- 1Department of Crop and Soil Sciences, Washington State University, Pullman, WA, United States
- 2Department of Crop Health, University of Rostock, Rostock, Germany
- 3Department of Crop and Soil Science, Oregon State University, Corvallis, OR, United States
- 4United States Department of Agriculture, Agricultural Research Service, Cereal Disease Laboratory, University of Minnesota, St. Paul, MN, United States
Cereal rust diseases, including leaf, stem, and stripe rust, are some of the most devastating and economically important diseases of barley. However, host–pathogen genetic interaction research for each pathosystem is typically conducted independently and in isolation. Examples of host resistance/susceptibility genes functioning sympathetically to multiple pathogens or antagonistically to additional pathogens have been reported. Therefore, consolidation of loci that have been reported in multiple studies and across pathosystems is useful for variety development to maximize resistance to multiple pathogens and avoid inadvertent incorporation of susceptibility loci that act antagonistically to other pathogens. This review summarizes loci reported in three key biotrophic pathosystems of barley, including leaf, stem, and stripe rust. In conjunction with previously consolidated net blotch loci, this review lays the foundation for a wider barley rust resistance/susceptibility atlas. This review aims to inform breeders and researchers in rapidly identifying accessions and loci that need further characterization and which loci would be most useful to introgress into elite varieties.
Introduction
Cereals are staple crops the world over, providing calories and protein to the growing global population (Hubbard et al., 2015; Kearney, 2010). Currently, barley ranks as the second most-produced temperate cereal in the world after wheat (Thiel et al., 2021). Barley is primarily used as animal feed yet is the key ingredient in brewing beer and distilling premium whiskey by providing the necessary nutrients and enzymes for alcohol production (Sayre-Chavez et al., 2022). Beer ranks third for the most consumed beverage worldwide behind water and tea (Salanță et al., 2020), and its consumption plays a major role in the social fabric in many parts of the world. Thus, a secure supply of malting quality barley is essential to support the multibillion-dollar added value industry. Barley is also reemerging as a food crop due to its human health benefits (Tosh and Bordenave, 2020), and efforts are underway to develop modern biofortified and heart-healthy hulless barley varieties.
A major constraint to barley yield and quality is disease outbreaks. Due to climate change, environmental conditions have led to more severe epidemics in important barley production regions, preventing the crops from reaching their full potential and requiring additional inputs for disease management and mitigation (Dean et al., 2012; Xie et al., 2018). Rusts have been a major bane of cereal production and a part of human history since the beginning of agriculture. The Romans performed sacrificial ceremonies of Robigalia to appease the god of cereal rust diseases, Robigus (Saunders et al., 2019). In addition, all rust species mentioned in this review are macrocyclic and heteroecious (Bolton et al., 2008; Jin et al., 2010; Leonard and Szabo, 2005). Thus, countries have performed extensive eradication programs of barberry, the alternative host to multiple rust species, in an attempt to prevent sexual recombination and stabilize rust populations to slow the evolution of new races of rust species (Saunders et al., 2019). Although not calculated for barley, the economic impact of rusts on wheat is estimated to be $4.3–5.0 billion, 3.5% of the $145 billion economic value of worldwide wheat (Figueroa et al., 2018).
Cereal rust species are members of the genus Puccinia from the basidiomycota division of fungi that are obligate biotrophs (Bolton et al., 2008; Jin et al., 2010; Jin and Steffenson, 1999; Leonard and Szabo, 2005). There are four primary species of rust known to infect barley (Table 1): Puccinia coronata var. hordei (Pch), the causal agent of crown rust; Puccinia hordei (Ph), the causal agent of the leaf (brown) rust; Puccinia striiformis f. sp. hordei (Psh), the causal agent of the stripe (yellow) rust; and Puccinia graminis f. sp. tritici (Pgt), the causal agent of the stem (black) rust, all showing close phylogenetic relationships with other cereal rusts (Brueggeman et al., 2020; Dracatos et al., 2019b; Park et al., 2015). The notion of formae speciales (ff. spp.) separating specialized forms of a species that infect specific host species was developed by Eriksson (1894). However, this specialization does not hold true for some formae speciales, as the wheat stem rust pathogen Pgt also infects barley. Depending on local fluctuations in pathogen virulence and the deployment of inadequate resistance loci, leaf, stem, and stripe rust of barley all have the potential to cause incredibly damaging epidemics (Dean et al., 2012; Kleinhofs et al., 2009). In comparison, crown rust does not cause significant yield loss (Jin et al., 1992; Jin and Steffenson, 1999), and therefore is relatively understudied.
Two types of resistance are described within cereal rust pathosystems: seedling/all-stage resistance (ASR) effective throughout the plant’s life cycle and adult plant resistance (APR) effective at the postseedling stage. ASR genes are often considered race-specific and associated with the hypersensitive response, whereas APR genes are often incomplete, providing additive resistance (Dracatos et al., 2015a), that can result in near-immunity when sufficiently stacked (Huerta-Espino et al., 2020). Some resistance/susceptibility genes are involved in interactions with multiple pathogens, whether as sympathetic resistance to multiple diseases or antagonistic relationships of resistance/susceptibility to biotrophic and necrotrophic pathogens. Therefore, the ability to report colocalization of newly reported loci currently requires intimate knowledge of respective pathosystems or an extensive and therefore time-consuming literature review. Previously, the Barley Genetics Newsletters provided an excellent resource for this purpose; however, with high-resolution mapping and the sheer abundance of loci reported in the current barley community, this easily becomes unmanageable. Similar consolidations have been achieved for various pathogens of wheat (Amo and Soriano, 2022; Jan et al., 2021; Pal et al., 2022; Peters Haugrud et al., 2022; Soriano and Royo, 2015; Yu et al., 2014; Zheng et al., 2021). This review consolidates resistance/susceptibility loci for the three highly devastating pathogens of barley leaf, stem, and stripe rust and the lesser pathogen crown rust against the Morex V3 reference genome (Mascher et al., 2021) to lay a foundation for a larger resistance/susceptibility atlas incorporating the net blotch consensus maps (Clare et al., 2020).
Genetics to crown rust
Crown rust was first identified in 1991 in the USA (Jin et al., 1992) and designated a variety of P. coronata in 1999 (Jin and Steffenson, 1999). Barley crown rust has currently only been documented in the USA, Hungary, and China (Tian et al., 2021). To date, only the Reaction to Puccinia coronata 1 (Rpc1) locus has been reported against Pch on chromosome 3H in HOR 2596 (Agrama et al., 2004). Rpc1 was reported to colocalize with Rph5, Rph6, and Rph7 (leaf rust); Rp1-D (stem rust); Run6 (loose smut); rym4 and rym5 (barley mild mosaic virus); and Ryd2 (barley yellow dwarf virus) at the time (Agrama et al., 2004; Jin and Steffenson, 2002). However, refinement of Rph5, Rph6, and Rph7 loci has shown that Rpc1 colocalizes with Rph10 (Martin et al., 2020). Candidate gene analysis for Rpc1 is limited due to the use of proprietary markers that encompass a large 314 Mb (175.45 to 489.01 Mb) genomic interval (Supplementary Table 1).
Genetics to leaf rust
Leaf rust is often reported as one of the most devastating barley diseases, with yield losses up to 62% and routinely contributing up to 30% yield loss under conditions conducive to epidemic formation (Cotterill et al., 1992; Griffey et al., 1994; King and Polley, 1976). Despite barley infections primarily being Ph, leaf rust can also be caused by P. triticina (Pt), the causal agent of wheat leaf rust (Kleinhofs et al., 2009). The genetics of leaf rust resistance have been studied since the 1920s (Fazlikhani et al., 2019), with at least 80 Leaf rust (Lr) (Prasad et al., 2020) and 28 Reaction to Puccinia hordei (Rph; Table 2) loci (Mehnaz et al., 2021a) identified within wheat and barley, respectively. Additionally, there are a series of Rphq partial resistance loci with minor effects (Marcel et al., 2007, 2008; Qi et al., 1998, 1999, 2000; Yeo et al., 2017), not to be confused with the set of RphQ loci (Ziems et al., 2014). Rphq1-10 and Rphq20-21 were first identified in L94 × Vada, Rphq11-13 in L94 × 116-5, Rphq14-15 and Rphq22-23 in Steptoe × Morex, and Rphq16-19 in Oregon Wolfe Barley (Marcel et al., 2007, 2008; Qi et al., 1998, 1999, 2000; Yeo et al., 2017).
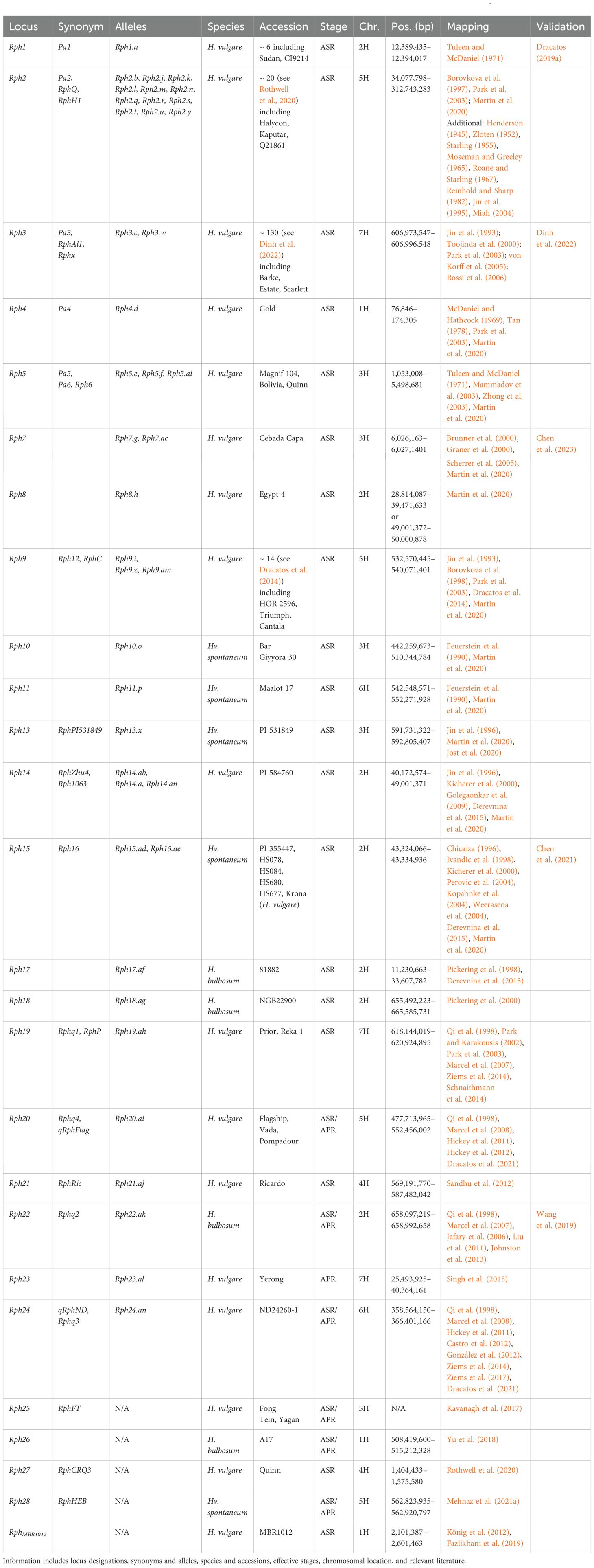
Table 2. Summary information of all designated resistance/susceptibility loci in barley to leaf rust pathogen Puccinia hordei.
Due to the number of reported loci, barley Rph loci appear complex; however, further research has revealed multiple Rph genes are allelic variants. In addition, Rphq loci are often elevated and reclassified into Rph loci. Therefore, the true number of Rph loci currently stands at 25 (Martin et al., 2020). The RphD locus is not assigned to any chromosome (Marcel et al., 2008). Additionally, two collections of diverse barley lines were found to carry uncharacterized leaf rust resistance (Mehnaz et al., 2021b; Verma et al., 2018). There are also three QTL detected within a population between Blenheim and E224/3, but marker intervals were not reported (Thomas et al., 1995).
Chromosome 1H
Rph4 was first mapped to the short arm of chromosome 1H (McDaniel and Hathcock, 1969; Qi et al., 1998; Tan, 1978). Additionally, RphMBR1012, when first mapped, was not suspected to be Rph4 (König et al., 2012). Further work delimited RphMBR1012 to a 500 kb interval (2.1–2.6 Mb) in close proximity to Rph4 (Fazlikhani et al., 2019). Subsequent work delimited Rph4 to a 97-kb interval (0.08–0.17 Mb), distal to RphMBR1012, and therefore is a distinct locus (Martin et al., 2020). Rph26 was identified in the wild bulbous barley line A17 (Yu et al., 2018), mapping to a 7.2-Mb interval (508.42–515.21 Mb) that was also mapped in previous studies (Gutiérrez et al., 2015; Hickey et al., 2011). Additional loci identified on chromosome 1H include RphQ1, RphQ3, and RphQ4 (Ziems et al., 2014); qGH_PBIC_3.91 (Dracatos et al., 2019b); Qlr.HeB-5-1H (Schnaithmann et al., 2014), Rphq14 (Yeo et al., 2017); Rphq21 (Marcel et al., 2008); and Rphq22 (Yeo et al., 2017).
Chromosome 2H
The Rph1 gene was first reported in the experimental line Minn. II 12.15 and cultivar Sudan (Roane and Starling, 1967; Watson and Butler, 1947) on chromosome 2H (Tuleen and McDaniel, 1971). Rph1 is highly likely to have been mapped as qField_PBIC_2016_3.14 (Dracatos et al., 2019b), QPh.2H-1 (Vatter et al., 2017), and QRph5 (Ziems et al., 2014). Rph1 encodes a coiled-coil (CC) nucleotide binding site leucine-rich repeat (NLR) receptor protein (Dracatos et al., 2019a). Phylogenetic classification of RPH1 places the NLR in the C9 clade, which includes the rice NLR Pik-2 that functions with the NLR Pik-1 (C12 clade) to confer resistance to Magnaporthe oryzae (Ashikawa et al., 2008; Bailey et al., 2018). Sequencing of rph1 mutants determined five mutants carry Rph1 mutations, whereas two mutants lacked mutations in Rph1 (Dracatos et al., 2019a). This observation supports the hypothesis that additional loci, such as a second NLR, are required for Rph1-mediated resistance. Rph8 was first described in Egypt 4 as an ASR locus (Tan, 1977), despite Egypt 4 previously being used as a susceptible check (Martin et al., 2020). The Rph8 locus remains elusive and understudied due to the lack of avirulent isolates on Rph8 (Jin et al., 1996). As Egypt 4 is the only known source of Rph8, introgression into Bowman delimited the locus to a 10.6-Mb (28.81–39.24 Mb) or 1.0-Mb (49.00–50.00 Mb) interval under the assumption that Rph8 is not allelic to Rph14/15/16/17 (Martin et al., 2020). This region is highly complex, with current mapping efforts delimiting Rph8, Rph14, Rph15, Rph16, and Rph17 to the same region (Derevnina et al., 2015; Martin et al., 2020). Rph14 was identified in barley accession PI584760 (Jin et al., 1996) and mapped to an 8.8-Mb interval (40.17–49.00 Mb) (Golegaonkar et al., 2009; Martin et al., 2020). In contrast, Rph15 was first identified in wild accession PI355447 (Chicaiza, 1996), and Rph16 in wild accessions HS078, HS084 (Ivandic et al., 1998), HS688 (Perovic et al., 2004), and potentially HS677 (Kopahnke et al., 2004), as well as landrace HOR1063 (Kicherer et al., 2000). Subsequently, Rph15 and Rph16 were confirmed to be allelic (Weerasena et al., 2004), whereas Rph14 was believed to be an independent locus based on segregation ratios (Derevnina et al., 2015). More recent studies dispute this, suggesting Rph14 may be allelic to Rph15/16 with the single susceptible individual separating Rph14 from Rph15/16 as a potential admixture (Derevnina et al., 2015). Therefore, Rph14, Rph15, and Rph16 are predicted to be an allelic series (Derevnina et al., 2015), of which Rph15/16 has already been validated as encoding a CC-NLR with an integrated zinc finger BED domain (Chen et al., 2021), located at 43.32–43.33 Mb. RPH15 is found in clade C24 within a subclade of related NLRs with N-terminal integrated zinc finger BED domains, including the resistance proteins Xa1, Xo1, Yr5, Yr7, and YrSP (Bailey et al., 2018; Marchal et al., 2018; Read et al., 2020; Yoshimura et al., 1998). These results suggest five potential alleles: Rph14.ab and Rph14.am (previously RphZhu4); Rph14.an (previously Rph1063) (Kicherer et al., 2000); and Rph15.ad and Rph15.ae (previously Rph16) (Derevnina et al., 2015; Martin et al., 2020). The allele designations.am and.an had already been applied to Rph9.am (Dracatos et al., 2014) and Rph24.an (Ziems et al., 2017) and therefore need to be addressed in the future. Rph17 is confirmed to be independent of Rph14/15/16 (Derevnina et al., 2015), mapping to a 22.4-Mb interval (11.23–33.61 Mb) using H. bulbosum × H. vulgare hybrids (Pickering et al., 1998). However, Rph17 currently encompasses Rph1 (Dracatos et al., 2019a) and the proximal end of Rph8 (Martin et al., 2020). Additionally, Rph17 is known to cosegregate with Mildew locus from Hordeum bulbosum (Mlhb), a powdery mildew resistance locus (Pickering et al., 1998). Lastly, RphQ7 maps in close proximity to this complex region (Ziems et al., 2014). Rph18 was also mapped using H. bulbosum × H. vulgare hybrids; however, it mapped to a 10.1-Mb interval (655.49–665.59 Mb) (Pickering et al., 2000), encompassing the validated Rph22 gene (Wang et al., 2019). Rph22 was originally mapped as Rphq2 (Qi et al., 1998) but was subsequently high-resolution mapped (Johnston et al., 2013) and identified as a lectin receptor-like kinase (Wang et al., 2019). Rph22 was also most likely mapped by association mapping studies as QPh.2H-2 (Vatter et al., 2017) and two unnamed QTL (Czembor et al., 2022; Gutiérrez et al., 2015). Both the wild bulbous barley and cultivated barley alleles confer stronger resistance responses to bulbous and cultivated barley leaf rust isolates, respectively (Wang et al., 2019). Additional loci mapping to chromosome 2H include Qlr.HeB-F23-2H (Schnaithmann et al., 2014); Rphq11 and Rphq12 (Qi et al., 2000; Yeo et al., 2017); QLr.S42-2H.a and QLr.S42-2H.b (von Korff et al., 2005); and two unnamed QTL (Amouzoune et al., 2022; Castro et al., 2012).
Chromosome 3H
Initially, the loci Rph5, Rph6, and Rph7 were believed to be dispersed along chromosome 3H (Zhong et al., 2003); however, Rph5 and Rph6 are currently hypothesized to be allelic variants, whereas Rph7 remains an independent locus. The Rph5/Rph6 and Rph7 loci are delimited to 4.5-Mb (1.05–5.50 Mb) and 134-kb intervals (5.98–6.12 Mb), respectively, using introgression mapping (Martin et al., 2020). Subsequently, Rph7 was identified as a NAC transcription factor at ~ 6.03 Mb (Chen et al., 2023). Four of eight mutants carried mutations in Rph7, indicating that additional loci are involved in Rph7-mediated resistance (Chen et al., 2023). Rph10 and Rph11 were originally mapped to chromosomes 3H and 6H within wild barley accessions Bar Giyyora 30 and Maalot 17, respectively (Feuerstein et al., 1990). Rph10 was subsequently mapped to an 18.1-Mb (442.26–510.34 Mb) interval (Martin et al., 2020). However, within the Rph10 backcross accession BW683, there is additional donor DNA in close proximity (~ 5.9 Mb) to Rph11 on chromosome 6H. Therefore, there is debate as to whether Rph10 was present in previous mapping studies due to the paucity of marker saturation at the time (Feuerstein et al., 1990) and that resistance may have been contributed by Rph11 (Martin et al., 2020).
Rph13 was identified in the experimental line PI531849, derived from a wild barley accession backcrossed to the British cultivar Berac (Martin et al., 2020). Rph13 was confirmed to be distinct from Rph1-12 (Jin et al., 1996) and mapped to a 1.1-Mb interval (591.73–592.81 Mb) (Jost et al., 2020). Additional loci mapping to chromosome 3H include qRphFra-3H (D., Singh et al., 2015), Qlr.HeB-F23-3H (Schnaithmann et al., 2014), qGH_PBIC_3.86 (Dracatos et al., 2019b), and three unnamed QTL (Czembor et al., 2022; Hickey et al., 2011). Lastly, several QTL map to chromosome 3H but have been unanchored, including Rphq17 and Rphq20 (Marcel et al., 2007); Rphq23 (Yeo et al., 2017); QLr.S42-3H.a (von Korff et al., 2005); and three unnamed QTL (Castro et al., 2012; Rossi et al., 2006; Thomas et al., 1995).
Chromosome 4H
Rph21 (RphRic) was identified in Ricardo (Sandhu et al., 2012), mapping to an 18.2-Mb interval (569.19–587.48 Mb). Unnamed QTL (Hickey et al., 2011) and QPh.4H-1 and QPh.4H-2 (Vatter et al., 2017) also map in close proximity to Rph21. Rph27 identified in cultivar Quinn provides limited value, only being effective against two pathotypes. In addition, due to the fact that DArTseq markers without sequence or positions were utilized, Rph27 was positioned using the gene HORVU.MOREX.r3.4HG0331680 and the full-length cDNA clone AK250035.1 (Rothwell et al., 2020) to a 350-kb interval between 1.40 and 1.76 Mb. As 20 DArTseq markers were determined to be in complete linkage with Rph27 (Rothwell et al., 2020), the Rph27 region is most likely larger, and further high-resolution mapping will be required before validation. Additional loci mapping to chromosome 4H include Qlr.HeB-5-4H and Qlr.HeB-F23-4H (Schnaithmann et al., 2014); Rphq10 (Qi et al., 1999); Rphq19 (Marcel et al., 2007); Rphq20 (Marcel et al., 2008); and QLr.S42-4H.a (von Korff et al., 2005). Lastly, two loci map to chromosome 4H but remain unanchored in Rphq5 (Qi et al., 1998) and Rphq8 (Ziems et al., 2014).
Chromosome 5H
Rph2 (RphQ) was first identified in Halycon, Kaputar, and Q21861 (Borovkova et al., 1997; Park et al., 2003) and is currently delimited to a large 278-Mb interval (34.07–312.74 Mb) (Martin et al., 2020). Currently, there are at least 12 allelic variants of the Rph2 locus present in at least 20 barley accessions (Table 2). However, as this locus is further refined, allelic variants may in fact be independent genes separating Rph2 into multiple loci due to the paucity of markers and small population size. Due to the large interval currently encompassing Rph2, it may also correspond to qField_PBIC_2018_rep1_11.79, qField_PBIC_2016_3.99, qField_PBIC_2018_rep2_17.45, and qGH_PBIC_9.67 (Dracatos et al., 2019b); RphQ9 (Ziems et al., 2014); and two unnamed QTL (Czembor et al., 2022; Schnaithmann et al., 2014).
The Rph9 locus identified in Ethiopian landrace HOR 2596 was designated after allelism tests with Rph1-8 donor accessions (Tan, 1977). The Rph12 locus was subsequently mapped to chromosome 5HL using Triumph and believed to be distinct from Rph9 based on both different reaction types and segregation with allelism tests with HOR 2596 (Rph9) (Jin et al., 1993). However, using a population of 3,858 F2 lines of Triumph × HOR 2596, no segregation could be identified and determined Rph9 (Rph9.i) and Rph12 (Rph9.z) to be allelic (Borovkova et al., 1998). Rph12 has also been identified in Franklin and Tallon (Park et al., 2003). The cultivar Cantala was subsequently identified to have an additional allele of Rph9 (Rph9.am) (Dracatos et al., 2014). The Rph9/12 locus is delimited to a 7.5-Mb interval (532.57–540.07 Mb) using introgression mapping within Bowman (Martin et al., 2020).
Rph20 (qRphFlag) and Rph24 (qRphND) were simultaneously mapped (Dracatos et al., 2021; Hickey et al., 2011); however, since intervals were not reported, determining the localization is troublesome. In addition, the DArT markers mapping Rph20 do not have significant homology to chromosome 5H and align to chromosome 4H in all Morex assemblies (Mascher et al., 2017, 2021; Monat et al., 2019). However, using lower-quality BLAST hits, Rph20 could be anchored to a 74.7-Mb interval (477.71–552.46 Mb) on chromosome 5H. Rph20 is believed to have been sourced from H. laevigatum or Gull that is subsequently present in derived accession Vada (Golegaonkar et al., 2009; Hickey et al., 2011; Hickey et al., 2012) and previously mapped as Rphq4 (Liu et al., 2011; Qi et al., 1998). Diagnostic markers developed to track Rph20 (Dracatos et al., 2021), map to chromosome 4H in all Morex assemblies (Mascher et al., 2017, 2021; Monat et al., 2019), which also may explain the inability to locate markers used by Hickey et al. (2011). Whether this region is misassembled in all Morex versions or other phenomena have occurred, such as a translocation relative to Morex, should be investigated. Based on this current information, Rph20 may also correspond to QPh.5H-1 (Vatter et al., 2017), RphQ10 (Ziems et al., 2014), and an unnamed QTL (Gutiérrez et al., 2015).
Rph25 (RphFT) was identified in the Chinese cultivar Fong Tein and Australian cultivar Yagan (Kavanagh et al., 2017). However, due to the use of reporting cM positions within a population and lack of sequence availability, Rph25 cannot be anchored to the Morex assembly. Rph28 (RphHEB) identified in wild barley was the most recently designated locus (Mehnaz et al., 2021a), mapping to an ~ 100-kb interval (562.28–562.92 Mb). Rph28 was most likely previously identified as an unnamed QTL (Schnaithmann et al., 2014).
Chromosome 6H
Rph11 was mapped to a 9.7-Mb interval (542.55–552.27 Mb) using introgression mapping but may also be allelic to Rph10 currently assigned to chromosome 3H (Martin et al., 2020), which was previously discussed. Rph24 (qRPhND) was mapped to a large 351 Mb region (Hickey et al., 2011) and subsequently, a 7.9-Mb region (358.56–366.40 Mb) using DArT markers (Ziems et al., 2017). Rph24 was also previously mapped as Rphq3 (González et al., 2012); QPh.6H-2 and QPh.6H-3 (Vatter et al., 2017); RphQ11 (Ziems et al., 2014); qRphYer2-6H (D., Singh et al., 2015); and multiple unnamed QTL (Castro et al., 2012; Czembor et al., 2022; González et al., 2012; Gutiérrez et al., 2015; Hickey et al., 2011; Qi et al., 1998; Ziems et al., 2014). Diagnostic markers targeting indels were subsequently developed to track Rph24 (Dracatos et al., 2021) in close proximity to the distal flank of the 7.9 Mb region. However, the fact that three distinct loci, QPh.6H-2, QPh.6H-3, and QPh.6H-4 (Sallam et al., 2017), were identified within the previously 351 Mb Rph24 interval (Hickey et al., 2011), suggests there may be additional resistance loci along the chromosome. Further investigation will be required to determine the true number of resistance loci in the region. Additional loci mapping to chromosome 6H includes QLr.S42-5H.a (von Korff et al., 2005), Rphq16 (Yeo et al., 2017), QPh.6H-1 (Vatter et al., 2017), qRphYer1-6H (Singh et al., 2015), three unanchored QTL (Castro et al., 2012), and unanchored Rphq15 (Marcel et al., 2007).
Chromosome 7H
The Rph3 gene was first reported in the cultivar Aim and subsequently Estate (Henderson, 1945; Roane and Starling, 1967). Rph3 was mapped to chromosome 7HL (Jin et al., 1993) and cloned as a small, unique, avirulence-dependent inducible membrane protein, reminiscent of TALE-activated executor resistance genes (Dinh et al., 2022). Rph3 exhibits increased diversity in wild accessions compared to domesticated germplasm (Dinh et al., 2022), most likely due to Rph3 being sourced from a wild accession. Rph3 can confer a strong hypersensitive response or incomplete resistance in the cultivar Ribari and barley accession L94, respectively (Martin et al., 2020). Rph19 was first identified in cultivar Prior, exhibiting the same resistance specificity as Reka 1 and mapped to a 2.8-Mb interval (618.14–620.92 Mb) (Park and Karakousis, 2002). Rph23 (qRphYer-7H) was identified within the Australian cultivar Yerong and believed to have originated from Russian landrace LV-Taganrog (D., Singh et al., 2015), mapping to a 14.9-Mb interval (25.49–40.36 Mb). Additional loci mapping to chromosome 7H include Rphq1, Rphq8, Rphq9, Rphq13, and Rphx (Qi et al., 1998); RphP/RphQ15 (Park and Karakousis, 2002; Ziems et al., 2014); RphQ2, RphQ12, RphQ13, and RphQ14 (Ziems et al., 2014); QPh.7H-1, QPh.7H-2, and QPh.7H-3 (Vatter et al., 2017); Qlr.HeB-F23-7H (Schnaithmann et al., 2014); and QLr.S42-7H.a (von Korff et al., 2005). Furthermore, five unnamed QTL map to chromosome 7H (Czembor et al., 2022; Gutiérrez et al., 2015; Rossi et al., 2006; Thomas et al., 1995), and one unnamed QTL remains unanchored to chromosome 7H (Hickey et al., 2011).
Association mapping
There have been six association mapping studies identifying leaf rust loci. The first association mapping study was conducted on 360 elite barley lines from Australia. A total of 11 of the 15 reported QTL were previously identified loci and four deemed novel (Ziems et al., 2014). The second association mapping study was performed in the Halle Exotic Barley (HEB)-5 nested association mapping (NAM) panel (Schnaithmann et al., 2014). At the time, only one novel QTL was identified with remaining loci corresponding to previously characterized Rph loci (Schnaithmann et al., 2014). The third association mapping study was conducted in Latin American barley, identifying two novel QTL out of six loci detected (Gutiérrez et al., 2015). The fourth was conducted with the HEB-25 NAM panel, a significant expansion of the HEB-5 population, identifying a total of two novel loci out of 11 (Vatter et al., 2018). The fifth association mapping study identified six QTL (Czembor et al., 2022), with the most significant marker-trait association (MTA) mapping within 1.2 Mb of RphQ11 identified on chromosome 6H (Ziems et al., 2014) and therefore is unlikely to be novel. The last association mapping conducted ASR and APR, claiming 58 MTA, 32 of which were novel. However, based on the low LOD thresholds utilized, it could be argued that all but seven of the MTA detected should be considered significant. In addition, these seven MTA are within a 7-Mb interval, suggesting all markers may form a single locus (Amouzoune et al., 2022).
Genetics to stem rust
The stem rust pathogen Pgt contains the largest host range within Puccinia with 28 hosts and is the primary cause of barley and wheat stem rust (Dracatos et al., 2015b). Stem rust is unique in that a barley-specific ff. spp. has not been identified. In addition, other ff. spp. are known to infect barley, including Puccinia graminis f. sp. secalis (Pgs), Puccinia graminis f. sp. avenae (Pga) (Brueggeman et al., 2020; Dracatos et al., 2015b), and a Pgt × Pgs hybrid known as Scabrum rust arising on triticale (Park, 2007). Barley stem rust epidemics in North America during the 1920s–1930s resulted in yield losses between 15% and 20% (Steffenson, 1992); however, 100% yield loss has been reported in wheat to the Digalu (Pgt race TKTTF) lineage (Singh et al., 2015). Due to the widespread deployment of Rpg1, barley stem rust epidemics were largely controlled until the late 1980s when Pgt race QCCJB overcame Rpg1 (Sharma Poudel et al., 2018). In addition, the Ug99 (race TTKSK) lineage of Pgt isolates overcame Sr31 in wheat and is seen as a major threat to global food security with 80%–95% of worldwide acreage of wheat considered susceptible (Singh et al., 2015). In addition, over 95% of barley cultivars and wild accessions surveyed are considered susceptible (Hatta et al., 2021; Prins et al., 2020; Steffenson et al., 2017). Due to effective resistance gene deployment and adequate disease management over the past 75 years, stem rust was not considered a major threat despite its enormous potential to devastate barley and wheat crops (Singh et al., 2011). The first reported epidemics in over 60 years in the UK and the first in decades across Germany and Sicily have caused devastating damage (Edae and Rouse, 2020; Lewis et al., 2018). Reports have also found Pgt isolates becoming more aggressive at both warmer and cooler temperatures (Lewis et al., 2018). Alarmingly, the first isolates with virulence on Rpg1 and rpg4/Rpg5 when stacked together in barley line Q21861 were recently reported in the Pacific Northwestern region of North America (Upadhaya et al., 2021).
Genetic studies describing stem rust resistance have been reported since the 1930s (Powers and Hines, 1933). Over 60 Stem rust (Sr) resistance loci have been designated within wheat, with at least nine identified and validated (Chen et al., 2018; Hatta et al., 2021). Multiple wheat stem rust resistance genes can remain functional in a susceptible barley background to provide resistance against Pgt. For example, the wheat genes Sr22, Sr33, Sr35, and Sr45 all provide resistance to the Pgt race TTKSK when transformed into the susceptible barley background of Golden Promise (Hatta et al., 2021). In comparison, only nine Reaction to Puccinia graminis (Rpg, Table 3) loci have been identified within barley (Hatta et al., 2021). Rpg1-rpg4, rpg6-Rpg7, and RpgU were first identified against Pgt, whereas Rpg5 and rpgBH were identified against Pgs (Zhou et al., 2014). The RpgU locus was identified in Peatland, Husky, and Diamond (Fox and Harder, 1995) and potentially SB90585 (Harder and Legge, 2000); however, the locus was never mapped. Based on segregation ratios, RpgU is not Rpg1, and most likely not Rpg2 or Rpg3 (Fox and Harder, 1995). However, further investigation is required to determine the location of RpgU and whether RpgU is allelic to any other Rpg loci. The recessive locus rpgBH was identified in Black Hulless, effective against Pgs (Steffenson et al., 1984). As allelism tests or mapping experiments have not been conducted, it cannot be ruled out that rpgBH is not a different and/or less effective allele of previously mapped Rpg loci. Up to two other loci were reported in Purple Nudum and Skinless (Babriwala, 1954; Luig, 1957), but were not investigated further. A total of nine loci were identified against Pga (Rgpaq1-9); however, only Rpgaq7 and Rpgaq9 did not colocalize with previously reported Rpg loci and were considered novel (Dracatos et al., 2016a). Lastly, a total of 17 required for P. graminis resistance (rpr) mutants have been obtained using fast neutron irradiation of Morex (Rpr1-7) and Q21861 (Rpr8-17), three of which have been mapped as Rpr1, Rpr2, and Rpr9 (Gill et al., 2016; Solanki et al., 2019; Zhang et al., 2006).
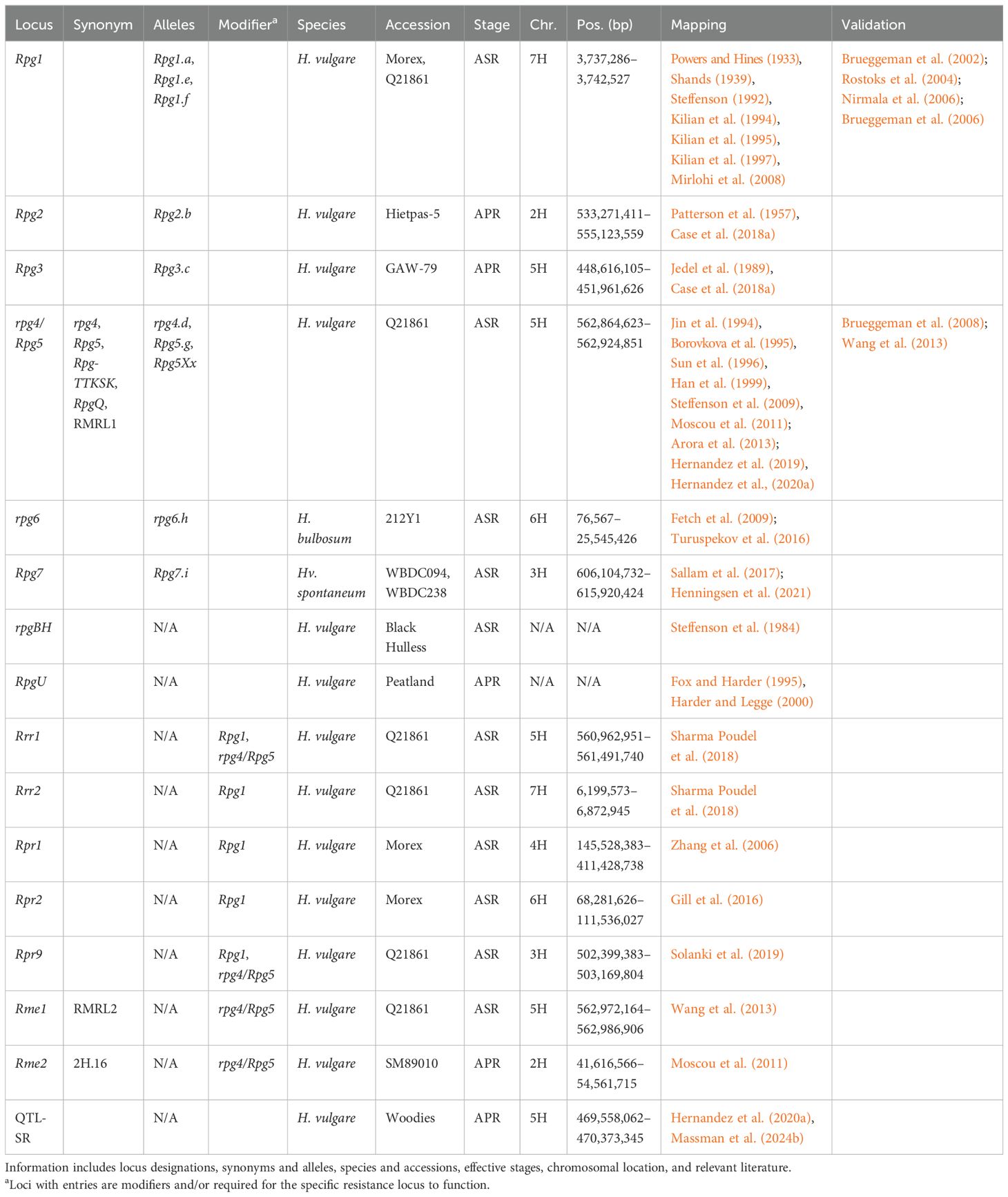
Table 3. Summary information of all designated resistance/susceptibility loci in barley to stem rust pathogen Puccinia graminis.
Chromosome 1H
Currently, only association mapping studies have identified stem rust resistance loci on chromosome 1H. These include Rpgaq1 (Dracatos et al., 2016a), Rpg-qtl-1H-11_11277 and Rpg-qtl-1H-12_20613 (Case et al., 2018b), and 10 unnamed QTL (Czembor et al., 2022; Sallam et al., 2017; Turuspekov et al., 2016). Current evidence suggests the 10 unnamed QTL do not overlap and therefore chromosome 1H may harbor untapped potential for stem rust resistance.
Chromosome 2H
Rpg2 was first described in Heitpas-5 (Patterson et al., 1957) and mapped within a 21.9-Mb interval (533.27–555.12 Mb) (Case et al., 2018a); however, it remains largely under investigation due to the low level of resistance it provides (Kleinhofs et al., 2009). Additionally, a trans-eQTL regulator hotspot was identified that results in the suppression of hundreds of genes after Pgt inoculation and is colocalized with an enhancer of rpg4/Rpg5-mediated resistance to Pgt race TTKSK at the adult stage (Moscou et al., 2011). Both the enhancer and regulator are hypothesized to be the same gene, and therefore for this manuscript, this locus has been designated Rpg4-modifier element 2 (Rme2) and mapped to a 12.9-Mb interval (41.6–51.6 Mb). Additional loci mapping to chromosome 2H include Rpg-qtl-PH-PI38-2H (258.46–264.39 Mb) nested within Rpg-qtl-HH-Hie-2H.1 (221.89–26830 Mb) (Case et al., 2018a) that was also identified with association mapping (Hernandez et al., 2020a). Other loci include Rpg-qtl-HH-Hie-2H.2, Rpg-qtl-HH-Hie-2H.3, and Rpg-qtl-HH-Hie-2H.4 (Case et al., 2018a); Rpg-qtl-2H-12_11278 (Case et al., 2018b); Rpg-qtl-2H_SCRI_RS_115905 and Rpg-qtl-2H_SCRI_RS_109266 (Mamo, 2013); Rpgaq9 and unanchored Rpgaq2 (Dracatos et al., 2016a); and nine unnamed QTL (Czembor et al., 2022; Sallam et al., 2017).
Chromosome 3H
Rpg7 is the most recently designated Rpg locus identified in wild barley accessions WBDC094 and WBDC238, mapping to a 9.8-Mb interval (606.10–615.92 Mb) (Henningsen et al., 2021). Association mapping identified the MTA Rpg-qtl-3H_SCRI_RS_180847 approximately 0.98 Mb from the boundary of Rpg7 (Mamo, 2013). Rpr9, which was a gene identified by mutant analysis of line Q21861, is required for both Rpg1- and rpg4/Rpg5-mediated resistance, and mapped to a 770-kb interval (502.40–503.17 Mb) (Solanki et al., 2019). Rpr9 is hypothesized to function by facilitating ubiquitination and degradation of proteins required for resistance, based on previous research on Rpg1 (Solanki et al., 2019). In addition, rpr9 mutants resulted in a stunted root phenotype due to the hypothesis that Rpr9 is involved in hormone signaling (Solanki et al., 2019). Rpr9 appears to be encompassed by Rpg-qtl-HH-Hip-3H (Case et al., 2018a) and potentially mapped in association mapping (Czembor et al., 2022). Additional loci mapping to chromosome 3H include Rpg-qtl-PH-Hip-3H and Rpg-qtl-HH-Hip-3H (Case et al., 2018a); Rpg-qtl-3H-SCRI_RS_199887 (Case et al., 2018b); and qGH_PBIC_3.11 (Dracatos et al., 2019b). A total of 11 unnamed QTL have also been mapped to chromosome 3H (Hernandez et al., 2020a; Mamo et al., 2014; Sallam et al., 2017; Turuspekov et al., 2016), whereas Rpgaq6 remains unanchored (Dracatos et al., 2016a). Rpg-qtl-PH-Hip-3H and Rpg-qtl-HH-Hip-3H show partial overlap and encompass six MTA identified via association mapping (Czembor et al., 2022; Mamo et al., 2014; Sallam et al., 2017), suggesting less unique loci on chromosome 3H and will require further investigation.
Chromosome 4H
The Rpr1 locus was mapped to a 265.9-Mb interval (145.53–411.43 Mb) in the Morex V3 genome (Zhang et al., 2006). Rpr1 was identified as a suppressor of Rpg1 and therefore required for Rpg1-mediated resistance; however, Rpr1 is not involved in rpg4/Rpg5-mediated resistance (Zhang et al., 2006). Six MTA were reported within the Rpr1 region using association mapping (Sallam et al., 2017; Turuspekov et al., 2016); however, considering the size of the Rpr1 region, it may encompass different resistance loci. Additional loci mapped to chromosome 4H include Rpg-qtl-PH-PI38-4H, Rpg-qtl-HH-Hie-4H (Case et al., 2018a), Rpg-qtl-4H_12_30995 (Mamo, 2013), seven MTA that most likely form a single QTL based on cM positions (Sallam et al., 2017), and a further four unnamed QTL (Czembor et al., 2022; Turuspekov et al., 2016).
Chromosome 5H
Rpg3 was first reported in GAW-79 (Jedel et al., 1989) and mapped to chromosome 5H (Case et al., 2018a). Using markers reported to be significant, Rpg3 maps to a 323.5-Mb interval (94.22–417.72 Mb) using Morex V3. However, these significant markers are not present within the linkage map reported. Utilizing the genetic marker information of the map, Rpg3 maps to a 2.3-Mb interval (448.62–450.88 Mb) (Case et al., 2018a), outside of the original Rpg3 interval. Rpg3 is more likely to be located within the 2.3-Mb interval based on Rpg3 being localized to chromosome 5HL and the higher-quality genome assembly of Morex V3. However, as Rpg3 remains under investigated due to low-level resistance (Kleinhofs et al., 2009), this discrepancy should be addressed for both tracking Rpg3 for breeding purposes and future validation.
The rpg4 locus was first identified as providing resistance to Pgt isolates, whereas Rpg5 provided resistance to Pgs isolates and later Pga isolates (Dracatos et al., 2015b; Sun et al., 1996; Sun and Steffenson, 2005). The rpg4 gene was originally suspected as HvAdf2 (Brueggeman et al., 2009; Kleinhofs et al., 2009), whereas the Rpg5 gene was validated as HvRga2, encoding an NLR with an integrated protein kinase (functional haplotype) or protein phosphatase 2C domain (PP2C, nonfunctional haplotype) (Brueggeman et al., 2008). However, increasing evidence found that rpg4-mediated resistance was not a result of HvAdf2 and required Rpg5 (HvRga2) and two additional genes located at the locus, HvRga1 and HvAdf3, to be functional (Arora et al., 2013; Wang et al., 2013). The HvAdf3 gene within the locus was also deemed to be a candidate susceptibility gene (Moscou et al., 2011). Therefore, the rpg4/Rpg5 complex was renamed to the rpg4-Mediated Resistance Locus (RMRL) that includes RMRL1 containing HvRga1, Rpg5 (HvRga2), and HvAdf3; and RMRL2 containing the yet unidentified Rpg4-modifier element 1 (Rme1), which is required for rpg4-mediated wheat stem rust resistance but not required for Rpg5-mediated rye stem rust resistance (Wang et al., 2013). RPG5 (HvRGA2) and HvRGA1 belong to MIC1 (C16) and C7 clades, respectively (Bailey et al., 2018). NLRs in the MIC1 and C7 clades were found to be in head-to-head orientation in grasses such as the paired NLRs RGA5/RGA4 in rice that confer resistance to M. oryzae through recognition of the effectors AVR-Pia and AVR1-CO39 (Bailey et al., 2018; Cesari et al., 2013). The MIC1 clade is unique in the grasses, as NLRs in the clade have diverse C-terminal integrated domains with RGA5 carrying an integrated heavy metal-associated domain (Bailey et al., 2018; Cesari et al., 2013).
Using six highly diverse barley accessions, only one allele of rpg4/Rpg5 was identified, suggesting cultivated barley could be extremely vulnerable to a lack of diversity at rpg4/Rpg5 (Mamo et al., 2014). In addition, the rpg4/Rpg5 complex was found to be present in nearly every resistant landrace from Switzerland when assessing resistance to Pgt races TTKSK and QCCJB (Steffenson et al., 2016). Additional studies mapped rpg4/Rpg5 using association mapping, identifying a novel allele of rpg4/Rpg5 designated Rpg5Xx (Hernandez et al., 2019, 2020a). Previous work found that the region containing Rme1 is approximately 220 kb in size, proximal to RMRL1, containing a heat shock protein, a zinc finger SEC14 protein, and an actin depolymerization-like protein (HvAdf1) (Wang et al., 2013). Further investigation into the Morex V3 found that Rme1 encompasses 321 kb. Leaf-expressed candidate genes include those previously identified as well as a PP2C protein. Further work is needed to establish sequence and structural variation within the Rme1 region relative to diverse barley accessions.
The rpg4/Rpg5 locus was the only locus to provide resistance to Pgt race TTKSK in barley (Brueggeman et al., 2009); however, an additional locus, required for rpg4-mediated resistance 1 (Rrr1), is required to facilitate rpg4/Rpg5-mediated resistance when stacked with Rpg1 (Sharma Poudel et al., 2018). The Rrr1 locus was mapped to a 529-kb interval (560.96–561.49 Mb) on chromosome 5H (Sharma Poudel et al., 2018). Similar phenomena were observed with Rpr9 (required for Rpg1- and rpg4/Rpg5-mediated resistance) described earlier on chromosome 3H (Solanki et al., 2019), and the identification of two novel loci on chromosomes 5H (not Rrr1) and 7H that were additive to Rpg4/Rpg5 resistance, i.e., rpg4/Rpg5 is required for resistance but not sufficient (Hernandez et al., 2019). These complex interactions have made the introgression of rpg4/Rpg5-mediated resistance to elite cultivars more complicated than originally anticipated (Hernandez et al., 2019).
For further complexity, the recently released Woodies germplasm (Woody-1, DH160733 and Woody-2, DH160754) designed to systematically stack stripe and stem rust resistance have a null allele of Rpg5, yet remain highly resistant to stem rust (Hernandez et al., 2020a, b; Massman et al., 2024a). The high level of resistance present in the Woodies has been attributed to the QTL-SR locus, encompassing a 1.8-Mb region (469.6–470.4 Mb) in close vicinity to the rpg4/Rpg5 complex (Massman et al., 2024b). Candidate gene analysis of the Morex V3 region revealed 10 candidate genes within the region of interest, and a further three NLR genes within 10 Mb that may underlie QTL-SR due to structural rearrangements. Further comparative analysis revealed that large regions of Morex are absent from the Woody-1 genome assembly (Massman et al., 2024b). Therefore, instead, the Woodies may lack a susceptibility gene that is present in the susceptible barley accession Morex. The identification of this novel locus further complicates the resistance puzzle; however, Woody-2 has been identified to be highly amenable to transformation, which will aid in gene validation.
Additional loci mapping to chromosome 5H include Rpg-qtl-PH-PI38-5H (Case et al., 2018a), Rpg-qtl-5H-11_11355 (Case et al., 2018b; Zhou et al., 2014), Rpg-qtl-5H-SCRI_RS_10929 (Mamo, 2013), and three unnamed QTL (Czembor et al., 2022; Sallam et al., 2017). Multiple MTA are deemed to delimit Rpg-qtl-5H-11_11355 as a QTL due to linkage disequilibrium (Case et al., 2018b; Zhou et al., 2014) and were also mapped in earlier and subsequent association mapping studies (Hernandez et al., 2020a; Mamo, 2013). The boundary of Rpg-qtl-5H-11_11355 is approximately 14 Mb distal to Rpg3 and therefore is likely to be a distinct locus.
Chromosome 6H
The rpg6 locus is the only locus identified within H. bulbosum to stem rust, providing recessive resistance (Fetch et al., 2009) and located within a 25.5-Mb interval (0.08–25.55 Mb). The rpg6 locus has also been mapped in three association mapping studies (Czembor et al., 2022; Sallam et al., 2017; Turuspekov et al., 2016). Rpr2 mapped to a 43.3-Mb (68.28–111.54 Mb) interval and hypothesized to function as a stabilizer of Rpg1, as the RPG1 protein was degraded faster than that of highly resistant stem rust-resistant lines (Gill et al., 2016). An MTA embedded within the Rpr2 interval was also reported in association mapping (Czembor et al., 2022). Additional loci that map to chromosome 6H include a further eight unnamed QTL (Czembor et al., 2022; Sallam et al., 2017), eight MTA that most likely form a single QTL (Turuspekov et al., 2016), and unanchored Rpgaq3, Rpgaq7, and Rpgaq8 (Dracatos et al., 2016a).
Chromosome 7H
Rpg1 was the first Rpg locus identified and provided durable, broad-spectrum resistance to Pgt against all North American Pgt isolates for approximately 70 years (Roelfs et al., 1993). Rpg1 was first reported in 1933, with sources found in Peatland, Chevron, and Kindred (Powers and Hines, 1933; Shands, 1939; Steffenson, 1992). Cloning of Rpg1 identified a gene encoding a protein with two tandem serine/threonine protein kinase domains (Brueggeman et al., 2002, 2006) located at 3.74 Mb. Transcript analysis found that Rpg1 has the highest expression in leaf epidermal cells (Rostoks et al., 2004) and is predominantly located in the cytosol (Nirmala et al., 2006). Both protein kinase domains are required for resistance; however, the second kinase domain is required for autophosphorylation (Nirmala et al., 2006). In addition, two effectors are required for autophosphorylation with RIN4, and subsequent degradation of RPG1 is required for resistance (Chai et al., 2012; Gill et al., 2012; Horvath et al., 2003; Nirmala et al., 2007, 2010, 2011). Either Rrr1 or another locus designated required for Rpg1-mediated resistance 2 (Rrr2) is required for Rpg1-mediated resistance in the presence of rpg4/Rpg5 (Sharma Poudel et al., 2018). Rrr2 was mapped to a 673-kb interval (6.20–6.87 Mb) (Sharma Poudel et al., 2018), but is not the 7H locus reported by Hernandez et al. (2019). Additional loci mapping to chromosome 7H include Rpg-qtl-PH-PI38-7H and Rpg-qtl-HH-Hie-7H (Case et al., 2018a); qGH_BPIC_4.6 (Dracatos et al., 2019b); 19 unnamed QTL (Czembor et al., 2022; Hernandez et al., 2020a; Sallam et al., 2017; Zhou et al., 2014); and unanchored Rpgaq4 and Rpgaq5 (Dracatos et al., 2016a).
Association mapping
To date, there have been eight association mapping studies used to characterize stem rust resistance. The first identified up to 15 MTAs in wild barley, two of which were associated with rpg4/Rpg5 (Steffenson et al., 2007). The second association mapping study was conducted in US breeding material against Pgt race TTKSK effectively identifying two novel QTL on chromosomes 5H and 7H, respectively (Zhou et al., 2014). These were both mapped again in subsequent association mapping studies (Case et al., 2018b; Hernandez et al., 2020a). In the third association mapping study, 17 MTAs were reported using Kazakh spring barley in two environments (Turuspekov et al., 2016). However, one marker misassigned to chromosome 2H and four markers assigned to unknown chromosomal loci can be consolidated with other MTAs into a total of eight distinct loci. After consolidating these loci, only markers located at the proximal end of chromosome 6H colocalize with the previously identified locus rpg6 (Fetch et al., 2009). While the other loci can be deemed novel within stem rust, four colocalize with the leaf rust resistance loci Rph13, Rph20, Rph21, and Rph26. The fourth association mapping study was conducted in wild barley, identifying 45 MTAs, many of which were novel at the time (Sallam et al., 2017). The fifth association mapping study identified seven QTL from the barley core collection; one locus of notable interest was on chromosome 5H, as it provided APR not conferred by the rpg4/Rpg5 complex (Case et al., 2018b). The sixth and seventh association mapping studies were conducted on a double haploid population designed to increase resistance to stem rust race TTKSK (Hernandez et al., 2019, 2020a). These association mapping studies identified eight and six MTAs, respectively (Hernandez et al., 2019, 2020a), none of which were claimed to be novel; however, a new allele of rpg4/Rpg5 was identified as Rpg5Xx (Hernandez et al., 2020a). The last association mapping study identified 48 significant MTAs using European barley accessions (Czembor et al., 2022).
Genetics to stripe rust
Barley stripe rust is predominantly caused by Psh; however, barley can also be infected by P. striiformis f. sp. tritici (Pst), the causal agent of wheat stripe rust. Yield losses exceeding 70% have been reported but typically cause approximately 40% yield loss under environmental conditions conducive to disease in a susceptible variety (Marshall and Sutton, 1995). Psh is therefore potentially the most damaging rust, despite arguably being the most understudied of the three major barley rusts. Barley stripe rust was first described by European workers in the late 1800s, causing particular issues in winter barley in the UK and the Netherlands (Wellings, 2011). Subsequently, Psh was first reported in Columbia in 1975, spreading throughout South America by 1982 (Dubin and Stubbs, 1986), Mexico by 1987, and in the USA by 1991 (Chen et al., 1995; Marshall and Sutton, 1995). Due to stripe rust prevalence in colder, wetter climates, often at higher altitudes, stripe rust is often regarded as cold-temperature rust (Dracatos et al., 2019b). Around 88% of worldwide wheat production is susceptible to stripe rust (Beddow et al., 2015) and is considered a major pathogen of barley, with 60%–70% of Australian barley considered susceptible (Dracatos et al., 2019b; Gyawali et al., 2021).
The genetics of stripe rust resistance have been studied since the 1940s (Murty, 1942). To date, there are 78 Yellow rust (Yr) loci (Jamil et al., 2020) and at least 50 Resistance to Puccinia striiformis (Rps; Table 4) loci identified in wheat and barley, respectively (Bettgenhaeuser et al., 2021; Chełkowski et al., 2003; Chen and Line, 1999, 2001, 2003; Nover and Scholz, 1969; Pahalawatta and Chen, 2005). However, only 10 Rps loci have been formally designated within the barley-P. striiformis pathosystem (Clare et al., 2016), and even fewer have been mapped. Earlier mapping efforts made use of restriction fragment length polymorphisms, simple sequence repeats, amplified fragment length polymorphisms, and resistance gene analog polymorphisms, which make anchoring loci to the Morex V3 troublesome (Castro et al., 2002, 2003a, 2003b; Chen et al., 1994; Thomas et al., 1995; Toojinda et al., 1998, 2000; Vales et al., 2005).
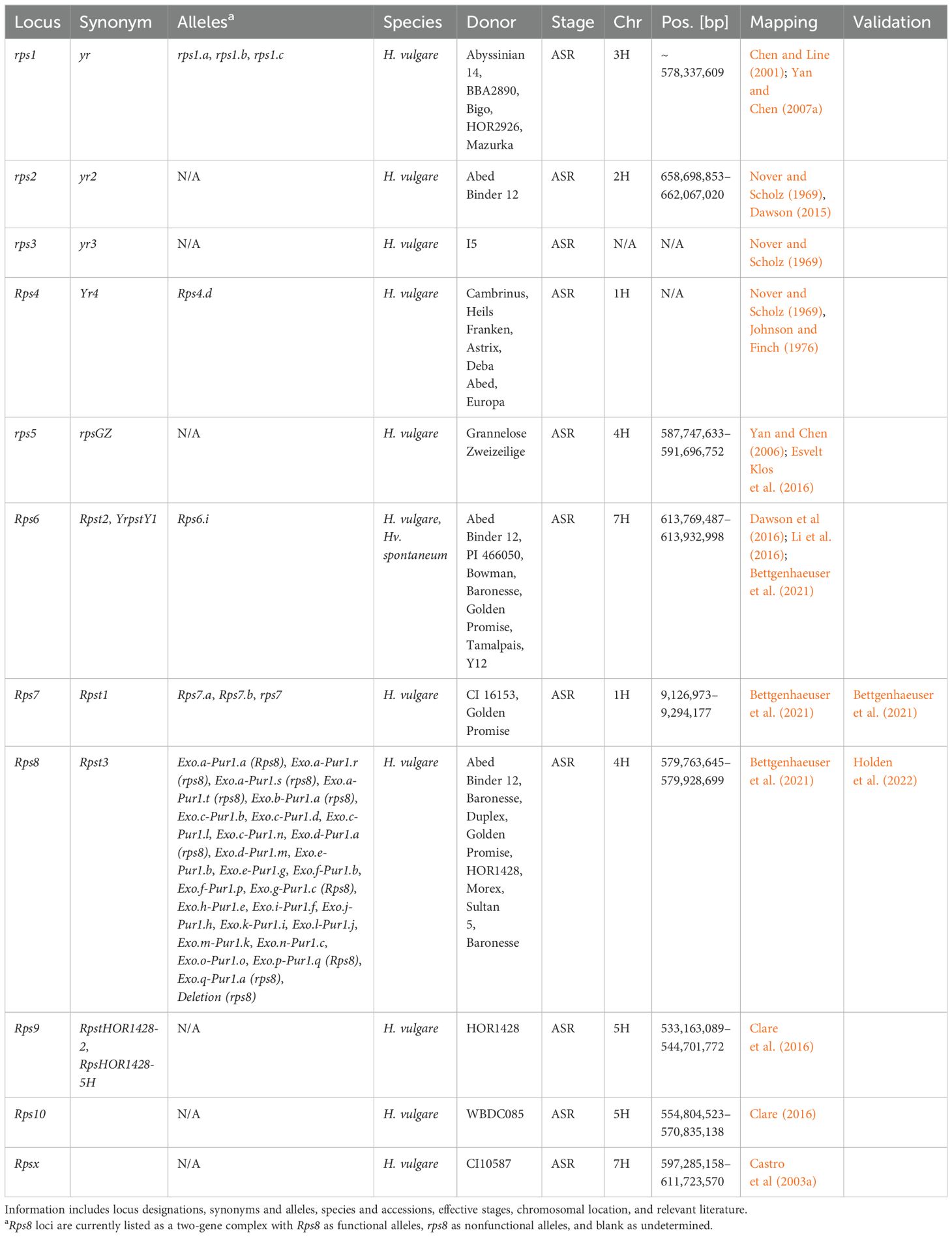
Table 4. Summary information of all designated resistance/susceptibility loci in barley to stripe rust pathogen Puccinia striiformis.
Another caveat is that these loci are functional against P. striiformis sensu lato, meaning they can function against both or only Psh or Pst isolates. The first four Rps loci were originally designated Yr loci; however, Rps4 has only been associated with chromosome 1H, and rps3 was never mapped (Johnson, 1968; Nover and Scholz, 1969). In addition, Yr4 through Yr13 were identified in India but were not consistent with international nomenclature (Verma et al., 2018). There were also at least eight additional loci with Puccinia striiformis (Ps) nomenclature (Chen and Line, 1999; Luthra and Chopra, 1990). Many of the original Rps loci function under a recessive mode of inheritance, with only five out of 26 under a dominant mode of inheritance (Chełkowski et al., 2003; Chen and Line, 1999). The first loci identified to govern nonadapted resistance to Pst were RpstS1 and RpstS2 in Steptoe (Pahalawatta and Chen, 2005). More recently, nonadapted resistance to Pst has seen a renewed research focus with the mapping of Rps6 (Dawson et al., 2016; Li et al., 2016) and subsequent cloning of Rps7 and Rps8 (Bettgenhaeuser et al., 2021; Holden et al., 2022).
Chromosome 1H
Rps7 was the first Rps gene to be identified and validated within the barley-P. striiformis pathosystem (Bettgenhaeuser et al., 2021). Rps7 encodes an NLR that was previously identified as the barley immune receptor Mildew locus a (Mla), conferring resistance to Blumeria graminis f. sp. hordei, and M. oryzae (Brabham et al., 2023; Inukai et al., 2006), and susceptibility to Cochliobolus sativus (Leng et al., 2018). The authors warned that due to different haplotypes of Mla providing different specificities, often to different pathogens, breeders should be careful not to inadvertently remove resistance to nonadapted pathogens (Bettgenhaeuser et al., 2021). Rps7 was also mapped as QPsh-DP-2R-1.1 and QPsh-rM-6R-1.1 (Gyawali et al., 2021); qGH_WUR_rep1_3.26 and qGH_WUR_rep2_5.78 (Dracatos et al., 2019b); RPsh-1H (Belcher et al., 2018); and unnamed QTL (Hernandez et al., 2020a). The genetic mapping of Rps4 using protein and phenotypic markers placed the gene ~ 6.2 cM proximal to Mla (Johnson et al., 1969). Additional loci mapping to chromosome 1H include qField_Mex2015_3.30 and qGH_PBIC_5.29 (against Psph, Dracatos et al., 2019b); QPsh-r24-6R-1.1, QPsh-rQ-6R-1.2, QPsh-r24-6R-1.2, and QPsh-DP-2R-1.2 (Gyawali et al., 2021); QPs.1H-1 (Vatter et al., 2018); QPsh.FW6-1H (Belcher et al., 2018); and two unnamed QTL (Belcher et al., 2018; Dracatos et al., 2016b). Furthermore, eight unnamed unanchored MTA/QTL are present on chromosome 1H (Thomas et al., 1995; Visioni et al., 2018).
Chromosome 2H
The rps2 locus was originally identified as a recessive resistance locus against Psh in Abed Binder 12 (Nover and Scholz, 1969); however, it is currently only one of two Rps loci to be functional against Psh and Pst. Subsequent high-resolution mapping determined that rps2 was additive rather than completely recessive and localizing to a 3.4-Mb (658.7–662.1 Mb) interval (Dawson, 2015). Additional loci mapping to chromosome 2H include QPs.2H-1, QPs.2H-2, and QPs.2H-3 (Vatter et al., 2018); Qpsh.316A.2Ha and Qpsh.316A.2Hb (Esvelt Klos et al., 2020); QPsh-rQ-6R-2.1, QPsh-r7S0-2R-2.1, QPsh-DP-6R-2.1, QPsh-DP-6R-2.2, and QPsh-DP-2R-2.1 (Gyawali et al., 2021); qField_Mex2015_5.13, qField_Mex2015_5.30, and qField_Ecuad2017_3.97 (Dracatos et al., 2019b); and QPsh.FW6-2H.1 (Belcher et al., 2018). Furthermore, a total of four unnamed QTL from 10 MTA (Belcher et al., 2018) and three unnamed QTL (Dracatos et al., 2016b; Gutiérrez et al., 2015) were mapped to chromosome 2H, while 15 MTA/QTL remain unanchored (Rao et al., 2007; Rossi et al., 2006; Visioni et al., 2018).
Chromosome 3H
The recessive rps1 resistance locus was identified in Bigo and Abyssinian 14 and mapped in BBA2890 using resistance gene analog polymorphisms (RGAP); however, due to the use of these markers, an interval cannot be reported (Nover and Scholz, 1969; Yan and Chen, 2007a). One marker used to track rps1 places the causal gene in the vicinity of 578.33 Mb (Yan and Chen, 2007b). Loci with markers in close proximity to rps1 have included Qpsh.316A.3H (Esvelt Klos et al., 2020); RPsh-3H, QPsh.FW6-3H.2, and two unnamed QTL (Belcher et al., 2018); QPs.3H-2 and QPs.3H-3 (Vatter et al., 2018); and QPsh-rQ-6R-3.1 (Gyawali et al., 2021). Additional loci mapping to chromosome 3H include QPsh-rQ-2R-3.1, QPsh.FW6-3H.1, QPsh-r7S0-6R-3.1, QPsh-DP-6R-3.1, and QPsh-r57-2R-3.1 (Gyawali et al., 2021); QPs.3H-1 and QPs.3H-4 (Vatter et al., 2018); RpsHOR1428-3H, also mapped by association mapping (Clare, 2016; Gutiérrez et al., 2015); and qGH_PBIC_3.14 (Dracatos et al., 2019b). A further three unnamed QTL were mapped to chromosome 3H (Belcher et al., 2018; Gutiérrez et al., 2015), while 11 MTA/QTL remain unanchored (Rao et al., 2007; Rossi et al., 2006; Visioni et al., 2018).
Chromosome 4H
The rps5 (rpsGZ) locus identified in Grannenlose Zweizeilige was mapped to chromosome 4H using RGAPs (Yan and Chen, 2006) and subsequently to a 3.9-Mb interval (587.75–591.70 Mb) (Esvelt Klos et al., 2016). The nonadapted Rps8 resistance locus, functional against Pst, was the second validated Rps locus as a two-gene complex encoding a receptor kinase and an Exo70. Both the receptor kinase and Exo70 are required for resistance (Holden et al., 2022) and are located within a 170-kb interval (579.76–579.92 Mb). Due to the two-gene complex of Rps8, there are at least 27 unique alleles/haplotypes currently reported (Holden et al., 2022). Rps8 was first mapped by Bettgenhaeuser et al. (2021); however, it was also most likely mapped with association mapping as QPsh-rQ-2R-4.1 and QPsh-rM-6R-4.1 (Gyawali et al., 2021) and an unnamed MTA (Hernandez et al., 2020a). Additional loci mapping to chromosome 4H include QPsh-r57-6R-4.1, QPsh-r57-6R-4.2, QPsh-r57-6R-4.3 QPsh-rG-2R-4.1, QPsh-rG-2R-4.2, QPsh-DP-6R-4.1, QPsh-DP-6R-4.2, QPsh-r7S0-2R-4.1, QPsh-r7S0-6R-4.1, and QPsh-rG-2R-4.3 (Gyawali et al., 2021); Qpsh4Ha and Qpsh4Hb (Esvelt Klos et al., 2020); and qGH_WUR_rep1_3.86 and qGH_WUR_rep2_3.28 (Dracatos et al., 2019b). A further three unnamed QTL (Belcher et al., 2018; Dracatos et al., 2016b; Gutiérrez et al., 2015) and 10 unanchored MTA/QTL (Rao et al., 2007; Rossi et al., 2006; Visioni et al., 2018) are mapped to chromosome 4H.
Chromosome 5H
The final two most recently designated nonadapted resistance loci are Rps9 and Rps10. Rps9 was identified as the second locus functional against Psh and Pst, utilizing a backcrossing scheme to isolate the locus from HOR 1428 in a Manchuria background and delimited to an 11.5-Mb interval (533.16–544.07 Mb) (Clare et al., 2016). Rps9 has also been identified as QPs.5H-1 (Vatter et al., 2018), QPsh-r57-2R-5.1 (Gyawali et al., 2021), and an unnamed QTL (Gutiérrez et al., 2015). The same strategy was used to isolate Rps10, functional against Pst, from WBDC085. A marker in complete coupling with Rps10 was not found; however, using the two peak markers at the BC2 and BC2F2 stages, Rps10 maps to a 16-Mb interval (554.80–570.84 Mb) (Clare, 2016). Rps10 was also mapped as QPsh-DP-2R-5.1 and QPsh-DP-2R-5.2 (Gyawali et al., 2021) and in close proximity to QPsh.FW6-5H.2 (Belcher et al., 2018) and QPsh-rM-6R-5.1 (Gyawali et al., 2021; Vatter et al., 2018) on the proximal flank and QPsh-r24-6R-5.2, QPsh-DP-2R-5.3, QPsh-rG-6R-5.2, and QPsh-rG-2R-5.1 (Gyawali et al., 2021) on the distal flank. Additional loci were mapped to chromosome 5H including the following: QPsh-rG-6R-5.1, QPsh-r24-6R-5.1, QPsh-r7S0-6R-5.1, QPsh-rM/Q-2R-5.1, QPsh-rM/Q-2R-5.2, QPsh-rM-2R-5.1, and QPsh-r24-2R-5.1 (Gyawali et al., 2021); QPsh.FW6-5H.1, QPsh.FW6-5H.3, six MTA most likely forming a single unnamed QTL, and another MTA (Belcher et al., 2018); one unnamed MTA (Hernandez et al., 2020a); qGH_PBIC_4.26 effective against Psph (Dracatos et al., 2019b); Qpsh.316A.5H (Esvelt Klos et al., 2020); five MTA that may form a single QTL (Dracatos et al., 2016b); and 14 unanchored MTA/QTL (Rao et al., 2007; Thomas et al., 1995; Visioni et al., 2018).
Chromosome 6H
Currently, only association mapping studies have identified stripe rust resistance loci on chromosome 6H. These include QPsh-rM-6R-6.1, QPsh-rM-6R-6.2, QPsh-rM-6R-6.3, QPsh.FW6-6H.4, QPsh-r57-6R-6.2, QPsh-DP-6R-6.1, QPsh-r7S0-6R-6.1, QPsh-r7S0-6R-6.2, QPsh-r7S0-6R-6.3, QPsh-r7S0-6R-6.4, QPsh-r24/57-6R-6.1, QPsh-rQ-6R-6.1 (Gyawali et al., 2021), QPsh.FW6-6H.1, QPsh.FW6-6H.2, QPsh.FW6-6H.3, one unnamed QTL (Belcher et al., 2018), Qpsh6H (Esvelt Klos et al., 2016), Qpsh.316A.6H (Esvelt Klos et al., 2020), and QPs.6H-1 (Vatter et al., 2018). Furthermore, seven MTA/QTL remain unanchored to chromosome 6H (Rao et al., 2007; Rossi et al., 2006; Visioni et al., 2018).
Chromosome 7H
Rps6 is functional only against Pst and was concurrently high-resolution mapped by two independent studies to a 267.6-kb (613.67–613.94 Mb) and 163.5-kb (613.77–613.93 Mb) interval (Dawson et al., 2016; Li et al., 2016). Rps6 was mapped in wild barley accessions and the German barley accession Abed Binder 12 (Dawson et al., 2016; Li et al., 2016) and potentially mapped in Franklin (Dracatos et al., 2016b). Rps6 was also identified using association mapping (Dracatos et al., 2016b; Gutiérrez et al., 2015). Rpsx was mapped to a 14.4-Mb (597.28–611.72 Mb) interval using CI10587 (Castro et al., 2003a). Rpsx has also been mapped as QPsh.FW6-7H (Belcher et al., 2018), two unnamed MTA forming a single QTL (Dracatos et al., 2016b), qGH_PBIC_4.51 against Psph (Dracatos et al., 2019b) and QPs.7H-1 (Vatter et al., 2018). Additional loci mapped to chromosome 7H include QPsh-r24/G-2R-7.1, QPsh-r24-2R-7.2, QPsh-rQ-6R-7.1, QPsh-r57-2R-7.1, QPsh-r7S0-2R-7.2, QPsh-rG-6R-7.1, and QPsh-rG-6R-7.2 (Gyawali et al., 2021); Rpsh-7H (Belcher et al., 2018); Qphs7H (Esvelt Klos et al., 2016); and Qpsh.316A.7H (Esvelt Klos et al., 2020). A single unnamed QTL also maps to chromosome 4H (Gutiérrez et al., 2015), whereas a further 10 MTA/QTL (Rao et al., 2007; Rossi et al., 2006; Visioni et al., 2018) remain unanchored to chromosome 4H.
Association mapping
To date, six association mapping studies have been used to characterize resistance to stripe rust in barley. The first used a Latin American barley population and identified a total of seven QTL, three of which were deemed novel (Gutiérrez et al., 2015). The second study, using the HEB-25 NAM panel, identified eight novel loci out of the 12 identified (Vatter et al., 2018). The third association study assessed Oregon and Minnesotan breeding material, identifying three ASR and 14 APR QTL, respectively, five of which were novel (Belcher et al., 2018). The fourth study assessed a global population of 261 barley accessions identifying 45 ASR and 18 APR QTL (Visioni et al., 2018); however, this cannot be verified as the DaRTseq markers could not be anchored to the Morex genome. The fifth study identified four QTL, none of which were novel; however, one locus was mapped for both stem and stripe rust (Hernandez et al., 2020a) in close proximity to Rph9. The last association mapping study assessed 336 ICARDA accessions with 42 ASR and 13 APR MTA identified, 33 of which were deemed novel (Gyawali et al., 2021).
Notable changes to net blotch consensus map
Marker positions for consolidated net blotch loci were identified using the Morex V3 genome assembly, further refining multiple loci after incorporating additional studies (Adhikari et al., 2020; Afanasenko et al., 2022; Alhashel et al., 2021, 2023; Clare et al., 2021; Czembor and Czembor, 2023; Esmail et al., 2023; Mazinani et al., 2020; Muria-Gonzalez et al., 2023; Skiba et al., 2022) and additional unanchored markers (Table 5; Supplementary Table 2). The total number of consensus loci for net blotch has been lowered from 73 to 72, despite the separation of SPN1 from Rpt5/Spt1, the identification of Rpt9 (Franckowiak and Platz, 2021), and the addition of two novel MTA (Czembor and Czembor, 2023). As noted, a major change in the net blotch consensus maps was the identification of Rpt9 (598.8–611.7 Mb), dividing Rpt4 (now 415.1–596.6 Mb) (Franckowiak and Platz, 2021) into two loci on the long arm of chromosome 7H. The Rpt4/Rpt9 region has been subsequently further characterized with two overlapping resistance loci identified at 592.2–602.0 Mb (Alhashel et al., 2021) and 587.1–598.8 Mb (Skiba et al., 2022), and therefore potentially mapping the same underlying gene of either Rpt4 or Rpt9. However, within this interval, a susceptibility locus was identified and high-resolution mapped (592.6–593.0 Mb) with the proposed name of Sptm1 (Alhashel et al., 2023). Whether Sptm1 is Rpt4, Rpt9, or a separate locus is currently unknown and requires further investigation. This is further supported by association mapping that identified two significant markers within the region (596.7 and 611.7 Mb) separated by multiple insignificant SNPs (Clare et al., 2021). Further work will be required to tease apart the true number of loci present in the Rpt4/Rpt9/Sptm1 region.
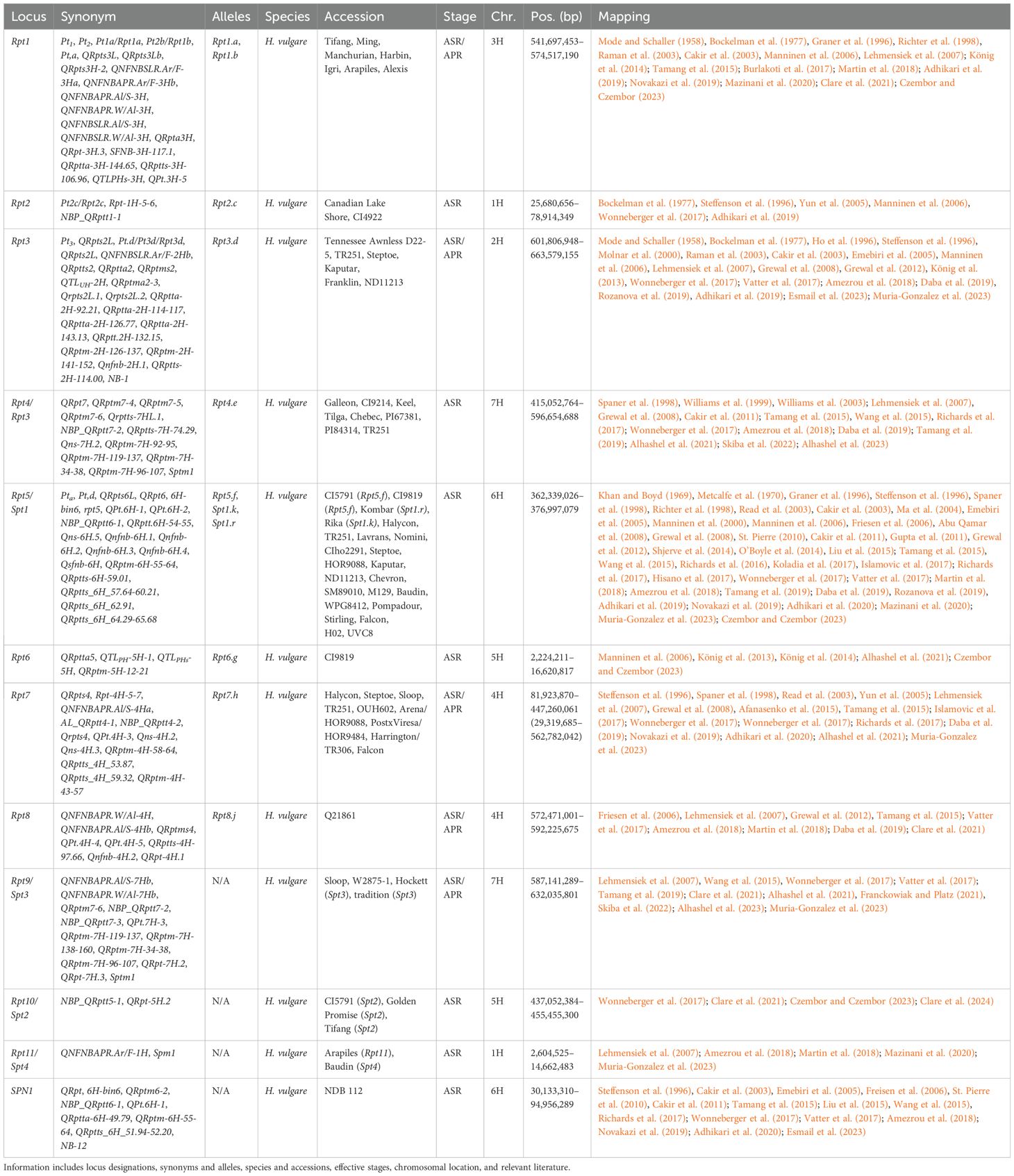
Table 5. Summary information of all designated resistance/susceptibility loci in barley net blotch to net blotch pathogen Pyrenophora teres.
The most broad and effective resistance gene, Rpt5, against Pyrenophora teres f. teres (Ptt) has been validated as a RLP at 364.7 Mb, whereas Spt1 remains under investigation (Effertz, 2023; Effertz et al., 2024). Due to the centromeric location of Rpt5/Spt1, the majority of markers identified on chromosome 5H are most likely in linkage disequilibrium currently spanning from 111.9 to 466.6 Mb. However, this separates the SPN1 locus originally delimited to 46.0–90.3 Mb (Liu et al., 2015). Additionally, further work will be required to determine if any additional loci are present within the large linkage block that currently delimits Rpt5/Spt1. Unfortunately, Rpt5 has been broken by Canadian (Akhavan et al., 2016), French (Arabi et al., 1992), Turkish (Çelik Oğuz and Karakaya, 2017), and Moroccan isolates (Li et al., 2023; Richards et al., 2024). Interestingly, an association mapping study into Egyptian germplasm did not identify Rpt5, instead identified seven significant markers on 3H (Esmail et al., 2023). These are reported as seven separate MTA; however, NB-2, NB-3, NB-4 colocalize with Rpt3H-4 (Afanasenko et al., 2022; Clare et al., 2021; Daba et al., 2019; Islamovic et al., 2017; König et al., 2014; Lehmensiek et al., 2007; Novakazi et al., 2019; Richards et al., 2017; Tamang et al., 2015, 2021; Wonneberger et al., 2017; Yun et al., 2005), and NB-5, NB-6, NB-7, and NB-8 colocalize with QRpts3La (Burlakoti et al., 2017; Cakir et al., 2011; Daba et al., 2019; Lehmensiek et al., 2007; Raman et al., 2003; Richards et al., 2017; Tamang et al., 2015, 2019; Vatter et al., 2017; Wonneberger et al., 2017).
Lastly, multiple susceptibility loci have been identified in the barley-P. teres f. maculata (Ptm) pathosystem, including Sptm1, described earlier. Firstly, Spt2 was been high-resolution mapped to a single pentatricopeptide repeat-containing protein candidate gene on chromosome 5H in two independent mapping populations that share CI5791 as a common parent (Clare et al., 2024). The Spt2 locus is unique in that all parents are considered resistance to the Ptm isolate 13IM8.3; however, all F1 individuals are considered hypersusceptible. This locus was previously mapped as resistance loci: NBP_QRptt5-1 (Wonneberger et al., 2017), QRpt-5H.2 (Clare et al., 2021), and potentially an unnamed QTL (Clare et al., 2020; Williams et al., 2003) raising interesting questions about whether the locus can function as a resistance and/or susceptibility target. Another locus designated Spm1 was identified within a 190-kb interval (9.12-9.31 Mb) on chromosome 1H (Muria-Gonzalez et al., 2023), that has previously been identified as resistance loci: QRptta-1H-4.11 (Amezrou et al., 2018), QRpta1H-2 (Mazinani et al., 2020), QNFNBAPR.Ar/F-1H (Lehmensiek et al., 2007) and a QTL identified between 19.4 and 25.7 cM (Martin et al., 2018). The Susceptibility to P. tritici-repentis 1 (Spr1) locus facilitates susceptibility to the pathogen P. tritici-repentis (Ptr), which primarily causes tan spots of wheat (Wei et al., 2020). The Spr1 locus to 9.4–12.0 Mb has also been included in the P. teres maps (Figure 1) and colocalizes with SFNB-2H-8-10 (Burlakoti et al., 2017), NBP_QRptt2-1 (Wonneberger et al., 2017), QRptts-2H-7.44 (Amezrou et al., 2018), QRptts-2H-9.00, QRptts-2H-161.70 (Adhikari et al., 2019), and unnamed QTL (Liu et al., 2015; Skiba et al., 2022; Tamang et al., 2015, 2019).
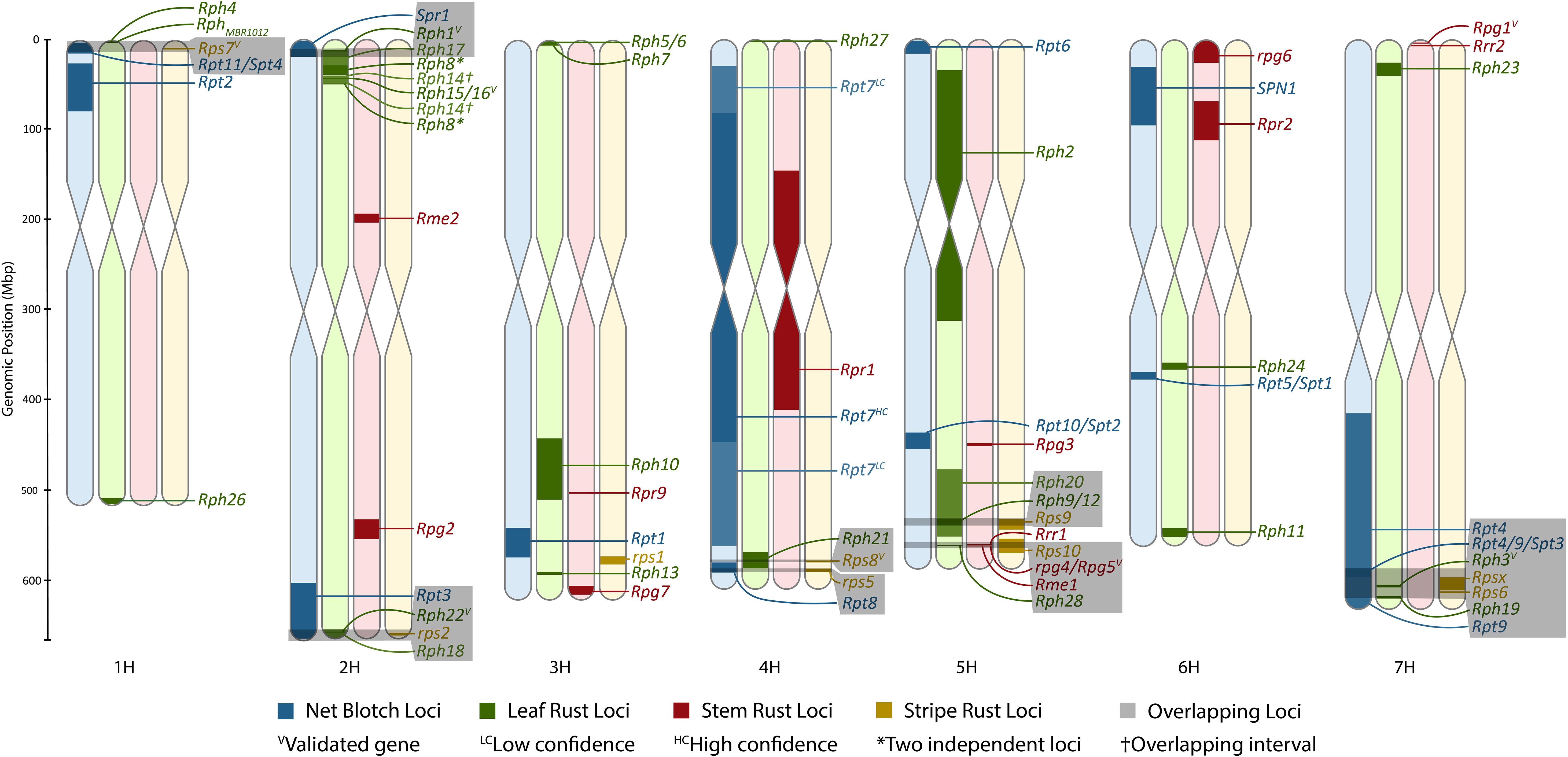
Figure 1. Karyotype density map of all rust pathosystems in comparison to previously anchored net blotch loci. The figure was made using Rideogram and restricted to only formally designated loci that could be anchored to the Morex V3 genome. The size of intervals may be increased to allow for visualization if an interval is sufficiently refined.
A trend of identifying susceptibility loci within the barley-P. teres pathosystem appears to be common after the initial identification of the Spt1 susceptibility locus (Richards et al., 2016), followed by Spt2 (Clare, 2022; Clare et al., 2024), Sptm1 (Alhashel et al., 2023), Spm1 (Muria-Gonzalez et al., 2023), and resistant accessions contributing susceptibility alleles at QRptm-3H-45-52, QRptm-5H-12-21, QRptm-5H-81-88, and QRptm-6H-60-64 (Alhashel et al., 2021). Loci nomenclature needs to be addressed urgently, considering multiple loci are identified to be effective against Ptt and Ptm. Despite recent research suggesting incipient speciation between Ptt and Ptm (Yuzon et al., 2023), there is considerable overlap (Clare et al., 2020) and the custom of utilizing three letters and numbers to designate barley loci (Bockelman et al., 1977). The designations Rpt and Spt provide no discrimination between Ptt and Ptm, unlike Sptm and Spm, which suggest these loci are only implicated within Ptm interactions, despite the fact these have been previously identified within the Ptt interaction but not formally designated. We therefore propose Sptm1 is renamed to Rpt4/Rpt9/Spt3 and Spm1 to Rpt11/Spt4. In addition, with the increased number of alleles being discovered at identified designated loci, e.g., Rph2 and Rps8 with 12 and 27 alleles, respectively, we also propose the removal of the current convention to designate alleles with a letter across all loci within the pathosystem and instead restart with each locus, i.e., Rpt2.b becomes Rpt2.a.
Genomic resources and locus colocalization
Currently, there are over 25 full pseudomolecule chromosome assemblies of barley to assess for allelic diversity, ranging from wild accessions, landraces, and cultivars (Jayakodi et al., 2020; Jiang et al., 2022; Mascher et al., 2021; Sakkour et al., 2022; Sato et al., 2021; Schreiber et al., 2020; Xu et al., 2021), that will no doubt expand with the ever-decreasing cost of long-read sequencing. These resources will allow for the rapid identification of new alleles once genes underlying resistance or susceptibility loci have been validated. In summary, chromosomes 6H and 5H contain the least and most amount of formally designated rust resistance genes, respectively (Figure 1; Table 6); however, this may not hold true as additional loci are added to the portfolio. A limitation of this atlas is that frequently raw data are not readily available to determine the nearest insignificant markers, to precisely delimit the MTA/QTL interval, and ultimately identify overlaps between loci. This is particularly a problem with association mapping studies and would therefore implore all marker data to be published in the future. With numerous studies remapping the same locus and the dearth of novel loci, research should focus on refining the genomic intervals underlying these intervals and gene validation.
A total of eight colocalizations of formally designated loci were identified, including Rps7 with Rpt11/Spt4; Spr1 with Rph1 or Rph17; rps2 with Rph18 or Rph22 and potentially Rpt3; Rps8 and Rph21; Rpt8 and rps5; Rph20 with Rph9/12 and Rps9; rpg4/Rpg5 with Rps10 and Rph28; and Rpt4/Rpt9/Spt3 with Rph3, Rph19, Rps6, and/or Rpsx on chromosomes 1H, 2H, 2H, 4H, 4H, 5H, 5H, and 7H, respectively (Figure 1). Rpg3 appears to colocalize with Rpt10/Spt2; however, the refined Spt2 region does not overlap with Rpg3, and therefore further investigation will be required to determine if Rpt10 colocalizes with Rpg3. The most notable colocalization is Rph28 and Rps10 with rpg4/Rpg5 on chromosome 5H. The Rph28 locus encompasses two of the three genes within the validated rpg4/Rpg5 complex, including the two NLRs of rpg4/Rpg5, Adf2, and two additional zinc fingers. In addition, Rps10 colocalizes with rpg4/Rpg5 and Rph28, although with a larger overlapping interval. Therefore, there is a high likelihood that the validated dual NLR genetic architecture of rpg4/Rpg5 and/or novel alleles of the rpg4/Rpg5 complex is functional against leaf, stripe, and stem rust. Both reports of Rph28 and Rps10 lack mention of the rpg4/Rpg5 complex, despite being validated over a decade ago and providing the most widespread resistance to stem rust, therefore showcasing the importance of high-resolution genetic mapping and developing a barley gene atlas.
Conclusion
This work was initiated due to the fact plant defense responses are highly coordinated and interconnected, with recent research showcasing sympathetic or antagonistic relationships of pathogen recognition mechanisms. Therefore, without a comprehensive resource collating all known resistance/susceptibility loci, identifying previously reported loci within additional accessions against different pathogens, or both, becomes burdensome. We therefore began the process of consolidating loci that confer resistance or susceptibility to three of the most important diseases of barley in leaf, stem, and stripe rust. In addition, previously reported net blotch loci were updated and included in the colocalization analysis. There is often difficulty determining which loci colocalize with each other due to asynchronous marker technologies and the larger mapping intervals of early mapping studies. However, researchers will be able to use this resource to quickly identify previously reported loci without intimate knowledge of these pathosystems, as we show with rpg4/Rpg5, Rph28, and Rps10 loci. Ideally, this atlas will be expanded to include additional fungal diseases as well as bacterial and viral diseases to identify conserved resistance mechanisms or pathogen-specific resistance to inform breeders in the development of highly resistant cultivars.
Author contributions
SC: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Software, Visualization, Writing – original draft, Writing – review & editing, Project administration. FN: Data curation, Investigation, Writing – original draft, Writing – review & editing, Formal analysis, Validation. PH: Writing – review & editing, Formal analysis, Validation. MM: Formal analysis, Investigation, Methodology, Resources, Writing – original draft, Writing – review & editing, Data curation, Software, Validation. RB: Funding acquisition, Resources, Supervision, Writing – review & editing, Validation.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. The research presented in this manuscript was supported by the USDA National Institute of Food and Agriculture Hatch project 1014919, Crop Improvement and Sustainable Production Systems (WSU reference 00011), and United States Department of Agriculture-Agricultural Research Service CRIS No. 5062-21220-025-000D.
Acknowledgments
The authors would like to thank Andrzej Kilian for DArT and DArTseq’s assistance. DArT marker sequences can be found at https://www.diversityarrays.com/technology-and-resources/sequences/. Authors would also like to acknowledge the maintainers of T3 Barley: Legacy and BreedBase, GrainGenes, HarvEST Barley, NCBI, Enslembl Plants and Geneious Prime to determine new genomic positions.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fagro.2024.1451281/full#supplementary-material
Supplementary Table 1 | All markers reported as intervals of surrounding barley disease resistance genes for cereal rust and their inferred loci designation based on consolidation.
Supplementary Table 2 | Upgraded marker positions for all reported net blotch resistance markers and their new loci consolidation.
Abbreviations
APR, adult plant resistance; ASR, seedling/all-stage resistance; f., form (different forms of a species with the same host); f. sp., forma specialis (same species with different host specificity); ff. spp., formae speciales (plural of f. sp.); MTA, marker-trait association; Pch, Puccinia coronata var. hordei; Ph, Puccinia hordei; Pt, Puccinia triticina; Psh, Puccinia striiformis f. sp. hordei; Pst, Puccinia striiformis f. sp. tritici; Pga, Puccinia graminis f. sp. avenae; Pgs, Puccinia graminis f. sp. secalis; Pgt, Puccinia graminis f. sp. tritici; Ptm, Pyrenophora teres f. maculata; Ptt, Pyrenophora teres f. teres; Ptr, Pyrenophora tritici-repentis; QTL, quantitative trait locus/loci; Rpc#, Reaction to Puccinia coronata #; Rpg#, Reaction to Puccinia graminis #; Rph#, Reaction to Puccinia hordei #; Rps#, Reaction to Puccinia striiformis #; Rpt#, Reaction to Pyrenophora teres #; SNP, single-nucleotide polymorphism; Spt#, Susceptibility to Pyrenophora teres #; Spr#, Susceptibility to Pyrenophora tritici-repentis #.
References
Abu Qamar M., Liu Z. H., Faris J. D., Chao S., Edwards M. C., Lai Z., et al. (2008). A region of barley chromosome 6H harbors multiple major genes associated with net type net blotch resistance. Theor. Appl. Genet. 117. doi: 10.1007/s00122-008-0860-x
Adhikari A., Steffenson B. J., Smith M. J., Dill-Macky R. (2019). Genome-wide association mapping of seedling net form net blotch resistance in an Ethiopian and Eritrean barley collection. Crop Sci. 59, 1625–1638. doi: 10.2135/cropsci2019.01.0003
Adhikari A., Steffenson B. J. J., Smith K. P. P., Smith M., Dill-Macky R. (2020). Identification of quantitative trait loci for net form net blotch resistance in contemporary barley breeding germplasm from the USA using genome-wide association mapping. Theor. Appl. Genet. 133, 1019–1037. doi: 10.1007/s00122-019-03528-5
Afanasenko O., Rozanova I., Gofman A., Lashina N., Novakazi F., Mironenko N., et al. (2022). Validation of molecular markers of barley net blotch resistance loci on chromosome 3H for marker-assisted selection. Agriculture 12, 439. doi: 10.3390/agriculture12040439
Afanasenko O. S., Koziakov A. V., Hedlay P. E., Lashina N. M., Anisimova A. V., Manninen O., et al. (2015). Mapping of the loci controlling the resistance to Pyrenophora teres f. teres and Cochliobolus sativus in two double haploid barley populations. Russian J. Genetics: Appl. Res. 5, 242–253. doi: 10.1134/S2079059715030028
Agrama H. A., Dahleen L., Wentz M., Jin Y., Steffenson B. (2004). Molecular mapping of the crown rust resistance gene rpc1 in barley. Phytopathology® 94, 858–861. doi: 10.1094/PHYTO.2004.94.8.858
Akhavan A., Turkington T. K., Askarian H., Tekauz A., Xi K., Tucker J. R., et al. (2016). Virulence of Pyrenophora teres populations in western Canada. Can. J. Plant Pathol. 38, 183–196. doi: 10.1080/07060661.2016.1159617
Alhashel A. F., Fiedler J. D., Nandety R. S., Skiba R. M., Bruggeman R. S., Baldwin T., et al. (2023). Genetic and physical localization of a major susceptibility gene to Pyrenophora teres f. maculata in barley. Theor. Appl. Genet. 136, 118. doi: 10.1007/s00122-023-04367-1
Alhashel A. F., Sharma Poudel R., Fiedler J., Carlson C. H., Rasmussen J., Baldwin T., et al. (2021). Genetic mapping of host resistance to the Pyrenophora teres f. maculata isolate 13IM8.3. G3 Genes|Genomes|Genetics 11, jkab341. doi: 10.1093/g3journal/jkab341
Amezrou R., Verma R. P. S., Chao S., Brueggeman R. S., Belqadi L., Arbaoui M., et al. (2018). Genome-wide association studies of net form of net blotch resistance at seedling and adult plant stages in spring barley collection. Mol. Breed. 38, 58. doi: 10.1007/s11032-018-0813-2
Amo A., Soriano J. M. (2022). Unravelling consensus genomic regions conferring leaf rust resistance in wheat via meta-QTL analysis. Plant Genome 15, e20185. doi: 10.1002/tpg2.20185
Amouzoune M., Rehman S., Benkirane R., Verma S., Gyawali S., Al-Jaboobi M., et al. (2022). Genome-wide association study of leaf rust resistance at seedling and adult plant stages in a global barley panel. Agriculture 12, 1829. doi: 10.3390/agriculture12111829
Arabi M. I., Barrault G., Sarrafi A., Albertini L. (1992). Variation in the resistance of barley cultivars and in the pathogenicity of Drechslera teres f. sp. maculata and D. teres f. sp. teres isolates from France. Plant Pathol. 41, 180–186. doi: 10.1111/j.1365-3059.1992.tb02336.x
Arora D., Gross T., Brueggeman R. (2013). Allele characterization of genes required for rpg4-mediated wheat stem rust resistance identifies rpg5 as the R gene. Phytopathology® 103, 1153–1161. doi: 10.1094/PHYTO-01-13-0030-R
Ashikawa I., Hayashi N., Yamane H., Kanamori H., Wu J., Matsumoto T., et al. (2008). Two adjacent nucleotide-binding site–leucine-rich repeat class genes are required to confer pikm-specific rice blast resistance. Genetics 180, 2267–2276. doi: 10.1534/genetics.108.095034
Bailey P. C., Schudoma C., Jackson W., Baggs E., Dagdas G., Haerty W., et al. (2018). Dominant integration locus drives continuous diversification of plant immune receptors with exogenous domain fusions. Genome Biol. 19, 23. doi: 10.1186/s13059-018-1392-6
Beddow J. M., Pardey P. G., Chai Y., Hurley T. M., Kriticos D. J., Braun H.-J., et al. (2015). Research investment implications of shifts in the global geography of wheat stripe rust. Nat. Plants 1, 15132. doi: 10.1038/nplants.2015.132
Belcher A. R., Cuesta-Marcos A., Smith K. P., Mundt C. C., Chen X., Hayes P. M. (2018). TCAP FAC-WIN6 elite barley GWAS panel QTL. I. Barley stripe rust resistance QTL in facultative and winter six-rowed malt barley breeding programs identified via GWAS. Crop Sci. 58, 103–119. doi: 10.2135/cropsci2017.03.0206
Bettgenhaeuser J., Hernández-Pinzón I., Dawson A. M., Gardiner M., Green P., Taylor J., et al. (2021). The barley immune receptor Mla recognizes multiple pathogens and contributes to host range dynamics. Nat. Commun. 12, 6915. doi: 10.1038/s41467-021-27288-3
Bockelman H. E., Sharp E. L., Eslick R. F. (1977). Trisomic analysis of genes for resistance to scald and net blotch in several barley cultivars. Can. J. Bot. 55, 2142–2148. doi: 10.1139/b77-242
Bolton M. D., Kolmer J. A., Garvin D. F. (2008). Wheat leaf rust caused by Puccinia triticina. Mol. Plant Pathol. 9, 563–575. doi: 10.1111/j.1364-3703.2008.00487.x
Borovkova I. G., Jin Y., Steffenson B. J. (1998). Chromosomal location and genetic relationship of leaf rust resistance genes rph9 and rph12 in barley. Phytopathology® 88, 76–80. doi: 10.1094/PHYTO.1998.88.1.76
Borovkova I. G., Steffenson B. J., Jin Y., Kilian A., Kleinhofs A., Blake T. K. (1997). Identification and mapping of a leaf rust resistance gene in barley line Q21861. Genome 40, 236–241. doi: 10.1139/g97-033
Borovkova I. G., Steffenson B. J., Jin Y., Rasmussen J. B., Kilian A., Kleinhofs A., et al. (1995). Identification of molecular markers linked to the stem rust resistance gene rpg4 in barley. Phytopathology® 85, 181–185.
Brabham H. J., Gómez de la Cruz D., Were V., Shimizu M., Saitoh H., Hernández-Pinzón I., et al. (2023). Barley MLA3 recognizes the host-specificity effector Pwl2 from Magnaporthe oryzae. Plant Cell 36, 447–470. doi: 10.1093/plcell/koad266
Brueggeman R., Drader T., Kleinhofs A. (2006). The barley serine/threonine kinase gene Rpg1 providing resistance to stem rust belongs to a gene family with five other members encoding kinase domains. Theor. Appl. Genet. 113, 1147–1158. doi: 10.1007/s00122-006-0374-3
Brueggeman R., Druka A., Nirmala J., Cavileer T., Drader T., Rostoks N., et al. (2008). The stem rust resistance gene Rpg5 encodes a protein with nucleotide-binding-site, leucine-rich, and protein kinase domains. Proc. Natl. Acad. Sci. 105, 14970 LP–14975. doi: 10.1073/pnas.0807270105
Brueggeman R., Rostoks N., Kudrna D., Kilian A., Han F., Chen J., et al. (2002). The barley stem rust-resistance gene Rpg1 is a novel disease-resistance gene with homology to receptor kinases. Proc. Natl. Acad. Sci. 99, 9328–9333. doi: 10.1073/pnas.142284999
Brueggeman R., Steffenson B. J., Kleinhofs A. (2009). The rpg4/Rpg5 stem rust resistance locus in barley; resistance genes and cytoskeleton dynamic. Cell Cycle 8, 977–981. doi: 10.4161/cc.8.7.8079
Brueggeman R. S., Solanki S., Ameen G., Effertz K., Poudel R. S., Karakaya A. (2020). “Fungal diseases affecting barley,” in Achieving sustainable cultivation of barley. Eds. Fox G., Li C. (Burleigh Dodds Science Publishing Limited, Cambridge, UK), 1–58.
Brunner S., Keller B., Feuillet C. (2000). Molecular mapping of the Rph7.g leaf rust resistance gene in barley (Hordeum vulgare l.). Theor. Appl. Genet. 101, 783–788. doi: 10.1007/s001220051544
Burlakoti R. R., Gyawali S., Chao S., Smith K. P., Horsley R. D., Cooper B., et al. (2017). Genome-wide association study of spot form of net blotch resistance in the Upper Midwest barley breeding programs. Phytopathology 107, 100–108. doi: 10.1094/PHYTO-03-16-0136-R
Cakir M., Gupta S., Li C., Hayden M., Mather D. E., Ablett G. A., et al. (2011). Genetic mapping and QTL analysis of disease resistance traits in the barley population Baudin × AC Metcalfe. Crop Pasture Sci. 62, 152–161. doi: 10.1071/CP10154
Cakir M., Gupta S., Platz G. J., Ablett G. A., Loughman R., Emebiri L. C., et al. (2003). Mapping and validation of the genes for resistance to Pyrenophora teres f. teres in barley (Hordeum vulgare L.). Aust J Agric Res. 54, 1369–1377. doi: 10.1071/AR02229
Case A. J., Bhavani S., Macharia G., Pretorius Z., Coetzee V., Kloppers F., et al. (2018a). Mapping adult plant stem rust resistance in barley accessions Hietpas-5 and GAW-79. Theor. Appl. Genet. 131, 2245–2266. doi: 10.1007/s00122-018-3149-8
Case A. J., Bhavani S., Macharia G., Steffenson B. J. (2018b). Genome-wide association study of stem rust resistance in a world collection of cultivated barley. Theor. Appl. Genet. 131, 107–126. doi: 10.1007/s00122-017-2989-y
Castro A. J., Capettini F., Corey A. E., Filichkina T., Hayes P. M., Kleinhofs A., et al. (2003a). Mapping and pyramiding of qualitative and quantitative resistance to stripe rust in barley. Theor. Appl. Genet. 107, 922–930. doi: 10.1007/s00122-003-1329-6
Castro A. J., Chen X., Hayes P. M., Johnston M. (2003b). Pyramiding quantitative trait locus (QTL) alleles determining resistance to barley stripe rust. Crop Sci. 43, 651–659. doi: 10.2135/cropsci2003.6510
Castro A. J., Chen X., Hayes P. M., Knapp S. J., Line R. F., Toojinda T., et al. (2002). Coincident QTL which determine seedling and adult plant resistance to stripe rust in barley. Crop Sci. 42, 1701–1708. doi: 10.2135/cropsci2002.1701
Castro A. J., Gamba F., German S., Gonzalez S., Hayes P. M., Pereyra S., et al. (2012). Quantitative trait locus analysis of spot blotch and leaf rust resistance in the BCD47 × Baronesse barley mapping population. Plant Breed. 131, 258–266. doi: 10.1111/j.1439-0523.2011.01930.x
Çelik Oğuz A., Karakaya A. (2017). Pathotypes of Pyrenophora teres on barley in Turkey. Phytopathol. Mediterr. 56, 224–234. doi: 10.14601/Phytopathol_Mediterr-20267
Cesari S., Thilliez G., Ribot C., Chalvon V., Michel C., Jauneau A., et al. (2013). The rice resistance protein pair RGA4/RGA5 recognizes the magnaporthe oryzae effectors AVR-pia and AVR1-CO39 by direct binding. Plant Cell 25, 1463–1481. doi: 10.1105/tpc.112.107201
Chai Y., Nirmala J., Kleinhofs A., Steffenson B. (2012). Failure of RPG1 protein to degrade in high-copy Rpg1 transgenic barley lines results in susceptibility to stem rust. Physiol. Mol. Plant Pathol. 80, 10–18. doi: 10.1016/j.pmpp.2012.07.003
Chełkowski J., Tyrka M., Sobkiewicz A. (2003). Resistance genes in barley (Hordeum vulgare L.) and their identification with molecular markers. J. Appl. Genet. 44, 291–309.
Chen C., Jost M., Clark B., Martin M., Matny O., Steffenson B. J., et al. (2021). BED domain-containing NLR from wild barley confers resistance to leaf rust. Plant Biotechnol. J. 19, 1206–1215. doi: 10.1111/pbi.13542
Chen C., Jost M., Outram M. A., Friendship D., Chen J., Wang A., et al. (2023). A pathogen-induced putative NAC transcription factor mediates leaf rust resistance in barley. Nat. Commun. 14, 5468. doi: 10.1038/s41467-023-41021-2
Chen X., Line R. F. (1999). Recessive Genes for Resistance to Puccinia striiformis f. sp. hordei in Barley. Phytopathology® 89, 226–232. doi: 10.1094/PHYTO.1999.89.3.226
Chen X., Line R. F. (2001). Genes for Resistance to Barley Stripe Rust (Puccinia striiformis f. sp. hordei). Barley Genet. Newslett. 31, 31–36.
Chen X., Line R. F. (2003). Identification of genes for resistance to Puccinia striiformis f. sp. hordei in 18 barley genotypes. Euphytica 129, 127–146. doi: 10.1023/A:1021585907493
Chen X., Line R. F., Leung H. (1995). Virulence and Polymorphic DNA Relationships of Puccinia striiformis f. sp. hordei to Other Rusts. Phytopathology® 85, 1335–1342. doi: 10.1094/Phyto-85-1335
Chen F. Q., Prehn D., Hayes P. M., Mulrooney D., Corey A., Vivar H. (1994). Mapping genes for resistance to barley stripe rust (Puccinia striiformis f. sp. hordei). Theor. Appl. Genet. 88, 215–219. doi: 10.1007/BF00225900
Chen S., Zhang W., Bolus S., Rouse M. N., Dubcovsky J. (2018). Identification and characterization of wheat stem rust resistance gene Sr21 effective against the Ug99 race group at high temperature. PloS Genet. 14, e1007287. doi: 10.1371/journal.pgen.1007287
Clare S. (2016). Development of a near-isogenic line panel in barley to stripe rust (Norwich: University of East Anglia). doi: 10.1186/s12870-024-05303-1
Clare S. J. (2022). Genetic Characterization of Barley and Pyrenophora teres Host Pathogen Interactions. (Pullman: Washington State University).
Clare S. J., Alhashel A. F., Li M., Effertz K. M., Poudel R. S., Zhang J., et al. (2024). High resolution mapping of novel non-transgressive hybrid susceptibility in barley exploited by P. teres f. maculata maps to a single pentatricopeptide repeat-containing protein. BMC Plant Biol 24, 622. doi: 10.1186/s12870-024-05303-1
Clare S. J., Çelik Oğuz A., Effertz K., Sharma Poudel R., See D., Karakaya A., et al. (2021). Genome-wide association mapping of Pyrenophora teres f. maculata and Pyrenophora teres f. teres resistance loci utilizing natural Turkish wild and landrace barley populations. G3 Genes|Genomes|Genetics 11, 1–13. doi: 10.1093/g3journal/jkab269
Clare S., Kitcher W., Gardiner M., Green P., Hubbard A., Moscou M. J. (2016). Defining the genetic architecture of stripe rust resistance in the barley accession HOR 1428. bioRxiv, 93773. doi: 10.1101/093773
Clare S. J., Wyatt N. A., Brueggeman R. S., Friesen T. L. (2020). Research advances in the Pyrenophora teres–barley interaction. Mol. Plant Pathol. 21, 272–288. doi: 10.1111/mpp.12896
Cotterill P. J., Rees R. G., Platz G. J., Dill-Macky R. (1992). Effects of leaf rust on selected Australian barleys. Aust. J. Exp. Agric. 32, 747–751. doi: 10.1071/EA9920747
Czembor J. H., Czembor E. (2023). Barley Sources of Resistance to the Net Form of Net Blotch (Pyrenophora teres f. teres). Biol. Life Sci. Forum 27, 9. doi: 10.3390/IECAG2023-15517
Czembor J. H., Czembor E., Suchecki R., Watson-Haigh N. S. (2022). Genome-wide association study for powdery mildew and rusts adult plant resistance in European spring barley from polish gene bank. Agronomy 7, 1–24. doi: 10.3390/agronomy12010007
Daba S. D., Horsley R., Brueggeman R., Chao S., Mohammadi M. (2019). Genome-wide association studies and candidate gene identification for leaf scald and net blotch in barley (Hordeum vulgare L.). Plant Dis. 103, 880–889. doi: 10.1094/PDIS-07-18-1190-RE
Dawson A. M. (2015). Elucidating the molecular genetics of host and nonhost resistance in barley to stripe rust. (Norwich: University of East Anglia).
Dawson A. M., Ferguson J. N., Gardiner M., Green P., Hubbard A., Moscou M. J. (2016). Isolation and fine mapping of Rps6: an intermediate host resistance gene in barley to wheat stripe rust. Theor. Appl. Genet. 129, 831–843. doi: 10.1007/s00122-015-2659-x
Dean R., Van Kan J., Pretorius Z. A., Hammond-Kosack K., Di Pietro A., Spanu P. D., et al. (2012). The Top 10 fungal pathogens in molecular plant pathology. Mol. Plant Pathol. 13, 414–430. doi: 10.1111/j.1364-3703.2011.00783.x
Derevnina L., Singh D., Park R. F. (2015). The genetic relationship between barley leaf rust resistance genes located on chromosome 2HS. Euphytica 203, 211–220. doi: 10.1007/s10681-014-1315-x
Dinh H. X., Singh D., Gomez de la Cruz D., Hensel G., Kumlehn J., Mascher M., et al. (2022). The barley leaf rust resistance gene Rph3 encodes a predicted membrane protein and is induced upon infection by avirulent pathotypes of Puccinia hordei. Nat. Commun. 13, 2386. doi: 10.1038/s41467-022-29840-1
Dracatos P. M., Ahmed M., Berlin A., Park R. F., Niks R. E. (2016a). Isolate Specificity and Polygenic Inheritance of Resistance in Barley to the Heterologous Rust Pathogen Puccinia graminis f. sp. avenae. Phytopathology® 106, 1029–1037. doi: 10.1094/PHYTO-10-15-0264-R
Dracatos P. M., Bartoš J., Elmansour H., Singh D., Karafiátová M., Zhang P., et al. (2019a). The coiled-coil NLR rph1, confers leaf rust resistance in barley cultivar Sudan. Plant Physiol. 179, 1362–1372. doi: 10.1104/pp.18.01052
Dracatos P. M., Haghdoust R., Singh R. P., Huerta Espino J., Barnes C. W., Forrest K., et al. (2019b). High-density mapping of triple rust resistance in barley using DArT-Seq markers. Front. Plant Sci. 10. doi: 10.3389/fpls.2019.00467
Dracatos P. M., Khatkar M. S., Singh D., Park R. F. (2014). Genetic mapping of a new race specific resistance allele effective to Puccinia hordei at the Rph9/Rph12 locus on chromosome 5HL in barley. BMC Plant Biol. 14, 1598. doi: 10.1186/s12870-014-0382-4
Dracatos P. M., Khatkar M., Singh D., Stefanato F., Park R. F., Boyd L. A. (2016b). Resistance in Australian barley (Hordeum vulgare) germplasm to the exotic pathogen Puccinia striiformis f. sp. hordei, causal agent of stripe rust. Plant Pathol. 65, 734–743. doi: 10.1111/ppa.12448
Dracatos P. M., Park R. F., Singh D. (2021). Validating molecular markers for barley leaf rust resistance genes rph20 and rph24. Plant Dis. 105, 743–747. doi: 10.1094/PDIS-08-20-1735-SC
Dracatos P. M., Singh D., Bansal U., Park R. F. (2015a). Identification of new sources of adult plant resistance to Puccinia hordei in international barley (Hordeum vulgare L.) germplasm. Eur. J. Plant Pathol. 141, 463–476. doi: 10.1007/s10658-014-0556-9
Dracatos P. M., Singh D., Fetch T., Park R. F. (2015b). Resistance to Puccinia graminis f. sp. avenae in Barley Is Associated with the Rpg5 Locus. Phytopathology® 105, 490–494. doi: 10.1094/PHYTO-08-14-0224-R
Dubin H. J., Stubbs R. W. (1986). Epidemic spread of barley stripe rust in south america. Plant Disease1 70, 141–144. doi: 10.1094/PD-70-141
Edae E. A., Rouse M. N. (2020). Association mapping of resistance to emerging stem rust pathogen races in spring wheat using genotyping-by-sequencing. Plant Genome 13, e20050. doi: 10.1002/tpg2.20050
Effertz K. (2023). Genetic and Functional Characterization of the Host-Pathogen Intereactions Underlying the Barley-Net Form Net Blotch Pathosystem. (Pullman: Washington State University).
Effertz K., Richards J. K., Clare S. J., Castillo M. D., Poudel R. S., Li M., et al. (2024). Rpt5 encodes a receptor-like protein that provides broad and effective net form net blotch (Pyrenophora teres f. teres) resistance in barley. bioRxiv. doi: 10.1101/2024.08.09.607401
Emebiri L. C., Platz C. G., Moody D. B. (2005). Disease resistance genes in a doubled haploid population of two-rowed barley segregating for malting quality attributes. Aust. J. Agric. Res. 56, 49–56. doi: 10.1071/AR04102
Eriksson J. (1894). Ueber die Specialisirung des Parasitismus bei den Getreiderostpilzen Vol. 12 (Berichte der Deutschen Botanischen Gesellschaft).
Esmail S. M., Jarquín D., Börner A., Sallam A. (2023). Genome-wide association mapping highlights candidate genes and immune genotypes for net blotch and powdery mildew resistance in barley. Comput. Struct. Biotechnol. J. 21, 4923–4932. doi: 10.1016/j.csbj.2023.10.014
Esvelt Klos K., Gordon T., Bregitzer P., Hayes P., Chen X. M., del Blanco I. A., et al. (2016). Barley Stripe Rust Resistance QTL: Development and Validation of SNP Markers for Resistance to Puccinia striiformis f. sp. hordei. Phytopathology® 106, 1344–1351. doi: 10.1094/PHYTO-09-15-0225-R
Esvelt Klos K., Hayes P., del Blanco I. A., Chen X., Filichkin T., Helgerson L., et al. (2020). Quantitative trait loci for field resistance to barley stripe rust derived from malting line 95SR316A. Crop Sci. 60, 1844–1853. doi: 10.1002/csc2.20154
Fazlikhani L., Keilwagen J., Kopahnke D., Deising H., Ordon F., Perovic D. (2019). High Resolution Mapping of Rph (MBR1012) Conferring Resistance to Puccinia hordei in Barley (Hordeum vulgare L.). Front. Plant Sci. 10. doi: 10.3389/fpls.2019.00640
Fetch T., Johnston P. A., Pickering R. (2009). Chromosomal Location and Inheritance of Stem Rust Resistance Transferred from Hordeum bulbosum into Cultivated Barley (H. vulgare). Phytopathology® 99, 339–343. doi: 10.1094/PHYTO-99-4-0339
Feuerstein U., Brown A. H. D., Burdon J. J. (1990). Linkage of rust resistance genes from wild barley (Hordeum spontaneum) with isozyme markers. Plant Breed. 104, 318–324. doi: 10.1111/j.1439-0523.1990.tb00442.x
Figueroa M., Hammond-Kosack K. E., Solomon P. S. (2018). A review of wheat diseases—a field perspective. Mol. Plant Pathol. 19, 1523–1536. doi: 10.1111/mpp.12618
Fox S. L., Harder D. E. (1995). Resistance to stem rust in barley and inheritance of resistance to race QCC. Can. J. Plant Science1 75, 781–788. doi: 10.4141/cjps95-132
Franckowiak J. D., Platz G. J. (2021). “Breeding barley for durable resistance to net and spot forms of net blotch,” in Achieving durable disease resistance in cereals (Cambridge, UK: Burleigh Dodds Science Publishing Limited), 567–586.
Friesen T. L., Faris J. D., Lai Z., Steffenson B. J. (2006). Identification and chromosomal location of major genes for resistance to Pyrenophora teres in a doubled-haploid barley population. Genome 49, 855–859. doi: 10.1139/g06-024
Gill U., Brueggeman R., Nirmala J., Chai Y., Steffenson B., Kleinhofs A. (2016). Molecular and genetic characterization of barley mutants and genetic mapping of mutant rpr2 required for Rpg1-mediated resistance against stem rust. Theor. Appl. Genet. 129, 1519–1529. doi: 10.1007/s00122-016-2721-3
Gill U., Nirmala J., Brueggeman R., Kleinhofs A. (2012). Identification, characterization and putative function of HvRin4, a barley homolog of Arabidopsis Rin4. Physiol. Mol. Plant Pathol. 80, 41–49. doi: 10.1016/j.pmpp.2012.08.002
Golegaonkar P. G., Karaoglu H., Park R. F. (2009). Molecular mapping of leaf rust resistance gene Rph14 in Hordeum vulgare. Theor. Appl. Genet. 119, 1281–1288. doi: 10.1007/s00122-009-1132-0
González A. M., Marcel T. C., Niks R. E. (2012). Evidence for a minor gene–for–minor gene interaction explaining nonhypersensitive polygenic partial disease resistance. Phytopathology® 102, 1086–1093. doi: 10.1094/PHYTO-03-12-0056-R
Graner A., Foroughi-Wehr B., Tekauz A. (1996). RFLP mapping of a gene in barley conferring resistance to net blotch (Pyrenophora teres). Euphytica 91, 229–234. doi: 10.1007/BF00021075
Graner A., Streng S., Drescher A., Jin Y., Borovkova I., Steffenson B. J. (2000). Molecular mapping of the leaf rust resistance gene Rph7 in barley. Plant Breed. 119, 389–392. doi: 10.1046/j.1439-0523.2000.00528.x
Grewal T. S., Rossnagel B. G., Pozniak C. J., Scoles G. J. (2008). Mapping quantitative trait loci associated with barley net blotch resistance. Theor. Appl. Genet. 116, 529–539. doi: 10.1007/s00122-007-0688-9
Grewal T. S., Rossnagel B. G., Scoles G. J. (2012). Mapping quantitative trait loci associated with spot blotch and net blotch resistance in a doubled-haploid barley population. Mol. Breed. 30, 267–279. doi: 10.1007/s11032-011-9616-4
Griffey C. A., Das M. K., Baldwin R. E., Waldenmaier C. M. (1994). Yield losses in winter barley resulting from a new race of puccinia hordei in North America. Plant Dis. 78, 256–259. doi: 10.1094/PD-78-0256
Gupta S., Li C., Loughman R., Cakir M., Westcott S., Lance R. (2011). Identifying genetic complexity of 6H locus in barley conferring resistance to Pyrenophora teres f. teres. Plant Breed. 130, 423–429. doi: 10.1111/j.1439-0523.2011.01854.x
Gutiérrez L., Germán S., Pereyra S., Hayes P. M., Pérez C. A., Capettini F., et al. (2015). Multi-environment multi-QTL association mapping identifies disease resistance QTL in barley germplasm from Latin America. Theor. Appl. Genet. 128, 501–516. doi: 10.1007/s00122-014-2448-y
Gyawali S., Mamidi S., Chao S., Bhardwaj S. C., Shekhawat P. S., Selvakumar R., et al. (2021). Genome-wide association studies revealed novel stripe rust resistance QTL in barley at seedling and adult-plant stages. Euphytica 217, 3. doi: 10.1007/s10681-020-02728-1
Han F., Kilian A., Chen J. P., Kudrna D., Steffenson B., Yamamoto K., et al. (1999). Sequence analysis of a rice BAC covering the syntenous barley Rpg1 region. Genome 42, 1071–1076. doi: 10.1139/g99-060
Harder D. E., Legge W. G. (2000). Effectiveness of different sources of stem rust resistance in barley. Crop Sci. 40, 826–833. doi: 10.2135/cropsci2000.403826x
Hatta M. A. M., Arora S., Ghosh S., Matny O., Smedley M. A., Yu G., et al. (2021). The wheat Sr22, Sr33, Sr35 and Sr45 genes confer resistance against stem rust in barley. Plant Biotechnol. J. 19, 273–284. doi: 10.1111/pbi.13460
Henderson M. T. (1945). Studies of sources of resistance and inheritance of reaction to leaf rust, Puccinia anomala Rostr., in barley. (Twin Cities, Minnesota: University of Minnesota).
Henningsen E. C., Omidvar V., Della Coletta R., Michno J.-M., Gilbert E., Li F., et al. (2021). Identification of Candidate Susceptibility Genes to Puccinia graminis f. sp. tritici in Wheat. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.657796
Hernandez J., del Blanco A., Filichkin T., Fisk S., Gallagher L., Helgerson L., et al. (2020a). A Genome-Wide Association Study of Resistance to Puccinia striiformis f. sp. hordei and P. graminis f. sp. tritici in Barley and Development of Resistant Germplasm. Phytopathology® 110, 1082–1092. doi: 10.1094/PHYTO-11-19-0415-R
Hernandez J., Meints B., Hayes P. (2020b). Introgression breeding in barley: perspectives and case studies. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.00761
Hernandez J., Steffenson B. J., Filichkin T., Fisk S. P., Helgerson L., Meints B., et al. (2019). Introgression of rpg4/Rpg5 Into Barley Germplasm Provides Insights Into the Genetics of Resistance to Puccinia graminis f. sp. tritici Race TTKSK and Resources for Developing Resistant Cultivars. Phytopathology® 109, 1018–1028. doi: 10.1094/PHYTO-09-18-0350-R
Hickey L. T., Lawson W., Platz G. J., Dieters M., Arief V. N., Germán S., et al. (2011). Mapping Rph20: a gene conferring adult plant resistance to Puccinia hordei in barley. Theor. Appl. Genet. 123, 55–68. doi: 10.1007/s00122-011-1566-z
Hickey L. T., Lawson W., Platz G. J., Dieters M., Franckowiak J. (2012). Origin of leaf rust adult plant resistance gene Rph20 in barley. Genome 55, 396–399. doi: 10.1139/g2012-022
Hisano H., Sakamoto K., Takagi H., Terauchi R., Sato K. (2017). Exome QTL-seq maps monogenic locus and QTLs in barley. BMC Genomics 18, 125. doi: 10.1186/s12864-017-3511-2
Ho K. M., Choo T. M., Tekauz A., Martin R. A. (1996). Genetic studies on net blotch resistance in a barley cross. Can. J. Plant Sci. 76, 715–719. doi: 10.4141/cjps96-123
Holden S., Bergum M., Green P., Bettgenhaeuser J., Hernández-Pinzón I., Thind A., et al. (2022). A lineage-specific Exo70 is required for receptor kinase–mediated immunity in barley. Sci. Adv. 8, eabn7258. doi: 10.1126/sciadv.abn7258
Horvath H., Rostoks N., Brueggeman R., Steffenson B., von Wettstein D., Kleinhofs A. (2003). Genetically engineered stem rust resistance in barley using the Rpg1 gene. Proc. Natl. Acad. Sci. 100, 364–369. doi: 10.1073/pnas.0136911100
Hubbard A., Lewis C. M., Yoshida K., Ramirez-Gonzalez R. H., Vallavieille-Pope C., Thomas J., et al. (2015). Field pathogenomics reveals the emergence of a diverse wheat yellow rust population. Genome Biol. 16, 23. doi: 10.1186/s13059-015-0590-8
Huerta-Espino J., Singh R., Crespo-Herrera L. A., Villaseñor-Mir H. E., Rodriguez-Garcia M. F., Dreisigacker S., et al. (2020). Adult plant slow rusting genes confer high levels of resistance to rusts in bread wheat cultivars from Mexico. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.00824
Inukai T., Vales M. I., Hori K., Sato K., Hayes P. M. (2006). RMo1 confers blast resistance in barley and is located within the complex of resistance genes containing mla, a powdery mildew resistance gene. Mol. Plant-Microbe Interactions® 19, 1034–1041. doi: 10.1094/MPMI-19-1034
Islamovic E., Bregitzer P., Friesen T. L. (2017). Barley 4H QTL confers NFNB resistance to a global set of P. teres f. teres isolates. Mol. Breed. 37, 29. doi: 10.1007/s11032-017-0621-0
Ivandic V., Walther U., Graner A. (1998). Molecular mapping of a new gene in wild barley conferring complete resistance to leaf rust (Puccinia hordei Otth). Theor. Appl. Genet. 97, 1235–1239. doi: 10.1007/s001220051015
Jamil S., Shahzad R., Ahmad S., Fatima R., Zahid R., Anwar M., et al. (2020). Role of genetics, genomics, and breeding approaches to combat stripe rust of wheat. Front. Nutr. 7. doi: 10.3389/fnut.2020.580715
Jan I., Saripalli G., Kumar K., Kumar A., Singh R., Batra R., et al. (2021). Meta-QTLs and candidate genes for stripe rust resistance in wheat. Sci. Rep. 11, 22923. doi: 10.1038/s41598-021-02049-w
Jafary H., Szabo L. J., Niks R. E. (2006). Innate nonhost immunity in barley to different heterologous rust fungi is controlled by sets of resistance genes with different and overlapping specificities. MPMI 19, 1270–1279. doi: 10.1094/MPMI-19-1270
Jayakodi M., Padmarasu S., Haberer G., Bonthala V. S., Gundlach H., Monat C., et al. (2020). The barley pan-genome reveals the hidden legacy of mutation breeding. Nature 588, 284–289. doi: 10.1038/s41586-020-2947-8
Jedel P. E., Metcalfe D. R., Martens J. W. (1989). Assessment of barley accessions PI 382313, PI 382474, PI 382915, and PI 382976 for stem rust resistance. Crop Sci. 29, 1473–1477. doi: 10.2135/cropsci1989.0011183X002900060030x
Jiang C., Lei M., Guo Y., Gao G., Shi L., Jin Y., et al. (2022). A reference-guided TILLING by amplicon-sequencing platform supports forward and reverse genetics in barley. Plant Commun. 3, 100317. doi: 10.1016/j.xplc.2022.100317
Jin Y., Cui G. H., Steffenson B. J., Franckowiak J. D. (1996). New leaf rust resistance genes in barley and their allelic and linkage relationships with other rph genes. Phytopathology 86, 887–890. doi: 10.1094/Phyto-86-887
Jin Y., Staler G. D., Franckowiak J. D., Steffenson B. J. (1993). Linkage between leaf rust resistance genes and morphological markers in barley. Phytopathology 83, 230–233. doi: 10.1094/Phyto-83-230
Jin Y., Steffenson B. J. (1999). Puccinia coronata var. hordei var. nov.: morphology and pathogenicity. Mycologia 91, 877–884. doi: 10.1080/00275514.1999.12061093
Jin Y., Steffenson B. J. (2002). Sources and genetics of crown rust resistance in barley. Phytopathology® 92, 1064–1067. doi: 10.1094/PHYTO.2002.92.10.1064
Jin Y., Steffenson B. J., Oberthur L. E., Baenziger P. S. (1992). Puccinia coronata on barley. Plant Dis. 76, 1283. doi: 10.1094/PD-76-1283C
Jin Y., Szabo L. J., Carson M. (2010). Century-old mystery of puccinia striiformis life history solved with the identification of berberis as an alternate host. Phytopathology® 100, 432–435. doi: 10.1094/PHYTO-100-5-0432
Jin Y., Steffenson B. J., Bockelman H. E. (1995). Evaluation of cultivated and wild barley for resistance to pathotypes of Puccinia hordei with wide virulence. Genet. Resour Crop Evol. 42, 1–6. doi: 10.1007/BF02310678
Jin Y., Steffenson B. J., Miller J. D. (1994). Inheritance of resistance to pathotypes QCC and MCC of Puccinia graminis f. sp. tritici in barley line Q21861 and temperature effects on the expression of resistance. Pythopathology 84, 452–455.
Johnson R. (1968). “Genetics of resistance of barley to stripe rust,” in 1st International Congress of Plant Pathology (University of California Press, Berkeley), 99.
Johnson R., Wolfe M. S., Scott P. R. (1969). Annual Report 1968 (Cambridge: Plant Breeding Institute).
Johnston P. A., Niks R. E., Meiyalaghan V., Blanchet E., Pickering R. (2013). Rph22: mapping of a novel leaf rust resistance gene introgressed from the non-host Hordeum bulbosum L. into cultivated barley (Hordeum vulgare L.). Theor. Appl. Genet. 126, 1613–1625. doi: 10.1007/s00122-013-2078-9
Johnson R., Finch R. A. (1976). Resistant to Puccinia striiformis west (stripe rust). Barley Genet. Newslett. 6, 130.
Jost M., Singh D., Lagudah E., Park R. F., Dracatos P. (2020). Fine mapping of leaf rust resistance gene Rph13 from wild barley. Theor. Appl. Genet. 133, 1887–1895. doi: 10.1007/s00122-020-03564-6
Kavanagh P. J., Singh D., Bansal U. K., Park R. F. (2017). Inheritance and characterization of the new and rare gene Rph25 conferring seedling resistance in Hordeum vulgare against Puccinia hordei. Plant Breed. 136, 908–912. doi: 10.1111/pbr.12535
Kearney J. (2010). Food consumption trends and drivers. Philos. Trans. R. Soc. London. Ser. B Biol. Sci. 365, 2793–2807. doi: 10.1098/rstb.2010.0149
Khan T. N., Boyd W. J. R. (1969). Inheritance of resistance to net blotch in barley. II. genes conditioning resistance against race W.A.-2. Can. J. Genet. Cytology 11, 592–597. doi: 10.1139/g69-069
Kicherer S., Backes G., Walther U., Jahoor A. (2000). Localising QTLs for leaf rust resistance and agronomic traits in barley (Hordeum vulgare L.). Theor. Appl. Genet. 100, 881–888. doi: 10.1007/s001220051365
Kilian A., Chen J., Han F., Steffenson B., Kleinhofs A. (1997). Towards map-based cloning of the barley stem rust resistance genes Rpg1 and rpg4 using rice as an intergenomic cloning vehicle. Plant Mol. Biol. 35, 187–195. doi: 10.1023/A:1005768222615
Kilian A., Kudrna D. A., Kleinhofs A., Yano M., Kurata N., Steffenson B., et al. (1995). Rice-barley synteny and its application to saturation mapping of the barley Rpg1 region. Nucleic Acids Res. 23, 2729–2733. doi: 10.1093/nar/23.14.2729
Kilian A., Steffenson B. J., Saghai Maroof M. A., Kleinhofs A. (1994). RFLP markers linked to the durable stem rust resistance gene Rpg1 in barley. Mol. Plant-Microbe Interact. 7, 298–301.
King J. E., Polley R. W. (1976). Observations on the epidemiology and effect on grain yield of brown rust in spring barley. Plant Pathol. 25, 63–73. doi: 10.1111/j.1365-3059.1976.tb01926.x
Kleinhofs A., Brueggeman R., Nirmala J., Zhang L., Mirlohi A., Druka A., et al. (2009). Barley stem rust resistance genes: structure and function. Plant Genome 2, 109–120. doi: 10.3835/plantgenome2009.02.0011
Koladia V. M., Faris J. D., Richards J. K., Brueggeman R. S., Chao S., Friesen T. L. (2017). Genetic analysis of net form net blotch resistance in barley lines CIho 5791 and tifang against a global collection of P. teres f. teres isolates. Theor. Appl. Genet. 130, 163–173. doi: 10.1007/s00122-016-2801-4
König J., Perovic D., Kopahnke D., Ordon F. (2013). Development of an efficient method for assessing resistance to the net type of net blotch (Pyrenophora teres f. teres) in winter barley and mapping of quantitative trait loci for resistance. Mol. Breed. 32, 641–650. doi: 10.1007/s11032-013-9897-x
König J., Kopahnke D., Steffenson B. J., Przulj N., Romeis T., Röder M. S., et al. (2012). Genetic mapping of a leaf rust resistance gene in the former Yugoslavian barley landrace MBR1012. Mol. Breed. 30, 1253–1264. doi: 10.1007/s11032-012-9712-0
König J., Perovic D., Kopahnke D., Ordon F. (2014). Mapping seedling resistance to net form of net blotch (Pyrenophora teres f. teres) in barley using detached leaf assay. Plant Breed. 133, 356–365. doi: 10.1111/pbr.12147
Kopahnke D., Nachtigal M., Ordon F., Steffenson B. J. (2004). Evaluation and Mapping of a Leaf Rust Resistance Gene Derived from Hordeum vulgare subsp. spontaneum. Czech J. Genet. Plant Breed. 40, 86–90. doi: 10.17221/3704-CJGPB
Lehmensiek A., Platz G. J., Mace E., Poulsen D., Sutherland M. W. (2007). Mapping of adult plant resistance to net form of net blotch in three Australian barley populations. Aust. J. Agric. Res. 58, 1191–1197. doi: 10.1071/AR07141
Leng Y., Zhao M., Wang R., Steffenson B. J., Brueggeman R. S., Zhong S. (2018). The gene conferring susceptibility to spot blotch caused by Cochliobolus sativus is located at the Mla locus in barley cultivar Bowman. Theor. Appl. Genet. 131, 1531–1539. doi: 10.1007/s00122-018-3095-5
Leonard K. J., Szabo L. E. S. J. (2005). Stem rust of small grains and grasses caused by Puccinia graminis. Mol. Plant Pathol. 6, 99–111. doi: 10.1111/j.1364-3703.2005.00273.x
Lewis C. M., Persoons A., Bebber D. P., Kigathi R. N., Maintz J., Findlay K., et al. (2018). Potential for re-emergence of wheat stem rust in the United Kingdom. Commun. Biol. 1, 13. doi: 10.1038/s42003-018-0013-y
Li K., Hegarty J., Zhang C., Wan A., Wu J., Guedira G. B., et al. (2016). Fine mapping of barley locus Rps6 conferring resistance to wheat stripe rust. Theor. Appl. Genet. 129, 845–859. doi: 10.1007/s00122-015-2663-1
Li J., Wyatt N. A., Skiba R. M., Kariyawasam G. K., Richards J. K., Effertz K., et al. (2023). Pathogen genetics identifies avirulence/virulence loci associated with barley chromosome 6H resistance in the Pyrenophora teres f. teres – barley interaction. doi: 10.1101/2023.02.10.527674
Liu Z., Holmes D. J., Faris J. D., Chao S., Brueggeman R. S., Edwards M. C., et al. (2015). Necrotrophic effector-triggered susceptibility (NETS) underlies the barley–Pyrenophora teres f. teres interaction specific to chromosome 6H. Mol. Plant Pathol. 16, 188–200. doi: 10.1111/mpp.12172
Liu F., Gupta S., Zhang X.-Q., Jones M., Loughman R., Lance R., et al. (2011). PCR markers for selection of adult plant leaf rust resistance in barley (Hordeum vulgare l.). Mol. Breed. 28, 657–666. doi: 10.1007/s11032-010-9517-y
Luig N. H. (1957). Inheritance studies of barley in relation to disease resistance (Sydney: University of Sydney).
Luthra J. K., Chopra V. L. (1990). Genetics of stripe rust resistance in barley. Indian J. Genet. Plant Breed. 50, 390–395.
Ma Z. Q., Lapitan N. L. V., Steffenson B. (2004). QTL mapping of net blotch resistance genes in a doubled-haploid population of six-rowed barley. Euphytica 137, 291–296. doi: 10.1023/B:EUPH.0000040441.36990.58
Mammadov J. A., Zwonitzer J. C., Biyashev R. M., Griffey C. A., Jin Y., Steffenson B. J., et al. (2003). Molecular mapping of leaf rust resistance gene Rph5 in barley. Crop Sci. 43, 388–393. doi: 10.2135/cropsci2003.3880
Mamo B. (2013). Genetic characterization of multiple disease resistance and agronomical and nutritional traits in Hordeum. (Twin Cities: University of Minnesota). PhD.
Mamo B. E., Smith K. P., Brueggeman R. S., Steffenson B. J. (2014). Genetic characterization of resistance to wheat stem rust race TTKSK in landrace and wild barley accessions identifies the rpg4/rpg5 locus. Phytopathology® 105, 99–109. doi: 10.1094/PHYTO-12-13-0340-R
Manninen O. M., Jalli M., Kalendar R., Schulman A., Afanasenko O., Robinson J. (2006). Mapping of major spot-type and net-type net-blotch resistance genes in the ethiopian barley line CI 9819. Genome 49, 1564–1571. doi: 10.1139/g06-119
Manninen O., Kalendar R., Robinson J., Schulman A. H. (2000). Application of BARE-1 retrotransposon markers to the mapping of a major resistance gene for net blotch in barley. Mol. Gen. Genet. MGG 264, 325–334. doi: 10.1007/s004380000326
Marcel T. C., Gorguet B., Ta M. T., Kohutova Z., Vels A., Niks R. E. (2008). Isolate specificity of quantitative trait loci for partial resistance of barley to Puccinia hordei confirmed in mapping populations and near-isogenic lines. New Phytol. 177, 743–755. doi: 10.1111/j.1469-8137.2007.02298.x
Marcel T. C., Varshney R. K., Barbieri M., Jafary H., de Kock M. J. D., Graner A., et al. (2007). A high-density consensus map of barley to compare the distribution of QTLs for partial resistance to Puccinia hordei and of defence gene homologues. Theor. Appl. Genet. 114, 487–500. doi: 10.1007/s00122-006-0448-2
Marchal C., Zhang J., Zhang P., Fenwick P., Steuernagel B., Adamski N. M., et al. (2018). BED-domain-containing immune receptors confer diverse resistance spectra to yellow rust. Nat. Plants 4, 662–668. doi: 10.1038/s41477-018-0236-4
Marshall D., Sutton R. L. (1995). Epidemiology of Stripe Rust, Virulence of Puccinia striiformis f. sp. hordei, and Yield Loss in Barley. Plant Dis. 79, 732–737. doi: 10.1094/PD-79-0732
Martin M. J., Chicaiza O., Caffarel J. C., Sallam A. H., Druka A., Waugh R., et al. (2020). Development of barley introgression lines carrying the leaf rust resistance genes Rph1 to Rph15. Crop Sci. 60, 282–302. doi: 10.1002/csc2.20057
Martin A., Platz G. J., Klerk D., Fowler R. A., Smit F., Potgieter F. G., et al. (2018). Identification and mapping of net form of net blotch resistance in South African barley. Mol. Breed. 38, 53. doi: 10.1007/s11032-018-0814-1
Mascher M., Gundlach H., Himmelbach A., Beier S., Twardziok S. O., Wicker T., et al. (2017). A chromosome conformation capture ordered sequence of the barley genome. Nature 544, 427–433. doi: 10.1371/journal.pgen.1005767
Mascher M., Wicker T., Jenkins J., Plott C., Lux T., Koh C. S., et al. (2021). Long-read sequence assembly: a technical evaluation in barley. Plant Cell 33, 1888–1906. doi: 10.1093/plcell/koab077
Massman C., Hernandez J., Clare S. J., Brooke M., Filichkin T., Fisk S., et al. (2024a). Registration of the “Woodies” multi–rust-resistant barley germplasm. J. Plant Registrations 18, 393–401. doi: 10.1002/plr2.20373
Massman C., Maughan P. J., Nandety R. S., Clare S. J., Fiedler J. D., Hayes P. M. (2024b). Exploratory genomic sequence analysis reveals structural differences at key loci for growth habit, seed dormancy, and rust resistance in barley. Genet. Resour. Crop Evol. doi: 10.1007/s10722-024-01875-x
Mazinani M. A., Aghnoum R., Navabpour S., Zaynali Nezhad K., Dehghan M. A. (2020). Identification of genomic regions associated with net blotch resistance in barley. Modern Genetics Journal 15, 235–247.
McDaniel M. E., Hathcock B. R. (1969). Linkage of the pa4 and mla, loci in barley. Crop Sci. 9, 822. doi: 10.2135/cropsci1969.0011183X000900060047x
Mehnaz M., Dracatos P. M., Park R. F., Singh D. (2021b). Mining middle eastern and central asian barley germplasm to understand diversity for resistance to puccinia hordei, causal agent of leaf rust. Agronomy 11, 2146. doi: 10.3390/agronomy11112146
Mehnaz M., Dracatos P., Pham A., March T., Maurer A., Pillen K., et al. (2021a). Discovery and fine mapping of Rph28: a new gene conferring resistance to Puccinia hordei from wild barley. Theor. Appl. Genet. 134, 2167–2179. doi: 10.1007/s00122-021-03814-1
Metcalfe D. R., Buchannon K. W., McDonald W. C., Reinbergs E. (1970). Relationships between the ‘Jet’ and “Milton” genes for resistance to loose smut genes for reistance to other barley dieases. Can. J. Plant Sci. 50, 423–427. doi: 10.4141/cjps70-077
Mirlohi A., Brueggeman R., Drader T., Nirmala J., Steffenson B. J., Kleinhofs A. (2008). Allele sequencing of the barley stem rust resistance gene Rpg1 identifies regions relevant to disease resistance. Phytopathology® 98, 910–918. doi: 10.1094/PHYTO-98-8-0910
Mode C. J., Schaller C. W. (1958). Two additional factors for host resistance to net blotch in barley. Agron. J. 50, 15–18.
Molnar S. J., James L. E., Kasha K. J. (2000). Inheritance and RAPD tagging of multiple genes for resistance to net blotch in barley. Genome 43, 224–231. doi: 10.1139/g99-111
Monat C., Padmarasu S., Lux T., Wicker T., Gundlach H., Himmelbach A., et al. (2019). TRITEX: chromosome-scale sequence assembly of Triticeae genomes with open-source tools. Genome Biol. 20, 284. doi: 10.1186/s13059-019-1899-5
Moscou M. J., Lauter N., Steffenson B., Wise R. P. (2011). Quantitative and qualitative stem rust resistance factors in barley are associated with transcriptional suppression of defense regulons. PloS Genet. 7, e1002208. doi: 10.1371/journal.pgen.1002208
Moseman J. G., Greeley L. W. (1965). New physiological strains of Puccinia hordei among physiological races identified in the united states from 1959 through 1964. Plant Dis. Rep. 49, 575–578.
Muria-Gonzalez M. J., Lawrence J. A., Palmiero E., D’Souza N. K., Gupta S., Ellwood S. R. (2023). Major susceptibility gene epistasis over minor gene resistance to spot form net blotch in a commercial barley cultivar. Phytopathology®. 113, 1058–1065. doi: 10.1094/PHYTO-10-22-0376-R
Murty S. S. (1942). Segregation and correlated inheritance of rust resistance and epidermal characters in a barley cross. Indian J. Genet. Plant Breed. 2, 73–75.
Nirmala J., Brueggeman R., Maier C., Clay C., Rostoks N., Kannangara G. C., et al. (2006). Subcellular localization and functions of the barley stem rust resistance receptor-like serine/threonine-specific protein kinase Rpg1. Proc. Natl. Acad. Sci. 103, 7518–7523. doi: 10.1073/pnas.0602379103
Nirmala J., Dahl S., Steffenson B. J., Kannangara C. G., von Wettstein D., Chen X., et al. (2007). Proteolysis of the barley receptor-like protein kinase RPG1 by a proteasome pathway is correlated with Rpg1-mediated stem rust resistance. Proc. Natl. Acad. Sci. 104, 10276–10281. doi: 10.1073/pnas.0703758104
Nirmala J., Drader T., Chen X., Steffenson B., Kleinhofs A. (2010). Stem rust spores elicit rapid RPG1 phosphorylation. Mol. Plant-Microbe Interactions® 23, 1635–1642. doi: 10.1094/MPMI-06-10-0136
Nirmala J., Drader T., Lawrence P. K., Yin C., Hulbert S., Steber C. M., et al. (2011). Concerted action of two avirulent spore effectors activates Reaction to Puccinia graminis 1 (Rpg1)-mediated cereal stem rust resistance. Proc. Natl. Acad. Sci. 108, 14676–14681. doi: 10.1073/pnas.1111771108
Novakazi F., Afanasenko O., Anisimova A., Platz G. J. J., Snowdon R., Kovaleva O., et al. (2019). Genetic analysis of a worldwide barley collection for resistance to net form of net blotch disease (Pyrenophora teres f. teres). Theor. Appl. Genet. 132, 2633–2650. doi: 10.1007/s00122-019-03378-1
Nover I., Scholz F. (1969). Genetische Untersuchungen zur Resistenz der Gerste gegen Gelbrost (Puccinia striiformis West.). Theor. Appl. Genet. 39, 150–155. doi: 10.1007/BF00272523
O’Boyle P. D., Brooks W. S., Barnett M. D., Berger G. L., Steffenson B. J., Stromberg E. L., et al. (2014). Mapping net blotch resistance in ‘Nomini’ and CIho 2291 barley. Crop Sci. 54, 2596–2602. doi: 10.2135/cropsci2013.08.0514
Pahalawatta V., Chen X. (2005). Inheritance and molecular mapping of barley genes conferring resistance to wheat stripe rust. Phytopathology® 95, 884–889. doi: 10.1094/PHYTO-95-0884
Pal N., Jan I., Saini D. K., Kumar K., Kumar A., Sharma P. K., et al. (2022). Meta-QTLs for multiple disease resistance involving three rusts in common wheat (Triticum aestivum L.). Theor. Appl. Genet. 135, 2385–2405. doi: 10.1007/s00122-022-04119-7
Park R. F. (2007). Stem rust of wheat in Australia. Crop Pasture Sci. 58, 558–566. doi: 10.1071/AR07117
Park R. F., Golegaonkar P. G., Derevnina L., Sandhu K. S., Karaoglu H., Elmansour H. M., et al. (2015). Leaf rust of cultivated barley: pathology and control. Annu. Rev. Phytopathol. 53, 565–589. doi: 10.1146/annurev-phyto-080614-120324
Park R. F., Karakousis A. (2002). Characterization and mapping of gene Rph19 conferring resistance to Puccinia hordei in the cultivar ‘Reka 1’ and several Australian barleys. Plant Breed. 121, 232–236. doi: 10.1046/j.1439-0523.2002.00717.x
Park R. F., Poulsen D., Barr A. R., Cakir M., Moody D. B., Raman H., et al. (2003). Mapping genes for resistance to Puccinia hordei in barley. Aust. J. Agric. Res. 54, 1323–1333. doi: 10.1071/AR02244
Patterson F., Shands R., Dickson J. (1957). Temperature and seasonal effects on seedling reactions of barley varieties to three races of Puccinia graminis f. sp. tritici. Phytopathology® 47, 395–402.
Perovic D., Stein N., Zhang H., Drescher A., Prasad M., Kota R., et al. (2004). An integrated approach for comparative mapping in rice and barley with special reference to the Rph16 resistance locus. Funct. Integr. Genomics 4, 74–83. doi: 10.1007/s10142-003-0100-z
Peters Haugrud A. R., Zhang Z., Friesen T. L., Faris J. D. (2022). Genetics of resistance to septoria nodorum blotch in wheat. Theor. Appl. Genet. 135, 3685–3707. doi: 10.1007/s00122-022-04036-9
Pickering R. A., Malyshev S., Künzel G., Johnston P. A., Korzun V., Menke M., et al. (2000). Locating introgressions of Hordeum bulbosum chromatin within the H. vulgare genome. Theor. Appl. Genet. 100, 27–31. doi: 10.1007/PL00002904
Pickering R. A., Steeffenson B. J., Hill A. M., Borovkova I. (1998). Association of leaf rust and powdery mildew resistance in a recombinant derived from a Hordeum vulgare × Hordeum bulbosum hybrid. Plant Breed. 117, 83–84. doi: 10.1111/j.1439-0523.1998.tb01453.x
Powers L., Hines L. (1933). Inheritance of reaction to stem rust and barbing of awns in barley crosses. J. Agric. Research1 46, 1121–1129.
Prasad P., Savadi S., Bhardwaj S. C., Gupta P. K. (2020). The progress of leaf rust research in wheat. Fungal Biol. 124, 537–550. doi: 10.1016/j.funbio.2020.02.013
Prins R., Steffenson B. J., Case A. J., Boshoff W. H. P., Agenbag G. M., Pretorius Z. A. (2020). Assessments and perspectives on stem rust resistance in South African malting barley. Australas. Plant Pathol. 49, 679–690. doi: 10.1007/s13313-020-00744-2
Read B. J., Raman H., McMichael G., Chalmers K. J., Ablett G. A., Platz G. J., et al. (2003). Mapping and QTL analysis of the barley population sloop × halcyon. Aust. J. Agric. Res. 54, 1145–1153. doi: 10.1071/AR03037
Reinhold M., Sharp E. L. (1982). Virulence types of Puccinia hordei from north america, north africa and the middle east. Plant Dis. 66, 1009–1011.
Richter K., Schondelmaier J., Jung C. (1998). Mapping of quantitative trait loci affecting Drechslera teres resistance in barley with molecular markers. Theor. Appl. Genet. 97, 1225–1234. doi: 10.1007/s001220051014
Rozanova I. V., Lashina N. M., Mustafin Z. S., Gorobets S. A., Efimov V. M., Afanasenko O. S., et al. (2019). SNPs associated with barley resistance to isolates of Pyrenophora teres f. teres. BMC Genomics 20, 292. doi: 10.1186/s12864-019-5623-3
Qi X., Fufa F., Sijtsma D., Niks R. E., Lindhout P., Stam P. (2000). The evidence for abundance of QTLs for partial resistance to Puccinia hordei on the barley genome. Mol. Breed. 6, 1–9. doi: 10.1023/A:1009649903383
Qi X., Jiang G., Chen W., Niks R. E., Stam P., Lindhout P. (1999). Isolate-specific QTLs for partial resistance to Puccinia hordei in barley. Theor. Appl. Genet. 99, 877–884. doi: 10.1007/s001220051308
Qi X., Niks R. E., Stam P., Lindhout P. (1998). Identification of QTLs for partial resistance to leaf rust (Puccinia hordei) in barley. Theor. Appl. Genet. 96, 1205–1215. doi: 10.1007/s001220050858
Raman H., Platz G. J., Chalmers K. J., Raman R., Read B. J., Barr A. R., et al. (2003). Mapping of genomic regions associated with net form of net blotch resistance in barley. Aust. J. Agric. Res. 54, 1359–1367. doi: 10.1071/AR03026
Rao H. S., Basha O. P., Singh N. K., Sato K., Dhaliwal H. S. (2007). Frequency distributions and composite interval mapping for QTL analysis in ‘Steptoe’ x ‘Morex’ barley mapping population. Barley Genet. Newslett. 37, 5–20.
Read A. C., Hutin M., Moscou M. J., Rinaldi F. C., Bogdanove A. J. (2020). Cloning of the rice xo1 resistance gene and interaction of the xo1 protein with the defense-suppressing xanthomonas effector tal2h. Mol. Plant-Microbe Interactions® 33, 1189–1195. doi: 10.1094/MPMI-05-20-0131-SC
Richards J., Chao S., Friesen T., Brueggeman R. (2016). Fine mapping of the barley chromosome 6H net form net blotch susceptibility locus. G3 Genes|Genomes|Genetics 6, 1809–1818. doi: 10.1534/g3.116.028902
Richards J. K., Friesen T. L., Brueggeman R. S. (2017). Association mapping utilizing diverse barley lines reveals net form net blotch seedling resistance/susceptibility loci. Theor. Appl. Genet. 130, 915–927. doi: 10.1007/s00122-017-2860-1
Richards J., Li J., Koladia V., Wyatt N., Rehman S., Brueggeman R. S., et al. (2024). A Moroccan Pyrenophora teres f. teres population defeats Rpt5, the broadly effective resistance on barley chromosome 6H. Phytopathology® 114, 193–199. doi: 10.1094/PHYTO-04-23-0117-R
Roane C. W., Starling T. M. (1967). Inheritance of reaction to Puccinia hordei in barley. II. Gene symbols for loci in differential cultivars. Phytopathology 57, 66–68.
Roelfs A. P., Long D. L., Roberts J. J. (1993). Races of Puccinia graminis in the United States during 1990. Plant Dis. 77, 125–128. doi: 10.1094/PD-77-0125
Rossi C., Cuesta-Marcos A., Vales I., Gomez-Pando L., Orjeda G., Wise R., et al. (2006). Mapping multiple disease resistance genes using a barley mapping population evaluated in Peru, Mexico, and the USA. Mol. Breed. 18, 355–366. doi: 10.1007/s11032-006-9043-0
Rostoks N., Steffenson B. J., Kleinhofs A. (2004). Structure and expression of the barley stem rust resistance gene Rpg1 messenger RNA. Physiol. Mol. Plant Pathol. 64, 91–101. doi: 10.1016/j.pmpp.2004.05.006
Rothwell C. T., Singh D., Dracatos P. M., Park R. F. (2020). Inheritance and characterization of rph27: A third race-specific resistance gene in the barley cultivar quinn. Phytopathology® 110, 1067–1073. doi: 10.1094/PHYTO-12-19-0470-R
Sakkour A., Mascher M., Himmelbach A., Haberer G., Lux T., Spannagl M., et al. (2022). Chromosome-scale assembly of barley Cv. ‘Haruna Nijo’ as a resource for barley genetics. DNA Res. 29, dsac001. doi: 10.1093/dnares/dsac001
Salanță L. C., Coldea T. E., Ignat M. V., Pop C. R., Tofană M., Mudura E., et al. (2020). Non-alcoholic and craft beer production and challenges. Processes 1382, 10–23. doi: 10.3390/pr8111382
Sallam A. H., Tyagi P., Brown-Guedira G., Muehlbauer G. J., Hulse A., Steffenson B. J. (2017). Genome-Wide Association Mapping of Stem Rust Resistance in Hordeum vulgare subsp. spontaneum. G3 Genes|Genomes|Genetics 7, 3491–3507. doi: 10.1534/g3.117.300222
Sandhu K. S., Forrest K. L., Kong S., Bansal U. K., Singh D., Hayden M. J., et al. (2012). Inheritance and molecular mapping of a gene conferring seedling resistance against Puccinia hordei in the barley cultivar Ricardo. Theor. Appl. Genet. 125, 1403–1411. doi: 10.1007/s00122-012-1921-8
Sato K., Mascher M., Himmelbach A., Haberer G., Spannagl M., Stein N. (2021). Chromosome-scale assembly of wild barley accession “OUH602. G3 Genes|Genomes|Genetics 11, jkab244. doi: 10.1093/g3journal/jkab244
Saunders D. G. O., Pretorius Z. A., Hovmøller M. S. (2019). Tackling the re-emergence of wheat stem rust in Western Europe. Commun. Biol. 2, 51. doi: 10.1038/s42003-019-0294-9
Sayre-Chavez B., Bettenhausen H., Windes S., Aron P., Cistué L., Fisk S., et al. (2022). Genetic basis of barley contributions to beer flavor. J. Cereal Sci. 104, 103430. doi: 10.1016/j.jcs.2022.103430
Scherrer B., Isidore E., Klein P., Kim J., Bellec A., Chalhoub B., et al. (2005). Large intraspecific haplotype variability at the Rph7 locus results from rapid and recent divergence in the barley genome. Plant Cell 17, 361–374. doi: 10.1105/tpc.104.028225
Schnaithmann F., Kopahnke D., Pillen K. (2014). A first step toward the development of a barley NAM population and its utilization to detect QTLs conferring leaf rust seedling resistance. Theor. Appl. Genet. 127, 1513–1525. doi: 10.1007/s00122-014-2315-x
Schreiber M., Mascher M., Wright J., Padmarasu S., Himmelbach A., Heavens D., et al. (2020). A genome assembly of the barley ‘Transformation reference’ Cultivar golden promise. G3 Genes|Genomes|Genetics 10, 1823–1827. doi: 10.1101/2020.02.12.945550
Shands R. G. (1939). Chevron, a barley variety resistant to stem rust and other diseases. Phytopathology® 29, 209–211.
Sharma Poudel R., Al-Hashel A. F., Gross T., Gross P., Brueggeman R. (2018). Pyramiding rpg4- and Rpg1-mediated stem rust resistance in barley requires the Rrr1 gene for both to function (Fargo, ND, United States: Department of Plant Pathology, North Dakota State University).
Shjerve R. A., Faris J. D., Brueggeman R. S., Yan C., Zhu Y., Koladia V., et al. (2014). Evaluation of a Pyrenophora teres f. teres mapping population reveals multiple independent interactions with a region of barley chromosome 6H. Fungal Genet. Biol. 70, 104–112. doi: 10.1016/j.fgb.2014.07.012
Singh D., Dracatos P., Derevnina L., Zhou M., Park R. F. (2015). Rph23: A new designated additive adult plant resistance gene to leaf rust in barley on chromosome 7H. Plant Breed. 134, 62–69. doi: 10.1111/pbr.12229
Singh R. P., Hodson D. P., Huerta-Espino J., Jin Y., Bhavani S., Njau P., et al. (2011). The emergence of ug99 races of the stem rust fungus is a threat to world wheat production. Annu. Rev. Phytopathol. 49, 465–481. doi: 10.1146/annurev-phyto-072910-095423
Singh R. P., Hodson D. P., Jin Y., Lagudah E. S., Ayliffe M. A., Bhavani S., et al. (2015). Emergence and spread of new races of wheat stem rust fungus: continued threat to food security and prospects of genetic control. Phytopathology® 105, 872–884. doi: 10.1094/PHYTO-01-15-0030-FI
Skiba R. M., Wyatt N. A., Kariyawasam G. K., Fiedler J. D., Yang S., Brueggeman R. S., et al. (2022). Host and pathogen genetics reveal an inverse gene-for-gene association in the P. teres f. maculata–barley pathosystem. Theor. Appl. Genet. 135, 3597–3609. doi: 10.1007/s00122-022-04204-x
Solanki S., Richards J., Ameen G., Wang X., Khan A., Ali H., et al. (2019). Characterization of genes required for both Rpg1 and rpg4-mediated wheat stem rust resistance in barley. BMC Genomics 20, 495. doi: 10.1186/s12864-019-5858-z
Soriano J. M., Royo C. (2015). Dissecting the genetic architecture of leaf rust resistance in wheat by QTL meta-analysis. Phytopathology® 105, 1585–1593. doi: 10.1094/PHYTO-05-15-0130-R
Spaner D., Shugar L. P., Choo T. M., Falak I., Briggs K. G., Legge W. G., et al. (1998). Mapping of disease resistance loci in barley on the basis of visual assessment of naturally occurring symptoms. Crop Sci. 38, 843–850. doi: 10.2135/cropsci1998.0011183X003800030037x
Starling T. M. (1955). Sources, inheritance, and linkage relationships of resistance to race 4 of leaf rust (Puccinia hordei otth), race 9 of powdery mildew (Erysiphe graminis hordei el marchal), and certain agronomic characters in barley (Ames: Iowa State University). doi: 10.31274/rtd-180813-14395
Steffenson B. J., Hayes P. M., Kleinhofs A. (1996). Genetics of seedling and adult plant resistance to net blotch (Pyrenophora teres f. teres) and spot blotch (Cochliobolus sativus) in barley. Theor. Appl. Genet. 92, 552–558. doi: 10.1007/BF00224557
Steffenson B. J., Jin Y., Brueggeman R. S., Kleinhofs A., Sun Y. (2009). Resistance to stem rust race TTKSK maps to the rpg4/Rpg5 complex of chromosome 5H of barley. Phytopathology® 99, 1135–1141. doi: 10.1094/PHYTO-99-10-1135
Steffenson B. J. (1992). Analysis of durable resistance to stem rust in barley. Euphytica 63, 153–167. doi: 10.1007/BF00023920
Steffenson B. J., Case A. J., Pretorius Z. A., Coetzee V., Kloppers F. J., Zhou H., et al. (2017). Vulnerability of Barley to African Pathotypes of Puccinia graminis f. sp. tritici and Sources of Resistance. Phytopathology® 107, 950–962. doi: 10.1094/PHYTO-11-16-0400-R
Steffenson B. J., Olivera P., Roy J. K., Jin Y., Smith K. P., Muehlbauer G. J. (2007). A walk on the wild side: mining wild wheat and barley collections for rust resistance genes. Aust. J. Agric. Res. 58, 532–544. doi: 10.1071/AR07123
Steffenson B. J., Solanki S., Brueggeman R. S. (2016). Landraces from mountainous regions of Switzerland are sources of important genes for stem rust resistance in barley. Alpine Bot. 126, 23–33. doi: 10.1007/s00035-015-0161-3
Steffenson B. J., Wilcoxson R. D., Roelfs A. P. (1984). Inheritance of Resistance to Puccinia graminis f. sp. secalis in Barley. Plant Dis. 68, 762–763. doi: 10.1094/PD-69-762
St. Pierre S., Gustus C., Steffenson B., Dill-Macky R., Smith K. P. (2010). Mapping net form net blotch and septoria speckled leaf blotch resistance loci in barley. Phytopathology 100, 80–84. doi: 10.1094/PHYTO-100-1-0080
Sun Y., Steffenson B. J. (2005). Reaction of barley seedlings with different stem rust resistance genes to Puccinia graminis f. sp. tritici and Puccinia graminis f. sp. secalis. Can. J. Plant Pathol. 27, 80–89. doi: 10.1080/07060660509507198
Sun Y., Steffenson B. J., Jin Y. (1996). Genetics of Resistance to Puccinia graminis f. sp. secalis in Barley Line Q21861. Pythopathology 86, 1299–1302. doi: 10.1094/Phyto-86-1299
Tamang P., Neupane A., Mamidi S., Friesen T., Brueggeman R. (2015). Association mapping of seedling resistance to spot form net blotch in a worldwide collection of barley. Phytopathology 105, 500–508. doi: 10.1094/PHYTO-04-14-0106-R
Tamang P., Richards J. K., Alhashal A., Sharma Poudel R., Horsley R. D., Friesen T. L., et al. (2019). Mapping of barley susceptibility/resistance QTL against spot form net blotch caused by Pyrenophora teres f. maculata using RIL populations. Theor. Appl. Genet. 132, 1953–1963. doi: 10.1007/s00122-019-03328-x
Tamang P., Richards J. K., Solanki S., Ameen G., Sharma Poudel R., Deka P., et al. (2021). The Barley HvWRKY6 Transcription Factor Is Required for Resistance Against Pyrenophora teres f. teres. Front. Genet. 11. doi: 10.3389/fgene.2020.601500
Tan B. H. (1977). A new gene for resistance to Puccinia hordei in certain Ethiopian barleys. Cereal Rust Bull. 5, 39–43.
Tan B. H. (1978). Verifying the genetic relationships between three leaf rust resistance genes in barley. Euphytica 27, 317–323. doi: 10.1007/BF00039148
Thiel J., Koppolu R., Trautewig C., Hertig C., Kale S. M., Erbe S., et al. (2021). Transcriptional landscapes of floral meristems in barley. Sci. Adv. 7, eabf0832. doi: 10.1126/sciadv.abf0832
Thomas W. T. B., Powell W., Waugh R., Chalmers K. J., Barua U. M., Jack P., et al. (1995). Detection of quantitative trait loci for agronomic, yield, grain and disease characters in spring barley (Hordeum vulgare L.). Theor. Appl. Genet. 91, 1037–1047. doi: 10.1007/BF00223917
Tian X., Yao Q., Zhang Z., Cheng X., Qin J., Kang Z., et al. (2021). Evidence of Occurrence of Barley Crown Rust Caused by Puccinia coronata var. hordei and Sexual Reproduction of the Pathogen Under Field Conditions in China. Plant Dis. 105, 2990–2999. doi: 10.1094/PDIS-09-20-2029-RE
Toojinda T., Baird E., Booth A., Broers L., Hayes P., Powell W., et al. (1998). Introgression of quantitative trait loci (QTLs) determining stripe rust resistance in barley: an example of marker-assisted line development. Theor. Appl. Genet. 96, 123–131. doi: 10.1007/s001220050718
Toojinda T., Broers L. H., Chen X. M., Hayes P. M., Kleinhofs A., Korte J., et al. (2000). Mapping quantitative and qualitative disease resistance genes in a doubled haploid population of barley (Hordeum vulgare). Theor. Appl. Genet. 101, 580–589. doi: 10.1007/s001220051519
Tosh S. M., Bordenave N. (2020). Emerging science on benefits of whole grain oat and barley and their soluble dietary fibers for heart health, glycemic response, and gut microbiota. Nutr. Rev. 78, 13–20. doi: 10.1093/nutrit/nuz085
Tuleen N. A., McDaniel M. E. (1971). “Location of genes pa and pa5,” in Barley Newsletter (Milwaukee, Wisconsin: American Malting Barley Association), vol. 15. , 106–107.
Turuspekov Y., Ormanbekova D., Rsaliev A., Abugalieva S. (2016). Genome-wide association study on stem rust resistance in Kazakh spring barley lines. BMC Plant Biol. 16, 6. doi: 10.1186/s12870-015-0686-z
Upadhaya A., Upadhaya G. C. S., Brueggeman R. S. (2021). The wheat stem rust (Puccinia graminis f. sp. tritici) population from Washington contains the most virulent isolates reported on barley. Plant Dis. 106, 223–230. doi: 10.1094/PDIS-06-21-1195-RE
Vales M. I., Schön C. C., Capettini F., Chen X. M., Corey A. E., Mather D. E., et al. (2005). Effect of population size on the estimation of QTL: a test using resistance to barley stripe rust. Theor. Appl. Genet. 111, 1260–1270. doi: 10.1007/s00122-005-0043-y
Vatter T., Maurer A., Kopahnke D., Perovic D., Ordon F., Pillen K. (2017). A nested association mapping population identifies multiple small effect QTL conferring resistance against net blotch (Pyrenophora teres f. teres) in wild barley. PloS One 12, e0186803. doi: 10.1371/journal.pone.0186803
Vatter T., Maurer A., Perovic D., Kopahnke D., Pillen K., Ordon F. (2018). Identification of QTL conferring resistance to stripe rust (Puccinia striiformis f. sp. hordei) and leaf rust (Puccinia hordei) in barley using nested association mapping (NAM). PloS One 13, e0191666.
Verma R. P. S., Selvakumar R., Gangwar O. P., Shekhawat P. S., Bhardwaj S. C., Rehman S., et al. (2018). Identification of additional sources of resistance to Puccinia striiformis f. sp. hordei (PSH) in a collection of barley genotypes adapted to the high input condition. J. Phytopathol. 166, 355–364. doi: 10.1111/jph.12693
Visioni A., Gyawali S., Selvakumar R., Gangwar O. P., Shekhawat P. S., Bhardwaj S. C., et al. (2018). Genome Wide Association Mapping of Seedling and Adult Plant Resistance to Barley Stripe Rust (Puccinia striiformis f. sp. hordei) in India. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.00520
von Korff M., Wang H., Léon J., Pillen K. (2005). AB-QTL analysis in spring barley. I. Detection of resistance genes against powdery mildew, leaf rust and scald introgressed from wild barley. Theor. Appl. Genet. 111, 583–590. doi: 10.1007/s00122-005-2049-x
Wang X., Mace E. S., Platz G. J., Hunt C. H., Hickey L. T., Franckowiak J. D., et al. (2015). Spot form of net blotch resistance in barley is under complex genetic control. Theor. Appl. Genet. 128, 489–499. doi: 10.1007/s00122-014-2447-z
Wang X., Richards J., Gross T., Druka A., Kleinhofs A., Steffenson B., et al. (2013). The rpg4-Mediated Resistance to Wheat Stem Rust (Puccinia graminis) in Barley (Hordeum vulgare) Requires Rpg5, a Second NBS-LRR Gene, and an Actin Depolymerization Factor. Mol. Plant-Microbe Interactions® 26, 407–418. doi: 10.1094/MPMI-06-12-0146-R
Wang Y., Subedi S., Vries H., Doornenbal P., Vels A., Hensel G., et al. (2019). Orthologous receptor kinases quantitatively affect the host status of barley to leaf rust fungi. Nat. Plants 5, 1129–1135. doi: 10.1038/s41477-019-0545-2
Watson I. A., Butler F. C. (1947). Resistance to barley leaf rust (Puccinia anomala Rost.). Proc. Linn. Soc. New South Wales 72, 379–386.
Weerasena J. S., Steffenson B. J., Falk A. B. (2004). Conversion of an amplified fragment length polymorphism marker into a co-dominant marker in the mapping of the Rph15 gene conferring resistance to barley leaf rust, Puccinia hordei Otth. Theor. Appl. Genet. 108, 712–719. doi: 10.1007/s00122-003-1470-2
Wei B., Moscou M. J., Sato K., Gourlie R., Strelkov S., Aboukhaddour R. (2020). Identification of a locus conferring dominant susceptibility to pyrenophora tritici-repentis in barley. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.00158
Wellings C. R. (2011). Global status of stripe rust: a review of historical and current threats. Euphytica 179, 129–141. doi: 10.1007/s10681-011-0360-y
Williams K. J., Platz G. J., Barr A. R., Cheong J., Willsmore K., Cakir M., et al. (2003). A comparison of the genetics of seedling and adult plant resistance to the spot form of net blotch (Pyrenophora teres f. maculata). Aust. J. Agric. Res. 54, 1387–1394. doi: 10.1071/AR03028
Williams K. J., Lichon A., Gianquitto P., Kretschmer J. M., Karakousis A., Manning S., et al. (1999). Identification and mapping of a gene conferring resistance to the spot form of net blotch (Pyrenophora teres f. maculata) in barley. Theor. Appl. Genet. 99, 323–327. doi: 10.1007/s001220051239
Wonneberger R., Ficke A., Lillemo M. (2017). Identification of quantitative trait loci associated with resistance to net form net blotch in a collection of Nordic barley germplasm. Theor. Appl. Genet. 130, 2025–2043. doi: 10.1007/s00122-017-2940-2
Xie W., Xiong W., Pan J., Ali T., Cui Q., Guan D., et al. (2018). Decreases in global beer supply due to extreme drought and heat. Nat. Plants 4, 964–973. doi: 10.1038/s41477-018-0263-1
Xu W., Tucker J. R., Bekele W. A., You F. M., Fu Y.-B., Khanal R., et al. (2021). Genome assembly of the canadian two-row malting barley cultivar AAC synergy. G3 Genes|Genomes|Genetics 11, jkab031. doi: 10.1093/g3journal/jkab031
Yan G. P., Chen X. M. (2006). Molecular mapping of a recessive gene for resistance to stripe rust in barley. Theor. Appl. Genet. 113, 529–537. doi: 10.1007/s00122-006-0319-x
Yan G., Chen X. (2007a). Identification of a Quantitative Trait Locus for High-Temperature Adult-Plant Resistance Against Puccinia striiformis f. sp. hordei in ‘Bancroft’ Barley. Phytopathology® 98, 120–127. doi: 10.1094/PHYTO-98-1-0120
Yan G., Chen X. (2007b). Molecular mapping of the rps1.a recessive gene for resistance to stripe rust in BBA 2890 barley. Phytopathology® 97, 668–673. doi: 10.1094/PHYTO-97-6-0668
Yeo F. K. S., Bouchon R., Kuijken R., Loriaux A., Boyd C., Niks R. E., et al. (2017). High-resolution mapping of genes involved in plant stage-specific partial resistance of barley to leaf rust. Mol. Breed. 37, 45. doi: 10.1007/s11032-017-0624-x
Yoshimura S., Yamanouchi U., Katayose Y., Toki S., Wang Z.-X., Kono I., et al. (1998). Expression of Xa1, a bacterial blight-resistance gene in rice, is induced by bacterial inoculation. Proc. Natl. Acad. Sci. 95, 1663–1668. doi: 10.1073/pnas.95.4.1663
Yu L.-X., Barbier H., Rouse M. N., Singh S., Singh R. P., Bhavani S., et al. (2014). A consensus map for Ug99 stem rust resistance loci in wheat. Theor. Appl. Genet. 127, 1561–1581. doi: 10.1007/s00122-014-2326-7
Yu X., Kong H. Y., Meiyalaghan V., Casonato S., Chng S., Jones E. E., et al. (2018). Genetic mapping of a barley leaf rust resistance gene Rph26 introgressed from Hordeum bulbosum. Theor. Appl. Genet. 131, 2567–2580. doi: 10.1007/s00122-018-3173-8
Yun S. J., Gyenis L., Hayes P. M., Matus I., Smith K. P., Steffenson B. J., et al. (2005). Quantitative trait loci for multiple disease resistance in wild barley. Crop Sci. 45, 2563–2572. doi: 10.2135/cropsci2005.0236
Yuzon J. D., Wyatt N. A., Vasighzadeh A., Clare S., Navratil E., Friesen T. L., et al. (2023). Hybrid inferiority and genetic incompatibilities drive divergence of fungal pathogens infecting the same host. Genetics 224, iyad037. doi: 10.1093/genetics/iyad037
Zhang L., Fetch T., Nirmala J., Schmierer D., Brueggeman R., Steffenson B., et al. (2006). Rpr1, a gene required for Rpg1-dependent resistance to stem rust in barley. Theor. Appl. Genet. 113, 847–855. doi: 10.1007/s00122-006-0342-y
Zheng T., Hua C., Li L., Sun Z., Yuan M., Bai G., et al. (2021). Integration of meta-QTL discovery with omics: Towards a molecular breeding platform for improving wheat resistance to Fusarium head blight. Crop J. 9, 739–749. doi: 10.1016/j.cj.2020.10.006
Zhong S., Effertz R. J., Jin Y., Franckowiak J. D., Steffenson B. J. (2003). Molecular mapping of the leaf rust resistance gene rph6 in barley and its linkage relationships with rph5 and rph7. Phytopathology 93, 604–609. doi: 10.1094/PHYTO.2003.93.5.604
Zhou H., Steffenson B. J., Muehlbauer G., Wanyera R., Njau P., Ndeda S. (2014). Association mapping of stem rust race TTKSK resistance in US barley breeding germplasm. TAG. Theor. Appl. Genet. Theoretische und angewandte Genetik 127, 1293–1304. doi: 10.1007/s00122-014-2297-8
Ziems L. A., Hickey L. T., Hunt C. H., Mace E. S., Platz G. J., Franckowiak J. D., et al. (2014). Association mapping of resistance to Puccinia hordei in Australian barley breeding germplasm. Theor. Appl. Genet. 127, 1199–1212. doi: 10.1007/s00122-014-2291-1
Ziems L. A., Hickey L. T., Platz G. J., Franckowiak J. D., Dracatos P. M., Singh D., et al. (2017). Characterization of rph24: A gene conferring adult plant resistance to puccinia hordei in barley. Phytopathology® 107, 834–841. doi: 10.1094/PHYTO-08-16-0295-R
Keywords: barley, leaf rust, stripe rust, stem rust, net blotch, atlas, Hordeum, Puccinia
Citation: Clare SJ, Novakazi F, Hayes PM, Moscou MJ and Brueggeman RS (2024) Colocalization of genetic regions that confer resistance/susceptibility against Puccinia species and association with Pyrenophora teres loci within the barley genome. Front. Agron. 6:1451281. doi: 10.3389/fagro.2024.1451281
Received: 18 June 2024; Accepted: 26 August 2024;
Published: 01 October 2024.
Edited by:
Karthikeyan Adhimoolam, Jeju National University, Republic of KoreaReviewed by:
Sujan Mamidi, HudsonAlpha Institute for Biotechnology, United StatesSudhir Navathe, Agharkar Research Institute, India
Bharathi Raja Ramadoss, Plenty Unlimited Inc., United States
Copyright © 2024 Clare, Novakazi, Hayes, Moscou and Brueggeman. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shaun J. Clare, c2hhdW4uY2xhcmVAd3N1LmVkdQ==
 Shaun J. Clare
Shaun J. Clare Fluturë Novakazi
Fluturë Novakazi Patrick M. Hayes
Patrick M. Hayes Matthew J. Moscou
Matthew J. Moscou Robert S. Brueggeman
Robert S. Brueggeman