
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
SYSTEMATIC REVIEW article
Front. Bioeng. Biotechnol. , 17 March 2021
Sec. Cell and Gene Therapy
Volume 9 - 2021 | https://doi.org/10.3389/fbioe.2021.626315
This article is part of the Research Topic Non-Viral Gene Therapy Systems for Orthopaedics View all 6 articles
 Piers Wilkinson1,2
Piers Wilkinson1,2 Ilya Y. Bozo3
Ilya Y. Bozo3 Thomas Braxton1,2
Thomas Braxton1,2 Peter Just4
Peter Just4 Elena Jones5
Elena Jones5 Roman V. Deev6
Roman V. Deev6 Peter V. Giannoudis7,8
Peter V. Giannoudis7,8 Georg A. Feichtinger1*
Georg A. Feichtinger1*Bone defects and improper healing of fractures are an increasing public health burden, and there is an unmet clinical need in their successful repair. Gene therapy has been proposed as a possible approach to improve or augment bone healing with the potential to provide true functional regeneration. While large numbers of studies have been performed in vitro or in vivo in small animal models that support the use of gene therapy for bone repair, these systems do not recapitulate several key features of a critical or complex fracture environment. Larger animal models are therefore a key step on the path to clinical translation of the technology. Herein, the current state of orthopedic gene therapy research in preclinical large animal models was investigated based on performed large animal studies. A summary and an outlook regarding current clinical studies in this sector are provided. It was found that the results found in the current research literature were generally positive but highly methodologically inconsistent, rendering a comparison difficult. Additionally, factors vital for translation have not been thoroughly addressed in these model systems, and the risk of bias was high in all reviewed publications. These limitations directly impact clinical translation of gene therapeutic approaches due to lack of comparability, inability to demonstrate non-inferiority or equivalence compared with current clinical standards, and lack of safety data. This review therefore aims to provide a current overview of ongoing preclinical and clinical work, potential bottlenecks in preclinical studies and for translation, and recommendations to overcome these to enable future deployment of this promising technology to the clinical setting.
While bone is highly competent at regeneration (Hadjidakis and Androulakis, 2006), a variety of situations can lead to damage that cannot be fully repaired by endogenous mechanisms. One of the most challenging examples is a major traumatic event resulting in significant bone loss, fragmentation, substantial damage to the surrounding soft tissue, or some combination of the above. Alternatively, other common etiologies for impaired healing and indications for augmentation of bone regeneration are surgical resection of a tumor or osteomyelitis (chronic bone infection), spinal fusion, and alveolar ridge atrophy caused by edentulism. Large bone defects that cannot be repaired by endogenous mechanisms leaving a permanent gap in the bone are termed “critical defects” (see Figure 1). Exactly how critical defects are defined is controversial, though sometimes a ratio of defect size to bone length is used. Typically, in humans, a defect of >1–2 cm in length where 50% of the bone circumference is lost will be critical (Lindsey et al., 2006; Spicer et al., 2012; Schemitsch, 2017). It is important to note that defect site and other factors have a major influence and may lead to defects that do not fit these parameters becoming critical or those that do fit them healing fully (Sanders et al., 2014; Schemitsch, 2017).
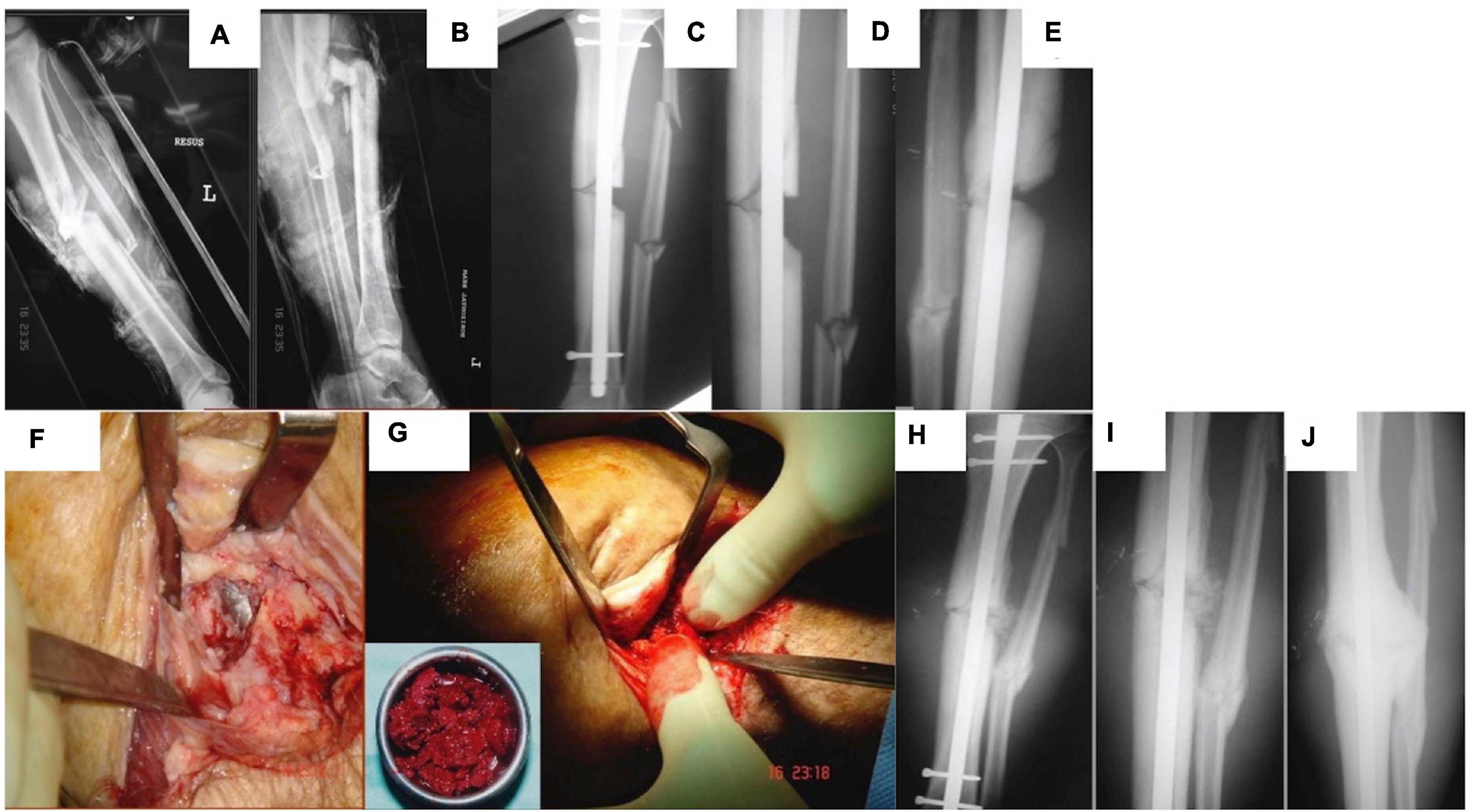
Figure 1. A male patient 29 years of age sustained a left tibial open fracture. (A) Initial anterior–posterior (AP). (B) Lateral radiographs of left tibia showing the midshaft fracture. (C) AP. (D) Lateral tibial radiographs showing that the fracture was stabilized with an intramedullary nail (note the tibial small defect laterally). (E) Lateral tibial radiograph at 4 months demonstrating lack of healing activity. (F) Intraoperative picture at 6 months showing non-union/bone loss and visualization of the nail. (G) Intraoperative picture showing bone grafting of the tibia with autologous bone graft. (H) AP postoperative. (I) Closer AP postoperative radiograph showing the area of small bone defect filled with the autologous bone graft. (J) AP radiograph 3 months lateral showing osseous healing of the tibial small defect.
Complications in small fracture healing are commonly seen in older patients or those suffering with comorbidities, for example, disorders associated with systemic inflammation such as diabetes (Claes et al., 1999, 2012). This is typically manifested as delayed union, while cessation of fracture repair without full defect closure is often termed a “non-union” (Panagiotis, 2005; Tall, 2018). Non-unions are typically filled with fibrotic tissue and have some superficial similarities to a joint (see Figure 1), leading to the alternative name of pseudoarthrosis. With the increasing age of the general population and prevalence of chronic conditions (Anderson and Horvath, 2004; van Oostrom et al., 2016), the clinical burden of impaired fracture healing is likely to increase in the future. Consequently, there has been a great deal of interest in the use of regenerative medicine and tissue engineering to encourage impaired bone repair. Numerous combinations of genes, vectors, proteins, cells, scaffolds, and methods to apply them have been proposed or investigated (Grol and Lee, 2018).
When considering orthopedic bone repair, gene therapy has several advantages over competing methods. The present standard of regenerative, intraoperative care for such defects is a bone autograft. Bone is removed from a healthy donor site on the patient (typically the iliac crest or fibula) and then used to fill the original defect (see Figure 1) or space prefabricated with the Masquelet technique. While this approach now has a high success rate, it is a complex surgical procedure with a risk of donor site morbidity (Kuik et al., 2016). Alternative approaches include synthetic bone substitutes and allo- or xenografts; however, these are limited in osteoinductive potential (Buser et al., 2016; Wang and Yeung, 2017; Haugen et al., 2019; Sohn and Oh, 2019). Recombinant growth factors to encourage endogenous repair have seen use in the clinic; however, their effectiveness is impacted by short biological half-life, immunogenicity in some patients, and a host of other side effects (Talwar et al., 2001; Hwang et al., 2009). To ensure a physiologically relevant level of protein is present long enough to induce an effect of a supraphysiological initial dose (which may have adverse effects) (Aspenberg, 2013; James et al., 2016), repeat applications (clinically challenging), or a sustained release system (technically challenging) is required. Some attempts have been made to develop small molecules to encourage bone regeneration; however, these approaches face similar problems to those of growth factors (Paralkar et al., 2003; Laurencin et al., 2014). Gene therapy can potentially avoid all of these issues by delivering genetic blueprints. A single treatment or application gene therapy can lead to targeted, sustained, and controlled expression of therapeutic gene/s of interest, all of which can be tuned using vector and expression cassette design.
Various gene therapy approaches are now available and intensely studied for clinical translation. An important part of any gene therapy is the vector or method by which genes of interest are introduced to the target cells. Traditionally, a host of different viruses have been used for this purpose, making use of their natural adaptions for cell targeting and entry to deliver genetic information (Lundstrom, 2018). However, viral vectors can be cytotoxic (Büning and Schmidt, 2015) and have the potential to induce an immune response, possibly rendering the initial therapy or follow-up treatments ineffective (Nayak and Herzog, 2010). Furthermore, certain vectors encourage transgene integration in the host genome leading to potentially dangerous insertional mutagenesis effects (Knight et al., 2013; David and Doherty, 2017). Because of this, there is an increasing push toward non-viral gene therapies as a safer and therefore more easily translatable alternative in situations where only short-term transgene expression is required. In such methods, nucleic acids are introduced either “naked” or as part of a synthetic carrier such as a capsule or nanoparticle (Yin et al., 2014). While these methods are deemed to be often safer, the advantageous aspects of viral gene delivery are lost. Non-viral approaches typically show low efficiency and limited transgene persistence, and the host immune responses to both the carrier and its nucleic acid contents are still a concern in terms of safety and efficacy (Al-Dosari and Gao, 2009). A possible approach to mitigate the problems of both approaches is ex vivo gene therapy, a combination of cell and gene therapy (CGT). In such approaches, cells are removed from the patient or donor, transduced or transfected in the lab, screened for successful modification, and then reintroduced to the patient (Gregory-Evans et al., 2012) (see Figure 2). Such approaches are theoretically attractive and have already seen some success in humans in a variety of conditions (Kumar et al., 2016; Hirsch et al., 2017). However, they are expensive, time-consuming, and labor-intensive and introduce a host of new safety and regulatory issues. These limitations act as a major barrier to widespread clinical translation.
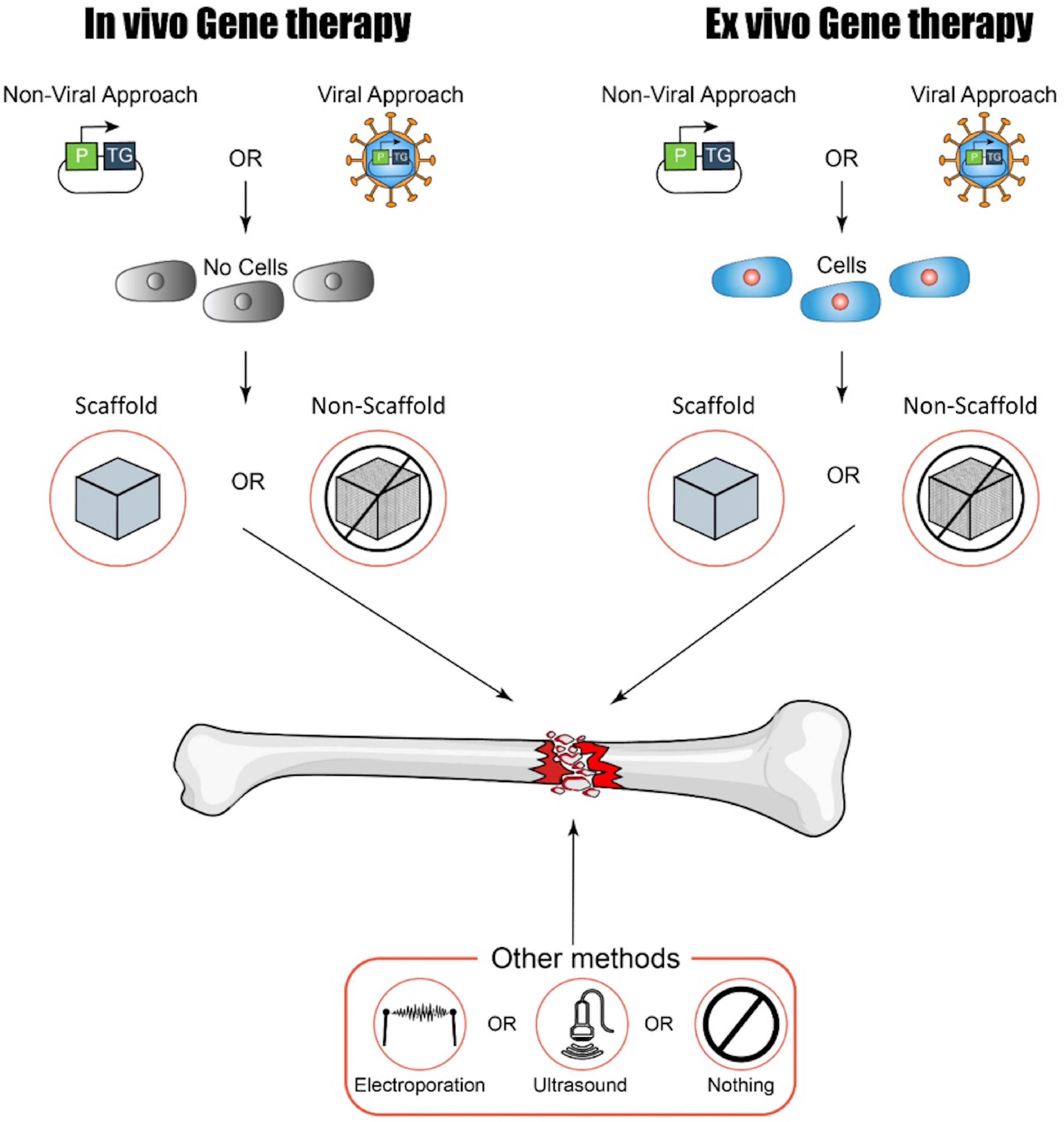
Figure 2. A summary of possible approaches to gene therapy for bone regeneration (kindly provided by D. Ilas). P, promoter; TG, transgene.
Another vital consideration is the therapeutic gene to be delivered. Many potential options have been proposed to encourage bone repair. Perhaps the most commonly used options to date are members of the bone morphogenetic protein (BMP) family of growth factors, due to their vital role in bone development, homeostasis, and repair (Bragdon et al., 2011; Wang et al., 2014). A popular alternative is vascular endothelial growth factor (VEGF) to encourage angiogenesis, vital in such a highly vascularized tissue as bone. These and numerous other options have been reviewed extensively elsewhere (Betz et al., 2018; Grol and Lee, 2018; Shapiro et al., 2018).
Each combination of delivery method, target cell population, and vector has a unique combination of positives and negatives, but common issues include targeting the correct cell population and establishing a suitable dose (Waehler et al., 2007; Yin et al., 2014). New approaches to vector and expression cassette design have been attempted to address these problems, but novel delivery methods may also be able to contribute to technological advancement and translatability. Scaffold or gene-activated matrix (GAM)-based delivery has drawn a great deal of interest from the tissue engineering community. In this approach, the gene therapy is immobilized on a 3D tissue engineering scaffold, allowing precise localization of the gene therapy to site of interest (Raisin et al., 2016; D’Mello et al., 2017). This technology has been developed further with the use of microbubble carriers, which can be sheared using ultrasound, providing unprecedented spatiotemporal control of gene delivery (Zhou et al., 2008; Nomikou et al., 2018). However, such approaches are still in their infancy and require further development.
Research and development of regenerative approaches for bone repair does not differ in terms of its iterative preclinical approach from other biomedical research fields; in fact, the large majority of preclinical animal research has been performed in small rodent models. While small rodent models have many advantages, they often fail to recapitulate key aspects of human biology (Chong et al., 2013; Seok et al., 2013; von Scheidt et al., 2017; King, 2018). Consequently, they can be poor predictors of the behavior of a treatment or therapy in humans. In the case of bone repair, the sheer difference in size, biomechanical loading, and biomechanics is particularly important, but mechanisms and rates of bone remodeling and biomechanics also vary widely between species (Reichert et al., 2009; Wancket, 2015; McGovern et al., 2018). Larger animal models are more similar to humans with regard to many of these factors, having the potential to be better models for orthopedic treatments. Rabbits (Inui et al., 1998; Murakami et al., 2002), sheep (Kirker-Head et al., 1998; den Boer et al., 2003), goats (Xu et al., 2005; Lian et al., 2009), dogs (Cook et al., 1994; Sumner et al., 2003), pigs (Lin et al., 2015; Bez et al., 2017), horses (Backstrom et al., 2004; Ishihara et al., 2008), and non-human primates (Andersson et al., 1978; Cook et al., 2002) have all been used as animal models for orthopedic implants or regenerative approaches to bone repair. Porcine bone has similar morphology and microstructure to that of humans; however, pigs are large and difficult to handle (Reichert et al., 2009; Wancket, 2015; Perleberg et al., 2018). Dogs also possess relevant bone physiology in several regards, but their status as a companion animal has prompted public concern over their use (O’Loughlin et al., 2008; McGovern et al., 2018). Sheep display favorable bone qualities, relative ease of handling, and lack of public objection to their use in research (Malhotra et al., 2014) but are still more expensive and difficult to house than are small rodents. There is no animal model that is unambiguously superior for bone research, and practical considerations such as animal handling, housing, and cost, which typically favor smaller models, cannot be ignored.
While preclinical animal experiments are required for the large majority of new drugs and treatments developed in Europe and the United States (Agency, 2008), regulators are currently highly flexible with regard to species choice. Industry guidelines published by the United States Food and Drug Administration (FDA) state that the species should demonstrate a biological response similar to that of humans and that the comparability of physiology and anatomy to humans should play a role (Center for Biologics Evaluation and Research, 2013). The European Medicines Agency (EMA) provides similar guidelines. It remains for the investigator to demonstrate that their species of choice is appropriate with regard to these and several additional factors (Agency, 2015). Testing in a second species is encouraged but sometimes not required, and there is no requirement to use larger animal models even in situations where they may be more representative of human biology.
The PICO (Problem, Intervention, Comparison, and Outcome) question that the review attempts to answer is as follows: Has gene therapy been successfully applied to regenerate bone in large animal models? We also aimed to assess the various methodologies and approaches used across the field, making a thorough survey of methods that have been employed in large animal models. Finally, we assessed the risk of bias (RoB) of all of the publications using an established framework.
The information gathered to answer these questions was then used to conduct a technology readiness assessment (TRA). A TRA aims to estimate the technology readiness level (TRL) of a technology, in this case gene therapy for orthopedic bone regeneration. TRAs were first developed by the National Aeronautics and Space Administration (NASA) to allow consistent discussion of technical maturity across various technologies but have now become a popular assessment technique in many diverse fields. Simply, TRAs rank a technology on a 9- or 10-point scale of TRLs, with higher values indicating technological maturity. In bioscience/medicine, these scores correspond to closeness to clinical translation. TRAs are still uncommon in the fields of biomedicine and biotechnology, and consequently, there is no widely accepted framework for their use. In this case, a slightly modified version of the United States Department of Defense TRA Deskbook guidelines for biomedical TRLs (Office of the Director of Defense Research and Engineering, 2009) has been applied, a highly simplified version of which can be seen in Table 1. More detailed versions of the frameworks for biologics and medical devices are provided in Supplementary Table 4.

Table 1. A simplified biomedical technology readiness level (TRL) framework, based on the United States government DoD TRA Deskbook (Office of the director of defense and engineering, 2009).
The databases MEDLINE, EMBASE, and BIOSIS were searched in August 2019 for terms relating to animal models, gene therapy, and bone. Controlled or curated vocabulary was used for all databases, with an additional topic search added to the BIOSIS search due to a lack of sufficiently specific curated terms. The initial search returned 1,483 publications, which were then taken for filtering (see Figure 3).
See the Supplementary Information for a detailed description of the literature search.
References were exported from the databases into the reference management software EndNote X7. EndNote’s duplicate detection function and a manual search were used to filter for duplicates. A relevance search was then conducted based on the listed inclusion and exclusion criteria.
Inclusion criteria were as follows:
• Use of gene therapy,
• Focus on bone repair,
• Use of large animal models, and
• English language.
Exclusion criteria were as follows:
• Exclusive use of small animal models (rabbits and smaller);
• Exclusively in vitro; and
• Review articles, letters to the editor, and other non-research article types.
A first pass was made by searching the titles for terms associated with exclusion criteria (e.g., “mouse”). Identified titles were manually examined and excluded if a reasonable assumption that exclusion criteria were met could be made (e.g., the publication “Combination therapy with BMP-2 and a systemic RANKL inhibitor enhances bone healing in a mouse critical-sized femoral defect” was excluded at this stage, as there was no indication that a large animal model was used). The remaining publications were manually screened, with both the titles and abstracts consulted. Finally, an availability check was conducted. See Figure 3 for a summary of the search and filtering process.
In order to provide a summary overview of the most common and recurring topics shared by the selected systematic review publications, we have developed a Python script, which automatically creates a visual representation of the most common key phrases, involved authors, journals, and publication years. We provide and maintain the current version as Jupyter Notebook under the following GitHub Repository: https://github.com/intonumbers/pubmed-insights.
First, we have queried the Entrez query and database system at the National Center for Biotechnology Information (NCBI) via their Entrez Programming Utilities (E-utilities) API for the corresponding 33 selected publications (Sayers, 2009).
To extract the most frequent key terms, we have included the values of the following keys per fetched publication dataset:
• Title,
• Abstract,
• Results,
• Keywords, and
• Conclusions.
Our data munging process’s first step was to remove all punctuation marks, special characters, and figures for each of the mentioned key–value pairs and replace all capital letters with their corresponding lowercase letters. We then removed common stop words (detailed stop-words list see GitHub repository) and lemmatized the remaining words using NLTK WordNet’s1 built-in Morphy function. Next, we have formed all n-grams between word-size 2 and 5 for all key–value pairs per publication dataset except for the keywords key–value pair, where we have defined each entry as n-gram regardless of their size.
Afterward, we have removed all n-gram duplicates per publication dataset.
For the final word cloud visualization, we have implemented a weighting score by multiplying the number of publications in which the n-gram occurs by the size of the corresponding n-gram and manually removed less meaningful n-grams (detailed ignore word list, see Supplementary Information). The program was applied to the dataset using the following parameters available in the graphical interface: Cloud Size = 100, Min Grams = 2, Max Grams = 5, Top Journals = 10, Long Gram Weight = ON, Remove incomplete author names = ON, and Remove Isolated Numbers = ON.
The size of each resulting word cloud item represents the magnitude of its n-gram’s score, taking into account the top 100 of the remaining entries. The size of each word cloud item represents the number of publications that the corresponding author name was found in the authors-key of the fetched publication datasets.
A bar chart was generated to depict the number of publications per year of the analyzed publication dataset. Another visualization was generated to show the distribution of the selected publications per journal. Only the top 10 journals based on the number of publications within the current dataset were considered.
The following study characteristics were extracted from each publication: model species, number of animals, defect site/s, defect size and type, fixation method, inclusion and type of cells, inclusion and type of a scaffold/construct, vector, nature of therapy (i.e., in vivo or ex vivo), therapeutic gene/s, promoter of therapeutic gene/s, therapy dose (including carrier information, cell number, and modification efficiency, if available), therapy delivery site, time between creation of defect and administration of therapy, length of experiment, methods used to investigate bone regeneration, and the methods used to investigate immune response to, persistence of, and localization of gene therapy.
A RoB assessment was performed using the Systematic Review Centre for Laboratory animal Experimentation (SYRCLE) RoB tool for animal studies (Hooijmans et al., 2014). The tool is a modified version of the Cochrane group RoB tool, initially developed to standardize assessment of study biases in clinical randomized controlled trials (RCTs). The SYRCLE RoB tool has been modified to account for differences between clinical RCTs and preclinical animal experiments. Briefly, the tool provides a framework to assess if the study took adequate steps to avoid bias through randomization and investigator blinding at various stages, and other factors such as full data reporting. Relevant animal baseline characteristics were decided to be age and weight.
All publications were assessed independently by two reviewers (PW and TB) for both extracted study characteristics and the SYRCLE RoB tool. Results were then compared and discussed, with the consensus view presented here.
Automated machine reading data analysis of all 33 publication abstracts of the manuscripts selected for systematic review illustrates that the most common converging topic areas are centered around the use of bone marrow stromal cells (BMSCs) and BMP genes (Figure 4A). Furthermore, there is an indication of the relatively common use of adenoviral vectors for human BMP (hBMP) gene delivery in several studies. The author word cloud (Figure 4B) illustrates the most prolific authors found in the dataset but is not a representation of overall publication activity or leadership in the gene therapy field, as it might represent a skewed dataset in this regard due to the performed preselection. It gives, however, a suitable overview of the most active authors investigating orthopedic gene therapeutics in large animal models. Finally, publication activity in this field peaked around 2009 (Figure 4C) in the current dataset with a current stagnation since 2013, indicating a potential need for more intense preclinical research activity in this field in order to accelerate the translation of gene therapies for orthopedic indications. The most common journals identified in the current dataset (Figure 4D) are Gene Therapy, the Journal of Orthopaedic Research, and Biomaterials.
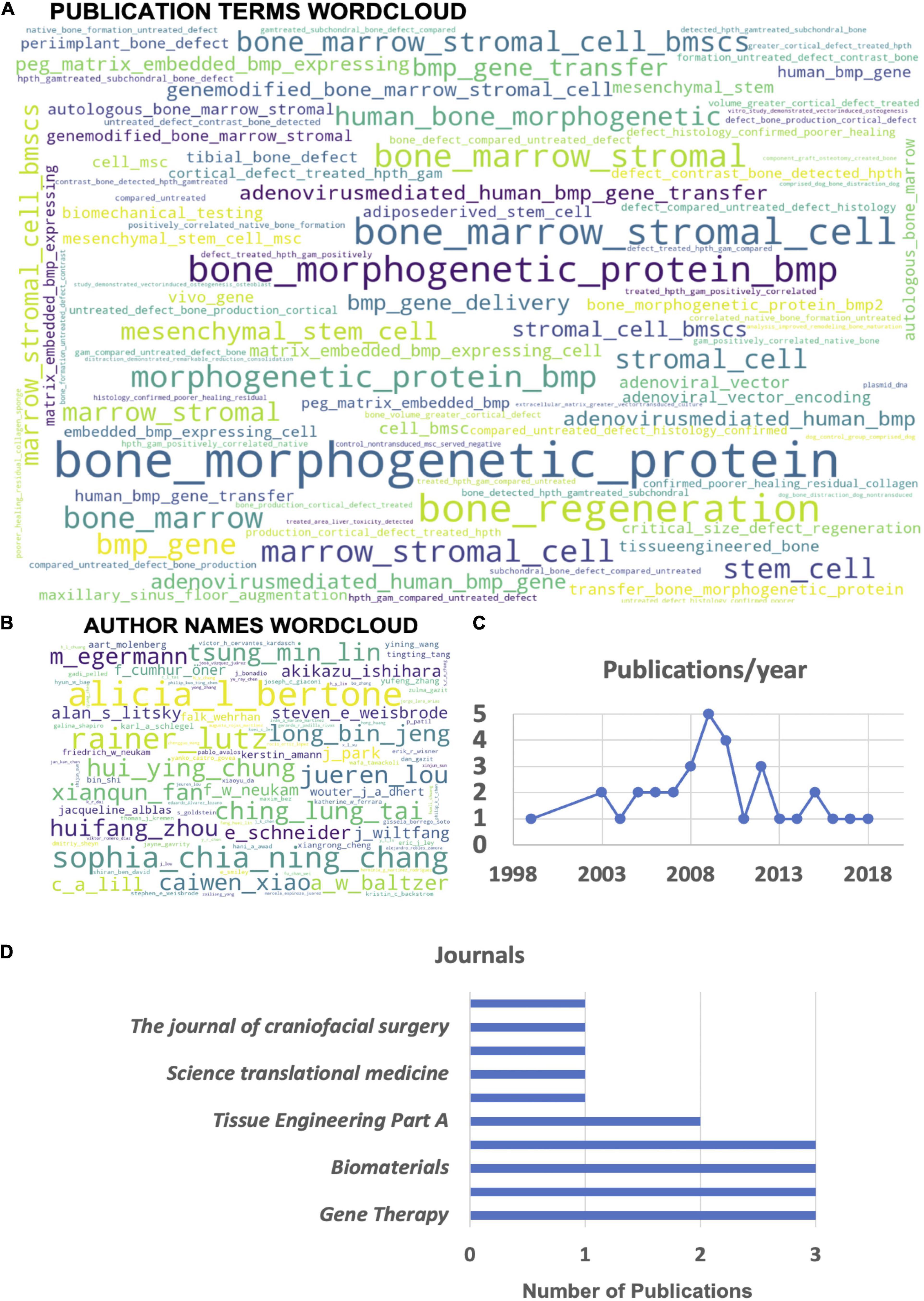
Figure 4. Automated machine reading analysis of the systematic review manuscript dataset (33 publications). (A) Word cloud depicting frequency of different n-grams across the abstracts of all selected publications. (B) Author name word cloud depicting the most common authors appearing on the highest frequency of publications within the analyzed dataset. (C) Overview of the yearly distribution of publication numbers within the dataset. (D) List of the top 10 journals with the most publications in the topic area within the analyzed dataset.
Thirty-three publications were identified as relevant during the search. These publications were found to have used five different species. Dogs were used by eight publications (Bonadio et al., 1999; Zhang et al., 2007, 2009; Xiao et al., 2010; Castro-Govea et al., 2012; Deng et al., 2014; Liu et al., 2016; Kim et al., 2018), goats by five publications (Dai et al., 2005; Xu et al., 2005; Lian et al., 2009; Wegman et al., 2012; Loozen et al., 2015), sheep by three publications (Egermann et al., 2006a, b; Santoni et al., 2008), pigs by four publications (Park et al., 2007; Lutz et al., 2008; Wehrhan et al., 2012, 2013), mini-pigs by eight publications (Chang et al., 2003a, b, 2009, 2010; Chen et al., 2010; Kroczek et al., 2010; Lin et al., 2015; Bez et al., 2017), and horses by five publications (Backstrom et al., 2004; Ishihara et al., 2008, 2009, 2010; Southwood et al., 2012) (see Figure 5).
A wide variety of defect sites were investigated. Eight publications created defects in the maxillofacial region (not including the frontal bone). The specific sites investigated were the mandible (Zhang et al., 2007, 2009; Kroczek et al., 2010; Castro-Govea et al., 2012), inner orbit (Xiao et al., 2010; Deng et al., 2014), sinus (Liu et al., 2016), and infraorbital rim of the maxilla (Chang et al., 2003a). Seven publications created defects in the cranial or calvarial region (including the frontal bone) (Chang et al., 2003b, 2009, 2010; Park et al., 2007; Lutz et al., 2008; Wehrhan et al., 2012, 2013). Eleven publications created defects in the long bones of the limbs. The bones investigated were the tibia (Bonadio et al., 1999; Dai et al., 2005; Xu et al., 2005; Egermann et al., 2006a, b; Santoni et al., 2008; Bez et al., 2017), femur (Bonadio et al., 1999; Lian et al., 2009; Lin et al., 2015), ulna (Chen et al., 2010), and radius (Kim et al., 2018). Eight publications used a handful of other sites. These were the iliac crest (Egermann et al., 2006b; Ishihara et al., 2010; Loozen et al., 2015), various metacarpals and metatarsals (Backstrom et al., 2004; Ishihara et al., 2008; Ishihara et al., 2009; Southwood et al., 2012), ribs (Ishihara et al., 2010), and L1 vertebra (Wegman et al., 2012). Various different types of defect were created. Full-thickness osteotomies of long bones were made by 14 publications (Bonadio et al., 1999; Dai et al., 2005; Xu et al., 2005; Egermann et al., 2006a, b; Ishihara et al., 2008, 2009; Santoni et al., 2008; Lian et al., 2009; Chen et al., 2010; Southwood et al., 2012; Lin et al., 2015; Bez et al., 2017; Kim et al., 2018), with defect sizes ranging from 1 to 50 mm (see Figure 6). Drill holes were used by 11 publications (Bonadio et al., 1999; Backstrom et al., 2004; Egermann et al., 2006b; Park et al., 2007; Lutz et al., 2008; Chang et al., 2009; Ishihara et al., 2010; Wehrhan et al., 2012, 2013; Loozen et al., 2015). Cuboid and oval defects were used by six publications (Chang et al., 2003a, b, 2010; Lian et al., 2009; Zhang et al., 2009; Liu et al., 2016). A full-thickness osteotomy followed by distraction osteogenesis was used by two publications (Kroczek et al., 2010; Castro-Govea et al., 2012). One publication employed a decortication approach (Wegman et al., 2012).
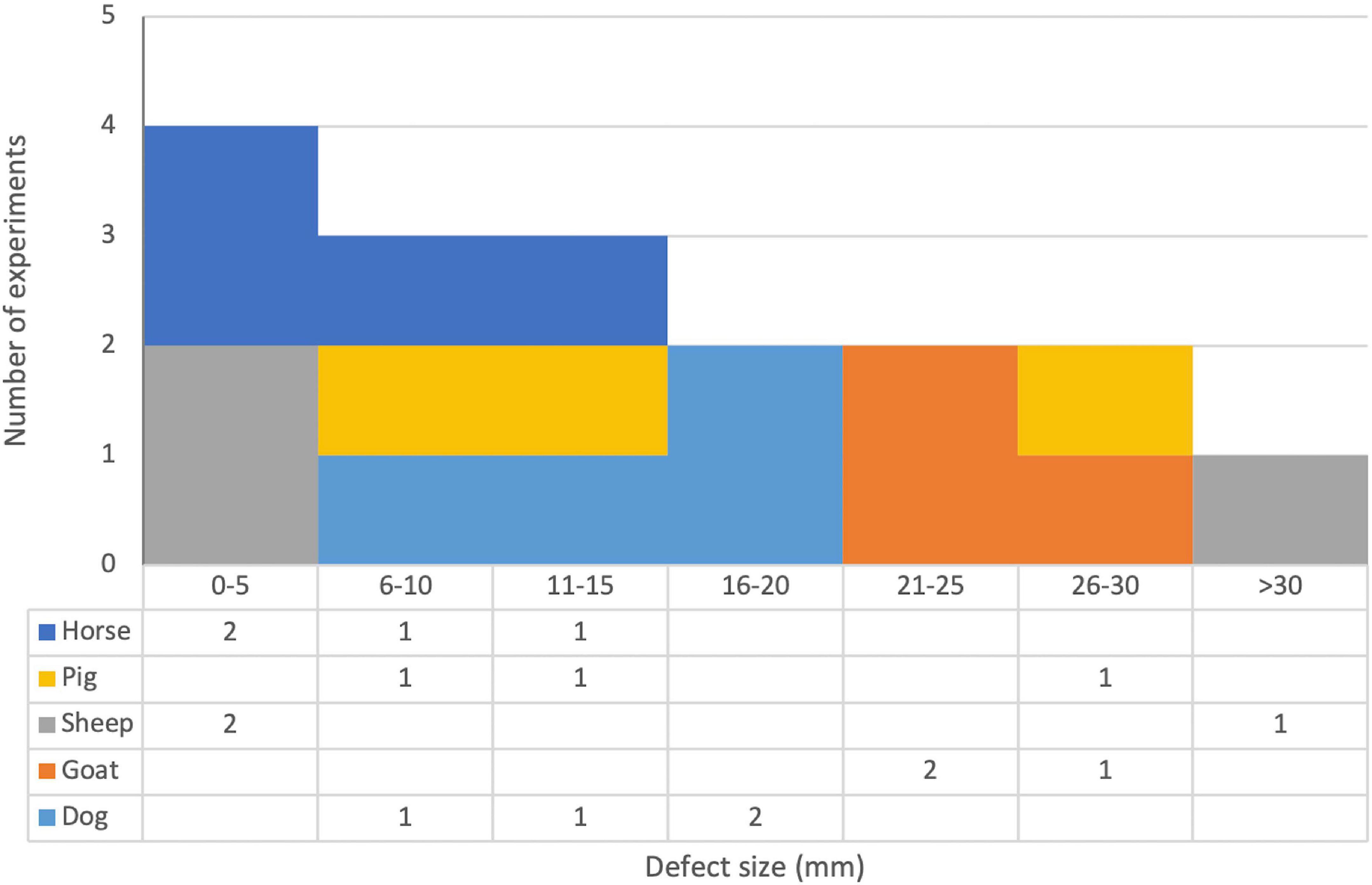
Figure 6. Stacked histogram and data table of defect sizes and species for publications that used full-thickness osteotomy defects in long bones. Note that publications that used multiple different defect sizes are included in all appropriate brackets and are counted twice if they used two different defect sizes that fell within the same bracket. See Table 3 for full details.
In long-bone studies where fixation was potentially necessary, six used internal fixation methods; five used plates (Santoni et al., 2008; Southwood et al., 2012; Lin et al., 2015; Bez et al., 2017; Kim et al., 2018); one used intramedullary rods (Lian et al., 2009); and five used external approaches, with two using plates (Bonadio et al., 1999; Egermann et al., 2006a), two using circular/Ilizarov frames (Dai et al., 2005; Xu et al., 2005), and one using a custom approach (Egermann et al., 2006b). Three studies in long bones did not state if they used fixation; however, these studies all used sites where stability could be provided by other nearby bones (Ishihara et al., 2008, 2009; Chen et al., 2010).
All publications delivered their gene therapy directly into the defect. There was little variability in the length of time between the creation of the defect and the application of gene therapy. The large majority of publications applied their therapy immediately after defect creation, but five left several days between defect creation and therapy application. No chronic models were used. One publication applied their therapy 5 days post defect creation (Kroczek et al., 2010), while four applied their therapies 14 days post defect creation (Ishihara et al., 2008, 2009, 2010; Bez et al., 2017). The length of experiments (measured as the number of days between defect creation and sacrifice of the last experimental group) varied substantially, ranging from 28 (Park et al., 2007; Lutz et al., 2008) to 182 days (Dai et al., 2005) (Mean 93.2, SD 40.7; see Figure 7). The full results for species and model information are presented in Table 2.
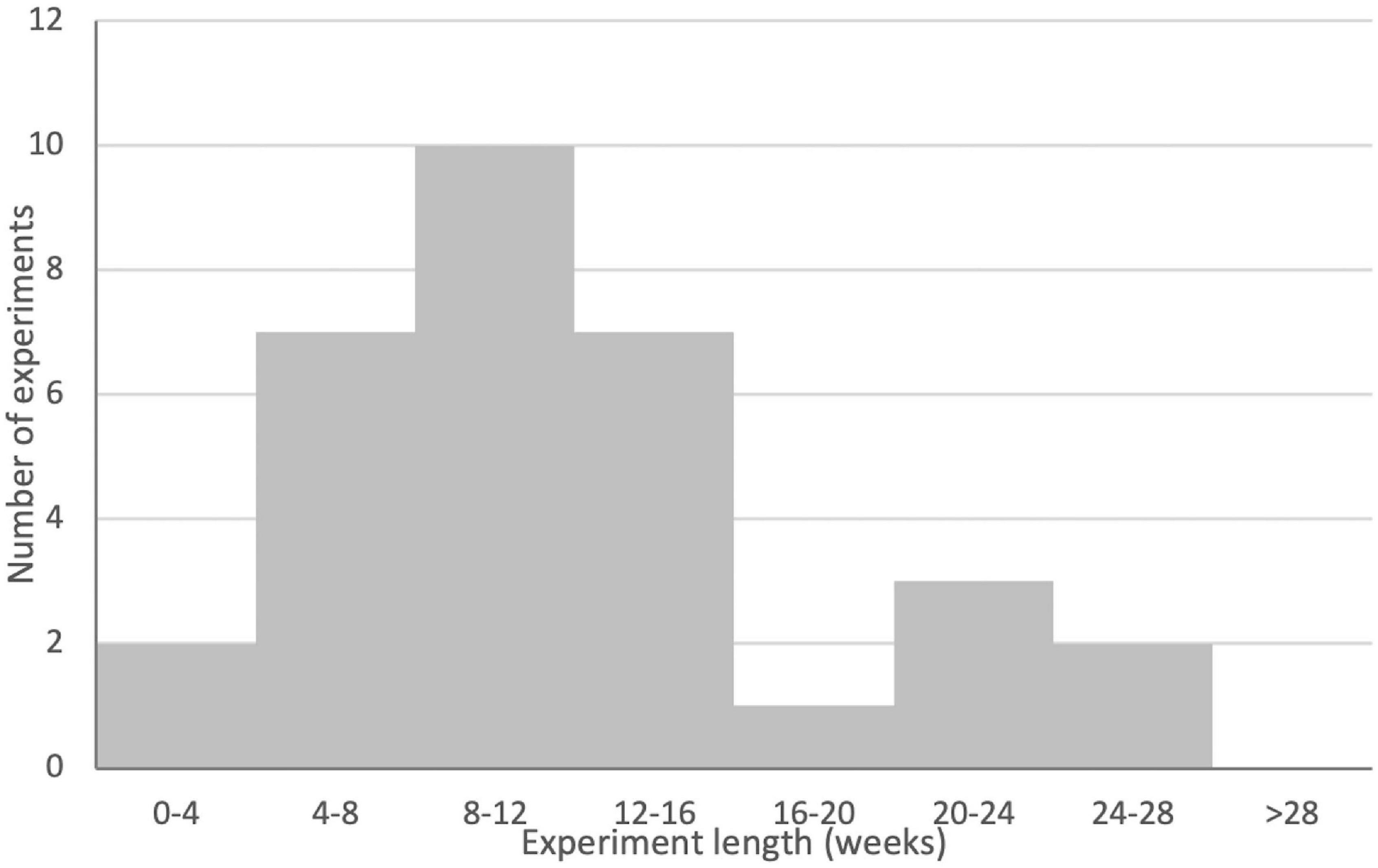
Figure 7. Histogram of experimental lengths for gene therapy groups across the publication set. Note that publications where experimental lengths were not clear or where different gene therapy treatment groups were sacrificed at different times are not included in this figure. Note for binning that experiments that landed on bin boundaries are included in the smaller bin (e.g., a 28-day experiment would be included in the 0- to 4-week bin). See Table 5 for full details.
Twenty-five publications used some form of scaffold or construct to fill the defect they had created, with a wide variety of scaffolds being employed (Figure 8) (Bonadio et al., 1999; Chang et al., 2003a, b, 2009, 2010; Backstrom et al., 2004; Dai et al., 2005; Park et al., 2007; Zhang et al., 2007, 2009; Lutz et al., 2008; Santoni et al., 2008; Lian et al., 2009; Chen et al., 2010; Xiao et al., 2010; Castro-Govea et al., 2012; Wegman et al., 2012; Wehrhan et al., 2012, 2013; Deng et al., 2014; Lin et al., 2015; Loozen et al., 2015; Liu et al., 2016; Bez et al., 2017; Kim et al., 2018). Ten publications used scaffolds that contained multiple substances or were multiphasic (Chang et al., 2003a, 2009; Dai et al., 2005; Zhang et al., 2007, 2009; Wegman et al., 2012; Wehrhan et al., 2012, 2013; Loozen et al., 2015; Kim et al., 2018). Seven publications used scaffolds that were entirely composed of collagen (Bonadio et al., 1999; Chang et al., 2003b, 2010; Backstrom et al., 2004; Park et al., 2007; Lutz et al., 2008; Bez et al., 2017), and four publications used scaffolds that contained it in combination with other materials (Chang et al., 2003a; Dai et al., 2005; Zhang et al., 2007, 2009). One publication used tricalcium phosphate (TCP) alone (Deng et al., 2014), and five used some form of calcium phosphate in combination with other materials (Wegman et al., 2012; Wehrhan et al., 2012, 2013; Loozen et al., 2015; Kim et al., 2018). Two publications used a scaffold solely composed of treated bone matrix (Castro-Govea et al., 2012; Liu et al., 2016), and three used bone matrix in combination with other materials (Dai et al., 2005; Chen et al., 2010; Kim et al., 2018). Two publications employed coral scaffolds (Lian et al., 2009; Xiao et al., 2010), and none used coral in combination with another material. Alginate was used by three publications, two investigated alginate only scaffolds (Chang et al., 2010; Wegman et al., 2012), and two investigated it in combination with other materials (Wegman et al., 2012; Loozen et al., 2015) (note that Wegman et al., 2012 tried both). Chitosan was employed by two publications, both times in combination with collagen (Zhang et al., 2007, 2009). One publication used gelatin in combination with various other materials in a complex multiphasic scaffold (Chang et al., 2009). A variety of biodegradable polymers were used by a small number of publications, largely in combination with other materials. Poly(ε-caprolactone) (PCL) (Kim et al., 2018), poly(L-lactide acid) (PLLA) (Chang et al., 2003a), and poly(ethylene glycol) (PEG) (Wehrhan et al., 2012, 2013) were used with other materials, while one publication used poly(lactic-co-glycolic acid) (PLGA)-only scaffold (Lin et al., 2015). Two studies used autologous bone grafts (Park et al., 2007; Lutz et al., 2008). Eight publications did not use a scaffold or construct (Xu et al., 2005; Egermann et al., 2006a, b; Ishihara et al., 2008, 2009, 2010; Kroczek et al., 2010; Southwood et al., 2012). See Table 3 for full details.
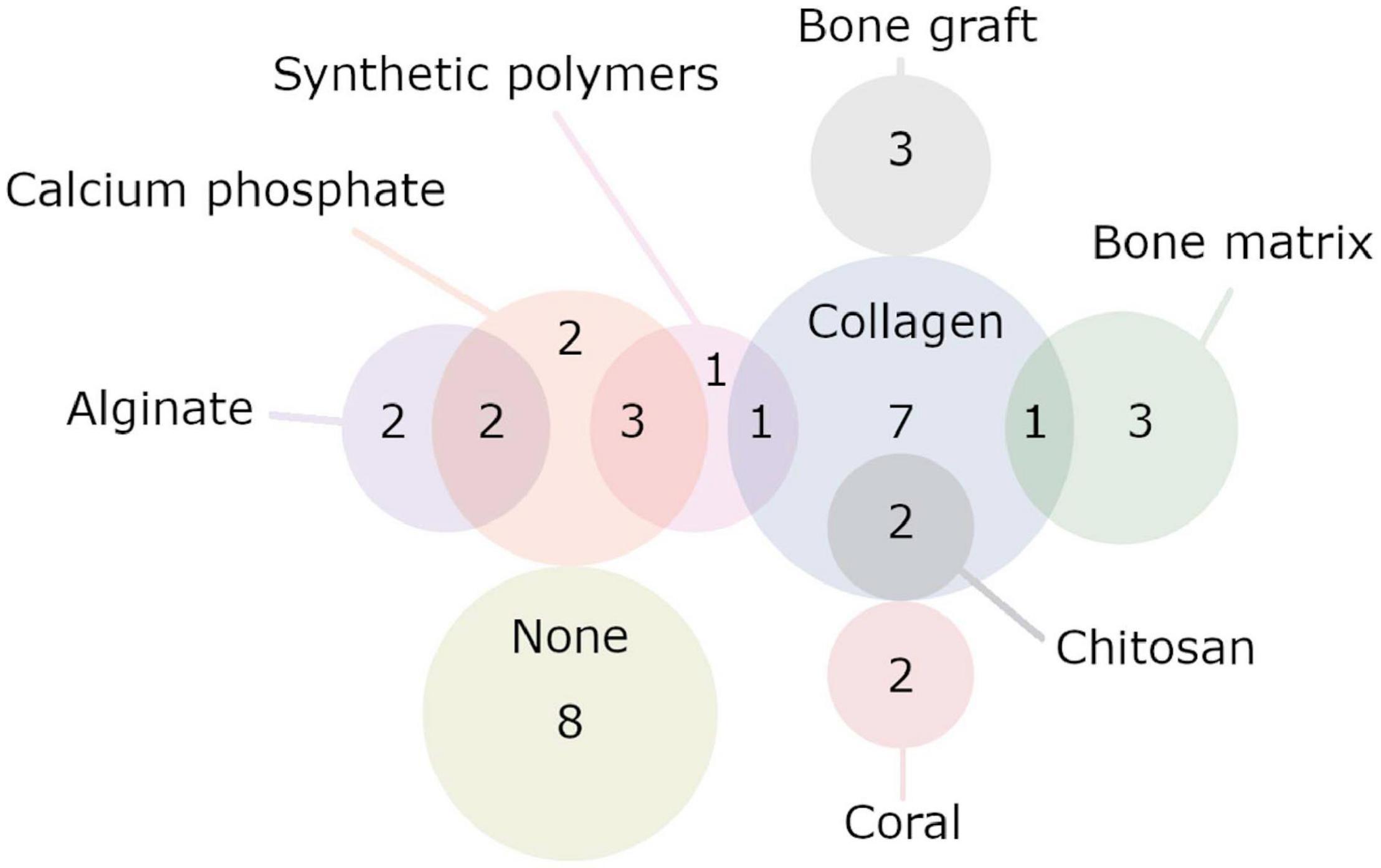
Figure 8. Set diagram representing the various biomaterials used by the publications. Here, only publications that used a single biomaterial or a combination of two are displayed. Two publications that used more complicated scaffolds/constructs made up of three or more materials are not included here. Note that publications were counted multiple times if they used multiple different experimental groups with different biomaterials. Publications included in the “None” group never used a biomaterial; it does not include publications that used a biomaterial but included a no-biomaterial group. See Table 4 for full details.
The vector systems used were less variable (see Figure 9). Approximately two thirds of the publications used viral vectors, accounting for 21 of the 33 publications in the set. Eighteen publications used adenovirus, with 12 clearly indicating that they used serotype 5 (Chang et al., 2003a, b, 2009, 2010; Dai et al., 2005; Xu et al., 2005; Egermann et al., 2006a, b; Ishihara et al., 2008, 2010; Castro-Govea et al., 2012; Southwood et al., 2012). The remaining six did not clearly state which form of adenovirus they used (Zhang et al., 2007, 2009; Ishihara et al., 2009; Lian et al., 2009; Xiao et al., 2010; Deng et al., 2014). Two publications used lentivirus (Liu et al., 2016; Kim et al., 2018), and a single publication used baculovirus (Lin et al., 2015). The remaining 12 publications used non-viral gene delivery methods. Seven used “naked” plasmids (Bonadio et al., 1999; Backstrom et al., 2004; Chen et al., 2010; Wegman et al., 2012; Wehrhan et al., 2012, 2013; Loozen et al., 2015), and five used plasmids with some form of carrier (Park et al., 2007; Lutz et al., 2008; Santoni et al., 2008; Kroczek et al., 2010; Bez et al., 2017). Eighteen publications used ex vivo approaches (Chang et al., 2003a, b, 2009, 2010; Dai et al., 2005; Xu et al., 2005; Ishihara et al., 2009, 2010; Lian et al., 2009; Chen et al., 2010; Xiao et al., 2010; Castro-Govea et al., 2012; Wehrhan et al., 2012, 2013; Deng et al., 2014; Lin et al., 2015; Liu et al., 2016; Kim et al., 2018), 14 used in vivo approaches (Bonadio et al., 1999; Backstrom et al., 2004; Egermann et al., 2006a, b; Park et al., 2007; Zhang et al., 2007, 2009; Ishihara et al., 2008; Lutz et al., 2008; Santoni et al., 2008; Kroczek et al., 2010; Southwood et al., 2012; Loozen et al., 2015; Bez et al., 2017), and one used a hybrid approach, where cells were sometimes included in a gene-activated scaffold (Wegman et al., 2012). See Table 3 for full details.
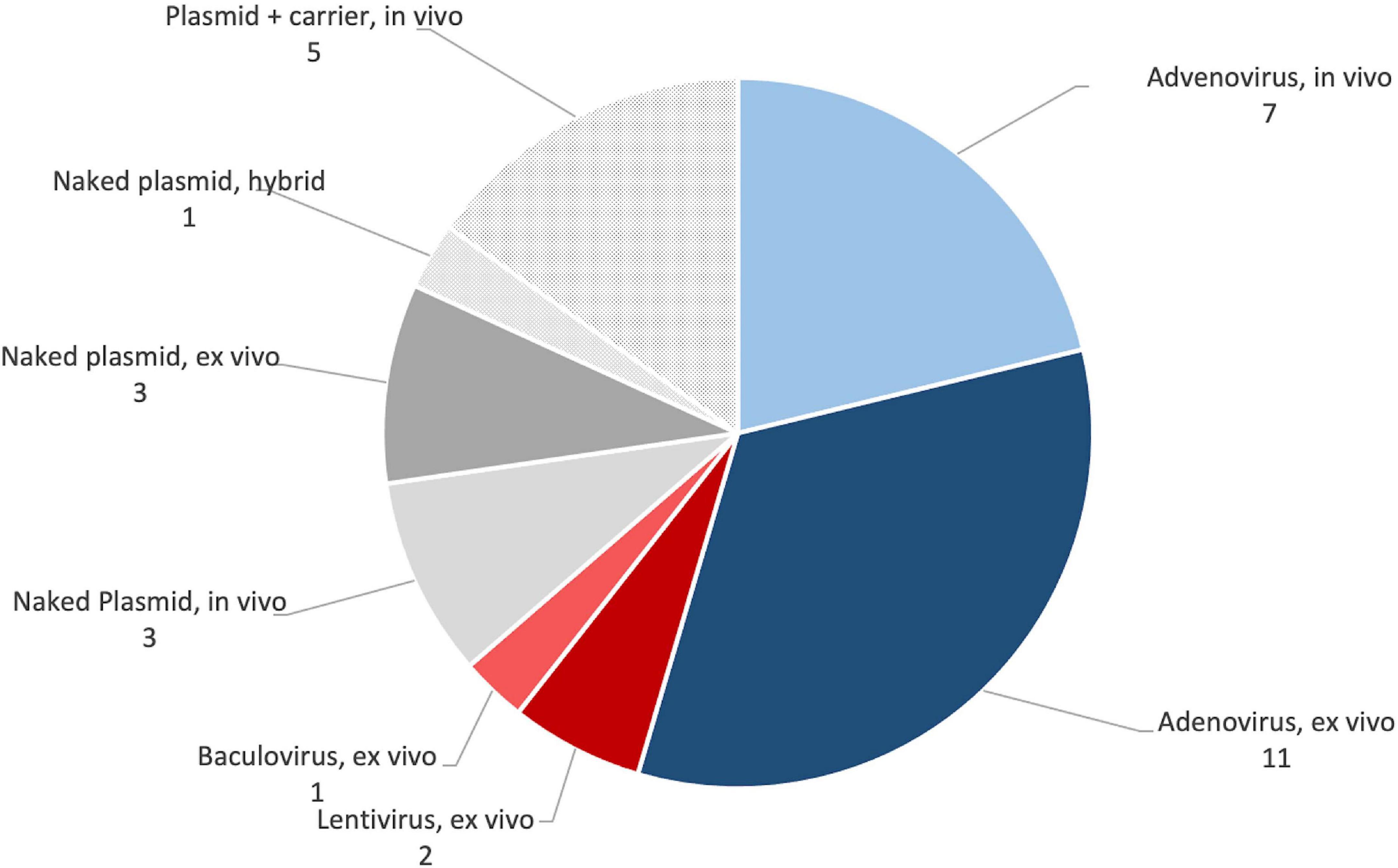
Figure 9. Summary of the popularity of different gene transfer approaches in preclinical studies. See Table 5 for full details.
Therapeutic genes were similarly consistent. Twenty-five publications used BMP-2 (Chang et al., 2003a, b, 2009, 2010; Dai et al., 2005; Xu et al., 2005; Egermann et al., 2006a, b; Park et al., 2007; Ishihara et al., 2008, 2009, 2010; Lutz et al., 2008; Santoni et al., 2008; Chen et al., 2010; Kroczek et al., 2010; Xiao et al., 2010; Castro-Govea et al., 2012; Southwood et al., 2012; Wegman et al., 2012; Wehrhan et al., 2012, 2013; Deng et al., 2014; Lin et al., 2015; Loozen et al., 2015), in three cases in combination with another gene (Chen et al., 2010; Southwood et al., 2012; Deng et al., 2014). Of these, 19 used hBMP-2 (Chang et al., 2003a, b, 2009, 2010; Dai et al., 2005; Xu et al., 2005; Egermann et al., 2006a, b; Park et al., 2007; Ishihara et al., 2008; Lutz et al., 2008; Santoni et al., 2008; Chen et al., 2010; Xiao et al., 2010; Castro-Govea et al., 2012; Southwood et al., 2012; Deng et al., 2014; Lin et al., 2015; Loozen et al., 2015), with six not indicating the species of origin of their gene (Ishihara et al., 2009, 2010; Kroczek et al., 2010; Wegman et al., 2012; Wehrhan et al., 2012, 2013). Five publications used BMP-7, with three using hBMP-7 (Zhang et al., 2007; Lian et al., 2009; Southwood et al., 2012), one using canine BMP-7 (Kim et al., 2018) (in a canine defect model, the only clearly indicated case where the model species and the species of origin of the therapeutic gene matched) and one not indicating the species of origin (Zhang et al., 2009). Three publications used VEGF (Chen et al., 2010; Deng et al., 2014; Lin et al., 2015), in all cases human. In all cases, VEGF was used either in comparison with or simultaneously to BMP-2. Two publications used parathyroid hormone (PTH), in both cases human and not in combination with other genes (Bonadio et al., 1999; Backstrom et al., 2004). BMP-6 was used by two publications (Ishihara et al., 2008; Bez et al., 2017), in both cases human and used in isolation, although compared with BMP-2 in one case. Individual publications used DMP1 (Liu et al., 2016) (alone) and PDGF-B (Zhang et al., 2009) (with or without BMP-7). In both cases, the species of origin of the genes was not indicated. See Table 4 for full details of the therapeutic genes used.
Promoter choice for expression cassettes was highly consistent. Twenty-three publications used cytomegalovirus (CMV) promoter or variants such as immediate-early (CMV-IE) (Bonadio et al., 1999; Chang et al., 2003a, b, 2009, 2010; Dai et al., 2005; Xu et al., 2005; Egermann et al., 2006a, b; Park et al., 2007; Zhang et al., 2007; Ishihara et al., 2008, 2010; Lutz et al., 2008; Kroczek et al., 2010; Castro-Govea et al., 2012; Southwood et al., 2012; Wehrhan et al., 2012, 2013; Deng et al., 2014; Lin et al., 2015; Loozen et al., 2015; Bez et al., 2017). One publication used the human elongation factor-1 alpha (EF-1α) promoter (Kim et al., 2018). Nine publications did not clearly state which promoter was used (Backstrom et al., 2004; Santoni et al., 2008; Ishihara et al., 2009; Lian et al., 2009; Zhang et al., 2009; Chen et al., 2010; Xiao et al., 2010; Wegman et al., 2012; Liu et al., 2016). See Table 4 for full details.
Twenty publications used cells as part of their therapy. Autologous BMSCs were overwhelmingly the most popular option, being used by 11 publications (Chang et al., 2003a, b, 2009, 2010; Dai et al., 2005; Xu et al., 2005; Lian et al., 2009; Xiao et al., 2010; Castro-Govea et al., 2012; Deng et al., 2014; Liu et al., 2016). Various other cell types were used by two publications each. These were autologous fibroblasts (Ishihara et al., 2009, 2010), allogeneic BMSCs (Wegman et al., 2012; Loozen et al., 2015), allogeneic adipose-derived mesenchymal stem cells (ADSCs) (Lin et al., 2015; Kim et al., 2018), and human fetal osteoblasts (Wehrhan et al., 2012, 2013). Autologous ADSCs were used by one publication (Chen et al., 2010). Thirteen publications did not use cells as a part of their gene therapy (Bonadio et al., 1999; Backstrom et al., 2004; Egermann et al., 2006a, b; Park et al., 2007; Zhang et al., 2007, 2009; Ishihara et al., 2008; Lutz et al., 2008; Santoni et al., 2008; Kroczek et al., 2010; Southwood et al., 2012; Bez et al., 2017). See Table 3 for details of the cells used by the publications. Doses were highly variable and reported in several different and non-comparable ways depending on several factors including vector, use of cells, use of scaffolds/constructs, and in vivo or ex vivo application. Frequently, scaffold-based applications (largely ex vivo) did not provide information regarding multiplicity of infection and/or gene transfer efficiency. Sixteen publications failed to provide one or both of these pieces of information (Chang et al., 2003a, b, 2010; Dai et al., 2005; Zhang et al., 2007, 2009; Lian et al., 2009; Chen et al., 2010; Xiao et al., 2010; Castro-Govea et al., 2012; Wegman et al., 2012; Wehrhan et al., 2012, 2013; Lin et al., 2015; Loozen et al., 2015; Kim et al., 2018). The picture is further confused by several publications that formally used in vivo approaches but added unmodified ex vivo cells to their gene-activated scaffolds shortly before implantation. In some cases, the number of cells was provided, but the quantity of vector in the scaffold was not (Zhang et al., 2007; Zhang et al., 2009). In a handful of cases, the number of cells applied to each scaffold was either not measured or not clearly provided (Castro-Govea et al., 2012; Loozen et al., 2015; Liu et al., 2016; Kim et al., 2018). Non-viral in vivo approaches accounted for five publications. The two publications that used naked DNA used quantities measured in tens of milligrams (Bonadio et al., 1999; Backstrom et al., 2004), while the three publications that used carrier agents used either 12 μg (Park et al., 2007; Lutz et al., 2008) or 1 mg (Bez et al., 2017). See Table 4 for dosage information. Unfortunately, it was not possible to make accurate estimates of the dose in terms comparable with human clinical doses (mg DNA/kg bodyweight) due to lack of information regarding the animals used. Often, only the range of weights of animals was provided with no measurement of central tendency, and furthermore, authors frequently did not provide information regarding animal breed or variety to allow a more accurate estimation. Estimated doses varied hugely, from 5.33 × 10–5 to 7.69 mg/kg (data available on request). All four scaffold-free viral in vivo approaches used roughly similar amounts of vector, between 1 × 1011 and 5 × 1011 particles (Egermann et al., 2006a, b; Ishihara et al., 2008; Southwood et al., 2012). Dosage estimates in particles/kg body weight were calculated based on estimated animal weight. Again, measures of central tendency were not provided by three of these publications, so rough estimates were made from the range of animal weights. Estimated viral doses were much more consistent than those of non-viral approaches, ranging between ∼4.7 × 108 and ∼1.6 × 109 particles/kg (data available on request).
Four families of techniques were commonly used to investigate regenerated tissue. These were 2D radiograph, CT and μCT, histology and biomechanical testing (see Figure 10). Two-dimensional radiographs were used by 12 publications (Bonadio et al., 1999; Backstrom et al., 2004; Dai et al., 2005; Xu et al., 2005; Egermann et al., 2006b; Ishihara et al., 2008, 2009; Santoni et al., 2008; Lian et al., 2009; Chen et al., 2010; Castro-Govea et al., 2012; Southwood et al., 2012) and not used by 21 (Chang et al., 2003a, b, 2009, 2010; Egermann et al., 2006a; Park et al., 2007; Zhang et al., 2007, 2009; Lutz et al., 2008; Ishihara et al., 2010; Kroczek et al., 2010; Xiao et al., 2010; Wegman et al., 2012; Wehrhan et al., 2012, 2013; Deng et al., 2014; Lin et al., 2015; Loozen et al., 2015; Liu et al., 2016; Bez et al., 2017; Kim et al., 2018). CT or μCT was used by 16 publications (Chang et al., 2003a, b, 2009, 2010; Backstrom et al., 2004; Dai et al., 2005; Egermann et al., 2006a, b; Ishihara et al., 2008, 2009, 2010; Xiao et al., 2010; Deng et al., 2014; Lin et al., 2015; Bez et al., 2017; Kim et al., 2018) and not used by 17 (Bonadio et al., 1999; Xu et al., 2005; Park et al., 2007; Zhang et al., 2007, 2009; Lutz et al., 2008; Santoni et al., 2008; Lian et al., 2009; Chen et al., 2010; Kroczek et al., 2010; Castro-Govea et al., 2012; Southwood et al., 2012; Wegman et al., 2012; Wehrhan et al., 2012, 2013; Loozen et al., 2015; Liu et al., 2016). Histology was used by all 33 publications. Various forms of biomechanical testing were used by 13 publications (Chang et al., 2003a, b, 2009, 2010; Dai et al., 2005; Xu et al., 2005; Egermann et al., 2006a, b; Ishihara et al., 2008, 2009; Lian et al., 2009; Lin et al., 2015; Bez et al., 2017), and not used by 20 (Bonadio et al., 1999; Backstrom et al., 2004; Park et al., 2007; Zhang et al., 2007, 2009; Lutz et al., 2008; Santoni et al., 2008; Chen et al., 2010; Ishihara et al., 2010; Kroczek et al., 2010; Xiao et al., 2010; Castro-Govea et al., 2012; Southwood et al., 2012; Wegman et al., 2012; Wehrhan et al., 2012, 2013; Deng et al., 2014; Loozen et al., 2015; Liu et al., 2016; Kim et al., 2018). A handful of other analysis techniques were used. These were microradiography by four publications (Park et al., 2007; Lutz et al., 2008; Kroczek et al., 2010; Southwood et al., 2012); calcein staining by three publications (Zhang et al., 2007, 2009; Xiao et al., 2010); immunohistochemistry for osteogenic markers by three publications (Bonadio et al., 1999; Wehrhan et al., 2013; Loozen et al., 2015); and PET/CT (Lin et al., 2015), radionuclide imaging (Chen et al., 2010), and single-photon imaging (Chen et al., 2010) by one publication each. The remaining 24 publications did not employ any additional methods. Outcome parameter information is provided in full in Supplementary Table 2.
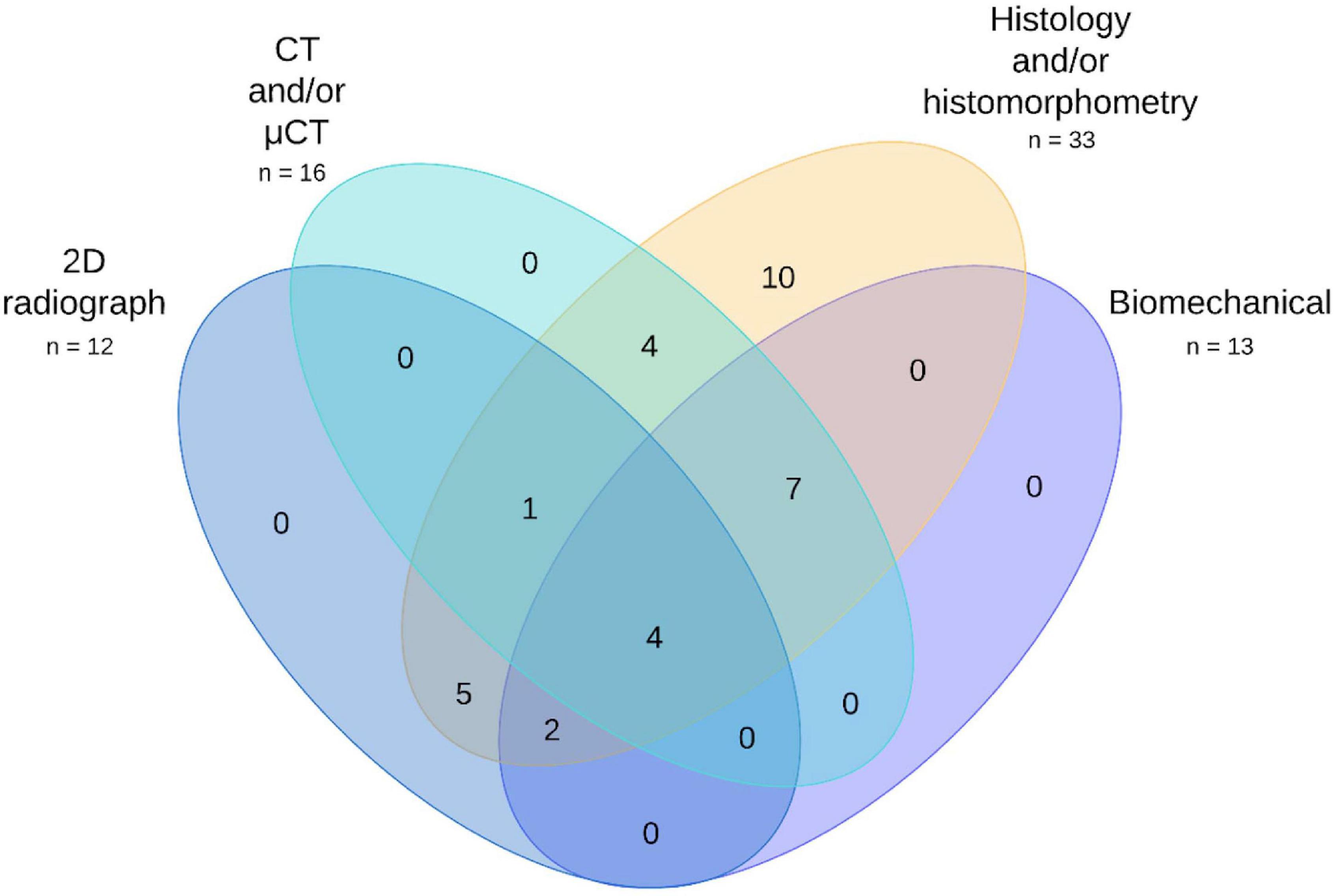
Figure 10. Venn diagram of the most commonly used preclinical evaluation techniques across the publication set. See Supplementary Data for full details.
The investigation of three additional factors was taken as a yardstick for closeness to clinical translation. These factors were immunological response, therapy persistence, and vector biodistribution. Note that many publications investigated vector persistence in vitro; however, only attempts to investigate persistence and the other factors in vivo were included at this stage. Eleven publications made some attempt to investigate the immune response to their therapy (Backstrom et al., 2004; Xu et al., 2005; Egermann et al., 2006a, b; Ishihara et al., 2008; Chen et al., 2010; Castro-Govea et al., 2012; Southwood et al., 2012; Wehrhan et al., 2012, 2013); however, this was often only an assessment of histology. Twenty-two publications made no reference to the immune response to their therapy whatsoever. Nine publications attempted to investigate transgene or vector persistence (Bonadio et al., 1999; Chang et al., 2003a, b; Egermann et al., 2006a, b; Santoni et al., 2008; Ishihara et al., 2010; Wegman et al., 2012; Bez et al., 2017), while 23 did not. Only five publications investigated vector biodistribution (Bonadio et al., 1999; Egermann et al., 2006a, b; Ishihara et al., 2008; Bez et al., 2017), with the remaining 28 failing to do so. Information regarding factors vital for translation is provided in Supplementary Table 1.
All 33 manuscripts used histological techniques that are expensive and difficult to perform. See Table 5 for a summary of methods used. Histological analyses were carried out according to different protocols; therefore, the obtained results have to be considered to be of different reliability. Six of the 33 publications (18%) used only H&E staining (with an additional two using Giemsa–eosin staining), and another two studies used only toluidine blue staining. Multicolor techniques allowing more accurate differentiation of elements in the newly formed tissues—Van Gieson bichrome, Masson trichrome, Masson–Goldner trichrome, and Gomori and Sanderson trichrome—were used in 13 publications (39.4%).
Morphometry and statistical analysis were performed only in 60.6% (n = 20) of studies (Backstrom et al., 2004; Dai et al., 2005; Xu et al., 2005; Egermann et al., 2006a, b; Park et al., 2007; Zhang et al., 2007, 2009; Ishihara et al., 2008, 2009, 2010; Lutz et al., 2008; Santoni et al., 2008; Xiao et al., 2010; Wegman et al., 2012; Wehrhan et al., 2012, 2013; Deng et al., 2014; Loozen et al., 2015; Liu et al., 2016), and in five cases, it did not assess the bone tissue but additional parameters such as the number of blood vessels, the severity of inflammation, and the linear sizes of bone callus. Nineteen studies (57.6%) used a complex technique for sectioning non-decalcified bone that had been previously placed in methyl methacrylate or its analogs, Technovit 9100 and Osteo-Bed (Polysciences) (Backstrom et al., 2004; Egermann et al., 2006a, b; Park et al., 2007; Zhang et al., 2007, 2009; Ishihara et al., 2008, 2009, 2010; Lutz et al., 2008; Santoni et al., 2008; Xiao et al., 2010; Wegman et al., 2012; Wehrhan et al., 2012, 2013; Deng et al., 2014; Loozen et al., 2015; Liu et al., 2016; Bez et al., 2017), but only eight of them (24.2% of all publications) then used the methods for assessment of dynamic indicators of bone formation [measure mineralizing surface (MS), mineral apposition rate (MAR), and bone formation rate (BFR)] (Backstrom et al., 2004; Zhang et al., 2007, 2009; Santoni et al., 2008; Ishihara et al., 2009, 2010; Xiao et al., 2010; Loozen et al., 2015), the main reason for using this technique. Static morphometric indicators were evaluated in all experiments where morphometry was performed, and these included the following: the proportion of bone tissue (BT/TV); the percentage of cartilage and connective tissue in the newly formed tissues; the areas of remaining bone substitute; and the length of bone tissue in direct contact with bone substitute (the percentage of available scaffold perimeter in contact to bone) or to a metal implant (bone-to-implant contact).
Von Kossa staining, a method commonly used to characterize mineralization of tissues, was used in 12% of the publications (n = 4) (Chang et al., 2003a, b, 2010; Castro-Govea et al., 2012). It is important to note that this method cannot be used to quantify mineralization and may mask important features of tissue reactions due to black color on histological slides. For these reasons, it is of limited value when assessing regeneration.
Immunohistochemical technique for detecting viral particles, expression of the protein encoded by transgene, and markers of cell differentiation within the newly formed tissues were used in 33.3% of all studies (n = 11) (Bonadio et al., 1999; Chang et al., 2003a, b; Park et al., 2007; Santoni et al., 2008; Kroczek et al., 2010; Wegman et al., 2012; Wehrhan et al., 2012, 2013; Lin et al., 2015; Loozen et al., 2015).
The SYRCLE RoB assessment revealed that a large majority of the publications had not considered or mentioned most factors covered by the tool (detailed analysis available on request). Sequence generation (that is, the assignment of animals to groups) was only randomized by six publications, with none of these stating which randomization method was used. Six additional publications indicated they had randomized treatment sites within animals or used a blocking strategy, although again none stated which randomization method was used. The remaining 21 publications made no indication that randomization had been considered. Only one publication clearly indicated that animal baseline characteristics had been considered during group allocation. Two others stated that they had tried to balance groups based on only a subset of our defined baseline characteristics or additional factors. The remaining 30 publications did not address group balance. The assessment factors of group allocation concealment, random animal housing, investigator blinding at the treatment stage, and random assessment at the outcome stage were not considered by any publications. Investigator blinding at the analysis stage was performed by two publications for portions of their data. No publications blinded investigators for all of their data. The remaining 31 papers did not indicate that blinding at the data analysis stage had been considered. Thirteen publications clearly addressed the subject of incomplete experimental outcomes, either stating none had occurred or listing their number and nature. Only one publication clearly stated that there had been incomplete outcomes during the study but did not provide the number or nature of these events. Nineteen publications did not mention incomplete outcomes. While it is possible none occurred in these studies, it cannot be safely assumed. Selective outcome reporting (not providing data that the methods section indicates was collected, or clear gaps in the methods section) was common, with 29 publications identified to have failed to provide some information. The large majority of these cases were minor, with common omissions including failing to provide non-significant p-values or histology or radiographic images for every time point and group. More serious failings included not clearly indicating numbers of animals used or group sizes, providing no radiographic images despite extensive use of the technique and providing no histology images despite extensive use of the technique. Only four publications were found to have provided all expected data and information. Several other sources of bias not directly covered by the assessment were identified across the publication set. Fourteen publications did not indicate that they had tested if their data were normally distributed prior to applying a parametric statistical test. Rarer issues included exceptionally small group sizes (n = 1 or 2) and use of animals for simultaneous experiments not included in the publication. The full results of the RoB assessment are available on request.
The publications reviewed here cover a wide range of approaches to gene therapy for bone regeneration in large animal models. This is both a blessing and a curse. It is clearly beneficial to attempt different approaches to establish which methods may be the most effective. However, there is currently such little consistency between studies that it is very difficult to compare effectiveness. Some simple trends across the literature can be clearly seen, for example, aspects of vector design, the use of orthotopic test sites, and bone analysis methods. However, almost all other elements of experimental design are highly variable, including crucially model species, defect sites, and specifics of analysis techniques.
While it is difficult to make generalizations, it seems that species choice was primarily influenced by practicality and researcher familiarity rather than advantages of the model system. Defects in horses were typically small and never larger than 15 mm (Southwood et al., 2012), while defects in goats were never smaller than 21 mm (Xu et al., 2005) despite being a substantially smaller species. A similar situation seemed to have occurred in choice of defect site, with little rationale provided as to why various locations where chosen. If we consider a model system to be a species, bone, and defect type, no particular model was used by more than a handful of publications. Porcine cranial or calvarial defects were the most popular, with seven publications (Chang et al., 2003b, 2009, 2010; Park et al., 2007; Wehrhan et al., 2012, 2013); however, even this group was split between pigs and mini-pigs, and defect locations were not described in a consistent manner (clear indications of where defects were created were not provided, with the terms skull, cranium, calvarium, frontal bone, and frontal skull all used by various publications). Frequently, publications used a combination of species, bone, and defect type that was unique. Similarities further break down when experimental techniques are considered. Even within a defined model system, publications used unique combinations of defect size, fixation method, vector, gene/s, cells, scaffold, dose, experimental length, data collection methods, and time points. The importance of defect fixation should not be overlooked, as fixation method and quality influence fracture healing (O’Sullivan et al., 1989; Claes et al., 1999; Hak et al., 2010). This will likely influence experimental results, and the performance of novel approaches likely should be tested using various different fixation approaches prior to translation. Additionally, the precise biomechanics of each defect location are unique and highly dependent on surgical technique. Even in areas that seem to be consistent, there were differences that lead to difficulty in comparing results. For example, approximately one third of publications used 2D radiographs to investigate rates of defect closure, but time points and measurement methods varied considerably between publications. A similar situation was seen with histology and histomorphometry, used by all the publications in the set, where different authors often used completely different and often highly qualitative evaluation parameters. Furthermore, there were additional experimental procedures that were not reviewed (though most often not provided by the publications) that are likely to have some influence on results. The most important was thought to be the specifics of the surgical methods used (e.g., treatment of the periosteum), details of vector design other than transgene and promoter, and the length of time between modification and implantation of cells for ex vivo approaches. Consequently, a direct comparison of results was not possible between the large majority of publications in the set, and even among those that were relatively similar, it was thought to be unproductive due to the numerous differences in experimental design. This made it impossible to thoroughly answer the sub-questions posed at the start of the review.
Despite difficulties in the comparison, there is a general trend of positive results among the set. Near universally, some form of improvement versus control groups was seen, with only two publications showing unambiguously negative results (Santoni et al., 2008; Southwood et al., 2012). It is of course possible that this could be due to positive publication bias, which has been shown to be problematic across a wide swathe of fields and at both investigative and editorial levels (Olson et al., 2002; Hopewell et al., 2009; Mlinarić et al., 2017). Despite this, it seems possible to say that at a very broad level gene therapy does seem able to improve bone regeneration at a proof-of-concept level in these model species. This does provide an answer to the primary question of the review but is admittedly rather unsatisfying.
The generally positive results found by the first portion of the review are called into question by the RoB assessment, which found that investigators are universally doing a poor job of avoiding bias. Of the nine core factors covered by the assessment, no publication satisfactorily addressed more than two, and there was no indication that many of the factors were considered by any of the publications at all. The only two factors considered by a substantial fraction of the set were incomplete outcome reporting and group randomization; and even so, no publications reported randomization methods satisfactorily. In terms of additional factors, the most serious was thought to be potential statistical errors. Though many publications used parametric tests, many failed to check if such approaches were appropriate for their data. Overall, it would seem that researchers are either unaware of or unconcerned by the risks of bias, which can potentially lead to erroneous positive results and cumulatively do great harm to the progress of the field if widespread. For example, in the field of stroke, it has been reported that apparent substantial improvements in outcome can be largely attributed to bias, leading to a great deal of wasted time and resources pursuing treatments that were actually much less effective than first thought (Macleod et al., 2009). Bias is also likely an important factor in the present reproducibility crisis seen throughout biological science and biomedicine (Nuzzo, 2015). Based on the results of this review, it seems that the field of gene therapy for bone regeneration is likely suffering from the same problems as many others. A widespread and concerted push to improve study design in regard to bias is clearly required.
In terms of technology readiness and closeness to translation, it seems that there is still some way to go. While results were generally positive, factors vital for gene therapy translation were routinely ignored. For the purposes of this review, the three factors of immune response, persistence of transgene expression, and biodistribution were taken as a yardstick for closeness to translation. Very few publications attempted to investigate these factors in any capacity, and those that did typically did not perform a thorough investigation (see Supplementary Table 1). Consequently, the reviewed approaches fit within TRL 4 (see Table 1 and Supplementary Table 4). TRL 5 would require in-depth safety studies, which have not been performed in these models. At the very least, more effort needs to be taken to fully characterize where and for how long therapies are working and possible immune responses. Alongside these, several additional factors should be more thoroughly investigated. While many publications demonstrated improved rates of union when compared with controls, the quality of the bone was highly variable or not adequately investigated. Biomechanical and μCT results were most useful in this regard, as they produce quantitative and potentially more comparable data. Several publications showed that their regenerated bone had comparable mechanical properties to native tissue (Chang et al., 2003a, b; Ishihara et al., 2008; Lian et al., 2009; Bez et al., 2017). Other publications found that despite improvements compared with no treatment controls or scaffold-only treatments, the regenerated bone showed significantly worse properties than native tissue (Dai et al., 2005; Xu et al., 2005; Lin et al., 2015). One publication used these methods but failed to compare with native tissue (Ishihara et al., 2009), while most failed to use them at all. A further criticism is the lack of comparison with existing and alternative treatments. Few studies investigated how gene therapy compared with autologous bone graft or recombinant growth factors (Park et al., 2007; Lutz et al., 2008; Kroczek et al., 2010). Only one publication, Bez et al. (2017) demonstrated that the tissue regenerated in their experiment had properties comparable with those of autograft controls across a variety of techniques including crucially mechanical testing. This is likely the clearest demonstration of non-inferiority yet provided in the field; however, it is currently a lone outlier. Others should follow the example of this publication when attempting to demonstrate the non-inferiority of their own approaches.
To improve the comparability of research in the field and help bring gene therapy for bone regeneration closer to translation, we suggest that several principles of experimental design, large animal models, and investigative approaches should be defined and broadly adhered to by the research community. This would help to reduce bias and allow comparisons to be more easily drawn between different studies without restricting investigators to particular therapeutic approaches. In the following section, we propose potential standards in these areas. Our recommendations here are summarized in Supplementary Figure 1. Furthermore, as discussed by Iglesias-López et al. (2019) and in Outlook and Current Status Regarding Clinical Studies and Authorized Clinical Gene Therapeutics for Orthopedics section, it is essential to clearly classify the type of drug developed and applicable translational pathway as well as to perform early engagement with regulatory authorities to inform translation-enabling preclinical studies, ensuring that study design and data procurement would either support or directly feed into preclinical data packages required for investigational new drug (IND)-enabling studies prior to clinical trials.
We encourage investigators to adhere to the “Animal Research: Reporting of In Vivo Experiments” (ARRIVE) guidelines laid out by the United Kingdom National Centre for the replacement, refinement, and reduction of animals in research (NC3Rs) (Kilkenny et al., 2010). Additional guidance should be taken from the “Planning Research and Experimental Procedures on Animals: Recommendations for Excellence” (PREPARE) guidelines from Norecopa, Norway’s National Consensus Platform for the advancement of the 3Rs. We would also recommend that researchers consult the SYRCLE RoB tool to further inform themselves of reporting expectations (Hooijmans et al., 2014). While ARRIVE and the SYRCLE RoB tools are not formally experimental design guides, they can both act as useful sources of information for method and data reporting expectations and therefore inform design considerations required to meet them. We would also recommend investigators examine the relevant standards from the International Organization for Standardization (ISO) to inform themselves as to the knowledge required before beginning the process of translating an approach. While there are presently no ISO standards specifically focusing on gene therapies, many from the ICS 11.100.20: “Biological evaluation of medical devices” family are relevant to GAMs. ISO 10993-1:2018 “Evaluation and testing within a risk management process,” ISO 10993-2:2006 “Animal welfare requirements,” ISO 10993-6:2016 “Tests for local effects after implantation,” and ISO 10993-11:2017 “Tests for systemic toxicity” all contain relevant information. Investigators pursuing cell-based therapies should also consult ISO 13022:2012 “Medical products containing viable human cells—Application of risk management and requirements for processing practices.”
To investigate non-inferiority in comparison with current treatment approaches, we recommend that investigators always include a group treated with the current clinical gold standard, autologous bone graft. Investigators could also include a group treated with a recombinant version of their transgene as an additional positive control.
We propose two different model systems, for load-bearing and non-load-bearing defects.
Load bearing: Sheep, femoral mid-diaphysis full-thickness osteotomy/segmental defect.
Non-load bearing: Mini-pig, calvarial full-thickness oval defect.
For load-bearing defects, we have selected the sheep femoral defect. While it could be argued that pigs are more representative of human bone in some regard (Reichert et al., 2009; Li et al., 2015; Wancket, 2015), sheep are of a comparable weight and have bones of a similar size to humans and are generally considered easier to work with. Additionally, they are a well-established model for long bone load-bearing defects in the regeneration of critical bone defects using other approaches (McGovern et al., 2018). Indeed, other investigators have already suggested sheep for standardized testing of biomaterials in large animals, although using a different defect location and type (Ferguson et al., 2018). For non-load-bearing defects, we have selected mini-pigs for a standard model species. The similarities between pig and human bone have been previously mentioned, and mini-pigs are easier to handle than their larger cousins. Additionally, this review has revealed that porcine calvarial defects are currently the most popular model system in the field; therefore, continued use of the model presents practical advantages.
As we have previously stated in this review, there is no clearly superior animal model for bone regeneration research, and as such, our recommendations here are fairly arbitrary. We are aware of the irony of suggesting our own arbitrary guidelines after criticizing authors for not providing adequate justification for model choice; however, we hope that the very act of standardization could prove highly beneficial to the field. If these suggestions prove to be impractical, we hope they will at least spur discussion of more appropriate standardized options.
We recommend authors place a greater emphasis on quantitative techniques such as biomechanical testing and μCT imaging and reduce their reliance on qualitative and semi-quantitative approaches such as histology and 2D radiographs. Quantitative techniques are vital for formally demonstrating the non-inferiority of novel approaches to the existing clinical gold standard and would allow a more valuable comparison of data between studies. Qualitative and semi-quantitative methods still have value as supporting information, and standardization of approaches would further improve their usefulness. Histology can provide extremely useful information that is not provided by other methods if good standards of practice, blinding, and evaluation are maintained. Novel, automated methods in digital histology and machine learning approaches certainly have the potential to revolutionize this approach and provide reliable results in the future (Aeffner et al., 2019). Undoubtedly, these techniques provide useful information, and we do not want to discourage investigators from their use; however, we advise against over-reliance. A combination of various quantitative and qualitative techniques will provide the best picture of examined tissue, adding real value to the study.
In summary, we would like to suggest possible standards for improving the quality, comparability, and reproducibility of outcome measures and procedures for future preclinical work in the field of orthopedic gene therapy (Supplementary mindmap in Supplementary Figure 1) in addition to the recommended use of model systems previously suggested.
To date, gene therapy for orthopedics has seen little use in the clinic, with only a single case of clinical translation. While numerous clinical trials have been carried out for osteoarthritis (Evans et al., 2018) and different types of myodystrophies, only two clinical studies have been initiated for bone regeneration (trial numbers NCT02293031 and NCT03076138), both in the Russian Federation. The first of these, for a gene-activated bone substitute based on collagen–hydroxyapatite scaffold containing a VEGFA plasmid, was started in November 2014. Despite the positive results, the study was withdrawn for commercial reasons (Bozo et al., 2016). The same group proceeded with another clinical trial aiming to investigate another gene-activated bone substitute based on an octacalcium phosphate scaffold and a VEGFA plasmid. The results of this study were reported at the TERMIS-EU Meeting in Rhodes 2019 (Bozo et al., 2019). Twenty patients with alveolar ridge atrophy and or mandible defects were successfully treated within 6 months, and no adverse events were observed. Approval for clinical use of this approach in the Russian Federation was granted in April 2019 and is now available for oral and maxillofacial surgeons. Between these two clinical trials, another group based in Kazan, Russia, published a clinical case report of a successful treatment of a patient with ulnar pseudarthrosis using the GAM based on demineralized bone allograft and a dual cassette plasmid encoding VEGFA and BMP2 (Masgutov et al., 2017).
Gene therapy for bone regeneration is now clearly on its way to clinical practice; however, the clinical data are currently limited, and no human trials have been performed outside the Russian Federation. Continued successful clinical translation of regenerative orthopedic gene therapy products is limited by several factors closely associated with the particular complexities of different regulatory environments. An excellent review of EMA and FDA frameworks for advanced therapy medicinal products (ATMPs) has been provided by Iglesias-López et al. (2019). This diversity in definitions, legal recommendations, and requirements for the translation of gene therapies for regenerative medicine applications across different regulatory framework increases risk and cost and leads to extensive translational timeframes for gene therapy developers in this space. In both jurisdictions, all types of advanced therapies discussed in this review would fall under the classification of medicines/biological products with differences in terms of further granularity and classification [e.g., gene therapy medicinal product (GTMP), combined ATMP (cATMP), and CGT products]. The extent and effect on translation of global regulatory differences and the diversity in interpretation and preclinical and clinical study requirements of orthopedic gene therapy approaches can be illustrated using the specific example of GAM translation using a biomaterial in conjunction with a gene therapy vector. In both EMA and FDA jurisdiction, this approach would be considered an ATMP. GAMs, for example, are considered to be combination products under FDA regulations. Similarly, under EMA regulations, they fulfill the definition of a GTMP as defined in Article 2(1) of Regulation (EC) No. 1394/2007 and a cATMP as defined in Article 2(1) of Regulation (EC) No. 1394/2007 due to the incorporation of a scaffold. The strict regulatory environments in the United States and EU have led to relatively slow clinical translation pathways, although novel expedited pathways/classifications are available for accelerated translation (e.g., RMAT in the United States). Furthermore, differences in terms of definitions, classification, and regulatory requirements between EMA and FDA regulatory legislation mostly require separate, independent clinical trials and preclinical data (e.g., IND submission requirements in the United States) procurement for each jurisdiction. This is further complicated by the oversight of clinical trials by national regulatory authorities [e.g., Medicines and Healthcare products Regulatory Agency (MHRA) in the United Kingdom] in individual member states in the EU. In EMA regulatory space, a centralized procedure for market authorization is mandatory for ATMPs. Therefore, parallel deployment of a novel gene therapy in the global market within the United States and EU requires significant resources and time and cannot be expedited, as for now no harmonized procedure is available covering both regulatory spaces. The regulatory requirements for translation of somatic gene therapeutics are, however, substantially different inside the Russian Federation. The advantage for rapid, accelerated translation of a novel gene therapeutic, in particular for regenerative applications if developed as a combination device with a biomaterial, is that if a material contains biologically active components, the combination product is still defined as a medical device. Only the biological component, in this case the plasmid DNA for GAMs, needs to be separately registered as a drug. The first treatment to be approved in this way was a gene therapy drug comprising plasmid DNA encoding VEGFA (“Neovasculgen®,” HSCI, Russia) for the treatment of chronic lower limb ischemia. After three clinical trials (trial numbers NCT02369809, NCT02538705, and NCT03068585), the drug was approved in the Russian Federation in 2011 (Deev et al., 2017) as a gene therapy. This approval subsequently allowed the use of the authorized Neovasculgen® product in the Nucleostim GAM combination device product, which is now undergoing clinical trials in the Russian Federation or has already completed trials depending on the indication (NCT03076138 and NCT02293031). This peculiarity of the Russian regulatory environment for combination products allowed a phased approach wherein an authorized gene therapy drug can be subsequently combined with a biomaterial and repurposed as a GAM. Thereby, for the new indication of orthopedic gene therapy, an accelerated path to translation can be pursued wherein the medical device pathway can be followed even if the resulting product is a cATMP. This means, however, that for new approaches that are developed, all components would need to follow separate translational pathways (i.e., the gene therapeutic biologic component used follows drug translation pathway and the materials have to follow the device route). Therefore, this can only be regarded as an advantage for accelerated translation if there is already a separate gene therapeutic that can be repurposed and integrated to a biomaterial for the subsequent indication.
This example illustrates that there are still significant worldwide differences in regulatory approaches to ATMP translation and that orthopedic gene therapy in particular encompasses a complex array of different approaches that can be classed as either GTMPs/CGTs or combination approaches with different terminologies, definitions, and regulatory requirements. Even relatively closely aligned regulatory frameworks such as EMA and FDA exhibit significant differences that result in the requirement of independent translational regulatory pathways to market authorization if one desires to bring an orthopedic gene therapy to the clinic for patient benefit. Ultimately, it would be highly desirable to facilitate harmonization and convergence (Iglesias-López et al., 2019) of different regulatory environments dealing with ATMPs to enable accelerated translation of these promising approaches in the future and facilitate cross-jurisdiction accreditation of preclinical and clinical study data. This would not only facilitate the successful deployment of these future therapies within the orthopedic space but also eliminate or at least ameliorate the major bottleneck of complex regulatory requirements and highly diverse global environments. Due to the nature of the complex matter of ATMPs and the current trend of global political compartmentalization and fragmentation, it is difficult to predict whether this would be a far-fetched possibility or an illusion; it would, however, be a clear driver to make clinical gene therapy a more widespread routine intervention in the future.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
PW wrote the major part of the manuscript and performed the systematic review. IB wrote the clinical trials part and edited and reviewed the manuscript. TB was the second reviewer performing the systematic review. PJ wrote the program and performed the machine reading analysis. EJ contributed to the manuscript text and edited and reviewed. RD performed the systematic analysis of histological methods, contributed to the manuscript text, and reviewed and edited the manuscript. PG participated in conceptualization, contributed to the clinical aspects of the manuscript and reviewed and edited the manuscript. GF proposed the manuscript, conceptualized its contents, supervised PW and TB, wrote the translational part of the manuscript, and reviewed and edited and handled submission. All authors contributed to the article and approved the submitted version.
IB is the CEO of the company Histograft, LLC. PJ is the CEO of the company Into Numbers Data Science GmbH. GF works as pro bono, non-salaried consultant for Histograft, LLC.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fbioe.2021.626315/full#supplementary-material
Aeffner, F., Zarella, M. D., Buchbinder, N., Bui, M. M., Goodman, M. R., Hartman, D. J., et al. (2019). Introduction to digital image analysis in whole-slide imaging: a white paper from the digital pathology association. J. Pathol. Inf. 10:9. doi: 10.4103/jpi.jpi_82_18
Agency, E. M. (2008). Non-Clinical Studies Required Before First Clinical use of Gene Therapy Medicinal Products. Amsterdam: European Medicines Agency.
Agency, E. M. (2015). Guideline on the Quality, Non-Clinical and Clinical Aspects of Gene Therapy Medicinal Products. Amsterdam: European Medicines Agency.
Al-Dosari, M. S., and Gao, X. (2009). Nonviral gene delivery: principle, limitations, and recent progress. AAPS J. 11, 671–681.
Anderson, G., and Horvath, J. (2004). The growing burden of chronic disease in America. Public Health Rep. 119, 263–270. doi: 10.1016/j.phr.2004.04.005
Andersson, G. B., Gaechter, A., Galante, J. O., and Rostoker, W. (1978). Segmental replacement of long bones in baboons using a fiber titanium implant. J. Bone Joint Surg. Am. 60, 31–40. doi: 10.2106/00004623-197860010-00005
Aspenberg, P. (2013). Special review: accelerating fracture repair in humans: a reading of old experiments and recent clinical trials. Bonekey. Rep. 2, 244–244.
Backstrom, K. C., Bertone, A. L., Wisner, E. R., and Weisbrode, S. E. (2004). Response of induced bone defects in horses to collagen matrix containing the human parathyroid hormone gene. Am. J. Vet. Res. 65, 1223–1232. doi: 10.2460/ajvr.2004.65.1223
Betz, V. M., Kochanek, S., Rammelt, S., Muller, P. E., Betz, O. B., and Messmer, C. (2018). Recent advances in gene-enhanced bone tissue engineering. J. Gene Med. 20:e3018. doi: 10.1002/jgm.3018
Bez, M., Sheyn, D., Tawackoli, W., Avalos, P., Shapiro, G., Giaconi, J. C., et al. (2017). In situ bone tissue engineering via ultrasound-mediated gene delivery to endogenous progenitor cells in mini-pigs. Sci. Transl. Med. 9:eaal3128. doi: 10.1126/scitranslmed.aal3128
Bonadio, J., Smiley, E., Patil, P., and Goldstein, S. (1999). Localized, direct plasmid gene delivery in vivo: prolonged therapy results in reproducible tissue regeneration. Nat. Med. 5, 753–759. doi: 10.1038/10473
Bozo, I. Y., Deev, R. V., Drobyshev, A. Y., Isaev, A. A., and Eremin, I. I. (2016). World’s first clinical case of gene-activated bone substitute application. Case Rep. Dent. 2016:8648949.
Bozo, I. Y., Drobyshev, A. Y., Komlev, V. S., Isaev, A. A., and Deev, R. V. (2019). Gene-activated matrix based on octacalcium phosphate and plasmid DNA carrying VEGF-A gene in clinical trial: safe and effective for bone grafting. (Rhodes, Greece).
Bragdon, B., Moseychuk, O., Saldanha, S., King, D., Julian, J., and Nohe, A. (2011). Bone morphogenetic proteins: a critical review. Cell Signal. 23, 609–620.
Büning, H., and Schmidt, M. (2015). Adeno-associated vector toxicity-to be or not to be? Mol. Ther. 23, 1673–1675. doi: 10.1038/mt.2015.182
Buser, Z., Brodke, D. S., Youssef, J. A., Meisel, H.-J., Myhre, S. L., Hashimoto, R., et al. (2016). Synthetic bone graft versus autograft or allograft for spinal fusion: a systematic review. J. Neurosurg. 25, 509–516. doi: 10.3171/2016.1.spine151005
Castro-Govea, Y., Cervantes-Kardasch, V. H., Borrego-Soto, G., Martínez-Rodríguez, H. G., Espinoza-Juarez, M., and Romero-Díaz, V. (2012). Human bone morphogenetic protein 2–transduced mesenchymal stem cells improve bone regeneration in a model of mandible distraction surgery. J. Craniofac. Surg. 23, 392–396. doi: 10.1097/scs.0b013e318240fe9b
Center for Biologics Evaluation and Research (2013). Preclinical Assessment of Investigational Cellular and Gene Therapy Products. Silver Spring, MA: FDA.
Chang, S. C. N., Chuang, H. L., Chen, Y. R., Chen, J. K., Chung, H. Y., Lu, Y. L., et al. (2003a). Ex vivo gene therapy in autologous bone marrow stromal stem cells for tissue-engineered maxillofacial bone regeneration. Gene Ther. 10, 2013–2019. doi: 10.1038/sj.gt.3302106
Chang, S. C.-N., Wei, F. C., Chuang, H., Chen, Y. R., Chen, J. K., Lee, K. C., et al. (2003b). Ex vivo gene therapy in autologous critical-size craniofacial bone regeneration. Plastic Reconstr. Surg. 112, 1841–1850. doi: 10.1097/01.prs.0000091168.73462.1a
Chang, S. C. N., Chung, H.-Y., Tai, C.-L., Chen, P. K. T., Lin, T.-M., and Jeng, L.-B. (2010). Repair of large cranial defects by hBMP-2 expressing bone marrow stromal cells: comparison between alginate and collagen type I systems. J. Biomed. Mater. Res. Part A 94, 433–441.
Chang, S. C.-N., Lin, T.-M., Chung, H.-Y., Chen, P. K.-T., Lin, F.-H., Lou, J., et al. (2009). Large-scale bicortical skull bone regeneration using ex vivo replication-defective adenoviral-mediated bone morphogenetic protein—2 gene—transferred bone marrow stromal cells and composite biomaterials. Operat. Neurosurg. 65(6 Suppl.), 75–81.
Chen, Q., Yang, Z., Sun, S., Huang, H., Sun, X., Wang, Z., et al. (2010). Adipose-derived stem cells modified genetically in vivo promote reconstruction of bone defects. Cytotherapy 12, 831–840. doi: 10.3109/14653249.2010.495980
Chong, A. S., Alegre, M. L., Miller, M. L., and Fairchild, R. L. (2013). Lessons and limits of mouse models. Cold Spring Harb. Perspect. Med. 3:a015495. doi: 10.1101/cshperspect.a015495
Claes, L., Recknagel, S., and Ignatius, A. (2012). Fracture healing under healthy and inflammatory conditions. Nat. Rev. Rheumatol. 8, 133–143. doi: 10.1038/nrrheum.2012.1
Claes, L., Heitemeyer, U., Krischak, G., Braun, H., and Hierholzer, G. (1999). Fixation technique influences osteogenesis of comminuted fractures. Clin. Orthop. Relat. Res. 365, 221–229. doi: 10.1097/00003086-199908000-00027
Cook, S. D., Salkeld, S. L., Patron, L. P., Sargent, M. C., and Rueger, D. C. (2002). Healing course of primate ulna segmental defects treated with osteogenic protein-1. J. Invest. Surg. 15, 69–79. doi: 10.1080/08941930290085822
Cook, S. D., Baffes, G. C., Wolfe, M. W., Sampath, T. K., and Rueger, D. C. (1994). Recombinant human bone morphogenetic protein-7 induces healing in a canine long-bone segmental defect model. Clin. Orthop. Relat. Res. 301, 302–312.
Dai, K. R., Xu, X. L., Tang, T. T., Zhu, Z. A., Yu, C. F., Lou, J. R., et al. (2005). Repairing of goat tibial bone defects with BMP-2 gene–modified tissue-engineered bone. Calcif. Tissue Int. 77, 55–61. doi: 10.1007/s00223-004-0095-z
David, R. M., and Doherty, A. T. (2017). Viral vectors: the road to reducing genotoxicity. Toxicol. Sci. 155, 315–325. doi: 10.1093/toxsci/kfw220
Deev, R., Plaksa, I., Bozo, I., and Isaev, A. (2017). Results of an international postmarketing surveillance study of pl-VEGF165 safety and efficacy in 210 patients with peripheral arterial disease. Am.J. Cardiovasc. Drugs 17, 235–242. doi: 10.1007/s40256-016-0210-3
den Boer, F. C., Wippermann, B. W., Blokhuis, T. J., Patka, P., Bakker, F. C., and Haarman, H. J. (2003). Healing of segmental bone defects with granular porous hydroxyapatite augmented with recombinant human osteogenic protein-1 or autologous bone marrow. J. Orthop. Res. 21, 521–528. doi: 10.1016/s0736-0266(02)00205-x
Deng, Y., Zhou, H., Yan, C., Wang, Y., Xiao, C., Gu, P., et al. (2014). In vitro osteogenic induction of bone marrow stromal cells with encapsulated gene-modified bone marrow stromal cells and in vivo implantation for orbital bone repair. Tissue Eng. Part A 20, 2019–2029. doi: 10.1089/ten.tea.2013.0604
D’Mello, S., Atluri, K., Geary, S. M., Hong, L., Elangovan, S., and Salem, A. K. (2017). Bone regeneration using gene-activated matrices. AAPS J. 19, 43–53. doi: 10.1208/s12248-016-9982-2
Egermann, M., Baltzer, A. W., Adamaszek, S., Evans, C., Robbins, P., Schneider, E., et al. (2006a). Direct adenoviral transfer of bone morphogenetic protein-2 cdna enhances fracture healing in osteoporotic sheep. Hum. Gene Ther. 17, 507–517. doi: 10.1089/hum.2006.17.507
Egermann, M., Lill, C. A., Griesbeck, K., Evans, C. H., Robbins, P. D., Schneider, E., et al. (2006b). Effect of BMP-2 gene transfer on bone healing in sheep. Gene Ther. 13, 1290–1299. doi: 10.1038/sj.gt.3302785
Evans, C. H., Ghivizzani, S. C., and Robbins, P. D. (2018). Gene delivery to joints by intra-articular injection. Hum. Gene Ther. 29, 2–14. doi: 10.1089/hum.2017.181
Ferguson, J. C., Tangl, S., Barnewitz, D., Genzel, A., Heimel, P., Hruschka, V., et al. (2018). A large animal model for standardized testing of bone regeneration strategies. BMC Vet. Res. 14:330. doi: 10.1186/s12917-018-1648-0
Gregory-Evans, K., Bashar, A. M., and Tan, M. (2012). Ex vivo gene therapy and vision. Curr. Gene Ther. 12, 103–115. doi: 10.2174/156652312800099607
Grol, M. W., and Lee, B. H. (2018). Gene therapy for repair and regeneration of bone and cartilage. Curr. Opin. Pharmacol. 40, 59–66. doi: 10.1016/j.coph.2018.03.005
Hadjidakis, D. J., and Androulakis, I. (2006). Bone remodeling. Ann. N. Y. Acad. Sci. 1092, 385–396.
Hak, D. J., Toker, S., Yi, C., and Toreson, J. (2010). The influence of fracture fixation biomechanics on fracture healing. Orthopedics 33, 752–755. doi: 10.3928/01477447-20100826-20
Haugen, H. J., Lyngstadaas, S. P., Rossi, F., and Perale, G. (2019). Bone grafts: which is the ideal biomaterial? J. Clin. Periodontol. 46, 92–102. doi: 10.1111/jcpe.13058
Hirsch, T., Rothoeft, T., Teig, N., Bauer, J. W., Pellegrini, G., De Rosa, L., et al. (2017). Regeneration of the entire human epidermis using transgenic stem cells. Nature 551, 327–327. doi: 10.1038/nature24487
Hooijmans, C. R., Rovers, M. M., De Vries, R. B. M., Leenaars, M., Ritskes-Hoitinga, M., and Langendam, M. W. (2014). SYRCLE’s risk of bias tool for animal studies. BMC Med. Res. Methodol. 14:43. doi: 10.1186/1471-2288-14-43
Hopewell, S., Loudon, K., Clarke, M. J., Oxman, A. D., and Dickersin, K. (2009). Publication bias in clinical trials due to statistical significance or direction of trial results. Cochrane Database Syst. Rev. 21:MR000006.
Hwang, C. J., Vaccaro, A. R., Lawrence, J. P., Hong, J., Schellekens, H., Alaoui-Ismaili, M. H., et al. (2009). Immunogenicity of bone morphogenetic proteins. J. Neurosurg. Spine 10, 443–451.
Iglesias-López, C., Agustí, A., Obach, M., and Vallano, A. (2019). Regulatory framework for advanced therapy medicinal products in Europe and United States. Front. Pharmacol. 10:921. doi: 10.3389/fphar.2019.00921
Inui, K., Maeda, M., Sano, A., Fujioka, K., Yutani, Y., Sakawa, A., et al. (1998). Local application of basic fibroblast growth factor minipellet induces the healing of segmental bony defects in rabbits. Calcif. Tissue Int. 63, 490–495. doi: 10.1007/s002239900563
Ishihara, A., Shields, K. M., Litsky, A. S., Mattoon, J. S., Weisbrode, S. E., Bartlett, J. S., et al. (2008). Osteogenic gene regulation and relative acceleration of healing by adenoviral-mediated transfer of human BMP-2 or -6 in equine osteotomy and ostectomy models. J. f Orthop. Res. 26, 764–771. doi: 10.1002/jor.20585
Ishihara, A., Zekas, L. J., Litsky, A. S., Weisbrode, S. E., and Bertone, A. L. (2009). Dermal fibroblast-mediated BMP2 therapy to accelerate bone healing in an equine osteotomy model. J. f Orthop. Res. 28, 403–411.
Ishihara, A., Zekas, L. J., Weisbrode, S. E., and Bertone, A. L. (2010). Comparative efficacy of dermal fibroblast-mediated and direct adenoviral bone morphogenetic protein-2 gene therapy for bone regeneration in an equine rib model. Gene Ther. 17, 733–744. doi: 10.1038/gt.2010.13
James, A. W., Lachaud, G., Shen, J., Asatrian, G., Nguyen, V., Zhang, X., et al. (2016). A review of the clinical side effects of bone morphogenetic protein-2. Tissue Eng. Part B Rev. 22, 284–297.
Kilkenny, C., Browne, W. J., Cuthill, I. C., Emerson, M., and Altman, D. G. (2010). Improving bioscience research reporting: the ARRIVE guidelines for reporting animal research. PLoS Biol. 8:e1000412. doi: 10.1371/journal.pbio.1000412
Kim, Y., Kang, B.-J., Kim, W., Yun, H.-S., and Kweon, O.-K. (2018). Evaluation of mesenchymal stem cell sheets overexpressing BMP-7 in canine critical-sized bone defects. Int. J. Mol. Sci. 19, 2073–2073. doi: 10.3390/ijms19072073
Kirker-Head, C. A., Gerhart, T. N., Armstrong, R., Schelling, S. H., and Carmel, L. A. (1998). Healing bone using recombinant human bone morphogenetic protein 2 and copolymer. Clin. Orthop. Relat. Res. 349, 205–217. doi: 10.1097/00003086-199804000-00026
Knight, S., Collins, M., and Takeuchi, Y. (2013). Insertional mutagenesis by retroviral vectors: current concepts and methods of analysis. Curr. Gene Ther. 13, 211–227. doi: 10.2174/1566523211313030006
Kroczek, A., Park, J., Birkholz, T., Neukam, F. W., Wiltfang, J., and Kessler, P. (2010). Effects of osteoinduction on bone regeneration in distraction: results of a pilot study. J. Cranio Maxillofac. Surg. 38, 334–344. doi: 10.1016/j.jcms.2009.10.010
Kuik, K., Putters, T. F., Schortinghuis, J., Van Minnen, B., Vissink, A., and Raghoebar, G. M. (2016). Donor site morbidity of anterior iliac crest and calvarium bone grafts: a comparative case-control study. J. Cranio Maxillofac. Surg. 44, 364–368. doi: 10.1016/j.jcms.2015.12.019
Kumar, S. R., Markusic, D. M., Biswas, M., High, K. A., and Herzog, R. W. (2016). Clinical development of gene therapy: results and lessons from recent successes. Mol. Ther. Methods Clin. Dev. 3:16034. doi: 10.1038/mtm.2016.34
Laurencin, C. T., Ashe, K. M., Henry, N., Kan, H. M., and Lo, K. W. H. (2014). Delivery of small molecules for bone regenerative engineering: preclinical studies and potential clinical applications. Drug. Dis. Today 19, 794–800. doi: 10.1016/j.drudis.2014.01.012
Li, Y., Chen, S.-K., Li, L., Qin, L., Wang, X.-L., and Lai, Y.-X. (2015). Bone defect animal models for testing efficacy of bone substitute biomaterials. J. Orthop. Transl. 3, 95–104. doi: 10.1016/j.jot.2015.05.002
Lian, Z., Chuanchang, D., Wei, L., Yilin, C., and Jiasheng, D. (2009). Enhanced healing of goat femur-defect using BMP7 gene-modified BMSCs and load-bearing tissue-engineered bone. J. f Orthop. Res. 28, 412–418. doi: 10.1002/jor.20973
Lin, C.-Y., Wang, Y.-H., Li, K.-C., Sung, L.-Y., Yeh, C.-L., Lin, K.-J., et al. (2015). Healing of massive segmental femoral bone defects in minipigs by allogenic ASCs engineered with FLPo/Frt-based baculovirus vectors. Biomaterials 50, 98–106. doi: 10.1016/j.biomaterials.2015.01.052
Lindsey, R. W., Gugala, Z., Milne, E., Sun, M., Gannon, F. H., and Latta, L. L. (2006). The efficacy of cylindrical titanium mesh cage for the reconstruction of a critical-size canine segmental femoral diaphyseal defect. J. Orthop. Res. 24, 1438–1453. doi: 10.1002/jor.20154
Liu, X., Li, Q., Wang, F., and Wang, Z. (2016). Maxillary sinus floor augmentation and dental implant placement using dentin matrix protein-1 gene-modified bone marrow stromal cells mixed with deproteinized boving bone: a comparative study in beagles. Arch. Oral Biol. 64, 102–108. doi: 10.1016/j.archoralbio.2016.01.004
Loozen, L. D., Van Der Helm, Y. J. M., Öner, F. C., Dhert, W. J. A., Kruyt, M. C., and Alblas, J. (2015). Bone morphogenetic protein-2 nonviral gene therapy in a goat iliac crest model for bone formation. Tissue Eng. Part A 21, 1672–1679. doi: 10.1089/ten.tea.2014.0593
Lutz, R., Park, J., Felszeghy, E., Wiltfang, J., Nkenke, E., and Schlegel, K. A. (2008). Bone regeneration after topical BMP-2-gene delivery in circumferential peri-implant bone defects. Clin. Oral Implants Res. 19, 590–599. doi: 10.1111/j.1600-0501.2007.01526.x
Macleod, M. R., Fisher, M., O’collins, V., Sena, E. S., Dirnagl, U., Bath, P. M. W., et al. (2009). Good laboratory practice: preventing introduction of bias at the bench. Stroke 40, e50–e52.
Malhotra, A., Pelletier, M. H., Yu, Y., Christou, C., and Walsh, W. R. (2014). A sheep model for cancellous bone healing. Front. Surg. 1:37. doi: 10.3389/fsurg.2014.00037
Masgutov, R., Chekunov, M., Zhuravleva, M., Masgutova, G., Teplov, O., Salikhov, R., et al. (2017). Use of gene-activated demineralized bone allograft in the therapy of ulnar pseudarthrosis. Case Report. BioNanoScience 7, 194–198. doi: 10.1007/s12668-016-0325-7
McGovern, J. A., Griffin, M., and Hutmacher, D. W. (2018). Animal models for bone tissue engineering and modelling disease. Dis. Model. Mech. 11:dmm033084. doi: 10.1242/dmm.033084
Mlinarić, A., Horvat, M., and Šupak Smolčić, V. (2017). Dealing with the positive publication bias: why you should really publish your negative results. Biochem. Med. 27:30201.
Murakami, N., Saito, N., Horiuchi, H., Okada, T., Nozaki, K., and Takaoka, K. (2002). Repair of segmental defects in rabbit humeri with titanium fiber mesh cylinders containing recombinant human bone morphogenetic protein-2 (rhBMP-2) and a synthetic polymer. J. Biomed. Mater. Res. 62, 169–174. doi: 10.1002/jbm.10236
Nayak, S., and Herzog, R. W. (2010). Progress and prospects: immune responses to viral vectors. Gene Ther. 17, 295–304. doi: 10.1038/gt.2009.148
Nomikou, N., Feichtinger, G. A., Saha, S., Nuernberger, S., Heimel, P., Redl, H., et al. (2018). Ultrasound-responsive gene-activated matrices for osteogenic gene therapy using matrix-assisted sonoporation. J. Tissue Eng. Regen. Med. 12, e250–e260.
Nuzzo, R. (2015). How scientists fool themselves - and how they can stop. Nat. Int. Week J. Sci. 526, 182–185.
Office of the Director of Defense Research and Engineering (2009). Technology Readiness Assessment (TRA) Deskbook, (Washington, DC: US Department of Defense (DoD)).
O’Loughlin, P. F., Morr, S., Bogunovic, L., Kim, A. D., Park, B., and Lane, J. M. (2008). Selection and development of preclinical models in fracture-healing research. JBJS 90, 79–84. doi: 10.2106/jbjs.g.01585
Olson, C. M., Rennie, D., Cook, D., Dickersin, K., Flanagin, A., Hogan, J. W., et al. (2002). Publication bias in editorial decision making. JAMA 287, 2825–2828. doi: 10.1001/jama.287.21.2825
O’Sullivan, M. E., Chao, E. Y., and Kelly, P. J. (1989). The effects of fixation on fracture-healing. JBJS 71, 306–310. doi: 10.2106/00004623-198971020-00022
Paralkar, V. M., Borovecki, F., Ke, H. Z., Cameron, K. O., Lefker, B., Grasser, W. A., et al. (2003). An EP2 receptor-selective prostaglandin E2 agonist induces bone healing. Proc. Natl. Acad. Sci. U.S.A. 100, 6736–6740. doi: 10.1073/pnas.1037343100
Park, J., Lutz, R., Felszeghy, E., Wiltfang, J., Nkenke, E., Neukam, F. W., et al. (2007). The effect on bone regeneration of a liposomal vector to deliver BMP-2 gene to bone grafts in peri-implant bone defects. Biomaterials 28, 2772–2782. doi: 10.1016/j.biomaterials.2007.02.009
Perleberg, C., Kind, A., and Schnieke, A. (2018). Genetically engineered pigs as models for human disease. Dis. Model Mech. 11:dmm030783. doi: 10.1242/dmm.030783
Raisin, S., Belamie, E., and Morille, M. (2016). Non-viral gene activated matrices for mesenchymal stem cells based tissue engineering of bone and cartilage. Biomaterials 104, 223–237. doi: 10.1016/j.biomaterials.2016.07.017
Reichert, J. C., Saifzadeh, S., Wullschleger, M. E., Epari, D. R., Schutz, M. A., Duda, G. N., et al. (2009). The challenge of establishing preclinical models for segmental bone defect research. Biomaterials 30, 2149–2163. doi: 10.1016/j.biomaterials.2008.12.050
Sanders, D. W., Bhandari, M., Guyatt, G., Heels-Ansdell, D., Schemitsch, E. H., Swiontkowski, M., et al. (2014). Critical-sized defect in the tibia: is it critical? Results from the SPRINT trial. J. Orthop. Trauma 28, 632–635. doi: 10.1097/bot.0000000000000194
Santoni, B. G., Simon Turner, A., Wheeler, D. L., Nicholas, R. W., Anchordoquy, T. J., and Ehrhart, N. (2008). Gene therapy to enhance allograft incorporation after host tissue irradiation. Clin. Orthop. Relat. Res. 466, 1921–1929. doi: 10.1007/s11999-008-0297-1
Sayers, E. (2009). “The E-utilities in-depth: parameters, syntax and More,” in Entrez Programming Utilities Help, (Bethesda (MD): National Center for Biotechnology Information (US)). Available online at: https://www.ncbi.nlm.nih.gov/books/NBK25499/
Schemitsch, E. H. (2017). Size matters: defining critical in bone defect size! J. Orthop. Trauma 31(Suppl. 5), S20–S22.
Seok, J., Warren, H. S., Cuenca, A. G., Mindrinos, M. N., Baker, H. V., Xu, W., et al. (2013). Genomic responses in mouse models poorly mimic human inflammatory diseases. Proc. Natl. Acad. Sci. U.S.A. 112, 1167–1172.
Shapiro, G., Lieber, R., Gazit, D., and Pelled, G. (2018). Recent advances and future of gene therapy for bone regeneration. Curr. Osteop. Rep. 16, 504–511. doi: 10.1007/s11914-018-0459-3
Sohn, H.-S., and Oh, J.-K. (2019). Review of bone graft and bone substitutes with an emphasis on fracture surgeries. Biomater. Res. 23:9.
Southwood, L. L., Kawcak, C. E., Hidaka, C., Mcilwraith, C. W., Werpy, N., Macleay, J., et al. (2012). Evaluation of direct in vivo gene transfer in an equine metacarpal IV ostectomy model using an adenoviral vector encoding the bone morphogenetic Protein-2 and Protein-7 Gene. Vet. Surg. 41, 345–354.
Spicer, P. P., Kretlow, J. D., Young, S., Jansen, J. A., Kasper, F. K., and Mikos, A. G. (2012). Evaluation of bone regeneration using the rat critical size calvarial defect. Nat. Protoc. 7, 1918–1929. doi: 10.1038/nprot.2012.113
Sumner, D. R., Turner, T. M., Cohen, M., Losavio, P., Urban, R. M., Nichols, E. H., et al. (2003). Aging does not lessen the effectiveness of TGFbeta2-enhanced bone regeneration. J. Bone Miner Res. 18, 730–736. doi: 10.1359/jbmr.2003.18.4.730
Tall, M. (2018). Treatment of aseptic tibial shaft non-union without bone defect. Orthop. Traumatol. Surg. Res. 104, S63–S69.
Talwar, R., Di Silvio, L., Hughes, F. J., and King, G. N. (2001). Effects of carrier release kinetics on bone morphogenetic protein-2-induced periodontal regeneration in vivo. J. Clin. Periodontol. 28, 340–347. doi: 10.1034/j.1600-051x.2001.028004340.x
van Oostrom, S. H., Gijsen, R., Stirbu, I., Korevaar, J. C., Schellevis, F. G., Picavet, H. S. J., et al. (2016). Time trends in prevalence of chronic diseases and multimorbidity not only due to aging: data from general practices and health surveys. PLoS One 11:e0160264. doi: 10.1371/journal.pone.0160264
von Scheidt, M., Zhao, Y., Kurt, Z., Pan, C., Zeng, L., Yang, X., et al. (2017). Applications and limitations of mouse models for understanding human atherosclerosis. Cell Metab. 25, 248–261. doi: 10.1016/j.cmet.2016.11.001
Waehler, R., Russell, S. J., and Curiel, D. T. (2007). Engineering targeted viral vectors for gene therapy. Nat. Rev. Genet. 8, 573–587.
Wancket, L. M. (2015). Animal models for evaluation of bone implants and devices. Vet. Pathol. 52, 842–850. doi: 10.1177/0300985815593124
Wang, R. N., Green, J., Wang, Z., Deng, Y., Qiao, M., Peabody, M., et al. (2014). Bone Morphogenetic Protein (BMP) signaling in development and human diseases. Genes Dis. 1, 87–105.
Wang, W., and Yeung, K. W. K. (2017). Bone grafts and biomaterials substitutes for bone defect repair: a review. Bioact. Mater. 2, 224–247. doi: 10.1016/j.bioactmat.2017.05.007
Wegman, F., Geuze, R. E., Van Der Helm, Y. J., Cumhur Oner, F., Dhert, W. J., and Alblas, J. (2012). Gene delivery of bone morphogenetic protein-2 plasmid DNA promotes bone formation in a large animal model. J. Tissue Eng. Regen. Med. 8, 763–770. doi: 10.1002/term.1571
Wehrhan, F., Amann, K., Molenberg, A., Lutz, R., Neukam, F. W., and Schlegel, K. A. (2012). PEG matrix enables cell-mediated local BMP-2 gene delivery and increased bone formation in a porcine critical size defect model of craniofacial bone regeneration. Clin. Oral Implants Res. 23, 805–813. doi: 10.1111/j.1600-0501.2011.02223.x
Wehrhan, F., Amann, K., Molenberg, A., Lutz, R., Neukam, F. W., and Schlegel, K. A. (2013). Critical size defect regeneration using PEG-mediated BMP-2 gene delivery and the use of cell occlusive barrier membranes - the osteopromotive principle revisited. Clin. Oral Implants Res. 24, 910–920. doi: 10.1111/j.1600-0501.2012.02489.x
Xiao, C., Huifang, Z., Ge, S., Tang, T., Hou, H., Luo, M., et al. (2010). Repair of orbital wall defects using biocoral scaffolds combined with bone marrow stem cells enhanced by human bone morphogenetic protein-2 in a canine model. Int. J. Mol. Med. 26, 517–525.
Xu, X. L., Tang, T., Dai, K., Zhu, Z. A., Guo, X. E., Yu, C., et al. (2005). Immune response and effect of adenovirus-mediated human BMP-2 gene transfer on the repair of segmental tibial bone defects in goats. Acta Orthop. 76, 637–646. doi: 10.1080/17453670510041709
Yin, H., Kanasty, R. L., Eltoukhy, A. A., Vegas, A. J., Dorkin, J. R., and Anderson, D. G. (2014). Non-viral vectors for gene-based therapy. Nat. Rev. Genet. 15, 541–555.
Zhang, Y., Shi, B., Li, C., Wang, Y., Chen, Y., Zhang, W., et al. (2009). The synergetic bone-forming effects of combinations of growth factors expressed by adenovirus vectors on chitosan/collagen scaffolds. J. Control. Release 136, 172–178. doi: 10.1016/j.jconrel.2009.02.016
Zhang, Y., Song, J., Shi, B., Wang, Y., Chen, X., Huang, C., et al. (2007). Combination of scaffold and adenovirus vectors expressing bone morphogenetic protein-7 for alveolar bone regeneration at dental implant defects. Biomaterials 28, 4635–4642. doi: 10.1016/j.biomaterials.2007.07.009
Keywords: bone regeneration, gene therapy, preclinical models, translational medical research, orthopedic and trauma, regenerative medicine
Citation: Wilkinson P, Bozo IY, Braxton T, Just P, Jones E, Deev RV, Giannoudis PV and Feichtinger GA (2021) Systematic Review of the Preclinical Technology Readiness of Orthopedic Gene Therapy and Outlook for Clinical Translation. Front. Bioeng. Biotechnol. 9:626315. doi: 10.3389/fbioe.2021.626315
Received: 05 November 2020; Accepted: 12 January 2021;
Published: 17 March 2021.
Edited by:
Aiming Yu, University of California, Davis, United StatesReviewed by:
Ye Tian, Northwestern Polytechnical University, ChinaCopyright © 2021 Wilkinson, Bozo, Braxton, Just, Jones, Deev, Giannoudis and Feichtinger. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Georg A. Feichtinger, Zy5mZWljaHRpbmdlckBsZWVkcy5hYy51aw==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.