
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Anim. Sci. , 05 June 2024
Sec. Animal Physiology and Management
Volume 5 - 2024 | https://doi.org/10.3389/fanim.2024.1399337
This article is part of the Research Topic Understanding the Female Reproductive Microbiome in Livestock View all 9 articles
 Molly S. Smith
Molly S. Smith Dallas R. Soffa
Dallas R. Soffa Brooke E. McAnally
Brooke E. McAnally Kyle J. Hickman-Brown
Kyle J. Hickman-Brown Erin L. Stockland
Erin L. Stockland Rebecca K. Poole*
Rebecca K. Poole*Prior studies in beef cattle have shown shifts in the reproductive microbiome prior to artificial insemination (AI), yet few have characterized both the reproductive microbiome and immune responses prior to AI, particularly in purebred Bos indicus. Therefore, the aim of this study was to characterize the circulating cytokines and the vaginal microbiome of Bos indicus females prior to fixed-time artificial insemination (FTAI). Bos indicus females (n = 79) on four separate ranch operations within a 20-mile radius in East Texas were subjected to the 7-day CO-Synch + controlled intervaginal drug-releasing (CIDR) protocol beginning on day (d)-9 with FTAI on d0. Blood samples were collected on d-9 (CIDR IN), d-2 (CIDR OUT), and d0 (AI DAY) for cytokine concentration analyses using the RayBiotech Quantibody Bovine Cytokine Array Q1 kit per the manufacturer’s instructions. Sterile vaginal swabs were inserted past the vulva, rotated, and stored at -80°C for microbiome analysis. Bacterial community analyses targeted the V4 hypervariable region of the 16S rRNA gene. Pregnancy status was determined by transrectal ultrasonography approximately 60 days after FTAI for resulting open females (n = 45) and pregnant females (n = 34). Regardless of pregnancy status, the vaginal relative abundance of Firmicutes differed between CIDR IN, CIDR OUT, and AI DAY (63.74% vs. 28.31% vs. 60.86% ± 3.66%, respectively; P < 0.01). Genera with phylum Firmicutes including Ruminococcus, Clostridium, Blautia, Butyrvibrio, and Mogibacterium followed a similar trend (P < 0.05). Butyrvibrio tended to have greater relative abundance in the vaginal samples of Cows than Heifers (4.17% ± 0.75% vs. 3.26% ± 0.77%; P = 0.07). Concentrations of the interferon (IFN)γ (2005.98 ± 471.94 pg/mL vs. 1185.40 ± 482.65 pg/mL; P < 0.01), interleukin (IL)1F5 (153.89 ± 141.07 pg/mL vs. 627.30 ± 149.28 pg/mL; P < 0.01), and interferon gamma-induced protein (IP)10 (9363.26 ± 2929.83 pg/mL vs. 5905.53 ± 2983.60 pg/mL; P = 0.05) were greater in Cows than Heifers. There was a parity-by-status interaction for IP10, with Open Cows having the greatest concentration compared all other groups (P < 0.05). These results indicate differences in circulating cytokines and shifts in the vaginal microbiome for in Bos indicus cattle prior to FTAI.
With demand for the global food supply continuing to increase with a growing world population, reproductive efficiency of cattle on cow/calf operations will need to adapt. When a female fails to establish a pregnancy, yet remains in the breeding herd, the rancher experiences an economic loss (Cooke et al., 2020). While infertility in cattle can be attributed to many factors, such as nutrition, genetics, or disease; the immune system plays an active role in reproduction and may influence reproductive efficiency. Additionally, with Bos indicus-type or Brahman cattle consisting of around 30% of the beef population in the United States, it is important to understand how these factors impact reproductive efficiency in this subspecies. Their adaptivity and hardiness in sub-tropical and tropical climates make them ideal for beef production on the Gulf Coast (NASS, 2017; Cooke et al., 2020).
The innate immune system in an animal is the initial defense in the reproductive tract against invasive pathogens. It plays an active role in pregnancy, especially in preventing the onset of infection (Singh et al., 2008). Upon identification of an infection or pathogenic bacteria in the reproductive tract, the innate immune system will release small communication proteins called cytokines. Cytokines can initiate an inflammatory response in an environment by communicating with immune cells (Wira et al., 2005; Azawi, 2008; Murphy et al., 2022). There are two major classifications of cytokines: pro- inflammatory or anti-inflammatory. Pro-inflammatory cytokines may cause a more sustained and severe immune reaction and can lead to detrimental effects on tissues. In contrast, anti-inflammatory cytokines may aid in healing and returning the immune system back to homeostasis after an immune response (Dinarello, 2000; Cavaillon, 2001). Particularly during the rebreeding period in postpartum cattle, the innate immune system plays an active role in preventing an infection from occurring (Singh et al., 2008). Previous work has demonstrated certain bacterial genera are associated with cytokine concentrations prior to breeding in beef cattle (Smith et al., 2023). Additionally, the immune system’s first reaction for pregnancy establishment appears at insemination (Fair, 2015). During insemination, a variety of inflammatory cells are introduced at the site of semen deposition, potentially affecting the overall inflammatory response and subsequent fertility (Robertson, 2005).
Sheldon et al. (2002) and Williams et al. (2004) postulated that bacterial contamination in the uterus could disrupt ovulation. Other studies have indicated that changes in hormone concentrations lead to shifts in bacterial relative abundances (Sandrini et al., 2015; Org et al., 2016; Poole et al., 2023; Smith et al., 2023). Ault et al. (2019a) observed greater relative abundances of various commensal and pathogenic bacterial phyla in cattle in response to increased or reduced concentrations of progesterone (P4). These shifts in bacterial communities of the reproductive microbiome could impact fertility (Swartz et al., 2014; Laguardia-Nascimento et al., 2015; Luecke et al., 2022; Smith et al., 2023). Evaluating certain factors, such as the reproductive microbiome, provides an opportunity to develop methods that may increase efficiency in animal agricultural practices (Sheldon et al., 2002).
Characterization of the vaginal microbiome and circulating cytokine concentrations on a purebred Bos indicus beef cattle operation remains to be determined. As such, a unique opportunity became available to collect samples from several purebred Bos indicus beef cattle operations subjected to an estrus synchronization protocol. Therefore, the first objective of the current study was to determine the concentrations of specific pro- and anti-inflammatory cytokines in purebred Bos indicus beef cattle in association with a fixed-time artificial insemination (FTAI) protocol on-farm. The second objective was to evaluate the vaginal bacterial relative abundances in the vagina of Bos indicus beef cattle on-farm subjected to a FTAI protocol. The hypothesis was that concentrations of pro- and anti-inflammatory cytokines as well as the taxonomic composition of bacteria within the vagina would shift throughout during the estrus synchronization protocol.
All animal procedures were conducted in accordance with Texas A&M-approved protocols. Purebred Bos indicus females from cow-calf seedstock operations (RANCH 1, n = 11; RANCH 2, n = 20; RANCH 3, n = 24; RANCH 4, n = 24) in Wharton County, Texas were used for data collection from September to December 2022. Of the 79 cattle, both Heifers (n = 30) and Cows (n = 49) were utilized in this study. Cows were at least 60 days postpartum and assumed to be free of any known reproductive, health, or physical ailments. Heifers had not previously been artificially inseminated (AI), or pasture exposed to a bull. Females were subjected to the 7-day CO-Synch + controlled intervaginal drug-releasing (CIDR) protocol, where only two animal handlings were required prior to FTAI. The sampling and FTAI protocol is depicted in Figure 1. In brief, gonadotropin-releasing hormone (GnRH; Factrel, 100 μg; Zoetis Animal Health, Troy Hills, NJ) was administered, and CIDR devices (Eazi Breed CIDR; Zoetis Animal Health, Troy Hills, NJ) inserted 9 days (d-9; CIDR IN) prior to FTAI, which occurred 60–66 hours after CIDR removal, on d0 (AI DAY). On d-2 (CIDR OUT), an injection of prostaglandin F2α (PGF2α; Lutalyse, 5 mL; 5 mg/mL; Zoetis Animal Health, Troy Hills, NJ) was administered, and CIDRs were removed.
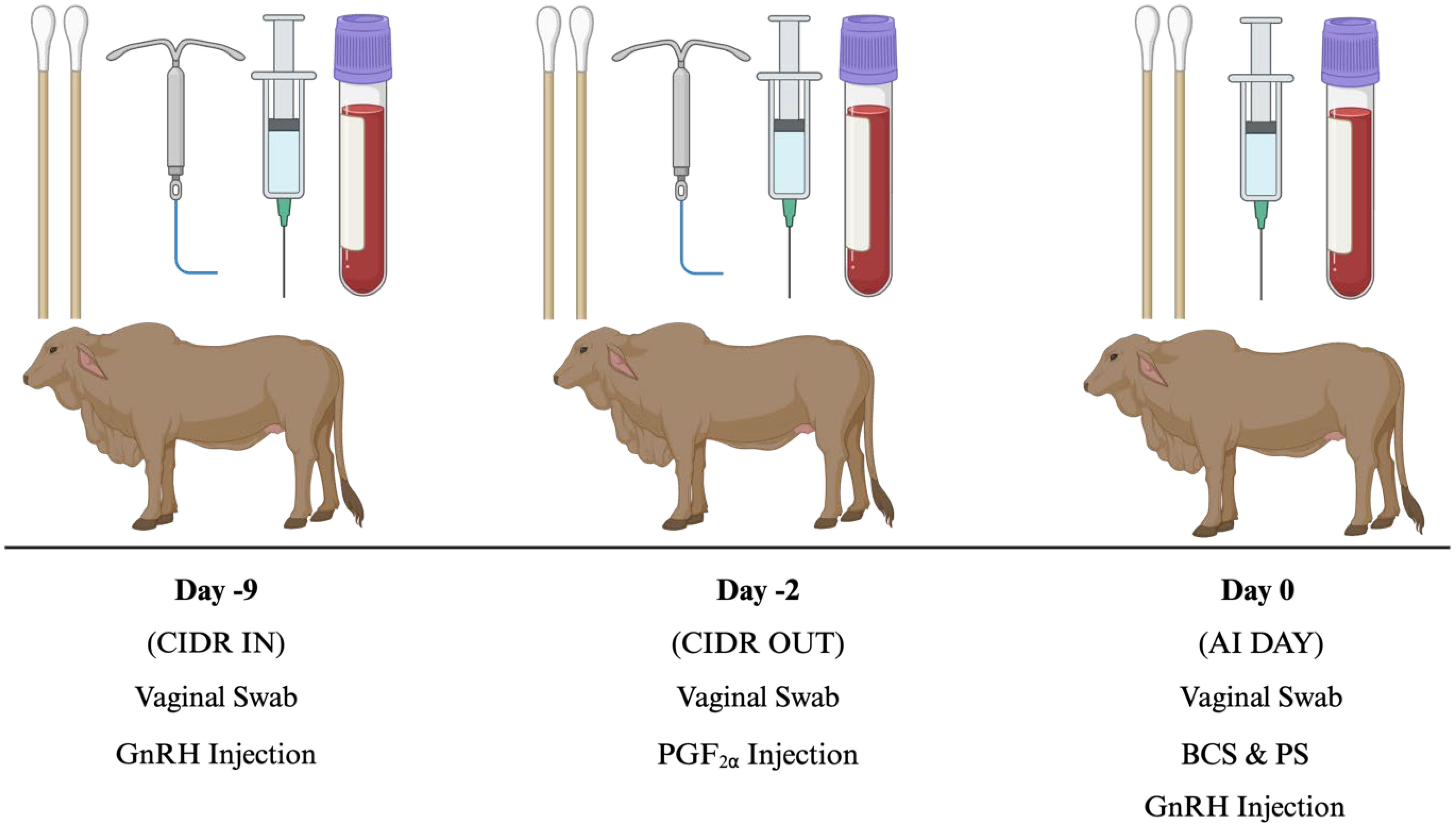
Figure 1 7-day CO-SYNCH + CIDR protocol experimental timeline used in Bos indicus cows for estrus synchronization. AI, Artificial Insemination; BCS, Body Condition Score; CIDR, Controlled Intervaginal Drug Releasing Device; PS, Patch Score; PGF2α, Prostaglandin injection; GnRH, Gonadotropin-Releasing Hormone. Created with BioRender.com.
Groups were assigned post-hoc as determined by pregnancy status via transrectal ultrasonography, resulting in Open (n = 45) and Pregnant (n = 34) females. Criteria for animal inclusion in this study the: females were cycling at the time of estrus synchronization and female responded to the estrus synchronization protocol based on P4 concentrations (described in subsequent sections).
Cattle body condition scores (BCS) and patch/paint scores were recorded on d0 (AI DAY). Vaginal swabs were collected on d-9 (CIDR IN), d-2 (CIDR OUT), and d0 (AI DAY) for bacterial DNA extraction and sequencing. During vaginal sampling, the perineal region was cleaned and spread prior to insertion of the sterile swabs to minimize fecal contamination. Swabs were inserted past the vulva and rotated six times against the vaginal wall. In addition, when sampling on-farm, a set of swabs was opened in between collections to document environmental conditions (control samples). Swab samples were collected in duplicate and stored in microcentrifuge tubes at -80°C until sequencing. Blood samples were collected on CIDR IN, CIDR OUT, and AI DAY via coccygeal venipuncture into 10 mL sterile vacutainer K2 EDTA collection tubes (BD Vacutainer, Becton, Dickinson and Company, NJ). Samples were immediately placed on ice and centrifuged at 1,500 g for 20 minutes. Plasma was transferred into 1.5 mL tubes and stored at -20°C until P4 and cytokine analysis. For blood and swab samples collected, on CIDR IN, samples were taken prior to GnRH injection and CIDR insertion, CIDR OUT swab sampling was performed following CIDR removal, and AI DAY sampling occurred prior to AI rod and lubricant insertion.
Progesterone concentrations were quantified per the manufacturer’s instructions using a commercial double-antibody radioimmunoassay (RIA) kit (MP Biomedicals, Santa Ana, CA), as previously described (Pohler et al., 2016). Concentrations were calculated using a calculated standard curve, including high/low reference samples, for quality control. The intra- and inter-assay coefficients of variation (CVs) were 4.30% and 14.24%, respectively.
Concentrations of interleukin (IL)13, IL1α, IL1F5, IL21, interferon (IFN)α, IFNγ, tumor necrosis factor (TNF)α, chemokine ligand (CXCL)9 (MIG), CXCL10 (IP10), and chemokine ligand 4 (MIP1b) were measured in the Bovine Cytokine Array Q1 kit (RayBiotech Life, Inc., Peachtree Corners, GA). Plasma samples were diluted 1:1 as suggested by the manufacturer, and cytokine concentrations for CIDR IN, CIDR OUT, and AI DAY samples were quantified. Assays were run per the manufacturer’s instructions, with slides being stored in a dark box at 4°C before shipment to the manufacturer for laser scanning and data extraction. These assays have been previously validated by Poole et al. (2019); Poole et al. (2021) and Smith et al. (2023).
Microbiome analysis was performed on the vaginal swab samples from CIDR IN, CIDR OUT and AI DAY as previously described in Smith et al. (2023). Briefly, samples were delivered to FERA Diagnostics and Biologicals Corp. (College Station, TX) for DNA extraction and 16S rRNA sequencing. Samples were transferred to 96-well plates, and DNA extraction was performed per the manufacturer’s instructions using the Mag-Bind Universal Pathogen 96 Kit (Omega Bio-Tek, Norcross, GA). As previously described, the 16S amplicons were amplified by PCR for individual metagenomic DNA samples (Bicalho et al., 2017). The V4 hypervariable region of the bacterial 16S rRNA bacterial genome was amplified with 515F (5′-GTGCCAGCMGCCGCGGTAA-3′) and 806R (5′-GGACTACHVGGGTWTCTAAT-3′) primers using methods for the Illumina MiSeq platform (Caporaso et al., 2012). The generated 16S rRNA gene sequences were assembled using the standard MiSeq pipeline as previously described (Coil et al., 2015; Bringhenti et al., 2021; Tomazi et al., 2023). The representative sequences for each operational taxonomic unit were compared against the Greengenes database (https://greengenes2.ucsd.edu/) for taxonomy assignment, and only full-length, high-quality reads were used. The MiSeq reporter classification was based on the Greengenes database, and the output used from this workflow was a classification of reads at multiple taxonomic levels (kingdom, phylum, class, order, family, genus, and species). However, for the purposes of this study, only the phylum and genus levels were reported.
Pregnancy status was analyzed using the MIXED procedure in Statistical Analysis Software (SAS; version 9.4, SAS Inst. Inc., Cary, NC) and included ranch as a random effect and parity and sire as a fixed effect. For each ranch, there was only one inseminator. Body condition scores and P4 concentrations were analyzed using PROC MIXED in SAS 9.4 with ranch as a random effect and parity, day, and pregnancy status as fixed effects and all interactions were evaluated. Bacterial abundances and cytokine concentrations were analyzed using PROC MIXED in SAS 9.4 and included ranch as a random effect and parity, day, and pregnancy status as fixed effects and all interactions were evaluated. All variables were tested for normality and homogeneity of variance using the HOVTEST=LEVENE option in PROC GLM in SAS 9.4. All bacterial abundance data and cytokine concentrations were normally distributed. Terms with a significance value of P > 0.10 were removed from the complete model in a stepwise manner to derive the final reduced model for each variable. Results were reported as least square means ± standard error of the mean. A statistical significance was reported at P ≤ 0.05. A tendency was reported at P > 0.05 and ≤ 0.10.
Between parity status (Cow vs. Heifer), no difference was observed in pregnancy rate (P > 0.10). There was no difference in pregnancy rate by sire (P > 0.10). No differences in P4 concentrations were seen by parity (P > 0.10) or by pregnancy status (Open vs. Pregnant; P > 0.10). A difference in P4 concentrations were observed by day (P < 0.01). Specifically, AI DAY exhibited reduced P4 concentrations versus CIDR IN (0.38 vs. 2.66 ± 0.57 ng/mL) and CIDR OUT (0.38 vs. 4.76 ± 0.57 ng/mL). Additionally, a difference in P4 concentrations were observed between CIDR IN and CIDR OUT days (2.66 vs. 4.76 ± 0.57 ng/mL). A difference was found between the BCS of Heifers and Cows, with Cows having greater body condition (5.45 ± 0.09 vs 5.76 ± 0.08; P < 0.01). Between pregnancy status, a difference was observed with Pregnant females having a greater BCS than Open females (5.75 ± 0.09 vs 5.50 ± 0.08; P < 0.01). Overall, there was no interaction between parity and pregnancy status for BCS (P > 0.10).
Differences in cytokine concentrations are displayed in Table 1. There was no difference observed by day regarding cytokine concentrations. By the main effect of parity, concentrations of IFNγ (2005.98 ± 471.94 pg/mL vs. 1185.40 ± 482.65 pg/mL; P < 0.01), IL1F5 (153.89 ± 141.07 pg/mL vs. 627.30 ± 149.28 pg/mL; P < 0.01), and IP10 (9363.26 ± 2929.83 pg/mL vs. 5905.53 ± 2983.60 pg/mL; P = 0.05) were greater in Cows when compared to Heifers. By the main effect of pregnancy status, there were greater concentrations of MIP1b in Open females (463.29 ± 97.47 pg/mL vs. 278.41 ± 97.91 pg/mL; P < 0.01) than in Pregnant females. Furthermore, there was a tendency to have greater concentrations of IL13 (8345.18 ± 1693.93 pg/mL vs. 6322.70 ± 1703.07 pg/mL; P = 0.06) in Open females compared with Pregnant females. There was a parity-by-status interaction for IP10, with Open Cows having the greatest concentration compared all other groups (Pregnant Cows, Pregnant Heifers, Open Heifers; P < 0.05).
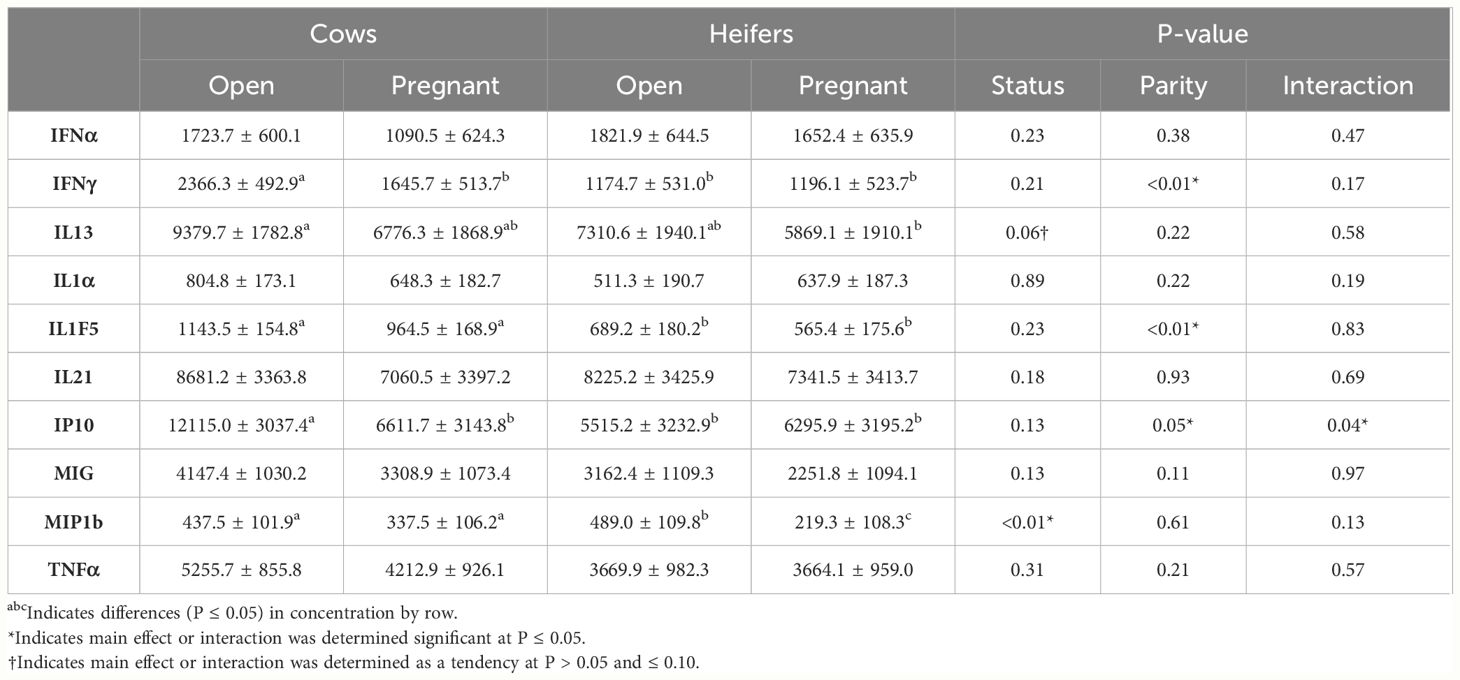
Table 1 Differences in cytokine concentrations by main effects of parity (Cow and Heifer), resulting pregnancy status (Open and Pregnant), and the interaction.
Within the vagina, Firmicutes, Proteobacteria, Bacteroidetes, Fusobacteria, Actinobacteria, Tenericutes, and Euryarchaeota were the most abundant phyla observed (greater than 1% relative abundance). Cows tended to have a greater relative abundance of the phylum Firmicutes than Heifers (53.39% ± 3.50% vs. 48.55% ± 3.50%; P = 0.07). No differences were observed by pregnancy status for phyla greater than 1% abundance in the vagina (P > 0.10).
Shifts in phyla by day are displayed in Figure 2. For the phylum of Firmicutes, there was a decrease in relative abundance from CIDR IN to CIDR OUT (63.74% ± 3.66% vs. 28.31% ± 3.66%) and an increase in relative abundance from CIDR OUT to AI DAY (28.31% ± 3.66% vs. 60.86% ± 3.66%; P < 0.01). Both Proteobacteria (P < 0.01) and Bacteroidetes (P = 0.045) had an increase in relative abundance from CIDR IN to CIDR OUT (7.79% ± 1.79% vs. 26.39% ± 1.79%; 12.25% ± 1.30% vs. 15.68% ± 1.30%, respectively) and then experienced a decrease in relative abundance from CIDR OUT to AI DAY (26.39% ± 1.79% vs. 9.63% ± 1.79%; 15.68% ± 1.30% ± 12.73% ± 1.30%, respectively). Furthermore, CIDR OUT had the greatest relative abundance of Fusobacteria, followed by CIDR IN, and then AI DAY (15.38% ± 3.58% vs. 5.71% ± 3.58% vs. 4.64% ± 3.58%, respectively; P < 0.01). Additionally, a difference was observed for Actinobacteria between CIDR IN and CIDR OUT (5.15% ± 1.31% vs. 10.19% ± 1.31%; P < 0.01), as well as between AI DAY and CIDR OUT (3.93% ± 1.31% vs. 10.19% ± 1.31%; P < 0.01). Tenericutes relative abundance had a difference by day (P = 0.05) between AI DAY and CIDR IN (4.50% ± 1.28% vs. 1.41% ± 1.28%). The phylum of Euryarchaeota saw a difference by day between AI DAY and CIDR OUT (1.47% ± 0.14% vs. 0.35% ± 0.14%; P < 0.01), as well as CIDR IN and CIDR OUT (1.73% ± 0.14% vs. 0.35% ± 0.14%; P < 0.01).
No differences were detected between pregnancy status for genera greater than 1% relative abundance in the vagina (P > 0.10). Butyrvibrio tended to have greater relative abundance in the vaginal samples of Cows than Heifers (4.17% ± 0.75% vs. 3.26% ± 0.77%; P = 0.07). Additionally, Mannheimia tended to differ between Cows and Heifers (2.43% ± 1.07% vs. 4.93% ± 1.22%; P = 0.08).
Furthermore, there was a parity-by-day interaction for Mannheimia (P = 0.02) with CIDR OUT for Heifers having the greatest relative abundance (14.12% ± 1.92%) and CIDR IN for Cows and Heifers having the least relative abundance (0.29% ± 1.58% and 0.20% ± 1.92%, respectively). Additionally, there was a tendency for Blautia to have a parity-by-day interaction (P = 0.07) with CIDR IN day for Cows having the greatest relative abundance (7.14% ± 1.02%) and CIDR OUT day for Cows and Heifers having the least relative abundance (2.36% ± 1.02% and 2.36% ± 1.06%, respectively).
Relative abundance of significant vaginal genera on CIDR IN, CIDR OUT, and AI DAY are displayed in Table 2. A difference was seen in Ruminococcus, Clostridium, Blautia, Butyrvibrio, and Mogibacterium, with CIDR IN having greater relative abundance than CIDR OUT. Furthermore, when comparing CIDR OUT to AI DAY, Ruminococcus, Clostridium, Blautia, Butyrvibrio, and Mogibacterium had increased relative abundance. Additionally, there was a difference by day for Mannheimia, with CIDR OUT exhibiting a greater relative abundance than CIDR IN and AI DAY.
It has been previously established that the immune system plays a role in beef cattle fertility; in addition to the relationship between different bacterial communities and the immune system (Smith et al., 2023). The first aim of this study was to determine concentrations of pro- and anti-inflammatory cytokines in purebred Bos indicus beef cattle plasma samples on producer operations before FTAI. Furthermore, Ault et al. (2019a) stated that relative abundances of bacteria appear to shift during estrus synchronization and may influence fertility. Therefore, an additional objective was to evaluate the fluctuations in vaginal bacteria relative abundance associated with the 7-day CO-Synch synchronization protocol in purebred Bos indicus beef cattle.
The immune system will respond to microbial invasion by releasing cytokines or chemokines into the bloodstream to heal or mitigate the effects of infection (Cavaillon, 2001; Murphy et al., 2022). In addition, cytokines are essential for pregnancy establishment and pregnancy maintenance (Yockey and Iwasaki, 2019). In the current study, the pro-inflammatory cytokine IFNγ was observed to be greater in Cows compared to Heifers. A greater concentration of pro-inflammatory cytokines in the reproductive tract is typically associated with uterine involution (Balkwill, 2006; Galvao et al., 2011), which potentially explains the greater concentrations in Cows, who have undergone the involution process, as compared to Heifers in the current study. The pleiotropic cytokine IP10 also had greater concentrations measured in Cows than in Heifers as well as a parity-by-status interaction for Open Cows. Interferon gamma-induced protein 10 is regulated by IFNγ signaling and nuclear factor kappa B (NF-κB) activity, thus it makes sense that both IFNγ and IP10 concentrations would be greater in Cows compared to Heifers. Pleiotropic cytokines can display both pro- and anti-inflammatory behavior and stimulate a wide variety of cells (Zhang and An, 2007). As a biomarker, IP10 has been shown as an indicator of infection prior to onset of clinical symptoms in humans (Moreno et al., 2022; Wang et al., 2023). Collectively, greater IP10 concentrations in Open Cows prior to AI could indicate inflammation hindering the establishment of a pregnancy. The anti-inflammatory IL1F5 was also observed in greater concentration in Cows than in Heifers; and in one study, it was found to have a correlation with bacteria that induce cell inflammation (Smith et al., 2023). Additionally, IL1F5 inhibits the activation of NF-κB, which is important for regulating pro-inflammatory immune responses (Abdulrahman Alrabiah et al., 2021). Therefore, the increased concentrations of pro-inflammatory immune responses (i.e., IFNγ) would also be associated with an increase in an anti-inflammatory immune response (i.e., IL1F5) in Cows.
Concentrations of the anti-inflammatory cytokine IL13 tended to be greater in concentration for Open females compared to Pregnant females. Ma et al. (2013) and Dickinson et al. (2016) concluded that IL13 inhibits cell breakdown of old and damaged cell components. Interleukin 13 will also increase the production of an IL1 receptor antagonist, which can bind with IL1 and reduce its pro-inflammatory properties. Typically, IL13 is involved in downregulating pro-inflammatory cytokine and chemokine production (de Vries, 1998); however, some studies investigating the pathogenesis of asthma and the lungs have demonstrated that IL13 can induce chemokine responses (Zhu et al., 2002). Interestingly, concentrations of the chemokine MIP1b were greater in Open cattle than in Pregnant cattle. The increased concentrations of MIP1b may be attributed to its function as a macrophage-attracting chemokine that plays a role in controlling macrophage influx to tissues, such as the vagina (von Stebut et al., 2003). In addition, cytokine concentrations did not differ by day of the synchronization protocol in the current study, which is different from a previous study by Smith et al. (2023), where it was noted that hormone changes were associated with alterations in cytokine concentrations by day in plasma, vaginal flushes, and uterine flushes.
Bacterial phyla such as Firmicutes, Bacteroidetes, Proteobacteria, Actinobacteria, and Tenericutes have been discovered to dominate the reproductive tract of beef cattle which aligns with the findings of the current study (Swartz et al., 2014; Clemmons et al., 2017; Messman et al., 2020; Pickett et al., 2022). Additionally, some studies have shown that bacterial abundances will shift due to hormonal alterations in Bos taurus females (Ault et al., 2019a, Ault et al., 2019b; Poole et al., 2023). In Bos indicus cattle specifically, some studies have shown that Firmicutes, Proteobacteria, and Bacteroidetes are frequently detected (Laguardia-Nascimento et al., 2015; Giannattasio-Ferraz et al., 2019). The current study identified that the relative abundance of Firmicutes was more abundant at CIDR IN than CIDR OUT. These results coincide with Ault et al. (2019b), where Bos taurus females subjected to an estrus synchronization protocol had decreased relative abundances of Firmicutes two days prior to AI (synonymous with CIDR OUT in the current study). In addition, the relative abundance of Firmicutes increased from CIDR OUT to AI DAY, where removal of the CIDR device and administration of a PGF2α injection caused the corpus luteum to undergo luteolysis and P4 concentrations to decrease. More specifically, as P4 decreased the relative abundance of Firmicutes appeared to increase. Notably, the tendency for Cows to have greater relative abundances of Firmicutes compared to Heifers was also observed in a study completed by Moore et al. (2017). Oppositely, the relative abundances of Proteobacteria and Bacteroidetes were reduced on CIDR IN compared to CIDR OUT days. The shift in the relative abundance of Proteobacteria was also correlated with P4 (Ault et al., 2019b), further demonstrating that as P4 concentrations increase, Proteobacteria relative abundance increases. Interestingly, an increased abundance of Bacteroidetes and decreased relative abundance of Proteobacteria has been found in Bos taurus metritic cows (Jeon et al., 2015), although no incidences of uterine inflammation were noted in this study.
The genus of Butyrvibrio, within phylum Firmicutes, was seen in greater relative abundance in the vagina of Cows compared to Heifers. Furman et al. (2020) reported that Butyrvibrio had greater abundance in the rumen of Bos taurus calves that were delivered vaginally versus via cesarean section, indicating this may be a common genus found in the vaginal microbiome. Though not typically categorized as pathogenic, Butyrivibrio has been previously identified within the uterus of Bos taurus Open cows (Clemmons et al., 2017; Ault et al., 2019b). Ruminococcus, Blautia, and Mogibacterium, all genera under the phylum of Firmicutes, had greater relative abundance at CIDR IN day than CIDR OUT day. This indicates that the introduction of a CIDR may have caused shifts in the vaginal microbiome. Such alterations to the microbiome have been shown to cause a rise in vaginal pH which may then influence fertility (Dias, 2022). While Butyrvibrio and Mogibacterium have been associated with compromised fertility (Clemmons et al., 2017; Ault et al., 2019a), no differences by fertility outcomes were observed in this study. The genus Mannheimia, under the phylum of Proteobacteria, had greatest relative abundance on CIDR OUT day and is known to be pleomorphic, which means it can be irregular and vary in its form (Laishevtsev, 2020). The occurrence of Mannheimia in the vagina of Bos indicus beef cows is not surprising as it has also been found in the vagina of dairy cows (Bicalho et al., 2017), but the association between the greater relative abundance of Mannheimia and its relation to CIDR OUT day needs to be further assessed.
In conclusion, cytokine concentrations may be associated with parity or fertility outcomes. This study also suggests that bacterial relative abundances can shift through an estrus synchronization protocol in Bos indicus cattle. Additional research may evaluate hormone concentrations with or without the introduction of a foreign object (e.g., CIDR) on bacterial shifts. Future research is necessary to determine the factors that can positively affect the immune system and vaginal microbiome to improve reproductive efficiency in Bos indicus cattle.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
The animal studies were approved by Texas A&M University Institutional Animal Care and Use Committee. The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent was obtained from the owners for the participation of their animals in this study.
MS: Investigation, Methodology, Writing – original draft, Writing – review & editing. DS: Methodology, Writing – review & editing. BM: Methodology, Writing – review & editing. KH-B: Methodology, Writing – review & editing. ES: Methodology, Writing – review & editing. RP: Conceptualization, Formal Analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the USDA-NIFA Hatch/Multistate Project W4112-TEX09926: Reproductive performance in domestic ruminants. The Walter and Wilma Bush Graduate Assistantship from the Texas A&M College of Agriculture and Life Sciences provided assistantship funding for MS.
Special appreciation is extended to J.D. Hudgins Inc. – Forgason Division, J.D. Hudgins Inc. – Locke Division, B.R. Cutrer Inc., and Heritage Cattle Company for generously providing access to their ranch facilities and cattle.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abdulrahman Alrabiah N., Evans A. C. O., Fahey A. G., Cantwell N., Lonergan P., Mccormack J., et al. (2021). Immunological aspects of ovarian follicle ovulation and corpus luteum formation in cattle. Reproduction. 162, 209–225. doi: 10.1530/REP-21-0165
Ault T. B., Clemmons B. A., Reese S. T., Dantas F. G., Franco G. A., Smith T. P. L., et al. (2019a). Bacterial taxonomic composition of the postpartum cow uterus and vagina prior to artificial insemination. J. Anim. Sci. 97, 4305–4313. doi: 10.1093/jas/skz212
Ault T. B., Clemmons B. A., Reese S. T., Dantas F. G., Franco G. A., Smith T. P. L., et al. (2019b). Uterine and vaginal bacterial community diversity prior to artificial insemination between Pregnant and Non-Pregnant postpartum cows. J. Anim. Sci. 97 (10): 4298–4304. doi: 10.1093/jas/skz210
Azawi O. I. (2008). Postpartum uterine infection in cattle. Anim. Reprod. Sci. 105, 187–208. doi: 10.1016/j.anireprosci.2008.01.010
Balkwill F. (2006). TNF-α in promotion and progression of cancer. Cancer Metastasis Rev. 25, 409–416. doi: 10.1007/s10555-006-9005-3
Bicalho M. L. S., Santin T., Rodrigues M. X., Marques C. E., Lima S. F., Bicalho R. C. (2017). Dynamics of the microbiota found in the vaginas of dairy cows during the transition period: Associations with uterine diseases and reproductive outcome. J. Dairy Sci. 100, 3043–3058. doi: 10.3168/jds.2016-11623
Bringhenti L., Pallu M., Silva J. C., Tomazi T., Tomazi A. C. C. H., Rorigues M. X., et al. (2021). Effect of treatment of pneumonia and otitis media with tildipirosin or florfenicol + flunixin meglumine on health and upper respiratory tract microbiota of pre-weaned Holstein dairy heifers. J. Dairy Sci. 104, 10291–10309. doi: 10.3168/jds.2020-19945
Caporaso J. G., Lauber C. L., Walters W. A., Berg-Lyons D., Huntley J., Fierer N., et al. (2012). Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 6, 1621–1624. doi: 10.1038/ismej.2012.8
Cavaillon J. M. (2001). Pro- versus anti-inflammatory cytokines: myth or reality. Mol. Cell. Biol. 47, 695–702.
Clemmons B. A., Reese S. T., Dantas F. G., Franco G. A., Smith T. P. L., Adeyosoye O. I., et al. (2017). Vaginal and uterine bacterial communities in postpartum lactating cows. Front. Microbiol. 8. doi: 10.3389/fmicb.2017.01047
Coil D., Jospin G., Darling A. E. (2015). A5-miseq: an updated pipeline to assemble microbial genomes from Illumina MiSeq data. Bioinformatics 31, 587–589. doi: 10.1093/bioinformatics/btu661
Cooke R. F., Cardoso R. C., Cerri R. L. A., Lamb G. C., Pohler K. G., Riley D. G., et al. (2020). Cattle adapted to tropical and subtropical environments: genetic and reproductive considerations. J. Anmi. Sci. 98, skaa015. doi: 10.1093/jas/skaa015
de Vries J. E. (1998). The role of IL-13 and its receptor in allergy and inflammatory responses. J. Allergy Clin. Immunol. 102, 165–169. doi: 10.1016/S0091-6749(98)70080-6
Dias N. (2022). Changes in vaginal microbiome of beef cows enrolled in estrous synchronization protocols and its relation to fertility (Blacksburg, Virginia: Dissertation. Virginia Polytechnic Institute and State University).
Dickinson J. D., Alevy Y., Malvin N. P., Patel K. K., Gunsten S. P., Holtzman M. J., et al (2016). IL13 activates autophagy to regulate secretion in airway epithelial cells. Autophagy. 12, 397–409. doi: 10.1080/15548627.2015.1056967
Dinarello C. A. (2000). Proinflammatory cytokines. Chest. 118, 503–508. doi: 10.1378/chest.118.2.503
Fair T. (2015). The contribution of the maternal immune system to the establishment of pregnancy in cattle. Frontiers. 6. doi: 10.3389/fimmu.2015.00007
Furman O., Shenhav L., Sasson G., Kokou F., Honig H., Jacoby S., et al. (2020). Stochasticity constrained by deterministic effects of diet and age drive rumen microbiome assembly dynamics. Nat. Commun. 11, 1–13. doi: 10.1038/s41467-020-15652-8
Galvao K. N., Santos N. R., Galvao J. S., Gilbert R. O. (2011). Association between endometritis and endometrial cytokine expression in postpartum Holstein cows. Theriogenology. 76, 290–299. doi: 10.1016/j.theriogenology.2011.02.006
Giannattasio-Ferraz S., Laguardia-Nascimento M., Gasparini M. R., Leite L. R., Araujo F. M. G., de Matos Salim A. C., et al. (2019). A common vaginal microbiota composition among breeds of Bos taurus indicus (Gyr and Nellore). Braz. J. Microbiol. 50, 1115–1124. doi: 10.1007/s42770-019-00120-3
Jeon S. J., Vieira-Neto A., Gobikrushanth M., Daetz R., Mingoti R. D., Parize A. C. B., et al. (2015). Uterine microbiota progression from calving until establishment of metritis in dairy cows. Appl. Environ. Microbiol. 81, 6324–6332. doi: 10.1128/AEM.01753-15
Laguardia-Nascimento M., Branco K. M. G. R., Gasparini M. R., Giannattasio-Ferraz S., Leite L. R., Araujo F. M. G., et al. (2015). Vaginal microbiome characterization of Nellore cattle using metagenomic analysis. PloS One 10, e0143294. doi: 10.1371/journal.pone.0143294
Laishevtsev A. I. (2020). “Mannheimiosis of cattle, sheep, and goats,” in IOP Conf. Ser.: Earth environ. Sci, Vol. 548. doi: 10.1088/1755–1315/548/7/072038
Luecke S. M., Webb E. M., Dahlen C. R., Reynolds L. P., Amat S. (2022). Seminal and vagino-uterine microbiome and their individual and interactive effects on cattle fertility. Front. Microbiol. 13. doi: 10.3389/fmicb.2022.1029128
Ma Y., Galluzzi L., Zitvogel L., Kroemer G. (2013). Autophagy and cellular immune responses. Immunity. 39, 211–227. doi: 10.1016/j.immuni.2013.07.017
Messman R. D., Contreras-Correa Z. E., Paz H. A., Perry G., Lemley C. O. (2020). Vaginal bacterial community composition and concentrations of estradiol at the time of artificial insemination in Brangus heifers. J. Anim. Sci. 98, skaa178. doi: 10.1093/jas/skaa178
Moore S. G., Ericsson A. C., Poock S. E., Melendez P., Lucy M. C. (2017). Hot topic: 16S rRNA gene sequencing reveals the microbiome of the virgin and pregnant bovine uterus. J. Dairy Sci. 100, 4953–4960. doi: 10.3168/jds.2017-12592
Moreno B., Hueso L., Ortega R., Benito E., Martinez-Hervas S., Peiro M., et al. (2022). Association of chemokines IP-10/CXCL10 and I-TAC/CXCL11 with insulin resistance and enhance leukocyte endothelial arrest in obesity. Microvascular Res. 139, 104254. doi: 10.1016/j.mvr.2021.104254
Murphy K., Weaver C., Berg L. (2022). Janeway’s immunobiology. 10th ed (New York, NY: W.W. Norton & Company).
NASS. (2017). National Agricultural Statistics Service, agricultural statistics board. USDA, Washington, DC.
Org E., Mehrabian M., Parks B. W., Shipkova P., Liu X., Drake T. A., et al. (2016). Sex differences and hormonal effects on gut microbiota composition in mice. Gut Microbes 7, 312–322. doi: 10.1080/19490976.2016.1203502
Pickett A. T., Cooke R. F., Mackey S. J., Brandao A. P., Columbo E. A., Oliviera Filho R. V., et al. (2022). Shifts in bacterial communities in the rumen, vagina, and uterus of beef heifers receiving different levels of concentrate. J. Anim. Sci. 100, 1–11. doi: 10.1093/jas/skac338
Pohler K. G., Pereira M. H. C., Lopes F. R., Lawrence J. C., Keisler D. H., Smith M. F., et al. (2016). Circulating concentrations of bovine pregnancy-associated glycoproteins and late embryonic mortality in lactating dairy herds. J. Dairy Sci. 99, 1584–1594. doi: 10.3168/jds.2015-10192
Poole R. K., Ault-Seay T. B., Payton R. R., Myer P. R., Lear A. S., Pohler K. G. (2021). Evaluation of reproductive tract cytokines in post-partum beef cows relating to reproductive microbiota and fertility outcomes. Front. Anim. Sci. 2. doi: 10.3389/fanim.2021.704714
Poole R. K., Brown A. R., Poore M. H., Pickworth C. L., Poole D. H. (2019). Effects of endophyte-infected tall fescue seed and protein supplementation on stocker steers: II. Adaptive and innate immune function. J. Anim. Science. 97, 4160–4170. doi: 10.1093/jas/skz250
Poole R. K., Pickett A. T., Oliveira Filho R. V., de Melo G. D., Palanisamy V., Chitlapilly Dass S., et al. (2023). Shifts in uterine bacterial communities associated with endogenous progesterone and 17β-estradiol concentrations in beef cattle. Domest. Anim. Endocrinol. 82, 106766. doi: 10.1016/j.domaniend.2022.106766
Robertson S. A. (2005). Seminal plasma and male factor signaling in the female reproductive tract. Cell Tissue Res. 322, 43–52. doi: 10.1007/s00441-005-1127-3
Sandrini S., Aldriwesh M., Alruways M., Freestone P. (2015). Microbial endocrinology: host-bacteria communication within the gut microbiome. J. Endocrinol. 225, R21–R34. doi: 10.1530/JOE-14-0615
Sheldon I. M., Noakes. D. E., Rycroft A. N., Pfeiffer D. U., Dobson H. (2002). Influence of uterine bacterial contamination after parturition on ovarian dominant follicle selection and follicle growth and function in cattle. Reproduction 123, 837–845. doi: 10.1530/rep.0.1230837
Singh J., Murray R. D., Mshelia G., Woldehiwet Z. (2008). The immune status of the bovine uterus during the peripartum period. Vet. J. 175, 301–309. doi: 10.1016/j.tvjl.2007.02.003
Smith M. S., Hickman-Brown K. J., McAnally B. E., Oliveira Filho R. V., De Melo G. D., Pohler K. G., et al. (2023). Reproductive microbiome and cytokine profiles associated with fertility outcomes of postpartum beef cows. J. Anim. Sci. 101:skad219. doi: 10.1093/jas/skad219
Swartz J. D., Lachman M., Westveer K., O’Neill T., Geary T., Kott R. W., et al. (2014). Characterization of the vaginal microbiota of ewes and cows reveals a unique microbiota with low levels of lactobacilli and near-neutral pH. Front. Vet. Sci. 1. doi: 10.3389/fvets.2014.00019
Tomazi A. C. C. H., Tomazi T., Bringhenti L., Vinhal A. P. A., Rorigues M. X., Bilby T. R., et al. (2023). Treatment with 2 commercial antibiotics reduced clinical and systemic signs of pneumonia and the abundance of pathogenic bacteria in the upper respiratory tract of preweaning dairy calves. J. Dairy Sci. 106, 2750–2771. doi: 10.3168/jds.2022-22451
von Stebut E., Mets M., Milon G., Knop J., Maurer M. (2003). Early macrophage influx to sites of cutaneous granuloma formation is dependent on MIP-1 / released from neutrophils recruited by mast cell-derived TNF. Blood. 101, 210–215. doi: 10.1182/blood-2002-03-0921
Wang P., Xu Y., Liu M., Li H., Wang H., Liu Y., et al. (2023). Risk factors and early markers for echovirus type 11 associated hemorrhage-hepatitis syndrome in neonates, a retrospective cohort study. Front. Pediatr. 11. doi: 10.3389/fped.2023.1063558
Williams E. J., Fischer D. P., Pfeiffer D. U., England G. C. W., Noakes D. E., Dobson H., et al. (2004). Clinical evaluation of postpartum vaginal mucus reflects uterine bacterial infection and the immune response in cattle. Theriogenology. 63, 102–117. doi: 10.1016/j.theriogenology.2004.03.017
Wira C. R., Grant-Tschudy K. S., Crane-Godreau M. A. (2005). Epithelial cells in the female reproductive tract: a central role as sentinels of immune protection. Am. J. Reprod. Immunol. 53, 65–76. doi: 10.1111/j.1600-0897.2004.00248.x
Yockey L. J., Iwasaki A. (2019). Role of interferons and cytokines in pregnancy and fetal development. J. Immunol. 49, 397–412. doi: 10.1016/j.immuni.2018.07.017
Zhang J.-M., An J. (2007). Cytokines, inflammation, and pain. In Anesthesiol Clin. 45, 27–37. doi: 10.1097/AIA.0b013e318034194e
Keywords: beef, Bos indicus, cytokines, fertility, microbiome, on-farm
Citation: Smith MS, Soffa DR, McAnally BE, Hickman-Brown KJ, Stockland EL and Poole RK (2024) On-farm study: cytokine profiles and vaginal microbiome of Bos indicus cattle before artificial insemination. Front. Anim. Sci. 5:1399337. doi: 10.3389/fanim.2024.1399337
Received: 11 March 2024; Accepted: 20 May 2024;
Published: 05 June 2024.
Edited by:
Mohan Mondal, ICAR-National Dairy Research Institute, Eastern Regional Station, Kalyani, IndiaReviewed by:
Kyle McLean, The University of Tennessee, Knoxville, United StatesCopyright © 2024 Smith, Soffa, McAnally, Hickman-Brown, Stockland and Poole. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rebecca K. Poole, cmtwb29sZUB0YW11LmVkdQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.