- 1Institute of Sericulture and Apiculture, College of Animal Sciences, Zhejiang University, Hangzhou, China
- 2Max Planck Fellow Group on Plankton Community Interaction, Max Planck Institute for Chemical Ecology, Jena, Germany
- 3Analysis Center of Agrobiology and Environmental Sciences, Zhejiang University, Hangzhou, China
- 4Department of Natural Product Biosynthesis, Max Planck Institute for Chemical Ecology, Jena, Germany
- 5Department of Bioorganic Chemistry, Max Planck Institute for Chemical Ecology, Jena, Germany
- 6Key Laboratory for Molecular Animal Nutrition, Ministry of Education, Hangzhou, China
Mulberry (Morus) is an economically important woody tree that is suitable for use in sericulture as forage and in medicine. However, this broad-leaved tree is facing multiple threats ranging from phytopathogens to insect pests. Here, a Gram-positive, endospore-forming bacterium (ZJU1) was frequently isolated from healthy mulberry plants by screening for foliar endophytes showing antagonism against pathogens and pests. Whole-genome sequencing and annotation resulted in a genome size of 4.06 Mb and classified the bacterium as a novel strain of Bacillus amyloliquefaciens that has rarely been identified from tree leaves. An integrative approach combining traditional natural product chemistry, activity bioassays, and high-resolution mass spectrometry confirmed that strain ZJU1 uses a blend of antimicrobials including peptides and volatile organic compounds to oppose Botrytis cinerea, a major phytopathogenic fungus causing mulberry gray mold disease. We showed that the inoculation of endophyte-free plants with ZJU1 significantly decreased both leaf necrosis and mortality under field conditions. In addition to the direct interactions of endophytes with foliar pathogens, in planta studies suggested that the inoculation of endophytes also induced plant systemic defense, including high expression levels of mulberry disease resistance genes. Moreover, when applied to the generalist herbivore Spodoptera litura, ZJU1 was sufficient to reduce the pest survival rate below 50%. A previously undiscovered crystal toxin (Cry10Aa) could contribute to this insecticidal effect against notorious lepidopteran pests. These unique traits clearly demonstrate that B. amyloliquefaciens ZJU1 is promising for the development of successful strategies for biocontrol applications. The search for new plant-beneficial microbes and engineering microbiomes is therefore of great significance for sustainably improving plant performance.
Introduction
Microorganisms colonizing plant surfaces (the rhizosphere and phyllosphere) and inner tissues (the endosphere) play an important role in plant host health and productivity (Barra et al., 2018; Cordovez et al., 2019). Recent high-throughput approaches targeting the entire microbiota, such as whole-metagenome sequencing, have shown that bacteria dominate all plant compartments and perform various beneficial activities (Remus-Emsermann et al., 2012; Leach et al., 2017). For instance, common phyllosphere bacteria belonging to the genus Sphingomonas protect Arabidopsis against phytopathogens by reducing pathogen growth (Innerebner et al., 2011). Furthermore, the transplantation of rhizosphere bacterial communities from resistant tomato suppresses disease symptoms in susceptible cultivars (Kwak et al., 2018), demonstrating the potential for microbiome engineering to increase crop yield (Foo et al., 2017; Dini-Andreote and Raaijmakers, 2018).
In contrast to well-studied phyllosphere and rhizosphere bacteria, only a few studies have specifically examined foliar endophytic bacteria (Ding et al., 2013; Müller et al., 2015). In general, foliar bacterial endophytes occur at low population densities inside plants; however, they can establish a mutualistic relationship with their hosts (Rosenblueth and Martínez-Romero, 2006). For instance, several endophytic Bacillus species produce highly diverse antimicrobials against a variety of phytopathogens, insects and nematodes, indicating potential application in biocontrol (Lopes et al., 2018). Therefore, a more thorough examination of endophytes in diverse plants may result in numerous additional microbial resources that may be used in bioengineering and biotechnology to improve agricultural productivity.
Mulberry (Morus spp.) is a woody perennial tree that is widely cultivated throughout subtropical and temperate regions (Berg, 2001). Mulberry is well-known as an economically important feed crop for the domesticated silkworm (Bombyx mori) in sericulture but also attracts farmers because of its delicious fruit and multiple uses in traditional medicine (He et al., 2013). However, mulberry plants are unfortunately facing multiple threats from phytopathogens and herbivores. For instance, gray mold caused by the fungal pathogen Botrytis cinerea is a widespread and destructive disease of mulberry trees (Elad et al., 2004). Necrotrophic B. cinerea is difficult to control or prevent because it exhibits multifarious modes of attack and a wide host range (Droby and Lichter, 2007), and it can persist as mycelia and/or conidia for extended periods as sclerotia in plant debris (Williamson et al., 2007). In particular, B. cinerea has already developed multiple resistance to several or even all currently used fungicides, which is probably promoted by their excessive use (Weber and Hahn, 2019). Moreover, mulberry is often attacked by a number of insect pests, mostly belonging to the Lepidoptera (Chen B. et al., 2018; Chen et al., 2019). Since mulberry leaves are used to feed silkworms or are processed into pharmaceutical products, the improper use of agrochemicals to treat these diseases and herbivores could be hazardous to silkworms and even humans. Biocontrol appears to offer a valid alternative to solve this problem. An endophytic bacterium, Burkholderia cepacia, that has previously been isolated from mulberry leaves, inhibits the plant-pathogenic fungus Colletotrichum dematium (Ji et al., 2010). However, B. cepacia is an opportunistic human pathogen and attacks young plants such as tobacco, limiting its possible use as a biocontrol agent (Shommu et al., 2015). Considering that diverse bacteria may live inside a leaf, the investigation of new endophytes from mulberry plants has become essential.
In the present study, we screened leaf endophytes associated with healthy trees in a mulberry-planting field and identified the Gram-positive, endospore-forming bacterial strain Bacillus amyloliquefaciens ZJU1, showing antagonism against both B. cinerea and the generalist insect herbivore Spodoptera litura (Chen et al., 2016b). Endophytic B. amyloliquefaciens has been isolated from the seeds and stems of grass plants such as rice (Oryza sativa) and the herb Bacopa monnieri and is known to provide benefits to host plants (Lopes et al., 2018). However, B. amyloliquefaciens has rarely been isolated from the leaves of woody plants. Thus, B. amyloliquefaciens ZJU1 represents a novel strain for examining the potential of biocontrol and other bioapplications. Currently, whole-genome sequencing is poised to provide substantial information for understanding key bacterial characteristics (Liang et al., 2018). Together with sequencing efforts, comparative genomic analysis of this strain with other strains isolated from various environments provided further insight into the pan- and core genomes, suggesting the existence of functional diversity in Bacillus species. Based on both genetic and chemical characterization, we aimed to elucidate the possible mechanisms of this antagonism to lay a foundation for future biocontrol applications.
Materials and Methods
Sample Collection and DNA Extraction
Five healthy mulberry trees were randomly selected from a mulberry-planting field in Hangzhou, China (30°18′6.31″ N, 120°05′9.25″ E), and leaves were collected randomly from different branches of each sampled tree in October 2017. For sampling, surface-disinfected gloves and razor blades as well as sterile bags were used. The individual leaves were severed aseptically from the petioles by using a sterile razor blade, placed a sterile bag, and kept on ice until further processing. The collected leaves were washed with sterile water to remove dust and other debris before being placed in 0.25% NaOCl and 80% EtOH (3 min each step) to remove leaf surface microorganisms. The samples were then rinsed three times in sterile water (1 min each time) again and dried on sterile filter paper to remove excess water. To assess the efficacy of surface disinfection, 100 μL of the last rinse was plated on a PDA plate and incubated at 30°C for 4 days. The plates were examined for the absence or presence of microorganism colony growth. Leaf samples that showed any microbial growth from the last rinse water (the control) were removed from further analyses. The endophytic bacteria were isolated from the surface-sterilized mulberry leaves using the fragmentation technique (Liotti et al., 2018) as follows: the cleaned leaves were aseptically cut into 1 × 1 cm pieces, placed on PDA plates and incubated at 30°C for 4 days. The resulting bacterial colonies were randomly picked and subcultured at least three times before identification. The genomic DNA of pure cultures was extracted using the MasterPure™ Complete DNA and RNA Purification Kit (Epicenter, Madison, USA) according to the manufacturer's instructions.
Genome Sequencing, Assembly and Annotation
The whole genome of B. amyloliquefaciens ZJU1 was sequenced using PacBio RSII in Majorbio Bio-pharm Technology Co., Ltd. (Shanghai, China). 1,154 Mbp high-quality reads were generated, and filtered by following 5′ end containing non-A, G, C, and T bases, sequences shorter than 25 bps, sequences contained with 10% “N” bases, adaptor sequences and low quality reads with quality scores <20. SOAPdenovo V1.05 was used to generate contigs and scaffolds using k-mer sizes and gaps were filled closed by PCR amplification method (Chen et al., 2016a; Jia et al., 2017). The raw dataset, including both single and paired end reads (average read length of 7,483 bp) is deposited at DDBJ/EMBL/GenBank under BioProject ID: PRJNA544619, SRR9125050. Genes of strain ZJU1 were predicted by Glimmer V3.02 software (http://www.cbcb.umd.edu/software/glimmer/). The predicted protein sequences were compared against the Nr, genes, string, and GO databases using BLAST 2.2.28+ and a cut off value 1e−5 (Altschul et al., 1990; Ashburner et al., 2000; Li et al., 2002), respectively. To obtain annotation information, sequences were further compared against the Clusters of Orthologous Group sequence database (COG, http://www.ncbi.nlm.nih.gov/COG/) and Kyoto Encyclopedia of Genes and Genomes database (KEGG, https://www.genome.jp/kegg/) using cutoff value 1e−5. GI island sequences were predicted by three methods: IslandPATH-DIMOB, SIGI-HMM, and IslandPick, and visualized on the chromosome. Transfer RNA (tRNA) and ribosome RNA (rRNA) genes were detected by tRNAscan-SE V1.3.1 (http://lowelab.ucsc.edu/tRNAscan-SE/index.html) and Barrnap V0.4.2 (http://www.vicbioinformatics.com/software.barrnap.shtml), respectively (Lowe and Eddy, 1997). Interspersed repetitive sequences were identified from genomic sequences using RepeatMasker (http://www.repeatmasker.org), and tandem repeats using Tandem Repeats Finder (TRF) (Benson, 1999; Saha et al., 2008). Prediction of prophage was performed by PHAST (http://phast.wishartlab.com) (Zhou et al., 2011). Furthermore, CRISPRFinder online tool (http://crispr.i2bc.paris-saclay.fr/) was used for CRISPR identification (Grissa et al., 2007). Genomic circle map was visualized by Circos V0.64 (http://circos.ca/).
Multi-Genome Analysis
A range of complete genome sequences with protein functions of genus Bacillus were downloaded from NCBI database (12th March 2019, https://www.ncbi.nlm.nih.gov/genome/). MUMER V3.0 was applied for identification of homology regions and for collinearity analysis by default parameters (Kurtz et al., 2004). The inferred collinear genes were used for further phylogenetic and evolutionary analyses. Pan-genome analysis was performed using PGAP, and changes in the number of core genes were predicted by fitting the exponential decay function: . Meanwhile, the pan-genome size was predicted by power function fitting: F(n) = knγ. When γ < 0, it is considered to be a closed pan-genome, and γ ≥ 0, an open pan-genome. Bacillus genomes were selected for homologous gene analysis by OrthoMCL (http://orthomcl.org/common/downloads/software/v2.0/) with thresholds as follows: E-Value, 1e-5; Markov Inflation Index, 1.5. Based on homologous gene clustering analysis, single copy homologous genes were selected for multiple sequence alignment by MAFFT (https://mafft.cbrc.jp/alignment/software/), and Gblocks (http://molevol.cmima.csic.es/castresana/Gblocks.html) was used for quality control. Phylogenetic tree based on single copy genes was constructed using RAxML (https://github.com/stamatak/standard-RAxML). antiSMASH was employed to identify secondary metabolite gene clusters in the ZJU1 genome (Blin et al., 2019).
Antagonism Test Against the Pathogen B. cinerea and the Insect Pest S. litura
To test the biocontrol ability of strain ZJU1 against pathogenic fungi, ZJU1 and B. cinerea cultures were inoculated simultaneously on PDA plates. Bacillus cereus was used as a negative control. Furthermore, a fresh ZJU1 culture was inoculated into PDB medium (200 g/L potato, 20 g/L glucose, pH 5.6 ± 0.2) and grown at 30°C. The bioassay was performed according to previous studies (Shao et al., 2017). Briefly, a 1 mL aliquot of a culture collected after 24 h of incubation was centrifuged at 13,000 rpm for 5 min to collect the supernatant. A 50 μL aliquot of the resulting supernatant was used in the well diffusion assay to observe antagonistic activity against B. cinerea. For in planta infestation experiments, fresh mulberry leaves were inoculated with strain ZJU1, the negative control B. cereus and the challenge pathogen B. cinerea (mycelial plug excised from a PDA plate) and then covered with a polythene bag for 48 h to maintain humidity and avoid any disturbance. Disease incidence and lesion sizes were surveyed. Subsequently, the inoculated leaves were excised and immersed in 80% acetone to extract the chlorophyll contents as previously described (Ritchie, 2006). The absorbance was measured at wavelengths of 645 and 663 nm, respectively. Total chlorophyll content was determined by the following equations: total chlorophyll (μg/mg) = 8.02 (A663) + 20.2 (A645) (Arnon, 1949).
Total RNA from the infested leaves was extracted using the MasterPure™ Complete DNA and RNA Purification Kit (Epicenter, Madison, USA) to investigate the expression of plant disease resistance genes. The extracted RNA was further reverse transcribed to cDNA using random primers (Vazyme Biotech, Nanjing, China). Quantitative RT-PCR (qRT-PCR) was carried out in a Roche LightCycler 480 system (Roche, Basel, Switzerland) using SYBR® qPCR master mix (Vazyme Biotech, Nanjing, China). The primer sequences for all test genes are shown in Table S1. RT-PCR was conducted using the following program: 5 min of denaturation at 95°C, followed by 40 cycles of 10 s at 95°C, 10 s of annealing at an appropriate temperature, and 10 s of elongation at 72°C, followed by a final melting curve step (from 65 to 92°C, 0.5°C/s). Each sample included at least five replicates.
To investigate the insecticidal properties of strain ZJU1, different bacterial cultures were prepared and mixed with artificial feed (pinto bean-based diets) at a dose of 7 x 103 CFU per gram of feed. S. litura larvae were maintained under three regimes: (i) no treatment, (ii) feed containing strain ZJU1, and (iii) feed containing B. cereus as a negative control. The diet was changed every 24 h. The disease symptoms and survival rates were recorded until pupation.
For protein structure prediction, the Cry10Aa sequences (Gene ID: 9779107; 5759939) were searched against BlastN and further analyzed by Geneious V5.5.7 (Kearse et al., 2012). The resulting amino acid sequences were aligned with the CLUSTALW MUSCLE algorithm. The secondary and tertiary structures of Cry10Aa were predicted by Phyre2 using structure templates and default parameters extracted from the Protein Data Bank (PDB) (Kelley and Sternberg, 2009).
In situ Metabolite Extraction, UHPLC-HR-MS and GC-MS Analyses
For the cocultivation of the endophyte bacterium ZJU1 and the pathogenic fungus B. cinerea, each strain was first precultivated for 2 days at 27°C under ambient light/dark conditions. Then, both strains were inoculated onto LB agar plates by striking living cells on a line with an inoculation loop, while the controls consisted of incubating each organism alone with no other stimulus. The experiment was conducted in biological triplicates. After 8 days of incubation at 27°C, the plates were photographed, and the antibiosis areas and bands of the same size located at the edge of the axenic cultures were recovered, cut into small pieces and extracted with 100% methanol under overnight static conditions, followed by 30 min of sonication in an ultrasonic bath. The organic filtrates were passed through folded Whatman® filters (quality 595 ½, diameter 110 mm) and dried under a nitrogen flow for 2 h. The samples were then diluted in methanol:water (1:1, dilution 1/10, UHPLC-grade CHEMSOLUTE®), and 1 μL was injected into a C-18 column (Kinetex, 100 × 2.1 mm, 2.6 μm, Thermo Accucore) and analyzed with a UHPLC-HR-MS (UltiMate 3000 UHPLC, Dionex, USA) coupled to a Q-Exactive Plus Orbitrap mass spectrometer (Thermo Fisher Scientific, Dreieich, Germany). The mass spectrometry analysis was performed in both positive- and negative-ion mode with a scan range of m/z 100–1,500 for both the full-scan and the targeted data-dependent mass spectrometry experiment (ddMS, Top N). The peak resolutions were set at 70,000 and 17,500 for the full-scan and ddMS analyses, respectively. The MS/MS experiments were conducted with an isolation window of 0.4 m/z at a peak resolution of 35,000 (average NCE 15, 30, 45). The maximum ion time was set to 200 ms, and the AGC target was set to 3e6. The analytical standard was prepared in methanol:water (1:1), and a 10 μL aliquot was injected under similar conditions to those mentioned above. The raw MS data and MS/MS spectra were uploaded to the open data repository Dryad (https://www.datadryad.org/) under the https://doi.org/10.5061/dryad.0rxwdbrv4.
For GC-MS analysis, 250 mL Erlenmeyer flasks filled with 100 mL sterile LB medium were inoculated with 50 μL of a ZJU1 culture, then covered with aluminum foil and incubated at 30°C for 48 h. Volatile organic compounds were collected via solid-phase micro extraction (SPME) using 6 biological replicates, and every replicate was sampled for 1 h at room temperature. The SPME fibers and the holders were obtained from Supelco (divinylbenzene-carboxen-polydimethylsiloxane fiber, 50/30 μm DVB/CAR/PDMS, Bellefonte, PA, USA). The fibers were first conditioned according to the manufacturer's instructions. The needles were inserted into the volatile collection equipment through a hole in the aluminum foil cover. After 1 h of exposure, the fiber was introduced into the GC injector for thermal desorption and analysis of the volatile organic compounds. An ISQ LT and Trace 1310 (Thermo Fisher Scientific, Dreieich, Germany) device equipped with a ZB5 column (30 m, 0.25 mm I.D., 0.25 μm film thickness) was run connected to a guard column (10 m, Phenomenex, Aschaffenburg, Germany) using helium (1.2 mL min−1) as the carrier gas. Mass spectra were measured in electron impact (EI) mode at 70 eV, 41–450 m/z. Volatiles were eluted under the following programmed conditions: 40°C (2 min isotherm), followed by heating at 10°C min−1 to 200°C and at 40°C min−1 to 280°C (1 min isotherm). The GC injector (split ratio 1:10), transfer line and ion source were set at 230, 280, and 250°C, respectively. Compounds were identified using standards.
Data Processing and Metabolomics Analysis
The raw data were analyzed in Compound Discoverer software 2.1 (Thermo Fisher Scientific, Dreieich, Germany) for peak picking, deconvolution and identification of the metabolites. The mass tolerance for fragment matching and composition prediction was set to 5 ppm, while the intensity tolerance and threshold were 30 and 0.1%, respectively. Peak detection was performed with a 30% intensity tolerance, signal-to-noise threshold of 3 and mass tolerance of 5 ppm. The signal-to-noise ratio was 3 for fragment matching and composition predictions. Peaks were filtered using a signal-to-noise ratio of 1.5. Raw MS/MS profiles were compared with open data repositories (Metabolika, KEGG, mzCloud) within Compound Discoverer without changing the suggested parameters of the program. Mass tolerance was set to 5 ppm for all tools used for spectrum similarity searched. The identification of surfactin was confirmed on the basis of the MS/MS fragments and retention times of the purchased standard (Sigma-Aldrich, Darmstadt, Germany). The identification of bacillibactin, bacillaene and bacilysin was accomplished by accurate mass interpretation and database matching based on previous literature (Phister et al., 2004; Hertlein et al., 2014).
Results
Isolation and Identification of the Endophytic Bacterium B. amyloliquefaciens From Mulberry Leaves
B. cinerea is the most frequent fungal pathogen isolated from the mulberry-planting fields (Table S2), and causes severe anthracnose infections of mulberry. The symptoms of this mulberry gray mold disease consist of brown to black necrotic spots on the leaves of the tree, leading to yield loss of leaves for silkworm feeding and other uses. The infected leaves that fall in the field are a source of primary inoculum in the following year. To develop potential biocontrol strategies against gray mold, we screened endophytes in a mulberry field where gray mold has never been reported. Isolates from mulberry foliage were tested for their capacity to suppress B. cinerea. Based on colony and cell morphology, endospore formation and Gram staining, a Gram-positive, rod-shaped, endospore-forming bacterium was frequently isolated from healthy mulberry leaves (Figure 1A) and showed high efficacy in controlling gray mold caused by B. cinerea on agar plates under laboratory conditions (Figures 1B,C).
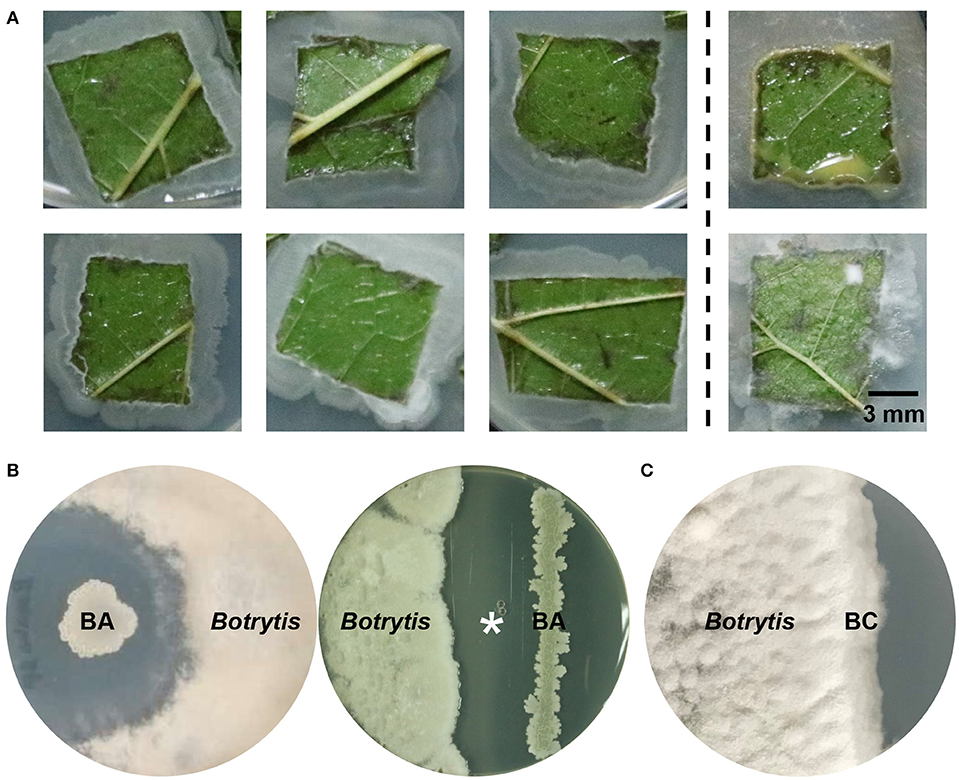
Figure 1. Isolation and identification of the mulberry leaf endophytic bacterium B. amyloliquefaciens. (A) Survey of indigenous bacterial endophytes from healthy mulberry leaves. Left panel, endophytes identified as Bacillus amyloliquefaciens; right panel, other bacterial and fungal endophytes. (B) Antagonistic effect of a newly isolated strain, B. amyloliquefaciens ZJU1 (BA), on the growth of the pathogenic fungus Botrytis cinerea. (C) The negative control Bacillus cereus (BC) did not show any antagonistic activity against B. cinerea. The asterisk indicates the competition zone extracted for the chemical detection and identification of antimicrobials.
Strain ZJU1 is a cultured representative that was initially identified by 16S rRNA analysis as Bacillus amyloliquefaciens. It is well-established that the species B. amyloliquefaciens is associated with many plants (Belbahri et al., 2017), whereas to the best of our knowledge, this is the first report of the presence of B. amyloliquefaciens inside woody tree leaves. Phylogenetic reconstruction based on 16S rRNA sequences also showed that ZJU1 forms a phylogenetic lineage distinct from other B. amyloliquefaciens strains (Figure 2A). Thus, B. amyloliquefaciens ZJU1 represents a novel leaf endophyte that is closely associated with healthy mulberry trees.
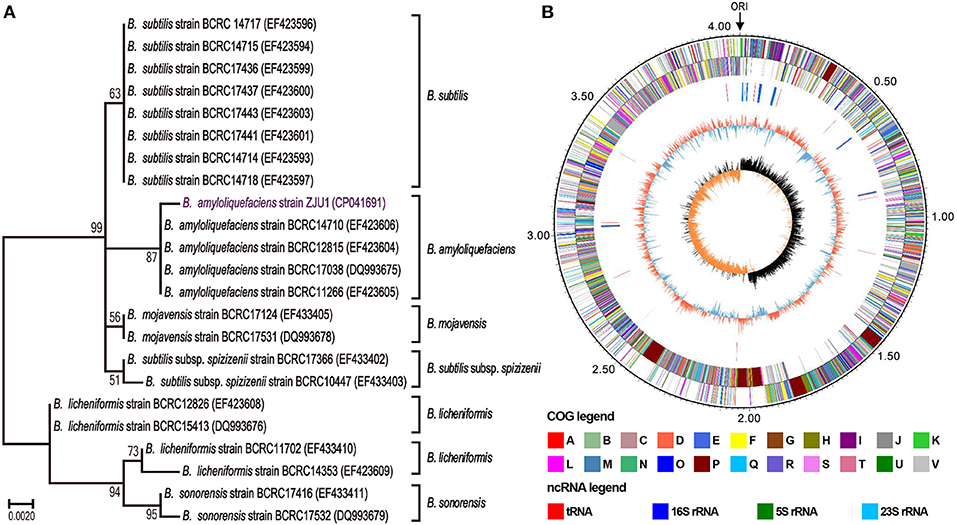
Figure 2. Phylogenetic analysis and circular chromosome of strain ZJU1. (A) Phylogenetic tree of B. amyloliquefaciens ZJU1 based on the 16S rRNA gene sequences of 23 representative Bacillus strains. BCRC, the Bioresource Collection and Research Center, Japan. The tree was built using the construct/test neighbor-joining method with bootstrap test values (based on 500 replications) expressed as a percentage of 100 at the branch points. Only bootstrap percentages above 50% are shown. (B) The circular genome map revealing the genetic basis of ZJU1. From outside to inside, the map shows the (1) size of the complete genome (M); (2–3) sequences encoding the amino acids in proteins on the + and – strands, with different colors representing different COG functional classifications; (4) rRNA and tRNA; (5) GC content, where the outward red portion indicates that the GC content in this region is higher than the average GC content of the whole genome, and the inward blue portion indicates that the GC content in this region is lower than the whole genome average, where the higher the peak value, the greater the difference from the average GC content; and (6) GC skew value (G–C/G+C). When the value is positive, the CDS is more likely to be transcribed from the positive chain; otherwise, the CDS is more likely to be transcribed from the negative chain.
Whole-Genome Sequencing of B. amyloliquefaciens ZJU1 and Comparative Genomic Analyses
We sequenced the ZJU1 genome to investigate the general characteristics of this Bacillus strain. A total of 1.15 billion high-quality bases (284-fold genome coverage) were generated via the whole-genome shotgun sequencing approach. De novo assembly of the sequences successfully generated a 4,064,151 bp circular chromosome with 4,144 genes, and an average gene length of 876 bp (Figure 2B), and no plasmids were detected. The total length of the genes was estimated to be 3,630,891 bp, with a 47.22% GC content, accounting for 89.34% of the entire genome (Table S3). Genome component analyses, including the prediction of genomic islands, CRISPR sequences, tRNA and rRNA, prophages, and repetitive sequences, are also shown in Figure 2B, Figure S1 and Tables S4–S7. Among these components, 9 genomic islands, 86 tRNA genes, and 27 rRNA operons were predicted in the ZJU1 chromosome.
The functional annotation of the resulting genomic sequences was based on a whole-genome BLAST search against several commonly used databases. In the COG analysis, 2,947 COGs were classified into 24 functional categories, and amino acid transport and metabolism constituted the most enriched metabolic category, including 283 related genes. The second highest percentage of COGs was in the transcription category, with 251 genes involved (Figure S2). Gene ontology (GO) analysis indicated that the genes related to metabolic process, catalytic activity and cell parts accounted for the highest proportions among the biological process, molecular function and cellular component categories, respectively (Figure S3). In the KEGG pathways, the number of genes involved in the metabolic pathways (583 unigenes), the biosynthesis of secondary metabolites (277 unigenes) and microbial metabolism in diverse environments (158 unigenes) accounted for 30.68% of the predicted genes (Table S8). Whole-genome-based phylogenetic analysis of ZJU1 revealed that it may be more appropriately identified as a B. amyloliquefaciens subsp. plantarum strain, and the most closely related strain to ZJU1 with an available genome sequence is within B. amyloliquefaciens Y2, which was isolated from wheat (Figure S4).
For comparative genomic analysis, the global gene repertoire of the genus Bacillus was first determined by profiling 42 representative genome assemblies (Table S9). In total, 82,950 genes were clustered from these different species into 8,804 orthologous groups (OGs) by OrthoMCL. Among these clusters, 2,093 contained only a single gene copy (singletons); 667 contained two gene copies (doublets); and 6,044 contained three or more gene copies. We found that these Bacillus genomes shared 2,043 gene family clusters, and strain ZJU exhibited the second largest number (317) of unique clusters after Bacillus velezensis SRCM101413 (413 clusters) (Figure 3A). The gene numbers corresponding to the core genome of genus Bacillus decreased with the addition of new strains, and the accumulation curve for the number of genes in common tended to reach a plateau with the inclusion of 16 genomes (in green, Figure 3B). In contrast to the core genome estimates, the total number of the possible genes—the pangenome—of Bacillus species appeared not to have been saturated, and the gene accumulation curve continued to rise, as depicted in Figure 3B (in red). Based on these genome sequences, the Bacillus core genome consisted of 1,263 genes, representing approximately 28.34% of the average genome size (N = 21) in the same genus. The pangenome for all lineages consisted of 12,889 genes, corresponding to about 3-fold the average size of these selected genomes.
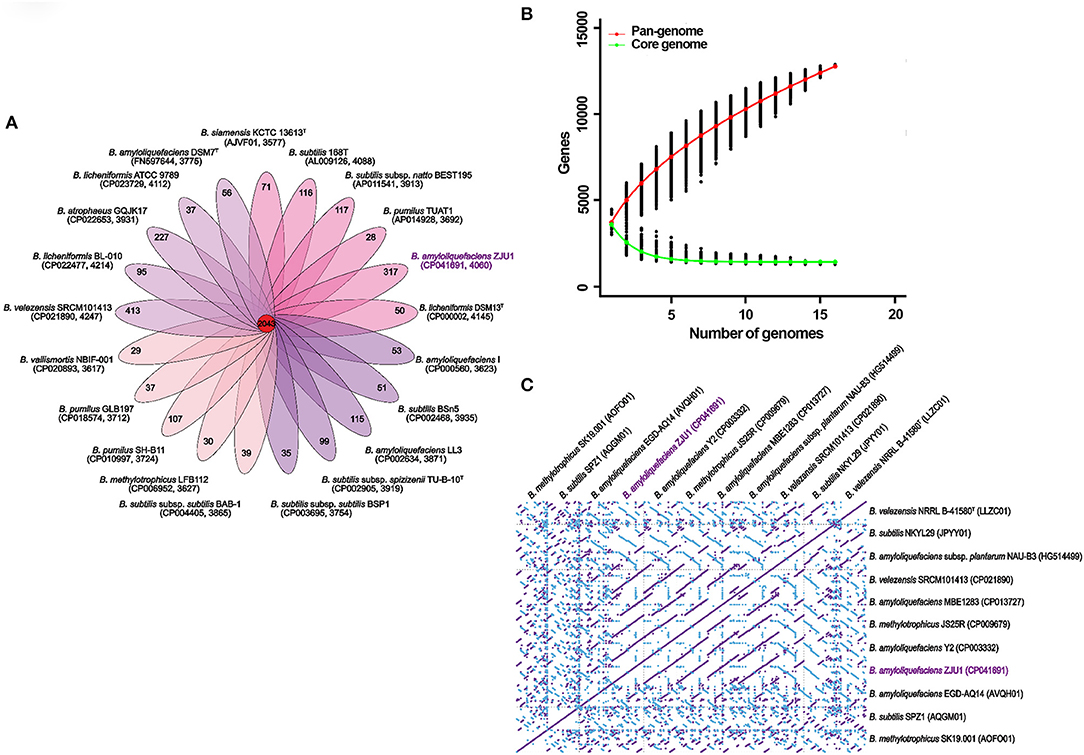
Figure 3. Multigenomic analysis revealing the pangenome of Bacillus and genome synteny. (A) Flower plot illustrating the number of shared (core) and specific (accessory/dispensable) genes based on clusters of orthologs. Each petal displays the number of strain-specific genes found in each genome, with the core orthologous gene number in the center. Inside the brackets, the accession number of the strain and its total number of gene families are shown. (B) Pan and core genome gene numbers in representative Bacillus taxa. The red continuous curve represents the total number of genes (pangenome) for a given number of sequentially added genomes; green indicates the number of ubiquitous genes (core genome) as a function of the number of sequentially added genomes. The vertical bars correspond to standard deviations after repeating random combinations of the genomes. (C) Synteny plot of Bacillus chromosomes based on whole-genome alignments. The diagonal lines calculated with MUMmer display the homologous regions in pairwise genome comparisons. Forward and complimentary strands between the genomic sequences are represented in purple and blue, respectively.
To better understand the correlated gene arrangements among taxa, we examined synteny and collinearity in the Bacillus genomes by aligning the gene loci. Chromosomal collinearity assessed through whole-genome alignments revealed a striking level of conservation in the same direction between the genome sequence of ZJU1 and that of strain B. amyloliquefaciens Y2. As expected for two strains with very close phylogenetic relationships, nearly all regions of sequence similarity fell along the diagonals of the forward strand, except for the only two gene sites, indicating a generally similar gene and sequence order (Figure 3C). Similarly, the longest stretches of conserved syntenic blocks were observed between the Bacillus amyloliquefaciens NAU-B3 and ZJU1 genomes along the diagonals of the complimentary strand. Therefore, B. amyloliquefaciens NAU-B3 is most similar to ZJU1 on the reverse strand. Notably, the two strains were the same as ZJU1 belonging to the B. amyloliquefaciens species, but both were isolated from grass plants. Some other environmental and food product strains also showed some similarities to the ZJU1 genome, such as JPYY01 (soil), LLZC01 (river), and CP021890 (fermentation food).
The Bacterial Endophyte ZJU1 Limits Fungal Pathogen Damage in Mulberry Tree
Genome analysis further revealed major traits of strain ZJU1 in the antagonistic effects on the growth of B. cinerea. There were 31 gene clusters related to secondary metabolite biosynthesis in this strain (Table S10). Among these clusters, 10 candidates covering over 689 kb in total were identified as antimicrobials, including four polyketides, three non-ribosomal peptides (NRPS), one ribosomally synthesized and posttranslationally modified peptide (RiPP), and two others (Table 1). In particular, using high-resolution mass spectrometry, we were able to characterize whether these molecules were truly secreted by ZJU1. The chemical identification of active fractions in the competition zone indicated that surfactin, bacillaene, bacillibactin, and bacilysin were specifically produced by B. amyloliquefaciens ZJU1 to inhibit the growth of the phytopathogenic fungus B. cinerea (Table 1). Moreover, gas chromatography-mass spectrometry (GC-MS) analysis showed that strain ZJU1 also emitted a cocktail of diffusible and volatile organic compounds (VOCs) with antifungal activity, including 2-heptanone, 2-nonanone, and 2-undecanone (Figure S5). Therefore, the bacterial endophyte ZJU1 presents an inherent ability to produce diverse secondary metabolites directed against B. cinerea and other phytopathogens.
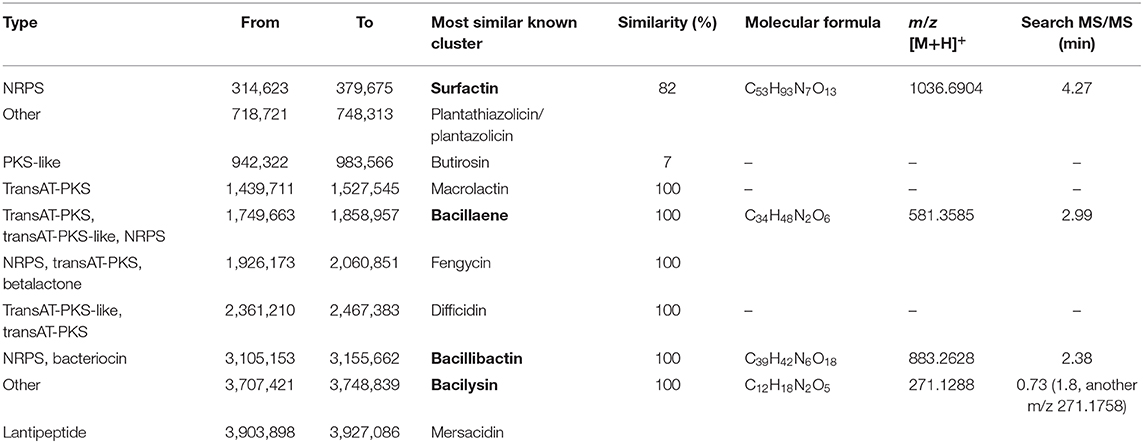
Table 1. Gene clusters directing antimicrobial synthesis in B. amyloliquefaciens ZJU1 and the in situ determination of metabolites.
To validate its use in controlling plant disease under field conditions, ZJU1 was tested in planta on mulberry leaves together with the negative control bacterium Bacillus cereus (Figure 4A). The negative control B. cereus, a common commensal bacillus species isolated from the same field, appeared not to increase plant performance or affect the pathogenicity of B. cinerea; the infection of plants with B. cinerea caused typical disease symptoms. By contrast, the endophyte ZJU1 strongly inhibited the mycelial growth and conidial germination of the fungal pathogen and reduced the incidence or severity of leaf disease in mulberry. Since leaf chlorophyll content provides valuable information about the physiological status of plants, we investigated the chlorophyll content of mulberry leaves after pathogen challenge. The measurement of chlorophyll content showed that chlorophyll was lost more quickly in B. cinerea-infected and B. cereus coinoculated plants; however, ZJU1-coinoculated plants suffered less, thereby maintaining their photosynthetic capacity and preventing pathogen-induced leaf senescence (Figure 4B). These results demonstrated that the bacterial endophyte ZJU1 is sufficient to limit fungal pathogen damage in mulberry plants.
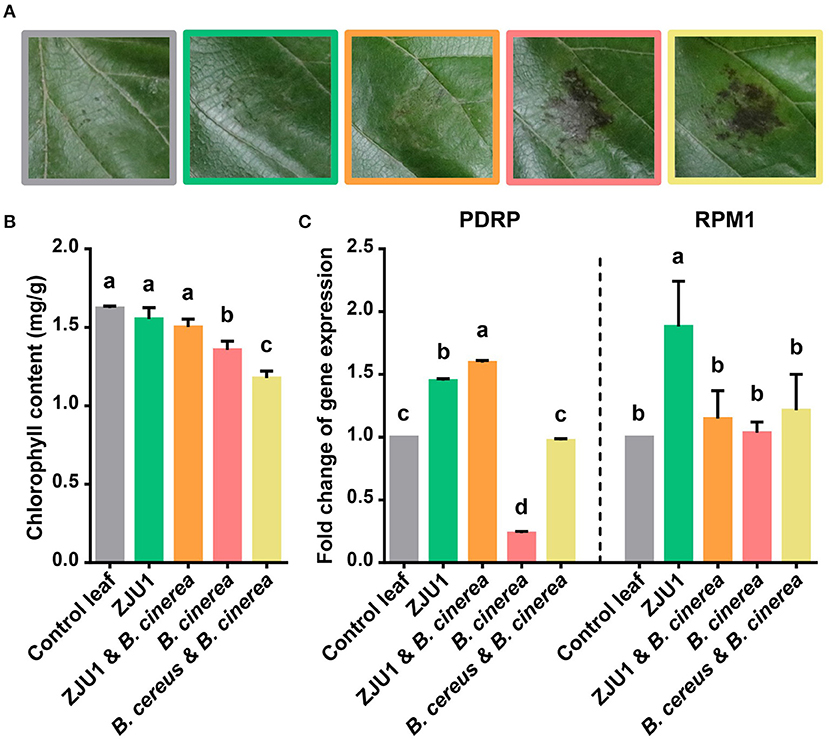
Figure 4. The bacterial endophyte ZJU1 limits fungal pathogen damage in mulberry. (A) Anthracnose disease inhibition by strain ZJU1 on mulberry leaves. Representative results of at least five independent experiments are shown. Same color key as in (B,C). (B) Leaf chlorophyll content indicating the physiological status of plants under different treatments. (C) Expression of putative disease resistance protein-encoding genes of Morus. PDRP, putative disease resistance protein SUMM2; RPM1, disease resistance protein RPM1. Different letters indicate a significant (P < 0.05) difference between treatments by one-way analysis of variance. Error bars indicate ±SD of five replicates.
In addition to the direct antagonism of pathogens, other mechanisms of protection may also exist, such as the stimulation of systemic host responses. We further characterized the mulberry disease resistance gene expression profiles from the in planta experiments. Consistent with previous reports, the plant pathogen B. cinerea reduced the expression level of the putative Morus disease resistance protein (PDRP) after infection. In contrast, the transcript abundance of pathogen defense marker genes (PDRP and RPM1) increased significantly in the presence of ZJU1 (Figure 4C). B. cereus did not significantly enhance the expression level of resistance genes.
Taken together, these results suggest that the production of diverse antagonistic metabolites and activation of the plant defense pathway jointly contribute to the control of the fungal pathogen in mulberry trees.
Strain ZJU1 Antagonizes the Herbivorous Insect Pest S. litura
In addition to the diverse gene clusters involved in the production of antimicrobial metabolites, the in silico analysis of genomic sequences revealed that B. amyloliquefaciens ZJU1 also harbors genes related to the biosynthesis of toxins, particularly the crystal (Cry) proteins that target herbivorous insects. Various Bacillus spp. have evolved specific Cry toxins to colonize insects, which are now being been widely used as a biocontrol strategy in transgenic and organic farming (Castillo-Esparza et al., 2019). Since the main insect pests of mulberry are lepidopteran herbivores, a biocontrol trial with the ZJU1 strain has been performed in the devastating agricultural pest S. litura (Lepidoptera: Noctuidae). The insecticidal activity of ZJU1 was proven via the oral infection of S. litura larvae. The feeding assay showed that the average survival rate of the Spodoptera larvae was below 50% after exposure to B. amyloliquefaciens ZJU1 (Figure 5A). Meanwhile, the untreated control larvae and B. cereus-treated group exhibited higher survival rates (90 and 84%, respectively) during development into pupae. ZJU1 resulted in disease symptoms, pupal malformation, and high mortality (Figure 5B), as described in other lepidopterans infected by Gram-positive entomopathogens (Shao et al., 2017).
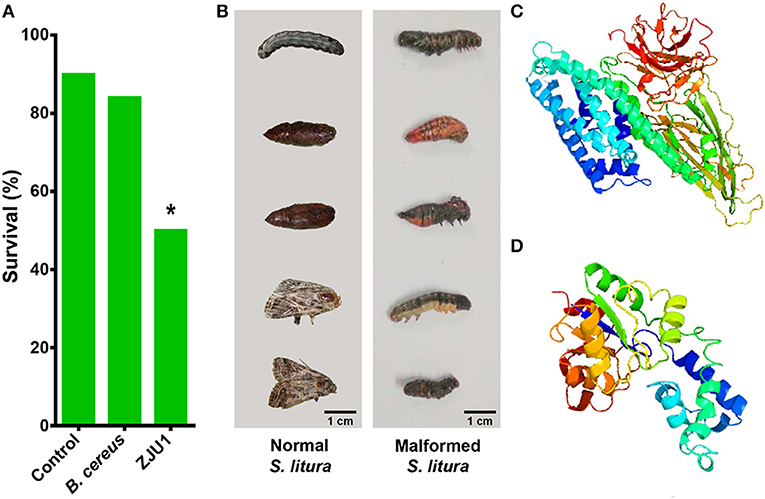
Figure 5. Strain ZJU1 antagonizes the herbivorous insect pest S. litura. (A) Oral insecticidal activity of strain ZJU1. (B) Left panel, normal S. litura not exposed to strain ZJU1; right panel, larval and pupal deformities caused by ZJU1. (C) Tertiary structure prediction for Cry10Aa of B. thuringiensis and (D) B. amyloliquefaciens by Phyre2. Image colored by the rainbow N/C terminus. *Represents a significant difference.
We compared the sequence of the Cry toxin gene in B. amyloliquefaciens ZJU1 with all sequences in the NCBI database, which revealed that this gene is most similar to the Cry10Aa gene from B. thuringiensis. A further comparison of the stereo structures of the Cry10Aa protein from the two Bacillus species provided insight into some important characteristics of this kind of pesticidal crystal toxin. We found that both Cry10Aa proteins are mostly composed of α-helix and β-sheet structures (Figures 5C,D). Although the protein of B. thuringiensis (Figure 5C) has a more complex structure than that in B. amyloliquefaciens ZJU1 (Figure 5D), interestingly, a higher proportion of α-helix (53%) structures was found in the Cry10Aa of B. amyloliquefaciens ZJU1 compared to that of B. thuringiensis (only 32%). Considering that Cry toxins have been successfully used as a bioinsecticide against caterpillars, beetles, mosquitoes, and flies, the new Cry toxin identified from the mulberry endophyte ZJU1 not only offers another positive effect on host plants but also represents a novel biocontrol agent in biotechnology.
Discussion
The development and application of microbial agents for the biocontrol of diseases and pests has received a significant amount of interest in recent years (Xie et al., 2019). In particular, native isolates offer environmentally friendly alternatives to chemical germicides and pesticides and are commonly accepted compared to genetically modified biocontrols. Endophytes living inside plants exhibit an intimate and often symbiotic interaction with their hosts, thus representing a valuable resource for the screening microbial agents with biotechnological potential. To date, only a few plants have ever been completely studied relative to their endophytic biology; consequently, the opportunity to find new and beneficial bacterial endophytes is considerable among the diverse plants in different ecosystems. In this study, we systematically investigated the leaf endophytes of mulberry, an ecologically, economically, and medicinally important plant, and identified a novel Bacillus strain ZJU1 that is a promising candidate for the development of biocontrol agents and biotechnology innovation.
The genus Bacillus comprises a physiologically versatile group of bacteria that includes strains isolated from diverse habitats such as food products, soil, rhizosphere and plant tissue (Vallet et al., 2017). Bacillus has been recognized as a good option for biocontrol applications. Most of their registered products (e.g., in the EU pesticide database) are based on Bacillus (Berg et al., 2017). There are a number of different reasons for their prevalent use: they present advantages over Gram-negative bacteria due to spores they form, allowing them to survive unfavorable conditions; they are easy to formulate and exhibit a prolonged shelf-life and high temperature stability (Wu et al., 2018; Xu et al., 2018; Zhang et al., 2018). Bacillus species such as Bacillus licheniformis, Bacillus cereus, Bacillus subtilis, Bacillus coagulans, and Bacillus clausii are already used as bacterial antagonists of plant pathogens and as plant growth-promoting bacteria (Cisternas-Jamet et al., 2019; Gautam et al., 2019). To better understand the relationship between B. amyloliquefaciens ZJU1 and other Bacillus strains, we performed comparative genomic analysis of the 21 representative strains in this work, which revealed a high level of conservation among the genomes. The genes of the core genome present in all strains are essential to the Bacillus life cycle, traits for habitat adaptation and biotechnological potential. Despite the limited number of strains employed here, this result is also consistent with previous studies on a broad range of Bacillus taxa (Alcaraz et al., 2010; Kim et al., 2017). Notably, ZJU1, showing the highest degree of collinearity with Bacillus amyloliquefaciens Y2, exhibits relatively greater differences in gene contents (317 non-shared family clusters) that may herald its unusual biological potential. Rarefaction analysis of pangenomes further reflected a tremendous increase in strain-specific new traits, indicating the wide diversity of biological functions harbored by Bacillus species. Clearly, more sequencing efforts of new strains are necessary to better understand the genomic structures and diversity of these agriculturally and industrially important bacteria.
Genome sequencing and analysis of B. amyloliquefaciens ZJU1 revealed that it encodes an impressive arsenal of antimicrobial compounds, among which four products exhibiting highly antifungal activity were detected in situ (Loeffler et al., 1986; Chen et al., 2009; Um et al., 2013; Li et al., 2014; Chen K. et al., 2018). In particular, non-ribosomally synthesized lipopeptides and polyketides have been demonstrated to make a significant contribution to plant disease protection (Molinatto et al., 2017). In addition, ZJU1 can produce a variety of bioactive VOCs to antagonize phytopathogens. Among these compounds, 2-heptanone and 2-nonanone were shown to exhibit strong antifungal properties (Wu et al., 2019), suggesting that VOCs produced by ZJU1 also play a role in the process of biocontrol. Our in planta experiments finally verified that the occurrence of gray mold disease caused by the major mulberry pathogen B. cinerea could be successfully suppressed through the application of ZJU1. In addition to controlling diseases directly through its antimicrobial activity, ZJU1 stimulates the expression of immune-related genes in plants and prevents its hosts from continuing to deteriorate under the action of B. cinerea. At the same time, B. cinerea significantly suppressed the host immune response, a well-known virulence strategy for pathogen invasion (Xin et al., 2018). In the interaction between Arabidopsis leaves and the plant-beneficial Bacillus cereus strain AR156, genes involved in several defense pathways, such as salicylate- and jasmonate/ethylene-dependent signaling pathways, were upregulated (Lopes et al., 2018), indicating that beneficial microorganisms can manipulate host immunity to establish a successful relationship with the host. Altogether, these results demonstrate that ZJU1 protects plants against pathogens not only by directly reducing pathogen growth but also by indirectly inducing plant systemic resistance.
A variety of Bacillus species, particularly B. thuringiensis, have been reported to exhibit insecticidal potential involving several mechanisms, including the production of crystal toxins such as the Cry and Cyt proteins (Berry and Crickmore, 2017). However, to the best of our knowledge, no studies have reported any B. amyloliquefaciens strain Cry toxin genes to date. Here, a Cry gene was detected in the strain ZJU1 for the first time. This Cry10Aa gene encodes a completely different insecticidal protein compared to those encoded by other entomopathogenic Bacillus spp., and notably, the Cry10Aa toxin of ZJU1 has a compact structure and, hence, a more stable configuration (53% alpha helix ratio) than its close relative in B. thuringiensis (Pardo-López et al., 2013). Furthermore, our feeding experiment validated that B. amyloliquefaciens ZJU1 had negative effects on the generalist lepidopteran pest S. litura growing from larva to pupa, thereby positively affecting plant development. These results, taken together, highlight that the antagonistic bacterium ZJU1 can also serve as a new microbial insecticide against widespread lepidopteran pests and can broaden the application of crystal toxins in genetically modified crops to support sustainable agriculture.
In conclusion, the leaf endophyte B. amyloliquefaciens ZJU1 isolated from mulberry tree not only represents a novel plant-beneficial bacterium but also possesses great potential for use in bioengineering and biotechnology. Our systemic genomic and functional characterization provides valuable and comprehensive information and will facilitate its wider effective application. Clearly, the production of all of these antibiotic and insecticidal compounds suggests that B. amyloliquefaciens ZJU1 is a good candidate for the development of biocontrol strategies against emerging gray mold-related infectious diseases and notorious lepidopteran pests, for instance, through artificial inoculation of the soil/rhizosphere or seeds/seedlings and direct injection into plant tissues. Considering the complete genome sequence reported here and its amenability to genetic manipulation, ZJU1 could be further engineered to increase plant performance, as it was recently shown that engineering of the banana endosphere microbiome improved Fusarium wilt resistance in banana (Anderson et al., 2019; Liu et al., 2019). Our study therefore highlights that expanding the investigation of the plant microbiome could lead to additional successful discoveries of unexplored novel microbial symbionts with biotechnological potential.
Data Availability Statement
The raw dataset, including both single and paired end reads (average read length of 7,482.9 bp) is deposited at DDBJ/EMBL/GenBank under BioProject ID: PRJNA544619, SRR9125050.
Author Contributions
YS and XL designed the experiments. SX, XZ, and BC performed the genomic and biological experiments. MV, MK, AD, and WB performed the chemical work and analyzed the data. CS assisted with the bacterial isolation and chlorophyll content measurement. SX, MV, MK, and YS wrote the manuscript with contributions from all authors.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Prof. Dr. Georg Pohnert for support with the mass spectrometry. This project was funded by the National Natural Science Foundation of China (Grant Nos. 31970483, 31601906), China Agriculture Research System (Grant No. CARS-18-ZJ0302), Zhejiang province analysis and testing science and technology project (Grant No. 2018C37060), and Max Planck Society, Germany. We thank editors at Nature Research Editing Service from Springer Nature for editorial assistance.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fbioe.2019.00488/full#supplementary-material
References
Alcaraz, L. D., Moreno-Hagelsieb, G., Eguiarte, L. E., Souza, V., Herrera-Estrella, L., and Olmedo, G. (2010). Understanding the evolutionary relationships and major traits of Bacillus through comparative genomics. BMC Genomics 11:332. doi: 10.1186/1471-2164-11-332
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local aligment search tool. J. Mol. Biol. 215, 403–410. doi: 10.1016/S0022-2836(05)80360-2
Anderson, J. A., Ellsworth, P. C., Faria, J. C., Head, G. P., Owen, M. D. K., Pilcher, C. D., et al. (2019). Genetically engineered crops: importance of diversified integrated pest management for agricultural sustainability. Front. Bioeng. Biotechnol. 7:24. doi: 10.3389/fbioe.2019.00024
Arnon, D. I. (1949). Copper enzymes in isolated chloroplasts. Polyphenoloxidase in Beta vulgaris. Plant Physiol. 24, 1–15. doi: 10.1104/pp.24.1.1
Ashburner, M., Ball, C. A., Blake, J. A., Botstein, D., Butler, H., Cherry, J. M., et al. (2000). Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 25, 25–29. doi: 10.1038/75556
Barra, P. J., Viscardi, S., Jorquera, M. A., Duran, P. A., Valentine, A. J., and de la Luz Mora, M. (2018). Understanding the strategies to overcome phosphorus-deficiency and aluminum-toxicity by ryegrass endophytic and rhizosphere phosphobacteria. Front. Microbiol. 9:1155. doi: 10.3389/fmicb.2018.01155
Belbahri, L., Chenari Bouket, A., Rekik, I., Alenezi, F. N., Vallat, A., Luptakova, L., et al. (2017). Comparative genomics of Bacillus amyloliquefaciens strains reveals a core genome with traits for habitat adaptation and a secondary metabolites rich accessory genome. Front. Microbiol. 8:1438. doi: 10.3389/fmicb.2017.01438
Benson, G. (1999). Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 27, 573–580. doi: 10.1093/nar/27.2.573
Berg, C. C. (2001). Moreae, Artocarpeae, and Dorstenia (Moraceae), With Introductions to the Family and Ficus and With Additions and Corrections to Flora Neotropica Monograph 7. New York, NY: New York Botanical Garden Press, 346.
Berg, G., Köberl, M., Rybakova, D., Müller, H., Grosch, R., and Smalla, K. (2017). Plant microbial diversity is suggested as the key to future biocontrol and health trends. FEMS Microbiol. Ecol. 93:fix050. doi: 10.1093/femsec/fix050
Berry, C., and Crickmore, N. (2017). Structural classification of insecticidal proteins - towards an in silico characterisation of novel toxins. J. Invertebr. Pathol. 142, 16–22. doi: 10.1016/j.jip.2016.07.015
Blin, K., Shaw, S., Steinke, K., Villebro, R., Ziemert, N., Lee, S. Y., et al. (2019). antiSMASH 5.0: updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res. 47, W81–W87. doi: 10.1093/nar/gkz310
Castillo-Esparza, J. F., Hernández-González, I., and Ibarra, J. E. (2019). Search for cry proteins expressed by Bacillus spp. genomes, using hidden Markov model profiles. 3 Biotech. 9:13. doi: 10.1007/s13205-018-1533-3
Chen, B., Du, K., Sun, C., Vimalanathan, A., Liang, X., Li, Y., et al. (2018). Gut bacterial and fungal communities of the domesticated silkworm (Bombyx mori) and wild mulberry-feeding relatives. ISME J. 12, 2252–2262. doi: 10.1038/s41396-018-0174-1
Chen, B., Sun, C., Liang, X., Lu, X., Gao, Q., Alonso-Pernas, P., et al. (2016a). Draft genome sequence of Enterococcus mundtii SL 16, an indigenous gut bacterium of the polyphagous pest Spodoptera littoralis. Front. Microbiol. 7:1676. doi: 10.3389/fmicb.2016.01676
Chen, B., Teh, B. S., Sun, C., Hu, S., Lu, X., Boland, W., et al. (2016b). Biodiversity and activity of the gut microbiota across the life history of the insect herbivore Spodoptera littoralis. Sci. Rep. 6:29505. doi: 10.1038/srep29505
Chen, B., Xie, S., Zhang, X., Zhang, N., Feng, H., Sun, C., et al. (2019). Gut microbiota metabolic potential correlates with body size between mulberry-feeding lepidopteran pest species. Pest Manag. Sci. doi: 10.1002/ps.5642. [Epub ahead of print].
Chen, K., Tian, Z., Luo, Y., Cheng, Y., and Long, C. A. (2018). Antagonistic activity and the mechanism of Bacillus amyloliquefaciens DH-4 against citrus green mold. Phytopathology 108, 1253–1262. doi: 10.1094/PHYTO-01-17-0032-R
Chen, X. H., Koumoutsi, A., Scholz, R., and Borriss, R. (2009). More than anticipated - production of antibiotics and other secondary metabolites by Bacillus amyloliquefaciens FZB42. J. Mol. Microbiol. Biotechnol. 16, 14–24. doi: 10.1159/000142891
Cisternas-Jamet, J., Salvatierra-Martínez, R., Vega-Gálvez, A., Uribe, E., Goñi, M. G., and Stoll, A. (2019). Root inoculation of Green bell pepper (Capsicum annum) with Bacillus amyloliquefaciens BCC047: effect on biochemical composition and antioxidant capacity. J. Sci. Food Agric. 99, 5131–5139. doi: 10.1002/jsfa.9758
Cordovez, V., Dini-Andreote, F., Carrión, V. J., and Raaijmakers, J. M. (2019). Ecology and evolution of plant microbiomes. Annu. Rev. Microbiol. 73, 69–88. doi: 10.1146/annurev-micro-090817-062524
Ding, T., Palmer, M. W., and Melcher, U. (2013). Community terminal restriction fragment length polymorphisms reveal insights into the diversity and dynamics of leaf endophytic bacteria. BMC Microbiol. 13:1. doi: 10.1186/1471-2180-13-1
Dini-Andreote, F., and Raaijmakers, J. M. (2018). Embracing community ecology in plant microbiome research. Trends Plant Sci. 23, 467–469. doi: 10.1016/j.tplants.2018.03.013
Droby, S., and Lichter, A. (2007). “Post-harvest botrytis infection: etiology, development and management,” in Botrytis: Biology, Pathology and Control, eds Y. Elad, B. Williamson, P. Tudzynski, and Delen, N (Dordrecht: Springer Netherlands), 349–367.
Elad, Y., Williamson, B., Tudzynski, P., and Delen, N. (2004). Botrytis: Biology, Pathology and Control. Dordrecht: Springer.
Foo, J. L., Ling, H., Lee, Y. S., and Chang, M. W. (2017). Microbiome engineering: current applications and its future. Biotechnol. J. 12:1600099. doi: 10.1002/biot.201600099
Gautam, S., Chauhan, A., Sharma, R., Sehgal, R., and Shirkot, C. K. (2019). Potential of Bacillus amyloliquefaciens for biocontrol of bacterial canker of tomato incited by Clavibacter michiganensis ssp. michiganensis. Microb. Pathog. 130, 196–203. doi: 10.1016/j.micpath.2019.03.006
Grissa, I., Vergnaud, G., and Pourcel, C. (2007). CRISPRFinder: a web tool to identify clustered regularly interspaced short palindromic repeats. Nucleic Acids Res. 35, W52–57. doi: 10.1093/nar/gkm360
He, N., Zhang, C., Qi, X., Zhao, S., Tao, Y., Yang, G., et al. (2013). Draft genome sequence of the mulberry tree Morus notabilis. Nat. Commun. 4:2445. doi: 10.1038/ncomms3445
Hertlein, G., Müller, S., Garcia-Gonzalez, E., Poppinga, L., Süssmuth, R. D., and Genersch, E. (2014). Production of the catechol type siderophore bacillibactin by the honey bee pathogen Paenibacillus larvae. PLoS ONE 9:e108272. doi: 10.1371/journal.pone.0108272
Innerebner, G., Knief, C., and Vorholt, J. A. (2011). Protection of Arabidopsis thaliana against leaf-pathogenic Pseudomonas syringae by Sphingomonas strains in a controlled model system. Appl. Environ. Microbiol. 77, 3202–3210. doi: 10.1128/AEM.00133-11
Ji, X., Lu, G., Gai, Y., Gao, H., Lu, B., Kong, L., et al. (2010). Colonization of Morus alba L. by the plant-growth-promoting and antagonistic bacterium Burkholderia cepacia strain Lu10-1. BMC Microbiol. 10:243. doi: 10.1186/1471-2180-10-243
Jia, B., Chun, B. H., Cho, G. Y., Kim, K. H., Moon, J. Y., Yeo, S. H., et al. (2017). Complete genome sequences of two acetic acid-producing Acetobacter pasteurianus strains (Subsp. ascendens LMG 1590(T) and Subsp. paradoxus LMG 1591(T)). Front. Bioeng. Biotechnol. 5:33. doi: 10.3389/fbioe.2017.00033
Kearse, M., Moir, R., Wilson, A., Stones-Havas, S., Cheung, M., Sturrock, S., et al. (2012). Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647–1649. doi: 10.1093/bioinformatics/bts199
Kelley, L. A., and Sternberg, M. J. (2009). Protein structure prediction on the web: a case study using the phyre server. Nat. Protoc. 4, 363–371. doi: 10.1038/nprot.2009.2
Kim, Y., Koh, I., Young Lim, M., Chung, W. H., and Rho, M. (2017). Pan-genome analysis of Bacillus for microbiome profiling. Sci. Rep. 7:10984. doi: 10.1038/s41598-017-11385-9
Kurtz, S., Phillippy, A., Delcher, A. L., Smoot, M., Shumway, M., Antonescu, C., et al. (2004). Versatile and open software for comparing large genomes. Genome Biol. 5:R12. doi: 10.1186/gb-2004-5-2-r12
Kwak, M. J., Kong, H. G., Choi, K., Kwon, S.-K., Song, J. Y., Lee, J., et al. (2018). Rhizosphere microbiome structure alters to enable wilt resistance in tomato. Nat. Biotechnol. 36, 1100–1109. doi: 10.1038/nbt.4232
Leach, J. E., Triplett, L. R., Argueso, C. T., and Trivedi, P. (2017). Communication in the phytobiome. Cell 169, 587–596. doi: 10.1016/j.cell.2017.04.025
Li, B., Li, Q., Xu, Z., Zhang, N., Shen, Q., and Zhang, R. (2014). Responses of beneficial Bacillus amyloliquefaciens SQR9 to different soilborne fungal pathogens through the alteration of antifungal compounds production. Front. Microbiol. 5:636. doi: 10.3389/fmicb.2014.00636
Li, W., Jaroszewski, L., and Godzik, A. (2002). Tolerating some redundancy significantly speeds up clustering of large protein databases. Bioinformatics 18, 77–82. doi: 10.1093/bioinformatics/18.1.77
Liang, X., Sun, C., Chen, B., Du, K., Yu, T., Luang-In, V., et al. (2018). Insect symbionts as valuable grist for the biotechnological mill: an alkaliphilic silkworm gut bacterium for efficient lactic acid production. Appl. Microbiol. Biotechnol. 102, 4951–4962. doi: 10.1007/s00253-018-8953-1
Liotti, R. G., da Silva Figueiredo, M. I., da Silva, G. F., de Mendonça, E. A. F., and Soares, M. A. (2018). Diversity of cultivable bacterial endophytes in Paullinia cupana and their potential for plant growth promotion and phytopathogen control. Microbiol. Res. 207, 8–18. doi: 10.1016/j.micres.2017.10.011
Liu, Y., Zhu, A., Tan, H., Cao, L., and Zhang, R. (2019). Engineering banana endosphere microbiome to improve Fusarium wilt resistance in banana. Microbiome 7:74. doi: 10.1186/s40168-019-0690-x
Loeffler, W., Tschen, J. S. M., Vanittanakom, N., Kugler, M., Knorpp, E., Hsieh, T.-F., et al. (1986). Antifungal effects of bacilysin and fengymycin from Bacillus-subtilis F-29-3 a comparison with activities of other Bacillus antibiotics. J. Phytopathol. Phytopathol. Zeitschrift 115, 204–213. doi: 10.1111/j.1439-0434.1986.tb00878.x
Lopes, R., Tsui, S., Gonçalves, P. J. R. O., and de Queiroz, M. V. (2018). A look into a multifunctional toolbox: endophytic Bacillus species provide broad and underexploited benefits for plants. World J. Microbiol. Biotechnol. 34:94. doi: 10.1007/s11274-018-2479-7
Lowe, T. M., and Eddy, S. R. (1997). tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25, 955–964. doi: 10.1093/nar/25.5.955
Molinatto, G., Franzil, L., Steels, S., Puopolo, G., Pertot, I., and Ongena, M. (2017). Key impact of an uncommon plasmid on Bacillus amyloliquefaciens subsp. plantarum S499 developmental traits and lipopeptide production. Front. Microbiol. 8:17. doi: 10.3389/fmicb.2017.00017
Müller, H., Berg, C., Landa, B. B., Auerbach, A., Moissl-Eichinger, C., and Berg, G. (2015). Plant genotype-specific archaeal and bacterial endophytes but similar Bacillus antagonists colonize Mediterranean olive trees. Front. Microbiol. 6:138. doi: 10.3389/fmicb.2015.00138
Pardo-López, L., Soberón, M., and Bravo, A. (2013). Bacillus thuringiensis insecticidal three-domain Cry toxins: mode of action, insect resistance and consequences for crop protection. FEMS Microbiol. Rev. 37, 3–22. doi: 10.1111/j.1574-6976.2012.00341.x
Phister, T. G., O'Sullivan, D. J., and McKay, L. L. (2004). Identification of bacilysin, chlorotetaine, and iturin a produced by Bacillus sp. strain CS93 isolated from pozol, a Mexican fermented maize dough. Appl. Environ. Microbiol. 70, 631–634. doi: 10.1128/AEM.70.1.631-634.2004
Remus-Emsermann, M. N., Tecon, R., Kowalchuk, G. A., and Leveau, J. H. (2012). Variation in local carrying capacity and the individual fate of bacterial colonizers in the phyllosphere. ISME J. 6, 756–765. doi: 10.1038/ismej.2011.209
Ritchie, R. J. (2006). Consistent sets of spectrophotometric chlorophyll equations for acetone, methanol and ethanol solvents. Photosyn. Res. 89, 27–41. doi: 10.1007/s11120-006-9065-9
Rosenblueth, M., and Martínez-Romero, E. (2006). Bacterial endophytes and their interactions with hosts. Mol. Plant Microbe Interact. 19, 827–837. doi: 10.1094/MPMI-19-0827
Saha, S., Bridges, S., Magbanua, Z. V., and Peterson, D. G. (2008). Empirical comparison of ab initio repeat finding programs. Nucleic Acids Res. 36, 2284–2294. doi: 10.1093/nar/gkn064
Shao, Y., Chen, B., Sun, C., Ishida, K., Hertweck, C., and Boland, W. (2017). Symbiont-derived antimicrobials contribute to the control of the lepidopteran gut microbiota. Cell Chem. Biol. 24, 66–75. doi: 10.1016/j.chembiol.2016.11.015
Shommu, N. S., Vogel, H. J., and Storey, D. G. (2015). Potential of metabolomics to reveal Burkholderia cepacia complex pathogenesis and antibiotic resistance. Front. Microbiol. 6:668. doi: 10.3389/fmicb.2015.00668
Um, S., Fraimout, A., Sapountzis, P., Oh, D. C., and Poulsen, M. (2013). The fungus-growing termite Macrotermes natalensis harbors bacillaene-producing Bacillus sp. that inhibit potentially antagonistic fungi. Sci. Rep. 3:3250. doi: 10.1038/srep03250
Vallet, M., Vanbellingen, Q. P., Fu, T., Le Caer, J. P., Della-Negra, S., Touboul, D., et al. (2017). An integrative approach to decipher the chemical antagonism between the competing endophytes Paraconiothyrium variabile and Bacillus subtilis. J. Nat. Prod. 80, 2863–2873. doi: 10.1021/acs.jnatprod.6b01185
Weber, R. W. S., and Hahn, M. (2019). Grey mould disease of strawberry in northern Germany: causal agents, fungicide resistance and management strategies. Appl. Microbiol. Biotechnol. 103, 1589–1597. doi: 10.1007/s00253-018-09590-1
Williamson, B., Tudzynski, B., Tudzynski, P., and van Kan, J. A. (2007). Botrytis cinerea: the cause of grey mould disease. Mol. Plant Pathol. 8, 561–580. doi: 10.1111/j.1364-3703.2007.00417.x
Wu, S., Zhuang, G., Bai, Z., Cen, Y., Xu, S., Sun, H., et al. (2018). Mitigation of nitrous oxide emissions from acidic soils by Bacillus amyloliquefaciens, a plant growth-promoting bacterium. Glob. Chang. Biol. 24, 2352–2365. doi: 10.1111/gcb.14025
Wu, Y., Zhou, J., Li, C., and Ma, Y. (2019). Antifungal and plant growth promotion activity of volatile organic compounds produced by Bacillus amyloliquefaciens. Microbiologyopen 8:e813. doi: 10.1002/mbo3.813
Xie, S., Lan, Y., Sun, C., and Shao, Y. (2019). Insect microbial symbionts as a novel source for biotechnology. World J. Microbiol. Biotechnol. 35:25. doi: 10.1007/s11274-019-2599-8
Xin, X. F., Kvitko, B., and He, S. Y. (2018). Pseudomonas syringae: what it takes to be a pathogen. Nat. Rev. Microbiol. 16, 316–328. doi: 10.1038/nrmicro.2018.17
Xu, W. F., Ren, H. S., Ou, T., Lei, T., Wei, J. H., Huang, C. S., et al. (2018). Genomic and functional characterization of the endophytic Bacillus subtilis 7PJ-16 strain, a potential biocontrol agent of mulberry fruit sclerotiniose. Microb. Ecol. 77, 651–663. doi: 10.1007/s00248-018-1247-4
Zhang, Q. X., Zhang, Y., He, L. L., Ji, Z. L., and Tong, Y. H. (2018). Identification of a small antimycotic peptide produced by Bacillus amyloliquefaciens 6256. Pestic. Biochem. Physiol. 150, 78–82. doi: 10.1016/j.pestbp.2018.07.006
Keywords: endophyte, fungal pathogen, lepidopteran pest, mulberry, symbiosis
Citation: Xie S, Vallet M, Sun C, Kunert M, David A, Zhang X, Chen B, Lu X, Boland W and Shao Y (2020) Biocontrol Potential of a Novel Endophytic Bacterium From Mulberry (Morus) Tree. Front. Bioeng. Biotechnol. 7:488. doi: 10.3389/fbioe.2019.00488
Received: 26 September 2019; Accepted: 30 December 2019;
Published: 23 January 2020.
Edited by:
Paola Duran, University of La Frontera, ChileReviewed by:
Ashok K. Dubey, Netaji Subhas Institute of Technology, University of Delhi, IndiaM. Sudhakara Reddy, Thapar University, India
Copyright © 2020 Xie, Vallet, Sun, Kunert, David, Zhang, Chen, Lu, Boland and Shao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yongqi Shao, eXNoYW9Aemp1LmVkdS5jbg==
 Sen Xie
Sen Xie Marine Vallet
Marine Vallet Chao Sun3
Chao Sun3 Bosheng Chen
Bosheng Chen Wilhelm Boland
Wilhelm Boland Yongqi Shao
Yongqi Shao