- 1Department of Mathematics, Morgan State University, Baltimore, MD, United States
- 2Department of Mathematics, Southern University of Science and Technology, Shenzhen, Guangdong, China
Introduction: Biot's consolidation model in poroelasticity describes the interaction between the fluid and the deformable porous structure. Based on the fixed-stress splitting iterative method proposed by Mikelic et al. (Computat Geosci, 2013), we present a network approach to solve Biot's consolidation model using physics-informed neural networks (PINNs).
Methods: Two independent and small neural networks are used to solve the displacement and pressure variables separately. Accordingly, separate loss functions are proposed, and the fixed stress splitting iterative algorithm is used to couple these variables. Error analysis is provided to support the capability of the proposed fixed-stress splitting-based PINNs (FS-PINNs).
Results: Several numerical experiments are performed to evaluate the effectiveness and accuracy of our approach, including the pure Dirichlet problem, the mixed partial Neumann and partial Dirichlet problem, and the Barry-Mercer's problem. The performance of FS-PINNs is superior to traditional PINNs, demonstrating the effectiveness of our approach.
Discussion: Our study highlights the successful application of PINNs with the fixed-stress splitting iterative method to tackle Biot's model. The ability to use independent neural networks for displacement and pressure offers computational advantages while maintaining accuracy. The proposed approach shows promising potential for solving other similar geoscientific problems.
1. Introduction
Biot's consolidation model in poroelasticity describes the interaction between fluid flow and the porous structure it saturates. This model was first proposed by Biot [1], and has a wide range of applications, including biomechanics [2] and petroleum engineering [3]. The partial differential equations (PDEs) for the quasi-static Biot system in a bounded domain Ω ⊂ ℝd (where d = 2 or 3) over the time interval (0, T] are as follows:
Here, u is the displacement of solid, p is the fluid pressure, g is the body force, f is a source or sink term, σ(u) = 2με(u) + λdivuI with being the strain tensor. The Lamé constants λ and μ are expressed in terms of the Young's modulus E and the Poisson ratio ν as
Other physical parameters are the Biot-Willis constant α > 0, which is close to 1, Biot's modulus M > 0, and hydraulic conductivity K. Equation (1) represents the mass conservation and Equation (2) means forces balance. For ease of presentation, we assume the following pure Dirichlet conditions.
The initial conditions are
The discussion of the existence and uniqueness of the solution of Biot's system (1)–(5) can be found in [4–6]. In this work, we focus on the algorithm aspect.
Several classical numerical methods have been proposed to solve this problem, including finite volume methods [7], virtual element methods [8], and mixed finite element methods [9]. Biot's model is a multiphysics problem involving both linear elasticity and porous media flow. Numerical difficulties such as elastic locking and pressure oscillations can arise, especially for models based on two-field formulations [10–13]. To overcome these difficulties, various methods have been proposed, such as the discontinuous Galerkin method [14], stabilized finite element methods [11, 15], and three-field or four-field reformulations using inf-sup stable finite element pairs [12, 16–18]. These methods may face challenges in terms of large computational overhead. The fixed-stress splitting iterative method [19, 20] is proposed to address this issue. This method breaks down the original problem into two subproblems and solves them in an iterative manner, rather than solving the entire system at once. The method relies on the contraction mapping principle to prove its convergence [19] and has been shown to be efficient through various studies. Further studies have been carried out based on the fixed-stress splitting iterative method, such as the analysis of the relationship between the convergence rate and the stabilization parameter [21–23] and the implementation of a parallel-in-time strategy to speed up the computations [24].
In recent years, deep neural networks (DNNs) have demonstrated impressive potential in solving partial differential equations (PDEs) with a wide range of applications in various domains. Among DNNs, physics-informed neural networks (PINNs) have become a popular class due to their ability to solve PDEs without meshing. PINNs have proven successful in solving high-dimensional problems and interface problems, and they can tackle inverse problems with slight modifications of the loss function [25–27]. Several studies have used PINNs to tackle the Biot's model [28–30], where the key advantage of PINNs over traditional methods such as finite element methods is their ability to avoid numerical difficulties arising from meshing and not requiring inf-sup stability. Therefore, from a flexibility perspective, deep neural network methods are preferable. However, most of these studies employ a monolithic approach to train the neural network solution, with only one study using a sequential training approach [27], which lacks theoretical analysis and only presents numerical experiments. In this paper, we propose a combination of physics-informed neural networks with fixed-stress splitting method (FS-PINNs) to solve Biot's equations. We employ two PINNs and incorporate them into the fixed-stress splitting iterative method. Our method involves two separate neural networks, one for solid displacement and another for fluid pressure, leading to faster convergence and lower computational cost than classical PINNs [31, 32]. Through a detailed analysis of the monotonic convergence of the fixed-stress splitting method, we present an error estimate for the solution of the proposed FS-PINNs. Future work could include refining the neural network architecture to improve the accuracy and further reduce the computational cost. In addition, the potential application of our approach to other PDE problems and multiphysics problems could be explored. Overall, the proposed FS-PINNs represent a promising approach for solving poroelastic models with faster convergence and lower computational cost.
The remaining sections of the paper are organized as follows. Section 2 provides an overview of the fixed-stress splitting iterative method for Biot's model. In Section 3, we introduce the fixed-stress splitting iterative PINNs (FS-PINNs) and present theoretical analyses to demonstrate their approximation properties. Section 4 presents numerical experiments to demonstrate the effectiveness of the proposed method. Finally, conclusions are drawn in Section 5.
2. Fixed-stress splitting method for Biot's equations
In this section, we introduce an iterative scheme proposed in [19], called the fixed-stress splitting method. Instead of solving Biot's model (1)–(5) in a monolithic way, this method decouples the original problem into two subproblems and solves them in an iterative manner. Given a large enough stabilization parameter βFS and an initial guess (p0, u0), the standard fixed-stress splitting method computes a sequence of approximations as follows:
The first step: Given pn and un, we solve for pn+1 satisfying
The second step: Using pn+1, we solve for un+1 satisfying
To derive the variational formulation of the fixed-stress problem (6)–(11), we define the proper functional spaces with H1(Ω) denoting the Hilbert subspace as follows.
Multiplying (6), (9) by test functions, and applying the integration by parts, we obtain the variational problems: for a given t ≥ 0, find {(pn + 1, un + 1)} ⊂ W × V such that
Assume that (p, u) ∈ W × V is the unique solution of Biot's system (1)–(5). Given a large enough stabilization parameter βFS, the sequence {(pn+1, un+1)} ⊂ W × V generated by Equations (12, 13) converges to the solution (p, u). More precisely, we can state the following:
THEOREM 2.1. The sequence {(pn+1, un+1)} ⊂ W × V generated by Equations (6)–(11) converges to (p, u) ∈ W × V for any . If one denotes and , then there holds
where is a positive constant strictly smaller than 1.
REMARK 2.2. We have included a convergence analysis of the fixed-stress splitting iterative method in the Appendix. In our proof, we demonstrate that the differences and approach zeros as n tends to infinity. Our proof helps to illustrate the convergence of the fixed-stress splitting method.
3. The classical PINNs and the FS-PINNs for solving Biot's model
In this section, we introduce the training procedure of the classical PINNs for solving Biot's model, and propose an iterative deep learning method combining the fixed-stress splitting method, called FS-PINNs. The proposed FS-PINNs consist of two independent PINNs, one for pressure, and the other for displacement. We then present the training procedure and a theoretical analysis of the proposed FS-PINNs.
3.1. Training procedure of the classical PINNs
The idea of using the classical PINNs to solve PDEs is from [25], which can be easily extended to solve Biot's model. The main idea is to design loss functions for fully connected neural networks. In this context, we provide a brief overview of the training procedure for solving problem (1)–(5), which is illustrated in Figure 1.
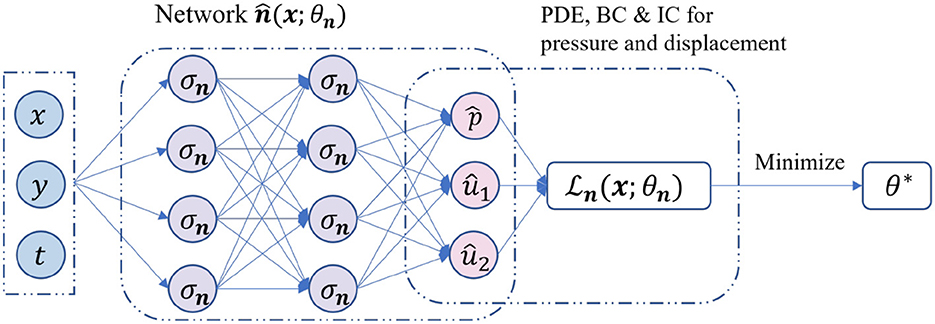
Figure 1. The training procedure of the classical PINNs using unified activation functions for a 2D Biot's model.
The functional form of a classical PINN is given as follows.
Here, is the network approximation of the exact solution (p; u) with Ln hidden layers with the input . The collection of parameter θn is given by
The mapping function is defined as
Here, represent the weights, biases, and activation functions of the i-th layer for network . It is assumed that the number of neurons in each hidden layer is set to be the same Nn and that all activation functions are identical.
After setting up the architecture, we can begin training the parameter θn using the procedure:
where is the loss function. The loss function for the classical PINNs generally consists of three components: the PDEs loss, the initial condition loss, and the boundary condition loss. The PDEs loss represents the residual of the governing equations, while the initial and boundary condition losses ensure that the predicted solution satisfies the initial and boundary conditions. The loss function is denoted by and has the following representation.
The above expression uses the mean square error (MSE) to measure errors. In this expression, represents the initial and boundary training data, while represents the collocation points in the domain. We then optimize θn to obtain θ*. For cases involving mixed boundary conditions, one can refer to [28] for more details.
3.2. Training procedure of the proposed FS-PINNs
The primary innovation of our approach is incorporating the fixed-stress strategy into the training procedure of PINNs. The proposed method includes two independent PINNs to solve for displacement and fluid pressure separately. We can then train our FS-PINNs iteratively with respect to their corresponding loss functions. Figure 2 illustrates the training procedure.
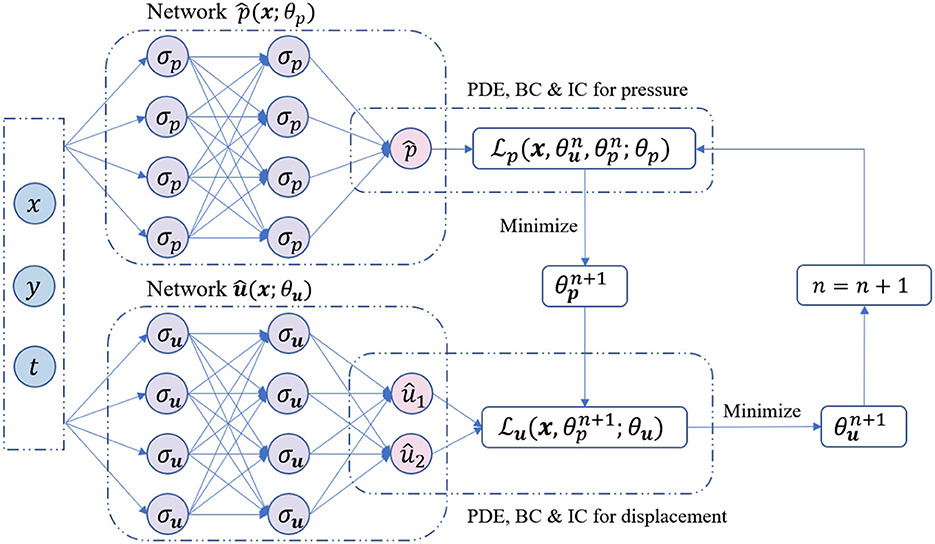
Figure 2. The training procedure of the proposed FS-PINNs. Two independent networks are trained iteratively with respect to the corresponding loss functions.
Similar to the classical PINNs, the functional forms of the two independent PINNs are as follows.
Here, refers to the network approximation of pressure p with Lp hidden layers, while refers to the network approximation of displacement u with Lu hidden layers. The collection of parameters of and are denoted by θp and θu, respectively, and are given by:
The mapping functions and are defined as
In this context, and represent the weights, biases, and activation functions of the i-th layer for and , respectively. It is also assumed that the number of neurons in each hidden layer of and is set to Np and Nu, respectively. Furthermore, all activation functions and are assumed to be identical.
From now, our attention turns to developing the loss functions. By utilizing the fixed-stress splitting method detailed in Equations (6)–(8) and Equations (9)–(11), we can impose constraints on the networks and , respectively. We begin by setting n = 0 and . We can then formulate the optimization problem as follows. In the first step, we obtain by solving for the minimum of :
In the second step, we obtain by solving for the minimum of :
To be clear, and denote the loss functions for networks and , respectively. These loss functions are defined as follows:
In the above expressions, and denote the initial and boundary training data on p(x) and u(x), respectively. Additionally, and represent the collocation points for f(x) and g(x) in the domain, respectively. By repeatedly updating θp and θu using (25) and (26), we can get the network approximations of Biot's model: and .
3.3. Analysis of the proposed FS-PINNs
The classical PINNs have shown great potential in solving PDEs, but quantifying their errors remains an open problem [26]. The accuracy of neural network approximation is influenced by various factors, such as network architecture, training data, etc. In this section, we assume that PINNs can solve the two subproblems within a certain error tolerance. We then employ an iterative strategy for the proposed FS-PINNs. It is crucial to consider the possibility of error accumulation and divergence since network approximations can not guarantee convergence of the fixed-stress iteration. To address this issue, we observe that the fixed-stress splitting method, which is essentially equivalent to the fixed-point iterative scheme, has been shown to mitigate error accumulation. Therefore, we present a mathematical analysis to demonstrate the robustness of our proposed FS-PINNs. Given an initial guess , we describe the procedures of (25) and (26) as follows.
Step 1: Given and , we employ (25) to obtain a network solution for the following problem.
The corresponding loss function for imposes the initial condition and the boundary condition in the mean square sense.
Step 2: using , we employ (26) to obtain a network solution for the following problem.
The corresponding loss function for imposes the initial condition and the boundary condition in the mean square sense.
To analyze the numerical errors and , we can express them as the sum of two terms: and , which can be dealt with separately. We assume that the Physics-Informed Neural Networks (PINNs) used to solve Equations (29) and (32) can achieve a certain error tolerance, denoted by δ, for the approximations and obtained in each step. This is expressed as
Such assumptions are reasonable if the neural networks are sufficiently deep and wide [33]. For estimating the terms (pn+1−p) and (un+1−u), we utilize integration by parts and employ the fixed-stress iterative method to analyze convergence. We then build on Theorem 2.1 to perform mathematical analysis.
THEOREM 3.1. Assume that (p, u) ∈ W × V is the unique solution of the problem (1)–(5). Let be the network approximations of Equations (29)–(34), and (pn+1, un+1) ∈ W × V be the exact solution of (6)–(11). We define and . Under the assumption (35) with a positive δ, if βFS is large enough, then there exists a positive number such that the following estimate holds for all n ≥ 0.
where C is a positive constant. Moreover, the limit superior exists and satisfies:
Proof. Firstly, we start with the second step of FS-PINNs. We subtract the original Equation (2) from the n-th iteration of (32). For the resulted equation, taking the derivative with respect to t, we obtain
Multiplying (38) by , applying the divergence theorem, the Cauchy-Schwarz inequality, and the Poincaré inequality, we shall derive the following inequality with any ϵ1 > 0.
On the other hand, there holds [23]
Based on the above inequality and (35), we can deduce the following inequality from (39).
Similar to the derivation of (38), subtracting the (n+1)-th iteration Equation (32) from (2) and taking the derivative with respect to t, we obtain
Subtracting (38) from (41) yields
We multiply (42) by , and repeat the same argument as that for (40) to derive
Next, we consider the entire system of the FS-PINNs. Subtracting the original Equation (1) from the (n+1)-th iteration Equation (29) yields
Multiplying (44) by , and (41) by , respectively, we then take a summation to derive the following equation.
We recall the following algebraic identities [“(·, ·)” means the l2 inner product].
Applying (46) to the LHS of (45), and discarding some positive terms, we shall obtain
We then deduce the following inequality based on (43).
For estimating an upper bound of the RHS term in (45), we use the Cauchy-Schwarz inequality with any ϵ2 > 0, and (40). We see that
We let , by combining (48) and (49), we see that
where
We choose ϵ1 and ϵ2 small enough, such that the contraction coefficient , defined as follows, is strictly smaller than 1.
Then, we can rewrite (50) in the following form.
This leads to the limit supremum.
This completes our proof.
4. Benchmark tests
In this section, the efficacy of the proposed FS-PINNs is presented through three case studies. The first study involves a pure Dirichlet boundary value problem, while the second study considers a problem with mixed Dirichlet-Neumann boundary conditions. Finally, the third study investigates Barry-Mercer's problem. All experiments were implemented in the open-source machine learning tool PyTorch [34] on a single NVIDIA GeForce RTX 3060 GPU. Our code is available at https://github.com/newpolarbear/FS-PINNs. We evaluate the computational efficiency of our proposed method by comparing its true errors and runtime overheads with those of classical PINNs.
4.1. Models generation
In practice, the training procedure of classical PINNs is heavily influenced by several factors. One of these factors is the design of the neural network architecture, which does not have a universal standard. It's well-known that the network's representational capability is affected by its depth and width. While deeper and wider networks have greater representation capacity, they may suffer from slower training, compromises can be made to reduce runtime overhead. Another factor is the choice of nonlinear activation functions and optimization methods. In this work, we used as the activation function and Adam optimizer with a fixed learning rate of 0.0001. In addition, some factors are specific to the problem being solved, such as the number of epochs, collocation points, and initial and boundary training data. In this study, we randomly select 10, 000 points as collocation points within the domain. The initial training data and each boundary training data are set to 500 and 200, respectively.
In addition to the standard configurations of classical PINNs, there are other aspects that come into play when training FS-PINNs. One FS-PINN is composed of two separate networks, which directly impact the performance. Moreover, training FS-PINNs involves networks optimization for each fixed-stress iteration, as shown in Figure 2. In the n-th fixed-stress iteration, we carry out the optimization procedure of network parameters θp and θu. For simplicity, we assume that the number of epochs for optimizing θp equals that for optimizing θu. Since the fixed-stress splitting method is an iterative algorithm, achieving accurate solutions at each step is not so necessary. For simplicity, we set the fixed-stress stabilization parameter to .
The training procedure of FS-PINNs is dynamic, loss functions and can only show the performance in the current fixed-stress iteration. To evaluate the proposed method, we consider the true errors between network solutions and exact solutions as follows.
Here, , û1, and û2 represent the neural network solutions of pressure, x-displacement, and y-displacement, respectively, while p, u1, and u2 denote their corresponding exact solutions. The total mean squared error can be computed by taking the summation of these errors:
4.2. Example 1
The first numerical experiment is based on the example in [16]. The domain is Ω = [0, 1]2 and the final time is T = 0.5, where pure Dirichlet boundary conditions are applied. In this experiment, the source or sink term f, body force g, initial conditions, and boundary conditions are chosen such that the exact solutions are provided as follows:
The model parameters are set to E = 1.0, ν = 0.3, α = 1.0, M = 1.0, and K = 1.0.
4.2.1. Robustness of the FS-PINNs architecture
The performance of FS-PINNs is highly dependent on its architecture. We fixed the total width to Np+Nu = 30 and depths to Lp = Lu = 3, where Np, Nu are the number of neurons, and Lp, Lu are the number of layers. We then designed an experiment with five sets of parameter selection for Np:Nu. To ensure that the networks are well-trained, we applied 20 fixed-stress iterations and 3,000 epochs for each fixed-stress step. The experiment is tested repeatedly to ensure the reliability of the results.
Table 1 shows the runtime overheads for various parameter selections, revealing that a more uniform separation exhibits lower runtime overhead. Notably, the case Np = 5:Nu = 25 performs poorly compared to other cases, because wider networks experience slower training, which leads to a significant increase in the runtime overhead of network û. Conversely, the other four cases exhibit similar performance, suggesting that the proposed FS-PINNs with reasonable architecture are resilient to runtime overhead.
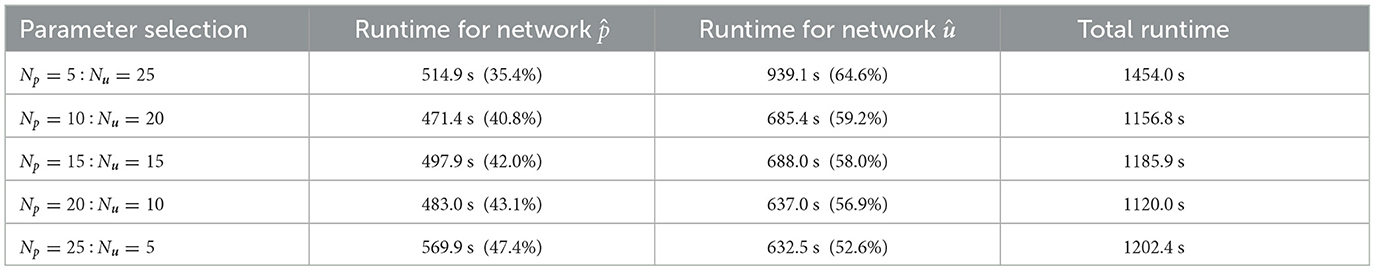
Table 1. Runtime overheads of five different parameter selections for FS-PINNs architecture of Example 1.
Figure 3 tracks the trends of all MSE terms during different parameter selections training procedures, revealing that the case Np = 25:Nu = 5 performs worst, and the case Np = 5:Nu = 25 is less stable. The inadequate representation ability of tiny networks leads to inaccurate solutions. Network approximations and depend on the interdependent loss functions for FS-PINNs. On the other hand, the trends of the remaining three cases are quite similar, which implies that FS-PINNs with reasonable architecture can withstand accuracy issues.
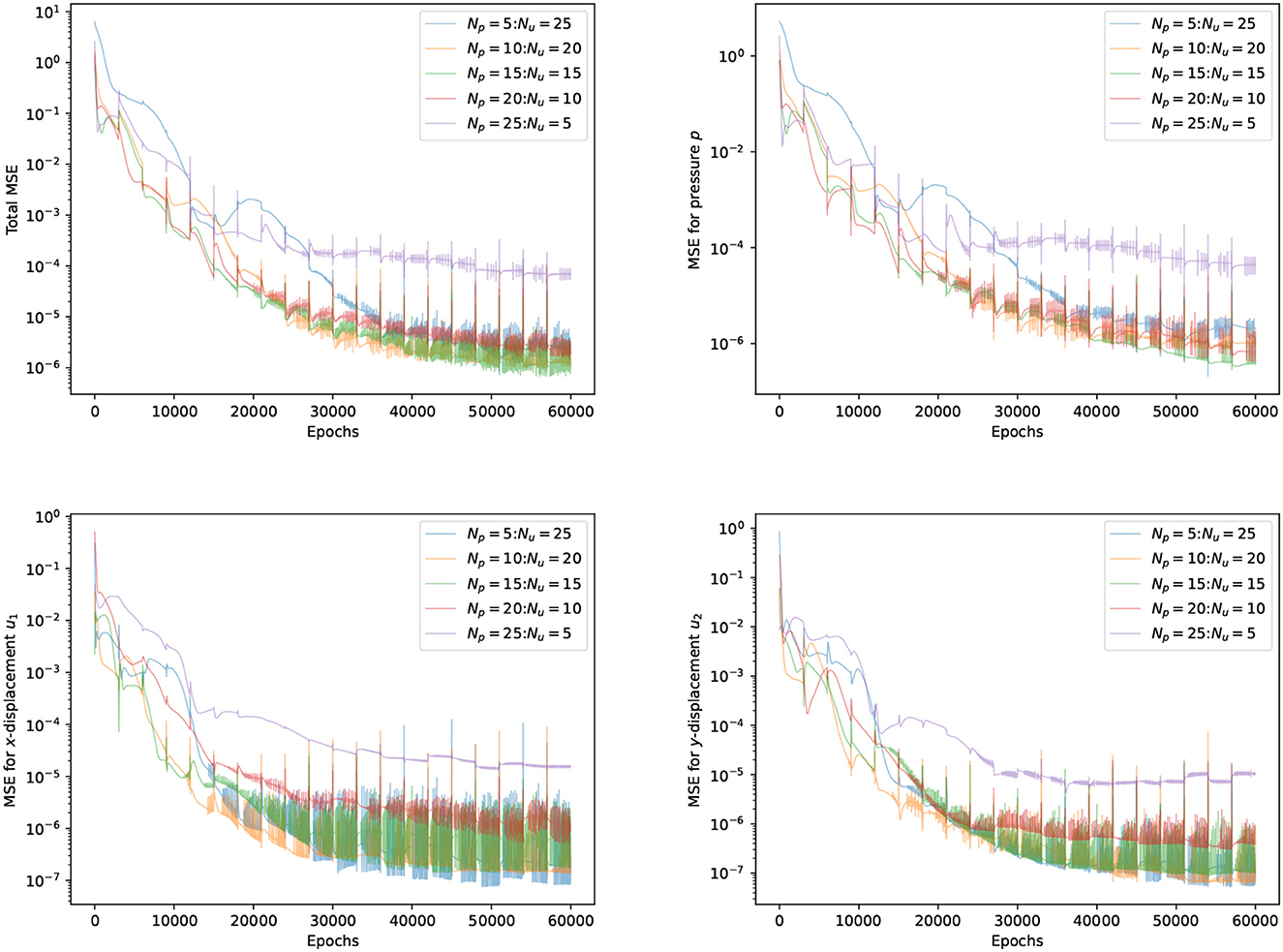
Figure 3. Trends of all the MSE terms at T = 0.5 for five different FS-PINNs architecture during training with respect to epoch.
4.2.2. Acceleration effect of the FS-PINNs training
This section demonstrates the acceleration effect of the FS-PINNs over the classical PINNs. Biot's equations are solved using both methods with the same setup as in Example 1. The number of neurons for the two networks in FS-PINNs is fixed to Np = Nu = 15 (as indicated by Table 1), while the total number of neurons in classical PINNs is set to Nn = 30 with the number of layers Ln = 3. To examine the effect of the number of epochs optimizing networks and the number of fixed-stress iterations, we then conduct the test with a constant number of the multiplication of the above two number, which is set to 20, 000.
Figure 4 shows the trends of MSE terms for solving the problem in Example 1. We present the results generated by one classical PINN and four FS-PINNs under identical conditions, which demonstrates the acceleration effect of FS-PINNs. By decoupling the original optimization problem into two small sub-problems, together with the fixed-stress coefficient L < 1, FS-PINNs increase efficiency and maintain certain accuracy. From this perspective, FS-PINNs offer more flexibility in solving the problem through different setting of optimizing epoch and fixed-stress iteration. In Figure 4, we can see that a smaller number of epochs lead to faster convergence initially but lower accuracy eventually. Since the fixed-stress splitting method is a numerical approach, high accuracy is not essential for each fixed-stress step. It implies that a large number of optimizing epochs is not so helpful.
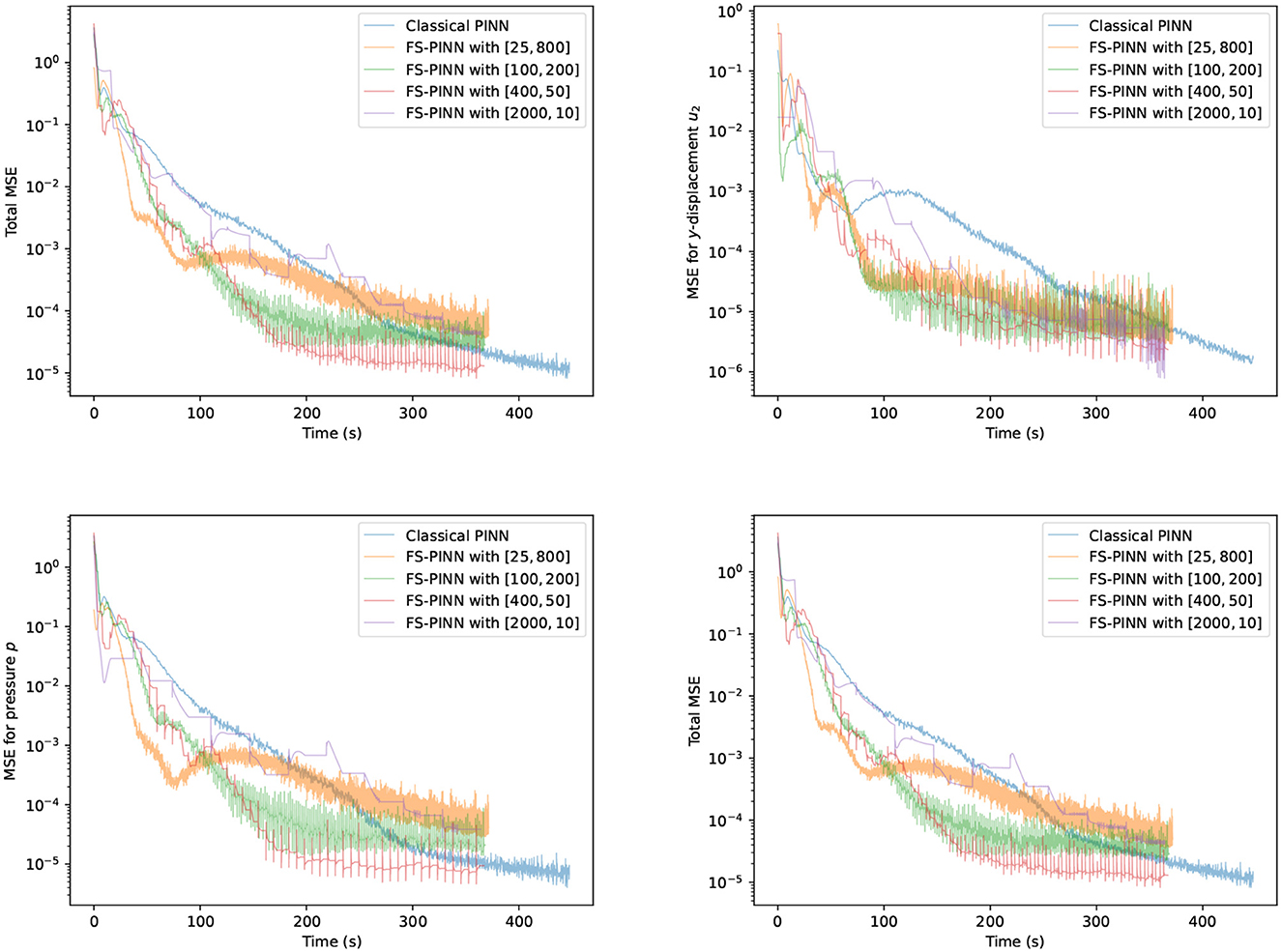
Figure 4. Trends of all the MSE terms at T = 0.5 for one classical PINN and four FS-PINNs with [the number of epochs training networks, the number of fixed-stress iterations] during training with respect to runtime overhead.
Among all the results generated by FS-PINNs, here we consider in-depth observations for the case of [400, 50]. Here, 400 means the number of epochs, and 50 means the number of fixed-stress iterations. To test the cases with a larger parameter λ, we also include the testing result when the Poisson ratio ν = 0.49 for Example 1. Figure 5 provides a comparison between the analytical solution and the numerical solution given by FS-PINNs with [400, 50]. Theses results show that the proposed FS-PINNs with ν = 0.3 and ν = 0.49 accurately predict the pressure p and displacement u with errors within an acceptable tolerance range. However, it is true that when λ is large, to obtain a more accurate solution, we need to put a relatively large weight for the first equation in the total loss function. And also, we need to increase the number of epochs and set a finer learning rate for the training of the network for u.
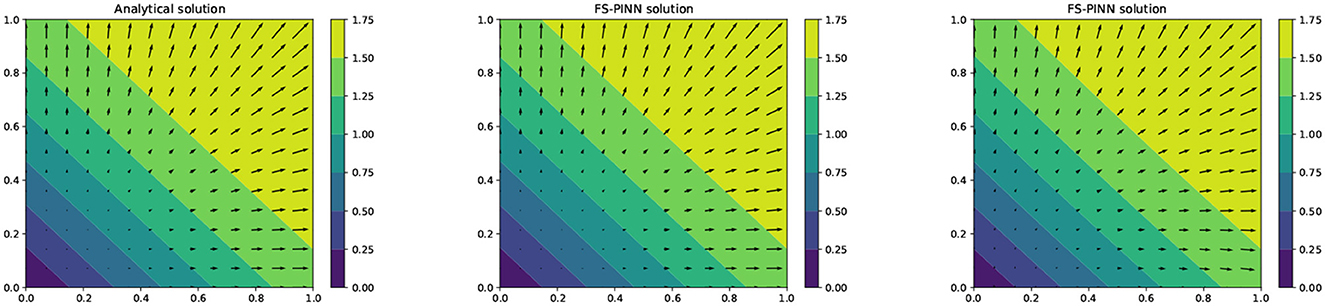
Figure 5. A comparison of the analytical solution (left), the FS-PINN solution with ν = 0.3 (middle), and the FS-PINN solution ν = 0.49 (right) at T = 0.5 for Example 1.
4.3. Example 2
The Biot's problem with mixed boundary conditions presented in [2] is tested. We consider the square domain Ω = [0, 1]2, with Dirichlet boundary Γ1∪Γ3 and Neumann boundary Γ2∪Γ4. Specifically, Γ1 = (1, y);0 ≤ y ≤ 1, Γ2 = (x, 0);0 ≤ x ≤ 1, Γ3 = (0, y);0 ≤ y ≤ 1, and Γ4 = (x, 1);0 ≤ x ≤ 1. Setting the final time to T = 0.2, we choose the body force g, the source or sink term f, as well as the initial and boundary conditions such that exact solutions are as follows.
The parameters of the model are E = 1.0, ν = 0.3, α = 1.0, M = 1.0, K = 1.
In dealing with mixed boundary conditions, it is necessary to select suitable loss functions capable of handling both Dirichlet and Neumann boundary conditions. To this end, we follow the approach taken in [28, 32] and employ the corresponding loss functions and for pressure and displacement, respectively. In this test, following the result in Section 4.2.2., we employ the FS-PINN with [400, 50] using the same architecture to solve Example 2.
In Figure 6, we compare the analytical solution with our FS-PINN solution and observe that our approach can approximate solutions for problems with mixed boundary conditions. However, its performance is not as good as that of solving the pure Dirichlet problem in Example 1. The network approximations in the boundary region are sub-optimal because all the parameters used were selected to solve Example 1. Hence, the complexity of Example 2 presents a challenge to our network, which could potentially be overcome by optimizing the parameters, such as by using additional collocation points in the boundary region.
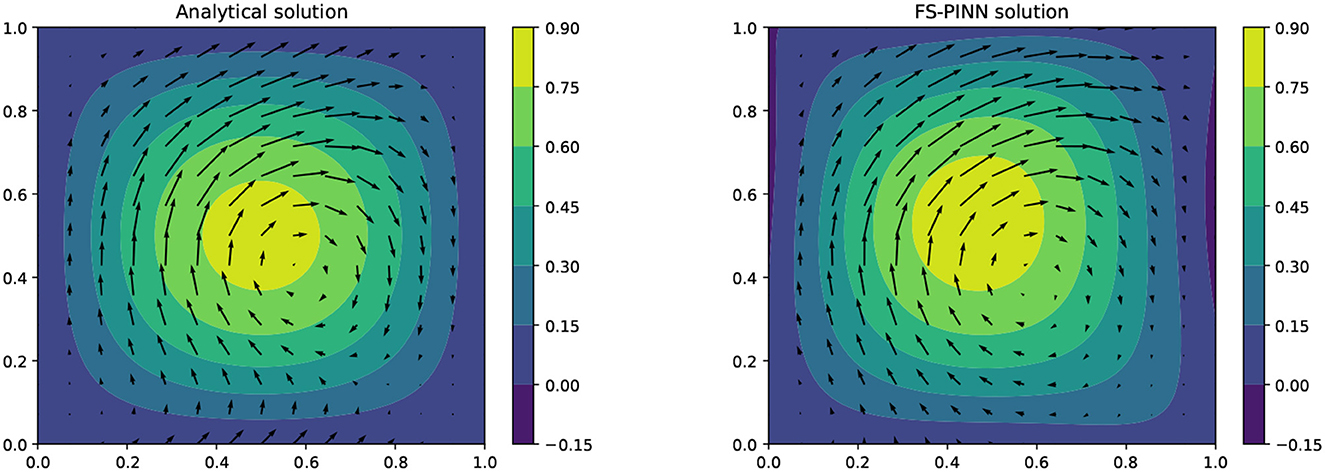
Figure 6. A comparison of the analytical solution (left) and the FS-PINN solution with [400, 50] (right) at T = 0.2 for Example 2.
4.4. Example 3
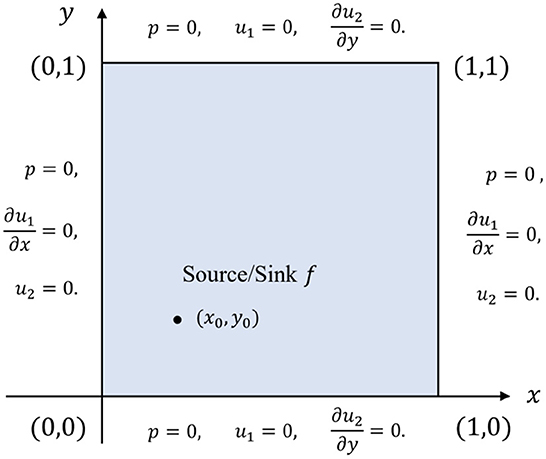
The Barry-Mercer's problem is a classical benchmark problem of porous media involving injection of fluids [14, 31, 35, 36]. It is a two-dimensional problem in the square domain Ω = [0, 1]2 that has a point source f located at (x0, y0) = (0.25, 0.25), which makes it challenging to solve numerically. Figure 7 depicts the boundary conditions under consideration.
As for the initial conditions, we set
To define the body force term g and the source/sink term f, we use the following expressions:
where δ denotes the Dirac function. We approximate the Dirac function using the Gaussian distribution [31], that is, with cg = 0.04. The physical parameters are specified as follows:
Denote λn = nπ, λq = qπ, and , then the analytical solution is as follows:
where
In this case, we use the same architecture as in Section 4.2.2, which is the FS-PINN with [400, 50], to solve Barry-Mercer's problem. As mentioned in the literature [37], networks training relies on the propagation of information from initial and boundary points to the interior points. To improve the performance of our FS-PINN, we apply weighted loss functions proposed in [32], and increase the weights of the initial and boundary loss terms. Specifically, we replace , with , in the loss functions (27), (28), respectively. The solution obtained from our FS-PINN closely matches the analytic solution at T = π/4 for Example 3, as demonstrated in Figure 8. This indicates the effectiveness of our approach in approximating the analytic solution.
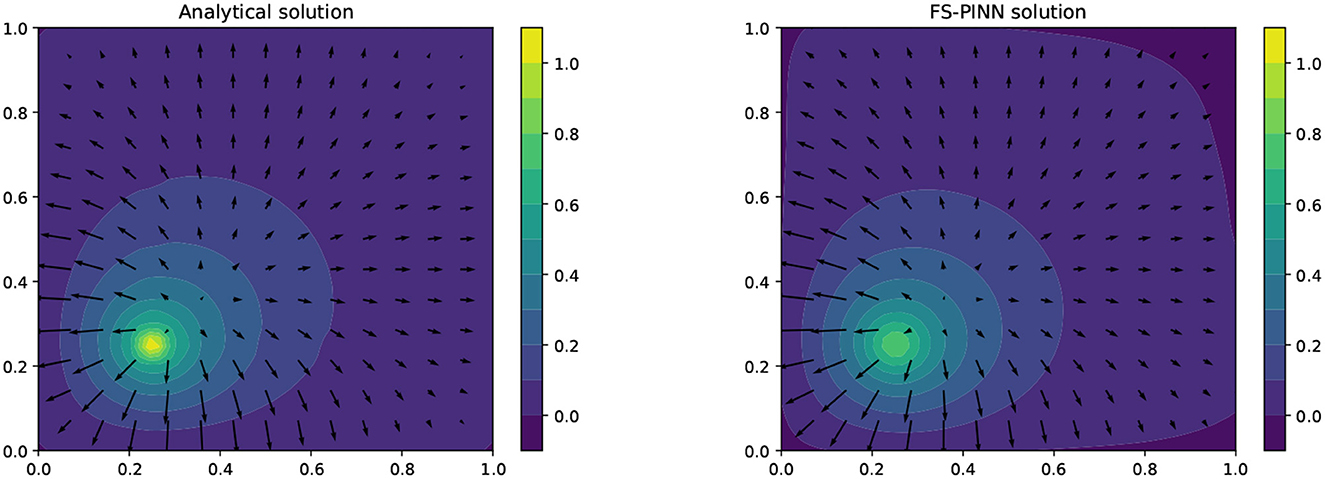
Figure 8. A comparison of the analytical solution (left) and the FS-PINN solution (right) at T = π/4 for Example 3.
5. Conclusion
This paper presents a new approach to solving Biot's consolidation model by combining Physics-informed neural networks (PINNs) with the fixed-stress splitting method. We demonstrate that our method achieves faster convergence and lower computational costs than the neural network methods which solve Biot's problem in a monolithic way. We provide a theoretical analysis to show the convergence of our proposed FS-PINNs. The numerical experiments illustrate the effectiveness of our approach on various test cases with different boundary conditions and physical parameters.
While our approach has several advantages over some traditional methods for solving poroelastic models, such as being meshless, not requiring inf-sup stability, there are also some limitations and challenges that need to be addressed in future work. These include choosing optimal network architectures and parameters for different problems, improving accuracy and reducing computational costs even further, and extending the approach to other PDE problems and multi-physics problems. We hope that our work can inspire more research on combining deep learning techniques with iterative methods for solving PDEs.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author.
Author contributions
MC: conceptualization, investigation, writing—original draft, validation, methodology, writing—review and editing, supervision, formal analysis, and funding acquisition. HG: investigation, writing—original draft, validation, visualization, writing—review and editing, formal analysis, and data curation. PH: investigation, writing—original draft, validation, visualization, writing—review and editing, software, and data curation. JL: conceptualization, investigation, funding acquisition, validation, methodology, project administration, and supervision. All authors contributed to the article and approved the submitted version.
Funding
The work of MC was partially supported by the NIH-RCMI grant through 347 U54MD013376, the affiliated project award from the Center for Equitable Artificial Intelligence and Machine Learning Systems (CEAMLS) at Morgan State University (project ID 02232301), and the National Science Foundation awards (1831950 and 2228010). The work of HG, PH, and JL were partially supported by the NSF of China No. 11971221, Guangdong NSF Major Fund No. 2021ZDZX1001, the Shenzhen Sci-Tech Fund Nos. RCJC20200714114556020, JCYJ20200109115422828, and JCYJ20190809150413261, National Center for Applied Mathematics Shenzhen, and SUSTech International Center for Mathematics.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Biot MA. General theory of three-dimensional consolidation. J Appl Phys. (1941) 12:155–64. doi: 10.1063/1.1712886
2. Ju G, Cai M, Li J, Tian J. Parameter-robust multiphysics algorithms for Biot model with application in brain edema simulation. Math Comput Simul. (2020) 177:385–403. doi: 10.1016/j.matcom.2020.04.027
3. Kim J, Tchelepi HA, Juanes R. Stability and convergence of sequential methods for coupled flow and geomechanics: fixed-stress and fixed-strain splits. Comput Methods Appl Mech Eng. (2011) 200:1591–606. doi: 10.1016/j.cma.2010.12.022
4. Ženíšek A. The existence and uniqueness theorem in Biot's consolidation theory. Aplik Matem. (1984) 29:194–211. doi: 10.21136/AM.1984.104085
5. Showalter RE. Diffusion in poro-elastic media. J Math Anal Appl. (2000) 251:310–40. doi: 10.1006/jmaa.2000.7048
6. Phillips PJ, Wheeler MF. A coupling of mixed and continuous Galerkin finite element methods for poroelasticity I: the continuous in time case. Comput Geosci. (2007) 11:131–44. doi: 10.1007/s10596-007-9045-y
7. Nordbotten JM. Stable cell-centered finite volume discretization for Biot equations. SIAM J Numer Anal. (2016) 54:942–68. doi: 10.1137/15M1014280
8. Coulet J, Faille I, Girault V, Guy N, Nataf F. A fully coupled scheme using virtual element method and finite volume for poroelasticity. Comput Geosci. (2020) 24:381–403. doi: 10.1007/s10596-019-09831-w
9. Yi SY. A coupling of nonconforming and mixed finite element methods for Biot's consolidation model. Numer Methods Part Diff Equat. (2013) 29:1749–77. doi: 10.1002/num.21775
10. Yi SY. A study of two modes of locking in poroelasticity. SIAM J Numer Anal. (2017) 55:1915–36. doi: 10.1137/16M1056109
11. Rodrigo C, Gaspar F, Hu X, Zikatanov L. Stability and monotonicity for some discretizations of the Biot's consolidation model. Comput Methods Appl Mech Eng. (2016) 298:183–204. doi: 10.1016/j.cma.2015.09.019
12. Lee JJ, Mardal KA, Winther R. Parameter-robust discretization and preconditioning of Biot's consolidation model. SIAM J Sci Comput. (2017) 39:A1–24. doi: 10.1137/15M1029473
13. Cai M, Zhang G. Comparisons of some iterative algorithms for Biot equations. Int J Evol Equat. (2017) 10:267–82.
14. Phillips PJ, Wheeler MF. Overcoming the problem of locking in linear elasticity and poroelasticity: an heuristic approach. Comput Geosci. (2009) 13:5–12. doi: 10.1007/s10596-008-9114-x
15. Rodrigo C, Hu X, Ohm P, Adler JH, Gaspar FJ, Zikatanov L. New stabilized discretizations for poroelasticity and the Stoke's equations. Comput Methods Appl Mech Eng. (2018) 341:467–84. doi: 10.1016/j.cma.2018.07.003
16. Feng X, Ge Z, Li Y. Analysis of a multiphysics finite element method for a poroelasticity model. IMA J Numer Anal. (2018) 38:330–59. doi: 10.1093/imanum/drx003
17. Oyarzúa R, Ruiz-Baier R. Locking-free finite element methods for poroelasticity. SIAM Journal on Numerical Anal. (2016) 54:2951–73. doi: 10.1137/15M1050082
18. Yi SY, Bean ML. Iteratively coupled solution strategies for a four-field mixed finite element method for poroelasticity. Int J Numer Analyt Methods Geomech. (2017) 41:159–79. doi: 10.1002/nag.2538
19. Mikelić A, Wheeler MF. Convergence of iterative coupling for coupled flow and geomechanics. Comput Geosci. (2013) 17:455–61. doi: 10.1007/s10596-012-9318-y
20. Mikelić A, Wang B, Wheeler MF. Numerical convergence study of iterative coupling for coupled flow and geomechanics. Comput Geosci. (2014) 18:325–41. doi: 10.1007/s10596-013-9393-8
21. Both JW, Borregales M, Nordbotten JM, Kumar K, Radu FA. Robust fixed stress splitting for Biot's equations in heterogeneous media. Appl Math Lett. (2017) 68:101–8. doi: 10.1016/j.aml.2016.12.019
22. Bause M, Radu FA, Köcher U. Space-time finite element approximation of the Biot poroelasticity system with iterative coupling. Comput Methods Appl Mech Eng. (2017) 320:745–68. doi: 10.1016/j.cma.2017.03.017
23. Storvik E, Both JW, Kumar K, Nordbotten JM, Radu FA. On the optimization of the fixed-stress splitting for Biot's equations. Int J Numer Methods Eng. (2019) 120:179–94. doi: 10.1002/nme.6130
24. Borregales M, Kumar K, Radu FA, Rodrigo C, Gaspar FJ. A partially parallel-in-time fixed-stress splitting method for Biot's consolidation model. Comput Math Appl. (2019) 77:1466–78. doi: 10.1016/j.camwa.2018.09.005
25. Raissi M, Perdikaris P, Karniadakis GE. Physics-informed neural networks: a deep learning framework for solving forward and inverse problems involving nonlinear partial differential equations. J Comput Phys. (2019) 378:686–707. doi: 10.1016/j.jcp.2018.10.045
26. Lu L, Meng X, Mao Z, Karniadakis GE. DeepXDE: a deep learning library for solving differential equations. SIAM Rev. (2021) 63:208–28. doi: 10.1137/19M1274067
27. Amini D, Haghighat E, Juanes R. Inverse modeling of nonisothermal multiphase poromechanics using physics-informed neural networks. arXiv preprint arXiv:220903276. (2022). doi: 10.1016/j.jcp.2023.112323
28. Kadeethum T, Jørgensen TM, Nick HM. Physics-informed neural networks for solving nonlinear diffusivity and Biot's equations. PLoS ONE. (2020) 15:e0232683. doi: 10.1371/journal.pone.0232683
29. Millevoi C, Spiezia N, Ferronato M. On Physics-Informed Neural Networks Architecture for Coupled Hydro-Poromechanical Problems. (2022). doi: 10.2139/ssrn.4074416
30. Bekele YW. Physics-informed deep learning for flow and deformation in poroelastic media. arXiv preprint arXiv:201015426. (2020).
31. Haghighat E, Amini D, Juanes R. Physics-informed neural network simulation of multiphase poroelasticity using stress-split sequential training. Comput Methods Appl Mech Eng. (2022) 397:115141. doi: 10.1016/j.cma.2022.115141
32. Amini D, Haghighat E, Juanes R. Physics-informed neural network solution of thermo-hydro-mechanical (THM) processes in porous media. arXiv preprint arXiv:220301514. (2022). doi: 10.1061/(ASCE)EM.1943-7889.0002156
33. De Ryck T, Jagtap AD, Mishra S. Error estimates for physics informed neural networks approximating the Navier-Stokes equations. arXiv preprint arXiv:220309346. (2022). doi: 10.1093/imanum/drac085
34. Paszke A, Gross S, Massa F, Lerer A, Bradbury J, Chanan G, et al. PyTorch: An imperative style, high-performance deep learning library. In:Wallach H, Larochelle H, Beygelzimer A, Alche-Buc F, Fox E, Garnett R, , editors. Advances in Neural Information Processing Systems, Vol. 32. Curran Associates (2019). Available online at: https://proceedings.neurips.cc/paper_files/paper/2019/file/bdbca288fee7f92f2bfa9f7012727740-Paper.pdf
35. Barry S, Mercer G. Exact solutions for two-dimensional time-dependent flow and deformation within a poroelastic medium. J Appl Mech. (1999) 66:536–40. doi: 10.1115/1.2791080
36. Phillips PJ. Finite Element Methods in Linear Poroelasticity: Theoretical and Computational Results. Austin, TX: The University of Texas at Austin (2005).
37. Daw A, Bu J, Wang S, Perdikaris P, Karpatne A. Mitigating propagation failures in pinns using evolutionary sampling, conference paper at ICLR 2023?. (Under review).
Appendix
Proof of Theorem 2.1
Proof. Subtracting (6), (9) from (1), (2), respectively, we shall derive
After differentiating (2) with respect to t, one has
We multiply (1), (3) by , , respectively, and use integration by parts to obtain
Combining Equations (4) and (5) results in the following equation.
Applying the algebraic identities (46) to (6), we derive
The difference of two successive iterations based on (2) will yield
Multiplying (8) by , integration by parts, and then applying Cauchy-Schwarz inequality, we derive that
Discarding the second, third, and fourth terms in (7), we choose to obtain
which directly implies (14).
Keywords: physics-informed neural networks, the fixed-stress method, Biot's model, iterative algorithm, separated networks
Citation: Cai M, Gu H, Hong P and Li J (2023) A combination of physics-informed neural networks with the fixed-stress splitting iteration for solving Biot's model. Front. Appl. Math. Stat. 9:1206500. doi: 10.3389/fams.2023.1206500
Received: 15 April 2023; Accepted: 14 July 2023;
Published: 03 August 2023.
Edited by:
Haizhao Yang, Purdue University, United StatesReviewed by:
Kent-Andre Mardal, University of Oslo, NorwayQifeng Liao, ShanghaiTech University, China
Copyright © 2023 Cai, Gu, Hong and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mingchao Cai, TWluZ2NoYW8uQ2FpJiN4MDAwNDA7bW9yZ2FuLmVkdQ==
 Mingchao Cai
Mingchao Cai Huipeng Gu
Huipeng Gu Pengxiang Hong2
Pengxiang Hong2 Jingzhi Li
Jingzhi Li