- 1Department of Animal Science, University of Tennessee, Knoxville, TN, United States
- 2Department of Animal Science, Texas A&M University, College Station, TX, United States
- 3United States Meat Animal Research Center, Agricultural Research Service, United States Department of Agriculture, Clay Center, NE, United States
Bacterial communities play major roles in rumen and uterine function toward optimal animal performance and may be affected by changes occurring during heifer development such as nutritional supplementation for optimal growth and the attainment of puberty. The effect of different levels of protein supplementation on ruminal and uterine bacterial communities following weaning was examined through first breeding of heifers. Angus heifers (n = 39) were blocked by initial body weight (BW) and randomly assigned to one of three 163-day (d) crude protein (CP) supplementation diets including control (10% CP, n = 14), 20% CP (n = 11), or 40% CP (n = 14) treatment groups. Growth and development were monitored by body weight, with blood progesterone concentration determined every 14 d to determine pubertal status. Uterine flush and rumen fluid were collected on d 56, 112, and 163 relative to the start of supplementation. Bacterial DNA was extracted from fluid samples, the V1–V3 hypervariable region of the 16S rRNA gene was amplified, and amplicons were sequenced then processed in R 4.1. Statistical analyses were performed in SAS 9.4 with a GLIMMIX procedure utilizing fixed effects of protein, month, pubertal status, and interactions, with random effects including BW, interaction of BW and protein, and heifer within the interaction, and repeated measures of day. In the uterus, pubertal status and day of supplementation affected the observed amplicon sequence variants (ASVs) and led to clustering of samples in a principal coordinate analysis (PCoA; P < 0.05), but no effect of protein supplementation was observed. Ruminal samples clustered in PCoA (P = 0.001), and observed ASVs were impacted over time (P < 0.0001), but no effect of protein supplementation was detected. In contrast, protein supplementation, pubertal status, and day of supplementation affected the abundance of multiple phyla and genera in the uterus and rumen (P < 0.05). Temporal and pubertal status effects on the heifer’s uterine bacterial communities potentially indicate a maturing uterine microbiome. Protein supplementation did not impact microbial diversity measures but did affect the abundance of individual bacterial phyla and genera that may provide future opportunities to manipulate bacterial community composition and maximize productivity.
Introduction
The development of replacement heifers is crucial for their incorporation into a breeding cow herd. The nutrition provided to heifers during the time of development can affect reproductive efficiency as it influences age at puberty, future success as a breeding cow, and longevity in the herd (Bellows and Short, 1971). Therefore, to support future reproductive performance, it is crucial to provide the required nutrition to meet the demands of the growing heifer. Heifers should ideally reach puberty early, attain approximately 60% of their mature body weight by the first breeding, and achieve a successful pregnancy with as few inseminations as possible to calve by 2 years of age (Larson, 2007; Kuehn et al., 2011; Perry, 2016). Weaned heifers are often developed during a time of dormant forages with lower quality in operations that employ defined breeding seasons. Protein is one of the most vital nutrients for development and reproduction but is often the first limiting nutrient in dormant forages (Marston et al., 1995). Previous studies have shown that supplementing heifers with protein increases their breeding success, muscle growth, and weight gain (Lalman et al., 1993; Martin et al., 2007; Dickinson et al., 2019). Therefore, supplemental feeds high in crude protein are often provided to heifers for continued growth and development for future reproductive success.
The rumen microbiome has been well established as the major producer of energy for the ruminant through the conversions of indigestible feed stuffs to provide volatile fatty acids (VFA), amino acids, metabolites, vitamins, and other useful nutrients (Bergman, 1990). Ruminal bacterial communities can be affected by different feedstuffs that vary in amount and quality of protein, and other components such as fiber and starches (Belanche et al., 2012). Shifts in the rumen bacterial community composition may alter the nutrients metabolized and absorbed for use by the ruminant, which has been shown to have effects systemically and in the reproductive tract through immune system responses (Zebeli et al., 2012; Bilal et al., 2016). Providing supplemented protein in excess has been shown to potentially impact the pH of the reproductive tract by increased plasma urea and ammonia reaching the uterus through histotrophic secretions of the endometrium (Elrod and Butler, 1993; Dawuda et al., 2002). Histotroph contains a variety of nutrients and other factors that contribute to overall uterine health and embryo development (Bazer et al., 2015). Therefore, differences in the nutrient profiles of variable feedstuffs, which impact the rumen microbial communities, could impact future reproductive success by altering the composition of endometrial secretions and the uterine environment via nutrient transporters.
The uterine microbiome of ruminants has been recently determined to be an important contributor to the uterine environment. The reproductive tract was originally thought to be essentially sterile, with the presence of bacteria indicating infection. The emergence of next-generation sequencing has provided the opportunity to recognize the reproductive tract as harboring its own distinct microbiome (Swartz et al., 2014; Laguardia-Nascimento et al., 2015; Clemmons et al., 2017). The composition of the uterine bacterial communities prior to breeding has been associated with differences in breeding outcomes (Ault et al., 2019a; Ault et al., 2019b). For maintenance of healthy bacterial communities in the uterus, the environment must be favorable for microorganisms to perform their metabolic processes, which can benefit reproductive success. However, there has been limited research on the factors that can impact the composition of the uterine bacterial community, potentially compromising the establishment and maintenance of pregnancy. Previous studies have suggested that excess protein and non-protein nitrogen in the diet can result in increased circulating urea and ammonia that can reach the uterus and decrease pH (Elrod and Butler, 1993; Dawuda et al., 2002; Ocon and Hansen, 2003). The observed impacts of diet on the uterine environment in cattle suggests that nutrition may influence the bacterial communities in the uterus. Additionally, differences in uterine bacterial communities have been detected between different phases of the estrous cycle and throughout estrus synchronization protocols (Quereda et al., 2020; Ault et al., 2019a; Ault et al., 2019b). Thus, the physiological changes that occur through the attainment of puberty and changes in hormone response in heifers may affect the uterine bacterial communities present. Through heifer development, the impact of nutrition on the uterine environment may provide the opportunity to influence uterine bacterial communities toward optimal reproductive health and efficiency.
The objective of the current study was to evaluate the effects of different levels of protein supplement on ruminal and uterine bacterial communities following weaning (pre-pubertal) through the first breeding (pubertal) of beef heifers. We hypothesized that 1) protein supplementation will affect developing heifer’s ruminal and uterine bacterial communities during development, and 2) bacterial communities will differ between prepubertal and pubertal heifers.
Materials and Methods
All animal experimental procedures for the current study were approved by the Institutional Animal Care and Use Committee at the University of Tennessee, Knoxville.
Experimental Design
Commercial Angus heifers (n = 60) housed at the Middle Tennessee Research and Education Center were enrolled in the study 23 days (d) following weaning. Figure 1 depicts the following study timeline with sampling and estrus synchronization days. On d 0, heifers were evaluated for baseline measurements including body weight (BW), body condition scores (BCS), and blood samples via the jugular vein for determination of pubertal status. Heifers were blocked by BW and assigned to a treatment group (n = 20 heifers per treatment): control supplement (CON; 1.59 kg corn per day), 20% crude protein (CP) supplement (P20; 1.59 kg per day: 75% dried distillers’ grains 25% corn), or 40% CP supplement (P40; 1.59 kg per day: 75% soybean meal 25% dried distillers’ grains). Heifers were maintained in 1.21-hectare pens with five heifers per pen resulting in four pens per treatment group. Supplements were provided four times each week for 140 d, beginning in early October through late March. Body weight, BCS, and blood samples were collected once every 2 weeks to monitor development and pubertal onset. Blood samples were centrifuged at 5,000 × g and 4°C for 20 min to collect serum. Serum samples were stored at -80°C until further analysis. Circulating progesterone (P4) samples were quantified in serum using the Double Antibody RIA Kit (MP Biomedicals, Irvine, CA) validated previously by Pohler et al. (2016). The progesterone concentration of ≥1 ng/ml in two consecutive samplings that corresponded with a normal estrous cycle indicated a functional corpus luteum, representing puberty (Polat et al., 2009). The heifer was considered pubertal at the time of the second sample of ≥1 ng/ml P4. Serum P4 concentrations at puberty, age at puberty, BW, and BCS for all heifers throughout development, with supplement and total diet feed analyses, were reported in Brandt (2020). Cytokine concentrations of the uterine flush leading up to breeding for all heifers were reported in Ault-Seay et al. (2021).
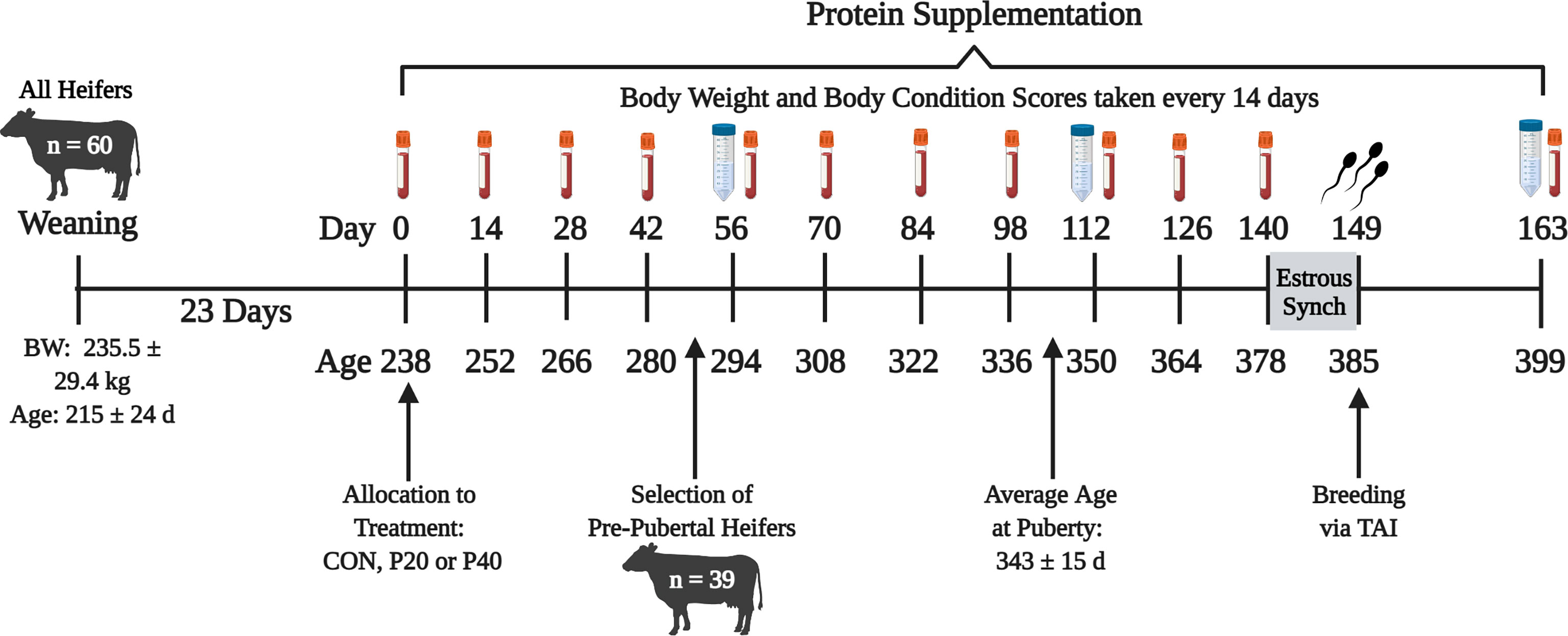
Figure 1 Timeline of experiment beginning at weaning of all heifers until 2 weeks after breeding. Red tubes indicate blood samples every 14 days. Blue tubes represent uterine flush and rumen fluid collection every 56 days. BW, body weight; CON, control 10% crude protein supplement; P20, 20% crude protein supplement; P40, 40% crude protein supplement; TAI, timed artificial insemination.
Only prepubertal heifers (n = 39) were selected to continue through the remainder of the study based on serum P4 concentrations from the four blood collections between d 0 through 42 (Figure 1), resulting in 14 heifers from the CON and P40 treatment groups and 11 heifers from the P20 treatment group. Uterine flushes and rumen fluid were collected every other month on d 56, 112, and 163, relative to the start of protein supplementation, to evaluate the bacterial communities present. For uterine flushes, a Foley catheter was inserted through the vagina and cervix into the uterus to deliver 20 ml of sterile saline, manipulated throughout the uterus by rectal massage, then collected by syringe (Clemmons et al., 2017). Rumen fluid (~50 ml) was collected by oroesophageal tubing into the rumen attached to a hand pump with resulting fluid collected into a flask (Guan et al., 2008). Foley catheters and oroesophageal tubing were disinfected and washed between each heifer to eliminate contamination. Uterine flush and rumen fluid were immediately placed on ice for transport then stored at -80°C until bacterial community analysis.
Prior to the final uterine flush and rumen fluid collections on d 163, heifers were subjected to estrus synchronization utilizing an industry standard 7-Day Co-Synch + CIDR protocol beginning on d 140. Controlled intravaginal drug release devices (CIDR; 1.38 g P4; Zoetis Animal Health, Florham Park, NJ) were inserted, and gonadotropin-releasing hormone (100 µg; Cystorelin, Boehringer Ingelheim, Duluth, GA) was administered i.m. on d 140. An injection of prostaglandin F2α (500 µg; cloprostenol sodium; Synchsure, Boehringer Ingelheim, Duluth, GA) was administered i.m. at CIDR removal on d 147. Approximately 52 h after CIDR removal, heifers were administered GnRH, followed by artificial insemination on d 149.
Bacterial DNA Extraction, Library Preparation, and Sequencing
Uterine flush bacterial DNA extraction was performed according to methods from Clemmons et al. (2017) and recommendations for low-biomass samples (Weinroth et al., 2022). Samples were thawed to room temperature (22°C), and 5-ml aliquots were placed in 15-ml conical tubes for centrifugation at 5,000 × g and 4°C for 10 min. The supernatant was removed, and the resulting pellet was resuspended in enzymatic lysis buffer (4% (w/v) sodium dodecyl sulfate (SDS), 500 mM NaCl, and 50 mM EDTA) for incubation at 37°C. Bacterial DNA was extracted using the DNEasy Blood and Tissue kit (Qiagen, Hilden, Germany) according to manufacturer protocol. Extracted DNA concentrations were determined using a spectrophotometer (DeNovix, Wilmington, DE, USA) then stored at -20°C.
Bacterial DNA was extracted from rumen fluid using a modified method validated by Yu and Morrison (2004). Samples were thawed to room temperature (22°C), and approximately 0.2 g of rumen fluid was transferred to a ZR BashingBead Lysis Tube (Zymo Research Corp., Santa Ana, CA, USA) with lysis buffer for chemical lysis. Samples were placed in the TissueLyser II system (Qiagen, Hilden, Germany) for mechanical lysis at 21 Hz for 3 min. Cell debris was removed using 10 M ammonium acetate, then nucleic acids were precipitated using isopropanol. Proteinase K and RNase were used to remove all proteins and RNA. Lastly, DNA samples were purified by centrifugation in QIAmp columns from the QIAamp DNA Stool Mini Kit (Qiagen, Hilden, Germany). Resulting DNA concentrations were quantified via using a spectrophotometer (DeNovix, Wilmington, Delaware) then stored at -20°C.
Library preparation and sequencing were performed at the US Meat Animal Research Center (Clay Center, NE, USA). Bacterial DNA was amplified using polymerase chain reaction (PCR) with AccuPrime Taq high-fidelity DNA polymerase (Life Technologies, Carlsbad, CA, USA) for library preparation. Primers 27F (5′-Adapter/Index/AGAGTTTGATCCTGGCTCAG) and 519R (5′-Adapter/Index/GTATTACCGCGGCTGCTG) with TruSeq indices were used to target the V1–V3 hypervariable region of the bacterial 16S rRNA gene (Myer et al., 2015). The resulting rumen and uterine bacterial DNA libraries were sequenced using the 2 × 300, v3 600-cycle kit on the Illumina MiSeq platform (Illumina, Inc., San Diego, CA, USA).
Sequence Processing and Statistical Analyses
Sequence quality filtering, trimming, and taxonomic assignment were performed in R 4.1 using the dada2 pipeline (Callahan et al., 2016). Forward sequences were filtered with standard dada2 parameters and a truncation length of 270. Resulting sequences were used to construct an amplicon sequence variant (ASV) table. Chimeric sequences were identified and removed, then taxa were assigned using the SILVA database version 138 (Quast et al., 2013). Metadata were imported and merged with the taxa table for alpha and beta diversity analyses using the phyloseq package (McMurdie and Holmes, 2013) in R 4.1. Unassigned reads were classified to their last known taxonomic classification, and sequences assigned as Eukaryota or Cyanobacteria were removed for all downstream analyses. Alpha diversity indices were calculated for observed species by observed ASVs, and richness and evenness using the Shannon Diversity Index. Beta diversity analyses utilized the principle coordinate analysis (PCoA) method with Bray Curtis distances. Visualizations were generated with ggplot2 including ellipses representing a 95% confidence interval for each cluster. Relative abundance plots were generated for taxa using ggplot2 for the top 10 phyla and genera with a relative abundance ≥1%. Genera with a relative abundance <1% were combined to one group for visualization.
For beta diversity, the vegan package (Oksanen et al., 2017) with the adonis function in R 4.1 was used to perform a PERMANOVA with 999 permutations to determine the significance of each PCoA. Statistical analyses to determine differences in alpha diversity indices and bacterial taxa abundances were performed in SAS 9.4 (SAS Institute, Cary, NC, USA). Normality of observed ASVs and bacterial taxa raw abundance values were evaluated by the univariate procedure. Data were determined to be non-normal with a Shapiro–Wilk value of W < 0.80. Non-normal data were normalized by log transformation to achieve a normal distribution and Shapiro–Wilk value of W > 0.85. A completely randomized block design with repeated measures was utilized with the GLIMMIX procedure including fixed effects of day of sampling, protein supplementation treatment, and pubertal status, and interactions of day × protein and protein × pubertal status. Random effects included the block of BW, interaction of BW × protein, and heifer within the interaction, with repeated measures of day. For bacterial taxa abundance data, means are reported as the mean raw abundance value × 100 to indicate percent relative abundance out of 100%. Significance was determined by P ≤ 0.05 and trends by 0.05 ≤ P ≤ 0.10 for all analyses.
Results
Cluster analysis of 16S rRNA amplicon sequences by PCoA among all samples indicated distinct separation in beta diversity of microbial communities between the uterus and rumen (R2 = 0.13, P = 0.001, Figure 2). Rumen samples clustered tightly indicating a high degree of similarity of bacterial community composition among all rumen samples. Uterine samples clustered loosely indicating higher dissimilarity of microbial communities among uterine samples (Figure 2). A total of 39 phyla and 1,633 genera were detected among all uterine and ruminal samples. Supplementary Figure 1 illustrates the overall relative abundance of the top 10 phyla detected among all samples between the uterus and rumen. Firmicutes and Bacteroidota were the most abundant phyla in the uterus and rumen, respectively, independent of protein supplementation treatment, pubertal status, or day of sampling. Supplementary Figure 2 depicts genera with a relative abundance ≥1% in the uterus and rumen, with all genera present in <1% grouped. Twenty-three genera in the uterus and 25 genera in the rumen were detected with relative abundances ≥1%. The total average relative abundance of genera with <1% abundance was 30.27% in the uterus, and 20.01% in the rumen. Prevotella was the most abundant genus in both uterine and ruminal samples across all groups.
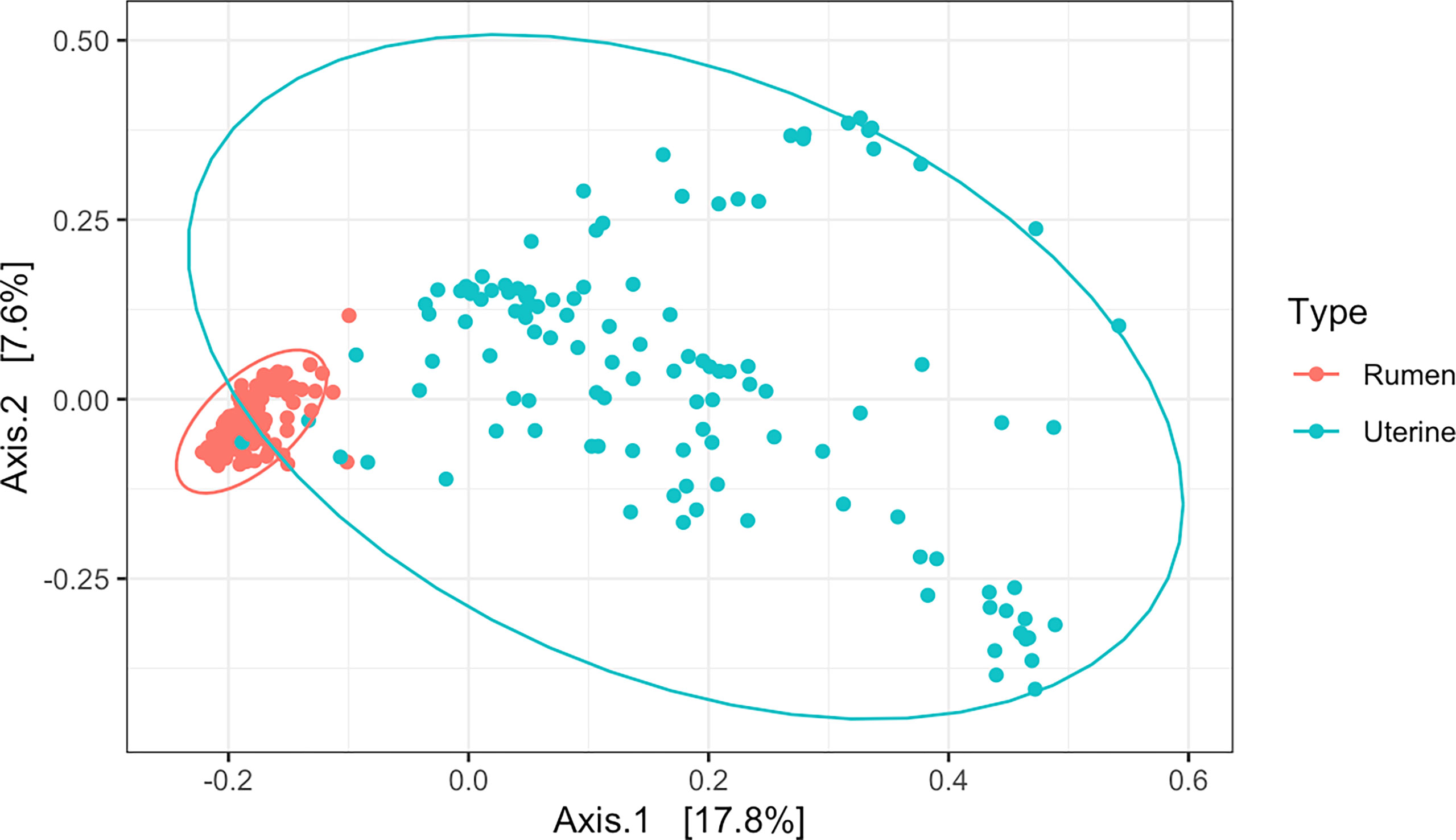
Figure 2 Beta diversity by principal coordinate analysis with Bray Curtis distances between all uterine and rumen samples (R2 = 0.13, P = 0.001). Significance was determined by PERMANOVA with 999 permutations. Ellipses represent 95% confidence interval.
Protein Supplementation Level
The level of protein supplementation did not affect alpha diversity metrics, observed ASVs, or Shannon’s Diversity Index, in the uterus or rumen (Table 1). Similarly, no clear clustering pattern by protein supplementation level was observed by the beta diversity PCoA in uterine (R2 = 0.02, P = 0.06, Figure 3A) or ruminal (R2 = 0.02, P < 0.01, Figure 3B) samples, despite calculated significance in the rumen. However, multiple phylum and genus relative abundances were affected in both the uterus and rumen by protein supplementation level.
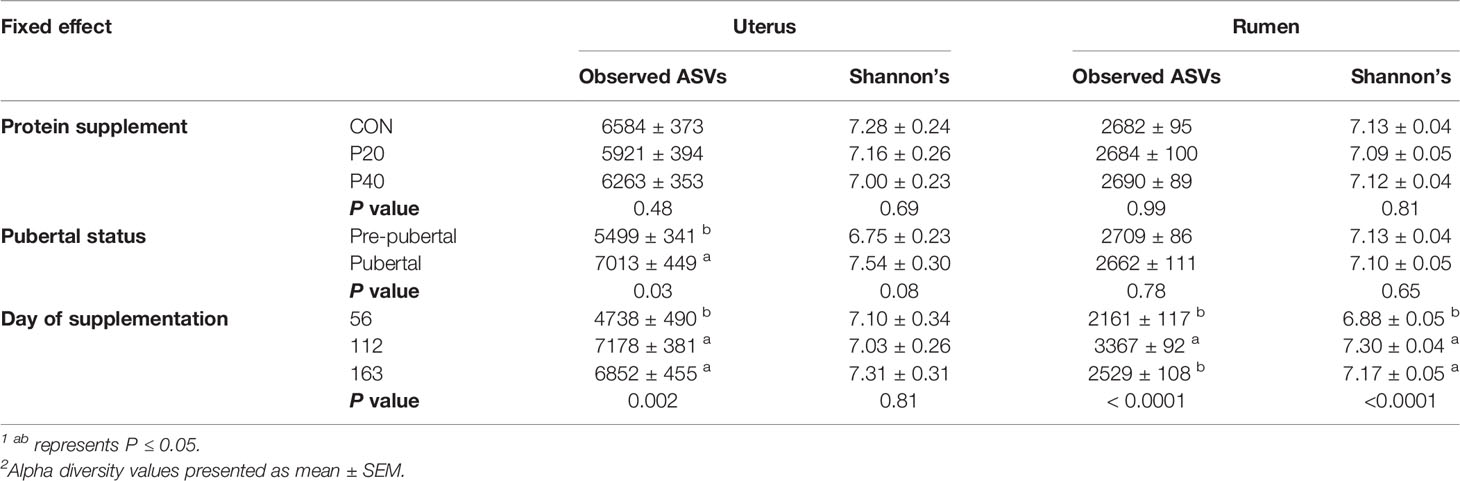
Table 1 Impact of protein supplement, pubertal status, and day of supplementation on alpha diversity metrics observed ASVs and Shannon’s Diversity Index in the uterus and rumen1,2.
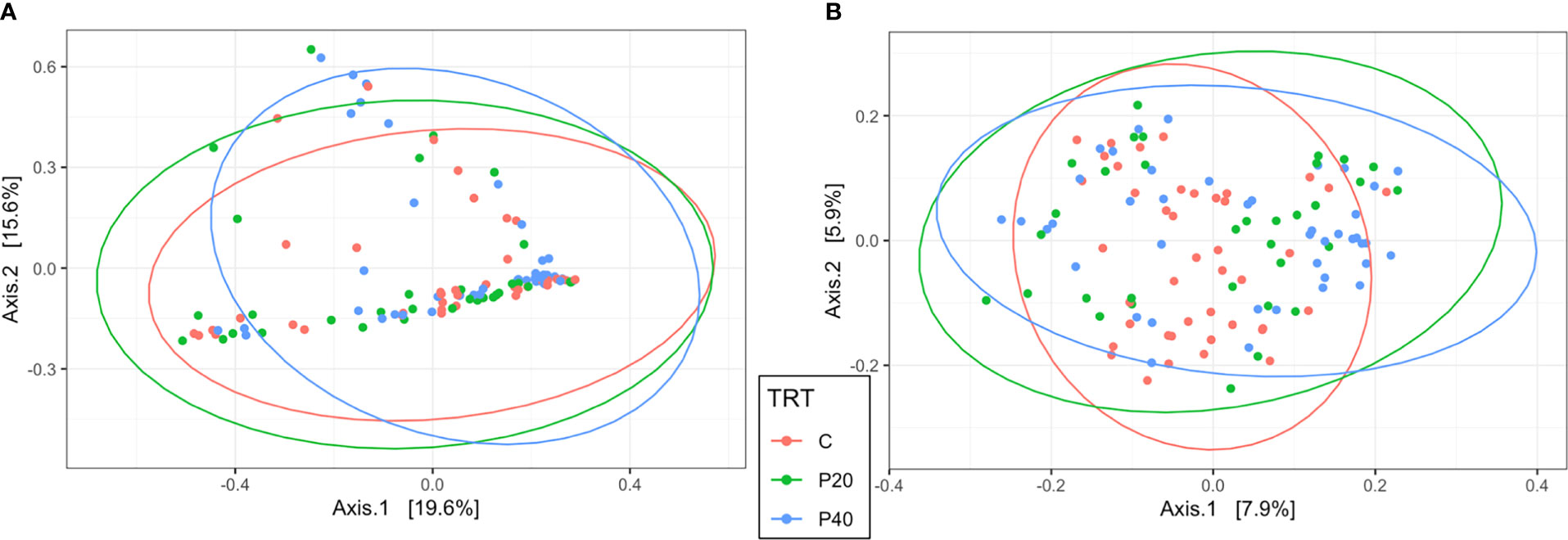
Figure 3 Beta diversity by principal coordinate analysis with Bray Curtis distances for protein supplementation treatment in the uterus (A; R2 = 0.02, P = 0.06) and rumen (B; R2 = 0.02, P = 0.008). Significance was determined by PERMANOVA with 999 permutations. Ellipses represent 95% confidence interval. CON, control, 10% crude protein supplement; P20, 20% crude protein supplement; P40, 40% crude protein supplement.
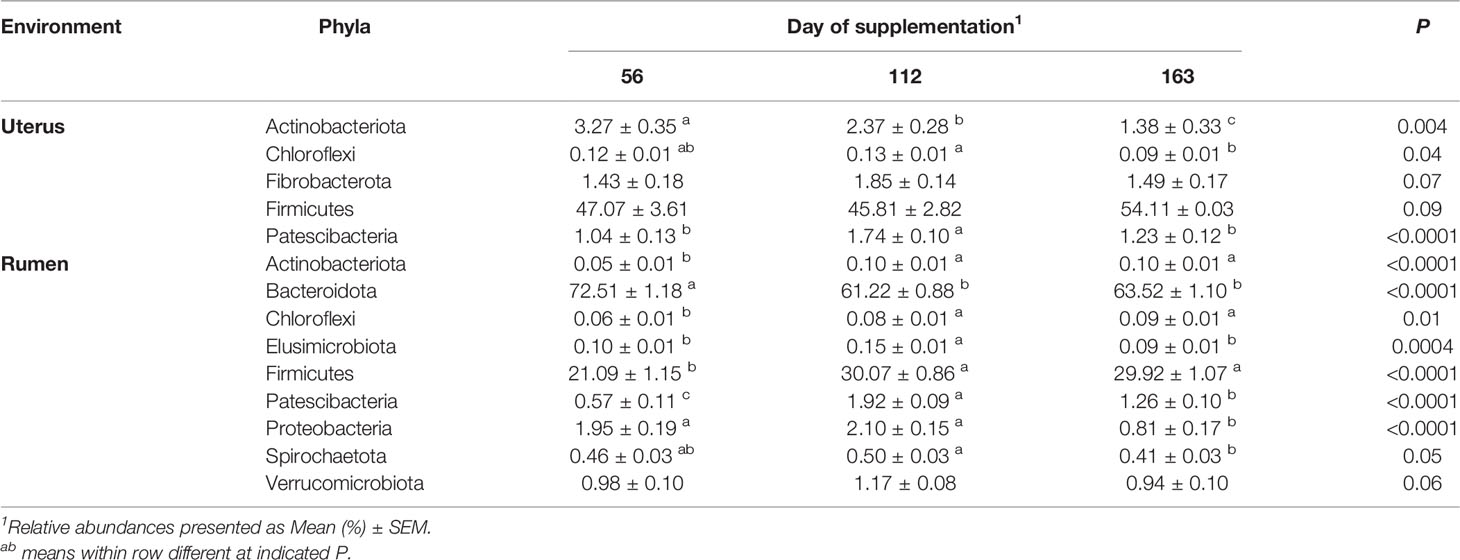
In the uterus, Firmicutes was the most abundant phyla and was affected by protein treatment, with P20 having the greatest and P40 having the least abundance (P = 0.05, Table 2). Fibrobacterota and Proteobacteria abundances were affected by protein supplementation (P ≤ 0.01) and tended to affect the abundance of Chloroflexi (P = 0.09; Table 2). In the rumen, Bacteroidota abundance increased with increasing crude protein as P40 heifers had the greatest abundance, and control heifers with the least (P = 0.02; Table 2). Actinobacteriota abundance in the rumen was also affected by protein supplementation, with P20 heifers having the greatest abundance (P = 0.02; Table 2). Additionally, Firmicutes, Proteobacteria, and Spirochaetota abundances tended to be affected by protein supplementation (P < 0.10; Table 2).
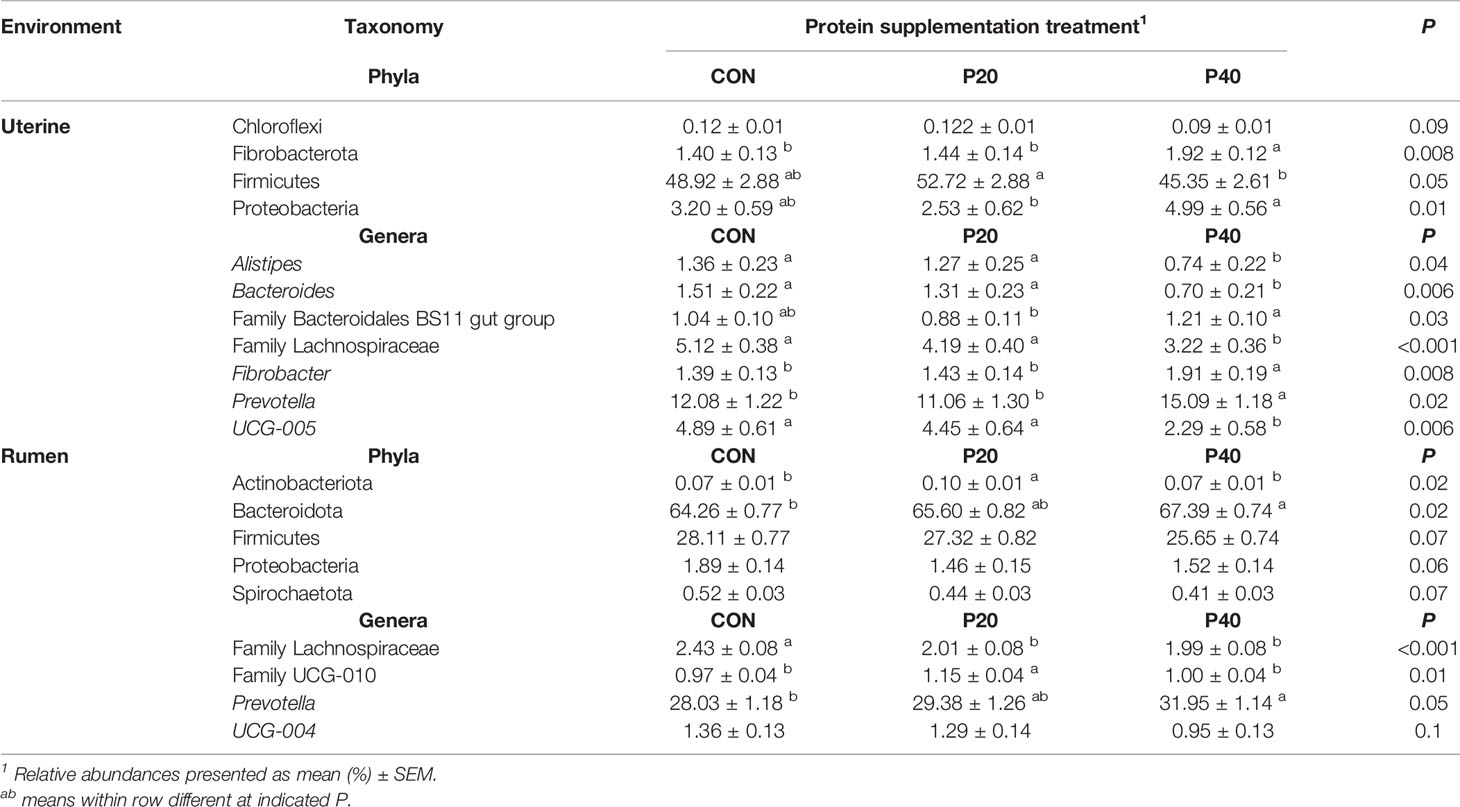
Table 2 Bacterial phyla and genera with relative abundance ≥1% in the uterus and rumen impacted by protein supplementation treatment.
Relative abundances of genera Alistipes, Bacteroides, Fibrobacter, Prevotella, UCG-005, and unclassified genera from Family Bacteroidales BS11 and Family Lachnospiraceae were affected by protein supplementation in the uterus (P < 0.05; Table 2). The most abundant uterine genus, Prevotella, had the greatest abundance in the P40 supplement group, with lesser, similar abundances in the CON and P20 groups (P = 0.02; Table 2). Similarly in the rumen, Prevotella was the most abundant genus and increased in abundance with increasing crude protein level of the supplement (P = 0.05; Table 2). The protein supplementation level also affected relative abundances of the unclassified genera from Family Lachnospiraceae and Family UCG-010 (P ≤ 0.01) in the rumen and tended to affect UCG-004 (P = 0.10; Table 2).
Pubertal Status
In the uterus, observed ASVs and Shannon’s Diversity Index were affected by pubertal status. Pubertal heifers had a greater number of observed ASVs (P = 0.03) and tended to have a higher Shannon Diversity Index value (P = 0.08), than prepubertal heifers (Table 1). Clustering was observed in the PCoA by pubertal status in uterine samples (R2 = 0.03, P = 0.001), depicting high variation in prepubertal heifers and tighter clustering of pubertal heifers (Figure 4A). However, pubertal status did not impact rumen alpha diversity metrics or beta diversity PCoA clustering (R2 = 0.02, P > 0.05; Figure 4B).
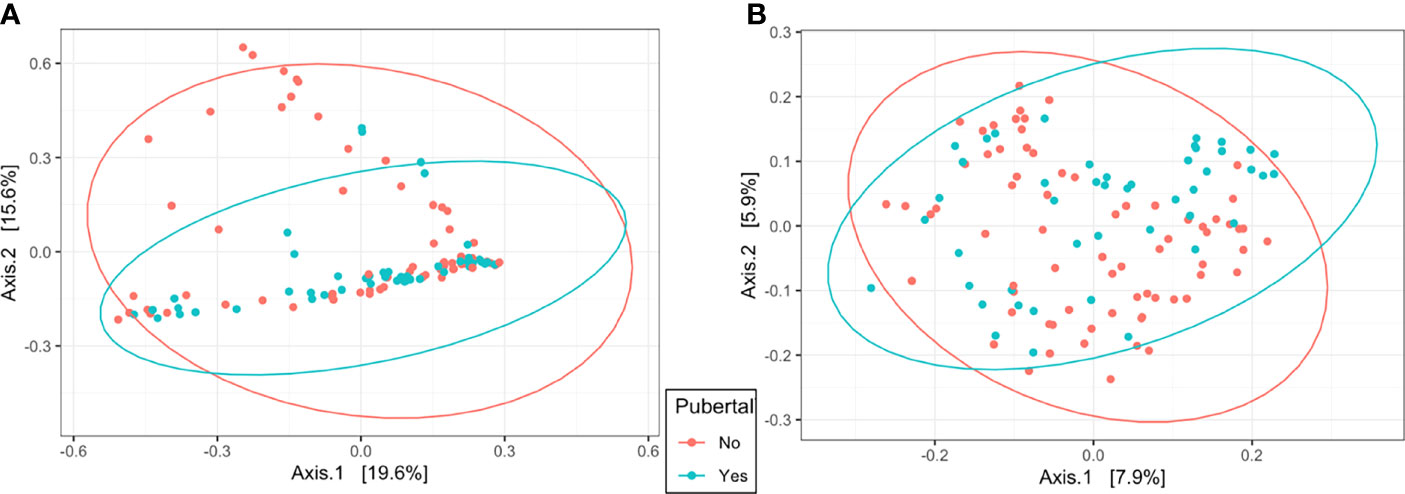
Figure 4 Beta diversity by principal coordinate analysis with Bray Curtis distances for pubertal status in the uterus (A; R2 = 0.03, P = 0.001) and rumen (B; R2 = 0.02, P = 0.01). Significance was determined by PERMANOVA with 999 permutations. Ellipses represent 95% confidence interval.
Pubertal status affected the abundance of the most abundant uterine bacterial phyla, Firmicutes, with prepubertal heifers having the greatest abundance compared to pubertal heifers (P = 0.04 Table 3). The abundance of Firmicutes was less in pubertal heifers, although multiple phyla were greater or tended to be greater in pubertal heifers including Bacteroidota (P = 0.06), Elusimicrobiota (P = 0.05), Proteobacteria (P = 0.04), and Verrucomicrobiota (P = 0.07; Table 3). In the rumen, pubertal status did not affect the abundance of Bacteroidota, but did affect Actinobacteria, Firmicutes, and Proteobacteria (P < 0.05; Table 3). Similar to the uterus, Firmicutes was also decreased in the rumen, while Proteobacteria was increased in pubertal heifers compared to prepubertal heifers (Table 3).
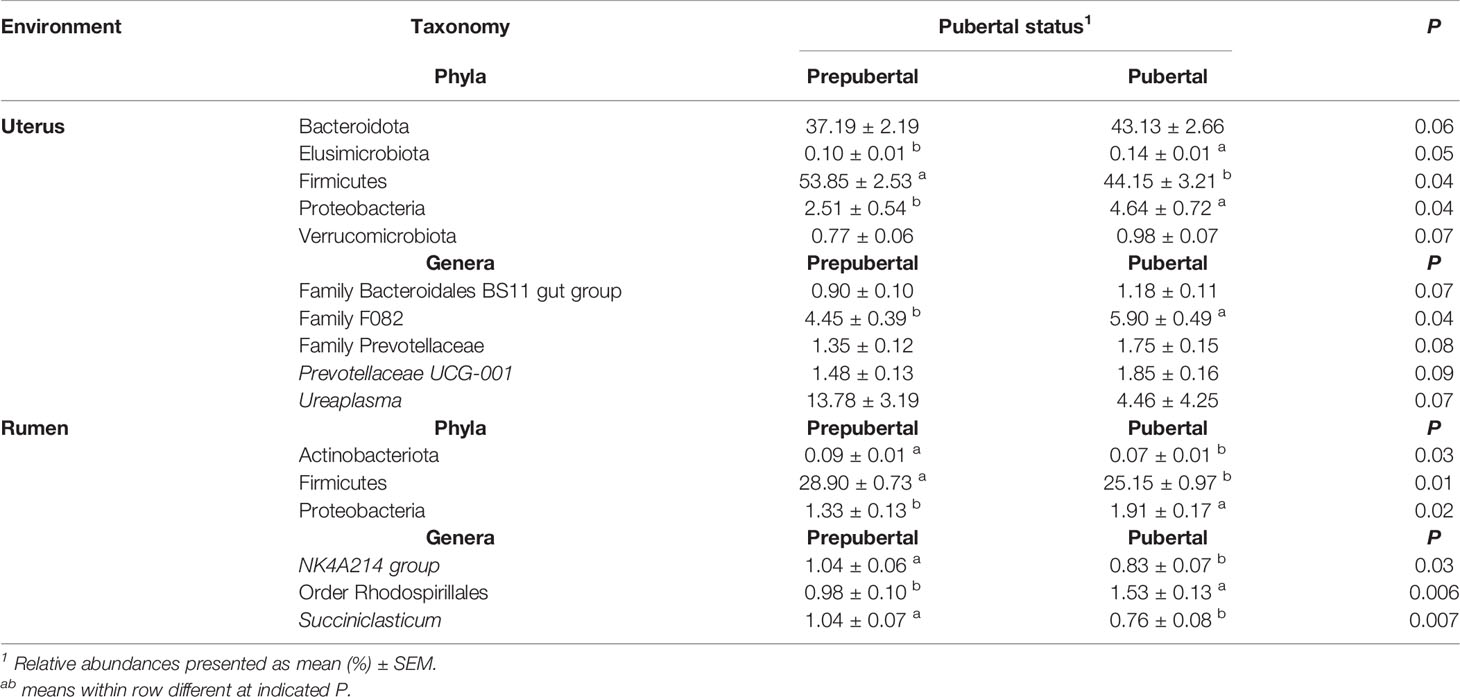
Table 3 Bacterial phyla and genera with relative abundance ≥1% in the uterus and rumen impacted by pubertal status.
The relative abundance of unclassified genera from Family F082 (P = 0.04) was affected in the uterus by pubertal status and tended to affect Prevotellaceae-UCG001, Ureaplasma, and unclassified genera from Family Bacteroidales BS11 and Family Prevotellaceae (P < 0.10; Table 3). All of these genera had a greater relative abundance in pubertal heifers, except Ureaplasma (Table 3). Puberty affected the ruminal relative abundance of NK4A214 and Succiniclasticum, which were greater in prepubertal heifers and unclassified genera from Order Rhodospirillales which was greater in pubertal heifers (P < 0.05; Table 3).
Day of Supplementation
Observed ASVs in the uterus were affected by day of supplementation. The least number of observed ASVs was detected on d 56 then significantly increased to the greatest observed ASVs by d 112 and remained similar on d 163 (P < 0.001; Table 1). The Shannon Diversity Index followed a similar trend but was not significant (Table 1). Ruminal observed ASVs and Shannon’s Diversity were affected by day, with the greatest number detected on d 112 and less observed ASVs on d 56 and d 163 (P < 0.0001; Table 1).
Beta diversity analysis by PCoA indicated uterine samples clustered by day with overlap (R2 = 0.10, P = 0.001, Figure 5A). Samples on d 56 had the greatest variation overlapping with d 112 and 163 uterine samples. However, d 112 and 163 samples clustered tightly and depicted the greatest separation between days, with slight overlap (Figure 5A). Rumen samples had similar PCoA clustering by day as the uterus by day with overlap (R2 = 0.08, P = 0.001, Figure 5B). The greatest separation was observed between d 56 and 112 indicating the greatest difference in overall community composition.
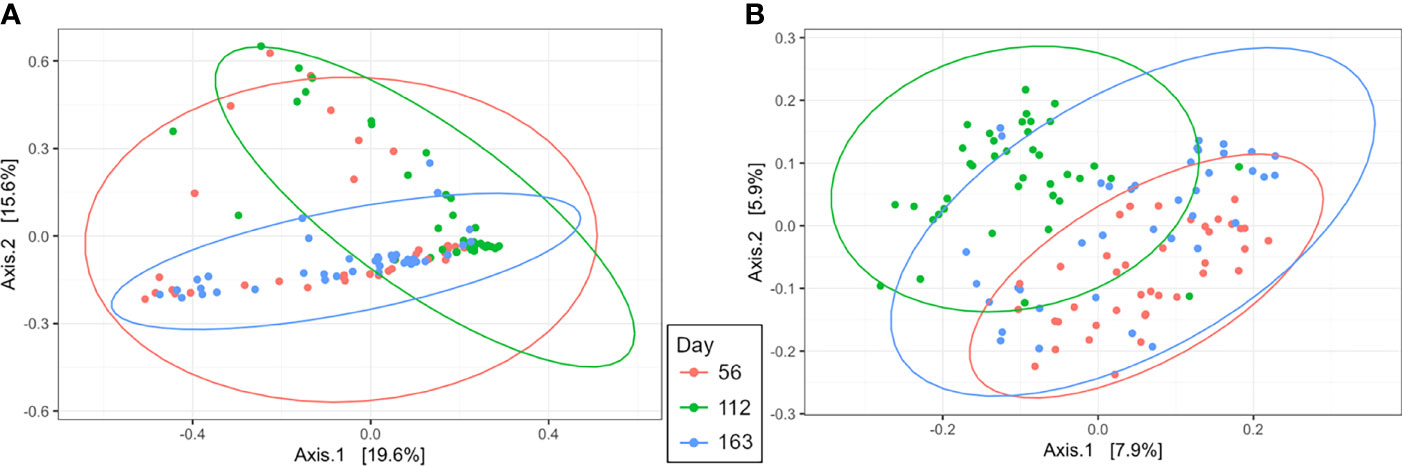
Figure 5 Beta diversity by principal coordinate analysis with Bray Curtis distances for day of supplementation in the uterus (A; R2 = 0.10, P = 0.001) and rumen (B; R2 = 0.08, P = 0.001). Significance was determined by PERMANOVA with 999 permutations. Ellipses represent 95% confidence interval.
In the uterus, day of supplementation tended to affect the abundance of Firmicutes (P = 0.09, Table 4). Multiple additional phyla also shifted over time including Actinobacteriota, Chloroflexi, and Patescibacteria (P < 0.05; Table 4), with a tendency for Fibrobacterota (P = 0.07). All top 10 phyla detected in the rumen were affected by the day of sampling (P ≤ 0.05; Table 4), except Fibrobacterota. The most abundant phyla in the rumen, Bacteroidota, had the greatest abundance on d 56, with lesser, similar abundances on d 112 and 163 (P < 0.0001; Table 4). The majority of genera in the rumen and uterus had shifts in their abundance over time; therefore, Table 5 indicates the top 5 most abundant genera that shifted over time in each environment, with all other affected genera listed in Supplementary Table 1. There were 23 genera detected in the uterus with a relative abundance ≥1%, of which 16 shifted over time (P ≤ 0.10). All rumen genera detected with relative abundance ≥1% shifted over time (P ≤ 0.10), except for unclassified genera from Family Prevotellaceae.
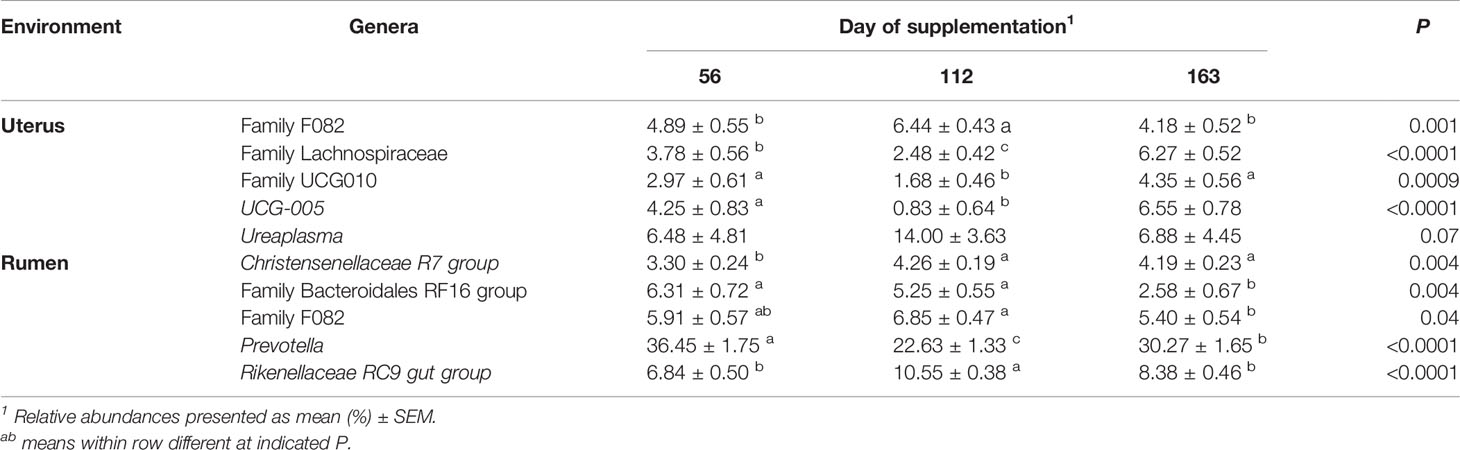
Table 5 Top 5 most abundant bacterial genera in the uterus and rumen impacted by day of supplementation.
Interactions of Protein Supplementation, Day of Supplementation, and Pubertal Status
The triple interaction of protein supplementation level, day of supplementation, and pubertal status, and the interaction of day of supplementation × pubertal status, was not able to be analyzed in the current study due to all heifers being prepubertal at the first sampling on d 56. Alpha diversity metrics in the uterus and rumen had no significant interaction of protein supplementation level × day of supplementation, or for any of the top 10 uterine bacterial phyla relative abundances. For uterine genera, however, a protein supplementation × day interaction was observed for UCG-005 (P = 0.04) and unclassified genera from Family Lachnospiraceae (P < 0.01). In the rumen, a supplement × day interaction was detected for phyla Bacteroidota (P = 0.01), Firmicutes (P = 0.02), and Spirochaetota (P = 0.04), and tendencies for Actinobacteria (P = 0.10) and Chloroflexi (P = 0.09). Ruminal genera Prevotella, UCG-004, and unclassified genera from Family Lachnospiraceae (P ≤ 0.05) were affected by the interaction. Neither alpha diversity metrics nor relative abundance of genera showed significant effects for the interaction of protein supplementation level × pubertal status in the rumen or uterus. The only significant interaction detected was the phyla Actinobacteria in the rumen (P = 0.03).
Discussion
Ruminal and uterine bacterial communities have been established as playing important roles in feed efficiency and breeding outcome success, respectively (Myer et al., 2015; Ault et al., 2019a). Therefore, bacterial communities of these environments may be affected during heifer development and potentially influence the long-term productivity of the heifer. Physiological changes associated with attainment of puberty, and nutritional supplementation commonly applied to support heifer growth and development, may influence the composition and effects of these microbial communities. The first objective of the current study was to evaluate the impact of differing levels of supplemented crude protein throughout heifer development on ruminal and uterine bacterial communities. Ruminal bacterial communities are well known to shift with changes in dietary feedstuffs or nutrient composition (McCann et al., 2014; Clemmons et al., 2019a; Gruninger et al., 2019). As expected, we observed that ruminal bacterial communities were affected as a result of differences in protein supplementation treatment consistent with previous studies (Table 2). Prevotella, the most abundant genus detected in the rumen, and its respective phylum Bacteroidota were observed to have greater abundances as supplement crude protein level increased. Prevotella spp. are commonly the dominant genus in the rumen, although abundances may differ by the type of diet such as a greater concentrate compared to fiber diet (Stevenson and Weimer, 2007; Pitta et al., 2014). Additionally, abundance of Prevotella has been greater in heifers than mature cows (Liu et al., 2017) and different between cattle of differing feed efficiencies (Carberry et al., 2012; Myer et al., 2015). Prevotella spp. are functionally diverse but are well established as major proteolytic bacterial species in the rumen and capable of proteolysis via a variety of mechanisms (Wallace, 1996; Griswold et al., 1999). Multiple studies have observed a greater abundance of Prevotella spp. in diets with greater crude protein content (Belanche et al., 2012; Pitta et al., 2014), as observed in the current study. Due to the high proteolytic activity of Prevotella, increases in its abundance potentially impact nitrogen utilization and amino acids reaching the lower gastrointestinal tract for absorption and use by the host (Bach et al., 2005). However, additional data from the current study did not observe changes in uterine pH based on protein supplementation treatment, and only two amino acid concentrations in the uterine luminal fluid were impacted (Brandt, 2020).
Potential impacts of nutrition on uterine bacterial communities in bovine have not been previously studied. The current study applied a common base diet across all heifer groups with only crude protein level of the supplement modified. With the protein supplement, overall diet crude proteins were 11%, 15%, and 19% among groups. Regardless, the relative abundance of multiple bacterial phyla and genera was affected in the uterus by the level of crude protein supplement (Table 2). Firmicutes was the most abundant bacterial phylum detected in the uterus, but Prevotella was the most abundant genus and increased in abundance with increasing supplemented crude protein, similar to ruminal Prevotella. The relative abundance of Prevotella spp. in the uterus of healthy cows has been previously observed to range between <2% and 12% on average (Clemmons et al., 2017; Ault et al., 2019a). Interestingly, increased Prevotella spp. in the uterus is typically associated with uterine disease in postpartum cows (Sheldon et al., 2009; Galvão et al., 2019) and was one of eight bacteria that differed in abundance between successful (<1% abundance) and failed (>1% abundance) pregnancies in apparently healthy postpartum cows (Ault et al., 2019a), indicating potential to influence breeding success. However, limited research exists on the uterine microbiome of heifers between weaning and breeding. The presence of Prevotella, and other uterine disease-associated bacteria, have been found on endometrial biopsies from the uterus of virgin, pubertal heifers (Moore et al., 2017). The current study also found common uterine pathogen Ureaplasma to also be highly abundant in the uterus of heifers, but relative abundance was not affected by protein supplementation level and did not shift over time. Other bacteria not associated as uterine pathogens were also affected by protein supplementation, such as Alistipes and Bacteroides, with the greatest abundance detected in control and P40 groups. The abundance of these bacteria was detected in >1% relative abundance in uterine samples but not rumen samples. Alistipes and Bacteroides have been previously associated with greater abundances in the lower gastrointestinal tract and feces of ruminants (Holman and Gzyl, 2019). Vaginal bacterial communities commonly share genera with gastrointestinal and fecal bacteria, which can enter into the uterus (Clemmons et al., 2017), or possibly enter the uterine environment through blood (Jeon et al., 2017). Alistipes and Bacteroides have been recognized as core vaginal bacteria (Rodrigues et al., 2015), and proposed as potential predictors of reproductive success based on abundance in fecal samples (McClure, 2018). Further research is needed to understand the role of potentially pathogenic bacteria from gastrointestinal sources, and their effects on the uterine environment and endometrial tissue. Despite the minimal differences in overall diet crude protein, differences in uterine bacterial communities by protein supplementation treatments were detected. Greater differences in diet such as different forage to concentrate ratios, energy supply, or other overall nutrient levels may have a larger impact on additional bacteria in the uterus. Therefore, the ability of protein supplementation to affect the abundances of bacteria in the uterus may allow for manipulation of the microbiome toward an optimal uterine environment.
The attainment of puberty is a significant milestone in heifer development. The activity of endometrial tissues and the uterine environment changes with the onset of puberty including tissue gene expression, hormonal signal response, and uterine histotrophic secretions (Cánovas et al., 2014; Fortes et al., 2018). Therefore, an additional objective of the current study was to evaluate the impact of pubertal status on bacterial communities, beginning with prepubertal heifers and ending 2 weeks following the first breeding. Alpha diversity metrics indicated differences in the bacterial community of the uterus during pubertal attainment, as observed ASVs increased over time and were greater in pubertal than prepubertal heifers (Table 1). In a study of the vaginal microbiome between cows and heifers, cows had a greater number of observed bacterial species than heifers (Laguardia-Nascimento et al., 2015). Pubertal heifers also had increased similarity of their overall uterine microbiome than prepubertal heifers according to beta diversity analyses of the current study (Figure 4A). When evaluating specific bacterial taxa in the uterus of our study, five phyla, including the two most abundant of Firmicutes and Bacteroidota, were affected by pubertal status (Table 3). Firmicutes was greater in prepubertal than pubertal heifers, but the remaining four were observed to have greater relative abundances in pubertal heifers compared to prepubertal heifers. Similar results were observed previously in cows during estrus synchronization leading up to breeding (Ault et al., 2019a) where Firmicutes was highly abundant (61%–74% abundance) and dominated the uterine bacterial communities at the beginning of estrous synchronization during low P4 concentrations. Firmicutes then decreased to 36% on average, with other phyla significantly increasing, 2 days before breeding during higher P4 concentrations (Ault et al., 2019a). Luteal function and resulting P4 concentrations are the defining parameter of pubertal animals, indicative of their ability to successfully ovulate and establish and maintain a pregnancy (Kinder et al., 1995; Atkins et al., 2013; Fortes et al., 2018). Progesterone influences uterine function through changes in endometrial gene expression related to activities such as immune response, nutrient exchange to the lumen, and other processes related to the establishment of pregnancy (Forde et al., 2010; Forde et al., 2014; Fortes et al., 2018). Companion data from the current study reported by Brandt (2020) found that 18 amino acids, regardless of protein supplementation treatment, reached their highest concentration in the uterus at d 163 indicating changes to the uterine environment through development to puberty attainment. These changes occurring with pubertal onset from P4 stimulation to the uterine environment and endometrial tissue may influence the bacterial communities of the uterus, leading to a more similar overall uterine microbiome of pubertal animals.
Furthermore, there was no difference in observed ASVs and fewer bacterial phyla and genera were affected by pubertal status in the rumen in contrast to the uterus (Tables 1, 3). Steroid hormone receptors associated with reproduction, such as estrogen and androgen receptors, have been reportedly expressed in the rumen, but P4 nuclear receptor expression has not been detected (Pfaffl et al., 2003). Therefore, minimal direct impact on the bacterial communities is expected in the rumen due to the changes specifically associated with puberty and increased P4 secretion. Temporal shifts in bacterial diversity and relative abundances of taxa, however, were observed in the current study of both the rumen and uterus (Tables 4, 5). Alpha diversity analyses indicated changes in the number of ASVs, and beta diversity indicated a slight shift in the rumen overall rumen bacterial community composition over time (Table 1). Ruminal bacterial communities have been shown to undergo shifts according to a change in diet (McCann et al., 2014; Clemmons et al., 2019a; Gruninger et al., 2019). However, previous research has indicated stability of the rumen microbiome by 9 weeks following a diet change (Clemmons et al., 2019b). Sampling of the rumen bacterial communities in the current study did not occur until 56 days following the beginning of supplementation. Therefore, the rumen microbiome had likely reached any diet-induced equilibrium, following the addition of supplements to the heifer’s diet, prior to sampling. The development of the rumen microbiome from birth to adulthood is not an immediate process, with the number of species shown to increase from birth up to 2 years of age (Guo et al., 2020), along with shifts in bacterial taxa composition (Jami et al., 2013). The first few days of life and weaning are likely the two most influential periods on rumen microbiome development (Guo et al., 2020; Amin and Seifert, 2021). Therefore, the rumen microbiome of heifers in the current study was likely continuing to develop following weaning into its mature state.
In conclusion, weaning through breeding during heifer development is a crucial period to evaluate the uterine and ruminal bacterial communities and factors that may influence their composition, such as nutrition and puberty attainment. Despite relatively minor changes in overall diet crude protein, the current study was able to detect differences in the uterine and rumen bacterial communities with varying levels of protein supplementation. Pubertal status had a greater impact on uterine bacterial communities than in the rumen. The effects observed over time and by pubertal status on bacterial communities of the heifer uterus potentially indicates progression of a maturing uterine microbiome through development. Future studies are needed to understand how nutrition and the overall diet may be used to manipulate uterine bacterial communities, and how the changing uterine microbiome through development may impact the uterine environment. Together, such studies would contribute to a greater understanding of the developing uterine bacterial communities and how they might be modified to improve future reproductive success and lifetime reproductive efficiency.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA819438.
Ethics Statement
The animal study was reviewed and approved by Institutional Animal Care and Use Committee at the University of Tennessee, Knoxville.
Author Contributions
TA-S was responsible for primary manuscript preparation, sampling, DNA extraction, and data analyses. KB assisted with sample collections and manuscript preparation. MH assisted with data analyses and manuscript preparation. RP assisted with laboratory analyses and manuscript preparation. TS was responsible for DNA library preparation and sequencing. DM, SM, FS, and KP were responsible for experimental design development and manuscript preparation. JR, LS, KM, and PM were responsible for sampling, experimental design development, and manuscript preparation. All authors contributed to the article and approved the submitted version.
Funding
The authors would like to thank the state of Tennessee through UT AgResearch, the Department of Animal Science, and the USDA National Institute of Food and Agriculture, Hatch Project No. 1019048, for providing funding and support for this research.
Author Disclaimer
Mention of a trade name, proprietary product, or specific equipment does not constitute a guarantee or warranty by the USDA and does not imply approval to the exclusion of other products that may be suitable. USDA is an equal opportunity provider and employer.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors would like to thank Kevin Thompson and the Middle Tennessee Research and Education Center for assistance in collecting samples and providing care for the animals on study, and Dr. J. Lannett Edwards for assistance in development of the uterine flush technique and experimental design. Thanks are given to Kristen Kuhn, Kelsey McClure, and Bob Lee for technical support of 16S amplification and sequencing at USDA-MARC.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fanim.2022.903909/full#supplementary-material
References
Amin N., Seifert J. (2021). Dynamic Progression of the Calf’s Microbiome and its Influence on Host Health. Comput. Struct. Biotechnol. J. 19, 989–1001. doi: 10.1016/j.csbj.2021.01.035
Atkins J. A., Pohler K. G., Smith M. F. (2013). Physiology and Endocrinology of Puberty in Heifers. Vet. Clin. North Am. Food Anim. 29 (3), 479–492. doi: 10.1016/j.cvfa.2013.07.008
Ault T. B., Clemmons B. A., Reese S. T., Dantas F. G., Franco G. A., Smith T. P. L., et al. (2019a). Bacterial Taxonomic Composition of the Postpartum Cow Uterus and Vagina Prior to Artificial Insemination. J. Anim. Sci. 97 (10), 4305–4313. doi: 10.1093/jas/skz212
Ault T. B., Clemmons B. A., Reese S. T., Dantas F. G., Franco G. A., Smith T. P. L., et al. (2019b). Uterine and Vaginal Bacterial Community Diversity Prior to Artificial Insemination Between Pregnant and Nonpregnant Postpartum Cows. J. Anim. Sci. 97 (10), 4298–4304. doi: 10.1093/jas/skz210
Ault-Seay T. B., Harrison T. D., Brandt K. J., Payton R. R., Schneider L. G., Myer P. R., et al. (2021). Short Communication: The Effects of Protein Level on Cytokines and Chemokines in the Uterine Environment of Beef Heifers During Development. J. Anim. Sci. 99, 1–7. doi: 10.1093/jas/skab105
Bach A., Calsamiglia S., Stern M. D. (2005). Nitrogen Metabolism in the Rumen. J. Dairy Sci. 88, E9–E21. doi: 10.3168/jds.S0022-0302(05)73133-7
Bazer F. W., Wang X., Johnson G. A., Wu G. (2015). Select Nutrients and Their Effects on Conceptus Development in Mammals. Anim. Nutr. 1 (3), 85–95. doi: 10.1016/j.aninu.2015.07.005
Belanche A., Pinloche E., Edwards J. E., Moorby J. M., Newbold C. J., Doreau M. (2012). Shifts in the Rumen Microbiota Due to the Type of Carbohydrate and Level of Protein Ingested by Dairy Cattle are Associated With Changes in Rumen Fermentation. J. Nutr. 142 (9), 1684–1692. doi: 10.3945/jn.112.159574
Bellows R. A., Short R. E. (1971). Relationships Among Weight Gains, Age at Puberty and Reproductive Performance in Heifers. J. Anim. Sci. 32 (1), 127–131. doi: 10.2527/jas1971.321127x
Bergman E. N. (1990). Energy Contributions of Volatile Fatty Acids From the Gastrointestinal Tract in Various Species. Physiol. Rev. 70 (2), 567–590. doi: 10.1152/physrev.1990.70.2.567
Bilal M. S., Abaker J. A., ul Aabdin Z., Xu T., Dai H., Zhang K., et al. (2016). Lipopolysaccharide Derived From the Digestive Tract Triggers an Inflammatory Response in the Uterus of Mid-Lactating Dairy Cows During SARA. BMC Vet. Res. 12 (1), 284. doi: 10.1186/s12917-016-0907-1
Brandt K. J. (2020). The Effect of Supplemental Protein on the Uterine Environment During Heifer Development (Knoxville, TN: University of Tennessee).
Callahan B. J., McMurdie P. J., Rosen M. J., Han A. W., Johnson A. J. A., Holmes S. P. (2016). DADA2: High-Resolution Sample Inference From Illumina Amplicon Data. Nat. Methods 13 (7), 581–583. doi: 10.1038/nmeth.3869
Cánovas A., Reverter A., DeAtley K. L., Ashley R. L., Colgrave M. L., Fortes M. R. S., et al. (2014). Multi-Tissue Omics Analyses Reveal Molecular Regulatory Networks for Puberty in Composite Beef Cattle. PloS One 9 (7), e102551. doi: 10.1371/journal.pone.0102551
Carberry C. A., Kenny D. A., Han S., McCabe M. S., Waters S. M. (2012). Effect of Phenotypic Residual Feed Intake and Dietary Forage Content on the Rumen Microbial Community of Beef Cattle. Appl. Environ. Microbiol. 78 (14), 4949–4958. doi: 10.1128/AEM.07759-11
Clemmons B. A., Martino C., Schneider L. G., Lefler J., Embree M. M., Myer P. R. (2019b). Temporal Stability of the Ruminal Bacterial Communities in Beef Steers. Sci. Rep. 9 (1), 9522. doi: 10.1038/s41598-019-45995-2
Clemmons B. A., Reese S. T., Dantas F. G., Franco G. A., Smith T. P. L., Adeyosoye O. I., et al. (2017). Vaginal and Uterine Bacterial Communities in Postpartum Lactating Cows. Front. Microbiol. 8. doi: 10.3389/fmicb.2017.01047
Clemmons B. A., Voy B. H., Myer P. R. (2019a). Altering the Gut Microbiome of Cattle: Considerations of Host-Microbiome Interactions for Persistent Microbiome Manipulation. Microb. Ecol. 77 (2), 523–536. doi: 10.1007/s00248-018-1234-9
Dawuda P. M., Scaramuzzi R. J., Leese H. J., Hall C. J., Peters A. R., Drew S. B., et al. (2002). Effect of Timing of Urea Feeding on the Yield and Quality of Embryos in Lactating Dairy Cows. Theriogenology 58 (8), 1443–1455. doi: 10.1016/S0093-691X(02)00973-1
Dickinson S. E., Elmore M. F., Kriese-Anderson L., Elmore J. B., Walker B. N., Dyce P. W., et al. (2019). Evaluation of Age, Weaning Weight, Body Condition Score, and Reproductive Tract Score in Pre-Selected Beef Heifers Relative to Reproductive Potential. J. Anim. Sci. Biotechnol. 10 (1), 18. doi: 10.1186/s40104-019-0329-6
Elrod C. C., Butler W. R. (1993). Reduction of Fertility and Alteration of Uterine pH in Heifers Fed Excess Ruminally Degradable Protein. J. Anim. Sci. 71, 694–701. doi: 10.2527/1993.713694x
Forde N., Beltman M. E., Duffy G. B., Duffy P., Mehta J. P., Ó'Gaora P., et al. (2010). Changes in the Endometrial Transcriptome During the Bovine Estrous Cycle: Effect of Low Circulating Progesterone and Consequences for Conceptus Elongation. Biol. Reprod. 84 (2), 266–278. doi: 10.1095/biolreprod.110.085910
Forde N., Simintiras C. A., Sturmey R., Mamo S., Kelly A. K., Spencer T. E., et al. (2014). Amino Acids in the Uterine Luminal Fluid Reflects the Temporal Changes in Transporter Expression in the Endometrium and Conceptus During Early Pregnancy in Cattle. PloS One 9 (6), e100010. doi: 10.1371/journal.pone.0100010
Fortes M. R. S., Zacchi L. F., Nguyen L. T., Raidan F., Weller M.M.D.C.A., Choo J. J. Y., et al. (2018). Pre- and Post-Puberty Expression of Genes and Proteins in the Uterus of Bos Indicus Heifers: The Luteal Phase Effect Post-Puberty. Anim. Genet. 49 (6), 539–549. doi: 10.1111/age.12721
Galvão K. N., Bicalho R. C., Jeon S. J. (2019). Symposium Review: The Uterine Microbiome Associated With the Development of Uterine Disease in Dairy Cows. J. Dairy Sci. 76, 290–299. doi: 10.3168/jds.2019-17106
Griswold K. E., White B. A., Mackie R. I. (1999). Diversity of Extracellular Proteolytic Activities Among Prevotella Species From the Rumen. Curr. Microbiol. 39 (4), 187–194. doi: 10.1007/s002849900443
Gruninger R. J., Ribeiro G. O., Cameron A., McAllister T. A. (2019). Invited Review: Application of Meta-Omics to Understand the Dynamic Nature of the Rumen Microbiome and How it Responds to Diet in Ruminants. Animal 13 (9), 1843–1854. doi: 10.1017/S1751731119000752
Guan L. L., Nkrumah J. D., Basarab J. A., Moore S. S. (2008). Linkage of Microbial Ecology to Phenotype: Correlation of Rumen Microbial Ecology to Cattle's Feed Efficiency. FEMS Microbiol. Lett. 288 (1), 85–91. doi: 10.1111/j.1574-6968.2008.01343.x
Guo C. Y., Ji S. K., Yan H., Wang Y. J., Liu J. J., Cao Z. J., et al. (2020). Dynamic Change of the Gastrointestinal Bacterial Ecology in Cows From Birth to Adulthood. Microbiol. Open 9 (11), e1119–e1119. doi: 10.1002/mbo3.1119
Holman D. B., Gzyl K. E. (2019). A Meta-Analysis of the Bovine Gastrointestinal Tract Microbiota. FEMS Microbiol. Ecol. 95 (6), 1–9. doi: 10.1093/femsec/fiz072
Jami E., Israel A., Kotser A., Mizrahi I. (2013). Exploring the Bovine Rumen Bacterial Community From Birth to Adulthood. ISME J. 7 (6), 1069–1079. doi: 10.1038/ismej.2013.2
Jeon S. J., Cunha F., Vieira-Neto A., Bicalho R. C., Lima S., Bicalho M. L., et al. (2017). Blood as a Route of Transmission of Uterine Pathogens From the Gut to the Uterus in Cows. Microbiome 5 (1), 109. doi: 10.1186/s40168-017-0328-9
Kinder J., Bergfeld E., Wehrman M., Peters K., Kojima F. (1995). Endocrine Basis for Puberty in Heifers and Ewes. J. Reprod. Fertil. Suppl (49), 393–408. doi: 10.1530/biosciprocs.3.030
Kuehn L. A., Cundiff L. V., Freetly H. C. (2011). Growth Curves of Crossbred Cows Sired by Hereford, Angus, Belgian Blue, Brahman, Boran, and Tuli Bulls, and the Fraction of Mature Body Weight and Height at Puberty. J. Anim. Sci. 89 (8), 2373–2379. doi: 10.2527/jas.2011-3847
Laguardia-Nascimento M., Branco K. M., Gasparini M. R., Giannattasio-Ferraz S., Leite L. R., Araujo F. M., et al. (2015). Vaginal Microbiome Characterization of Nellore Cattle Using Metagenomic Analysis. PloS One 10 (11), e0143294. doi: 10.1371/journal.pone.0143294
Lalman D. L., Petersen M. K., Ansotegui R. P., Tess M. W., Clark C. K., Wiley J. S. (1993). The Effects of Ruminally Undegradable Protein, Propionic Acid, and Monensin on Puberty and Pregnancy in Beef Heifers. J. Anim. Sci. 71 (11), 2843–2852. doi: 10.2527/1993.71112843x
Larson R. L. (2007). Heifer Development: Reproduction and Nutrition. Vet. Clin. North Am. Food Anim. 23 (1), 53–68. doi: 10.1016/j.cvfa.2006.11.003
Liu C., Meng Q., Chen Y., Xu M., Shen M., Gao R., et al. (2017). Role of Age-Related Shifts in Rumen Bacteria and Methanogens in Methane Production in Cattle. Front. Microbiol. 8 (1563). doi: 10.3389/fmicb.2017.01563
Marston T. T., Lusby K. S., Wettemann R. P. (1995). Effects of Postweaning Diet on Age and Weight at Puberty and Milk Production of Heifers. J. Anim. Sci. 73 (1), 63–68. doi: 10.2527/1995.73163x
Martin J. L., Cupp A. S., Rasby R. J., Hall Z. C., Funston R. N. (2007). Utilization of Dried Distillers Grains for Developing Beef Heifers. J. Anim. Sci. 85 (9), 2298–2303. doi: 10.2527/jas.2007-0076
McCann J. C., Wickersham T. A., Loor J. J. (2014). High-Throughput Methods Redefine the Rumen Microbiome and its Relationship With Nutrition and Metabolism. Bioinform. Biol. Insights 8, 15389. doi: 10.4137/bbi.S15389
McClure M. W. (2018). The Vaginal Microbiome Related to Reproductive Traits in Beef Heifers (Ann Arbor: M.S., University of Arkansas).
McMurdie P. J., Holmes S. (2013). Phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PloS One 8 (4), e61217. doi: 10.1371/journal.pone.0061217
Moore S. G., Ericsson A. C., Poock S. E., Melendez P., Lucy M. C. (2017). Hot Topic: 16S rRNA Gene Sequencing Reveals the Microbiome of the Virgin and Pregnant Bovine Uterus. J. Dairy Sci. 100 (6), 4953–4960. doi: 10.3168/jds.2017-12592
Myer P. R., Smith T. P. L., Wells J. E., Kuehn L. A., Freetly H. C. (2015). Rumen Microbiome From Steers Differing in Feed Efficiency. PloS One 10 (6), e0129174. doi: 10.1371/journal.pone.0129174
Ocon O. M., Hansen P. J. (2003). Disruption of Bovine Oocytes and Preimplantation Embryos by Urea and Acidic pH. J. Dairy Sci. 86 (4), 1194–1200. doi: 10.3168/jds.S0022-0302(03)73703-5
Oksanen J., Blanchet F. G., Friendly M., Kindt R., Legendre P., McGlinn D., et al. (2017) Vegan: Community Ecology Package. Available at: https://cran.r-project.org/web/packages/vegan/index.html.
Perry G. A. (2016). Factors Affecting Puberty in Replacement Beef Heifers. Theriogenology 86 (1), 373–378. doi: 10.1016/j.theriogenology.2016.04.051
Pfaffl M. W., Lange I. G., Meyer H. H. D. (2003). The Gastrointestinal Tract as Target of Steroid Hormone Action: Quantification of Steroid Receptor mRNA Expression (AR, Erα, Erβ and PR) in 10 Bovine Gastrointestinal Tract Compartments by Kinetic RT-PCR. J. Steroid Biochem. Mol. 84 (2), 159–166. doi: 10.1016/S0960-0760(03)00025-6
Pitta D. W., Parmar N., Patel A. K., Indugu N., Kumar S., Prajapathi K. B., et al. (2014). Bacterial Diversity Dynamics Associated With Different Diets and Different Primer Pairs in the Rumen of Kankrej Cattle. PloS One 9 (11), e111710. doi: 10.1371/journal.pone.0111710
Polat B., Colak A., Kaya M., Ucar O. (2009). Stimulation of Delayed Puberty in Heifers by Using a PRID Regime. Rev. Med. Vet. 160 (3), 149–153.
Pohler K. G., Pereira M. H. C., Lopes F. R., Lawrence J. C., Keisler D. H., Smith M. F. , et al. (2016). Circulating concentrations of bovine pregnancy-associated glycoproteins and late embryonic mortality in lactating dairy herds. J. Dairy Sci. 99 (2), 1584–1594. doi: 10.3168/jds.2015-10192
Quast C., Pruesse E., Yilmaz P., Gerken J., Schweer T., Yarza P., et al. (2013). The SILVA Ribosomal RNA Gene Database Project: Improved Data Processing and Web-Based Tools. Nucleic Acids Res. 41 (1), D590–D596. doi: 10.1093/nar/gks1219
Quereda J. J., Barba M., Mocé M. L., Gomis J., Jiménez-Trigos E., García-Muñoz Á., et al. (2020). Vaginal Microbiota Changes During Estrous Cycle in Dairy Heifers. Front. Vet. Sci. 7 (371). doi: 10.3389/fvets.2020.00371
Rodrigues N. F., Kästle J., Coutinho T. J., Amorim A. T., Campos G. B., Santos V. M., et al. (2015). Qualitative Analysis of the Vaginal Microbiota of Healthy Cattle and Cattle With Genital-Tract Disease. Genet. Mol. Res. 14 (2), 6518–6528. doi: 10.4238/2015.June.12.4
Sheldon I. M., Cronin J., Goetze L., Donofrio G., Schuberth H. J. (2009). Defining Postpartum Uterine Disease and the Mechanisms of Infection and Immunity in the Female Reproductive Tract in Cattle. Biol. Reprod. 81 (6), 1025–1032. doi: 10.1095/biolreprod.109.077370
Stevenson D. M., Weimer P. J. (2007). Dominance of Prevotella and Low Abundance of Classical Ruminal Bacterial Species in the Bovine Rumen Revealed by Relative Quantification Real-Time PCR. Appl. Microbiol. Biotechnol. 75 (1), 165–174. doi: 10.1007/s00253-006-0802-y
Swartz J. D., Lachman M., Westveer K., O’Neill T., Geary T., Kott R. W., et al. (2014). ). Characterization of the Vaginal Microbiota of Ewes and Cows Reveals a Unique Microbiota With Low Levels of Lactobacilli and Near-Neutral pH. Front. Vet. Sci. 1 (19). doi: 10.3389/fvets.2014.00019
Wallace R. (1996). The proteolytic systems of ruminal microorganisms. Annales de zootechnie, INRA/EDP Sciences. 45 (1), 301–308.
Weinroth M. D., Belk A. D., Dean C., Noyes N., Dittoe D. K., Rothrock M. J. Jr., et al. (2022). Considerations and Best Practices in Animal Science 16S Ribosomal RNA Gene Sequencing Microbiome Studies. J. Anim. Sci. 100 (2), 1–18. doi: 10.1093/jas/skab346
Yu Z., Morrison M.. (2004). Improved extraction of PCR-quality community DNA from digesta and fecal samples. Biotechniques. 36 (5), 808–812. doi: 10.2144/04365st04
Keywords: bacteria, beef heifer, development, nutrition, protein, rumen, uterus
Citation: Ault-Seay TB, Brandt KJ, Henniger MT, Payton RR, Mathew DJ, Moorey SE, Schrick FN, Pohler KG, Smith TPL, Rhinehart JD, Schneider LG, McLean KJ and Myer PR (2022) Bacterial Communities of the Uterus and Rumen During Heifer Development With Protein Supplementation. Front. Anim. Sci. 3:903909. doi: 10.3389/fanim.2022.903909
Received: 24 March 2022; Accepted: 13 June 2022;
Published: 07 July 2022.
Edited by:
Mohan Mondal, ICAR-National Dairy Research Institute, Eastern Regional Station, IndiaReviewed by:
Hatem Eltahan, Agricultural Research Center, EgyptWeinan Zhou, University of Illinois at Urbana-Champaign, United States
Rajwali Khan, University of Agriculture, Pakistan
Medhavi Sudarshan, Patliputra University, India
Shengru Wu, Northwest A&F University, China
Copyright © 2022 Ault-Seay, Brandt, Henniger, Payton, Mathew, Moorey, Schrick, Pohler, Smith, Rhinehart, Schneider, McLean and Myer. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Phillip R. Myer, cG15ZXJAdXRrLmVkdQ==; Kyle J. McLean, a21jbGVhMTBAdXRrLmVkdQ==
†Present Address: Kiernan J. Brandt, South Dakota State University, Brookings, SD, United States
 Taylor B. Ault-Seay
Taylor B. Ault-Seay Kiernan J. Brandt1†
Kiernan J. Brandt1† Madison T. Henniger
Madison T. Henniger Rebecca R. Payton
Rebecca R. Payton Ky G. Pohler
Ky G. Pohler Liesel G. Schneider
Liesel G. Schneider Kyle J. McLean
Kyle J. McLean Phillip R. Myer
Phillip R. Myer