
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Anim. Sci., 22 November 2021
Sec. Animal Welfare and Policy
Volume 2 - 2021 | https://doi.org/10.3389/fanim.2021.756101
This article is part of the Research TopicBiology Meets Technology: Aquatic Animals in Novel and New Aquaculture Production SystemsView all 6 articles
Microbial gill diseases caused by either opportunistic or specific pathogens are an emerging area of concern for aquaculture producers in part due to their sometimes complex and/or cryptic nature. Many antimicrobial treatments used in aquacultural settings are broad spectrum in nature. The effect of such therapeutics upon reduction and recolonization of commensal or pathogenic microbiota post-treatment has received little attention to date. Commensal bacteria are an integral component of the barrier function of mucosal surfaces in animals. This study evaluated the effect of several commercially relevant antimicrobial treatments upon the diversity and composition of branchial bacteria of Atlantic salmon. Here we exposed Atlantic salmon smolt to a number of commercially relevant antimicrobial treatments including chemotherapeutants (chloramine-t and hydrogen peroxide) and antibiotics (oxytetracycline and florfenicol) in vivo. Subsequently we examined the change in bacterial load, 16S rRNA gene expression, and taxonomic diversity post-treatment upon the gills. Results revealed a decrease in cultivable bacterial colonies after antimicrobial treatment, and a downstream decrease in bacterial richness and abundance post-treatment, with colonization of several prominent pathogenic taxa including Vibrio and Tenacibaculum. Temporal tracing over a 14-day period demonstrated that the bacteriome of gill mucus is sensitive to change, and altered by antimicrobial treatment and handling. This study identified candidate antimicrobial treatments which could be implemented in future studies to illustrate the effect of dysbiosis on microbial gill diseases.
Global finfish aquaculture continues to increase rapidly to meet market demands and the need for a sustainable, high yield protein source for a burgeoning global population. Production stressors, life cycle stages, adverse water quality, diet, and disease are some factors affecting the overall health of intensive aquaculture systems (Beck and Peatman, 2015). Compromise and infection on the gill can lead to reduced productivity and economic losses (Rozas-Serri, 2019). The increasing use of high-density animal production systems producing high levels of waste effluent can lead to pathogen proliferation and impinge on production success if not correctly processed or recycled (Noga, 2010; de Bruijn et al., 2018). Pathogen infiltration of fish often occurs through mucosal barriers, including the gill, skin and gastrointestinal tract (Merrifield and Rodiles, 2015). Fish gill surfaces are in constant contact with the aquatic external environment containing an abundance of microbes including pathogenic agents. Mucosal gill surfaces can be a portal for pathogens to colonize and infiltrate leading to localized or systemic disease, and compromise to the gill can impact upon physiological processes due to the multifunctional nature of the organ. Diseases and disorders of the gill are therefore often multifactorial and complex in nature (Mitchell and Rodger, 2011). Some gill conditions such as complex gill disease (CGD) have multiple known aetiological agents (e.g., Neoparamoeba perurans, Candidatus Piscichlamydia salmonis, Desmozoon lepeophtherii, salmon gill poxvirus and Candidatus Branchiomonas cysticola) (Gjessing et al., 2019) which interact in a co-infection. Other presumed single-agent gill conditions such as columnaris disease (Flavobacterium columnare) and yellow mouth (Tenacibaculum maritimum) have strong environmental influences, including temperature and salinity, which can affect infection severity (Bandilla et al., 2006; Wynne et al., 2020).
The concept of an innate immune benefit in-part provisioned by commensal bacteria within the mucus layer is known colloquially as barrier health (Beck and Peatman, 2015). The commensal bacterial community inhabiting these areas represent a component of the defense barrier against pathogens (Cabillon and Lazado, 2019), and are thought to be most effective when the microbial community is highly diverse (Wilson and Laurent, 2002; Merrifield and Rodiles, 2015). These microbes colonize gill mucus and provide competitive exclusion toward opportunistic pathogens, synthesis of antimicrobial compounds (e.g., bacteriocins, antimicrobial peptides, hydrogen peroxide) and assist with immune functions such as phagocytic activity (Gómez and Balcázar, 2008; Cabillon and Lazado, 2019). The diversity of the commensal microbiota can be profiled using next generation sequencing techniques providing an indicator of health for a given aquaculture species (Derome et al., 2016).
Disease can occur within intensive aquaculture systems, and treatment of animals in both a prophylactic and therapeutic nature is essential to maintain welfare and promote optimal growth. Treatment options include the use of chemotherapeutics (e.g., oxidative compounds), antibiotics as well as manipulation of water quality parameters (i.e., transfer to salt/freshwater, change in temperature). Chemotherapeutic treatment is well-documented for a range of prominent bacterial and fungal infections, an example of this is the use of chloramine-t (Cl-T) as an immersion bath treatment for salmonid diseases such as bacterial gill disease and columnaris disease (caused by Flavobacterium spp.) (Bullock et al., 1991; Genaro Sanchez et al., 1996; Bowker et al., 2008). Ecoparasites are also commonly treated in this fashion. Hydrogen peroxide (H2O2) is an oxidative chemical which breaks down into environmentally friendly by products (water and oxygen gas). It has been used within the salmonid industry for decades as one of the most reliable treatments of sea lice (Lepeoptheirus salmonis and Caligus spp.), and to a lesser extent amoebic gill disease (AGD) (Kiemer and Black, 1997; Adams et al., 2012; Powell et al., 2015). Antibiotic treatments are often utilized to treat more systemic or internal bacterial infection events, which can be administered via immersion bath or oral feed delivery. Commonly used compounds include florfenicol and oxytetracycline, which have a bacteriostatic killing action used to effectively treat conditions such as furunculosis (Aeromonas salmonicida), vibriosis (Vibrio spp.), piscirickettsiosis (Piscirickettsia salmonis) (Nordmo et al., 1994; Lundén et al., 1999; Noga, 2010; Henríquez et al., 2016; Schmidt et al., 2017). However, due to the broad-spectrum killing action of many antimicrobial agents, non-target taxa can be affected by the treatment process (Noga, 2010). This may inadvertently lead to a microbial imbalance, often termed as “dysbiosis” (Egan and Gardiner, 2016; Francino, 2016), which can lead to further health issues and susceptibility to infection. Therapeutic Cl-T treatment in rainbow trout (Oncorhynchus mykiss) left the skin in an infection prone state, and was colonized by secondary opportunist agents including Tenacibaculum and Pseudomonas (Genaro Sanchez et al., 1996). Mohammed and Arias (2015) demonstrated that microbial dysbiosis of channel catfish Ictalurus punctatus induced by antibacterial bath immersion was associated with increased susceptibility to experimentally induced columnaris disease. Similarly, Schmidt et al. (2017) demonstrated that after challenging black molly (Poecilia sphenops) with Vibrio anguillarum, treatment with streptomycin led to significant mortality. Survival in the antibiotic treated fish which were also provided a probiotic additive treatment showed a much higher survival rate, indicating that the subtle microbiome supplementation lessened the impact of microbial dysbiosis on the fish. Despite these examples, there is a limited understanding of the effect broad spectrum antimicrobial compounds have in the context of bacterial dysbioses upon mucosal surfaces, especially those of the gills. Furthermore, the temporal response of gill microbiota post-treatment is largely undescribed past the initial treatment window, along with quantification of the impact of topical (immersion bathing) vs. systemic (in feed) antimicrobial treatments, and sources of community recolonization.
In this study we used a combination of microbiological, molecular and amplicon sequencing techniques to determine the impact of several antimicrobial treatment regimes. The major aims of the study were to determine if broad spectrum antimicrobial compounds affect the commensal gill community, and to evaluate the post-treatment response of the bacterial community following such treatments. Results revealed a downstream decrease in bacterial richness and abundance over time post-treatment, and prominent colonization of several known pathogenic taxa. This area of research is still largely uncovered, and hence our findings are pertinent to inform the greater aquaculture field of the potential outcomes arising from antimicrobial usage in the context of commensal bacteria. This research will have implications for future studies investigating the intricacies of microbial gill disease, and offer a model process to induce microbial dysbioses in an applied scenario. It will also provide insights regarding the significance of commensal bacteria as a component of the mucosal health barrier.
All activities relating to fish maintenance and sampling in this trial were reviewed and approved by CSIRO Queensland Animal Ethics Committee (permit 2018-18) under the guidelines of the Australian Code of Practice for the Care and Use of Animals in Research.
A cohort of 200 Atlantic salmon (Salmo salar) parr were on-reared at the Bribie Island Research Centre in purpose-built recirculation systems for a period of ~9 months. Fish were fed daily to satiation on a commercial pelleted feed, maintained at a mean rearing temperature of 12°C ±1 and fixed photoperiod of (14L: 10D). Fish were prepared for smoltification after attaining a mean weight of 150 g. Fish were exposed to a constant photoperiod (24L: 0D) at an intensity of 3300 lumen for a period of 5 weeks and then transferred from the freshwater RAS to marine flowthrough water via constant system water exchange within the 5,000 L tank. Post smolt were then allowed to acclimate to marine water over a period of two weeks prior to trial commencement, with water temperature held at 15°C ±1, dissolved oxygen 90–110% sat, TA-N <0.50 ppm, and salinity 35–36 ppt.
Upon trial commencement 147 Atlantic salmon post-smolt (229.4 ± 0.4 g) were anesthetized, individually weighed and stocked into an array of seven identical 500 L tanks (n = 21) which were provided with identical flowthrough seawater and had independent drainage systems. During the trial, the water temperature was maintained at 15 ± 0.5°C and the dissolved oxygen 90–110% saturation by chilling infrastructure and aeration with oxygen monitoring probes and automated oxygen modulation. The system was operated as flow-through, with seawater pumped from approximately 300 m off the beach adjacent to the research station then through a series of 16 spin disk filters (40 μm) and 10 multimedia filters (~10–15 μm), after which received ozone treatment from two 100 gO3.h generator units (Wedeco OCS-GSO30). The ozone treated seawater was then pumped via ultra violet sterilizers, providing 80 mJ.cm2 dosed to two (~8 m3) granular activated carbon vessels for a contact time of >9 min to remove unwanted by-products from the ozone treatment. Finally, the seawater was pumped to a header tank, which fed directly into a pipe system delivering treated seawater to this experiment. The array was light-controlled and maintained a 14L:10D photoperiod throughout.
Antimicrobial treatments in the form of in-feed antibiotics and immersion therapeutic baths were conducted to reduce branchial bacteria loads. Antibiotic coated feeds were prepared for this work by adding pelleted feed (3 mm Spectra, Skretting P/L, Cambridge, TAS) into a Hobart mixer bowl (Hobart, Ohio, USA) with the required addition of pre-warmed fish oil (60°C) combined with an emulsion of concentration of either 79 ppm.kg−1 oxytetracycline hydrochloride (OTC) (CCD, NSW, Australia) or 10 ppm.kg−1 of florfenicol (FF) (Abbey Labs, NSW, Australia) respectively. Each compound emulsion was then poured into separate pellet bowls, where a sealed lid was added and the chamber was evacuated of air using a vacuum pump at 350 P.S.I for 5 min, until visible air escaping the pellets was no longer observed. Antibiotic coated pellets were stored in the dark at −20°C, and the daily ration was taken from the freezer to be loaded into the autofeeder hopper. Antibiotics pellets were offered daily to fish in OTC and FF tanks, completing a 10-day course duration.
At the completion of the antibiotic course, the therapeutic chemical bath treatments were carried out to synchronize sample timing (Figure 1). The three oxidative immersion bath treatments used were as follows; chloramine-tihydrate (Cl-T, Sigma Aldrich, USA) in saltwater at 25 ppm for 60 min, hydrogen peroxide (H2O2, Solvay Interox, Australia) divided into both a saltwater treatment at 1,250 ppm for 15 min, and a freshwater treatment at 500 ppm for 20 min. Fish from each stocked tank (n = 21) were transferred to four identical static baths made up the three aforementioned bath treatment concentrations, along with a sham bath (bath control) containing only filtered saltwater for 60 min. Fish behavior and welfare was visually monitored throughout the duration of the bath treatments, and upon completion, 18 fish were netted back into their holding tank and three fish were sampled from each treatment. After initial treatment fish from all groups were held in their respective experimental tanks for the duration of the trial. All tanks including antibiotic feed treatments were offered a daily ration of 1% bodyweight (BW) (3 mm Skretting Spectra) via autofeeder system (Arvotec wolf controller, Arvotec-Oy). Daily maintenance included recording water quality (temperature and dissolved oxygen), observing fish for irregular behavior, cleaning tank systems and collection of any uneaten feed at the conclusion of the autofeeder activity period. This collected feed was retained into a mesh sieve (1 mm aperture), where it could be transferred to individual trays and dried overnight at 105°C to obtain dry weight and allow total uneaten feed to be calculated.
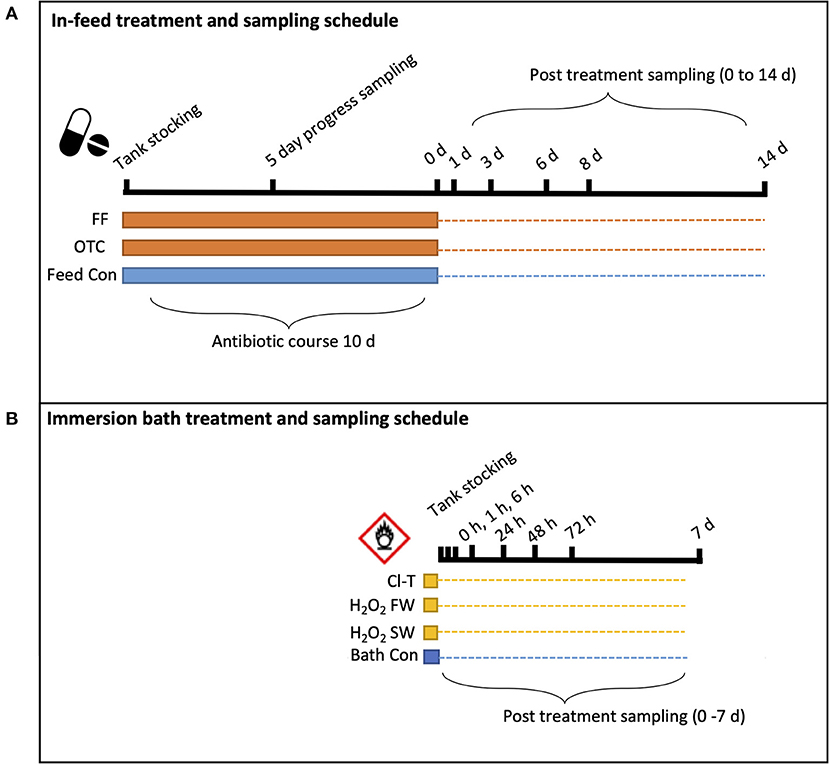
Figure 1. Schematic illustration of experimental sampling regime for each group of (A) infeed antibiotics and (B) oxidative bath treatments. Solid banding indicates the disinfection “period” for each group, while the dotted line represents the longitudinal trial sampling period to its completion. Black ticked lines and respective times indicate the sampling timepoint when fish were sampled.
To account for the systemic nature of in-feed antibiotic administration, the sampling schedule for feed treatments was extended to a 14-day timeframe, whilst oxidative bath treatments were expected to have shorter term impact and recovery and thus sampling was over a shorter duration (7 days) targeting the immediate post-bath period.
Gill mucus was sampled by taking a swab of the first right hand side (R1) hemibranch (three rotations along the length of the arch) and placing the swab into 1 ml of filtered, autoclaved seawater in a 1.5 ml tube. Each tube was then vortexed for 15 s, before a 500 μl aliquot was pipetted onto an aerobic count film (3M petri-film®) and incubated at 35 ± 0.1°C for 48 h before colonies were visually counted in and recorded (calculated as CFU.ml−1).
On each nominated sampling timepoint (Figure 1), three individual fish from each group were humanely killed (by immersion in 100 ppm AQUI-S™) and then sampled. A mucosal gill sample was taken by swabbing all anterior and posterior hemibranch from the entire right-hand side of the gill basket (eight surfaces). This was achieved using a sterile cotton swab (Westlabs), where the swab was rotated three times on each of the eight hemibranch surfaces. Swabs were then transferred to a 1.5 ml screw cap tube containing 1 ml of RNAlater solution, and stored at 4°C for 24 h before being frozen and stored at −80°C until further processing could occur.
Tank water samples were collected by filling 3 sterile HDPE collection bottles with 500 ml volume from each individual tank, and passaging the contents of each bottle through a 0.22 μm Sterivex™ (Millipore) filter membrane unit using a peristaltic pump (RP-100 series, Lachat Instruments) to retain bacterial cells. The Sterivex™ filter chamber was then flooded with 2 mL of RNAlater solution and then stored at −20°C prior to DNA extraction.
Bacterial DNA was extracted from both mucosal cotton swab samples as well as from 0.22 μm sterivex water filter units. Mucosal swab samples were extracted using the Qiagen DNeasy spin column extraction kit, using a modified protocol to the standard blood and tissue documentation. Briefly, swab samples were agitated and centrifuged, with excess RNAlater fixative pipetted off to waste. The process then followed the blood and tissue documentation until completion. Sterivex filter samples were extracted using the Qiagen Sterivex DNA extraction kit, as per manufacturers protocols. Genomic DNA quality and concentration was verified using a nanodrop ND1000 spectrophotometer (Life Technologies).
Target gene for the assay was the V3–V4 hypervariable region of the 16S rRNA bacterial gene, defined by a 174 bp fragment using the following primers 341f 5′-CCTACGGGAGGCAGCAG-3′ and 515r 5′-ATTCCGCGGCTGGCA-3′ as described in López-Gutiérrez et al. (2004). The Atlantic salmon elongation factor gene EL1-a (ELF) described in Bland et al. (2012) was used as the reference gene in this assay, amplifying a 66 bp fragment using the primer set S-ELF.f 5'-GGCCAGATCTCCCAGGGCTAT-3' and S-ELF.r 5'-TGAACTTGCAGGCGATGTGA-3'.
Extracted DNA was diluted to a working concentration of 10 ng.μl for all samples. Real time PCRs were carried out in a ViiA™ 7 Real-Time PCR Machine (Applied Biosystems). qPCR was performed in a single-plex 25ul reaction containing 1.25 μl of 10 uM forward and reverse primer, 8 μl of RNase-free H2O, and 12.5 μl of SYBR Green qPCR Mastermix including hotstart Taq polymerase (Bioline). Each reaction contained 2 μl of normalized template DNA (30 ng.ul−1). PCR reactions were subjected to the following thermal cycling: 95°C for 10 min, then 35 cycles of 95°C for 30 s, 72°C for 40 s and 76°C for 35 s. A melt curve was also included in the assay, set at 95°C for 15 s, 60°C for 1 min and 95°C for 1 s. PCR reaction volumes were loaded into a 384-well plate in triplicate for both the target and reference gene on each biological sample.
After gDNA was extracted, it was amplified by PCR using Illumina fused primers which targeted the V1–V3 hypervariable region of the 16S rRNA gene. The sequences for the forward and reverse primers were as described in Table 1.

Table 1. Primer sequences used in the current study for PCR submission amplifying the V1–V3 region of the 16S rRNA gene.
DNA concentration of each sample was quantified using a nanodrop spectrophotometer (ND-1000) and were diluted to 10 ng.μl−1 as template for the PCR reactions, using Platinum Taq Hi fidelity mastermix (Thermo Fisher). Cycling was completed as per the following conditions; 94 °C for 90 s; 25 cycles of 94 °C for 30 s, 52 °C for 30 s, 72 °C for 90 s; and a final extension of 72 °C for 10 min. Sequencing was carried out on an Illumina Miseq platform at the Ramaciotti Center for Genomics (UNSW, Sydney), generating forward and reverse reads of 300 bp in length.
A negative control of ultrapure analytical grade water was included within the PCR reaction. After cycling was completed, PCR product amplification was verified via gel electrophoresis for the target amplicon. A negative process control (unused cotton swab opened and placed into a tube with RNAlater) and a mock community standard (ZymoBIOMICS Microbial Community Standard, Zymo Research) containing a known composition of 8 bacterial species was sequenced to validate sequencing effort and quality. Sequencing reads from the demultiplexed samples analyzed in this study have been deposited in the NCBI Sequence Read Archive (SRA) under the BioProject accession PRJNA718152.
Raw Illumina amplicon sequencing data files were processed using the open-source software pipeline “Quantitative Insights into Microbial Ecology 2” QIIME2 (Caporaso et al., 2010). Paired end sequences from the forward and reverse reads were merged for each sample and were denoised using the q2-dada2 plugin (Callahan et al., 2016) with default parameters. Quality control including chimeric sequence removal from the dataset was completed during dada2 processing, along with subsequent removal of host DNA and exclusion of chloroplast and mitochondrial sequences. Amplicon Sequence Variants (ASV's) were classified taxonomically using the classify-sklearn method in the QIIME2 q2-feature-classifier plugin using default parameters (Bokulich et al., 2018). The SILVA 16S rRNA 99% taxonomy database release 132 (Quast et al., 2012), was used as reference sequences for taxonomic classification.
All statistics were performed in R version 3.6.0 (R Core Team, 2020). Daily feed consumption was calculated as % bodyweight consumed per day by subtracting collected, dried uneaten pellets from the total ration fed to each tank. The mean ± SD of this metric was compared between treatments using a one-way ANOVA and Tukey post-hoc testing. Bacterial count plate data was arcsine-transformed and the CFU mL values analyzed using a one-way ANOVA with Tukey PSD post-hoc testing. Real time qPCR data were analyzed as log-fold change between treatment groups of the ratio of the gene of interest (16S rRNA) after against the housekeeping (salmon Ef1α) control, and assessed using two-way ANOVA with treatment and timepoint as factors (padj < 0.05). Samples from the 16S NGS data were rarefied using R package QsRutils (Quensen, 2020) performed on a maximum subsampling depth of 7,171 sequences per sample (Supplementary Figure S1). Obvious contaminant artifact present in the negative control sequences was identified and subsetted from biological samples via the Decontam package (Davis et al., 2018). Using the Phyloseq package (McMurdie and Holmes, 2013) taxonomic assignments were generated and alpha diversity indices calculated (Observed ASV's, Shannon diversity, Faith's phylogenetic distance). The alpha diversity metrics were analyzed via non-parametric means (Kruskal-Wallis test) and further pairwise comparisons using a Wilcoxon Test (Rank Sum Test). Beta-diversity comparisons were made via NMDS using Bray Curtis pairwise distances. Differences between groups was analyzed using PERMANOVA via the pairwise Adonis package (Martinez Arbizu, 2019). Relative taxonomic abundance was analyzed using the DeSeq2 package (Love et al., 2014) to test for differentially abundance bacterial taxa between groups. The origin of gill mucus samples was investigated by comparing the core branchial bacterial community against the source tank water using the FEAST package (Shenhav et al., 2019). All figures were produced using the R package ggplot2 (Wickham, 2016).
In the current study we aimed to characterize the effect that known antimicrobial agents would have on the mucosal bacterial communities of the Atlantic salmon gill surface. To achieve this, several known antimicrobial treatments administered were in line with FDA recommended delivery (bath or in-feed) and dosage. Feed intakes for antibiotic treatment were examined daily to ensure that fish consumed enough feed to receive the correct dosage listed in Table 2.
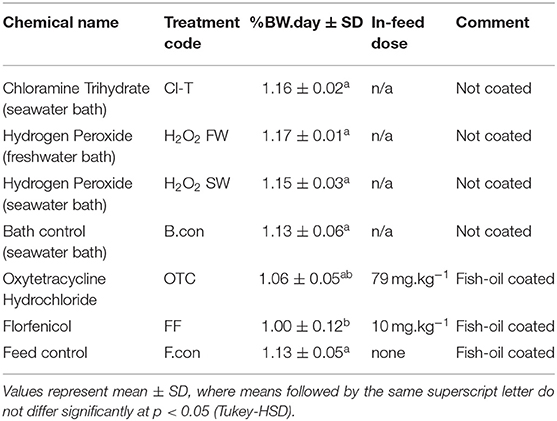
Table 2. Feed intake measured as percent bodyweight per day consumption over the total antibiotic course timeframe.
Mean feed consumption across all tanks remained at or above 1% bodyweight per day for the duration of the antibiotic course (or habituation period for other groups). Mass specific intake (% BW.day) was significantly lower for the florfenicol coated diet comparative to the fish-oil coated commercial pellets (ANOVA, F(6, 63) = 4.788, p < 0.001). There was no difference in intake values between the two antibiotic coated diets (OTC and FF), or between commercial pellet and fish-oil coated commercial pellets.
Total bacterial counts (CFU.ml−1) from the R1 hemibranch surface varied within the trial and across sampling dates, with the time by treatment interaction significant (ANOVA, F(32, 96) = 2.135, p < 0.001). This was characterized by a decline in viable count plate colonies directly after all antimicrobial (bath and in-feed) treatments. Immersion bath treatments including Cl-T, H2O2 FW and H2O2 SW recorded lowest count numbers directly post-bath (0 h) which largely increased over the 7-day trial period (Figure 2A). The seawater bath control also showed an initial decrease in CFU.ml at the start of the trial, increasing significantly at the 1 hr timepoint (p < 0.05) followed by relative stabilization toward the end of the trial period. Antibiotic treatments (Figure 2B; OTC, FF) were observed at significantly lower levels to the control at the mid-course (p < 0.01), completion of the course (0 h; p < 0.001), and 1-day post treatment (p < 0.05). All three groups remained relatively consistent between subsequent timepoints, with the feed control group being significantly higher at 14 days (p < 0.01).
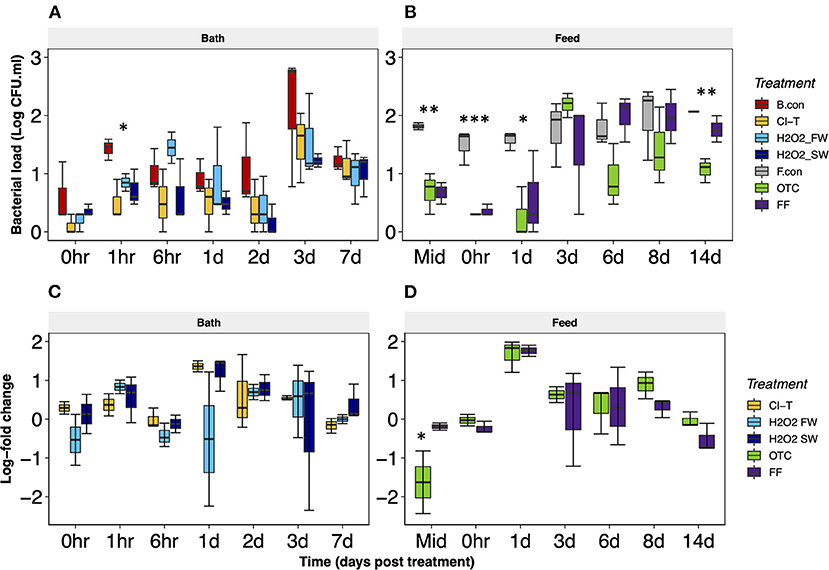
Figure 2. Log transformed bacterial count plate data + SE for bath (A) and in-feed (B) treatments. Data are presented as estimated marginal means, with the error bars representing the standard error. At each timepoint a comparison of different treatments (between means of CFU.ml counts) were compared and differences are indicated by *p ≤ 0.05, **p < 0.01, or ***p ≤ 0.001. (C,D) shows quantitative PCR log-fold 16S rRNA gene expression of gill mucosa samples from immersion bath (C) and in-feed treatments (D). An asterisk *indicates a significant difference from the control group at that particular timepoint (p < 0.05).
Quantitative PCR data were assessed as the log-fold change of the 16S rRNA gene against a reference housekeeping gene (ELF), and compared back to the respective control from the bath and in-feed groups (Figures 2C,D). Immersion bath treatments were variable throughout the 7-day period. The H2O2 FW group demonstrated lower 16S gene expression of the at 0, 6 h and 1-day post-bath to the reference control, but this was not statistically significant. At the day 3 timepoint, Cl-T and H2O2 SW groups had a higher log-fold expression, although this was not significant. At the 2-, 3- and 7-day sample points, all three bath treatments expressed positive log-fold increase in 16S gene expression. OTC treated gill mucus at the mid-course point in Figure 2D showed a significant decrease in 16S gene expression (padj < 0.05). Both OTC and FF treatments were characterized by a decrease in 16S expression between the mid-course sample to 1-day post-treatment. Log-fold expression was slightly higher between day 3 and day 8, before values stabilized close to the control reference point for the 14-day timepoint.
From the 147 gill swabs and 28 tank water samples we obtained a total of 7,939,968 raw sequence reads. QC and merging sequences resulted in an average of 43,152 reads per sample, with only one sample below 7,000 reads. The subsequent ASV table generated 7,296 bacterial taxa from which the diversity and taxonomic analyses were computed.
Alpha diversity metrics were used to assess bacterial richness (Observed ASVs) and diversity (Shannon index) and compare community structure within antibacterial treatments across timepoints (Figure 3). Timepoints within each group were analyzed to determine the magnitude of change post-treatment. Over a 7 day period post-bath treatment period, bacterial richness and diversity in fish gills exposed to Cl-T and filtered seawater (bath control) remained static. There were no significant interactions over time (p > 0.05). Hydrogen peroxide treated fish in both freshwater and seawater did differ longitudinally in both observed ASVs and Shannon index, and was deemed statistically significant (p < 0.05; Figures 3A,C). Both groups were characterized by an increase in richness and diversity at 1 and 6 h, before values decreased and remained relatively stable. Within the in-feed groups, ASV richness differed with both antibiotic treatments starting and finishing at similar levels, while the feed control slightly increased over time. Shannon diversity was low in the initial timepoints for all groups, and increased consistently to the 14 day timepoint. Kruskal Wallis testing demonstrated that none of the longitudinal changes in both observed ASVs or Shannon diversity were statistically significant for OTC, FF or the feed control (Figures 3B,D; p > 0.05).
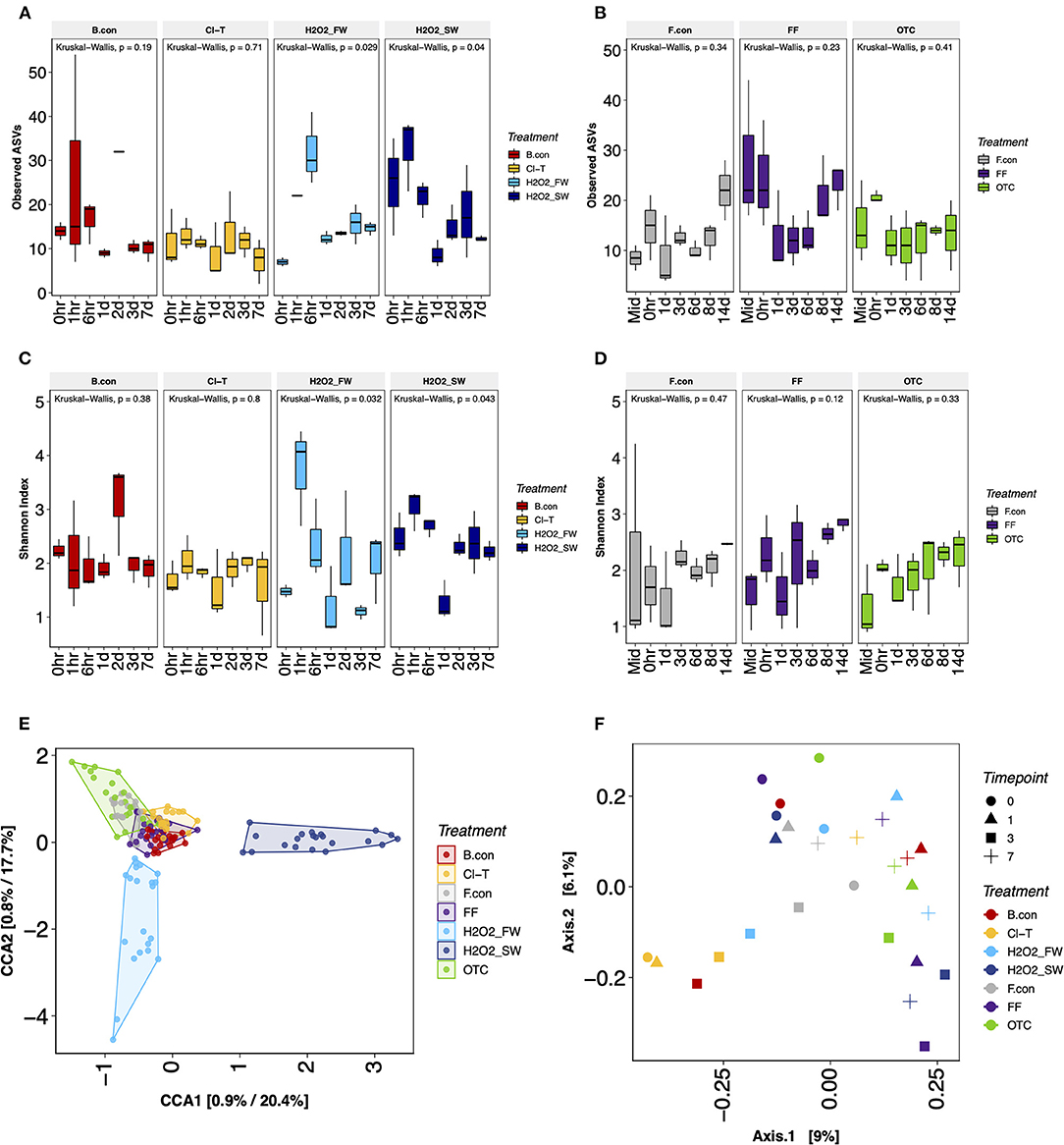
Figure 3. Alpha richness expressed as ASV richness for gill mucus samples in bath (A) and in-feed (B) antimicrobial treatments, and Shannon diversity (C,D) for each respective group. Beta diversity visualized through CCA ordinations for gill mucus (E) and tank water (F) samples.
Beta diversity was visualized using ordination of the gill mucus samples via canonical correspondence analysis (CCA) demonstrated that treatments grouped together strongly throughout the experimental period (Figure 3E). PERMANOVA comparing treatment group and timepoint indicated a significant interaction (p < 0.001) for both factors, but the Treatment*Time interaction was not significant (p > 0.05). Pairwise adonis for each comparison of gill mucus samples revealed significant differences between H2O2 FW and all treatments (p < 0.001), as well as H2O2 SW and all other treatments (p < 0.001). Cl-T was significantly different to the bath control, feed control, FF at OTC treatments (p < 0.001). Tank water sample groups (Figure 3F) were also significantly different from one another when grouped by treatment (PERMANOVA, p < 0.001), with the difference being between the Cl-T and OTC tank (p < 0.05).
To further understand differences in bacterial richness and diversity observed in alpha and beta metrics, we examined the relative abundance of bacterial taxa at the phylum level to assess community change within each treatment across the time course. The most abundant phylum associated with gill mucus samples were Proteobacteria (37.30%), Verrucomicrobia (26.05%), Actinobacteria (24.69%), Bacteriodetes (9.30%) and Firmicutes (2.43%). Actinobacteria appeared present in all timepoints, but was most apparent within the Cl-T and H2O2 SW bath treatments. Bacteriodetes and Firmicutes were inconsistent in abundance throughout most treatments, with no distinct trends apparent. The phylum Verrucomicrobia was also highly abundant, but appeared to decrease within the in-feed treatments over the duration of the trial, and was in lower levels in the H2O2 SW group (decreasing longitudinally after bath), and completely absent in the H2O2 FW group until 1-day post bath before returning in high abundance (Figure 4A). DeSeq2 analysis indicated that both Verrucomicrobia and Proteobacteria were statistically different between the bath treatment groups (padj < 0.001). Only the phylum Verrucomicrobia was deemed significantly different for in-feed treatment groups (padj < 0.01). The 100 most prevalent ASVs were classified to genus level assignments and compared to identify key genera involved in the post perturbation period. Further examination of the 10 most abundant genus revealed that the genus Rubritalea was more prevalent at the beginning of the trial for all treatments excluding H2O2 FW, and that the abundance of Pseudoaltermonas increased markedly at the final sampling points (Supplementary Figure S2). When compared to the reference bath control, the dominant genera Rubritalea and Cutibacterium were differentially expressed in Cl-T, H2O2 FW and H2O2 SW groups (padj < 0.001; Figure 4B). The same taxa were not statistically different for in-feed treatments compared to the feed control (padj > 0.05). Other prominent taxa included Pseudoaltermonas, Vibrio and Tenacibaculum. These genera were at higher abundance toward the later stages of the sampling period.
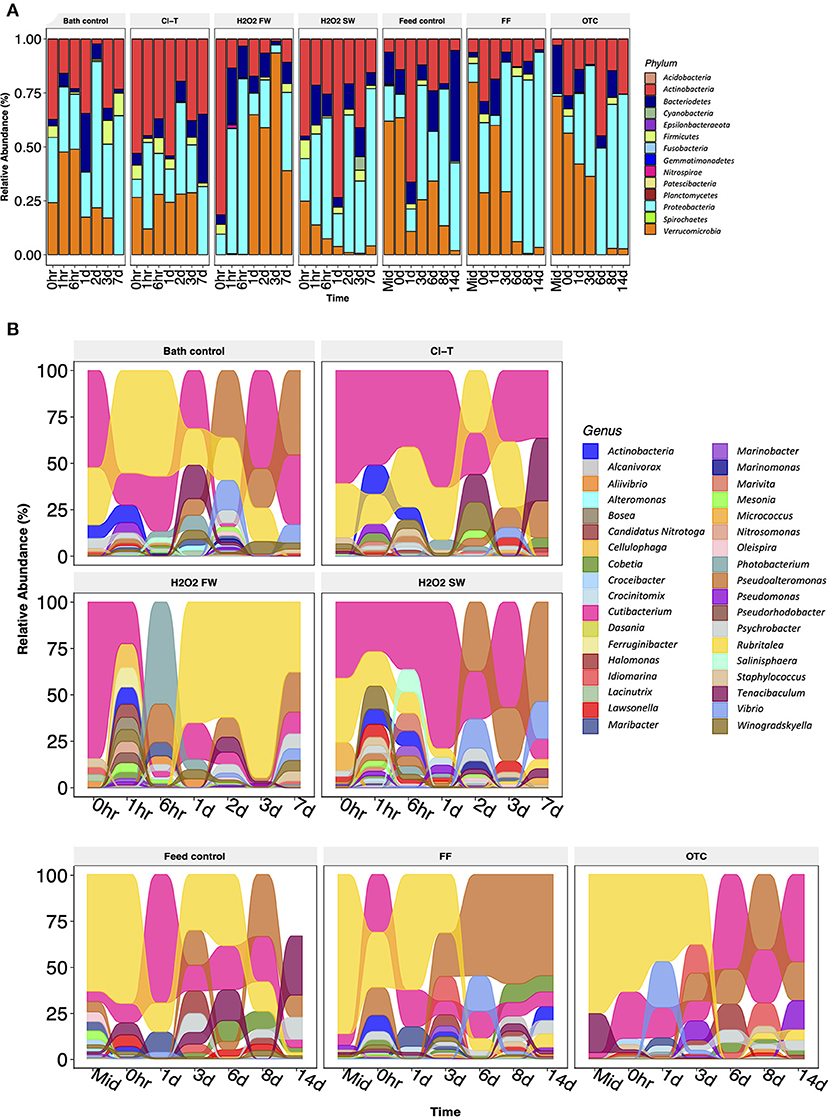
Figure 4. (A) Relative abundance of phylum level assignments obtained from gill mucus samples. Samples are grouped by timepoint in longitudinal fashion (each bar n = 3). (B) Alluvial plot of genus level assignments from the top 100 most prevalent ASVs in the study. These data indicate the bacterial community was likely in a dynamic state, which changed rapidly over a short-term period, characterized by large changes in abundance between dominant taxa.
In addition to profiling the bacterial community of gill mucus, we also examined the bacteria of tank water in which the fish reside. Taxonomically, the dominant tank water derived taxa at the genus level include Alteromonas, Pseudoalteromonas, Crocinitomix, Tenacibaculum and Winogradskyella (Supplementary Figure S3). Overall, many ASVs were common between samples obtained from the gill mucus and the tank environment. The FEAST package was used to determine the source origin of the gill mucus samples, by assessing the contribution from tank water microbiota. A large proportion of tank water-based bacteria were present on the gill for all bath (Figure 5A) and feed (Figure 5B) groups. FEAST demonstrated that tank water contributed a significantly higher proportion of the gill mucus community post treatment with hydrogen peroxide. Both H2O2 FW and H2O2 SW directly post bath and at 1-day sample points were significantly influenced by tank water communities (t-test, p < 0.001). Conversely, in-feed OTC had the lowest contribution of tank water-based bacteria on the gill surface, which decreased over time.
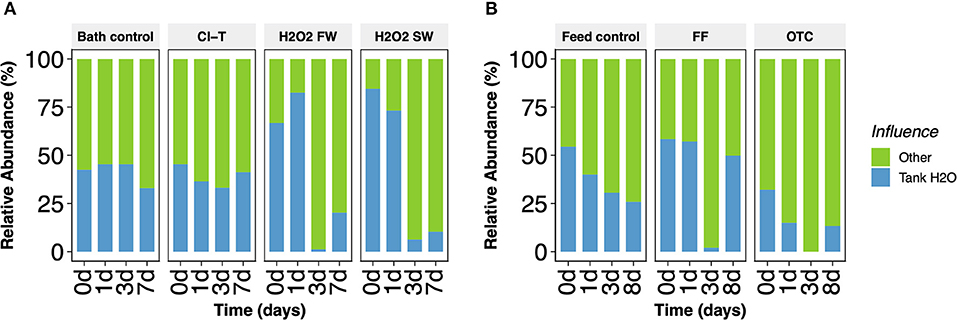
Figure 5. Relative abundance of bacterial sources obtained in gill mucus samples from (A) bath and (B) feed treatment groups. Core bacterial microbiota originating from tank water derived origins (blue) are shown comparative to other sources (green).
The bacterial community upon gill surfaces provide protection to the host by maintaining a diverse range of taxa which deter against pathogenic opportunist microbes. The beneficial action of commensal microbiota is integral to the non-specific immunity of fish species, assisting in the overall defense against infection and disease. In aquaculture and many other primary production industries, the control of disease sometimes involves therapeutic treatments to be administered to animals to prevent substantive stock losses. However, the effect that these treatments have on the commensal microbiota and thus barrier health is not fully understood. The antibacterial treatment options used in this study dramatically reduced bacterial counts derived from the anterior holobranch. Interestingly, this effect was relatively brief for fish treated either with dietary antibiotics or a bath immersion. A rapid increase in bacterial numbers following bath treatments was observed, with an increase in cultivable colonies at the 1- and 6-h timepoints, which was generally lower than the bath control samples. Very low numbers of bacterial colonies detected upon the gills treated with OTC and FF persisted slightly longer, with some evidence of recolonization at 1-day post treatment and peaking at day 3. This was a similar result to that observed by Carlson et al. (2015), where rifampicin treated Gambusia were rapidly recolonized 2.6 days after treatment. Bacterial counts taken in this study however were limited to culturable heterotrophic species, and therefore may not entirely reflect total richness or diversity. Nonetheless, this result does demonstrate that the culturable bacterial load can be significantly lowered using antibacterial treatments. Fish that underwent a sham treatment of filtered seawater displayed lower viable counts compared to unbathed control fish that were not fed antibiotics. It is possible that multiple netting and handling events during the immersion bath process may have impacted upon the gill bacterial community. A study completed by Minniti et al. (2017) demonstrated that the Atlantic salmon skin microbiome was greatly altered after fish were netted from a holding tank and their skin bacterial community differed markedly to unhandled fish for at least 24 h. Handling fish induces an acute stress response in salmonids (Demers and Bayne, 1997) which is typically confluent with increased ventilatory action and shedding of gill mucus (Roberts and Powell, 2005), which may reduce bacterial load within the mucosa. Subsequent post-transfer into a tank of ozonated and filtered seawater may also be attributable to the lower overall CFU.ml counts (and slower recruitment) than what was recorded in the in-feed treatment groups, where handling had only occurred 10 days prior for that cohort. Irrespective of handling effect, the overall antimicrobial action resulted in more marked reductions in cultivable colonies in fish that were exposed to antimicrobial treatments.
Quantitative PCR of the fish gill mucus in this study also demonstrated that there was a brief but significant reduction of 16S rRNA gene copies noted within the OTC and to lesser extent the FF antibiotic treatments at the completion of the 10-day course. Despite being a very different assessment technique to the aerobic count plates for evaluating bacterial load, there appears to be some agreement between results. Previous studies have correlated 16S rDNA assays with reasonable agreement to colony count methodologies (Bach et al., 2002), a result that is logical given the high proportion of aerobic heterotrophic bacteria present in aquatic environs (Cole et al., 1988). These results in the current study demonstrate that both methods show the required sensitivity to capture differences in the bacterial load on the gill surface after antimicrobial treatment in antibiotic fed fish. The interpretation of SYBR green 16S qPCR data for immersion bath groups appeared to remain relatively static in comparison. A possible limitation of this assay for oxidative bath treatments is an inability to distinguish between bacterial DNA that originates from killed or live cells at the time of sampling. It is possible that a reduction in 16S gene expression may not be observed in the oxidative bath treatments, but the treatments may have still had a significant bactericidal effect. A process such as screening samples with propidium-monoazide (PMA) to remove DNA from lysed cells and measure only live bacterial DNA would be useful to attain further accuracy in future studies, as demonstrated in previous studies (Li et al., 2017). Shannon diversity indices also reflected the post-treatment results observed in the count plate and qPCR data for the antibiotic groups. Both OTC and FF began at a very low Shannon index, which gradually increased throughout the trial period, presumably as bacteria were able to recolonize the gill surface. Richness of the bacterial communities were largely unchanged throughout this early post-treatment period. It may be possible that taxa that were naturally resistant to specific antimicrobial treatments were able to proliferate easily with a lack of competition and substantial nutrient resource (Noga, 2010).
Taxonomically, largescale changes in the bacterial community were noted immediately after bath treatments. This indicates that the composition of the bacterial community was likely impacted by the treatment applications. Most groups appeared to share a low number of dominant taxa (predominantly Cutibacterium and Rubritalea) from 0–6 h, but this began to diverge into more disparate communities. The taxonomic analysis did however show a significant shift in phylum and genus from the initial samples and the bath control fish. Interestingly, the movement of fish from freshwater back to marine water in the H2O2 FW group resulted in total removal of Rubritalea, which colonized again one day post-bath. This is not surprising given that the Rubritaleaceae are generally psychrophilic marine based bacterium (Song et al., 2019), however the function of this taxon may require more investigation as a colonizer of the gill surface in prior studies (Schmidt et al., 2017; Wilkes Walburn et al., 2019; Slinger et al., 2020). This taxon appeared to colonize the gill mucus in the highest proportion during both antibiotic treated groups. Source-tracking analysis (FEAST) compared the gill mucus community to the tank water sources collected in this study to determine the proportion of the gill community that is directly influenced by the water environment. Commonality between the 100 most abundant ASVs derived from gill mucus and tank water was very high. The unique species found on the gill surface only related to several known nitrogen synthesizing taxa (Marivita, Nitrotoga and Nitrosomonas) (Yoon et al., 2013; Dang et al., 2017), along with chemoautotrophs which may occupy favorable niche habitats on the gill surface (e.g., Marinomonas, Pseudorhodobacter and Micrococcus) (Nierychlo et al., 2020). Interestingly, this analysis demonstrates that fish bathed in hydrogen peroxide (both freshwater and saltwater) were initially more likely to recruit gill microbiota from the source tank water, and lacked the enrichment from these functional gill-based taxa. The dominance of tank water sourced gill bacteria was reduced 3 days post-treatment, but suggested that gill mucus recolonization may at least initially be reflective of microbiota from the external milieu. Water samples obtained at four timepoints from experimental tanks were largely consistent, but showed a significant group effect between OTC and Cl-T tank water. This may be due to the poor oral bioavailability of oxytetracycline in marine fish, where complexation is likely responsible for a lack of effective absorption when given as medicated feed in seawater. It is thought that around 90% of the drug passes into the receiving environment in the form of uneaten feed, feaces and urine (Noga, 2010).
The diversity of bacterial taxa were greatly reduced in antibiotic treated fish in the current study. Shannon diversity indices were progressively higher in the OTC and FF groups post-course over a 14 day period, with the respective taxonomic analyses demonstrating that gill mucus was dominated by Rubritalea, Cutibacterium and Tenacibaculum at the initial timepoints. Treatment with such broad-spectrum antibiotics have been known to remove a significant proportion of the host commensal community, which can cause a range of detrimental effects including increased disease susceptibility (Gupta et al., 2019; Rosado et al., 2019). Studies within mammalian biology have demonstrated that broad spectrum antibiotic application has significant impacts on the host microbiota. In mice, a distinct decrease in species diversity and subsequently higher rates of pathogen colonization have been observed (Sekirov et al., 2008). Channel catfish (I. punctatus) treated with in-feed florfenicol at 20 mg.kg demonstrated a decrease in diversity compared to non-treated fish, and a significant dysbiosis dominated by the genus Plesiomonas for 10 days post treatment (Wang et al., 2019). Some consideration has also been given to mitigating the effects of antibiotic usage with an addition of a commensal or functional probiotic. For example, Schmidt et al. (2017) demonstrated that Phaeobacter inhibens S4Sm and Bacillus pumilus RI06-95Sm could effectively colonize fish gills and alleviate the impacts of antibiotic usage post challenge with V. anguillarum.
An increase in known pathogen-associated bacteria was observed after treatment in this study, although it appears several taxa are present in low numbers as part of a normal bacterial community. Taxa including Vibrio and Pseudoalteromonas were most abundant toward the end of the post-treatment period for OTC, FF and bathing in H2O2 with saltwater. These taxa, in combination with Tenacibaculum, Staphylococcus, Aliivibrio, Pseudomonas and Photobacterium made up a high proportion of the 100 most prevalent ASVs detected in this study. This assemblage of known pathogenic microbiota is known as the “pathobiome,” with proliferation of this clade leading to negative impacts to the host by promoting multifocal health issues (Sweet and Bulling, 2017; Bass et al., 2019). These data indicate that antimicrobial treatment and associated husbandry stressors of apparently healthy stock could potentially lead to an increase in abundance of potentially harmful clades of bacterium. It is known that pathogenic threats can be effectively neutralized by a functioning and diverse commensal bacterial layer (Cabillon and Lazado, 2019). The reduction in commensal bacteria from antimicrobial treatment may have resulted in nutrient rich areas of low bacterial density or mucus layer coverage where infiltration of such opportunistic bacterium could occur. It is possible that branchial gill damage from the various treatment options may have removed or partially impacted the mucosal layer initially (Powell and Perry, 1997), which may have also played a role in the subsequent colonization. Both antibiotics used in the current study have been observed to cause immunosuppressive effects to the specific and innate salmonid immune systems (Lundén et al., 1999; Noga, 2010; Enis Yonar et al., 2011). Similarly, the oxidative action of both chloramine-T and hydrogen peroxide are known to cause innate immune suppression in Atlantic salmon and rainbow trout (Yavuzcan Yildiz et al., 2009; Vera and Migaud, 2016). Such suppression to the immune function of fish gills may provide scope for pathogenic species to more readily colonize these areas, increasing host susceptibility to infection. Such host susceptibility extends to the terrestrial environment, where honey bees exposed to the herbicide glyphosate suffer microbial imbalances to the gut microbiome. Animals challenged with the known pathogen Serratia marcescens suffer higher mortality as a result of this antimicrobial action (Motta et al., 2018). Further substantiation is required to determine if direct impacts of the antimicrobial treatment, possible immunosuppression on the host (from treatment), or the absence of the commensal microbiota after microbial reduction/alteration can influence pathogenic susceptibility and colonization of the gill.
In this study we examined several antimicrobial treatment applications that successfully reduced cultivable gill bacteria, and caused significant post-treatment impacts on branchial bacteriomic diversity and taxonomic composition. The ability to confidently infer treatment effects here may be limited as the study was restricted to sampling biological replicates within a single experimental unit for each treatment group. However, the results from this study support previous research suggesting that antimicrobial treatments may have significant and lasting effects on the composition of branchial microbiota. The antimicrobial treatments identified in this study would be suitable for future studies of branchial dysbiosis and its potential impacts upon microbial gill diseases or more broadly the role of commensal microbiota in fish health.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA718152.
The animal study was reviewed and approved by CSIRO Queensland Animal Ethics Committee.
JS, JW, and MA prepared the original project concept, formulated the study design and project objectives, performed data interpretation, and wrote the manuscript, with input from all co-authors. JS completed the trial and provided samples and performed nucleic acid extractions. JS and JW performed bioinformatic analysis. All authors contributed to the article and approved the submitted version.
Funding was provided by the Institute for Marine and Antarctic Studies (University of Tasmania), via provision of a PhD scholarship awarded to JS.
This study received funding from Tassal Group Ltd. The funder was not involved in the study design, collection, analysis, interpretation of data, the writing of this article or the decision to submit it for publication. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The authors wish to thank Dr. Richard Taylor for review of this manuscript, and the CSIRO salmon health facility technical staff (Chris Stratford and Lambertus Koster) for their contribution to husbandry of salmon stocks used in this study.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fanim.2021.756101/full#supplementary-material
Supplementary Figure S1. Rarefaction curve of gill mucus and water filter samples in the current study. Samples were subsetted to an even sampling depth of 7,171, removing one failed H2O sample and one failed swab sample, along with the swab blank and NTC controls. Water filter samples demonstrated a consistently higher species richness compared to gill mucus samples.
Supplementary Figure S2. Heatmap of the 10 most abundant genus level assignments obtained from gill mucus samples, depicting relative abundance percentage of taxa temporally for each treatment group, for ease of visualization of the three dominant taxon in this dataset.
Supplementary Figure S3. Relative abundance of genus level ASV assignments within tank water samples collected, displayed within treatment groups. Samples were collected at four timepoints throughout the trial (n = 1 for each bar).
Adams, M. B., Crosbie, P. B. B., and Nowak, B. F. (2012). Preliminary success using hydrogen peroxide to treat Atlantic salmon, Salmo salar L., affected with experimentally induced amoebic gill disease (AGD). J. Fish Dis. 35, 839–848. doi: 10.1111/j.1365-2761.2012.01422.x
Bach, H. J., Tomanova, J., Schloter, M., and Munch, J. C. (2002). Enumeration of total bacteria and bacteria with genes for proteolytic activity in pure cultures and in environmental samples by quantitative PCR mediated amplification. J. Microbiol. Methods 49, 235–245. doi: 10.1016/S0167-7012(01)00370-0
Bandilla, M., Valtonen, E. T., Suomalainen, L. R., Aphalo, P. J., and Hakalahti, T. (2006). A link between ectoparasite infection and susceptibility to bacterial disease in rainbow trout. Int. J. Parasitol. 36, 987–991. doi: 10.1016/j.ijpara.2006.05.001
Bass, D., Stentiford, G. D., Wang, H. C., Koskella, B., and Tyler, C. R. (2019). The pathobiome in animal and plant diseases. Trends Ecol. Evol. 34, 996–1008. doi: 10.1016/j.tree.2019.07.012
Beck, B., and Peatman, E. (2015). Mucosal Health in Aquaculture. Waltham, MA: Academic Press. doi: 10.1016/B978-0-12-417186-2.00013-3
Bland, F., McIntosh, R., Bain, N., and Snow, M. (2012). Development and validation of a range of endogenous controls to support the implementation of practical Taqman real-time PCR-based surveillance for fish diseases within aquaculture. J. Fish Dis. 35, 447–454. doi: 10.1111/j.1365-2761.2012.01363.x
Bokulich, N. A., Kaehler, B. D., Rideout, J. R., Dillon, M., Bolyen, E., Knight, R., et al. (2018). Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2's q2-feature-classifier plugin. Microbiome 6:90. doi: 10.1186/s40168-018-0470-z
Bowker, J. D., Carty, D. G., Telles, L., David, B., and Oviedo, D. (2008). Efficacy of chloramine-T to control mortality in freshwater-reared salmonids diagnosed with bacterial gill disease. N. Am. J. Aquac. 70, 20–26. doi: 10.1577/A06-042.1
Bullock, G. L., Herman, R. L., and Waggy, C. (1991). Hatchery efficacy trials with chloramine-t for control of bacterial gill disease. J. Aquat. Anim. Health 3, 48–50. doi: 10.1577/1548-8667(1991)003<0048:HETWCT>2.3.CO;2
Cabillon, N., and Lazado, C. (2019). Mucosal barrier functions of fish under changing environmental conditions. Fishes 4:2. doi: 10.3390/fishes4010002
Callahan, B. J., McMurdie, P. J., Rosen, M. J., Han, A. W., Johnson, A. J. A., and Holmes, S. P. (2016). DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583. doi: 10.1038/nmeth.3869
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336. doi: 10.1038/nmeth.f.303
Carlson, J. M., Hyde, E. R., Petrosino, J. F., Manage, A. B. W., and Primm, T. P. (2015). The host effects of Gambusia affinis with an antibiotic-disrupted microbiome. Comp. Biochem. Physiol. Part - C Toxicol. Pharmacol. 178, 163–168. doi: 10.1016/j.cbpc.2015.10.004
Cole, J., Findlay, S., and Pace, M. (1988). Bacterial production in fresh and saltwater ecosystems: a cross-system overview. Mar. Ecol. Prog. Ser. 43, 1–10. doi: 10.3354/meps043001
Dang, H., Jose Alava, J., Zhuo Jia, L., Huang, Z., He, J., Hou, D., et al. (2017). Environmental factors shape water microbial community structure and function in shrimp cultural enclosure ecosystems. Front. Microbiol 8, 2359. doi: 10.3389/fmicb.2017.02359
Davis, N. M., Proctor, D. M., Holmes, S. P., Relman, D. A., and Callahan, B. J. (2018). Simple statistical identification and removal of contaminant sequences in marker-gene and metagenomics data. Microbiome 6, 1–14. doi: 10.1186/s40168-018-0605-2
de Bruijn, I., Liu, Y., Wiegertjes, G. F., and Raaijmakers, J. M. (2018). Exploring fish microbial communities to mitigate emerging diseases in aquaculture. FEMS Microbiol. Ecol. 94, 1–12. doi: 10.1093/femsec/fix161
Demers, N. E., and Bayne, C. J. (1997). The immediate effects of stress on hormones and plasma lysozyme in rainbow trout. Dev. Comp. Immunol. 21, 363–373. doi: 10.1016/S0145-305X(97)00009-8
Derome, N., Gauthier, J., Boutin, S., and Llewellyn, M. (2016). “Bacterial opportunistic pathogens of fish,” in The Rasputin Effect: When Commensals and Symbionts Become Parasitic. Advances in Environmental Microbiology, Vol. 3, ed C. Hurst (Cham: Springer), 81–108. doi: 10.1007/978-3-319-28170-4_4
Egan, S., and Gardiner, M. (2016). Microbial dysbiosis: rethinking disease in marine ecosystems. Front. Microbiol. 7:991. doi: 10.3389/fmicb.2016.00991
Enis Yonar, M., Mişe Yonar, S., and Silici, S. (2011). Protective effect of propolis against oxidative stress and immunosuppression induced by oxytetracycline in rainbow trout (Oncorhynchus mykiss, W.). Fish Shellfish Immunol. 31, 318–325. doi: 10.1016/j.fsi.2011.05.019
Francino, M. P. (2016). Antibiotics and the human gut microbiome: dysbioses and accumulation of resistances. Front. Microbiol. 6:1543. doi: 10.3389/fmicb.2015.01543
Genaro Sanchez, J., Speare, D. J., MacNair, N., and Johnson, G. (1996). Effects of a prophylactic chloramine-T treatment on growth performance and condition indices of rainbow trout. J. Aquat. Anim. Health 8, 278–284. doi: 10.1577/1548-8667(1996)008<0278:EOAPCT>2.3.CO;2
Gjessing, M. C., Steinum, T., Olsen, A. B., Lie, K. I., Tavornpanich, S., Colquhoun, D. J., et al. (2019). Histopathological investigation of complex gill disease in sea farmed Atlantic salmon. PLoS ONE 14:e0222926. doi: 10.1371/journal.pone.0222926
Gómez, G. D., and Balcázar, J. L. (2008). A review on the interactions between gut microbiota and innate immunity of fish. FEMS Immunol. Med. Microbiol. 52, 145–154. doi: 10.1111/j.1574-695X.2007.00343.x
Gupta, S., Fernandes, J., and Kiron, V. (2019). Antibiotic-induced perturbations are manifested in the dominant intestinal bacterial phyla of Atlantic salmon. Microorganisms 7:233. doi: 10.3390/microorganisms7080233
Henríquez, P., Kaiser, M., Bohle, H., Bustos, P., and Mancilla, M. (2016). Comprehensive antibiotic susceptibility profiling of Chilean Piscirickettsia salmonis field isolates. J. Fish Dis. 39, 441–448. doi: 10.1111/jfd.12427
Kiemer, M. C. B., and Black, K. D. (1997). The effects of hydrogen peroxide on the gill tissues of Atlantic salmon, Salmo salar L. Aquaculture 153, 181–189. doi: 10.1016/S0044-8486(97)00037-9
Lane, D. J., Pace, B., Olsen, G. J., Stahl, D. A., Sogin, M. L., and Pace, N. R. (1985). Rapid determination of 16S ribosomal RNA sequences for phylogenetic analyses. Proc. Natl. Acad. Sci. U. S. A. 82, 6955–6959. doi: 10.1073/pnas.82.20.6955
Li, R., Tun, H. M., Jahan, M., Zhang, Z., Kumar, A., Fernando, D., et al. (2017). Comparison of DNA-, PMA-, and RNA-based 16S rRNA Illumina sequencing for detection of live bacteria in water. Sci. Rep. 7, 1–11. doi: 10.1038/s41598-017-02516-3
López-Gutiérrez, J. C., Henry, S., Hallet, S., Martin-Laurent, F., Catroux, G., and Philippot, L. (2004). Quantification of a novel group of nitrate-reducing bacteria in the environment by real-time PCR. J. Microbiol. Methods 57, 399–407. doi: 10.1016/j.mimet.2004.02.009
Love, M. I., Huber, W., and Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550. doi: 10.1186/s13059-014-0550-8
Lundén, T., Miettinen, S., Lönnström, L. G., Lilius, E. M., and Bylund, G. (1999). Effect of florfenicol on the immune response of rainbow trout (Oncorhynchus mykiss). Vet. Immunol. Immunopathol. 67, 317–325. doi: 10.1016/S0165-2427(98)00232-3
Martinez Arbizu, P. (2019). pairwiseAdonis: Pairwise Multilevel Comparison Using Adonis. R Package Version 0.3. Available online at: https://github.com/pmartinezarbizu/pairwiseAdonis (accessed January 04, 2020).
McMurdie, P. J., and Holmes, S. (2013). phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 8:1217. doi: 10.1371/journal.pone.0061217
Merrifield, D. L., and Rodiles, A. (2015). “Mucosal health in aquaculture,” in Mucosal Health in Aquaculture, eds B. Beck and E. Peatman (Plymouth: Elsevier Inc.), 273–295. doi: 10.1016/B978-0-12-417186-2.00010-8
Minniti, G., Hagen, L. H., Porcellato, D., Jørgensen, S. M., Pope, P. B., and Vaaje-Kolstad, G. (2017). The skin-mucus microbial community of farmed Atlantic salmon (Salmo salar). Front. Microbiol. 8:2043. doi: 10.3389/fmicb.2017.02043
Mitchell, S. O., and Rodger, H. D. (2011). A review of infectious gill disease in marine salmonid fish. J. Fish Dis. 34, 411–432. doi: 10.1111/j.1365-2761.2011.01251.x
Mohammed, H. H., and Arias, C. R. (2015). Potassium permanganate elicits a shift of the external fish microbiome and increases host susceptibility to columnaris disease. Vet. Res. 46, 1–13. doi: 10.1186/s13567-015-0215-y
Motta, E. V. S., Raymann, K., and Moran, N. A. (2018). Glyphosate perturbs the gut microbiota of honey bees. Proc. Natl. Acad. Sci. U. S. A. 115, 10305–10310. doi: 10.1073/pnas.1803880115
Nierychlo, M., Andersen, K. S., Xu, Y., Green, N., Jiang, C., Albertsen, M., et al. (2020). MiDAS 3: An ecosystem-specific reference database, taxonomy and knowledge platform for activated sludge and anaerobic digesters reveals species-level microbiome composition of activated sludge. Water Res. 182:115955. doi: 10.1016/j.watres.2020.115955
Nordmo, R., Varma, K. J., Sutherland, I. H., and Brokken, E. S. (1994). Florfenicol in Atlantic salmon, Salmo salar L.: field evaluation of efficacy against furunculosis in Norway. J. Fish Dis. 17, 239–244. doi: 10.1111/j.1365-2761.1994.tb00219.x
O'Farrell, H. E., Shaw, J. G., Goh, F., Bowman, R. V., Fong, K. M., Krause, L., et al. (2019). Potential clinical utility of multiple target quantitative polymerase chain reaction (qPCR) array to detect microbial pathogens in patients with chronic obstructive pulmonary disease (COPD). J. Thorac. Dis. 11, S2254–S2265. doi: 10.21037/jtd.2019.10.39
Powell, M. D., and Perry, S. F. (1997). Respiratory and acid-base disturbances in rainbow trout blood during exposure to chloramine-T under hypoxia and hyperoxia. J. Fish Biol. 50, 418–428. doi: 10.1006/jfbi.1996.0312
Powell, M. D., Reynolds, P., and Kristensen, T. (2015). Freshwater treatment of amoebic gill disease and sea-lice in seawater salmon production: Considerations of water chemistry and fish welfare in Norway. Aquaculture 448, 18–28. doi: 10.1016/j.aquaculture.2015.05.027
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., et al. (2012). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596. doi: 10.1093/nar/gks1219
Quensen, J. (2020). QsRutils: R Functions Useful for Community Ecology. Available online at: https://github.com/jfq3/QsRutils (accessed January 04, 2020).
R Core Team (2020). R: A language and environment for statistical computing. Vienna: R Foundation for Statistical Computing. Available online at: https://www.R-project.org/ (accessed August 01, 2020).
Roberts, S. D., and Powell, M. D. (2005). The viscosity and glycoprotein biochemistry of salmonid mucus varies with species, salinity and the presence of amoebic gill disease. J. Comp. Physiol. B Biochem. Syst. Environ. Physiol. 175, 1–11. doi: 10.1007/s00360-005-0473-5
Rosado, D., Xavier, R., Severino, R., Tavares, F., Cable, J., and Pérez-Losada, M. (2019). Effects of disease, antibiotic treatment and recovery trajectory on the microbiome of farmed seabass (Dicentrarchus labrax). Sci. Rep. 9, 1–11. doi: 10.1038/s41598-019-55314-4
Rozas-Serri, M. (2019). Gill diseases in marine salmon aquaculture with an emphasis on amoebic gill disease. CAB Rev. Perspect. Agric. Vet. Sci. Nutr. Nat. Resour. 14, 1–15. doi: 10.1079/PAVSNNR201914032
Schmidt, V., Gomez-Chiarri, M., Roy, C., Smith, K., and Amaral-Zettler, L. (2017). Crossm probiotics reduces antibiotic-associated mortality in fish. Am. Soc. Microbiol. 2, 1–13. doi: 10.1128/mSystems.00133-17
Sekirov, I., Tam, N. M., Jogova, M., Robertson, M. L., Li, Y., Lupp, C., et al. (2008). Antibiotic-induced perturbations of the intestinal microbiota alter host susceptibility to enteric infection. Infect. Immun. 76, 4726–4736. doi: 10.1128/IAI.00319-08
Shenhav, L., Thompson, M., Joseph, T. A., Briscoe, L., Furman, O., Bogumil, D., et al. (2019). FEAST: fast expectation-maximization for microbial source tracking. Nat. Methods 16, 627–632. doi: 10.1038/s41592-019-0431-x
Slinger, J., Adams, M. B., and Wynne, J. W. (2020). Comparison of bacterial diversity and distribution on the gills of Atlantic salmon (Salmo salar L.): an evaluation of sampling techniques. J. Appl. Microbiol., jam.14969. doi: 10.1111/jam.14969
Song, J., Kang, I., Joung, Y., Yoshizawa, S., Kaneko, R., Oshima, K., et al. (2019). Genome analysis of Rubritalea profundi SAORIC-165 T, the first deep-sea verrucomicrobial isolate, from the northwestern Pacific Ocean. J. Microbiol. 57, 413–422. doi: 10.1007/s12275-019-8712-8
Sweet, M. J., and Bulling, M. T. (2017). On the importance of the microbiome and pathobiome in coral health and disease. Front. Mar. Sci. 4:9. doi: 10.3389/fmars.2017.00009
Vera, L. M., and Migaud, H. (2016). Hydrogen peroxide treatment in Atlantic salmon induces stress and detoxification response in a daily manner. Chronobiol. Int. 33, 530–542. doi: 10.3109/07420528.2015.1131164
Wang, E., Yuan, Z., Wang, K., Gao, D., Liu, Z., and Liles, M. R. (2019). Consumption of florfenicol-medicated feed alters the composition of the channel catfish intestinal microbiota including enriching the relative abundance of opportunistic pathogens. Aquaculture 501, 111–118. doi: 10.1016/j.aquaculture.2018.11.019
Wickham, H. (2016). ggplot2: Elegant Graphics for Data Analysis. New York, NY: Springer-Verlag New York Available online at: https://ggplot2.tidyverse.org (accessed January 12, 2019).
Wilkes Walburn, J., Wemheuer, B., Thomas, T., Copeland, E., O'Connor, W., Booth, M., et al. (2019). Diet and diet-associated bacteria shape early microbiome development in Yellowtail Kingfish (Seriola lalandi). Microb. Biotechnol. 12, 275–288. doi: 10.1111/1751-7915.13323
Wilson, J. M., and Laurent, P. (2002). Fish gill morphology: inside out. J. Exp. Zool. 293, 192–213. doi: 10.1002/jez.10124
Wynne, J. W., Thakur, K. K., Slinger, J., Samsing, F., Milligan, B., Powell, J. F. F., et al. (2020). Microbiome profiling reveals a microbial dysbiosis during a natural outbreak of tenacibaculosis (Yellow Mouth) in Atlantic Salmon. Front. Microbiol. 11:586387. doi: 10.3389/fmicb.2020.586387
Yavuzcan Yildiz, H., Meric, I., Mehmet, G., and Ergonul, B. (2009). Changes of non-specific immune parameters in rainbow trout, oncorhynchus mykiss, after exposure to antimicrobial agents used in aquaculture. J. Appl. Aquac. 21, 139–150. doi: 10.1080/10454430903113529
Yoon, J. H., Kang, S. J., and Lee, U. S. (2013). Marivita geojedonensis sp. nov., isolated from seawater. Int. J. Syst. Evol. Microbiol. 63, 423–427. doi: 10.1099/ijs.0.039065-0
Keywords: dysbiosis, Atlantic salmon, antimicrobial, treatment, pathobiome, mucosal health
Citation: Slinger J, Wynne JW and Adams MB (2021) Profiling Branchial Bacteria of Atlantic Salmon (Salmo salar L.) Following Exposure to Antimicrobial Agents. Front. Anim. Sci. 2:756101. doi: 10.3389/fanim.2021.756101
Received: 10 August 2021; Accepted: 01 November 2021;
Published: 22 November 2021.
Edited by:
Carlo C. Lazado, Norwegian Institute of Food, Fisheries and Aquaculture Research (Nofima), NorwayCopyright © 2021 Slinger, Wynne and Adams. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Joel Slinger, am9lbC5zbGluZ2VyQGNzaXJvLmF1
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.