- 1State Key Laboratory of Microbial Metabolism, Joint International Research Laboratory of Metabolic and Developmental Sciences, School of Life Sciences and Biotechnology, Shanghai Jiao Tong University, Shanghai, China
- 2State Key Lab of Bioreactor Engineering, East China University of Science and Technology, Shanghai, China
Filamentous fungal strains of Trichoderma reesei have been widely used for cellulase production, and great effort has been devoted to enhancing their cellulase titers for the economic biorefinery of lignocellulosic biomass. In our previous studies, artificial zinc finger proteins (AZFPs) with the Gal4 effector domain were used to enhance cellulase biosynthesis in T. reesei, and it is of great interest to modify the AZFPs to further improve cellulase production. In this study, the endogenous activation domain from the transcription activator Xyr1 was used to replace the activation domain of Gal4 of the AZFP to explore impact on cellulase production. The cellulase producer T. reesei TU-6 was used as a host strain, and the engineered strains containing the Xyr1 and the Gal4 activation domains were named as T. reesei QS2 and T. reesei QS1, respectively. Compared to T. reesei QS1, activities of filter paper and endoglucanases in crude cellulase produced by T. reesei QS2 increased 24.6 and 50.4%, respectively. Real-time qPCR analysis also revealed significant up-regulation of major genes encoding cellulase in T. reesei QS2. Furthermore, the biomass hydrolytic performance of the cellulase was evaluated, and 83.8 and 97.9% more glucose was released during the hydrolysis of pretreated corn stover using crude enzyme produced by T. reesei QS2, when compared to the hydrolysis with cellulase produced by T. reesei QS1 and the parent strain T. reesei TU-6. As a result, we proved that the effector domain in the AZFPs can be optimized to construct more effective artificial transcription factors for engineering T. reesei to improve its cellulase production.
Introduction
Lignocellulosic biomass is abundantly available as a renewable resource, which is mainly composed of cellulose, hemicelluloses, and lignin. Degradation of the cellulose component by cellulase into glucose to produce biofuels and biobased chemicals has attracted extensive research attention (De Bhowmick et al., 2018). However, high production cost of cellulase makes the bioconversion process too expensive for practical applications, which ultimately limits the utilization of lignocellulosic biomass for biorefinery (Klein-Marcuschamer et al., 2012).
Filamentous fungi are commonly used as cellulase producers, among which Trichoderma reesei has been widely studied for cellulase production (Liu et al., 2013). Without doubt, enhancing cellulase production by T. reesei is of great importance for developing lignocellulosic biorefinery.
The cellulase enzymatic complex of T. reesei has been shown to consist of at least two cellobiohydrolases (CBHs), eight endo-β-1,4-glucanases (EGs), and seven β-glucosidases (BGLs) that act synergistically upon insoluble cellulose substrate (Druzhinina and Kubicek, 2017). The synthesis of these cellulase components is strictly controlled by various regulators, including at least six positive transcriptional activators (Xyr1, Ace2, Ace3, Vib1, BglR, and the Hap2/3/5 complex) as well as three repressors (Ace1, Rce1, and the carbon catabolite repressor Cre1) (Aro et al., 2001, 2003; Cao et al., 2017; Zhang F. et al., 2018). Recently, great effort has been made to genetically modify these endogenous transcription factors to reprogram the transcriptional regulation network to improve cellulase production in T. reesei (Derntl et al., 2013; Lv et al., 2015; Zhang et al., 2017; Rassinger et al., 2018; Wang et al., 2019). On the other hand, artificial transcription factors (ATFs) have also been studied to genetically engineer cellulase production. For example, the repressor Cre1 could be changed to transcriptional activator for cellulases gene expression by replacement of the VP16 activation domain (AD) (Zhang J. et al., 2018). In the previous studies, a library of artificial zinc finger proteins (AZFPs) was explored for expression in bacteria and yeast (Park et al., 2003; Ma et al., 2015), and screened for mutants with changed phenotypes. The AZFPs were designed to be composed of four zinc fingers as a DNA-binding domain (DBD) followed by an AD of Gal4p (Gal4AD). We have modified the library and successfully obtained mutants with enhanced cellulase production in T. reesei Rut-C30 (Zhang et al., 2016). However, Gal4AD in the AZFPs originates from budding yeast Saccharomyces cerevisiae, and it remains unknown whether exogenous transcriptional regulation domains can efficiently recruit protein complex to initiate transcription. Therefore, it is of great interest to investigate the effect of endogenous effector domains in the AZFPs on regulation of cellulase production in T. reesei.
In T. reesei, Xyr1 is the Gal4 family transcription activator that is essential for cellulase/hemicellulase gene transcription (Dos Santos Castro et al., 2016). We therefore designed a new transcription factor AZFPM2-Xyr1AD harboring the native effector domain of Xyr1 in this study. Production of cellulase, transcription of key genes related to cellulase biosynthesis, and degradation of lignocellulosic biomass in the recombinant strain carrying AZFPM2-Xyr1AD were compared with the parent strain and the control strain with the exogenous Gal4AD. The results provide basis for further optimizing the effector domains of AZFPs to enhance their stimulating effects on cellulase production by T. reesei.
Materials and Methods
Strains, Culture Media, and Culture Conditions
Escherichia coli GB05-dir was used for vector construction and propagation, which was cultivated in lysogeny broth (LB) medium with 4 μg/mL tetracycline (Wang et al., 2016).
T. reesei TU-6 (ATCC MYA-256), a non-homologous end joining pathway deficient, uridine auxotrophic derivative of QM9414 (Gruber et al., 1990), was used as the parent strain in this study. The strain and its derivatives were cultured on PDA plate for 5–7 days at 28°C to produce conidia. For fermentation experiment, T. reesei strains were inoculated with 106 spores/mL into 250 mL Erlenmeyer flasks containing 50 mL minimal medium (MM) supplemented with 0.1% uridine, 0.2% peptone, and 2% glucose at 28°C and 200 rpm for 48 h. Then, mycelia were harvested by filtration and washed twice with MM solution without any carbon source. Equal amounts (0.4 g cell wet weight) of mycelia were transferred into 250 mL Erlenmeyer flask containing 50 mL MM supplemented with 2% (w/v) microcrystalline cellulose and 2% (w/v) wheat bran, and were cultivated at 28°C, shaking at 200 rpm. The composition of the MM solution is described in the previous report (Liu et al., 2016).
Plasmid Construction and Fungal Transformation
Firstly, the AD of Xyr1 was amplified from T. reesei TU-6 cDNA and fused with the AZFPM2-Gal4 DBD amplified from T. reesei M2 genomic DNA by overlap extension PCR using primers AZFP-F and Xyr1-R, generating AzfpM2-xyr1AD coding sequence. To overexpress the AZFPs (AzfpM2-gal4AD and AzfpM2-xyr1AD) at the xyn3 loci in T. reesei TU-6, the AzfpM2-gal4AD coding sequence was amplified from T. reesei M2 genomic DNA, and then the two AZFPs coding sequences were ligated into the Nco I and Xba I sites of pCB303 (Zhang et al., 2016) to obtain the plasmids pCB310 and pCB311, respectively. Subsequently, the two AZFPs (AZFPM2-Gal4AD and AZFPM2-Xyr1AD) expression cassettes which contained the AZFPs coding sequence and the terminator TtrpC were amplified from pCB310 and pCB311, respectively. Additionally, Two DNA fragments containing approximately 1.5 kb of up- and downstream the xyn3 non-coding region and the pyr4 selection marker cassette were amplified from T. reesei QM9414 genomic DNA, respectively. Finally, five fragments including the AZFP (AZFPM2-Gal4AD or AZFPM2-Xyr1AD) expression cassette, the pyr4 expression cassette, the up- and downstream xyn3 non-coding region, and the pUG6 fragment amplified from the pUG6 vector were joined into the pUG6-AZFPM2-Gal4AD or pUG6-AZFPM2- Xyr1AD vector by RecET direct cloning technology (Wang et al., 2016). All primers used in this study were listed in Supplementary Table S1.
The protoplast transformation protocol follows previous report (Derntl et al., 2015). Transformants were cultivated and screened on MM plates containing 2% glucose without uridine. Southern blot was performed to verify the correct mutants. T. reesei QS1 and QS2 strain are the engineered strains overexpression AzfpM2-gal4AD and AzfpM2-xyr1AD at the xyn3 loci, respectively.
Southern-Blot Analysis
Chromosomal DNA was isolated from mycelia by grinding in liquid nitrogen as described previously (Derntl et al., 2015). For analysis of T. reesei QS1 and QS2 genome, Nde I was used as the restriction endonuclease for genome digestion. Probes were amplified from the genomic DNA with the primers xyn3-probe-F/R in Supplementary Table S1. Then, the Nde I-digested genomic DNA was hybridized by the probe. Finally, the probe-hybridized DNA fragments were detected and visualized with the DIG high prime DNA Labeling and Detection Starter kit I (Roche Diagnostics, Mannheim, Germany), respectively.
Western Blot Analysis
The conidia of the parent strain T. reesei TU-6 and the transformants were cultivated in MM medium containing 2% cellulose as a carbon source and 0.1% uridine to support growth. The mycelia at 72 h were collected by filtration and used to extract intracellular protein after grinding in liquid nitrogen. For detection of the His-tagged AZFP, 30 μg cell protein extracts were separated in 12% SDS-PAGE, and then the proteins were electro-transferred to PVDF membrane (Millipore, United States). An anti-His antibody (GenScript, Nanjing, China) was used to incubate with the membrane, washed, and subsequently incubated with HRP-conjugated goat anti-rabbit IgG as a secondary antibody. Bands on the blotting membrane were visualized using the DAB kit (CWBIO, Shanghai, China) according the manufacturer’s instructions.
Biochemical Assays
The activities of filter paper (FPase), endo-β-glucanase (CMCase), exo-β-glucanase (pNPCase), β-glucosidase (pNPGase), and Xylanase were determined as described elsewhere (Wood and Bhat, 1988; Gao et al., 2017). Total extracellular proteins were assayed using the BCA Kit (Beyotime, Shanghai, China).
Transcription Analysis by RT-qPCR
The T. reesei strains were cultured, and harvested at 24 and 48 h. Total RNA was extracted using the Spin Column Plant Total RNA Purification Kit (Sangon Biotech, China), and 1μg RNA was reverse transcribed to cDNA using the PrimeScript® RT Reagent Kit with gDNA Eraser (Takara Japan). RT-qPCR analysis was carried out with iQ SYBR Green Supermix Kit (Bio-Rad, United States) and the CFX Connect Real-Time PCR Detection 96 System (Bio-Rad, United States) using the primers listed in Supplementary Table S2. Three biological replicates for all reactions were carried out, and the relative transcription of genes was calculated according to the 2–ΔΔCT method using the reference gene tef1 for normalization (Livak and Schmittgen, 2001; Steiger et al., 2010).
Saccharification of Pretreated Lignocellulosic Biomass
Alkaline-pretreated corn stover (APCS) and Jerusalem artichoke stalk (APJAS) were used as substrates in the saccharification process and the chemical compositions of APCS and APJAS were described before (Meng et al., 2018). The crude enzyme was placed in 30 mL citrate buffer (50 mM, pH4.8) containing 5% (W/V) substrate and the reaction mixture was incubated at 150 rpm, 50°C. Enzyme loading was adjusted to the same protein dosage (30 mg/g substrate). Glucose released was detected by HPLC at interval of 12 h. Cellulose conversion was calculated as follows:
Results
Construction of the Recombinant T. reesei Containing the Novel Artificial Transcription Factor AZFPM2-Xyr1AD
In our previous work, transformants containing AZFPs coding sequences driven by the constitutive promoter pki were screened (Zhang et al., 2016), and one hyper-cellulolytic mutant T. reesei M2 was selected by detecting FPase activity during liquid fermentation for cellulase production in flasks (Meng et al., 2020). Subsequently, the AZFP coding sequence in T. reesei M2 was amplified and analyzed. As shown in Supplementary Figure S1, the AZFP coding sequence is composed of four zinc fingers acting as a DBD, followed by a Gal4AD, and was named AZFPM2-Gal4AD. Considering the heterologous origin of Gal4AD, we are interested in whether cellulase production could be further improved when Gal4AD was replaced by the endogenous Xyr1AD. In addition, the pki promoter was changed to a relatively stronger promoter of xyn3. Therefore, a new artificial transcription factor AZFPM2-Xyr1AD was constructed, and was constitutively expressed at the xyn3 locus in T. reesei TU-6, yielding the strain T. reesei QS2. For comparison purposes, the AZFPM2-Gal4AD was also inserted in the same locus, and the resultant strain was named T. reesei QS1 (Figure 1A). Southern blot analysis confirmed correct integration at the xyn3 locus in T. reesei QS1 and QS2 strains, respectively (Figures 1B,C). On the other hand, the expression of AZFPs in the cell lysate of T. reesei QS1 and QS2 mutants was confirmed by Western blot analysis (Figure 1D), indicating the observed phenotypic changes of transformants in cellulase production were resulted from the expression of the integrated AZFPs.
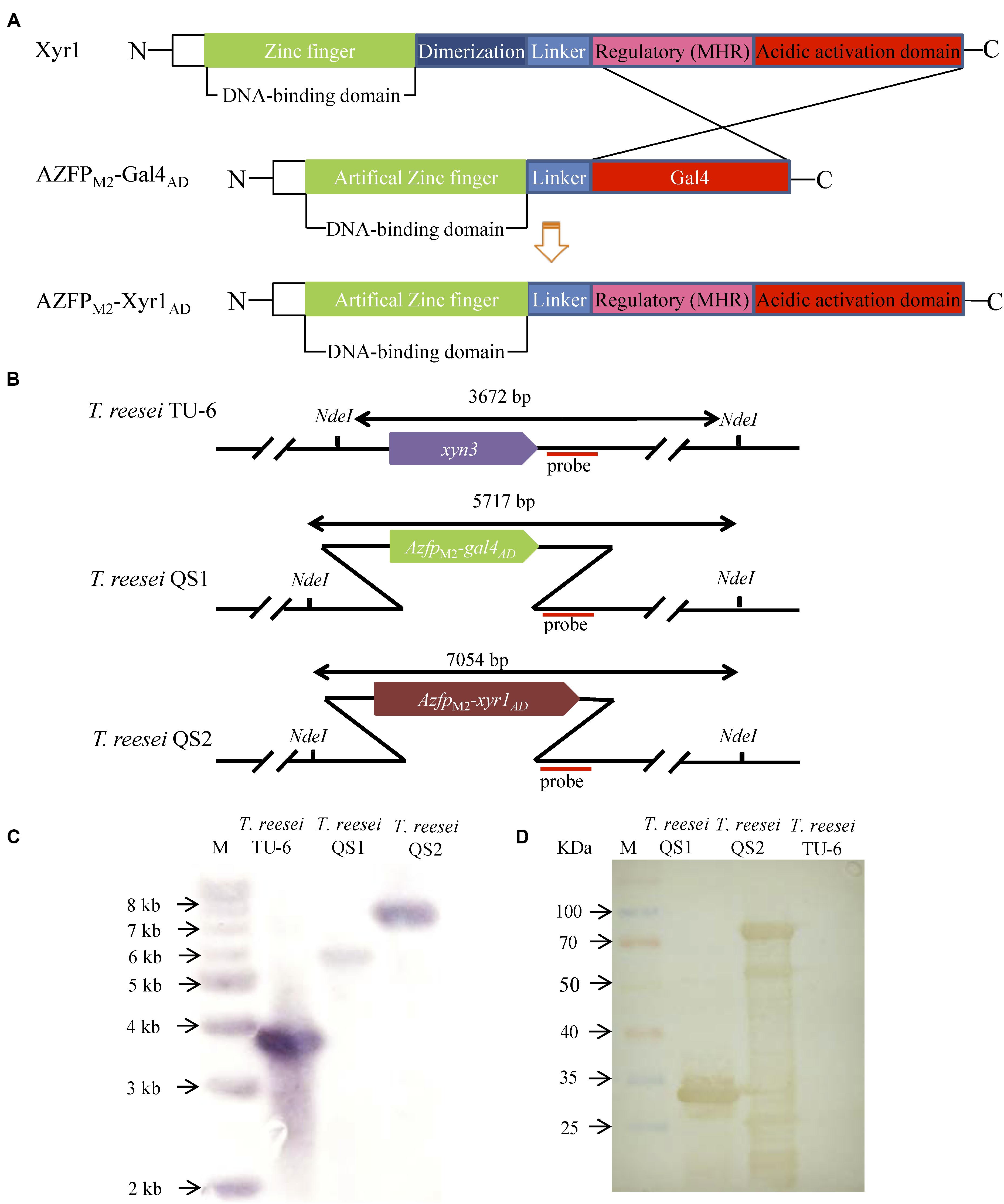
Figure 1. Construction of fusion transcription factor AZFPM2-Xyr1AD in T. reesei. (A) Schematic map of AZFPM2-Xyr1AD. (B) Schematic diagram of Southern bolt analysis. (C) Southern blot analysis of transformants QS1 and QS2 with AZFPs at xyn3 loci, respectively. Southern blot analysis of the genome digested with Nde I. A fragment of 3.7 kb is present in the parent strain, and the 5.7 and 7.1 kb bands are shown in the mutant strains QS1 and QS2, respectively. (D) Western blot analysis for AZFPM2-Gal4AD/Xyr1AD in T. reesei QS1 and QS2, respectively. T. reesei TU-6 represent the parent strain.
Influence of AZFPM2-Xyr1AD Overexpression on Cellulolytic Activity
The corresponding activities of cellulolytic enzymes from T. reesei QS2, QS1, and the parent strain T. reesei TU-6 were further evaluated. As shown in Figure 2A, T. reesei QS2 displayed the highest cellulase activity (FPase) of 2.4 IU/mL at the 5th day among these strains, 24.2 and 73.7% higher than that secreted by T. reesei QS1 and the parent strain TU-6, respectively. In addition, the endoglucanase activity (CMCase) of T. reesei QS2 improved 50.4 and 132.4%, respectively, compared to that of T. reesei QS1 and TU-6 strain (Figure 2B). In case of exoglucanase activity, T. reesei QS2 produced a pNPCase activity of 0.68 IU/mL, which displayed a 25.8 and 52.0% increase compared to the respective strains. However, we found that the activity of β-glucosidase (pNPGase) varied among these strains (Figure 2C). Surprisingly, T. reesei QS2 exhibited an obviously 48.4% higher β-glucosidase activity than T. reesei QS1 mutant, while 24.4% lower β-glucosidase activity than that of the parent strain TU-6 at the 7th day fermentation in flask (Figure 2D). To compare the roles of the two AZFPs in regulation of cellulase synthesis, the xyn3 gene locus was chosen for insertion of the AZFPs. As expected, xylanase produced by T. reesei QS2 was similar to that of T. reesei QS1, but lower than that of the parent strain (Figure 2E), which was caused by the disruption of xyn3 ORF in these two mutants. However, the extracellular proteins secreted by T. reesei QS2 showed a 21.5 and 20.2% increase in comparison with that of T. reesei QS1 and TU-6 strain, respectively (Figure 2F). These results suggest that optimization of artificial transcription factors with endogenous ADs is a viable strategy for increasing cellulase production.
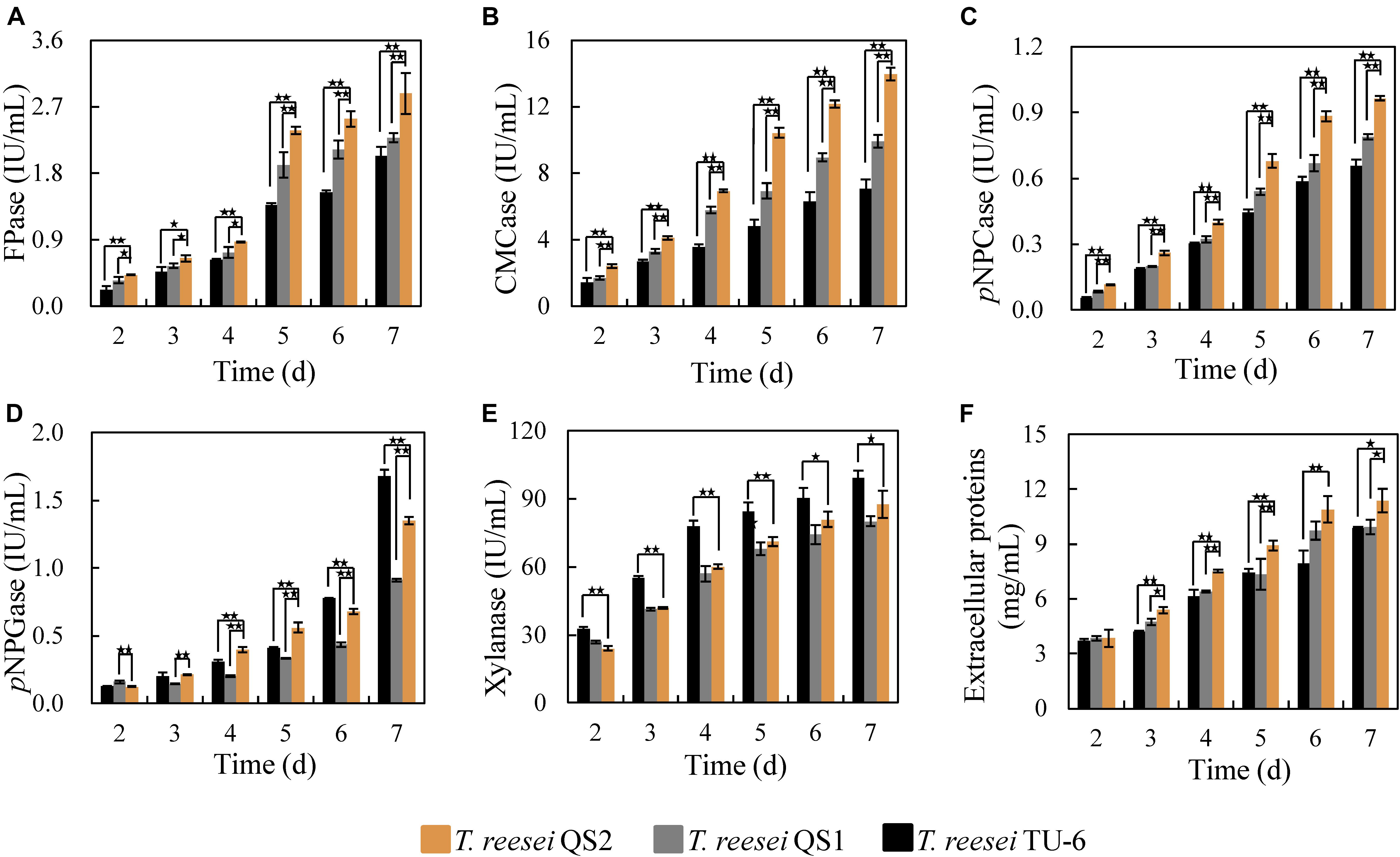
Figure 2. Cellulase production by T. reesei QS2, QS1 and TU-6. (A–E) The activities for FPase, CMCase, pNPCase, pNPGase and xylanase, respectively. (F) Total extracellular proteins content. The strains were cultured at 28°C and 180 rpm in flasks using minimal medium supplemented with 2% cellulose and 2% wheat bran as carbon source. Error bar denotes the standard deviations (SD) of three replicates (*p < 0.05, **p < 0.01).
Influence of AZFPM2-Xyr1AD Overexpression on Transcript Levels of Cellulase Encoding Genes
To gain further insight into how the overexpression of AZFPM2-Xyr1AD influences the regulation of the cellulase biosynthesis in T. reesei QS2, the transcriptional levels of major cellulase and related transcription factor genes were measured. For T. reesei QS2, higher expression of the major cellulase genes (cbh1, cbh2, egl1, egl2, and bgl1), 2. 3-, 2. 9-, 2. 5-, 2. 4-, and 3.2-folds compared with that of T. reesei QS1, was observed early at 24 h, which may consequently lead to more efficient production of cellulase characterized by the increased FPase activity. In addition, the expression of cellulolytic enzyme genes (cbh2, egl1, egl2, and bgl1) was also significantly up-regulated and a dramatic down-regulation of xyn2 at 24 h was observed in T. reesei QS2 compared with the parent strain T. reesei TU-6, which was in accordance with the activities of cellulolytic enzymes produced by T. reesei QS2 (Figure 3A). On the other hand, the transcription of accessory protein encoding genes in T. reesei QS2 also exhibited significant elevation when compared with T. reesei QS1 and the parent strain (Figure 3B). Among these genes, the transcription of cel61a and cip2 in T. reesei QS2 were 4.1- and 2.7-fold as well as 19.7- and 2.3-fold higher than that from T. reesei QS1 and TU-6 at 24 h, respectively, suggesting that the crude enzyme secreted by T. reesei QS2 is more suitable for further hydrolysis of lignocellulosic biomass.
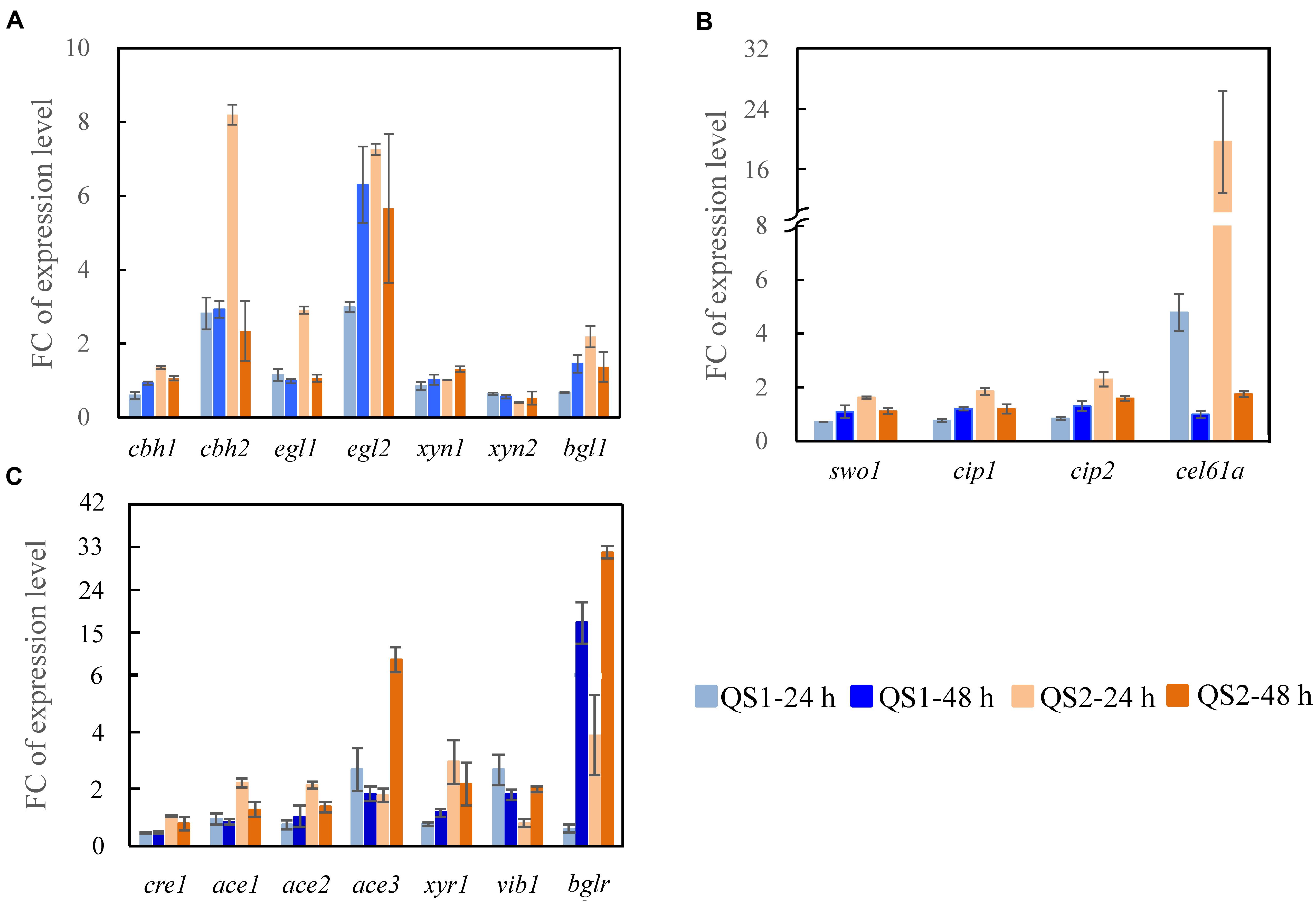
Figure 3. Transcription analysis of genes encoding major cellulolytic enzymes and regulators. Genes encoding (A) cellulolytic enzymes, (B) regulators, and (C) accessory proteins were analyzed for T. reesei QS1 and QS2 mutants with T. reesei TU-6 as the reference strain. Strains were cultured at 28°C and 180 rpm in flasks using minimal medium supplemented with 2% cellulose and 2% wheat bran as a carbon source for 24 and 48 h, respectively. Expression levels of the reference gene tef1 were used as an endogenous control. The value is the mean of three biological replicates with SD as the error bars. FC represents fold change of the transcription levels with targeted genes detected in the mutants over that detected in the control.
In addition to the enzyme-encoding genes mentioned above, the transcriptional levels of the most known cellulase transcription factor genes were also significantly altered in both T. reesei QS1 and QS2 (Figure 3C). Among these transcription factor genes, xyr1, which encodes the major transcriptional activator for cellulase biosynthesis (Stricker et al., 2006), was significantly up-regulated by 3.8- and 2.9- fold in T. reesei QS2 at 24 compared with that of T. reesei QS1 and the parent strain, respectively. Similar trend was observed at 48 h. Even more, an enhanced expression of another transcription activator gene ace3, approximately 5.0- and 9.2-folds in T. reesei QS2 compared with that of T. reesei QS1 and the parent strain, was also observed at 48 h, indicating that the production of cellulase might be specifically regulated by these transcription factors. As for bglr encoding an activator specific for regulating β-glucosidases except bgl1, its expression in T. reesei QS2 was substantially enhanced to 6.5-folds at the early stage of 24 h, and then attenuated to 1.9-folds at 48 h compared with that of T. reesei QS1. Whereas, T. reesei QS2 and QS1 showed highly similar transcription profile across the tested genes in comparison to the parent strain, but the magnitude of gene expression change was more pronounced in T. reesei QS2. Conclusively, we assume that the AZFPM2-Xyr1AD is more advantageous for regulation of cellulase biosynthesis in T. reesei than the AZFPM2-Gal4AD.
Saccharification of Lignocellulosic Substrates by Cellulolytic Enzyme Preparations From the T. reesei Strains
In order to evaluate the hydrolysis ability of the cellulase produced by the T. reesei strains on lignocellulosic biomass, the crude enzyme complexes were used to saccharify APCS and APJAS. At the same enzyme loading (30 mg/g substrate), the glucose release (28.3 g/L corresponding to 86.9% cellulose conversion) using the T. reesei QS2 enzyme in the saccharification of APCS increased 83.8 and 97.9% compared with that from T. reesei QS1 (15.4 g/L corresponding to 47.3% cellulose conversion) and TU-6 (14.3 g/L corresponding to 43.9% cellulose conversion) at 60 h (Figure 4A). In contrast, similar as described in the previous reports, less glucose was released using APJAS as a substrate, and less difference was observed using crude enzymes from different strains, which would be caused by different composition and structure of APCS and APJAS (Meng et al., 2018). However, we still found slightly more glucose release by cellulase produced with T. reesei QS2 among the three strains. The glucose concentrations are 15.5, 14.4, and 13.6 g/L (corresponding to 58.7, 54.5, and 51.4% cellulose conversion), for T. reesei QS2, QS1 and the parent strain, respectively (Figure 4B). These results revealed that the optimized enzyme system of T. reesei QS2 benefits scarification of lignocellulosic biomass.
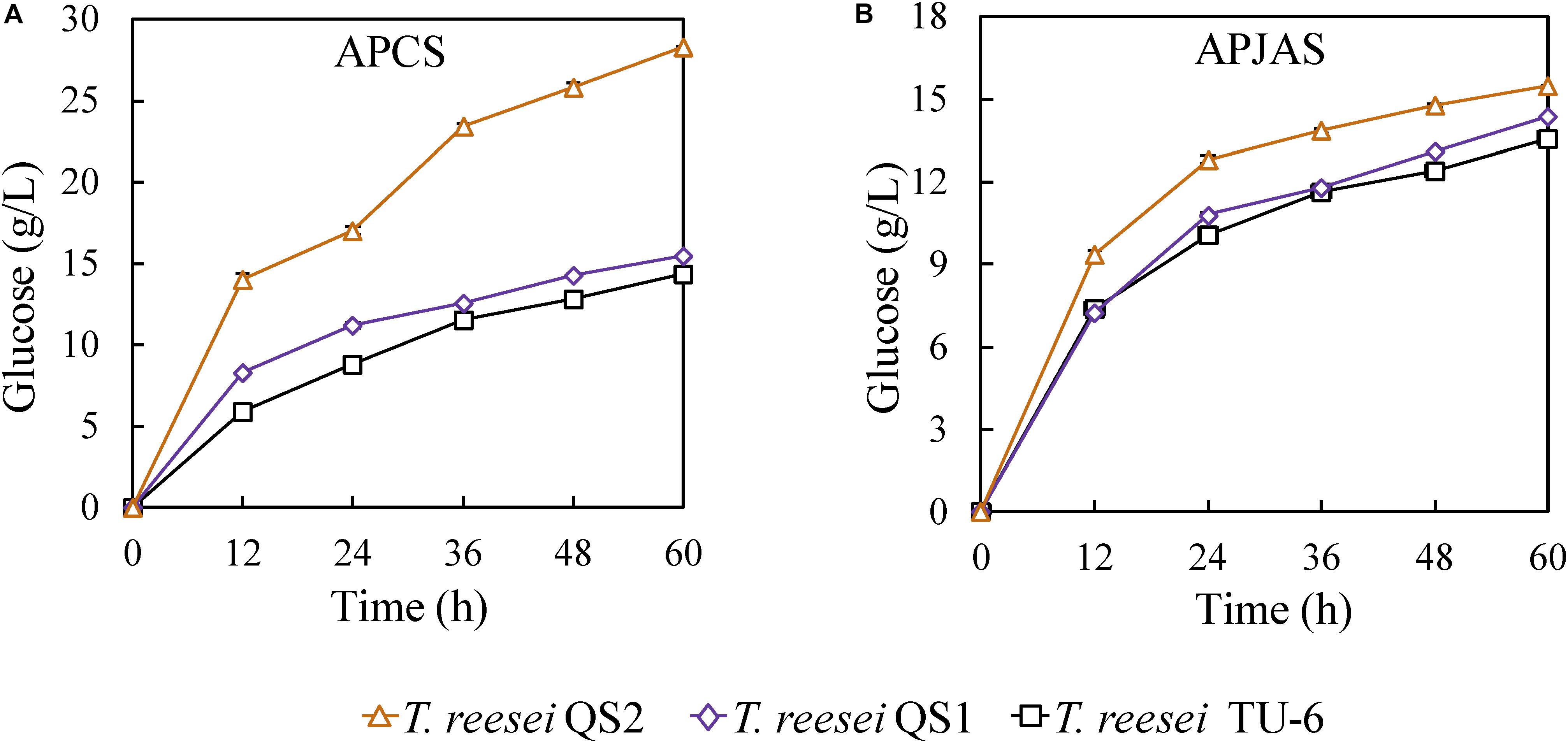
Figure 4. Hydrolysis of alkali pretreated corn stover (APCS) and jerusalem artichoke stalk (APJAS) by cellulases produced by T. reesei QS2, QS1, and TU-6. Sugar release from (A) alkali pretreated corn stover (APCS) and (B) alkali pretreated jerusalem artichoke stalk (APJAS) was examined. The same protein dosage (30 mg/g substrate) was used for enzymatic hydrolysis experiments. Data are represented as the mean of triplicate with standard deviation (SD) as the error bars.
Discussion
Based on the existing knowledge about the regulatory network of cellulase biosynthesis in T. reesei, great efforts have been made to reprogram cellulase transcription with the aim to enhance cellulase production by engineering endogenous transcription factors. However, the regulatory mechanism of cellulase biosynthesis is still not fully clear, and the cross-regulation of transcription factors involved in cellulase biosynthesis is very complicated, which all limit the development of robust strains (Liu and Qu, 2019). Taking advantage of the diversity of zinc finger DBDs, AZFPs with the yeast Gal4AD have been developed to increase cellulase production in T. reesei (Zhang et al., 2016). Yeast Gal4p has been identified as a transcription activator for regulation of genes involved in galactose catabolims, which contains an N-terminal DBD (1∼147aa) and a C-terminal AD (768∼881aa) (Hidalgo et al., 2001). The acidic part of the C-terminal activation region functions to stimulate transcription via the recruitment of SAGA (Spt-Ada-Gcn5-Acetyltransferase) in vivo (Larschan and Winston, 2001). The yeast Gcn5 orthologous protein encoding gene in T. reesei has also been identified, which plays a crucial role in regulation of cellulase gene expression (Xin et al., 2013). However, no further evidences showed that the exogenous Gal4AD activates transcription by recruiting SAGA complex for the regulation of cellulase biosynthesis in T. reesei at present. Xyr1 (Xylanase regulator) is a Gal4-like binuclear Zn-cluster protein, which can activate the expression of cellulase and hemicellulase gene expression in T. reesei. Lack of Xyr1 resulted in not only the absence of lignocellulosic enzyme production, but also down-regulation of genes encoding MFS (Major facilitator superfamily) transporters and ABC (Adenosine triphosphate (ATP)-binding cassette) transporters in inducing medium (Dos Santos Castro et al., 2016). As shown in Figure 1A, the highly conserved domain of this protein contains the DBD (91∼131aa) recognizing the nucleotide binding motif [GGC(T/A)4] (Furukawa et al., 2009), as well as a middle homology region (MHR, 314∼632aa) and a short acidic AD (767∼940aa) within the distal portion of the C-terminus. The missing 140 C-terminal amino acids of Xyr1 abolished cellulase production (Lichius et al., 2015). Recent studies have revealed the transcription factor Xyr1AD could recruit SWI/SNF to remodel nucleosomes positioned in cellulase gene promoters for activating their transcription (Cao et al., 2019), suggesting the important role of this AD in regulation of cellulase gene expression. Therefore, we speculated that Xyr1AD may function better than Gal4AD to regulate cellulase gene expression in T. reesei. Moreover, it was reported that the mating type locus protein MAT1-2-1 could directly interact with the MHR and AD of Xyr1 to regulate expression of cellulase genes in response to light, which suggest that the MHR might be critical for enhancing transcription of target genes (Zheng et al., 2017). Herein, we used the endogenous Xyr1AD including MHR instead of the original Gal4AD to rebuild a new AZFP, and the results demonstrated that this optimization of AZFP not only improves cellulase production, but also balances cellulase system for efficient biomass conversion. In our recent work, it was found that the AZFPM2-Gal4AD could down-regulate the expression of a newly identified cellulase repressor Ctf1, resulting in relief of the repression of cellulase activator Vib-1 and Ace3 by Ctf1 for improvement of cellulase production (Meng et al., 2020), which was consistent with transcription analysis in Figure 3C (Fold change > 2). Here, the expression of ctf1 was also significantly down-regulated by AZFPM2-Gal4AD, but not by AZFPM2-Xyr1AD (Supplementary Figure S2), which suggest that the regulatory mechanisms of these two AZFPs on cellulase biosynthesis are different due to the different effector domains. Taken together, our results proved the important roles of the effector domains in the AZFP regulating cellulase production in filamentous fungi. Using synthetic biology tools, other effector domains as well as synthetic effector domains can be created, and the numbers of effector domains can also be modulated in the AZFPs to improve cellulase production.
In order to compare the roles of the AZFPM2-Gal4AD and AZFPM2-Xyr1AD in cellulase biosynthesis, the xyn3 gene locus was chosen for foreign DNA insertion, since the xyn3 promoter was optimal for the expression of a specific gene and deletion of xyn3 gene did not affect the cellulase activity (Rahman et al., 2009). The results showed that the activities of CMCase and pNPCase were improved significantly, which was consistent with transcription analysis in Figure 3. In the future work, various other integration sites can be evaluated to optimize the cellulase enzymatic complex. The cellulase produced by T. reesei QS2 displayed better performance in saccharification of lignocellulosic biomass comparing with that of T. reesei QS1. Besides the increased β-glucosidase activity in cellulase mixture of T. reesei QS2 in comparison to that of T. reesei QS1 (Figure 2D), another possible reason for enhanced hydrolysis efficiency is the enhancement of accessary protein components, such as Swo1, Cip1, Cip2, and Cel61A, the encoding genes of which were up-regulated in T. reesei QS2 (Figure 3). The protein Cip2 exhibits synergistic activity with the lytic polysaccharide monooxygenase (Cel61A) to break down the linkage present in the hemicellulose-lignin matrix (Duranová et al., 2009; Pierce et al., 2017). It will be important to explore regulation of accessory protein biosynthesis by the AZFP developed in this study, so that further synthetic biology design can be performed to increase hydrolysis efficiency of lignocellulosic biomass.
Conclusion
In this study, we designed a new artificial transcription factor AZFPM2-Xyr1AD in T. reesei, and found that AZFPM2-Xyr1AD could significantly improve the expression of major cellulase genes. Moreover, the cellulase produced by the T. reesei QS2 carrying AZFPM2-Xyr1AD was proven to be highly effective in the saccharification of the pretreated lignocellulosic biomass. As a result, we proved that the endogenous effector domain Xyr1AD is more advantageous for construction of AZFP not only in improving cellulase production, but also in optimizing cellulase complex for biomass conversion.
Data Availability Statement
All datasets generated for this study are included in the article/Supplementary Material.
Author Contributions
X-QZ and Q-SM conceived the project and designed the experiments. Q-SM carried out experiments and measurements, and interpreted experimental data. WW, FZ, C-GL, X-QZ, and F-WB revised the manuscript. X-QZ supported the research funding. All authors read and approved the final manuscript.
Funding
This work was financially funded by the Natural Science Foundation of China (No. 21536006).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We acknowledge the final support from the Open Funding Project of the State Key Laboratory of Bioreactor Engineering and the Fundamental Research Funds for the Central Universities (No. 222201714053) and State Key Laboratory of Microbial Technology Open Projects Fund (No. M2017-10). We are grateful to Dr. Jin-Soo Kim in ToolGen Inc., South Korea and Prof. Xu Fang in Shandong University for kindly providing the plasmids containing artificial zinc finger protein genes and T. reesei TU-6 strain, respectively.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fbioe.2020.00675/full#supplementary-material
References
Aro, N., Ilmén, M., Saloheimo, A., and Penttilä, M. (2003). ACEI of Trichoderma reesei is a repressor of cellulase and xylanase expression. Appl. Environ. Microbiol. 69, 56–65. doi: 10.1128/AEM.69.1.56-65.2003
Aro, N., Saloheimo, A., Ilmen, M., and Penttilä, M. (2001). ACEII, a novel transcriptional activator involved in regulation of cellulase and xylanase genes of Trichoderma reesei. J. Biol. Chem. 276, 24309–24314. doi: 10.1074/jbc.M003624200
Cao, Y., Zheng, F., Wang, L., Zhao, G., Chen, G., Zhang, W., et al. (2017). Rce1, a novel transcriptional repressor, regulates cellulase gene expression by antagonizing the transactivator Xyr1 in Trichoderma reesei. Mol. Microbiol. 105, 65–83. doi: 10.1111/mmi.13685
Cao, Y., Zheng, F., Zhang, W., Meng, X., and Liu, W. (2019). Trichoderma reesei XYR1 recruits SWI/SNF to facilitate cellulase gene expression. Mol. Microbiol. 112, 1145–1162. doi: 10.1111/mmi.14352
De Bhowmick, G., Sarmah, A. K., and Sen, R. (2018). Lignocellulosic biorefinery as a model for sustainable development of biofuels and value added products. Bioresour. Technol. 247, 1144–1154. doi: 10.1016/j.biortech.2017.09.163
Derntl, C., Gudynaite-Savitch, L., Calixte, S., White, T., Mach, R. L., and Mach-Aigner, A. R. (2013). Mutation of the Xylanase regulator 1 causes a glucose blind hydrolase expressing phenotype in industrially used Trichoderma strains. Biotechnol. Biofuels. 6:62. doi: 10.1186/1754-6834-6-62
Derntl, C., Kiesenhofer, D. P., Mach, R. L., and Mach-Aigner, A. R. (2015). Novel strategies for genomic manipulation of Trichoderma reesei with the purpose of strain engineering. Appl. Environ. Microbiol. 81, 6314–6323. doi: 10.1128/AEM.01545-15
Dos Santos Castro, L., de Paula, R. G., Antonieto, A. C., Persinoti, G. F., and Silva-Rocha, R. (2016). Understanding the role of the master regulator XYR1 in Trichoderma reesei by global transcriptioanl analysis. Front. Microbiol. 7:175. doi: 10.3389/fmicb.2016.00175
Druzhinina, I. S., and Kubicek, C. P. (2017). Genetic engineering of T. reesei cellulases and their production. Microb. Biotechnol. 10, 1485–1499. doi: 10.1111/1751-7915.12726
Duranová, M., Hirsch, J., Kolenová, K., and Biely, P. (2009). Fungal glucuronoyl esterases and substrate uronic acid recognition. Biosci. Biotechnol. Biochem. 73, 2483–2487. doi: 10.1271/bbb.90486
Furukawa, T., Shida, Y., Kitagami, N., Mori, K., Kato, M., Kobayashi, T., et al. (2009). Identification of specific binding sites for XYR1, a transcriptional activator of cellulolytic and xylanolytic genes in Trichoderma reesei. Fungal Genet. Biol. 46, 564–574. doi: 10.1016/j.fgb.2009.04.001
Gao, L., Li, Z., Xia, C., Qu, Y., Liu, M., Yang, P., et al. (2017). Combining manipulation of transcription factors and overexpression of the target genes to enhance lignocellulolytic enzyme production in Penicillium oxalicum. Biotechnol. Biofuels 10:100. doi: 10.1186/s13068-017-0783-3
Gruber, F., Visser, J., Kubicek, C. P., and de Graaff, L. H. (1990). The development of a heterologous transformation system for the cellulolytic fungus Trichdoerma reesei based on a pyrG-negative mutant strian. Curr. Genet. 18, 71–76. doi: 10.1007/bf00321118
Hidalgo, P., Ansari, A. Z., Schmidt, P., Hare, B., Simkovich, N., Farrell, S., et al. (2001). Recruitment of the transcriptional machinery through GAL11P: structure and interactions of the GAL4 dimerization domain. Genes Dev. 15, 1007–1020. doi: 10.1101/gad.873901
Klein-Marcuschamer, D., Oleskowicz-Popiel, P., Simmons, B. A., and Blanch, H. W. (2012). The challenge of enzyme cost in the production of lignocellulosic biofuels. Biotechnol. Bioeng. 109, 1083–1087. doi: 10.1002/bit.24370
Larschan, E., and Winston, F. (2001). The S. cerevisiae SAGA complex functions in vivo as a coactivator for transcriptional activation by Gal4. Genes Dev. 15, 1946–1956. doi: 10.1101/gad.911501
Lichius, A., Bidard, F., Buchholz, F., Le Crom, S., Martin, J., Schackwitz, W., et al. (2015). Genome sequencing of the Trichoderma reesei QM9136 mutant identifies a truncation of the transcriptional regulator XYR1 as the cause for its cellulase-negative phenotype. BMC Genomics 16:326. doi: 10.1186/s12864-015-1526-0
Liu, G., Qin, Y., Li, Z., and Qu, Y. (2013). Development of highly efficient, low-cost lignocellulolytic enzyme systems in the post-genomic ear. Biotechnol. Adv. 31, 962–975. doi: 10.1016/j.biotechadv.2013.03.001
Liu, G., and Qu, Y. (2019). Engineering of filamentous fungi for efficient conversion of lignocellulose: tools, recent advances and prospects. Biotechnol. Adv. 37, 519–529. doi: 10.1016/j.biotechadv.2018.12.004
Liu, K., Dong, Y., Wang, F., Jiang, B., Wang, M., and Fang, X. (2016). Regulation of cellulase expression, sporulation, and morphogenesis by velvet family proteins in Trichoderma reesei. Appl. Microbiol. Biotechnol. 100, 769–779. doi: 10.1007/s00253-015-7059-2
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lv, X., Zheng, F., Li, C., Zhang, W., Chen, G., and Liu, W. (2015). Characterization of a copper responsive promoter and its mediated overexpression of the xylanase regulator 1 results in an induction-independent production of cellulases in Trichoderma reesei. Biotechnol. Biofuels 8:67. doi: 10.1186/s13068-015-0249-4
Ma, C., Wei, X., Sun, C., Zhang, F., Xu, J., Zhao, X., et al. (2015). Improvement of acetic acid tolerance of Saccharomyces cerevisiae using a zinc-finger-based artificial transcription factor and identification of novel genes involved in acetic acid tolerance. Appl. Microbiol. Biotechnol. 99, 2441–2449. doi: 10.1007/s00253-014-6343-x
Meng, Q. S., Liu, C. G., Zhao, X. Q., and Bai, F. W. (2018). Engineering Trichoderma reesei Rut-C30 with the overexpression of egl1 at the ace1 locus to relieve repression on cellulase production and to adjust the ratio of cellulolytic enzymes for more efficient hydrolysis of lignocellulosic biomass. J. Biotechnol. 285, 56–63. doi: 10.1016/j.jbiotec.2018.09.001
Meng, Q. S., Zhang, F., Liu, C. G., Zhao, X. Q., and Bai, F. W. (2020). Identification of a novel repressor encoded by the putative gene ctf1 for cellulase biosynthesis in Trichoderma reesei through artificial zinc finger engineering. Biotechnol. Bioeng 117, 1747–1760. doi: 10.1002/bit.27321
Park, K. S., Lee, D. K., Lee, H., Lee, Y., Jang, Y. S., Kim, Y. H., et al. (2003). Phenotypic alteration of eukaryotic cells using randomized libraries of artificial transcription factors. Nat. Biotechnol. 21, 1208–1214. doi: 10.1038/nbt868
Pierce, B. C., Agger, J. W., Wichmann, J., and Meyer, A. S. (2017). Oxidative cleavage and hydrolytic boosting of cellulose in soybean spent flakes by Trichoderma reesei Cel61A lytic polysaccharide monooxygenase. Enzyme Microb. Technol. 98, 58–66. doi: 10.1016/j.enzmictec.2016.12.007
Rahman, Z., Shida, Y., Furukawa, T., Suzuki, Y., Okada, H., Ogasawara, W., et al. (2009). Application of Trichoderma reesei cellulase and xylanase promoters through homologous recombination for enhanced production of extracellular β-glucosidase I. Biosci. Biotechnol. Biochem. 73, 1083–1089. doi: 10.1271/bbb.80852
Rassinger, A., Gacek-Matthews, A., Strauss, J., Mach, R. L., and Mach-Aigner, A. R. (2018). Truncation of the transcriptional repressor protein Cre1 in Trichoderma reesei Rut-C30 turns it into an activator. Fungal Biol. Biotechnol. 5:15. doi: 10.2286/s40694-018-0059-0
Steiger, M. G., Mach, R. L., and Mach-Aigner, A. R. (2010). An accurate normalization strategy for RT-qPCR in Hypocrea jecorina (Trichoderma reesei). J. Biotechnol. 145, 30–37. doi: 10.1016/j.jbiotec.2009.10.012
Stricker, A. R., Grosstessner-Hain, K., Würleitner, E., and Mach, R. L. (2006). Xyr1 (xylanase regulator 1) regulates both the hydrolytic enzyme system and D-xylose metabolism in Hypocrea jecorina. Eukaryot. Cell 5, 2128–2137. doi: 10.1128/EC.00211-06
Wang, F., Zhang, R., Han, L., Guo, W., Du, Z., Niu, K., et al. (2019). Use of fusion transcription factors to reprogram cellulase transcription and enable efficient cellulase production in Trichoderma reesei. Biotechnol. Biofuels 12:244. doi: 10.1186/s13068-019-1589-2
Wang, H., Li, Z., Jia, R., Hou, Y., Yin, J., Bain, X., et al. (2016). RecET direct cloning and Redαβ recombineering of biosynthetic gene clusters, large operons or single genes for heterologous expression. Nat. Protoc. 11, 1175–1190. doi: 10.1038/nprot.2016.054
Wood, T. M., and Bhat, K. M. (1988). Methods for measuring cellulase activities. Methods Enzymol. 160, 87–112. doi: 10.1016/0076-6879(88)60109-1
Xin, Q., Gong, Y., Lv, X., Chen, G., and Liu, W. (2013). Trichoderma reesei histone acetyltransferase Gcn5 regulates fungal growth, condidiation, and cellulase gene expression. Curr. Microbiol. 67, 580–589. doi: 10.1007/s00284-013-0396-4
Zhang, F., Bai, F., and Zhao, X. (2016). Enhanced cellulase production from Trichoderma reesei Rut-C30 by engineering with an artificial zinc finger protein library. Biotechnol. J. 11, 1282–1290. doi: 10.1002/biot.201600227
Zhang, F., Zhao, X., and Bai, F. (2018). Improvement of cellulase production in Trichoderma reesei Rut-C30 by overexpression of a novel regulatory gene Trvib-1. Bioresour. Technol. 247, 676–683. doi: 10.1016/j.biortech.2017.09.126
Zhang, J., Zhang, G., Wang, W., and Wei, D. (2018). Enhanced cellulase production in Trichoderma reesei RUT-C30 via constitution of minimal transcriptional activators. Microb. Cell Fact 17:75. doi: 10.1186/s12934-018-0926-7
Zhang, X., Li, Y., Zhao, X., and Bai, F. (2017). Constitutive cellulase production from glucose using the recombinant Trichoderma reesei strain overexpressing an artificial transcription activator. Bioresour. Technol. 223, 317–322. doi: 10.1016/j.biortech.2016.10.083
Keywords: Trichoderma reesei, cellulase production, artificial transcription factors, effector domain, lignocellulosic biomass
Citation: Meng Q-S, Zhang F, Wang W, Liu C-G, Zhao X-Q and Bai F-W (2020) Engineering the Effector Domain of the Artificial Transcription Factor to Improve Cellulase Production by Trichoderma reesei. Front. Bioeng. Biotechnol. 8:675. doi: 10.3389/fbioe.2020.00675
Received: 01 March 2020; Accepted: 01 June 2020;
Published: 25 June 2020.
Edited by:
Jean Marie François, Institut Biotechnologique de Toulouse (INSA), FranceReviewed by:
Fernando Segato, University of São Paulo, BrazilMingfeng Cao, University of Illinois at Urbana-Champaign, United States
Copyright © 2020 Meng, Zhang, Wang, Liu, Zhao and Bai. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xin-Qing Zhao, eHF6aGFvQHNqdHUuZWR1LmNu
 Qing-Shan Meng
Qing-Shan Meng Fei Zhang1
Fei Zhang1 Wei Wang
Wei Wang Chen-Guang Liu
Chen-Guang Liu Xin-Qing Zhao
Xin-Qing Zhao Feng-Wu Bai
Feng-Wu Bai