
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Bioeng. Biotechnol. , 30 July 2020
Sec. Bioprocess Engineering
Volume 8 - 2020 | https://doi.org/10.3389/fbioe.2020.00873
This article is part of the Research Topic CAZymes in Biorefinery: From Genes to Application View all 8 articles
 Birgitte Zeuner1
Birgitte Zeuner1 Thore Bach Thomsen1
Thore Bach Thomsen1 Mary Ann Stringer2
Mary Ann Stringer2 Kristian B. R. M. Krogh2
Kristian B. R. M. Krogh2 Anne S. Meyer1*
Anne S. Meyer1* Jesper Holck1
Jesper Holck1Fungal genomes often contain several copies of genes that encode carbohydrate active enzymes having similar activity. The copies usually have slight sequence variability, and it has been suggested that the multigenecity represents distinct reaction optima versions of the enzyme. Whether the copies represent differences in substrate attack proficiencies of the enzyme have rarely been considered. The genomes of Aspergillus species encode several pectin lyases (EC 4.2.2.10), which all belong to polysaccharide lyase subfamily PL1_4 in the CAZy database. The enzymes differ in terms of sequence identity and phylogeny, and exhibit structural differences near the active site in their homology models. These enzymes catalyze pectin degradation via eliminative cleavage of the α-(1,4) glycosidic linkages in homogalacturonan with a preference for linkages between methyl-esterified galacturonate residues. This study examines four different pectin lyases (PelB, PelC, PelD, and PelF) encoded by the same Aspergillus sp. (namely A. luchuensis), and further compares two PelA pectin lyases from two related Aspergillus spp. (A. aculeatus and A. tubingensis). We report the phylogeny, enzyme kinetics, and enzymatic degradation profiles of the enzymes’ action on apple pectin, citrus pectin, and sugar beet pectin. All the pectin lyases exerted highest reaction rate on apple pectin [degree of methoxylation (DM) 69%, degree of acetylation (DAc) 2%] and lowest reaction rate on sugar beet pectin (DM 56%, DAc 19%). Activity comparison at pH 5–5.5 produced the following ranking: PelB > PelA > PelD > PelF > PelC. The evolution of homogalacturonan-oligomer product profiles during reaction was analyzed by liquid chromatography with mass spectrometry (LC-MS) detection. This analyses revealed subtle differences in the product profiles indicating distinct substrate degradation preferences amongst the enzymes, notably with regard to acetyl substitutions. The LC-MS product profiling analysis thus disclosed that the multigenecity appears to provide the fungus with additional substrate degradation versatility. This product profiling furthermore represents a novel approach to functionally compare pectin-degrading enzymes, which can help explain structure-function relations and reaction properties of disparate copies of carbohydrate active enzymes. A better understanding of the product profiles generated by pectin modifying enzymes has significant implications for targeted pectin modification in food and biorefinery processes.
Pectin lyases (EC 4.2.2.10) catalyze cleavage of the α-(1,4) glycosidic linkages between methyl-esterified galacturonic acid (GalA) units in the homogalacturonan backbone of pectin through a β-elimination reaction (Figure 1). The β-elimination leads to formation of a 4,5-unsaturated 6-O-methylated galacturonide molecule in the non-reducing end of one of the cleavage products, which has absorbance maximum at 235 nm. Pectin lyases thus prefer linkages between methyl-esterified (methoxylated) GalA units, but also catalyze bond cleavage at sites where only the GalA moiety in the +1 subsite is methoxylated, albeit with lower specific activity (Mutenda et al., 2002; Van Alebeek et al., 2002). Accordingly, pectin lyase activity is heavily dependent on the degree of methoxylation (DM) of the substrate, and also on the methyl ester distribution: A blockwise distribution of methoxylated GalA units results in higher pectin lyase activity than a random distribution (Mutenda et al., 2002; Van Alebeek et al., 2002). Detailed studies on Aspergillus niger pectin lyase A have further revealed requirement for methoxylated GalA in subsites +1 and +3, whereas subsites +2, +4, and -1 to -4 appear to allow accommodation of unesterified GalA units (Van Alebeek et al., 2002).
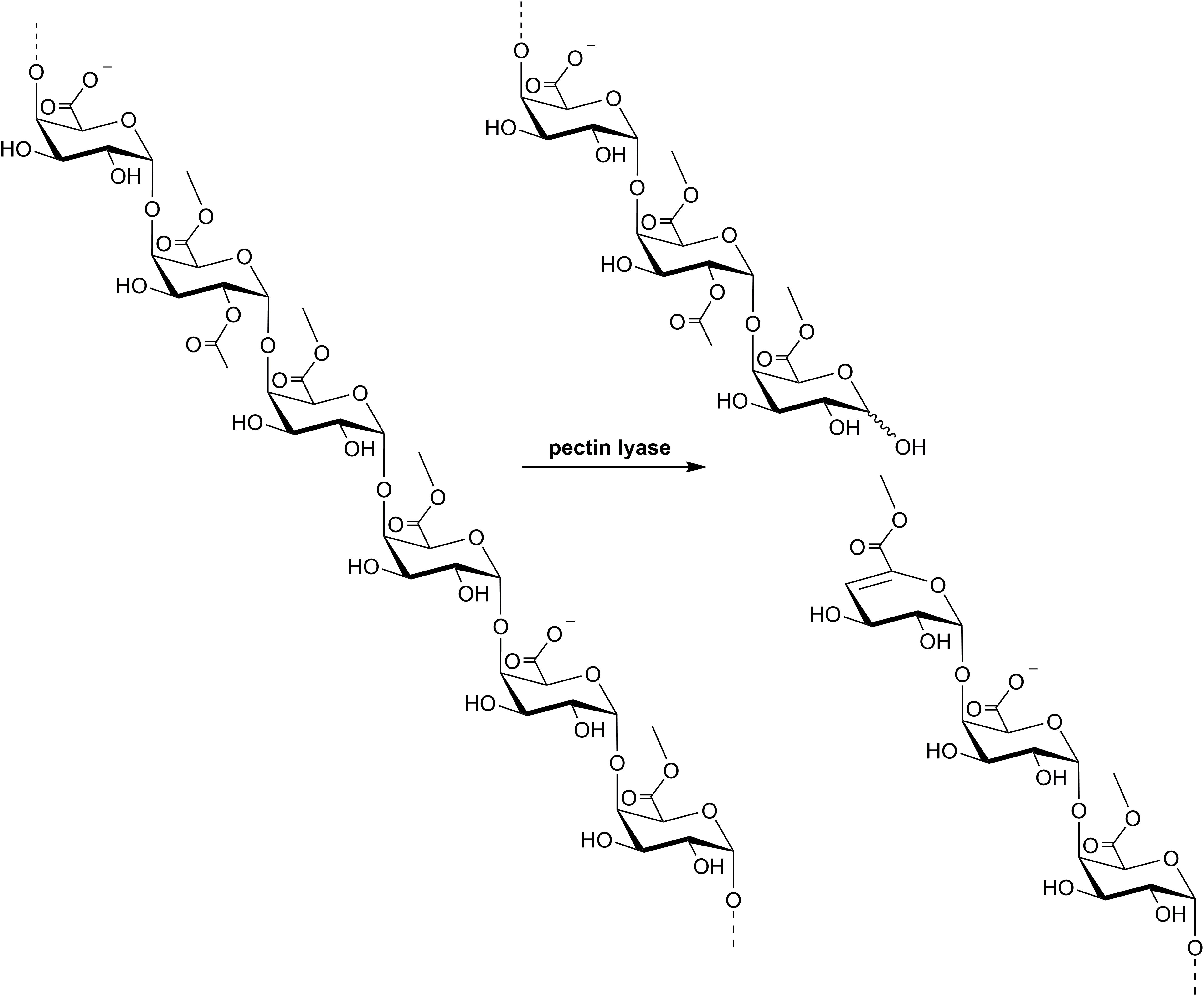
Figure 1. Stylized illustration of a pectin lyase-catalyzed β-elimination reaction on pectin. GalA can be methoxylated on O-6 and acetylated on O-2 and/or O-3. The illustration depicts a pectin fragment substrate with a degree of methoxylation (DM) of 67% and a degree of acetylation (DAc) of 17%, whereas the true substitution degrees vary between pectins of different sources (Table 2). Note the preference for cleavage between methylesterified GalA moieties as outlined in more detail in the text.
Important industrial applications of pectin lyases [usually employed in mixtures with other pectin depolymerizing enzymes, notably endo-polygalacturonases (EC 3.2.1.15)] include wine and juice pre-press maceration as well as juice clarification (Mantovani et al., 2005; Kassara et al., 2019). Uses of enzymes, including pectinase blends and pectin lyases, in food and beverage processing constitute about 30% of the total enzymes market, and a conservative estimate is that the global market value for pectin modifying enzymes is above US$ 100 million and growing. Pectin lyases are also actively investigated for special new uses such as bio-degumming and cleaning of bast fibers (Kohli and Gupta, 2019), and for production of distinct pectic oligosaccharides having prebiotic or anti-inflammatory properties (Chung et al., 2017; Tan et al., 2018). Most commercial pectinases originate from filamentous fungi, notably several derive from the Aspergillus section Nigri such as Aspergillus niger and A. aculeatus (Sandri et al., 2011). These fungi are specialized in secreting a broad spectrum of enzymes, including pectin-degrading enzymes, which can degrade and utilize the surrounding biomass (Benoit et al., 2012; de Vries et al., 2017; Kowalczyk et al., 2017).
Genome sequencing of the industrial enzyme-producing strain A. niger CBS 513.88 has revealed that this fungus contains multiple genes encoding for the same enzymatic activity (Pel et al., 2007; Andersen et al., 2012). Of the 131 genes encoding secreted polysaccharide-active enzymes, five (3.8%) encode pectin lyases; these five are denominated pelA, pelB, pelC, pelD, and pelF (Pel et al., 2007). Expression profiling of strains derived from A. niger CBS 120.49 has shown that the genes are active at different stages of pectin degradation, suggesting different regulatory mechanisms either related to extent of substrate degradation or to the increased acidification of the medium during pectinase action (de Vries et al., 2002). From studies on closely related Aspergillus species, it has furthermore become evident that the different species have different enzymatic strategies for efficient degradation of the same substrate (Benoit et al., 2015; Barrett et al., 2020b). Characterization of the individual gene products can therefore be used to efficiently select enzymes for a specific application, e.g., optimal degradation of a given substrate under a certain set of reaction conditions.
We hypothesized that the multiple copies (multigenecity) of genes encoding for pectin lyases in Aspergilli might be related to small differences in substrate attack preferences of the enzymes. Such differences could reflect adaptation to particular substitution patterns on the pectin backbone and materialize as minor variations in the active site region of the enzymes. This study was undertaken to examine PelB, PelC, PelD, and PelF from A. luchuensis in terms of substrate specificity, oligomer product profiles, pH-temperature optima, kinetic constants, active site and thermal stability. Two PelA pectin lyases from A. aculeatus and A. tubingensis were included for comparison. A. tubingensis and A. luchuensis both belong to the A. niger clade (Varga et al., 2011; Hong et al., 2013) and the pectin lyases studied here show > 90% sequence identity to the corresponding enzyme proteins from A. niger CBS 513.88 (Table 1). A. aculeatus is less closely related, yet all three Aspergillus spp. belong to section Nigri (Varga et al., 2011; Hong et al., 2013).
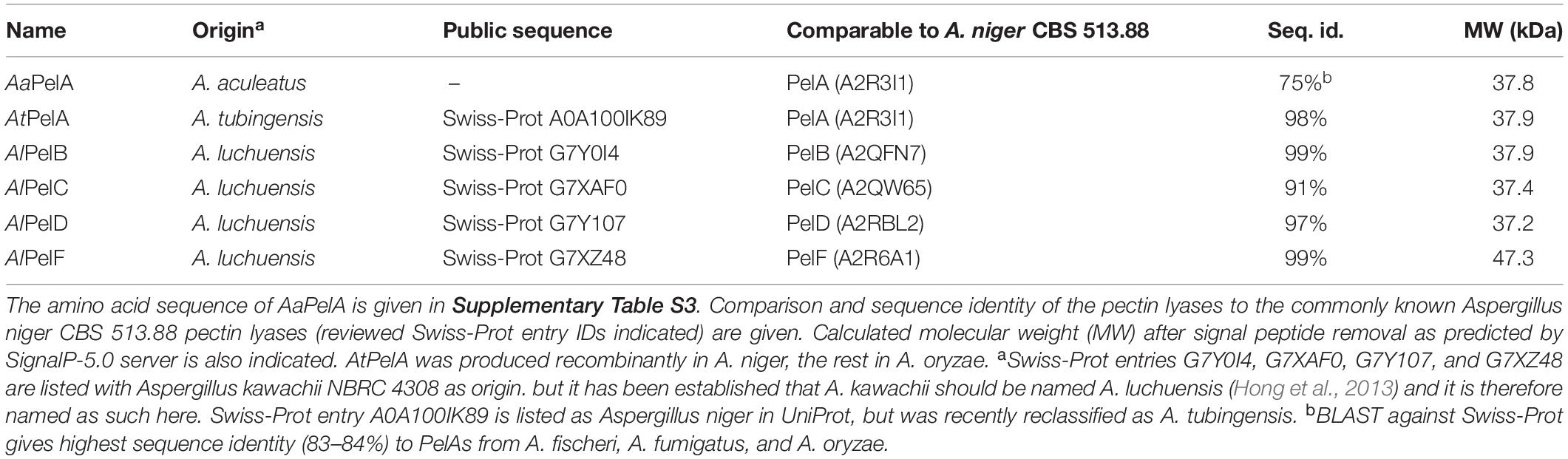
Table 1. Name, origin, and publicly available sequence entries of the Aspergillus sp. pectin lyases used in the current work.
Sugar beet pectin was provided by CP Kelco ApS (Lille Skensved, Denmark). Pectin from apple with 70–75% esterification (apple pectin), pectin from citrus peel (citrus pectin), and polygalacturonic acid and all other chemicals were purchased from Sigma-Aldrich (Merck) (Darmstadt, Germany).
Genes encoding the full length native pectin lyases (Table 1) were PCR amplified using gene specific primers from either a cDNA pool from Aspergillus tubingensis or genomic DNA from Aspergillus luchuensis and Aspergillus aculeatus. The amplified full-length coding sequences were cloned into Aspergillus expression vectors and recombinantly expressed in Aspergillus niger for AtPelA and Aspergillus oryzae for the remaining pectin lyases. The enzymes were purified to SDS-PAGE electrophoretic purity (Supplementary Figure S1) using hydrophobic interaction chromatography followed by ion exchange chromatography.
All pectin lyases are classified in the CAZy polysaccharide lyase family 1 (PL1). PL1 comprises pectin lyases (EC 4.2.2.10), pectate lyases (EC 4.2.2.2), and exo-pectate lyases (EC 4.2.2.9). Based on phylogenetic analysis of the catalytic domain, PL1 is currently divided into 13 subfamilies, which to some extent reflects substrate specificity (Lombard et al., 2010). Following CAZy nomenclature, PL1 subfamily X is written PL1_X. Characterized pectin lyases are found in PL1_4 and PL1_8, both of which are monospecific subfamilies comprising pectin lyases only. The six Aspergillus pectin lyases studied here all belong to PL1_4.
The sequences of the six pectin lyases studied in the current work were compared to the reviewed Swiss-Prot entries by BLAST analysis from UniProt. Phylogenetic trees were made in CLC Main Workbench (QIAGEN, Hilden, Germany) using all reviewed Swiss-Prot entries for pectin lyases and the sequences of the pectin lyases studied in the current work. Homology models of the enzymes were generated using SWISS-MODEL (Waterhouse et al., 2018). Conserved amino acids in the substrate binding site were identified and colored using PyMOL (Schrödinger, New York, NY, United States). Homology models and crystal structures were aligned in PyMOL using the super function. Multiple sequence alignments were generated with ClustalW in MUSCLE (Madeira et al., 2019). Signal peptides were predicted using the SignalP 5.0 web server (Almagro Armenteros et al., 2019), and the theoretical molecular weight of the secreted enzymes were calculated from the amino acid sequence in CLC Main Workbench. Secondary structure prediction was performed with PSIPRED (Buchan and Jones, 2019). The CUPP webserver for functional annotation was accessed at cupp.info (Barrett and Lange, 2019; Barrett et al., 2020a).
Apple pectin, citrus pectin, and sugar beet pectin substrates were subjected to acid hydrolysis with 4% sulfuric acid following the NREL method (Sluiter et al., 2012). The resulting monosaccharide composition was determined by HPAEC-PAD using a CarboPac PA1 column (Dionex, Sunnyvale, CA, United States) as described previously (Zeuner et al., 2018).
In order to determine degree of methoxylation (DM) and degree of acetylation (DAc) of the pectin substrates, methyl and acetyl substitutions were released by saponification as described previously (Voragen et al., 1986). The released methanol and acetate were quantified by HPLC (Shimadzu, Kyoto, Japan) essentially as described previously (Agger et al., 2010) using an Aminex HPX-87H ion exclusion column (BioRad, Hercules, CA, United States). Elution was performed over 40 min with 4 mM sulfuric acid at a flow rate of 0.6 mL/min and 63°C. Compounds were detected by refractive index detection using methanol and acetate as external standards. The DM is defined as the percentage of methoxylated GalA moieties out of the total amount of GalA moieties. Similarly, the DAc is defined as the percentage of acetyl substituted GalA units.
The initial reaction rates of the pectin lyases were determined on four different pectin substrates: apple pectin, citrus pectin, sugar beet pectin, and polygalacturonic acid using a substrate concentration of 1 g/L in 100 mM sodium acetate buffer at pH 5. The enzyme dosage was 0.4% (w/w) E/S for AaPelA, AtPelA, and AlPelB, 12.5% (w/w) E/S for AlPelC, 0.625% (w/w) E/S for AlPelD, and 1.25% (w/w) E/S for AlPelF. The pectin lyase activity was monitored at 25°C for 10 min by measuring the change in absorbance at 235 nm using an Infinite 200Pro plate reader (Tecan, Männedorf, Switzerland). This change was converted to product formation using an extinction coefficient of 5500 M–1 cm–1 for the 4,5-unsaturated galacturonide product (Kester and Visser, 1994). One unit (U) is defined as the amount of enzyme which catalyzes the formation of one μmol of 4,5-unsaturated galacturonide product per min at pH 5.0 and 25°C. In turn, the reported specific activity is given as μmol product released per min per mg enzyme.
The pH-temperature optima of AaPelA, AtPelA, AlPelB, and AlPelD were determined by response surface methodology (RSM) using a two-factor face-centered central composite design of experiments (CCF) with three center points. Each sample was analyzed in duplicate. Statistical analysis, model generation, and graphic illustration was made in JMP 14 Pro (SAS, Cary, NC, United States). All reactions were performed with pectin solutions of 2 g/L where pH was adjusted by NaOH and HCl. For AtPelA and AlPelD a pH range of 3.5–5.5 was employed for all substrates. For AaPelA and AlPelB the pH range was 5.5–7.5, except for AaPelA on sugar beet pectin, where it was 4.5–6.5. The tested temperature range was 45–65°C for AaPelA, 50–70°C for AtPelA, 40–60°C for AlPelB, and 40–70°C for AlPelD. Enzyme dosages ranged from 0.0125 to 0.05% E/S depending on the initial substrate screening results. Reactions took place in preheated substrate solution for 5 min. The reaction was stopped by heating at 95°C. Finally, the extent of reaction was determined from the difference in absorbance at 235 nm when compared to an enzyme-free solution of the same substrate.
Kinetic constants were determined using the same reaction setup at the pH-temperature optima predicted for each enzyme on each substrate based on the RSM results; for AaPelA on sugar beet pectin where no model was generated, center point values (pH 5.5, 55°C) were used instead. Eight different pectin concentrations in the range from 0.25 to 10 g/L were used. The substrate concentration was converted to molar concentration of bonds between GalA moieties based on the monosaccharide composition (Table 2). Reactions were run in triplicates. Kinetic constants were determined from linear regression in a Hanes-Woolf plot of the data.
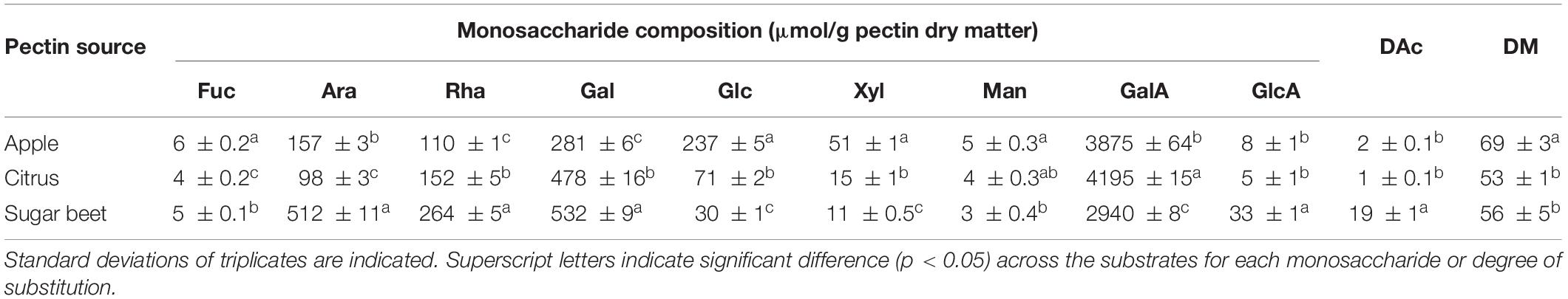
Table 2. Average monosaccharide composition in μmol/g pectin dry matter, degree of acetylation (DAc), and degree of methoxylation (DM) of pectin from apple, citrus, and sugar beet.
A thermal shift assay (TSA) (Lo et al., 2004) was used to determine the melting point (Tm) of the pectin lyases at pH 5. Purified pectin lyase samples were prepared for TSA by diluting to a standard concentration of 0.24 mg/mL in Milli-Q water. SYPRO Orange dye (S6650; Life Technologies, Carlsbad, CA, United States) was diluted 1:200 in assay buffer. The assay buffer was 0.1 M succinic acid, 0.1 M HEPES, 0.1 M CHES, 0.1 M CAPS, 0.15 M KCl, 1 mM CaCl2, and 0.01 % Triton X100, adjusted to pH 5. For measurement, 10 μl of diluted enzyme sample was combined with 20 μl of diluted dye in the well of a TSA assay plate (Roche LightCycler 480 Multiwell plate 96, white; Roche, Basel, Switzerland) and the plate was covered with optic sealing foil. Thermal ramping and fluorescence measurements were run in a Roche Lightcycler 480 II machine (Roche, Basel, Switzerland) running with a ramp from 25 to 99°C at a rate of 200°C/h. The data collected was analyzed by Roche LightCycler 480 software (release 1.5.0 SP4). All samples were analyzed in duplicate and averaged.
For AlPelF, a different analysis method was used for determining Tm, namely nano-differential scanning fluorimetry (nDSF). The enzyme was diluted to 0.5 mg/mL and buffer exchanged to 50 mM acetic acid (pH 5.0) using a PD SpinTrap G-25 column (GE Healthcare, Uppsala, Sweden). The nDSF experiments were done utilizing a Prometheus NT.48 (NanoTemper, Munich, Germany). The experiments were conducted from 20 to 95°C with a temperature gradient of 200°C/h. The transition temperatures (Tm values) were obtained from peak values derived from the first-derivative of the signal trace (350/330 nm fluorescence ratio or 330 nm fluorescence) using PR.ThermControl software (NanoTemper, Munich, Germany).
The pectin degradation product profiles were studied for all six pectin lyases using 10 g/L pectin (apple, citrus, or sugar beet) in acetate buffer at pH 5.5 and 40°C. An equimolar enzyme dosage of 60 nM (corresponding to 0.023% E/S for AtPelA) was used for all enzymes, but due to low activity enzyme dosages of 600 nM were also included for AlPelC and AlPelF. After 5 and 20 min, and 2 and 24 h, samples were taken out and the enzymes were inactivated by heating at 95°C for 10 min. The samples were centrifuged (14,000 g, 5 min) to remove insoluble particles.
For size exclusion chromatography (SEC) analysis of the pectin degradation profiles, the samples were diluted 4 times in the eluent buffer and filtered (0.22 μm). The SEC analysis was performed by injecting on a TSKgel G3000PW column (300 mm × 7.5 mm) equipped with a TSKgel PWH guard column (7.5 mm × 7.5 mm) (Tosoh Bioscience, Tokyo, Japan) using an UltiMate iso-3100 SD pump and an RI-101 refractive index detector. Elution took place at 40°C using 0.1 M NaNO3 with 0.02% NaN3 as eluent and a flow rate of 1 mL/min. Pullulan standards (180 Da to 110 kDa) were used as reference.
Identification and relative quantification of pectic oligosaccharides was performed by liquid chromatography electrospray ionization mass spectrometry (LC-ESI-MS) on an Amazon SL iontrap (Bruker Daltonics, Bremen Germany) coupled to an UltiMate 3000 UHPLC from Dionex (Sunnyvale, CA, United States). Samples of 10 μL were injected on a porous graphitized carbon column (Hypercarb PGC, 150 mm × 2.1 mm, 3 μm; Thermo Fisher Scientific, Waltham, MA, United States). The chromatography was performed at 0.4 mL/min at 70 °C on a three-eluent system with eluent A (water), eluent B (acetonitrile), and eluent C (100 mM ammonium acetate pH 5). The elution profile was as follows: 0–1 min, 0% B; 1–30 min, linear gradient to 30% B; 30–35 min, isocratic 30% B; 35–40 min, isocratic 0% B. In addition, a constant level of 10% C was maintained throughout the elution. For every four samples, a cleaning procedure of 0–15 min, isocratic 50% B and C; 15–40 min, isocratic 10% C was performed to avoid build-up of polymeric pectin. The electrospray was operated in negative or positive mode with UltraScan mode and a scan range from 100 to 2000 m/z, smart parameter setting of 500 m/z, capillary voltage at 4.5 kV, end plate off-set 0.5 kV, nebulizer pressure at 3.0 bar, dry gas flow at 12.0 L/min, and dry gas temperature at 280°C. Identification of observed compounds by m/z and MS2 fragmentation pattern was performed in DataAnalysis 4.2 and relative quantification based on intensity peaks in positive mode was performed in Compass QuantAnalysis 2.2 (Bruker Daltonics, Bremen Germany).
All samples were investigated with respect to their content of all confirmed and putative compounds. The intensity of each compound was defined as the area under the curve of a given extracted ion chromatogram. In order to investigate the product profile, or fingerprint, of each enzyme rather than their overall activity/productivity, all intensities were normalized with respect to the most abundant compound in a given enzyme-substrate-time combination. Data was clustered by hierarchical clustering with complete linkage on the euclidian distance matrix and visualized in the pheatmap-package using R version 3.6.0.
A representative sample (AtPelA, sugar beet pectin, 24 h) was selected for validation of the method. Three consecutive injections were made for technical replicates. The average coefficient of variation (CV) of the normalized values for all compounds was 9.0% (±5.5%) with no observed linearity between CV% and compound average (data not shown).
One-way ANOVA for determination of statistical significance was performed with JMP 14 Pro (SAS, Cary, NC, United States). Statistical significance was established at p < 0.05.
The multigenecity of pectinolytic enzymes from Aspergilli has been the subject of several studies over the last three decades (Harmsen et al., 1990; Kusters-van Someren et al., 1992; de Vries et al., 2002; Martens-Uzunova and Schaap, 2009; Benoit et al., 2012; Kowalczyk et al., 2017; He et al., 2018). Differences in pH optima (Supplementary Table S1) and expression inducers may partially explain the large gene families observed for several different classes of pectinolytic enzymes (Martens-Uzunova and Schaap, 2009; Andersen et al., 2012), but there is a need for rigorous biochemical characterization of the gene products including specificity toward pectins of different origin and in particular determination of product profiles in order to fully understand this multigenecity. In fact, several cases exist where one of the genes in a pectinolytic gene family is present in one Aspergillus niger strain, but absent from another (Martens-Uzunova and Schaap, 2009). For instance, the genome of A. niger ATCC 1015 harbors a putatively pectin lyase-encoding pelE gene, but there is no evidence of such a gene in the genome of A. niger CBS 513.88 (Martens-Uzunova and Schaap, 2009). The presence of pelE in A. niger CBS 120.49 was described 30 years ago, although some doubt was raised about its actual function (Harmsen et al., 1990). The genomes of the A. tubingensis and A. luchuensis strains from which pectin lyases were sourced in the current work also contain pelE genes (UniProt entries A0A100I604 and G7XZW9, respectively), but no reports on PelE expression or activity of the pectin lyases from these strains exist. In a recent study, all five pectin lyase genes of A. niger CBS 513.88 – pelA, pelB, pelC, pelD, and pelF – were cloned and overexpressed in A. niger SH-2 (He et al., 2018). Of these, the pelA-recombinant strain exhibited the highest pectin lyase activity on apple pectin (DM 50–75%), and thus only PelA was selected for characterization.
In the current work, we aimed to express and systematically characterize representatives of Aspergillus sp. PelA, PelB, PelC, PelD, and PelF. The PL1_4 pectin lyases selected for the current work originated from three different strains which all belong to the Aspergillus section Nigri: A. aculeatus, A. tubingensis, and A. luchuensis (Table 1). The two latter strains were originally both assigned as A. niger, but subsequently determined via sequence analysis to in fact be the very closely related species A. tubingensis and A. luchuensis. Indeed, A. tubingensis and A. luchuensis both belong to the A. niger clade (Varga et al., 2011; Hong et al., 2013). We attempted to express AlPelA recombinantly in A. oryzae, but did not succeed. The PelA from A. tubingensis (which we named AtPelA for the current work) is 99% identical to the PelA of A. luchuensis (AlPelA; UniProt G7XV55) and is therefore useful as a PelA representative to compare to PelB, PelC, PelD, and PelF from A. luchuensis. The UniProt entries list different Aspergillus species as origins of the employed pectin lyases, e.g., A. kawachii which has since been replaced by A. luchuensis (Hong et al., 2013). Together, these shifting taxonomic designations indicate that assigning exact Aspergillus species classification is a complex matter. In the current work, we have named the pectin lyases according to their most recent species classification as well as to their identity with the A. niger CBS 513.88 pectin lyases named PelA, PelB, PelC, PelD, and PelF (Table 1).
In order to allow comparison of the current work to literature, the six PL1_4 pectin lyases were compared to Swiss-Prot pectin lyase sequences in terms of sequence identity (Table 1) and phylogeny (Figure 2). A clear distinction between bacterial and fungal pectin lyases is observed (Figure 2). AtPelA, AaPelA, AlPelD, and AlPelB cluster in the same fungal cluster (FC1), whereas AlPelC and AlPelF cluster in each their fungal cluster (FC2 and FC3, respectively, Figure 2). This division is also evident from their pairwise sequence comparisons: AlPelC and AlPelF are clearly different from the members of FC1 and also different from each other (sequence identities ≤ 50%; Supplementary Figure S2). The putative PelE from A. niger CBS 120.49 (SwissProt B3GQR3) clusters at the edge of FC2 containing the PelC representatives, which have not been deeply characterized either (Figure 2 and Supplementary Table S1). The phylogenetic tree emphasizes the close relatedness of the pectin lyases used in the current work to those of A. niger CBS 513.88 and CBS 120.49 Figure 2); this is also apparent from their high sequence identity (Table 1). Furthermore, the high similarity between AtPelA and PelA from A. luchuensis (AlPelA) is also clear from the phylogenetic analysis (Figure 2). As expected, AaPelA clusters with AtPelA and PelA (Figure 2, Table 1, and Supplementary Figure S2). The decision to include AaPelA and no other A. aculeatus PL1_4 enzymes in the study reflects the fact that PelA is the most studied enzyme of the five, possibly due to its high activity and expression levels (He et al., 2018).
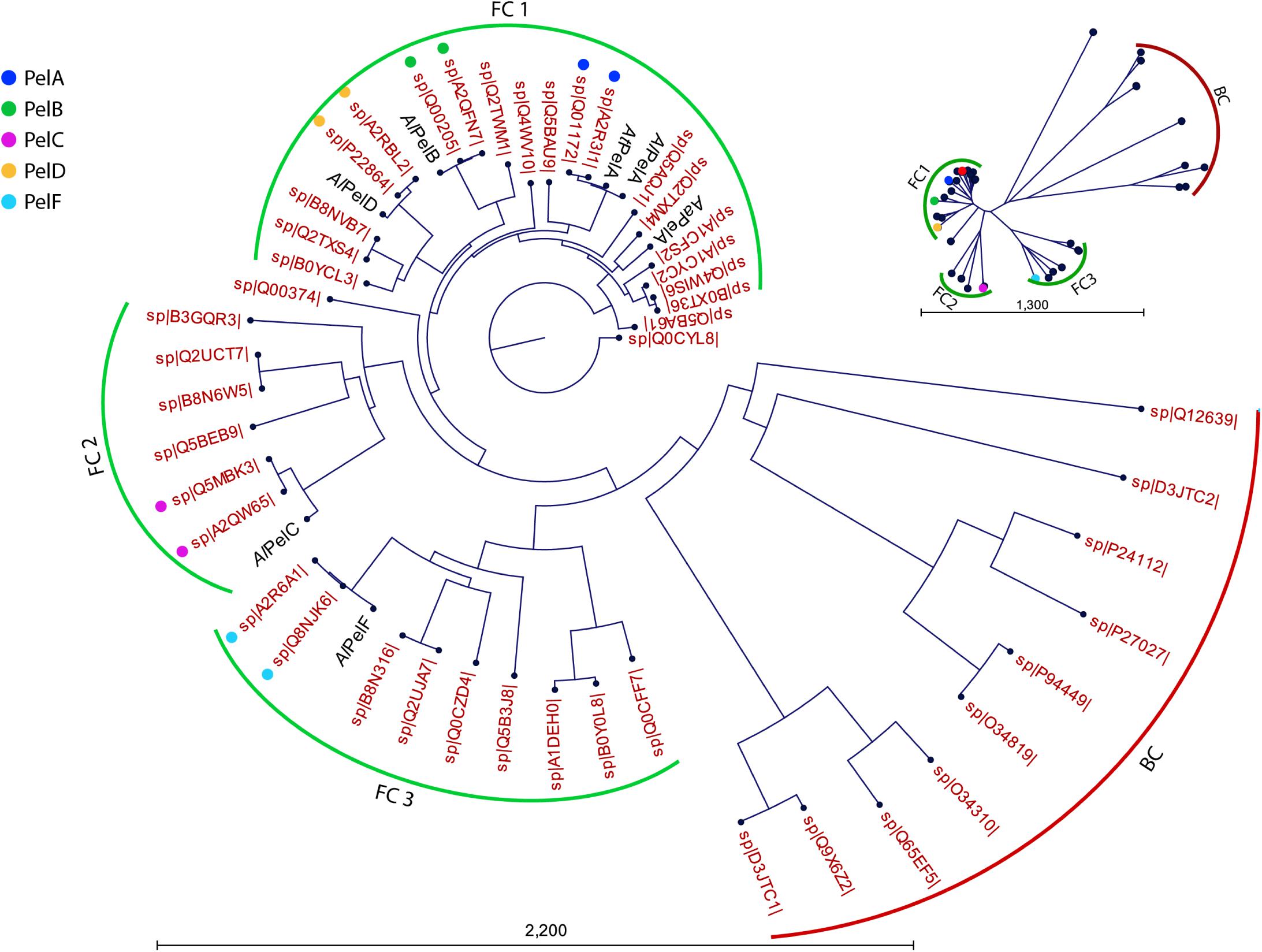
Figure 2. Phylogenetic tree of pectin lyases included in the current study shown versus all reviewed Swiss-Prot entries (red labels indicating Swiss-Prot accession numbers). From the radial tree (top right), three clusters were identified: a bacterial cluster (BC, red) and three fungal clusters (FC1-FC3, green). Pectin lyases PelA, PelB, PelC, PelD, and PelF from A. niger strains CBS 513.88 and CBS 120.49 are indicated with colored dots. The UniProt database does not indicate CBS 120.49 in these entries, but tracking the submissions they all originate from A. niger CBS 120.49. AlPelA from the same A. luchuensis strain as AlPelB, AlPelC, AlPelD, and AlPelF is indicated to visualize its close relatedness to AtPelA studied here.
Two PL1_4 subfamily members have resolved crystal structures: A. niger PelA (PDB IDs 1IDK and 1IDJ; Mayans et al., 1997), which has 99% sequence identity to AtPelA, and A. niger PelB (PDB ID 1QCX; Vitali et al., 1998), which has 98% sequence identity to AlPelB. Structurally, the two pectin lyases appear highly similar. Neither contain ligand structures, but several crystal structures of bacterial PL1 pectate lyases from other subfamilies do. Together, this enables homology modeling and identification of the active site in the PL1_4 pectin lyases studied here. From the homology models it is evident that the three-dimensional structures of AaPelA, AtPelA, AlPelB, and AlPelD are indeed very similar (Figure 3). In contrast, AlPelC and AlPelF appear to exhibit a more open conformation around the active site and the homology models suggest that AlPelC and AlPelF have a more disordered and open loop in this position instead of the short α-helix observed in the crystal structures of PelA and PelB as well as in the models of AaPelA, AtPelA, AlPelB, and AlPelD (Figure 3). From the multiple sequence alignment it is evident that AlPelC and AlPelF have shorter loops in this region (marked in blue in Supplementary Figure S3). This loop contains a conserved substrate-interacting Trp residue corresponding to Trp81 in A. niger PelA and PelB (Mayans et al., 1997; Vitali et al., 1998; Herron et al., 2002). In the AlPelC homology model this Trp is completely aligned with that of AtPelA, whereas the Trp side chain points in the opposite direction in the AlPelF model (Figure 3). For a neighboring loop above the active site (marked in gray in Supplementary Figure S3), AlPelC is similar to the other pectin lyases, whereas AlPelF exhibits a longer, disordered loop in this position (Figure 3). Importantly, this loop contains a Trp residue (Trp66 in A. niger PelA and PelB), which interacts with the methoxyl group of GalA in subsite +3 (Mayans et al., 1997; Herron et al., 2002). This Trp residue is generally conserved, but absent from AlPelF (Figure 3 and Supplementary Figure S3). Furthermore, AlPelF is larger than the other five pectin lyases, which are almost identical in predicted size (glycosylations excluded; Table 1). This larger size is due to a C-terminal stretch found in AlPelF (Supplementary Figure S3), which according to a BLAST analysis appears to be unique to PelF from various Aspergillus species. The function of this C-terminal stretch is unknown; it could not be modeled by homology modeling, and secondary structure prediction suggested the entire structure to be a random coil. In conclusion, AlPelF differs most from the generally similar structures of the other pectin lyases. This assessment of the six pectin lyases show that there is good agreement between the phylogenetic analysis and the three-dimensional enzyme structure (Figures 2, 3).
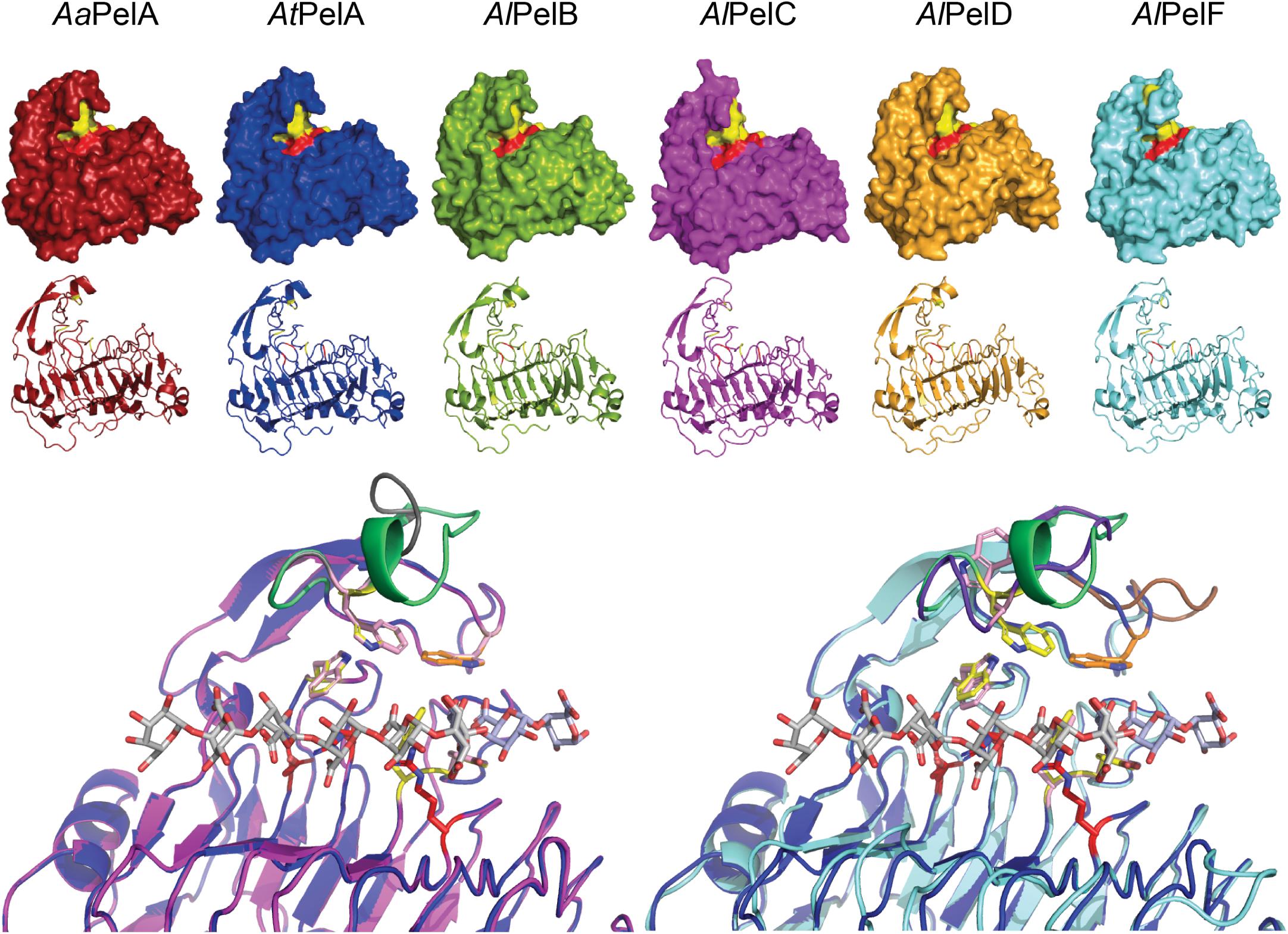
Figure 3. Top: Homology models of the six PL1_4 pectin lyases studied here. Three charged residues believed to hold the catalytic function (one Asp and two Arg residues) are indicated in red, while aromatic residues which interact with the substrate and maintain the active site architecture are indicated in yellow (Mayans et al., 1997; Herron et al., 2002; Van Alebeek et al., 2002). The C-terminals of AlPelD and AlPelF could not be modeled and are thus not visible. Bottom: Active site zoom in structural alignments of homology models of AtPelA (blue) and AlPelC (magenta) and of AtPelA and AlPelF (cyan), respectively. The pectate oligomer ligands (gray and light blue) are derived from alignment with Bacillus subtilis pectate lyase (PDB IDs: 3KRG and 5AMV) (Seyedarabi et al., 2010; Ali et al., 2015). Assumed catalytic residues (red) are indicated alongside substrate interacting residues (yellow in AtPelA, light pink in AlPelC and AlPelF). The substrate-interacting Trp86 in AtPelA (corresponding to Trp66 in crystal structures of A. niger PelA and PelB (Mayans et al., 1997; Vitali et al., 1998; Herron et al., 2002), which were numbered without the 20-amino acid signal peptide) is shown in orange to highlight its absence in AlPelF, where this differing loop region is shown in brown (Supplementary Figure S3). The loop above the active site, which is four amino acids shorter in AlPelC and AlPelF and contains the substrate-interacting Trp residue corresponding to Trp81 in A. niger PelA and PelB (Mayans et al., 1997; Vitali et al., 1998; Herron et al., 2002), is highlighted in green in AtPelA, gray in AlPelC, and purple in AlPelF. Alignment of AlPelF and AlPelC with AlPelB is highly similar to that with AtPelA (not shown).
Using the CUPP webserver for functional annotation (Barrett and Lange, 2019; Barrett et al., 2020a) confirmed that all the included enzymes are predicted to belong to PL1_4. AaPelA, AtPelA, AlPelB, AlPelD, and AlPelF were all assigned EC 4.2.2.10 and CUPP group PL1:12.11 (Barrett et al., 2020a). In contrast, AlPelC could not be assigned to a CUPP group due to a low score. Although AlPelF differs most in terms of structure (Figure 3), CUPP singled out AlPelC as being more different. The detailed grouping provided by CUPP relies on an unsupervised peptide-based clustering algorithm that can provide systematic grouping and functional annotation of carbohydrate-active enzymes (CAZymes) based on comparison of peptide motifs in the enzyme amino acid sequences (Barrett and Lange, 2019). This emphasizes that AlPelC may be functionally different from the other pectin lyases studied here.
Quantitatively, the major constituent of pectin is homogalacturonan followed by rhamnogalacturonan I (RG-I). Homogalacturonan consists of an unbranched chain of α-(1,4)-linked GalA units, which may be methoxylated and/or acetylated. Homogalacturonan can also be further substituted to form other less abundant pectin components, namely xylogalacturonan, apiogalacturonan, and rhamnogalacturonan II (RG-II). RG-I is made up of a backbone of alternating α-(1,2)-linked rhamnose and α-(1,4)-linked GalA units, where the rhamnose units are substituted with neutral sidechains of galactan, arabinan, and/or different arabino-galactan sidechains (Bonnin et al., 2014).
Compared to apple and citrus pectins, sugar beet pectin is known to have shorter homogalacturonan stretches, more RG-I, and a higher degree of acetylation (DAc) (Buchholt et al., 2004). This was reflected in the monosaccharide composition observed in the current work: sugar beet pectin contained more rhamnose, arabinose, and galactose (indicators of RG-I) and less GalA than apple and citrus pectins (Table 2). The DAc was 19% for sugar beet pectin and only 1% and 2% for citrus and apple pectins, respectively (Table 2). Comparing apple and citrus pectins, citrus pectin had slightly more GalA. Since pectin lyase prefers methylesterified GalA, the degree of methoxylation (DM) is an important parameter: Apple pectin had the highest DM of 69%, whereas there was no significant difference between sugar beet pectin (56%) and citrus pectin (53%; Table 2). This is in agreement with previous reports on DM for these pectin sources (Müller-Maatsch et al., 2016). Not accounting for other factors such as methyl group distribution, the pectin lyases were thus expected to have the highest affinity for apple pectin due to the high DM. Similar DMs were observed for citrus pectin and sugar beet pectin, but due to the significantly higher DAc observed in sugar beet pectin, it was expected that the pectin lyases would exhibit the lowest affinity for sugar beet pectin.
The specific activity of the six pectin lyases was determined as initial rates obtained at 25°C in 1 g/L solutions of apple pectin (DM 69%, DAc 2%), citrus pectin (DM 53%, DAc 1%), sugar beet pectin (DM 56%, DAc 19%), and polygalacturonic acid at pH 5.0 (Table 3). The highest initial rate was obtained with AlPelB on apple pectin. Indeed, all the pectin lyases exhibited highest activity on apple pectin, which is likely linked to the high DM of this pectin substrate. In general, the following substrate preference was observed: apple pectin > citrus pectin > sugar beet pectin > polygalacturonic acid, which reflects both DM and DAc of the substrates (Table 3). As expected, the difference in DM appeared to determine the difference in preference between apple and citrus pectins (Tables 2, 3), whilst the markedly higher DAc of sugar beet pectin perhaps in combination with a higher amount of RG-I (Table 2) impaired pectin lyase activity on sugar beet pectin. Finally, the consistently low activities of all pectin lyases on polygalacturonic acid (Table 3) confirm the requirement for methoxylation to achieve pectin lyase activity. The same trend was recently reported for PelA from A. niger CBS 513.88, which had highest activity on citrus pectin (DM ≥ 85%), a relative activity of 59% compared to citrus pectin on a sample of apple pectin with a DM of 50–75%, and almost no activity on polygalacturonic acid (He et al., 2018).
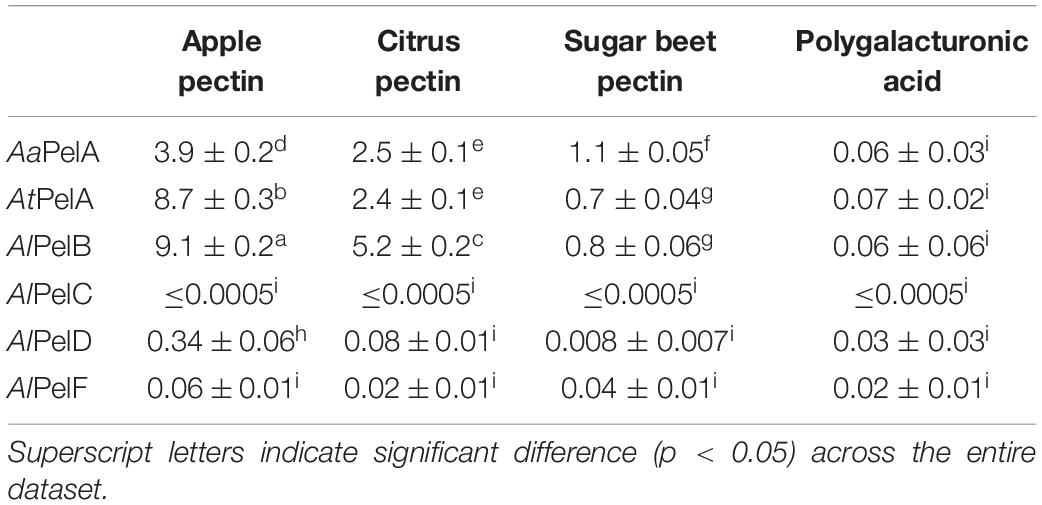
Table 3. Specific enzyme activity (μmol product released per min per mg enzyme) determined from initial rates of the six pectin lyases on four different pectin substrates at pH 5 and 25°C: Apple pectin, citrus pectin, sugar beet pectin, and polygalacturonic acid; c.f. Table 2 for composition of apple, citrus, and sugar beet pectins.
Among the six pectin lyases, AlPelB had the highest initial rate on both apple pectin and citrus pectin, whereas AaPelA had highest activity on sugar beet pectin (Table 3). AtPelA had higher activity on apple pectin than AaPelA, but the opposite was observed for sugar beet pectin (Table 3), and the two PelA pectin lyases exhibited similar activity on citrus pectin and polygalacturonic acid. For apple and citrus pectins, the following general ranking in specific activity was observed: PelB > PelA > > PelD > PelF > PelC (Table 3). The activity of AlPelC was at the detection limit despite using a high enzyme dosage. For polygalacturonic acid there was no significant difference between the very low specific activities, indicating that the enzymes do not have pectate lyase activity.
In a recent study, the fermentation broths of A. niger SH-2 used for recombinant expression of A. niger CBS 513.88 PelA, PelB, PelC, PelD, and PelF were tested on apple pectin (DM 50–75%) at pH 5.2. The enzyme activities in U/mL was PelA > PelD > > PelF > PelB, while PelC could not be expressed. However, these activities also reflect the expression levels, and especially PelB was expressed in lower amounts than the other pectin lyases (He et al., 2018). It was previously reported that A. niger CBS 120.49 PelB was more prone to protease degradation than PelA of the same strain when expressed homologously (Kusters-van Someren et al., 1992). As a consequence, it may not be possible to detect the high specific activity of PelB without controlling the enzyme dosage.
Data on pH and temperature optima of pectin lyases from Aspergillus sp. are scarce (Supplementary Table S1). Recently, the optima for PelA from A. niger CBS 513.88 were determined separately on citrus pectin (DM ≥ 85%) to pH 4.5 and 50°C (He et al., 2018). Previously, PelA from A. niger (identified as PLII in the commercial Ultrazym preparation) was found to have a slightly substrate concentration-dependent optimum around pH 5.5–6.5 when determined at 25°C on apple pectin with a DM of 94% (van Houdenhoven, 1975; Kester and Visser, 1994). The same was true for Ultrazym PLI, which was found to be PelD from A. niger (van Houdenhoven, 1975; Gysler et al., 1990; Kester and Visser, 1994). PelF from A. niger ZJF also exhibited an acidic pH optimum of 5.0 and a temperature optimum of 43°C on citrus pectin (Xu et al., 2015). In contrast, PelB from A. niger CBS 120.49 had an alkaline optimum at pH 8.5 when determined at 25°C (Kester and Visser, 1994).
As pH optima are temperature dependent, we set out to estimate combined pH-temperature optima for the four most active pectin lyase in the current study using response surface methodology. Based on preliminary studies, acidic pH ranges (pH 3.5–5.5) were selected for AtPelA and AlPelD, whereas a range from slightly acidic to neutral pH (pH 5.5–7.5) was selected for AaPelA and AlPelB. Temperatures ranged from 40°C to 70°C (Table 4) and the reaction was monitored for 5 min. All the generated models had R2 above 0.92, and most of them above 0.98, indicating that the modeled response surfaces described the data well (Supplementary Table S2 and Supplementary Figure S4). For most models, the lack of fit was statistically significant (Supplementary Table S2). This indicates that the generated models are not sufficient for predicting activity at the identified optima, and is a result of the fact that the model assumes that the effect of pH and temperature can be described by quadratic polynomials. This is, however, not always the case for pH and temperature optima, which often follow non-symmetrical bell-shaped curves. For a few of the models, one of the main factors (T or pH) was not significant, but they were kept in the model in order to predict a pH-temperature optimum for the four pectin lyases on three different pectins (Table 4, Supplementary Table S2, and Supplementary Figure S4). For AaPelA on sugar beet pectin, no model was obtained as the differences between the data points in the design were not statistically significant within the chosen ranges (Supplementary Table S2 and Supplementary Figure S4).
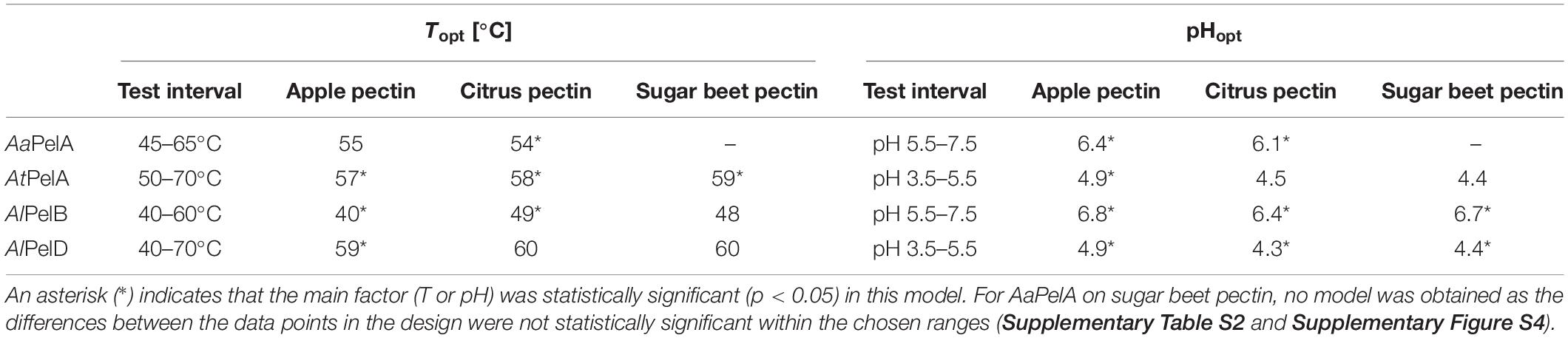
Table 4. Combined pH-temperature optima on apple pectin, citrus pectin, and sugar beet pectin predicted from the regression models generated by the two-factor face-centered central composite experimental design (Supplementary Table S2).
In general, AtPelA and AlPelD exhibited pH-temperature optima, which were a combination of high temperature and low pH: pH 4.4–4.9 and 57–59°C for AtPelA and pH 4.3–4.4 and 59–60°C for AlPelD (Table 4). However, for AlPelD on citrus and sugar beet pectins, T was not a significant factor, indicating low variability across the range from 40 to 70°C (Supplementary Figure S4). The same was true for pH for AtPelA (Table 4 and Supplementary Table S2). For AaPelA and AtPelB, the pH-temperature optima were a combination of lower temperature and higher pH: pH 6.1–6.4 and 54–55°C for AaPelA and pH 6.4–6.8 and ≤ 40–49°C for AlPelB (Table 4). These trends are fairly well in line with previously reported optima for A. niger pectin lyases (Supplementary Table S1), and can partially explain the pectin lyase multigenecity in Aspergilli. For AlPelB we observe a lower (yet high in comparison with the other pectin lyases) pH optimum than found for PelB from A. niger CBS 120.49 (Kester and Visser, 1994). This is most likely tied to the fact that we have included an elevated temperature (40–60°C), whereas the previously established pH optimum of 8.5 was determined at 25°C, where PelB is more stable. Remarkably, the fact that the two PelA pectin lyases included here differ in their pH-temperature optima emphasize that results obtained with a given pectin lyase from one Aspergillus strain is not directly transferable to one of another strain. However, this effect may not be as dramatic within the A. niger clade where sequence similarities above 90% are observed for the enzymes discussed here, whereas the sequence similarity between AaPelA and AtPelA is only 75% (Table 1 and Supplementary Figure S2).
Generally, pH values of the optima were higher on apple pectin than on citrus or sugar beet pectins (Table 4). The pKa of pectin decreases with increasing pH (Plaschina et al., 1978). Possibly, the higher DM of the apple pectin entails that a higher pH is required to reach the net charge preferred by the pectin lyases as compared to the lower-DM pectins.
Thermal stability of the pectin lyases was assessed by determining the melting temperatures (Tm) at pH 5. The highest Tm was found for AaPelA (75°C), followed by AtPelA (68°C), AlPelD (67°C), and AlPelB (65°C). The lowest Tm values were found for AlPelF (61°C) and AlPelC (57°C). Significant difference (p < 0.05) was observed between all values, indicating that the PelA pectin lyases were the most thermally stably, whereas the enzymes of low specific activity, i.e., AlPelF and AlPelC, also had the lowest thermal stability.
The kinetic parameters of the four most active pectin lyases were determined at the estimated pH-temperature optima (Table 4) on apple pectin, citrus pectin, and sugar beet pectin (Table 5). The enzymes differed in terms of Km values on the different substrates except for AaPelA where no significant difference in Km was observed across the three pectins (11–14 mM GalA). For AtPelA the Km was significantly lower on apple pectin (8 mM GalA) than on citrus pectin (19 mM GalA), and highest on sugar beet pectin (25 mM GalA). For both AlPelB and AlPelD, no significant difference in Km was observed between citrus and sugar beet pectins, indicating that acetylations had little effect on affinity for these enzymes (Table 5). The effect of methoxylation differed between the two: while Km decreased with increasing DM for AlPelB, an increase was observed for AlPelD (Table 5). The lowest Km values were observed for AtPelA on apple pectin and for AlPelD on citrus and sugar beet pectins (Table 5). The latter were at the same level as that observed previously for A. niger PelD on high-methoxylated apple pectin (Supplementary Table S1). However, for A. niger PelA and PelB, previously reported Km values are lower than the values observed here; this could be linked to the use of chemically esterified pectins in previous studies, where the pectins were esterified to a much higher DM than we used here, namely a very high DM of around 95% (Supplementary Table S1) (van Houdenhoven, 1975; Kusters-van Someren et al., 1991; Kester and Visser, 1994).
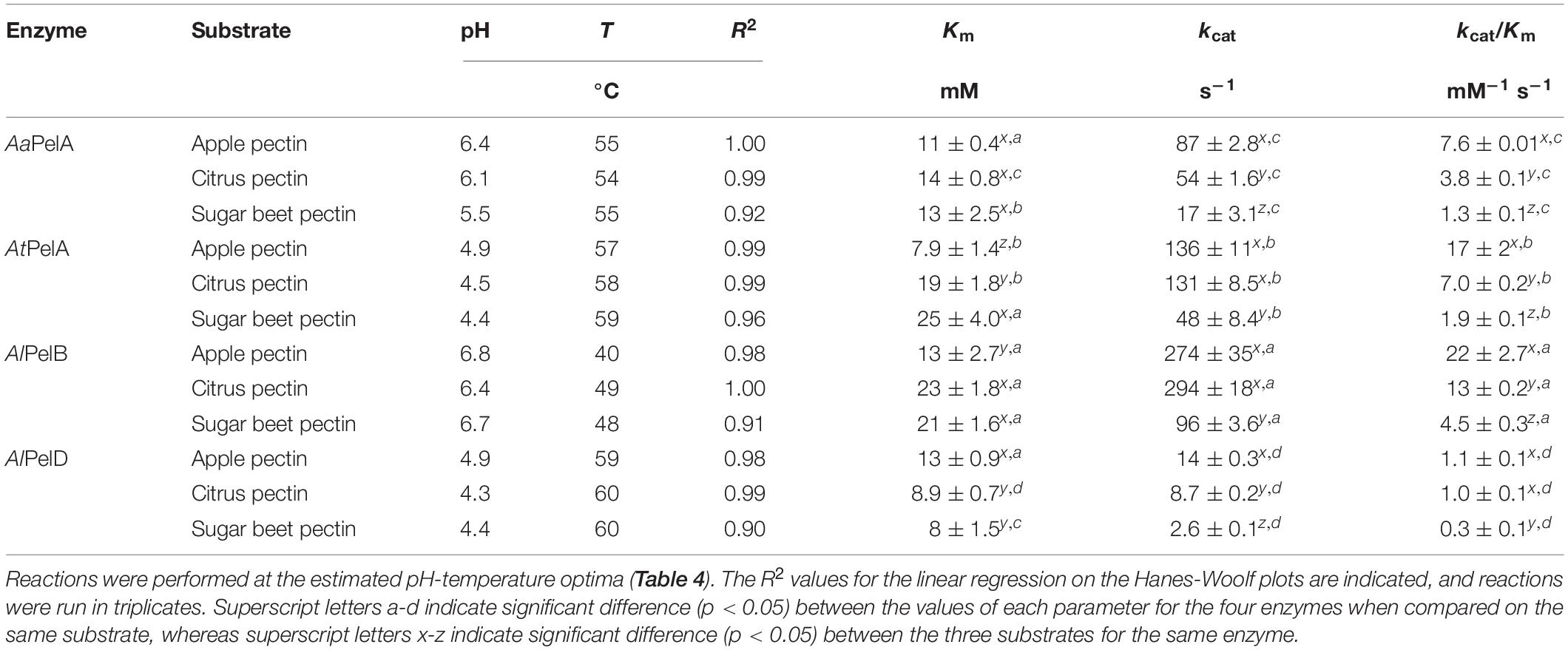
Table 5. Kinetic parameters Km (in mM GalA) and kcat (in mol product released per mol enzyme per second) for AaPelA, AtPelA, AlPelB, and AlPelD on apple pectin (DM 69 ± 3%, DAc 2 ± 0.1%), citrus pectin (DM 53 ± 1%, DAc 1 ± 0.1%), and sugar beet pectin (DM 56 ± 5%, DAc 19 ± 1%).
For all enzymes, kcat was significantly lower on sugar beet pectin than on the other pectin substrates (Table 5). For AaPelA and AlPelD, kcat was highest on apple pectin, whereas there was no significant difference between kcat values on apple and citrus pectins for AtPelA and AlPelB (Table 5). For AaPelA, AtPelA, and AlPelB kcat/Km was significantly higher on apple pectin. For AlPelD the difference to citrus pectin was not statistically significant, but the value for apple pectin was higher. For all enzymes, kcat/Km was significantly lower on sugar beet pectin (Table 5). Again, this directly reflects the preference of pectin lyases for a highly methoxylated substrate and suggests a preference for substrates with low DAc. Regardless of the substrate, large differences in kcat/Km were observed between the enzymes, which ranked in the following order with respect to kcat/Km: AlPelB > AtPelA > AlPelA > AlPelD (Table 5). The finding that AlPelB was the most efficient enzyme is in good agreement with literature data on A. niger pectin lyases, where the highest kcat/Km has indeed been observed for PelB (Supplementary Table S1). In that previous study (Kester and Visser, 1994), the kcat/Km was higher than observed in the current work, which could be due to the use of a highly methylesterified substrate, and that the reaction took place at pH 8.5. However, such a high pH is close to the pH where pectin spontaneously decomposes by β-elimination (Renard and Thibault, 1996). Analogously, significantly lower kcat/Km values were obtained for AaPelA and AlPelD as compared to the A. niger PelA and PelD, which for AaPelA was linked to an eight times higher Km and for AlPelD to a five times higher kcat (Table 5 and Supplementary Table S1). For AtPelA the kcat/Km of 17 mM–1 s–1 on apple pectin was comparable to that of the A. niger PelA (18–22 mM–1 s–1); the use of a higher reaction temperature in the current work to increase kcat balanced out the negative effect of the lower DM on Km in this particular case (Table 5 and Supplementary Table S1).
To compare the pectin degradation profiles of all six pectin lyases, they were dosed in equimolar ratios on apple pectin, citrus pectin, and sugar beet pectin. For simplicity, a single set of reaction conditions was selected, reflecting the best compromise based on the obtained pH-temperature optima (Table 4): pH 5.5 and 40°C. The reactions were monitored over 24 h and analyzed by size exclusion chromatography (SEC) and liquid chromatography coupled to mass spectrometry with electrospray ionization (LS-ESI-MS). From the SEC chromatograms, the concentration of pectin molecules below ~100 kDa could be monitored over time (Figure 4). The SEC analysis of pectin degradation products reflect the enzyme activity and substrate specificity data well: Apple pectin is degraded faster than citrus pectin, which is in turn degraded faster than sugar beet pectin (Figure 4). In a comparison of the six pectin lyases, the specific activities (Table 3) are directly reflected in the development of the SEC profiles over time: PelB > PelA > PelD > PelF > PelC (Figure 4 and Supplementary Figure S5). Comparing the two PelA pectin lyases, they were again found to be equally fast on citrus pectin, while AtPelA was faster on apple pectin, and AaPelA was faster on sugar beet pectin (Figure 4 and Table 3). As a consequence of their low activity, AlPelC and AlPelF were dosed 10 times higher than the other pectin lyases in order to better capture their product profiles in the LC-MS-based product profiling. For AlPelF the increased dosage had a large effect on pectin degradation (Supplementary Figure S5). For AlPelC the very low activity was only detectable after 24 h, with a noticeable difference between the two enzyme dosages (Figure 4 and Supplementary Figure S5).
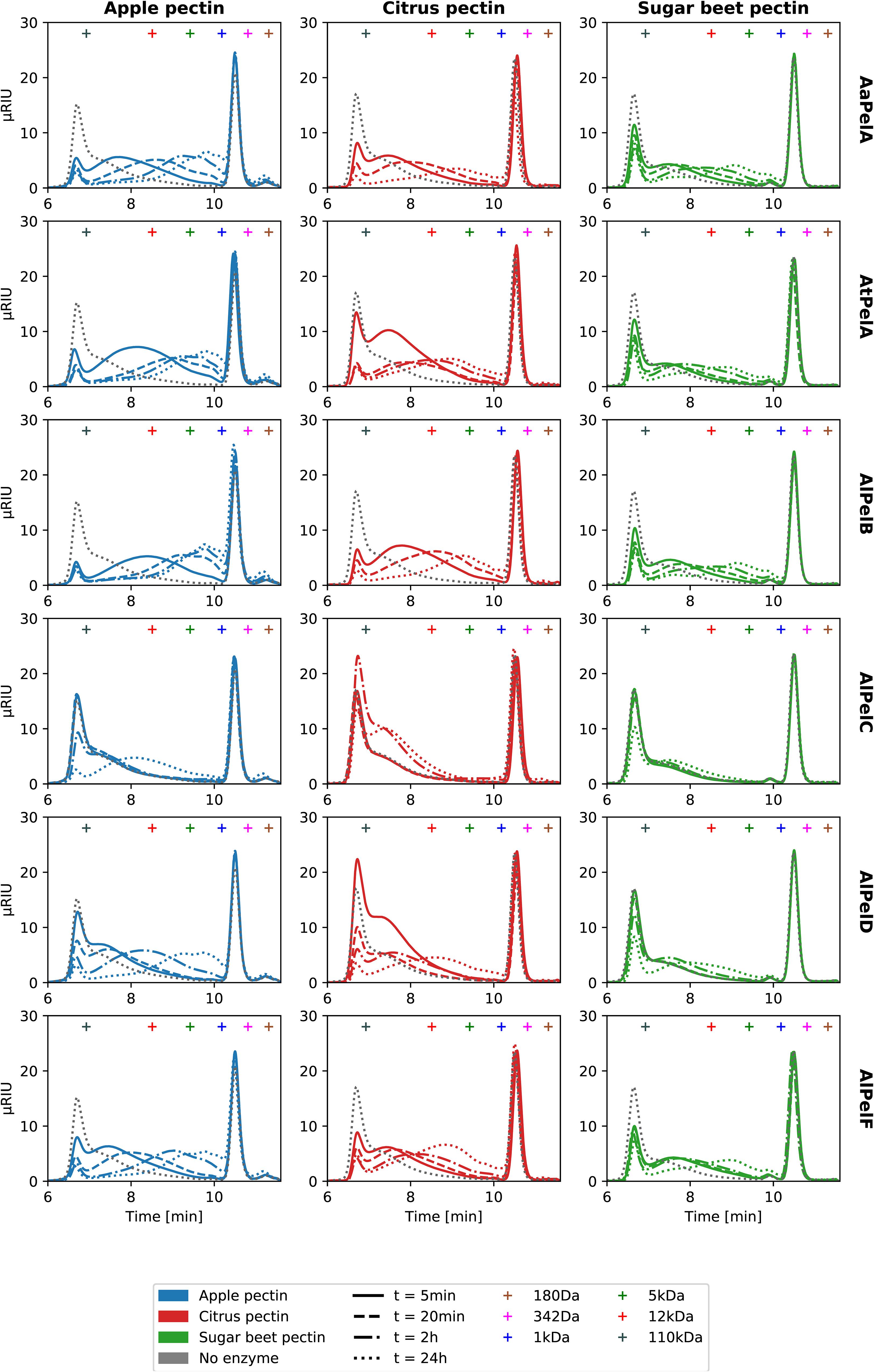
Figure 4. Size exclusion chromatography (SEC) chromatograms for degradation of 10 g/L apple pectin (blue), citrus pectin (red), and sugar beet pectin (green) by 60 nM AaPelA, AtPelA, AlPelB, or AlPelD (corresponding to 0.023% E/S for AtPelA) and 600 nM AlPelC or AlPelF at pH 5.5 and 40°C. For chromatograms of AlPelC and AlPelF at 60 nM, refer to Supplementary Figure S5. Retention times of pullulan standard (180 Da to 110 kDa) are indicated by crosses.
The product profiles obtained by LC-ESI-MS were expected to contain both partly and fully methoxylated structures. Partly methoxylated pectin-derived oligosaccharides can easily be identified by MS2 in negative mode (Quéméner et al., 2003a; Ralet et al., 2005; Leijdekkers et al., 2011; Remoroza et al., 2012), whereas fully methoxylated pectin derived oligosaccharides can be identified in positive mode (Van Alebeek et al., 2000; Mutenda et al., 2002; Quéméner et al., 2003b). Manual inspection of the chromatograms obtained in negative mode revealed 28 unique compounds and 5 isomers all present in at least one enzyme-substrate-time combination (Table 6). The nomenclature of the identified compounds are gAxmyaz, meaning x GalA residues, y methyl substitutions, and z acetyl substitutions, e.g., a trimer with three GalA, one methyl substitution, and one acetyl substitution is denoted gA3m1a1. All compounds observed in negative mode appeared as [M-H], except gA3m3 and gA4m4 which appeared as formate [M+FA] – adducts and with a disproportionally low intensity. For all compounds observed in single charge state, the molecular formula could be confirmed by MS2 fragmentation with almost exclusively Ci (or Zj due to isomeric masses) and 0,2Ai fragments observed, according to the nomenclature of Domon and Costello (Domon and Costello, 1988), including the occasional loss of 32 Da that can be attributed to methoxylation at the galacturonic acid residue in the reducing end (Supplementary Figure S6; Quéméner et al., 2003b). For larger structures, e.g., molecular masses larger than ~1100 Da, a tendency toward double charging was observed (Table 6), making full identification from the fragmentation pattern difficult. Hence, for these compounds it was necessary to rely on the m/z values only. All compounds carried a 4,5 unsaturation in the non-reducing end due to the lyase reaction. For some masses, two distinct peaks were observed with two different retention times, indicating two different molecular structures (denoted #1 or #2; Table 6). The different retention times of the compounds are assumed to derive from different positions of the methyl and/or acetyl substitutions. The porous graphite column in use has previously shown the capability to separate different isomers of compounds with identical molecular composition, but with different linkages and/or planarity (Jamek et al., 2018; Mosbech et al., 2018; Zeuner et al., 2018).
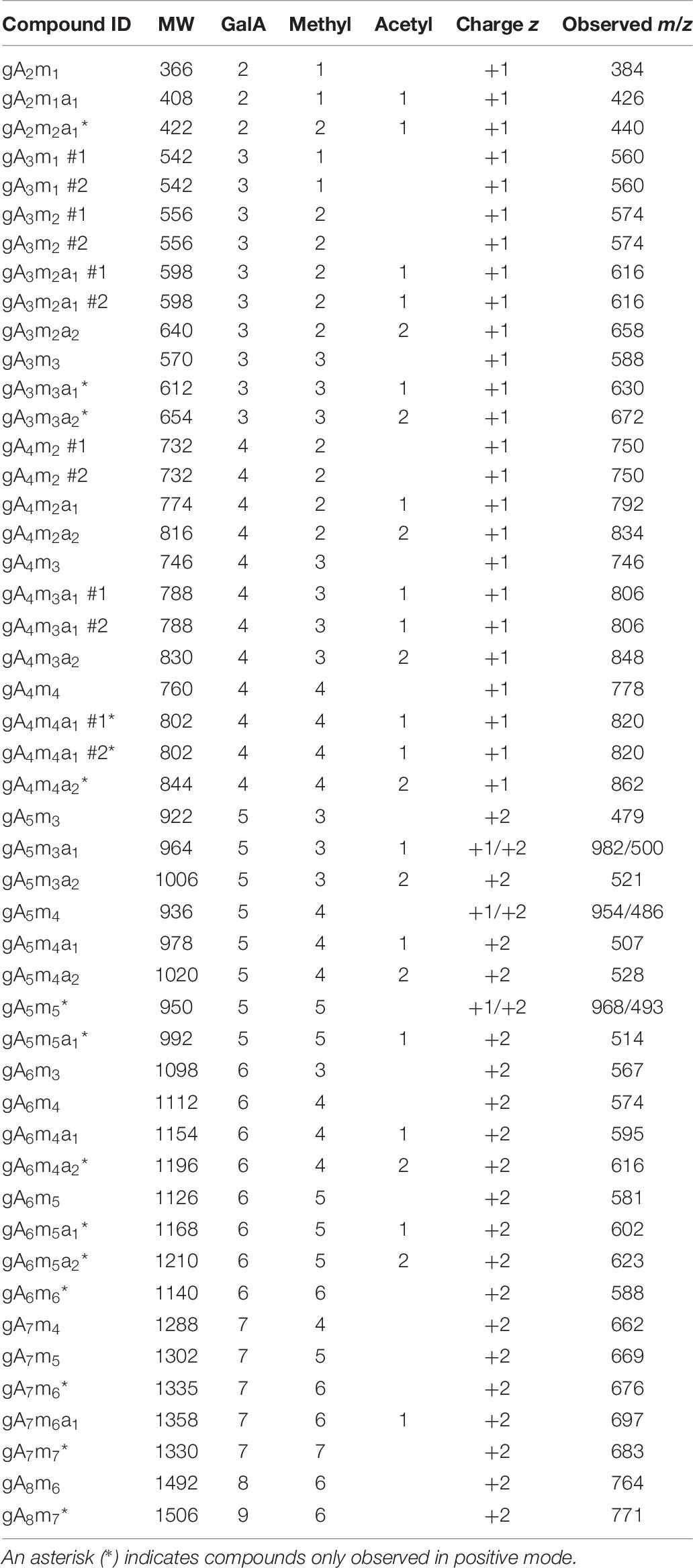
Table 6. Compound ID, molecular weight (MW), number of GalA residues, number of methyl- and acetyl substitutions, charge state, and observed m/z in positive mode of all identified pectic oligosaccharides.
Investigation of the chromatograms obtained in positive mode revealed another 14 unique compounds and 1 isomer all present in at least one enzyme-substrate-time combination (Table 6), with the majority being fully methoxylated compounds. All identified compounds were observed as single charge [M+NH4]+ or double charge [½M + NH4]2+ ammonium adducts due to the constant presence of ammonium formate in the eluent. The structural verification by MS2 fragmentation was somewhat complex due to an unexpected fragmentation pattern. From literature, fully methoxylated pectin derived oligosaccharides are expected to form Ci/Zj ions upon fragmentation in positive mode (Mutenda et al., 2002), although a mixed pattern including all possible ion types can be observed for pectic oligosaccharides in general (Körner et al., 1999). In the present case, assuming Bi and Ci fragments and the NH4+ ion retained on all fragments, all observed fragments had and m/z value 7 Da lower than expected. For example, this pattern was observed in positive mode for gA4m3, for which the structure could be verified in negative mode (Supplementary Figures S6, S7A). A number of putative, fully methoxylated structures were observed in positive mode, including gA3m3, gA4m4, and gA5m5. They all shared the “Bi/Ci -7 Da” pattern (Supplementary Figures S7B–D). The compound gA5m5 was observed both as a single charge and a double charge ion, with the fragmentation pattern of the latter giving more information (Supplementary Figure S7E). Despite not being the most dominant fragments, true Ci ions were observed. Since this pattern is observed for compounds where additional structural verification in negative mode is available, it is assumed that the proposed structures with full methoxylation are correct.
Contrary to fragmentation in negative mode, where only Ci type (and isomeric Zj type) ions are observed, fragmentation in positive mode revealed additional information on acetylation patterns for different isomers sharing the same mass such as gA4m4a1 #1 and #2. The extracted ion chromatogram with the corresponding mass 820 m/z clearly indicates at least two different compounds with baseline separation (Figure 5A). The fragmentation pattern of gA4m4a1 #1 (Figure 5B) shows B3 and C3 fragments that have lost the acetyl group, indicating that the acetyl substitution is on the reducing end galacturonic acid unit. In contrast, the acetyl group is retained on B3 and C3 fragments and lost on B2 and C2 fragments in the fragmentation pattern of gA4m4a1 #2 (Figure 5C), indicating that the acetyl group is on the third galacturonic acid moiety when counting from the non-reducing end. No cross ring fragmentation was observed in either cases, leaving the position (O2 or O3) of the acetyl substitution unsolved.
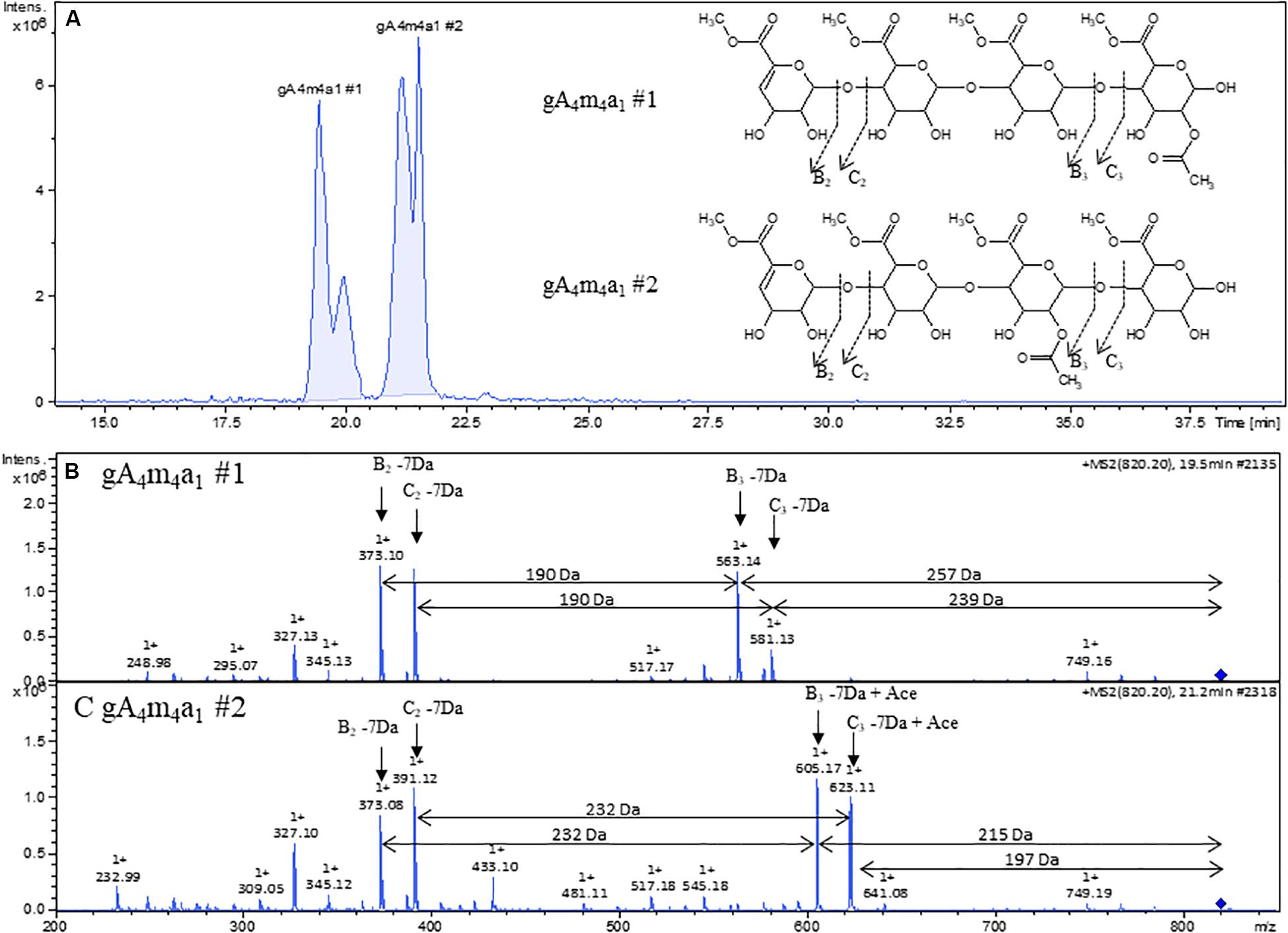
Figure 5. (A) Extracted ion chromatogram of 820 m/z with proposed structures inserted. (B) Positive ion mode ESI-MS2 of gA4m4a1 #1. (B) Positive ion mode of ESI-MS2 of gA4m4a1.
The observed peak intensities showed that relatively few compounds, primarily gA3m2 and gA3m3, and to some extend gA2m1, gA4m3, gA4m4, and gA6m4, were the dominant products for the Aspergillus sp. pectin lyases acting on apple and citrus pectins, with a more disperse profile when acting on sugar beet pectin (Supplementary Figure S8). An increased level of most of the identified compounds was observed over time, although with different rates dependent on type of compound, substrate and enzyme. The levels of larger structures such as gA6m5, gA6m6, gA7m7, and gA8m7 exhibited transient maxima during the 24 h reaction, indicating that these pectic oligomers were substrates for the pectin lyases (Supplementary Figure S8). The large variations in production rates among the enzymes are coherent with activity data (Table 3, Figure 4, and Supplementary Figure S8). Because of the large variation in activity among the enzymes, it conceals the details of the profiles for enzymes with lower activity, such as AlPelC and AlPelD. Hence, the data was normalized in order to facilitate an in-depth investigation and comparison of the product profiles.
For all enzymes acting on apple pectin, gA3m3 was generally the most dominant product throughout the time course (Figure 6A). In the early stages (5, 20 min) some of the longer compounds were more highly represented (degree of polymerization (DP) of 5–7, especially gA6m5), but the relative abundance of these went down in favor of shorter compound like gA3m2 during the extended reaction (120 min, 24 h). The relative levels of gA4m3 and gA4m4 were steady throughout the time course, and there was a tendency to build-up of gA6m4 (Figure 6A). The shift from longer to shorter compounds was most predominantly observed for AlPelB, which has the highest specific activity (Table 3) and the highest kcat/Km on apple pectin (Table 5). The product profilse after 24 h were quite similar for AaPelA, AtPelA, AlPelB, and AlPelF with gA3m2 and gA3m3 as the main products. An even narrower product profile was observed for AlPelD, clearly favoring gA3m3 and indicating a preference for fully methoxylated pectin. In contrast, a more dispersed profile was observed for AlPelC (Figure 6A). This more dispersed profile could be caused by the low activity of AlPelC (Table 3 and Figure 4), since the profile after 24 h looked more like the profiles of AlPelD and AlPelF after 20 min. However, it may also reflect a more selective substrate specificity, where the enzyme failed to degrade the longer compounds. Indeed, a vast increase in gA6m5 levels was observed for AlPelC during 24 h, whereas slight decreases in the observed peak intensities were observed for the other pectin lyases over the course of the reaction (Supplementary Figure S8). The clustering analysis of the product profiles was most clear after 24 h, with the profiles of AlPelB and AlPelA clustering closest together, and the profiles of AlPelD and AlPelC clustering second-most and most apart from the other pectin lyases, respectively.
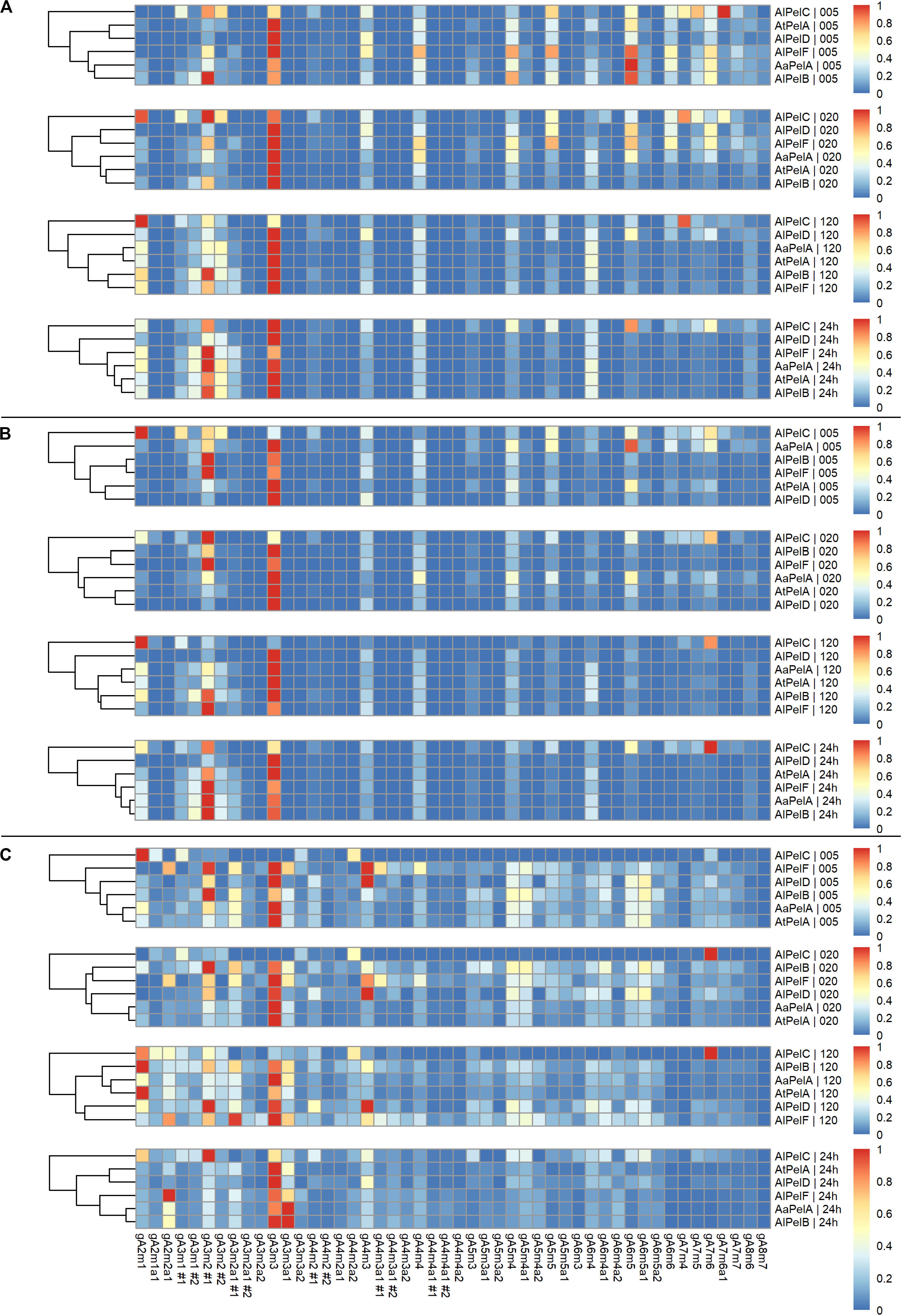
Figure 6. Heat maps of normalized relative intensities for pectin lyases acting on apple pectin (A), citrus pectin (B), and sugar beet pectin (C). Panels top to bottom: 5, 20, 120 min, and 24 h of reaction. For each substrate the pectin lyases are clustered according to product profiles within each time point.
In general, the product profile for all pectin lyases acting on citrus pectin looked similar to the profile on apple pectin (Figure 6B). This was expected due to the similarities of the two substrates in terms of fairly high methylation, low acetylation, and a low content of RGI. Similar product profile patterns were observed with gA3m2 and gA3m3 as the main end products for AaPelA, AtPelA, AlPelB and AlPelF, almost solely gA3m3 for AlPelD, and a more dispersed profile for AlPelC (Figure 6B). A notable difference compared to the profiles of enzymes acting on apple pectin was the lower relative levels of DP6-7 observed for citrus pectin. For apple pectin, especially gA6m5 was major contributor to the profiles particularly in the early stages of the reaction; a similarly, important contribution from this compound to the product profile was not observed on citrus pectin (Figures 6A,B). This might be due to the different distributions of methyl substitutions in the two substrates. The clustering analysis was almost identical to that observed for apple pectin, with the product profiles of AlPelB and AaPelA clustering most, and the product profiles of AlPelD and AlPelC being the most different from the other enzymes after 24 h (Figure 6B).
From activity measurements (Tables 3, 5 and Figure 4) as well as the peak intensities of the individual compounds (Supplementary Figure S8), it is evident that sugar beet pectin is an inferior substrate compared to apple and citrus pectins, but decent degradation was nevertheless observed. The product profile of pectin lyases acting on sugar beet pectin was more differentiated than the profiles of apple and citrus pectins, due to the relatively higher presence of acetyl substituted compounds (Figure 6C). For all enzymes, except AlPelC, the major compound throughout the time course was gA3m3, with relatively high levels of DP5-6. The low levels of gA6m6 and longer compounds could be due to the nature of the sugar beet pectin substrate, where the structures necessary for releasing these products are present in limited amounts. The tendency of the longer compounds (DP5-6) to be outnumbered by the shorter compounds over time was also evident for the enzymes acting on sugar beet pectin. Remarkably, for AaPelA and AlPelB one of the most dominating compounds was the fully methoxylated gA3m3a1, a compound that features a galacturonic acid residue with both a methyl and an acetyl substitution (Figure 5). These double substitutions has previously been reported as rare in sugar beet pectin (Ralet et al., 2005). The relative contribution of gA3m3a1 to the product profile compared to gA3m3 was most pronounced after 24 h, indicating that while especially AaPelA, AlPelB, and AlPelF accommodate the acetylated, fully methoxylated substrate, they release the non-acetylated variant first (Figure 6C and Figure Supplementary S8). Again, a more dispersed profile was observed for AlPelC, with gA3m2 being the main product. The cluster analyses after 24 h were similar to the ones of apple and citrus pectins with the product profiles of AlPelB and AaPelA clustering most, and AlPelC being most different (Figure 6C).
Examining the product profiles in a broader perspective with the intensities summed up according to DP with and without acetyl substitutions it was evident that DP3 is the most preferred product throughout the reactions (Figure 7). AlPelC showed a more differentiated profile compared to the other enzymes during the early phases of the reaction, but after 2 h of reaction DP3 was also dominant for this enzyme (Figure 7). In general, only negligible amounts of acetyl substituted compounds were observed in reactions on apple and citrus pectins, which was highly expected due to the low DAc of these substrates (Table 2), whereas slightly more differentiated profiles were observed for sugar beet reactions due to substrate acetylation. Hence, the division is more applicable for the profiles of enzymes acting on sugar beet pectin and the segregation revealed a division of the enzymes in two groups. AaPelA, AlPelB, and AlPelF showed equal preference/tolerance for DP3 with and without acetyl substitutions as a product, and even a higher preference for releasing acetyl substituted DP4-6 compared to non-substituted DP4-6 (Figure 7). In contrast, AtPelA, AlPelC, and AlPelD showed a notable preference for releasing non-acetylated DP3 compared to acetylated DP3. However, while AlPelC specifically preferred release of non-acetylated pectic oligosaccharides regardless of DP, the result was more blurred for AtPelA and AlPelD, where the tolerance to acetylation changed across the DP range (Figure 7).
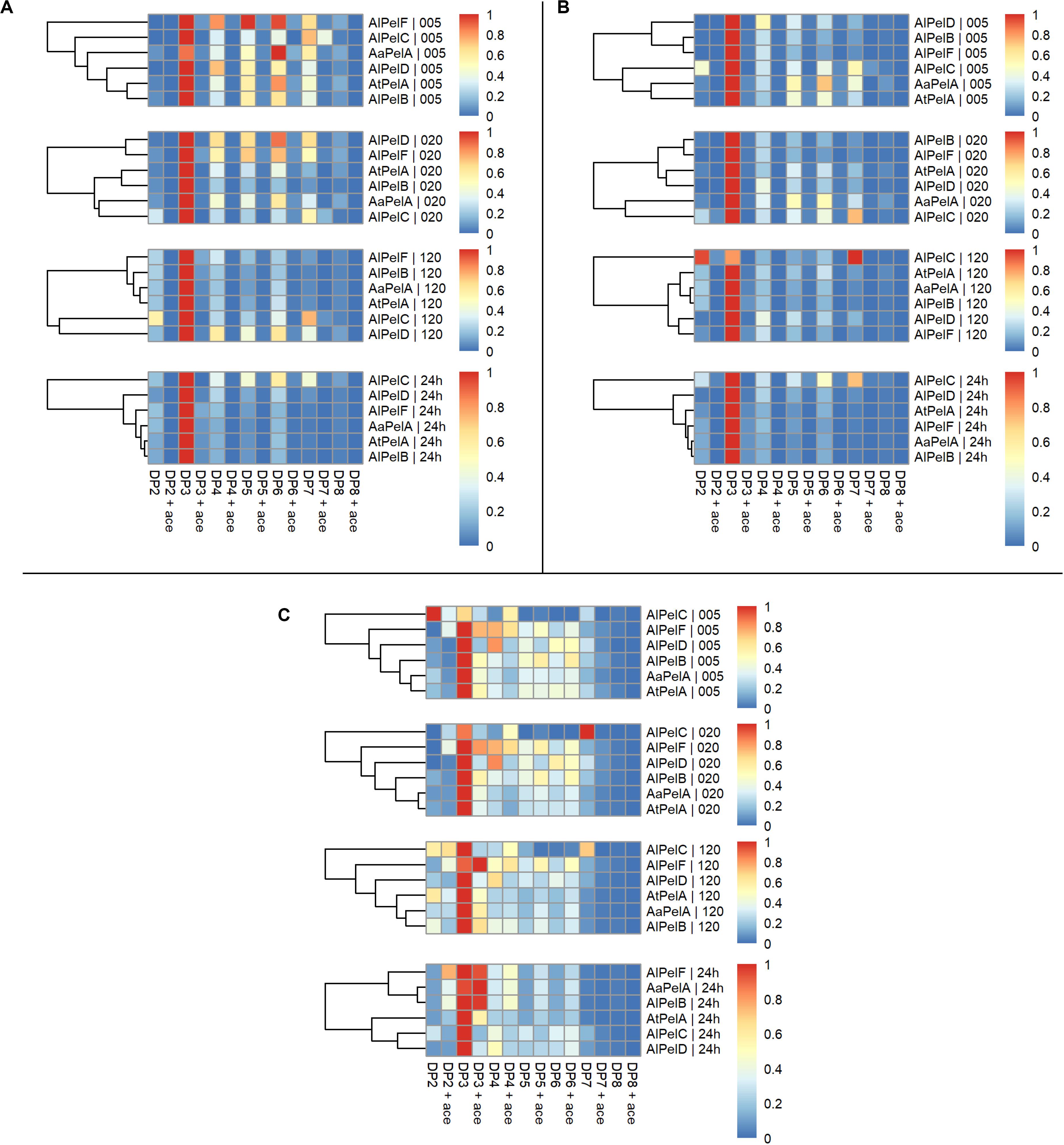
Figure 7. Heat maps of normalized summed relative intensities for individual degrees of polymerization divided non-acetyl substituted (DP columns) and acetyl substituted (DP ace columns) of pectin lyases acting on apple pectin (A), citrus pectin (B), and sugar beet pectin (C). Panels top to bottom: 5, 20, 120 min, and 24 h. For each substrate the pectin lyases are clustered according to product profiles within each time point.
A broad product profile was observed after 5 and 20 min for all enzymes, with a shift toward the narrow profile of mainly DP3 over the 24 h of reaction. Even though a limited decrease in the levels of the longer DPs was observed (Supplementary Figure S8), this shift in the profiles can be explained by the production rates of the different compounds. The apparent production rate of DP3 is the sum of products originating from degradation of longer oligomers/polymers not accounted for in the profile analysis. It seems likely that DP3 does not serve as a substrate for any of the pectin lyases, which is in agreement with previous studies (Van Alebeek et al., 2002). In contrast, the apparent formation rate of longer DP, e.g., DP6, is the net sum of the positive production rate and the negative degradation rate. If the latter is lower than the former, the net result is a positive apparent production rate of DP6, albeit lower than the production rate of DP3. In all cases, a shift in the profiles from higher DP (DP5-7) to DP3 was observed, with the exception of AlPelC that had a more differentiated profile throughout the reactions, regardless of substrate (Figures 6, 7). For the cluster analyses on citrus and sugar beet pectins after 24 h reaction, AaPelA and AlPelB clustered closest together, whereas AtPelA and AlPelB clustered closest together in reactions on apple pectin (Figures 6, 7). Furthermore, AlPelD and AlPelC had the second and most different product profile, when compared to the rest of the enzymes after 24 h. Phylogenetically, AlPelF clustes far from PelA and PelB (Figure 2) and has a different profile than these enzymes at short reaction times (Figures 6, 7). However, at extended reaction times (and high enzyme dosage), the AlPelF profile became increasingly similar to that of the PelAs and AlPelB. Thus, the differences observed for AlPelF could merely be a result of low activity, which could in turn be explained by lack of an otherwise conserved substrate-interacting Trp residue (Figure 3).
Major differences in specific activity were observed among the different pectin lyases (Table 3), and through systematic experiments, we also observed differences in pH-temperature optima for PelA, PelB, and PelD (Table 4). Clearly, from a competitive growth perspective, the ability of the fungus to produce a battery of similar enzymes having optimal activity at different pH and temperature represents a major advantage, and this fitness theory provides an explanation for the multigenecity phenomenon also for the pectin lyases. Interestingly, the ranking of kcat/Km across the three pectin substrates was the same for all of the four most active enzymes, but some discrepancies were observed for the Km values, suggesting different propensity toward methyl and acetyl substitutions among the pectin lyases (Table 4). This interpretation was further substantiated by the detailed LC-MS product profiling, where differences in substrate degradation proficiency as well as in the end product profiles were observed between the enzymes (Figures 6, 7). In particular, the discriminative substrate degradation pattern analysis revealed a significant differential activity sensibility to acetyl substitutions.
A. niger regulates the expression of carbohydrate-degrading enzymes at the transcriptional level. At least three different transcriptional activators are linked to pectin degradation, namely GaaR, RhaR, and AraR, which control the production of enzymes responsible for degradation of homogalacturonan, RG-I, and RG-I arabinan and arabinogalactan side chains, respectively (Kowalczyk et al., 2017). Moreover, the GalA-dependent transcriptional repressor GaaX and the general carbon catabolite repressor CreA also take part in controlling expression of pectinolytic enzymes in A. niger including the pectin lyases (Kowalczyk et al., 2017; Niu et al., 2017). When grown on sugar beet pectin, PelA was found to be regulated by GaaR, whereas PelD was regulated by both GaaR, RhaR, and possibly also AraR (Alazi et al., 2016; Kowalczyk et al., 2017). These transcription regulators are proteins which are activated by the presence and concentration of certain monosaccharides or a catabolic product thereof (Kowalczyk et al., 2017). Indeed, GaaR is induced by 2-keto-3-deoxy-L-galactonate, an intermediate of the GalA catabolic pathway (Alazi et al., 2016, 2017). Signal molecules such as GalA may be released from the complex carbohydrates by enzymes which are constitutively expressed at low levels. Indeed, A. niger PelB, PelC, and PelF and several other pectinolytic enzymes were found not to be regulated by GaaR. Data indicated that they may instead be regulated by the general carbon catabolite repressor CreA and could therefore play a role in release of GalA to induce expression of PelA and PelD (de Vries et al., 2002; Niu et al., 2015; Kowalczyk et al., 2017). For PelE from A. niger CBS 120.49 transcription levels were very low, and it is therefore hard to conclude whether PelE is also regulated by the GalA-induced GaaX-GaaR system (Alazi et al., 2018). Expression profiling and transcriptomics on A. niger grown on sugar beet pectin or GalA revealed that PelA and PelD (both regulated by GaaR) were predominantly expressed/transcribed after short incubation times. In contrast, levels of PelB and PelF (mRNA) increased throughout 24 h of incubation (de Vries et al., 2002; Alazi et al., 2016). The mRNA levels of PelC were very low, again questioning its role in pectin degradation (Alazi et al., 2016, 2018), in agreement with the very low activity observed in the current work. Comparing mRNA levels a general tendency was observed: PelA > PelF > PelD > PelB, and PelC and PelE were hardly transcribed (Martens-Uzunova and Schaap, 2009; Alazi et al., 2016, 2018; Kowalczyk et al., 2017).
Based on this, PelA appears to remain the major pectin lyase responsible for pectin degradation by Aspergilli in nature with high activity and high expression levels. However, the fact that AlPelB was observed to be more efficient enzyme in the current work, indicates a possible important role of this enzyme in releasing GalA to induce the transcription of PelA despite low transcription levels, especially at elevated pH. Indeed, varying expression levels may be the reason for the presence of the much less efficient AlPelF to release GalA for GaaR activation. From an industrial perspective, where enzymes are produced recombinantly, AlPelB thus appears to be promising candidate for an efficient enzyme with a broad pH range. In contrast, the reason for maintaining pelC and pelE in the chromosomes is more elusive, given the inferior activity of AlPelC and low transcription levels of both enzymes. The fact that pelE could not be identified in A. niger CBS 513.88 (Pel et al., 2007; Martens-Uzunova and Schaap, 2009) may suggest evolutionary pressure to lose such genes from the multigene family.
In order to study the multigenecity of pectin lyases in Aspergilli, representatives of PelA, PelB, PelC, PelD, and PelF were recombinantly expressed and characterized. A comparison of the pectin lyases in terms of activity at acidic pH, which is relevant for e.g., juice processing applications, the following ranking materialized: PelB > PelA > PelD > PelF > PelC. Interestingly, PelB was consistently the most efficient pectin lyase even at acidic conditions, despite having a higher pH optimum than PelA. In general, the highest extent of pectin degradation was observed for PelA and PelB. A systematic assessment of pH-temperature optima revealed significant differences in reaction optima among the different types of the pectin lyases, and homology modeling of the different versions of the pectin lyases supported that the slight sequence disparity amongst the copies, fosters structural differences in the active site region of the enzymes. Together with differences in thermal stability amongst the pectin lyases, these differences and the activity optimum variation easily supports the prevailing theory that this multigenecity is related to competitive fitness evolution providing the fungus with optimal tools for survival in different environments.
By providing a compendium of the degradation product profiles for the available Aspergillus pectin lyases on three different types of pectin (from apple, citrus, and sugar beet), which varied in degrees of methoxylation and acetylation, the present work added a new dimension to pectin lyase diversity characterization, and moreover provided a new angle to understand why fungi possess multiple copies of similar CAZymes. The discriminative LC-MS based substrate degradation pattern analysis provides support to the idea that the multigenecity is a prerequisite for the sophisticated biological “plasticity” that grants the fungus the ability to degrade an array of almost similar structural moieties, in this case pectin having different methoxylation and acetylation patterns. This interpretation agrees with the idea that the evolution of the CAZymes-secretome is an integral part of fungal speciation (Benoit et al., 2015; Barrett et al., 2020b). The product profiling method presented here may furthermore be useful as a novel tool in enzyme characterization as it enables discrimination between different enzymes with identical catalytic activity, and thus provides a new approach to continued exploration of the action of CAZymes on complex biomass substrates.
All datasets generated for this study are included in the article/Supplementary Material.
JH, AM, MS, and KK conceptualized and supervised the study. MS and KK expressed and purified the enzymes and determined their melting temperatures. TT performed experiments to determine substrate monosaccharide composition, enzyme pH-temperature optima, and kinetic constants. BZ determined initial rates on different pectin substrates and performed SDS-PAGE analysis. BZ, TT, and MS performed all in silico analyses. JH performed experiments and LC-MS and SEC analyses for product profiling and degradative pattern display. JH, AM, BZ, and TT analyzed all data. BZ, TT, and JH drafted the manuscript. AM, JH, MS, KK, and BZ revised the final manuscript. All authors contributed to the article and approved the submitted version.
This work was part of the BioValue SPIR Industrial Inovation Platform funded Innovation Fund Denmark, case no. 0603-00522B.
MS and KK are employed by the company Novozymes A/S.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Mikael Lenz Strube, Department of Biotechnology and Biomedicine, Technical University of Denmark, is thanked for assistance in data handling and data analysis.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fbioe.2020.00873/full#supplementary-material
Agger, J., Viksø-Nielsen, A., and Meyer, A. S. (2010). Enzymatic xylose release from pretreated corn bran arabinoxylan: differential effects of deacetylation and deferuloylation on insoluble and soluble substrate fractions. J. Agric. Food Chem. 58, 6141–6148. doi: 10.1021/jf100633f
Alazi, E., Khosravi, C., Homan, T. G., du Pré, S., Arentshorst, M., Di Falco, M., et al. (2017). The pathway intermediate 2-keto-3-deoxy-L-galactonate mediates the induction of genes involved in D-galacturonic acid utilization in Aspergillus niger. FEBS Lett. 591, 1408–1418. doi: 10.1002/1873-3468.12654
Alazi, E., Knetsch, T., Di Falco, M., Reid, I. D., Arentshorst, M., Visser, J., et al. (2018). Inducer-independent production of pectinases in Aspergillus niger by overexpression of the D-galacturonic acid-responsive transcription factor gaaR. Appl. Microbiol. Biotechnol. 102, 2723–2736. doi: 10.1007/s00253-018-8753-7
Alazi, E., Niu, J., Kowalczyk, J. E., Peng, M., Aguilar Pontes, M. V., van Kan, J. A. L., et al. (2016). The transcriptional activator GaaR of Aspergillus niger is required for release and utilization of d-galacturonic acid from pectin. FEBS Lett. 590, 1804–1815. doi: 10.1002/1873-3468.12211
Ali, S., Søndergaard, C. R., Teixeira, S., and Pickersgill, R. W. (2015). Structural insights into the loss of catalytic competence in pectate lyase activity at low pH. FEBS Lett. 589, 3242–3246. doi: 10.1016/j.febslet.2015.09.014
Almagro Armenteros, J. J., Tsirigos, K. D., Sønderby, C. K., Petersen, T. N., Winther, O., Brunak, S., et al. (2019). SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 37, 420–423. doi: 10.1038/s41587-019-0036-z
Andersen, M. R., Giese, M., de Vries, R. P., and Nielsen, J. (2012). Mapping the polysaccharide degradation potential of Aspergillus niger. BMC Genomics 13:313. doi: 10.1186/1471-2164-13-313
Barrett, K., Hunt, C. J., Lange, L., and Meyer, A. S. (2020a). Conserved unique peptide patterns (CUPP) online platform: peptide-based functional annotation of carbohydrate active enzymes. Nucleic Acids Res. 48, W110–W115. doi: 10.1093/nar/gkaa375
Barrett, K., Jensen, K., Meyer, A. S., Frisvad, J. C., and Lange, L. (2020b). Fungal secretome profile categorization of CAZymes by function and family corresponds to fungal phylogeny and taxonomy: example Aspergillus and Penicillium. Sci. Rep. 10:5158. doi: 10.1038/s41598-020-61907-1
Barrett, K., and Lange, L. (2019). Peptide-based functional annotation of carbohydrate-active enzymes by conserved unique peptide patterns (CUPP). Biotechnol. Biofuels 12, 1–21. doi: 10.1186/s13068-019-1436-5
Benoit, I., Coutinho, P. M., Schols, H. A., Gerlach, J. P., Henrissat, B., and de Vries, R. P. (2012). Degradation of different pectins by fungi: correlations and contrasts between the pectinolytic enzyme sets identified in genomes and the growth on pectins of different origin. BMC Genomics 13:321. doi: 10.1186/1471-2164-13-321
Benoit, I., Culleton, H., Zhou, M., DiFalco, M., Aguilar-Osorio, G., Battaglia, E., et al. (2015). Closely related fungi employ diverse enzymatic strategies to degrade plant biomass. Biotechnol. Biofuels 8:107. doi: 10.1186/s13068-015-0285-0
Bonnin, E., Garnier, C., and Ralet, M. C. (2014). Pectin-modifying enzymes and pectin-derived materials: applications and impacts. Appl. Microbiol. Biotechnol. 98, 519–532. doi: 10.1007/s00253-013-5388-6
Buchan, D. W. A., and Jones, D. T. (2019). The PSIPRED protein analysis workbench: 20 years on. Nucleic Acids Res. 47, W402–W407. doi: 10.1093/nar/gkz297
Buchholt, H. C., Christensen, T. M. I. E., Fallesen, B., Ralet, M. C., and Thibault, J. F. (2004). Preparation and properties of enzymatically and chemically modified sugar beet pectins. Carbohydr. Polym. 58, 149–161. doi: 10.1016/j.carbpol.2004.06.043
Chung, W. S. F., Meijerink, M., Zeuner, B., Holck, J., Louis, P., Meyer, A. S., et al. (2017). Prebiotic potential of pectin and pectic oligosaccharides to promote anti-inflammatory commensal bacteria in the human colon. FEMS Microbiol. Ecol. 93:fix127. doi: 10.1093/femsec/fix127
de Vries, R. P., Jansen, J., Aguilar, G., Paøenicová, L., Joosten, V., Wülfert, F., et al. (2002). Expression profiling of pectinolytic genes from Aspergillus niger. FEBS Lett. 530, 41–47. doi: 10.1016/S0014-5793(02)03391-4
de Vries, R. P., Riley, R., Wiebenga, A., Aguilar-Osorio, G., Amillis, S., Uchima, C. A., et al. (2017). Comparative genomics reveals high biological diversity and specific adaptations in the industrially and medically important fungal genus Aspergillus. Genome Biol. 18:28. doi: 10.1186/s13059-017-1151-0
Domon, B., and Costello, C. E. (1988). A systematic nomenclature for carbohydrate fragmentations in FAB-MS/MS spectra of glycoconjugates. Glycoconj. J. 5, 397–409. doi: 10.1007/BF01049915
Gysler, C., Harmsen, J. A. M., Kester, H. C. M., Visser, J., and Heim, J. (1990). Isolation and structure of the pectin lyase D-encoding gene from Aspergillus niger. Gene 89, 101–108. doi: 10.1016/0378-1119(90)90211-9
Harmsen, J. A. M., Kusters-van Someren, M. A., and Visser, J. (1990). Cloning and expression of a second Aspergillus niger pectin lyase gene (pelA): indications of a pectin lyase gene family in A. niger. Curr. Genet. 18, 161–166. doi: 10.1007/BF00312604
He, Y., Pan, L., and Wang, B. (2018). Efficient over-expression and application of high-performance pectin lyase by screening Aspergillus niger pectin lyase gene family. Biotechnol. Bioprocess Eng. 23, 662–669. doi: 10.1007/s12257-018-0387-1
Herron, S. R., Benen, J. A. E., Scavetta, R. D., Visser, J., and Jurnak, F. (2002). Structure and function of pectic enzymes: virulence factors of plant pathogens. Proc. Natl. Acad. Sci.U.S.A. 97, 8762–8769. doi: 10.1073/pnas.97.16.8762
Hong, S. B., Lee, M., Kim, D. H., Varga, J., Frisvad, J. C., Perrone, G., et al. (2013). Aspergillus luchuensis, an industrially important black Aspergillus in East Asia. PLoS One 8:e0063769. doi: 10.1371/journal.pone.0063769
Jamek, S. B., Mikkelsen, J. D., Busk, P. K., Meyer, A. S., Holck, J., Zeuner, B., et al. (2018). Loop protein engineering for improved transglycosylation activity of a β-N-Acetylhexosaminidase. ChemBioChem 19, 1858–1865. doi: 10.1002/cbic.201800181
Kassara, S., Li, S., Smith, P., Blando, F., and Bindon, K. (2019). Pectolytic enzyme reduces the concentration of colloidal particles in wine due to changes in polysaccharide structure and aggregation properties. Int. J. Biol. Macromol. 140, 546–555. doi: 10.1016/j.ijbiomac.2019.08.043
Kester, H. C. M., and Visser, J. (1994). Purification and characterization of pectin lyase B, a novel pectinolytic enzyme from Aspergillus niger. FEMS Microbiol. Lett. 120, 63–68. doi: 10.1016/0378-1097(94)00176-6
Kohli, P., and Gupta, R. (2019). Application of calcium alginate immobilized and crude pectin lyase from Bacillus cereus in degumming of plant fibres. Biocatal. Biotransformation 37, 341–348. doi: 10.1080/10242422.2018.1564745
Körner, R., Limberg, G., Christensen, T. M. I. E., Mikkelsen, J. D., and Roepstorff, P. (1999). Sequencing of partially methyl-esterified oligogalacturonates by tandem mass spectrometry and its use to determine pectinase specificities. Anal. Chem. 71, 1421–1427. doi: 10.1021/ac981240o
Kowalczyk, J. E., Lubbers, R. J. M., Peng, M., Battaglia, E., Visser, J., and De Vries, R. P. (2017). Combinatorial control of gene expression in Aspergillus niger grown on sugar beet pectin. Sci. Rep. 7, 1–12. doi: 10.1038/s41598-017-12362-y
Kusters-van Someren, M., Flipphi, M., de Graaff, L., van den Broeck, H., Kester, H., Hinnen, A., et al. (1992). Characterization of the Aspergillus niger pelB gene: structure and regulation of expression. MGG Mol. Gen. Genet. 234, 113–120. doi: 10.1007/BF00272352
Kusters-van Someren, M. A., Harmsen, J. A. M., Kester, H. C. M., and Visser, J. (1991). Structure of the Aspergillus niger pelA gene and its expression in Aspergillus niger and Aspergillus nidulans. Curr. Genet. 20, 293–299. doi: 10.1007/BF00318518
Leijdekkers, A. G. M., Sanders, M. G., Schols, H. A., and Gruppen, H. (2011). Characterizing plant cell wall derived oligosaccharides using hydrophilic interaction chromatography with mass spectrometry detection. J. Chromatogr. A 1218, 9227–9235. doi: 10.1016/j.chroma.2011.10.068
Lo, M. C., Aulabaugh, A., Jin, G., Cowling, R., Bard, J., Malamas, M., et al. (2004). Evaluation of fluorescence-based thermal shift assays for hit identification in drug discovery. Anal. Biochem. 332, 153–159. doi: 10.1016/j.ab.2004.04.031
Lombard, V., Bernard, T., Rancurel, C., Brumer, H., Coutinho, P. M., and Henrissat, B. (2010). A hierarchical classification of polysaccharide lyases for glycogenomics. Biochem. J. 432, 437–444. doi: 10.1042/bj20101185
Madeira, F., Park, Y. M., Lee, J., Buso, N., Gur, T., Madhusoodanan, N., et al. (2019). The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 47, W636–W641. doi: 10.1093/nar/gkz268
Mantovani, C. F., Geimba, M. P., and Brandelli, A. (2005). Enzymatic clarification of fruit juices by fungal pectin lyase. Food Biotechnol. 19, 173–181. doi: 10.1080/08905430500316284
Martens-Uzunova, E. S., and Schaap, P. J. (2009). Assessment of the pectin degrading enzyme network of Aspergillus niger by functional genomics. Fungal Genet. Biol. 46(Suppl. 1), S170–S179. doi: 10.1016/j.fgb.2008.07.021
Mayans, O., Scott, M., Connerton, I., Gravesen, T., Benen, J., Visser, J., et al. (1997). Two crystal structures of pectin lyase A from Aspergillus reveal a pH driven conformational change and striking divergence in the substrate-binding clefts of pectin and pectate lyases. Structure 5, 677–689. doi: 10.1016/S0969-2126(97)00222-0
Mosbech, C., Holck, J., Meyer, A. S., and Agger, J. W. (2018). The natural catalytic function of CuGE glucuronoyl esterase in hydrolysis of genuine lignin-carbohydrate complexes from birch. Biotechnol. Biofuels 11:71. doi: 10.1186/s13068-018-1075-2
Müller-Maatsch, J., Bencivenni, M., Caligiani, A., Tedeschi, T., Bruggeman, G., Bosch, M., et al. (2016). Pectin content and composition from different food waste streams in memory of Anna Surribas, scientist and friend. Food Chem. 201, 37–45. doi: 10.1016/j.foodchem.2016.01.012
Mutenda, K. E., Körner, R., Christensen, T. M. I. E., Mikkelsen, J., and Roepstorff, P. (2002). Application of mass spectrometry to determine the activity and specificity of pectin lyase A. Carbohydr. Res. 337, 1217–1227. doi: 10.1016/s0008-6215(02)00127-1
Niu, J., Alazi, E., Reid, I. D., Arentshorst, M., Punt, P. J., Visser, J., et al. (2017). An evolutionarily conserved transcriptional activator-repressor module controls expression of genes for D-galacturonic acid utilization in Aspergillus niger. Genetics 205, 169–183. doi: 10.1534/genetics.116.194050
Niu, J., Homan, T. G., Arentshorst, M., de Vries, R. P., Visser, J., and Ram, A. F. J. (2015). The interaction of induction and repression mechanisms in the regulation of galacturonic acid-induced genes in Aspergillus niger. Fungal Genet. Biol. 82, 32–42. doi: 10.1016/j.fgb.2015.06.006
Pel, H. J., de Winde, J. H., Archer, D. B., Dyer, P. S., Hofmann, G., Schaap, P. J., et al. (2007). Genome sequencing and analysis of the versatile cell factory Aspergillus niger CBS 513.88. Nat. Biotechnol. 25, 221–231. doi: 10.1038/nbt1282
Plaschina, I. G., Braudo, E. E., and Tolstoguzov, V. B. (1978). Circular dichroism studies of pectin solutions. Carbohydr. Res. 60, 1–8. doi: 10.1016/S0008-6215(00)83459-X
Quéméner, B., Cabrera Pino, J. C., Ralet, M. C., Bonnin, E., and Thibault, J. F. (2003a). Assignment of acetyl groups to O-2 and/or O-3 of pectic oligogalacturonides using negative electrospray ionization ion trap mass spectrometry. J. Mass Spectrom. 38, 641–648. doi: 10.1002/jms.478
Quéméner, B., Désiré, C., Debrauwer, L., and Rathahao, E. (2003b). Structural characterization by both positive and negative electrospray ion trap mass spectrometry of oligogalacturonates purified by high-performance anion-exchange chromatography. J. Chromatogr. A 984, 185–194. doi: 10.1016/S0021-9673(02)01729-6
Ralet, M. C., Cabrera, J. C., Bonnin, E., Quéméner, B., Hellìn, P., and Thibault, J. F. (2005). Mapping sugar beet pectin acetylation pattern. Phytochemistry 66, 1832–1843. doi: 10.1016/j.phytochem.2005.06.003
Remoroza, C., Cord-Landwehr, S., Leijdekkers, A. G. M., Moerschbacher, B. M., Schols, H. A., and Gruppen, H. (2012). Combined HILIC-ELSD/ESI-MS n enables the separation, identification and quantification of sugar beet pectin derived oligomers. Carbohydr. Polym. 90, 41–48. doi: 10.1016/j.carbpol.2012.04.058
Renard, C. M. G. C., and Thibault, J.-F. (1996). “Pectins in mild alkaline conditions: beta-elimination and kinetics of demethylation,” in Pectins and Pectinases. Book Series: Progress in Biotechnology, eds J. Visser and A. G. J. Voragen, (Amsterdam: Elsevier), 603–608. doi: 10.1016/s0921-0423(96)80292-9
Sandri, I. G., Fontana, R. C., Barfknecht, D. M., and da Silveira, M. M. (2011). Clarification of fruit juices by fungal pectinases. LWT Food Sci. Technol. 44, 2217–2222. doi: 10.1016/j.lwt.2011.02.008
Seyedarabi, A., To, T. T., Ali, S., Hussain, S., Fries, M., Madsen, R., et al. (2010). Structural insights into substrate specificity and the anti β-elimination mechanism of pectate lyase. Biochemistry 49, 539–546. doi: 10.1021/bi901503g
Sluiter, A., Hames, B., Ruiz, R., Scarlata, C., Sluiter, J., Templeton, D., et al. (2012). Laboratory Analytical Procedure (LAP): Determination of Structural Carbohydrates and Lignin in Biomass. Golden, CO: National Renewable Energy Laboratory.
Tan, H., Chen, W., Liu, Q., Yang, G., and Li, K. (2018). Pectin oligosaccharides ameliorate colon cancer by regulating oxidative stress- and inflammation-activated signaling pathways. Front. Immunol. 9:1504. doi: 10.3389/fimmu.2018.01504
Van Alebeek, G. J. W. M., Christensen, T. M. I. E., Schols, H. A., Mikkelsen, J. D., and Voragen, A. G. J. (2002). Mode of action of pectin lyase A of Aspergillus niger on differently C6-substituted oligogalacturonides. J. Biol. Chem. 277, 25929–25936. doi: 10.1074/jbc.M202250200
Van Alebeek, G. J. W. M., Zabotina, O., Beldman, G., Schols, H. A., and Voragen, A. G. J. (2000). Structural analysis of (methyl-esterified) oligogalacturonides using post-source decay matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. J. Mass Spectrom. 35, 831–840. doi: 10.1002/1096-9888(200007)35:7<831::aid-jms7>3.0.co;2-4
van Houdenhoven, F. E. A. (1975). Studies on Pectin Lyase. PhD thesis, Agricultural University, Wageningen.
Varga, J., Frisvad, J. C., Kocsubé, S., Brankovics, B., Tóth, B., Szigeti, G., et al. (2011). New and revisited species in Aspergillus section Nigri. Stud. Mycol. 69, 1–17. doi: 10.3114/sim.2011.69.01
Vitali, J., Schick, B., Kester, H. C. M., Visser, J., and Jurnak, F. (1998). The three-dimensional structure of Aspergillus niger pectin lyase B at 1.7-Å resolution. Plant Physiol. 116, 69–80. doi: 10.1104/pp.116.1.69
Voragen, A. G. J., Schols, H. A., and Pilnik, W. (1986). Determination of the degree of methylation and acetylation of pectins by h.p.l.c. Top. Catal. 1, 65–70. doi: 10.1016/S0268-005X(86)80008-X
Waterhouse, A., Bertoni, M., Bienert, S., Studer, G., Tauriello, G., Gumienny, R., et al. (2018). SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 46, W296–W303. doi: 10.1093/nar/gky427
Xu, S. X., Qin, X., Liu, B., Zhang, D. Q., Zhang, W., Wu, K., et al. (2015). An acidic pectin lyase from Aspergillus niger with favourable efficiency in fruit juice clarification. Lett. Appl. Microbiol. 60, 181–187. doi: 10.1111/lam.12357
Keywords: pectin lyase, multigenecity, sugar beet pectin, apple pectin, citrus pectin, product profiling
Citation: Zeuner B, Thomsen TB, Stringer MA, Krogh KBRM, Meyer AS and Holck J (2020) Comparative Characterization of Aspergillus Pectin Lyases by Discriminative Substrate Degradation Profiling. Front. Bioeng. Biotechnol. 8:873. doi: 10.3389/fbioe.2020.00873
Received: 11 April 2020; Accepted: 07 July 2020;
Published: 30 July 2020.
Edited by:
André Damasio, State University of Campinas, BrazilReviewed by:
Noppol Leksawasdi, Chiang Mai University, ThailandCopyright © 2020 Zeuner, Thomsen, Stringer, Krogh, Meyer and Holck. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Anne S. Meyer, YXNtZUBkdHUuZGs=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.