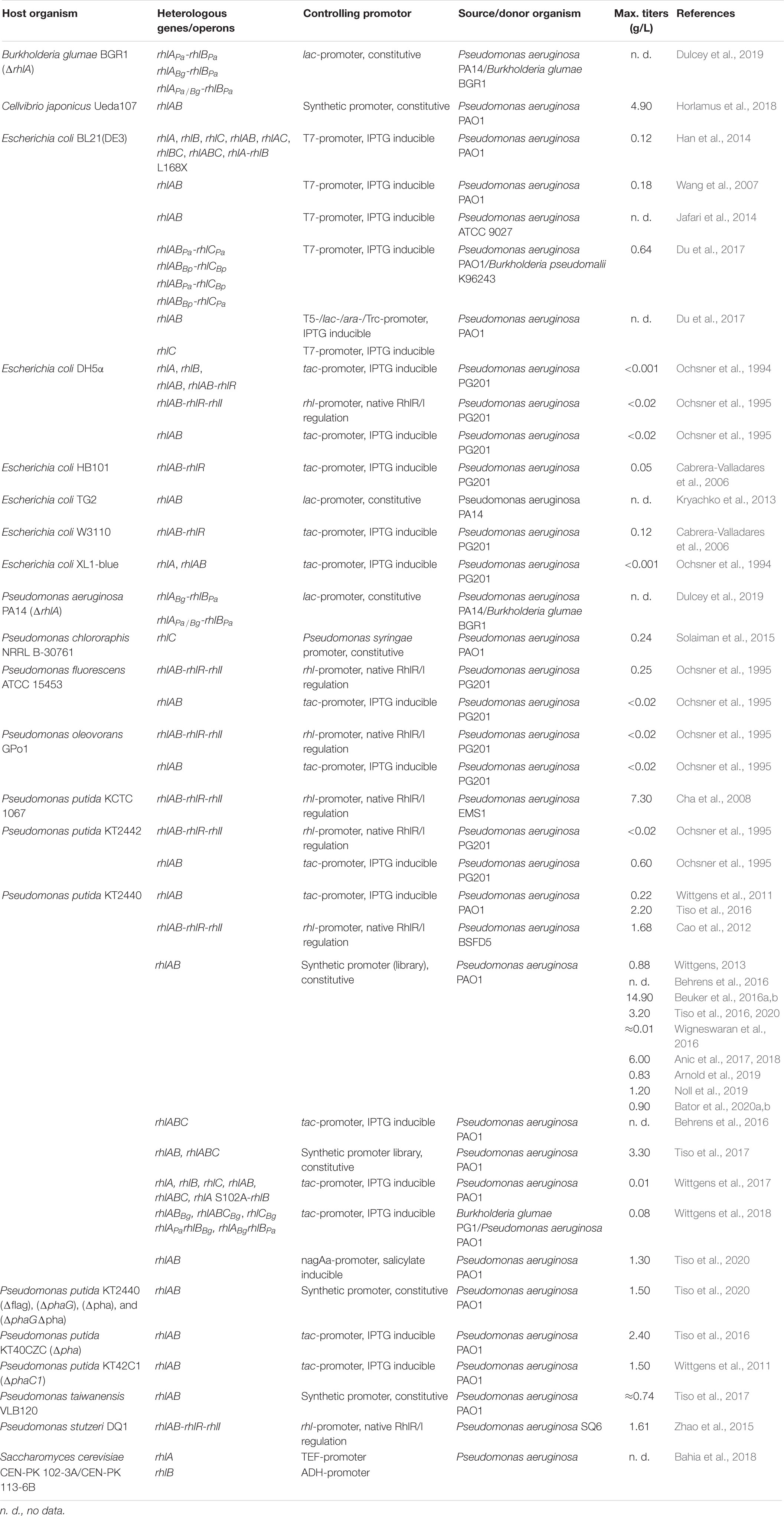
- 1Institute of Pharmaceutical Biotechnology, Ulm University, Ulm, Germany
- 2Ulm Center for Peptide Pharmaceuticals (U-PEP), Ulm University, Ulm, Germany
- 3Department Synthesis of Macromolecules, Max-Planck-Institute for Polymer Research Mainz, Mainz, Germany
The first heterologous expression of genes responsible for the production of rhamnolipids was already implemented in the mid-1990s during the functional identification of the rhlAB operon. This was the starting shot for multiple approaches to establish the rhamnolipid biosynthesis in different host organisms. Since most of the native rhamnolipid producing organisms are human or plant pathogens, the intention for these ventures was the establishment of non-pathogenic organisms as heterologous host for the production of rhamnolipids. The pathogenicity of producing organisms is one of the bottlenecks for applications of rhamnolipids in many industrial products especially foods and cosmetics. The further advantage of heterologous rhamnolipid production is the circumvention of the complex regulatory network, which regulates the rhamnolipid biosynthesis in wild type production strains. Furthermore, a suitable host with an optimal genetic background to provide sufficient amounts of educts allows the production of tailor-made rhamnolipids each with its specific physico-chemical properties depending on the contained numbers of rhamnose sugar residues and the numbers, chain length and saturation degree of 3-hydroxyfatty acids. The heterologous expression of rhl genes can also enable the utilization of unusual carbon sources for the production of rhamnolipids depending on the host organism.
Introduction
Rhamnolipids are among the best characterized biosurfactants and possess outstanding properties to replace or complement conventional surfactants based on petrochemistry in industrial and biotechnological applications. Rhamnolipids are considered as eco-friendly and sustainable, show a very low toxicity, are highly or even perfectly biocompatible and biodegradable (Desai and Banat, 1997; van Hamme et al., 2006; Hirata et al., 2009; Lima et al., 2011; Johann et al., 2016). As an important additional aspect, rhamnolipids exclusively originate from microbiological hosts and can be produced from renewable substrates in contrast to their synthetical counterparts, which primary base on fossil resources (Müller et al., 2010b). Combined with their better foaming properties and remarkable stabilities against extreme pH-values, temperatures and salt concentrations, rhamnolipids offer a broad application potential (Lang and Wullbrandt, 1999; Nitschke and Pastore, 2006; Banat et al., 2010). Rhamnolipids are used in traditional surfactant applications like washing detergents and cleaning agents (Nguyen and Sabatini, 2011), but also in cosmetics and foods (Maier and Soberón-Chávez, 2000; Nitschke and Costa, 2007), bioremediation (Nguyen et al., 2008; Liu et al., 2018) and (microbial) enhanced oil recovery (Wang et al., 2007; Sharma et al., 2018).
The amphiphilic rhamnolipids belong to the class of glycolipids and are composed of a hydrophilic molecule domain consisting of one or two L-rhamnoses β-glycosidically bound to a hydrophobic counterpart consisting of one or two 3-hydroxyfatty acids (Figure 1; Déziel et al., 1999; Abdel-Mawgoud et al., 2010). Based on the number of rhamnose residues, they are separated into mono- and di-rhamnolipids. The number of 3-hydroxyfatty acids they contain allows a further sub-classification into the four known species of natively occurring rhamnolipids, namely the predominant mono- and di-rhamno-di-lipids (mRdL and dRdL) and the rarer to find mono-and di-rhamno-mono-lipids (mRmL and dRmL) (Syldatk et al., 1985a,b; Déziel et al., 1999; Abdel-Mawgoud et al., 2010). The chain length of the fatty acids can vary within the species ranging typically from 8 to 16 carbon atoms depending on the producing wild type. Short-chain rhamnolipids containing a predominant C10-C10 hydrophobic moiety are produced in the highest concentrations known so far by the human-pathogenic organism Pseudomonas aeruginosa (Giani et al., 1997; Müller et al., 2010a) and were first described by Jarvis and Johnson (1949). Among a few other organisms, especially bacteria from the genus Burkholderia, are able to produce rhamnolipids with long-chain fatty acids and predominant C14-C14 congener (Häußler et al., 1998; Andrä et al., 2006; Funston et al., 2016). In organisms naturally producing rhamnolipids are essential for swarming motility, involved in biofilm formation and act as hemolysins (Köhler et al., 2000; Davey et al., 2003; Tremblay et al., 2007). In addition, they enhance the uptake of hydrophobic substrates (Zhang and Miller, 1995; Al-Tahhan et al., 2000; Noordman and Janssen, 2002) and play a role in shielding the producing cells from host defense (McClure and Schiller, 1996; van Gennip et al., 2009; Alhede et al., 2009).
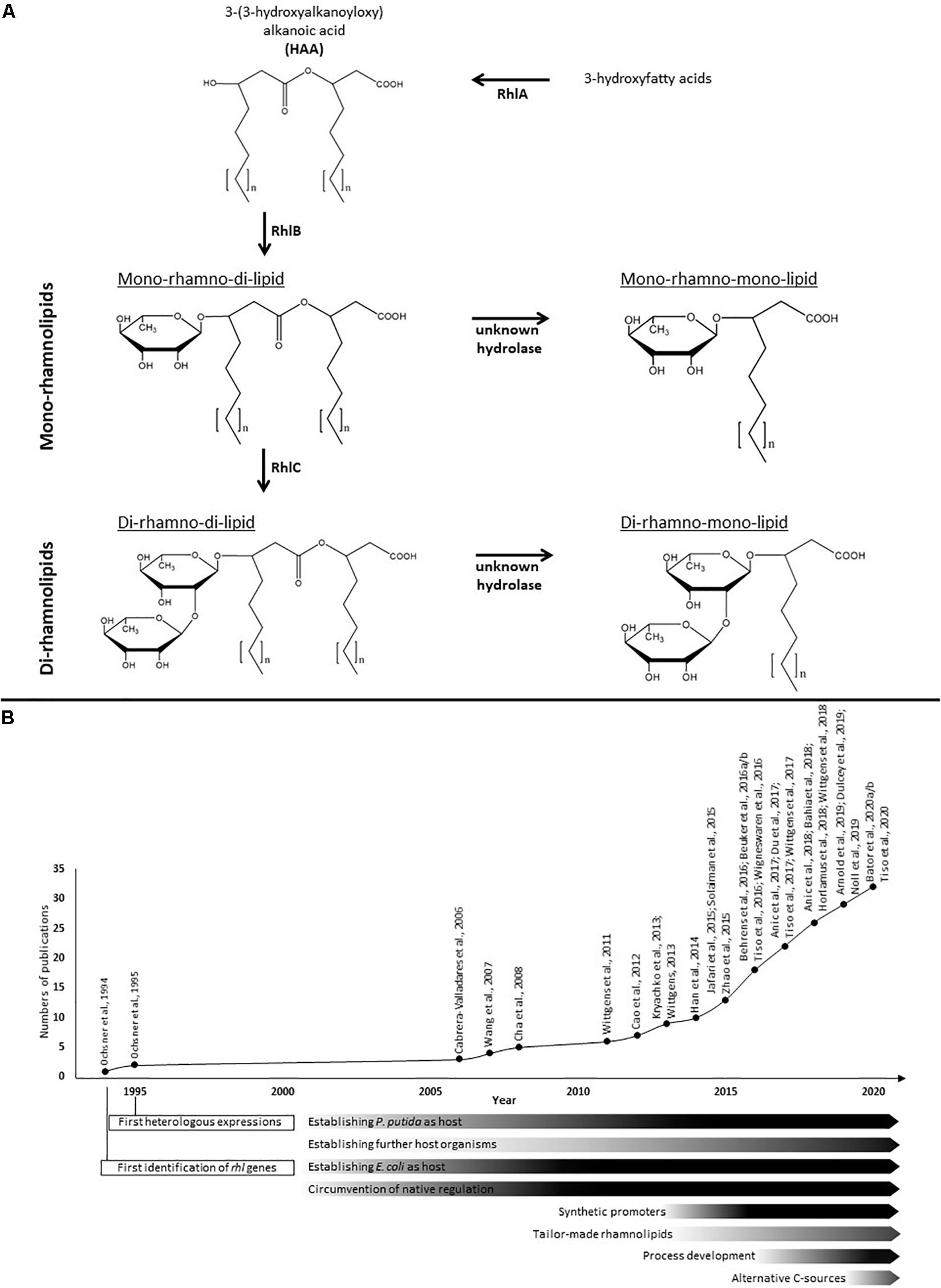
Figure 1. Rhamnolipid biosynthesis and research efforts in heterologous rhamnolipid production. (A) The biosynthesis of rhamnolipids occurs in consecutive enzymatic reactions. The esterification of two 3-hydroxyfatty acids is catalyzed by the acyltransferase RhlA and generates HAAs as rhamnolipid precursors. The rhamnosyltransferase I (RhlB) generates mono-rhamnolipids by adding a dTDP-L-rhamnose to the HAAs. Di-rhamnolipids are synthesized by the rhamnosyltransferase II (RhlC) by linking a second dTDP-L-rhamnose to the mono-rhamnolipids. Both rhamnolipid species can be further processed by hydrolases to create mono-rhamno-mono-lipids and di-rhamno-mono-lipids containing only one fatty acid chain. The fatty acids chain lengths of rhamnolipids typically vary between C8 and C16. (B) Time course of efforts and milestones in the research of heterologous rhamnolipid production.
The biosynthesis of rhamnolipids is initiated by the esterification of two 3-hydroxyfatty acids to form the rhamnolipid precursor molecules 3-(3-hydroxyalkanoyloxy)alcanoic acids (HAAs), which typically form the hydrophobic moiety of the final rhamnolipids. This dimerization is catalyzed by the acyltransferase RhlA (Déziel et al., 2003; Zhu and Rock, 2008). It is controversially discussed, whether the 3-hydroxyfatty acids are descending from the fatty acid de novo synthesis while bound to the acyl carrier protein (Rehm et al., 2001), as it has been described earlier to be the exclusively accepted substrate of RhlA (Zhu and Rock, 2008), or if the β-oxidation is the main provider of 3-hydroxyfatty acids for the HAA and rhamnolipid biosynthesis (Zhang et al., 2012; Abdel-Mawgoud et al., 2014). An HAA molecule together with one molecule of dTDP-L-rhamnose descending from glucose-6-phosphate (Olvera et al., 1999; Rahim et al., 2000) are used as substrates for the biosynthesis of mono-rhamnolipids by the rhamnosyltransferase I (RhlB) (Ochsner et al., 1994; Wittgens et al., 2017). Finally, di-rhamnolipids are synthesized by adding a second dTDP-L-rhamnose molecule to the mono-rhamnolipids by the rhamnosyltransferase II (RhlC) (Rahim et al., 2001). Subsequently, the biosynthesis of mono-rhamno-mono-lipids and di-rhamno-mono-lipids can occur through hydrolysis of the second 3-hydroxyacyl chain probably by two specific but yet unknown α/β-hydrolases (Wittgens et al., 2017). In P. aeruginosa the rhlA and rhlB genes are organized within an operon (Ochsner et al., 1994), while rhlC is part of a second operon encoded together with PA1131, which shares similarities with transport proteins of the major facilitator superfamily (MFS) (Rahim et al., 2001). However, PA1131 appeared to be not involved in rhamnolipid biosynthesis or secretion (Wittgens et al., 2017). In contrast, in Burkholderia species all rhl genes are located within a single gene cluster (Dubeau et al., 2009; Lim et al., 2009).
Apart from the natural rhamnolipid producer organisms an increasing number of different recombinant hosts was generated by introduction of rhl genes. In this review, we will give an update of different approaches for the heterologous production of diverse rhamnolipids from the functional identification of the rhlAB operon in the 1990s to efficient modern production strains. We explain the reasons for this concept as well as the challenges to find a suitable host organism and the current strategies to synthesize tailor-made rhamnolipids. Thereby, we focus on real heterologous expression of foreign genes in recombinant hosts rather than metabolic engineering of wild type rhamnolipid producers.
Heterologous Production of Rhamnolipids—Advantages and Challenges
The overall arguments for a heterologous expression of target genes—in this case the rhl genes responsible for the biosynthesis of rhamnolipids—are numerous. The first implementation was performed to finally proof the responsibility of the rhlAB operon for the mono-rhamnolipid biosynthesis. After the successful identification of the rhlAB operon in studies with P. aeruginosa mutant strains Ochsner et al. (1994) expressed single rhl genes and operons heterologously in Escherichia coli DH5α (Table 1). While the evidence for a rhamnosyltransferase activity using 3-hydroxydecanoyl-3-hydroxydecanoate and TDP-rhamnose as substrate was positive, they could not detect any production of rhamnolipids in E. coli.
A further reason for heterologous expression is to overcome the pathogenicity of most of the native producing organisms, which is disadvantageous for many industrial applications especially in foods and cosmetics (Toribio et al., 2010; Müller and Hausmann, 2011). The best known rhamnolipid producer P. aeruginosa as well as some rhamnolipid producing species of the genus Burkholderia like B. pseudomallei (Häußler et al., 1998, 2003) are human-pathogens, others like B. plantarii and B. glumae are at least plant-pathogens (Manso Pajarron et al., 1993; Andrä et al., 2006; Hörmann et al., 2010; Costa et al., 2011). Therefore, already in 1995 Ochsner et al. established the heterologous expression of rhlAB in four different organisms (Table 1). The operon was either controlled by an inducible tac-promoter or transcriptionally regulated by RhlR/I being part of a recombinant rhlABRI gene cluster. However, a reliable induction of rhlAB especially through the native regulation could not be achieved in all of the hosts, e.g., in E. coli. Nevertheless, the successful production of considerable amounts of mono-rhamnolipids after expression of rhlABRI was demonstrated for Pseudomonas putida (Cha et al., 2008; Cao et al., 2012) and Pseudomonas stutzeri (Zhao et al., 2015) as well (Table 1). This strategy certainly avoided only a part of the complex regulation system for the rhamnolipid biosynthesis. In P. aeruginosa the cell density depending quorum sensing system consisting primarily of LasR/I and RhlR/I as its central components is involved in the transcriptional regulation of both rhl operons (Williams and Cámara, 2009; Reis et al., 2011). Although they seem to be hierarchical organized, both systems are able to induce the expression of several genes including the rhl genes independent from each other (Latifi et al., 1996; Schuster and Greenberg, 2006). Experiments using P. aeruginosa mutant strains revealed a reduction of rhamnolipids of about 55%, when RhlI was absent, but even of almost 80% in the absence of LasI (Pearson et al., 1997), because its synthesized autoinducer bound to LasR is one of at least four regulators, which can induce the rhlR expression (Medina et al., 2003). In contrast, in LasR deficient mutants, the rhamnolipid biosynthesis is delayed, but finally reached concentrations like the wild type (Dekimpe and Déziel, 2009). Aside, further systems are existing being responsible to modulate the quorum sensing response and subsequent the rhamnolipid biosynthesis, e.g., the PQS system as the so-called third quorum sensing system (Pesci et al., 1999), the quorum quenching (Sio et al., 2006), the global regulators RsaL and Vfr (Rampioni et al., 2007, 2009; Croda-García et al., 2011) and further signaling systems (Wilhelm et al., 2007; Rosenau et al., 2010; Henkel et al., 2013), which is often depending on the cultivation conditions (Duan and Surette, 2007). Similar complex regulatory systems were also described for rhamnolipid producing Burkholderia (Nickzad et al., 2015; Nickzad and Déziel, 2016).
Since their regulation is one of the bottlenecks for the high-yield production of rhamnolipids (Toribio et al., 2010; Müller and Hausmann, 2011), it seems to be very challenging to enhance rhamnolipid production also in specific native producers, which are characterized as non-pathogenic like the P. aeruginosa strain ATCC 9027 (Grosso-Becerra et al., 2016) or species like P. chlororaphis (Gunther et al., 2005, 2006), B. thailandensis (Dubeau et al., 2009; Funston et al., 2016; Elshikh et al., 2017) and B. kururiensis (Tavares et al., 2012). Noteworthy, the non-pathogenic P. chlororaphis produce only mono-rhamnolipids and was complemented for di-rhamnolipid biosynthesis by heterologous expression of rhlC (Solaiman et al., 2015). However, almost none of these components exist in E. coli and, therefore, cannot positively affect the expression of rhlR/I or rhlAB. On the other hand, this gene cluster is not repressed by any influence and its successful sole native expression was repeatedly demonstrated (Table 1).
More recent studies focused on complete decoupling of heterologous rhlAB and rhlC expression from the native regulation. Especially for applications in the (microbial) enhanced oil recovery, rhamnolipids were produced in E. coli using the common T7 expression system (Wang et al., 2007; Han et al., 2014; Jafari et al., 2014; Du et al., 2017), while others established a constitutive expression of rhlAB in E. coli (Kryachko et al., 2013). More frequently, P. putida KT2440 wild type or engineered strains were used as heterologous host for rhamnolipid biosynthesis using an inducible tac-promoter or constitutive expressed and partly synthetic promoters (Wittgens et al., 2011, 2017; Wittgens, 2013; Behrens et al., 2016; Beuker et al., 2016a,b; Tiso et al., 2016, 2017, 2020; Anic et al., 2017, 2018; Noll et al., 2019; Table 1). Besides well-known short-chain rhamnolipids from P. aeruginosa also the heterologous production of long chain rhamnolipids was established in this organism by expressing rhlAB and rhlC from B. glumae (Wittgens et al., 2018).
Except typical strategies to enable heterologous expression in a specific host, e.g., codon usage optimization, application of compatible shuttle vectors and promoters, the availability of educts for the rhamnolipid biosynthesis is a challenging bottleneck. Thereby, especially the amount of dTDP-L-rhamnose seems to limit the productivity of rhamnolipids as it was shown in recombinant E. coli strains. Coexpression of the rmlBDAC operon, which converts glucose-1-phosphate into dTDP-L-rhamnose solved this problem and increased the rhamnolipid titers (Cabrera-Valladares et al., 2006).
Other researchers left the domain of bacteria and expressed codon-optimized genes responsible for the biosynthesis of mono-rhamnolipids in Saccharomyces cerevisiae (Bahia et al., 2018). As described for E. coli, they also had to use coexpression of the rmlBDAC operon to enable sufficient amounts of dTDP-L-rhamnose.
Recent studies are more focused on establishing unusual carbon sources or cultivation conditions to make the rhamnolipid biosynthesis more economical, e.g., by using cheap raw materials from waste streams. These studies were based on well-established host organisms like P. putida KT2440 engineered to utilize ethanol, pyrolysis oil or alternative sugars like xylose and arabinose as part of lignocellulosic hydrolysates or from agricultural residues (Arnold et al., 2019; Horlamus et al., 2019; Wang et al., 2019; Bator et al., 2020a,b) or they were cultivated in biofilms to avoid foaming as in conventional bioreactors (Wigneswaran et al., 2016). Moreover, new heterologous hosts for rhamolipid production were exploited, e.g., Cellvibrio japonicus (Horlamus et al., 2018), which can even utilize polymeric substrates like xylan and cellulose (Gardner, 2016), or Pseudomonas stutzeri, which was used to produce rhamnolipids under anaerobic conditions (Zhao et al., 2015; Table 1).
Tailor-Made Rhamnolipids—Future Perspectives
The terms “tailor-made” or “designer rhamnolipids” initially were introduced to claim the possibility to freely choose the numbers of L-rhamnose sugar and 3-hydroxyfatty acid residues in the desired rhamnolipids. Mono-rhamnolipids are exclusively produced by expression of rhlAB, but expression of rhlABC typically results in a mixture of mono- and di-rhamnolipids. Several strategies from enzyme design and pathway modifications to congener enrichment by purification can be used to increase the purity of di-rhmnolipids (Manso Pajarron et al., 1993; Tiso et al., 2017; Wittgens and Rosenau, 2018). Moreover, the identification of the α/β-hydrolase(s), which synthesize mRmL and dRmL using typical mRdL and dRdL as precursors, would open up the possibility to specifically produce these rare rhamnolipid species. This would enlarge the portfolio of different available rhamnolipid species with specific physico-chemical properties for several applications. The so far only other known rhamnolipid modifying enzyme is the naringinase from Aspergillus niger, which removes a single L-rhamnose residue from di- and mono-rhamnolipids and finally generates HAAs (Trummler et al., 2003).
Another purpose for tailor-made rhamnolipids is to define the fatty acid chain lengths for each of the four species and possibly their degree of saturation customized for any type of specific applications. So far most of the publications used rhl genes from P. aeruginosa for the heterologous production of rhamnolipids in different hosts and only a few reports about the successful heterologous expression of rhl genes from Burkholderia (Wittgens et al., 2018; Dulcey et al., 2019). However, it has been described for Pseudomonas desmolyticum to produce rhamnolipids with predominant C6-C8 fatty acids (Jadhav et al., 2011), while other bacteria of the genus Thermus produce much longer rhamnolipids with up to 24 carbon atoms and a predominant C16-C16 congener (Řezanka et al., 2011), but none of them were produced heterologously so far. Germer et al. (2020) identified additional and partly unknown rhlA variants from different organisms by homology search. The heterologous expression of five of these rhlA variants in E. coli resulted in the production of two novel HAA congener compositions different from the typical composition of Pseudomonas and Burkholeria with its predominant C10-C10 and C14-C14 congeners. RhlA from P. fluorescens LMG 05825 produces HAA with 49% of a predominant C10-C14 congener and a slightly less C10-C12 congener of about 38%, while HAAs produced by RhlA from Dickeya dadantii Dd586_2334 contains a C10-C14 congener with 73%. These HAA mixtures could easily processed subsequently into mono- and di-rhamnolipids by additional expressions of appropriate rhlB and rhlC. Especially hybrid operons combining rhl genes from different organisms could give novel insights into the responsibility for defining rhamnolipid chain length. Such an approach using hybrid rhlAB operons from P. aeruginosa and B. glumae verified, that RhlA seems to mainly determine the fatty acid lengths used for the biosynthesis first of HAAs, but subsequent also for rhamnolipids (Wittgens et al., 2018). Similar experiments using the two originating species as hosts for such hybrid operons further indicated an influence of the host organism and its provided 3-hydroxyfatty acids (Dulcey et al., 2019).
A further possibility to achieve novel chain lengths in rhamnolipids is to manipulate the involved enzymes by single amino acid or whole domain exchanges to alter the accepted length of 3-hydroxyfatty acids as it was recently done. Thereby, Dulcey et al. (2019) created a chimeric RhlA variant using different original enzyme domains for RhlA originating form P. aeruginosa and B. glumae. Combined in an operon with rhlB from P. aeruginosa, the newly designed enzyme produced rhamnolipids with a predominant C12-C12 congener when heterologous expressed in B. glumae, which is the average between the typical short-chain rhamnolipids from Pseudomnas and the long-chain rhamnolipids from Burkholderia. In contrast, if the same operon was expressed in P. aeruginosa as host, the amount of C12-C12 congener was only increased in a mixture with a still predominant C10-C10 congener. It was further demonstrated, that mutagenized RhlA reached activities more than twice as much as the wild type RhlA (Dulcey et al., 2019) and amino acid exchanges of RhlB resulted in a shifted pattern of the rhamnolipid composition from C10-C10 to C10-C8 congeners indicating a further specificity of RhlB to specific HAAs containing certain fatty acid length (Han et al., 2014). Further enzyme modifications could result in a much more specific congener enriched composition through improved specificity of responsible enzymes to specific fatty acid chain lengths.
These strategies will help to develop rhamnolipids with their structural diversity as a platform molecule, which harbors an enormous potential to adopt tailor-made properties to meet a huge variety of demands of surfactants for various applications.
Discussion
A specific heterologous host provides a given genetic and metabolic background to investigate the influence of different rhl genes by introducing specific or synthetic biosynthesis pathways for the production of various rhamnolipid species and congeners. Many different recombinant host organisms were established in the past belonging typically and with only a few exceptions to beta- or gamma-proteobacteria like the wild type producers.
A fundamental requirement to any rhamnolipid producing organism is its resistance against high rhamnolipid concentrations. This is most probably the reason, why none of the rhamnolipid producers is a Gram-positive organism, because rhamnolipids possess antimicrobial properties especially against Gram-positives (Abalos et al., 2001; Haba et al., 2003). Obviously, their single cytoplasmic membrane surrounded by the peptidoglycan layer does not represent an effective barrier against the influence of rhamnolipids (Sotirova et al., 2008, 2012). Recombinant P. putida appeared to be a good choice for heterologous rhamnolipid production, since with this organism the highest rhamnolipid titers of about 15 g/L could be achieved (Beuker et al., 2016a), while without using a profound bioprocess strategy titers of only a few hundred mg/L were reported for E. coli (Cabrera-Valladares et al., 2006; Wang et al., 2007; Du et al., 2017). However, none of the heterologous rhamnolipid producers so far reached the at least 40 g/L, which can reproducible be achieved using P. aeruginosa (Müller et al., 2011).
The most critical demand on heterologous rhamolipid producer so far is to provide sufficient amounts of educts. Gram-negative bacteria typically synthesize dTDP-L-rhamnose as they are part of the lipopolysaccharides in the other membrane of these bacteria (Rahim et al., 2000; Poon et al., 2008). However, the amount of this educt appears to limit heterologous rhamnolipid production especially in E. coli (Cabrera-Valladares et al., 2006) and probably other host organisms, but through suitable strategies like the coexpression of the rmlBDAC operon the amount of this educt can be increased.
The introduction of foreign and especially synthetic promoters enabled not only the circumvention of native quorum-sensing dependent regulation of rhamnolipid biosynthesis, but also the possibility to optimize and fine-tune the expression of responsible genes. This strategy will also allow to uncouple additional genes from their native regulation, e.g., responsible for the educt synthesis, which are usually strongly regulated in the wild types (Aguirre-Ramírez et al., 2012), to further improve the rhamnolipid biosynthesis. Metabolic engineering was also successfully used to lower the intrinsic metabolic burden by deleting high energy or resource-demanding side activities, e.g., the biosynthesis of polyhydroxyalkanoate (PHA) and the formation of flagella, which strongly increased the amounts of rhamnolipids (Wittgens et al., 2011; Tiso et al., 2016, 2020). This strain engineering in conjunction with strategies for bioprocess development (feeding strategies, media compositions, downstream processing, etc.) will contribute to lower the production cost for rhamnolipids and make them a more economical alternative in the future.
Moreover, the use of different rhamnolipid producer strains or more preferable the heterologous expression of the responsible genes already allows the production of several rhamnolipid species and congeners. Furthermore, real tailor-made rhamnolipids will probably become available soon through the identification and expression of novel rhamnolipid processing enzymes or enzyme modifications leading to a considerable enlargement of the portfolio of diverse rhamnolipids with the possibility to freely choose the chemical entities they consist of. This can assign a decisive role to rhamnolipids not only as the first class of true “designer” biosurfactant per se, but also as the first group of biosurfactants that can be produced exclusively in biotechnological processes at the same time.
Author Contributions
AW and FR created figures and drafted the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by grants from the Ministry of Science, Research and the Arts of Baden-Württemberg (MWK; Förderkennzeichen: 7533-10-5-186A and 7533-10-5-190). Furthermore, the authors are grateful to the EU project Horizon 2020 AD GUT (ID: 686271) for providing financial support.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Abalos, A., Pinazo, A., Infante, M. R., Casals, M., García, F., and Manresa, A. (2001). Physicochemical and antimicrobial properties of new rhamnolipids produced by Pseudomonas aeruginosa AT10 from soybean oil refinery wastes. Langmuir 17, 1367–1371. doi: 10.1021/la0011735
Abdel-Mawgoud, A. M., Lépine, F., and Déziel, E. (2010). Rhamnolipids: diversity of structures, microbial origins and roles. Appl. Microbiol. Biotechnol. 86, 1323–1336. doi: 10.1007/s00253-010-2498-2
Abdel-Mawgoud, A. M., Lépine, F., and Déziel, E. (2014). A stereospecific pathway diverts β-oxidation intermediates to the biosynthesis of rhamnolipid biosurfactants. Chem. Biol. 21, 156–164. doi: 10.1016/j.chembiol.2013.11.010
Aguirre-Ramírez, M., Medina, G., González-Valdez, A., Grosso-Becerra, V., and Soberón-Chávez, G. (2012). The Pseudomonas aeruginosa rmlBDAC operon, encoding dTDP-L-rhamnose biosynthetic enzymes, is regulated by the quorum-sensing transcriptional regulator RhlR and the alternative sigma factor σS. Microbiology 158, 908–916. doi: 10.1099/mic.0.054726-0
Alhede, M., Bjarnsholt, T., Jensen, P. Ø, Phipps, R. K., Moser, C., Christophersen, L., et al. (2009). Pseudomonas aeruginosa recognizes and responds aggressively to the presence of polymorphonuclear leukocytes. Microbiology 155, 3500–3508. doi: 10.1099/mic.0.031443-0
Al-Tahhan, R. A., Sandrin, T. R., Bodour, A. A., and Maier, R. M. (2000). Rhamnolipid-induced removal of lipopolysaccharide from Pseudomonas aeruginosa: effect on cell surface properties and interaction with hydrophobic substrates. Appl. Environ. Microbiol. 66, 3262–3268. doi: 10.1128/AEM.66.8.3262-3268.2000
Andrä, J., Rademann, J., Howe, J., Koch, M. H. J., Heine, H., Zähringer, U., et al. (2006). Endotoxin-like properties of a rhamnolipid exotoxin from Burkholderia (Pseudomonas) plantarii: immune cell stimulation and biophysical characterization. Biol. Chem. 387, 301–310. doi: 10.1515/BC.2006.040
Anic, I., Apolonia, I., Franco, P., and Wichmann, R. (2018). Production of rhamnolipids by integrated foam adsorption in a bioreactor system. AMB Express 8:122. doi: 10.1186/s13568-018-0651-y
Anic, I., Nath, A., Franco, P., and Wichmann, R. (2017). Foam adsorption as an ex situ capture step for surfactants produced by fermentation. J. Biotechnol. 258, 181–189. doi: 10.1016/j.jbiotec.2017.07.015
Arnold, S., Henkel, M., Wanger, J., Wittgens, A., Rosenau, F., and Hausmann, R. (2019). Heterologous rhamnolipid biosynthesis by P. putida KT2440 on bio-oil derived small organic acids and fractions. AMB Express 9:80. doi: 10.1186/s13568-019-0804-7
Bahia, F. M., de Almeida, G. C., de Andrade, L. P., Campos, C. G., Queiroz, L. R., da Silva, R. L. V., et al. (2018). Rhamnolipids production from sucrose by engineered Saccharomyces cerevisiae. Sci. Rep. 8:2905. doi: 10.1038/s41598-018-21230-2
Banat, I., Franzetti, A., Gandolfi, I., Bestetti, G., Martinotti, M., Fracchia, L., et al. (2010). Microbial biosurfactants production, applications and future potential. Appl. Microbiol. Biotechnol. 87, 427–444. doi: 10.1007/s00253-010-2589-0
Bator, I., Karmainski, T., Tiso, T., and Blank, L. M. (2020a). Killing two birds with one stone – strain engineering facilitates the development of a unique rhamnolipid production process. Front. Bioeng. Biotechnol. 8:899. doi: 10.3389/fbioe.2020.00899
Bator, I., Wittgens, A., Rosenau, F., Tiso, T., and Blank, L. M. (2020b). Comparison of three xylose pathways in Pseudomonas putida KT2440 for the synthesis of valuable products. Front. Bioeng. Biotechnol. 7:480. doi: 10.3389/fbioe.2019.00480
Behrens, B., Engelen, J., Tiso, T., Blank, L. M., and Hayen, H. (2016). Characterization of rhamnolipids by liquid chromatography/mass spectrometry after solid-phase extraction. Anal. Bioanal. Chem. 408, 2505–2514. doi: 10.1007/s00216-016-9353-y
Beuker, J., Barth, T., Steier, A., Wittgens, A., Rosenau, F., Henkel, M., et al. (2016a). High titer heterologous rhamnolipid production. AMB Express 6:124. doi: 10.1186/s13568-016-0298-5
Beuker, J., Steier, A., Wittgens, A., Rosenau, F., Henkel, M., and Hausmann, R. (2016b). Integrated foam fractionation for heterologous rhamnolipid production with recombinant Pseudomonas putida in a bioreactor. AMB Express 6:11. doi: 10.1186/s13568-016-0183-2
Cabrera-Valladares, N., Richardson, A. P., Olvera, C., Treviño, L. G., Déziel, E., Lépine, F., et al. (2006). Monorhamnolipids and 3-(3-hydroxyalkanoyloxy)alkanoic acids (HAAs) production using Escherichia coli as a heterologous host. Appl. Microbiol. Biotechnol. 73, 187–194. doi: 10.1007/s00253-006-0468-5
Cao, L., Wang, Q., Zhang, J., Li, C., Yan, X., Lou, X., et al. (2012). Construction of a stable genetically engineered rhamnolipid-producing microorganism for remediation of pyrene-contaminated soil. World. J. Microbiol. Biotechnol. 28, 2783–2790. doi: 10.1007/s11274-012-1088-0
Cha, M., Lee, N., Kim, M., Kim, M., and Lee, S. (2008). Heterologous production of Pseudomonas aeruginosa EMS1 biosurfactant in Pseudomonas putida. Bioresour. Technol. 99, 2192–2199. doi: 10.1016/j.biortech.2007.05.035
Costa, S. G. V. A. O., Déziel, E., and Lépine, F. (2011). Characterization of rhamnolipid production by Burkholderia glumae. Lett. Appl. Microbiol. 53, 620–627. doi: 10.1111/j.1472-765X.2011.03154.x
Croda-García, G., Grosso-Becerra, V., Gonzalez-Valdez, A., Servín-González, L., and Soberón-Chávez, G. (2011). Transcriptional regulation of Pseudomonas aeruginosa rhlR: role of the CRP orthologue Vfr (virulence factor regulator) and quorum-sensing regulators LasR and RhlR. Microbiology 157, 2545–2555. doi: 10.1099/mic.0.050161-0
Davey, M. E., Caiazza, N. C., and O’Toole, G. A. (2003). Rhamnolipid surfactant production affects biofilm architecture in Pseudomonas aeruginosa PAO1. J. Bacteriol. 185, 1027–1036. doi: 10.1128/JB.185.3.1027-1036.2003
Dekimpe, V., and Déziel, E. (2009). Revisiting the quorum-sensing hierarchy in Pseudomonas aeruginosa: the transcriptional regulator RhlR regulates LasR specific factors. Microbiology 155, 712–723. doi: 10.1099/mic.0.022764-0
Desai, J. D., and Banat, I. M. (1997). Microbial production of surfactants and their commercial potential. Microbiol. Mol. Biol. Rev. 61, 47–64. doi: 10.1128/.61.1.47-64.1997
Déziel, E., Lépine, F., Dennie, D., Boismenu, D., Mamer, O. A., and Villemur, R. (1999). Liquid chromatography/mass spectrometry analysis of mixtures of rhamnolipids produced by Pseudomonas aeruginosa strain 57RP grown on mannitol or naphthalene. Biochim. Biophys. Acta. 1440, 244–252. doi: 10.1016/S1388-1981(99)00129-8
Déziel, E., Lépine, F., Milot, S., and Villemur, R. (2003). rhlA is required for the production of a novel biosurfactant promoting swarming motility in Pseudomonas aeruginosa: 3-(3-hydroxyalkanoyloxy)alkanoic acids (HAAs), the precursors of rhamnolipids. Microbiology 149, 2005–2013. doi: 10.1099/mic.0.26154-0
Du, J., Zhang, A., Hao, J., and Wang, J. (2017). Biosynthesis of di-rhamnolipids and variations of congeners composition in genetically-engineered Escherichia coli. Biotechnol. Lett. 39, 1041–1048. doi: 10.1007/s10529-017-2333-2
Duan, K., and Surette, M. G. (2007). Environmental regulation of Pseudomonas aeruginosa PAO1 Las and Rhl quorum-sensing systems. J. Bacteriol. 189, 4827–4836. doi: 10.1128/JB.00043-07
Dubeau, D., Déziel, E., Woods, D. E., and Lépine, F. (2009). Burkholderia thailandensis harbors two identical rhl gene clusters responsible for the biosynthesis of rhamnolipids. BMC Microbiol. 9:263. doi: 10.1186/1471-2180-9-263
Dulcey, C. E., López de Los Santos, Y., Létourneau, M., Déziel, E., and Doucet, N. (2019). Semi-rational evolution of the 3-(3-hydroxyalkanoyloxy)alkanoate (HAA) synthase RhlA to improve rhamnolipid production in Pseudomonas aeruginosa and Burkholderia glumae. FEBS J. 286, 4036–4059. doi: 10.1111/febs.14954
Elshikh, M., Funston, S., Ahmed, S., Marchant, R., and Banat, I. B. (2017). Rhamnolipids from non-pathogenic Burkholderia thailandensis E264: physicochemical characterization, antimicrobial and antibiofilm efficacy against oral hygiene related pathogens. N. Biotechnol. 36, 26–36. doi: 10.1016/j.nbt.2016.12.009
Funston, S. J., Tsaousi, K., Rudden, M., Smyth, T. J., Stevenson, P. S., Marchant, R., et al. (2016). Characterising rhamnolipid production in Burkholderia thailandensis E264, a non-pathogenic producer. Appl. Microbiol. Biotechnol. 100, 7945–7956. doi: 10.1007/s00253-016-7564-y
Gardner, J. G. (2016). Polysaccharide degradation systems of the saprophytic bacterium Cellvibrio japonicus. World J. Microbiol. Biotechnol. 32:121. doi: 10.1007/s11274-016-2068-6
Germer, A., Tiso, T., Müller, C., Behrens, B., Vosse, C., Scholz, K., et al. (2020). Exploiting the natural diversity of RhlA acyltransferases for the synthesis of the rhamnolipid precursor 3-(3-Hydroxyalkanoyloxy)alkanoic acid. Appl. Environ. Microbiol. 86:e002317-19. doi: 10.1128/AEM.02317-19
Giani, C., Wullbrandt, D., Rothert, R., and Meiwes, J. (1997). Pseudomonas Aeruginosa and Its Use in a Process for the Biotechnological Preparation of L-Rhamnose. US005658793A. Frankfurt: Hoechst AG.
Grosso-Becerra, M. V., González-Valdez, A., Granados-Martínez, M. J., Morales, E., Servín-González, L., Méndez, J. L., et al. (2016). Pseudomonas aeruginosa ATCC 9027 is a non-virulent strain suitable for mono-rhamnolipids production. Appl. Microbiol. Biotechnol. 100, 9995–10004. doi: 10.1007/s00253-016-7789-9
Gunther, N. W. IV, Nuñez, A., Fett, W., and Solaiman, D. K. Y. (2005). Production of rhamnolipids by Pseudomonas chlororaphis, a nonpathogenic bacterium. Appl. Environ. Microbiol. 71, 2288–2293. doi: 10.1128/AEM.71.5.2288-2293.2005
Gunther, N. W. IV, Nuñez, A., Fortis, L., and Solaiman, D. K. Y. (2006). Proteomic based investigation of rhamnolipid production by Pseudomonas chlororaphis strain NRRL B-30761. J. Ind. Microbiol. Biotech. 33, 914–920. doi: 10.1007/s10295-006-0169-1
Haba, E., Pinazo, A., Jauregui, O., Espuny, M. J., Infante, M. R., and Manresa, A. (2003). Physicochemical characterization and antimicrobial properties of rhamnolipids produced by Pseudomonas aeruginosa 47T2 NCBIM 40044. Biotechnol. Bioeng. 81, 316–322. doi: 10.1002/bit.10474
Han, L., Liu, P., Peng, Y., Lin, J., Wang, Q., and Ma, Y. (2014). Engineering the biosynthesis of novel rhamnolipids in Escherichia coli for enhanced oil recovery. J. Appl. Microbiol. 117, 139–150. doi: 10.1111/jam.12515
Häußler, S., Nimtz, M., Domke, T., Wray, V., and Steinmetz, I. (1998). Purification and characterization of a cytotoxic exolipid of Burkholderia pseudomallei. Infect. Immun. 66, 1588–1593. doi: 10.1128/IAI.66.4.1588-1593.1998
Häußler, S., Rohde, M., von Neuhoff, N., Nimtz, M., and Steinmetz, I. (2003). Structural and functional cellular changes induced by Burkholderia pseudomallei rhamnolipid. Infect. Immun. 71, 2970–2975. doi: 10.1128/IAI.71.5.2970-2975.2003
Henkel, M., Schmidberger, A., Kühnert, C., Beuker, J., Bernard, T., Schwartz, T., et al. (2013). Kinetic modeling of the time course of N-butyryl-homoserine lactone concentration during batch cultivations of Pseudomonas aeruginosa PAO1. Appl. Microbiol. Biotechnol. 97, 7607–7616. doi: 10.1007/s00253-013-5024-5
Hirata, Y., Ryu, M., Oda, Y., Igarashi, K., Nagatsuka, A., Furuta, T., et al. (2009). Novel characteristics of sophorolipids, yeast glycolipid biosurfactants, as biodegradable low-foaming surfactants. J. Biosci. Bioeng. 108, 142–146. doi: 10.1016/j.jbiosc.2009.03.012
Horlamus, F., Wang, Y., Steinbach, D., Vahidinasab, M., Wittgens, A., Rosenau, F., et al. (2019). Potential of biotechnological conversion of lignocellulose hydrolyzates by Pseudomonas putida KT2440 as a model organism for a bio-based economy. Glob. Change Biol. Bioenergy 11:12. doi: 10.1111/gcbb.12647
Horlamus, F., Wittgens, A., Noll, P., Michler, J., Müller, I., Weggenmann, F., et al. (2018). One-step bioconversion of hemicellulose polymers to rhamnolipids with Cellvibrio japonicus: a proof-of-concept for a potential host strain in future bioeconomy. Glob. Change Biol. Bioenergy 11:1. doi: 10.1111/gcbb.12542
Hörmann, B., Müller, M. M., Syldatk, C., and Hausmann, R. (2010). Rhamnolipid production by Burkholderia plantarii DSM 9509T. Eur. J. Lipid Sci. Tech. 112, 674–680. doi: 10.1002/ejlt.201000030
Jadhav, M., Kalme, S., Tamboli, D., and Govindwar, S. (2011). Rhamnolipid from Pseudomonas desmolyticum NCIM-2112 and its role in the degradation of Brown 3REL. J. Basic Microbiol. 51, 385–396. doi: 10.1002/jobm.201000364
Jafari, A., Raheb, J., Bardania, H., and Rasekh, B. (2014). Isolation, cloning and expression of rhamnolipid operon from Pseudomonas aeroginosa ATCC 9027 in logarithmic phase in E. coli BL21. Am. J. Life Sci. 2, 22–30.
Jarvis, F. G., and Johnson, M. J. (1949). A glycolipide produced by Pseudomonas aeruginosa. J. Am. Chem. Soc. 71, 4124–4126. doi: 10.1021/ja01180a073
Johann, S., Seiler, T. B., Tiso, T., Bluhm, K., Blank, L. M., and Hollert, H. (2016). Mechanism-specific and whole-organism ecotoxicity of mono-rhamnolipids. Sci. Total Environ. 548-549, 155–163. doi: 10.1016/j.scitotenv.2016.01.066
Köhler, T., Curty, L. K., Barja, F., van Delden, C., and Pechère, J. C. (2000). Swarming of Pseudomonas aeruginosa is dependent on cell-to-cell signaling and requires flagella and pili. J. Bacteriol. 182, 5990–5996. doi: 10.1128/JB.182.21.5990-5996.2000
Kryachko, Y., Nathoo, S., Lai, P., Voordouw, J., Prenner, E. J., and Voordouw, G. (2013). Prospects for using native and recombinant rhamnolipid producers for microbially enhanced oil recovery. Int. Biodeter. Biodegr. 81, 133–140. doi: 10.1016/j.ibiod.2012.09.012
Lang, S., and Wullbrandt, D. (1999). Rhamnose lipids – biosynthesis, microbial production and application potential. Appl. Microbiol. Biotechnol. 51, 22–32. doi: 10.1007/s002530051358
Latifi, A., Foglino, M., Tanaka, K., Williams, P., and Lazdunski, A. (1996). A hierarchical quorum sensing cascade in Pseudomonas aeruginosa links the transcriptional activators LasR and RhIR (VsmR) to expression of the stationary-phase sigma factor RpoS. Mol. Microbiol. 21, 1137–1146. doi: 10.1046/j.1365-2958.1996.00063.x
Lim, J. Y., Lee, T.-H., Nahm, B. H., Choi, Y. D., Kim, M., and Hwang, I. (2009). Complete genome sequence of Burkholderia glumae BGR1. J. Bacteriol. 191, 3758–3759. doi: 10.1128/JB.00349-09
Lima, T. M. S., Procópio, L. C., Brandão, F. D., Carvalho, A. M. X., Tótola, M. R., and Borges, A. C. (2011). Biodegradability of bacterial surfactants. Biodegradation 22, 585–592. doi: 10.1007/s10532-010-9431-3
Liu, G., Zhong, H., Yang, X., Liu, Y., Shao, B., and Liu, Z. (2018). Advances in applications of rhamnolipids biosurfactant in environmental remediation: a review. Biotechnol. Bioeng. 115, 796–814. doi: 10.1002/bit.26517
Maier, R. M., and Soberón-Chávez, G. (2000). Pseudomonas aeruginosa rhamnolipids: biosynthesis and potential applications. Appl. Microbiol. Biotechnol. 54, 625–633. doi: 10.1007/s002530000443
Manso Pajarron, A., de Koster, C. G., Heerma, W., Schmidt, M., and Haverkamp, J. (1993). Structure identification of natural rhamnolipid mixtures by fast atom bombardment tandem mass spectrometry. Glycoconj. J. 10, 219–226. doi: 10.1007/BF00702203
McClure, C. D., and Schiller, N. L. (1996). Inhibition of macrophage phagocytosis by Pseudomonas aeruginosa rhamnolipids in vitro and in vivo. Curr. Microbiol. 33, 109–117. doi: 10.1007/s002849900084
Medina, G., Juarez, K., Diaz, R., and Soberón-Chávez, G. (2003). Transcriptional regulation of Pseudomonas aeruginosa rhlR, encoding a quorum-sensing regulatory protein. Microbiology 149, 3073–3081. doi: 10.1099/mic.0.26282-0
Müller, M. M., and Hausmann, R. (2011). Regulatory and metabolic network of rhamnolipid biosynthesis: traditional and advanced engineering towards biotechnological production. Appl. Microbiol. Biotechnol. 91, 251–264. doi: 10.1007/s00253-011-3368-2
Müller, M. M., Hörmann, B., Kugel, M., Syldatk, C., and Hausmann, R. (2011). Evaluation of rhamnolipid production capacity of Pseudomonas aeruginosa PAO1 in comparison to the rhamnolipid over-producer strains DSM 7108 and DSM 2874. Appl. Microbiol. Biotechnol. 89, 585–592. doi: 10.1007/s00253-010-2901-z
Müller, M. M., Hörmann, B., Syldatk, C., and Hausmann, R. (2010a). Pseudomonas aeruginosa PAO1 as a model for rhamnolipid production in bioreactor systems. Appl. Microbiol. Biotechnol. 87, 167–174. doi: 10.1007/s00253-010-2513-7
Müller, M. M., Kügler, J. H., Henkel, M., Gerlitzki, M., Hörmann, B., Pöhnlein, M., et al. (2010b). Rhamnolipids - next generation surfactants? J. Biotechnol. 162, 366–380. doi: 10.1016/j.jbiotec.2012.05.022
Nguyen, T. T., and Sabatini, D. A. (2011). Characterization and emulsification properties of rhamnolipid and sophorolipid biosurfactants and their applications. Int. J. Mol. Sci. 12, 1232–1244. doi: 10.3390/ijms12021232
Nguyen, T. T., Youssef, N. H., McInerney, M. J., and Sabatini, D. A. (2008). Rhamnolipid biosurfactant mixtures for environmental remediation. Water Res. 42, 1735–1743. doi: 10.1016/j.watres.2007.10.038
Nickzad, A., and Déziel, E. (2016). Adaptive significance of quorum sensing-dependent regulation of rhamnolipids by integration of growth rate in burkholderia glumae: a trade-off between survival and efficiency. Front. Microbiol. 7:1215. doi: 10.3389/fmicb.2016.01215
Nickzad, A., Lépine, F., and Déziel, E. (2015). Quorum sensing controls swarming motility of Burkholderia glumae through regulation of rhamnolipids. PLoS One 10:e0128509. doi: 10.1371/journal.pone.0128509
Nitschke, M., and Costa, S. G. V. A. O. (2007). Biosurfactants in food industry. Trends Food Sci. Technol. 18, 252–259. doi: 10.1016/j.tifs.2007.01.002
Nitschke, M., and Pastore, G. M. (2006). Production and properties of a surfactant obtained from Bacillus subtilis grown on cassava wastewater. Bioresour. Technol. 97, 336–341. doi: 10.1016/j.biortech.2005.02.044
Noll, P., Treinen, C., Müller, S., Senkalla, S., Lilge, L., Hausmann, R., et al. (2019). Evaluating temperature-induced regulation of a ROSE-like RNA-thermometer for heterologous rhamnolipid production in Pseudomonas putida KT2440. AMB Express 9:154. doi: 10.1186/s13568-019-0883-5
Noordman, W. H., and Janssen, D. B. (2002). Rhamnolipid stimulates uptake of hydrophobic compounds by Pseudomonas aeruginosa. Appl. Environ. Microbiol. 68, 4502–4508. doi: 10.1128/AEM.68.9.4502-4508.2002
Ochsner, U. A., Fiechter, A., and Reiser, J. (1994). Isolation, characterization, and expression in Escherichia coli of the Pseudomonas aeruginosa rhlAB genes encoding a rhamnosyltransferase involved in rhamnolipid biosurfactant synthesis. J. Biol. Chem. 269, 19787–19795.
Ochsner, U. A., Reiser, J., Fiechter, A., and Witholt, B. (1995). Production of Pseudomonas aeruginosa rhamnolipid biosurfactants in heterologous hosts. Appl. Environ. Microbiol. 61, 3503–3506. doi: 10.1128/AEM.61.9.3503-3506.1995
Olvera, C., Goldberg, J. B., Sánchez, R., and Soberón-Chávez, G. (1999). The Pseudomonas aeruginosa algC gene product participates in rhamnolipid biosynthesis. FEMS Microbiol. Lett. 179, 85–90. doi: 10.1111/j.1574-6968.1999.tb08712.x
Pearson, J. P., Pesci, E. C., and Iglewski, B. H. (1997). Roles of Pseudomonas aeruginosa las and rhl quorum-sensing systems in control of elastase and rhamnolipid biosynthesis genes. J. Bacteriol. 179, 5756–5767. doi: 10.1128/JB.179.18.5756-5767.1997
Pesci, E. C., Milbank, J. B. J., Pearson, J. P., McKnight, S., Kende, A. S., Greenberg, E. P., et al. (1999). Quinolone signaling in the cell-to-cell communication system of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U.S.A. 96, 11229–11234. doi: 10.1073/pnas.96.20.11229
Poon, K. K., Westman, E. L., Vinogradov, E., Jin, S., and Lam, J. S. (2008). Functional characterization of MigA and WapR: putative rhamnosyltransferases involved in outer core oligosaccharide biosynthesis of Pseudomonas aeruginosa. J. Bacteriol. 190, 1857–1865. doi: 10.1128/JB.01546-07
Rahim, R., Burrows, L. L., Monteiro, M. A., Perry, M. B., and Lam, J. S. (2000). Involvement of the rml locus in core oligosaccharide and O polysaccharide assembly in Pseudomonas aeruginosa. Microbiology 146, 2803–2814. doi: 10.1099/00221287-146-11-2803
Rahim, R., Ochsner, U. A., Olvera, C., Graninger, M., Messner, P., Lam, J. S., et al. (2001). Cloning and functional characterization of the Pseudomonas aeruginosa rhlC gene that encodes rhamnosyltransferase 2, an enzyme responsible for di-rhamnolipid biosynthesis. Mol. Microbiol. 40, 708–718. doi: 10.1046/j.1365-2958.2001.02420.x
Rampioni, G., Schuster, M., Greenberg, E. P., Bertani, I., Grasso, M., Venturi, V., et al. (2007). RsaL provides quorum sensing homeostasis and functions as a global regulator of gene expression in Pseudomonas aeruginosa. Mol. Microbiol. 66, 1557–1565. doi: 10.1111/j.1365-2958.2007.06029.x
Rampioni, G., Schuster, M., Greenberg, E. P., Zennaro, E., and Leoni, L. (2009). Contribution of the RsaL global regulator to Pseudomonas aeruginosa virulence and biofilm formation. FEMS Microbiol. Lett. 301, 210–217. doi: 10.1111/j.1574-6968.2009.01817.x
Rehm, B. H., Mitsky, T. A., and Steinbüchel, A. (2001). Role of fatty acid de novo biosynthesis in polyhydroxyalkanoic acid (PHA) and rhamnolipid synthesis by pseudomonads: establishment of the transacylase (PhaG)-mediated pathway for PHA biosynthesis in Escherichia coli. Appl. Environ. Microbiol. 67, 3102–3109. doi: 10.1128/AEM.67.7.3102-3109.2001
Reis, R. S., Pereira, A. G., Neves, B. C., and Freire, D. M. G. (2011). Gene regulation of rhamnolipid production in Pseudomonas aeruginosa – A review. Bioresour. Technol. 102, 6377–6384. doi: 10.1016/j.biortech.2011.03.074
Řezanka, T., Siristova, L., and Sigler, K. (2011). Rhamnolipid-producing thermophilic bacteria of species thermus and Meiothermus. Extremophiles 15, 697–709. doi: 10.1007/s00792-011-0400-5
Rosenau, F., Isenhardt, S., Gdynia, A., Tielker, D., Schmidt, E., Tielen, P., et al. (2010). Lipase LipC affects motility, biofilm formation and rhamnolipid production in Pseudomonas aeruginosa. FEMS Microbiol. Lett. 309, 25–34. doi: 10.1111/j.1574-6968.2010.02017.x
Schuster, M., and Greenberg, E. P. (2006). A network of networks: quorum-sensing gene regulation in Pseudomonas aeruginosa. Int. J. Med. Microbiol. 296, 73–81. doi: 10.1016/j.ijmm.2006.01.036
Sharma, R., Singh, J., and Verma, N. (2018). Optimization of rhamnolipid production from Pseudomonas aeruginosa PBS towards application for microbial enhanced oil recovery. 3 Biotech 8:20. doi: 10.1007/s13205-017-1022-0
Sio, C. F., Otten, L. G., Cool, R. H., Diggle, S. P., Braun, P. G., Bos, R., et al. (2006). Quorum quenching by an N-acyl-homoserine lactone acylase from Pseudomonas aeruginosa PAO1. Infect. Immun. 74, 1673–1682. doi: 10.1128/IAI.74.3.1673-1682.2006
Solaiman, D. K. Y., Ashby, R. D., Gunther, N. W. IV, and Zerkowski, J. A. (2015). Dirhamnose-lipid production by recombinant nonpathogenic bacterium Pseudomonas chlororaphis. Appl. Microbiol. Biotechnol. 99, 4333–4342. doi: 10.1007/s00253-015-6433-4
Sotirova, A., Avramova, T., Stoitsova, S., Lazarkevich, I., Lubenets, V., Karpenko, E., et al. (2012). The importance of rhamnolipid-biosurfactant-induced changes in bacterial membrane lipids of Bacillus subtilis for the antimicrobial activity of thiosulfonates. Curr. Micrbiol. 65, 534–541. doi: 10.1007/s00284-012-0191-7
Sotirova, A. V., Spasova, D. I., Galabova, D. N., Karpenko, E., and Shulga, A. (2008). Rhamnolipid-biosurfactant permeabilizing effects on Gram-positive and Gram-negative bacterial strains. Curr. Microbiol. 56, 639–644. doi: 10.1007/s00284-008-9139-3
Syldatk, C., Lang, S., Matulovic, U., and Wagner, F. (1985a). Production of four interfacial active rhamnolipids from n-alkanes or glycerol by resting cells of Pseudomonas species DSM 2874. Z. Naturforsch. C 40, 61–67. doi: 10.1515/znc-1985-1-213
Syldatk, C., Lang, S., Wagner, F., Wray, V., and Witte, L. (1985b). Chemical and physical characteritation of four interfacial-active rhamnolipids from Pseudomonas spec. DSM 2874 grown on n-alkanes. Z. Naturforsch. C 40, 51–60. doi: 10.1515/znc-1985-1-212
Tavares, L. F. D., Silva, P. M., Junqueira, M., Mariano, D. C. O., Nogueira, F. C. S., Domont, G. B., et al. (2012). Characterization of rhamnolipids produced by wild-type and engineered Burkholderia kururiensis. Appl. Microbiol. Biotechnol. 97, 1909–1921. doi: 10.1007/s00253-012-4454-9
Tiso, T., Ihling, N., Kubicki, S., Biselli, A., Schonhoff, A., Bator, I., et al. (2020). Integration of genetic and process engineering for optimized rhamnolipid production using Pseudomonas putida. Front. Bioeng. Biotechnol. 8:976. doi: 10.3389/fbioe.2020.00976
Tiso, T., Sabelhaus, A., Behrens, B., Wittgens, A., Rosenau, F., Hayen, H., et al. (2016). Creating metabolic demand as an engineering strategy in Pseudomonas putida – Rhamnolipid synthesis as an example. Metab. Eng. Commun. 3, 234–244. doi: 10.1016/j.meteno.2016.08.002
Tiso, T., Zauter, R., Tulke, H., Leuchtle, B., Li, W. J., Behrens, B., et al. (2017). Designer rhamnolipids by reduction of congener diversity: production and characterization. Microb. Cell Fact. 16:225. doi: 10.1186/s12934-017-0838-y
Toribio, J., Escalante, A. E., and Soberón-Chávez, G. (2010). Rhamnolipids: production in bacteria other than Pseudomonas aeruginosa. Eur. J. Lipid Sci. Technol. 112, 1082–1087. doi: 10.1002/ejlt.200900256
Tremblay, J., Richardson, A. P., Lépine, F., and Déziel, E. (2007). Self-produced extracellular stimuli modulate the Pseudomonas aeruginosa swarming motility behaviour. Environ. Microbiol. 9, 2622–2630. doi: 10.1111/j.1462-2920.2007.01396.x
Trummler, K., Effenberger, F., and Syldatk, C. (2003). An integrated microbial/enzymatic process for production of rhamnolipids and L-(+)-rhamnose from rapeseed oil with Pseudomonas sp. DSM 2874. Eur. J. Lipid Sci. Technol. 105, 563–571. doi: 10.1002/ejlt.200300816
van Gennip, M., Christensen, L. D., Alhede, M., Phipps, R., Jensen, P. Ř, Christophersen, L., et al. (2009). Inactivation of the rhlA gene in Pseudomonas aeruginosa prevents rhamnolipid production, disabling the protection against polymorphonuclear leukocytes. APMIS 117, 537–546. doi: 10.1111/j.1600-0463.2009.02466.x
van Hamme, J. D., Singh, A., and Ward, O. P. (2006). Physiological aspects. Part 1 in a series of papers devoted to surfactants in microbiology and biotechnology. Biotechnol. Adv. 24, 604–620. doi: 10.1016/j.biotechadv.2006.08.001
Wang, Q., Fang, X., Bai, B., Liang, X., Shuler, P. J., and Goddard, W. A. III, et al (2007). Engineering bacteria for production of rhamnolipid as an agent for enhanced oil recovery. Biotechnol. Bioeng. 98, 842–853. doi: 10.1002/bit.21462
Wang, Y., Horlamus, F., Henkel, M., Kovacic, F., Schläfle, S., Hausmann, R., et al. (2019). Growth of engineered Pseudomonas putida KT2440 on glucose, xylose, and arabinose: hemicellulose hydrolysates and their major sugars as sustainable carbon sources. Glob. Change Biol. Bioenergy 11, 249–259. doi: 10.1111/gcbb.12590
Wigneswaran, V., Nielsen, K. F., Sternberg, C., Jensen, P. R., Folkesson, A., and Jelsbak, L. (2016). Biofilm as a production platform for heterologous production of rhamnolipids by the non-pathogenic strain Pseudomonas putida KT2440. Microb. Cell Fact. 15:181. doi: 10.1186/s12934-016-0581-9
Wilhelm, S., Gdynia, A., Tielen, P., Rosenau, F., and Jaeger, K. E. (2007). The autotransporter esterase EstA of Pseudomonas aeruginosa is required for rhamnolipid production, cell motility, and biofilm formation. J. Bacteriol. 189, 6695–6703. doi: 10.1128/JB.00023-07
Williams, P., and Cámara, M. (2009). Quorum sensing and environmental adaptation in Pseudomonas aeruginosa: a tale of regulatory networks and multifunctional signal molecules. Curr. Opin. Microbiol. 12, 182–191. doi: 10.1016/j.mib.2009.01.005
Wittgens, A. (2013). Konstruktion Neuer Produktionsstämme Für Die Heterologe Rhamnolipidsynthese in dem nicht-pathogenen Wirt Pseudomonas Putida KT2440. Dissertation, Ulm University, Ulm.
Wittgens, A., Kovacic, F., Müller, M. M., Gerlitzki, M., Santiago-Schübel, B., Hofmann, D., et al. (2017). Novel insights into biosynthesis and uptake of rhamnolipids and their precursors. Appl. Microbiol. Biotechnol. 101, 2865–2878. doi: 10.1007/s00253-016-8041-3
Wittgens, A., and Rosenau, F. (2018). On the road towards tailor-made rhamnolipids: current state and perspectives. Appl. Microbiol. Biotechnol. 102, 8175–8185. doi: 10.1007/s00253-018-9240-x
Wittgens, A., Santiago-Schuebel, B., Henkel, M., Tiso, T., Blank, L. M., Hausmann, R., et al. (2018). Heterologous production of long-chain rhamnolipids from Burkholderia glumae in Pseudomonas putida-a step forward to tailor-made rhamnolipids. Appl. Microbiol. Biotechnol. 102, 1229–1239. doi: 10.1007/s00253-017-8702-x
Wittgens, A., Tiso, T., Arndt, T. T., Wenk, P., Hemmerich, J., Müller, C., et al. (2011). Growth independent rhamnolipid production from glucose using the non-pathogenic Pseudomonas putida KT2440. Microb. Cell Fact. 10:80. doi: 10.1186/1475-2859-10-80
Zhang, L., Veres-Schalnat, T. A., Somogyi, A., Pemberton, J. E., and Maier, R. M. (2012). Fatty acid cosubstrates provide β-oxidation precursors for rhamnolipid biosynthesis in Pseudomonas aeruginosa, as evidenced by isotope tracing and gene expression assays. Appl. Environ. Microbiol. 78, 8611–8622. doi: 10.1128/AEM.02111-12
Zhang, Y., and Miller, R. M. (1995). Effect of rhamnolipid (biosurfactant) structure on solubilization and biodegradation of n-alkanes. Appl. Environ. Microbiol. 61, 2247–2251. doi: 10.1128/AEM.61.6.2247-2251.1995
Zhao, F., Shi, R., Zhao, J., Li, G., Bai, X., Han, S., et al. (2015). Heterologous production of Pseudomonas aeruginosa rhamnolipid under anaerobic conditions for microbial enhanced oil recovery. J. Appl. Microbiol. 118, 379–389. doi: 10.1111/jam.12698
Keywords: rhamnolipids, biosurfactants, Pseudomonas putida, Pseudomonas aeruginosa, Burkholderia glumae, heterologous production, quorum sensing
Citation: Wittgens A and Rosenau F (2020) Heterologous Rhamnolipid Biosynthesis: Advantages, Challenges, and the Opportunity to Produce Tailor-Made Rhamnolipids. Front. Bioeng. Biotechnol. 8:594010. doi: 10.3389/fbioe.2020.594010
Received: 12 August 2020; Accepted: 07 October 2020;
Published: 22 October 2020.
Edited by:
Gloria Soberón-Chávez, National Autonomous University of Mexico, MexicoReviewed by:
Fengjie Cui, Jiangsu University, ChinaVivek Sharma, Chandigarh University, India
Roberta Barros Lovaglio, Federal University of São Carlos, Brazil
Copyright © 2020 Wittgens and Rosenau. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Andreas Wittgens, YW5kcmVhcy53aXR0Z2Vuc0B1bmktdWxtLmRl
 Andreas Wittgens
Andreas Wittgens Frank Rosenau
Frank Rosenau