- 1Department of Science, University Roma Tre, Rome, Italy
- 2Department of Biological and Environmental Sciences and Technologies (DiSTeBA), University of Salento, Lecce, Italy
The bottom-up branch of synthetic biology includes—among others—innovative studies that combine cell-free protein synthesis with liposome technology to generate cell-like systems of minimal complexity, often referred to as synthetic cells. The functions of this type of synthetic cell derive from gene expression, hence they can be programmed in a modular, progressive and customizable manner by means of ad hoc designed genetic circuits. This experimental scenario is rapidly expanding and synthetic cell research already counts numerous successes. Here, we present a review focused on the exchange of chemical signals between liposome-based synthetic cells (operating by gene expression) and biological cells, as well as between two populations of synthetic cells. The review includes a short presentation of the “molecular communication technologies,” briefly discussing their promises and challenges.
Molecular Communications and Synthetic Cells (SCs)
Natural organisms coordinate their activities through communication. Isolated cells, tissue cells, as well as higher organisms, share their environment with other living forms. Tactile, physical, and especially chemical signals define in unique and complex manner the sensory world of living beings. Communications in the chemical domain are ubiquitous intercellular processes, and play important roles in all organisms.
Inspired by the already mentioned capabilities of natural organisms, a new branch of biomimetic technology has been proposed which focuses on molecular communications (Nakano et al., 2011, 2013). Network engineers have envisioned the exploitation of chemical exchanges as the basis for developing new types of Information and Communication Technologies (the so-called bio-chem-ICTs, Figure 1A). This is an exciting new arena for engineers and biologists that aims at the construction of well-characterized biological parts, devices, and systems that will process chemical information in a controlled and programmable manner, as it happens with classical electric signals. The challenge, here, relies on the ability of managing communication and information processing through chemical signals with the same mastery as nature has done for billions of years. Such a broad and innovative territory of research offers several opportunities for various approaches to synthetic biology, which needs adequate theoretical frameworks, numerical modeling strategies, and experimental methodologies. More generally, bio-chem-ICTs refers to radically new forms of computation, communication, and information processing approaches—at the nano- and micro-scale levels—based on chemical and biochemical systems (Amos et al., 2011).
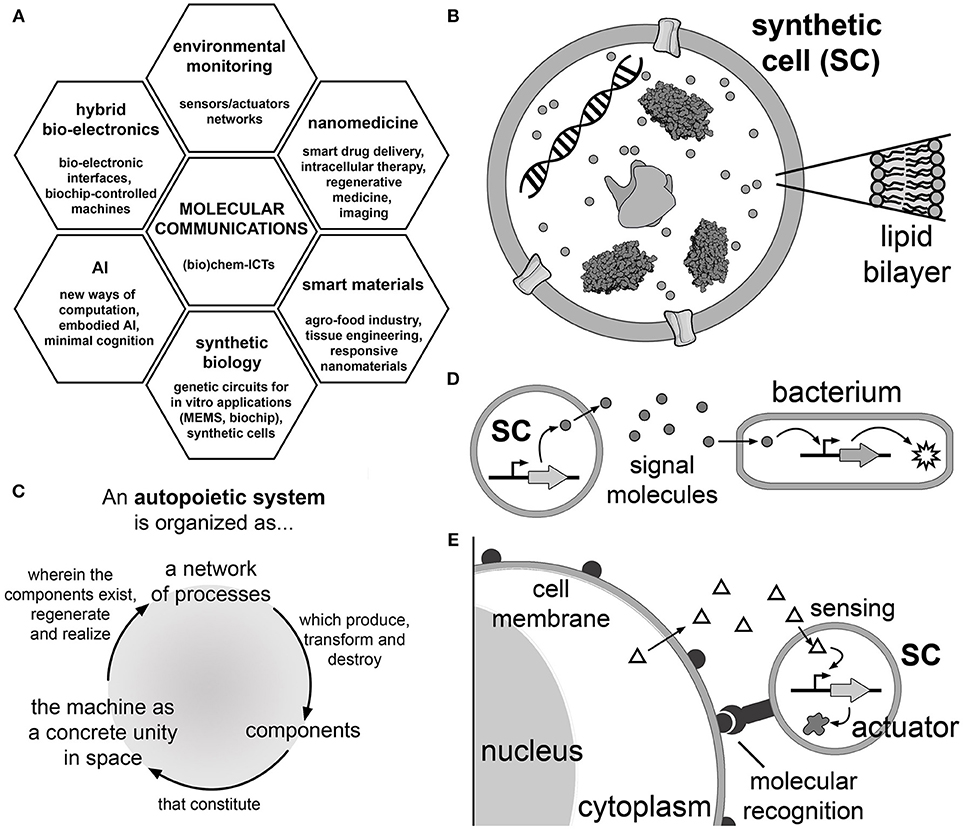
Figure 1. Molecular communications based on synthetic cell (SC) technology. (A) Application areas of molecular communication research. Molecular communication is a (bio)chem-information and communication technology that can be applied to nanomedicine (smart drug delivery systems), smart responsive materials, synthetic biology (construction of biochips), artificial intelligence (AI), hybrid bio-electronic systems and for sensors in environmental monitoring (Nakano et al., 2013). (B) Synthetic cells are cell-like systems, generally built by encapsulating a number of (bio)molecular components into artificial micro-compartments. One of the possible designs focuses on liposome-based SCs operating by gene expression (Luisi, 2002; Luisi et al., 2006). With this aim, TX-TL kits produce the protein(s) of interest starting from the corresponding DNA sequence. The SC membrane can be functionalized with membrane proteins as pores (Noireaux and Libchaber, 2004) and receptors (Hamada et al., 2014); cytoskeletal proteins have been implemented as well (Maeda et al., 2012). (C) The principles of autopoiesis (self-production) (Varela et al., 1974), which guides the long-term goal of constructing SCs that produce all their components. Autopoiesis provides insights into the spatial and dynamical organization that a chemical system should be endowed with in order to display self-maintenance, organizational closure, homeostasis and reproduction achieved by the internal processes of manufacturing and assembling its components. (D) Schematic representation of a SC which produces and releases a signal molecule into the environment. The signal is perceived by a natural cell (e.g., a bacterium) that consequently activates a response (for example, a reporter protein, an enzyme operating as an actuator to perform a certain operation, including a reply signaling) (Nakano et al., 2011; Stano et al., 2012). Table 1 reports several cases of unidirectional or bidirectional molecular communications between SCs, or between SCs and natural cells. (E) The vision of using SCs as smart drug delivery systems or for enzyme replacement therapy (Leduc et al., 2007). SCs, intended as a biotechnological evolution of current liposomes for drug delivery, reach and bind to the target cells by a molecular recognition mechanism and activate their internal circuits responding to chemical stimuli and consequently act, in a programmable manner, for a certain task (e.g., producing a therapeutic or diagnostic agent Ding et al., 2018; Krinsky et al., 2018, or a secondary easy-to-detect signal, etc.). The chemical stimulus can be an endogenous chemical that derives from the target cell itself (as shown in the cartoon) or from other tissues (not shown), as well as purposely-added exogenous chemicals (not shown).
Owing to our direct involvement in the field (Stano et al., 2012; Rampioni et al., 2014, 2018), and considering recent exciting reports, in this review we present and discuss the intersection between the bio-chem-ICT idea of exchanging chemical signals in a programmable way, and the bottom-up synthetic biology approach focused on the construction of cell-like systems based on gene expression inside liposomes (Luisi, 2002; Noireaux and Libchaber, 2004; Luisi et al., 2006; Ichihashi et al., 2010; Stano et al., 2011; Nourian and Danelon, 2013; Spencer et al., 2013). For simplicity, we will shortly refer to these systems simply as “synthetic cells” (SCs, Figure 1B), keeping in mind that these are rather simple mimics of biological cells.
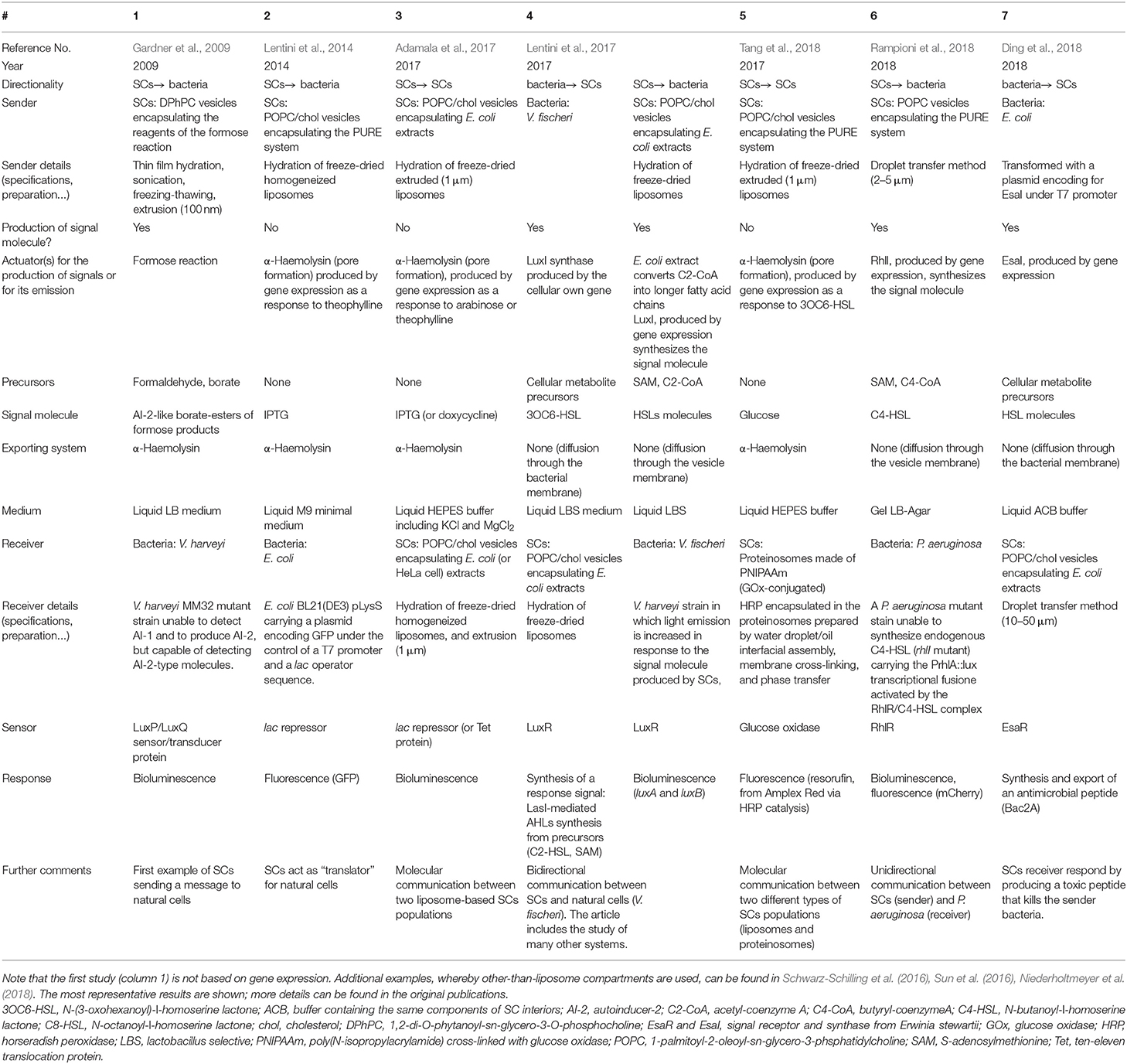
Table 1. Comparative analysis of six experimental reports based on liposome SCs that send/receive chemical signals (2009–2018).
In this mini-review, the principles on which liposome-based SCs operate will be summarized, together with an explanation of the reason why they could contribute significantly to molecular communication technologies on account of their inherent possibilities in terms of design, modeling, control, programmability, and modularity. Next, recent experimental reports focused on chemical communication between SCs and natural cells (or with other SCs) will be reviewed (see also Lentini et al., 2016), while the opportunities and challenges facing this novel research arena will be discussed in the final section.
Before advancing in the discussion, two notes of warning are intended for readers unfamiliar with this research field. Firstly, the term “synthetic cell” is also used in synthetic biology to indicate living cells generated either by engineering biological cells (e.g., metabolic engineering, genetic optimization, or reprogramming), as well as by the transplantation of an entire synthetic genome in a living cell deprived of its own genome. Second, bottom-up synthetic biology approaches aiming at constructing cell-like systems are not restricted to liposome-based SCs. No less interesting are systems based on other types of compartments (Walde et al., 1994; Martino et al., 2012; Huang et al., 2014; Karzbrun et al., 2014; Dora Tang et al., 2015; Rideau et al., 2018), nor those based on new artificial molecules (Kurihara et al., 2011; Marguet et al., 2013; Taylor et al., 2015). Interested readers can refer to recent reviews for a broader discussion (Buddingh and van Hest, 2017; Salehi-Reyhani et al., 2017; Göpfrich et al., 2018; Schwille et al., 2018). The current review will focus only on SCs based on gene expression inside liposomes.
Basic Principles on Liposome-Based SCs Operating Via Gene Expression
SCs based on gene expression inside liposomes find their origin in early studies on cell models aiming at achieving minimal life-like behaviors (Morowitz et al., 1988; Luisi and Varela, 1989; Schmidli et al., 1991; Oberholzer et al., 1995a,b, 1999; Szostak et al., 2001; Luisi, 2002; Pohorille and Deamer, 2002; Mansy and Szostak, 2009). Born within the origins-of-life community, this research was intended as a means of investigating the emergence of life on Earth, more precisely by demonstrating the emergence of life as a system-level phenomenon due to a particular type of organization (the autopoietic one). Hence, the autopoietic (self-production) (Varela et al., 1974; Luisi and Varela, 1989; Luisi, 2003) (Figure 1C) and the chemoton theories (chemical automaton) (Gánti, 1975) are two valuable theoretical frameworks for the construction of SCs which display features of biological organisms. Starting in the first years of 2000, SCs and similar constructs became highly relevant also in the context of synthetic biology, either as tools for generating basic knowledge, or as systems designed for applied research, i.e., biotechnology and nanomedicine.
The SCs discussed in this review are liposomes, with a size ranging typically from 0.1 to 10–100 μm: they contain DNA and a cell-free gene expression system. They are made by assembling liposomes in an aqueous phase which contains all the molecules needed to be encapsulated for accomplishing protein synthesis from a DNA template (e.g., enzymes, ribosomes, tRNAs, nucleotides, amino acids etc.). The protein synthesis machinery can derive from a cell extract or from a reconstituted system [such as the PURE system (Shimizu et al., 2001)]. Accordingly, it can be noted that SC technology is based on liposome technology (including microfluidics) and cell-free systems (including biochemical reconstitution approaches). As a result of the reactions occurring in their aqueous lumen and/or on their boundary surface, SCs can display behavior(s) typical of living cells. For example, SCs produce proteins from a corresponding gene; in turn, the synthesized protein can be an enzyme that converts substrates into products, or it can be a pore-forming protein, creating pores on the liposome membrane, or it can be a receptor that binds a signal molecule, etc. More in general, SCs can be functionalized with any chemical network of biological relevance that is functional in vitro.
Several reactions different from gene expression have been successfully performed inside liposomes, confirming the potentiality of SCs in terms of scope, programmability, and functionality. Some examples are: PCR and RT-PCR (Oberholzer et al., 1995a; Shohda et al., 2011; Lee et al., 2014; Tsugane and Suzuki, 2018), DNA replication (Sakatani et al., 2018; van Nies et al., 2018), and several enzymatic reactions. Moreover cytoskeletal elements have been reconstituted inside SCs (Cabré et al., 2013; Furusato et al., 2018; Litschel et al., 2018). Ad hoc designed gene circuits lead to SCs that can perform useful operations in a programmable way, including communication, as discussed below. SCs with the capacity of self-producing all their own constitutive components, and which possibly grow-and-divide as living cells do, are still missing, although interesting reports that show progress in this directions have been published (Kurihara et al., 2011).
This mini-review focuses on SCs capable of communicating with biological cells and with each other. However, other interesting research directions are under current development, including the construction of SCs with nested design (Deng et al., 2017; York-Duran et al., 2017; Hindley et al., 2018), the production of ATP inside SCs (Feng et al., 2016; Altamura et al., 2017; Lee et al., 2018), the attempts of self-producing SC parts (Schmidli et al., 1991; Kuruma et al., 2009; Scott et al., 2016; Li et al., 2017; Exterkate et al., 2018), and the shift from isolated SCs to “SC communities”, including tissue-like structures (Carrara et al., 2012; Hadorn et al., 2013; Booth et al., 2016).
SCs That Exchange Chemical Signals: a Bottom-up Synthetic Biology Platform for Molecular Communications
SCs based on gene expression inside liposomes can be useful tools for developing molecular communication technologies (Stano et al., 2012). Current SC technology allows building simple systems capable of exchanging chemical signals, and therefore performing elementary signal processing. The idea is to design SCs capable of communicating with each other or with biological cells in a programmable manner (Figure 1D). This innovative perspective has multifold theoretical and practical consequences. From the theoretical viewpoint, SCs that can regulate their internal mechanisms in response to external perturbations (the chemical signaling) are de facto experimental tools for investigating minimal cognitive systems (Damiano and Stano, 2018a,b). Considering the proposed extension of the Turing imitation game to the SC realm (Cronin et al., 2006), molecular communication can contribute to the determination of life-likeness criteria as referred to SCs, as recently investigated by the Sheref Mansy group (Lentini et al., 2017). In a more practical perspective, an expansion of actual drug delivery strategies can be proposed. Inspired by the scenario depicted by Leduc and collaborators (Figure 1E) (Leduc et al., 2007), SCs could activate internal mechanisms upon perception of chemical signals, thus acting as “intelligent” drug carriers. As an example, SCs could be targeted to specific cells (e.g., tumoural cells) by exploiting antigen-antibody recognition. Once localized, their internal genetic circuit could be activated by chemical stimuli produced by the target cell itself or by other endogenous or exogenous chemical signals. These “smart” SCs could produce and release therapeutics (or drugs) in situ. Note that a recent study has reported SCs (injected into the tumor) that constitutively produce a toxin against breast cancer cells (Krinsky et al., 2018). The therapeutic (or diagnostic) use of SCs is, today, still a hypothetic scenario. Nevertheless, continuous improvements in SC design and construction is expected to favor a more rapid prototyping, thus accelerating the path toward applicative purposes.
Sensors, Actuators, Controllers, and Molecular Diffusion
Like hardware robots or conventional communication devices, SCs are embodied systems composed of molecular elements that perform specific operations. Hardware components, such as sensors, controllers, and actuators (Mataric, 2007; Wang et al., 2013) have their molecular counterparts in SCs.
In the context of SCs operating by gene expression, sensors can be protein receptors or RNA aptamers that bind to a signal molecule and consequently change their conformation. This event directly or indirectly affects the “controller system,” which is based on the regulation of gene expression by protein receptors or RNA aptamers (riboswitches) at the transcriptional or translational level, respectively. These mechanisms are well-understood (Alberts et al., 2014). Depending on its design, the regulatory circuit can involve a single gene or multiple genes. As a result of this sensing-and-regulation system, the synthesis of an actuator (a protein) is promoted or inhibited. In turn, the actuator operates on some further step (e.g., producing a signal molecule, catalyzing a useful reaction, creating a pore on the SC membrane, acting as a controller/regulator of another circuit, etc.). Key examples of this general mechanism will be commented on in section A Survey of Published Reports and listed in Table 1.
To provide SCs with communication capability, water-soluble proteins (sensors, regulators, signal-producing elements, or components of the gene expression machinery) should be either encapsulated, or synthesized in the SC lumen. This has become a standard practice, somehow, at least for some prokaryotic proteins (Stano et al., 2011). It is not trivial, instead, dealing with membrane-associated and integral membrane sensors/receptors, even if reports have shown that this is a feasible goal in SCs technology (strategies as membrane protein reconstitution Yanagisawa et al., 2011; Altamura et al., 2017; Jørgensen et al., 2017 or synthesis-from-within Kuruma et al., 2009; Hamada et al., 2014; Soga et al., 2014 have been employed). Genetic circuits of distinctive complexity have already been proven to be functional, also inside liposomes (Noireaux et al., 2003; Shin and Noireaux, 2012; Siegal-Gaskins et al., 2014).
In addition to molecular elements, in order to establish an intercellular communication channel, diffusion of the signal molecule in the outer aqueous environment should be taken into account. The signal molecule cannot be directed toward the communication partner, but it spreads in all direction, guided by the concentration gradient. Although the average behavior of many signal molecules can be foreseen, individual molecules follow an erratic path. In addition to free diffusion, for closely packed SCs, communication through gap junctions (reconstituted in liposomes) has been proposed (Ramundo-Orlando et al., 2005; Moritani et al., 2010).
A Survey of Published Reports
The pioneer experimental report on a simple cell-like system sending a signal molecule to biological cells was published by the Ben Davis group (Gardner et al., 2009). The authors encapsulated the precursors of the formose reaction inside liposomes, and observed that one class of products of the intra-vesicular reaction escaped the liposomes through a channel formed by α-haemolysin and spontaneously reacted with the borate ions present in the external medium to generate furanosyl-boronates structurally similar to the quorum sensing (QS) signal molecule AI-2, that naturally triggers bioluminescence in Vibrio harveyi. Remarkably, the “synthetic” signal released by the liposome was able to induce natural behavior (i.e., light emission) in this bacterium.
Despite its great interest as proof of the concept study, the SCs used by Ben Devis and co-workers were not based on gene expression, therefore they lacked those aspects of programmability and control that are peculiar to synthetic biology. Being a novel research area, literature on the liposome-based SCs which operate by gene expression to interface with natural cells (or with other SCs) is, to the best of our knowledge, limited to the six studies that are summarized in Table 1 together with the already cited study by Gardner et al. (2009). Additional cases involving non-liposome compartments are also available (Gupta et al., 2013; Schwarz-Schilling et al., 2016; Sun et al., 2016; Niederholtmeyer et al., 2018), but these will not be discussed in this mini-review.
In 2014, Sheref Mansy and collaborators designed SCs acting as “translators” for the bacterium Escherichia coli, using theophylline as trigger and isopropyl β-D-1-thiogalactopyranoside (IPTG) as signal molecule (Lentini et al., 2014). These SCs are liposomes containing IPTG, the PURE system as the transcription-translation (TX-TL) machinery, and a DNA template coding for a riboswitch that, after binding to the free-diffusible molecule theophylline, activated the expression of the pore forming protein α-haemolysin. The authors demonstrated that only in the presence of theophylline, did IPTG escape the liposomes through α-haemolysin, and activate the expression of the green fluorescent protein (GFP) gene in receiver E. coli cells. In this way, SCs acted as chemical translators allowing E. coli to sense theophylline (the latter molecule cannot be normally sensed by E. coli).
Adamala et al. (2017) built SCs containing engineered genetic circuits and regulatory cascades. These SCs can be controlled/triggered by external signals, and can be fused together in order to bring together products of incompatible reactions. In particular, the group lead by Edward Boyden showed that by using cell lysates with transcriptional-translational activity, DNA vectors encoding genes for IPTG (or doxycycline) detection and permeable chemical inducers, as arabinose or theophylline, the arabinose (or theophylline) activates the α-haemolysin production in the first SC population, so that pre-encapsulated impermeable IPTG (or doxycycline) could be released, and thus activate a response in a second SC population.
The group of Sheref Mansy recently reported two-way chemical communication between SCs and bacteria (Lentini et al., 2017). They exploited cell extracts to generate SCs able to synthesize molecules perceived by Vibrio fischeri, V. harveyi, E. coli, and Pseudomonas aeruginosa. In particular, the expression of LuxI-like synthases inside liposomes, in the presence of acetyl coenzyme A and S-adenosylmethionine (SAM), resulted in the production of molecules able to activate E. coli and V. fischeri-based biosensor strains for acyl-homoserine lactone (AHLs) detection. Cell extracts operated both for TX-TL reactions and for the synthesis of some AHL precursors. Moreover, it was shown that SCs containing ad hoc designed genetic circuits could express QS signal molecule receptors able to trigger the expression of reporter and QS signal synthase genes (e.g., gfp and luxI), upon perception of QS signal molecules produced by bacteria. The extent to which SCs could “imitate” natural cells in term of their response to the investigated QS signal molecule was estimated by a sort of cellular Turing test (Cronin et al., 2006).
The signaling between liposome-based SCs and proteinosomes (cell-like particles made of proteins) mediated by glucose, has been recently reported by a joint work of the groups of Sheref Mansy and Stephen Mann (Tang et al., 2018). In this study, the unidirectional signaling pathway was based on: (i) liposome transmitters, containing the PURE system, a DNA plasmid carrying a chemically inducible repression switch (EsaR), a gene coding for α-haemolysin, and glucose; (ii) proteinosome receivers, consisting of a cross-linked enzymatically active glucose oxidase (Gox)-poly(N-isopropylacrylamide) (PNIPAAm) membrane and encapsulated horseradish peroxidase (HRP). The addition of the permeable AHL molecule N-(3-oxohexanoyl)-L-homoserine lactone (3OC6-HSL) triggered intravesicular α-hemolysin expression and consequent membrane pore formation in liposome-based SCs, which allowed the release of glucose contained in the aqueous lumen. Glucose oxidation on the proteinosome membrane produced hydrogen peroxide, which in turn converted a molecule into a fluorescent output by reacting with the HRP encapsulated in proteinosome. This study provides an example of molecular communication between two different types of artificial cell-like systems.
A recent report comes from our laboratory, and it deals with unidirectional SC–P. aeruginosa communication, based on the QS AHL signal molecule C4-HSL (Rampioni et al., 2018). In particular, SCs were prepared by encapsulating the PURE system inside GVs prepared by the droplet transfer method (Pautot et al., 2003; Fujii et al., 2014), together with butyryl coenzyme A and SAM as precursors, and a plasmid encoding for RhlI, the synthase for C4-HSL production. SCs produced C4-HSL (a natural QS signal molecule), which was perceived by P. aeruginosa both in liquid medium and in gel. In particular, P. aeruginosa modified its gene expression pattern in response to the C4-HSL-produced by SCs, demonstrating that reprogramming of gene expression in the bacterial cell is similar when interacting with other bacteria or with SCs. The entire TX-TL mechanism was assessed by rhlI mRNA and RhlI protein quantification, as well as by chemical identification of the C4-HSL signal produced by the SCs. The experimental results interestingly match with previously published numerical modeling (Rampioni et al., 2014), confirming the predictive power of in silico simulations in SCs research.
Finally, the Tan group reported an interesting study where SCs and bacteria engaged unidirectional communication in various ways (SCs to SCs, bacteria to SCs, and SCs to bacteria) (Ding et al., 2018). In this case, the QS signal molecule was produced via the EsaI synthase, and was perceived by the cognate EsaR receptor. Gene expression in SCs was triggered when binding of the QS signal molecule to EsaR led to derepression of an EsaR-controlled promoter region. Quite interestingly, the authors designed SCs that produce an antimicrobial peptide (Bac2A) in response to QS signal molecules sent by bacteria—a proof of principle of the use of signal processing and actuation dynamics for the generation of SCs interfacing with natural cells. Moreover, SCs embedded in biofilms were also reported.
Directions and Challenges for Future Work
The works compared in Table 1 represent proof-of-concept pioneer works that will likely stimulate further research to expand SC capabilities related to molecular communications. In this context, several challenges and open questions can be envisaged. Some refers to mechanistic, biochemical and biological aspects, others to the capability of engineering molecular communications.
With respect to the mechanisms of molecular communication, “sender” and “receiver” SCs mainly relied on transmembrane diffusion of signal molecules. This simple approach has been effective because some QS signal molecules, such as short-tail AHLs, can cross the lipid bilayer (Pearson et al., 1999). The generation of α-haemolysin pores is a drastic (yet effective) solution that has been used to bypass the low permeability of SC membranes when non free-diffusible signal molecules have been used (e.g., IPTG or glucose), but this causes the release of all the low-MW compounds contained inside SCs (the cut-off molecular weight value for the α-haemolysin pore is 3 kDa; Song et al., 1996). An alternative could be the use of DNA nanopores, whose properties are tunable by design (Krishnan et al., 2016). The future employment of more sophisticated import/export mechanisms based on membrane proteins will allow expanding the chemical repertoire of signal molecules secreted or perceived by SCs (e.g., peptides), thus increasing the communication capability and specificity. In this respect, ongoing progress on the functionalization of SC envelopes with integral membrane proteins is promising (see section Basic Principles on Liposome-Based SCs Operating via Gene Expression).
Looking at the biological partners of SCs for molecular communications, early studies focused on bacteria, since they are prone to genetic engineering and their intercellular communication systems have been thoroughly studied at the molecular level, especially in the case of QS systems. From a practical viewpoint, SC/bacteria communication is a technological platform for the long-term goal of interfering with bacterial populations and for therapeutic strategies that could be devised against infections. Indeed, the ability of SCs to drive gene expression in response to external cues envisages the generation of injectable SCs endowed with the ability to produce or release an antimicrobial compound only in response to a signal molecule produced by a bacterial pathogen. The study reported in Table 1 by the Tan group (Ding et al., 2018) has provided a proof-of-principle that SCs can be generated which are able to kill bacteria by a mechanism triggered by the bacteria themselves.
Proving that SCs can communicate with eukaryotic cells is one of the next milestones, especially when nanomedicine applications are devised. This complex task could require the generation of SCs with internal operations that rely on eukaryotic signal synthesis or more complex signal reception machineries. The relevance of these approaches is that SCs could be employed as intelligent drug-delivery systems that perform a therapeutic action by extracting information from their microenvironment. As mentioned, the generation of SCs constitutively producing a tumor-killing protein (the Pseudomonas exotoxin A) has been recently described (Krinsky et al., 2018). Another task would involve enzyme replacement therapy (Itel et al., 2017). For example, SCs that consume excess phenylalanine could play a therapeutic role in phenylketonuria (Leduc et al., 2007). Notably, Thomas M. S. Chang proposed in a pre-liposome age the therapeutic use of enzyme-containing semi-permeable collodion capsules circulating in the bloodstream (Chang, 1964, 1972). The generation of SC interfacing via molecular communication with neural cells can also be imagined. The resulting hybrid bio/synthetic cell networks could also be exploited for innovative investigations of neural functions (Pinato et al., 2011).
Considering the engineering plan of networking SCs (or SCs and biological cells), the rigorous design of molecular communication channels requires a proper modeling of the physical and information levels. At the physical level stochastic diffusion plays a central role. This peculiar aspect is the ultimate limit of molecular communication (when compared to traditional electro-magnetic systems) because it is essentially a random process. Intercellular molecular communications rely on diffusion of chemical signals under a concentration gradient. They are, therefore, slow stochastic processes; their success depends on a number of factors, like the sender/receiver ratio, their spatial arrangement, the viscosity of the medium, and the temperature. Numerical models can be useful to understand the limiting factors and the constraints operating at this (inescapable) physical level (Nakano et al., 2011, 2013). The stochastic dimension of molecular communications affects its reliability. Facing with it represents an engineering challenge. The second aspect refers to the amount of information transmitted in the molecular communication “channel,” and this is a theoretical issue. To apply classical information and communication theory to such a novel scenario, “information” should be defined with respect to the type of signal molecules, number of sent/received molecules, and time-dependent concentration profile (switch-like, pulse-like, etc.). Control theory for bottom-up synthetic biology should be delineated (Del Vecchio et al., 2016). Its peculiarity stems from molecular discreteness, random timing of sending/receiving, nature of “noise,” etc.
In conclusion, SCs could significantly contribute to the origin of a very novel research field based on communication with biological cells. Thanks to their modular constructive principle, their biocompatibility and programmability, SCs of the type discussed in this review have the unique ability to act as passive carriers of hydrophilic and hydrophobic drugs, and to actively drive gene expression in response to chemical stimuli from other cells and from the environment.
At present, main challenges in this field rely on our capacity of (i) designing and build multi-functional SCs based on a proper genetic circuit and auxiliary molecular parts/devices, (ii) building homogeneous populations of SCs that are stable in biological fluids, (iii) and being able to control SC behavior even in a complex and fluctuating environment, such as a human host. All these challenges will probably be solved in the near future thanks to constant improvements in SC technology (in a broad sense, i.e., not necessarily restricted to liposomes). Along this path, there will be room for developing various systems in which in vitro usage will generate opportunities for understanding principles of biological systems and constructing short-term devices (e.g., biosensors).
Author Contributions
PS conceived the research, all authors wrote the paper.
Funding
The authors acknowledge the Italian Ministry for Education, University and Research, MIUR-Italy for the FIRB2010 project No. RBFR10LHD1_002, and for the Grant of Excellence Departments (Art. 1, c. 314-337, Legge 232/2016).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Adamala, K. P., Martin-Alarcon, D. A., Guthrie-Honea, K. R., and Boyden, E. S. (2017). Engineering genetic circuit interactions within and between synthetic minimal cells. Nat. Chem. 9, 431–439. doi: 10.1038/nchem.2644
Alberts, B., Johnson, A. D., Lewis, J., Morgan, D., Raff, M., Roberts, K., et al. (2014). Molecular Biology of the Cell. 6th Edn. New York, NY: W. W. Norton & Company.
Altamura, E., Milano, F., Tangorra, R. R., Trotta, M., Omar, O. H., Stano, P., et al. (2017). Highly oriented photosynthetic reaction centers generate a proton gradient in synthetic protocells. Proc. Natl. Acad. Sci. U.S.A. 114, 3837–3842. doi: 10.1073/pnas.1617593114
Amos, M., Dittrich, P., McCaskill, J., and Rasmussen, S. (2011). Biological and chemical information technologies. Procedia Computer Sci. 7, 56–60. doi: 10.1016/j.procs.2011.12.019
Booth, M. J., Schild, V. R., Graham, A. D., Olof, S. N., and Bayley, H. (2016). Light-activated communication in synthetic tissues. Sci. Adv. 2:e1600056. doi: 10.1126/sciadv.1600056
Buddingh, B. C., and van Hest, J. C. M. (2017). Artificial cells: synthetic compartments with life-like functionality and adaptivity. Acc. Chem. Res. 50, 769–777. doi: 10.1021/acs.accounts.6b00512
Cabré, E. J., Sánchez-Gorostiaga, A., Carrara, P., Ropero, N., Casanova, M., Palacios, P., et al. (2013). Bacterial division proteins FtsZ and ZipA induce vesicle shrinkage and cell membrane invagination. J. Biol. Chem. 288, 26625–26634. doi: 10.1074/jbc.M113.491688
Carrara, P., Stano, P., and Luisi, P. L. (2012). Giant vesicles “colonies”: a model for primitive cell communities. ChemBioChem 13, 1497–1502. doi: 10.1002/cbic.201200133
Chang, T. M. S. (1964). Semipermeable microcapsules. Science 146, 524–525. doi: 10.1126/science.146.3643.524
Cronin, L., Krasnogor, N., Davis, B. G., Alexander, C., Robertson, N., Steinke, J. H. G., et al. (2006). The imitation game—a computational chemical approach to recognizing life. Nat. Biotechnol. 24, 1203–1206. doi: 10.1038/nbt1006-1203
Damiano, L., and Stano, P. (2018a). Synthetic biology and artificial intelligence: grounding a cross-disciplinary approach to the synthetic exploration of (embodied) cognition. Complex Syst. 27, 199–228. doi: 10.25088/complexsystems.27.3.199
Damiano, L., and Stano, P. (2018b). Understanding embodied cognition by building models of minimal life. Preparatory steps and a preliminary autopoietic framework. Commun. Computer Info. Sci. 830, 73–87. doi: 10.1007/978-3-319-78658-2_6
Del Vecchio, D., Dy, A. J., and Qian, Y. (2016). Control theory meets synthetic biology. J. R. Soc. Interface 13:20160380. doi: 10.1098/rsif.2016.0380
Deng, N.-N., Yelleswarapu, M., Zheng, L., and Huck, W. T. S. (2017). Microfluidic assembly of monodisperse vesosomes as artificial cell models. J. Am. Chem. Soc. 139, 587–590. doi: 10.1021/jacs.6b10977
Ding, Y., Contreras-Llano, L. E., Morris, E., Mao, M., and Tan, C. (2018). Minimizing context dependency of gene networks using artificial cells. ACS Appl. Mater. Interfaces 10, 30137–30146. doi: 10.1021/acsami.8b10029
Dora Tang, T.-Y., van Swaay, D., deMello, A., Ross Anderson, J. L., and Mann, S. (2015). In vitro gene expression within membrane-free coacervate protocells. Chem. Commun. 51, 11429–11432. doi: 10.1039/c5cc04220h
Exterkate, M., Caforio, A., Stuart, M. C. A., and Driessen, A. J. M. (2018). Growing membranes in vitro by continuous phospholipid biosynthesis from free fatty acids. ACS Synth. Biol. 7, 153–165. doi: 10.1021/acssynbio.7b00265
Feng, X., Jia, Y., Cai, P., Fei, J., and Li, J. (2016). Coassembly of photosystem II and ATPase as artificial chloroplast for light-driven ATP synthesis. ACS Nano 10, 556–561. doi: 10.1021/acsnano.5b05579
Fujii, S., Matsuura, T., Sunami, T., Nishikawa, T., Kazuta, Y., and Yomo, T. (2014). Liposome display for in vitro selection and evolution of membrane proteins. Nat. Protoc. 9, 1578–1591. doi: 10.1038/nprot.2014.107
Furusato, T., Horie, F., Matsubayashi, H. T., Amikura, K., Kuruma, Y., and Ueda, T. (2018). De novo synthesis of basal bacterial cell division proteins FtsZ, FtsA, and ZipA inside giant vesicles. ACS Synth. Biol. 7, 953–961. doi: 10.1021/acssynbio.7b00350
Gánti, T. (1975). Organization of chemical reactions into dividing and metabolizing units: the chemotons. Biosystems 7, 15–21. doi: 10.1016/0303-2647(75)90038-6
Gardner, P. M., Winzer, K., and Davis, B. G. (2009). Sugar synthesis in a protocellular model leads to a cell signalling response in bacteria. Nat. Chem. 1, 377–383. doi: 10.1038/nchem.296
Göpfrich, K., Platzman, I., and Spatz, J. P. (2018). Mastering complexity: towards bottom-up construction of multifunctional eukaryotic synthetic cells. Trends Biotechnol. 36, 938–951. doi: 10.1016/j.tibtech.2018.03.008
Gupta, A., Terrell, J. L., Fernandes, R., Dowling, M. B., Payne, G. F., Raghavan, S. R., et al. (2013). Encapsulated fusion protein confers “sense and respond” activity to chitosan-alginate capsules to manipulate bacterial quorum sensing. Biotechnol. Bioeng. 110, 552–562. doi: 10.1002/bit.24711
Hadorn, M., Boenzli, E., Sørensen, K. T., De Lucrezia, D., Hanczyc, M. M., and Yomo, T. (2013). Defined DNA-mediated assemblies of gene-expressing giant unilamellar vesicles. Langmuir 29, 15309–15319. doi: 10.1021/la402621r
Hamada, S., Tabuchi, M., Toyota, T., Sakurai, T., Hosoi, T., Nomoto, T., et al. (2014). Giant vesicles functionally expressing membrane receptors for an insect pheromone. Chem. Commun. 50, 2958–2961. doi: 10.1039/c3cc48216b
Hindley, J. W., Elani, Y., McGilvery, C. M., Ali, S., Bevan, C. L., Law, R. V., et al. (2018). Light-triggered enzymatic reactions in nested vesicle reactors. Nat. Commun. 9:1093. doi: 10.1038/s41467-018-03491-7
Huang, X., Li, M., and Mann, S. (2014). Membrane-mediated cascade reactions by enzyme-polymer proteinosomes. Chem. Commun. 50, 6278–6280. doi: 10.1039/c4cc02256d
Ichihashi, N., Matsuura, T., Kita, H., Sunami, T., Suzuki, H., and Yomo, T. (2010). Constructing partial models of cells. Cold Spring Harb. Perspect. Biol. 2:a004945. doi: 10.1101/cshperspect.a004945
Itel, F., Schattling, P. S., Zhang, Y., and Städler, B. (2017). Enzymes as key features in therapeutic cell mimicry. Adv. Drug Deliv. Rev. 118, 94–108. doi: 10.1016/j.addr.2017.09.006
Jørgensen, I. L., Kemmer, G. C., and Pomorski, T. G. (2017). Membrane protein reconstitution into giant unilamellar vesicles: a review on current techniques. Eur. Biophys. J. 46, 103–119. doi: 10.1007/s00249-016-1155-9
Karzbrun, E., Tayar, A. M., Noireaux, V., and Bar-Ziv, R. H. (2014). Programmable on-chip DNA compartments as artificial cells. Science 345, 829–832. doi: 10.1126/science.1255550
Krinsky, N., Kaduri, M., Zinger, A., Shainsky-Roitman, J., Goldfeder, M., Benhar, I., et al. (2018). Synthetic cells synthesize therapeutic proteins inside tumors. Adv. Healthc. Mater. 7:e1701163. doi: 10.1002/adhm.201701163
Krishnan, S., Ziegler, D., Arnaut, V., Martin, T. G., Kapsner, K., Henneberg, K., et al. (2016). Molecular transport through large-diameter DNA nanopores. Nat. Commun. 7:12787. doi: 10.1038/ncomms12787
Kurihara, K., Tamura, M., Shohda, K.-I., Toyota, T., Suzuki, K., and Sugawara, T. (2011). Self-reproduction of supramolecular giant vesicles combined with the amplification of encapsulated DNA. Nat. Chem. 3, 775–781. doi: 10.1038/nchem.1127
Kuruma, Y., Stano, P., Ueda, T., and Luisi, P. L. (2009). A synthetic biology approach to the construction of membrane proteins in semi-synthetic minimal cells. Biochim. Biophys. Acta 1788, 567–574. doi: 10.1016/j.bbamem.2008.10.017
Leduc, P. R., Wong, M. S., Ferreira, P. M., Groff, R. E., Haslinger, K., Koonce, M. P., et al. (2007). Towards an in vivo biologically inspired nanofactory. Nat. Nanotechnol. 2, 3–7. doi: 10.1038/nnano.2006.180
Lee, K. Y., Park, S.-J., Lee, K. A., Kim, S.-H., Kim, H., Meroz, Y., et al. (2018). Photosynthetic artificial organelles sustain and control ATP-dependent reactions in a protocellular system. Nat. Biotechnol. 36, 530–535. doi: 10.1038/nbt.4140
Lee, S., Koo, H., Na, J. H., Lee, K. E., Jeong, S. Y., Choi, K., et al. (2014). DNA amplification in neutral liposomes for safe and efficient gene delivery. ACS Nano 8, 4257–4267. doi: 10.1021/nn501106a
Lentini, R., Santero, S. P., Chizzolini, F., Cecchi, D., Fontana, J., Marchioretto, M., et al. (2014). Integrating artificial with natural cells to translate chemical messages that direct E. coli behaviour. Nat. Commun. 5:4012. doi: 10.1038/ncomms5012
Lentini, R., Yeh Martín, N., Forlin, M., Belmonte, L., Fontana, J., Cornella, M., et al. (2017). Two-way chemical communication between artificial and natural cells. ACS Central Sci. 3, 117–123. doi: 10.1021/acscentsci.6b00330
Lentini, R., Yeh Martín, N., and Mansy, S. S. (2016). Communicating artificial cells. Curr. Opin. Chem. Biol. 34, 53–61. doi: 10.1016/j.cbpa.2016.06.013
Li, J., Haas, W., Jackson, K., Kuru, E., Jewett, M. C., Fan, Z. H., et al. (2017). Cogenerating synthetic parts toward a self-replicating system. ACS Synth. Biol. 6, 1327–1336. doi: 10.1021/acssynbio.6b00342
Litschel, T., Ramm, B., Maas, R., Heymann, M., and Schwille, P. (2018). Beating vesicles: encapsulated protein oscillations cause dynamic membrane deformations. Angew. Chem. Int. Ed. 57, 16286–16290. doi: 10.1002/anie.201808750
Luisi, P. L. (2002). Toward the engineering of minimal living cells. Anat. Rec. 268, 208–214. doi: 10.1002/ar.10155
Luisi, P. L. (2003). Autopoiesis: a review and a reappraisal. Naturwissenschaften 90, 49–59. doi: 10.1007/s00114-002-0389-9
Luisi, P. L., Ferri, F., and Stano, P. (2006). Approaches to semi-synthetic minimal cells: a review. Naturwissenschaften 93, 1–13. doi: 10.1007/s00114-005-0056-z
Luisi, P. L., and Varela, F. J. (1989). Self-replicating micelles—A chemical version of a minimal autopoietic system. Origins Life Evol. Biosphere 19, 633–643. doi: 10.1007/BF01808123
Maeda, Y. T., Nakadai, T., Shin, J., Uryu, K., Noireaux, V., and Libchaber, A. (2012). Assembly of MreB filaments on liposome membranes: a synthetic biology approach. ACS Synth. Biol. 1, 53–59. doi: 10.1021/sb200003v
Mansy, S. S., and Szostak, J. W. (2009). Reconstructing the emergence of cellular life through the synthesis of model protocells. Cold Spring Harb. Symp. Quan. Biol. 74, 47–54. doi: 10.1101/sqb.2009.74.014
Marguet, M., Bonduelle, C., and Lecommandoux, S. (2013). Multicompartmentalized polymeric systems: towards biomimetic cellular structure and function. Chem. Soc. Rev. 42, 512–529. doi: 10.1039/c2cs35312a
Martino, C., Kim, S.-H., Horsfall, L., Abbaspourrad, A., Rosser, S. J., Cooper, J., et al. (2012). Protein expression, aggregation, and triggered release from polymersomes as artificial cell-like structures. Angew. Chem. Int. Edit. 51, 6416–6420. doi: 10.1002/anie.201201443
Mataric, M. J. (2007). The Robotics Primer. Cambridge, MA: MIT Press. Available online at: https://mitpress.mit.edu/books/robotics-primer
Moritani, Y., Nomura, S. M., Morita, I., and Akiyoshi, K. (2010). Direct integration of cell-free-synthesized connexin-43 into liposomes and hemichannel formation. FEBS J. 277, 3343–3352. doi: 10.1111/j.1742-4658.2010.07736.x
Morowitz, H. J., Heinz, B., and Deamer, D. W. (1988). The chemical logic of a minimum protocell. Orig. Life Evol. Biosph. 18, 281–287.
Nakano, T., Eckford, A. W., and Haraguchi, T. (2013). Molecular Communications. Cambridge: Cambridge University Press.
Nakano, T., Moore, M., Enomoto, A., and Suda, T. (2011). “Molecular communication technology as a biological ICT,” in Biological Functions for Information and Communication Technologies Studies in Computational Intelligence, ed H. Sawai (Berlin; Heidelberg: Springer), 49–86.
Niederholtmeyer, H., Chaggan, C., and Devaraj, N. K. (2018). Communication and quorum sensing in non-living mimics of eukaryotic cells. Nat. Commun. 9, 5027. doi: 10.1038/s41467-018-07473-7
Noireaux, V., Bar-Ziv, R., and Libchaber, A. (2003). Principles of cell-free genetic circuit assembly. Proc. Natl. Acad. Sci. U.S.A. 100, 12672–12677. doi: 10.1073/pnas.2135496100
Noireaux, V., and Libchaber, A. (2004). A vesicle bioreactor as a step toward an artificial cell assembly. Proc. Natl. Acad. Sci. U.S.A. 101, 17669–17674. doi: 10.1073/pnas.0408236101
Nourian, Z., and Danelon, C. (2013). Linking genotype and phenotype in protein synthesizing liposomes with external supply of resources. ACS Synth. Biol. 2, 186–193. doi: 10.1021/sb300125z
Oberholzer, T., Albrizio, M., and Luisi, P. (1995a). Polymerase chain-reaction in liposomes. Chem. Biol. 2, 677–682. doi: 10.1016/1074-5521(95)90031-4
Oberholzer, T., Nierhaus, K. H., and Luisi, P. L. (1999). Protein expression in liposomes. Biochem. Biophys. Res. Commun. 261, 238–241. doi: 10.1006/bbrc.1999.0404
Oberholzer, T., Wick, R., Luisi, P. L., and Biebricher, C. K. (1995b). Enzymatic RNA replication in self-reproducing vesicles: an approach to a minimal cell. Biochem. Biophys. Res. Commun. 207, 250–257. doi: 10.1006/bbrc.1995.1180
Pautot, S., Frisken, B. J., and Weitz, D. A. (2003). Production of unilamellar vesicles using an inverted emulsion. Langmuir 19, 2870–2879. doi: 10.1021/la026100v
Pearson, J. P., Van Delden, C., and Iglewski, B. H. (1999). Active efflux and diffusion are involved in transport of Pseudomonas aeruginosa cell-to-cell signals. J. Bacteriol. 181, 1203–1210.
Pinato, G., Raffaelli, T., D'Este, E., Tavano, F., and Cojoc, D. (2011). Optical delivery of liposome encapsulated chemical stimuli to neuronal cells. J. Biomed. Opt. 16:095001. doi: 10.1117/1.3616133
Pohorille, A., and Deamer, D. (2002). Artificial cells: prospects for biotechnology. Trends Biotechnol. 20, 123–128. doi: 10.1016/S0167-7799(02)01909-1
Rampioni, G., D'Angelo, F., Messina, M., Zennaro, A., Kuruma, Y., Tofani, D., et al. (2018). Synthetic cells produce a quorum sensing chemical signal perceived by Pseudomonas aeruginosa. Chem. Commun. 54, 2090–2093. doi: 10.1039/C7CC09678J
Rampioni, G., Mavelli, F., Damiano, L., D'Angelo, F., Messina, M., Leoni, L., et al. (2014). A synthetic biology approach to bio-chem-ICT: first moves towards chemical communication between synthetic and natural cells. Nat Comput. 13, 333–349. doi: 10.1007/s11047-014-9425-x
Ramundo-Orlando, A., Serafino, A., Schiavo, R., Liberti, M., and d'Inzeo, G. (2005). Permeability changes of connexin32 hemi channels reconstituted in liposomes induced by extremely low frequency, low amplitude magnetic fields. Biochim. Biophys. Acta 1668, 33–40. doi: 10.1016/j.bbamem.2004.11.003
Rideau, E., Dimova, R., Schwille, P., Wurm, F. R., and Landfester, K. (2018). Liposomes and polymersomes: a comparative review towards cell mimicking. Chem. Soc. Rev. 47, 8572–8610. doi: 10.1039/c8cs00162f
Sakatani, Y., Yomo, T., and Ichihashi, N. (2018). Self-replication of circular DNA by a self-encoded DNA polymerase through rolling-circle replication and recombination. Sci. Rep. 8:13089. doi: 10.1038/s41598-018-31585-1
Salehi-Reyhani, A., Ces, O., and Elani, Y. (2017). Artificial cell mimics as simplified models for the study of cell biology. Exp. Biol. Med. 242, 1309–1317. doi: 10.1177/1535370217711441
Schmidli, P. K., Schurtenberger, P., and Luisi, P. L. (1991). Liposome-mediated enzymatic synthesis of phosphatidylcholine as an approach to self-replicating liposomes. J. Am. Chem. Soc. 113, 8127–8130. doi: 10.1021/ja00021a043
Schwarz-Schilling, M., Aufinger, L., Mückl, A., and Simmel, F. C. (2016). Chemical communication between bacteria and cell-free gene expression systems within linear chains of emulsion droplets. Integr. Biol. 8, 564–570. doi: 10.1039/C5IB00301F
Schwille, P., Spatz, J., Landfester, K., Bodenschatz, E., Herminghaus, S., Sourjik, V., et al. (2018). MaxSynBio: avenues towards creating cells from the bottom up. Angew. Chem. Int. Ed. 57, 3382–13392. doi: 10.1002/anie.201802288
Scott, A., Noga, M. J., de Graaf, P., Westerlaken, I., Yildirim, E., and Danelon, C. (2016). Cell-Free Phospholipid Biosynthesis by Gene-Encoded Enzymes Reconstituted in Liposomes. PLoS ONE 11:e0163058. doi: 10.1371/journal.pone.0163058
Shimizu, Y., Inoue, A., Tomari, Y., Suzuki, T., Yokogawa, T., Nishikawa, K., et al. (2001). Cell-free translation reconstituted with purified components. Nat. Biotechnol. 19, 751–755. doi: 10.1038/90802
Shin, J., and Noireaux, V. (2012). An E. coli cell-free expression toolbox: application to synthetic gene circuits and artificial cells. ACS Synth. Biol. 1, 29–41. doi: 10.1021/sb200016s
Shohda, K., Tamura, M., Kageyama, Y., Suzuki, K., Suyama, A., and Sugawara, T. (2011). Compartment size dependence of performance of polymerase chain reaction inside giant vesicles. Soft Matter 7, 3750–3753. doi: 10.1039/C0SM01463J
Siegal-Gaskins, D., Tuza, Z. A., Kim, J., Noireaux, V., and Murray, R. M. (2014). Gene circuit performance characterization and resource usage in a cell-free “breadboard.” ACS Synth. Biol. 3, 416–425. doi: 10.1021/sb400203p
Soga, H., Fujii, S., Yomo, T., Kato, Y., Watanabe, H., and Matsuura, T. (2014). In vitro membrane protein synthesis inside cell-sized vesicles reveals the dependence of membrane protein integration on vesicle volume. ACS Synth. Biol. 3, 372–379. doi: 10.1021/sb400094c
Song, L., Hobaugh, M. R., Shustak, C., Cheley, S., Bayley, H., and Gouaux, J. E. (1996). Structure of staphylococcal alpha-hemolysin, a heptameric transmembrane pore. Science 274, 1859–1866.
Spencer, A. C., Torre, P., and Mansy, S. S. (2013). The encapsulation of cell-free transcription and translation machinery in vesicles for the construction of cellular mimics. J. Vis. Exp. 80:e51304. doi: 10.3791/51304
Stano, P., Carrara, P., Kuruma, Y., de Souza, T. P., and Luisi, P. L. (2011). Compartmentalized reactions as a case of soft-matter biotechnology: synthesis of proteins and nucleic acids inside lipid vesicles. J. Mater. Chem. 21, 18887–18902. doi: 10.1039/c1jm12298c
Stano, P., Rampioni, G., Carrara, P., Damiano, L., Leoni, L., and Luisi, P. L. (2012). Semi-synthetic minimal cells as a tool for biochemical ICT. BioSystems 109, 24–34. doi: 10.1016/j.biosystems.2012.01.002
Sun, S., Li, M., Dong, F., Wang, S., Tian, L., and Mann, S. (2016). Chemical signaling and functional activation in colloidosome-based protocells. Small 12, 1920–1927. doi: 10.1002/smll.201600243
Szostak, J. W., Bartel, D. P., and Luisi, P. L. (2001). Synthesizing life. Nature 409, 387–390. doi: 10.1038/35053176
Tang, T.-Y. D., Cecchi, D., Fracasso, G., Accardi, D., Coutable-Pennarun, A., Mansy, S. S., et al. (2018). Gene-mediated chemical communication in synthetic protocell communities. ACS Synth. Biol. 7, 339–346. doi: 10.1021/acssynbio.7b00306
Taylor, A. I., Pinheiro, V. B., Smola, M. J., Morgunov, A. S., Peak-Chew, S., Cozens, C., et al. (2015). Catalysts from synthetic genetic polymers. Nature 518, 427–430. doi: 10.1038/nature13982
Tsugane, M., and Suzuki, H. (2018). Reverse transcription polymerase chain reaction in giant unilamellar vesicles. Sci. Rep. 8:9214. doi: 10.1038/s41598-018-27547-2
van Nies, P., Westerlaken, I., Blanken, D., Salas, M., Mencía, M., and Danelon, C. (2018). Self-replication of DNA by its encoded proteins in liposome-based synthetic cells. Nat. Commun. 9:1583. doi: 10.1038/s41467-018-03926-1
Varela, F. G., Maturana, H. R., and Uribe, R. (1974). Autopoiesis: the organization of living systems, its characterization and a model. Biosystems 5, 187–196. doi: 10.1016/0303-2647(74)90031-8
Walde, P., Goto, A., Monnard, P., Wessicken, M., and Luisi, P. (1994). Oparins reactions revisited—enzymatic-synthesis of poly(adenylic acid). J. Am. Chem. Soc. 116, 7541–7547. doi: 10.1021/ja00096a010
Wang, Y.-H., Wei, K. Y., and Smolke, C. D. (2013). Synthetic biology: advancing the design of diverse genetic systems. Annu. Rev. Chem. Biomol. Eng. 4, 69–102. doi: 10.1146/annurev-chembioeng-061312-103351
Yanagisawa, M., Iwamoto, M., Kato, A., Yoshikawa, K., and Oiki, S. (2011). Oriented reconstitution of a membrane protein in a giant unilamellar vesicle: experimental verification with the potassium channel KcsA. J. Am. Chem. Soc. 133, 11774–11779. doi: 10.1021/ja2040859
York-Duran, M. J., Godoy-Gallardo, M., Labay, C., Urquhart, A. J., Andresen, T. L., and Hosta-Rigau, L. (2017). Recent advances in compartmentalized synthetic architectures as drug carriers, cell mimics and artificial organelles. Coll. Surf. B Biointerf. 152, 199–213. doi: 10.1016/j.colsurfb.2017.01.014
Keywords: synthetic cells, bottom-up synthetic biology, molecular communications, quorum sensing, lipid vesicles (liposomes), cell-free protein synthesis
Citation: Rampioni G, D'Angelo F, Leoni L and Stano P (2019) Gene-Expressing Liposomes as Synthetic Cells for Molecular Communication Studies. Front. Bioeng. Biotechnol. 7:1. doi: 10.3389/fbioe.2019.00001
Received: 30 September 2018; Accepted: 02 January 2019;
Published: 17 January 2019.
Edited by:
Francesca Ceroni, Imperial College London, United KingdomReviewed by:
Yo Suzuki, J. Craig Venter Institute, United StatesKerstin Göpfrich, Max Planck Institute for Medical Research (MPIMF), Germany
Copyright © 2019 Rampioni, D'Angelo, Leoni and Stano. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Pasquale Stano, cGFzcXVhbGUuc3Rhbm9AdW5pc2FsZW50by5pdA==
 Giordano Rampioni
Giordano Rampioni Francesca D'Angelo
Francesca D'Angelo Livia Leoni
Livia Leoni Pasquale Stano
Pasquale Stano