
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Bioeng. Biotechnol. , 21 August 2020
Sec. Synthetic Biology
Volume 8 - 2020 | https://doi.org/10.3389/fbioe.2020.01013
This article is part of the Research Topic Mining, Deciphering and Engineering of Bacterial Natural Products View all 6 articles
Chlorothricin (CHL), produced by Streptomyces antibioticus DSM 40725 (wild-type strain, WT), belongs to a growing family of spirotetronate antibiotics that have biological activities inhibiting pyruvate carboxylase and malate dehydrogenase. ChlF2, a cluster-situated SARP regulator, can activate the transcription of chlJ, chlC3, chlC6, chlE1, chlM, and chlL to control CHL biosynthesis. Co-expression of chlF2 and chlK encoding type II thioesterase in WT strain under the control of Pkan led to high production of chlorothricin by 840% in comparison with that of WT. Since the inhibitory activity of CHL against several Gram-positive bacteria is higher than des-CHL, combinatorial strategies were applied to promote the conversion of des-CHL to CHL. Over-expression of chlB4, encoding a halogenase, combining with the supplementation of sodium chloride led to further 41% increase of CHL production compared to that of F2OE, a chlF2 over-expression strain. These findings provide new insights into the fine-tuned regulation of spirotetronate family of antibiotics and the construction of high-yield engineered strains.
Streptomycetes are the most abundant source of antibiotics widely applied in clinical treatments, animal husbandry, and plant crop protection (Chen et al., 2008; Guo et al., 2010; Zhang et al., 2016). High-producing strains are demanded to meet the needs of industrial production (Li D. et al., 2019; Zheng et al., 2019). It is common that antibiotic biosynthesis is specified by the corresponding gene cluster, usually including pathway-specific regulatory genes (Liu et al., 2013; Li and Tan, 2017; Guan et al., 2019). In recent years, spirotetronate polyketides have been defined as a new family of microbial metabolites with potent antitumor properties or antibacterial activity, of which chlorothricin (CHL) was discovered as the first member of the spirotetronate family in 1969 (Lacoske and Theodorakis, 2015). Of the spirotetronate/spirotetramate natural products, members of this family usually possess a characteristic pentacyclic aglycone comprising a trans-decalin system and a tetronate or tetramate spiro-conjugate, e.g., kijanimicin, pyrrolosporin, tetrocarcin, lobophorin, versipelostatin, nomimicin, and abyssomicin (Igarashi et al., 2012; Li S. et al., 2013; Lacoske and Theodorakis, 2015; Tian et al., 2015). For instance, abyssomicin C and tetrocarcin A exhibit significant activity against antibiotic-resistant Staphylococcus aureus and Mycobacterium tuberculosis, as well as for the treatment of various lymphomas and solid tumors (Braddock and Theodorakis, 2019).
Chlorothricin possesses a pentacyclic aglycone that is conjugated with a modified methylsalicylic acid (MSA) moiety through a disaccharide linkage (Supplementary Figure S1) and shows good anti-inflammation activity, anti-cholesterol activity. Both CHL aglycone (chlorothricolide) and MSA are polyketide products assembled by iterative type I polyketide synthases, while MSA needs to be further functionalized by C2-O-methylation and C5-chlorination through ChlB5 and ChlB4, respectively (Yi et al., 2019). Disruption of chlB4 and chlB5 led to the accumulation of deschloro-chlorothricin (des-CHL) and desmethylsalicyl-CHL, respectively (Yi et al., 2019). The antibacterial activity is highly dependent on the glycosylation of chlorothricolide and halogenation of MSA. The glycosylation of chlorothricolide is also crucial for stability (Jia et al., 2006; Shao et al., 2006; Ding et al., 2010).
The regulation of biosynthesis of the secondary metabolites usually involves a complex regulatory network that responds to environmental or nutritional factors, signaling molecules, even the antibiotics themselves or their intermediates (Liu et al., 2013). The biosynthesis of most antibiotics is typically catalyzed by a series of enzymes that are encoded by structural genes and modulated by relevant pathway-specific regulators (Liu et al., 2013). Our previous work showed that ChlF1 is an essential activator and directly activates the transcription of the chlJ, which encodes an acyl-CoA carboxyl transferase involved in the CHL biosynthesis, while represses the transcription of chlF1, chlK, and chlG (Li Y. et al., 2016). We have revealed that the glycosylated end product CHL and its intermediates can act as signaling molecules of the regulator ChlF1. Also, the antibacterial activity of these molecules is correlated with their binding affinity to ChlF1 (Li Y. et al., 2016). But the role of another cluster-situated regulator (CSR) ChlF2 has not been reported up to now.
ChlF2 belongs to the family of Streptomyces antibiotic regulatory proteins (SARPs), which have a typical N-terminal OmpR-type DNA-binding domain and an additional bacterial transcriptional activation domain (Tanaka et al., 2007). Representative examples of SARPs include ActII-ORF4 and RedD from Streptomyces coelicolor A3 (2), which have function in the regulation of actinorhodin and undecylprodigiosin biosynthesis, respectively. A SARP-type regulator TylS is essential for tylosin production and controls the expression of another global activator gene tylR (Bate et al., 2002). Heterologous expression of SARP-type regulator gene papR2 from Streptomyces pristinaespiralis in Streptomyces lividans resulted in the activation of the silent undecylprodigiosin (Red) gene cluster (Krause et al., 2020).
In this article, we demonstrate that ChlF2 is an essential regulator for CHL biosynthesis. It positively controls the transcription of chlJ, chlC3, chlC6, chlE1, chlM, and chlL. Over-expression of chlF2 and its co-transcribed type II thioesterase coding gene chlK effectively increased CHL production by 840% in comparison to WT production levels. We provide some evidences that des-CHL and CHL have inhibitory activity against cancer cells. Meanwhile, CHL exhibits higher biological activities than des-CHL against several Gram-positive bacteria. Since ChlB4 is responsible for the C5-chlorination of MSA, over-expression of chlB4 was performed to promote the production of CHL, which is further enhanced with the addition of sodium chloride. These findings provide new insights into the fine-tuned regulation of the biosynthesis of spirotetronate antibiotics and lay foundation for constructing high-yield engineered strains.
Streptomyces were grown on MS agar (2% soybean flour, 2% mannitol, 2% agar) for spore preparation and conjugal transfer, and in liquid YEME medium (0.3% yeast extract, 0.5% tryptone, 0.3% malt extract, 20% sucrose, 5 mM MgCl2, 1% glucose, 0.5% glycine) for total DNA isolation. Streptomyces coelicolor M1146 and its derivatives were cultured on AS-1 agar (0.1% yeast extract, 0.02% L-alanine, 0.02% L-arginine, 0.02% L-asparagine, 0.5% soluble starch, 0.25% NaCl, and 1% Na2SO4) for phenotypical analysis. Escherichia coli JM109 was used as a general host for propagating plasmids at 37°C. E. coli ET12567/pUZ8002 was used for E. coli–Streptomyces conjugal transfer. The temperature-sensitive shuttle plasmid pKC1139 was used to construct recombinant plasmids for gene disruption and over-expression. The integrative plasmids pSET152 and pIJ10500, which can integrate into the chromosome by site-specific recombination at the phage ΦC31 or ΦBT1 attachment site (attB), respectively, were used for gene complementation, over-expression, or gusA reporter system (Supplementary Table S1). When necessary, antibiotics were used at the following final concentrations: 100 μg/ml of apramycin, ampicillin, kanamycin, or hygromycin and 50 μg/ml of chloramphenicol in LB for E. coli; 25 μg/ml of nalidixic acid or apramycin, 50 μg/ml of kanamycin, and 12.5 μg/ml of hygromycin in MS for Streptomyces antibioticus; 25 μg/ml of nalidixic acid and 50 μg/ml of apramycin, kanamycin, or hygromycin in MS for Streptomyces coelicolor.
All the plasmids, strains, and primers used in this work are listed in Supplementary Tables S1–S3, respectively, and the detailed procedures for plasmids and strains construction are described in Supplementary Material.
The spore suspension was inoculated into a 250-ml flask containing 50 ml YEME medium, incubated for 48 h in a rotary shaker at 220 rpm and 28°C as the seed culture. The seed culture was inoculated into fermentation medium (2% cold pressed soybean flour, 2% mannitol, 0.2% CaCO3, pH 6.8) for CHL and des-CHL production. Cultures of S. antibioticus grown at 28°C for 7 days were harvested and extracted with methanol. The samples were analyzed on Agilent 1100 HPLC (Zorbax, SB-C18, 4.6 × 250 mm, 5 μm) at detection wavelength 222 nm. des-CHL and CHL were identified by LC-MS as previously described (Li Y. et al., 2016).
This experiment was performed as previously described (Li Y. et al., 2016). Total RNA was isolated from cultures of S. antibioticus grown at several time points (48, 72, and 96 h); 500 ng of pure DNA-free RNA was used to prepare cDNA, and each 20 μl of PCR reaction mixture contained 10 μl of 2× SuperReal PreMix, 6 pmol of each primer, and 1 μl of threefold diluted cDNA (1000-fold diluted for amplification of 16S rRNA as internal control). The primers used for qRT-PCR were listed in Supplementary Table S3. RT-qPCR was performed in a Rotor-Gene Q with FastFire qPCR PreMix (SYBR Green) kit according to the manufacturer’s instructions.
To detect the biological activities of des-CHL and CHL against Bacillus subtilis CGMCC 1.1630 (B. subtilis) and Staphylococcus aureus CGMCC 1.89 (S. aureus), the purified des-CHL and CHL dissolved in 30 μl methanol were added into the holes with 0.8 cm diameter in LB agar medium containing 1% (v/v) B. subtilis or S. aureus culture, respectively. The plate was incubated at 37°C for 8 h, and the antibacterial activity was estimated by measuring the diameter of the inhibition zones.
To determine the minimum inhibitory concentrations (MICs) of des-CHL and CHL on bacteria, antibacterial activity was measured according to the method described previously (Lu et al., 2015). Purified CHL and des-CHL were dissolved in DMSO to form a stock solution (100 mg/ml) and serially diluted with LB prior to mixing with indicator strains, which were pre-incubated in LB on a rotary shaker at 37°C for overnight. Assays for determining MIC were performed on 96-well microtiter plates containing diluted compounds, indicator strains (0.5%) and 0.5% DMSO. The growth of indicator strains was measured after 12 h of incubation for Staphylococcus epidermidis ATCC 35984 (S. epidermidis), S. aureus, B. subtilis, and Bacillus cereus CGMCC 1.1626 (B. cereus), and 18 h for Streptococcus pneumoniae 010 (S. pneumoniae) and Streptococcus pyogenes #2 (S. pyogenes) on a microplate reader (Epoch, Biotech) at wavelength of 600 nm (Supplementary Table S2). Strains grown in LB medium containing 0.5% DMSO without test compounds were used as positive controls, and LB medium containing 0.5% DMSO was used as blank. All tests were carried out in triplicate. The 50% inhibitory concentration (IC50) against cancer cell lines was performed as described previously (Xu et al., 2017), in which human lung carcinoma cell line A549, human lung adenocarcinoma cell line Calu-3, human hepatocellular carcinoma cell line HepG2, and human breast adenocarcinoma cell line MCF-7 were used.
Streptomyces antibiotic regulatory protein family regulators are widespread in Streptomycetes (Krause et al., 2020). The deduced product of chlF2 of S. antibioticus is a protein of 262 aa, containing a typical DNA binding domain at the N-terminus and two other functional domains at the C-terminus: a BTA (bacterial transcriptional activator) domain and a tetratricopeptide repeat (TPR) domain usually mediating protein-protein interactions and assembly of multi-protein complex (Supplementary Figure S2A). ChlF2 was deduced to be a SARP-type regulator, which shows 68% amino acid sequence identity with PlaR1 involved in phenalinolactone biosynthesis of Streptomyces sp. TÜ6071 (Myronovskyi et al., 2011), 59% identity with PyrF2 involved in pyrroindomycin biosynthesis of Streptomyces rugosporus (Wu et al., 2012), and 55% and 53% identity with NanR2 and NanR1 involved in nanchangmycin biosynthesis of Streptomyces nanchangensis NS3226 (Sun et al., 2003), respectively (Supplementary Figure S2B).
In order to determine the role of ChlF2 in CHL biosynthesis, a chlF2 disruption mutant (ΔchlF2) was constructed via homologous recombination. In ΔchlF2, a 708 bp internal fragment of chlF2 was replaced by a kanamycin resistance gene (kan) in its parent strain S. antibioticus DSM 40725. Southern blot experiment was carried out to confirm the correctness of ΔchlF2 mutant. Genomic DNAs of seven conjugants and WT were digested with NcoI, and 1 kb DNA fragment containing kan from pUC119:neo was used as probe. As expected, all these preliminarily selected transformants exhibited positive signals at 2363 bp and 794 bp, whereas WT strain did not exhibit signal due to the absence of kan, indicating that the resulting disruption strains were correct (Supplementary Figures S3A,B).
The fermentation of ΔchlF2 and WT strain was carried out to compare the change of CHL production. HPLC analysis revealed that a characteristic peak corresponding to CHL standard in the extract of WT disappeared in the ΔchlF2 mutant (Figures 1A,B). Also, an inhibition zone was observed with the fractions from WT but not with that from ΔchlF2 in the bioassays against S. aureus and B. subtilis, suggesting that CHL biosynthesis was abolished in ΔchlF2. To verify whether this was due to chlF2 disruption, the chlF2 complementary strain ΔchlF2/pSET152:PupchlF2-chlF2, in which the transcription of chlF2 was driven by a deduced promoter sequence (a short stretch of upstream chlF2 coding sequence), was constructed based on the integrative plasmid pSET152. Unexpectedly, there was no peak corresponding to CHL detected in the fermentation broth of ΔchlF2/pSET152:PupchlF2-chlF2 (data not shown). Further experiment demonstrated that CHL production could not be restored unless chlF2 and its adjacent gene chlK were co-expressed under the control of the native promoter PchlK, as shown by HPLC analysis and the bioassays against S. aureus and B. subtilis (Figures 1A,B), implying that chlF2 transcription may depend on the promoter of the adjacent gene chlK. The above results indicated that ChlF2 is an activator involved in CHL biosynthesis.
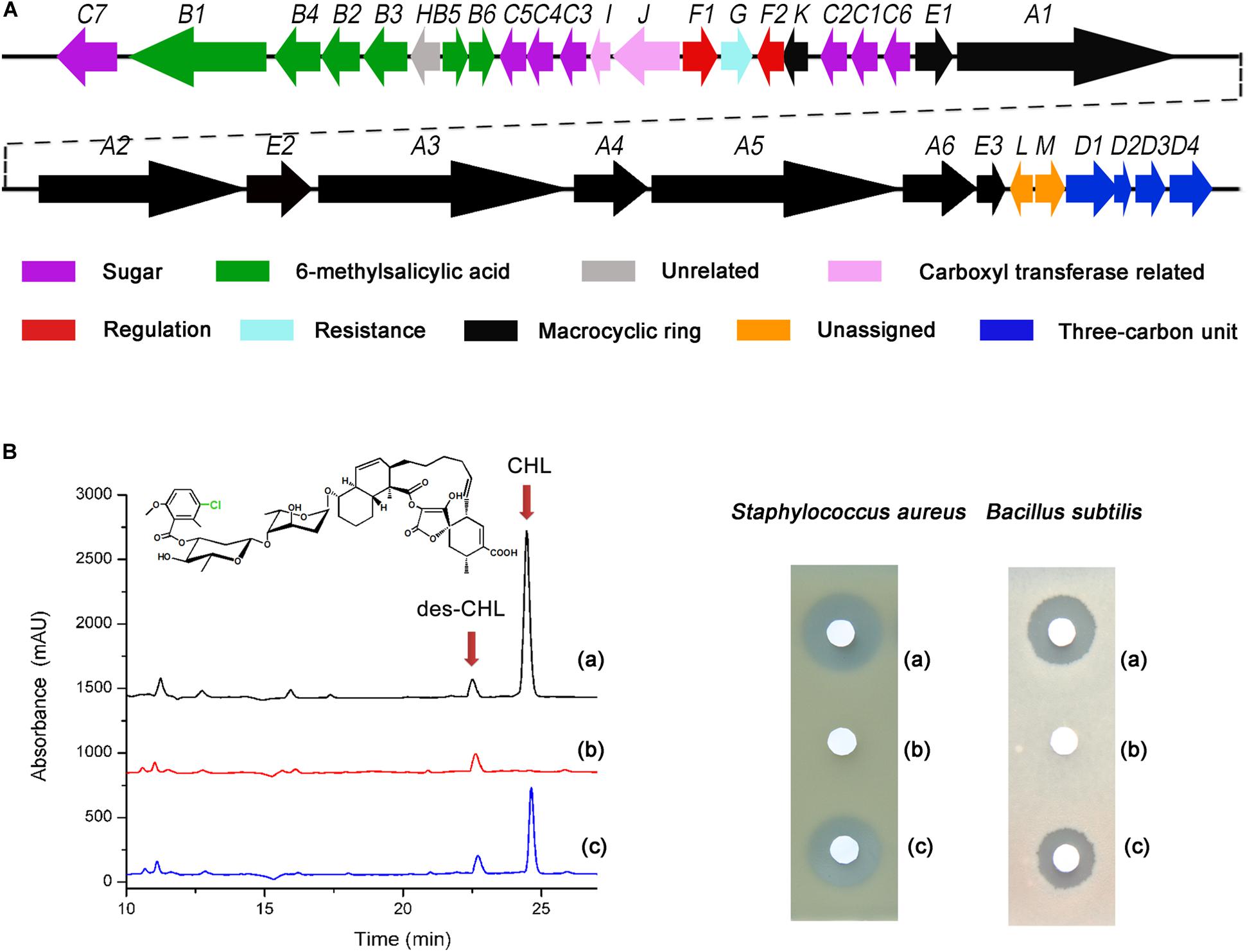
Figure 1. Effect of chlF2 disruption on CHL production. (A) Genetic organization of the chl cluster. Each arrow indicates a separate ORF and orientation of the transcription. (B) HPLC analysis and bioassays of CHL produced by WT and its derivatives. (a) Wild-type S. antibioticus DSM 40725; (b) ΔchlF2, chlF2 disruption mutant; (c) ΔchlF2/pSET152:PchlK-chlK-chlF2, chlF2 complementary strain. des-CHL indicates deschloro-chlorothricin, CHL indicates chlorothricin. The chemical structure of CHL is presented, and the position of Cl is indicated with green color.
In order to confirm whether chlK and chlF2 are co-transcribed, RT-PCR experiment was carried out. Primer pairs PF2-F/PK-R and B16S-F/B16S-R were used to confirm the transcriptional profile of chlKF2 and 16S rRNA coding gene. The results showed that the expected sizes of PCR fragments (429 bp for 16s rRNA coding gene and 1398 bp for chlKF2) were observed in accordance with the positive control (genome DNA of WT as template), but not visible in the negative control (RNA without reverse transcription as template), indicating that chlK and chlF2 were located in the same operon (Supplementary Figures S4A,B). Overall, these results confirmed that chlF2 is an indispensable activator gene for CHL production.
To find the direct targets of ChlF2 in the chl cluster, we first tried to purify ChlF2 prior to the electrophoretic mobility shift assay (EMSA), but unfortunately, we could not get the soluble protein after over-expression of chlF2 in vitro. Therefore, the in vivo reporting system of gusA was taken into account (Sherwood and Bibb, 2013). Several promoters (PchlB4, PchlB5, PchlC3, PchlJ, PchlF1, PchlK, PchlC1, PchlC6, PchlE1, PchlA1, PchlL, and PchlM) were chosen and individually inserted upstream of the gusA coding region (encoding β-glucuronidase, GUS) (Figure 2A). The gusA-fused plasmids were respectively introduced into S. coelicolor M1146 to generate recombinant strains MB4 (S. coelicolor M1146/pIJ10500:PchlB4-gusA), MB5 (M1146/pIJ10500:PchlB5-gusA), MC3 (M1146/pIJ10500:PchlC3-gusA), MJ (M1146/pIJ10500:PchlJ-gusA), MF1 (M1146/pIJ10500:PchlF1-gusA), MK (M1146/pIJ10500:PchlK-gusA), MC1 (M1146/pIJ10500:PchlC1-gusA), MC6 (M1146/pIJ10500:PchlC6-gusA), ME1 (M1146/pIJ10500:PchlE1-gusA), MA1 (M1146/pIJ10500:PchlA1-gusA), ML (M1146/pIJ10500:PchlL-gusA), and MM (M1146/pIJ10500:PchlM-gusA), and GUS activity (for hydrolyzing 5-bromo-4-chloro-3-indolyl-β-d-glucuronide to the blue product 5,5′-dibromo-4,4′-dichloro-indigo) was subsequently observed via color change. A chlF2 over-expression plasmid pKC1139-Pkan-kan-chlF2 was integrated into the φC31 attB site of the above-mentioned constructed recombinant strains to get the corresponding reporter strains FMB4 (S. coelicolor M1146/pIJ10500:PchlB4-gusA and pKC1139-Pkan-kan-chlF2), FMB5 (M1146/pIJ10500:PchlB5-gusA and pKC1139-Pkan-kan-chlF2), FMC3 (M1146/pIJ10500:PchlC3-gusA and pKC1139-Pkan-kan-chlF2), FMJ (M1146/pIJ10500:PchlJ-gusA and pKC1139-Pkan-kan-chlF2), FMF1 (M1146/pIJ10500:PchlF1-gusA and pKC1139-Pkan-kan-chlF2), FMK (M1146/pIJ10500:PchlK-gusA and pKC1139-Pkan-kan-chlF2), FMC1 (M1146/pIJ10500:PchlC1-gusA and pKC1139-Pkan-kan-chlF2), FMC6 (M1146/pIJ10500:PchlC6-gusA and pKC1139-Pkan-kan-chlF2), FME1 (M1146/pIJ10500:PchlE1-gusA and pKC1139-Pkan-kan-chlF2), FMA1 (M1146/pIJ10500:PchlA1-gusA and pKC1139-Pkan-kan-chlF2), FML (M1146/pIJ10500:PchlL-gusA and pKC1139-Pkan-kan-chlF2), and FMM (M1146/pIJ10500:PchlM-gusA and pKC1139-Pkan-kan-chlF2) accordingly. If ChlF2 could bind to the target genes, the expression of gusA would be activated to show blue color. The results showed that GUS activity of FMC3, FMJ, FMC6, FME1, FML, and FMM strains were obviously observed, revealing that the promoters of chlC3, chlJ, chlC6, chlE1, chlL, and chlM are probably the targets of ChlF2 (Figure 2B). It was noteworthy that GUS activity was detected in strains MB5, MB4, MF1, and MK, in which ChlF2 is absent, implying that chlB5, chlB4, chlF1, and chlK were constitutively expressed but not regulated by ChlF2. Neither FMC1 nor FMA1 activity was detectable under both conditions, indicating that ChlF2 could not bind to the promoters of chlC1 and chlA1 (Figure 2B).
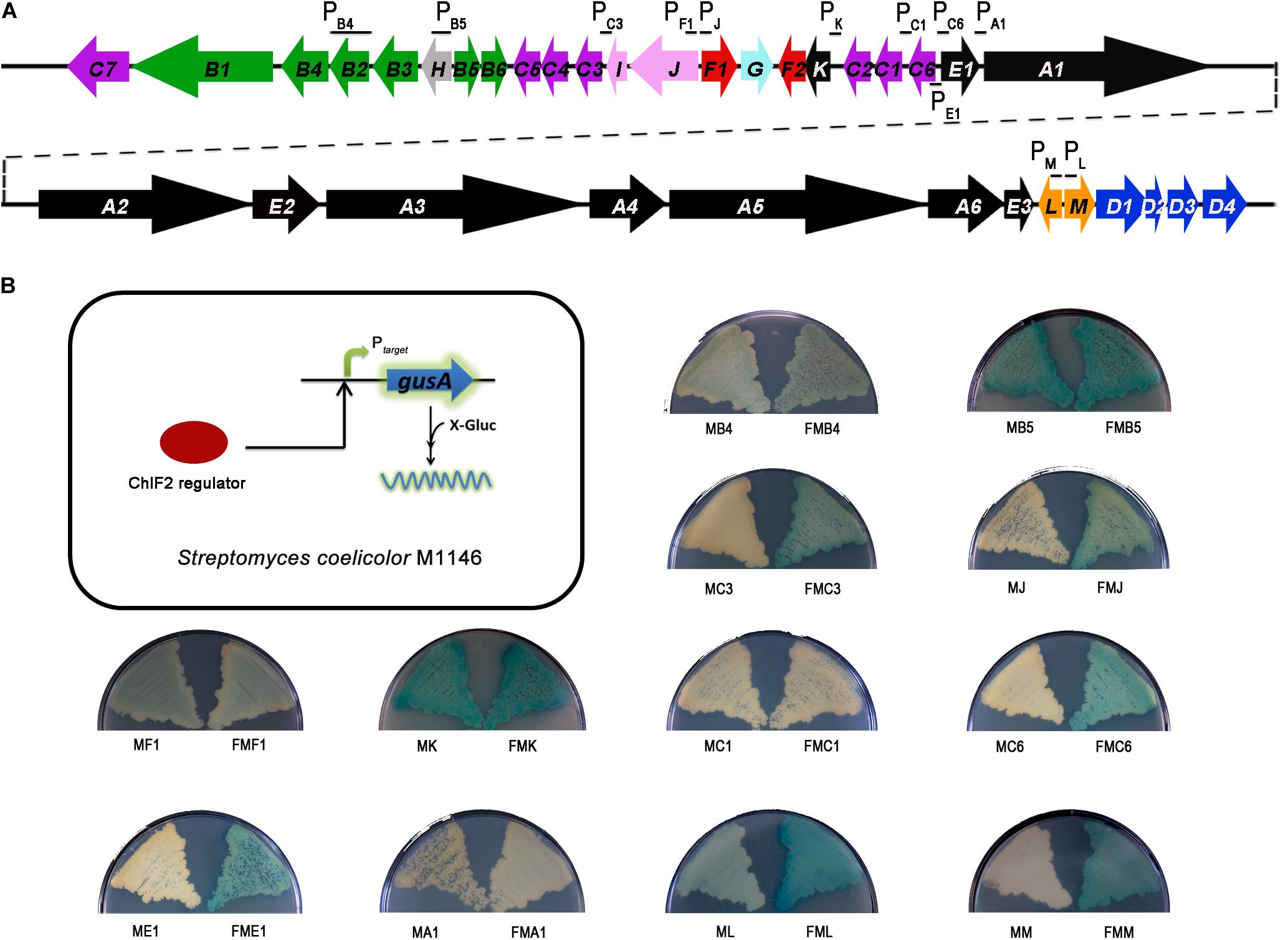
Figure 2. In vivo determination of the interaction between ChlF2 and its targets. (A) Promoter regions selected for detecting the binding activity of ChlF2 were indicated by short black lines in chl cluster. (B) Schematic diagram of gusA transcriptional fusion assays and chromogenic assays of S. coelicolor M1146 derivatives. MB4, MB5, MC3, MJ, MF1, MK, MC1, MC6, ME1, MA1, ML, and MM strains were used as controls. FMB4, FMB5, FMC3, FMJ, FMF1, FMK, FMC1, FMC6, FME1, FMA1, FML, and FMM strains were used to identify the binding regions of ChlF2.
To determine the effect of chlF2 disruption on its target genes, quantitative real-time PCR (RT-qPCR) analysis was performed to assess the transcriptional profiles of target genes chlM, chlL, chlE1, chlJ, chlC3, chlC6, and non-target gene chlF2 as a negative control. Total RNA was prepared from the cultures of WT and ΔchlF2 at different time points (48 h, 72 h, 96 h). The results showed that the transcripts of chlM, chlL, chlE1, chlC3, chlJ, and chlC6 were significantly decreased in ΔchlF2 at each time point in comparison with those in WT strain (Figure 3), revealing that ChlF2 can activate the transcription of these genes directly. The transcription of non-target gene chlF2 was not affected (Figure 3). These data indicated that ChlF2 positively regulates its target genes.
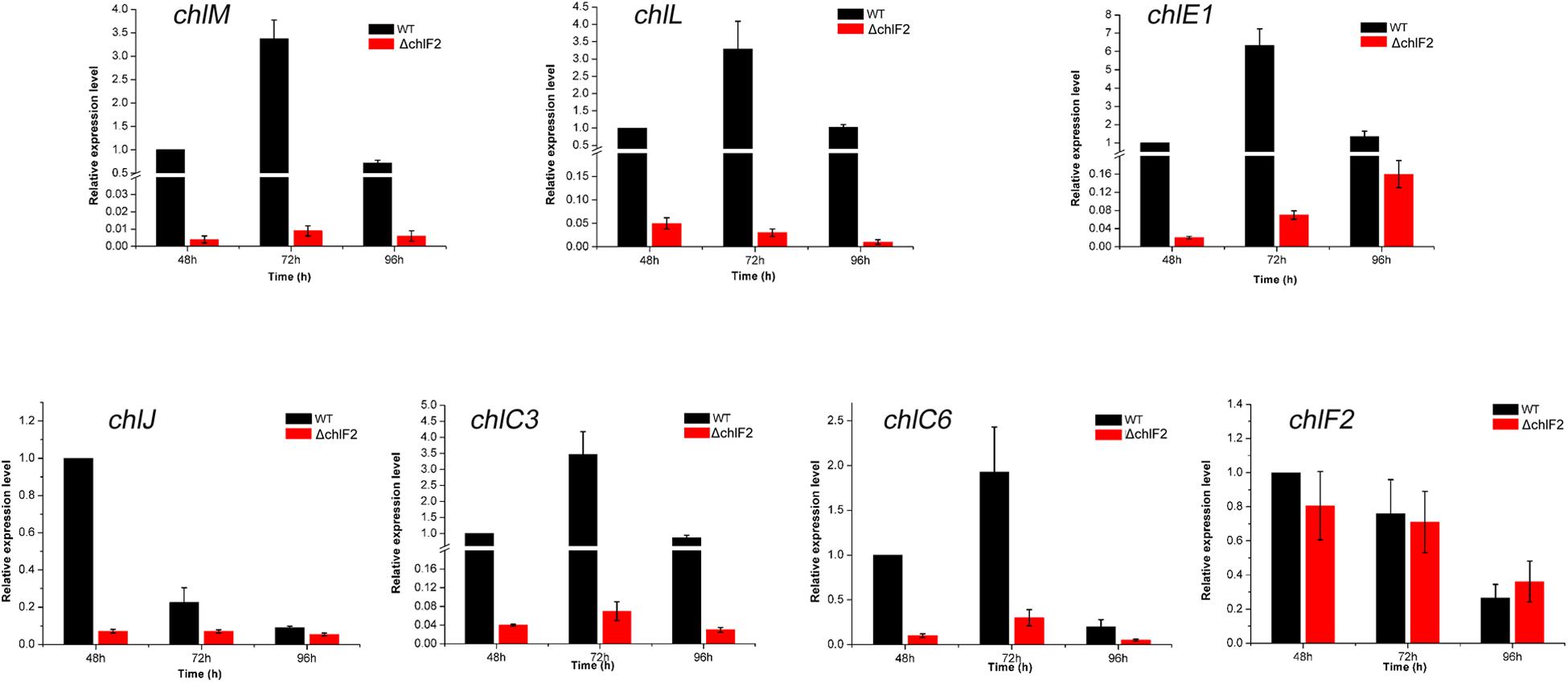
Figure 3. RT-qPCR transcriptional analysis of selected chl genes in WT and ΔchlF2. RNA samples were isolated from fermentation cultures after incubation for 48, 72, and 96 h. The data are presented as the averages of three independent experiments conducted in triplicate. Error bars show standard deviations.
ChlF2 is an important activator of CHL biosynthesis, and chlF2 complementary experiments showed that chlK and chlF2 are located in the same transcriptional unit under the control of PchlK (Figure 1A and Supplementary Figure S4). Previous studies on the role of ChlK showed that in-frame deletion of chlK led to partial decrease of CHL (Li Y. et al., 2016), whereas remarkable enhancement was observed in mutant F2OE, in which chlK was replaced by a resistance gene kan and its promoter Pkan via homologous recombination. Actually chlF2 and kan are co-transcribed under the control of strong promoter Pkan (Figures 4A,B). Bioassays were performed against B. subtilis with the fractions corresponding to CHL and des-CHL from WT and F2OE based on HPLC analysis. Inhibition zones with the fractions of des-CHL and CHL from F2OE were much bigger than those from WT, which were consistent with the HPLC data (Figure 4C). These results strongly supported that ChlF2 is a crucial activator for CHL biosynthesis, and ChlK exerts some functions but is not decisive. The transcriptional levels of all six target genes in F2OE remarkably increased at each time point compared with those in WT. A more than 100-fold transcriptional enhancement of the non-target gene chlF2 in F2OE was likely attributed to the exogenous insertion of constitutive promoter Pkan (Figure 4D).
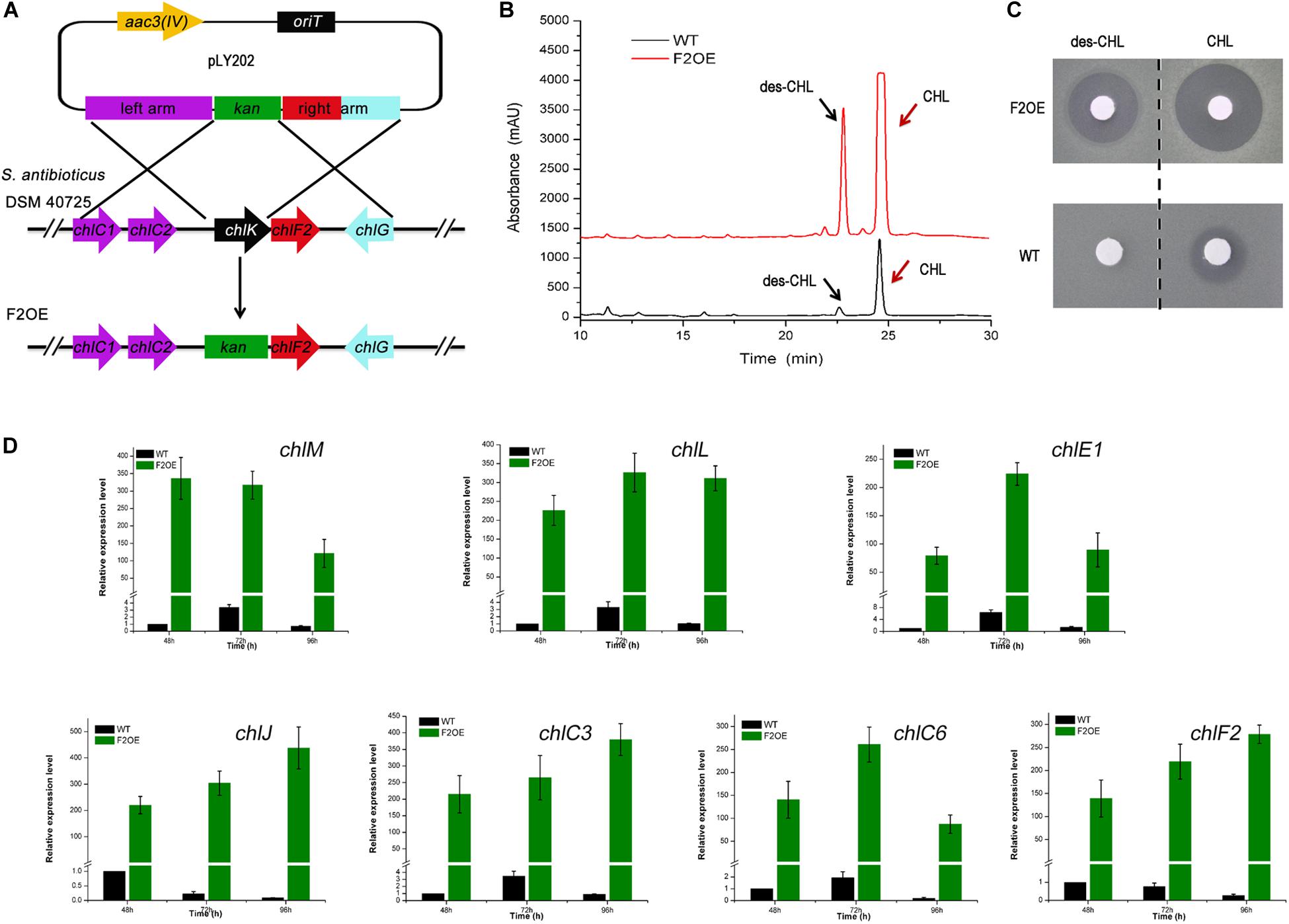
Figure 4. Metabolite analysis of WT and F2OE. (A) Construction of chlF2 over-expression engineered strain (S. antibioticus F2OE) via homologous recombination. chlK was replaced by kan and its promoter Pkan on the chromosome. (B) HPLC analysis of the extracts of WT and S. antibioticus F2OE. des-CHL indicates deschloro-chlorothricin, CHL indicates chlorothricin. (C) Bioassays of the extracts of WT and S. antibioticus F2OE against B. subtilis. (D) RT-qPCR transcriptional analysis of selected chl genes in WT and F2OE. The data are presented as the averages of three independent experiments conducted in triplicate. Error bars show standard deviations.
To enhance CHL production effectively, several strategies toward chlF2 and chlK were attempted to understand their role. Firstly, the integrative plasmids pSET152:PchlK-chlK-chlF2, pSET152:Pkan-chlF2, pSET152:Pkan-kan-chlF2, and pSET152:Pkan-chlK-chlF2 were constructed and introduced into WT, respectively. The resulting recombinant strains were named as YL01, YL02, YL03, and YL04 accordingly (Figure 5A). After fermentation and HPLC analysis, in comparison with WT, the production of CHL and des-CHL was increased by 300% and 310% in YL01, and 750% and 900% in YL02, respectively (Figure 5B). The CHL production in YL03 and YL04 was increased by 760% and 840% compared with WT, respectively (Figure 5B). Furthermore, des-CHL was significantly increased by 980% and 1100%, respectively. These results revealed the importance of promoter Pkan for chlF2 and chlK expression, and which in turn led to high-production of CHL and des-CHL.
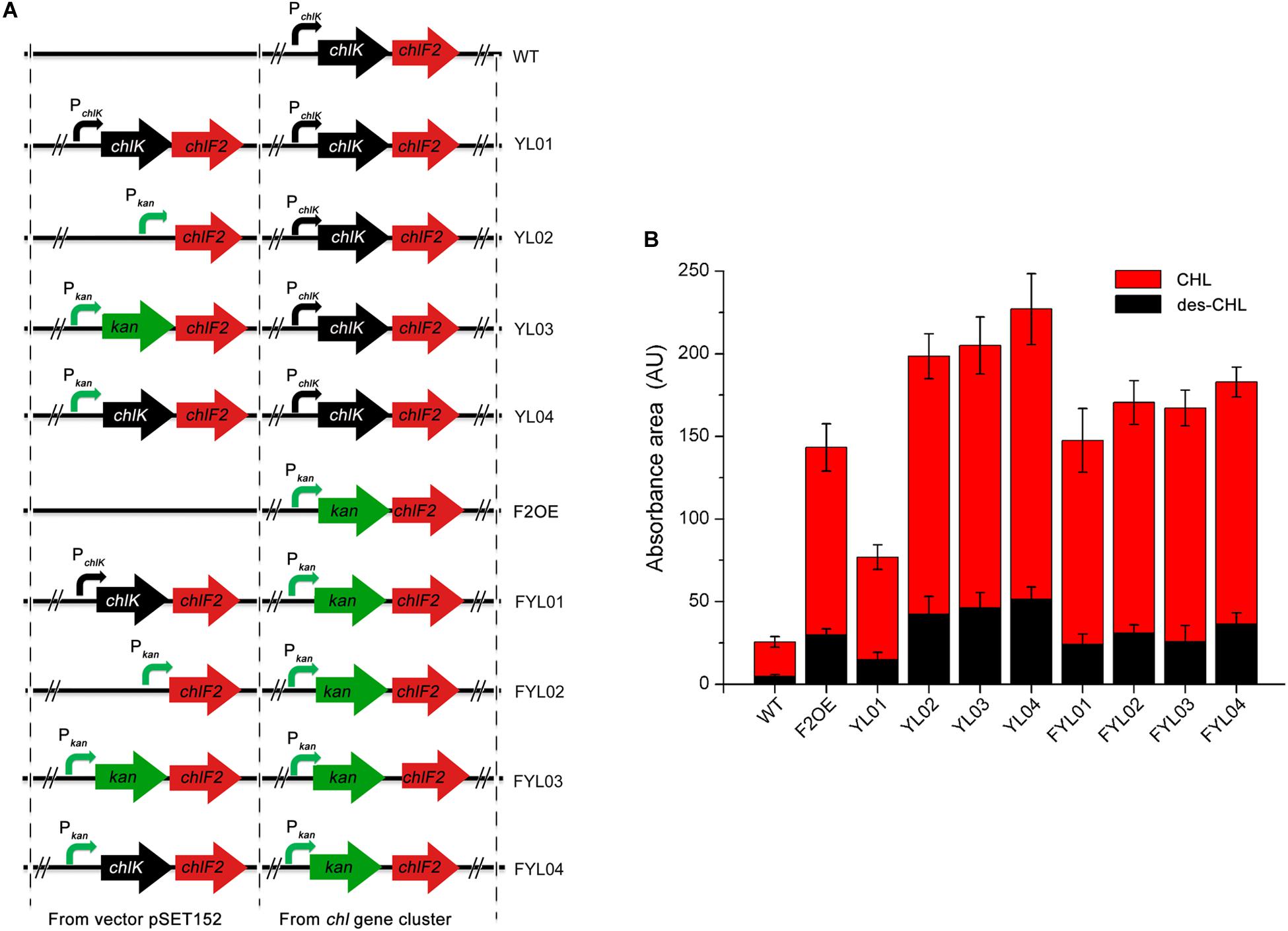
Figure 5. Comparison of antibiotics production between WT and its recombinant over-expression strains. (A) The constructions of chlF2 over-expression strains. (B) Absorbance area of des-CHL and CHL detected by HPLC in WT and its derivatives. CHL, chlorothricin; des-CHL, deschloro-chlorothricin.
Since F2OE exhibited much higher production of CHL than WT, it was consequently used as an alternative host strain to further improve the antibiotics production. The following four integrative plasmids pSET152:PchlK-chlK-chlF2, pSET152:Pkan-chlF2, pSET152:Pkan-kan-chlF2, and pSET152:Pkan-chlK-chlF2 were separately introduced into F2OE to generate strains FYL01, FYL02, FYL03 and FYL04 (Figure 5A). HPLC analysis indicated that a range of 590%∼700% improvement of CHL production was observed in these strains compared to WT strain, even though FYL02, FYL03, and FYL04 did not exhibit higher production of des-CHL and CHL than YL02, YL03, and YL04 (Figure 5B), implying strong promoter and cellular tolerance are crucial elements. Thus, YL04 was selected as the high-yield producing strains of CHL and des-CHL for further studies.
Chlorothricin and des-CHL are two major spirotetronate components. To determine their bioactivities, CHL and des-CHL were purified from YL04 high-producing strain, and their antibacterial and anticancer activities were evaluated. MIC values were determined against several Gram-positive bacteria, which ranged from 31.25 to 100.00 μg/ml. CHL showed improved antibacterial activity in comparison with des-CHL against B. subtilis, B. cereus and S. aureus (Table 1). For instance, MIC of CHL was 31.25 μg/ml against the above three indicator strains, while MIC of des-CHL was 62.5 μg/ml against B. subtilis and B. cereus (Table 1). CHL and des-CHL showed less than 80% inhibitory activity at the maximum tested concentration 100 μg/ml against S. epidermidis, S. pyogenes, and S. pneumoniae. In addition, we could not detect any biological activity against P. aeruginosa PA14, A. longipes CGMCC 3.2946, and C. albicans CGMCC 2.4159 (Table 1). Meanwhile, the CHL and des-CHL showed inhibitory activity against A549 (non-small-cell lung cancer) and Calu-3 (human lung adenocarcinoma cancer), HepG2 (liver cancer), and MCF-7 (breast cancer) with IC50 ranging from 22.5 μM to 181.3 μM (Figure 6). Overall, both compounds exhibited wide antibacterial or anticancer cell line spectrum, indicating their potential for drug development.
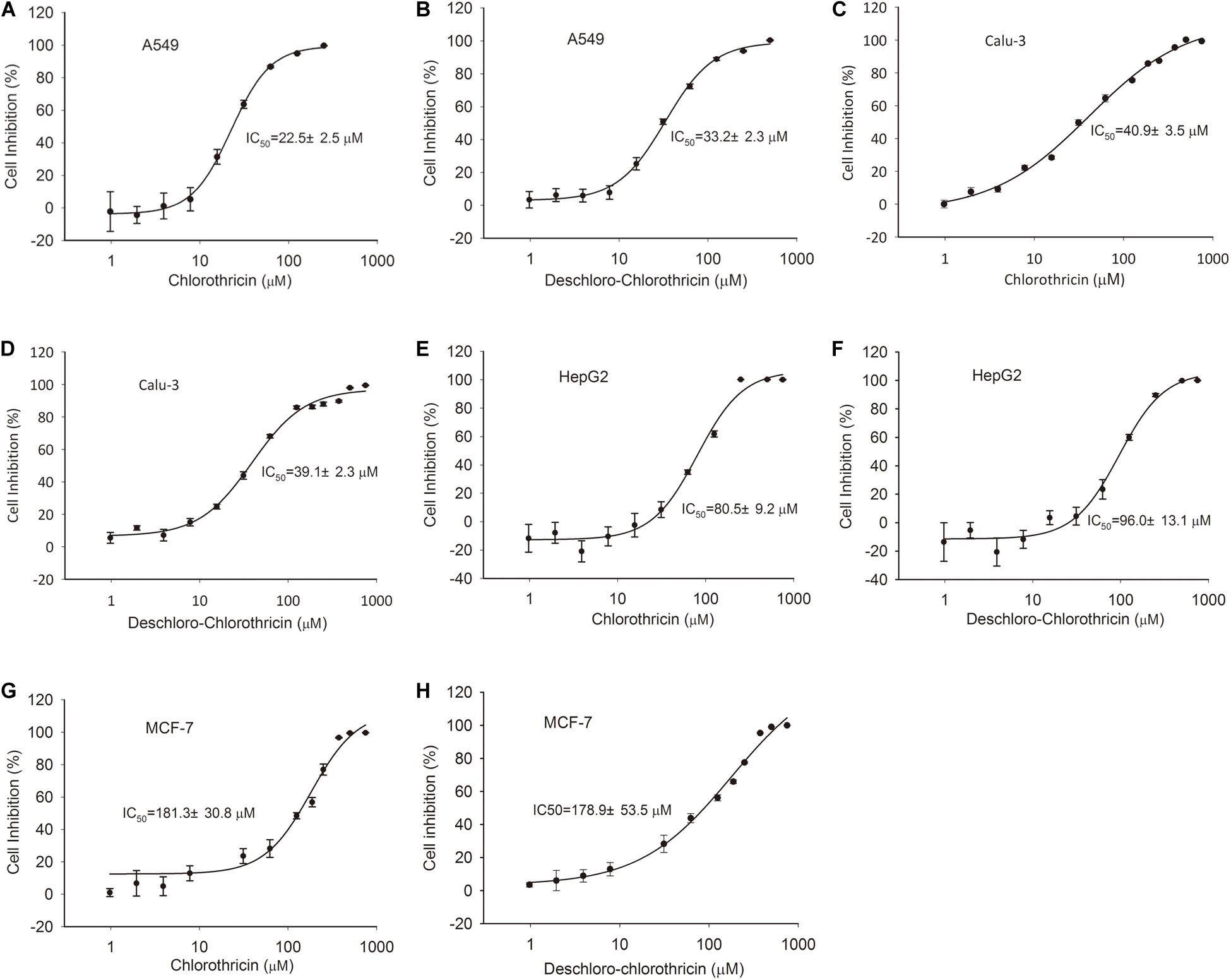
Figure 6. Cytotoxic assays of CHL and des-CHL. Cytotoxic assays of CHL against A549 (A), Calu-3 (C), HepG2 (E), and MCF-7 (G). Cytotoxic assays of des-CHL against A549 (B), Calu-3 (D), HepG2 (F), and MCF-7 (H). CHL, chlorothricin; des-CHL, deschloro-chlorothricin. A549 indicates human lung carcinoma cell line, Calu-3 indicates human lung adenocarcinoma cell line, HepG2 indicates human hepatocellular carcinoma cell line, MCF-7 indicates human breast adenocarcinoma cell line. IC50 (50% inhibitory concentration) values were labeled. The error bars indicated the standard deviations (SD) of three independent experiments.
As aforementioned, over-expression of chlK-chlF2 or kan-chlF2 under the control of Pkan led to accumulation of des-CHL. Because CHL exhibits higher activity than des-CHL against B. cereus, B. subtilis, and S. aureus, it is necessary to promote the conversion of des-CHL to CHL. chlB4 encoding halogenase is a key cluster-situated gene that responsible for the halogenation of MSA to form CHL. We speculated that the possible insufficient expression of halogenase ChlB4 or shortage of chloride ions in the medium could be a limiting factor for the accumulation of des-CHL. Thus, in order to further improve the production of CHL, the chlB4 was first over-expressed under the control of Pkan in F2OE to generate B4F2OE strain (Figure 7A). The result showed that both des-CHL and CHL of B4F2OE increased 7% and 29% compared with that of F2OE, respectively (Figure 7B). When optimal concentration of 0.05% NaCl as the chloride reservoir was added into the medium before fermentation, des-CHL production in B4F2OEN was markedly decreased about 52% in comparison to that in F2OE, while CHL increased 41% (Figure 7B). These results indicated that combination of the two strategies together can efficiently promote the conversion of des-CHL to CHL. To investigate the transcriptional level of chlB4 in F2OE, F2OEN, B4F2OE, and B4F2OEN, RT-qPCR experiment showed that the transcripts of chlB4 in F2OEN, B4F2OE, and B4F2OEN were all upregulated in varying degrees at different time points (Figure 7C).
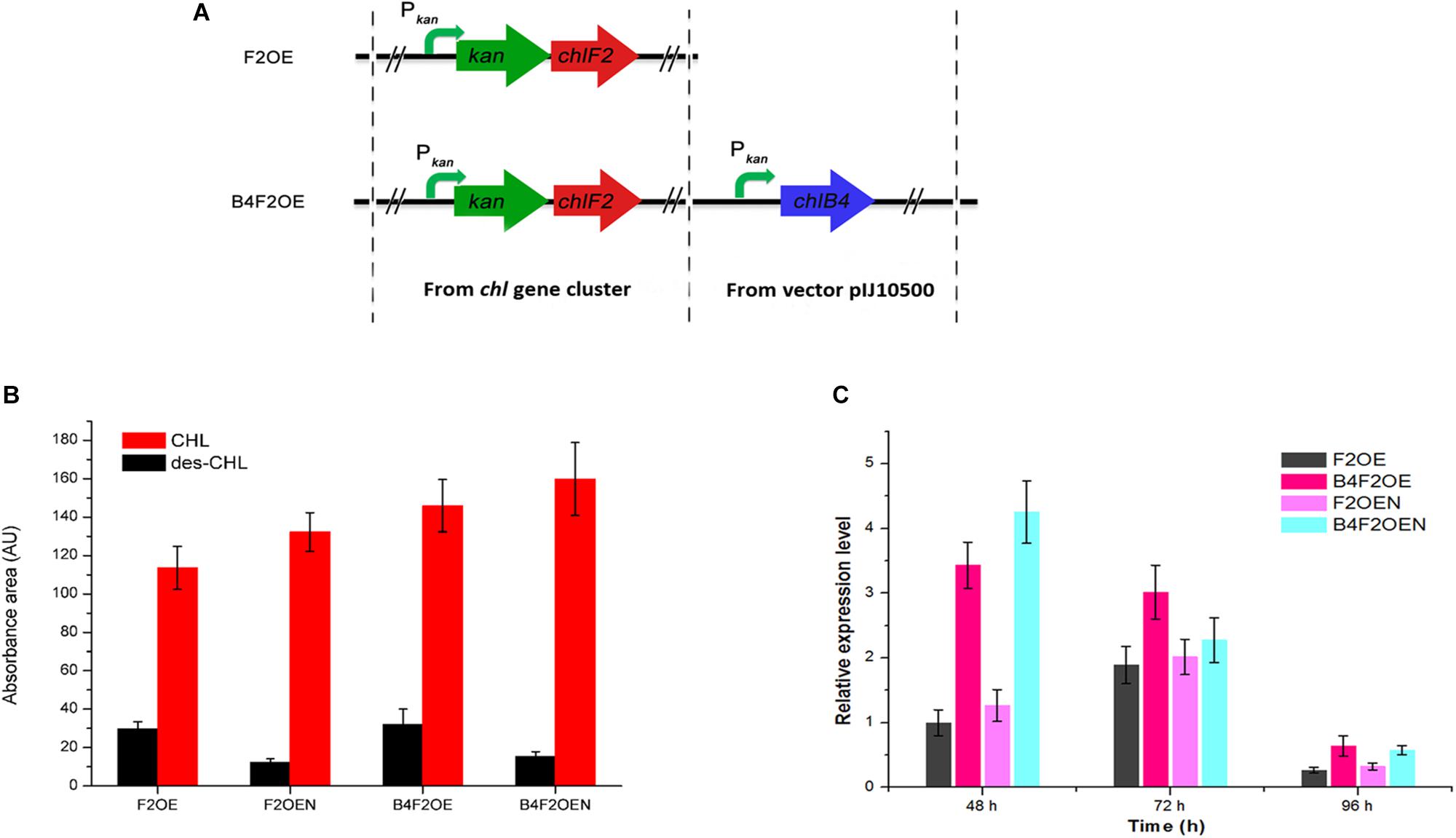
Figure 7. Metabolites analysis of WT and its recombinant strains. (A) The construction of B4F2OE strain. (B) HPLC analysis of the extracts of WT and its recombinant strains. des-CHL indicates deschloro-chlorothricin, CHL indicates chlorothricin. (C) RT-qPCR analysis of chlB4 in F2OE, F2OEN, B4F2OE and B4F2OEN. WT, S. antibioticus DSM 40725; WTN, WT with the addition of 0.05% NaCl when fermentation; F2OE, chlF2 over-expression strain; F2OEN, F2OE with the addition of 0.05% NaCl when fermentation; B4F2OE, halogenase coding gene chlB4 and chlF2 co-expression strain, B4F2OEN, B4F2OE with the addition of 0.05% NaCl when fermentation.
Streptomyces antibiotic regulatory proteins (SARPs) are a specific family of paralogous proteins that are associated with antibiotic biosynthesis in many streptomycetes. SARPs are widespread in Streptomyces with a varying length (from less than 300 aa to more than 1000 aa) and have three major functional domains (a N-terminal DNA binding domain, a central AAA domain and a conserved C-terminal domain of unknown function) (Liu et al., 2013). As we know, the biosynthesis of antibiotics is often hierarchically controlled by multi-level regulators in Streptomyces (Li C. et al., 2019), some of which are cascade regulation. For example, SARP family regulator PolY can activate PolR, and the latter can regulate the transcription of the whole polyoxin biosynthetic gene cluster to control polyoxin biosynthesis in Streptomyces cacaoi subsp. asoensis (Li R. et al., 2009, 2010). Some regulators exercise synergistic regulation with other proteins, such as OvmZ, its interplay with OvmW can activate the transcription of ovmOI-ovmH operon to control oviedomycin production (Xu et al., 2017). Two pathway-specific regulatory genes chlF1 and chlF2 are included in chl gene cluster, so we are curious to know the relationship between ChlF1 and ChlF2. First of all, GUS activity assays demonstrated that the expression of chlF1 was not regulated by ChlF2, while our previous EMSA experiments had illustrated that ChlF1 directly represses the transcription of chlK (Li Y. et al., 2016). Since chlK and chlF2 are co-transcribed, chlF2 was speculated to be regulated by ChlF1 through controlling the PchlK promoter. Subsequent RT-qPCR analysis confirmed that the transcription of chlF2 was negatively controlled by ChlF1 (Supplementary Figure S5). However, unlike chlF2, the over-expression of chlF1 driven by its own promoter did not lead to remarkable improvement of CHL production. Therefore, we speculated that the increase of CHL production in F2OE or YL04 is ascribed to the transcriptional enhancement of several structural genes activated by ChlF2 directly. Meanwhile, replacing PchlK with stronger promoter Pkan could help relieve the repression of ChlF1 on ChlF2.
As aforementioned, co-expression of both chlK and chlF2 or co-expression of both kan and chlF2 significantly increased the yield of des-CHL and CHL. It is likely that the ChlK or KanP may act as ‘helper’ of ChlF2 or form heterodimer with ChlF2. To assess if ChlK and ChlF2 or KanP and ChlF2 interacted, bacterial two-hybrid system based on the reconstitution of activity of adenylate cyclase in E. coli BTH101 (cya-) was performed as described in the Supplementary Material. chlK and kan were respectively fused to the T25 domain of an adenylate cyclase coding gene to generate plasmids pKT25-chlK and pKT25-kan, and chlF2 was fused to the T18 domain of the adenylate cyclase coding gene to generate plasmid pUT18-chlF2. Once the two proteins interacted, adenylate cyclase activity would be restored due to its T25 domain and T18 domain getting closer and then the strain would exhibit blue color on LB plate containing X-Gal (Supplementary Figure S6A). As a result, neither SZ-kanF2 nor SZ-KF2 strain exhibited blue color compared to the positive control SZ-zip, implying that ChlF2 and ChlK or ChlF2 and KanP don’t interact with each other in vivo (Supplementary Figure S6B). So it is unlikely that ChlK and KanP could facilitate CHL production via forming heterodimer with ChlF2. However, an alternative hypothesis about ChlK coordination with ChlF2 may be proposed based on the enzymatic activity. ChlK is a type II thioesterase and plays an ‘editor’ role to ensure the specific loading efficiency via hydrolyzing aberrantly loaded substrates. Our previous study showed that in-frame deletion of chlK resulted in declined CHL production, but not abolished completely (Li Y. et al., 2016), while over-expression of chlK along with chlF2 in YL-series or FYL-series strains did improve CHL production. One of the underlying mechanisms about co-expression of ChlK and ChlF2 could be considered to enhance substrate loading efficiency and accuracy during the chlorothricolide or MSA (methylsalicylic acid) biosynthesis, and increase the enzyme activity of several target genes of ChlF2 which in turn enhanced the CHL production.
Since multiple components with different potency are usually co-produced as a mixture in the manufacture process of most microbial antibiotics. The selective enhancement of the desired components and reduction of the undesired components are always expected in metabolic engineering (Cropp et al., 2001). Thus, it is very important to confirm the crucial switch that controls the metabolic flux toward the desired antibiotics and then genetic manipulation of the corresponding genes. In WT, chlB4 encoding halogenase is a key cluster-situated gene for catalyzing C5-chlorination of MSA. Its over-expression combined with the addition of NaCl facilitated the conversion of des-CHL to CHL. It can be perceived that more copies of chlB4 and optimization of the medium component may lead to an even higher yield. Moreover, since a high proportion of CHL is accumulated intracellularly rather than being exported out of the cells, the overproduced CHL may cause feedback regulation as described in our previous study (Li Y. et al., 2016), enhancing the cellular tolerance in combination with the above strategies would be promising to further improve CHL yield.
Taken together, the clarification of the regulatory mechanisms paves a road to further optimize the corresponding genes for strain improvement beneficial for commercial application. The findings also provide basis for investigating the biosynthesis and regulation of other spirotetronate family of antibiotics.
All datasets generated for this study are included in the article/Supplementary Material.
YL carried out most of the experiments, analyzed the primary data, and wrote the draft manuscript. JHZ performed the anticancer experiment and revised the manuscript. JZZ and HG constructed part of recombinant plasmids and strains related to GUS assays. WL conceived the partial research work. HT conceived the whole research work and revised the manuscript. All authors read and approved the final manuscript.
This work was supported by the National Natural Science Foundation of China (Grant Nos. 31800029 and 31771378) and National Key Research and Development Program of China (2018YFA0901900).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We are grateful to Dr. Zhoujie Xie, and Professor Luyan Ma for kindly providing strains (Streptococcus pneumoniae 010, Streptococcus pyogenes #2, Staphylococcus epidermidis ATCC 35984, Pseudomonas aeruginosa PA14).
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fbioe.2020.01013/full#supplementary-material
Bate, N., Stratigopoulos, G., and Cundliffe, E. (2002). Differential roles of two SARP-encoding regulatory genes during tylosin biosynthesis. Mol. Microbiol. 43, 449–458. doi: 10.1046/j.1365-2958.2002.02756.x
Braddock, A. A., and Theodorakis, E. A. (2019). Marine spirotetronates: biosynthetic edifices that inspire drug discovery. Mar. Drugs 17:232. doi: 10.3390/md17040232
Chen, Y., Deng, W., Wu, J., Qian, J., Chu, J., Zhuang, Y., et al. (2008). Genetic modulation of the overexpression of tailoring genes eryK and eryG leading to the improvement of erythromycin A purity and production in Saccharopolyspora erythraea fermentation. Appl. Environ. Microbiol. 74, 1820–1828. doi: 10.1128/AEM.02770-07
Cropp, A., Chen, S., Liu, H., Zhang, W., and Reynolds, K. A. (2001). Genetic approaches for controlling ratios of related polyketide products in fermentation processes. J. Ind. Microbiol. Biotechnol. 27, 368–377. doi: 10.1038/sj.jim.7000206
Ding, W., Lei, C., He, Q., Zhang, Q., Bi, Y., and Liu, W. (2010). Insights into bacterial 6-methylsalicylic acid synthase and its engineering to orsellinic acid synthase for spirotetronate generation. Chem. Biol. 17, 495–503. doi: 10.1016/j.chembiol.2010.04.009
Guan, H., Li, Y., Zheng, J., Liu, N., Zhang, J., and Tan, H. (2019). Important role of a LAL regulator StaR in the staurosporine biosynthesis and high-production of Streptomyces fradiae CGMCC 4.576. Sci. China Life Sci. 62, 1638–1654. doi: 10.1007/s11427-019-1597-6
Guo, J., Zhao, J., Li, L., Chen, Z., Wen, Y., and Li, J. (2010). The pathway specific regulator AveR from Streptomyces avermitilis positively regulates avermectin production while it negatively affects oligomycin biosynthesis. Mol. Genet. Genomics 283, 123–133. doi: 10.1007/s00438-009-0502-2
Igarashi, Y., Iida, T., Oku, N., Watanabe, H., Furihata, K., and Miyanouchi, K. (2012). Nomimicin, a new spirotetronate-class polyketide from an actinomycete of the genus Actinomadura. J. Antibiot. 65, 355–359. doi: 10.1038/ja.2012.30
Jia, X. Y., Tian, Z. H., Shao, L., Qu, X. D., Zhao, Q. F., Tang, J., et al. (2006). Genetic characterization of the chlorothricin gene cluster as a model for spirotetronate antibiotic biosynthesis. Chem. Biol. 13, 575–585. doi: 10.1016/j.chembiol.2006.03.008
Krause, J., Handayani, I., Blin, K., Kulik, A., and Mast, Y. (2020). Disclosing the potential of the SARP-type regulator PapR2 for the activation of antibiotic gene clusters in Streptomycetes. Front. Microbiol. 18:225. doi: 10.3389/fmicb.2020.00225
Lacoske, M. H., and Theodorakis, E. A. (2015). Spirotetronate polyketides as leads in drug discovery. J. Nat. Prod. 78, 562–575. doi: 10.1021/np500757w
Li, C., He, H., Wang, J., Liu, H., Wang, H., Zhu, Y., et al. (2019). Characterization of a LAL-type regulator NemR in nemadectin biosynthesis and its application for increasing nemadectin production in Streptomyces cyaneogriseus. Sci. China Life Sci. 62, 394–405. doi: 10.1007/s11427-018-9442-9
Li, D., Zhang, J., Tian, Y., and Tan, H. (2019). Enhancement of salinomycin production by ribosome engineering in Streptomyces albus. Sci. China Life Sci. 62, 276–279. doi: 10.1007/s11427-018-9474-7
Li, R., Liu, G., Xie, Z., He, X., Chen, W., Deng, Z., et al. (2010). PolY, a transcriptional regulator with ATPase activity, directly activates transcription of polR in polyoxin biosynthesis in Streptomyces cacaoi. Mol. Microbiol. 75, 349–364. doi: 10.1111/j.1365-2958.2009.06968.x
Li, R., Xie, Z., Tian, Y., Yang, H., Chen, W., You, D., et al. (2009). polR, a pathway-specific transcriptional regulatory gene, positively controls polyoxin biosynthesis in Streptomyces cacaoi subsp. asoensis. Microbiology 155(Pt 6), 1819–1831. doi: 10.1099/mic.0.028639-0
Li, S., Xiao, J., Zhu, Y., Zhang, G., Yang, C., Zhang, H., et al. (2013). Dissecting glycosylation steps in lobophorin biosynthesis implies an iterative glycosyltransferase. Org. Lett. 15, 1374–1377. doi: 10.1021/ol400342e
Li, Y., Li, J., Tian, Z., Xu, Y., Zhang, J., Liu, W., et al. (2016). Coordinative modulation of chlorothricin biosynthesis by binding of the glycosylated intermediates and end product to a responsive regulator ChlF1. J. Biol. Chem. 291, 5406–5417. doi: 10.1074/jbc.M115.695874
Li, Y., and Tan, H. (2017). Biosynthesis and molecular regulation of secondary metabolites in microorganisms. Sci. China Life Sci. 60, 935–938. doi: 10.1007/s11427-017-9115-x
Liu, G., Chater, K. F., Chandra, G., Niu, G., and Tan, H. (2013). Molecular regulation of antibiotic biosynthesis in Streptomyces. Microbiol. Mol. Biol. Rev. 77, 112–143. doi: 10.1128/MMBR.00054-12
Lu, C., Liao, G., Zhang, J., and Tan, H. (2015). Identification of novel tylosin analogues generated by a wblA disruption mutant of Streptomyces ansochromogenes. Microb. Cell Fact 14:173. doi: 10.1186/s12934-015-0359-5
Myronovskyi, M., Welle, E., Fedorenko, V., and Luzhetskyy, A. (2011). Beta-glucuronidase as a sensitive and versatile reporter in Actinomycetes. Appl. Environ. Microbiol. 77, 5370–5383. doi: 10.1128/AEM.00434-11
Shao, L., Qu, X. D., Jia, X. Y., Zhao, Q. F., Tian, Z. H., Wang, M., et al. (2006). Cloning and characterization of a bacterial iterative type I polyketide synthase gene encoding the 6-methylsalicyclic acid synthase. Biochem. Biophys. Res. Commun. 345, 133–139. doi: 10.1016/j.bbrc.2006.04.069
Sherwood, E. J., and Bibb, M. J. (2013). The antibiotic planosporicin coordinates its own production in the actinomycete Planomonospora alba. Proc. Natl. Acad. Sci. U.S.A. 110, E2500–E2509. doi: 10.1073/pnas.1305392110
Sun, Y., Zhou, X., Dong, H., Tu, G., Wang, M., Wang, B., et al. (2003). A complete gene cluster from Streptomyces nanchangensis NS3226 encoding biosynthesis of the polyether ionophore nanchangmycin. Chem. Biol. 10, 431–441. doi: 10.1016/s1074-5521(03)00092-9
Tanaka, A., Takano, Y., Ohnishi, Y., and Horinouchi, S. (2007). AfsR recruits RNA polymerase to the afsS promoter: a model for transcriptional activation by SARPs. J. Mol. Biol. 369, 322–333. doi: 10.1016/j.jmb.2007.02.096
Tian, Z., Sun, P., Yan, Y., Wu, Z., Zheng, Q., Zhou, S., et al. (2015). An enzymatic [4+2] cyclization cascade creates the pentacyclic core of pyrroindomycins. Nat. Chem. Biol. 11, 259–265. doi: 10.1038/nchembio.1769
Wu, Q., Wu, Z., Qu, X., and Liu, W. (2012). Insights into pyrroindomycin biosynthesis reveal a uniform paradigm for tetramate/tetronate formation. J. Am. Chem. Soc. 134, 17342–17345. doi: 10.1021/ja304829g
Xu, J., Zhang, J., Zhuo, J., Li, Y., Tian, Y., and Tan, H. (2017). Activation and mechanism of a cryptic oviedomycin gene cluster via the disruption of a global regulatory gene, adpA, in Streptomyces ansochromogenes. J. Biol. Chem. 292, 19708–19720. doi: 10.1074/jbc.M117.809145
Yi, X., Zhao, Q., Tian, Z., Jia, X., Cao, W., Liu, W., et al. (2019). Insights into the functionalization of the methylsalicyclic moiety during the biosynthesis of chlorothricin by comparative kinetic assays of the activities of two KAS III-like acyltransferases. Chin. J. Chem. 37, 821–826. doi: 10.1002/cjoc.201900134
Zhang, Y., He, H., Liu, H., Wang, H., Wang, X., and Xiang, W. (2016). Characterization of a pathway-specific activator of milbemycin biosynthesis and improved milbemycin production by its overexpression in Streptomyces bingchenggensis. Microb. Cell Fact 15:152. doi: 10.1186/s12934-016-0552-1
Keywords: Streptomyces antibioticus, gene regulation, chlorothricin, high production, halogenase
Citation: Li Y, Zhang J, Zheng J, Guan H, Liu W and Tan H (2020) Co-expression of a SARP Family Activator ChlF2 and a Type II Thioesterase ChlK Led to High Production of Chlorothricin in Streptomyces antibioticus DSM 40725. Front. Bioeng. Biotechnol. 8:1013. doi: 10.3389/fbioe.2020.01013
Received: 11 June 2020; Accepted: 03 August 2020;
Published: 21 August 2020.
Edited by:
Xu-Ming Mao, Zhejiang University, ChinaReviewed by:
Chin-Yuan Chang, National Chiao Tung University, TaiwanCopyright © 2020 Li, Zhang, Zheng, Guan, Liu and Tan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Huarong Tan, dGFuaHJAaW0uYWMuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.