
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Bioeng. Biotechnol. , 03 March 2021
Sec. Industrial Biotechnology
Volume 9 - 2021 | https://doi.org/10.3389/fbioe.2021.627082
This article is part of the Research Topic Microbial Production of Biopolyesters and their Building Blocks: Opportunities and Challenges View all 7 articles
Polyhydroxyalkanoate (PHA) synthase is an enzyme that polymerizes the acyl group of hydroxyacyl-coenzyme A (CoA) substrates. Aeromonas caviae PHA synthase (PhaCAc) is an important biocatalyst for the synthesis of a useful PHA copolymer, poly[(R)-3-hydroxybutyrate-co-(R)-3-hydroxyhexanoate] [P(3HB-co-3HHx)]. Previously, a PhaCAc mutant with double mutations in asparagine 149 (replaced by serine [N149S]) and aspartate 171 (replaced by glycine [D171G]) was generated to synthesize a 3HHx-rich P(3HB-co-3HHx) and was named PhaCAc NSDG. In this study, to further increase the 3HHx fraction in biosynthesized PHA, PhaCAc was engineered based on the three-dimensional structural information of PHA synthases. First, a homology model of PhaCAc was built to target the residues for site-directed mutagenesis. Three residues, namely tyrosine 318 (Y318), serine 389 (S389), and leucine 436 (L436), were predicted to be involved in substrate recognition by PhaCAc. These PhaCAc NSDG residues were replaced with other amino acids, and the resulting triple mutants were expressed in the engineered strain of Ralstonia eutropha for application in PHA biosynthesis from palm kernel oil. The S389T mutation allowed the synthesis of P(3HB-co-3HHx) with an increased 3HHx fraction without a significant reduction in PHA yield. Thus, a new workhorse enzyme was successfully engineered for the biosynthesis of a higher 3HHx-fraction polymer.
Polyhydroxyalkanoates (PHAs) are bio-based polyesters produced by a wide range of microorganisms as carbon and energy storage materials. The wild-type strain H16 of Ralstonia eutropha (or Cupriavidus necator) is one of the best-known PHA-producing bacteria (Sudesh et al., 2000; Steinbüchel and Hein, 2001). There has been long-standing interest in using PHAs as biodegradable bioplastics that could serve as alternatives to petrochemical plastics. Recently, PHAs have attracted attention as biodegradable and biocompatible thermoplastics for use in a wide range of agricultural, marine, and medical applications because of their excellent biodegradability (Akiyama et al., 2003).
Polyhydroxyalkanoates mainly consist of short-chain length (SCL; C3 to C5) and/or medium-chain-length (MCL; C6 and longer) monomers (Rehm, 2003). Among the SCL-PHAs, poly[(R)-3-hydroxybutyrate] [P(3HB)] is the most common bacterial PHA in nature. Although P(3HB) is a highly crystalline, hard, and brittle polymer, it begins to decompose at a temperature close to its melting point, making it difficult to process this polymer (Lehrle and Williams, 1994). Copolymerization of MCL monomers with a 3HB unit leads to notable changes in the physical properties of PHA, depending on the molecular structure and copolymer composition (Noda et al., 2005). The best-studied SCL/MCL-PHA copolymer is poly(3HB-co-3-hydroxyhexanoate) [P(3HB-co-3HHx)]. In this polymer, an important aspect is to control the level of the 3HHx monomer fraction for practical application in many fields. For example, the elongation at break increases from 5 to 760% by increasing the 3HHx fraction from 0 to 15 mol% (Doi et al., 1995; Chen et al., 2000; Andreeßen et al., 2014). P(3HB-co-3HHx) with 10–15 mol% 3HHx fraction can be used as an alternative to conventional plastics such as polypropylene and polyethylene (Shimamura et al., 1994; Chen et al., 2000; Andreeßen et al., 2014). However, it is difficult to efficiently produce P(3HB-co-3HHx) with such a high 3HHx fraction. Thus, significant efforts have been made to increase the 3HHx fraction in P(3HB-co-3HHx) biosynthesis (Jian et al., 2010; Budde et al., 2011; Arikawa and Matsumoto, 2016a).
The bacterium Aeromonas caviae is an original strain that can produce P(3HB-co-3HHx) from plant oils and fatty acids (Shimamura et al., 1994). Aeromonas caviae PHA synthase (PhaCAc) shows substrate specificity toward 3HB and 3-hydroxyvalerate monomers, as well as the 3HHx monomer (Kobayashi et al., 1994). From this point of view, PhaCAc is a valuable biocatalyst for production of P(3HB-co-3HHx). However, the polymer production capacity of A. caviae is not superior to that of other PHA producers. With the help of genetic engineering, recombinant R. eutropha expressing PhaCAc was generated, which demonstrated remarkable enhancement of P(3HB-co-3HHx) production from plant oils (Fukui and Doi, 1997, 1998; Kahar et al., 2004).
Additionally, to increase the 3HHx fraction in P(3HB-co-3HHx), various strategies have been developed. One effective approach is to increase the expression of (R)-specific enoyl-coenzyme A (CoA) hydratase (PhaJ4bRe), which provides R-3-hydroxyacyl-CoA precursors for PHA synthesis from the β-oxidation cycle, to reinforce the supply of the 3HHx monomer (Arikawa and Matsumoto, 2016a). In contrast, the 3HHx fraction in the polymer was increased by deleting the gene for the 3HB supplier acetoacetyl-CoA reductase (PhaBRe) to suppress the 3HB monomer supply; however, the PHA yield decreased (Budde et al., 2011).
Another approach to increase the 3HHx fraction in PHA is the engineering of PHA synthase (Kichise et al., 2002; Tsuge et al., 2004, 2007a,b; Watanabe et al., 2012). In previous studies, PhaCAc was modified via evolutionary engineering approaches, and several mutation sites (e.g., asparagine 149, aspartate 171, valine 214, and phenylalanine 518) enhanced the 3HHx polymerization capacity (Amara et al., 2002; Kichise et al., 2002; Tsuge et al., 2004). Furthermore, a double mutant of PhaCAc, termed the NSDG mutant, which has two amino acid substitutions of asparagine 149 by serine (N149S) and aspartate 171 by glycine (D171G), was generated as a superior enzyme capable of synthesizing P(3HB-co-3HHx) with a higher 3HHx fraction than the wild-type enzyme (Tsuge et al., 2007b). However, since then, no PhaCAc mutant with further high 3HHx polymerization ability has been created.
The three-dimensional structure of a protein provides important information for understanding its biochemical function and catalytic mechanism. Homology modeling aims to build three-dimensional protein structure models using experimentally determined structures of related family members as templates. Thus, homology modeling is a powerful tool for understanding and predicting the three-dimensional structure of unknown proteins to determine beneficial mutation sites and improve protein properties (Stoilov et al., 1998; Lee et al., 2011). Recently, some research groups have determined the partial crystal structure of R. eutropha PHA synthase (PhaCRe), which is classified into the same group (class I) as PhaCAc based on its substrate specificity and subunit structure (Wittenborn et al., 2016; Kim et al., 2017). According to their crystal structures, three active residues, Cys319, Asp480, and His508, in PhaCRe are in close proximity. Additionally, amino acid residues that make up the substrate pocket have been identified (Wittenborn et al., 2016; Kim et al., 2017). Moreover, structural information on the available PHA synthases has been increasing (Chek et al., 2017, 2019, 2020).
In this study, using a newly constructed homology model of PhaCAc, three amino acid residues were predicted to be constituents of the substrate pocket and involved in substrate recognition. Based on this prediction, site-specific mutagenesis was conducted on PhaCAc NSDG to introduce an additional third mutation. The resulting triple mutants were expressed in the strain 005dC1Z126TRCB, an engineered R. eutropha strain, grown on palm kernel oil as a carbon source for PHA biosynthesis. It was found that the triple mutant PhaCAc NSDG/S389T is capable of synthesizing P(3HB-co-3HHx) with a higher 3HHx fraction than the parental PhaCAc NSDG. Furthermore, the selected PhaCAc triple mutants were isolated as PHA granule-associated enzymes from R. eutropha and characterized through enzyme kinetic analysis to understand how the catalytic function changed.
Bacterial strains and gene expression plasmids used are listed in Table 1. All Escherichia coli strains were grown in Luria-Bertani (LB) medium. The E. coli strains JM109 and S17-1 were used for plasmid construction and as donors in the intergeneric conjugation experiments, respectively. All R. eutropha strains were grown in nutrient broth (Difco Laboratories, Detroit, MI, United States).
To delete the phaCAcNSDG gene in the R. eutropha CnTRCB strain (Arikawa and Matsumoto, 2016a), the gene deletion plasmid pNS2X-sacB-phaC1AdS (Sato et al., 2013) was introduced into the CnTRCB strain by conjugation from the donor strain E. coli S17-1. The deletion of phaC was confirmed through PCR. The resulting strain was named 005dC1Z126TRCB, which retained phaA and phaB involved in the 3HB monomer supply and provided greater proportions of 3HHx than the H16 strain, by enhancing the expression of phaJ4bRe.
A template-based modeling method using HyperChem (HYPERCUBE, INC., Gainsville, FL, United States) (Froimowitz, 1993) was used to predict the structure of PhaCAc using PDB:5T6O (residues 201–589) from PhaCRe as a template.
Plasmids expressing wild-type PhaCAc, the double mutant NSDG, and the triple mutants NSDG-Y318/S389/L436X were constructed based on the pCUP3 vector, which is stably maintained in R. eutropha (Sato et al., 2013). The wild-type phaCAc (WT-phaCAc) and phaCAc NSDG genes were obtained through PCR with MunI_PhaCAc_F and SpeI_PhaCAc_R as primers, using the plasmid pColdI::phaCAc and the genomic DNA of the R. eutropha strain KNK005 as a template, respectively (Sato et al., 2013; Ushimaru et al., 2014). These fragments were digested by MunI and SpeI, and then cloned into the same sites of the pCUP3 vector. The Ptrp fragment, which was amplified by PCR using pKK388-1 (Clontech, Palo Alto, CA, United States) as a template (Arikawa and Matsumoto, 2016a), was digested with MunI and ligated with MunI-digested pCUP3 vectors carrying WT-phaCAc and phaCAc NSDG genes to yield pCUP3-Ptrp-WT-phaCAc and pCUP3-Ptrp-phaCAc NSDG, respectively. Site-directed mutagenesis of phaCAcNSDG gene was performed by overlap extension PCR (Ho et al., 1989). Reverse primers containing a point mutation were designed as listed in Supplementary Table S1, and primers containing a restriction enzyme site were designed as (pCUP3_IF_MunI_trp_F) 5′-ACA TTGCGCTGAAAGAAGGGCCAATTGTGCTTCTGGCGTC-3′ and (pCUP3_SpeI_IF_R) 5′-GCTCGGATCCACTAGTCGGCT GCCGACTGGT-3′ (the underlined sequences indicate the MunI and SpeI sites, and the bold sequences indicate in-fusion alignment). Using the corresponding primers in phaCAc_Y318/S389/L436X_R and phaCAc_Y318/S389/L436X_F (Supplementary Table S1), the DNA fragments were amplified. The resulting fragments after one round of PCR were used as templates, and PCR was performed again using the outside primers with MunI and SpeI sites. The resulting phaCAcNSDG fragments with point mutations were digested using MunI and SpeI, and then inserted into the corresponding restriction sites in the pCUP3 vector. The resulting pCUP3-Ptrp-NSDG-Y318/S389/L436X plasmids were introduced into an engineered strain of R. eutropha 005dC1Z126TRCB strain, in which phaC gene was disrupted. Transformation was performed through electroporation, as described previously (Sato et al., 2013; Arikawa et al., 2016b).
Polyhydroxyalkanoate production was performed in 50 mL of mineral salt (MS) medium (Kato et al., 1996) with 1.29 g/L (NH4)2SO4 and 1.5 w/v% palm kernel oil as a sole carbon source for 72 h. Kanamycin was added to the medium at a concentration of 50 mg/L to maintain the plasmid in the cells. After cultivation, the collected cells were washed with water and ethanol to remove the remaining carbon sources and then lyophilized (Arikawa et al., 2016b). The PHA content in the cells was determined by gas chromatography (GC) after methanolysis of approximately 15 mg of lyophilized cells in the presence of 15% (v/v) sulfuric acid, as previously described (Lakshman and Shamala, 2006).
The PHA synthase activity assay was performed, wherein the amount of CoA released was measured using 5,5-dithiobis(2-nitrobenzoic acid) (DTNB) with the following modifications: PHA synthase assay was initiated by adding the granule-associated PhaCAc, which was obtained from 24 h of R. eutropha culture broth by ultracentrifugation as previously described (Valentin and Steinbüchel, 1994; Harada et al., 2019). After incubation with the substrate, R-3HB-CoA (0.1–20 mM) or R-3HHx-CoA (0.1–20 mM) (synthesized by China Suli Co., Ltd., China) at 30°C for 30 s, the reaction was stopped by adding trichloroacetic acid (2.5% (v/v) final concentration). A portion of the reaction mixture was diluted with DTNB solution (10 mM DTNB in 500 mM potassium phosphate buffer, pH 8.0) to obtain a final concentration of 1 mM DTNB. The reaction mixtures were incubated for 30 min in the dark and then centrifuged. The absorbance of the supernatant was measured at 412 nm. The molar absorbance coefficient ε412 = 13,600 M–1cm–1 was used to determine the concentration of the thiol group of free CoA.
The concentration of the granule-associated PhaCAc was determined as previously described (Harada et al., 2019), after incubation with rabbit antiserum against a peptide from the C-terminus of PhaCAc, followed by incubation with a goat anti-rabbit antibody conjugated with horseradish peroxidase (HRP; Santa Cruz Biotechnology, CA, United States). Proteins were visualized using the ECL Plus Western Blotting Detection Reagent (Bio-Rad, Hercules, CA, United States). Data were recorded using a CCD camera FAS-1000 (Toyobo, Osaka, Japan). Quantitative analysis of PhaCAc concentration on PHA granules was performed using calibration curves prepared using purified PhaCAc (130–520 ng). Band intensities were quantified using the ImageJ software1.
To identify the beneficial mutation site for increasing the 3HHx fraction, a homology model of PhaCAc was first built by targeting PhaCRe. The PhaCRe template had 38.3% (220/575) sequence identity and 75.7% (435/575) similarity with PhaCAc, while in residues 201–589, sequence identity was 43.2% (164/380) and similarity was 81.8% (311/380). The constructed homology model is shown in Figures 1 and 2. Compared to the structure of PhaCRe, PhaCAc has a partly wide and deep cavity space around the catalytic domain (Figures 2A,B). This is in good agreement with the experimental observation that PhaCAc has a broader substrate specificity than PhaCRe (Fukui and Doi, 1997). From the comparison of these structural models, two amino acid residues adjacent to the active center (PhaCAc vs. PhaCRe: Y318 vs. F318, S389 vs. T393) were found to be different. It was presumed that Y318 and S389 determine the depth and width of the substrate pocket of PhaCAc, respectively. The substrate entrance tunnel of these models was further compared (Figures 2C,D), and additional differences were found (PhaCAc vs. PhaCRe: L436 vs. Y440). In PhaCAc, L436 mainly contributes to expanding the substrate entrance tunnel, because there is a significant difference in the amino acid size at the homologous positions in these structural models.
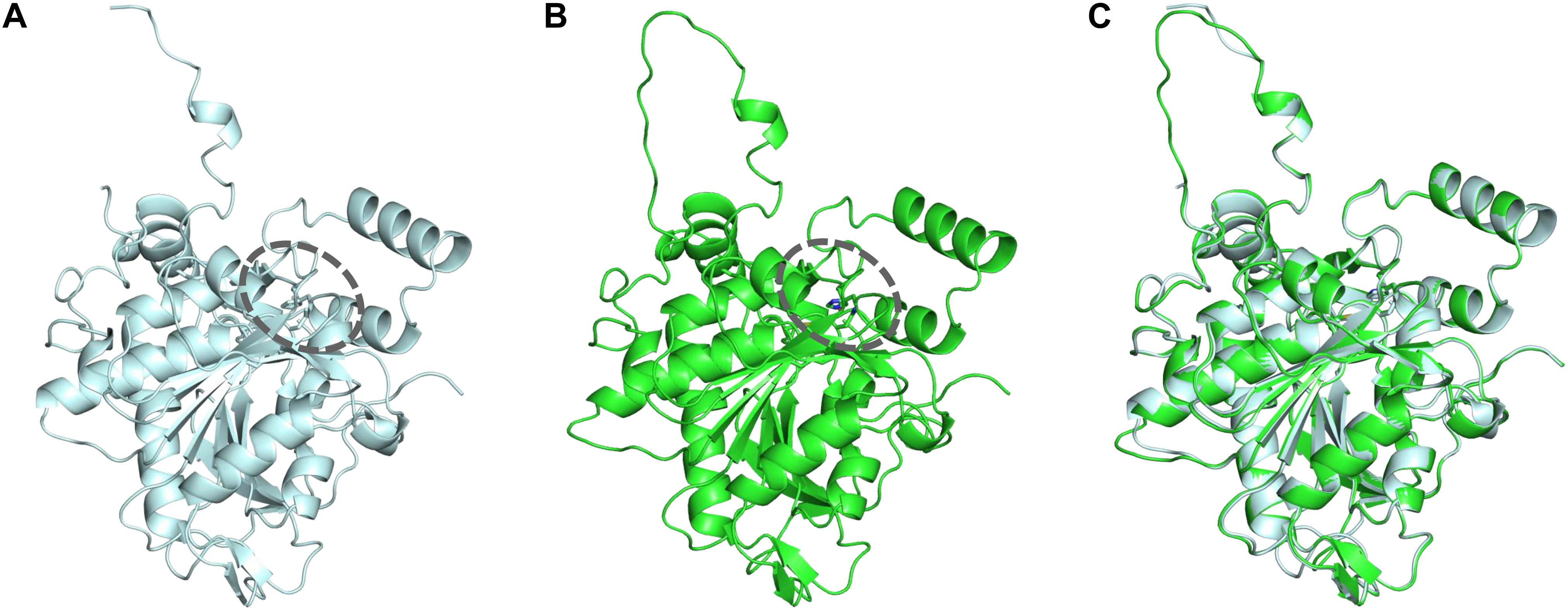
Figure 1. (A) Crystal structure of the PhaCRe catalytic domain (amino acid sequence 201–589). (B) Homology model of PhaCAc catalytic domain (amino acid sequence 201–581). (C) Overlay of PhaCRe (light blue) with the homology model PhaCAc (green). Dotted circle indicates the substrate pocket.
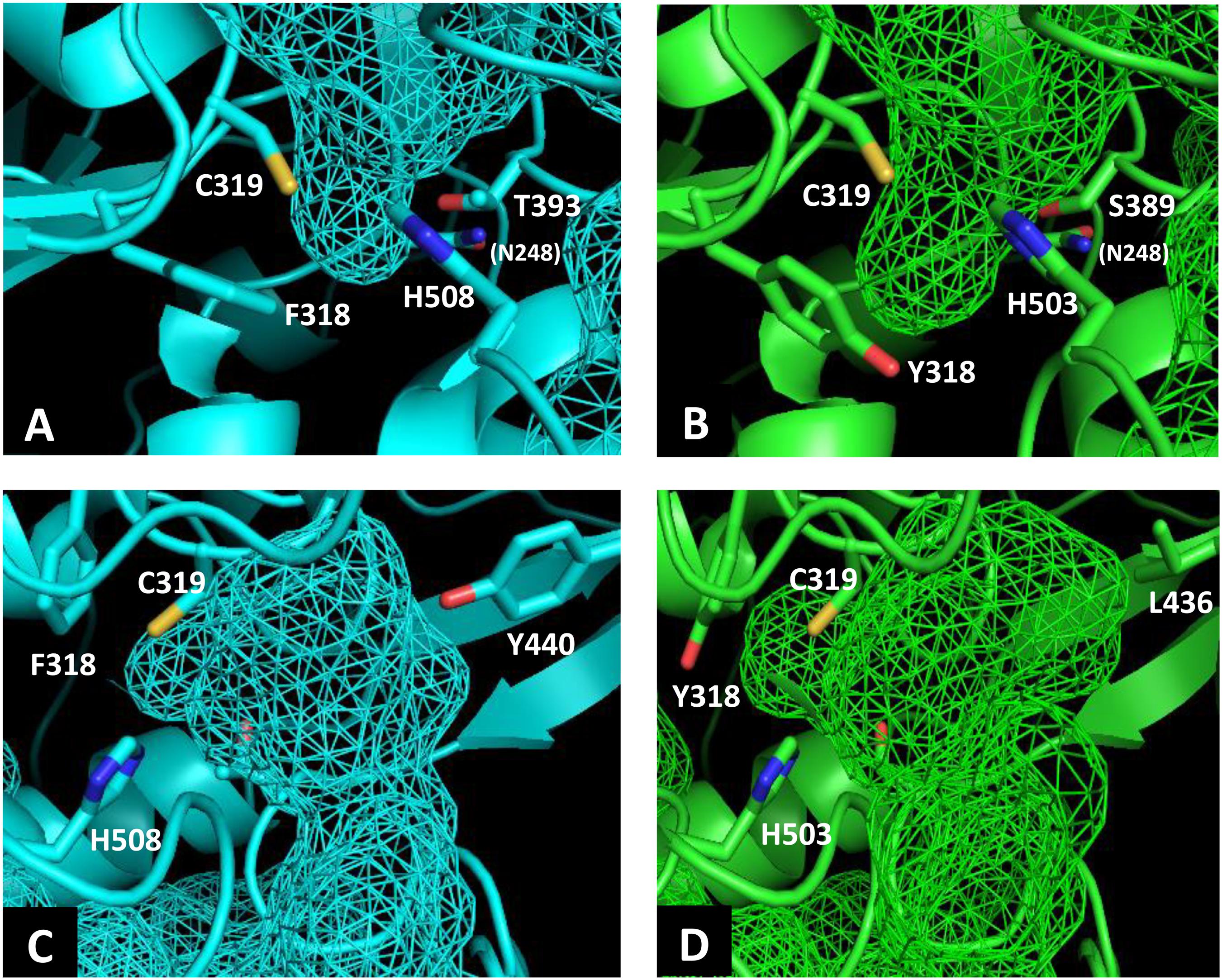
Figure 2. Comparison of the substrate pocket around the active site between PhaCRe (A) and PhaCAc (B). Comparison of the substrate entrance tunnel between (C) PhaCRe and (D) PhaCAc. Cavity spaces are indicated with a mesh.
As the Y318 of PhaCAc was predicted to determine the depth of the substrate pocket based on the homology model, we investigated the effect of the amino acid size at this position on 3HHx polymerization ability. To replace Y318, we selected Leu, Ile, and Met, which are smaller than Tyr, with the aim of expanding the substrate pocket space. The three PhaCAc mutants with NSDG mutations and either Y318L/I/M mutations were generated by site-directed mutagenesis and expressed in the engineered R. eutropha strain 005dC1Z126TRCB to induce P(3HB-co-3HHx) biosynthesis from palm kernel oil. The results are presented in Table 2. The strain expressing the wild-type enzyme accumulated 80.3 wt% P(3HB-co-3HHx) of dried cells, with 7.4 mol% of 3HHx fraction. Meanwhile, the strain expressing PhaCAc NSDG accumulated 85.7 wt% P(3HB-co-3HHx) of dried cells with 13.1 mol% of 3HHx fraction. A very small amount (less than 0.1 mol%) of 3-hydroxyoctanoate (3HO) was also detected, which is consistent with previous study (Tsuge et al., 2007b). PhaCAc NSDG was confirmed to have the ability to synthesize P(3HB-co-3HHx) with a higher 3HHx fraction than the wild-type enzyme. Compared to NSDG and NSDG/Y318X, a slight increase in the 3HHx fraction was observed in the strain expressing the NSDG/Y318I mutant, whereas the other two strains showed a considerable decrease in the 3HHx fraction. As for the NSDG/Y318L mutant, it showed a slight increase (0.8 mol%) in the 3HO fraction. On the contrary, expression of the NSDG/Y318I mutant notably decreased polymer accumulation (11.7 wt%) in the cells compared to the parental NSDG strain (85.7 wt%). Thus, additional mutagenesis of Y318 was not beneficial.
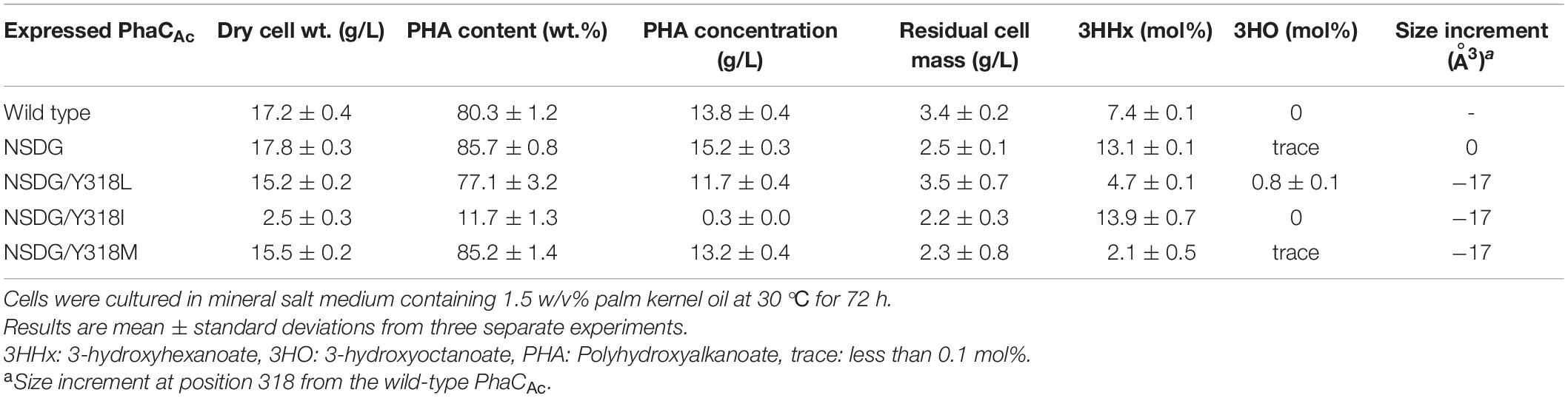
Table 2. Polyhydroxyalkanoate accumulation in Ralstonia eutropha strain 005dC1Z126TRCB by expressing PhaCAc NSDG with Y318X mutation.
S389 in PhaCAc contributes to cavity formation near the active center. It is homologous to T393 in PhaCRe, and the cavity space in PhaCAc is larger due to the volume of one methyl group. To further expand the cavity space, the amino acid residue at position 389 was replaced with Ala (S389A), which is a smaller amino acid. To examine the opposite effect on the amino acid size, this residue was also replaced with the larger amino acid Thr (S389T) with the aim of narrowing the space. The two PhaCAc mutants with NSDG mutations and either S389A/T mutations were generated by site-directed mutagenesis and evaluated for P(3HB-co-3HHx) biosynthesis. The results are presented in Table 3. The additional S389A mutation did not alter the 3HHx fraction. However, the S389T mutation in PhaCAc NSDG increased the 3HHx fraction to 14.9 mol% without a significant decrease in PHA yield. Since the 3HHx fraction increased due to replacement with the bulkier amino acid in the mutant, further replacements were conducted using Val, Leu, Ile, and Cys, which have bulkier side chains than Ser based on their van der Waals volumes (Darby and Creighton, 1995; Tsuge et al., 2009). As a result, a slight increase in the 3HHx fraction up to 13.8 mol% was observed by introducing S389V/L/I/C mutations in PhaCAc NSDG. Of the mutations tested, the S389T mutation was the most effective in increasing the 3HHx fraction, followed by S389V. It was found that mutagenesis at position 318 in PhaCAc may enhance the 3HHx polymerization ability, although replacement with relatively bulky amino acids was effective.
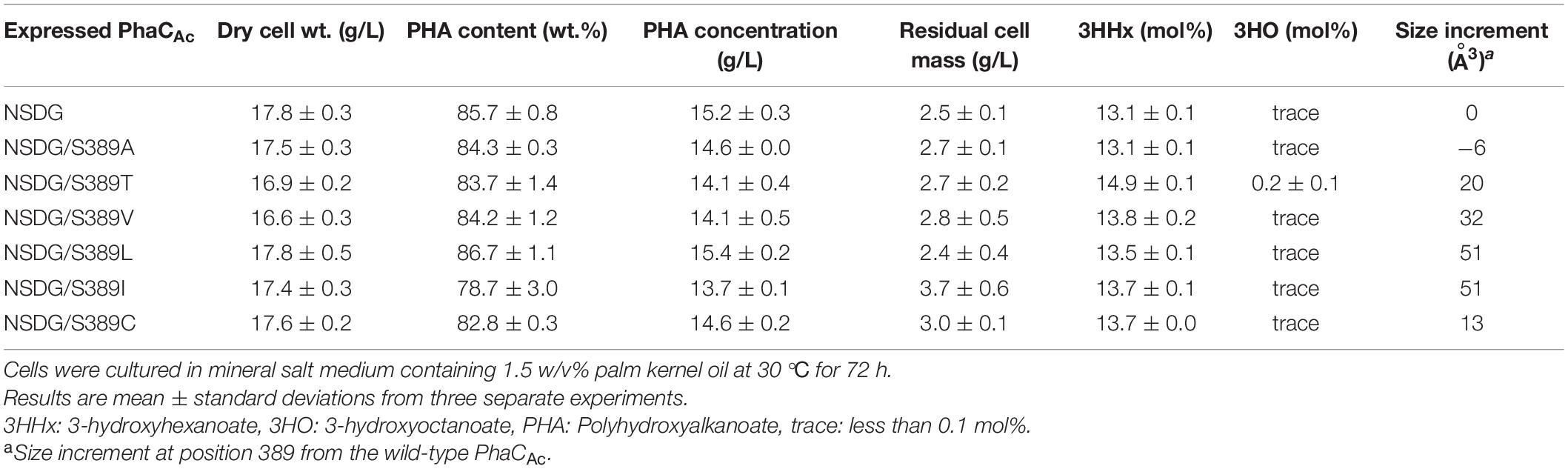
Table 3. Polyhydroxyalkanoate accumulation in Ralstonia eutropha strain 005dC1Z126TRCB by expressing PhaCAc NSDG with S389X mutation.
L436 is an amino acid located slightly outside the active center, which corresponds to Y440 in PhaCRe. As predicted by homology modeling, the cavity of PhaCAc is larger than that of PhaCRe because of the difference in the amino acid side size at this position. To examine the effect of mutagenesis for L436 on the 3HHx polymerization ability of PhaCAc NSDG, site-directed mutagenesis was performed. To examine the expanding effect of the pocket space, L436A/V mutations were introduced into PhaCAc NSDG. In addition, L436Y/I mutations were introduced to examine the opposite narrowing effect (Darby and Creighton, 1995; Tsuge et al., 2009). The results are listed in Table 4. PHA accumulation was observed for all strains with polymer contents greater than 80 wt%. However, these mutations showed a decrease in the 3HHx fraction; The L436A and L436Y mutations showed 21% and 66% reductions in the 3HHx fraction, respectively, when compared to the parental NSDG strain. Based on this observation, the residue at position 436 may be involved in substrate recognition, but mutagenesis at this position did not result in an increase in the 3HHx fraction of the polymer.
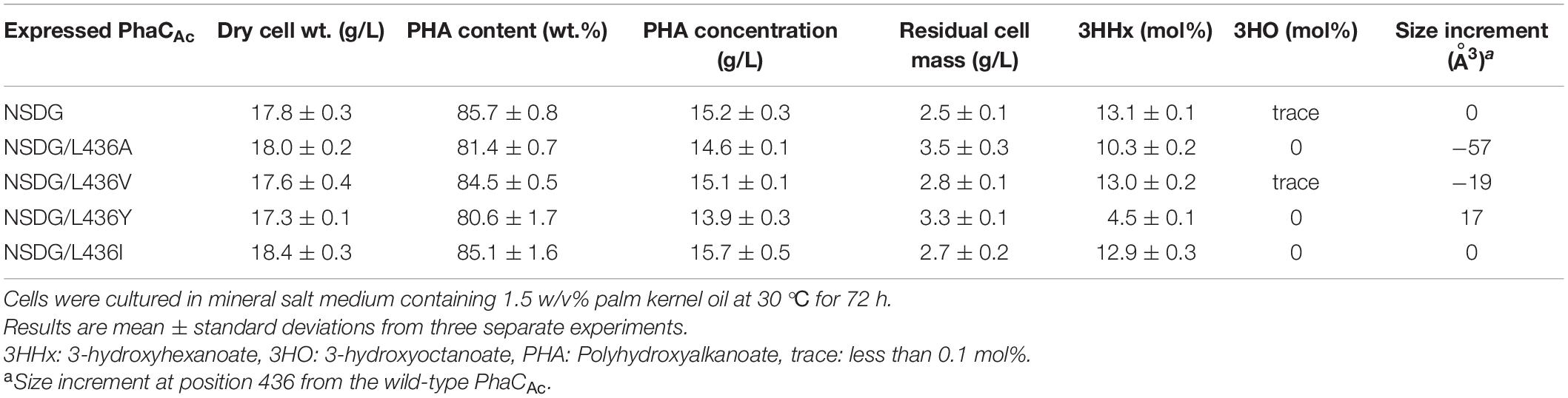
Table 4. Polyhydroxyalkanoate accumulation in Ralstonia eutropha strain 005dC1Z126TRCB by expressing PhaCAc NSDG with L436X mutation.
To obtain a better understanding of the polymerization ability of the 3HHx monomer of PhaCAc, granule-associated PHA synthases were prepared and used for enzyme kinetic analysis. The granule-associated PHA synthase does not exhibit a lag phase (Gerngross et al., 1994; Taguchi et al., 2002) because the enzyme is already activated and thus is suitable for use in accurate kinetic analysis. To determine the PhaCAc concentrations on the surface of the isolated PHA granules, western blotting was performed using an antibody against PhaCAc. The kinetic parameters determined for wild-type PhaCAc, NSDG mutant, and NSDG/S389X mutants are listed in Table 5. The NSDG mutant and NSDG/S389X showed a lower Michaelis constant (Km) for the R-3HHx-CoA substrate than the wild-type PhaCAc but was not significant for the R-3HB-CoA substrate. In addition, the NSDG mutant and NSDG/S389X mutants showed a higher turnover number (kcat) for both substrates than the wild-type PhaCAc, except for NSDG/S389V toward R-3HHx-CoA. Kinetic analysis revealed that the substrate affinity and turnover number, especially for R-3HHx-CoA, increased in the NSDG mutant. Among the mutants tested, the Km values of S389V/C mutants for R-3HHx-CoA, which were 0.46 mM and 0.53 mM, respectively, showed smaller values than that of the parental NSDG strain (0.73 mM). The decrease in Km value indicates the increased affinity between enzyme and substrate, thus providing evidence of the reinforced ability of 3HHx polymerization by these mutations. In contrast, by introducing the S389T mutation into PhaCAc NSDG, the Km value slightly increased for both R-3HB-CoA and R-3HHx-CoA. Furthermore, the kcat value significantly increased for both substrates by up to 3.4-fold compared to the parental NSDG enzyme. Thus, the increase in the 3HHx fraction caused by the S389T mutation could be attributed to the increased catalytic turnover of the enzyme, rather than the increased affinity between the substrate and the enzyme.
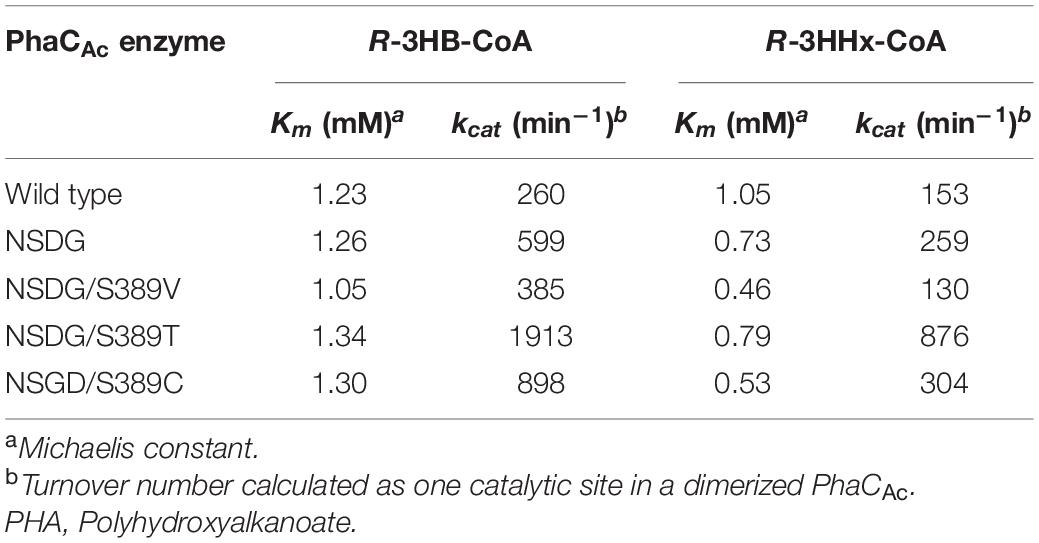
Table 5. Kinetic parameters of PHA granule-associated PhaCAc for R-3HB-CoA and R-3HHx-CoA substrates.
This study aimed to increase the 3HHx fraction in P(3HB-co-3HHx) by engineering PhaCAc. Based on evolutionary engineering, we had already generated a PhaCAc NSDG mutant as a workhorse to synthesize a high 3HHx-fraction polymer. The mutation positions of NSDG are at the N-terminal region of PhaCAc, and these amino acid residues are predicted to not be involved in the formation of the substrate pocket. Thus, to further modify the PhaCAc NSDG for higher 3HHx-fraction polymer synthesis, we attempted to change the cavity space of the substrate pocket by replacing certain amino acids. Recently, two research groups have published the partial crystal structure of PhaCRe (Wittenborn et al., 2016; Kim et al., 2017). PhaCRe can polymerize up to C5 monomers, whereas PhaCAc is capable of polymerizing up to C6 monomers. The difference in substrate specificity may be caused by the size of the substrate pocket near the active center (Kim et al., 2017). From this viewpoint, the three-dimensional structures around the cavity pocket space of PhaCRe and the homology model of PhaCAc were compared, mainly focusing on the difference in the spread of amino acid side chains. As possible determining residues for the pocket size of PhaCAc, three amino acids, namely Y318, S389, and L436, were identified in this study.
Our homology model suggests that Y318 may be an important residue that determines the pocket size (Figure 2B). Interestingly, this position is Ala in PHA synthases from Pseudomonas spp. (class II) that can polymerize MCL monomers up to C14. Therefore, it is reasonable to hypothesize that a mutation at this position has a significant influence on the pocket depth. The amino acid at this position in PhaCRe (F318) has been suggested to stabilize the structure of the substrate pocket (Kim et al., 2017). Indeed, mutagenesis at this position of PhaCRe led to a decrease in 75% of the synthase activity (Kim et al., 2017). In our study, mutation of Y318 of PhaCAc also resulted in a significant reduction in polymer synthesis (Table 2). Y318 maintains the structure of the substrate pocket and is strongly related to the polymerization ability in the same manner as PhaCRe.
The docking simulation using the crystal structure of PhaCRe suggested that Y440 is located in the substrate entrance tunnel and contributes to the structural stabilization of the β-mercaptoethylamine/pantothenate (β-MP) moiety of R-3HB-CoA (Kim et al., 2017). Y440 stabilizes the substrate orientation by interacting with neighboring amino acids to efficiently catalyze the polymerization reaction. In PhaCAc, the corresponding L436 was considered to regulate the space of the substrate entrance tunnel based on the homology model (Figure 2D). In fact, mutagenesis of L436 limited the substrate specificity of PhaCAc and reduced the 3HHx fraction in the biosynthesized polymer (Table 4). Among the NSDG/L436X mutants examined, the most remarkable reduction in the 3HHx fraction was observed for the NSDG/L436Y mutant, probably due to the narrowest pocket space by replacement with the largest amino acid Tyr.
However, the effect of 3HHx polymerization ability cannot always be explained by the reduction and expansion of pocket space due to amino acid replacement. In this study, we found that the 3HHx fraction in PHA increased after narrowing the substrate pocket by mutagenesis of S389 (Table 3). However, this observation was opposite to our hypothesis.
To better understand the effect of S389 mutagenesis, the kinetics of the enzymes with the S389X mutation were investigated. Kinetic analysis provided new information on the changes in catalytic function due to S389X mutations. It was revealed that substrate affinity for R-3HHx-CoA was increased by S389V/C mutations, whereas the catalytic turnover of the enzyme was increased by the S389T mutation. Thus, the increase in the 3HHx fraction caused by the S389T mutation may be due to the increased catalytic turnover of the enzyme, rather than the change in binding affinity between the enzyme and substrate. The relationship between pocket size narrowing and 3HHx polymerization ability may be explained by stabilization of the substrate orientation when the substrate accesses the active site. The proper orientation of the substrate may increase the efficiency of the catalytic reaction. However, further studies are required to elucidate the underlying mechanisms of mutagenesis.
In conclusion, by comparing the substrate pocket structures of PhaCRe and PhaCAc, a new beneficial mutation position at S389 was found to enhance the 3HHx polymerization ability of PhaCAc NSDG. Since the discovery of the NSDG mutation, additional mutations conferring a superior ability of 3HHx polymerization have not been found by an evolutionary engineering approach. Thus, this is a successful example of PHA synthase engineering by effectively exploiting the findings from the three-dimensional structure of proteins.
The raw data supporting the conclusion of this article will be made available by the authors, without undue reservation.
KH, SK, KO, SY, TT, and SS jointly conceived the study. KH, SK, KO, and SY performed the experiments. KH wrote the manuscript in consultation with SK, KO, SY, TT, and SS. All authors read and approved the final manuscript.
This research was supported by the Adaptable and Seamless Technology Transfer Program through Target-driven R&D (A-Step), JST, Japan.
KH, SK, KO, SY, and SS are employees of Kaneka, Co., Ltd.
The remaining author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fbioe.2021.627082/full#supplementary-material
Akiyama, M., Tsuge, T., and Doi, Y. (2003). Environmental life cycle comparison of polyhydroxyalkanoates produced from renewable carbon resources by bacterial fermentation. Polym. Degrad. Stabil. 80, 183–194. doi: 10.1016/s0141-3910(02)00400-7
Amara, A. A., Steinbuchel, A., and Rehm, B. H. A. (2002). In vivo evolution of the Aeromonas punctata polyhydroxyalkanoate (PHA) synthase: isolation and characterization of modified PHA synthases with enhanced activity. Appl. Microbiol. Biotechnol. 59, 477–482. doi: 10.1007/s00253-002-1035-3
Andreeßen, B., Taylor, N., and Steinbüchel, A. (2014). Poly(3-hydroxypropionate): A promising alternative to fossil fuel-based materials. Appl. Environ. Microbiol. 80, 6574–6582. doi: 10.1128/aem.02361-14
Arikawa, H., and Matsumoto, K. (2016a). Evaluation of gene expression cassettes and production of poly(3-hydroxybutyrate-co-3-hydroxyhexanoate) with a fine modulated monomer composition by using it in Cupriavidus necator. Microb. Cell Fact. 15:184. doi: 10.1186/s12934-016-0583-7
Arikawa, H., Sato, S., Fujiki, T., and Matsumoto, K. (2016b). A study on the relation between poly(3-hydroxybutyrate) depolymerases or oligomer hydrolases and molecular weight of polyhydroxyalkanoates accumulating in Cupriavidus necator H16. J. Biotechnol. 227, 94–102. doi: 10.1016/j.jbiotec.2016.04.004
Budde, C. F., Riedel, S. L., Willis, L. B., Rha, C. K., and Sinskey, A. J. (2011). Production of poly(3-hydroxybutyrate-co-3-hydroxyhexanoate) from plant oil by engineered Ralstonia eutropha strains. Appl. Environ. Microbiol. 77, 2847–2854. doi: 10.1128/aem.02429-10
Chek, M. F., Hiroe, A., Hakoshima, T., Sudesh, K., and Taguchi, S. (2019). PHA synthase (PhaC): Interpreting the functions of bioplastic-producing enzyme from a structural perspective. Appl. Microbiol. Biotechnol. 103, 1131–1141. doi: 10.1007/s00253-018-9538-8
Chek, M. F., Kim, S. Y., Mori, T., Arsad, H., Samian, M. R., Sudesh, K., et al. (2017). Structure of polyhydroxyalkanoate (PHA) synthase PhaC from Chromobacterium sp. USM2, producing biodegradable plastics. Sci. Rep. 7:5312. doi: 10.1038/s41598-017-05509-4
Chek, M. F., Kim, S., Mori, T., Tan, H. T., and Sudesh, K. (2020). Asymmetric open-closed dimer mechanism of polyhydroxyalkanoate synthase PhaC of polyhydroxyalkanoate synthase PhaC. iScience 23:101084. doi: 10.1016/j.isci.2020.101084
Chen, G. Q., Wu, Q., Zhao, K., and Yu, P. H. (2000). Functional polyhydroxyalkanoates synthesized by microorganisms. Chinese J. Polym. Sci. 18, 389–396.
Darby, N. J., and Creighton, T. E. (1995). Protein structure: In Focus. Biochem. Educ. 23:46. doi: 10.1016/0307-4412(95)90200-7
Doi, Y., Kitamura, S., and Abe, H. (1995). Microbial synthesis and characterization of poly(3-hydroxybutyrate-co-3-hydroxyhexanoate). Macromolecules 28, 4822–4828. doi: 10.1021/ma00118a007
Froimowitz, M. (1993). HyperChem: a software package for computational chemistry and molecular modeling. Biotechniques 14, 1010–1013.
Fukui, T., and Doi, Y. (1997). Cloning and analysis of the poly(3-hydroxybutyrate-co-3- hydroxyhexanoate) biosynthesis genes of Aeromonas caviae. J. Bacteriol. 179, 4821–4830. doi: 10.1128/jb.179.15.4821-4830.1997
Fukui, T., and Doi, Y. (1998). Efficient production of polyhydroxyalkanoates from plant oils by Alcaligenes eutrophus and its recombinant strain. Appl. Microbiol. Biotechnol. 49, 333–336. doi: 10.1007/s002530051178
Gerngross, T. U., Snell, K. D., Peoples, O. P., Sinskey, A. J., Csuhai, E., Masamune, S., et al. (1994). Overexpression and purification of the soluble polyhydroxyalkanoate synthase from Alcaligenes eutrophus: Evidence for a required posttranslational modification for catalytic activity. Biochemistry 33, 9311–9320. doi: 10.1021/bi00197a035
Jian, J., Li, Z. J., Ye, H. M., Yuan, M. Q., and Chen, G. Q. (2010). Metabolic engineering for microbial production of polyhydroxyalkanoates consisting of high 3-hydroxyhexanoate content by recombinant Aeromonas hydrophila. Bioresour. Technol. 101, 6096–6102. doi: 10.1016/j.biortech.2010.02.089
Harada, K., Nambu, Y., Mizuno, S., and Tsuge, T. (2019). In vivo and in vitro characterization of hydrophilic protein tag-fused Ralstonia eutropha polyhydroxyalkanoate synthase. Int. J. Biol. Macromol. 138, 379–385. doi: 10.1016/j.ijbiomac.2019.07.095
Ho, S. N., Hunt, H. D., Horton, R. M., Pullen, J. K., and Pease, L. R. (1989). Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77, 51–59. doi: 10.1016/0378-1119(89)90358-2
Kahar, P., Tsuge, T., Taguchi, K., and Doi, Y. (2004). High yield production of polyhydroxyalkanoates from soybean oil by Ralstonia eutropha and its recombinant strain. Polym. Degrad. Stabil. 83, 79–86. doi: 10.1016/s0141-3910(03)00227-1
Kato, M., Bao, H. J., Kang, C. K., Fukui, T., and Doi, Y. (1996). Production of a novel copolyester of 3-hydroxybutyric acid and medium-chain-length 3-hydroxyalkanoic acids by Pseudomonas sp. 61-3 from sugars. Appl. Microbiol. Biotechnol. 45, 363–370. doi: 10.1007/s002530050697
Kichise, T., Taguchi, S., and Doi, Y. (2002). Enhanced accumulation and changed monomer composition in polyhydroxyalkanoate (PHA) copolyester by in vitro evolution of Aeromonas caviae PHA synthase. Appl. Environ. Microbiol. 68, 2411–2419. doi: 10.1128/aem.68.5.2411-2419.2002
Kim, J., Kim, Y. J., Choi, S. Y., Lee, S. Y., and Kim, K. J. (2017). Crystal structure of Ralstonia eutropha polyhydroxyalkanoate synthase C-terminal domain and reaction mechanisms. Biotechnol. J. 12:1600648. doi: 10.1002/biot.201600648
Kobayashi, G., Shiotani, T., Shima, Y., Doi, Y. (1994). “Biosynthesis and Characterization of poly(3-hydroxybutyrate-co-3-hydroxyhexanoate) from oils and fats by Aeromonas sp. OL-338 and Aeromonas sp. FA-440”, in Biodegradable Plastics and Polymers. Y. Doi and K. Fukuda eds. Amsterdam: Elsevier. 12, 410–416. doi: 10.1016/b978-0-444-81708-2.50044-0
Lakshman, K., and Shamala, T. R. (2006). Extraction of polyhydroxyalkanoate from Sinorhizobium meliloti cells using Microbispora sp. culture and its enzymes. Enzyme Microb. Technol. 39, 1471–1475. doi: 10.1016/j.enzmictec.2006.03.037
Lee, J. H., Lee, Y., Ryu, H., Kang, D. W., Lee, J., Lazar, J., et al. (2011). Structural insights into transient receptor potential vanilloid type 1 (TRPV1) from homology modeling, flexible docking, and mutational studies. J. Comput. Aided. Mol. Des. 25, 317–327. doi: 10.1007/s10822-011-9421-5
Lehrle, R. S., and Williams, R. J. (1994). Thermal degradation of bacterial poly(hydroxybutyric acid): Mechanisms from the dependence of pyrolysis yields on sample thickness. Macromolecules 27, 3782–3789. doi: 10.1021/ma00092a017
Noda, I., Green, P. R., Satkowski, M. M., and Schechtman, L. A. (2005). Preparation and properties of a novel class of polyhydroxyalkanoate copolymers. Biomacromolecules 6, 580–586. doi: 10.1021/bm049472m
Rehm, B. H. A. (2003). Polyester synthases: natural catalysts for plastics. Biochem. J. 376, 15–33. doi: 10.1042/bj20031254
Sato, S., Fujiki, T., and Matsumoto, K. (2013). Construction of a stable plasmid vector for industrial production of poly(3-hydroxybutyrate-co-3-hydroxyhexanoate) by a recombinant Cupriavidus necator H16 strain. J. Biosci. Bioeng. 116, 677–681. doi: 10.1016/j.jbiosc.2013.05.026
Shimamura, E., Kasuya, K., Kobayashi, G., Shiotani, T., Shima, Y., and Doi, Y. (1994). Physical properties and biodegradability of microbial poly(3-hydroxybutyrate-co-3-hydroxyhexanoate). Macromolecules 27, 878–880. doi: 10.1021/ma00081a041
Steinbüchel, A., and Hein, S. (2001). Biochemical and molecular basis of microbial synthesis of polyhydroxyalkanoates in microorganisms. Adv. Biochem. Eng. Biotechnol. 71, 81–123. doi: 10.1007/3-540-40021-4_3
Stoilov, I., Akarsu, A. N., Alozie, I., Child, A., Barsoum-Homsy, M., Turacli, M. E., et al. (1998). Sequence analysis and homology modeling suggest that primary congenital glaucoma on 2p21 results from mutations disrupting either the hinge region or the conserved core structures of cytochrome P4501B1. Am. J. Hum. Genet. 62, 573–584. doi: 10.1086/301764
Sudesh, K., Abe, H. Y., and Doi, Y. (2000). Synthesis, structure and properties of polyhydroxyalkanoates: biological polyesters. Prog. Polym. Sci. 25, 1503–1555. doi: 10.1016/s0079-6700(00)00035-6
Taguchi, S., Nakamura, H., Hiraishi, T., Yamato, I., and Doi, Y. (2002). In vitro evolution of a polyhydroxybutyrate synthase by intragenic suppression-type mutagenesis. J. Biochem. 131, 801–806. doi: 10.1093/oxfordjournals.jbchem.a003168
Tsuge, T., Saito, Y., Kikkawa, Y., Hiraishi, T., and Doi, Y. (2004). Biosynthesis and compositional regulation of poly[(3-hydroxybutyrate)-co-(3-hydroxyhexanoate)] in recombinant Ralstonia eutropha expressing mutated polyhydroxyalkanoate synthase genes. Macromol. Biosci. 4, 238–242. doi: 10.1002/mabi.200300077
Tsuge, T., Watanabe, S., Sato, S., Hiraishi, T., Abe, H., Doi, Y., et al. (2007a). Variation in copolymer composition and molecular weight of polyhydroxyalkanoate generated by saturation mutagenesis of Aeromonas caviae PHA synthase. Macromol. Biosci. 7, 846–854. doi: 10.1002/mabi.200700023
Tsuge, T., Watanabe, S., Shimada, D., Abe, H., Doi, Y., and Taguchi, S. (2007b). Combination of N149S and D171G mutations in Aeromonas caviae polyhydroxyalkanoate synthase and impact on polyhydroxyalkanoate biosynthesis. FEMS Microbiol. Lett. 277, 217–222. doi: 10.1111/j.1574-6968.2007.00958.x
Tsuge, T., Yamamoto, T., Yano, K., Abe, H., Doi, Y., and Taguchi, S. (2009). Evaluating the ability of polyhydroxyalkanoate synthase mutants to produce P(3HB−co−3HA) from soybean oil. Macromol. Biosci. 9, 71–78. doi: 10.1002/mabi.200800118
Ushimaru, K., Motoda, Y., Numata, K., and Tsuge, T. (2014). Phasin proteins activate Aeromonas caviae polyhydroxyalkanoate (PHA) synthase but not Ralstonia eutropha PHA synthase. Appl. Environ. Microbiol. 80, 2867–2873. doi: 10.1128/aem.04179-13
Valentin, H. E., and Steinbüchel, A. (1994). Application of enzymatically synthesized short-chain-length hydroxy fatty acid coenzyme A thioesters for assay of polyhydroxyalkanoic acid synthases. Appl. Microbiol. Biotechnol. 40, 699–709. doi: 10.1007/s002530050053
Watanabe, Y., Ichinomiya, Y., Shimada, D., Saika, A., Abe, H., Taguchi, S., et al. (2012). Development and validation of an HPLC-based screening method to acquire polyhydroxyalkanoate synthase mutants with altered substrate specificity. J. Biosci. Bioeng. 113, 286–292. doi: 10.1016/j.jbiosc.2011.10.015
Keywords: polyhydroxyalkanoate, PHA synthase, copolymer composition, site-directed mutagenesis, homology modeling
Citation: Harada K, Kobayashi S, Oshima K, Yoshida S, Tsuge T and Sato S (2021) Engineering of Aeromonas caviae Polyhydroxyalkanoate Synthase Through Site-Directed Mutagenesis for Enhanced Polymerization of the 3-Hydroxyhexanoate Unit. Front. Bioeng. Biotechnol. 9:627082. doi: 10.3389/fbioe.2021.627082
Received: 08 November 2020; Accepted: 28 January 2021;
Published: 03 March 2021.
Edited by:
Huibin Zou, Qingdao University of Science and Technology, ChinaReviewed by:
Yanning Zheng, Institute of Microbiology, Chinese Academy of Sciences, ChinaCopyright © 2021 Harada, Kobayashi, Oshima, Yoshida, Tsuge and Sato. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Takeharu Tsuge, dHN1Z2UudC5hYUBtLnRpdGVjaC5hYy5qcA==; Shunsuke Sato, U2h1bnN1a2UuU2F0b0BrYW5la2EuY28uanA=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.