- 1Department of Oncology, Faculty of Science and Medicine, Immunology and Microbiology, University of Fribourg, Fribourg, Switzerland
- 2Laboratory of Medical and Molecular Biology, Centre National de la Recherche Scientifique, Reims, France
- 3Swiss Integrative Center for Human Health, Fribourg, Switzerland
Colorectal cancer (CRC) is the third cause of cancer-related mortality in industrialized countries. Local invasion and metastasis formation are events associated with poor prognosis for which today there are no effective therapeutic options. Invasion and metastasis are strongly modulated by cells of the tumor microenvironment (TME), in particular fibroblasts and endothelial cells. Unraveling interactions between tumor cells and cells of the TME may identify novel mechanisms and therapeutic targets to prevent or treat metastasis. We report here the development and in vivo validation of a 3D tumor spheroid model to study the interactions between CRC cells, fibroblasts and endothelial cells in vitro. Co-cultured fibroblasts promoted SW620 and HCT116 CRC spheroid invasion, and this was prevented by the SRC and FGFR kinase inhibitors Dasatinib and Erdafitinib, respectively. To validate these findings in vivo, we injected SW620 cells alone or together with fibroblasts orthotopically in the caecum of mice. Co-injection with fibroblasts promoted lung metastasis growth, which was fully reversed by treatment with Dasatinib or Erdafitinib. Co-culture of SW620 or HCT116 CRC spheroids with endothelial cells suppressed spheroid growth while it had no effect on cancer cell migration or invasion. Consistent with this in vitro effect, co-injected endothelial cells significantly inhibited primary tumor growth in vivo. From these experiments we conclude that effects on cancer cell invasion and growth induced by co-cultured TME cells and drug treatment in the 3D spheroid model in vitro, are predictive of in vivo effects. The 3D spheroid model may be considered as an attractive model to study the effect of heterotypic cellular interactions and drug activities on cancer cells, as animal testing alternative. This model may be adapted and further developed to include different types of cancer and host cells and to investigate additional functions and drugs.
Introduction
After decades of experimental and molecular cancer research, drug development and testing, cancer remains a leading cause of death worldwide (Siegel et al., 2017b). Colorectal cancer (CRC) is the third cause of cancer-related mortality in industrialized countries (Brenner et al., 2014; Siegel et al., 2017a). Next generation DNA sequencing has allowed the identification of recurrent CRC mutations thereby opening the way to a better detection, classification and treatment of CRC (Dienstmann and Tabernero, 2016). Four consensus molecular subtypes (CMS) of clinical relevance have been recently reported based on RNA expression profiling (Guinney et al., 2015). At the cellular level, however, many questions remain unsolved, in particular those concerning mechanisms of invasion and metastasis, two of the main hallmarks of cancer (Hanahan and Weinberg, 2011; Valastyan and Weinberg, 2011; Sleeman et al., 2012). This is clinically highly relevant as cancer cell migration and local invasion are the first steps toward metastatic dissemination, which will eventually determine patient outcome (Brenner et al., 2014; McAllister and Weinberg, 2014).
Invasion and metastasis are not fully cell autonomous events. Multiple elements of the tumor microenvironment (TME) play critical roles not only in supporting local tumor growth but also in promoting invasion and metastasis in several cancer types, including CRC (Gout and Huot, 2008; Lorusso and Ruegg, 2008; Joyce and Pollard, 2009; Jeon et al., 2015; Malandrino et al., 2018). Growth factors production, metabolic changes, extracellular matrix (ECM) remodeling, activation of host cells such as fibroblasts, mesenchymal stem cells and endothelial cells as well as recruitment and polarization of immune and inflammatory cells, contribute to cancer growth, local invasion and distant metastasis formation (Lorusso and Ruegg, 2008; Quail and Joyce, 2013; Kalluri, 2016). As there are no robust, curative treatment for metastatic disease, the understanding of the mechanisms used by cells of the TME and their interaction with tumor cells to promote invasion and metastasis is essential for the rational development of new treatments impinging on local invasion and metastasis formation. The knowledge about the complex signaling circuits occurring between the tumor cells and the different cell populations of the TME is growing but still incomplete (Quail and Joyce, 2013; Bhome et al., 2015). Studying heterotypic cellular interaction in vivo is limited due to constrains in accessing the tissue, the simultaneous presence of multiple cell types, and the difficulty in selectively modulating specific cell types or intercellular interactions. In addition, in vivo monitoring requires invasive procedures and time-course experiments necessitate large amounts of animals (Taketo, 2006; Clarke, 2007; Golovko et al., 2015).
In vitro 2D co-culture models mimicking cancer-stromal cell interaction are widely used to identify new therapeutic targets and study new drugs. However, 2D tissue culture conditions do not mimic well in vivo heterotypic interactions, leaving a wide gap between in vitro and in vivo models (Bartlett et al., 2014). It is now generally accepted that 3D tissue culture is the preferred way of investigating cancer cells in vitro to bridge this gap. 3D tissue culture represents a more physiological setting to study morphology, cell cycle progression, cellular interactions, gene and protein expression, invasion, migration, and tumor metabolism. This is particular relevant to drug discovery and testing of anti-cancer agents as cells have different sensitivities in 3D vs. 2D conditions, including CRC cells (Stadler et al., 2015; Weiswald et al., 2015; Pereira et al., 2016; Penfornis et al., 2017; Ravi et al., 2017; Jin et al., 2018; Langhans, 2018). In addition, in vitro 3D co-culture models constitute invaluable tools to interrogate the role of individual cells of the TME and their interactions with cancer cells in tumor progression (Herrmann et al., 2014; Thoma et al., 2014; Horie et al., 2015; Ravi et al., 2015, 2017). We previously reported a 3D spheroid in vitro model of CRC to study multicellular interactions between tumor cells and fibroblasts and used it to decipher mechanisms by which fibroblasts promote CRC invasion (Knuchel et al., 2015). We showed that cell surface presentation of fibroblasts-derived FGF-2 to cancer cells, leads to integrin αvβ5-dependent and SRC-mediated adhesion of cancer cells to fibroblasts, and contact-dependent tumor cell elongation, migration and invasion. Here we report the in vivo validation of results obtained with co-cultured fibroblasts and SRC and fibroblast growth factor receptor (FGFR) inhibitors in this 3D model in vitro. We also tested the effects of endothelial cells as additional cells to reconstitute the multicellular interactions in the TME.
Results
Dasatinib or Erdafitinib Treatment Prevents Fibroblast-Promoted CRC Cell Migration and Invasion in vitro
We previously developed a 3D CRC cell-fibroblasts co-culture model and used it to demonstrate that fibroblasts promote contact-dependent cancer cell motility and invasion. Treatment with the SRC inhibitor CGP77675 and the FGFR inhibitor PD161570 prevented these in vitro effects (Knuchel et al., 2015). These results raised the question whether fibroblasts would also promote CRC invasion/metastasis in vivo in a SCR and FGFR-dependent manner. To test this hypothesis, we used two drugs in clinical practice or clinical development: Dasatinib, a BCR/ABL and SRC family tyrosine kinases inhibitor used to treat chronic myelogenous leukemia (CML) and acute lymphoblastic leukemia (ALL) (Lindauer and Hochhaus, 2014), and Erdafitinib, a potent pan-FGFR inhibitor (Perera et al., 2017) in clinical testing in advanced solid tumors, including breast, prostate, colon, bladder, esophageal and non-small-cell lung cancers (www.clinicaltrials.gov). Dasatinib reduced SRC phosphorylation (Figures 1A–C) in cancer cells and or Erdafitinib inhibited FGF-2 production in fibroblasts (Supplementary Figure S1). In drug titration experiments we identified non-toxic Dasatinib or Erdafitinib concentrations to use in the in vitro experiments (50 nM and nM, respectively, Figures 1D–F). Dasatinib or Erdafitinib treatment of SW620 and HCT116 CRC cells co-cultured with fibroblasts reduced fibroblast-induced cancer cell elongation, motility and invasion under 2D (Figure 2 and Supplementary Figure S2) and 3D conditions in vitro (Figure 3).
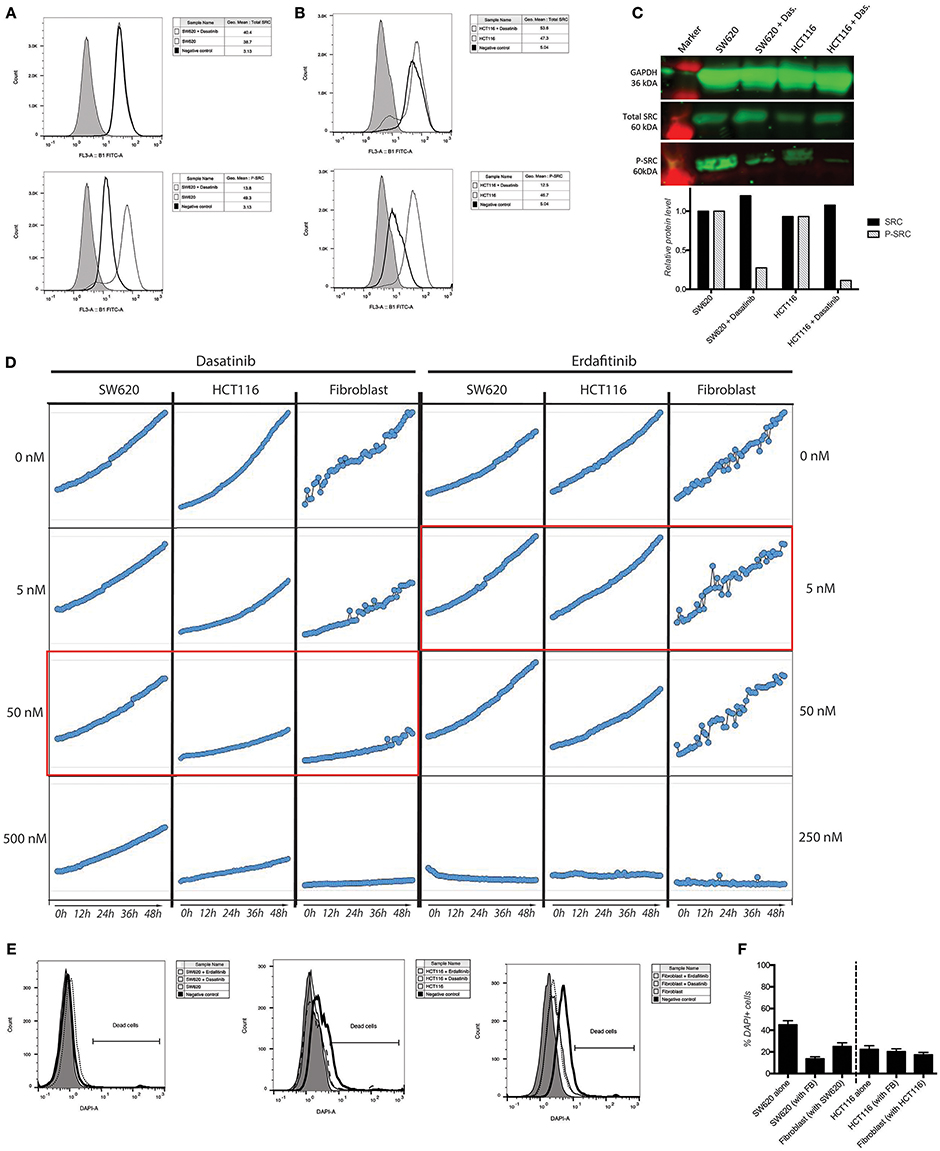
Figure 1. Activity and toxicity of Dasatinib and Erdafitinib. (A,B) Intracellular detection of total and phospho-SRC in SW620 (A) and HCT116 (B) show that Dasatinib inhibits SRC phosphorylation. (C) Western blot analysis confirms that Dasatinib suppresses SRC phosphorylation in cancer cells. (D) Growth curve of SW620 and HCT116 over 48 h in presence or absence of the described drugs at the described concentration. In red the in vitro used concentration for the two drugs. (E) Quantification of cell dead by flow cytometry after 7 days in 3D assay conditions. (F) Viability measurements of the different cell lines cultured in 2D conditions in the presence or absence of the corresponding inhibitor for 48 h using DAPI staining.
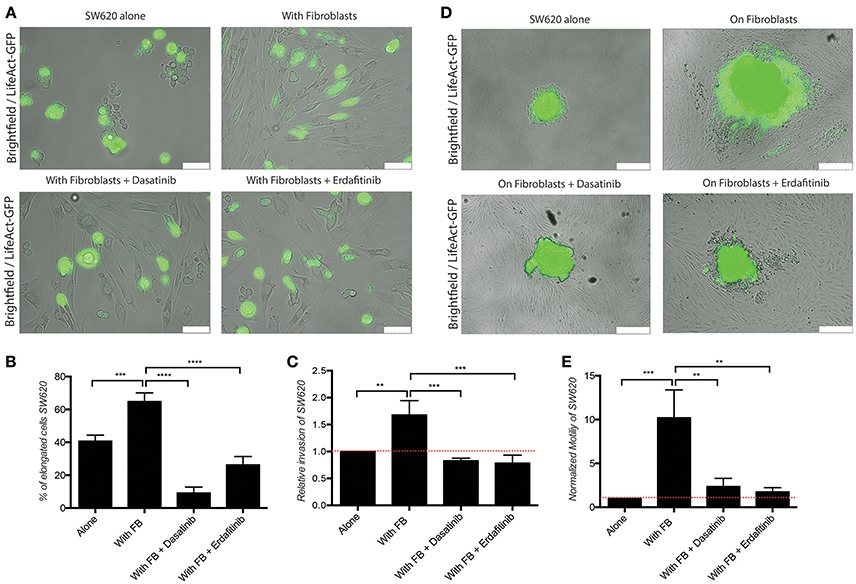
Figure 2. Dasatinib and Erdafitinib reduce fibroblasts-induced SW620 cancer cell elongation, migration and invasion in vitro. (A) Representative images of SW620-LifeAct-GFP cells cultured under 2D conditions with and without fibroblasts in the absence or presence of Dasatinib and Erdafitinib for 48 h. White bars represent 100 μm. (B) Quantification of elongation of SW620 cells of experiment in (A), cultured as indicated, at day 4. (C) Quantification of SW620 cell spheroid 2D invasion under the indicated conditions after 4 days of culture. (D) Representative images of SW620-LifeAct-GFP cells of experiment in (C) cultured as indicated, at day 4. White bars represent 500 μm. (E) Quantification of motility of SW620 cancer cells cultured as indicated for 48 h. Both inhibitors block fibroblasts-induced SW620 elongation, migration and invasion. All quantification data represent mean values ± SD. **p ≤ 0.01, ***p ≤ 0.001, and ****p ≤ 0.0001. Red line represent control value at 1.
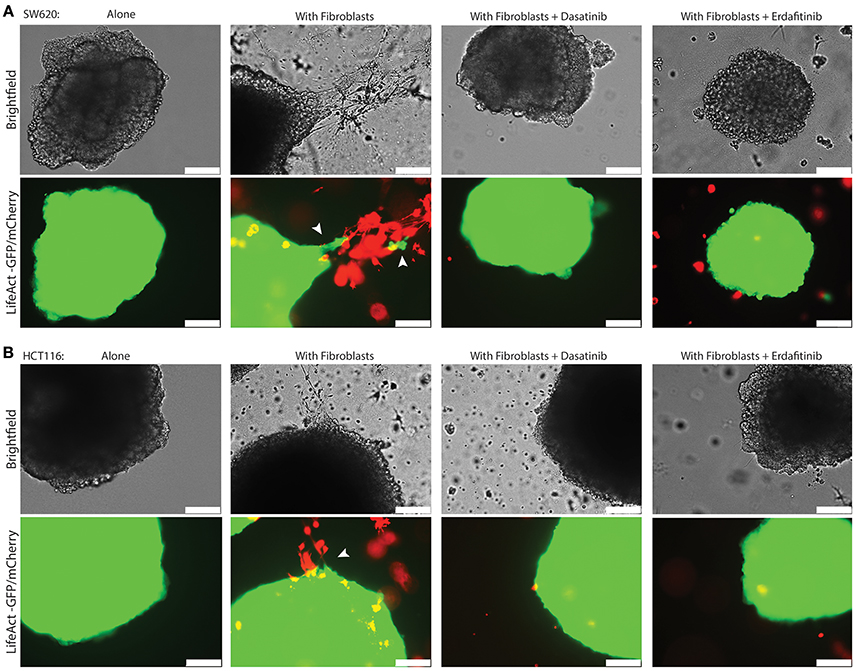
Figure 3. Dasatinib and Erdafitinib inhibitors reduce fibroblasts-induced SW620 cancer cell invasion in vitro under 3D condition. Representative images of (A) SW620-LifeAct-GFP and (B) HCT116-LifeAct-GFP 3D spheroid invasion with and without LifeAct-mCherry labeled fibroblasts in the absence or presence of Dasatinib and Erdafitinib inhibitors after 7 days. Both inhibitors are blocking fibroblasts-induced SW620 3D invasion. Arrows are indication the invasion area. White bars represent 250 μm.
Dasatinib or Erdafitinib Treatment Prevents Fibroblast-Promoted CRC Cell Migration and Invasion in vivo
The pro-invasive effect of co-cultured fibroblasts in vitro raised the question whether fibroblasts co-injected with CRC cells would promote cancer metastasis in vivo. To this end SW620 cells expressing luciferase were orthotopically injected alone or together with fibroblasts in the caecum of immunocompromised (NSG) mice. Fibroblasts and tumor cells were mixed and injected at a 1:1 ratio which is a realistic approximation of the average Cancer Associated Fibroblasts (CAF): epithelial cell ratio observed in human CRC, considering variability observed in different CRC subtypes, stages and intertumoral heterogeneity (Henry et al., 2007; Isella et al., 2015; Nishishita et al., 2018). Fibroblasts co-injected with tumor cells did not impact primary tumor growth (Supplementary Figure S3). However, they induced a significant increase in the lung metastatic burden as detected by luciferase activity measured ex vivo (Figure 4A), and by immunohistochemistry (IHC) for human vimentin of the lungs (Figure 4D). Fibroblasts co-injection promoted the formation of larger metastatic nodules (Figures 4C,D), while it did not increase the number of the nodules themselves (Figure 4B).
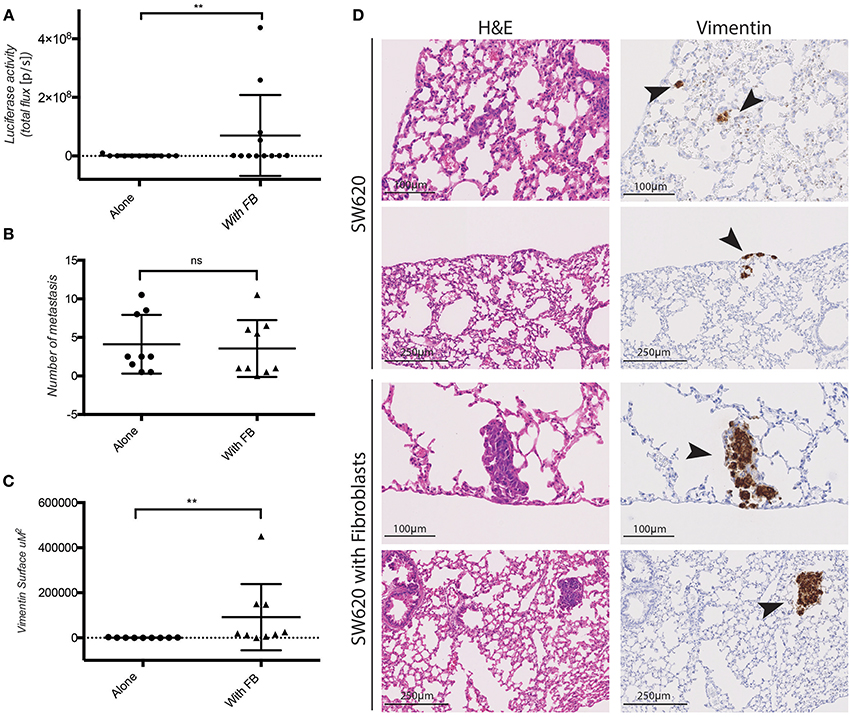
Figure 4. Fibroblasts promote SW620 colorectal cancer cell metastasis. (A) Ex-vivo Luciferase activity in the lung of mice injected with SW620-A299 ± fibroblasts. (B) Quantification of the number of metastatic nodules in the lung of mice orthotopically injected with SW620-A299 cells ± fibroblasts. (C) Quantification of the metastatic lung surface positive for vimentin by IHC of mice orthotopically injected with SW620-A299 cells ± fibroblasts. (D) Representatives images of consecutive sections of lungs of mice injected with SW620 ± fibroblasts stained by H&E (left) and for human vimentin by IHC (right). Arrows indicate metastases. Twelve mice per group were used. Quantification data represent mean values ± SD. Scale bars are given on the images. **p ≤ 0.01; Circles represent cells injected alone and triangles in presence of fibroblasts.
Next, we tested whether Dasatinib or Erdafitinib treatment would impinge on fibroblast-promoted metastasis formation in vivo, as it did for cancer cell migration and invasion in vitro. Treatments were started 2 weeks after primary tumor development was confirmed by In Vivo Bio Luminescence imaging (IBL; data not shown). Animal treated with Dasatinib or Erdafitinib showed a significant and homogeneous reduction of fibroblast-induced lung metastasis burden as detected by IBL imaging (Figure 5A). Immunohistochemical analysis of the lungs confirmed that Dasatinib and Erdafitinib treatment nearly completely reduced metastatic burden in the lungs as detected by IBL or by vimentin IHC of the lungs (Figures 5B,C). The number of metastatic lesions in the lungs, however, was not affected by the treatment (Supplementary Figures S4B–D). From week 4 on under treatment, animals were losing weight with both treatments (Supplementary Figure S4A), known side effect of kinase inhibitors due to intestinal toxicity (Galinsky and Buchanan, 2009), and therefore experiments were prematurely terminated at 6 weeks. As at this time point lung metastases in the treated animals are still microscopic, and therefore no significant increase in IBL signal was detected compared to untreated control animals in which lung metastases are already well established (Supplementary Figures S4B–D).
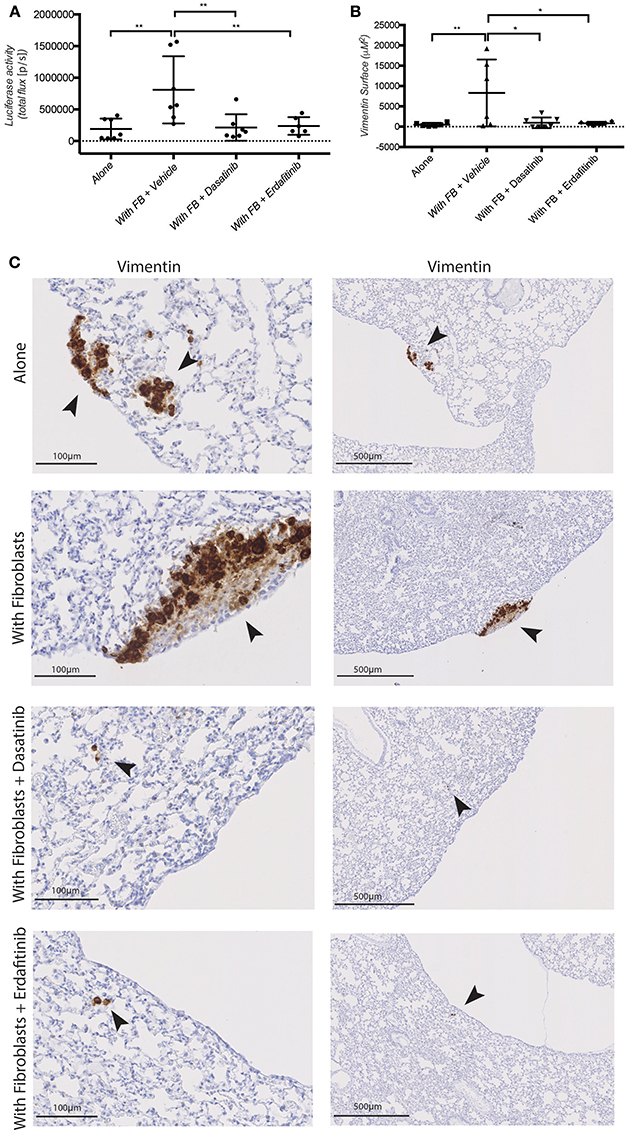
Figure 5. Dasatinib and Erdafitinib suppress fibroblasts-induced metastasis in vivo. (A) Ex-vivo Luciferase activity in the lung of mice orthotopically injected with SW620-A299 cells ± fibroblasts treated with Dasatinib or Erdafitinib or vehicle only as indicated. (B) Quantification of the metastatic lung surface positive by vimentin by IHC of mice of the same experiment. (C) Representatives images of consecutive sections of lungs of mice of the same experiment stained by IHC for human vimentin. Arrows are indication the metastases. Seven mice per group were used. Quantification data represent mean values ± SD. Scale bars are given on the images. *p ≤ 0.05, **p ≤ 0.01; Circles represent cells injected alone and triangles in presence of fibroblasts.
These data demonstrate that fibroblasts orthotopically co-injected with CRC cell promoted metastatic outgrowth (increased volume of metastatic lesions) in the lung, without however affecting the number of lesions. Conversely, treatment with the SRC and pan-FGFR inhibitors Dasatinib and Erdafitinib, respectively, prevented the fibroblasts-induced increase in lung metastasis. These in vivo results are consistent with in vitro results obtained with the 3D co-culture model, and further validate our previously in vitro-only study (Knuchel et al., 2015).
Endothelial Cells Suppress CRC Cell Spheroid Growth in the in vitro 3D Co-Culture Culture Model
Based on these results we reasoned that this 3D model might be further developed to higher complexity by adding endothelial cells to mimic the vascular compartment of the TME. To this end we used an immortalized human umbilical vein endothelial cells (HUVEC)-derived cell line, Ea.hy296, previously reported to retain characteristics and functions of differentiated endothelial, such as angiogenesis, homeostasis/thrombosis, blood pressure and inflammation and are able to form a long lasting tube-like network in vitro (Edgell et al., 1983, 1990; Bauer et al., 1992; Rieber et al., 1993). We confirmed expression of the endothelial marker CD31 at similar level as in HUVEC (Supplementary Figure S5A). Ea.hy296 cells were genetically modified to express the life dye Azurite to assure visualization in the co-cultures (Figure 6A). As endothelial cells need a flat surface to form a visible tube-like network, the 3D model has been adapted accordingly by using a thicker second gel layer. 3D Co-culture experiments with CRC cells, fibroblasts and Ea.hy296 endothelial cells show that neither fibroblasts nor cancer cells directly interact with endothelial cells (Figures 6B,C). However, the presence of Ea.hy296 endothelial cells consistently reduced SW620 and HCT116 cell 3D spheroid growth (Figures 6C,D).
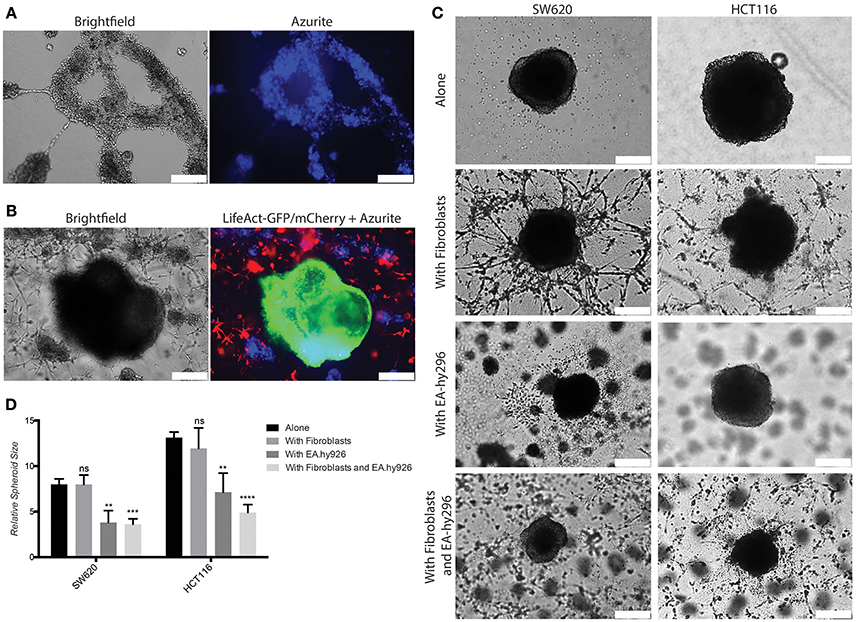
Figure 6. Co-culture with endothelial cells under 3D conditions inhibits SW620 and HCT116 spheroid growth in vitro. (A) Representative images of Ea.hy296 cells expressing Azurite cultured for 8 days under 3D conditions. (B) Representative images of HCT116-LifeAct-GFP cells cultured under 3D conditions with Ea.hy296-Azurite-cells and fibroblasts-LifeAct-mCherry cells. (C) Representatives brightfield images of SW620 and HCT116 colon cancer cells cultured for 7 days under 3D conditions in presence or absence of fibroblasts ± Ea.hy296 endothelial cells. (D) Quantification of relative size of SW620 and HT116 spheroids after 7 days of 3D culture. Quantification data represent mean values ± SD. White bars represent 500 μm. **p ≤ 0.01, ***p ≤ 0.001, and ****p ≤ 0.0001.
Co-Injected Endothelial Cells Suppress Cancer Cell Growth in vivo
To validate these in vitro 3D results, Luciferase expressing SW620 and HCT116 CRC cells were injected subcutaneously in the back of NSG mice, alone and together with Ea.hy296 or HUVEC. Tumor growth was monitored over time by IBL and animals were sacrificed after 10 days. Co-injection of cancer cells with either Ea.hy296 or HUVEC significantly reduced the growth of both SW620 and HCT116 CRC cells (Figure 7 and Supplementary Figures S5B–D).
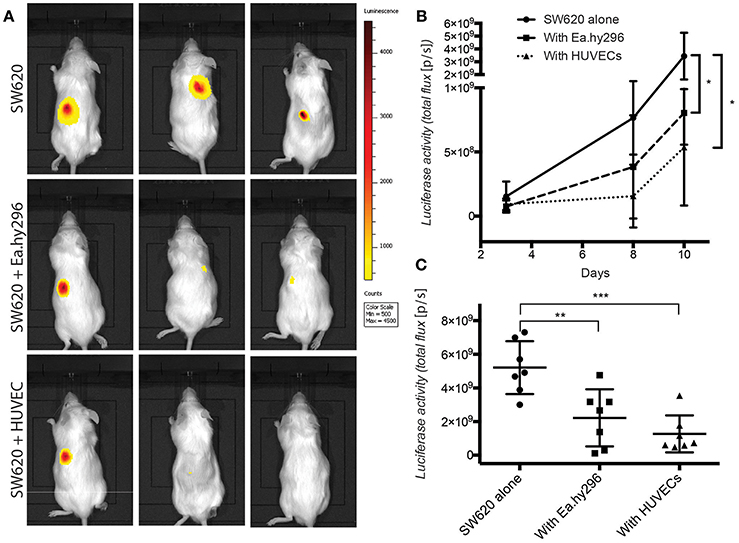
Figure 7. Co-injected endothelial cells reduce SW620 colon cancer growth in vivo. (A) Representative images of luciferase activity in mice subcutaneously injected with SW620-A299 in the presence or absence of EA.hy296 or HUVEC after 8 days. (B) Quantification of luciferase activity by IBL in mice subcutaneously injected at the indicated conditions over time. (C) Quantification of ex-vivo luciferase activity in tumors recovered 10 days after injection at the indicated conditions. Seven mice per group were used. Quantification data represent mean values ± SD. *p ≤ 0.1, **p ≤ 0.01 and ***p ≤ 0.001.
Taken together these results demonstrate that, in contrast to fibroblasts, which promoted spheroids invasion in vitro and metastasis formation in vivo, co-cultured and co-injected endothelial cells suppressed spheroids growth in vitro and tumor growth in vivo, respectively.
Discussion
Local invasion and metastasis development in distant organs are the leading cause of cancer-related death (Siegel et al., 2017b). Mechanisms governing invasion and metastasis are only in part understood and the contribution of host cells at the primary and metastatic sites is being progressively unraveled (Quail and Joyce, 2013). Further understanding of these processes is essential in order to identify new mechanism and targets to develop novel therapeutic strategies to prevent tumor cell dissemination and outgrowth at distant sites. The multi-cellular nature of the TME and its multitude of soluble factors contributing to heterotypic communication, the changes in the extracellular matrix and the dynamic evolution of tumor-host complicate the in vivo study of the TME. In addition, animal models are expensive, time consuming, and subjected to higher variability (Heijstek et al., 2005; Clarke, 2007). Based on these considerations, we set up to develop a 3D heterotypic co-culture model allowing the study of the interactions between cancer cells and cells of the TME. Using this model, we previously reported that co-cultured fibroblasts promote CRC cells motility and invasion through a FGF-2/FGFR, αvβ5-integrin, and SRC -dependent mechanism (Knuchel et al., 2015). The in vivo relevance of this pathway and its therapeutic implication were not assessed.
To prove that our 3D in vitro model is relevant to in vivo conditions, we performed in vivo experiments in mice in which we orthotopically injected SW620 and HCT116 CRC cells with or without fibroblasts. As a clinically relevant readout we monitored metastasis formation. Indeed, we observed that co-injected fibroblasts promoted the formation of larger metastatic nodules in the lungs, without, however, increasing the number of metastatic foci. Importantly, the larger metastatic burden induced by co-injected fibroblasts was prevented by inhibiting SRC and FGFR using Dasatinib and Erdafitinib, respectively. Consistent with the fibroblasts promoting effect, Dasatinib and Erdafitinib decrease the size of the lesions but not their numbers. Clinically, Dasatinib was tested in metastatic CRC patients, alone or in combination therapies, but showed no effect on disease progression (Sharma et al., 2012; Parseghian et al., 2017). These results are not comparable with our settings, as patients had already established metastases, while we tested the efficacy of SRC inhibition on the de novo formation of metastases. Our results indicate that SRC inhibition blocks metastasis formation when given early during disease progression. This setting would be comparable to an adjuvant treatment, for which to our knowledge, a clinical trial has not been reported yet. Through a gene-silencing approach, we have previously demonstrated that inhibition of SRC in tumor cells, but not in fibroblasts, is essential for the effect observed with pharmacological inhibition of SRC in the co-culture setting (Knuchel et al., 2015).
Taken together these in vivo results validate the in vitro results obtained with the 3D model at the cellular (i.e., fibroblasts promoted CRC cell invasion) and molecular (i.e., SRC and FGFR-depended fibroblast mechanism) levels. Although the endpoints of the two assays (i.e., motility and invasion in vitro vs. metastasis in vivo) are different, they are functionally linked as local invasion is considered as one of the first step of the metastatic cascade (Valastyan and Weinberg, 2011; Sleeman et al., 2012).
Next, we added endothelial cells to the 3D model to mimic the presence of a tumor-associated vasculature. Endothelial cells did not physically interact with fibroblasts or cancer cells and did not promote invasion but, unexpectedly, significantly decreased tumor spheroids growth. Consistent with these results in vivo, we indeed observed that tumor growth was decreased in animal co-injected with tumor cells and endothelial cells compared to tumor cells alone. Three dimensional models of tumor cells (spheroids)—endothelial cell interaction in vitro, have been largely used to study morphological and functions effect of tumor cells on endothelial cells including gene expression and sprouting angiogenesis, effect or angiogenic factors or anti-angiogenic drugs (Chopra et al., 1997; Khodarev et al., 2003; Van Moorst and Dass, 2011; Correa de Sampaio et al., 2012; Szot et al., 2013; Chiew et al., 2017; Wan et al., 2017). Co-culture with endothelial cells was shown to promote cancer cell (melanoma) invasion along tube likes structures (Yamamoto et al., 2014) or tumor cell growth when mixed inside spheroids (Upreti et al., 2011). However, in an on-top Matrigel model of ovarian cancer cells co-cultured with a tubular network of HUVEC, the presence of HUVEC clearly reduced tumor cell growth (Wan et al., 2017). No in vivo validation was performed in that work. The reason for this inhibitory effect remains elusive at this point and therefore these results should be carefully considered in their context. It is however tempting to speculate that quiescent, non-angiogenic endothelial cells may exert paracrine anti-proliferative/quiescence promoting effects to neighboring cells in contrast to pro-inflammatory and stimulatory effects of angiogenic endothelial cells (Potente et al., 2011). Furthermore, an important point to consider is that we used endothelial cells of macrovascular origin (HUVEC and a HUVEC-derived line) rather than microvascular endothelial cells from organs relevant to CRC, such as the colon itself (for the primary tumor) or the lung or liver for metastases. Considering the growing interest in understanding the role and activities of organotypic endothelial cells (Augustin and Koh, 2017; Potente and Makinen, 2017), this is an experimental aspect to include in follow-up experiments.
There are many different 3D assays described in the literature or available commercially (Kimlin et al., 2013b; Hoarau-Véchot et al., 2018), in the context of CRC (Nietzer et al., 2016; Pereira et al., 2016), vasculature modeling (Bersini et al., 2016; Haase and Kamm, 2017) and cancer invasion (Berens et al., 2015; De Jaeghere et al., 2018). The model that we proposed has been specifically developed to study the effect of cells of the TME, and in particular fibroblasts, on tumor cell motility and invasion. Globally, 3D in vitro models differ at two levels: firstly, on the preparation of the cancer spheroids. Hanging drop technique is the standard in creating spheroid either using manual pipetting or Bioprinting (Albritton and Miller, 2017). We used manual pipetting with a single spheroid per well. Secondly, models differ on the nature of the 3D matrix used. Both natural and synthetic matrices have been proposed and used for 3D culture conditions and invasion studies. For example, basement membrane extracts and hyaluronic acid are commonly used as biologically-derived matrixes; Polyethylene glycol (PEG), polyvinyl alcohol (PVA), polylactide-co-glycolide (PLG), and polycaprolactone (PLA) are common materials used to form synthetic scaffolds (Asghar et al., 2015; Chung et al., 2017). These matrices face similar limitations and potential criticisms, as they do not faithfully replicate the extracellular matrix of the TME. We opted for Matrigel, as there is ample literature on it and it is still the most widely used matrix in 3D assays, thereby allowing for direct for comparisons with other published reports (Benton et al., 2014).
In conclusion, we have developed a 3D in vitro heterotypic co-culture model to study CRC cell—stroma cell interactions. The model was validated for two CRC cell lines (i.e., SW620 and HCT116), two TME cell types (i.e., fibroblasts and endothelial cells) and two functions (i.e., growth and invasion/metastasis). This model may be adapted to different cancer types, such as breast or prostate, to include additional host cells, such as immune/inflammatory cells, microvascular endothelial cells of diverse organ origins, to test for additional functions, such as metabolism (Zecchin et al., 2015) or gene expression and under different conditions (e.g., hypoxia or starvation) (Bersini et al., 2016). This will help accelerating project progression and reducing the number of in vivo experiments and animal used (Kimlin et al., 2013a; Thoma et al., 2014).
Materials and Methods
Cell Culture
The human colorectal carcinoma cell lines SW620, HCT116, endothelial cells EA.hy296 and HEK-293T cells were purchased from ATCC (LGC Standards). Dermal fibroblasts were isolated from human neonatal foreskin as previously described (Knuchel et al., 2015). HUVEC were isolated from fresh umbilical cords dissociation and cultured as previously described (Yilmaz et al., 1998; Dormond et al., 2001). SW620 were cultured in RPMI GlutaMAXTM, HCT116, EA.hy296, fibroblasts and HEK-293T in DMEM GlutaMAXTM, all supplemented with 10% FCS, 100 U/ml Penicillin and 100 μg/ml Streptomycin. Cell detachment was performed using Trypsin-EDTA. Cell counting and viability determination was performed by trypan blue exclusion using Neubauer Counting Chamber. For co-culture experiments, fibroblasts and colon cancer cells were grown for 48 h at a 1:1 ratio in DMEM GlutaMAXTM. Growth curve to assess viability were monitored by Incucyte 10x. All cell culture reagents were purchased from Life Technologies. Protocols for collection and use of human samples were approved by the Ethic committees of Cantons Fribourg, Vaud, Berne and Ticino, Switzerland.
Inhibitors of Signaling
Dasatinib (Medkoo) was used in vitro at 50 nM in DMSO and in vivo at 30 mg/kg in 80 mM sodium citrate buffer pH 3.1 orally daily for 20 days. Erdafitinib (Medkoo) was used in vitro at 5 nM in DMSO and in vivo at 25 mg/kg in water with 1% Tween 80 orally daily for 20 days.
Vectors and Infections
LifeAct lentiviral vectors in GFP and mCherry were kindly provided by Dr. Olivier Pertz, Basel. A299 lentiviral vectors for Luciferase in GFP was kindly provided by Dr. Albert SantaMaria, Fribourg. pLV-Azurite plasmid was obtained from Addgene. Lentiviral particles were generated in HEK-293T cells by transducing the vector of interest with pMD2G (pSD11) and pMDLgpRRE (pSD16) plasmids using calcium phosphate transfection (Dull et al., 1998), followed, for LifeAct vectors, by an antibiotic-based selection of the infected cells and, for Azurite and A299 vectors, by a cell sorting using BD FACS ARIA Fusion based on GFP or blue fluorescent expression.
2D and 3D Culture Assay
Cancer cells spheroids were prepared as previously described (Kelm et al., 2003), using the hanging drop technic in a Terazaki plate with 500 cells per well in 20 μl medium for 72 h. For 2D spheroids assay, cancer cell spheroids were placed on top of a confluent fibroblasts layer. 3D spheroid assays were performed according to the 3D-On-Top method (Lee et al., 2007). Seven mg/ml Matrigel growth factor reduced (Corning) was used as matrix (Benton et al., 2014). Briefly, a first matrix layer was seeded on the well bottom of a non-treated multi-well plate to polymerize. A single spheroid was deposed on the layer and incubated 30 min at 37°C to adhere. A second matrix layer diluted 1:5 in culture medium was deposed on top of the spheroid. For 3D assay with fibroblasts, 75,000 fibroblasts/ml were added to the gel. For 3D assay with endothelial cells, the second layer on top of the spheroid was replaced by a 7 mg/ml Matrigel growth factors reduced matrix, mixed or not with 75,000 fibroblasts/ml. After 45 min polymerization 150,000 Ea.hy296 were seeded on top in DMEM. Time course of the in vitro experiments is given in Supplementary Figure S6A.
In Vivo Experiments
Male NOD/SCID common gamma chain (IL-2Rγ−/−) ko (NSG) animals between 7 and 8 weeks old were used for in vivo experimentation and were purchased from the University of Lausanne. Twelve or 7 mice per group were used, depending on the expected difference of 40 or 50% or more for a SD of 30%, alpha = 0.05, and beta = 0.8. Orthotopic injection in the caecum was performed as previously described (Céspedes et al., 2007; Ragusa et al., 2014). The number of tumor cells was kept constrain in all conditions (i.e., ± fibroblasts) in accordance with widely accepted protocols (Picard et al., 1986; Erez et al., 2010; Li et al., 2012; Shintani et al., 2013; Cohen et al., 2017; Nakaoka et al., 2017; Wang et al., 2017). Briefly one million SW620-A299 cells, mixed or not with one million fibroblasts, resuspended in 50 μl DMEM solution containing 50% Matrigel were injected using a 30G needle in the submucosa of the caecum under a binocular loupe. The 1:1 fibroblasts: tumor cell ration was chosen as a realistic approximation of the average CAF: epithelial cell ratio reported for human CRC (Henry et al., 2007; Isella et al., 2015; Nishishita et al., 2018). Tumor growth was monitored by in vivo bioluminescence after i.p. injection of 1.5mg/g luciferin (Biosynth), using IVIS Lumina II (Xenogen). Sub-cutaneous tumor transplantation model were performed by injecting subcutaneously in the back a cell suspension of one million SW620-A299 cells or HCT116-A299, mixed or not with one million Ea.hy296 in 50 μl DMEM solution containing 50% Matrigel. Animal procedures were performed in accordance with the Swiss and French legislations on animal experimentation and approved by the Cantonal Veterinary Service of the Canton Fribourg (2016_06_FR). A schematic of the experiment is represented in the Supplementary Figure s6B.
Immunohistochemistry
Slide staining's were performed on Leica BOND RX system following manufacturer's instructions. Paraffin embedded organ cuts were pre-treated 15 min with 1% H2O2 in Methanol for vimentin staining. Primary antibody (Dako) was incubated for 40 min at 1:100.
Flow Cytometry
For viability assay, cultures were harvested on ice and resuspended in PBS containing 3% FCS and 5 mM EDTA. DAPI staining (Miltenyi) was used at 1/1,000 dilutions 5 min before analysis. For 3D recovery and viability testing, cells were dissociated from recovered spheroids using the BD Recovery Solution (BD Biosciences) following manufacturer instructions.
For endothelial cells characterization an anti-CD31-Alexafluor 647 (BD Biosciences) was used at 1/100 dilution.
For intracellular staining, cells were fixed and permeabilized using a BD Bioscience Fix/Perm assay following manufacturer instructions. Anti-SRC, phospho-SRC and FGF-2 unlabeled primary antibodies (Cell Signaling) were used 30 min at 1/100 dilution and secondary anti rabbit-Alexa Fluor 488 (Cell Signaling) was used at 1/300 for 30 min. A MACSQuant instrument was used to perform experiments and FlowJo 10.4 (Treestar Inc.) software was used to analyze all data.
Western Blotting
Cells were lysed using RIPA lysis buffer (Cell Signaling) supplemented with protease Cocktail inhibitor (Sigma), 2 mM PMSF, 2 mM BGE (Sigma) and 0.2 mM Orthovanadate (Sigma) on ice and re-suspended in SDS buffer containing 10% Glycerol, 5% β-mercatoethanol, 60 mM Tris-Cl, trace of bromophenol blue and 2% SDS (Sigma-Aldrich). SDS-PAGE, blotting, and detection were performed as previously described using a Lumina Odyssey instrument.
Imaging and Data Analysis
Imaging of 2D and 3D spheroids assay were performed with 5x and 40x objectives with Leica AF6000 inverted fluorescence microscope. For 3D assay, spheroid growth was quantified using Photoshop CS6 (Adobe) by calculating the spheroid size area on the 7 days image, normalized on initial spheroid size. For 2D assay, invasion was quantified using Photoshop CS6 (Adobe) by calculating the invaded area on image taken at day 4 and normalized on the initial spheroid size. For co-culture experiments images were taken every hour at different magnifications with an inverted fluorescence microscope (Leica AF6000). Cancer cell elongation was quantified manually by scoring individual cells using Image J “Cell Counter” plugin on taken images at 48 h co-culture. Elongated cells were defined as single cells with a minimal length-width rapport of 2:1, and at least one sharp-ending extremity. Cancer cell motility was quantified based on single cell track follow for 24 h on time-lapse movies using Image J “Manual Tracking” and “Chemotaxis Tool” plugins. Western Blot images were quantified for protein level using Image J “Gel Analyze Tool” plugin. For immunohistochemistry images, slides were scanned using a NanoZoomer 2.0 HT (Hamamatsu) and images were extracted and analyzed using NDP.view2 analysis software (Hamamatsu). Metastases were counted based on the presence of vimentin positive staining in the lung. Each isolated positive cell or cell cluster (touching) was considered as a metastasis.
Statistical Analysis
Each experiment was repeated independently a minimum of 3 times in triplicate conditions. Acquired data were analyzed using Prism Software (GraphPad). Statistical comparisons were performed by un-paired two-tailed Student's t-test or by two-way ANOVA with Bonferroni post-test. Results were considered to be significantly from p < 0.05.
Author Contributions
SC planned and designed the project; designed and performed experiments; analyzed data; wrote the manuscript. LR designed and performed part of the experiments on endothelial cells; analyzed data. CR planned and designed the project; evaluated experiments; wrote the manuscript.
Funding
This work was supported by grants from the 3R Research Foundation Switzerland (Project n°144-15), the Medic Foundation and the Swiss National Science Foundation (31003A_135738) to CR.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors wish to thank Dr. Oliver Pertz (University of Basel) for providing lentiviral plasmids expressing LifeAct and Dr. Albert SantaMaria (University of Fribourg) for providing lentiviral plasmids A299.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fbioe.2018.00097/full#supplementary-material
References
Albritton, J. L., and Miller, J. S. (2017). 3D bioprinting: improving in vitro models of metastasis with heterogeneous tumor microenvironments. Dis. Model Mech. 10, 3–14. doi: 10.1242/dmm.025049
Asghar, W., El Assal, R., Shafiee, H., Pitteri, S., Paulmurugan, R., and Demirci, U. (2015). Engineering cancer microenvironments for in vitro 3-D tumor models. Mater. Today 18, 539–553. doi: 10.1016/j.mattod.2015.05.002
Augustin, H. G., and Koh, G. Y. (2017). Organotypic vasculature: from descriptive heterogeneity to functional pathophysiology. Science 357:eaal2379. doi: 10.1126/science.aal2379
Bartlett, R., Everett, W., Lim, S., G, N., Loizidou, M., Jell, G., et al. (2014). Personalized in vitro cancer modeling - fantasy or reality? Transl. Oncol. 7, 657–664. doi: 10.1016/j.tranon.2014.10.006
Bauer, J., Margolis, M., Schreiner, C., Edgell, C. J., Azizkhan, J., Lazarowski, E., et al. (1992). In vitro model of angiogenesis using a human endothelium-derived permanent cell line: contributions of induced gene expression, G-proteins, and integrins. J. Cell. Physiol. 153, 437–449.
Benton, G., Arnaoutova, I., George, J., Kleinman, H. K., and Koblinski, J. (2014). Matrigel: from discovery and ECM mimicry to assays and models for cancer research. Adv. Drug Deliv. Rev. 79–80, 3–18. doi: 10.1016/j.addr.2014.06.005
Berens, E. B., Holy, J. M., Riegel, A. T., and Wellstein, A. (2015). A cancer cell spheroid assay to assess invasion in a 3D setting. J. Vis. Exp. 105:e53409 doi: 10.3791/53409
Bersini, S., Gilardi, M., Arrigoni, C., Talò, G., Zamai, M., Zagra, L., et al. (2016). Human in vitro 3D co-culture model to engineer vascularized bone-mimicking tissues combining computational tools and statistical experimental approach. Biomaterials 76, 157–172. doi: 10.1016/j.biomaterials.2015.10.057
Bhome, R., Bullock, M. D., Al Saihati, H. A., Goh, R. W., Primrose, J. N., Sayan, A. E., et al. (2015). A top-down view of the tumor microenvironment: structure, cells and signaling. Front. Cell. Dev. Biol. 3:33. doi: 10.3389/fcell.2015.00033
Brenner, H., Kloor, M., and Pox, C. P. (2014). Colorectal cancer. Lancet 383, 1490–1502. doi: 10.1016/S0140-6736(13)61649-9
Céspedes, M. V., Espina, C., García-Cabezas, M. A., Trias, M., Boluda, A., Gomez Del Pulgar, M. T., et al. (2007). Orthotopic microinjection of human colon cancer cells in nude mice induces tumor foci in all clinically relevant metastatic sites. Am. J. Pathol. 170, 1077–1085. doi: 10.2353/ajpath.2007.060773
Chiew, G. G. Y., Wei, N., Sultania, S., Lim, S., and Luo, K. Q. (2017). Bioengineered three-dimensional co-culture of cancer cells and endothelial cells: a model system for dual analysis of tumor growth and angiogenesis. Biotechnol. Bioeng. 114, 1865–1877. doi: 10.1002/bit.26297
Chopra, V., Dinh, T. V., and Hannigan, E. V. (1997). Three-dimensional endothelial-tumor epithelial cell interactions in human cervical cancers. In Vitro Cell. Dev. Biol. Anim. 33, 432–442. doi: 10.1007/s11626-997-0061-y
Chung, M., Ahn, J., Son, K., Kim, S., and Jeon, N. L. (2017). Biomimetic model of tumor microenvironment on microfluidic platform. Adv. Healthc. Mater. 6:1700196. doi: 10.1002/adhm.201700196
Clarke, A. R. (2007). Cancer genetics: mouse models of intestinal cancer. Biochem. Soc. Trans. 35, 1338–1341. doi: 10.1042/BST0351338
Cohen, N., Shani, O., Raz, Y., Sharon, Y., Hoffman, D., Abramovitz, L., et al. (2017). Fibroblasts drive an immunosuppressive and growth-promoting microenvironment in breast cancer via secretion of Chitinase 3-like 1. Oncogene 36, 4457–4468. doi: 10.1038/onc.2017.65
Correa de Sampaio, P., Auslaender, D., Krubasik, D., Failla, A. V., Skepper, J. N., Murphy, G., et al. (2012). A heterogeneous in vitro three dimensional model of tumour-stroma interactions regulating sprouting angiogenesis. PLoS ONE 7:e30753. doi: 10.1371/journal.pone.0030753
De Jaeghere, E., De Vlieghere, E., Van Hoorick, J., Van Vlierberghe, S., Wagemans, G., Pieters, L., et al. (2018). Heterocellular 3D scaffolds as biomimetic to recapitulate the tumor microenvironment of peritoneal metastases in vitro and in vivo. Biomaterials 158, 95–105. doi: 10.1016/j.biomaterials.2017.12.017
Dienstmann, R., and Tabernero, J. (2016). Spectrum of gene mutations in colorectal cancer: implications for treatment. Cancer J. 22, 149–155. doi: 10.1097/PPO.0000000000000191
Dormond, O., Foletti, A., Paroz, C., and Rüegg, C. (2001). NSAIDs inhibit alpha V beta 3 integrin-mediated and Cdc42/Rac-dependent endothelial-cell spreading, migration and angiogenesis. Nat. Med. 7, 1041–1047. doi: 10.1038/nm0901-1041
Dull, T., Zufferey, R., Kelly, M., Mandel, R. J., Nguyen, M., Trono, D., et al. (1998). A third-generation lentivirus vector with a conditional packaging system. J. Virol. 72, 8463–8471.
Edgell, C. J., Haizlip, J. E., Bagnell, C. R., Packenham, J. P., Harrison, P., Wilbourn, B., et al. (1990). Endothelium specific Weibel-Palade bodies in a continuous human cell line, EA.hy926. In Vitro Cell. Dev. Biol. 26, 1167–1172.
Edgell, C. J., Mcdonald, C. C., and Graham, J. B. (1983). Permanent cell line expressing human factor VIII-related antigen established by hybridization. Proc. Natl. Acad. Sci. U.S.A. 80, 3734–3737. doi: 10.1073/pnas.80.12.3734
Erez, N., Truitt, M., Olson, P., Arron, S. T., and Hanahan, D. (2010). Cancer-associated fibroblasts are activated in incipient neoplasia to orchestrate tumor-promoting inflammation in an NF-kappaB-dependent manner. Cancer Cell 17, 135–147. doi: 10.1016/j.ccr.2009.12.041
Galinsky, I., and Buchanan, S. (2009). Practical management of dasatinib for maximum patient benefit. Clin. J. Oncol. Nurs. 13, 329–335. doi: 10.1188/09.CJON.329-335
Golovko, D., Kedrin, D., Yilmaz, Ö. H., and Roper, J. (2015). Colorectal cancer models for novel drug discovery. Expert Opin. Drug Discov. 10, 1217–1229. doi: 10.1517/17460441.2015.1079618
Gout, S., and Huot, J. (2008). Role of cancer microenvironment in metastasis: focus on colon cancer. Cancer Microenviron. 1, 69–83. doi: 10.1007/s12307-008-0007-2
Guinney, J., Dienstmann, R., Wang, X., de Reyniès, A., Schlicker, A., Soneson, C., et al. (2015). The consensus molecular subtypes of colorectal cancer. Nat. Med. 21, 1350–1356. doi: 10.1038/nm.3967
Haase, K., and Kamm, R. D. (2017). Advances in on-chip vascularization. Regen. Med. 12, 285–302. doi: 10.2217/rme-2016-0152
Hanahan, D., and Weinberg, R. A. (2011). Hallmarks of cancer: the next generation. Cell 144, 646–674. doi: 10.1016/j.cell.2011.02.013
Heijstek, M. W., Kranenburg, O., and Borel Rinkes, I. H. (2005). Mouse models of colorectal cancer and liver metastases. Dig. Surg. 22, 16–25. doi: 10.1159/000085342
Henry, L. R., Lee, H. O., Lee, J. S., Klein-Szanto, A., Watts, P., Ross, E. A., et al. (2007). Clinical implications of fibroblast activation protein in patients with colon cancer. Clin. Cancer Res. 13, 1736–1741. doi: 10.1158/1078-0432.CCR-06-1746
Herrmann, D., Conway, J. R., Vennin, C., Magenau, A., Hughes, W. E., Morton, J. P., et al. (2014). Three-dimensional cancer models mimic cell-matrix interactions in the tumour microenvironment. Carcinogenesis 35, 1671–1679. doi: 10.1093/carcin/bgu108
Hoarau-Véchot, J., Rafii, A., Touboul, C., and Pasquier, J. (2018). Halfway between 2D and animal models: are 3D cultures the ideal tool to study cancer-microenvironment interactions? Int. J. Mol. Sci. 19:E181. doi: 10.3390/ijms19010181
Horie, M., Saito, A., Yamaguchi, Y., Ohshima, M., and Nagase, T. (2015). Three-dimensional Co-culture model for tumor-stromal interaction. J. Vis. Exp. 96:e52469 doi: 10.3791/52469
Isella, C., Terrasi, A., Bellomo, S. E., Petti, C., Galatola, G., Muratore, A., et al. (2015). Stromal contribution to the colorectal cancer transcriptome. Nat. Genet. 47, 312–319. doi: 10.1038/ng.3224
Jeon, J. S., Bersini, S., Gilardi, M., Dubini, G., Charest, J. L., Moretti, M., et al. (2015). Human 3D vascularized organotypic microfluidic assays to study breast cancer cell extravasation. Proc. Natl. Acad. Sci. U.S.A. 112, 214–219. doi: 10.1073/pnas.1417115112
Jin, M. Z., Han, R. R., Qiu, G. Z., Ju, X. C., Lou, G., and Jin, W. L. (2018). Organoids: an intermediate modeling platform in precision oncology. Cancer Lett. 414, 174–180. doi: 10.1016/j.canlet.2017.11.021
Joyce, J. A., and Pollard, J. W. (2009). Microenvironmental regulation of metastasis. Nat. Rev. Cancer 9, 239–252. doi: 10.1038/nrc2618
Kalluri, R. (2016). The biology and function of fibroblasts in cancer. Nat. Rev. Cancer 16, 582–598. doi: 10.1038/nrc.2016.73
Kelm, J. M., Timmins, N. E., Brown, C. J., Fussenegger, M., and Nielsen, L. K. (2003). Method for generation of homogeneous multicellular tumor spheroids applicable to a wide variety of cell types. Biotechnol. Bioeng. 83, 173–180. doi: 10.1002/bit.10655
Khodarev, N. N., Yu, J., Labay, E., Darga, T., Brown, C. K., Mauceri, H. J., et al. (2003). Tumour-endothelium interactions in co-culture: coordinated changes of gene expression profiles and phenotypic properties of endothelial cells. J. Cell Sci. 116, 1013–1022. doi: 10.1242/jcs.00281
Kimlin, L. C., Casagrande, G., and Virador, V. M. (2013b). In vitro three-dimensional (3D) models in cancer research: an update. Mol. Carcinog. 52, 167–182. doi: 10.1002/mc.21844
Kimlin, L., Kassis, J., and Virador, V. (2013a). 3D in vitro tissue models and their potential for drug screening. Expert Opin. Drug Discov. 8, 1455–1466. doi: 10.1517/17460441.2013.852181
Knuchel, S., Anderle, P., Werfelli, P., Diamantis, E., and Rüegg, C. (2015). Fibroblast surface-associated FGF-2 promotes contact-dependent colorectal cancer cell migration and invasion through FGFR-SRC signaling and integrin alphavbeta5-mediated adhesion. Oncotarget 6, 14300–14317. doi: 10.18632/oncotarget.3883
Langhans, S. A. (2018). Three-dimensional in vitro cell culture models in drug discovery and drug repositioning. Front. Pharmacol. 9:6. doi: 10.3389/fphar.2018.00006
Lee, G. Y., Kenny, P. A., Lee, E. H., and Bissell, M. J. (2007). Three-dimensional culture models of normal and malignant breast epithelial cells. Nat. Methods 4, 359–365. doi: 10.1038/nmeth1015
Li, H. J., Reinhardt, F., Herschman, H. R., and Weinberg, R. A. (2012). Cancer-stimulated mesenchymal stem cells create a carcinoma stem cell niche via prostaglandin E2 signaling. Cancer Discov. 2, 840–855. doi: 10.1158/2159-8290.CD-12-0101
Lindauer, M., and Hochhaus, A. (2014). Dasatinib. Recent Results Cancer Res. 201, 27–65. doi: 10.1007/978-3-642-54490-3_2
Lorusso, G., and Rüegg, C. (2008). The tumor microenvironment and its contribution to tumor evolution toward metastasis. Histochem. Cell Biol. 130, 1091–1103. doi: 10.1007/s00418-008-0530-8
Malandrino, A., Kamm, R. D., and Moeendarbary, E. (2018). In vitro modeling of mechanics in cancer metastasis. ACS Biomater. Sci. Eng. 4, 294–301. doi: 10.1021/acsbiomaterials.7b00041
McAllister, S. S., and Weinberg, R. A. (2014). The tumour-induced systemic environment as a critical regulator of cancer progression and metastasis. Nat. Cell Biol. 16, 717–727. doi: 10.1038/ncb3015
Nakaoka, H. J., Tanei, Z., Hara, T., Weng, J. S., Kanamori, A., Hayashi, T., et al. (2017). Mint3-mediated L1CAM expression in fibroblasts promotes cancer cell proliferation via integrin alpha5beta1 and tumour growth. Oncogenesis 6:e334. doi: 10.1038/oncsis.2017.27
Nietzer, S., Baur, F., Sieber, S., Hansmann, J., Schwarz, T., Stoffer, C., et al. (2016). Mimicking metastases including tumor stroma: a new technique to generate a three-dimensional colorectal cancer model based on a biological decellularized intestinal scaffold. Tissue Eng. C Methods 22, 621–635. doi: 10.1089/ten.tec.2015.0557
Nishishita, R., Morohashi, S., Seino, H., Wu, Y., Yoshizawa, T., Haga, T., et al. (2018). Expression of cancer-associated fibroblast markers in advanced colorectal cancer. Oncol. Lett. 15, 6195–6202. doi: 10.3892/ol.2018.8097
Parseghian, C. M., Parikh, N. U., Wu, J. Y., Jiang, Z. Q., Henderson, L., Tian, F., et al. (2017). Dual inhibition of EGFR and c-Src by cetuximab and dasatinib combined with FOLFOX chemotherapy in patients with metastatic colorectal cancer. Clin. Cancer Res. 23, 4146–4154. doi: 10.1158/1078-0432.CCR-16-3138
Penfornis, P., Vallabhaneni, K. C., Janorkar, A. V., and Pochampally, R. R. (2017). Three dimensional tumor models for cancer studies. Front. Biosci. 9, 162–173.
Pereira, J. F., Awatade, N. T., Loureiro, C. A., Matos, P., Amaral, M. D., and Jordan, P. (2016). The third dimension: new developments in cell culture models for colorectal research. Cell. Mol. Life Sci. 73, 3971–3989. doi: 10.1007/s00018-016-2258-2
Perera, T. P. S., Jovcheva, E., Mevellec, L., Vialard, J., De Lange, D., Verhulst, T., et al. (2017). Discovery and pharmacological characterization of JNJ-42756493 (Erdafitinib), a functionally selective small-molecule FGFR family inhibitor. Mol. Cancer Ther. 16, 1010–1020. doi: 10.1158/1535-7163.MCT-16-0589
Picard, O., Rolland, Y., and Poupon, M. F. (1986). Fibroblast-dependent tumorigenicity of cells in nude mice: implication for implantation of metastases. Cancer Res. 46, 3290–3294.
Potente, M., Gerhardt, H., and Carmeliet, P. (2011). Basic and therapeutic aspects of angiogenesis. Cell 146, 873–887. doi: 10.1016/j.cell.2011.08.039
Potente, M., and Mäkinen, T. (2017). Vascular heterogeneity and specialization in development and disease. Nat. Rev. Mol. Cell Biol. 18, 477–494. doi: 10.1038/nrm.2017.36
Quail, D. F., and Joyce, J. A. (2013). Microenvironmental regulation of tumor progression and metastasis. Nat. Med. 19, 1423–1437. doi: 10.1038/nm.3394
Ragusa, S., Cheng, J., Ivanov, K. I., Zangger, N., Ceteci, F., Bernier-Latmani, J., et al. (2014). PROX1 promotes metabolic adaptation and fuels outgrowth of Wnt(high) metastatic colon cancer cells. Cell Rep. 8, 1957–1973. doi: 10.1016/j.celrep.2014.08.041
Ravi, M., Paramesh, V., Kaviya, S. R., Anuradha, E., and Solomon, F. D. (2015). 3D cell culture systems: advantages and applications. J. Cell. Physiol. 230, 16–26. doi: 10.1002/jcp.24683
Ravi, M., Ramesh, A., and Pattabhi, A. (2017). Contributions of 3D cell cultures for cancer research. J. Cell. Physiol. 232, 2679–2697. doi: 10.1002/jcp.25664
Rieber, A. J., Marr, H. S., Comer, M. B., and Edgell, C. J. (1993). Extent of differentiated gene expression in the human endothelium-derived EA.hy926 cell line. Thromb. Haemost. 69, 476–480.
Sharma, M. R., Wroblewski, K., Polite, B. N., Knost, J. A., Wallace, J. A., Modi, S., et al. (2012). Dasatinib in previously treated metastatic colorectal cancer: a phase II trial of the University of Chicago Phase II Consortium. Invest. New Drugs 30, 1211–1215. doi: 10.1007/s10637-011-9681-x
Shintani, Y., Abulaiti, A., Kimura, T., Funaki, S., Nakagiri, T., Inoue, M., et al. (2013). Pulmonary fibroblasts induce epithelial mesenchymal transition and some characteristics of stem cells in non-small cell lung cancer. Ann. Thorac. Surg. 96, 425–433. doi: 10.1016/j.athoracsur.2013.03.092
Siegel, R. L., Miller, K. D., Fedewa, S. A., Ahnen, D. J., Meester, R. G. S., Barzi, A., et al. (2017a). Colorectal cancer statistics, 2017. CA Cancer J. Clin. 67, 177–193. doi: 10.3322/caac.21395
Siegel, R. L., Miller, K. D., and Jemal, A. (2017b). Cancer statistics, 2017. CA Cancer J. Clin. 67, 7–30. doi: 10.3322/caac.21387
Sleeman, J. P., Christofori, G., Fodde, R., Collard, J. G., Berx, G., Decraene, C., et al. (2012). Concepts of metastasis in flux: the stromal progression model. Semin. Cancer Biol. 22, 174–186. doi: 10.1016/j.semcancer.2012.02.007
Stadler, M., Walter, S., Walzl, A., Kramer, N., Unger, C., Scherzer, M., et al. (2015). Increased complexity in carcinomas: analyzing and modeling the interaction of human cancer cells with their microenvironment. Semin. Cancer Biol. 35, 107–124. doi: 10.1016/j.semcancer.2015.08.007
Szot, C. S., Buchanan, C. F., Freeman, J. W., and Rylander, M. N. (2013). In vitro angiogenesis induced by tumor-endothelial cell co-culture in bilayered, collagen I hydrogel bioengineered tumors. Tissue Eng. C Methods 19, 864–874. doi: 10.1089/ten.tec.2012.0684
Taketo, M. M. (2006). Mouse models of gastrointestinal tumors. Cancer Sci. 97, 355–361. doi: 10.1111/j.1349-7006.2006.00190.x
Thoma, C. R., Zimmermann, M., Agarkova, I., Kelm, J. M., and Krek, W. (2014). 3D cell culture systems modeling tumor growth determinants in cancer target discovery. Adv. Drug Deliv. Rev. 69–70, 29–41. doi: 10.1016/j.addr.2014.03.001
Upreti, M., Jamshidi-Parsian, A., Koonce, N. A., Webber, J. S., Sharma, S. K., Asea, A. A., et al. (2011). Tumor-endothelial cell three-dimensional spheroids: new aspects to enhance radiation and drug therapeutics. Transl. Oncol. 4, 365–376. doi: 10.1593/tlo.11187
Valastyan, S., and Weinberg, R. A. (2011). Tumor metastasis: molecular insights and evolving paradigms. Cell 147, 275–292. doi: 10.1016/j.cell.2011.09.024
Van Moorst, M., and Dass, C. R. (2011). Methods for co-culturing tumour and endothelial cells: systems and their applications. J. Pharm. Pharmacol. 63, 1513–1521. doi: 10.1111/j.2042-7158.2011.01352.x
Wan, X., Bovornchutichai, P., Cui, Z., O'neill, E., and Ye, H. (2017). Morphological analysis of human umbilical vein endothelial cells co-cultured with ovarian cancer cells in 3D: an oncogenic angiogenesis assay. PLoS ONE 12:e0180296. doi: 10.1371/journal.pone.0180296
Wang, Y., Gan, G., Wang, B., Wu, J., Cao, Y., Zhu, D., et al. (2017). Cancer-associated fibroblasts promote irradiated cancer cell recovery through autophagy. EBioMedicine 17, 45–56. doi: 10.1016/j.ebiom.2017.02.019
Weiswald, L. B., Bellet, D., and Dangles-Marie, V. (2015). Spherical cancer models in tumor biology. Neoplasia 17, 1–15. doi: 10.1016/j.neo.2014.12.004
Yamamoto, S., Hotta, M. M., Okochi, M., and Honda, H. (2014). Effect of vascular formed endothelial cell network on the invasive capacity of melanoma using the in vitro 3D co-culture patterning model. PLoS ONE 9:e103502. doi: 10.1371/journal.pone.0103502
Yilmaz, A., Bieler, G., Spertini, O., Lejeune, F. J., and Rüegg, C. (1998). Pulse treatment of human vascular endothelial cells with high doses of tumor necrosis factor and interferon-gamma results in simultaneous synergistic and reversible effects on proliferation and morphology. Int. J. Cancer 77, 592–599.
Keywords: colorectal cancer, three-dimensional model (3D), in vitro, invasion, tumor microenvironment, heterotypic interactions, animal use alternatives, 3R principles
Citation: Cattin S, Ramont L and Rüegg C (2018) Characterization and In Vivo Validation of a Three-Dimensional Multi-Cellular Culture Model to Study Heterotypic Interactions in Colorectal Cancer Cell Growth, Invasion and Metastasis. Front. Bioeng. Biotechnol. 6:97. doi: 10.3389/fbioe.2018.00097
Received: 12 March 2018; Accepted: 25 June 2018;
Published: 17 July 2018.
Edited by:
Ivan Martin, Universität Basel, SwitzerlandReviewed by:
Christian Hirt, ETH Zürich, SwitzerlandMichela Pozzobon, Università degli Studi di Padova, Italy
Copyright © 2018 Cattin, Ramont and Rüegg. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Curzio Rüegg, Y3VyemlvLnJ1ZWdnQHVuaWZyLmNo
 Sarah Cattin
Sarah Cattin Laurent Ramont
Laurent Ramont Curzio Rüegg
Curzio Rüegg