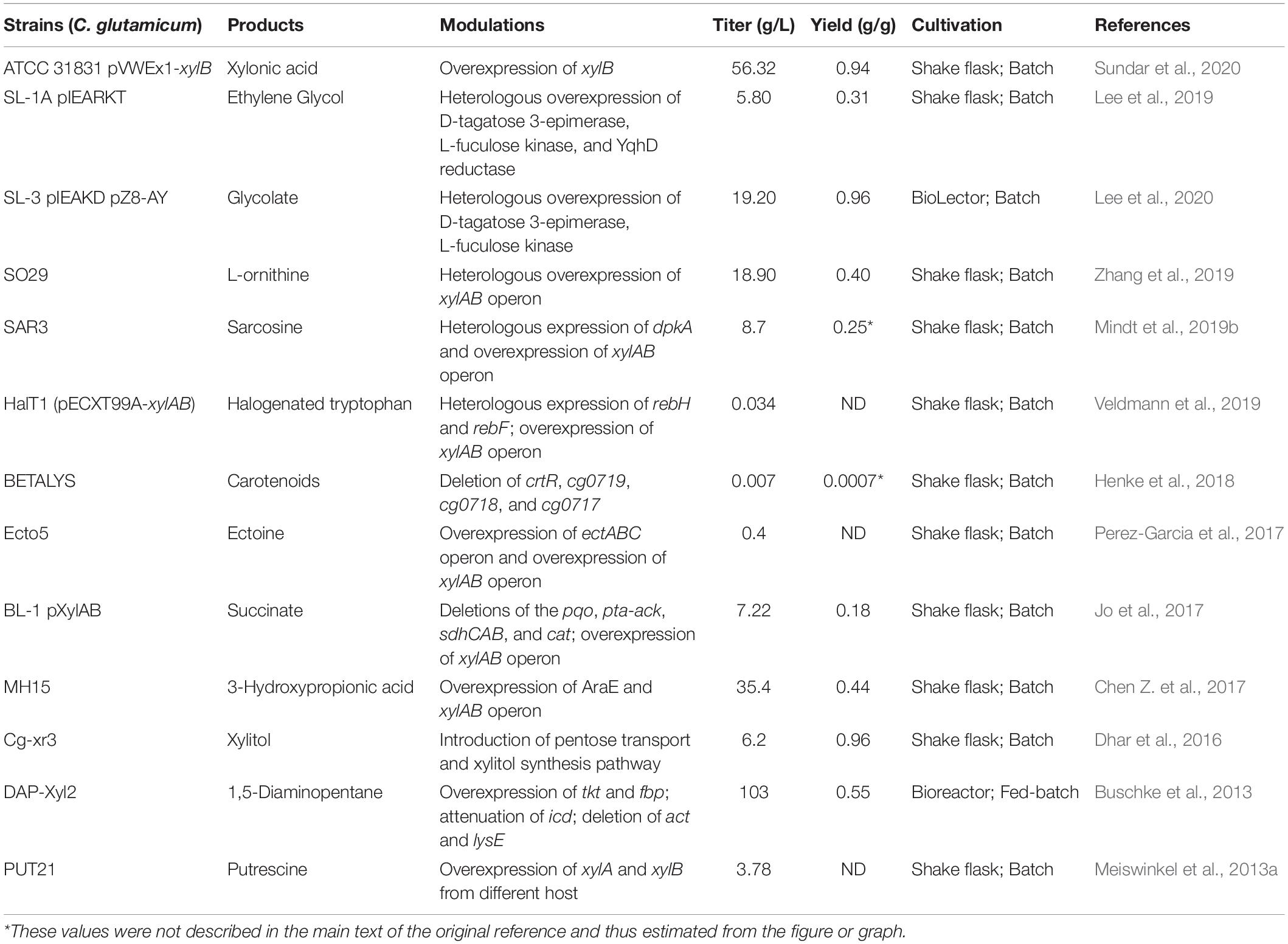
- 1College of Bioscience and Bioengineering, Jiangxi Agricultural University, Nanchang, China
- 2Jiangxi Engineering Laboratory for the Development and Utilization of Agricultural Microbial Resources, Jiangxi Agricultural University, Nanchang, China
Due to the non-renewable nature of fossil fuels, microbial fermentation is considered a sustainable approach for chemical production using glucose, xylose, menthol, and other complex carbon sources represented by lignocellulosic biomass. Among these, xylose, methanol, arabinose, glycerol, and other alternative feedstocks have been identified as superior non-food sustainable carbon substrates that can be effectively developed for microbe-based bioproduction. Corynebacterium glutamicum is a model gram-positive bacterium that has been extensively engineered to produce amino acids and other chemicals. Recently, in order to reduce production costs and avoid competition for human food, C. glutamicum has also been engineered to broaden its substrate spectrum. Strengthening endogenous metabolic pathways or assembling heterologous ones enables C. glutamicum to rapidly catabolize a multitude of carbon sources. This review summarizes recent progress in metabolic engineering of C. glutamicum toward a broad substrate spectrum and diverse chemical production. In particularly, utilization of lignocellulosic biomass-derived complex hybrid carbon source represents the futural direction for non-food renewable feedstocks was discussed.
Introduction
Since isolation in 1957, Corynebacterium glutamicum has been intensively applied for biobased chemical production due to its ability to secrete amino acids, which are traditionally used as drugs or health products (Kogure and Inui, 2018; Wu et al., 2019). These amino acids and their derived chemicals are worth billions of dollars per year. Development of engineered strains with higher production performance is an active field that has attracted numerous researchers (Lee and Wendisch, 2017b; Li et al., 2017; D’Este et al., 2018; Zhang et al., 2018a,b). In particular, most amino acids, including L-glutamate, L-lysine, L-arginine, L-valine, and L-ornithine, have achieved industrial-scale production due to rapid development of gene-editing and fermentation-manipulation techniques. These amino acids are used in human nutrition, food additives, and drug preparation, applications that benefit from the use of biologically safe C. glutamicum (Lee and Wendisch, 2017a). In addition to amino acids, C. glutamicum has also been extensively modulated to produce a multitude of valuable products, including bulk chemicals, natural products, polymers, proteins, and biofuels (Becker et al., 2018b; Shanmugam et al., 2018, 2019; Wendisch et al., 2018). This vigorous development has benefited from fossil fuel depletion and anthropogenic climate change caused by the emission of toxic gases generated from oil decomposition, which traditionally served as the major source of manufactured chemicals (Sun et al., 2018). However, biobased production of metabolites using C. glutamicum consumes large amounts of glucose, obtained from the hydrolysis reaction of starch, creating competition for food with humans. Hence, it is critical to exploit alternative renewable carbon sources, such as agricultural wastes, industrial wastes, and others for the cultivation of industrial model strains. In the past few years, research efforts have shifted toward biobased production of metabolites from non-food renewable feedstocks. Here are summarized recent advances in the utilization of alternative C-resources, including xylose, arabinose, methanol, glycerol, and mannitol, to produce high-value chemicals using C. glutamicum (Figure 1).
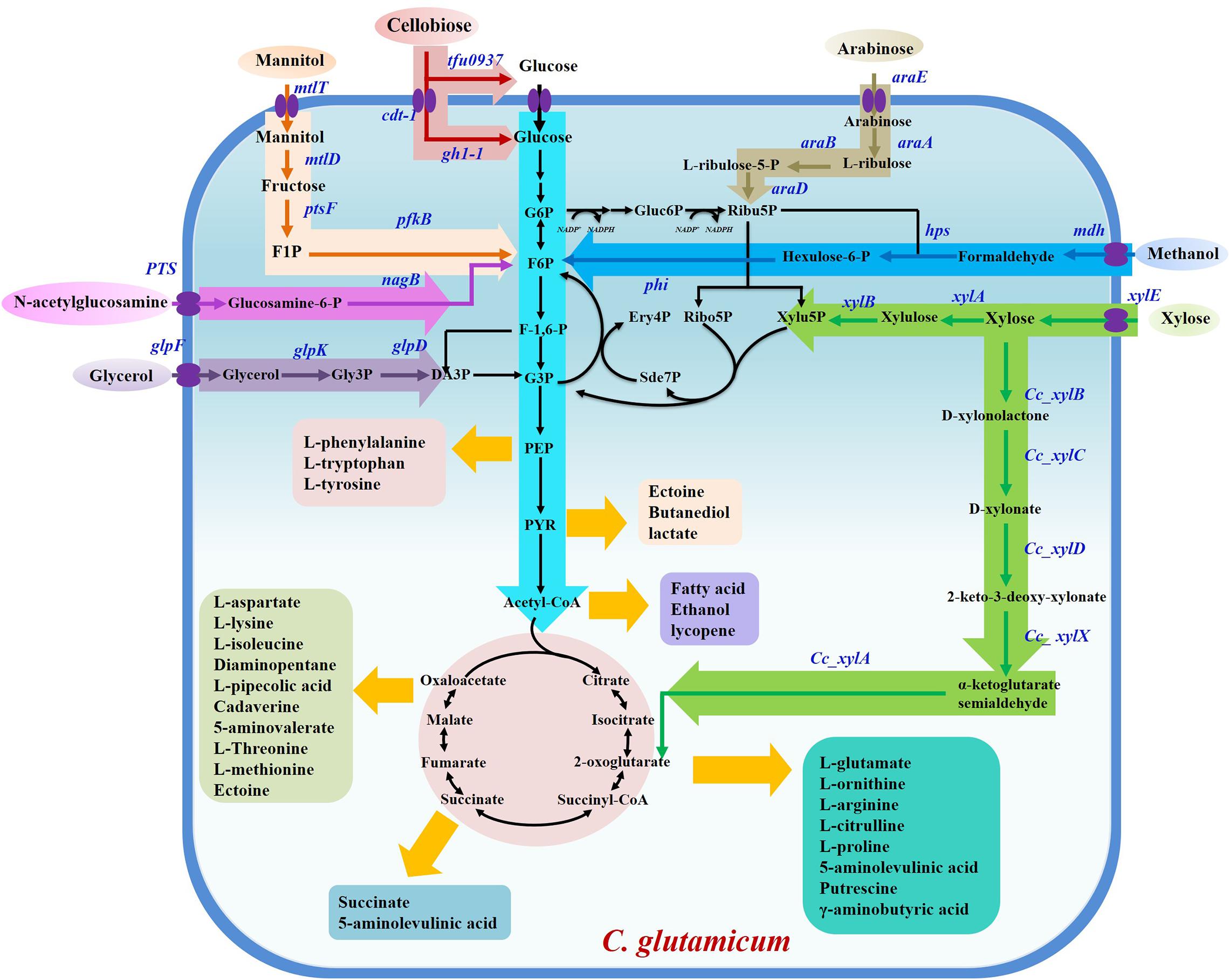
Figure 1. Overview of non-food renewable carbon resource utilization and chemicals production in C. glutamicum. G6P, glucose-6-phosphate; F6P, fructose-6-phosphate; F-1,6-P, fructose-1,6-diphosphate; G3P, glyceraldehyde-3-phosphate; DA3P, dihydroxyacetone-3-phosphate; PEP, phosphoenolpyruvate; PYR, pyruvate; Gluc6P, gluconate-6-phosphate; Ribu5P, L-ribulose-5-phosphate; Ribo5P, ribose-5-phosphate; Xylu5P, xylulose-5-phosphate; Ery4P, erythritol-4-phosphate; Sde7P, sedoheptulose-7-phosphate; F1P, fructose-1-phosphate; Gly3P, glycerol-3-phosphate.
Bioconversion of Single-Carbon Sources to Valuable Chemicals
Xylose
Xylose, a pentose, is the major constituent of lignocellulose biomass, and was deemed to be the second most abundant biological resource, after glucose (Elmore et al., 2020). High-value utilization of xylose is shifting from bioenergy to a broader range of chemicals. Engineering microbes to produce biobased chemicals from xylose provides a potential alternative to threatening human food security by continuing to use glucose. In C. glutamicum, the absence of xylose isomerase means there is no established endogenous metabolic pathway for xylose assimilation. To overcome this obstacle, xylA, which encodes xylose isomerase and originates from various microorganisms, has been amplified and employed to reconstruct the xylose utilization metabolic pathway in C. glutamicum (Zhao et al., 2018). Heterologous expression of xylA from Xanthomonas campestris converts imported xylose into xylulose, demonstrating better performance than xylA from other microbial species. Tandem expression of X. campestris xylAB operon by employing a plasmid or chromosomal insertion of gene copy approach enables impressive conversion of xylose to chemicals in C. glutamicum (Table 1). For instance, we previously demonstrated plasmid-based expression of X. campestris xylAB operon in the L-ornithine-producing strain C. glutamicum SO26 that significantly improved the yield of L-ornithine. By adjusting the expression of the xylAB operon, we achieved an L-ornithine titer of 18.9 g/L, representing the highest L-ornithine production titer from xylose recorded to date (Zhang et al., 2019). In contrast to the xylose isomerase (XI) pathway, the Weimberg (WMB) pathway, consisting of multistep reactions catalyzed by enzymes including xylose dehydrogenase, xylonolactonase, xylonate dehydratase, 2-keto-3-deoxy-D-xylonate dehydratase, and α-ketoglutarate semialdehyde dehydrogenase, was introduced into C. glutamicum to progressively convert xylose to α-ketoglutarate (Figure 1; Zhao et al., 2018; Choi et al., 2019). The enzymes involved in the WMB pathway are encoded by genes listed as belonging to the xylXABCD operon in Caulobacter crescentus. Heterologous expression of the xylXABCD operon allowed C. glutamicum to utilize xylose without loss of carbon; however, cell growth was hindered by inferior metabolic flux. To overcome this growth inhibition, the mutated strain C. glutamicum WMB2evo was selected by adaptive laboratory evolution, providing a 3.7-fold increase in growth rate compared to the original strain (Radek et al., 2017). Results from comparative genomics analysis suggest that mutations in IolR, which upregulated the expression of iolT1, significantly improved xylose transport and contributed to increased cell growth (Radek et al., 2017). It is universally acknowledged that reinforcing the xylose uptake system markedly increases conversion from xylose to valuable chemicals. Currently, two transmembrane proteins capable of transporting xylose, XylE (Yim et al., 2016) and AraE (Chen Z. et al., 2017), are applied to boost xylose utilization, which promotes the accumulation of metabolites. C. glutamicum ATCC 31831, possessing the endogenic arabinose transporter AraE, and C. glutamicum ATCC 13032, which lacks this transporter, were simultaneously modulated to produce xylonic acid from xylose; as a result, engineered C. glutamicum ATCC 31831 produced approximately 10% more xylonic acid than engineered C. glutamicum ATCC 13032 (Sundar et al., 2020). During 120 h of fermentation, 75% xylose consumption in the engineered C. glutamicum ATCC 31831 strain likewise outstripped the 60% xylose consumption observed in the AraE-absent C. glutamicum ATCC 13032 strain. It can be widely accepted that C. glutamicum ATCC 31831 is a superior host for the conversion from xylose to valued chemicals. A robust uptake system exerts the crucial role in xylose utilization by using C. glutamicum. Although xylose is the second abundant carbon source next to glucose, the fermentation of pure xylose to produce compounds has been little investigated. This probably due to xylose generally derives from complex carbon sources, frequently obtained by the pretreatment of cellulose, rather than pure sugars like glucose.
Methanol
Methanol is an attractive alternative feedstock that can be easily obtained from the oxidation of methane. Ongoing development of natural gas, the main source of methane, and use of exploitation technology enable the steady conversion from methane to methanol, continuously reducing the price of methanol (Zhao et al., 2018). Consequently, microbes engineered to convert methanol to valuable metabolites have received considerable attention regarding their comparable cost to glucose-based processes. In microbes, the ribulose monophosphate (RuMP) pathway demonstrates advantages in generating ATP and NAD(P)H, which are frequently used for methanol assimilation, a process in which methanol is converted to fructose 6-phosphate, catalyzed by methanol dehydrogenase (encoded by mdh), 3-hexulose-6-phosphate synthase (encoded by hps), and 6-phospho-3-hexuloisomerase (encoded by phi) (Figure 1; Zhao et al., 2018). In addition to natural occurrence in methylotrophic microorganisms, the RuMP pathway has been completely assembled in synthetic methylotrophs and is feasible for the conversion of methanol to metabolites (Antoniewicz, 2019). In C. glutamicum, despite the existence of an endogenous metabolic pathway in which the four reactions catalyzed by AdhE, Ald, FdhF, and MshC convert methanol to CO2, this strain requires an auxiliary carbon source to maintain cell growth (Witthoff et al., 2013). In order to continuously utilize methanol, the RuMP pathway was introduced to C. glutamicum by heterogeneous expression of Mdh from Bacillus methanolicus and Hps and Phi from Bacillus subtilis that produced an average methanol consumption rate of 1.7 mM/h in a glucose-methanol-containing medium (Witthoff et al., 2015). Simultaneously, a non-methylotrophic strain was successfully modulated to produce 13C-label cadaverine from methanol, though high amounts of methanol added to the fermentation medium were toxic to C. glutamicum cells (Lessmeier et al., 2015). Accordingly, to enhance the methanol tolerance of C. glutamicum, adaptive laboratory evolution was applied to screen mutant strains that exhibited tolerance to high methanol content (Lessmeier and Wendisch, 2015; Wang et al., 2020). Comparative genome analysis of these mutant strains indicated that the amino acid substitution of A165T in MetY and a shortened Cat are crucial factors for improving methanol tolerance (Lessmeier and Wendisch, 2015). In addition, adaptive laboratory evolution was also applied to screen mutant strains capable of increased methanol-dependent cell growth in methanol-xylose-containing medium (Tuyishime et al., 2018). In these mutant strains, the carbon atom in methanol was used to synthesize cell materials, co-factors, and intermediates, which were subsequently converted to L-glutamate through interference with the biosynthesis of the cell wall (Tuyishime et al., 2018). Results from transcriptome analysis illustrate that increased methanol concentrations readjusted the metabolic flux distribution of the glycolysis pathway, amino acid biosynthesis pathway, oxidative phosphorylation reactions, ribosome biosynthesis, and parts of the tricarboxylic acid cycle in C. glutamicum (Wang et al., 2020). Moreover, mechanistic analysis of mutant strains obtained by adaptive laboratory evolution of methanol-glucose co-utilizing strains demonstrated that improved expression of mdh-hxlAB, improved supplementation of riboflavin, and S288N mutation in MetY contribute to the distinct methanol-dependent growth of C. glutamicum (Hennig et al., 2020). Inspired by research on methanol bioconversion, a similar study claimed that methyl acetate can be easily obtained from the carbonylation of methanol and CO and that this was utilized by C. glutamicum through the introduction of a highly active esterase (Choo et al., 2016). In practice, current progress consistently indicated that cell growth of C. glutamicum was hindered by the cytotoxicity of methanol and formaldehyde, which restricted the conversion from methanol to chemicals. In addition to using engineered C. glutamicum to improve utilization efficiency and tolerance of methanol, the range of synthetic chemicals produced should also be further expanded.
Glycerol
Glycerol, a carbon-containing waste resulting from the biodiesel production process, has been widely applied in microbial fermentation owing to its high-reduction property, which frequently leads to higher maximum theoretical yields for producing chemicals (Xiberras et al., 2019). Recently, global markets witnessed remarkable growth in the biodiesel industry that subsequently stimulated research toward utilization of byproduct glycerol (Vivek et al., 2017). Due to its abundance, high-reduction ability, non-toxicity, and low cost, glycerol is regarded as a favorable carbon substrate for numerous industrial microbes, including Escherichia coli (Westbrook et al., 2019), Saccharomyces cerevisiae (Xiberras et al., 2019), B. subtilis (Fan et al., 2018), and Pseudomonas species (Pobletecastro et al., 2020) to produce various types of metabolites. Glycerol is typically not treated as a carbon resource for C. glutamicum-based bioprocesses due to the absence of glycerol oxidation pathway enzymes in this strain. In practice, glpK-encoded glycerol kinase and glpD-encoded glycerol-3-phosphate dehydrogenase from E. coli are introduced into C. glutamicum to obtain engineered strains capable of metabolizing glycerol (Rittmann et al., 2008). In order to further improve glycerol utilization, heterogeneous expression of glpF, which encodes a glycerol facilitator from E. coli, was developed, accelerating the growth of C. glutamicum on glycerol. These strategies have been intensively applied to developing engineered C. glutamicum to convert glycerol into chemicals. For instance, overexpression of E. coli_glpFKD in corresponding C. glutamicum strains produced 3.5 ± 0.8 mM, 23.0 ± 2.3 mM, 17.9 ± 0.4 mM, and 23.6 ± 0.9 mM of L-glutamate, L-lysine, L-ornithine, and L-arginine, respectively (Rittmann et al., 2008; Meiswinkel et al., 2013b). Additionally, plasmid-based overexpression of E. coli_glpFKD in engineered C. glutamicum produced 11 ± 1 mM L-pipecolic acid at a yield of 0.14 ± 0.02 g/g glycerol (Pérez-García et al., 2017), 0.6 ± 0.0 g/L ectoine at a yield of 0.055 ± 0.003 g/g glycerol (Perez-Garcia et al., 2017), and 38.4 g/L succinate at a yield of 1.02 g/g glycerol (Wang et al., 2016), indicating that the majority of chemicals can be produced from glycerol in manipulated strains. Although rapid progress has been made in modulation of C. glutamicum to convert glycerol to chemicals, there are still uncertainties: At first, crude glycerol obtained from biodiesel production bioprocesses contains growth inhibitors that limit its application for the majority of industrial microorganisms (Vivek et al., 2017). Second, poor metabolic flux to the pentose phosphate pathway, using glycerol as carbon resource, results in inadequate cofactor NADPH regeneration, hindering the biosynthesis of chemicals including amino acids, fatty acids, and others, which requires NADPH to drive multistep enzyme reactions (Xiberras et al., 2019). In conclusion, if these obstacles can be addressed, microbial fermentation will be a promising approach for converting glycerol to metabolites.
Arabinose
Arabinose is the second most abundant pentose, following xylose, in plant cellulose biomass, and is intensively used as a carbon feedstock for industrial microbes. Assimilation of arabinose requires three enzyme actions and a membrane protein. First, arabinose isomerase (encoded by araA) catalyzes L-arabinose to synthesize L-ribulose. Second, ribulokinase (encoded by araB) catalyzes L-ribulose to synthesize L-ribulose-5-P. Third, ribulose-5-phosphate 4-epimerase (encoded by araD) catalyzes L-ribulose-5-P to synthesize D-xylulose-5-P, and this terminal metabolite is able to enter the pentose phosphate pathway (Gopinath et al., 2012; Zhao et al., 2018; Choi et al., 2019). AraE, a membrane channel protein that functions as a xylose transporter, was able to simultaneously transport L-arabinose. C. glutamicum strains, with the exception of C. glutamicum ATCC31831, cannot utilize arabinose due to lack of this metabolic pathway (Zhao et al., 2018; Choi et al., 2019). By heterogeneously assembling the arabinose metabolic pathway from E. coli, researchers engineered C. glutamicum to produce succinic acid from arabinose (Kawaguchi et al., 2008). Subsequently, C. glutamicum ATCC31831 with the ability to grow on L-arabinose was discovered by random screening. Analysis of the genome of this strain suggested that genes associated with arabinose utilization were included in the araBAD operon, which is negatively controlled by the transcription factor AraR (Kuge et al., 2015). In addition, simultaneous utilization of L-arabinose and D-glucose in C. glutamicum ATCC31831 indicated that carbon metabolism repression was ineffective against arabinose in this strain (Kawaguchi et al., 2009). Hence, development of recombinant C. glutamicum strains for converting arabinose to chemicals has received extensive attention. Heterologous expression of the E. coli araBAD operon in C. glutamicum HalT1 enabled fermentation production of 7-Cl-Trp and Trp at a titer of 52 ± 1 mg/L and 2.4 ± 0.1 g/L, respectively (Veldmann et al., 2019). Co-expression of the E. coli araBAD operon and X. campestris xylA endowed engineered C. glutamicum with the ability to simultaneously utilize arabinose and xylose as well as improve the production titers of L-lysine, L-glutamate, L-ornithine, and putrescine (Meiswinkel et al., 2013a). Despite the existence of endogenous arabinose metabolic pathways in C. glutamicum ATCC31831, researchers prefer heterologous expression of the araBAD operon from E. coli to construct engineered C. glutamicum strains (Meiswinkel et al., 2013a; Pérez-García et al., 2017). Because hydrolyzed cellulose is composed of glucose, xylose, arabinose, and other sugars, it can be acknowledged that the arduous feasibility of mixed sugar fermentation is the main bottleneck for the utilization of arabinose (Jojima et al., 2015; Mindt et al., 2019b).
Mannose or Mannitol
In addition to xylose and arabinose, mannose is another feedstock, making up approximately 20% of the sugar composition in lignocellulose hydrolyzate. Engineering recombinant strains to convert mannose to valuable chemicals has captured widespread attention. Mannose catabolism relies largely on the branching metabolism of the glycolytic pathway, similar to other sugars. In this metabolic pathway, mannose is transported into the cytoplasm, accompanied by acquisition of the phosphoryl group from phosphoenolpyruvate to generate mannose-6-phosphate. Subsequently, phosphomannose isomerase is employed to catalyze mannose-6-phosphate to synthesize fructose-6-phosphate, which is an intermediate in the glycolytic pathway. In C. glutamicum, a well-established native mannose metabolic pathway consists of glucose or fructose permeases (encoded by ptsG or ptsF), as well as phosphomannose isomerase (encoded by manA), which has been applied for the biosynthesis of various organic acids from mannose (Sasaki et al., 2011). Under anaerobic conditions, co-overexpression of manA and ptsF not only accelerated utilization of mannose but also broke down carbon metabolite repression, enabling simultaneous metabolism of glucose and mannose in C. glutamicum (Sasaki et al., 2011). Currently, engineered C. glutamicum to utilize mannose requires deforestation to prepare raw materials that are unsustainable; therefore, attention has been transferred to its reductive format, mannitol, which can be easily obtained from hydrolysis of marine plants.
Similar to mannose, the catabolic pathway of mannitol involves two transmembrane transport systems and two enzymatic reactions; mannitol can also be processed in C. glutamicum (Figure 1). First, it is transported from extracellular to intracellular space using the transmembrane protein MtlT and converted to fructose by mannitol dehydrogenase (encoded by mtlD) (Peng et al., 2011). Subsequently, fructose is transported twice across the cell membrane to produce fructose-1-phosphate by employing a non-specific PTS transport system composed of an EI (encoded by ptsIH) and an EII (encoded by ptsF) membrane protein. Moreover, fructose-1-phosphate is applied to generate fructose-1,6-bisphosphate under the catalysis of phosphofructokinase (encoded by pfkB), which enters the glycolysis pathway (Laslo et al., 2012). Therefore, it is reasonable to speculate that sufficient expression of mannitol transporter and mannitol dehydrogenase is a critical factor for the utilization of mannitol in C. glutamicum. In general, C. glutamicum is unable to utilize mannitol until the negative regulator MtlR is removed; this regulator severely represses the expression of mtlTD operon (Peng et al., 2011). Accordingly, deletion of MtlR was performed in the lysine-producing strain C. glutamicum Lys12 to generate the recombinant strain C. glutamicum SEA-1, which is capable of producing 8.5 mM L-lysine from mannitol (Hoffmann et al., 2018). However, the recombinant C. glutamicum SEA-1 strain could only metabolically convert mannitol as long as the cell growth rate and biosynthesis of intracellular cofactor NADPH were much lower than in the parent C. glutamicum Lys12 strain cultured on glucose (Hoffmann et al., 2018). Fructose was found in the fermentation broth of strain C. glutamicum SEA-1, along with secondary growth observed in the fermentation process, indicating that fructose was incompletely converted to fructose-1-phosphate and secreted into the extracellular space. Thus, there are important factors, such as low expression of related enzymes, inefficient fructose uptake system, and insufficient supply of cofactor NADPH, that restrict biotransformation from mannitol to chemicals in MtlR-stripped C. glutamicum. Further regulatory mechanism analyses of mannitol metabolism, rational design and modification of related gene targets, and adoption of appropriate metabolic engineering approaches to optimize the metabolic flux of mannitol catabolism and fructose metabolism pathways are crucial for improving mannitol utilization efficiency.
N-Acetylglucosamine
N-Acetylglucosamine is an amino sugar that serves as a monomer for chitin, a polymer widely present in the exoskeleton of crustaceans. Cooked shrimp serves as food for humans, particularly in China, producing several million tons of shellfish waste, of which chitin makes up approximately half of the dry weight. Simple disposal of these wastes not only wastes biological resources but also causes environmental pollution, detracting from sustainable development of the national economy. Engineering microbes to utilize abundant polysaccharides from crustacean exoskeletons in order to produce chemicals has attracted extensive attention from researchers. Generally, the metabolic pathway of N-acetylglucosamine consists of a specific transport system and two enzyme actions, catalyzed by N-acetylglucosamine-6-phosphatedeacetylase (encoded by nagA) and glucosamine-6P deaminase (encoded by nagB), converting N-acetylglucosamine to fructose-6-phosphate that enters the glycolytic pathway (Matano et al., 2016). In C. glutamicum, the absence of a specific uptake system prevents the utilization of extracellular N-acetylglucosamine, requiring heterogeneous expression of nagE from Corynebacterium glycinophilum; this expression enables prompt transportation and feasible assimilation of N-acetylglucosamine. Co-expression of exogenous nagA, nagB, and nagE generated C. glutamicum capable of producing various chemicals, including L-lysine (Sgobba et al., 2018), L-citrulline (Eberhardt et al., 2014), lycopene (Matano et al., 2014), putrescine (Uhde et al., 2013), 7-chloro-L-tryptophan (Veldmann et al., 2019), 5-aminovalerate (Jorge et al., 2017b), gamma-aminobutyric acid (Jorge et al., 2017a), ectoine (Perez-Garcia et al., 2017), and L-pipecolic acid (Pérez-García et al., 2017) from N-acetylglucosamine. Consequently, N-acetylglucosamine is a promising alternative carbon source for C. glutamicum, although further investigation is required to accelerate the conversion from N-acetylglucosamine to bulk chemicals at the industrial scale. Additionally, the solid nature of chitin is unfavorable in preprocessing, as it hinders industrial application of amino sugars.
Cellobiose
Cellobiose, a disaccharide formed by the connection of two glucose molecules, is a carbon feedstock generated by degradation of cellulose through a synergistic reaction between endoglucanase and cellobiohydrolase (Li et al., 2020). Currently, direct utilization of cellobiose requires a β-glucosidase that catalyzes breakage of β-1,4-glucoside bonds and converts cellobiose to glucose. Due to the absence of a cellobiose transporter and low permeability of cellobiose, expression of β-glucosidase in microorganisms invariably requires secretion or surface display in order for the enzyme to contact the substrate. Traditionally, secreted expression or surface display of β-glucosidase has been applied for biorefinery chemical production in a multitude of industrial strains, including Clostridium thermocellum (Dash et al., 2019), S. cerevisiae (Yun et al., 2018), Pseudomonas putida (Dvoøák and de Lorenzo, 2018), E. coli (Satowa et al., 2020), and C. glutamicum (Adachi et al., 2013). C. glutamicum displays high β-glucosidase activity derived from Saccharophagus degradans, successfully displaying β-glucosidase fused with the C-terminus of the anchor protein PorC. On the surface, C. glutamicum enables simultaneous saccharification and fermentation, producing three-fold more L-lysine than a β-glucosidase-secretory C. glutamicum strain (Adachi et al., 2013). Co-expression of endoglucanase and β-glucosidase, either secretory or surface-displaying, in C. glutamicum DM1729 directly converts cellulose to L-lysine (Anusree et al., 2016). However, due to insufficient β-glucosidase activity, the yield of L-lysine and the cellobiose consumption rate of this engineered C. glutamicum were underdeveloped. To address limitations on enzyme activity, Tfu0937 from Thermobifida fusca, which shares high β-glucosidase activity with E. coli, was codon-optimized and introduced into C. glutamicum. Fusion with the CgR0949 signal sequence resulted in a Tfu0937-secreting strain that produced 9.7 g/L lysine, an improvement of approximately eight-fold compared to the original β-glucosidase-displaying strains (1.08 g/L) (Matsuura et al., 2019). Additionally, intracellular utilization of cellobiose in C. glutamicum is feasible through heterologous expression of codon-optimized cdt-1, which encodes a cellobiose transporter, and the gh1-1 gene from Neurospora crassa (Lee et al., 2016). By adaptive evolution, this mutant strain was also applied for the co-utilization of cellobiose and xylose.
Direct Bioconversion From Plant Biomass Hydrolyzates to Chemicals
Lignocellulosic biomass is generally regarded as agricultural waste, stored in huge amounts of organic carbon sources. Although many cellulosic ethanol factories are gradually coming into operation, lignocellulosic biomass is still underutilized and poses major environmental problems in some economically disadvantaged areas. Thus, there is an urgent need to improve the utilization of lignocellulosic biomass, which is a promising alternative raw material for the biobased production of chemicals. Pretreated by high-temperature acid hydrolysis, lignocellulosic materials such as corn stalk, rice straw, cassava bagasse, and wheat bran can be converted into corresponding hydrolyzates containing various monosaccharides (glucose, xylose, arabinose, etc.) that can be used to provide nutrition for microbes.
Corn Straw Hydrolyzate
Corn is an economic and food crop that is widely planted in numerous countries. Billions of tons of corn straw biomass are generated annually by harvesting corn. Most corn straw is incinerated, and only a fraction is applied for feeding animals, causing an extreme waste of resources and environmental pollution. To address these problems, corn straw has been preprocessed by mixing with dilute sulfuric acid and mild treatment at high temperature to generate exploitable corn straw hydrolyzate, which contains multitudinous carbohydrates, including glucose, xylose, arabinose, and mannose, which are favorable substrates for microbial fermentation (Choi et al., 2019). However, hydrolyzate generated from dilute-acid pretreatment process invariably contains fermentation inhibitors, including furfural, 5-hydroxymethylfurfural (HMF), and phenolic aldehydes, that affect the growth of microbes such as C. glutamicum. Hence, improving the tolerance of C. glutamicum to these inhibitors was investigated by adaptive evolutionary analysis to obtain a mutant strain capable of rapid growth on corn straw hydrolyzate (Wang et al., 2018). Exploration of tolerance mechanisms using a transcriptome analysis approach found that overexpression of CGS9114_RS01115, which encodes an alcohol dehydrogenase, as well as numerous oxidoreductase genes, accelerated conversion of these aldehyde inhibitors in C. glutamicum (Zhou et al., 2019). Meanwhile, excess biotin, which is unavoidable in lignocellulosic hydrolyzate produced by the pretreatment process, results in a rigid cell membrane that hampers secretion of L-glutamate, restricting cellulosic L-glutamate production (Wen et al., 2018). This unfavorable phenomenon can be reversed by extrinsic addition of osmotic agents, such as Tween 40, ethambutol, and penicillin, a step that not only improves the yield of L-glutamate but also promotes accumulation of L-arginine and L-ornithine during fermentation (Chen M. et al., 2015; Jiang et al., 2020). In addition, the uptake system of biotin is reinforced by overexpression of the bioYMN operon, which encodes biotin transporter, reducing biotin content in corn straw hydrolyzate and stimulating faster cell growth and multiplicative cellulosic L-glutamate production (Han et al., 2019). Moreover, to increase the transport of L-glutamate, MscCG, the primary L-glutamate transporter, was truncated, which stimulated L-glutamate production in biotin-rich corn stover hydrolyzates (Wen and Bao, 2019; Kawasaki and Martinac, 2020). Reducing biotin concentration in corn stover hydrolyzates is indispensable for cellulosic L-glutamate production. In addition to L-glutamate, L-lysine is a bulk amino acid that is in tremendous global demand and can be produced from corn straw hydrolyzate. For instance, fermentation cultivation of C. glutamicum SIIM B253 in high-glucose corn straw hydrolyzate resulted in L-lysine accumulation at a titer of 7.4 g/L. By modulating nutrient concentration, fermentation conditions, and simultaneous saccharification and fermentation processes, the yield of L-lysine was further elevated to 33.8 g/L, representing the highest yield of cellulosic L-lysine (Chen et al., 2019). As illustrated in these examples, C. glutamicum-based cellulosic biomass utilization has focused on the assimilation of glucose in corn straw hydrolyzate. Xylose serves as the second most abundant carbon feedstock in corn straw hydrolyzate, though C. glutamicum does not contain its own xylose metabolic machinery. Heterologous assimilation of xylose metabolic pathways from X. campestris were processed in strain SAZ3 that then produced 98.6 g/L of succinate by two-stage fermentations of glucose and xylose in corn straw hydrolyzate (Mao et al., 2018). However, despite extensive investigations aims at engineering C. glutamicum strains to produce valuable chemicals from corn straw hydrolyzate, multitude of problems, including the complexity of nutrients, generation of fermentation inhibitors, complex pretreatment process, and limited sugar yield, still restrict the large-scale application of corn straw hydrolyzate. In summary, corn straw hydrolyzate is an alternative renewable feedstock with huge potential in the field of microbial fermentation if those drawbacks could be addressed.
Other Cellulose Hydrolyzates
There are many food crops grown on Earth. In addition to corn straw, cellulose hydrolyzate prepared from crop residues such as cassava bagasse, straw stalk, and wheat bran are also applied for the cultivation of industrial microbes. Among these, cassava bagasse is a residuum generated from the starch extraction process and frequently identified as widespread waste, occupying a high proportion of disposal capacity (Padi and Chimphango, 2020). Engineering microbes for bioconversion of cassava bagasse has aroused intense interest in the past few decades (Sugumaran et al., 2014). Currently, the enzymatic hydrolysis products of cassava bagasse contains amounts of glucose and trace amounts of xylose, arabinose, and acids has been resoundingly used for the bioproduction of various bulk chemicals, including alcohols (Huang et al., 2019), organic acids (Wang and Yang, 2013), and fatty acids (Chen J. et al., 2015). For example, an immobilized C. glutamicum cell device, using a mixture of substrates and alkalis, has been discovered and can produce succinic acid from cassava bagasse (Shi et al., 2014). After immobilization in a porous polyurethane filler, C. glutamicum can be used for cyclic utilization of hydrolyzate generated by the two-step enzymatic hydrolysis of cassava bagasse, producing 22.5 g/L succinic acid for each round of fermentation (Shi et al., 2014). Additionally, enzymatic hydrolysis of cassava bagasse (contains (w/w) 50.3% starch, and 12.2% fiber, and 6.5% moisture) recovers approximately half the mass of glucose (0.53 g glucose/g cassava bagasse), which can then be utilized by engineered C. glutamicum. This produced 18.5 g/L of 5-aminolevulinic acid during fed-batch fermentation, exhibiting 90.1% thrift on the cost of carbon material (Chen et al., 2020). In addition to cassava, rice is another widely planted food crop that generates a hundred billion tons of waste straw in the harvested phase. Traditionally, incineration disposal of rice straw has resulted in extensive haze and waste of resources. Acid treatment of rice straw generates 42 g/L of carbohydrate, containing 40 mM glucose, 166 mM xylose, and 66 mM arabinose, which can be utilized by engineered C. glutamicum containing a heterogeneously assembled xylose and arabinose metabolic pathway and produced 96 mM of L-glutamate during 100 h of fermentation cultivation (Gopinath et al., 2011). Similarly, acid treatment of wheat bran generates 41 g/L of carbohydrate, containing 125 mM glucose, 62 mM xylose, and 64 mM arabinose, was also can be utilized by engineered C. glutamicum to produce L-glutamate (Gopinath et al., 2011). Inspired by this example, biobased production of N-ethylglycine from rice straw hydrolyzate was enabled in engineered C. glutamicum by introducing a mutant DpkA from P. putida (Mindt et al., 2019a). Moreover, small quantities of plant biomass hydrolyzates, such as those derived from sorghum, softwood lignin, and Miscanthus, also deserve attention in the fermentation field. Biobased production of 5-aminovaleric acid (Joo et al., 2017) and biogasoline isopentenol (Sasaki et al., 2019) from Miscanthus hydrolyzate, as well as cis-muconic acid from depolymerized small aromatics of softwood lignin (Becker et al., 2018a), is feasible. Biomass hydrolyzates provide an alternative to glucose, creating enormous potential to alleviate the problem of industrial fermentation competing with humans for food.
Conclusion and Perspectives
In this review, non-food carbon sources for the fermentation cultivation of C. glutamicum and chemical production were summarized. These feedstocks, including xylose, methanol, arabinose, glycerol, mannitol, N-acetylglucosamine, cellobiose, and cellulose hydrolyzates, provide alternative and renewable substrates to produce biobased chemicals. Microbial fermentation using these substrates is expected to alleviate the problem of food competition between industrial fermentation and human nutrition. However, there are still many technical bottlenecks in the practical application of these carbon sources. First, the tolerance of C. glutamicum to toxic materials, such as methanol and cellulose hydrolyzate, requires additional improvement to meet the demands of industrial fermentation. Fermentation inhibitors, including furfurals and phenolic aldehydes generated in the dilute-acid hydrolysis process of cellulose, are crucial factors restricting cellulose conversion. In addition to fermentation inhibitors, some nutrients may themselves hinder biosynthesis and secretion of target chemicals. Second, the instability of allogenically assembled metabolic pathways frequently restricts the biosynthesis of chemicals. These metabolic pathways disrupt the original metabolic balance, resulting in low growth and relatively low substrate utilization rates. Dynamic regulation of allochthonous metabolic pathways and chromosome insertion of genes involved in substrate utilization provide an effective solution for unstable pathway engineering. Third, the abundance and outlook of the alternative feedstock are important factors to consider when evaluating the technological index. As sea levels rise, land desertifies, the total human population of the earth increases, and the area of arable land decreases; biomass from terrestrial plants will no longer be the ideal substrate source. Utilization of marine biomass in the form of mannitol, mannose, and trehalose is one alternative, owing to the heavy specific proportion of these substrates in oceans. Meanwhile, stimulated by the development of sea-rice (Chen R. et al., 2017) and sand rice (Genievskaya et al., 2017), the utilization of rice straw hydrolyzate will also play an important role in industrial fermentation in the future.
Author Contributions
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication. BZ wrote and submitted this manuscript. YJ and ZL revised the “Bioconversion of Single-Carbon Sources to Valuable Chemicals” section. FW and X-YW revised the “Direct Bioconversion from Plant Biomass Hydrolysates to Chemicals” section.
Funding
This work was supported by the National Natural Science Foundation of China (No. 32000057) and Jiangxi Provincial Natural Science Foundation (No. 20202BAB213023), and Jiangxi Key R&D Program (No. 20171ACF60006).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Editage (www.editage.cn) for English language editing.
References
Adachi, N., Takahashi, C., Onomurota, N., Yamaguchi, R., Tanaka, T., and Kondo, A. (2013). Direct L-lysine production from cellobiose by Corynebacterium glutamicum displaying beta-glucosidase on its cell surface. Appl. Microbiol. Biotechnol. 97, 7165–7172. doi: 10.1007/s00253-013-5009-4
Antoniewicz, M. R. (2019). Synthetic methylotrophy: strategies to assimilate methanol for growth and chemicals production. Curr. Opin. Biotechnol. 59, 165–174. doi: 10.1016/j.copbio.2019.07.001
Anusree, M., Wendisch, V. F., and Nampoothiri, K. M. (2016). Co-expression of endoglucanase and β-glucosidase in Corynebacterium glutamicum DM1729 towards direct lysine fermentation from cellulose. Bioresour. Technol. 213, 239–244. doi: 10.1016/j.biortech.2016.03.019
Becker, J., Kuhl, M., Kohlstedt, M., Starck, S., and Wittmann, C. (2018a). Metabolic engineering of Corynebacterium glutamicum for the production of cis, cis-muconic acid from lignin. Microb. Cell Fact. 17:115. doi: 10.1186/s12934-018-0963-2
Becker, J., Rohles, C. M., and Wittmann, C. (2018b). Metabolically engineered Corynebacterium glutamicum for bio-based production of chemicals, fuels, materials, and healthcare products. Metab. Eng. 50, 122–141. doi: 10.1016/j.ymben.2018.07.008
Buschke, N., Becker, J., Schafer, R., Kiefer, P., Biedendieck, R., and Wittmann, C. (2013). Systems metabolic engineering of xylose-utilizing Corynebacterium glutamicum for production of 1,5-diaminopentane. Biotechnol. J. 8, 557–570. doi: 10.1002/biot.201200367
Chen, J., Liu, X., Wei, D., and Chen, G. (2015). High yields of fatty acid and neutral lipid production from cassava bagasse hydrolysate (CBH) by heterotrophic Chlorella protothecoides. Bioresour. Technol. 191, 281–290. doi: 10.1016/j.biortech.2015.04.116
Chen, J., Wang, Y., Guo, X., Rao, D., Zhou, W., Zheng, P., et al. (2020). Efficient bioproduction of 5-aminolevulinic acid, a promising biostimulant and nutrient, from renewable bioresources by engineered Corynebacterium glutamicum. Biotechnol. Biofuels 13:41. doi: 10.1186/s13068-020-01685-0
Chen, M., Chen, X., Wan, F., Zhang, B., Chen, J., and Xiong, Y. (2015). Effect of Tween 40 and DtsR1 on l-arginine overproduction in Corynebacterium crenatum. Microb. Cell Fact. 14:119. doi: 10.1186/s12934-015-0310-9
Chen, R., Cheng, Y., Han, S., Van Handel, B., Dong, L., Li, X., et al. (2017). Whole genome sequencing and comparative transcriptome analysis of a novel seawater adapted, salt-resistant rice cultivar - sea rice 86. BMC Genomics 18:655. doi: 10.1186/s12864-017-4037-3
Chen, Z., Huang, J., Wu, Y., Wu, W., Zhang, Y., and Liu, D. (2017). Metabolic engineering of Corynebacterium glutamicum for the production of 3-hydroxypropionic acid from glucose and xylose. Metab. Eng. 39, 151–158. doi: 10.1016/j.ymben.2016.11.009
Chen, Z., Liu, G., Zhang, J., and Bao, J. (2019). A preliminary study on l-lysine fermentation from lignocellulose feedstock and techno-economic evaluation. Bioresour. Technol. 271, 196–201. doi: 10.1016/j.biortech.2018.09.098
Choi, J. W., Jeon, E. J., and Jeong, K. J. (2019). Recent advances in engineering Corynebacterium glutamicum for utilization of hemicellulosic biomass. Curr. Opin. Biotechnol. 57, 17–24. doi: 10.1016/j.copbio.2018.11.004
Choo, S., Um, Y., Han, S. O., and Woo, H. M. (2016). Engineering of Corynebacterium glutamicum to utilize methyl acetate, a potential feedstock derived by carbonylation of methanol with CO. J. Biotechnol. 224, 47–50. doi: 10.1016/j.jbiotec.2016.03.011
D’Este, M., Alvarado-Morales, M., and Angelidaki, I. (2018). Amino acids production focusing on fermentation technologies - a review. Biotechnol. Adv. 36, 14–25. doi: 10.1016/j.biotechadv.2017.09.001
Dash, S., Olson, D. G., Chan, S. H. J., Amadornoguez, D., Lynd, L. R., and Maranas, C. D. (2019). Thermodynamic analysis of the pathway for ethanol production from cellobiose in Clostridium thermocellum. Metab. Eng. 55, 161–169. doi: 10.1016/j.ymben.2019.06.006
Dhar, K. S., Wendisch, V. F., and Nampoothiri, K. M. (2016). Engineering of Corynebacterium glutamicum for xylitol production from lignocellulosic pentose sugars. J. Biotechnol. 230, 63–71. doi: 10.1016/j.jbiotec.2016.05.011
Dvoøák, P., and de Lorenzo, V. (2018). Refactoring the upper sugar metabolism of Pseudomonas putida for co-utilization of cellobiose, xylose, and glucose. Metab. Eng. 48, 94–108. doi: 10.1016/j.ymben.2018.05.019
Eberhardt, D., Jensen, J. V., and Wendisch, V. F. (2014). L-citrulline production by metabolically engineered Corynebacterium glutamicum from glucose and alternative carbon sources. AMB Express 4:85. doi: 10.1186/s13568-014-0085-0
Elmore, J. R., Dexter, G. N., Salvachúa, D., O’Brien, M., and Guss, A. M. J. M. E. (2020). Engineered Pseudomonas putida simultaneously catabolizes five major components of lignocellulosic biomass: glucose, xylose, arabinose, coumaric acid, and acetic acid. Metab. Eng. 62, 62–71. doi: 10.1016/j.ymben.2020.08.001
Fan, X., Wu, H., Jia, Z., Li, G., Li, Q., Chen, N., et al. (2018). Metabolic engineering of Bacillus subtilis for the co-production of uridine and acetoin. Appl. Microbiol. Biotechnol. 102, 8753–8762. doi: 10.1007/s00253-018-9316-7
Genievskaya, Y., Abugalieva, S., Zhubanysheva, A., and Turuspekov, Y. (2017). Morphological description and DNA barcoding study of sand rice (Agriophyllum squarrosum, Chenopodiaceae) collected in Kazakhstan. BMC Plant Biol. 17(Suppl. 1):177. doi: 10.1186/s12870-017-1132-1
Gopinath, V., Meiswinkel, T. M., Wendisch, V. F., and Nampoothiri, K. M. (2011). Amino acid production from rice straw and wheat bran hydrolysates by recombinant pentose-utilizing Corynebacterium glutamicum. Appl. Microbiol. Biotechnol. 92, 985–996. doi: 10.1007/s00253-011-3478-x
Gopinath, V., Murali, A., Dhar, K. S., and Nampoothiri, K. M. (2012). Corynebacterium glutamicum as a potent biocatalyst for the bioconversion of pentose sugars to value-added products. Appl. Microbiol. Biotechnol. 93, 95–106. doi: 10.1007/s00253-011-3686-4
Han, X., Li, L., and Bao, J. (2019). Microbial extraction of biotin from lignocellulose biomass and its application on glutamic acid production. Bioresour. Technol. 288:121523. doi: 10.1016/j.biortech.2019.121523
Henke, N. A., Wiebe, D., Perezgarcia, F., Peterswendisch, P., and Wendisch, V. F. (2018). Coproduction of cell-bound and secreted value-added compounds: simultaneous production of carotenoids and amino acids by Corynebacterium glutamicum. Bioresour. Technol. 247, 744–752. doi: 10.1016/j.biortech.2017.09.167
Hennig, G., Haupka, C., Brito, L. F., Ruckert, C., Cahoreau, E., Heux, S., et al. (2020). Methanol-essential growth of Corynebacterium glutamicum: adaptive laboratory evolution overcomes limitation due to methanethiol assimilation pathway. Int. J. Mol. Sci. 21:3617. doi: 10.3390/ijms21103617
Hoffmann, S. L., Jungmann, L., Schiefelbein, S., Peyriga, L., Cahoreau, E., Portais, J. C., et al. (2018). Lysine production from the sugar alcohol mannitol: design of the cell factory Corynebacterium glutamicum SEA-3 through integrated analysis and engineering of metabolic pathway fluxes. Metab. Eng. 47, 475–487. doi: 10.1016/j.ymben.2018.04.019
Huang, J., Du, Y., Bao, T., Lin, M., Wang, J., and Yang, S. (2019). Production of n-butanol from cassava bagasse hydrolysate by engineered Clostridium tyrobutyricum overexpressing adhE2: kinetics and cost analysis. Bioresour. Technol. 292:121969. doi: 10.1016/j.biortech.2019.121969
Jiang, Y., Huang, M., Chen, X., and Zhang, B. (2020). Proteome analysis guided genetic engineering of Corynebacterium glutamicum S9114 for Tween 40-triggered improvement in L-ornithine production. Microb. Cell Fact. 19:2. doi: 10.1186/s12934-019-1272-0
Jo, S., Yoon, J., Lee, S., Um, Y., Han, S. O., and Woo, H. M. (2017). Modular pathway engineering of Corynebacterium glutamicum to improve xylose utilization and succinate production. J. Biotechnol. 258, 69–78. doi: 10.1016/j.jbiotec.2017.01.015
Jojima, T., Noburyu, R., Sasaki, M., Tajima, T., Suda, M., Yukawa, H., et al. (2015). Metabolic engineering for improved production of ethanol by Corynebacterium glutamicum. Appl. Microbiol. Biotechnol. 99, 1165–1172. doi: 10.1007/s00253-014-6223-4
Joo, J. C., Oh, Y. H., Yu, J. H., Hyun, S. M., Khang, T. U., Kang, K. H., et al. (2017). Production of 5-aminovaleric acid in recombinant Corynebacterium glutamicum strains from a miscanthus hydrolysate solution prepared by a newly developed miscanthus hydrolysis process. Bioresour. Technol. 245, 1692–1700. doi: 10.1016/j.biortech.2017.05.131
Jorge, J. M. P., Nguyen, A. Q., Pérez-García, F., Kind, S., and Wendisch, V. F. (2017a). Improved fermentative production of gamma-aminobutyric acid via the putrescine route: systems metabolic engineering for production from glucose, amino sugars, and xylose. Biotechnol. Bioeng. 114, 862–873. doi: 10.1002/bit.26211
Jorge, J. M. P., Pérez-García, F., and Wendisch, V. F. (2017b). A new metabolic route for the fermentative production of 5-aminovalerate from glucose and alternative carbon sources. Bioresour. Technol. 245(Pt B), 1701–1709. doi: 10.1016/j.biortech.2017.04.108
Kawaguchi, H., Sasaki, M., Vertes, A. A., Inui, M., and Yukawa, H. (2008). Engineering of an l-arabinose metabolic pathway in Corynebacterium glutamicum. Appl. Microbiol. Biotechnol. 77, 1053–1062. doi: 10.1007/s00253-007-1244-x
Kawaguchi, H., Sasaki, M., Vertes, A. A., Inui, M., and Yukawa, H. (2009). Identification and functional analysis of the gene cluster for l-arabinose utilization in Corynebacterium glutamicum. Appl. Environ. Microbiol. 75, 3419–3429. doi: 10.1128/AEM.02912-08
Kawasaki, H., and Martinac, B. (2020). Mechanosensitive channels of Corynebacterium glutamicum functioning as exporters of l-glutamate and other valuable metabolites. Curr. Opin. Chem. Biol. 59, 77–83. doi: 10.1016/j.cbpa.2020.05.005
Kogure, T., and Inui, M. (2018). Recent advances in metabolic engineering of Corynebacterium glutamicum for bioproduction of value-added aromatic chemicals and natural products. Appl. Microbiol. Biotechnol. 102, 8685–8705. doi: 10.1007/s00253-018-9289-6
Kuge, T., Teramoto, H., and Inui, M. (2015). AraR, an l-arabinose-responsive transcriptional regulator in Corynebacterium glutamicum ATCC 31831, exerts different degrees of repression depending on the location of its binding sites within the three target promoter regions. J. Bacteriol. 197, 3788–3796. doi: 10.1128/jb.00314-15
Laslo, T., von Zaluskowski, P., Gabris, C., Lodd, E., Ruckert, C., Dangel, P., et al. (2012). Arabitol metabolism of Corynebacterium glutamicum and its regulation by AtlR. J. Bacteriol. 194, 941–955. doi: 10.1128/Jb.06064-11
Lee, J., Saddler, J. N., Um, Y., and Woo, H. M. (2016). Adaptive evolution and metabolic engineering of a cellobiose- and xylose- negative Corynebacterium glutamicum that co-utilizes cellobiose and xylose. Microb. Cell Fact. 15:20. doi: 10.1186/s12934-016-0420-z
Lee, J. H., and Wendisch, V. F. (2017a). Biotechnological production of aromatic compounds of the extended shikimate pathway from renewable biomass. J. Biotechnol. 257, 211–221. doi: 10.1016/j.jbiotec.2016.11.016
Lee, J. H., and Wendisch, V. F. (2017b). Production of amino acids - genetic and metabolic engineering approaches. Bioresour. Technol. 245(Pt B), 1575–1587. doi: 10.1016/j.biortech.2017.05.065
Lee, S. S., Choi, J. I., and Woo, H. M. (2019). Bioconversion of xylose to ethylene glycol and glycolate in engineered Corynebacterium glutamicum. ACS Omega 4, 21279–21287. doi: 10.1021/acsomega.9b02805
Lee, S. S., Park, J., Heo, Y. B., and Woo, H. M. (2020). Case study of xylose conversion to glycolate in Corynebacterium glutamicum: current limitation and future perspective of the CRISPR-Cas systems. Enzyme Microb. Technol. 132:109395. doi: 10.1016/j.enzmictec.2019.109395
Lessmeier, L., Pfeifenschneider, J., Carnicer, M., Heux, S., Portais, J. C., and Wendisch, V. F. (2015). Production of carbon-13-labeled cadaverine by engineered Corynebacterium glutamicum using carbon-13-labeled methanol as co-substrate. Appl. Microbiol. Biotechnol. 99, 10163–10176. doi: 10.1007/s00253-015-6906-5
Lessmeier, L., and Wendisch, V. F. (2015). Identification of two mutations increasing the methanol tolerance of Corynebacterium glutamicum. BMC Microbiol. 15:216. doi: 10.1186/s12866-015-0558-6
Li, J., Zhang, Y., Li, J., Sun, T., and Tian, C. (2020). Metabolic engineering of the cellulolytic thermophilic fungus Myceliophthora thermophila to produce ethanol from cellobiose. Biotechnol. Biofuels 13:23. doi: 10.1186/s13068-020-1661-y
Li, Y., Wei, H., Wang, T., Xu, Q., Zhang, C., Fan, X., et al. (2017). Current status on metabolic engineering for the production of l-aspartate family amino acids and derivatives. Bioresour. Technol. 245(Pt B), 1588–1602. doi: 10.1016/j.biortech.2017.05.145
Mao, Y., Li, G., Chang, Z., Tao, R., Cui, Z., Wang, Z., et al. (2018). Metabolic engineering of Corynebacterium glutamicum for efficient production of succinate from lignocellulosic hydrolysate. Biotechnol. Biofuels 11:95. doi: 10.1186/s13068-018-1094-z
Matano, C., Kolkenbrock, S., Hamer, S. N., Sgobba, E., Moerschbacher, B. M., and Wendisch, V. F. (2016). Corynebacterium glutamicum possesses β-N-acetylglucosaminidase. BMC Microbiol. 16:177. doi: 10.1186/s12866-016-0795-3
Matano, C., Uhde, A., Youn, J. W., Maeda, T., Clermont, L., Marin, K., et al. (2014). Engineering of Corynebacterium glutamicum for growth and L-lysine and lycopene production from N-acetyl-glucosamine. Appl. Microbiol. Biotechnol. 98, 5633–5643. doi: 10.1007/s00253-014-5676-9
Matsuura, R., Kishida, M., Konishi, R., Hirata, Y., Adachi, N., Segawa, S., et al. (2019). Metabolic engineering to improve 1,5-diaminopentane production from cellobiose using ß-glucosidase-secreting Corynebacterium glutamicum. Biotechnol. Bioeng. 116, 2640–2651. doi: 10.1002/bit.27082
Meiswinkel, T. M., Gopinath, V., Lindner, S. N., Nampoothiri, K. M., and Wendisch, V. F. (2013a). Accelerated pentose utilization by Corynebacterium glutamicum for accelerated production of lysine, glutamate, ornithine and putrescine. Microb. Biotechnol. 6, 131–140. doi: 10.1111/1751-7915.12001
Meiswinkel, T. M., Rittmann, D., Lindner, S. N., and Wendisch, V. F. (2013b). Crude glycerol-based production of amino acids and putrescine by Corynebacterium glutamicum. Bioresour. Technol. 145, 254–258. doi: 10.1016/j.biortech.2013.02.053
Mindt, M., Hannibal, S., Heuser, M., Risse, J. M., Sasikumar, K., Nampoothiri, K. M., et al. (2019a). Fermentative production of N-alkylated glycine derivatives by recombinant Corynebacterium glutamicum using a mutant of imine reductase DpkA from Pseudomonas putida. Front. Bioeng. Biotechnol. 7:232. doi: 10.3389/fbioe.2019.00232
Mindt, M., Heuser, M., and Wendisch, V. F. (2019b). Xylose as preferred substrate for sarcosine production by recombinant Corynebacterium glutamicum. Bioresour. Technol. 281, 135–142. doi: 10.1016/j.biortech.2019.02.084
Padi, R. K., and Chimphango, A. F. A. (2020). Feasibility of commercial waste biorefineries for cassava starch industries: techno-economic assessment. Bioresour. Technol. 297:122461. doi: 10.1016/j.biortech.2019.122461
Peng, X., Okai, N., Vertes, A. A., Inatomi, K., Inui, M., and Yukawa, H. (2011). Characterization of the mannitol catabolic operon of Corynebacterium glutamicum. Appl. Microbiol. Biotechnol. 91, 1375–1387. doi: 10.1007/s00253-011-3352-x
Pérez-García, F., Max Risse, J., Friehs, K., and Wendisch, V. F. (2017). Fermentative production of L-pipecolic acid from glucose and alternative carbon sources. Biotechnol. J. 12:1600646. doi: 10.1002/biot.201600646
Perez-Garcia, F., Ziert, C., Risse, J. M., and Wendisch, V. F. (2017). Improved fermentative production of the compatible solute ectoine by Corynebacterium glutamicum from glucose and alternative carbon sources. J. Biotechnol. 258, 59–68. doi: 10.1016/j.jbiotec.2017.04.039
Pobletecastro, I., Wittmann, C., and Nikel, P. I. (2020). Biochemistry, genetics and biotechnology of glycerol utilization in Pseudomonas species. Microb. Biotechnol. 13, 32–53. doi: 10.1111/1751-7915.13400
Radek, A., Tenhaef, N., Muller, M. F., Brusseler, C., Wiechert, W., Marienhagen, J., et al. (2017). Miniaturized and automated adaptive laboratory evolution: evolving Corynebacterium glutamicum towards an improved d-xylose utilization. Bioresour. Technol. 245, 1377–1385. doi: 10.1016/j.biortech.2017.05.055
Rittmann, D., Lindner, S. N., and Wendisch, V. F. (2008). Engineering of a glycerol utilization pathway for amino acid production by Corynebacterium glutamicum. Appl. Environ. Microbiol. 74, 6216–6222. doi: 10.1128/AEM.00963-08
Sasaki, M., Teramoto, H., Inui, M., and Yukawa, H. (2011). Identification of mannose uptake and catabolism genes in Corynebacterium glutamicum and genetic engineering for simultaneous utilization of mannose and glucose. Appl. Microbiol. Biotechnol. 89, 1905–1916. doi: 10.1007/s00253-010-3002-8
Sasaki, Y., Eng, T., Herbert, R. A., Trinh, J., Chen, Y., Rodriguez, A., et al. (2019). Engineering Corynebacterium glutamicum to produce the biogasoline isopentenol from plant biomass hydrolysates. Biotechnol. Biofuels 12:41. doi: 10.1186/s13068-019-1381-3
Satowa, D., Fujiwara, R., Uchio, S., Nakano, M., Otomo, C., Hirata, Y., et al. (2020). Metabolic engineering of E. coli for improving mevalonate production to promote NADPH regeneration and enhance acetyl-CoA supply. Biotechnol. Bioeng. 117, 2153–2164. doi: 10.1002/bit.27350
Sgobba, E., Blobaum, L., and Wendisch, V. F. (2018). Production of food and feed additives from non-food-competing feedstocks: valorizing N-acetylmuramic acid for amino acid and carotenoid fermentation with Corynebacterium glutamicum. Front. Microbiol. 9:2046. doi: 10.3389/fmicb.2018.02046
Shanmugam, S., Sun, C., Chen, Z., and Wu, Y. R. (2019). Enhanced bioconversion of hemicellulosic biomass by microbial consortium for biobutanol production with bioaugmentation strategy. Bioresour. Technol. 279, 149–155. doi: 10.1016/j.biortech.2019.01.121
Shanmugam, S., Sun, C., Zeng, X., and Wu, Y. R. (2018). High-efficient production of biobutanol by a novel Clostridium sp. strain WST with uncontrolled pH strategy. Bioresour. Technol. 256, 543–547. doi: 10.1016/j.biortech.2018.02.077
Shi, X., Chen, Y., Ren, H., Liu, D., Zhao, T., Zhao, N., et al. (2014). Economically enhanced succinic acid fermentation from cassava bagasse hydrolysate using Corynebacterium glutamicum immobilized in porous polyurethane filler. Bioresour. Technol. 174, 190–197. doi: 10.1016/j.biortech.2014.09.137
Sugumaran, K. R., Jothi, P., and Ponnusami, V. (2014). Bioconversion of industrial solid waste-Cassava bagasse for pullulan production in solid state fermentation. Carbohydr. Polym. 99, 22–30. doi: 10.1016/j.carbpol.2013.08.039
Sun, C., Zhang, S., Xin, F., Shanmugam, S., and Wu, Y. R. (2018). Genomic comparison of Clostridium species with the potential of utilizing red algal biomass for biobutanol production. Biotechnol. Biofuels 11:42. doi: 10.1186/s13068-018-1044-9
Sundar, M. S. L., Susmitha, A., Rajan, D., Hannibal, S., Sasikumar, K., Wendisch, V. F., et al. (2020). Heterologous expression of genes for bioconversion of xylose to xylonic acid in Corynebacterium glutamicum and optimization of the bioprocess. AMB Express 10:68. doi: 10.1186/s13568-020-01003-9
Tuyishime, P., Wang, Y., Fan, L., Zhang, Q., Li, Q., Zheng, P., et al. (2018). Engineering Corynebacterium glutamicum for methanol-dependent growth and glutamate production. Metab. Eng. 49, 220–231. doi: 10.1016/j.ymben.2018.07.011
Uhde, A., Youn, J. W., Maeda, T., Clermont, L., Matano, C., Krämer, R., et al. (2013). Glucosamine as carbon source for amino acid-producing Corynebacterium glutamicum. Appl. Microbiol. Biotechnol. 97, 1679–1687. doi: 10.1007/s00253-012-4313-8
Veldmann, K. H., Minges, H., Sewald, N., Lee, J. H., and Wendisch, V. F. (2019). Metabolic engineering of Corynebacterium glutamicum for the fermentative production of halogenated tryptophan. J. Biotechnol. 291, 7–16. doi: 10.1016/j.jbiotec.2018.12.008
Vivek, N., Sindhu, R., Madhavan, A., Anju, A. J., Castro, E., Faraco, V., et al. (2017). Recent advances in the production of value added chemicals and lipids utilizing biodiesel industry generated crude glycerol as a substrate – metabolic aspects, challenges and possibilities: an overview. Bioresour. Technol. 239, 507–517. doi: 10.1016/j.biortech.2017.05.056
Wang, C., Cai, H., Chen, Z., and Zhou, Z. (2016). Engineering a glycerol utilization pathway in Corynebacterium glutamicum for succinate production under O2 deprivation. Biotechnol. Lett. 38, 1791–1797. doi: 10.1007/s10529-016-2166-4
Wang, X., Khushk, I., Xiao, Y., Gao, Q., and Bao, J. (2018). Tolerance improvement of Corynebacterium glutamicum on lignocellulose derived inhibitors by adaptive evolution. Appl. Microbiol. Biotechnol. 102, 377–388. doi: 10.1007/s00253-017-8627-4
Wang, Y., Fan, L., Tuyishime, P., Liu, J., Zhang, K., Gao, N., et al. (2020). Adaptive laboratory evolution enhances methanol tolerance and conversion in engineered Corynebacterium glutamicum. Commun. Biol. 3:217. doi: 10.1038/s42003-020-0954-9
Wang, Z., and Yang, S. (2013). Propionic acid production in glycerol/glucose co-fermentation by Propionibacterium freudenreichii subsp. shermanii. Bioresour. Technol. 137, 116–123. doi: 10.1016/j.biortech.2013.03.012
Wen, J., and Bao, J. (2019). Engineering Corynebacterium glutamicum triggers glutamic acid accumulation in biotin-rich corn stover hydrolysate. Biotechnol. Biofuels 12:86. doi: 10.1186/s13068-019-1428-5
Wen, J., Xiao, Y., Liu, T., Gao, Q., and Bao, J. (2018). Rich biotin content in lignocellulose biomass plays the key role in determining cellulosic glutamic acid accumulation by Corynebacterium glutamicum. Biotechnol. Biofuels 11:132. doi: 10.1186/s13068-018-1132-x
Wendisch, V. F., Mindt, M., and Pérez-García, F. (2018). Biotechnological production of mono- and diamines using bacteria: recent progress, applications, and perspectives. Appl. Microbiol. Biotechnol. 102, 3583–3594. doi: 10.1007/s00253-018-8890-z
Westbrook, A. W., Miscevic, D., Kilpatrick, S., Bruder, M., Mooyoung, M., and Chou, C. P. (2019). Strain engineering for microbial production of value-added chemicals and fuels from glycerol. Biotechnol. Adv. 37, 538–568. doi: 10.1016/j.biotechadv.2018.10.006
Witthoff, S., Muhlroth, A., Marienhagen, J., and Bott, M. (2013). C1 metabolism in Corynebacterium glutamicum: an endogenous pathway for oxidation of methanol to carbon dioxide. Appl. Environ. Microbiol. 79, 6974–6983. doi: 10.1128/AEM.02705-13
Witthoff, S., Schmitz, K., Niedenfuhr, S., Noh, K., Noack, S., Bott, M., et al. (2015). Metabolic engineering of Corynebacterium glutamicum for methanol metabolism. Appl. Environ. Microbiol. 81, 2215–2225. doi: 10.1128/AEM.03110-14
Wu, X. Y., Guo, X. Y., Zhang, B., Jiang, Y., and Ye, B. C. (2019). Recent advances of L-ornithine biosynthesis in metabolically engineered Corynebacterium glutamicum. Front. Bioeng. Biotechnol. 7:440. doi: 10.3389/fbioe.2019.00440
Xiberras, J., Klein, M., and Nevoigt, E. (2019). Glycerol as a substrate for Saccharomyces cerevisiae based bioprocesses - Knowledge gaps regarding the central carbon catabolism of this ‘non-fermentable’ carbon source. Biotechnol. Adv. 37:107378. doi: 10.1016/j.biotechadv.2019.03.017
Yim, S. S., Choi, J. W., Lee, S. H., and Jeong, K. J. (2016). Modular optimization of a hemicellulose-utilizing pathway in Corynebacterium glutamicum for consolidated bioprocessing of hemicellulosic biomass. ACS Synth. Biol. 5, 334–343. doi: 10.1021/acssynbio.5b00228
Yun, E. J., Oh, E. J., Liu, J. J., Yu, S., Kim, D., Kwak, S., et al. (2018). Promiscuous activities of heterologous enzymes lead to unintended metabolic rerouting in Saccharomyces cerevisiae engineered to assimilate various sugars from renewable biomass. Biotechnol. Biofuels 11:140. doi: 10.1186/s13068-018-1135-7
Zhang, B., Gao, G., Chu, X. H., and Ye, B. C. (2019). Metabolic engineering of Corynebacterium glutamicum S9114 to enhance the production of L-ornithine driven by glucose and xylose. Bioresour. Technol. 284, 204–213. doi: 10.1016/j.biortech.2019.03.122
Zhang, B., Ren, L. Q., Yu, M., Zhou, Y., and Ye, B. C. (2018a). Enhanced L-ornithine production by systematic manipulation of L-ornithine metabolism in engineered Corynebacterium glutamicum S9114. Bioresour. Technol. 250, 60–68. doi: 10.1016/j.biortech.2017.11.017
Zhang, B., Yu, M., Wei, W. P., and Ye, B. C. (2018b). Optimization of L-ornithine production in recombinant Corynebacterium glutamicum S9114 by cg3035 overexpression and manipulating the central metabolic pathway. Microb. Cell Fact. 17:91. doi: 10.1186/s12934-018-0940-9
Zhao, N., Qian, L., Luo, G., and Zheng, S. (2018). Synthetic biology approaches to access renewable carbon source utilization in Corynebacterium glutamicum. Appl. Microbiol. Biotechnol. 102, 9517–9529. doi: 10.1007/s00253-018-9358-x
Keywords: Corynebacterium glutamicum, renewable feedstocks, metabolic engineering, chemical, fermentation
Citation: Zhang B, Jiang Y, Li Z, Wang F and Wu X-Y (2020) Recent Progress on Chemical Production From Non-food Renewable Feedstocks Using Corynebacterium glutamicum. Front. Bioeng. Biotechnol. 8:606047. doi: 10.3389/fbioe.2020.606047
Received: 14 September 2020; Accepted: 31 October 2020;
Published: 10 December 2020.
Edited by:
Yi-Rui Wu, Shantou University, ChinaReviewed by:
Wenming Zhang, Nanjing Tech University, ChinaK. Madhavan Nampoothiri, National Institute for Interdisciplinary Science and Technology (CSIR), India
Copyright © 2020 Zhang, Jiang, Li, Wang and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bin Zhang, emhhbmdiaW4yOTE5QGp4YXUuZWR1LmNu
 Bin Zhang
Bin Zhang Yan Jiang1,2
Yan Jiang1,2 Zhimin Li
Zhimin Li Xiao-Yu Wu
Xiao-Yu Wu