- 1State Key Laboratory for Diagnosis and Treatment of Infectious Diseases, Collaborative Innovation Center for Diagnosis and Treatment of Infectious Diseases, The First Affiliated Hospital, College of Medicine, Zhejiang University, Hangzhou, China
- 2Department of Internal Medicine, Section of Infectious Diseases, Yale University School of Medicine, New Haven, CT, United States
- 3Intensive Care Unit, The First Affiliated Hospital, College of Medicine, Zhejiang University, Hangzhou, China
Background
Coagulase-negative staphylococci (CNS) are key players in the majority of food fermentation ecosystems, which are commonly found in the production of fermented meat and milk products (Blaiotta et al., 2005; Resch et al., 2008). Strains of CNS have been implicated in exerting desirable effects as components of a fermentation flora, such as color formation, aroma development, and shelf-life enhancement, and may therefore have the potential for future application as starter cultures (Zell et al., 2008). Staphylococcus condimenti is one of the most prominent species and has the potential for use in starter cultures for the production of fermented sausage and cured ham (Zell et al., 2008). S. condimenti DSM 11674 was originally isolated from fermenting soy sauce mash and suggested to be a new species in 1998 (Probst et al., 1998). However, S. condimenti has been found in a few clinical samples (Argemi et al., 2015; Misawa et al., 2015). Therefore, some concerns have been raised with regard to the safety of this species for use in food production (Zell et al., 2008; Seitter et al., 2011a,b). To further understand the biochemical and genetic characteristics of DSM 11674 and advance the potential biotechnological applications of this strain, we constructed the complete genome sequence of S. condimenti DSM 11674.
Materials and Methods
Bacterial Strain and Biochemical Characterization
Staphylococcus condimenti DSM 11674 (= JCM 6074 = CIP 105760) was obtained from the Deutsche Sammlung von Mikroorganismen undZellkulturen. The isolate was identified by 16S rRNA sequencing. The sequence was then compared against NCBI database and EzTaxon-e database. To further explore its potential application in food fermentation, we calculated the nitrate reductase activity and catalase activity of S. condimenti DSM 11674 as described previously (Herrero et al., 1996; Miralles et al., 1996). Nitrite reductase activity was determined as described previously (Neubauer et al., 1999; Gotterup et al., 2007).
Minimum Inhibitory Concentrations (MICs) and DNA Purification
Minimum inhibitory concentrations were established by the Vitek 2 Compact system with AST-GP67 card (bioMe’rieux, France). The MICs were interpreted according to Clinical and Laboratory Standards Institute (CLSI, 2016). Genomic DNA was extracted from 3-ml overnight cultures using a Gentra Puregene Yeast/Bact Kit (Qiagen, Hilden, Germany). Bacteria were treated with lysis buffer containing Proteinase K and RNaseA for 2 h at 65°C, and DNA purification was performed according to the manufacturer’s recommended protocols.
Genome Sequencing and Assembly
The genome of S. condimenti DSM 11674 was sequenced on the PacBio RS II single-molecule real-time (SMRT) system. Raw sequence data were de novo assembled using the hierarchical genome-assembly process (HGAP) protocol (Chin et al., 2013) and RS HGAP Assembly 2.1
Genome Annotation
The genome was annotated using the Rapid Annotation using Subsystem Technology server (Aziz et al., 2008) and the NCBI Prokaryotic Genome Annotation Pipeline. Ribosomal RNAs were detected by RNAmmer (Lagesen et al., 2007) and transfer RNAs by tRNAscan-SE (Lowe and Eddy, 1997). CRISPRFinder was used to screen for the presence of CRISPR elements (Grissa et al., 2007). Coding sequences were analyzed to detect toxin genes by using VirulenceFinder2 and by comparing the protein sequences using BLASTP with sequences in virulence factor database (Chen et al., 2005). The Antibiotic Resistance Genes Database was applied to classify antibiotic resistance genes (Liu and Pop, 2009).
Comparative Genomic Analysis
The core genome alignment module in the rapid large-scale prokaryote pan genome analysis (Roary) pipeline was used to extract predicted coding regions from 21 complete Staphylococci genome sequences (Page et al., 2015). Core genes were defined as those present in all isolates with default parameters. Common and unique orthologous groups identified among the genomes were defined as previously described (Zheng et al., 2014). Full chromosome alignments were performed using progressive MAUVE (Darling et al., 2010).
Results and Discussion
Biochemical and Antimicrobial Characteristics
In our study, the strain of S. condimenti DSM 11674 has the highest capacity to reduce nitrate (13.67 mM nitrate reduced to nitrite per milligram of dry weight) and exhibits a high catalase activity compared to Staphylococcus aureus ATCC 25923 and the clinical isolate of S. condimenti CJ1628 (Figure S1A in Supplementary Material). Moreover, the strain of S. condimenti DSM 11674 exhibited the enhanced nitrite reductase activity when cultured with nitrite (2 mM) and nitrate (20 mM) under anaerobic condition (Table S1 in Supplementary Material). Antimicrobial susceptibility tests show that S. condimenti DSM 11674 is susceptible to all antibiotics tested, including amikacin, ampicillin/sulbactam, cefazolin, cefepime, ceftazidime, ceftriaxone, ciprofloxacin, ertapenem, gentamicin, imipenem, levofloxacin, tobramycin, and trimethoprim/sulfamethoxazole. These data are consistent with that of traditional starter culture Staphylococcus carnosus (Landeta et al., 2013) and indicate that S. condimenti is suitable as fermented meat starter.
Genome Features
The complete circular chromosome was 2,659,676 bp with a G + C content of 34.7%. A total of 2,516 protein coding genes, 18 rRNA genes, 58 tRNA genes, 46 pseudogenes, and 2 CRISPR arrays were identified in the genome (Table S2 and Figure S2 in Supplementary Material).
Comparison of Staphylococci Genomes
A total of 28,680 gene clusters and 22 core genes were defined, and a phylogenetic tree was constructed based on the core gene alignment (Figure 1A; Table S3 in Supplementary Material). On this tree, Staphylococcus saprophyticus ATCC 15305, three Staphylococcus xylosus isolates, S. carnosus TM300, Staphylococcus hyicus ATCC11249, Staphylococcus schleiferi 1360-13, and S. condimenti DSM 11674 formed a monophyletic branch, providing strong evidence for the taxonomic relatedness of these isolates (Figure 1A). Of note, S. condimenti DSM 11674 has the closest relationship with S. carnosus TM300. On the basis of this, we further identified unique and shared gene content in S. condimenti, with commercial meat starter culture bacteria S. carnosus TM300 (Rosenstein et al., 2009) and S. xylosus SMG-121 (El Haddad et al., 2014), which are widely used in the food industry. A Venn diagram of the unique/shared gene content was generated with a custom R script using the VennDiagram package (Figure 1B). These three strains share 1,743 CDS in their genome. In addition, a noticeable overlap between DSM 11674 and TM300 became evident, and these two strains shared 493 orthologous CDS. Moreover, 280 CDS from the DSM 11674 genome were classified as unique. The MAUVE analysis revealed a significant portion of the genetic information has been conserved among DSM 11674 and TM300, as the majority of the local collinear blocks are shared by these two strains (Figure S3 in Supplementary Material).
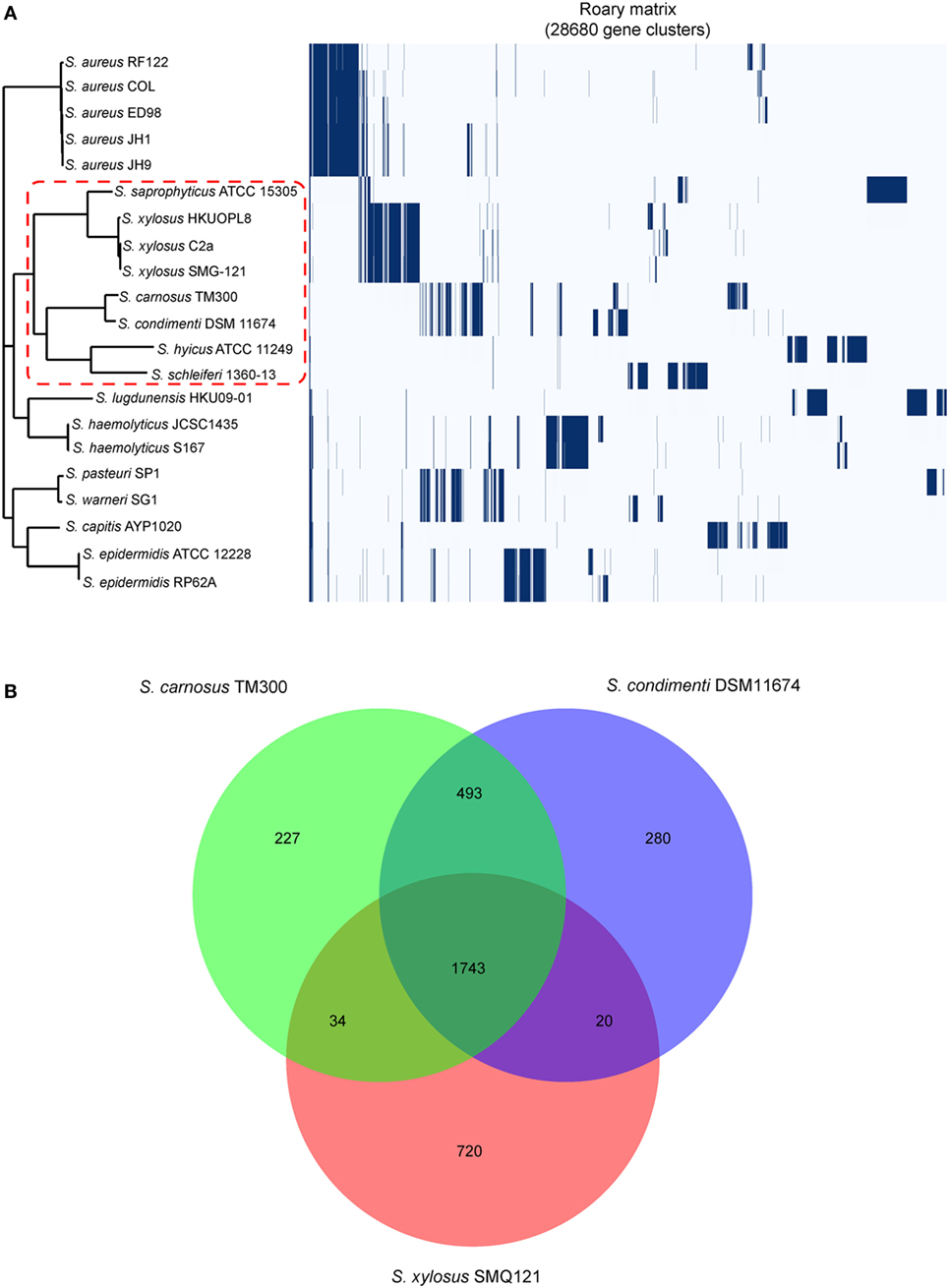
Figure 1. Genomic comparison of the Staphylococcus condimenti DSM 11674 with other staphylococci. (A) Phylogenetic tree based on all core gene sequences of 21 staphylococci complete genomes. Multiple sequence alignments of concatenated core gene sequences were calculated within Roary pipeline. The branch of the members of the S. condimenti is delimited by a red dash line. The isolates used in this study include Staphylococcus schleiferi 1360-13 (CP009470), Staphylococcus epidermidis RP62A (CP000029), S. epidermidis ATCC 12228 (AE015929), Staphylococcus capitis AYP1020 (CP007601), Staphylococcus warneri SG1 (CP003668), Staphylococcus pasteuri SP1 (CP004014), Staphylococcus haemolyticus S167 (CP013911), S. haemolyticus JCSC1435 (AP006716), Staphylococcus lugdunensis HKU09-01 (CP001837), Staphylococcus hyicus (CP008747), S. condimenti DSM 11674 (CP015114), Staphylococcus carnosus TM300 (AM295250), Staphylococcus xylosus SMG-121 (CP008724), S. xylosus C2a (LN554884), S. xylosus HKUOPL8 (CP007208.1), Staphylococcus saprophyticus ATCC 15305 (AP008934), Staphylococcus aureus RF122 (AJ938182), S. aureus COL (CP000046), S. aureus ED98 (CP001781), S. aureus JH1 (CP000736), and S. aureus JH9 (CP000703). (B) Core genome analysis of S. condimenti DSM 11674, S. carnosus TM300, and S. xylosus SMG-121. Numbers inside the Venn diagrams indicate the number of genes found to be shared among the indicated genomes.
Fermentative Activity-Associated Genes
In silico analyses revealed that complete pathways involved in the reduction of nitrate to nitrite (nitrate reductase, WP_047131530) and further to ammonia (nitrite reductase, WP_047131535) were found in the genome of DSM 11674. Two catalases (WP_047130958, WP_047132101) were also identified in genome. These data are in agreement with our enzyme activity results and provide clues to explain the production of both nitrate reductase and catalase in DSM 11674. In addition, two l-lactate dehydrogenase (WP_047131934, WP_047132743) and two d-lactate dehydrogenase (WP_047132560, WP_047131604) were encoded, which match with the phenotypic trait that both l-lactate and d-lactate are produced in this strain (Probst et al., 1998). Interestingly, lactate dehydrogenase has been reported to play a role in the improvement of starter fermentative activity (Cheng et al., 2014). Therefore, these results indicated that DSM 11674 has strong potential for use as a novel starter culture.
Salt-Dependent and Salt Acclimation Genes
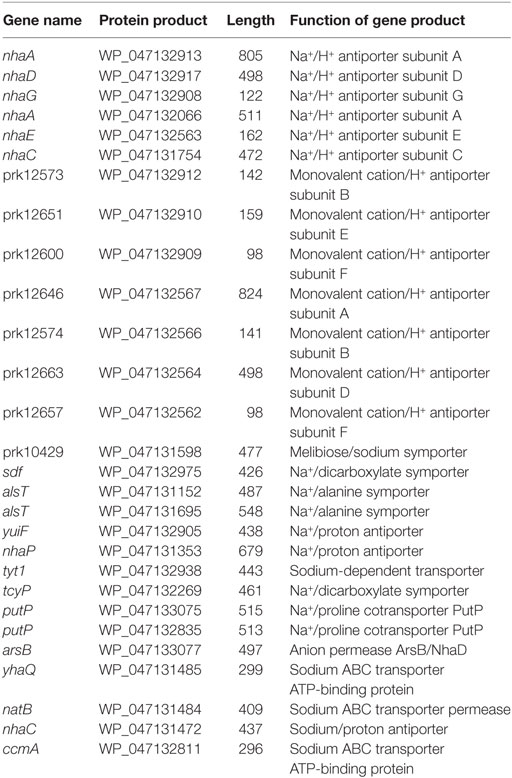
During soy sauce mash fermentation, the DSM 11674 strain experiences significant osmotic stress. To explain the genetic determinants involved in the acclimation of this strain to high salt conditions, we identified several genes known to be important for survival under saline stress (Table 1). The strain contains six Na+/H+ antiporter subunits and seven monovalent cation/H+ antiporter subunits, which are homologs of the antiporter genes of S. carnosus TM300. Furthermore, we identified 15 additional salt-dependent and salt acclimation genes in the DSM 11674 strain. This high content of osmoprotective factors in the genome is consistent well with the ability of this species to grow readily in the presence of 15% NaCl (Probst et al., 1998).
Stress Response and Antimicrobial Resistance Genes
The DSM 11674 genome also possesses genes encoding an ATP synthase complex (WP_047132344-WP_047132350), which enables regulation of the internal pH and could confer the ability to adapt to stressful conditions (Cotter and Hill, 2003). Moreover, we identified two cold-shock protein-enconding (CspA and CspC) genes in the chromosome, which are involved in stress responses (Katzif et al., 2003). Also, the heat-shock regulon (hrcA-grpE-dnaK-dnaJ and groESL) (Singh et al., 2007; Rossi et al., 2017) and several other heat-shock protein encoding genes were found in DSM 11674. Finally, the screening of antimicrobial resistance genes revealed a putative β-lactamase encoding gene; this was consistent with susceptibility testing results. Thus, DSM 11674 strain shows technological characteristics that makes it a good candidate for biotechnical application.
In summary, this study reports the complete genome sequence of S. condiment, a bacterial strain that is potentially useful in a variety of food preparation applications. Genomics-based analysis of this functional staphylococci starter culture candidate revealed important insights into its metabolic capacities and niche adaptations. This is also the first comparative genome sequence analysis of staphylococci starter culture strains, revealing their core genome and pan genome. Finally, the biochemical and genetic characteristics of S. condimenti DSM 11674 revealed in this study are essential to generate further insights into the functional role of staphylococci in general and S. condimenti in particular during the food fermentation process.
Ethics Statement
This article does not contain any studies with human participants or animals performed by any of the authors.
Data Access
The complete genome sequence of Staphylococcus condimenti DSM 11674 has been deposited at DDBJ/EMBL/GenBank under the accession number CP015114.
Author Contributions
BZ conceived and designed the research; HD and JC performed experiments and analyzed data; LG and AH analyzed data; BZ, HD, and AH wrote the manuscript; and all authors commented on the manuscript and approved the contents.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Dr. Tiancheng Zhang of Tianjin Lakeside Pangugene Development co., Ltd (TLPC) for PacBio SMRT sequencing.
Funding
This work was supported by the National Key R&D Program of China (No. 2016YFD0501105); National Natural Science Foundation of China (81301461); and Zhejiang Provincial Natural Science Foundation of China (No. LY17H190003).
Supplementary Material
The Supplementary Material for this article can be found online at http://journal.frontiersin.org/article/10.3389/fbioe.2017.00056/full#supplementary-material.
Figure S1. Biochemical characteristics of Staphylococcus condimenti DSM 11674. (A) Nitrate reductase activity of S. condimenti DSM 11674. (B) Catalase activity of S. condimenti DSM 11674. Enzyme activity was measured as spectrophotometric methods. Staphylococcus aureus ATCC 25923 and clinical isolate S. condimenti CJ1628 were used for comparison.
Figure S2. Genome atlas of Staphylococcus condimenti DSM 11674. The circles represent (from the outside to the inside): circle 1, reverse CDS (cyan); circle 2, forward CDS (yellow); circle 3, tRNAs (blue); circle 4, rRNAs (red); circle 5, GC plot; and circle 6, GC skew.
Figure S3. Genomic comparison of the Staphylococcus condimenti DSM 11674 with starter culture strains Staphylococcus carnosus TM300 and Staphylococcus xylosus SMQ121 by Mauve. Alignment is represented as local collinear blocks (LCBs) filled with a similarity plot. LCBs of conserved sequences among the strains are represented by rectangles of the same color. Connecting lines can be used to visualize synteny or rearrangement. LCBs positioned above or under the chromosome (black line) correspond to the forward and reverse orientation, respectively. The level of conservation is equivalent to the level of vertical color filling within the LCBs. Sequences not placed within an LCB are unique for the particular strain.
Footnotes
References
Argemi, X., Riegel, P., Lavigne, T., Lefebvre, N., Grandpre, N., Hansmann, Y., et al. (2015). Implementation of matrix-assisted laser desorption ionization-time of flight mass spectrometry in routine clinical laboratories improves identification of coagulase-negative staphylococci and reveals the pathogenic role of Staphylococcus lugdunensis. J. Clin. Microbiol. 53, 2030–2036. doi: 10.1128/JCM.00177-15
Aziz, R. K., Bartels, D., Best, A. A., Dejongh, M., Disz, T., Edwards, R. A., et al. (2008). The RAST server: rapid annotations using subsystems technology. BMC Genomics 9:75. doi:10.1186/1471-2164-9-75
Blaiotta, G., Casaburi, A., and Villani, F. (2005). Identification and differentiation of Staphylococcus carnosus and Staphylococcus simulans by species-specific PCR assays of sodA genes. Syst. Appl. Microbiol. 28, 519–526. doi:10.1016/j.syapm.2005.03.007
Chen, L., Yang, J., Yu, J., Yao, Z., Sun, L., Shen, Y., et al. (2005). VFDB: a reference database for bacterial virulence factors. Nucleic Acids Res. 33, D325–D328. doi:10.1093/nar/gki008
Cheng, X., Dong, Y., Su, P., and Xiao, X. (2014). Improvement of the fermentative activity of lactic acid bacteria starter culture by the addition of Mn(2)(+). Appl. Biochem. Biotechnol. 174, 1752–1760. doi:10.1007/s12010-014-1156-z
Chin, C. S., Alexander, D. H., Marks, P., Klammer, A. A., Drake, J., Heiner, C., et al. (2013). Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat. Methods 10, 563–569. doi:10.1038/nmeth.2474
CLSI. (2016). Performance Standards for Antimicrobial Susceptibility Testing: Twenty-Six Informational Supplement M100-S26. Wayne, PA: Clinical and Laboratory Standards Institute.
Cotter, P. D., and Hill, C. (2003). Surviving the acid test: responses of Gram-positive bacteria to low pH. Microbiol. Mol. Biol. Rev. 67, 429–453. doi:10.1128/MMBR.67.3.429-453.2003
Darling, A. E., Mau, B., and Perna, N. T. (2010). progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS ONE 5:e11147. doi:10.1371/journal.pone.0011147
El Haddad, L., Ben Abdallah, N., Plante, P. L., Dumaresq, J., Katsarava, R., Labrie, S., et al. (2014). Improving the safety of Staphylococcus aureus polyvalent phages by their production on a Staphylococcus xylosus strain. PLoS ONE 9:e102600. doi:10.1371/journal.pone.0102600
Gotterup, J., Olsen, K., Knochel, S., Tjener, K., Stahnke, L. H., and Moller, J. K. (2007). Relationship between nitrate/nitrite reductase activities in meat associated staphylococci and nitrosylmyoglobin formation in a cured meat model system. Int. J. Food Microbiol. 120, 303–310. doi:10.1016/j.ijfoodmicro.2007.08.034
Grissa, I., Vergnaud, G., and Pourcel, C. (2007). CRISPRFinder: a web tool to identify clustered regularly interspaced short palindromic repeats. Nucleic Acids Res. 35, W52–W57. doi:10.1093/nar/gkm360
Herrero, M., Mayo, B., Gonzalez, B., and Suarez, J. E. (1996). Evaluation of technologically important traits in lactic acid bacteria isolated from spontaneous fermentations. J. Appl. Bacteriol. 81, 565–570. doi:10.1111/j.1365-2672.1996.tb03548.x
Katzif, S., Danavall, D., Bowers, S., Balthazar, J. T., and Shafer, W. M. (2003). The major cold shock gene, cspA, is involved in the susceptibility of Staphylococcus aureus to an antimicrobial peptide of human cathepsin G. Infect. Immun. 71, 4304–4312. doi:10.1128/IAI.71.8.4304-4312.2003
Lagesen, K., Hallin, P., Rodland, E. A., Staerfeldt, H. H., Rognes, T., and Ussery, D. W. (2007). RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 35, 3100–3108. doi:10.1093/nar/gkm160
Landeta, G., Curiel, J. A., Carrascosa, A. V., Munoz, R., and De Las Rivas, B. (2013). Characterization of coagulase-negative staphylococci isolated from Spanish dry cured meat products. Meat Sci. 93, 387–396. doi:10.1016/j.meatsci.2012.09.019
Liu, B., and Pop, M. (2009). ARDB – antibiotic resistance genes database. Nucleic Acids Res. 37, D443–D447. doi:10.1093/nar/gkn656
Lowe, T. M., and Eddy, S. R. (1997). tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25, 955–964. doi:10.1093/nar/25.5.0955
Miralles, M. C., Flores, J., and Perezmartinez, G. (1996). Biochemical tests for the selection of Staphylococcus strains as potential meat starter cultures. Food Microbiol. 13, 227–236. doi:10.1006/fmic.1996.0028
Misawa, Y., Yoshida, A., Okugawa, S., and Moriya, K. (2015). First reported case of Staphylococcus condimenti infection associated with catheter-related bacteraemia. New Microbes New Infect. 3, 18–20. doi:10.1016/j.nmni.2014.10.002
Neubauer, H., Pantel, I., and Gotz, F. (1999). Molecular characterization of the nitrite-reducing system of Staphylococcus carnosus. J. Bacteriol. 181, 1481–1488.
Page, A. J., Cummins, C. A., Hunt, M., Wong, V. K., Reuter, S., Holden, M. T., et al. (2015). Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics 31, 3691–3693. doi:10.1093/bioinformatics/btv421
Probst, A. J., Hertel, C., Richter, L., Wassill, L., Ludwig, W., and Hammes, W. P. (1998). Staphylococcus condimenti sp. nov., from soy sauce mash, and Staphylococcus carnosus (Schleifer and Fischer 1982) subsp. utilis subsp. nov. Int. J. Syst. Bacteriol. 48(Pt 3), 651–658. doi:10.1099/00207713-48-3-651
Resch, M., Nagel, V., and Hertel, C. (2008). Antibiotic resistance of coagulase-negative staphylococci associated with food and used in starter cultures. Int. J. Food Microbiol. 127, 99–104. doi:10.1016/j.ijfoodmicro.2008.06.013
Rosenstein, R., Nerz, C., Biswas, L., Resch, A., Raddatz, G., Schuster, S. C., et al. (2009). Genome analysis of the meat starter culture bacterium Staphylococcus carnosus TM300. Appl. Environ. Microbiol. 75, 811–822. doi:10.1128/AEM.01982-08
Rossi, C. C., De Oliveira, L. L., De Carvalho Rodrigues, D., Urmenyi, T. P., Laport, M. S., and Giambiagi-Demarval, M. (2017). Expression of the stress-response regulators CtsR and HrcA in the uropathogen Staphylococcus saprophyticus during heat shock. Antonie Van Leeuwenhoek 110, 1105–1111. doi:10.1007/s10482-017-0881-z
Seitter, M., Geng, B., and Hertel, C. (2011a). Binding to extracellular matrix proteins and formation of biogenic amines by food-associated coagulase-negative staphylococci. Int. J. Food Microbiol. 145, 483–487. doi:10.1016/j.ijfoodmicro.2011.01.026
Seitter, M., Nerz, C., Rosenstein, R., Gotz, F., and Hertel, C. (2011b). DNA microarray based detection of genes involved in safety and technologically relevant properties of food associated coagulase-negative staphylococci. Int. J. Food Microbiol. 145, 449–458. doi:10.1016/j.ijfoodmicro.2011.01.021
Singh, V. K., Utaida, S., Jackson, L. S., Jayaswal, R. K., Wilkinson, B. J., and Chamberlain, N. R. (2007). Role for dnaK locus in tolerance of multiple stresses in Staphylococcus aureus. Microbiology 153, 3162–3173. doi:10.1099/mic.0.2007/009506-0
Zell, C., Resch, M., Rosenstein, R., Albrecht, T., Hertel, C., and Gotz, F. (2008). Characterization of toxin production of coagulase-negative staphylococci isolated from food and starter cultures. Int. J. Food Microbiol. 127, 246–251. doi:10.1016/j.ijfoodmicro.2008.07.016
Keywords: Staphylococcus condimenti, complete genome, comparative genomic analysis, starter culture, fermentation
Citation: Dong H, Chen J, Hastings AK, Guo L and Zheng B (2017) Complete Genome Sequence and Comparative Analysis of Staphylococcus condimenti DSM 11674, a Potential Starter Culture Isolated from Soy Sauce Mash. Front. Bioeng. Biotechnol. 5:56. doi: 10.3389/fbioe.2017.00056
Received: 01 July 2017; Accepted: 11 September 2017;
Published: 06 October 2017
Edited by:
Xiao-Jun Ji, Nanjing Tech University, ChinaCopyright: © 2017 Dong, Chen, Hastings, Guo and Zheng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Beiwen Zheng, emhlbmdidyYjeDAwMDQwO3pqdS5lZHUuY24=
†These authors have contributed equally to this work.
 Huihui Dong1,2†
Huihui Dong1,2† Andrew K. Hastings
Andrew K. Hastings Lihua Guo
Lihua Guo Beiwen Zheng
Beiwen Zheng