
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Bioeng. Biotechnol. , 10 January 2020
Sec. Synthetic Biology
Volume 7 - 2019 | https://doi.org/10.3389/fbioe.2019.00446
This article is part of the Research Topic 4th Applied Synthetic Biology in Europe View all 14 articles
The global warming conjugated with our reliance to petrol derived processes and products have raised strong concern about the future of our planet, asking urgently to find sustainable substitute solutions to decrease this reliance and annihilate this climate change mainly due to excess of CO2 emission. In this regard, the exploitation of microorganisms as microbial cell factories able to convert non-edible but renewable carbon sources into biofuels and commodity chemicals appears as an attractive solution. However, there is still a long way to go to make this solution economically viable and to introduce the use of microorganisms as one of the motor of the forthcoming bio-based economy. In this review, we address a scientific issue that must be challenged in order to improve the value of microbial organisms as cell factories. This issue is related to the capability of microbial systems to optimize carbon conservation during their metabolic processes. This initiative, which can be addressed nowadays using the advances in Synthetic Biology, should lead to an increase in products yield per carbon assimilated which is a key performance indice in biotechnological processes, as well as to indirectly contribute to a reduction of CO2 emission.
The global warming is caused by the anthropogenic emission of greenhouse gases (GHGS), among which emission of carbon dioxide (CO2) is broadly accepted by the scientific community as the most contributing factor to this climate change. While Nature has been orchestrated with a natural carbon cycle, the industrial revolution that begun in the early 19th century resulted in a rise of CO2 emission that slowly but definitively exceeded the natural geochemical carbon cycle. According to the Keeling curve, which reports the daily carbon dioxide measurements at Mauna Loa observatory, the CO2 emission has dramatically increased over the last 50 years, mostly due to deforestation and burning of fossil fuels (https://scripps.ucsd.edu/programs/keelingcurve/). These issues are now becoming critical for the human being, which urged the scientific community to find solutions to decrease our reliance on fossil fuels that should eventually change our life style. Biofuels and more generally biorefinery have emerged as promising solution whose purpose is to exploit microorganisms as cell factories to convert non-edible but renewable carbon sources such as lignocellulosic sugars into bioethanol as alternative energy (Duwe et al., 2019; Rosales-Calderon and Arantes, 2019) or into commodity chemicals that can replace those obtained from petrochemistry (Clark et al., 2012; Chen and Dou, 2016; Singhvi and Gokhale, 2019). In spite of the apparent sustainability of this solution, the use of chemoorganotrophic microbes such as yeast or E. coli present a caveat, as CO2 is released during the process of carbon metabolism (Figure 1). A wonderful alternative would be to exploit autotrophic microorganisms such as acetogenic anaerobic bacteria or microalgae (Schiel-Bengelsdorf and Durre, 2012; Scaife et al., 2015) as they have the capability to capture atmospheric CO2. However, these biological systems are at the moment industrially inefficient, due to their slow growth, poor productivity and low energy conversion yield (Claassens, 2017). The purpose of this mini-review is to expose and discuss original strategies that have emerged recently and that have employed synthetic biology tools to rewire the carbon metabolism of heterotrophic microorganisms to achieve maximal carbon conservation during their metabolism. Basically, this endeavor can be reached by either redesigning carbon metabolism network to optimize carbon conservation or recapturing carbon loss using CO2 fixing or carboxylating systems. Autotrophic CO2 fixation by reductive pentose phosphate cycle (rPP also known as Calvin-Benson cycle) as well as other natural metabolic pathways that perform carbon fixation including reductive TCA cycle, 3-hydroxypropionate cycle, 3-hydroxypropionate/4-hydroxybutyrate cycle, and dicarboxylate/4-hydroxybutyrate cycle are not considered in this mini-review, since they have been extensively reviewed in previous excellent papers (Berg et al., 2010; Fuchs and Berg, 2014; Erb and Zarzycki, 2018).
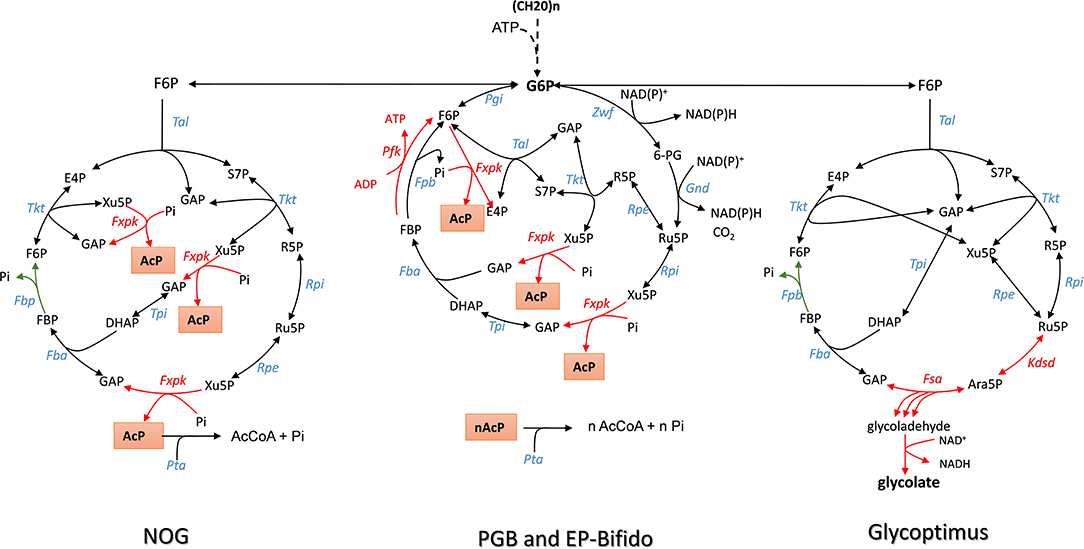
Figure 1. The scheme of non–oxidative glycolysis (NOG), Embden-Pentose-Bifido (EP-Bifido), and Glycoptimus synthetic pathways. Enzymes abbreviation are Fxpk, F6P/Xu5P-phosphoketolase; Tal, transaldolase; Tkt, transketolase; Fbp, fructose-1,6- bisphosphatase; Fba, fructose1,6 bisphosphate aldolase; tpi, triose phosphate isomerase; Rpe, ribulose 5-P epimerase; Rpi, ribose 5-P isomerase; Kdsd, arabinose-5-P isomerase; Fsa, fructose-6P aldolase; Alda, glycolaldehyde dehydrogenase. Metabolites: F6P, fructose-6P; PEP, phosphoenolpyruvate; Pyr, pyruvate; E4P, erythrose-4P; S7P, sedoheptulose-7P; R5P, ribose-5P; Ru5P, ribulose-5P; X5P, xylulose-5P; GAP, glyceraldehyde-3P; DHAP, dihydroxyacetone-P; FBP, fructose-1,6-bisphosphate; AcP, acetyl-Pi, AcCoA, acetyl-CoA.
Glycolysis or the Embden-Meyerhof-Parnas pathway (EMP) is a fundamental metabolic pathway in most living systems that decomposes sugars into pyruvate and recovers energy of this breakdown into ATP and reducing equivalents NADH. To fuel the cell with some essential anabolic precursors, pyruvate has to be decarboxylated into acetyl-CoA. This decarboxylation step releases carbon dioxide in the environment, resulting in 33% loss of carbon yield. This wasted CO2 may have major impact on the overall economy of bio-based products derived from fermentable carbon sources. This 33% carbon loss due to the decarboxylation of pyruvate has been challenged recently by Liao's group (Bogorad et al., 2013) who constructed a synthetic pathway termed “the non-oxidative glycolysis (NOG)” that can overcome this carbon loss, leading to a conversion of one mole of glucose to 3 moles of acetyl-moieties (Figure 1 and Table 1 for the stoichiometric equation). The logic of NOG relies on the phosphorylating cleavage of sugar phosphates by a phosphoketolase (Fxpk), which, combined with a carbon rearrangement cycles, creates a cyclic pathway with F6P as the input molecule (Figure 1). The irreversibility of this pathway is ensured by the phosphoketolase reaction that cleaves Xu5P into glyceraldehyde-3-P (GAP) and AcP and by the fructose-1,6-bisphosphatase reaction (Fbp) which recycles back F6P from FBP. The carbon rearrangement also involves transaldolase (Tal) and transketolase (Tkt) and the net result of this FBP-dependent cycle is the irreversible formation of three acetyl-phosphate (AcP) molecules from one F6P molecule. As phosphoketolase also displays phosphorylating cleavage activity on F6P (Tittmann, 2014), cleaving the latter molecule into AcP and E4P, a sedoheptulose bisphosphate (SBP)-dependent network can be conceived yielding to the same solution. In this case, this cycle will not need transaldolase (tal gene) but will require a SBP-aldolase to condense DHAP and E4P into SBP, and a sedoheptulose phosphatase (Sbp) to recycle back S7P. Such cycle can potentially exist since Sbp is essential in the Calvin-Benson cycle, and has been identified in yeast as being implicated in ribogenesis (Clasquin et al., 2011) and in methylotrophic bacteria where it participates in the ribulose 5-monophosphate (RuMP) cycle (Stolzenberger et al., 2013).
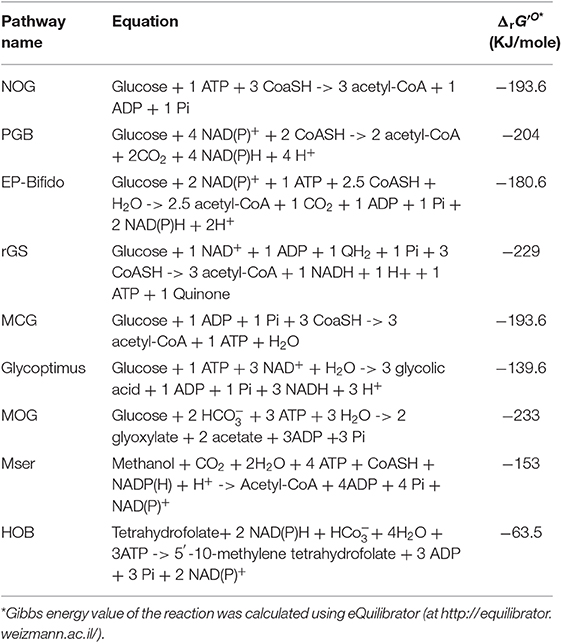
Table 1. Stoichiometric equation of carbon conservative pathways described in Figures 1–5, and their calculated thermodynamic value*.
In their Nature report, Liao and coworkers succeeded in demonstrating the in vitro and in vivo functioning of NOG (Bogorad et al., 2013). The in vitro system required the core of eight enzymes, which were either purchased when commercially available or produced by expression in E. coli. This was the case for phosphoketolase which is encoded by fxpk gene in Bifidobacterium adolescensis (Yin et al., 2005) and for some enzymes of the pentose phosphate pathway (PPP). In accordance with the pathway model, F6P initially added at 10 mM was readily and completely converted into 30 mM AcP. To validate the in vivo pathway, they chose to use xylose as the initial carbon substrate, as the phosphotransferase-dependent uptake and phosphorylation of glucose is not compatible with NOG and also because xylose does not cause repression of the fbp encoding fructose-1,6-bisphosphatase (Sedivy et al., 1984), which is needed together with the overexpression of fxpk. Other enzymes of the NOG pathway were natively expressed on a high copy plasmid, including pta and ackA which codes for phosphate acetyl transferase and acetate kinase, respectively, and which convert AcP into acetate. In addition, competitive fermentative pathways were disabled (i.e., deletion of ldhA, pflB, adhE, and frdBC to prevent production of lactate, formate, ethanol, and succinate) and the experiment was carried out under anaerobic condition to avoid further oxidation of acetate in the TCA cycle. Under this condition, 2.2 moles of acetate per mole of xylose were produced by the engineered E. coli equipped with the NOG pathway. In theory, the maximal yield that can be obtained by this conversion process should be 2.5. This yield termed the thermodynamic feasibility “YE” can be calculated as the ratio between the degree of reduction of the substrate γs to that of the product γp. YE is independent of the stoichiometry of the pathway and provides yield limits of the thermochemical process whatsoever the followed pathway or the catalyst used (Dugar and Stephanopoulos, 2011). In this case, YE of acetate from xylose (γs = 20) is 2.5 since γp of acetate = 6, whereas the stoichiometric yield “Yst” is 1,66. Therefore, the efficiency of the NOG pathway which can be calculated as the ratio of YE to Yst is 50% better than the classical EMP pathway. The fact that the in vivo conversion of xylose into acetate by the engineered E. coli expressing the NOG pathway only reached 88% of YE was due to some by-products formation, notably succinate.
According to the stoichiometric equation (Table 1), each mole of acetyl-CoA produced from glucose by NOG can be converted into acetate, which is accompanied by the production of one ATP. Therefore, a net balance of 2 ATP and 3 acetate molecules can be obtained from glucose by NOG. However, this pathway is not compatible for growth as it can generate neither the 12 metabolic building blocks nor the reducing power needed for synthetizing all cellular constituents. These features may explain why NOG has not been retained as a viable pathway during evolution. Nevertheless, in a second seminal paper published in PNAS, Liao's group succeeded in evolving a E. coli strain that could grow while relying on NOG for carbon catabolism (Lin et al., 2018). To achieve this challenge, they blocked glycolysis at the level of glyceraldehyde-3-P dehydrogenase and phosphoglycerate kinase by deleting the corresponding genes as well as the methylglyoxal and the Entner-Doudoroff pathways to prevent bypass of glucose into pyruvate. The glyoxylate shunt and gluconeogenesis was then upregulated to enable conversion of acetyl-CoA to metabolic intermediates required for growth, whereas the reducing power and ATP were derived from TCA, which could be done only under aerobic condition. Since these genetic interventions could potentially generate futile cycles, notably at the level of F6P/FBP and pyruvate/ acetate, pfkA gene encoding the major phosphofructokinase and poxB encoding pyruvate oxidase were deleted. The final strain termed PHL13 contained 10 genes deletion and the 2 overexpressed genes pck and fxpk encoding PEP carboxykinase and phosphoketolase. This engineered strain was still unable to grow on glucose unless acetate was present. It was therefore subjected to evolution by serial transfer to selective media and meanwhile, a quest for the limiting enzymes in the NOG cycle was carried out by devising a clever whole-pathway assay in crude extract of the engineered strains. Practically, the assay was to measure the rate of AcP production from F6P in a crude extract augmented with a mix of all purified NOG enzymes and determine the effect of removing a particular enzyme from this mix on AcP production. This study identified phosphoketolase as the most limiting enzyme followed by transketolase and transaldolase. Therefore, these genes were expressed on a high copy plasmid that also contained genes encoding the glucose permease (glf) from Zymomonas mobilis, the glucokinase (glk), and a AMP-insensitive PEP-activated FBPase (glpX) from E. coli (Donahue et al., 2000). Strain PHL13 that carried out this constructed plasmid was then subjected to further adaptive laboratory evolution (Mattanovich et al., 2014; Jang et al., 2019) to get rid of its dependence to acetate and rely only on glucose for growth. After further improvement of transketolase activity by the whole-pathway assay, these authors could isolate a slow growth colony NOG21 on glucose minimal medium, which was further evolved to yield a faster growing colony NOG26. Use of 13C glucose labeled on C3 and C4 confirmed that the growth was accomplished by NOG since acetate was labeled on its C1 and C2, as expected. Genome sequencing of this evolved strain unexpectedly unraveled a reduced expression of fxpk and pck. This result could be explained by a fitness response of the strain to the growth condition in order to avoid “kinetic traps” that would cause imbalance of the metabolic flux, as for instance by draining out all oxaloacetate from the TCA cycle or Xu5P from the NOG cycle. Other mutations were identified in the metabolic network such as a deletion of pykF encoding the major pyruvate kinase or mutation in pts genes, which likely reduced the PEP dependent phosphorylation of glucose, leaving thus more PEP for gluconeogenesis. All these genomic modifications likely resulted in a fine-tuning of pathway regulation that would be difficult to predict a priori.
From a biotechnological perspective, the potential of NOG is very limited unless this pathway is coupled with another to make compounds other than acetate. An example would be to establish a NOG- based reductive fermentation in which additional reducing agents such as molecular hydrogen is provided together with sugars. In this condition, 3 moles ethanol can be produced per mole of glucose which would increase by 50% the maximal yield from natural sugar fermentation pathway. The challenge here will be to express hydrogenase to allow the input of additional reducing equivalents. Another practical use of NOG shall be to combine with the Calvin-Benson-Bassham (CBB) cycle (Calvin, 1962) for the biosynthesis of acetyl-coA derived products, such as 1-butanol and fatty acids by autotrophic organisms (Liu et al., 2011). This metabolic rewiring could represent a 50% increase of CO2 into acetyl-CoA over the native pathway, concomitantly reducing the requirement of RuBisCO activity to reach the same carbon yield (Tcherkez et al., 2006).
A serious drawback of the NOG pathway is its inability to generate reducing power that is essential for the biosynthesis of added-value molecules from acetyl-CoA such as isoprene, fatty acids or polyhydroxybutyrate. A first approach to overcome this limitation was proposed by Opgenorth et al. (2016) who designed a synthetic in vitro pathway termed “Pentose-Bifido-Glycolysis” cycle (PBG) for the synthetic conversion of glucose into PHB via acetyl-CoA. This PBG cycle is divided in three metabolic parts. The first part is the phosphorylation of glucose into G6P which enters the cycle through the oxidative branch of the PPP and breaks down into Xu5P while producing NADPH. In a second phase, Xu5P is split into AcP and GAP by the phosphoketolase Fxpk. While AcP can be converted into acetyl-CoA by the CoA-phosphotransferase enzyme (Pta), GAP is recycled in a third phase into G6P. This occurs by the condensation of GAP and DHAP into FBP by the aldolase Fba, followed by a process involving a unique phosphofructokinase that catalyzes the ATP-dependent formation of F6P from FBP. At variance to the traditional phosphofructo-1-kinase reaction (Uyeda, 1979), this E. coli PFK encoded by pfkB was found to work reversibly and thus to regenerate ATP needed for glucose phosphorylation. This last phase enables to complete the cycle in ATP neutral manner. Accordingly, for each glucose entering the PBG cycle, two moles of acetyl-CoA, 2 moles of CO2 and 4 moles of NADPH is produced at zero cost of ATP (Table 1). However, there are two caveats with this synthetic pathway design. First the phosphoketolase Fxpk can also split F6P into E4P and AcP, which could abrogate the functioning of the cycle. Secondly, there is large excess of NADPH, as the synthesis of one PHB monomer only requires one NADPH, which therefore could lead to a feedback inhibition of the pathway. To solve the first issue, these authors (Opgenorth et al., 2016) complemented the pathway with PPP enzymes, namely transketolase (Tkt), transaldolase (Tal) and ribose-5-P isomerase (Rpi) in order to recycle E4P into the cycle (Figure 2B). The second problem was disentangled by introducing a “NAD(P)H purge valve” to regulate the build-up of NADPH. The outcome of this sophisticated purge valve is to uncouple carbon flux from NADPH production enabling a non-stoichiometric pathway production of NADPH. The construction of this purge valve requires the presence of NAD+-dependent glucose-6-P dehydrogenase (Zwf) and NAD+-dependent phosphogluconate dehydrogenase (Gnd) enzymes together with their native NADP+ dependent counterparts and a H2O-forming NADH oxidase (NoxE). While NADPH is reoxidized during PHB production, high level of NADPH would not penalize the cycle since it is taken over by the NAD+-dependent enzymes which continue to provide the precursor acetyl-CoA while NADH is readily reoxidized by NoxE. This original cell-free system production of bio-based chemicals from glucose turned out to be quite efficient as the authors reported a maximum productivity of the PBG cycle of 0.7 g PHB/L/h with a yield of 90%. There is great hope in developing such a process at an industrial scale since cell-free system can overcome common problems afflicting in vivo biological systems, such as building-up toxic intermediates, low productivities due to competing pathways and undesirable byproducts (Dudley et al., 2015). However, cell-free systems require to solve major technical and economic problems including production, purification, and stability of enzymes or their potential recycling.
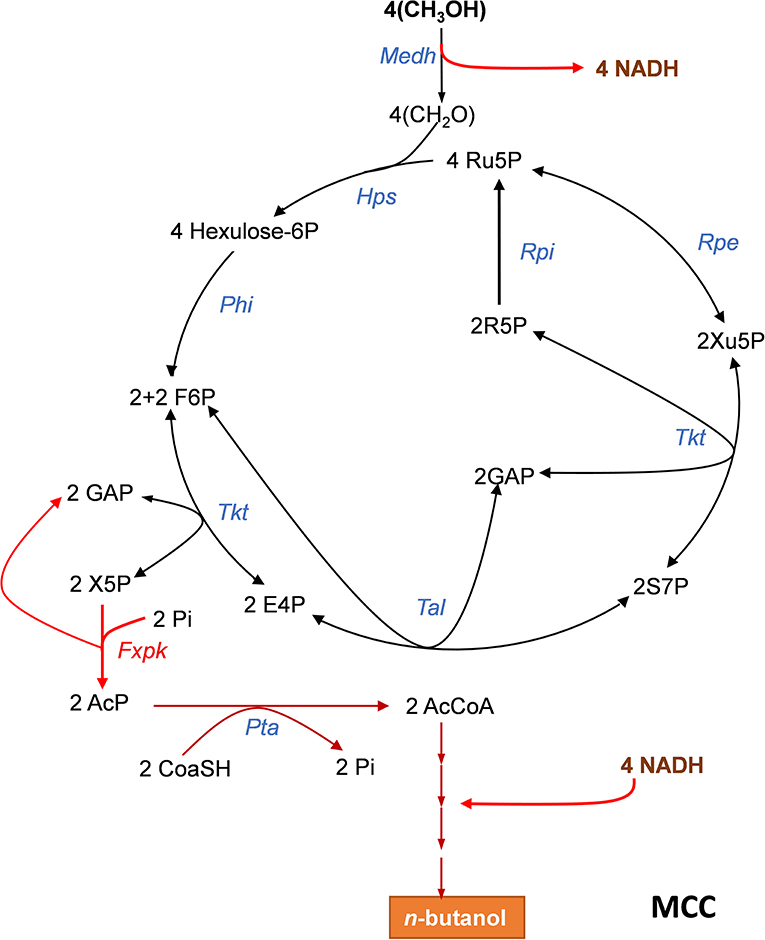
Figure 2. The methanol condensation cycle (MCC) for assimilation of methanol into higher alcohol with maximal carbon conservation. The MCC is a combination of the RuMP with part of the NOG allowing to avoid pyruvate decarboxylation and to bypass ATP dependency. It results in the net production of acetyl-CoA (AcCoA) from 2 methanol. Here, it is represented the metabolic pathway leading to the production of n-butanol which requires 4 methanol. Enzymes abbreviations are: Medh, NAD+-dependent methanol dehydrogenase; Hps, hexulose-6P synthase; Phi, phosphohexuloseisomerase; Tkt, transketolase, Fxkp, F6P/Xu5P phosphoketolase; Tal, transaldolase; Rpe, ribulose-5-P epimerase; Rpi, ribose-5-isomerase. Abbreviation for metabolites as in Figure 1.
Another approach to disentangle the failure of the NOG pathway to generate redox power was proposed by Wang et al. (2019) through the in vivo construction of an “EP-Bifido” pathway. This synthetic pathway is very similar to PGB as it combines EMP and PPP pathway with the expression of phosphoketolase enzymes acting on F6P and Xu5P, but it does not employ PFK reaction to regenerate the ATP that is needed to phosphorylate glucose (Figure 2B). As indicated in Figure 1, the major difference between NOG and the EP-Bifido is that the latter implicates the oxidative portion of the PPP, which leads to the production of NADPH. However, this metabolic process occurs at the expense of one CO2. The remaining reaction steps share those of the NOG with notably Fbp that provides the second driving force and recycles back F6P into the cycle. In theory, this pathway design should yield 2.5 acetyl CoA per mole glucose with two moles of NAD(P)H at the cost of 1 ATP (see Table 1). However, it may have the capacity to achieve an even higher yield of acetyl-CoA, as this will depend on favoring G6P into PPP with respect to EMP. In theory, 2.66 moles of acetyl-CoA could be produced from 1 glucose if 66% of G6P goes through PPP. To create this EP-Bifido pathway and expect to attain this optimal carbon yield, the following genetic modifications were carried out in E. coli. On the one hand, zwf and gnd were overexpressed to direct glucose into the PPP together with fxpk encoding F6P/Xu5P phosphoketolase from Bifidus adolescentis. On the other hand, the engineered strain was deleted for pfkA encoding the major phosphofructokinase, edd/eda encoding enzymes of the Entner-Doudoroff and ackA encoding acetate kinase to avoid, respectively, ATP wasteful by FBP/F6Pcycle, the production of pyruvate that could bypass the EP-Bifido pathway and the loss of AcP into acetate. Altogether, the engineered E. coli strain equipped with the EF-Bifido pathway grew 3 times slower than the wild type strain, did not produce any acetate and released 50 to 70% less CO2 than the wild type strain. The performance of the EF-Bifido pathway in acyl-CoA derived polydroxybutyrate (PHB), fatty acids or mevalonate was moreover demonstrated by showing that the yield of these products was improved by 145, 56, and 48% respectively in engineered strains as compared to the control strains (Wang et al., 2019), arguing for a biotechnological relevance of reengineering the carbon metabolism of E. coli with this pathway.
While the refactoring of central carbon metabolic pathway was devoted to maximal acetyl-moieties from glucose, the glycoptimus pathway deals with a rewiring of the carbon metabolism to achieve maximal yield of glycolic acid (GA) from C5 and C6 sugars without carbon loss (Figure 1). According to thermodynamic calculation (Dugar and Stephanopoulos, 2011), the YE of glycolic acid (γp = 6) from glucose (γs = 24) or from pentose (γp = 20) should be 4 and 3.3, respectively, whereas the maximal yield YS based on the pathway stoichiometry is 2 from glucose and 1.66 from pentose (Dolan and Welch, 2018). Using the natural pathway, no <13 genetic modifications were executed to achieve a production of 52 g/L glycolate from glucose at a yield of 50 % of the Yst (Soucaille, 2007). Further genetic modifications were proposed by Deng et al. (2018), by combining Dahms pathway with the glyoxylate shunt, enabling a yield close to 90% of the stoichiometric yield. The problem to reach the thermodynamic YE lies at two levels. A first one is to avoid the loss of CO2 at the pyruvate decarboxylation step, while the second is more challenging as it requires to capture one carbon mole as CO2. Since direct CO2 fixation by heterotrophic system is still very complicated as experienced by Milo and coworkers in E. coli (Antonovsky et al., 2016), we rewired the carbon central metabolism of E. coli in order to overcome the loss of CO2 during the conversion of sugars into glycolic acid. Theoretically, the proposed pathway should lead to a maximal yield of 3 GA per glucose (Table 1) or 2.5 GA per pentose, which is 50% higher than obtained by the natural pathway (Figure 1). This pathway design implicates the overexpression of kdsD and fsaA of E. coli which codes for an arabinose-5P (Ara5P) isomerase and a class I aldolase, respectively. Even though these two genes are endogenous to E. coli, this pathway is not natural in this bacteria because the Kdsd enzyme is naturally implicated in the synthesis of 2-keto-3-deoxy-octulosonate (KDO), a constituent of the outer member of cell wall lipopolysaccharide (Lim and Cohen, 1966), whereas Fsa has been initially reported as a F6P aldolase that catalyzes the aldol cleavage of F6P into dihydroxyacetone (DHA) and glyceraldehyde-3-P (GAP) (Schurmann and Sprenger, 2001). The bacterium E. coli also harbors an ortholog of fsaA termed talC or fsaB (Reizer et al., 1995) but the physiological function of these two genes is still completely unknown. In particular they are not expressed under standard—LB- culture condition (Schurmann and Sprenger, 2001). Nevertheless, Clapes' team demonstrated the remarkable originality of Fsa enzyme as a unique biocatalyst for asymmetric cross-aldol addition of glycoladehyde. They also reported that Fsa can cleave Ara5P into glycolaldehyde and GAP with a 10 fold higher affinity than F6P (Garrabou et al., 2009), which was confirmed in Lachaux et al. (2019). Therefore, the consecutive action of Kdsd and Fsa leads to the conversion of pentose phosphate intermediate Ru5P into glycolaldehyde and GAP (Figure 1). While glycolaldehyde can be oxidized into glycolic acid by an aldehyde dehydrogenase encoded by aldA (Caballero et al., 1983), GAP is recycled into Ru5P through the non-oxidative pentose phosphate pathway (Figure 1). This cycling scheme may result in theory in the production of 3 moles of GA per mole of glucose, only if the lower part of the glycolysis at the level of GAP is blocked. It also cost one ATP but generates 3 moles of NADH (Table 1) that needs to be reoxidized to allow the continuous functioning of the cycle. The reoxidation of NADH requires an active respiratory chain which is coupled to the synthesis of ATP. This ATP provision will be useful to regenerate ATP used in glucose phosphorylation, for uptake of sugars as well as for cellular maintenance. The in vitro and in vivo function of the glycoptimus pathway was validated although the yield of GA from glucose and xylose did not exceed 30% of the maximal yield expected. Possible explanation of this poor efficiency of the pathway could be that (i) the blockage of the lower part of glycolysis by deletion of gapA encoding GADPH was detrimental for cell viability; (ii) all competitive pathways using the same intermediaries have not yet been dismantled in the engineered strains; and (iii) the carbon flux in the kdsD-fsaA pathway is not optimized yet (Lachaux et al., 2019). Finally, the reoxidation of NADH is likely to generate more ATP than needed for the functioning of the glycoptimus cycle, which may result in feedback inhibition due to accumulation of intermediates and eventually depletion of cofactors. Solving these various issues together with the development of an appropriate fermentation practice should lead to an economically viable process of glycolic acid synthesis.
Abundant carbon sources other than sugars, such as methanol can be also exploited for the biosynthesis of added-value molecules. However, assimilation of these alternative resources also poses problem with carbon conversation. As an example, the ribulose-5- monophosphate pathway (RuMP) that is present in methylotrophic bacteria and in some yeasts such as Pichia pastoris (Yurimoto et al., 2002; Muller et al., 2015; Russmayer et al., 2015) is one of the three natural methanol assimilatory pathways known so far (Zhang et al., 2017) which can produce one acetyl-CoA from three formaldehydes at the expense of the decarboxylation of pyruvate. Liao's team (Bogorad et al., 2014) showed that the decarboxylation step could be avoided by combining the RuMP with phosphoketolase enzymes, leading to the production of acetyl-CoA from 2 formaldehydes through a methanol condensation cycle (MCC) as depicted in Figure 2. However, this MCC can only work if the thermodynamically unfavorable NAD+-dependent methanol dehydrogenase is coupled to the reoxidation of NADH into reduced products such as ethanol, n-butanol or even higher alcohols (Bogorad et al., 2014). Remarkably, the MCC reaction is completely redox balanced and independent of ATP, and thus does not involve the phosphofructokinase enzyme of the RuMP, as well as the triose phosphate isomerase (Tpi), fructose 1,6-bisphosphate aldolase (Fba) and fructose-1,6-bisphosphatase (Fbp) of the NOG pathway. This property has been exploited to demonstrate the in vitro functioning of MCC to the production of ethanol or n-butanol. Therefore, a cell-free system could be potentially exploitable for larger scale production of higher alcohols with maximal yield and at high productivity if conditions for enzymes and intermediates stability are assured. In addition, this in vitro study pointed out the role of phosphoketolase in the robustness of the MCC, showing that its optimal running requires well-balanced levels of this enzyme with respect to the others, as a low or excessive phosphoketolase activity could trigger kinetic traps that significantly diminish rate of end-product production, due to either accumulation or depletion of intermediates metabolites in the cycle.
The TCA or citric acid cycle elucidated by Krebs and Johnsson in 1937 (see Krebs and Johnson, 1980 for an historical perspective or Akram, 2014 for a recent review of TCA role in intermediary metabolism) is considered as the central metabolic hub of the cell whose amphibolic nature provides intermediates for numerous metabolic functions. This cycle generates energy and reducing power needed for anabolic activities of the cell at the expense of complete oxidation of pyruvate into carbon dioxide. The glyoxylate shunt, discovered by Krebs and Kornberg in 1957 (see Dolan and Welch, 2018) for an excellent review on this pathway) avoids the decarboxylation steps of the TCA cycle and allows acetyl-CoA to be converted into a C4 carbon without carbon loss. This shunt is therefore key for organisms that grow on C2 to C4 carbon such as acetate, glycerol and malate, or on ketogenic amino acids (i.e., aspartate, glutamate). The glyoxylate shunt takes carbon away from the TCA cycle at the level of isocitrate, just before the commencement of the first decarboxylation step. This requires two specific enzymes that are the hallmark of this cycle: isocitrate lyase that cleaves isocitrate into succinate (C4) and glyoxylate (C2), and malate synthase, which condenses glyoxylate with acetyl-CoA to malate. Due to the large negative Gibbs value of the malate synthase reaction (ΔrG'0 = −44 kJ/mol), the glyoxylate cycle can only run in the acetyl-CoA condensation. However, running the cycle clockwise directly could be beneficial to supply acetyl-CoA from sugars by bypassing the decarboxylation step of pyruvate. As illustrated in Figure 3, a reversal of the glyoxylate shunt (rGS) in which the thermodynamically favorable PEP carboxylating reaction is implemented could generate 2 acetyl-CoA from TCA intermediates. This challenge was raised by the Liao' group according to the following strategy (Mainguet et al., 2013). At first, they demonstrated that isocitrate lyase (ICL) is reversible in vivo by showing that a E. coli strain made auxotroph for glutamate (glu− strain) by deletion of gltA and prpC encoding citrate synthase was able to grow on glucose mineral medium supplemented with glyoxylate and succinate. In contrast, this glu− strain was unable to recover growth when glyoxylate was replaced by malate, even if dctA gene encoding a non-glucose repressible malate importer from B. subtilis had been overexpressed. These results confirmed that malate synthase reaction was not reversible in vivo. To get around this obstacle, they wished to express an ATP-dependent malate thiokinase (Mtk) encoded by mtkA and mtkB from Methylobacterium extorquens. In this methanotrophic bacterium, this enzyme contributes to C1 assimilation through the serine cycle (Fei et al., 2014). This reaction step is then followed by the cleavage of malyl-CoA into glyoxylate and acetyl-CoA by a malyl-coA lyase (Mcl) naturally present in microorganisms that use the 3-hydroxypropionate cycle for autotrophic carbon dioxide fixation (Berg et al., 2007). Unfortunately, the expression of mtkAB and mcl from Methylobacterium extorquens expressed in a glu− strain did not rescue growth on malate and succinate likely because Mtk was not functional. They overcame this problem by screening various putative malate thiokinase and discovered that sucCD-2 from Methylococcus capsulatus annotated as encoding a succinyl-CoA synthetase (Ward et al., 2004) had very good Mtk activity. Expression of sucCD-2 gene with mcl restored the capacity of growth of a glu− strain on glucose supplemented with malate and succinate. The second important action was to recycle OAA from isocitrate that is accompanied by the release of a second molecule of acetyl-coA. This involves to reverse the aconitase and the citrate synthase reaction. While the first one is readily reversible, the conversion of citrate to OAA by citrate synthase is strongly unfavorable (ΔrG'°~ + 34 KJ/mol). It can nevertheless be obtained by expressing an ATP-dependent citrate lyase encoded by acl which is present in several eukaryotic cells and in some archeabacteria (Fatland et al., 2000; Verschueren et al., 2019). The in vivo functionality of the Acl reaction was tested in E. coli strain made auxotroph to aspartate (asp− strain) by deleting genes encoding all enzymes that produce its precursor OAA (ie ppc, mdh, mqo), as well as citrate synthase and citrate lyase (gltA and citE). As expected, the asp− strain recovered growth on a glucose minimal medium when supplemented with citrate. Although the thermodynamically unfavorable reactions were solved to potentially create a reverse glyoxylate cycle, it was also necessary to remove isocitrate dehydrogenase (icd) to avoid siphoning isocitrate obtained by condensation of glyoxylate and succinate into α-ketoglutarate. Then, the assembly of the complete pathway in asp− strain which was otherwise deleted for mdh, mqo, ppc, icd, and aceB consisted in the overexpression of BsdctA, McsuccD-2, Rsmcl, and Ctacl genes. This strain was shown to grow on glucose supplemented with malate and succinate, demonstrating the in vivo function of this rGS. Until now, the complete demonstration of the “in vivo” reversal of the glyoxylate cycle has not been fully demonstrated. It will require to connect OAA to malate which can be easily carried out by the reversible NAD+-dependent malate dehydrogenase encoded by mdh. However, cycling malate shall require also expression of a fumarase (Fum) and a fumarate reductase (Frd), both enzymes being preferentially active under anaerobic conditions (Lin and Iuchi, 1991; Tseng, 1997).
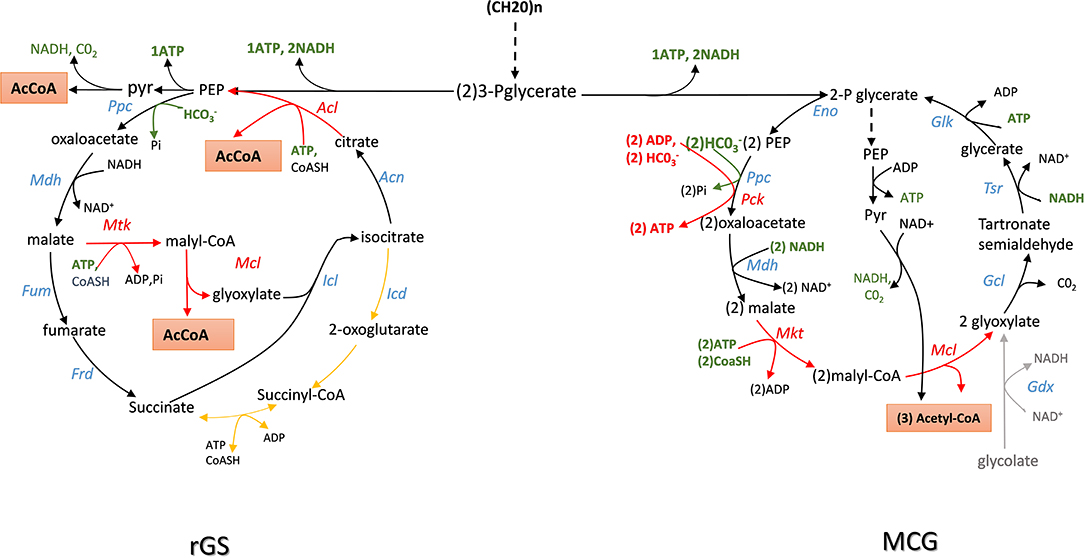
Figure 3. Scheme of the reversal of glyxoylate shunt (rGS) and maly-CoA Glycerate (MGC) cycling pathway for the production of acetyl-CoA from sugars with maximal carbon conservation. Enzymes abbreviation are: Ppc, PEP carboxylase; Mdh, malate dehydrogenase; Fum, fumarase; Frd, fumarate reductase; AceA, isocitrate lyase; Aco, aconitase; Acl, acyl-CoA lyase: Mtk, malate thiokinase; Mcl, malyl-CoA lyase; Gdh, glycolate dehydrogenase; Gcl, glyoxylate carboxyligase; Tsr, tartronate semialdehyde reductase; Glk, glycerate kinase; Eno, enolase.
In theory, the integration of rGS with the central carbon metabolic pathway via the PEP carboxylase encoded by ppc could lead to the conversion of one mole glucose into 3 moles of acetyl-CoA, thus achieving a 50% yield increase over the native pathway. Contrary to the NOG pathway that converts glucose to 3 AcCOA at the expense of one ATP, the conversion of glucose into AcCOA by the rGS generates one extra ATP. In addition, while NOG is redox neutral, the rGS pathway results in the net production of one NADH and one reduced quinone per glucose (Table 1). This indicates that rGS is more energetically efficient than NOG which could be an advantage with respect to the production of acetyl-CoA derived products such as isoprenoids, fatty acids, long chain alcohols as they required reduced power and ATP (Tabata and Hashimoto, 2004; Kondo et al., 2012; Robles-Rodriguez et al., 2018).
Like the rGS pathway, the synthetic Malyl-CoA-Glycerate (MCG) cycling pathway allows the conversion of glucose into acetyl-CoA. As depicted in Figure 3, this synthetic pathway relies on 6 natural reaction steps that are catalyzed by endogenous enzymes in E. coli, completed by the heterologous expression of mtkAB and mcL from methylotrophic bacteria. The importance of these two reaction steps in the in vivo functionality of the MCG was demonstrated by showing that the growth on glucose of an acetyl-COA auxotroph E. coli strain (ΔaceE ΔpoxB ΔplfB) can be rescued upon the expression of mtkAB and mcl genes. Like NOG and rGS, the theoretical molar yield of acetyl-COA per glucose consumed is 3 (Table 1). Another interesting feature of the MCG is to be able to assimilate C2 compound such as glycolate or glyoxylate (Figure 3). Hence, in photosynthetic organisms, expression of MCG could improve carbon fixation in combination with the CBB cycle by re-assimilating glycolate produced by photorespiration. Also, the conversion of glucose into acetyl-CoA by the MCG is redox neutral and costs one ATP for glucose phosphorylation, unless the glycolytic PEP carboxykinase as it exists in the rumen bacterium Actinobacillus succinogenes (Kim et al., 2004; Leduc et al., 2005) is used instead of PPC, which would result in the production of one extra ATP.
In an elegant paper published in 2010, Bar-Even et al. (2010) developed a constrained-based modeling approach that explored all possibilities that could be devised from a repertoire of ~5,000 known metabolic enzymes reported in the KEGG database (https://www.genome.jp/kegg/pathway.html) to generate carbon fixation pathways that are alternative to the 5 known to date. Criteria used in their modeling were (i) the enzyme's specific activity is equal or superior to that of RuBisCO which is used as the benchmark activity; (ii) a minimal energy cost of the pathway, which corresponds to the cost in NAD(P)H, ferredoxins, FADH2, and ATP in the production of one mole of products from CO2; (iii) thermodynamic feasibility of the pathway under a plausible physiological range of metabolites concentrations; (iv) topology of the pathway, which incorporates the number of enzymatic reactions the carbon fixation pathway can make as an independent unit and (v) the compatibility or integration of this synthetic pathway with the endogenous metabolic network. According to these criteria, they identified several cycling pathways containing four to six enzymatic steps leading to the production of the C2 compound glyoxylate. Although most of them were unrealistic because thermodynamically unfeasible or using oxygen-sensitive ferredoxin-oxidoreductase enzyme, these authors found that kinetically efficient pathways absolutely require a carboxylating enzyme with high activity and affinity toward CO2 or whose reaction must be physiologically irreversible. PEP carboxylase (ppc gene) and pyruvate carboxylase (pyc gene) came at first in their listing, followed by acetyl-CoA and propionyl-CoA carboxylase. Accordingly, they designed a pathway family termed Malonyl-CoA-Oxaloacetate-Glyoxylate (MOG) cycle employing either PEP carboxylase or pyruvate carboxylase as the sole carboxylating enzyme and that has a higher carboxylating activity than the reductive pentose pathway (rPP). This MOG pathway resembles the natural C4 cycle in which the recuperation of CO2 arising from malate decarboxylation to pyruvate by RuBisCO is replaced by a PEP or pyruvate carboxylating enzyme. The net product of MOG is the C2-carbon glyoxylate, which can be converted into GAP by the bacterial-like glycerate pathway (Eisenhut et al., 2006; Igamberdiev and Kleczkowski, 2018) (Figure 4). Interestingly, this pathway turns out to be thermodynamically feasible and more efficient than the rPP, although it has not been found in Nature yet. Moreover, combined with the plant natural C4-cycle, it could allow to overcome the futile CO2 cycling that is taking place in the bundle sheath cell and which is due to malate decarboxylation coupled to reassimilation of CO2 by RuBisCO. In addition, this coupling could result in an extra CO2 fixation with the release of glyoxylate. Therefore, it can be anticipated that the expression of MOG in autotrophic cells may provide great advantages from a biotechnological point of view, with faster growth and increased crop yields because of additional carbon dioxide input and absence of competing oxygenation reaction that reduces carbon fixation by plants. Whether the implementation of this MOG pathway in a heterotrophic cell like E. coli is biotechnologically relevant is less obvious. Indeed, this implementation will require the expression of heterologous genes encoding six out of the nine reactions of this cyclic pathway (Figure 4), namely the malate thiokinase (mtkAB), the malyl-CoA lyase (mcl), the methylmalonyl-COA carboxytransferase (mct), the malonyl-CoA reductase (mcr), the β-alanine-pyruvate transaminase (bapta), and the alanine 2,3 aminomutase (aam). More importantly, the stoichiometry evaluation of this route from glucose indicated that it can produce 2 moles of glyoxylate and 2 moles of acetate, with the requirement of two mole of CO2. However, while globally thermodynamically favorable and redox balanced, this reaction can occur at the expense of 3 ATP, which can be obtained by consumption of acetate, and hence resulting in the reemission of 2 moles of CO2. Therefore, this route has likely no meaning to be expressed in a chemoorganotrophic system.
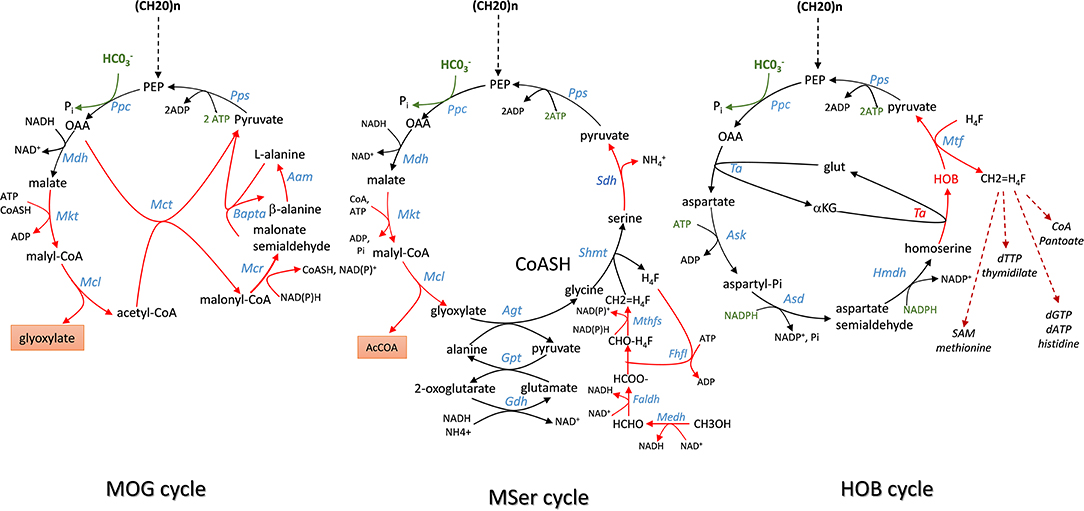
Figure 4. Scheme of Malonyl-coA-Oxaloacetate- Glyoxylate (MOG), Modified Serine (MSer) and 4-hydroxy-2-oxo-butyrate (HOB) cycling pathway employing the efficient PEP carboxylase to capture Co2 into either product or biomass. Enzymes abbreviation are: Ppc, PEP carboxylase; Mdh, malate dehydrogenase; Mtk, malate thiokinase; Mcl, malyl -CoA lyase; Mct, malonyl-CoA acetyl transferase; Mcr, malonyl CoA reductase; Bapta, β-alanine: pyruvate transaminase; Aam, alanine 2,3 aminomutase; Pps, PEP synthase; Ta, transaminase; Ask, aspartate kinase; Asd, aspartyl-Pi semialdehyde dehydrogenase; HmdH, homoserine dehydrogenase; Hmtf, hydroxymethyl transferase.
In methylotrophic bacteria, the serine cycle is a natural pathway to assimilate C1 carbon such as methanol or methane into acetyl-CoA intermediate without loss of carbon (Smejkalova et al., 2010). A modified serine cycle pathway has been designed in E. coli to allow co-assimilation of C1-carbon such as methanol or formate with bicarbonate to generate acetyl-COA as a precursor for several bio-based products (Yu and Liao, 2018). This synthetic pathway encompasses nine enzymatic reactions for the cycle in which CO2 is captured by the reaction catalyzed by PEP carboxylase, and four additional reaction enabling assimilation into 5–10 methylene tetrahydrofolate (CH2 = H4F). The methyl unit is afterwards transferred on serine cycle by a reaction involving glycine and catalyzed by serine hydroxymethyl transferase (Shmt). From these 13 enzymatic steps, seven (highlighted in red in Figure 5) had to be implemented using heterologous genes from various organisms, which included S. cerevisiae for the glyoxylate:alanine transaminase (Agt) encoded by AGX1, serine dehydratase (Sdh) from Cupriavidus necator encoded by SdaA, malate thiokinase (Mkt), and malyl-CoA lyase(Mcl) from M.extorquens. For methanol oxidation and its conversion to 5–10 methylene tetrahydrofolate, a variant of the Cupriavidus necator methanol dehydrogenase (Medh) with higher specific activity was used together with formate-H4F ligase (Fhlfl) and 5,10-methylene tetrahydrofolase synthase (Mthfs) from Moorella thermoacetica. The NAD+-linked formadehyde dehydrogenase (Faldh) from Pseudomonas putida was employed to oxidize formaldehyde to formate. All these genes were carried on 3 different plasmids and transformed into E. coli strain HY106, which was deleted for genes to avoid byproducts formation and shortage of intermediates in the cycle. Accordingly, aceB and glcB both encoding malate synthase were deleted to avoid competitive reverse reaction catalyzed by Mkt and Mcl. The gcl gene encoding glyoxylate carboxyligase was deleted to prevent shortage of glyoxylate into tartronate semialdehyde, as well as gcvP to avoid loss of glycine by decarboxylation. Genes encoding lactate dehydrogenase (ldhA) and fumarate reductase (frABCD) were also deleted in this strain to avoid shortage of pyruvate into D-lactate and malate into succinate. As indicated in Table 1, co-assimilation of methanol with CO2 needs 3 moles ATP and 1 mole reduced equivalent per mole of acetyl-CoA produced. Hence, it cannot proceed without addition of another carbon source to provide this energy and cofactors. Thus, the in vivo function of the complete cycle was demonstrated using C13-methanol together with xylose to supply energy and PEP. In addition the pyruvate to malate flux was increased upon overexpression of pyc and mdh encoding pyruvate carboxylase and malate dehydrogenase in the engineered E. coli equipped with the Mser pathway. The engineered strain could produce acetate from which 33% came from methanol. When the same experiment was carried out with both C13 methanol and C13 bicarbonate, the amount of acetate produced was the same but the labeling of both C1 and C2- carbon of acetyl-moieties was enriched by 2-fold, indicating that bicarbonate has been also incorporated into acetyl moieties. To conclude, the construction of this synthetic serine cycle in E. coli enables the assimilation of C1 compounds such as methanol to increase the production of C2-compounds as precursor of bio-based products such as PHB, isoprenoid, fatty acids, etc. In addition, this cycle, combined with the glyoxylate shunt, is capable to support growth from pyruvate that is formed from two C1-carbon (formate or methanol) and one bicarbonate, unless the mole excess of NAD(P)H is reoxidized by respiratory chain coupled with ATP.
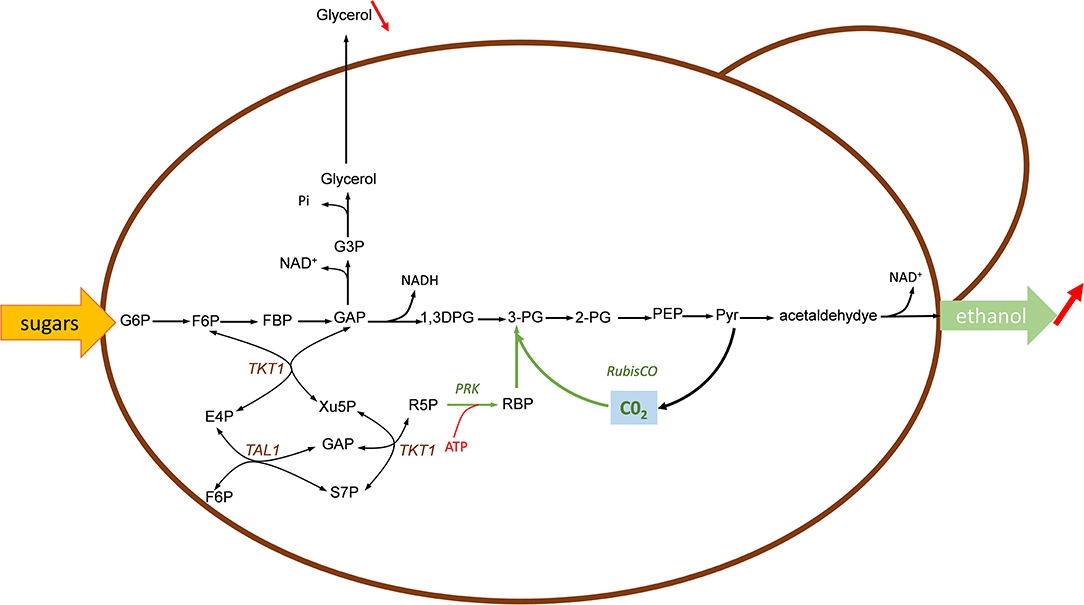
Figure 5. Engineering sugars fermentation with Calvin-cycle enzymes for fixing in-situ CO2 to improve bioethanol yield in yeast. In green are shown the Calvin-cycle enzymes corresponding to RuBisCO and phosphoribulose-5-P kinase (PRK). The expression of this Calvin-cycle enzymes enables the use of CO2 as alternative electron acceptor for reoxidation of NADH, thereafter reducing glycerol production under anaerobic condition. Enzymes: RuBisCO, ribulose 1,5 bisphosphate carboxylase; PRK, phosphoribulose 5-P kinase; Tkt, transketolase; Tal, transaldolase. Metabolites abbreviation, as in Figure 1 except: 1,3-DPG, 1,3 diphosphoglycerate; 3-PG, 3-phosphoglycerate; 2-PG, 2-phosphoglycerate; G3P, glycerol-3-P.
Another illustration about the potential function of carboxylating enzyme to incorporate CO2 into biomass comes from the work of Bouzon et al. (2017). These authors built a synthetic pathway that is alternative to the ubiquitous one carbon pathway transferring C1-moities (i.e., formyl, hydroxymethyl, or methyl group) from tetrahydrofolate for the synthesis of purine nucleotides, thymidylate, methionine, or coenzyme A (Figure 4). These C1-moities are obtained from the pivotal intermediate 5,10-methylene tetrathydrofolate (CH2=H4F) produced by the condensation of tetrahydrofolate with formaldehyde that originates from serine or glycine. To replace these amino acids as C1-donor, these authors devised a cyclic pathway in which the C1 compound (in this case formaldehyde) is obtained from the aldolytic cleavage of the non-natural intermediate 2-keto-4-hydroxybutyrate (HOB). As shown in Figure 4, the complete cycle encompasses 8 enzymatic steps in which the C1- source is that is captured by PEP carboxylase to yield OAA. Through a set of 4 natural enzymatic step, OAA is converted into homoserine which gives rise to HOB through a transaminase reaction. The best candidate for this non-natural transaminase reaction was obtained by employing an automated evolutionary technology originally developed by Marlière et al. for evolving E. coli possessing DNA in which thymine was replaced by the synthetic building block 5-chlorouracil (Marliere et al., 2011). The evolved transaminase was found to correspond to a variant of the alanine: pyruvate transaminase encoded by alaC having amino change at position 142 (Ala to Pro) and 245 (Tyr to Glu). The HOB is then cleaved into pyruvate and formaldehyde which is readily transferred into H4F by the promiscuous hydroxymethytransferase (Mtf) encoded by panB. The cycle is then closed by converting back pyruvate into PEP, which implicates PEP synthase. Overall, the functioning of this cycle costs two moles of NADPH and three moles of ATP per mole of CO2 incorporated (Table 1). To implement this synthetic pathway that completely rewires the C1-canonical metabolism by imposing HOB as the essential metabolic intermediate, the authors banked on an evolutionary trajectory of a E. coli strain lacking all enzymes necessary for the natural C1- transfer moieties using an original automated evolutionary technology that is now commercialized as “Heurisko” by the company Altar (http://www.altar.bio/). The authors estimated that the CO2 assimilated in the biomass of E. coli expressing this HOB pathway is about 2 times more than in wild type strain (Bouzon et al., 2017). However, being quite energy consuming, the relevance of this pathway in the production of biotechnological molecules methionine is questionable.
Under anaerobic condition, sugar fermentation by the yeast Saccharomyces cerevisiae into ethanol generated excess of NADH, which must be reoxidized to warrant redox-cofactor balancing (Van Dijken and Scheffers, 1986). This reoxidation diverts between 4 and 10% of the sugar into glycerol, which has a significant impact on the economy of large scale yeast-based bioethanol production. To reduce or even eliminate this by-product and in the same time, to increase the ethanol yield, the group of Jack Pronk in TU Delft proposed to use CO2 as electron acceptor for the reoxidation of this excess of NADH. A theoretical analysis indicates that the replacement of glycerol by CO2 can increase the ethanol yield from sugar by 14% if the CO2 is incorporated through phosphoribulokinase (PRK) and ribulose-1,5-bisphosphate carboxylase/oxygenase (RuBisCO) onto ribulose 5-P (Ru5P) (Figure 5). In their design, Ru5P must be produced from the non-oxidative pentose phosphate route in order to reoxidize all biosynthetic NADH and not NADPH through a transhydrogenase-type conversion NADPH-> NADH (Guadalupe-Medina et al., 2013). This clever idea was demonstrated in two elegant papers published in Biotechnology for Biofuels (Guadalupe-Medina et al., 2013; Papapetridis et al., 2018). In the first paper, the authors expressed the codon-optimized prokaryotic form II RuBisCO-encoding cbbM gene from T. denitrificans in a centromeric plasmid under the strong TDH3 promoter and showed that RuBisCO was functional only upon co-expression of E. coli groEL/groES encoding chaperones whereas surprisingly T denitrificans chaperones encoded by cbb02/cbbQ2 were ineffective. The PRK from Spinachia oleracea was integrated together with the E. coli groEL/groES gene in the yeast genome at the CAN1 locus under the galactose-inducible GAL1 promoter. Remarkably, expression of this minimal Calvin cycle in this engineered yeast cultivated under anaerobic chemostat condition showed a 68% reduction of glycerol production accompanied by a 10% increase of ethanol yield. Saturating the culture with CO2 resulted in a 90% decrease of glycerol, which could be explained in part by the low affinity of form II RuBisCO for CO2 (KC02 = 0.26 mM). In a second paper, the authors carried out several targeted metabolic engineering to optimize the fermentation kinetic, improving ethanol yield and reducing glycerol production. To achieve this goal, nine copies of the cbbM overexpression cassette along with a single expression cassette of E. coli groEL/groES chaperones were integrated at the SGA1 locus using CRISPR-Cas9 tool followed by the integration at X-2 locus of the PRK1 under the inducible anaerobic DAN1 promoter. The resulting engineered strain expressing a more robust Calvin-Benson cycle showed significant (31%) reduction of glycerol under glucose anaerobic condition. However, the contribution of the PRK/RuBisCO pathway to NADH reoxidation decreased with increased growth rate, with a concurrent higher contribution of the glycerol pathway. Therefore, a complementary strategy was to abrogate this pathway by removal of glycerol-3-P dehydrogenase encoded by GPD1 and GPD2. While GPD1 is essential for growth of Saccharomyces cerevisiae under osmotic stress (Albertyn et al., 1994), loss of GPD2 strongly affects growth under anaerobic condition (Ansell et al., 1997). Accordingly, the deletion of GPD2 in the engineered strain bearing the Calvin cycle resulted in a 62 % decrease of glycerol and 13% increase of ethanol yield as compared to a reference strain. An additional engineering strategy was dedicated to overexpress genes encoding the transketolase and transaldolase with the aim to increase the availability of Ru5P. Indeed, this intermediate is not solely the substrate of PRK, but it is also indirectly implicated in the biosynthesis of nucleic acid, aromatic amino acids and vitamins. Thus, its depletion through PRK could be in part responsible for the poor growth of the strain expressing the Calvin cycle. In accordance with this idea, overexpression of non-oxidative PPP genes RPEI, TKL1, TKL2, TAL1, RKI1 in the engineered strain also deleted for GPD2 resulted in a specific growth rate that was virtually the same as the reference strain under anaerobic batch condition on glucose. Furthermore, this engineered strain achieved the best fermentation performance with an increased ethanol yield of 15% and a 90% decrease of glycerol production (Papapetridis et al., 2018).
A comparable strategy has been applied by two other research groups which have equipped a xylose-utilizing Saccharomyces cerevisiae with the two Calvin-cycle enzymes PRK and RuBisCO to enhance xylose fermentation with in situ CO2 fixation (Li et al., 2017; Xia et al., 2017). As S. cerevisiae cannot naturally ferment xylose, fungal pathway consisting of a xylose reductase (XR) and xylulose dehydrogenase (XDH) or the bacterial xylose isomerase (XI) have been heterologously expressed, together with upregulation of the native pentose phosphate pathway (PPP) to achieve efficient and rapid xylose fermentation (Matsushika et al., 2009; Young et al., 2010; Oreb et al., 2012). While the XR/XDH pathway appears to be more effective for xylose fermentation rate and ethanol productivity than the XI pathway (Karhumaa et al., 2007), it leads to cofactor imbalance with a surplus of NADH that is regenerated at the cost of by-products formation such as glycerol and xylitol. Expression of the PRK-RuBisCO module should exploit this NADH surplus for recycling CO2 that is generated during the fermentation. This strategy was developed by Xia et al. (2017) which used a previously evolutionary engineered S. cerevisiae SR8 strain that was able to efficiently co-ferment xylose and glucose. This engineered strain harbored the XR/XDH together with an upregulated PPP and was deleted for PHO13 and ALD6 as beneficial targets for xylose fermentation (Kim et al., 2013). Similarly to the work of Guadalupe-Medina et al. (2013), two copies of the cbbM gene encoding form-II RuBisCO from R. rubrum under the strong TDH3 promoter and groEL/groES cassette under TEF1 promoter were integrated in the yeast genome (notably at ALD6 and PHO13 locus, respectively) leading to a stable platform SR8C+ strain. Only RuBisCO activity was detected in yeast strain co-expressing cbbM and groEL/ES. Then, while the overexpression of PRK from Spinacia oleracea alone was toxic for growth on xylose but not on glucose, likely because of ATP depletion or consecutive hyperaccumulation of RuBP, expression of the PRK-RuBisCO module rescued complete growth on xylose, which was accompanied by a 24% reduction of xylitol and 10% increase of ethanol yield while glycerol yield was similar to the control strain. Moreover, it was demonstrated that this engineered S. cerevisiae carrying the Calvin-cycle enzymes released 7% less CO2 than the control strain during anaerobic xylose fermentation. In a more recent publication, Li et al. (2017) engineered a xylose-utilizing S. cerevisiae for co-utilization of maltose, xylose and CO2. The rationale behind this strategy was to provide metabolite precursors, energy and reduced cofactor useful for xylose reduction by XR from maltose, which otherwise does not repress xylose fermentation. In addition, a mXR variant that preferred NADH over NADPH was co-expressed together with XR and XDH on a high copy plasmid pRS425. At variance to previous works, genes encoding form-I RuBisCO of Ralstonia eutropha or form-II of R. rubrum, PRK from Spinacia oleracea or Ralstonia eutrophia together with E. coli groEL and groES encoding chaperone were cloned in a low copy plasmid YcpLac33 and transformed into the xylose-utilizing yeast. They reported that these engineered strains grew better on a mix maltose-xylose medium than control strain. They exhibited higher ethanol productivity and yield, with the highest increase of ethanol yield (+ 15%) and sugar consumption rate (+ 63%) with the engineered strain expressing heterotrophic form-I RuBisCO. Using 13C-labeled CO2 and a metabolic flux index MFI h−C02 (Gong et al., 2015) for relative quantification of flux ratio between the CO2-fixing by-pass pathway and the central carbon metabolic pathway, an incorporation of 8% CO2 into Ru5P was estimated at a rate of 390 ± 50 mg CO2/L/h, which is a rate that is in the range of CO2 fixation by natural autotrophic microbes (Gong et al., 2015). Altogether, these results indicated that the beneficial effect of implementing the CO2-fixation pathway on growth and fermentation largely overcome the additional cost of ATP that is required when this pathway is expressed.
The RuBisCO-based pathway was also investigated in E. coli initially as a screen to find out variants of RuBisCO enzyme with higher catalytic activity and better affinity to CO2 (Parikh et al., 2006). It was then exploited for in situ CO2 assimilation in bioproduction (Zhuang and Li, 2013; Gong et al., 2015). For this purpose, the RuBisCO-encoding genes rbcl-rbcX-rbcS from the cyanobacterium Synechococcus sp PC7002 and PRK-encoding gene from Synechococcus elongatus were expressed on a high copy plasmid pET30a and arabinose was provided to the medium as an extra carbon source to enhance C5-phosphorylated sugar required for RuBisCO enzyme. Under this condition, the authors reported that the expression of RuBisCO alone promoted faster and complete consumption of arabinose, whereas the overexpression of PRK alone caused some growth retardation, which was explained by accumulation of RuBP. It was shown that the engineered strain expressing the Calvin-cycle was able to assimilate under anaerobic growth condition CO2 at a rate comparable to that of the autotrophic cyanobacteria and microalgae (Gong et al., 2015). However, to overcome the co-cultivation condition, the same authors decided to enhance the non-oxidative pentose phosphate pathway by either overexpressing transketolase (tktAB) or by deleting glucose-6-P dehydrogenase (zwf) encoding gene. These genetic actions resulted in a significant decrease of CO2 yield per glucose consumed. Genes encoding D-lactate dehydrogenase (ldh) and fumarate reductase (frd) were further deteled in this zwf mutant in order to favor recycling of in situ CO2 into fermentation products ethanol or acetate. However, while this reduced CO2 production proved the Calvin cycle pathway to work in vivo, and the expression of this cycle did not impede the growth on glucose, only 70% of carbon initially fed in the medium was recovered at the end of the fermentation. Moreover, there was no significant increase in C2-related compounds (Li et al., 2015; Yang et al., 2016). It turned out that this fermentation profile was explained by a huge accumulation of pyruvate that likely resulted from an excessive glycolytic flux at the sugar uptake and phosphorylation step leading to accumulation of NADH and concurrent reduction of pyruvate-formate lyase activity. This activity was inhibited by two concurrent actions, namely a direct inhibition by NADH and a downregulation of pflB encoding this enzyme due to the repression of its transcriptional activator encoded by arcA (Yang et al., 2016). Intriguingly, these transcriptional effects were already observed upon the simple overexpression of RuBisCO, suggesting a direct impact of the protein on the central carbon metabolism. To overcome these inhibitory problems, pyruvate decarboxylase and alcohol dehydrogenase encoding genes from Zymomonas mobilis were overexpressed in the Δzwf Δldh Δfrbd mutant strain equipped with the Calvin-cycle enzymes PRK and RuBisCO. This genetic intervention resulted in a yield of C-2 compounds from glucose 20% higher than that of theoretical fermentation value, which was due to a direct in situ CO2 recycling into these products by the RuBisCO system. In addition, a rate of 53 mg/L/h of CO2 consumption was evaluated, which was in the range of rate measured for CO2 fixation by microalgae (Gonzalez Lopez et al., 2009; Ho et al., 2012), arguing that mixotrophic fermentation can be a competitive alternative for reducing loss of CO2 and converting it into bio-based products.
The carbon management during the metabolic conversion of renewable carbon sources such as sugars by heterotrophic organisms is an attractive, yet partial, solution to combat the acute problem of continuous CO2 emission associated with human activities. Since carbon dioxide is an intrinsic by-product of carbon metabolism ensuring the irreversibility of metabolic pathways in which it is involved, the rewiring of carbon metabolism aiming to circumvent this carbon loss must take into account this cost. However, in most of the case that were presented here, this cost can be overridden by the benefit of a higher carbon yield achieved by these refractory or synthetic pathways. This higher carbon yield is the second advantage of this carbon management since it can in principle increase the practical performance of a product per carbon consumed, which is a decisive parameter for the economic evaluation of a biotechnology process. Nonetheless, the synthetic pathways aiming at reducing carbon loss or assimilate CO2 are still in their infancy. Substantial work is required to evaluate the potential for industrial application of some of them, such as NOG, MCG, rGS or glycoptimus, whereas others such as the MOG, MSer, and HOB will likely remain as elegant intellectual models illustrating the power of Synthetic Biology in its ability to reformat metabolic pathways.
While heterotrophic organisms are not naturally able to assimilate carbon dioxide, an attractive and complementary strategy to those developed elsewhere to capture CO2 is to equip these organisms with the two key Calvin cycle enzymes in order to capture CO2 produced during sugar fermentation, and meanwhile use it as an electron acceptor for NADH reoxidation. This dual function has been developed in yeast and E. coli and was shown to increase C2- products such as ethanol (Papapetridis et al., 2018; Tseng et al., 2018). The high concentration of carbon dioxide that prevails in industrial fermentations and their usual anaerobic to micro-aeration conditions are factors that should support the proper functioning of this capture system since RuBisCO has low affinity to CO2 and its carboxylation activity is antagonized by oxygen. Therefore, this engineering strategy could be implemented without major investment at the industrial scale owing to the fact that GMO's legislation is modified.
Another promising strategy that is already attracting much interest is to exploit the high catalytic capacity of carboxylating enzymes such as the PEP carboxylase to capture “CO2” in vivo. In addition of engineering specific pathways allowing this capture such as rGS or MCG, a great advantage of using heterotrophic organisms such as yeast or E. coli for CO2 fixation is their fast growth rates and capability to reach high cell density in bioreactors, both criteria contributing to CO2 assimilation rate that are higher than that of auxotrophic organisms. Therefore, engineering heterotrophic microbes for CO2 fixation can be a promising avenue to both reduce CO2 emission and in the meantime increase carbon yield as the fixed CO2 can be easily integrated into the central carbon metabolism. To be even more efficient, the next step will be to employ other energy source than sugar for CO2 fixation.
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors wish to acknowledge Agence Nationale de la Recherche (ANR), Ministry of Research and Education (MENRT), and TWB for technical and financial support.
Akram, M. (2014). Citric acid cycle and role of its intermediates in metabolism. Cell Biochem. Biophys. 68, 475–478. doi: 10.1007/s12013-013-9750-1
Albertyn, J., Hohmann, S., Thevelein, J. M., and Prior, B. A. (1994). GPD1, which encodes glycerol-3-phosphate dehydrogenase, is essential for growth under osmotic stress in Saccharomyces cerevisiae, and its expression is regulated by the high-osmolarity glycerol response pathway. Mol. Cell. Biol. 14, 4135–4144. doi: 10.1128/MCB.14.6.4135
Ansell, R., Granath, K., Hohmann, S., Thevelein, J. M., and Adler, L. (1997). The two isoenzymes for yeast NAD+-dependent glycerol 3-phosphate dehydrogenase encoded by GPD1 and GPD2 have distinct roles in osmoadaptation and redox regulation. EMBO J 16, 2179–2187.
Antonovsky, N., Gleizer, S., Noor, E., Zohar, Y., Herz, E., Barenholz, U., et al. (2016). Sugar synthesis from CO2 in Escherichia coli. Cell 166, 115–125. doi: 10.1016/j.cell.2016.05.064
Bar-Even, A., Noor, E., Lewis, N. E., and Milo, R. (2010). Design and analysis of synthetic carbon fixation pathways. Proc. Natl. Acad. Sci. U.S.A. 107, 8889–8894. doi: 10.1073/pnas.0907176107
Berg, I. A., Kockelkorn, D., Buckel, W., and Fuchs, G. (2007). A 3-hydroxypropionate/4-hydroxybutyrate autotrophic carbon dioxide assimilation pathway in Archaea. Science 318, 1782–1786. doi: 10.1126/science.1149976
Berg, I. A., Kockelkorn, D., Ramos-Vera, W. H., Say, R. F., Zarzycki, J., Hugler, M., et al. (2010). Autotrophic carbon fixation in archaea. Nat. Rev. Microbiol. 8, 447–460. doi: 10.1038/nrmicro2365
Bogorad, I. W., Chen, C. T., Theisen, M. K., Wu, T. Y., Schlenz, A. R., Lam, A. T., et al. (2014). Building carbon-carbon bonds using a biocatalytic methanol condensation cycle. Proc. Natl. Acad. Sci. U.S.A. 111, 15928–15933. doi: 10.1073/pnas.1413470111
Bogorad, I. W., Lin, T. S., and Liao, J. C. (2013). Synthetic non-oxidative glycolysis enables complete carbon conservation. Nature 502, 693–697. doi: 10.1038/nature12575
Bouzon, M., Perret, A., Loreau, O., Delmas, V., Perchat, N., Weissenbach, J., et al. (2017). A synthetic alternative to canonical one-carbon metabolism. ACS Synth. Biol. 6, 1520–1533. doi: 10.1021/acssynbio.7b00029
Caballero, E., Baldoma, L., Ros, J., Boronat, A., and Aguilar, J. (1983). Identification of lactaldehyde dehydrogenase and glycolaldehyde dehydrogenase as functions of the same protein in Escherichia coli. J. Biol. Chem. 258, 7788–7792.
Calvin, M. (1962). The path of carbon in photosynthesis. Science 135, 879–889. doi: 10.1126/science.135.3507.879
Chen, R., and Dou, J. (2016). Biofuels and bio-based chemicals from lignocellulose: metabolic engineering strategies in strain development. Biotechnol. Lett. 38, 213–221. doi: 10.1007/s10529-015-1976-0
Claassens, N. J. (2017). A warm welcome for alternative CO2 fixation pathways in microbial biotechnology. Microb. Biotechnol. 10, 31–34. doi: 10.1111/1751-7915.12456
Clark, J. H., Luque, R., and Matharu, A. S. (2012). Green chemistry, biofuels, and biorefinery. Annu. Rev. Chem. Biomol. Eng. 3, 183–207. doi: 10.1146/annurev-chembioeng-062011-081014
Clasquin, M. F., Melamud, E., Singer, A., Gooding, J. R., Xu, X., Dong, A., et al. (2011). Riboneogenesis in yeast. Cell 145, 969–980. doi: 10.1016/j.cell.2011.05.022
Deng, Y., Ma, N., Zhu, K., Mao, Y., Wei, X., and Zhao, Y. (2018). Balancing the carbon flux distributions between the TCA cycle and glyoxylate shunt to produce glycolate at high yield and titer in Escherichia coli. Metab. Eng. 46, 28–34. doi: 10.1016/j.ymben.2018.02.008
Dolan, S. K., and Welch, M. (2018). The glyoxylate shunt, 60 years on. Annu. Rev. Microbiol. 72, 309–330. doi: 10.1146/annurev-micro-090817-062257
Donahue, J. L., Bownas, J. L., Niehaus, W. G., and Larson, T. J. (2000). Purification and characterization of glpX-encoded fructose 1, 6-bisphosphatase, a new enzyme of the glycerol 3-phosphate regulon of Escherichia coli. J. Bacteriol. 182, 5624–5627. doi: 10.1128/JB.182.19.5624-5627.2000
Dudley, Q. M., Karim, A. S., and Jewett, M. C. (2015). Cell-free metabolic engineering: biomanufacturing beyond the cell. Biotechnol. J. 10, 69–82. doi: 10.1002/biot.201400330
Dugar, D., and Stephanopoulos, G. (2011). Relative potential of biosynthetic pathways for biofuels and bio-based products. Nat. Biotechnol. 29, 1074–1078. doi: 10.1038/nbt.2055
Duwe, A., Tippkotter, N., and Ulber, R. (2019). Lignocellulose-biorefinery: ethanol-focused. Adv. Biochem. Eng. Biotechnol. 166, 177–215. doi: 10.1007/10_2016_72
Eisenhut, M., Kahlon, S., Hasse, D., Ewald, R., Lieman-Hurwitz, J., Ogawa, T., et al. (2006). The plant-like C2 glycolate cycle and the bacterial-like glycerate pathway cooperate in phosphoglycolate metabolism in cyanobacteria. Plant Physiol. 142, 333–342. doi: 10.1104/pp.106.082982
Erb, T. J., and Zarzycki, J. (2018). A short history of RubisCO: the rise and fall (?) of Nature's predominant CO2 fixing enzyme. Curr. Opin. Biotechnol. 49, 100–107. doi: 10.1016/j.copbio.2017.07.017
Fatland, B., Anderson, M., Nikolau, B. J., and Wurtele, E. S. (2000). Molecular biology of cytosolic acetyl-CoA generation. Biochem. Soc. Trans. 28, 593–595. doi: 10.1042/bst0280593
Fei, Q., Guarnieri, M. T., Tao, L., Laurens, L. M., Dowe, N., and Pienkos, P. T. (2014). Bioconversion of natural gas to liquid fuel: opportunities and challenges. Biotechnol. Adv. 32, 596–614. doi: 10.1016/j.biotechadv.2014.03.011
Fuchs, G., and Berg, I. A. (2014). Unfamiliar metabolic links in the central carbon metabolism. J. Biotechnol. 192(Pt B), 314–322. doi: 10.1016/j.jbiotec.2014.02.015
Garrabou, X., Castillo, J. A., Guerard-Helaine, C., Parella, T., Joglar, J., Lemaire, M., et al. (2009). Asymmetric self- and cross-aldol reactions of glycolaldehyde catalyzed by D-fructose-6-phosphate aldolase. Angew. Chem. Int. Ed. Engl. 48, 5521–5525. doi: 10.1002/anie.200902065
Gong, F., Liu, G., Zhai, X., Zhou, J., Cai, Z., and Li, Y. (2015). Quantitative analysis of an engineered CO2-fixing Escherichia coli reveals great potential of heterotrophic CO2 fixation. Biotechnol. Biofuels 8:86. doi: 10.1186/s13068-015-0268-1
Gonzalez Lopez, C. V., Acien Fernandez, F. G., Fernandez Sevilla, J. M., Sanchez Fernandez, J. F., Ceron Garcia, M. C., and Molina Grima, E. (2009). Utilization of the cyanobacteria Anabaena sp. ATCC 33047 in CO2 removal processes. Bioresour. Technol. 100, 5904–5910. doi: 10.1016/j.biortech.2009.04.070
Guadalupe-Medina, V., Wisselink, H. W., Luttik, M. A., De Hulster, E., Daran, J. M., Pronk, J. T., et al. (2013). Carbon dioxide fixation by Calvin-Cycle enzymes improves ethanol yield in yeast. Biotechnol. Biofuels 6:125. doi: 10.1186/1754-6834-6-125
Ho, S. H., Chen, C. Y., and Chang, J. S. (2012). Effect of light intensity and nitrogen starvation on CO2 fixation and lipid/carbohydrate production of an indigenous microalga Scenedesmus obliquus CNW-N. Bioresour. Technol. 113, 244–252. doi: 10.1016/j.biortech.2011.11.133
Igamberdiev, A. U., and Kleczkowski, L. A. (2018). The glycerate and phosphorylated pathways of serine synthesis in plants: the branches of plant glycolysis linking carbon and nitrogen metabolism. Front. Plant Sci. 9:318. doi: 10.3389/fpls.2018.00318
Jang, S., Kim, M., Hwang, J., and Jung, G. Y. (2019). Tools and systems for evolutionary engineering of biomolecules and microorganisms. J. Ind. Microbiol. Biotechnol. 46, 1313–1326. doi: 10.1007/s10295-019-02191-5
Karhumaa, K., Garcia Sanchez, R., Hahn-Hagerdal, B., and Gorwa-Grauslund, M. F. (2007). Comparison of the xylose reductase-xylitol dehydrogenase and the xylose isomerase pathways for xylose fermentation by recombinant Saccharomyces cerevisiae. Microb. Cell Fact. 6:5. doi: 10.1186/1475-2859-6-5
Kim, P., Laivenieks, M., Vieille, C., and Zeikus, J. G. (2004). Effect of overexpression of Actinobacillus succinogenes phosphoenolpyruvate carboxykinase on succinate production in Escherichia coli. Appl. Environ. Microbiol. 70, 1238–1241. doi: 10.1128/AEM.70.2.1238-1241.2004
Kim, S. R., Skerker, J. M., Kang, W., Lesmana, A., Wei, N., Arkin, A. P., et al. (2013). Rational and evolutionary engineering approaches uncover a small set of genetic changes efficient for rapid xylose fermentation in Saccharomyces cerevisiae. PLoS ONE 8:e57048. doi: 10.1371/journal.pone.0057048
Kondo, T., Tezuka, H., Ishii, J., Matsuda, F., Ogino, C., and Kondo, A. (2012). Genetic engineering to enhance the Ehrlich pathway and alter carbon flux for increased isobutanol production from glucose by Saccharomyces cerevisiae. J. Biotechnol. 159, 32–37. doi: 10.1016/j.jbiotec.2012.01.022
Krebs, H. A., and Johnson, W. A. (1980). The role of citric acid in intermediate metabolism in animal tissues. FEBS Lett. 117 (Suppl), K1–10. doi: 10.1016/0014-5793(80)80564-3
Lachaux, C., Frazao, C. R., Krauser, F., Morin, N., Walther, T., and Francois, J. (2019). A new synthetic pathway for the bioproduction of glycolic acid from lignocellulosic sugars aimed at maximal carbon conservation Front. Bioeng. Biotechnol. 7:359. doi: 10.3389/fbioe.2019.00359
Leduc, Y. A., Prasad, L., Laivenieks, M., Zeikus, J. G., and Delbaere, L. T. (2005). Structure of PEP carboxykinase from the succinate-producing Actinobacillus succinogenes: a new conserved active-site motif. Acta Crystallogr. D Biol. Crystallogr. 61, 903–912. doi: 10.1107/S0907444905008723
Li, Y. H., Ou-Yang, F. Y., Yang, C. H., and Li, S. Y. (2015). The coupling of glycolysis and the Rubisco-based pathway through the non-oxidative pentose phosphate pathway to achieve low carbon dioxide emission fermentation. Bioresour. Technol. 187, 189–197. doi: 10.1016/j.biortech.2015.03.090
Li, Y. J., Wang, M. M., Chen, Y. W., Wang, M., Fan, L. H., and Tan, T. W. (2017). Engineered yeast with a CO2-fixation pathway to improve the bio-ethanol production from xylose-mixed sugars. Sci. Rep. 7:43875. doi: 10.1038/srep43875
Lim, R., and Cohen, S. S. (1966). D-phosphoarabinoisomerase and D-ribulokinase in Escherichia coli. J. Biol. Chem. 241, 4304–4315.
Lin, E. C., and Iuchi, S. (1991). Regulation of gene expression in fermentative and respiratory systems in Escherichia coli and related bacteria. Annu. Rev. Genet. 25, 361–387. doi: 10.1146/annurev.ge.25.120191.002045
Lin, P. P., Jaeger, A. J., Wu, T. Y., Xu, S. C., Lee, A. S., Gao, F., et al. (2018). Construction and evolution of an Escherichia coli strain relying on non-oxidative glycolysis for sugar catabolism. Proc. Natl. Acad. Sci. U.S.A. 115, 3538–3546. doi: 10.1073/pnas.1802191115
Liu, X., Sheng, J., and Curtiss, R III. (2011). Fatty acid production in genetically modified cyanobacteria. Proc. Natl. Acad. Sci. U.S.A. 108, 6899–6904. doi: 10.1073/pnas.1103014108
Mainguet, S. E., Gronenberg, L. S., Wong, S. S., and Liao, J. C. (2013). A reverse glyoxylate shunt to build a non-native route from C4 to C2 in Escherichia coli. Metab. Eng. 19, 116–127. doi: 10.1016/j.ymben.2013.06.004
Marliere, P., Patrouix, J., Doring, V., Herdewijn, P., Tricot, S., Cruveiller, S., et al. (2011). Chemical evolution of a bacterium's genome. Angew. Chem. Int. Ed Engl. 50, 7109–7114. doi: 10.1002/anie.201100535
Matsushika, A., Inoue, H., Kodaki, T., and Sawayama, S. (2009). Ethanol production from xylose in engineered Saccharomyces cerevisiae strains: current state and perspectives. Appl. Microbiol. Biotechnol. 84, 37–53. doi: 10.1007/s00253-009-2101-x
Mattanovich, D., Sauer, M., and Gasser, B. (2014). Yeast biotechnology: teaching the old dog new tricks. Microb. Cell Fact. 13:34. doi: 10.1186/1475-2859-13-34
Muller, J. E., Meyer, F., Litsanov, B., Kiefer, P., and Vorholt, J. A. (2015). Core pathways operating during methylotrophy of Bacillus methanolicus MGA3 and induction of a bacillithiol-dependent detoxification pathway upon formaldehyde stress. Mol. Microbiol. 98, 1089–1100. doi: 10.1111/mmi.13200
Opgenorth, P. H., Korman, T. P., and Bowie, J. U. (2016). A synthetic biochemistry module for production of bio-based chemicals from glucose. Nat. Chem. Biol. 12, 393–395. doi: 10.1038/nchembio.2062
Oreb, M., Dietz, H., Farwick, A., and Boles, E. (2012). Novel strategies to improve co-fermentation of pentoses with D-glucose by recombinant yeast strains in lignocellulosic hydrolysates. Bioengineered 3, 347–351. doi: 10.4161/bioe.21444
Papapetridis, I., Goudriaan, M., Vazquez Vitali, M., De Keijzer, N. A., Van Den Broek, M., Van Maris, A. J. A., et al. (2018). Optimizing anaerobic growth rate and fermentation kinetics in Saccharomyces cerevisiae strains expressing Calvin-cycle enzymes for improved ethanol yield. Biotechnol. Biofuels 11:17. doi: 10.1186/s13068-017-1001-z
Parikh, M. R., Greene, D. N., Woods, K. K., and Matsumura, I. (2006). Directed evolution of RuBisCO hypermorphs through genetic selection in engineered E. coli. Protein Eng. Des. Sel. 19, 113–119. doi: 10.1093/protein/gzj010
Reizer, J., Reizer, A., and Saier, M. H. Jr. (1995). Novel phosphotransferase system genes revealed by bacterial genome analysis–a gene cluster encoding a unique Enzyme I and the proteins of a fructose-like permease system. Microbiology 141(Pt 4), 961–971. doi: 10.1099/13500872-141-4-961
Robles-Rodriguez, C. E., Munoz-Tamayo, R., Bideaux, C., Gorret, N., Guillouet, S. E., Molina-Jouve, C., et al. (2018). Modeling and optimization of lipid accumulation by Yarrowia lipolytica from glucose under nitrogen depletion conditions. Biotechnol. Bioeng. 115, 1137–1151. doi: 10.1002/bit.26537
Rosales-Calderon, O., and Arantes, V. (2019). A review on commercial-scale high-value products that can be produced alongside cellulosic ethanol. Biotechnol. Biofuels 12:240. doi: 10.1186/s13068-019-1529-1
Russmayer, H., Buchetics, M., Gruber, C., Valli, M., Grillitsch, K., Modarres, G., et al. (2015). Systems-level organization of yeast methylotrophic lifestyle. BMC Biol. 13:80. doi: 10.1186/s12915-015-0186-5
Scaife, M. A., Nguyen, G. T., Rico, J., Lambert, D., Helliwell, K. E., and Smith, A. G. (2015). Establishing Chlamydomonas reinhardtii as an industrial biotechnology host. Plant J. 82, 532–546. doi: 10.1111/tpj.12781
Schiel-Bengelsdorf, B., and Durre, P. (2012). Pathway engineering and synthetic biology using acetogens. FEBS Lett. 586, 2191–2198. doi: 10.1016/j.febslet.2012.04.043
Schurmann, M., and Sprenger, G. A. (2001). Fructose-6-phosphate aldolase is a novel class I aldolase from Escherichia coli and is related to a novel group of bacterial transaldolases. J. Biol. Chem. 276, 11055–11061. doi: 10.1074/jbc.M008061200
Sedivy, J. M., Daldal, F., and Fraenkel, D. G. (1984). Fructose bisphosphatase of Escherichia coli: cloning of the structural gene (fbp) and preparation of a chromosomal deletion. J. Bacteriol. 158, 1048–1053.
Singhvi, M. S., and Gokhale, D. V. (2019). Lignocellulosic biomass: hurdles and challenges in its valorization. Appl. Microbiol. Biotechnol. 103, 9305–9320. doi: 10.1007/s00253-019-10212-7
Smejkalova, H., Erb, T. J., and Fuchs, G. (2010). Methanol assimilation in Methylobacterium extorquens AM1: demonstration of all enzymes and their regulation. PLoS ONE 5:e13001. doi: 10.1371/journal.pone.0013001
Soucaille, P. (2007). Glycolic Acid Production by Fermentation from Renewable Resources. France Patent Application: WO/2007/141316.
Stolzenberger, J., Lindner, S. N., Persicke, M., Brautaset, T., and Wendisch, V. F. (2013). Characterization of fructose 1,6-bisphosphatase and sedoheptulose 1,7-bisphosphatase from the facultative ribulose monophosphate cycle methylotroph Bacillus methanolicus. J. Bacteriol. 195, 5112–5122. doi: 10.1128/JB.00672-13
Tabata, K., and Hashimoto, S. (2004). Production of mevalonate by a metabolically-engineered Escherichia coli. Biotechnol. Lett. 26, 1487–1491. doi: 10.1023/B:BILE.0000044449.08268.7d
Tcherkez, G. G., Farquhar, G. D., and Andrews, T. J. (2006). Despite slow catalysis and confused substrate specificity, all ribulose bisphosphate carboxylases may be nearly perfectly optimized. Proc. Natl. Acad. Sci. U.S.A. 103, 7246–7251. doi: 10.1073/pnas.0600605103
Tittmann, K. (2014). Sweet siblings with different faces: the mechanisms of FBP and F6P aldolase, transaldolase, transketolase and phosphoketolase revisited in light of recent structural data. Bioorg. Chem. 57, 263–280. doi: 10.1016/j.bioorg.2014.09.001
Tseng, C. P. (1997). Regulation of fumarase (fumB) gene expression in Escherichia coli in response to oxygen, iron and heme availability: role of the arcA, fur, and hemA gene products. FEMS Microbiol. Lett. 157, 67–72. doi: 10.1111/j.1574-6968.1997.tb12754.x
Tseng, I. T., Chen, Y. L., Chen, C. H., Shen, Z. X., Yang, C. H., and Li, S. Y. (2018). Exceeding the theoretical fermentation yield in mixotrophic Rubisco-based engineered Escherichia coli. Metab. Eng. 47, 445–452. doi: 10.1016/j.ymben.2018.04.018
Uyeda, K. (1979). Phosphofructokinase. Adv. Enzymol. Relat. Areas Mol. Biol. 48, 193–244. doi: 10.1002/9780470122938.ch4
Van Dijken, J. P., and Scheffers, W. A. (1986). Redox balances in the metabolism of sugars by yeast. FEMS Microbiol. Rev. 32, 199–224. doi: 10.1111/j.1574-6968.1986.tb01194.x
Verschueren, K. H. G., Blanchet, C., Felix, J., Dansercoer, A., De Vos, D., Bloch, Y., et al. (2019). Structure of ATP citrate lyase and the origin of citrate synthase in the Krebs cycle. Nature 568, 571–575. doi: 10.1038/s41586-019-1095-5
Wang, Q., Xu, J., Sun, Z., Luan, Y., Li, Y., Wang, J., et al. (2019). Engineering an in vivo EP-bifido pathway in Escherichia coli for high-yield acetyl-CoA generation with low CO2 emission. Metab. Eng. 51, 79–87. doi: 10.1016/j.ymben.2018.08.003
Ward, N., Larsen, O., Sakwa, J., Bruseth, L., Khouri, H., Durkin, A. S., et al. (2004). Genomic insights into methanotrophy: the complete genome sequence of Methylococcus capsulatus (Bath). PLoS Biol. 2:e303. doi: 10.1371/journal.pbio.0020303
Xia, P. F., Zhang, G. C., Walker, B., Seo, S. O., Kwak, S., Liu, J. J., et al. (2017). Recycling carbon dioxide during xylose fermentation by engineered Saccharomyces cerevisiae. ACS Synth. Biol. 6, 276–283. doi: 10.1021/acssynbio.6b00167
Yang, C. H., Liu, E. J., Chen, Y. L., Ou-Yang, F. Y., and Li, S. Y. (2016). The comprehensive profile of fermentation products during in situ CO2 recycling by Rubisco-based engineered Escherichia coli. Microb. Cell Fact. 15:133. doi: 10.1186/s12934-016-0530-7
Yin, X., Chambers, J. R., Barlow, K., Park, A. S., and Wheatcroft, R. (2005). The gene encoding xylulose-5-phosphate/fructose-6-phosphate phosphoketolase (xfp) is conserved among Bifidobacterium species within a more variable region of the genome and both are useful for strain identification. FEMS Microbiol. Lett. 246, 251–257. doi: 10.1016/j.femsle.2005.04.013
Young, E., Lee, S. M., and Alper, H. (2010). Optimizing pentose utilization in yeast: the need for novel tools and approaches. Biotechnol. Biofuels 3:24. doi: 10.1186/1754-6834-3-24
Yu, H., and Liao, J. C. (2018). A modified serine cycle in Escherichia coli coverts methanol and CO2 to two-carbon compounds. Nat. Commun. 9:3992. doi: 10.1038/s41467-018-06496-4
Yurimoto, H., Hirai, R., Yasueda, H., Mitsui, R., Sakai, Y., and Kato, N. (2002). The ribulose monophosphate pathway operon encoding formaldehyde fixation in a thermotolerant methylotroph, Bacillus brevis S1. FEMS Microbiol. Lett. 214, 189–193. doi: 10.1111/j.1574-6968.2002.tb11345.x
Zhang, W., Zhang, T., Wu, S., Wu, A. L., Xin, F., Dong, W., et al. (2017). Guidance for engineering of synthetic methylotrophy based on methanol metabolism in methylotrophy. RSC Adv. 7, 4083–4091. doi: 10.1039/C6RA27038G
Keywords: microbial physiology, metabolic engineering, synthetic biology, carbon dioxide, bio-based products, chemicals
Citation: François JM, Lachaux C and Morin N (2020) Synthetic Biology Applied to Carbon Conservative and Carbon Dioxide Recycling Pathways. Front. Bioeng. Biotechnol. 7:446. doi: 10.3389/fbioe.2019.00446
Received: 27 August 2019; Accepted: 11 December 2019;
Published: 10 January 2020.
Edited by:
Jens O. Krömer, Helmholtz Centre for Environmental Research (UFZ), GermanyReviewed by:
S. Venkata Mohan, Indian Institute of Chemical Technology (CSIR), IndiaCopyright © 2020 François, Lachaux and Morin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jean Marie François, ZnJhbl9qbUBpbnNhLXRvdWxvdXNlLmZy; amVhbi5tYXJpZS1mcmFuY29pc0BpbnJhLmZy
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.