
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Bioeng. Biotechnol., 25 May 2020
Sec. Nanobiotechnology
Volume 8 - 2020 | https://doi.org/10.3389/fbioe.2020.00433
 Reham Samir Hamida1*
Reham Samir Hamida1* Mohamed Abdelaal Ali2
Mohamed Abdelaal Ali2 Doaa A. Goda3
Doaa A. Goda3 Mahmoud Ibrahim Khalil1,4*
Mahmoud Ibrahim Khalil1,4* Mayasar Ibrahim Al-Zaban5*
Mayasar Ibrahim Al-Zaban5*Emerging antibiotic-resistant bacteria result in increased mortality and have negative economic impacts. It is necessary to discover new strategies to create alternative antibacterial agents that suppress the bacterial resistance mechanism and limit the spread of serious infectious bacterial diseases. Silver nanoparticles may represent a new medicinal agents as alternative antibiotics affect different bacterial mechanisms such as virulence and resistance. In addition to that of silver nitrate (AgNO3) and ampicillin, for the first time, the inhibitory effect of silver nanoparticles synthesized using Desertifilum sp. (D-SNPs) was evaluated against five pathogenic bacteria using the agar well diffusion method. Also, the influence of D-SNPs and AgNO3 on bacterial antioxidant and metabolic activities was studied. The antibacterial activity of D-SNPs and AgNO3 against methicillin-resistant Staphylococcus aureus (MRSA) strains was studied at the morphological and molecular level. D-SNPs and AgNO3 have the ability to inhibit the growth of the five bacterial strains and resulted in an imbalance in the CAT, GSH, GPx and ATPase levels. MRSA treated with D-SNPs and AgNO3 showed different morphological changes such as apoptotic bodies formation and cell wall damage. Moreover, both caused genotoxicity and denaturation of MRSA cellular proteins. Additionally, TEM micrographs showed the distribution of SNPs synthesized by MRSA. This result shows the ability of MRSA to reduce silver nitrate into silver nanoparticles. These data indicate that D-SNPs may be a significant alternative antibacterial agent against different bacteria, especially MDR bacteria, by targeting the virulence mechanism and biofilm formation, leading to bacterial death.
Many pathogenic bacteria have become a principle cause of serious infectious diseases, and with the evolution of multi-drug-resistant (MDR) bacteria, the problem has become more complicated (Brown and Wright, 2016). For example, gram-positive Streptococcus mutans is the most well-known bacterium causing dental caries (Metwalli et al., 2013). Among gram-negative bacteria, Salmonella typhimurium is one of the most common bacterial strains causing NTS infections related to human illnesses such as acute gastroenteritis (Scallan et al., 2011), while Escherichia coli is one of the most common bacteria responsible for a wide range of infections such as cholecystitis and urinary tract infection (Lim et al., 2010). After E. coli, Klebsiella pneumonia is the second most common disease-causing gram-negative bacterium, causing invasive infections such as pneumonia and meningitis (Podschun and Ullmann, 1998; Vading et al., 2018).
The multi-drug-resistant bacteria problem is growing rapidly due to the uncontrolled and indiscriminate usage of antibiotics for the treatment of bacterial infections (Yah and Simate, 2015). The overloading usage of antibiotics can result in issues such as MDR bacterial spread, prolonged infection treatment and high mortality risk (Yah and Simate, 2015). Both gram-negative and gram-positive bacteria include members that are distinguished as MDR bacteria or those sensitive to antibiotics (Cillóniz et al., 2019; Koulenti et al., 2019). S. aureus is considered a leading cause of morbidity and mortality (World Health Organization, 2017), especially with the rapid emergence of methicillin-resistant Staphylococcus aureus (MRSA) strains, which are resistant to all known β-lactam antibiotics (Boucher et al., 2009), as well as reports that clinical MRSA strains are also resistant to vancomycin, linezolid, and daptomycin (Nannini et al., 2010). Endocarditis, osteomyelitis, necrotizing pneumonia and sepsis infections occur when these bacteria disseminate into the blood stream (Lowy, 1998).
The appearance of the phenomenon of MDR bacteria has motivated researchers to search for solutions to explain and solve this problem as well as produce efficient alternatives to antibiotics (Gurunathan et al., 2018). These alternatives are distinguished by their potential to decrease bacterial resistance mechanisms and/or to serve as anti-virulence agents to act on pathogen virulence (Wright and Sutherland, 2007; Maura et al., 2016). One of these strategies to overcome the spread of MDR bacteria is nanotechnology, which aims to synthesize nanoparticles (NPs) featuring unique physicochemical and biological properties including anticancer (Bin-Meferij and Hamida, 2019), anti-inflammatory, and antimicrobial activity (Yallappa et al., 2016; Jagielski et al., 2018; Jalal et al., 2019). Green nanotechnology is modern science that depends on natural sources such as plants (Raut et al., 2010), bacteria (Saravanan et al., 2017), mushrooms (Debnath et al., 2019), and cyanobacteria to fabricate NPs from metals (Bin-Meferij and Hamida, 2019). The green synthesis of nanoparticles is an environmentally friendly process with low cost and energy savings (Cicci et al., 2017). Biogenic NPs are applied in different fields such as medicine, biotechnology and industry (Kubik et al., 2005). For example, gold NPs are used in photothermal therapy for cancer diseases (Ali et al., 2019); silicon nanoparticles are used in cell imaging (Guo et al., 2019); and titanium dioxide NPs are applied in photodynamic and sonodynamic therapy (Bogdan et al., 2017).
Silver nanoparticles (SNPs) are one of the most important NPs that play significant roles in biomedical applications such as wound healing, cell imaging, diagnosis, disease treatment, and contraceptive devices (Naidu Krishna et al., 2015). Furthermore, SNPs have been used in sensing, drug discovery and tissue engineering (Narducci, 2007). In addition, as SNPs are potent anticancer agents (Murugesan et al., 2019), they are considered significant antimicrobial agents with particularly high potential to effectively treat antibiotic-resistant bacteria (Leid et al., 2011; Yah and Simate, 2015). Many publications have reported that SNPs are unique antimicrobial agents against bacteria, fungi and viruses (Lu et al., 2013; Mori et al., 2013; Ouda, 2014). SNPs significantly suppress the growth of various antibiotic-resistant bacteria such as Staphylococcus aureus and Pseudomonas aeruginosa that cause mastitis infection (Leid et al., 2011).
The mechanism of action of SNPs against bacteria is still poorly understood. Yuan et al. (2017) showed that the lethal activity of SNPs against bacteria is due to the reactivity of SNPs with bacterial biocompounds such as proteins, sugars, DNA and enzymes, leading to genetic mutation and structural alterations to the bacterial cell wall and membrane as well as proteins, ultimately causing bacterial inhibition of growth and cell death.
Thus, developing and synthesizing novel biogenic SNPs as alternative antibacterial materials that enhance bacterial death is a promising strategy. In the current investigation, for the first time, we aimed to screen the inhibitory activity of biogenic SNPs, which were synthesized previously by our team by using a novel cyanobacteria strain Desertifilum IPPAS B-1220 (Hamida et al., 2020), against five pathogenic gram-negative and gram-positive bacteria, namely, Escherichia coli ATCC 25922, Salmonella typhimurium ATCC 14028, Klebsiella pneumoniae (clinical isolate), Streptococcus mutans and methicillin-resistant Staphylococcus aureus (MRSA) clinical isolates. Moreover, for the first time, we demonstrated the ability of MRSA bacteria to fabricate SNPs from silver nitrate (AgNO3). Finally, we added a new mechanism to explain the molecular basis of action of D-SNPs and silver ions against MRSA.
Silver nitrate (AgNO3), bacterial culture materials, and chemicals related to evaluating enzyme activities were purchased from Sigma-Aldrich (St. Louis, MO, United States), and PiBind resin was purchased from Expedeon, San Diego, United States.
To prepare stock solutions of D-SNPs and silver nitrate powders, 1.54 mg of the powder was dissolved in 1 ml of distilled water.
Five pathogenic bacterial strains included gram-negative and gram-positive bacteria were obtained from City of Scientific Research and Technological Applications and Almery University Hospital (Alexandria, Egypt). These groups of bacteria included Escherichia coli ATCC 25922, Salmonella typhimurium ATCC 14028, drug-resistance Klebsiella pneumoniae (clinical isolate), Streptococcus mutans and methicillin-resistant Staphylococcus aureus (MRSA) (clinical isolates). Ten microliters were taken from glyceride bacteria stocks and seeded into Luria-Bertani (LB) broth for 24 h at 37°C. Fresh bacterial cultures were obtained at a concentration of 0.5 McFarland scale (104 CFU/mL). Then, fresh LB agar plates were prepared, and 50 μL of each bacterial strain was poured and gently spread on the agar plates. After spreading the bacteria on the plates, four 8 mm wells were made in all plates for further experiments with the treatments.
To assess the inhibitory activity of D-SNPs and AgNO3 against five pathogenic bacteria, 100 μL of D-SNPs, AgNO3 (1.54 mg/mL), distilled water as a negative control and ampicillin antibiotic as a positive control were added to each agar plate well. The treated plates were incubated at 37°C for 24 h. At the end of the incubation time, the diameter of the inhibition zone (IZ) in mm was evaluated.
The minimum inhibition concentration (MIC) refers to the lowest concentration of antimicrobial agent at which bacterial growth is inhibited. To determine the MIC, the serial dilution method was used. In brief, different concentrations of D-SNPs and AgNO3 were prepared (1.8, 1.5, 1.2, 0.9, 0.6, and 0.3 mg/mL) and (2.4, 2.1, 1.8, 1.5, 1.2, 0.9, 0.6, and 0.3 mg/mL) respectively. In 96-well plates, 100 μL/well of bacterial suspension (104 CFU/mL) was subjected to 100 μL of different concentrations of both D-SNPs and AgNO3 and then incubated for 24 h at 37°C. After 24 h, the bacterial turbidity in comparison with the 0.5 McFarland media turbidity was checked with the naked eye. To ensure the previous step, 10 μL of the resultant MIC values determined was tested against each bacterial strain using the agar well diffusion method.
The minimum bactericidal concentration (MBC) is termed the lowest concentration of antimicrobial drugs that kill bacteria. To assess the MBC values, different concentrations of D-SNPs and AgNO3 (1.8, 1.5, 1.2, 0.9, 0.6, and 0.3 mg/mL) and (2.4, 2.1, 1.8, 1.5, 1.2, 0.9, 0.6, and 0.3 mg/mL) respectively were prepared, and 100 μL of each treatment concentration was added to 100 μL of each bacterial strain (104 CFU/mL) and incubated for 24 h at 37°C. After 24 h, the bacterial turbidity in comparison with the 0.5 McFarland media turbidity was checked with the naked eye. To ensure the previous step, the resultant MBC values determined were tested against each bacterial strain using the agar well diffusion method.
To evaluate the effect of D-SNPs and AgNO3 on the antioxidant activity, the bacterial cells before and after treatment with D-SNPs and silver nitrate were incubated at ambient temperature for 24 h. Then, the cells were centrifuged at 10,000 rpm for 5 min, and the pellets were washed with phosphate-buffered saline (PBS) and lysed using a sonicator (IKAT 10 basic sonicator, United States). Oxidative stress markers such as GPx, GSH, and CAT were assayed as follows:
Catalase enzyme activity in the supernatants was evaluated spectrophotometrically (UV 2505 spectrophotometer, Los Angeles, United States) by measuring the drop in hydrogen peroxide (H2O2) concentration at 240 nm. In a quartz cuvette, 10 μL of supernatant was added to 2.89 mL of PBS (pH 7.4) at time 0. After adding 0.1 mL of 300 mM hydrogen peroxide to the previous mixture, the absorbance was measured at 240 nm for 5 min at 1-min intervals. The catalase activity was expressed in terms of mmol/min mg protein (Mal et al., 2013).
The method mentioned by Paglia and Valentine (1967) was applied to measure the activity of GPx antioxidant. In brief, the samples were added to the reaction mixture (50 mM potassium phosphate buffer (pH 7.0), 1 mM EDTA, 1 mM sodium azide, 0.2 mM NADPH, 1 U glutathione reductase, and 1 mM reduced glutathione) and allowed to equilibrate for 5 min at 25°C. The reaction was started by adding 0.1° mL of 2.5 mM H2O2. Absorbance was measured at 340 nm for 5 min. Values were expressed as n mol of NADPH oxidized to NADP by using the extinction coefficient of 6.2 × 103 M–1 cm–1 at 340 nm. The activity of GPx was expressed in terms of n mol NADPH consumed/min/mg protein (Chakraborty et al., 2012).
To evaluate the GSH level in samples with or without D-SNP and AgNO3 treatments, the supernatant was mixed with a solution of cold (4°C) 320 mM sulfosalicylic acid, 28 mM L-ascorbic acid, and 4 mM EDTA for protein precipitation. Then, the mixture was centrifuged at 27,000 × g (Hettich Zentrifugen, mikro 200, Germany) for 15 min at 4°C to remove the precipitated proteins, and the clear supernatants were used to evaluate the GSH concentration through a GSH assay kit based on the colorimetric method described by Beutler (1963). This reaction depends on the reduction of 5,5′-dithiobis (2-nitrobenzoic acid) (DTNB, dissolved in 25 mM PBS, pH 7.0) by the reduced glutathione to give a yellow product measured at 405 nm. The GSH content was expressed as μM GSH per milligram.
The influence of D-SNPs and silver nitrate on the ATPase activity of five selected bacteria was determined using a colorimetric ATPase assay according to the method described by Andrés and Fierro (2010). Ten microliters of supernatant was incubated with 10 μL of PiBind resin to remove the free inorganic phosphate (Pi). The amount of Pi released was calculated by spectrophotometry (UV 2505 spectrophotometer, Los Angeles, United States) at A650. For all experiments, calibration was performed using a standard range of Pi concentrations, and data were determined for a minimum of three independent assays.
Bacterial specimens were fixed by immersing the cells immediately in ice-cold 4F1G [formalin-glutaraldehyde mixture (pH 7.2)] (10 mL formaldehyde 40% + 2 mL glutaraldehyde 50% + 1.16 gm sodium phosphate monobasic + 0.27 gm NaOH + distilled water up to 100 mL) and were rinsed in 0.1 M phosphate buffer for 2 h at 4°C. This step was followed by post-fixation using 1% osmium tetroxide (OsO4) for 2 h at 4°C. Then, the specimens were washed with 0.1 M phosphate buffer several times for 10 min. After fixation, the specimens were serially dehydrated with graded ethanol, infiltrated in propylene oxide and embedded in an Araldite Epon mixture. Ultrathin (70 nm) sections from the selected area were cut with a glass knife on an LKB Ultramicrotome and placed on 200-mesh copper grids. The ultrathin sections after double staining with 2% uranyl acetate and lead citrate were examined using a JEOL 100 CX Electron Microscope (JEOL, Tokyo, Japan) operating at 80 kV.
In this investigation, the expression levels of transcription-repair-coupling factor (mfD), α-Hemolysin (hly) and Ag43 phase-variable biofilm formation autotransporter CP4-44 prophage (flu) genes were used as molecular markers to evaluate the influence of D-SNPs and silver nitrate via quantitative RT-PCR. Total RNA was purified from samples using TRIzol Reagent (15596026, Life Technologies, United States). The yield and quality of total RNA were determined spectrophotometrically (UV 2505 spectrophotometer, Los Angeles, United States) based on the absorbance at 260 nm and the 260/280 nm ratio, respectively. Mfd, hly, and flu mRNA were evaluated using Maximas SYBR Green/Fluorescein qPCR Master Mix by Rotor-Gene Q (Qiagen, United States). One microgram of total RNA was reverse-transcribed into single-stranded complementary DNA by using a QuantiTects Reverse Transcription Kit (Qiagen, United States) with a random primer hexamer in a two-step RT-PCR reaction in which any genomic DNA (gDNA) contamination was eliminated using gDNA Wipeout buffer. Total cDNA (30 ng) was used as a template for amplification with the specific primer pair used at a final concentration of 300 nM (Table 1). Each sample was subjected to real-time PCR in duplicate, and the mean values of the duplicates were used for subsequent analysis. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was utilized as a housekeeping gene. Rotor-Gene Q automatically collected data and analyzed the value of the threshold cycle (Ct), which was normalized to an average Ct value of the housekeeping genes (ΔCt), and the relative expression of each representative was calculated as 2–ΔCt.
To estimate the influence of D-SNPs and silver nitrate on the structure of cellular proteins of MRSA, protein electrophoresis was performed and monitored via SDS-PAGE. Total soluble proteins of MRSA were extracted and purified using TriFast (Peqlab, VWR company) (isolation of RNA, DNA and protein simultaneously). The OmniPAGE Mini vertical electrophoresis unit included Power Pro 5 as a power supply (Cleaver Scientific, United Kingdom) and was used to fractionate the bacterial protein content via SERVAGelTM TG PRiMETM 10% (SERVA, Germany). This step was followed by staining the gel with 0.1% Coomassie blue R-250 for 2 h. Then, the gel was distained with a solution (1:3:6) of glacial acetic acid: methanol: water. Finally, a gel documentation system (Geldoc-it, UVP, United Kingdom) was applied for data analysis using Totallab analysis software version 1.0.1 (Newcastle-Upon-Tyne, United Kingdom).
All experiments were repeated at least three times, and the data are presented as the mean ± SEM. Prism 8.3 software (GraphPad Software Inc., La Jolla, CA, United States) was used to determine the statistically significant difference between treatments and the control through one-way analysis of variance (ANOVA). The significance of the data is represented at P < 0.00001, P < 0.0001, P < 0.001, and P < 0.01. For SNP measurement, ImageJ (National Institutes of Health, Bethesda, MD, United States) was performed.
Biogenic SNPs and silver ions are well known as potent antimicrobial agents against different microbes and do not exhibit any cross-resistance with antibiotics (Guggenbichler et al., 1999; Durán et al., 2016). In our recently published article (Hamida et al., 2020), we synthesized and characterized silver nanoparticles (D-SNPs) using the novel cyanobacteria species Desertifilum IPPAS B-1220. These nanoparticles were spherical in shape with a nano-size range from 4.5 to 26 nm (Figure 1). In the current study, in addition to AgNO3, D-SNPs were screened for the first time against five different gram-positive and gram-negative bacteria.

Figure 1. TEM micrograph of SNPs synthesized by Desertifilum IPPAS B-1220 demonstrating the spherical shape and distribution of D-SNPs with a size range from 4.5 to 26 nm. Scale bar, 100 nm.
The D-SNPs showed inhibitory effects against all selected pathogens. The inhibitory activity of D-SNPs was found to be higher (MRSA > S. typhimurium > K. pneumoniae > E. coli > S. mutans) than that of silver ions (Figure 2). However, D-SNPs resulted in significant toxicity against both gram-positive bacteria and gram-negative bacteria. MRSA was the most sensitive bacterial strain to D-SNPs and silver nitrate, with IZ diameters equal to 23.7 ± 0.08 and 18 ± 0.58, respectively; however, MRSA subjected to ampicillin showed an IZ diameter of 26.7 ± 0.33. S. mutans was the most resistant microbe toward both D-SNPs and silver nitrate, with the smallest IZ diameters of 13.3 ± 0.03 and 11.4 ± 0.12, respectively (Table 2). The convergence of IZ diameter values of D-SNPs and ampicillin antibiotic against MRSA indicates that D-SNPs act as a promising antibacterial agent, especially against MDR bacteria. The higher inhibitory capacity of D-SNPs against MRSA than that of AgNO3 may be because their small size and large surface area allowed more surface contact with microorganisms (Logeswari et al., 2015). Another important reason for the enhanced antibacterial activity of D-SNPs may be attributed to the Desertifilum biocompounds that coated SNPs (Durán et al., 2016).
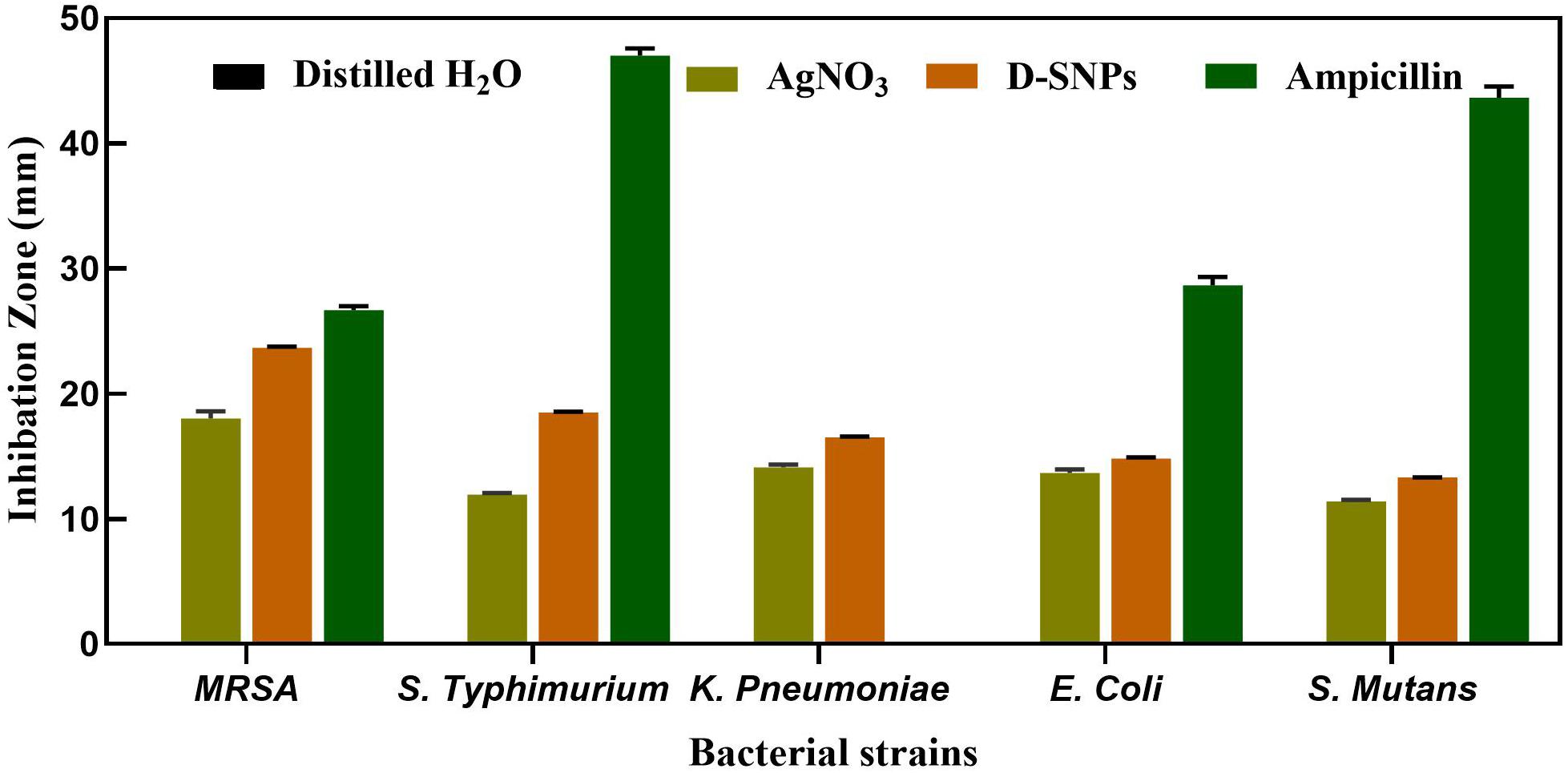
Figure 2. Inhibitory effect of D-SNPs, silver nitrate and ampicillin against five medically pathogenic bacteria including Escherichia coli, Salmonella typhimurium, and Klebsiella pneumoniae, Streptococcus mutans, and Methicillin-resistant Staphylococcus aureus.
Many reports have shown that SNPs are more toxic against gram-negative bacteria than against gram-positive bacteria due to the thickness of the cell wall of the latter (Slavin et al., 2017). Gram-negative bacteria are characterized by thin cell walls that allow SNPs to invade bacterial cells, while gram-positive bacteria have thick peptidoglycan cell walls that decrease the efficiency of SNPs to penetrate the cell wall (Dorobantu et al., 2015). These findings do not completely agree with the current data demonstrating the ability of D-SNPs to inhibit both gram-positive and gram-negative bacterial growth. However, our data related to S. typhimurium, K. pneumoniae, E. coli, and S. mutans agree with the results of Dorobantu et al. (2015) and Slavin et al. (2017) in which the gram-positive S. mutans bacterium showed the least response toward D-SNPs in comparison with that of S. typhimurium, K. pneumoniae and E. coli, with an IZ diameter of 13.3 ± 0.03. The lower activity of D-SNPs and AgNO3 toward S. mutans may be due to the structure of its cell wall. Based on all previous findings, the variable values of the IZ diameter after treating five bacteria with D-SNPs may not only depend on the thickness of the bacterial cell wall, as occurs in the case of MRSA, considering its resistance potential and the fact that E. coli exhibits the lowest IZ diameter value (14.8 ± 0.07) among the other tested gram-negative bacteria; the variable IZ diameter values may thus also depend on the reactivity between D-SNPs and bacteria, the mechanism of defense used by bacteria against D-SNPs, the functional groups surrounding the surface of D-SNPs and the opposite charge attraction between the surfaces of bacteria and D-SNPs. Romero-Urbina et al. (2015) showed that positively charged SNPs were more potent antibacterial agents owing to the greater affinity between these positively charged SNPs and the bacterial cell walls of MRSA. Salas-Orozco et al. (2019) showed that the response of bacteria toward silver nanoparticles depends on bacterial mechanisms that may be intrinsic, such as efflux pumps, downregulation of porins, and chromosomal resistance genes, or extrinsic, such as point and adaptive mutations and plasmids with resistance genes. Recent publications have reported that the enhanced inhibitory effect of biogenic silver nanoparticles may be due to neutral compounds surrounding the nanoparticles (Durán et al., 2016; Qais et al., 2019).
With the exception of K. pneumoniae, Table 3 shows that the MIC and MBC values of D-SNPs were lower than those of AgNO3. K. pneumoniae showed the same MIC and MBC values (1.2 and 1.5 mg/mL, respectively) for D-SNPs and AgNO3. These data confirm the results of the agar well diffusion method and indicate that D-SNPs were the most potent antibacterial material in comparison with AgNO3. Previous data have reported that plant-derived SNPs exhibited MIC values (10 mg/mL) against E. coli, K. pneumoniae, S. typhimurium, and MRSA of 2.5, 5, 2.5, and 2.5 mg/mL, respectively (Punjabi et al., 2018). These results may provide researchers with a new direction, that is, the synthesis of new alternative antibacterial drugs using cyano-nanotechnology.
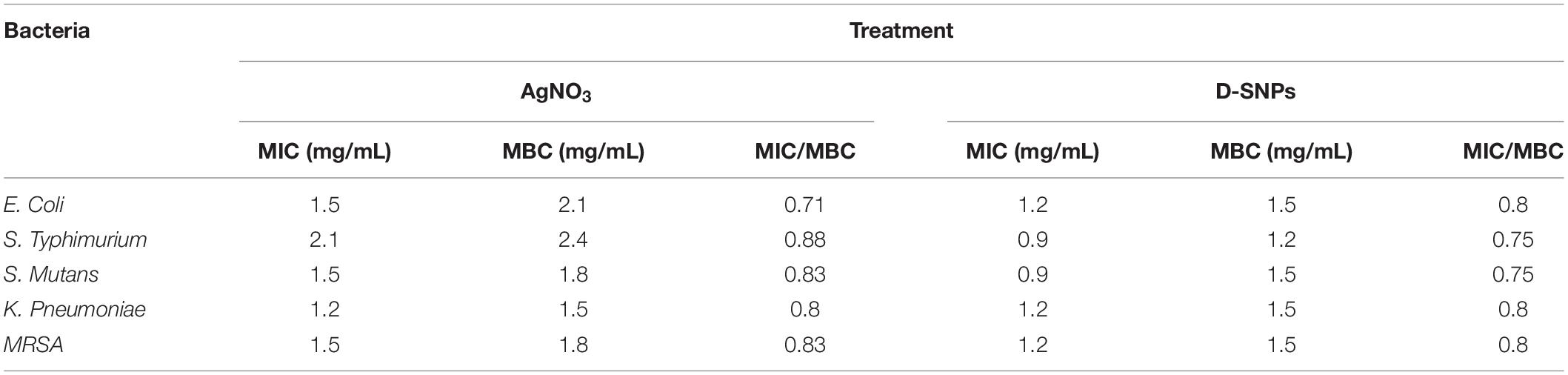
Table 3. Minimum inhibition concentration (MIC) and Minimum bactericidal concentration (MBC) of silver nitrate and D-SNPs against five pathogenic bacteria.
Many publications have tried to explain the mechanism of action of SNPs against bacteria, and some of these reports attributed the lethal effect of SNPs to their ability to induce the formation of reactive oxygen species (ROS) and metabolic toxicity (Yuan et al., 2017; Gurunathan et al., 2018). The current study screened the relationship between D-SNPs and AgNO3 and the levels of antioxidants such as GPx, GSH and catalase as well as the activity of ATPase.
Glutathione (GSH) is an antioxidant that exists in all living organisms such as plants, animals, bacteria, fungi and archaea. GSH has the ability to protect cells from damage caused by ROS such as free radicals, peroxides, and lipid peroxides as well as those from heavy metals (Pompella et al., 2003). With the exception of that in S. mutans, the GSH activity in all tested bacteria was decreased after D-SNP treatment in comparison with the control and silver nitrate treatments (Figure 3A). A similar result was observed for AgNO3; the GSH level was decreased in the five selected bacteria, however, the effect of D-SNPs on the GSH level was stronger than that of silver nitrate. The decrease in GSH levels may be due to the D-SNPs causing intense oxidative stress by inducing ROS formation inside bacteria. This finding is consistent with data reported by Gurunathan et al. (2018), who showed that after treatment of pathogenic bacteria with silver nanoparticles, the level of antioxidant GSH was sharply decreased. Banerjee et al. (2010) reported that treatment of Pseudomonas putida with SNPs led to increased peroxidation with a simultaneous weakening of the antioxidant defense system.

Figure 3. Effect of D-SNPs and silver nitrate on antioxidants and enzymes activities. (A) GSH. (B) GPx. (C) CAT. (D) ATPase. Data from at least three independent experiments performed at least in triplicate are presented as the mean SEM; P-values were calculated versus untreated bacterial cells: ****P < 0.00001, ***P < 0.0001, **P < 0.001, and *P < 0.01.
With respect to untreated bacteria, S. mutans showed increased GSH levels after treatment with D-SNPs. This result indicated that S. mutans may be more resistant to D-SNPs by increasing the GSH antioxidant level. This finding is consistent with data obtained from the agar well diffusion assay, in which S. mutans was the pathogen least affected by D-SNPs. Barros et al. (2019) mentioned that some bacteria detoxify heavy metals and nanoparticles by increasing the antioxidant levels. E. coli showed a greater decrease in the GSH levels after treatment with silver nitrate than that after treatment with D-SNPs. This result may indicate that oxidative stress caused by silver nitrate may be more than that caused by D-SNPs. Additionally, the defense mechanism of bacteria toward heavy metals and nanoparticles depends on many factors such as the type of bacteria, their neutral habitats, the stressors in their habitat and their capacity to react to external stress (Japenga et al., 1990; Fabrega et al., 2011).
Glutathione peroxidase (GPx) is a member of the enzyme family characterized by peroxidase activity. These enzymes protect the cells from oxidative stress through the reduction of lipid hydroperoxides to their corresponding alcohols and the reduction of free hydrogen peroxide to water (Muthukumar and Nachiappan, 2010; Muthukumar et al., 2011). The GPx enzyme activity of the five bacterial strains after administration of D-SNPs and AgNO3 was increased in comparison with that of the control (Figure 3B); however, in comparison with AgNO3, the D-SNPs led to a greater increase in the GPx enzyme activity. The increase in the GPx level may be due to D-SNPs increasing the production of ROS, and the bacteria fight this stress by scavenging hydroperoxides through increased expression of the GPx enzyme. These data are in accordance with that of Barros et al., who demonstrated that exposure of Pseudomonas sp. M1 to both silver ions and SNPs resulted in increased GPx antioxidant levels (Barros et al., 2019). The current data corroborate data from GSH level analysis, in which the higher activity of the GPx enzyme is related to the low GSH level as GPx converts the GSH substrate into GSSG (oxidized form), affecting the GSH stores required for cellular protection against oxidative stress and inducing bacterial cell death (Yuan et al., 2017).
Catalase is a common enzyme that exists in approximately all living organisms exposed to oxygen; it catalyzes the decomposition reaction of hydrogen peroxide to water and oxygen. This enzyme plays an important role in protecting cells from oxidative stress resulting from ROS (Chelikani et al., 2004). The five bacterial strains after 24 h of treatment with D-SNPs and AgNO3 showed a reduction in catalase enzyme activity (Figure 3C). Barros et al. showed that the catalase enzyme activity in P. aeruginosa cells after a short exposure (1 h) to SNPs was increased, while after a long exposure (12–24 h), the activity of catalase was decreased (Barros et al., 2019). This finding may explain why after a short exposure to silver ions and D-SNPs, the catalase activity increases as a response to external stress in the experiment to mitigate the external danger, while with increasing exposure time, the bacteria lose their ability to detoxify the silver nanoparticles and silver ions, so the catalase activity drops. Additionally, Gurunathan et al. (2018) reported that after being treated with SNPs, Prevotella melaninogenica and Arcanobacterium pyogenes showed low catalase activity after 12 h.
In all tested bacteria with the exception of S. typhimurium, D-SNPs were the agent most responsible for reducing the catalase activity compared to that of AgNO3 and untreated bacteria. S. typhimurium showed a greater decrease in catalase levels after treatment with silver nitrate than that with D-SNPs. These results showed that D-SNPs may be responsible for a significant drop in antioxidant levels as a result of causing intense oxidative damage in the bacterial cell by increasing the production of ROS (Banerjee et al., 2010; Gurunathan et al., 2018). Additionally, these findings are consistent with data reported by Yuan et al. (2017) showing that biogenic SNPs caused a decrease in the catalase level of drug-resistant strains of Staphylococcus aureus and Pseudomonas aeruginosa. The observed positive association between GPx and catalase activities under D-SNP exposure suggested the contribution of these enzymes to the scavenging of peroxides/hydroperoxides. The more pronounced activities of antioxidant enzymes under D-SNP exposure were consistent with the higher toxicity of the nano-form (Barros et al., 2019). Our results showed the involvement of different antioxidants such as GPx, GSH (part of the ascorbate–glutathione cycle) and catalase in coping with the oxidative stress induced by both silver nitrate and D-SNPs. However, the greatest imbalance in antioxidant activities was related to exposure to D-SNPs, which may be due to their small size, charge and coating with Desertifilum biocompounds. These factors may increase the potential of D-SNPs to penetrate bacterial cells and interfere with bacterial biomolecules.
ATPase is an enzyme responsible for the decomposition of adenosine triphosphate (ATP) into adenosine diphosphate (ADP) and the release of free phosphate ions or the inverse reaction. The dephosphorylation reaction results in liberating energy, which the enzyme harnesses to drive other biochemical reactions such as anabolic processes (Riley and Peters, 1981). The five bacterial strains treated with D-SNPs and silver nitrate exhibited decreased ATPase activity compared with that of the control (Figure 3D). However, D-SNP caused a greater decrease in ATPase activity than silver nitrate did. The decrease in ATPase activity may be because D-SNPs interfere with the ATPase enzyme, causing dysfunction of this enzyme. This finding is in agreement with the results of Cui et al., who stated that gold NPs inhibit ATPase activity, leading to a decreased ATP content in E. coli (Cui et al., 2012).
Based on the results from the analysis of enzyme activities, the mechanism by which D-SNPs and silver ions kill bacteria may be explained by an increase in ROS formation, leading to increased oxidative stress and metabolic toxicity. This toxicity leads to damage to biomolecules such as enzymes, proteins and DNA, resulting in bacterial cell death (Gurunathan et al., 2018).
TEM micrographs showed that untreated MRSA cells were generally intact and closely packed, with typical cytoplasmic membranes, and their cell walls were rigid, with thickened peptidoglycan (PG) layers (Figures 4A,B). In contrast, MRSA cells treated with silver nitrate lost their coccal appearance and appeared to undergo lysis, leading to liberation of the cellular contents into the surrounding environment and ultimately resulting in cell disruption (Figures 4C,D). Moreover, the morphological changes after treating MRSA with silver nitrate included disintegration of the bacterial cells, a decrease in the electron density of the cytoplasm and in their sizes (Figure 5A). D-SNPs resulted in deformation of the MRSA shape, reduction in bacterial density, cell bursting, scattering of cell debris and vacuoles (Figures 4E,F). Additionally, the D-SNPs led to an increase in cell size (Figure 5A), disruption of the cell wall, damage to PG layers and induced the formation of many pores, holes and folds and cytoplasmic filaments; all these signs confirm that D-SNPs induce bacterial cell death (Figure 4F). Figure 5B demonstrates the affinity between D-SNPs and bacterial cell walls, in which clusters of D-SNPs bind to the cell walls, resulting in cytoplasmic leakage and discharge of cytoplasmic components that aid in bacterial cell lysis and induce apoptotic body formation. These findings are in agreement with those of Romero-Urbina et al. (2015), who studied the effect of positively charged SNPs on the morphology of MRSA and showed that SNPs induce peptidoglycan layer damage, porosity of the cell membrane and discharge of cytoplasmic material. The D-SNP and silver nitrate treatments resulted in morphological changes of MRSA; however, the acute changes were caused by D-SNPs. It was found that D-SNPs inside bacterial cells have a smaller size, with a size range from 2 to 14.6 nm, than that of D-SNPs outside bacterial cells, with a size range equal to 2.5–26.8 nm (Figures 6A,B). The larger size of D-SNPs outside the bacterial cells than inside the cells may be due to the agglomeration of D-SNPs outside the bacteria and indicated that the larger particles may be trapped outside while small D-SNPs easily enter the bacteria (Romero-Urbina et al., 2015). On the other hand, the TEM micrographs also showed the existence of electron dense spherical particles that may be SNPs resulting from the reduction of silver nitrate by MRSA (Figures 4C,D). The silver nanoparticles synthesized by MRSA (M-SNPs) were distributed at the plasma membrane, cytoplasm and genetic material and have sizes ranging from 3.6 to 20.6 nm, and their average size is 9.6 ± 0.4 nm (Figure 6C). It is interesting to note that this investigation represents the first report showing the ability of MRSA to fabricate small SNPs from silver nitrate. Jung et al. mentioned in their publication that it is common to observe the presence of electron-dense particles or precipitates around damaged bacterial cells of S. aureus treated with silver ions (Jung et al., 2008).
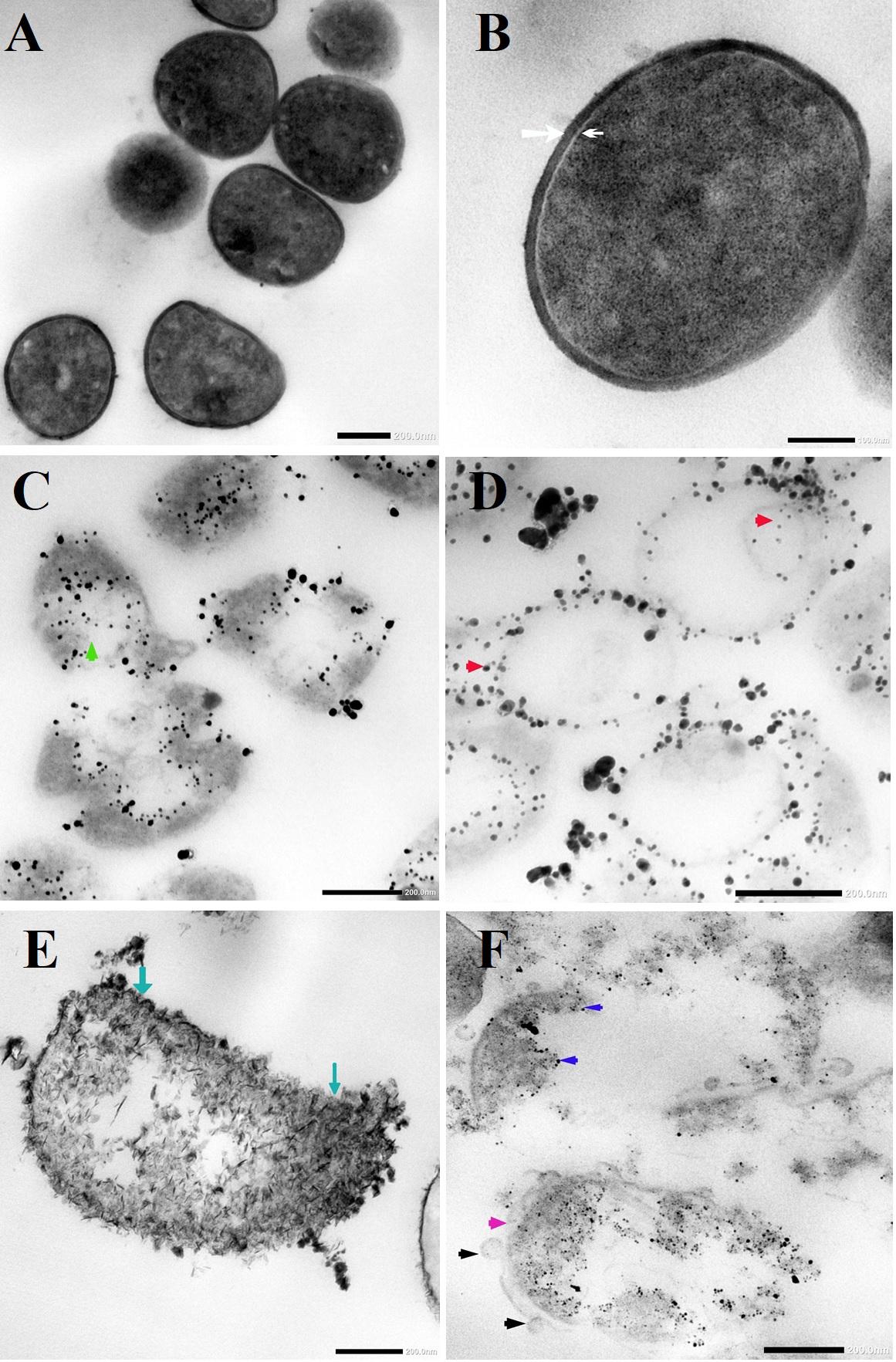
Figure 4. (A,B) TEM micrographs of untreated MRSA showing intact closely packed cells with thicken peptidoglycan layers cell walls (white arrow) and bacterial membrane (head arrow). (C) MRSA treated with silver nitrate appeared to undergo lysis (green arrow). (D) Distribution of SNPs synthesized by MRSA (red arrow). (E) MRSA treated with D-SNPs showing cell wall disruption with folds formation (blue arrow). (F) MRSA subjected with D-SNPs showing separation plasma membrane from the cell wall (pink arrow), secretory vacuoles (black arrow), D-SNPs inside cells (dark blue arrow) causing cytoplasmic lashing. Scale bar, 200 nm.
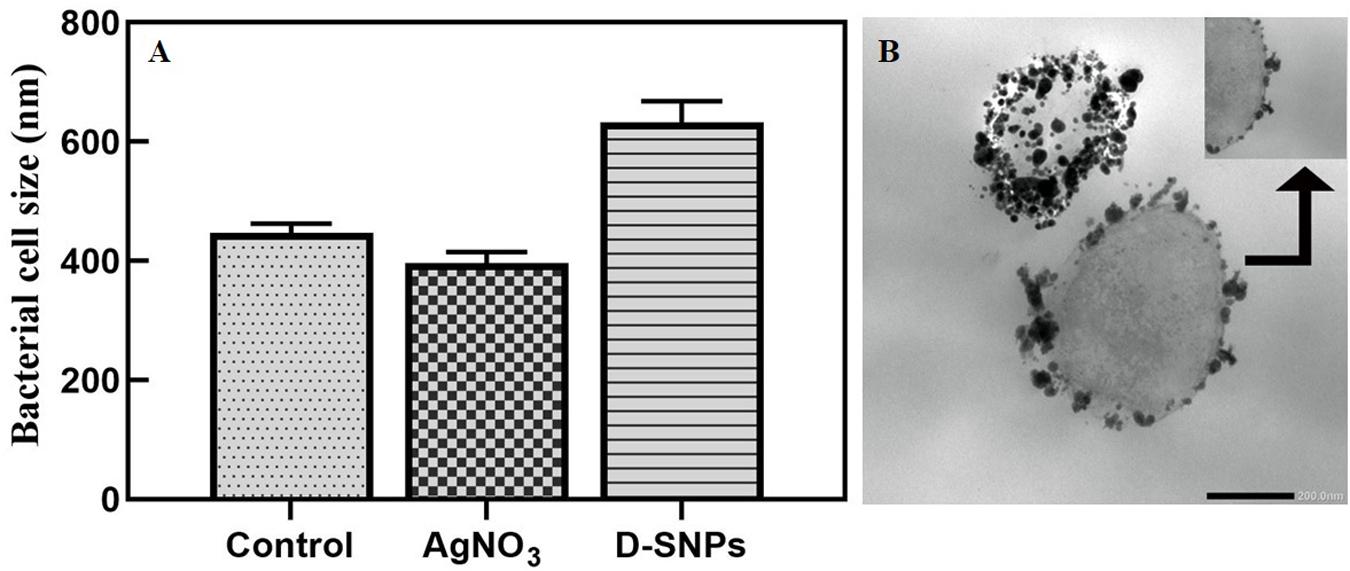
Figure 5. (A) MRSA cell size after treating with silver nitrate and D-SNPs. (B) TEM image showing the D-SNPs attached on MRSA cell wall. Scale bar, 200 nm.

Figure 6. (A) D-SNPs size distribution outside MRSA cell. (B) D-SNPs size distribution inside MRSA cell. (C) Size distribution of SNPs synthesized by MRSA.
Our consideration of this result is that MRSA may be attempting to overcome the external stress of a heavy metal (silver nitrate); the bacteria convert the bulk material into small nanoparticles (NPs), but these NPs kill the bacteria. Wakshlak et al. (2015) called this phenomenon a “zombie effect,” in which bacteria synthesize NPs that kill them.
It was found that not all bacteria are able to perform this action, suggesting that the ability of MRSA to fabricate the smaller SNPs from silver nitrate may be a resistance feature that aids the bacterium. Pandian et al. (2010) reported that there are two possible mechanisms for killing bacteria with silver nitrate. First, at lower concentrations of silver nitrate, silver ions are converted to silver nanoparticles by bacterial cells, and these synthesized NPs kill the bacteria from within. However, at higher concentrations, silver nitrate induces apoptosis by increasing the hydrogen peroxide levels through inactivation of thiol group-containing proteins such as NADH dehydrogenase II or by direct binding with DNA (Pandian et al., 2010).
The change in gene expression of MRSA mfD, hly and flu genes was evaluated using qRT-PCR before and after treatment of MRSA with D-SNPs and silver nitrate at a concentration of 1.54 mg/mL. Total RNA was extracted, cDNA was created, and q real-time PCR was performed using specific primers of mfD, hly, and flu, with GAPDH as a housekeeping gene control. The data showed that the exposure of MRSA to D-SNPs and silver nitrate caused upregulation of mfD gene expression but downregulation of the expression of hly and flu genes (Figure 7). Moreover, the higher increase in the mRNA levels of the mfD gene may be a bacterial response to genotoxic damage caused by D-SNPs as a result of intense oxidative stress. This finding is consistent with that of Ashmore et al. (2018), who treated E. coli with polymer-coated SNPs and reported that upregulation of the mfD gene was a response of the bacteria to damage caused by the NPs. On the other hand, the decrease in the mRNA level of the flu gene may be due to the stress induced by D-SNPs causing loss or disruption of bacterial function such as biofilm formation, as represented here by the flu gene. The inhibitory effect of zinc NPs on the flu gene of E. coli was reported by Shakerimoghaddam et al. (2017). The authors showed that zinc NPs reduced the expression of the flu and fimH genes, resulting in a reduction in biofilm formation (Shakerimoghaddam and Ghaemi, 2017; Shakerimoghaddam et al., 2017).
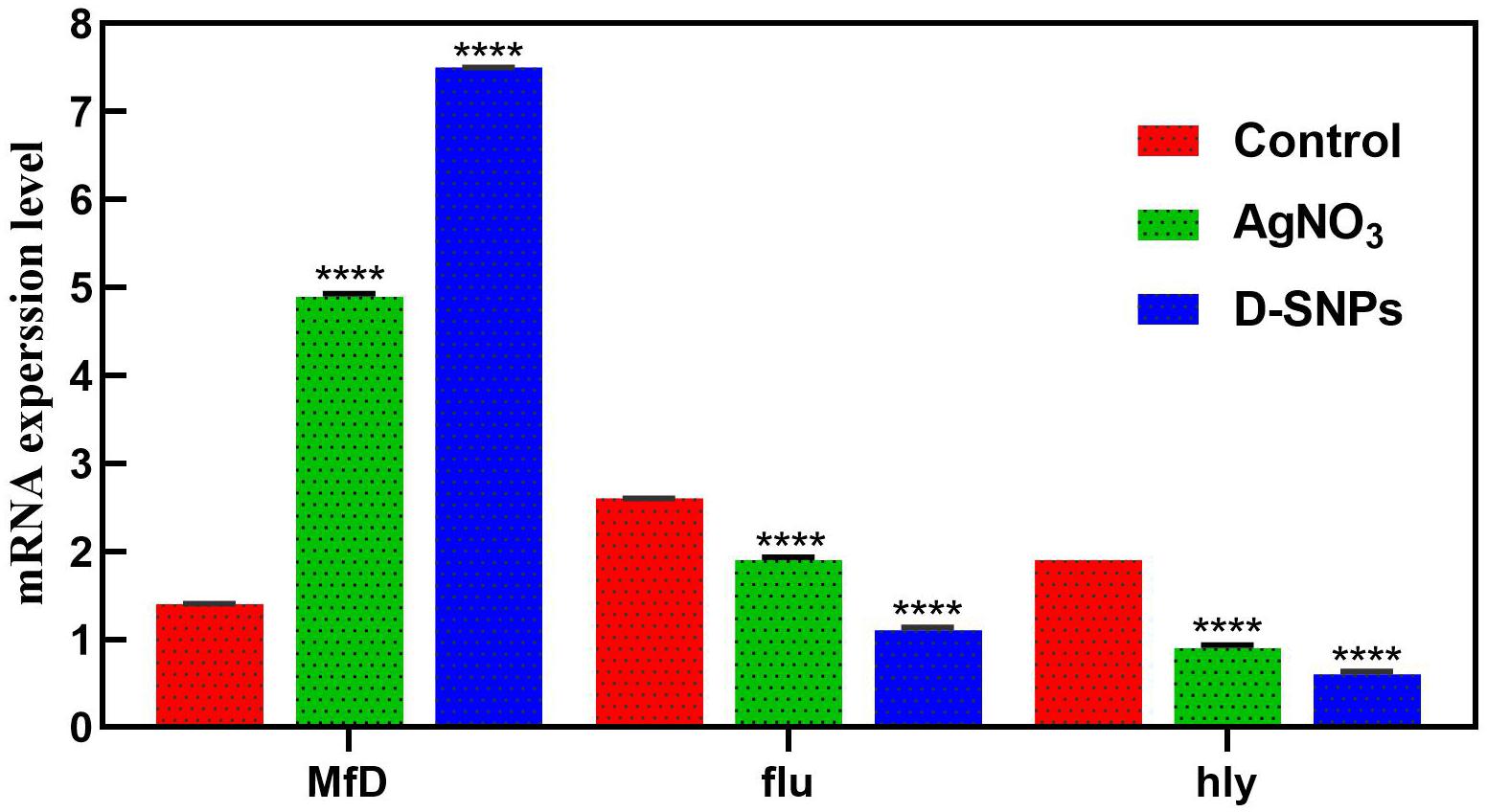
Figure 7. mRNA expression level of mfD, flu, hly genes of MRSA before and after treatment with AgNO3 or D-SNPs. Data from at least three independent experiments performed at least in triplicate are presented as the mean ± SEM; P-values were calculated versus untreated bacterial cells: ****P < 0.00001.
Additionally, the downregulation of the alpha-hemolysin gene as a crucial virulence factor of the pathogen may indicate that D-SNPs affect bacterial defense mechanisms by reducing virulence and invasiveness (Soleimani and Habibi-Pirkoohi, 2017). This result showed that D-SNPs may be a promising anti-virulence agent. Das et al. (2016) showed that multi-drug-resistant Pseudomonas aeruginosa strains treated with SNPs exhibited downregulation of the hly gene. A recent publication reported that biogenic SNPs synthesized using Shewanella oneidensis act as potent anti-virulence agents by downregulating the hly gene of E. coli (Tanhaeian and Ahmadi, 2018).
Similarly, the upregulation of the mfD gene and downregulation of flu and hly genes of MRSA after exposure to silver nitrate and/or M-SNPs may be due to silver nitrate and/or M-SNPs causing genotoxicity by inducing ROS formation (Pandian et al., 2010; Barros et al., 2019). It was found that the effect of D-SNPs on the gene expression of mfD, hly and flu of MRSA was greater than that of silver nitrate and/or M-SNPs. This finding may be due to the features of the D-SNPs including the Desertifilum-derived materials that coated the particles, the particle size and the charge of the D-SNPs. These factors may facilitate the penetration through the MRSA plasma membrane, and attraction between D-SNPs and genetic material allowed these NPs to exhibit the highest genotoxicity as a result of intense oxidative stress (Romero-Urbina et al., 2015). Based on previous data, we supposed that D-SNPs and silver nitrate and/or M-SNPs caused genotoxicity and imbalance in the gene expression of MRSA through induction of oxidative stress.
The SDS-PAGE results of proteins extracted from MRSA before and after treatment with D-SNPs and silver nitrate are shown in Figures 8A,B. Considering that of the control, the results showed that the band intensity of proteins of MRSA treated with D-SNPs was very thin, and new protein bands appeared. Additionally, in comparison with those of untreated bacteria, the large molecular weight protein bands from the treated bacteria were largely prevented from entering the electrophoresis gel. This result reflects that ROS formed after treating MRSA with D-SNPs may induce significant covalent modifications to the proteins that either caused the formation of insoluble aggregates that were too large to enter the electrophoresis gel or degradation of proteins into fragments that were too small to be detected by the technique (Wang et al., 2013). Moreover, these results may be attributed to the D-SNPs inducing the expression of new proteins as a stress response and/or preventing the synthesis of other proteins by inhibiting many translation factors. These data are in accordance with the results of Soliman et al. (2018) who reported that biogenic SNPs lead to the presence of new protein bands and increasing in band intensity of samples treated with SNPs were more than in untreated samples at the two separation speed of gel electrophoresis (4000, 14000 rpm). They mentioned that SNPs induced bacterial and fungal cells (Bacillus sp. and Candida sp. respectively) to produce new proteins as a response to this stress. Moreover, in case E. coli spiked with SNPs, the protein profile remain unchanged in comparing with untreated cells, however, band intensities were comparatively less than those of the control (Soliman et al., 2018). This finding implies that the affinity of D-SNPs to the thiol groups of proteins causing unfolding of the protein chain might cause cellular protein modification and degradation of the proteins (Gogoi et al., 2006; Velusamy et al., 2016). Tamiyakul et al. (2019) showed that SDS-PAGE of treated S. aureus and E. coli with SNPs coated with poly (4-styrenesulfonic acid-co-maleic acid) polymer exhibited more protein expression in comparison with untreated bacteria. They suggested that protein pattern modification results in bacterial death through interacting with metabolic function and synthesis of cell wall, nucleic acid, and protein (Tamiyakul et al., 2019). Tiwari et al. (2017) studied the effect of chemically synthesized polyvinylpyrrolidone-SNPs on sRS-307 bacterial proteins using SDS-PAGE. They showed that SNPs resulted in overexpression of 43 kDa protein while a protein with 35 kDa undergoes downregulation. Similar results were observed for silver nitrate and/or M-SNPs, but the protein bands with large electrophoretic molecular weights were slightly heavier than those from the D-SNP treatment. Additionally, the number of protein bands after treatment with silver nitrate was 12, while that in the case of D-SNP treatment was 14 (Table 4), indicating that the D-SNPs caused more fragmentation and denaturation of proteins.
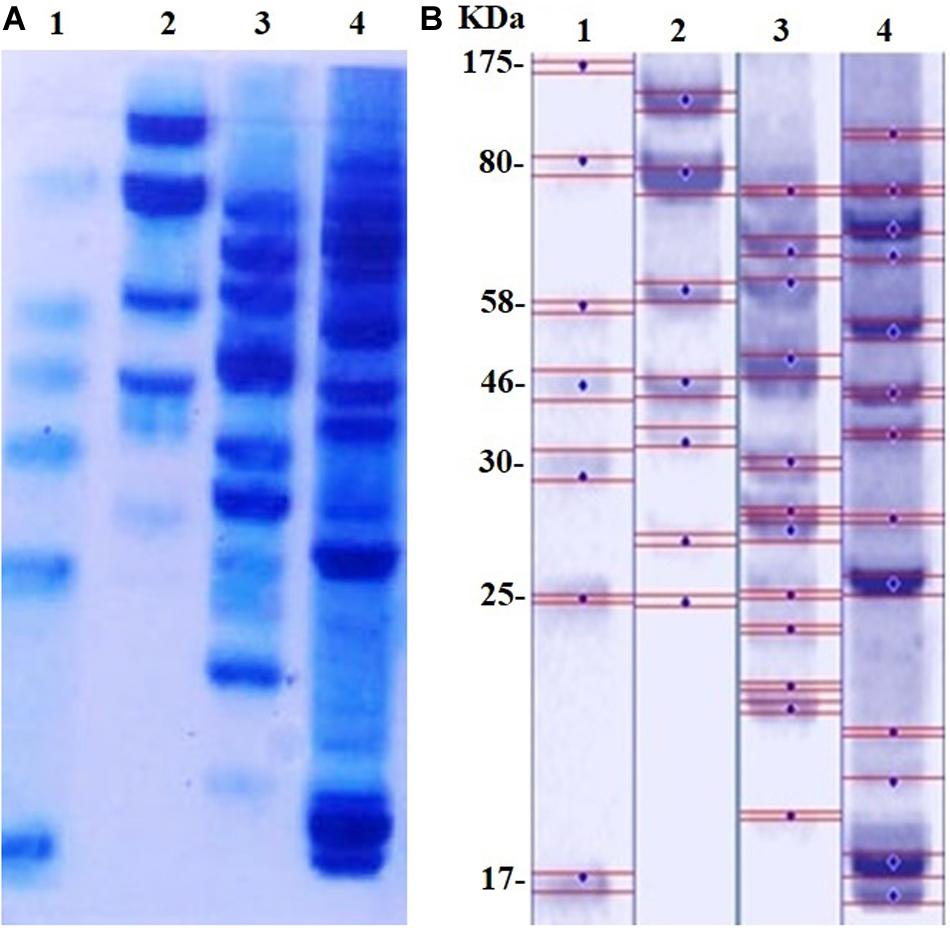
Figure 8. (A) SDS-PAGE of MRSA cells (104 CFU/mL) incubated with D-SNPs or silver nitrate for 24 h. (B) Computerized analysis of band intensities. (1) Marker, (2) untreated MRSA, (3) MRSA treated with AgNO3, (4) MRSA treated with D-SNPs.
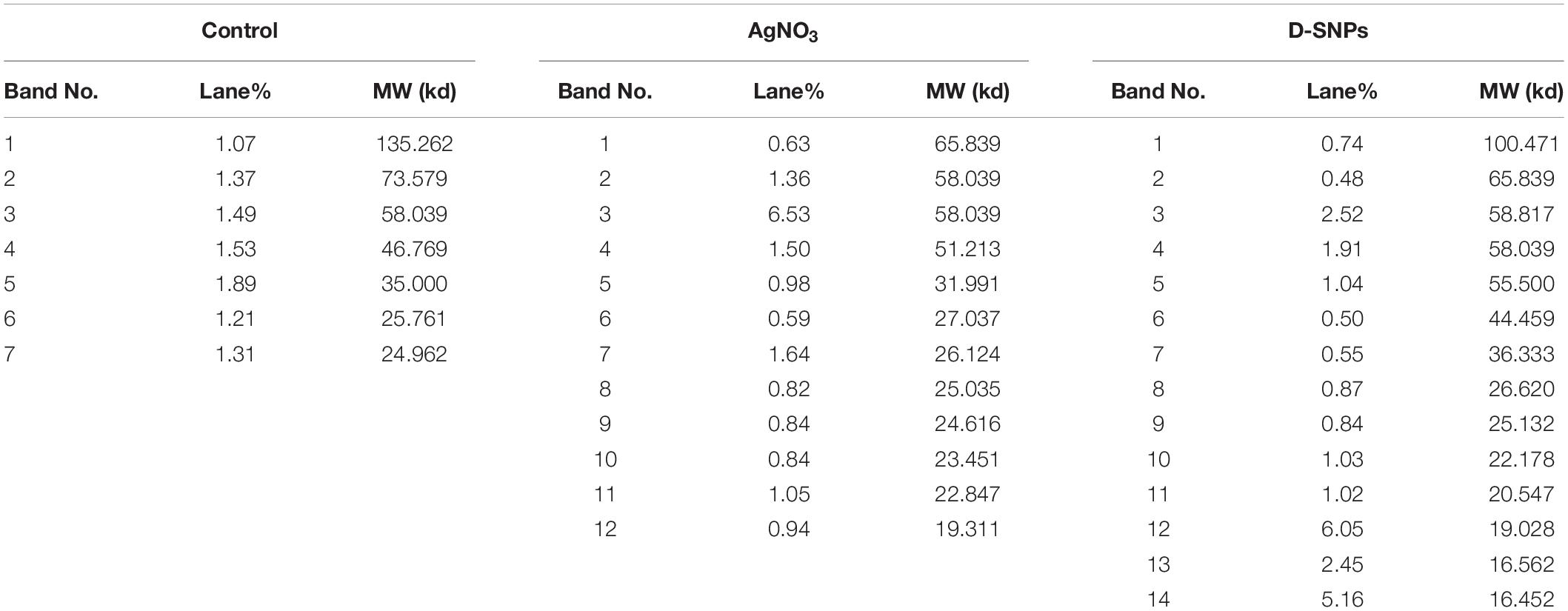
Table 4. Number and intensity of protein bands of MRSA cells before and after treatment with D-SNPs or AgNO3.
The higher toxicity of D-SNPs to MRSA than that of silver nitrate and/or M-SNPs may be due to the existence of Desertifilum biomolecules that surround the SNPs, preventing the agglomeration of SNPs, improving the ability of NPs to contact bacterial cells and promoting their antimicrobial properties (Chen et al., 2016). It has been suggested that D-SNPs cause inactivation of cellular proteins and loss of DNA replication ability by inducing ROS formation, leading to intense oxidative stress and bacterial cell death (Figure 9).
Biogenic silver nanoparticles may represent a new chance in the medical fields because they can act as alternative antibiotics that affect different bacterial mechanisms such as virulence and resistance. We explored the potential of novel D-SNPs and AgNO3 to inhibit the growth of different pathogenic bacteria including multi-drug-resistant bacteria such as MRSA. It was possible to conclude that D-SNPs were the most effective compound, showing potent antibacterial, anti-biofilm formation and anti-virulence activities against bacteria. Additionally, this investigation examined for the first time the ability of MRSA to biofabricate smaller SNPs from silver nitrate. Furthermore, the insights of this study into how novel D-SNPs impact bacterial metabolism could provide key information on the mechanisms of action, resulting in the discovery of new therapeutic methodologies for the development of better antibacterial agents. We suggest for the first time that the mechanism of action of D-SNPs and AgNO3 against MRSA is mainly related to oxidative stress induced by the generation of ROS, which results in an imbalance of antioxidant activity leading to the oxidation of biomolecules such as DNA, enzymes, RNA and proteins as well as bacterial death. This work demonstrates that green D-SNPs are efficient antibacterial agents that may serve as alternative drugs to antibiotics, opening the door to create synergetic nano-antibiotics to address multi-drug-resistant bacteria. Other findings are needed to complete the study of D-SNPs against different bacteria and fungi and the targeting of other genes to elucidate the specific molecular mechanism of these nanoparticles against bacteria.
The data supporting this article are available in Figures 1–9 and Tables 1–4. The data sets analyzed in the present study are available from the corresponding author upon reasonable request.
RH, MA, DG, MK, and MA-Z contributed conception and design of the study. RH, DG, and MA-Z downloaded and organized datasets. RH performed the statistical and result analysis and wrote the first draft of the manuscript. All authors contributed to manuscript revision, read, and approved the submitted version.
This research was included in a request for patent 120410334 from King Abdulaziz City for Science and Technology, Saudi Arabia, submitted on 07/01/2020, titled “Antibacterial activity of biosynthesized silver nanoparticles” https://epatentsso.saip.gov.sa/.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
This research was funded by the Deanship of Scientific Research at Princess Nourah bint Abdulrahman University through the Fast-track Research Funding Program.
μ g, microgram; μ L, microliter; AgNO3, silver nitrate; D-SNPs, silver nanoparticles synthesized by Desertifilum sp.; h, hour; IZ, inhibition zone; kb, kilobase; kV, kilovolt; mg, milligram; min, minute; mL, milliliter; mM, millimolar; nm, nanometer; NPs, nanoparticles; PCR, polymerase chain reaction; rpm, revolutions per minute; SNPs, silver nanoparticles; TEM, transmission electron microscope; UV, Ultraviolet-visible spectroscopy.
Ali, M. R., Wu, Y., and El-Sayed, M. A. (2019). Gold nanoparticle-assisted plasmonic photothermal therapy advances towards clinical application. J. Phys. Chem. C 123, 15375–15393.
Andrés, M. T., and Fierro, J. F. (2010). Antimicrobial mechanism of action of transferrins: selective inhibition of H+-ATPase. Antimicrob. Agents Chemother. 54, 4335–4342. doi: 10.1128/AAC.01620-09
Ashmore, D. A., Chaudhari, A., Barlow, B., Barlow, B., Harper, T., Vig, K., et al. (2018). Evaluation of E. coli inhibition by plain and polymer-coated silver nanoparticles. Rev. Inst. Med. Trop. São Paulo 60, 1–11. doi: 10.1590/s1678-9946201860018
Banerjee, M., Mallick, S., Paul, A., Chattopadhyay, A., and Ghosh, S. S. (2010). Heightened reactive oxygen species generation in the antimicrobial activity of a three component iodinated chitosan- silver nanoparticle composite. Langmuir 26, 5901–5908. doi: 10.1021/la9038528
Barros, D., Pradhan, A., Mendes, V. M., Manadas, B., Santos, P. M., Pascoal, C., et al. (2019). Proteomics and antioxidant enzymes reveal different mechanisms of toxicity induced by ionic and nanoparticulate silver in bacteria. Environ. Sci. Nano 6, 1207–1218.
Beutler, E. (1963). Improved method for the determination of blood glutathione. J. Lab. Clin. Med. 61, 882–888.
Bin-Meferij, M. M., and Hamida, R. S. (2019). Biofabrication and antitumor activity of silver nanoparticles utilizing novel Nostoc sp. Bahar M. Int. J. Nanomedicine 14, 9019–9029. doi: 10.2147/IJN.S230457
Bogdan, J., Pławińska-Czarnak, J., and Zarzyńska, J. (2017). Nanoparticles of titanium and zinc oxides as novel agents in tumor treatment: a review. Nanoscale Res. Lett. 12, 225–240. doi: 10.1186/s11671-017-2007-y
Boucher, H. W., Talbot, G. H., Bradley, J. S., Edwards, J. E., Gilbert, D., Rice, L. B., et al. (2009). Bad bugs, no drugs: no ESKAPE! An update from the Infectious Diseases Society of America. Clin. Infect. Dis. 48, 1–12. doi: 10.1086/595011
Brown, E. D., and Wright, G. D. (2016). Antibacterial drug discovery in the resistance era. Nature 529, 336–343. doi: 10.1038/nature17042
Chakraborty, S. P., Pramanik, P., and Roy, S. (2012). Staphylococcus aureus infection induced oxidative imbalance in neutrophils: possible protective role of nanoconjugated vancomycin. ISRN Pharmacol. 2012:435214. doi: 10.5402/2012/435214
Chelikani, P., Fita, I., and Loewen, P. C. (2004). Diversity of structures and properties among catalases. Cell. Mol. Life Sci. 61, 192–208. doi: 10.1007/s00018-003-3206-5
Chen, J., Li, S., Luo, J., Wang, R., and Ding, W. (2016). Enhancement of the antibacterial activity of silver nanoparticles against phytopathogenic bacterium Ralstonia solanacearum by stabilization. J. Nanomater. 2016, 1–16.
Cicci, A., Sed, G., Tirillò, J., Stoller, M., and Bravi, M. (2017). Production and characterization of silver nanoparticles in cultures of the cyanobacterium A. platensis (Spirulina). Chem. Eng. Trans. 57, 1405–1410.
Cillóniz, C., Dominedò, C., and Torres, A. (2019). Multidrug resistant gram-negative bacteria in community-acquired pneumonia. Crit. Care 23, 79–88. doi: 10.1186/s13054-019-2371-3
Cui, Y., Zhao, Y., Tian, Y., Zhang, W., Lü, X., and Jiang, X. (2012). The molecular mechanism of action of bactericidal gold nanoparticles on Escherichia coli. Biomaterials 33, 2327–2333. doi: 10.1016/j.biomaterials.2011.11.057
Das, B., Dash, S. K., Mandal, D., Adhikary, J., Chattopadhyay, S., Tripathy, S., et al. (2016). Green-synthesized silver nanoparticles kill virulent multidrug-resistant Pseudomonas aeruginosa strains: a mechanistic study. BLDE Univ. J. Health Sci. 1, 89–101.
Debnath, G., Das, P., and Saha, A. K. (2019). Green synthesis of silver nanoparticles using mushroom extract of Pleurotus giganteus: characterization, antimicrobial, and α-amylase inhibitory activity. BioNanoScience 9, 611–619.
Dorobantu, L. S., Fallone, C., Noble, A. J., Veinot, J., Ma, G., Goss, G. G., et al. (2015). Toxicity of silver nanoparticles against bacteria, yeast, and algae. J. Nanopart. Res. 17, 172–185.
Durán, N., Durán, M., De Jesus, M. B., Seabra, A. B., Fávaro, W. J., and Nakazato, G. (2016). Silver nanoparticles: a new view on mechanistic aspects on antimicrobial activity. Nanomedicine 12, 789–799. doi: 10.1016/j.nano.2015.11.016
Fabrega, J., Luoma, S. N., Tyler, C. R., Galloway, T. S., and Lead, J. R. (2011). Silver nanoparticles: behaviour and effects in the aquatic environment. Environ. Int. 37, 517–531. doi: 10.1016/j.envint.2010.10.012
Gogoi, S. K., Gopinath, P., Paul, A., Ramesh, A., Ghosh, S. S., and Chattopadhyay, A. (2006). Green fluorescent protein-expressing Escherichia coli as a model system for investigating the antimicrobial activities of silver nanoparticles. Langmuir 22, 9322–9328. doi: 10.1021/la060661v
Guggenbichler, J.-P., Böswald, M., Lugauer, S., and Krall, T. (1999). A new technology of microdispersed silver in polyurethane induces antimicrobial activity in central venous catheters. Infection 27, S16–S23. doi: 10.1007/bf02561612
Guo, D., Ji, X., Peng, F., Zhong, Y., Chu, B., Su, Y., et al. (2019). Photostable and biocompatible fluorescent silicon nanoparticles for imaging-guided co-delivery of siRNA and doxorubicin to drug-resistant cancer cells. Nano Micro Lett. 11, 27–40.
Gurunathan, S., Choi, Y.-J., and Kim, J.-H. (2018). Antibacterial efficacy of silver nanoparticles on endometritis caused by Prevotella melaninogenica and Arcanobacterum pyogenes in dairy cattle. Int. J. Mol. Sci. 19, 1210–1230. doi: 10.3390/ijms19041210
Hamida, R. S., Abdelmeguid, N. E., Ali, M. A., and Mohammed, M. (2020). Synthesis of silver nanoparticles using a novel cyanobacteria Desertifilum sp. extract: their antibacterial and cytotoxicity effects. Int. J. Nanomedicine 15, 49–63. doi: 10.2147/IJN.S238575
Jagielski, T., Bakuła, Z., Pleń, M., Kamiński, M., Nowakowska, J., Bielecki, J., et al. (2018). The activity of silver nanoparticles against microalgae of the Prototheca genus. Nanomedicine 13, 1025–1036. doi: 10.2217/nnm-2017-0370
Jalal, M., Ansari, M. A., Alzohairy, M. A., Ali, S. G., Khan, H. M., Almatroudi, A., et al. (2019). Anticandidal activity of biosynthesized silver nanoparticles: effect on growth, cell morphology, and key virulence attributes of Candida species. Int. J. Nanomedicine 14, 4667–4679. doi: 10.2147/IJN.S210449
Japenga, J., Zschuppe, K., De Groot, A., and Salomons, W. (1990). Heavy metals and organic micropollutants in floodplains of the river Waal, a distributary of the river Rhine, 1958-1981. Neth. J. Agric. Sci. 38, 381–397.
Jung, W. K., Koo, H. C., Kim, K. W., Shin, S., Kim, S. H., and Park, Y. H. (2008). Antibacterial activity and mechanism of action of the silver ion in Staphylococcus aureus and Escherichia coli. Appl. Environ. Microbiol. 74, 2171–2178. doi: 10.1128/AEM.02001-07
Koulenti, D., Xu, E., Yin Sum Mok, I., Song, A., Karageorgopoulos, D. E., Armaganidis, A., et al. (2019). Novel antibiotics for multidrug-resistant gram-positive microorganisms. Microorganisms 7, 270–294. doi: 10.3390/microorganisms7100386
Kubik, T., Bogunia-Kubik, K., and Sugisaka, M. (2005). Nanotechnology on duty in medical applications. Curr. Pharm. Biotechnol. 6, 17–33. doi: 10.2174/1389201053167248
Leid, J. G., Ditto, A. J., Knapp, A., Shah, P. N., Wright, B. D., Blust, R., et al. (2011). In vitro antimicrobial studies of silver carbene complexes: activity of free and nanoparticle carbene formulations against clinical isolates of pathogenic bacteria. J. Antimicrob. Chemother. 67, 138–148. doi: 10.1093/jac/dkr408
Lim, J. Y., Yoon, J. W., and Hovde, C. J. (2010). A brief overview of Escherichia coli O157: H7 and its plasmid O157. J. Microbiol. Biotechnol. 20, 5–14.
Logeswari, P., Silambarasan, S., and Abraham, J. (2015). Synthesis of silver nanoparticles using plants extract and analysis of their antimicrobial property. J. Saudi Chem. Soc. 19, 311–317.
Lu, Z., Rong, K., Li, J., Yang, H., and Chen, R. (2013). Size-dependent antibacterial activities of silver nanoparticles against oral anaerobic pathogenic bacteria. J. Mater. Sci. Mater. Med. 24, 1465–1471. doi: 10.1007/s10856-013-4894-5
Mal, P., Dutta, K., Bandyopadhyay, D., Basu, A., Khan, R., and Bishayi, B. (2013). Azithromycin in combination with riboflavin decreases the severity of Staphylococcus aureus infection induced septic arthritis by modulating the production of free radicals and endogenous cytokines. Inflamm. Res. 62, 259–273. doi: 10.1007/s00011-012-0574-z
Maura, D., Ballok, A. E., and Rahme, L. G. (2016). Considerations and caveats in anti-virulence drug development. Curr. Opin. Microbiol. 33, 41–46. doi: 10.1016/j.mib.2016.06.001
Metwalli, K. H., Khan, S. A., Krom, B. P., and Jabra-Rizk, M. A. (2013). Streptococcus mutans, Candida albicans, and the human mouth: a sticky situation. PLoS Pathog. 9:e1003616. doi: 10.1371/journal.ppat.1003616
Mori, Y., Ono, T., Miyahira, Y., Nguyen, V. Q., Matsui, T., and Ishiharam, M. (2013). Antiviral activity of silver nanoparticle/chitosan composites against H1N1 influenza A virus. Nanoscale Res. Lett. 8, 93–99. doi: 10.1186/1556-276X-8-93
Murugesan, K., Koroth, J., Srinivasan, P. P., Singh, A., Mukundan, S., Karki, S. S., et al. (2019). Effects of green synthesised silver nanoparticles (ST06-AgNPs) using curcumin derivative (ST06) on human cervical cancer cells (HeLa) in vitro and EAC tumor bearing mice models. Int. J. Nanomedicine 14, 5257–5270. doi: 10.2147/IJN.S202404
Muthukumar, K., and Nachiappan, V. (2010). Cadmium-induced oxidative stress in Saccharomyces cerevisiae. Indian J. Biochem. Biophys. 47, 383–390.
Muthukumar, K., Rajakumar, S., Sarkar, M. N., and Nachiappan, V. (2011). Glutathione peroxidase3 of Saccharomyces cerevisiae protects phospholipids during cadmium-induced oxidative stress. Antonie Van Leeuwenhoek 99, 761–771. doi: 10.1007/s10482-011-9550-9
Naidu Krishna, S., Govender, P., and Adam, J. K. (2015). Nano silver particles in biomedical and clinical applications. J. Pure Appl. Microbiol. 9, 103–112.
Nannini, E., Murray, B. E., and Arias, C. A. (2010). Resistance or decreased susceptibility to glycopeptides, daptomycin, and linezolid in methicillin-resistant Staphylococcus aureus. Curr. Opin. Pharmacol. 10, 516–521. doi: 10.1016/j.coph.2010.06.006
Narducci, D. (2007). An introduction to nanotechnologies: What’s in it for us? Vet. Res. Commun. 31, 131–137. doi: 10.1007/s11259-007-0082-8
Ouda, S. M. (2014). Antifungal activity of silver and copper nanoparticles on two plant pathogens, Alternaria alternata and Botrytis cinerea. Res. J. Microbiol. 9, 34–42.
Paglia, D. E., and Valentine, W. N. (1967). Studies on the quantitative and qualitative characterization of erythrocyte glutathione peroxidase. J. Lab. Clin. Med. 70, 158–169.
Pandian, S. R. K., Deepak, V., Kalishwaralal, K., Viswanathan, P., and Gurunathan, S. (2010). Mechanism of bactericidal activity of Silver Nitrate-a concentration dependent bi-functional molecule. Braz. J. Microbiol. 41, 805–809. doi: 10.1590/S1517-83822010000300033
Podschun, R., and Ullmann, U. (1998). Klebsiella spp. as nosocomial pathogens: epidemiology, taxonomy, typing methods, and pathogenicity factors. Clin. Microbiol. Rev. 11, 589–603.
Pompella, A., Visvikis, A., Paolicchi, A., De Tata, V., and Casini, A. F. (2003). The changing faces of glutathione, a cellular protagonist. Biochem. Pharmacol. 66, 1499–1503. doi: 10.3389/fphar.2015.00098
Punjabi, K., Mehta, S., Chavan, R., Chitalia, V., Deogharkar, D., and Deshpande, S. (2018). Efficiency of biosynthesized silver and zinc nanoparticles against multi drug resistant pathogens. Front. Microbiol. 9:2207. doi: 10.3389/fmicb.2018.02207
Qais, F. A., Shafiq, A., Khan, H. M., Husain, F. M., Khan, R. A., Alenazi, B., et al. (2019). Antibacterial effect of silver nanoparticles synthesized using Murraya koenigii (L.) against multidrug-resistant pathogens. Bioinorg. Chem. Appl. 2019:4649506. doi: 10.1155/2019/4649506
Raut, R. W., Kolekar, N. S., Lakkakula, J. R., Mendhulkar, V. D., and Kashid, S. B. (2010). Extracellular synthesis of silver nanoparticles using dried leaves of Pongamia pinnata (L) pierre. Nano Micro Lett. 2, 106–113.
Riley, M., and Peters, M. (1981). The localization of the anion-sensitive ATPase activity in corneal endothelium. Biochim. Biophys. Acta 644, 251–256. doi: 10.1016/0005-2736(81)90382-5
Romero-Urbina, D. G., Lara, H. H., Velázquez-Salazar, J. J., Arellano-Jiménez, M. J., Larios, E., Srinivasan, A., et al. (2015). Ultrastructural changes in methicillin-resistant Staphylococcus aureus induced by positively charged silver nanoparticles. Beilstein J. Nanotechnol. 6, 2396–2405. doi: 10.3762/bjnano.6.246
Salas-Orozco, M., Nińo-Martínez, N., Martínez-Castańón, G.-A., Méndez, M. F. T., Jasso, E. C., and Ruiz, F. (2019). Mechanisms of resistance to silver nanoparticles in endodontic bacteria: a literature review. J. Nanomater. 2019:7630316.
Saravanan, C., Rajesh, R., Kaviarasan, T., Muthukumar, K., Kavitake, D., and Shetty, P. H. (2017). Synthesis of silver nanoparticles using bacterial exopolysaccharide and its application for degradation of azo-dyes. Biotechnol. Rep. 15, 33–40. doi: 10.1016/j.btre.2017.02.006
Scallan, E., Hoekstra, R. M., Angulo, F. J., Tauxe, R. V., Widdowson, M.-A., Roy, S. L., et al. (2011). Foodborne illness acquired in the United States—major pathogens. Emerg. Infect. Dis 17, 7–15. doi: 10.3201/eid1701.p11101
Shakerimoghaddam, A., and Ghaemi, E. A. (2017). Effects of Zno nanoparticles on initial adhesion and fimH gene expression level of uropathogenic Eschercia coli. J. Clin. Basic Res. 1, 25–28.
Shakerimoghaddam, A., Ghaemi, E. A., and Jamalli, A. (2017). Zinc oxide nanoparticle reduced biofilm formation and antigen 43 expressions in uropathogenic Escherichia coli. Iran. J. Basic Med. Sci. 20, 451–456. doi: 10.22038/IJBMS.2017.8589
Slavin, Y. N., Asnis, J., Häfeli, U. O., and Bach, H. (2017). Metal nanoparticles: understanding the mechanisms behind antibacterial activity. J. Nanobiotechnol. 15, 65–85. doi: 10.1186/s12951-017-0308-z
Soleimani, M., and Habibi-Pirkoohi, M. (2017). Biosynthesis of silver nanoparticles using Chlorella vulgaris and evaluation of the antibacterial efficacy against Staphylococcus aureus. Avicenna J. Med. Biotechnol. 9, 120–125.
Soliman, H., Elsayed, A., and Dyaa, A. (2018). Antimicrobial activity of silver nanoparticles biosynthesised by Rhodotorula sp. strain ATL72. Egyptian J. Basic Appl. Sci. 5, 228–233.
Tamiyakul, H., Roytrakul, S., Jaresitthikunchai, J., Phaonakrop, N., Tanasupawat, S., and Warisnoicharoen, W. (2019). Changes in protein patterns of Staphylococcus aureus and Escherichia coli by silver nanoparticles capped with poly (4-styrenesulfonic acid-co-maleic acid) polymer. Asian Biomed. 13, 39–47.
Tanhaeian, A., and Ahmadi, F. S. (2018). Inhibitory effect of biologically synthesized silver nanoparticle on growth and virulence of E. coli. Zahedan J. Res. Med. Sci. 20, 1–6.
Tiwari, V., Tiwari, M., and Solanki, V. (2017). Polyvinylpyrrolidone-capped silver nanoparticle inhibits infection of carbapenem-resistant strain of Acinetobacter baumannii in the human pulmonary epithelial cell. Front. Immunol. 8:973. doi: 10.3389/fimmu.2017.00973
Vading, M., Nauclér, P., Kalin, M., and Giske, C. (2018). Invasive infection caused by Klebsiella pneumoniae is a disease affecting patients with high comorbidity and associated with high long-term mortality. PLoS One 13:e0195258. doi: 10.1371/journal.pone.0195258
Velusamy, P., Kumar, G. V., Jeyanthi, V., Das, J., and Pachaiappan, R. (2016). Bio-inspired green nanoparticles: synthesis, mechanism, and antibacterial application. Toxicol. Res. 32, 95–102. doi: 10.5487/TR.2016.32.2.095
Wakshlak, R. B.-K., Pedahzur, R., and Avnir, D. (2015). Antibacterial activity of silver-killed bacteria: the” zombies” effect. Sci. Rep. 5:9555. doi: 10.1038/srep09555
Wang, Y., Jett, S. D., Crum, J., Schanze, K. S., Chi, E. Y., and Whitten, D. G. (2013). Understanding the dark and light-enhanced bactericidal action of cationic conjugated polyelectrolytes and oligomers. Langmuir 29, 781–792. doi: 10.1021/la3044889
World Health Organization (2017). Prioritization of Pathogens to Guide Discovery, Research and Development of New Antibiotics for Drug-Resistant Bacterial Infections, Including Tuberculosis. Geneva: World Health Organization.
Wright, G. D., and Sutherland, A. D. (2007). New strategies for combating multidrug-resistant bacteria. Trends Mol. Med. 13, 260–267.
Yah, C. S., and Simate, G. S. (2015). Nanoparticles as potential new generation broad spectrum antimicrobial agents. DARU J. Pharm. Sci. 23, 43–57. doi: 10.1186/s40199-015-0125-6
Yallappa, S., Manjanna, J., Dhananjaya, B., Vishwanatha, U., Ravishankar, B., Gururaj, H., et al. (2016). Phytochemically functionalized Cu and Ag nanoparticles embedded in MWCNTs for enhanced antimicrobial and anticancer properties. Nano Micro Lett. 8, 120–130. doi: 10.1007/s40820-015-0066-0
Yuan, Y.-G., Peng, Q.-L., and Gurunathan, S. (2017). Effects of silver nanoparticles on multiple drug-resistant strains of Staphylococcus aureus and Pseudomonas aeruginosa from mastitis-infected goats: an alternative approach for antimicrobial therapy. Int. J. Mol. Sci. 18, 569–591. doi: 10.3390/ijms18030569
Keywords: MRSA, inhibitory activity, Desertifilum sp., bacteria, silver ion, silver nanoparticles
Citation: Hamida RS, Ali MA, Goda DA, Khalil MI and Al-Zaban MI (2020) Novel Biogenic Silver Nanoparticle-Induced Reactive Oxygen Species Inhibit the Biofilm Formation and Virulence Activities of Methicillin-Resistant Staphylococcus aureus (MRSA) Strain. Front. Bioeng. Biotechnol. 8:433. doi: 10.3389/fbioe.2020.00433
Received: 07 February 2020; Accepted: 15 April 2020;
Published: 25 May 2020.
Edited by:
Amir A. Zadpoor, Delft University of Technology, NetherlandsReviewed by:
Nanasaheb D. Thorat, University of Limerick, IrelandCopyright © 2020 Hamida, Ali, Goda, Khalil and Al-Zaban. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Reham Samir Hamida, cmVoYW0uaHVzc2VpbkBhbGV4dS5lZHUuZWc=; cmVoYW1zYW1pcjg1NEBnbWFpbC5jb20=; Mahmoud Ibrahim Khalil, bS5raGFsaWxAYmF1LmVkdS5sYg==; Mayasar Ibrahim Al-Zaban, TWlhbHphYmFuQHBudS5lZHUuc2E=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.