
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
TECHNOLOGY AND CODE article
Front. Bioinform., 25 July 2022
Sec. Integrative Bioinformatics
Volume 2 - 2022 | https://doi.org/10.3389/fbinf.2022.835805
This article is part of the Research TopicTowards Integrating Protein Sequence, Structure, Function & Therapeutics Discovery for Collaborative ResearchView all 5 articles
 Elizabeth V. Castro1,2
Elizabeth V. Castro1,2 John W. Shepherd1
John W. Shepherd1 Ryan S. Guggenheim1,2
Ryan S. Guggenheim1,2 Manimone Sengvoravong1
Manimone Sengvoravong1 Bailey C. Hall1
Bailey C. Hall1 McKenzie K. Chappell1,3
McKenzie K. Chappell1,3 Jessica A. Hearn1,3
Jessica A. Hearn1,3 Olivia N. Caraccio1
Olivia N. Caraccio1 Cora Bissman1,3
Cora Bissman1,3 Sydney Lantow1
Sydney Lantow1 Damian Buehner1
Damian Buehner1 Harry R. Costlow1
Harry R. Costlow1 David M. Prather4
David M. Prather4 Abigail M. Zonza3
Abigail M. Zonza3 Mallory Witt1
Mallory Witt1 Jeffrey A. Zahratka1,3*
Jeffrey A. Zahratka1,3*Ion channels are integral membrane protein complexes critical for regulation of membrane potential, cell volume, and other signaling events. As complex molecular assemblies with many interacting partners, ion channels have multiple structural and functional domains. While channel sequence and functional data are readily available across multiple online resources, there is an unmet need for functional annotation directly relating primary sequence information, 2D interactions, and three-dimensional protein structure. To this end, we present ChanFAD (Channel Functional Annotation Database), to provide the research community with a centralized resource for ion channel structure and functional data. ChanFAD provides functional annotation of PDB structures built on the National Center for Biotechnology Information’s iCn3D structure viewing tool while providing additional information such as primary sequence, organism, and relevant links to other databases. Here we provide a brief tour of ChanFAD functionality while showing example use cases involving drug-channel interactions and structural changes based on mutation. ChanFAD is freely available and can be accessed at https://www.chanfad.org/.
Ion channels are a broad family of integral membrane proteins characterized by the ability to transport ions and other small molecules across the phospholipid bilayer. Ubiquitously expressed and evolutionarily ancient, ion channels are critical regulators of cell volume, membrane potential, and gene expression (Berridge, 2014). Structurally, ion channels are made from multiple protein subunits that integrate into complex assemblies of scaffold proteins, enzymes, and other regulators to mediate localized intracellular signaling (Catterall, 2010; Dai et al., 2009). As multi-subunit protein complexes, ion channels have multiple structural and functional domains necessary for transmembrane localization, post-translational modification, and protein-protein interactions giving rise to channel function. Studying ion channel structure, function, and regulation has been a rewarding endeavor in diverse biological disciplines, but there is a notable lack of detailed annotation data mapping functional domains and post-translational modification sites to three-dimensional protein structure. To address this need, we introduce the Channel Functional Annotation Database (ChanFAD), a centralized community resource for relating genomic, biophysical, and bioinformatic information to map ion channel function to three-dimensional protein structure. ChanFAD entries are manually curated using a combination of primary literature, previously validated annotations, and available PDB structures in the National Center for Biotechnology Information’s iCn3D model structure viewer (Wang et al., 2020). Importantly, ChanFAD aims to build upon known annotation information found in general resources such as Pfam and Gene Ontology by focusing on functional domains specific to each ion channel complex in more detail. Each ChanFAD page contains the annotated PDB structure displayed in iCn3D accompanied by a downloadable selection file for external use, a link to the primary sequence in FASTA format, a “two-dimensional” representation of functional domains using the Swiss Institute of Bioinformatics neXtProt feature viewer, and links to relevant databases related to ion channel structure, function, or both. Specifically, ChanFAD provides links to the structural databases of RCSB PDB, UniProt, and predicted structures from AlphaFold; functional data from AmiGO. InterPro, and KEGG; and ion channel-specific databases of Channelpedia and IUPHAR entries for a given protein (Mao et al., 2005; Carbon et al., 2009, 2021; Blum et al., 2021; Jumper et al., 2021; Harding et al., 2022; Varadi et al., 2022). Additionally, the ChanFAD website provides links to other relevant resources to the ion channel community not specific to individual channel complexes, such as computational models provided by ModelDB and Ion Channel Genealogy, HOLE and ChannelsDB for exploring channel pore structure, and more generalized databases such as Pfam and the Neuroscience Information Framework (Smart et al., 1993; Gardner et al., 2008; McDougal et al., 2017; Podlaski et al., 2017; Pravda et al., 2018; Mistry et al., 2021). By providing functional annotation information for ion channels and grouping relevant links together, we envision multidisciplinary teams, including geneticists, structural biologists, and electrophysiologists, collaboratively working together to decipher ion channel structure-function relationships. Our aim is to provide an intuitive interface for bridging the gap between the 3D structural properties often studied by biophysicists and electrophysiologists with the functional domains probed by geneticists and molecular biologists. Here, we briefly detail the annotation process and demonstrate three examples of ChanFAD functionality highlighting the importance of structure-function relationships in ion channel activity and disease.
ChanFAD aims to be a comprehensive resource for ion channel functional annotation data. The defining feature of the database is its annotations, which are manually curated from a combination of previously annotated functional domains found on Pfam and NCBI’s Protein database, existing iCn3D data, and primary literature. ChanFAD annotations are represented in iCn3D selection sets, which allow for convenient 3D representation of a given domain. Annotations are created by finding an ion channel with a PDB structure on the RCSB PDB database (Berman, 2000). Next, the curator performs a thorough literature search, looking for descriptions of other domains not previously represented. Each domain is then organized and mapped to a PDB file using iCn3D’s Defined Sets feature, which is then freely available as part of an embedded iCn3D instance for the public on ChanFAD. The annotation process is then replicated for each unique subunit found in the PDB file, when applicable. One potential limitation of long annotations is the iCn3D shared link limit of 4,000 characters. As large protein complexes can easily result in detailed annotations that generate long links, we made the decision to annotate only the largest subunits of the PDB file when our annotations reach this limit, and only label each unique protein once, even if multiple identical chains are present. However, extremely large channels such as ryanodine receptors still exceed this limit, and possibilities for full annotation are currently being implemented for visualization.
The initial release of ChanFAD is built on the web using the Django framework (v2.1.2) in Python (v3.9.9) and can be accessed at https://www.chanfad.org/.
The most straightforward usage of ChanFAD is simple viewing of annotated domains, which can be especially useful for studying emerging topics or mapping genetic regions of interest onto a 3D protein structure. The example we will cover in this manuscript is the open reading frame 3a viral ion channel from SARS-CoV-2, the causative agent of COVID-19 (PDB ID: 7KJR). While ORF3a function is still being characterized at the time of writing, one of the channel’s possible quaternary structures has been solved using cryo-electron microscopy in lipid nanodiscs to reveal a peculiar homodimer configuration (Kern et al., 2020). While the initial RCSB entry shows a SARS-CoV-like ORF3a-like domain covering the entire length of each protein subunit, further exploration of the literature demonstrates multiple functional domains, including those for caveolin-1 interaction, clathrin-mediated endocytosis, RNA-binding, and TRAF3-binding ultimately resulting in recruitment of the NLRP3 inflammasome potentially resulting in apoptosis (Hassan et al., 2020; Issa et al., 2020; Ren et al., 2020). Each of these domains are represented and available to view in ChanFAD, as in the example screenshot shown in Figure 1. Also notable in the screenshot are direct links to the RCSB PDB database (by clicking on the PDB ID), reference to the publication characterizing the structure, UniProt entry, primary sequence in FASTA format, structure prediction using AlphaFold, and entries on AmiGO and InterPro. Notably, links for Channelpedia, KEGG, and IUPHAR are not available as information on ORF3a structure and function is still emerging.
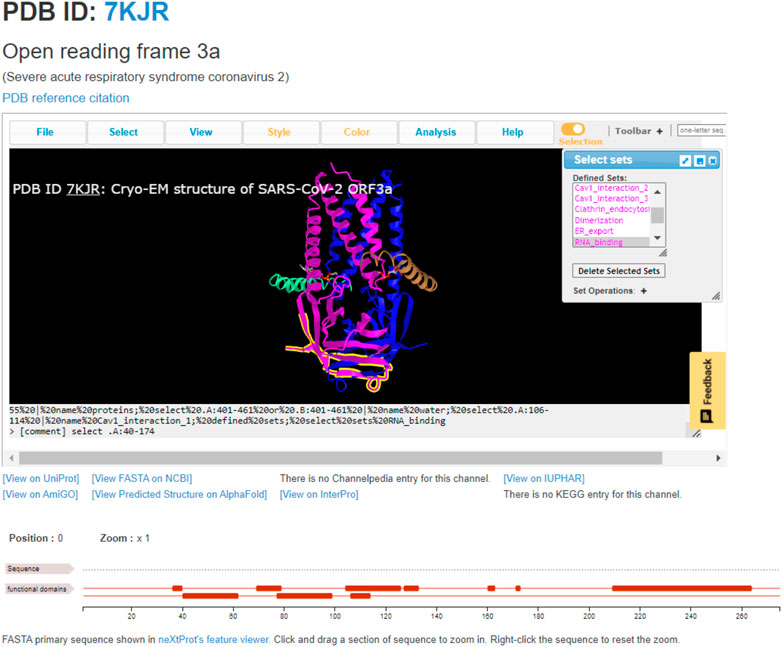
FIGURE 1. The annotated SARS-CoV-2 open reading frame 3a ion channel (PDB ID: 7KJR) with RNA-binding domain highlighted. This domain is the cytoplasmic side of the cell membrane, where it may be involved in binding viral RNA and apoptosis.
ChanFAD can also leverage the strength of iCn3D’s viewing capabilities for easy visualization of ligand-channel interactions. In this example, we will examine the binding of the rabbit (Oryctolagus cuniculus) voltage-gated calcium channel alpha-1 subunit (CaV1.1) with the benzothiazepine pore blocker diltiazem (PDB ID: 6JPB) (Zhao et al., 2019). Diltiazem has a straightforward mechanism of action: it physically blocks the channel pore on the intracellular side of the selectivity filter, disrupting ion flow (Tang et al., 2019). Structurally, the selectivity filter is a set of amino acids in the pore-forming domains of voltage-gated ion channels which confer ion specificity to the channels (Jiang et al., 2002). Annotation of the critical residues for diltiazem binding (Y1365, A1369, and I1372) are present in ChanFAD, and can be visualized in the iCn3D panel for view along with the previously mentioned RCSB PDB and Channelpedia database links (Figure 2) (Zhao et al., 2019). In contrast to the ORF3a entry discussed in Figure 1, the 6JPB entry shown in Figure 2 does have available links for the Channelpedia, KEGG, and IUPHAR databases. We foresee having this information in a single entry will facilitate studies of mapping genetic mutations to channel function both in research and classroom settings.
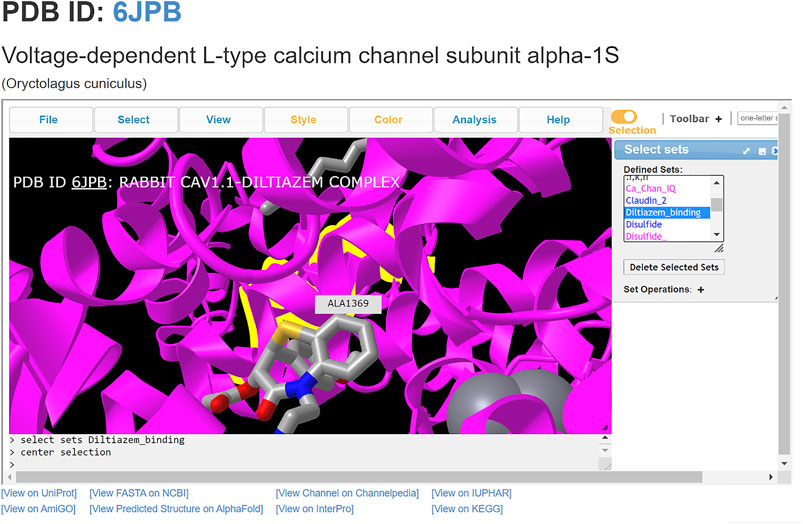
FIGURE 2. Visualization of diltiazem blocking the CaV1.1 channel pore (PDB: 6JPB). Diltiazem interacts with three conserved residues (Y1365, A1369, and I1372) on the intracellular side of the channel to block calcium conductance. Ca2+ ions shown as gray spheres. The alanine residue (A1369) is centered, with the diltiazem molecule visibly blocking the channel pore.
ChanFAD annotations can also be leveraged with iCn3D’s full capabilities for visualizing mutations. One powerful capability of iCn3D is the ability to make point mutations to PDB structures and render them in the viewer using scap (Jacobson et al., 2002; Xiang & Honig, 2001). Although ChanFAD automatically loads annotation files directly into iCn3D when the database is accessed, the annotation files are available for public download for further downstream analysis. For example, we will look at mutation of a human calcium-gated potassium channel subunit (SK4, PDB ID: 6CNN) where specific point mutations can result in dehydrated hereditary stomatocytosis type 2 (Lee & MacKinnon, 2018). Affected patients have a heterozygous substitution in a conserved calmodulin interaction domain (R352H), resulting in increased sensitivity to calcium ions and frequent channel opening resulting in altered red blood cell shape, cellular fragility, and anemia (Rapetti-Mauss et al., 2015). First, we obtain the mutated PDB by using iCn3D’s built-in mutation function (Analysis → Mutation) and specifying the R352H mutation (e.g., 6CNN_A_352_H) and opening a new iCn3D browser instance. Loading the ChanFAD annotation into the mutated PDB, we can highlight the calmodulin binding domain and see the change in the protein side chain, permitting visualization of structural changes for downstream analysis (Figure 3). In this way, we envision ChanFAD annotations can be used in tandem with shareable iCn3D links and PDB files for reproducible structural studies of ion channels.
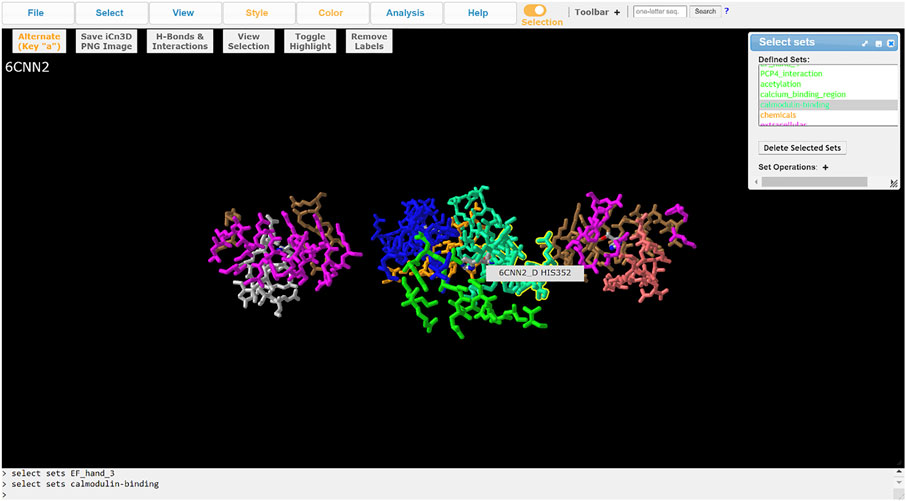
FIGURE 3. Visualization of the SK4 calcium-activated potassium channel (PDB ID: 6CNN) in iCn3D with ChanFAD annotation and the R352H mutation introduced using scap. R352H is in the calmodulin interaction domain, which is responsible for calcium binding and channel opening. The figure only shows the four calmodulin binding domains (one for each protein subunit) for readability.
As a database of manual annotations, the primary future direction of ChanFAD is the addition of new entries available for view. However, there are a few other features our team would like to add in the future. First, providing a table of annotated domains with associated references would be a valuable reference for structural and genetic studies, even if the full three-dimensional representation found in iCn3D is not needed. Second, we wish to add more robust literature curation to each database entry to help researchers and integrate peer-reviewed experiments with structural and functional domains, which synergizes well with the tabular representation of annotated domains mentioned previously. Third, we are actively working to implement iCn3D functionality for large annotations resulting in shared links of greater than 4,000 characters. One potential solution is the use of iCn3D PNG files or state files, which should be integrated into ChanFAD at a later date. We are also exploring the possibility of using natural language processing methods, such as BioBERT, to perform text mining on published literature to speed up the annotation process (Lee et al., 2020). Finally, as a community resource, we are working to implement both a community submission process for new annotation data through our Django-based web framework which would then be reviewed for accuracy and made available, and a convenient and secure way to directly load the annotation files from ChanFAD directly into other applications. Our ultimate goal is to easily facilitate collaborative research about ion channel structure and function.
Publicly available datasets were analyzed in this study. This data can be found here: https://github.com/jzahratka/ChanFAD.
EVC, JWS, RSG, MS, BCH, MKC, JAH, ONC, CB, SL, DB, HRC, DMP, AMZ, MW, and JAZ all created annotations for the database. JAZ designed the project, built the database, and wrote the manuscript, while each of the other authors (EVC, JWS, RSG, MS, BCH, MKC, JAH, ONC, CB, SL, DB, HRC, DMP, MW, and AMZ) all provided feedback for editing the manuscript.
Baldwin Wallace University Faculty Development Funds—Used in part to cover the publication fees.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We would like to thank the Baldwin Wallace University departments of Neuroscience, Psychology, Biology and Geology, and Chemistry for their support of this project. We would also like to thank all of the hard work put in by the iCn3D team, the RCSB PDB team, and individual submitters of PDB structural data without which this project could not be possible.
Berman, H. M., Bhat, T. N., Bourne, P. E., Feng, Z., Gilliland, G., Weissig, H., et al. (2000). The Protein Data Bank and the Challenge of Structural Genomics. Nat. Struct. Biol. 7 (Suppl. 1), 957–959. doi:10.1093/nar/28.1.23510.1038/80734
Berridge, M. J. (2014). Module 10: Neuronal Signalling. Cell. Signal. Biol. 6, csb0001010. doi:10.1042/csb0001010
Blum, M., Chang, H. Y., Chuguransky, S., Grego, T., Kandasaamy, S., Mitchell, A., et al. (2021). The InterPro Protein Families and Domains Database: 20 Years on. Nucleic Acids Res. 49 (D1), D344–D354. doi:10.1093/nar/gkaa977
Carbon, S., Ireland, A., Mungall, C. J., Shu, S., Marshall, B., Lewis, S., et al. (2009). AmiGO: Online Access to Ontology and Annotation Data. Bioinformatics 25 (2), 288–289. doi:10.1093/bioinformatics/btn615
Carbon, S., Douglass, E., Good, B. M., Unni, D. R., Harris, N. L., Mungall, C. J., et al. (2021). The Gene Ontology Resource: Enriching a GOld Mine. Nucleic Acids Res. 49 (D1), D325–D334. doi:10.1093/nar/gkaa1113
Catterall, W. A. (2010). Signaling Complexes of Voltage-Gated Sodium and Calcium Channels. Neurosci. Lett. 486 (2), 107–116. doi:10.1016/j.neulet.2010.08.085
Dai, S., Hall, D. D., and Hell, J. W. (2009). Supramolecular Assemblies and Localized Regulation of Voltage-Gated Ion Channels. Physiol. Rev. 89 (2), 411–452. doi:10.1152/physrev.00029.2007
Gardner, D., Akil, H., Ascoli, G. A., Bowden, D. M., Bug, W., Donohue, D. E., et al. (2008). The Neuroscience Information Framework: A Data and Knowledge Environment for Neuroscience. Neuroinformatics 6 (3), 149–160. doi:10.1007/s12021-008-9024-z
Harding, S. D., Armstrong, J. F., Faccenda, E., Southan, C., Alexander, S. P. H., Davenport, A. P., et al. (2022). The IUPHAR/BPS Guide to PHARMACOLOGY in 2022: Curating Pharmacology for COVID-19, Malaria and Antibacterials. Nucleic Acids Res. 50 (D1), D1282–D1294. doi:10.1093/nar/gkab1010
Hassan, S. S., Choudhury, P. P., Basu, P., and Jana, S. S. (2020). Molecular Conservation and Differential Mutation on ORF3a Gene in Indian SARS-CoV2 Genomes. Genomics 112 (5), 3226–3237. doi:10.1016/j.ygeno.2020.06.016
Issa, E., Merhi, G., Panossian, B., Salloum, T., and Tokajian, S. (2020). SARS-CoV-2 and ORF3a: Nonsynonymous Mutations, Functional Domains, and Viral Pathogenesis. MSystems 5 (3), e00266–20. doi:10.1128/mSystems.00266-20
Jacobson, M. P., Friesner, R. A., Xiang, Z., and Honig, B. (2002). On the Role of the Crystal Environment in Determining Protein Side-Chain Conformations. J. Mol. Biol. 320 (3), 597–608. doi:10.1016/S0022-2836(02)00470-9
Jiang, Y., Lee, A., Chen, J., Cadene, M., Chait, B. T., and MacKinnon, R. (2002). The Open Pore Conformation of Potassium Channels. Nature 417 (6888), 523–526. doi:10.1038/417523a
Jumper, J., Evans, R., Pritzel, A., Green, T., Figurnov, M., Ronneberger, O., et al. (2021). Highly Accurate Protein Structure Prediction with AlphaFold. Nature 596 (7873), 583–589. doi:10.1038/s41586-021-03819-2
Kern, D. M., Sorum, B., Hoel, C. M., Sridharan, S., Remis, J. P., Toso, D. B., et al. (2020). Cryo-EM Structure of the SARS-CoV-2 3a Ion Channel in Lipid Nanodiscs. BioRxiv 2020, 156554. doi:10.1101/2020.06.17.156554
Lee, C. H., and MacKinnon, R. (2018). Activation Mechanism of a Human SK-Calmodulin Channel Complex Elucidated by Cryo-EM Structures. Science 360 (6388), 508–513. doi:10.1126/science.aas9466
Lee, J., Yoon, W., Kim, S., Kim, D., Kim, S., So, C. H., et al. (2020). BioBERT: A Pre-trained Biomedical Language Representation Model for Biomedical Text Mining. Bioinformatics 36 (4), 1234–1240. doi:10.1093/bioinformatics/btz682
Mao, X., Cai, T., Olyarchuk, J. G., and Wei, L. (2005). Automated Genome Annotation and Pathway Identification Using the KEGG Orthology (KO) as a Controlled Vocabulary. Bioinformatics 21 (19), 3787–3793. doi:10.1093/bioinformatics/bti430
McDougal, R. A., Morse, T. M., Carnevale, T., Marenco, L., Wang, R., Migliore, M., et al. (2017). Twenty Years of ModelDB and beyond: Building Essential Modeling Tools for the Future of Neuroscience. J. Comput. Neurosci. 42 (1), 1–10. doi:10.1007/s10827-016-0623-7
Mistry, J., Chuguransky, S., Williams, L., Qureshi, M., Salazar, G. A., Sonnhammer, E. L. L., et al. (2021). Pfam: The Protein Families Database in 2021. Nucleic Acids Res. 49 (D1), D412–D419. doi:10.1093/nar/gkaa913
Podlaski, W. F., Seeholzer, A., Groschner, L. N., Miesenböck, G., Ranjan, R., and Vogels, T. P. (2017). Mapping the Function of Neuronal Ion Channels in Model and Experiment. ELife 6, 1–22. doi:10.7554/elife.22152
Pravda, L., Sehnal, D., Svobodová Vareková, R., Navrátilová, V., Toušek, D., Berka, K., et al. (2018). ChannelsDB: Database of Biomacromolecular Tunnels and Pores. Nucleic Acids Res. 46 (D1), D399–D405. doi:10.1093/nar/gkx868
Rapetti-Mauss, R., Lacoste, C., Picard, V., Guitton, C., Lombard, E., Loosveld, M., et al. (2015). A Mutation in the Gardos Channel Is Associated with Hereditary Xerocytosis. Blood 126 (11), 1273–1280. doi:10.1182/blood-2015-04-642496
Ren, Y., Shu, T., Wu, D., Mu, J., Wang, C., Huang, M., et al. (2020). The ORF3a Protein of SARS-CoV-2 Induces Apoptosis in Cells. Cell. Mol. Immunol. 17 (8), 881–883. doi:10.1038/s41423-020-0485-9
Smart, O. S., Goodfellow, J. M., and Wallace, B. A. (1993). The Pore Dimensions of Gramicidin A. Biophys. J. 65 (6), 2455–2460. doi:10.1016/S0006-3495(93)81293-1
Tang, L., Gamal El-Din, T. M., Lenaeus, M. J., Zheng, N., and Catterall, W. A. (2019). Structural Basis for Diltiazem Block of a Voltage-Gated Ca2+ Channel. Mol. Pharmacol. 96 (4), 485–492. doi:10.1124/mol.119.117531
Varadi, M., Anyango, S., Deshpande, M., Nair, S., Natassia, C., Yordanova, G., et al. (2022). AlphaFold Protein Structure Database: Massively Expanding the Structural Coverage of Protein-Sequence Space with High-Accuracy Models. Nucleic Acids Res. 50 (D1), D439–D444. doi:10.1093/nar/gkab1061
Wang, J., Youkharibache, P., Zhang, D., Lanczycki, C. J., Geer, R. C., Madej, T., et al. (2020). ICn3D, a Web-Based 3D Viewer for Sharing 1D/2D/3D Representations of Biomolecular Structures. Bioinformatics 36 (1), 131–135. doi:10.1093/bioinformatics/btz502
Xiang, Z., and Honig, B. (2001). Extending the Accuracy Limits of Prediction for Side-Chain Conformations. J. Mol. Biol. 311 (2), 421–430. doi:10.1006/jmbi.2001.4865
Keywords: ion channels, functional annotation, protein structure, database, proteomics
Citation: Castro EV, Shepherd JW, Guggenheim RS, Sengvoravong M, Hall BC, Chappell MK, Hearn JA, Caraccio ON, Bissman C, Lantow S, Buehner D, Costlow HR, Prather DM, Zonza AM, Witt M and Zahratka JA (2022) ChanFAD: A Functional Annotation Database for Ion Channels. Front. Bioinform. 2:835805. doi: 10.3389/fbinf.2022.835805
Received: 14 December 2021; Accepted: 17 May 2022;
Published: 25 July 2022.
Edited by:
Raul Cachau, Frederick National Laboratory for Cancer Research (NIH), United StatesReviewed by:
Pierre Tuffery, Institut National de la Santé et de la Recherche Médicale (INSERM), FranceCopyright © 2022 Castro, Shepherd, Guggenheim, Sengvoravong, Hall, Chappell, Hearn, Caraccio, Bissman, Lantow, Buehner, Costlow, Prather, Zonza, Witt and Zahratka. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jeffrey A. Zahratka, anphaHJhdGtAYncuZWR1
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.