
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Appl. Math. Stat., 02 November 2022
Sec. Mathematical Biology
Volume 8 - 2022 | https://doi.org/10.3389/fams.2022.1023310
This article is part of the Research TopicData science in neuro- and onco-biologyView all 6 articles
In neuroscience, the structural connectivity matrix of synaptic weights between neurons is one of the critical factors that determine the overall function of a network of neurons. The mechanisms of signal transduction have been intensively studied at different time and spatial scales and both the cellular and molecular levels. While a better understanding and knowledge of some basic processes of information handling by neurons has been achieved, little is known about the organization and function of complex neuronal networks. Experimental methods are now available to simultaneously monitor the electrical activity of a large number of neurons in real time. The analysis of the data related to the activities of individual neurons can become a very valuable tool for the study of the dynamics and architecture of neural networks. In particular, advances in optical imaging techniques allow us to record up to thousands of neurons nowadays. However, most of the efforts have been focused on calcium signals, that lack relevant aspects of cell activity. In recent years, progresses in the field of genetically encoded voltage indicators have shown that imaging signals could be well suited to record spiking and synaptic events from a large population of neurons. Here, we present a methodology to infer the connectivity of a population of neurons from their voltage traces. At first, putative synaptic events were detected. Then, a multi-class logistic regression was used to fit the putative events to the spiking activities and a penalization term was allowed to regulate the sparseness of the inferred network. The proposed Multi-Class Logistic Regression with L1 penalization (MCLRL) was benchmarked against data obtained from in silico network simulations. MCLRL properly inferred the connectivity of all tested networks, as indicated by the Matthew correlation coefficient (MCC). Importantly, MCLRL was accomplished to reconstruct the connectivity among subgroups of neurons sampled from the network. The robustness of MCLRL to noise was also assessed and the performances remained high (MCC > 0.95) even in extremely high noise conditions (> 95% noisy events). Finally, we devised a procedure to determine the optimal MCLRL regularization term, which allows us to envision its application to experimental data.
Emergent dynamics in neural networks are primarily determined by the inter-connectivity among neurons. The connectivity patterns can be studied using mathematical tools derived from graph theory [1]. During the maturation process, brain areas undergo profound changes that are reflected in refinements of neuron properties, synapse specificity, and the rearrangement of neural connections. The latter is accompanied by changes in emergent activities. For instance, while transiting to the mature stage, the hippocampus display synchronous events while the Purkinje cells' activities of the cerebellum manifest as traveling waves. It has been shown that the underlying network topology, i.e., the distribution of connections, differs, with the presence of hub neurons, i.e., scale-free organization, in the hippocampus and local connections, i.e., regular network, in the cerebellum [2]. Since the set of connections determines how network wide activities emerge from neurons' properties there is a great interest in determining the connectome (i.e., all connection patterns) adopting the most efficient technique.
We point out that, given the interest in studying neural connectomes, different definitions of connectivity exist [2]. The anatomical (or structural) connectivity is defined by the set of synaptic connections that exist among the neurons. Instead, functional connectivity considers the statistical interrelation between the neuron's activities. A typical example of functional connectivity is given by the spike-cross-correlation that is computed across pairs of spike trains. There are also several types of algorithms used to estimate the strength of functional connectivity, that depend also on the type of signals considered for the analysis. Instead, effective connectivity is related to the direct (or causal) influence of one node on another. The latter can be estimated by stimulating one node and investigating the information flow to the downstream neurons. As an alternative approach, one can gain insights into the anatomical connectivity setting a causal mathematical model relating neuron's activities. Then, statistical inference of the model parameters allows an understanding of how the neurons influence each other. The design of functional and especially effective connectivity algorithms is much dependent on the brain scale considered. Here we restrict our attention to the meso-scale, that is how the neurons are wired together and how they transmit information throughout the network.
Most of the existing connectivity algorithms rely on spike train analysis as the gold standard technique to record from the population of neurons, i.e., extra-cellular and genetic-encoded calcium fluorescence, allowing access to high signal-to-noise ratio signals. Since the analysis of spike trains considers only the time point at which a spike is generated this reduces the amount of data to store and process for each unit. However, this approach comes also with some drawbacks. For instance, the use of standard spike cross-correlation analysis involves numerous false positives, connections inferred by the method that, however, do not exist in the structural connectome [3, 4]. To circumvent this problem [4] combined the spike cross-correlation analysis by incorporating synaptic-kernels in a generalized linear model named GLMCC. The study showed that GLMCC reduced significantly the number of false positives with respect to standard approaches. However, since GLMCC infers the impact of mono-synaptic contacts in a noisy cross-correlogram, it also requires large recording time windows (refer to Table 1 in Kobayashi et al. [4]), that can be out of reach in standard experimental conditions. Moreover, long recordings are hardly stationary and it is often difficult to disentangle the contribution of non-stationary fluctuations on the results.
Here, we make the hypothesis we can record spikes and post-synaptic-potentials from all neurons of interest. The gold standard to record such types of signals, in experiments, is the patch clamp technique. However, the latter is well suited to record the activities of up to a maximum of around ten neurons [5]. Instead, a technique based on genetically expressed molecules with fluorescent signals proportional to voltage (Genetically Encoded Voltage Indicator, GEVI) appears to be more indicated here. Recent works have shown that sub-threshold signals can reliably be recorded together with action potentials [6]. In cell culture experiments, it has been shown that post-synaptic-potentials (PSPs) can be sensed with the GEVI [7]. Importantly, the most distal synaptic inputs that reach a neuron might be overlooked as noise because their amplitude can be very low (order of 0.5 mV [8]). However, as we will see, the proposed inference algorithm is quite robust to noisy PSPs. Thus, despite there are still some experimental issues to solve [9] it can be envisioned that the GEVIs, or a similar technique, will allow to record spikes and PSP signals at the population level. Interestingly, spiking activities and PSP events could also be obtained by combining different optical recording techniques [10], e.g., calcium (i.e., spikes) together with neurotransmitter (i.e., synaptic events) imaging recordings. As anticipated, a fundamental step of the proposed framework consists of the detection of spiking and PSP events. While spikes are easy to detect because of the high voltage excursion, the same can be much more challenging for PSPs. A distinctive property of PSPs is the onset phase, which is characterized by a marked change in the voltage derivative. The family of Perona-Malik (PM) filters represent a proper tool to denoise data by preserving the onset of a signal. Interestingly, we have contributed to improving the performances of standard PM filters [11–13] involving a new non-local diffusive term. A description of the improved algorithm is given in the Supplementary material.
In this study, we skip the detection phase and extract all events directly from the simulations. Then, to investigate the robustness of the proposed methodology, we increased the amount of falsely detected events (named 'instrumental noise' here). A key ingredient of biological networks is sparseness. We recognize a population sparsity (how many neurons are active right now), a time sparsity (how often the cells fire), and a connection sparsity (each neuron connects to a narrow subset of neurons). Here, we focus our attention on the latter type of sparseness. A logistic model is introduced linking the spiking activities to the PSPs and an L1-penalization term is included in the likelihood function to promote connectivity sparseness in the inferred network. Here, we extended a previous study [14], to include both excitatory and inhibitory neurons. The inference worked well on small networks even considering the mixed populations. The paper also analyses the impact of noise on these networks, showing that the inference is quite robust to extremely high noise levels. Then, the size of the networks was increased to 100 neurons and different network organizations (random, distance based, and clustered) were analyzed, again yielding good results. It is worth noting that, in most of the experiments, neurons are highly under-sampled, i.e., only a very small number of neurons can be recorded from a given neuronal population (refer to e.g., Pillow et al. [15], Stevenson et al. [16], and Vidne et al. [17]). This problem was addressed here by randomly selecting 20 out of 100 neurons running the inference on these subsets. Again the results were very good in terms of different metrics. Finally, we designed a simple data-driven approach to find a proxy of the optimal penalization term from experimental data. The proxy was defined as the smallest penalization term satisfying Dale's principle (i.e., neurons are either excitatory or inhibitory), and it worked optimally on small and medium sized networks made of 20 and 100 neurons, respectively.
In all analyses, except if stated otherwise, the errors and their error bars correspond to one standard deviation computed over the samples. For the boxplots, we adopted the following convention. The extremes of the boxes are relative to the first and third quartiles, the whiskers to the 5th and 95th percentiles and the red line corresponds to the mean.
The network model was implemented in the NEST simulation ambient [18]. We simulated the activity of each node using the single compartment neurons proposed by Hill and Tononi [19]. The latter aimed to simulate the temporal dynamics of the membrane potential by a biophysical plausible set of equations. With respect to the fully-fledged Hodgkin and Huxley model, the “Hill-Tononi” modeling allowed a realistic simulation of the sub-threshold potential while the generation of a spike was replaced by an integrate and fire mechanisms. In the network, each node/neuron had the same set of default parameters taken from the class of “cortical excitatory” neurons (refer to Table 2 in Hill and Tononi [19]). The nodes were split into two populations, relative to the excitatory and inhibitory neurons. In this study, the ratio between excitatory and inhibitory neurons was kept at 4:1 (80% excitatory, 20% inhibitory). Each spike of a neuron determined a post synaptic potential (PSP) in the contacted neuron(s) with a fixed transmission delay (0.1 ms). In particular, excitatory neurons caused excitatory post synaptic potentials (EPSPs) described by the ‘AMPA' model and the inhibitory neurons caused inhibitory post synaptic potentials (IPSPs) described by the “GABA-A” model (refer to Table 3 in Hill and Tononi [19]). In order to facilitate the spread of activities throughout the network, we increased all synaptic strengths to 4, decreased all recovery time constants to 10 ms, injected a fixed current of 10 pA to each neuron, and included additional “AMPA” synaptic inputs randomly distributed in time and governed by a Poisson process with an average frequency of 30 Hz. The simulations were typically run for 5–10 s (Tmax).
A graph G is a pair G = (V, E), where V is the set of nodes or vertices and E is the set of links or edges made by paired vertices: the link e = {v1, v2} belongs to E if and only the two vertices v1 and v2 are connected. The vertices are often labeled with the natural numbers, so that V = {1, …, n}.
A directed graph is a graph G = (V, E) where the links have a direction, also called arcs. Accordingly, E becomes a set of ordered pairs of vertices, so that (v1, v2)∈E means that there exists an arc starting from v1 and ending in v2, while (v2, v1)∈E ensures the presence of the opposite arc.
The adjacency matrix A = (ai, j) of a graph of n nodes is defined with |ai, j| is either 1 or 0, respectively when a connection exists or not from the node i to j. The sparseness of a directed graph G = (E, V) is given by the fraction of existing connections with respect to the possible ones:
A weighted graph is a directed graph in which each arc is given a numerical weight, typically positive. A weighted graph may be described by the square n×n matrix Θ = (θi, j) of the weights of the directed edges. Summing up, V = {1, …, n}, E = {(i, j):θi, j ≠ 0}, and for the arc (i, j) ∈ E, θi, j is its weight.
In a random graph, each node is connected to a fraction of the other neurons of the network without any specific constraint. The amount of arcs in a random graph is typically determined by the connectivity probability (Pconn). In our study, Pconn = 0.3 in the 20-neuron networks indicates that each neuron, on average, connects to ≃6 neurons of the network. Instead, in a distance based graph, the connectivity probability is not constant anymore and depends on the reciprocal distances between nodes so that 0 ≤ pij ≤ 1 represents the probability to connect node i to node j. In this study, we defined the connectivity probability in terms of a Gaussian function: where σ modulates the probability to connect neurons (i.e., a lower sigma yields smaller patches of connected neurons), while di, j stands for the distance between nodes i and j. In all simulations, the neurons were placed in a unity square and σ = 0.2. In the clustered 100-neurons networks, 25 neurons (excitatory or inhibitory, randomly selected) were placed in each of the four clusters located in the circles of centers (0.2,0.2), (0.2,0.8), (0.8, 0.2), (0.8, 0.8), and radius = 0.2. Then, the connectivity was determined by the Gaussian function with σ = 0.2. For each network type (random, Gauss, cluster), we tested 10 different connectomes changing the seed of the generative algorithm.
Similar to our previous report [14], once the detection of spikes and PSPs have been performed, the definition of the event-based model was obtained with a discretization of the temporal events. The discretization time is chosen based on considerations about the signals of interest. The conduction delay (i.e., the time it takes for an action potential to propagate throughout an axon) is typically small as the longest axons in the central nervous system are on the order of 1 mm and the conduction velocity in myelinated nerve fibers is ≃100 mm/ms, resulting in a propagation delay of ≃0.01 ms. Then, synaptic events are subject to temporal jitters that are on the order of ≤ 1ms (i.e., asynchronous release [20]). The discretization time was therefore set to dt = 1 ms and the external processes were defined as:
and the internal processes for neuron i at time step t:
where EPSP and IPSP correspond to the excitatory and inhibitory post-synaptic potentials, respectively. A Markovian property was assumed, such that for any i = 1, …, n the internal processes of neuron i were linked to the external processes of the neurons at the proceeding time-step, so that, for any internal activity a∈{−1, 0, +1} and t∈{1, …, Tmax}
To model the network activity, we use a weighted directed graph (V, Θ) of the excitatory/inhibitory synaptic links between neurons, so that, for j, i∈{1, …, n}, the null weight Θj→i = 0 means that there is no synaptic link from neuron j to i, while Θj→i > 0 modeled a synaptic link that started from neuron j to i. When Θj→i > 0, an action potential influenced the internal process according to the nature of the neuron j (excitatory/inhibitory).
Therefore, we divided the set of neurons V into two sets V+ and V−, the sets of the excitatory and inhibitory neurons, respectively, so that V = V+ ∪ V−. The excitatory and inhibitory property was then applied to the synaptic links by superscribing + and − to Θj→i, so that if the neuron j was excitatory, while when j was inhibitory.
A multi-class (or multinomial) logistic regression model was used to compare the excitatory/inhibitory activity to the baseline (no-PSP observed) with the weights Θ of the excitatory/inhibitory synaptic links:
which implied time-homogeneity in the window of observation: for any internal activity a∈{−1, 0, +1} and t1, t2∈{1, …, Tmax}
This is a well defined process when probabilities are positive for each class [21], as in our context (i.e., , since the neurons are silent, , most of the time).
The explicit conditional probabilities were then found by solving the system Equation (4), for any i = 1, …, n,
that allowed us to write the log-likelihood of the internal process for each neuron i as
Since a direct estimation of Θ suffered from an unbalanced number of internal events, we proceeded as in King and Zeng [22] by counting with the number of internal events that differed from a (a∈{0, +1, −1}) and weighted the data to obtain the modified log-likelihood function:
The goal of our methodology is to find a sparse geometry of the network. In terms of parameter estimation, this corresponds to determining whether or not the Θj→i are different from 0. A robust approach to this problem is the classical LASSO penalization, which introduces a penalty term for the 1-norm of Θj→i.
Summing up, the problem can be solved by minimizing the penalized weighted log-likelihood function
The penalization factor λ allowed for regulation of the number of connections. Indeed, at low λ, the inferred graph had numerous arcs while the increase of λ caused a decrease in the number of admissible arcs/links. In order to test the robustness of MCLRL, we introduced the concept of “instrumental” noise (νstrum). For a given simulation, the idea was to increase the detection noise by randomly flipping some of the “no PSP is observed” into either an EPSP or an IPSP (refer to Equation 3). The amount of “instrumental” noise was made proportional to the endogenous Poisson noise (30 Hz). For instance, in Figure 3, (0,1) indicates that no excitatory noise is introduced (0) while the inhibitory one (1) has the same rate (30 Hz) as the endogenous Poisson noise. In this specific case (νstrum=(0,1)), the performances of the MCLRL algorithm were assessed keeping the same (overall) noise level on the excitatory and inhibitory connections.
The Dale's precision index (Dpi) of neuron i was defined as:
where wrong represents the incorrect synaptic events assigned to neuron i (e.g., inhibitory event to an excitatory neuron) and total the overall amount of events. The mean Dale precision index (Dpi) of the excitatory or the inhibitory population was defined as:
where n is the number of neurons.
The performances of MCLRL can be quantified in terms of some synthetic indexes [23] computed on the confusion matrices obtained by comparing the inferred connectome (infer-conn) to the structural one (struct-conn). Following a standard convention, we call the true positive (TP) as the amount of connections part of infer-conn and struct-conn, the false positive (FP) as the amount part of infer-conn but not of struct-conn, the true negative (TN) the amount neither part of infer-conn nor of struct-conn and the false negative the amount absent in infer-conn but part of struct-conn. In this study, we defined three types of confusion matrices, for the excitatory, inhibitory, and overall connections (all).
Simple quantities can be defined in terms of the confusion matrices. For instance, the true positive rate (TPR), the false positive rate (FPR) and the Youden index were defined as:
The receiver operative curve (ROC), a gold standard in binary classification, was obtained by varying the penalization term λ (Section 2.4) and reporting TPR vs. FPR (refer to Figure 2A). The “goodness” of a ROC can be evaluated using different synthetic measures. For instance, the maximum of the Youden index is frequently used as a reference point as it corresponds to the maximum sensitivity and specificity. The maximum of the Youden index is typically found at the “top-left” coordinates of the ROC plot. Instead, the Area Under the Curve (AUC) corresponds to the integral of TPR vs. FPR. However, an accurate estimation of it would have required spanning the entire x-axis up to FPR close to 1 that, however, is obtained for very low λ values and is quite time consuming to compute. Three different ROCs could be defined for excitatory, inhibitory, and overall connections.
In this study, we relied on the Matthew correlation coefficient (MCC) [4, 23] because it better weights the number of false positives (FP) and false negatives (FN) with respect to other indexes. MCC is defined as:
Again, three different types of MCC were defined and named MCC(exc), MCC(inh), and MCC(all), respectively, for excitatory, inhibitory, and overall connections. As a function of λ, MCC was low for small/big values of the penalization term λ. Indeed, at small λ many false connections enter the connectome (e.g., gray neurons in Figure 1B) causing an increase in FP and a decrease in MCC. On the other side, at high λ many correct connections are lost causing an increase in FN and a consequent decrease in MCC.
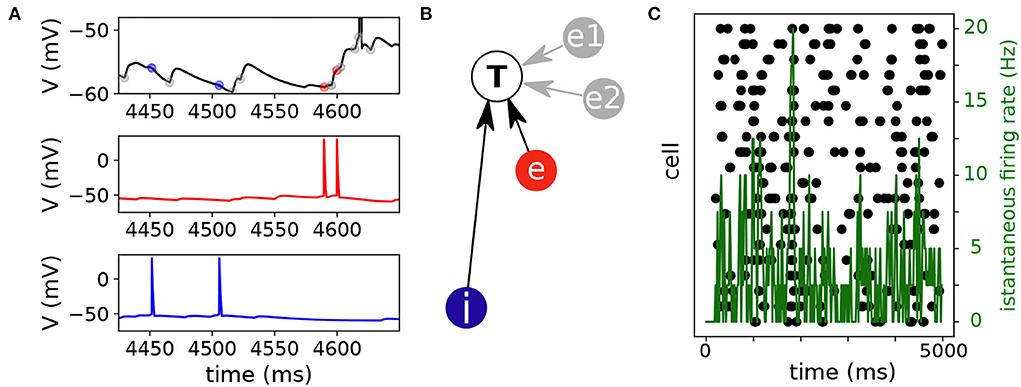
Figure 1. Network activity and inference. (A) Representative voltage traces of three neurons of the network. The voltage trace of an illustrative neuron (top, black trace) was modulated by excitatory and inhibitory inputs. An excitatory neuron (middle, red trace) had two spikes and elicited excitatory post synaptic potentials in the target neuron (top, red marks). Similarly, an inhibitory neuron (blue trace) had also two spikes and elicited inhibitory post synaptic potentials in the target neuron (top, blue marks). The rest of the synaptic inputs (gray marks) come either from endogenous noise or from other neurons of the network. (B) The inference algorithm associated the excitatory and inhibitory events with spikes of the corresponding neurons (excitatory, e and inhibitory, i). Two additional connections from neurons e1 and e2 (gray circles) were inferred by the algorithm to explain the excitatory inputs [gray marks in panel (A)]. (C) The raster plot shows that spiking activity in the network was rather sparse. Black symbols indicate spike events (x-axis spike timing, y-axis cell number). The mean firing rate (green trace) was computed with a 20 ms binsize.
Simulations were performed for network activity with 16 excitatory and 4 inhibitory neurons (4:1 ratio). The goal here was to verify that in small networks with mixed excitatory and inhibitory neurons the proposed multi-class logistic regression with lasso penalization performed similarly to the binary logistic regression used in our previous report [14]. The connectivity probability was set at 0.3., which yielded an average of 6 connections per neuron (114 connections, on average, per network, range: 101 − 133). At the single cell level, the neuron's activity was modulated by noisy events (black/gray symbols) intermingled by excitatory and inhibitory inputs (respectively red and blue symbols) originating from the network (Figure 1A). The multi-class logistic regression with lasso penalization associates the excitatory and inhibitory spiking activities to the synaptic events observed in the target cell (T, Figure 1B). Additional connections (and possibly wrong ones, refer to Section 2.6) from other neurons (e1 and e2) could be inferred to explain the putative synaptic events (gray marks, Figure 1A) occurring in the target cell. At the network level spiking activity (black marks, Figure 1C) was sparse in time and space. Across simulations, the population means firing rates (i.e., averaged mean firing rates) were 2.75 ± 0.19 Hz and 2.77 ± 0.41 Hz, respectively, for the excitatory and inhibitory neuron populations (n = 10 simulations). Given this premise, the inference algorithm has to infer the right connections in a highly noisy context. Indeed, each neuron receives a net synaptic input from the network ≃16.5 Hz (≃6 synaptic contacts x 2.75 Hz) while the endogenous Poisson noise comes at 30 Hz (Section 2.2).
In order to verify how the proposed inference algorithm performed, we spanned the penalization weight λ (refer to Section 2.4) from 0.1 to 10. In a representative example, with respect to the Receiver Operative Curve (ROC) (Figure 2A), the Youden index peak was close to one for λ≃2.5. Similarly, the Matthew correlation coefficient (MCC) reached the peak (≃1) for the same λ value (Figure 2B). Note that MCC has the characteristic bell shape, with lower values at the boundary of the domain where low/high λ values imply a high/low sparseness of the network (Section 2.6). For instance, when the penalization term λ was ‘too small', MCLRL associated most of the synaptic events with any neurons of the network. In the latter case, the inference returned several wrong connections (e.g., neurons e1 and e2 in Figure 1B). These observations were consistent across simulations with peak MCC values falling above 0.98 in all cases (n = 10, Figure 2C). The peak MCC was reached at small but different λ (range: 1.43 to 2.5, λmax, Figure 2D) and the λ at which MCC started to decay (i.e., measured with respect to 0.99 of the peak, λknee) was quite variable (range: 2.5 to 10). It was found that neither λmax nor λknee correlated with network sparseness (not shown). As reported in Figures 2C,D, the Youden index provided similar results to MCC. In all inferred networks (n = 10), the MCC curves of inhibition were systematically higher than the ones relative to excitation.
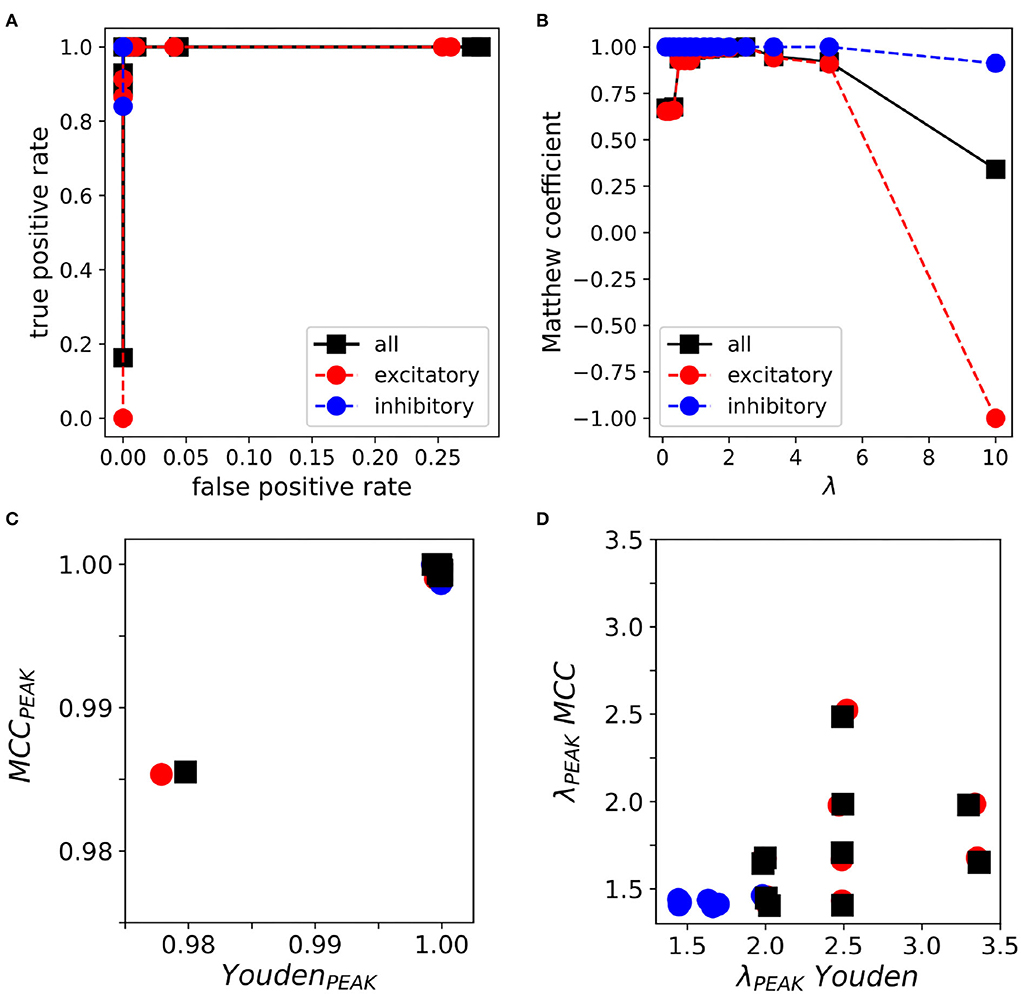
Figure 2. Multi-Class Logistic Regression with L1 (MCLRL) inference on 20-neuron networks. (A) Receiver operative curve relative to a representative simulation. The true positive rate reaches the value one with a false positive rate close to zero. (B) Matthew correlation coefficient (MCC) relative to the same simulation. MCC reaches a peak value close to one and decays steeply for λ ≥ 3. (C) The peak values of MCC and the Youden index are comparably high in a set of simulations (n = 10). In 9 out of 10 simulations, MCC and Youden index reach the value 1. (D) The peak MCC and Youden indexes are reached at small λ values. The λPEAK relative to the Youden indexes is much more spread with respect to the ones obtained with MCC. Note, a small jitter (≤ 0.1%) is added to each data point for clarity. In all panels, the colors are relative to excitation (red), inhibition (blue), and all connections (black).
In order to clarify the impact of the detection noise on the inference process, we tested different paradigms. As a first test, we checked the MCLRL robustness to noise. To this aim, we increased the “instrumental noise” (Section 2.4) for the different networks (n = 10) of the previous Section. Figure 3A shows that noise mostly affected the shape of the Matthew curves while the peaks remained close to one. Note that different repetitions of the same ‘instrumental noise' on the same network resulted in small deviations from the average MCC curves (i.e., the error bars—1 standard deviation—are barely visible). The latter result was somehow expected as different repetitions of the same noise level determine flips of the synaptic events at different time points, that do not affect significantly the conditional probabilities relating spikes to synaptic events (Section 2.4). Figure 3B shows that the MCC peaks were high (>0.96) up to the instrumental noise (2,4), which corresponded to an additional noise of 60 Hz (excitation) and 120 Hz (inhibition). Even when the noise frequency was brought to 240 Hz (excitation) and 480 Hz (inhibition) the performances were still good (condition (8,16)). Importantly, the resilience to the noise of MCLRL was quite impressive as in the latter condition the ratio between the genuine and noisy events is on the order of 2% [i.e., 16.5/(480 + 240 + 30 + 16.5)]. In short, the latter tests suggested that high noise levels did not affect the MCC peak but it caused a decrease in the left and right arms of the MCC parabola and reduced the set of optimal λ values. These results also explained why the MCC(inh) curve was systematically higher than MCC(exc) (Section 3.1). Indeed, since the inhibitory noise was not introduced in the simulations, MCC(inh) remained high for any λ. Another source of noise is relative to the incorrect detection of internal events. There are different such cases we can analyze. The most trivial one consists of missed detection of either excitatory and/or inhibitory events. The latter includes the relevant physiological case of synaptic failures (i.e., when a fraction of the presynaptic spikes do not elicit PSPs). Another case is when synaptic events are assigned to the wrong class. For instance, this can occur when inhibitory events are misinterpreted as excitatory ones. The latter happens when the IPSPs occur at voltages below the reversal potential of the GABA-A receptor. We investigated this on the same 20 neuron networks used before and we tested different amounts of wrong detections, spanning from 20, 50to 90%. Again the results were pretty good in terms of MCC peak (Figures 3C,D), though, when 90% of the inhibitory events were misinterpreted as excitatory ones, a decrease in the MCC peaks was observed. Note that, on average, the latter 90% of noise had a greater impact on the MCC peak than the (8, 16) instrumental noise tested before. The findings indicate that MCLRL was robust with respect to different types of noises. One might wonder in what conditions the performances of MCLRL would significantly deteriorate. A key ingredient of MCLRL is the proper estimation of the conditional probabilities linking the external (i.e., spikes) to the internal (i.e., synaptic signal) events (Section 2.4). The performances of MCLRL worsened when, for example, numerous neurons fired synchronously (i.e., within the discretization time dt = 1 ms) that weakened the causal relationship between spikes and synaptic events.
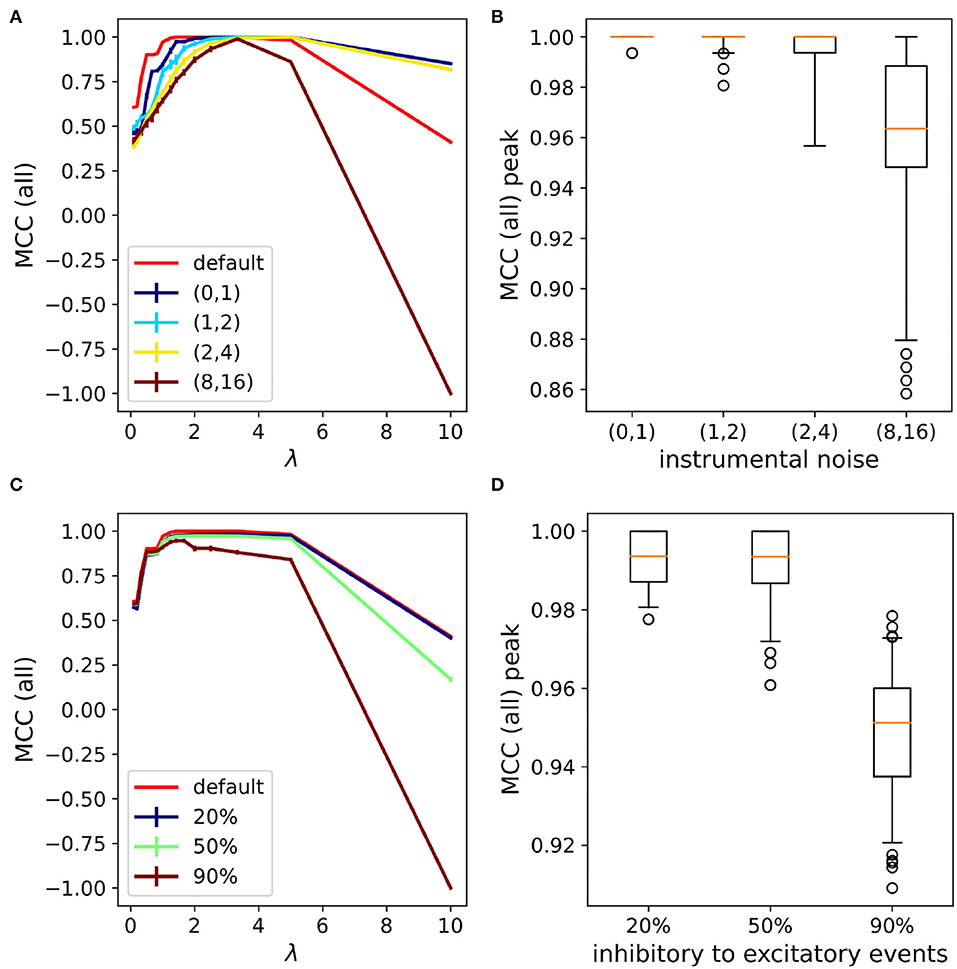
Figure 3. MCLRL performances with varying levels of noise. (A) MCC curves (all) with the increasing noise level. The numbers enclosed in the parenthesis indicate the scaling factor with respect to the endogenous Poisson noise rate. Note that the default curve (red) corresponds to the black curve reported in Figure 2B. (B) The distribution of MCC peaks obtained across different networks (n = 10, the same as Section 3.1) with different seeds (m=10 per network) of the given noise levels stay above 0.86 (mean > 0.96). (C) MCC curves (all) with incorrect detection of the inhibitory events. The percentages 20, 50, and 90% indicated the fraction of inhibitory events that have been wrongly assigned to excitatory events. (D) The distribution of MCC peaks obtained across different networks (n = 10, the same as Section 3.1) and repetitions (m=10) of the given percentage of inhibitory events flipped to excitatory events (mean > 0.95).
In order to verify the inference also works on a larger network we simulated the activity of 100 neurons with the same 4:1 ratio between excitatory and inhibitory neurons. The connectivity probability was lowered to 0.2 and the recording time window was set to 5 s. Ten different networks were simulated with the given parameters. In Figure 4A, the activity of a representative network has been reported. After an initial synchronous event, the network settled down to a sparse activity regime. Again, the performances were optimal with a peak Matthew correlation coefficient close to one (Figure 4B). In addition, the decay of MCC at high λ (λ ≥ 5) was smoother compared to the case of 20 neurons (Figure 2). Overall, the simulated networks performances were close to optimal, with peak Matthew correlation coefficient MCCPEAK = (0.996 ± 0.004, 1 ± 0) and the corresponding λPEAK = (2.58 ± 0.26, 0.23 ± 0.09).
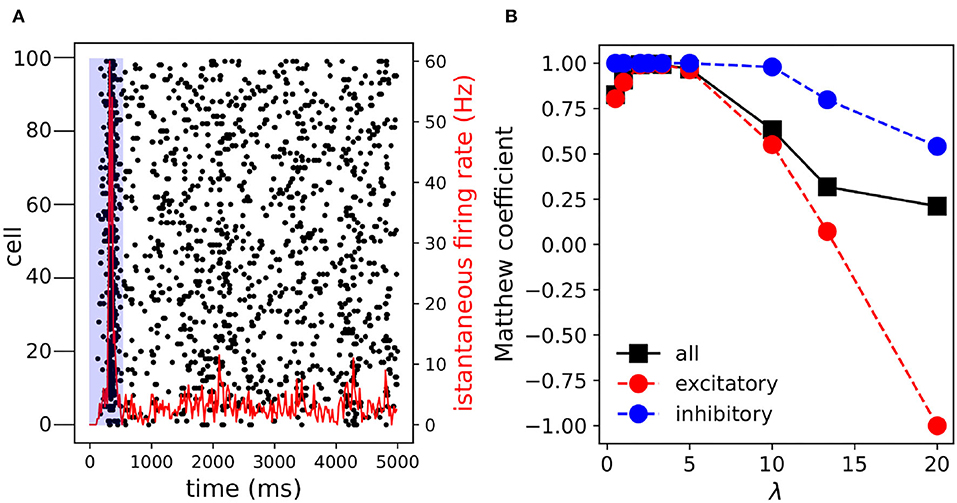
Figure 4. Multi-Class Logistic Regression with L1 inference on 100 neurons. (A) Raster plot. After an initial transient synchronous event (t≃200 ms) the network activity is pretty sparse. The activities falling in the blue shaded area are discarded from the inference process. (B) The Matthew correlation coefficient increases and reaches the peak value MCCMAX = (0.995, 1, 0.996) and the corresponding λMAX = (2.5, 0.5, 2.5). In the parenthesis the arguments are relative to excitation, inhibition and all connections. Note, when the inference is performed on the entire time interval (0–5,000 ms) the peak MCC slightly decreases MCCMAX = (0.991, 1, 0.993).
The inference of network connections from node activities faced a set of problems that are also found in related scientific questions when data sampling was far from optimal. For instance, multi-unit recordings, nowadays, allow us to simultaneously record signals from thousands of neurons. Still, their activities are likely regulated by several neurons that have not been recorded. The latter involves a set of multifaceted problems. The unobserved neurons could act as common drivers of the observed population and affect the inference process. In addition, unobserved neurons in a feed-forward chain may also affect the outcome of the inference process. Disentangling the partial contributions of the unobserved neurons may, therefore, represent an intricate problem to solve. Here, we addressed the question by running the MCLRL over groups of 20 neurons randomly sampled from networks of 100 neurons of the previous Section. We found that also in this case MCLRL managed to accurately infer the connectivity of the 20 neurons networks (m = 10 repetitions) and the performances were comparable to the previous results (refer to Section 3.1) with MCCMAX = (0.995 ± 0.010, 1 ± 0, 0.995 ± 0.009) and the corresponding λMAX = (2.05 ± 0.16, 0.5 ± 0, 0.5 ± 0, 2.05 ± 0.16). Since the neurons were randomly selected the fraction of excitatory vs. inhibitory neurons varied from (65%, 35%) to (90%, 10%), indicating that the MCLRL performance was not affected by the type of connection.
The previous results showed that MCLRL is robust to noise and it scales well with the network size. So far we have considered medium sized networks with excitatory and inhibitory neurons. In order to simulate network activities with a higher realism, one should also consider that neurons connect with a higher probability to the nearby ones. In this case, the inference will have to find more connections among neurons located close to each other. At first, we randomly distributed 100 neurons (80/20 % E/I ratio) and the connection probability was determined by a Gaussian function. Then, in order to exacerbate the concept, we considered networks characterized by the presence of neural assemblies and hubs. As in other fields, network organization can play an important role in emergent dynamics [2]. For instance, in cell cultures network bursts (i.e., events that involve a large fraction of the network) originate from specific regions of the network characterized by recurrent connections among neurons [24]. We considered four clusters (Figure 5A) with inter-cluster connectivity (i.e., the percentage of connections across clusters) of (16.2 ± 2.7%) (m = 10 networks). The link-length distribution was concentrated on small values compared to the Gauss connectivity (Figure 5B). Interestingly, the peak MCC (all) reached the value ≃1 for λ≃2 in all considered networks (Figure 5C). In addition, the MCC curves of the 4-clusters networks had a larger plateau with a peak MCC≃1. Overall, the MCC curves relative to the random and Gaussian networks were much more comparable (the former slightly higher at small and large λ). These results indicate that the MCC peak was not much affected by network sparseness. Instead, the MCC plateau does depend on network sparsity (4 clusters 6.2 ± 0.3, random 19.7 ± 0.5, Gauss 15.2 ± 0.5, error is one standard deviation). Indeed, for the four clusters, the number of connections of the anatomical connectome was considerably lower than in the other ones. As seen for inhibition (e.g., Figures 1B, 4B), the inference performs much better in terms of MCC when there are lesser connections to detect. Note that, the spiking activity was reduced in the 4-clusters networks (MFR = 2.70 ± 0.09 Hz) with respect to the Gauss networks (MFR = 3.12 ± 0.09 Hz), presumably because of the lower sparseness with respect to the Gauss networks. Finally, we performed an additional analysis (similar to Section 3.3) by sampling 20 out of the 100 neurons. Here, for each network, we selected two groups of 20 neurons with opposite properties. In the first (second) group, we selected the neurons that were less (most) connected to the other neurons of the network. This choice resulted in 20-neuron networks with sparseness in the 4-clusters 3.2 ± 0.8 (less) and 9.4 ± 2.5 (most). Instead, in the Gauss networks sparseness was 8.9 ± 1.6 (less) and 26.8 ± 2.9 (most). Interestingly, the subsampled networks had an MCC peak close to one in all cases (Figure 5D). The performances of the “less” connected sub-networks (triangle down) were only slightly worse compared to the “most” connected sub-networks (triangle up). Moreover, note that also λPEAK was in the same range as the values obtained for the 100-neuron networks (Figure 5C).
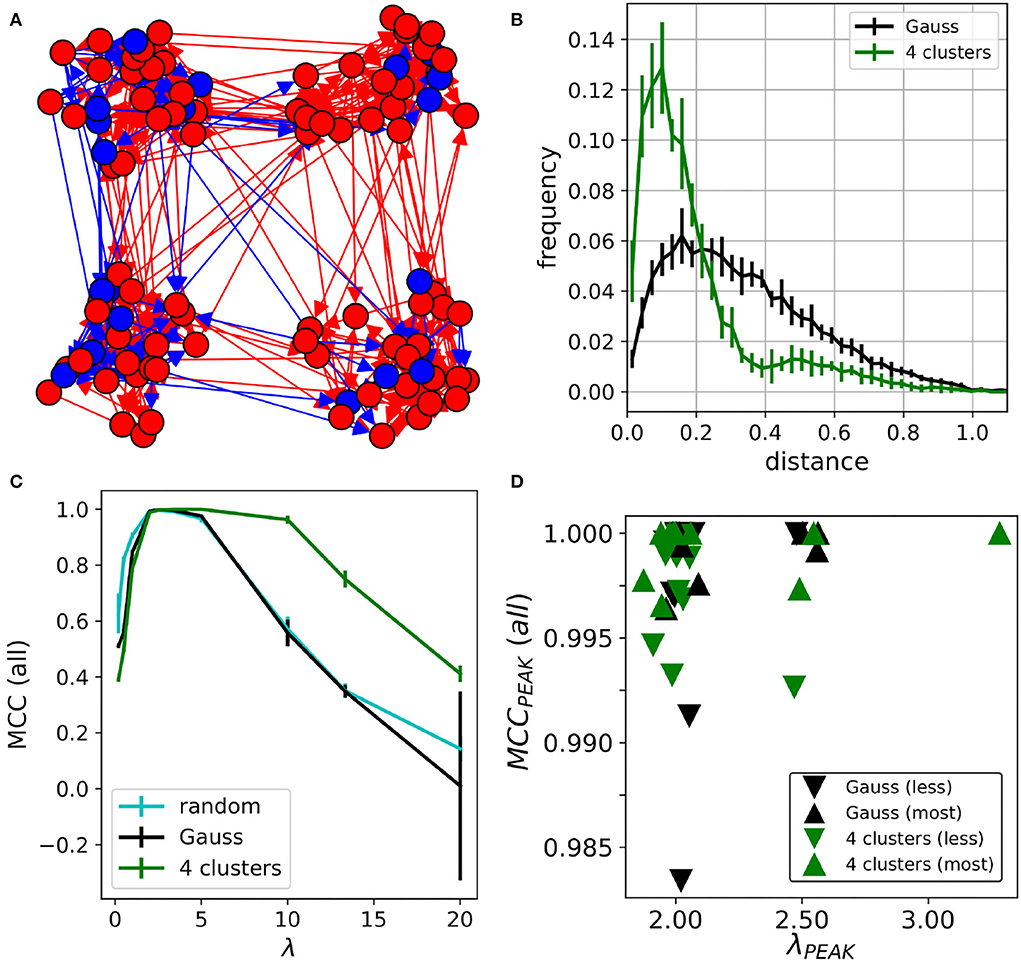
Figure 5. Networks with topological constraints. (A) 4-clusters networks. Red/blue circles indicate excitatory/inhibitory neurons. The arrows point to the connected neurons. (B) Link-length distribution of the Gauss and 4-clusters networks (C) MCC curves relative to the random (cyan), Gauss (black) and the 4-clusters (green). Error bars are one standard deviation computed over 10 realizations of the same type of network. (D) The MCC (all) of the sampled 20 neurons is high for the “less” and the “most” connected neurons in the network. Note that data points were slightly jittered (≃0.1%) to increase readability.
The results obtained so far indicate that MCLRL was quite effective in inferring the connectivity of small to medium sized networks. However, in the concrete context of experimental data, it is not clear what λ should be used to infer the network connectivity. Based on the previous results (Section 3.1), the optimal λ fell in the range of 1.43–2.5 but we also found that the optimal λ could be located in a tiny interval (Figure 3A). There are different possible routes in determining the right λ inspecting how the inferred graph changes at different λ. For instance, one could monitor the link length distribution of the inferred links and fit it to a theoretical distribution. Unfortunately, the latter distribution might be hard to define for a generic neural network, which prevents its usage. Another possible approach consists in monitoring the nature of synaptic connections. At the mature stage, a neuron makes either excitatory or inhibitory synapses with other neurons of the network, a rule known as Dale's principle in the Neuroscience field [25]. At small λ, the inference returns many more connections with respect to the structural connectome and the neurons will have connections with mixed signs (i.e., excitatory and inhibitory), thus violating Dale's principle. The increase of λ selects the most reliable connections and it will gradually satisfy Dale's principle. Indeed, we found that Dale's precision index (Dpi, Section 2.5) increased with λ and reached the plateau =1 for λ∈[1, 3] (Figure 6A), a range of values compatible with the optimal MCC λ values (Figure 6B). In order to properly compute the Dalton precision index (Dpi), each neuron has to be labeled according to its category (either excitatory or inhibitory). From an experimental point of view, the neurons can be labeled based on their firing patterns (e.g., inhibitory neurons typically fire more robustly than excitatory ones). Alternatively, the neurons could be labeled using different fluorescent reporters. Importantly, one can also rely on our data driven approach to label the neurons. Indeed, at high enough λ, when MCC starts to decrease, MCLRL does not infer all existing connections but all neurons are unambiguously labeled as excitatory or inhibitory. Dale's principle can thus be invoked to estimate the right λ and we defined λDale as the first point of the Dpi plateau (= 1 in panel A). At first, we tested the idea on the 20-neuron networks presented before [Section 3.2 with νstrum = (0, 1)]. Interestingly, λDale was comparable to the one maximizing the MCC index (Figure 6B). Now, since the Dpi plateaus of the excitatory and the inhibitory sub-populations occur at different λDale (Figure 6A) we can either choose one or the other. We found that adopting the maximum λDale yielded high values of the MCC (exc) and MCC (inh) indexes (Figure 6C). Then, we repeated the same simulations on the 100-neuron networks (Section 3.3) with νstrum = (0, 1) and obtained slightly lower, but still high MCC values (Figure 6D).
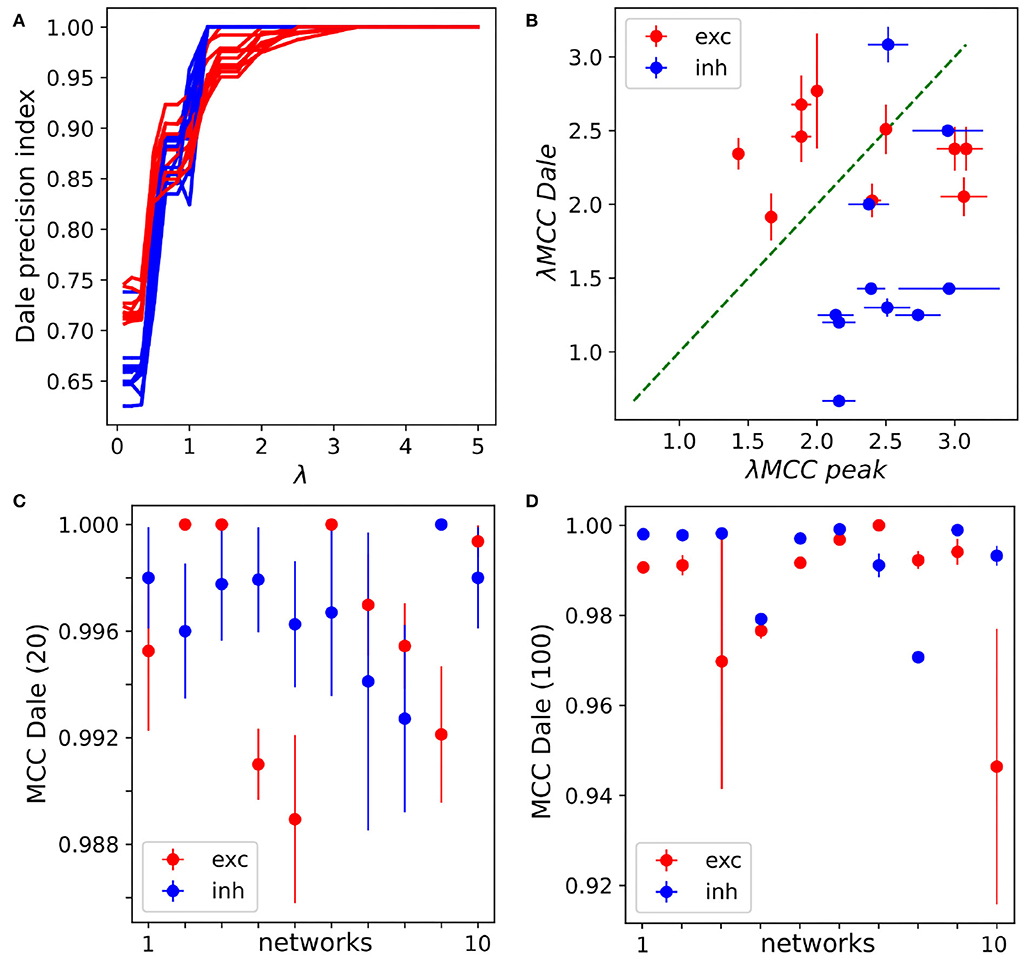
Figure 6. Network inference with the Dale precision index. (A) The Dale precision index of excitation and inhibition increases with increasing λ. In the example, a specific 20-neurons network is considered and different seeds of the instrumental noise [νstrum = (0, 1)] have been tested (m=10 repetitions). (B) The λs relative to the peak MCC are comparable to the one obtained with the Dpi plateau in 20-neurons networks. Each point is relative to the excitation/inhibition lambdas of a different network (n = 10, see Section 3.1), the mean λ and standard error of the mean are obtained using 10 different seeds for νstrum = (0, 1) [see panel (A)]. (C) For each network (1 to 10, x-axis) the max λDale between excitation and inhibition is used. The inference works very well on all 20-neurons networks, with MCC Dale(20) > 0.98. The standard error of the mean is obtained varying the seed of the instrumental noise. (D) The max λDale between excitation and inhibition is used for inference in the 100-neurons networks yielding MCC Dale(100) > 0.94) in all tested networks.
Thus, the penalization term provided a means to properly weigh sparseness toward causality and thereby select the most reliable links in the network. Interestingly, we found that MCLRL recovers the correct connectome using very small samples of activity (5–10 s). This result is of interest as neural signals are not stationary. In addition, the method may allow to track the changes in network connectivity across time. We have demonstrated that MCLRL is quite robust to noise, indeed it achieves to reconstruct with high fidelity the connectome, in 20 neurons networks, even when the fraction of genuinely detected signals is less than 5% of the total events. Interestingly, when scaling the network size to 100 neurons we found that MCLRL still worked well on networks with different topologies. Finally, we designed a simple data-driven procedure to determine the penalization term λ. We introduced Dale's precision index (Dpi) which quantified how well the MCLRL inference properly detects the excitatory and inhibitory nature of the connections (Dale's principle). Since, Dpi increases with increasing λ and reaches a plateau (=1) we empirically selected the optimal λ based on the first point of the plateau. Our results, on 20-neurons and 100-networks, showed that this simple criterion allowed us to ascertain a λ that was close to the optimal one obtained by the maximization of MCC. Importantly, the latter procedure yielded simple and effective criteria to select an appropriate λ when working with experimental data.
Although we have characterized many aspects of MCLRL, several others can be further explored. We found that a 5–10 s time window was enough to obtain good performances. Evidently, when the time window is shortened too much (e.g., 1 s) the MCC curves will decrease. Interestingly, one could investigate the interplay between noise level and recording time windows. Preliminary results (not shown) indicate that larger time windows can widen the MCC parabola, thus increasing the region of “optimal” λ values. In this work, all neurons were endowed with the same parameters (i.e., “cortical excitatory,” Section 2.2), consequently they also fired almost at the same rate. However, in more realistic networks there is a larger heterogeneity in the firing properties of the neurons (e.g., the inhibitory fast spiking inter-neurons fire at a much higher rate than excitatory neurons), which could be worth investigating.
Concerning the realism of the simulation framework, we foresee an ambitious application of the MCLRL algorithm to the study of realistic in silico neural networks (e.g., cerebellum [26], striatum [27], hippocampus [28], and the thalamo-cortical circuit [29]). This research line will presumably require a complete rewriting of the current code. Indeed, the MCLRL algorithm has been implemented based on the sklearn library for machine learning in Python [30], which however is not well suited to address large scale problems. Instead, the PMLS library1 has been optimized for these types of problems. PMLS manages to solve a 100 million dimensional sparse problem that corresponds to inferring the connectivity of networks made by 10,000 neurons.
In addition it would be interesting to apply the proposed MCLRL algorithm to experimental data. In this case, we could test the full pipeline of analysis, using the refined Perona-Malik algorithm (see Supplementary material) to preprocess the data (gathering spikes and synaptic events) and run the MCLRL algorithm with the penalization term λ optimized using the Dale's principle reported before. To this regard we have already demonstrated the efficacy of our refined Perona-Malik algorithm applied to calcium signals [11].
We also foresee significant applications of the proposed MCLRL algorithm to the study of neural diseases. Nowadays, there is an increasing interest in studying the impact of neural diseases on brain function through computational models of the brain. In Dyhrfjeld-Johnsen et al. [31], the consequences of cell loss and axonal sprouting, that occur in temporal lobe epilepsy, were studied in a computational model of the dentate gyrus. As the simulated disease progressed, the authors found that the network properties shifted from a small world topology to a more regular one. It would be interesting to parallel such computational studies running the proposed MCLRL on experimental data to verify the model predictions. A recent review [32] covered the implication of graph changes in Parkinson's and Alzheimer's diseases, which are both characterized by a progressive weakening of network connectivity. Other works addressed the study of bipolar/unipolar depression [33] and cognitive aging [34]. Importantly, most of the studies correlating brain diseases to changes in the properties of a graph are based on macro-scale recordings (e.g., EEG). The MCLRL algorithm however takes explicitly into account spiking and synaptic activities, which prevents a straightforward application of the proposed methodology to macro-scale recordings. However, there have been some efforts toward defining causal models relating, for example, the EEG responses to trans-magnetic-stimulation across brain states [35], it would be interesting to investigate whether the same ideas of the MCLRL can be adapted to such modeling approaches.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation. The latest version of the simulation and analysis code is available at: https://github.com/thierrynieus/Lasso.
TN performed simulations and analysis and wrote the first draft of the manuscript. DB contributed to an early version of the study. TN, SD, GN, and GA finalized the manuscript. GN and GA supervised the project. All authors contributed to the article and approved the submitted version.
Funded in part by a grant from the Italian Ministry of Foreign Affairs and International Cooperation, Multi-Scale Brain Function (MSBFIINE) India-Italy Network of Excellence (DST-MAE). TN was supported by fundings from the BigMath Project (H2020-MSCA-ITN-2018, grant agreement no. 812912), the Human Brain Project (grant agreement no. 945539), and from Core Facility INDACO-Università degli Studi di Milano.
Computational resources provided by the Core Facility INDACO, which is a project of High Performance Computing at the Università degli Studi di Milano http://www.unimi.it.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fams.2022.1023310/full#supplementary-material
1. Bullmore E, Sporns O. Complex brain networks: graph theoretical analysis of structural and functional systems. Nat Rev Neurosci. (2009) 10:186–98. doi: 10.1038/nrn2575
2. Feldt S, Bonifazi P, Cossart R. Dissecting functional connectivity of neuronal microcircuits: experimental and theoretical insights. Trends Neurosci. (2011) 34:225–36. doi: 10.1016/j.tins.2011.02.007
3. Garofalo M, Nieus T, Massobrio P, Martinoia S. Evaluation of the performance of information theory-based methods and cross-correlation to estimate the functional connectivity in cortical networks. PLoS ONE. (2009) 4:e6482. doi: 10.1371/journal.pone.0006482
4. Kobayashi R, Kurita S, Kurth A, Kitano K, Mizuseki K, Diesmann M, et al. Reconstructing neuronal circuitry from parallel spike trains. Nat Commun. (2019) 10:4468. doi: 10.1038/s41467-019-12225-2
5. Perin R, Markram H. A computer-assisted multi-electrode patch-clamp system. J Vis Exp. (2013) 18:50630. doi: 10.3791/50630
6. Gong Y, Huang C, Li JZ, Grewe BF, Zhang Y, Eismann S, et al. High-speed recording of neural spikes in awake mice and flies with a fluorescent voltage sensor. Science. (2015) 350:1361–6. doi: 10.1126/science.aab0810
7. Jin L, Han Z, Platisa J, Wooltorton JRA, Cohen LB, Pieribone VA. Single action potentials and subthreshold electrical events imaged in neurons with a fluorescent protein voltage probe. Neuron. (2012) 75:779–85. doi: 10.1016/j.neuron.2012.06.040
8. Williams SR, Stuart GJ. Dependence of EPSP efficacy on synapse location in neocortical pyramidal neurons. Science. (2002) 295:1907–10. doi: 10.1126/science.1067903
9. Platisa J, Pieribone VA. Genetically encoded fluorescent voltage indicators: are we there yet? Curr Opin Neurobiol. (2018) 50:146–53. doi: 10.1016/j.conb.2018.02.006
10. Brondi M, Bruzzone M, Lodovichi C, dal Maschio M. Optogenetic methods to investigate brain alterations in preclinical models. Cells. (2022) 11:1848. doi: 10.3390/cells11111848
11. Palazzolo G, Moroni M, Soloperto A, Aletti G, Naldi G, Vassalli M, et al. Fast wide-volume functional imaging of engineered in vitro brain tissues. Sci Rep. (2017) 7:8499. doi: 10.1038/s41598-017-08979-8
12. Aletti G, Moroni M, Naldi G. A new nonlocal nonlinear diffusion equation for data analysis. Acta Appl Math. (2020) 168:109–35. doi: 10.1007/s10440-019-00281-1
13. Aletti G, Benfenati A, Naldi G. A new nonlocal nonlinear diffusion equation: the one-dimensionale case. Bull Aust Math Soc. (2022) 106:333–9. doi: 10.1017/S0004972722000363
14. Aletti G, Lonardoni D, Naldi G, Nieus T. From dynamics to links: a sparse reconstruction of the topology of a neural network. Commun Appl Ind Math. (2019) 10:2–11. doi: 10.2478/caim-2019-0002
15. Pillow JW, Shlens J, Paninski L, Sher A, Litke AM, Chichilnisky EJ, et al. Spatio-temporal correlations and visual signalling in a complete neuronal population. Nature. (2008) 454:995–99. doi: 10.1038/nature07140
16. Stevenson IH, Rebesco JM, Miller LE, Körding KP. Inferring functional connections between neurons. Curr Opin Neurobiol. (2008) 18:582–8. doi: 10.1016/j.conb.2008.11.005
17. Vidne M, Ahmadian Y, Shlens J, Pillow JW, Kulkarni J, Litke AM, et al. Modeling the impact of common noise inputs on the network activity of retinal ganglion cells. J Comput Neurosci. (2012) 33:97–121. doi: 10.1007/s10827-011-0376-2
18. Marc-Oliver G, Markus D. NEST (NEural simulation tool). Scholarpedia. (2007). 2:1430. doi: 10.4249/scholarpedia.1430
19. Hill S, Tononi G. Modeling sleep and wakefulness in the thalamocortical system. J Neurophysiol. (2005) 93:1671–98. doi: 10.1152/jn.00915.2004
20. Silver RA. Estimation of nonuniform quantal parameters with multiple-probability fluctuation analysis: theory, application and limitations. J Neurosci Methods. (2003) 130:127–41. doi: 10.1016/j.jneumeth.2003.09.030
21. Menard SW. Applied Logistic Regression Analysis. Quantitative Applications in the Social Sciences. Thousand Oaks, CA: SAGE University Paper (2002).
22. King G, Zeng L. Logistic regression in rare events data. Polit Anal. (2001) 9:137–63. doi: 10.1093/oxfordjournals.pan.a004868
23. Tharwat A. Classification assessment methods. Appl Comput Inform. (2021) 17:168–92. doi: 10.1016/j.aci.2018.08.003
24. Lonardoni D, Amin H, Di Marco S, Maccione A, Berdondini L, Nieus T. Recurrently connected and localized neuronal communities initiate coordinated spontaneous activity in neuronal networks. PLoS Comput Biol. (2017) 13:e1005672. doi: 10.1371/journal.pcbi.1005672
25. Eccles JC. From electrical to chemical transmission in the central nervous system: the closing address of the sir henry dale centennial symposium Cambridge, 19 September 1975. Notes Rec R Soc Lond. (1976) 30:219–30. doi: 10.1098/rsnr.1976.0015
26. Solinas S, Nieus T, D'Angelo E. A realistic large-scale model of the cerebellum granular layer predicts circuit spatio-temporal filtering properties. Front Cell Neurosci. (2010) 4:12. doi: 10.3389/fncel.2010.00012
27. Hjorth JJJ, Kozlov A, Carannante I, Frost Nylén J, Lindroos R, Johansson Y, et al. The microcircuits of striatum in silico. Proc Natl Acad Sci USA. (2020) 117:9554–65. doi: 10.1073/pnas.2000671117
28. Romani A, Schürmann F, Markram H, Migliore M. Reconstruction of the hippocampus. In:Giugliano M, Negrello M, Linaro D, , editors. Computational Modelling of the Brain, Vol. 1359. Cham: Springer International Publishing (2022). p. 261–83.
29. Iavarone E, Yi J, Shi Y, Zandt BJ, O'Reilly C, Van Geit W, et al. Experimentally-constrained biophysical models of tonic and burst firing modes in thalamocortical neurons. PLoS Comput Biol. (2019) 15:e1006753. doi: 10.1371/journal.pcbi.1006753
30. Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, et al. Scikit-learn: machine learning in python. J Mach Learn Res. (2011) 12:2825–30. Available online at: https://www.jmlr.org/papers/v12/pedregosa11a.html
31. Dyhrfjeld-Johnsen J, Santhakumar V, Morgan RJ, Huerta R, Tsimring L, Soltesz I. Topological determinants of epileptogenesis in large-scale structural and functional models of the dentate Gyrus derived from experimental data. J Neurophysiol. (2007) 97:1566–87. doi: 10.1152/jn.00950.2006
32. Miraglia F, Vecchio F, Pappalettera C, Nucci L, Cotelli M, Judica E, et al. Brain connectivity and graph theory analysis in Alzheimer's and Parkinson's disease: the contribution of electrophysiological techniques. Brain Sci. (2022) 12:402. doi: 10.3390/brainsci12030402
33. Cha J, Spielberg JM, Hu B, Altinay M, Anand A. Differences in network properties of the structural connectome in bipolar and unipolar depression. Psychiatry Res Neuroimaging. (2022) 321:111442. doi: 10.1016/j.pscychresns.2022.111442
34. Nordin K, Gorbach T, Pedersen R, Panes Lundmark V, Johansson J, Andersson M, et al. DYNAMIC: a prospective longitudinal study of dopamine and brain connectomes: a new window into cognitive aging. J Neurosci Res. (2022) 100:1296–320. doi: 10.1002/jnr.25039
Keywords: network inference, effective connectivity, multi-class logistic regression, lasso penalization, genetic encoded voltage indicator, patch clamp recording, in silico simulation
Citation: Nieus T, Borgonovo D, Diwakar S, Aletti G and Naldi G (2022) A multi-class logistic regression algorithm to reliably infer network connectivity from cell membrane potentials. Front. Appl. Math. Stat. 8:1023310. doi: 10.3389/fams.2022.1023310
Received: 19 August 2022; Accepted: 12 October 2022;
Published: 02 November 2022.
Edited by:
Sara Sommariva, Università degli Studi di Genova, ItalyReviewed by:
Roberto Guidotti, University of Studies G. d'Annunzio Chieti and Pescara, ItalyCopyright © 2022 Nieus, Borgonovo, Diwakar, Aletti and Naldi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Thierry Nieus, dGhpZXJyeS5uaWV1c0B1bmltaS5pdA==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.