- 1Jiangxi Engineering Laboratory for the Development and Utilization of Agricultural Microbial Resources, College of Bioscience and Engineering, Jiangxi Agricultural University, Nanchang, China
- 2Laboratory of Biosystems and Microanalysis, State Key Laboratory of Bioreactor Engineering, East China University of Science and Technology, Shanghai, China
L-ornithine, a valuable non-protein amino acid, has a wide range of applications in the pharmaceutical and food industries. Currently, microbial fermentation is a promising, sustainable, and environment-friendly method to produce L-ornithine. However, the industrial production capacity of L-ornithine by microbial fermentation is low and rarely meets the market demands. Various strategies have been employed to improve the L-ornithine production titers in the model strain, Corynebacterium glutamicum, which serves as a major indicator for improving the cost-effectiveness of L-ornithine production by microbial fermentation. This review focuses on the development of high L-ornithine-producing strains by metabolic engineering and reviews the recent advances in breeding strategies, such as reducing by-product formation, improving the supplementation of precursor glutamate, releasing negative regulation and negative feedback inhibition, increasing the supply of intracellular cofactors, modulating the central metabolic pathway, enhancing the transport system, and adaptive evolution for improving L-ornithine production.
Introduction
The amino acid market is worth several billion dollars. Hence, the production of amino acids has been an active field of biotechnology research in recent years (Lee and Wendisch, 2017; Li et al., 2017; D'Este et al., 2018). Currently, most amino acids are produced by microbial fermentation, a technique adopted in Japan several decades ago. Microbial fermentation is an eco-friendly technology, which can address the growing concerns about environmental issues and can be used to establish a sustainable economy independent of fossil fuels (Becker et al., 2018; Kogure and Inui, 2018). L-ornithine, a non-protein amino acid, is widely used to improve human health as it is reported to have beneficial effects on the liver and the heart (Acharya et al., 2009; Rathi and Taneja, 2018; Butterworth and McPhail, 2019). L-ornithine is also produced by microbial fermentation using Escherichia coli, Saccharomyces cerevisiae, or Corynebacterium glutamicum as the microbial cell factory (Mitsuhashi, 2014; Becker and Wittmann, 2015). E. coli is a widely used microbe for the development of engineered strains to produce chemicals as it has several advantages, such as rapid propagation, availability of a completely sequenced genome, and ease of genetic manipulation (Sarria et al., 2017; Li et al., 2018). E. coli could be engineered for producing L-ornithine by rational modulation of the urea cycle and optimizing the fermentation process (Lee and Cho, 2006). However, E. coli synthesizes endotoxin, which is banned in the food and pharmaceutical industries (Okuda et al., 2016). This limits the application of E. coli as an ideal host to produce L-ornithine. S. cerevisiae, an alternative host, has been identified as a generally recognized as safe (GRAS) strain. S. cerevisiae exhibits strong robustness or tolerance to harsh growing conditions and is used for the production of various food and pharmaceutical compounds (Guo et al., 2018; Yu et al., 2018; Luo et al., 2019). Modular metabolic engineering strategies were used to engineer the carbon source transport system, central metabolic pathway, ammonia metabolism, energy supply, and transport of small molecular weight compounds in S. cerevisiae to produce L-ornithine (Qin et al., 2015). However, the production titer of L-ornithine was low when S. cerevisiae was used as a chassis microorganism and hence, the process could not be scaled up to industrial production. As shown in Table 1, C. glutamicum is predominantly used to produce L-ornithine. C. glutamicum, a gram positive bacterium, was intensively engineered by mutation breeding to enable utilization of a broad spectrum of carbon sources to produce desired chemical compounds (Jeandet et al., 2018; Becker and Wittmann, 2019; Kim et al., 2019). Recent developments in genetic manipulation tools have enabled further genetic modifications, which can further improve L-ornithine production in C. glutamicum by metabolic engineering. In this review, we have summarized the current advances in the metabolic engineering strategies for C. glutamicum. Comprehensive information on the modification of C. glutamicum metabolic pathways has been provided to improve the production of L-ornithine.
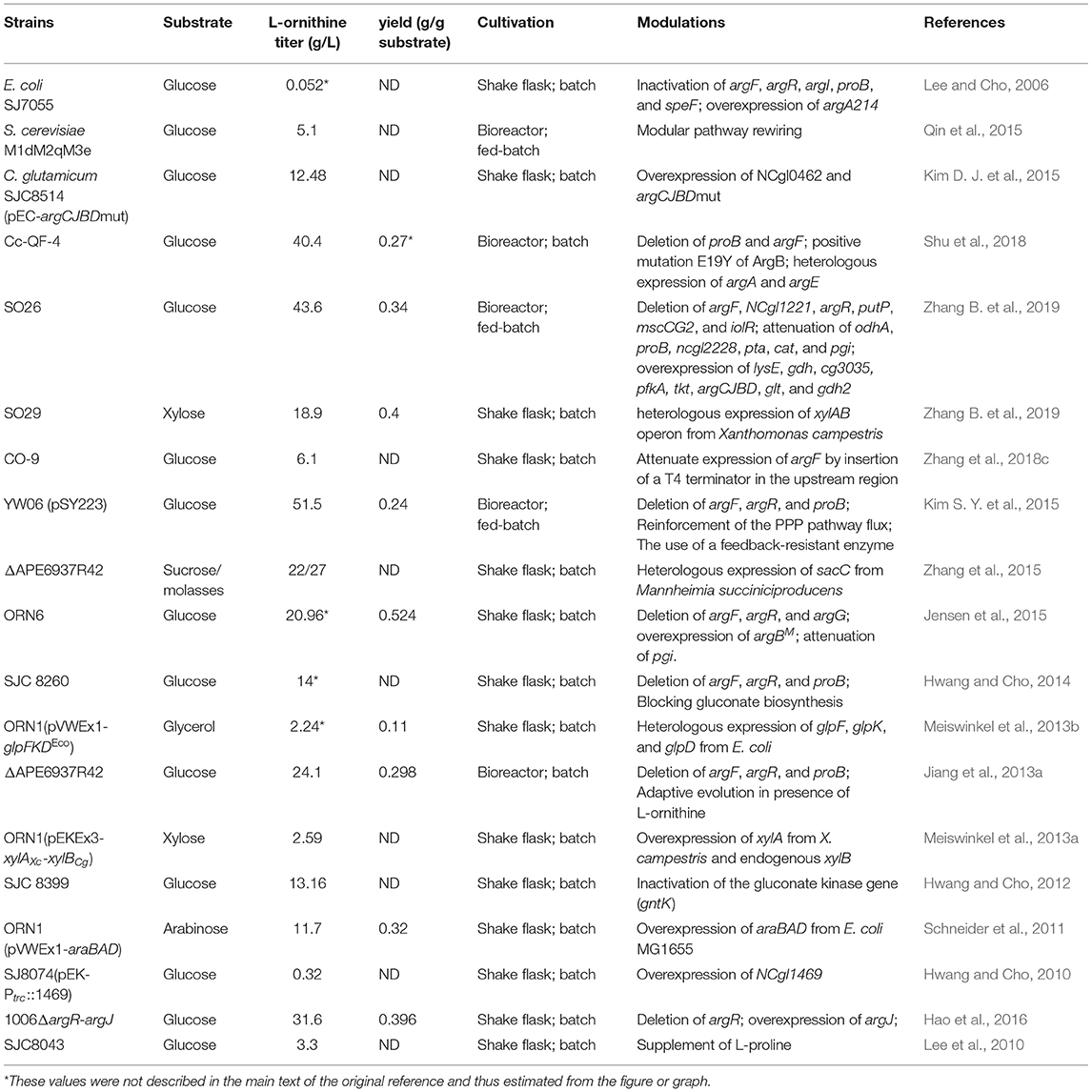
Table 1. Characteristic of L-ornithine producing strains developed by metabolic engineered strategies.
Main Biosynthesis Pathways Of L-Ornithine
In C. glutamicum, the biosynthesis of L-ornithine occurs through the urea cycle coupled with the tricarboxylic acid cycle. As described in Figure 1, α-oxoglutarate, an intermediate metabolite in the tricarboxylic acid cycle pathway, can be converted to glutamate by the reversible enzyme, glutamate dehydrogenase (encoded by gdh/NCgl0181). Further, glutamate is converted to L-ornithine through the enzymatic activities of N-acetylglutamate synthase (encoded by cg3035/NCgl2644), N-acetylglutamate kinase (encoded by argB/NCgl1342), N-acetyl-gamma-glutamyl-phosphate reductase (encoded by argC/NCgl1340), acetylornithine aminotransferase (encoded by argD/NCgl1343), and ornithine acetyltransferase (encoded by argJ/NCgl1341). Additionally, L-ornithine can be converted to L-citrulline and L-arginine through the enzymatic activities of ornithine carbamoyltransferase (encoded by argF/NCgl1344), argininosuccinate synthase (encoded by argG/NCgl1346), and argininosuccinate lyase (encoded by argH/NCgl1347). The genes encoding these enzymes are arranged in two operons, argCJBDFR and argGH. The two operons are repressed by a negative regulatory protein, ArgR/NCgl1345 (Ikeda et al., 2009; Stäbler et al., 2011). The generation of 1 mole of L-ornithine requires 2 moles of NADPH during the conversion of α-oxoglutarate to glutamate and N-acetyl-γ-glutamyl-phosphate to N-acetylglutamate semialdehyde. The enzyme activity of N-acetylglutamate kinase is regulated by L-arginine and L-citrulline via a feedback regulation mechanism. To select and engineer high L-ornithine-producing C. glutamicum strains, it is necessary to release the feedback regulation, improve supplementation of cofactor NADPH, and modulate metabolic pathways (Figure 2).
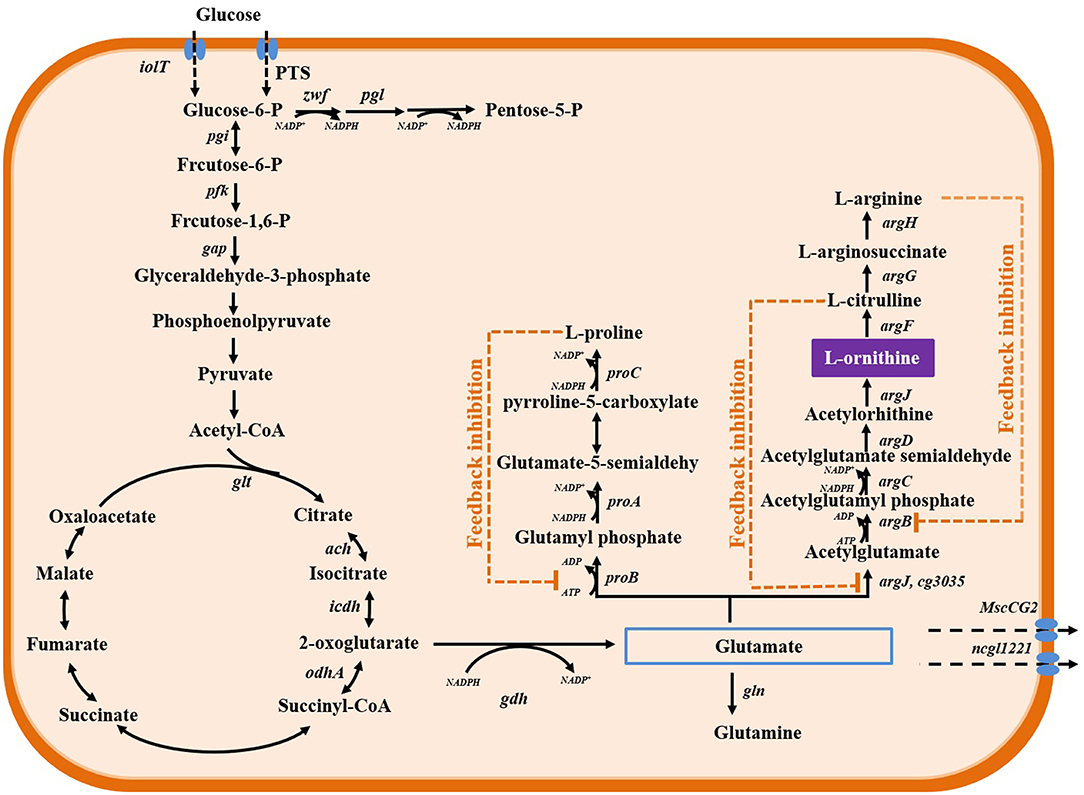
Figure 1. L-ornithine biosynthesis metabolic pathways in C. glutamicum. The genes annotation of enzymes as describe in previous study (Zhang B. et al., 2019). The dotted orange line represents feedback inhibition.
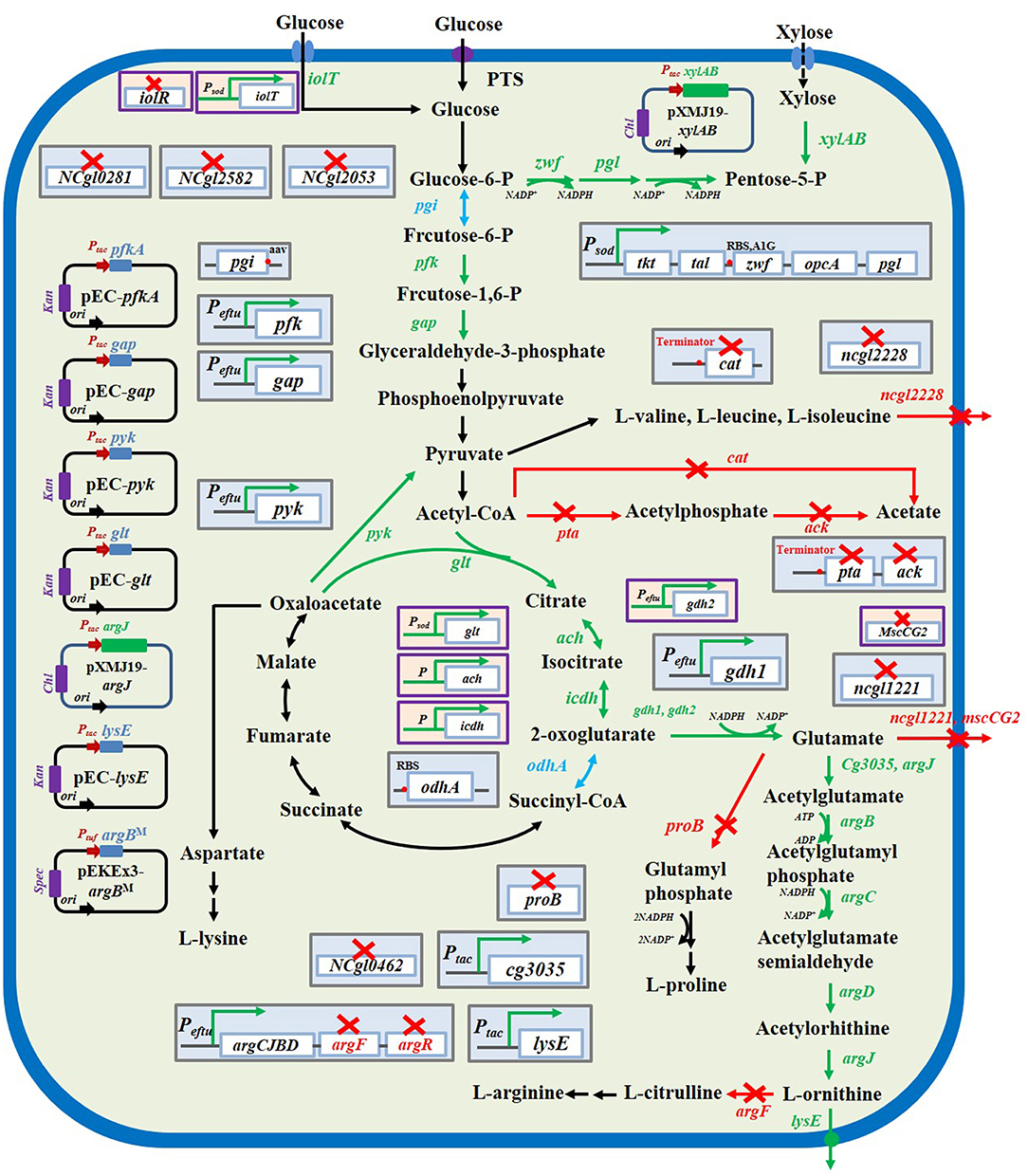
Figure 2. L-ornithine biosynthesis metabolic pathways in C. glutamicum and strategies to improve L-ornithine accumulation. The red× represented this pathway were inactivated. The blue font and arrows represented that pathways were attenuated. The green font and arrows indicated that pathways were overexpressed. The genes encoding enzymes involved in catalytically relevant reactions.
Blocking The BY-Product Pathways Via Gene Disruption
Under anaerobic conditions, C. glutamicum strains are reported to exhibit enhanced glutamate production and decreased L-ornithine production (Gourdon and Lindley, 1999; Lapujade et al., 1999; Hirasawa and Wachi, 2017; Tuyishime et al., 2018). L-ornithine, an intermediate compound in the L-arginine biosynthetic pathway, was synthesized using glutamate as a precursor, which was tightly regulated by feedback inhibition of the terminal metabolites, such as L-arginine and L-citrulline (Ikeda et al., 2009). The enzymatic activity of N-acetylglutamate kinase, which is involved in the main synthetic pathway of L-ornithine, was inhibited by L-arginine and L-citrulline at a concentration of <1 mM (Xu et al., 2012a,b). Thus, the biosynthetic pathways of L-arginine and L-citrulline not only consume L-ornithine, but also hinder its production. To promote the accumulation of L-ornithine, argF (encodes ornithine carbamoyltransferase) was disrupted to block the metabolic pathway that converts L-ornithine to L-citrulline and L-arginine. This markedly inhibited the production of L-citrulline and L-arginine in all the L-ornithine-producing C. glutamicum strains. However, the deletion of argF also resulted in a major hindrance in cell growth as the cells lacked the ability to synthesize arginine. To address this limitation, L-arginine was added to the fermentation medium to meet the demand for cell growth. However, the supplementation of L-arginine increases the cost and the operational complexity. Therefore, it is necessary to adopt appropriate strategies to attenuate the expression of argF. Metabolic engineering strategies, such as changing the ribosome binding site (Xiao et al., 2019), replacing the promoter (Xu et al., 2019), changing the translation initiation codon (Otten et al., 2015), and transcription interference by clustered regular interspaced short palindromic repeats (CRISPR) (Cleto et al., 2016; Yoon and Woo, 2018; Park et al., 2019) were successfully applied to optimize the flux of the metabolic pathway. The expression of argF was attenuated by changing the ribosome binding site and the translation initiation codon. It was difficult to attenuate the catabolism of L-ornithine owing to the enhanced transcription level of the argCJBDF operon. Additionally, the expression of argF was attenuated by introducing a strong terminator in the upstream region of argF. The L-ornithine titer in the engineered C. glutamicum was 6.1 g/L, which was 11-fold higher than that in the original strain and 42.8% higher than that in the mutant strain with argF deletion (Zhang et al., 2018c). It must be noted that insertion of a strong terminator in the upstream region of argF can not only attenuate the expression of argF, but also stimulate the expression of argCJBD gene clusters, which are required for the synthesis of ornithine. Therefore, attenuation of argF expression is a promising strategy for constructing an efficient L-ornithine-producing C. glutamicum.
Glutamate is also utilized to form L-proline via three steps of enzymatic reactions (Lee et al., 2010). The biosynthesis of a compete metabolite of L-proline not only consumes carbon sources, but also the NADPH cofactor, which limits L-ornithine production (Wendisch et al., 2016). In various studies, the proB/NCgl2274 (encodes γ-glutamyl kinase) gene has been deleted to construct high L-ornithine-producing strains by blocking the L-proline biosynthetic pathway. The engineered L-ornithine-producing strains that exhibit enhanced L-ornithine production include YW06 (pSY223) (Kim S. Y. et al., 2015) and Cc-QF-1 (Shu et al., 2018). However, the deletion of argF in C. glutamicum and inactivation of proB negatively affected cell growth due to L-proline deficiency. Thus, proline must be added to the culture medium to maintain normal cell growth. However, the addition of proline to the culture medium increases the operational cost and complexity. To address these limitations, recent studies have attenuated the expression of proB by inserting a terminator, which improved L-ornithine production without disturbing the cell growth (Zhang et al., 2018a). As suggested in the modulation of argF, attenuating the expression of proB by inserting a terminator is an efficient strategy for constructing a L-ornithine-producing C. glutamicum strain. In addition to the biosynthetic pathway of proline, the biosynthetic pathways of other amino acids, such as valine, isoleucine, and leucine also consume carbon sources, which limits L-ornithine production. Therefore, a recent study reported that deleting the ncgl2228 gene (encodes a putative branched amino acid transporter) in the engineered C. glutamicum orn8 strain decreased the yield of L-leucine (from 1.90 to 0.13 g/L), L-valine (from 2.16 to 0.22 g/L), and L-isoleucine (from 1.60 to 0.04 g/L) and increased the yield of L-ornithine (from 19 to 22 g/L) (Zhang et al., 2018a). Since the accumulation of by-products in the fermentation prevents the utilization of carbon source and cofactors for producing valuable compounds, it has been concluded that the inactivation of a corresponding transporter is an efficient strategy to reduce accumulation of by-products and increase production of the desired compound.
Removal Of Feedback Repression And Feedback Inhibition
In the C. glutamicum genome, the genes encoding the enzymes involved in the biosynthesis of L-ornithine are present in the argCJBD operon, which is repressed by the negative regulatory protein, ArgR. The expression of argCJBD operon is repressed at low level, which is an important rate-limiting factor for the biosynthesis of L-ornithine. Hence, the argR gene was deleted in all the engineered L-ornithine-producing C. glutamicum strains, which provided a powerful strategy for improving the L-ornithine production titer. Inactivation of argR in the C. glutamicum 1006 strain improved the yield of L-ornithine (from 20.5 to 28.3 g/L) and yield from 0.256 to 0.354 g/g glucose (Hao et al., 2016). However, the yield of L-ornithine did not improve further upon overexpression of the argCJBD operon by employing a plasmid or by inserting a strong promoter as the ArgR repressor was absent (Zhang B. et al., 2017; Zhang et al., 2018a). This suggested that the production of L-ornithine limited by the low expression of the argCJBD operon can be addressed by removing the ArgR repressor, which concurred with the results described in the construction of the L-arginine-producing strain (Chen et al., 2014; Park et al., 2014).
As described above, the deletion of argR removed the transcription control on the main biosynthetic pathway of L-ornithine in C. glutamicum. However, stimulation of the argCJBD operon expression is not enough for releasing the negative control of the L-ornithine biosynthetic pathway. This is because of the presence of dual control systems in C. glutamicum: feedback repression and feedback inhibition. For feedback inhibition of L-ornithine, the enzyme activity of ornithine acetyltransferase (encoded by argJ) was inhibited by 50% by using 5 mM L-ornithine. The overexpression of argJ increased the yield of L-ornithine from 28.3 to 31.6 g/L in the engineered C. glutamicum 1006 strain (Hao et al., 2016). However, the mechanism underlying the inhibition of ornithine acetyltransferase by L-ornithine has not been elucidated. This mechanism can be an effective target for improving L-ornithine production. The gene encoding the ornithine acetyltransferase enzyme was subjected to site-directed mutagenesis, which did not result in the generation of valuable mutants (Shu et al., 2018). Additionally, the fermentation medium for producing L-ornithine was manually supplemented with a certain amount of arginine that resulted in the feedback inhibition of the rate-limiting enzyme, N-acetylglutamate kinase (encoded by argB), which is unfavorable for the accumulation of L-ornithine. Several studies have extensively evaluated the release of the negative effect mediated by the addition of L-arginine. The heterologous expression of argCJBD derived from the L-arginine-producing strain, C. glutamicum ATCC 21831, in the engineered C. glutamicum YW03 strain resulted in the production of 7.2 g/L L-ornithine, which was 2.6-fold higher than the L-ornithine titer produced upon expression of argCJBD operon derived from C. glutamicum ATCC 13032 in the C. glutamicum YW03 strain (2.0 g/L) (Kim S. Y. et al., 2015). These results suggest that the argB sequences are inconsistent between C. glutamicum ATCC 21831 and C. glutamicum ATCC 13032, which resulted in differential L-ornithine production. Furthermore, several studies have been performed recently to discover arginine-insensitive N-acetylglutamate kinase in the engineered L-arginine-producing strains (Ikeda et al., 2009; Xu et al., 2012a,b; Zhang J. et al., 2017). The production of L-ornithine was markedly improved upon overexpression of mutant argB, such as argBE19R, argBH268N, argBG287D, argBA49V, M54V, argBA49V, M54V, G287D, argBE.coli in the engineered C. glutamicum ORN2B strain. The L-ornithine titers in the C. glutamicum ORN2B (pEKEx3-argBE19R), ORN2B (pEKEx3-argBA49V, M54V), and ORN2B (pEKEx3argBE.col) strains were 0.307, 0.3, and 0.3 g/g glucose, respectively. These titers were higher than those in the C. glutamicum ORN2B (pEKEx3-argB) strain (0.257 g/g glucose) (Jensen et al., 2015).
Improving The Supplementation Of Glutamate By Overexpression Of gdh And Blocking Glutamate Secretion
In addition to the optimization of L-ornithine biosynthetic pathway by blocking other competitive metabolic pathways and releasing feedback regulation, adequate supply of precursor glutamate was reported to be another key factor for developing high L-ornithine-producing strains. Currently, many powerful strategies, such as overexpression of the endogenous or exogenous glutamate dehydrogenase, inhibition of the glutamate secretion system, and attenuation of α-ketoglutarate dehydrogenase, have been employed to engineer C. glutamicum for improving the flux of glutamate supplementation. The plasmid-based overexpression of gdh (encodes glutamate dehydrogenase) in the C. glutamicum ΔAP strain increased L-ornithine production by 16% (from 8.6 to 10.0 g/L). Simultaneously, individual chromosomal integration of the rocG/BSU37790 gene (encodes NADH-coupled glutamate dehydrogenase) derived from Bacillus subtilis improved L-ornithine concentration from 12.12 ± 0.57 to 14.84 ± 0.57 g/L in the engineered C. glutamicum ΔAPER strain (Jiang et al., 2013b). Additionally, glutamate was synthesized from α-ketoglutarate, which is an intermediate metabolite of the tricarboxylic acid cycle. However, the tricarboxylic acid cycle is closely related to cell growth and cannot be blocked. To balance the cell growth and L-ornithine production, the expression of odhA (NCgl1084), which encodes an enzyme component of the α-ketoglutarate dehydrogenase complex, was attenuated to reduce the metabolic flux of the tricarboxylic acid cycle and provide more glutamate for the biosynthesis of L-ornithine. When the native ribosome binding site (predicted translation initial intensity was 1,613 au) of odhA was replaced with a synthetic ribosome binding site with a predicted translation initial intensity of 837 au on the chromosome, the L-ornithine production titer increased from 13.7 to 16 g/L in the engineered C. glutamicum Sorn4 strain (Zhang B. et al., 2017). Furthermore, the overexpression of pyruvate carboxylase and deletion of phosphoenolpyruvate carboxykinase also improved glutamate concentration in the fermentation medium, but did not promote the accumulation of L-ornithine (Hwang et al., 2008). The authors claimed that the supply of glutamate is not a rate-limiting step for L-ornithine production. However, the major reason for the unchanged concentration of L-ornithine may be the extracellular secretion of glutamate, which increased the cost of downstream reaction and precursor wastage. The secretion of glutamate can be inhibited by deleting the NCgl1221 gene (encodes a glutamate transporter). This was first discovered for developing the L-arginine-producing C. glutamicum strain. The deletion of NCgl1221 markedly improved the L-arginine production titer (Park et al., 2014; Chen M. et al., 2015). Inspired by these reports, inactivation of NCgl1221 was introduced into the C. glutamicum Sorn1 strain, which dramatically decreased the secretion of glutamate and improved L-ornithine production by 22.7% (from 7.97 to 9.8 g/L). However, the deletion of NCgl1221 is not enough to interrupt the glutamate secretion due to the presence of another glutamate transporter MscCG2 (encoded by mscCG2) in C. glutamicum. Hence, both NCgl1221 and mscCG2 genes were deleted in the engineered C. glutamicum S9114 strain, which exhibited negligible glutamate production and enhanced L-ornithine accumulation (Zhang B. et al., 2019). Hence, improving the glutamate availability is an efficient strategy for L-ornithine accumulation in C. glutamicum.
Modulating The Central Metabolic Pathway
Upon consumption of glucose for L-ornithine production by C. glutamicum strains, the metabolic flux of central metabolic pathways, including the glycolytic pathway and the tricarboxylic acid cycle pathway, may be a limiting factor as these pathways provide the carbon skeleton. Various strategies have been explored for increasing the metabolic flux of central metabolic pathways for producing L-ornithine. Proteomic studies indicated differential proteomes in the parent strain and L-ornithine-producing strain. The effect of individual plasmid-based overexpression of pgi (NCgl0817), pfkA (NCgl1202), gap (NCgl0900), pyk (NCgl2008), pyc (NCgl0659), and glt (NCgl0795) on L-ornithine production was investigated in the C. glutamicum ΔAP strain. Modulating the expression of enzymes involved in the glycolytic pathway, such as Pgi, PfkA, GapA, and Pyk, improved L-ornithine production (Jiang et al., 2013b). A common feature in the enhanced expression of these genes is the improved metabolic flux of glycolysis. Additionally, a recent study focused on strengthening the glycolytic pathway by inserting a strong Peftu promoter in the upstream region of pfkA in the engineered C. glutamicum SO1 strain, which improved the yield of L-ornithine from 23.8 to 26.5 g/L (Zhang et al., 2018b). These results suggested the importance of the glycolytic pathway in L-ornithine production.
The tricarboxylic acid cycle is a source of the carbon skeleton for the biosynthesis of amino acids, such as L-lysine, L-isoline, L-threonine, L-glutamate, L-arginine, L-proline, L-ornithine, and L-citrulline. A recent study reported that rationally engineering the tricarboxylic acid cycle by overexpressing the pyc, ppc (NCgl1523), and gdh genes (encoding pyruvate carboxylase, phosphoenolpyruvate carboxylase, and glutamate dehydrogenase, respectively) and deleting the P1 promoter of glt (encodes citrate synthase) improved the lysine yield (from 14.47 ± 0.41 to 23.86 ± 2.16 g/L) and carbon yield (from 36.18 to 59.65%) in the C. glutamicum JL68 strain (Xu et al., 2018a). Hence, the tricarboxylic acid cycle pathway was also modulated by overexpressing the glt, icdh (NCgl0634), and ach (NCgl1482) genes in the L-ornithine-producing C. glutamicum SO16 strain. The insertion of a strong Psod promoter in the upstream region of glt improved the yield of L-ornithine from 30.8 to 34.1 g/L. Simultaneously, rational modulation of the tricarboxylic acid cycle pathway by overexpressing the glt gene improved the glucose consumption in the C. glutamicum SO16 strain (Zhang B. et al., 2019).
Enhanced Supply Of Intracellular Cofactor NADPH And ACETYL-CoA
Efficient supply of the cofactor is important in preventing obstruction of the metabolic pathways to enable production of valuable chemicals using the microbial cell factory (Xu et al., 2018b). NADPH, an important cofactor, is widely used in the biosynthesis of various amino acids, such as lysine (Wu et al., 2019), valine (Zhang H. et al., 2018), methionine (Li et al., 2016), ornithine (Hwang and Cho, 2012), and arginine (Chen M. et al., 2015). Intracellular supplementation of NADPH can be increased by various strategies, such as improving the metabolic flux of the pentose phosphate pathway (Siedler et al., 2013), overexpression of NAD kinase (Lindner et al., 2010; Xu et al., 2016), and developing a NADP-dependent glyceraldehyde 3-phosphate dehydrogenase (Takeno et al., 2010, 2016; Hoffmann et al., 2018). NADPH is a rate-limiting factor in the biosynthesis of metabolites in C. glutamicum. In the metabolic pathway of L-ornithine, 2 moles of NADPH are consumed in two steps of enzymatic reactions: conversion from α-oxoglutarate to glutamate catalyzed by glutamate dehydrogenase and from N-acetyl-glutamyl-phosphate to N-acetyl-glutamyl-semialdehyde catalyzed by N-acetyl-γ-glutamyl-phosphate reductase. Ornithine production is frequently limited by the supply of NADPH. Hence, the metabolic flux was redirected toward the pentose phosphate pathway by attenuating pgi, replacing the native promoter of tkt operon with a strong Psod promoter, and changing the translation initiation codon (G1A) of zwf (NCgl1514) in the YW03 strain, which improved the yield of L-ornithine from 261.3 to 417.2 mg/L (Kim S. Y. et al., 2015). Additionally, the carbon flux of the pentose phosphate pathway was enhanced by deleting the gluconate kinase gene in the C. glutamicum SJC8039 strain, which increased the intracellular NADPH concentration by 51.8%. L-Ornithine production yield in the engineered C. glutamicum SJC8399 strain with double deletion of NCgl2399 and NCgl2905 was 13.16 g/L, which is higher than that (8.78 g/L) in the parent C. glutamicum SJC8039 strain (Hwang and Cho, 2012). Furthermore, inactivation of NCgl0281, NCgl2582, and NCgl2053 genes, which encode putative NADP+-dependent oxidoreductases, enhanced the intracellular NADPH content by 72.4%. Inactivation of these genes inhibited glucose dehydrogenase activity and improved L-ornithine yield by 66.3% in the engineered SJC8039 strain (Hwang and Cho, 2014). Moreover, NADPH availability for L-ornithine production in the C. glutamicum ΔAPER strain was improved by heterologous expression of NADP-dependent glyceraldehyde-3-phosphate dehydrogenase derived from Clostridium acetobutylicum, which improved L-ornithine production (Jiang et al., 2013b). Therefore, increased intracellular NADPH supply is a major limiting factor for developing high L-ornithine-producing C. glutamicum strains.
In addition to NADPH, the supply of acetyl-CoA cofactor is required to produce L-ornithine. The acetyl-CoA co-factor is consumed during the conversion of oxaloacetate to citrate and glutamate to N-acetylglutamate. Recently, overexpression of cg3035 (encodes an N-acetylglutamate synthetase) was reported to be an efficient strategy for improving L-ornithine accumulation. This was the first study to report the role of acetyl-CoA in L-ornithine production. The intracellular concentration of acetyl-CoA was improved by attenuating the acetate biosynthesis pathway through insertion of a terminator in the upstream region of pta-ack (NCgl2657-NCgl2656) operon or cat (NCgl2480) in the C. glutamicum SO1 strain. Modulation of the acetate biosynthesis pathway improved the yield of L-ornithine from 23.8 to 26 g/L (Zhang et al., 2018b).
Enhanced Transport System
One of the efficient strategies for developing engineered strains to produce amino acids is overexpression of a specific transport system, such as Ncgl1221 (Nakamura et al., 2007; Nakayama et al., 2012) and MscCG2 (Wang Y. et al., 2018) glutamate transporters, lysine transporter (LysE/NCgl1214) (Vrljic et al., 1996; Kind et al., 2011), phenylalanine transporter (AroP) (Shang et al., 2013), and branched chain amino acid transporters (BrnFE) (Kennerknecht et al., 2002; Chen C. et al., 2015). Currently, there are no reported ornithine-specific transporters. The effect of plasmid-based overexpression of lysE (encodes a lysine transporter) on L-ornithine production was investigated in an engineered C. glutamicum ATCC 13032 strain containing inactivated argF. The intracellular and extracellular concentrations of L-ornithine were measured in short-term fermentation. The analysis indicated that LysE is not an ornithine transporter (Bellmann et al., 2001). However, there was not much progress on the ornithine transport system until the overexpression of lysE was studied in the L-ornithine-producing C. glutamicum Sorn8 strain. In this strain, the yield of L-ornithine increased by 21.8% when compared to the parent strain after 72 h of fermentation (Zhang B. et al., 2017). Subsequently, the effect of LysE on L-ornithine production was evaluated by deleting the lysE gene in the engineered C. glutamicum orn1 strain, which reduced the L-ornithine yield by 41.7%. Furthermore, overexpression of LysE by insertion of a strong Ptac promoter in the upstream region of lysE also improved the L-ornithine production titer from 15.3 to 19 g/L. This suggested that LysE promotes L-ornithine production (Zhang et al., 2018a). Although the direct transporter of L-ornithine is not known, the LysE transporter may contribute to developing high L-ornithine-producing strains.
Adaptive Evolution
The strategies discussed so far for increasing the L-ornithine production titer were based on genetic modifications. However, direct manipulations of one or more genes may negatively affect cell growth or metabolism. Therefore, the combination of irrational metabolic engineering methods, such as adaptive laboratory evolution (Stella et al., 2019) and genetic modification strategies will facilitate the development of high L-ornithine-producing strains. Adaptive laboratory evolution is widely applied for the construction of microbial strains, which not only stimulate latent metabolic pathways but also promote the phenotype and environmental adaptation of engineered strains (Garst et al., 2017; Yu et al., 2018). Compared to rational genetic modifications, adaptive laboratory evolution has provided extensive genetic manipulation targets to improve the strain phenotype. Adaptive laboratory evolution was applied for improving the strain phenotype of C. glutamicum ΔAPE. The L-ornithine production titer in the resultant C. glutamicum ΔAPE6937 strain was 13.6 ± 0.5 g/L, which was 20% higher than that in the parent strain (11.3 ± 0.3 g/L) (Jiang et al., 2013a). The accelerated cell growth caused by adaptive laboratory evolution was the main factor that contributed to the enhanced L-ornithine production titer in the C. glutamicum ΔAPE strain.
Alternative Substrates
Currently, majority of L-ornithine is produced from glucose, which is also used for human nutrition and animal feed industries. To minimize the competition for glucose between food security and industrial biotechnology, several studies have investigated the production of L-ornithine using non-edible feedstocks, such as xylose and arabinose.
Xylose is the second most abundant sustainable raw material for fermentation. Xylose is obtained from the hydrolysis of lignocellulosic biomasses and is widely used for the production of valuable chemicals, such as succinate (Mao et al., 2018), sarcosine (Mindt et al., 2019), 5-aminovalerate (Jorge et al., 2017b), L-pipecolic acid (Perez-Garcia et al., 2017), 3-hydroxypropionic acid (Chen et al., 2017), and γ-aminobutyric acid (Jorge et al., 2017a) in engineered C. glutamicum strains. The utilization of xylose can be improved by heterologous expression of xylose isomerase (Jo et al., 2017), overexpression of myo-inositol/proton symporter IolT1 (Brusseler et al., 2018), and adaptive laboratory evolution (Radek et al., 2017) of C. glutamicum. Previously, L-ornithine was produced from xylose by engineering the xylose metabolic pathways in the C. glutamicum ORN1 strain. The yield of L-ornithine was further enhanced by overexpressing xylA (encodes xylose isomerase) derived from Xanthomonas campestris and endogenous xylB (encodes xylulokinase) in the C. glutamicum ORN1 strain, which improved the L-ornithine yield from 9.4 ± 1.4 to 19.6 ± 1.9 mM (Meiswinkel et al., 2013a). Recently, heterologous expression of xylAB operon derived from X. campestris in the engineered C. glutamicum SO26 strain resulted in the production of 16.32 g/L L-ornithine with a yield of 0.14 g/g xylose. The production titer of L-ornithine was further improved to 18.9 g/L by optimizing the concentration of the inducer, isopropyl β-D-1-thiogalactopyranoside (IPTG) (Zhang B. et al., 2019). Arabinose is another component of lignocellulosic biomass hydrolysate and is widely used for producing valuable compounds (Zahoor et al., 2012; Zhao et al., 2018). Several studies have developed superior microbial cell factories, such as C. glutamicum that can efficiently utilize arabinose (Henke et al., 2018; Kawaguchi et al., 2018; Mindt et al., 2019). The heterologous expression of araBAD operon derived from E. coli enabled the C. glutamicum ORN1 strain to utilize arabinose. This engineered strain produced 89 ± 3 mM L-ornithine with a yield of 0.37 M/M arabinose (Schneider et al., 2011).
Conclusions And Perspectives
Currently, several comprehensive and rational genetic strategies have been employed for engineering C. glutamicum strains to produce L-ornithine. There is rapid progress in improving strain performance by combining the optimization of the metabolic pathways in C. glutamicum. For example, Kim S. Y. et al. (2015) reported the construction of a recombinant C. glutamicum by disrupting proB, argR, and argF, and overexpressing the operon of argCJBD from C. glutamicum ATCC 21831. Fed-batch culture of the engineered strain in a 6.6-L fermenter yielded 51.5 g/L of L-ornithine production titer from glucose. In our previous work, a recombinant C. glutamicum with modulation in the deletion of argF, NCgl1221, argR, putP, mscCG2, and iolR, attenuation of odhA, proB, ncgl2228, pta, cat, and pgi, and overexpression of lysE, gdh, cg3035, pfkA, tkt, argCJBD, glt, and gdh2 produced 43.6 g/L of L-ornithine during a 72 h fed-batch cultivation (Zhang B. et al., 2019). Although the production of L-ornithine by microbial fermentation is a promising and attractive strategy, very few microbes have been used for industrial production due to low yield and low productivity. Metabolic engineering of C. glutamicum is an efficient strategy to improve L-ornithine production. The mechanisms underlying the transportation of L-ornithine must be further investigated by new engineering techniques, such as genomics (Teusink and Smid, 2006), transcriptomics (Kim et al., 2017), and proteomics (Shen et al., 2016; Varela et al., 2018) to aid the development of high L-ornithine-producing strains. CRISPR/Cas9, a novel gene-editing technology, is a rapid and efficient method that can be applied for genetic modifications, such as gene deletion, large fragment DNA assembly, or site-directed gene mutation (Cho et al., 2017; Jiang et al., 2017; Wang B. et al., 2018; Zhang J. et al., 2019) in C. glutamicum, which can accelerate the pathway engineering in C. glutamicum strains. Irrational breeding strategies such as induced mutation breeding, or adaptive evolution have enabled rapid progress in improving the yield of L-ornithine. However, random mutations and labor-intensive screening methods remain as bottleneck issues. Recently, high-throughput screening of mutant strains was developed using a metabolite biosensor, which can measure the metabolite concentrations based on the fluorescence signals or antibiotic selective pressure (Hammer and Avalos, 2016; Lin et al., 2017; Yeom et al., 2018). Due to an improvement in the screening efficiency using a metabolite biosensor, the traditional irrational breeding strategy is considered a promising method for the construction of engineered strains (Zheng et al., 2018). Future studies must focus on developing an efficient intracellular L-ornithine biosensor, which can activate the irrational breeding of high L-ornithine-producing strain.
Author Contributions
X-YW planned and wrote the first manuscript. X-YG, YJ, and B-CY modified this manuscript. BZ supervised and finalized the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by grants from the National Natural Science Foundation of China (Grant No. 31730004) and Jiangxi Key R&D Program (Grant No. 20171ACF60006).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The handling Editor declared a shared affiliation, though no other collaboration, with one of the authors B-CY.
Acknowledgments
We thank Dr. Xue-Lan Chen for providing pk18mobsacB. We also thank Dr. Zhong-Gui Mao and Dr. Li-Ming Liu for providing Corynebacterium glutamicum S9114. We would like to thank Editage (www.editage.cn) for English language editing.
References
Acharya, S. K., Bhatia, V., Sreenivas, V., Khanal, S., and Panda, S. K. (2009). Efficacy of L-ornithine L-aspartate in acute liver failure: a double-blind, randomized, placebo-controlled study. Gastroenterology 136, 2159–2168. doi: 10.1053/j.gastro.2009.02.050
Becker, J., Rohles, C. M., and Wittmann, C. (2018). Metabolically engineered Corynebacterium glutamicum for bio-based production of chemicals, fuels, materials, and healthcare products. Metab. Eng. 50, 122–141. doi: 10.1016/j.ymben.2018.07.008
Becker, J., and Wittmann, C. (2015). Advanced biotechnology: metabolically engineered cells for the bio-based production of chemicals and fuels, materials, and health-care products. Angew. Chem. Int. Ed. Engl. 54, 3328–3350. doi: 10.1002/anie.201409033
Becker, J., and Wittmann, C. (2019). A field of dreams: lignin valorization into chemicals, materials, fuels, and health-care products. Biotechnol. Adv. 37:107360. doi: 10.1016/j.biotechadv.2019.02.016
Bellmann, A., Vrljic, M., Patek, M., Sahm, H., Kramer, R., and Eggeling, L. (2001). Expression control and specificity of the basic amino acid exporter LysE of Corynebacterium glutamicum. Microbiology 147, 1765–1774. doi: 10.1099/00221287-147-7-1765
Brusseler, C., Radek, A., Tenhaef, N., Krumbach, K., Noack, S., and Marienhagen, J. (2018). The myo-inositol/proton symporter IolT1 contributes to d-xylose uptake in Corynebacterium glutamicum. Bioresour. Technol. 249, 953–961. doi: 10.1016/j.biortech.2017.10.098
Butterworth, R. F., and McPhail, M. J. W. (2019). L-Ornithine L-Aspartate (LOLA) for hepatic encephalopathy in cirrhosis: results of randomized controlled trials and meta-analyses. Drugs 79, 31–37. doi: 10.1007/s40265-018-1024-1
Chen, C., Li, Y., Hu, J., Dong, X., and Wang, X. (2015). Metabolic engineering of Corynebacterium glutamicum ATCC13869 for L-valine production. Metab. Eng. 29, 66–75. doi: 10.1016/j.ymben.2015.03.004
Chen, M., Chen, X., Wan, F., Zhang, B., Chen, J., and Xiong, Y. (2015). Effect of tween 40 and dtsR1 on L-arginine overproduction in Corynebacterium crenatum. Microb. Cell. Fact. 14:119. doi: 10.1186/s12934-015-0310-9
Chen, X. L., Zhang, B., Tang, L., Jiao, H. T., Xu, H. Y., Xu, F., et al. (2014). Expression and characterization of ArgR, an arginine regulatory protein in Corynebacterium crenatum. Biomed. Environ. Sci. 27, 436–443. doi: 10.3967/bes2014.072
Chen, Z., Huang, J., Wu, Y., Wu, W., Zhang, Y., and Liu, D. (2017). Metabolic engineering of Corynebacterium glutamicum for the production of 3-hydroxypropionic acid from glucose and xylose. Metab. Eng. 39, 151–158. doi: 10.1016/j.ymben.2016.11.009
Cho, J. S., Choi, K. R., Prabowo, C. P. S., Shin, J. H., Yang, D., Jang, J., et al. (2017). CRISPR/Cas9-coupled recombineering for metabolic engineering of Corynebacterium glutamicum. Metab. Eng. 42, 157–167. doi: 10.1016/j.ymben.2017.06.010
Cleto, S., Jensen, J. V., Wendisch, V. F., and Lu, T. K. (2016). Corynebacterium glutamicum metabolic engineering with CRISPR interference (CRISPRi). ACS Synth. Biol. 5, 375–385. doi: 10.1021/acssynbio.5b00216
D'Este, M., Alvarado-Morales, M., and Angelidaki, I. (2018). Amino acids production focusing on fermentation technologies - a review. Biotechnol. Adv. 36, 14–25. doi: 10.1016/j.biotechadv.2017.09.001
Garst, A. D., Bassalo, M. C., Pines, G., Lynch, S. A., Halweg-Edwards, A. L., Liu, R., et al. (2017). Genome-wide mapping of mutations at single-nucleotide resolution for protein, metabolic and genome engineering. Nat. Biotechnol. 35, 48–55. doi: 10.1038/nbt.3718
Gourdon, P., and Lindley, N. D. (1999). Metabolic analysis of glutamate production by Corynebacterium glutamicum. Metab. Eng. 1, 224–231. doi: 10.1006/mben.1999.0122
Guo, J., Suastegui, M., Sakimoto, K. K., Moody, V. M., Xiao, G., Nocera, D. G., et al. (2018). Light-driven fine chemical production in yeast biohybrids. Science 362, 813–816. doi: 10.1126/science.aat9777
Hammer, S. K., and Avalos, J. L. (2016). Metabolic engineering: biosensors get the green light. Nat. Chem. Biol. 12, 894–895. doi: 10.1038/nchembio.2214
Hao, N., Mu, J., Hu, N., Xu, S., Shen, P., Yan, M., et al. (2016). Implication of ornithine acetyltransferase activity on l-ornithine production in Corynebacterium glutamicum. Biotechnol. Appl. Biochem. 63, 15–21. doi: 10.1002/bab.1353
Henke, N. A., Wiebe, D., Perez-Garcia, F., Peters-Wendisch, P., and Wendisch, V. F. (2018). Coproduction of cell-bound and secreted value-added compounds: simultaneous production of carotenoids and amino acids by Corynebacterium glutamicum. Bioresour. Technol. 247, 744–752. doi: 10.1016/j.biortech.2017.09.167
Hirasawa, T., and Wachi, M. (2017). Glutamate fermentation-2: mechanism of L-glutamate overproduction in Corynebacterium glutamicum. Adv. Biochem. Eng. Biotechnol. 159, 57–72. doi: 10.1007/10_2016_26
Hoffmann, S. L., Jungmann, L., Schiefelbein, S., Peyriga, L., Cahoreau, E., Portais, J. C., et al. (2018). Lysine production from the sugar alcohol mannitol: design of the cell factory Corynebacterium glutamicum SEA-3 through integrated analysis and engineering of metabolic pathway fluxes. Metab. Eng. 47, 475–487. doi: 10.1016/j.ymben.2018.04.019
Hwang, G. H., and Cho, J. Y. (2010). Identification of a suppressor gene for the arginine-auxotrophic argJ mutation in Corynebacterium glutamicum. J. Ind. Microbiol. Biotechnol. 37, 1131–1136. doi: 10.1007/s10295-010-0760-3
Hwang, G. H., and Cho, J. Y. (2012). Implication of gluconate kinase activity in L-ornithine biosynthesis in Corynebacterium glutamicum. J. Ind. Microbiol. Biotechnol. 39, 1869–1874. doi: 10.1007/s10295-012-1197-7
Hwang, G. H., and Cho, J. Y. (2014). Enhancement of L-ornithine production by disruption of three genes encoding putative oxidoreductases in Corynebacterium glutamicum. J. Ind. Microbiol. Biotechnol. 41, 573–578. doi: 10.1007/s10295-013-1398-8
Hwang, J. H., Hwang, G. H., and Cho, J. Y. (2008). Effect of increased glutamate availability on L-ornithine production in Corynebacterium glutamicum. J. Microbiol. Biotechnol. 18, 704–710.
Ikeda, M., Mitsuhashi, S., Tanaka, K., and Hayashi, M. (2009). Reengineering of a Corynebacterium glutamicum L-arginine and L-citrulline producer. Appl. Environ. Microbiol. 75, 1635–1641. doi: 10.1128/AEM.02027-08
Jeandet, P., Sobarzo-Sanchez, E., Clement, C., Nabavi, S. F., Habtemariam, S., Nabavi, S. M., et al. (2018). Engineering stilbene metabolic pathways in microbial cells. Biotechnol. Adv. 36, 2264–2283. doi: 10.1016/j.biotechadv.2018.11.002
Jensen, J. V., Eberhardt, D., and Wendisch, V. F. (2015). Modular pathway engineering of Corynebacterium glutamicum for production of the glutamate-derived compounds ornithine, proline, putrescine, citrulline, and arginine. J. Biotechnol. 214, 85–94. doi: 10.1016/j.jbiotec.2015.09.017
Jiang, L. Y., Chen, S. G., Zhang, Y. Y., and Liu, J. Z. (2013a). Metabolic evolution of Corynebacterium glutamicum for increased production of L-ornithine. BMC Biotechnol. 13:47. doi: 10.1186/1472-6750-13-47
Jiang, L. Y., Zhang, Y. Y., Li, Z., and Liu, J. Z. (2013b). Metabolic engineering of Corynebacterium glutamicum for increasing the production of L-ornithine by increasing NADPH availability. J. Ind. Microbiol. Biotechnol. 40, 1143–1151. doi: 10.1007/s10295-013-1306-2
Jiang, Y., Qian, F., Yang, J., Liu, Y., Dong, F., Xu, C., et al. (2017). CRISPR-Cpf1 assisted genome editing of Corynebacterium glutamicum. Nat. Commun. 8:15179. doi: 10.1038/ncomms15179
Jo, S., Yoon, J., Lee, S. M., Um, Y., Han, S. O., and Woo, H. M. (2017). Modular pathway engineering of Corynebacterium glutamicum to improve xylose utilization and succinate production. J. Biotechnol. 258, 69–78. doi: 10.1016/j.jbiotec.2017.01.015
Jorge, J. M., Nguyen, A. Q., Perez-Garcia, F., Kind, S., and Wendisch, V. F. (2017a). Improved fermentative production of gamma-aminobutyric acid via the putrescine route: Systems metabolic engineering for production from glucose, amino sugars, and xylose. Biotechnol. Bioeng. 114, 862–873. doi: 10.1002/bit.26211
Jorge, J. M. P., Perez-Garcia, F., and Wendisch, V. F. (2017b). A new metabolic route for the fermentative production of 5-aminovalerate from glucose and alternative carbon sources. Bioresour. Technol. 245, 1701–1709. doi: 10.1016/j.biortech.2017.04.108
Kawaguchi, H., Yoshihara, K., Hara, K. Y., Hasunuma, T., Ogino, C., and Kondo, A. (2018). Metabolome analysis-based design and engineering of a metabolic pathway in Corynebacterium glutamicum to match rates of simultaneous utilization of D-glucose and L-arabinose. Microb. Cell. Fact. 17:76. doi: 10.1186/s12934-018-0927-6
Kennerknecht, N., Sahm, H., Yen, M. R., Patek, M., Saier, M. H. Jr., and Eggeling, L. (2002). Export of L-isoleucine from Corynebacterium glutamicum: a two-gene-encoded member of a new translocator family. J. Bacteriol. 184, 3947–3956. doi: 10.1128/JB.184.14.3947-3956.2002
Kim, D. J., Hwang, G. H., Um, J. N., and Cho, J. Y. (2015). Increased L-ornithine production in Corynebacterium glutamicum by overexpression of a gene encoding a putative aminotransferase. J. Mol. Microbiol. Biotechnol. 25, 45–50. doi: 10.1159/000375124
Kim, H. T., Khang, T. U., Baritugo, K. A., Hyun, S. M., Kang, K. H., Jung, S. H., et al. (2019). Metabolic engineering of Corynebacterium glutamicum for the production of glutaric acid, a C5 dicarboxylic acid platform chemical. Metab. Eng. 51, 99–109. doi: 10.1016/j.ymben.2018.08.007
Kim, S., Jeong, H., Kim, E. Y., Kim, J. F., Lee, S. Y., and Yoon, S. H. (2017). Genomic and transcriptomic landscape of Escherichia coli BL21(DE3). Nucleic. Acids Res. 45, 5285–5293. doi: 10.1093/nar/gkx228
Kim, S. Y., Lee, J., and Lee, S. Y. (2015). Metabolic engineering of Corynebacterium glutamicum for the production of L-ornithine. Biotechnol. Bioeng. 112, 416–421. doi: 10.1002/bit.25440
Kind, S., Kreye, S., and Wittmann, C. (2011). Metabolic engineering of cellular transport for overproduction of the platform chemical 1,5-diaminopentane in Corynebacterium glutamicum. Metab. Eng. 13, 617–627. doi: 10.1016/j.ymben.2011.07.006
Kogure, T., and Inui, M. (2018). Recent advances in metabolic engineering of Corynebacterium glutamicum for bioproduction of value-added aromatic chemicals and natural products. Appl. Microbiol. Biotechnol. 102, 8685–8705. doi: 10.1007/s00253-018-9289-6
Lapujade, P., Goergen, J. L., and Engasser, J. M. (1999). Glutamate excretion as a major kinetic bottleneck for the thermally triggered production of glutamic acid by Corynebacterium glutamicum. Metab. Eng. 1, 255–261. doi: 10.1006/mben.1999.0129
Lee, J. H., and Wendisch, V. F. (2017). Production of amino acids-genetic and metabolic engineering approaches. Bioresour. Technol. 245, 1575–1587. doi: 10.1016/j.biortech.2017.05.065
Lee, S. Y., Cho, J. Y., Lee, H. J., Kim, Y. H., and Min, J. (2010). Enhancement of ornithine production in proline-supplemented Corynebacterium glutamicum by ornithine cyclodeaminase. J. Microbiol. Biotechnol. 20, 127–131. doi: 10.4014/jmb.0907.07034
Lee, Y. J., and Cho, J. Y. (2006). Genetic manipulation of a primary metabolic pathway for L-ornithine production in Escherichia coli. Biotechnol. Lett. 28, 1849–1856. doi: 10.1007/s10529-006-9163-y
Li, S., Li, Y., and Smolke, C. D. (2018). Strategies for microbial synthesis of high-value phytochemicals. Nat. Chem. 10, 395–404. doi: 10.1038/s41557-018-0013-z
Li, Y., Cong, H., Liu, B., Song, J., Sun, X., Zhang, J., et al. (2016). Metabolic engineering of Corynebacterium glutamicum for methionine production by removing feedback inhibition and increasing NADPH level. Antonie Van Leeuwenhoek 109, 1185–1197. doi: 10.1007/s10482-016-0719-0
Li, Y., Wei, H., Wang, T., Xu, Q., Zhang, C., Fan, X., et al. (2017). Current status on metabolic engineering for the production of l-aspartate family amino acids and derivatives. Bioresour. Technol. 245, 1588–1602. doi: 10.1016/j.biortech.2017.05.145
Lin, J. L., Wagner, J. M., and Alper, H. S. (2017). Enabling tools for high-throughput detection of metabolites: metabolic engineering and directed evolution applications. Biotechnol. Adv. 35, 950–970. doi: 10.1016/j.biotechadv.2017.07.005
Lindner, S. N., Niederholtmeyer, H., Schmitz, K., Schoberth, S. M., and Wendisch, V. F. (2010). Polyphosphate/ATP-dependent NAD kinase of Corynebacterium glutamicum: biochemical properties and impact of ppnK overexpression on lysine production. Appl. Microbiol. Biotechnol. 87, 583–593. doi: 10.1007/s00253-010-2481-y
Luo, X., Reiter, M. A., d'Espaux, L., Wong, J., Denby, C. M., Lechner, A., et al. (2019). Complete biosynthesis of cannabinoids and their unnatural analogues in yeast. Nature 567, 123–126. doi: 10.1038/s41586-019-0978-9
Mao, Y., Li, G., Chang, Z., Tao, R., Cui, Z., Wang, Z., et al. (2018). Metabolic engineering of Corynebacterium glutamicum for efficient production of succinate from lignocellulosic hydrolysate. Biotechnol. Biofuels 11:95. doi: 10.1186/s13068-018-1094-z
Meiswinkel, T. M., Gopinath, V., Lindner, S. N., Nampoothiri, K. M., and Wendisch, V. F. (2013a). Accelerated pentose utilization by Corynebacterium glutamicum for accelerated production of lysine, glutamate, ornithine and putrescine. Microb. Biotechnol. 6, 131–140. doi: 10.1111/1751-7915.12001
Meiswinkel, T. M., Rittmann, D., Lindner, S. N., and Wendisch, V. F. (2013b). Crude glycerol-based production of amino acids and putrescine by Corynebacterium glutamicum. Bioresour. Technol. 145, 254–258. doi: 10.1016/j.biortech.2013.02.053
Mindt, M., Heuser, M., and Wendisch, V. F. (2019). Xylose as preferred substrate for sarcosine production by recombinant Corynebacterium glutamicum. Bioresour. Technol. 281, 135–142. doi: 10.1016/j.biortech.2019.02.084
Mitsuhashi, S. (2014). Current topics in the biotechnological production of essential amino acids, functional amino acids, and dipeptides. Curr. Opin. Biotechnol. 26, 38–44. doi: 10.1016/j.copbio.2013.08.020
Nakamura, J., Hirano, S., Ito, H., and Wachi, M. (2007). Mutations of the Corynebacterium glutamicum NCgl1221 gene, encoding a mechanosensitive channel homolog, induce L-glutamic acid production. Appl. Environ. Microbiol. 73, 4491–4498. doi: 10.1128/AEM.02446-06
Nakayama, Y., Yoshimura, K., and Iida, H. (2012). A gain-of-function mutation in gating of Corynebacterium glutamicum NCgl1221 causes constitutive glutamate secretion. Appl. Environ. Microbiol. 78, 5432–5434. doi: 10.1128/AEM.01310-12
Okuda, S., Sherman, D. J., Silhavy, T. J., Ruiz, N., and Kahne, D. (2016). Lipopolysaccharide transport and assembly at the outer membrane: the PEZ model. Nat. Rev. Microbiol. 14, 337–345. doi: 10.1038/nrmicro.2016.25
Otten, A., Brocker, M., and Bott, M. (2015). Metabolic engineering of Corynebacterium glutamicum for the production of itaconate. Metab. Eng. 30, 156–165. doi: 10.1016/j.ymben.2015.06.003
Park, J., Yu, B. J., Choi, J. I., and Woo, H. M. (2019). Heterologous production of squalene from glucose in engineered Corynebacterium glutamicum using multiplex CRISPR interference and high-throughput fermentation. J. Agric. Food. Chem. 67, 308–319. doi: 10.1021/acs.jafc.8b05818
Park, S. H., Kim, H. U., Kim, T. Y., Park, J. S., Kim, S. S., and Lee, S. Y. (2014). Metabolic engineering of Corynebacterium glutamicum for L-arginine production. Nat. Commun. 5:4618. doi: 10.1038/ncomms5618
Perez-Garcia, F., Max Risse, J., Friehs, K., and Wendisch, V. F. (2017). Fermentative production of L-pipecolic acid from glucose and alternative carbon sources. Biotechnol. J. 12:1600646. doi: 10.1002/biot.201600646
Qin, J., Zhou, Y. J., Krivoruchko, A., Huang, M., Liu, L., Khoomrung, S., et al. (2015). Modular pathway rewiring of Saccharomyces cerevisiae enables high-level production of L-ornithine. Nat. Commun. 6:8224. doi: 10.1038/ncomms9224
Radek, A., Tenhaef, N., Muller, M. F., Brusseler, C., Wiechert, W., Marienhagen, J., et al. (2017). Miniaturized and automated adaptive laboratory evolution: Evolving Corynebacterium glutamicum towards an improved d-xylose utilization. Bioresour. Technol. 245, 1377–1385. doi: 10.1016/j.biortech.2017.05.055
Rathi, S., and Taneja, S. (2018). Terminating and episode of overt hepatic encephalopathy: L-ornithine-L-aspartate may have some role. Hepatology 67:797. doi: 10.1002/hep.29570
Sarria, S., Kruyer, N. S., and Peralta-Yahya, P. (2017). Microbial synthesis of medium-chain chemicals from renewables. Nat. Biotechnol. 35, 1158–1166. doi: 10.1038/nbt.4022
Schneider, J., Niermann, K., and Wendisch, V. F. (2011). Production of the amino acids l-glutamate, l-lysine, l-ornithine and l-arginine from arabinose by recombinant Corynebacterium glutamicum. J. Biotechnol. 154, 191–198. doi: 10.1016/j.jbiotec.2010.07.009
Shang, X., Zhang, Y., Zhang, G., Chai, X., Deng, A., Liang, Y., et al. (2013). Characterization and molecular mechanism of AroP as an aromatic amino acid and histidine transporter in Corynebacterium glutamicum. J.Bacteriol. 195, 5334–5342. doi: 10.1128/JB.00971-13
Shen, H. J., Cheng, B. Y., Zhang, Y. M., Tang, L., Li, Z., Bu, Y. F., et al. (2016). Dynamic control of the mevalonate pathway expression for improved zeaxanthin production in Escherichia coli and comparative proteome analysis. Metab. Eng. 38, 180–190. doi: 10.1016/j.ymben.2016.07.012
Shu, Q., Xu, M., Li, J., Yang, T., Zhang, X., Xu, Z., et al. (2018). Improved L-ornithine production in Corynebacterium crenatum by introducing an artificial linear transacetylation pathway. J. Ind. Microbiol. Biotechnol. 45, 393–404. doi: 10.1007/s10295-018-2037-1
Siedler, S., Lindner, S. N., Bringer, S., Wendisch, V. F., and Bott, M. (2013). Reductive whole-cell biotransformation with Corynebacterium glutamicum: improvement of NADPH generation from glucose by a cyclized pentose phosphate pathway using pfkA and gapA deletion mutants. Appl. Microbiol. Biotechnol. 97, 143–152. doi: 10.1007/s00253-012-4314-7
Stäbler, N., Oikawa, T., Bott, M., and Eggeling, L. (2011). Corynebacterium glutamicum as a host for synthesis and export of D-amino acids. J. Bacteriol. 193, 1702–1709. doi: 10.1128/JB.01295-10
Stella, R. G., Wiechert, J., Noack, S., and Frunzke, J. (2019). Evolutionary engineering of Corynebacterium glutamicum. Biotechnol. J. 14:e1800444. doi: 10.1002/biot.201800444
Takeno, S., Hori, K., Ohtani, S., Mimura, A., Mitsuhashi, S., and Ikeda, M. (2016). l-Lysine production independent of the oxidative pentose phosphate pathway by Corynebacterium glutamicum with the Streptococcus mutans gapN gene. Metab. Eng. 37, 1–10. doi: 10.1016/j.ymben.2016.03.007
Takeno, S., Murata, R., Kobayashi, R., Mitsuhashi, S., and Ikeda, M. (2010). Engineering of Corynebacterium glutamicum with an NADPH-generating glycolytic pathway for L-lysine production. Appl. Environ. Microbiol. 76, 7154–7160. doi: 10.1128/AEM.01464-10
Teusink, B., and Smid, E. J. (2006). Modelling strategies for the industrial exploitation of lactic acid bacteria. Nat. Rev. Microbiol. 4, 46–56. doi: 10.1038/nrmicro1319
Tuyishime, P., Wang, Y., Fan, L., Zhang, Q., Li, Q., Zheng, P., et al. (2018). Engineering Corynebacterium glutamicum for methanol-dependent growth and glutamate production. Metab. Eng. 49, 220–231. doi: 10.1016/j.ymben.2018.07.011
Varela, C., Schmidt, S. A., Borneman, A. R., Pang, C. N. I., Krömerx, J. O., Khan, A., et al. (2018). Systems-based approaches enable identification of gene targets which improve the flavour profile of low-ethanol wine yeast strains. Metab. Eng. 49, 178–191. doi: 10.1016/j.ymben.2018.08.006
Vrljic, M., Sahm, H., and Eggeling, L. (1996). A new type of transporter with a new type of cellular function: L-lysine export from Corynebacterium glutamicum. Mol. Microbiol. 22, 815–826. doi: 10.1046/j.1365-2958.1996.01527.x
Wang, B., Hu, Q., Zhang, Y., Shi, R., Chai, X., Liu, Z., et al. (2018). A RecET-assisted CRISPR-Cas9 genome editing in Corynebacterium glutamicum. Microb. Cell. Fact. 17:63. doi: 10.1186/s12934-018-0910-2
Wang, Y., Cao, G., Xu, D., Fan, L., Wu, X., Ni, X., et al. (2018). A novel Corynebacterium glutamicum l-glutamate exporter. Appl. Environ. Microbiol. 84:e02691-17. doi: 10.1128/AEM.02691-17
Wendisch, V. F., Jorge, J. M., Pérez-García, F., and Sgobba, E. (2016). Updates on industrial production of amino acids using Corynebacterium glutamicum. World J. Microbiol. Biotechnol. 32:105. doi: 10.1007/s11274-016-2060-1
Wu, W., Zhang, Y., Liu, D., and Chen, Z. (2019). Efficient mining of natural NADH-utilizing dehydrogenases enables systematic cofactor engineering of lysine synthesis pathway of Corynebacterium glutamicum. Metab. Eng. 52, 77–86. doi: 10.1016/j.ymben.2018.11.006
Xiao, F., Wang, H., Shi, Z., Huang, Q., Huang, L., Lian, J., et al. (2019). Multi-level metabolic engineering of Pseudomonas mutabilis ATCC31014 for efficient production of biotin. Metab. Eng. doi: 10.1016/j.ymben.2019.05.005. [Epub ahead of print].
Xu, J. Z., Wu, Z. H., Gao, S. J., and Zhang, W. (2018a). Rational modification of tricarboxylic acid cycle for improving L-lysine production in Corynebacterium glutamicum. Microb. Cell. Fact. 17:105. doi: 10.1186/s12934-018-0958-z
Xu, J. Z., Yang, H. K., and Zhang, W. G. (2018b). NADPH metabolism: a survey of its theoretical characteristics and manipulation strategies in amino acid biosynthesis. Crit. Rev. Biotechnol. 38, 1061–1076. doi: 10.1080/07388551.2018.1437387
Xu, M., Qin, J., Rao, Z., You, H., Zhang, X., Yang, T., et al. (2016). Effect of Polyhydroxybutyrate (PHB) storage on L-arginine production in recombinant Corynebacterium crenatum using coenzyme regulation. Microb. Cell. Fact. 15:15. doi: 10.1186/s12934-016-0414-x
Xu, M., Rao, Z., Dou, W., Jin, J., and Xu, Z. (2012a). Site-directed mutagenesis studies on the L-arginine-binding sites of feedback inhibition in N-acetyl-L-glutamate kinase (NAGK) from Corynebacterium glutamicum. Curr. Microbiol. 64, 164–172. doi: 10.1007/s00284-011-0042-y
Xu, M., Rao, Z., Dou, W., Yang, J., Jin, J., and Xu, Z. (2012b). Site-directed mutagenesis and feedback-resistant N-acetyl-L-glutamate kinase (NAGK) increase Corynebacterium crenatum L-arginine production. Amino Acids 43, 255–266. doi: 10.1007/s00726-011-1069-x
Xu, N., Wei, L., and Liu, J. (2019). Recent advances in the applications of promoter engineering for the optimization of metabolite biosynthesis. World J. Microbiol. Biotechnol. 35:33. doi: 10.1007/s11274-019-2606-0
Yeom, S. J., Kim, M., Kwon, K. K., Fu, Y., Rha, E., Park, S. H., et al. (2018). A synthetic microbial biosensor for high-throughput screening of lactam biocatalysts. Nat. Commun. 9:5053. doi: 10.1038/s41467-018-07488-0
Yoon, J., and Woo, H. M. (2018). CRISPR interference-mediated metabolic engineering of Corynebacterium glutamicum for homo-butyrate production. Biotechnol. Bioeng. 115, 2067–2074. doi: 10.1002/bit.26720
Yu, T., Zhou, Y. J., Huang, M., Liu, Q., Pereira, R., David, F., et al. (2018). Reprogramming yeast metabolism from alcoholic fermentation to lipogenesis. Cell 174, 1549–1558. doi: 10.1016/j.cell.2018.07.013
Zahoor, A., Lindner, S. N., and Wendisch, V. F. (2012). Metabolic engineering of Corynebacterium glutamicum aimed at alternative carbon sources and new products. Comput. Struct. Biotechnol. J. 3:e201210004. doi: 10.5936/csbj.201210004
Zhang, B., Gao, G., Chu, X. H., and Ye, B. C. (2019). Metabolic engineering of Corynebacterium glutamicum S9114 to enhance the production of l-ornithine driven by glucose and xylose. Bioresour. Technol. 284, 204–213. doi: 10.1016/j.biortech.2019.03.122
Zhang, B., Ren, L. Q., Yu, M., Zhou, Y., and Ye, B. C. (2018a). Enhanced l-ornithine production by systematic manipulation of l-ornithine metabolism in engineered Corynebacterium glutamicum S9114. Bioresour. Technol. 250, 60–68. doi: 10.1016/j.biortech.2017.11.017
Zhang, B., Yu, M., Wei, W. P., and Ye, B. C. (2018b). Optimization of L-ornithine production in recombinant Corynebacterium glutamicum S9114 by cg3035 overexpression and manipulating the central metabolic pathway. Microb. Cell. Fact. 17:91. doi: 10.1186/s12934-018-0940-9
Zhang, B., Yu, M., Zhou, Y., Li, Y., and Ye, B. C. (2017). Systematic pathway engineering of Corynebacterium glutamicum S9114 for L-ornithine production. Microb. Cell. Fact. 16:158. doi: 10.1186/s12934-017-0776-8
Zhang, B., Yu, M., Zhou, Y., and Ye, B. C. (2018c). Improvement of L-ornithine production by attenuation of argF in engineered Corynebacterium glutamicum S9114. AMB Express 8:26. doi: 10.1186/s13568-018-0557-8
Zhang, H., Li, Y., Wang, C., and Wang, X. (2018). Understanding the high L-valine production in Corynebacterium glutamicum VWB-1 using transcriptomics and proteomics. Sci. Rep. 8:3632. doi: 10.1038/s41598-018-21926-5
Zhang, J., Xu, M., Ge, X., Zhang, X., Yang, T., Xu, Z., et al. (2017). Reengineering of the feedback-inhibition enzyme N-acetyl-L-glutamate kinase to enhance L-arginine production in Corynebacterium crenatum. J. Ind. Microbiol. Biotechnol. 44, 271–283. doi: 10.1007/s10295-016-1885-9
Zhang, J., Yang, F., Yang, Y., Jiang, Y., and Huo, Y. X. (2019). Optimizing a CRISPR-Cpf1-based genome engineering system for Corynebacterium glutamicum. Microb. Cell. Fact. 18:60. doi: 10.1186/s12934-019-1109-x
Zhang, Y. Y., Bu, Y. F., and Liu, J. Z. (2015). Production of L-ornithine from sucrose and molasses by recombinant Corynebacterium glutamicum. Folia Microbiol. 60, 393–398. doi: 10.1007/s12223-014-0371-x
Zhao, N., Qian, L., Luo, G., and Zheng, S. (2018). Synthetic biology approaches to access renewable carbon source utilization in Corynebacterium glutamicum. Appl. Microbiol. Biotechnol. 102, 9517–9529. doi: 10.1007/s00253-018-9358-x
Keywords: L-ornithine, Corynebacterium glutamicum, metabolic engineering, genetic engineering, fermentation
Citation: Wu X-Y, Guo X-Y, Zhang B, Jiang Y and Ye B-C (2020) Recent Advances of L-ornithine Biosynthesis in Metabolically Engineered Corynebacterium glutamicum. Front. Bioeng. Biotechnol. 7:440. doi: 10.3389/fbioe.2019.00440
Received: 16 October 2019; Accepted: 11 December 2019;
Published: 09 January 2020.
Edited by:
Hui Wu, East China University of Science and Technology, ChinaReviewed by:
Xixian Xie, Tianjin University of Science and Technology, ChinaTsutomu Tanaka, Kobe University, Japan
Copyright © 2020 Wu, Guo, Zhang, Jiang and Ye. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bin Zhang, emhhbmdiaW4yOTE5QDE2My5jb20=
 Xiao-Yu Wu1
Xiao-Yu Wu1 Bin Zhang
Bin Zhang Bang-Ce Ye
Bang-Ce Ye