
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Bioeng. Biotechnol., 03 March 2021
Sec. Synthetic Biology
Volume 9 - 2021 | https://doi.org/10.3389/fbioe.2021.622835
This article is part of the Research TopicLactic Acid Bacteria: Microbial Metabolism and Expanding ApplicationsView all 15 articles
 Lieke A. van Gijtenbeek†
Lieke A. van Gijtenbeek† Thomas H. Eckhardt†‡
Thomas H. Eckhardt†‡ Lucía Herrera-Domínguez†
Lucía Herrera-Domínguez† Elke Brockmann
Elke Brockmann Kristian Jensen
Kristian Jensen Asger Geppel
Asger Geppel Kristian Fog Nielsen
Kristian Fog Nielsen Jannik Vindeloev
Jannik Vindeloev Ana Rute Neves‡
Ana Rute Neves‡ Gunnar Oregaard*
Gunnar Oregaard*Lactococcus lactis cheese starter cultures typically contain a mix of many strains and may include variants that produce and/or tolerate the antimicrobial bacteriocin nisin. Nisin is well-established as an effective agent against several undesirable Gram-positive bacteria in cheese and various other foods. In the current study, we have examined the effect of nisin on 710 individual L. lactis strains during milk fermentations. Changes in milk acidification profiles with and without nisin exposure, ranging from unaltered acidification to loss of acidification, could be largely explained by the type(s) and variants of nisin immunity and nisin degradation genes present, but surprisingly, also by genotypic lineage (L. lactis ssp. cremoris vs. ssp. lactis). Importantly, we identify that nisin degradation by NSR is frequent among L. lactis and therefore likely the main mechanism by which dairy-associated L. lactis strains tolerate nisin. Insights from this study on the strain-specific effect of nisin tolerance and degradation during milk acidification is expected to aid in the design of nisin-compatible cheese starter cultures.
Lactococcus lactis is a lactic acid bacterium widely used in the dairy industry for milk fermentation. Some L. lactis strains produce the Class I bacteriocin nisin, a 34 amino-acid-long peptide that shows potent antimicrobial activity against a broad range of Gram-positive bacteria (Mattick and Hirsch, 1947; Gross and Morell, 1971). Nisin is heat-resistant and acid-tolerant, largely because of a set of stable post-translational modifications (Delves-Broughton, 1996). Nisin in powder format was first introduced to the market in the 1950's as a natural product to contribute to shelf-life extensions of cheese by preventing growth of food spoilage organisms including Clostridia, Propionibacteria and Listeria (Hirsch, 1951; Delves-Broughton, 1996; Molloy et al., 2011). Nisin-producing strains can be added or are naturally present in cheese starter cultures (Delves-Broughton, 1996; de Arauz et al., 2009; Ávila et al., 2014).
To match the increasing demands of the dairy industry, new starter cultures are constantly being developed by either blending traditional undefined cultures or targeted culture design (Ayad et al., 2001). Starter cultures for semi-hard cheeses like Gouda or Edam are typically composed of a mixture of lactococcal strains, including Lactococcus lactis ssp. lactis and Lactococcus lactis ssp. cremoris, and Leuconostoc strains (Erkus et al., 2013; Düsterhöft et al., 2017). Differences in strain composition and diversity of the starter culture impact robustness toward phage attack, acidification, eye-formation, flavor development, and rheological properties of the cheese. Hence, it is crucial to understand how the starter culture composition and subsequently the cheese quality is affected by different factors. L. lactis strains devoid of nisin immunity or degradation machineries are highly susceptible to nisin. We therefore hypothesized that presence of nisin may inhibit L. lactis cheese culture strains to different extents depending on the genetic blueprint of the strains, e.g., presence of nisin immunity and/or nisin degradation genes in each strain.
Nisin exerts multiple modes of action against bacteria. Small amounts of nisin can be sufficient to hinder cell division by binding to and subsequent displacement of lipid II molecules, which are essential precursors required for bacterial cell wall synthesis (Brötz et al., 1998; Breukink et al., 1999; Wiedemann et al., 2001; Hasper et al., 2006). When nisin concentrations increase, it assembles together with lipid II into membrane pore-forming entities (Hasper et al., 2004). At even higher concentrations, nisin molecules are also known to self-assemble into pores without the requirement for lipid II (Sahl et al., 1987; Breukink et al., 1999). Pore formation results in the dissipation of proton motive force, cytoplasm leakage including the release of autolysins, and eventually cell death (Bierbaum and Sahl, 1985; Breukink et al., 1999; MartÃnezCuesta et al., 2000). Nisin also prevents the outgrowth of bacterial spores, presumably by lipid II binding and pore formation (Egan et al., 2016 and references therein). Since nisin targets any bacterial cytoplasmic membrane including that of L. lactis itself, nisin-producing L. lactis strains co-express proteins that confer nisin autoimmunity. These are NisI, a membrane associated lipoprotein, and NisFEG, an ABC transporter (Kuipers et al., 1993; Engelke et al., 1994; Qiao et al., 1995; Siegers and Entian, 1995). Extracellular NisI binds nisin thereby abolishing pore formation (Takala et al., 2004; AlKhatib et al., 2014a; Hacker et al., 2015). Recently, it was shown that NisI also functions in cell aggregation thereby reducing accessibility to lipid II even more (AlKhatib et al., 2014a). NisFEG forms an efflux pump that translocates nisin from the membrane into the extracellular space (Stein et al., 2003). NisI and NisFEG have a synergistic effect and enable L. lactis cells to tolerate high levels of nisin (up to 700 nM, ~2.35 mg L−1), which drops to 10–30% when expressed separate from each other (Kuipers et al., 1993; Qiao et al., 1995; Duan et al., 1996; Ra et al., 1996, 1999; Stein et al., 2003; AlKhatib et al., 2014a,b).
Nisin production and immunity by L. lactis strains are established by a conserved biosynthesis gene cassette consisting of four transcriptional units, nisABTCIPRK, nisI, nisRK, and nisFEG (Kuipers et al., 1993; Ra and Saris, 1995; de Ruyter et al., 1996; Ra et al., 1996; Li and O'Sullivan, 2006; Trmčić et al., 2011) The nisA gene encodes the nisin bacteriocin of which several variants have been reported (A, Z, F, and Q) in L. lactis (Mulders et al., 1991; De Kwaadsteniet et al., 2008; Fukao et al., 2008). The transcription of the nisin biosynthesis and autoimmunity operons (nisABTCIPRK and nisFEG) is activated through a nisin-mediated positive feedback loop conducted by the two-component system NisRK (Kuipers et al., 1995; de Ruyter et al., 1996; Kleerebezem et al., 1997). Due to this positive autoregulation, strains that carry the complete nisin biosynthesis cassette are locked in a nisin-producing state. The two other transcriptional units, nisI and nisRK, are driven by a weak and a relatively strong constitutive promoter, respectively, that function in the absence of nisin (de Ruyter et al., 1996; Li and O'Sullivan, 2006). However, both are believed to be trivial in nisin-producing strains as transcription from the nisA promoter overrules the effect of the internal operator sites.
In addition to nisI and nisFEG, a third nisin resistance determinant (nsr) was mapped to certain plasmids in non-nisin producing lactococci (McKay and Baldwin, 1984; Froseth and McKay, 1991; Liu et al., 1997). Already in 1984, it was suggested that the product of this gene might function as a nisinase, a nisin degrading enzyme previously discovered in Streptococcus thermophilus (Alifax and Chevalier, 1962). Over a quarter of a century later, it was confirmed that NSR is a tail-specific membrane-bound protease that cleaves nisin Z after the 28th position (Sun et al., 2009). This renders the remaining peptide close to inactive through a significant reduction in membrane affinity and pore-forming capacity (Sun et al., 2009). Over the years, similar membrane-associated proteases have been identified in Gram-positive species (Khosa et al., 2013; Draper et al., 2015). One of these is the well-characterized Streptococcus agalactiae nisin resistance protein SaNSR of which its structure has been resolved (Khosa et al., 2015). While the IC50 of a nisin-sensitive L. lactis strain overexpressing NisI is 5-to-10-fold greater than the wild type, SaNSR overexpression in the same cells enabled cells to withstand up to 18-to-20-fold more nisin (Khosa et al., 2013).
To investigate the potential effect of nisin on L. lactis strains, we initiated a study directed at evaluating the effect of nisin on the individual acidification profiles of 710 individual L. lactis strains, from which many are derived or used in common starter cultures. By genome sequencing and PCR, we correlated the observed acidification profiles with the presence or absence of nisin immunity and degradation genes, concurrently giving insights in the collection-wide distribution of such elements. Furthermore, nisin production and degradation was measured using a newly developed high-throughput HPLC-MS/MS method, which enabled us to elucidate the nisin degradation capacity of each strain during milk fermentations.
A total of 710 proprietary L. lactis strains originating from the Chr. Hansen Culture Collection were used in this study, for details see Supplementary Table 1. The strains were routinely grown as standing cultures in Oxoid M17 (Thermo Fisher Scientific, Waltham, MA, USA) with glucose 0.5% (w/v) (GM17) or a mixture of 1% (w/v) glucose and 1% (w/v) lactose (GLM17) at 30°C for 18 h.
All 710 strains were subjected to whole-genome sequencing on an Illumina MiSeq, yielding reads of 250 bases. The reads were assembled into contigs using CLC Genomics Workbench (Qiagen, Århus, Denmark). Contigs with a mean depth of coverage <0.25 of the total mean depth of coverage were discarded as contaminants. A gene search was conducted in all 710 genomes to map the presence of genes encoding nisin-related proteins. The query sequences used in the gene search can be found in Supplementary Table 2. The genomes were searched using blastn and tblastn for nucleotide sequence and protein sequence queries, respectively, with an E-value cut-off of 0.01. A gene was considered present in a genome if a hit with more than 90% query coverage and 80% identity was found. After observing nisin degradation in strains where no nsr gene was detected, it was found that some strains had shorter versions of nsr. To identify such variants, we repeated the tblastn search for the nsr protein sequence while reducing the query coverage threshold to 20% (Schliep, 2011). Whole genome k-mer trees [K-mer length: 16, prefix: AT, distance function: Feature Frequency Profile via Jensen-Shannon divergences (FFP)] of the 710 draft genomes and 219 RefSeq genomes were calculated using the Microbial Genomics module of the CLC Genomics workbench. A newly developed MLST scheme was employed including the household genes dnaK, fusA, groEL, gyrA, gyrB, ileS, lep, pheS, recA, rpoA, rpoB, and rpoC (see Supplementary File 1 for sequences of indicated genes for all strains). Concatenated sequences of the twelve genes were used to calculate a maximum likelihood tree with the Phangorn package I (Schliep, 2011) in R software. Due to quality requirements, a total of 206 instead of 219 L. lactis RefSeq genomes were taken along in the MLST analysis. Both k-mer and MLST trees were plotted with iTOLv5.5 (Letunic and Bork, 2019).
Cell-free supernatants (CFS) with or without nisin were obtained from 500 mL GLM17 cultures of L. lactis well-studied strains ATCC11454 or Wg2, respectively, by centrifugation for 5 min at 5,000×g. Collected supernatant was adjusted to pH 6.0 by the addition of 0.25 M NaOH, filter-sterilized using a Minisart® 0.22 μm filter (Sartorius, Göttingen, Germany) and stored at −80°C in 10 mL aliquots. CFS of Wg2 yielded 0.0 μg ml−1 nisin A, whereas CFS of ATCC 11454 yielded 6.1 ± 0.8 μg ml−1 nisin A, as measured using the HPLC-MS/MS method detailed below. The two CFS types were subsequently used to evaluate the milk acidification profiles of the 710 Lactococcus strains as follows. A volume of 20 μl fresh overnight culture of each L. lactis strain grown in GLM17 in a 96-wells microtiter plate was used to inoculate 1,980 μl prewarmed (30°C) pasteurized semi-skimmed milk supplemented with 0.2% (w/v) yeast extract and 5% (v/v) pH indicator solution (1 g L−1 Bromocresol Purple sodium salt (Sigma Aldrich, St. Louis, MS, United States); 1 g L−1 Bromocresol Green sodium salt (Sigma Aldrich), pH 7.0, filtered-sterilized). One hundred and fifty microliter of each of the 710 inoculated milk samples were then mixed with 50 μl of one of the two CFS types described above, resulting in a final nisin A concentration of 0.0 or 1.5 μg ml−1, and incubated for 18 h at 30°C on flatbed scanners (HP ScanJet G4010) to obtain HUE-values every 6 min. Acidification profiles were obtained by converting HUE-values into pH values as described previously (Poulsen et al., 2019). Experiments with CFS from Wg2 and ATCC 11454 were always run in parallel and the experiments were performed in triplicates: one technical and two biological replicates.
As mentioned above, inoculation material for the milk acidifications originated from overnight cultures with different biomass and pH, which caused small changes in both initial and end pH, as well as the time for the acidification to start. Therefore, in order to define robust thresholds for phenotypic classification, the acidification curves were first normalized to the same maximum (initial) pH of 6.4. The pH values were re-scaled by setting the minimum and maximum pH for each pair of curves per strain to 0 and 1, respectively. Next, curves were also normalized in time so that starting timepoint of each pair of acidification curves was the same across replicates. The starting time was calculated per replicate using the milk acidification curve that had no nisin. This starting point was then subtracted across all timepoints from each pair of curves. Finally, absolute change in pH, area under the curve and starting timepoint for each curve and condition were calculated and averaged across replicates. These parameters were used to define the four different phenotypes. For detailed description on the data processing steps see Supplementary Table 3. Data and statistical analyses were performed using the computing environment R. Raincloud plots were made according to Allen et al. (2019). The central matrix layout was used using the UpSet package (Gu et al., 2016).
To set up a detection method for full-length and NSR-degraded nisin A, CFS was collected from L. lactis strains CH-1 (nisA–, nsr+) and ATCC 11454 (nisA+, nsr–) grown in GM17 spiked with 0.9 μg ml−1 nisin A (Chrisin®, Chr. Hansen A/S, Denmark). Chrisin® was shown to be 2.26% pure when compared to nisin A from Sigma-Aldrich (defined as 2.5% by FAO, 2013). Sterile GM17 spiked with 0.9 μg/ml nisin A was taken along as a blank control sample. A 30 μL sample from each CFS was then mixed in 1.5 mL Eppendorf tubes with 870 μL extraction buffer to which nisin Q was introduced as an internal standard (IS) to improve the precision of the assay. Nisin Q was chosen to get: (i) an as similar molecule to nisin A as possible; (ii) an IS not interfering with nisin A measurements; and (iii) an IS also susceptible to NSR degradation. The extraction buffer containing nisin Q was made as follows: CFS collected from a culture of the nisin Q-producing CH-5 strain grown in GM17 was quenched by adding 3% (v/v) acetonitrile (ACN) and 1% (v/v) formic acid (FA). 1.5 ml of this extract was then added to 100 ml of extraction buffer consisting of 100 mg/L Bovine Serum Albumin (BSA; A-2153, Sigma), 20% (v/v) ACN and 0.5% (v/v) FA dissolved in MilliQ water. The extracts were transferred to polypropylene HPLC vials, and nisin A, nisin Q, and nisin1−28 (nisin A degraded by NSR) levels were analyzed on a binary ACQUITY UPLC system (Waters, Milford, MA, USA) equipped with a sample organizer (held at 8°C) and connected to a Xevo TQ-XS Triple Quadrupole Mass Spectrometry (MS) instrument (Waters) equipped with an Electrospray source operated in positive mode. One microliter subsamples were injected onto a PLRP-S 300 Å, 2.1 × 150 mm, 3 μm HPLC column (Agilent Technologies, Waldbronn, Germany) held at 60°C and eluted using a linear gradient of 25–40% (v/v) ACN with 0.1% (v/v) FA using a flow rate of 0.5 mL min−1. The following MS conditions were applied: Capillary voltage: 3 kV, collision gas (Ar), desolvation temperature: 550°C, desolvation gas flow: 1,100 L/h, cone flow: 150 L/h, nebulizer: 6 bar. For all analytes the cone was held at 40 V and a fragmentation energy of 16 eV. Waters TargetLynx Software was used for data analysis and peak integration. The following multiple reaction monitoring (MRM) fragmentations, with a dwell time of 20 ms, were used for detection: (1) the [M+5H]5+ ion of nisin A from m/z 671.7 to 811.2, (2) the [M+5H]5+ ion of nisin Q from m/z 666.2 to 804.4, and (3) the [M+4H]4+ ion of nisin1−28 m/z 680.55–869.7). Nisin1−28 eluted ~0.35 min later than nisin A but 0.29 min earlier than nisin Q. From calibrants with constant nisin Q and variable nisin A concentrations, the nisin A peak areas, divided by the peak area of the nisin Q peak area, was used to determine the nisin A concentrations. From a degradation of nisin A with cell free NSR culture extract, the response factor difference between nisin A and nisin1−28 was estimated to approximate nisin1−28 concentrations in ATCC 11454 and CH-1 CFS. This method was also used to obtain the nisin A concentrations in the CFS of ATCC 11454 and Wg2 added during the milk acidification experiment.
Nisin degradation and nisin A production by each of the 710 L. lactis strains was assessed by measuring the decrease or increase of nisin in samples spiked with known concentrations of nisin, using a high-throughput HPLC-MS/MS as follows. Two milliliter of skimmed milk supplemented with 0.9 μg ml−1 nisin A and 0.2% (w/v) yeast extract was inoculated with 20 μl of a L. lactis GLM17 overnight culture and incubated in 2.5 mL deep well plates for 18 h at 30°C and stored at −20°C until further analysis. As a control, non-inoculated skimmed milk samples, to which 0.9 μg ml−1 nisin A was added, were taken along. Thirty microliter from each of the thawed samples was extracted with 870 μL extraction buffer in a 1 mL microtiter deep well plate. The plates were shaken for 1 h on an orbital mixing table, left overnight at 5°C and centrifuged at 6,000×g for 30 min, after which the samples were loaded for HPLC-MS/MS as described in the method above, except that a MRM for nisin Z was also included at m/z 667.2–739. Due to variations in the control samples from plate to plate, nisin concentrations in control samples were set to 1 AU mL−1 and nisin concentrations in fermented milk samples were converted to ratios of this arbitrary control value. Since we added a known concentration of nisin A, a decrease or increase of nisin A levels was used as a proxy for degradation and production, respectively. Degradation of nisin was confirmed in a subset of samples by measuring the accumulation of the NSR degradation product with the method described in the section above (Supplementary Table 1).
Strains with a nsr– genotype that displayed a nisin degrading phenotype, as well as strains where the nsr gene was found on the edge of a contig, were individually examined for the presence of the nsr gene using a colony PCR approach with REDTaq DNA polymerase Master Mix (VWR, Radnor, PA, USA) and primers annealing to two conserved regions within the nsr gene of L. lactis strains (oLIGI021 and oLIGI022), ranging from nucleotide 112–901 of the 958-nts long nsr gene. A total of 46 strains were tested by colony PCR using oLIGI021 and oLIGI022 to confirm the presence of a copy of the nsr gene. Of the 15 strains in which the nsr gene was detected at the edge of a contig in the genome sequence, 14 strains resulted in a PCR product validating the presence of nsr, while one strain contained an estimated insertion of roughly 6 kB in its nsr gene. The 31 remaining strains, corresponding to those in which the nsr gene could not be detected in the genome sequence but that showed signs of nisin degradation, 25 were confirmed to carry the nsr gene. Primer sets to confirm several genotypes were oLIGI023/24 for nisA, oLIGI043/44 for nisI, oLIGI045/46 for nisF. An overview of primers used in this study can be found in Supplementary Table 4.
For targeted gene deletions in strain CH-2, the pCS1966/oroP system was employed (Solem et al., 2008). Standard molecular cloning techniques were performed essentially as described (Sambrook et al., 1989). Flanking regions of either nisA or nsr gene were amplified using Q5 DNA polymerase (NEB, Ipswich, MA, USA) from CH-2 chromosomal DNA with primer pairs oLIGI003/oLIGI004 and oLIGI005/oLIGI006 or primer pairs oLIGI014/oLIGI015 and oLIGI016/oLIGI017, respectively. The backbone of pCS1966 was amplified using oLIGI001 and oLIGI002. Fragments were assembled using NEBuilder® HiFi DNA Assembly Master Mix (NEB) and transformed into competent E. coli DH5α cells (NEB), yielding pLIGI001 and pLIGI003. Purification of DNA fragments was done using the Monarch® PCR & DNA Cleanup and DNA Gel Extraction Kits (NEB) and plasmids were isolated using the NucleoSpin® Plasmid kit (Macherey-Nagel, Düren, Germany). pLIGI001 was then used to delete nisA, whereas pLIGI003 was used to delete nsr, both as described previously (Solem et al., 2008). Electrocompetent L. lactis cells were transformed using electroporation with a Bio-Rad Gene Pulser (Bio-Rad Laboratories, Richmond, CA). PCR amplification on colonies with REDTaq DNA Polymerase Master Mix (VWR) was used for routine checks for correct DNA constructs.
For well diffusion assays, strains ATCC 11454, CH-1, CH-2, CH-3, CH-4, mutant strains CH-2 ΔnisA and CH-2 Δnsr, and the nisin-sensitive MG1363 strain were grown to stationary phase in 16 h from single colonies in GM17. CFS of all strains except MG1363 was collected by centrifugation for 10 min at 4,000×g and subsequent filter-sterilization of the supernatant. The MG1363 culture was diluted 1,000 times in fresh GM17 broth, mixed 1:1 with GM17 agar and poured as a soft agar layer while leaving out 10 mm holes that were filled with 200 μL of the collected CFS samples. The assay was incubated at room temperature until halos were visible. For measurements of nisin A and the nisin1−28 fragment in GM17, the cultures were grown as described above and stored for 24 h at −80°C before CFS was collected using 0.22 μm 96w filter plates (Acroprep, Pall laboratory, Ann Arbor, MI). These samples were then subjected to HPLC-MS/MS analysis using the extraction and analysis procedure as described above.
A total of 710 L. lactis strains were monitored, in a time-resolved fashion, to evaluate their ability to acidify milk in the presence of nisin (CFS collected from the nisin producing L. lactis strain ATCC 11454, CFSnisin) or in absence of nisin (CFS collected from the non-nisin producing L. lactis strain Wg2, CFScontrol). Without nisin, all strains were able to acidify milk in <18 h: the majority (88%) of the strains were able to acidify milk from a start pH of 6.4 down to a pH value below 5.0, while the remaining strains acidified milk to an end pH between 5.0 and 6.0 instead (Supplementary Table 1). The addition of CFSnisin (final concentration of nisin A in milk of 1.5 μg ml−1), led to a wide variation of acidification profiles, ranging from unaltered acidification to loss of acidification. In order to get an impartial comparison of the obtained milk acidification profiles in the presence or absence of nisin for each separate strain, the start and end pH were normalized to correspond to values 1 and 0, respectively, and the curves were synchronized so that the control curves would start acidifying at the same timepoint. From each normalized acidification curve the absolute change in pH (ΔpH) was derived, and the difference in area under the curve (ΔAUC) and acidification starting time points (Δtime0) between CFSnisin and CFScontrol treated conditions for every strain were calculated. After multiple attempts for classification, we noticed that ΔAUC alone could not allow for a clear separation of the phenotypes. Instead, combining these three parameters (ΔpH, ΔAUC, Δtime0) allowed us to classify strains into four distinct phenotypic groups (rules are described in Supplementary Table 3). We define the resulting phenotypic groups as (A) loss of acidification (LA), (B) highly delayed acidification (HDA), (C) mildly delayed acidification (MDA), and (D) unaltered acidification (UA) (Figure 1 and Supplementary Figure 1).
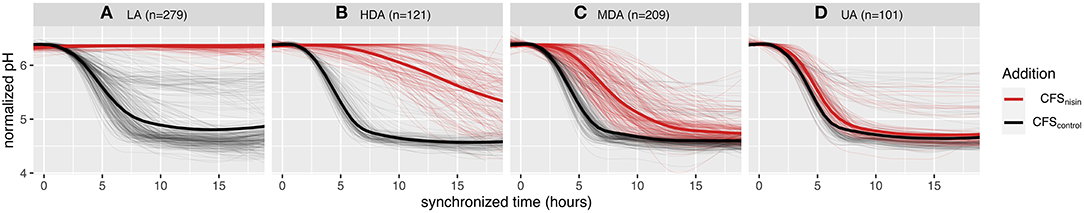
Figure 1. Effect of nisin on milk acidification profiles and phenotypic classification of 710 L. lactis strains. Milk acidification curves of strains grown at 30°C without nisin (CFScontrol) and with 1.5 μg ml−1 nisin addition (CFSnisin) The thin lines depict the averaged milk acidification data of three replicates while the thicker line depicts the averaged curves per group. The nisin-containing milk phenotypes are classified to four groups; (A) LA, loss of acidification; (B) HDA, highly delayed acidification; (C) MDA, mildly delayed acidification; and (D) UA, unaltered acidification. All acidification curves are synchronized based on acidification onset of each respective CFScontrol profile and normalized to a start pH of 6.4.
When milk fermentations were carried out in the presence of CFSnisin, 279 L. lactis strains clustered to the LA phenotype, indicating that these strains are sensitive to nisin (Figure 1A). A total of 101 L. lactis strains grouped to the UA phenotype, for which both the acidification rate and depth remained mostly unchanged upon the addition of CFSnisin compared to addition of CFScontrol (Figure 1D). For the remaining strains, the effect of nisin resulted in a range of delays in the onset of acidification. Of these, 121 strains clustered to the HDA and 209 to the MDA phenotypes (Figures 1B,C). We identified a small set of strains that did not show a delay but a change in acidification depth as a result of nisin addition, which therefore also clustered to the mild (MDA) phenotypic group, see also Supplementary Figure 2A.
In order to assess the genotypic diversity, the strains in the collection were whole-genome sequenced and their genomic relatedness assessed by a k-mer-based and a MLST-based tree construction (Figure 2 and Supplementary Figure 3). From this analysis, 64% of the strains (n = 457) belong to L. lactis ssp. lactis and the remaining 36% (n = 253) of the strains belong to L. lactis ssp. cremoris. We compared our genomes to 206 unique lactococcal assemblies publicly available (July 2020) in NCBI (National Center for Biotechnology Information) RefSeq database of which 73% and 26% map to L. lactis ssp. lactis and sspcremoris, respectively (Figure 2 and Supplementary Figure 3). Furthermore, the comparison shows that our strain collection spans the full spectrum of genomic diversity represented by the publicly available L. lactis genomes with some smaller lineages exclusively represented in our collection and to a lesser extent vice versa. We subsequently searched the genome sequences of our collection for orthologous groups of protein sequences (OGs) to capture ssp. cremoris with a reversed phenotype e.g., strains that display a ssp. lactis phenotype while having a ssp. cremoris genotype, as previously reported (Kelleher et al., 2017; Wels et al., 2019). This analysis showed that 25% of the ssp. cremoris strains in our collection carry sequences corresponding to OGs specific for ssp. cremoris strains with a ssp. lactis phenotype (Figure 2). Accordingly, these strains cluster in one of the two ssp. cremoris lineages, together with previously identified ssp. cremoris strains with a ssp. lactis phenotype such as KW2, N41, V4, MG1363, and NCDO763 (Wels et al., 2019).
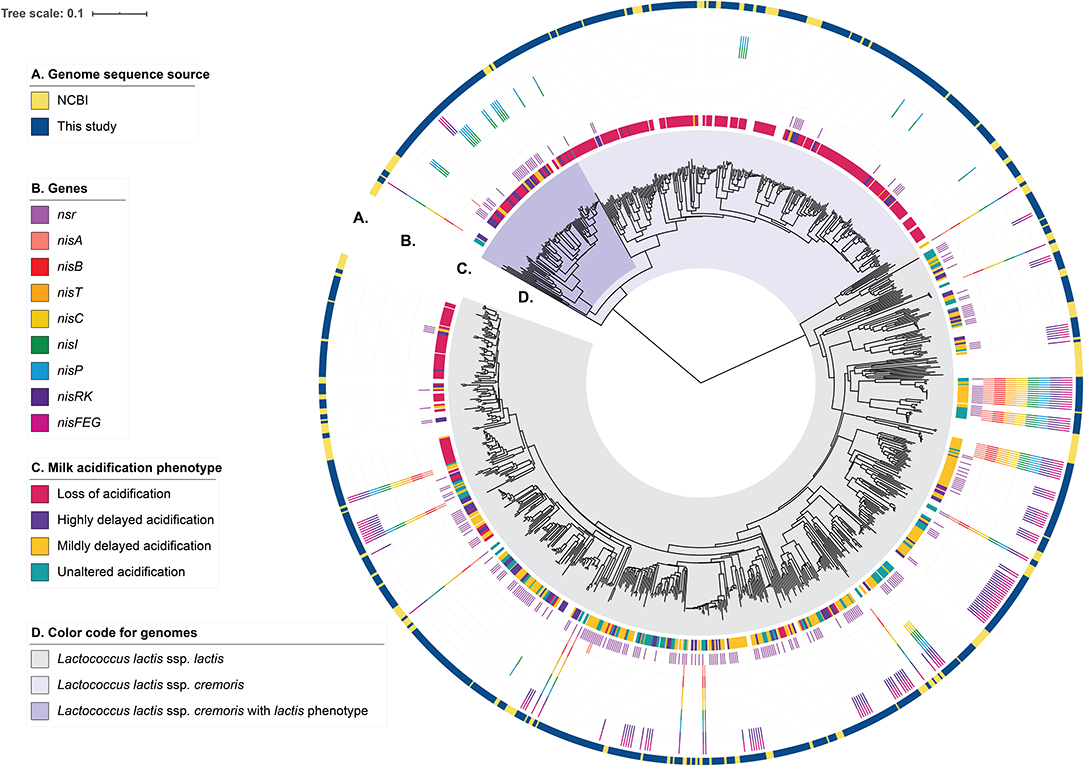
Figure 2. Genotypic diversity of the L. lactis strains. A k-mer tree depicting the genotypic relatedness of 929 L. lactis ssp. cremoris and ssp. lactis strains. Indicated from outer to inner ring are: (A) Strain-specific genome sequence origin, obtained either from the 710 draft genomes from this study or the publicly available 219 L. lactis RefSeq genomes. (B) Strain-specific presence of nisin-related genes or gene clusters. (C) Strain-specific milk acidification phenotypes. (D) Subspecies clusters of L. lactis ssp. lactis and L. lactis ssp. cremoris. The latter is further divided into whether ssp. cremoris strain is predicted to have a cremoris or a lactis phenotype.
To better understand how the presence of genes for nisin biosynthesis, nisin immunity and/or nisin degradation correlate to the observed variability in milk acidification upon addition of nisin, we performed gene-trait matching of nisin-related genes to observed acidification phenotypes. The distribution of genes encoding key proteins involved in nisin biosynthesis (a nisin structural gene: nisA; the nisin biosynthetic machinery: nisB, nisT, nisC, and nisP; nis gene regulation: nisR and nisK), nisin immunity (nisI and nisFEG) and nisin degradation (nsr) within the 710 strains along with the milk acidification phenotypes (LA, HDA, MDA, UA) are visualized in Figure 2 and Supplementary Figure 1. It is apparent that most ssp. cremoris have the LA phenotype, whereas the ssp. lactis have more variable phenotypes, with many strains belonging to UA, MDA, and HDA.
A total of 45 strains (6.3%) contain the full nisin gene cluster required for nisin biosynthesis (nisABTCIPRK-FEG). In fact, 24 of these strains acidified milk without any delay in the presence of CFSnisin, and were classified to the UA phenotypic group, while 21 do so with only a mild delay or higher end pH and therefore classified to the MDA phenotypic group (Figure 3 and Supplementary Figure 2). Out of 45 strains, 44 belong to the ssp. lactis group. The single ssp. cremoris strain that contains the nisin gene cassette groups to the genotype cremoris/phenotype lactis clade and is one out of only three ssp. cremoris strains that are not affected by the addition of nisin (Wels et al., 2019) (see also Figure 2).
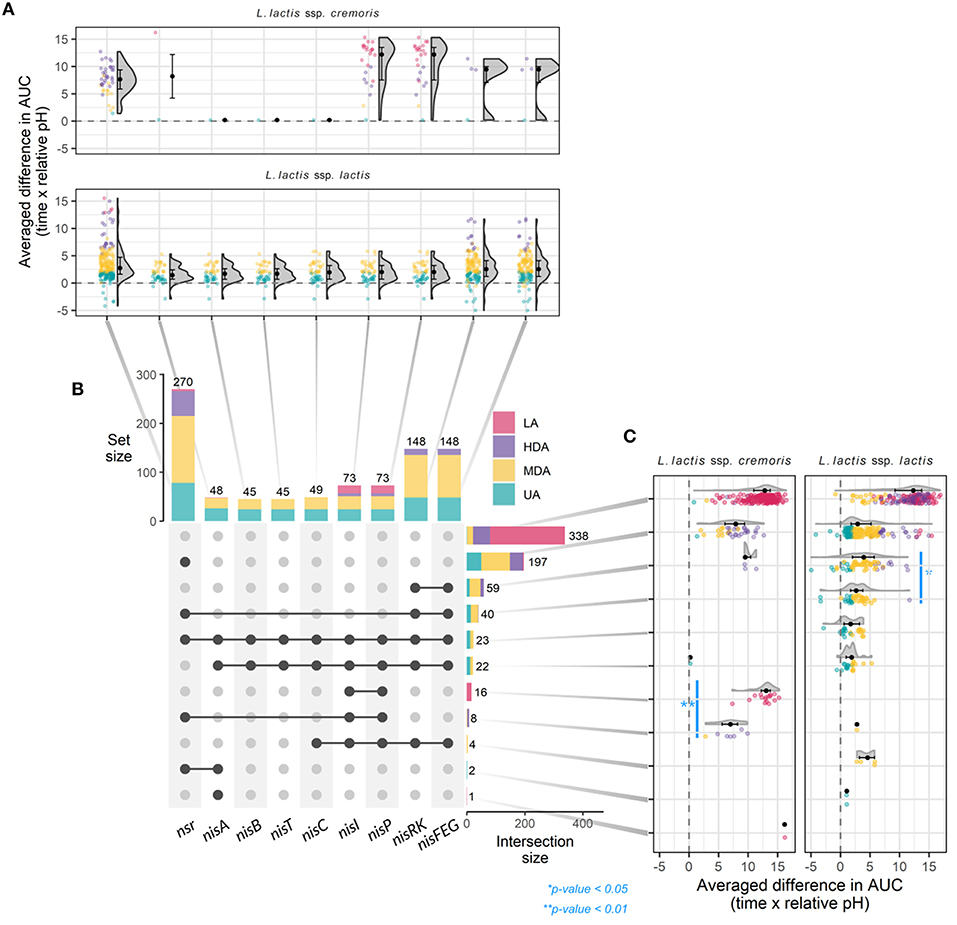
Figure 3. Gene-trait matching of nisin tolerance genotypes with milk acidification patterns. (A) Raincloud plots depicting the averaged ΔAUC of every strain for milk acidification curves with and without nisin, grouped by individual nisin-related gene and subspecies. (B) Matrix layout showing individual presence (vertical) and overlap in (horizontal) nisin-related genes in 710 L. lactis strain and the respective distributions of phenotypes and total number of strains that contain the indicated gene or set of genes. Dark dots connected with solid lines indicate genes present in a group, while gray dots indicate gene absence. (C) Raincloud plots depicting ΔAUC values as in (A), but now grouped by genotype for nisin-related genes and subspecies. For all raincloud plots, black dots represent the median values whereas black lines mark the boundaries for the center 50% of the distribution. P-values were calculated using the Mann-Whitney-Wilcoxon test.
Interestingly, of all strains that contain a full nisin biosynthesis gene cluster, 23 also contain nsr (Figure 3 and Supplementary Figure 1). We therefore wondered if the presence of both nsr and immunity genes would allow for faster acidification than that of only the immunity genes. We found a bimodal distribution in the acidification delays for the former group of strains indicating that two subgroups exist, one containing strains that acidify better than the strains with just immunity and a second, less tolerant group (Figure 3). Interestingly, the two subgroups appeared to have different variations of the nsr sequence (variant 25 for the UA group and variant 17 for the MDA group), putatively yielding proteases with different activities (Supplementary Table 1 and Supplementary Figure 6D).
To summarize, the current data confirms previous reports that the presence of nisin immunity genes nisI and nisFEG together constitute a strong mechanism in establishing nisin tolerance. Moreover, we find that the full nisin gene cluster is a feature mainly present in ssp. lactis.
Many strains exist that do not possess the full subset of genes for nisin biosynthesis and immunity but, instead, only contain one nisin immunity component (nisI and/or nisFEG) (Figure 3 and Supplementary Figure 1) (Wels et al., 2019). Four ssp. lactis were identified that contain genes for full nisin immunity (nisIP-RK-FEG) but have lost the corresponding synthesis genes. All show an MDA phenotype. A total of 59 strains contain nisRK-FEG without the rest of the nisin gene cassette. Like reported previously, we find that nisFEG genes always co-occur with nisRK (Wels et al., 2019). The three ssp. cremoris strains that possess unaccompanied nisRK-FEG display a HDA phenotype, while the 56 ssp. lactis strains appear divided over HDA (9), MDA (37), and UA (10) phenotypes (Figure 3 and Supplementary Figure 1). This again indicates that ssp. lactis strains with only nisRK-FEG are coping better with nisin than ssp. cremoris strains with the same set of immunity genes. Furthermore, nisRK-FEG genes are more prevalent in ssp. lactis than in ssp. cremoris. As reported previously, the nisI gene, when found without the remainder of the nisin biosynthesis cassette, typically still co-occurs with nisP (Tarazanova et al., 2016; Wels et al., 2019). In our collection, nisIP is uniquely present in 16 ssp. cremoris, all displaying LA (Figure 3). In contrast to these findings, NisI was previously reported to deliver substantial, but not full immunity against nisin in the absence of NisFEG (AlKhatib et al., 2014a; Tarazanova et al., 2016). NisI is believed to interact in an equimolar stoichiometry with nisin molecules (Hacker et al., 2015). This could lead to a significant surplus of nisin molecules under the tested conditions here in which a high concentration of nisin was employed. In order to test if NisI alone has the capacity to protect the cells against nisin, albeit at lower levels, the acidification experiments were repeated for these strains in the presence of 0.2 μg ml−1 nisin (Supplementary Figure 4). We did not detect an improvement in all but one of the acidification profiles in the presence of lower levels of nisin.
In short, strains with NisFEG alone are mostly ssp. lactis and can cope relatively well in the presence of nisin, whereas strains that only possess NisI are uniquely ssp. cremoris and do not. Interestingly, the average difference in AUC is slightly less in nsr+ strains compared to nisFEG+ strains, and even more so in nsr+/nisFEG+ strains, indicating that the presence of the nsr gene confers better tolerance toward nisin, and that NSR thus is a stronger protective force than the individual immunity systems from the nisin gene cassette (Figure 3).
A striking 270 L. lactis strains (38%) possess a gene encoding the nisin protease NSR (Figure 3) of which the majority (98%) were able to acidify milk, albeit with a wide window of delays (UA: 78, MDA: 137, and HDA: 51 and LA: 4). Of these, 197 of the strains (73%) contain only nsr and none of the immunity genes (Figure 3). The fact that 149 strains with nsr+ genotype and no nisin immunity genes display a UA or MDA phenotype clearly demonstrates the importance of NSR for nisin tolerance. The milk acidification profiles of the strains that only have the nsr+ genotype differ greatly between ssp. lactis and ssp. cremoris strains, with the former showing generally better acidification in the presence of nisin (Figure 3C and Supplementary Figure 5). In summary, it is evident that there is large phenotypic variation between nsr+ strains in terms of nisin tolerance. The general trend is that strains with only nsr are able to acidify milk in the presence of nisin, albeit with different delays in the onset of acidification. It could be that the underlying differences in genetic make-up and/or post-transcriptional regulation between ssp. cremoris and ssp. lactis play an important role in further determining nisin tolerance.
A total of 338 strains did neither contain nsr, nisI nor nisFEG. As expected, the majority of these strains (258; 76%) did not acidify milk in the presence of nisin. However, 1, 21, and 58 strains mapped to the UA, MDA, and HDA phenotypic group, respectively (Figure 3 and Supplementary Table 1). As these phenotypes deviate from the predicted LA phenotype of a strain with a nsr- nisI- nisFEG- genotype, we confirmed the absence of nisin immunity and degradation genes using a colony PCR. Therefore, the reason behind the UA, MDA, and HDA phenotypes of these strains remains elusive.
Since a high fraction of nsr+ strains were tolerant to nisin in milk, we were interested to find out to what extent nisin can be degraded by distinct nsr+ strains. To study the impact of each strain on nisin degradation in more detail, we first validated the role of lactococcal NSR in nisin A degradation. To accurately measure nisin A and detect its NSR degradation product, an assay based on high resolution mass spectrometry was developed. It was previously shown that nisin Z is cleaved after position 28 by the NSR enzyme thereby creating a large and a small fragment, which for nisin A should result in two fragments with monoisotopic masses of 2718.1854 Da (nisin1−28) and 869.3752 Da (nisin29−34), respectively (Sun et al., 2009). We initially identified nisin1−28 in CFS of CH-1(nis–, nsr+) incubated with nisin A by high resolution mass spectrometry on a HPLC-QTOF instrument (data not shown). Using a HPLC-MS/MS Triple Quadrupole Mass spectrometer, MS/MS fragmentation of CFS eluates collected from ATCC 11454 (nisA+, nsr–) and CH-1 (nsr–, nsr+) strains grown in the presence of nisin A and later on spiked with nisin Q as an internal standard led to the identification of quantification ions specific for nisin A, nisin Q, and nisin1−28 (Figure 4). In CFS of ATCC 11454, an increase in nisin A can be detected over the nisin A-spiked control sample. In contrast, no nisin A could be detected in the CFS derived from the nsr+ strain CH-1. However, in the latter, a fragment corresponding to the large NSR degradation product nisin1−28 could be accurately detected. This confirms that the lactococcal NSR is able to degrade nisin A. This was then used to measure nisin A and detect its NSR degradation in all further samples.
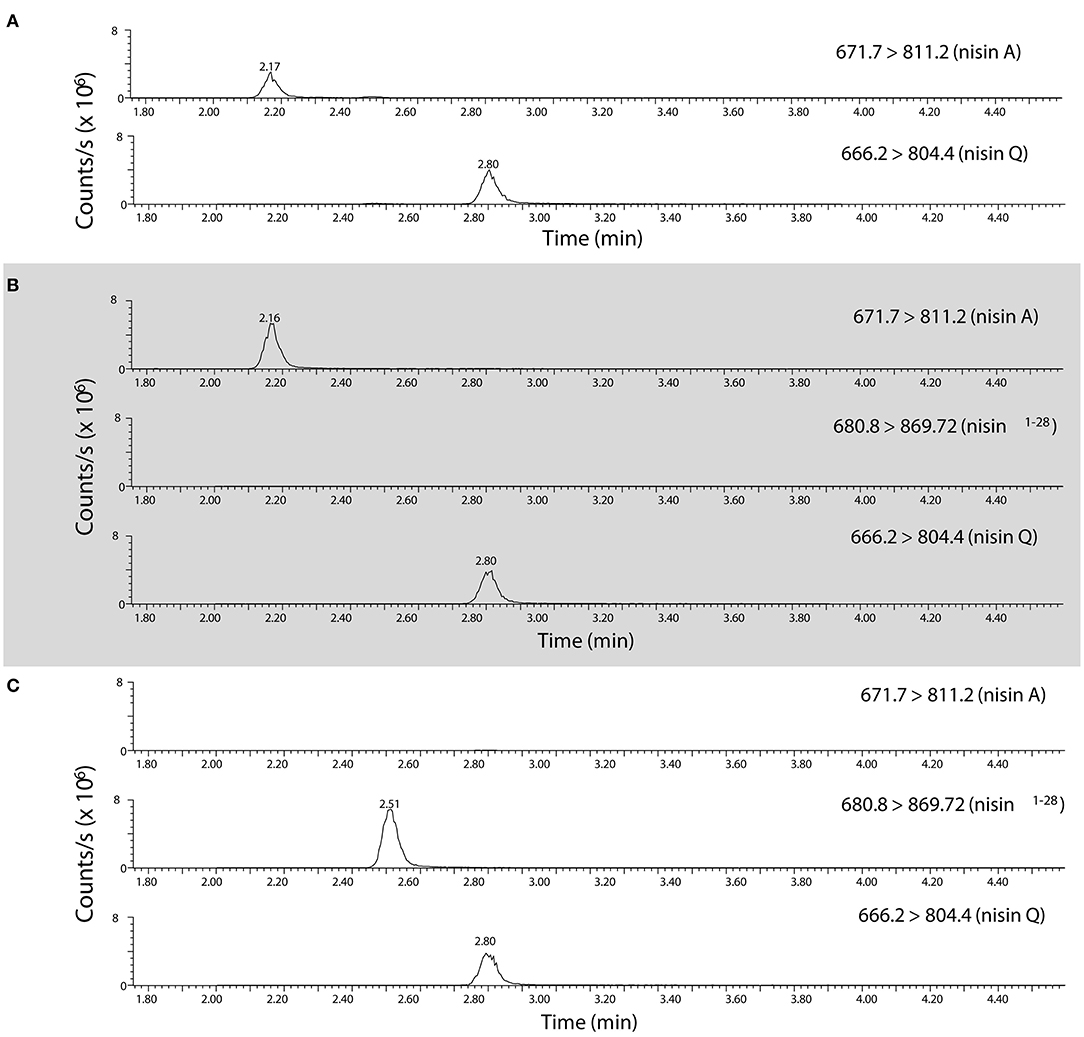
Figure 4. Identification of nisin A, nisin Q, and nisin1−28 in CFS using high resolution mass spectrometry. HPLC-MS/MS MRM transitions of quantification ions for nisin A [the [M+5H]5+ ion from m/z 671.7 to 811.2], nisin Q [the [M+5H]5+ ion of nisin Q from m/z 666.2 to 804.4,] and nisin1−28 [the [M+4H]4+ ion from m/z 680.55 to 869.7] as detected in (A) a control mix of nisin A and nisin Q, (B) CFS collected from CHCC 11454 (nisA+, nsr-) and (C), CFS collected from CH-1 (nsr–, nsr+). Note that the parent isolation window of the [M+4H]4+ ion of nisin1−28 (m/z 680.55) was moved m/z 0.25 as the A+1 isotopomer was the most abundant, creating the MRM transition 680.8 → 869.7.
We next aimed to study how much nisin A is broken down into nisin1−28 in a co-culture containing both a nisin-degrading and a nisin-producing strain. To do so, we employed ATCC 11454 and CH-1 as control as well as three different nisA+ nsr+ strains. CH-2 (nisA+ nsr+) has the same genetic make-up as CH-1 but has received the sucrose-nisin transposable element from ATCC 11454 via conjugation. CH-3 (nisA+ nsr+) also possesses an intact nsr gene, while CH-4 was found to naturally contain a point mutation in the nsr gene, introducing a stop codon at amino acid position 58 yielding a truncated NSR protein. To validate that the possession of nisA or nsr alone is responsible for nisin synthesis or nisin degradation, a clean deletion of either nisA or nsr was generated in CH-2, resulting in strains CH-2ΔnisA and CH-2Δnsr, respectively. CFS of the seven L. lactis strains grown to stationary phase in a rich nutrient broth (GM17) was collected and tested for antibacterial activity by means of well diffusion assays using a nisin-sensitive L. lactis strain as indicator. In addition, the presence of nisin A and nisin1−28 was assessed using the HPLC-MS/MS method described above. As expected, CFS of strains producing both nisin and NSR displayed a reduced activity against the nisin-sensitive indicator strain (Figure 5). In agreement, large amount of nisin1−28 degradation products and no full-length nisin peptides were detected in the CFS of CH-2 and CH-3 (Figure 5). CFS of CH-4 (nisA+ nsr1−57) showed the same level of inhibition as ATCC 11454 (Figure 5). In addition, nisin1−28 could not be detected by HPLC-MS/MS, confirming the loss in function of truncated NSR1−57. Deleting nisA in CH-2 effectively ceased the production of nisin A and thereby also any degraded nisin1−28, while deleting nsr in CH-2 abolished the presence of nisin1−28 in the CFS (Figure 5). However, nisin levels were similar to those detected for ATCC 11454 in CH-2Δnsr, confirming that NSR breaks down nisin produced by nisA+ nsr+ strains. Together, this data confirms that full-length NSR suffices for nisin degradation.
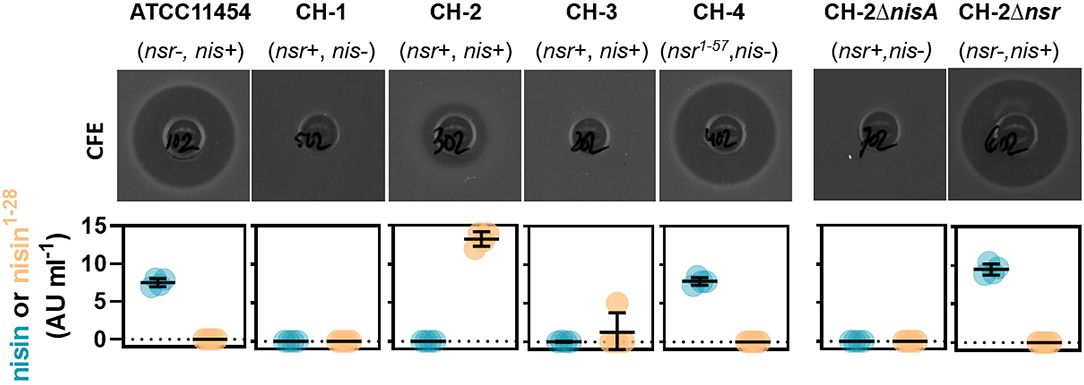
Figure 5. Impact of nsr genotype on bactericidal activity and nisin production. Top row: Well diffusion assays showing the antibacterial activity of CFS collected from indicated strains grown in GM17 for 16 h against the nisin-sensitive L. lactis strain Wg2. Bottom row: Graphs displaying nisin A and nisin1−28 concentrations in CFS collected from indicated strains grown in GM17 for 24 h as measured using HPLC-MS/MS.
After establishing that functional NSR is responsible for nisin degradation, we spiked milk with 0.9 μg mL−1 nisin A and determined changes in its concentration after fermentation for 18 h with each of the 710 L. lactis strains. This allowed us to map the impact of each strain on nisin: Either nisin concentrations increased, remained unaltered or decreased. We used this information to further divide the established phenotypic groups based on acidification profiles into a 3-by-4 phenotypic matrix (Figure 6). In addition, we plotted nisin degradation per variation of the NSR protein sequence to screen for possible inactive variants (Supplementary Figure 6).
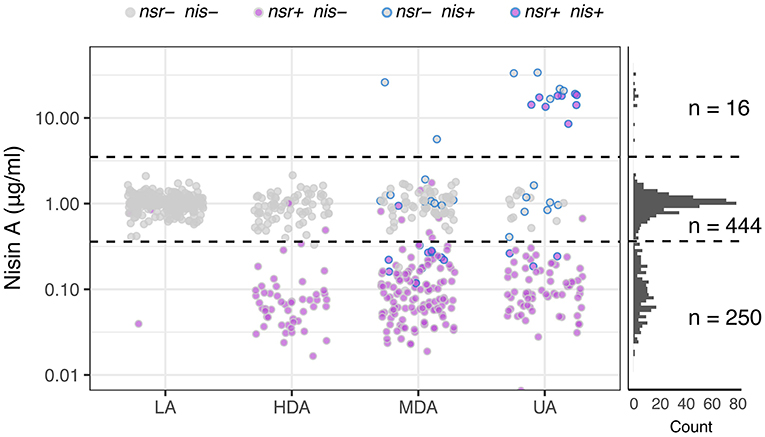
Figure 6. Nisin degradation by individual L. lactis strains. Dot plots depicting nisin A concentrations as quantified with HPLC-MS/MS after incubating 0.9 μg ml−1 of nisin A in milk inoculated with L. lactis strains belonging to one of the four phenotypic groups LA, HDA, MDA and UA. Dots are further color-coded to indicate the presence or absence of nsr and/or nisin biosynthesis genes (nis) in the tested strain. The right panel displays the distribution of resultant nisin A concentrations derived from the complete set of tested strains (n = 710). Horizontal dashed lines are drawn to indicate the boundaries between observations showing an increase of nisin A (16 datapoints above 3.5—upper dashed line), an unchanged nisin A concentration (444 datapoints between dashed lines) or a decreased nisin concentration (250 datapoints below 0.36—lower dashed line).
The top row of the matrix displayed in Figure 6 shows those strains that led to an increase in nisin A levels above the initial nisin A concentration. As expected, all 16 strains in this group contain the complete nisin A biosynthesis gene cassette and have either an UA or an MDA phenotype (Figure 6). Interestingly, we observe a >5-fold increase in the levels of nisin A of the eight nsr+ strains with a nisin biosynthesis cassette, indicating that the nisin production rate by these strains is higher than their nisin degradation rate (Figure 6). It should be noted that, out of 45 strains that contain a full nisin biosynthesis cassette, 29 encode versions other than nisin A that are not detected in the employed HPLC-MS/MS method. As a consequence, these will show up in the middle or bottom row instead.
In the middle row of Figure 6, the largest number of strains can be found. In milk fermentations carried out with these strains, the nisin A concentration remained stable. These largely correspond to strains that do not have the nsr gene (430 out of 444). Note that strains mapping to the LA phenotype typically fail to degrade nisin A (Figure 6). Fourteen nsr+ strains did not show nisin-degrading activity. Further examination of the NSR protein sequences of these strains revealed 13 different NSR variants (Supplementary Figure 6). Six of these variants were also present in strains with degradation activity, while strains possessing the remaining seven variants showed no indication of nisin degradation. The NSR protein sequences of these seven strains were compared to variant 25, the most abundant form in strains that degrade nisin (also present in CH-1) and a homolog of S. aureus NSR of which the structure has been resolved (Khosa et al., 2016) (Supplementary Figure 6). One strain had a N-terminal truncated NSR (variant 21), which likely renders the protease incapable of being transported out of the cell where nisin degradation takes place. For the remaining variants, we found several amino acid substitutions in domains that we speculate abolishes NSR activity (Supplementary Figure 6C) (Khosa et al., 2016).
Strains that map to the bottom row in Figure 6 cause a decline in nisin A, indicative of nisin degradation. Almost all of the 250 milk fermentations (99%) in which a decrease in nisin was observed were carried out by nsr+ strains, further strengthening the view that NSR is the fundamental contributor to nisin degradation and hence allows strains to grow and acidify milk in the presence of nisin (Figure 6). We noted in the acidification experiments that strains containing nsr as well as nisin immunity showed a bimodal distribution with respect to delay in acidification (Figure 3C). Because NSR variant 17 or 25 were overrepresented in the subgroups with either more or less delay in acidification, respectively, we evaluated their respective nisin degradation capacities when situated in non-nisin producers and found that variant 25 reduced nisin A levels more than variant 17 (Supplementary Figure 6D).
In summary, it is evident from these nisin measurements that nisin degradation by NSR is very common and a mechanism utilized by L. lactis strains to protect against nisin.
Nisin-producing strains are added to milk fermentations for cheese production, but how genetic variation in L. lactis strains contributes to differences regarding milk acidification remains largely unexplored. We therefore investigated the effect of nisin on milk fermentations from 710 individual L. lactis strains. We found that changes in milk acidification by nisin can largely be explained by the presence or absence of nisin immunity (nisI, nisFEG) or degradation (nsr) genes, in a subspecies dependent manner. Still, within each group a range of strain-specific acidification profiles occurs, which reveal nuances that are easily overlooked in single strain studies focusing on the singular or combinatorial contribution of nisin degradation and/or immunity to nisin tolerance.
Our study identifies nisin degradation by NSR as a very common feature present in 38% of the L. lactis strain collection examined (710 strains). In view of these results, nisin degradation via NSR appears to be the main mechanism by which lactococci tolerate nisin. Based on this prevalence, we find it likely that one or more nsr+ L. lactis strains would occur both in natural milk fermentations and undefined starter cultures. In agreement, we have found NSR-specific breakdown products in milk fermented with L. lactis-based starter cultures, and in cheese (data not shown). The immunity factors NisFEG and NisI only deliver self-protection, while NSR offers a community-level nisin resistance, cross-protecting also nisin-sensitive strains through the degradation of nisin in the environment. Importantly, the data shows that ssp. lactis nsr+ strains are better equipped to degrade nisin in milk than ssp. cremoris nsr+ strains.
We believe that NSR activity in a given starter culture is an important determinant for the level by which nisin impacts mixed-strain milk fermentations. On the one hand, NSR presents an advantage by protecting the nisin-sensitive members of the starter culture. On the other hand, a reduction of nisin levels might allow for the growth of undesirable organisms, such as Clostridia, which will compete for nutrients and may lead to spoilage of the cheese (Meijer et al., 1998; Sallami et al., 2004). These antitheses pose an important challenge on starter culture design.
Nisin production and autoimmunity is a trait naturally encountered in strains isolated from raw milk and natural bulk starter cultures (Alegría et al., 2010; Cosentino et al., 2012). Of the L. lactis strains examined, 6.3% of the strains contain a full nisin biosynthesis cassette. Thus, this trait is significantly less common than the nsr genotype. Nisin synthesis is a property typically assigned to ssp. lactis strains, which is further supported by our dataset in which all but one of the nisin-producing strains belong to ssp. lactis (De Vuyst, 1994; Alegría et al., 2010; Virolainen et al., 2012). Strikingly, we have identified several nisin producers that also carry the nsr gene. At this stage we are unable to put forward an explanation for the role of NSR in nisin-producing strains and we question whether it has a different function that confers a competitive advantage under the right conditions.
Tolerance to nisin can be achieved via two mechanisms: degradation with proteases or activity of the immunity proteins NisI and NisFEG. A close inspection of the L. lactis collection revealed that about 9% of the strains possessed nisRK-FEG, but no synthesis genes. In general, these strains show mild delays in the onset of acidification. The expression of nisFEG genes is dependent on nisin-induced activation of NisRK (de Ruyter et al., 1996; Ra et al., 1996). Since nisRK-FEG strains do not produce nisin, it is likely that NisFEG proteins are absent in cells of pre-cultures but build up after inoculation in nisin-containing milk. The time that elapses before a sufficient amount of NisFEG is produced for cells to effectively remove nisin from the membrane and resume growth might dictate the severity of the milk acidification lag phase of nisRK-FEG strains. Interestingly, nisRK-FEG is almost uniquely found in ssp. lactis and only present in three ssp. cremoris strains. In fact, the unaccompanied nisIP operon is the only nisin element specific for ssp. cremoris strains. This plasmid-localized operon was shown to confer protection against 20 ng ml−1 nisin in ssp. cremoris NCDO712 (Tarazanova et al., 2016; Wels et al., 2019). However, the results presented in the current study do not support that NisI alone gives nisin immunity under application-relevant conditions in which nisin concentrations typically range from 1.25 to 7.5 μg ml−1 (Davies et al., 1997; Sallami et al., 2004; Aly et al., 2012). Because nisIP is only expressed at a low constitutive rate in the absence of nisin-induced nisABTCIPRK transcription (Kuipers et al., 1995; de Ruyter et al., 1996; Li and O'Sullivan, 2006; Trmčić et al., 2011), NisI levels are likely to remain lower in nisIP strains than in those with the full nisin cassette. NisI quantity directly impacts nisin tolerance due to the fixed 1:1 molar stoichiometry with which it interacts with nisin molecules (Takala et al., 2004; Hacker et al., 2015; Jeong and Ha, 2018). Once nisin outnumbers NisI proteins exposed on the cell surface, the latter can no longer protect nisin from reaching lipid II (AlKhatib et al., 2014a). Taken together, NisI in nisIP strains seems to constitute a functional defense mechanism only against low levels of nisin, for instance, when NSR is produced by the same or another strain in the culture. NisFEG in nisRK-FEG strains, on the other hand, allows cells to grow in the presence of higher levels of nisin. Like strains that have the full set of immunity genes in combination with NSR, NisFEG or NisI with NSR generally results in an additive effect in nisin tolerance. Given the lack of correlation between the presence of nisIP and tolerance to nisin in ssp. cremoris, it is reasonable to speculate that NisIP plays a different role in this subspecies.
Unlike ssp. lactis, nisin-tolerant ssp. cremoris strains appear to be rare, as substantiated by the following observations derived from our dataset. First, effective nisin immunity and/or resistance seem to be less prevalent in ssp. cremoris. Second, ssp. cremoris strains that contain nisFEG and/or nsr generally acidify nisin-containing milk with more delay than ssp. lactis of the same genotype. Third, those ssp. cremoris strains capable of acidifying milk with a delay have genotypes corresponding to a lactis phenotype (Kelleher et al., 2017; Wels et al., 2019). L. lactis ssp. cremoris strains are used for flavor development of specific cheeses such as cheddar (Broadbent et al., 1998; Børsting et al., 2015). This has been attributed to their ability to lyse, a process that can be accelerated by bacteriocins, thereby releasing intracellular enzymes like peptidases contributing to the flavor during ripening (Morgan et al., 1997). It can thus be argued that the nisin sensitivity of ssp. cremoris is desirable and might even have been selected for during its domestication. In general, ssp. cremoris are less tolerant than ssp. lactis to stress such as elevated temperature, low pH, salt and reactive oxygen species (Kim et al., 1999; Sanders et al., 1999; Dijkstra et al., 2014, 2016). Based on our results, we find that ssp. cremoris strains are also less tolerant to nisin.
We identified 20 ssp. lactis and two ssp. cremoris strains that are tolerant (UA or MDA) to nisin, but do not encode any obvious nisin immunity or degradation machineries. We performed a search for homologs of multidrug transporters such as the CprABC-type and BceAB-type systems that are known to confer resistance to nisin in other Gram-positive bacteria, including pathogens and spoilers (Clemens et al., 2018 and references therein), but the results were inconclusive (data not shown). At this stage, we cannot put forward an explanation to why nisin failed to impair acidification of milk by L. lactis strains devoid of nisin immunity and degradation pathways. It is well-known that Gram-positive bacteria can adapt to tolerate more nisin (Kramer et al., 2004, 2006, 2008; Giaouris et al., 2008; Bergholz et al., 2013). For instance, adaptive laboratory evolution yielded a ssp. lactis strain that could withstand over 75-fold more nisin than its mother strain through temporary transcriptomic alterations (Kramer et al., 2006). Changes in expression were mapped to genes involved in cell wall and phospholipid composition, drug transport and the cell envelope stress response (Kramer et al., 2006; Giaouris et al., 2008). Similar transcriptional responses were also observed in Staphylococcus aureus, Streptococcus pneumoniae, Listeria monocytogenes, and Bacillus subtilis challenged with nisin (Ming and Daeschel, 1995; Mazzotta and Montville, 1997; Peschel et al., 1999; Hansen et al., 2009; Collins et al., 2010a,b, 2012; Majchrzykiewicz et al., 2010). Moreover, L. lactis cells treated with nisin showed overlapping physiological responses to cells challenged with other membrane-perturbing compounds such as Lcn972, another lipid II-binding bacteriocin produced by L. lactis, c2 lytic phages, and lysozyme, or by the recombinant production of heterologous membrane proteins (Kramer et al., 2006; Martínez et al., 2007; Veiga et al., 2007; Fallico et al., 2011; Pinto et al., 2011; Roces et al., 2012a,b). In many cases, pre-activating expression of the identified genes improved tolerance toward membrane perturbing compounds including nisin. In order to pinpoint factors involved in the observed subspecies-dependent deviations in innate nisin tolerance, it would be of importance to study nisin-induced changes in gene expression profiles of ssp. lactis and ssp. cremoris strains that do not have any immunity or degradation machineries in place.
In conclusion, we have characterized a large set of L. lactis strains for which we mapped the contribution of nisin immunity and/or degradation genes to nisin tolerance and degradation during milk fermentations. The prevalence of nsr and associated nisin degradation is a more common trait than previously reported, albeit the degree of protection established by NSR seems to be highly variable. Full immunity (nisI plus nisFEG), especially in combination with nsr, conferred the best protection. Unlike nisFEG, nisI alone hardly delivered immunity under application-relevant conditions. Particularly, nisin impacts ssp. cremoris milk acidification more than ssp. lactis. Strains that do not produce nisin but have immunity are of great interest for the design of nisin-compatible starter cultures as these would circumvent the requirement for NSR to protect starter culture composition. Such strains have been identified previously and are also found in the current study (Tarazanova et al., 2016; Wels et al., 2019). We believe that the generated dataset lays a new foundation toward understanding how nisin influences cheese fermentation processes that typically involve starter cultures containing multiple nisin-related genotypic variants. Furthermore, the knowledge of how individual strains react to nisin is highly relevant for the design of compatible culture compositions to help fermentation deliver the desired flavor and characteristic properties together with a strong bioprotective effect in the cheese.
All data is available, except the whole genome sequences, because these are proprietary strains.
Experiments were designed by TE, JV, GO, LG, and KN. Experiments were performed by TE, LG, LH-D, AG, and KN. Data was analyzed by LH-D, LG, KN, KJ, EB, TE, AN, and GO. All authors contributed to the first draft with main contributions by LG and TE. Editing and finalization of manuscript was done by LG, LH-D, TE, AN, and GO.
The authors declare that this study was funded entirely by Chr. Hansen A/S. The funder was not involved in the study design, collection, analysis, interpretation of data, the writing of this article or the decision to submit it for publication.
All authors were employed by CHR HANSEN A/S during the work presented here. CHR HANSEN A/S is a company that develops and commercializes dairy starter cultures.
We acknowledge the input and effort made by Martin Karkov Kristensen to study the effect of nisin degrading strains in complex cultures and for inspirational scientific discussions. The large amount of data made available in this work could only be composed after a great effort done by Birgit Albrecht Svendsen, Elahe Ghanei Moghadam, Rikke Dollerup Bech, Helle Simonsen and our Chr. Hansen NGS team.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fbioe.2021.622835/full#supplementary-material
Supplementary Figure 1. Effect of nisin on milk acidification profiles by 710 L. lactis strains co-plotted according to subspecies, phenotypic classification, and genetic make-up of nisin-related gene sets. Milk acidification curves at 30°C of (A) L. lactis ssp. lactis strains and (B) L. lactis ssp. cremoris strains, in the absence (CFScontrol) or presence of 1.5 μg ml−1 nisin (CFSnisin) and further grouped according to acidification phenotype (rows) and presence or absence of genes for nisin immunity, synthesis, and/or degradation (columns). Thin lines depict averaged milk acidification data of three replicates while thick lines depict the averaged curves per group. The nisin-containing milk phenotypes are: LA, loss of acidification; HDA, highly delayed acidification; MDA, mildly delayed acidification; and UA, unaltered acidification. All acidification curves are synchronized based on acidification onset of each respective CFScontrol profile and normalized to a start pH of 6.4.
Supplementary Figure 2. Effect of nisin on milk acidification of strains containing a full nisin biosynthesis gene cassette. Graphs depicting milk acidification at 30°C in the absence (CFScontrol) or presence of 1.5 μg ml−1 nisin (CFSnisin) by strains with nisABTCIPRK-FEG genotypes mapping to the (A) MDA or (B) UA phenotypic group. Each graph depicts the averaged curve of three replicates. Strain IDs as further described in Supplementary Table 1 and subspecies (lactis or cremoris) are indicated above each pair of curves. All acidification curves are synchronized based on acidification onset of each respective CFScontrol profile and normalized to a start pH of 6.4.
Supplementary Figure 3. Genotypic diversity of the L. lactis strains as revealed by a MLST tree. A MLST-tree depicting the genotypic relatedness of 916 L. lactis ssp. cremoris and ssp. lactis strains. Indicated from outer to inner ring are: (A) Strain-specific genome sequence origin, obtained either from the 710 draft genomes from this study or the publicly available 206 L. lactis RefSeq genomes. (B) Strain-specific presence of nisin-related genes or gene clusters. (C) Strain-specific milk acidification phenotypes. (D) Subspecies clusters of L. lactis ssp. lactis and L. lactis ssp. cremoris.
Supplementary Figure 4. The presence of nisI as the sole nisin tolerance factor is not sufficient to deliver protection of ssp. cremoris strains to dairy-relevant nisin concentrations. Single replicates of milk acidification profiles of L. lactis ssp. cremoris strains with nsr- nisIP+ nisFEG- genotypes in the absence (CFScontrol), presence of 0.2 μg ml−1 nisin or presence of 1.5 μg ml−1 (CFScontrol). All acidification curves are synchronized based on acidification onset of each respective CFScontrol profile and normalized to a start pH of 6.4.
Supplementary Figure 5. The presence of nsr leads to greater nisin tolerance in ssp. lactis strains than in ssp. cremoris strains. Upper panels: Milk acidification profiles of ssp. cremoris and ssp. lactis nsr+ strains in the absence (CFScontrol) or presence (CFSnisin) of 1.5 μg ml−1 nisin. Thin lines depict the averaged curve between replicates of the same strains while thick lines depict the averaged acidification of each genotypic group. All acidification curves are synchronized based on acidification onset of each respective CFScontrol profile and normalized to a start pH of 6.4. Lower panels: Subspecific distributions of nsr+ strains over acidification phenotypes LA, HDA, MDA, and UA per subspecies.
Supplementary Figure 6. Nisin A levels by NSR variant and subspecies show different protection efficiency. Dot plots depicting resultant nisin A concentrations as quantified with HPLC-MS/MS after incubating 0.9 μg ml−1 of nisin A in milk inoculated with (A) L. lactis ssp. cremoris or (B) L. lactis ssp. lactis, as a function of the NSR variant detected in each strain. Dots are further coded according to the phenotypic group (LA, HDA, MDA, and UA) each strain belongs (colors) and if the nsr gene is present alone or accompanied by nisin biosynthesis genes (shapes). NSR variants highlighted in green or blue/red indicate most common or non-nisin-degrading NSR variants, respectively. (C) Protein sequence alignments of the second-most common NSR variant 17 and non-degrading NSR variants with the most common and functional NSR variant 25 from CH-2. Color-shaded regions of the query sequence of NSR variant 25 indicate domains important for NSR catalytic activity and/or nisin binding reported for the structure function of NSR from S. agalactiae (Khosa et al., 2016). (D) Box and raincloud plot depicting the difference in resultant nisin A levels in milk fermentations performed with non-nisin A producing strains containing either NSR variant 17 or 25 (Mann-Whitney U, P < 0.01).
Supplementary Table 1. List of strains, their genotypes and milk acidification phenotypes, used in this study.
Supplementary Table 2. Query sequences used for gene blasts to identify nisin biosynthesis, immunity and degradation genes.
Supplementary Table 3. Details on data processing steps.
Supplementary Table 4. Primers used in this study.
Supplementary Table 5. Separate milk acidification profiles for all 710 L. lactis strains in the presence and absence of nisin.
Supplementary File 1. Sequences of the 12 conserved genes (dnaK, fusA, groEL, gyrA, gyrB, ileS, lep, pheS, recA, rpoA, rpoB, and rpoC) for each of the 710 L. lactis strains, used for the MLST tree presented in Supplementary Figure 3.
Alegría, Á., Delgado, S., Roces, C., López, B., and Mayo, B. (2010). Bacteriocins produced by wild Lactococcus lactis strains isolated from traditional, starter-free cheeses made of raw milk. Int. J. Food Microbiol. 143, 61–66. doi: 10.1016/j.ijfoodmicro.2010.07.029
Alifax, P. R., and Chevalier, R. (1962). Etude de la nisinase produite par Streptococcus thermophilus. J. Dairy Res. 29, 233–240. doi: 10.1017/S0022029900011043
AlKhatib, Z., Lagedroste, M., Fey, I., Kleinschrodt, D., Abts, A., and Smits, S. H. J. (2014a). Lantibiotic immunity: inhibition of nisin mediated pore formation by NisI. PLoS ONE 9:e102246. doi: 10.1371/journal.pone.0102246
AlKhatib, Z., Lagedroste, M., Zaschke, J., Wagner, M., Abts, A., Fey, I., et al. (2014b). The C-terminus of nisin is important for the ABC transporter NisFEG to confer immunity in Lactococcus lactis. Microbiol. Open 3, 752–763. doi: 10.1002/mbo3.205
Allen, M., Poggiali, D., Whitaker, K., Marshall, T. R., and Kievit, R. A. (2019). Raincloud plots: a multi-platform tool for robust data visualization. Wellcome Open Res. 4:63. doi: 10.12688/wellcomeopenres.15191.1
Aly, S., Floury, J., Piot, M., Lortal, S., and Jeanson, S. (2012). The efficacy of nisin can drastically vary when produced in situ in model cheeses. Food Microbiol. 32, 185–190. doi: 10.1016/j.fm.2012.06.001
Ávila, M., Gómez-Torres, N., Hernández, M., and Garde, S. (2014). Inhibitory activity of reuterin, nisin, lysozyme and nitrite against vegetative cells and spores of dairy-related Clostridium species. Int. J. Food Microbiol. 172, 70–75. doi: 10.1016/j.ijfoodmicro.2013.12.002
Ayad, E. H. E., Verheul, A., Wouters, J. T. M., and Smit, G. (2001). Population dynamics of lactococci from industrial, artisanal and non-dairy origins in defined strain starters for Gouda-type cheese. Int. Dairy J. 11, 51–61. doi: 10.1016/S0958-6946(01)00041-3
Bergholz, T. M., Tang, S., Wiedmann, M., and Boor, K. J. (2013). Nisin resistance of Listeria monocytogenes is increased by exposure to salt stress and is mediated via LiaR. Appl. Environ. Microbiol. 79, 5682–5688. doi: 10.1128/AEM.01797-13
Bierbaum, G., and Sahl, H.-G. (1985). Induction of autolysis of staphylococci by the basic peptide antibiotics Pep 5 and nisin and their influence on the activity of autolytic enzymes. Arch. Microbiol. 141, 249–254. doi: 10.1007/BF00408067
Børsting, M. W., Stallknecht, M. K., Vogensen, F. K., and Ardö, Y. (2015). Influence of proteolytic Lactococcus lactis subsp. cremoris on ripening of reduced-fat Cheddar cheese made with camel chymosin. Int. Dairy J. 41, 38–45. doi: 10.1016/j.idairyj.2014.09.002
Breukink, E., Wiedemann, I., van Kraaij, C., Kuipers, O. P., Sahl, H. G., and de Kruijff, B. (1999). Use of the cell wall precursor lipid II by a pore-forming peptide antibiotic. Science 286, 2361–2364. doi: 10.1126/science.286.5448.2361
Broadbent, J. R., Strickland, M., Weimer, B. C., Johnson, M. E., and Steele, J. L. (1998). Peptide accumulation and bitterness in Cheddar cheese made using single-strain Lactococcus lactis starters with distinct proteinase specificities. J. Dairy Sci. 81, 327–337. doi: 10.3168/jds.S0022-0302(98)75581-X
Brötz, H., Josten, M., Wiedemann, I., Schneider, U., Götz, F., Bierbaum, G., et al. (1998). Role of lipid-bound peptidoglycan precursors in the formation of pores by nisin, epidermin and other lantibiotics. Mol. Microbiol. 30, 317–327. doi: 10.1046/j.1365-2958.1998.01065.x
Clemens, R., Zaschke-Kriesche, J., Khosa, S., and Smits, S. H. J. (2018). Insight into two ABC transporter families involved in lantibiotic resistance. Front. Mol. Biosci. 4:91. doi: 10.3389/fmolb.2017.00091
Collins, B., Curtis, N., Cotter, P. D., Hill, C., and Ross, R. P. (2010a). The ABC transporter AnrAB contributes to the innate resistance of Listeria monocytogenes to nisin, bacitracin, and various β-lactam antibiotics. Antimicrob. Agents Chemother. 54, 4416–4423. doi: 10.1128/AAC.00503-10
Collins, B., Guinane, C. M., Cotter, P. D., Hill, C., and Paul Ross, R. (2012). Assessing the contributions of the LiaS histidine kinase to the innate resistance of Listeria monocytogenes to nisin, cephalosporins, and disinfectants. Appl. Environ. Microbiol. 78, 2923–2929. doi: 10.1128/AEM.07402-11
Collins, B., Joyce, S., Hill, C., Cotter, P. D., and Ross, R. P. (2010b). TelA contributes to the innate resistance of Listeria monocytogenes to nisin and other cell wall-acting antibiotics. Antimicrob. Agents Chemother. 54, 4658–4663. doi: 10.1128/AAC.00290-10
Cosentino, S., Fadda, M. E., Deplano, M., Melis, R., Pomata, R., and Pisano, M. B. (2012). Antilisterial activity of nisin-like bacteriocin-producing Lactococcus lactis subsp. lactis isolated from traditional Sardinian dairy products. J. Biomed. Biotechnol. 2012:376428. doi: 10.1155/2012/376428
Davies, E. A., Bevis, H. E., and Delves-Broughton, J. (1997). The use of the bacteriocin, nisin, as a preservative in ricotta-type cheeses to control the food-borne pathogen Listeria monocytogenes. Lett. Appl. Microbiol. 24, 343–346. doi: 10.1046/j.1472-765X.1997.00145.x
de Arauz, L. J., Jozala, A. F., Mazzola, P. G., and Vessoni Penna, T. C. (2009). Nisin biotechnological production and application: a review. Trends Food Sci. Technol. 20, 146–154. doi: 10.1016/j.tifs.2009.01.056
De Kwaadsteniet, M., Ten Doeschate, K., and Dicks, L. M. T. (2008). Characterization of the structural gene encoding nisin F, a new lantibiotic produced by a Lactococcus lactis subsp. lactis isolate from freshwater catfish (Clarias gariepinus). Appl. Environ. Microbiol. 74, 547–549. doi: 10.1128/AEM.01862-07
de Ruyter, P. G., Kuipers, O. P., Beerthuyzen, M. M., van Alen-Boerrigter, I., and de Vos, W. M. (1996). Functional analysis of promoters in the nisin gene cluster of Lactococcus lactis. J. Bacteriol. 178, 3434–3439. doi: 10.1128/JB.178.12.3434-3439.1996
De Vuyst, L. (1994). Nisin production variability between natural Lactococcus lactis subsp. lactis strains. Biotechnol. Lett. 16, 287–292. doi: 10.1007/BF00134627
Delves-Broughton, J. (1996). Applications of the bacteriocin, nisin. Antonie van Leeuwenhoek 69, 193–202. doi: 10.1007/BF00399424
Dijkstra, A. R., Alkema, W., Starrenburg, M. J. C., Hugenholtz, J., Van Hijum, S. A. F. T., and Bron, P. A. (2016). Strain-dependent transcriptome signatures for robustness in Lactococcus lactis. PLoS ONE 11:e0167944. doi: 10.1371/journal.pone.0167944
Dijkstra, A. R., Setyawati, M. C., Bayjanov, J. R., Alkema, W., Van Hijum, S. A. F. T., Bron, P. A., et al. (2014). Diversity in robustness of Lactococcus lactis strains during heat stress, oxidative stress, and spray drying stress. Appl. Environ. Microbiol. 80, 603–611. doi: 10.1128/AEM.03434-13
Draper, L. A., Cotter, P. D., Hill, C., and Ross, R. P. (2015). Lantibiotic resistance. Microbiol. Mol. Biol. Rev. 79, 171–191. doi: 10.1128/MMBR.00051-14
Duan, K., Harvey, M. L., Liu, C.-Q., and Dunn, N. W. (1996). Identification and characterization of a mobilizing plasmid, pND300, in Lactococcus lactis M189 and its encoded nisin resistance determinant. J. Appl. Bacteriol. 81, 493–500. doi: 10.1111/j.1365-2672.1996.tb03538.x
Düsterhöft, E. M., Engels, W., and Huppertz, T. (2017). “Chapter 34: gouda and related cheeses,” in Cheese, 4th Edn, eds P. L. H. McSweeney, P. F. Fox, P. D. Cotter, and D. W. Everett (San Diego, CA: Academic Press), 865–888. doi: 10.1016/B978-0-12-417012-4.00034-X
Egan, K., Field, D., Rea, M. C., Ross, R. P., Hill, C., and Cotter, P. D. (2016). Bacteriocins: Novel solutions to age old spore-related problems? Front. Microbiol. 7:461. doi: 10.3389/fmicb.2016.00461
Engelke, G., Gutowski-Eckel, Z., Kiesau, P., Siegers, K., Hammelmann, M., and Entian, K. D. (1994). Regulation of nisin biosynthesis and immunity in Lactococcus lactis 6F3. Appl. Environ. Microbiol. 60, 814–825. doi: 10.1128/AEM.60.3.814-825.1994
Erkus, O., De Jager, V. C. L., Spus, M., Van Alen-Boerrigter, I. J., Van Rijswijck, I. M. H., Hazelwood, L., et al. (2013). Multifactorial diversity sustains microbial community stability. ISME J. 7, 2126–2136. doi: 10.1038/ismej.2013.108
Fallico, V., Ross, R. P., Fitzgerald, G. F., and McAuliffe, O. (2011). Genetic response to bacteriophage infection in Lactococcus lactis reveals a four-strand approach involving induction of membrane stress proteins, D-alanylation of the cell wall, maintenance of proton motive force, and energy conservation. J. Virol. 85, 12032–12042. doi: 10.1128/JVI.00275-11
FAO (2013). Compendium of Food Additive Specifications. Joint FAO/WHO Expert Committee on Food Additives (JECFA), 77th Meeting 2013. FAO JECFA Monographs 14. Available online at: http://www.fao.org/3/a-at879e.pdf
Froseth, B. R., and McKay, L. L. (1991). Molecular characterization of the nisin resistance region of Lactococcus lactis subsp. lactis biovar diacetylactis DRC3. Appl. Environ. Microbiol. 57, 804–811. doi: 10.1128/AEM.57.3.804-811.1991
Fukao, M., Obita, T., Yoneyama, F., Kohda, D., Zendo, T., Nakayama, J., et al. (2008). Complete covalent structure of nisin Q, new natural nisin variant, containing post-translationally modified amino acids. Biosci. Biotechnol. Biochem. 72, 1750–1755. doi: 10.1271/bbb.80066
Giaouris, E., Briandet, R., Meyrand, M., Courtin, P., and Chapot-Chartier, M. P. (2008). Variations in the degree of D-alanylation of teichoic acids in Lactococcus lactis alter resistance to cationic antimicrobials but have no effect on bacterial surface hydrophobicity and charge. Appl. Environ. Microbiol. 74, 4764–4767. doi: 10.1128/AEM.00078-08
Gross, E., and Morell, J. L. (1971). The structure of nisin. J. Am. Chem. Soc. 93, 4634–4635. doi: 10.1021/ja00747a073
Gu, Z., Eils, R., and Schlesner, M. (2016). Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 32, 2847–2849. doi: 10.1093/bioinformatics/btw313
Hacker, C., Christ, N. A., Duchardt-Ferner, E., Korn, S., Göbl, C., Berninger, L., et al. (2015). The solution structure of the lantibiotic immunity protein NisI and its interactions with nisin. J. Biol. Chem. 290, 28869–28886. doi: 10.1074/jbc.M115.679969
Hansen, M. E., Wangari, R., Hansen, E. B., Mijakovic, I., and Jensen, P. R. (2009). Engineering of Bacillus subtilis 168 for increased nisin resistance. Appl. Environ. Microbiol. 75, 6688–6695. doi: 10.1128/AEM.00943-09
Hasper, H. E., De Kruijff, B., and Breukink, E. (2004). Assembly and stability of nisin-Lipid II pores. Biochemistry 43, 11567–11575. doi: 10.1021/bi049476b
Hasper, H. E., Kramer, N. E., Smith, J. L., Hillman, J. D., Zachariah, C., Kuipers, O. P., et al. (2006). An alternative bactericidal mechanism of action for lantibiotic peptides that target lipid II. Science 313, 1636–1637. doi: 10.1126/science.1129818
Hirsch, A. (1951). Growth and nisin production of a strain of Streptococcus lactis. J. Gen. Microbiol. 5, 208–221. doi: 10.1099/00221287-5-1-208
Jeong, J. H., and Ha, S. C. (2018). Crystal structure of NisI in a lipid-free form, the nisin immunity protein, from Lactococcus lactis. Antimicrob. Agents Chemother. 62. doi: 10.1128/AAC.01966-17
Kelleher, P., Bottacini, F., Mahony, J., Kilcawley, K. N., and van Sinderen, D. (2017). Comparative and functional genomics of the Lactococcus lactis taxon; insights into evolution and niche adaptation. BMC Genomics 18:267. doi: 10.1186/s12864-017-3650-5
Khosa, S., AlKhatib, Z., and Smits, S. H. J. (2013). NSR from Streptococcus agalactiae confers resistance against nisin and is encoded by a conserved nsr operon. Biol. Chem. 394, 1543–1549. doi: 10.1515/hsz-2013-0167
Khosa, S., Frieg, B., Mulnaes, D., Kleinschrodt, D., Hoeppner, A., Gohlke, H., et al. (2016). Structural basis of lantibiotic recognition by the nisin resistance protein from Streptococcus agalactiae. Sci. Rep. 6:18679. doi: 10.1038/srep18679
Khosa, S., Hoeppner, A., Kleinschrodt, D., and Smits, S. H. J. (2015). Overexpression, purification, crystallization and preliminary X-ray diffraction of the nisin resistance protein from Streptococcus agalactiae. Acta Crystallogr. Sect. FStructural Biol. Commun. 71(Pt 6), 671–675. doi: 10.1107/S2053230X15006226
Kim, W. S., Ren, J., and Dunn, N. W. (1999). Differentiation of Lactococcus lactis subspecies lactis and subspecies cremoris strains by their adaptive response to stresses. FEMS Microbiol. Lett. 171, 57–65. doi: 10.1111/j.1574-6968.1999.tb13412.x
Kleerebezem, M., Quadri, L. E. N., Kuipers, O. P., and De Vos, W. M. (1997). Quorum sensing by peptide pheromones and two-component signal-transduction systems in Gram-positive bacteria. Mol. Microbiol. 24, 895–904. doi: 10.1046/j.1365-2958.1997.4251782.x
Kramer, N. E., Hasper, H. E., van den Bogaard, P. T. C., Morath, S., de Kruijff, B., Hartung, T., et al. (2008). Increased D-alanylation of lipoteichoic and a thickened septum are main determinants in the nisin resistance mechanism of Lactococcus lactis. Microbiology 154, 1755–1762. doi: 10.1099/mic.0.2007/015412-0
Kramer, N. E., Smid, E. J., Kok, J., De Kruijff, B., Kuipers, O. P., and Breukink, E. (2004). Resistance of Gram-positive bacteria to nisin is not determined by Lipid II levels. FEMS Microbiol. Lett. 239, 157–161. doi: 10.1016/j.femsle.2004.08.033
Kramer, N. E., Van Hijum, S. A. F. T., Knol, J., Kok, J., and Kuipers, O. P. (2006). Transcriptome analysis reveals mechanisms by which Lactococcus lactis acquires nisin resistance. Antimicrob. Agents Chemother. 50, 1753–1761. doi: 10.1128/AAC.50.5.1753-1761.2006
Kuipers, O. P., Beerthuyzen, M. M., De Ruyter, P. G. G. A., Luesink, E. J., and De Vos, W. M. (1995). Autoregulation of nisin biosynthesis in Lactococcus lactis by signal transduction. J. Biol. Chem. 270, 27299–27304. doi: 10.1074/jbc.270.45.27299
Kuipers, O. P., Beerthuyzen, M. M., Siezen, R. J., and De Vos, W. M. (1993). Characterization of the nisin gene cluster nisABTCIPR of Lactococcus lactis: requirement of expression of the nisA and nisI genes for development of immunity. Eur. J. Biochem. 216, 281–291. doi: 10.1111/j.1432-1033.1993.tb18143.x
Letunic, I., and Bork, P. (2019). Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 47, W256–W259. doi: 10.1093/nar/gkz239
Li, H., and O'Sullivan, D. J. (2006). Identification of a nisI promoter within the nisABCTIP operon that may enable establishment of nisin immunity prior to induction of the operon via signal transduction. J. Bacteriol. 188, 8496–8503. doi: 10.1128/JB.00946-06
Liu, C. Q., Harvey, M. L., and Dunn, N. W. (1997). Cloning of a gene encoding nisin resistance from Lactococcus lactis subsp. lactis M189 which is transcribed from an extended−10 promoter. J. Gen. Appl. Microbiol. 43, 67–73. doi: 10.2323/jgam.43.67
Majchrzykiewicz, J. A., Kuipers, O. P., and Bijlsma, J. J. E. (2010). Generic and specific adaptive responses of Streptococcus pneumoniae to challenge with three distinct antimicrobial peptides, bacitracin, LL-37, and nisin. Antimicrob. Agents Chemother. 54, 440–451. doi: 10.1128/AAC.00769-09
MartÃnezCuesta, M. C., Kok, J., Herranz, E., Peláez, C., Requena, T., and Buist, G. (2000). Requirement of autolytic activity for bacteriocin-induced lysis. Appl. Environ. Microbiol. 66, 3174–3179. doi: 10.1128/AEM.66.8.3174-3179.2000
Martínez, B., Zomer, A. L., Rodríguez, A., Kok, J., and Kuipers, O. P. (2007). Cell envelope stress induced by the bacteriocin Lcn972 is sensed by the lactococcal two-component system CesSR. Mol. Microbiol. 64, 473–486. doi: 10.1111/j.1365-2958.2007.05668.x
Mattick, A. T., and Hirsch, A. (1947). Further observations on an inhibitory substance (nisin) from lactic streptococci. Lancet 2, 5–8. doi: 10.1016/s0140-6736(47)90004-4
Mazzotta, A. S., and Montville, T. J. (1997). Nisin induces changes in membrane fatty acid composition of Listeria monocytogenes nisin-resistant strains at 10°C and 30°C. J. Appl. Microbiol. 82, 32–38. doi: 10.1111/j.1365-2672.1997.tb03294.x
McKay, L. L., and Baldwin, K. A. (1984). Conjugative 40-megadalton plasmid in Streptococcus lactis subsp. diacetylactis DRC3 is associated with resistance to nisin and bacteriophage. Appl. Environ. Microbiol. 47, 68–74. doi: 10.1128/AEM.47.1.68-74.1984
Meijer, W., van de Bunt, B., Twigt, M., de Jonge, B., Smit, G., and Hugenholtz, J. (1998). Lysis of Lactococcus lactis subsp. cremoris SK110 and its nisin-immune transconjugant in relation to flavor development in cheese. Appl. Environ. Microbiol. 64, 1950–1953. doi: 10.1128/AEM.64.5.1950-1953.1998
Ming, X., and Daeschel, M. A. (1995). Correlation of cellular phospholipid content with nisin resistance of Listeria monocytogenes Scott A. J. Food Prot. 58, 416–420. doi: 10.4315/0362-028X-58.4.416
Molloy, E. M., Hill, C., Cotter, P. D., and Ross, R. P. (2011). “Bacteriocins,” in Encyclopedia of Dairy Sciences, 2nd Edn, eds J. W. Fuquay, P. F. Fox, and P. L. H. McSweeney (London: Elsevier Inc; Academic Press), 420–429. doi: 10.1016/B978-0-12-374407-4.00049-2
Morgan, S., Ross, R. P., and Hill, C. (1997). Increasing starter cell lysis in Cheddar cheese using a bacteriocin-producing adjunct. J. Dairy Sci. 80, 1–10. doi: 10.3168/jds.S0022-0302(97)75906-X
Mulders, J. W. M., Boerrigter, I. J., Rollema, H. S., Siezen, R. J., and de Vos, W. M. (1991). Identification and characterization of the lantibiotic nisin Z, a natural nisin variant. Eur. J. Biochem. 201, 581–584. doi: 10.1111/j.1432-1033.1991.tb16317.x
Peschel, A., Otto, M., Jack, R. W., Kalbacher, H., Jung, G., and Götz, F. (1999). Inactivation of the dlt operon in Staphylococcus aureus confers sensitivity to defensins, protegrins, and other antimicrobial peptides. J. Biol. Chem. 274, 8405–8410. doi: 10.1074/jbc.274.13.8405
Pinto, J. P. C., Kuipers, O. P., Marreddy, R. K. R., Poolman, B., and Kok, J. (2011). Efficient overproduction of membrane proteins in Lactococcus lactis requires the cell envelope stress sensor/regulator couple CesSR. PLoS ONE 6:e21873. doi: 10.1371/journal.pone.0021873
Poulsen, V. K., Derkx, P., and Oregaard, G. (2019). High-throughput screening for texturing Lactococcus strains. FEMS Microbiol. Lett. 366. doi: 10.1093/femsle/fnz001
Qiao, M., Immonen, T., Koponen, O., and Saris, P. E. J. (1995). The cellular location and effect on nisin immunity of the NisI protein from Lactococcus lactis N8 expressed in Escherichia coli and L. lactis. FEMS Microbiol. Lett. 131, 75–80. doi: 10.1111/j.1574-6968.1995.tb07757.x
Ra, R., Beerthuyzen, M. M., De Vos, W. M., Saris, P. E. J., and Kuipers, O. P. (1999). Effects of gene disruptions in the nisin gene cluster of Lactococcus lactis on nisin production and producer immunity. Microbiology 145, 1227–1233. doi: 10.1099/13500872-145-5-1227
Ra, R. S., and Saris, P. E. J. (1995). Characterization of prokaryotic mRNAs by RT-PCR. Biotechniques 18, 792–795.
Ra, S. R., Qiao, M., Immonen, T., Pujana, I., and Saris, P. E. J. (1996). Genes responsible for nisin synthesis, regulation and immunity form a regulon of two operons and are induced by nisin in Lactoccocus lactis N8. Microbiology 142, 1281–1288. doi: 10.1099/13500872-142-5-1281
Roces, C., Courtin, P., Kulakauskas, S., Rodríguez, A., Chapot-Chartier, M. P., and Martínez, B. (2012a). Isolation of Lactococcus lactis mutants simultaneously resistant to the cell wall-active bacteriocin Lcn972, lysozyme, nisin, and bacteriophage c2. Appl. Environ. Microbiol. 78, 4157–4163. doi: 10.1128/AEM.00795-12
Roces, C., Pérez, V., Campelo, A. B., Blanco, D., Kok, J., Kuipers, O. P., et al. (2012b). The putative lactococcal extracytoplasmic function anti-sigma factor Llmg2447 determines resistance to the cell wall-active bacteriocin Lcn972. Antimicrob. Agents Chemother. 56, 5520–5527. doi: 10.1128/AAC.01206-12
Sahl, H. G., Kordel, M., and Benz, R. (1987). Voltage-dependent depolarization of bacterial membranes and artificial lipid bilayers by the peptide antibiotic nisin. Arch. Microbiol. 149, 120–124. doi: 10.1007/BF00425076
Sallami, L., Kheadr, E. E., Fliss, and Vuillemard, J. C. (2004). Impact of autolytic, proteolytic, and nisin-producing adjunct cultures on biochemical and textural properties of cheddar cheese. J. Dairy Sci. 87, 1585–1594. doi: 10.3168/jds.S0022-0302(04)73312-3
Sambrook, J., Fritsch, E. F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual. Cold Spring Harbour, NY: Cold Spring Harbor Laboratory Press.
Sanders, J. W., Venema, G., and Kok, J. (1999). Environmental stress responses in Lactococcus lactis. FEMS Microbiol. Rev. 23, 483–501. doi: 10.1111/j.1574-6976.1999.tb00409.x
Schliep, K. P. (2011). phangorn: phylogenetic analysis in R. Bioinformatics 27, 592–593. doi: 10.1093/bioinformatics/btq706
Siegers, K., and Entian, K. D. (1995). Genes involved in immunity to the lantibiotic nisin produced byLactococcus lactis 6F3. Appl. Environ. Microbiol. 61:1082. doi: 10.1128/AEM.61.3.1082-1089.1995
Solem, C., Defoor, E., Jensen, P. R., and Martinussen, J. (2008). Plasmid pCS1966, a new selection/counterselection tool for lactic acid bacterium strain construction based on the oroP gene, encoding an orotate transporter from Lactococcus lactis. Appl. Environ. Microbiol. 74, 4772–4775. doi: 10.1128/AEM.00134-08
Stein, T., Heinzmann, S., Solovieva, I., and Entian, K. D. (2003). Function of Lactococcus lactis nisin immunity genes nisI and nisFEG after coordinated expression in the surrogate host Bacillus subtilis. J. Biol. Chem. 278, 89–94. doi: 10.1074/jbc.M207237200
Sun, Z., Zhong, J., Liang, X., Liu, J., Chen, X., and Huan, L. (2009). Novel mechanism for nisin resistance via proteolytic degradation of nisin by the nisin resistance protein NSR. Antimicrob. Agents Chemother. 53, 1964–1973. doi: 10.1128/AAC.01382-08
Takala, T. M., Koponen, O., Qiao, M., and Saris, P. E. J. (2004). Lipid-free NisI: interaction with nisin and contribution to nisin immunity via secretion. FEMS Microbiol. Lett. 237, 171–177. doi: 10.1111/j.1574-6968.2004.tb09693.x
Tarazanova, M., Beerthuyzen, M., Siezen, R., Fernandez-Gutierrez, M. M., de Jong, A., van der Meulen, S., et al. (2016). Plasmid complement of Lactococcus lactis NCDO712 reveals a novel pilus gene cluster. PLoS ONE 11:e0167970. doi: 10.1371/journal.pone.0167970
Trmčić, A., Monnet, C., Rogelj, I., and Bogovič Matijašić, B. (2011). Expression of nisin genes in cheese: a quantitative real-time polymerase chain reaction approach. J. Dairy Sci. 94, 77–85. doi: 10.3168/jds.2010-3677
Veiga, P., Bulbarela-Sampieri, C., Furlan, S., Maisons, A., Chapot-Chartier, M. P., Erkelenz, M., et al. (2007). SpxB regulates O-acetylation-dependent resistance of Lactococcus lactis peptidoglycan to hydrolysis. J. Biol. Chem. 282, 19342–19354. doi: 10.1074/jbc.M611308200
Virolainen, N., Guglielmetti, S., Arioli, S., and Karp, M. (2012). Bioluminescence-based identification of nisin producers - a rapid and simple screening method for nisinogenic bacteria in food samples. Int. J. Food Microbiol. 158, 126–132. doi: 10.1016/j.ijfoodmicro.2012.07.007
Wels, M., Siezen, R., Van Hijum, S., Kelly, W. J., and Bachmann, H. (2019). Comparative genome analysis of Lactococcus lactis indicates niche adaptation and resolves genotype/phenotype disparity. Front. Microbiol. 10:4. doi: 10.3389/fmicb.2019.00004
Wiedemann, I., Breukink, E., van Kraaij, C., Kuipers, O. P., Bierbaum, G., de Kruijff, B., et al. (2001). Specific binding of nisin to the peptidoglycan precursor lipid II combines pore formation and inhibition of cell wall biosynthesis for potent antibiotic activity. J. Biol. Chem. 276, 1772–1779. doi: 10.1074/jbc.M006770200
Keywords: high-throughput screening, milk acidification, nisin degradation, nisin tolerance, nisin biosynthesis, NSR, Lactococcus lactis, gene-trait matching
Citation: van Gijtenbeek LA, Eckhardt TH, Herrera-Domínguez L, Brockmann E, Jensen K, Geppel A, Nielsen KF, Vindeloev J, Neves AR and Oregaard G (2021) Gene-Trait Matching and Prevalence of Nisin Tolerance Systems in Lactococus lactis. Front. Bioeng. Biotechnol. 9:622835. doi: 10.3389/fbioe.2021.622835
Received: 29 October 2020; Accepted: 22 January 2021;
Published: 03 March 2021.
Edited by:
Jian-Ming Liu, Technical University of Denmark, DenmarkReviewed by:
Per Erik Joakim Saris, University of Helsinki, FinlandCopyright © 2021 van Gijtenbeek, Eckhardt, Herrera-Domínguez, Brockmann, Jensen, Geppel, Nielsen, Vindeloev, Neves and Oregaard. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gunnar Oregaard, ZGtnb3JAY2hyLWhhbnNlbi5jb20=
†These authors have contributed equally to this work
‡Present address: Thomas H. Eckhardt, DSM Food Specialties, Wageningen, Netherlands
Ana Rute Neves, Arla Foods Ingredients P/S, Videbaek, Denmark
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.