- 1Department of Agricultural Research for Northern Sweden, Swedish University of Agricultural Sciences, Umeå, Sweden
- 2Department of Animal Nutrition and Management, Swedish University of Agricultural Sciences, Uppsala, Sweden
- 3Department of Animal Nutrition and Management, Swedish University of Agricultural Sciences, Umeå, Sweden
- 4Department of Agricultural Sciences, University of Helsinki, Helsinki, Finland
- 5Department of Molecular Sciences, Swedish University of Agricultural Sciences, Uppsala, Sweden
The objective was to determine the effects on rumen microbiome and milk quality of reducing the methane (CH4) produced from enteric fermentation by the addition of Asparagopsis taxiformis (AT) to the diets of dairy cows. Six Nordic Red cows at 122 ± 13.7 (mean ± SD) days in milk, of parity 2.7 ± 0.52 and producing 36 kg ± 2.5 kg milk per day at the start of the trial were divided into three blocks by milk yield and assigned to an extra-period Latin-square change-over design comprising two dietary treatments. An extra period of observation was added to the Latin-square change-over design to control for carry-over effects. The dietary treatments were a diet consisting of grass silage and a commercial concentrate mixture (60:40) either not supplemented or supplemented with 0.5% AT on an organic matter intake basis. On average, daily CH4 production, CH4 yield, and CH4 intensity decreased by 60%, 54%, and 58%, respectively, in cows fed the diet supplemented with AT. Furthermore, hydrogen gas emitted by cows fed diets supplemented with AT increased by more than five times compared with cows fed a non-AT-supplemented diet. Feed intake was decreased and milk production altered, reflecting a decreased yield of milk fat in cows fed an AT-supplemented diet, but feed efficiency increased. Rumen fermentation parameters were changed to promote propionate rather than acetate and butyrate fermentation. The most prominent change in milk quality was an increase in bromine and iodine when the diet was supplemented with AT. The reduction in CH4 was associated with a shift from Methanobrevibacter to Methanomethylophilaceae in the archaeal population and a lower relative abundance of Prevotella in the bacterial population. Changes in milk fat odd-numbered and branched-chain fatty acids in the current study of AT supplementation support observed differences in ruminal archaeal and bacterial populations.
Introduction
The climate commitments made in the Paris Agreement aim to protect planet habitability by limiting global warming to 1.5°C above the pre-industrial baseline (European Commission, 2022). The environmental goals of carbon neutrality are directly related to the increased concentrations of greenhouse gases (GHGs) in the atmosphere (IPCC, 2021). Food systems contribute up to 30% of global GHG emissions (Clark et al., 2020). Emissions of methane (CH4) from ruminant livestock (via enteric fermentation and anaerobic manure storage) account for 33% of all anthropogenic CH4 sources and represent the second greatest anthropogenic CH4 source after fossil fuels (Jackson et al., 2020; Crippa et al., 2021). Methane in the atmosphere produces around 28 times more heat on a 100-year time scale than carbon dioxide (CO2). However, when CH4 decomposes to CO2 in the atmosphere, it will be bound to vegetation in the near future and will not contribute to further warming. This implies that reducing CH4 emissions will have an immediate effect on the global temperature (Sterner and Johansson, 2017). Demands on food production are increasing, and the global cattle population is predicted to increase as agricultural land in the world becomes dominated by pastures and permanent meadows used for livestock (FAO, 2019).
Short-term and efficient reductions in enteric emissions of CH4 from ruminants primarily involve feed manipulation mitigation strategies. Only a few of these feed manipulation strategies have been shown to be efficient and applicable in real farming situations. The inhibitor 3-nitroxypropanol has demonstrated CH4 reductions of only approximately 30%, when given in very low doses to dairy cows (Hristov et al., 2015). Recently, the application of macroalgae as antimethanogenic feed additives has been thoroughly reviewed by Abbott et al. (2020). Despite green and brown macroalgae potentially possessing CH4-mitigating effects, which can be attributed to several bioactive compounds (e.g., peptides, carbohydrates, lipids, and phlorotannins), the effects have been small in vitro (Krizsan et al., 2022) and are likely negligible in vivo. The tropical macroalga Asparagopsis taxiformis has recently shown the greatest potential CH4-reducing effect, amounting to an almost 100% reduction in CH4 production in vitro (Chagas et al., 2019), between a 55% and 65% reduction in vivo when added to the diet of dairy cows (Stefenoni et al., 2021), and at most a 98% reduction in vivo when added to the diet of steers (Roque et al., 2021), all at an inclusion level of 0.5% of organic matter (OM). A. taxiformis is a natural supplement, and the inhibitory effect is caused by the volatile compound bromoform in the last step of methanogenesis by ruminal archaea (Machado et al., 2016). Chagas et al. (2019) suggested a strong dose-dependent CH4-mitigating effect of A. taxiformis at the inclusion level of 0.5% of OM with the least impact on rumen fermentation parameters.
This study aimed to assess the effects on rumen microbiome and milk quality when dairy cow diets are supplemented with A. taxiformis at the inclusion level of 0.5% of OM to achieve an optimal antimethanogenic effect in dairy cows. Additionally, the study contributes to our understanding of the effects on dairy cow production performance and metabolism when applying A. taxiformis as a feed manipulation CH4 mitigation strategy.
Materials and methods
The feeding trial was conducted at Röbäcksdalen experimental farm of the Swedish University of Agricultural Sciences in Umeå (63°45′N, 20°17′E) from 8 October to 9 December 2020.
Cows, experimental design, and diets
Six Nordic Red dairy cows weighing (mean ± SD) 611 kg ± 62.1 kg, being 122 ± 13.7 days in milk, of parity 2.7 ± 0.52, and producing 36.3 kg ± 2.47 kg of milk per day pretrial, were blocked by milk yield (MY) and used in the experiment. The cows were kept in an insulated free-stall barn, offered a total mixed ration (TMR) ad libitum, mixed with a feed mixer (Nolan A/S, Viborg, Denmark), and delivered with a Mullerup M2000 Free Feeder automatic feeding wagon (GEA Farm Technologies Mullerup A/S, Ullerslev, Denmark) in individual feed troughs three times per day at 07:00, 13:00, and 20:00. The cows had free access to drinking water and were milked twice per day, at 06:00 and 16:30, throughout the experiment.
The cows were randomly allocated to a dietary treatment within block, i.e., square, and assigned to an extra-period Latin-square change-over design consisting of three equal squares (Lucas, 1957). The experimental periods lasted 21 days, with the last 7 days used for data recording and sampling. All cows were fed a control diet composed of 600 g/kg dry matter (DM) of grass silage, 390 g/kg DM of a commercial concentrate (Komplett Amin 180; Lantmännen Lantbruk AB, Malmö, Sweden), and 10 g/kg DM of mineral mix (Mixa Optimal; Lantmännen Lantbruk AB, Malmö, Sweden). Dietary treatments comprised either no supplementation (control) or supplementation with the macroalga Asparagopsis taxiformis (AT) at 0.5% of organic matter (OM) intake. The daily supplementation of AT was in line with an earlier dose–response in vitro experiment conducted by Chagas et al. (2019).
The grass silage was harvested as a first cut at Röbäcksdalen experimental farm on 13 June 2020 from a second year ley of timothy grass (Phleum pratense) and some red clover (Trifolium pratense) (seed ratio 80:20; botanical analysis not made). The grass was cut with a mower conditioner (GMT 3605 FlexP; JF-Stoll A/S, Sønderborg, Denmark), wilted overnight, and harvested with a precision chop forage wagon (ES 5000 MetaQ ProTec; JF-Stoll A/S, Sønderborg, Denmark). A formic acid-based additive (ProMyr XR 630; Perstorp Holding AB, Perstorp, Sweden) was applied at a rate of approximately 3.5 L/Mg and the forage material was ensiled in bunker silos until use in the experiment.
Asparagopsis taxiformis in the gametophyte life cycle stage was harvested at Angústias, Faial Island, Azores, Portugal (38°31′45″N, 28°37′09″W), between April and May 2020 by SeaExperts (Feteira, Ilha Do Faial, Portugal). Biomass was collected at a depth of 3 to 6 m by scuba divers and stored in a dark cooled container in a supporting boat before reaching land, where it was rinsed and immediately frozen and stored at –25°C. The frozen material was shipped to European Freeze Dry ApS (Kirke Hyllinge, Denmark) and the material was freeze-dried. The dried AT was vacuum stored in light-excluding bags and transported to the Swedish University of Agricultural Sciences in Umeå. During the experiment, the bags containing AT were stored in a dark basement storage holding room temperature, and bags were brought up and processed successively. The freeze-dried macroalga was ground using an electric hand mixer before being manually mixed with the TMR in the individual feedbunks (60% of the total daily AT dose at 07:00 and 40% of the total daily AT dose at 13:00). The delivery of the TMR was monitored and adjusted in the afternoon daily to avoid feed residues and achieve the intended provision of AT to each cow daily.
Data recording, sampling, and calculations
Individual feed intake was recorded daily by Roughage Intake Control feeders (Insentec B.V., Marknesse, The Netherlands) and daily MY was recorded using a gravimetric milk recorder (SAC, S.A. Christensen and Co Ltd, Kolding, Denmark). The body weight (BW) of the cows was measured after morning milking for 3 days before the experiment and for the last 3 days of each period.
Mass fluxes of CH4, carbon dioxide (CO2), and oxygen (O2) were recorded daily by the GreenFeed emission monitoring (GEM) system (GreenFeed; C-Lock., Rapid City, SD, USA) as described by Huhtanen et al. (2015). Gas calibrations (N2 and a mixture of CH4, O2, and CO2) were performed once a week and CO2 recovery tests were conducted every second week throughout the entire experiment. The average recovery (mean ± SE) was 100% ± 0.6%. The air filters of the GEM units were cleaned twice a week in order to maintain the airflow above 26 L/s. The same commercial concentrate (Komplett Amin 180; Lantmännen Lantbruk AB, Malmö, Sweden) as in the TMR was given to the cows in order to ensure regular visits by the cows to the GEM system. The program was set to allow each cow to visit at a minimum of 5-h intervals and to give eight drops of 50 g of concentrate during each visit. The interval between drops was set at 40 s.
The DM concentration was determined twice a week for the silages and once a week for the concentrates. All feed samples were oven-dried at 60°C for 48 h. The diets were adjusted twice weekly to account for changes in DM content. The dried feed samples were ground to pass through a 1-mm sieve in a cutter mill (SM 2000; Retsch Ltd., Haan, Germany) to provide samples for chemical analysis, or through a 2-mm sieve to provide samples for indigestible neutral detergent fiber (iNDF) analysis. Additionally, silages were sampled for 3 days in the last week of each period to obtain a composite sample for fermentation quality analysis. The silage samples were stored frozen at –20°C before and after grinding. Frozen silage samples were ground in a cutter mill (SM 2000) to pass through a 20-mm sieve.
Milk samples were collected during days 19–21. Pooled milk samples were made on day 19, weighted according to MY from morning and afternoon milking, for sensory testing and bromoform analysis. The samples for sensory testing were stored at 5°C in glass bottles that were provided by the dairy co-operative (Norrmejerier, Umeå, Sweden). The sensory analysis was conducted by the same two trained persons throughout the whole trial directly after the last milk sample collection. Sensory taste was compared with an equal test of the milk from cows in the experiment before the start of the experimental diets. The milk sample for bromoform analysis was stored frozen at –20°C until analysis. Samples for analysis of fat, protein and lactose, and urea composition of the milk were taken during four consecutive milking occasions in each period, and stored in plastic bottles with preservative (Bronopol) at 5°C until analysis. From the same four milking occasions a pooled sample of milk, weighted according to morning and afternoon MY, was made for each cow in each period for analysis of fatty acid composition, and stored at –20°C until analysis.
Rumen fluid was collected from all cows on day 19 in each period. The collection and preparation of rumen fluid was carried out in accordance with Chagas et al. (2021). In addition, 5-mL subsamples of filtered rumen fluid were transferred to Falcon tubes and preserved with 0.5 mL of sulfuric acid for ammonia determination and 1 mL of metaphosforic acid (25%) for volatile fatty acid (VFA) analysis. Samples were immediately frozen on dry ice and kept at –80°C in a freezer until analysis.
Fecal spot samples (approximately 250 g) from all cows were collected in connection with morning and afternoon milking on days 19–21 in each period. After the last collection of each period, the fecal samples were oven dried at 60°C for 48 h and milled to pass through a 1-mm sieve in a cutter mill. Fecal samples used for the analysis of iNDF were milled using mortar and pestle to pass through a 2.5-mm sieve. Fecal samples were pooled by cow within period.
The chemical composition of diets was calculated based on the intake, dietary ingredient composition determined from fresh weight proportions, and ingredient chemical composition. Total intake was calculated as TMR intake plus concentrate intake from the GEM system. The apparent digestibility of nutrients was calculated using iNDF as an internal marker in feeds and feces (Huhtanen et al., 1994). Potentially digestible neutral detergent fiber (pdNDF) was calculated as neutral detergent fiber (NDF) – iNDF. The metabolizable energy content was calculated based on coefficients from feed tables (LUKE, 2019). Milk constituent concentrations were calculated as a weighted mean of the combined morning and afternoon milk yields. Daily energy-corrected milk (ECM) yield was calculated in accordance with Sjaunja (1990). Feed efficiency was calculated as the daily yield of ECM/daily amount of dry matter intake (DMI), and milk nitrogen efficiency (MNE) as the ratio of nitrogen (N) milk yield in grams to N intake in kilograms.
Compositional analysis
The DM, ash, and NDF concentrations of feed and fecal samples, and fermentation quality of silages, were analyzed as described by Pang et al. (2021). Total N in the samples was analyzed with the Kjeldahl method (Nordic Committee on Food Analysis, 1976), degrading the sample in a 2520 Digestor adding Kjeltabs CK (Foss Analytical A/S Hilleröd, Denmark), and distilling and titrating using a Kjeltec 8400 Analyser unit equipped with an 8460 sampler unit (Foss Analytical A/S Hilleröd, Denmark). Crude protein (CP) concentration was calculated as N × 6.25. The concentration of indigestible NDF (iNDF) in feed ingredients and feces was analyzed as described by Krizsan et al. (2015).
The milk samples were analyzed for fat, protein, urea, and lactose concentration at the laboratory of Eurofins Milk Testing Sweden AB (Jönköping, Sweden) using infrared reflectance spectroscopy (CombiFoss 6000, Foss Electric, Hillerod, DK). Furthermore, milk samples were analyzed for fatty acid (FA) composition according to Lamminen et al. (2019). In brief, milk samples of 1 mL were extracted using a mixture of ammonia, ethanol, diethylether, and hexane. Lipid extracts were combined and evaporated to dryness at 40°C under N. Samples were dissolved in hexane and methyl acetate and methylated by methanolic sodium methoxide at laboratory room temperature for 5 min (Shingfield et al., 2003). Fatty acid methyl esters (FAMEs) were analyzed using a gas chromatograph (GC2010 Plus, Shimadzu, Kyoto, Japan) equipped with an autosampler (AOC-20s, Shimadzu), a split-splitless auto-injector (AOC-20i, Shimadzu), a flame ionization detector, and a 100-m fused silica capillary column (i.d. 0.25 mm) coated with a 0.2-μm film of cyanopropyl polysiloxane (CP-SIL 88, Agilent J&W, Santa Clara, CA, USA). Peaks were identified by comparison of retention times with FAME standards (GLC463, Nu-Chek Prep, Elysian, MN, USA; 10-2100-9, 10-2600-12, 10-2800-7, 21-1412-7, 21-1413-7, 21-1600-8, 21-1614-7, 21-1615–7, Larodan, Malmö, Sweden) and by cross-referring to published isomeric profiles of milk fat (Shingfield et al., 2003). The identification was further validated with mass spectrometry (GCMS-QP2010 Ultra, Shimadzu). Milk FA composition was expressed as a weight percentage of total FA using theoretical relative response factors (Wolff et al., 1995).
Microbial analysis
DNA isolation and 16S rRNA gene amplicon sequencing
Bacterial library preparation and sequencing were performed by Novogene with total DNA extracted using TIANamp Stool DNA Kit and primer pair 515F (GTGBCAGCMGCC GCGGTAA)/805R (GACTACHVGGGTATCTAATCC). For the archaeal library, total DNA was extracted with the FastDNA™ Spin kit for soil (MP Biomedicals) with an additional wash step with 500 μL of 5.5 M guanidine thiocyanate (Sigma-Aldrich, Missouri, USA) for humic acid removal (Singh, 2020). A two-step PCR protocol with the primer pair 516F (TGYCAGCCGCCGCGGTAAHACCVGC)/915R (GTGCTCCCCCGCCAATTCCT) was used as described by Müller et al. (2016). Archaeal amplicon sequencing was performed using the SNP&SEQ Platform (National Genomics Infrastructure, Uppsala, Sweden). Illumina MiSeq sequencing with v3 chemistry and paired end reads (2 × 300 bp) was used for both bacterial and archaeal amplicon libraries.
Sequence analysis
Analyses of 16S rRNA sequencing data was performed on the demultiplexed fastq reads. BBDuk in BBMap (Bushnell, 2014) was used for the adapter and primer removal and quality filtering. Amplicon sequence variants (ASVs), abundance, and taxonomy tables were prepared using package dada2 in R/RStudio (v3.4.0/2021.09.0+351). Taxonomic annotations were performed using the 16S rRNA database formatted for dada2 with Genome Taxonomy Database taxonomies (v202) (Alishum et al., 2021). Abundance, taxonomy tables, and sample metadata were merged and used for visualization and statistical analysis using packages phyloseq (v) (1.38.0) (McMurdie and Holmes, 2013), vegan (v2.6-2) (Oksanen et al., 2019), and ggplot2 (v3.3.6) (Wickham, 2016). Raw reads have been deposited in SRA at NCBI under accession number PRJNA911204 for archaea and PRJNA911215 for bacteria.
Statistical analysis
Experimental data were subjected to analysis of variance using the generalized linear model (GLM) procedure in SAS (SAS Inc. 2002–2003, Release 9.4 SAS Inst., Inc., Cary, NC, USA) by applying the following model:
where Yijkl is the dependent variable, µ is the mean of all observations, Pj is the fixed effect of period i, Cj(S)k is the fixed effect of cow j within square k, Tl is the fixed effect of experimental dietary treatment l, and ϵijkl ~ N(0, ) is the random residual error.
Potential carry-over effects on all traits were evaluated by including a residual effect in the model described above. There were no significant (p ≥ 0.22) residual effects for any production traits evaluated, and the effect was not included in the final analysis. The residual effect was only significant (P = 0.05) for cis-11-octadecenoic acid (cis-vaccenic acid) among all milk fatty acids analyzed.
Results
Ingredient and diet composition
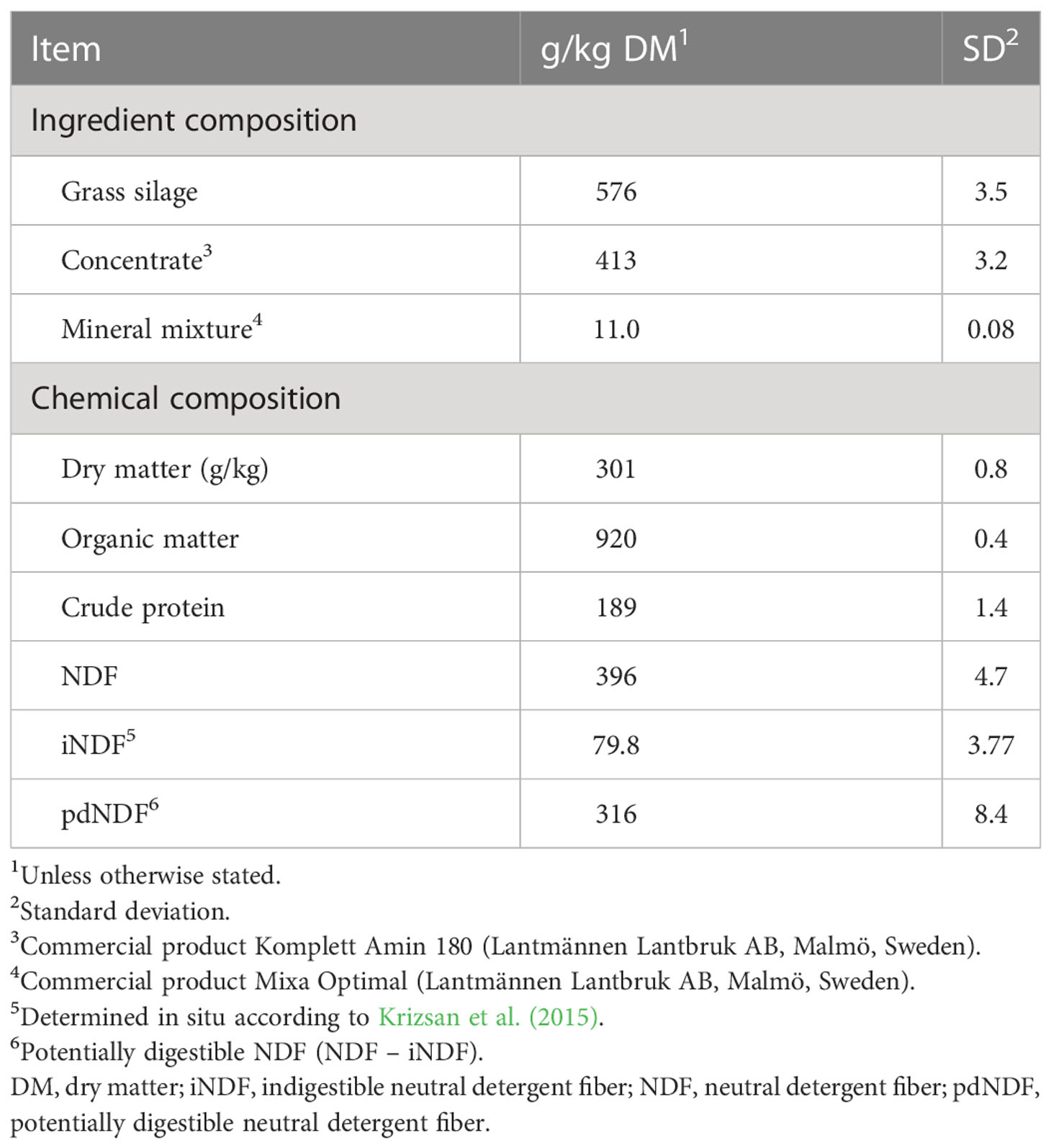
The grass silage displayed a low DM content and high lactic acid concentration, but was well preserved, with low pH and low levels of ammonia-N and butyric acid (Table 1). The concentrations of metabolizable energy and indigestible NDF indicated a relatively early harvest, and grass silage CP was high. The grass silage was fed with a commercial concentrate product, which resulted in a diet composition reflecting the high CP of both feed ingredients (Table 2).
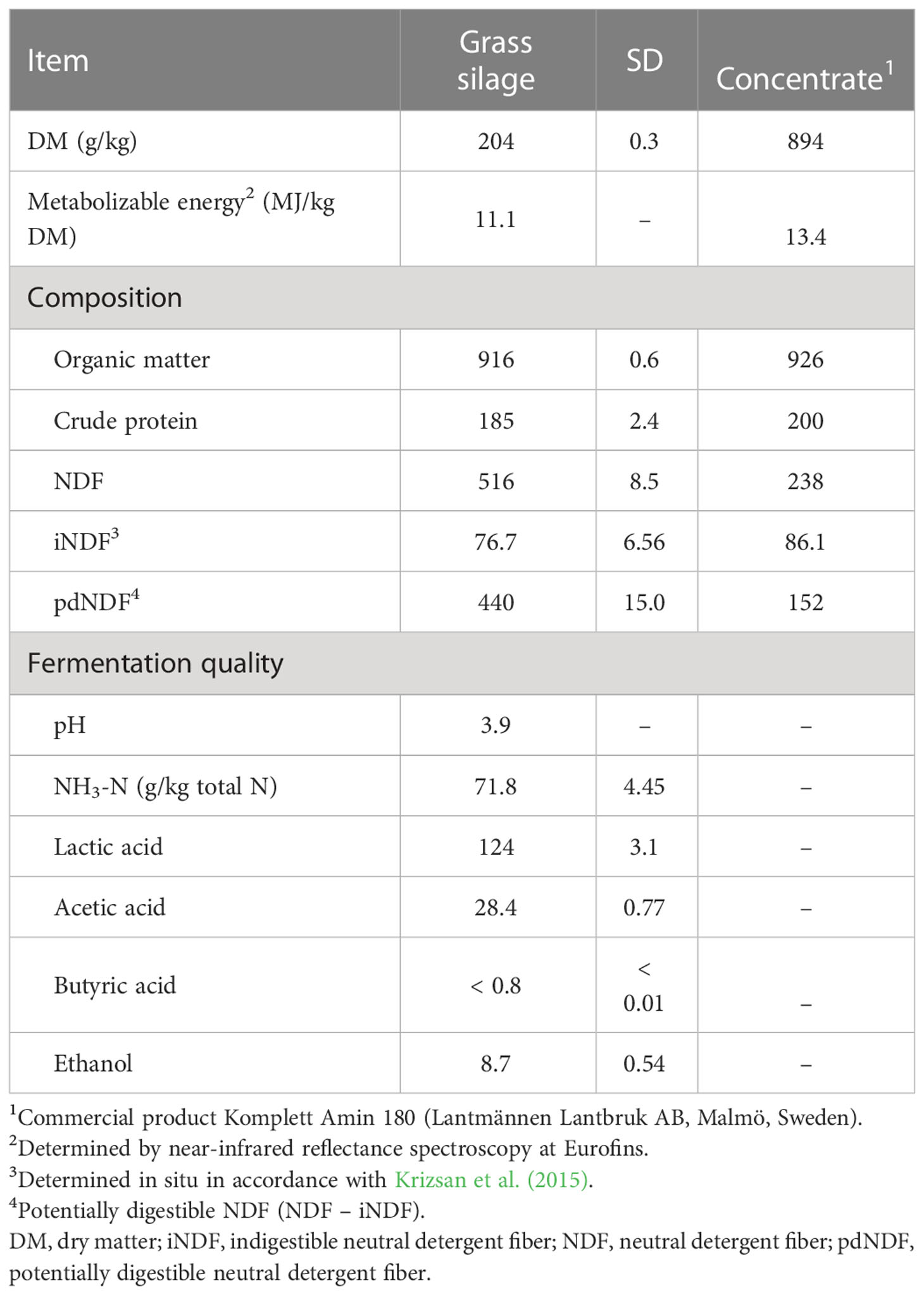
Table 1 Dietary ingredient composition and grass silage fermentation quality [g/kg dry matter (DM) unless otherwise stated].
Intake, milk production, and digestibility
Total intake of DM and silage decreased (p< 0.01) by 2.8 and 1.8 kg per day during the experiment for the cows that were supplemented with AT (Table 3). Yield of ECM decreased (P = 0.05) by 1.9 kg per day as a result of decreased (p = 0.03) fat yield for cows fed AT. However, the most prominent change in quality composition was the far greater (p< 0.01) concentrations of bromine (more than an eightfold increase) and iodine (more than a 15-fold increase) in the milk of the cows that were fed antimethanogenic diets with AT. Generally, feed efficiency increased (p< 0.01) and cows were smaller (p = 0.02) when their diet was supplemented with AT. The sensory panel could not detect any odd flavors or changes in sensory quality from pre-trial milk samples in the milk samples from cows fed diets with AT (results not presented). Feeding AT had no effect on the total concentration of saturated fatty acids (SFAs) or monounsaturated fatty acids in milk fat, averaging 74 and 22 g per 100 g of fatty acids, respectively (Supplementary Table S1). However, the proportion of several short- and medium-chain SFAs of 6–12 carbon atoms in milk fat was increased by AT supplementation (p ≤ 0.01). Feeding AT also affected the concentration of several odd-numbered and branched-chain FAs of 5–15 carbon atoms (p< 0.05). Treatment had no effect on milk fat cis-9 18:1, 18:2n-6, or 18:3n-3 concentrations, but AT enriched trans-11 18:1, cis-9,trans-11 18:2, and trans-11, cis-15 18:2 in milk fat at the expense of 18:0 (p = 0.01).
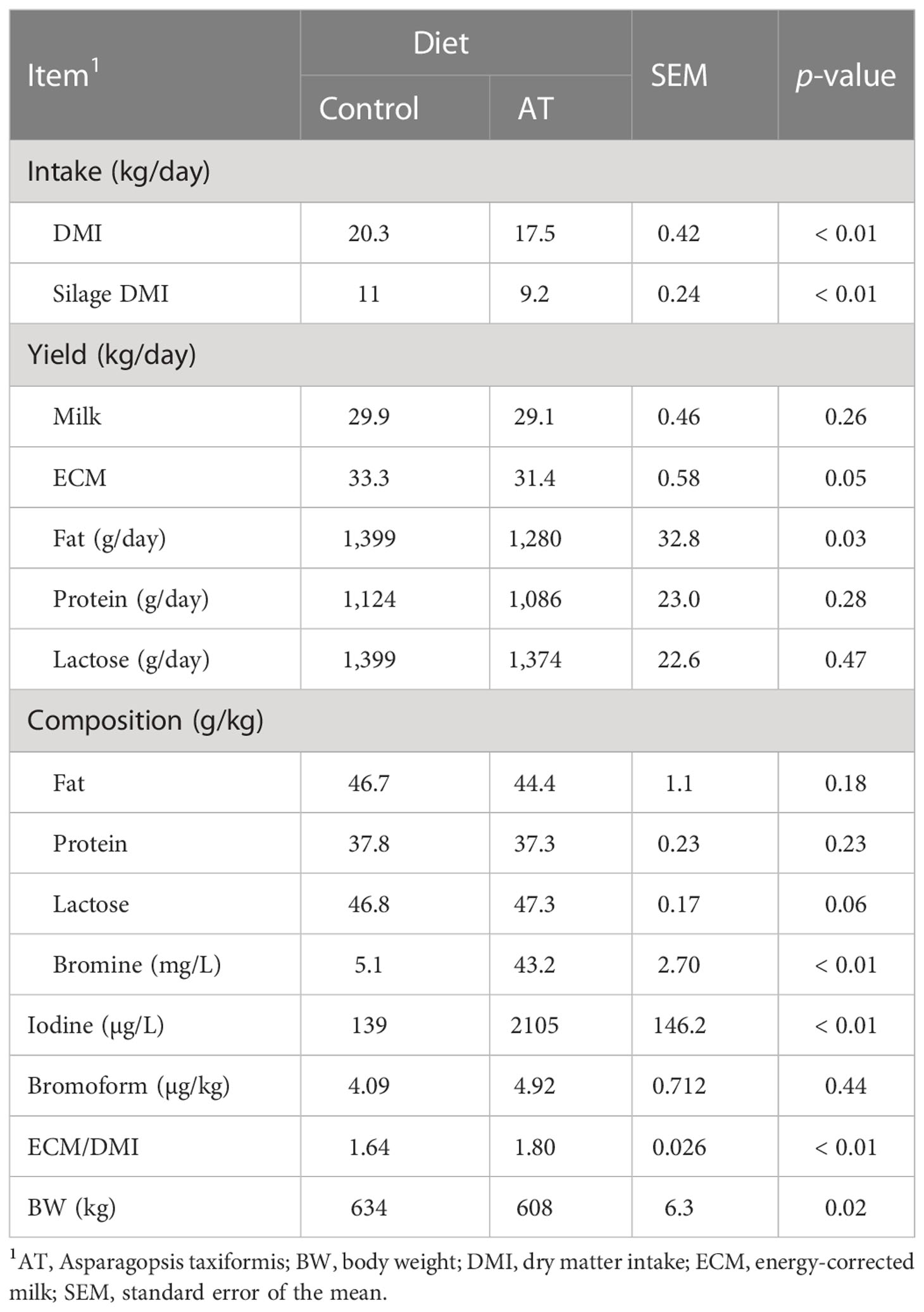
Table 3 Intake and production data for cows fed a grass silage-based diet without (control) or with supplementation with Asparagopsis taxiformis (AT).
Daily intake of OM and NDF was 2.6 kg and 1.2 kg lower, respectively (p< 0.01), when cows were supplemented AT, but no measures of total tract diet digestibility were affected by the antimethanogenic feed supplementation (Table 4). There were no differences (p = 0.19) in fecal bromoform concentration between the cows fed diets supplemented with AT and those fed non-supplemented diets (results not presented).
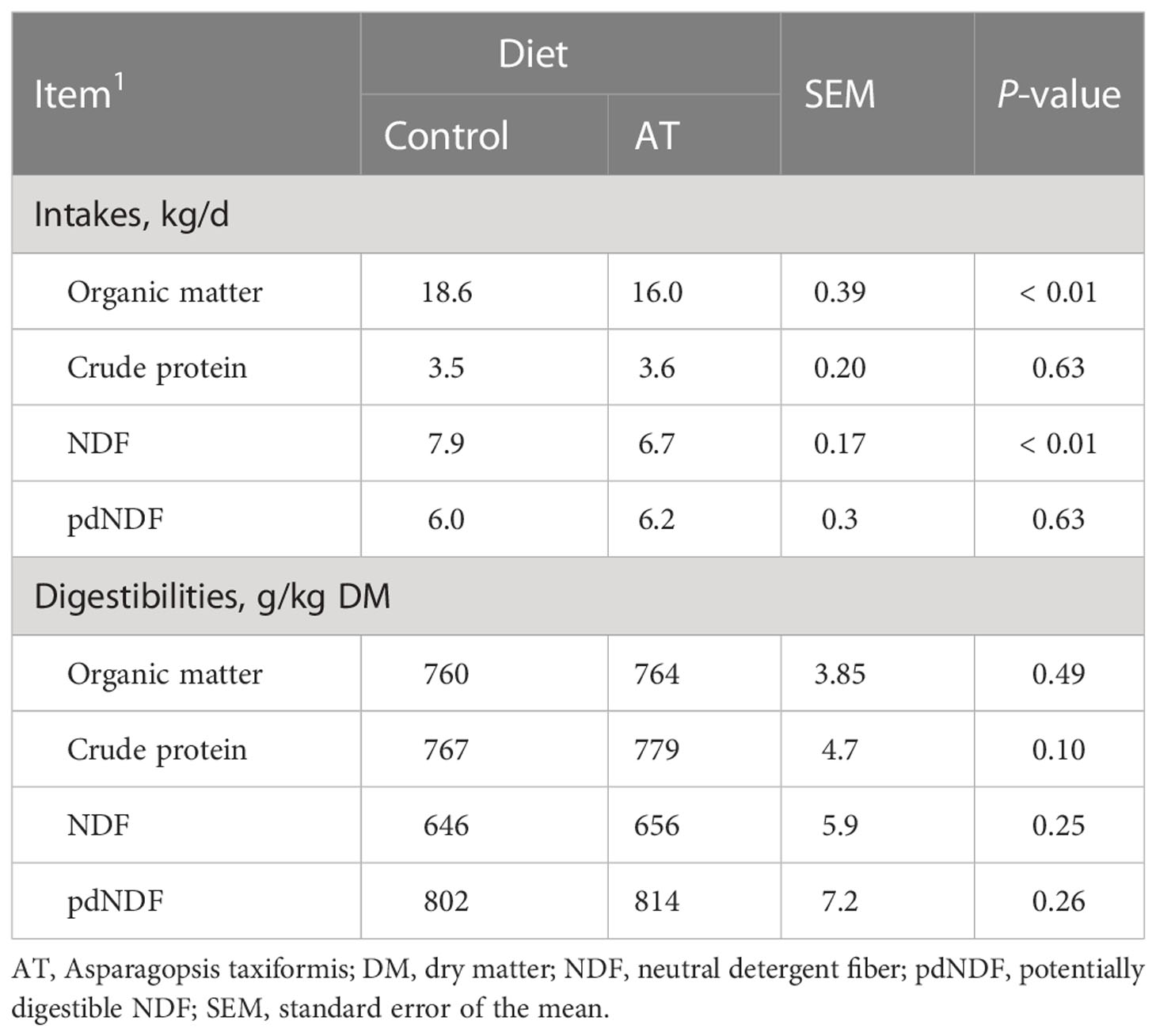
Table 4 Intakes and digestibilities of dietary chemical components for cows fed a grass silage-based diet without (control) or with supplementation with Asparagopsis taxiformis (AT).
Gases emissions and rumen fermentation
Feeding the cows the antimethanogenic AT supplementation decreased (p ≤ 0.04) total emissions of CH4 and CO2, CH4 yield (g CH4/kg DM), CH4 intensity (g CH4/kg ECM), and (Table 5) CH4/CO2 ratio, but increased (p< 0.01) CO2 yield and H2 production.
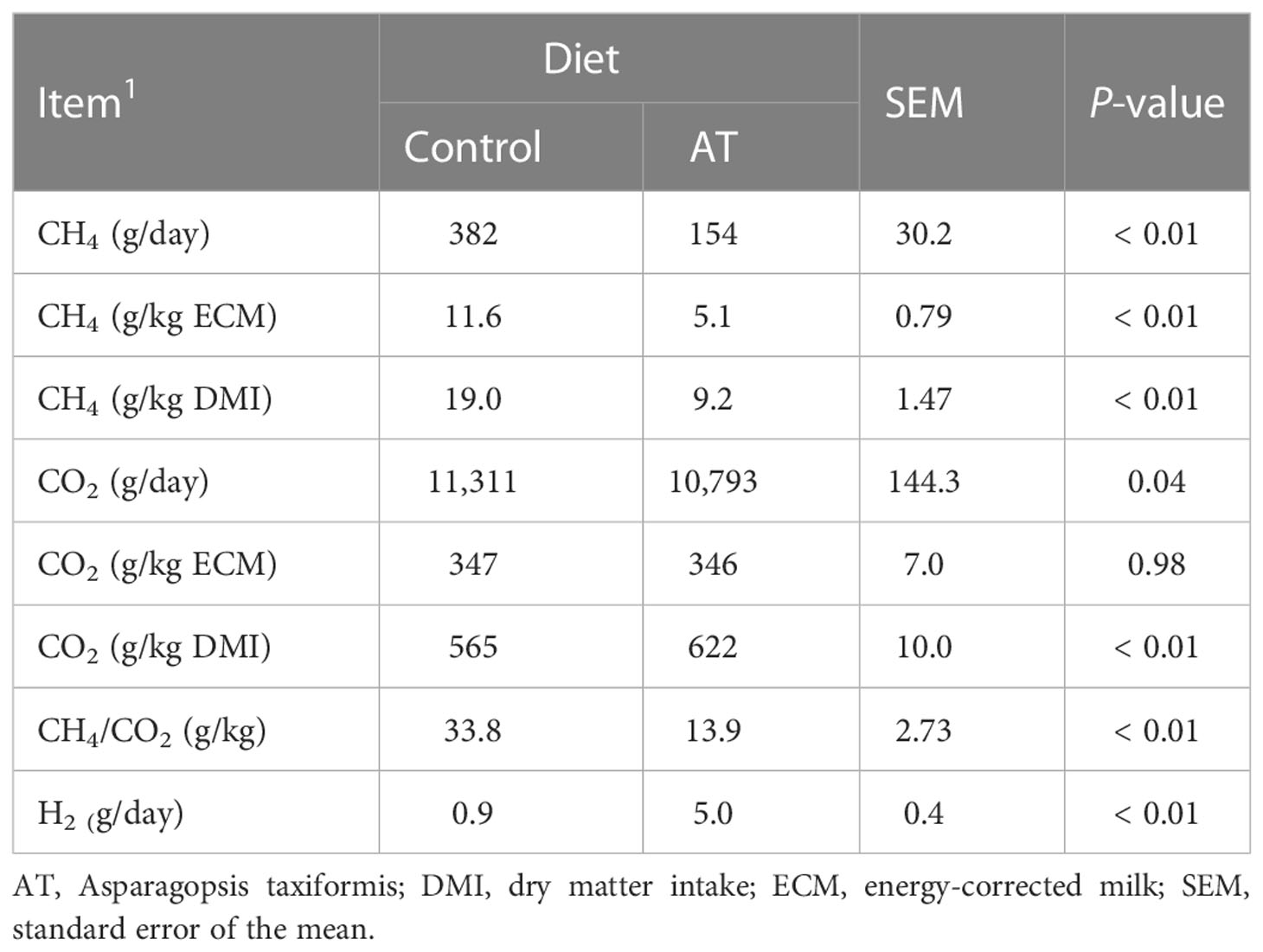
Table 5 Methane (CH4), carbon dioxide (CO2), and hydrogen (H2) emissions for cows fed a grass silage-based diet without (control) or with supplementation with Asparagopsis taxiformis (AT).
(Table 6) Total VFA decreased (p = 0.03) when AT was included in the diets. Molar proportions of individual VFAs were altered (p ≤ 0.04) toward more propionate and valerate, and less acetate and butyrate for cows fed AT-supplemented diets.
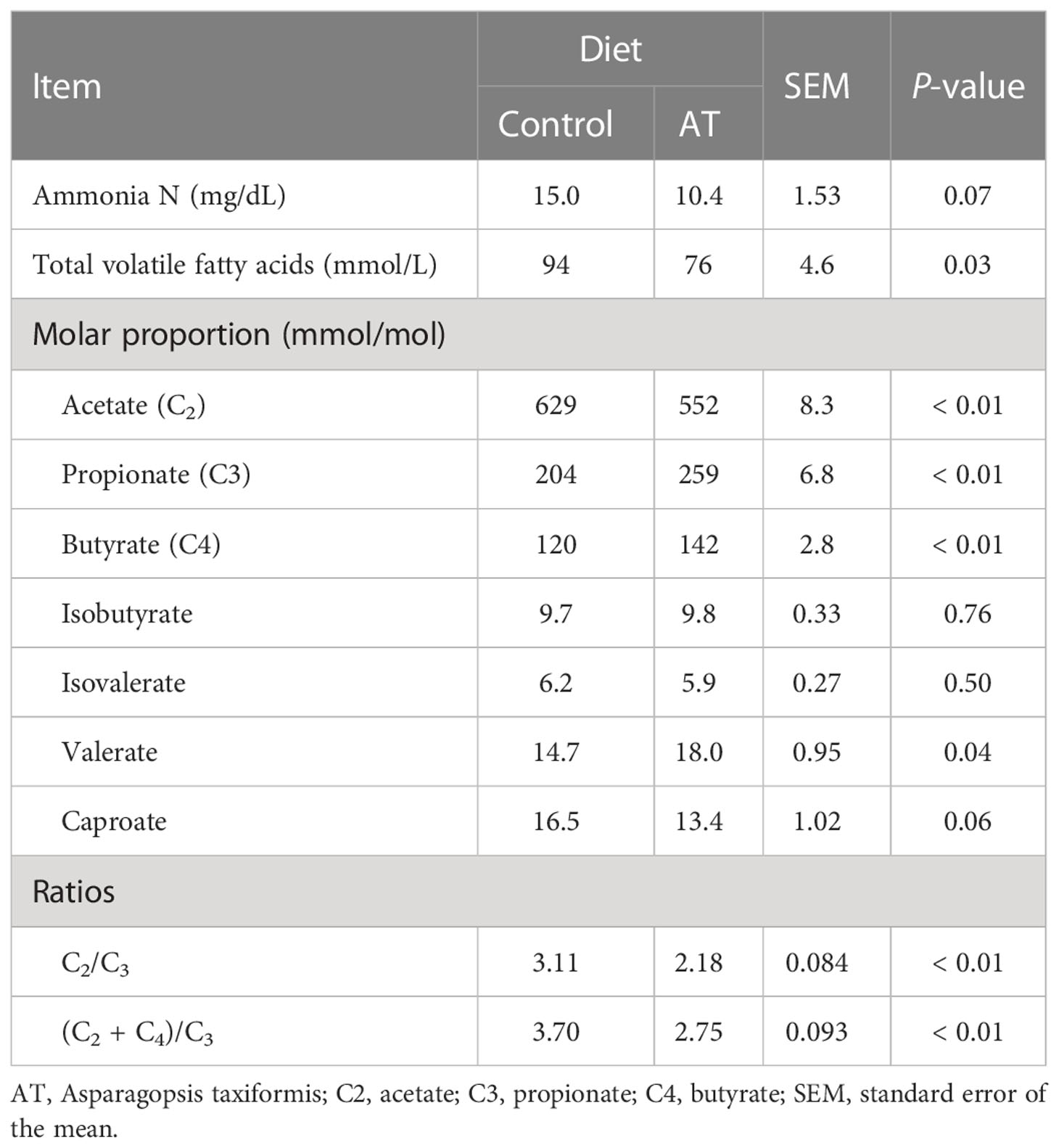
Table 6 Ruminal fermentation for cows fed a grass silage-based diet without (control) or with supplementation with Asparagopsis taxiformis (AT) .
All rumen fluid samples were all below detection level (0.09 ug bromoform/g sample; results not presented).
Microbial data
The structure of the rumen archaeal and bacterial community in the dairy cows was characterised by sequencing the V4 region of archaeal and bacterial 16S rRNA gene, respectively. After quality control of raw data and filtering of untargeted taxa (“NA” at kingdom level, bacterial reads from archaeal data and vice versa) a total of 362,798 and 1,896,528 readsrepresented the 1,349 and 8,139 ASVs of archaeal and bacterial community, respectively.
Archaea
The archaeal community was represented by two different phyla, Thermoplasmatota and Methanobacteriota, where Thermoplasmatota and Methanobacteriota represented on average 53% ± 32% (mean ± SE) and 44% ± 33% in relative abundance, respectively. Orders were represented by Methanomassiliicoccales (53% ± 33%) and Methanobacteriales (44% ± 33%). Methanobacteriota was represented only by the genus Methanobrevibacter and Thermoplasmatota mainly by unclassified Methanomethylophilaceae with genus identity UBA71 (43% ± 29%) and MX-02 (6.0% ± 10%). Analysis using the GLM procedure to evaluate any differences between archaeal sequences was performed and revealed no significant differences at phyla or genus level. The AT-treated group had a numerically lower relative abundance of Methanobacteriota (29.9%) than the control group (58.8%) with a tendency to significance [p = 0.05, standard error of the mean (SEM) = 8.80] (Figure 1). Thermoplasmatota was numerically higher in the AT group (66.2%) than in the control group (40.3%), with a tendency to significance (p = 0.08, SEM = 9.10). At genus level, Methanobrevibacter was numerically higher in the control group (p = 0.05, SEM = 8.87), and unclassified UBA 71 belonging to the family Methanomethylophilaceae had a tendency of difference in higher relative abundance for AT treatment (p = 0.09). Differences at species level between treatments were observed only in species UBA71_sp900767505_RS_GCF_9007675 (Table 7), which was mainly present in the AT group, and there was a tendency (p = 0.06) for the relative abundance of the species Methanobrevibacter_A_sp001548675 to be higher in the control group.
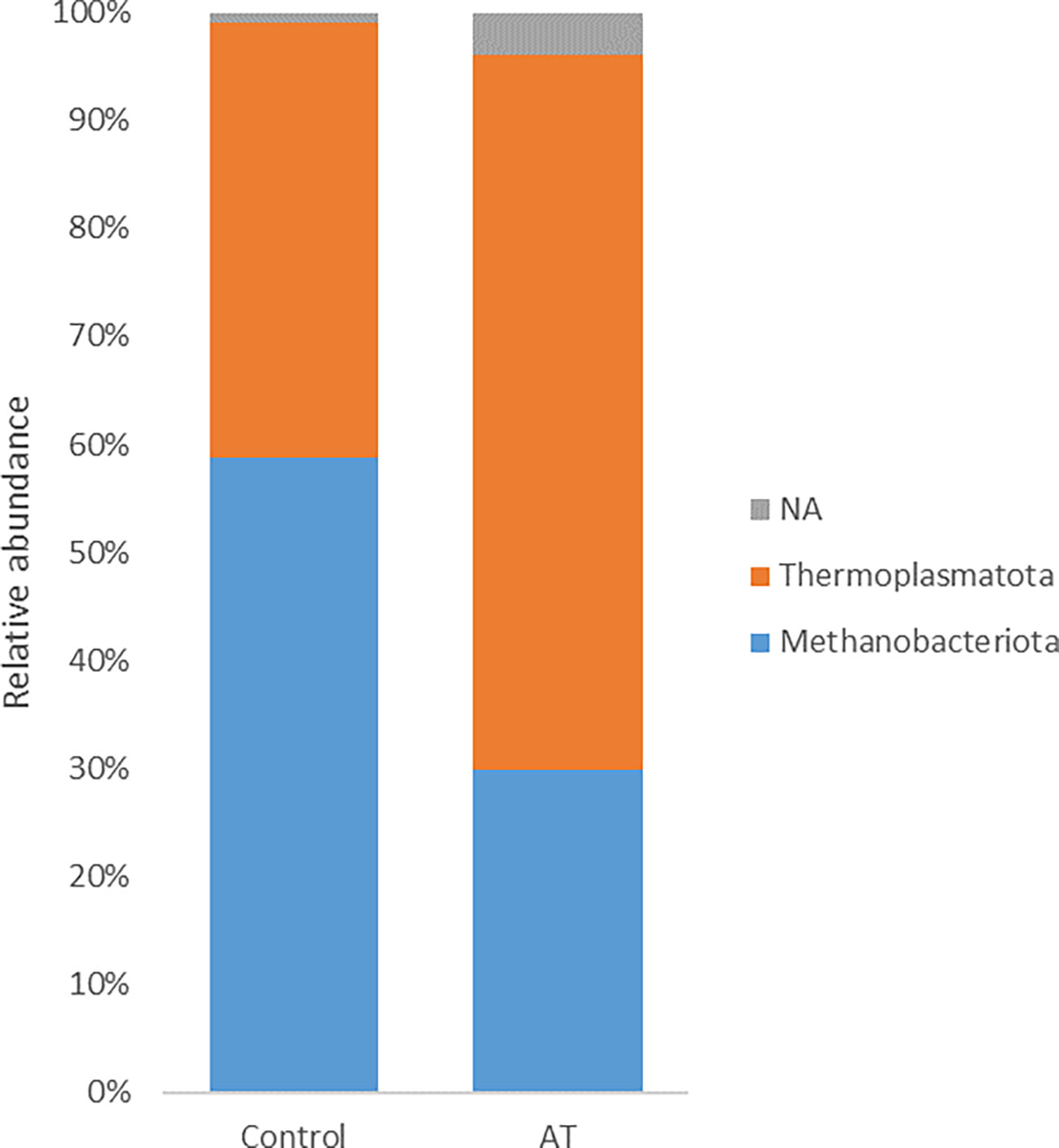
Figure 1 Least-squares means of relative abundance of archaea at phylum level for cows with a diet without (control) or with supplementation with Asparagopsis taxiformis (AT). NA, not applicable.
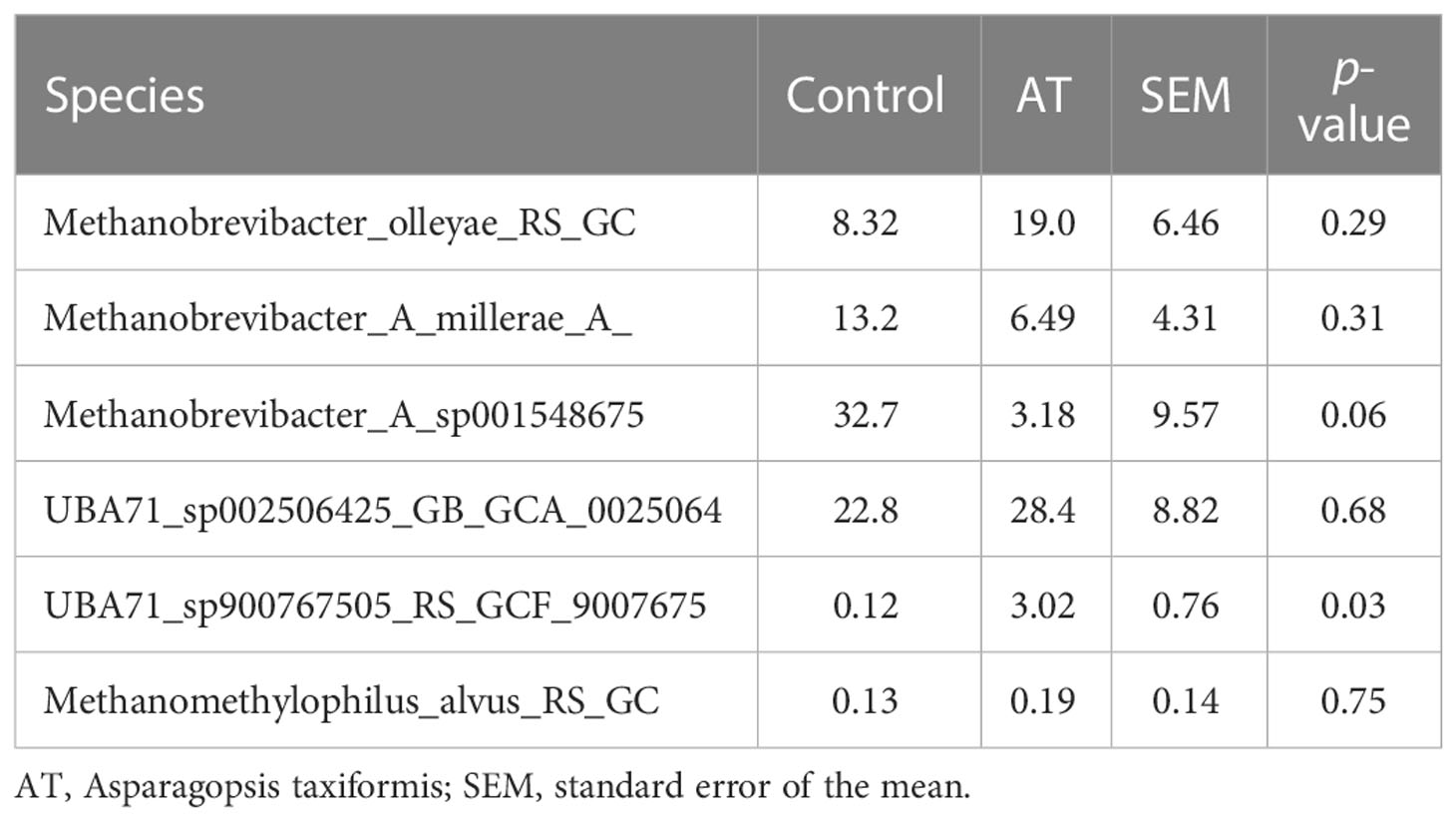
Table 7 Mean abundance of archaeal species for cows with a diet without (control) or with supplementation with Asparagopsis taxiformis (AT).
Bacteria
The bacterial population was represented by 44 different phyla, with 17 phyla found across all samples. The bacterial composition was dominated by the Firmicutes phylum, representing on average 50.5% ± 10.0% of all sequences, followed by Bacteroidota (38.8% ± 12.0%), Proteobacteria (4.1% ± 2.2%), Actinobacteriota (1.7% ± 1.3%), and Verrucomicrobiota (1.7% ± 1.4%). The remaining phyla represented less than 1% of all sequences. At genus level, Prevotella represented on average 34.3% ± 13.8% of all sequences. Other abundant genera were unidentified Clostridia (CAG-492) (4.4% ± 2.2%), Saccharofermentans (3.3% ± 2.0%), Prevotellamassilia (3.3% ± 0.2%) and Ruminococcus_E (2.4% ± 0.9%).
The 10 most abundant phyla accounted for > 98% of the sequences generated from the bacterial communities of both the control and AT-supplemented groups. Analysis using the GLM procedure revealed statistically significant differences in the relative abundance of Bacteroidota (p = 0.04, SEM = 3.35), with a higher abundance in the control treatment group than in the AT treatment group, with abundances of 45.6% and 33.9%, respectively (Figure 2). Firmicutes had a tendency to difference (p = 0.07, SEM = 6.65), with a numerically higher abundance in the AT group than in the control group, with abundances of 54.5% and 46.5% respectively. Proteobacteria was more abundant in the AT group (5.1%) than in the control group (3.1%) (p = 0.04, SEM = 0.58).
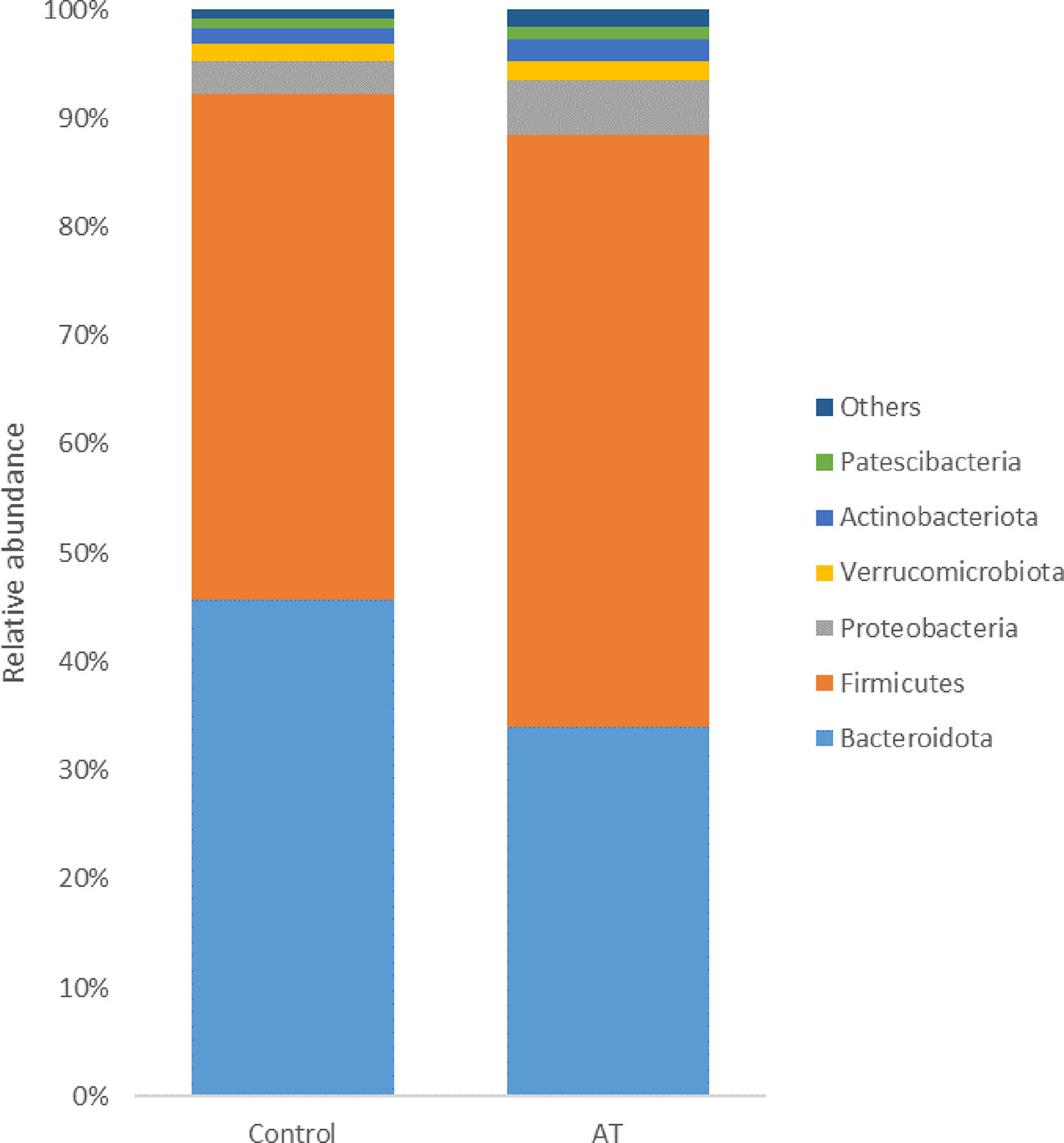
Figure 2 Least-squares means of relative abundance of bacteria at phylum level for cows with a diet without (control) or with supplementation with Asparagopsis taxiformis (AT). Phyla with a lower abundance than 1% are summarized as “others”.
Discussion
In line with the results of this study, several in vivo experiments have confirmed the considerable potential of AT as an antimethanogenic feed supplement across several ruminant species (Li et al., 2016; Muizelaar et al., 2021; Roque et al., 2021; Stefenoni et al., 2021). However, studies investigating the effect of feeding AT to cows on the rumen microbial community are rare. It is known that the most abundant antimethanogenic compound found in AT is bromoform (CHBr3). A common feature that is specific to all methanogens is the use of methyl coenzyme M-reductase (Mcr) (Luton et al., 2002). In the last step in methanogenesis, the methyl group in methyl coenzyme M is reduced to CH4 by Mcr and coenzyme M is regenerated. Bromoform acts as an inhibitor of coenzyme-M, and so methanogenesis is inhibited (Machado et al., 2016). As previously mentioned, the inhibition of methanogenesis is known, but the effect on the microbial community is not well understood and rarely studied, especially in in vivo studies. In this study, we did not find any significant differences in the abundance of archaeal communities between cows fed with or without AT. At the phylum level, it was shown that Thermoplasmota was numerically more abundant in the AT treatment group and Methanobacteriota were numerically more abundant in the control group. Methanobacteriota was dominated by the genus Methanobrevibacter, and Thermoplasmatota was mainly represented by genus UBA71, which belongs to the family Methanomethylophilaceae. In most studies on rumen fluid from dairy cows, Methanobrevibacter, mainly using H2 as a substrate to reduce CO2 to CH4, is the dominant genus in dairy cows (Henderson et al., 2015). Little is currently known about the Methanomethylophilaceae family, but some members within the family use methyl groups for CH4 production, and these methylotrophic members have been associated with reduced CH4 emissions from the rumen (Poulsen et al., 2013), which is in line with the findings in this study. 16S rRNA data offer limited mechanistic insight, and for future studies to clarify the mechanisms within the methanogenic community shotgun metagenomics or metatranscriptomics could be used.
In the bacterial community, Fimicutes and Bacteroidota (previously named as Bacteroidetes) were the most abundant phyla in our study, which is in line with most studies on the rumen bacterial community (Henderson et al., 2015; Williamson et al., 2022). However, there was a difference in the abundance of the phyla Bacteroidota, which had a higher abundance in cows not fed AT than in those receiving AT treatment. That difference was also found at genus level for Prevotella, which is the main genus within Bacteroidota, with a higher abundance in the control group than in the AT treatment group. These results differ from what was found in the in vitro study by O'Hara et al. (2022), where Prevotella abundance was increased in the AT treatment group in the intermediate phase of the incubation. Prevotella species are Gram-negative anaerobes and produce a variety of extracellular degradative enzymes, which degrade starch and hemicellulose in plant cell walls and also exhibit proteolytic activity, although this varies greatly between Prevotella species (Stevenson and Weimer, 2007). It has been speculated that Prevotella spp. may redirect excess H2 to propionate when CH4 is inhibited in the rumen (Aguilar-Marin et al., 2020). However, propionate levels were reduced by AT supplementation, which is in line with the inhibition of Prevotella in this study. Propionate and acetate are both fermentation products of Prevotella, which makes the relationship between enteric CH4 emission and Prevotella more complex to interpret. To obtain a holistic understanding of the effect on the microbial population, metagenomics or metatranscriptomics studies are required.
Stefenoni et al. (2021) reported decreased feed intake, milk yield, and yield of milk fat and protein when dairy cows’ diets were supplemented with AT at the same inclusion level as in this study. The induced changes in milk fat yield in this study could be explained by the altered rumen fermentation pattern, promoting lower proportions of lipogenic VFA. Milk fatty acid composition was similar to previous reports on grass silage-based diets (Lindmark Månsson, 2008). However, the enrichment of the biohydrogenation intermediates of 18-carbon unsaturates in milk fat at the expense of the final product, 18:0 (Shingfield et al., 2010), suggests that ruminal biohydrogenation of feed fatty acids was less complete in cows fed AT. Milk fat odd-numbered and branched-chain fatty acids are largely synthesized by rumen microbiota and reflect their relative abundance (Vlaeminck et al., 2006). Subtle but systematic changes in milk fat odd-numbered and branched-chain fatty acids in the current study of AT supplementation support observed differences in ruminal archaeal population.
The most prominent change in milk quality was the increase in bromine and iodine when the cows’ diets were supplemented with AT, in line with the results of Stefenoni et al. (2021). Stefenoni et al. (2021) observed an increase in bromine of equal magnitude in the milk of cows given AT supplementation and those not given AT supplementation, but a much lower change in milk iodine than in our study. However, in absolute values, Stefenoni et al. (2021) reported a concentration of milk bromine comparable to that in our study, but a far higher concentration of milk iodine. Excess consumption of iodine by humans can potentially cause thyroid dysfunction (Leung and Braverman, 2014), but harmful amounts would depend on total milk consumption, assuming that all retail milk was from AT supplemented cows (Antaya et al., 2015), and on age (Zimmermann et al., 2005). The recommended daily intake (RDA) of iodine differs according to age, but in general ranges from 90 to 150 μg/day, with additional intake recommended for those who are pregnant or lactating (Russell et al., 2001; EFSA, 2017). The upper limit for iodine consumption varies from two to four times higher than the RDA, i.e., between 200 and 600 μg per day (EFSA, 2017). From the results of our study, intake of a cup of milk (225 mL would provide approximately 470 μg of iodine per day, which would exceed the upper limit for children aged up to either 8 years (Russell et al., 2001) or 14 years (EFSA, 2017). For bromine, there are fewer recommendations available. However, in a study by Van Leeuwen et al. (1987), the provisional acceptable daily intake for bromine was 0.4 mg/kg body weight. This means that if milk contained 43.2 mg/L of bromine, as in our study, a child that weighs 10 kg should not exceed a daily intake of 10 mL of milk while an adult weighing 60 kg could consume 0.5 L of milk without exceeding the acceptable daily intake.
Data availability statement
The datasets presented in this study can be found in online repositories. The name of the repository and accession numbers can be found below: NCBI; PRJNA911204 and PRJNA911215.
Ethics statement
The use and handling of animals in this experiment was approved by the Swedish Ethics Committee on Animal Research (Dnr A 32–16), represented by the Court of Appeal for Northern Norrland in Umeå, and the experiment was carried out in accordance with laws and regulations governing experiments performed with live animals in Sweden.
Author contributions
SK: conceived and supervised the study and acquired funding. SK and JC: inputs to data analysis. All authors: methodology. JC, SK, and MR: conducted the experiment. All authors: wrote the full paper. All authors contributed to the article and approved the submitted version.
Funding
The study was part of the RumenPredict project within the FACCE ERA-GAS funding scheme as part of the EU Horizon 2020 Research and Innovation Program. This research was also supported by the Swedish Research Council for Environment, Agricultural Sciences, and Spatial Planning (FORMAS).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fanim.2023.1112969/full#supplementary-material
Supplementary Table 1 | Milk fatty acid (FA) composition for cows with a diet without (control) or with supplementation with Asparagopsis taxiformis (AT)
References
Abbott D. W., Aasen I. M., Beauchemin K. A., Gröndahl F., Gruninger R., Hayes M., et al. (2020). Seaweed and seaweed bioactives for mitigation of enteric methane: Challenges and opportunities. Animals 10, 2432. doi: 10.3390/ani10122432
Aguilar-Marin S. B., Betancur-Murillo C. L., Isaza G. A., Mesa H., Jovel J. (2020). Lower methane emissions were associated with higher abundance of ruminal prevotella in a cohort of Colombian buffalos. BMC Microbiol. 20, 1–13. doi: 10.1186/s12866-020-02037-6
Alishum A., Greenfield P., Seersholm F., Christophersen C. (2021). DADA2 formatted 16S RRNA gene sequences for both bacteria & archaea. Zenodo. doi: 10.5281/zenodo.4735821
Antaya N. T., Soder K. J., Kraft J., Whitehouse N. L., Guindon N. E., Erickson P. S., et al. (2015). Incremental amounts of ascophyllum nodosum meal do not improve animal performance but do increase milk iodine output in early lactation dairy cows fed high-forage diets. J. Dairy Sci. 98, 1991–2004. doi: 10.3168/jds.2014-8851
Chagas J. C. C., Ramin M., Exposito R. G., Smidt H., Krizsan S. J. (2021). Effect of a low-methane diet on performance and microbiome in lactating dairy cows accounting for individual pre-trail methane emissions. Animals 11, 2597. doi: 10.3390/ani11092597
Chagas J. C. C., Ramin M., Krizsan S. J. (2019). In vitro evaluation of different dietary methane mitigation strategies. Animals 9, 1120. doi: 10.3390/ani9121120
Clark M. A., Domingo N. G., Colgan K., Thakrar S. K., Tilman D., Lynch J., et al. (2020). Global food system emissions could preclude achieving the 1.5 and 2 c climate change targets. Science 370, 705–708. doi: 10.1126/science.aba7357
Crippa M., Solazzo E., Guizzardi D., Monforti-Ferrario F., Tubiello F. N., Leip A. (2021). Food systems are responsible for a third of global anthropogenic GHG emissions. Nat. Food 2, 198–209. doi: 10.1038/s43016-021-00225-9
EFSA (European Food Safety Authority) (2017). Dietary Reference Valuesfor nutrients. Summary Report. EFSA supporting publication 2017:e15121.98pp. doi: 10.2903/sp.efsa.2017.e15121
European Commission (2022). Available at: https://ec.europa.eu/clima/eu-action/climate-strategies-targets/2050-long-term-strategy_sv. (Accessed September 12, 2022).
FAO (2019). The State of Food and Agriculture 2019. Moving forward on food loss and waste reduction. Rome. Licence: CC BY-NC-SA 3.0 IGO.
Henderson G., Cox F., Ganesh S., Jonker A., Young W., Collaborators, G.R.C, et al. (2015). Rumen microbial community composition varies with diet and host, but a core microbiome is found across a wide geographical range. Sci. Rep. 5, 1–15. doi: 10.1038/srep14567
Hristov A. N., Oh J., Giallongo F., Frederick T. W., Harper M. T., Weeks H. L., et al. (2015). An inhibitor persistently decreased enteric methane emission from dairy cows with no negative effect on milk production. Proc. Natl. Acad. Sci. 112, 10663–10668. doi: 10.1073/pnas.1504124112
Huhtanen P., Cabezas-Garcia E. H., Utsumi S., Zimmerman S. (2015). Comparison of methods to determine methane emissions from dairy cows in farm conditions. J. dairy Sci. 98 (5), 3394–3409. doi: 10.3168/jds.2014-9118
Huhtanen P., Kaustell K., Jaakkola S. (1994). The use of internal markers to predict total digestibility and duodenal flow of nutrients in cattle given six different diets. Anim. Feed Sci. Technol. 48, 211–227. doi: 10.1016/0377-8401(94)90173-2
IPCC (2021). Climate Change 2021: The Physical Science Basis. Contribution of Working Group I to the Sixth Assessment Report of the Intergovernmental Panel on Climate Change [Masson-Delmotte V., Zhai P., Pirani A., Connors S. L., Péan C., Berger S., et al. (eds.)]. Cambridge University Press, Cambridge, United Kingdom and New York, NY, USA, 2391 pp. doi: 10.1017/9781009157896
Jackson R. B., Saunois M., Bousquet P., Canadell J. G., Poulter B., Stavert A. R., et al. (2020). Increasing anthropogenic methane emissions arise equally from agricultural and fossil fuel sources. Environ. Res. Lett. 15, 071002. doi: 10.1088/1748-9326/ab9ed2
Krizsan S. J., Rinne M., Nyholm L., Huhtanen P. (2015). New recommendations for the ruminal in situ determination of indigestible neutral detergent fibre. Anim. Feed Sci.Technol. 205, 31–41. doi: 10.1016/j.anifeedsci.2015.04.008
Krizsan S. J., Hayes M., Gröndahl F., Ramin M., O´Hara P., Owen K. (2022). Characterization and in vitro assessment of seaweed bioactives with potential to reduce methane production. Front. Anim. Sci. Sec. Animal Nutrition. doi: 10.3389/fanim.2022.1062324
Lamminen M., Halmemies-Beauchet-Filleau A., Kokkonen T., Jaakkola S., Vanhatalo A. (2019). Different microalgae species as a substitutive protein feed for soya bean meal in grass silage based dairy cow diets. Anim. Feed Sci. Technol. 247, 112–126. doi: 10.1016/j.anifeedsci.2018.11.005
Leung A. M., Braverman L. E. (2014). Consequences of excess iodine. Nat. Rev. Endocrinol. 10, 136–142. doi: 10.1038/nrendo.2013.251
Li X., Norman H. C., Kinley R. D., Laurence M., Wilmot M., Bender H., et al. (2016). Asparagopsis taxiformis decreases enteric methane production from sheep. Anim. Prod. Sci. 58, 681–688. doi: 10.1071/AN15883
Lindmark Månsson H. (2008). Fatty acids in bovine milk fat. Food Nutr. Res. 52 (1), p.1821. doi: 10.3402/fnr.v52i0.1821
Lucas H. L. (1957). Extra-period Latin-square change-over designs. J. Dairy Sci. 40, 225–239. doi: 10.3168/jds.S0022-0302(57)94469-7
LUKE (2019). Finnish Feed tables. Available at: https://portal.mtt.fi/portal/page/portal/Rehutaulukot/feed_tables_english.
Luton P. E., Wayne J. M., Sharp R. J., Riley P. W. (2002). The mcrA gene as an alternative to 16S rRNA in the phylogenetic analysis of methanogen populations in landfill. Microbiol. 148, 3521–3530. doi: 10.1099/00221287-148-11-3521
Machado L., Magnusson M., Paul N. A., Kinley R., de Nys R., Tomkins N. (2016). Identification of bioactives from the red seaweed asparagopsis taxiformis that promote antimethanogenic activity in vitro. J. Appl. Phycol. 28, 3117–3126. doi: 10.1007/s10811-016-0830-7
McMurdie P. J., Holmes S. (2013). Phyloseq: An r package for reproducible interactive analysis and graphics of microbiome census data. PloS One 8, e61217. doi: 10.1371/journal.pone.0061217
Muizelaar W., Groot M., van Duinkerken G., Peters R., Dijkstra J. (2021). Safety and transfer study: Transfer of bromoform present in asparagopsis taxiformis to milk and urine of lactating dairy cows. Foods 10 (3), 584. doi: 10.3390/foods10030584
Müller B., Sun L., Westerholm M., Schnürer A. (2016). Bacterial community composition and fhs profiles of low- and high-ammonia biogas digesters reveal novel syntrophic acetate-oxidising bacteria biotechnol. Biofuels 9, 1–18.
Nordic Committee on Food Analysis. (1976). Nitrogen de-termination in foods and feeds according to Kjeldahl. Method No 6. Statens Teknologiska Forskningscentral. Esbo, Finland.
O'Hara E., Moote P. E., Terry S., Beauchemin K., McAllister T., Abbott W., et al. (2022). Comparative analysis of macroalgae supplementation on the rumen microbial community: Asparagopsis taxiformis inhibits major ruminal methanogenic, fibrolytic, and volatile fatty acid-producing microbes in vitro. bioRxiv preprint. doi: 10.1101/2022.09.08.507231
Oksanen J., Blanchet F., Friendly M., Kindt R., Legendre P., McGlinn D., et al. (2019). Vegan: Community ecology package (Comprehensive R Archive Network (CRAN).
Pang D., Yan T., Krizsan S. J. (2021). Effect of strategy for harvesting regrowth grass silage on performance in dairy cows. J. Dairy Sci. 104, 367–380. doi: 10.3168/jds.2020-18888
Poulsen M., Schwab C., Jensen B. B., Engberg R. M., Spang A., Canibe N., et al. (2013). Methylotrophic methanogenic thermoplasmata implicated in reduced methane emissions from bovine rumen. Nat. Commun. 4, 1428. doi: 10.1038/ncomms2432
Roque B. M., Venegas M., Kinley R. D., de Nys R., Duarte T. L., Yang X., et al. (2021). Red seaweed (Asparagopsis taxiformis) supplementation reduces enteric methane by over 80 percent in beef steers. PloS One 16, e0247820. doi: 10.1371/journal.pone.0247820
Russell R. M., Beard J. L., Cousins R. J., Dunn J. T., Ferland G., Hambidge K. M., et al. (2001). Dietary reference intakes for vitamin a, vitamin K, arsenic, boron, chromium, copper, iodine, iron, manganese, molybdenum, nickel, silicon, vanadium, and zinc; a report of the panel on micronutrients, subcommittees on upper reference levels of nutrients and of interpretation and uses of dietary reference intakes, and the standing committee on the scientific evaluation of dietary reference intakes food and nutrition board institute of medicine (Washington, DC, USA: National Academies Press).
Shingfield K. J., Ahvenjärvi S., Toivonen V., Ärölä A., Nurmela K. V. V., Huhtanen P., et al. (2003). Effect of dietary fish oil on biohydrogenation of fatty acids and milk fatty acid content in cows. Anim. Sci. 77, 165–179. doi: 10.1017/S1357729800053765
Shingfield K. J., Bernard L., Leroux C., Chilliard Y. (2010). Role of trans fatty acids in the nutritional regulation of mammary lipogenesis in ruminants. Animal 4, 1140–1166. doi: 10.1017/S1751731110000510
Sjaunja L. O. (1990). “A Nordic proposal for an energy-corrected milk (ECM) formula,” in 27th Session International Committee for Recording and Productivity of Milk Animals; 2-6 July 1990, Paris, France.
Stefenoni H. A., Räisänen S. E., Cueva S. F., Wasson D. E., Lage C. F. A., Melgar A., et al. (2021). Effects of the macroalga asparagopsis taxiformis and oregano leaves on methane emission, rumen fermentation, and lactational performance of dairy cows. J. Dairy Sci. 104, 4157–4173. doi: 10.3168/jds.2020-19686
Sterner E. O., Johansson D. J. (2017). The effect of climate–carbon cycle feedbacks on emission metrics. Environ. Res. Lett. 12, 034019. doi: 10.1088/1748-9326/aa61dc
Stevenson D. M., Weimer P. J. (2007). Dominance of prevotella and low abundance of classical ruminal bacterial species in the bovine rumen revealed by relative quantification real-time PCR. Appl. Microbiol. Biotechnol. 75, 165–174. doi: 10.1007/s00253-006-0802-y
Van Leeuwen F. R., Sangster B., Hildebrandt A. G. (1987). The toxicology of bromide ion. CRC Crit. Rev. toxicol. 18, 189–213. doi: 10.3109/10408448709089861
Vlaeminck B., Fievez V., Tamminga S., Dewhurst R. J., Van Vuuren A., De Brabander D., et al. (2006). Milk odd-numbered and branched-chain fatty acids in relation to the rumen fermentation pattern. J. Dairy Sci. 89, 3954–3964. doi: 10.3168/jds.S0022-0302(06)72437-7
Wickham H. (2016). Ggplot2: Elegant graphics for data analysis. 2nd ed (Cham: Springer International Publishing). doi: 10.1007/978-3-319-24277-4
Williamson J. R., Callaway T. R., Lourenco J. M., Ryman V. E. (2022). Characterization of rumen, fecal, and milk microbiota in lactating dairy cows. Front. Microbiol. 13. doi: 10.3389/fmicb.2022.984119
Wolff R. L., Bayard C. C., Fabien R. J. (1995). Evaluation of sequential methods for the determination of butterfat fatty acid composition with emphasis ontrans-18: 1 acids. application to the study of seasonal variations in french butters. J. Am. Oil Chem. So. 72, 1471–1483. doi: 10.1007/BF02577840
Keywords: archaea, bacteria, milk quality, seaweed, ruminant, methane
Citation: Krizsan SJ, Ramin M, Chagas JCC, Halmemies-Beauchet-Filleau A, Singh A, Schnürer A and Danielsson R (2023) Effects on rumen microbiome and milk quality of dairy cows fed a grass silage-based diet supplemented with the macroalga Asparagopsis taxiformis. Front. Anim. Sci. 4:1112969. doi: 10.3389/fanim.2023.1112969
Received: 30 November 2022; Accepted: 30 January 2023;
Published: 27 February 2023.
Edited by:
Alexandros Mavrommatis, Agricultural University of Athens, GreeceReviewed by:
Andres Alfredo Pech-Cervantes, Fort Valley State University, United StatesStefanie Santos, Federal University of Bahia, Brazil
Copyright © 2023 Krizsan, Ramin, Chagas, Halmemies-Beauchet-Filleau, Singh, Schnürer and Danielsson. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sophie J. Krizsan, c29waGllLmtyaXpzYW5Ac2x1LnNl
 Sophie J. Krizsan
Sophie J. Krizsan Mohammad Ramin
Mohammad Ramin Juana C. C. Chagas
Juana C. C. Chagas Anni Halmemies-Beauchet-Filleau
Anni Halmemies-Beauchet-Filleau Abhijet Singh
Abhijet Singh Anna Schnürer
Anna Schnürer Rebecca Danielsson
Rebecca Danielsson