- 1Department of Computer Science, Ben-Gurion University of the Negev, Beer-Sheva, Israel
- 2Department of EOAS, The University of British Columbia, Vancouver, BC, Canada
Recent advancements in machine learning techniques for protein structure prediction motivate better results in its inverse problem–protein design. In this work we introduce a new graph mimetic neural network, MimNet, and show that it is possible to build a reversible architecture that solves the structure and design problems in tandem, allowing to improve protein backbone design when the structure is better estimated. We use the ProteinNet data set and show that the state of the art results in protein design can be met and even improved, given recent architectures for protein folding.
1 Introduction
Protein structure prediction (PSP) and protein backbone design are two related tasks. The former, aims at deriving three dimensional models of proteins from their residue (amino-acid) sequences. The latter, protein backbone design, aka the inverse protein folding problem, focus on suggesting sequences that are likely to fold to a given structure. These two scientific problems have remained open for more than 4 decades already, with unprecedented recent successes (Sun and Kim, 2017; Strokach et al., 2020; Ben-Sasson et al., 2021; Jumper et al., 2021). They have common underlying physical principles, and consequently the same computational infrastructure may cope with both (Jones, 1994; Leaver-Fay et al., 2011; Norn et al., 2021). Yet, these problems are typically considered separately, with many scientific endeavours focusing on either prediction or design but not on both. Thus, the current breakthrough in PSP does not directly affect the design filed, which seems to somewhat lag behind. Here we present a unifying framework that handles both problems simultaneously offering a conceptual way to harness progress in PSP to push the design field forward. Our method, dubbed MimNet, is inspired by the reversibility of the folding/unfolding physical processes, and the ability of molecular dynamics simulations to reproduce this behaviour. We use essentially the same mathematical formulation to mimic (hence the name) the physical processes by non-physical, alchemy, processes that reversibly transform “coordinates” between sequence and Cartesian spaces. To this end we leverage bi-directional Graph Convolutional Neural Networks (GCNs) that solve both problems, PSP and design, using the same learned weights.
Evolutionary inference, mainly in the form of multiple sequence alignments (MSAs) of homologous proteins, has long been recognized as a major source of structural insight. First order statistics of MSAs including conservation profiles, position-specific score matrices (PSSMs) (Gribskov et al., 1987; Altschul et al., 1997) and hidden Markov models (Remmert et al., 2012) allow the identification of remote homologs as modeling templates (Bates et al., 2001; Schwede et al., 2003; Hildebrand et al., 2009; Waterhouse et al., 2018), and reliable prediction of one-dimensional structural features like secondary structure (Rost and Sander, 1993), transmembrane segments (Rost et al., 1995), and solvent accessibility (Adamczak et al., 2004). Second order statistics, like direct coupling (Morcos et al., 2011) and inverted covariance matrices (Jones et al., 2012; Li et al., 2019), are used to predict spatial contacts between residues (Vassura et al., 2008; Kamisetty et al., 2013; Tetchner et al., 2014). The use of second order statistics require “deep” MSAs, that is alignments of many diverged homologous sequences. Searching for these homologous sequences, in large and rapidly growing datasets, becomes a major task in PSP that consumes much time and computational resources. Further, the sheer size of “deep” MSA datasets makes them hard to share.
In recent years, deep learning techniques boost the efforts to solve the PSP problem (AlQuraishi, 2019a; Drori et al., 2019; Hou et al., 2019; Kandathil et al., 2019; Xu, 2019; Zheng et al., 2019; Senior et al., 2020; Baek et al., 2021), reaching remarkable unprecedented performance, as was demonstrated in the CASP13 and CASP14 competitions (Abriata et al., 2019; Kryshtafovych et al., 2019; Jumper et al., 2021). Currently, all the state-of-the-art, high performance, methods rely on second order statistics derived from “deep” MSAs. In this study we chose an alternative rout to PSP, namely using first order information only (Li et al., 2017; Gao et al., 2018; AlQuraishi, 2019a; Torrisi et al., 2020; Xu et al., 2021). Giving up on the structural clues provided by second order information, this approach has received less attention in recent literature. Nevertheless, by using only first-order statistics we are able to focus on the development of new prediction strategies with reasonable resources. To this end, we follow the example of (AlQuraishi, 2019a) and benchmark on the relatively light-weight ProteinNet (AlQuraishi, 2019b), in which the only inputs to PSP are sequences and PSSMs (formulated as a per-position probability density function). We further adopt a lean protein representation with a single interaction point, the Cα atom, per residue. Finer grained protein representations (i.e., more interaction points per residue) are more expressive, and thus likely to achieve better results. Yet, when developing a novel methodology we do not aim at state-of-art performance but rather at simplicity and ease of experimentation. The Cα representation for that matter is expressive enough. Yet, our MimNet is on-par with previous studies that used only first order data, and it outperforms (AlQuraishi, 2019a), which uses the same training and test sets, and thus can be directly compared (see Section 3.3).
PSP gained much more attention over the years compared with its inverse, protein design. One apparent reason, for the more central role of PSP, is that progress in this field is monitored and accelerated by two accepted and objective benchmarks: Critical Assessment of Structure Prediction (CASP) (Moult et al., 1997; Kryshtafovych et al., 2019) and Continuous Automated Model EvalutiOn (CAMEO) (Haas et al., 2018). The protein design field lacks benchmarks like CASP and CAMEO, and a reliable evidence of success requires expensive synthesis and functional characterization in the laboratory (Dahiyat and Mayo, 1996; Basanta et al., 2020; Ben-Sasson et al., 2021). When laboratory validation is infeasible (e.g., this study), researchers resort to re-designing proteins of known structures.
Notwithstanding the evident importance of CASP and CAMEO they compare the “end product” (i.e., prediction) of elaborate pipelines. A mediocre implementation of one stage of the pipeline may overshadow other, better implemented stages. Most importantly, data collection and preprocessing may dominate the performance of data driven methods. The PSP data-set of this study, ProteinNet, offers an alternative type of benchmark. In the spirit of machine learning data-sets such as ImageNet (Deng et al., 2009), it provides predefined training and test sets (based on the PDB and on CASP validation) and allows researchers to focus on the development of novel machine learning schemes. Using a reversible network, this study begs to use the same data-set also as a benchmark for its design side. Specifically, we aim to reproduce the residue probabilities of ProteinNet’s PSSMs based on the Cα Coordinates. Indeed, a network can learn to generate sequences (Strokach et al., 2020) as the “natural” output of a design algorithm. However, sequences are in a sense arbitrary, as homologous proteins share similar if not identical (up to measurement errors) structures, while their sequences are diverged. Thus, the design problem of some structure does not have a unique solution, but rather a family of sequences that we represent by a PSSM that associates each position with a residue probability distribution function. Individual sequences may then be sampled form the PSSM or, if the target structure exists in nature, retrieved from sequence data-sets with a search algorithm (e.g., BLAST). The former is similar in spirit to the veteran back-to-consensus strategy of more traditional protein design approaches (Bershtein et al., 2008; Chandler et al., 2020).
Employing deep learning for protein design task is a relatively new idea (O’Connell et al., 2018; Ingraham et al., 2019; Anand-Achim et al., 2021; Norn et al., 2021). A similar approach to ours was recently presented in (Strokach et al., 2020) where graph methods were proposed for protein design, reporting promising results by treating the problem as a graph node classification problem, surpassing other de-novo design codes. While (Strokach et al., 2020) and our method share some similarities, ours is largely different as we use reversible architectures which offer numerous advantages, discussed in the following. As we show in Section 3.2, our approach obtains better results on a large data-set derived from the Protein Data Bank (PDB).The main part of this work is the introduction of a framework that unifies the treatment of protein folding and design. Our framework mimics the physical formulation of protein folding using a neural network. Hence, we coin the term Mimetic Deep Neural Networks (MimNet), which we apply to graphs, describing protein structures. While our work focuses on protein folding and design, the proposed network can be applied with any node or edge data that is available, thus it suits using both first or second order statistics.
The main idea is to generate a reversible transformation from the structure to the sequence and vice versa by using reversible neural network architectures (Chang et al., 2018; Ruthotto and Haber, 2019). These networks are bi-directional (can propagate forward and backward), and hence allow us to jointly train them to solve both the folding and the design problems, utilizing both the sequence and the structure of the protein simultaneously. This effectively doubles the amount of the data with respect to the network parameters. Such networks can utilize any type of layer, from structured to graph convolution or attention, harnessing recent advances in the understanding of protein folding architectures. Another important advantage of such a network is its memory footprint. Since the network is reversible, it is possible to train an arbitrarily long network without storing the activations, at the cost of double the computation of the backward pass (Chang et al., 2018). This enables the use of very deep networks that are impossible to use otherwise.
Furthermore, the physical folding process can be described by a second order differential equation derived from Hamiltonian dynamics. Therefore, we consider reversible architectures that are inspired by Hamiltonian dynamics hyperbolic differential equations (Chang et al., 2018; Ruthotto and Haber, 2019), which can be used to simulate this process. One can therefore claim that such a mimetic network is more faithful to the physics of the protein folding problem compared to a standard deep network like a ResNet (He et al., 2016). In particular, using such a dynamics has conservation properties that avoid the well known phenomenon of over-smoothing in graphs (Ming Chen et al., 2020; Zhao and Akoglu, 2020; Chamberlain et al., 2021; Eliasof et al., 2021). In this paper, we particularly explore the use of reversible GCNs, which are graph-based deep learning methods (Gao and Ji, 2019; Wang et al., 2019). Such networks are inspired by molecular dynamics, and their pairwise interactions are reminiscent of the residue interactions in a three dimensional physical system.
The rest of the paper is organized as follows. In Section 2 we discuss the problem and introduce the key mathematical ideas which constitute the building blocks of our network. In particular, we discuss multiscale reversible networks and different types of graph convolution techniques that are used to solve the problem. We then define our MimNet and its objective functions. In Section 3 we perform numerical experiments with data obtained from ProteinNet (AlQuraishi, 2019b). ProteinNet is a publicly available data set that contains both sequences and PSSMs and thus allows for the training of a folding network with first order statistics as done in the recurrent geometric network (RGN) (AlQuraishi, 2019a). The size of the data set, its structure, and the division into training validating and testing subsets, were carefully selected, allowing one to rigorously test the design problem as well. Finally, in Section 4 we discuss the results and summarize the paper.
2 Methods
Before discussing the particular network and architecture, we define the data and the functions of folding and design problems. Specifically, assume that
We now review the concept of a mimetic deep neural network, that is, a neural network whose functional form mimics simulations of folding dynamics. To this end, a deep network can be thought of as a time discretization of a differential equation (Chen et al., 2018; Ruthotto and Haber, 2019). According to this interpretation, each layer represents the state of the system at some particular pseudo-time. The mimetic properties are first discussed in pseudo-time, namely, how the network propagates from one layer to the other. The second mimetic property considers the spatial domain, meaning, how a particular residue in the protein interacts with another residue. These properties are put together to generate a mimetic deep neural network that imitates Verlet integration in a high dimensional space, using network architectures that are derived by discretized differential operators in time and space (Ruthotto and Haber, 2019; Eliasof and Treister, 2020). The treatment in both space and time are put together within a network optimization procedure to train the system and yield a network that can solve both the folding and the design problems.
2.1 Reversible Networks and Dynamical Systems
In this subsection we show how to build a mimetic network in time by using reversible dynamics. Reversible systems play a major role in physics for applications that range from Hamiltonian dynamics to wave equations. Broadly speaking, a reversible system is one that can propagate forward in time without information loss and therefore, can propagate backwards in time. Simple physical examples are a pendulum or a wave. These systems (in their idealized form) do not change their entropy, and therefore allow for forward or backward integration in time. Typical molecular dynamics is solved using reversible methods (Allen, 2004) (that is, integrating Hamiltonian dynamics) and therefore, it is natural to explore neural network architectures with similar properties.
To be more specific, given the input for the folding task [S, S+] (e.g., the concatenation of the one hot encoding sequence design and PSSM matrices) we first apply
where Y0 contains nf channels of n-length sequence features, embedded by the transformation q(⋅, ⋅), parameterized by the weights θe. This layer transforms the input to the latent space of the network. Here we use a 1D convolution for q, but other transformations may also be suitable.
The initial state Y0 and its velocity vector V0 are then pushed forward by a deep residual neural network. In particular, we consider a network with the following structure
where j = 0, … , T is the layer index. h is a parameter that represents a time step size and θj are learnt parameters that characterize the j-th layer. The system in Eq. 2.2a can be interpreted as a Verlet type discretization of a dynamical system with learnable forces that are the gradients of some potential function. A particular case of such dynamics is obtained by setting g = Id (the identity transformation) yielding the second order dynamics
This scheme is reversible, regardless of the choice f (which we discuss Section 2.3), since we can express Yj−1 as a function of Yj and Yj+1. The propagation forward (and backward) is not complete without defining the boundary conditions Y−1 and YT+1. Here we arbitrarily choose Y−1 = Y0 and YT−1 = YT, that is, initializing the network with zero velocity, i.e., V0 = 0. An illustration of the dynamics is plotted in Figure 1. Note, that this formulation is a slight (but important) modification of the standard ResNet (He et al., 2016) that reads
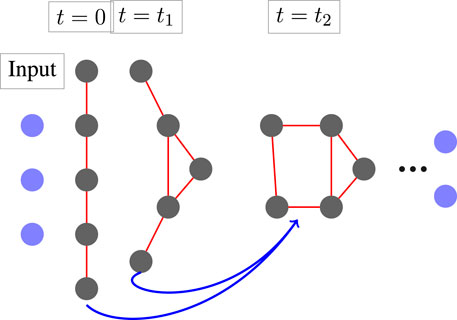
FIGURE 1. The architecture of MimNet with graph convolution layers. An embedding layer transforms the input into a latent space which then propagates through a GCN layer fed with the outputs of two previous layers. A graph that represents the protein structure is computed after each layer. The final layer is then projected back to obtain residue coordinates.
Given the final state of the system YT, we predict the coordinates X by projecting YT onto a three dimensional space
where
constraining the distance between every two residues to c = 3.8 Å. We have found that when the data is noisy implementing this constraint is needed in order to obtain physically feasible results (see Section 2.5.3).
In the forward pass, described above, the folding problem was solved, where we march from the protein design attributes (as in Section 2.1) to its coordinates. In the backward pass, we solve the design problem, where our goal is to predict the sequence given its coordinates. We start the backward pass by embedding the coordinates into the network feature space, i.e.,
where
where
2.2 Graph Convolutional Networks
Section 2.1 considers the propagation of the network from its initial condition (a sequence) to its final one (3D structure) and vice versa. The discussion was agnostic to the choice of the function f(⋅, ⋅) in Eq. 2.3 that realizes the network in hand. In this section we review the concept of a graph network and discuss its computation.
The idea behind a graph based method is rooted in the physics of the problem. Energy based simulations can be thought of as pairwise interactions on a graph based on the L2 distance between the residues. Indeed, as the distance between residues is smaller, the interaction between them is stronger. This motivates us to use machine learning techniques that mimic this property. As the dynamical system is evolving, the interactions between pairs of close residues is significantly larger compared to far ones.
One of the most successful techniques for image and speech processing is Convolution Neural Networks (CNN) (Krizhevsky et al., 2012; Goodfellow et al., 2016). The method relays on the structured grids on which sequences and images are defined. That is, every element has neighbouring elements in a structured manner. In recent years, similar ideas were extended to more complex geometries and manifolds, which can be conveniently represented by a graph (Ranjan et al., 2018; Hanocka et al., 2019; Wang et al., 2019). The main idea is to replace the structured convolution with a graph based convolution. That is, rather than convolving each location in the feature map with its near neighbours, define the distance between each location based on node or edge features, and then convolve the residues that are close on the graph.
To be more specific, we let Yj be the state at the jth layer. Then, we define a graph convolution block as follows:
where
Many graph based networks employ a graph convolution with fixed connectivity (Ranjan et al., 2018; Bouritsas et al., 2019). This is reasonable if the final topology is known. However, for protein folding we start with an unknown structure and it is evolving (learnt) from the data. Therefore, rather than using a fixed graph for the network we let the graph evolve throughout network. We thus recompute a weighted graph Laplacian at each layer, or, for computational saving, every few layers. To this end, we compute the weighted distance matrix between each two residues
where α is a scaling parameter (we set α = 10) and D is the L2 distance between each two residues
The vector 1 is a vector of ones of appropriate size. Using the distance matrix we define the graph Laplacian as
The approach of dynamically updating the connectivity of the graph was also suggested in (Wang et al., 2019), where k nearest neighbors are equally chosen per node, regardless of their distances. However, this imposes a non-smooth transition of the graph, which may result in optimization difficulties. Thus, we employ a weighted fully connected graph that can smoothly strengthen or weaken the connectivity of the residues. Since the weighting of the edges makes the graph Laplacian continuously differentiable with respect to the network parameter - a smoother optimization trajectory is obtained, leading to better results in our experience.
2.3 Multiscale Graph Networks
The limitation of graph based networks, similar to other convolution methods is that they generate strong local interactions only. Hence, spatially-distant connections may suffer from weak interactions (due to small weights), and information will be spread slowly within the network—requiring more layers to compensate for. An elegant way to introduce long-range interactions and pass information between far-away parts of the graph is to consider a multiscale framework.
To this end, instead of a standard graph convolution
Our graph U-net is comprised of nLevels graph scales. At each level we perform a GCN block where we use both the graph and sequence neighbors in our convolutions:
where Kj is a 1D convolution with kernel of size 9, connecting nodes on the protein sequence, and Lj is the graph Laplacian operator from Eq. 2.11.
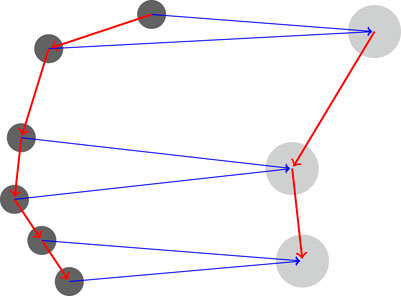
FIGURE 2. Coarsening a protein. The features of each two residues in the chain (left graph) are averaged together and a new coarse graph is computed (right) for the coarse protein. The graph Laplacian is computed directly from the rediscretized coarse protein.
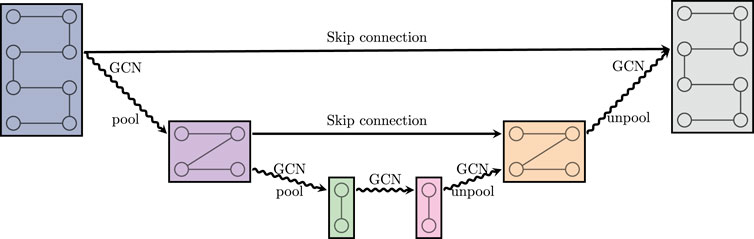
FIGURE 3. A graph U-net. GCN is defined in (2.12). Pool and unpool denote graph coarsening and refinement, respectively. Skip connection denotes a summation of the respective feature maps.
2.4 The MimNet Architecture
Combining our building blocks together, we now define our bi-directional mimetic architecture, called MimNet. The network consists of three main components - the opening embedding layer, a stacked graph U-net modules, and a closing embedding layer.
At the start and end of MimNet we use the embedding layers Eq. 2.1 and Eq. 2.5, both of which are implemented using a simple 1 × 1 convolution of appropriate sizes (n = 40). At the core of our network we employ a series of T graph U-nets modules. Each graph U-net is defined according to section 2.3, and all of them are of identical dimensions. That is, each has nf channels of equal dimensions on the finest level.
2.5 Training MimNet
Our MimNet allows us to build the physics of the problem into the neural network. This needs to be followed by a thoroughly thought training process. In particular, care needs be taken when choosing the appropriate problem to minimize and the appropriate choice of loss functions and regularization. We now discuss these choices for our training.
2.5.1 The Optimization Problem
Since we have a bi-directional network, we use both directions to train the network. We define the objective function
Here θ are all the parameters of the network, F is the forward network from sequence to coordinates and F† is the backward mapping from coordinates to sequence. The loss functions ℓfold and ℓdesign are chosen to measure the discrepancy between the estimated and true coordinates and between the predicted and true sequence design, respectively. The choice of these functions is to be discussed next. Finally, R(⋅) is a regularization term that ensures stability of the network and is described below.
2.5.2 Loss Function for the Design Problem
The loss function for the design problem aims at an accurate PSSM and sequence prediction by the network. Noting
We interpret both
where ⊘ denotes element-wise division. Having a PSSM as an output allows for greater flexibility when designing a protein, since there is not necessarily a unique answer to the design process, and the PSSM represents this ambiguity. As we show later, utilizing the PSSM-based loss in Eq. 2.15 has a significant contribution to the accuracy of the network compared to a loss based on the sequence alone used in (Strokach et al., 2020). Thus, as we demonstrate later, the coupling of PSSM and sequence estimation yields favorable performance.
2.5.3 Loss Function for the Folding Problem
We turn our attention for the loss function for the folding problem. Clearly, one cannot simply compare the coordinates obtained by the network, F(S) to the observed coordinates of the sequence, as they are invariant with respect to rotation and translation. Similar to the work (AlQuraishi, 2019a) one can compare the distance matrices obtained from the coordinates. Let
Where M is a masking matrix, and nM is the number of non-zeros in M. Zero elements in M correspond to either missing data in the native structure or to large distances. In proteins residue distances may range from a few to dozens Ångstroms. Naively minimizing the L2 distance would therefore focused on the large scale structure of the protein and might neglect the small scale structures as they would contribute remarkably less to the loss function. This motivated previous studies to use a threshold value and ignore distances larger than that threshold. For example, AlphaFold (Senior et al., 2020) and RGN (AlQuraishi, 2019a) use a 22.8Å cutoff, which correspond to six times the distance between consecutive Cα atoms. During the training phase, we used a slightly more conservative value of 7 residues, which translates to 7 × 3.8Å = 26.6Å. For evaluation and comparison purposes, we report our results using the same cutoff distance of 22.8 Å as RGN.
2.5.4 Regularization
The last component in our optimization scheme is the regularization, R(θ). We rewrite θ = [θ0, … , θL] where θj are the parameters used for the j-th layer. Then, smooth dynamics are obtained if the total variation of the dynamical system’s parameters is small (Ruthotto and Haber, 2019). Thus we choose the following regularization function
We note, that since the parameters θi denote the weights of the i-th UNet in our network, our regularization term considers the correspondingly-sized layers across subsequent layers of UNets. An example of the rather smooth folding process throughout the network is given in Figure 4. Note that we do not use the standard Tikhonov regularization (so called weight decay) on the weights as they do not guarantee smoothness in time which is crucial for reversible networks and integration in time (Celledoni et al., 2020).
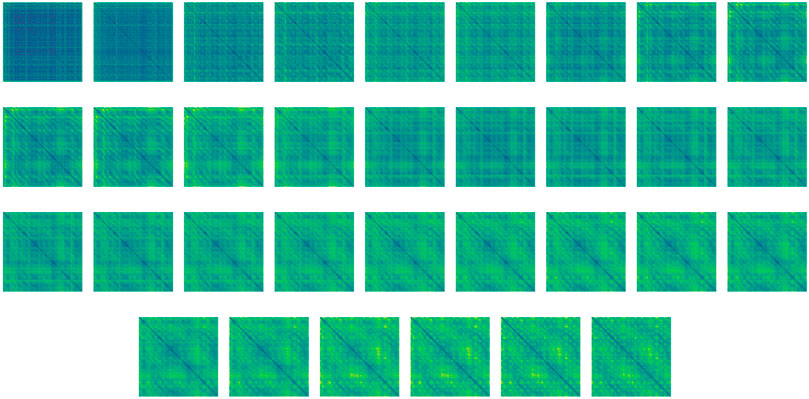
FIGURE 4. Distance maps of the high-dimensional feature evolution from unstructured (top left) to structured (bottom right) throughout the bi-directional network, during structure prediction of T0783. For a better demonstration of the process smoothness, the network includes 32 layers instead of the six layers used throughout the numerical study. Step size was adjusted to match the integration time. The idea of layer interpolation is discussed in Ruthotto and Haber (2019); Chen et al. (2018); Han et al. (2018).
2.6 Full Atom Model Reconstruction
While Cα models of protein structures are simple and allow fast structure estimate, they are not useful in practice. This is because it is possible to obtain a low dRMSD score for models that are not physically feasible. Such models can have too short distances between non-consecutive residues, and their geometry may be inverted as the loss function is invariant to mirror symmetry. While coping with this task in an end-to-end fashion is our long-term goal, we currently cope with it with a post processing stage based on our in-house molecular modeling Package MESHI (Kalisman et al., 2005) and scwrl4 (Krivov et al., 2009). An example of a post-processed reconstruction appears in Figure 5. This reconstruction stage consists of four steps:
• Random assignment of Cβ and backbone atoms close to their corresponding Cα atoms
• Several rounds of energy minimization, gradually reducing constraint on the Cα atom positions. The energy function includes standard bonded (bond, angle, plane out-of-plane, and torsion-angle (Amir et al., 2008) pairwise non-bonded term (Summa and Levitt, 2007) and hydrogen-bonds terms (Levy-Moonshine et al., 2009).
• Assigning side-chain atoms to their most common rotamer
• Side-chain repacking with scwrl4.
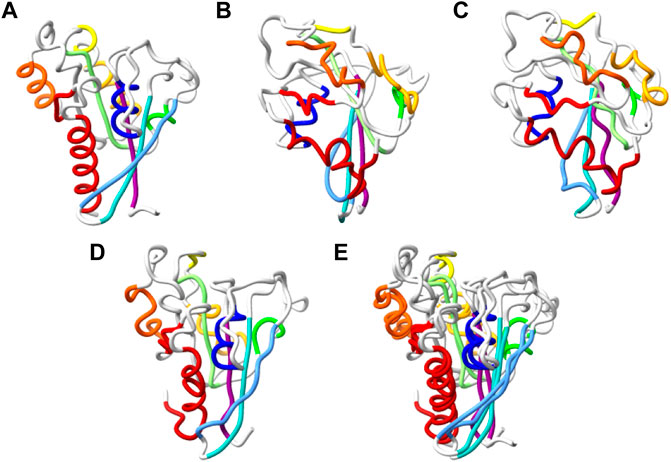
FIGURE 5. Reconstruction of chirally accurate full atom model. (A–D). Alpha trace representation of: the native structure of T0798 (A), MimNet output with dRMSD 5.194Å, GDTTS 0.19, LDDT 0.424 (B), and two energy minimize models of the MimNet output (C) and its mirror image (D). The secondary structure elements of the native structure are colored from first (dark blue) to last (dark red). The atoms of the other models are colored the same, regardless of their conformation. The energy of the minimized model (C) is −7838 (arbitrary units) and a dRMSD/GDTTS/LDDT of 5.076Å/0.20/0.426, while the energy of the minimized mirror image (D) is −18474, and a dRMSD/GDTTS/LDDT of 4.866Å/0.57/0.4358. (E). A ribbon representation of minimized mirror image, superimposed on the native structure.
3 Numerical Experiments
We verify our method by performing three sets of experiments—protein folding, design, and an ablation study to quantify the contribution of the reversible learning.
3.1 Dataset and Settings
3.1.1 Dataset
For the experiments we used the data set supplied by ProteinNet (AlQuraishi, 2019b). The data contains proteins processed from the PDB data set, and is organized to hold training, validation and testing splits specifically for CASP 7–12. The ProteinNet data for CASP 11, for instance, contains 42,338 proteins that are less than 1,000 residue long for training, 224 proteins for validation and 81 test proteins. All training and validation proteins were publicly available before CASP11, and the test set proteins are CASP11 targets. This data set was used in (AlQuraishi, 2019a) and more recently in (Drori et al., 2019). We use the 90% thinning version of the data, as reported in (AlQuraishi, 2019a). While the first order statistics is available, second order statistics cannot be downloaded freely and requires complex and expensive pre-processing. We therefore use only first order statistics in this work. We compare the results to two other recent methods RGN, which uses identical information, and proteinSolver. Note that the recent success of the AlphaFold2, as well as other methods, in CASP 14 were achieved using second order statistics.
3.1.2 Network and Optimization Settings
Throughout our experiments, we use our MimNet as described in Section 2.4 with nf = 128 with nLevels = 3 and T = 6. Other hyper-parameters, e.g., deeper networks using T > 6, may yield better performance and are worthy of consideration at corresponding computational cost. We use the Adam optimizer with a learning rate of 0.0001 and a batch size of 1, trained for 250 epochs. For the protein folding loss described in Eq. 2.5.3, we impose a dynamic mask M such that it begins from considering all distances, and linearly decreasing to the cutoff distance used of our training phase, which is 26.6Å. This way, our optimization scheme starts by capturing the global structure of the protein and gradually optimizing local interactions (distances) between residues. Our experiments are carried on an Nvidia Titan RTX. Our code is implemented in PyTorch (Paszke et al., 2019).
3.1.3 Comparisons
To evaluate our results we compare them to two recent studies, in the fields of protein design and PSP. First, for protein design, we compare our results with those of (Strokach et al., 2020). Their ProteinSolver is a graph neural network for the solution of the design problem. Using a sophisticated graph representation of the protein and its features, ProteinSolver obtains state of the art results, and a remarkable improvement over previous design studies. Thus, we consider it a good benchmark. Second, the results of the PSP side of our study are compared to the work of (AlQuraishi, 2019a). While that work did not achieve state of the art results (compared with methods that use second-order statistics), it is the only recent PSP study, known to us, that uses first order information from the ProteinNet dataset only.
3.2 Protein Design
The design experiments used Cα models, obtained from ProteinNet, to predict the corresponding PSSMs. As discussed in Section 2.5.2, the PSSMs are soft-assignments of the designed sequences, in accordance with the multiple design possibilities given a structure. The KL-divergence between the predicted PSSM and the ground-truth PSSM as supplied by the data in proteinNet is thus a natural measure, which we use for both training and performance evaluation. To compare our results with the recent work of ProteinSolver (Strokach et al., 2020) we employed the code and weights that are supplied with the paper (based on the 2015 version of the PDB) and used this pre-trained model to generate PSSMs for the proteinNet test sets data (CASP 7–12). The PSSMs predicted by ProteinSolver are then compared to the true PSSMs using KL-divergence. Table 1 depicts the design performances of proteinSolver and our MimNet on the various ProteinNet test sets. It shows a major improvement in MimNet, although ProteinSolver utilizes a superset of both the training and test sets of (CASP 7–11).
3.3 Protein Folding
Our folding experiment used first-order data (PSSM and a one hot sequence encoding), obtained from the ProteinNet test sets, to generate Cα models of the corresponding native structures. The accuracies of these models are evaluated by dRMSD to make the results comparable to those of RGN that used the same data sets (AlQuraishi, 2019a). Table 2 depicts the average performances of each method on the different test sets, suggest that the use of MimNet can significantly improve the accuracy of protein folding.

TABLE 2. dRMSD (Å) comparison of recent Protein-Folding methods. Averages of FM (novel folds)/TBM (known folds) are shown.
3.4 Additional Experimental Setup
To further test the method’s performance, we deviated from the ProteinNet scheme by creating new test sets. To this end, performance of a model based on CASPi training set, is tested by the union of all CASP test sets CASPi, … ,CASP12. This way, all but the last model were tested on larger and somewhat different test sets. The performances, presented in Figure 6, are consistent with results of the standard test sets reported in Tables 2, 4. We show the dRMSD and Accuracy, which is consistent with the KL divergence score.
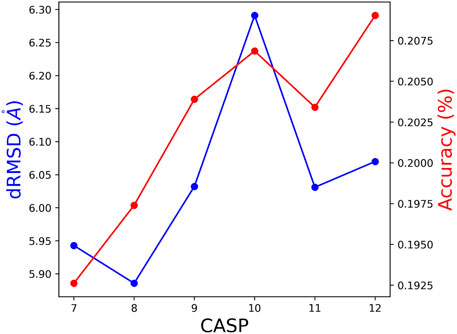
FIGURE 6. The obtained dRMSD and residue accuracy on the union of test of subsequent CASP editions. (e.g., the test set for CASP 8 is a union of CASP 8 until 12).
3.5 Per-Target Performance
Per-target performance in both folding and design is presented in Figure 7, and is consistent with the average values presented the Table 3. Interestingly, we can see a negative correlation between the obtained dRMSD and accuracy. That is, where our network yields lower (better) dRMSD, a higher sequence prediction accuracy is obtained.
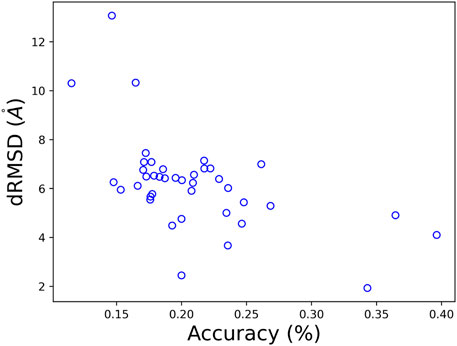
FIGURE 7. A comparison of the obtained dRMSD and residue accuracy on CASP 12 test set with our MimNet.
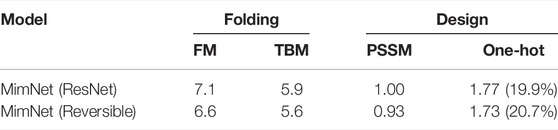
TABLE 3. A comparison of the reversible and standard ResNet architectures on folding and design tasks on CASP 11. For the folding task, we report both FM and TBM proteins dRMSD (Å) scores. For the design task, the KL-divergence of the PSSM and sequence (one-hot labels) are reported, along with the sequence prediction accuracy.
3.6 The Significance of Reversibility
One of the key contributions of our work is the introduction of reversible network architecture that can jointly learn both protein folding and design, which have not been done until now. We therefore delve on the significance of the reversibility scheme. There are three advantages for a reversible network architecture over a one directional network. First, and most important, typical one directional graph networks (e.g., ResNet like) are known to have smoothing properties that do not recover high frequencies well (Ming Chen et al., 2020; Zhao and Akoglu, 2020), a problem that can be alleviated by using second-order dynamics networks (Eliasof et al., 2021), which are also reversible, and behave like our dynamics in 2.3. Second, although we have not used this property in this work, reversible network allow to use very deep networks without limitation on memory (Gomez et al., 2017). Finally, in some cases, reversible architecture yields better results in training and generalizing compared to their one directional counterparts (Zhu et al., 2017; Chang et al., 2018; Yang et al., 2019). Most importantly it is a common approach in Language models (Devlin et al., 2018).
To validate the observations above on the considered data-set, we compare the reversible architecture to a standard ResNet (as in Eq. 2.4) architecture using the same settings. Namely, we employ the same input and output as well as number of layers and channels (as described in Section 3.1.2), yielding an identical number of parameters. The results for this experiment are provided in Table 3. The results suggest that using the reversible architecture is favored.
We now examine the importance of training the network in a bi-directional manner. We compare the behavior of our network when trained for both directions, versus the case of optimizing it only one direction (from sequence to coordinates and vice versa). Our results are summarized in Tables 4, 5 , suggesting that in some cases coupling the learning of folding and design problems can lead to better results in both folding and design, however, the effect of training on both sets of data at the same time, was marginal. Curiously, the reversible training experiments were our original motivation for the inclusion of PSSM in the design loss function (for symmetry considerations). In retrospect it turned out that this has a considerable positive effect on the design performance. Notably, the accuracy of one-hot prediction improves when the network is trained using PSSM (Table 4).

TABLE 4. Comparison between Coordinates to Design (C → D) and reversible learning (C ↔ D) on CASP 7–12. The letters in the brackets represent the data considered by the loss function. P denotes PSSM, O denotes one-hot encoding of the sequence and P/O implies both data are utilized. We report the KL-divergence scores of the PSSM, the sequence or both. The accuracy of the sequence is depicted in brackets where applicable.
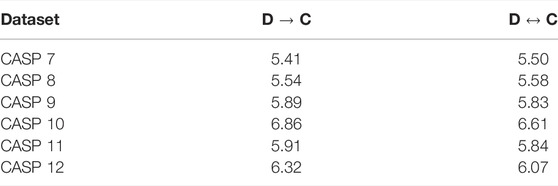
TABLE 5. Comparison between Design to Coordinates (D → C) and reversible learning (C ↔ D) on CASP 7–12. Results are reported in dRMSD (Å).
4 Conclusion
In this work we introduce a novel approach that unifies the treatment of protein folding and protein design. Our methodology is based on a combination of two recently studied techniques developed in deep learning. The first is a reversible architecture. Such an architecture allows us to propagate forward and backward and therefore have a network that can propagate sequence information into coordinates information and, more importantly for the protein design, propagate backward from a structure to a sequence. In the context of employing neural networks for molecular dynamics, it is natural to couple the reversible architecture with a graph representation, since our network models the pairwise interactions between amino acids as edges in the graph. Furthermore, to allow far field interactions, a multiscale structure is used.
In this scheme the input of one direction is the output of the other. The folding direction starts from a sequence and a PSSM formulated as a matrix of residue probabilities, and its output is a protein conformation. The loss function for training is the standard dRMSD metric applied to the output conformation and the native structure. Beyond the scope of this study, the scores of general PSSMs may not represent probabilities. In such cases however one can employ the SoftMax scheme to convert the scores to residue probabilities. The native structure in turn is the input to the design direction and the output is a PSSM and sequence, with their KL-divergence serving as the loss function for training. Previous studies only derived a single output from the PSSM. We note that the design problem does not have a unique answer and therefore we follow a more probabilistic approach.
We have performed extensive numerical experiments that compares both folding and design on the CASP 7–12 data sets. These data sets contain tens of thousands of proteins that we trained both on folding and design tasks. We compared the results of the protein folding to a recent work that uses only first order information. We have shown that our network performs on par or better than that network for the folding task. More importantly however, we have shown a significant improvement on the protein design task, achieving a KL-divergence loss that is less than half of a recently published work. We attribute this success to the use of recent protein folding architectures as well as using extensive data sets that allow better training of the proposed architecture. We note that our network is generic and can use both first order statistics, as in this study, and second order statistics.
Finally, we believe that the ProteinNet data-set constitutes a great leap forward as it allows scientists to compare methods on the same footings, similarly to the impact of ImageNet (Deng et al., 2009) on the computer vision community.
Data Availability Statement
Publicly available datasets were analyzed in this study. This data can be found here: ProteinNet at github.com/aqlaboratory/proteinnet.
Author Contributions
ET, CK, and EH conceived the project. ME, TB, EH, and ET developed the new DL scheme. ME and EH wrote the DL code and run the experiments. CK developed the full atom reconstruction. ET, CK, EH, and ME wrote the manuscript. All authors read the manuscript and agreed on its submission.
Funding
The research reported in this paper was funded by Genomica.ai and MITACS, and by grant no. 2018209 from the United States—Israel Binational Science Foundation (BSF), Jerusalem, Israel. ME is supported by Kreitman High-tech scholarship.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abriata, L. A., Tamò, G. E., and Dal Peraro, M. (2019). A Further Leap of Improvement in Tertiary Structure Prediction in Casp13 Prompts New Routes for Future Assessments. Proteins 87, 1100–1112. doi:10.1002/prot.25787
Adamczak, R., Porollo, A., and Meller, J. (2004). Accurate Prediction of Solvent Accessibility Using Neural Networks-Based Regression. Proteins 56, 753–767. doi:10.1002/prot.2017
Allen, M. P. (2004). Introduction to Molecular Dynamics Simulation. Comput. soft matter: Synth. Polym. proteins 23, 1–28.
AlQuraishi, M. (2019a). End-to-end Differentiable Learning of Protein Structure. Cell Syst 8 (4), 292–e3. doi:10.1016/j.cels.2019.03.006
AlQuraishi, M. (2019b). Proteinnet: a Standardized Data Set for Machine Learning of Protein Structure. BMC Bioinformatics 20, 311. doi:10.1186/s12859-019-2932-0
Altschul, S. F., Madden, T. L., Schäffer, A. A., Zhang, J., Zhang, Z., Miller, W., et al. (1997). Gapped BLAST and PSI-BLAST: a New Generation of Protein Database Search Programs. Nucleic Acids Res. 25, 3389–3402. doi:10.1093/nar/25.17.3389
Amir, E. D., Kalisman, N., and Keasar, C. (2008). Differentiable, Multi-Dimensional, Knowledge-Based Energy Terms for Torsion Angle Probabilities and Propensities. Proteins 72, 62–73. doi:10.1002/prot.21896
Anand-Achim, N., Eguchi, R. R., Mathews, I. I., Perez, C. P., Derry, A., Altman, R. B., et al. (2021). Protein Sequence Design with a Learned Potential. bioRxiv. doi:10.1101/2020.01.06.895466
Baek, M., DiMaio, F., Anishchenko, I., Dauparas, J., Ovchinnikov, S., Lee, G. R., et al. (2021). Accurate Prediction of Protein Structures and Interactions Using a Three-Track Neural Network. Science 373, 871–876. doi:10.1126/science.abj8754
Basanta, B., Bick, M. J., Bera, A. K., Norn, C., Chow, C. M., Carter, L. P., et al. (2020). An Enumerative Algorithm for De Novo Design of Proteins with Diverse Pocket Structures. Proc. Natl. Acad. Sci. U S A. 117, 22135–22145. doi:10.1073/pnas.2005412117
Bates, P. A., Kelley, L. A., MacCallum, R. M., and Sternberg, M. J. (2001). Enhancement of Protein Modeling by Human Intervention in Applying the Automatic Programs 3d-Jigsaw and 3d-Pssm. Proteins 5, 39–46. doi:10.1002/prot.1168
Ben-Sasson, A. J., Watson, J. L., Sheffler, W., Johnson, M. C., Bittleston, A., Somasundaram, L., et al. (2021).Design of Biologically Active Binary Protein 2D Materials. Nature 589. 468–473. doi:10.1038/s41586-020-03120-8
Bershtein, S., Goldin, K., and Tawfik, D. S. (2008). Intense Neutral Drifts Yield Robust and Evolvable Consensus Proteins. J. Mol. Biol. 379, 1029–1044. doi:10.1016/j.jmb.2008.04.024
Bouritsas, G., Bokhnyak, S., Ploumpis, S., Bronstein, M., and Zafeiriou, S. (2019). “Neural 3d Morphable Models: Spiral Convolutional Networks for 3d Shape Representation Learning and Generation,” in Proceedings of the IEEE/CVF International Conference on Computer Vision, 7213–7222. doi:10.1109/iccv.2019.00731
Celledoni, E., Ehrhardt, M. J., Etmann, C., McLachlan, R. I., Owren, B., Schönlieb, C.-B., et al. (2020). Structure Preserving Deep Learning. arXiv preprint arXiv:2006.03364.
Chamberlain, B. P., Rowbottom, J., Gorinova, M., Webb, S., Rossi, E., and Bronstein, M. M. (2021). Grand: Graph Neural Diffusion. arXiv preprint arXiv:2106.10934.
Chandler, P. G., Broendum, S. S., Riley, B. T., Spence, M. A., Jackson, C. J., McGowan, S., et al. (2020). Strategies for Increasing Protein Stability. New York, NY: Springer US, 163–181. doi:10.1007/978-1-4939-9869-2_10
Chang, B., Meng, L., Haber, E., Ruthotto, L., Begert, D., and Holtham, E. (2018). Reversible Architectures for Arbitrarily Deep Residual Neural Networks. In Proceedings of the AAAI Conference on Artificial Intelligence, 32.
Chen, R. T. Q., Rubanova, Y., Bettencourt, J., and Duvenaud, D. (2018). Neural Ordinary Differential Equations. 32nd Conference On Neural Information Processing Systems (NeurIPS).
Dahiyat, B. I., and Mayo, S. L. (1996). Protein Design Automation. Protein Sci. 5, 895–903. doi:10.1002/pro.5560050511
Deng, J., Dong, W., Socher, R., Li, L.-J., Li, K., and Fei-Fei, L. (2009). “ImageNet: A Large-Scale Hierarchical Image Database,” in CVPR09. doi:10.1109/cvpr.2009.5206848
Devlin, J., Chang, M.-W., Lee, K., and Toutanova, K. (2018). Bert: Pre-training of Deep Bidirectional Transformers for Language Understanding. arXiv preprint arXiv:1810.04805.
Drori, I., Thaker, D., Srivatsa, A., Jeong, D., Wang, Y., Nan, L., et al. (2019). Accurate Protein Structure Prediction by Embeddings and Deep Learning Representations. Machine Learn. Comput. Biol. (Mlcb).
Eliasof, M., Haber, E., and Treister, E. (2021). Pde-gcn: Novel Architectures for Graph Neural Networks Motivated by Partial Differential Equations. Adv. Neural Inf. Process. Syst. 34.
Eliasof, M., and Treister, E. (2020). Diffgcn: Graph Convolutional Networks via Differential Operators and Algebraic Multigrid Pooling. 34th Conference On Neural Information Processing Systems (NeurIPS 2020). Vancouver, Canada.
Gao, H., and Ji, S. (2019). “Graph U-Nets,” in international conference on machine learning (PMLR), 2083–2092.
Gao, Y., Wang, S., Deng, M., and Xu, J. (2018). Raptorx-angle: Real-Value Prediction of Protein Backbone Dihedral Angles through a Hybrid Method of Clustering and Deep Learning. BMC Bioinformatics 19, 100. doi:10.1186/s12859-018-2065-x
Gomez, A. N., Ren, M., Urtasun, R., and Grosse, R. B. (2017). “The Reversible Residual Network: Backpropagation without Storing Activations,” in Proceedings of the 31st International Conference on Neural Information Processing Systems, 2211–2221.
Goodfellow, I., Bengio, Y., and Courville, A. (2016). Deep Learning. MIT Press. Available at: http://www.deeplearningbook.org.
Gribskov, M., McLachlan, A. D., and Eisenberg, D. (1987). Profile Analysis: Detection of Distantly Related Proteins. Proc. Natl. Acad. Sci. U S A. 84, 4355–4358. doi:10.1073/pnas.84.13.4355
Haas, J., Barbato, A., Behringer, D., Studer, G., Roth, S., Bertoni, M., et al. (2018). Continuous Automated Model Evaluation (Cameo) Complementing the Critical Assessment of Structure Prediction in Casp12. Proteins 86, 387–398. doi:10.1002/prot.25431
Han, J., Jentzen, A., and E, W. (2018). Solving High-Dimensional Partial Differential Equations Using Deep Learning. Proc. Natl. Acad. Sci. U S A. 115, 8505–8510. doi:10.1073/pnas.1718942115
Hanocka, R., Hertz, A., Fish, N., Giryes, R., Fleishman, S., and Cohen-Or, D. (2019). MeshCNN. ACM Trans. Graph. 38, 1–12. doi:10.1145/3306346.3322959
He, K., Zhang, X., Ren, S., and Sun, J. (2016). “Deep Residual Learning for Image Recognition,” in Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, 770–778. doi:10.1109/cvpr.2016.90
Hildebrand, A., Remmert, M., Biegert, A., and Söding, J. (2009). Fast and Accurate Automatic Structure Prediction with HHpred. Proteins 77, 128–132. doi:10.1002/prot.22499
Hou, J., Wu, T., Cao, R., and Cheng, J. (2019). Protein Tertiary Structure Modeling Driven by Deep Learning and Contact Distance Prediction in Casp13. Proteins 87, 1165–1178. doi:10.1002/prot.25697
Ingraham, J., Garg, V. K., Barzilay, R., and Jaakkola, T. (2019). “Generative Models for Graph-Based Protein Design,” in Advances in Neural Information Processing Systems.
Jones, D. T., Buchan, D. W., Cozzetto, D., and Pontil, M. (2012). PSICOV: Precise Structural Contact Prediction Using Sparse Inverse Covariance Estimation on Large Multiple Sequence Alignments. Bioinformatics 28, 184–190. doi:10.1093/bioinformatics/btr638
Jones, D. T. (1994). De Novo protein Design Using Pairwise Potentials and a Genetic Algorithm. Protein Sci. 3, 567–574. doi:10.1002/pro.5560030405
Jumper, J., Evans, R., Pritzel, A., Green, T., Figurnov, M., Ronneberger, O., et al. (2021). Highly Accurate Protein Structure Prediction with Alphafold. Nature, 1–11. doi:10.1038/s41586-021-03819-2
Kalisman, N., Levi, A., Maximova, T., Reshef, D., Zafriri-Lynn, S., Gleyzer, Y., et al. (2005). MESHI: a New Library of Java Classes for Molecular Modeling. Bioinformatics 21, 3931–3932. doi:10.1093/bioinformatics/bti630
Kamisetty, H., Ovchinnikov, S., and Baker, D. (2013). Assessing the Utility of Coevolution-Based Residue-Residue Contact Predictions in a Sequence- and Structure-Rich Era. Proc. Natl. Acad. Sci. U S A. 110, 15674–15679. doi:10.1073/pnas.1314045110
Kandathil, S. M., Greener, J. G., and Jones, D. T. (2019). Recent Developments in Deep Learning Applied to Protein Structure Prediction. Proteins 87, 1179–1189. doi:10.1002/prot.25824
Krivov, G. G., Shapovalov, M. V., and Dunbrack, R. L. (2009). Improved Prediction of Protein Side-Chain Conformations with SCWRL4. Proteins 77, 778–795. doi:10.1002/prot.22488
Krizhevsky, A., Sutskever, I., and Hinton, G. E. (2012). “Imagenet Classification with Deep Convolutional Neural Networks,” in Advances in neural information processing systems, 1097–1105.
Kryshtafovych, A., Schwede, T., Topf, M., Fidelis, K., and Moult, J. (2019). Critical Assessment of Methods of Protein Structure Prediction (CASP)-Round XIII. Proteins 87, 1011–1020. doi:10.1002/prot.25823
Leaver-Fay, A., Tyka, M., Lewis, S. M., Lange, O. F., Thompson, J., Jacak, R., et al. (2011). “Rosetta3,” in Methods in Enzymology. Editors M. L. Johnson, and L. Brand (Academic Press), 487, 545–574. of Computer Methods, Part C. doi:10.1016/B978-0-12-381270-4.00019-6
Levy-Moonshine, A., Amir, el-A. D., and Keasar, C. (2009). Enhancement of Beta-Sheet Assembly by Cooperative Hydrogen Bonds Potential. Bioinformatics 25, 2639–2645. doi:10.1093/bioinformatics/btp449
Li, H., Hou, J., Adhikari, B., Lyu, Q., and Cheng, J. (2017). Deep Learning Methods for Protein Torsion Angle Prediction. BMC Bioinformatics 18, 417–426. doi:10.1186/s12859-017-1834-2
Li, Y., Hu, J., Zhang, C., Yu, D. J., and Zhang, Y. (2019). ResPRE: High-Accuracy Protein Contact Prediction by Coupling Precision Matrix with Deep Residual Neural Networks. Bioinformatics 35, 4647–4655. doi:10.1093/bioinformatics/btz291
Ming Chen, Z. W., Zengfeng Huang, B. D., and Li, Y. (2020). Simple and Deep Graph Convolutional Networks.
Morcos, F., Pagnani, A., Lunt, B., Bertolino, A., Marks, D. S., Sander, C., et al. (2011). Direct-coupling Analysis of Residue Coevolution Captures Native Contacts across many Protein Families. Proc. Natl. Acad. Sci. U S A. 108, E1293–E1301. doi:10.1073/pnas.1111471108
Moult, J., Hubbard, T., Bryant, S. H., Fidelis, K., and Pedersen, J. T. (1997). Critical Assessment of Methods of Protein Structure Prediction (Casp): Round Ii. Proteins Suppl 1, 2–6. doi:10.100210.1002/(sici)1097-0134(1997)1+<2::aid-prot2>3.0.co;2-t
Norn, C., Wicky, B. I. M., Juergens, D., Liu, S., Kim, D., Tischer, D., et al. (2021). Protein Sequence Design by Conformational Landscape Optimization. Proc. Natl. Acad. Sci. 118. doi:10.1073/pnas.2017228118
O'Connell, J., Li, Z., Hanson, J., Heffernan, R., Lyons, J., Paliwal, K., et al. (2018). Spin2: Predicting Sequence Profiles from Protein Structures Using Deep Neural Networks. Proteins 86, 629–633. doi:10.1002/prot.25489
Paszke, A., Gross, S., Massa, F., Lerer, A., Bradbury, J., Chanan, G., et al. (2019). “Pytorch: An Imperative Style, High-Performance Deep Learning Library,” in Advances in Neural Information Processing Systems 32. Editors H. Wallach, H. Larochelle, A. Beygelzimer, F. d'Alché-Buc, E. Fox, and R. Garnett (Curran Associates, Inc.), 8024–8035.
Ranjan, A., Bolkart, T., Sanyal, S., and Black, M. J. (2018). “Generating 3d Faces Using Convolutional Mesh Autoencoders,” in Proceedings of the European Conference on Computer Vision (ECCV), 704–720. doi:10.1007/978-3-030-01219-9_43
Remmert, M., Biegert, A., Hauser, A., and Söding, J. (2012). HHblits: Lightning-Fast Iterative Protein Sequence Searching by HMM-HMM Alignment. Nat. Methods 9, 173–175. doi:10.1038/nmeth.1818
Ronneberger, O., Fischer, P., and Brox, T. (2015). U-net: Convolutional Networks for Biomedical Image Segmentation.
Rost, B., Casadio, R., Fariselli, P., and Sander, C. (1995). Transmembrane Helices Predicted at 95% Accuracy. Protein Sci. 4, 521–533. doi:10.1002/pro.5560040318
Rost, B., and Sander, C. (1993). Prediction of Protein Secondary Structure at Better Than 70% Accuracy. J. Mol. Biol. 232, 584–599. doi:10.1006/jmbi.1993.1413
Ruthotto, L., and Haber, E. (2019). Deep Neural Networks Motivated by Partial Differential Equations. J. Math. Imaging Vis., 1–13. doi:10.1007/s10851-019-00903-1
Schwede, T., Kopp, J., Guex, N., and Peitsch, M. C. (2003). SWISS-MODEL: an Automated Protein Homology-Modeling Server. Nucleic Acids Res. 31, 3381–3385. doi:10.1093/nar/gkg520
Senior, A. W., Evans, R., Jumper, J., Kirkpatrick, J., Sifre, L., Green, T., et al. (2020). Improved Protein Structure Prediction Using Potentials from Deep Learning. Nature 577, 706–710. doi:10.1038/s41586-019-1923-7
Shah, S., Ghosh, P., Davis, L. S., and Goldstein, T. (2018). Stacked U-Nets: A No-Frills Approach to Natural Image Segmentation
Silverman, B. W. (1986). Density Estimation for Statistics and Data Analysis. London: Chapman & Hall.
Strokach, A., Becerra, D., Corbi-Verge, C., Perez-Riba, A., and Kim, P. M. (2020). Fast and Flexible Protein Design Using Deep Graph Neural Networks. Cel Syst 11, 402–e4. doi:10.1016/j.cels.2020.08.016
Summa, C. M., and Levitt, M. (2007). Near-native Structure Refinement Using In Vacuo Energy Minimization. Proc. Natl. Acad. Sci. U S A. 104, 3177–3182. doi:10.1073/pnas.0611593104
Sun, M. G. F., and Kim, P. M. (2017). Data Driven Flexible Backbone Protein Design. Plos Comput. Biol. 13, e1005722. doi:10.1371/journal.pcbi.1005722
Tetchner, S., Kosciolek, T., and Jones, D. T. (2014). Opportunities and Limitations in Applying Coevolution-Derived Contacts to Protein Structure Prediction. Bio-Algorithms and Med-Systems 10, 243–254. doi:10.1515/bams-2014-0013
Torrisi, M., Pollastri, G., and Le, Q. (2020). Deep Learning Methods in Protein Structure Prediction. Comput. Struct. Biotechnol. J. 18, 1301–1310. doi:10.1016/j.csbj.2019.12.011
Vassura, M., Margara, L., Di Lena, P., Medri, F., Fariselli, P., and Casadio, R. (2008). Reconstruction of 3d Structures from Protein Contact Maps. Ieee/acm Trans. Comput. Biol. Bioinform 5, 357–367. doi:10.1109/TCBB.2008.27
Wang, Y., Sun, Y., Liu, Z., Sarma, S. E., Bronstein, M. M., and Solomon, J. M. (2019). Dynamic Graph Cnn for Learning on point Clouds. ACM Trans. Graph. 38, 1–12. doi:10.1145/3326362
Waterhouse, A., Bertoni, M., Bienert, S., Studer, G., Tauriello, G., Gumienny, R., et al. (2018). SWISS-MODEL: Homology Modelling of Protein Structures and Complexes. Nucleic Acids Res. 46, W296–W303. doi:10.1093/nar/gky427
Xu, J. (2019). Distance-based Protein Folding Powered by Deep Learning. Proc. Natl. Acad. Sci. U S A. 116, 16856–16865. doi:10.1073/pnas.1821309116
Xu, J., Mcpartlon, M., and Li, J. (2021). Improved Protein Structure Prediction by Deep Learning Irrespective of Co-evolution Information. Nat. Machine Intelligence 1–9. doi:10.1038/s42256-021-00348-5
Yang, G., Huang, X., Hao, Z., Liu, M.-Y., Belongie, S., and Hariharan, B. (2019). Pointflow: 3d point Cloud Generation with Continuous Normalizing Flows.
Zhao, L., and Akoglu, L. (2020). “Pairnorm: Tackling Oversmoothing in \{gnn\}s,” in International Conference on Learning Representations.
Zheng, W., Li, Y., Zhang, C., Pearce, R., Mortuza, S. M., and Zhang, Y. (2019). Deep-learning Contact-Map Guided Protein Structure Prediction in Casp13. Proteins 87, 1149–1164. doi:10.1002/prot.25792
Keywords: graph neural networks, protein design, protein folding, deep learning, protein sructure prediction
Citation: Eliasof M, Boesen T, Haber E, Keasar C and Treister E (2022) Mimetic Neural Networks: A Unified Framework for Protein Design and Folding. Front. Bioinform. 2:715006. doi: 10.3389/fbinf.2022.715006
Received: 26 May 2021; Accepted: 29 March 2022;
Published: 05 May 2022.
Edited by:
Liam James McGuffin, University of Reading, United KingdomReviewed by:
Liang-Tsung Huang, Tzu Chi University, TaiwanBjörn Wallner, Linköping University, Sweden
Silvia Crivelli, Lawrence Berkeley National Laboratory, United States
Steven Farrell, Lawrence Berkeley National Laboratory, United States, in collaboration with reviewer SC
Rafael Zamora-Resendiz, Lawrence Berkeley National Laboratory, United States, in collaboration with reviewer SC
Copyright © 2022 Eliasof, Boesen , Haber , Keasar and Treister . This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Eldad Haber , ZWxkYWRoYWJlckBnbWFpbC5jb20=
 Moshe Eliasof
Moshe Eliasof Tue Boesen 2
Tue Boesen 2 Chen Keasar
Chen Keasar  Eran Treister
Eran Treister 