- Guangdong Province Key Laboratory of Improved Variety Reproduction in Aquatic Economic Animals, Biomedical Center, School of Life Sciences, Institute of Synthetic Biology, Sun Yat-sen University, Guangzhou, China
Aromatic compounds derived from aromatic amino acids are an important class of diverse chemicals with a wide range of industrial and commercial applications. They are currently produced via petrochemical processes, which are not sustainable and eco-friendly. In the past decades, significant progress has been made in the construction of microbial cell factories capable of effectively converting renewable carbon sources into value-added aromatics. Here, we systematically and comprehensively review the recent advancements in metabolic engineering and synthetic biology in the microbial production of aromatic amino acid derivatives, stilbenes, and benzylisoquinoline alkaloids. The future outlook concerning the engineering of microbial cell factories for the production of aromatic compounds is also discussed.
Introduction
Aromatic compounds are an important class of diverse chemicals. They have extensive applications in the production of various solvents, plastics, fine chemicals, food and feed additives, nutraceuticals, and pharmaceuticals (Averesch and Kromer, 2018; Wang J. et al., 2018). Moreover, many aromatic products have various biological activities and are widely used in pharmaceutical industries. Typically, aromatic compounds are chemically synthesized from petroleum-derived feedstocks, such as benzene, xylene, and toluene. With the progressive exhaustion of traditional feedstocks and the continuous deterioration of the environment, the biotechnological production of aromatic chemicals from renewable sugar feedstock in an eco-friendly manner has received considerable attention as a promising alternative for the synthesis of aromatic compounds.
Aromatic compounds can be produced by various plants, algae, fungi, and bacteria via the shikimate pathway. The shikimate pathway begins with the condensation of phosphoenolpyruvate (PEP) in the Embden-Meyerhof-Parnas pathway (EMP) and that of D-erythrose 4-phosphate (E4P) in the pentose phosphate pathway (PPP) to form 3-deoxy-D-arabino-heptulosonate-7-phosphate (DAHP). Six additional enzymatic steps are necessary for the synthesis of chorismite, which is converted to phenylpyruvate (PP) or 4-hydroxyphenylpyruvate (4HPP) via prephenate, through the Phe-Tyr branch that is catalyzed by chorismate mutase/prephenate dehydratase (PheA) or the chorismate mutase/prephenate dehydrogenase (TyrA) complex. Chorismate is also converted to anthranilate (AA) by the Trp branch in a reaction catalyzed by the anthranilate synthase (TrpED) complex. Finally, Phe, Tyr, and Trp are synthesized via their own biosynthetic pathways using PP, 4HPP, or anthranilate, respectively, as precursors (Figure 1). The enzymes and regulations involved in the aromatic amino acid biosynthetic pathway have been well-studied and characterized. Cao et al. (2020) well-summarized the transcriptional regulation mechanism. Herein, we only outline the feedback inhibition mechanism in Escherichia coli and Saccharomyces cerevisiae (Figure 1). In E. coli, DAHP synthase, which is encoded by aroF, aroG, and aroH is feedback-inhibited by L-tyrosine (L-Tyr), L-phenylalanine (L-Phe), and L-tryptophan (L-Trp), respectively. Additionally, aroF, aroG, and aroH are negatively regulated by TyrR, a transcriptional regulatory protein. Chorismate mutase or prephenate dehydrogenase encoded by tyrA or pheA is also regulated through feedback inhibition by L-Tyr or L-Phe, respectively. DAHP synthase, 3-dehydroquinate (DHQ) synthase (AroB), shikimate kinase (AroKL), 5-enolpyruvoylshikimate 3-phosphate (EPSP) synthase (AroA), and choirmate synthase (AroC) are the rate-limiting enzymes in the shikimate pathway (Dell and Frost, 1993). In S. cerevisiae, DAHP synthase is encoded by ARO4 and ARO3, and regulated through feedback inhibition by L-Tyr and L-Phe. ARO1 is a pentafunctional enzyme that catalyzes steps 2 through 6 during the biosynthesis of chorismate.
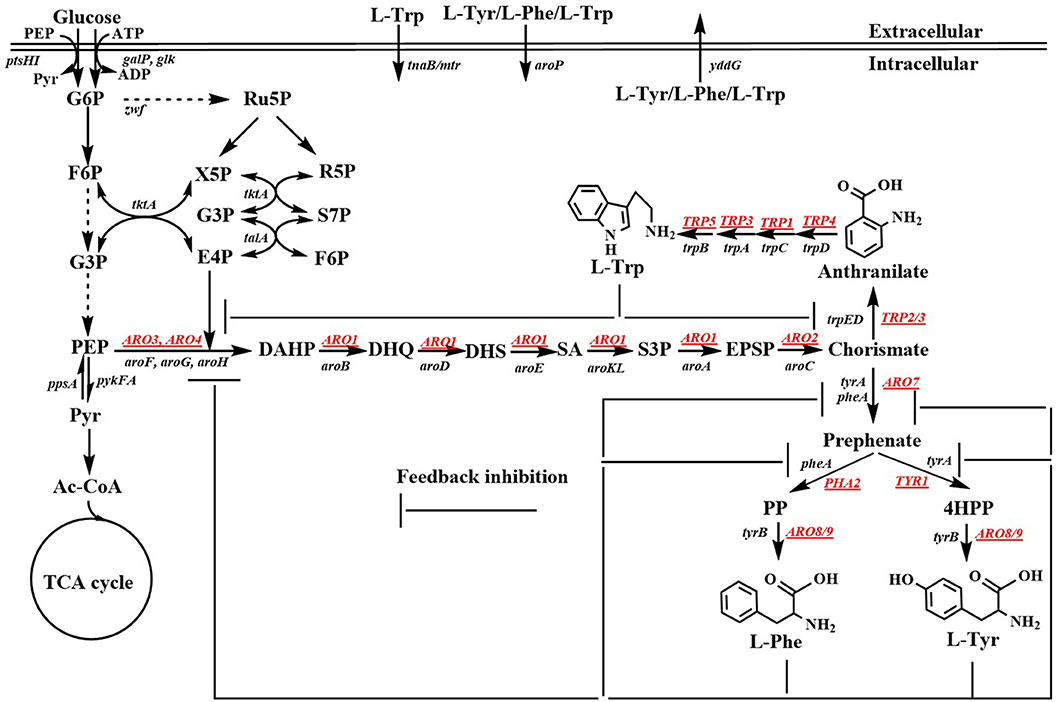
Figure 1. Biosynthetic pathway of aromatic amino acids. Black, Escherichia coli genes; Underlined genes, Saccharomyces cerevisiae genes; G6P, glucose-6-phosphate; F6P, fructose-6-phosphate; G3P, glyceraldehde-3-phosphate; PEP, phosphoenolpyruvate; Pyr, pyruvate; Ac-CoA, acetyl-CoA; TCA cycle, tricarboxylic acid cycle; Ru5P, ribulose-5-phosphate; X5P, xylulose-5-phosphate; R5P, ribose-5-phosphate; S7P, sedoheptulose-7-phosphate; E4P, D-erythrose 4-phosphate; DAHP, 3-deoxy-D-arabino-heptulosonate-7-phosphate; DHQ, 3-dehydroquinate; DHS, 3-dehydroshikimate; SA; shikimic acid; S3P, shikimate-3-phosphate; EPSP, 5-enolpyruvylshikimate 3-phosphate; PP, phenylpyruvate; 4HPP, 4-hydroxyphenylpyruvate; L-Phe, L-phenylanine; L-Tyr, L-tyrosine; L-Trp, L-tryptophan. ptsHI, phosphocarrier protein HPr and PTS enzyme I genes; galP, galactose, H (+) symporter gene; glk, glucokinase gene; zwf, glucose-6-phosphate dehydrogenase gene; tkt, transketolase 1 gene; tal, transaldolase A gene; ppsA, phosphoenolpyruvate synthase; pykFA, pyruvate kinase I and II genes; aroF/aroG/aroH, 3-deoxy-D-arabino-heptulosonate-7-phosphate synthase genes; aroB, 3-dehydroquinate synthase gene; aroD, 3-dehydroquinate dehydratase gene; aroE, dehydroshikimate reductase gene; aroK/aroL, shikimate kinase genes; aroA, EPSP synthase gene; aroC, chorismate synthase gene; pheA/tyrA, chorismate mutase/prephenate dehydrogenase gene; tyrB, aromatic-amino-acid transaminase gene; trpD/trpE, anthranilate synthase gene; trpC, phosphoribosylanthranilate isomerase gene; trpA, indoleglycerol phosphate aldolase gene; trpB, tryptophan synthase gene; tnaB/mtr, tryptophan: H (+) symporter gene; aroP, aromatic amino acid: H (+) symporter gene; yddG, aromatic amino acid exporter gene; ARO3/ARO4, 3-deoxy-D-arabino-heptulosonate-7-phosphate synthase genes; ARO2, chorismate synthase; ARO7, chorismate mutase gene; TYR1, prephenate dehydrogenase gene; ARO8/ARO9, aromatic acid aminotransferase gene; PHA2, prephenate dehydratase gene; TRP2/TRP3, anthranilate synthase gene; TRP4, anthranilate phosphoribosyl transferase gene; TRP1, N-(5'-phosphoribosyl)-anthranilate isomerase gene; TRP3, indole-3-glycerol phosphate synthase gene; TRP5, tryptophan synthetase gene.
With the developments of metabolic engineering and synthetic biology, significant advances have been made by engineering the shikimate pathway to produce natural or non-natural aromatic compounds at the gram level in microorganisms, especially in E. coli and S. cerevisiae (Averesch and Kromer, 2018; Wang J. et al., 2018; Wu et al., 2018; Huccetogullari et al., 2019). Here, we provide a comprehensive and systematic overview of the recent advancements in metabolic engineering and synthetic biology used to achieve the microbial production of aromatic compounds derived from aromatic amino acids. These aromatic compounds include aromatic amino acid derivatives, stilbenes, and benzylisoquinoline alkaloids. More aromatic amino acid derivatives were reviewed in this study. Some groups gave some good reviews about the metabolic engineering strategies and the progress for the production of aromatic amino acid derivatives before 2019 (Averesch and Kromer, 2018; Huccetogullari et al., 2019; Cao et al., 2020). Overexpression of the feedback-resistant mutants and transketolase are the common strategies for engineering microorganisms to produce aromatic ammonic acids. These strategies will not be specially mentioned. To avoid repeating, we will focus on the production of aromatic compounds using metabolic engineering microorganisms.
Production of Aromatic Amino Acid Derivatives
Aromatic amino acid derivatives are a class of important aromatic compounds derived from aromatic amino acids.
Phenylalanine Derivatives
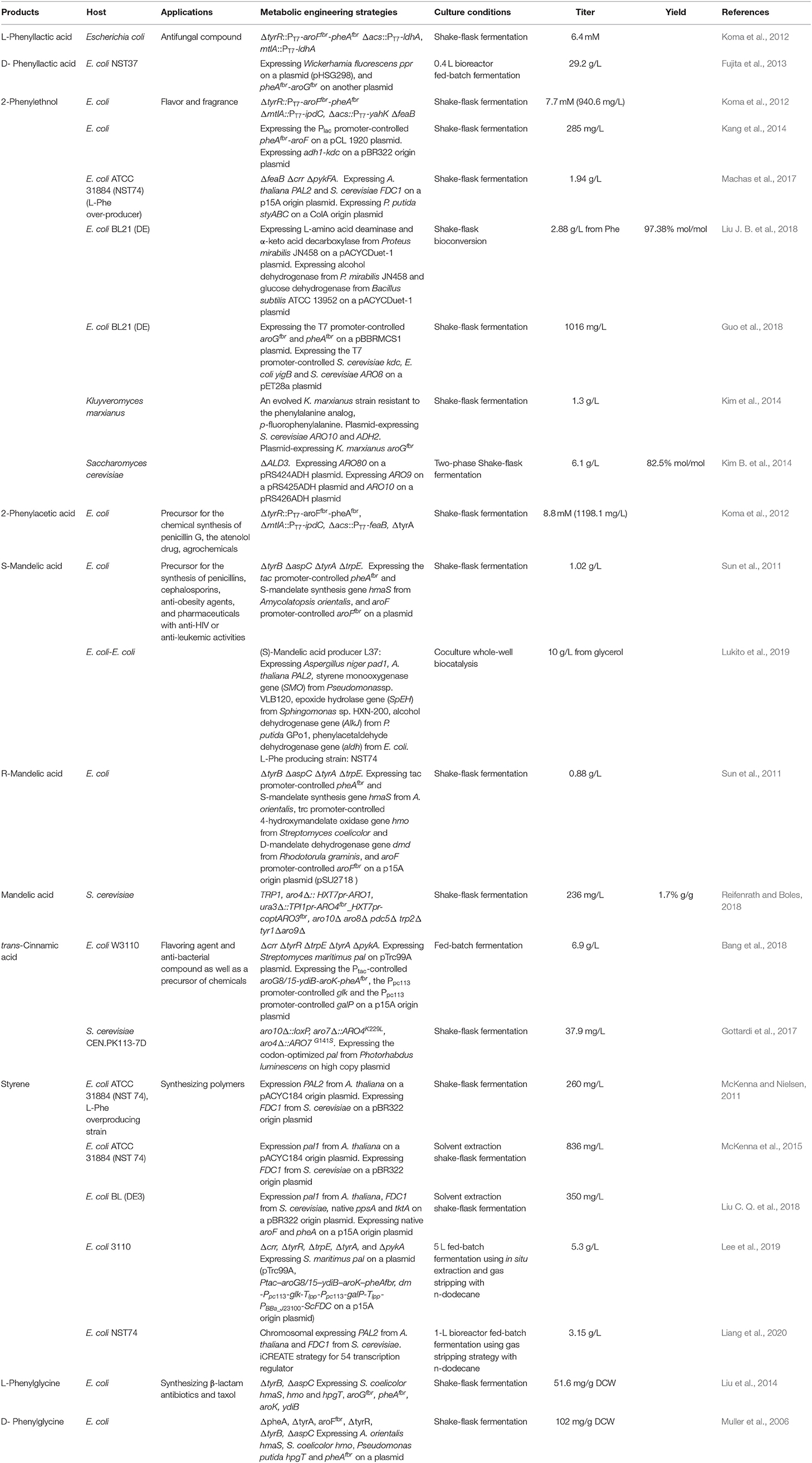
Many aromatic chemicals can be derived from L-Phe or its precursor phenylpyruvate by a suitable biosynthetic pathway (Figure 2, Table 1). Phenylpyruvate derivatives, such as phenyllactic acid (PLA), 2-phenylethanol (2PE), 2-phenylacetic acid and mandelic acid, are synthesized using phenylpyruvate as a common precursor (Figure 2, Table 1). PLA is an antifungal compound. The replacement of the acs and mtlA with the Cupriavidus necator JCM20644 lactate dehydrogenase gene ldhA under the control of the T7 promoter in the L-Phe producing E. coli strain resulted in the production of 6.4 mM of L-PLA from glucose in shake flask cultures (Koma et al., 2012). Overexpression of Wickerhamia fluorescens phenylpyruvate reductase gene (ppr), native feedback-resistant aroGfbr and pheAfbr in the L-Phe producing strain E. coli NST37 resulted in the production of 29.2 g/L of D-PLA from glucose in 0.4 L bioreactor fed-batch fermentations (Fujita et al., 2013). They also found that the replacement of W. fluorescens ppr with lactate dehydrogenase gene (L-ldh) from Pediococcus acidilactici generated L-PLA. Recently, whole-cell biocatalysis has been successfully used to produce L-PLA from L-Phe or phenylpyruvate. An E. coli expressing Lactobacillus plantarum lactate dehydrogenase gene (L-ldh), Proteus mirabilis L-amino acid deaminase gene (L-aad), and Candida boidinii formate dehydrogenase gene (fdh) was constructed for the production of L-PLA by using whole-cell biocatalysis (Hou et al., 2019). The resulting strain produced 54.0 g/L of L-PLA from L-Phe with the aid of glucose, which was used as a co-substrate for cofactor regeneration.
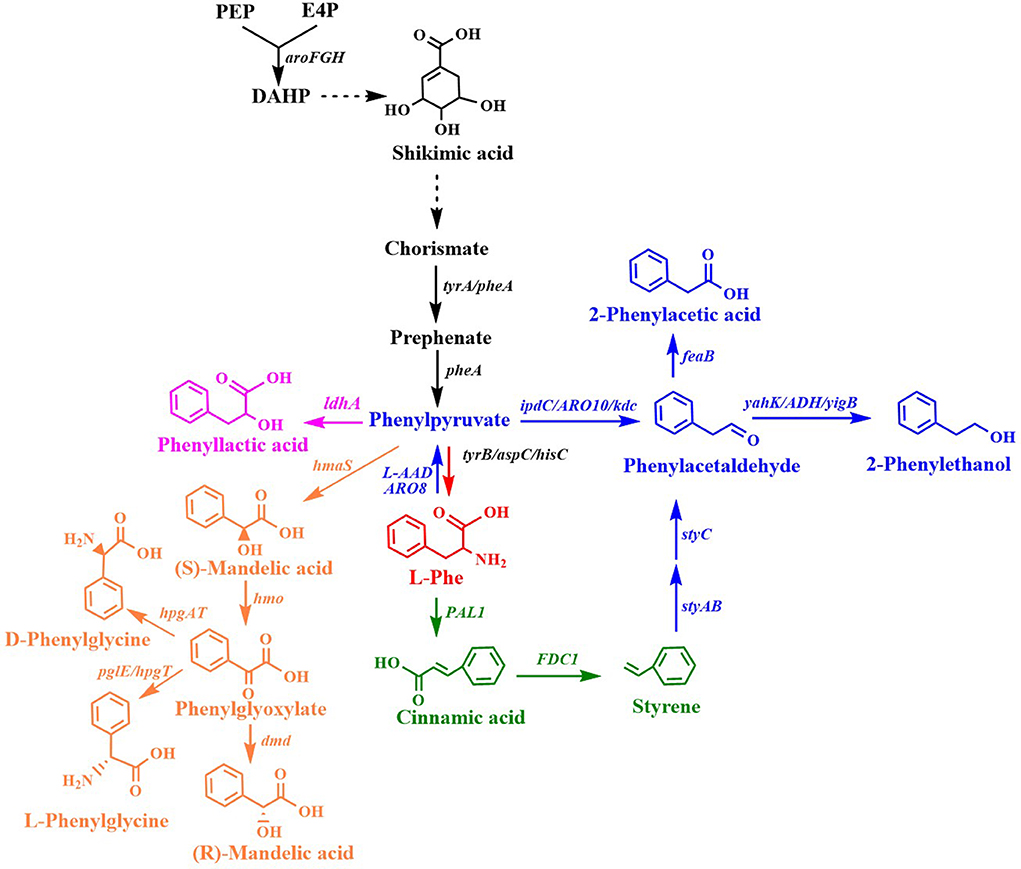
Figure 2. Biosynthesis of L-phenylalanine derivatives. ldhA, lactate dehydrogenase gene; ipdC/ARO10/kdc, phenylpyruvate decarboxylase gene; yahK/ADH6/yigB/adh, aldehyde reductase; feaB, phenylacetaldehyde dehydrogenase gene; L-AAD, L-amino acid deaminase gene; ARO8: PAL1, phenylalanine ammonia lyase gene; FDC1, ferulic acid decarboxylase 1 gene; styAB, styrene monooxygenase genes; styC, styrene oxide isomerase gene; hmaS, S-mandelate synthesis gene; hmo, l-4-hydroxymandelate oxidase gene; hpgT, l-4-hydroxyphenylglycine transaminase gene; pglE, phenylglycine aminotransferase; hpgAT, D-(4-Hydroxy)phenylglycine aminotransferase gene; dmd, D-mandelate dehydrogenase gene.
2PE, which has high-value as flavor and fragrance compound, has a wide range of applications in the cosmetic, perfumery, and food industries. The indol-3-pyruvate/phenylpyruvate decarboxylase gene ipdC from Azospirillum brasilense NBRC102289 was introduced into a Phe overproducing E. coli strain, resulting in the production of 2PE (Figure 2) (Koma et al., 2012). The chromosomal overexpression of A. brasilense ipdC and E. coli yahK, and the deletion of feaB resulted in the production of 7.7 mM (940.6 mg/L) of 2PE from glucose in shake flask cultures (Koma et al., 2012). The overexpression of the phenylpyruvate decarboxylase gene kdc from Pichia pastoris GS115 and alcohol dehydrogenase (ADH) gene adh1 from S. cerevisiae S288c in E. coli resulted in the production of 2PE (Kang et al., 2014). The co-overexpression of the four genes kdc, adh1, pheAfbr, and aroF improved 2PE production to 285 mg/L from glucose in shake flask cultures. A novel route (styrene-derived pathway) has been established for 2PE production in E. coli (Machas et al., 2017). The styrene-derived pathway comprised PAL2 from Arabidopsis thaliana, FDC1 from S. cerevisiae, and styABC from Pseudomonas putida S12. The introduction of the styrene-derived pathway, as well as the deletion of the competing pathways (feaB, crr, and pfkFA) resulted in the production of 1.94 g/L of 2PE from glucose in shake flask cultures. The results also demonstrated that the styrene-derived pathway more efficiently produced 2PE than the Ehrlich pathway. The pathway for 2PE production, which involves L-amino acid deaminase, α-keto acid decarboxylase, and ADH, was mimicked from the 2PE natural producer P. mirabilis JN458 (Liu J. B. et al., 2018). The recombinant E. coli harboring this P. mirabilis pathway produced 2.88 g/L 2PE from Phe with a molar yield of 97.38%. The plasmid-based overexpression of S. cerevisiae kdc, E. coli yigB, and S. cerevisiae aro8, as well as the enhancement of the expression of the feedback-resistant aroGfbr and pheAfbr in E. coli resulted in the production of 1,016 mg/L 2PE from glucose (Guo et al., 2018). After the introduction of alcohol acetyltransferase gene ATF1 from S. cerevisiae into the 2PE producer, 2-phenylethyl acetate was produced and achieved 687 mg/L. Kiuyveromyces marxianus, S. cerevisiae, and P. pastoris are typical yeast systems that have been engineered to synthesize 2PE. The phenylpyruvate decarboxylase (ARO10) and ADH (ADH2) genes of S. cerevisiae were expressed in an evolved K. marxianus that was resistant to the phenylalanine analog, p-fluorophenylalanine, which resulted in 2PE production (Kim et al., 2014). The overexpression of the feedback-resistant aroGfbr from K. marxianus further increased the production of 2PE from glucose to 1.3 g/L. In S. cerevisiae, the overexpression of ARO9, ARO10, and ARO80 (the latter is a transcription activator of ARO9 and ARO10), and the deletion of ALD3 improved 2PE production (Kim B. et al., 2014). After optimizing the fermentation conditions, the engineered S. cerevisiae produced 6.1 g/L of 2PE from glucose with a molar yield of 82.5%. ARO8, which encodes aromatic aminotransferase I, was identified to be a transcriptional regulator of ARO10 (Romagnoli et al., 2015). The deletion of ARO8 improved 2PE production in S. cerevisiae (Romagnoli et al., 2015). P. pastoris, a type of the methylotrophic yeast, can rapidly grow to extremely high cell densities in very simple and defined media. P. pastoris can naturally synthesize low levels of 2PE. Overexpression of the biosynthetic pathway of 2PE from S. cerevisiae (ARO10-ADH6) in P. pastoris enhanced the 2PE titer to 416 mg/L from 26 mg/L (Kong et al., 2020). Increasing the availability of the precursor of phenylpyruvate furtherly improved the 2PE titer to 1,169 mg/L by overexpressing S. cerevisiae ARO8, E. coli aroGfbr and pheAfbr.
2-Phenylacetic acid and 4-hydroxyphenylacetic acid (4HPAA) are intermediates in the chemical synthesis of penicillin G, atenolol, agrochemicals, and other compounds. The production of 2-phenylacetic acid was achieved by the overexpression of endogenous feaB and ipdC from A. brasilense NBRC102289 in E. coli using the Ehrlich pathway intermediates phenylacetaldehyde as precursors (Figure 2) (Koma et al., 2012). The overexpression of A. brasilense ipdC and E. coli feaB, as well as the deletion of tyrA in the Phe overproducing E. coli strain resulted in the de novo production of 8.8 mM (1,198.1 mg/L) of 2-phenylacetic acid in shake flask cultures (Koma et al., 2012).
Mandelic acid and 4-hydroxymandelic acid (4HMA) are valuable aromatic fine chemicals used as precursors for the production of pharmaceuticals, flavors, and cosmetics. Mandelic acid has been widely used in the synthesis of semisynthetic penicillins, cephalosporins, anti-obesity agents, and pharmaceuticals with anti-HIV or anti-leukemic activities, as well as for the resolution of racemic alcohols and amines (Reifenrath and Boles, 2018). The biosynthetic pathway for mandelic acid production has been successfully established in E. coli (Sun et al., 2011) and S. cerevisiae (Reifenrath and Boles, 2018). The biosynthetic pathway for mandelic acid (Figure 2) was first established in E. coli by introducing the S-mandelate synthesis gene hmaS from Amycolatopsis orientalis (Sun et al., 2011). The overexpression of hmaS from A. orientalis and the feedback-resistant pheAfbr and aroFfbr, along with the deletion of the competing pathways resulted in the de novo production of 1.02 g/L of S-mandelic acid in shake flask cultures. The co-expression of the 4-hydroxymandelate oxidase gene hmo from Streptomyces coelicolor and D-mandelate dehydrogenase gene dmd from Rhodotorula graminis in the aforementioned S-mandelic acid producing strain led to the de novo production of 0.88 g/L of R-mandelic acid in shake flask cultures. S-Mandelate synthase HmaS from A. orientalis can convert 4-hydroxyphenylpyruvate (4HPP) to 4HMA, but also phenylpyruvate to mandelic acid in E. coli (Sun et al., 2011; Li et al., 2016). It was also found that the introduction of codon-optimized hmaS from A. orientalis in S. cerevisiae produced mandelic acid (Reifenrath and Boles, 2018). Mandelic acid production was increased by the replacement of hmaS from A. orientalis with the corresponding gene from Nocardia uniformis. The strategies used for the enhancement of the expression of the aromatic amino acid pathway as well as the deletion of the competing pathways were used to improve the production of mandelic acid. The resulting strain produced 236 mg/L mandelic acid with a mass yield of 1.7% in shake flask cultures (Reifenrath and Boles, 2018). Lukito et al. (2019) developed a whole cell-based cascade biotransformation for the production of (S)-mandelic acid from styrene, L-Phe, glucose, or glycerol. The recombinant E. coli strain LZ37 expressing the 6-step enzyme cascades produced 160 mM (S)-mandelic acid from L-Phe with a yield of 80%. Coupling the E. coli strain LZ37 with the L-Phe producing E. coli strain NST74 enabled the de novo production of 63 mM (10 g/L) or 52 mM (8 g/L) (S)-mandelic acid from glycerol and glucose, respectively.
Cinnamic acid is used as a flavoring agent and antibacterial compound. The overexpression of Streptomyces maritimus L-phenylalanine ammonia lyase gene pal in an E. coli strain overproducing L-Phe resulted in the production of 6.9 g/L of trans-cinnamic acid in a fed-batch fermentation process (Figure 2) (Bang et al., 2018). It was demonstrated that the bacterial phenylalanine ammonia lyase from Photorhabdus luminescens was superior to the plant enzyme from A. thaliana for the production of trans-cinnamic acid in yeast (Gottardi et al., 2017). After optimizing the biosynthetic pathway, the overexpression of ARO4K229L, ARO7G141S, and codon-optimized pal from P. luminescens, and deletion of ARO10 in S. cerevisiae CEN.PK113-7D resulted in the production of 37.9 mg/L of trans-cinnamic acid (Gottardi et al., 2017).
Styrene and its derivatives act as monomers and petroleum-based feedstocks, which is valuable because they can be used as raw materials in industrial processes. They have many uses, including in the manufacture of polystyrenes, plastics, and styrene-butadiene. Styrene can be synthesized from glucose by the co-expression of phenylalanine ammonia lyase and trans-cinnamate decarboxylase (Figure 2). Mckenna and Nielsen screened candidate isoenzymes for the two steps from bacterial, yeast, and plant (McKenna and Nielsen, 2011). Finally, the overexpression of PAL2 from A. thaliana and FDC1 from S. cerevisiae in an L-Phe overproducing E. coli strain led to the accumulation of 260 mg/L of styrene in shake flask cultures. To avoid the effects of the toxicity of styrene, solvent extraction fermentation was used to improve the amount of styrene produced, which achieved 836 mg/L (McKenna et al., 2015). A similar strategy was also used for achieving styrene production in E. coli BL (DE3), which yielded 350 mg/L of styrene in an extraction fermentation process using isopropyl myristate (Liu C. Q. et al., 2018). This titer was achieved by the overexpression of PAL2 from A. thaliana, FDC1 from S. cerevisiae, native ppsA, tktA, aroF, and pheA, using a two-plasmid system. An L-Phe overproducing E. coli strain overexpressing S. maritimus pal and FDC1 from S. cerevisiae was engineered for the production of styrene (Lee et al., 2019). The resulting strain produced 5.3 g/L of styrene in a 5-L fed-batch fermentation process using in situ extraction and gas stripping with n-dodecane. The styrene synthetic pathway containing PAL2 from A. thaliana and FDC1 from S. cerevisiae was integrated into the SS9 site of an L-Phe overproducing strain E. coli NST 74 to obtain a styrene producing strain (Liang et al., 2020). After optimization of the expression levels of the two genes PAL2 and FDC1, iCREATE strategy for 54 transcription regulator genes was used to improve styrene-tolerance and the production of styrene. The resulting strain produced 3.15 g/L in a 1-L bioreactor fed-batch fermentation using gas stripping strategy with n-dodecane. Although S. cerevisiae (McKenna et al., 2014) and S. lividans (Fujiwara et al., 2016b) have been used as host strains for styrene production, the titer (about 30 mg/L) is much lower than that obtained using engineered E. coli.
Phenylglycine, as one of the unnatural amino acids, has been widely used in the synthesis of penicillin, virginiamycin S, pristinamycin I, and the antitumor compound taxol (Liu et al., 2014). Phenylglycine is synthesized from phenylpyruvate via phenylglyoxylate (Figure 2). An artificial biosynthetic pathway, which consists of HmaS (l-4-hydroxymandelate synthase), Hmo (l-4-hydroxymandelate oxidase), and HpgT (l-4-hydroxyphenylglycine transaminase) from S. coelicolor, was constructed for the production of L-phenylglycine in E. coli (Liu et al., 2014). Deletions of both tyrB and aspC as well as increasing the copies of both hmo and hpgT further increased the production of L-phenylglycine to 51.6 mg/gDCW (Liu et al., 2014). A novel L-phenylglycine biosynthetic pathway via the PglE reaction was reported in Streptomyces pristinaespiralis (Osipenkov et al., 2018). Zhou et al. (2016) constructed an artificial eight-step biosynthetic pathway containing 10 different enzymes for the production of L-phenylglycine from L-Phe. E. coli expressing the pathway produced 34 mM (5.1 g/L) L-phenylglycine with a yield of 85% from L-Phe. An artificial biosynthetic pathway was created for the production of D-phenylglycine in E. coli (Muller et al., 2006). The pathway consists of HmaS from A. orientalis, Hmo from S. coelicolor, and HpgAT from P. putida. This pathway was introduced into the L-Phe producing E. coli strain KB532 for the de novo production of D-phenylglycine. The deletion of both tyrB and aspC further improved the production of D-phenylglycine, which reached 102 mg/g DCW (Muller et al., 2006). Zhou Y. et al. (2017) created a biosynthetic pathway for the production of D-phenylglycine from mandelic acid, styrene, or L-Phe. The biosynthetic pathway has eight steps catalyzed by nine enzymes: A. thaliana PAL2, Aspergillus niger phenylacrylic acid decarboxylase (PAD1), styrene monooxygenase (SMO) from Pseudomonas sp. VLB120, epoxide hydrolase (SpEH) from Sphingomonas sp. HXN-200, alcohol dehydrogenase gene (AlKJ) from P. putida GPo1, phenylacetaldehyde dehydrogenase (Aldh) from E. coli, (S)-mandelate dehydrogenase (SMDH) from P. putida ATCC 12633, D-phenylglycine aminotransferase (DpgAT) from Pseudomonas stutzeri ST-201 and glutamate dehydrogenase (GluDH) from E. coli. E. coli expressing the nine enzymes of the eight-step pathway produced 50 mM D-phenylglycine from 60 mM L-Phe with a yield of 83% using whole-cell catalysis (Zhou Y. et al., 2017).
Tyrosine Derivatives
L-Tyr derivatives were synthesized from 4-hydroxyphenylpyruvate (4HPP) and L-Tyr (Figure 3). Many L-Tyr derivatives were also used as a precursor for the production of a wide range of valuable aromatic compounds with pharmaceutical value (Table 2).
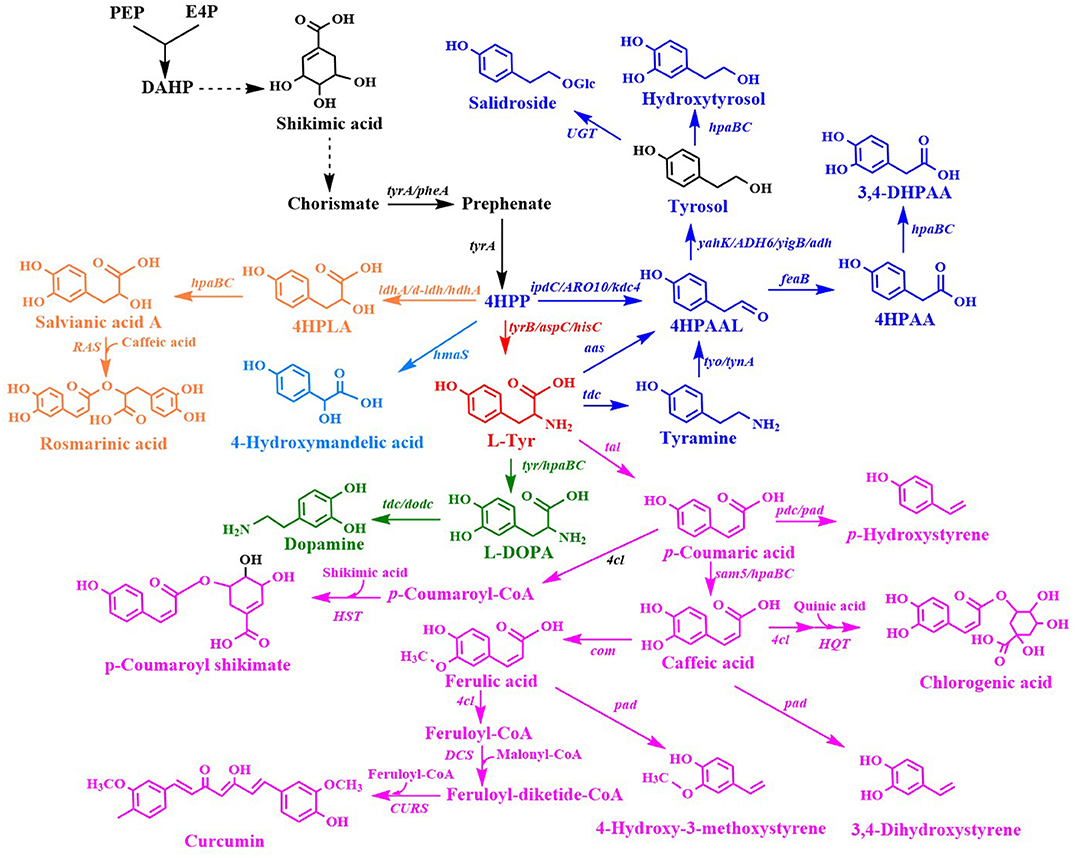
Figure 3. Biosynthesis of L-tyrosine derivatives. 4HPLA, 4-hydroxyphenyllactic acid; 4HPAAL, 4-hydroxyphenylacetaldehyde; 4HPAA, 4-hydroxyphenylacetic acid; ldhA/d-ldh, D-lactate dehydrogenase gene; hdhA, 2-hydroxyacid dehydrogenase gene; hpaBC, p-hydroxyphenylacetate 3-hydroxylase genes; ipdC/ARO10/kdc, phenylpyruvate decarboxylase gene; yahK/ADH6, alcohol dehydrogenase gene; UGT, uridine diphosphate dependent glycosyltransferase gene; aas, aromatic aldehyde synthase gene; RAS, rosmarinic acid synthase gene; tyr, tyrosinase gene; tdc, tyrosine decarboxylase gene; tyo/tynA, tyramine oxidase gene; tal, tyrosine ammonia lyase gene; dodc, L-DOPA decarboxylase; pdc, p-coumaric acid decarboxylase gene; 4cl, 4-coumaroyl-coenzyme A ligases gene; sam5, 4-coumarate hydroxylase gene; com, caffeic acid methyltransferase gene; HQT, hydroxycinnamate-CoA quinate transferase gene; HST, hydroxycinnamate-CoA shikimate transferase gene; pad, phenolic acid decarboxylase gene; CURS, curcumin synthase gene.
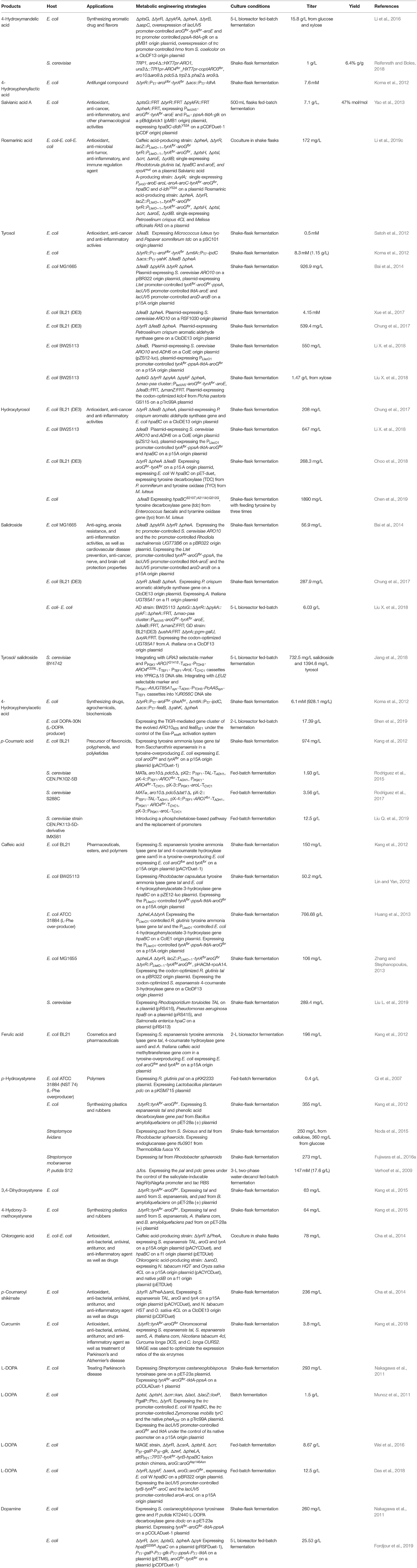
4HMA serves as a building block for the synthesis of aromatic drugs and flavors. The biosynthetic pathway for 4HMA production has been successfully established in E. coli and S. cerevisiae (Li et al., 2016; Reifenrath and Boles, 2018). The overexpression of the codon-optimized hmaS from A. orientalis in the tyrosine-producing strain, E. coli BKT5, resulted in the formation of 4HMA (Figure 3) (Li et al., 2016). After optimizing the expression of hmaS by fine-tuning four promoters of different strengths which were combined with three plasmids with different copy numbers, and blocking the competing pathway by deleting tyrB and aspC, 4HMA production was improved. The resulting strain produced 15.8 g/L 4HMA from a mixture of glucose and xylose in a 5-L bioreactor fed-batch fermentation process. It was also found that the introduction of the codon-optimized hmaS from A. orientalis in S. cerevisiae produced 4HMA (Reifenrath and Boles, 2018). The production of 4HMA was increased by the replacement of hmaS from A. orientalis with the corresponding gene from N. uniformis. The strategies used for the enhancement of the expression of the aromatic amino acid pathway as well as the deletion of the competing pathways were used to improve the production of 4HMA. The strain exhibiting the best performance produced 1 g/L of 4HMA with a mass yield of 6.4% in shake flask cultures (Reifenrath and Boles, 2018).
4-Hydroxyphenyllactic acid (4HPLA) is known to be an antifungal compound. The replacement of acs with C. necator JCM20644 ldhA under the control of the T7 promoter in the L-Tyr producing E. coli strain resulted in the production of 7.6 mM 4HPLA in shake flask cultures (Figure 3) (Koma et al., 2012).
Salvianic acid A (3,4-dihydroxyphenllactic acid, also known as danshensu), is a naturally occurring plant polyphenolic acid that is recognized for its superior antioxidant activities. It has a variety of other pharmacological activities, including improved cerebral blood flow, inhibition of platelet activation and arterial thrombosis, as well as anti-cancer and anti-inflammatory activities. Yao et al. developed an artificial biosynthetic pathway for salvianic acid A production in E. coli (Yao et al., 2013). 4-Hydroxyphenylpyruvate was converted to salvianic acid A via D-lactate dehydrogenase (encoded by d-ldh from Lactobacillus pentosus) and the hydroxylase complex (encoded by hpaBC from E. coli) (Figure 3). After optimizing the pathway using a modular engineering approach and deleting the genes involved in regulatory and competing pathways, the metabolically engineered E. coli strain produced 7.1 g/L of salvianic acid A with a yield of 0.47 mol/mol glucose. To overcome the drawbacks caused by plasmid system, the salvianic acid A biosynthetic pathway was integrated into the chromosomal of the L-Tyr producing E. coli strain BAK5 to obtain a plasmid-free strain for the production of salvianic acid A (Zhou L. et al., 2017). The final strain BKD13 produced 5.6 g/L salvianic acid A by fed-batch fermentation. A cell-free system was established for the production of salvianic acid A from phenylpyruvate. The cell-free system contained D-mandelate dehydrogenase from Thermococcus barophilus, phenylalanine 4-hydroxylase from Thermomonospora curvata, and Hydroxyphenylacetate 3-hydroxylase from Sulfobacillus acidophilus TPY. The resulting cell-free system produced 392 mM of salvianic acid A with a yield of 85% (Li et al., 2019a).
Rosmarinic acid is a plant-derived natural compound belong to the family of polyphenolic compounds. It is an ester of caffeic acid and salvianic acid A. It has diverse important nutraceutical and pharmaceutical values, including antioxidant, anti-inflammatory, anti-microbial, anti-tumor and anti-viral, and immune regulation activities. After comparing pathway enzymes, Bloch and Schmidt-Dannert (2014) constructed an artificial biosynthetic pathway of rosmarinic acid (Figure 3), which consisted of 2-hydroxyacid dehydrogenase (HdhA) from Lactobacillus delbrueckii, 4-hydroxyphenylacetate 3-hydroxylase complex (HpaBC) from E. coli W, Tal from Rhodobacter sphaeroides, 4-coumarate-CoA ligase 2 (4CL2) from A. thaliana and rosmarinic acid synthase from Melissa officinalis. The introduction of this pathway into E. coli by using three plasmids resulted in the de novo production of rosmarinic acid. Jiang et al. (2016) established another biosynthetic pathway of rosmarinic acid using a different set of enzymes. The recombinant E. coli strain harboring this pathway produced 130 mg/L rosmarinic acid from caffeic acid using whole-cell biocatalysis. The biosynthetic pathway of rosmarinic acid from 4-hydroxyphenylpyruvate is a diverging and converging pathway. Modular co-culture engineering with three E. coli strains was used to balance the different pathway modules for the production of rosmarinic acid (Li et al., 2019c). The three-strain co-culture produced 172 mg/L rosmarinic acid, exhibiting 38-fold improvement over the mono-culture. Yan et al. (2019) established a cell-free system with ATP and CoA double regeneration for the production of rosmarinic acid, which reached 320.04 mg/L/h.
Tyrosol and hydroxytyrosol are two major ingredients in olive oil, and both are considered bioactive ingredients that have various biological activities. It has been reported that tyrosol plays an important role in the prevention of cardiovascular diseases, osteopenia, melanin pigmentation, as well as anti-inflammatory responses (Kim et al., 2018). In addition, tyrosol can lower the risk of developing Alzheimer's disease (Kim et al., 2018). Three biosynthetic pathways have been developed for tyrosol production (Figure 3). In the first pathway, tyrosine is converted into tyramine by tyrosine decarboxylase (TDC), which is followed by the further consecutive transformation of tyramine into tyrosol by tyramine oxidase (TYO) and ADH (Satoh et al., 2012). The second pathway is known as the yeast Ehrlich pathway, wherein 4HPP is decarboxylated to 4-hydroxyphenylacetaldehyde (4HPAAL), which is acted on by the endogenous enzyme ADH (Hazelwood et al., 2008; Bai et al., 2014; Xue et al., 2017). In the third pathway, tyrosine is directly converted to 4HPAAL by aromatic aldehyde synthase (AAS), and then converted to tyrosol by ADH (Chung et al., 2017; Jiang et al., 2018). The plasmid-mediated overexpression of the tyo gene from Micrococcus luteus and tyrosine decarboxylase gene from Papaver somniferum in the feaB-knockout mutant of E. coli BW25113 resulted in the production of 0.50 mM of tyrosol (Satoh et al., 2012) in shake flask cultures. The yeast Ehrlich pathway for tyrosol production was established by the overexpression of indol-3-pyruvate/phenylpyruvate decarboxylase gene ipdC from A. brasilense NBRC102289 (Koma et al., 2012). The co-overexpression of A. brasilense ipdC and E. coli yahK, as well as the deletions of feaB and pheA in a Tyr-overproducing E. coli strain resulted in the production of 8.3 mM (1.15 g/L) tyrosol in shake flask cultures (Koma et al., 2012). The yeast Ehrlich pathway for tyrosol production was also established by the overexpression of S. cerevisiae pyruvate decarboxylase gene ARO10 (Bai et al., 2014). A combination of the knock out of the competitive pathways and negative regulation, along with the enhanced expression of the pathway genes enhanced the level of tyrosol produced to 926.9 mg/L. In another study, ARO10 from S. cerevisiae was overexpressed in the pheA/feaB double knockouts of E. coli BL21 (DE3), which resulted in the synthesis of 4.15 mM (573.4 mg/L) tyrosol from glucose during 48-h shake flask culture (Xue et al., 2017). Recently, it was demonstrated that the overexpression of the S. cerevisiae ADH gene ADH6 further increased tyrosol production (Li X. et al., 2018). The resulting E. coli strain containing the yeast Ehrlich pathway produced 1,469 mg/L of tyrosol from tyrosine and 550 mg/L of tyrosol from a simple carbon source, such as glucose. The overexpression of E. coli hpaBC in the tyrosol-overproducing strain led to the production of 647 mg/L hydroxytyrosol from a simple carbon source, such as glucose (Figure 3) (Li X. et al., 2018). In another recent study, the kdc4 gene from P. pastoris GS115 was more efficient for the biosynthesis of tyrosol than the previously reported ARO10 gene from S. cerevisiae (Liu X. et al., 2018). After the deletion of genes involved in regulatory and competing pathways, the resulting E. coli strain produced 1.47 g/L tyrosol from xylose in shake flask cultures, which is the highest titer produced so far. An AAS-derived pathway was first established for tyrosol and hydroxytyrosol production by the overexpression of the aas gene from Petroselinum crispum (Chung et al., 2017). The three different aas genes from A. thaliana, Petunia hybrid, and P. crispum were compared, and it was found that the P. crispum aas gene was most suitable for the production of tyrosol and hydroxytyrosol in E. coli. The deletion of tyrR, pheA, and feaB further increased tyrosol production. The overexpression of the aas gene from P. crispum in the tyrR/pheA/feaB knockouts of E. coli BL21 (DE3) resulted in the production of 539.4 mg/L of tyrosol in shake flask cultures (Chung et al., 2017). The authors also found that the titer of the product generated using the AAS-derived pathway was higher than that generated using the TDC-TYO-derived pathway. Hydroxytyrosol can be synthesized from tyrosol by hydroxylation (Figure 3). Chung et al. (2017) compared E. coli hpaBC and Saccharothrix espanaensis sam5 and found that the co-overexpression of E. coli hpaBC and P. crispum aas in E. coli produced greater levels of hydroxytyrosol. The plasmid overexpression of E. coli hpaBC and P. crispum aas in the tyrosol-overproducing strain resulted in the production of 208 mg/L of hydroxytyrosol in shake flask cultures (Chung et al., 2017). Judging from the reported titers of tyrosol, the yeast Ehrlich pathway has the highest efficiency among the three pathways, subsequent to the AAS-derived pathway. Salidroside is a glucoside of tyrosol and one of the major ingredients of the medicinal herb Rhodiola. It has various biological properties, including anti-aging, anoxia resistance, and anti-inflammation activities, as well as cardiovascular disease prevention, anti-cancer, nerve protection, and brain cell protection properties (Bai et al., 2014). It can be synthesized from tyrosol by a uridine diphosphate dependent glycosyltransferase (UGT) (Figure 3). Salidroside was first produced by introducing the glycosyltransferase UGT73B6 from Rhodiola sachalinensis into the tyrosol-overproducing E. coli strain (Bai et al., 2014). The resulting E. coli strain produced 56.9 mg/L of salidroside in shake flask cultures. Twelve UGTs from A. thaliana were screened, and UGT85A1 was found to be the most suitable enzyme for the production of salidroside from tyrosol (Chung et al., 2017). The overexpression of A. thaliana UGT85A1 in the tyrosol-overproducing E. coli strain with the AAS-derived pathway resulted in the production of 287.9 mg/L salidroside in shake flask cultures. The results of the two studies described above showed that most of the tyrosol was not glycosylated into salidroside (Bai et al., 2014; Chung et al., 2017). To overcome the drawbacks observed in E. coli monocultures, a syntrophic E. coli-E. coli co-culture system was developed for salidroside production (Liu X. et al., 2018). The co-culture system composed of the aglycone (AG) and the glycoside (GD) strains, which convergently accommodated the biosynthetic pathways for the production of tyrosol and salidroside, respectively. The co-culture of the AG and DG strains resulted in the production of 6.03 g/L salidroside in a 5-L bioreactor fed-batch fermentation process (Liu X. et al., 2018). Tyrosol is a native metabolite of yeast that is derived from endogenous Ehrlich pathway. Thus, S. cerevisiae has been successfully used as a host strain for tyrosol production. An AAS-derived pathway has been successfully introduced into S. cerevisiae for tyrosol and salidroside production (Jiang et al., 2018). Plasmid overexpressing of aas from P. crispum in S. cerevisiae improved the production tyrosol to 398.9 mg/L from 170.8 mg/L. Chromosomal overexpression of P. crispum aas, A. thaliana UGT85A1 as well as the enhancement of the expressions of ARO4K229L, ARO7G141S, and AROL resulted in the production of tyrosol and salidroside to 1394.6 and 732.5 mg/L, respectively (Jiang et al., 2018). An AAS-derived pathway containing P. crispum Aas and E. coli Adh was overexpressed for the production of tyrosol in S. cerevisiae (Guo et al., 2019). The replacement of PDC1 with E. coli tyr mutant tyrAM53I/A354V further improved the production of tyrosol by 440 times. The introduction of tyrosine decarboxylase from P. somniferum, tyrosine oxidase from M. luteus, and 4-hydroxyphenylacetate 3-monooxygenase from E. coli in a tyrosine-overproducing E. coli strain resulted in the production of 268.3 mg/L of hydroxytyrosol (Choo et al., 2018). A HpaBC mutant (HpaBCS210T/A211M/Q212G) active on both tyrosol and tyramine was obtained by directed divergent evolution (Chen et al., 2019). Then, they constructed a multiple pathway network to convert tyrosine to hydroxytyrosol using the same set of enzymes. Overexpression of the hpaBC mutant, tyrosine decarboxylase gene (tdc) from Enterococcus faecalis and tyramine oxidase gene (tyo) from M. luteus in the feaB deletion E. coli strain enabled the production of hydroxytyrosol of 1,890 mg/L (Chen et al., 2019). A whole-cell catalytic method was developed for the production of hydroxytyrosol from L-DOPA (Li et al., 2019b). Aromatic amino acid aminotransferase (TyrB) from E. coli and aldehyde reductase (YahK) from E. coli, a-keto acid decarboxylase (Kdc) from P. mirabilis JN458, and L-glutamate dehydrogenase (Gdh) were co-expressed in E. coli to catalyze the production of hydroxytyrosol from L-DOPA. The yield of hydroxytyrosol reached 36.33 mM (5.59 g/L) using whole-cell catalysis of the recombinant E. coli (Li et al., 2019b).
The production of 4HPAA was achieved by the overexpression of endogenous feaB and ipdC from A. brasilense NBRC102289 in E. coli, using the Ehrlich pathway intermediates 4-hydroxyphenylacetaldehyde as precursors (Koma et al., 2012). The overexpression of A. brasilense ipdC and E. coli feaB, as well as the deletion of yahK/pheA in the Tyr-overproducing E. coli strain resulted in the production of 6.1 mM (928.1 mg/L) of 2-phenylacetic acid in shake flask cultures (Figure 3) (Koma et al., 2012). Our lab constructed an E. coli strain for the production of 4HPAA by applying a combinatorial strategy of directed evolution of pathway enzymes and quorum-sensing (QS)-based dynamic regulation of the pathway. The resulting strain produced 17.39 g/L 4HPAA in 2-L bioreactor fed-batch fermentation (Shen et al., 2019). The 4HPAA titer is the highest value reported to date.
Hydroxycinnamic acids are a class of hydroxylate aromatic acids synthesized via the phenylpropanoid pathway. They mainly include cinnamic acid, caffeic acid, p-coumaric acid, and ferulic acid. The biotechnological production of p-coumaric acid was first achieved in S. cerevisiae from phenylalanine by phenylalanine ammonia lyase (PAL)-catalyzed formation of cinnamic acid and sequential hydroxylation of cinnamic acid via the cytochrome P450-dependent enzyme 4-cinnamic acid hydroxylase (C4H) (Ro and Douglas, 2004). However, the lower expression of C4H led to the production of an extremely low level of p-coumaric acid. To improve p-coumaric acid production, a more straightforward route that employed tyrosine as the direct precursor was established in E. coli. The overexpression of the tyrosine ammonia lyase gene tal from S. espanaensis in a tyrosine-overproducing E. coli strain resulted in the production of 974 mg/L of p-coumaric acid in shake flask cultures (Figure 3) (Kang et al., 2012). The overexpression of the heterologous biosynthetic pathway of caffeic acid (including S. espanaensis tyrosine ammonia lyase gene tal and 4-coumarate hydroxylase gene sam5) or ferulic acid (including tyrosine ammonia lyase gene tal, 4-coumarate hydroxylase gene sam5 from S. espanaensis, and caffeic acid methyltransferase gene com from A. thaliana) (Figure 3) led to the production of 150 mg/L of caffeic acid or 196 mg/L ferulic acid, respectively (Kang et al., 2012). This pathway was also introduced in S. cerevisiae to enable the production of 1.9 g/L of p-coumaric acid in a deep-well plate fed-batch fermentation process (Rodriguez et al., 2015). This titer was obtained by the inactivation of phenylpyruvate decarboxylase and pyruvate decarboxylase, in conjunction with the overexpression of genes encoding for feedback-resistant DAHP synthase, chorismate mutase, and shikimate kinase (Rodriguez et al., 2015). Then, this research group further enhanced p-coumaric acid titer to 3.56 g/L by deleting the amino acid transporter gene tat1 (Rodriguez et al., 2017). Recently, this group further enhanced the production of p-coumaric acid to 12.5 g/L by introducing a phosphoketalose-based pathway to divert glycolytic flux toward erythrose 4-phosphate synthesis, and optimizing carbon distribution between glycolysis and the aromatic amino acid biosynthetic pathway by promoter replacement (Liu Q. et al., 2019). The p-coumaric acid titer is the highest value reported to date. This pathway has also been introduced into P. putida KT2440 (Calero et al., 2016) and Synechocystis PCC 6803 (Xue et al., 2014; Tantong et al., 2018) to achieve the production of p-coumaric acid. However, the p-coumaric acid titer obtained with these engineered microorganisms was lower than that obtained with S. cerevisiae. Caffeic acid exhibits antioxidant, anti-viral, anti-cancerous, and anti-inflammatory activities. Caffeic acid is commonly synthesized by a cytochrome P450 enzyme p-coumarate 3-hydroxylase (C3H), which catalyzes the hydroxylation of p-coumaric acid (Figure 3). The overexpression of E. coli 4-hydroxyphenylacetate 3-hydroxylase gene hpaBC and Rhodobacter capsulatus tyrosine ammonia lyase gene tal in a tyrosine-overproducing E. coli strain expressing the PLlacO1-controlled tyrAfbr-ppsA-tktA-aroGfbr resulted in the production of 50.2 mg/L of caffeic acid (Lin and Yan, 2012). The titer was further enhanced to 766.68 mg/L by increasing the availability of tyrosine (Huang et al., 2013). Inclusion of the coumaroyl-CoA/caffeic -CoA biosynthesis route in the tyrosine ammonia lyase-4-coumarate 3-hydroxylase pathway did not improve caffeic acid production (Zhang and Stephanopoulos, 2013). The overexpression of Rhodotorula glutinis tal and S. espanaensis 4-coumarate 3-hydroxylase gene in a tyrosine-overproducing E. coli strain yielded 106 mg/L of caffeic acid in a 2-L bioreactor batch fermentation process (Zhang and Stephanopoulos, 2013). HpaB from Pseudomonas aeruginosa and HpaC from Salmonella enterica along with Tal from Rhodosporidium toruloides was overexpressed in S. cerevisiae for the production of caffeic acid, which reached 289.4 mg/L (Liu L. et al., 2019). This is the first report on engineering S. cerevisiae for the production of caffeic acid. The highest level of caffeic acid (10.2 g/L) could be produced from p-coumaric acid by using whole-cell catalysis of engineered E. coli (Furuya and Kino, 2014). This level was much higher than that obtained from glucose, indicating that the availability of p-coumaric acid and tyrosine might be key factors influencing caffeic acid production in E. coli. From the caffeic acid yield reported in the literature, the titer using whole-cell catalysis is much higher than that obtained by using fermentation. This reason may be the high toxicity of caffeic acid on host strain.
p-Hydroxystyrene is a derivative of styrene. It has the same applications as styrene. The de novo biosynthesis of p-hydroxystyrene from glucose was achieved in E. coli by the decarboxylation of p-coumaric acid (Figure 3). The L-Phe overproducing strain E. coli ATCC 31884 expressing R. glutinis pal and the p-coumaric acid decarboxylase gene pdc from L. plantarum produced 0.40 g/L of p-hydroxystyrene in a 14-L fermenter (Qi et al., 2007). Recently, an artificial biosynthetic pathway was established in E. coli to produce p-hydroxystyrene using the tyrosine ammonia lyase gene tal from S. espanaensis and phenolic acid decarboxylase gene pad from Bacillus amyloliquefaciens (Kang et al., 2015). The overexpression of this pathway in the L-Tyr producing E. coli strain (chromosome-expressing aroGfbr and tyrAfbr in the tyrA deletion strain) resulted in the production of 355 mg/L of p-hydroxystyrene in shake flask cultures after 36 h. The introduction of the tal, 4-coumarate 3-hydroxylase gene sam5 from S. espanaensis and pad genes or the tal, sam5, caffeic acid methyltransferase gene com from A. thaliana and pad genes into the above L-Tyr producing E. coli strain led to the production of 63 mg/L of 3,4-dihydroxystyrene or 64 mg/L of 4-hydroxy-3-methoxystyrene in shake flask cultures after 36 h, respectively (Kang et al., 2015). Since phenolic acid decarboxylase genes are widely distributed in various Streptomyces species, Streptomyces strains have been successfully used as the hosts for p-hydroxystyrene production. After comparing the phenolic acid decarboxylase gene pad obtained from S. sviceus, S. hygroscopicus, and S. cattleya, pad from S. sviceus and tal from R. sphaeroides were used to create the biosynthetic pathway of p-hydroxystyrene (Noda et al., 2015). The introduction of this pathway into the endoglucanase-screening S. lividans species directly yielded 250 mg/L p-hydroxystyrene from cellulose. This research group also engineered S. mobaraense for p-hydroxystyrene production, which yielded 273 mg/L of the product from glucose (Fujiwara et al., 2016a). The titer was obtained by introducing a single gene that encodes Tal from R. sphaeroides. P. putida S12 has also been used as the host strain for p-hydroxystyrene production. The overexpression of the bifunctional enzyme gene pal/tal from R. toruloides and pdc from L. plantarum in the pcs knockout strain of P. putida S12 resulted in the production of 147 mM (17.6 g/L) of p-hydroxystyrene in a two-phase fed-batch fermentation process (Verhoef et al., 2009).
In plants, hydroxycinnamic acids are commonly conjugated with quinic acid, shikimic acid, malic acid, and glycerol (Kim B. G. et al., 2013). Hydroxycinnamic acid conjugates have stronger anti-oxidative activity than hydroxycinnamic acid. Hydroxycinnamic acid conjugates are synthesized by hydroxycinnamoyl transferase (Figure 3). It has been demonstrated that the activity of hydroxycinnamate-CoA shikimate transferase (HST) from Nicotiana tabacum to p-coumaroyl-CoA is higher than that to other acyl donors (such as caffeoyl-CoA and feruloyl-CoA) (Kim B. G. et al., 2013). They also found that that activity of hydroxycinnamate-CoA quinate transferase (HQT) from N. tabacum to caffeoyl-CoA is higher than that to other acyl donors (such as coumaroyl-CoA and feruloyl-CoA). This group constructed an E. coli strain overexpressing HQT from N. tabacum and 4CL from Oryza sativa for the production of chlorogenic acid from caffeic acid for the first time (Figure 3). The recombinant E. coli strain produced 450 mg/L chlorogenic acid from caffeic acid (Kim B. G. et al., 2013). Then, this group constructed an E. coli-E. coli culture system. The chlorogenic acid producing E. coli strain was co-cultured with a caffeic acid producing E. coli strain for the de novo production of chlorogenic acid, which reached 78 mg/L (Cha et al., 2014). They also engineered an E. coli strain for the de novo synthesis of p-coumaroyl shikimate, which reached 236 mg/L.
Curcumin is a nature phenylpropanoid from the plant Curcuma longa. It has diverse therapeutic properties including anti-oxidant, anti-cancer, anti-inflammatory, anti-Alzeimer's, Anti-HIV, anti-Parkinson, and cholesterol-lowering activities. Rodrigues's group constructed an engineered E. coli expressing A. thaliana 4CL, C. longa diketide-CoA synthase gene (DCS) and C. longa curcumin synthase gene (CURS1) for the production of curcumin from ferulic acid (Figure 3) (Rodrigues et al., 2015). In this study, they also constructed an artificial biosynthetic pathway from L-tyrosine via caffeic acid by caffeoyl-CoA 3-O-methyl-transferase (CCoAOMT) from Medicago sativo. Curcumin can also be produced from caffeic acid, p-coumaric acid or tyrosine, respectively. Then, after the optimization of culture conditions, the resulting strain produced 959.3 mM (353 mg/L) curcumin from ferulic acid in shake flask cultures (Couto et al., 2017). Recently, it was demonstrated that the curcumin titer obtained through the ferulic acid pathway by caffeic acid O-methyltransferase (COMT) was higher than that obtained through the caffeic acid by CCoAOMT (Rodrigues et al., 2020). In this study, they established a two-strain coculture system to further improve the production of curcumin from L-Tyr, which reached 43.2 mM (15.9 mg/L). This titer is the highest titer of curcumin obtained from L-Tyr to date. The coculture increased the production of curcumin by 6.6-fold compared to the mono-culture. An artificial curcumin biosynthetic pathway was integrated into the chromosome of the engineered L-Tyr overproducing E. coli ΔCOS1 strain for the de novo production of curcumin from glucose (Kang et al., 2018). Then, multiplex automated genome engineering (MAGE) was applied for balancing the expression of the six enzymes of the biosynthetic pathway. The resulting strain produced 3.8 mg/L curcumin from glucose in shake flask cultures.
L-DOPA is an aromatic compound derived from L-tyrosine (Figure 3). L-DOPA has been used to treat Parkinson's disease, which is caused by deficiency of the neurotransmitter dopamine. Nakagawa et al. (2011) constructed an E. coli expressing the Streptomyces castaneoglobisporus tyrosinase gene tyr and native tyrAfb, aroGfbr, tktA, and ppsA, which could produce 293 mg/L of L-DOPA from glucose. The authors also co-expressed tyr from S. castaneoglobisporus and L-DOPA decarboxylase gene dodc from P. putida KT2440 in the L-Tyr producing E. coli strain, resulting in the production of 260 mg/L of dopamine. Munoz et al. (2011) reported an engineered E. coli with HpaBC activity, which could produce 1.5 g/L of L-DOPA from glucose. This titer was achieved by the overexpression of E. coli W hpaBC, native tktA, atoGfbr, pheA, and Zymomonas mobilis tyrC in the phosphotransferase system and tyrR knockout strain. Our group first used the singleplex genome engineering approach to create an L-DOPA-producing strain, E. coli DOPA-1 (Wei et al., 2016). MAGE based on 23 targets was then used to further improve L-DOPA production. The resulting strain, E. coli DOPA-30N, produced 8.67 g/L of L-DOPA within 60 h in a 5 L fed-batch fermentation process (Wei et al., 2016). Overexpression of E. coli W hpaBC along with the enhancement of the expression level of the downstream tyrosine biosynthetic pathway of shikimate in the tyrR/pheA/serA knockout E. coli strain resulted in producing 12.5 g/L L-DOPA in a 5-L bioreactor fed-batch fermentation (Das et al., 2018). To increase the catalytic efficiency of HpaBC, HpaB from E. coli was directly evolved and three mutants with higher activity were obtained (Fordjour et al., 2019). Overexpression of the mutant hpaBG295RC in an L-Tyr producing E. coli strain produced 25.53 g/L L-DOPA in a 5-L bioreactor fed-batch fermentation.
Tryptophan Derivatives
L-Trp is a common precursor for the production of many important aromatic chemicals (Figure 4). A few L-Trp derivatives that have been synthesized by engineering microorganisms are listed in Table 3.
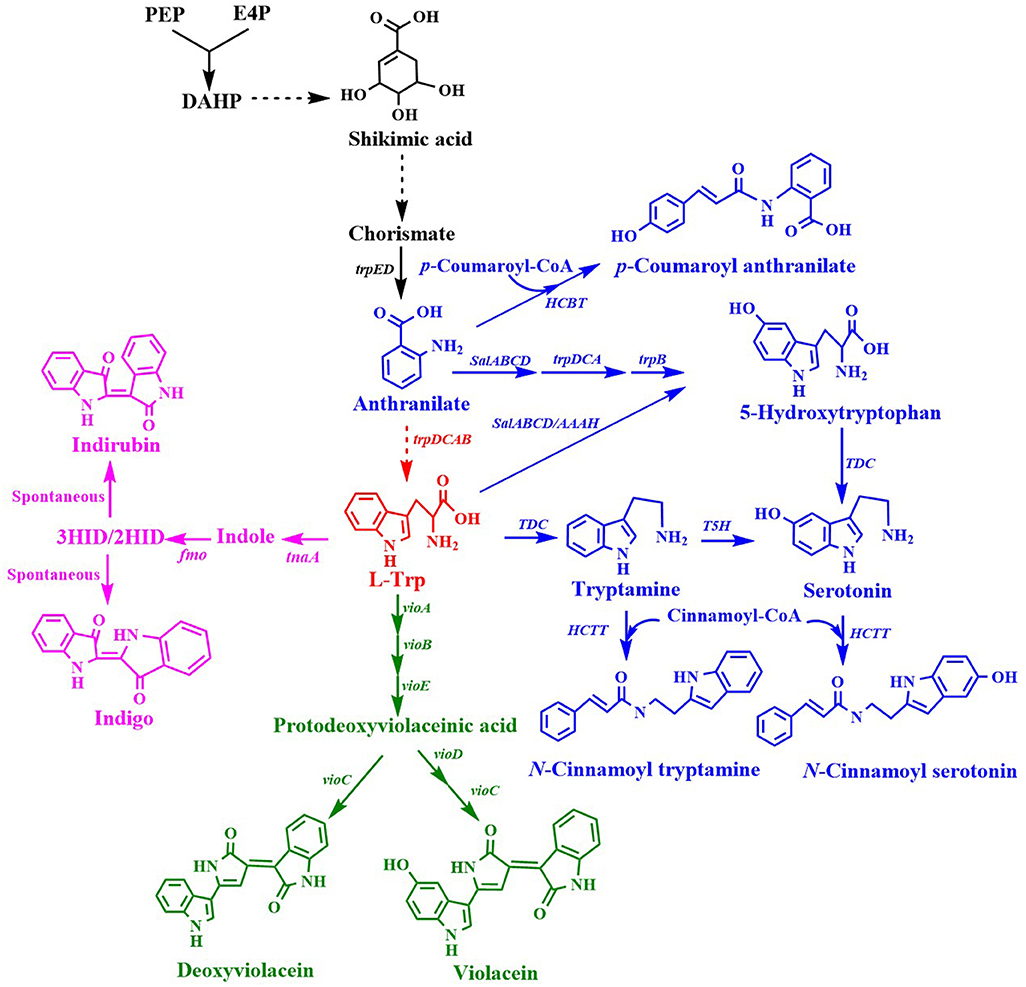
Figure 4. Biosynthesis of L-tryptophan derivatives. HCBT, anthranilate N-hydroxycinnamoyl/benzoyltransferase gene; salABCD, salicylate 5-hydroxylase gene; AAAH, aromatic amino acid hydroxylase gene; TDC, tryptophan decarboxylase gene; T5H, tryptamine 5-hydroxylase gene; tnaA, tryptophanase gene; fmo, flavin-containing monooxygenase gene; HCTT, hydroxycinnamoyl-CoA:tryptamine N-hydroxycinnamoyl transferase gene; vioA, tryptophan 2-monooxygenase/L-tryptophan oxidase gene; vioB, 2-imino-3-(indol-3-yl)propanoate dimerase gene; vioE, violacein biosynthesis protein gene; vioC, violacein synthase gene; vioD, protodeoxyviolaceinate monooxygenase gene.
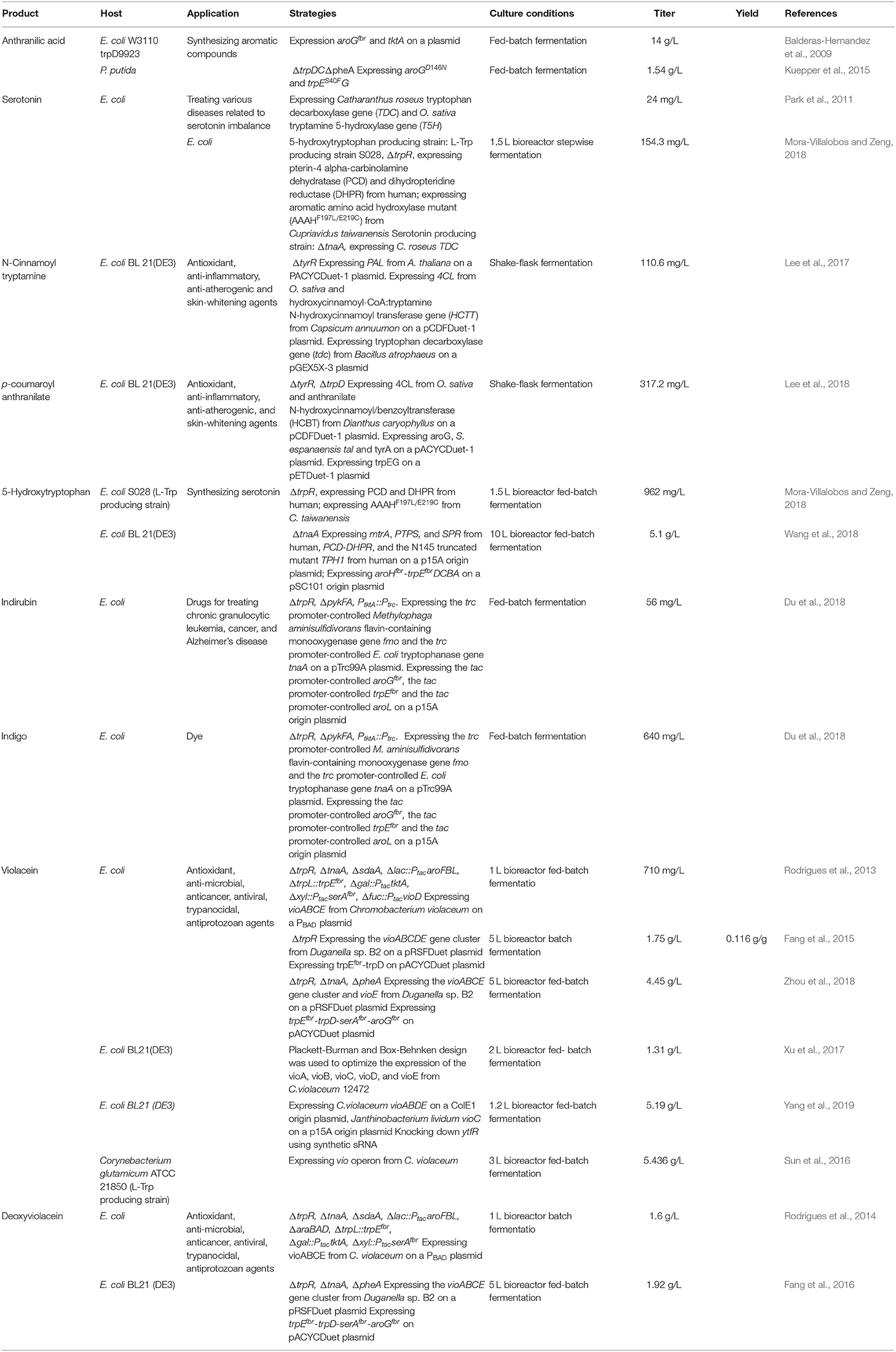
Indirubin, also known as couraupitine B, is an indole alkaloid that is a major bioactive ingredient of a traditional oriental medicine called Danggui Longhui Wan. It is currently used as a drug to treat chronic granulocytic leukemia and may have therapeutic value in the treatment of cancer and Alzheimer's disease. Indirubin is derived from L-Trp, which is converted into indole by native tryptophanase, then reduced to 2-hydroxyindole (2HID) and 3-hydroxyindole (3HID) by heterologous monooxygenase or dioxygenase, and finally undergoes spontaneous dimerization with 3HID and 2HID to form indirubin (Figure 4). Recently, an artificial de novo biosynthetic pathway was developed for the production of 56 mg/L indirubin and 640 mg/L indigo from glucose (Du et al., 2018). The titer was achieved by the overexpression of Methylophaga aminisulfidivorans flavin-containing monooxygenase gene fmo and E. coli tryptophanase gene tnaA as well as by enhancing the expression of the feedback-resistant aroGfbr and trpEfbr in the trpR/pykFA knockout E. coli strain. This was the first report about the direct production of indirubin from glucose in a shake flask culture.
Violacein and deoxyviolacein are secondary metabolites that receives high interest due to their biological activities such as anti-microbial, anti-cancer, anti-viral, trypanocidal, anti-protozoan, and anti-oxidant activities (Fang et al., 2015). E. coli expressing the vioABCE cluster from Chromobacterium violaceum under control of the inducible araC system produced 180 mg/L deoxyviolacein from glucose (Figure 4) (Rodrigues et al., 2013). System-level analysis for the serine, chorismate and L-Trp biosynthesis and the non-oxidative pentose phosphate pathway revealed the bottlenecks in L-Trp biosynthesis. After eliminating the bottlenecks in L-Trp supply, the deoxyviolacein titer was further improved to 320 mg/L. Moreover, co-expression of the vioD from C. violaceum in the above deoxyviolacein producing strain dVio-6 led to the production of 710 mg/L violacein from glucose in a fed-batch fermentation (Rodrigues et al., 2013). In their further study (Rodrigues et al., 2014), the L-arabinose metabolism of the above deoxyviolacein producing strain dVio-6 was eliminated to prevent catabolism of the inducer L-arabinose by knocking the araBAD genes. The resulting E. coli strain dVio-8 produced 1.6 g/L deoxyviolacein from glycerol in a fed-batch fermentation (Rodrigues et al., 2014). Using another vioABCE cluster from Duganella sp. B2, an engineered E. coli strain was used to produce violacein (Fang et al., 2015). Co-overexpression of this gene cluster, trpEM293T/S40L and trpD in the trpR deletion E. coli strain led to the production of 1.75 g/L violacein in a 5 L bioreactor batch fermentation (Fang et al., 2015). This group then developed an intermediate sensor-assisted push-pull strategy (InterSPPS) (Fang et al., 2016). In this study, an L-Trp biosensor was first developed. Then the L-Trp biosensor was used to sequentially enhance the upstream and downstream modular of L-Trp in the deoxyviolacein biosynthetic pathway. By this means, the deoxyviolacein titer was increased by 4.4-fold (1.92 g/L) (Fang et al., 2016). This group's study also demonstrated that VioE is the rate-limiting enzyme for the violacein biosynthesis (Zhou et al., 2018). Overexpression of the vioE further improved the production of violacein. The final recombinant E. coli strain produced 4.45 g/L violacein in 5-L bioreactor fed-batch fermentation (Zhou et al., 2018). A statistical model-based multivariate regulator strategy was developed to improve metabolic pathway efficiency (Xu et al., 2017). A combination of Plackett-Burman design with Box-Behnken design was used to optimize the expression levels of the violacein biosynthetic pathway genes by using different strength promoters. The final E. coli strain produced 1.31 g/L of violacein in a 2 L bioreactor fed-batch fermentation (Xu et al., 2017). Constraints-based flux balance analysis (FBA) identified tryptophan biosynthesis and NADPH availability as limiting (Immanuel et al., 2018). The recombinant E. coli strain based on FBA produced a 6-fold increase in violacein yield. The synthetic small regulatory RNA (sRNA) technology has been successfully used for genome-scale screening of beneficial knockdown gene targets in a violacein producing E. coli strain (Yang et al., 2019). As a result, knocking down ytfR (encoding sugar ABC transporter) increased 600% violacein titer. The ytfR knockdown strain produced 5.19 g/L violacein in fed-batch fermentation. Corynebacterium glutamicum has been successfully used as a host strain for the production of violacein. Introduction of the vio operon from C. violaceum into the L-Trp producer C. glutamicum ATCC 2185 led to the production of 5.436 g/L violacein in a 3 L bioreactor fed-batch fermentation (Sun et al., 2016). The value is the highest to date.
Production of Stilbenes
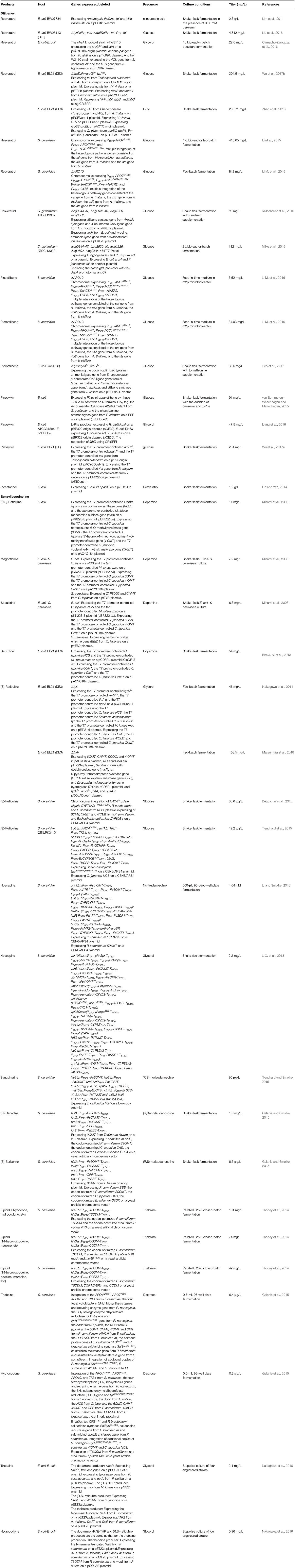
Stilbenes are composed of a skeleton with two aromatic rings joined by a methylene bridge. They are biosynthesized through the condensation of Co-A-linked cinnamic acids with three molecules of malonyl-CoA catalyzed by stilbene synthase (Figure 5, Table 4). Pinosylvin, resveratrol, and piceatannol are the three stilbenoids derived from cinnamic acid, p-coumaric acid, and caffeic acid, respectively. Recently, some 4-coumaroyl-coenzyme A ligases (4CLs) and stilbene synthase (STS) were introduced into microorganisms for the production of resveratrol, pterostilbene, piceatannol, and pinosylvin (Shrestha et al., 2019; Thapa et al., 2019). In most studies, resveratrol was produced from p-coumaric acid by the co-expression of 4CL and STS (Beekwilder et al., 2006; Watts et al., 2006; Katsuyama et al., 2007b; Lim et al., 2011), or from L-Tyr (or L-Phe) by the co-expression of Tal (or Pal), 4CL, and STS (Katsuyama et al., 2007a; Wu et al., 2013; Wang S. Y. et al., 2015). The highest titer (2.3 g/L) of resveratrol was obtained from p-coumaric acid by engineered E. coli BW27784 containing the 4cl1 gene from A. thaliana and sts gene from Vitis vinifera in the presence of 0.05 mM cerulenin (Lim et al., 2011). The authors also demonstrated that the combination of 4CL from A. thaliana and STS from V. vinifera was the most suitable combination for resveratrol production. The de novo biosynthesis of resveratrol from glucose was reported for the first time by Liu et al. (2016). The tal gene from R. glutinis, 4cl gene from P. crispum, and sts gene from V. vinifera were site-specifically integrated into the loci of the genes tyrR and trpED within the chromosome of E. coli BW25113 (DE3). The engineered strain produced 4.612 mg/L of resveratrol from glucose in shake flask cultures (Liu et al., 2016). A co-culture of p-coumaric acid producing E. coli and resveratrol producing E. coli was used to improve resveratrol production from glycerol to 22.6 mg/L in a 1-L bioreactor batch fermentation process (Camacho-Zaragoza et al., 2016). Recently, the CRISPRi system was used to repress fabFBID involved in the fatty acid biosynthetic pathway, which improved the amount of resveratrol produced from glucose to 304.5 mg/L (Wu et al., 2017b). The titer was obtained by the introduction of the malonate assimilation pathway from Rhizobium trifolii and the repression of the fatty acid biosynthetic pathway using CRISPRi. Since 2003, S. cerevisiae has been successfully used as the host strain for stilbene production from p-coumaric acid, by the introduction of the 4CL216 gene from a hybrid poplar and the vst1 gene from V. vinifera (Becker et al., 2003). Li et al. (2015) first reported an engineered S. cerevisiae capable of the de novo biosynthesis of resveratrol from glucose. The introduction of the tal gene from Herpetosiphon aurantiacus, 4cl gene from A. thaliana, and sts gene from V. vinifera in an engineered S. cerevisiae overexpressing the feedback-insensitive alleles of ARO4 and ARO7 resulted in the de novo production of resveratrol from glucose (Li et al., 2015). The overexpression of the inactivation-resistant variant of acetyl-CoA carboxylase (Acc1pS659A/S1157A) to abolish its phosphorylation, and the increase in the copy number of the heterologous resveratrol biosynthetic pathway via tyrosine further improved the amount of resveratrol produced from glucose to 415.65 mg/L, or that produced from ethanol to 532.42 mg/L, in a fed-batch fermentation process. The authors then applied a strategy involving pull-push-block stain engineering to further improve the amount of resveratrol produced from glucose in a fed-batch fermentation process to 812 mg/L (Li M. et al., 2016). The titer was obtained by the multiple integrations of the heterologous pathway genes via phenylalanine, enhancement of P450 activity, upregulation of the expression levels of the feedback-insensitive alleles of ARO4 and ARO7, and the inactivation-resistant variant of acetyl-CoA carboxylase (Acc1pS659A/S1157A), along with blockage of the decarboxylation of phenylpyruvate. Kallscheuer et al. (2016) constructed a C. glutamicum platform strain for the production of resveratrol from glucose. The heterologous biosynthetic pathway genes containing the sts gene from Arachis hypogaea, 4cl gene from P. crispum and tal gene from Flavobacterium johnsoniae, and the aroH gene from E. coli were introduced into the four gene cluster knockout C. glutamicum strain. The final strain produced 59 mg/L of resveratrol from glucose in the presence of 25 μM of cerulenin in shake flask cultures (Kallscheuer et al., 2016). Then, this group found that increasing the intracellular acetyl-CoA availability through the reduction of citrate synthase activity further improved the production of resveratrol to 112 mg/L (Milke et al., 2019). A RppA-coupled malonyl-CoA biosensor was developed for high-throughput screening of targets increasing the malonyl-CoA pool (Yang et al., 2018). The authors applied the biosensor to screen an 1858 synthetic sRNA library and found 14 knockdown targets that generally enhanced malonyl-CoA level in E. coli. The pabA knockdown using sRNA increased resveratrol production by 4.2-fold, which achieved 51.8 mg/L (Yang et al., 2018). After screening pathway genes from various species and exploring their expression pattern, an artificial resveratrol biosynthetic pathway was created and was introduced into E. coli strain for the production of resveratrol (Zhao et al., 2018). Co-overexpression of chaperone protein genes (groeS-groEL), transport gene (ompF), and malony-CoA biosynthetic pathway genes (accBC-dtsR1) from C. glutamicum along with antisense inhibiting fabD significantly increased the production of resveratrol from L-Tyr, which reached 238.71 mg/L.
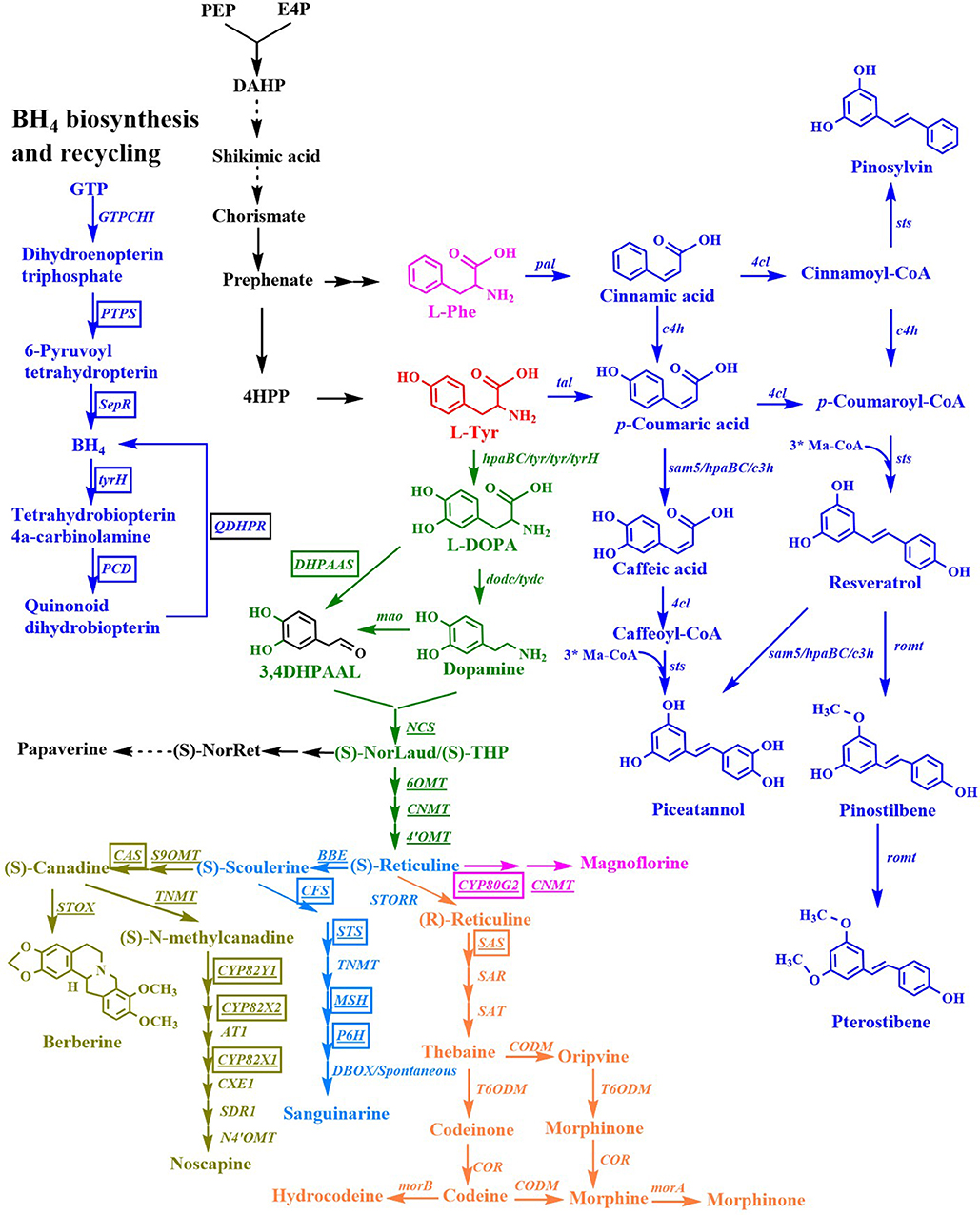
Figure 5. Biosynthesis of stilbenes, benzylisoquinoline alkaloids. Black, microorganism genes; Boxed genes, mammalian (Rattus norvegicus/ Bombyx mori) genes; Underlinded, plant genes; Boxed and underlined genes, cytochrome P450 genes; Ma-CoA, malonyl-CoA; (S)-NorLaud, (S)-norlaudanosoline; (S)-THP, (S)-tetrahydropapaveroline; (S)-NorRet, (S)-norreticuline; BH4, tetrahydrobiopterin; pal, phenylalanine ammonia lyase gene; 4cl, 4-coumaroyl-coenzyme A ligase gene; sts, stilbene synthase gene; c4h, 4-cinnamic acid hydroxylase gene; sam5, 4-coumarate hydroxylase gene; c3h, p-coumarate 3-hydroxylase gene; romt, resveratrol O-methyltransferase; tyr/tyrH, tyrosinase gene; DHPAAS, 3,4-dihydroxyphenylacetaldehyde synthase gene; dodc, DOPA decarboxylase gene; tydc, tyrosine decarboxylase gene; mao, monoamine oxidase gene; NCS, norcoclaurine synthase gene; 6OMT, norcoclaurine 6-O-methyltransferase gene; CNMT, coclaurine-N-methyltransferase gene; 4'-OMT, 3'-hydroxy-N-methylcoclaurine-4'-O-methyltransferase gene; CYP80G2, corytuberine synthase gene; BBE, berberine bridge enzyme gene; GTPCHI, GTP cyclohydrolase I gene; PTPS, 6-pyruvoyl-tetrahydropterin synthase gene; SepR, sepiapterin reductase gene; PCD, pterin-4α-carbinolamine dehydratase gene; QDHPR, quinonoid dihydropteridine reductase gene; S9OMT, scoulerine 9-O-methyltransferase gene; CAS, canadine synthase gene; CPR, cytochrome P450 reductase gene; TNMT, tetrahydroprotoberberine cis-N-methyltransferase gene; CYP82Y1, 1-hydroxy-N-methylcanadine synthase gene; CYP82X2, 1- hydroxy-N-methylcanadine 13-hydroxylase gene; AT1, 1,13-dihydroxy-N-methylcandine 13-O-acetyltransferase gene; CYP82X1, 1-hydroxy-13-O-acetyl-N-methylcanadine 8-hydroxylase gene; CXE1, 3-O-acetylpapaveroxine carboxylesterase gene; SDR1, short-chain dehydrogenase/reductase gene; N4'-OMT, narcotoline-4′-O-methyltransferase gene; CFS, cheilanthifoline synthase gene; STS, stylopine synthase gene; TNMT, tetrahydroprotoberberine N-methyltransferase gene; MSH, cis-N-methylstylopine 14-hydroxylase gene; P6H, protopine 6-hydroxylase gene; DBOX, dihydrobenzophenanthridine oxidase gene; STOX, (S)-tetrahydroprotoberberine oxidase gene; STORR, (S)-reticuline isomerase gene; SAS, salutaridine synthase gene; SAR, salutaridinol reductase gene; SAT, salutaridinol acetyltransferase gene; CODM, codeine-O-demethylase gene; T6OMT, thebaine 6-O-demethylase gene; COR, codeinone reductase gene; morA, morphine dehydrogenase; morB, morphine reductase.
Resveratrol is sensitive to light and oxygen, which limits its bioavailability and bioactivity. Both properties can be enhanced by the substitution of hydroxyl groups with methoxy groups. Resveratrol methyltransferase from A. thaliana (Heo et al., 2017), O. sativa (Katsuyama et al., 2007a), Sorghum bicolor (Jeong et al., 2014; Li M. et al., 2016), Vitis riparia (Jeong et al., 2014), and V. vinifera (Wang Y. C. et al., 2015; Li M. et al., 2016; Kallscheuer et al., 2017; Wang et al., 2018) were used to convert resveratrol to pinostilbene/pterostilbene (Figure 5). The methyltransferase from O. sativa methylated the hydroxy groups at positions 3 and 5 of resveratrol to form pinostilbene/pterostilbene (Katsuyama et al., 2007a). The methylation of resveratrol by the methyltransferase from S. bicolor yielded pinostilbene as the major product and a very small amount of pterostilbene, whereas the methyltransferase from V. riparia showed very low methylation activity when resveratrol was used as a substrate (Jeong et al., 2014). The expression of the methyltransferase enzyme from V. vinifera or A. thaliana led to the production of the major product pterostilbene from resveratrol, and a very small amount of pinostilbene (Wang Y. C. et al., 2015; Li M. et al., 2016; Heo et al., 2017; Wang et al., 2018). The titer of pterostilbene (50 mg/L) produced from p-coumaric acid by the methyltransferase from V. vinifera (Wang Y. C. et al., 2015) was higher than that (10 mg/L) produced by the methyltransferase from A. thaliana (Heo et al., 2017), indicating that the methyltransferase from V. vinifera is more beneficial for the production of pterostilbene. The de novo biosynthesis of pinostilbene or pterostilbene from glucose was achieved for the first time by the introduction of the methyltransferase from S. bicolor or V. vinifera, respectively, into resveratrol producing yeast (Li M. et al., 2016). The engineered S. cerevisiae containing the methyltransferase from S. bicolor produced 5.52 mg/L of pinostilbene from glucose. The engineered S. cerevisiae containing the methyltransferase from V. vinifera produced 34.93 mg/L of pterostilbene and 1.96 mg/L of pinostilbene from glucose. Heo et al. (2017) also engineered E. coli for the de novo biosynthesis of pterostilbene from glucose by the overexpression of caffeic acid O-methyltransferase from A. thaliana, tyrosine ammonia lyase from S. espanaensis, p-coumarate: CoA ligase from N. tabacum, and stilbene synthase from V. vinifera in the L-Tyr-overproducing E. coli strain. The resulting E. coli produced 33.6 mg/L of pterostilbene from glucose in shake flask cultures containing additional L-methionine medium.
Pinosylvin is a natural resveratrol analog that lacks a hydroxyl group at the C-4 position. It is synthesized from L-Phe in three enzymatic steps (Figure 5). L-Phe is first transformed to cinnamic acid by L-Phe ammonia lyase. Subsequently, the enzyme 4-coumarate/cinnamate:coenzyme A ligase converts the cinnamic acid to cinnamoyl-CoA. Finally, the enzyme stilbene synthase catalyzes the stepwise condensation of three molecules of malonyl-CoA with one molecule of cinnamoyl-CoA, yielding one molecule of pinosylvin. The production of pinosylvin was achieved from L-Phe for the first time by the co-expression of the phenylalanine ammonia lyase gene from Rhodotorula rubra, the 4-coumarate:CoA ligase gene from Lithospermum erythrorhizon, the stilbene synthase gene from A. hypogaea, and the acetyl-CoA carboxylase gene from C. glutamicum (Katsuyama et al., 2007a). The resulting E. coli strain produced 20 mg/L pinosylvin from L-Phe. De novo biosynthesis of pinosylvin was achieved for the first time by the co-expression of the stilbene synthase gene from Pinus strobus, the 4-coumarate: CoA ligase gene from S. coelicolor, and the phenylalanine ammonia lyase gene from P. crispum (van Summeren-Wesenhagen and Marienhagen, 2015). The addition of cerulenin to increase the intracellular malonyl-CoA pools and the directed evolution of stilbene synthase from P. strobus further enhanced pinosylvin production. The final engineered E. coli strain produced 91 mg/L of pinosylvin from glucose after the addition of cerulenin and L-Phe (van Summeren-Wesenhagen and Marienhagen, 2015). Cerulenin is expensive and its use is not cost-effective for a large-scale fermentation process. In seeking a better alternative, the CRISPRi system was used to repress the fadD gene, which increased the availability of malonyl-CoA and resulted in the production of 47.5 mg/L of pinosylvin from glycerol (Liang et al., 2016). A rational modular design approach was used to improve pinosylvin production from glucose in E. coli (Wu et al., 2017a). The overall pinosylvin synthetic pathway was divided into two modules that were separated at cinnamic acid. The combined optimization of the transcriptional and translational levels of these two modules by modifying the plasmid copy numbers, promoter strength, and 5' region of the mRNA secondary structure increased the titer of pinosylvin by 16 times. The CRISPRi system was also used to repress fabB, fabF, adhE, eno, fumC, and sucC to improve pinosylvin production. The resulting E. coli produced 281 mg/L of pinosylvin, which was the highest titer directly obtained from glucose without the provision of supplementation with any additional precursors (Wu et al., 2017a). The metabolic engineering strategy based on the post-translational modification of proteins was used to optimize the lysine acylation of the stilbene synthase and to improve pinosylvin production by 220% (Xu et al., 2018).
Piceatannol is another natural resveratrol analog with an additional phenolic hydroxyl group at the C-3 position. It can be synthesized from caffeic acid or resveratrol (Figure 5). E. coli W endogenous non-P450 monooxygenase (HpaBC) catalyzed the ortho-hydroxylation of resveratrol to piceatannol, which resulted in the maximal titer of 1.2 g/L from resveratrol with a nearly 100% molar yield (Lin and Yan, 2014). An engineered C. glutamicum was constructed for the production of pinosylvin or piceatannol from cinnamic acid or caffeic acid, respectively, by the introduction of stilbene synthase from A. hypogaea (Kallscheuer et al., 2016).
Production of Benzylisoquinoline Alkaloids
Benzylisoquinoline alkaloids (BIAs) are a large family of L-tyrosine-derived plant-specialized compounds with a variety of therapeutic uses. This class of compounds includes the opioid analgesics morphine and codeine, the antibiotics sanguinarine and berberine, the muscle relaxants (+)-tubocurarine and papaverine, and the cough suppressant noscapine. BIA synthesis begins with the condensation of dopamine and 4-hydroxyphenylacetaldehyde or 3,4-di hydroxyphenylacetaldehyde to form (S)-norcoclaurine or (S)-norlaudanosoline ((S)-NorLaud, also known as (S)-tetrahydropapaveroline, (S)-THP) in engineered microorganisms (Figure 5, Table 4) (Diamond and Desgagne-Penix, 2016; Narcross et al., 2016b). Both (S)-norcoclaurine and (S)-norlaudanosoline are unstable end-products, because they are subject to enzymatic oxidation as well as spontaneous oxidation at alkaline pHs. Thus, reticuline is frequently used as the key branch-point intermediate for the de novo synthesis of BIAs. Reticuline was produced in E. coli for the first time by the overexpression of monoamine oxidase (Mao) from M. luteus, norcoclaurine synthase (NCS) from Coptis japonica, norcoclaurine 6-O-methyltransferase (6OMT) from C. japonica, coclaurine-N-methyltransferase (CNMT) from C. japonica, and 3'-hydroxy-N-methylcoclaurine-4'-O-methyltransferase (4'OMT) from C. japonica (Figure 5) (Minami et al., 2008). Since some plant enzymes are not well-expressed in E. coli in an active form, various types of BIAs were then synthesized from reticuline using engineered S. cerevisiae. Co-cultures of E. coli expressing the reticuline biosynthetic genes and S. cerevisiae expressing C. japonica CYP80G2 and CNMT, or the C. japonica berberine bridge enzyme (BBE) were used to produce magnoflorine or scoulerine, respectively, from dopamine (Figure 5) (Minami et al., 2008). The overall yield of magnoflorine or scoulerine from dopamine was 1.9 or 2.2%, respectively. After optimizing the fermentation conditions and the gene copy numbers, the reticuline titer generated from dopamine was increased by 5-fold to 54 mg/L (Kim J. S. et al., 2013). Recently, this group comparatively analyzed the effect of the three methyltransferases on the biosynthesis of reticuline and found that the combination of 6OMT from P. somniferum, CNMT from P. somniferum, and 4'OMT from C. japonica was the most suitable combination for reticuline production (Matsumura et al., 2017). The de novo biosynthesis of reticuline was achieved for the first time by the co-expression of tyrosinase from Ralstonia solanacearum, DOPA decarboxylase (Dodc) from P. putida strain KT2440, Mao from M. luteus, NCS from C. japonica, 6OMT from C. japonica, CNMT from C. japonica, and the 4'OMT from C. japonica in the L-Tyr overproducing E. coli strain (Nakagawa et al., 2011). The final strain produced 46 mg/L (S)-reticuline from glycerol in a fed-batch fermentation process. Then, this group further improved the (S)-reticuline titer to 163.5 Mg/L by an engineered E. coli strain EM353, which possessed a tyrosine-overproducing pathway, a pathway producing dopamine from L-Tyr along with the BH4-synthesis pathway, and a pathway producing (S)-reticuline from dopamine (Matsumura et al., 2018). In the above studies, tetrahydropapaveroline (THP), a reticuline precursor, is synthesized by Mao and Dodc. Recently, an insect Bombyx mori 3,4-dihydroxyphenylacetaldehyde synthase (DHPAAS) and its variants with Phe79Tyr, Tyr80Phe, and Asn192His were identified to replace Mao and Dodc for direct production (R,S)-THP in a single enzyme system from L-DOPA (Vavricka et al., 2019).
S. cerevisiae is a more attractive host for BIA synthesis, due to its superior ability to express the plant enzymes and the cytochrome P450s, which are prevalent in BIA biosynthesis. The synthesis of reticuline in S. cerevisiae was initially achieved with a yield of 10% from norlaudanosoline by the over-expressions of P. somniferum 6OMT, CNMT, and 4'OMT (Hawkins and Smolke, 2008). S. cerevisiae strains were also engineered for the production of the protoberberine intermediate (S)-canadine and the morphinan intermediate salutaridine from (R,S)-norlaudanosoline (Hawkins and Smolke, 2008). The yield of reticuline obtained from norlaudanosoline in S. cerevisiae was then increased to 20% (Fossati et al., 2014). These two research groups constructed a de novo biosynthetic pathway for producing reticuline from glucose via norcoclaurine and reported their results at the same time (DeLoache et al., 2015; Trenchard et al., 2015). To solve the issue of the poor activity of tyrosine hydroxylase in yeast, an enzyme-coupled biosensor of L-DOPA was developed. Using this sensor, a mutant (W13L/F309L) of CYP76AD1 from Bete vilgaris was obtained to improve its L-DOPA yield by 2.8-fold via directed evolution (DeLoache et al., 2015). The co-expression of the feedback-insensitive Aro4, the CYP76AD1 variant, DOPA decarboxylase (DODC) from P. putida, a newly identified NCS from P. somniferum, 6OMT, CNMT, and 4′OMT from P. somniferum, and the cytochrome P450 N-methylcoclaurine hydroxylase (NMCH) from Eschscholzia californica in S. cerevisiae resulted in the de novo production of 80.6 μg/L of reticuline from glucose (DeLoache et al., 2015). In the study by the research group led by Smolke, the mammalian tyrosine hydroxylase mutant (W166Y/R37E/R38E) from Rattus norvegicus, the four tetrahydrobiopterin (BH4) biosynthesis and recycling enzymes from R. norvegicus, and bacterial DOPA decarboxylase from P. putida were introduced in the L-Tyr overproducing S. cerevisiae strain for the production of norcoclaurine (Trenchard et al., 2015). The co-expression of optimized NCS from C. japonica, the three methyltransferases (6OMT, CNMT, and 4′OMT) from P. somniferum, the cytochrome P450 (CYP80B1) from E. californica, and its reductase partner (CPR) from P. somniferum in the aforementioned engineered S. cerevisiae strain resulted in the de novo production of 19.2 mg/L of reticuline from glucose in a shake flask fermentation process (Trenchard et al., 2015).
Noscapine has long been used as a cough suppressant. Due to its long-established safety record, noscapine is a compelling candidate anti-cancer drug and has been used off-label to treat cancers in some countries. Noscapine is synthesized from L-Tyr through scoulerine (Figure 5). The synthetic pathway of noscapine with the 10-gene cluster was discovered in P. somniferum (Winzer et al., 2012). A 14-step biosynthetic pathway for the production of noscapine from norlaudanosoline was engineered for the first time in yeast (Li and Smolke, 2016). After the optimization of the expression of 16 plant enzymes involved in this biosynthetic pathway, the engineered S. cerevisiae strain produced 1.64 mM of noscapine. This group subsequently reported the de novo production of noscapine in S. cerevisiae, through the reconstruction of a biosynthetic pathway comprising 37 enzymes from plants, bacteria, mammals, and yeasts (Li Y. et al., 2018). After engineering rate-limiting pathway enzymes, optimizing enzyme expression levels, introducing modifications to the endogenous yeast metabolism to enhance NADPH availability, and optimizing fermentation conditions, the noscapine titer was improved by over 18,000-fold to ~2.2 mg/L.
Sanguinarine is a BIA with recognized antimicrobial activities. It may have potential as an anti-neoplastic drug. Fossati et al. constructed a 10-gene plant pathway in S. cerevisiae that allowed the production of the sanguinarine precursor dihydrosanguinarine from (R,S)-norlaudanosoline, which resulted in a yield of 1.5% (Figure 5) (Fossati et al., 2014). The spontaneous oxidation of dihydrosanguinarine to sanguinarine was observed in this study. The 10 enzymatic steps included four reactions catalyzed by plant cytochrome P450s (Fossati et al., 2014). After optimizing the expression of the cytochrome P450s (including P450 enzymatic variants, the pairing of CPRs with P450 enzymatic variants, gene number copy and promoter) and culture conditions, 80 μg/L sanguinarine was produced from (R, S)-norlaudanosoline in the engineered S. cerevisiae (Trenchard and Smolke, 2015). This titer was achieved by the expression of P. somniferum 6OMT, CNMT, 4'OMT, BBE, CPR, TNMT, and MSH, E. californica CFS, STS, and P6H, and A. thaliana ATR1. Mining of transcriptome libraries identified the superior cytochrome P450 enzymes CFS and SPS (Narcross et al., 2016a). They improved the level of molar conversion of norlaudanosoline to dihydrosanguinarine to 10%. This study demonstrated that the mining of transcriptome libraries combined with gene synthesis is an effective strategy for pathway engineering.
Hawkins and Smolke reconstructed the biosynthetic pathway for the berberine precursor (S)-canadine in S. cerevisiae to achieve the microbial biosynthesis of (S)-canadine from (R,S)-norlaudanosoline (Figure 5) (Hawkins and Smolke, 2008). Then, this group optimized cytochrome P450 oxidoreductase (canadine synthase CAS), scoulerine 9-O-methyltransferase S9OMT, and culture conditions, which improved the canadine and berberine production from (R,S)-norlaudanosoline to 1.8 and 6.5 mg/L, respectively (Galanie and Smolke, 2015). Their studies also demonstrated that canadine can be easily and spontaneously converted to berberine. The de novo synthesis of berberine has not been achieved in microorganisms to date. The abovementioned strategies as reported by Li Y. et al. (2018), which were used for the de novo synthesis of noscapine, can be used for berberine production, because berberine and noscapine have the same precursor, i.e., (S)-canadine.
Thodey et al. (2014) engineered S. cerevisiae strains to convert thebaine to codeine, morphine, hydromorphone, hydrocodone, and oxycodone, by expressing the thebaine 6-O-demethylase (T6ODM), codeine-O-demethylase (CODM), and codeinone reductase (COR1.3) from P. somniferum and MorAB from P. putida M10. The authors improved the total opioid titers to 130 mg/L by titrating gene copy number, applying a spatial engineering strategy, and optimizing culture conditions. In another study, a seven-gene pathway was constructed in S. cerevisiae for the production of codeine from (R)-reticuline (Fossati et al., 2015). These seven genes included salutaridine synthase gene (SAS), salutaridine reductase gene (SAR), salutaridinol acetyltransferase gene (SAT), and T6ODM, COR1.3, and CODM from P. somniferum. Another research group engineered 21-gene and 23-gene pathways in S. cerevisiae for the de novo production of thebaine and hydrocodone, respectively, from dextrose (Galanie et al., 2015). The genes of the 21-gene pathway for thebaine synthesis composed 5 modules: (1) precursor overproduction modular, (2) tetrahydrobiopterin (BH4) modular, (3) (S)-norcoclaurine modular, (4) (S)-reticuline modular, (5) thebaine modular (Figure 5, Table 4). And they also demonstrated that the (S)-reticuline modular is the key pathway for opioid production (Galanie et al., 2015). Integrating additional gene copies of the (S)-reticuline modular increased reticuline production to 82 μg/L from 20 μg/L. The integration of 21 genes of the thebaine synthetic pathway and additional copies of the (S)-reticuline modular (TyrH, 4'OMT, and NCS) in S. cerevisiae resulted in the production of 6.4 μg/L thebaine. The authors also described that the overexpression of T6ODM from P. somniferum and MorB from P. putida M10 in a yeast artificial chromosome vector in the above thebaine producing yeast stain resulted in the production of ~0.3 μg/L of hydrocodone (Galanie et al., 2015). A stepwise culture strategy was used for the de novo production of opiates from glycerol using four E. coli strains (Nakagawa et al., 2016). These four E. coli strains included dopamine, (R,S)-THP, (R,S)-reticuline, and thebaine producing E. coli strains. The stepwise culture of the four engineered E. coli strains yielded 2.1 mg/L of thebaine from glycerol, which corresponded to a 300-fold increase from the yield observed with the developed yeast system (Nakagawa et al., 2016). After the introduction of T6ODM obtained from P. somniferum and morB from P. putida into the thebaine producing E. coli strain, 0.36 mg/L of hydrocodone was produced using the four-step culture system (Nakagawa et al., 2016). A neopinone isomerase gene (NISO) was recently identified from opium poppy (P. somniferum) (Dastmalchi et al., 2019b). Overexpression of this NISO in an engineered yeast significantly improved the production of codeine and hydrocodone, respectively. Recently, nine benzylisoquinoline alkaloid uptake permease genes (BUPs) were also identified from P. somniferum (Dastmalchi et al., 2019a). Overexpression of BUP in an engineered yeast further enhanced the production of (S)-reticuline, thebaine, codeine, and morphine, respectively.
Concluding Remarks and Future Prospects
With the rapid development of metabolic engineering and synthetic biology, as well as the increased knowledge regarding the native biosynthetic pathways of aromatic natural compounds, heterologous biosynthesis of aromatic products in microorganisms has become a promising alternative for the production of aromatics and their derivatives. However, their titers and yields are still too low for industrial production. The following may be new directions that can enhance the production of aromatic amino acid derivatives.
First, many aromatic compounds have anti-microbial activity and are highly toxic to microbial host cells, thus limiting the microbial production of aromatic chemicals. Adaptive laboratory evolution (ALE) is a general approach for improving the tolerance of host cells. However, t improvements in tolerance cannot guarantee an increase in yield. The biosensor-driven ALE approaches can rapidly screen the highly efficient evolved strain from the ALE libraries tolerant to the aromatic compounds. Some biosensors of aromatic compounds have been developed (Li et al., 2017; Wang et al., 2017). Shikimic acid and resveratrol biosensors have been developed and applied to increase the production of shikimic acid (Li et al., 2017) and resveratrol (Xiong et al., 2017), respectively. Liu et al. (2017) developed an L-Phe biosensor and applied the biosensor to improve L-Phe production, by screening hyperproducing strains from a ribonucleotide binding site library and random mutagenesis library.
Second, whole-cell biocatalysis may be a powerful strategy for the production of aromatic chemicals. The whole-cell biocatalysis processes consist of two stages: cell culture and the conversion of the substrates (Lin and Tao, 2017). After cells are cultured, cells are harvested and then suspended in the desired buffer to convert substrates into products. Because the whole-cell biocatalysis process separates the cell growth from the formation of target products, it is unnecessary to consider the cytotoxicity of products. It has been successfully used for the production of L-PLA from L-Phe (Hou et al., 2019), D-phenylglycine from L-Phe (Zhou Y. et al., 2017), rosmarinic acid from caffeic acid (Jiang et al., 2016), L-DOPA from hydroxytyrosol (Li et al., 2019b), and caffeic acid from p-coumaric acid (Furuya and Kino, 2014). It can also be used for the de novo biosynthesis of products when cells harbor the de novo biosynthetic pathway. Using whole-cell biocatalysis of the engineered C. glutamicum resulted in the highest shikimic acid titer and yield attained by microbial production reported so far (141 g/L and 0.49 g of shikimic acid per g of glucose (Kogure et al., 2016).
Third, because regulation mechanisms of aromatic amino acid formation have been quite well-known and thus do no longer present obstacles in increasing the aromatic amino acid availability. The identification of better enzymes of the heterogeneous pathway and the further improvement of these enzymes will play important roles in further improving the production of aromatic compounds. A mutant of A. thaliana 4CL1 exhibiting a higher catalytic efficiency was obtained by directed evolution via error-prone PCR (Xiong et al., 2017). The use of this mutant improved resveratrol production by 4.7-fold in E. coli.
Fourth, modular co-cultures might be a useful approach for improving the production titer of aromatic compounds. This approach divides a complete biosynthetic pathway into separate serial modules and uses selected microbial strains to accommodate individual modules for biosynthesis. There are a number of main benefits of using this approach. These include: (1) reduction of the metabolic burden on each host strain; (2) allowing the selection of a suitable host strain and culture conditions for each module that meets the requirements of specific biosynthetic steps; (3) reduction in the undesirable interference from different pathways; (4) maintenance of an easy balance of the biosynthetic pathway among individual pathway modules, by simply changing the strain-to-strain ratio; (5) high-efficiency utilization of complex materials containing multiple active substrates; and (6) support provided for the plug-and-play biosynthesis of various target products (Zhang and Wang, 2016). Co-culture engineering strategies can significantly improve the production of salidroside (Liu X. et al., 2018) and resveratrol (Camacho-Zaragoza et al., 2016). BIAs are tyrosine derivatives. Strategies for improving the production of tyrosine in E. coli and S. cerevisiae are similar, but the highest published titers of aromatic amino acids in E. coli are currently an order of magnitude higher than that in S. cerevisiae. Moreover, plant enzymes and cytochrome P450s are more efficiently expressed in S. cerevisiae than in E. coli. Thus, the modular co-culture of E. coli and S. cerevisiae—during which E. coli expresses the microbial genes for synthesizing (S)-THP and S. cerevisiae express the plant enzymes and cytochrome P450s required for synthesizing BIAs—might be an efficient strategy for the biotechnological production of BIAs. Minami et al. (2008) constructed a co-culture system involving E. coli and S. cerevisiae for the production of magnoflorine or scoulerine from dopamine.
Fifth, most of the studies that have been discussed used the plasmid system to achieve the expression of heterologous pathways. Drawbacks that include the instability and the necessity of use of antibiotics in plasmid expression systems limit the industrial use of plasmid expression systems. To overcome these drawbacks associated with plasmid expression systems, a chemically inducible chromosomal evolution (CIChE) system (Tyo et al., 2009) and an auxotrophic system (Shukal et al., 2019) were developed to achieve high gene copy expression. However, the constructed CIChE strains contained an antibiotic resistance marker (chloramphenicol resistance) (Tyo et al., 2009). Subsequently, we modified CIChE strains and developed an approach that involves triclosan inducible chromosomal evolution (Chen et al., 2013). We applied this approach to engineer a lycopene (Chen et al., 2013) and shikimic acid (Cui et al., 2014) hyperproducing E. coli strains that do not carry a plasmid or antibiotic marker.
The final, as in the cases for developing strains overproducing other compounds, systems metabolic engineering will also be used for developing microbial strains efficiently producing various aromatic chemicals. This approach, which integrates metabolic engineering with systems biology, might represent another efficient strategy to improve the production of aromatic compounds. Comparative analyses using omics might lead to the identification of unknown bottlenecks and help overcome them, and might enable the use of reverse engineering, i.e., the application of novel targets in existing aromatic-producing strains. Although systems metabolic engineering has successfully been applied to engineer microorganisms as a way of improving the production of chemicals (Cho et al., 2015; Shen et al., 2016; Li and Liu, 2017), there are few reports regarding the production of aromatic compounds.
Author Contributions
Y-PS, F-XN, and Y-BH provided draft text for the section Production of Aromatic Amino Acid Derivatives. Z-BY provided draft text for the section Production of Stilbens. LF provided draft text for the section Production of Benzylisoquinoline Alkaloids. J-ZL conceived, reviewed the literature and extracted data, plotted figures and wrote the manuscript. All authors read and approved the final manuscript.
Funding
This work was funded by the National Natural Science Foundation of China (Grant No. 21808248 and 31901024), the Natural Science Foundation of Guangdong Province (No. 2018A030310255), the Open Fund Project of Shenzhen Innovation Institute of Synthetic Biology (DWKF20190003), and the Fundamental Research Funds for the Central Universities (19lgpy133) for their financial support.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Averesch, N. J. H., and Kromer, J. O. (2018). Metabolic engineering of the shikimate pathway for production of aromatics and derived compounds-present and future strain construction strategies. Front. Bioeng. Biotechnol. 6:32. doi: 10.3389/fbioe.2018.00032
Bai, Y. F., Bi, H. P., Zhuang, Y. B., Liu, C., Cai, T., Liu, X. N., et al. (2014). Production of salidroside in metabolically engineered Escherichia Coli. Sci. Rep. 4:6640. doi: 10.1038/srep06640
Balderas-Hernandez, V. E., Sabido-Ramos, A., Silva, P., Cabrera-Valladares, N., Hernandez-Chavez, G., Baez-Viveros, J. L., et al. (2009). Metabolic engineering for improving anthranilate synthesis from glucose in Escherichia coli. Microb. Cell Fact. 8:19. doi: 10.1186/1475-2859-8-19
Bang, H. B., Lee, K., Lee, Y. J., and Jeong, K. J. (2018). High-level production of trans-cinnamic acid by fed-batch cultivation of Escherichia coli. Process Biochem. 68, 30–36. doi: 10.1016/j.procbio.2018.01.026
Becker, J. V. W., Armstrong, G. O., Van der Merwe, M. J., Lambrechts, M. G., Vivier, M. A., and Pretorius, I. S. (2003). Metabolic engineering of Saccharomyces cerevisiae for the synthesis of the wine-related antioxidant resveratrol. FEMS Yeast Res. 4, 79–85. doi: 10.1016/S1567-1356(03)00157-0
Beekwilder, J., Wolswinkel, R., Jonker, H., Hall, R., de Vos, C. H. R., and Bovy, A. (2006). Production of resveratrol in recombinant microorganisms. Appl. Environ. Microb. 72, 5670–5672. doi: 10.1128/AEM.00609-06
Bloch, S. E., and Schmidt-Dannert, C. (2014). Construction of a chimeric biosynthetic pathway for the de novo biosynthesis of rosmarinic acid in Escherichia coli. Chem. Biol. Chem. 15, 2393–2401. doi: 10.1002/cbic.201402275
Calero, P., Jensen, S. I., and Nielsen, A. T. (2016). Broad-host-range ProUser vectors enable fast characterization of inducible promoters and optimization of p-coumaric acid production in Pseudomonas Putida KT2440. ACS Synth. Biol. 5, 741–753. doi: 10.1021/acssynbio.6b00081
Camacho-Zaragoza, J. M., Hernandez-Chavez, G., Moreno-Avitia, F., Ramirez-Iniguez, R., Martinez, A., Bolivar, F., et al. (2016). Engineering of a microbial coculture of Escherichia coli strains for the biosynthesis of resveratrol. Microb. Cell Fact. 15:163. doi: 10.1186/s12934-016-0562-z
Cao, M. F., Gao, M. R., Suastegui, M., Mei, Y. Z., and Shao, Z. Y. (2020). Building microbial factories for the production of aromatic amino acid pathway derivatives: from commodity chemicals to plant-sourced natural products. Metab. Eng. 58, 94–132. doi: 10.1016/j.ymben.2019.08.008
Cha, M. N., Kim, H. J., Kim, B. G., and Ahn, J. H. (2014). Synthesis of chlorogenic acid and p-coumaroyl shikimates from glucose using engineered Escherichia coli. J. Microbiol. Biotechnol. 24, 1109–1117. doi: 10.4014/jmb.1403.03033
Chen, W., Yao, J., Meng, J., Han, W. J., Tao, Y., Chen, Y. H., et al. (2019). Promiscuous enzymatic activity-aided multiple-pathway network design for metabolic flux rearrangement in hydroxytyrosol biosynthesis. Nat. Commun. 10:960. doi: 10.1038/s41467-019-08781-2
Chen, Y. Y., Shen, H. J., Cui, Y. Y., Chen, S. G., Weng, Z. M., Zhao, M., et al. (2013). Chromosomal evolution of Escherichia coli for the efficient production of lycopene. BMC Biotechnol. 13:6. doi: 10.1186/1472-6750-13-6
Cho, C., Choi, S. Y., Luo, Z. W., and Lee, S. Y. (2015). Recent advances in microbial production of fuels and chemicals using tools and strategies of systems metabolic engineering. Biotechnol. Adv. 33, 1455–1466. doi: 10.1016/j.biotechadv.2014.11.006
Choo, H. J., Kim, E. J., Kim, S. Y., Lee, Y., Kim, B. G., and Ahn, J. H. (2018). Microbial synthesis of hydroxytyrosol and hydroxysalidroside. Appl. Biol. Chem. 61, 295–301. doi: 10.1007/s13765-018-0360-x
Chung, D., Kim, S. Y., and Ahn, J. H. (2017). Production of three phenylethanoids, tyrosol, hydroxytyrosol, and salidroside, using plant genes expressing in Escherichia coli. Sci. Rep. 7:2578. doi: 10.1038/s41598-017-02042-2
Couto, M. R., Rodrigue, J. L., and Rodrigues, L. R. (2017). Optimization of fermentation conditions for the production of curcumin by engineered Escherichia coli. J. R. Soc. Interface 14:20170470. doi: 10.1098/rsif.2017.0470
Cui, Y. Y., Ling, C., Zhang, Y. Y., Huang, J., and Liu, J. Z. (2014). Production of shikimic acid from Escherichia coli through chemically inducible chromosomal evolution and cofactor metabolic engineering. Microb. Cell Fact. 13:21. doi: 10.1186/1475-2859-13-21
Das, A., Tyagi, N., Verma, A., Akhtar, S., and Mukherjee, K. J. (2018). Metabolic engineering of Escherichia coli w3110 strain by incorporating genome-level modifications and synthetic plasmid modules to enhance L-dopa production from glycerol. Prep. Biochem. Biotech. 48, 671–682. doi: 10.1080/10826068.2018.1487851
Dastmalchi, M., Chang, L. M., Chen, R. J., Yu, L. S., Chen, X., Hagel, J. M., et al. (2019a). Purine permease-type benzylisoquinoline alkaloid transporters in opium poppy. Plant Physiol. 181, 916–933. doi: 10.1104/pp.19.00565
Dastmalchi, M., Chen, X., Hagel, J. M., Chang, L. M., Chen, R. J., Ramasamy, S., et al. (2019b). Neopinone isomerase is involved in codeine and morphine biosynthesis in opium poppy. Nat. Chem. Biol. 15, 384–390. doi: 10.1038/s41589-019-0247-0
Dell, K., and Frost, J. (1993). Identification and removal of impediments to biocatalytic synthesis of aromatics from D-glucose: rate-limiting enzymes in the common pathway of aromatic amino acid biosynthesis. J. Am. Chem. Soc. 115, 11581–11589. doi: 10.1021/ja00077a065
DeLoache, W. C., Russ, Z. N., Narcross, L., Gonzales, A. M., Martin, V. J. J., and Dueber, J. E. (2015). An enzyme-coupled biosensor enables (s)-reticuline production in yeast from glucose. Nat. Chem. Biol. 11, 465–471. doi: 10.1038/nchembio.1816
Diamond, A., and Desgagne-Penix, I. (2016). Metabolic engineering for the production of plant isoquinoline alkaloids. Plant Biotechnol. J. 14, 1319–1328. doi: 10.1111/pbi.12494
Du, J., Yang, D., Luo, Z. W., and Lee, S. Y. (2018). Metabolic engineering of Escherichia coli for the production of indirubin from glucose. J. Biotechnol. 267, 19–28. doi: 10.1016/j.jbiotec.2017.12.026
Fang, M. Y., Wang, T. M., Zhang, C., Bai, J. L., Zheng, X., Zhao, X. J., et al. (2016). Intermediate-sensor assisted push-pull strategy and its application in heterologous deoxyviolacein production in Escherichia coli. Metab. Eng. 33, 41–51. doi: 10.1016/j.ymben.2015.10.006
Fang, M. Y., Zhang, C., Yang, S., Cui, J. Y., Jiang, P. X., Lou, K., et al. (2015). High crude violacein production from glucose by Escherichia coli engineered with interactive control of tryptophan pathway and violacein biosynthetic pathway. Microb. Cell Fact. 14:8. doi: 10.1186/s12934-015-0192-x
Fordjour, E., Adipah, F. K., Zhou, S. H., Du, G. C., and Zhou, J. W. (2019). Metabolic engineering of Escherichia coli BL21 (DE3) for de novo production of L-DOPA from D-glucose. Microb. Cell Fact. 18:74.
Fossati, E., Ekins, A., Narcross, L., Zhu, Y., Falgueyret, J. P., Beaudoin, G. A. W., et al. (2014). Reconstitution of a 10-gene pathway for synthesis of the plant alkaloid dihydrosanguinarine in Saccharomyces cerevisiae. Nat. Commun. 5:3283. doi: 10.1038/ncomms4283
Fossati, E., Narcross, L., Ekins, A., Falgueyret, J. P., and Martin, V. J. J. (2015). Synthesis of morphinan alkaloids in Saccharomyces cerevisiae. PLoS ONE 10:e0124459. doi: 10.1371/journal.pone.0124459
Fujita, T., Nguyen, H. D., Ito, T., Zhou, S. M., Osada, L., Tateyama, S., et al. (2013). Microbial monomers custom-synthesized to build true bio-derived aromatic polymers. Appl. Microbiol. Biot. 97, 8887–8894. doi: 10.1007/s00253-013-5078-4
Fujiwara, R., Noda, S., Kawai, Y., Tanaka, T., and Kondo, A. (2016a). 4-vinylphenol production from glucose using recombinant Streptomyces mobaraense expressing a tyrosine ammonia lyase from Rhodobacter sphaeroides. Biotechnol. Lett. 38, 1543–1549. doi: 10.1007/s10529-016-2126-z
Fujiwara, R., Noda, S., Tanaka, T., and Kondo, A. (2016b). Styrene production from a biomass-derived carbon source using a coculture system of phenylalanine ammonia lyase and phenylacrylic acid decarboxylase-expressing Streptomyces lividans transformants. J. Biosci. Bioeng. 122, 730–735. doi: 10.1016/j.jbiosc.2016.05.005
Furuya, T., and Kino, K. (2014). Catalytic activity of the two-component flavin-dependent monooxygenase from Pseudomonas aeruginosa toward cinnamic acid derivatives. Appl. Microbiol. Biol. 98, 1145–1154. doi: 10.1007/s00253-013-4958-y
Galanie, S., and Smolke, C. D. (2015). Optimization of yeast-based production of medicinal protoberberine alkaloids. Microb. Cell Fact. 14:144. doi: 10.1186/s12934-015-0332-3
Galanie, S., Thodey, K., Trenchard, I. J., Interrante, M. F., and Smolke, C. D. (2015). Complete biosynthesis of opioids in yeast. Science 349, 1095–1100. doi: 10.1126/science.aac9373
Gottardi, M., Grun, P., Bode, H. B., Hoffmann, T., Schwab, W., Oreb, M., et al. (2017). Optimisation of trans-cinnamic acid and hydrocinnamyl alcohol production with recombinant Saccharomyces cerevisiae and identification of cinnamyl methyl ketone as a by-product. FEMS Yeast Res. 17:fox091. doi: 10.1093/femsyr/fox091
Guo, D. Y., Zhang, L. H., Kong, S. J., Liu, Z. J., Li, X., and Pan, H. (2018). Metabolic engineering of Escherichia coli for production of 2-phenylethanol and 2-phenylethyl acetate from glucose. J. Agr. Food Chem. 66, 5886–5891. doi: 10.1021/acs.jafc.8b01594
Guo, W., Huang, Q. L., Liu, H., Hou, S. L., Niu, S. H., Jiang, Y., et al. (2019). Rational engineering of chorismate-related pathways in Saccharomyces Cerevisiae for improving tyrosol production. Front. Bioeng. Biotechnol. 7:152. doi: 10.3389/fbioe.2019.00152
Hawkins, K. M., and Smolke, C. D. (2008). Production of benzylisoquinoline alkaloids in Saccharomyces cerevisiae. Nat. Chem. Biol. 4, 564–573. doi: 10.1038/nchembio.105
Hazelwood, L. A., Daran, J. M., van Maris, A. J. A., Pronk, J. T., and Dickinson, J. R. (2008). The ehrlich pathway for fusel alcohol production: a century of research on Saccharomyces cerevisiae metabolism. Appl. Environ. Microb. 74, 2259–2266. doi: 10.1128/AEM.02625-07
Heo, K. T., Kang, S. Y., and Hong, Y. S. (2017). De novo biosynthesis of pterostilbene in an Escherichia coli strain using a new resveratrol o-methyltransferase from Arabidopsis. Microb. Cell Fact. 16:30. doi: 10.1186/s12934-017-0644-6
Hou, Y., Gao, B., Cui, J. D., Tan, Z. L., Qiao, C. S., and Jia, S. R. (2019). Combination of multi-enzyme expression fine-tuning and co-substrates addition improves phenyllactic acid production with an Escherichia coli whole-cell biocatalyst. Bioresour. Technol. 287:121423. doi: 10.1016/j.biortech.2019.121423
Huang, Q., Lin, Y. H., and Yan, Y. J. (2013). Caffeic acid production enhancement by engineering a phenylalanine over-producing Escherichia coli strain. Biotechnol. Bioeng. 110, 3188–3196. doi: 10.1002/bit.24988
Huccetogullari, D., Luo, Z. W., and Lee, S. Y. (2019). Metabolic engineering of microorganisms for production of aromatic compounds. Microb. Cell Fact. 18:41. doi: 10.1186/s12934-019-1090-4
Immanuel, S. R. C., Banerjee, D., Rajankar, M. P., and Raghunathan, A. (2018). Integrated constraints based analysis of an engineered violacein pathway in Escherichia coli. Biosystems 171, 10–19. doi: 10.1016/j.biosystems.2018.06.002
Jeong, Y. J., An, C. H., Woo, S. G., Jeong, H. J., Kim, Y. M., Park, S. J., et al. (2014). Production of pinostilbene compounds by the expression of resveratrol o-methyltransferase genes in Escherichia coli. Enzyme Microb. Tech. 54, 8–14. doi: 10.1016/j.enzmictec.2013.09.005
Jiang, J., Bi, H., Zhuang, Y., Liu, S., Liu, T., and Ma, Y. (2016). Engineered synthesis of rosmarinic acid in Escherichia coli resulting production of a new intermediate, caffeoyl-phenyllactate. Biotechnol. Lett. 38, 81–88. doi: 10.1007/s10529-015-1945-7
Jiang, J. J., Yin, H., Wang, S. A., Zhuang, Y. B., Liu, S. W., Liu, T., et al. (2018). Metabolic engineering of Saccharomyces cerevisiae for high-level production of salidroside from glucose. J. Agr. Food Chem. 66, 4431–4438. doi: 10.1021/acs.jafc.8b01272
Kallscheuer, N., Vogt, M., Bott, M., and Marienhagen, J. (2017). Functional expression of plant-derived o-methyltransferase, flavanone 3-hydroxylase, and flavonol synthase in Corynebacterium glutamicum for production of pterostilbene, kaempferol, and quercetin. J. Biotechnol. 258, 190–196. doi: 10.1016/j.jbiotec.2017.01.006
Kallscheuer, N., Vogt, M., Stenzel, A., Gatgens, J., Bott, M., and Marienhagen, J. (2016). Construction of a Corynebacterium glutamicum platform strain for the production of stilbenes and (2s)-flavanones. Metab. Eng. 38, 47–55. doi: 10.1016/j.ymben.2016.06.003
Kang, S. Y., Choi, O., Lee, J. K., Ahn, J. O., Ahn, J. S., Hwang, B. Y., et al. (2015). Artificial de novo biosynthesis of hydroxystyrene derivatives in a tyrosine overproducing Escherichia coli strain. Microb. Cell Fact. 14:78. doi: 10.1186/s12934-015-0268-7
Kang, S. Y., Choi, O., Lee, J. K., Hwang, B. Y., Uhm, T. B., and Hong, Y. S. (2012). Artificial biosynthesis of phenylpropanoic acids in a tyrosine overproducing Escherichia coli strain. Microb. Cell Fact. 11:153. doi: 10.1186/1475-2859-11-153
Kang, S. Y., Heo, K. T., and Hong, Y. S. (2018). Optimization of artificial curcumin biosynthesis in E. coli by randomized 5'-UTR sequences to control the multienzyme pathway. ACS Synth. Biol. 7, 2054–2062. doi: 10.1021/acssynbio.8b00198
Kang, Z., Zhang, C. Z., Du, G. C., and Chen, J. (2014). Metabolic engineering of Escherichia coli for production of 2-phenylethanol from renewable glucose. Appl. Biochem. Biotechnol. 172, 2012–2021. doi: 10.1007/s12010-013-0659-3
Katsuyama, Y., Funa, N., and Horinouchi, S. (2007a). Precursor-directed biosynthesis of stilbene methyl ethers in Escherichia coli. Biotechnol. J. 2, 1286–1293. doi: 10.1002/biot.200700098
Katsuyama, Y., Funa, N., Miyahisa, I., and Horinouchi, S. (2007b). Synthesis of unnatural flavonoids and stilbenes by exploiting the plant biosynthetic pathway in Escherichia coli. Chem. Biol. 14, 613–621. doi: 10.1016/j.chembiol.2007.05.004
Kim, B., Cho, B. R., and Hahn, J. S. (2014). Metabolic engineering of Saccharomyces cerevisiae for the production of 2-phenylethanol via ehrlich pathway. Biotechnol. Bioeng. 111, 115–124. doi: 10.1002/bit.24993
Kim, B. G., Jung, W. D., Mok, H., and Ahn, J. H. (2013). Production of hydroxycinnamoyl-shikimates and chlorogenic acid in Escherichia coli: production of hydroxycinnamic acid conjugates. Microb. Cell Fact. 12:15. doi: 10.1186/1475-2859-12-15
Kim, J. S., Nakagawa, A., Yamazaki, Y., Matsumura, E., Koyanagi, T., Minami, H., et al. (2013). Improvement of reticuline productivity from dopamine by using engineered Escherichia coli. Biosci. Biotechnol. Biochem. 77, 2166–2168. doi: 10.1271/bbb.130552
Kim, S. Y., Song, M. K., Jeon, J. H., and Ahn, J. H. (2018). Current status of microbial phenylethanoid biosynthesis. J. Microbiol. Biotechnol. 28, 1225–1232. doi: 10.4014/jmb.1805.05021
Kim, T. Y., Lee, S. W., and Oh, M. K. (2014). Biosynthesis of 2-phenylethanol from glucose with genetically engineered Kluyveromyces marxianus. Enzyme Microb. Tech. 61–62:44–47. doi: 10.1016/j.enzmictec.2014.04.011
Kogure, T., Kubota, T., Suda, M., Hiraga, K., and Inui, M. (2016). Metabolic engineering of Corynebacterium glutamicum for shikimate overproduction by growth-arrested cell reaction. Metab. Eng. 38, 204–216. doi: 10.1016/j.ymben.2016.08.005
Koma, D., Yamanaka, H., Moriyoshi, K., Ohmoto, T., and Sakai, K. (2012). Production of aromatic compounds by metabolically engineered Escherichia coli with an expanded shikimate pathway. Appl. Environ. Microb. 78, 6203–6216. doi: 10.1128/AEM.01148-12
Kong, S., Pan, H., Liu, X., Li, X., and Guo, D. (2020). De novo biosynthesis of 2-phenylethanol in engineered Pichia pastoris. Enzyme Microb. Technol. 133:109459. doi: 10.1016/j.enzmictec.2019.109459
Kuepper, J., Dickler, J., Biggel, M., Behnken, S., Jager, G., Wierckx, N., et al. (2015). Metabolic engineering of Pseudomonas putida kt2440 to produce anthranilate from glucose. Front. Microbiol. 6:1310. doi: 10.3389/fmicb.2015.01310
Lee, K., Bang, H. B., Lee, Y. H., and Jeong, K. J. (2019). Enhanced production of styrene by engineered Escherichia coli and in situ product recovery (ISPR) with an organic solvent. Microb. Cell Fact. 18:79. doi: 10.1186/s12934-019-1129-6
Lee, S. J., Sim, G. Y., Kang, H., Yeo, W. S., Kim, B. G., and Ahn, J. H. (2018). Synthesis of avenanthramides using engineered Escherichia coli. Microb. Cell Fact. 17:46. doi: 10.1186/s12934-018-0896-9
Lee, S. J., Sim, G. Y., Lee, Y., Kim, B. G., and Ahn, J. H. (2017). Engineering of Escherichia coli for the synthesis of n-hydroxycinnamoyl tryptamine and serotonin. J. Ind. Microbiol. Biot. 44, 1551–1560. doi: 10.1007/s10295-017-1975-3
Li, C., Jia, P., Bai, Y. J., Fan, T. P., Zheng, X. H., and Cai, Y. J. (2019b). Efficient synthesis of hydroxytyrosol from l-3,4-dihydroxyphenylalanine using engineered Escherichia coli whole cells. J. Agric. Food Chem. 67, 6867–6873. doi: 10.1021/acs.jafc.9b01856
Li, C., Zhang, Z., and Wang, J. (2019a). A thermophilic biofunctional multienzyme cascade reaction for cell-free synthesis of salvianic acid a and 3,4-dihydroxymandelic acid. ACS Sustain. Chem. Eng. 7, 18247–18253. doi: 10.1021/acssuschemeng.9b05496
Li, F. F., Zhao, Y., Li, B. Z., Qiao, J. J., and Zhao, G. R. (2016). Engineering Escherichia coli for production of 4-hydroxymandelic acid using glucose-xylose mixture. Microb. Cell Fact. 15:90. doi: 10.1186/s12934-016-0489-4
Li, H., Liang, C. N., Chen, W., Jin, J. M., Tang, S. Y., and Tao, Y. (2017). Monitoring in vivo metabolic flux with a designed whole-cell metabolite biosensor of shikimic acid. Biosens. Bioelectron. 98, 457–465. doi: 10.1016/j.bios.2017.07.022
Li, M., Kildegaard, K. R., Chen, Y., Rodriguez, A., Borodina, I., and Nielsen, J. (2015). De novo production of resveratrol from glucose or ethanol by engineered Saccharomyces cerevisiae. Metab. Eng. 32, 1–11. doi: 10.1016/j.ymben.2015.08.007
Li, M., Schneider, K., Kristensen, M., Borodina, I., and Nielsen, J. (2016). Engineering yeast for high-level production of stilbenoid antioxidants. Sci. Rep. 6:36827. doi: 10.1038/srep36827
Li, X., Chen, Z., Wu, Y., Yan, Y., Sun, X., and Yuan, Q. (2018). Establishing an artificial pathway for efficient biosynthesis of hydroxytyrosol. ACS Synth. Biol. 7, 647–654. doi: 10.1021/acssynbio.7b00385
Li, Y., Li, S., Thodey, K., Trenchard, I., Cravens, A., and Smolke, C. D. (2018). Complete biosynthesis of noscapine and halogenated alkaloids in yeast. Proc. Natl. Acad. Sci. U.S.A. 115, E3922–E3931. doi: 10.1073/pnas.1721469115
Li, Y. R., and Smolke, C. D. (2016). Engineering biosynthesis of the anticancer alkaloid noscapine in yeast. Nat. Commun. 7:12137. doi: 10.1038/ncomms12137
Li, Z., and Liu, J. Z. (2017). Transcriptomic changes in response to putrescine production in metabolically engineered Corynebacterium glutamicum. Front. Microbiol. 8:1987. doi: 10.3389/fmicb.2017.01987
Li, Z., Wang, X., and Zhang, H. (2019c). Balancing the non-linear rosmarinic acid biosynthetic pathway by modular co-culture engineering. Metab. Eng. 54, 1–11. doi: 10.1016/j.ymben.2019.03.002
Liang, J. L., Guo, L. Q., Lin, J. F., He, Z. Q., Cai, F. J., and Chen, J. F. (2016). A novel process for obtaining pinosylvin using combinatorial bioengineering in Escherichia coli. World J. Microb. Biol. 32:102. doi: 10.1007/s11274-016-2062-z
Liang, L. Y., Liu, R. M., Foster, K. E. O., Alakshchoudhury, Cook, S., Cameron, J. C., et al. (2020). Genome engineering of E. coli for improved styrene production. Metab. Eng. 57, 74–84. doi: 10.1016/j.ymben.2019.09.007
Lim, C. G., Fowler, Z. L., Hueller, T., Schaffer, S., and Koffas, M. A. G. (2011). High-yield resveratrol production in engineered Escherichia coli. Appl. Environ. Microb. 77, 3451–3460. doi: 10.1128/AEM.02186-10
Lin, B. X., and Tao, Y. (2017). Whole-cell biocatalysts by design. Microb. Cell Fact. 16:106. doi: 10.1186/s12934-017-0724-7
Lin, Y. H., and Yan, Y. J. (2012). Biosynthesis of caffeic acid in Escherichia coli using its endogenous hydroxylase complex. Microb. Cell Fact. 11:42. doi: 10.1186/1475-2859-11-42
Lin, Y. H., and Yan, Y. J. (2014). Biotechnological production of plant-specific hydroxylated phenylpropanoids. Biotechnol. Bioeng. 111, 1895–1899. doi: 10.1002/bit.25237
Liu, C. Q., Men, X., Chen, H. L., Li, M. J., Ding, Z. R., Chen, G. Q., et al. (2018). A systematic optimization of styrene biosynthesis in Escherichia coli BL21(DE3). Biotechnol. Biofuels 11:14. doi: 10.1186/s13068-018-1017-z
Liu, J. B., Jiang, J., Bai, Y. J., Fan, T. P., Zhao, Y., Zheng, X. H., et al. (2018). Mimicking a new 2-phenylethanol production pathway from Proteus mirabilis JN458 in Escherichia coli. J. Agric. Food. Chem. 66, 3498–3504. doi: 10.1021/acs.jafc.8b00627
Liu, L., Liu, H., Zhang, W., Yao, M. D., Li, B. Z., Liu, D., et al. (2019). Engineering the biosynthesis of caffeic acid in Saccharomyces cerevisiae with heterologous enzyme combinations. Engineering 5, 287–295. doi: 10.1016/j.eng.2018.11.029
Liu, Q., Yu, T., Li, X. W., Chen, Y., Campbell, K., Nielsen, J., et al. (2019). Rewiring carbon metabolism in yeast for high level production of aromatic chemicals. Nat. Commun. 10:4976. doi: 10.1038/s41467-019-12961-5
Liu, S. P., Liu, R. X., El-Rotail, A. A., Ding, Z. Y., Gu, Z. H., Zhang, L., et al. (2014). Heterologous pathway for the production of L-phenylglycine from glucose by E. coli. J. Biotechnol. 186, 91–97. doi: 10.1016/j.jbiotec.2014.06.033
Liu, X., Li, X. B., Jiang, J. L., Liu, Z. N., Qiao, B., Li, F. F., et al. (2018). Convergent engineering of syntrophic Escherichia coli coculture for efficient production of glycosides. Metab. Eng. 47, 243–253. doi: 10.1016/j.ymben.2018.03.016
Liu, X. L., Lin, J., Hu, H. F., Zhou, B., and Zhu, B. Q. (2016). De novo biosynthesis of resveratrol by site-specific integration of heterologous genes in Escherichia coli. FEMS Microbiol. Lett. 363:fnw061. doi: 10.1093/femsle/fnw061
Liu, Y. F., Zhuang, Y. Y., Ding, D. Q., Xu, Y. R., Sun, J. B., and Zhang, D. W. (2017). Biosensor-based evolution and elucidation of a biosynthetic pathway in Escherichia coli. ACS Synth. Biol. 6, 837–848. doi: 10.1021/acssynbio.6b00328
Lukito, B. R., Sekar, B. S., Wu, S., and Li, Z. (2019). Mandelic acid-whole cell-based cascade biotransformation for the production of (S)-mandelic acid from styrene, L-phenylalanine, glucose, or glycerol. Adv. Synth. Catal. 361, 3560–3568. doi: 10.1002/adsc.201900373
Machas, M. S., McKenna, R., and Nielsen, D. R. (2017). Expanding upon styrene biosynthesis to engineer a novel route to 2-phenylethanol. Biotechnol. J. 12:1700310. doi: 10.1002/biot.201700310
Matsumura, E., Nakagawa, A., Tomabechi, Y., Ikushiro, S., Sakaki, T., Katayama, T., et al. (2018). Microbial production of novel sulphated alkaloids for drug discovery. Sci. Rep. 8:7980. doi: 10.1038/s41598-018-26306-7
Matsumura, E., Nakagawa, A., Tomabechi, Y., Koyanagi, T., Kumagai, H., Yamamoto, K., et al. (2017). Laboratory-scale production of (s)-reticuline, an important intermediate of benzylisoquinoline alkaloids, using a bacterial-based method. Biosci. Biotechenol. Biochem. 81, 396–402. doi: 10.1080/09168451.2016.1243985
McKenna, R., Moya, L., McDaniel, M., and Nielsen, D. R. (2015). Comparing in situ removal strategies for improving styrene bioproduction. Bioproc. Biosyst. Eng. 38, 165–174. doi: 10.1007/s00449-014-1255-9
McKenna, R., and Nielsen, D. R. (2011). Styrene biosynthesis from glucose by engineered E. coli. Metab. Eng. 13, 544–554. doi: 10.1016/j.ymben.2011.06.005
McKenna, R., Thompson, B., Pugh, S., and Nielsen, D. R. (2014). Rational and combinatorial approaches to engineering styrene production by Saccharomyces cerevisiae. Microb. Cell Fact. 13:123. doi: 10.1186/s12934-014-0123-2
Milke, L., Ferreira, P., Kallscheuer, N., Braga, A., Vogt, M., Kappelmann, J., et al. (2019). Modulation of the central carbon metabolism of Corynebacterium glutamicum improves malonyl-CoA availability and increases plant polyphenol synthesis. Biotechnol. Bioeng. 116, 1380–1391. doi: 10.1002/bit.26939
Minami, H., Kim, J. S., Ikezawa, N., Takemura, T., Katayama, T., Kumagai, H., et al. (2008). Microbial production of plant benzylisoquinoline alkaloids. Proc. Natl. Acad. Sci. U.S.A. 105, 7393–7398. doi: 10.1073/pnas.0802981105
Mora-Villalobos, J. A., and Zeng, A. P. (2018). Synthetic pathways and processes for effective production of 5-hydroxytryptophan and serotonin from glucose in Escherichia coli. J. Biol. Eng. 12:3. doi: 10.1186/s13036-018-0094-7
Muller, U., van Assema, F., Gunsior, M., Orf, S., Kremer, S., Schipper, D., et al. (2006). Metabolic engineering of the E-coli L-phenylalanine pathway for the production of D-phenylglycine (D-Phg). Metab. Eng. 8, 196–208. doi: 10.1016/j.ymben.2005.12.001
Munoz, A. J., Hernandez-Chavez, G., de Anda, R., Martinez, A., Bolivar, F., and Gosset, G. (2011). Metabolic engineering of Escherichia coli for improving L-3,4-dihydroxyphenylalanine (L-DOPA) synthesis from glucose. J. Ind. Microbiol. Biot. 38, 1845–1852. doi: 10.1007/s10295-011-0973-0
Nakagawa, A., Matsumura, E., Koyanagi, T., Katayama, T., Kawano, N., Yoshimatsu, K., et al. (2016). Total biosynthesis of opiates by stepwise fermentation using engineered Escherichia coli. Nat. Commun. 7:10390. doi: 10.1038/ncomms10390
Nakagawa, A., Minami, H., Kim, J. S., Koyanagi, T., Katayama, T., Sato, F., et al. (2011). A bacterial platform for fermentative production of plant alkaloids. Nat. Commun. 2:326. doi: 10.1038/ncomms1327
Narcross, L., Bourgeois, L., Fossati, E., Burton, E., and Martin, V. J. J. (2016a). Mining enzyme diversity of transcriptome libraries through DNA synthesis for benzylisoquinoline alkaloid pathway optimization in yeast. ACS Synth. Biol. 5, 1505–1518. doi: 10.1021/acssynbio.6b00119
Narcross, L., Fossati, E., Bourgeois, L., Dueber, J. E., and Martin, V. J. J. (2016b). Microbial factories for the production of benzylisoquinoline alkaloids. Trends Biotechnol. 34, 228–241. doi: 10.1016/j.tibtech.2015.12.005
Noda, S., Kawai, Y., Tanaka, T., and Kondo, A. (2015). 4-Vinylphenol biosynthesis from cellulose as the sole carbon source using phenolic acid decarboxylase- and tyrosine ammonia lyase-expressing Streptomyces lividans. Bioresour.Technol. 180, 59–65. doi: 10.1016/j.biortech.2014.12.064
Osipenkov, N., Kulik, A., and Mast, Y. (2018). Characterization of the phenylglycine aminotransferase PglE from Streptomyces pristinaespiralis. J. Biotechnol. 278, 34–38. doi: 10.1016/j.jbiotec.2018.05.007
Park, S., Kang, K., Lee, S. W., Ahn, M. J., Bae, J. M., and Back, K. (2011). Production of serotonin by dual expression of tryptophan decarboxylase and tryptamine 5-hydroxylase in Escherichia coli. Appl. Microbiol. Biot. 89, 1387–1394. doi: 10.1007/s00253-010-2994-4
Qi, W. W., Vannelli, T., Breinig, S., Ben-Bassat, A., Gatenby, A. A., Haynie, S. L., et al. (2007). Functional expression of prokaryotic and eukaryotic genes in Escherichia coli for conversion of glucose to p-hydroxystyrene. Metab. Eng. 9, 268–276. doi: 10.1016/j.ymben.2007.01.002
Reifenrath, M., and Boles, E. (2018). Engineering of hydroxymandelate synthases and the aromatic amino acid pathway enables de novo biosynthesis of mandelic and 4-hydroxymandelic acid with Saccharomyces cerevisiae. Metab. Eng. 45, 246–254. doi: 10.1016/j.ymben.2018.01.001
Ro, D. K., and Douglas, C. J. (2004). Reconstitution of the entry point of plant phenylpropanoid metabolism in yeast (Saccharomyces cerevisiae) - implications for control of metabolic flux into the phenylpropanoid pathway. J. Biol. Chem. 279, 2600–2607. doi: 10.1074/jbc.M309951200
Rodrigues, A. L., Becker, J., Lima, A. O. D. S., Porto, L. M., and Wittmann, C. (2014). Systems metabolic engineering of Escherichia coli for gram scale production of the antitumor drug deoxyviolacein from glycerol. Biotechnol. Bioeng. 111, 2280–2289. doi: 10.1002/bit.25297
Rodrigues, A. L., Trachtmann, N., Becker, J., Lohanatha, A. F., Blotenberg, J., Bolten, C. J., et al. (2013). Systems metabolic engineering of Escherichia coli for production of the antitumor drugs violacein and deoxyviolacein. Metab. Eng. 20, 29–41. doi: 10.1016/j.ymben.2013.08.004
Rodrigues, J. L., Araujo, R. G., Prather, K. L. J., Kluskens, L. D., and Rodrigues, L. R. (2015). Production of curcuminoids from tyrosine by a metabolically engineered Escherichia coli using caffeic acid as an intermediate. Biotechnol. J. 10, 599–609. doi: 10.1002/biot.201400637
Rodrigues, J. L., Gomes, D., and Rodrigues, L. R. (2020). A combinatorial approach to optimize the production of curcuminoids from tyrosine in Escherichia coli. Front. Bioeng. Biotechnol. 8:59. doi: 10.3389/fbioe.2020.00059
Rodriguez, A., Chen, Y., Khoomrung, S., Ozdemir, E., Borodina, I., and Nielsen, J. (2017). Comparison of the metabolic response to over-production of p-coumaric acid in two yeast strains. Metab. Eng. 44, 265–272. doi: 10.1016/j.ymben.2017.10.013
Rodriguez, A., Kildegaard, K. R., Li, M. J., Borodina, I., and Nielsen, J. (2015). Establishment of a yeast platform strain for production of p-coumaric acid through metabolic engineering of aromatic amino acid biosynthesis. Metab. Eng. 31, 181–188. doi: 10.1016/j.ymben.2015.08.003
Romagnoli, G., Knijnenburg, T. A., Liti, G., Louis, E. J., Pronk, J. T., and Daran, J. M. (2015). Deletion of the Saccharomyces cerevisiae ARO8 gene, encoding an aromatic amino acid transaminase, enhances phenylethanol production from glucose. Yeast 32, 29–45. doi: 10.1002/yea.3015
Satoh, Y., Tajima, K., Munekata, M., Keasling, J. D., and Lee, T. S. (2012). Engineering of a tyrosol-producing pathway, utilizing simple sugar and the central metabolic tyrosine, in Escherichia coli. J. Agric. Food Chem. 60, 979–984. doi: 10.1021/jf203256f
Shen, H. J., Cheng, B. Y., Zhang, Y. M., Tang, L., Li, Z., Bu, Y. F., et al. (2016). Dynamic control of the mevalonate pathway expression for improved zeaxanthin production in Escherichia coli and comparative proteome analysis. Metab. Eng. 38, 180–190. doi: 10.1016/j.ymben.2016.07.012
Shen, Y. P., Fong, L. S., Yan, Z. B., and Liu, J. Z. (2019). Combining directed evolution of pathway enzymes and dynamic pathway regulation using a quorum-sensing circuit to improve the production of 4-hydroxyphenylacetic acid in Escherichia coli. Biotechnol. Biofuels 12:94. doi: 10.1186/s13068-019-1438-3
Shrestha, A., Pandey, R. P., and Sohng, J. K. (2019). Biosynthesis of resveratrol and piceatannol in engineered microbial strains: achievements and perspectives. Appl. Microbiol. Biotechnol. 103, 2959–2972. doi: 10.1007/s00253-019-09672-8
Shukal, S., Chen, X. X., and Zhang, C. Q. (2019). Systematic engineering for high-yield production of viridiflorol and amorphadiene in auxotrophic Escherichia coli. Metab. Eng. 55, 170–178. doi: 10.1016/j.ymben.2019.07.007
Sun, H. N., Zhao, D. D., Xiong, B., Zhang, C. Z., and Bi, C. H. (2016). Engineering Corynebacterium glutamicum for violacein hyper production. Microb. Cell Fact. 15:148. doi: 10.1186/s12934-016-0545-0
Sun, Z., Ning, Y., Liu, L., Liu, Y., Sun, B., Jiang, W., et al. (2011). Metabolic engineering of the l-phenylalanine pathway in Escherichia coli for the production of s- or r-mandelic acid. Microb. Cell Fact. 10:71. doi: 10.1186/1475-2859-10-71
Tantong, S., Nuengchamnong, N., Kumphune, S., Incharoensakdi, A., Lindblad, P., and Sirikantaramas, S. (2018). Synechocystis pcc 6803 cells heterologously expressing bacterial tyrosine ammonia lyase can use exogenous tyrosine for p-coumaric acid production. J. Plant Biochem. Biot. 27, 118–122. doi: 10.1007/s13562-017-0416-8
Thapa, S. B., Pandey, R. P., Park, Y. I., and Sohng, J. K. (2019). Biotechnological advances in resveratrol production and its chemical diversity. Molecules 24:2571. doi: 10.3390/molecules24142571
Thodey, K., Galanie, S., and Smolke, C. D. (2014). A microbial biomanufacturing platform for natural and semisynthetic opioids. Nat. Chem. Biol. 10, 837–844. doi: 10.1038/nchembio.1613
Trenchard, I. J., Siddiqui, M. S., Thodey, K., and Smolke, C. D. (2015). De novo production of the key branch point benzylisoquinoline alkaloid reticuline in yeast. Metab. Eng. 31, 74–83. doi: 10.1016/j.ymben.2015.06.010
Trenchard, I. J., and Smolke, C. D. (2015). Engineering strategies for the fermentative production of plant alkaloids in yeast. Metab. Eng. 30, 96–104. doi: 10.1016/j.ymben.2015.05.001
Tyo, K. E. J., Ajikumar, P. K., and Stephanopoulos, G. (2009). Stabilized gene duplication enables long-term selection-free heterologous pathway expression. Nat. Biotechnol. 27, 760–U115. doi: 10.1038/nbt.1555
van Summeren-Wesenhagen, P. V., and Marienhagen, J. (2015). Metabolic engineering of Escherichia coli for the synthesis of the plant polyphenol pinosylvin. Appl. Environ. Microb. 81, 840–849. doi: 10.1128/AEM.02966-14
Vavricka, C. J., Yoshida, T., Kuriya, Y., Takahashi, S., Ogawa, T., Ono, F., et al. (2019). Mechanism-based tuning of insect 3,4-dihydroxyphenylacetaldehyde synthase for synthetic bioproduction of benzylisoquinoline alkaloids. Nat. Commun. 10:2015. doi: 10.1038/s41467-019-09610-2
Verhoef, S., Wierckx, N., Westerhof, R. G. M., de Winde, J. H., and Ruijssenaars, H. J. (2009). Bioproduction of p-hydroxystyrene from glucose by the solvent-tolerant bacterium Pseudomonas putida s12 in a two-phase water-decanol fermentation. Appl. Environ. Microb. 75, 931–936. doi: 10.1128/AEM.02186-08
Wang, H. J., Liu, W. Q., Shi, F., Huang, L., Lian, J. Z., Qu, L., et al. (2018). Metabolic pathway engineering for high-level production of 5-hydroxytryptophan in Escherichia coli. Metab. Eng. 48, 279–287. doi: 10.1016/j.ymben.2018.06.007
Wang, J., Shen, X. L., Rey, J., Yuan, Q. P., and Yan, Y. J. (2018). Recent advances in microbial production of aromatic natural products and their derivatives. Appl. Microbiol. Biot. 102, 47–61. doi: 10.1007/s00253-017-8599-4
Wang, Q. Z., Tang, S. Y., and Yang, S. (2017). Genetic biosensors for small-molecule products: design and applications in high-throughput screening. Front. Chem. Sci. Eng. 11, 15–26. doi: 10.1007/s11705-017-1629-z
Wang, S. Y., Zhang, S. W., Xiao, A. F., Rasmussen, M., Skidmore, C., and Zhan, J. X. (2015). Metabolic engineering of Escherichia coli for the biosynthesis of various phenylpropanoid derivatives. Metab. Eng. 29, 153–159. doi: 10.1016/j.ymben.2015.03.011
Wang, Y. C., Bhuiya, M. W., Zhou, R., and Yu, O. (2015). Pterostilbene production by microorganisms expressing resveratrol o-methyltransferase. Ann. Microbiol. 65, 817–826. doi: 10.1007/s13213-014-0922-z
Watts, K. T., Lee, P. C., and Schmidt-Dannert, C. (2006). Biosynthesis of plant-specific stilbene polyketides in metabolically engineered Escherichia coli. BMC Biotechnol. 6:22. doi: 10.1186/1472-6750-6-22
Wei, T., Cheng, B. Y., and Liu, J. Z. (2016). Genome engineering Escherichia coli for L-DOPA overproduction from glucose. Sci. Rep. 6:30080. doi: 10.1038/srep30080
Winzer, T., Gazda, V., He, Z., Kaminski, F., Kern, M., Larson, T. R., et al. (2012). A papaver somniferum 10-gene cluster for synthesis of the anticancer alkaloid noscapine. Science 336, 1704–1708. doi: 10.1126/science.1220757
Wu, F. L., Cao, P., Song, G. T., Chen, W. J., and Wang, Q. H. (2018). Expanding the repertoire of aromatic chemicals by microbial production. J. Chem. Technol. Biot. 93, 2804–2816. doi: 10.1002/jctb.5690
Wu, J. J., Liu, P. R., Fan, Y. M., Bao, H., Du, G. C., Zhou, J. W., et al. (2013). Multivariate modular metabolic engineering of Escherichia coli to produce resveratrol from l-tyrosine. J. Biotechnol. 167, 404–411. doi: 10.1016/j.jbiotec.2013.07.030
Wu, J. J., Zhang, X., Zhu, Y. J., Tan, Q. Y., He, J. C., and Dong, M. S. (2017a). Rational modular design of metabolic network for efficient production of plant polyphenol pinosylvin. Sci. Rep. 7:1459. doi: 10.1038/s41598-017-01700-9
Wu, J. J., Zhou, P., Zhang, X., and Dong, M. S. (2017b). Efficient de novo synthesis of resveratrol by metabolically engineered Escherichia coli. J. Ind. Microbiol. Biot. 44, 1083–1095. doi: 10.1007/s10295-017-1937-9
Xiong, D. D., Lu, S. K., Wu, J. Y., Liang, C. N., Wang, W., Wang, W. Z., et al. (2017). Improving key enzyme activity in phenylpropanoid pathway with a designed biosensor. Metab. Eng. 40, 115–123. doi: 10.1016/j.ymben.2017.01.006
Xu, J. Y., Xu, Y., Chu, X. H., Tan, M. J., and Ye, B. C. (2018). Protein acylation affects the artificial biosynthetic pathway for pinosylvin production in engineered E. coli. ACS Chem. Biol. 13, 1200–1208. doi: 10.1021/acschembio.7b01068
Xu, P., Rizzoni, E. A., Sul, S. Y., and Stephanopoulos, G. (2017). Improving metabolic pathway efficiency by statistical model-based multivariate regulatory metabolic engineering. ACS Synth. Biol. 6, 148–158. doi: 10.1021/acssynbio.6b00187
Xue, Y., Zhang, Y., Cheng, D., Daddy, S., and He, Q. F. (2014). Genetically engineering Synechocystis sp pasteur culture collection 6803 for the sustainable production of the plant secondary metabolite p-coumaric acid. Proc. Natl. Acad. Sci. U.S.A. 111, 9449–9454. doi: 10.1073/pnas.1323725111
Xue, Y. X., Chen, X. Z., Yang, C., Chang, J. Z., Shen, W., and Fan, Y. (2017). Engineering Eschericha coli for enhanced tyrosol production. J. Agric. Food Chem. 65, 4708–4714. doi: 10.1021/acs.jafc.7b01369
Yan, Y., Jia, P., Bai, Y. J., Fan, T. P., Zheng, X. H., and Cai, Y. J. (2019). Production of rosmarinic acid with ATP and CoA double regenerating system. Enzyme Microb. Technol. 131:109392. doi: 10.1016/j.enzmictec.2019.109392
Yang, D., Kim, W. J., Yoo, S. M., Choi, J. H., Ha, S. H., Lee, M. H., et al. (2018). Repurposing type III polyketide synthase as a malonyl-CoA biosensor for metabolic engineering in bacteria. Proc. Natl. Acad. Sci. U.S.A. 115, 9835–9844. doi: 10.1073/pnas.1808567115
Yang, D., Yoo, S. M., Gu, C., Ryu, J. Y., Lee, J. E., and Lee, S. Y. (2019). Expanded synthetic small regulatory RNA expression platforms for rapid and multiplex gene expression knockdown. Metab. Eng. 54, 180–190. doi: 10.1016/j.ymben.2019.04.003
Yao, Y. F., Wang, C. S., Qiao, J. J., and Zhao, G. R. (2013). Metabolic engineering of Escherichia coli for production of salvianic acid a via an artificial biosynthetic pathway. Metab. Eng. 19, 79–87. doi: 10.1016/j.ymben.2013.06.001
Zhang, H. R., and Stephanopoulos, G. (2013). Engineering E. coli for caffeic acid biosynthesis from renewable sugars. Appl. Microbiol. Biot. 97, 3333–3341. doi: 10.1007/s00253-012-4544-8
Zhang, H. R., and Wang, X. N. (2016). Modular co-culture engineering, a new approach for metabolic engineering. Metab. Eng. 37, 114–121. doi: 10.1016/j.ymben.2016.05.007
Zhao, Y., Wu, B. H., Liu, Z. N., Qiao, J. J., and Zhao, G. R. (2018). Combinatorial optimization of resveratrol production in engineered E-coli. J. Agric. Food Chem. 66, 13444–13453. doi: 10.1021/acs.jafc.8b05014
Zhou, L., Ding, Q., Jiang, G. Z., Liu, Z. N., Wang, H. Y., and Zhao, G. R. (2017). Chromosome engineering of Escherichia coli for constitutive production of salvianic acid A. Microb. Cell Fact. 16:84. doi: 10.1186/s12934-017-0700-2
Zhou, Y., Wu, S. K., and Li, Z. (2016). Cascade biocatalysis for sustainable asymmetric synthesis: from biobased L-phenylalanine to high-value chiral chemicals. Angew. Chem. Int. Ed. 55, 11647–11650. doi: 10.1002/anie.201606235
Zhou, Y., Wu, S. K., and Li, Z. (2017). One-pot enantioselective synthesis of D-phenylglycines from racemic mandelic acids, styrenes, or biobased L-phenylalanine via cascade biocatalysis. Adv. Synth. Catal. 359, 4305–4316. doi: 10.1002/adsc.201700956
Keywords: aromatic amino acid derivatives, stilbenes, benzylisoquinoline alkaloids, metabolic engineering, microorganism, strain construction
Citation: Shen Y-P, Niu F-X, Yan Z-B, Fong LS, Huang Y-B and Liu J-Z (2020) Recent Advances in Metabolically Engineered Microorganisms for the Production of Aromatic Chemicals Derived From Aromatic Amino Acids. Front. Bioeng. Biotechnol. 8:407. doi: 10.3389/fbioe.2020.00407
Received: 28 February 2020; Accepted: 14 April 2020;
Published: 05 May 2020.
Edited by:
Shihui Yang, Hubei University, ChinaReviewed by:
Mingfeng Cao, University of Illinois at Urbana-Champaign, United StatesCongqiang Zhang, Agency for Science, Technology and Research (A*STAR), Singapore
Copyright © 2020 Shen, Niu, Yan, Fong, Huang and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jian-Zhong Liu, bHNzbGp6JiN4MDAwNDA7bWFpbC5zeXN1LmVkdS5jbg==
 Yu-Ping Shen
Yu-Ping Shen Fu-Xing Niu
Fu-Xing Niu Jian-Zhong Liu
Jian-Zhong Liu