- 1Department of Applied Biology, Adama Science and Technology University, Adama, Ethiopia
- 2Bio and Emerging Technology Institute, Agricultural Biotechnology, Addis Ababa, Ethiopia
Non-conservative K232A substitution in the diacylglycerol acyl-CoA acyltransferase 1 (DGAT1) gene has been reported to explain variation in milk traits in cattle. The objective of this study was to estimate allele and genotype frequencies and to assess associations between K232A variants and milk yield and composition in cattle populations of Ethiopia. Blood samples for genomic DNA extraction and milk samples for analysis of milk components were collected from 92 randomly selected cattle of five Ethiopian breeds. Gene-specific primers were used to amplify 278 bp of the exon 8 region of DGAT1. Allele and genotype frequencies were calculated using Power Marker, and the GLM function of SAS software was used to assess the associations of detected genetic variation with milk traits. Boran * Holstein Friesian (HF) crosses produced a higher daily milk yield than the other breeds (p< 0.05), whereas the Boran and Begait breeds produced milk with higher fat and protein contents (p< 0.05), and Horro produced milk with a higher content of lactose, than the other breeds studied (p< 0.05). Alleles K and A and genotypes AA, KA, and KK were detected. The frequency of K232 ranged from 0.50 in Boran * HF crosses to 0.97 in the Horro population. The frequency of the KK and KA genotypes in the zebu population ranged from 0.50 to 0.94 and from 0.03 to 0.50, respectively. The AA genotype was associated with higher milk yield in Boran * HF crosses, whereas the KA genotype was associated with higher milk yield in the zebu populations (p< 0.05). The fat and lactose contents of milk produced with the KA genotype were lower than those of milk produced with the KK genotype in all genetic groups (p< 0.05). Substitution of one copy of the K allele led to a significant (p< 0.05) increase in fat content, of up to 0.81%, a decrease in daily milk yield of up to 3 L, and a decrease in lactose content of 0.58% in the sampled populations. The association study confirmed that the DGAT1 K232A marker had significant effects on daily milk yield, milk fat and lactose contents in the investigated cattle. These results suggested that the DGAT1 K232A marker may be utilized to accelerate future molecular breeding of dairy cattle, tropical zebu, their crossbreeds, after validation in a larger population.
Introduction
The cattle agriculture sector in the tropics faces the challenges of growing productivity and profitability, in order to meet increased demand from a growing human population. Ethiopia is home to a large variety of cattle populations that experience an assortment of harsh ecological conditions, but interventions aimed at animal genetic improvement are scarce.
Increasing milk volume increment is the principal goal in the breeding of dairy cattle worldwide (Meredith et al., 2012). However, newer breeding objectives, notably to improve milk composition traits, are a response to the demand for a healthier human diet, and the basis of the milk-pricing system has shifted from a standard amount to milk composition, with a direct effect on farms’ financial performance (Krovvidi et al., 2013).
Local breeds’ performance levels can be increased through genetic improvement in milk performance traits, but efficient genetic improvement requires information on genetic variation and its effects on milk yield. It is known that genetic selection increases the frequency of alleles with a positive effect on a given attribute (Dekkers, 2004).
Improving productivity through genetic selection is a common goal of many animal breeding programs around the world (Meredith et al., 2012; Narayana et al., 2017; Heimes et al., 2019). Several genes affecting economically important traits in livestock species have been localized within quantitative trait loci (QTL) dispersed throughout the genome.
In cattle breeds, the diacylglycerol O-acyl transferase 1 (DGAT1) gene encodes acyl coenzyme A:diacylglycerol acyltransferase, a protein involved in fat metabolism (Grisart et al., 2004; Yu and Ginsberg, 2004; Coleman and Mashek, 2011; Yen et al., 2015). The important role of this gene in the metabolism of milk fat makes it an interesting candidate for the genetic manipulation of milk characteristics. The mutation, which results in a lysine to alanine substitution at position 232 (K232A), has been linked to differences in the kinetics of the enzymes encoded by the two allelic variants (Grisart et al., 2004). Grisart et al. (2004) demonstrated that the lysine variant, which represents the “wild type” and is designated the K allele, is distinguished by a higher maximum rate of reaction of the enzyme involved in triglycerides synthesis than the alanine variant, thereby increasing the fat percentage in the animal’s milk. The lysine variant has been linked to higher fat yield and fat and protein percentages, whereas the alanine variant has been linked to higher milk and protein yields (Grisart et al., 2002; Krovvidi et al., 2021; Elzaki et al., 2022; Gothwal et al., 2022).
The distribution of the allele frequencies of the DGAT1 K232A mutation has been studied in various Holstein populations and other exotic breeds, and the effects of DAGT1 polymorphisms on milk yield traits have been researched (Bovenhuis et al., 2016; Krovvidi et al., 2021; Li et al., 2021; Gothwal et al., 2022). Information on the effect of DGAT1 K232A on the daily milk yield and milk composition of different cattle breeds is scarce in tropical zebu and their crossbred population. The K232A genotypes of DGAT1 have been shown to have significant effects on milk composition traits in some tropical zebu populations, including Sudanese Kenana, Butana, Butana–Holstein, and Benin cattle breeds (Rahamattalla et al., 2015; Houaga et al., 2017; Houaga et al., 2018; Elzaki et al., 2022). However, whether or not the known DGAT1 K232A mutations segregate and are associated with milk yield and milk quality traits in zebu and their crossbred cattle populations in Ethiopia has not been well studied. Before the aforementioned marker is used for the genetic improvement of Ethiopian cattle productivity, its effect should be clearly examined.
Ethiopia is endowed with indigenous cattle, such as the Boran, Begait, Horro, and Fogera breeds, that have promising yield potential (Rege and Tawah, 1999; Zerabruk et al., 2007). A recent study revealed the polymorphic nature of the DGAT1 gene in cattle populations in Ethiopia (Samuel et al., 2022), but further study on the influence of DGAT1 K232A on milk traits is required. The detection of mutations affecting dairy traits will facilitate the application of marker-assisted selection and provides insights into the genetics of milk traits in tropically adapted cattle breeds. Therefore, the present study aimed to estimate the milk composition variation as well as allele and genotypic frequencies of the DGAT1 K232A mutation and to assess the effects of the two allelic variants on daily milk yield and milk composition in cattle populations in Ethiopia.
Materials and methods
Sample collection and DNA extraction
The animal study was reviewed and approved by the Ethical Review Board of Adama Science and Technology University (certificate reference number RECSoANS/BIO/07/2021). Written informed consent was obtained from the owners for the participation of their animals in this study.
A total of 92 animals from five cattle populations (17 Boran, 16 Begait, 18 Fogera, 17 Horro, and 24 Boran * HF cross (75% F1 HF** (HF * Boran) were sampled from state-owned farms in Ethiopia.
Boran cattle are predominantly distributed in the semi-arid and arid areas of southern Ethiopia, northern Kenya, and south-western Somalia, and are large East African zebu cattle known for their fast growth, high milk yield, and heavy muscling. They are known to need a calm environment (Rege and Tawah, 1999). The Horro cattle breed is widely distributed in south-western and west Ethiopia. It is one of the sanga * zebu intermediate types and is used for milk and meat yield, and to provide draft power (Rege and Tawah, 1999; Edea et al., 2012; Mwai et al., 2015). The Fogera cattle breed is found in the north-west highlands of Ethiopia and is reared mainly for dairy yield and for its draft power. It belongs to the zenga group, which is a cross between sanga and zebu (Rege and Tawah, 1999; Zerabruk et al., 2007). The Begait breed is grouped under large East African zebu (Rege and Tawah, 1999) and is used primarily for milk and meat yield (Zerabruk et al., 2007; Assefa and Hailu, 2018).
Blood and milk samples were obtained only from randomly selected and sampled lactating cows. The studied animals were raised on natural grazing without concentrate supplementation and were sampled at the same time, eliminating the effect of season. Blood samples (4 mL)were collected from the tail head of each animal under aseptic conditions and gently mixed with ethylene diamine tetraacetic acid (EDTA) anticoagulant placed into an ice box containing ice. Extraction of genomic DNA was carried out using the salting-out extraction procedure (Nasiri et al., 2005). The quality of the DNA and its concentration were assessed using a NanoDrop1000 spectrophotometer and electrophoresis in 1.5% agarose gels. Those DNA samples with good quality and quantity were used for amplification and sequencing.
PCR amplification and sequencing of the DGAT1 gene region
Based on the reference sequence (AJ318490) the primers were designed to amplify a 278-bp fragment enclosing the p.Lys232Ala mutation (forward: 5′-AAGGCCAAGGCTGGTGAG-3′; reverse: 5′-GGCGAAGAGGAAGTAGTAG-3′) using Primer3Plus software (Rozen and Skaletsky, 2000). PCR was carried out in a total volume of 25 μL containing, 5* PCR buffer (5 μL), 1.5 mM MgCl2 (3 μL), 10 mM dNTP mix (1 μL), forward primer 10 pmol/μL (0.5 μL), reverse primer 10 pmol/μL (0.5 μL), genomic DNA 25 ng/μL (2 μL), Taq DNA polymerase 5 U/μL (0.3 μL) and DNAase-free water (12.7 μL). The optimized thermal profile comprised an initial denaturation at 94°C for 3 min, 30 cycles of denaturation at 94°C for 1 min, annealing at 57°C for 45 s, elongation at 72°C for 1 min, and a final extension at 72°C for 7 min. Finally, the PCR products were visualized post electrophoresis on 1.7% agarose gel in TAE buffer (Tris base, acetic acid, and EDTA) followed by GelRed staining. The PCR products were sequenced on an ABI 3730 sequencer (Applied Biosystems, Foster City, CA, USA) using the Sanger dideoxy chain termination method at Konkuk University, Seoul, South Korea. The sequences were analyzed and deposited in GenBank with the accession numbers ON262825–ON262849.
In the early morning and late afternoon, milk samples (50 mL) were collected from each animal in clean, dry, grease-free, and labeled milk collection vials. The milk samples were collected in the mid-milking stage to avoid sampling fluctuations in the milk composition data. No milk was collected from the animals that were in the terminal or early stage of lactation. Milk samples were stored at 4°C and then sent to the Holeta dairy research laboratory for milk composition analyses. The percentages of milk fat, protein, lactose, and non-fat solids were determined using a Lactoscan ultrasonic milk analyzer (Milkotronic Ltd, Nova Zagora, Bulgaria).
Data management and statistical analysis
Prior to analysis, all chromatograms were visualized and sequence fragments were edited using Bio-edit version 7.0.5.3 and aligned using the ClustalX2 software package (Larkin et al., 2007). Genotype and allele frequencies were calculated using Power Marker (version 3.25) (Liu and Muse, 2005). The standard error (SE) of allelic frequency was calculated as [p(1 – p)/2n]1/2, where n is the sample size and p is the frequency of allele (Spiess, 1977).
To investigate the effect of breed type and DGAT1 genotypes on milk components, we used a generalized linear model (GLM), and the difference between means was calculated using the Tukey honestly significant different (HSD) assess with the statistical software package SAS version 9.3 (SAS, 2011). Breed type and genotype were fitted as fixed independent variables, while the observed traits (daily milk yield, fat percentage, protein percentage, non-fat solids percentage, and lactose percentage) were fitted as dependent variables. The effects of fixed factors were fitted in the statistical model below:
where Yij is the observed trait (daily milk yield, fat percentage, protein percentage, non-fat solids percentage, or lactose percentage), μ is the population mean, Bi is the fixed effect of ith breed type (i = 5), Gj is the fixed effect of DGAT1 genotype (KK, KA, or AA), (B × G)ij is the fixed interaction effect between breed and DGAT1 genotype, and ϵijk is the random residual associated with each record.
The quantitative effects of one copy of the K allele of DGAT1 K232A were also calculated. The linear model was used, with the replacement of genotype effect by the linear regression on the number of desired alleles: Yijk = μ + b*xi + eij
where Yij is a phenotypic observation (milk traits) of the animal, μ is the overall mean, xi is the number of desired alleles (0, 1, or 2), b is the regression coefficient representing the allele substitution effect, and eij is a random residual effect. The results of breed and DGAT1 genotype effects are presented as least-squares means ± SE. A probability of less than 0.05 was considered to indicate a significant difference.
Results
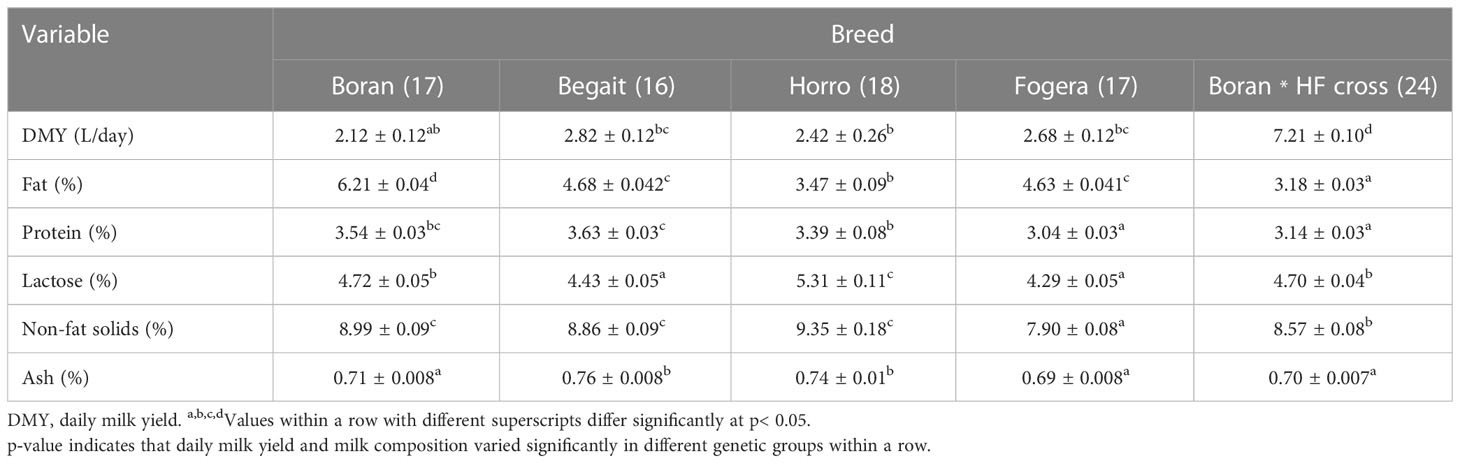
Effect of breed on milk yield and composition
Statistically significant differences in milk components were observed among the studied breeds (Table 1). The Boran lowland breed produced milk with a significantly higher fat content than the other breeds (p< 0.05), while the other lowland breed, Begait, produced milk with a significantly higher protein content (p< 0.05). On the other hand, Horro produced milk with higher lactose content than the other breeds (p< 0.05). The Boran, Begait, and Horro breeds all produced milk with a higher content of non-fat solids than other breeds.
Least-squares means of milk components across breeds are reported in Table 1. Boran * HF crosses produced a higher daily milk yield than the other breeds (p< 0.05).
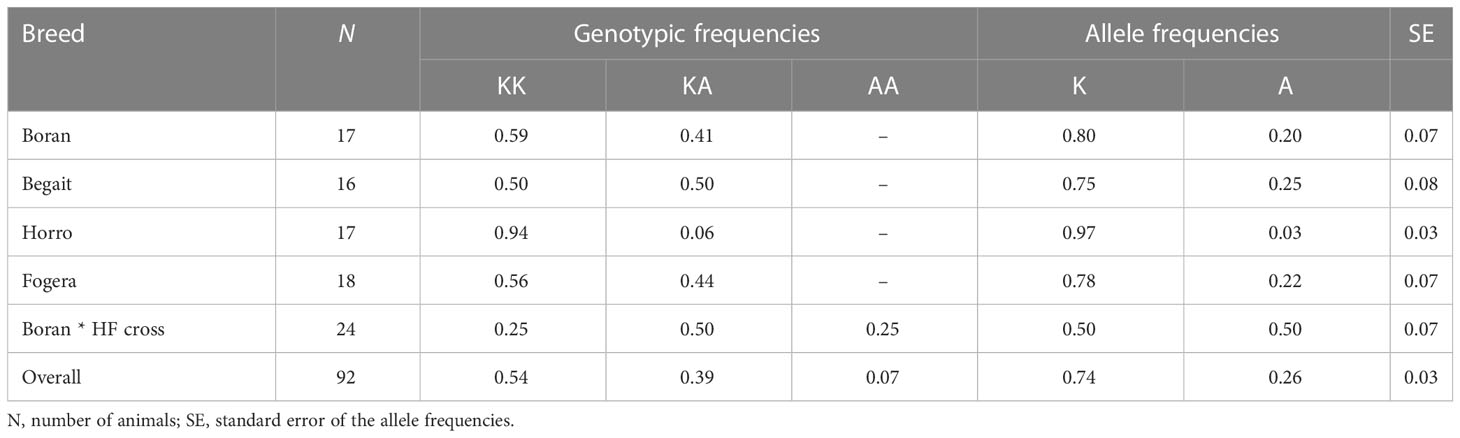
Allele and genotype frequencies of the K232A protein variant
The genotypic and allelic frequencies of the DGAT1 gene in Boran, Begait, Horro, Fogera, and Boran * HF cross cattle are presented in Table 2.
The DGAT1 K allele, corresponding to lysine substitution, was the predominant allele in all zebu populations. All three possible genotypes of DGAT1 K232A were observed in Boran * HF crosses, although the KA genotype predominated. The KK genotype predominated in the Boran and Fogera populations, but was even more prevalent in Horro. In Begait, the KK and KA genotypes have identical frequency distributions (Figure 1; Table 2).
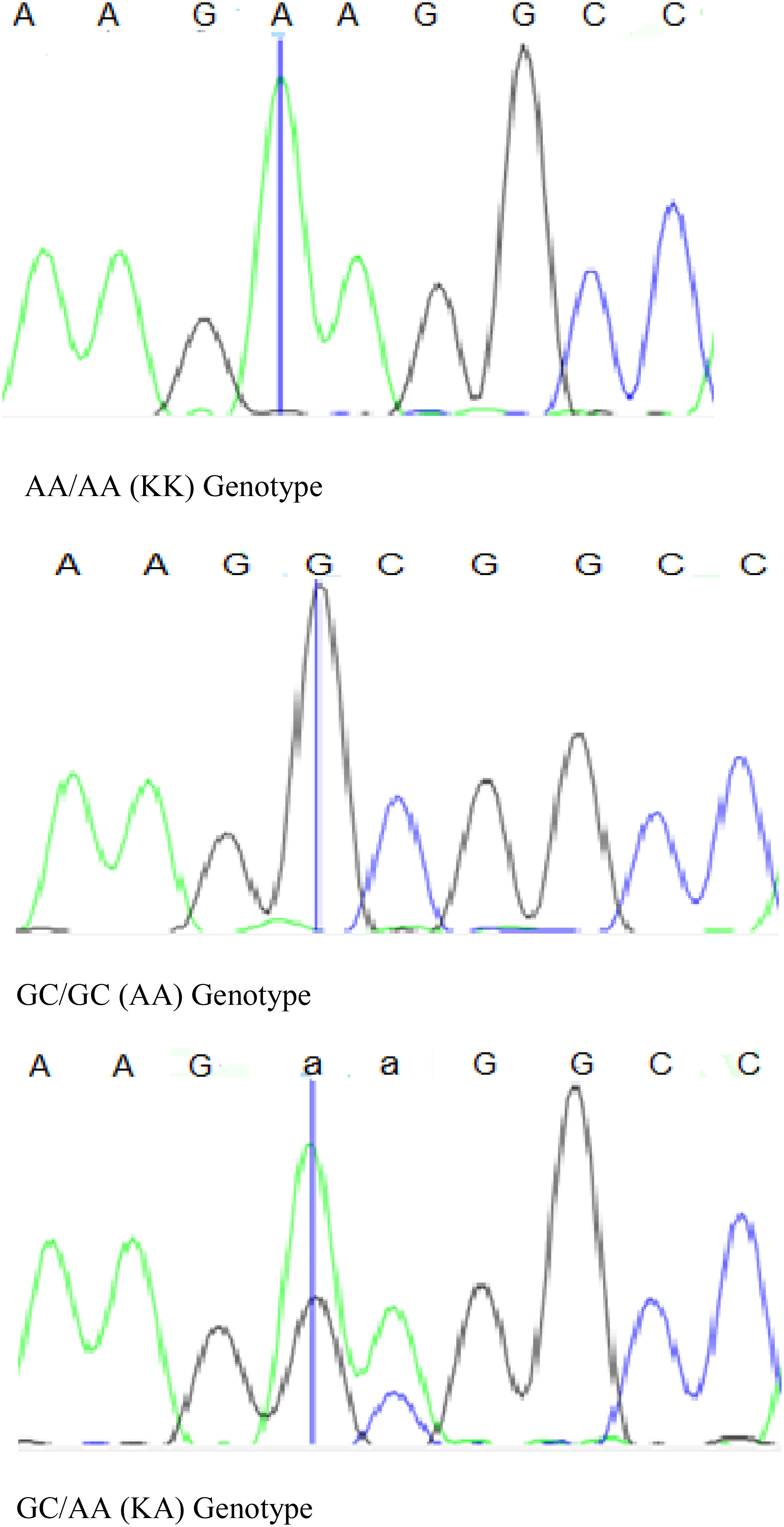
Figure 1 Sequence trace views of sequencing traces for positions 10,433 and 10,434. The vertical line indicates nucleotide position 10,433. Positions 10,433 and 10,434 are responsible for the K232A substitution. The positions of individual mutations were named according to the sequence available in GenBank (AJ318490).
Effect of DGAT1 K232A variants on daily milk yield and composition
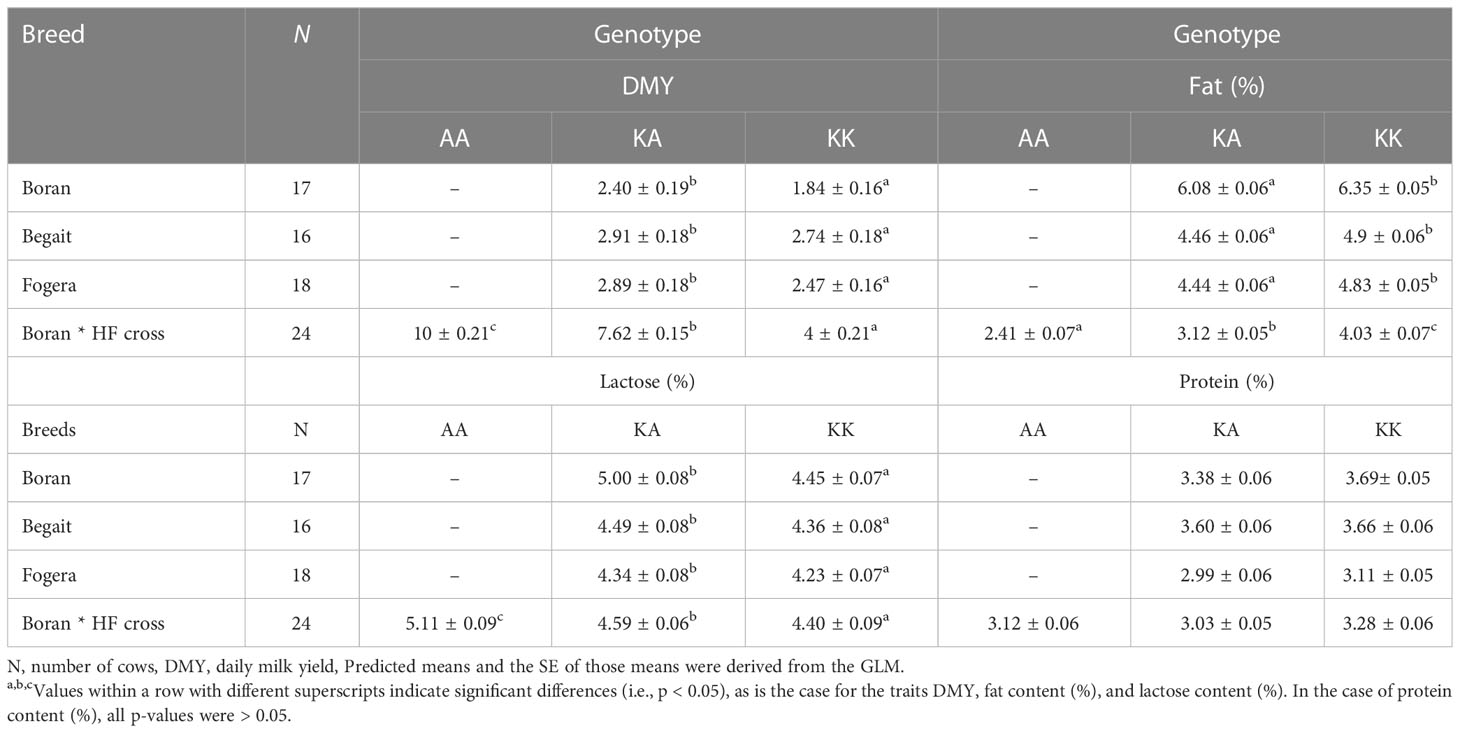
Variability at the DGAT1 K232A polymorphic site indicated that this locus can be considered for association studies only in Boran, Begait, Fogera, and Boran * HF crosses. In Horro populations, the minor allele frequency is less than 0.1 and, hence, does not qualify for association studies (Table 3).
The AA genotype was associated with greater milk yield in Boran * HF crosses, whereas the KA genotype was associated with greater milk yield in the zebu populations (p< 0.05). The fat and lactose contents of milk produced with the KA genotype were lower than those of milk produced with the KK genotype in all genetic groups (p< 0.05). The percentages of milk protein and non-fat solids were lower in milk produced with the KA genotype than in milk produced with the KK genotype in all the genetic groups (p > 0.05).
Substitution of the K allele showed that even one copy of this allele leads to a significant (p< 0.05) increase in fat content in Boran * HF crosses (of up to 0.81%) and a significant increase of protein content (of up to 0.32%) in Boran breeds. In contrast, substitution of the K allele leads to a decrease in daily milk yield of up to 3 L in Boran * HF crosses and a decrease in milk lactose content of 0.58% in Horro cattle.
Discussion
Effect of breed on milk yield and composition
The milk fat content of the studied breeds was within the range of the standard milk composition requirement for cows (3.18%–6.21%). Zebu cows can give milk containing up to 7% fat (O’Mahony, 1988). Significant differences among the studied breeds were observed for all milk components. Similar studies have reported the effect of breed on milk composition in South African (Myburgh et al., 2012) and Benin cattle breeds (Houaga et al., 2017). Boran cows in Ethiopia presented a higher fat content (6.21%) than Boran cows in South Africa (2.68%) and Borgou and White Fulani cows in Benin (4.78% and 4.85%, respectively) (Myburgh et al., 2012; Houaga et al., 2017). Thus, according to the present study, Boran * HF cross cows are to be favored for daily milk yield, whereas Boran cows are best in terms of milk fat percentage. The observed differences between breeds are therefore likely to be due to differences in genetic background. However, another the reason for the variation in milk components among cattle breeds could be differences in the forage nutrient composition of natural grazing. The investigated cattle breeds are reared in different agro-ecological zones with different floristic compositions. Cow’s milk composition could be affected by forage species and variety, climate, and growth stage of forage plants.
Allele and genotype frequencies of the K232A protein variant
The variants of the analyzed gene were found in all the five breeds, and therefore it is the allelic distribution that characterizes the differences between breeds. The present study revealed that the lysine variant of the DGAT1 gene, i.e., the K232 allele, was common in Horro, Boran, and Fogera cattle. This is in agreement with studies in other Bos indicus breeds, in which the frequency of the lysine variant K232 ranged from 0.80 to 1 in four types African of zebu (Banyo Gudali, Kenana, Butana, and White Fulani) (Abu et al., 2015; Rahamattalla et al., 2015; Elzaki et al., 2022) and three types of Indian cattle (Sahiwal, Gir, and Kankrej) (Ganguly et al., 2013; Patel and Chauhan, 2017; Gothwal et al., 2022), and was comparable to the frequencies of K232 in Borgou (0.77) and White Fulani (0.92) cattle of Benin (Houaga et al., 2018). The higher frequency of the K allele seen in those three Ethiopian breeds might reflect the exceptional adaptive value of these breeds. The overall immunity of zebu cattle to diseases, and their ability to adapt to and survive on a coarse feed diet, could be due to increased acyltransferase activity of the DGAT1 gene, as DGAT catalyzes the synthesis of retinol esters and thus regulates the synthesis of vitamin A. Manga and Řiha (2011) reported a favorable association of the DGAT1 K allele with low somatic cell count in lactating cows that partly explains the exceptional genetic resistance of native cattle to mastitis (Liu et al., 2007).
In the present study, the observed frequencies of the K allele (0.50–0.97) are higher than those observed (0.210–0.380) in the native Turkish and European Bos taurus breeds (Kaupe et al., 2004), with the exception of the Modicana and Jersey breeds (Valenti et al., 2019). The finding in the present study indicated the tendency of fixation of DGAT1 K allele in Horro cattle did not support its use as a reliable universal marker for milk yield and composition traits for the population. Fixation of the DGAT1 K allele in ongole and buffalo breeds in India was noted by Krovvidi et al. (2021), whereas Kaupe et al. (2004) found fixation of the DGAT1 A allele in five Bos taurus breeds (Belgian Blue beef, Gelbvieh, Hereford, Pinzgaurer, and Slavonian Syrmian). The high proportion of the lysine variant K232 observed in the current study might contribute to the higher fat content in native cattle in Ethiopia and other Bos indicus breeds than in HF cross cattle. The frequency of the lysine variant K232 in the investigated Boran * HF cross cattle is similar to that found in studies of other HF populations or their crosses, which have reported a range from 0.21 to 0.63 (Ahani et al., 2015; Akyüz et al., 2015; Komisarek and Kolenda, 2016; Bhat et al., 2017; Li et al., 2020; Krovvidi et al., 2021). The differences in the frequency of the lysine variant K232 among breeds might result from different breeding goals and the considerable Holstein contribution to the blood level of Boran * HF cross cattle.
Allele A is a rare allele detected in Boran, Begait, Fogera and Boran * HF crosses. A locus is considered polymorphic if the most common allele has a frequency of ≤ 0.95 and therefore the less common allele (at a locus with two alleles) has a frequency of ≥ 0.05 (Hartl and Clark, 1997). Accordingly, in the present study, the reason why the DGAT1 locus was not found to be polymorphic in Horro cattle populations may be that one animal among the sampled population had the A allele. The A allele frequency observed in the zebu populations was relatively similar to that found in the indigenous cattle of Sudan (0.037–0.15) (Abu et al., 2015). The A allele frequency observed in Boran * HF cross cattle was higher than reported by Li et al. (2021) in Kiwi cross cattle in New Zealand and lower than in Butana–Holstein crossbred cattle in Sudan (Elzaki et al., 2022). To reiterate, the low A allele frequency observed in native breeds of Ethiopia might be attributed to Bos taurus cattle introgression.
The KK and KA genotypic frequencies observed in in the zebu populations in the present study ranged from 0.50 to 0.94 and from 0.03 to 0.50, respectively, and are comparable to the frequencies observed in most Bos indicus cattle (Abu et al., 2015; Rahamattalla et al., 2015; Houaga et al., 2017; Elzaki et al., 2022). An excess of heterozygotes over homozygotes in Boran * HF cross breeds indicates that heterozygotes are preferred, which could be the effect of natural selection over the years of evolution and would be effective in selecting for or against polymorphism.
Effect of DGAT1 K232A on daily milk yield and composition
The association study confirmed that the DGAT1 K232A marker had significant effects on daily milk yield and milk fat and lactose contents in the investigated cattle. Similar effects of the KK genotype on milk composition content traits, and an even greater effect of the AA and KA genotypes on average milk yield, have been observed in Holstein, Holstein crosses, Jersey and its local Kashmiri crosses, Holstein *Jersey crosses, Gir, and Kankrej cattle populations in different parts of the world (Molee et al., 2012; Akyüz et al., 2015; Bhat et al., 2017; Patel and Chauhan, 2017; Li et al., 2021).
In contrast, Manga and Řiha (2011) have reported that the DGAT1K allele (lysine variant) is associated with higher milk yield in Holstein cows.
Substitution of the K allele showed that even one copy of this allele led to a significant (p< 0.01) increase in fat and protein content and a decrease in milk yield and lactose and non-fat solids content in all breeds considered. Similar effects of allele substitution were reported by Grisart et al. (2002). They observed that substitution of one copy of A allele by K allele reduced average milk yield by 130 L, increased average milk fat content by 5.76 kg, and reduced milk protein by 2.45 kg in HF cattle, and reduced milk yield by 110 L, increased milk fat by 3.30 kg, and reduced milk protein by 2.48 kg in a Jersey cattle population. Moreover, an increase in milk yield of 548 kg and in protein yield of 12.6 kg, accompanied by a decrease in milk fat of 15.4 kg, has been reported in HF animals homozygous for the A allele of the DGAT1 gene (Hradecká et al., 2008).
Conclusion
Our results show that Boran * HF cross cows are the best breed in terms of daily milk yield, whereas Boran cows are best in terms of milk fat percentage. Allele K and genotype KK were prevalent in the zebu cattle populations. The overall diversity indices showed that the DGAT1 locus was polymorphic in the Boran, Begait, Fogera, and Boran * HF cross populations. The association study confirmed that the DGAT1 K232A marker has significant effects on daily milk yield, milk fat and lactose content in Boran, Begait, Fogera and Boran * HF cross populations. The DGAT1 K232A marker could be utilized for breeding and improvement of milk yield and composition traits in local cattle populations in Ethiopia after validation in a larger population.
Data availability statement
The datasets presented in this study can be found online at https://www.ncbi.nlm.nih.gov/genbank/, under the accession numbers ON262825–ON262849.
Ethics statement
The animal study was reviewed and approved by the Ethical Review Board of Adama Science and Technology University (certificate reference number RECSoANS/BIO/07/2021). Written informed consent was obtained from the owners for the participation of their animals in this study. Animal samples were obtained in compliance with local/national laws in force at the time of sampling. Data exchange was in accordance with national and international regulations, and approved by the owners. The procedure involving sample collection followed the recommendation of directive 2010/63/EU. All methods were carried out in accordance with relevant guidelines and regulations. All authors gave their informed consent prior to their inclusion in the study.
Author contributions
Conceptualization: BS, HDi, and HDa. Data curation: BS. Formal analysis: BS. Funding acquisition: BS. Investigation: BS. Methodology: BS, HDi, and HDa. Project administration: BS. Resources: BS, HDi, and HDa. Supervision: HDi and HDa; Visualization: BS. Writing – original draft: BS. Writing – review and editing: BS, HDi, and HDa. All authors contributed to the article and approved the submitted version.
Acknowledgments
This research work was supported by the Ethiopian Ministry of Education for data collection and Konkuk University of South Korea for sequencing.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abu S. M., Said Ahmed A., Ahmed M. K. A., Reissmann M., Brockmann G. A., Rahmatalla S. A. (2015). Variants of the diacylglycerol acyltransferase 1 (DGAT 1) gene in Sudanese dairy cattle (Kenana and butana). J. Mol. Genet. 7 (1), 5–9. doi: 10.36478/jmolgene.2015.5.9
Ahani S., Mashhadi M. H., Nassiri M. R., Aminafshar M., Haddadi M. (2015). Characterization of single nucleotide polymorphism in diacylglycerol acyltransferase (DGAT1) gene loci of Iranian Holstein cattle. Res. Opin. Anim. Veterinary Sci. 5 (5), 231–236.
Akyüz B., Agaoglu O. K., Akçay A., Agaoglu A. R. (2015). Effects of DGAT1 and GH1 polymorphism on milk yield in Holstein cows reared in Turkey. Slovenian Veterinary Res. 52 (4), 185–191.
Assefa A., Hailu A. (2018). Ethiopian Indigenous cattle breed’s diversity, distribution, purpose of keeping, and their potential threats. J. Biodiv. Innovativ. Assoc. 7 (5), 770–789.
Bennewitz J., Reinsch N., Paul S., Looft C., Kaupe B., Weimann C., et al. (2004). The DGAT1 K232A mutation is not solely responsible for the milk yield quantitative trait locus on the bovine chromosome 14. J. Dairy Sci. 87 (2). doi: 10.3168/jds.S0022-0302(04)73182-3
Bhat S. A., Ahmad S. M., Ganai N. A., Khan S. M., Malik A., Shah R. A., et al. (2017). Association of DGAT1, beta-casein and leptin gene polymorphism with milk quality and yield traits in Jersey and its cross with local kashmiri cattle. J. Entomology Zoology Stud. 5 (6), 557–561.
Bovenhuis H., Visker M. H. P. W., Poulsen N. A., Sehested J., van Valenberg H. J. F., van Arendonk J. A. M., et al. (2016). Effects of the diacylglycerol o-acyltransferase 1 (DGAT1) K232A polymorphism on fatty acid, protein, and mineral composition of dairy cattle milk. J. Dairy Sci. 99 (4), 3113–23. doi: 10.3168/jds.2015-10462
Coleman R. A., Mashek D. G. (2011). Mammalian triacylglycerol metabolism: synthesis, lipolysis, and signaling. Chem. Rev. 111 (10), 6359–86. doi: 10.1021/cr100404w
Da Silva M. V. G. B., Sonstegard T. S., Thallman R. M., Connor E. E., Schnabel R. D., Van Tassell C. P. (2010). Characterization of dgat1 allelic effects in a sample of north American holstein cattle. Anim. Biotechnol. 21 (2). doi: 10.1080/10495390903504625
Dekkers J. C. (2004). Commercial application of marker- and gene-assisted selection in livestock: strategies and lessons. J. Anim. Sci. 82 (E-Suppl):313–28. doi: 10.2527/2004.8213_supplE313x
Edea Z., Dadi H., Kim S. W., Dessie T., Kim K.-S. (2012). Comparison of SNP variation and distribution in indigenous Ethiopian and Korean cattle (Hanwoo) populations. Genomics Inf. 10 (3), 200–5. doi: 10.5808/gi.2012.10.3.200
Elzaki S., Korkuć P., Arends D., Reissmann M., Brockmann G. A. (2022). Effects of DGAT1 on milk performance in Sudanese butana * Holstein crossbred cattle. Trop. Anim. Health yield 54 (2), 142–7. doi: 10.1007/s11250-022-03141-7
Ganguly I., Kumar S., Gaur G. K., Singh U., Kumar A., Kumar S., et al. (2013). DGAT1 polymorphism K232A in sahiwal (Indian zebu) and frieswal (Holstein friesian x sahiwal crossbred) cattle. Indian J. Anim. Res. 47 (4), 360–3.
Gautier M., Capitan A., Fritz S., Eggen A., Boichard D., Druet T. (2007). Characterization of the DGAT1 K232A and variable number of tandem repeat polymorphisms in French dairy cattle. J. Dairy Sci. 90 (6), 2980–2988. doi: 10.3168/jds.2006-707
Gothwal A., Magotra A., Bangar Y. C., Malik B. S., Yadav A. S., Garg A. R. (2022). Candidate K232A mutation of DGAT1 gene associated with yield and reyield traits in Indian dairy cattle. Anim. Biotechnol. 33:1–9. doi: 10.1080/10495398.2022.2109041
Grisart B., Coppieters W., Farnir F., Karim L., Ford C., Berzi P., et al. (2002). Positional candidate cloning of a QTL in dairy cattle: identification of a missense mutation in the bovine DGAT1 gene with major effect on milk yield and composition. Genome Res. 12 (2), 222–231. doi: 10.1101/gr.224202
Grisart B., Farnir F., Karim L., Cambisano N., Kim J. J., Kvasz A., et al. (2004). Genetic and functional confirmation of the causality of the DGAT1 K232A quantitative trait nucleotide in affecting milk yield and composition. Proc. Natl. Acad. Sci. U.S.A. 101 (8), 2398–2403. doi: 10.1073/pnas.0308518100
Hartl D. L., Clark A. G. (1977). Principles of Population Genetics 3rd edition (Sunderland USA: Sinauer Associates Inc.), 542.
Heimes A., Brodhagen J., Weikard R., Hammon H. M., Meyerholz M. M., Petzl W., et al. (2019). Characterization of functional traits with focus on udder health in heifers with divergent paternally inherited haplotypes on BTA18. BMC Veterinary Res. 15 (1), 241–251. doi: 10.1186/s12917-019-1988-4
Houaga I., Muigai A. W. T., Kyallo M., Githae D., Youssao I. A. K., Stomeo F. (2017). Effect of breed and diacylglycerol acyltransferase 1 gene polymorphism on milk yield traits in beninese white Fulani and borgou cows. Global J. Anim. Breed. Genet. 5 (5), 403–412.
Houaga I., Muigai A. W. T., Ng’ang’a F. M., Ibeagha-Awemu E. M., Kyallo M., Youssao I. A. K., et al. (2018). Milk fatty acid variability and association with polymorphisms in SCD1 and DGAT1 genes in white Fulani and borgou cattle breeds. Mol. Biol. Rep. 45 (6), 1849–1862. doi: 10.1007/s11033-018-4331-4
Hradecká E., Čítek J., Panicke L., Řehout V., Hanusová L. (2008). The relation of GH1, GHR and DGAT1 polymorphisms with estimated breeding values for milk yield traits of German Holstein sires. Czech J. Anim. Sci. 53 (6), 238–245. doi: 10.17221/362-cjas
Kaupe B., Winter A., Fries R., Erhardt G. (2004). DGAT1 polymorphism in bos indicus and bos taurus cattle breeds. J. Dairy Res. 71 (2), 182–187. doi: 10.1017/S0022029904000032
Komisarek J., Kolenda M. (2016). The effect of DGAT1 polymorphism on milk yield traits in dairycows depending on environmental temperature. Turkish J. Veterinary Anim. Sci. 40 (2), 251–254. doi: 10.3906/vet-1508-7
Krovvidi S., Sudhakar K., Panneerselvam S., Thiruvenkadan A. K., Abraham J., Vinodkumar G. (2013). Factors effecting milk composition of crossbred dairy cattle in southern India. Int. J. Food Agric. Veterinary Sci. 3 (1), 229–232. doi: 10.1007/s11250-021-02560-2
Krovvidi S., Thiruvenkadan A. K., Murali N., Saravanan R., Vinoo R., Metta M. (2021). Evaluation of non-synonym mutation in DGAT1 K232A as a marker for milk yield traits in ongole cattle and murrah buffalo from southern India. Trop. Anim. Health yield 53 (118), 1–7. doi: 10.1007/s11250-021-02560-2
Larkin M. A., Blackshields G., Brown N. P., Chenna R., Mcgettigan P. A., McWilliam H., et al. (2007). Clustal W and clustal X version 2.0. Bioinformatics 23 (21), 2947–2948. doi: 10.1093/bioinformatics/btm404
Li F., Cai C., Qu K., Liu J., Jia Y., Hanif Q., et al. (2020). DGAT1 K232A polymorphism is associated with milk yield traits in Chinese cattle. Anim. Biotechnol 32(1-5). doi: 10.1080/10495398.2020.1711769
Li Y., Zhou H., Cheng L., Edwards G. R., Hickford J. G. H. (2021). Effect of DGAT1 variant (K232A) on milk traits and milk fat composition in outdoor pasture-grazed dairy cattle. New Z. J. Agric. Res. 64 (1), 1–13. doi: 10.1080/00288233.2019.1589537
Liu Z., Chen N., Shi X., Tsang B., Yu Y. H. (2007). Upregulation of myocellular DGAT1 augments triglyceride synthesis in skeletal muscle and protects against fat-induced insulin resistance. J. Clin. Invest. 117 (6), 1679–1689. doi: 10.1172/JCI30565
Liu K., Muse S. V. (2005). PowerMarker: an integrated analysis environment for genetic marker analysis. Bioinformatics 21 (9), 2128–2129. doi: 10.1093/bioinformatics/bti282
Manga I., Řiha H. (2011). The DGAT1 gene K232A mutation is associated with milk fat content, milk yield and milk somatic cell count in cattle (Short communication). Arch. Anim. Breed. 54 (3), 257–263. doi: 10.5194/aab-54-257-2011
Meredith B. K., Kearney F. J., Finlay E. K., Bradley D. G., Fahey A. G., Berry D. P., et al. (2012). Genome-wide associations for milk yield and somatic cell score in Holstein-friesian cattle in Ireland. BMC Genet. 13:1–11. doi: 10.1186/1471-2156-13-21
Molee A., Duanghaklang N., Na-Lampang P. (2012). Effects of acyl-CoA: diacylglycerol acyl transferase 1 (DGAT1) gene on milk yield traits in crossbred Holstein dairy cattle. Trop. Anim. Health yield 44 (4), 751–755. doi: 10.1007/s11250-011-9959-1
Mwai O., Hanotte O., Kwon Y. J., Cho S. (2015). Invited review - African indigenous cattle: unique genetic resources in a rapidly changing world. Asian-Australasian J. Anim. Sci. 28 (7), 911–921. doi: 10.5713/ajas.15.0002R
Myburgh J., Osthoff G., Hugo A., de Wit M., Nel K., Fourie D. (2012). Comparison of the milk composition of free-ranging indigenous African cattle breeds. South Afr. J. Anim. Sci. 42 (1), 1–13. doi: 10.4314/sajas.v42i1.1
Narayana S. G., Schenkel F. S., Fleming A., Koeck A., Malchiodi F., Jamrozik J., et al. (2017). Genetic analysis of groups of mid-infrared predicted fatty acids in milk. J. Dairy Sci. 100 (6), 4731–4744. doi: 10.3168/jds.2016-12244
Nasiri H., Forouzandeh M., Rasaee M. J., Rahbarizadeh F. (2005). Modified salting-out method: high-yield, high-quality genomic DNA extraction from whole blood using laundry detergent. J. Clin. Lab. Anal. 19 (6), 229–232. doi: 10.1002/jcla.20083
O’Mahony F. (1988). Rural dairy technology: experiences in Ethiopia (No. 4; dairy technology unit) (ILRI).
Patel J., Chauhan J. (2017). Evaluation of DGAT1-exon 8 K232A substitution in gir and kankrej (Bos indicus), Indian origin cattle and its association with milk yield traits. Genetika 49 (2), 627–634. doi: 10.2298/GENSR1702627P
Rahamattalla S., ReiBmann M., Mueller U., Brockmann G. A. (2015). Indentification of genetic variants influencing milk yield traits in Sudanese dairy cattle. Res. J. Anim. Sci. 9 (2-4), 12–22).
Rege J. E. O., Tawah C. L. (1999). The state of African cattle genetic resources II. geographical distribution, characteristics and uses of present-day breeds and strains. Anim. Genet. Resour. Inf. 26:1–25. doi: 10.1017/s1014233900001152
Rozen S., Skaletsky H. (2000). Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. (Clifton N.J.) 132:365–286. doi: 10.1385/1-59259-192-2:365
Samuel B., Mengistie D., Assefa E., Kang M., Park C., Dadi H., et al. (2022). Genetic diversity of DGAT1 gene linked to milk yield in cattle populations of Ethiopia. BMC Genomic Data 23 (1), 1–10. doi: 10.1186/s12863-022-01080-8
Valenti B., Criscione A., Moltisanti V., Bordonaro S., De Angelis A., Marletta D., et al. (2019). Genetic polymorphisms at candidate genes affecting fat content and fatty acid composition in modicana cows: effects on milk yield traits in different feeding systems. Animal 13 (6), 1332–1340. doi: 10.1017/S1751731118002604
Yen C. L. E., Nelson D. W., Yen M. I. (2015). Inassessinal triacylglycerol synthesis in fat absorption and systemic energy metabolism. J. Lipid Res. 56 (3), 489–501. doi: 10.1194/jlr.R052902
Yu Y. H., Ginsberg H. N. (2004). The role of acyl-CoA:diacylglycerol acyltransferase (DGAT) in energy metabolism. Ann. Med. 36 (4), 252–261. doi: 10.1080/07853890410028429
Keywords: DGAT1 K232A, Ethiopian cattle, frequency distribution, milk yield, milk quality
Citation: Samuel B, Dadi H and Dinka H (2023) Effect of the DGAT1 K232A mutation and breed on milk traits in cattle populations of Ethiopia. Front. Anim. Sci. 4:1096706. doi: 10.3389/fanim.2023.1096706
Received: 12 November 2022; Accepted: 03 April 2023;
Published: 01 May 2023.
Edited by:
Geoffrey E. Dahl, University of Florida, United StatesReviewed by:
Ludmila Zavadilová, Institute of Animal Science, CzechiaYogesh Bangar, Lala Lajpat Rai University of Veterinary and Animal Sciences, India
Copyright © 2023 Samuel, Dadi and Dinka. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Behailu Samuel, YmVoYWlsdXMyMDAxQGdtYWlsLmNvbQ==
 Behailu Samuel
Behailu Samuel Hailu Dadi
Hailu Dadi Hunduma Dinka1
Hunduma Dinka1