- 1Institute of Biology, Federal University of Uberlândia, Uberlândia, Minas Gerais, Brazil
- 2Illinois Natural History Survey, University of Illinois at Urbana-Champaign, Champaign, IL, United States
- 3Pontifical Catholic University of Rio Grande do Sul, Porto Alegre, Rio Grande do Sul, Brazil
- 4Institute for Genomics and Evolutionary Medicine, Temple University, Philadelphia, PA, United States
- 5Department of Biology, University of Louisiana at Lafayette, Lafayette, LA, United States
Neotropical Freshwater Fish (NFF) fauna exhibits the greatest phenotypic disparity and species richness among all continental aquatic vertebrate faunas, with more than 6,345 species distributed across the mostly tropical regions of Central and South America. The last two decades have seen a proliferation of molecular phylogenies, often at the species level, covering almost all 875 valid NFF genera. This study presents the most comprehensive genome-wide, time-calibrated phylogenetic hypothesis of NFF species to date, based on DNA sequences generated over decades through the collaborative efforts of the multinational ichthyological research community. Our purpose is to build and curate an extensive molecular dataset allowing researchers to evaluate macroevolutionary hypotheses in the NFF while facilitating continuous refinement and expansion. Using thousands of DNA sequences from dozens of studies, we compiled a supermatrix of 51 markers for 5,984 taxa, representing 3,167 NFF species. Based on this dataset, we built the most species-rich time-calibrated phylogeny of the NFF taxa to date, summarizing the collective efforts of the ichthyological research community since the midpoint of the last century. We provide a summary review of this remarkable evolutionary history and hope this dataset provides a framework for forthcoming studies of the NFF fauna, documenting compelling, emergent patterns in the world’s most diverse continental vertebrate fauna.
Introduction
The Neotropics is a vast biological realm encompassing over 17 million km2 from Central Mexico to southern Argentina, home to some of the world’s largest rivers, including the Amazon, Orinoco, and La Plata (Lévêque et al., 2008; Van der Sleen and Albert, 2022; Dagosta et al., 2024). These rivers harbor the most diverse continental aquatic vertebrate fauna on Earth, an astonishing diversity of freshwater fishes totaling over 6,345 valid species at present, with more discovered every year (Reis et al., 2016; Albert et al., 2020; Chamon et al., 2022; Becker et al., 2023; Dagosta et al., 2024). Among these are many families found nowhere else on earth, such as the piranha (Serrasalmidae) and electric eel (Gymnotidae), lending the Neotropical Freshwater Fish (NFF) fauna a readily identifiable character and a unique place among the freshwater faunas of the world (Albert et al., 2011).
A large majority (77%) of NFF species diversity is concentrated in just five taxonomic orders (Albert et al., 2011); Characiformes, Cichliformes, Cyprinodontiformes, Gymnotiformes, and Siluriformes. The NFF fauna evolved in isolation for tens of millions of years when South America was an island continent, between its final separation from Africa c. 100 Ma and Antarctica (c. 34 Ma) to its connection with Central America c. 12–3.5 Ma. Over these millions of years, the evolutionary history of NFF taxa was shaped by dynamic processes of river capture (Tagliacollo et al., 2015; Albert et al., 2018; Boschman et al., 2023) and marine incursions (Lovejoy et al., 1998; Bloom et al., 2011; Abreu et al., 2020) at continental scales. This process gave rise to the greatest variety of phenotypes and ecological traits of any continental fish fauna on Earth (Su et al., 2019; Albert et al., 2020). The macroevolutionary patterns in the NFF fauna therefore serve as a compelling example of how continental faunas diversify over evolutionary time periods (Miller and Román-Palacios, 2021), driven by the interactions between intrinsic organismal traits and environmental factors like plate tectonics and climate variability (Silva et al., 2016; Cassemiro et al., 2023; Val et al., 2022).
Yet studying this evolutionary history is challenging as, with more than 6,345 species, inferring comprehensive and accurate relationships among NFF clades is expensive in time and computational resources regardless of the datasets or methods used for phylogeny estimation. This challenge is compounded by the phenotypic diversity of these fishes, which exhibit a myriad of morphological, physiological and behavioral adaptations (Crampton, 2011; Val and Almeida-Val, 2012; Albert et al., 2020; Soares et al., 2023). Nevertheless, substantial molecular data have been produced by the ichthyological research community across hundreds of studies and are now available in public repositories (e.g., Musilová et al., 2008; Cramer, et al., 2011; Oliveira et al., 2011; Thomaz et al., 2015; Tagliacollo et al., 2016; Bragança and Costa, 2018). These data give us an opportunity to build a large-scale, time-calibrated phylogeny as a framework for future evolutionary studies documenting macro-scale patterns in the world’s most diverse continental vertebrate fauna.
Such a phylogeny will help us disentangle the complexity that makes the Neotropics the most species-rich region for freshwater fish on Earth in several ways (Cerezer et al., 2023; Cassemiro et al., 2023). A comprehensive phylogenetic tree is essential for studying evolutionary patterns, offering insights into speciation (Albert et al., 2020), adaptive radiation (López-Fernández, 2021), and lineage diversification (Miller and Román-Palacios, 2021). Considering a phylogenetic tree in the context of major earth history events further reveals the historical distributions of lineages coexisting under similar geological or trophic conditions (Emerson and Gillespie, 2008; Baumsteiger et al., 2012). A phylogeny will also enable large-scale macroevolutionary analyses by facilitating the investigation of rate changes over time (Graham et al., 2018), morphological variations across distantly related lineages (Winemiller, 1991; Jablonski, 2017), and other macroevolutionary patterns (Henao Diaz et al., 2019). Comprehensive phylogenetic trees can even inform conservation strategies by clarifying diversity patterns and identifying unique lineages within regions of high evolutionary diversity (Albert et al., 2020). These insights are invaluable for prioritizing conservation efforts (Dagosta et al., 2021).
To this end, we build on the work of dozens of ichthyologists spanning over three decades of taxonomic, phylogenetic and biogeographical research to present a comprehensive, time-calibrated phylogeny of NFF diversity. The purpose of this study is not primarily to introduce new evolutionary hypotheses or propose new taxonomic classifications, but to compile and systematically organize an extensive, reproducible, and extensible dataset of DNA sequences, alignments, matrices, and phylogenetic trees into a single resource. This is a collective effort, integrating genetic alignments, phylogenetic trees, and divergence time estimates to summarize our current understanding of the evolutionary history of the most diverse continental ichthyofauna on Earth. We aim to continue refining and expanding the dataset, enabling future studies to test alternative evolutionary hypotheses in the NFF. By elucidating NFF relationships on a broad scale, this study establishes a framework for further research and opens new avenues in historical biogeography, macroecology, and macroevolution within the Neotropical ichthyofauna.
Methodology
NFF species
The discovery of new species and the reclassification of existing ones are essential processes for refining our understanding of biodiversity and species boundaries. This taxonomic evolution inevitably makes constructing an up-to-date phylogeny challenging. We compiled the list of all valid NFF species described up to December 2021 using information available from original descriptions (Thimotheo et al., 2020; Malabarba et al., 2021), taxonomic reviews (e.g., Crampton et al., 2016; Villa-Navarro et al., 2017; Ferrer and Malabarba, 2020), checklists (Ferraris et al., 2017; Beltrão et al., 2019; Meza-Vargas et al., 2021), and online catalogs (e.g., Eschmeyer’s Catalog of Fishes) (Fricke, 2021). In total, we compiled a list of 6,345 species, 870 genera and 97 families.
Acquiring DNA sequences
We used the R package PhylotaR (Bennett et al., 2018) to extract published DNA sequences from metadata repositories such as GenBank. PhylotaR is a pipeline designed to automate the retrieval of orthologous DNA sequences, enabling the construction of large-scale phylogenetic trees. It clusters sequences into operational taxonomic units (OTUs) based on predefined taxonomic ranks, such as families, genera, or species, and organizes them by gene region. We used PhylotaR to retrieve sequences for each OTU across all loci from GenBank, downloading them at the family level, using the GenBank Taxonomy Database as a reference (Schoch et al., 2020). To mitigate potential errors from taxonomic misidentification or low-quality sequencing, we retrieved a maximum of three sequences (individuals) per species, when available, and visually inspected the alignments. The vast majority (over 95%) of sequences ascribed to the same species exhibited near-zero nucleotide variation, strongly indicating that the individual sequences were correctly identified, and we therefore retained these in the alignment. In cases where multiple sequences purporting to represent the same species showed high nucleotide variation, suggesting issues related to taxonomic misidentification or sequencing errors, an outlier could be easily identified on the basis of poor homology or imprecise identification (e.g., names including “cf.”, “sp.” and “aff.”), and these were excluded. This approach allowed us to detect and exclude outliers, mitigating aberrant phylogenetic signals due to poor data curation. A similar approach has been applied successfully in other phylogenetic studies (e.g., Arce-H et al., 2017; Mirande, 2019).
PhylotaR retrieved DNA sequences from GenBank through BLAST searches, identifying 51 orthologous clusters, resulting in 43,850 bp, 21,223 sequences, 5,984 terminals, and 3,167 species representing 741 genera (or 86% of NFF genera). The retrieved DNA markers consisted of 61% mitochondrial and 39% nuclear genomic sequences, primarily comprising protein-coding genes and a few non-coding genomic regions (e.g., tRNAs; rRNAs) many of which have been commonly sequenced for phylogenetic research (e.g., Li et al., 2007; Oliveira et al., 2011; Tagliacollo et al., 2012; Ochoa et al., 2017; Melo et al., 2018; Fontenelle et al., 2021). Since orthologous sequences were independently retrieved at the family level, some markers were available for certain families but not for others. Nonetheless, all families showed some degree of marker overlap, ensuring reliable empirical estimates of species evolutionary relationships. To elucidate relationships at finer taxonomic scales, we split the D-LOOP marker into alignments for three specific clades: Siluriformes, Poecilidae, and Cichlidae. This strategy guaranteed a more reliable hypothesis of homologies for this rapidly evolving genetic marker. The complete list of accessions can be found in the Supplementary Material, located in the “csv” folder, under the filename “Annex_1 - accession_numbers.xlsx”.
Alignment, trimming and partitioning scheme
After downloading the DNA sequences from GenBank, we individually aligned each genetic marker using MAFFT v. 7 (Katoh and Toh, 2008) with default parameters. To enhance data quality, we conducted trimming with GBlocks (Castresana, 2000; Talavera and Castresana, 2007) to remove gap-rich sites at both ends of the alignments, thereby avoiding subjective manual trimming. We achieved this by setting the “Allowed Gap Positions” parameter to -b5 = h, which permits gaps in internal positions but removes them from the flanking regions. This approach ensures that Gblocks eliminates gaps in non-conserved areas while retaining gaps in more conserved regions of the alignment. We concatenated the trimmed alignments into a supermatrix, assigning each marker to an independent partition and GTR evolutionary model with gamma (G) in four rate categories to account for rate heterogeneity among sites. With PartitionFinder2 (Lanfear et al., 2017), we concurrently estimated an optimal partitioning scheme and identified the best-fitting models of molecular evolution for selected partitions. The best-fitting scheme for the supermatrix estimated 30 independent partitions, with each adhering to a GTR + G model. While the partitions share the GTR + G models, each has an independent set of parameter estimates. Alignments both before and after GBocks analysis and partition information are available in the Supplementary Material in the “mtx” and “pfinder” folders respectively.
Tree inferences
We carried out a maximum likelihood tree search using RAxML-HPC v.8 (Stamatakis, 2014). We incorporated a backbone tree with some node constraints, especially at the family level, based on other evolutionary hypotheses available in the literature (e.g., (Armbruster et al., 2015; Tagliacollo et al., 2016; Lujan, et al., 2018 Melo et al., 2022; Reis and Lehmann, 2022). The backbone tree and its respective node constraints are available in the Supplementary Material in the “mtx” folder. These constraints were necessary due to the low phylogenetic signal for resolving deep node relationships. Unfortunately, alternative genomic markers like UCEs (Faircloth et al., 2012) that are more suitable for inferring deeper node relationships were not available for enough NFF taxa to be included in the matrix. We designated the two lamprey (Agnatha) species Geotria australis and Mordacia lapicida as outgroup taxa and rooted the tree at their most recent common ancestral node. RAxML estimated all free model parameters and optimized the likelihood of the final tree under the GAMMA model using its rapid hill-climbing mode. To assess tree support, we conducted 100 bootstrap replicates on the resulting likelihood tree topology. RAxML analyses were performed on the CIPRES Gateway portal (Miller et al., 2010). Since some analyses are compatible with fully bifurcating trees exclusively and will not be possible given trees with polytomies, we include both fully resolved trees and trees with poorly supported nodes (bootstrap values below 70%) collapsed in our Supplementary Material in the “phy” folder. We advise researchers pursuing downstream work to use the high-confidence trees where possible, and only use the fully resolved trees if their given approach is incompatible with polytomies.
Divergence time estimates
Estimating divergence times concurrently with topology using Bayesian inference would be challenging due to the large size of the matrix and the number of terminals. Additionally, given the geological conditions of the Neotropics, fish fossils are typically quite rare, leaving us with few reliable primary calibrations. As an alternative, we used the congruification method implemented in the R package geiger v2 (Pennell et al., 2014) to estimate divergence times using a reference time-calibrated tree as a source of secondary calibrations (Eastman et al., 2013; Pennell et al., 2014). In brief, divergence times of a target tree (with branch lengths in units of nucleotide substitutions) can be estimated by taking any divergence times also found in the reference tree as secondary calibrations. For this, we selected a published global time-calibrated phylogeny of ray-finned fishes based on 1,105 selected phylogenomic markers from 303 extant species representing 66 or 72 recognized ray-finned fish orders, and including 31 fossil calibrations (Hughes et al., 2018). As an additional source of secondary calibrations among cartilaginous fish, we used the shark tree of life based on 10 fossil calibrations (Stein et al., 2018). Divergences shared between our tree and these two reference trees were identified using the congruify.phylo () function in geiger (Pennell et al., 2014) in R (R Core TeamR, 2023). These divergence times were then used to calibrate nodes in our tree using treePL (Smith and O'Meara, 2012). We first initialized the treePL optimization by setting thorough and prime to TRUE, and then used the default priming step to optimize parameters when estimating divergence times. The suggested optimized parameters included opt = 1, optad = 1, and optcvad = 4. The treePL input files with all calibration points and parameter settings can be found in our Supplementary Materials in the “phy” folder.
Results
Species relationships inferred from RAxML
Here we present the most species-rich NFF tree to date, comprising 5,984 terminals and 3,167 species across 741 genera (Figure 1). The majority of the phylogenetic relationships we recover are consistent with published molecular studies (Oliveira et al., 2011; Near et al., 2012; Sullivan et al., 2013; Tagliacollo et al., 2016; Betancur-R et al., 2017; Hughes et al., 2018; Ilves et al., 2018; Betancur-R et al., 2019), as expected given that much of the data underlying our novel supermatrix was submitted to GenBank as part of numerous past phylogenetic efforts. Relationships among the four largest clades of the NFF, Characiformes, Siluriformes, Cichliformes, and Cyprinodontiformes corroborate the current consensus in fish classification (Dornburg and Near, 2021). Our tree estimates positions Lepisosteiformes (i.e., gars) as the sister group to Teleostei, with the clade Euteleostei as the sister group to Elopiformes (Anguillidae, Megalopidae), Osteoglossiformes (Arapaimidae, Osteoglossidae), and Otocephala (Figure 2). Within Otocephala, our findings support a close relationship between the Clupeiformes and Characiphysae, with Characiformes forming the sister group to Siluriformes and Gymnotiformes (Figure 2).
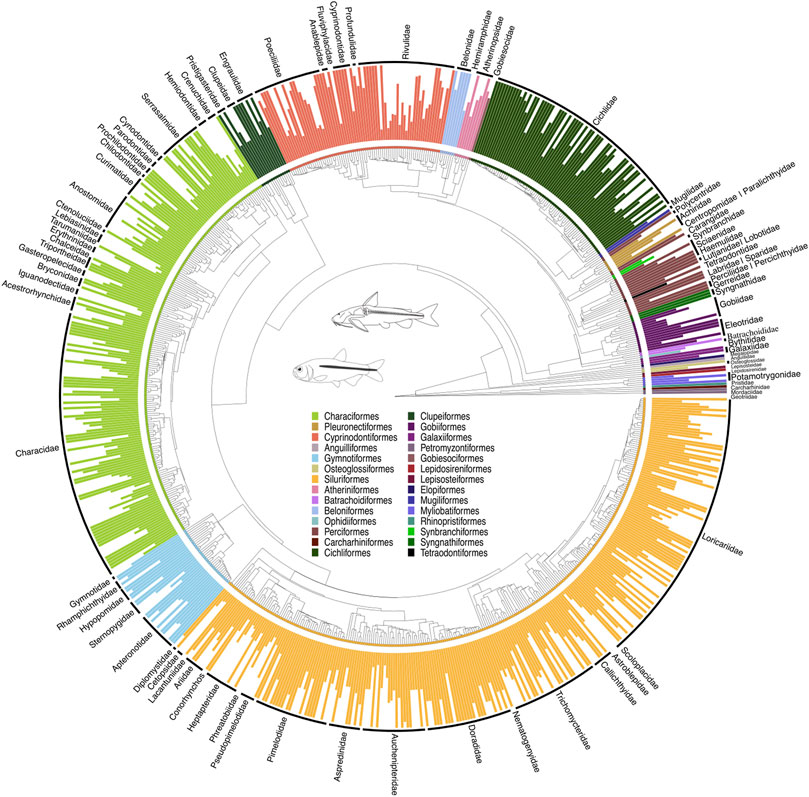
Figure 1. A time-calibrated phylogeny of 3,167 NFF species (or 51% of NFF) allocated to 741 genera (or 86% of NFF), illustrating evolutionary relationships among genera across major lineages. This tree was estimated using RAxML-HPC and time-calibrated through congruification (Eastman et al., 2013) employing divergence tree estimates from Hughes et al. (2018). The tree resulted from alignments of 51 orthologous markers, totaling 43,850 bp, with 21,223 sequences and 5,984 terminals. Vertical bars indicate the complete species per genus represented on the tree. All GenBank accession numbers, alignments, species-level relationships, divergence clade estimates and species representativeness percentages are provided in the Supplementary Material. This phylogeny, as well as one with nodes bearing bootstrap values below 70% collapsed, is available as a Newick string in the Supplementary Material and can be found in the “phy” folder.
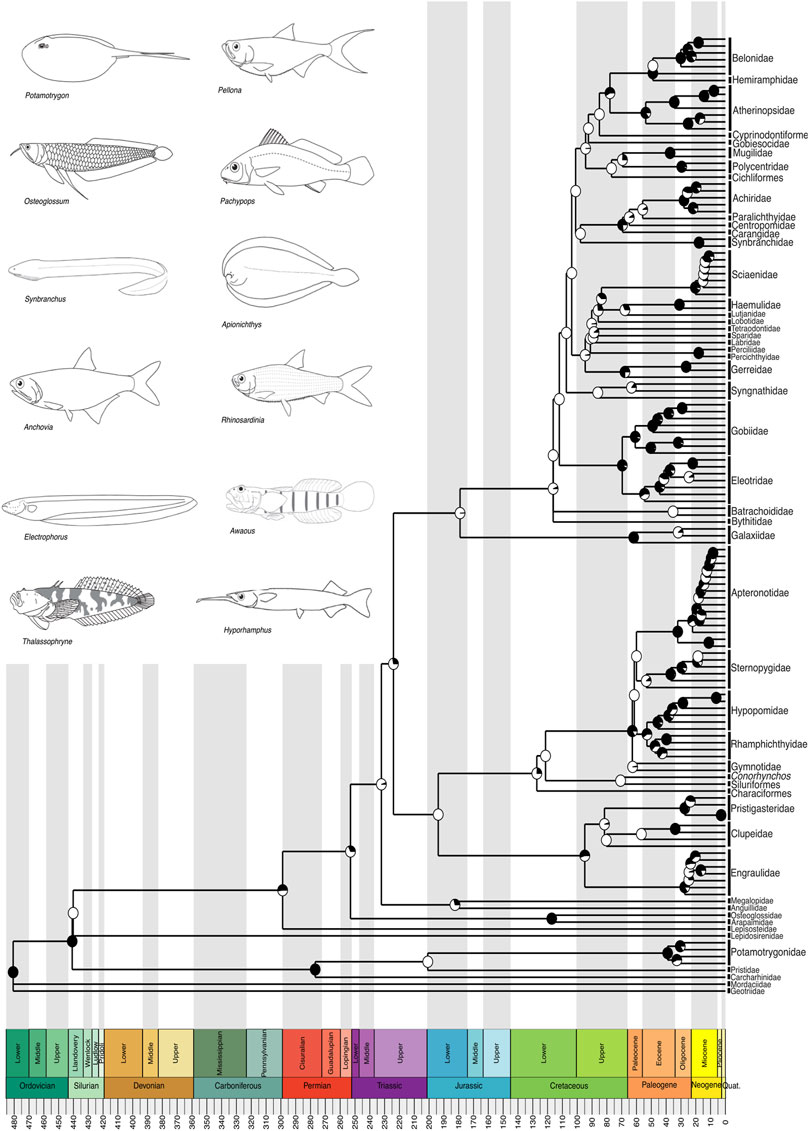
Figure 2. A time-calibrated phylogeny of Neotropical Freshwater Fishes (NFF) illustrating evolutionary relationships among genera from all recognized families, except within Characiformes (shown in detail in Figure 3), Siluriformes (shown in detail in Figure 4), Cichliformes (shown in detail in Figure 5), and Cyprinodontiformes (shown in detail in Figure 6). Divergence time estimates indicate that the evolutionary history of the NFF has its roots in the Ordovician, around 480 million years ago (mya), a period of high diversification in the oceans, particularly among agnathans. Pie charts on nodes indicate bootstrap support, with the proportion of black space corresponding to the percentage of bootstrap replicates in support. Full black circles represent 100% support. All GenBank accession numbers, alignments, species-level relationships, divergence clade estimates and species representativeness percentages are provided in the Supplementary Material. This phylogeny, as well as one with nodes bearing bootstrap values below 70% collapsed, is available as a Newick string in the Supplementary Material and can be found in the “phy” folder.
Within the first major NFF clade, Characiformes, which includes the piranha as well as over a thousand small tetras and darters, our tree includes only representatives of the suborder Characoidei, recapitulating the monophyly of families and several well-established relationships at genus level (Oliveira et al., 2011; Tagliacollo et al., 2012; Abe et al., 2014; Thomaz et al., 2015; Melo et al., 2022). These relationships include Crenuchidae as the sister group to all other characiforms, the monophyly of Anostomoidea (sensu Vari, 1983; Vari, 1983; i.e., Curimatidae, Prochilodontidae, Anostomidae, and Chilodontidae, but see Betancur et al., 2019), a sister-group relationship between Ctenoluciidae and Lebiasinidae, and the monophyly of Characidae (sensu Oliveira et al., 2011, but see Melo et al., 2024), including the genus Amazonspinther as the sister group to all remaining characids (Figure 3).
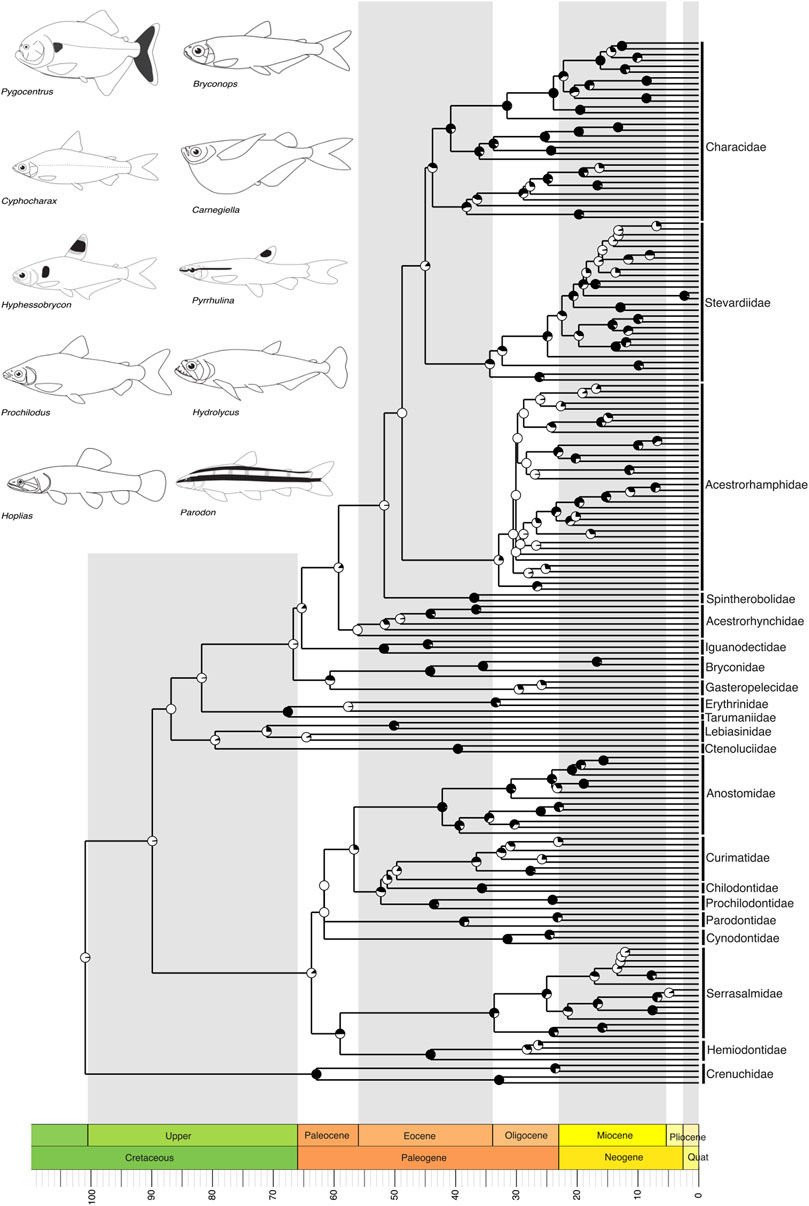
Figure 3. A time-calibrated phylogeny of Characiformes, the second-largest clade in the Neotropics, illustrating evolutionary relationships among genera of all currently recognized families. Divergence times place the most recent common ancestor of Neotropical characiforms at the boundary of the Upper Cretaceous, around 100 million years ago (mya). Pie charts on nodes indicate bootstrap support, with the proportion of black space corresponding to the percentage of bootstrap replicates in support. Full black circles represent 100% support. All GenBank accession numbers, alignments, species-level relationships, divergence clade estimates and species representativeness percentages are provided in the Supplementary Material. This phylogeny, as well as one with nodes bearing bootstrap values below 70% collapsed, is available as a Newick string in the Supplementary Material and can be found in the “phy” folder.
Within the second major clade, Siluriformes, comprising over a thousand catfishes ranging from miniatures such as Scoloplax to the six-foot riverine predator Brachyplatystoma, our tree indicates that Diplomystidae forms the sister group to all other catfishes, which are further divided into two primary clades (sensu Sullivan et al., 2006) Loricarioidei and Siluroidei (Figure 4). Within Siluroidei, several well-established relationships were corroborated, such as the close relationships between Doradidae, Aspredinidae, and Auchenipteridae, as well as the grouping of Pseudopimelodidae, Pimelodidae, Phreatobiidae, Heptapteridae and Conorhynchos (Figure 4). Within Loricarioidei, our tree indicates that Nematogenyidae is the sister group to the remaining families (Figure 4), and Trichomycteridae is sister to remaining loricarioids, with Callichthyidae being sister to advanced loricarioids and Loricariidae, and Astroblepidae forming a sister group to Scoloplacidae (Figure 4). A point to note is that within Trichomycteridae, the subfamily Vandelliinae is not closely related to Tridentinae and Stegophilinae. This result must be interpreted with caution. Several independent studies have shown a close relationship between these three clades including through genomic data (de Pinna, 1998; Ochoa et al., 2017; Ochoa et al., 2020), which are important to our understanding of the origins of parasitism in the Trichomycteridae clade.
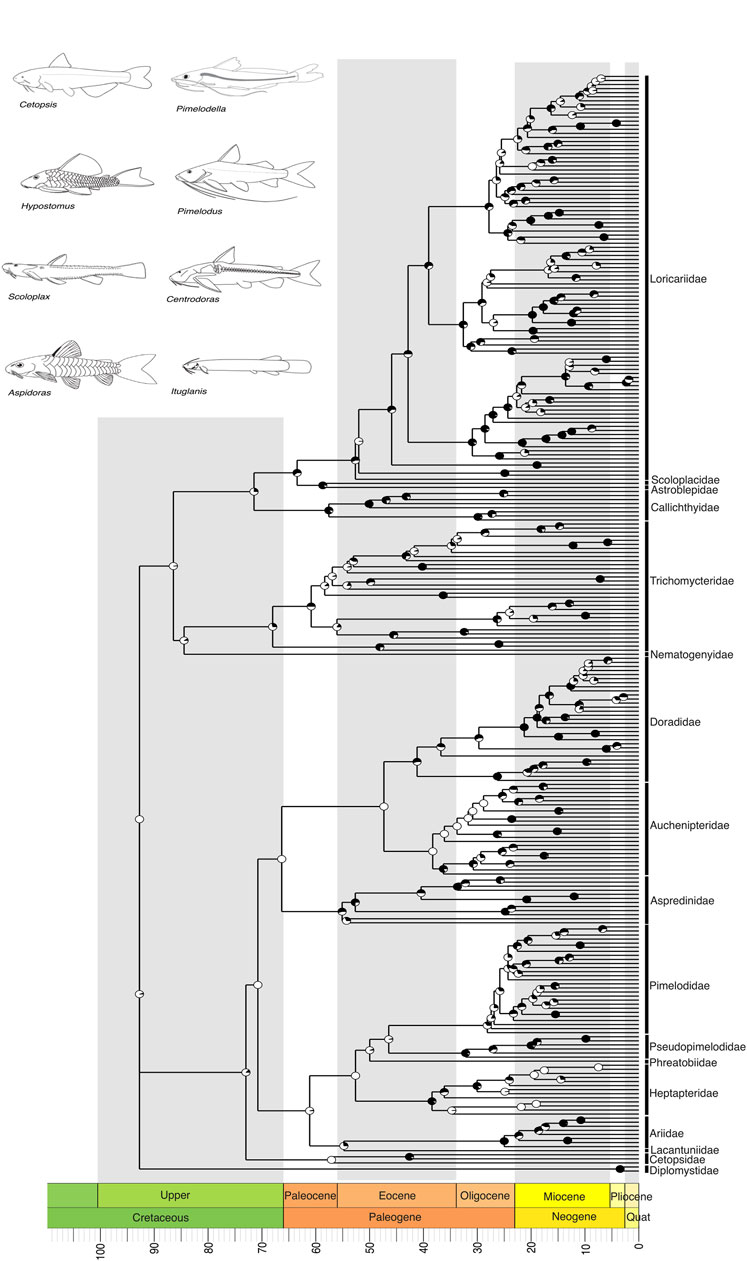
Figure 4. A time-calibrated phylogeny of siluriformes, the largest clade of Neotropical Freshwater Fishes (NFF), illustrating evolutionary relationships among genera of all currently recognized Neotropical families. Divergence time estimates suggest that the most recent common ancestor of Neotropical siluriformes and some of its families (e.g., stem clades: Diplomystidae, Cetopsidae, Nematogenyidae, Trichomycteridae) arose in the Upper Cretaceous, around 100 to 66 million years ago (mya). Pie charts on nodes indicate bootstrap support, with the proportion of black space corresponding to the percentage of bootstrap replicates in support. Full black circles represent 100% support. All GenBank accession numbers, alignments, species-level relationships, divergence clade estimates and species representativeness percentages are provided in the Supplementary Material. This phylogeny, as well as one with nodes bearing bootstrap values below 70% collapsed, is available as a Newick string in the Supplementary Material and can be found in the “phy” folder.
Within the third major clade, Cichliformes, including cichlid fishes like the peacock bass and the freshwater angelfish and discus, our tree supports the monophyly of the seven proposed tribes (sensu López-Fernández et al., 2010; Ilves et al., 2018). The relationships we recover suggest that Cichlini is the sister group to Retroculini, and together they represent the earliest-diverging lineages among Neotropical cichlids (Figure 5). Astronotini and Chaetobranchini were recovered as sister groups, collectively forming a sister clade to Geophagini (Figure 5). This latter clade is related to a large group consisting of Cichlasomatini, which is sister to Heroini, a lineage that includes representatives in Central America (Figure 5). Within Heroini, the biogeographical distribution patterns of species suggest that the tribe originated in South America, with subsequent colonization of Central America and the Caribbean occurring through more than one independent event during the Neogene (Figure 5).
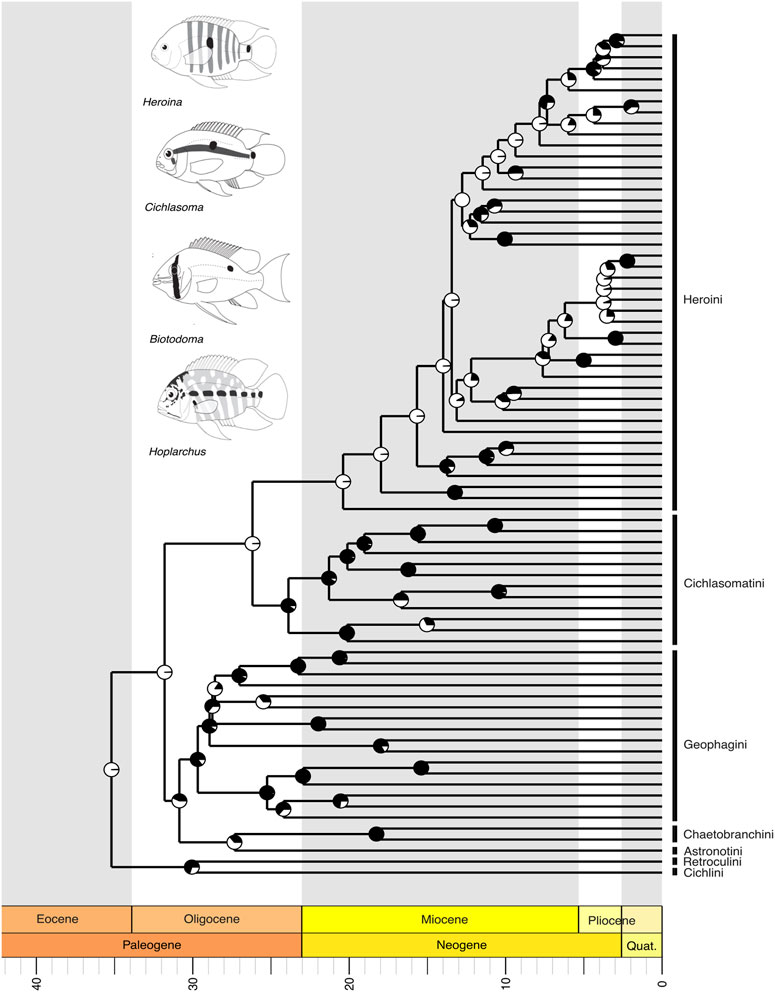
Figure 5. A time-calibrated phylogeny of the Neotropical Cichlidae, showing evolutionary relationships among genera of all currently recognized Neotropical families. Divergence time estimates indicate that Cichlidae is the youngest of the four largest clades of Neotropical Freshwater Fishes (NFF), with the most recent common ancestor evolving in the Cenozoic, during the Eocene, around 40 million years ago (mya). Pie charts on nodes indicate bootstrap support, with the proportion of black space corresponding to the percentage of bootstrap replicates in support. Full black circles represent 100% support. All GenBank accession numbers, alignments, species-level relationships, divergence clade estimates and species representativeness percentages are provided in the Supplementary Material. This phylogeny, as well as one with nodes bearing bootstrap values below 70% collapsed, is available as a Newick string in the Supplementary Material and can be found in the “phy” folder.
Within the fourth major NFF clade, Cyprinodontiformes, including the livebearing guppies and killifish, our tree revealed a close relationship between two major clades: one comprising the family Rivulidae and another consisting of Profundulidae, Cyprinodontidae, Fluviphylacidae, Anablepidae, and Poeciliidae (Figure 6). In this latter clade, we recovered Profundulidae as the sister group to all other families, followed by Cyprinodontidae as sister to the remaining clades (Figure 6). Poeciliidae, the largest group within this clade, was recovered as the sister group to Anablepidae, and together they formed a sister group relationship with Fluviphylacidae (Figure 6). As seen in Cichliformes, the biogeographical distribution patterns suggest that this group originated in South America, with subsequent colonization of Central America.
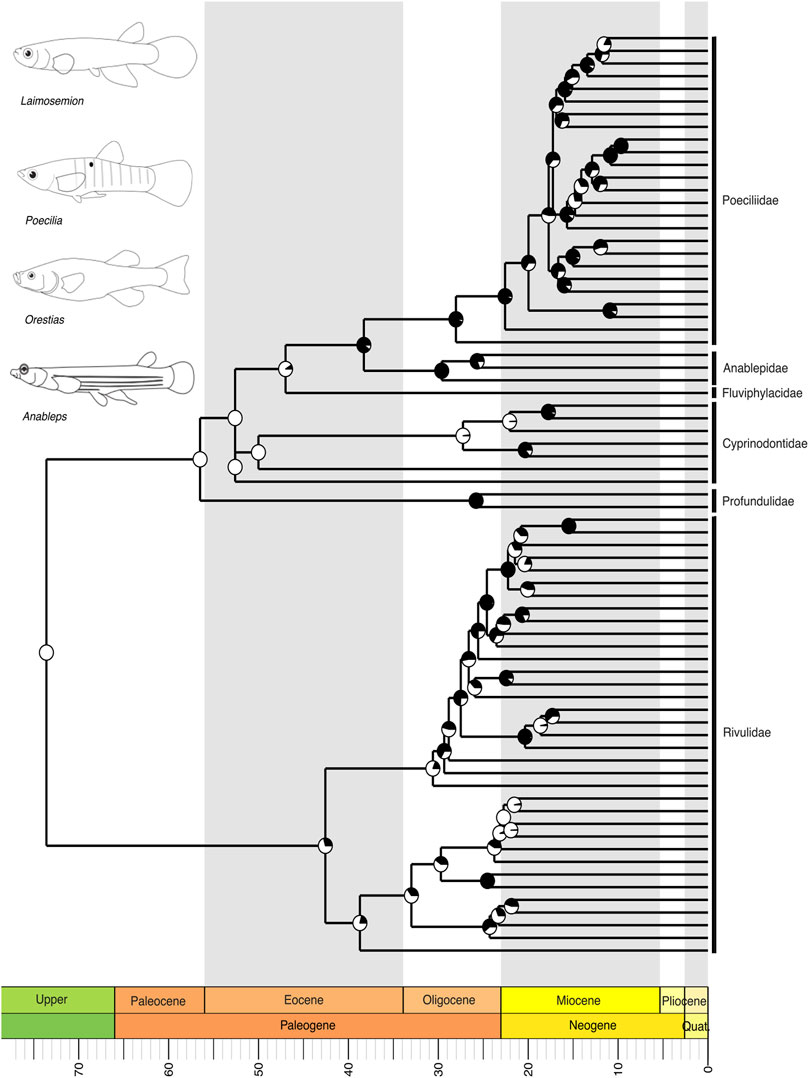
Figure 6. A time-calibrated phylogeny of Cyprinodontiformes illustrates the evolutionary relationships among Neotropical clades including Rivulidae, Profundulidae, Cyprinodontidae, Poeciliidae, Anablepidae, and Fluviphylacidae, as well as their respective genera. Divergence estimates place the origin of this clade in the Upper Cretaceous, with the most recent common ancestor of Rivulidae, Cyprinodontidae, and Fluviphylacidae appearing in the Eocene, and that of Profundulidae, Poeciliidae, and Anablepidae in the Oligocene. Pie charts on nodes indicate bootstrap support, with the proportion of black space corresponding to the percentage of bootstrap replicates in support. Full black circles represent 100% support. All GenBank accession numbers, alignments, species-level relationships, divergence clade estimates and species representativeness percentages are provided in the Supplementary Material. This phylogeny, as well as one with nodes bearing bootstrap values below 70% collapsed, is available as a Newick string in the Supplementary Material and can be found in the “phy” folder.
Time-calibrated tree
Our divergence time estimates align closely with the established timeline of NFF evolution found in our reference phylogenies, Hughes et al. (2018) and Stein et al. (2018), and other published independently-estimated divergence times accessible through TimeTree.org (McMahan et al., 2013; Kappas et al., 2016; Fontenelle et al., 2021; Melo et al., 2022). The NFF earliest lineages to diversify, the jawless lamprey (Agnatha) species, diverged from other clades in the Paleozoic, specifically during the Ordovician, approximately 480 million years ago (mya) (Figure 2). This divergence aligns with the Great Ordovician Biodiversification (GOBI) event, during which vertebrates, particularly fishes, emerged and reached their peak diversification in the Devonian (Servais et al., 2010). The three gnathostome vertebrate lineages with extant freshwater species in the Neotropics—Chondrichthyes, Sarcopterygii, and Actinopterygii—share a most recent common ancestor from the Silurian, with a subsequent split in the Permian, between 299 and 252 mya (Figure 2). The Permian period was marked by the formation of Pangaea and one of the largest mass extinction events in Earth’s history (Erwin et al., 2002), creating new evolutionary opportunities for surviving lineages to diversify, including the Actinopterygian ray-finned fishes, which today dominate freshwater environments worldwide.
The Actinopterygii, encompassing all four major clades of the NFF (Characiformes, Siluriformes, Cichliformes, and Cyprinodontiformes) as well as the electric eel and other electric fishes (Gymnotiformes), diverged from their sister group the Sarcopterygian lobe-finned fishes during the Silurian, with most families within Actinopterygii diversifying during the Paleogene, specifically between 66 and 23 mya (Figure 2). This period was characterized by significant geomorphological changes in South America, including the uplift of the Andes, which rearranged river basins, particularly in the Western Amazon (Hoorn et al., 2010; Wilkinson et al., 2010; Albert et al., 2018) and triggered marine incursions along the sub-Andean Foreland (Bloom et al., 2011). At 23 mya, the Miocene epoch saw the formation of the Pebas system, a network of long-lived lakes and wetlands in western Amazonia (Hoorn et al., 2010; Boschman et al., 2023). Most modern lineage diversification within the NFF occurred during this interval, suggesting that the Pebas system, along with the shift in the Amazon River from its original Northward drainage to its current mouth at Belém in Eastern Brazil, played an important role in shaping one of the richest freshwater vertebrate faunas on Earth (Albert et al., 2018; Albert et al., 2021).
We recover the crown of the Characiformes in the Upper Cretaceous, with most families evolving during the Paleogene, particularly in the Eocene (55–33 mya) (Figure 3). The Siluriformes also originated in the Upper Cretaceous, with the majority of families diversifying during the Paleogene, primarily in the Eocene (Figure 4). Fossil records indicate that both Characiformes and Siluriformes were already established in the continental waters of South America during the Upper Cretaceous (Alves et al., 2019). This is further supported by the modern distribution of Characiformes in Africa and South America, and Siluriformes globally, except in Antarctica (although an Eocene–Oligocene fossil has been reported from Antarctica; (Grande and Eastman, 1986), suggesting that dispersal likely occurred during the era of the Pangaea supercontinent.
The Neotropical cichlids of the subfamily Cichlinae (Cichlidae), the youngest of the major NFF clades, evolved approximately 40 million years ago during the Cenozoic (Figure 5). Within this clade, all South American genera originated in the Miocene, while a few Central American genera emerged later during the Pliocene-Pleistocene (Figure 5). The origins of Central American cichlids remain a point of debate (Holden and Dietz, 1972; MacPhee, 1999; Bacon et al., 2015), likely involving multiple colonization and diversification events associated with the Caribbean plate margins (Tagliacollo et al., 2017). The Cyprinodontiformes of the Neotropics also diverged in the Upper Cretaceous, with the most recent common ancestors of Rivulidae, Cyprinodontidae, and Fluviphylacidae diversifying in the Eocene, while those of Profundulidae, Poeciliidae, and Anablepidae diverged in the Oligocene (Figure 6). All phylogeny files are available in easily viewable and editable Newick format in the Supplementary Material in the “phy” folder. Divergence estimates for NFF families are provided in Supplementary Table S1.
Discussion
Constructing a comprehensive phylogenetic tree of NFF species from public data repositories such as GenBank is important in both scientific and conservation contexts (Winter et al., 2013; Tucker et al., 2017). Scientifically, the tree presented here provides a holistic view of the NFF biodiversity, unraveling evolutionary relationships and establishing a comparative framework to test macroevolutionary hypotheses in the Neotropics. It will also form a baseline for continuous updates as new sequences become available through the collective efforts of taxonomists and evolutionary biologists, advancing our understanding of one of the Earth’s most diverse biotas (e.g., Antonelli et al., 2017). Maintaining an accurate timed phylogeny also benefits databases such as TimeTree.org, allowing researchers who are not phylogenetic experts to export phylogenies or summaries of divergence times for their own downstream analyses (Kumar et al., 2022).
This phylogeny also plays an important role in conservation efforts. It will inform targeted initiatives based on evolutionary distinctiveness and phylogenetic shape and rate metrics, and will contribute to threat assessments among the NFF species (Weber et al., 2017; Albert et al., 2020). Given that continental freshwaters harbor diverse yet fragile ecosystems threatened by human activities, prioritizing geographic areas with the highest number of coexisting evolutionary lineages, including those with threatened species, can significantly enhance conservation efforts for NFF taxa (Tagliacollo et al., 2021; Dagosta et al., 2021).
Maintaining this comprehensive NFF tree presents challenges due to the ongoing discovery of new species, taxonomic revisions, and advancements in genomic sequencing technologies (Remec et al., 2021). Consequently, community-driven efforts are crucial for updating the tree, encompassing the inclusion of newly-described taxa, review of taxonomic changes, and the integration of new DNA markers generated by next-generation sequencing technologies such as UCEs (Faircloth et al., 2012) and exons (Bragg et al., 2016). Over the years, new DNA sequences will be generated as methodologies improve, allowing for the continuous refinement of evolutionary hypotheses. Future studies will also incorporate phylogenomic data, improving node support and removing some tree constraints that were necessary due to the lack of sufficient phylogenetic signal to recover deeper node evolutionary relationships.
Using publicly available sequences for research comes with inherent limitations, including misidentifications of taxa, where sequences may be attributed to the wrong species due to errors in specimen identification, and incomplete coverage in the alignments, which can affect the accuracy and comprehensiveness of analyses. While misidentified sequences are common on public sequence repositories like GenBank (Li et al., 2018), we mitigated their impact through rigorous manual evaluation by multiple experts in Neotropical ichthyology, and we will continue to review public data by the community and evaluate the reliability of sequences during future updates to the tree. As these updates unfold, the tree will expand in the number of both taxa and molecular markers, requiring the utilization of bioinformatic tools to standardize and streamline such updating processes. Because public databases can often contain sequences that are incomplete or represent only certain regions of the genome, which can impact the accuracy and comprehensiveness of analyses (Li et al., 2018), researchers should always be aware of these limitations and biases when using publicly available DNA sequences and exercise caution in interpreting and generalizing results derived from such data. Incorporating multiple data sources and rigorous validation processes can help mitigate these issues to some extent.
One possible alternative is to continue developing methodologies of phylogenetic placement which allow us to determine the phylogenetic position of new species without remaking the entire tree (e.g., Matsen et al., 2010). Importantly, phylogenetic placement techniques require less time and computational power than full Bayeseian or likelihood-based tree inferences and can be implemented collaboratively on platforms connected to computer servers across different locations. While this strategy provides a more efficient means of performing small, localized updates to the phylogenetic tree, it will still periodically be necessary to rebuild the entire tree to incorporate all new taxa and genomic markers, which will be a computationally intensive process. This only underscores the importance of investing in high powered computational infrastructure as the field evolves.
The ichthyological research community is invited to embark on a collaborative journey to enhance the phylogenetic tree of NFF species. With a dedication to continuous data contribution, available online in repositories such as GenBank, periodic taxonomic reviews, and methodological advancements, the community can collectively refine and expand this initial effort to elucidate the evolutionary relationships among NFF taxa. Emphasizing data sharing, interdisciplinary collaboration, and community engagement will foster a clear understanding and application of these phylogenetic insights in future studies. The curation of public repositories, development of new methods, and interdisciplinary collaboration ensure a robust and accessible framework for ongoing research, aiming to reveal the intricate processes and patterns of formation of the diverse NFF taxa. A proactive approach to address data gaps solidifies the commitment to advancing our knowledge of the evolutionary relationships of NFF species, supporting empirical means for biodiversity conservation plans, and contributing to global scientific endeavors uncovering the origins and diversification of freshwater vertebrate faunas in the Neotropics.
Conclusion
This study presents the first comprehensive, time-calibrated phylogeny of NFF species, based on a supermatrix of 51 genes with a total of 43,850 bp for 5,984 specimens in 3,167 species representing 50% of all valid NFF species and 741 or 86% of all NFF genera. This supermatrix reflects three decades of molecular research by the ichthyological community by drawing on sequence data deposited in GenBank in hundreds of separate studies. While standing out as the most species-rich dataset of NFF taxa assembled to date, the accuracy of this phylogeny is affected by the representation of genes and taxa included in previous investigations, with known biases in the representations of fish species by clade (e.g., family), traits (e.g., adult body size), geographic and ecological ranges (e.g., investigator access), and conservation status. Despite these shortcomings, this study establishes an initial framework for fauna-wide investigations of macroevolutionary and macroecological patterns, acknowledging the need for continual updates as science progresses. Our commitment as a scientific community to regular updates ensures its increasing accuracy and relevance, aligning with advancing ichthyological knowledge. This study underscores the importance of collaborative efforts and unlocks avenues for future research in historical biogeography, macroevolution, and macroecology. By fostering active collaboration and embracing scientific inquiry, we hope this assembled tree serves as a start point for more efforts to protect freshwater fishes, marking a stride toward sustainable biodiversity conservation.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author contributions
VT: Conceptualization, Data curation, Formal Analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing–original draft, Writing–review and editing. MT: Formal Analysis, Writing–original draft, Writing–review and editing. RR: Conceptualization, Project administration, Writing–original draft, Writing–review and editing. RG: Visualization, Writing–review and editing. VC: Visualization, Writing–review and editing. MR: Visualization, Writing–review and editing. JC: Conceptualization, Investigation, Project administration, Writing–review and editing. JA: Conceptualization, Data curation, Formal Analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing–original draft, Writing–review and editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. VT is supported by Fundação de Amparo à Pesquisa do Estado de Minas Gerais (FAPEMIG #BPD-00201-22, #APQ-01493-24). RG, VC and MR are supported by Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES). JA is supported by US National Science Foundation DEB 0614334, 0741450, and 1354511. RR is supported by Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq, processes # 302760/2022-9 and # 402665/2023-6). JC is supported by NSF 2318917. This study was financed in part by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior–Brasil (CAPES) – Finance Code 001.
Acknowledgments
We acknowledge Peter van der Sleen for the line artwork.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fbinf.2024.1433995/full#supplementary-material
References
Abe, K. T., Mariguela, T. C., Avelino, G. S., Foresti, F., and Oliveira, C. (2014). Systematic and historical biogeography of the Bryconidae (Ostariophysi: Characiformes) suggesting a new rearrangement of its genera and an old origin of Mesoamerican ichthyofauna. BMC Evol. Biol. 14, 152–215. doi:10.1186/1471-2148-14-152
Abreu, J. M. S., Saraiva, A. C. S., Albert, J. S., and Piorski, N. M. (2020). Paleogeographic influences on freshwater fish distributions in northeastern Brazil. J. S. Am. Earth Sci. 102, 102692. doi:10.1016/j.jsames.2020.102692
Albert, J. S., Bernt, M. J., Fronk, A. H., Fontenelle, J. P., Kuznar, S. L., and Lovejoy, N. R. (2021). Late Neogene megariver captures and the Great Amazonian biotic interchange. Glob. Planet. Change 205, 103554. doi:10.1016/j.gloplacha.2021.103554
Albert, J. S., Craig, J. M., Tagliacollo, V. A., and Petry, P. (2018). Upland and lowland fishes: a test of the river capture hypothesis. Mt. Clim. Biodivers., 273–294.
Albert, J. S., Petry, P., and Reis, R. E. (2011). “Major biogeographic and phylogenetic patterns,” in Historical biogeography of neotropical freshwater fishes. Editors J. S. Albert, and R. E. Reis (Univ. California Press), 21–57.
Albert, J. S., Tagliacollo, V. A., and Dagosta, F. (2020). Diversification of Neotropical freshwater fishes. Annu. Rev. Ecol. Evol. Syst. 51, 27–53. doi:10.1146/annurev-ecolsys-011620-031032
Alves, Y. M., Bergqvist, L. P., and Brito, P. M. (2019). The dorsal and pectoral fin spines of catfishes (Ostariophysi: siluriformes) from the Bauru Group (Late Cretaceous), Brazil: a comparative and critical analysis. J. S. Am. Earth Sci. 92, 32–40. doi:10.1016/j.jsames.2019.02.016
Antonelli, A., Hettling, H., Condamine, F. L., Vos, K., Nilsson, R. H., Sanderson, M. J., et al. (2017). Toward a self-updating platform for estimating rates of speciation and migration, ages, and relationships of taxa. Syst. Biol. 66, 152–166. doi:10.1093/sysbio/syw066
Arce-H, M., Lundberg, J. G., and O'Leary, M. A. (2017). Phylogeny of the North American catfish family Ictaluridae (Teleostei: siluriformes) combining morphology, genes and fossils. Cladistics 33, 406–428. doi:10.1111/cla.12175
Armbruster, J. W., Werneke, D. C., and Tan, M. (2015). Three new species of saddled loricariid catfishes, and a review of Hemiancistrus, Peckoltia, and allied genera (Siluriformes). Zookeys 480, 97–123. doi:10.3897/zookeys.480.6540
Bacon, C. D., Silvestro, D., Jaramillo, C., Smith, B. T., Chakrabarty, P., and Antonelli, A. (2015). Biological evidence supports an early and complex emergence of the Isthmus of Panama. Proc. Natl. Acad. Sci. 112, 6110–6115. doi:10.1073/pnas.1423853112
Baumsteiger, J., Kinziger, A. P., and Aguilar, A. (2012). Life history and biogeographic diversification of an endemic western North American freshwater fish clade using a comparative species tree approach. Mol. Phylogenetics Evol. 65, 940–952. doi:10.1016/j.ympev.2012.08.015
Becker, F. G., Ferrer, J., Loureiro, M., dos Reis, R. B., and Malabarba, L. R. (2023). “Fish diversity and conservation in a neotropical grassland region,” in South Brazilian grasslands: ecology and conservation of the Campos Sulinos (Cham: Springer International Publishing), 319–348. doi:10.1007/978-3-031-42580-6_12
Beltrão, H., Zuanon, J., and Ferreira, E. (2019). Checklist of the ichthyofauna of the rio negro basin in the Brazilian Amazon. ZooKeys 881, 53–89. doi:10.3897/zookeys.881.32055
Bennett, D. J., Hettling, H., Silvestro, D., Zizka, A., Bacon, C. D., Faurby, S., et al. (2018). phylotaR: an automated pipeline for retrieving orthologous DNA sequences from GenBank in R. Life 8, 20. doi:10.3390/life8020020
Betancur-R, R., Arcila, D., Vari, R. P., Hughes, L. C., Oliveira, C., Sabaj, M. H., et al. (2019). Phylogenomic incongruence, hypothesis testing, and taxonomic sampling: the monophyly of characiform fishes. Evolution 73, 329–345. doi:10.1111/evo.13649
Betancur-R, R., Wiley, E. O., Arratia, G., Acero, A., Bailly, N., Miya, M., et al. (2017). Phylogenetic classification of bony fishes. BMC Evol. Biol. 17, 162. doi:10.1186/s12862-017-0958-3
Bloom, D. D., Lovejoy, N. R., Albert, J. S., and Reis, R. E. (2011). The biogeography of marine incursions in South America. Hist. Biogeogr. Neotropical Freshw. fishes, 137–144. doi:10.1525/california/9780520268685.003.0008
Boschman, L. M., Carraro, L., Cassemiro, F. A., de Vries, J., Altermatt, F., Hagen, O., et al. (2023). Freshwater fish diversity in the western Amazon basin shaped by Andean uplift since the Late Cretaceous. Nat. Ecol. and Evol. 7, 2037–2044. doi:10.1038/s41559-023-02220-8
Bragança, P. H., and Costa, W. J. (2018). Time-calibrated molecular phylogeny reveals a Miocene–Pliocene diversification in the Amazon miniature killifish genus Fluviphylax (Cyprinodontiformes: cyprinodontoidei). Org. Divers. and Evol. 18, 345–353. doi:10.1007/s13127-018-0373-7
Bragg, J. G., Potter, S., Bi, K., and Moritz, C. (2016). Exon capture phylogenomics: efficacy across scales of divergence. Mol. Ecol. Resour. 16, 1059–1068. doi:10.1111/1755-0998.12449
Cassemiro, F. A., Albert, J. S., Antonelli, A., Menegotto, A., Wüest, R. O., Cerezer, F., et al. (2023). Landscape dynamics and diversification of the megadiverse South American freshwater fish fauna. Proc. Natl. Acad. Sci. 120, e2211974120. doi:10.1073/pnas.2211974120
Castresana, J. (2000). Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 17, 540–552. doi:10.1093/oxfordjournals.molbev.a026334
Cerezer, F. O., Dambros, C. S., Coelho, M. T., Cassemiro, F. A., Barreto, E., Albert, J. S., et al. (2023). Accelerated body size evolution in upland environments is correlated with recent speciation in South American freshwater fishes. Nat. Commun. 14, 6070. doi:10.1038/s41467-023-41812-7
Chamon, C. C., Serra, J. P., Camelier, P., Zanata, A. M., Fichberg, I., and Marinho, M. M. F. (2022). Building knowledge to save species: 20 years of ichthyological studies in the Tocantins-Araguaia River basin. Biota Neotropica 22, e20211296. doi:10.1590/1676-0611-BN-2021-1296
Cramer, C. A., Bonatto, S. L., and Reis, R. E. (2011). Molecular phylogeny of the neoplecostominae and hypoptopomatinae (siluriformes: Loricariidae) using multiple genes. Mol. phylogenetics Evol. 59, 43–52. doi:10.1016/j.ympev.2011.01.002
Crampton, W. G. (2011). An ecological perspective on diversity and distributions. Hist. Biogeogr. Neotropical Freshw. fishes, 165–189. doi:10.1525/california/9780520268685.003.0010
Crampton, W. G., de Santana, C. D., Waddell, J. C., and Lovejoy, N. R. (2016). Phylogenetic systematics, biogeography, and ecology of the electric fish genus Brachyhypopomus (Ostariophysi: Gymnotiformes). PLoS One 11, e0161680. doi:10.1371/journal.pone.0161680
Dagosta, F. C., de Pinna, M., Peres, C. A., and Tagliacollo, V. A. (2021). Existing protected areas provide a poor safety-net for threatened Amazonian fish species. Mar. Freshw. Ecosyst. 31, 1167–1189. doi:10.1002/aqc.3461
Dagosta, F. C. P., Monção, M. S., Nagamatsu, B. A., Pavanelli, C. S., Carvalho, F. R., Lima, F. C., et al. (2024). Fishes of the upper rio Paraná basin: diversity, biogeography and conservation. Neotropical Ichthyol. 22, e230066. doi:10.1590/1982-0224-2023-0066
de Pinna, M. D. (1998). Phylogenetic relationships of Neotropical Siluriformes (Teleostei: ostariophysi): historical overview and synthesis of hypotheses. Phylogeny Classif. Neotropical fishes.
Dornburg, A., and Near, T. J. (2021). The emerging phylogenetic perspective on the evolution of actinopterygian fishes. Annu. Rev. Ecol. Evol. Syst. 52, 427–452. doi:10.1146/annurev-ecolsys-122120-122554
Eastman, J. M., Harmon, L. J., and Tank, D. C. (2013). Congruification: support for time scaling large phylogenetic trees. Methods Ecol. Evol. 4, 688–691. doi:10.1111/2041-210X.12051
Emerson, B. C., and Gillespie, R. G. (2008). Phylogenetic analysis of community assembly and structure over space and time. Trends Ecol. and Evol. 23, 619–630. doi:10.1016/j.tree.2008.07.005
Erwin, D. H., Bowring, S. A., Yugan, J., Koeberl, C., and MacLeod, K. G. (2002). End-Permian mass extinctions: a review. Special Papers-Geological Soc. Am., 363–384.
Faircloth, B. C., McCormack, J. E., Crawford, N. G., Harvey, M. G., Brumfield, R. T., and Glenn, T. C. (2012). Ultraconserved elements anchor thousands of genetic markers spanning multiple evolutionary timescales. Syst. Biol. 61, 717–726. doi:10.1093/sysbio/sys004
Ferraris, C. J., de Santana, C. D., and Vari, R. P. (2017). Checklist of Gymnotiformes (osteichthyes: ostariophysi) and catalogue of primary types. Neotropical Ichthyol. 15. doi:10.1590/1982-0224-20160067
Ferrer, J., and Malabarba, L. R. (2020). Systematic revision of the Neotropical catfish genus Scleronema (Siluriformes: Trichomycteridae), with descriptions of six new species from Pampa grasslands. Neotropical Ichthyol. 18, e190081. doi:10.1590/1982-0224-2019-0081
Fontenelle, J. P., Portella Luna Marques, F., Kolmann, M. A., and Lovejoy, N. R. (2021). Biogeography of the Neotropical freshwater stingrays (Myliobatiformes: potamotrygoninae) reveals effects of continent-scale paleogeographic change and drainage evolution. J. Biogeogr. 48, 1406–1419. doi:10.1111/jbi.14086
Fricke, R. (2021). Genera/Species by family/subfamily in Eschmeyer's catalog of fishes. California: California Academy of Sciences.
Graham, C. H., Storch, D., and Machac, A. (2018). Phylogenetic scale in ecology and evolution. Glob. Ecol. Biogeogr. 27, 175–187. doi:10.1111/geb.12686
Grande, L., and Eastman, J. T. (1986). A review of Antarctic ichthyofaunas in the light of new fossil discoveries. Palaeontology 29, 113–137.
Henao Diaz, L. F., Harmon, L. J., Sugawara, M. T. C., Miller, E. T., and Pennell, M. W. (2019). Macroevolutionary diversification rates show time dependency. Proc. Natl. Acad. Sci. U.S.A. 116, 7403–7408. doi:10.1073/pnas.1818058116
Holden, J. C., and Dietz, R. S. (1972). Galapagos gore, NazCoPac triple junction and Carnegie/Cocos ridges. Nature 235, 266–269. doi:10.1038/235266a0
Hoorn, C., Wesselingh, F. P., Ter Steege, H., Bermudez, M. A., Mora, A., Sevink, J., et al. (2010). Amazonia through time: andean uplift, climate change, landscape evolution, and biodiversity. science 330, 927–931. doi:10.1126/science.1194585
Hughes, L. C., Ortí, G., Huang, Y., Sun, Y., Baldwin, C. C., Thompson, A. W., et al. (2018). Comprehensive phylogeny of ray-finned fishes (Actinopterygii) based on transcriptomic and genomic data. Proc. Natl. Acad. Sci. 115, 6249–6254. doi:10.1073/pnas.1719358115
Ilves, K. L., Torti, D., and López-Fernández, H. (2018). Exon-based phylogenomics strengthens the phylogeny of Neotropical cichlids and identifies remaining conflicting clades (Cichliformes: Cichlidae: Cichlinae). Mol. Phylogenetics Evol. 118, 232–243. doi:10.1016/j.ympev.2017.10.008
Jablonski, D. (2017). Approaches to macroevolution: 1. General concepts and origin of variation. Evol. Biol. 44, 427–450. doi:10.1007/s11692-017-9420-0
Kappas, I., Vittas, S., Pantzartzi, C. N., Drosopoulou, E., and Scouras, Z. G. (2016). A time-calibrated mitogenome phylogeny of catfish (Teleostei: siluriformes). PLoS One 11, e0166988. doi:10.1371/journal.pone.0166988
Katoh, K., and Toh, H. (2008). Recent developments in the MAFFT multiple sequence alignment program. Briefings Bioinforma. 9, 286–298. doi:10.1093/bib/bbn013
Kumar, S., Suleski, M., Craig, J. M., Kasprowicz, A. E., Sanderford, M., Li, M., et al. (2022). TimeTree 5: an expanded resource for species divergence times. Mol. Biol. Evol. 39, msac174. doi:10.1093/molbev/msac174
Lanfear, R., Frandsen, P. B., Wright, A. M., Senfeld, T., and Calcott, B. (2017). PartitionFinder 2: new methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol. Biol. Evol. 34, 772–773. doi:10.1093/molbev/msw260
Lévêque, C., Oberdorff, T., Paugy, D., Stiassny, M. L. J., and Tedesco, P. A. (2008). “Global diversity of fish (Pisces) in freshwater,” in Freshwater animal diversity assessment, developments in hydrobiology. Editors E. V. Balian, C. Lévêque, H. Segers, and K. Martens (Netherlands, Dordrecht: Springer), 545–567. doi:10.1007/978-1-4020-8259-7_53
Li, C., Ortí, G., Zhang, G., and Lu, G. (2007). A practical approach to phylogenomics: the phylogeny of ray-finned fish (Actinopterygii) as a case study. BMC Evol. Biol. 7, 44–11. doi:10.1186/1471-2148-7-44
Li, X., Shen, X., Chen, X., Xiang, D., Murphy, R. W., and Shen, Y. (2018). Detection of potential problematic Cytb gene sequences of fishes in GenBank. Front. Genet. 9, 30. doi:10.3389/fgene.2018.00030
López-Fernández, H. (2021). “Neotropical riverine cichlids: adaptive radiation and macroevolution at continental scales,” in The behavior, ecology and evolution of cichlid fishes. Editors M. E. Abate, and D. L. G. Noakes (Netherlands, Dordrecht: Springer), 135–173. doi:10.1007/978-94-024-2080-7_5
López-Fernández, H., Winemiller, K. O., and Honeycutt, R. L. (2010). Multilocus phylogeny and rapid radiations in Neotropical cichlid fishes (Perciformes: Cichlidae: Cichlinae). Mol. Phylogenetics Evol. 55, 1070–1086. doi:10.1016/j.ympev.2010.02.020
Lovejoy, N. R., Bermingham, E., and Martin, A. P. (1998). Marine incursion into South America. Nature 396, 421–422. doi:10.1038/24757
Lujan, N. K., Armbruster, J. W., and Lovejoy, N. R. (2018). Multilocus phylogeny, diagnosis and generic revision of the Guiana Shield endemic suckermouth armoured catfish tribe Lithoxini (Loricariidae: hypostominae). Zoological J. Linn. Soc. 184, 1169–1186. doi:10.1093/zoolinnean/zly025
MacPhee, R. D. E. (1999). Paleogeography of the Caribbean region: implications for Cenozoic biogeography. Am. Mus. Nat. Hist. 238, 1–95.
Malabarba, L. R., Chuctaya, J., Hirschmann, A., De Oliveira, E. B., and Thomaz, A. T. (2021). Hidden or unnoticed? MULTIPLE lines of evidence support the recognition of a new species of Pseudocorynopoma (Characidae: corynopomini). J. Fish Biol. 98, 219–236. doi:10.1111/jfb.14572
Matsen, F. A., Kodner, R. B., and Armbrust, E. V. (2010). pplacer: linear time maximum-likelihood and Bayesian phylogenetic placement of sequences onto a fixed reference tree. BMC Bioinforma. 11, 538. doi:10.1186/1471-2105-11-538
McMahan, C. D., Chakrabarty, P., Sparks, J. S., Smith, W. L., and Davis, M. P. (2013). Temporal patterns of diversification across global cichlid biodiversity (Acanthomorpha: Cichlidae). PloS one 8, e71162. doi:10.1371/journal.pone.0071162
Melo, B. F., Ota, R. P., Benine, R. C., Carvalho, F. R., Lima, F. C., Mattox, G. M., et al. (2024). Phylogenomics of Characidae, a hyper-diverse Neotropical freshwater fish lineage, with a phylogenetic classification including four families (Teleostei: Characiformes). Zoological J. Linn. Soc. 202, zlae101. doi:10.1093/zoolinnean/zlae101
Melo, B. F., Sidlauskas, B. L., Hoekzema, K., Vari, R. P., Dillman, C. B., and Oliveira, C. (2018). Molecular phylogenetics of neotropical detritivorous fishes of the family Curimatidae (Teleostei: Characiformes). Mol. Phylogenetics Evol. 127, 800–812. doi:10.1016/j.ympev.2018.06.027
Melo, B. F., Sidlauskas, B. L., Near, T. J., Roxo, F. F., Ghezelayagh, A., Ochoa, L. E., et al. (2022). Accelerated diversification explains the exceptional species richness of tropical characoid fishes. Syst. Biol. 71, 78–92. doi:10.1093/sysbio/syab040
Meza-Vargas, V., Faustino-Fuster, D. R., Chuctaya, J., Hidalgo, M., and Torres, H. O. (2021). Checklist of freshwater fishes from Loreto, Peru. Rev. Peru. Biol. 28, e21911. doi:10.15381/rpb.v28iespecial.21911
Miller, E. C., and Román-Palacios, C. (2021). Evolutionary time best explains the latitudinal diversity gradient of living freshwater fish diversity. Glob. Ecol. Biogeogr. 30, 749–763. doi:10.1111/geb.13253
Miller, M. A., Pfeiffer, W., and Schwartz, T. (2010). “Creating the CIPRES Science Gateway for inference of large phylogenetic trees,” in Gateway computing environments workshop (GCE) (Ieee), 1–8. doi:10.1109/GCE.2010.5676129
Mirande, J. M. (2019). Morphology, molecules and the phylogeny of Characidae (Teleostei, Characiformes). Cladistics 35, 282–300. doi:10.1111/cla.12345
Musilová, Z., Říčan, O., Janko, K., and Novák, J. (2008). Molecular phylogeny and biogeography of the neotropical cichlid fish tribe Cichlasomatini (Teleostei: Cichlidae: cichlasomatinae). Mol. Phylogenetics Evol. 46, 659–672. doi:10.1016/j.ympev.2007.10.011
Near, T. J., Eytan, R. I., Dornburg, A., Kuhn, K. L., Moore, J. A., Davis, M. P., et al. (2012). Resolution of ray-finned fish phylogeny and timing of diversification. Proc. Natl. Acad. Sci. 109, 13698–13703. doi:10.1073/pnas.1206625109
Ochoa, L. E., Datovo, A., DoNascimiento, C., Roxo, F. F., Sabaj, M. H., Chang, J., et al. (2020). Phylogenomic analysis of trichomycterid catfishes (Teleostei: siluriformes) inferred from ultraconserved elements. Sci. Rep. 10, 2697. doi:10.1038/s41598-020-59519-w
Ochoa, L. E., Roxo, F. F., Do Nascimiento, C., Sabaj, M. H., Datovo, A., Alfaro, M., et al. (2017). Multilocus analysis of the catfish family Trichomycteridae (Teleostei: ostariophysi: siluriformes) supporting a monophyletic trichomycterinae. Mol. Phylogenetics Evol. 115, 71–81. doi:10.1016/j.ympev.2017.07.007
Oliveira, C., Avelino, G. S., Abe, K. T., Mariguela, T. C., Benine, R. C., Ortí, G., et al. (2011). Phylogenetic relationships within the speciose family Characidae (Teleostei: ostariophysi: Characiformes) based on multilocus analysis and extensive ingroup sampling. BMC Evol. Biol. 11, 275. doi:10.1186/1471-2148-11-275
Pennell, M. W., Eastman, J. M., Slater, G. J., Brown, J. W., Uyeda, J. C., FitzJohn, R. G., et al. (2014). Geiger v2. 0: an expanded suite of methods for fitting macroevolutionary models to phylogenetic trees. Bioinformatics 30, 2216–2218. doi:10.1093/bioinformatics/btu181
R Core Team, R. (2023). _R: a language and environment for statistical. Computing_. Vienna, Austria: R Foundation for Statistical Computing. Available at: https://www.R-project.org/.
Reis, R. E., Albert, J. S., Di Dario, F., Mincarone, M. M., Petry, P., and Rocha, L. A. (2016). Fish biodiversity and conservation in South America. J. Fish Biol. 89, 12–47. doi:10.1111/jfb.13016
Reis, R. E., and Lehmann, A. P. (2022). A new genus of armored catfish (Siluriformes: Loricariidae) from the Greater Amazon, with a review of the species and description of five new species. Neotropical Ichthyol. 20, e220002. doi:10.1590/1982-0224-2022-0002
Remec, Z. I., Trebusak Podkrajsek, K., Repic Lampret, B., Kovac, J., Groselj, U., Tesovnik, T., et al. (2021). Next-generation sequencing in newborn screening: a review of current state. Front. Genet. 12, 662254. doi:10.3389/fgene.2021.662254
Schoch, C. L., Ciufo, S., Domrachev, M., Hotton, C. L., Kannan, S., Khovanskaya, R., et al. (2020). NCBI Taxonomy: a comprehensive update on curation, resources and tools. Database 2020, baaa062. doi:10.1093/database/baaa062
Servais, T., Owen, A. W., Harper, D. A., Kröger, B., and Munnecke, A. (2010). The great Ordovician biodiversification event (GOBE): the palaeoecological dimension. Palaeogeogr. Palaeoclimatol. Palaeoecol. 294, 99–119. doi:10.1016/j.palaeo.2010.05.031
Silva, G. S., Roxo, F. F., Lujan, N. K., Tagliacollo, V. A., Zawadzki, C. H., and Oliveira, C. (2016). Transcontinental dispersal, ecological opportunity and origins of an adaptive radiation in the Neotropical catfish genus Hypostomus (Siluriformes: Loricariidae). Mol. Ecol. 25, 1511–1529. doi:10.1111/mec.13583
Smith, S. A., and O'Meara, B. C. (2012). treePL: divergence time estimation using penalized likelihood for large phylogenies. Bioinformatics 28, 2689–2690. doi:10.1093/bioinformatics/bts492
Soares, B. E., Benone, N. L., Leitão, R. P., Leal, C. G., Santos, L. L., de Assis Montag, L. F., et al. (2023). The ecomorphological diversity of Amazonian stream fishes is constrained by phylogenetic relationships. doi:10.32942/X2N59N
Stamatakis, A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. doi:10.1093/bioinformatics/btu033
Stein, R. W., Mull, C. G., Kuhn, T. S., Aschliman, N. C., Davidson, L. N., Joy, J. B., et al. (2018). Global priorities for conserving the evolutionary history of sharks, rays and chimaeras. Nat. Ecol. and Evol. 2, 288–298. doi:10.1038/s41559-017-0448-4
Su, G., Villéger, S., and Brosse, S. (2019). Morphological diversity of freshwater fishes differs between realms, but morphologically extreme species are widespread. Glob. Ecol. Biogeogr. 28, 211–221. doi:10.1111/geb.12843
Sullivan, J. P., Lundberg, J. G., and Hardman, M. (2006). A phylogenetic analysis of the major groups of catfishes (Teleostei: siluriformes) using rag1 and rag2 nuclear gene sequences. Mol. phylogenetics Evol. 41, 636–662. doi:10.1016/j.ympev.2006.05.044
Sullivan, J. P., Muriel-Cunha, J., and Lundberg, J. G. (2013). Phylogenetic relationships and molecular dating of the major groups of catfishes of the Neotropical superfamily Pimelodoidea (Teleostei, Siluriformes). Proc. Acad. Nat. Sci. Phila. 162, 89–110. doi:10.1635/053.162.0106
Tagliacollo, V. A., Bernt, M. J., Craig, J. M., Oliveira, C., and Albert, J. S. (2016). Model-based total evidence phylogeny of Neotropical electric knifefishes (Teleostei, Gymnotiformes). Mol. Phylogenetics Evol. 95, 20–33. doi:10.1016/j.ympev.2015.11.007
Tagliacollo, V. A., Dagosta, F. C. P., Pinna, M. de, Reis, R. E., and Albert, J. S. (2021). Assessing extinction risk from geographic distribution data in Neotropical freshwater fishes. Neotropical Ichthyol. 19. doi:10.1590/1982-0224-2021-0079
Tagliacollo, V. A., Duke-Sylvester, S. M., Matamoros, W. A., Chakrabarty, P., and Albert, J. S. (2017). Coordinated dispersal and pre-isthmian assembly of the central american ichthyofauna. Syst. Biol. 66, 183–196. doi:10.1093/sysbio/syv064
Tagliacollo, V. A., Roxo, F. F., Duke-Sylvester, S. M., Oliveira, C., and Albert, J. S. (2015). Biogeographical signature of river capture events in Amazonian lowlands. J. Biogeogr. 42, 2349–2362. doi:10.1111/jbi.12594
Tagliacollo, V. A., Souza-Lima, R., Benine, R. C., and Oliveira, C. (2012). Molecular phylogeny of Aphyocharacinae (Characiformes, Characidae) with morphological diagnoses for the subfamily and recognized genera. Mol. Phylogenetics Evol. 64, 297–307. doi:10.1016/j.ympev.2012.04.007
Talavera, G., and Castresana, J. (2007). Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst. Biol. 56, 564–577. doi:10.1080/10635150701472164
Thimotheo, M. G. R., Benine, R. C., Oliveira, C., and Silva, G. de S. da C. (2020). New species of the Corumbataia cuestae group (Siluriformes: Loricariidae) from the Rio Tocantins basin, with comments on its phylogenetic relationships. Neotropical Ichthyol. 18. doi:10.1590/1982-0224-2020-0060
Thomaz, A. T., Arcila, D., Ortí, G., and Malabarba, L. R. (2015). Molecular phylogeny of the subfamily Stevardiinae Gill, 1858 (Characiformes: Characidae): classification and the evolution of reproductive traits. BMC Evol. Biol. 15, 146–225. doi:10.1186/s12862-015-0403-4
Tucker, C. M., Cadotte, M. W., Carvalho, S. B., Davies, T. J., Ferrier, S., Fritz, S. A., et al. (2017). A guide to phylogenetic metrics for conservation, community ecology and macroecology. Biol. Rev. 92, 698–715. doi:10.1111/brv.12252
Val, A. L., and de Almeida-Val, V. M. (2012). Fishes of the Amazon and their environment: physiological and biochemical aspects. Springer Sci. and Bus. Media 32.
Val, P., Lyons, N. J., Gasparini, N., Willenbring, J. K., and Albert, J. S. (2022). Landscape evolution as a diversification driver in freshwater fishes. Front. Ecol. Evol. 9, 788328. doi:10.3389/fevo.2021.788328
Van der Sleen, P., and Albert, J. S. (2022). Patterns in freshwater fish diversity. Reference Module Earth Syst. Environ. Sci. 26, 894–907.
Vari, R. P. (1983). Phylogenetic relationships of the families Curimatidae. Prochilodontidae, Anostomidae, Chilodontidae (Pisces, Characiformes).
Villa-Navarro, F. A., Acero, A., and Cala, P. (2017). Taxonomic review of Trans-Andean species of Pimelodus (Siluriformes: Pimelodidae), with the descriptions of two new species. Zootaxa 4299, 337–360. doi:10.11646/zootaxa.4299.3.2
Weber, M. G., Wagner, C. E., Best, R. J., Harmon, L. J., and Matthews, B. (2017). Evolution in a community context: on integrating ecological interactions and macroevolution. Trends Ecol. and Evol. 32, 291–304. doi:10.1016/j.tree.2017.01.003
Wilkinson, M. J., Marshall, L. G., Lundberg, J. G., and Kreslavsky, M. H. (2010). Megafan environments in northern South America and their impact on Amazon Neogene aquatic ecosystems. Amaz. Landsc. species Evol. a look into past 1, 162–184. doi:10.1002/9781444306408.ch10
Winemiller, K. O. (1991). Ecomorphological diversification in lowland freshwater fish assemblages from five biotic regions. Ecol. Monogr. 61, 343–365. doi:10.2307/2937046
Keywords: neotropics, freshwater fishes, phylogeny, divergence times, macroevolution
Citation: Tagliacollo VA, Tan M, Reis RE, Gaia R, Carrijo V, Ranuzi M, Craig JM and Albert JS (2024) Time-calibrated phylogeny of neotropical freshwater fishes. Front. Bioinform. 4:1433995. doi: 10.3389/fbinf.2024.1433995
Received: 16 May 2024; Accepted: 18 November 2024;
Published: 06 December 2024.
Edited by:
Sudhindra R. Gadagkar, Midwestern University, United StatesReviewed by:
Soham Sengupta, St. Jude Children’s Research Hospital, United StatesLarry Page, Florida Museum of Natural History, United States
Copyright © 2024 Tagliacollo, Tan, Reis, Gaia, Carrijo, Ranuzi, Craig and Albert. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Victor A. Tagliacollo, dmF0YWdsaWFjb2xsb0BnbWFpbC5jb20=
†ORCID: Victor A. Tagliacollo, https://orcid.org/0000-0003-1493-9409; Milton Tan, https://orcid.org/0000-0002-9803-0827; Roberto E. Reis, https://orcid.org/0000-0003-3746-6894; Ronielson Gaia, https://orcid.org/0000-0001-6507-747X; Virgilio Carrijo, https://orcid.org/0009-0008-3138-6352; Marcelo Ranuzi, https://orcid.org/0000-0002-3887-493X; Jack M. Craig, https://orcid.org/0000-0003-0590-5944; James S. Albert, https://orcid.org/0000-0001-5477-1749
 Victor A. Tagliacollo
Victor A. Tagliacollo Milton Tan
Milton Tan Roberto E. Reis
Roberto E. Reis Ronielson Gaia
Ronielson Gaia Virgilio Carrijo
Virgilio Carrijo Marcelo Ranuzi
Marcelo Ranuzi Jack M. Craig
Jack M. Craig James S. Albert
James S. Albert