- 1UMR Transfrontalière BioEcoAgro N° 1158, Univ. Lille, INRAE, Univ. Liège, UPJV, YNCREA, Univ. Artois, Univ. Littoral Côte d'Opale, ICV – Institut Charles Viollette, Lille, France
- 2UR Risques Microbiens, Normandie Univ, UNICAEN, U2RM, Caen, France
Enterocin DD14 (EntDD14) is a two-peptide leaderless bacteriocin produced by Enterococcus faecalis 14, a strain previously isolated from meconium. EntDD14 has a strong antibacterial activity against Gram-positive bacteria. Leaderless bacteriocins, unlike bacteriocins with leader peptides, are immediately active after their translation, and a producing strain has then to develop specific mechanisms to protect both intra and extracellular compartments. The in silico analysis of Ent. faecalis 14 genome allowed to locate downstream of structural ddAB genes, 8 other adjacent genes, designed ddCDEFGHIJ, which collectively may form three operons. To gain insights on immunity mechanisms of Ent. faecalis 14, mutant strains knocked out in ddAB genes encoding bacteriocin precursor peptides (Δbac) and/or ABC transporter (ΔddI) of EntDD14 were constructed and characterized. Importantly, Δbac mutant strains, from which structural ddAB genes were deleted, resulted unable to produce EntDD14 and sensitive to exogenous EntDD14 showing their involvement in the Ent. faecalis 14 immunity system. Moreover, the sensitivity of Δbac mutants appeared not to be associated with the down-regulation of ddCDEFGHIJ gene expression since they were similarly expressed in both Δbac and wild-type strains during the log phase while they were found significantly down-regulated in the Δbac mutant strain after 24 h of growth. Data gathered from this study suggest also the implication of the ABC transporter (ddHIJ) in the active export of EntDD14 but ruled-out its involvement in the primary self-immunity system. Interestingly, non-bacteriocin producing Ent. faecalis JH2-2 cells transformed with ddAB, or ddAB plus genes encoding the ABC transporter (ddAB-HIJ) did not produce EntDD14 and remained sensitive to its action. Of note, trans-complementation of the Δbac mutant strain with these constructions allowed to recover the WT phenotype. To the best of our knowledge, this is the first study delineating the role of the intracellular two-peptide leaderless bacteriocins in their self-immunity.
Introduction
Enterococci are members of the lactic acid bacteria (LAB) group that are known to produce potent bacteriocins, named enterocins. Bacteriocins are ribosomally synthesized, antimicrobial peptides active against a group of closely related species (Klaenhammer, 1993; Nes et al., 1996). Many studies have reported the promising potential of bacteriocins for use in in different areas like food industry (Perez et al., 2014; Juturu and Wu, 2018), and both clinical and veterinary applications (Papo and Shai, 2005; Diez-Gonzalez, 2007; van Heel et al., 2011; Hu et al., 2018). According to their physicochemical properties, bacteriocins can cause perturbations of cell membranes (pore formation, followed by efflux of metabolites and ions), depolarization of membranes, inhibition of cell wall synthesis or septum formation, disruption of DNA replication and transcription, or interference at the ribosomal level preventing protein synthesis (Cotter et al., 2013; Verma et al., 2014; Johnson et al., 2018).
Different classifications of bacteriocins have been proposed. Cotter et al. (2013) subdivided bacteriocins into two main classes. Class I contains bacteriocins that undergo significant post-translational modifications and class II contains unmodified peptides but that can undergo only slight modification such as the formation of disulphide bridges or circularization. Recently, Alvarez-Sieiro et al. proposed a classification constituted of three classes, with emphasis on modified bacteriocins (Alvarez-Sieiro et al., 2016). Class I contains small post-translationally modified peptides, designed RiPPs (ribosomally synthesized, post-translationally modified peptides), and are <10 kDa; class II contains unmodified bacteriocins smaller than 10 KDa; and class III contains unmodified bacteriocins larger than 10 KDa and endowed with bacteriolytic or non-lytic mechanisms.
Class II bacteriocins of the lactic acid bacteria can be divided into four groups; Class IIa pediocin-like bacteriocins are peptides that containing a YGNGV motif; Class IIb contain two-peptide bacteriocins whose antimicrobial activity depends on the complementarity of the two peptide partners; Class IIc are circular bacteriocins; and class IId groups unmodified linear peptides, non-pediocin-like, single-peptide bacteriocins that do not fit into the three other groups (Cotter et al., 2013). This latter group is relatively diverse and includes a leaderless bacteriocin subgroup whose members are different from other bacteriocins. While most bacteriocins are synthesized with an N-terminal leader peptide, leaderless bacteriocins do not involve any N-terminal leader sequence for their export outside of the cell (Cotter et al., 2013).
Many leaderless bacteriocins with different characteristics have been recently described in the literature, overall constituting a large group. Leaderless bacteriocins can be composed of from one to four peptides. The leaderless single peptide bacteriocins include enterocin Q, aureocin A53, BHT-B, LsbB, lacticin Q, lacticin Z, weisselicin Y, weissellicin M, enterocin EJ97, enterocin K1, epidermicin NI01, lactolisterin BU, for review see Perez et al. (2018). The leaderless two-peptide bacteriocins group enterocin L50 (A, B), enterocin MR10 (A, B), enterocin DD14 (A, B), enterocin Ent7 (A, B) and enterocin A5-11 (A, B) (Cintas et al., 1998; Batdorj et al., 2006; Martín-Platero et al., 2006; Liu et al., 2011; Caly et al., 2017). The leaderless multi-peptide bacteriocins include aureocin A70 (A, B, C, D) (Netz et al., 2001) and garvicin KS (A, B, C), cereucin X (A, B, C), cereucin V (A, B, C), and cereucin H (A, B, C, D) (Ovchinnikov et al., 2016).
Enterocin DD14 is a two-peptide leaderless bacteriocin produced by Ent. faecalis 14 isolated from meconium at the Roubaix Hospital, in the north of France (Al Atya et al., 2015). The antagonistic activity of EntDD14 was reported against a panel of Gram-positive bacteria such as Staphylococcus aureus, Listeria monocytogenes, Ent. faecalis, Bacillus subtilis and Clostridium perfringens.
The scale-up production of bacteriocins requires an understanding of their genetic regulation and expression. Thus, it would be pertinent to identify the mechanisms involved in the secretion of EntDD14, a bacteriocin which lacks a transport signal domain. It has been reported that bacteriocin production is generally associated with the expression of specific immunity proteins, allowing protection of the bacteriocinogenic strain from the toxicity of its own bacteriocin (Bierbaum and Sahl, 2009; Nissen-Meyer et al., 2009). The leaderless bacteriocins are active immediately after their translation process (Perez et al., 2018). Their intracellular toxicity has been reported for lacticin Q (Iwatani et al., 2012), but many questions related to this topic remain to be solved. The self-immunity system would have to protect the bacteriocinogenic strain from both intra- and extracellular bacteriocins. To date there are little data in the literature reporting biosynthetic pathways of leaderless bacteriocins. Genes implicated in the secretion and immunity of aureocin A70 (Netz et al., 2001; Coelho et al., 2014), aureocin A53 (Nascimento et al., 2012) and lacticins Q and Z (Iwatani et al., 2012, 2013) have been experimentally confirmed. Interestingly, the protein LmrB, an ABC-type multi-drug transporter, has been reported to play a role in secretion and immunity of the leaderless bacteriocin, LsbB, and that of a non-leaderless bacteriocin, LsbA (Gajic et al., 2003). Here, the two-peptide leaderless bacteriocin EntDD14 has been investigated at the molecular level to decipher and better understand its role in the self-immunity system.
Materials and Methods
Bacterial Strains, Plasmids, and Growth Conditions
Bacterial strains, plasmids and oligonucleotides used in this study are listed in Tables 1, 2. Cultures of Ent. faecalis, Listeria innocua and C. perfringens strains were performed in M17 medium supplemented with 0.5% (w/v) of glucose (GM17) or in brain heart infusion (BHI) at 37°C under semi-aerobic or anaerobic conditions. Where necessary, media were supplemented with erythromycin (Em) (150 μg/mL) or chloramphenicol (Cm) (15 μg/mL). Growth of the cultures was followed by measuring the optical density at a wavelength of 600 nm (OD600) with a spectrophotometer (Aquoalabo, France) and by determining cfu counts on agar plates. Escherichia coli strains were cultivated at 37°C in Luria-Bertani (LB) broth with vigorous shaking (160 rpm) or on LB agar medium. Where appropriate, ampicillin (100 μg/mL), erythromycin (150 μg/mL) or chloramphenicol (10 μg/mL) was added to the medium. Bacterial stocks were stored at −80°C in GM17, BHI or LB broth supplemented with 50% (v/v) glycerol.
General Molecular Methods
Molecular cloning and other standard techniques were performed essentially as previously described (Sambrook and Russell, 2001). Antibiotics, chemicals, and enzymes were of reagent grade. Restriction endonucleases and T4 ligase were obtained from ThermoFicher Scientific and used in accordance with the manufacturer's instructions. Plasmids and PCR products were purified using NucleoSpin kits (Macherey-Nagel, Dünen, Germany) and the final plasmid constructions and the resulting deletion mutants were verified by PCR and sequencing (Eurofins, Ebersberg, Germany). The resulting sequences were analyzed using the SnapGenes tool (GSL Biotech LLC, CA). The strains were transformed by heat shock for E. coli and by electroporation for Ent. faecalis using Gene Pulser Apparatus (Bio-Rad Laboratories).
Antibacterial Activity Assays
The screening of antimicrobial activity of WT Ent. faecalis 14 and its isogenic mutant strains against the target bacteria was performed using well-diffusion and the spot-on-lawn methods (Caly et al., 2017). Briefly, BHI plates (1% agar) were inoculated with L. innocua, C. perfringens, Ent. faecalis JH2-2 and Ent. faecalis V583 strains and were allowed to dry. Then, 4 μL of a bacterial culture or 50 μL of culture supernatant/pure EntDD14 of the tested strain was spotted on the plate and incubated overnight at 37°C in appropriate conditions. The radius of the inhibition zone was measured from the edge of the well/spot to the edge of the inhibition halo. Antibacterial activity was quantified using the Arbitrary Units (AU) method (Cintas et al., 2000).
EntDD14 Purification and Quantification
A volume of 200 mL culture of Ent. faecalis 14 was grown for 24 h in GM17 medium at 37°C without shaking. The culture supernatant was harvested by centrifugation (8,000 rpm) and was incubated for 24 h at 4°C with CM Sephadex® C-25 (GE Healthcare Life Sciences, Milwaukee, WI). The resin was then washed with 5 bed volumes (BV) of 50 mM sodium phosphate (pH 6.3) and 5 BV of 0.5 M NaCl. The resin-bound bacteriocin was then eluted with 2 BV of 1.5 M NaCl. The EntDD14 was then further purified by gel filtration using PD MidiTrap G-10 columns (GE Healthcare Life Sciences) in Milli-Q water. The eluted solution was dried by miVac Sample Concentrators (SP Scientific NY-USA) and the dried samples were then resuspended in appropriate volumes of Milli-Q water to give the desired concentration. After each purification step, protein concentration was measured using a bicinchoninic acid assay (Sigma-Aldrich). The purity of the EntDD14 was verified by MALDI-TOF/MS analyses (Bruker Daltonics, Bremen, Germany) according to the recommendations of the manufacturer.
Sensitivity Assays to Pure EntDD14
To analyze the sensitivity of the Ent. faecalis 14 mutant strains, and other targets to the purified EntDD14, kinetics at 37°C in GM17 medium, supplemented with purified EntDD14 at a final concentration of 10 μg/mL, were performed in a 96-well microplate using a SpectraMax i3 spectrophotometer (Molecular Devices, CA, USA). The cultures were inoculated with the same bacterial charge thus giving the same initial OD600 of 0.2. Measurements were taken every 30 min at OD600 for 12 H. In parallel, the activity of EntDD14 (60 μg/mL) or serial dilutions of culture supernatant was also tested using the well-diffusion assay and the bacteriocin activity was expressed in AU/mL, which corresponds to the inverse of the last active dilution multiplied by 100 (Batdorj et al., 2006).
Construction of the Double Mutant Strain Δbac
The double mutant strain Δbac was constructed by allelic exchange using a method based on the conditional replication of the pLT06 vector (Thurlow et al., 2009). For this purpose, we used the Ent. faecalis 14 strain (Al Atya et al., 2015) to prepare the DNA template for PCR amplification using the Phusion™ High-Fidelity DNA Polymerase Mix (Thermo Fisher Scientific, Strasbourg, France). First, we amplified separately the upstream (900 bp) and downstream (1,057 bp) regions of the ddA/ddB genes using the primer pairs Bac1F-PstI/Bac2R-Stop and Bac3F-Stop/ Bac4R-NcoI, respectively (Table 2). The Bac3F-Stop and Bac2R-Stop primers share a complementary sequence of 24 bp, where four stop codons and the BamHI restriction site were inserted (Figure S1). To generate the entire DNA fragment harboring the mutated ddA and ddB genes, a second PCR was performed using the primer pair Bac1F-PstI/Bac4R-NcoI and the mix of the two previous amplified fragments as template DNA. The resulting DNA fragment of 1,981 bp was excised from the electrophoresis agarose gel and purified. After digestion with appropriate restriction enzymes (PstI and NcoI), this fragment was cloned into the pLT06 plasmid to generate pLT06ΔddAB using the E. coli JM109 strain. This recombinant plasmid was used to transform Ent. faecalis 14 and clones were obtained on GM17 with 15 μg/mL of Cm at 30°C (the permissive temperature of the plasmid). To trigger the first cross-over (CO), cultures were grown for 2½ h at 30°C then shifted to 42°C for an additional 4 h in the presence of Cm. After serial dilutions, agar plates supplemented with 15 μg/mL Cm and X-Gal 80 μg/ml were inoculated. The larger blue colonies were verified by PCR using the outer primers Bac5F with one of the primers located on the plasmid (ORIf or KS05seqR). A colony from the 1st CO was cultured in the absence of selection pressure until the culture had reached stationary phase (~2 × 109 cfu/mL). Serial dilutions were prepared and a new culture of ~100 cfu/mL was inoculated and subsequently grown to stationary phase overnight. At this step, the chloramphenicol-susceptible clones underwent the 2nd CO and were screened for the deletion of the ddA and ddB genes. The BacFscreen primer (Table 2) was designed on the basis of the introduced modified sequence in place of the ddA and ddB sequence (Figure S1) thereby allowing the screening by PCR of the chloramphenicol-susceptible clones obtained after the second crossing-over event. When BacFscreen primer was used in combination with the Bac6R primer (located outside of the construct; Figure S1), only the double mutant strain Δbac clones were able to produce a 1,132 pb DNA product. Moreover, PCR with Bac5F/Bac6R amplified a shorter fragment than that amplified in the wild-type, confirming that the deletion occurred in the structure of the ddA and ddB genes. Finally, the EntDD14 genetic environment of the obtained mutant clones was also verified by sequencing.
Construction of the ΔddI Mutant Strain
To delete the gene ddI of Ent. faecalis14 corresponding to an ATP-binding protein of the ABC transporter, a strategy identical to that used to generate the Δbac mutant strain was followed. Briefly, the upstream (1,128 bp) and downstream (987 bp) regions of the ddI gene were amplified using the primer pairs ddI 1F-PstI/ddI 2R-Stop and ddI 3F-Stop/ddI 4R-NcoI, respectively (Table 2). A second PCR using the primer pair ddI 1F-PstI/ddI 4R-NcoI was performed using the mix of the two previous amplified fragments as DNA template in order to obtain the entire DNA fragment harboring the mutated ddI gene. After that, the resulting DNA fragment was cloned into the pLT06 vector and the resulting plasmid pLT06ΔddI was transformed into E. coli JM109 then into Ent. faecalis 14. After the 1st and 2nd CO events, the chloramphenicol-susceptible clones were screened for the deletion of ddI gene by PCR using BacFscreen/ddI 6R and ddI 5F/ddI 6R primer pairs. Finally, the genetic environment of the resulting mutant strains was verified by sequencing.
Construction of the Triple Mutant Strain ΔIΔbac
The ΔddI mutant strain was used to generate the Ent. faecalis 14 ΔIΔbac triple mutant strain by using a double homologous recombination as described above. Competent Ent. faecalis 14 ΔddI cells were transformed by the recombinant pLT06ΔddAB plasmid and the same method was then used to obtain the triple mutant strain ΔIΔbac. All these mutant strains and their required genetic correctness were checked by PCR and sequencing.
Complementation of Ent. Faecalis 14 Mutant Strains and Construction of Ent. Faecalis JH2-2 Harboring Structural and ABC Transporter Genes
For the complementation assays, a DNA fragment containing the entire ddA, ddB and the promoter region was obtained by PCR using Bac-compF-KpnI and Bac-compR-BamHI primers and cloned into pAT18 (Table 1). The recombinant plasmid pAT18:ddAB was used to transform competent cells of the Ent. faecalis 14 Δbac mutant and Ent. faecalis JH2-2 after passing through E. coli XL-1 blue selected on LB, Em (150 μg/mL), X-gal (40 μg/mL) and IPTG (40 μg/mL). The colonies obtained on GM17 agar Em (150 μg/mL) after 48 h at 37°C, (Ent. faecalis 14Δbac comp and JH2-2 pAT18:ddAB) were analyzed by PCR and sequencing for the presence of the recombinant plasmid. In the same way we complemented the ΔddI mutant strain using the primers ddI-compF-BamHI and ddI-compR-PstI. To complement the ΔIΔbac triple mutant strain, we constructed a pAT18:ddAB-HIJ containing the structural genes ddA and ddB and the ABC transporter module ddH, ddI, and ddJ intact genes. For this, a first PCR with the OpAF-KpnI/OpBR primer pair was used to amplify the ddAB genes including the promoter region. A second PCR with the Op HF/Op JR-BamHI primers amplified the ddHIJ genes of the ABC transporter. As the Op BR and Op HF primers share a complemented sequence of 24 bp (see Table 2), a PCR using the AF-KpnI/Op JR-BamHI primers was able to amplify the ddAB-HIJ genes. After digestion by BamHI/KpnI, the obtained fragment was cloned into the pAT18 vector to obtain pAT18:ddAB-HIJ (3,7 kb). This construction was used to transform the ΔIΔbac mutant and Ent. faecalis JH2-2 strains leading to ΔIΔbac-Comp and Ent. faecalis JH2-2-ddAB-HIJ strains, respectively.
RNA Isolation and Microarrays Analysis
For microarray analysis, three distinct cultures of Ent. faecalis 14Δbac were used (Δbac1, Δbac2 and Δbac3) in order to compare with the Ent. faecalis 14 wild-type strain (WT1, WT2 and WT3), after 6 and 24 h of growth in GM17 medium. Analyses were performed with total RNA isolated using NucleoSpinTM RNA Plus columns (Macherey-Nagel, Hoerdt, France). RNA quality was verified with Nanodrop and absorbance ratios A260/280 and A260/230 were between 2.0 and 2.2. RNA quality was also examined with Bioanalyzer 2100 (Agilent, Les Ulis, France) and a minimal RNA integrity number (RIN) of 0.8 was required for all samples.
Agilent G2509F Ent. faecalis custom oligo-based DNA microarray (8 × 15 K) containing spots of 60-mer oligonucleotide probes (in duplicate) were used to study the gene expression profile. RNA amplification, staining, hybridization and washing were conducted according to the manufacturer's instructions. Slides were scanned at 5 μm/pixel resolution using the GenePix 4000B scanner (Molecular Devices Corporation, Sunnyvale, CA, USA). Images were used for grid alignment and expression data digitization with GenePix Pro 6.0 software. Expression data were normalized by Quantile algorithm. To ascertain the quality of normalized data, filtering of data was mandatory for flagged signals. Expression data from the 3 wild-type samples were filtered for p < 0.05 and the average was calculated for each gene. Genes of interest were selected for this study (ddABCDEFGHIJ) and the log2 ratio from individual Δbac samples and the mean of WT samples was calculated. The corresponding probes designed for these genes are shown in Table 3.
Bioinformatics Analysis
The Ent. faecalis 14 genome sequence was obtained from GenBank under accession number CP021161. Here, different databases and bioinformatics tools were used to analyze the DNA sequences and predict the EntDD14 operon organization, with location of putative promoters and terminators. This analysis included the Basic Local Alignment Search Tool of the National Center for Biotechnology Information (BLAST-NCBI: https://blast.ncbi.nlm.nih.gov/Blast.cgi), the SnapGene®4.3.7 tool, the Softberry software for analysis of bacterial genomes (http://www.softberry.com), the ARNold program (http://rssf.i2bc.paris-saclay.fr/toolbox/arnold/index.php#Results) and the RibEx program (Abreu-Goodger and Merino, 2005)
Statistical Analysis
Differences between samples were calculated using the Student test. P ≤ 0.05 were considered to be significant.
Results
Genetic Environment of EntDD14 Encoding DNA
Previous sequencing of the Ent. faecalis 14 genome showed that the EntDD14 encoding genes are chromosomal determinants (Belguesmia et al., 2017). In silico analysis of the genetic neighborhood of the EntDD14 structural genes revealed a cluster of 10 open reading frames (ORFs) ddABCDEFGHIJ (Figure 1). This cluster encodes two-peptide leaderless bacteriocins (DdA and DdB), five unknown proteins (DdC, D, E, F, and G) and a putative ABC transporter (DdH, I and J) (Table 4). The sequence alignment of this 6.5kb region with the BLAST-NCBI showed a high homology with the translated two-peptide leaderless bacteriocin regions of other known enterococci such as Ent. faecalis DBH18 (GenBank ID: EF502034.2) (Arbulu et al., 2016), Ent. faecalis KB1 (GenBank ID: CP022712.1), Ent. faecalis plasmid Efsorialis-p1 (GenBank ID: CP015884.1), Ent. faecium SRCM103341 plasmid (GenBank ID: CP035137.1), Ent. faecium strain FA3 plasmid (GenBank ID: CP042833.1), Ent. faecium strain 6T1a plasmid pEF1 (GenBank ID: DQ198088.1), Ent. faecium strain HB-1 plasmid (GenBank ID: CP040877.1), Ent. faecium strain SRCM103470 plasmid (GenBank ID: CP035221.1), Ent. faecium strain Gr17 plasmid pGR17 (GenBank ID: CP033377.1) and Ent. faecium strain 4928STDY7387800 (GenBank ID: LR607382.1). Noteworthy, the sequence analysis by the Bacterial Operon and Gene Prediction (http://www.softberry.com) predicted three operons. The first operon consisted of the ddAB genes, the second contained the ddCDEFGH genes and the third consisted of the two ddIJ genes (Figure 1). The structural genes ddA and ddB are expected to be co-transcribed as only one promoter has been detected upstream of the ddA gene and a clear processing site motif of 48 bp was detected between the ddB and ddC genes (Figure 2). The alignment of the DdA and DdB peptides with all other known two-peptide leaderless bacteriocins displayed a very high degree of identity. In this sense, EntDD14 was identical to MR-10 and Ent7 produced by Ent. faecalis MRR 10-3 (Martín-Platero et al., 2006) and Ent. faecalis 710C (Liu et al., 2011), respectively (Figure 3). Moreover, DdA and DdB were 98 and 95% identical to the corresponding peptide of L50 and A5-11 bacteriocins produced, respectively, by Ent. faecium L50 (Cintas et al., 2000) and Ent. durans (Batdorj et al., 2006; Belguesmia et al., 2013). Interestingly, all the bacteriocinogenic strains above-cited share at least a set of 3 genes, which are highly homologous to the second ABC transport system (As-48FGH) described for the cyclic bacteriocin AS-48 (Diaz et al., 2003) (Figure 1).
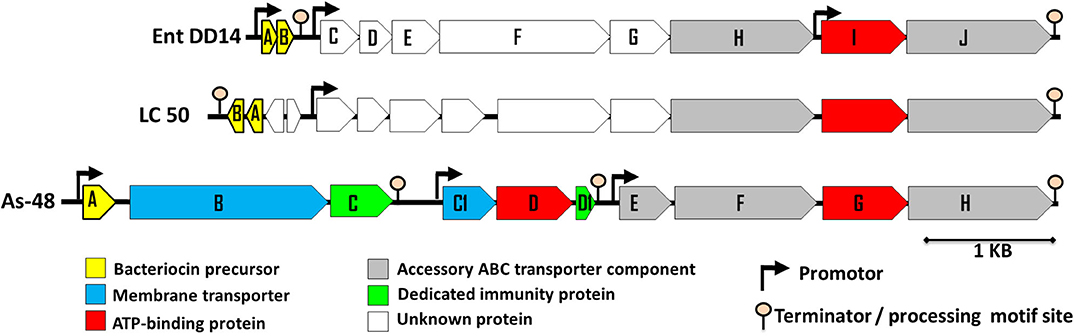
Figure 1. Genetic organization of Ent. faecalis 14 predicted ORFs. The genetic organization of Ent. faecalis 14 is compared to that of Ent. faecalis LC50 and to that of Ent. faecalis AS48 that are shown on the figure and produce enterocins L50AB and AS-48, respectively. The predicted promoters and ORFs were obtained using Softberry software.

Figure 2. Genetic characterization of the ddA and ddB structural genes. The processing site motif has been detected by using both ARNold and RibEX programs.

Figure 3. Alignment of the EntDD14 peptides with other leaderless two-peptide bacteriocins. The EntDD14 peptides (A and B) are aligned with the corresponding peptides of Ent 7, MR-10, L50, and A5-11 produced by Ent. faecalis 710C, Ent. faecalis MRR 10-3, Ent. faecium L50, and Ent. Durans, respectively.
Antimicrobial Activity of Ent. Faecalis 14 Is Solely Due to EntDD14
The antibacterial activity of Ent. faecalis 14 against L. innocua, C. perfringens, Ent. faecalis JH2-2 and Ent. faecalis V583 was solely attributable to EntDD14, and not to any other potential antibacterial agent, which could be produced by the WT strain. Indeed, the Δbac mutant strain, deleted in ddA and ddB genes, was completely devoid of any antibacterial activity despite the comparable pH values of the supernatants obtained from the mutant and the WT strains. Noteworthy, after 24 h of culture, the supernatant final pH measured was 5.12 for the WT strain and 4.87 for the Δbac mutant strain. This slight difference in pH values could be explained by the basic nature of the two peptides of EntDD14 produced by the WT strain. Similar data were obtained with the WT vs. the ΔddIΔbac triple mutant strain (Figure 4), which strengthens this argument. Trans-complementation assays conducted upon cloning the ddA and ddB genes into the Gram-positive replicative plasmid pAT18 recovered the WT phenotype in the Δbac mutant strain as shown by the complemented strain Ent. faecalis 14 Δbac-Comp phenotype. Therefore, production of EntDD14 was comparable between the WT and the Ent. faecalis 14 Δbac-Comp complemented strains (Figure 4). Thus, the inhibition potency of Ent. faecalis 14 was exclusively due to EntDD14 production, and not to any other factor such as pH decrease or organic acid production, which occur in LAB during glucose metabolism.
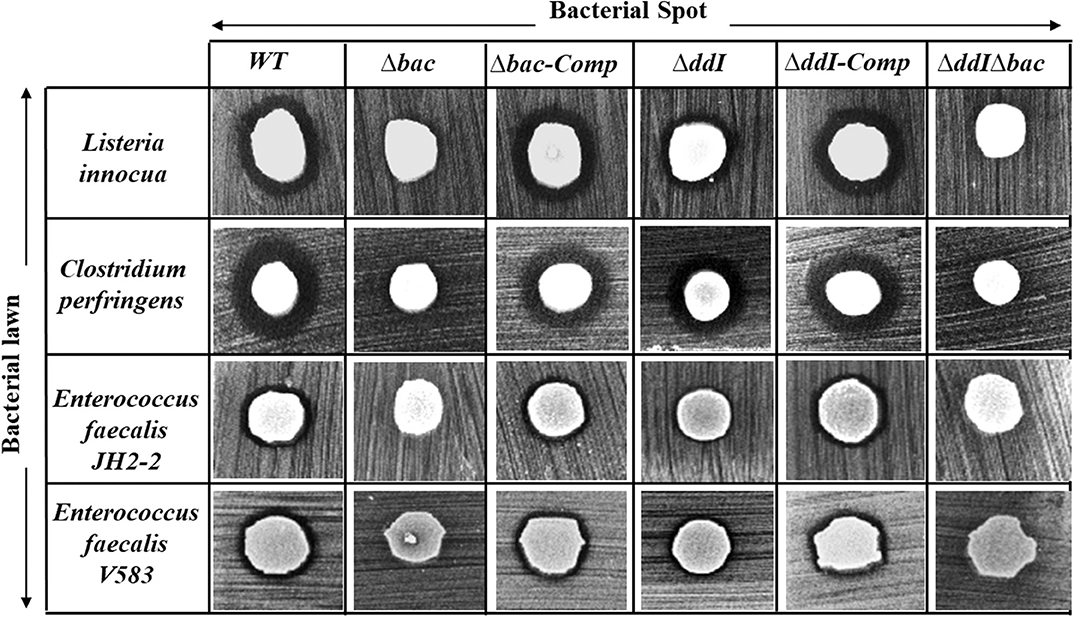
Figure 4. Antimicrobial activity of Ent. faecalis 14 wild-type, its null and complemented mutant strains against Listeria innocua, Clostridium perfringens, Enterococcus faecalis JH2-2 and Enterococcus faecalis V583. Δbac: Ent. faecalis 14 mutant deleted in ddA and ddB bacteriocin structural genes, Δbac-Comp: the Δbac complemented strain, ΔddI: Ent. faecalis 14 mutant deleted in ddI gene, ΔddI-Comp: the ΔddI complemented strain, ΔddIΔbac: Ent. faecalis 14 mutant deleted in ddI, ddA, and ddB genes. If present, the inhibition zone indicated the susceptibility of the bacterial lawn (target bacteria) to the produced EntDD14 bacteriocin. The data are representative of at least three independent experiments.
The Role of the ABC Transporter in the Transport of EntDD14
Bacterial ABC transporters are often composed of an ATP-binding protein and one or more accessory proteins (Beis and Rebuffat, 2019). In the EntDD14 locus, the ABC transporter is located downstream of the structural genes encoding for the EntDD14 enterocin. As shown in Figure 1 and Table 4, the ABC transporter associated with EntDD14 production contained at least three genes named dd-H, I, and J. The gene ddH encodes a putative efflux system protein YvrP. The ddI gene encodes an ABC transporter ATP-binding protein of the ATPase family, and the ddJ gene encodes a putative macrolide export ATP-binding/permease protein (MacB). Our results show that this ABC transporter is involved in the active transport of intracellular EntDD14 peptides to the outside of the membrane. Indeed, the mutant deleted in the ddI gene encoding an ATP binding cassette of the ABC transporter has considerably reduced the production of EntDD14, comparative to the WT strain (Figure 4). Indeed, the ΔddI mutant strain produced only 25% of the EntDD14 produced by the WT strain. This rate was based on the anti-Listeria activity of the ΔddI mutant strain (200 AU/mL) compared to the WT (800 AU/mL). The levels of EntDD14 production were restored when the ΔddI mutant strain was complemented with a functional ddI gene cloned into pAT18 vector (Ent. faecalis ΔddI-Comp) (Figure 4), arguing the role of this putative transporter in the active export of EntDD14. Nevertheless, this result suggested that additional systems are also exporting EntDD14 outside of the cell, as this mutant strain, was still producing about 25% of EntDD14 compared to the WT strain. To check whether other non-specific enterococcal transporters were involved in EntDD14 export, we transformed the non-bacteriocinogenic and sensitive Ent. faecalis JH2-2 strain with the EntDD14 precursor including its promoter region and structural ddA and ddB genes cloned into the pAT18 vector. No production of EntDD14 was observed in the resulting strains (data not shown).
The Contribution of the ABC Transporter to the Immunity System Against Exogenous EntDD14
ABC transporters can contribute to bacteriocin immunity by pumping bacteriocins to the outside of the cell membrane (Diaz et al., 2003; Sánchez-Hidalgo et al., 2003; Beis and Rebuffat, 2019). To better understand the role of the ABC transporter in immunity to EntDD14, we performed sensitivity tests to EntDD14 by the well-diffusion method and also by growth kinetics in the presence of purified EntDD14 (10 μg/mL). Unexpectedly, the ΔddI mutant strain did not exhibit any sensitivity to EntDD14 even at highest concentration (60 μg/ml) nor to the cell-free supernatant (CFS) obtained from the WT strain (Figure 5). Notably, EntDD14's MIC values were similar to those obtained for the WT (resistant phenotype). Growth kinetics in the presence of EntDD14 at 10 μg/mL showed no significant differences between the WT strain, the ΔddI mutant strain and the complemented ddI-Comp strain (Figure 6A). These results refuted therefore the involvement of an ATP-binding protein/ATPase transporter in the self-immunity system of EntDD14. In order to unravel the role of the entire ABC transporter (ddHIJ) in the mechanism of immunity, we constructed a recombinant plasmid pAT18:ddAB-HIJ which we used to transform the sensitive Ent. faecalis JH2-2 strain. In this case, if the ABC transporter played a dual transport and immunity function, then this construction would be sufficient to produce EntDD14 and to immunize the host strain against the inhibitory action of EntDD14. Unexpectedly, the obtained clones of Ent. faecalis JH2-2-ddAB-HIJ were unable to produce EntDD14. In addition, growth kinetics in the presence of 10 μg/mL of EntDD14 showed the sensitive phenotype of Ent. faecalis JH2-2 compared to the WT strain. Overall, no significant difference was observed between Ent. faecalis JH2-2 and the transformed strain, JH2-2-ddAB-HIJ (Figure 6B). To verify the integrity of the pAT18:ddAB-HIJ construction, we used it in order to complement the Δbac double mutant strain, which is deleted in both ddA and ddB, and also to transform the ΔddIΔbac triple mutant strain, which is deleted in genes ddA, ddB and ddI of the ABC transporter. Remarkably, the WT phenotype was restored in both cases. Indeed, the pAT18:ddAB-HIJ construction transformed the Δbac mutant strain to a producer strain and the resulting transformed strain ΔddIΔbac produced as much EntDD14 as the WT (data not shown).
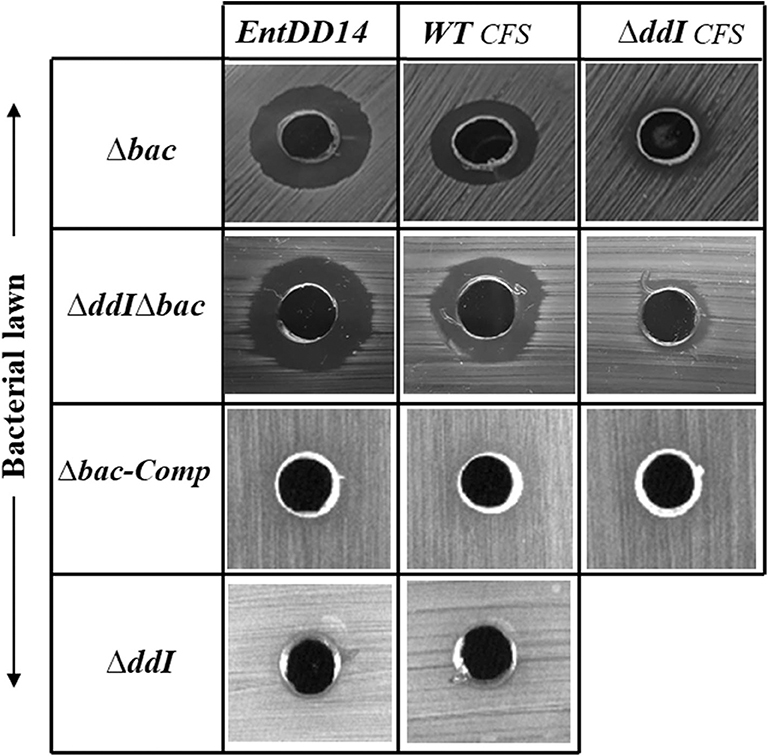
Figure 5. Sensitivity of the Ent. faecalis 14 mutant and complemented strains to pure EntDD14 at 60 μg/mL, cell free supernatant (CFS) of the wild-type strain and that of the ΔddI mutant. Δbac: Ent. faecalis 14 mutant deleted in ddA and ddB bacteriocin structural genes, ΔddIΔbac: Ent. faecalis 14 mutant deleted in ddI, ddA, and ddB genes, Δbac-Comp: the Δbac complemented strain and ΔddI: Ent. faecalis 14 mutant deleted in ddI gene. If present, the inhibition zone indicates the susceptibility of the bacterial lawn (target bacteria) to EntDD14. The data are representative of at least three independent experiments.
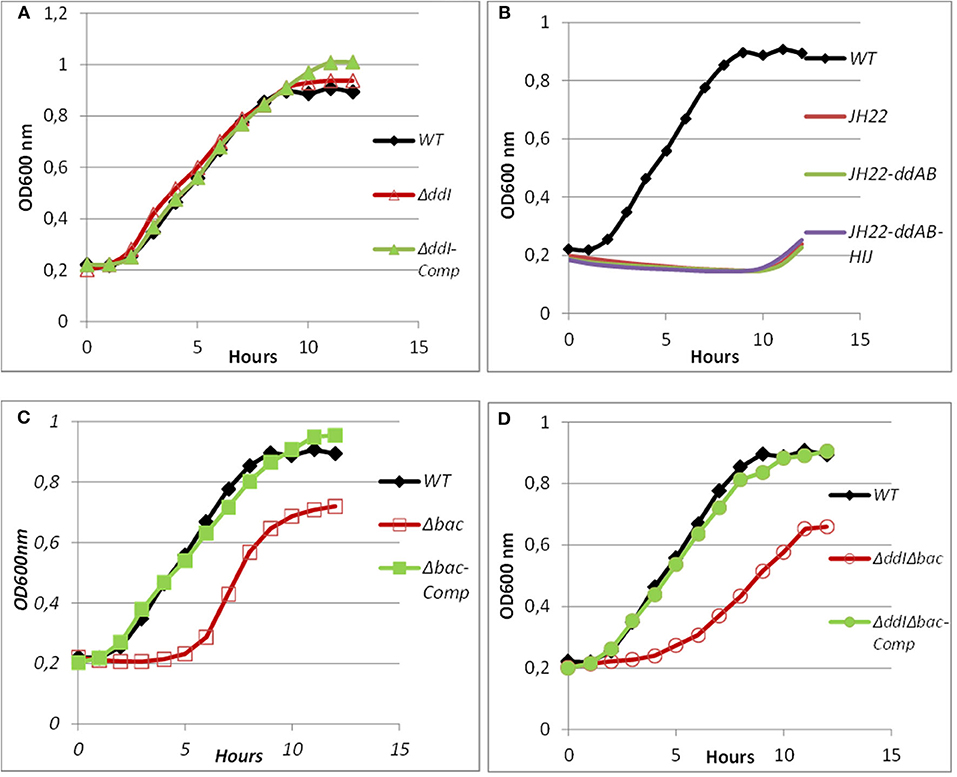
Figure 6. Growth curves of WT, mutated and complemented Ent. faecalis 14 strains in the presence of pure EntDD14 at 10 μg/mL. Ent. faecalis 14 WT (♦) (A): ΔddI mutant (Δ) and the ΔddI complemented strain (▴); (B): Ent. faecalis JH2-2 (red line) Ent. faecalis JH2-2-ddAB (green line) and Ent. faecalis JH2-2-ddAB-HIJ (purple line); (C): Δbac mutant (□) and the Δbac complemented strain (■); and (D): ΔddIΔbac mutant (°) and the ΔddIΔbac complemented strain (•).The data are the means of at least three independent experiments.
All these data ruled out the involvement of the ABC transporter in the primary self-immunity system of the two peptides leaderless bacteriocin EntDD14. Moreover, the precursor genes ddAB and ABC transporter genes ddHIJ in themselves seem to be insufficient to produce EntDD14.
Intracellular EntDD14 Is Involved in the Self-Immunity System
Mutants obtained during this study were screened for their susceptibility to bacteriocin EntDD14. Of importance, the Δbac mutant strain appeared to be sensitive to its own bacteriocin. This was observed not only when the mutant strain was treated with pure EntDD14 at 60 μg/mL but also when it was exposed to the supernatant of the WT strain, or to that of the less producing strain ΔddI (Figure 5). During growth in the presence of 10 μg/ml of EntDD14, the Δbac mutant strain extended its lag phase and entered the exponential growth phase only after 5 h of incubation (Figure 6C). The OD600 values obtained after 12 h of growth were significantly different between the WT strain and the Δbac mutant strain (P = 0.0001). The trans-complementation experiment using the pAT18-ddAB construction allowed the Δbac mutant strain to recover its resistance to EntDD14 (Figures 5, 6C). To confirm this result, we constructed a Δbac mutant in a host strain other than Ent. faecalis 14, and thus selected the Ent. faecalis 14 ΔddI mutant strain. This strain was resistant to EntDD14 and produced about 25% of this bacteriocin compared to the WT strain. As expected, ΔddIΔbac did not produce EntDD14 and was found to be sensitive to this bacteriocin, like the Δbac mutant strain (Figures 5, 6D). The complementation of this triple mutant with the pAT18:ddAB-HIJ construction fully restored the phenotype of the WT strain, which displayed almost identical kinetics in the presence of EntDD14 at 10 μg/mL (Figure 6C). Taken together, these results highlight the clear role of intracellular EntDD14 in the mechanism of its own immunity.
Gene Expression Analysis of EntDD14 Cluster
To explain the sensitive phenotype of the Δbac mutant strain to EntDD14, we checked whether the genes ddCDEFGHIJ were expressed in this strain. To this end, we used data generated from a comparative transcriptomic analysis of the Δbac mutant strain vs. the WT, after 6 and 24 h of growth in GM17 medium. The gene expression analysis did not reveal any significant differences between these strains after 6 h of growth except for the ddF gene which was slightly down-regulated, as the log2 ratio Δbac/WT ranged from −1 to 1 (Figure 7 and Table S1). Interestingly, all analyzed ddCDEFGHIJ genes were found to be significantly down-regulated in the Δbac mutant strain after 24 h of growth in GM17 (Figure 7 and Table S1). Moreover, mean gene expression showed that ddI, ddE and ddJ genes were the most deregulated with log2 Δbac/WT ratios of − 2.86, − 1.78, and − 1.70, respectively (Table S1). Finally, as expected, ddA and ddB encoding the structural genes were highly down-regulated in the Δbac mutant strain at both 6 and 24 h since they were deleted in this host (Figure 7 and Table S1).
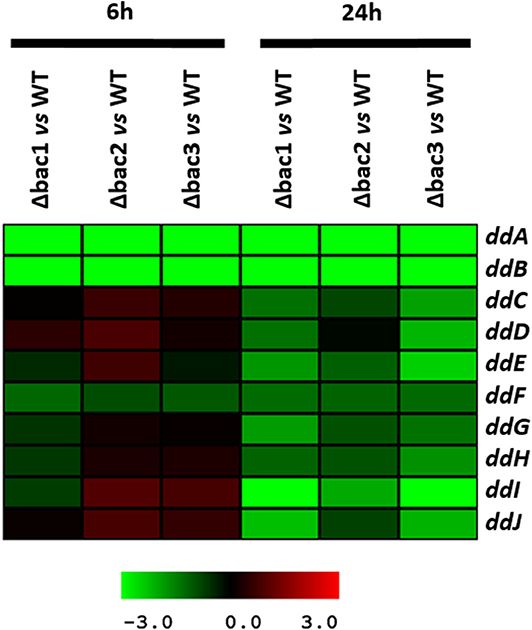
Figure 7. Profile of the EntDD14 predicted ORFs in Δbac vs. WT strain. Log2 ratios were registered for three biological repetitions after 6 and 24 h of growth in GM17 medium.
Discussion
Bacteriocins are known for their effectiveness in fighting and eradicating microbial pathogens (Quintana et al., 2014; Graham et al., 2017; Johnson et al., 2018). Interactions of these charged antimicrobial peptides with cell membrane leads to the formation of ion-permeable channels, resulting in perturbation of ion gradients by passive efflux of vital intracellular metabolites, such as ions, amino acids and ATP, which causes serious metabolic disruption and finally cell death (Karpinski et al., 2016). At the molecular level, bacteriocin production requires complex machinery consisting of the transcription-translation processes, transport out of the cell, and also immunity mechanisms to protect the strain from the toxicity of its own products (Bastos et al., 2015; Bountra et al., 2017).
Leaderless bacteriocins are active after their translation, and do not undergo significant post-translational modifications or processing. Therefore, bacteria producing such bacteriocins have certainly specific immunity systems that ensure their protection from the toxicity of their own peptides. This protection is expected to be ascertained from the translation step until their export outside of the cell. Of note, no-dedicated immunity proteins have been reported for leaderless two-peptide bacteriocins. Only the role of export and immunity of the ABC transporter has been described. Several mechanisms of self-immunity have been described for many bacteriocins. These involve for the well-known nisin, a lipoprotein, designed NisI, which is able to bind and thereby prevent nisin to access its predicted target lipid II through direct interactions (Hacker et al., 2015). In direct line, Abi proteins, which are putative membrane-bound metalloproteases, have been reported to play a role in the self-immunity of bacteriocins (Kjos et al., 2010). Using EntDD14 as a representative model of this category of bacteriocins, based on its high homology with other known leaderless two peptides bacteriocins (Figure 3), we have undertaken a genetic study to obtain relevant information on the immunity system of two-peptide leaderless bacteriocin-producers. First, an in silico genetic analysis of the EntDD14 bacteriocin region predicted a least 10 ORFs (ddABCDEFGHIJ), which could be organized into three closed operons. As shown in Figure 2, predictions of mRNA folding, using ARNold and RibEX algorithms permitted the location of a potential secondary structure between ddB and ddC, suggesting a possible post-transcriptional regulation, in which the synthesis of peptides A and B can be dissociated from the translation of other genes. To the best of our knowledge regulation and expression of bacteriocins at the post-transcriptional level has been reported only for the cyclic Enterocin AS-48 (Fernández et al., 2008).
Here, a BLAST analysis of the proteins encoded by the EntDD14 gene cluster did not reveal any significant homology with other known dedicated immunity proteins. However, sequence alignment established that the ddHIJ genes share significant similarities with several of the ABC transporters found in most leaderless, and cyclic bacteriocins, despite the absence of sequence similarities for the structural genes encoding these bacteriocins (Cintas et al., 2000; Diaz et al., 2003; Iwatani et al., 2012; Nascimento et al., 2012). These data support a potential relationship in the mediated mechanisms (production and/or immunity) and a close structural feature between leaderless and cyclic bacteriocins (Towle and Vederas, 2017; Perez et al., 2018). In the case of EntDD14, the ABC transporter was found to be involved in bacteriocin export, as a ΔddI mutant strain, affected in an ATP-binding protein of the ATPase family, could produce only 25% of bacteriocin compared to the wild-type strain. Similar results were reported for the leaderless single peptide aureocin 53. Indeed, the aureocin 53 bacteriocinogenic strain could produce only 24% of this bacteriocin when the ABC transporter aucEFG was affected in the first gene aucE (Nascimento et al., 2012). Here, we established that in the ΔddI mutant, production of EntDD14 was not completely abolished, indicating that another export pathway is also used by the bacteriocinogenic strain. Unexpectedly, the susceptible Ent. faecalis JH2-2 strain was unable to produce EntDD14 when transformed by the ddAB genes. In terms of the bacteriocin export pathways, it was established that enterocin L50 can be successfully expressed heterologously in two yeast expression systems through the general secretory pathways (Basanta et al., 2009, 2010), suggesting a possible use of another transport pathway. Here, the ddF gene, which encodes a putative pH domain protein, was found to be down-regulated after 6 h and 24 h of growth in the Δbac mutant but not in the wild-type strain (Figure 7, Table S1). The ddF gene displayed 23% identity (and 42% of positive residue) with orf8 of the pRJ9 plasmid of S. aureus (Netz et al., 2002). Remarkably, in S. aureus, the disruption of this gene abolished bacteriocin production (Nascimento et al., 2012). Therefore, we can hypothesize that ddF gene might play a role in the export of EntDD14.
The three-component ABC transporters, which are similar to those of ddHIJ, were previously shown to contribute to bacteriocin immunity by active extrusion of the peptide, most likely by reducing the concentration of the bacteriocin in direct contact with the cytoplasmic membrane (Klein and Entian, 1994; Siegers and Entian, 1995; Yarmus et al., 2000). Similarly, Rincé et al. (1997) reported that the self-immunity to the lantibiotic lacticin 481 was abolished when any lctE, lctF, or lctG gene was deleted (Rincé et al., 1997).
Taking into consideration all these published data, and in order to gain novel insights into the immunity systems of two-peptide leaderless bacteriocins, we looked at the role of the ABC transporter in the synthesis and immunity of EntDD14. To this end, we tested the sensitivity of the ΔddI mutant strain to EntDD14, and also by cloning ddAB-HIJ genes and studying their effects in the sensitive Ent. faecalis JH2-2 strain. Consequently our data were in discordance with those formerly reported. Indeed, the ΔddI mutant strain, as well as the WT strain were resistant to EntDD14 (up to 60 μg/mL) (Figures 5, 6A). Furthermore, cloning and expression of the full ABC transporter in Ent. faecalis JH2-2 strain did not grant any resistance to EntDD14 even at low concentrations (Figure 6B). Although cloning of the entire ABC transporter did not allow the Ent. faecalis JH2-2 strain to gain resistance to exogenous EntDD14, this does not necessarily rule out the role of this ABC transporter in the detoxification of intracellular EntDD14. This has already been attributed to the ABC transporter for aureocin 53, which contains two other immunity proteins named AucIA and AucIB (Nascimento et al., 2012). AucIA and AuIB exhibited, respectively, 28 and 29% identity with the proteins encoded by ddD and ddC. These homologies were even higher, 52 and 60%, when compared to the products of DdC and DdD, and their aureocin counterparts AucIA and AucIB, suggesting therefore that the DdC and DdD proteins could be good candidates for studying the immunity of bacteriocinogenic Ent. faecalis 14.
Considering the different genetic constructions, their expression and phenotypes, we assume that intracellular EntDD14 plays a role in its own immunity system. This original finding was strengthened by the phenotypes of the Δbac and ΔddIΔbac mutant strains, which exhibited sensitivity to EntDD14, as opposed to the WT and the complemented strains (Figure 5). At low EntDD14 concentration, the Δbac and ΔddIΔbac mutant strains showed an extended lag phase and a final OD600 significantly below that of the WT strain (Figures 6C,D), pointing again to the role of the intracellular EntDD14 in its own immunity system. The sensitivity of null-mutant strains was not ascribed to shut-down of expression of the ddCDEFGHIJ genes, as mostly these genes were expressed during the log phase of the Δbac mutant strain (Figure 7, Table S1).
The comparative study of the gene expression of the Δbac mutant strain vs. WT strain underpinned the role of EntDD14 in the positive regulation of the ddCDEFGHIJ genes since all of them were significantly down-regulated in the Δbac mutant strain, mainly after 24 h of growth (Figure 7, Table S1). Recent studies have reported the implication of transcriptional regulators in the synthesis of lacticin Q and aureocin A70 (Coelho et al., 2016; Iwatani et al., 2016). Environmental factors such as temperature or nutrition-adaptation were reported to control biosynthesis of enterocin L50 and enterocin Q in the multiple bacteriocin producing strains Ent. faecium L50 (Criado et al., 2006) and Weissella hellenica QU 13 (Masuda et al., 2016). In the case of non-leaderless bacteriocins, regulation by quorum sensing and leader peptide seemed to be involved (Kleerebezem, 2004; Kuipers et al., 2004; Rink et al., 2007; Abts et al., 2013). Related to that, Perez et al. (2017) reported that mutations in the leader peptide altered the overall conformation of the precursor peptide, which reduced, or enhanced its ability to fit and interact with the substrate-binding cleft of its processing enzyme(s).
As a conclusion, we report that to the best of our knowledge, an intracellular two-peptide leaderless bacteriocin plays a role in its own self-immunity system. The characterization of other genes of the EntDD14 operon (ddCDEFG) and their role in overall EntDD14 expression and immunity constitutes our next goal.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
DD, AB, and RL conceived the ideas, designed the experiments, discussed the data throughout the project, and wrote the article. RL performed the experiments and AL-D did the transcriptomic analysis. DD, RL, AB, and AL-D revised and approved the manuscript dissertation. All authors contributed to the article and approved the submitted version.
Funding
This work has been supported by the CPER/FEDER grant awarded by la Région des Hauts-de-France (2016-2021).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The MALDI-TOF/MS profiling experiments to verify the purity of EntDD14 were performed on the REALCAT platform issued from French governmental subvention administrated by the French National Research Agency (ANR) program [ANR-11-EQPX-0037]. The authors are grateful to Nacim Barache for his assistance in some experiments of this work. The authors would like to thank Prof. Leon Dicks and Dr. Steve W. Elson for their critical reading of the manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fbioe.2020.00644/full#supplementary-material
References
Abreu-Goodger, C., and Merino, E. (2005). RibEx: a web server for locating riboswitches and other conserved bacterial regulatory elements. Nucleic Acids Res 33, W690–W692. doi: 10.1093/nar/gki445
Abts, A., Montalban-Lopez, M., Kuipers, O. P., Smits, S. H., and Schmitt, L. (2013). NisC binds the FxLx motif of the nisin leader peptide. Biochemistry 52, 5387–5395. doi: 10.1021/bi4008116
Al Atya, A. K., Drider-Hadiouche, K., Ravallec, R., Silvain, A., Vachee, A., and Drider, D. (2015). Probiotic potential of Enterococcus faecalis strains isolated from meconium. Front. Microbiol. 6:227. doi: 10.3389/fmicb.2015.00227
Alvarez-Sieiro, P., Montalbán-López, M., Mu, D., and Kuipers, O. P. (2016). Bacteriocins of lactic acid bacteria: extending the family. Appl. Microbiol. Biotechnol. 100, 2939–2951. doi: 10.1007/s00253-016-7343-9
Arbulu, S., Jimenez, J. J., Borrero, J., Sánchez, J., Frantzen, C., Herranz, C., et al. (2016). Draft genome sequence of the bacteriocinogenic strain Enterococcus faecalis DBH18, isolated from mallard ducks (Anas platyrhynchos). Genome Announc. 4:e00663-16. doi: 10.1128/genomeA.00663-16
Basanta, A., Gómez-Sala, B., Sánchez, J., Diep, D. B., Herranz, C., Hernández, P. E., et al. (2010). Use of the yeast Pichia pastoris as an expression host for secretion of enterocin L50, a leaderless two-peptide (L50A and L50B) bacteriocin from Enterococcus faecium L50. Appl. Environ. Microbiol. 76, 3314–3324. doi: 10.1128/AEM.02206-09
Basanta, A., Herranz, C., Gutiérrez, J., Criado, R., Hernández, P. E., and Cintas, L. M. (2009). Development of bacteriocinogenic strains of Saccharomyces cerevisiae heterologously expressing and secreting the leaderless enterocin L50 peptides L50A and L50B from Enterococcus faecium L50. Appl. Environ. Microbiol. 75, 2382–2392. doi: 10.1128/AEM.01476-08
Bastos, M. do. C. de. F., Coelho, M. L. V., and Santos, O. C. da S. (2015). Resistance to bacteriocins produced by gram-positive bacteria. Microbiology 161, 683–700. doi: 10.1099/mic.0.082289-0
Batdorj, B., Dalgalarrondo, M., Choiset, Y., Pedroche, J., Métro, F., Prévost, H., et al. (2006). Purification and characterization of two bacteriocins produced by lactic acid bacteria isolated from Mongolian airag. J. Appl. Microbiol. 101, 837–848. doi: 10.1111/j.1365-2672.2006.02966.x
Beis, K., and Rebuffat, S. (2019). Multifaceted ABC transporters associated to microcin and bacteriocin export. Res. Microbiol. 170, 399–406. doi: 10.1016/j.resmic.2019.07.002
Belguesmia, Y., Choiset, Y., Rabesona, H., Baudy-Floc'h, M., Le Blay, G., Haertlé, T., et al. (2013). Antifungal properties of durancins isolated from Enterococcus durans A5-11 and of its synthetic fragments. Lett. Appl. Microbiol. 56, 237–244. doi: 10.1111/lam.12037
Belguesmia, Y., Leclère, V., Duban, M., Auclair, E., and Drider, D. (2017). Draft genome sequence of Enterococcus faecalis DD14, a bacteriocinogenic lactic acid bacterium with anti-clostridium activity. Genome Announc. 5, e00695-17. doi: 10.1128/genomeA.00695-17
Bierbaum, G., and Sahl, H.-G. (2009). Lantibiotics: mode of action, biosynthesis and bioengineering. Curr. Pharm. Biotechnol. 10, 2–18. doi: 10.2174/138920109787048616
Bougherra, F., Dilmi-Bouras, A., Balti, R., Przybylski, R., Adoui, F., Elhameur, H., et al. (2017). Antibacterial activity of new peptide from bovine casein hydrolyzed by a serine metalloprotease of Lactococcus lactis subsp lactis BR16. J. Funct. Foods 32, 112–122. doi: 10.1016/j.jff.2017.02.026
Bountra, K., Hagelueken, G., Choudhury, H. G., Corradi, V., El Omari, K., Wagner, A., et al. (2017). Structural basis for antibacterial peptide self-immunity by the bacterial ABC transporter McjD. EMBO J. 36, 3062–3079. doi: 10.15252/embj.201797278
Caly, D. L., Chevalier, M., Flahaut, C., Cudennec, B., Al Atya, A. K., Chataigné, G., et al. (2017). The safe enterocin DD14 is a leaderless two-peptide bacteriocin with anti-Clostridium perfringens activity. Int. J. Antimicrob. Agents 49, 282–289. doi: 10.1016/j.ijantimicag.2016.11.016
Cintas, L. M., Casaus, P., Herranz, C., Hâvarstein, L. S., Holo, H., Hernández, P. E., et al. (2000). Biochemical and genetic evidence that Enterococcus faecium L50 produces enterocins L50A and L50B, the sec-dependent enterocin P, and a novel bacteriocin secreted without an N-terminal extension termed enterocin Q. J. Bacteriol. 182, 6806–6814. doi: 10.1128/jb.182.23.6806-6814.2000
Cintas, L. M., Casaus, P., Holo, H., Hernandez, P. E., Nes, I. F., and Håvarstein, L. S. (1998). Enterocins L50A and L50B, Two novel bacteriocins from Enterococcus faecium L50, are related to staphylococcal hemolysins. J. Bacteriol. 180, 1988–1994.
Coelho, M. L. V., Coutinho, B. G., Cabral da Silva Santos, O., Nes, I. F., and Bastos, M. do. C. de. F. (2014). Immunity to the Staphylococcus aureus leaderless four-peptide bacteriocin aureocin A70 is conferred by AurI, an integral membrane protein. Res. Microbiol. 165, 50–59. doi: 10.1016/j.resmic.2013.11.001
Coelho, M. L. V., Fleming, L. R., and Bastos, M. do. C. de. F. (2016). Insights into aureocin A70 regulation: participation of regulator AurR, alternative transcription factor σ(B) and phage φ11 regulator cI. Res. Microbiol. 167, 90–102. doi: 10.1016/j.resmic.2015.10.004
Cotter, P. D., Ross, R. P., and Hill, C. (2013). Bacteriocins - a viable alternative to antibiotics? Nat. Rev. Microbiol. 11, 95–105. doi: 10.1038/nrmicro2937
Criado, R., Gutiérrez, J., Martín, M., Herranz, C., Hernández, P. E., and Cintas, L. M. (2006). Immunochemical characterization of temperature-regulated production of enterocin L50 (EntL50A and EntL50B), enterocin P, and enterocin Q by Enterococcus faecium L50. Appl. Environ. Microbiol. 72, 7634–7643. doi: 10.1128/AEM.00983-06
Diaz, M., Valdivia, E., Martínez-Bueno, M., Fernández, M., Soler-González, A. S., Ramírez-Rodrigo, H., et al. (2003). Characterization of a new operon, as-48EFGH, from the as-48 gene cluster involved in immunity to enterocin AS-48. Appl. Environ. Microbiol. 69, 1229–1236. doi: 10.1128/aem.69.2.1229-1236.2003
Diez-Gonzalez, F. (2007). Applications of bacteriocins in livestock. Curr. Issues Intest. Microbiol. 8, 15–23
Fernández, M., Sánchez-Hidalgo, M., García-Quintáns, N., Martínez-Bueno, M., Valdivia, E., López, P., et al. (2008). Processing of as-48ABC RNA in AS-48 enterocin production by Enterococcus faecalis. J. Bacteriol. 190, 240–250. doi: 10.1128/JB.01528-07
Gajic, O., Buist, G., Kojic, M., Topisirovic, L., Kuipers, O. P., and Kok, J. (2003). Novel mechanism of bacteriocin secretion and immunity carried out by lactococcal multidrug resistance proteins. J. Biol. Chem. 278, 34291–34298. doi: 10.1074/jbc.M211100200
Graham, C. E., Cruz, M. R., Garsin, D. A., and Lorenz, M. C. (2017). Enterococcus faecalis bacteriocin EntV inhibits hyphal morphogenesis, biofilm formation, and virulence of Candida albicans. Proc. Natl. Acad. Sci. U.S.A. 114, 4507–4512. doi: 10.1073/pnas.1620432114
Hacker, C., Christ, N. A., Duchardt-Ferner, E., Korn, S., Göbl, C., Berninger, L., et al. (2015). The solution structure of the lantibiotic immunity protein NisI and its interactions with nisin. J. Biol. Chem. 290, 28869–28886. doi: 10.1074/jbc.M115.679969
Hu, J., Ma, L., Nie, Y., Chen, J., Zheng, W., Wang, X., et al. (2018). A Microbiota-derived bacteriocin targets the host to confer diarrhea resistance in early-weaned piglets. Cell Host Microbe. 24, 817–832.e8. doi: 10.1016/j.chom.2018.11.006
Iwatani, S., Horikiri, Y., Zendo, T., Nakayama, J., and Sonomoto, K. (2013). Bifunctional gene cluster lnqBCDEF mediates bacteriocin production and immunity with differential genetic requirements. Appl. Environ. Microbiol. 79, 2446–2449. doi: 10.1128/AEM.03783-12
Iwatani, S., Ishibashi, N., Flores, F. P., Zendo, T., Nakayama, J., and Sonomoto, K. (2016). LnqR, a TetR-family transcriptional regulator, positively regulates lacticin Q production in Lactococcus lactis QU 5. FEMS Microbiol. Lett. 363:fnw200. doi: 10.1093/femsle/fnw200
Iwatani, S., Yoneyama, F., Miyashita, S., Zendo, T., Nakayama, J., and Sonomoto, K. (2012). Identification of the genes involved in the secretion and self-immunity of lacticin Q, an unmodified leaderless bacteriocin from Lactococcus lactis QU 5. Microbiology 158, 2927–2935. doi: 10.1099/mic.0.062943-0
Johnson, M. E. M., Jung, D. Y.-G., Jin, D. Y.-Y., Jayabalan, D. R., Yang, D. S. H., and Suh, P. J. W. (2018). Bacteriocins as food preservatives: challenges and emerging horizons. Crit. Rev. Food Sci. Nutr. 58, 2743–2767. doi: 10.1080/10408398.2017.1340870
Juturu, V., and Wu, J. C. (2018). Microbial production of bacteriocins: latest research development and applications. Biotechnol. Adv. 36, 2187–2200. doi: 10.1016/j.biotechadv.2018.10.007
Karpinski, T., Szkaradkiewicz, A., Caballero, B., Finglas, P. M., and Toldrá, F. (2016). “Bacteriocins,” in Encyclopedia of Food and Health, eds B. Caballero, P. M. Finglas, and F. Toldrá (Amsterdam: Academic Press), 312–419.
Kjos, M., Snipen, L., Salehian, Z., Nes, I. F., and Diep, D. B. (2010). The abi proteins and their involvement in bacteriocin self-immunity. J. Bacteriol. 192, 2068–2076. doi: 10.1128/JB.01553-09
Klaenhammer, T. R. (1993). Genetics of bacteriocins produced by lactic acid bacteria. FEMS Microbiol. Rev. 12, 39–85. doi: 10.1111/j.1574-6976.1993.tb00012.x
Kleerebezem, M. (2004). Quorum sensing control of lantibiotic production; nisin and subtilin autoregulate their own biosynthesis. Peptides 25, 1405–1414. doi: 10.1016/j.peptides.2003.10.021
Klein, C., and Entian, K. D. (1994). Genes involved in self-protection against the lantibiotic subtilin produced by Bacillus subtilis ATCC 6633. Appl. Environ. Microbiol. 60, 2793–2801.
Kuipers, A., Boef, E., de Rink, R., Fekken, S., Kluskens, L. D., Driessen, A. J. M., et al. (2004). NisT, the transporter of the lantibiotic nisin, can transport fully modified, dehydrated, and unmodified prenisin and fusions of the leader peptide with non-lantibiotic peptides. J. Biol. Chem. 279, 22176–22182. doi: 10.1074/jbc.M312789200
Liu, X., Vederas, J. C., Whittal, R. M., Zheng, J., Stiles, M. E., Carlson, D., et al. (2011). Identification of an N-terminal formylated, two-peptide bacteriocin from Enterococcus faecalis 710C. J. Agric. Food Chem. 59, 5602–5608. doi: 10.1021/jf104751v
Martín-Platero, A. M., Valdivia, E., Ruíz-Rodríguez, M., Soler, J. J., Martín-Vivaldi, M., Maqueda, M., et al. (2006). Characterization of antimicrobial substances produced by Enterococcus faecalis MRR 10-3, isolated from the uropygial gland of the hoopoe (upupa epops). Appl. Environ. Microbiol. 72, 4245–4249. doi: 10.1128/AEM.02940-05
Masuda, Y., Perez, R. H., Zendo, T., and Sonomoto, K. (2016). Nutrition-adaptive control of multiple-bacteriocin production by Weissella hellenica QU 13. J. Appl. Microbiol. 120, 70–79. doi: 10.1111/jam.12997
Nascimento, J., dos, S., Coelho, M. L. V., Ceotto, H., Potter, A., Fleming, L. R., et al. (2012). Genes involved in immunity to and secretion of aureocin A53, an atypical class II bacteriocin produced by Staphylococcus aureus A53. J. Bacteriol. 194, 875–883. doi: 10.1128/JB.06203-11
Nes, I. F., Diep, D. B., Håvarstein, L. S., Brurberg, M. B., Eijsink, V., and Holo, H. (1996). Biosynthesis of bacteriocins in lactic acid bacteria. Antonie Van Leeuwenhoek 70, 113–128. doi: 10.1007/BF00395929
Netz, D. J., Sahl, H. G., Marcelino, R., dos Santos Nascimento, J., de Oliveira, S. S., Soares, M. B., et al. (2001). Molecular characterisation of aureocin A70, a multi-peptide bacteriocin isolated from Staphylococcus aureus. J. Mol. Biol. 311, 939–949. doi: 10.1006/jmbi.2001.4885
Netz, D. J. A., Pohl, R., Beck-Sickinger, A. G., Selmer, T., Pierik, A. J., Bastos, M., et al. (2002). Biochemical characterisation and genetic analysis of aureocin A53, a new, atypical bacteriocin from Staphylococcus aureus. J. Mol. Biol. 319, 745–756. doi: 10.1016/S0022-2836(02)00368-6
Nissen-Meyer, J., Rogne, P., Oppegard, C., Haugen, H., and Kristiansen, P. (2009). Structure-function relationships of the non-lanthionine-containing peptide (class II) bacteriocins produced by gram-positive bacteria. CPB 10, 19–37. doi: 10.2174/138920109787048661
Ovchinnikov, K. V., Chi, H., Mehmeti, I., Holo, H., Nes, I. F., and Diep, D. B. (2016). Novel group of leaderless multipeptide bacteriocins from gram-positive bacteria. Appl. Environ. Microbiol. 82, 5216–5224. doi: 10.1128/AEM.01094-16
Papo, N., and Shai, Y. (2005). Host defense peptides as new weapons in cancer treatment. Cell. Mol. Life Sci. 62, 784–790. doi: 10.1007/s00018-005-4560-2
Perez, R. H., Sugino, H., Ishibashi, N., Zendo, T., Wilaipun, P., Leelawatcharamas, V., et al. (2017). Mutations near the cleavage site of enterocin NKR-5-3B prepeptide reveal new insights into its biosynthesis. Microbiology 163, 431–441. doi: 10.1099/mic.0.000435
Perez, R. H., Zendo, T., and Sonomoto, K. (2014). Novel bacteriocins from lactic acid bacteria (LAB): various structures and applications. Microb. Cell Fact. 13:S3. doi: 10.1186/1475-2859-13-S1-S3
Perez, R. H., Zendo, T., and Sonomoto, K. (2018). Circular and leaderless bacteriocins: biosynthesis, mode of action, applications, and prospects. Front. Microbiol. 9:2085. doi: 10.3389/fmicb.2018.02085
Quintana, V. M., Torres, N. I., Wachsman, M. B., Sinko, P. J., Castilla, V., and Chikindas, M. (2014). Antiherpes simplex virus type 2 activity of the antimicrobial peptide subtilosin. J. Appl. Microbiol. 117, 1253–1259. doi: 10.1111/jam.12618
Rincé, A., Dufour, A., Uguen, P., Le Pennec, J. P., and Haras, D. (1997). Characterization of the lacticin 481 operon: the Lactococcus lactis genes lctF, lctE, and lctG encode a putative ABC transporter involved in bacteriocin immunity. Appl. Environ. Microbiol. 63, 4252–4260. doi: 10.1128/AEM.63.11.4252-4260.1997
Rink, R., Kluskens, L. D., Kuipers, A., Driessen, A. J. M., Kuipers, O. P., and Moll, G. N. (2007). NisC, the cyclase of the lantibiotic nisin, can catalyze cyclization of designed nonlantibiotic peptides. Biochemistry 46, 13179–13189. doi: 10.1021/bi700106z
Sahm, D. F., Kissinger, J., Gilmore, M. S., Murray, P. R., Mulder, R., Solliday, J., et al. (1989). In vitro susceptibility studies of vancomycin-resistant Enterococcus faecalis. Antimicrob. Agents Chemother. 33, 1588–1591. doi: 10.1128/AAC.33.9.1588
Sambrook, J., and Russell, D. W. (2001). Molecular Cloning: A Laboratory Manual. New York, NY: Cold Spring Harbor Laboratory Press.
Sánchez-Hidalgo, M., Maqueda, M., Gálvez, A., Abriouel, H., Valdivia, E., and Martínez-Bueno, M. (2003). The genes coding for enterocin EJ97 production by enterococcus faecalis EJ97 are located on a conjugative plasmid. Appl. Environ. Microbiol. 69, 1633–1641. doi: 10.1128/AEM.69.3.1633-1641.2003
Siegers, K., and Entian, K. D. (1995). Genes involved in immunity to the lantibiotic nisin produced by Lactococcus lactis 6F3. Appl. Environ. Microbiol. 61, 1082–1089. doi: 10.1128/AEM.61.3.1082-1089.1995
Thurlow, L. R., Thomas, V. C., and Hancock, L. E. (2009). Capsular polysaccharide production in Enterococcus faecalis and contribution of CpsF to capsule serospecificity. J. Bacteriol. 191, 6203–6210. doi: 10.1128/JB.00592-09
Towle, K. M., and Vederas, J. C. (2017). Structural features of many circular and leaderless bacteriocins are similar to those in saposins and saposin-like peptides. Medchemcomm 8, 276–285. doi: 10.1039/C6MD00607H
Trieu-Cuot, P., Carlier, C., Poyart-Salmeron, C., and Courvalin, P. (1991). Shuttle vectors containing a multiple cloning site and a lacZ alpha gene for conjugal transfer of DNA from Escherichia coli to gram-positive bacteria. Gene 102, 99–104. doi: 10.1016/0378-1119(91)90546-N
van Heel, A. J., Montalban-Lopez, M., and Kuipers, O. P. (2011). Evaluating the feasibility of lantibiotics as an alternative therapy against bacterial infections in humans. Expert. Opin. Drug Metab. Toxicol. 7, 675–680. doi: 10.1517/17425255.2011.573478
Verma, A. K., Banerjee, R., Dwivedi, H. P., Juneja, V. K., and Carl, A. B. (2014). “BACTERIOCINS | potential in food preservation,” in Encyclopedia of Food Microbiology, eds C. A. Batt and M. L. Tortorello (Cambridge, MA: Elsivier Ltd, Academic Press), 180–206. doi: 10.1016/B978-0-12-384730-0.00029-X
Yagi, Y., and Clewell, D. B. (1980). Recombination-deficient mutant of Streptococcus faecalis. J. Bacteriol. 143, 966–970. doi: 10.1128/JB.143.2.966-970.1980
Yanisch-Perron, C., Vieira, J., and Messing, J. (1985). Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mpl8 and pUC19 vectors. Gene 33, 103–119. doi: 10.1016/0378-1119(85)90120-9
Keywords: leaderless peptides, two-peptide enterocin DD14, bacterial self-immunity, regulation, antimicrobial peptide, ABC transporter
Citation: Ladjouzi R, Lucau-Danila A, Benachour A and Drider D (2020) A Leaderless Two-Peptide Bacteriocin, Enterocin DD14, Is Involved in Its Own Self-Immunity: Evidence and Insights. Front. Bioeng. Biotechnol. 8:644. doi: 10.3389/fbioe.2020.00644
Received: 01 April 2020; Accepted: 26 May 2020;
Published: 26 June 2020.
Edited by:
Csaba Fehér, Budapest University of Technology and Economics, HungaryReviewed by:
Sylvie Françoise Rebuffat, Muséum National d'Histoire Naturelle, FranceThomas Dandekar, Julius Maximilian University of Würzburg, Germany
Copyright © 2020 Ladjouzi, Lucau-Danila, Benachour and Drider. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Djamel Drider, ZGphbWVsLmRyaWRlciYjeDAwMDQwO3VuaXYtbGlsbGUuZnI=
 Rabia Ladjouzi
Rabia Ladjouzi Anca Lucau-Danila
Anca Lucau-Danila Abdellah Benachour
Abdellah Benachour Djamel Drider1*
Djamel Drider1*