- Université Paris Saclay, Université Paris Cité, ENS Paris Saclay, CNRS, SSA, INSERM, Centre Borelli, Gif-sur-Yvette, France
This article describes a non-parametric clustering algorithm with an outlier removal step. Our method is based on tools from topological data analysis: we define a new filtration on metric spaces which is a variant of the Vietoris–Rips filtration that adds information about the points' nearest neighbor to the persistence diagram. We prove a stability theorem for this filtration, and evaluate our method on point cloud and graph datasets, showing that it can compete with state-of-the-art methods while being non-parametric.
1 Introduction
Agglomerative hierarchical clustering is one of the main classes of clustering algorithms [1, 2]. It considers each data point as its own cluster and iteratively merges clusters. All state-of-the-art clustering algorithms require to choose parameters such as the number of clusters (K-means, spectral clustering, etc.), some thresholds (hierarchical clustering), or a density parameter for density-based clustering [DBSCAN [3, 4] and ToMATo [5]]. The choice of parameters can be problematic in some cases, especially when clustering is only one step in a larger algorithm.
Topological data analysis (TDA), and more specifically persistent homology [6, 7], has been a popular research subject over the last decades, with many applications [8]. The idea is to construct a sequence of simplicial complexes (a filtration) from given data and to study how its structure evolves when going through the filtration, using a tool called persistence diagram. The success of TDA is partly due to stability theorems, which state that close datasets will have close persistence diagrams, according to some distance between datasets/diagrams, ensuring in particular that TDA results are robust to noise. One of the most used filtrations is the Vietoris–Rips filtration, for which a stability theorem has been proved in [9].
Concretely, 0-dimensional persistent homology links data points until only one connected component is left. It thus naturally induces a hierarchical clustering algorithm as the one described in [10–12], that use the Vietoris–Rips filtration to study brain networks or co-occurrence networks, or the ToMATo algorithm [5], which combines TDA with a density-based approach. Persistent homology highlights important components of the data and is thus a suitable tool for threshold choice in hierarchical clustering. In this study, we introduce a new filtration for metric spaces1 that adds information about the points' nearest neighbor to the persistence diagram which makes it easier to identify clusters on the diagram and to remove outliers. We prove its stability and use it to define a non-parametric algorithm to perform clustering with outlier removal.2
Section 2 is an introduction to TDA. In Section 3, we introduce our filtration and proved its stability. In Section 4, we present the algorithm and some experiments on point cloud and graph datasets.
2 Topological data analysis background
In this section, we introduce some notions of topological data analysis.
2.1 Simplicial complexes and filtrations
Definition 1. A k-simplex on a set X is an unordered tuple σ = [x0, ..., xk] of k+1 distinct elements of X. The elements x0, ..., xk are called the vertices of σ. If each vertex of a simplex τ is also a node of σ, then τ is called a face of σ. A simplicial complex K is a set of simplices such that any face of a simplex of K is a simplex of K. We call the set of all simplices on X.
Definition 2. An increasing filtration3 of a simplicial complex K is a family of simplicial complexes (Kα)α≥0 such that K0 = ∅, and ⋃α≥0Kα = K.
This definition is useful for theory, but in practice, the number of simplices is finite, in which case, we will only use a finite set of indices αi such that and αi ≤ α < αi+1⇒Kαi = Kα. Without loss of generality, we can also assume that for all i, there exists a simplex σi+1∈K such that . See Figure 1 and [6, 7, 13] for examples of filtrations.
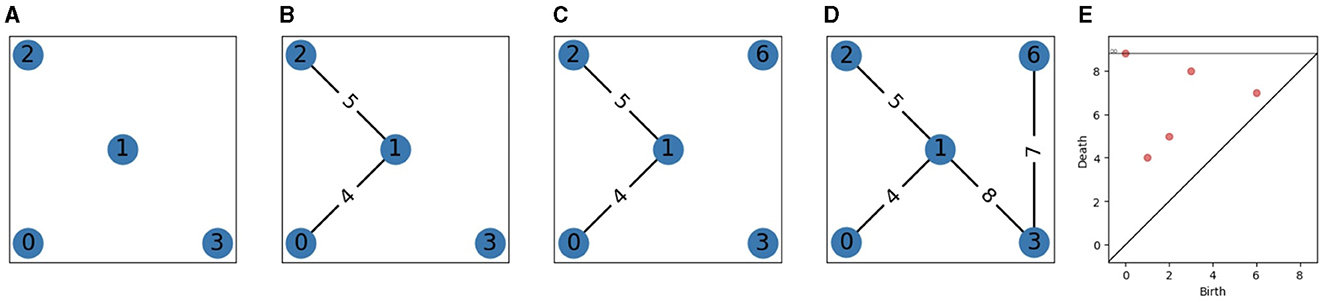
Figure 1. A filtration and its persistence diagram. (A) Steps 0 to 3 (add vertices 0, 1, 2, and 3). (B) Steps 4 and 5 (add edges 4 and 5). (C) Step 6 (add vertex 6). (D) Steps 7 and 8 (add edges 7 and 8). (E) 0-dimensional persistence diagram.
In clustering applications, data are a (finite) metric space (X, d). A common and easily computable filtration on X is the Vietoris–Rips filtration VR(X) = (VR(X, α))α≥0 defined for all simplices σ by:
The main idea of TDA is to build a filtration on top of the data and study how the structure of the simplicial complexes evolves while increasing the filtration parameter using persistent homology. Our clustering algorithm only requires 0-dimensional persistent homology, which we will explain in terms of connected components. Persistent homology is defined for higher dimensions in Section 2.3 (we define it for the sake of completeness, as Theorem 10 is about all dimensions).
2.2 0-dimensional persistent homology
Let be a filtration. For each index i, we go from to by adding a simplex σi+1 to . If σi+1 is a vertex, then it creates a new connected component in , and we say that this component is born at αi+1. If σi+1 is an edge, it links two vertices that where already in (by definition of a simplicical complex). If those vertices belonged to different connected components in , they become one component in : we say that one of the two components (the one with the lowest birth date, by convention) absorbed the younger one, and that the younger one died at αi+1. In all other cases, there is no change in connected components when adding σi+1 (changes are made in higher dimensional homology, see Section 2.3). At the end of the filtration, all remaining components die at α = ∞. The 0-dimensional persistence diagram of a filtration is defined as the multiset of all pairs (birthdateanddeathdate) obtained by the above construction, counted with multiplicity, along with all pairs (x, x) with infinite multiplicity.
See Figures 1A–E for an example of persistence diagram. If (b, d) is a point of a persistence diagram, we call d−b its persistence (or the persistence of the corresponding connected component).
A distance between persistence diagrams can be defined as follows:
Definition 3. The bottleneck distance between diagrams D and D′ is defined as:
where Γ(D, D′) is the set of bijections from D to D′, where a point of multiplicity m is treated as m points, and where ||p−q||∞ = |xp−xq| when p = (xp, yp), q = (xq, yq) and yp = yq = +∞.
Note that it is necessary to include the diagonal in persistence diagrams so there exists bijections between diagrams that do not have the same number of persistent pairs.
2.3 Persistent homology in any dimension
2.3.1 Simplicial homology
Let K be a simplicial complex with maximal simplex dimension d, 𝔽 be a field and 0 ≤ k ≤ d.
Definition 4. The space Ck(K) of k-chains is defined as the set of formal sums of k-simplices of K with coefficients in F, that is to say, if all the k-simplices of K are σ1, …, σnk, all the elements of the form:
Ck(K) is a vector space whose addition and scalar multiplication are naturally defined as follows: if and , then:
and for λ ∈ 𝔽,
Using a field F gives the more general definition, but from now on, we will only consider 𝔽 = ℤ/2ℤ, so the coefficients are modulo 2, which allows to avoid orientation considerations.
Definition 5. Let σ = [v1, …, vk] be a k-simplex with vertices v1, …, vk, and be the (k−1)-simplex spanned by those points minus vi. The boundary operator ∂ is defined as:
We have the following sequence of linear maps:
They satisfy ∂°∂ = 0 : we call such a sequence of maps a chain complex. This constitutes the setup for homology. We can now define cycle and boundaries, homology groups, and Betti numbers.
Definition 6. We define the set Zk(K) of k-cycles of K as :
and the set Bk(K) of k-boundaries of K as :
We have Bk(K) ⊂ Zk(K) ⊂ Ck(K), so we can define the kth homology group as:
and the kth Betti number:
βk represents the number of k-dimensional “holes”. For example, β0 is the number of connected components of K, β1 is the number of loops, and β2 is the number of voids. Betti numbers of simplicial complexes are computable using a filtration, see [7] for the algorithm.
2.3.2 Persistent homology
Persistent homology considers studying the evolution of homology groups while increasing the filtration parameter.
Let be a filtration such that for each index i, we go from to by adding a simplex σi+1 to .
We call the respective spaces of k-chains, k-cycles, k-boundaries, kth homology group and kth Betti number of Ki. The goal is to follow the evolution of as i increases. It can be shown [7] that when a k-simplex σi+1 (k>0) is added, it either creates a new homology class in (i.e., a new k-cycle that is independent of those of ) or it closes a k−1-dimensional hole of , so has one less homology class than , in that case, we say that σi+1 killed a homology class (by convention, we always consider that when two classes merge, the younger class gets killed). If k = 0, each new vertex creates a homology class in H0.
The final result of persistent homology is the set of all so-called persistent pairs (σl(j), σj) such that for each j, σl(j) creates a component and σj kills it. We say that the persistence (or lifetime) of such a pair is j−l(j)−1. The algorithms to compute them are described in detail in [7]. The k-dimensional persistence diagram is the set of points of coordinates (αl(j), αj) such that σl(j) is a k-simplex (counted with multiplicity). The points of the diagonal y = x are added with infinite multiplicity (it is useful to define distances). The bottleneck distance between two k-dimensional persistence diagrams is defined exactly as in Definition 3.
3 Nearest Neighbor Vietoris-Rips filtration and stability
In this section, we introduce a new filtration: the Nearest Neighbor Vietoris-Rips (NNVR) filtration, and prove its stability.
Definition 7. Let (X, d) be a metric space. The Nearest Neighbor Vietoris-Rips filtration NNVR(X, α)α≥0 is defined by:
and for all k-simplices σ with k ≥ 1:
This means that the NNVR filtration is the same as the VR filtration except for 0-simplices, which all enter the VR filtration at α = 0, whereas they enter the NNVR filtration when α is equal to the distance to their nearest neighbor. It is easy to verify that NNVR is indeed a filtration. Figure 2 shows an example of NNVR filtration.
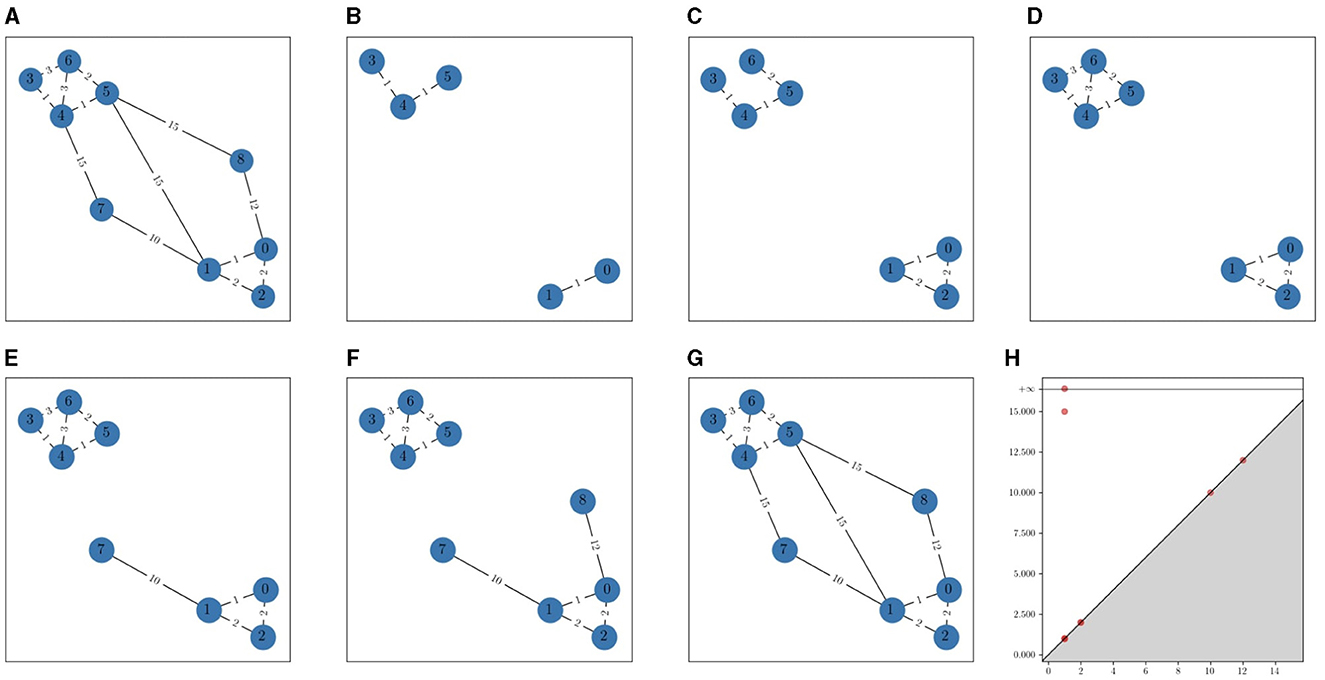
Figure 2. An example of NNVR filtration. The edge weights represent the distance between vertices. For each value of α, vertices and edges with filtration value equal to α are added. (A) Full graph. (B) α = 1. (C) α = 2. (D) α = 3. (E) α = 10. (F) α = 12. (G) α = 15. (H) α = Persistence diagram.
The stability theorem for the VR filtration is proved in [9], we use the same framework as they did to adapt the proof to the NNVR filtration. The theorem states that the NNVR filtration is stable with respect to the Gromov–Hausdorff distance [14], which we define below, along with the notions required to prove the theorem.
Definition 8. Let X, Y be two sets and C ⊂ X × Y. For any A ⊂ X and B ⊂ Y, we note C(A) = {y ∈ Y:∃x ∈ A, (x, y) ∈ C} and C⊤(B) = {x ∈ X:∃y ∈ B, (x, y) ∈ C}. C is called a correspondence between X and Y if C(X) = Y and
C⊤(Y) = X.
Definition 9. Let (X, dX) and (Y, dY) be metric spaces and C be a correspondence between X and Y. The distortion of C is defined as
The Gromov–Hausdorff distance between X and Y is:
We now state the stability theorem.
Theorem 10. Let (X, dX) and (Y, dY) be totally bounded metric spaces. Let DX (resp. DY) be the persistence diagram of NNVR(X) (resp. NNVR(Y)). Then:
To prove this theorem, we introduce simplicial correspondences and use results from [9].
Definition 11. Let (resp. ) be a filtration of a simplicial complex on a set X (resp. Y). A correspondence C is called ε-simplicial, if
• For any α ≥ 0 and simplex , any finite subset of C(σ) is a simplex of .
• For any α ≥ 0 and simplex , any finite subset of C⊤(σ) is a simplex of .
Proposition 4.2 from [9] states that if there is an ε-simplicial correspondence between two filtered metric spaces X and Y, then there exists a so-called ε-interleaving between the persistent homology groups of X and Y which is a family of linear maps between homology groups and for all α, that commute with the persistence structure. Theorem 2.3 from [9] states that if there exists such an ε-interleaving then ε is an upper bound on the bottleneck distance.4 The following lemma synthesizes those results.
Lemma 12. Let (resp. ) be a filtration of a finite simpicial complex on a totally bounded metric space X (resp. Y), with persistence diagrams DX and DY. If there exists an ε-simplicial correspondence, then dB(DX, DY) ≤ ε.
Proof of Theorem 10: Let ε > 2dGH(X, Y). There exists a correspondence C such that (C) ≤ ε. Let us show that C is ε-simplicial.
Let α ≥ 0, σ be a simplex of NNVR(X, α) and τ be a finite subset of C(σ).
Case 1: τ only has one element: τ = [y]. There exists x∈σ such that y∈C(x) and a sequence such that 0 < dX(x,xn) and (dX(x,xn))n∈ℕ converges to a value lower than or equal to α. Moreover, there exists a sequence such that for all n, yn ∈ C(xn). Then:
So: and τ = [y] ∈ NNVR(Y, α + ε).
Case 2: τ = [y0, ..., yk], k≥1. For all i, j, there exists xi, xj ∈ σ such that yi ∈ C(xi) and yj ∈ C(xj). Then :
where the last inequality holds because [xi, xj]∈NNVR(X, α).
The same proof shows that if σ is a simplex in NNVR(Y, α), then all finite subsets of C⊤(σ) are simplices of NNVR(X, α+ε). So, C is ε-simplicial and Lemma 12 ends the proof.
4 Outlier-removing clustering
4.1 Algorithm
Our clustering algorithm relies on the 0-dimensional persistent homology construction described in Section 2.2. The idea is to construct the 0D persistence diagram of the NNVR filtration of the data, then use it to choose a birth threshold (which tells us which points are outliers) and a persistence threshold (which tells us what the clusters are). The thresholds can be chosen manually or automatically. If they are chosen using a non-parametric procedure, then the whole clustering algorithm is non-parametric (see next section for more details). The clustering algorithm works as follows:
• Input: a finite metric space (X, d).
• Compute the filtration NNVR(X) and its 0-dimensional persistence diagram.
• Choose a birth threshold and a persistence threshold.
• Mark all points whose birth dates are above the birth threshold as outliers. Let Y be the remaining points.
• Compute 0-dimensional persistent homology on NNVR(Y), but do not add edges that would merge components whose persistence is above the persistence threshold.
• Output: a simplicial complex, where each connected component is a cluster.
The benefit of using the NNVR filtration is that, compared to the VR filtration, it separates outliers from essential components on the persistence diagram. Components corresponding to outliers indeed have late death dates in both filtrations as they are far away from other points. Thus, they give very persistent points with the VR filtration (where every birth date is 0), which can be hard to distinguish from other persistent points (as illustrated below). The NNVR filtration moves them to the right of the persistence diagram. Note that, if there are no outliers, the same algorithm can be used with the VR filtration and no birth threshold.
Seeing (X, d) as a graph with n vertices and m edges, the filtration can be computed in O(m) (the filtration value of each vertex is the distance to its nearest neighbor, and the filtration value of each edge is its weight, so reading the m edge weights is enough to compute the filtration). Computing 0D persistent homology requires to compute a minimal spanning tree, which can be done in O(m(m, n))) using Kruskall's algorithm. An algorithm to choose the thresholds (such as max-jump, described below) should work in O(n), as there are n points on the persistence diagram. So, the overall complexity of the method is O(m(m, n))), so O(n2(n)) in the worst case, as m ≤ n2, with equality if the whole distance matrix is computed (in Euclidean spaces, for example).
4.2 Choice of thresholds
After computing the persistence diagram D, two thresholds are needed: a birth threshold, which is a threshold on the list of birth dates of points on the diagram, and a persistence threshold, which is a threshold on the list of persistence of points on the diagram. The choice of the birth threshold determines which points are considered as outliers: if a point on the persistence diagram has a birth date above the threshold, we exclude the corresponding point in X and detect it as an outlier. The choice of the persistence threshold determines which components are significant enough to be considered as clusters. The number of points whose persistence is above the threshold is the number of detected clusters.
A natural method to choose the birth and persistence thresholds is to apply the max-jump algorithm to the list of birth dates and to the list of persistence values (without the point of infinite persistence). The algorithm is defined in Algorithm 1.
The idea behind this algorithm is to separate significant values from non-significant ones by using the biggest variation. It is fully non-parametric, and will always output at least two clusters (one corresponding to the infinite persistence point, and at least one point above the persistence threshold). A drawback of this method is that it can output a persistence threshold that is too low if clusters are not at the same distance from one another (see point cloud 5 in section 4.3. for an example).
We propose two possible solutions to this problem, which introduce the choice of a parameter. The first one is to apply a sigmoid function to the normalized list before using max-jump. The normalization makes the data vary from 0 to 1, which makes it easier to choose a unique parameter λ for a whole dataset, and the sigmoid function pushes significant values close to 1, which can lead to a lower threshold than the one obtained with max-jump. The second alternative we propose is using OTSU's method [15] for gray-level threshold selection. This requires to choose a number of bins for the histograms, which we set to 1,024 by default. As the persistence distribution is skewed, we apply the algorithm twice and remove values under the threshold after each iteration. The point cloud experiments on Section 4.3 illustrate how those alternatives can improve clustering. For every other dataset, we will always use max-jump by default. Other methods can be preferred depending on the application, to make use of prior knowledge on the data (for example, knowledge about the expected number of clusters or their size).
Figure 3 shows the thresholded persistence diagram (with max-jump) and the results of our algorithm on the graph from Figure 2.
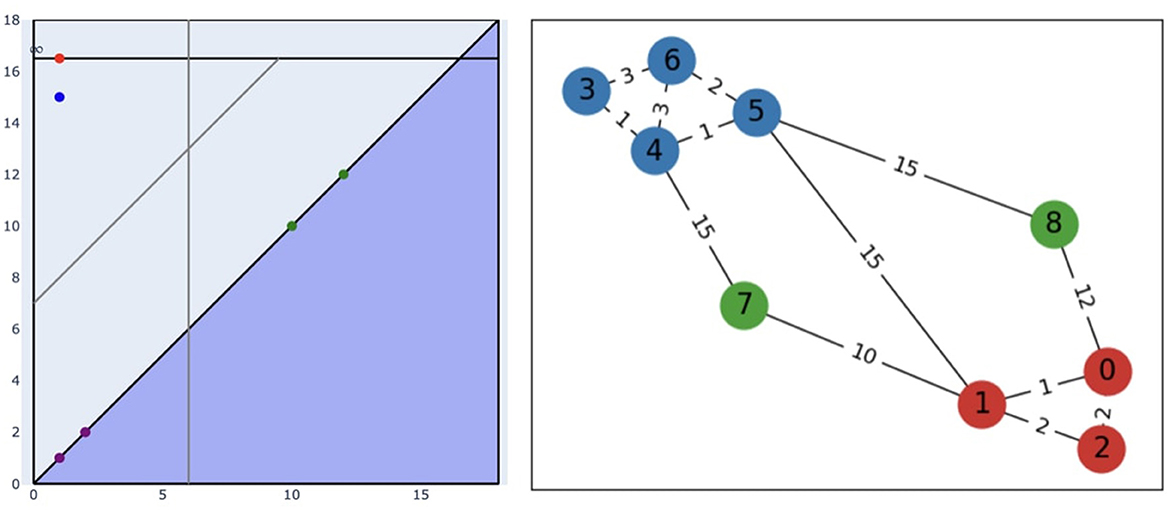
Figure 3. (Left) The persistence diagram from Figure 2 with both thresholds chosen with max-jump. The vertical gray line represents the birth threshold, the gray line parallel to the diagonal represents the persistence threshold (as the persistence of a point is proportional to its distance to the diagonal). (Right) Clusters (blue and red) and outliers (green) obtained with our algorithm. A point of a given color on the diagram represents the component that contains the nodes of this color on the graph.
4.3 Experiments
We applied our algorithm to the karate club graph, which is a classical example for community detection in networks [16], to point clouds from the scikit-learn documentation5 to which we added four outliers (one in each corner), and to synthetic graph datasets. Results were evaluated using the Rand index [17], which is the ratio of the number of correctly labeled points over the total number of points, ignoring label permutations (so perfect clustering gives a Rand index of 1 and the index is close to 0 for bad clustering).
4.3.1 Karate club graph
The karate club graph shows the interactions between the 34 members of a karate club (nodes 0–33). Each interaction is represented by an edge with a weight between 1 and 7 (a high weight means more interaction). After a conflict between the instructor (node 0) and administrator (node 33), the club split up into two groups: one with the instructor and one with the administrator. The goal is to predict those groups based on the interaction. We added 10 outliers (nodes 34–43, that could represent people outside the club), each one with two edges randomly linking it to other nodes with a random weight between 0.5 and 1. We inverted the weights of the edges to get a new weighted graph and used the shortest-path distance.
The results are shown in Figure 4 along with persistence diagrams for the NNVR and VR filtrations, and results obtained with the Louvain Community Detection Algorithm [18], which is a modularity optimization method [we used the NetworkX [19] implementation with default parameters]. The Rand indices are 0.95 for the NNVR method, 0.65 for the VR method, and 0.73 for the Louvain method. Note that points from both diagrams have the same death dates, so outliers have a higher persistence than relevant points on the VR diagram, which is not the case on the NNVR diagram. The algorithm exactly recovers the true clusters for all nodes, except nodes 5, 6, 16, and 10, that form a cluster. This can be explained by the fact that node 16 is not linked to node 0 and only has two links to 5 and 6, and the edges (5,6) and (5,10) are, respectively, heavier than (5,0) and (10,0). This point is more persistent than the blue one on Figure 4 because of the proximity between nodes 8 and 33.
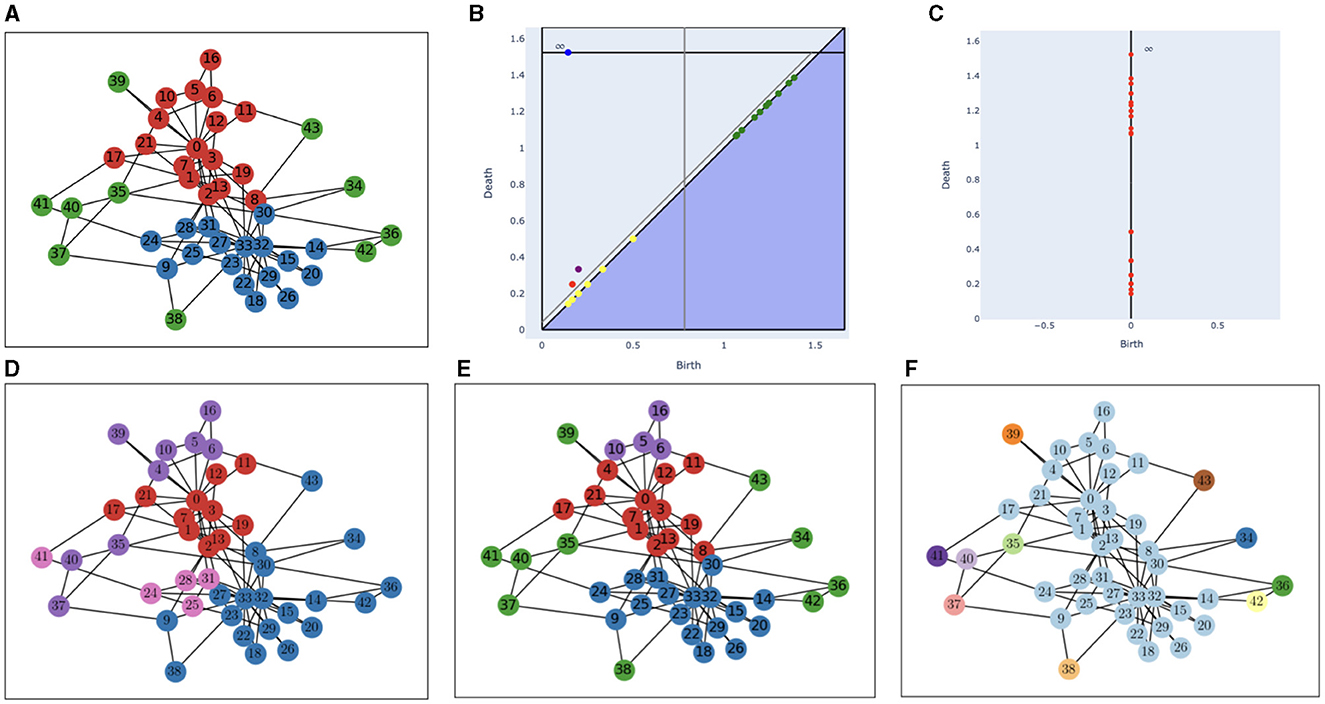
Figure 4. (A) Ground truth for the karate club graph with additional outliers, (B) persistence diagram of the NNVR filtration, (C) persistence diagram of the VR filtration, (D) clusters obtained with the Louvain Community Detection Algorithm, (E) clusters obtained with the NNVR algorithm, and (F) clusters obtained with the VR algorithm. On (B), the two gray lines represent the two thresholds, the blue (resp. red, purple) point corresponds to the blue (resp. red, purple) cluster on the graph, green points correspond to outliers, and yellow points are below the persistence threshold.
4.3.2 Point clouds
In the case of point clouds, the metric space is a set of points in 2D equipped with the Euclidean distance. Figures 5, 6 and Table 1 show the results of 10 clustering algorithms [3, 20–27], and three versions of our algorithm that all use max-jump to choose the birth threshold but different methods (described above) for the persistence threshold: one using max-jump, one using it after applying a sigmoid with parameter 4, and one using OTSU. We also test our algorithm with the VR filtration (and max-jump persistence threshold), on three point clouds. For all algorithms, we used the same parameters as the ones used in the scikit-learn documentation.
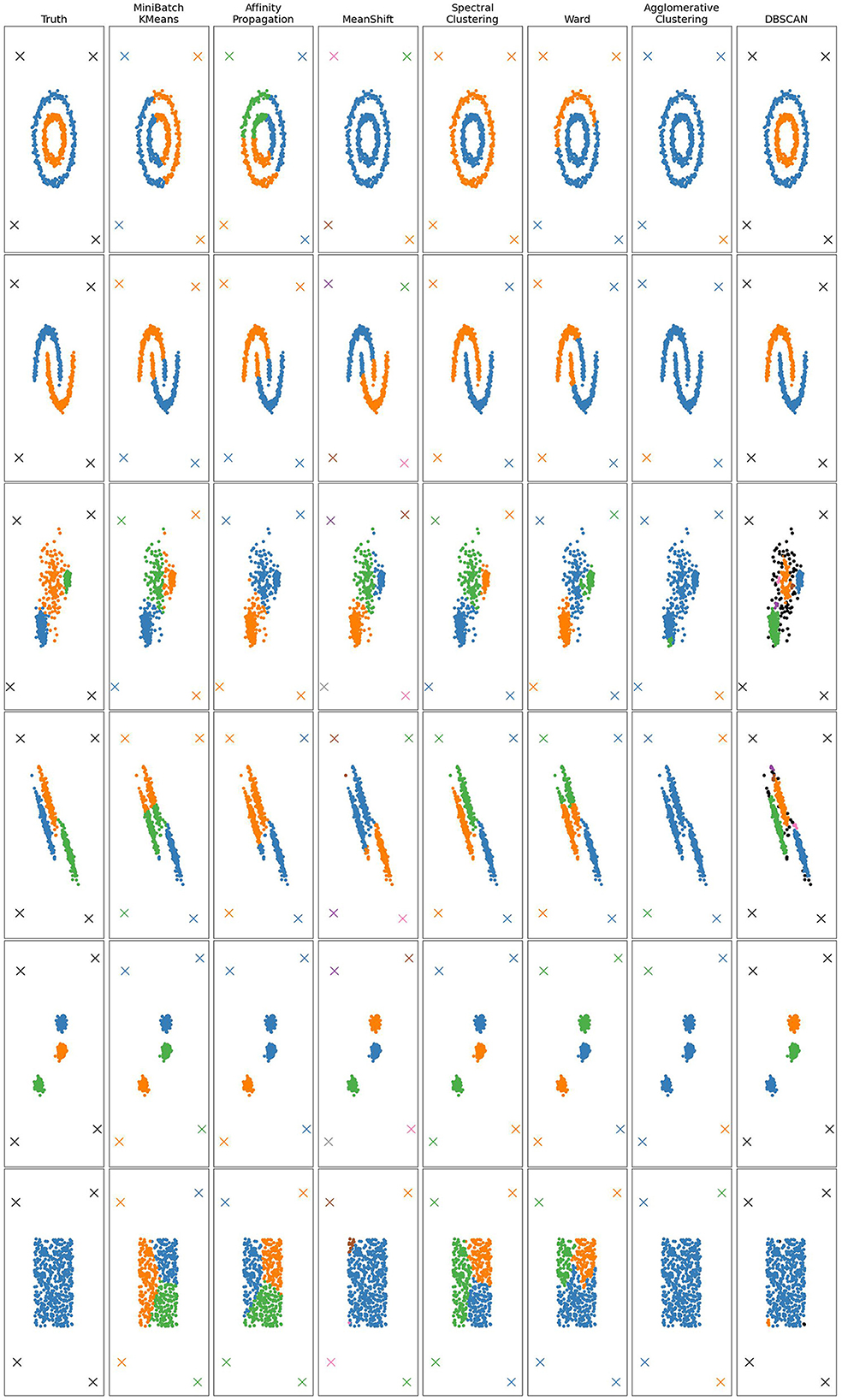
Figure 5. Clustering results for six point clouds obtained with several methods. Points marked with an X are outliers.
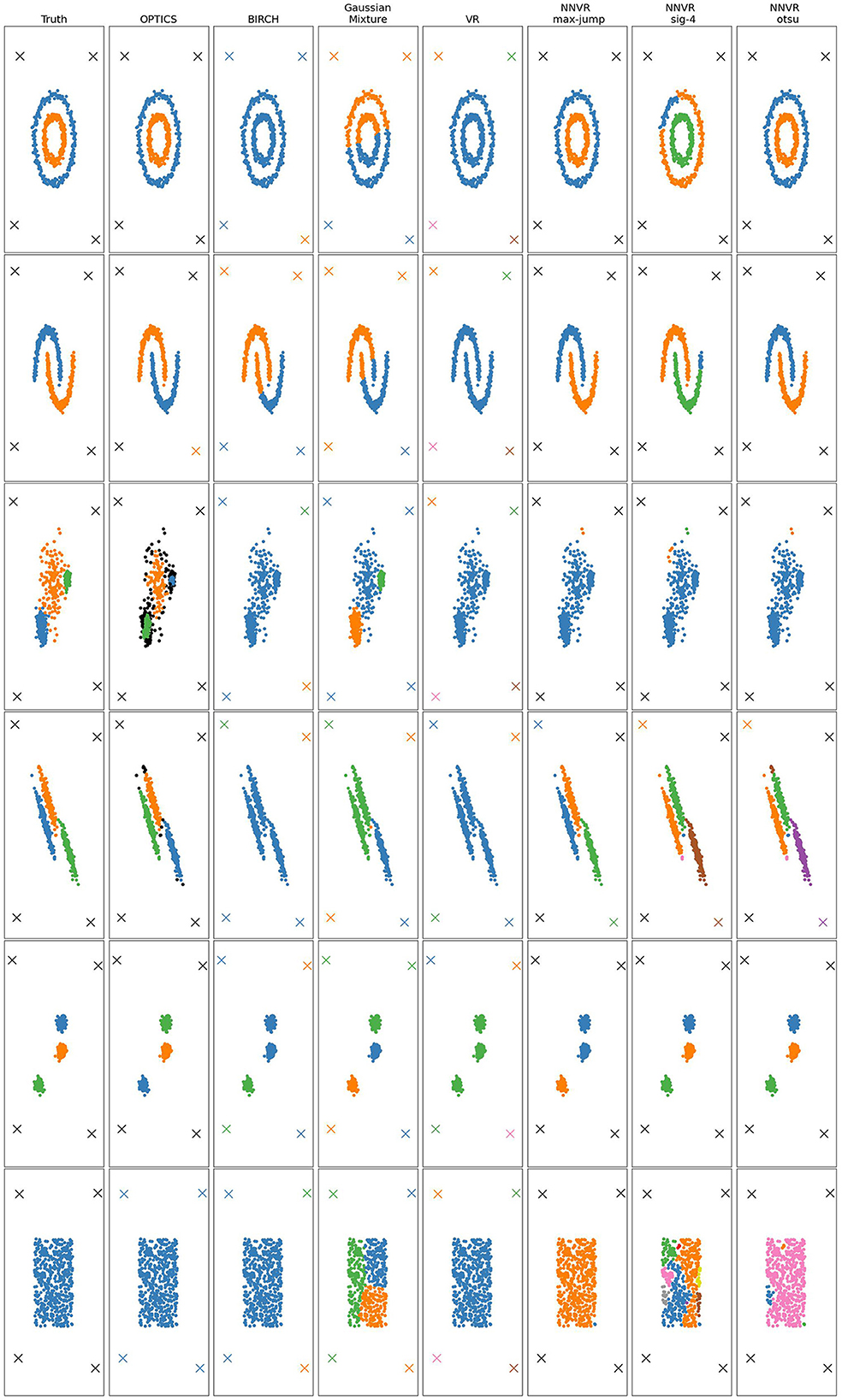
Figure 6. Clustering results for six point clouds obtained with several methods, including the NNVR filtration with max-jump, sigmoid of parameter 4 and OTSU, and VR filtration. Points marked with an X are outliers.
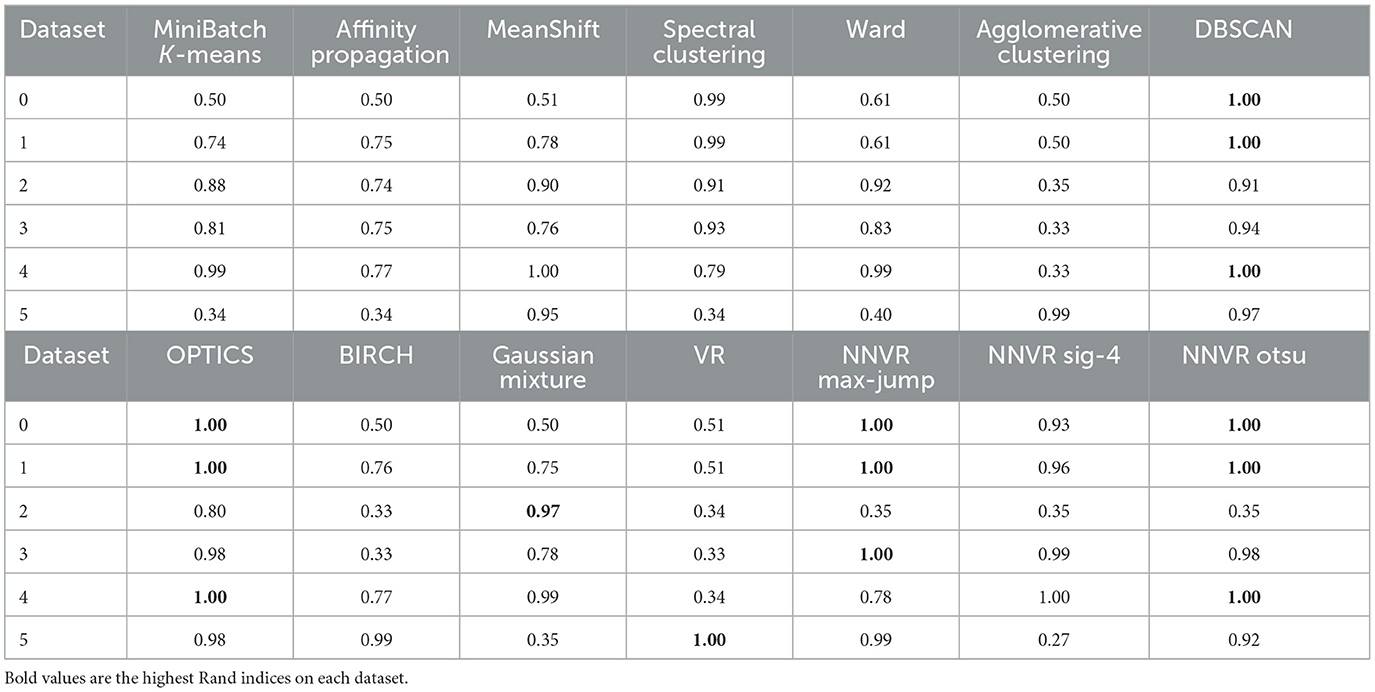
Table 1. Rand indices for the 14 clustering algorithms applied to the six datasets represented on Figures 5, 6.
The NNVR algorithm with max-jump performs almost perfectly on datasets 1, 2, 4, and 6. On dataset 5, the persistence threshold is too high so the algorithm detects two clusters instead of three, which is not the case for the sigmoid and OTSU methods, which work perfectly on this dataset. The drawback of the sigmoid and OTSU methods is that they tend to choose more clusters than max-jump and can thus choose too many such as for datasets 4 and 6 (and 1 and 2 for the sigmoid). Density-based methods outperform the others on dataset 3. Dataset 6 shows that our algorithm cannot output only one cluster. The only methods capable of detecting outliers are DBSCAN, OPTICS, and our NNVR method. Our method only fails to detect two of the four outliers on dataset 4. DBSCAN and OPTICS detect too many outliers on datasets 3 and 4.
4.3.3 Synthetic graphs
We used the NetworkX's [19] function to generate synthetic graph datasets with a number of clusters varying from 2 to 10 and containing outliers. This functions takes as input the size of each cluster, a probability pin, and a probability pout. It outputs a random graph where each edge has a probability pin of existing between two nodes from the same cluster, and pout if they are from different clusters. The data were generated as follows:
• Fix a number of iterations n and a number of clusters k, pin = 0.3 and pout = 0.01.
• Set the size of each cluster to 5 plus a random integer between −3 and 3.
• Add five clusters of size 1 (the outliers).
• Generate n graphs with the above procedure, and add their adjacency matrices together. The resulting matrix is the adjacency matrix of a weighted graph Gn, k.
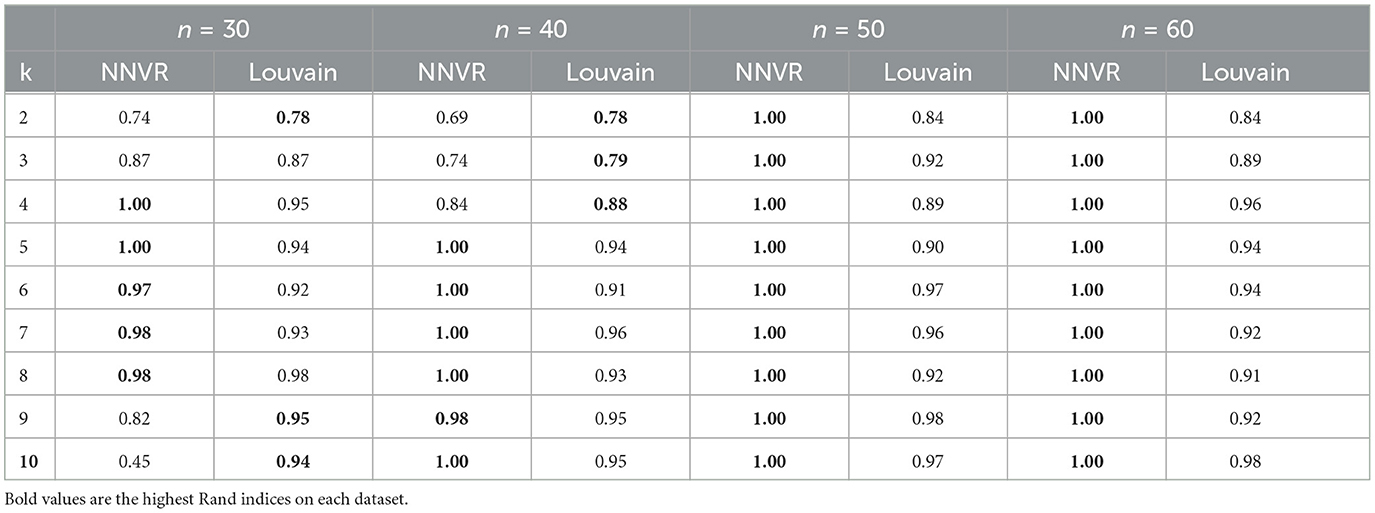
See Figure 7 for examples. Table 2 shows the results obtained with our algorithm (using max-jump for both thresholds) and with the Louvain method [18], for n = 30, 40, 50 and k varying from 2 to 10. Our method performs better than Louvain's on 5 and 6 graphs out of 9 for n = 30 and 40, respectively, and for higher values of n, when clusters are denser, our method always detects the right clusters and outliers.
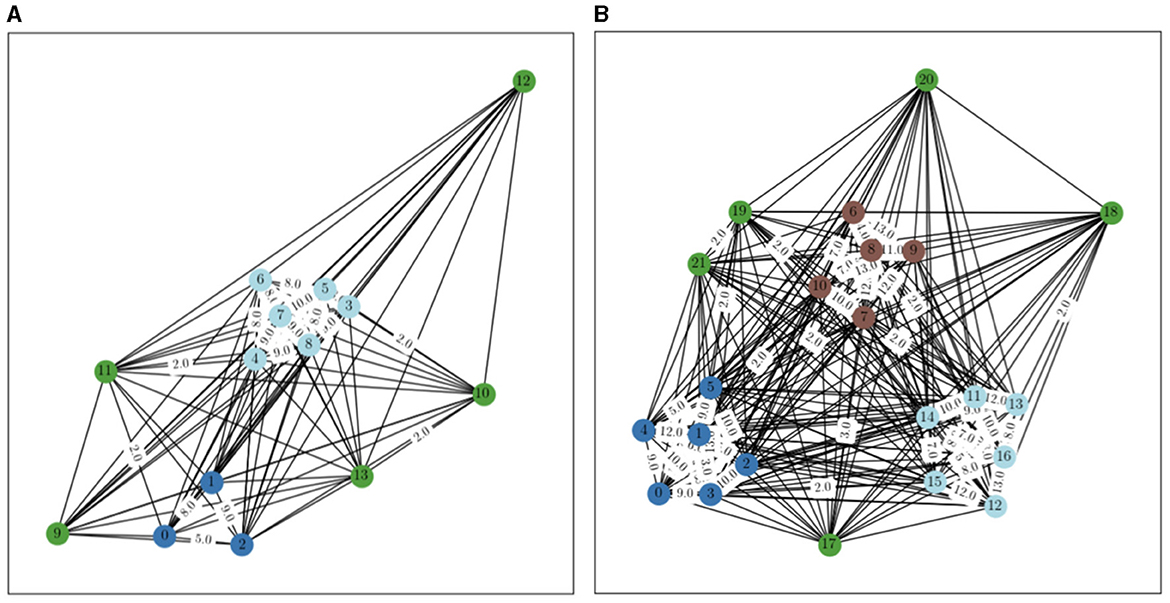
Figure 7. Generated graphs with n = 30, with 2 clusters on (A) and 3 on (B). Edges without a label have a weight of 1.
5 Conclusion
In this study, we have defined the NNVR filtration, proved its stability, and integrated it in a non-parametric hierarchical clustering algorithm. The NNVR filtration contains more information than the VR filtration, which makes it possible to add an outlier detection step to persistence-based clustering. Our experiments show that our algorithm can perform well despite being fully non-parametric, and can thus be useful in many applications. Future research could explore more advanced methods to choose thresholds on the persistence diagram that could for example select both thresholds simultaneously or incorporate some knowledge depending on the application.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
Author contributions
AB: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Software, Validation, Visualization, Writing – original draft, Writing – review & editing. BT: Supervision, Validation, Writing – review & editing. LO: Supervision, Validation, Writing – review & editing.
Funding
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Acknowledgments
The authors thank Thibaut Germain for helpful discussions.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fams.2024.1260828/full#supplementary-material
Footnotes
1. ^For a review on filtrations over graphs, see [13]. Note that a metric space can be seen as a non-directed weighted graph.
2. ^Here, we define an outlier as an isolated point, and consider that two close points form a cluster.
3. ^In this study, we will simply use the term filtration, as all filtrations considered will be increasing.
4. ^This theorem requires a tameness hypothesis to be verified. Here, it is the case because Proposition 5.1 from [9] applies to the NNVR filtration with the exact same proof as for the VR filtration.
References
1. Saxena A, Prasad M, Gupta A, Bharill N, Patel OP, Tiwari A, et al. A review of clustering techniques and developments. Neurocomputing. (2017) 267:664–81. doi: 10.1016/j.neucom.2017.06.053
2. Murtagh F, Contreras P. Algorithms for hierarchical clustering: an overview. Wiley Interdiscipl Rev Data Mining Knowl Discov. (2017) 7:e1219. doi: 10.1002/widm.1219
3. Ester M, Kriegel HP, Sander J, Xu X. A density-based algorithm for discovering clusters in large spatial databases with noise. KDD. (1996) 96:226–31.
4. Schubert E, Sander J, Ester M, Kriegel HP, Xu X, DBSCAN. revisited, revisited: why and how you should (still) use DBSCAN. ACM Transact Database Syst. (2017) 42:1–21. doi: 10.1145/3068335
5. Chazal F, Guibas LJ, Oudot SY, Skraba P. Persistence-based clustering in Riemannian manifolds. J. ACM. (2013) 60:1–38. doi: 10.1145/2535927
6. Edelsbrunner H, Harer JL. Computational Topology: An Introduction. Providence, RI: American Mathematical Society (2022).
7. Boissonnat JD, Chazal F, Yvinec M. Geometric and Topological Inference. Vol. 57. Cambridge: Cambridge University Press (2018).
8. Chazal F, Michel B. An introduction to topological data analysis: fundamental and practical aspects for data scientists. Front Artif Intell. (2021) 4:667963. doi: 10.3389/frai.2021.667963
9. Chazal F, De Silva V, Oudot S. Persistence stability for geometric complexes. Geometriae Dedicata. (2014) 173:193–214. doi: 10.1007/s10711-013-9937-z
10. Lee H, Kang H, Chung MK, Kim BN, Lee DS. Persistent brain network homology from the perspective of dendrogram. IEEE Trans Med Imaging. (2012) 31:2267–77. doi: 10.1109/TMI.2012.2219590
11. Kim H, Hahm J, Lee H, Kang E, Kang H, Lee DS. Brain networks engaged in audiovisual integration during speech perception revealed by persistent homology-based network filtration. Brain Connect. (2015) 5:245–58. doi: 10.1089/brain.2013.0218
12. Rieck B, Fugacci U, Lukasczyk J, Leitte H. Clique community persistence: a topological visual analysis approach for complex networks. IEEE Trans Vis Comput Graph. (2017) 24:822–31. doi: 10.1109/TVCG.2017.2744321
13. Aktas ME, Akbas E, Fatmaoui AE. Persistence homology of networks: methods and applications. Appl Netw Sci. (2019) 4:1–28. doi: 10.1007/s41109-019-0179-3
14. Burago D, Burago Y, Ivanov S. A Course in Metric Geometry. Vol. 33. Providence, RI: American Mathematical Society (2022).
15. Otsu N. A threshold selection method from gray-level histograms. IEEE Trans Syst Man Cybern. (1979) 9:62–6. doi: 10.1109/TSMC.1979.4310076
16. Newman ME. Detecting community structure in networks. Eur Phys J B. (2004) 38:321–30. doi: 10.1140/epjb/e2004-00124-y
17. Rand WM. Objective criteria for the evaluation of clustering methods. J Am Stat Assoc. (1971) 66:846–50. doi: 10.1080/01621459.1971.10482356
18. Blondel VD, Guillaume JL, Lambiotte R, Lefebvre E. Fast unfolding of communities in large networks. J. Stat. Mech. (2008) 2008:P10008. doi: 10.1088/1742-5468/2008/10/P10008
19. Hagberg A, Swart PS, Chult D. Exploring Network Structure, Dynamics, and Function Using NetworkX. Los Alamos, NM: Los Alamos National Lab (LANL) (2008).
20. Arthur D, Vassilvitskii S. K-means++ the advantages of careful seeding. In: Proceedings of the Eighteenth Annual ACM-SIAM Symposium on Discrete Algorithms. Philadelphia, PA: SIAM (2007). p. 1027–35.
21. Frey BJ, Dueck D. Clustering by passing messages between data points. Science. (2007) 315:972–6. doi: 10.1126/science.1136800
22. Comaniciu D, Meer P. Mean shift: a robust approach toward feature space analysis. IEEE Trans Pattern Anal Mach Intell. (2002) 24:603–19. doi: 10.1109/34.1000236
23. Shi. Multiclass spectral clustering. In: Proceedings Ninth IEEE International Conference on Computer Vision. New York, NY: IEEE (2003). p. 313–9.
24. Nielsen F, Nielsen F. Hierarchical Clustering. Introduction to HPC With MPI for Data Science. New York, NY: Springer (2016). p. 195–211.
25. Ankerst M, Breunig MM, Kriegel HP, Sander J. Optics: ordering points to identify the clustering structure. ACM Sigmod Record. (1999) 28:49–60. doi: 10.1145/304181.304187
26. Zhang T, Ramakrishnan R, Livny M. BIRCH: an efficient data clustering method for very large databases. ACM Sigmod Record. (1996) 25:103–14. doi: 10.1145/235968.233324
Keywords: hierarchical clustering, topological data analysis, filtration, stability, outlier removal
Citation: Bois A, Tervil B and Oudre L (2024) Persistence-based clustering with outlier-removing filtration. Front. Appl. Math. Stat. 10:1260828. doi: 10.3389/fams.2024.1260828
Received: 18 July 2023; Accepted: 13 March 2024;
Published: 04 April 2024.
Edited by:
Lixin Shen, Syracuse University, United StatesReviewed by:
Mehmet Aktas, Georgia State University, United StatesUlderico Fugacci, National Research Council (CNR), Italy
Copyright © 2024 Bois, Tervil and Oudre. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Alexandre Bois, YWxleGFuZHJlLmJvaXNAZW5zLXBhcmlzLXNhY2xheS5mcg==
 Alexandre Bois
Alexandre Bois Brian Tervil
Brian Tervil Laurent Oudre
Laurent Oudre