- 1Institute of Mathematical Sciences, Claremont Graduate University, Claremont, CA, United States
- 2Johann Radon Institute, Linz, Austria
- 3Department of Mathematics, Vanderbilt University, Nashville, TN, United States
We consider the question of 30-min prediction of blood glucose levels measured by continuous glucose monitoring devices, using clinical data. While most studies of this nature deal with one patient at a time, we take a certain percentage of patients in the data set as training data, and test on the remainder of the patients; i.e., the machine need not re-calibrate on the new patients in the data set. We demonstrate how deep learning can outperform shallow networks in this example. One novelty is to demonstrate how a parsimonious deep representation can be constructed using domain knowledge.
1. Introduction
Deep Neural Networks, especially of the convolutional type (DCNNs), have started a revolution in the field of artificial intelligence and machine learning, triggering a large number of commercial ventures and practical applications. There is therefore a great deal of theoretical investigations about when and why deep (hierarchical) networks perform so well compared to shallow ones. For example, Montufar et al. [1] showed that the number of linear regions that can be synthesized by a deep network with ReLU nonlinearities is much larger than by a shallow network. Examples of specific functions that cannot be represented efficiently by shallow networks have been given very recently by Telgarsky [2] and Safran and Shamir [3].
It is argued in Mhaskar and Poggio [4] that from a function approximation point of view, deep networks are able to overcome the so-called curse of dimensionality if the target function is hierarchical in nature; e.g., a target function of the form
where each function has a bounded gradient, can be approximated by a deep network comprising n units, organized as a binary tree, up to an accuracy . In contrast, a shallow network that cannot take into account this hierarchical structure can yield an accuracy of at most . In theory, if samples of the functions h, h1,2, … are known, one can construct the networks explicitly, without using any traditional learning algorithms such as back propagation.
One way to think of the function in Equation (1) is to think of the inner functions as the features of the data, obtained in a hierarchical manner. While classical machine learning with shallow neural networks requires that the relevant features of the raw data should be selected by using domain knowledge before the learning can start, deep learning algorithms appear to select the right features automatically. However, it is typically not clear how to interpret these features. Indeed, from a mathematical point of view, it is easy to show that a structure such as Equation (1) is not unique, so that the hierarchical features cannot be defined uniquely, except perhaps in some very special examples.
In this paper, we examine how a deep network can be constructed in a parsimonious manner if we do allow domain knowledge to suggest the compositional structure of the target function as well as the values of the constituent functions. We study the problem of predicting, based on the past few readings of a continuous glucose monitoring (CGM) device, both the blood glucose (BG) level and the rate at which it would be changing 30 min after the last reading. From the point of view of diabetes management, a reliable solution to this problem is of great importance. If a patient has some warning that that his/her BG will rise or drop in the next half hour, the patient can take certain precautionary measures to prevent this (e.g., administer an insulin injection or take an extra snack) [5, 6].
Our approach is to first construct three networks based on whether a 5-min prediction, using ideas in Mhaskar et al. [7], indicates the trend to be in the hypoglycemic (0–70 mg/dL), euglycemic (70–180 mg/dL), or hyperglycemic (180–450 mg/dL) range. We then train a “judge” network to get a final prediction based on the outputs of these three networks. Unlike the tacit assumption in the theory in Mhaskar and Poggio [4], the readings and the outputs of the three constituent networks are not dense in a Euclidean cube. Therefore, we use diffusion geometry ideas in Ehler et al. [8] to train the networks in a manner analogous to manifold learning.
From the point of view of BG prediction, a novelty of our paper is the following. Most of the literature on this subject which we are familiar with does the prediction patient-by-patient; for example, by taking 30% of the data for each patient to make the prediction for that patient. In contrast, we consider the entire data for 30% of the patients as training data, and predict the BG level for the remaining 70% of patients in the data set. Thus, our algorithm transfers the knowledge learned from one data set to another, although it does require the knowledge of both the data sets to work with. From this perspective, the algorithm has similarity with the meta-learning approach by Naumova et al. [5], but in contrast to the latter, it does not require a meta-features selection.
We will explain the problem and the evaluation criterion in Section 2. Some prior work on this problem is reviewed briefly in Section 3. The methodology and algorithm used in this paper are described in Section 4. The results are discussed in Section 5. The mathematical background behind the methods described in Section 4 is summarized in Appendices A.1 and A.2.
2. Problem Statement and Evaluation
We use a clinical data set provided by the DirectNet Central Laboratory [9], which lists BG levels of different patients taken at 5-min intervals with the CGM device; i.e., for each patient p in the patient set P, we are given a time series {sp(tj)}, where sp(tj) denotes the BG level at time tj. Our goal is to predict for each j, the level sp(tj+m), given readings sp(tj), …, sp(tj−d+1) for appropriate values of m and d. For a 30-min prediction, m = 6, and we took d = 7 (a sampling horizon tj − tj−d+1 of 30 min has been suggested as the optimal one for BG prediction in Mhaskar et al. [7] and Hayes et al. [10].
In this problem, numerical accuracy is not the central objective. To quantify the clinical accuracy of the considered predictors, we use the Prediction Error-Grid Analysis (PRED-EGA) [11], which has been designed especially for glucose predictors and which, together with its predecessors and variations, is by now a standard metric in the blood glucose prediction problem (see for example, [5, 12–14]). This assessment methodology records reference BG estimates paired with the BG estimates predicted for the same moments, as well as reference BG directions and rates of change paired with the corresponding estimates predicted for the same moments. As a result, the PRED-EGA reports the numbers (in percent) of Accurate (Acc.), Benign (Benign) and Erroneous (Error) predictions in the hypoglycemic, euglycemic and hyperglycemic ranges separately. This stratification is of great importance because consequences caused by a prediction error in the hypoglycemic range are very different from ones in the euglycemic or the hyperglycemic range.
3. Prior Work
Given the importance of the problem, many researchers have worked on it in several directions. Relevant to our work is the work using a linear predictor and work using supervised learning.
The linear predictor method estimates based on the previous d readings, and then predicts
Perhaps the most classical of these is the work [15] by Savitzky and Golay, that proposes an approximation of by the derivative of a polynomial of least square fit to the data (tk, sp(tk)), k = j, …, j − d + 1. The degree n of the polynomial acts as a regularization parameter. However, in addition to the intrinsic ill–conditioning of numerical differentiation, the solution of the least square problem as posed above involves a system of linear equations with the Hilbert matrix of order n, which is notoriously ill–conditioned. Therefore, it is proposed in Lu et al. [16] to use Legendre polynomials rather than the monomials as the basis for the space of polynomials of degree n. A procedure to choose n is given in Lu et al. [16], together with error bounds in terms of n and the estimates on the noise level in the data, which are optimal up to a constant factor for the method in the sense of the oracle inequality. A substantial improvement on this method was proposed in Mhaskar et al. [7], which we summarize in Appendix A.1. As far as we are aware, this is the state of the art in this approach in short term blood glucose prediction using linear prediction technology.
There exist several BG prediction algorithms in the literature that use a supervised learning approach. These can be divided into three main groups.
The first group of methods employ kernel-based regularization techniques to achieve prediction (for example, [5, 17] and references therein), where Tikhonov regularization is used to find the best least square fit to the data (tk, sp(tk)), k = j, …, j − d + 1, assuming the minimizer belongs to a reproducing kernel Hilbert space (RKHS). Of course, these methods are quite sensitive to the choice of kernel and regularization parameters. Therefore, the authors in Naumova et al. [5, 17] develop methods to choose both the kernel and regularization parameter adaptively, or through meta-learning (“learning to learn”) approaches.
The second group consists of artificial neural network models (such as [13, 18]). In Pappada et al. [13], for example, a feed-forward neural network is designed with eleven neurons in the input layer (corresponding to variables such as CGM data, the rate of change of glucose levels, meal intake and insulin dosage), and nine neurons with hyperbolic tangent transfer function in the hidden layer. The network was trained with the use of data from 17 patients and tested on data from 10 other patients for a 75-min prediction, and evaluated using the classical Clarke Error-Grid Analysis (EGA) [19], which is a predecessor of the PRED-EGA assessment metric. Classical EGA differs from PRED-EGA in the sense that it only compares absolute BG concentrations with reference BG values (and not rates of change in BG concentrations as well). Although good results are achieved in the EGA grid in this paper, a limitation of the method is the large amount of additional input information necessary to design the model, as described above.
The third group consists of methods that utilize time-series techniques such as autoregressive (AR) models (for example, [14, 20]). In Reifman et al. [14], a tenth-order AR model is developed, where the AR coefficients are determined through a regularized least square method. The model is trained patient-by-patient, typically using the first 30% of the patient's BG measurements, for a 30 or 60-min prediction. The method is tested on a time series containing glucose values measured every minute, and evaluation is again done through the classical EGA grid. The authors in Sparacino et al. [20] develop a first-order AR model, patient-by-patient, with time-varying AR coefficients determined through weighted least squares. Their method is tested on a time series containing glucose values measured every 3 min, and quantified using statistical metrics such as measuring the mean square of the errors. As noted in Naumova et al. [5], these methods seem to be sensitive to gaps in the input data.
4. Methodology in the Current Paper
Our proposed method represents semi-supervised learning that follows an entirely different approach from those described above. It is not a classical statistics/optimization based approach; instead, it is based on function approximation on data defined manifolds, using diffusion polynomials. In this section, we describe our deep learning method, which consists of two layers, in details.
Given the time series {sp(tj)} of BG levels at time tj for each patient p in the patient set P, where tj − tj−1 = 5 min, we start by formatting the data into the form {(xj, yj)}, where
We will use the notation
We also construct the diffusion matrix from . This is done by normalizing the rows of the weight matrix in Equation (A6), following the approach in Lafon [21, pp. 33–34].
Having defined the input data and the corresponding diffusion matrix, our method proceeds as follows.
4.1. First Layer: Three Networks in Different Clusters
To start our first layer training, we form the training patient set TP by randomly selecting (according to a uniform probability distribution) M% of the patients in P. The training data are now defined to be all the data (xj, yj) corresponding to the patients in TP. We will use the notations
and
Next, we make a short term prediction Lxj(tj+1) of the BG level sp(tj+1) after 5 min, for all the given measurements , by applying the linear predictor method (summarized in Section 3 and Appendix A.1). Based on these 5-min predictions, we divide the measurements in into three clusters and ,
with
The motivation of this step is to gather more information concerning the training set to ensure more accurate predictions in each BG range—as noted previously, consequences of prediction error in the separate BG ranges are very different.
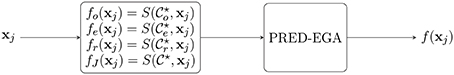
In the sequel, let S(Γ, xj) denote the result of the method of Appendix A.2 [that is, S(Γ, xj) is defined by Equation (A5)], used with training data Γ and evaluated at a point . After obtaining the three clusters and , we compute the three predictors
as well as the “judge” predictor, based on the entire training set ,
using the summability method of Appendix A.2 [specifically, Equation (A4)]. We remark that, as discussed in Mhaskar [22], our approximations in Equation (A5) can be computed as classical radial basis function networks, with exactly as many neurons as the number of eigenfunctions used in Equation (A3). (As mentioned in Appendix A.2, this number is determined to ensure that the system Equation (A4) is still well conditioned.)
The motivation of this step is to decide which one of the three predictions (fo, fe or fr) is the best prediction for each datum . Since we do not know in advance in which blood glucose range a particular datum will result, we need to use all of the training data for the judge predictor fJ to choose the best prediction.
4.2. Second Layer (Judge): Final Output
In the last training layer, a final output is produced based on which fℓ, ℓ ∈ {o, e, r} gives the best placement in the PRED-EGA grid, using fJ as the reference value. The PRED-EGA grid is constructed by using comparisons of fo (resp., fe and fr) with the reference value fJ—specifically, it involves comparing
as well as the rates of change
for all . Based on these comparisons, PRED-EGA classifies fo(xj) (resp., fe(xj) and fr(xj)) as being Accurate, Benign or Erroneous. As our final output f(xj) of the target function at , we choose the one among fℓ(xj), ℓ ∈ {o, e, r} with the best classification, and if there is more than one achieving the best grid placement, we output the one among fℓ(xj), ℓ ∈ {o, e, r} that has value closest to fJ(xj). For the first and last xj for each patient p ∈ P (for which the rate of change Equation (2) cannot be computed), we use the one among fℓ(xj), ℓ ∈ {o, e, r} that has value closest to fJ(xj) as the final output.
4.3. Evaluation
Lastly, to evaluate the performance of the final output we use the actual reference values yj to place f(xj) in the PRED-EGA grid.
We repeat the process described in Sections 4.1–4.3 for a fixed number of trials, after which we report the average of the PRED-EGA grid placements, over all and over all trials, as the final evaluation.
A summary of the method is given in Algorithm 1. A flowchart diagram of the algorithm is shown in Figure 1.
4.4. Remarks
1. An optional smoothing step may be applied before the first training layer step in Section 4.1 to remove any large spikes in the given time series {sp(tj)}, p ∈ P, that may be caused by patient movement, for example. Following ideas in Sparacino et al. [20], we may apply flat low-pass filtering (for example, a first-order Butterworth filter). In this case, the evaluation of the final output in Section 4.3 is done by using the original, unsmoothed measurements as reference values.
2. We remark that, as explained in Section 1, the training set may also be implemented in a different way: instead of drawing M% of the patients in P and using their entire data sets for training, we could construct the training set for each patient p ∈ P separately by drawing M% of a given patient's data for training, and then construct the networks fℓ, ℓ ∈ {o, e, r} and fJ for each patient separately. This is a different problem, studied often in the literature (see for example, [14, 20, 23]). We do not pursue this approach in the current paper.
5. Results and Discussion
As mentioned in Section 2, we apply our method to data provided by the DirecNet Central Laboratory. Time series for 25 patients are considered. This specific data set was designed to study the performance of CGM devices in children with Type I diabetes; as such, all of the patients are less than 18 years old. Each time series {sp(tj)} contains more than 160 BG measurements, taken at 5-min intervals. Our method is a general purpose algorithm, where these details do not play any significant role, except in affecting the outcome of the experiments.
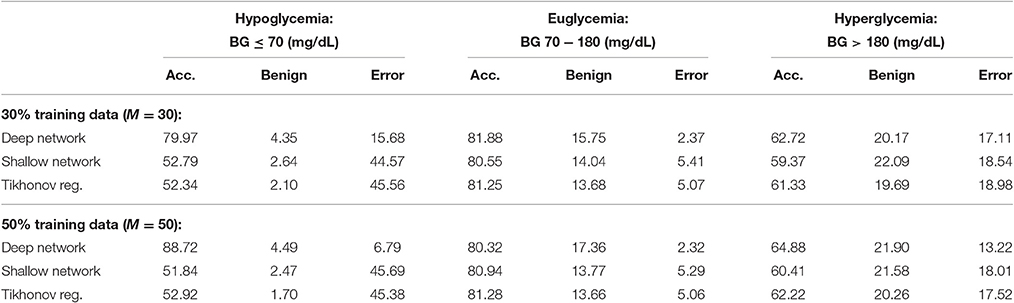
We provide results obtained by implementing our method in Matlab, as described in Algorithm 1. For our implementation, we employ a sampling horizon tj − tj−d+1 of 30 min (d = 7), a prediction horizon tj+m − tj of 30 min (m = 6), and a total of 100 trials (T = 100). We provide results for both 30% training data (M = 30) and 50% training data (M = 50) (which is comparable to approaches followed in for example [13, 14]). After testing our method on all 25 patients, the average PRED-EGA scores (in percent) are displayed in Table 1. For 30% training data, the percentage of accurate predictions and predictions with benign consequences is 84.32% in the hypoglycemic range, 97.63% in the euglycemic range, and 82.89% in the hyperglycemic range, while for 50% training data, we have 93.21% in the hypoglycemic range, 97.68% in the euglycemic range, and 86.78% in the hyperglycemic range.
For comparison, Table 1 also displays the PRED-EGA scores obtained when implementing a shallow (ie, one layer) feed-forward network with 20 neurons using Matlab's Neural Network Toolbox, using the same parameters d, m, M and T as in our implementation, with Matlab's default hyperbolic tangent sigmoid activation function. The motivation for using 20 neurons is the following. As mentioned in Section 4.1, each layer in our deep network can be viewed as a classical neural network with exactly as many neurons as the number of eigenfunctions used in Equations (A9) and (A11). This number is determined to ensure that the system in Equation (A10) is well conditioned; in our experiments, it turned out to be at most 5. Therefore, to compare our two-layer deep network (where the first layer consists of three separate networks) with a classical shallow neural network, we use 20 neurons. It is clear that our deep network performs substantially better than the shallow network; in all three BG ranges, our method produces a lower percentage of erroneous predictions.
For a further comparison, we also display in Table 1 the PRED-EGA scores obtained when training through supervised learning using a standard Tikhonov regularization to find the best least square fit to the data; specifically, we implemented the method described in Poggio and Smale [24, pp. 3–4] using a Gaussian kernel with σ = 100 and regularization constant γ = 0.0001. Again, our method produces superior results, especially in the hypoglycemic range.
Figures 2, 3 display boxplots for the percentage accurate predictions in each BG range for each method over the 100 trials.
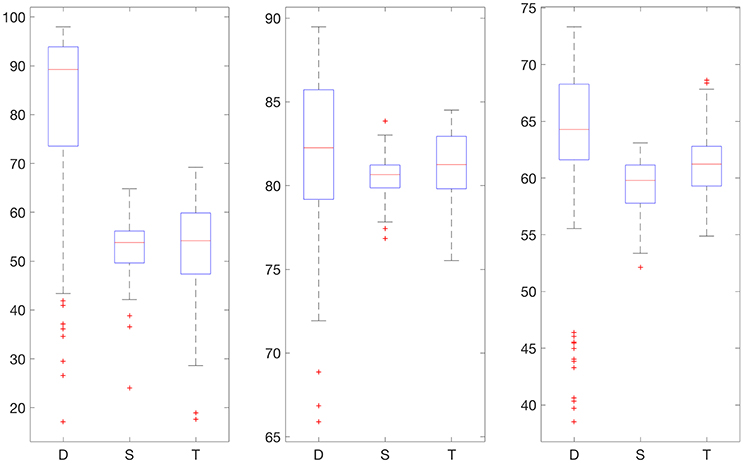
Figure 2. Boxplot for the 100 experiments conducted with no smoothing and 30% training data for each prediction method (D, deep network; S, shallow network; T, Tikhonov regularization). The three graphs show the percentage accurate predictions in the hypoglycemic range (left), euglycemic range (middle), and hyperglycemic range (right).
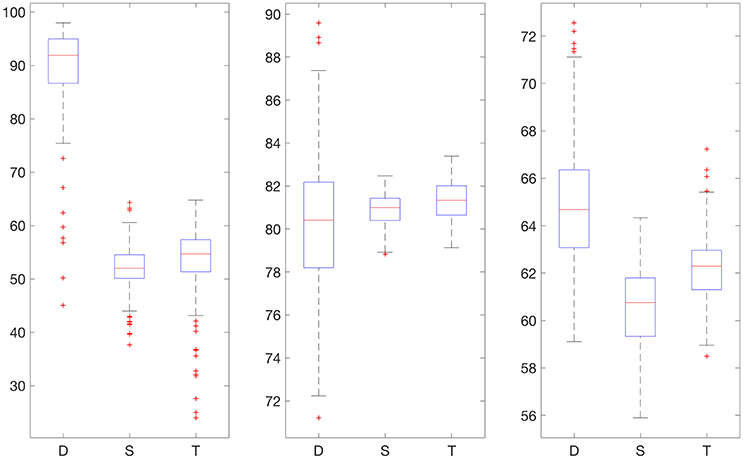
Figure 3. Boxplot for the 100 experiments conducted with no smoothing and 50% training data for each prediction method (D, deep network; S, shallow network; T, Tikhonov regularization). The three graphs show the percentage accurate predictions in the hypoglycemic range (left), euglycemic range (middle) and hyperglycemic range (right).
We also provide results obtained when applying smoothing through flat low-pass filtering to the given time series, as explained in Section 4.4. For our implementations, we used a first-order Butterworth filter with cutoff frequency 0.8, with the same input parameters as before. The results are given in Table 2. For 30% training data, the percentage of accurate predictions and predictions with benign consequences is 88.92% in the hypoglycemic range, 97.64% in the euglycemic range, and 77.58% in the hyperglycemic range, while for 50% training data, we have 96.43% in the hypoglycemic range, 97.96% in the euglycemic range, and 85.29% in the hyperglycemic range. Figures 4, 5 display boxplots for the percentage accurate predictions in each BG range for each method over the 100 trials.
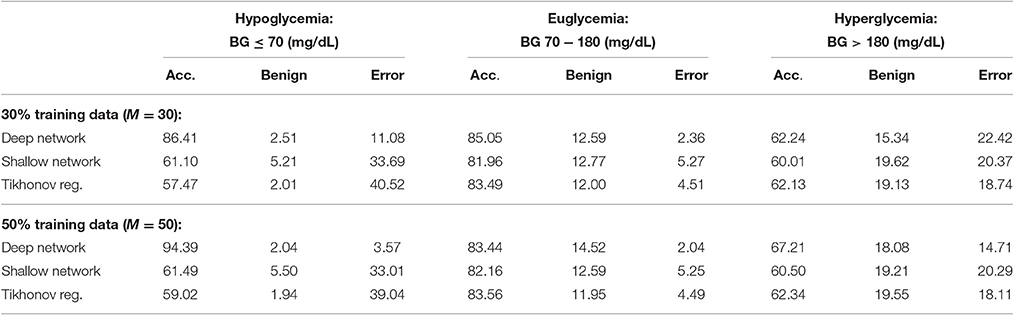
Table 2. Average PRED-EGA scores (in percent): M% of patients used for training with flat low-pass filtering.
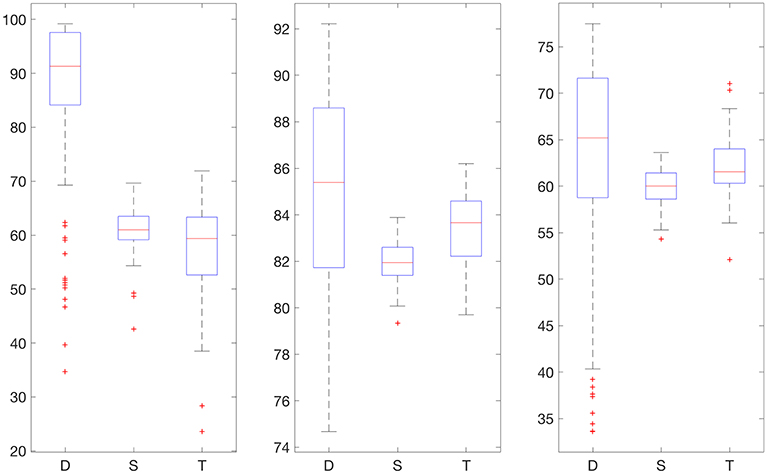
Figure 4. Boxplot for the 100 experiments conducted with flat low-pass filtering and 30% training data for each prediction method (D, deep network; S, shallow network; T, Tikhonov regularization). The three graphs show the percentage accurate predictions in the hypoglycemic range (left), euglycemic range (middle) and hyperglycemic range (right).
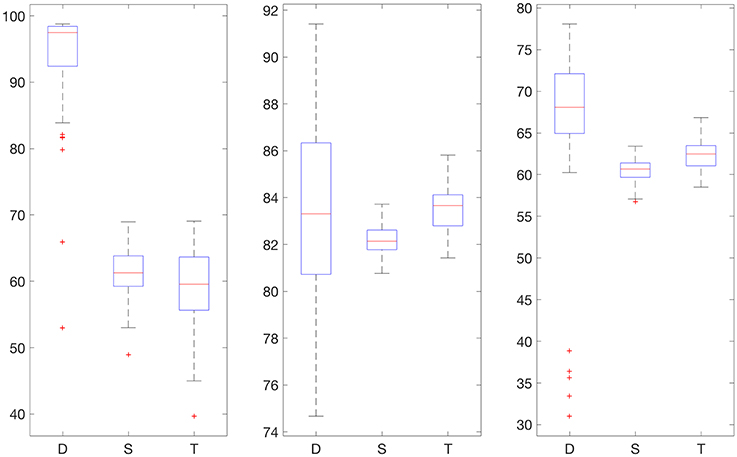
Figure 5. Boxplot for the 100 experiments conducted with flat low-pass filtering and 50% training data for each prediction method (D, deep network; S, shallow network; T, Tikhonov regularization). The three graphs show the percentage accurate predictions in the hypoglycemic range (left), euglycemic range (middle) and hyperglycemic range (right).
In all our experiments the percentage of erroneous predictions is substantially smaller with deep networks than the other two methods we have tried, with the only exception of hyperglycemic range with 30% training data and flat low pass filtering. Our method's performance also improves when the amount of the training data increases from 30% to 50%, because the percentage of erroneous predictions decreases in all of the BG ranges in all experiments.
Moreover, from this viewpoint, our deep learning method outperforms the considered competitors, except in the hyperglycemic BG range in Table 2. A possible explanation for this is that the DirecNet study was done on children (less than 18 years of age) with type 1 diabetes, for a period of roughly 26 h. It appears that children usually have prolonged hypoglycemic periods as well as profound postprandial hyperglycemia (high blood sugar “spikes” after meals)—according to Boland et al. [25], more than 70% of children display prolonged hypoglycemia while more than 90% of children display significant postprandial hyperglycemia. In particular, many of the patients in the data set only exhibit very limited hyperglycemic BG spikes—for the patient labeled 43, for example, there exists a total of 194 BG measurements, of which only 5 measurements are greater than 180 mg/dL. This anomaly might have affected the performance of our algorithm in the hyperglycemic range. Obviously, it performs remarkably better than the other techniques we have tried, including fully supervised training, while ours is only semi-supervised.
6. Conclusion
The prediction of blood glucose levels based on continuous glucose monitoring system data 30 min ahead is a very important problem with many consequences for the health care industry. In this paper, we suggest a deep learning paradigm based on a solid mathematical theory as well as domain knowledge to solve this problem accurately as assessed by the PRED-EGA grid, developed specifically for this purpose. It is demonstrated in Mhaskar and Poggio [4] that deep networks perform substantially better than shallow networks in terms of expressiveness for function approximation when the target function has a compositional structure. Thus, the blessing of compositionality cures the curse of dimensionality. However, the compositional structure is not unique, and it is open problem to decide whether a given target function has a certain compositional structure. In this paper, we have demonstrated an example where domain knowledge can be used to build an appropriate compositional structure, leading to a parsimonious deep learning design.
Ethics Statement
The clinical data set used in this publication is publicly available online [9]. The data were collected during the study that was carried out in accordance with the recommendations of DirecNet, Jaeb Center for Health Research, with written informed consent from all 878 subjects. All subjects gave written informed consent in accordance with the Declaration of Helsinki. The protocol was approved by DirecNet, Jaeb Center for Health Research.
Author Contributions
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The research of HM is supported in part by ARO Grant W911NF-15-1-0385. The research of SP is partially supported by Austrian Science Fund (FWF) Grant I 1669-N26.
References
1. Montufar GF, Pascanu R, Cho K, Bengio Y. On the number of linear regions of deep neural networks. In: Advances in Neural Information Processing Systems. Red Hook, NY: Curran Associates, Inc. (2014) p. 2924–32.
2. Telgarsky M. Representation benefits of deep feedforward networks. arXiv preprint (2015) arXiv:150908101
3. Safran I, Shamir O. Depth separation in relu networks for approximating smooth non-linear functions. arXiv preprint (2016) arXiv:161009887
4. Mhaskar HN, Poggio T. Deep vs. shallow networks: an approximation theory perspective. Anal Appl. (2016) 14:829–48. doi: 10.1142/S0219530516400042
5. Naumova V, Pereverzyev SV, Sivananthan S. A meta-learning approach to the regularized learning - Case study: blood glucose prediction. Neural Netw. (2012) 33:181–93. doi: 10.1016/j.neunet.2012.05.004
6. Snetselaar L. Nutrition Counseling Skills for the Nutrition Care Process. Sudbury, MA: Jones & Bartlett Learning (2009).
7. Mhaskar HN, Naumova V, Pereverzyev SV. Filtered Legendre expansion method for numerical differentiation at the boundary point with application to blood glucose predictions. Appl Math Comput. (2013) 224:835–47. doi: 10.1016/j.amc.2013.09.015
8. Ehler M, Filbir F, Mhaskar HN. Locally learning biomedical data using diffusion frames. J Comput Biol. (2012) 19:1251–64. doi: 10.1089/cmb.2012.0187
9. DirecNet Central Laboratory. (2005). Available online at: http://direcnet.jaeb.org/Studies.aspx
10. Hayes AC, Mastrototaro JJ, Moberg SB, Mueller JC Jr, Clark HB, Tolle MCV, et al. Algorithm sensor augmented bolus estimator for semi-closed loop infusion system. Google Patents (2009). US Patent 7,547,281.
11. Sivananthan S, Naumova V, Man CD, Facchinetti A, Renard E, Cobelli C, et al. Assessment of blood glucose predictors: the prediction-error grid analysis. Diabet Technol Ther. (2011) 13:787–96. doi: 10.1089/dia.2011.0033
12. Naumova V, Pereverzyev SV, Sivananthan S. Adaptive parameter choice for one-sided finite difference schemes and its application in diabetes technology. J Complex. (2012) 28:524–38. doi: 10.1016/j.jco.2012.06.001
13. Pappada SM, Cameron BD, Rosman PM, Bourey RE, Papadimos TJ, Olorunto W, et al. Neural network-based real-time prediction of glucose in patients with insulin-dependent diabetes. Diabet Technol Ther. (2011) 13:135–41. doi: 10.1089/dia.2010.0104
14. Reifman J, Rajaraman S, Gribok A, Ward WK. Predictive monitoring for improved management of glucose levels. J Diabet Sci Technol. (2007) 1:478–86. doi: 10.1177/193229680700100405
15. Savitzky A, Golay MJE. Smoothing and differentiation of data by simplified least squares procedures. Anal Chem. (1964) 36:1627–39. doi: 10.1021/ac60214a047
16. Lu S, Naumova V, Pereverzev SV. Legendre polynomials as a recommended basis for numerical differentiation in the presence of stochastic white noise. J Inverse ILL Posed Prob. (2013) 21:193–216. doi: 10.1515/jip-2012-0050
17. Naumova V, Pereverzyev SV, Sivananthan S. Extrapolation in variable RKHSs with application to the blood glucose reading. Inverse Prob. (2011) 27:075010. doi: 10.1088/0266-5611/27/7/075010
18. Pappada SM, Cameron BD, Rosman PM. Development of a neural network for prediction of glucose concentration in type 1 diabetes patients. J Diabet Sci Technol. (2008) 2:792–801. doi: 10.1177/193229680800200507
19. Clarke WL, Cox D, Gonder-Frederick LA, Carter W, Pohl SL. Evaluating clinical accuracy of systems for self-monitoring of blood glucose. Diabetes Care (1987) 10:622–8. doi: 10.2337/diacare.10.5.622
20. Sparacino G, Zanderigo F, Corazza S, Maran A, Facchinetti A, Cobelli C. Glucose concentration can be predicted ahead in time from continuous glucose monitoring sensor time-series. IEEE Trans Biomed Eng. (2007) 54:931–7. doi: 10.1109/TBME.2006.889774
22. Mhaskar HN. Eignets for function approximation on manifolds. Appl Comput Harmon Anal. (2010) 29:63–87. doi: 10.1016/j.acha.2009.08.006
23. Eren-Oruklu M, Cinar A, Quinn L, Smith D. Estimation of future glucose concentrations with subject-specific recursive linear models. Diabet Technol Ther. (2009) 11:243–53. doi: 10.1089/dia.2008.0065
24. Poggio T, Smale S. The mathematics of learning: dealing with data. Notices AMS (2003) 50:537–44. Available online at: http://www.ams.org/notices/200305/fea-smale.pdf
25. Boland E, Monsod T, Delucia M, Brandt CA, Fernando S, Tamborlane WV. Limitations of conventional methods of self-monitoring of blood glucose lessons learned from 3 days of continuous glucose sensing in pediatric patients with type 1 diabetes. Diabetes Care (2001) 24:1858–62. doi: 10.2337/diacare.24.11.1858
26. Maggioni M, Mhaskar HN. Diffusion polynomial frames on metric measure spaces. Appl Comput Harmon Anal. (2008) 24:329–53. doi: 10.1016/j.acha.2007.07.001
27. Belkin M, Niyogi P. Towards a theoretical foundation for Laplacian-based manifold methods. In: International Conference on Computational Learning Theory. Berlin: Springer (2005). p. 486–500. doi: 10.1007/11503415_33
28. Singer A. From graph to manifold Laplacian: the convergence rate. Appl Comput Harmon Anal. (2006) 21:128–34. doi: 10.1016/j.acha.2006.03.004
A. Appendix
A.1. Filtered Legendre Expansion Method
In this appendix, we review the mathematical background for the method developed in Mhaskar et al. [7] for short term blood glucose prediction. As explained in the text, the main mathematical problem can be summarized as that of estimating the derivative of a function at the end point of an interval, based on measurements of the function in the past. Mathematically, if f : [−1, 1] → ℝ is continuously differentiable, we wish to estimate f′(1) given the noisy values {yj = f(tj) + ϵj} at points . We summarize only the method here, and refer the reader to Mhaskar et al. [7] for the detailed proof of the mathematical facts.
For this purpose, we define the Legendre polynomials recursively by
Let h:ℝ → ℝ be an even, infinitely differentiable function, such that h(t) = 1 if t ∈ [0, 1/2] and h(t) = 0 if t ≥ 1. We define
Next, given the points , we use least squares to find n = nd such that
and the following estimates are valid for all polynomials of degree < 2n:
for some positive constant A. We do not need to determine A, but it is guaranteed to be proportional to the condition number of the system in Equation (A1).
Finally, our estimate of f′(1) based on the values yj = f(tj)+ϵj, j = 1, ⋯ , d, is given by
It is proved in Mhaskar et al. [7, Theorem 3.2] that if f is twice continuously differentiable,
and |ϵj| ≤ δ then
where
the minimum being over all polynomials P of degree < n/2.
A.2. Function Approximation on Data Defined Spaces
While classical approximation theory literature deals with function approximation based on data that is dense on a known domain, such as a cube, torus, sphere, etc., this condition is generally not satisfied in the context of machine learning. For example, the set of vectors (sp(tj), …, sp(tj − d + 1)) is unlikely to be dense in a cube in ℝd. A relatively recent idea is to think of the data as being sampled from a distribution on an unknown manifold, or more generally, a locally compact, quasi-metric measure space. In this appendix, we review the theoretical background that underlies our experiments reported in this paper. The discussion here is based mainly on Maggioni and Mhaskar [26], Mhaskar [22], and Ehler et al. [8].
Let 𝕏 be a locally compact quasi-metric measure space, with ρ being the quasi-metric and μ* being the measure. In the context of machine learning, μ* is a probability measure and 𝕏 is its support. The starting point of this theory is a non-decreasing sequence of numbers , such that λ0 = 0 and λk → ∞ as k → ∞, and a corresponding sequence of bounded functions that forms an orthonormal sequence in L2(μ*). Let Πλ = span{ϕk : λk < λ}, and analogously to Equation (A2), we define for a uniformly continuous, bounded function f : 𝕏 → ℝ,
With the function h as defined in Appendix A.1, we define
Next, let be the “training data”. Our goal is to learn a function f : 𝕏 → ℝ based on the values . Toward this goal, we solve an under-determined system of equations
for the largest possible λ for which this system is still well conditioned. We then define an approximation, analogous to the classical radial basis function networks, by
It is proved in Maggioni and Mhaskar [26], Mhaskar [22], and Ehler et al. [8] that under certain technical conditions, for a uniformly continuous, bounded function f : 𝕏 → ℝ, we have
where c > 0 is a generic positive constant. Thus, the approximation σλ(f) is guaranteed to yield a “good approximation” that is asymptotically best possible given the training data.
In practice, one has a point cloud rather than the space 𝕏. The set is a subset of some Euclidean space ℝD for a possibly high value of D, but is assumed to lie on a compact sub-manifold 𝕏 of ℝD, with this manifold having a low dimension. We build the graph Laplacian from as follows: With a parameter ε > 0, induces the weight matrix defined by
We build the diagonal matrix and define the (unnormalized) graph Laplacian as
Various other versions of this Laplacian are also used in practice. For M → ∞ and ε → 0, the eigenvalues and interpolations of the eigenvectors of converge toward the “interesting” eigenvalues λk and eigenfunctions ϕk of a differential operator Δ𝕏. This behavior has been studied in detail by many authors, e.g., Belkin and Niyogi [27], Lafon [21], and Singer [28]. When are uniformly distributed on 𝕏, this operator is the Laplace-Beltrami operator on 𝕏. If are distributed according to the density p, then the graph Laplacian approximates the elliptic Schrödinger type operator , whose eigenfunctions ϕk also form an orthonormal basis for L2(𝕏, μ).
Keywords: deep learning, deep neural network, diffusion geometry, continuous glucose monitoring, blood glucose prediction
Citation: Mhaskar HN, Pereverzyev SV and van der Walt MD (2017) A Deep Learning Approach to Diabetic Blood Glucose Prediction. Front. Appl. Math. Stat. 3:14. doi: 10.3389/fams.2017.00014
Received: 12 April 2017; Accepted: 28 June 2017;
Published: 14 July 2017.
Edited by:
Dabao Zhang, Purdue University, United StatesReviewed by:
Polina Mamoshina, Insilico Medicine, Inc., United StatesPing Ma, University of Georgia, United States
Copyright © 2017 Mhaskar, Pereverzyev and van der Walt. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Maria D. van der Walt, bWFyeWtlLnRob21AZ21haWwuY29t
 Hrushikesh N. Mhaskar
Hrushikesh N. Mhaskar Sergei V. Pereverzyev
Sergei V. Pereverzyev Maria D. van der Walt
Maria D. van der Walt