- 1State Key Laboratory of Biocatalysis and Enzyme Engineering, Environmental Microbial Technology Center of Hubei Province, College of Life Science, Hubei University, Wuhan, China
- 2Wuhan Junan Biotechnology Co., Ltd., Wuhan, China
Poly-γ-glutamic acid (γ-PGA) is an emerging biopolymer with various applications and γ-PGAs with different molecular weights exhibit distinctive properties. However, studies on the controllable molecular weights of biopolymers are limited. The purpose of this study is to achieve production of γ-PGAs with a wide range of molecular weights through manipulating the expression of γ-PGA depolymerase (PgdS) in Bacillus licheniformis WX-02. Firstly, the expression and secretion of PgdS were regulated through engineering its expression elements (four promoters and eight signal peptides), which generated γ-PGAs with molecular weights ranging from 6.82 × 104 to 1.78 × 106 Da. Subsequently, through combination of promoters with signal peptides, the production of γ-PGAs with a specific molecular weight could be efficiently obtained. Interestingly, the γ-PGA yield increased with the reduced molecular weight in flask cultures (Pearson correlation coefficient of −0.968, P < 0.01). Finally, in batch fermentation, the highest yield of γ-PGA with a weight-average molecular weight of 7.80 × 104 Da reached 39.13 g/L under glutamate-free medium. Collectively, we developed an efficient strategy for one-step production of γ-PGAs with specific molecular weights, which have potential application for industrial production of desirable γ-PGAs.
Introduction
Poly-γ-glutamic acid (γ-PGA), an anionic polymer composed of repeated D- and L-glutamic acid units via γ-amide linkages, is mainly produced by Bacillus species (e.g., B. licheniformis, B. subtilis, B. amyloliquefaciens) (Sirisansaneeyakul et al., 2017). Due to its versatile physical properties, γ-PGA has been used in various fields, including agriculture, food, cosmetics, and pharmaceutical industries (Cao et al., 2018). Notably, depending on the producers, the molecular weight (Mw) of γ-PGA varied with a range from 1.0 × 104 to over 2.0 × 106 Da (Sirisansaneeyakul et al., 2017). The γ-PGAs with different Mws are exploited in different applications (Ogunleye et al., 2015). For example, γ-PGAs with high-Mws are used as superior flocculants in the wastewater treatment (Yokoi et al., 1996). The medium-Mw γ-PGAs (∼9.9 × 105 Da) can efficiently remove basic dyes from solution (Inbaraj et al., 2006). Low-Mw γ-PGAs (2.0 × 104–2.7 × 105 Da) can be used as the drug carrier and tissue engineering nanocomposite in the biomedical industry (Shu et al., 2014). Thus, the controlled Mw is critical for the development and application of γ-PGA.
Previously, efforts have been made to change the Mws of γ-PGAs through changing fermentation conditions (Zeng et al., 2016; Feng et al., 2017). Also, several physical methods such as ultrasonication and heating (Pérez−Camero et al., 1999), as well as chemical methods like acidic and alkaline hydrolysis (Kubota et al., 1996), have been applied to depolymerize γ-PGA. However, since those methods lack precise control of Mws, the increasing polydispersity leads to a complicated purification process, which in turn limits the application of γ-PGA. By contrast, the enzymatic depolymerization via γ-PGA depolymerase is an alternative way to produce γ-PGA with desirable Mws with several superiorities, including mild reaction conditions and non-pollution (Yao et al., 2009). However, the extraction and purification of the depolymerase PgdS and subsequent hydrolysis processes are time-consuming and comparatively tedious, hampering the application of the enzymatic method for tailor-made γ-PGA production.
Recently, one-step fermentation is becoming an attractive method for biorefinery. For example, γ-PGA with a wider range of Mws (4.0 × 104–8.5 × 106 Da) were achieved in B. subtilis via expression of different γ-PGA synthetases (Halmschlag et al., 2019). Several endo-type PgdS depolymerases have been characterized (Suzuki and Tahara, 2003; Tian et al., 2014; Sha et al., 2018). Tian et al. (2014) characterized the PgdS hydrolase from B. licheniformis WX-02 and expressed this enzyme to achieve efficient production of low-Mw γ-PGAs. Moreover, Sha et al. (2018) achieved production of γ-PGA with different Mws through expressing four PgdS hydrolases with different hydrolytic activity. These results indicate that Mws of γ-PGA are correlated with PgdS hydrolase activity.
In this study, we develop a simple system for one-step production of γ-PGA through regulating the expression of PgdS depolymerase in B. licheniformis WX-02, a glutamate independent producer. The promoter and signal peptide are the vital factors for expression and secretion of proteins (Zhang et al., 2016). The Mws of γ-PGA was systematically modified by manipulating the promoters and signal peptides independently and in combination. By generating PgdS with different hydrolytic activities, γ-PGAs with a broad range of Mws were efficiently produced. Therefore, the strategy of engineering PgdS expression cassette is feasible to produce γ-PGAs with specific Mws.
Materials and Methods
Strains, Plasmids and Culture Conditions
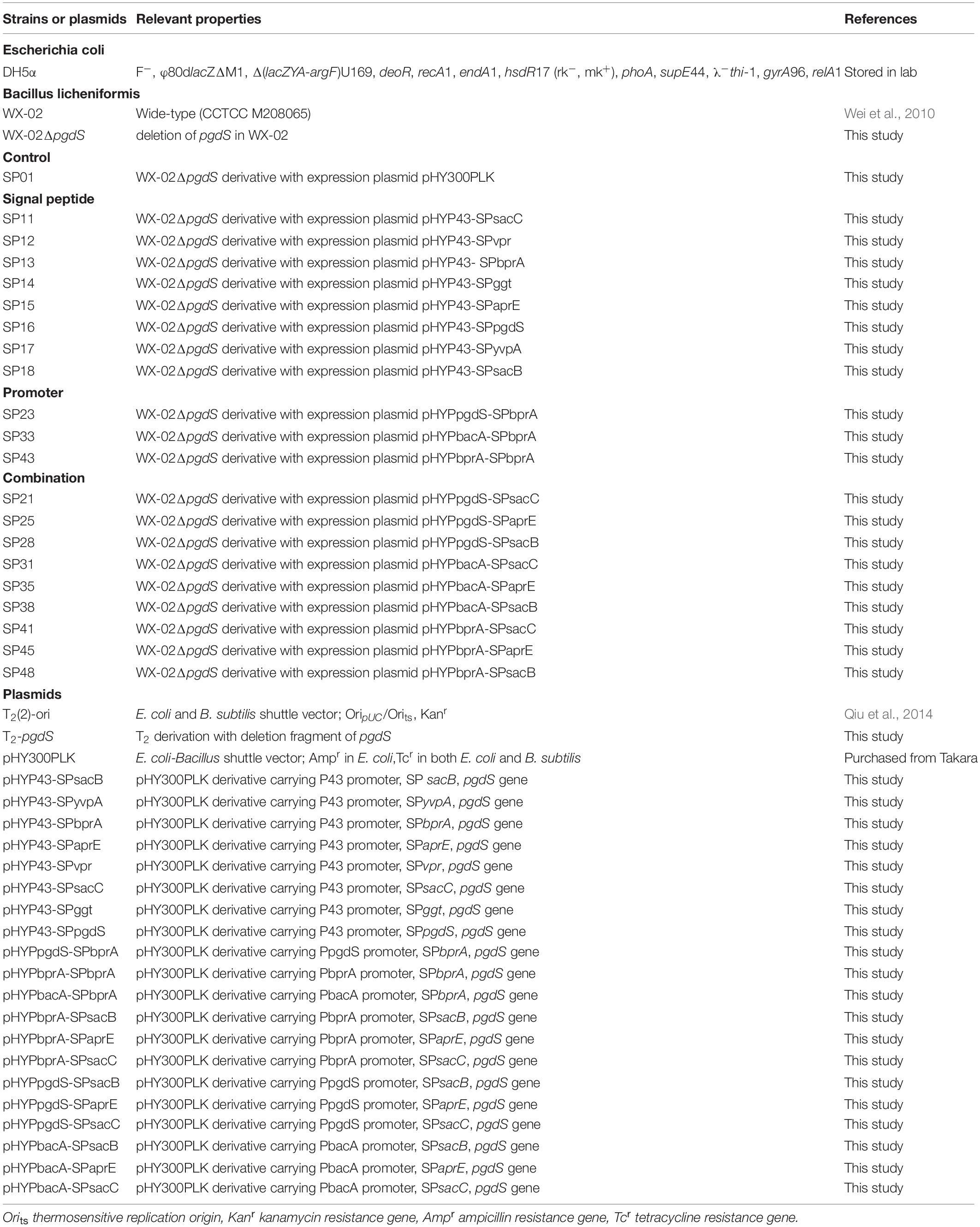
All strains and plasmids employed in this work were listed in Table 1. The pHY-300PLK plasmid and temperature-sensitive plasmid T2(2)-ori were used to construct the expression and deletion vectors, respectively. All strains were generally cultivated at 37°C in Luria Bertani (LB) medium. When necessary, antibiotics (20 μg/L kanamycin, or 20 μg/L tetracycline) were supplemented into the media.
For the seed cultures, B. licheniformis cells were precultured in LB medium and incubated at 37°C for 12 h. For γ-PGA fermentation, 250 mL flasks containing 50 mL medium (glucose 80.00 g/L, sodium citrate 30.00 g/L, NH4Cl 8.00 g/L, NaNO3 15.00 g/L, K2HPO4⋅3H2O 1.00 g/L, MgSO4⋅7H2O 1.00 g/L, ZnSO4⋅7H2O 1.00 g/L, CaCl2 1.00 g/L, MnSO4⋅H2O 0.15 g/L, pH7.2) were inoculated with 3% (v/v) seed cultures and cultivated at 37°C.
The B. licheniformis cells grow in 1-L bioreactor (T&J Bio-engineering Co., Ltd., Shanghai, China) with 0.60 L of media for 10 h. The aeration rate was 1.0 vvm, and the stirring speed was 300 rpm. The volume fractions of exhausted O2 and CO2 were measured by exhaust gas analyzer, and the volumetric mass transfer coefficient were calculated by the exhaust gas analysis system (T&J Bio−engineering Co., Ltd., Shanghai, China) (Cai et al., 2018).
Batch fermentation was carried out in the 3-L bioreactor (T&J Bio-engineering Co., Ltd., Shanghai, China) containing 1.8 L media with an aeration rate of 1.0 vvm. The agitation speed was starting to set at 300 rpm and increased to 600 rpm after 12 h. Samples were collected periodically to analyze the cell growth, glucose, and γ-PGA concentrations. ΔpgdS was further verified by diagnostic PCR and DNA sequencing.
Construction of Gene Expression Strains
As an example, the construction procedure of the plasmid pHYP43-SPsacB (containing P43 promoter and SPsacB) was described. Briefly, P43 promoter of B. subtilis 168, signal peptide of levansucrase SacB (SPsacB), gene pgdS (without its own signal peptide) and amyL terminator of B. licheniformis WX-02 were amplified and fused by overlapping PCR to obtain the expression cassettes (Figure 1). The expression cassettes were inserted into EcoRI/XbaI-cut pHY-300PLK, and the resulting plasmids were transformed into WX-02ΔpgdS to obtain recombinant strain SP18. Other PgdS expression strains were constructed by the similar method. Notably, all the recombinant vectors were verified by DNA sequencing. Moreover, the empty plasmid pHY300PLK was transformed into WX-02ΔpgdS to generate the control strain SP01.
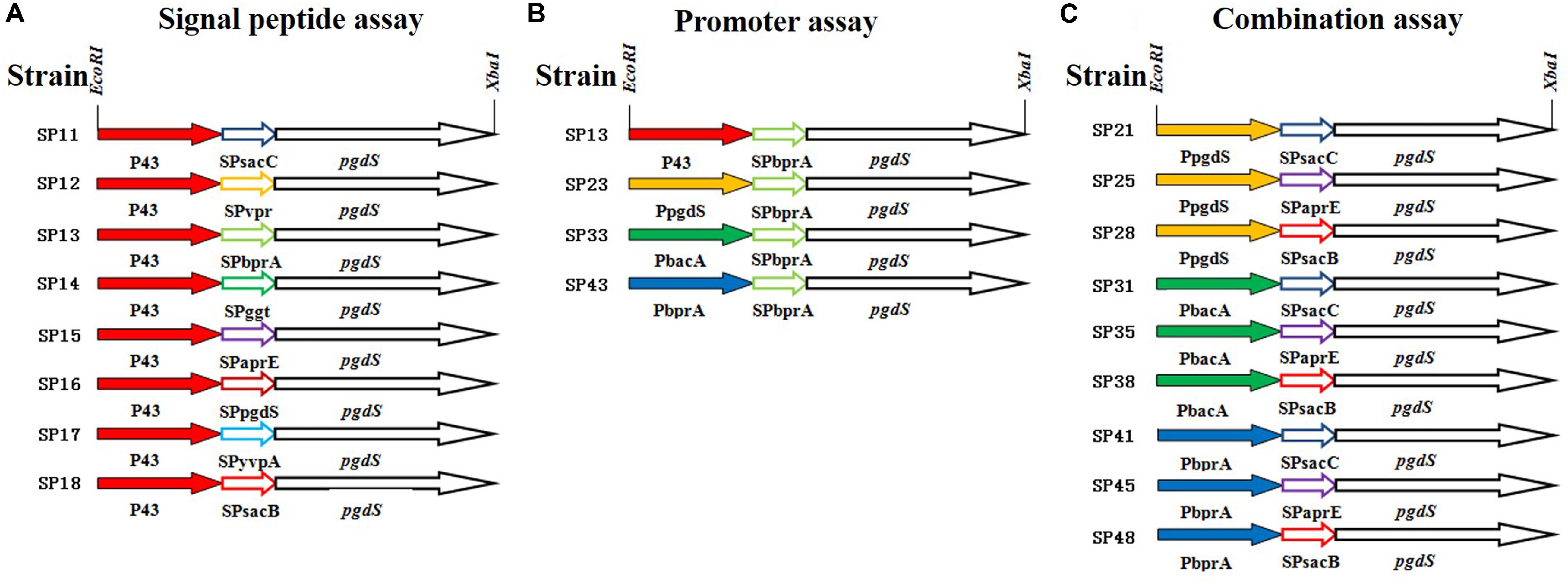
Figure 1. The details of PgdS expression cassettes constructed by manipulating its promoter and signal peptide independently and in combination. (A) Signal peptide assay containing SPsacB, SPaprE, SPggt, SPvpr, SPbprA, SPyvpA, SPpgdS, and SPsacC using P43 promoter; (B) promoter assay containing P43, PbacA, PpgdS, and PbprA using BprA signal peptide; (C) combination assay containing three promoters (PpgdS, PbacA, and PbprA) and three signal peptides (SPsacB, SPpgdS, and SPsacC). Different colors indicate different promoters or signal peptides, and the named rule of strains: first number means promoter, second number means signal peptide.
Enzymatic Assay for PgdS
Crude enzyme solution of PgdS was attained by removing the cells via centrifugation (13 700 × g, 20 min). Enzyme activity was measured as described by Ashiuchi et al. (2006). Briefly, the reaction contains 20 μM f potassium phosphate buffer (pH 6.0), 0.2 nM 1.0 × 106 Da γ-PGA substrate, 2 μM dithiothreitol and 10 μL of crude enzyme solution, and was incubated at 37°C for 4 h. After stopped, 100 μL of reaction solution was withdrawn and mixed with 100 μL of 0.2 M borate buffer (pH 8.5) and 20 μL of 10 mmol/L fluorodinitrobenzene (FDNB, in acetone). The hydrolysis of FDNB-modified fragments and the assay of dinitrophenyl glutamate monomers (DNPGlu) were performed as described previously (Ashiuchi et al., 2006). One unit of PgdS was defined as the amount of enzyme that generated 1 nmol of terminal glutamate group per minute.
SDS-PAGE Analysis
SDS-PAGE analysis was applied to determine the extracellular contents of PgdS in recombinant strains and the BSA (bovine serum albumin) was used as a protein standard. Briefly, 1 mL fermentation broth was centrifuged at 10000 × g for 10 min, and the supernatant was precipitated by 6.12 mol/L trichloroacetic acid (TCA). The precipitate was washed with absolute alcohol, and re-dissolved in a 100 uL solution which containing 2 mol/L thiourea and 8 mol/L urea. Protein concentrations were quantitated by Bradford assay (Hammond and Kruger, 1988) and BSA was used as a protein standard. Protein samples as while as BSA solution were mixed with 2 × SDS–PAGE loading buffer containing β-mercaptoethanol in 1:1 ratio, then boiling water bath for 5 min and then samples contain loading buffer were analyzed by the 15% (w/v) gel (Cai et al., 2016). The gels were then imaged with a Bio-Rad’s Gel Doc XR + system (Bio-Rad, United States). The target bands in the gels were visualized and quantified with ImageJ software (Rasband, W.S., ImageJ, United States National Institutes of Health, Bethesda, MD, United States) (Gallagher, 2014), and the BSA proteins were used as standards.
Quantitative PCR Analysis
When the cells grew into the mid-logarithmic growth phase, the cells were collected for RNA extraction according to Shi’s method (Shi et al., 2019), The HiScript®II Q RT SuperMix for qPCR (+gDNA wiper) (Vazyme, China) was employed for cDNA synthesis. The real-time PCR was performed using iTaqTM Universal SYBR® Green Supermix (Bio-Rad, United States). The experiments were performed in three replicates, and 16S rRNA was used as the reference gene (Rocha et al., 2015). The relative transcriptional level of pgdS gene was calculated using the 2–ΔΔCt method.
Analytical Methods
The cell density (OD600) was measured using the 752N spectrophotometer (Shanghai Opler Instrument Co., Ltd., Shanghai, China). The glucose concentration was determined by SBA-40C bioanalyzer (Academy of Sciences, Shandong, China).
The purification of γ –PGA was carried out by the method reported previously (Tian et al., 2014). Number average molecular weight (Mn), weight average molecular weight (Mw), and polydispersity index (Mw/Mn) of γ-PGA were measured using gel permeation chromatography (GPC) with a refractive index (RI) detector and a Shodex OHpak SB-806 HQ column (8.0 mm ID × 300 mm, 13 μm) (Birrer et al., 1994). Pullulan standards of narrow polydispersity (SHANGHAI ZZBIO CO., Ltd., Shanghai, China) were employed to establish a calibration curve. The concentration of γ-PGA was calculated from the peak area of the GPC measurements, with purified γ-PGA as a standard (Birrer et al., 1994; Xu et al., 2019).
Statistical Analysis
All data are represented as the means of three replicates and bars represent the standard deviations. Data were analyzed by SPSS Statistics software v. 19.0. Pearson correlation coefficient, t-test, and ANOVA test were carried out to compare means values and p < 0.05 were considered statistically significant.
Results
To systematically study the influence of PgdS expression level on the Mws of γ-PGAs, the native pgdS gene was deleted from the genome of B. licheniformis WX-02. PgdS depolymerase was expressed episomally and controlled by the promoter and signal peptide independently and in combination. We employed the strategy to construct twenty recombinant strains with different PgdS hydrolytic activities to generate γ-PGA with different Mws. In our previous study, the nattokinase was used as a reporter to test the effects of signal peptide on protein expression (Cai et al., 2016). According to the data, nattokinase protein with the eight signal peptides (SPsacB, SPaprE, SPpgdS, SPbprA, SPggt, SPsacC, SPyvpA, and SPvpr) expressed in various levels (nearly ten times between the highest level and the lowest level, due to the high copy number of plasmid harboring pgdS gene, we tend to choose the signal peptides with low expression level to avoid over expression of PgdS) (Supplementary Table S2). In order to get multiple expression levels of PgdS, those eight signal peptides and four promoters (P43, PpgdS, PbprA, and PbacA) were employed to construct pgdS expression vectors (Figure 1). According to the result of pre-experiment (Supplementary Figure S1A), the PGA titers of each strain were highest at 36 h. The molecular weight of PGA was decreased with the prolonged fermentation (Supplementary Figure S1B), while, at the later period of fermentation (30–42 h), the γ-PGA molecular weight values for the recombinants SP12, SP14, and SP18 were maintained. So the time of fermentation of all the strains were set as 36 h.
Modulating PgdS Secretion to Tune γ-PGA Molecular Weight
All PgdS expression vectors were transferred into the pgdS deletion strain WX-02ΔpgdS, and the resultant strains were designated B. licheniformis SP11–SP18, respectively (Table 1).
In our previous study, we have construct SP library in B. licheniformis using nattokinase as reporter (Cai et al., 2016). We choose eight different SP from B. licheniformis to expression PgdS. The SDS-PAGE analysis of the culture supernatants (Supplementary Figure S2) showed a band of the predicted molecular mass (4.2 × 104 Da) among the SP12–SP18 strains, except the control strain SP01 and SP11 (carrying cassette with the combination of P43-SPsacC). Among these strains, the secretion efficiency ratios were SPsacB > SPyvpA > SPpgdS > SPaprE > SPggt > SPbprA > SPvpr > SPsacC (Table 2). As indicated in Figure 2, overexpression of PgdS enabled efficient reduction of Mws, which was consistent with the results reported by Tian et al. (2014) and Sha et al. (2018). The PgdS overexpression strains also produced γ-PGA with lower Mns values and polydispersity index (Table 2). The Mws of γ-PGAs produced by recombinant strains were ranged from (6.82 ± 0.51) × 104 to (1.71 ± 0.08) × 106 Da, compared with the control strain (1.99 ± 0.11) × 106 Da. Interestingly, cell growth showed no significant differences between the recombinant strains and control strain SP01. Therefore, our results indicated that γ-PGA with a wide range of Mws (6.82 × 104–1.71 × 106 Da) could be achieved via modulating the secretory capability of PgdS.
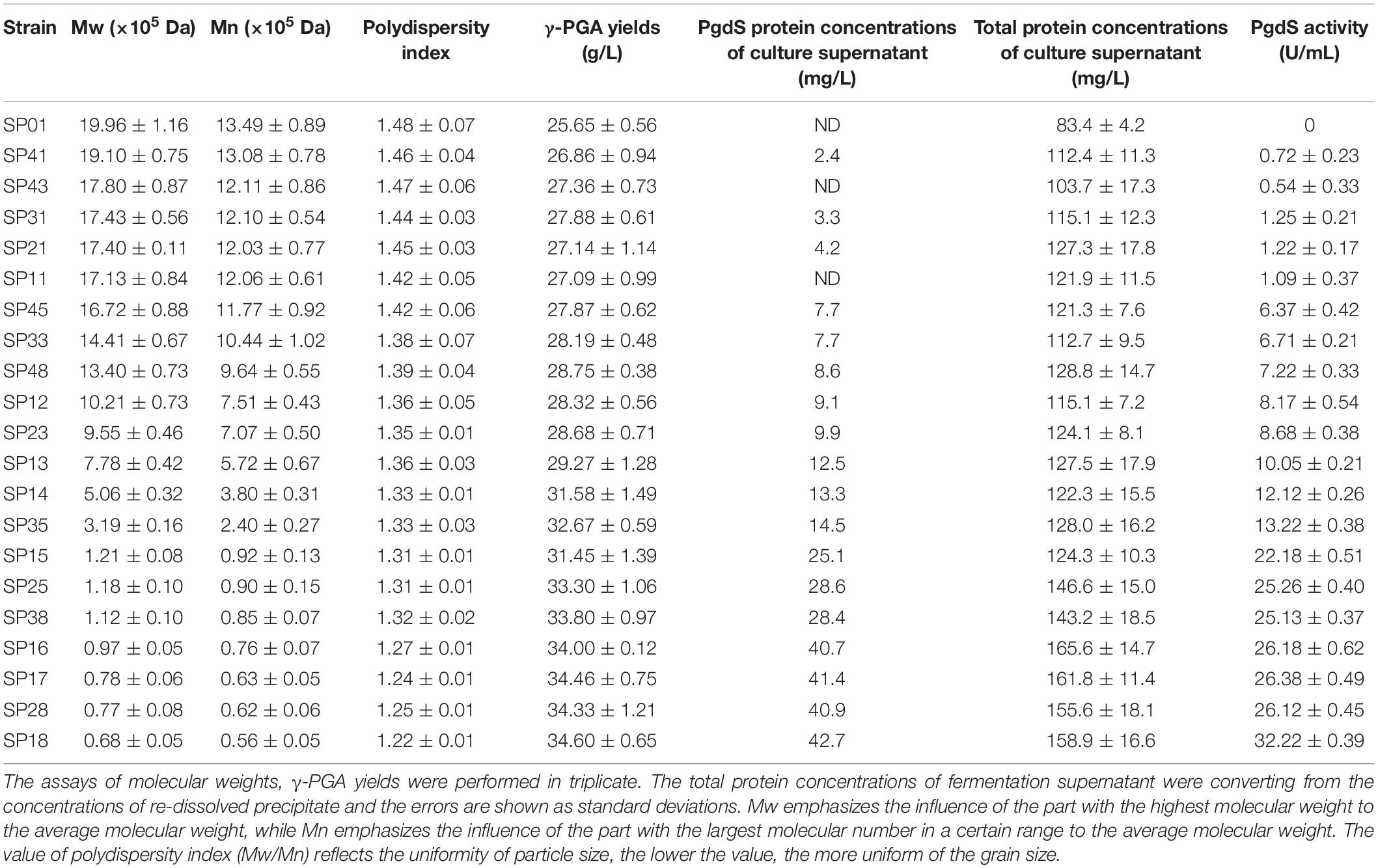
Table 2. The γ-PGA yields of recombinant strains and the PgdS concentrations of culture supernatants.
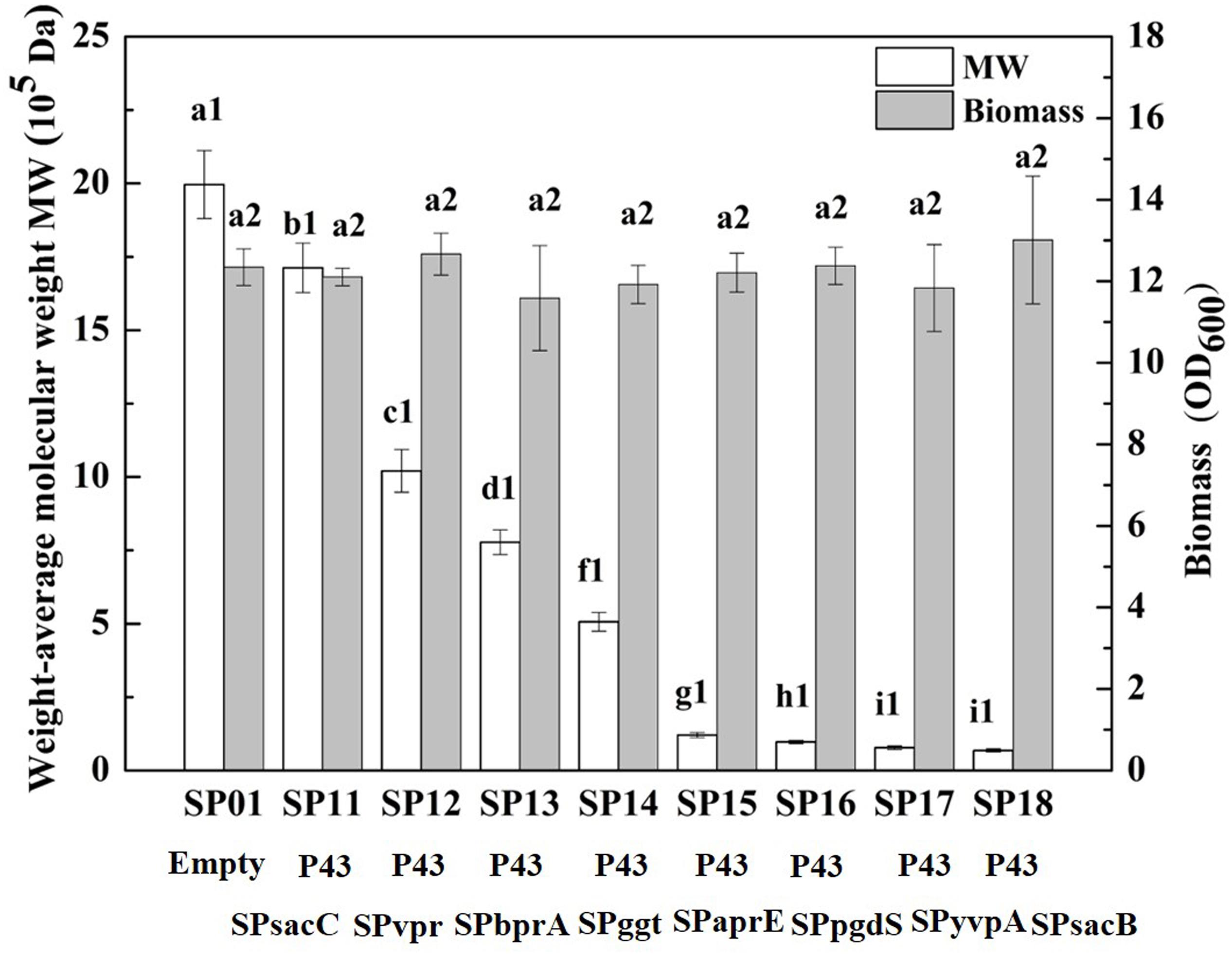
Figure 2. Comparison of Mws and biomass in B. licheniformis under different signal peptides. The data are presented as the mean ± SD of three replications. Different lowercase letters indicate significant differences among strains at 0.05 level (p < 0.05).
Modification of γ-PGA Molecular Weight by Regulating pgdS Transcriptional Level
According to the above results, the Mws of γ-PGAs could be tuned by PgdS secretion. We then evaluated the effect of the transcriptional levels of pgdS on Mws by using promoters with different strength. Four promoters P43, PpgdS, PbacA, and PbprA were applied to drive the transcription of pgdS. The four recombinant vectors harboring the signal peptide BprA were constructed and transformed into the WX-02ΔpgdS, generated four recombinant strains SP13, SP23, SP33, and SP43, respectively (Table 1).
To examine the expression efficiency of the four promoters, the transcriptional levels of pgdS driven by these promoters were determined by real-time PCR. The results showed that the transcription levels from P43, PpgdS, and PbacA were increased in different degrees compared with the PbprA, and P43 exhibited the highest transcription level (p < 0.05) (Figure 3A). The expression strength of different promoters was also determined by SDS-PAGE assay (Supplementary Figure S3), and the concentrations of PgdS were quantified (Table 2), the results confirmed that the expression strength of these four promoters was in a descending order of P43 > PpgdS > PbacA > PbprA.
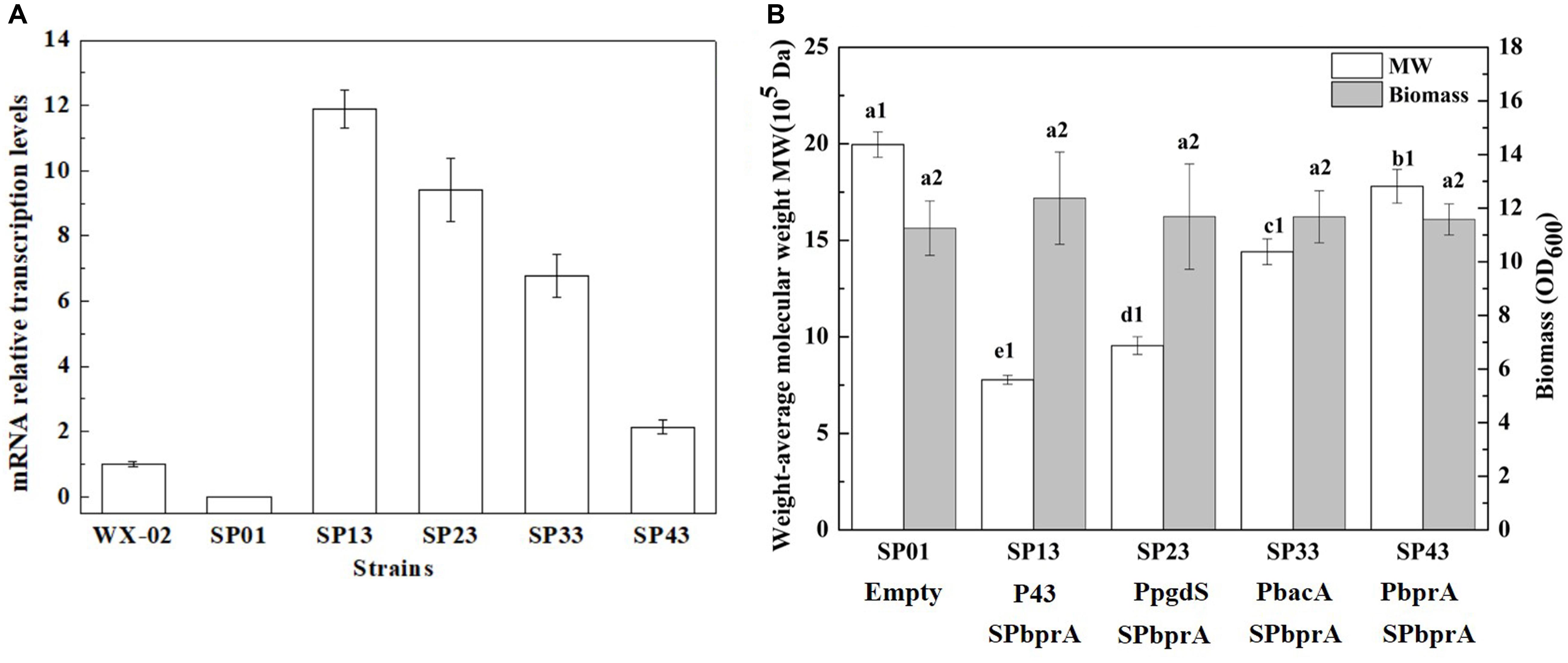
Figure 3. The transcriptional levels of gene pgdS among strains with different promoters (A). Data are expressed as mean ± SD of three replications. Comparison of Mws and biomass in B. licheniformis under different promoters (B). The data are presented as the mean ± SD of three replications. Different lowercase letters indicate significant differences among strains at 0.05 level (p < 0.05).
After 36 h cultivation, we measured the Mws and yields of γ-PGA produced by these recombinant strains. The Mws of the polymer produced by SP13, SP23, SP33, and SP43 were (7.78 ± 0.23) × 105, (9.55 ± 0.46) × 105, (14.41 ± 0.67) × 105, and (17.80 ± 0.87) × 105 Da, respectively (Figure 3B), which decreased by 61.02, 52.15, 27.81, and 10.82%, respectively, compared to the control strain SP01 (19.96 × 105 Da). However, the cell growth of these four recombinant strains were approximately the same as that of SP01. Collectively, our results showed that an apparent reduction of Mws along with the enhanced promoter strength.
Varying Levels of PgdS Expression Enable Controlling the Molecular Weights of γ-PGAs
In order to further boost the performance, the promoter and signal peptide were combined to regulate PgdS expression. Three promoters [PpgdS(H), PbacA(M) and PbprA(L)] and signal peptides [SPsacB(H), SPpgdS(M), SPsacC(L)] were selected and combined to form nine expression cassettes (Figure 1). Every expression cassette was cloned in pHY300PLK, and the resultant vectors were transformed into WX-02ΔpgdS. The transformed strains were designated as B. licheniformis SP21, SP25, SP28, SP31, SP35, SP38, SP41, SP45, and SP48, respectively.
As shown in Table 2, the activities of PgdS in strains SP28, SP25, and SP38 (26.12 ± 0.45, 25.26 ± 0.4, and 25.13 ± 0.37 U/mL) were significantly greater among all strains, and SP35 (13.22 ± 0.38 U/mL) and SP48 (7.22 ± 0.33 U/mL) exhibited moderate activity. In contrast, weak enzyme activities were detected in strains SP21, SP31, SP41, and SP45. The results of SDS-PAGE assay (Supplementary Figures S4, S5) and the concentrations of PgdS protein of each strains (Table 2) were consistent with PgdS activity assay. The Mws of γ-PGA produced by these combinations were measured at 36 h (Figure 4). The Mws of γ-PGA mediated by the nine combinations varied greatly from (7.74 ± 0.80) × 104 to (1.91 ± 0.08) × 106 Da. To assess the relationship between PgdS expression and γ-PGA Mws, the Pearson correlation coefficients were calculated (Weaver and Wuensch, 2013). A correlation coefficient of −0.945 indicated a strong inverse relation between extracellular PgdS activity and γ-PGA Mws (P < 0.01) (Supplementary Figure S6A). Taken together, engineering PgdS expression cassette was an efficient approach to produce γ-PGAs with specific Mws.
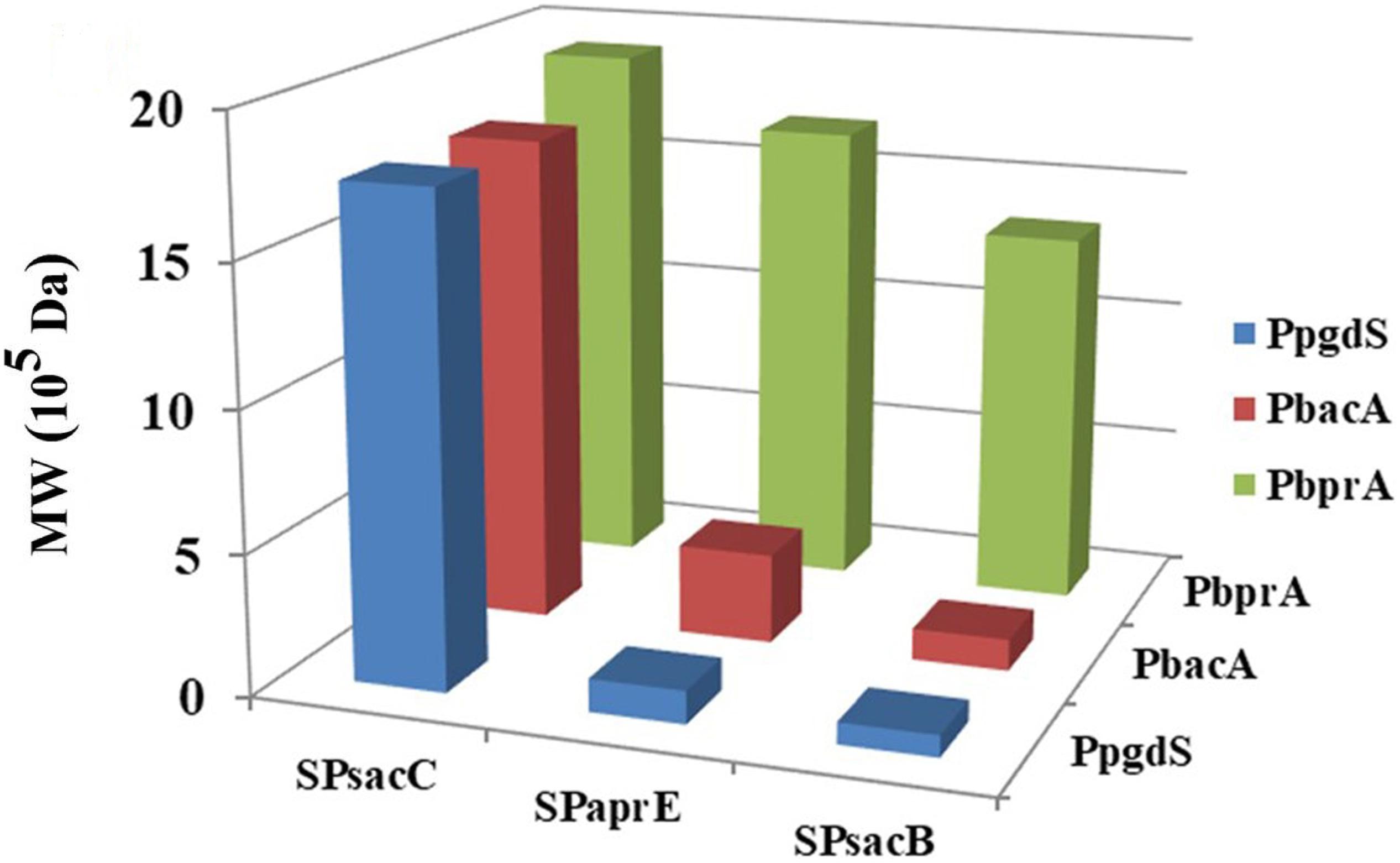
Figure 4. The Mws of γ-PGA in recombinant strains (SP01, SP41, SP45, SP48, SP31, SP35, SP38, SP21, SP25, and SP28). Data are expressed as mean ± SD of three replications, M: protein marker.
Increasing γ-PGA Yield Along With Reduced Molecular Weights
In this study, we achieved the production of low-, medium-, and high-Mw γ-PGA in the same microbial chassis. The effect of PgdS expression levels on γ-PGA yield was further studied (Table 2). We found that the γ-PGA yield in recombinant strains was obviously increased compared to the control strain when the Mws of γ-PGA decreased by over 50%. A dispersion diagram was obtained with the PgdS yields and logarithms of γ-PGA molecular weight (Supplementary Figure S6B), showing the increasing γ-PGA yield along with the reduction of molecular weights (Pearson correlation coefficient = −0.958). Furthermore, the strain SP18 produced 34.60 g/L γ-PGAs, which increased by 34.89% compared with the control strain (25.65 g/L).
The viscosity of fermentation broth affects the oxygen transfer rate (OTR), which is proportional to the volumetric mass transfer coefficient and substrate utilization. The OTR plays vital roles in the growth of strains and the production of target metabolites (Damiani et al., 2014). We hypothesized that improvement of γ-PGA yield in engineered strains may be due to the higher OTR. To prove this hypothesis, the OTRs of fermentation broths from the B. licheniformis SP01 (Mw, 1.99 × 106 Da), SP12 (Mw, 1.02 × 106 Da), SP14 (Mw, 5.06 × 105 Da), and SP18 (Mw, 6.82 × 104 Da) were compared in 1-L fermenter (Figure 5). All recombinant strains showed improved kLa compared to the control strain SP01 throughout the fermentation process. Among them, kLa and DO were consistent with the trend of values negatively correlated with the γ-PGA Mws in these recombinant strains (Figures 5A,B). These results confirmed that low viscosity of fermentation broth could improve γ-PGA yield by increasing the oxygen transfer rate.
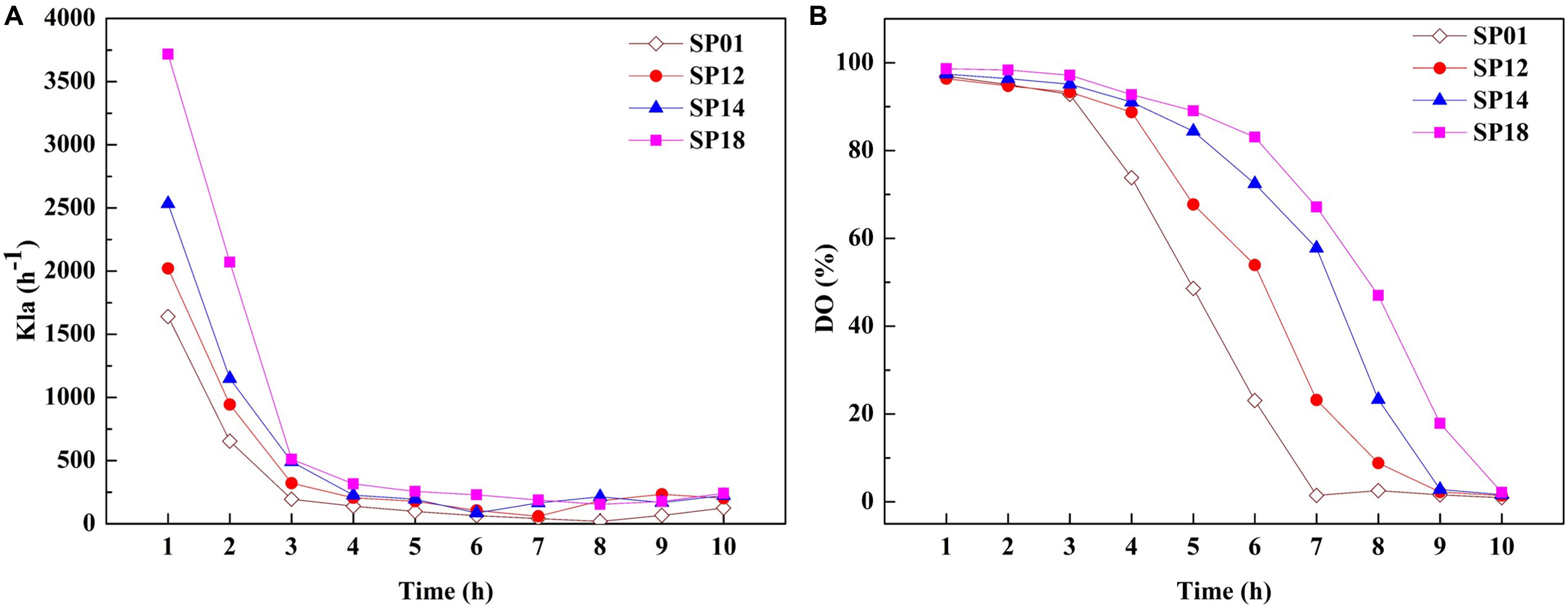
Figure 5. Comparison of kLa (A) and dissolved oxygen (DO) (B) between control strain SP01 and recombinant strains in a 1-L fermenter. Time courses of recombinant strains SP01, SP12, SP14, and SP18 in batch culture. The kLa and DO were measured and calculated at regular intervals.
Large-Scale Fermentation of Recombinant B. licheniformis in 3-L Fermenter
Based on the above results, an efficient system for one-step synthesis of γ-PGA with a wide range of Mws (6.82 × 104–1.99 × 106 Da) was developed by regulating PgdS expression. Therefore, in order to further explored the applicability of this system in large-scale γ-PGA production, the engineered strains SP01, SP12, SP14, and SP18 were carried out in a 3-L bioreactor, respectively (Figure 6). Compared to the control strain SP01, the DO values in cultures of SP12, SP14, and SP18 were maintained at higher levels, resulting in higher cell growth rates and biomass (Figure 6). The highest yield of γ-PGAs produced by the SP01 was 29.00 g/L at 30 h (Figure 5A). In contrast, the maximal γ-PGA yields in strains SP12, SP14, and SP18 reached 33.05, 35.91, and 39.13 g/L, respectively, increasing by 12.25, 23.83, and 34.93% (Figures 5B–D), and the Mws of γ-PGAs in corresponding strains were 1.42 × 106, 5.56 × 105, and 7.83 × 104 Da, respectively. Therefore, our results demonstrated that it was feasible and efficient to produce specific γ-PGAs using glutamate-free medium by the controllable expression of PgdS.
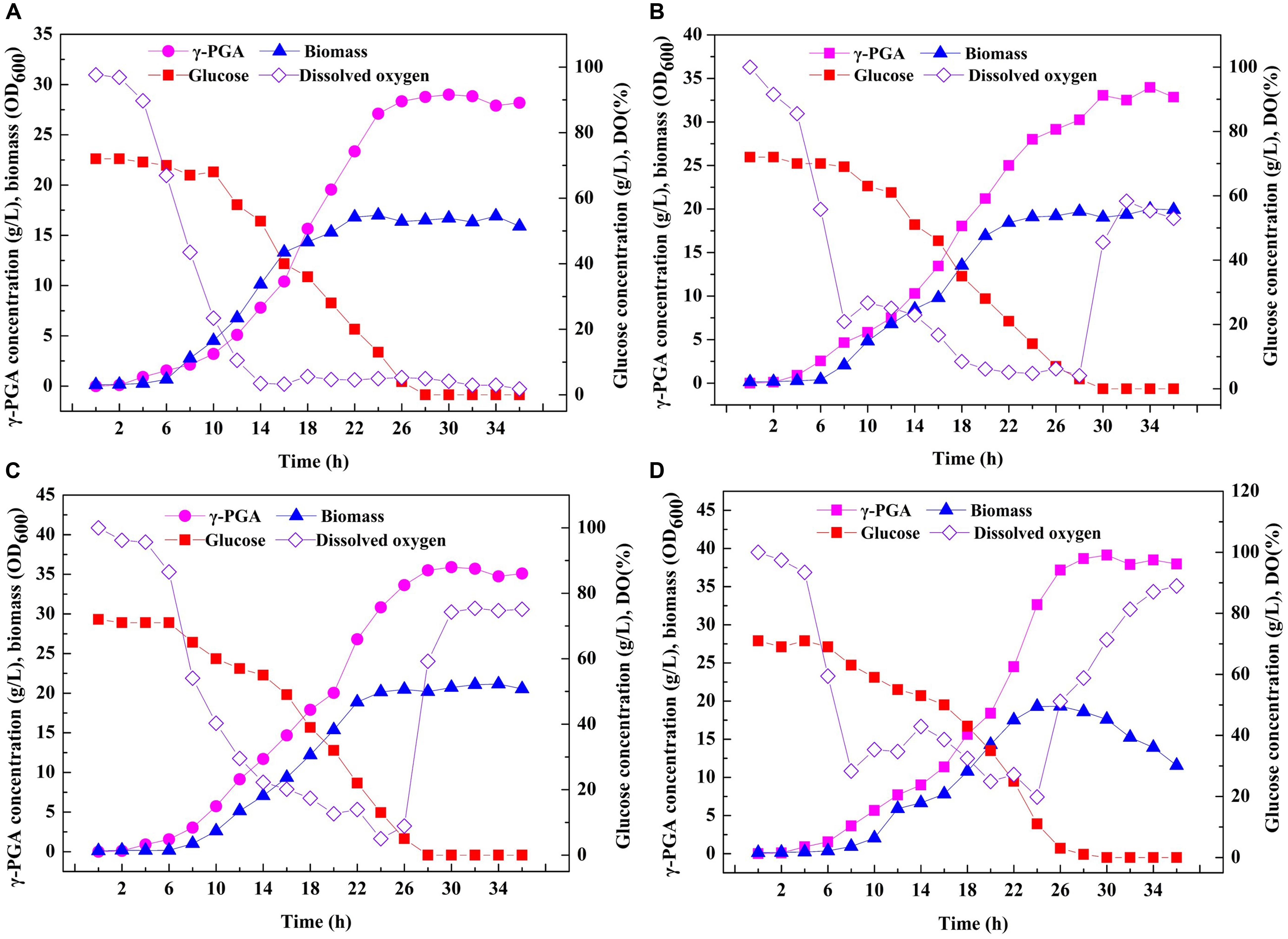
Figure 6. Large-scale fermentation of recombinant B. licheniformis strains in 3-L fermenter. Time courses of recombinant strains SP01 (A), SP12 (B), SP14 (C), and SP18 (D) in batch culture. γ-PGA concentration, biomass (OD600), glucose concentration and DO were measured and calculated at regular intervals.
Discussion
Since γ-PGAs with different Mws show specific applications, the Mws must be precisely controlled. γ-PGAs produced by a single strain was often difficult to meet the requirements of molecular weight differentiation (Supplementary Table S4). In this work, we aimed to develop a convenient system for one-step production of regulated-molecular-weight γ-PGAs through the controlled expression of PgdS depolymerase by manipulating the promoters and signal peptides independently and in combination.
Currently, many endo-type γ-PGA hydrolases (PgdS) has been characterized from various strains (Suzuki and Tahara, 2003; Tian et al., 2014) and used as a target to improve the yields of γ-PGAs (Scoffone et al., 2013; Sha et al., 2018). It has been proved that PgdS degraded γ-PGA in the extracellular region (Yao et al., 2009). The optimization of signal peptides is an efficient method to increase the secretion of desirable proteins (Zhang et al., 2016). According to Cai et al., 81 signal peptides were identified in B. licheniformis WX-02 by the SignalP tool1 (Cai et al., 2016). The effects of all signal peptides on the secretion of nattokinase from B. subtilis MBS 04-6 were investigated, and the strains with different signal peptides showed different activities ranging from 0 to 31.99 FU/mL (Cai et al., 2016). In this study, we investigated the effects of eight proteases signal peptides (SacC, Vpr, BprA, GGT, AprE, PgdS, YvpA, and SacB) on PgdS secretion and γ-PGA Mws. Our results showed that higher γ-PGA degradation capability was achieved with the increased PgdS secretion, which was consistent with the previous reports (Yuan et al., 2015; Jin et al., 2016). Besides, the optimal signal peptides varied for different proteins (Degering et al., 2010). In this study, the maximum activity of PgdS being obtained under the signal peptide of SPsacB in B. licheniformis among the selected signal peptides (Table 2), which is different with the optimal signal peptide (SPaprE) for nattokinase secretion (Cai et al., 2016).
The promoter is another critical element for controlling the protein expression (Wang et al., 2014). There are many classes of promoters that could be employed to dynamically regulate the expression of protein, such as inducible promoters, constitutive promoters of different strengths, condition-responsive promoters, and growth phase responsive promoters (Fontana et al., 2018; Maury et al., 2018; Tekel et al., 2019). In this work, we explore the effects of pgdS transcriptional levels on γ-PGA Mws using promoters with different strengths (P43, PbacA, PbprA, and PpgdS). P43 is considered as a strong promoter for protein expression (Zhang et al., 2007). PpgdS is the native promoter of PgdS depolymerase and regulated by sigma D factor from B. licheniformis (Mitsunaga et al., 2016). PbacA is the promoter of bacABC operon involved in bacitracin synthesis from B. licheniformis DW2 (Shu et al., 2018). PbprA, the promoter of bacillopeptidase F (BprA), is a weak promoter according to our transcriptome data (data not shown) and used as a control. Among the four promoters, P43 promoter is the most effective one to drive the expression of pgdS.
In order to further evaluate the effect of regulated PgdS expression on the Mws of γ-PGAs, we explored various levels of PgdS expression by combining promoters and signal peptides. Through this strategy, the combined strains provided γ-PGA with a wide range of Mws [(7.74 ± 0.80) × 104–(1.91 ± 0.08) × 106 Da]. Based on these results, regulating PgdS expression was an efficient and simple method to obtain γ-PGAs with specific Mws. Compared to other methods, we could achieve broader range of Mws of γ-PGAs, which is beneficial to broaden the industrial application of γ-PGAs. Previous studies have mainly focused on increasing the production of high-Mw γ-PGAs (Birrer et al., 1994; Scoffone et al., 2013), limiting the applications of γ-PGAs in other fields. However, one limitation of this study is that the determination of the correlations between the PgdS expression and γ-PGA Mws is based on small dataset (n = 9). We will achieve precise control of Mws via the titratable regulation of PgdS expression using the further development of tools.
Specially, all recombinant strains containing pgdS gene produced higher γ-PGA yields compared with the control strain SP01, which confirmed that overexpression of PgdS is an efficient approach to enhance γ-PGA production. Due to the high viscosity of high-Mw γ-PGAs, it severely decreases oxygen transfer in the fermentation process, resulting in inhibition of cell growth and limit of γ-PGA production (Su et al., 2010). In this paper, the kLa and DO of fermentation broths in recombinant strains were improved compared to the control strain SP01 with high-Mw γ-PGA. One possible explanation for the increased DO was that the reduced Mws decreased the viscosity of the fermentation broth, enabling higher oxygen transfer rate, further improving cell growth, substrate utilization and γ-PGA yields.
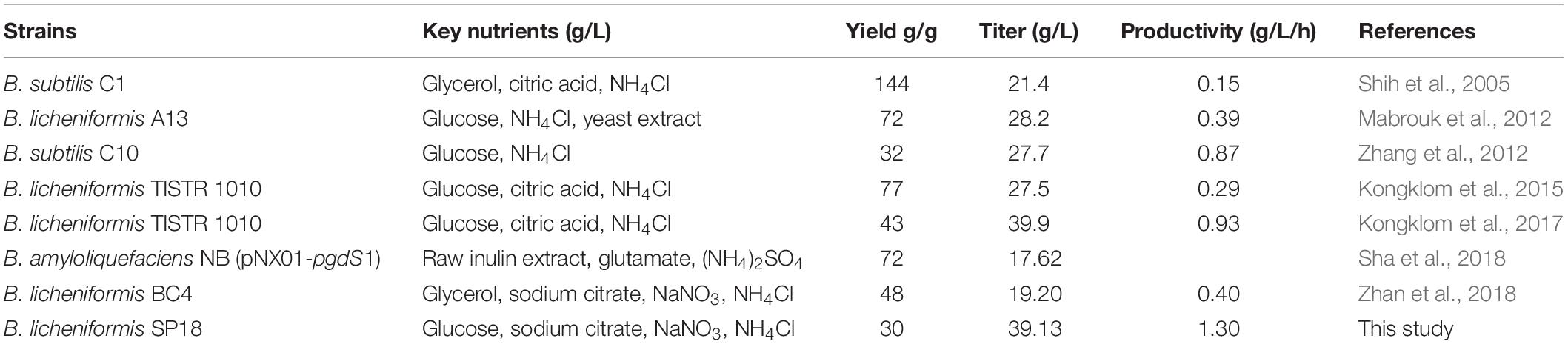
Ultimately, we investigated the applicability of the system in the large-scale production of specific-molecular-weight γ-PGAs. Our results indicated that the γ-PGA could be efficiently produced on a high level (29.00–39.13 g/L) without additional glutamic acid using the recombinant strains in 3-L fermenter. The results from this work and related studies were summarized in Table 3. In particular, the highest yield of γ-PGA (39.13 g/L) with molecular-weight value of 7.83 × 104 Da was obtained in SP18 strain, which was 34.93% higher than that of control strain SP01, and the yield was higher than those produced by most γ-PGA producers (Table 3). Moreover, the γ-PGA productivity was 1.30 g/L/h, which was the highest from glutamate-free medium to date. One possible explanation for the increased DO was that the reduced Mws decreased the viscosity of the fermentation broth, enabling higher oxygen transfer rate, further improving cell growth, substrate utilization and γ-PGA yields. Collectively, it is more applicable to industrially produce specific-molecular-weight γ-PGAs using our strategies than previously reported studies.
In conclusion, we developed an efficient system for one-step production of specific Mws γ-PGA through regulating the PgdS expression in B. licheniformis for the first time. The ability to produce γ-PGA with low, medium, and high Mws ranging between 6.82 × 104 Da and 1.99 × 106 Da was demonstrated by manipulating the promoter and signal peptide independently and in combination. The maximum production of γ-PGA (Mw, 7.83 × 104 Da) reached 39.13 g/L from glutamate-free medium in batch fermentation. Our study presented a potential method for commercial production of specific-molecular-weight γ-PGA, and this strategy could also be used to produce other biopolymers by precisely controlling corresponding depolymerase expression.
Data Availability Statement
All datasets generated for this study are included in the article/Supplementary Material.
Author Contributions
SC and FS designed and supervised the study. HW, DW, YX, JD, and JC performed the experiments. DW and HW analyzed the data and wrote the manuscript. YZ, QW, DC, SC, and FS revised the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (No. 31972849), the National Key Research and Development Program of China (2018YFA090039), and the Technical Innovation Special Fund of Hubei Province (No. 2018ACA149).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fbioe.2020.00728/full#supplementary-material
Footnotes
References
Ashiuchi, M., Nakamura, H., Yamamoto, M., and Misono, H. (2006). Novel poly -γ-glutamate-processing enzyme catalyzing γ-glutamyl DD-amidohydrolysis. J. Biosci. Bioeng. 102, 60–65. doi: 10.1263/jbb.102.60
Birrer, G. A., Cromwick, A. M., and Gross, R. A. (1994). Gamma-poly(glutamic acid) formation by Bacillus licheniformis 9945a: physiological and biochemical studies. Int. J. Biol. Macromol. 16, 265–275. doi: 10.1016/0141-8130(94)90032-9
Cai, D., Chen, Y., He, P., Wang, S., Mo, F., Li, X., et al. (2018). Enhanced production of poly−γ−glutamic acid by improving ATP supply in metabolically engineered Bacillus licheniformis. Biotechnol. Bioeng. 115, 2541–2553. doi: 10.1002/bit.26774
Cai, D., Wei, X., Qiu, Y., Chen, Y., Chen, J., Wen, Z., et al. (2016). High-level expression of nattokinase in Bacillus licheniformis by manipulating signal peptide and signal peptidase. J. Appl. Microbiol. 121, 704–712. doi: 10.1111/jam.13175
Cao, M., Feng, J., Sirisansaneeyakul, S., Song, C., and Chisti, Y. (2018). Genetic and metabolic engineering for microbial production of poly-gamma-glutamic acid. Biotechnol. Adv. 36, 1424–1433. doi: 10.1016/j.biotechadv.2018.05.006
Damiani, A. L., Kim, M. H., and Wang, J. (2014). An improved dynamic method to measure kLa in bioreactors. Biotechnol. Bioeng. 111, 2120–2125. doi: 10.1002/bit.25258
Degering, C., Eggert, T., Puls, M., Bongaerts, J., Evers, S., Maurer, K. H., et al. (2010). Optimization of protease secretion in Bacillus subtilis and Bacillus licheniformis by screening of homologous and heterologous signal peptides. Appl. Environ. Microbiol. 76, 6370–6376. doi: 10.1128/AEM.01146-10
Feng, J., Shi, Q., Zhou, G., Wang, L., Chen, A., Xie, X., et al. (2017). Improved production of poly-γ-glutamic acid with low molecular weight under high ferric ion concentration stress in Bacillus licheniformis ATCC 9945a. Process. Biochem. 56, 30–36. doi: 10.1016/j.procbio.2017.02.017
Fontana, J., Dong, C., Ham, J. Y., Zalatan, J. G., and Carothers, J. M. (2018). Regulated expression of sgRNAs tunes CRISPRi in E. coli. Biotechnol. J. 13:e1800069. doi: 10.1002/biot.201800069
Gallagher, S. R. (2014). Digital image processing and analysis with ImageJ. Curr Protoc. Essent Lab. Techni. 3, 36–42. doi: 10.1002/9780470089941.eta03cs9
Halmschlag, B., Steurer, X., Putri, S. P., Fukusaki, E., and Blank, L. M. (2019). Tailor-made poly-γ-glutamic acid production. Metab. Eng. 55, 239–248. doi: 10.1016/j.ymben.2019.07.009
Hammond, J. B. W., and Kruger, N. J. (1988). The Bradford method for protein quantitation. Methods Mol. Biol. 3, 25–32. doi: 10.1385/0-89603-126-8:25
Inbaraj, B. S., Chiu, C. P., Ho, G. H., Yang, J., and Chen, B. H. (2006). Removal of cationic dyes from aqueous solution using an anionic poly-gamma-glutamic acid-based adsorbent. J. Hazard. Mater. 137, 226–234. doi: 10.1016/j.jhazmat.2006.01.057
Jin, P., Kang, Z., Yuan, P., Du, G., and Chen, J. (2016). Production of specific-molecular-weight hyaluronan by metabolically engineered Bacillus subtilis 168. Metab. Eng. 35, 21–30. doi: 10.1016/j.ymben.2016.01.008
Kongklom, N., Luo, H., Shi, Z., Pechyen, C., Chisti, Y., and Sirisansaneeyakul, S. (2015). Production of poly-γ-glutamic acid by glutamic acid-independent Bacillus licheniformis TISTR 1010 using different feeding strategies. Biochem. Eng. J. 100, 67–75. doi: 10.1016/j.bej.2015.04.007
Kongklom, N., Shi, Z., Chisti, Y., and Sirisansaneeyakul, S. (2017). Enhanced production of Poly-gamma-glutamic Acid by Bacillus licheniformis TISTR 1010 with environmental controls. Appl. Biochem. Biotechno.l 182, 990–999. doi: 10.1007/s12010-016-2376-1
Kubota, H., Nambu, Y., and Endo, T. (1996). Alkaline hydrolysis of poly (γ−glutamic acid) produced by microorganism. J. Polym. Sci. Pol. Chem. 34, 1347–1351. doi: 10.1002/(SICI)1099-0518(199605)34:7<1347::AID-POLA24<3.0.CO;2-8
Mabrouk, M., Abou-Zeid, D., and Sabra, W. (2012). Application of Plackett–Burman experimental design to evaluate nutritional requirements for poly (γ-glutamic acid) production in batch fermentation by Bacillus licheniformis A13. Afr. J. Appl. Microbiol. Res 1, 6–18.
Maury, J., Kannan, S., Jensen, N. B., Oberg, F. K., Kildegaard, K. R., Forster, J., et al. (2018). Glucose-dependent promoters for dynamic regulation of metabolic pathways. Front. Bioeng. Biotechnol. 6:63. doi: 10.3389/fbioe.2018.00063
Mitsunaga, H., Meissner, L., Buchs, J., and Fukusaki, E. (2016). Branched chain amino acids maintain the molecular weight of poly(gamma-glutamic acid) of Bacillus licheniformis ATCC 9945 during the fermentation. J. Biosci. Bioeng. 122, 400–405. doi: 10.1016/j.jbiosc.2016.03.007
Ogunleye, A., Bhat, A., Irorere, V. U., Hill, D., Williams, C., and Radecka, I. (2015). Poly-gamma-glutamic acid: production, properties and applications. Microbiology. 161(Pt 1), 1–17. doi: 10.1099/mic.0.081448-0
Pérez−Camero, G., Congregado, F., Bou, J. J., and Muñoz−Guerra, S. (1999). Biosynthesis and ultrasonic degradation of bacterial poly (γ−glutamic acid). Biotechnol. Bioeng. 63, 110–115. doi: 10.1002/(sici)1097-0290(19990405)63:1<110::aid-bit11>3.0.co;2-t
Qiu, Y., Xiao, F., Wei, X., Wen, Z., and Chen, S. (2014). Improvement of lichenysin production in Bacillus licheniformis by replacement of native promoter of lichenysin biosynthesis operon and medium optimization. Appl. Microbiol. Biotechnol. 98, 8895–8903. doi: 10.1007/s00253-014-5978-y
Rocha, J., Santos, S., and Pacheco, G. (2015). Bacterial reference genes for gene expression studies by RT-qPCR: survey and analysis. Anton Van Leeuw. 108, 685–693. doi: 10.1007/s10482-015-0524-1
Scoffone, V., Dondi, D., Biino, G., Borghese, G., Pasini, D., Galizzi, A., et al. (2013). Knockout of pgdS and ggt genes improves gamma-PGA yield in B. subtilis. Biotechnol. Bioeng. 110, 2006–2012. doi: 10.1002/bit.24846
Sha, Y., Zhang, Y., Qiu, Y., Xu, Z., Li, S., Feng, X., et al. (2018). Efficient biosynthesis of low-molecular-weight poly-gamma-glutamic acid by stable overexpression of PgdS hydrolase in Bacillus amyloliquefaciens NB. J Agric. Food. Chem. 67, 282–290. doi: 10.1021/acs.jafc.8b05485
Shi, J., Zhan, Y., Zhou, M., He, M., Wang, Q., Li, X., et al. (2019). High-level production of short branched-chain fatty acids from waste materials by genetically modified Bacillus licheniformis. Bioresour. Technol. 271, 325–331. doi: 10.1016/j.biortech.2018.08.134
Shih, L., Wu, P. J., and Shieh, C. J. (2005). Microbial production of a poly (γ-glutamic acid) derivative by Bacillus subtilis. Process. Biochem. 40, 2827–2832. doi: 10.1016/j.procbio.2004.12.009
Shu, C., Wang, D., Guo, J., Song, J., Chen, S., Chen, L., et al. (2018). Analyzing AbrB-knockout effects through genome and transcriptome sequencing of Bacillus licheniformis DW2. Front. Microbiol. 9:307. doi: 10.3389/fmicb.2018.00307
Shu, X., Shi, Q., Feng, J., Xie, X., and Chen, Y. (2014). Design and in vitro evaluation of novel γ-PGA/hydroxyapatite nanocomposites for bone tissue engineering. J. Mater. Sci. 49, 7742–7749. doi: 10.1007/s10853-014-8484-9
Sirisansaneeyakul, S., Cao, M., Kongklom, N., Chuensangjun, C., Shi, Z., and Chisti, Y. (2017). Microbial production of poly-gamma-glutamic acid. World. J. Microbiol. Biotechnol. 33:173. doi: 10.1007/s11274-017-2338-y
Su, Y., Li, X., Liu, Q., Hou, Z., Zhu, X., Guo, X., et al. (2010). Improved poly-γ-glutamic acid production by chromosomal integration of the Vitreoscilla hemoglobin gene (vgb) in Bacillus subtilis. Bioresour. Technol. 101, 4733–4736. doi: 10.1016/j.biortech.2010.01.128
Suzuki, T., and Tahara, Y. (2003). Characterization of the Bacillus subtilis ywtD gene, whose product is involved in gamma-polyglutamic acid degradation. J. Bacteriol. 185, 2379–2382. doi: 10.1128/JB.185.7.2379-2382.2003
Tekel, S. J., Smith, C. L., Lopez, B., Mani, A., Connot, C., Livingstone, X., et al. (2019). Engineered orthogonal quorum sensing systems for synthetic gene regulation in Escherichia coli. Front. Bioeng. Biotechnol. 7:80. doi: 10.3389/fbioe.2019.00080
Tian, G., Fu, J., Wei, X., Ji, Z., Ma, X., Qi, G., et al. (2014). Enhanced expression of pgdS gene for high production of poly−γ−glutamic aicd with lower molecular weight in Bacillus licheniformis WX−02. J. Chem. Technol. Biotechnol. 89, 1825–1832. doi: 10.1002/jctb.4261
Wang, Y., Liu, Y., Wang, Z., and Lu, F. (2014). Influence of promoter and signal peptide on the expression of pullulanase in Bacillus subtilis. Biotechnol. Lett. 36, 1783–1789. doi: 10.1007/s10529-014-1538-x
Weaver, B., and Wuensch, K. L. (2013). SPSS and SAS programs for comparing Pearson correlations and OLS regression coefficients. Behav. Res. Methods. 45, 880–895. doi: 10.3758/s13428-012-0289-7
Wei, X., Ji, Z., and Chen, S. (2010). Isolation of halotolerant Bacillus licheniformis WX-02 and regulatory effects of sodium chloride on yield and molecular sizes of poly-gamma-glutamic acid. Appl. Biochem. Biotechnol. 160, 1332–1340. doi: 10.1007/s12010-009-8681-1
Xu, G., Zha, J., Cheng, H., Ibrahim, M. H. A., Yang, F., Dalton, H., et al. (2019). Engineering Corynebacterium glutamicum for the de novo biosynthesis of tailored poly-gamma-glutamic acid. Metab. Eng. 56, 39–49. doi: 10.1016/j.ymben.2019.08.011
Yao, J., Jing, J., Xu, H., Liang, J., Wu, Q., Feng, X., et al. (2009). Investigation on enzymatic degradation of γ-polyglutamic acid from Bacillus subtilis NX-2. J. Mol. Catal. B Enzym. 56, 158–164. doi: 10.1016/j.molcatb.2007.12.027
Yokoi, H., Arima, T., Hirose, J., Hayashi, S., and Takasaki, Y. (1996). Flocculation properties of poly (γ-glutamic acid) produced by Bacillus subtilis. J. Ferment. Bioeng. 82, 84–87. doi: 10.1016/0922-338X(96)89461-X
Yuan, P., Lv, M., Jin, P., Wang, M., Du, G., Chen, J., et al. (2015). Enzymatic production of specifically distributed hyaluronan oligosaccharides. Carbohydr. Polym. 129, 194–200. doi: 10.1016/j.carbpol.2015.04.068
Zeng, W., Liang, Z., Li, Z., Bian, Y., Li, Z., Tang, Z., et al. (2016). Regulation of poly-γ-glutamic acid production in Bacillus subtilis GXA-28 by potassium. J. Taiwan. Inst. Chem. E. 61, 83–89. doi: 10.1016/j.jtice.2015.12.026
Zhan, Y., Sheng, B., Wang, H., Shi, J., Cai, D., Yi, L., et al. (2018). Rewiring glycerol metabolism for enhanced production of poly-γ-glutamic acid in Bacillus licheniformis. Biotechnol. Biofuels. 11:306. doi: 10.1186/s13068-018-1311-9
Zhang, A. L., Liu, H., Yang, M. M., Gong, Y. S., and Chen, H. (2007). Assay and characterization of a strong promoter element from B. subtilis. Biochem. Biophys. Res. Commun. 354, 90–95. doi: 10.1016/j.bbrc.2006.12.137
Zhang, H., Zhu, J., Zhu, X., Cai, J., Zhang, A., Hong, Y., et al. (2012). High-level exogenous glutamic acid-independent production of poly-(γ-glutamic acid) with organic acid addition in a new isolated Bacillus subtilis C10. Bioresour. Technol. 116, 241–246. doi: 10.1016/j.biortech.2011.11.085
Keywords: Bacillus licheniformis, Poly-γ-glutamic acid, molecular weight, PgdS depolymerase, controllable degradation
Citation: Wang D, Wang H, Zhan Y, Xu Y, Deng J, Chen J, Cai D, Wang Q, Sheng F and Chen S (2020) Engineering Expression Cassette of pgdS for Efficient Production of Poly-γ-Glutamic Acids With Specific Molecular Weights in Bacillus licheniformis. Front. Bioeng. Biotechnol. 8:728. doi: 10.3389/fbioe.2020.00728
Received: 03 October 2019; Accepted: 09 June 2020;
Published: 09 July 2020.
Edited by:
Bernd Rehm, Griffith University, AustraliaReviewed by:
Jin-Song Gong, Jiangnan University, ChinaSamarthya Bhagia, Biosciences Division (BSD), Oak Ridge National Laboratory, United States
Copyright © 2020 Wang, Wang, Zhan, Xu, Deng, Chen, Cai, Wang, Sheng and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Feng Sheng, c2hlbmdmc2tAMTYzLmNvbQ==; Shouwen Chen, bWVsMjEyQDEyNi5jb20=
†These authors have contributed equally to this work
 Dong Wang1†
Dong Wang1† Qin Wang
Qin Wang Shouwen Chen
Shouwen Chen