- 1Agriculture and Agri-Food Canada, Lethbridge Research and Development Centre, Lethbridge, Alberta, AB, Canada
- 2Agriculture and Agri-Food Canada, Lacombe Research and Development Centre, Lacombe, Alberta, AB, Canada
Sustainable crop production systems should promote large and diverse soil microbial communities to enhance biological soil processes rather than depend solely on chemical interventions that include pesticide applications. Crop rotation increases above-ground temporal diversity which, relative to monoculture, usually increases soil microbial diversity. But comparisons between short and long crop rotations that also include pesticide effects are rare. A 5-yr (2013-2017) field study was conducted to investigate crop rotation and fungicide effects on the soil microbiome and activity. There were nine rotations, with or without fungicide applications, that included four 2-yr rotations (wheat preceded by canola, barley, pea, or flax), four 3-yr rotations where barley or canola were added to the 2-yr rotations, and one rotation where canola and wheat were stacked (canola-canola-wheat-wheat). In 2017, soil microbial biomass, composition, diversity and enzyme activities were measured in the rhizosphere of the final wheat crop in each rotation. Fungicides reduced fungal richness (the number of different fungal taxa) in the wheat rhizosphere (e.g., Chao1 indices of 64.0 vs. 79.9) especially in 2-yr rotations, but rotation length/type and the crops that preceded wheat had different effects on different taxa. Two of the three most predominant prokaryotic phyla, Proteobacteriota and Actinobacteriota, responded differently to rotation length: 3-yr rotations enriched the former (27.4% vs. 20.1% relative abundances), but 2-yr rotations enriched the latter (19.9% vs. 28.3% relative abundances). Relative to oilseed crops preceding the sampled wheat, a field pea preceding crop enriched Actinobacteriota (31.7% vs. 24.8% relative abundances) and the most abundant fungal class, Sordariomycetes (39.1% vs. 22.1% relative abundances), in addition to increasing microbial biomass carbon (MBC) and arylsulphatase activity by 33% and 57%, respectively. Correlations of the relative abundances of fungal or prokaryotic genera with β-glucosidase and arylsulphatase activities were similar (both positive or negative), but they were the opposite of correlations with acid phosphomonoesterase, suggesting a close link between C and S cycling. Besides the nutrient cycling implications of these soil microbial characteristics, there is need to study their biological disease control significance.
1 Introduction
Sustainable crop production should combine biological processes with chemical processes for crop nutrition and crop protection (pest control), instead of replacing the former with the latter. The chemical interventions that rely on chemical processes include inorganic fertilizer and pesticide applications. Biological processes in the soil are mostly mediated by soil microorganisms, and they include biological nitrogen fixation by rhizobia, nutrient uptake by mycorrhiza, cycling of nutrients through microbial decomposition of organic soil inputs, biological pest control through microbial predation and competition for resources, and detoxification of agro-chemicals. Therefore, sustainable crop production systems should sustain large and diverse soil microbial communities. Crop rotation increases above-ground temporal diversity, which should translate into below-ground diversity (Venter et al., 2016). In a global meta-analysis conducted on 76 studies, Liu et al. (2023b) reported that, relative to monoculture, crop rotation increased soil microbial biomass C (MBC) by 13.43% and bacterial Shannon’s diversity index by 7.68%. A similar meta-analysis used 20 studies and reported 15.11% and 3.36% increases in microbial evenness and diversity, respectively, with crop rotation (Venter et al., 2016).
The comparisons above were between crop rotations and monocultures. However, comparisons among different rotation lengths on their effects on the soil microbiome are not as many as rotation vs. monoculture comparisons. Some studies show that long (diverse) rotations result in greater soil microbial diversity than short (simple) rotations. In Ohio, Huo et al. (2023) identified eight bacterial amplicon sequence variants (ASVs) in a corn-soybean-wheat rotation that were not present in a corn-soybean rotation. In the North China Plain, Liu et al. (2023a) reported 15% and 20-23% greater relative abundances of Actinobacteriota and Ascomycota, respectively, in a sweet potato-winter wheat-summer corn rotation than in a winter wheat-summer corn rotation. One conclusion from a review of crop rotation effects on agroecosystem services and resilience was that diversified crop rotations increased resistance to pest/disease infestation and accelerated recovery from the infestations (Liu et al., 2022). The reasons that crop rotations (rotation vs. monoculture or long vs. short rotations) increase soil microbial diversity include soil physical, chemical and biological changes that rotational crops create, e.g., successive crops having different root actions that produce different niches for microbial exploitation, and build-up of different residual root exudates and crop residues (Venter et al., 2016). In Midwest USA, relative to a conventional corn-soybean rotation, a 4-yr corn-soybean-oat-alfalfa rotation reduced soil resistance to root growth by 8%, and increased cation exchange capacity, salt-extractable soil C and MBC by 16%, 157% and 62%, respectively (Baldwin-Kordick et al., 2022). More short vs. long rotation studies are required to build on existing knowledge.
Whereas crop rotations can foster large and diverse soil microbial communities, pesticides can have adverse effects on soil microbial communities because they are toxic by definition. In a review of agricultural input impacts on soil organisms, Bünemann et al. (2006) reported that the pesticide order of negative effects on the soil microbiome was: fungicides > insecticides > herbicides. Fungicides have direct negative effects on the soil microbiome because they are toxic to fungi, but they can also have indirect effects on other microorganisms through trophic relations, e.g., on the bacteria that feed on the fungi killed by the fungicide, by use of resources previously used by the killed fungi, and by attracting fungicide-degrading bacteria (Han et al., 2021; Meyer et al., 2024). Han et al. (2021) reported 42-3500% increase in tebuconazole-degrading bacterial genera in the tebuconazole fungicide treatment relative to the control. However, in a review of the toxic effects of tebuconazole to the soil microbiota, Roman et al. (2021) noted that most of the studies reported moderate, and sometimes transient, dose-dependent effects on soil microbial biomass and diversity. Riedo et al. (2023) reported similar results in a microcosm experiment evaluating three fungicides in three soils. Studies that combine crop rotation length and fungicide effects on the soil microbiome are required to discover any interactive effects.
The objective of this study was to assess how crop rotations and fungicide applications affected soil microbial biomass, the composition and diversity of prokaryotic (bacterial and archaeal) communities, and enzyme activities in the rhizosphere of wheat, which was the final crop in each rotation. For crop rotations, we particularly investigated how rotation length and the crops preceding the final wheat crop affected the soil microbial properties.
The setup of the trial was innovative because (a) it did not include monoculture crops, (b) it included a stacked rotation (canola-canola-wheat-wheat), and (c) the crops were no-till managed. All these three aspects of the rotations reflected common practices in the study area.
2 Materials and methods
2.1 Experimental site, setup and management
The five-year field trial was conducted from 2013 to 2017 at Lacombe Research and Development Centre (113°45’W, 52°27’N), Alberta, Canada, with 533 mm annual rainfall and 3.5°C average annual temperature. The soil was a loam (51% sand, 30% silt and 19% clay) Black Chernozem (Typic Haplustoll in Soil Taxonomy) with pH 6.6 and 45.2 g organic C kg-1. There were nine rotations, with or without fungicide applications, arranged in a randomized complete block design with four replications (Table 1). They included four 2-yr rotations (canola-wheat, barley-wheat, pea-wheat, and flax-wheat), four 3-yr rotations where barley or canola were added to the 2-yr rotations (barley-canola-wheat, canola-barley-wheat, barley-pea-wheat, and barley-flax-wheat), and one rotation where canola and wheat were stacked (canola-canola-wheat-wheat). In 2017, all the rotations were sown to wheat, the final crop that was sampled to evaluate each rotation. Therefore, in addition to evaluating the rotations in terms of length/type (2-yr, 3-yr, or stacked), the effects of the crop preceding wheat (canola, barley, pea, or flax) were also evaluated (Table 1).
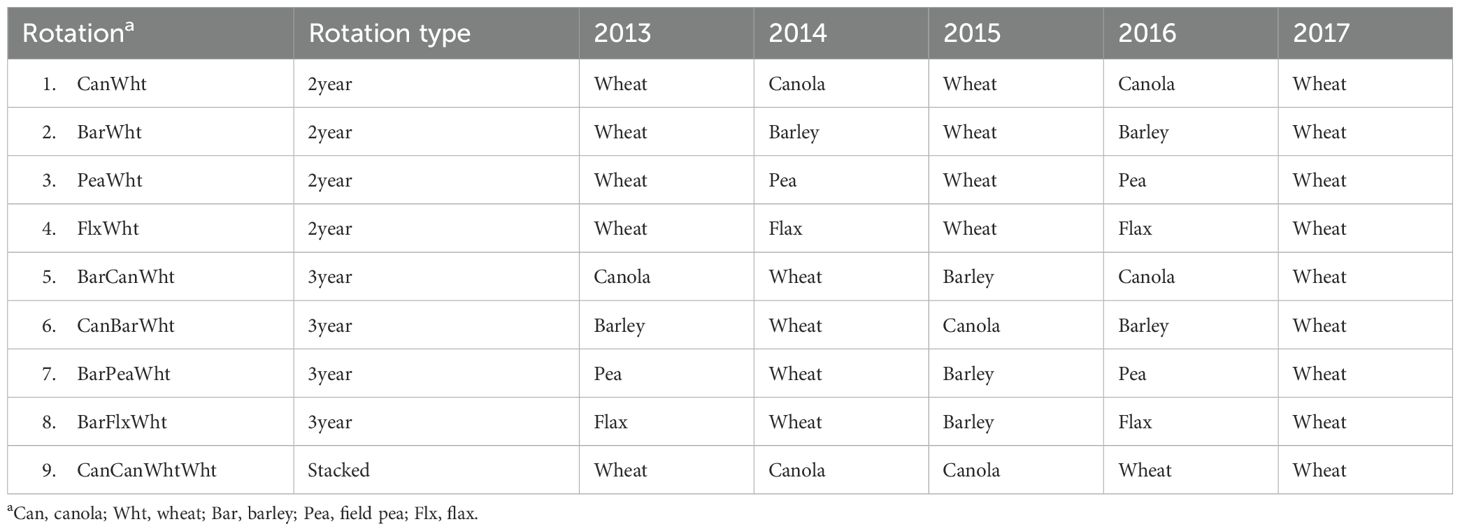
Table 1. List of crop rotations, each with or without fungicide applications. The 2017 wheat rhizosphere was sampled for microbiological analysis; the 2016 crops were the preceding crops.
For the fungicide treatment, the fungicides applied depended on the crop. In cereals, the fusarium head blight fungicide prothioconazole+tebucanzole was applied as Prosaro 250 EC (Bayer, 125 g prothioconazole L-1 + 125 g tebuconazole L-1) at 790 mL ha-1 with a water volume of 100 L ha-1. In barley, the fungicide was applied at 70-100% head emergence growth stage, and in wheat it was applied between 75% head emergence on the main stem and 50% flowering. This fungicide also targeted cereal leaf spots (tan spot, etc.) and rusts. Both actives are part of the triazole chemical group in the Fungicide Resistance Action Committee (FRAC) fungicide Group 3, and they affect sterol biosynthesis (https://www.frac.info/docs/default-source/publications/frac-code-list/frac-code-list-2024.pdf, accessed on 12 September, 2024). In canola, two fungicides were applied: one for blackleg and the other for sclerotinia. The blackleg fungicide was pyraclostrobin, which was applied as Headline EC (250 g pyraclostrobin L-1) in a tank mix with supported canola herbicides at the 2 to 6-leaf stage at 395 mL ha-1, with a water volume of 100 L ha-1. The fungicide for sclerotinia was prothioconazole, applied as Proline 480 SC (480 g prothioconazole L-1) at the early bloom to full bloom stage of canola at the rate of 363 mL ha-1, with a water volume of 100 L ha-1. Pyraclostrobin is part of the methoxy-carbamates chemical group (Quinone outside inhibitors) in the FRAC fungicide Group 11, and it affects mitochondrial respiration. In field pea, the fungicide was fluxapyroxad + pyraclostrobin, applied as Priaxor DS (250 g fluxapyroxad L-1 and 250 g pyraclostrobin L-1) at the beginning of flowering at a rate of 296 mL L ha-1, with a water volume of 100 L ha-1. In flax, the fungicide was pyraclostrobin, applied as Headline EC (250 g pyraclostrobin L-1) 7-10 days after initiation of flowering at a rate of 395 mL ha-1, with a water volume of 100 L ha-1. Fluxapyroxad affects respiration and is part of the pyrazole-4-carboxamides chemical group (Succinate dehydrogenase inhibitors) in the FRAC fungicide Group 7.
The crops were seeded in May each year and were managed according to standard agronomic practices, including no tillage. Fertilizer applications were based on soil-test results for non-legumes, and field peas were inoculated with Rhizobium inoculant.
2.2 Soil sampling
In 2017, we sampled soil from the rhizosphere of wheat, the final crop of each rotation, at the flag leaf growth stage. The wheat plants were excavated from four random 0.5 m lengths of row in each treatment, i.e., four locations per plot. We removed loose soil from the roots by shaking the plants by hand, and the remaining soil that had strongly adhered to the roots was carefully brushed off and recovered as rhizosphere soil. We combined the four rhizosphere soil samples from each plot to make a composite sample and passed it through a 2 mm sieve. The samples for DNA extraction were frozen at −20°C, those for enzyme analysis were stored at 4°C, and those for microbial biomass C (MBC) analysis were air-dried and stored at 4°C.
2.3 Microbial biomass C analysis
Soil MBC was measured using the substrate-induced respiration method (Horwath and Paul, 1994): 300 mg of glucose was dissolved in 9.0 mL water and added to 50 g of air-dry soil to bring it to 50% water-holding capacity (determined by measuring the gravimetric water content after draining a water-saturated column of soil until the water stopped dripping). After stir-mixing, the soil was incubated in a 1 L jar for 3 h at 22°C, and the amount of CO2 that accumulated in the head space was measured using gas chromatography.
2.4 Microbiome analysis: DNA extraction, sequencing and bioinformatics
Soil DNA was extracted using the Qiagen DNeasy Powerlyzer PowerSoil Kit (Quagen, Toronto, Ontario) following the manufacturer’s instructions. Sequencing was performed using the 515F/806R primer set (515F: 5`-ACACTGACGACATGGTTCTACAGTGCCAGCMGCCGCGGTAA – 3`; 806R: 5`-TACGGTAGCAGAGACTTGGTCTGGACTACHVGGGTWTCTAAT – 3`) to amplify the V4 hypervariable region of the prokaryotic (bacterial and archaeal) 16S rRNA gene (Caporaso et al., 2011). Samples were first barcoded and then pooled. To characterize soil fungi, we used the ITS primers ITS1F/ITS2R (ITS1F: 5` - CTTGGTCATTTAGAGGAAGTAA – 3`; ITS2R: 5`- GCTGCGTTCTTCATCGATGC – 3`) (Bellemain et al., 2010). Again, the samples were first barcoded and then pooled. The amplicon libraries were then purified and quantified separately. Each library was sequenced on an Illumina MiSeq platform at Génome Québec Innovation Centre at McGill University (Montréal, Canada).
The 16S sequence data was analyzed using the DADA2 package (Callahan et al., 2016) (version 3.14) of the R statistical computing language. Raw 16S sequences were filtered based on quality, errors were inferred, Amplicon Sequence Variants (ASVs) were selected, chimeras were removed and taxonomy assignment was performed with the SILVA_NR99_v138.1 database. For the ITS sequence analysis, we used the PIPITS pipeline (GitHub - hsgweon/pipits: Automated pipeline for analyses of fungal ITS from the Illumina) to produce OTU abundance and taxonomic assignment tables, after assigning the sequences with RDP Classifier against the UNITE fungal ITS reference dataset (Gweon et al., 2015). Raw sequences have been deposited in the NCBI Sequence Read Archive (SRA) repository under BioProject ID PRJNA1175403. For both 16S and ITS, the ASV, OTU and taxonomic assignment tables were exported to the Marker-gene Data Profiling (MDP) module of the online platform MicrobiomeAnalyst for further bioinformatics analysis (https://www.microbiomeanalyst.ca/) (Chong et al., 2020; Lu et al., 2023). The sequences were filtered for low counts and low variance before analysis. The relative microbial abundances at various classification levels (phylum to genus) were calculated. We calculated the following α-diversity indices at OTU level: ACE (abundance-based coverage estimator), Chao1, Fisher and Shannon, after rarifying the data to the minimum library size. β-diversity was visualized using Principal Coordinate Analysis (PCoA) on a Bray-Curtis dissimilarity index, and group differences in microbial community structures were assessed for statistical significance using permutational multivariate analysis of variance (PERMANOVA), all in MicrobiomeAnalyst.
2.5 Enzyme analysis
The activities of β-glucosidase (C cycling), N-acetyl-β-glucosaminidase (NAG) (N cycling), and acid phosphomonoesterase (P cycling) were measured using microplate fluorimetric assays (Deng et al., 2011) as described by Lupwayi et al. (2019). These assays were based on the detection of 4-methylumbelliferone (MUF) released by the enzymatic hydrolysis of MUF-labelled substrates (MUF-β-D-glucoside, MUF-β-N-acetyl-glucosaminide, or MUF-phosphate) incubated with soil at the optimal pH of each enzyme (pH 5.5 for NAG, and pH 6.0 for the other two enzymes). Arylsulphatase (S cycling) activity was measured in a bench-scale assay to colorimetrically measure the p-nitrophenol released by the enzyme after incubating 1 g soil with buffered (pH 6.0) p-nitrophenyl-β-D-sulphate (Dick et al., 1996).
2.6 Statistical analysis
Soil MBC, the relative abundances of the microbiome at selected classification levels, the α-diversity indices, and enzyme activity data were statistically analysed by analysis of variance (ANOVA) as a crop rotation x fungicide factorial in a RCBD. The 5% level of significance was used to determine statistical significance, and the means were separated by Tukey HSD test when statistical significance was detected. To further examine which rotation properties affected the results, the following orthogonal contrasts were constructed:-
A. 2-yr vs. 3-yr rotations: Rotations 1-4 vs. Rotations 5-8.
B. 2-yr vs. stacked rotations: Rotations 1-4 vs. Rotation 9.
C. Broadleaf vs. barley preceding crops within 2-yr rotations: Rotations 1, 3, and 4 vs. Rotation 2.
D. Pea vs. oilseed preceding crops within 2-yr rotations: Rotation 3 vs. Rotations 1 and 4.
We used only 2-yr rotations for contrasts C and D because including 3-yr rotations would, in some cases, confound the effects with the very crops that we were contrasting.
The relationships between the composition of the microbial genera and enzyme activities were assessed through Pearson correlation analysis, and some of the relationships were modeled with linear regression analysis when the correlations were significant at the 5% significance level.
3 Results
3.1 Microbial biomass C
Microbial biomass C in the wheat rhizosphere was highest in pea-wheat rotation and lowest in barley-canola-wheat rotation, but rotation length (2-yr vs. 3-yr or 2-yr vs. stacked rotations) had no effect (Table 2). Even though there was no difference in MBC between broadleaf preceding crops and barley preceding crop, MBC was 33% greater in the wheat preceded by pea (970 mg C kg-1 soil) than in wheat preceded by oilseed crops (an average of 729 mg C kg-1 soil). Fungicide applications did not affect MBC.
3.2 Fungal microbiome
The most abundant fungal class was Sordariomycetes (Supplementary Figure S1), and its relative abundance was greater when wheat was preceded by barley, particularly in barley-wheat rotation without fungicides, than by broadleaf crops (Table 3). At the genus level of this fungal class, the relative abundance of the genus Acremonium was also greater in barley-wheat and canola-barley-wheat rotations than in other rotations, but there was rotation x fungicide interaction (Figure 1A). Sordariomycetes were also more abundant when the preceding crop was pea relative to oilseed preceding crops (39.1% vs. 22.1% relative abundances) (Table 3), exemplified by the high relative abundance of the genus Clamydocillium in the pea-wheat rotation without fungicides (Figure 1B). The relative abundance of the class Leotiomycetes was highest in the flax-wheat rotation (Table 3), especially where no fungicides were applied. This flax effect was the main contributor to the greater relative abundance of this class in the wheat preceded by oilseed crops relative to pea preceding crop, as shown for the genus Entimonentora (Figure 1C). The genus Penicillium (from class Eurotiomycetes), which was the most predominant fungal genus (Supplementary Figure S2), was also enriched in the flax-wheat rotation (Figure 1D). No particular rotation type or preceding crop explained the differences in the relative abundances of Microbotromycetes, and fungicides had no effect (Table 3). Three-year rotations, particularly barley-flax-wheat and barley-pea-wheat, had greater relative abundances of Rhizophydiomycetes than 2-yr rotations, and fungicides had no effect. However, fungicides reduced the relative abundance of the genus Zymoseptoria (from class Dothideomycetes) (Figure 1F); this genus (Z. tritici) is associated with wheat speckled leaf blotch. Zoopagomycetes were more abundant in wheat preceded by pea, especially in barley-pea-wheat rotation with fungicides and pea-wheat rotation without fungicides, than by oilseed preceding crops as shown for the genus Syncephalis (Figure 1E).
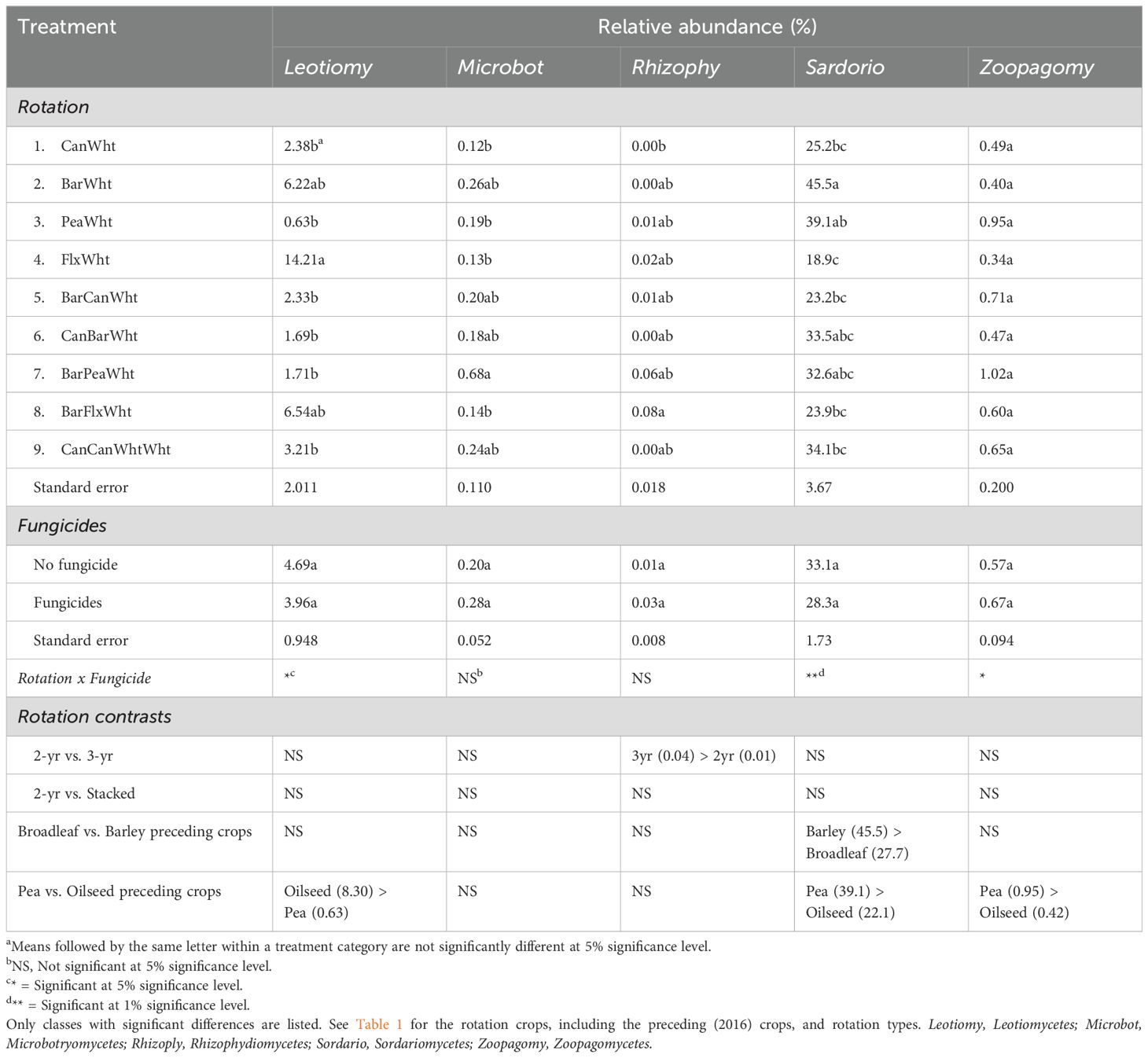
Table 3. Crop rotation and fungicide effects on the relative abundances of fungal classes in the wheat rhizosphere.
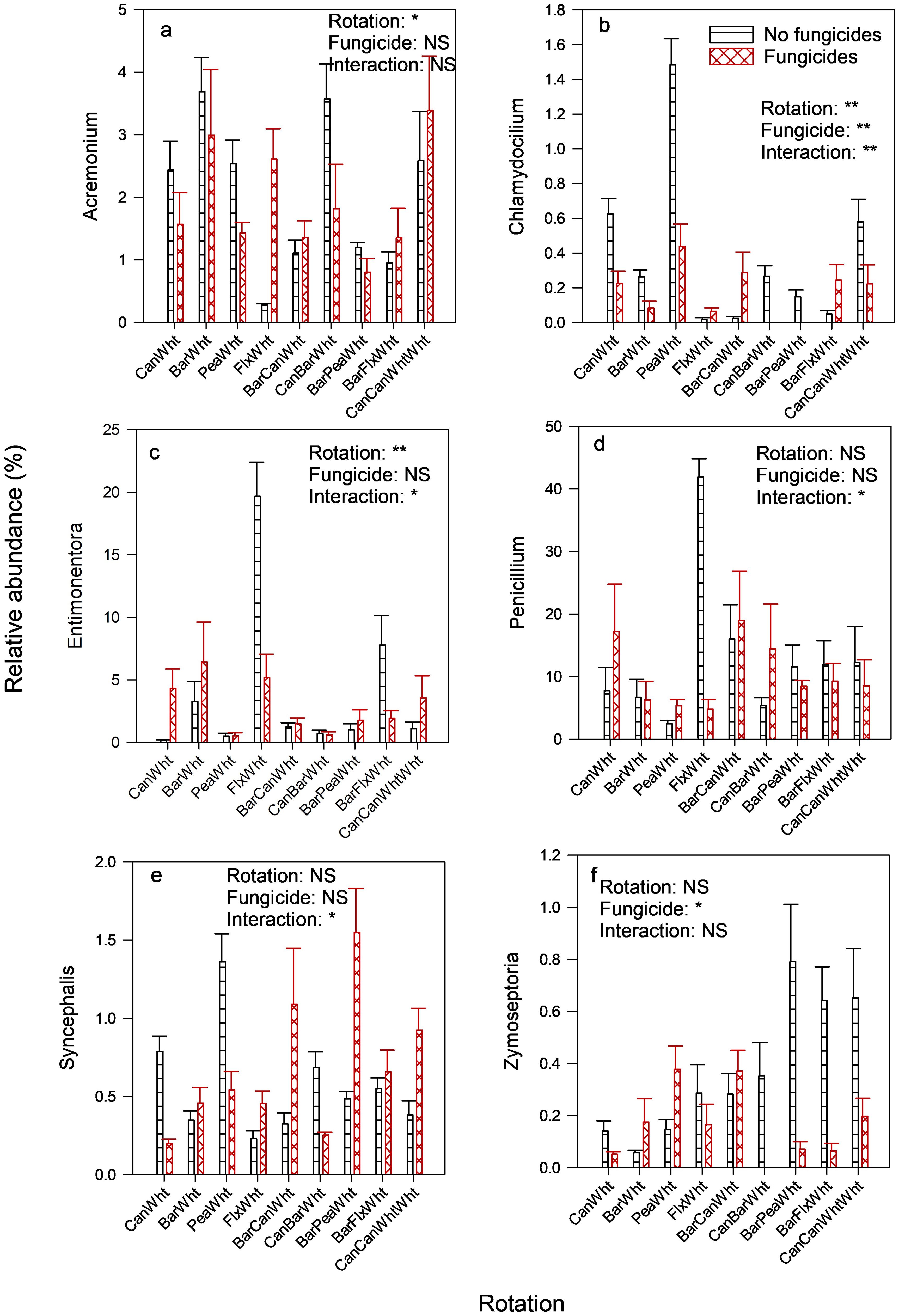
Figure 1. Crop rotation and fungicide effects on the relative abundances of fungal genera in the wheat rhizosphere. Only the genera (that had an average relative abundance of at least 0.20%) with significant differences are included. Can, Canola; Wht, Wheat; Bar, Barley; Pea, Field pea; Flx, Flax. Treatment effects: ** = significant at 1% significance level; * = significant at 5% significance level; NS, not significant at 5% significance level. Fungal genera depicted include (A) Acremonium, (B) Chlamydocilium, (C) Entimonentora, (D) Penicillium, (E) Syncephalis, and (F) Zymoseptoria.
The Chao 1, ACE, and Fisher indices of fungal α-diversity, which all denote species richness, were greater in 2-yr rotations than in 3-yr rotations or the stacked rotation (Figures 2A–C). These indices were also all reduced by fungicide applications (e.g., Chao1 indices of 64.0 vs. 79.9), especially in 2-yr rotations. The Shannon index of diversity, however, was greater in 3-yr rotations than in 2-yr rotations, and fungicide applications increased this index in 2-yr rotations (Figure 2D). Principal Coordinate Analysis (PCoA) indicated that fungal β-diversity was only affected by crop rotation (PERMANOVA F-value = 1.7801; R-squared = 0.18437; p-value = 0.007), and Figure 3 shows that the pea-wheat, barley-wheat, and barley-pea-wheat rotations had slightly different fungal community structures than the other rotations. Fungicides did not affect fungal β-diversity.
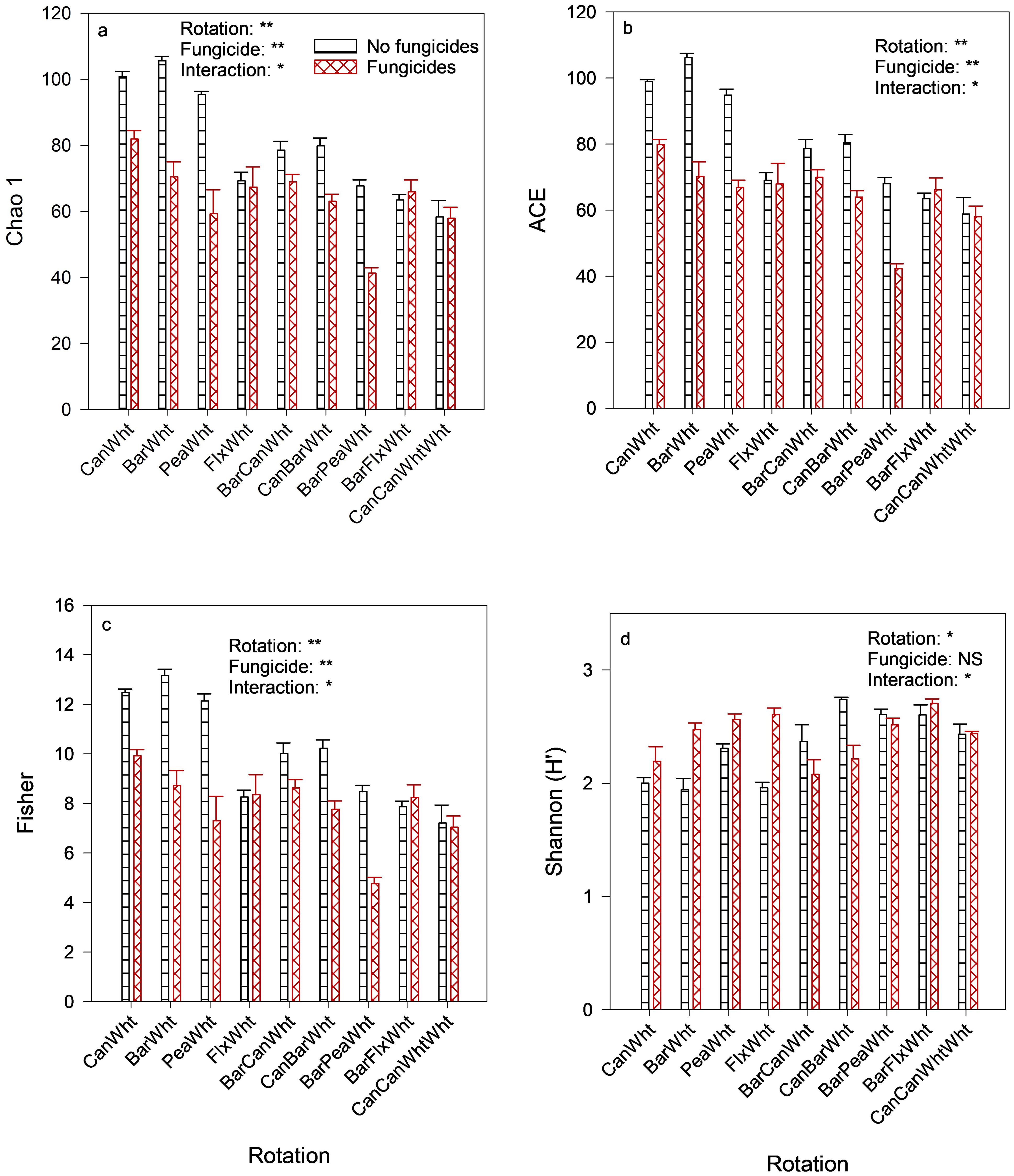
Figure 2. Crop rotation and fungicide effects on fungal α-diversity in the wheat rhizosphere. Can, Canola; Wht, Wheat; Bar, Barley; Pea, Field pea; Flx, Flax; ACE, Abundance-based coverage estimator. Treatment effects: ** = significant at 1% significance level; * = significant at 5% significance level; NS, not significant at 5% significance level. Indices depicted are (A) Chao 1, (B) Abundance-Based Coverage Estimator (ACE), (C) Fisher, and (D) Shannon.
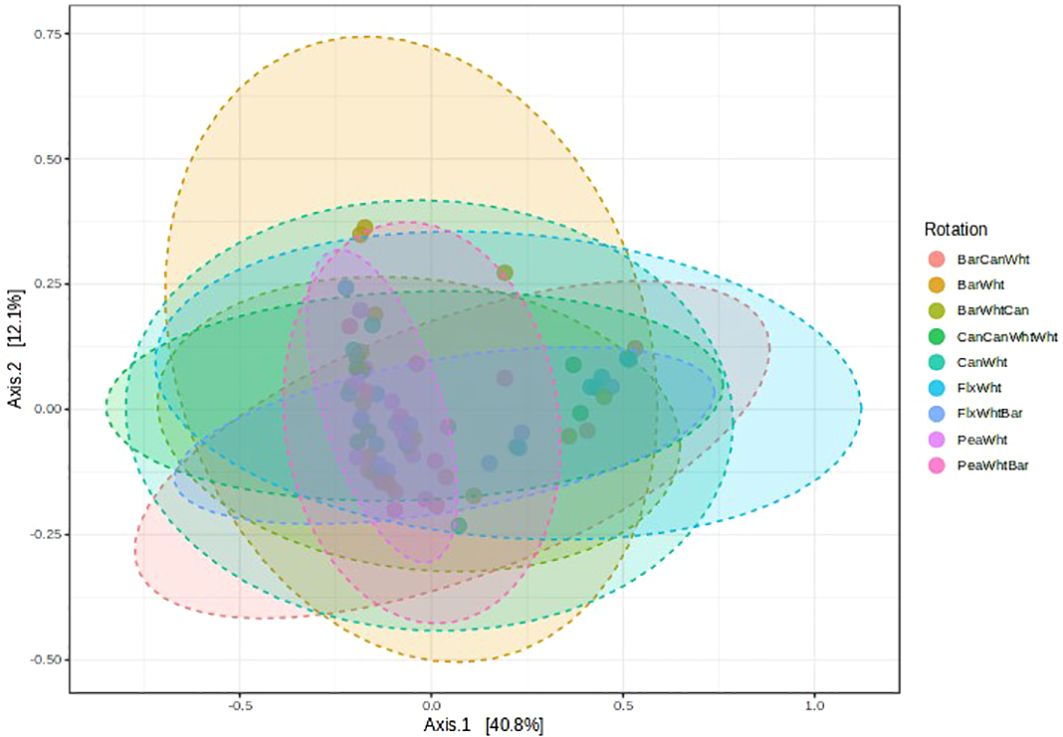
Figure 3. Principal coordinate analysis (PCoA) showing crop rotation effects on fungal β-diversity in the wheat rhizosphere. Can, Canola; Wht, Wheat; Bar, Barley; Pea, Field pea; Flx, Flax. PERMANOVA F-value = 1.7801; R-squared = 0.18437; p-value = 0.007.
3.3 Prokaryotic microbiome
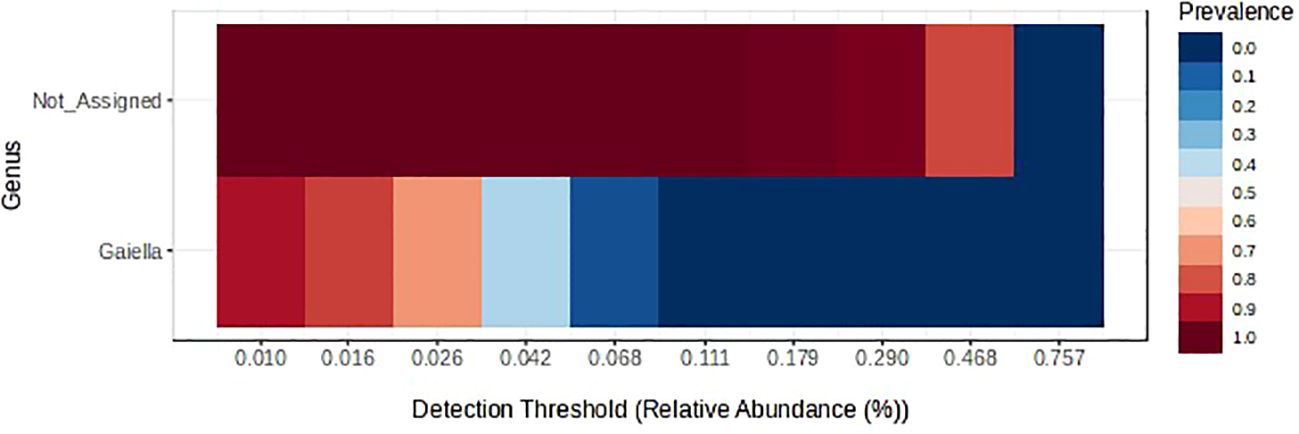
The most predominant prokaryotic phyla, each with about 24% relative abundance, were Chloroflexi, Actinobacteriota, and Proteobacteriota (Supplementary Figure S3). Crop rotation did not affect the relative abundance of the bacterial phylum Acidobacteriota, which was increased by fungicides (Table 4). These effects were also observed at genus level, as shown by the relative abundance of the genus RB41 (Figure 4A). The relative abundance of Actinobacteriota was greater in 2-yr than in 3-yr rotations, and was reduced by fungicides especially in 2-yr rotations (Table 4). These effects were also observed in the genus Pseudonocardia (Figure 4B), but the genus Gaiella only responded to the rotation (Figure 4C). The rotation effects on the phylum Chloroflexi could not be categorized by rotation type, and fungicides had no effect (Table 3). The relative abundance of the archaea phylum Crerchaeota was greater in 2-yr rotations than in 3-yr rotations (Table 4), and fungicides reduced its abundance in 2-yr rotations while increasing it in 3-yr rotations. Latecibacterota were more abundant in the wheat preceded by barley than that preceded by broadleaf crops, and fungicides increased its abundance in the barley-wheat rotation (Table 3). Proteobacteriota were more abundant in 3-yr than in 2-yr rotations (27.4% vs. 20.1% relative abundances, Table 4), as illustrated by the genus Luteimonas (Figure 4D), and in wheat preceded by oilseed crops than by pea crop as illustrated by the genus Rhodanobacter (Figure 4E). Rhodanobacter (from phylum Proteobacteriota) was the most predominant prokaryotic genus (Supplementary Figure S4). Fungicides increased the relative abundance of Proteobacteriota in 2-yr rotations (except flax-wheat rotation). Although there were no treatment effects on the relative abundances of the phylum Firmicutes, the genus Granulicella was enriched in the flax-wheat rotation without fungicides (Figure 4F). An un-assigned taxon and the genus Gaiella were the core prokaryotic taxa that were observed in at least 85% of the samples were (Figure 5).
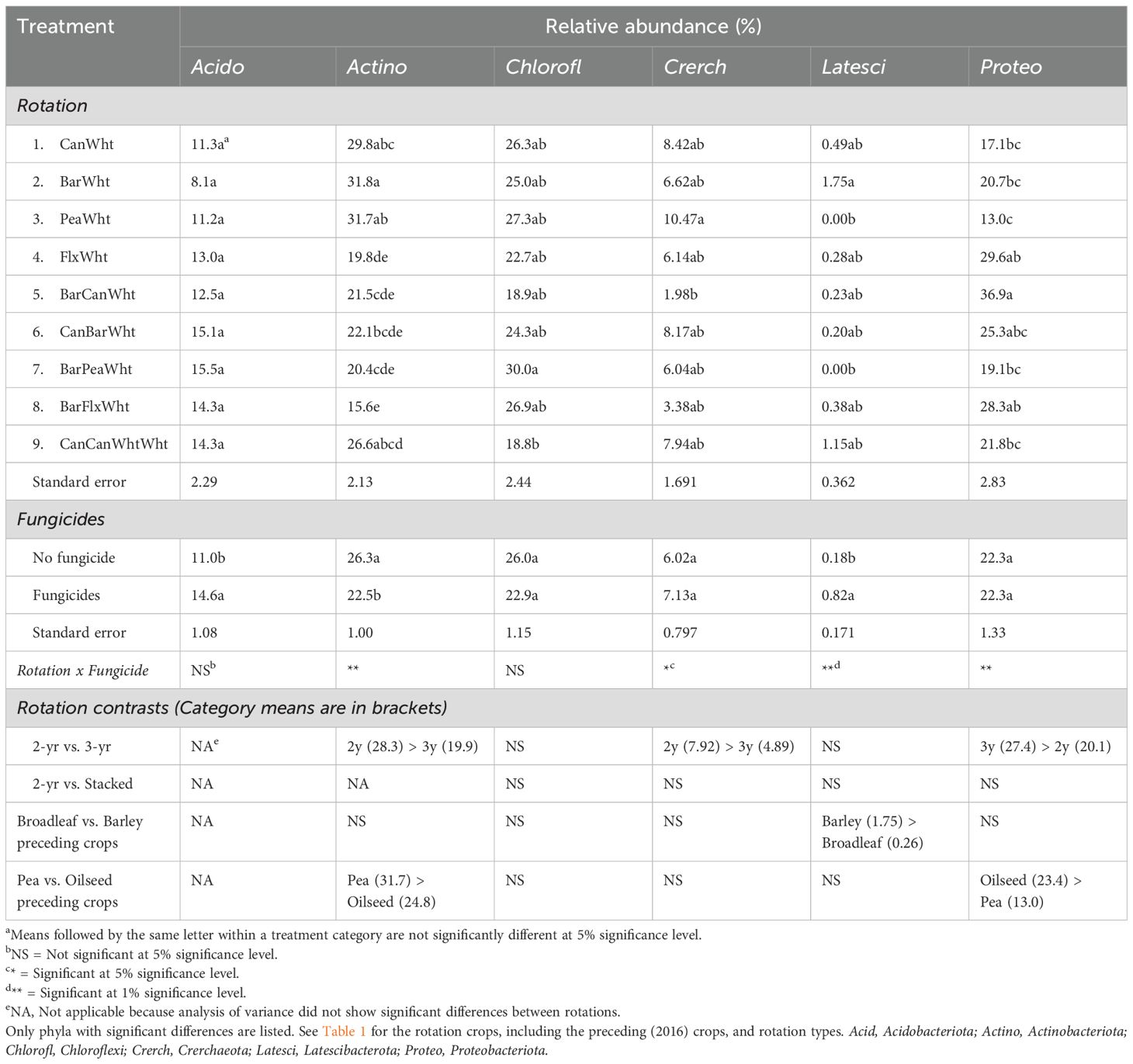
Table 4. Crop rotation and fungicide effects on the relative abundances of prokaryotic phyla in the wheat rhizosphere.
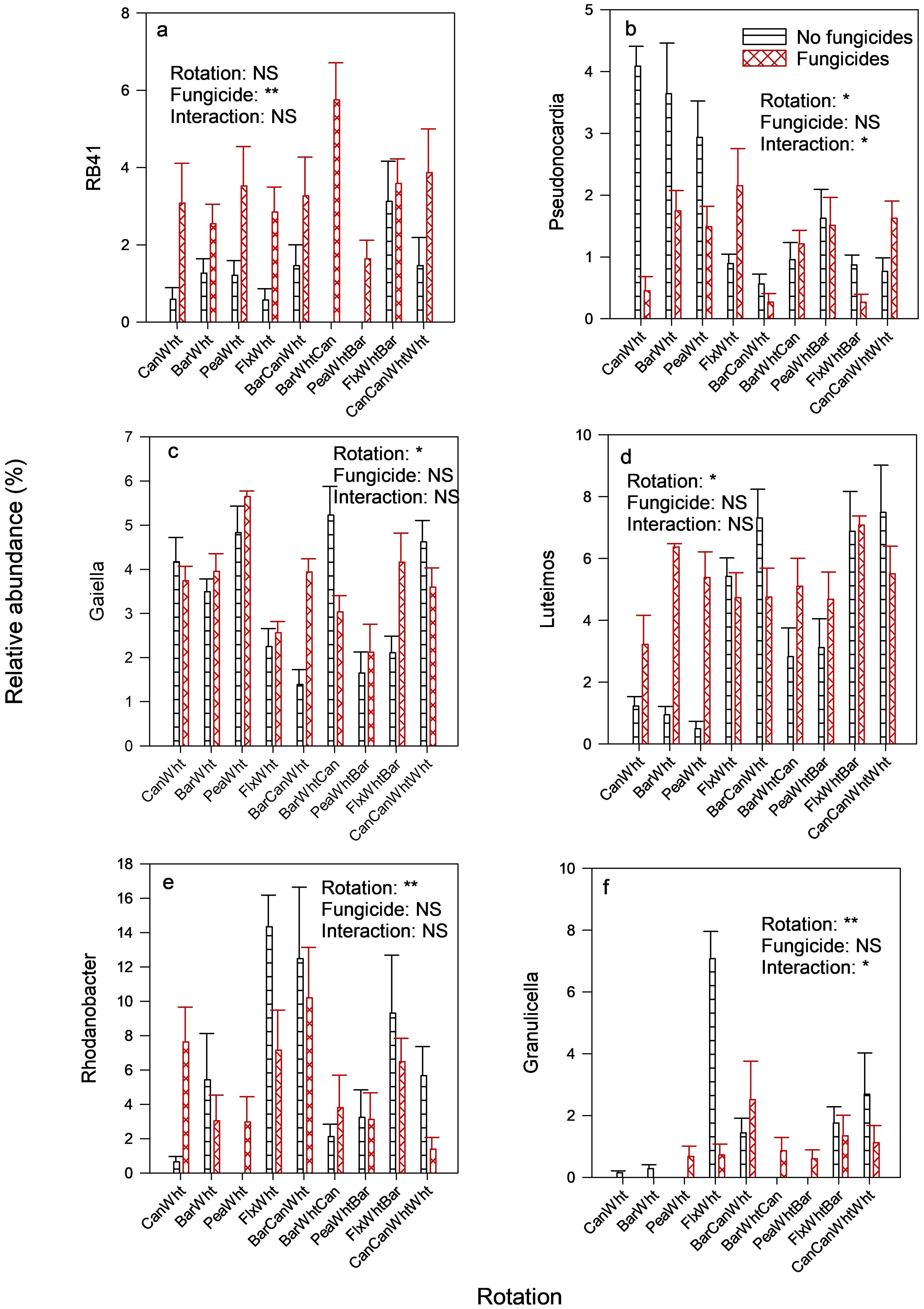
Figure 4. Crop rotation and fungicide effects on the relative abundances of prokaryotic genera in the wheat rhizosphere. Only the genera (that had an average relative abundance of at least 0.50%) with significant differences are included. Can, Canola; Wht, Wheat; Bar, Barley; Pea, Field pea; Flx, Flax. Treatment effects: ** = significant at 1% significance level; * = significant at 5% significance level; NS, not significant at 5% significance level. Prokaryotic genera depicted include (A) RB41, (B) Pseudonocardia, (C) Gaiella, (D) Luteimos, (E) Rhodanobacter and (F) Granulicella.
Neither the α-diversity nor the β-diversity of prokaryotic communities was affected by the crop rotations or fungicides.
3.4 Enzyme activities
There were no rotation or fungicide effects on the activity of N-acetyl-β-glucosaminidase (NAG), nor were there fungicide effects on the activity of any other enzyme (Table 5). The activity of β-glucosidase was highest in canola-wheat rotation, and lowest in barley-canola-wheat rotation, with no clear indications of the effects of rotation type or preceding crop. Similarly, acid phosphomonoesterase activity was highest in barley-canola-wheat rotation and lowest in canola-barley-wheat rotation, showing a crop sequence effect. The activity of arylsulphatase was 57% greater when pea was the preceding crop (107.7 mg C kg-1 soil) than where an oilseed crop preceded the wheat (68.8 mg C kg-1 soil).
3.5 Relationships between the microbiome composition and enzyme activities
None of the relative abundances of fungal genera were correlated with the activity of N-acetyl-β-glucosaminidase that mediates N cycling (Table 6). The relative abundances of six fungal genera, all from the classes Sordariomycetes (the genera Chlamydocillium, Chloridium, Marquandomycetes, and Trichoderma) or Dothideomycetes (the genera Sporormiella and Zymoseptoria) were positively correlated with β-glucosidase activity; and three genera, from the classes Eurotiomycetes (the genera Oidiodendron and Penicillium) or Leotiomycetes (the genus Entimomentora) had negative correlations. The genera that were negatively correlated with β-glucosidase activity were also negatively correlated with arylsulphatase, but positively correlated with acid phosphomonoesterase. Similarly, negative correlations with acid phosphomonoesterase were the opposite of correlations with β-glucosidase or arylsulphatase activities. Some of these relationships are depicted with linear regression in Figure 6.
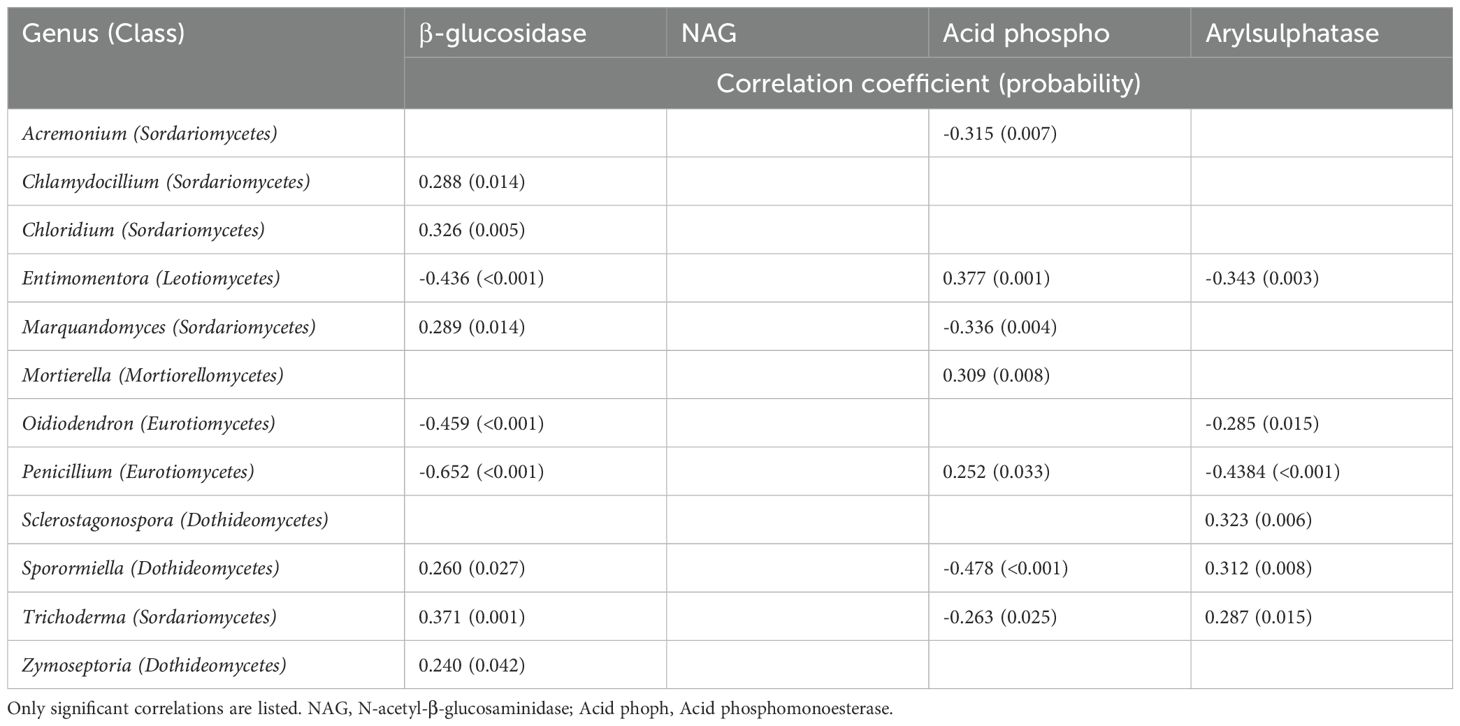
Table 6. Correlations between the relative abundances of fungal genera and enzyme activities in the wheat rhizosphere.
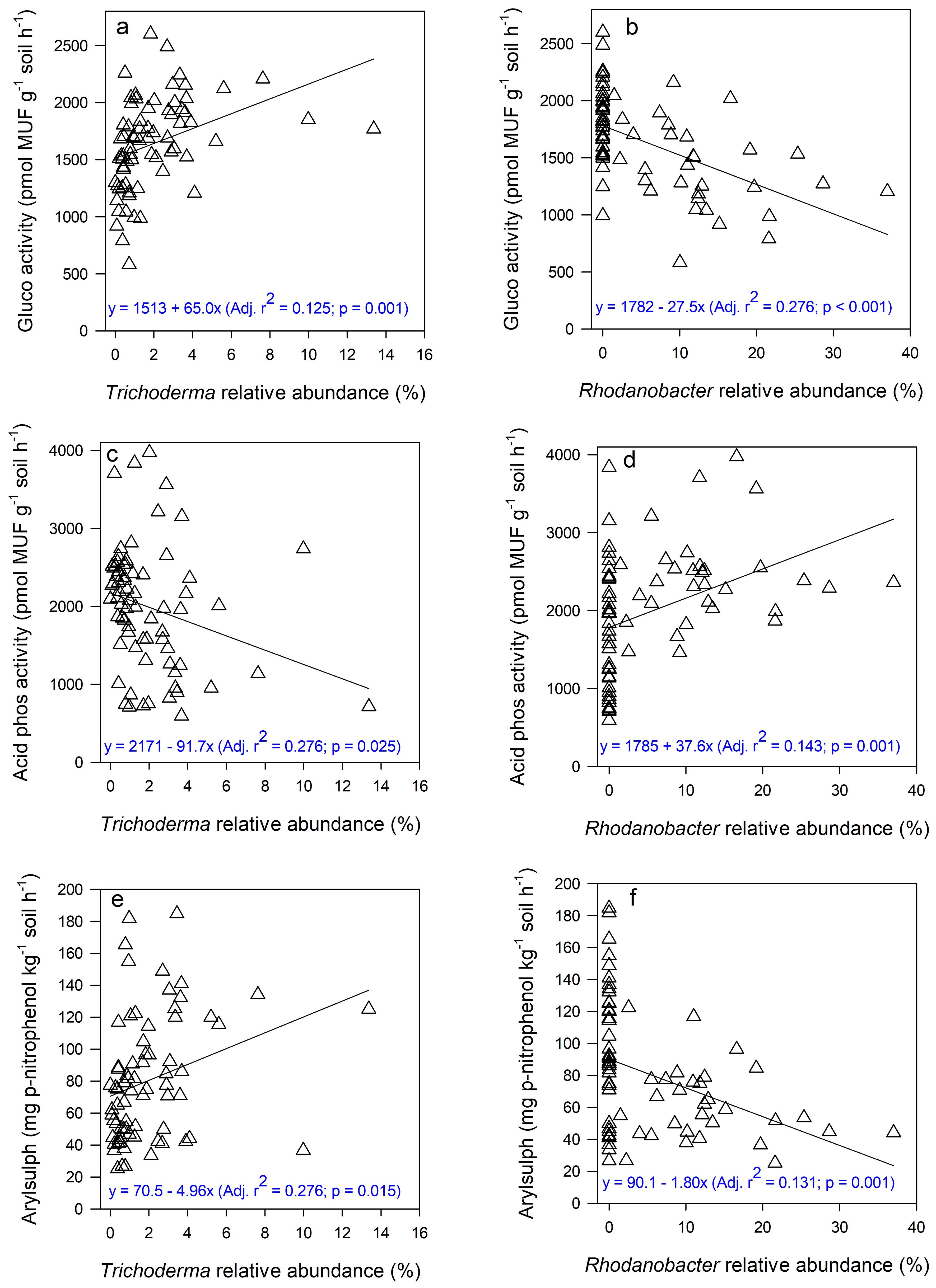
Figure 6. Relationships between the relative abundances of the fungal genus Trichoderma (class Sordariomycetes) and the activities of β-glucosidase (gluco), acid phosphomonoesterase (acid phos) and arylsulphatase (arylsulph) (A, C, E), and between the relative abundances of the bacterial genus Rhodanobacter (phylum Proteobacteriota) and the activities of the same enzymes (B, D, F).
As with fungal genera, correlations of the relative activities of prokaryotic genera with β-glucosidase and arylsulphatase activities were similar, i.e., both positive or negative, but the opposite of correlations with acid phosphomonoesterase. The relative abundances of prokaryotic genera from the phylum Proteobacteriota (the genera Masillia, Rhodonobacter, and Sphingomonas) that were negatively correlated with β-glucosidase activity were also negatively correlated with arylsulphatase activity, but positively correlated with acid phophomonoesterase activity (Supplementary Table S1). However, some genera from the phylum Actinobacteria (e.g., Marmoricola) had negative correlations, while others (e.g., Rubrobacter) had positive correlations, with the activities of the two enzymes. Some of these relationships are depicted with linear regression in Figure 6.
4 Discussion
4.1 Fungicides
The most consistent fungicide effect was the reduction of fungal richness indices of α-diversity, especially in 2-yr rotations (Figures 2A–C), but the Shannon index of α-diversity increased in these rotations (Figure 2D). Microbial diversity is a function of richness (number of different microbial taxa) and evenness (the relative distributions of those taxa) (Bo-Ra et al., 2017). Our results suggest that fungicides reduced the number of different fungal taxa in 2-yr rotations, but those taxa were more evenly distributed in those rotations than in 3-yr rotations, i.e., were not dominated by few taxa. It is possible that, without fungicides, there were more fungal taxa in 2-yr rotations than 3-yr rotations because of the fungal pathogens that were targeted by the fungicides. However, the fungicides applied were in-crop foliar fungicides that did not target soil fungal pathogens. Therefore, these effects on fungal diversity were probably non-target effects from the fungicides that reached the soil through unclosed crop canopies or by dripping from crop leaves and stems. Because different fungicides were applied to the different crops in each rotation, it is difficult to attribute their effects to particular fungicides. However, 2-year wheat-based rotations received a higher proportion of the triazole fungicides prothioconazole and tebuconazole (because wheat was grown every other year) than 3-year rotations (Table 1), suggesting that one or both of these fungicides could have reduced soil fungal richness. Lloyd et al. (2021) reported that prothioconazole applied to blueberries reduced soil fungal richness. By contrast, Vasilchenko et al. (2023) reported that fungicides reduced the Shannon indices of both fungal and bacterial communities in the soil but did not affect Chao 1 indices (of richness). The fungicide effects on the soil microbiome were only assessed once in our study, due to resource limitations. With one-time sampling, it is not possible to know if the effects were transient, as has been reported in some studies (reviewed by Roman et al., 2021). Nonetheless, even transient effects can have consequences. However, these fungicide effects on fungal diversity had no apparent effects on nutrient cycling in our study because enzyme activities were not affected.
Although the fungicides did not affect the diversity of the prokaryotes in our study, they reduced the relative abundance of Actinobacteriota while increasing those of Acidobacteriota and Latescibacterota (Table 4). Divergent fungicide effects on different soil prokaryotes have also been reported in other studies, and the increases in the relative abundances have mainly been attributed to enrichment of the prokaryotes that detoxify fungicides (Han et al., 2021; Vasilchenko et al., 2023).
4.2 Rotation length/type
Two of the three most predominant prokaryotic phyla, Actinobacteriota and Proteobacteriota, responded differently to rotation length. Relative to 2-yr rotations, 3-yr rotations increased the relative abundances of Proteobacteriota (Table 3) and the fungal class Rhizophydiomycetes (Table 2), but the opposite was true for the Actinobacteriota and archaeal phylum Crerchaeota (Table 3). Liu et al. (2023a) also reported that the relative abundance of Actinobacteriota was greater in a 3-yr rotation (spring sweet potato-winter wheat-summer corn) than a 2-yr rotation (winter wheat-summer corn).
Fungal α-diversity also responded differently to rotation length: the indices of species richness (Chao 1, ACE, and Fisher) were all greater in 2-yr rotations than in 3-yr rotations or the stacked rotation (Figures 2A–C), but the reverse was true for the index (Shannon) that combines richness and evenness (Figure 2D). The study period for this field trial was probably too short for the 3-year rotations to shape the soil microbiome. Between 2013 and 2017, the 2-yr rotations completed two cycles, but the 3-yr rotations completed only one cycle. The ideal study duration would be one in which each crop rotation completes at least two cycles, as was the case in a 12-yr soil conservation study that had 1-yr to 6-yr rotations (Larney et al., 2017). In a 16-yr field trial, Yang et al. (2023) reported that the soil microbial network complexity increased with increased crop rotational diversity, and that the network complexity had a stronger effect on soil multifunctionality (characterized by using functions related to soil properties, basal respiration, enzyme activities, and nitrogen cycling) than microbial community composition and diversity.
4.3 Preceding crops
The most abundant fungal class, Sordariomycetes, and one of the three most abundant prokaryotic phyla, Actinobacteriota, were both enriched by a pea preceding crop relative to oilseed preceding crops, and so was MBC (Table 2) and arylsulphatase activity (Supplementary Table S1). Borase et al. (2021) also reported that adding the legume mungbean to a rice-wheat rotation increase soil MBC and the activity of arylsulphatase. However, in our study, the reverse was true for the fungal class Leotiomycetes and bacterial phylum Proteobacteriota due to a flax effect (Tables 3, 4). The reason for the enrichment of Leotiomycetes by flax is unknown, but Tan et al. (2017) reported that some Leotiomycetes were potential pathogens for the medicinal crop sanchi (Panax notoginseng) in China. The main difference between pea and oilseed crops is biological nitrogen fixation by the former, and oil production by the latter. In field and greenhouse studies of crop residue decomposition, Lupwayi et al. (2004a, b) reported that soil MBC was greater under decomposing field pea residues than canola residues even though their decomposition pattens were not much different. Many studies have shown positive effects of legume-based crop rotations to the soil microbiome with regard to MBC and diversity (reviewed by Lupwayi and Kennedy, 2007; Schaedel et al., 2021; Liu et al., 2023b), but others have shown that adding many legumes to a rotation can have negative effects (Nayyar et al., 2009; Lupwayi et al., 2012; Bainard et al., 2017). Thus, Bainard et al. (2017) reported that adding more than two legume phases to a 4-yr rotation increased fungal pathogens that included Fusarium avenaceum, F. redolens, and Alternaria spp. One of the oilseed crops, canola, produces glucosinolates that release biocides which act as soil fumigants, thereby affecting the soil microbiome (McCully et al., 2008). In wheat-based cropping systems in Washington State (USA), Hansen et al. (2019) reported that canola reduced soil fungi and gram+ bacteria in some locations. Therefore, different factors can explain the differences between legume and oilseed crops in their effects on the soil microbiome.
Relative to oilseed preceding crops, a barley preceding crop enriched the predominant fungal class Sordariomycetes (Table 3) and one of the least predominant prokaryotic phyla, Latescibacterota (Table 4). These results suggest that these taxa thrive in low-N soil environments because Lupwayi et al. (2004b) reported barley and canola crop residue N concentrations of 4.9 and 10.9 mg g-1, respectively, resulting in C/N ratios of 93 and 41, respectively. The reduced abundance of Sordariomycetes relative to wheat could not be attributed to the soil fumigation effects of canola glucosinolates as the flax-wheat rotation had slightly lower relative abundance of Sordariomycetes than the canola-wheat rotation (Table 3).
4.4 Relationships between microbiome composition and functioning
There were two consistent trends in the relationships between the microbiome composition and enzyme activities. The first one was that the relative abundances of none of the fungal genera were correlated with the activities of N-acetyl-β-glucosaminidase that mediates N cycling. This observation does not necessarily suggest that fungi were not involved in N cycling, but rather that there probably was not much variation in the relative abundances of fungal genera and/or activities of N-acetyl-β-glucosaminidase to reveal relationships between them. Actually, there were no crop rotation or fungicide effects on the activity of this enzyme (Table 5). Lupwayi et al. (2024) reported a negative correlation between the relative abundance of Acremonium spp. and the activity of N-acetyl-β-glucosaminidase. That study involved the insecticide ivermectin which killed some soil microarthropods (but not others) that presumably increased soil chitin, a substrate for N-acetyl-β-glucosaminidase. The second observation was that correlations of the relative abundances of fungal or prokaryotic genera with β-glucosidase and arylsulphatase activities were similar but the opposite of correlations with acid phosphomonoesterase. The implication is that P cycling was less dependent on soil organic C than S cycling, probably because soil P exists in many chemical forms in addition to organic forms. In addition to microbial mineralization of organic P (Raguet et al., 2023), soil P can be made available to plants from chemical pools through chemical dissolution or P-solubilizing microorganisms (Islam et al., 2024).
The bacterial genus Gaiella was the core prokaryotic identifiable (assigned) taxon that was present in 85% of the soil samples (Figure 5) even though its relative abundance was highest in the pea-wheat rotation and lowest in the barley-pea-wheat rotation (Figure 4C). However, the nutrient-cycling role of this bacterium in these rotations is unclear because its relative abundance was not correlated with any enzyme activity. In rice-wheat and sugarcane-wheat rotations in India, this genus was considered as plant-growth-promoting because its relative abundance was positively correlated with availability of N, P, K, Fe, and S (Ansari et al., 2024). However, in corn-soybean rotations and monoculture soybean in China, its relative abundance was negatively correlated with soybean yield (Xu et al., 2024). Therefore, the ecological services provided by Gaiella spp. to cropping systems may be context-dependent.
The relative abundances of the fungal and prokaryotic genera were related to activities of nutrient-cycling enzymes to gauge their contributions to crop nutritional soil health. Other studies have related such abundances to crop protection soil health (Mazzola and Gu, 2002; Stirling et al., 2012; Siegel-Hertz et al., 2018). In a comparison of fusarium wilt suppressive soils with fusarium wilt conducive soils, Siegel-Hertz et al. (2018) reported that the following fungal genera (also found in our study, Table 6) were exclusively found in fusarium-suppressive soils: Acremonium, Mortierella, and Penicillium. The prokaryotic genera that were more abundant in fusarium-suppressive soils than in fusarium-conducive soils (also in our Supplementary Table S1) were: Massilia, Arthrobacter, and Rubrobacter. Therefore, some of the genera identified in our study were probably involved in nutrient (N, P, and S) cycling and biological disease control.
5 Conclusion
We obtained valuable information on crop rotation and fungicide effects on soil microbial biomass, the composition and diversity of prokaryotic communities, and enzyme activities even though the study period was probably not long enough for 3-yr rotations to shape the soil microbiome. Whereas fungicide applications reduced fungal richness in the wheat rhizosphere (e.g., Chao1 indices of 64.0 vs. 79.9), especially in 2-yr rotations, rotation length/type and the crops that preceded wheat had different effects on the relative abundances of different taxa. Two of the three most predominant prokaryotic phyla, Proteobacteriota and Actinobacteriota, responded differently to rotation length: 3-yr rotations enriched the former (27.4% vs. 20.1% relative abundances), but 2-yr rotations enriched the latter (19.9% vs. 28.3% relative abundances). Relative to oilseed crops preceding the sampled wheat, a pea preceding crop enriched Actinobacteriota (31.7% vs. 24.8% relative abundances) and the most abundant fungal class, Sordariomycetes (39.1% vs. 22.1% relative abundances), in addition to increasing microbial biomass carbon (MBC) and arylsulphatase activity by 33% and 57%, respectively. Correlations of the relative abundances of fungal or prokaryotic genera with β-glucosidase and arylsulphatase activities were both either positive or negative, but the opposite of correlations with acid phosphomonoesterase, suggesting that S cycling was more linked to C cycling than P cycling. Besides the crop nutritional (nutrient cycling) significance of these soil microbial characteristics, there is need top study their biological disease control aspects.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
NL: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Project administration, Supervision, Visualization, Writing – original draft, Writing – review & editing. TT: Conceptualization, Data curation, Funding acquisition, Investigation, Methodology, Project administration, Supervision, Writing – review & editing. BT: Data curation, Investigation, Project administration, Supervision, Writing – review & editing. HK: Data curation, Project administration, Resources, Writing – review & editing. RO: Data curation, Formal analysis, Methodology, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was financially supported by Western Grains Research Foundation and Agriculture and Agri-Food Canada.
Acknowledgments
We are grateful to Noryne Rauhala, Jackie Busaan, Deb Clark, and annual summer students for their technical assistance in managing the field crops, fungicide application, and soil sampling, as well as the assistance of Larry Michielsen, Elizabeth Sroka, Jennifer Zuidhof, and Patty Reid with plot seeding, herbicide application and harvesting. We also thank Andrea Brown and Derrick Kanashiro for assistance with soil analysis, and to Jackie Zorz and Mathew Richards for bioinformatics analysis. This study was financially supported by Western Grains Research Foundation and Agriculture and Agri-Food Canada.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fagro.2024.1455448/full#supplementary-material
References
Ansari W. A., Kumar M., Krishna R., Singh A., Zeyad M. T., Tiwari P., et al. (2024). Influence of rice-wheat and sugarcane-wheat rotations on microbial diversity and plant growth promoting bacteria: Insights from high-throughput sequencing and soil analysis. Microbiol. Res. 278, 127533. doi: 10.1016/j.micres.2023.127533
Bainard L. D., Navarro-Borrell A., Hamel C., Braun K., Hanson K., Gan Y. (2017). Increasing the frequency of pulses in crop rotations reduces soil fungal diversity and increases the proportion of fungal pathotrophs in a semiarid agroecosystem. Agric. Ecosyst. Environ. 240, 206–214. doi: 10.1016/j.agee.2017.02.020
Baldwin-Kordick R., De M., Lopez M. D., Liebman M., Lauter N., Marino J., et al. (2022). Comprehensive impacts of diversified cropping on soil health and sustainability. Agroecol. Sustain. Food Syst. 46, 331–363. doi: 10.1080/21683565.2021.2019167
Bellemain E., Carlsen T., Brochmann C., Coissac E., Taberlet P., Kauserud H. (2010). ITS as an environmental DNA barcode for fungi: an in silico approach reveals potential PCR biases. BMC Microbiol. 10, 189. doi: 10.1186/1471-2180-10-189
Bo-Ra K., Jiwon S., Robin B. G., Jun Hyung L., Doo Wan K., Kuk-Hwan S., et al. (2017). Deciphering diversity indices for a better understanding of microbial communities. J. Microbiol. Biotechnol. 27, 2089–2093. doi: 10.4014/jmb.1709.09027
Borase D. N., Murugeasn S., Nath C. P., Hazra K. K., Singh S. S., Kumar N., et al. (2021). Long-term impact of grain legumes and nutrient management practices on soil microbial activity and biochemical properties. Arch. Agron. Soil Sci. 67, 2015–2032. doi: 10.1080/03650340.2020.1819532
Bünemann E. K., Schwenke G. D., Van Zwieten L. (2006). Impact of agricultural inputs on soil organisms -a review. Australian J. Soil Res. 44, 379–406. doi: 10.1071/SR05125
Callahan B. J., McMurdie P. J., Rosen M. J., Han A. W., Johnson A. J. A., Holmes S. P. (2016). DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583. doi: 10.1038/nmeth.3869
Caporaso J. G., Lauber C. L., Walters W. A., Berg-Lyons D., Lozupone C. A., Turnbaugh P. J., et al. (2011). Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. U.S.A. 108, 4516–4522. doi: 10.1073/pnas.1000080107
Chong J., Liu P., Zhou G., Xia J. (2020). Using MicrobiomeAnalyst for comprehensive statistical, functional, and meta-analysis of microbiome data. Nat. Protoc. 15, 799–821. doi: 10.1038/s41596-019-0264-1
Deng S., Kang H., Freeman C. (2011). “Microplate fluorometric assay for soil enzymes,” in Methods in Soil Enzymology. Ed. Dick R. P. (Madison, WI: Soil Science Society of America), 311–318.
Dick R. P., Breakwell D. P., Turco R. F. (1996). “Soil enzyme activities and biodiversity measurements as integrative microbiological indicators,” in Methods for assessing soil quality. Eds. Doran J. W., Jones A. J. (Madison, WI: Soil Science Society of America), 247–271. Special Publication Number 49.
Gweon H. S., Oliver A., Taylor J., Booth T., Gibbs M., Read D. S., et al. (2015). PIPITS: an automated pipeline for analyses of fungal internal transcribed spacer sequences from the Illumina sequencing platform. Methods Ecol. Evol. 6, 973–980. doi: 10.1111/mee3.2015.6.issue-8
Han L., Kong X., Xu M., Nie J. (2021). Repeated exposure to fungicide tebuconazole alters the degradation characteristics, soil microbial community and functional profiles. Environ. pollut. 287, 117660. doi: 10.1016/j.envpol.2021.117660
Hansen J. C., Schillinger W. F., Sullivan T. S., Paulitz T. C. (2019). Soil microbial biomass and fungi reduced with canola introduced into long-term monoculture wheat rotations. Front. Microbiol. 10, 1488. doi: 10.3389/fmicb.2019.01488
Horwath W. R., Paul E. A. (1994). “Microbial biomass,” in Methods of Soil Analysis. Part 2. Microbiological and Biochemical Properties. Eds. Weaver R. W., Angle S., Bottomly P., Bezdicek D., Smith S., Tabatabai A., Wollum A. (Madison, WI: Soil Science Society of America), 753–773.
Huo D., Malacrinò A., Lindsey L. E., Benitez M.-S. (2023). Subtle responses of soil bacterial communities to corn-soybean-wheat rotation. Phytobiomes J. 7, 392–400. doi: 10.1094/PBIOMES-05-22-0032-R
Islam M., Siddique K. H. M., Padhye L. P., Pang J., Solaiman Z. M., Hou D., et al. (2024). A critical review of soil phosphorus dynamics and biogeochemical processes for unlocking soil phosphorus reserves. Adv. Agron. 185, 153–249. doi: 10.1016/bs.agron.2024.02.004
Larney F. J., Pearson D. C., Blackshaw R. E., Lupwayi N. Z. (2017). Soil changes over 12 yr of conventional vs. conservation management on irrigated rotations in southern Alberta. Can. J. Soil Sci. 97, 249–265. doi: 10.1139/cjss-2016-0141
Liu C., Plaza-Bonilla D., Coulter J. A., Kutcher H. R., Beckie H. J., Wang L., et al. (2022). Diversifying crop rotations enhances agroecosystem services and resilience. Adv. Agron. 173, 299–335. doi: 10.1016/bs.agron.2022.02.007
Liu Q., Li T., Chen L., Zhang S., Zhao Y., Chen Y., et al. (2023a). Taxonomic and functional diversity of the soil microbiome recruited from alternative crops in a rotation system. Eur. J. Soil Sci. 74, e13410. doi: 10.1111/ejss.13410
Liu Q., Zhao Y., Li T., Chen L., Chen Y., Sui P. (2023b). Changes in soil microbial biomass, diversity, and activity with crop rotation in cropping systems: A global synthesis. Appl. Soil Ecol. 186, 104815. doi: 10.1016/j.apsoil.2023.104815
Lloyd A. W., Percival D., Yurgel S. N. (2021). Effect of fungicide application on lowbush blueberries soil microbiome. Microorganisms 9, 1366. doi: 10.3390/microorganisms9071366
Lu Y., Zhou G., Ewald J., Pang Z., Shiri T., Xia J. (2023). MicrobiomeAnalyst 2.0: comprehensive statistical, functional and integrative analysis of microbiome data. Nucleic Acids Res. 51, W310–W318. doi: 10.1093/nar/gkad407
Lupwayi N. Z., Clayton G. W., O’Donovan J. T., Harker K. N., Turkington T. K., Rice W. A. (2004a). Decomposition of crop residues under conventional and zero tillage. Can. J. Soil Sci. 84, 403–410. doi: 10.4141/S03-082
Lupwayi N. Z., Clayton G. W., O’Donovan J. T., Harker K. N., Turkington T. K., Rice W. A. (2004b). Soil microbiological properties during decomposition of crop residues under conventional and zero tillage. Can. J. Soil Sci. 84, 411–419. doi: 10.4141/S03-083
Lupwayi N. Z., Floate K. D., Petri R. M. (2024). The veterinary parasiticide ivermectin increased the activity of an enzyme that mediates soil chitin degradation on a prairie grassland. Appl. Soil Ecol. 197, 105338. doi: 10.1016/j.apsoil.2024.105338
Lupwayi N. Z., Kennedy A. C. (2007). Grain legumes in Northern Great Plains: impacts on selected biological soil processes. Agron. J. 99, 1700–1709. doi: 10.2134/agronj2006.0313s
Lupwayi N. Z., Lafond G. P., May W. E., Holzapfel C. B., Lemke R. L. (2012). Intensification of field pea production: impact on soil microbiology. Agron. J. 104, 1189–1196. doi: 10.2134/agronj2012.0046
Lupwayi N. Z., Zhang Y., Hao X., Thomas B. W., Eastman A. H., Schwinghamer T. D. (2019). Linking soil microbial biomass and enzyme activities to long-term manure applications and their nonlinear legacy. Pedobiol 74, 34–42. doi: 10.1016/j.pedobi.2019.04.001
Mazzola M., Gu Y. H. (2002). Wheat genotype-specific induction of soil microbial communities suppressive to disease incited by Rhizoctonia solani anastomosis group AG-5 and AG-8. Phytopathol. 92, 1300–1307. doi: 10.1094/PHYTO.2002.92.12.1300
McCully M. E., Miller C., Sprague S. J., Huang C. X., Kirkegaard J. A. (2008). Distribution of glucosinolates and sulphur-rich cells in roots of field-grown canola (Brassica napus). New Phytol. 180, 193–205. doi: 10.1111/j.1469-8137.2008.02520.x
Meyer C., Jeanbille M., Breuil M.-C., Bru D., Höfer K., Screpanti C., et al. (2024). Soil microbial community fragmentation reveals indirect effects of fungicide exposure mediated by biotic interactions between microorganisms. J. Hazard. Mater. 470, 134231. doi: 10.1016/j.jhazmat.2024.134231
Nayyar A., Hamel C., Lafond G., Gossen B. D., Hanson K., Germida J. (2009). Soil microbial quality associated with yield reduction in continuous-pea. Appl. Soil Ecol. 43, 115–121. doi: 10.1016/j.apsoil.2009.06.008
Raguet P., Cade-Menun B., Mollier A., Abdi D., Ziadi N., Karam A., et al. (2023). Mineralization and speciation of organic phosphorus in a sandy soil continuously cropped and phosphorus-fertilized for 28 years. Soil Biol. Biochem. 178, 108938. doi: 10.1016/j.soilbio.2022.108938
Riedo J., Yokota A., Walther B., Bartolomé N., van der Heijden M. G. A., Bucheli T. D., et al. (2023). Temporal dynamics of total and bioavailable fungicide concentrations in soil and their effect upon nine soil microbial markers. Sci. Total Environ. 878, 162995. doi: 10.1016/j.scitotenv.2023.162995
Roman D. L., Voiculescu D. I., Filip M., Ostafe V., Isvoran A. (2021). Effects of triazole fungicides on soil microbiota and on the activities of enzymes found in soil: a review. Agriculture 11, 893. doi: 10.3390/agriculture11090893
Schaedel M., Hidrobo G., Grossman J. (2021). From microns to meters: exploring advances in legume microbiome diversity for agroecosystem benefits. Front. Sustain. Food Syst. 5, 668195. doi: 10.3389/fsufs.2021.668195
Siegel-Hertz K., Edel-Hermann V., Chapelle E., Terrat S., Raaijmakers J. M., Steinberg C. (2018). Comparative microbiome analysis of a fusarium wilt suppressive soil and a fusarium wilt conducive soil from the Châteaurenard Region. Front. Microbiol. 9, e568. doi: 10.3389/fmicb.2018.00568
Stirling G. R., Smith M. K., Smith J. P., Stirling A. M., Hamill S. D. (2012). Organic inputs, tillage and rotation practices influence soil health and suppressiveness to soilborne pests and pathogens of ginger. Aust. Plant Pathol. 41, 99–112. doi: 10.1007/s13313-011-0096-0
Tan Y., Cui Y., Li H., Kuang A., Li X., Wei Y., et al. (2017). Rhizospheric soil and root endogenous fungal diversity and composition in response to continuous Panax notoginseng cropping practices. Microbiol. Res. 194, 10–19. doi: 10.1016/j.micres.2016.09.009
Vasilchenko A. V., Poshvina D. V., Semenov M. V., Timofeev V. N., Iashnikov A. V., Stepanov A. A., et al. (2023). Triazoles and strobilurin mixture affects soil microbial community and incidences of wheat diseases. Plants 12, 660. doi: 10.3390/plants12030660
Venter Z. S., Jacobs K., Hawkins H.-J. (2016). The impact of crop rotation on soil microbial diversity: A meta-analysis. Pedobiologia 59, 215–223. doi: 10.1016/j.pedobi.2016.04.001
Xu L., Jin S., Su Y., Lyu X., Yan S., Wang C., et al. (2024). Combined metagenomics and metabolomic analysis of microbial community structure and metabolic function in continuous soybean cropping soils of Songnen Plain, China. Chem. Biol. Technol. Agric. 11, 46. doi: 10.1186/s40538-024-00569-x
Keywords: soil microbial diversity, soil nutrient cycling, soil enzyme activities, pesticides, fungicide
Citation: Lupwayi NZ, Turkington TK, Tidemann BD, Kubota H and Polo RO (2024) Crop rotation and fungicides impact microbial biomass, diversity and enzyme activities in the wheat rhizosphere. Front. Agron. 6:1455448. doi: 10.3389/fagro.2024.1455448
Received: 26 June 2024; Accepted: 11 October 2024;
Published: 12 November 2024.
Edited by:
Anna De Marco, University of Naples Federico II, ItalyReviewed by:
Qiongli Bao, Ministry of Agriculture and Rural Affairs, ChinaPasquale Napoletano, University of Molise, Italy
Copyright © 2024 His Majesty the King in Right of Canada, as represented by the Minister of Agriculture and Agri-Food Canada for the contribution of N.Z. Lupwayi, T.K. Turkington, B.D.Tidemann, H. Kubota, R.O. Polo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Breanne D. Tidemann, YnJlYW5uZS50aWRlbWFubkBhZ3IuZ2MuY2E=
 Newton Z. Lupwayi
Newton Z. Lupwayi T. Kelly Turkington2
T. Kelly Turkington2 Breanne D. Tidemann
Breanne D. Tidemann Hiroshi Kubota
Hiroshi Kubota