- 1Department of Molecular Science and Technology, Ajou University, Suwon, South Korea
- 2Bio R&D Center, Noroo Holdings Co., Ltd., Suwon, South Korea
- 3School of Energy and Chemical Engineering, Ulsan National Institute of Science and Technology (UNIST), Ulsan, South Korea
- 4Department of Applied Chemistry and Biological Engineering, Ajou University, Suwon, South Korea
Coenzyme B12-dependent glycerol dehydratase (GDHt) catalyzes the dehydration reaction of glycerol in the presence of adenosylcobalamin to yield 3-hydroxypropanal (3-HPA), which can be converted biologically to versatile platform chemicals such as 1,3-propanediol and 3-hydroxypropionic acid. Owing to the increased demand for biofuels, developing biological processes based on glycerol, which is a byproduct of biodiesel production, has attracted considerable attention recently. In this review, we will provide updates on the current understanding of the catalytic mechanism and structure of coenzyme B12-dependent GDHt, and then summarize the results of engineering attempts, with perspectives on future directions in its engineering.
Introduction
Glycerol dehydratase (GDHt; EC 4.2.1.30) catalyzes the dehydration reaction of glycerol yielding 3-hydroxypropanal (3-HPA) that can be biologically converted into 1,3-propanediol (1,3-PD) or 3-hydroxypropionic acid (3-HP) by additionally expressing 1,3-propanediol dehydrogenase or aldehyde dehydrogenase, respectively, in microorganisms (Pawelkiewicz, 1965; Schneider et al., 1970; Stroinski et al., 1974; Toraya, 2000a; Huang et al., 2002; Liu et al., 2016; Park et al., 2017). 1,3-PD is used as a monomer for synthesizing polyethers, polyurethanes, and polyesters (Koutinas et al., 2014; Garlapati et al., 2016; Liu et al., 2016). 3-HP is a versatile platform chemical that can be converted into acrylic acid, acrylonitrile, and malonic acid (Valdehuesa et al., 2013; Chen and Liu, 2016; Kalantari et al., 2017). In addition, 3-HPA has an inhibitory effect on the growth of a wide variety of bacteria and therefore, prevents food spoilage, making it a suitable food preservative (Vollenweider and Lacroix, 2004). In particular, owing to the increase in the demand for biofuels, biological processes based on glycerol, a byproduct of biodiesel production, have been a recent focus of research (Da Silva et al., 2009; Ganesh et al., 2012; Cremonez et al., 2015; Ferrero et al., 2015). Besides glycerol, GDHt is also involved in the dehydration reaction of 1,2-propanediol to produce propanal, and the dehydration of 2,3-butanediol to yield butanone (Toraya et al., 1976; Chen et al., 2015).
Glycerol dehydratase are categorized on the basis of their reliance on coenzyme B12 (adenosylcobalamin (AdoCbl): coenzyme B12-independent GDHt and coenzyme B12-dependent GDHt (Yamanishi et al., 2002; Liao et al., 2003a; O’Brien et al., 2004). They share low sequence homology and are structurally very distinct despite the surprisingly similar architecture of the substrate-binding pockets (Liu et al., 2010; Martins-Pinheiro et al., 2016). Only coenzyme B12-independent GDHt from Clostridium butyricum has been experimentally characterized (O’Brien et al., 2004). The coenzyme B12-independent GDHt is extremely sensitive to oxygen and requires strict anaerobic conditions for its activity (Raynaud et al., 2003). However, most industrial microorganisms are cultured in the presence of oxygen, and the enzyme has rarely been used for the production of biochemicals so far. On the other hand, coenzyme B12-dependent GDHts are relatively resistant to aerobic conditions (Jiang et al., 2016), and have been utilized for bioconversion of glycerol (Liu et al., 2016). However, the complex cofactor, coenzyme B12, often undergoes chemical modifications during the reaction, resulting in catalytically inactive forms, and needs to be added in media for maintaining the enzyme activity. The dehydration reaction catalyzed by coenzyme B12-dependent GDHt has been reported as the rate-limiting step for the bioconversion of glycerol into 1,3-PD or 3-HP (Ahrens et al., 1998; Yuanyuan et al., 2004).
Coenzyme B12-dependent GDHt and its applications have previously been reviewed in several papers (Jiang et al., 2016; Liu et al., 2016; Jers et al., 2019). With increasing interest in the enzyme, studies on its biochemical features and engineering of the biocatalyst have recently been reported. In this review, we will provide updates on the current understanding of the structure and catalytic mechanism of B12-dependent GDHt, and in particular, describe its catalytic mechanism obtained through computational studies. To date, only limited attempts have been made to engineer coenzyme B12-dependent GDHt probably because of its multimeric structure and complicated reaction mechanism involving radicals. We summarize the findings from site-directed mutagenesis studies as well as recently reported engineering attempts. We also provide perspectives on the future directions in engineering coenzyme B12-dependent GDHt.
Dehydration Reaction Catalyzed by Coenzyme B12-Dependent GDHt
The overall reaction of GDHt is shown in Figure 1. The binding of a substrate such as glycerol induces conformational changes in the enzyme, which lengthens the bond between the Co atom and the adenosyl moiety of AdoCbl from 1.95–2.2 Å to 2.5 Å, followed by breakage of the C-Co bond (Toraya et al., 1977; Mancia et al., 1996; Shibata et al., 1999; Liao et al., 2003a). The adenosyl radical rotates alongside its glycosidic bond and abstracts the hydrogen atom from the substrate, resulting in the formation of a substrate radical (Frey, 1990, 2001; Mancia and Evans, 1998; Frey and Reed, 2000). Next, the OH group in the second carbon migrates to the terminal carbon, and a new radical is formed at the second carbon. The potassium ion present at the active site of the enzyme plays an important role in the OH migration during the catalysis (Shibata et al., 1999; Kamachi et al., 2007). The resulting 1,1-diol is unstable and is readily converted into an aldehyde group by releasing H2O. Next, the hydrogen atom is abstracted back to the substrate radical from 5′-deoxyadenosine, resulting in an aldehyde product and adenosyl radical.
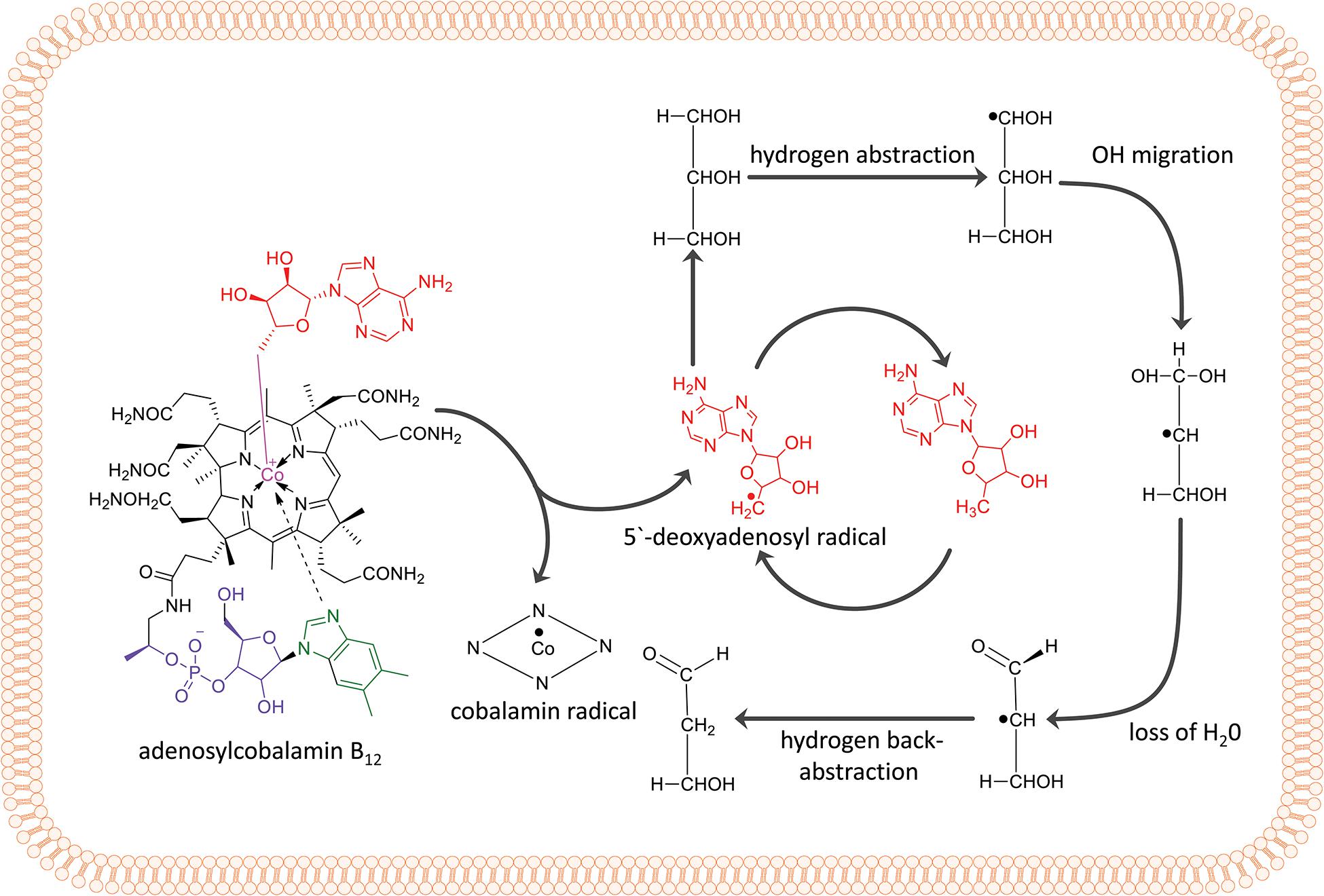
Figure 1. Adenylcobalamin B12 and the essential features of the dehydration reaction by coenzyme B12-dependent GDHt, including hydrogen abstraction, OH migration, loss of H2O, and hydrogen back-abstraction.
The Structure of Coenzyme B12-Dependent GDHt
The crystal structures of the coenzyme B12-dependent GDHt from Klebsiella pneumoniae (KpGDHt) in the presence or absence of a substrate have been reported (Yamanishi et al., 2002; Liao et al., 2003a). KpGDHt exists as a dimer of αβγ-heterotrimer, (αβγ)2, and the dimerization is induced by the interaction of two α-subunits (Figure 2A). The β- and γ-subunits are separately bound to the α-subunit. AdoCbl resides between the α- and β-subunits of each αβγ-heterotrimer.
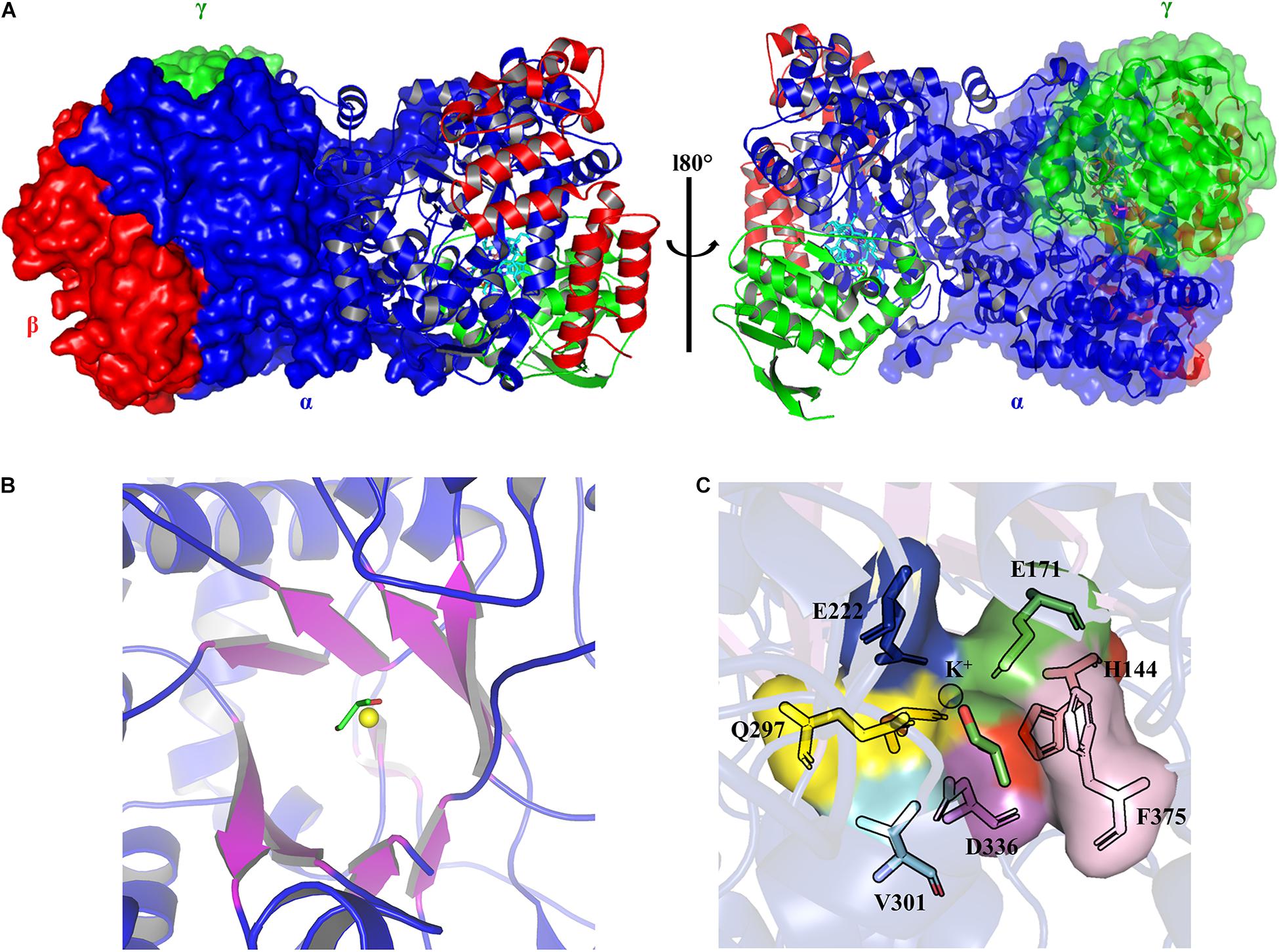
Figure 2. The three-dimensional structure models of KpGDHt (PDB ID 1IWP). (A) The dimeric form of KpGDHt. The α-, β-, and γ-subunits are shown in blue, red, and green color, respectively. The structures are shown by the ribbon or surface representation. (B) A topological diagram of the TIM barrel. The barrel β-sheets are shown in purple color. The substrate (shown by the stick model) and the K+ ion (yellow sphere) are present at the active site. (C) A zoom-in view of the active site residues interacting with the substrate and the K+ ion. The figures were created using PyMOL.
The α-subunit possesses the triosephosphate isomerase barrel (TIM) structure, where the substrate and the essential cofactor K+ are bound (Figure 2B). In the absence of a substrate, the potassium ion is hexacoordinated with amino acids at the active site and a water molecule (Liao et al., 2003a). The interaction is specific, and the K+ ion is unlikely to be exchanged with other monovalent cations such as (Shibata et al., 1999). Binding the substrate breaks the bond with water, and the K+ ion is instead heptacoordinated with two OH groups of the substrate and five oxygen atoms from residues at the active site (Gluα 171, Glnα 142, Gluα 222, Glnα 297, and Serα 363) (Figure 2C; Yamanishi et al., 2002). Structural changes take place during the dissociation of the product from the enzyme, which leads to the positioning of the cofactor in its apo conformation (Yamanishi et al., 2002; Liao et al., 2003a; Toraya, 2003, 2014).
A crack formed between the 10th and 11th β strands of the α-subunit has been assumed to be the path of substrate entry to the active site (Figure 3; Shibata et al., 1999). Three Asn and three Gln residues located around the crack possibly facilitate the entry of neutral hydrophilic compounds into the active site. The β-subunit forms the binding pocket for AdoCbl with the α-subunit, and the Rossmann fold-like structure in the central part of the β-subunit plays an important role in the interaction with the lower axial ligand of cobalamin (Liu et al., 2016). The γ-subunit is located far from the active site of the α-subunit and AdoCbl, and its role has been assumed to support the barrel structure of the α-subunit and the overall structure of coenzyme B12-dependent GDHt (Toraya, 2000a).
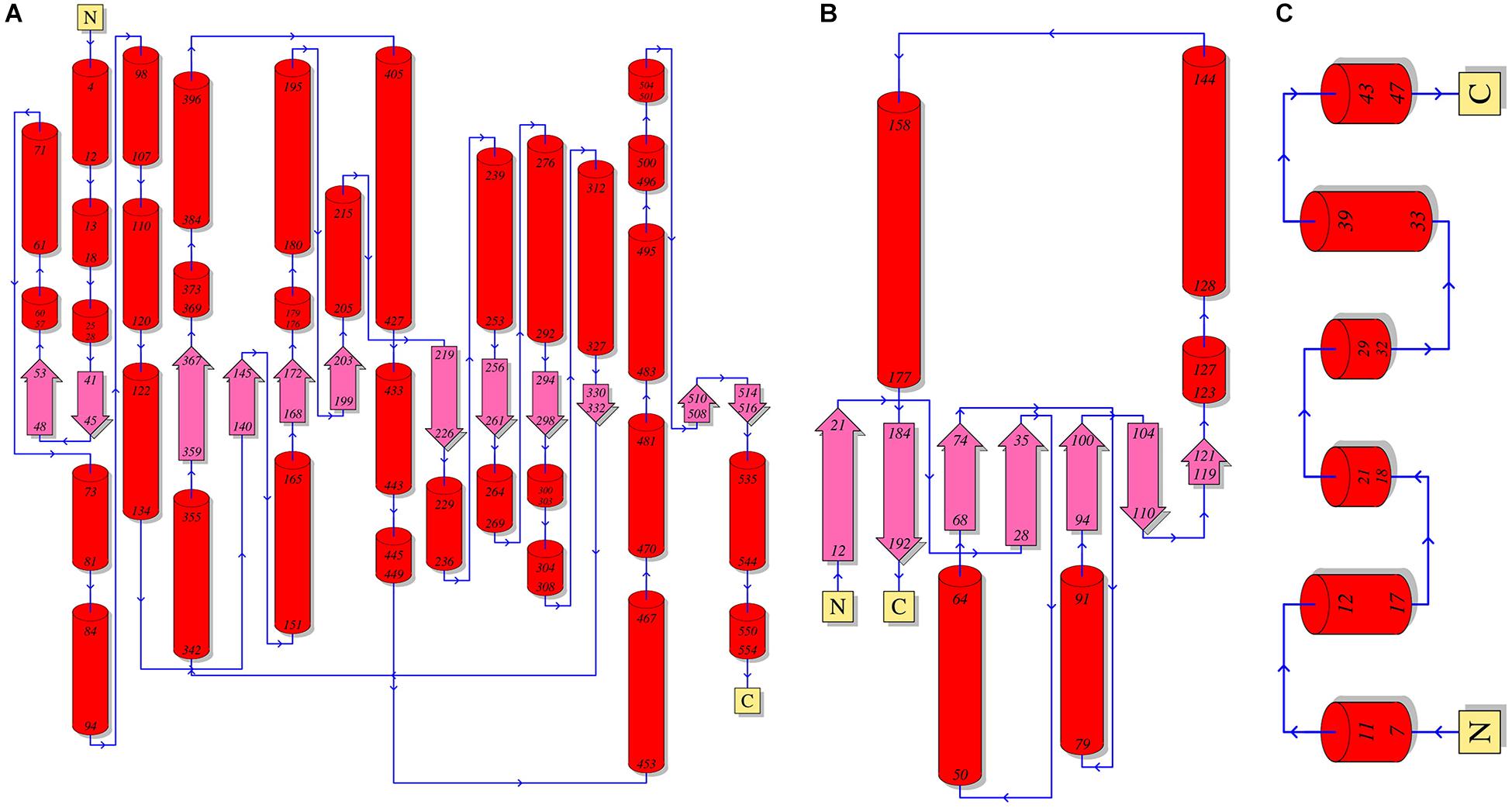
Figure 3. A schematic diagram of the secondary structures of the α-(A), β-(B), and γ-(C) subunits of coenzyme B12-dependent GDHt. The α-helices are shown as red cylinders and the β-strands as turquoise arrows.
Cobalamin derivatives are one of the most complex organic cofactors in nature. AdoCbl, the active form of cobalamin for coenzyme B12-dependent GDHt, resides in the pocket formed at the interface of the α- and β-subunits (Banerjee and Ragsdale, 2003; Maddock et al., 2015). Cobalamin has a complex structure consisting of cyclic tetrapyrroles, called corrin ring, with the cobalt atom at its center. The ring carries one nucleotide-derived tail comprising the dimethylbenzimidazole (DBI) group in addition to four propionamide, three acetamide, and eight methyl groups (Abeles and Dolphin, 1976; Banerjee, 1999; Figure 1). These peripheral groups interact with the residues in the α/β interface, which maintains the cofactor in a proper position for the radical-based catalysis (Figure 4; Shibata et al., 2018). The crystal structure of KpGDHt reported by Yamanishi et al. has cyanocobalamin, an analog of AdoCbl (Rétey, 1990; Yamanishi et al., 2002). The cyanocobalamin-KpGDHt complex structure provides information regarding how the peripheral groups of the corrin ring interact with the amino acid residues of the binding pocket (Shibata et al., 1999; Toraya, 2000a, 2014). Recently, the same group illustrated the complex structure of coenzyme B12-dependent diol dehydratase (DDHt) with AdoCbl (Shibata et al., 2018); coenzyme B12-dependent DDHt has nearly the same structure and catalytic mechanism as coenzyme B12-dependent GDHt (Toraya, 1994). This is the first complex structure including AdoCbl that shows how the native cofactor interacts with the enzyme. The adenosyl group of AdoCbl interacts with Thrα 172 and Serα 224 whereas the ribose moiety is stabilized by two hydrogen bonds through the acetamide group of corrin and Serα 224. The substrate-free and substrate-bound structures exhibited the outward and inward shifts of the α-acetamide group, which was suggested to be linked to the opening and closing of a plausible channel for the passage of substrate and product (Shibata et al., 2018).
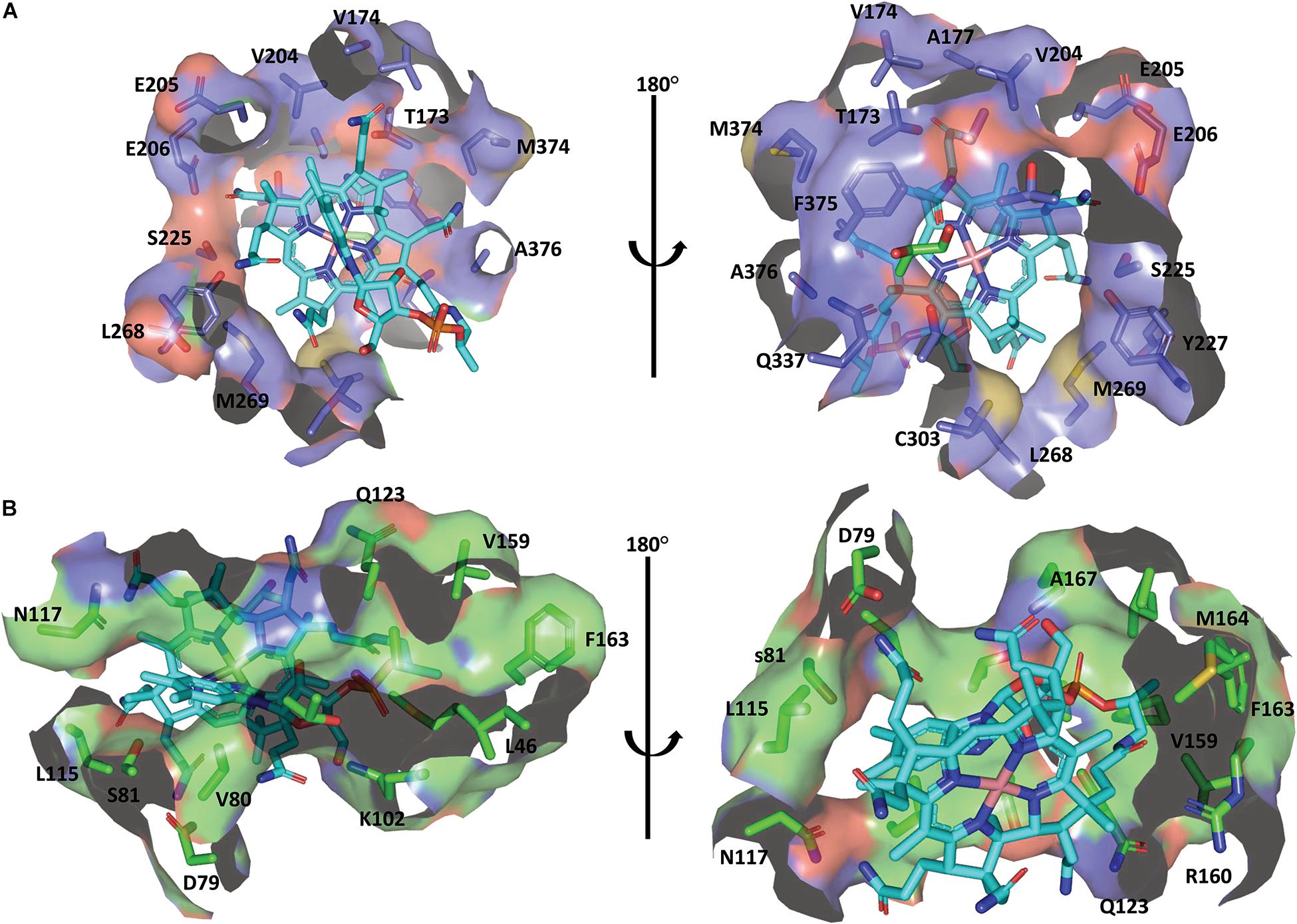
Figure 4. The binding mode of cobalamin in the KpGDHt active site. (A) Residues of the α-subunit interacting with the cobalamin molecule. (B) Residues of the β-subunit interacting with the cobalamin molecule. The B12 molecule is shown by the cyan stick model. The figures were created using PyMOL.
Inactivation and Reactivation of Coenzyme B12-Dependent GDHt
Studies on coenzyme B12-dependent GDHts have shown reaction inactivation by glycerol (substrate-bound) or oxygen (apo-form), both of which are involved in the failure to regenerate AdoCbl in the catalytic cycle, yielding a tightly bound catalytically incompetent cobalamin at the active site of the enzyme (Figure 5A). The latter inactivation, known as physiological inactivation, is due to the cleavage of the partially activated Co-C bond via binding of oxygen to the bond, whereas in the former case, known as mechanism-based inactivation, irreversible homolysis occurs during the glycerol dehydration reaction (Toraya et al., 1976; Bachovchin et al., 1977; Toraya and Abeles, 1980; Toraya, 2000a; Tobimatsu et al., 2000; Seifert et al., 2001; Yamanishi et al., 2012).
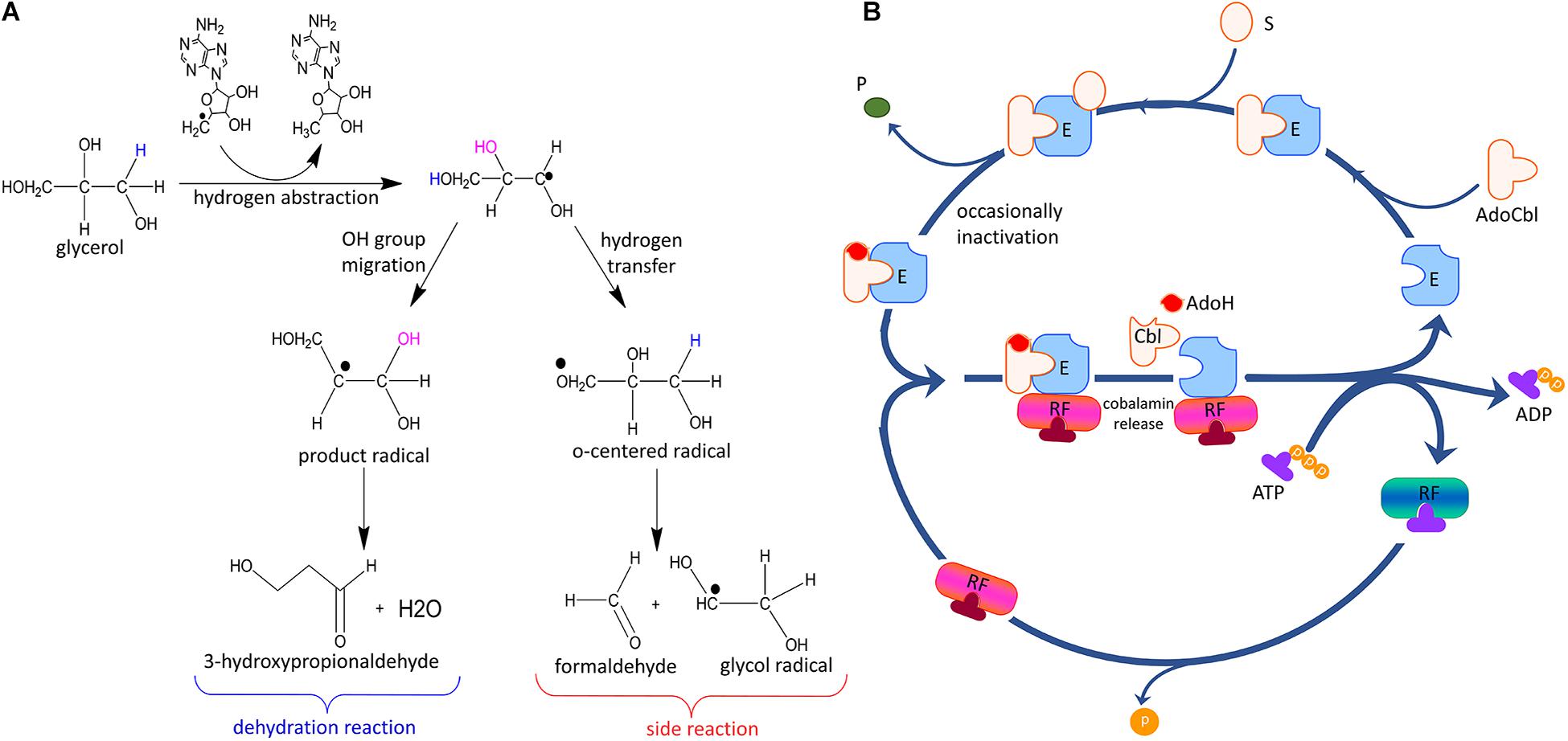
Figure 5. General mechanism of inactivation and reactivation of coenzyme B12-dependent GDHt. (A) Side reactions leading to the inactivation of coenzyme B12-dependent GDHt. (B) The mechanism of reactivation via cofactor exchange by GDHt reactivase. E: apoenzyme; AdoCbl: adenosylcobalamin; S: substrate; P: product; RF: reactivase; AdoH: 5′-deoxyadenosine; Cbl: cobalamin or damaged cofactor (Modified with permission from Bilicì et al., 2019).
The inactive cobalamin can be replaced with catalytically competent cobalamin by GDHt reactivase (Figure 5B) (Honda et al., 1980; Mori and Toraya, 1999). The structure of GDHt reactivase represents two αβ-heterodimers along with a hexacoordinated Mg2+ ion bound at the interface of the α- and β-subunits (Liao et al., 2003b). The α-subunit of GDHt reactivase contains four domains: the ATPase domain resembling those of molecular chaperons GroEL and Hsp70, the insert domain, the linker domain, and the swiveling domain (Figure 6A). Interestingly, the structure of the β-subunit of GDHt reactivase is similar to that of the β-subunit of GDHt (Figure 6B). The β-subunit swap hypothesis was proposed as a reactivation mechanism because of the structural features (Bennett et al., 1995; Liao et al., 2003b; Shibata et al., 2005). ATP hydrolysis by GDHt reactivase destabilizes the structure of the α-subunit, which facilitates its β-subunit dissociation from the swiveling domain of the α-subunit. Owing to their structural similarity, the β-subunit of GDHt reactivase binds to the α-subunit of GDHt replacing its β-subunit, during which the inactivated coenzyme B12 is released from GDHt (Yamanishi et al., 2002).
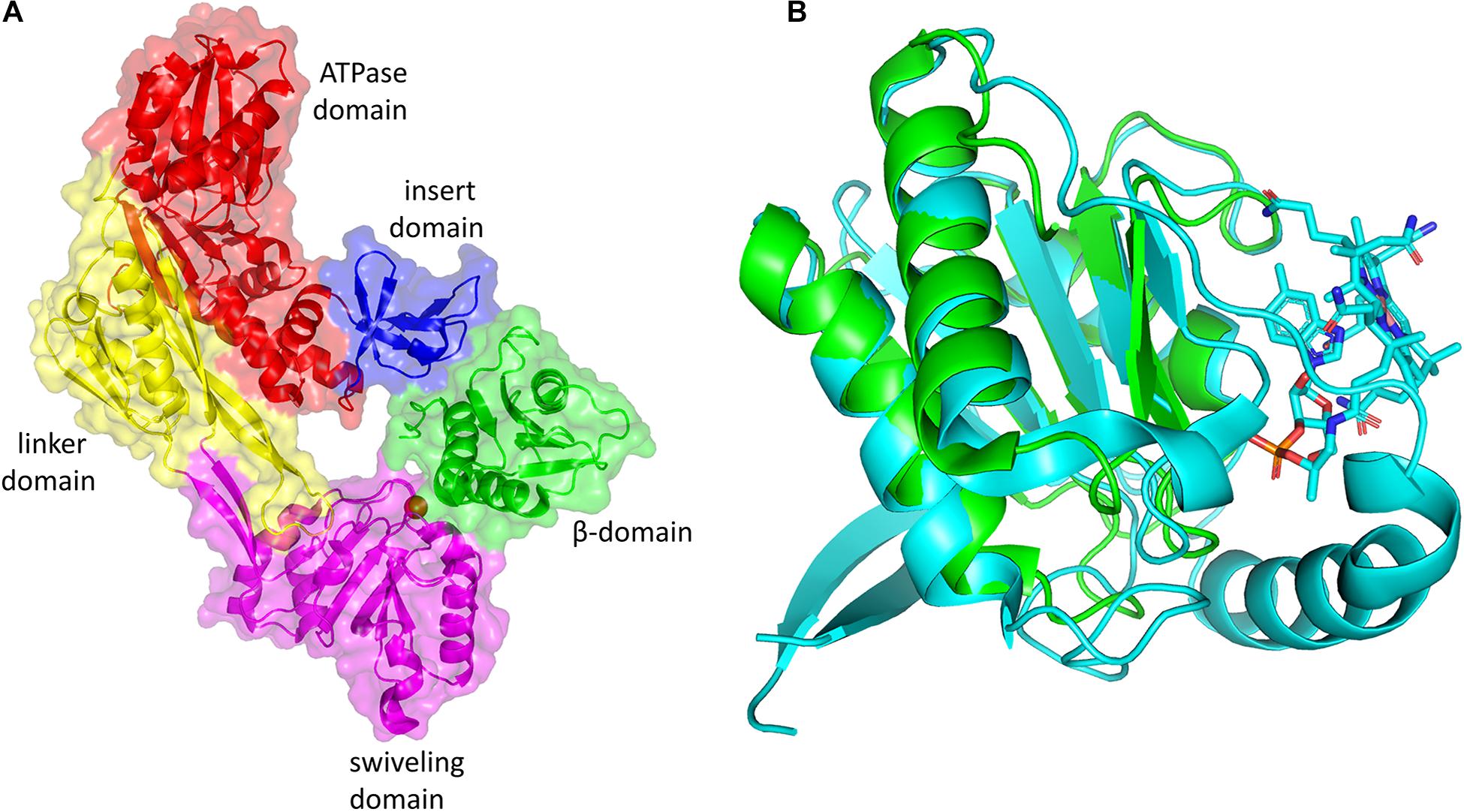
Figure 6. (A) Three-dimensional structure of GDHt reactivase αβ heterodimer (PDB ID: 1NBW). The ATPase, linker, swiveling, and insert domains of the α-subunit are colored red, yellow, magenta, and blue, respectively. The β- subunit is colored green. (B) Superimposition of the β-subunit of GDHt (cyan) and the β-subunit of GDHt reactivase (green). The B12 molecule is shown by the stick model. The figures were created using PyMOL.
Molecular Understanding of the Dehydration Reaction
Coenzyme B12-dependent GDHt and coenzyme B12-dependent DDHt are isofunctional enzymes; they have the same catalytic mechanisms and are very structurally similar with a slight difference in substrate specificities. These extreme similarities suggest that these two enzymes possibly evolved from a common ancestor (Poznanskaja et al., 1979; Toraya, 1994, 1999, 2000b; Yamanishi et al., 2002; Liao et al., 2003a; Liu et al., 2010). Thus, catalytic mechanisms are discussed based on previous studies on DDHt and GDHt hereinafter. In a pioneering study on coenzyme B12-dependent DDHt from K. pneumoniae (KpDDHt), Bachovchin et al. reported two binding modes of glycerol, the pro-S and pro-R conformations, depending on the position of the abstracted hydrogen (Figures 7A,B; Bachovchin et al., 1977). The dehydration reaction occurred dominantly when the substrate was bound in the pro-R conformation, whereas the inactivation reaction was preferable in the pro-S conformation. Doitomi et al. (2014) analyzed the three steps of substrate transformation (hydrogen abstraction, OH migration, and hydrogen re-abstraction) for both the pro-R and pro-S conformations using quantum mechanics/molecular mechanics methods. The C3-OH group in the pro-S conformation was oriented toward Ser301 of the α-subunit, and the hydrogen bond between them was suggested to increase the activation energy for the migration of the C2-OH group. Therefore, inactivation could take place prior to hydrogen recombination. In another computational study, Biliæ et al. also reported the interaction of the two conformations of glycerol at the active site of KpDDHt with respect to the orientation of the C3-OH group. The OH group in the pro-S conformation was oriented toward Ser301 of the α-subunit, and that in the pro-R conformation was oriented toward Asp335 (Figures 7C,D; Bilicì et al., 2019). An attempt was made to introduce mutations into coenzyme B12-dependent DDHt to favor its interaction with the pro-R conformation of glycerol (Yamanishi et al., 2012), which will be described later.
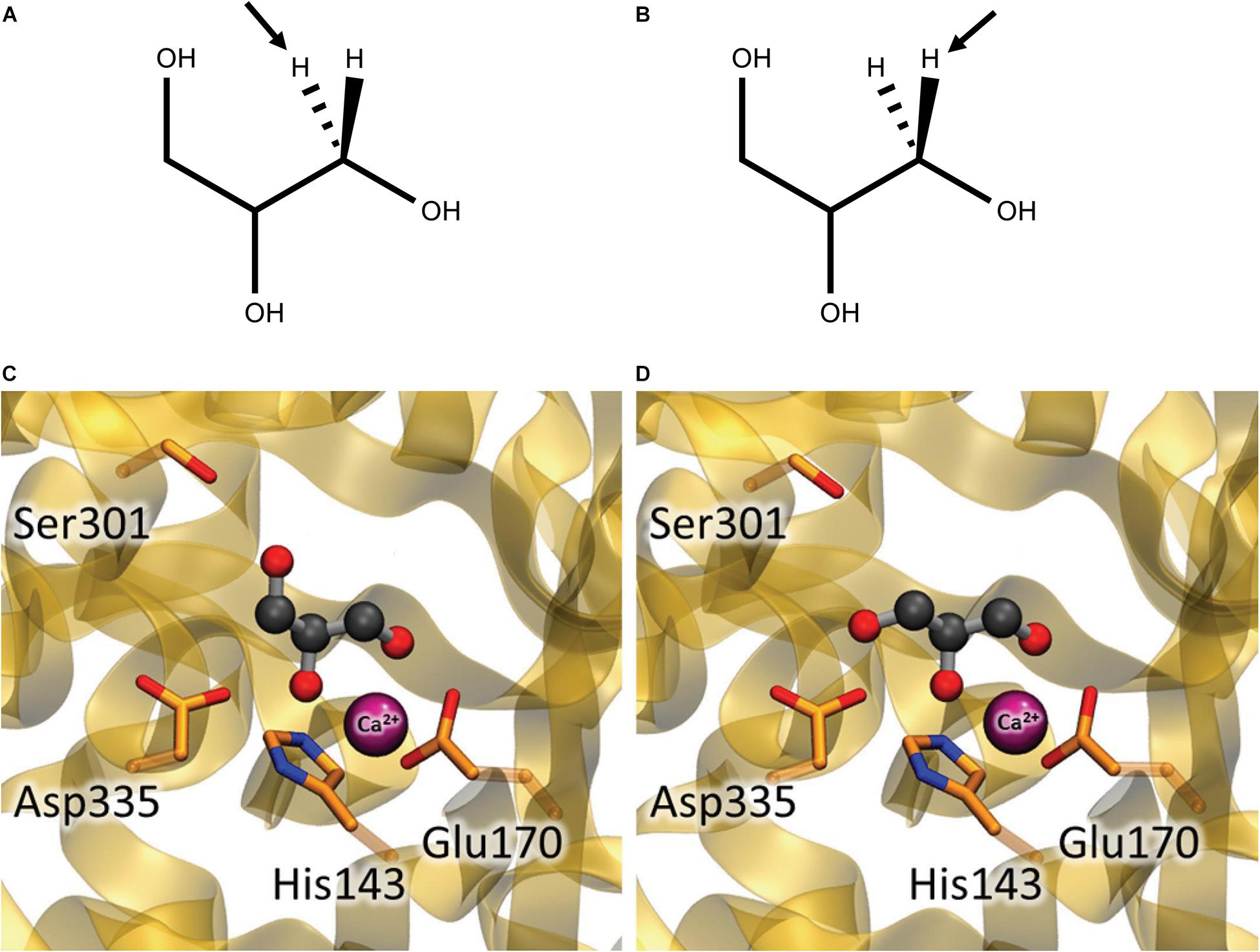
Figure 7. Glycerol in the pro-S (A,C) and pro-R (B,D) conformations at the active site of coenzyme B12-dependent DDHt. (A,B) Hydrogen abstracted in the first catalytic step are shown by arrows. (C,D) The C3-OH group of glycerol is oriented toward Ser301 in the pro-S conformation while toward Asp335 in the pro-R conformation (Modified with permission from Bilicì et al., 2019).
Engineering of Coenzyme B12-Dependent GDHt
Improvement in Catalytic Activity
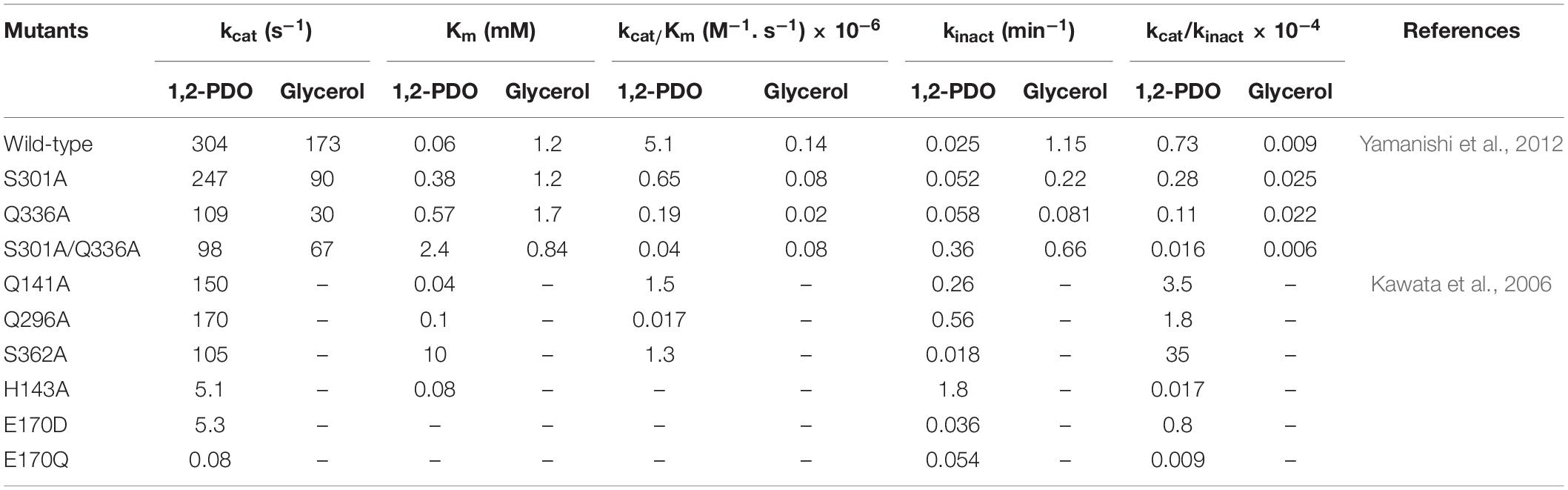
One approach to engineering enzymes is to introduce mutations at their active sites when the structures are available, in particular for the residues to interact with substrates (Yagonia et al., 2015; Lee et al., 2019). In addition, understanding the catalytic mechanism may provide useful insights into how a particular amino acid residue functions in enzymatic reactions. A study was reported on site-directed mutagenesis of the residues at the active site of coenzyme B12-dependent DDHt from K. oxytoca (KoDDHt) (Kawata et al., 2006). Substitution into Ala was made for Glnα 141, Glnα 296, Serα 362, Hisα 143, Gluα 170, and Gluα 221; Gluα 170 was further mutated to Asp, Gln, or His. Aspα 335 was mutated to Asp, Gln, and His, or Asn. All the mutations at Gluα 170, Gluα 221, and Aspα 335 abolished the activity of KoDDHt. Other variants also showed a decrease in activity compared to the wild-type (Table 1). These residues exhibited a lack of tolerance to mutagenesis, which demonstrates their important role in catalysis (Wilke et al., 2005). Yamanishi et al. identified two residues, Serα 301 and Glnα 335, of KoDDHt that play an important role in differentiating the two conformations of glycerol, the pro-S and pro-R conformations (Yamanishi et al., 2012); previous studies have demonstrated that glycerol in the pro-S conformation induces a mechanism-based inactivation (Toraya et al., 1976; Bachovchin et al., 1977). Substituting each of these residues with alanine showed a lower inactivation rate than the wild-type enzyme, but their enzyme activities decreased at the same time. This result suggests that the hydrogen bond interactions between the substrate 3-OH group and the active site residues have an important role in mechanism-based inactivation.
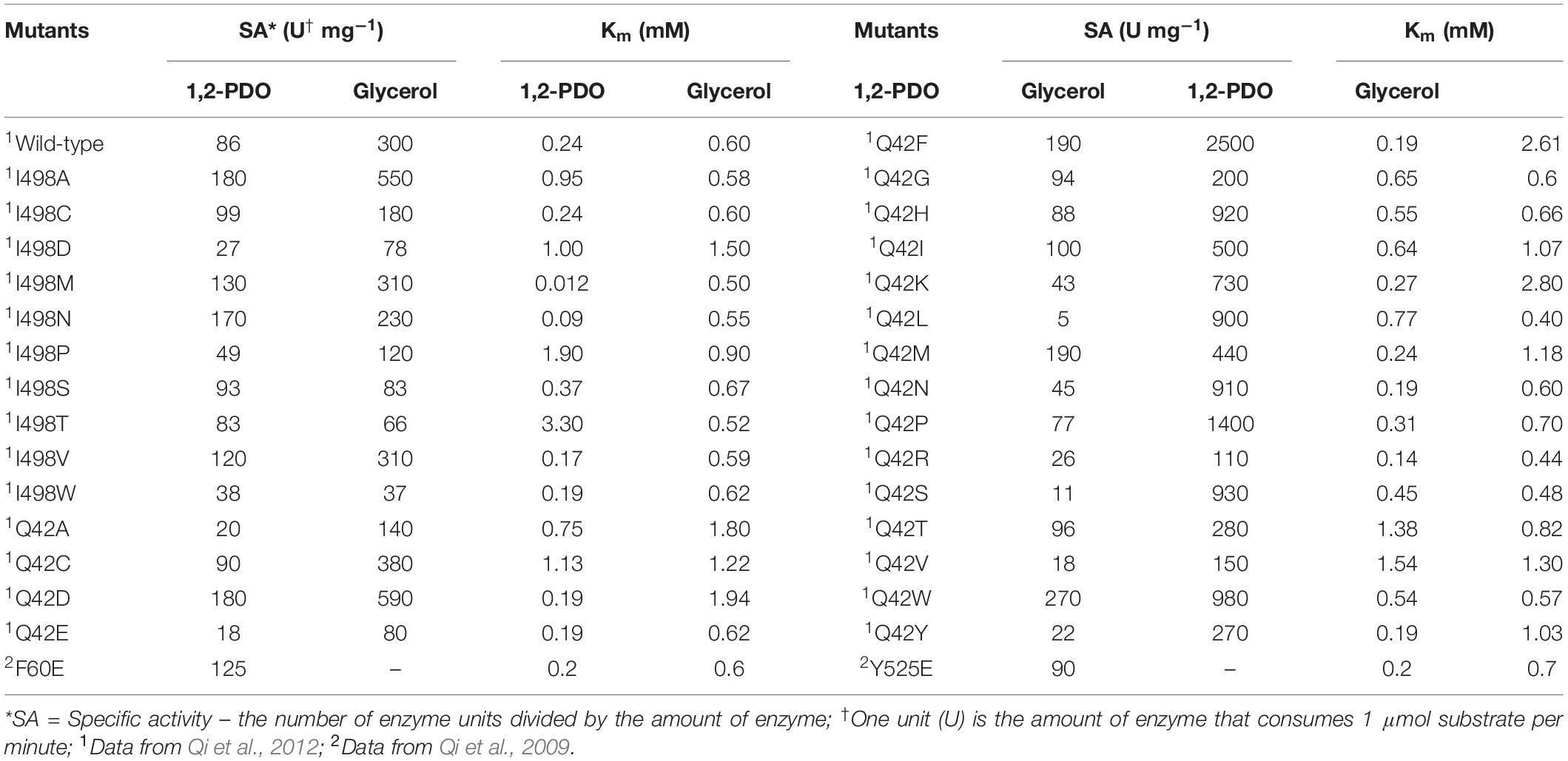
Mutations that are far from the active site sometimes result in improvement in enzyme activity, in addition to other physical properties such as stability and solubility (Guan et al., 2004; Morley and Kazlauskas, 2005; Shukla et al., 2017). These variants are usually detected by screening the libraries generated via random mutagenesis. The substitutions can induce subtle changes at the active sites, possibly via the interaction network of the residues or structural dynamics, which has been reported to be related to enzyme activities (Mesecar et al., 1997; Whittle and Shanklin, 2001; Han and Shin, 2019). Qi and colleagues applied a directed evolution approach to a KpGDHt library generated via a random mutagenesis method for improving catalytic activity and stability. The authors found two variants, Ile498Val of the α-subunit and Gln42Leu of the β-subunit, which demonstrated improved thermal and pH stability compared to the wild-type enzyme; the two positions were located far from the active site (Qi et al., 2009). Variants with a moderate increase in the catalytic efficiency toward glycerol were found in site-saturation libraries focusing on Ileα 498 and Glnβ 42. Ile498Ala and Gln42Phe mutations increased the activity of glycerol by 1.8- and 8-fold, respectively (Qi et al., 2009). The two residues were then subjected to saturation mutagenesis. Interestingly, three variants, all of which have mutations at position 42 of the β-subunit, exhibited substantially improved catalytic efficiency (kcat/Km) toward both glycerol and 1,2-propanediol (Table 2). Another attempt was made to engineer KpGDHt by introducing mutations using the PopMuSiC program (Kwasigroch et al., 2002), a computer-aided rational design program that predicts the thermodynamic stability changes caused by mutations (Qi et al., 2012). This study reported that the Tyr525Glu mutation in the α-subunit increased the catalytic activities of glycerol and 1,2-propanediol by 2- and 1.8-fold, respectively, whereas the α-Phe60Glu mutation showed opposite effects on the two substrates, increased the activity of 1,2-propanediol but decreased the activity of glycerol. These studies suggest that positions that are distant from the active site of the enzyme could be important targets for engineering.
The Fusion of α- and β-Subunits
The β-subunit of KpGDHt is prone to dissociate from the enzyme complex during purification (Yamanishi et al., 2002). This problem could be circumvented through the fusion of the α- and β-subunits via a peptide linker; the C-terminus of the α-subunit is located close to the N-terminus of the β-subunit (Figure 8; Wang et al., 2009; Maddock et al., 2017). Wang et al. fused the α- and the β-subunits of KpGDHt using a 20-residue linker of (Gly4Ser)4, and the engineered enzyme exhibited comparable catalytic activities (kcat/Km) to the wild-type enzyme (Wang et al., 2009). Maddock et al. adopted a linker of G(PT)4T(PT)7G from endoglucanase A of Cellulomonas fimi, and the enzyme unexpectedly showed a 20°C increase in the optimal temperature for the activity toward 1,2-propanediol (Maddock et al., 2017). In an attempt to engineer KpGDHt to improve its resistance to inactivation, an interesting variant was isolated which has a mutation at the stop codon (TAA) of the α-subunit to CAA (Gln), resulting in the fusion of the α- and β-subunits (Gibson et al., 2013). The fused enzyme showed a slower inactivation rate than the wild-type enzyme in an assay using the cell lysate.
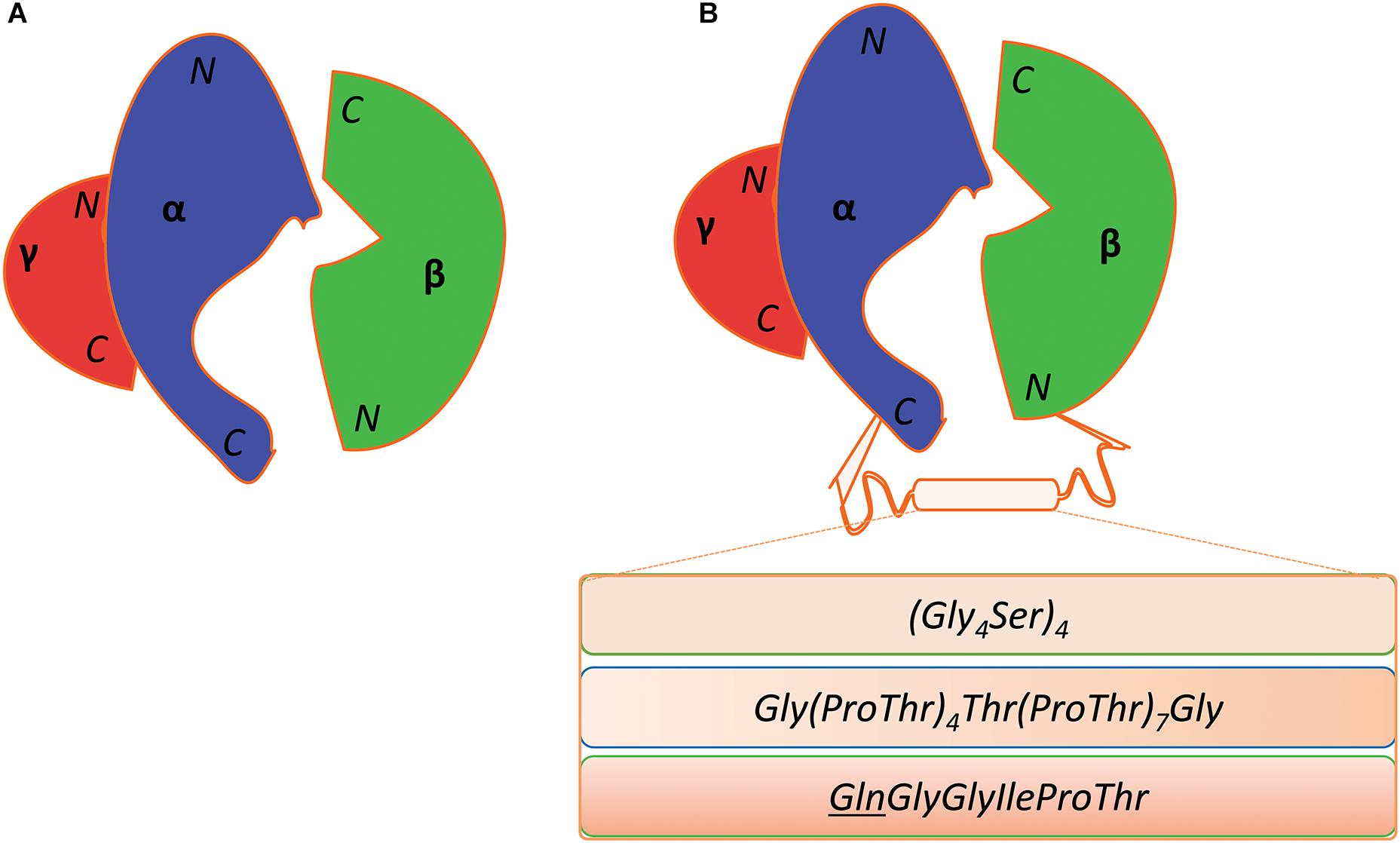
Figure 8. Fusion of the α- and β-subunit of coenzyme B12-dependent GDHt. (A) Diagrammatic representation of the wild-type coenzyme B12-dependent GDHt. Their N- and C-termini of the α-, β-, and γ-subunits are indicated. (B) The three different linkers are shown in the box between the C-terminus of the α-subunit and the N-terminus of the β-subunit. The mutation of the stop codon (TAA) of the α-subunit into a Gln codon (CAA) resulted in the third linker; the Gln position was underlined in the sequence.
Alternation of Substrate Specificity
KpGDHt has a promiscuous activity to dehydrate 2,3-butanediol to butanone, as there is no known enzyme for the reaction (Zhang et al., 2014; Chen et al., 2015). Butanone is an industrial solvent, used in the manufacture of paints, wood coatings, adhesives, inks, and pharmaceuticals. Maddock et al. reported that the catalytic efficiency (kcat/Km) of KpGDHt toward 2,3-butanediol was several hundred-fold lower than that toward 1,2-propanediol (Maddock et al., 2017). They applied strategies of combinatorial active site saturation and consensus-guided mutagenesis to improve the activity of KpGDHt toward meso-2,3-butanediol. It had been hypothesized that starting from a more stable protein would increase the rate of success in the protein engineering based on the observation that stable proteins are more resistant to mutations (Gummadi, 2003; Bloom et al., 2006). Maddock et al. (2017) used a fused enzyme, in which the α- and β-subunits of KpGDHt were linked via a Pro-rich linker showing much higher stability than the wild-type enzyme, as a template for generating libraries and found that a single point mutation (α-Thr200Ser) resulted in a four-fold increase in the catalytic efficiency of KpGDHt toward meso-2,3-butanediol by screening over 5,500 variants.
Perspectives on the Engineering of Coenzyme B12-Dependent GDHt
Coenzyme B12-dependent GDHt is the key enzyme in the biological conversion of glycerol into 1,3-PD or 3-HP, and it has been utilized in developing processes producing them. The enzyme, however, has a critical drawback of losing its activity resulting from the modification of AdoCbl, and its reactivation needs the action of GDHt reactivase involving the consumption of ATP. Supplementation of coenzyme B12 in media is necessary even for microorganisms naturally synthesizing the cofactor to attain high productivity. Therefore, engineering an enzyme that is resistant to inactivation should be an important research direction in applying coenzyme B12-dependent GDHts for industrial processes. Structural and computational studies have demonstrated that the conformation of glycerol plays a role in GDHt inactivation (Bachovchin et al., 1977; Bilicì et al., 2019). Mutations were introduced at the active site of coenzyme B12-dependent DDHt to favor its interaction with the pro-R conformation, and the variants showed slower inactivation rates than the wild-type enzyme even though their activities decreased (Yamanishi et al., 2012). The results suggest some possibilities to improve resistance to inactivation by engineering the substrate-binding site of coenzyme B12-dependent GDHts.
An engineered enzyme in which the α- and β-subunits were fused via a linker displayed a higher resistance to inactivation than the wild-type enzyme (Gibson et al., 2013). The result suggests that linking the two subunits is an alternative strategy to engineer the enzyme particularly for resisting inactivation. However, the result was demonstrated in an assay using cell lysate, and the fused enzyme needs further characterizations before a conclusion can be drawn. In particular, how fusion affects reactivation by GDHt reactivase remains unknown. Linkers to connect the two proteins can affect the properties of the fused proteins (Reddy Chichili et al., 2013; Morales-Luna et al., 2018; Rullán-Lind et al., 2018). A few linkers have been utilized so far, and only the case mentioned above was investigated with respect to inactivation. Systematic studies on the linkers to connect the two subunits can yield engineered variants more resistant to inactivation. The study by Liu and colleagues revealed that some bacterial species, such as Mesorhizobium loti and Mycobacterium smegmatis, express GDHt enzymes in the form of an αγ-heterodimer where the α-subunits have additional sequences homologous to the β-subunit of KpGDHt (Liu et al., 2010). These enzymes would be candidates for application in the bioconversion of glycerol.
Ethics Statement
This manuscript does not include any data from studies involving animal or human subjects.
Author Contributions
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
Funding
This research was supported by the National Research Foundation of Korea funded by the Ministry of Science and ICT (2015M3D3A1A01064878 and 2018R1A2B6001562) and the Ministry of Education (2019R1A6A1A11051471).
Conflict of Interest
SA is employed by the Noroo Holdings Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Abeles, R. H., and Dolphin, D. (1976). The vitamin B12 coenzyme. Acc. Chem. Res. 9, 114–120. doi: 10.1021/ar50099a006
Ahrens, K., Menzel, K., Zeng, A. P., and Deckwer, W. D. (1998). Kinetic, dynamic, and pathway studies of glycerol metabolism by Klebsiella pneumoniae in anaerobic continuous culture: III. Enzymes and fluxes of glycerol dissimilation and 1,3−propanediol formation. Biotechnol. Bioeng. 59, 544–552. doi: 10.1002/(sici)1097-0290(19980905)59:5<544::aid-bit3>3.0.co;2-a
Bachovchin, W. W., Eagar, R. G. Jr., Moore, K. W., and Richards, J. H. (1977). Mechanism of action of adenosylcobalamin: glycerol and other substrate analogs as substrates and inactivators for propanediol dehydratase-kinetics, stereospecificity, and mechanism. Biochemistry 16, 1082–1092. doi: 10.1021/bi00625a009
Banerjee, R., and Ragsdale, S. W. (2003). The many faces of vitamin B12: catalysis by cobalamin-dependent Enzymes. Annu. Rev. Biochem. 72, 209–247. doi: 10.1146/annurev.biochem.72.121801.161828
Bennett, M. J., Schlunegger, M. P., and Eisenberg, D. (1995). 3D domain swapping: a mechanism for oligomer assembly. Protein Sci. 4, 2455–2468. doi: 10.1002/pro.5560041202
Bilicì, L., Baricì, D., Banhatti, R. D., Smith, D. M., and Kovacevic, B. (2019). Computational study of glycerol binding within the active site of coenzyme B12-dependent diol dehydratase. J. Phys. Chem. B 123, 6178–6187. doi: 10.1021/acs.jpcb.9b04071
Bloom, J. D., Labthavikul, S. T., Otey, C. R., and Arnold, F. H. (2006). Protein stability promotes evolvability. Proc. Natl. Acad. Sci. U S A. 103, 5869–5874. doi: 10.1073/pnas.0510098103
Chen, Z., and Liu, D. (2016). Toward glycerol biorefinery: metabolic engineering for the production of biofuels and chemicals from glycerol. Biotechnol. Biofuels 9:205. doi: 10.1186/s13068-016-0625-8
Chen, Z., Wu, Y., Huang, J., and Liu, D. (2015). Metabolic engineering of Klebsiella pneumoniae for the de novo production of 2-butanol as a potential biofuel. Bioresour. Technol. 197, 260–265. doi: 10.1016/j.biortech.2015.08.086
Cremonez, P. A., Feroldi, M., Nadaleti, W. C., de Rossi, E., Feiden, A., de Camargo, M. P., et al. (2015). Biodiesel production in Brazil: current scenario and perspectives. Renew. Sustain. Energy Rev. 42, 415–428. doi: 10.1016/j.rser.2014.10.004
Da Silva, G. P., Mack, M., and Contiero, J. (2009). Glycerol: a promising and abundant carbon source for industrial microbiology. Biotechnol. Adv. 27, 30–39. doi: 10.1016/j.biotechadv.2008.07.006
Doitomi, K., Tanaka, H., Kamachi, T., Toraya, T., and Yoshizawa, K. (2014). Computational mutation design of diol dehydratase: Catalytic ability toward glycerol beyond the wild-type enzyme. Bull. Chem. Soc. Jpn. 87, 950–959. doi: 10.1246/bcsj.20140115
Ferrero, G. O., Almeida, M. F., Alvim-Ferraz, M. C., and Dias, J. M. (2015). Glycerol-enriched heterogeneous catalyst for biodiesel production from soybean oil and waste frying oil. Energy Convers. Manag. 89, 665–671. doi: 10.1016/j.enconman.2014.10.032
Frey, P. A. (1990). Importance of organic radicals in enzymic cleavage of unactivated carbon-hydrogen bonds. Chem. Rev. 90, 1343–1357. doi: 10.1021/cr00105a014
Frey, P. A. (2001). Radical mechanisms of enzymatic catalysis. Annu. Rev. Biochem. 70, 121–148. doi: 10.1146/annurev.biochem.70.1.121
Frey, P. A., and Reed, G. H. (2000). Radical mechanisms in adenosylmethionine-and adenosylcobalamin-dependent enzymatic reactions. Arch. Biochem. Biophys. 382, 6–14. doi: 10.1006/abbi.2000.2010
Ganesh, I., Ravikumar, S., and Hong, S. H. (2012). Metabolically engineered Escherichia coli as a tool for the production of bioenergy and biochemicals from glycerol. Biotechnol. Bioprocess Eng. 17, 671–678. doi: 10.1007/s12257-011-0446-3
Garlapati, V. K., Shankar, U., and Budhiraja, A. (2016). Bioconversion technologies of crude glycerol to value added industrial products. Biotechnol. Rep. 9, 9–14. doi: 10.1016/j.btre.2015.11.002
Gibson, K. J., Liao, D.-I., and Tang, X. S. (2013). B12-dependent dehydratases with improved reaction kinetics. U.S. Patent No 8,569,469. Washington, DC: U.S. Patent and Trademark Office.
Guan, C., Kumar, S., Kucera, R., and Ewel, A. (2004). Changing the enzymatic activity of T7 endonuclease by mutations at the β-bridge site: alteration of substrate specificity profile and metal ion requirements by mutation distant from the catalytic domain. Biochemistry 43, 4313–4322. doi: 10.1021/bi036033j
Gummadi, S. N. (2003). What is the role of thermodynamics on protein stability? Biotechnol. Bioprocess Eng. 8, 9–18. doi: 10.1007/bf02932892
Han, S.-W., and Shin, J.-S. (2019). Activity improvements of an engineered ω-transaminase for ketones are positively correlated with those for cognate amines. Biotechnol. Bioprocess Eng. 24, 176–182. doi: 10.1007/s12257-018-0377-3
Honda, S., Toraya, T., and Fukui, S. (1980). In situ reactivation of glycerol-inactivated coenzyme B12-dependent enzymes, glycerol dehydratase and diol dehydratase. J. Bacteriol. 143, 1458–1465.
Huang, H., Gong, C. S., and Tsao, G. T. (2002). Production of 1,3-propanediol by Klebsiella pneumoniae. Appl. Biochem. Biotechnol. 98-100, 687–698. doi: 10.1007/978-1-4612-0119-9_56
Jers, C., Kalantari, A., Garg, A., and Mijakovic, I. (2019). Production of 3-hydroxypropanoic acid from glycerol by metabolically engineered bacteria. Front. Bioeng. Biotechnol. 7:124. doi: 10.3389/fbioe.2019.00124
Jiang, W., Wang, S., Wang, Y., and Fang, B. (2016). Key enzymes catalyzing glycerol to 1,3-propanediol. Biotechnol. Biofuels 9:57.
Kalantari, A., Chen, T., Ji, B., Stancik, I. A., Ravikumar, V., Franjevic, D., et al. (2017). Conversion of glycerol to 3-hydroxypropanoic acid by genetically engineered Bacillus subtilis. Front. Microbiol. 8:638. doi: 10.3389/fmicb.2017.00638
Kamachi, T., Toraya, T., and Yoshizawa, K. (2007). Computational mutation analysis of hydrogen abstraction and radical rearrangement steps in the catalysis of coenzyme B12-dependent diol dehydratase. Chem. Eur. J. 13, 7864–7873. doi: 10.1002/chem.200601466
Kawata, M., Kinoshita, K., Takahashi, S., Ogura, K. I., Komoto, N., Yamanishi, M., et al. (2006). Survey of catalytic residues and essential roles of glutamate-α170 and aspartate-α335 in coenzyme B12-dependent diol dehydratase. J. Biol. Chem. 281, 18327–18334. doi: 10.1074/jbc.m601910200
Koutinas, A. A., Vlysidis, A., Pleissner, D., Kopsahelis, N., Garcia, I. L., Kookos, I. K., et al. (2014). Valorization of industrial waste and by-product streams via fermentation for the production of chemicals and biopolymers. Chem. Soc. Rev. 43, 2587–2627. doi: 10.1039/c3cs60293a
Kwasigroch, J. M., Gilis, D., Dehouck, Y., and Rooman, M. (2002). PoPMuSiC, rationally designing point mutations in protein structures. Bioinformatics 18, 1701–1702. doi: 10.1093/bioinformatics/18.12.1701
Lee, H.-S., Na, J. G., Lee, J., and Yeon, Y. J. (2019). Structure-based mutational studies of D-3-hydroxybutyrate dehydrogenase for substrate recognition of aliphatic hydroxy acids with a variable length of carbon chain. Biotechnol. Bioprocess Eng. 24, 605–612. doi: 10.1007/s12257-019-0135-1
Liao, D.-I., Dotson, G., Turner, I. Jr., Reiss, L., and Emptage, M. (2003a). Crystal structure of substrate free form of glycerol dehydratase. J. Inorg. Biochem. 93, 84–91. doi: 10.1016/s0162-0134(02)00523-8
Liao, D.-I., Reiss, L., Turner, I. Jr., and Dotson, G. (2003b). Structure of glycerol dehydratase reactivase: a new type of molecular chaperone. Structure 11, 109–119. doi: 10.1016/s0969-2126(02)00935-8
Liu, J.-z, Xu, W., Chistoserdov, A., and Bajpai, R. K. (2016). Glycerol dehydratases: biochemical structures, catalytic mechanisms, and industrial applications in 1,3-propanediol production by naturally occurring and genetically engineered bacterial strains. Appl. Biochem. Biotechnol. 179, 1073–1100. doi: 10.1007/s12010-016-2051-6
Liu, Y., Gallo, A. A., Bajpai, R. K., Chistoserdov, A., Nelson, A. T., Segura, L. N., et al. (2010). The diversity and molecular modelling analysis of B12-dependent and B12-independent glycerol dehydratases. Int. J. Boinform. Res. Appl. 6, 484–507. doi: 10.1504/ijbra.2010.037988
Maddock, D. J., Gerth, M. L., and Patrick, W. M. (2017). An engineered glycerol dehydratase with improved activity for the conversion of meso-2,3-butanediol to butanone. Biotechnol. J. 12:1700480. doi: 10.1002/biot.201700480
Maddock, D. J., Patrick, W. M., and Gerth, M. L. (2015). Substitutions at the cofactor phosphate-binding site of a clostridial alcohol dehydrogenase lead to unexpected changes in substrate specificity. Protein Eng. Des. Sel. 28, 251–258. doi: 10.1093/protein/gzv028
Mancia, F., and Evans, P. R. (1998). Conformational changes on substrate binding to methylmalonyl CoA mutase and new insights into the free radical mechanism. Structure 6, 711–720. doi: 10.1016/s0969-2126(98)00073-2
Mancia, F., Keep, N. H., Nakagawa, A., Leadlay, P. F., McSweeney, S., Rasmussen, B., et al. (1996). How coenzyme B12 radicals are generated: the crystal structure of methylmalonyl-coenzyme A mutase at 2 Å resolution. Structure 4, 339–350. doi: 10.1016/s0969-2126(96)00037-8
Martins-Pinheiro, M., Lima, W. C., Asif, H., Oller, C. A., and Menck, C. F. (2016). Evolutionary and functional relationships of the dha regulon by genomic context analysis. PLoS One 11:e0150772. doi: 10.1371/journal.pone.0150772
Mesecar, A. D., Stoddard, B. L., and Koshland, D. E. (1997). Orbital steering in the catalytic power of enzymes: small structural changes with large catalytic consequences. Science 277, 202–206. doi: 10.1126/science.277.5323.202
Morales-Luna, L., Serrano-Posada, H., González-Valdez, A., Ortega-Cuellar, D., Vanoye-Carlo, A., Hernández-Ochoa, B., et al. (2018). Biochemical characterization and structural modeling of fused glucose-6-phosphate dehydrogenase-phosphogluconolactonase from Giardia lamblia. Int. J. Mol. Sci. 19:2518. doi: 10.3390/ijms19092518
Mori, K., and Toraya, T. (1999). Mechanism of reactivation of coenzyme B12-dependent diol dehydratase by a molecular chaperone-like reactivating factor. Biochemistry 38, 13170–13178. doi: 10.1021/bi9911738
Morley, K. L., and Kazlauskas, R. J. (2005). Improving enzyme properties: when are closer mutations better? Trends Biotechnol. 23, 231–237. doi: 10.1016/j.tibtech.2005.03.005
O’Brien, J. R., Raynaud, C., Croux, C., Girbal, L., Soucaille, P., and Lanzilotta, W. N. (2004). Insight into the mechanism of the B12-independent glycerol dehydratase from Clostridium butyricum: preliminary biochemical and structural characterization. Biochemistry 43, 4635–4645. doi: 10.1021/bi035930k
Park, Y. S., Choi, U. J., Nam, N. H., Choi, S. J., Nasir, A., Lee, S.-G., et al. (2017). Engineering an aldehyde dehydrogenase toward its substrates, 3-hydroxypropanal and NAD+, for enhancing the production of 3-hydroxypropionic acid. Sci. Rep. 7, 1–12.
Pawelkiewicz, J. (1965). Enzymic conversion of glycerol into β-hydroxypropionaldehyde in a cell-free extract from Aerobacter aerogenes. Acta Biochim. Pol. 12, 207–218.
Poznanskaja, A. A., Tanizawa, K., Soda, K., Toraya, T., and Fukui, S. (1979). Coenzyme B12-dependent diol dehydrase: purification, subunit heterogeneity, and reversible association. Arch. Biochem. Biophys. 194, 379–386. doi: 10.1016/0003-9861(79)90630-1
Qi, X., Chen, Y., Jiang, K., Zuo, W., Luo, Z., Wei, Y., et al. (2009). Saturation-mutagenesis in two positions distant from active site of a Klebsiella pneumoniae glycerol dehydratase identifies some highly active mutants. J. Biotechnol. 144, 43–50. doi: 10.1016/j.jbiotec.2009.06.015
Qi, X., Guo, Q., Wei, Y., Xu, H., and Huang, R. (2012). Enhancement of pH stability and activity of glycerol dehydratase from Klebsiella pneumoniae by rational design. Biotechnol. Lett. 34, 339–346. doi: 10.1007/s10529-011-0775-5
Raynaud, C., Sarçabal, P., Meynial-Salles, I., Croux, C., and Soucaille, P. (2003). Molecular characterization of the 1,3-propanediol (1,3-PD) operon of Clostridium butyricum. Proc. Natl. Acad. Sci. U.S.A. 100, 5010–5015. doi: 10.1073/pnas.0734105100
Reddy Chichili, V. P., Kumar, V., and Sivaraman, J. (2013). Linkers in the structural biology of protein–protein interactions. Protein Sci. 22, 153–167. doi: 10.1002/pro.2206
Rétey, J. (1990). Enzymic reaction selectivity by negative catalysis or how do enzymes deal with highly reactive intermediates? Angew. Chem. 29, 355–361. doi: 10.1002/anie.199003551
Rullán-Lind, C., Pietri, R. B., Vázquez-Cintrón, M., and Baerga-Ortiz, A. (2018). Fused dimerization increases expression, solubility, and activity of bacterial dehydratase enzymes. Protein Sci. 27, 969–975. doi: 10.1002/pro.3404
Schneider, Z., Larsen, E. G., Jacobson, G., Johnson, B. C., and Pawelkiewicz, J. (1970). Purification and properties of glycerol dehydrase. J. Biol. Chem. 245, 3388–3396.
Seifert, C., Bowien, S., Gottschalk, G., and Daniel, R. (2001). Identification and expression of the genes and purification and characterization of the gene products involved in reactivation of coenzyme B12-dependent glycerol dehydratase of Citrobacter freundii. Eur. J. Biochem. 268, 2369–2378. doi: 10.1046/j.1432-1327.2001.02123.x
Shibata, N., Masuda, J., Tobimatsu, T., Toraya, T., Suto, K., Morimoto, Y., et al. (1999). A new mode of B12 binding and the direct participation of a potassium ion in enzyme catalysis: X-ray structure of diol dehydratase. Structure 7, 997–1008. doi: 10.1016/s0969-2126(99)80126-9
Shibata, N., Mori, K., Hieda, N., Higuchi, Y., Yamanishi, M., and Toraya, T. (2005). Release of a damaged cofactor from a coenzyme B12-dependent enzyme: X-ray structures of diol dehydratase-reactivating factor. Structure 13, 1745–1754. doi: 10.1016/j.str.2005.08.011
Shibata, N., Sueyoshi, Y., Higuchi, Y., and Toraya, T. (2018). Direct participation of a peripheral side chain of a corrin ring in coenzyme B12 catalysis. Angew. Chem. 130, 7956–7961. doi: 10.1002/ange.201803591
Shukla, H., Shukla, R., Sonkar, A., Pandey, T., and Tripathi, T. (2017). Distant Phe345 mutation compromises the stability and activity of Mycobacterium tuberculosis isocitrate lyase by modulating its structural flexibility. Sci. Rep. 7:1058. doi: 10.1038/s41598-017-01235-z
Stroinski, A., Pawelkiewicz, J., and Johnson, B. C. (1974). Allosteric interactions in glycerol dehydratase: purification of enzyme and effects of positive and negative cooperativity for glycerol. Arch. Biochem. Biophys. 162, 321–330. doi: 10.1016/0003-9861(74)90189-1
Tobimatsu, T., Kajiura, H., and Toraya, T. (2000). Specificities of reactivating factors for adenosylcobalamin-dependent diol dehydratase and glycerol dehydratase. Arch. Microbiol. 174, 81–88. doi: 10.1007/s002030000179
Toraya, T. (1994). Diol dehydrase and glycerol dehydrase, coenzyme B12-dependent isozymes. Met. Ions Biol. Syst. 30, 217–254.
Toraya, T. (1999). “Diol dehydratase and glycerol dehydratase,” in Chemistry and Biochemistry of B12, ed. R. Banerjee (New York: John Wiley and Sons), 783–809.
Toraya, T. (2000a). Radical catalysis of B12 enzymes: structure, mechanism, inactivation, and reactivation of diol and glycerol dehydratases. Cell. Mol. Life Sci. 57, 106–127. doi: 10.1007/s000180050502
Toraya, T. (2000b). The structure and the mechanism of action of coenzyme B12-dependent diol dehydratases. J. Mol. Catal. B: Enzym. 10, 87–106. doi: 10.1016/s1381-1177(00)00117-x
Toraya, T. (2003). Radical catalysis in coenzyme B12-dependent isomerization (eliminating) reactions. Chem. Rev. 103, 2095–2128. doi: 10.1021/cr020428b
Toraya, T. (2014). Cobalamin-dependent dehydratases and a deaminase: Radical catalysis and reactivating chaperones. Arch. Biochem. Biophys. 544, 40–57. doi: 10.1016/j.abb.2013.11.002
Toraya, T., and Abeles, R. H. (1980). Inactivation of dioldehydrase in the presence of a coenzyme-B12 analog. Arch. Biochem. Biophys. 203, 174–180. doi: 10.1016/0003-9861(80)90166-6
Toraya, T., and Mori, K. (1999). A reactivating factor for coenzyme B12-dependent diol dehydratase. J. Biol. Chem. 274, 3372–3377. doi: 10.1074/jbc.274.6.3372
Toraya, T., Shirakashi, T., Kosuga, T., and Fukui, S. (1976). Substrate specificity of coenzyme B12-dependent diol dehydrase: glycerol as both a good substrate and a potent inactivator. Biochem. Biophys. Res. Commun. 69, 475–480. doi: 10.1016/0006-291x(76)90546-5
Toraya, T., Ushio, K., Fukui, S., and Hogenkamp, P. (1977). Studies on the mechanism of the adenosylcobalamin-dependent diol dehydrase reaction by the use of analogs of the coenzyme. J. Biol. Chem. 252, 963–970.
Valdehuesa, K. N. G., Liu, H., Nisola, G. M., Chung, W. J., Lee, S. H., and Park, S. J. (2013). Recent advances in the metabolic engineering of microorganisms for the production of 3-hydroxypropionic acid as C3 platform chemical. Appl. Microbiol. Biotechnol. 97, 3309–3321. doi: 10.1007/s00253-013-4802-4
Vollenweider, S., and Lacroix, C. (2004). 3-hydroxypropionaldehyde: applications and perspectives of biotechnological production. Appl. Microbiol. Biotechnol. 64, 16–27. doi: 10.1007/s00253-003-1497-y
Wang, X. D., Zhang, X. E., Guo, Y. C., Zhang, Z. P., Cao, Z. A., and Zhou, Y. F. (2009). Characterization of glycerol dehydratase expressed by fusing its α- and β-subunits. Biotechnol. Lett. 31, 711–717. doi: 10.1007/s10529-009-9911-x
Whittle, E., and Shanklin, J. (2001). Engineering Δ9-16: 0-acyl carrier protein (ACP) desaturase specificity based on combinatorial saturation mutagenesis and logical redesign of the castor Δ9-18: 0-ACP desaturase. J. Biol. Chem. 276, 21500–21505. doi: 10.1074/jbc.m102129200
Wilke, C. O., Bloom, J. D., Drummond, D. A., and Raval, A. (2005). Predicting the tolerance of proteins to random amino acid substitution. Biophys. J. 89, 3714–3720. doi: 10.1529/biophysj.105.062125
Yagonia, C. F. J., Park, H. J., Hong, S. Y., and Yoo, Y. J. (2015). Simultaneous improvements in the activity and stability of Candida antarctica lipase B through multiple-site mutagenesis. Biotechnol. Bioprocess Eng. 20, 218–224. doi: 10.1007/s12257-014-0706-0
Yamanishi, M., Kinoshita, K., Fukuoka, M., Saito, T., Tanokuchi, A., Ikeda, Y., et al. (2012). Redesign of coenzyme B12 dependent diol dehydratase to be resistant to the mechanism-based inactivation by glycerol and act on longer chain 1,2-diols. FEBS J. 279, 793–804. doi: 10.1111/j.1742-4658.2012.08470.x
Yamanishi, M., Yunoki, M., Tobimatsu, T., Sato, H., Matsui, J., Dokiya, A., et al. (2002). The crystal structure of coenzyme B12-dependent glycerol dehydratase in complex with cobalamin and propane-1,2-diol. Eur. J. Biochem. 269, 4484–4494. doi: 10.1046/j.1432-1033.2002.03151.x
Yuanyuan, Z., Yang, C., and Baishan, F. (2004). Cloning and sequence analysis of the dhaT gene of the 1,3-propanediol regulon from Klebsiella pneumoniae. Biotechnol. Lett. 26, 251–255. doi: 10.1023/b:bile.0000013715.04456.0a
Keywords: glycerol dehydratase, coenzyme B12, glycerol, inactivation, reactivase, enzyme engineering
Citation: Nasir A, Ashok S, Shim JY, Park S and Yoo TH (2020) Recent Progress in the Understanding and Engineering of Coenzyme B12-Dependent Glycerol Dehydratase. Front. Bioeng. Biotechnol. 8:500867. doi: 10.3389/fbioe.2020.500867
Received: 27 September 2019; Accepted: 17 September 2020;
Published: 05 November 2020.
Edited by:
Spiros Nicolas Agathos, Catholic University of Louvain, BelgiumReviewed by:
Wu Xu, University of Louisiana at Lafayette, United StatesGyoo Yeol Jung, Pohang University of Science and Technology, South Korea
Won Jae Choi, Institute of Chemical and Engineering Sciences (A∗STAR), Singapore
Copyright © 2020 Nasir, Ashok, Shim, Park and Yoo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tae Hyeon Yoo, dGFlaHllb255b29AYWpvdS5hYy5rcg==
 Abdul Nasir
Abdul Nasir Somasundar Ashok
Somasundar Ashok Jeung Yeop Shim3
Jeung Yeop Shim3 Tae Hyeon Yoo
Tae Hyeon Yoo