- Department of Plant and Environmental Sciences, Faculty of Science, University of Copenhagen, Frederiksberg, Denmark
Microalgae offer a promising biological platform for sustainable biomanufacturing of a wide range of chemicals, pharmaceuticals, and fuels. The model microalga Chlamydomonas reinhardtii is thus far the most versatile algal chassis for bioengineering and can grow using atmospheric CO2 and organic carbons (e.g., acetate and pure cellulose). Ability to utilize renewable feedstock like lignocellulosic biomass as a carbon source could significantly accelerate microalgae-based productions, but this is yet to be demonstrated. We observed that C. reinhardtii was not able to heterotrophically grow using wheat straw, a common type of lignocellulosic biomass, likely due to the recalcitrant nature of the biomass. When the biomass was pretreated with alkaline, C. reinhardtii was able to grow using acetate that was released from the biomass. To establish an eco-friendly and self-sustained growth system, we engineered C. reinhardtii to secrete a fungal acetylxylan esterase (AXE) for hydrolysis of acetylesters in the lignocellulosic biomass. Two transgenic strains (CrAXE03 and CrAXE23) secreting an active AXE into culture media were isolated. Incubation of CrAXE03 with wheat straw resulted in an eight-fold increase in the algal cell counts with a concomitant decrease of biomass acetylester contents by 96%. The transgenic lines showed minor growth defects compared to the parental strain, indicating that secretion of the AXE protein imposes limited metabolic burden. The results presented here would open new opportunities for applying low-cost renewable feedstock, available in large amounts as agricultural and manufacturing by-products, for microalgal cultivation. Furthermore, acetylesters and acetate released from them, are well-known inhibitors in lignocellulosic biofuel productions; thus, direct application of the bioengineered microalga could be exploited for improving renewable biofuel productions.
Introduction
Microalgae offer a promising biological platform for sustainable biomanufacturing of a wide range of bioproducts from water, air, and light. C. reinhardtii is one of the best characterized algal species and has been developed as a robust expression platform for production of recombinant proteins, oils, and chemicals with broad industrial applications (Almaraz-Delgado et al., 2014; Rasala and Mayfield, 2015). Well-characterized molecular tools are also available and allow efficient and robust expressions of transgenes from the nuclear and chloroplast genomes, making this microalga by far the most powerful chassis for algal synthetic biology and bioengineering (Schroda et al., 2000; Eichler-Stahlberg et al., 2009; Jinkerson and Jonikas, 2015; Mussgnug, 2015; Díaz-Santos et al., 2016).
Chlamydomonas reinhardtii can grow photoautotrophically using CO2 as the sole carbon source and also heterotrophically or mixotrophically using organic carbons such as acetate (Harris, 2009). Furthermore, it was recently shown that C. reinhardtii can utilize pure cellulose through secretion of cellulolytic enzymes (Blifernez-Klassen et al., 2012), raising an exciting prospect of using plant biomass, such as agricultural residues and manufacturing by-products, as alternative carbon sources for improving the cost performance of microalgae-based productions. However, direct utilization of renewable biomass by C. reinhardtii is yet to be demonstrated. Thus far, evidence of direct utilization of lignocellulosic biomass is only reported for the oleaginous microalga Auxenochlorella protothecoides UTEX 25 (Vogler et al., 2018).
Over 30 recombinant proteins have been produced in C. reinhardtii, a small subset of which were secreted to culture media. Recombinant protein secretion into culture media has several advantages, including protein glycosylation, which can faciliate folding and increased protein stability (Lingg et al., 2012; Mathieu-Rivet et al., 2014; Ramos-Martinez et al., 2017), and simplification of downstream processing, which can circumvent costly cell harvesting steps (Hellwig et al., 2004; Nikolov and Woodard, 2004). Moreover, the algal biomass can be exploited as a co-product, adding more values to the process (Gangl et al., 2015). Among secreted recombinant proteins produced by C. reinhardii are luciferase (Laursen et al., 2013), fluorescent proteins (Lauersen et al., 2015; Ramos-Martinez et al., 2017), xylanase (Rasala et al., 2012), laccase (Chiaiese et al., 2011), human glycoprotein erythropoietin (Eichler-Stahlberg et al., 2009), an ice-binding protein (Lauersen et al., 2013), and human growth factors (Chávez et al., 2016; Baier et al., 2018). Glyconengineering was recently applied to enhance the product yield (Ramos-Martinez et al., 2017). However, current yields are still very low, making further enhancement of product yields and product recovery technologies critically important (Baier et al., 2018). Alternatively, exploration into new avenues of applications, where engineered microalgal cultures could be directly utilized without product purifications, could open new opportunities, as it has been explored in yeast (Sun et al., 2012; Kricka et al., 2014; Liang et al., 2014; Lee et al., 2017).
In this study, we investigated utilization of lignocellulosic biomass for cultivation of C. reinhardtii. Lignocellulosic biomass is widely used as renewable feedstock for productions of bioethanol and animal feeds are available in huge amounts as agricultural residues such as wheat straw. Our initial test, however, revealed that C. reinhardtii was not able to grow using wheat straw as the carbon source. We exploited the fact that hemicelluloses and pectin in plant biomass can be highly acetyl esterified and it was previously shown that acetate released from softwood biomass could be assimilated by C. reinhardtii, although this required prior removal of toxic substances (Liang et al., 2013; Hu et al., 2016). To establish an eco-friendly and self-sustained system of biomass acetylester utilization, C. reinhardtii was engineered to secrete an acetylxylan esterase (AXE), capable of hydrolyzing acetylesters in lignocellulosic biomass. Our results demonstrated that the AXE-secreting C. reinhardtii was able to directly utilize acetylesters in lignocellulosic biomass, leading to simultaneous reduction in biomass acetylester contents.
Materials and Methods
Strains, Media, and Culture Conditions
Chlamydomonas reinhardtii wild type, photosynthetic mutant FUD16 (Ketchner et al., 1995), the cell wall-deficient strain UVM4 (Neupert et al., 2009), and transgenic strains generated in this study were routinely cultivated mixotrophically in tris-acetate-phosphate (TAP) media supplemented with 1 g L−1 acetic acid (Gorman and Levine, 1965) or in minimal medium (MM) containing 0.1 g L−1 acetic acid, instead of 1 g L−1 (Blifernez-Klassen et al., 2012). The pH value of all the media were adjusted to 7.0. Liquid cultures of C. reinhardtii were cultivated under the standard conditions (at the photon flux of 120 μmol m−2 s−1, 25°C) at a constant shaking (120 rpm) in an orbital shaker. Cell numbers in cultures were determined by counting in a Neubauer hemocytometer under a bright-field microscope. MM medium containing wheat biomass (MM+Biomass) was prepared as follows. Straws from wheat (Triticum aestivum) were first ground to small pieces, washed in 96% (v/v) ethanol followed by wash in 100% (v/v) acetone, and dried in air. Ten milligrams of dried straw were resuspended in 1 mL of a 50-mM ammonium formate buffer, pH 6.5, into which thermostable α-amylase (Megazyme, https://www.megazyme.com) was added at the final enzyme concentration of 2 U mL−1 to remove starch. The mixture was incubated for 2 h at 85°C and was centrifuged at 3,500 × g for 5 min at room temperature (~23°C). The pellet was washed in water and then in 100% (v/v) acetone and dried in air. The de-starched wheat straw was then added to MM at the concentration of 15 g L−1 and the mixture was autoclaved. The acetylester content of the de-starch biomass was 2.5% (w/w). All cultures were supplemented with antibiotics (50 μg mL−1 ampicillin, 25 μg mL−1 kasugamycin) to reduce the risk of contamination.
Generation of the CrAXE Construct
All standard chemicals and reagents were purchased from SigmaAldrich (http://www.sigmaaldrich.com). Restriction enzymes were purchased from New England Biolabs (https://www.neb.com). Oligonucleotide primers were purchased from Integrated DNA Technologies Inc. (https://www.idtdna.com). The signal sequence in AXE protein from Aspergillus nidulans (AnAXE) was analyzed in silico analysis using the publicly available tool SignalP version 4.1 (http://www.cbs.dtu.dk/services/SignalP/). Codon optimization of the AXE gene was performed using the webtool OPTIMIZER (Puigbò et al., 2007). A codon-optimized AXE gene was synthesized (Integrated DNA Technologies, Inc.) and overhangs were added by PCR using primer AnAXEΔSP-F (5′-gcacagctcctgcgtgggccGTGAAGCTCCAGTACCTG) and AnAXE-R (5′-tcgaactgcgggtggctccaGCTCGTAAAACCGAACCAC). The uppercase letters indicate target specific sequences and the lowercase letters indicate overlapping sequences with a transformation vector (pERC-SSVENUS) (Ramos-Martinez et al., 2017). The PCR product was fused with the vector by overlap extension PCR (Bryksin and Matsumura, 2010) such that the codon-optimized AXE gene is in translational fusion with the gametolysin signal peptide (MSLATRRFGAAAALLVAACVLCTAPAWA) and a strep-tag (WSHPQFEK). The final expression gene construct was named CrAXE (Supplementary Material). The vector backbone contained the aphVIII gene encoding an aminoglycoside 3′-phosphotransferase from Streptomyces rimosus to provide resistance to paromomycin, which was used to select transformed C. reinhardtii (Sizova et al., 1996). The expression of the transgene and the resistance gene was controlled by the chimeric promoter Hsp70A-RbcS2 containing the first intron of RbscS2. The RbcS2 3′ UTR was placed downstream of the transgenes and the resistance cassette for correct termination. For selection in E. coli the ampicillin resistance gene, bla encoding β-lactamase is introduced into the plasmid under the control of the AmpR promoter.
Transformation of the UVM4 Strain of C. reinhardtii
The UVM4 strain was transformed by the glass bead method (Kindle, 1998) using 2 μg of the plasmid construct that has been linearized through ScaI restriction digestion. The mixture of the UVM4 cells and the linearized plasmid was incubated overnight and then harvested at 1,100 ×g for 5 min at room temperature (~23°C) and spread over TAP media containing 15 g L−1 agar and 15 mg L−1 paromomycin. After 7 days of incubation under the standard growth conditions, paromomycin resistant colonies were transferred to 96-well plates containing 250 μL of the liquid TAP media and 15 mg L−1 paromomycin in each well and were grown for 7 days. Ten microliters of each culture were transferred onto TAP agar media containing 15 mg L−1 paromomycin and further incubated under the standard conditions for 4 days. Each colony was transferred into 50 μL Chelex-100 5% (w/v) (Bio-Rad, https://www.bio-rad.com) in a well in 96-well PCR plate (VWR, https://www.vwr.com) and DNA was extracted as previously described (Cao et al., 2009). PCR was performed using gene specific primers (GSP3 5′-CTAGAACTAGTGCTGAGGCTTG and GSP4 5′-CGAAGGATCCCGCTTCAAATAC) and the HotMaster Taq DNA polymerase (ThermoFisher Scientific, https://www.thermofisher.com). PCR products were analyzed by gel electrophoresis in a tris-acetate-ethylenediaminetetraacetic acid buffer containing 1% (w/v) agarose.
Reverse Transcriptase-PCR
Total RNAs were isolated from transgenic lines grown for 5 days in liquid TAP media until the cell density reached approximately 1 × 107 cells mL−1 using the TRIzol reagent (Invitrogen, https://www.thermofisher.com) according to the manufacturer's instructions. Concentrations of purified RNAs were determined spectrophotometrically at 260 nm. One microgram of each sample was reverse transcribed into cDNA using iScript cDNA Synthesis kit (Bio-Rad) following the manufacturer's instruction. The following gene-specific forward primers were used: MR11 (5′-CTCATGCTGGACCGCCGC) and MR12 (5′-TTCTCGAACTGCGGGTGGCT). PCR was carried out using HotMaster Taq DNA Polymerase. PCR products were analyzed by gel electrophoresis as described above.
Immunoblotting
The UVM4 strain and transgenic lines were grown for 5 days in TAP media under the standard conditions and 2 mL of each culture were centrifuged at 3,500 ×g for 5 min at the room temperature. The cell pellet was resuspended in 200 μL of water. The supernatant was transferred to a fresh tube and proteins were precipitated by mixing with 4 mL of pre-cooled acetone, followed by centrifugation at 12,000 ×g for 10 min at 4°C. The precipitate was air dried and dissolved in 200 μL of water. The resulting sample was mixed with four-fold concentrated Laemmlii Sample Buffer (Bio-Rad) in a 3:1 ratio and incubated at 95°C for 5 min. Forty five microliters of the mixture were loaded in a lane of a 12% Criterion™ TGX Stain-Free™ Protein Gel (Bio-Rad), and, after electrophoresis, proteins were transferred to a poly(vinylidene fluoride) membrane. Immunodetection of CrAXE was carried out using a strep-tag antibody (https://www.qiagen.com) and polyclonal rabbit anti-mouse antibodies conjugated with a horse-radish peroxidase (Dako, https://www.agilent.com). Probed proteins were visualized using the chemiluminescence substrate SuperSignal™ (ThermoFisher Scientific, Inc.).
Acetylesterase Activity Assay
Microalgal strains were grown in liquid TAP media under the standard conditions. Acetylesterase activity was monitored as previously reported (Chung et al., 2002). One milliliter of each culture was centrifuged at 3,500 ×g for 5 min at the room temperature and 15 μL of the supernatant were transferred to a well in a 96-well plate. Subsequently, 285 μL of 100 mM potassium phosphate buffer, pH 7.0, containing 35 μM 4-nitrophenyl acetate was added. The mixture was incubated at 37°C and the changes in the absorbance at 410 nm was recorded using a Spectra MAX 190 plate reader (Molecular Devices, https://www.moleculardevices.com). One unit of enzyme activity is defined as the amount of enzyme catalyzing the hydrolysis of 1 μmol substrate per min per milliliter of sample.
Determination of Biomass-Bound Acetylester Contents
Ten milligrams of the biomass materials were suspended in 1 mL of 1 M NaOH and incubated for 1 h at 22°C. The mixture was centrifuged at 3,500 ×g for 5 min and the supernatant was filtered through a 0.22-μm pore-size filter hydrophobic membrane. Acetic acid was detected by HPLC featuring a Synergi Hydro-RP (Phenomenex, http://www.phenomenex.com) column equipped with SecurityGuard Guard Cartridge (Phenomenex) and the diode-array detector SPD-M20A (Shimadzu, http://www.shimadzu.com). To elute analytes, 20 mM K3PO4, pH 2.9, was used at the constant flow rate of 0.7 mL min−1 at 22°C. Quantification was performed by integrating 210-nm peak area and comparing it with a standard curve generated using varying concentrations of acetic acid.
Results
Non-engineered C. reinhardtii Cannot Directly Utilize Wheat Biomass as the Carbon Source
To test if lignocellulosic biomass can be used as a carbon source for C. reinhardtii, the wild-type C. reinhardtii was cultivated in minimal medium supplemented with 15 g l−1 of wheat straw biomass (MM+Biomass) in the dark. The wild type grew only slightly in MM+Biomass. No difference was observed between growth in MM+Biomass and that in MM (Figure 1A), indicating that wheat biomass did not support growth of the wild-type C. reinhardtii in the dark. Control experiments showed that the wild type grew robustly in TAP media in the dark. Next, the FUD16 mutant strain of C. reinhardtii was used to test heterotrophic growth using wheat biomass. This strain is defective in photoautotrophic growth and requires organic carbons for growth, thus allowing us to evaluate heterotrophic assimilation of wheat straw by the microalga (Blifernez-Klassen et al., 2012). Similar to the results described above, the FUD16 strain grew only slightly in MM and MM+Biomass. In contrast, when MM was supplemented with 15 g l−1 of carboxymethyl cellulose (MM+CMC), the FUD16 was able to grew as previously reported (Blifernez-Klassen et al., 2012) (Figure 2). These results indicate that C. reinhardtii is not capable of utilizing cellulose in lignocellulosic biomass as a carbon source, possibly because hemicelluloses and lignins occlude cellulolytic enzymes produced by the microalga.
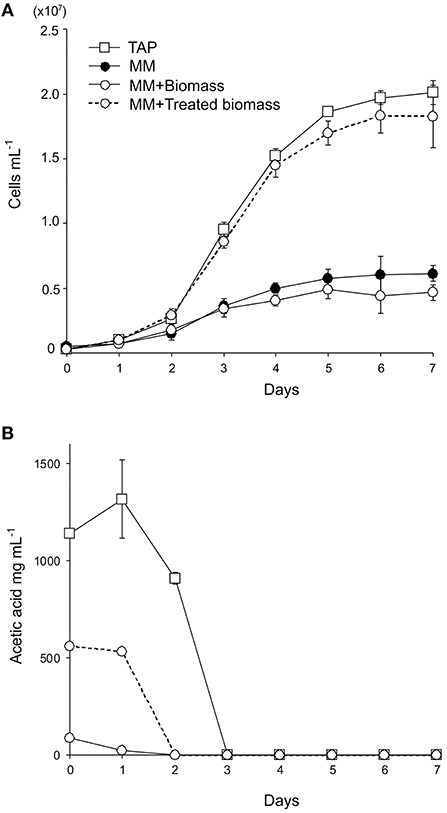
Figure 1. Heterotrophic cultivation of C. reinhardtii using wheat straw biomass. (A) Growth curves of the wild-type strain in enriched media (TAP media, square symbols with a solid line), minimal medium (MM, filled circles with a solid line), MM containing 15 g L−1 wheat straw biomass (MM+Biomass, open circles with a solid line), and MM containing 15 g L−1 alkaline-treated wheat straw biomass (MM+Treated biomass, open circles with a dotted line). (B) Acetate contents in the wild-type C. reinhardtii cultures in TAP, MM+Biomass, and MM+Treated biomass. Three independent cultures were analyzed for each conditions. Average values and standard deviations are shown.
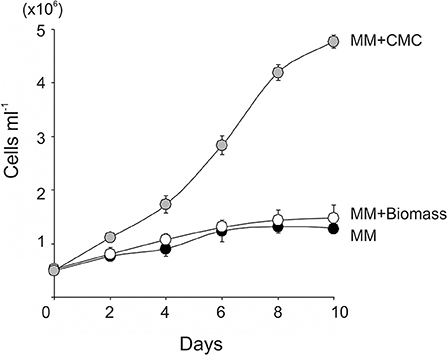
Figure 2. Growth of the photosynthesis deficient strain (FUD16) of C. reinhardtii using wheat biomass as a carbon source. The FUD16 strain was cultivated in the TAP, the MM containing 15 g L−1 of CMC (MM+CMC), and the MM containing 15 g L−1 of wheat biomass (MM+Biomass). Three independent cultures were analyzed for each condition. Average values and standard deviations are shown.
Previously, wild-type C. reinhardtii was shown to assimilate acetate that was released from softwood following fast pyrolysis treatments (Liang et al., 2013; Hu et al., 2016). We tested if acetate released from wheat straw biomass can support growth of the microalga. Wheat biomass was treated with alkaline (1 M NaOH) and added to MM to generate “MM+Treated biomass” at the biomass content of 15 g l−1. The pH value of the media was adjusted to 7.0. The wild-type C. reinhardtii grew similarly well in MM+Treated biomass to that in TAP media (Figure 1B). Acetates in the culture media were completely removed by C. reinhardtii by day 3 of the cultivation, indicating that the wild-type C. reinhardtii can assimilate acetates chemically released from wheat biomass.
Generation and Characterization of C. reinhardtii Secreting the CrAXE Enzyme
In order to circumvent uses of NaOH and to establish an eco-friendly and self-sustained growth system, we engineered C. reinhardtii to secrete an AXE enzyme for enzymatic hydrolysis of biomass acetylesters. An AXE from the fungus A. nidulans (AnAXE), belonging to the carbohydrate esterase family 1 in Carbohydrate Active Enzyme database (www.cazy.org), was chosen for expression in C. reinhardtii. This enzyme has been successfully expressed in a broad range of heterologous hosts (Bauer et al., 2006; Pogorelko et al., 2013; Mai-Gisondi et al., 2015). The codon usage of the AnAXE gene was optimized according to the genome codon usage of C. reinhardtii (Puigbò et al., 2007). The native signal sequence was predicted to be within the first 19 acid residues of the AnAXE protein by in silico analysis using SignalP (http://www.cbs.dtu.dk/services/SignalP/) (Petersen et al., 2011) and was replaced by the signal sequence of gametolysin from C. reinhardtii to ensure efficient secretion of the recombinant protein (Ramos-Martinez et al., 2017). The design of the expression gene construct, CrAXE, is shown in Figure 3A. The C. reinhardtii UVM4 strain was used for transformation because of its advantages in reduced gene silencing and high heterologous gene expression (Neupert et al., 2009).
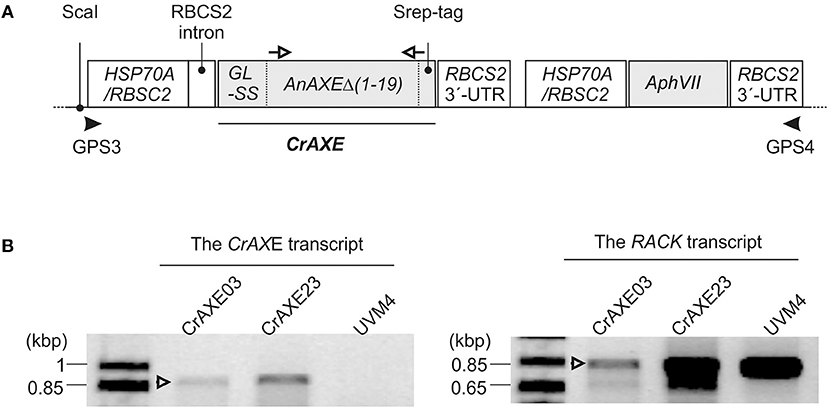
Figure 3. Generation of C. reinhardtii transgenic strains expressing the CrAXE transcript. (A) Schematic illustrations of the gene constructs. The following non-translated genetic elements were included in order to enhance transcription and translation of the recombinant protein: the HSP70A/RBSC2 promoter, the RBCS2 intron, and the RBCS2 3′ UTR, as previously described in Ramos-Martinez et al. (2017). GL-SS indicates the signal sequence derived from gametolysin of C. reinhardtii as described in the afore-mentioned reference and is fused to the truncated AnAXE lacking its native signal sequence and a strep tag for detection of the recombinant protein. Primers used for genotyping by PCR and gene specific primers use for RT-PCR are shown by black arrowheads and open arrows, respectively. (B) An agarose gel electrophoretic analysis of RT-PCR products derived from the transgenic strains CrAXE03 and CrAXE23. The binding sites of oligonucleotide primers used for RT-PCR analysis are indicated as white arrows in panel A. The expected product size is 859 bp. As a loading control, amplification of a house-keeping gene, RACK, was performed.
Following transformation with the DNA construct and selection in the presence of paromomycin, 288 colonies were screened by PCR for each construct for the presence of the full-length construct (Figure 3A). The presence of the entire expression cassette, spanning the promoter and the resistance gene, in the genome was confirmed for 158 independent colonies. The expression of the CrAXE transcript in 24 transgenic strains, and two transgenic strains, CrAXE03 and CrAXE23, were shown to express the CrAXE transcript (Figure 3B).
Secretion of the CrAXE Enzyme From Transgenic C. reinhardtii Strains
Expression of the CrAXE protein was tested for the transgenic strains CrAXE03 and CrAXE23 alongside the UVM4 strain as a control. Four-day cultures of these strains were separated into cells and cell-free media and immunoblotting was performed using a strep-tag antibody. Both transgenic strains showed a strong band at ~35 kDa in the media fraction and a very faint band at ~33 kDa in the cell fraction (Figure 4A). As expected, the UVM4 did not show any detectable products. The expected molecular weight of CrAXE following cleavage of the gametolysin signal sequence is 33.4 kDa. But, as previously reported, C. reinhardtii appears to perform yet uncharacterized post-translational modifications, causing increases of apparent molecular weights by ca. 2 kDa (Rasala et al., 2012; Ramos-Martinez et al., 2017). Thus, it was concluded that the 35 kDa product in the media fraction represents the secreted CrAXE protein with a post-translational modification and the 33 kDa product in the cell fractions represents nascent and unmodified CrAXE protein en route for secretion.
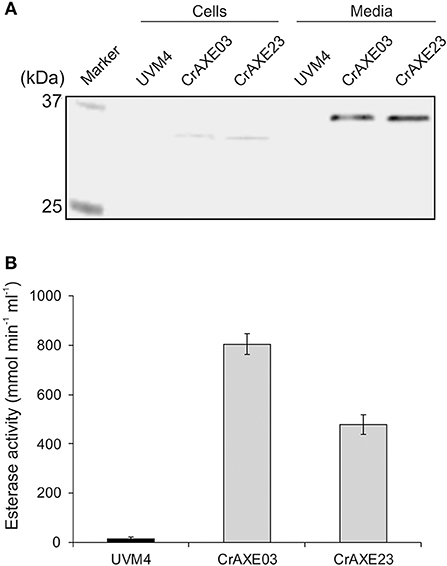
Figure 4. Secretion of the CrAXE protein by the transgenic strains of C. reinhardtii. (A) Immunoblot analysis of cell lysates (Cells) and cell-free media fractions (Media). (B) Acetylesterase activity in cell-free media fractions. The UVM strain and the transgenic strains CrAXE03 and CrAXE23 were grown for 4 days in TAP media under the standard growth conditions. Two milliliters of each culture were centrifuged and the cell pellet and the media fraction were recovered. For immunoblotting, the cell pellet was resuspended in 200 μL of water. Proteins in the media fraction were precipitated by cold acetone and resuspended in 200 μL of water. The equal volume of each material was loaded on a SDS-PAGE gel and immunoblotting was performed using a monoclonal anti-strep antibody. Activity assays were performed directly for the media fraction using 4-nitrophenyl acetate.
Acetylesterase activity in cell-free media fractions was measured as previously described (Chung et al., 2002). The transgenic strains CrAXE03 and CrAXE23 showed acetylesterase activity of 800 and 478 μmol min−1 mL−1, respectively (Figure 4B). The difference between the two lines may be caused by a difference in terms of genome integration sites and/or a number of integrated transgene (Eichler-Stahlberg et al., 2009; Ramos-Martinez et al., 2017). The media fraction derived from the UVM4 strain showed a minor activity close to the detection limit. Taken together, these results demonstrate that the CrAXE03 and CrAXE23 strains secrete active CrAXE enzyme into media.
Characterization of the Transgenic C. reinhardtii in TAP Media
Growth of the transgenic strains CrAXE03 and CrAXE23 were investigated alongside the UVM4 strain. The transgenic strains grew slower than the UVM4 strain by day 2 (one-way ANOVA, P < 0.05), but they reached similar cell counts to that of the UVM4 strain by day 3 (Figure 5A). Thus, the production of the CrAXE protein imposes a temporal burden for the growth of C. reinhardtii during the early growth phase but the transgenic lines can reach comparable cell counts as that of the UVM4 strain in the late and stationary growth phases. Acetylesterase activity in the culture media of the transgenic lines increased as the cell counts increased, with the CrAXE03 strain showing higher activity than the CrAXE23 strain (ANOVA, P < 0.05, Figure 5B). In contrast, the UVM4 strain showed activity that was marginally above the detection limit (Figure 5B).
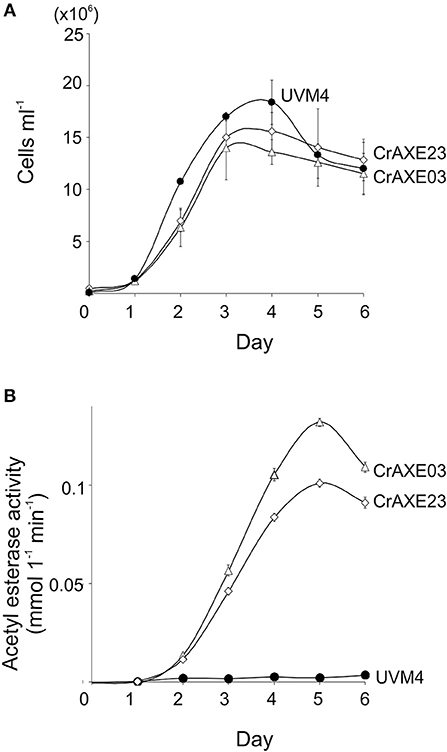
Figure 5. Characterization of the transgenic strains CrAXE03 and CrAXE23. (A) Growth curves. (B) Acetylesterase activity in the media as a function of time (days). Three independent cultures were analyzed for each genotype. Standard errors are shown. Two-way ANOVA showed that the growth curves and acetylesterase activities between the UVM4 strains and the transgenic strains (CrAXE02 and CrAXE23) were significantly different (P < 0.05).
Heterotrophic Growth of C. reinhardtii Using Wheat Straw Biomass
We then tested if the CrAXE03 strain could grow heterotrophically in MM+Biomass. The CrAXE03 line increased cell counts by eight-folds in the MM+Biomass media in the dark, whereas the UVM4 strain showed only marginal growth under the same conditions (Figure 6A). When the UVM4 strain was supplemented with a commercial acetylxylan esterase at the final concentration of 2 U mL−1, it was able to grow to the level similar to that of the CrAXE03 strain. Acetylester contents in biomass materials were determined at the end of the cultivation period (day 10) and compared to the level detected in untreated biomass. A small decrease of acetylesters, by 6%, was found for the biomass treated with the UVM4 strain, possibly due to a basal level of non-specific esterase activities that were observed for the UVM4 culture (Figure 4B). A significantly larger reduction, by 81%, was observed when the UVM4 culture was supplemented with the commercial acetylxylan esterase (Figure 6B). A greater reduction, by 96%, of acetylester contents was observed for the biomass treated with the CrAXE03 strain (Figure 6B). Taken together, these results demonstrate that secretion of an AXE enables C. reinhardtii to sequester acetate from acetylesters in wheat straw biomass and assimilate it for growth.
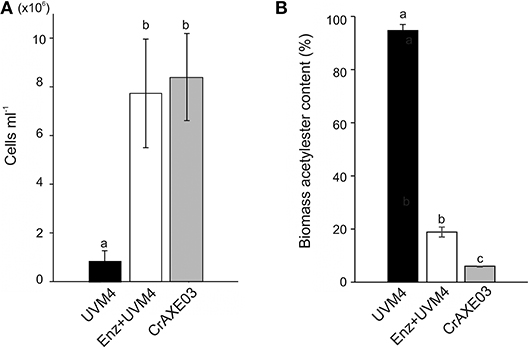
Figure 6. Cultivation of the UVM4 strain and the transgenic strain CrAXE03 of C. reinhardtii in the presence of wheat biomass. (A) Cell numbers. (B) Acetylester content in the wheat straw biomass. The strains were cultivated in the minimal medium containing wheat biomass (MM+Biomass) for 10 days. “Enz+” indicates that a commercial acetylxylan esterase at the concentration of 2 U mL−1 was added at the start of cultivation. a, b, and c indicate statistically significant difference (P < 0.05).
Discussion
In this study, we showed that secretion of an AXE by C. reinhardtii enabled the microalga to utilize acetylesters in wheat straw as a carbon source. Notably, non-engineered C. reinhardtii strains (the wild type, the FUD16 strain, and the UVM4 strain) were not able to grow under the same conditions, indicating that recalcitrance of wheat straw hinders assimilation of cellulose (Pauly and Keegstra, 2010). Direct utilization of lignocellulosic biomass has been reported for A. protothecoides UTEX 25, likely through assimilation of carbohydrates in the biomass (Vogler et al., 2018), but assimilation of acetylesters has not been. Several industrially important microalga, such as Chlorella vulgaris and Haematococcus pluviaris, utilize acetate as an organic carbon source (Kobayashi et al., 1993; Estévez-Landazábal et al., 2013). Thus, the approach presented in this study could be applied for a broader range of microalgae, paving a path toward more robust cultivation of microalga.
It is worth noting that treatment of wheat straw biomass by AXE-secreting C. reinhardtii caused decreases of acetylesters content in the biomass. It is widely known that acetylesters in lignocellulosic biomass impede saccharification of lignocellulose and acetates released from acetylesters significantly inhibit fermentation of saccharification products by microorganisms like yeast (Casal et al., 1996; Helle et al., 2003; Selig and Himmel, 2009). Notably, a reduction of 20% in the acetate concentration after biomass pretreatment would result increases in the ethanol yields by about 10% and corresponding reduction in the ethanol price (Klein-Marcuschamer et al., 2010). Several approaches have been exploited to solve the acetylester/acetate issues, including extraction by organic solvents (Aghazadeh and Engelberth, 2016) and genetic engineering of plants for reducing contents of acetylation (Lee et al., 2011; Manabe et al., 2011, 2013; Pogorelko et al., 2013; Xiong et al., 2013; Ratke et al., 2015; Schultink et al., 2015). While these approaches are beneficial, they also suffer from negative environmental impacts due to usage of chemical solvents or regulatory restrictions imposed on agricultural cultivation of genetically-modified plants. Thus, direct application of the engineered C. reinhardtii culture to lignocellulosic biomass could potentially offer an eco-friendly and flexible way for reducing biomass acetylester contents, thereby improving renewable biofuel productions.
For advancing the present approach to industrial applications, some issues need to be resolved, particularly those relating to host strains. The experiments in this study were carried out using the C. reinhardtii UVM4 strain because it offers low gene silencing activity (Neupert et al., 2009). But this strain also lacks the cell wall. The lack of the cell wall may be advantageous because the secreted protein is directly deposited into the culture media without having to pass through the cell wall matrix as in the case of the wild type, but it could also render the cell more prone to stresses imposed by mixing of cultures (i.e., shear stress) and possible toxic compounds in media. These aspects are yet to be rigorously examined in large-scale growth facilities relevant for industrial applications. As for uses of industrially important microalgae as the host organisms, such as those mentioned above, establishment of efficient transformation protocols and genetic tools are prerequisite and is still to come. Lastly and not least, genetic instability is a key issue and is widely observed for nuclear genome transformation in C. reinhardtii, where expression of transgenes typically diminishes over generations. Unfortunately, underlining causes of genetic instability are not fully understood. Thus, future research opportunities in microalgal biotechnology are abundant and will likely cover wide topics from carbon sources, development of robust transformation and genetic tools, optimization of growth facility, and discoveries of ways to overcome genetic instability.
Data Availability
All datasets generated for this study are included in the manuscript and the supplementary files.
Author Contributions
EMR-M, LF, and YS conceived the study. EMR-M, LF, and MKR performed experiments. EMR-M, LF, MKR, and YS performed data analysis. EMR-M and YS prepared the original draft. All authors contributed to the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fbioe.2019.00035/full#supplementary-material
References
Aghazadeh, M., and Engelberth, A. S. (2016). Techno-economic analysis for incorporating a liquid-liquid extraction system to remove acetic acid into a proposed commercial scale biorefinery. Biotechnol. Prog. 32, 971–977. doi: 10.1002/btpr.2325
Almaraz-Delgado, A. L., Flores-Uribe, J., Pérez-España, V. H., Salgado-Manjarrez, E., and Badillo-Corona, J. A. (2014). Production of therapeutic proteins in the chloroplast of Chlamydomonas reinhardtii. AMB Express 4:57. doi: 10.1186/s13568-014-0057-4
Baier, T., Kros, D., Feiner, R. C., Lauersen, K. J., Müller, K. M., and Kruse, O. (2018). Engineered fusion proteins for efficient protein secretion and purification of a human growth factor from the green microalga Chlamydomonas reinhardtii. ACS Synth. Biol. 7, 2547–2557. doi: 10.1021/acssynbio.8b00226
Bauer, S., Vasu, P., Persson, S., Mort, A. J., and Somerville, C. R. (2006). Development and application of a suite of polysaccharide-degrading enzymes for analyzing plant cell walls. Proc. Natl. Acad. Sci. U.S.A. 103, 11417–11422. doi: 10.1073/pnas.0604632103
Blifernez-Klassen, O., Klassen, V., Doebbe, A., Kersting, K., Grimm, P., Wobbe, L., et al. (2012). Cellulose degradation and assimilation by the unicellular phototrophic eukaryote Chlamydomonas reinhardtii. Nat. Commun. 3:1214. doi: 10.1038/ncomms2210
Bryksin, A. V., and Matsumura, I. (2010). Overlap extension PCR cloning: a simple and reliable way to create recombinant plasmids. Biotechniques 48, 463–465. doi: 10.2144/000113418
Cao, M., Fu, Y., Guo, Y., and Pan, J. (2009). Chlamydomonas (Chlorophyceae) colony PCR. Protoplasma 235, 107–110. doi: 10.1007/s00709-009-0036-9
Casal, M., Cardoso, H., and Leão, C. (1996). Mechanisms regulating the transport of acetic acid in Saccharomyces cerevisiae. Microbiology 142, 1385–1390. doi: 10.1099/13500872-142-6-1385
Chávez, M. N., Schenck, T. L., Hopfner, U., Centeno-Cerdas, C., Somlai-Schweiger, I., Schwarz, C., et al. (2016). Towards autotrophic tissue engineering: photosynthetic gene therapy for regeneration. Biomaterials 75, 25–36. doi: 10.1016/j.biomaterials.2015.10.014
Chiaiese, P., Palomba, F., Tatino, F., Lanzillo, C., Pinto, G., Pollio, A., et al. (2011). Engineered tobacco and microalgae secreting the fungal laccase POXA1b reduce phenol content in olive oil mill wastewater. Enzyme Microb. Technol. 49, 540–546. doi: 10.1016/j.enzmictec.2011.06.002
Chung, H. J., Park, S. M., Kim, H. R., Yang, M. S., and Kim, D. H. (2002). Cloning the gene encoding acetyl xylan esterase from Aspergillus ficuum and its expression in Pichia pastoris. Enzyme Microb. Technol. 31, 384–391. doi: 10.1016/S0141-0229(02)00122-9
Díaz-Santos, E., Vila, M., Vigara, J., and León, R. (2016). A new approach to express transgenes in microalgae and its use to increase the flocculation ability of Chlamydomonas reinhardtii. J. Appl. Phycol. 28, 1611–1621. doi: 10.1007/s10811-015-0706-2
Eichler-Stahlberg, A., Weisheit, W., Ruecker, O., and Heitzer, M. (2009). Strategies to facilitate transgene expression in Chlamydomonas reinhardtii. Planta 229, 873–883. doi: 10.1007/s00425-008-0879-x
Estévez-Landazábal, L. L., Barajas-Solano, A. F., Barajas-Ferreira, C., and Kafarov, V. (2013). Improvement of lipid productivity on Chlorella vulgaris using waste glycerol and sodium acetate. CT&F - Ciencia, Tecnología y Futuro. 5, 113–116.
Gangl, D., Zedler, J. A. Z., Rajakumar, P. D., Ramos-Martinez, E. M., Riseley, A., Włodarczyk, A., et al. (2015). Biotechnological exploitation of microalgae. J. Exp. Bot. 66, 6975–6990. doi: 10.1093/jxb/erv426
Gorman, D. S., and Levine, R. P. (1965). Cytochrome f and plastocyanin: their sequence in the photosynthetic electron transport chain of Chlamydomonas reinhardi. Proc. Natl. Acad. Sci. U.S.A. 54, 1665–1669. doi: 10.1073/pnas.54.6.1665
Harris, E. H. (ed.). (2009). “Introduction to Chlamydomonas and its laboratory use,” in The Chlamydomonas Sourcebook, (Oxford: Academic Press), 1–444.
Helle, S., Lam, J., White, B., Duff, S., and Cameron, D. (2003). Effect of inhibitory compounds found in biomass hydrolysates on growth and xylose fermentation by a genetically engineered strain of S. cerevisiae. Enzym. Microb. Tech. 33, 786–792. doi: 10.1016/S0141-0229(03)00214-X
Hellwig, S., Drossard, J., Twyman, R. M., and Fischer, R. (2004). Plant cell cultures for the production of recombinant proteins. Nat. Biotechnol. 22, 1415–1422. doi: 10.1038/nbt1027
Hu, J., Lin, Y., Zhang, Z., Xiang, T., Mei, Y., Zhao, S., et al. (2016). High-titer lactic acid production by Lactobacillus pentosus FL0421 from corn stover using fed-batch simultaneous saccharification and fermentation. Bioresour. Technol. 214, 74–80. doi: 10.1016/j.biortech.2016.04.034
Jinkerson, R. E., and Jonikas, M. C. (2015). Molecular techniques to interrogate and edit the Chlamydomonas nuclear genome. Plant J. 82, 393–412. doi: 10.1111/tpj.12801
Ketchner, S. L., Drapier, D., Olive, J., Gaudriault, S., Girard-Bascou, J., and Wollman, F. A. (1995). Chloroplasts can accommodate inclusion bodies. Evidence from a mutant of Chlamydomonas reinhardtii defective in the assembly of the chloroplast ATP synthase. J Biol Chem. 270, 15299–15306. doi: 10.1074/jbc.270.25.15299
Kindle, K. L. (1998). High-frequency nuclear transformation of Chlamydomonas reinhardtii. Methods Enzymol. 297, 27–38. doi: 10.1016/S0076-6879(98)97005-7
Klein-Marcuschamer, D., Oleskowicz-Popiel, P., Simmons, B. A., and Blanch, H. W. (2010). Technoeconomic analysis of biofuels: A wiki-based platform for lignocellulosic biorefineries. Biomass Bioenergy 34, 1914–1921. doi: 10.1016/j.biombioe.2010.07.033
Kobayashi, M., Kakizono, T., and Nagai, S. (1993). Enhanced carotenoid biosynthesis by oxidative stress in acetate-induced cyst cells of a green unicellular alga, Haematococcus pluvialis. Appl. Environ. Microbiol. 59, 897–873.
Kricka, W., Fitzpatrick, J., and Bond, U. (2014). Metabolic engineering of yeasts by heterologous enzyme production for degradation of cellulose and hemicellulose from biomass: a perspective. Front. Microbiol. 5:174. doi: 10.3389/fmicb.2014.00174
Lauersen, K. J., Kruse, O., and Mussgnug, J. H. (2015). Targeted expression of nuclear transgenes in Chlamydomonas reinhardtii with a versatile, modular vector toolkit. Appl. Microbiol. Biotechnol. 99, 3491–3503. doi: 10.1007/s00253-014-6354-7
Lauersen, K. J., Vanderveer, T. L., Berger, H., Kaluza, I., Mussgnug, J. H., Walker, V. K., et al. (2013). Ice recrystallization inhibition mediated by a nuclear-expressed and -secreted recombinant ice-binding protein in the microalga Chlamydomonas reinhardtii. Appl. Microbiol. Biotechnol. 97, 9763–9772. doi: 10.1007/s00253-013-5226-x
Laursen, T., Naur, P., and Møller, B. L. (2013). Amphipol trapping of a functional CYP system. Biotechnol. Appl. Biochem. 60, 119–127. doi: 10.1002/bab.1092
Lee, C., Teng, Q., Zhong, R., and Ye, Z. H. (2011). The four Arabidopsis reduced wall acetylation genes are expressed in secondary wall-containing cells and required for the acetylation of xylan. Plant Cell Physiol. 52, 1289–1301. doi: 10.1093/pcp/pcr075
Lee, C.-R., Sung, B. H., Lim, K.-M., Kim, M.-J., Sohn, M. J., Bae, J.-H., et al. (2017). Co-fermentation using recombinant Saccharomyces cerevisiae yeast strains hyper-secreting different cellulases for the production of cellulosic bioethanol. Sci. Rep. 7:4428. doi: 10.1038/s41598-017-04815-1
Liang, Y., Si, T., Ang, E. L., and Zhao, H. (2014). Engineered pentafunctional minicellulosome for simultaneous saccharification and ethanol fermentation in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 80, 6677–6684. doi: 10.1128/AEM.02070-14
Liang, Y., Zhao, X., Rover, M., Johnston, P., Brown, R., Jarboe, L., et al. (2013). Utilization of acetic acid-rich pyrolytic bio-oil by microalgae Chlamydomonas reinhardtii: reducing bio-oil toxicity and enhancing algal toxicity tolerance. Bioresour. Technol. 133, 500–506. doi: 10.1016/j.biortech.2013.01.134
Lingg, N., Zhang, P., Song, Z., and Bardor, M. (2012). The sweet tooth of biopharmaceuticals: importance of recombinant protein glycosylation analysis. Biotechnol. J. 7, 1462–1472. doi: 10.1002/biot.201200078
Mai-Gisondi, G., Turunen, O., Pastinen, O., Pahimanolis, N., and Master, E. R. (2015). Enhancement of acetyl xylan esterase activity on cellulose acetate through fusion to a family 3 cellulose binding module. Enzyme Microb. Technol. 79–80, 27–33. doi: 10.1016/j.enzmictec.2015.07.001
Manabe, Y., Nafisi, M., Verhertbruggen, Y., Orfila, C., Gille, S., Rautengarten, C., et al. (2011). Loss-of-function mutation of REDUCED WALL ACETYLATION2 in Arabidopsis leads to reduced cell wall acetylation and increased resistance to Botrytis cinerea. Plant Physiol. 155, 1068–1078. doi: 10.1104/pp.110.168989
Manabe, Y., Verhertbruggen, Y., Gille, S., Harholt, J., Chong, S. L., Pawar, P. M., et al. (2013). Reduced wall acetylation proteins play vital and distinct roles in cell wall O-acetylation in Arabidopsis. Plant Physiol. 163, 1107–1117. doi: 10.1104/pp.113.225193
Mathieu-Rivet, E., Kiefer-Meyer, M.-C., Vanier, G., Ovide, C., Burel, C., Lerouge, P., et al. (2014). Protein N-glycosylation in eukaryotic microalgae and its impact on the production of nuclear expressed biopharmaceuticals. Front. Plant Sci. 5:359. doi: 10.3389/fpls.2014.00359
Mussgnug, J. H. (2015). Genetic tools and techniques for Chlamydomonas reinhardtii. Appl. Microbiol. Biotechnol. 99, 5407–5418. doi: 10.1007/s00253-015-6698-7
Neupert, J., Karcher, D., and Bock, R. (2009). Generation of Chlamydomonas strains that efficiently express nuclear transgenes. Plant J. 57, 1140–1150. doi: 10.1111/j.1365-313X.2008.03746.x
Nikolov, Z. L., and Woodard, S. L. (2004). Downstream processing of recombinant proteins from transgenic feedstock. Curr. Opin. Biotechnol. 15, 479–486. doi: 10.1016/j.copbio.2004.08.006
Pauly, M., and Keegstra, K. (2010). Plant cell wall polymers as precursors for biofuels. Curr. Opin. Plant Biol. 13, 305–312. doi: 10.1016/j.pbi.2009.12.009
Petersen, T. N., Brunak, S., von Heijne, G., and Nielsen, H. (2011). SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods 8, 785–786. doi: 10.1038/nmeth.1701
Pogorelko, G., Lionetti, V., Fursova, O., Sundaram, R. M., Qi, M., Whitham, S. A., et al. (2013). Arabidopsis and Brachypodium distachyon transgenic plants expressing Aspergillus nidulans acetylesterases have decreased degree of polysaccharide acetylation and increased resistance to pathogens. Plant Physiol. 162, 9–23. doi: 10.1104/pp.113.214460
Puigbò, P., Guzmán, E., Romeu, A., and Garcia-Vallvé, S. (2007). OPTIMIZER: a web server for optimizing the codon usage of DNA sequences. Nucleic Acids Res. 35, 126–131. doi: 10.1093/nar/gkm219
Ramos-Martinez, E. M., Fimognari, L., and Sakuragi, Y. (2017). High-yield secretion of recombinant proteins from the microalga Chlamydomonas reinhardtii. Plant Biotechnol. J. 15, 1214–1224. doi: 10.1111/pbi.12710
Rasala, B. A., Lee, P. A., Shen, Z., Briggs, S. P., Mendez, M., and Mayfield, S. P. (2012). Robust expression and secretion of Xylanase1 in Chlamydomonas reinhardtii by fusion to a secretion gene and processing with the FMDV 2A peptide. PLoS ONE 7:e43349. doi: 10.1371/journal.pone.0043349
Rasala, B. A., and Mayfield, S. P. (2015). Photosynthetic biomanufacturing in green algae; production of recombinant proteins for industrial, nutritional, and medical uses. Photosynth. Res. 123, 227–239. doi: 10.1007/s11120-014-9994-7
Ratke, C., Pawar, P. M.-A., Balasubramanian, V. K., Naumann, M., Duncranz, M. L., Derba-Maceluch, M., et al. (2015). Populus GT43 family members group into distinct sets required for primary and secondary wall xylan biosynthesis and include useful promoters for wood modification. Plant Biotechnol. J. 13, 26–37. doi: 10.1111/pbi.12232
Schroda, M., Blocker, D., and Beck, C. F. (2000). The HSP70A promoter as a tool for the improved expression of transgenes in Chlamydomonas. Plant J. 21, 121–131. doi: 10.1046/j.1365-313x.2000.00652.x
Schultink, A., Naylor, D., Dama, M., and Pauly, M. (2015). The role of the plant-specific ALTERED XYLOGLUCAN9 protein in Arabidopsis cell wall polysaccharide O-acetylation. Plant Physiol. 167, 1271–1283. doi: 10.1104/pp.114.256479
Selig, A. W., and Himmel, M. M. (2009). The impact of cell wall acetylation on corn stover hydrolysis by cellulolytic and xylanolytic enzymes. Cellulose 16, 711–722. doi: 10.1007/s10570-009-9322-0
Sizova, I., Lapina, T., Frolova, O. N., Alexandrova, N. N., Akopiants, K. E., and Danilenko, V. N. (1996). Stable nuclear transformation of Chlamydomonas reinhardtii with a Streptomyces rimosus gene as the selective marker. Gene 181, 13–18.
Sun, J., Wen, F., Si, T., Xu, J. H., and Zhao, H. (2012). Direct conversion of xylan to ethanol by recombinant Saccharomyces cerevisiae strains displaying an engineered minihemicellulosome. Appl. Environ. Microbiol. 78, 3837–3845. doi: 10.1128/AEM.07679-11
Vogler, B. W., Starkenburg, S. R., Sudasinghe, N., Schambach, J. Y., Rollin, J. A., Pattathil, S., et al. (2018). Characterization of plant carbon substrate utilization by Auxenochlorella protothecoides. Algal Res. 34, 37–48. doi: 10.1016/j.algal.2018.07.001
Keywords: chlamydomonas reinhardtii, protein secretion, acetylxylan esterase, lignocellulosic biomass, direct application of engineered algae
Citation: Ramos-Martinez EM, Fimognari L, Rasmussen MK and Sakuragi Y (2019) Secretion of Acetylxylan Esterase From Chlamydomonas reinhardtii Enables Utilization of Lignocellulosic Biomass as a Carbon Source. Front. Bioeng. Biotechnol. 7:35. doi: 10.3389/fbioe.2019.00035
Received: 03 December 2018; Accepted: 08 February 2019;
Published: 28 February 2019.
Edited by:
Maurycy Daroch, Peking University, ChinaReviewed by:
Md. Rezaul Islam Khan, Pohang University of Science and Technology, South KoreaAnchalee Sirikhachornkit, Kasetsart University, Thailand
Klaas J. Jan Hellingwerf, University of Amsterdam, Netherlands
Copyright © 2019 Ramos-Martinez, Fimognari, Rasmussen and Sakuragi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yumiko Sakuragi, eXNhQHBsZW4ua3UuZGs=
†Present Address: Erick Miguel Ramos-Martinez, Aliga ApS, Hjørring, Denmark
Lorenzo Fimognari, Chr. Hansen A/S, Plant Health Innovation, Hørsholm, Denmark
‡These authors have contributed equally to this work
 Erick Miguel Ramos-Martinez
Erick Miguel Ramos-Martinez Lorenzo Fimognari†‡
Lorenzo Fimognari†‡ Maria K. Rasmussen
Maria K. Rasmussen Yumiko Sakuragi
Yumiko Sakuragi