
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Bioeng. Biotechnol., 05 November 2020
Sec. Bioprocess Engineering
Volume 8 - 2020 | https://doi.org/10.3389/fbioe.2020.598392
This article is part of the Research TopicDevelopment and Application of Clostridia as Microbial Cell-factories for Biofuels and Biochemicals ProductionView all 13 articles
N-Butanol, a valuable solvent and potential fuel extender, can be produced via acetone-butanol-ethanol (ABE) fermentation. One of the main drawbacks of ABE fermentation is the high toxicity of butanol to producing cells, leading to cell membrane disruption, low culture viability and, consequently, low produced concentrations of butanol. The goal of this study was to obtain mutant strains of Clostridium beijerinckii NRRL B-598 with improved butanol tolerance using random chemical mutagenesis, describe changes in their phenotypes compared to the wild-type strain and reveal changes in the genome that explain improved tolerance or other phenotypic changes. Nine mutant strains with stable improved features were obtained by three different approaches and, for two of them, ethidium bromide (EB), a known substrate of efflux pumps, was used for either selection or as a mutagenic agent. It is the first utilization of this approach for the development of butanol-tolerant mutants of solventogenic clostridia, for which generally there is a lack of knowledge about butanol efflux or efflux mechanisms and their regulation. Mutant strains exhibited increase in butanol tolerance from 36% up to 127% and the greatest improvement was achieved for the strains for which EB was used as a mutagenic agent. Additionally, increased tolerance to other substrates of efflux pumps, EB and ethanol, was observed in all mutants and higher antibiotic tolerance in some of the strains. The complete genomes of mutant strains were sequenced and revealed that improved butanol tolerance can be attributed to mutations in genes encoding typical stress responses (chemotaxis, autolysis or changes in cell membrane structure), but, also, to mutations in genes X276_07980 and X276_24400, encoding efflux pump regulators. The latter observation confirms the importance of efflux in butanol stress response of the strain and offers new targets for rational strain engineering.
Butanol can be produced from renewable feedstocks of different kinds, including agricultural waste materials, by acetone-butanol-ethanol (ABE) fermentation using solventogenic clostridia (Lee et al., 2008). The most famous species of the group of solventogenic clostridia are Clostridium acetobutylicum and Clostridium beijerinckii, both of which share a common process bottleneck – low tolerance to butanol.
Butanol is a toxic metabolite that tends to incorporate into the cell membrane, increases membrane fluidity and may disrupt membrane functions (Bowles and Ellefson, 1985; Lepage et al., 1987; Patakova et al., 2018; Vasylkivska and Patakova, 2020). In C. acetobutylicum, at inhibitory levels, effects of butanol on cell membrane results in lower ATP generation by the cell, a malfunction in nutrient uptake and an inability of the cell to maintain its internal pH (Bowles and Ellefson, 1985). In Gram-negative bacteria, butanol damages the inner and outer membranes and can also result in a change in cell shape (Fletcher et al., 2016). Such damage, at the cellular level, results in low culture viability and decreased growth rate and, as a consequence, low final butanol concentrations achieved after fermentation (Ingram, 1986).
Different methods have been used to obtain butanol-tolerant strains of solventogenic clostridia, including targeted modifications, e.g., overexpression of genes encoding heat-shock proteins or modification of fatty acids synthesis (Tomas et al., 2003; Zhao et al., 2003; Mann et al., 2012; Liao et al., 2017), serial transfer and adaptation (Lin and Blaschek, 1983; Baer et al., 1987; Soucaille et al., 1987; Xue et al., 2012; Liu et al., 2013; Yang and Zhao, 2013) or random mutagenesis (Hermann et al., 1985; Matta-el-Ammouri et al., 1986; Annous and Blaschek, 1991; Jain et al., 1994; Mao et al., 2010; Kong et al., 2016; Tanaka et al., 2017). Use of these methods resulted in improved survival in the presence of butanol at a concentration of 10–12 g/L for the wild-type strain (WTS) to 16–18 g/L for mutant strains (Vasylkivska and Patakova, 2020), sometimes up to 23 g/L, as in the case of mutant strain C. beijerinckii BA101 (Qureshi and Blaschek, 2001). Surprisingly, random and targeted mutagenesis have apparently produced very similar improvements in butanol tolerance, although this probably reflects our incomplete understanding of butanol tolerance mechanisms and their regulation (Patakova et al., 2018). In most cases, obtained mutant strains in addition to higher tolerance exhibited higher butanol production, usually an improvement from approximately 9–12 g/L for WTS to 13–16 g/L for mutant strains, sometimes up to 19–21 g/L for selected mutants (Vasylkivska and Patakova, 2020). However, some authors have reported increased tolerance, but not increased butanol production for obtained mutants, both for random (Baer et al., 1987; Gallardo et al., 2017; Máté de Gérando et al., 2018) and targeted (Zhao et al., 2003; Alsaker et al., 2004; Mann et al., 2012; Jones et al., 2016) mutagenesis. Thus, although it is commonly accepted that increased butanol tolerance leads to higher production, such data suggest strain-specific dependence between butanol tolerance and production or even the absence of any direct connection.
Efflux is one of the innate mechanisms of stress response in bacteria and it is based on active transport of the substances from the cell. This mechanism is mostly associated with antibiotic resistance, however, it has been shown that it also takes part in the butanol stress response of Pseudomonas putida and Escherichia coli (Fisher et al., 2014; Bui et al., 2015; Basler et al., 2018; Zhang et al., 2018). It was suggested that efflux can be very effective when cells are dealing with high solvent concentrations (Segura et al., 2012), and enhancement of efflux pump activity could possibly shift metabolic flux, resulting in higher production (Mukhopadhyay, 2015).
Efflux, particularly butanol efflux, is rarely studied in solventogenic clostridia (Vasylkivska and Patakova, 2020). Up-regulation of ATP-binding cassette transporters (Schwarz et al., 2012) and putative efflux pump regulators (Sedlar et al., 2019) observed under cultivation with butanol stress are the only evidence of innate butanol efflux in the group. Recently, a butanol efflux pump from P. putida S12 was expressed in Clostridium saccharoperbutylacetonicum, resulting in improved butanol tolerance (Jiménez-Bonilla et al., 2020) and demonstrating that butanol efflux studies in solventogenic Clostridium have potential for the development of butanol-tolerant production strains. As no native butanol efflux pumps have yet been reported for solventogenic clostridia, targeted engineering cannot be used for the study. Nevertheless, it was shown that efflux enhancement can be achieved even by a point mutation in the efflux pump gene sequence (Bohnert et al., 2007) or in the efflux pump promoter or regulator. To generate such mutations, ethyl methanesulfonate (EMS) or ethidium bromide (EB) can be used. EMS can alkylate guanine bases in DNA, resulting in unidirectional random transition mutations between GC and AT base pairs. Such a type of mutation can lead to an amino acid change and even loss of protein function. EMS was previously successfully used by Jain et al. (1994) to obtain a high butanol producing stable asporogenic mutant, C. beijerinckii ATCC 55025, formerly C. acetobutylicum ATCC 55025 (Jain et al., 1994). EB is a known substrate of different efflux pumps in both Gram-positive (Patel et al., 2010) and Gram-negative bacteria (Paixao et al., 2009) and also a chemical mutagen (Ohta et al., 2001). Use of EB may enhance efflux in the strain, resulting in improved butanol tolerance. Therefore, for this study, we have chosen three approaches to develop butanol-tolerant mutants of solventogenic C. beijerinckii NRRL B-598:
(1) Random chemical mutagenesis using EMS with selection on butanol (EMS + butanol mutants).
(2) Random chemical mutagenesis using EMS with selection on EB (EMS + EB mutants).
(3) Random chemical mutagenesis using EB as a mutagenic agent, where strains were selected directly on agar plates containing EB (EB mutants).
Phenotypic behavior of selected mutant strains was observed by testing tolerance to different substances such as butanol, EB, ethanol and antibiotics, and metabolite production. The complete genomes of mutant strains that exhibited improved butanol tolerance were sequenced to understand their changes in phenotype and to contribute to knowledge about mutations that lead to increased butanol tolerance. To the best of our knowledge, the genomic sequence of only a few butanol-tolerant strains obtained by random mutagenesis are available.
Clostridium beijerinckii NRRL B-598 (WTS), former Clostridium pasteurianum NRRL B-598 (Sedlar et al., 2017), obtained from the Agricultural Research Service Culture Collection (1815 N. University Street, Peoria, IL 61604) was used in this study. The culture was maintained in the form of a spore suspension [containing about 2.2⋅108 spores/ml, determined by method described by Branska et al. (2018)] in sterile distilled water at 4°C. For each cultivation experiment, 450 μl of spore preserve was used for 100 ml of cultivation medium.
Mutant strains described in this work were obtained using three different approaches of random chemical mutagenesis (Figure 1). Firstly, ethyl methanesulfonate (EMS) was used as a mutagenic agent in combination with selection in butanol (EMS + butanol mutants), secondly, EMS was used for mutagenesis but selection was carried out in ethidium bromide (EB) (EMS + EB mutants) and, finally, EB was used as a mutagenic agent and strains were directly selected on agar plates containing EB without exposition to EMS (EB mutants).
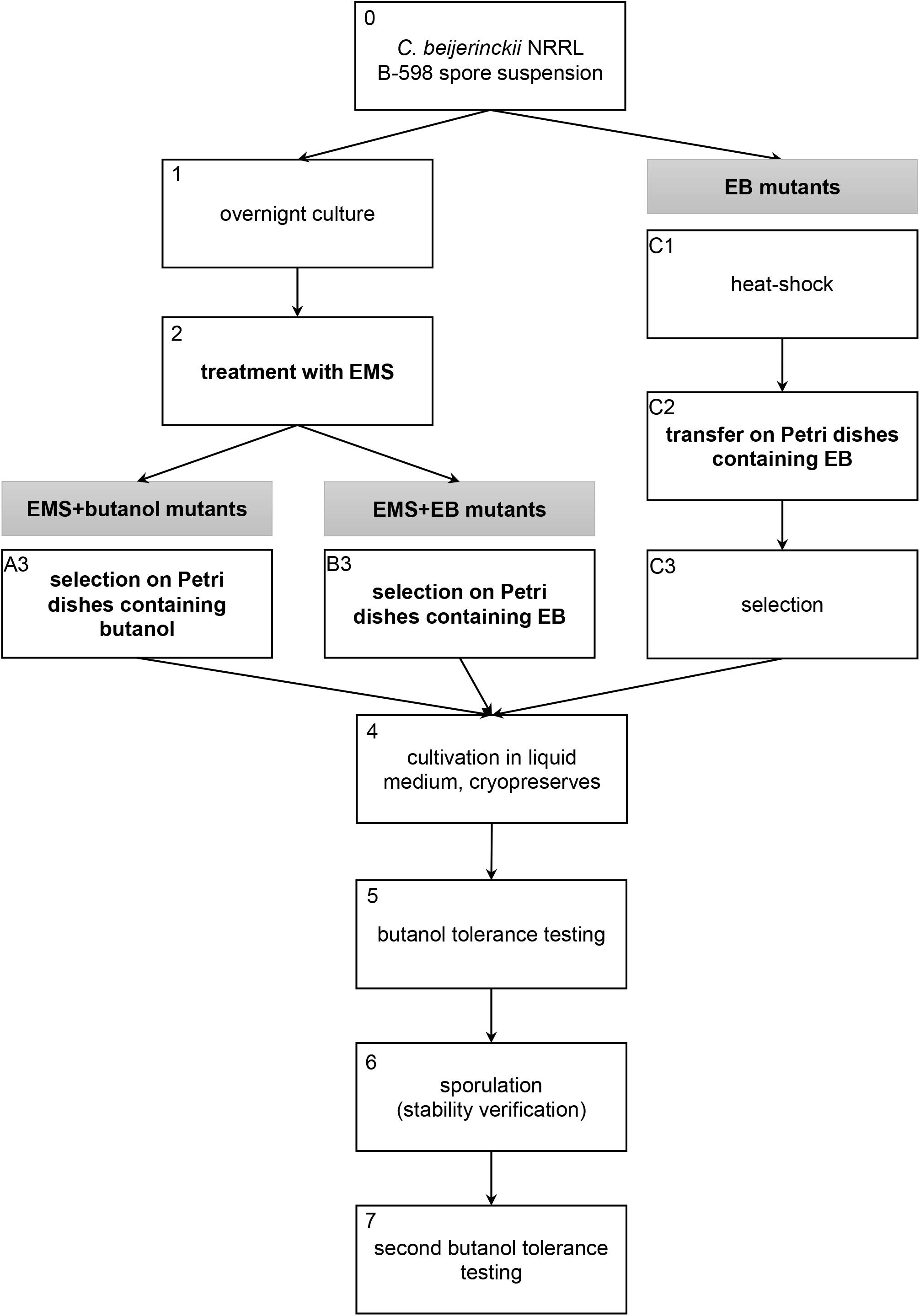
Figure 1. Scheme of three random chemical mutagenesis approaches that were used to obtain mutant strains of C. beijerinckii NRRL B-598 with increased butanol tolerance. EMS, ethyl methanesulfonate; EB, ethidium bromide.
For the first two approaches, when EMS was used as a mutagenic agent, a spore suspension of C. beijerinckii NRRL B-598 was heated to 80°C for 2 min, vortexed and transferred to Erlenmeyer flasks with TYA medium (Vasylkivska et al., 2019) containing 20 g/L of glucose (analytical reagent grade, PENTA, Chrudim, Czechia). The strain was cultivated in a Concept 400 anaerobic chamber (Ruskinn Technology) under a stable N2 atmosphere at 37°C (Figure 1, step 1). After 24 h of cultivation, 2 ml × 2 ml of cell suspension were transferred into sterile Eppendorf tubes, centrifuged and 0.5 ml of fresh sterile TYA medium were added to the cell pellet. Eppendorf tubes were vortexed and 20 μl/ml of EMS (pure, Sigma-Aldrich) were added (Figure 1, step 2). The exposition time was 40 min, during which Eppendorf tubes were placed in the anaerobic chamber. The cell suspension with mutagen was further centrifuged and washed twice with sterile physiological solution. The washed cell suspension was transferred to Petri dishes (250 μl of suspension on each plate) containing HPLC-grade butanol (Sigma-Aldrich) (Figure 1, step A3) or EB (for molecular biology, 10 mg/mL in H2O, Sigma-Aldrich) (Figure 1, step B3) for selection. Tolerance of WTS to butanol and EB was tested prior to mutagenesis procedure (details are described in section “Tolerance Testing”) and twice as high a concentration of butanol (for EMS + butanol mutants) or twice as high a concentration of EB (for EMS + EB mutants) than WTS was able to tolerate were used for mutants selection on Petri dishes (Figure 1, step 3). Inoculated petri dishes were cultivated for 48 h at 37°C in the anaerobic chamber. Colonies obtained were transferred to test tubes containing 10 ml of TYA medium with 20 g/L of glucose, and cultivated overnight in the anaerobic chamber (Figure 1, step 4). The cell suspension was cryopreserved in 30% (v/v) glycerol (analytical reagent grade, PENTA, Chrudim, Czechia) solution below −80°C.
EB mutant strains, prepared by direct cultivation on agar plates containing EB without exposition to EMS, were obtained as follows: the spore suspension of C. beijerinckii NRRL B-598 was heated to 80°C for 2 min (Figure 1, step C1), vortexed and 50 μl of the spore suspension were transferred onto Petri dishes containing either 0.5 mg/L or 2 mg/L of EB (Figure 1, step C2). Agar plates were cultivated in the anaerobic chamber for 24 h, chosen colonies (Figure 1, step C3) were cultivated in TYA medium and the cell culture was cryopreserved (Figure 1, step 4).
Mutant strains obtained by all methods were tested for butanol tolerance (details are described in section “Tolerance Testing) (Figure 1, step 5). Selected mutant strains with improved butanol tolerance compared to the wild-type strain (WTS) were cultivated in TYA medium (EB mutants) or on TYA agar plates (EMS + butanol and EMS + EB mutants) until sporulation was observed (Olympus BX51 microscope); spore preserves were prepared (Figure 1, step 6). Inocula were prepared from the spore preserves and mutant strains were tested one more time for butanol tolerance (Figure 1, step 7). After this step, the selected strains were further used in the experiments described in this article.
Inocula of the wild-type strain and of the mutant strains (except for the first butanol tolerance testing shown as step 5 in Figure 1) were prepared from spore suspensions, and inocula of the mutant strains for the first butanol tolerance testing were prepared from the cryopreserves. Prior to inoculation, the spore suspensions of strains were heated to 80°C for 2 min and vortexed (heat-shock); cryopreserves were thawed at room temperature and vortexed. TYA medium containing 20 g/L of glucose was used for the preparation of inocula and inoculated test tubes were transferred to the anaerobic chamber and cultivated at 37°C overnight.
Tolerance to various substances was tested in microtiter plates containing 120 μl of TYA medium and 10 μl of cell culture. Medium for the experiment additionally contained 0.02 g/L of acid base indicator, bromocresol purple (suitable for indicator, dye content 90%, Sigma-Aldrich), and each substance in different concentrations. Eight substances were tested: two metabolites [butanol in the range of 0 to 30 g/L and ethanol (analytical reagent grade, PENTA, Chrudim, Czechia) in the range of 0 to 65 g/L], efflux pump inducer ethidium bromide EB in the range of 0–6 mg/L and five antibiotics [chloramphenicol (≥98% (HPLC), Sigma-Aldrich] in the range of 0 to 150 mg/L, tetracycline [98.0–102.0% (HPLC), Sigma-Aldrich] in the range of 0–30 mg/L, streptomycin in a form of streptomycin sulfate [>95.0% (TN), Tokyo Chemical Industry] in the range of 0 to 35 mg/L, ampicillin in a form of sodium salt (pure Ph. EUr., AppliChem) in the range of 0–100 mg/L and erythromycin [(for microbiological assay, Sigma-Aldrich) in the range of 0–100 mg/L]. Inoculated microtiter plates were cultivated in the anaerobic chamber at 37°C for 24 h. Results of the testing were evaluated visually as a change in color of the medium, from purple to yellow due to acid production and medium pH shift indicating growth of the strain. For each strain and each substrate, butanol tolerance was tested in at least three repetitions and TYA medium without additions was used as a control.
For the determination of glucose consumption rate and metabolite production, WTS and mutant strains were cultivated in triplicates in non-shaken Erlenmeyer flasks. TYA medium containing 40 g/L of glucose was used for both inoculum preparation and cultivation experiment itself. The inoculum was prepared from the spore preserves after heat-shock (as described in section “Tolerance Testing”) and cultivated in an anaerobic chamber under a stable N2 atmosphere at 37°C overnight. For the cultivation experiment, flasks were inoculated with 10% (v/v) cell culture and cultivated in the anaerobic chamber for 72 h. At the end of cultivation, samples were taken for pH measurement and subsequent HPLC analysis (details of the analysis are described in section “Analytical Methods”).
Cultivation in triplicates in non-shaken Erlenmeyer flasks with pH control was performed the same way as described above, but TYA medium after sterilization prior to inoculation was supplemented with CaCO3 so that concentration 10 g/L of CaCO3 was achieved.
According to the result of experiment described in Section “Cultivation in Erlenmeyer Flasks,” one selected mutant strain was chosen for batch cultivation alongside with WTS in triplicates in parallel Multiforce 1L bioreactors (Infors HT).
TYA medium for the inoculum was prepared with glucose concentrations of 20 g/L and for cultivation in bioreactors with concentrations of 40 g/L. The inoculum was prepared from the spore preserves as described in Section “Cultivation in Erlenmeyer Flasks.”. Prior to inoculation of bioreactors, oxygen was exchanged with N2 and the pH of the medium was adjusted to 6.4. Bioreactors were inoculated with 10% (v/v) cell culture. Cultivation temperature was 37°C and agitation speed was set to 3.3 s–1 throughout the cultivation, the pH was not controlled. During the cultivation, samples were taken for OD measurements and subsequent HPLC analysis (details of the analysis are described in section “Analytical Methods”).
Culture growth was measured as the optical density OD of the culture broth at 600 nm (Varian Cary 50 UV-Vis spectrophotometer, Agilent) against TYA medium as a blank (Vasylkivska et al., 2019).
The concentrations of lactic acid (retention time tR 6.9 min), acetic acid (tR 8.1 min), ethanol (tR 11.7 min), acetone (tR 12.3 min), butyric acid (tR 13.5 min), butanol (tR 24.9 min), and glucose (tR 4.8 min) were measured using HPLC with refractive index detection (Agilent Series 1200 HPLC). A column with stationary phase of Polymer IEX H from 8 μm (Watrex) was used for the separation. Samples of culture broth were centrifuged and the supernatants were microfiltered. The sample injection volume was 20 μl, the column temperature was 60°C, and 5 mM H2SO4 was used as the mobile phase with a flow-rate of 1 ml/min. The concentration of substances was determined from calibration curves (Sedlar et al., 2018).
DNA was isolated from exponentially growing cultures prepared by inoculation of TYA medium with heat-shocked spores (as described above) using commercially available isolation kit DNeasy UltraClean Microbial Kit (Qiagen, Hilden, Germany), following recommended instructions. Library construction and sequencing of the sample was performed by CEITEC Genomics core facility (Brno, Czechia) on Illumina NextSeq 500, pair-end, 150 bp.
The quality assessment of particular steps of data processing were done using FastQC in combination with MultiQC to summarize the reports across all samples (Ewels et al., 2016). Adapter and quality trimming and filtering of singletons was performed with Trimmomatic v0.36 (Bolger et al., 2014). Remaining high quality paired reads were mapped to the genome sequence of the wild type strain with BWA-mem v0.7.15 (Li and Durbin, 2009). The latest genome assembly of the C. beijerinckii NRRL B-598 CP011966.3 (Sedlar et al., 2019) was used as a reference. Resulting SAM (Sequence Read Alignment/Map) files were indexed and transformed into more compact BAM (Binary Read Alignment/Map) format using SAMtools v1.9 (Li et al., 2009). Sequences in BAM files were cleaned, sorted, deduplicated and files were indexed with Picard Tools v2.21.6. Single nucleotide variants in genome sequences between wild type strain and particular mutant strains were called with GATK v4.1.4.1 (McKenna et al., 2010). Detected variants were further filtered in order to reduce the number of false positives. For this purpose, WTS resequencing data gathered within the same sequencing run, were used. All variants that were called simultaneously in the WTS and mutant were filtered out. Moreover, we used coverage of false detections in the WTS to set a threshold for filtering, and only variants covered by more than 25.9% of an average coverage of a strain were used. This threshold corresponded to the highest coverage of falsely detected mutations in the WTS. Moreover, not all variants were detected in the whole population. We again set a threshold using analysis of the WTS. Only variants that were called in at least 30.8% of the population were counted. Furthermore, we detected structural variations to the genome sequences, including copy number variations (CNV) with Pilon v 1.22 (Walker et al., 2014). All these analyses were performed with R/Bioconductor using functions from the genomeIntervals v1.42 (Gagneur et al., 2020), Biostrings v2.54 (Pagès et al., 2020), vcfR v1.11 (Knaus and Grünwald, 2017), and ggplot2 (Wickham, 2009) packages.
As a first step, WTS C. beijerinckii NRRL B-598 was tested for its tolerance to butanol, ethanol and EB. Testing revealed that the strain was able to grow in medium containing no more than 11 ± 1 g/L of butanol, 38.5 ± 2.1 g/L of ethanol or 1.75 ± 0.40 mg/L of EB. EMS + butanol and EMS + EB mutants were, therefore, picked (Figure 1, step 3) from agar plates containing approximately twice as high a concentration of butanol or EB, which were 20 g/L and 4 g/L, respectively, prior to both butanol tolerance testing in microtiter plates (Figure 1, steps 5 and 7).
Number of strains being reduced through screening process is shown in Table 1. For mutagenesis using EMS as mutagenic agent, total of 45 colonies were selected during step A3 (Figure 1) and 45 during B3 (Figure 1). The first butanol tolerance testing (Figure 1, step 5) revealed that 32 strains exhibited increased butanol tolerance compared to WTS. To ensure that increased butanol tolerance was a result of mutation and not adaptation, these 32 strains were cultivated on TYA agar plates until sporulation was observed (Figure 1, step 6) and then spores were germinated and the strains were once again tested for butanol tolerance (Figure 1, step 7). After the second round of butanol tolerance testing, six strains exhibiting increased butanol tolerance were selected for further experiments: EMS + butanol mutants B33 and B44 and EMS + EB mutants E15, E28, E32, and E33 (Table 1).
Using another approach, mutagenesis of WTS was carried out on agar plates containing EB with no exposition to EMS (EB mutants, Figure 1 steps C1–C3). 24 colonies (Table 1) were picked during C3 step (Figure 1). After the subsequent butanol tolerance test in microtiter plates (Figure 1, step 5), three EB mutants strains exhibited an increase in tolerance, strains A, B, and C. These strains were also cultivated until sporulation was observed (Figure 1, step 6) and then the spores were germinated and the strains were again tested for butanol tolerance (Figure 1, step 7). This confirmed that the acquired increase in tolerance was a stable phenotype.
All of mutant strains showed a significant increase in butanol and ethanol tolerance compared to WTS (p < 0.05, two-sample t-test) and were able to grow in a medium containing up to 25.0 g/L of butanol or up to 55.0 g/L of ethanol (Figure 2).
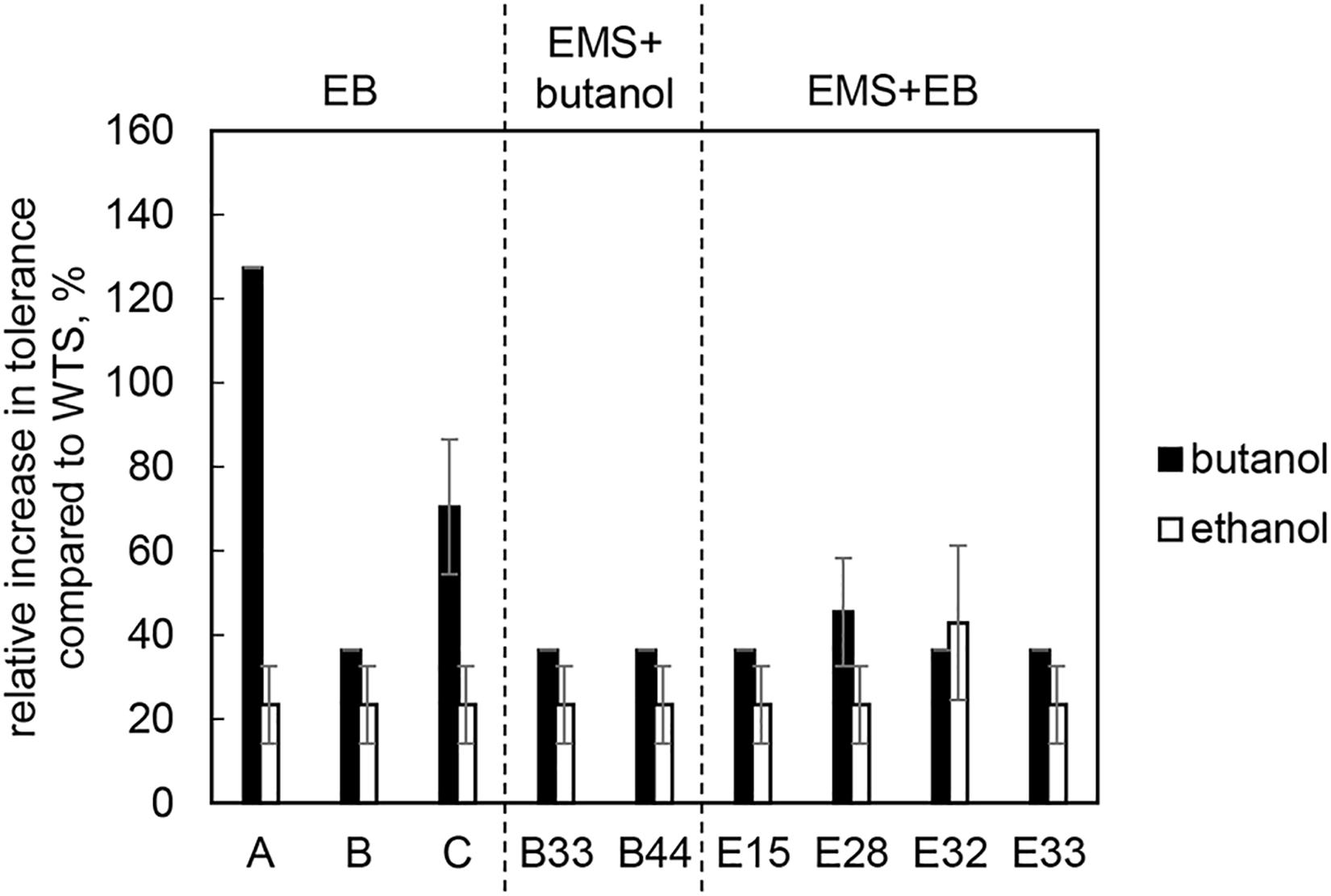
Figure 2. Relative increase in butanol and ethanol tolerance of mutant strains compared to C. beijerinckii NRRL B-598 (WTS). Tolerance levels are expressed as the maximum concentration of butanol or ethanol in TYA medium at which growth of the strains was still detectable. The original tolerance of WTS to butanol and ethanol was 11 ± 1 g/L and 38.5 ± 2.1 g/L, respectively. EB – strains obtained by random chemical mutagenesis using EB as a mutagenic agent, when strains were selected directly on the agar plates containing EB; EMS + butanol – strains obtained by random chemical mutagenesis using EMS with selection on butanol; EMS + EB – strains obtained by random chemical mutagenesis using EMS with selection on EB.
As the goal of the study was to increase the efflux capacity of mutant strains, the tolerance of mutant strains and the WTS to different substrates of efflux pumps, i.e., EB and antibiotics, was also tested. Mutants acquired increased EB (up to 5 mg/L) tolerance, see Figure 3.
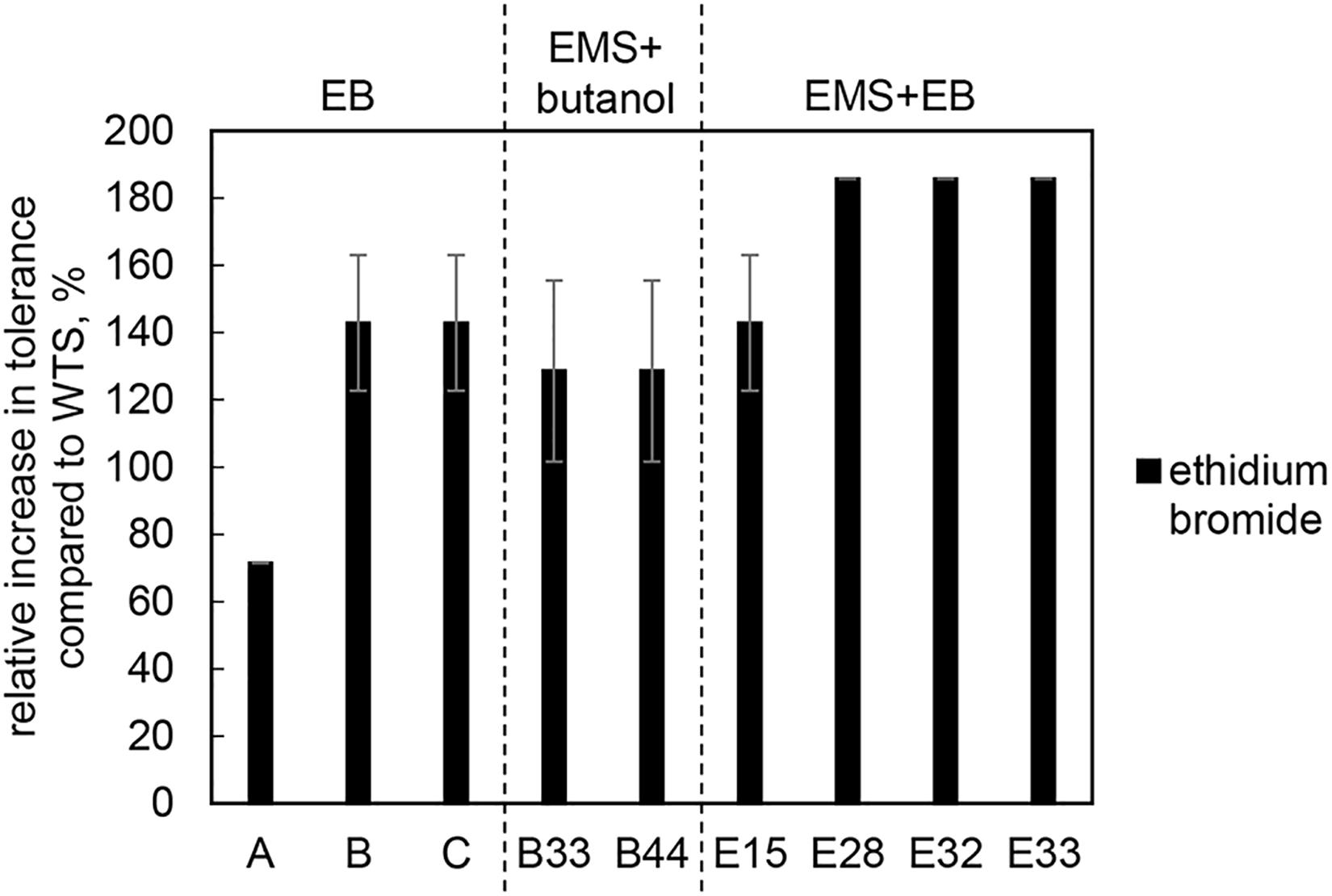
Figure 3. Relative increase in tolerance to EB of mutant strains compared to C. beijerinckii NRRL B-598 (WTS). Tolerance levels are expressed as the maximum concentration of substances in TYA medium at which growth of the strains was still detectable. Original tolerance of WTS to EB was 1.75 ± 0.4 mg/L. EB – strains obtained by random chemical mutagenesis using EB as a mutagenic agent, when strains were selected directly on agar plates containing EB; EMS + butanol - strains obtained by random chemical mutagenesis using EMS with selection on butanol; EMS + EB – strains obtained by random chemical mutagenesis using EMS with selection on EB.
Wild-type strain was tolerant to tetracycline, chloramphenicol and streptomycin at concentrations of 3.0 ± 0.0, 19.0 ± 1.4, and 20.0 ± 0.0 μg/ml, respectively, but was sensitive to ampicillin and erythromycin. The sensitivity toward ampicillin and erythromycin remained unchanged in all mutant strains, but tolerance to tetracycline and chloramphenicol was modified and, in some cases, increased. Surprisingly, tolerance to streptomycin decreased in all cases and strains B33 (EMS + butanol mutant), E15, E32, and E33 (EMS + EB mutants) exhibited complete growth inhibition in the presence of this antibiotic, see Figure 4.
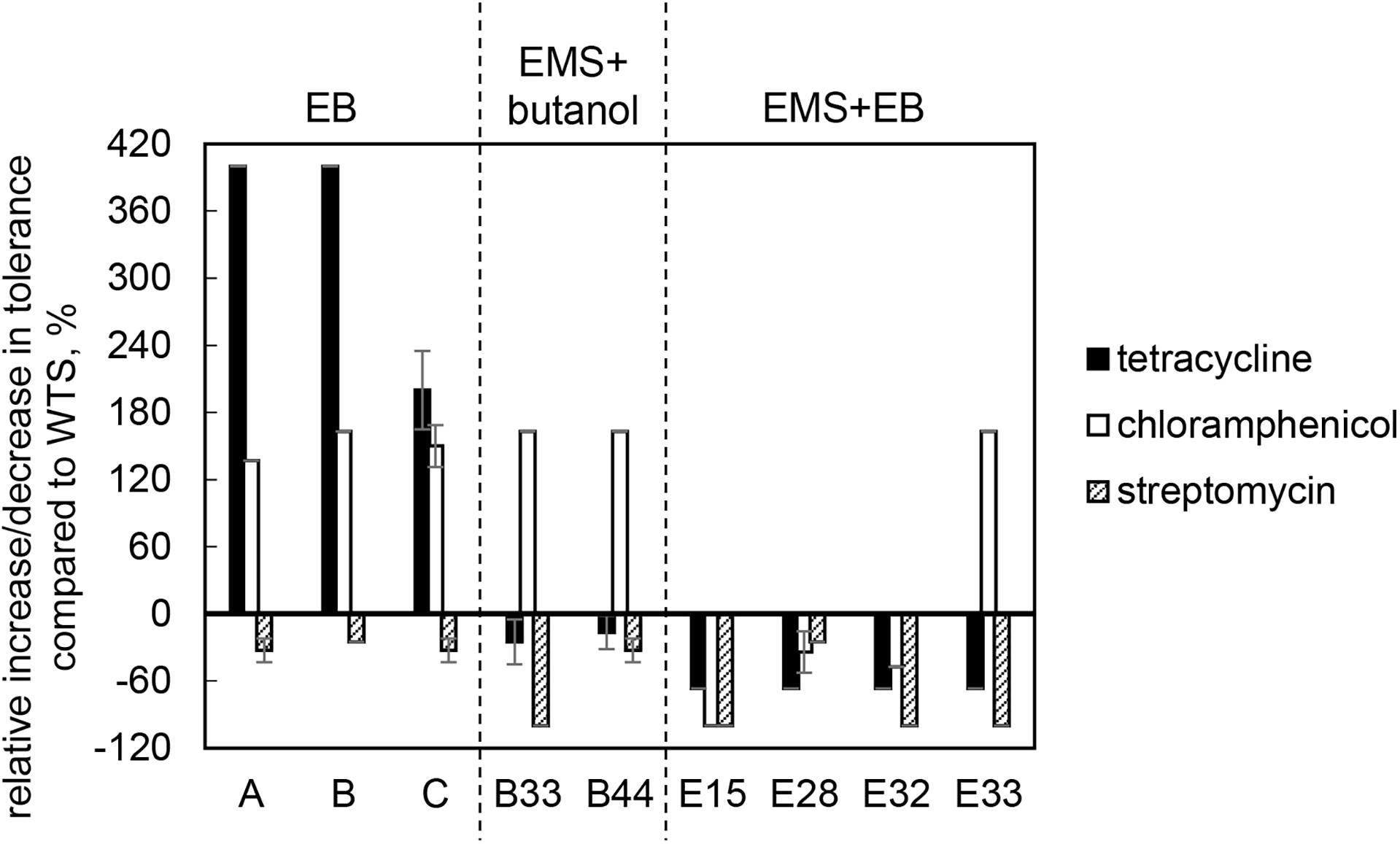
Figure 4. Relative increase/decrease in tolerance to antibiotics tetracycline, chloramphenicol and streptomycin in mutant strains compared to C. beijerinckii NRRL B-598 (WTS). Tolerance levels are expressed as the maximum concentration of substances in TYA medium at which growth of the strains was still detectable. Tolerance of WTS was 3.0 ± 0.0, 19.0 ± 1.4, and 20.0 ± 0.0 μg/ml of tetracycline, chloramphenicol and streptomycin, respectively. EB – strains obtained by random chemical mutagenesis using EB as a mutagenic agent, when strains were selected directly on agar plates containing EB; EMS + butanol – strains obtained by random chemical mutagenesis using EMS with selection on butanol; EMS + EB – strains obtained by random chemical mutagenesis using EMS with the selection on EB.
Production of solvents and acids was tested in flasks containing TYA medium. Mutant strains A and C exhibited similar fermentation profiles as WTS and produced butanol in similar concentrations under standard cultivation conditions (Supplementary Table 1). Other mutant strains, B (EB mutant), B33, B44 (EMS + butanol mutants), E15, E28, E32, and E33 (EMS + EB mutants), probably developed the so-called “acid crash” phenotype and their respective fermentation outputs achieved higher concentrations of butyric, acetic or lactic acids compared to WTS.
Inspired by the study of the mutant strain C. beijerinckii BA105, which at first was seem to only exhibit the acid crash phenotype but it was later revealed that significantly higher butanol production can be achieved by the strain under pH regulation (Seo et al., 2017), we tested TYA medium with the addition of 10 g/L CaCO3 for partial pH control (Table 2). WTS and mutant strains A and C (EB mutants) produced 11–16% higher concentrations of butanol and consumed the total amount of glucose present in the medium. Furthermore, mutant strain B (EB mutant) was able to produce butanol at a concentration of 4.7 ± 0.5 g/L, which is significantly (p < 0.05, two-sample t-test), almost 12 times, higher than that achieved in the medium without supplementation. However, EMS + butanol and EMS + EB stayed in the acidogenic phase of fermentation and the only difference compared to previous experiments was a significantly (p < 0.05, two-sample t-test) higher concentration of butyric acid at the end of fermentation.
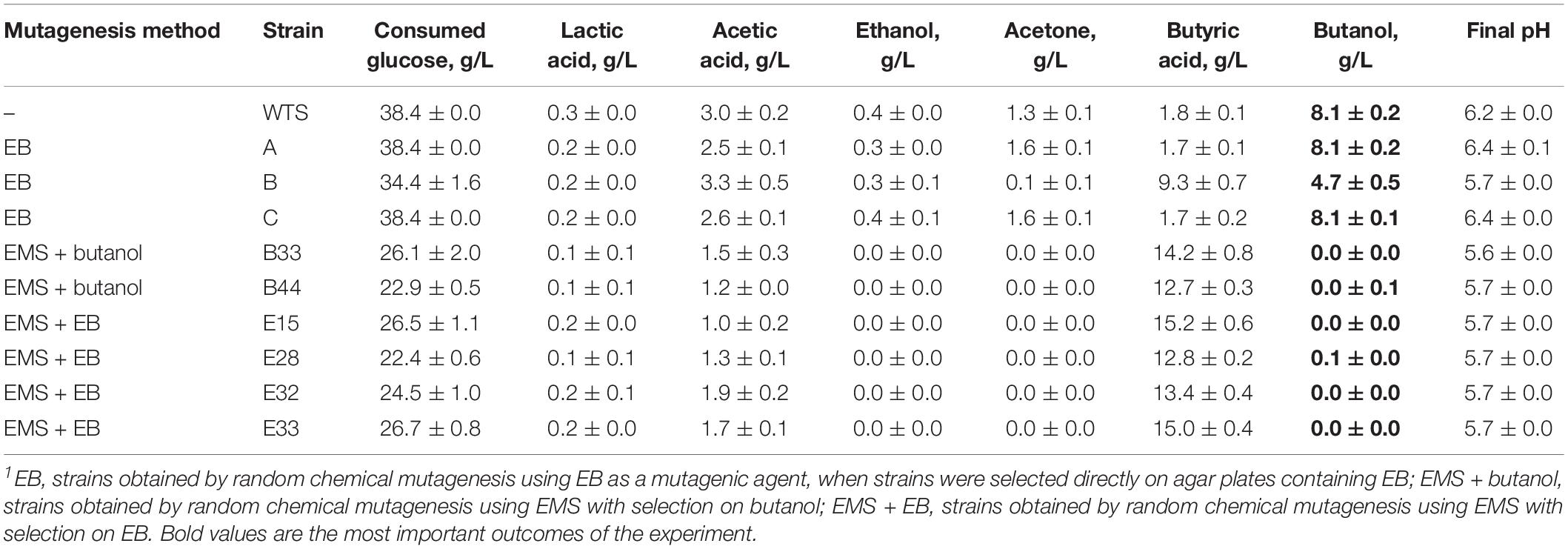
Table 2. Concentrations of glucose, acids and solvents and pH reached by C. beijerinckii NRRL B-598 (WTS) and its mutant strains in TYA medium containing 10 g/L of CaCO3 after 72 h cultivation1.
The behavior of the mutant strain with the overall highest butanol production, EB mutant C, was compared with WTS in parallel batch bioreactor fermentation (Supplementary Figure 1). At first sight, the fermentation profiles, growth curves and glucose consumption curves looked very similar, however, a thorough analysis of fermentation data revealed differences in the fermentation dynamics. The pH and growth curves show that the mutant strain C switched to solventogenesis earlier than the WTS and grew faster during the exponential phase. This was also confirmed by calculation of the specific growth rate (μ) for the exponential phase of growth (first 5 h of cultivation as no lag phase was observed), which was 0.32 ± 0.03 h–1 for the WTS and 0.48 ± 0.01 h–1 for the mutant strain C. Comparison of fermentation parameters for the first 24 h of cultivation and total fermentation time (48 h) is given in Supplementary Table 2. Although the strains reached the same butanol yield (Supplementary Table 2), productivity for mutant C was somewhat higher when calculated for the first 24 h of fermentation, which is in accordance with its higher glucose consumption rate during this period (Supplementary Table 2).
To identify the causes of phenotypic changes in mutant strains, their genomes were sequenced and compared with the WTS genome. After quality trimming and deduplication, 2.4–4.5 million high quality (average Phred score Q ≈ 35) paired sequences mapped to the reference genome suggesting coverage from 115 × to 221 × per mutant strain (see Supplementary Table 3). Sequencing of mutant strains revealed that most of the mutations were single-nucleotide polymorphisms (SNP). In unfiltered data, we detected from 21 to 51 SNPs or short indels per mutant strain and 18 false positive detections in the WTS (see Supplementary Files 4). Nevertheless, after filtering (see section “Materials and Methods”), the number of SNPs was reduced to the range from one to 17 per mutant strain (see Table 3). In total, 21 non-synonymous mutations were captured, including a mutation disrupting an open reading frame. SNPs in six genes were observed in at least two mutant strains (Figure 4). SNPs in X276_13415 encoding a putative S-layer family protein was detected in five mutant strains, SNPs in X276_14460 encoding cytochrome b5 was observed in four strains. EB mutants strain A, B and C included SNPs in the same genes, X276_22865 and X276_03000 encoding a carbohydrate ABC transporter permease and an AAA family ATPase, respectively (Figure 4 and Table 3), and no mutations were observed in these gene when EMS was used as the mutagen.
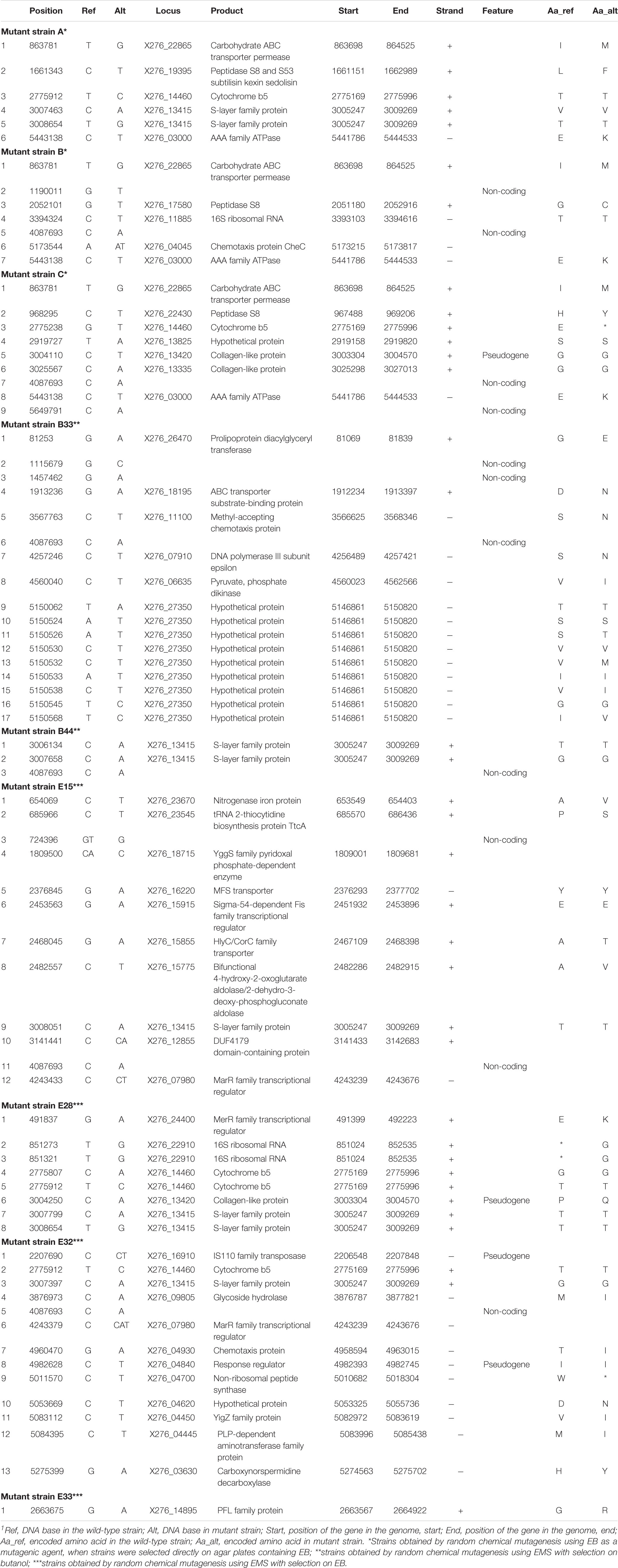
Table 3. List of single-nucleotide polymorphisms that occurred in mutant strains of C. beijerinckii NRRL B-598 exhibiting high butanol tolerance1.
In addition to SNPs, we detected several longer genome changes, including copy number variations (CNVs) (Tables 4, 5). No copy number variations were detected for mutant strain E32. CNVs were detected for similar positions in strains A, B. C, B44, E28, and E33 (Table 5).

Table 4. List of mutations that caused longer changes in the genomes of mutant strains of C. beijerinckii NRRL B-598 exhibiting high butanol tolerance1.
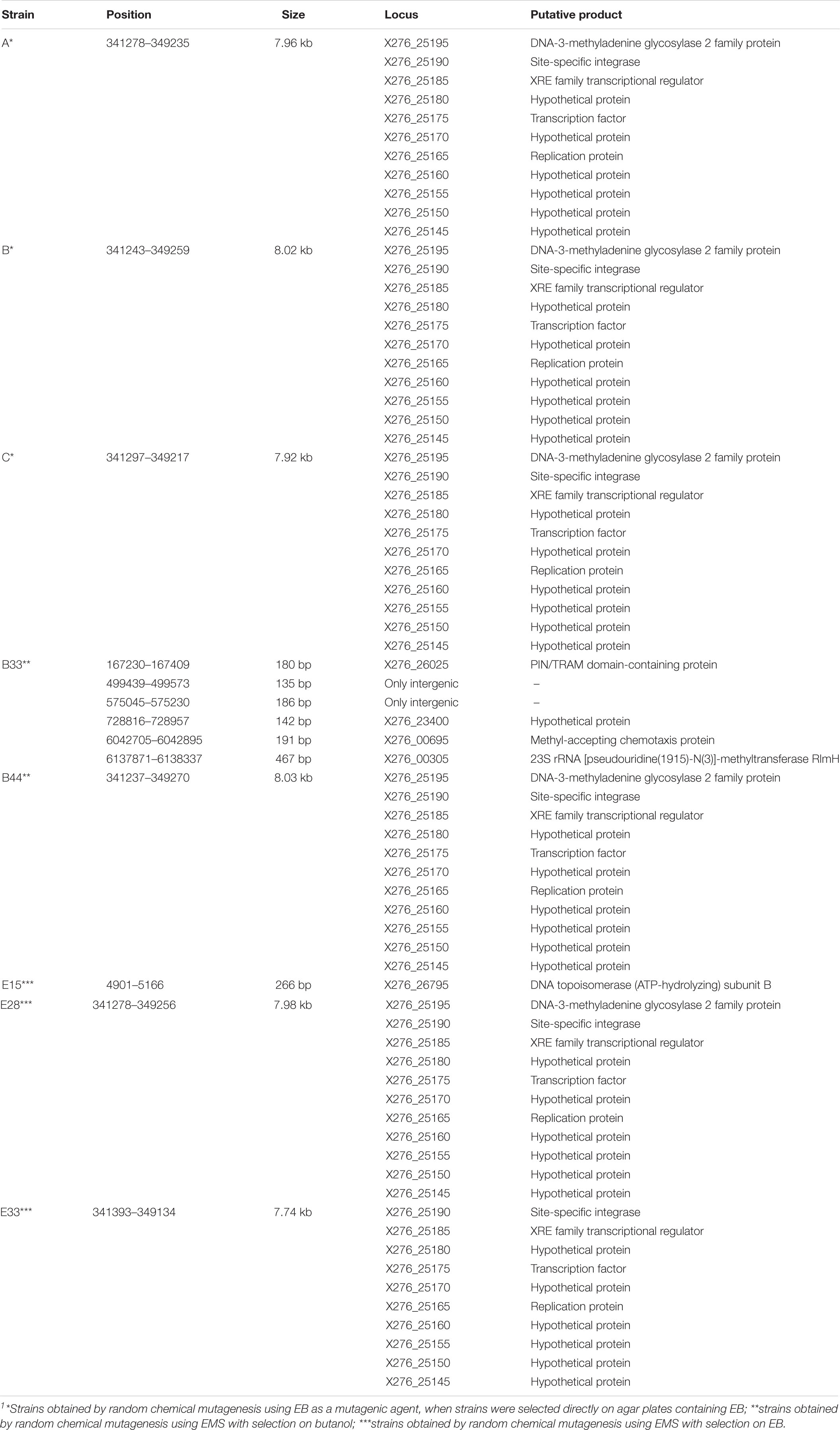
Table 5. List of copy number variations in the genomes of mutant strains of C. beijerinckii NRRL B-598 exhibiting high butanol tolerance1.
Using three different approaches of random chemical mutagenesis, we were able to obtain nine mutant strains of C. beijerinckii NRRL B-598 exhibiting from 36 to 127% increases in butanol tolerance (Figure 2). Similar increases in tolerance have been described for other solventogenic clostridia; in fact, the most well-studied and best-performing mutant strain, C. beijerinckii BA101, was likewise obtained by random chemical mutagenesis (Annous and Blaschek, 1991). C. beijerinckii BA101, an offspring of C. beijerinckii NCIMB 8052, exhibited around a 110% increase in butanol tolerance (Qureshi and Blaschek, 2001). Similarly, the asporogenic mutant C. beijerinckii ATCC 55025, an offspring of C. acetobutylicum ATCC 4259, was able to tolerate about 11.4 g/L of butanol while its WTS only 5 g/L (Jain et al., 1994). Interestingly, two of our nine mutant strains of C. beijerinckii NRRL B-598 with the highest increase in butanol tolerance were obtained by mutagenesis on agar plates containing EB (EB mutants) (Figure 2), a method that has not previously been used for this purpose in solventogenic Clostridium.
Despite increased butanol tolerance, no mutant strain exhibited an increase in butanol production compared with WTS (Supplementary Table 1 and Table 2), which is a divergence from well-known mutant strains obtained in a similar way. For example, C. beijerinckii BA101 was able to produce around 20 g/L of butanol during batch cultivation (an increase of over 100%) (Annous and Blaschek, 1991; Chen and Blaschek, 1999) and C. beijerinckii ATCC 55025 displayed an increase in butanol concentration between 22 and 38% (Jain et al., 1994). However, it has previously been shown in multiple studies, both for random (Baer et al., 1987; Gallardo et al., 2017; Máté de Gérando et al., 2018) and targeted (Zhao et al., 2003; Alsaker et al., 2004; Mann et al., 2012; Jones et al., 2016) mutagenesis, that increased butanol tolerance does not always result in improved butanol production.
EMS + butanol and EMS + EB mutant strains exhibited the acid crash phenotype under standard cultivation conditions and under pH regulation via CaCO3 supplementation (Supplementary Table 1 and Table 2), producing high concentrations of butyric acid. It was shown for the WTS that it is possible to cultivate the strain at larger scale under constant pH regulation to produce butyric acid as the main fermentation product (Drahokoupil and Patáková, 2020). Therefore, these mutant strains can be further studied as alternative butyric acid producers.
Along with higher butanol tolerance, mutant strains of C. beijerinckii NRRL B-598 exhibited improved tolerance to ethanol and EB (Figures 2, 3). Tolerance testing revealed that mutant strains obtained by different approaches behaved differently. While tolerance to ethanol increased similarly in all mutant strains (Figure 2), EMS + EB mutants exhibited higher tolerance to EB than other strains (Figure 3). In the case of antibiotics, EB mutants generally exhibited higher tolerance (Figure 4). On the other hand, antibiotic tolerance of EMS + EB strains was lower than the WTS, except for strain E33 when tested for chloramphenicol (Figure 4). These differences can be probably explained by different mutations that occurred in the strains, so to reveal the differences at the gene level, genomes of all mutant strains were sequenced and variant callings were analyzed.
Variant calling in bacteria is a neglected topic and mainly, attention is paid to eukaryotes. Nevertheless, one of the first benchmarking studies dealing with bacterial variant calling by Bush et al. (2020) showed that a combination of BWA for mapping and GATK HaplotypeCaller brought the best results for closely related strains. As this was the case in our study, we used these tools to capture SNPs (Supplementary Files 4). Moreover, we took advantage of our data containing resequencing of the WTS that was recently used to update its genome assembly (CP011966.3) (Sedlar et al., 2019). We used false positive detections of SNPs in the WTS sequencing data to infer our own filtering rules (see section “Materials and Methods”) (Table 3). While this approach is good for SNP detection, it is unable to call structural variants. Thus, we utilized Pilon to detect longer changes in genomic sequences (Tables 4, 5). The combination of various approaches for detection of short and longer changes is quite common for eukaryotes (Long et al., 2013). Utilization of Pilon is advantageous in our case as the reference sequence was constructed using this approach, and thus, we were again able to remove false positives by filtering variants that were falsely called in the WTS data. SNPs in individual genes of the mutants are discussed further in the text, however, it is difficult to discuss CNVs because the topic has been scarcely studied in bacteria. CNVs are usually studied in comparative analyses (Větrovský and Baldrian, 2013; Greenblum et al., 2015) and the approach cannot be applied in our case.
Only few genome sequences of butanol-tolerant mutant strains obtained by random mutagenesis are currently available [C. beijerinckii SA-1 (Sandoval-Espinola et al., 2013), C. pasteurianum M150B (Sandoval et al., 2015), C. acetobutylicum ATCC 55025 and C. acetobutylicum JB200 (Xu et al., 2017)]. Genomic sequences of C. acetobutylicum ATCC 55025 and C. acetobutylicum JB200, as well as C. pasteurianum M150B, included multiple mutations, for example, 143 SNPs and 67s SNP for C. acetobutylicum ATCC 55025 and C. pasteurianum M150B, respectively. On the other hand, similar to the case of C. beijerinckii SA-1, for which 10 genetic polymorphisms were confirmed, including eight SNPs, genomes of our mutant strains of C. beijerinckii NRRL B-598 included one to 17 SNPs (21–51 prior to filtering), one longer change in strains B33, E15, and E33 and one or more copy number variations in all of the strains, except E32 (Tables 3–5). Some of the mutations revealed in C. beijerinckii NRRL B-598 mutant strains with increased butanol tolerance can be connected to tolerance mechanisms, while others may play roles in metabolic enhancement rather than tolerance improvement. Many of the mutations were membrane-related, which correlates with the fact that normal cellular responses of the strains to the solvent was mainly at the membrane level (Patakova et al., 2018). A similar result was observed in E. coli when a genomic library enrichment strategy was used under butanol challenge (Reyes et al., 2011). Examples of membrane-related mutations in C. beijerinckii NRRL B-598 mutant strains are mutations in genes encoding peptidases, transporters or cytochrome b5 (Table 3). Interestingly, mutations in genes encoding peptidase S8 were observed not only in our EB mutants A (X276_19395), B (X276_17580), and C (X276_22430) (Table 3), but also in a mutant strain of C. beijerinckii SA-1, the offspring of C. beijerinckii NCIMB 8052 (Sandoval-Espinola et al., 2013). Additionally, it seems that EB mutants A, B and C not only exhibited different phenotypes from other mutants for which EMS was used as a mutagenic agent, but were also different in terms of mutations and formed a distinguishable cluster, as shown in Figure 5.

Figure 5. A bi-clustered binary heatmap showing the distribution of genes with single-nucleotide polymorphisms among mutant strains of C. beijerinckii NRRL B-598 with improved butanol tolerance. Complete linkage clustering is based on binary distance between genes and mutants, respectively. Strains A, B, and C were obtained by random chemical mutagenesis using EB as a mutagenic agent, when strains were selected directly on agar plates containing EB; Strains B33 and B44 were obtained by random chemical mutagenesis using EMS with selection on butanol; strains E15, E28, E32, and E33 were obtained by random chemical mutagenesis using EMS with selection on EB.
We analyzed changes in the genomes of mutant strains to determine whether some of the mutations occurring in EMS + butanol and EMS + EB strains could lead to the acid-crash phenotype. In the case of strain E15, the mutation occurred in gene X276_15915 encoding the sigma-54-dependent Fis family transcriptional regulator (Table 3). This may be causing the acid crash phenotype. Sigma-54 regulates sugar consumption and carbon metabolism in C. beijerinckii (Hocq et al., 2019). Thus, a higher glucose uptake rate and acid production rate, the reasons for the acid crash phenotype (Maddox et al., 2000), may occur due to mutations in regulatory genes such as sigma-54. A similar hypothesis has already been proposed for the degenerate strain C. beijerinckii DG-8052 (Lv et al., 2016). Unfiltered data contained a mutation that might be responsible for the phenotype; it is situated in gene X276_15350 encoding NADP-dependent glyceraldehyde-3-phosphate dehydrogenase in mutant strain E28 (Supplementary Files 4). The gene product plays a role in the formation of NADPH, which is necessary for the biosynthetic processes and modulation of redox potential (Liu et al., 2015). The importance of the gene for metabolite production was shown when expression of NADP-dependent glyceraldehyde-3-phosphate dehydrogenase of C. acetobutylicum in E. coli resulted in improved productivity of lycopene and ε-caprolactone (Martínez et al., 2008). As in the case of solventogenic strain C. saccharoperbutylacetonicum, where defects in genes encoding enzymes responsible for NADH formation resulted in strain degeneration (Hayashida and Yoshino, 1990), mutations in genes encoding NADPH formation in mutant strain E28 could affect the cells in a similar way. Nevertheless, this mutation (in gene X276_15350) was later filtered (Table 3) and, therefore, acid crash in the E28 strain probably happened due to some other changes. The reason for the acid crash phenotype in other mutant strains was not clear and needs further investigation.
The faster growth and somewhat higher butanol productivity of mutant C, observed during the first 24 h of cultivation (Supplementary Table 2), can probably be attributed to mutations in genes encoding a carbohydrate ABC transporter permease (X276_22865) and an AAA family ATPase (X276_03000), in which mutations were also detected for the other EB mutants, A and B (Figure 5). Use of non-PTS mechanisms of glucose uptake during the solventogenic phase, such as ABC transporters, resulted in more complete glucose utilization and increased butanol production in C. beijerinckii BA101 (Lee et al., 2005). A BLASTp (Gish and States, 1993) analysis of AAA family ATPase X276_03000 showed that it contained motifs similar to the PTS operon transcription anti-terminator in Clostridioides difficile 630, therefore it probably plays a part in PTS regulation. It was reported that the genome of the butanol-overproducing mutant C. beijerinckii SA-1 included mutations in PTS genes (Sandoval-Espinola et al., 2013) and that the mutant strain C. beijerinckii BA101 had a partially defective PTS (Lee and Blaschek, 2001; Lee et al., 2005). Therefore, a connection between PTS and regulation of solvent production, contributing to butanol overproduction, was hypothesized (Sandoval-Espinola et al., 2013).
Mutations in genes connected to butanol tolerance can also be identified in mutant strains of C. beijerinckii NRRL B-598. For example, in genes encoding chemotaxis proteins in strains B (X276_04045), B33 (X276_11100 and X276_00695), and E32 (X276_04930) (Tables 3, 5). Chemotaxis is one the important mechanisms of adaptation to environmental stress (Zhao et al., 2007), in our case, the presence of produced butanol in the medium. It was shown that butanol acts as a repellent for C. acetobutylicum, meaning that it induces negative chemotaxis in the strain (Gutierrez and Maddox, 1987). For strain E32, mutations were observed in the gene X276_09805 encoding glycoside hydrolase, which belongs to family 25 (Table 3). This family includes enzymes with lysozyme activity and ones connected to autolysin production, for example, lyc gene (CA_C0554) of C. acetobutylicum ATCC 824. It was reported that enzymes that take part in autolysis and cell wall recycling in solventogenic clostridia are also connected to butanol tolerance (Patakova et al., 2018). In mutant strain B33, mutations were detected in X276_26470 encoding prolipoprotein diacylglyceryl transferase (Table 3), which catalyzes attachments of lipoproteins to cell membrane. Lipoproteins take part in multiple processes in the cell, including membrane transport, modifications of the cell wall, and antibiotic tolerance (Nielsen and Lampen, 1982; Chimalapati et al., 2012); therefore, mutations in prolipoprotein diacylglyceryl transferase can also be connected to butanol tolerance mechanisms.
One of most interesting mutations connected to improved butanol tolerance was revealed in EMS + EB mutants E15, E32, and E33 in the gene X276_07980 encoding the MarR family transcriptional regulator (Tables 3, 4). Similarly to our result, mutations in genes encoding the MarR family transcriptional regulator occurred for the hyper butanol-tolerant and -producing strain, C. acetobutylicum JB200 and the authors hypothesized that this mutation contributed to enhanced butanol tolerance (Xu et al., 2017). Interestingly, MarR is a regulator that contributes to antibiotic resistance, as well as resistance to organic solvents and oxidative stress agents, by modulating the efflux pump and porin expression (Sharma et al., 2017). Thus, other regulators may also contribute to enhanced butanol tolerance. For example, mutations in gene X276_24400 encoding the MerR family transcriptional regulator in mutant strain E28 (Table 3). A BLASTp (Gish and States, 1993) analysis showed that the MerR gene shares similarity with the BmrR regulator from Bacillus subtilis, which controls the Bmr efflux pump for removal of antibiotics, dyes and disinfectants (Ahmed et al., 1994). Therefore, mutations in efflux pump regulators were found in all EMS + EB mutants.
These results suggest that the increased or decreased tolerance of mutant strains to antibiotics, EB or solvents (butanol and ethanol) (Figures 2–4) may also be due to altered activities of efflux pumps controlled by regulators. Transcriptional analysis of C. beijerinckii NRRL B-598 revealed upregulation of the TetR/AcrR family regulators putatively involved in efflux pump gene transcription after butanol addition to the medium (Sedlar et al., 2019), suggesting that efflux pumps might be involved in overcoming butanol stress in the strain. In the C. beijerinckii NRRL B-598 genome, 55 genes were identified as putatively encoding efflux pumps (Jureckova et al., 2018). Some of these pumps may be able to remove antibiotics, EB or solvents from a cell. For example, genes encoding Gram-positive efflux pumps capable of transporting tested antibiotics have been described for Streptococcus pneumoniae, B. subtilis, Enterococcus faecalis, Lactococcus lactis, and Corynebacterium glutamicum (Schindler and Kaatz, 2016). An efflux pump responsible for increased ethanol tolerance was reported for Saccharomyces cerevisiae BY4741 (Yang et al., 2013). Furthermore, native efflux pumps able to actively remove butanol were recently discovered in Pseudomonas putida and E. coli (Basler et al., 2018; Zhang et al., 2018), and heterologous expression of such a butanol efflux pump from P. putida in butanol-producing C. saccharoperbutylacetonicum led to increased butanol tolerance of the strain (Jiménez-Bonilla et al., 2020).
Our study shows that random chemical mutagenesis using EB can be successfully used for the generation of butanol-tolerant mutant strains in solventogenic Clostridium. While mutagenesis with EMS as a mutagenic agent, which is more commonly used for such a purpose, resulted in mutants exhibiting the acid-crash phenotype, use of EB alone did not disrupt solvent production. Moreover, use of EB for both mutagenesis and selection resulted in increased tolerance to several different substrates of efflux pumps.
We speculate that the acid crash phenotype in mutant strain E15 was acquired due to mutations in the gene encoding the sigma-54-dependent Fis family transcriptional regulator. Further investigations are needed to reveal the reasons for the phenotype in other EMS + butanol and EMS + EB strains.
Higher butanol tolerance of the strains may be connected to mutations in genes connected to the stress response, for example, glycoside hydrolase or prolipoprotein diacylglyceryl transferase. However, the most prominent change in tolerance to substrates of efflux pumps, including butanol, can be explained by mutations in genes encoding efflux pump regulators, which were found in EMS + EB mutants. These regulators can be further studied in research connected to butanol tolerance mechanisms using targeted mutagenesis.
The genome sequencing data have been deposited in the NCBI Sequence Read Archive (SRA) under the project accession number PRJNA229510 (https://www.ncbi.nlm.nih.gov/sra/?term=PRJNA229510). WTS data are available under accession number SRX6419139 and mutant strains data under accession number within the range from SRX8614691 to SRX8614699.
MV, KS, IP, and PP designed the study. MV and BB performed the experiments and analyzed the data. KS and KJ performed the bioinformatics analyses. MV wrote the original draft, KS, BB, and PP contributed to manuscript writing. All authors read and approved the final manuscript.
This work was supported by GACR (Grantova Agentura Ceske Republiky), Czechia, project number 17-00551S.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Computational resources were partially provided by the CESNET LM2015042 and the CERIT Scientific Cloud LM2015085, under the programme “Projects of Large Research, Development, and Innovations Infrastructures”. We acknowledge the CF Genomics of CEITEC supported by the NCMG research infrastructure (LM2015091 funded by MEYS CR) for their support in obtaining the scientific data presented in this paper.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fbioe.2020.598392/full#supplementary-material
Ahmed, M., Borsch, C. M., Taylor, S. S., Vázquez-Laslop, N., and Neyfakh, A. A. (1994). A protein that activates expression of a multidrug efflux transporter upon binding the transporter substrates. J. Biol. Chem. 269, 28506–28513.
Alsaker, K. V., Spitzer, T. R., and Papoutsakis, E. T. (2004). Transcriptional analysis of spo0A overexpression in Clostridium acetobutylicum and its effect on the cell’s response to butanol stress. J. Bacteriol. 186, 1959–1971. doi: 10.1128/JB.186.7.1959-1971.2004
Annous, B. A., and Blaschek, H. P. (1991). Isolation and characterization of Clostridium acetobutylicum mutants with enhanced amylolytic activity. Appl. Environ. Microbiol. 57, 2544–2548.
Baer, S. H., Blaschek, H. P., and Smith, T. L. (1987). Effect of butanol challenge and temperature on lipid composition and membrane fluidity of butanol-tolerant Clostridium acetobutylicum. Appl. Environ. Microbiol. 53, 2854–2861.
Basler, G., Thompson, M., Tullman-Ercek, D., and Keasling, J. (2018). A Pseudomonas putida efflux pump acts on short-chain alcohols. Biotechnol. Biofuels 11:136. doi: 10.1186/s13068-018-1133-9
Bohnert, J. A., Schuster, S., Fähnrich, E., Trittler, R., and Kern, W. V. (2007). Altered spectrum of multidrug resistance associated with a single point mutation in the Escherichia coli RND-type MDR efflux pump YhiV (MdtF). J. Antimicrob. Chemother. 59, 1216–1222. doi: 10.1093/jac/dkl426
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Bowles, L. K., and Ellefson, W. L. (1985). Effects of butanol on Clostridium acetobutylicum. Appl. Environ. Microbiol. 50, 1165–1170.
Branska, B., Pechacova, Z., Kolek, J., Vasylkivska, M., and Patakova, P. (2018). Flow cytometry analysis of Clostridium beijerinckii NRRL B-598 populations exhibiting different phenotypes induced by changes in cultivation conditions. Biotechnol. Biofuels 11:99. doi: 10.1186/s13068-018-1096-x
Bui, L. M., Lee, J. Y., Geraldi, A., Rahman, Z., Lee, J. H., and Kim, S. C. (2015). Improved n-butanol tolerance in Escherichia coli by controlling membrane related functions. J. Biotechnol. 204, 33–44. doi: 10.1016/j.jbiotec.2015.03.025
Bush, S. J., Foster, D., Eyre, D. W., Clark, E. L., De Maio, N., Shaw, L. P., et al. (2020). Genomic diversity affects the accuracy of bacterial single-nucleotide polymorphism-calling pipelines. Gigascience 9:giaa007. doi: 10.1093/gigascience/giaa007
Chen, C. K., and Blaschek, H. P. (1999). Acetate enhances solvent production and prevents degeneration in Clostridium beijerinckii BA101. Appl. Microbiol. Biotechnol. 52, 170–173. doi: 10.1007/s002530051504
Chimalapati, S., Cohen, J. M., Camberlein, E., MacDonald, N., Durmort, C., Vernet, T., et al. (2012). Effects of deletion of the Streptococcus pneumoniae lipoprotein diacylglyceryl transferase gene Lgt on ABC transporter function and on growth in vivo. PLoS One 7:e41393. doi: 10.1371/journal.pone.0041393
Drahokoupil, M., and Patáková, P. (2020). Production of butyric acid at constant pH by a solventogenic strain of Clostridium beijerinckii. Czech J. Food Sci. 38, 185–191. doi: 10.17221/95/2020-CJFS
Ewels, P., Magnusson, M., Lundin, S., and Kaller, M. (2016). MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics 32, 3047–3048. doi: 10.1093/bioinformatics/btw354
Fisher, M. A., Boyarskiy, S., Yamada, M. R., Kong, N., Bauer, S., and Tullman-Ercek, D. (2014). Enhancing tolerance to short-chain alcohols by engineering the Escherichia coli AcrB efflux pump to secrete the non-native substrate n-butanol. ACS Synth. Biol. 3, 30–40. doi: 10.1021/sb400065q
Fletcher, E., Pilizota, T., Davies, P. R., McVey, A., and French, C. E. (2016). Characterization of the effects of n-butanol on the cell envelope of E. coli. Appl. Microbiol. Biotechnol. 100, 9653–9659. doi: 10.1007/s00253-016-7771-6
Gagneur, J., Toedling, J., Bourgon, R., and Delhomme, N. (2020). genomeIntervals: Operations on Genomic Intervals. R package version 1.44.2. Available online at: https://bioconductor.org/packages/release/bioc/html/genomeIntervals.html (accessed June 13, 2020).
Gallardo, R., Alves, M., and Rodrigues, L. R. (2017). Influence of nutritional and operational parameters on the production of butanol or 1,3-propanediol from glycerol by a mutant Clostridium pasteurianum. N. Biotechnol. 34, 59–67. doi: 10.1016/j.nbt.2016.03.002
Gish, W., and States, D. J. (1993). Identification of protein coding regions by database similarity search. Nat. Genet. 3, 266–272. doi: 10.1038/ng0393-266
Greenblum, S., Carr, R., and Borenstein, E. (2015). Extensive strain-level copy-number variation across human gut microbiome species. Cell 160, 583–594. doi: 10.1016/j.cell.2014.12.038
Gutierrez, N. A., and Maddox, I. S. (1987). Role of chemotaxis in solvent production by Clostridium acetobutylicum. Appl. Environ. Microbiol. 53, 1924–1927.
Hayashida, S., and Yoshino, S. (1990). Degeneration of solventogenic Clostridium caused by a defect in NADH generation. Agric. Biol. Chem. 54, 427–435. doi: 10.1271/bbb1961.54.427
Hermann, M., Fayolle, F., Marchal, R., Podvin, L., Sebald, M., and Vandecasteele, J. P. (1985). Isolation and characterization of butanol-resistant mutants of Clostridium acetobutylicum. Appl. Environ. Microbiol. 50, 1238–1243.
Hocq, R., Bouilloux-Lafont, M., Lopes Ferreira, N., and Wasels, F. (2019). σ 54 (σ L) plays a central role in carbon metabolism in the industrially relevant Clostridium beijerinckii. Sci. Rep. 9:7228. doi: 10.1038/s41598-019-43822-2
Ingram, L. O. (1986). Microbial tolerance to alcohols: role of the cell membrane. Trends Biotechnol. 4, 40–44. doi: 10.1016/0167-7799(86)90152-6
Jain, M. K., Beacom, D., and Datta, R. (1994). Mutant strain of C. acetobutylicum and process for making butanol. Patent 12:242. doi: 10.1016/0734-9750(94)90895-8
Jiménez-Bonilla, P., Zhang, J., Wang, Y., Blersch, D., De-Bashan, L.-E., Guo, L., et al. (2020). Enhancing the tolerance of Clostridium saccharoperbutylacetonicum to lignocellulosic-biomass-derived inhibitors for efficient biobutanol production by overexpressing efflux pumps genes from Pseudomonas putida. Bioresour. Technol 312:123532. doi: 10.1016/j.biortech.2020.123532
Jones, A. J., Venkataramanan, K. P., and Papoutsakis, T. (2016). Overexpression of two stress-responsive, small, non-coding RNAs, 6S and tmRNA, imparts butanol tolerance in Clostridium acetobutylicum. FEMS Microbiol. Lett. 363:fnw063. doi: 10.1093/femsle/fnw063
Jureckova, K., Koscova, P., Sedlar, K., Kolek, J., Patakova, P., and Provaznik, I. (2018). “In silico prediction of genes coding efflux pumps in Clostridium beijerinckii NRRL B-598,” in Proceedings of the 6th International Conference on Chemical Technology, eds M. Vesely, Z. Hrdlicka, J. Hanika, and J. Lubojacky (Czech Republic: Czech Society of Industrial Chemistry), 86–90.
Knaus, B. J., and Grünwald, N. J. (2017). vcfr: a package to manipulate and visualize variant call format data in R. Mol. Ecol. Resour. 17, 44–53. doi: 10.1111/1755-0998.12549
Kong, X., He, A., Zhao, J., Wu, H., Ma, J., Wei, C., et al. (2016). Efficient acetone-butanol-ethanol (ABE) production by a butanol-tolerant mutant of Clostridium beijerinckii in a fermentation-pervaporation coupled process. Biochem. Eng. J. 105, 90–96. doi: 10.1016/j.bej.2015.09.013
Lee, J., and Blaschek, H. P. (2001). Glucose uptake in Clostridium beijerinckii NCIMB 8052 and the solvent-hyperproducing mutant BA101. Appl. Environ. Microbiol. 67, 5025–5031.
Lee, J., Mitchell, W. J., Tangney, M., and Blaschek, H. P. (2005). Evidence for the presence of an alternative glucose transport system in Clostridium beijerinckii NCIMB 8052 and the solvent-hyperproducing mutant BA101. Appl. Environ. Microbiol. 71, 3384–3387. doi: 10.1128/AEM.71.6.3384-3387.2005
Lee, S. Y., Park, J. H., Jang, S. H., Nielsen, L. K., Kim, J., and Jung, K. S. (2008). Fermentative butanol production by clostridia. Biotechnol. Bioeng. 101, 209–228. doi: 10.1002/bit.22003
Lepage, C., Fayolle, F., Hermann, M., and Vandecasteele, J. P. (1987). Changes in membrane lipid composition of Clostridium acetobutylicum during acetone-butanol fermentation: effects of solvents, growth temperature and pH. Microbiology 133, 103–110. doi: 10.1099/00221287-133-1-103
Li, H., and Durbin, R. (2009). Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760. doi: 10.1093/bioinformatics/btp324
Li, H., Handsaker, B., Wysoker, A., Fennell, T., Ruan, J., Homer, N., et al. (2009). The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079. doi: 10.1093/bioinformatics/btp352
Liao, Z., Zhang, Y., Luo, S., Suo, Y., Zhang, S., and Wang, J. (2017). Improving cellular robustness and butanol titers of Clostridium acetobutylicum ATCC824 by introducing heat shock proteins from an extremophilic bacterium. J. Biotechnol. 252, 1–10. doi: 10.1016/j.jbiotec.2017.04.031
Lin, Y. L., and Blaschek, H. P. (1983). Butanol production by a butanol-tolerant strain of Clostridium acetobutylicum in extruded corn broth. Appl. Environ. Microbiol. 45, 966–973. doi: 10.1128/aem.45.3.966-973.1983
Liu, J., Qi, H., Wang, C., and Wen, J. (2015). Model-driven intracellular redox status modulation for increasing isobutanol production in Escherichia coli. Biotechnol. Biofuels 8:108.
Liu, X. B., Gu, Q. Y., and Yu, X. B. (2013). Repetitive domestication to enhance butanol tolerance and production in Clostridium acetobutylicum through artificial simulation of bio-evolution. Bioresour. Technol. 130, 638–643. doi: 10.1016/j.biortech.2012.12.121
Long, Q., Rabanal, F. A., Meng, D., Huber, C. D., Farlow, A., Platzer, A., et al. (2013). Massive genomic variation and strong selection in Arabidopsis thaliana lines from Sweden. Nat. Genet. 45, 884–890. doi: 10.1038/ng.2678
Lv, J., Jiao, S., Du, R., Zhang, R., Zhang, Y., and Han, B. (2016). Proteomic analysis to elucidate degeneration of Clostridium beijerinckii NCIMB 8052 and role of Ca2+ in strain recovery from degeneration. J. Ind. Microbiol. Biotechnol. 43, 741–750. doi: 10.1007/s10295-016-1754-6
Maddox, I. S., Steiner, E., Hirsch, S., Wessner, S., Gutierrez, N. A., Gapes, J. R., et al. (2000). The cause of “acid crash” and “acidogenic fermentations” during the batch acetone-butanol-ethanol (ABE-) fermentation process. J. Mol. Microbiol. Biotechnol. 2, 95–100.
Mann, M. S., Dragovic, Z., Schirrmacher, G., and Lutke-Eversloh, T. (2012). Over-expression of stress protein-encoding genes helps Clostridium acetobutylicum to rapidly adapt to butanol stress. Biotechnol. Lett. 34, 1643–1649. doi: 10.1007/s10529-012-0951-2
Mao, S., Luo, Y., Zhang, T., Li, J., Bao, G., Zhu, Y., et al. (2010). Proteome reference map and comparative proteomic analysis between a wild type Clostridium acetobutylicum DSM 1731 and its mutant with enhanced butanol tolerance and butanol yield. J. Proteome Res. 9, 3046–3061. doi: 10.1021/pr9012078
Martínez, I., Zhu, J., Lin, H., Bennett, G. N., and San, K. Y. (2008). Replacing Escherichia coli NAD-dependent glyceraldehyde 3-phosphate dehydrogenase (GAPDH) with a NADP-dependent enzyme from Clostridium acetobutylicum facilitates NADPH dependent pathways. Metab. Eng. 10, 352–359. doi: 10.1016/j.ymben.2008.09.001
Máté de Gérando, H., Wasels, F., Bisson, A., Clement, B., Bidard, F., Jourdier, E., et al. (2018). Genome and transcriptome of the natural isopropanol producer Clostridium beijerinckii DSM6423. BMC Genom. 19:242. doi: 10.1186/s12864-018-4636-7
Matta-el-Ammouri, G., Janati-Idrissi, R., Rambourg, J.-M., Petitdemange, H., and Gay, R. (1986). Acetone butanol fermentation by a Clostridium acetobutylicum mutant with high solvent productivity. Biomass 10, 109–119. doi: 10.1016/0144-4565(86)90059-4
McKenna, A., Hanna, M., Banks, E., Sivachenko, A., Cibulskis, K., Kernytsky, A., et al. (2010). The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303. doi: 10.1101/gr.107524.110
Mukhopadhyay, A. (2015). Tolerance engineering in bacteria for the production of advanced biofuels and chemicals. Trends Microbiol. 23, 498–508. doi: 10.1016/j.tim.2015.04.008
Nielsen, J. B. K., and Lampen, J. O. (1982). Membrane-bound penicillinases in gram-positive bacteria. J. Biol. Chem. 257, 4490–4495.
Ohta, T., Tokishita, S. I., and Yamagata, H. (2001). Ethidium bromide and SYBR Green I enhance the genotoxicity of UV-irradiation and chemical mutagens in E. coli. Mutat. Res. Genet. Toxicol. Environ. Mutagen. 492, 91–97. doi: 10.1016/S1383-5718(01)00155-3
Pagès, H., Aboyoun, P., Gentleman, R., and DebRoy, S. (2020). Biostrings: Efficient Manipulation of Biological Strings. R package version 2.56.0. Available online at: https://bioconductor.org/packages/release/bioc/html/Biostrings.html (accessed June 13, 2020).
Paixao, L., Rodrigues, L., Couto, I., Martins, M., Fernandes, P., de Carvalho, C. C. C. R., et al. (2009). Fluorometric determination of ethidium bromide efflux kinetics in Escherichia coli. J. Biol. Eng. 3:18. doi: 10.1186/1754-1611-3-18
Patakova, P., Kolek, J., Sedlar, K., Koscova, P., Branska, B., Kupkova, K., et al. (2018). Comparative analysis of high butanol tolerance and production in clostridia. Biotechnol. Adv. 36, 721–738. doi: 10.1016/j.biotechadv.2017.12.004
Patel, D., Kosmidis, C., Seo, S. M., and Kaatz, G. W. (2010). Ethidium bromide MIC screening for enhanced efflux pump gene expression or efflux activity in Staphylococcus aureus. Antimicrob. Agents Chemother. 54, 5070–5073. doi: 10.1128/AAC.01058-10
Qureshi, N., and Blaschek, H. P. (2001). Recent advances in ABE fermentation: Hyper-butanol producing Clostridium beijerinckii BA101. J. Ind. Microbiol. Biotechnol. 27, 287–291. doi: 10.1038/sj.jim.7000114
Reyes, L. H., Almario, M. P., and Kao, K. C. (2011). Genomic library screens for genes involved in n-butanol tolerance in Escherichia coli. PLoS One 6:e17678. doi: 10.1371/journal.pone.0017678
Sandoval, N. R., Venkataramanan, K. P., Groth, T. S., and Papoutsakis, E. T. (2015). Whole-genome sequence of an evolved Clostridium pasteurianum strain reveals Spo0A deficiency responsible for increased butanol production and superior growth. Biotechnol. Biofuels 8:227. doi: 10.1186/s13068-015-0408-7
Sandoval-Espinola, W. J., Makwana, S. T., Chinn, M. S., Thon, M. R., Andrea Azcárate-Peril, M., and Bruno-Bárcena, J. M. (2013). Comparative phenotypic analysis and genome sequence of Clostridium beijerinckii SA-1, an offspring of NCIMB 8052. Microbiology 159, 2558–2570. doi: 10.1099/mic.0.069534-0
Schindler, B. D., and Kaatz, G. W. (2016). Multidrug efflux pumps of Gram-positive bacteria. Drug Resist. Updat. 27, 1–13. doi: 10.1016/j.drup.2016.04.003
Schwarz, K. M., Kuit, W., Grimmler, C., Ehrenreich, A., and Kengen, S. W. M. (2012). A transcriptional study of acidogenic chemostat cells of Clostridium acetobutylicum - Cellular behavior in adaptation to n-butanol. J. Biotechnol. 161, 366–377. doi: 10.1016/j.jbiotec.2012.03.018
Sedlar, K., Kolek, J., Gruber, M., Jureckova, K., Branska, B., Csaba, G., et al. (2019). A transcriptional response of Clostridium beijerinckii NRRL B-598 to a butanol shock. Biotechnol. Biofuels 12:243. doi: 10.1186/s13068-019-1584-7
Sedlar, K., Kolek, J., Provaznik, I., and Patakova, P. (2017). Reclassification of non-type strain Clostridium pasteurianum NRRL B-598 as Clostridium beijerinckii NRRL B-598. J. Biotechnol. 244, 1–3. doi: 10.1016/j.jbiotec.2017.01.003
Sedlar, K., Koscova, P., Vasylkivska, M., Branska, B., Kolek, J., Kupkova, K., et al. (2018). Transcription profiling of butanol producer Clostridium beijerinckii NRRL B-598 using RNA-Seq. BMC Genom. 19:415. doi: 10.1186/s12864-018-4805-8
Segura, A., Molina, L., Fillet, S., Krell, T., Bernal, P., Muñoz-Rojas, J., et al. (2012). Solvent tolerance in Gram-negative bacteria. Curr. Opin. Biotechnol. 23, 415–421. doi: 10.1016/j.copbio.2011.11.015
Seo, S.-O., Janssen, H., Magis, A., Wang, Y., Lu, T., Price, N. D., et al. (2017). Genomic, transcriptional, and phenotypic analysis of the glucose derepressed Clostridium beijerinckii mutant exhibiting acid crash phenotype. Biotechnol. J. 12, 1700182. doi: 10.1002/biot.201700182
Sharma, P., Haycocks, J. R. J., Middlemiss, A. D., Kettles, R. A., Sellars, L. E., Ricci, V., et al. (2017). The multiple antibiotic resistance operon of enteric bacteria controls DNA repair and outer membrane integrity. Nat. Commun. 8:1444. doi: 10.1038/s41467-017-01405-7
Soucaille, P., Joliff, G., Izard, A., and Goma, G. (1987). Butanol tolerance and autobacteriocin production by Clostridium acetobutylicum. Curr. Microbiol. 14, 295–299. doi: 10.1007/BF01568139
Tanaka, Y., Kasahara, K., Hirose, Y., Morimoto, Y., Izawa, M., and Ochi, K. (2017). Enhancement of butanol production by sequential introduction of mutations conferring butanol tolerance and streptomycin resistance. J. Biosci. Bioeng. 124, 400–407. doi: 10.1016/j.jbiosc.2017.05.003
Tomas, C. A., Welker, N. E., and Papoutsakis, E. T. (2003). Overexpression of groESL in Clostridium acetobutylicum results in increased solvent production and tolerance, prolonged metabolism, and changes in the cell’s transcriptional program. Appl. Environ. Microbiol. 69, 4951–4965. doi: 10.1128/AEM.69.8.4951-4965.2003
Vasylkivska, M., Jureckova, K., Branska, B., Sedlar, K., Kolek, J., Provaznik, I., et al. (2019). Transcriptional analysis of amino acid, metal ion, vitamin and carbohydrate uptake in butanol-producing Clostridium beijerinckii NRRL B-598. PLoS One 14:e0224560. doi: 10.1371/journal.pone.0224560
Vasylkivska, M., and Patakova, P. (2020). Role of efflux in enhancing butanol tolerance of bacteria. J. Biotechnol. 320, 17–27. doi: 10.1016/j.jbiotec.2020.06.008
Větrovský, T., and Baldrian, P. (2013). The variability of the 16S rRNA gene in bacterial genomes and its consequences for bacterial community analyses. PLoS One 8:e57923. doi: 10.1371/journal.pone.0057923
Walker, B. J., Abeel, T., Shea, T., Priest, M., Abouelliel, A., Sakthikumar, S., et al. (2014). Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9:e112963. doi: 10.1371/journal.pone.0112963
Xu, M., Zhao, J., Yu, L., and Yang, S. T. (2017). Comparative genomic analysis of Clostridium acetobutylicum for understanding the mutations contributing to enhanced butanol tolerance and production. J. Biotechnol. 263, 36–44. doi: 10.1016/j.jbiotec.2017.10.010
Xue, C., Zhao, J., Lu, C., Yang, S. T., Bai, F., and Tang, I. C. (2012). High-titer n-butanol production by Clostridium acetobutylicum JB200 in fed-batch fermentation with intermittent gas stripping. Biotechnol. Bioeng. 109, 2746–2756. doi: 10.1002/bit.24563
Yang, K. M., Woo, J. M., Lee, S. M., and Park, J. B. (2013). Improving ethanol tolerance of Saccharomyces cerevisiae by overexpressing an ATP-binding cassette efflux pump. Chem. Eng. Sci. 103, 74–78. doi: 10.1016/j.ces.2012.09.015
Yang, S.-T., and Zhao, J. (2013). Adaptive engineering of Clostridium for increased butanol production. U.S. Patent No US8450093B1. Washington, DC: U.S. Patent and Trademark Office.
Zhang, Y., Dong, R., Zhang, M., and Gao, H. (2018). Native efflux pumps of Escherichia coli responsible for short and medium chain alcohol. Biochem. Eng. J. 133, 149–156. doi: 10.1016/j.bej.2018.02.009
Zhao, K., Liu, M., and Burgess, R. R. (2007). Adaptation in bacterial flagellar and motility systems: from regulon members to ‘foraging’-like behavior in E. coli. Nucleic Acids Res. 35, 4441–4452. doi: 10.1093/nar/gkm456
Zhao, Y., Hindorff, L. A., Chuang, A., Monroe-Augustus, M., Lyristis, M., Harrison, M. L., et al. (2003). Expression of a cloned cyclopropane fatty acid synthase gene reduces solvent formation in Clostridium acetobutylicum ATCC 824. Appl. Environ. Microbiol. 69, 2831–2841. doi: 10.1128/AEM.69.5.2831-2841.2003
Keywords: butanol tolerance, random chemical mutagenesis, solventogenic Clostridium species, genome sequence, butanol efflux
Citation: Vasylkivska M, Branska B, Sedlar K, Jureckova K, Provaznik I and Patakova P (2020) Phenotypic and Genomic Analysis of Clostridium beijerinckii NRRL B-598 Mutants With Increased Butanol Tolerance. Front. Bioeng. Biotechnol. 8:598392. doi: 10.3389/fbioe.2020.598392
Received: 24 August 2020; Accepted: 20 October 2020;
Published: 05 November 2020.
Edited by:
Fu-Li Li, Qingdao Institute of Bioenergy and Bioprocess Technology (CAS), ChinaReviewed by:
Yi Wang, Auburn University, United StatesCopyright © 2020 Vasylkivska, Branska, Sedlar, Jureckova, Provaznik and Patakova. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Maryna Vasylkivska, dmFzeWxraW1AdnNjaHQuY3o=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.