
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Bioeng. Biotechnol. , 08 July 2020
Sec. Synthetic Biology
Volume 8 - 2020 | https://doi.org/10.3389/fbioe.2020.00691
This article is part of the Research Topic Development and Application of Novel Genome Engineering Tools in Microbial Biotechnology View all 22 articles
Filamentous fungi are important eukaryotic organisms crucial in substrate degradation and carbon cycle on the earth and have been harnessed as cell factories for the production of proteins and other high value-added products in recent decades. As cell factories, filamentous fungi play a crucial role in industrial protein production as both native hosts and heterologous hosts. In this review, the regulation strategies of carbohydrate active enzyme expression at both transcription level and protein level are introduced, and the transcription regulations are highlighted with induction mechanism, signaling pathway, and promoter and transcription factor regulation. Afterward, the regulation strategies in protein level including suitable posttranslational modification, protein secretion enhancement, and protease reduction are also presented. Finally, the challenges and perspectives in this field are discussed. In this way, a comprehensive knowledge regarding carbohydrate active enzyme production regulation at both transcriptional and protein levels is provided with the particular goal of aiding in the practical application of filamentous fungi for industrial protein production.
In recent years, green biomanufacturing is getting increasing attention because of the serious energy crisis and environment pollution. Searching for green and environmentally friendly ways for industrial production has gradually become a new theme for human beings, such as converting hardly degradable biomass into fermentable sugars and ethanol, as well as replacing fossil energy with environmentally friendly clean energy. However, the transformation of biomass requires a huge number of industrial enzymes, especially carbohydrate active enzymes (CAZymes), such as cellulases and amylases. It is crucial to attain better cost performance or improved properties for these industrial enzymes. Microorganisms show irreplaceable advantages in bioeconomy and green biomanufacturing for their fast growth, short culture period, and low culture cost. Filamentous fungi, which widely exist on earth, play a crucial role in global carbon cycle for their excellent performance in degrading organic matters and converting plant biomass into cost-effective fermentable sugar. In addition, filamentous fungi can also produce a lot of secondary metabolites that could be used as antibiotics, such as penicillin, or some other drugs that are applied in tumor therapy. Moreover, they can produce many organic acids and other chemical materials.
Altogether, the application of fungal biotechnology enables the development of many industrial fields, such as enzyme and pharmaceutical production, biofuels and biochemistry, food, agriculture, pulp and paper, detergents, textiles, and crop protection. As a consequence, filamentous fungi are getting increasing attention for their major role in industrial production (Dunford, 2012; Deacon, 2013; Benocci et al., 2017; Fang and Qu, 2018; Jiang et al., 2018). As eukaryotic cell factory, filamentous fungi can presumably serve as ideal hosts with rapid growth rate on simple and inexpensive media.
There are several kinds of hosts for protein expression, including both prokaryotic and eukaryotic protein expression system (Altmann et al., 1999; Balbas and Lorence, 2004; Kantardjieff and Zhou, 2013; Schmoll and Dattenböck, 2016; Vega, 2016), and their comparison is listed in Table 1. As eukaryotic cell factory, filamentous fungi have numerous advantages that cannot be replaced by other organisms. For instance, filamentous fungi can grow rapidly on simple and cheap media and even fermented or unfermented agroindustrial wastes. They also have a strong survivability, which make them an ideal cell factory for producing drugs, antibiotics, industrial enzymes, and other substances. Most importantly, filamentous fungi have a strong ability in protein expression and perform various posttranslational processing correctly, including glycosylation, peptide chain shearing, and disulfide bond formation, which are similar to mammal cells (Bergquist et al., 2002). Besides, filamentous fungi have a powerful secretory pathway, including signal recognition particle signaling and efficient function of the endoplasmic reticulum in protein modification, as well as rapid clearance of misfolded proteins, fusion between vesicles and target membranes, and apical secretion of the proteins, which conferred them the ability to produce eukaryotic proteins correctly (Kavanagh, 2011; Karagiosis and Baker, 2012; Fang and Qu, 2018).
The common hosts of filamentous fungi are Aspergillus species, Trichoderma species, and Penicillium species, such as Aspergillus niger, Aspergillus oryzae, Aspergillus nidulans, Trichoderma reesei, Penicillium oxalicum, and other model fungi such as Neurospora crassa, all of which can be used for both mechanism investigation and protein expression. Besides, T. reesei and P. oxalicum are likely to be applied in cellulases expression and plant biomass degradation, whereas A. niger and A. oryzae are often applied in food industry. What’s more, the genetic toolboxes of model filamentous fungi A. nidulans and N. crassa have been fully developed and used for the investigations of various mechanisms in filamentous fungi, especially for N. crassa, as the N. crassa Gene Knockout Library is available and brought great convenience for further studies. Filamentous fungi are not suitable for those proteins that are easily produced by other hosts with a considerable yield due to their complex and time-consuming genetic manipulations. As saprophytic fungi, most of the filamentous fungi possess the advantages of biomass degradation. Therefore, filamentous fungi are often used for the expression of CAZymes, which are responsible for the degradation of plant biomass in industrial field, thus providing clean energy by green manufacture.
This review elucidates the regulating strategies in enzyme expression at both transcription level and protein level. Filamentous fungal cell factories produce both endogenous and heterologous enzymes. The filamentous fungi–derived CAZymes, such as cellulases, are often expressed in their native hosts. The expression of these endogenous proteins is regulated at the transcription level to a large extent under the control of carbon catabolite repression (CCR). The promoter and transcription factor regulation mechanisms as well as signal pathways of protein expression in transcription level will be highlighted in detail, which brings a better understanding of the transcription regulation and further applications in the improvement of CAZyme expression. Furthermore, the conventional regulation strategies for improving heterologous expression, such as increasing copy number, codon optimization, protein fusion expressing, and protease reduction, are also introduced briefly. The summarized regulation strategies for enhancing protein expression in filamentous fungi are shown in Figure 1.
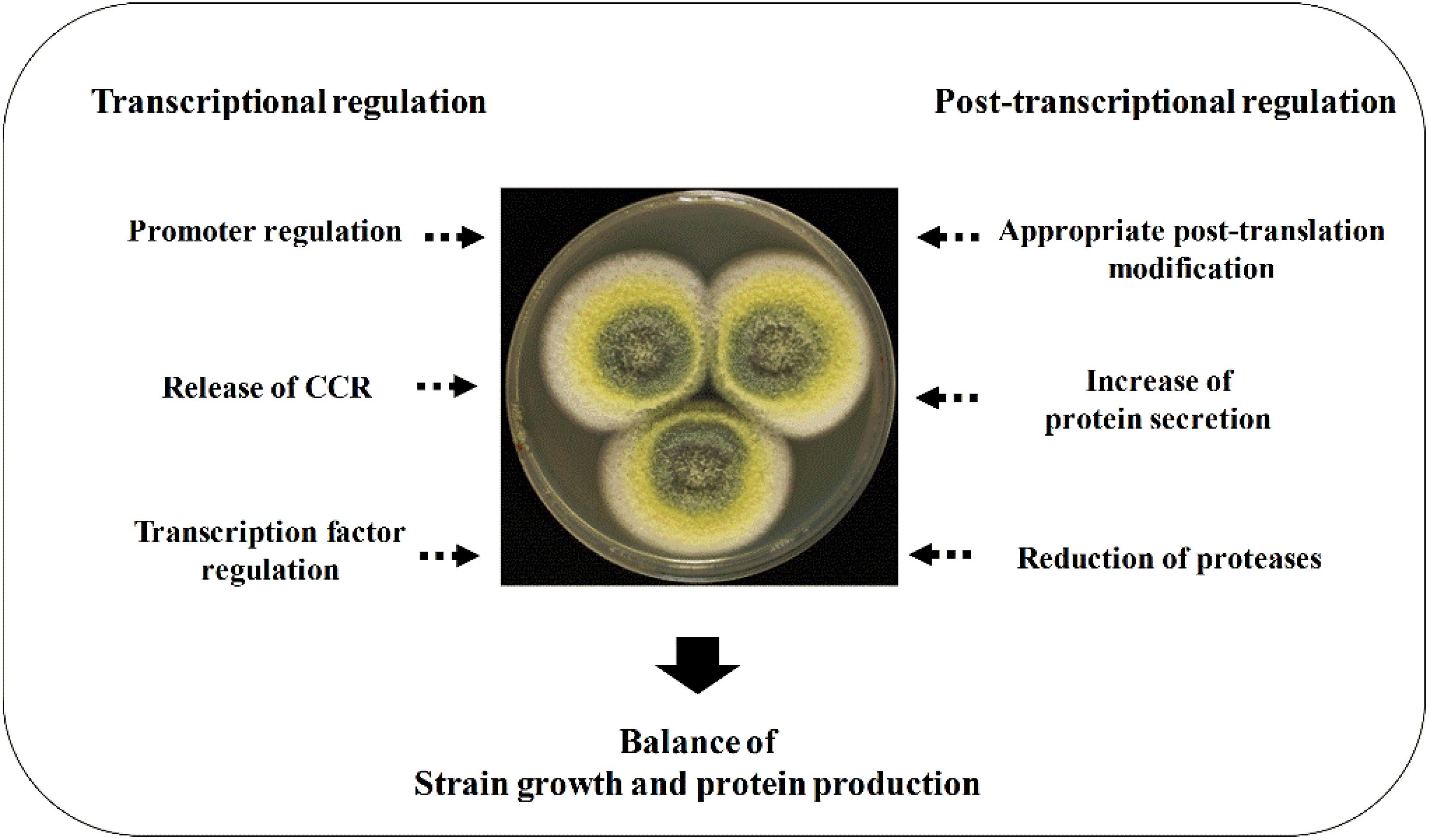
Figure 1. Regulation strategies for enhancing protein expression in filamentous fungi. The enhancement of protein expression at both transcriptional and translational levels is required, and a fermentation optimization is needed to balance the strain growth and protein production.
Most of protein production systems for filamentous fungi cell factories require transformation methods and gene editing strategies, such as vectors, insertion manner, and selection markers, and the present studies and advances of genetic manipulations of filamentous fungi are listed in Table 2. Meanwhile, the efficiency of mRNA production and protein expression mostly depends on transcriptional regulation, which includes promoter regulation, CCR regulation, and transcription factor regulation. Herein, various regulation strategies in transcription level for protein expression in filamentous fungi are introduced.
The strategy of using known regulatory elements for protein production is widely applied in fungi cell factory, and the utilization of strong promoters in filamentous fungi can efficiently improve the transcription level of target genes. The strong promoters such as the promoters of glucoamylase gene (glaA) in A. oryzae, glyceraldehyde-3-phosphate dehydrogenase gene (gpdA) in A. nidulans, α-amylase gene (amyB) in A. oryzae, and cellobiose hydrolysis enzyme gene (cbh1) in T. reesei are most commonly used strong promoters and have been successfully applied in the efficient expression of recombinant proteins.
The original promoters of CAZymes are often inducing promoters, which called for some specific induction conditions. Thus, the strategy of converting the inducing promoter to a strong constitutive promoter is efficient sometimes and has been commonly used in the transcriptional regulation of CAZymes (Su et al., 2012). However, constitutive promoters with strong transcription abilities are not always suitable for the enhancement of proteins as recombinant proteins may be toxic for the growth of the host strain. In this case, inducible promoters would be more preferable as their controllable ability to transfer to the protein expression phase from strain growth phase (Weinhandl et al., 2014).
As a consequence, the modification of the existing promoters and new promoters mining would be better choices. The promoter series were also applied for the overexpression of the target gene (Zhang and Xia, 2016). The modification of promoters greatly enhanced the expression of proteins when the binding sites of repressors were replaced by those of activators (Zou et al., 2012; Sun et al., 2020). Native promoters with different strength were mined and used when there were no suitable or enough promoters applied in some non-model strain with industrial value (Liu et al., 2018).
As we know, the factors that affect protein expression in filamentous fungi are mainly related to the hierarchy of carbon source utilization of the strain, the signaling sensing and transduction pathways that regulate catabolites, and expression of corresponding enzymes. CCR widely exists in various microorganisms with a regulation system of carbon source utilization, which determines the utilization hierarchy of a huge variety of carbon substrates. CCR ensures the utilization of preferential carbon sources, such as glucose, and inhibits the utilization of less preferred carbon sources by repressing the expression of CAZymes required for the catabolism of a wide range of alternative carbon sources (Kiesenhofer et al., 2016). A large number of genes involved in carbon catabolism are under the control of CCR, including the industrially important CAZymes such as cellulase, amylase, and xylanase (Kunitake et al., 2019). CCR energetically selects the preferential carbon sources, helps microorganisms adapt to the environment by absorbing favorable nutrients at maximum, and reduces the cost of CAZymes synthesis when the preferred carbon source is available, which represents an economical manner for carbon catabolism regulation (Adnan et al., 2017). However, CCR is caused not only by glucose, but also by other monosaccharides. It is reported that xylose also acts as a carbon catabolite repressor, whereas the expression of enzymes for xylose utilization can also be repressed in the presence of glucose (Prathumpai et al., 2004). Besides, the expression of alcohol dehydrogenase (ADH2) of Saccharomyces cerevisiae was shown to be inhibited by both glucose and the acetate (Simpson-Lavy and Kupiec, 2019).
The CCR regulation system exists in various fungi and involves several regulators. CCR is mediated by the Mig1 repressor (Kayikci and Nielsen, 2015) in yeast carbon metabolism, while it is often mediated by CreA/Cre1 in most of filamentous fungi. The transcriptional repressor CreA is a C2H2 finger domain DNA-binding protein and found to mediate CCR in A. nidulans with a transcript of 1.8 kb in length (Dowzer and Kelly, 1989), and the orthologs were identified as CRE1 in N. crassa and T. reesei with similar functions (Strauss et al., 1995; Serna et al., 1999). CreA mediates CCR with the help of CreB–CreC deubiquitination complex, which plays a crucial role in the function and stability of CreA (Todd et al., 2000; Lockington and Kelly, 2002; Ries et al., 2016). The subcellular localization of CreA is crucial for derepression of CCR and utilization of carbon source in A. nidulans (Ries et al., 2016), and the detailed introduction can be found in next part. CreA functions in repressing transcription of CAZymes via directly binding to 5’-SYGGRG-3’ on the promoters of target genes or the transcription activators (Benocci et al., 2017). Besides CCR mediation, CreA also functions in hyphal growth and metabolism in Aspergillus species, T. reesei, and Humicola insolens (Ries et al., 2016; Xu et al., 2019).
As CreA is a repressor for CAZyme expression, strategies of CreA deletion or modification have been used for improving the expression of genes related to carbon utilization. Xylose catabolism was activated in the CreA deletion strain even under high glucose concentration, whereas the major enzymes for xylose utilization were expressed only when glucose repression was relieved in the wild-type strain of A. nidulans (Prathumpai et al., 2004). The significantly enhanced expression of cellulase and hemicellulase in T. reesei was obtained when the cre1 gene of the mutant strain was either completely removed or partly truncated, which resulted in the derepressed cellulase expression even in the presence of glucose under both inducing and non-inducing conditions (Nakari-Setälä et al., 2009). However, persistent nuclear localization was obtained when domains of CreA in A. nidulans were deleted, which led to a repression of cellulase coding genes under cellulase-inducing conditions (Ries et al., 2016). More interestingly, a truncated Cre1 could turn into an activator, which functioned to activate and enhance the expression of cellulase and xylanase in T. reesei without causing any growth deficiencies. The truncated CreA, which served as an activator, exerted its function by locating to the nucleus and directly binding to the upstream regulatory regions of target genes under both inducing and repressing conditions, especially of the main transcription activator of the cellulases, Xyr1 (Rassinger et al., 2018).
As a consequence, the investigation of CCR is important for the regulation and enhancement of endogenous protein expression, whereas for foreign protein expression, choosing a pathway independent of CCR and a strong promoter to make the target protein expression in a constitutive manner might be a more efficient strategy for enhancing its expression level (Zhang et al., 2008).
There are two prerequisites for the expression of endogenous CAZymes in filamentous fungi; one is derepression of CCR, whereas the other one is the presence of inducers. Derepression would be introduced in the following paragraphs, whereas the mechanism of how inducers function in the signaling pathway and the interactions between inducers and elements or factors of the target genes still lack of full investigation. The derepression of CCR is an essential condition for the expression of CAZymes as they can only be induced by corresponding inducers when glucose was depleted in the wild-type strain without any genetic modifications (Gancedo, 1998; Sarma et al., 2007; de Souza et al., 2013; Brown et al., 2015; Wang et al., 2017).
Derepression of CCR is achieved by depletion of favorable carbon sources or a deletion of the crucial factors related to the CCR pathway, such as CreA and other repressors that mediated CCR or PKA pathway [cyclic adenosine monophosphate (cAMP); cAMP-dependent protein kinase A (PKA)], which are responsible for the nuclear location of repressors. The repression and derepression of CCR consist of two crucial pathways, which are the AMP-activated protein kinase (AMPK) pathway and the PKA pathway (Hardie, 2010; Adnan et al., 2017; Lin and Hardie, 2017; Kunitake et al., 2019).
AMPK is regarded as a sensor and regulator of nutritious conditions of the extracellular environment as it can switch on the expression of CAZymes for alternative carbon source utilization. It is activated in the condition of low energy by sensing the cellular adenine nucleotide level, and it was first discovered in mammalian cells for its crucial role in energy sensing to regulate the energy balance of the whole body in a way of inhibiting ATP consumption and accelerating ATP production by switching on alternative catabolic pathways (Lin and Hardie, 2017). G protein–coupled receptors (GPCRs), hexokinases, and hexose transporters are all involved in glucose sensing. When high concentration of glucose was sensed by the corresponding receptors, the glucose was transported into the cell and participated in glycolysis, leading to an increased ATP level and a reduced AMP/ATP ratio, which resulted in a silenced mode of AMPK. With the consumption of glucose and ATP, the AMP/ATP ratio was increased, and the AMPK pathway was activated, which further affected the subcellular localization of CAZyme repressors directly or indirectly by exporting repressors from the nucleus, followed by the degradation in cytoplasm (Tanaka et al., 2018). Thus, the regulatory sequences of target genes were released and further bound by transcription activators, which initiated the transcription of the corresponding CAZyme coding genes (Rubenstein, 2008; Brown et al., 2013).
AMPK always exists as heterotrimeric complexes and consists of the catalytic α subunit and the β and γ regulatory subunit. The α subunit of AMPK in S. cerevisiae, which is encoded by snf1 gene with the target of Ser/Thr site, is required for the derepression of CCR (Gancedo, 1998). The repressor Mig1 is regulated by phosphorylation with the protein kinase Snf1 in S. cerevisiae, which is the functional homolog of CreA. Snf1 is activated at a low glucose level by glucose sensing and signaling cascades. Mig1 is phosphorylated by Snf1 and removed to cytoplasm during glucose starvation, which led to a liberation of the regulatory sequences of target genes, whereas Snf1 is inactivated by a high extracellular glucose concentration and results in nucleus localization of Mig1. And as a consequence, Mig1 binds to the regulatory element upstream of the target genes and represses their transcription (Adnan et al., 2017). Similar to yeast, the derepression of CCR in filamentous fungi is controlled dominantly by the function of MAPK. Studies show that snfA in A. nidulans, which encodes Snf1 homolog protein kinases, is required for CreA derepression and cellulase production. The absence of SnfA led to an inactivation of removing CreA away from the nucleus under the inducing condition of growth on cellulose, which indicated that the subcellular localization was an important process responding to the nutrients in the environment (Brown et al., 2013). Besides, the presence of inducers (less favorable carbon sources) and carbon starvation are also crucial for the activation of nutrient sensing kinase pathways and the release of inducer binding sites for gene induction. And CreA derepression is necessary for the induction of CAZymes (Brown et al., 2013).
The PKA pathway, which plays an antagonistic role in the regulation of CAZymes compared with AMPK pathway, is also involved in the CCR. G protein–coupled receptors, the cellular second messenger cAMP, and adenylate cyclase are all involved in the PKA pathway and affected the downstream targets by phosphorylation cascades (Nogueira et al., 2015; Yang et al., 2018). PKA is essential for CCR as its deletion leads to a misfunction of CCR with impaired nucleus location effect of repressors and partial derepression of CAZymes, which is consistent with the result obtained by the PkaA deletion strain with a varied degree of derepression of cellulase genes under different culture conditions (Kunitake et al., 2019). PKAs are involved in the regulation of various physiological processes including growth, virulence, and metabolism, and responding to the extracellular nutrient status by a series of phosphorylation cascades (Ribeiro et al., 2019). PkaA in A. nidulans plays a curial role in the glucose signaling pathway, and the deletion of this protein kinase resulted in an increased secretion of CAZymes but a decreased growth in the presence of inducing carbon sources (Assis et al., 2015). A mutation of amino acid site of Cre1 led to a loss of phosphorylation and the function of DNA binding, which caused a carbon catabolite derepression, and suggested phosphorylation is required for recovering the function of Cre1 from an inactive conformation (Cziferszky et al., 2002).
When a high glucose level was sensed by GPCRs or other related receptors in Ras/cAMP pathway (Wang et al., 2004), Gα subunit (catalytic subunit of G protein) was activated, and further, adenylate cyclase was activated; cAMP level was thus increased via elevated adenylate cyclase activity. The generated cAMP would bind to the regulatory subunit of PKA, which led to the liberation of PKA catalytic subunit. PKA was activated by the release of catalytic subunit and then initiated phosphorylation cascades and transmitted signals to the downstream targets, leading to the nucleus location of repressors such as CreA (Brown et al., 2014; Ziv, 2020). Phosphorylation is essential for the subcellular localization of repressors, which is crucial for repression/derepression and the transcription of the majority of CAZymes. That is, repressors such as CreA would be imported to the nucleus and repress the expression of target genes when a high extracellular glucose level was sensed, while it would also be exported from nucleus into cytoplasm and finally degraded, which led to the derepression of CCR when the energy depletion is sensed by the cell (Tanaka et al., 2018).
The receptors sense the nutrients status and transmit the signal by phosphorylation cascades to the protein kinases including AMPK and PKA pathway, followed by the direct or indirect phosphorylation effects on the specific sites of the repressors with their final subcellular localization. Some results indicated that dephosphorylation of CreA was essential for CCR in T. reesei (Cziferszky et al., 2002), whereas other studies declared the dephosphorylation of CreA is also found in derepressing conditions in A. nidulans (Alam et al., 2016). Therefore, it cannot be concluded that the role of phosphorylation is repression or derepression of CAZymes. The derepression of CCR mediated by AMPK pathway plays a positive role in the expression of CAZymes by direct or indirect phosphorylation in different hosts (Gancedo, 1998; Rubenstein, 2008; Adnan et al., 2017), but it certainly functioned via mediating the nucleus exportation of repressors. However, PKA pathway–mediated CCR has negative effects on CAZyme expression, which also regulates the catabolism via nucleus location by indirect phosphorylation of repressors as no direct interaction was found between PKA and CreA, and it was deduced that the CreA was phosphorylated via the kinases controlled by PKA, which further mediated the nucleus import of CreA (Ribeiro et al., 2019). Both PKA and AMPK pathways are indispensably involved in nutrients sensing, signal transduction, catabolism, and growth and function synergistically with the counteract effects in the regulation of CAZyme expression. The possible pathway of carbon catabolism regulation is shown in Figure 2.
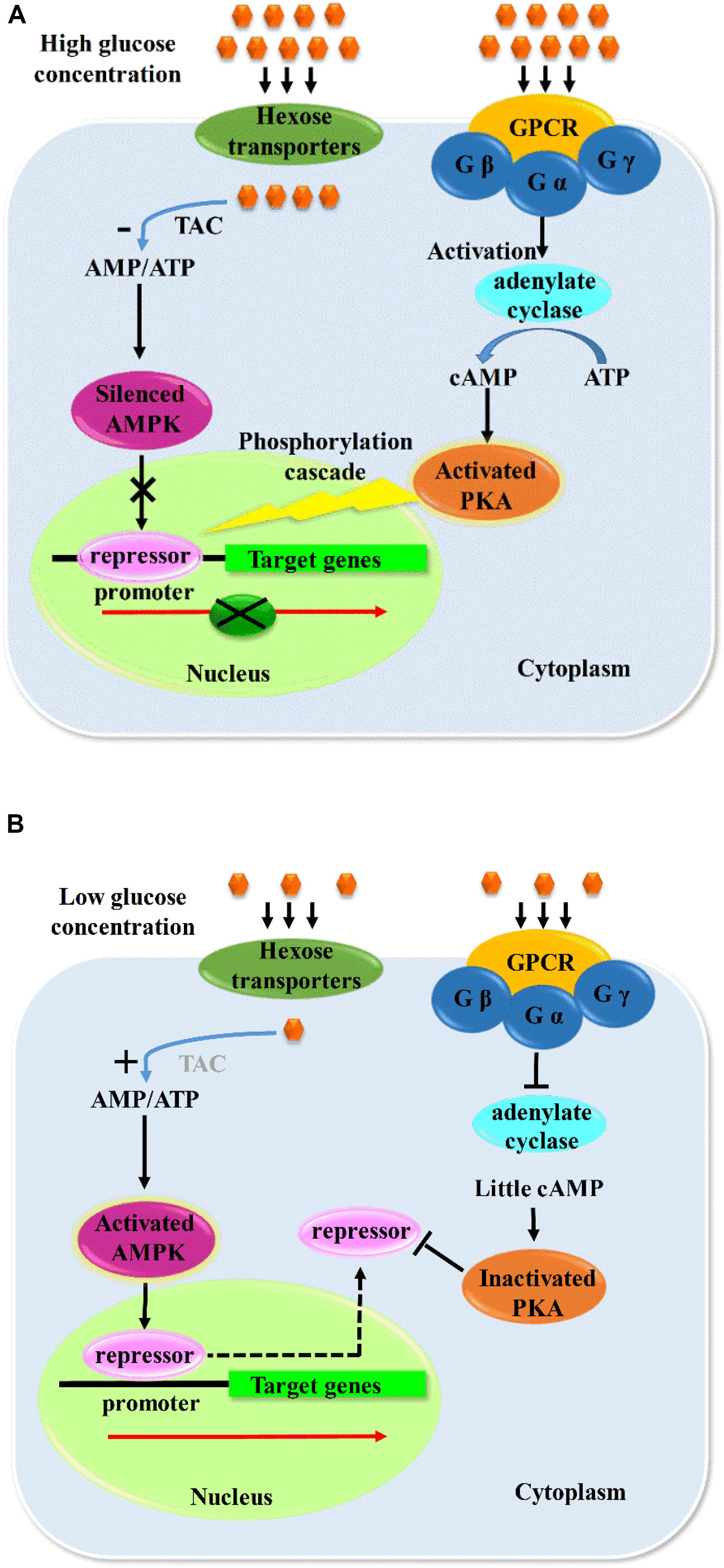
Figure 2. Probable model of carbon metabolism regulation. When the concentration of glucose is high outside the cell (A), G protein–coupled receptor would receive and activate G protein, and catalytic subunit of which would activate adenylate cyclase and result in the increased level of cAMP, and further activate PKA and a series of phosphorylation cascade, which would cause a nucleus location of repressors. In the same time, the high concentration of glucose would accelerate the tricarboxylic acid cycle and increase the concentration of ATP, and the decreased ratio of AMP/ATP would silence the AMPK and make it unable to mediate the export of repressors to cytoplasm from nucleus, thus forming a repression model of CAZymes, and vice versa (B). Gα, Gβ, and Gγ are the 3 subunits of G protein; Gα is the catalytic subunit of G protein. TAC, tricarboxylic acid cycle.
The transcription factors involved in CAZymes regulatory network are important regulatory elements in filamentous fungi. Besides CreA, which mediated CCR, there are many other transcription factors functioning as activators or repressors and playing a crucial role in the transcription regulation of CAZyme expression, most of which are Zn2Cys6-type transcription factors consisting of zinc finger DNA-binding domain and transcription activation/repression domain. The crucial activators functioning in CAZymes regulation are Xyr1 (xylanase regulator 1)/XlnR, CLR1/ClrA, and CLR2/ClrB, ClrC, and CLR-4 and also ACE2, ACE3, and AmyR, whereas the common repressors are ACE1, Rce1, and Xpp1 (xylanase promoter-binding protein 1) (Coradetti et al., 2012).
Xyr1 and its homologs are the main activators of cellulase gene expression, which play important roles in the expression of cellulolytic enzymes in many filamentous fungi, with a general consensus binding motif of the sequence GGCTRR (Castro et al., 2016; Benocci et al., 2017; Jiang et al., 2018). The overexpression of Xyr1 caused a relief from the CCR and thus produced cellulases in a constitutive expression manner, which resulted in the full expression of cellulases even on the non-inducing carbon sources in T. reesei, such as glucose and glycerol (Xinxing et al., 2015). AmyR is a key regulator in activating amylase expression and repressing cellulase gene expression in the meantime (Tani et al., 2001). The other activators also functioned together in the regulation of cellulases induction and positively regulated the expression of cellulases, such as CLR1/ClrA and CLR2/ClrB, ClrC, and CLR-4 and also ACE2 and ACE3 (Craig et al., 2015; Raulo et al., 2016; Liu et al., 2018; Zhang et al., 2019).
The factors such as ACE1, Rce1, and Xpp1 (xylanase promoter-binding protein 1) serve as transcriptional repressors of xylanase expression in T. reesei (Hypocrea jecorina), ACE1 can bind to the chb1 promoter of main cellulase gene, whose deletion led to an enhancement in the cellulase and xylanase expression (Saloheimo et al., 2003; Portnoy et al., 2011). The promotion of cellulase induction and extension of induction expression process were obtained by the disruption of repressor Rce1 coding gene in T. reesei (Cao et al., 2017). Xpp1 could only bind to the promoters of xylanases and regulate the expression of main xylanase, without affecting the expression of cellulases (Mach-Aigner et al., 2010; Derntl et al., 2015, 2017).
Transcription regulation based on transcription factors is a useful tool in improving the CAZyme expression, such as overexpressing the transcription activators or modulating the activators into constitutive expression, and deleting the repressors if their absence does not affect the growth of strain, or downregulating when they do affect the growth. What’s more, the use of fused transcription factors to release or attenuate CCR inhibition in cellulase transcription and the modification of the existing transcription repressors to inactive mode, which lose the repression function of target genes and have no influence on the strain growth in the meantime, are promising strategies for enhancing the production of CAZymes (Alazi and Ram, 2018; Wang et al., 2019).
Artificial transcriptional factors that are fused by DNA-binding domain and effectors of different transcription factors were used in cellulase production. The strategy of fusing the ACE2 effector domain with the DNA-binding domains of CRE1 and ACE1 was used to regulate the expression of cellulase (Su et al., 2009). The binding domain of CRE1 was fused to the effector and the binding domain of XYR1, which formed a constituted expression of cellulase based on glucose serving as the sole carbon source (Zhang X. et al., 2016). Enhanced cellulase production was also obtained when a strong transcriptional activation domain was fused to the C-terminus of the natural transcription factors (XYR1, ACE2, and ACE1), followed by the transfection into hypercellulolytic strain and the replacement of natural transcription factors by homologous recombination (Zhang et al., 2018b) or by replacing natural transcription factors with minimal transcriptional activators (Zhang et al., 2018a). Randomized artificial zinc finger protein library, which was constructed via linking multiple zinc finger domain by random shuffling (Park et al., 2003), was used in cellulase expression in T. reesei with a significant enhancement of cellulase expression (Zhang F. et al., 2016). An elevated cellulase production was obtained in T. reesei when 11 amino acids of the activator ACE3 were truncated, which was probably caused by increasing the interaction with the activator XYR1 (Chen et al., 2020). The modification of ClrB with middle region removal and fusing of DNA-binding/transcriptional activation domains together led to a derepression of CCR; as a consequence, that induction of cellulase in the presence of repression carbon sources such as glucose and glycerol was obtained in P. oxalicum without cellulose addition (Gao et al., 2019b). The transcription factor regulation strategies are shown in Table 3.
As mentioned previously, various transcription factors are involved in the complicated regulation network of CAZyme expression, but their mechanism investigations are still far from enough. The omics techniques play a crucial role in the investigation of transcription factors, followed by gene cloning and characterization, and their functions were finally identified by the knockout, complementation, overexpression, truncation, and even binding properties (Mäkinen et al., 2014; Li et al., 2015). Furthermore, transcriptome data are also necessary for novel regulator screening, and candidate genes were selected based on the combination of genome data and transcriptome data, which were further determined by experimental results, and finally, the underlying regulators that function in regulation of protein expression were uncovered and applied in the production of CAZymes (Derntl et al., 2017; Liao et al., 2018; Zhang et al., 2019). The flowchart of transcription factor mining is shown in Figure 3.
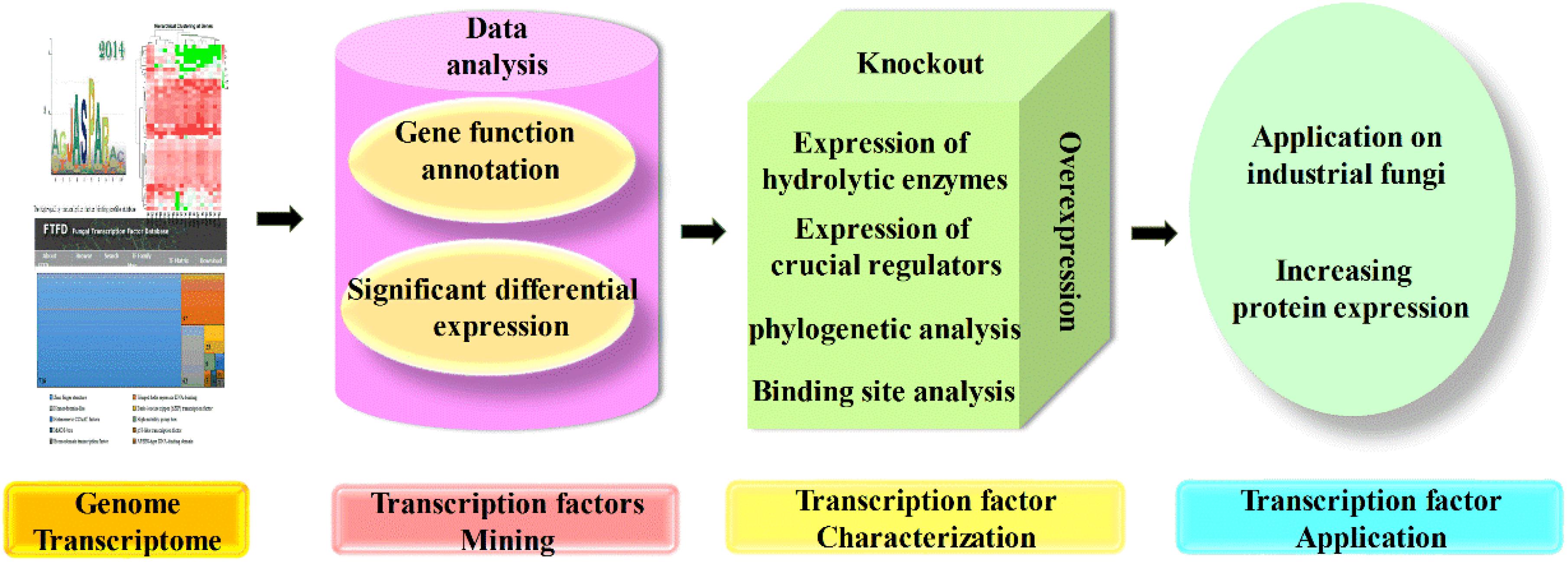
Figure 3. Flowchart of transcription factor mining. The mining of transcription factors often started by genome and transcriptome analysis, and candidate genes could be chosen for further characterization based on gene function and different expression level. The mutants with candidate genes knocked out; complementation and overexpression were constructed, and characterized by the expression of target regulated genes and crucial regulators, also the phylogenetic analysis and binding site analysis, which finally applied on other industrial fungi for the hyperproduction of proteins.
The transcription level of exogenous genes depends more on the transcription efficiency of insertion sites in genome than copy numbers in filamentous fungi when the genome integration strategy was applied (Zoglowek et al., 2014). Unlike heterologous gene expression with high copy number plasmids in Escherichia coli, the selective stress is required to ensure their stable inheritance when the extrachromosomal expression manner is used in fungi (Kavanagh, 2011), which caused a less application of increasing copy number in filamentous fungi.
The protein regulation includes translation and posttranslational modification, and protein secretion. The strategies during this process are mainly directed to heterologous expression, which are introduced in detail in previous studies (Sharma et al., 2009; Ward, 2011). Here are several strategies for improving protein expression in heterologous host strain.
Filamentous fungi can secrete protein out of the cell efficiently when endogenous genes were expressed, but the secretory ability significantly decreased when some heterologous genes were expressed. The strategy of codon optimization is crucial and useful for improvement of translation efficiency. Codon optimizing of heterologous genes according to the codon preference of host strain is an effective way and powerful tool to improve the expression efficiency of heterologous protein in filamentous fungi (Tanaka et al., 2014), which improves the translation efficiency of heterologous genes by the increase of steady-state mRNA level via the elimination of premature polyadenylation and avoidance of mRNA degradation (Li et al., 2007; Tokuoka et al., 2008). The improvement for heterologous protein focuses on the conventional strategies, which are gene fusion with a well-secreted protein and overexpression of foldases and chaperones (Archer et al., 1994). Constructing and selecting host strains with high secretory ability are also a way to improve the expression ability of foreign genes of filamentous fungi (Gombert et al., 2016). Inserting foreign genes into the downstream of the endogenous carrier protein with hyperproduction and high secretion ability by molecular manipulation, constructing the fusion expression system, and expressing the heterologous protein in a fusing manner followed by a cleavage of resulting proteins are also an effective way for improving the level of foreign gene secretion (Gustavsson et al., 2001). Human antibody fragment fused to a truncated endogenous enzyme was expressed in a fourfold protease deletion strain of N. crassa, which obtained a good secretion of 3 mg/L (Havlik et al., 2017). For facilitating the subsequent cleavage of the two fused proteins, a linker with proteolytic processing sites should be contained between the carrier protein and the target protein, which should be designed to allow the independent folding of the catalytic domain and the fused protein (Ward, 2011). Besides, the secretion of heterologous protein could also be increased by overexpression of foldases, signal peptides, and chaperones via facilitated protein folding (Goedegebuur et al., 2014; Nevalainen and Peterson, 2014). The application of heterologous signal peptides led to a high level of protein secretion; when enhanced green fluorescent protein served as a model, the heterologous protein human interferon β was finally successfully expressed in Aspergillus unguis (Madhavan et al., 2017). The porin B signal peptide in Corynebacterium glutamicum was used to improve the production of model protein endoxylanase with activity in high efficiency with a performance of 615 mg/L (An et al., 2013).
It is far from enough to express heterologous protein just in high transcription level and translation efficiency. The industrial and commercial values of the proteins are their structures and functional activities; therefore, appropriate posttranslation modification is crucial for the utilization of the protein produced by heterologous host strains. Unlike the simple prokaryotic expression systems, which are unable to carry out many of the posttranslation processing and lead to the secretion of inactive inclusion bodies, fungal cells have an outstanding performance of these processes. Among all the posttranslation modifications, glycosylation is very crucial for the biological activities of proteins; for example, cellobiohydrolases are typical glycoproteins, which undergo both N-linked glycosylation and O-linked glycosylation by the attachment of oligosaccharides (Stals et al., 2004; Ward, 2011). However, the overglycosylation of proteins would negatively influence enzyme activities, such as enzyme binding and protein stability. Choosing appropriate hosts for the expression of recombination protein is important because the level of glycosylation depends on host strain in a large extent. Generally speaking, the overglycosylation of filamentous fungi is less extensive than that in yeast, especially S. cerevisiae (Zoglowek et al., 2014). In some cases, strategies for reducing some extent of glycosylation are necessary for increasing the biology activities of proteins by site mutation, enzymatic deglycosylation, or even using glycosylation deficient strain (Kavanagh, 2011). In a word, an appropriate host strain and a reliable control system of glycosylation are both crucial during activation of heterologous protein.
The occurrence of some incorrect posttranslational processing in the heterologous protein expression during the cell growth of filamentous fungi is unavoidable, such as misfolding, impairment of intracellular transport, and proteolytic degradation. Proteolytic degradation is one of the most obvious reasons for the low yields of heterologous proteins (Chung et al., 2002). Proteolytic degradation occurs not only in intracellular but also extracellular by the function of endogenous proteases produced during fungal growth (Zoglowek et al., 2014).
There are several strategies in reducing the proteolysis in host cells. Deletion of the identified protease coding genes could bring a satisfactory cellulase production compared with the parent strain (Li et al., 2019). The application of protease-deficient strains for improving the production of heterologous proteins is efficient and commonly used, although it would also bring a reduction of industrial protein production in the meantime (Wang et al., 2005). In this case, the strategy of partially inactivating some of the more prominent extracellular proteases, such as alkaline proteases or metalloproteases, which were determined in the strain, could be chosen for improving the protein production (Ward, 2011). Protease inhibitors were also used to control protease activity and reduce the proteolysis, but they could only be applied in a small-scale protein expression system (Wiebe, 2003). Besides, disruption of the coding genes of some protease regulators could also work in reducing protease activity in Aspergillus species. What’s more, adjusting the pH of fungal culture away from optimal pH for proteases activities and inhibiting the proteolysis could reduce the degradation of recombination protein (Braaksma and Punt, 2008).
As stated above, there are so many advantages of filamentous fungal cell factory in producing industrial enzymes, and some achievements were obtained with a large amount of protein expression, which indicates filamentous fungi are a kind of efficient hosts for the production of CAZymes. The landmark studies of CAZymes are listed in Table 4. However, the mining of genome and the efforts for simplifying genetic technology are far from enough of non-model strains, such as lower transformation efficiency, multinuclear cells, lack of knowledge for available genetic engineering elements, exogenous gene insertion sites, and so on (Gupta et al., 2012; Nevalainen, 2020). It is difficult to improve the level of protein expression by a single genetic modification method. Fungal cells are always regulated by the CCR and could not express most of the CAZymes with the existence of readily available carbon sources such as glucose, whereas the lack of glucose cannot enrich the biomass of the strain, which leads to a low production of target proteins. Besides, the complex protease system of filamentous fungal cells makes them difficult to accumulate and secrete the expression products of foreign genes, resulting in low yields of target products. When some non-fungal genes from bacteria, mammals, or plants are expressed in fungal hosts, the level of protein expression would be much lower than that of fungi sourced genes (Broekhuijsen et al., 1993).
In addition, the process of protein expression often includes two stages of strain growth and protein induction, and the strain needs to reach the maximum biomass in the growth stage by utilizing some favorable carbon sources, whereas the favorable carbon sources should be depleted, and the corresponding inducers should be added for the initiation of protein induction stage. It is obvious that monitoring the status of carbon source consumption and control of fermentation process are inconvenient. There are also some filamentous fungi that form large mycelium pellets during fermentation, which would reduce the contact between fungal cells and the surrounding medium, hindering the efficient expression and secretion of proteins (Nevalainen, 2020). The transcription regulation of CAZymes in filamentous fungi mainly focuses on the modulation of signaling pathway or modification of transcription factors, to make CAZymes express in the presence of glucose and achieve a better growth of strain and substantial biomass, thus obtaining an elevated production of CAZymes.
The strategies such as downregulation or even deletion of genes involved in PKA pathway and repressors, upregulation of AMPK pathway, and overexpressing activators are often efficient in the enhancement of CAZyme expression. However, it should be noted that PKA functions in various physical process including cell growth and metabolism, as well as the repressor CreA; their downregulation might cause deficiency in strain growth and metabolism, which must lead to a weak production of enzymes far from best. The better choice for enhancing the expression of proteins is to keep the balance between cell growth and protein expression, such as modifying a repressor into factors without negative impacts or even positive effects for the target genes or modifying the promoter regions upstream of target genes by replacing the binding sites of repressors to those of activators, and modulate the PKA and AMPK pathway to make a perfect subcellular location of repressors and incapability for repressing the gene transcription without any negative impacts on cell growth.
Moreover, the growth of the strain would change the fluidity of the medium, resulting in changes in the conditions of nutrients, oxygen, and pH; in turn, the ability of secreting foreign protein also varied with the changes of growth status and conditions (Braaksma and Punt, 2008). Therefore, the improvement with multiple tolerances of the strain by metabolic engineering strategies is essential to enhance the stability of strain in various culture environments by the relief of multiple growth stresses and so to increase the production of proteins, which needs further efforts. A further conceptual point with regard to simplifying the genetic procedures and shortening the transformation time with an acceptable transformation efficiency, multiple tolerance host strains, and the balance between cell growth and protein expression would be the main goal for the development of fungi cell factory.
In summary, the present article systematically elucidated the strategies of protein expression in both transcription level and protein levels. Transcription regulations with both mechanism and signal pathway of protein expression were illustrated in detail. The strategies for producing industrial CAZymes in filamentous fungal cell factory of current studies were introduced, which are promoters and transcription factor regulation, protein expression and secretion regulation, and the balance of strain growth and protein expression. Although the catabolism regulatory network in filamentous fungal cells is really complex, our understanding of signaling pathways and mechanisms for CAZymes induction and also strategies of protein expression regulation improved the cognition of protein expression in filamentous fungi, which would benefit the investigations of CAZymes or filamentous fungi protein expression system.
TZ was responsible for the literature survey and the writing of whole manuscript. HL was responsible for the revise of manuscript. BL and CL were responsible for supervision. All authors contributed to the article and approved the submitted version.
This work was supported by the National Natural Science Foundation of China (Nos. 21736002 and 21706012) and the National Key Research and Development Program of China (2018YFA0901800).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Adnan, M., Zheng, W., Islam, W., Arif, M., Abubakar, Y., Wang, Z.-H., et al. (2017). Carbon catabolite repression in filamentous fungi. Int. J. Mol. Sci. 19:48. doi: 10.3390/ijms19010048
Alam, M. A., Kamlangdee, N., and Kelly, J. (2016). The CreB deubiquitinating enzyme does not directly target the CreA repressor protein in Aspergillus nidulans. Curr. Genet. doi: 10.1007/s00294-016-0666-3[Epub ahead of print].
Alazi, E., and Ram, A. (2018). Modulating transcriptional regulation of plant biomass degrading enzyme networks for rational design of industrial fungal strains. Front. Bioeng. Biotechnol. 6:133. doi: 10.3389/fbioe.2018.00133
Altmann, F., Staudacher, E., Wilson, I., and Maerz, L. (1999). Insect cells as hosts for the expression of recombinant glycoproteins. Glycoconj. J. 16, 109–123. doi: 10.1023/A:1026488408951
An, S. J., Yim, S. S., and Jeong, K. J. (2013). Development of a secretion system for the production of heterologous proteins in Corynebacterium glutamicum using the Porin B signal peptide. Protein Expr. Purif. 89, 251–257. doi: 10.1016/j.pep.2013.04.003
Archer, D., Jeenes, D., and MacKenzie, D. (1994). Strategies for improving heterologous protein production from filamentous fungi. Antonie Van Leeuwenhoek 65, 245–250. doi: 10.1007/BF00871952
Assis, L., Ries, L., Savoldi, M., Reis, T., Brown, N., and Goldman, G. (2015). Aspergillus nidulans protein kinase A plays an important role in cellulase production. Biotechnol. Biofuels 8:213. doi: 10.1186/s13068-015-0401-1
Balbas, P., and Lorence, A. (eds) (2004). Recombinant gene expression: Reviews and Protocols, 2nd Edn. Totowa, NJ: Humana Press. doi: 10.1385/1592597742
Benocci, T., Aguilar-Pontes, M. V., Zhou, M., Seiboth, B., and Vries, R. P. (2017). Regulators of plant biomass degradation in ascomycetous fungi. Biotechnol. Biofuels 10:152. doi: 10.1186/s13068-017-0841-x
Bergquist, P., Te’o, V., Gibbs, M., Cziferszky, A., de Faria, F., Azevedo, M., et al. (2002). Production of recombinant bleaching enzymes from thermophilic microorganisms in fungal hosts. Appl. Biochem. Biotechnol. 98-100, 165–176. doi: 10.1385/abab:98-100:1-9:165
Braaksma, M., and Punt, P. J. (2008). Aspergillus as a Cell Factory for Protein Production: Controlling Protease Activity in Fungal Production. Boca Raton, FL: CRC Press.
Broekhuijsen, M., Mattern, I., Contreras, R., Kinghorn, J., and Hondel, C. (1993). Secretion of heterologous proteins by Aspergillus niger: production of active human interleukin-6 in a protease-deficient mutant by KEX2-like processing of a glucoamylase-hIL6 fusion protein. J. Biotechnol. 31, 135–145. doi: 10.1016/0168-1656(93)90156-H
Brown, N., Gouvea, P., Krohn, N., Savoldi, M., and Goldman, G. (2013). Functional characterisation of the non-essential protein kinases and phosphatases regulating Aspergillus nidulans hydrolytic enzyme production. Biotechnol. Biofuels 6:91. doi: 10.1186/1754-6834-6-91
Brown, N., Reis, T., Reis, L., Caldana, C., Mah, J.-H., Yu, J.-H., et al. (2015). G-protein coupled receptor mediated nutrientsensing and developmental control in Aspergillus nidulans. Mol. Microbiol. 98, 420–439. doi: 10.1111/mmi.13135
Brown, N., Ries, L., and Goldman, G. (2014). How nutritional status signalling coordinates metabolism and lignocellulolytic enzyme secretion. Fungal Genet. Biol. 72, 48–63. doi: 10.1016/j.fgb.2014.06.012
Cao, Y., Zheng, F., Wang, L., Zhao, G., Chen, G., Zhang, W., et al. (2017). Rce1, a novel transcriptional repressor, regulates cellulase gene expression by antagonizing the transactivator Xyr1 in Trichoderma reesei. Mol. Microbiol. 105, 65–83. doi: 10.1111/mmi.13685
Castro, L., De Paula, R., Antoniêto, A., Persinoti, G., Silva-Rocha, R., and Silva, R. (2016). Understanding the role of the master regulator XYR1 in Trichoderma reesei by global transcriptional analysis. Front. Microbiol. 7:175. doi: 10.3389/fmicb.2016.00175
Chen, Y., Wu, C., Fan, X., Zhao, X., Zhao, X., Shen, T., et al. (2020). Engineering of Trichoderma reesei for enhanced degradation of lignocellulosic biomass by truncation of the cellulase activator ACE3. Biotechnol. Biofuels 13:62. doi: 10.1186/s13068-020-01701-3
Chung, H.-J., Park, S.-M., and Kim, D.-H. (2002). Characterization of Aspergillus niger mutants deficient of a protease. Mycobiology 30, 160–165. doi: 10.4489/MYCO.2002.30.3.160
Coradetti, S., Craig, J., Xiong, Y., Shock, T., Tian, C., and Glass, N. (2012). Conserved and essential transcription factors for cellulase gene expression in ascomycete fungi. Proc. Natl. Acad. Sci. U.S.A. 109, 7397–7402. doi: 10.1073/pnas.1200785109
Craig, J., Coradetti, S., Starr, T., and Glass, N. (2015). Direct target network of the Neurospora crassa plant cell wall deconstruction regulators CLR-1, CLR-2, and XLR-1. mBio 6:e1452-15. doi: 10.1128/mBio.01452-15
Cziferszky, A., Mach, R., and Kubicek, C. (2002). Phosphorylation positively regulates DNA binding of the carbon catabolite repressor Cre1 of Hypocrea jecorina (Trichoderma reesei). J. Biol. Chem. 277, 14688–14694. doi: 10.1074/jbc.M200744200
de Souza, W., Morais, E., Krohn, N., Savoldi, M., Goldman, M. H., Rodrigues, F., et al. (2013). Identification of metabolic pathways influenced by the G-protein coupled receptors GprB and GprD in Aspergillus nidulans. PLoS One 8:e62088. doi: 10.1371/journal.pone.0062088
Derntl, C., Kluger, B., Bueschl, C., Schuhmacher, R., Mach, R., and Mach-Aigner, A. (2017). Transcription factor Xpp1 is a switch between primary and secondary fungal metabolism. Proc. Natl. Acad. Sci. U.S.A. 114, E560–E569. doi: 10.1073/pnas.1609348114
Derntl, C., Rassinger, A., Srebotnik, E., Mach, R., and Mach-Aigner, A. (2015). Xpp1 regulates the expression of xylanases, but not of cellulases in Trichoderma reesei. Biotechnol. Biofuels 8:112. doi: 10.1186/s13068-015-0298-8
Dong, L., Lin, X., Yu, D., Huang, L., Wang, B., and Pan, L. (2019). High-level expression of highly active and thermostable trehalase from Myceliophthora thermophila in Aspergillus niger by using the CRISPR/Cas9 tool and its application in ethanol fermentation. J. Ind. Microbiol. Biotechnol. 47, 133–144. doi: 10.1007/s10295-019-02252-9
Dowzer, C., and Kelly, J. (1989). Cloning of the creA gene from Aspergillus nidulans: a gene involved in carbon catabolite repression. Curr. Genet. 15, 457–459. doi: 10.1007/BF00376804
Dunford, N. T. (2012). Food and Industrial Bioproducts and Bioprocessing. Hoboken, NJ: Wiley-Blackwell.
Fang, X., and Qu, Y. (2018). Fungal Cellulolytic Enzymes Microbial Production and Application: Microbial Production and Application. Berlin: Springer.
Gancedo, J. (1998). Yeast carbon catabolite repression. Microbiol. Mol. Biol. Rev. 62, 334–361. doi: 10.1128/MMBR.62.2.334-361.1998
Gao, J. (2017). Production of the versatile cellulase for cellulose bioconversion and cellulase inducer synthesis by genetic improvement of Trichoderma reesei. Biotechnol. Biofuels 10:272.
Gao, L., Li, S., Xu, Y., Xia, C., Xu, J., Liu, J., et al. (2019a). Mutation of a conserved alanine residue in transcription factor AraR leads to hyper-production of α-L-Arabinofuranosidases in Penicillium Oxalicum. Biotechnol. J. 14:e1800643. doi: 10.1002/biot.201800643
Gao, L., Xu, Y., Song, X., Li, S., Xia, C., Xu, J., et al. (2019b). Deletion of the middle region of the transcription factor ClrB in Penicillium oxalicum enables cellulase production in the presence of glucose. J. Biol. Chem. 294, 18685–18697. doi: 10.1074/jbc.RA119.010863
Goedegebuur, F., Neef-Kruithof, P., Pucci, J. P., and Ward, M. (2014). Over expression of foldases and chaperones improves protein production. U.S. Patent No 6,090,051. Washington, DC: U.S. Patent and Trademark Office. doi: 10.1074/jbc.ra119.010863
Gombert, A., Junior, J., Cerdán, M. E., and González-Siso, M.-I. (2016). Kluyveromyces marxianus as a host for heterologous protein synthesis. Appl. Microbiol. Biotechnol. 100, 6193–6208. doi: 10.1007/s00253-016-7645-y
Gupta, V. K., Tuohy, M., Ayyachamy, M., Turner, K., and O’Donovan, A. (2012). Laboratory Protocols in Fungal Biology: Current Methods in Fungal Biology. Berlin: Springer.
Gustavsson, M., Lehtiö, J., Denman, S., Teeri, T., Hult, K., and Martinelle, M. (2001). Stable linker peptides for a cellulose-binding domain-lipase fusion protein expressed in Pichia pastoris. Protein Eng. 14, 711–715. doi: 10.1093/protein/14.9.711
Hardie, D. (2010). “AMP-activated protein kinase,” in Handbook of Cell Signaling. Amsterdam: Elsevier.
Havlik, D., Brandt, U., Bohle, K., and Fleißner, A. (2017). Establishment of Neurospora crassa as a host for heterologous protein production using a human antibody fragment as a model product. Microb. Cell Fact. 16:128.
He, L., Guo, W., Li, J., Meng, Y., Wang, Y., Lou, H., et al. (2020). Two dominant selectable markers for genetic manipulation in Neurospora crassa. Curr. Genet. doi: 10.1007/s00294-020-01063-1 [Epub ahead of print].
Honda, Y., Tanigawa, E., Tsukihara, T., Nguyen Xuan, D., Kawabe, H., Sakatoku, N., et al. (2019). Stable and transient transformation, and a promoter assay in the selective lignin-degrading fungus, Ceriporiopsis subvermispora. AMB Express 9:92. doi: 10.1186/s13568-019-0818-1
Ichinose, S., Tanaka, M., Shintani, T., and Gomi, K. (2017). Increased production of biomass-degrading enzymes by double deletion of creA and creB genes involved in carbon catabolite repression in Aspergillus oryzae. J. Biosci. Bioeng. 125, 141–147. doi: 10.1016/j.jbiosc.2017.08.019
Istvan, W., Yang, L., Vang, J., Ahring, B., Lübeck, M., and Lübeck, P. S. (2017). A comparison of Agrobacterium-mediated transformation and protoplast-mediated transformation with CRISPR-Cas9 and bipartite gene targeting substrates, as effective gene targeting tools for Aspergillus carbonarius. J. Microbiol. Methods 135, 26–34. doi: 10.1016/j.mimet.2017.01.015
Jain, S., Durand, H., and Tiraby, G. (1992). Development of a transformation system for the thermophilic fungus Talaromyces sp. CL240 based on the use of phleomycin resistance as a dominant selectable marker. Mol. Gen. Genet. 234, 489–493. doi: 10.1007/bf00538710
Jiang, Y., Liu, K., Guo, W., Zhang, R., Liu, F., Zhang, N., et al. (2018). “Lignocellulase formation, regulation, and secretion mechanisms in Hypocrea jecorina (Trichoderma reesei) and other filamentous fungi: microbial production and application,” in Fungal Cellulolytic Enzymes, eds X. Fang, and Y. Qu, (Singapore: Springer), 43–59. doi: 10.1007/978-981-13-0749-2_3
Kantardjieff, A., and Zhou, W. (2013). Mammalian cell cultures for biologics manufacturing preface. Adv. Biochem. Eng. Biotechnol. 139, 1–9. doi: 10.1007/10_2013_255
Karagiosis, S. A., and Baker, S. E. (2012). “Fungal cell factories,” in Food and Industrial Bioproducts and Bioprocessing, ed. N. T. Dunford, (Oxford: Wiley-Blackwell), 205–219
Kavanagh, K. (2011). Fungi: Biology and Applications. Hoboken, NJ: Wiley. doi: 10.1002/9781119976950
Kayikci, Ö., and Nielsen, J. (2015). Glucose repression in Saccharomyces cerevisiae. FEMS Yeast Res. 15:fov068. doi: 10.1093/femsyr/fov068
Kiesenhofer, D., Mach-Aigner, A., and Mach, R. (2016). “Understanding the mechanism of carbon catabolite repression to increase protein production in filamentous fungi,” in Gene Expression Systems in Fungi: Advancements and Applications. Fungal Biology, eds M. Schmoll, and C. Dattenböck, (Cham: Springer), 275–288 doi: 10.1007/978-3-319-27951-0_12
Kunitake, E., Li, Y., Uchida, R., Nohara, T., Asano, K., Hattori, A., et al. (2019). CreA-independent carbon catabolite repression of cellulase genes by trimeric G-protein and protein kinase A in Aspergillus nidulans. Curr. Genet. 65, 941–952. doi: 10.1007/s00294-019-00944-4
Li, C., Lin, F., Zhou, L., Qin, L., Li, B.-Z., Zhou, Z., et al. (2017). Cellulase hyper-production by Trichoderma reesei mutant SEU-7 on lactose. Biotechnol. Biofuels 10:228. doi: 10.1186/s13068-017-0915-9
Li, D., Tang, Y., Lin, J., and Cai, W. (2017). Methods for genetic transformation of filamentous fungi. Microb. Cell Fact. 16:168. doi: 10.1186/s12934-017-0785-7
Li, F., Liu, Q., Li, X., Zhang, C., Li, J., Sun, W., et al. (2020). Construction of a new thermophilic fungus Myceliophthora thermophila platform for enzyme production using a versatile 2A peptide strategy combined with efficient CRISPR-Cas9 system. Biotechnol. Lett. 42, 1181–1191. doi: 10.1007/s10529-020-02882-5
Li, X., Liu, Q., Sun, W., He, Q., and Tian, C. (2019). Improving cellulases production by Myceliophthora thermophila through disruption of protease genes. Biotechnol. Lett. 42, 219–229. doi: 10.1007/s10529-019-02777-0
Li, X.-L., Skory, C., Ximenes, E., Jordan, D., Dien, B., and Hughes, S. (2007). Expression of an AT-rich xylanase gene from the anaerobic fungus Orpinomyces sp. strain PC-2 in and secretion of the heterologous enzyme by Hypocrea jecorina. Appl. Microbiol. Biotechnol. 74, 1264–1275. doi: 10.1007/s00253-006-0787-6
Li, Z., Yao, G., Wu, R., Gao, L., Kan, Q., Liu, M., et al. (2015). Synergistic and dose-controlled regulation of cellulase gene expression in Penicillium oxalicum. PLoS Genet. 11:e1005509. doi: 10.1371/journal.pgen.1005509
Liao, G.-Y., Zhao, S., Zhang, T., Li, C.-X., Liao, L.-S., Zhang, F.-F., et al. (2018). The transcription factor TpRfx1 is an essential regulator of amylase and cellulase gene expression in Talaromyces pinophilus. Biotechnol. Biofuels 11:276. doi: 10.1186/s13068-018-1276-8
Lin, L., Wang, S., Li, X., He, Q., Benz, J. P., and Tian, C. (2019). STK-12 acts as a transcriptional brake to control the expression of cellulase-encoding genes in Neurospora crassa. PLoS Genet. 15:e1008510. doi: 10.1371/journal.pgen.1008510
Lin, S.-C., and Hardie, D. (2017). AMPK: sensing glucose as well as cellular energy status. Cell Metab. 27, 299–313. doi: 10.1016/j.cmet.2017.10.009
Liu, Q., Gao, R., Li, J., Lin, L., Zhao, J., Sun, W., et al. (2017). Development of a genome-editing CRISPR/Cas9 system in thermophilic fungal Myceliophthora species and its application to hyper-cellulase production strain engineering. Biotechnol. Biofuels 10:1. doi: 10.1186/s13068-016-0693-9
Liu, Q., Li, J., Gao, R., Li, J., Ma, G., and Tian, C. (2018). CLR-4, a novel conserved transcription factor for cellulase gene expression in ascomycete fungi. Mol. Microbiol. 111, 373–394. doi: 10.1111/mmi.14160
Liu, Q., Zhang, Y., Li, F., Li, J., Sun, W., and Tian, C. (2019). Upgrading of efficient and scalable CRISPR–Cas-mediated technology for genetic engineering in thermophilic fungus Myceliophthora thermophila. Biotechnol. Biofuels 12:293. doi: 10.1186/s13068-019-1637-y
Liu, R., Chen, L., Jiang, Y., Zhou, Z., and Zou, G. (2015). Efficient genome editing in filamentous fungus Trichoderma reesei using the CRISPR/CAS9 system. Cell Discov. 1:15007. doi: 10.1038/celldisc.2015.7
Lockington, R., and Kelly, J. (2002). The WD40-repeat protein CreC interacts with and stabilizes the deubiquitinating enzyme CreB in vivo in Aspergillus nidulans: CreB and CreC interact in vivo. Mol. Microbiol. 43, 1173–1182. doi: 10.1046/j.1365-2958.2002.02811.x
Mach-Aigner, A., Grosstessner-Hain, K., Poças-Fonseca, M., Mechtler, K., and Mach, R. (2010). From an electrophoretic mobility shift assay to isolated transcription factors: a fast genomic-proteomic approach. BMC Genomics 11:644. doi: 10.1186/1471-2164-11-644
Madhavan, A., Pandey, A., and Sukumaran, R. (2017). Expression system for heterologous protein expression in the filamentous fungus Aspergillus unguis. Bioresour. Technol. 245, 1334–1342. doi: 10.1016/j.biortech.2017.05.140
Mäkinen, M., Valkonen, M., Westerholm-Parvinen, A., Aro, N., Arvas, M., Vitikainen, M., et al. (2014). Screening of candidate regulators for cellulase and hemicellulase production in Trichoderma reesei and identification of a factor essential for cellulase production. Biotechnol. Biofuels 7:14. doi: 10.1186/1754-6834-7-14
Michielse, C., Hooykaas, P., Hondel, C., and Ram, A. (2008). Agrobacterium-mediated transformation of the filamentous fungus Aspergillus awamori. Nat. Protoc. 3, 1671–1678. doi: 10.1038/nprot.2008.154
Nakari-Setälä, T., Paloheimo, M., Kallio, J., Vehmaanperä, J., Penttilä, M., and Saloheimo, M. (2009). Genetic modification of carbon catabolite repression in Trichoderma reesei for improved protein production. Appl. Environ. Microbiol. 75, 4853–4860. doi: 10.1128/AEM.00282-09
Navarrete, K., Roa, A., Vaca, Y. I, Espinoza, Navarro, C., and Chavez, R. (2009). Molecular characterization of the niaD and pyrG genes from Penicillium camemberti, and its uses as transformation markers. Cell. Mol. Biol. 14, 692–702.
Nevalainen, H., and Peterson, R. (2014). Making recombinant proteins in filamentous fungi- Are we expecting too much? Front. Microbiol. 5:75. doi: 10.3389/fmicb.2014.00075
Niu, J., Arentshorst, M., Seelinger, F., Ram, A., and Ouedraogo, J.-P. (2016). A set of isogenic auxotrophic strains for constructing multiple gene deletion mutants and parasexual crossings in Aspergillus niger. Arch. Microbiol. 198, 861–868. doi: 10.1007/s00203-016-1240-6
Nogueira, K., Nogueira, V., Costa, M., De Paula, R., Flávia, C., Mendonça Natividade, F., et al. (2015). Evidence of cAMP involvement in cellobiohydrolase expression and secretion by Trichoderma reesei in presence of the inducer sophorose. BMC Microbiol. 15:195. doi: 10.1186/s12866-015-0536-z
Park, K.-S., Lee, D.-K., Lee, H., Lee, Y., Jang, Y.-S., Kim, Y., et al. (2003). Corrigendum: phenotypic alteration of eukaryotic cells using randomized libraries of artificial transcription factors. Nat. Biotechnol. 21, 1208–1214. doi: 10.1038/nbt868
Portnoy, T., Margeot, A., Seidl-Seiboth, V., Le Crom, S., Chaabane, F., Linke, R., et al. (2011). Differential regulation of the cellulase transcription factors XYR1, ACE2, and ACE1 in Trichoderma reesei strains producing high and low levels of cellulase. Eukaryot. Cell 10, 262–271. doi: 10.1128/EC.00208-10
Prathumpai, W., Workman, M., and Nielsen, J. (2004). The effect of CreA in glucose and xylose catabolism in. Appl. Microbiol. Biotechnol. 63, 748–753. doi: 10.1007/s00253-003-1409-1
Randhawa, A., Ogunyewo, O., Eqbal, D., Gupta, M., and Yazdani, S. S. (2018). Disruption of zinc finger DNA binding domain in catabolite repressor Mig1 increases growth rate, hyphal branching, and cellulase expression in hypercellulolytic fungus Penicillium funiculosum NCIM1228. Biotechnol. Biofuels 11:15. doi: 10.1186/s13068-018-1011-5
Rassinger, A., Gacek- Matthews, A., Strauss, J., Mach, R., and Mach-Aigner, A. (2018). Truncation of the transcriptional repressor protein Cre1 in Trichoderma reesei Rut-C30 turns it into an activator. Fungal Biol. Biotechnol. 5:15. doi: 10.1186/s40694-018-0059-0
Raulo, R., Kokolski, M., and Archer, D. (2016). The roles of the zinc finger transcription factors XlnR, ClrA and ClrB in the breakdown of lignocellulose by Aspergillus niger. AMB Express 6:5. doi: 10.1186/s13568-016-0177-0
Ribeiro, L., Chelius, C., Boppidi, K., Naik, N., Hossain, S., Ramsey, J., et al. (2019). Comprehensive analysis of Aspergillus nidulans PKA phosphorylome identifies a novel mode of CreA regulation. mBio 10:e02825-18. doi: 10.1128/mBio.02825-18
Ries, L., Beattie, S., Espeso, E., Cramer, R., and Goldman, G. (2016). Diverse regulation of the CreA carbon catabolite repressor in Aspergillus nidulans. Genetics 203, 335–352. doi: 10.1534/genetics.116.187872
Rubenstein, E. (2008). Glucose Sensing and the Regulation of the AMP-Activated Protein Kinase in Yeast. Ph.D. thesis, University of Pittsburgh, Pittsburgh, PA.
Saloheimo, A., Aro, N., Ilmén, M., and Penttilä, M. (2003). ACEI of Trichoderma reesei is a repressor of cellulase and xylanase expression. Appl. Environ. Microbiol. 69, 56–65. doi: 10.1128/AEM.69.1.56-65.2003
Sarma, N., Haley, T., Haley, K., Buford, T., Willis, K., and Santangelo, G. (2007). Glucose-responsive regulators of gene expression in Saccharomyces cerevisiae function at the nuclear periphery via a reverse recruitment mechanism. Genetics 175, 1127–1135. doi: 10.1534/genetics.106.068932
Schmoll, M., and Dattenböck, C. (eds) (2016). Gene Expression Systems in Fungi: Advancements and Applications, 1st Edn. Berlin: Springer.
Serna, I., Ng, D., and Tyler, B. (1999). Carbon regulation of ribosomal genes in Neurospora crassa occurs by a mechanism which does not require Cre-1, the homologue of the aspergillus carbon catabolite repressor, CreA. Fungal Genet. Biol. 26, 253–269. doi: 10.1006/fgbi.1999.1121
Sharma, R., Katoch, M., Srivastava, P., and Qazi, G. (2009). Approaches for refining heterologous protein production in filamentous fungi. World J. Microbiol. Biotechnol. 25, 2083–2094. doi: 10.1007/s11274-009-0128-x
Shimizu, T., Ito, T., and Kanematsu, S. (2012). Transient and multivariate system for transformation of a fungal plant pathogen, Rosellinia necatrix, using autonomously replicating vectors. Curr. Genet. 58, 129–138. doi: 10.1007/s00294-012-0370-x
Simpson-Lavy, K., and Kupiec, M. (2019). Carbon catabolite repression: not only for glucose. Curr. Genet. 65, 1321–1323. doi: 10.1007/s00294-019-00996-6
Stals, I., Sandra, K., Geysens, S., Contreras, R., Beeumen, J., and Claeyssens, M. (2004). Factors influencing glycosylation of Trichoderma reesei cellulases. I: postsecretorial changes of the O- and N-glycosylation pattern of Ce17A. Glycobiology 14, 713–724. doi: 10.1093/glycob/cwh080
Strauss, J., Mach, R., Zeilinger, S., Hartler, G., Stöffler, G., Wolschek, M., et al. (1995). Cre1, the carbon catabolite repressor protein from Trichoderma reesei. FEBS Lett. 376, 103–107. doi: 10.1016/0014-5793(95)01255-5
Su, X., Chu, X., and Dong, Z. (2009). Identification of elevated transcripts in a Trichoderma reesei strain expressing a chimeric transcription activator using suppression subtractive hybridization. World J. Microbiol. Biotechnol. 25, 1075–1084. doi: 10.1007/s11274-009-9993-6
Su, X., Schmitz, G., Zhang, M., Mackie, R., and Cann, I. (2012). Heterologous gene expression in filamentous fungi. Adv. Appl. Microbiol. 81, 1–61. doi: 10.1016/B978-0-12-394382-8.00001-0
Sun, X., Zhang, X., Huang, H., Wang, Y., Tu, T., Bai, Y., et al. (2020). Engineering the cbh1 promoter of Trichoderma reesei for enhanced protein production by replacing the binding sites of a transcription repressor ACE1 to those of the activators. J. Agric. Food Chem. 68, 1337–1346. doi: 10.1021/acs.jafc.9b05452
Tamayo-Ramos, J. A., and Orejas, M. (2014). Enhanced glycosyl hydrolase production in Aspergillus nidulans using transcription factor engineering approaches. Biotechnol. Biofuels 7:103. doi: 10.1186/1754-6834-7-103
Tanaka, M., Ichinose, S., Shintani, T., and Gomi, K. (2018). Nuclear export-dependent degradation of the carbon catabolite repressor CreA is regulated by a region located near the C-terminus in Aspergillus oryzae. Mol. Microbiol. 110, 176–190. doi: 10.1111/mmi.14072
Tanaka, M., Tokuoka, M., and Gomi, K. (2014). Effects of codon optimization on the mRNA levels of heterologous genes in filamentous fungi. Appl. Microbiol. Biotechnol. 98, 3859–3867. doi: 10.1007/s00253-014-5609-7
Tani, S., Katsuyama, Y., Hayashi, T., Suzuki, H., Kato, M., Gomi, K., et al. (2001). Characterization of the amyR gene encoding a transcriptional activator for the amylase genes in Aspergillus nidulans. Curr. Genet. 39, 10–15. doi: 10.1007/s002940000175
Todd, R., Lockington, R., and Kelly, J. (2000). The Aspergillus nidulans creC gene involved in carbon catabolite repression encodes a WD40 repeat protein. Mol. Gen. Genet. 263, 561–570. doi: 10.1007/s004380051202
Tokuoka, M., Tanaka, M., Ono, K., Takagi, S., Shintani, T., and Gomi, K. (2008). Codon Optimization increases steady-state mRNA levels in Aspergillus oryzae heterologous gene expression. Appl. Environ. Microbiol. 74, 6538–6546. doi: 10.1128/AEM.01354-08
Vega, M. C. (2016). Advanced technologies for protein complex production and characterization. Anticancer Res. 36:4375.
Wang, B., Li, J., Gao, J., Cai, P., Han, X., and Tian, C. (2017). Identification and characterization of the glucose dual-affinity transport system in Neurospora crassa: pleiotropic roles in nutrient transport, signaling, and carbon catabolite repression. Biotechnol. Biofuels 10:17. doi: 10.1186/s13068-017-0705-4
Wang, F., Zhang, R., Han, L., Guo, W., Du, Z., Niu, K., et al. (2019). Use of fusion transcription factors to reprogram cellulase transcription and enable efficient cellulase production in Trichoderma reesei. Biotechnol. Biofuels 12:244. doi: 10.1186/s13068-019-1589-2
Wang, L., Ridgway, D., Gu, T., and Moo-Young, M. (2005). Bioprocessing strategies to improve heterologous protein production in filamentous fungal fermentations. Biotechnol. Adv. 23, 115–129. doi: 10.1016/j.biotechadv.2004.11.001
Wang, Y., Pierce, M., Schneper, L., Guldal, C., Zhang, X., Tavazoie, S., et al. (2004). Ras and Gpa2 mediate one branch of a redundant glucose signaling pathway in yeast. PLoS Biol. 2:E128. doi: 10.1371/journal.pbio.0020128
Ward, O. (2011). Production of recombinant proteins by filamentous fungi. Biotechnol. Adv. 30, 1119–1139. doi: 10.1016/j.biotechadv.2011.09.012
Weinhandl, K., Winkler, M., Glieder, A., and Camattari, A. (2014). Carbon source dependent promoters in yeasts. Microb. Cell Fact. 13:5. doi: 10.1186/1475-2859-13-5
Wiebe, M. (2003). Stable production of recombinant proteins in filamentous fungi - Problems and improvements. Mycologist 17, 140–144. doi: 10.1017/S0269915X03003033
Xinxing, L., Zheng, F., Li, C., Zhang, W., Chen, G., and Liu, W. (2015). Characterization of a copper responsive promoter and its mediated overexpression of the xylanase regulator 1 results in an induction-independent production of cellulases in Trichoderma reesei. Biotechnol. Biofuels 8:67. doi: 10.1186/s13068-015-0249-4
Xu, G., Li, J., Liu, Q., Sun, W., Jiang, M., and Tian, C. (2018). Transcriptional analysis of Myceliophthora thermophila on soluble starch and role of regulator AmyR on polysaccharide degradation. Bioresour. Technol. 265, 558–562. doi: 10.1016/j.biortech.2018.05.086
Xu, J.-R., and Bluhm, B. (eds) (2011). Fungal Genomics: Methods and Protocols, Vol. 722. Berlin: Springer.
Xu, X., Fan, C., Song, L., Li, J., Chen, Y., Zhang, Y., et al. (2019). A novel CreA-mediated regulation mechanism of cellulase expression in the thermophilic fungus Humicola insolens. Int. J. Mol. Sci. 20:3693. doi: 10.3390/ijms20153693
Yang, H., Liu, Y., Hao, X., Wang, D., Akhberdi, O., Xiang, B., et al. (2018). Regulation of the Gα-cAMP/PKA signaling pathway in cellulose utilization of Chaetomium globosum. Microb. Cell Fact. 17:160. doi: 10.1186/s12934-018-1008-6
Yao, G., Li, Z., Gao, L., Wu, R., Kan, Q., Liu, G., et al. (2015). Redesigning the regulatory pathway to enhance cellulase production in Penicillium oxalicum David Wilson. Biotechnol. Biofuels 8:71. doi: 10.1186/s13068-015-0253-8
Yao, G., Wu, R., Kan, Q., Gao, L., Liu, M., Yang, P., et al. (2016). Production of a high-efficiency cellulase complex via β-glucosidase engineering in Penicillium oxalicum. Biotechnol. Biofuels 9:78. doi: 10.1186/s13068-016-0491-4
Zhang, F., Bai, F., and Zhao, X. (2016). Enhanced cellulase production from Trichoderma reesei Rut-C30 by engineering with an artificial zinc finger protein library. Biotechnol. J. 11, 1282–1290. doi: 10.1002/biot.201600227
Zhang, G., Seiboth, B., Wen, C., Yaohua, Z., Xian, L., and Wang, T. (2009). A novel carbon source-dependent genetic transformation system for the versatile cell factory Hypocrea jecorina (anamorph Trichoderma reesei). FEMS Microbiol. Lett. 303, 26–32. doi: 10.1111/j.1574-6968.2009.01851.x
Zhang, J., Chen, Y., Wu, C., Liu, P., Wang, W., and Wei, D. (2019). The transcription factor ACE3 controls cellulase activities and lactose metabolism via two additional regulators in the fungus Trichoderma reesei. J. Biol. Chem. 294, 18435–18450. doi: 10.1074/jbc.RA119.008497
Zhang, J., Guoxiu, Z., Wang, W., and Wei, D. (2018a). Enhanced cellulase production in Trichoderma reesei RUT C30 via constitution of minimal transcriptional activators. Microb. Cell Fact. 17:75. doi: 10.1186/s12934-018-0926-7
Zhang, J., Pan, J., Guan, G., Ying, L., Xue, W., Tang, G., et al. (2008). Expression and high-yield production of extremely thermostable bacterial xylanaseB in Aspergillus niger. Enzyme Microb. Technol. 43, 513–516. doi: 10.1016/j.enzmictec.2008.07.010
Zhang, J., Wu, C., Wang, W., and Wei, D. (2018b). Construction of enhanced transcriptional activators for improving cellulase production in Trichoderma reesei RUT C30. Bioresour. Bioprocess. 5:40. doi: 10.1186/s40643-018-0226-4
Zhang, X., and Xia, L. (2016). Expression of Talaromyces thermophilus lipase gene in Trichoderma reesei by homologous recombination at the cbh1 locus. J. Ind. Microbiol. Biotechnol. 44, 377–385. doi: 10.1007/s10295-016-1897-5
Zhang, X., Li, Y., Zhao, X., and Bai, F. (2016). Constitutive cellulase production from glucose using the recombinant Trichoderma reesei strain overexpressing an artificial transcription activator. Bioresour. Technol. 223, 317–322. doi: 10.1016/j.biortech.2016.10.083
Ziv, C. (2020). Carbon source affects PKA-dependent polarity of Neurospora crassa in a CRE-1-dependent and independent manner. Fungal Genet. Biol. 45, 103–116. doi: 10.1016/j.fgb.2007.05.005
Zoglowek, M., Lübeck, P. S., Ahring, B., and Lübeck, M. (2014). Heterologous expression of cellobiohydrolases in filamentous fungi – An update on the current challenges, achievements and perspectives. Process Biochem. 50, 211–220. doi: 10.1016/j.procbio.2014.12.018
Keywords: filamentous fungal cell factory, protein expression, carbohydrate active enzymes, transcription factors, carbon catabolite repression, signal pathway, regulation strategies
Citation: Zhang T, Liu H, Lv B and Li C (2020) Regulating Strategies for Producing Carbohydrate Active Enzymes by Filamentous Fungal Cell Factories. Front. Bioeng. Biotechnol. 8:691. doi: 10.3389/fbioe.2020.00691
Received: 18 April 2020; Accepted: 03 June 2020;
Published: 08 July 2020.
Edited by:
Yuan Lu, Tsinghua University, ChinaReviewed by:
Shuobo Shi, Beijing University of Chemical Technology, ChinaCopyright © 2020 Zhang, Liu, Lv and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bo Lv, bHYtYkBiaXQuZWR1LmNu; Chun Li, bGljaHVuQHRzaW5naHVhLmVkdS5jbg==; bGljaHVuQGJpdC5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.