
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Bioeng. Biotechnol. , 17 January 2020
Sec. Synthetic Biology
Volume 7 - 2019 | https://doi.org/10.3389/fbioe.2019.00480
This article is part of the Research Topic 4th Applied Synthetic Biology in Europe View all 14 articles
Pseudomonas putida KT2440 is a well-established chassis in industrial biotechnology. To increase the substrate spectrum, we implemented three alternative xylose utilization pathways, namely the Isomerase, Weimberg, and Dahms pathways. The synthetic operons contain genes from Escherichia coli and Pseudomonas taiwanensis. For isolating the Dahms pathway in P. putida KT2440 two genes (PP_2836 and PP_4283), encoding an endogenous enzyme of the Weimberg pathway and a regulator for glycolaldehyde degradation, were deleted. Before and after adaptive laboratory evolution, these strains were characterized in terms of growth and synthesis of mono-rhamnolipids and pyocyanin. The engineered strain using the Weimberg pathway reached the highest maximal growth rate of 0.30 h−1. After adaptive laboratory evolution the lag phase was reduced significantly. The highest titers of 720 mg L−1 mono-rhamnolipids and 30 mg L−1 pyocyanin were reached by the evolved strain using the Weimberg or an engineered strain using the Isomerase pathway, respectively. The different stoichiometries of the three xylose utilization pathways may allow engineering of tailored chassis for valuable bioproduct synthesis.
For the establishment of a circular bioeconomy, the chemical industry has to overcome the massive and ever-increasing use of fossil resources and the concomitant production of environmental pollutions including greenhouse gases. The alternative is CO2 as carbon source, either directly or fixed via chemocatalysis or plants (Olah et al., 2009; Goeppert et al., 2012). CO2-fixation though cannot only proceed under natural conditions, there are also many synthetic approaches to convert CO2 to valuable products. The conversion of CO2 mainly results in the production of biomass in microbes and is well-studied. Hence, it immediately becomes clear that the knowledge about CO2-fixation pathways can be utilized to redirect the metabolic flow into the production of chemicals using the synthetic biology arsenal. On the one hand cell-free systems are used to fix CO2 via multienzyme cascades (Schwander et al., 2016; Satagopan et al., 2017), on the other hand engineered autotrophic microbes are used for the production of chemicals based on CO2 (Lan and Liao, 2011; Angermayr et al., 2012). If the usage of non-autotrophic microbes is desired for the production of multi-carbon compounds via CO2-fixation, either existing pathways (e.g., the Calvin cycle) or newly designed pathways have to be introduced into the cells (Guadalupe-Medina et al., 2013; Antonovsky et al., 2016; Bouzon et al., 2017; Schada von Borzyskowski et al., 2018), with the outcome of alternative stoichiometries (Liebal et al., 2018). CO2-fixation via plants is a common choice as lignocellulosic biomass is abundant and does not compete immediately with food applications. Lignocellulosic biomass contains about 25–50% hemicellulose (Saha, 2003). In addition to glucose, the pentose xylose is a dominant building block in hemicellulose. For example, bagasse fibers consist of ~23% xylose (Lee, 1997). While wheat straw and corn stover are industrially used as carbon source for bioethanol production (Larsen et al., 2008; Maas et al., 2008; Zhao et al., 2018), exemplifying the feasibility of the alternatives, most other microbes than yeast are still on the experimental scale.
The well-characterized chassis Pseudomonas putida KT2440 is known for its versatile metabolism, the richness of cofactors, and high solvent tolerance (Ramos et al., 1995; Nelson et al., 2002; Blank et al., 2010; Tiso et al., 2014). Further, the genome is sequenced and well-annotated (Nelson et al., 2002; Belda et al., 2016; Winsor et al., 2016; Nogales et al., 2019). The structure of the central carbon metabolism and the fluxes on different carbon sources are well-studied for P. putida KT2440 (Sudarsan et al., 2014). Further, many tools for genomic engineering are available (Martínez-García and de Lorenzo, 2011; Poblete-Castro et al., 2012; Nikel and de Lorenzo, 2018). Besides, its non-pathogenicity contributes to it being a chassis relevant for production of valuable products (Nelson et al., 2002; Tiso et al., 2014; Loeschcke and Thies, 2015; Wittgens and Rosenau, 2018).
However, P. putida KT2440 lacks the capability to metabolize xylose. Microbial assimilation of xylose has so far been observed by three different metabolic pathways: An isomerase and two oxidative pathways called Weimberg and Dahms. The Isomerase pathway is characterized by the presence of a xylose isomerase and xylulokinase. The resulting intermediate xylulose is subsequently phosphorylated to xylulose-5-phosphate and introduced into the pentose phosphate pathway. The Isomerase pathway is originally found in prokaryotes, such as Escherichia coli and Bacillus subtilis (David and Weismeyer, 1970; Wilhelm and Hollenberg, 1985). The oxidative pathways start with the oxidation of xylose to xylonate. This step is often catalyzed by a periplasmic dehydrogenase. A xylonate dehydratase then converts xylonate into 2-keto-3-deoxy-xylonate. The resulting intermediate is dehydrated to α-ketoglutaric semialdehyde in case of the Weimberg pathway. In case of the Dahms pathway, α-ketoglutaric semialdehyde is split by an aldolase into pyruvate and glycolaldehyde. While pyruvate is directly converted to acetyl-CoA in the central carbon metabolism, glycolaldehyde is further metabolized in several steps to 2-phosphoglycerate (Franden et al., 2018). 2-Phosphoglycerate is then converted in glycolysis. In the Weimberg pathway, an additional oxidation step from α-ketoglutaric semialdehyde to 2-oxoglutarate is present. The latter metabolite is converted in the tricarboxylic acid (TCA) cycle (Figure 1). The Weimberg pathway was reported for example to be present in P. fragi, Haloferax volcanii, and Caulobacter crescentus (Weimberg, 1961; Stephens et al., 2007; Johnsen et al., 2009). The genes of the latter species were already heterologously expressed in different organisms to establish the Weimberg pathway (Meijnen et al., 2009; Radek et al., 2014; Rossoni et al., 2018). Recently, also P. taiwanensis VLB120 was found to be a native xylose-consumer using the Weimberg pathway (Köhler et al., 2015). The Dahms pathway was first found to be present in an unclassified Pseudomonas strain (Dahms, 1974). Some of the genes of C. crescentus were also heterologously expressed to establish the Dahms pathway in E. coli, because E. coli harbors a gene coding for an aldolase, which catalyzes the last step of the Dahms pathway (Choi et al., 2017; Cabulong et al., 2018).
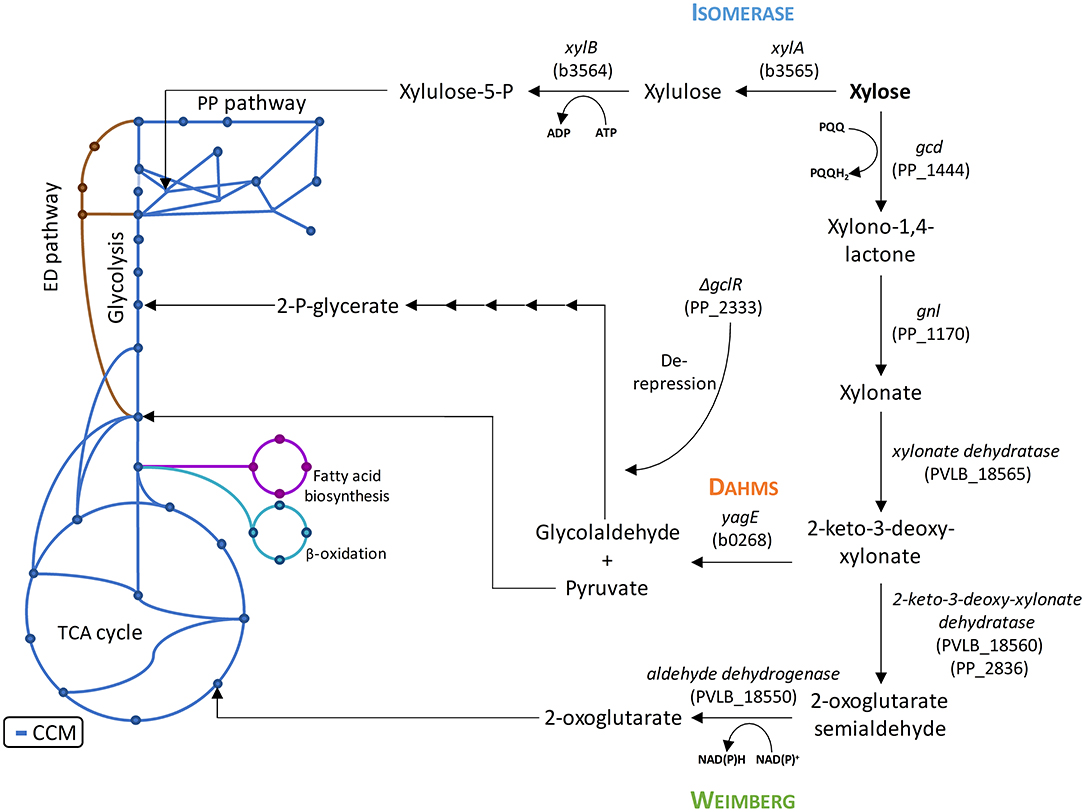
Figure 1. Introduction of xylose metabolism pathways into the central carbon metabolism of P. putida KT2440 with the presumed endogenous enzymes and heterologous enzymes from E. coli DH5α and P. taiwanensis VLB120. The blue lines indicate the central carbon metabolism based on (Sudarsan et al., 2014). The PP numbers represent the locus tag in P. putida KT2440, the PVLB numbers represent the locus tag in P. taiwanensis VLB120, and the b numbers represent the locus tag in E. coli K12, of which E. coli DH5α is derived from. ADP, adenosine diphosphate; ATP, adenosine triphosphate; NAD+, nicotinamide adenine dinucleotide; NADH, reduced nicotinamide adenine dinucleotide; PQQ, pyrroloquinoline quinone; PQQH2, reduced pyrroloquinoline quinone.
P. putida KT2440 has already been engineered to utilize xylose via the Isomerase pathway (Le Meur et al., 2012; Dvorák and de Lorenzo, 2018). Furthermore, P. putida S12 was equipped successfully with xylose metabolism genes to establish the Isomerase and the Weimberg pathway (Meijnen et al., 2008, 2009). In the studies mentioned above, growth via the Isomerase pathway was rather slow. This phenomenon occurred because most of the substrate was oxidized to the dead-end product xylonate. Further, xylose transport was reported to be inefficient in P. putida KT2440. By implementing adaptive laboratory evolution (ALE) growth on xylose could be improved (Meijnen et al., 2008). Using a different approach, Dvorák and de Lorenzo additionally inserted the proton-coupled symporter XylE, also achieving improved growth (Dvorák and de Lorenzo, 2018).
So far, metabolic engineering approaches were limited to metabolic alterations in order to synthesize a target compound based on a previously determined substrate. We here propose to exploit the metabolic potential of microbes for the production of chemicals by electing for the substrate of choice the most suited degradation pathway. Pseudomonads are suitable hosts for the heterologous production of several products, e.g., surfactants, aromatics, terpenoids, and phenazines besides others (Wittgens et al., 2011, 2018; Loeschcke and Thies, 2015; Schmitz et al., 2015b; Tiso et al., 2017; Wynands et al., 2018). However, the usage of xylose for the synthesis of valuable chemicals with P. putida KT2440 has not been reported so far.
In this work, we aimed at engineering a chassis, which can be used for the production of desired secondary metabolites using xylose. First, we investigated the theoretical potential for the synthesis of valuable products using the different xylose pathways via flux balance analysis (FBA). Second, we implemented the three different xylose pathways into P. putida KT2440 and optimized growth by ALE. Third, we used these strains for the synthesis of mono-rhamnolipids and pyocyanin, a derivative of phenazines. We demonstrate efficient growth on xylose via the oxidative pathways. The results show that using xylose pathways having different stoichiometries leads to differences in substrate consumption, growth, and production rates, as well as in biomass and product yields. We propose to use different xylose degradation pathways in dependence of product needs.
For strain maintenance and cloning experiments, the strains P. putida KT2440 (DSM6125, ATCC47054), E. coli DH5α (New England Biolabs, Ipswich, MA, USA), and E. coli PIR2 (ThermoFisher Scientific, Waltham, MA, USA) were routinely cultivated in LB medium containing 10 g L−1 peptone, 5 g L−1 yeast extract, and 10 g L−1 NaCl. P. putida was cultivated at 30°C and E. coli at 37°C. If required, 50 μg mL−1 kanamycin or 30 μg mL−1 gentamycin were added to the medium to avoid loss of plasmid. After mating procedures, P. putida strains were selected on cetrimide agar (Sigma-Aldrich, St. Louis, MO, USA). Growth and production experiments were performed using M9 minimal medium with a final composition (per L) of 8.5 g Na2HPO4x2H2O, 3 g KH2PO4, 0.5 g NaCl, 1 g NH4Cl, 2 mM MgSO4, 4.87 mg FeSO4x7H2O, 4.12 mg CaCl2x2H2O, 1.5 mg MnCl2x4H2O, 1.87 mg ZnSO4x7H2O, 0.3 mg H3BO3, 0.25 mg Na2MoO4x2H2O, 0.15 mg CuCl2x2H2O, 0.84 mg Na2EDTAx2H2O (Sambrook and Russell, 2001), and 10 g glucose for pre-cultures or 10 g xylose for main cultures. Growth experiments were performed in 500 mL shake flasks with 10% filling volume at 200 rpm and in 24-deep well-plates (SystemDuetz; Enzyscreen B.V., Heemstede, The Netherlands) with 1 mL filling volume at 300 rpm.
Strains for the isotope labeling experiments were grown under the conditions stated above. The medium contained 50% of 1-13C labeled xylose (99% purity, Sigma-Aldrich, St. Louis, MO, USA). When the culture reached pseudo-steady state, samples with defined amounts of biomass (0.3 mg) were taken, hydrolyzed, and derivatized according to Schmitz et al. (2015a). The MS data were processed with iMS2Flux (Poskar et al., 2012), which enables the correction of the data for the presence of naturally occurring isotopes and the determination of the fractional labeling of selected amino acids. The fractional labeling indicates the fraction of 13C-labeled carbon atoms.
The genes for xylose utilization were amplified from genomic DNA of E. coli DH5α and P. taiwanensis VLB120 (isolated with High Pure PCR Template Preparation Kit, Roche Holding, Basel, Switzerland). Plasmid construction of pBT-Isomerase, pBT-Weimberg, and pBT-Dahms was planned with NEBuilder Assembly online tool and performed with NEBuilder HiFi DNA Assembly (New England Biolabs, Ipswich, MA, USA) (Gibson et al., 2009). The expression vector pBT including the constitutive Ptac promoter (Koopman et al., 2010) was linearized with XbaI (New England Biolabs, Ipswich, MA, USA) prior to assembly. DNA fragments for Gibson Assembly were amplified using Q5 High-Fidelity DNA Polymerase (New England Biolabs, Ipswich, MA, USA) according to the manual. All used primers with their nucleotide sequences are listed in the supplemental information (Supplementary Table 1). For pBT-Isomerase, the operon xylAB (b3565 and b3564 from E. coli DH5α) was amplified using primers IB-27 and IB-28 and cloned into the linearized pBT vector by Gibson Assembly. To generate pBT-Weimberg, an operon from P. taiwanensis VLB120 was used. The first two genes (PVLB_18555 and PVLB_18550), originally on antisense strand, were amplified using primers IB-63 and IB-64 and the following two genes (PVLB_18560 and PVLB_18565) were amplified using primers IB-65 and IB-66. Afterwards, the linearized pBT vector and both amplified fragments were assembled in one reaction. After that, pBT-Weimberg was used as template for pBT-Dahms. The backbone including PVLB_18555 and PVLB_18550 was amplified from pBT-Weimberg with primers IB-67 and IB-74. Amplification of gene PVLB_18565 from P. taiwanensis VLB120 was performed using primers IB-68 and IB-124 and the gene for the aldolase yagE (b0268 from E. coli DH5α) was amplified using the primers IB-118 and IB-125 to create a synthetic operon. All fragments were assembled via Gibson reactions. In a final step to obtain pBT-Dahms, the putative dehydratase PVLB_18550 was deleted from the assembled vector. Therefore, the vector was amplified with primers IB-93 and IB-94. Subsequently, the amplified vector was self-ligated using T4 Polynucleotide Kinase, T4 DNA Ligase, and T4 DNA Ligase Reaction Buffer (all New England Biolabs GmbH, Ipswich, MA, USA) according to the manual.
The resulting plasmids, pBT-Isomerase, pBT-Weimberg, and pBT-Dahms, were transferred individually into chemically competent E. coli PIR2 cells using heat shock according to Hanahan (1983). Transformants were selected on LB agar with kanamycin. Positive colonies were verified via colony PCR using OneTaq 2x Master Mix with Standard Buffer (New England Biolabs, Ipswich, MA, USA). The efficiency of colony PCR was increased by lysis of cell material with alkaline polyethylene glycol according to Chomczynski and Rymaszewski (2006). Plasmids were validated by Sanger sequencing performed by Eurofins (Brussels, Belgium). Subsequently, plasmids were isolated with Monarch Plasmid Miniprep Kit (New England Biolabs, Ipswich, MA, USA) and transferred via electroporation in competent P. putida cells according to Choi et al. (2006). Electroporation was performed using a GenePulser Xcell (Biorad, Hercules, CA, USA) using a cuvette with a 2 mm gap and the settings 2.5 kV, 200 Ω, and 25 μF. Plasmids pBT-Isomerase and pBT-Weimberg were transferred individually in P. putida KT2440 and P. putida KT2440 Δgcd. Plasmid pBT-Dahms was transferred individually in P. putida KT2440, P. putida KT2440 ΔPP_2836, and P. putida KT2440 ΔgclR ΔPP_2836. Harboring of the plasmids was verified via colony PCR using primers IB-5 and IB-35 (as above).
Deletion mutants were obtained using the I-SceI-based system developed by Martínez-García and de Lorenzo (2011). Briefly, 700 bp upstream and downstream flanking regions of the target sites, named TS1 and TS2 regions, were amplified from the genomic DNA of P. putida KT2440 and cloned into the non-replicative pEMG vector by Gibson Assembly. The resulting plasmids, pEMG-gcd and pEMG-PP_2836, were transferred individually into chemically competent E. coli PIR2 cells using heat shock as described above. Afterwards, the plasmids were verified by Sanger sequencing and transferred from the E. coli PIR2 strains into the required Pseudomonas strains via triparental mating according to Wynands et al. (2018). The I-SceI-encoding plasmid pSW-2 was used and no 3-methylbenzoate for induction of I-SceI expression was needed according to Wynands et al. (2018). Positive colonies with loss of kanamycin resistance were verified for targeted deletion by colony PCR using OneTaq 2x Master Mix with Standard Buffer. To obtain marker-free and pure clones, the recombinant strains were cured of pSW-2 plasmid by re-inoculation in LB medium without gentamycin and verified again by colony PCR. In this study, the gene for glucose dehydrogenase (PP_1444) was deleted in the wild type, resulting in P. putida KT2440 Δgcd. The gene for the putative 2-keto-3-deoxy-xylonate dehydratase (PP_2836) was deleted in the wild type and P. putida KT2440 ΔgclR (Li et al., 2019), resulting in P. putida KT2440 ΔPP_2836 and P. putida KT2440 ΔgclR ΔPP_2836.
To demonstrate the production of secondary metabolites, two target molecules, rhamnolipids and phenazines, were chosen. For the production of mono-rhamnolipids, the mini-Tn7 delivery transposon vector (pBG) developed by Zobel et al. (2015), which inserts a synthesis module in a single genomic locus of the chromosome, was used. Therefore, plasmid pBG14ffg with a stronger constitutive, synthetic promoter than pBG14g (Sebastian Köbbing, RWTH Aachen University, personal communication) was used as backbone and amplified with primers SK06 and SK07. The genes enabling rhamnolipid production rhlAB from P. aeruginosa were amplified with primers SK08 and SK09 and cloned into the amplified backbone by Gibson Assembly. The resulting mini-Tn7 vector pSK02 was integrated into the genome of xylose consuming strains via homologous recombination according to Zobel et al. (2015). The integration of the rhamnolipid synthesis module into the attTn7 site was verified by colony PCR as described above. Mono-rhamnolipid producing clones were identified using cetrimide-blood agar plates (7.5% (v/v) sheep blood; Fiebig-Naehrstofftechnik, Idstein-Niederauroff, Germany). If rhamnolipid synthesis occurred, a halo around the colony was visible, because of their hemolytic activity. The 10 clones showing the largest halos were selected for experiments in minimal medium with 10 g L−1 xylose to quantitatively examine rhamnolipid production. Subsequently, the best three producers of each strain were subjected to replicate experiments and out of these the best was chosen for further characterization and is described in this study.
Phenazine production was achieved by plasmid-based expression. Plasmid pJNN_phzA1-G1,M,S (modified from Schmitz et al., 2015b) was transferred into competent xylose consuming strains via electroporation according to Choi et al. (2006). Harboring of the plasmid was confirmed via colony PCR as described above. Based on Schmitz et al. (2015b), five clones were used to inoculate 1 mL minimal medium containing antibiotics and 10 g L−1 xylose at a starting OD600 of 0.1 and induced with 0.1 mM salicylate to induce phenazine synthesis. If phenazine synthesis occurred, a blue color, which is typical for pyocyanin production, was observed. The best three producers of each strain were subjected to replicate experiments and out of these the best was chosen for further characterization and is described in this study.
All used strains in this study are listed in Table 1.
For adaptation to xylose, the different xylose-consuming strains were grown in M9 minimal medium containing 10 g L−1 xylose. OD600 was measured daily and the cells were sequentially transferred to fresh medium with a starting OD600 of 0.1. The sub-culturing was carried out 30 times for the isomerase-strain, 17 times for the Weimberg-strain, and 26 times for the Dahms-strain. The inhomogeneous culture was streaked out on LB-agar to obtain single isolates, which were subsequently tested for adaption to xylose in a 96-well-plate in a Growth Profiler 960 (Enzyscreen B.V., Heemstede, The Netherlands).
The optical density at 600 nm (OD600) was measured using an Ultrospec 10 cell density meter (Amersham Biosciences, UK). A correlation between OD600 and cell dry weight (CDW) was established. An OD600 of 1.0 corresponds with a cell dry weight of 369 mg L−1.
Xylose and xylonate concentrations in the supernatant were analyzed in a Beckmann Coulter System Gold High Performance Liquid Chromatography (HPLC) (Beckmann Coulter, Brea, CA, USA) with a Metab-AAC 300 × 7.8 mm separation column (particle size: 10 μm, ISERA GmbH, Düren, Germany), a UV detector 166 (Beckmann Coulter, Brea, CA, USA) at 210 nm and a refractory index detector (RI 2300, Knauer GmbH, Berlin, Germany). Elution was performed with 5 mM H2SO4 at a flow rate of 0.5 ml min−1 at 40°C.
Reversed-phase chromatography was performed for analyzing mono-rhamnolipid concentrations based on a method developed earlier (Behrens et al., 2016; Tiso et al., 2016). For sample preparation, the supernatant was mixed 1:1 with acetonitrile and stored at 4°C overnight. Subsequently, the mixture was centrifuged at 16,500 g for 2 min. All samples were filtered with Phenex RC syringe filters (0.2 μm, Ø 4 mm, Phenomenex, Torrance, USA). The HPLC system Ultimate 3000 with a Corona Veo Charged Aerosol Detector (Thermo Fisher Scientific, Waltham, MA, USA) was used. For separation, a NUCLEODUR C18 Gravity 150 × 4.6 mm column (particle size: 3 mm, Macherey-Nagel GmbH & Co. KG, Düren, Germany) was used. The flow rate was set to 1 ml min−1 and the column oven temperature was set to 40°C. Acetonitrile (A) and 0.2% (v/v) formic acid in ultra-pure water (B) were used as running buffers. The method started with a ratio of 70% buffer A: 30% buffer B and a linear gradient was applied to reach a ratio of 80:20% in 8 min. The acetonitrile fraction was increased linearly from 80 to 100% between 9 and 10 min and decreased linearly to 70% between 11 and 12.5 min. The measurement was stopped after 15 min.
The concentration of phenazines was determined in a spectrophotometer Cary 60 (Agilent, Santa Clara, CA, USA) by analyzing the blue phenazine derivative pyocyanin. Samples were centrifuged at 16,500 g for 2 min. Afterwards, the supernatant was vortexed and pyocyanin was measured at 691 nm. For quantification, Lambert-Beer's law with an extinction coefficient of 4.31 mM−1 cm−1 for pyocyanin was used (Filloux and Ramos, 2014).
13C-labeling pattern of proteinogenic amino acids were determined by gas chromatography coupled with mass spectrometry (GC-MS) as described in Schmitz et al. (2015a). A Trace GC Ultra coupled to an ISQ single quadrupole mass spectrometer with a TriPlus RSH autosampler (all Thermo Fisher Scientific, Waltham, MA, USA) was used. For separation, a TraceGOLD TG-5SilMS capillary column (length: 30 mm, inner diameter: 0.25 mm, film thickness: 0.25 μm, Thermo Fisher Scientific, Waltham, MA, USA) was used. A sample volume of 2 μL was injected at 270°C with a split ratio of 1:50. The flow rate of the carrier gas helium was set to 1 mL min−1 and the oven temperature was kept constant for 1 min at 140°C. The temperature was increased with a gradient of 10°C min−1 to 310°C and then kept constant for 1 min. The temperature of the transfer line and the ion source were both set to 280°C. Ionization was performed by electron impact ionization at −70 eV. All raw data were analyzed using Xcalibur (Thermo Fisher Scientific, Waltham, MA, USA).
For the prediction of product yields, the genome-scale model of P. putida KT2440, iJN1411, was used (Nogales et al., 2017). All simulations were carried out in MATLAB (version R2017b, the Mathworks, Inc., Natick, MA, USA) using the COBRA toolbox (Schellenberger et al., 2011), with the linear programming solver of Gurobi1. First, the existing model network was extended by the xylose utilization pathways as well as the biosynthesis routes to the desired products, e.g., mono-rhamnolipids, pyocyanin, and ethylene glycol, using reaction information from KEGG (Ogata et al., 1999). Subsequently, the evaluation of the respective product yields for the usage of the three xylose utilization pathways and their combination was performed. Here, the uptake of xylose for all investigated cases was set to 10 mmol h−1 and the uptake of other carbon sources (e.g., glucose) was set to zero. The production of the target molecule was used as objective function in the extended model.
With FBA one can compute the maximal product yield in different scenarios (e.g., growth, maintenance coefficients, aeration). Here, the genome-scale model P. putida KT2440 iJN1411 (Nogales et al., 2017) was modified by introducing the alternative xylose utilization pathways. In addition, various synthesis reactions for valuable products according to Werpy and Petersen (2004) and chemicals we are working on were introduced into iJN1411.
The maximal product yields of 12 of the 14 metabolites computed on xylose were produced by the Isomerase pathway (Table 2). The exceptions of maximal yields of metabolites are such products that are directly synthesized from intermediates of the Weimberg and Dahms pathway. For example, the intermediate 2-oxoglutarate of the Weimberg pathway is an interesting metabolite, because it is introduced into the TCA cycle without carbon loss. The direct conversion of 2-oxoglutarate into glutamate (or proline, OH-proline, etc.) would thus benefit from the Weimberg pathway. This was also computed by FBA with a maximal product yield of 1 mmol mmol−1 for the Weimberg pathway, and only 0.83 and 0.75 mmol mmol−1 for the Isomerase and the Dahms pathway, respectively. The intermediate glycolaldehyde, using the Dahms pathway for xylose degradation, itself is a relevant chemical for industrial applications and a precursor for ethylene glycol synthesis. Ethylene glycol can neither be synthesized via the Isomerase nor the Weimberg pathway, while the maximal product yield of the Dahms pathway is 1 mmol mmol−1. Consequently, the Dahms pathway is a favorable pathway to produce glycols out of xylose. In case of the Isomerase pathway, xylulose-5-phosphate is part of the pentose phosphate pathway and the intermediate erythrose-4-phosphate is a starting metabolite for aromatics synthesis via the shikimate pathway. This explains that the Isomerase pathway has the highest yield for the production of the here synthesized aromatic compound pyocyanin (0.27 mmol mmol−1). Moreover, the highest maximal mono-rhamnolipid yield is also possible via the Isomerase pathway (0.14 mmol mmol−1) and lower yields for the Weimberg and the Dahms pathway (0.08 and 0.12 mmol mmol−1) were computed. This is due to CO2 production using the TCA cycle for the Weimberg pathway and converting glycolaldehyde in several steps to 2-phosphoglycerate for the Dahms pathway, also releasing one CO2. In addition, the maximal product yields for all possible combinations of the xylose pathways (Isomerase + Weimberg, Isomerase + Dahms, Weimberg + Dahms, and Isomerase + Weimberg + Dahms) were computed. In general, it was observed that always one of the pathways in the combinations is favored and is thus exclusively used. Interestingly, and contrary to the above, the combination of the Weimberg and the Dahms pathways resulted in higher maximal product yields for nine of the 14 metabolites (Table 2) compared to the single pathway strains. Thus, in Table 2 only the combined activities of the latter pathways are also listed. In summary, the in silico study indicates that the strain equipped with the Isomerase pathway is best-suited for a range of chemicals including the production of the here chosen metabolites mono-rhamnolipids and pyocyanin. The Weimberg and Dahms pathways are favored in niche applications (Table 2), when xylose is the sole carbon and energy source.
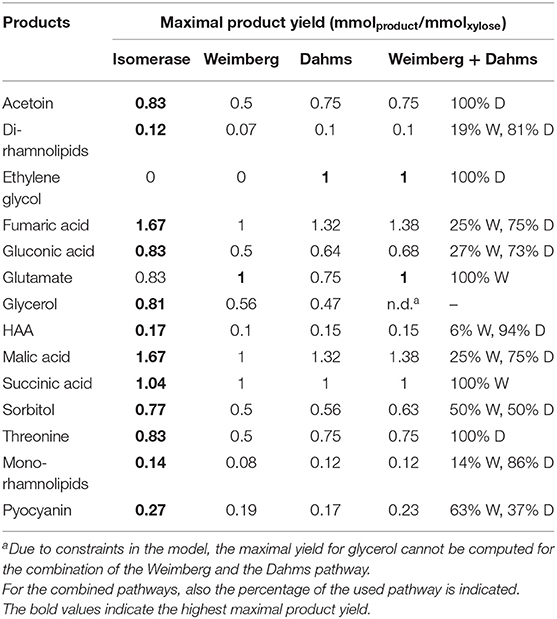
Table 2. FBA computed maximal product yield of valuable products for three different xylose utilization pathways (Isomerase, Weimberg, and Dahms) and the combination of the Weimberg (W) and the Dahms (D) pathway.
A genome analysis using BLAST showed that P. putida KT2440 possesses genes (Altschul et al., 1990), which might encode enzymes needed in xylose utilization, but a complete pathway seems to be absent, indeed correlating with no-growth on this sugar (Figure 2A). All genes required for the Isomerase, Weimberg, and Dahms pathways were compared to the genome of P. putida KT2440. No homologs for the genes xylA and xylB from E. coli for the Isomerase pathway are present. Additionally, no homologs for the genes PVLB_18565 from P. taiwanensis and yagE from E. coli for the Weimberg pathway and the Dahms pathway, respectively, are present. In contrast, homologous genes of the Weimberg pathway from P. taiwanensis are present (homologs to PVLB_18550, PVLB_18555, and PVLB_18560 with sequence identities of 68–72%). HPLC analysis revealed that xylose was consumed by non-growing P. putida KT2440, however xylose was converted into the dead-end product xylonate (Figure 2A). Xylonate synthesis proceeds likely via the gene products of gcd (PP_1444) and gnl (PP_1170) that are besides for glucose also active for xylose and its lactone derivative (Figure 1).
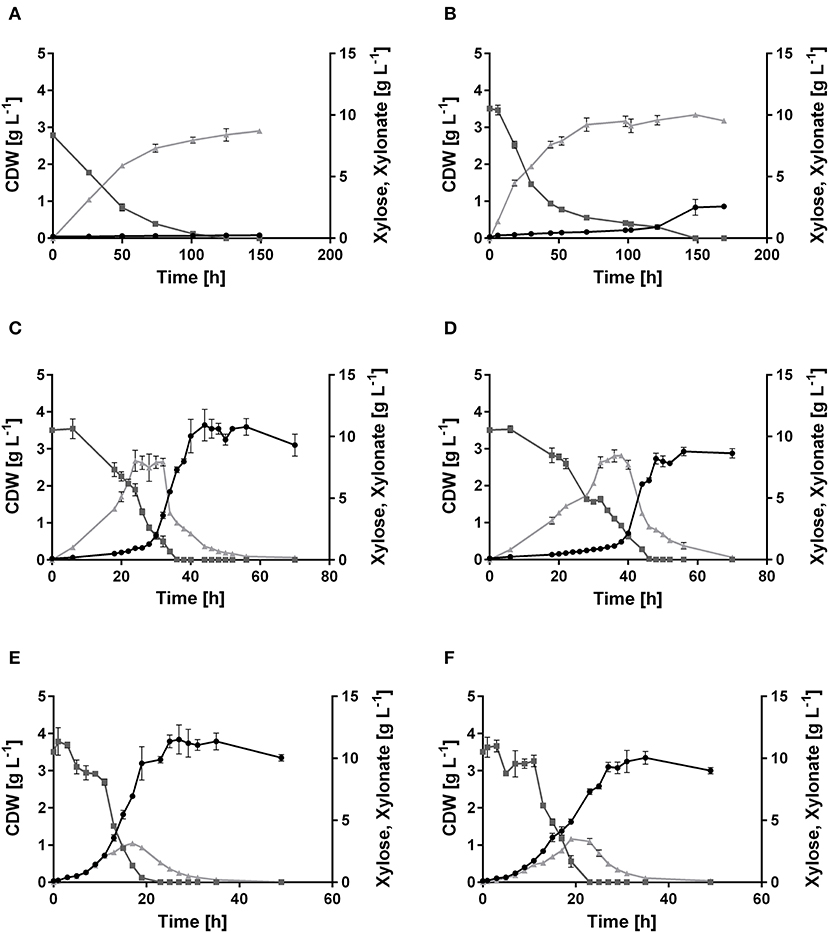
Figure 2. Growth of P. putida KT2440, its derivate xylose consuming strains, and the xylose-adapted strains in minimal medium containing 10 g L−1 xylose. Consumption of xylose ( , dark gray), formation of xylonate (
, dark gray), formation of xylonate ( , light gray), and cell dry weight (•, black) of (A) P. putida KT2440, (B) P. putida KT2440 pIso, (C) P. putida KT2440 pWeim, (D) P. putida KT2440ΔΔ pDahms, (E) P. putida KT2440 pWeim2, and (F) P. putida KT2440ΔΔ pDahms2. Error bars indicate deviation from the mean (n = 3).
, light gray), and cell dry weight (•, black) of (A) P. putida KT2440, (B) P. putida KT2440 pIso, (C) P. putida KT2440 pWeim, (D) P. putida KT2440ΔΔ pDahms, (E) P. putida KT2440 pWeim2, and (F) P. putida KT2440ΔΔ pDahms2. Error bars indicate deviation from the mean (n = 3).
In order to engineer P. putida for the utilization of xylose, P. putida was equipped with a plasmid for the Isomerase pathway and designated P. putida KT2440 pIso. After inoculation, strain P. putida KT2440 pIso showed little growth (Figure 2B) with a maximal growth rate of 0.02 h−1 and a substrate consumption rate of 0.47 mmol L−1 h−1 (Table 3). In addition, pBT-Isomerase was introduced into P. putida KT2440 Δgcd. This strain is unable to produce the dead-end product xylonate and thus more substrate should be available for biomass formation, as it was reported previously in a study with P. putida S12 (Meijnen et al., 2008). The resulting P. putida KT2440 Δgcd pIso showed no improved growth compared to P. putida KT2440 pIso (data not shown). Nevertheless, these results suggest that the cloned genes xylAB from E. coli are functionally expressed in P. putida KT2440 and allow slow growth of the recombinant strain.
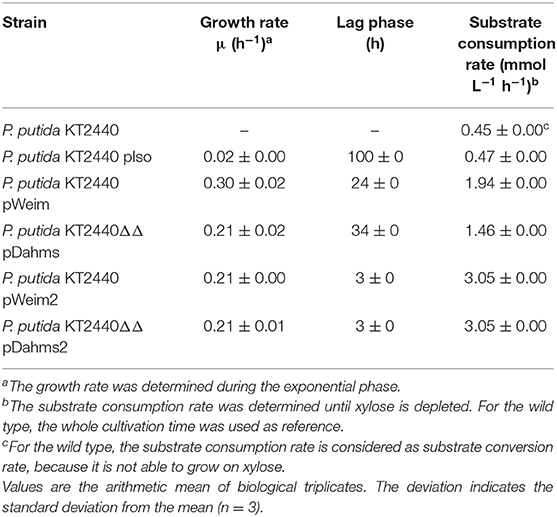
Table 3. Growth characteristics of wild type, recombinant, and evolved P. putida KT2440 strains on xylose.
In order to establish the oxidative Weimberg pathway in P. putida KT2440, the plasmid pBT-Weimberg was transferred into P. putida KT2440 resulting in P. putida KT2440 pWeim. Interestingly, P. putida KT2440 pWeim showed efficient growth with a maximal growth rate of 0.30 h−1 and a strong accumulation of xylonate (Figure 2C). This led to a long lag phase of ~24 h (Table 3). We define the lag phase as a phase before growth was observed due to homogenous adaptation. The growth of a small phenotypic subpopulation on the new carbon source due to heterogenous adaptation described by Kotte et al. (2014) is not considered. The observed lag phase indicates a bottleneck in the conversion of xylonate to 2-keto-3-deoxy-xylonate or in the transport from the periplasm to the cytoplasm. Further, the substrate consumption rate was about 4-fold higher (1.94 mmol L−1 h−1) in comparison to P. putida KT2440 pIso (Table 3). These results show that the xylose operon from P. taiwanensis is functionally expressed in P. putida KT2440 and results in efficient growth. To verify that the glucose dehydrogenase (Gcd) features a side-activity for xylose in P. putida KT2440, the plasmid pBT-Weimberg was introduced into P. putida KT2440 Δgcd yielding P. putida KT2440 Δgcd pWeim. After inoculation, no growth was observed (data not shown). This behavior confirms the assumption that Gcd is active for xylose because xylose needs to be converted to xylonate to establish the Weimberg pathway (Figure 1).
Moreover, the expression of genes encoding the Dahms pathway was targeted. Therefore, the plasmid for the Dahms pathway was introduced into wild type yielding P. putida KT2440 pDahms. As described before, genome analysis revealed that a homologous gene of PVLB_18560 is present (PP_2836). By comparison with genes expressing xylose-metabolizing enzymes from C. crescentus, PVLB_18560 is assumed to encode a dehydratase, which converts 2-keto-3-deoxy-xylonate to α-ketoglutaric semialdehyde (Figure 1). As a result, it is assumed that the product of the homologous gene PP_2836 in P. putida KT2440 is likely to catalyze this reaction. To verify this hypothesis, the plasmid pBT-Dahms was also transferred into P. putida KT2440 ΔPP_2836 yielding P. putida KT2440 ΔPP_2836 pDahms. P. putida KT2440 pDahms reached an OD600 of 8.2 after 74 h, while P. putida KT2440 ΔPP_2836 pDahms stopped growing after 48 h (final OD600 of 2.0). The latter strain showed cell clumping after 74 h, which implies stress. We speculated that low optical density and cell-clumping are caused by an accumulation of the toxic intermediate glycolaldehyde. These findings strongly indicate that PP_2836 encodes a 2-keto-3-deoxy-xylonate dehydratase. In order to overcome the toxic effect of glycolaldehyde, strain P. putida KT2440 ΔgclR was used. The glyoxylate carboligase oxidase pathway is active in this mutant, thus avoiding glycolaldehyde accumulation (Li et al., 2019). The plasmid pBT-Dahms was introduced into P. putida KT2440 ΔgclR ΔPP_2836 resulting in P. putida KT2440ΔΔ pDahms. In comparison to P. putida KT2440 ΔPP_2836 pDahms, strain P. putida KT2440ΔΔ pDahms indeed grew to higher cell densities, which led to the conclusion that the genes encoding the enzymes for the Dahms pathway are successfully expressed and the prevention of glycolaldehyde accumulation leads to efficient growth. The growth of P. putida KT2440ΔΔ pDahms was further examined and a maximal growth rate of 0.21 h−1 and a substrate consumption rate of 1.46 mmol L−1 h−1 was observed (Table 3). Further, accumulation of xylonate occurred as in the case of the Weimberg strain. Thereby, a lag phase of ~34 h results (Figure 2D).
As stated above, it was observed that not only a long lag phase occurs when the Isomerase pathway is used, but also when the oxidative pathways are used. Regulatory mechanisms are assumed as a cause, but in the case of the oxidative pathways, the used route for metabolization is different. There, the xylose gets oxidized to xylonate via xylono-1,4-lactone by the enzymes encoded by gcd and gnl. It is assumed that these enzymes are active for xylose, but with a lower specificity and thus, the conversion is slower than for glucose and a long lag phase is the result. Further, accumulation of xylonate was observed, which indicates a bottleneck in xylonate conversion or transport.
It was shown that all xylose metabolism plasmids are functionally expressed resulting in weak growth for the Isomerase pathway and efficient growth for the Weimberg and Dahms pathways, respectively. However, the lag phase of all strains was relatively long. To optimize growth, ALE was applied. For each of the strains, P. putida KT2440 pIso, P. putida KT2440 pWeim, and P. putida KT2440ΔΔ pDahms, two parallel cultures were sequentially transferred to fresh minimal medium with xylose as the sole carbon source (Figure 3A). The performance of P. putida KT2440 pWeim and P. putida KT2440ΔΔ pDahms was improved after 17 and 26 transfers corresponding to ~115 and 160 generations, respectively. The number of generations was calculated based on the starting OD600 and the end OD600 of every transfer. Afterwards, 24 isolates were obtained from the culture broth and up to six from each culture were randomly selected and tested for growth on xylose in comparison to the initial strains. In Figure 3B five isolates in comparison to the initial strain P. putida KT2440 pWeim are shown. The adaptation toward xylose is not similar for all isolates, as some isolates still had a longer lag phase, suggesting that an isolation step to obtain the best growing strain is required. Hereafter, the best isolates were designated as P. putida KT2440 pWeim2 and P. putida KT2440ΔΔ pDahms2. The adaptation of P. putida KT2440 pIso to xylose was problematic because the strain did not grow anymore after several transfers.
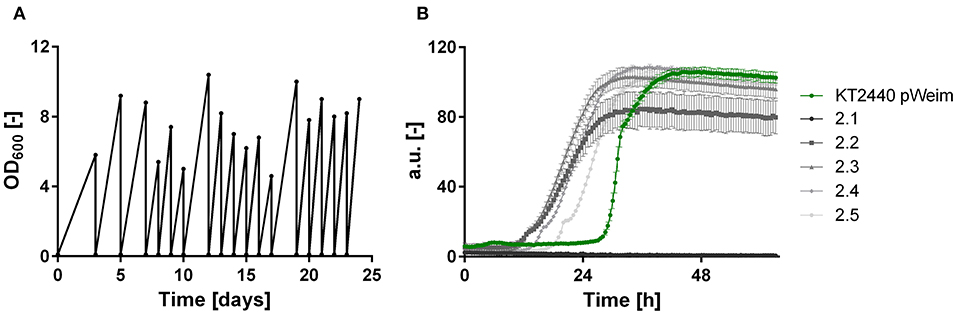
Figure 3. ALE and growth characteristics of isolates of P. putida KT2440 pWeim on xylose. (A) ALE of P. putida KT2440 pWeim on xylose as sole carbon source. A single representative culture is shown, (B) Comparison of growth for P. putida KT2440 pWeim (•, green) and five isolates from the second ALE culture on xylose. Error bars indicate deviation from the mean (n = 3).
The colonies showing the best performance (P. putida KT2440 pWeim2 and P. putida KT2440ΔΔ pDahms2) were characterized in detail regarding their growth behavior. Both showed improved growth compared to the initial strains. The lag phase of P. putida KT2440 pWeim2 was shortened from ~24 to 3 h (Figure 2E). However, the maximal growth rate of the strain was 0.21 h−1. Hence, the lag phase was 1.5-fold reduced compared to the initial strain (Table 3). For P. putida KT2440ΔΔ pDahms2, the lag phase was shortened from ~34 to 3 h (Figure 2F). The growth rate remained the same with a value of 0.21 h−1 (Table 3). Further, the substrate consumption rate was 3 mmol L−1 h−1 for both evolved strains, which is about 1.6- and 2-fold higher than the consumption rates of their initial strains.
To identify possible mutations, which are responsible for the reduced lag phase and xylonate accumulation, the plasmids of P. putida KT2440 pWeim2 and P. putida KT2440ΔΔ pDahms2 were isolated and sequenced. Mutations in the promoter, xylose genes, replication initiator protein, origin of replication, and antibiotic resistance of the plasmids carried by strains P. putida KT2440 pWeim2 and P. putida KT2440ΔΔ pDahms2 could be excluded. Hence, mutations in coding regions in the genome are expected and will be analyzed in the future.
To demonstrate that the isolated Weimberg or Dahms pathway are present in the engineered strains, growth experiments with 1-13C labeled xylose as substrate were performed. Afterwards, the labeling patterns of the proteinogenic amino acids were analyzed. Since xylose is metabolized via different routes using the Weimberg or the Dahms pathway, the labeling pattern is well-distinguishable (Figure 4). Xylose is converted in several steps to 2-oxoglutarate via the Weimberg pathway, which is a direct precursor of glutamate, glutamine, and proline. This would lead to 1-13C labeled glutamate, glutamine and proline. In the central carbon metabolism, 2-oxoglutarate is further processed via the TCA cycle and the labeling is removed via CO2 generation. Furthermore, in the case of the Dahms pathway, xylose is converted into pyruvate and glycolaldehyde. Since pyruvate is a precursor for the amino acids alanine, lysine, and valine, they are fractionally labeled. Pyruvate is also converted to acetyl-CoA in the central carbon metabolism, removing the labeled carbon atom by CO2 generation.
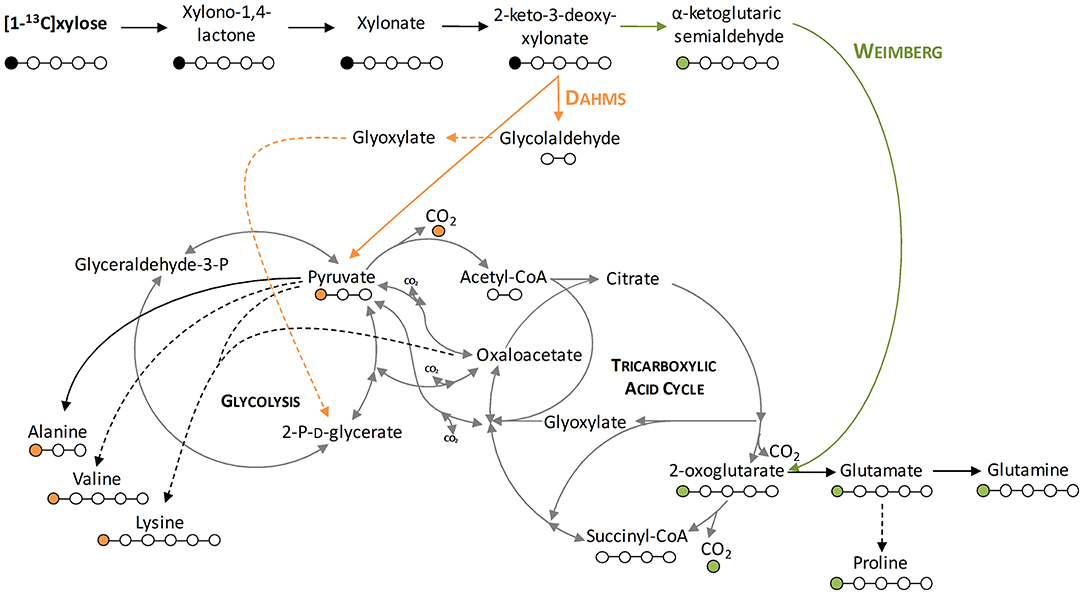
Figure 4. Depiction of the labeling of metabolites resulting from labeled xylose. Shown is the stable isotope introduction into the central carbon metabolism of P. putida KT2440. Orange lines indicate the Dahms pathway, green lines indicate the Weimberg pathway, and dashed lines indicate several enzymatic steps. Orange dots indicate the labeling resulting from the Dahms pathway and green dots indicate the labeling resulting from the Weimberg pathway.
For these experiments, P. putida KT2440 pWeim und P. putida KT2440ΔΔ pDahms were used. As seen before, deletion of the gene PP_2836 entails a completely different growth behavior, which indicates that the deletion of PP_2836 is necessary to establish the Dahms pathway. It is assumed that xylose is utilized via both pathways simultaneously (Weimberg and Dahms) in a strain that still harbors PP_2836 and is deficient in gclR (to activate degradation of the intermediate glycolaldehyde, see Figure 1). This assumption was experimentally tested by introducing the plasmid pBT-Dahms into P. putida ΔgclR. The resulting P. putida ΔgclR pDahms was also used in the labeling experiments.
Significant distinctions of the fractional labeling with labeled xylose were observed for all three strains (Figure 5). For the Weimberg-strain, alanine, lysine, and valine were almost not labeled at all while glutamate and proline showed a labeling about 0.1. This meets the expectations since 50% of 1-13C-labeled xylose was used (one atom out of ten carbon atoms from two xylose molecules should be labeled). For the Dahms-strain, the fractional labeling of the amino acids, which are derived from pyruvate (alanine, lysine, and valine), is about 0.13, 0.06, and 0.07, respectively, and for glutamate and proline below 0.05. Theoretically, a fractional labeling of about 0.17 for alanine, 0.08 for lysine, and 0.1 for valine would be assumed because one out of six, ten, and twelve carbon atoms from two xylose molecules should be labeled, respectively. The observed labeling is a little bit lower than expected, but the distribution meets the expectation since again 50% of 1-13C-labeled xylose was used. For more quantitative statements, metabolic flux analysis has to be performed in order to involve fluxes through the central carbon metabolism. Further, as described above the labeling is removed when CO2 is generated. However, via anaplerotic reactions the labeled CO2 can be reincorporated and ultimately result in labeled glutamate and proline. Nevertheless, both strains showed a distinct labeling pattern, which indicates that the Weimberg and Dahms pathways are truly used in isolation.
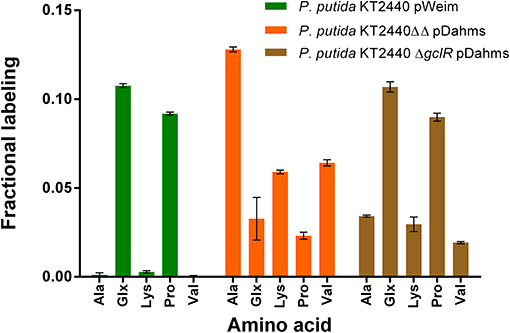
Figure 5. Fractional labeling of amino acids during growth in minimal medium containing 50% 1-13C-xylose. Error bars indicate deviation from the mean of three technical replicates from one growth experiment (n = 3). Ala, alanine; Glx, glutamate and deaminated glutamine; Lys, lysine; Pro, proline; Val, valine.
Moreover, for P. putida KT2440 ΔgclR pDahms it was determined that the fractional labeling of glutamate and proline was about 0.1 as in the case of the Weimberg-strain and that the labeling of alanine, lysine, and valine was in between the values of the Weimberg- and the Dahms-strain. These results show that the fractional labeling is a combination of the single fractional labelings of the Weimberg and the Dahms pathway. Thus, it is suggested that the Weimberg and the Dahms pathway are active simultaneously in this strain. Consequently, the above stated declaration that PP_2836 encodes a dehydratase is verified. The deletion of this gene is necessary to obtain a strain, which solely uses the Dahms pathway.
Different stoichiometries of the pathways have an influence on the production yield of specific metabolites. In order to show this dependency, two secondary metabolites (rhamnolipids and phenazines) were chosen for heterologous synthesis in efficient xylose metabolizers. Exemplarily, strains with the individual pathways (Isomerase, Weimberg, and Dahms) were chosen and the production of the initial strains and the adapted strains was investigated. For the production of rhamnolipids, the rhamnolipid synthesis module was integrated as single copy into the genome of all xylose utilizing strains. P. putida KT2440 EM42 Δgcd pSEVA2213_xylABE (Dvorák and de Lorenzo, 2018) was used as optimized strain using the Isomerase pathway because no evolved P. putida strain using the Isomerase pathway was generated. The rhamnolipid producers were designated P. putida KT2440 pIso_RL, P. putida KT2440 pWeim_RL, P. putida KT2440ΔΔ pDahms_RL, P. putida KT2440 pIso2_RL, P. putida KT2440 pWeim2_RL, and P. putida KT2440ΔΔ pDahms2_RL.
P. putida KT2440 pIso_RL, utilizing the Isomerase pathway for xylose degradation, showed only little rhamnolipid production (47 mg L−1) after 96 h, because most of the xylose was converted to the dead-end product xylonate. While P. putida KT2440 pIso2_RL produced seven times more mono-rhamnolipids after 54 h (Figure 6A), although the cells were clumped and did not reach high optical densities. On the contrary, the strains with the Weimberg and Dahms pathways reached higher rhamnolipid titers. P. putida KT2440 pWeim_RL produced 13 times more mono-rhamnolipids than P. putida KT2440 pIso_RL in less time (54 h) and the evolved strain with the rhamnolipid production module P. putida KT2440 pWeim2_RL produced even more mono-rhamnolipids after 30 h (720 mg L−1). Further, P. putida KT2440ΔΔ pDahms_RL and P. putida KT2440ΔΔ pDahms2_RL produced six times more mono-rhamnolipids after 54 h (290 mg L−1) and 13 times more mono-rhamnolipids after 30 h (620 mg L−1), respectively. Besides, differences in the substrate consumption were observed. While P. putida KT2440 pWeim2_RL and P. putida KT2440ΔΔ pDahms_RL consumed all of the provided xylose, xylose and xylonate in the cultures of the other rhamnolipid-producing strains was detected after growth stopped (Supplementary Figure 1). Hence, the ratio of the amount of product synthesized and the amount of substrate consumed (yield) is considered. P. putida KT2440 pIso2_RL had the highest yield (78 mg g−1), while P. putida KT2440 pWeim2_RL and P. putida KT2440ΔΔ pDahms2_RL reached comparable yields, however, 1.4-fold lower than P. putida KT2440 pIso2_RL (Figure 6C). As expected, the evolved and optimized strains showed a higher rhamnolipid concentration and yield than their initial xylose utilizing strains (Figures 6A,C).
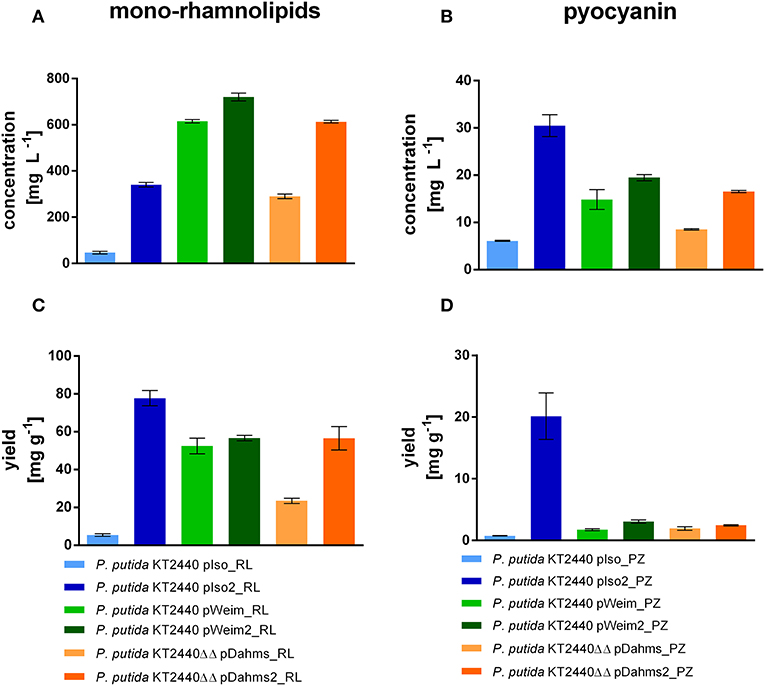
Figure 6. Mono-rhamnolipid and pyocyanin production from xylose by engineered P. putida strains. The minimal medium contained 10 g L−1 xylose. Shown are the product titers of mono-rhamnolipids (A), the product titers of pyocyanin (B), the yield of mono-rhamnolipids on xylose (C), and the yield of pyocyanin on xylose (D). Error bars indicate deviation from the mean (n = 3).
The synthesis of phenazines was achieved by transferring plasmid pJNN_phzA1-G1,S,M into all initial xylose utilizing strains and the optimized or evolved strains. The pyocyanin producers were named P. putida KT2440 pIso_PZ, P. putida KT2440 pWeim_PZ, P. putida KT2440ΔΔ pDahms_PZ, P. putida KT2440 pIso2_PZ, P. putida KT2440 pWeim2_PZ, and P. putida KT2440ΔΔ pDahms2_PZ. All strains produced pyocyanin in a similar range (~15–20 mg L−1, Figure 6B), but in a different period, except the strains using the Isomerase pathway. P. putida KT2440 pIso_PZ produced around three times less (6 mg L−1) pyocyanin and P. putida KT2440 pIso2_PZ produced roughly two times more (30 mg L−1) pyocyanin in comparison to the evolved strain using the Dahms pathway. Pyocyanin was harvested at the same time points as mono-rhamnolipids. Furthermore, as observed in rhamnolipid production experiments, differences in substrate consumption were recognized. P. putida KT2440 pIso_PZ converted even more xylose to xylonate than in the rhamnolipid production experiments, which could then not be used for pyocyanin production. Further, less xylose was consumed than in the rhamnolipid production experiment (Supplementary Figure 1). Consequently, the yield was determined and P. putida KT2440 pIso2_PZ showed the best yield (20 mg g−1) (Figure 6D). Furthermore, the evolved strains reached higher yields than their initial strains (P. putida KT2440 pWeim2_PZ: 3.2 mg g−1, P. putida KT2440ΔΔ pDahms2_PZ: 2.5 mg g−1). It was confirmed that the evolved and optimized strains could reach higher product titers and yields than their initial strains, also in case of pyocyanin production (Figures 6B,D). This can be explained by the adaptation toward the novel substrate xylose for P. putida KT2440 pWeim2 and P. putida KT2440ΔΔ pDahms2. In case of P. putida KT2440 pIso2 the production of xylonate is prevented by deletion of the glucose dehydrogenase in comparison to P. putida KT2440 pIso and consequently more carbon is available for the production of pyocyanin. Conspicuously, growth was slower for the phenazine-producing strains compared to the rhamnolipid-producing strains and consequently the total consumption of xylose was lower. This might be due to the metabolic burden through the usage of kanamycin to avoid loss of the plasmid harboring the phenazine synthesis genes during cultivation. Further, product toxicity might be a cause for the xylose leftovers. In accordance to the computed data in 3.1., P. putida KT2440 pIso2 harboring the rhamnolipid or phenazine genes showed in both scenarios the best yield (Figure 6).
In this study, we integrated three bacterial xylose utilization pathways—Isomerase, Weimberg, and Dahms—in P. putida KT2440, to compare in silico and in vivo the synthesis capacities of these alternative degradation pathways. While other studies focus on heterologous production with P. putida KT2440, we aimed at showing the production of various products (i.e., mono-rhamnolipids and pyocyanin) from xylose. Further, we wanted to emphasize that computational analysis can guide strain design, reaching higher yields.
FBA indicates strong preferences for alternative pathways depending on the product of choice due to different stoichiometries. It was determined that mainly the Isomerase pathway is preferred, however the Weimberg and Dahms pathways are favored in niche applications, when for example intermediates of the pathways are precursor for the products. To show this dependency, the xylose pathways and synthesis pathways were integrated in P. putida KT2440. In the course of isolating the Dahms pathway, two genes (PP_2836 and PP_4283) were deleted. After successful pathway implementation, the synthesis capacities for mono-rhamnolipids and pyocyanin were investigated. The resulting data matched the computed data. The approach of in silico metabolic network design driven by the product of choice, adds another degree of freedom for metabolic engineering.
The computed maximal product yield for both synthesized metabolites, mono-rhamnolipids and pyocyanin, was produced by the Isomerase pathway. In both cases, this was confirmed in vivo (Figure 6). While the highest rhamnolipid titer (700 mg L−1) was reached by the evolved strain using the Weimberg pathway, the highest yield (78 mg g−1) was reached by the strain using the Isomerase pathway. The highest pyocyanin titer (30 mg L−1) and yield (20 mg g−1) were also reached by the strain using the Isomerase pathway. Taken together, it was shown that the reached yields are dependent on the metabolization route and hence the stoichiometries of the pathway, which can be determined in silico.
Many studies deal with the heterologous production by P. putida KT2440. Production of biosurfactants, such as hydroxyalkanoyloxy alkanoates (HAA), mono-rhamnolipids, and di-rhamnolipids (Tiso et al., 2017; Wittgens et al., 2017, 2018), terpenoids (zeaxanthin and β-carotene) (Beuttler et al., 2011; Loeschcke et al., 2013), amino acid-derived compounds (e.g., phenazines) (Schmitz et al., 2015b), polyketides/non-ribosomal peptides (e.g., flaviolin, prodigiosin) (Gross et al., 2006; Loeschcke et al., 2013; Domröse et al., 2015), and N-methylglutamate (Mindt et al., 2018) was demonstrated. These examples show how diverse the production spectrum in P. putida KT2440 can be. To create sustainable production processes in times of high environmental pollution, one searches for alternative substrates (Vanholme et al., 2013). The concept of using xylose or other C5 sugars for platform chemicals was already considered before (Werpy and Petersen, 2004). For example, HAA and terpenoids are benefitting from the Isomerase pathway as acetyl-CoA is the precursor (Table 2). Prodigiosin synthesis involves the precursors pyruvate, proline, and malonyl-CoA (Williamson et al., 2006), favoring a combination of the Isomerase and the Weimberg pathway. The Weimberg pathway would be beneficial for synthesis of N-methylglutamate. The attempt of using xylose as renewable source for the production of metabolites was already considered in several studies, where P. taiwanensis VLB120 was used for the synthesis of mono-rhamnolipids, phenol, and 4-hydroxybenzoate (Tiso et al., 2017; Wynands et al., 2018; Lenzen et al., 2019). P. taiwanensis VLB120 utilizes xylose via the Weimberg pathway natively (Köhler et al., 2015), but engineering of the strain and enabling xylose utilization via the Isomerase pathway could enhance product yield on substrate due to the superior stoichiometry.
While the integration of the xylose pathways worked in vivo, we observed strong differences between the oxidative and the Isomerase pathway operations. Pseudomonades using the three xylose pathways natively were reported (Hochster, 1955; Dahms, 1974; Köhler et al., 2015), indicating that these pathways are compatible with the Pseudomonades metabolic network. Notably, we did not find any report suggesting the presence of two of the xylose pathways. The implementation of the Isomerase pathway in P. putida was shown before (Meijnen et al., 2008; Le Meur et al., 2012; Dvorák and de Lorenzo, 2018; Wang et al., 2019). In accordance to our results, only weak growth was observed in two studies (Meijnen et al., 2008; Dvorák and de Lorenzo, 2018), which was improved by rational or non-rational engineering. In another study, growth on xylose via the Weimberg pathway in P. putida was shown to be immediately efficient (Meijnen et al., 2009), matching our results. As already considered by Wang et al. (2019), xylose metabolization seems to be metabolically demanding for P. putida KT2440 using the isomerase route and might be the reason for the discrepancies. But how can this difference between the Isomerase pathway and the oxidative pathways be explained? The consideration of other carbon sources indicates that the usage of oxidative pathways is likely to have an advantage for Pseudomonas species. The metabolization of the C5 sugar arabinose proceeds in E. coli via the Isomerase pathway (Laikova, 2001), whereas in other Pseudomonas species the utilization of arabinose happens via oxidative steps to form the intermediate pentonic acid (Lockwood and Nelson, 1946; Weimberg and Doudoroff, 1955). In the case of galacturonic acid the utilization starts with an isomerase reaction in E. coli (Ashwell et al., 1960). Whereas, in Pseudomonas species the first enzyme is a dehydrogenase, which catalyzes an oxidation reaction (Kilgore and Starr, 1959; Richard and Hilditch, 2009). There might be two explanations why Pseudomonas favors oxidative pathways. First, the conversion of the substrate into an intermediate acid prevents other microbes from using this substrate, which is an advantage in terms of survival. Consequently, the environment is acidified due to the resulting intermediate acid, which creates a further advantage for propagation. P. putida KT2440 is a soil bacterium and is able to cope with extreme conditions, such as nutrient limitation, temperature shifts, and pH changes, in contrast to enterobacteria that occur in nutrient-rich niches (Martins Dos Santos et al., 2004; Reva et al., 2006). Second, the energy metabolism is more flexible, because the dehydrogenases have different redox cofactors. This can be explained using glucose as an example. In general, the redox metabolism is balanced and therefore, the rates of reduction and oxidation of the redox cofactors have to be highly similar (Blank et al., 2010). The electrons, which are released during oxidation of glucose and gluconate, are used to reduce PQQ and FAD+ and are feeding directly the electron transport chain. There, PQQ is directly reoxidized by transferring the electrons to ubiquinone in the inner membrane (Ebert et al., 2011; Tiso et al., 2014). The transport of glucose over the membrane costs two ATP per molecule, while only one proton and one sodium ion are necessary for the transport of gluconate and ketogluconate, respectively. Therefore, P. putida KT2440 saves energy using the oxidative pathway and does not require additional cofactor regeneration systems.
However, a prolonged lag phase was observed for all three engineered strains. P. putida KT2440 pIso had the longest lag phase with 100 h, P. putida KT2440 pWeim had a lag phase of 24 h, and P. putida KT2440ΔΔ pDahms had a lag phase of 34 h (Table 3). To identify possible bottlenecks, the plasmids were sequenced after ALE. Interestingly, no mutations in the xylose utilization genes and other encoding areas of the vectors could be detected. Different studies showed that mutations in the replication initiation protein or in the antibiotic resistance cassette could enhance growth by lowering the plasmid copy number, and subsequently reducing the need of resources for the synthesis of kanamycin resistance (Jakob et al., 2013; Mi et al., 2016). A second possible bottleneck could be the transport of xylonate from the periplasm to the cytoplasm. P. putida KT2440 harbors the transporter GntT (PP_3417), enabling the transport of gluconate from the periplasm into the cytoplasm (Porco et al., 1997). Such a transporter is characterized for E. coli. A conceivable possibility would be, that this transporter might be active for xylonate, but due to its high-affinity toward gluconate it might operate slower for xylonate. Another option could be that a slow, low-affinity transporter is used instead. To improve the growth performance and reduce the lag phase, ALE was implemented. Interestingly, the lag phase was reduced significantly, while the growth rates did not increase and in one case even decreased during ALE. This can be explained by positive selection for a reduced lag phase and negative selection for the growth rate. The fast-adapting cells does not have stringently the highest growth rate. After reduction of the lag phase, it can be selected for increased growth rates now. While laboratory evolution is an easy method to increase the overall fitness of a population in laboratory scale, the design of the ALE approach for adaptation of the population is not trivial. Conditions such as time point of transfer, passage size, and growth phase can alter in ALE studies (Charusanti et al., 2010; LaCroix et al., 2015). Batch cultivation and continuous (chemostat) cultivation are the mostly used ALE techniques (Dragosits and Mattanovich, 2013; Gresham and Dunham, 2014; LaCroix et al., 2017). Of these, regularly transferred batch cultures are more popular because effort and costs are rather low. However, this method has several limitations due to varying conditions (LaCroix et al., 2017). Further, this method is slower than an automated ALE because the transfer is usually done on a daily basis. While in automated ALE processes, several parameters, including the optical density and growth rate, are monitored online. If the growth rate increases over the course of the ALE, the passage frequency can automatically be increased. Additionally, cultures probably reach the stationary phase in batch cultivations, which then results in an improved survival in stationary phase or decreased lag phase (Wiser and Lenski, 2015). Thus, it is not surprising that the batch cultivations for adaptation used in this study resulted in a shortage of the lag phase.
The here presented alternative pathways for xylose utilization open another degree of freedom in the design and metabolic engineering of the production strain. Dependent on the product of interest, the experimenter can compute the best network design considering three different stoichiometries for xylose use. The general applicability of this approach is outlined here, while as an outlook the detailed single strain optimization and the co-consumption of carbon sources in silico and in vivo should be worked on.
All datasets generated for this study are included in the article/Supplementary Material.
IB performed all molecular engineering and characterization experiments, analyzed the data, prepared figures, conducted the in silico experiments, and wrote the manuscript. TT provided guidance on P. putida biotechnology and edited the manuscript. TT, AW, and FR discussed the data and critically revised the manuscript. LB advised on all experiments, analyzed and discussed data, and edited the manuscript.
The scientific activities of the Bioeconomy Science Center were financially supported by the Ministry of Culture and Science within the framework of the NRW Strategieprojekt BioSC (No. 313/323-400-00213). The GC-MS/MS was funded by the German Research Foundation DFG (Förderkennzeichen: INST 2221018-1 FUGG). Furthermore, the authors are grateful to the Fachagentur Nachwachsende Rohstoffe e.V. (FNR; Förderkennzeichen: 22013314), the Ministry of Science, Research and the Arts of Baden-Württemberg (MWK; Förderkennzeichen: 7533-10-5-186A and 7533-10-5-190) and the EU project Horizon 2020 AD GUT (ID: 686271) for providing financial support.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Prof. de Lorenzo [Systems and Synthetic Biology Program, Centro Nacional de Biotecnología (CNB-CSIC)] for providing P. putida EM42 Δgcd pSEVA2213_xylABE, Dr. Simone Schmitz (RWTH Aachen University) and Prof. Agler-Rosenbaum (Leibniz Institute for Natural Product Research and Infection Biology, Hans-Knöll-Institute) for providing plasmid pJNN_phzA1-G1,M,S, Sebastian Köbbing (RWTH Aachen University) for providing plasmid pBG14ffg, and Wing-Jin Li (RWTH Aachen University) for providing P. putida KT2440 ΔgclR. We kindly acknowledge Hans M. Böhne and Sebastian Kruth for their experimental help.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fbioe.2019.00480/full#supplementary-material
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. doi: 10.1016/S0022-2836(05)80360-2
Angermayr, S. A., Paszota, M., and Hellingwerf, K. J. (2012). Engineering a cyanobacterial cell factory for production of lactic acid. Appl. Environ. Microbiol. 78, 7098–7106. doi: 10.1128/AEM.01587-12
Antonovsky, N., Gleizer, S., Noor, E., Zohar, Y., Herz, E., Barenholz, U., et al. (2016). Sugar synthesis from CO2 in Escherichia coli. Cell 166, 115–125. doi: 10.1016/j.cell.2016.05.064
Ashwell, G., Wahba, A. J., and Hickman, J. (1960). Uronic acid metabolism in bacteria. I. Purification and properties of uronic acid isomerase in Escherichia coli. J. Biol. Chem. 235, 1559–1565.
Bagdasarian, M., Lurz, R., Rückert, B., Franklin, F. C. H., Bagdasarian, M. M., Frey, J., et al. (1981). Specific-purpose plasmid cloning vectors II. Broad host range, high copy number, RSF 1010-derived vectors, and a host-vector system for gene cloning in Pseudomonas. Gene 16, 237–247. doi: 10.1016/0378-1119(81)90080-9
Behrens, B., Baune, M., Jungkeit, J., Tiso, T., Blank, L. M., and Hayen, H. (2016). High performance liquid chromatography-charged aerosol detection applying an inverse gradient for quantification of rhamnolipid biosurfactants. J. Chromatogr. A 1455, 125–132. doi: 10.1016/j.chroma.2016.05.079
Belda, E., van Heck, R. G., José Lopez-Sanchez, M., Cruveiller, S., Barbe, V., Fraser, C., et al. (2016). The revisited genome of Pseudomonas putida KT2440 enlightens its value as a robust metabolic chassis. Environ. Microbiol. 18, 3403–3424. doi: 10.1111/1462-2920.13230
Beuttler, H., Hoffmann, J., Jeske, M., Hauer, B., Schmid, R. D., Altenbuchner, J., et al. (2011). Biosynthesis of zeaxanthin in recombinant Pseudomonas putida. Appl. Microbiol. Biotechnol. 89, 1137–1147. doi: 10.1007/s00253-010-2961-0
Blank, L. M., Ebert, B. E., Buehler, K., and Bühler, B. (2010). Redox biocatalysis and metabolism: molecular mechanisms and metabolic network analysis. Antioxid. Redox Signal. 13, 349–394. doi: 10.1089/ars.2009.2931
Bouzon, M., Perret, A., Loreau, O., Delmas, V., Perchat, N., Weissenbach, J., et al. (2017). A synthetic alternative to canonical one-carbon metabolism. ACS Synth. Biol. 6, 1520–1533. doi: 10.1021/acssynbio.7b00029
Cabulong, R. B., Lee, W. K., Bañares, A. B., Ramos, K. R. M., Nisola, G. M., Valdehuesa, K. N. G., et al. (2018). Engineering Escherichia coli for glycolic acid production from D-xylose through the Dahms pathway and glyoxylate bypass. Appl. Microbiol. Biotechnol. 102, 2179–2189. doi: 10.1007/s00253-018-8744-8
Charusanti, P., Conrad, T. M., Knight, E. M., Venkataraman, K., Fong, N. L., Xie, B., et al. (2010). Genetic basis of growth adaptation of Escherichia coli after deletion of pgi, a major metabolic gene. PLoS Genet. 6, 1–13. doi: 10.1371/journal.pgen.1001186
Choi, K. H., Gaynor, J. B., White, K. G., Lopez, C., Bosio, C. M., Karkhoff-Schweizer, R. A., et al. (2005). A Tn7-based broad-range bacterial cloning and expression system. Nat. Methods 2, 443–448. doi: 10.1038/nmeth765
Choi, K. H., Kumar, A., and Schweizer, H. P. (2006). A 10-min method for preparation of highly electrocompetent Pseudomonas aeruginosa cells: application for DNA fragment transfer between chromosomes and plasmid transformation. J. Microbiol. Methods 64, 391–397. doi: 10.1016/j.mimet.2005.06.001
Choi, S. Y., Kim, W. J., Yu, S. J., Park, S. J., Im, S. G., and Lee, S. Y. (2017). Engineering the xylose-catabolizing Dahms pathway for production of poly(d-lactate-co-glycolate) and poly(d-lactate-co-glycolate-co-d-2-hydroxybutyrate) in Escherichia coli. Microb. Biotechnol. 10, 1353–1364. doi: 10.1111/1751-7915.12721
Chomczynski, P., and Rymaszewski, M. (2006). Alkaline polyethylene glycol-based method for direct PCR from bacteria, eukaryotic tissue samples, and whole blood. Biotechniques 40, 454–458. doi: 10.2144/000112149
Dahms, S. A. (1974). 3-Deoxy-D-pentulosonic acid aldolase and its role in a new pathway of D-xylose degradation. Biochem. Biophys. Res. Commun. 60, 1433–1439. doi: 10.1016/0006-291X(74)90358-1
David, J. D., and Weismeyer, H. (1970). Control of xylose metabolism in Escherichia coli. BBA Gen. Subj. 201, 497–499. doi: 10.1016/0304-4165(70)90171-6
de Lorenzo, V., and Timmis, K. N. (1994). Analysis and construction of stable phenotypes in gram-negative bacteria with Tn5- and Tn10-derived minitransposons. Methods Enzymol. 235, 386–405. doi: 10.1016/0076-6879(94)35157-0
Ditta, G., Stanfield, S., Corbin, D., and Helinski, D. R. (1980). Broad host range DNA cloning system for Gram-negative bacteria: Construction of a gene bank of Rhizobium meliloti. Proc. Natl. Acad. Sci. U.S.A. 77, 7347–7351. doi: 10.1073/pnas.77.12.7347
Domröse, A., Klein, A. S., Hage-Hülsmann, J., Thies, S., Svensson, V., Classen, T., et al. (2015). Efficient recombinant production of prodigiosin in Pseudomonas putida. Front. Microbiol. 6, 1–10. doi: 10.3389/fmicb.2015.00972
Dragosits, M., and Mattanovich, D. (2013). Adaptive laboratory evolution - principles and applications for biotechnology. Microb. Cell Fact. 12, 1–17. doi: 10.1186/1475-2859-12-64
Dvorák, P., and de Lorenzo, V. (2018). Refactoring the upper sugar metabolism of Pseudomonas putida for co-utilization of cellobiose, xylose, and glucose. Metab. Eng. 48, 94–108. doi: 10.1016/j.ymben.2018.05.019
Ebert, B. E., Kurth, F., Grund, M., Blank, L. M., and Schmid, A. (2011). Response of Pseudomonas putida KT2440 to increased NADH and ATP demand. Appl. Environ. Microbiol. 77, 6597–6605. doi: 10.1128/AEM.05588-11
Franden, M. A., Jayakody, L. N., Li, W. J., Wagner, N. J., Cleveland, N. S., Michener, W. E., et al. (2018). Engineering Pseudomonas putida KT2440 for efficient ethylene glycol utilization. Metab. Eng. 48, 197–207. doi: 10.1016/j.ymben.2018.06.003
Gibson, D. G., Young, L., Chuang, R. Y., Venter, J. C., Hutchison, C. A., and Smith, H. O. (2009). Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6, 343–345. doi: 10.1038/nmeth.1318
Goeppert, A., Czaun, M., Surya Prakash, G. K., and Olah, G. A. (2012). Air as the renewable carbon source of the future: An overview of CO2 capture from the atmosphere. Energy Environ. Sci. 5, 7833–7853. doi: 10.1039/c2ee21586a
Gresham, D., and Dunham, M. J. (2014). The enduring utility of continuous culturing in experimental evolution. Genomics 104, 399–405. doi: 10.1016/j.ygeno.2014.09.015
Gross, F., Luniak, N., Perlova, O., Gaitatzis, N., Jenke-Kodama, H., Gerth, K., et al. (2006). Bacterial type III polyketide synthases: Phylogenetic analysis and potential for the production of novel secondary metabolites by heterologous expression in pseudomonads. Arch. Microbiol. 185, 28–38. doi: 10.1007/s00203-005-0059-3
Guadalupe-Medina, V., Wisselink, H. W., Luttik, M. A., De Hulster, E., Daran, J. M., Pronk, J. T., et al. (2013). Carbon dioxide fixation by Calvin-cycle enzymes improves ethanol yield in yeast. Biotechnol. Biofuels 6, 1–12. doi: 10.1186/1754-6834-6-125
Hanahan, D. (1983). Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166, 557–580. doi: 10.1016/S0022-2836(83)80284-8
Hanahan, D. (1985). “Techniques for transformation of E. coli,” in DNA Cloning: A Practical Approach, ed D. M. Glover (Oxford: IRL Press), 109–135.
Hochster, R. M. (1955). The formation of phosphorylated sugars from D-xylose by extracts of Pseudomonas hydrophila. Can. J. Microbiol. 1, 346–363. doi: 10.1139/m55-047
Jakob, F., Lehmann, C., Martinez, R., and Schwaneberg, U. (2013). Increasing protein production by directed vector backbone evolution. AMB Expr. 3:39. doi: 10.1186/2191-0855-3-39
Johnsen, U., Dambeck, M., Zaiss, H., Fuhrer, T., Soppa, J., Sauer, U., et al. (2009). D-xylose degradation pathway in the halophilic archaeon Haloferax volcanii. J. Biol. Chem. 284, 27290–27303. doi: 10.1074/jbc.M109.003814
Kilgore, W. W., and Starr, M. P. (1959). Uronate oxidation by phytopathogenic pseudomonads. Nature 183, 1412–1413. doi: 10.1038/1831412a0
Köhler, K. A. K., Blank, L. M., Frick, O., and Schmid, A. (2015). D-Xylose assimilation via the Weimberg pathway by solvent-tolerant Pseudomonas taiwanensis VLB120. Environ. Microbiol. 17, 156–170. doi: 10.1111/1462-2920.12537
Koopman, F., Wierckx, N., de Winde, J. H., and Ruijssenaars, H. J. (2010). Identification and characterization of the furfural and 5-(hydroxymethyl)furfural degradation pathways of Cupriavidus basilensis HMF14. Proc. Natl. Acad. Sci. U.S.A. 107, 4919–4924. doi: 10.1073/pnas.0913039107
Kotte, O., Volkmer, B., Radzikowski, J. L., and Heinemann, M. (2014). Phenotypic bistability in Escherichia coli's central carbon metabolism. Mol. Syst. Biol. 10, 1–11. doi: 10.15252/msb.20135022
LaCroix, R. A., Palsson, B. O., and Feist, A. M. (2017). A model for designing adaptive laboratory evolution experiments. Appl. Environ. Microbiol. 83, 1–14. doi: 10.1128/AEM.03115-16
LaCroix, R. A., Sandberg, T. E., O'Brien, E. J., Utrilla, J., Ebrahim, A., Guzman, G. I., et al. (2015). Use of adaptive laboratory evolution to discover key mutations enabling rapid growth of Escherichia coli K-12 MG1655 on glucose minimal medium. Appl. Environ. Microbiol. 81, 17–30. doi: 10.1128/AEM.02246-14
Laikova, O. (2001). Computational analysis of the transcriptional regulation of pentose utilization systems in the gamma subdivision of Proteobacteria. FEMS Microbiol. Lett. 205, 315–322. doi: 10.1111/j.1574-6968.2001.tb10966.x
Lan, E. I., and Liao, J. C. (2011). Metabolic engineering of cyanobacteria for 1-butanol production from carbon dioxide. Metab. Eng. 13, 353–363. doi: 10.1016/j.ymben.2011.04.004
Larsen, J., Øtergaard Petersen, M., Thirup, L., Li, H. W., and Iversen, F. K. (2008). The IBUS process - Lignocellulosic bioethanol close to a commercial reality. Chem. Eng. Technol. 31, 765–772. doi: 10.1002/ceat.200800048
Le Meur, S., Zinn, M., Egli, T., Thöny-Meyer, L., and Ren, Q. (2012). Production of medium-chain-length polyhydroxyalkanoates by sequential feeding of xylose and octanoic acid in engineered Pseudomonas putida KT2440. BMC Biotechnol. 12, 1–12. doi: 10.1186/1472-6750-12-53
Lee, J. (1997). Biological conversion of lignocellulosic biomass to ethanol. J. Biotechnol. 56, 1–24. doi: 10.1016/S0168-1656(97)00073-4
Lenzen, C., Wynands, B., Otto, M., Bolzenius, J., Mennicken, P., Wierckx, N., et al. (2019). High-yield production of 4-hydroxybenzoate from glucose or glycerol by an engineered Pseudomonas taiwanensis VLB120. Front. Bioeng. Biotechnol. 7:130. doi: 10.3389/fbioe.2019.00130
Li, W., Jayakody, L. N., Franden, M. A., Wehrmann, M., Daun, T., Hauer, B., et al. (2019). Laboratory evolution reveals the metabolic and regulatory basis of ethylene glycol metabolism by Pseudomonas putida KT2440. Environ. Microbiol. 21:3669–3682. doi: 10.1111/1462-2920.14703
Liebal, U. W., Blank, L. M., and Ebert, B. E. (2018). CO 2 to succinic acid – Estimating the potential of biocatalytic routes. Metab. Eng. Commun. 7, 1–10. doi: 10.1016/j.mec.2018.e00075
Lockwood, L. B., and Nelson, G. E. (1946). The oxidation of pentoses by Pseudomonas. J. Bacteriol. 52, 581–586.
Loeschcke, A., Markert, A., Wilhelm, S., Wirtz, A., Rosenau, F., Jaeger, K.-E., et al. (2013). TREX: a universal tool for the transfer and expression of biosynthetic pathways in bacteria. ACS Synth. Biol. 2, 22–33. doi: 10.1021/sb3000657
Loeschcke, A., and Thies, S. (2015). Pseudomonas putida- a versatile host for the production of natural products. Appl. Microbiol. Biotechnol. 99, 6197–6214. doi: 10.1007/s00253-015-6745-4
Maas, R. H., Bakker, R. R., Boersma, A. R., Bisschops, I., Pels, J. R., de Jong, E., et al. (2008). Pilot-scale conversion of lime-treated wheat straw into bioethanol: quality assessment of bioethanol and valorization of side streams by anaerobic digestion and combustion. Biotechnol. Biofuels 1, 1–13. doi: 10.1186/1754-6834-1-14
Martínez-García, E., and de Lorenzo, V. (2011). Engineering multiple genomic deletions in Gram-negative bacteria: analysis of the multi-resistant antibiotic profile of Pseudomonas putida KT2440. Environ. Microbiol. 13, 2702–2716. doi: 10.1111/j.1462-2920.2011.02538.x
Martins Dos Santos, V. A. P., Heim, S., Moore, E. R. B., Strätz, M., and Timmis, K. N. (2004). Insights into the genomic basis of niche specificity of Pseudomonas putida KT2440. Environ. Microbiol. 6:1264–1286. doi: 10.1111/j.1462-2920.2004.00734.x
Meijnen, J.-P., de Winde, J. H., and Ruijssenaars, H. J. (2008). Engineering Pseudomonas putida S12 for efficient utilization of D-xylose and L-arabinose. Appl. Environ. Microbiol. 74, 5031–5037. doi: 10.1128/AEM.00924-08
Meijnen, J.-P., de Winde, J. H., and Ruijssenaars, H. J. (2009). Establishment of oxidative D-xylose metabolism in Pseudomonas putida S12. Appl. Environ. Microbiol. 75, 2784–2791. doi: 10.1128/AEM.02713-08
Mi, J., Sydow, A., Schempp, F., Becher, D., Schewe, H., Schrader, J., et al. (2016). Investigation of plasmid-induced growth defect in Pseudomonas putida. J. Biotechnol. 231, 167–173. doi: 10.1016/j.jbiotec.2016.06.001
Mindt, M., Walter, T., Risse, J. M., and Wendisch, V. F. (2018). Fermentative production of N-methylglutamate from glycerol by recombinant Pseudomonas putida. Front. Bioeng. Biotechnol. 6:159. doi: 10.3389/fbioe.2018.00159
Nelson, K. E., Weinel, C., Paulsen, I. T., Dodson, R. J., Hilbert, H., Martins dos Santos, V. A. P., et al. (2002). Complete genome sequence and comparative analysis of the metabolically versatile Pseudomonas putida KT2440. Environ. Microbiol. 4, 799–808. doi: 10.1046/j.1462-2920.2002.00366.x
Nikel, P. I., and de Lorenzo, V. (2018). Pseudomonas putida as a functional chassis for industrial biocatalysis: From native biochemistry to trans-metabolism. Metab. Eng. 50, 142–155. doi: 10.1016/j.ymben.2018.05.005
Nogales, J., Gudmundsson, S., Duque, E., Ramos, J. L., and Palsson, B. O. (2017). Expanding the computable reactome in Pseudomonas putida reveals metabolic cycles providing robustness. bioRxiv [Preprint] 1–34. doi: 10.1101/139121
Nogales, J., Mueller, J., Gudmundsson, S., Canalejo, F. J., Duque, E., Monk, J., et al. (2019). High-quality genome-scale metabolic modelling of Pseudomonas putida highlights its broad metabolic capabilities. Environ. Microbiol. doi: 10.1111/1462-2920.14843
Ogata, H., Goto, S., Sato, K., Fujibuchi, W., Bono, H., and Kanehisa, M. (1999). KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 27, 29–34. doi: 10.1093/nar/27.1.29
Olah, G. A., Goeppert, A., and Prakash, G. K. (2009). Chemical recycling of carbon dioxide to methanol and dimethyl ether: from greenhouse gas to renewable, environmentally carbon neutral fuels and synthetic hydrocarbons. J. Org. Chem. 74, 487–498. doi: 10.1021/jo801260f
Panke, S., Witholt, B., Schmid, A., and Wubbolts, M. G. (1998). Towards a biocatalyst for (S)-styrene oxide production: Characterization of the styrene degradation pathway of Pseudomonas sp. strain VLB120. Appl. Environ. Microbiol. 64, 2032–2043. doi: 10.1128/AEM.64.6.2032-2043.1998
Poblete-Castro, I., Becker, J., Dohnt, K., dos Santos, V. M., and Wittmann, C. (2012). Industrial biotechnology of Pseudomonas putida and related species. Appl. Microbiol. Biotechnol. 93, 2279–2290. doi: 10.1007/s00253-012-3928-0
Porco, A., Peekhaus, N., Bausch, C., Tong, S., Isturiz, T., and Conway, T. (1997). Molecular genetic characterization of the Escherichia coli gntT gene of GntI, the main system for gluconate metabolism. J. Bacteriol. 179, 1584–1590. doi: 10.1128/jb.179.5.1584-1590.1997
Poskar, C. H., Huege, J., Krach, C., Franke, M., Shachar-Hill, Y., and Junker, B. H. (2012). iMS2Flux - a high-throughput processing tool for stable isotope labeled mass spectrometric data used for metabolic flux analysis. BMC Bioinformatics 13:295. doi: 10.1186/1471-2105-13-295
Radek, A., Krumbach, K., Gätgens, J., Wendisch, V. F., Wiechert, W., Bott, M., et al. (2014). Engineering of Corynebacterium glutamicum for minimized carbon loss during utilization of D-xylose containing substrates. J. Biotechnol. 192, 156–160. doi: 10.1016/j.jbiotec.2014.09.026
Ramos, J. L., Duque, E., Huertas, M. J., and Haïdour, A. (1995). Isolation and expansion of the catabolic potential of a Pseudomonas putida strain able to grow in the presence of high concentrations of aromatic hydrocarbons. J. Bacteriol. 177, 3911–3916. doi: 10.1128/jb.177.14.3911-3916.1995
Reva, O. N., Weinel, C., Weinel, M., Böhm, K., Stjepandic, D., Hoheisel, J. D., et al. (2006). Functional genomics of stress response in Pseudomonas putida KT2440. J. Bacteriol. 188, 4079–4092. doi: 10.1128/JB.00101-06
Richard, P., and Hilditch, S. (2009). D-galacturonic acid catabolism in microorganisms and its biotechnological relevance. Appl. Microbiol. Biotechnol. 82, 597–604. doi: 10.1007/s00253-009-1870-6
Rossoni, L., Carr, R., Baxter, S., Cortis, R., Thorpe, T., Eastham, G., et al. (2018). Engineering Escherichia coli to grow constitutively on D-xylose using the carbon-efficient Weimberg pathway. Microbiol. 164, 287–298. doi: 10.1099/mic.0.000611
Saha, B. C. (2003). Hemicellulose bioconversion. J. Ind. Microbiol. Biotechnol. 30, 279–291. doi: 10.1007/s10295-003-0049-x
Sambrook, J., and Russell, D. W. (2001). Molecular Cloning, 3rd Edn. New York, NY: Cold Spring Harbor Laboratory Press.
Satagopan, S., Sun, Y., Parquette, J. R., and Tabita, F. R. (2017). Synthetic CO2-fixation enzyme cascades immobilized on self-assembled nanostructures that enhance CO2/O2 selectivity of RubisCO. Biotechnol. Biofuels 10, 1–14. doi: 10.1186/s13068-017-0861-6
Schada von Borzyskowski, L., Carrillo, M., Leupold, S., Glatter, T., Kiefer, P., Weishaupt, R., et al. (2018). An engineered Calvin-Benson-Bassham cycle for carbon dioxide fixation in Methylobacterium extorquens AM1. Metab. Eng. 47, 423–433. doi: 10.1016/j.ymben.2018.04.003
Schellenberger, J., Que, R., Fleming, R. M., Thiele, I., Orth, J. D., Feist, A. M., et al. (2011). Quantitative prediction of cellular metabolism with constraint-based models: the COBRA Toolbox v2.0. Nat. Protoc. 6, 1290–1307. doi: 10.1038/nprot.2011.308
Schmitz, A., Ebert, B. E., and Blank, L. M. (2015a). “GC-MS-based determination of mass isotopomer distributions for 13C-based metabolic flux analysis,” in Hydrocarbon and Lipid Microbiology Protocols, eds. T. J. McGenity, K. N. Timmis, and B. N. Fernandez (Berlin: Springer, 223–243.
Schmitz, S., Nies, S., Wierckx, N., Blank, L. M., and Rosenbaum, M. A. (2015b). Engineering mediator-based electroactivity in the obligate aerobic bacterium Pseudomonas putida KT2440. Front. Microbiol. 6, 1–13. doi: 10.3389/fmicb.2015.00284
Schwander, T., Von Borzyskowski, L. S., Burgener, S., Cortina, N. S., and Erb, T. J. (2016). A synthetic pathway for the fixation of carbon dioxide in vitro. Science (80-) 354, 900–904. doi: 10.1126/science.aah5237
Stephens, C., Christen, B., Fuchs, T., Sundaram, V., Watanabe, K., and Jenal, U. (2007). Genetic analysis of a novel pathway for D-xylose metabolism in Caulobacter crescentus. J. Bacteriol. 189, 2181–2185. doi: 10.1128/JB.01438-06
Sudarsan, S., Dethlefsen, S., Blank, L. M., Siemann-Herzberg, M., and Schmid, A. (2014). The functional structure of central carbon metabolism in Pseudomonas putida KT2440. Appl. Environ. Microbiol. 80, 5292–5303. doi: 10.1128/AEM.01643-14
Tiso, T., Sabelhaus, P., Behrens, B., Wittgens, A., Rosenau, F., Hayen, H., et al. (2016). Creating metabolic demand as an engineering strategy in Pseudomonas putida – Rhamnolipid synthesis as an example. Metab. Eng. Commun. 3, 234–244. doi: 10.1016/j.meteno.2016.08.002
Tiso, T., Wierckx, N., and Blank, L. M. (2014). “Non-pathogenic Pseudomonas as platform for industrial biocatalysis,” in Industrial Bocatalysis, ed. P. Grunwald (Hoboken: Pan Stanford, 323–372.
Tiso, T., Zauter, R., Tulke, H., Leuchtle, B., Li, W.-J., Behrens, B., et al. (2017). Designer rhamnolipids by reduction of congener diversity: production and characterization. Microb. Cell Fact. 16:225. doi: 10.1186/s12934-017-0838-y
Vanholme, B., Desmet, T., Ronsse, F., Rabaey, K., Van Breusegem, F., De Mey, M., et al. (2013). Towards a carbon-negative sustainable bio-based economy. Front. Plant Sci. 4:174. doi: 10.3389/fpls.2013.00174
Wang, Y., Horlamus, F., Henkel, M., Kovacic, F., Schläfle, S., Hausmann, R., et al. (2019). Growth of engineered Pseudomonas putida KT2440 on glucose, xylose, and arabinose: hemicellulose hydrolysates and their major sugars as sustainable carbon sources. GCB Bioenergy 11, 249–259. doi: 10.1111/gcbb.12590
Weimberg, R., and Doudoroff, M. (1955). The oxidation of L-arabinose by Pseudomonas saccharophila. J. Biol. Chem. 217, 607–624.
Werpy, T., and Petersen, G. (2004). Top Value Added Chemicals From Biomass: Volume I - Results of Screening for Potential Candidates From Sugars and Synthesis Gas. Golden, CO: US-DoE (U.S. Department of Energy).
Wilhelm, M., and Hollenberg, C. P. (1985). Nucleotide sequence of the Bacillus subtilis xylose isomerase gene: extensive homology between the Bacillus and Escherichia coli enzyme. Nucleic Acids Res. 13, 5717–5722. doi: 10.1093/nar/13.15.5717
Williamson, N. R., Fineran, P. C., Leeper, F. J., and Salmond, G. P. (2006). The biosynthesis and regulation of bacterial prodiginines. Nat. Rev. Microbiol. 4, 887–899. doi: 10.1038/nrmicro1531
Winsor, G. L., Griffiths, E. J., Lo, R., Dhillon, B. K., Shay, J. A., and Brinkman, F. S. L. (2016). Enhanced annotations and features for comparing thousands of Pseudomonas genomes in the Pseudomonas genome database. Nucleic Acids Res. 44, 646–653. doi: 10.1093/nar/gkv1227
Wiser, M. J., and Lenski, R. E. (2015). A comparison of methods to measure fitness in Escherichia coli. PLoS ONE 10:e0126210. doi: 10.1371/journal.pone.0126210
Wittgens, A., Kovacic, F., Müller, M. M., Gerlitzki, M., Santiago-Schübel, B., Hofmann, D., et al. (2017). Novel insights into biosynthesis and uptake of rhamnolipids and their precursors. Appl. Microbiol. Biotechnol. 101, 2865–2878. doi: 10.1007/s00253-016-8041-3
Wittgens, A., and Rosenau, F. (2018). On the road towards tailor-made rhamnolipids: current state and perspectives. Appl. Microbiol. Biotechnol. 102, 8175–8185. doi: 10.1007/s00253-018-9240-x
Wittgens, A., Santiago-Schuebel, B., Henkel, M., Tiso, T., Blank, L. M., Hausmann, R., et al. (2018). Heterologous production of long-chain rhamnolipids from Burkholderia glumae in Pseudomonas putida—a step forward to tailor-made rhamnolipids. Appl. Microbiol. Biotechnol. 102, 1229–1239. doi: 10.1007/s00253-017-8702-x
Wittgens, A., Tiso, T., Arndt, T. T., Wenk, P., Hemmerich, J., Müller, C., et al. (2011). Growth independent rhamnolipid production from glucose using the non-pathogenic Pseudomonas putida KT2440. Microb. Cell Fact. 10, 1–17. doi: 10.1186/1475-2859-10-80
Wynands, B., Lenzen, C., Otto, M., Koch, F., Blank, L. M., and Wierckx, N. (2018). Metabolic engineering of Pseudomonas taiwanensis VLB120 with minimal genomic modifications for high-yield phenol production. Metab. Eng. 47, 121–133. doi: 10.1016/j.ymben.2018.03.011
Zhao, Y., Damgaard, A., and Christensen, T. H. (2018). Bioethanol from corn stover – a review and technical assessment of alternative biotechnologies. Prog. Energy Combust. Sci. 67, 275–291. doi: 10.1016/j.pecs.2018.03.004
Keywords: Pseudomonas putida, xylose, metabolic engineering, rhamnolipid, phenazine, pyocyanin, flux balance analysis, heterologous production
Citation: Bator I, Wittgens A, Rosenau F, Tiso T and Blank LM (2020) Comparison of Three Xylose Pathways in Pseudomonas putida KT2440 for the Synthesis of Valuable Products. Front. Bioeng. Biotechnol. 7:480. doi: 10.3389/fbioe.2019.00480
Received: 04 September 2019; Accepted: 23 December 2019;
Published: 17 January 2020.
Edited by:
Jean Marie François, UMS3582 Toulouse White Biotechnology (TWB), FranceReviewed by:
Antoine Danchin, INSERM U1016 Institut Cochin, FranceCopyright © 2020 Bator, Wittgens, Rosenau, Tiso and Blank. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lars M. Blank, bGFycy5ibGFua0Byd3RoLWFhY2hlbi5kZQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.