- NIAB East Malling, West Malling, United Kingdom
In the UK, strawberry is mostly grown in coconut coir substrate under protection. Coir substrate is usually used only for one or two cropping seasons because the continuous reuse of coir without any treatment leads to yield decline. In this study, we investigated the changes in bacterial and fungal communities in strawberry roots and bulk coir in relation to (i) the coir substrate age (cropping seasons) and (ii) oak or beech biochar amendment at planting. Coir age did not affect fungal/bacterial alpha (within-sample) diversity but affected beta (between-sample) diversity. Amendment with either oak or beech biochar did not lead to significant changes in either alpha or beta diversity for both fungi and bacteria, but it did alter the relative abundance of 13 fungal ASVs. This study identified six bacterial and 20 fungal ASVs with a significant positive linear relationship with coir age and also eight bacterial and 22 fungal ASVs with a significant negative linear relationship with coir age. Notably, the observed strawberry yield decline in reused coir substrate could be associated with a generalist root pathogen, Ilyonectria destructans (ex. Cylindrocarpon destructans), of which the abundance increased annually by 225% and 426% in strawberry root and bulk coir, respectively. Future research is needed to confirm the role of I. destructans in reused coir on strawberry plant health and fruit productivity and then to identify management strategies for yield decline mitigation.
1 Introduction
Strawberry is a beloved fruit known for its sweet and distinctive scent and is a high-value crop that supports both agriculture and local economies (Ulrich et al., 2018; Liu et al., 2023). Strawberries are a major fruit crop in more than 50 countries over five continents. In 2022, a total of 9.5 M t of strawberries was harvested from 0.4 M ha worldwide (FAOSTAT, https://www.fao.org/). Strawberry production in the UK in the past 20 years has shifted from open field toward substrate-based systems. These systems use substrates such as coconut coir, peat, and other materials to grow strawberries in protected (mostly polytunnel) environments (Philip, 2013; Robinson-Boyer et al., 2016; Fennimore et al., 2024). Coir, a waste fiber from coconut production, has been the most popular substrate for growing strawberries in soilless tabletop production in the UK for over a decade. Compared to peat, coir provides a more stable growing medium, with a higher level of production consistency—for example, coir has a lower acidity level, higher levels of calcium, and can be re-used for up to four seasons (Vidhana Arachchi and Somasiri, 1997; Abad et al., 2002; Hernández-Apaolaza et al., 2005). It is also a more sustainable option than peat-based media (Alexander et al., 2008).
Growing strawberries in coir bags allows for better management of soil-borne pests through the use of pesticides or natural parasites and limits the spread of soilborne pathogens to within the same bag, trough, or pot. Coir-grown table-top strawberries have improved fruit quality through intensive fertigation and crop protection, allowing out-of-season production, a much higher yield, and potentially better financial returns. Coir media is currently used mainly for one or a maximum of two growing seasons, and there is a growing environmental and economic need to extend the coir substrate lifetime. However, empirical evidence in the UK industry suggested significant reductions in yield associated with reusing coir substrate.
Biochar is a finely grained product similar to charcoal that can be produced from a variety of biomass feedstocks, including agricultural and industrial green wastes, and urban sludge (Lehmann and Rondón, 2006; Norah et al., 2015; Sheng et al., 2016). The application of biochar has been found to improve soil fertility (Nelson et al., 2011; Prendergast-Miller et al., 2014), increase soil microbial diversity (Rutigliano et al., 2014), and increase crop growth (Reynolds et al., 2003; Marris, 2006). When added to peat growing media in strawberry production, biochar has been reported to enhance root formation, increase fruit production, and improve the post-harvest resistance of fruit to gray mold (Botrytis cinerea) (De Tender et al., 2016). Two different biochars added to the potting medium of strawberry plants reduced the severity of gray mold, anthracnose (Colletrotrichum acutatum), and powdery mildew (Podosphaeria apahanis) (Meller Harel et al., 2012). However, the addition of biochar to soil-grown strawberry did not lead to a yield benefit in a UK study (Jay et al., 2015).
Recently, we reported a significant yield decline in strawberry equivalent to approximately 5% reduction for every year of coir use (Shuttleworth et al., 2021). Moreover, amending used coir with either oak or beech biochar did not have any discernible effects on fruit production. Yield reduction associated with continuous cropping is commonly observed in many crops (Mazzola and Manici, 2012; Xu et al., 2015; Wang et al., 2020). Among the important causes of yield decline in continuous cropping are the changes in the microbiome in growing media—for instance, apple replant disease is often associated with an increased inoculum of several plant pathogens and/or decreased number of beneficial microbes in soil (Mazzola and Manici, 2012; Cook et al., 2023). In addition to crop rotation, amending growing media with specific beneficial microbes or bioproducts directly is a strategy to alleviate yield decline.
To investigate the microbial causes of yield decline in reused coir, we sampled strawberry roots and bulk coir from the experiment where strawberry yield decline in reused coir was reported (Shuttleworth et al., 2021). We aimed to (i) determine changes in the overall root and coir microbiome as well as the specific taxa groups in relation to coir age (growing seasons) and in root samples and biochar amendment and (ii) identify specific microbial taxa associated with yield decline.
2 Materials and methods
2.1 Experimental design and plantation
The experimental design and strawberry fruit yield data were previously described by Shuttleworth et al. (2021). In short, the experiment was conducted in 2020 to 2021 using a fully randomized block design with two factors (12 treatments in total): reused coir age and biochar amendment. Reused coir age had four levels: (1) unused/virgin coir, (2) reused 1-year-old coir, (3) reused 2-year-old coir, and (4) reused 3-year-old coir. The biochar amendment factor had three levels, namely: (1) unamended control, (2) oak biochar (produced by ILVO, Belgium), and (3) beech biochar (produced by ECN-TNO, Netherlands). The key properties of the two biochar products used in the present study—oak (Amery et al., 2021) and beech (Vandecasteele et al., 2023)—are given in Supplementary Table S1 based on published studies.
In 2018, virgin and 1-year-old coir (obtained from a local commercial farm) were used to grow the June-bearer strawberry cv. ‘Malling Centenary’. Three biochar amendments were applied to both coir ages. In 2019, the coir used in 2018 was reused, becoming 1- and 2-year-old coir. A new batch of virgin coir was added as a control, and coir of all ages was amended with biochar. Similarly, the coir bags used in 2019 were reused in 2020 (becoming 1-, 2-, and 3-year-old coir) alongside additional virgin coir bags and all coir amended with biochar; this led to four coir ages. At the end of the 2018 and 2019 seasons, all the plants were removed from all the coir bags, and new plants were planted in the following season. In 2020, the randomized block design experiment had the four blocks with 12 plots, each randomly assigned to one of the 12 treatments. In each plot, there were four coir bags, each with six plants of everbearer cv. “prize” per bag, giving a total of 96 plants per treatment.
Before amendment, both biochar types were rehydrated in plastic containers using water to a biochar weight ratio of 1:2. An amendment of 50 mL of wet biochar was applied directly to each planting hole at planting time. Vegetative feed (YaraTera Kristalon Blue LB, Yara UK; 19% N, 6% P, 20% K, 3% Mg, 7.5% S, 0.025% B, 0.01% Cu, 0.07% Fe, 0.04% Mn, 0.004% Mo, and 0.025% Zn) or fruiting feed (YaraTera Kristalon Red LB, Yara UK; 12% N, 12% P, 36% K, 1% Mg, 2.5% S, 0.025% B, 0.01% Cu, 0.07% Fe, 0.04% Mn, 0.004% Mo, and 0.025% Zn) nutrient feeds were applied at 1% solution through irrigation lines (fertigation) at four drippers (2 L/H) per bag. Fertigation volume and concentration were adjusted according to plant development, with the conductivity (EC) kept between 1.6 and 2 mS/cm at pH 6. The fertigation frequency varied with plant growth stage, the weather forecast, and the substrate moisture which was determined weekly using a WET-2 Sensor (Delta-T Devices Ltd., UK).
2.2 Sampling roots and coir
The plants from 2020 were maintained for fruit production in 2021 as COVID-19 made it difficult to replant the trial with an additional virgin coir treatment. Furthermore, because of COVID-19 restrictions, we could not harvest ripe fruit twice or three times a week; we instead harvested all fruits at a monthly interval. Thus, fruit yield was represented by the total number of fruits per plant instead of class I marketable yield (Shuttleworth et al., 2021).
Both roots and coir were sampled at the end of the experiment in October 2021. Roots were collected from every plot. Two or three small pinches of fine roots were collected from six plants per plot (grown in one bag) and pooled into a single sample. The roots were gently washed in sterile water to remove coir and biochar and dried with a paper towel. The resulting root microbiome is a sum of rhizoplane and root endophytic microbiomes.
Bulk coir fibers were sampled from all biochar unamended plots only (all coir ages). Each sample consisted of a pool of coir fibers at four bags per plot. All samples were stored at -20°C until DNA extraction.
2.3 DNA extraction and sequencing
The root samples were freeze-dried, and dry weight (around 0.03 g) was recorded before homogenization with a Geno/Grinder 2010 (SPEX CertiPrep) for 4 min at 1,500 rpm using 50-mL tubes and 14 5-mm steel ball bearings. Phosphate-buffered saline (PBS) was prepared to 0.1 M, filter-sterilized through a 200-µM pore, added to the homogenized samples at 1:5 dry weight (mg) to volume (µL) ratio, and mixed by vortexing. DNA was extracted from 120 mL of PBS resuspended homogenate with the DNeasy Plant Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s protocol, including an optional Rnase A digestion step after lysis.
For coir extracts, the exact weights of freeze-dried coir samples (around 0.25 g) were recorded. Samples were then extracted by using DNeasy PowerSoil Kit (Qiagen) according to the manufacturer’s protocols with the following adjustment: at lysis stage, the samples were put in Geno/Grinder 2010 (SPEX CertiPrep) for 2 min at 1,750 rpm.
All samples were eluted in 100 µL, with the eluate passed through the column twice, and the yields were analyzed using a Nanodrop spectrophotometer (Thermo Scientific, Waltham, MA, USA) and a Qubit fluorometer (Invitrogen, Waltham, MA, USA). Some samples were also tested for amplification using endpoint PCR using ITS1/4 (White et al., 1990) and 16S 357-1492 (Muyzer et al., 1993) primers, and the products were run on 1.5% agarose gel with GelRed at 100 V for 1 h and visualized using a GelDoc imager (Bio-Rad, Hercules, CA, USA). The DNA samples were shipped to Novogene UK (Cambridge, UK) for PCR, library prep, and amplicon sequencing. The target primers were ITS1-1F [ITS1-1F-F: 5’-CTTGGTCATTTAGAGGAAGTAA-3’ (Gardes and Bruns, 1993)], ITS2 [5’-GCTGCGTTCTTCATCGATGC-’3 (White et al., 1990)], and 16S V5-V7 [799F: 5’-AACMGGATTAGATACCCKG-3’ (Chelius and Triplett, 2001) and 1193R: 5’-ACGTCATCCCCACCTTCC-3’ (Bodenhausen et al., 2013)]. The samples were sequenced on an Illumina NovaSeq platform in the 250-nt paired-end mode.
2.4 Sequencing processing and taxonomy assignment
Amplicon sequence variants (ASVs) were generated from a combined set of root and coir sample data but analyzed separately using a previously published pipeline (Papp-Rupar et al., 2022). Raw sequence reads with incorrect bases in the barcode or primer regions (whether forward or reverse) or which contain adapter contamination were discarded. The retained reads were then merged using the UPARSE pipeline V. 11.0 (Edgar, 2013) twice: (1) to produce reads for ASV generation using stringent criteria and (2) to produce reads for subsequent frequency table generation using much more permissive criteria. For ASV generation, the following settings were used: (1) a minimum read length of 250 bases, (2) zero different bases in the merged region, and (3) a minimum merged length of 400 (16S) or 185 (ITS) bases. These merged sequences were further filtered for quality with a maximum expected error (MEE) threshold of 0.2 (16S) and 0.1 (ITS) per sequence (Edgar and Flyvbjerg, 2015). The reads were then dereplicated, and sequences with less than eight replicates were discarded before the generation of denoised ASVs (UPARSE also removes suspected chimeral sequences). For frequency table generation, reads were merged using a “maximum number of different bases” set to an arbitrarily high number (100) to ensure that all reads were effectively merged. These unfiltered merged reads were aligned to the ASV representative sequences at the level of 97% similarity to produce an ASV frequency table. Finally, the SINTAX algorithm (https://www.drive5.com/usearch/manual/sintax_algo.html) was used to assign taxonomic ranks to each ASV with the Unite V8.3 (2021-05-10) fungal database (Köljalg et al., 2013) and “the RDP training set V18” database for the 16S rRNA gene (Cole et al., 2014). The SINTAX algorithm only resolves bacterial ASVs to the genus level but may resolve fungal ASVs to the species level. Taxonomy assignment confidence was set at the 50% level.
2.5 Statistical analysis of amplicon data
Only the most abundant ASVs that accounted for 99.9% of the total sequence reads were retained for statistical analysis. The ASV count data were normalized for library size by the median-of-ratios (MR) method implemented in DESeq2 (Love et al., 2014) before statistical analysis. Data from the two sample types (roots and coir) were analyzed separately. In all analyses, there were two factors: coir age and biochar amendment. All statistical analyses were carried out with R 4.1.3 (Team, 2019).
The rank of alpha diversity (Shannon and Simpson) indices, calculated with the R vegan 2.3–1 package (Dixon, 2003), was subjected to an analysis of variance (ANOVA) to assess the effects of treatment factors via a permutation test. The beta (Bray–Curtis) diversity indices were subjected to permutational multivariate ANOVA (PERMANOVA) to assess the effects of treatment factors (implemented as the Adonis function in the vegan package). ANOVA was applied to assess the treatment effects on the first four principal components (PCs) of both the bacterial and fungal microbiomes.
Although the within-sample (alpha) and between-sample (beta) diversity indices were analyzed, the present study focused on the differential abundance among coir of different ages and biochar amendments to identify microbes whose relative abundance was significantly influenced by coir age and biochar amendment. DESeq2 analyses were applied to assess the changes in the relative abundance of individual ASVs between specific biochar amendments: (1) oak biochar amendment vs. control, (2) beech biochar amendment vs. control, and (3) beech vs. oak biochar amendment. DESeq2 uses Wald test to assess for operational taxonomic units with a significant differential abundance (Anders and Huber, 2010; Love et al., 2014). To investigate the relative abundance of individual ASVs in relation to reused coir age, two analyses were used: (1) coir age as a factor with three degrees of freedom and (2) coir age as a continuous variable (hence, only the linear relationship with one degree of freedom was assessed). In all differential abundance (DESeq2) analyses, the full root sample model included the factorial design of biochar amendment and coir age as well as the block factor. For the coir samples, there was only a single treatment factor (coir age). Only ASVs with mean normalized counts above 100 were used in the differential abundance analysis with DESeq2.
Spearman and Pearson correlation coefficients were calculated for fruit yield in each plot with relative ASV abundance and all PC scores (based on ASV relative abundance). In addition, regression analysis was used to assess the effect of individual ASVs on yield using models where the block and biochar amendment were included. Only ASVs with average normalized counts greater than 100 were included in the correlation and regression analysis—normalized counts were log-transformed first before correlation and regression.
In all analyses with individual ASV abundance (DESeq2, correlation, and regression), the probability values were adjusted for multiple testing using the Benjamini–Hochberg (BH) method (Benjamin and Aikman, 1995).
3 Results
3.1 Root microbiome
3.1.1 General sequencing quality and taxonomy
All 48 strawberry root samples produced enough high-quality DNA for sequencing and achieved sufficient sequencing depth (Supplementary Figure S1). Table 1 gives the summary of sequencing reads per sample or per ASV. In total, there were 2,740 bacterial ASVs and 1,168 fungal ASVs used in the statistical analysis of the root microbiome.
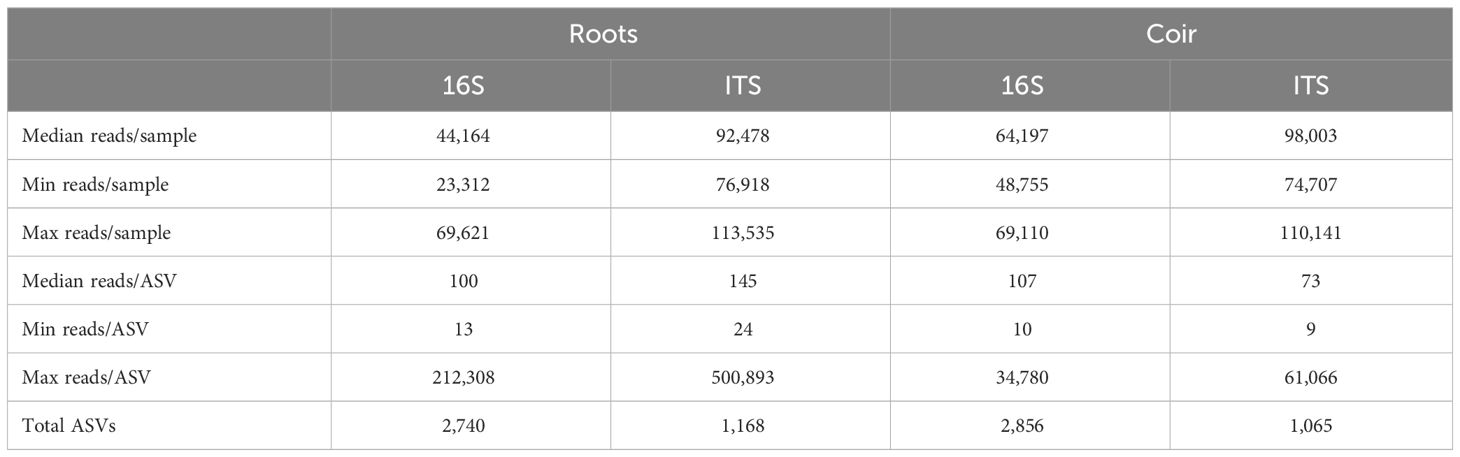
Table 1 Summary of sequencing reads per sample or ASV and total bacterial (16S) and fungal (ITS) ASVs found in strawberry root and coir samples.
There were 99.8%, 97.0%, 87.1%, 82.3%, and 73.9% of mapped bacterial reads that could be assigned to the rank of phylum, class, order, family, and genus, respectively, at the 50% confidence level. The two most common bacterial phyla were Actinobacteria and Proteobacteria, irrespective of treatments (Supplementary Figure S2A), accounting for approximately 50.5% and 39.4% of total mapped reads, respectively. Only 0.2% of reads failed to be assigned to a phylum with confidence. The top three bacterial ASVs in the root microbiome were from the Streptomyces genus, accounting for 26.1% of the total mapped reads.
There were 91.8%, 50.8%, 33.3%, 25.9%, 19.3%, and 14.6% of mapped fungal reads that could be assigned to the rank of phylum, class, order, family, genus, and species, respectively. Ascomycota and Basidiomycota accounted for 47.5% and 43.4% of total mapped reads, respectively (Supplementary Figure S2B). The top three most abundant fungal ASVs in the root microbiome could only be assigned to the phylum rank: one from Basidiomycota (13.2% mapped reads) and the other two from Ascomycota (8.7% and 5.5% mapped reads).
3.1.2 Within-sample (alpha) and between-sample (beta) diversity
Within-sample diversity indices varied greatly in root samples for both bacteria and fungi (Figure 1). However, neither biochar amendment nor coir age significantly affected the alpha diversity indices in roots. Overall, alpha diversity was higher for bacteria than fungi (Figure 1).
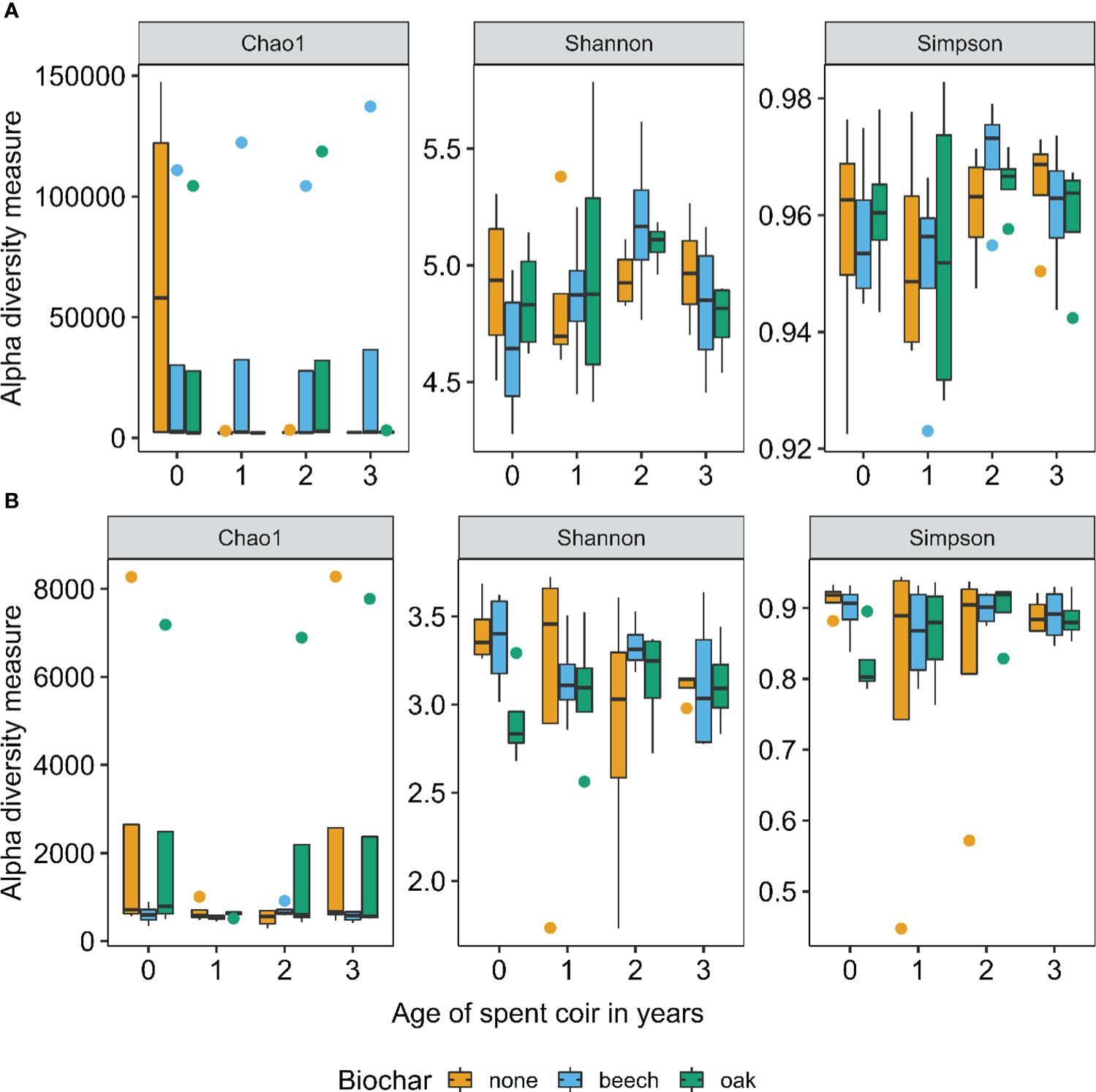
Figure 1 Within-sample (alpha) diversity indices for bacteria (A) and fungi (B) of 48 strawberry root samples in relation to biochar amendment and the age of reused coir (in years at planting).
The first principal components explained 16.4%, 5.9%, 5.3%, and 4.6% of the total variability in bacterial ASVs in roots, whereas the corresponding values for fungi were 10.7%, 9.3%, 7.0%, and 6.1%. The first four bacterial PC scores were not affected by biochar amendment. Only the second (P < 0.001) and third (P < 0.05) PC were significantly affected by the coir age. Furthermore, only the second PC was linearly related with coir age. As a factor, coir age explained about 10.0% of the total variability across all bacterial PCs from root samples.
Coir age, as a factor, significantly affected the first four fungal PC scores in root samples with P < 0.05 for the first and second, P < 0.01 for the third, and P < 0.001 for the fourth PC score. Furthermore, the first and fourth PCs were linearly related to the reused coir age. Summarized over all PCs, the coir age explained about 10.3% of the total fungal variability.
The non-parametric multivariate analysis of variance of the Bray–Curtis indices showed that only coir age was significantly (P < 0.001) affected between sample diversities, explaining 10.7% of the total variability in the β diversity indices. Similar results were obtained for fungi, except that coir age explained more variability (approximately 18.3%) in the β diversity indices. However, the sample separation due to reused coir age appeared to be clearer for bacteria (Figure 2A) than for fungi in the first two dimensions (Figure 2B).
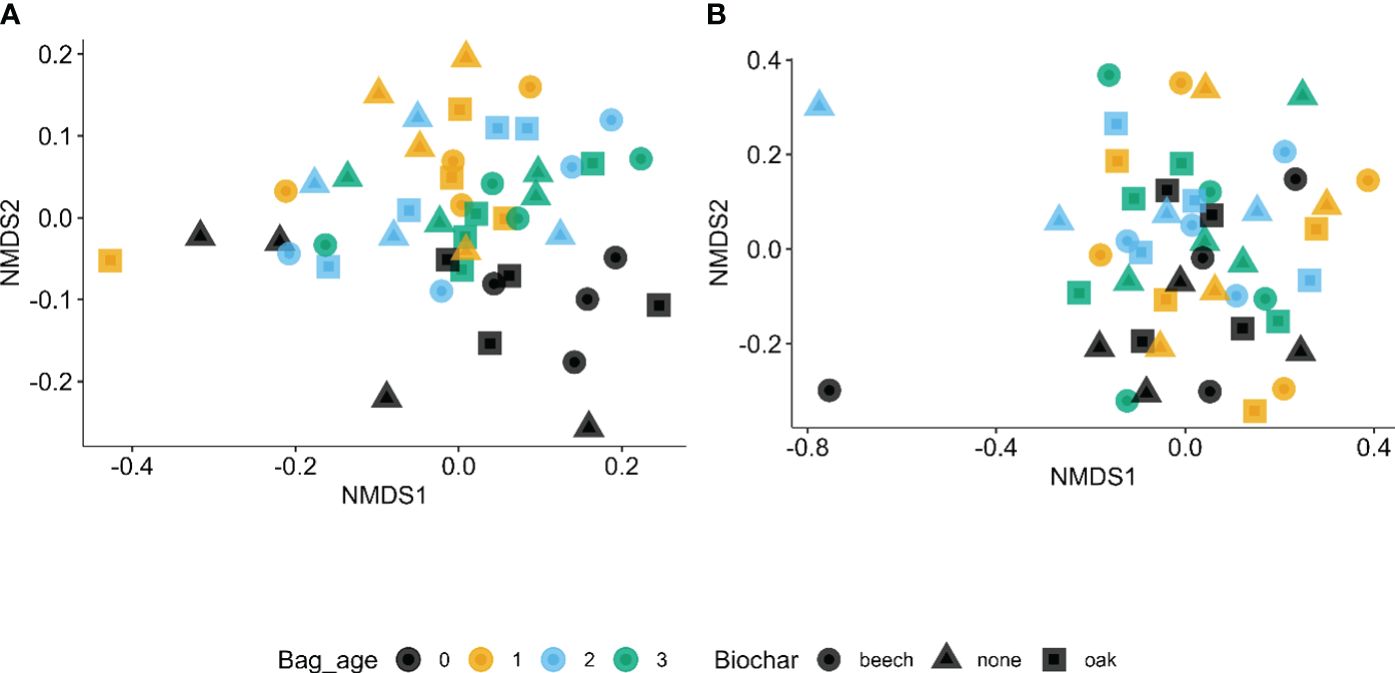
Figure 2 First two dimensions of non-metric multidimensional scaling of Bray–Curtis indices among the 48 strawberry root samples for bacteria (A) and fungi (B) in relation to the age of reused coir (in years at planting) and biochar amendment.
3.1.3 Differential abundance
Table 2 gives the summary of differential abundance analysis. The present focus was on those ASVs with an average count greater than 100. Of the 2,740 bacterial ASVs, the abundance of 20 bacterial ASVs was affected by reused coir age (as a factor), and the abundance of 14 bacterial ASVs had a significant linear relationship with reused coir age (Table 3): six increased with increasing reused coir age, including two Streptomyces ASVs. For the other eight ASVs, there was a decreasing relationship with increasing coir age, including two Novosphingobium ASVs and two Streptomyces ASVs. None of the bacterial ASVs with an average read count above 100 was affected significantly by biochar amendment.
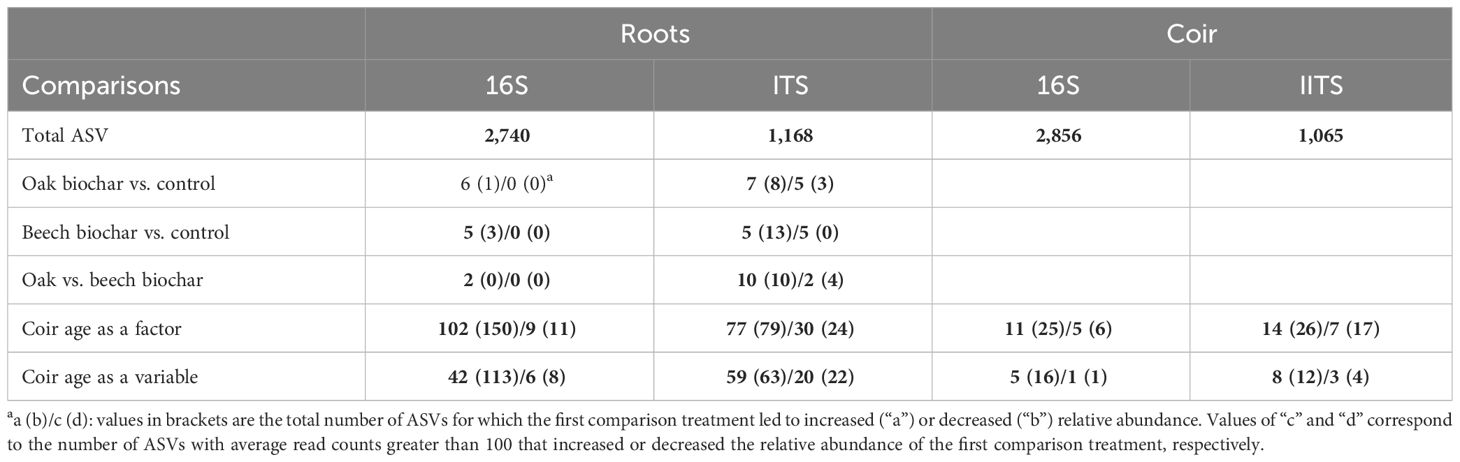
Table 2 Summary of differential abundance analysis of bacterial (16S) and fungal (ITS) communities in strawberry root and coir.
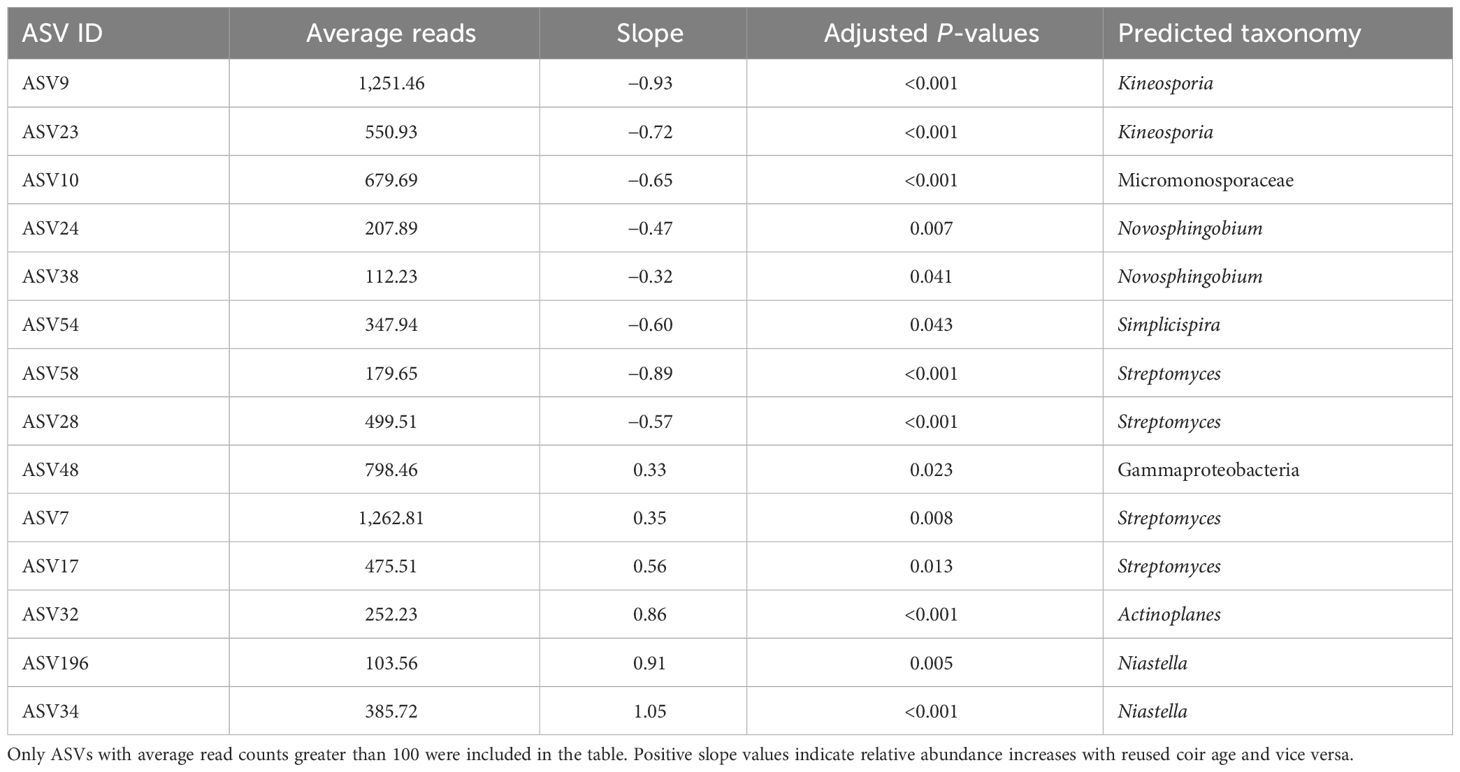
Table 3 Summary of bacterial ASV taxa in strawberry roots (endophytes and rhizoplane) that significantly linearly related to the reused coir age (years).
The abundance of 55 fungal ASVs was affected by reused coir age (as a factor), and the abundance of 41 fungal ASVs was in a linear relationship with reused coir age: 20 increased with increasing reused coir age, including Ilyonectria destructans and Flagelloscypha minutissima (Table 4). For the other 21 ASVs, there was a decreasing relationship with increasing coir age, including I. liriodendra, two Cadophora luteo-olivacea ASVs, two Zopfiella marina ASVs, and one Cadophora malorum ASV (Table 4).
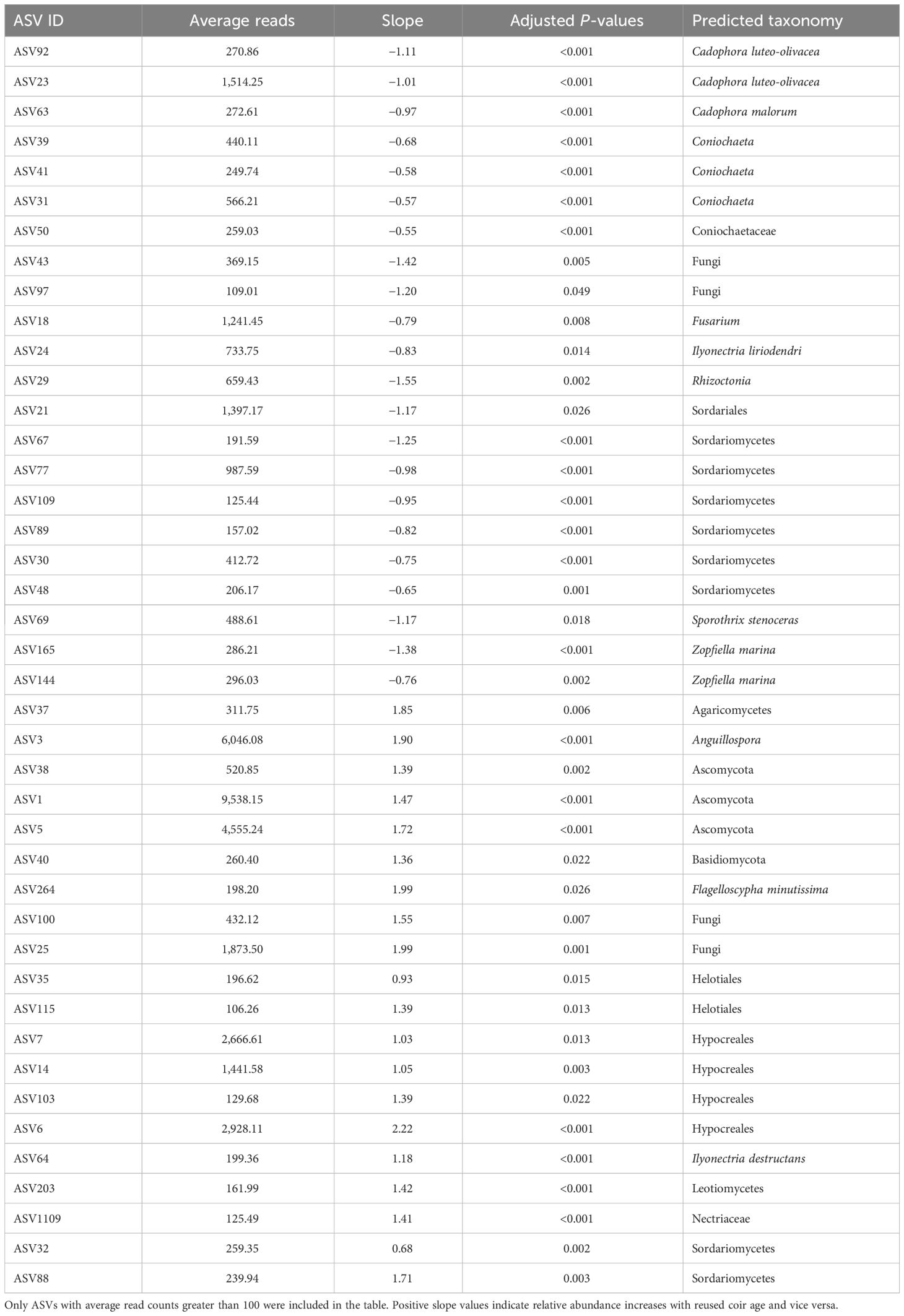
Table 4 Summary of fungal ASV in strawberry roots (endophytes and rhizoplane) that are significantly linearly related to the reused coir age (years).
Of the 1,168 fungal ASVs detected in the root microbiome, beech biochar amendment led to significant decreases in relative abundance of five ASVs compared to the control: one from F. minutissima, one from Rhizoctonia, and the other three ASVs could not be assigned to a rank below order (Table 5). Compared to the control, amendment with oak biochar led to significant increases and decreases in relative abundance for five and three ASVs, respectively. Of these eight ASVs, one was from F. minutissima (reduced abundance), and one Rhizoctonia, one Ceratobasidium, and one Dactylium (increased abundance) (Table 5).
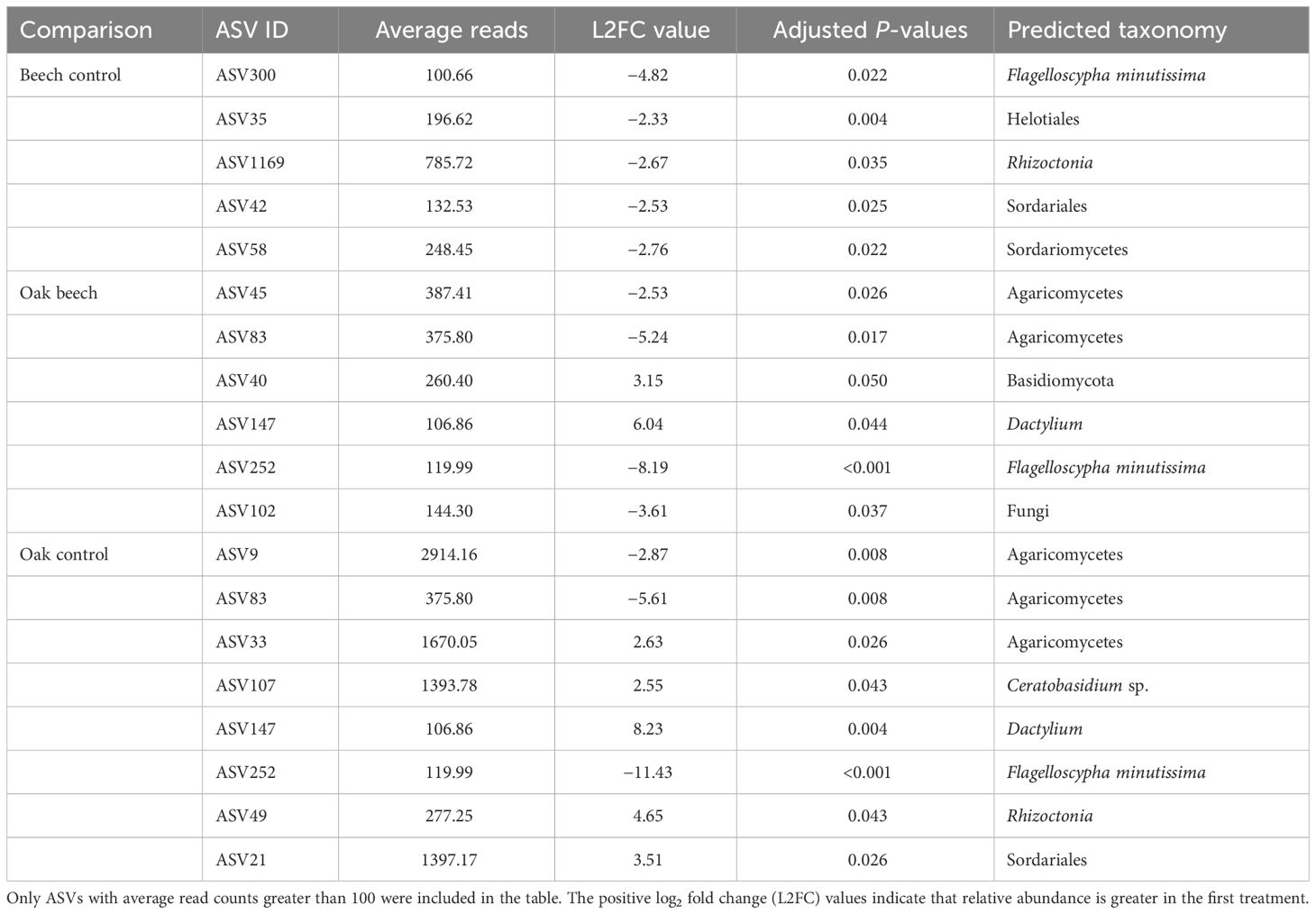
Table 5 Differential analysis summary of fungal ASVs in strawberry roots (endophytes and rhizoplane) with a significantly different abundance between biochar treatments.
3.1.4 Fruit yield in relation to biochar amendment, reused coir age, and microbial ASVs
The root microbiome consisted of 1,364 and 700 bacterial and fungal ASVs with an average read count greater than 100, respectively. Only one bacterial ASV (Bradyrhizobium) was correlated (Pearson coefficient) with fruit yield. None of the ASVs were significantly associated with fruit yield once the block and biochar factors were included in the regression.
Spearman and Pearson correlation indicated four and three fungal ASVs, respectively, with their relative abundance negatively correlated with fruit yield. Two were common ASVs, and the others could not be assigned to the phylum rank. When both block and biochar amendment were included in the linear regression, only the Linnemannia elongata ASV was negatively related to fruit yield. None of the bacterial or fungal PCs correlated with fruit yield.
3.2 Coir microbiome
3.2.1 General sequencing quality and taxonomy
Of the 16 coir samples, four failed to produce sufficiently high-quality DNA for sequencing: two from virgin coir and the other two from 1-year-old coir. A sufficient sequencing depth was achieved for the remaining 12 samples (Supplementary Figure S1). The summary of sequencing reads per sample or per ASV is given in Table 1. In total, the coir microbiome consisted of 2,856 bacterial and 1,066 fungal ASVs used for statistical analysis.
A total of 98.6%, 95.3%, 83.4%, 68.1%, and 58.5% of mapped bacterial reads could be assigned to the rank of phylum, class, order, family, and genus with confidence of at least 50%, respectively. The two most common bacterial phyla were Proteobacteria and Actinobacteria, irrespective of treatments (Supplementary Figure S3A), accounting for approximately 50.7% and 29.1% of the total mapped reads, respectively. Only 0.2% of reads failed to be assigned to a phylum level with confidence. The top four ASVs were all from the Streptomyces genus, accounting for 9.0% of the total mapped reads. One Bacillus ASV and two Rhizobiales ASVs were also among the top 10 most abundant ASVs.
A total of 94.3%, 85.9%, 69.0%, 51.9%, 46.2%, and 32.2% of mapped fungal reads could be assigned to the rank of phylum, class, order, family, genus, and species, respectively. Ascomycota and Basidiomycota accounted for 47.5% and 43.4% of total mapped reads, respectively (Supplementary Figure S2B). The top two most abundant fungal ASVs in coir were from Hypocreales (6.2% and 4.1%), and two Humicola fuscoatra ASVs were the third and fourth most abundant ASVs, jointly accounting for nearly 7.6% of the total mapped reads. Two Cladosporium ASVs were also among the top 10 most abundant fungal ASVs; one was classified as C. ramotenellum.
3.2.2 Within-sample (alpha) and between-sample (beta) diversity
Coir microbiome within-sample diversity indices varied greatly between samples of the same coir age for both bacteria and fungi, with the exception of bacteria in virgin coir and fungi in the 1-year-old coir, where much less variation was observed (Figure 3). The age of reused coir did not significantly affect the alpha diversity indices. Overall, alpha diversity was much higher for bacteria than fungi, particularly for the Chao1 and Shannon indices (Figure 3).
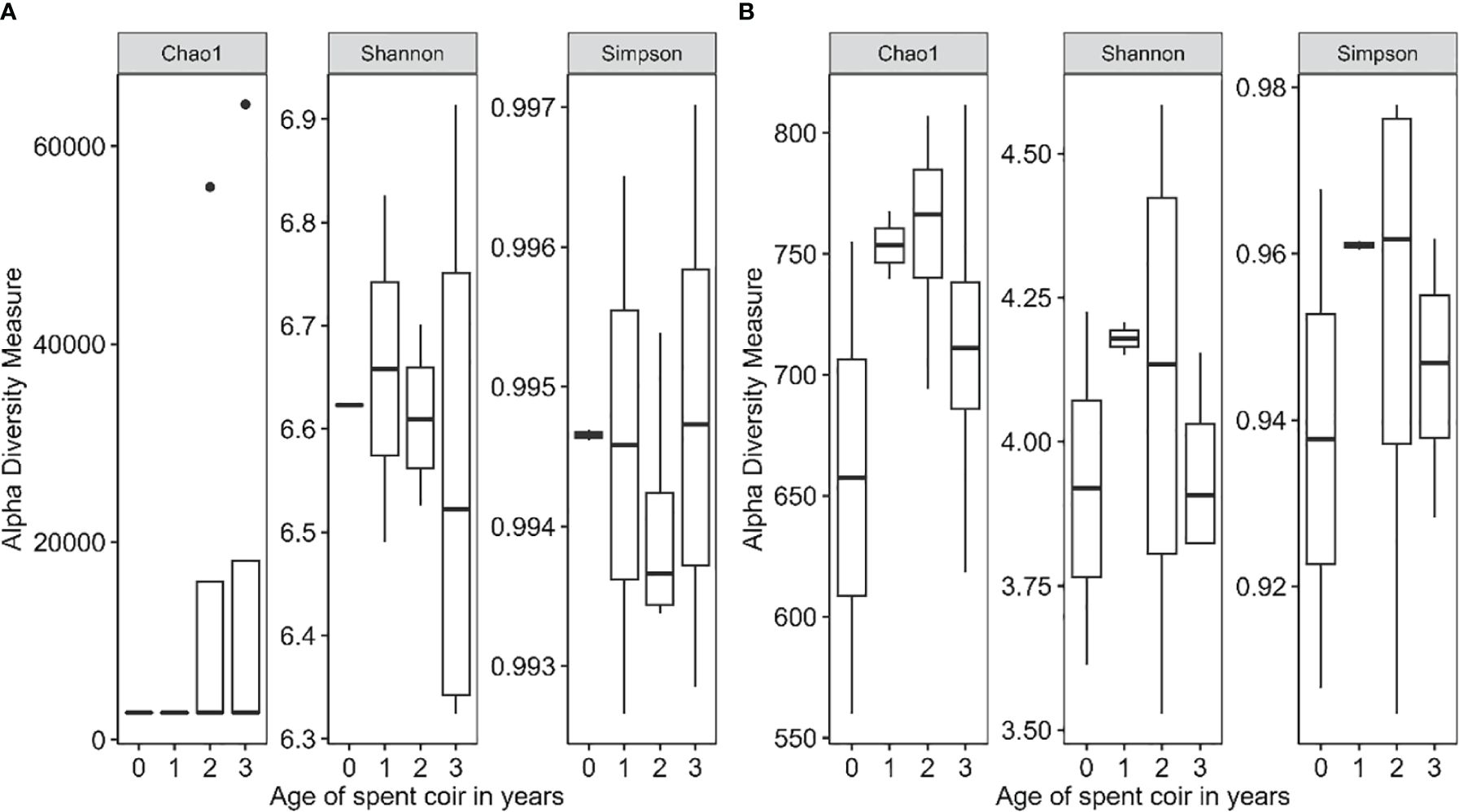
Figure 3 Within-sample (alpha) diversity indices for bacteria (A) and fungi (B) of 12 coir samples in relation to the age of reused coir (in years at planting).
The first principal components explained 24.4%, 16.1%, 12.5%, and 10.3% of the total variability in bacterial ASVs in coir, whereas the corresponding values for fungi were 22.9%, 14.3%, 12.9%, and 9.7%. The bacterial PC scores were not significantly affected by reused coir age. Coir age significantly affected the fourth fungal PC scores only (P < 0.01). Summarized over all fungal PCs, the coir age explained about 28.1% of the total variability.
A non-parametric multivariate analysis of variance of the Bray–Curtis indices suggested that coir age significantly affected the β diversities for both bacteria (P < 0.05) and fungi (P < 0.001), explaining about 33.1% and 36.7% of the total variability in the β diversity indices, respectively. Samples from virgin and 1-year-old coir appeared to be closer together in β diversity (Figure 4).
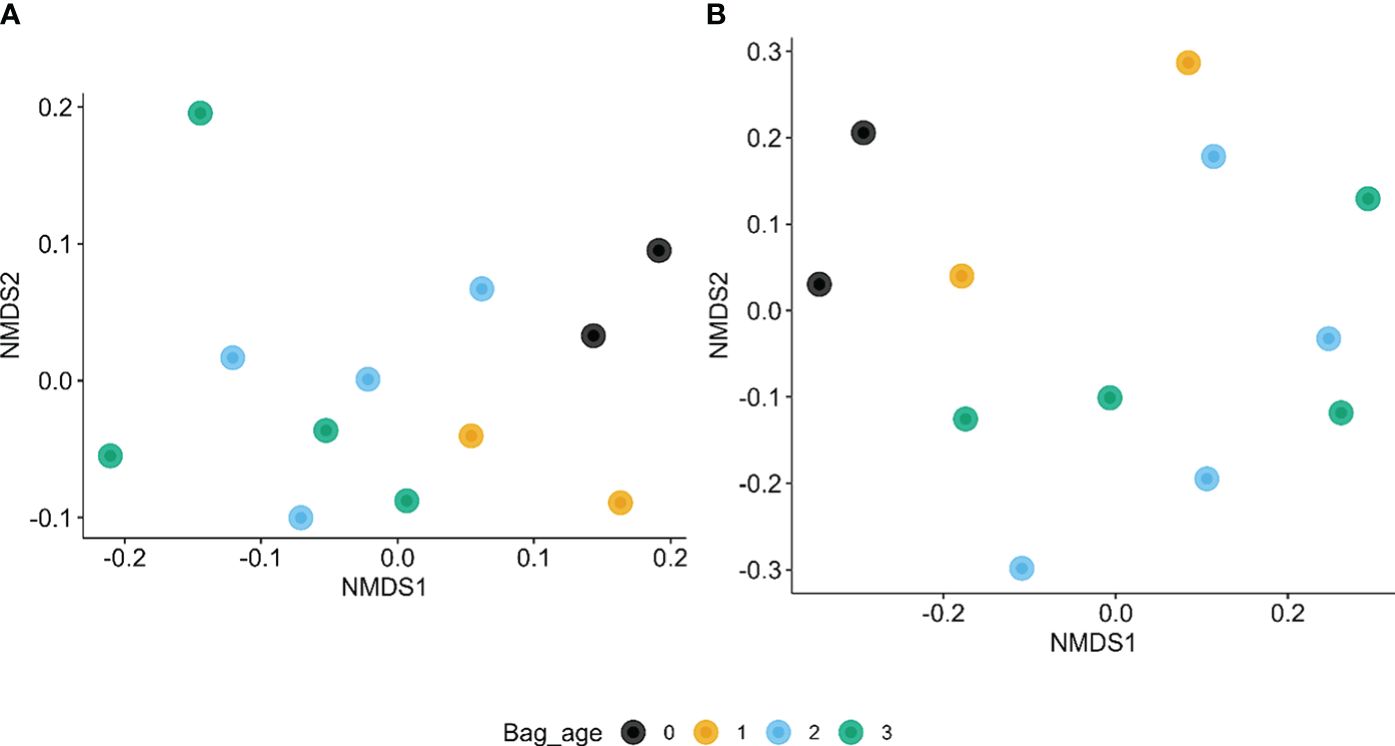
Figure 4 First two dimensions of non-metric multidimensional scaling of Bray–Curtis indices among the 12 coir samples for bacteria (A) and fungi (B) in relation to the age of reused coir (in years at planting) and biochar amendment.
3.2.3 Differential abundance
A summary of the results is given in Table 6. For the coir microbiome, only two comparisons were made: when coir age was treated as a factor and as a continuous variable. The present focus was on the comparison when coir age was treated as a continuous variable. The abundance of 11 out of 2,856 bacterial ASVs was significantly affected by coir age (as a factor), but only two ASVs had their relative abundance linearly related to coir age after block was taken into account—one was a Chlorophyta ASV (increasing with increasing coir age) and the other a Streptomyces (decreasing with increasing coir age) (Table 5). Similarly, the relative abundance of 24 out of 1,065 fungal ASVs was affected by reused coir age (as a factor), seven of which had their abundance linearly related with coir age (Table 5). Noticeably, an I. destructans ASV and a Penicillium ASV increased their relative abundance with increasing coir age.
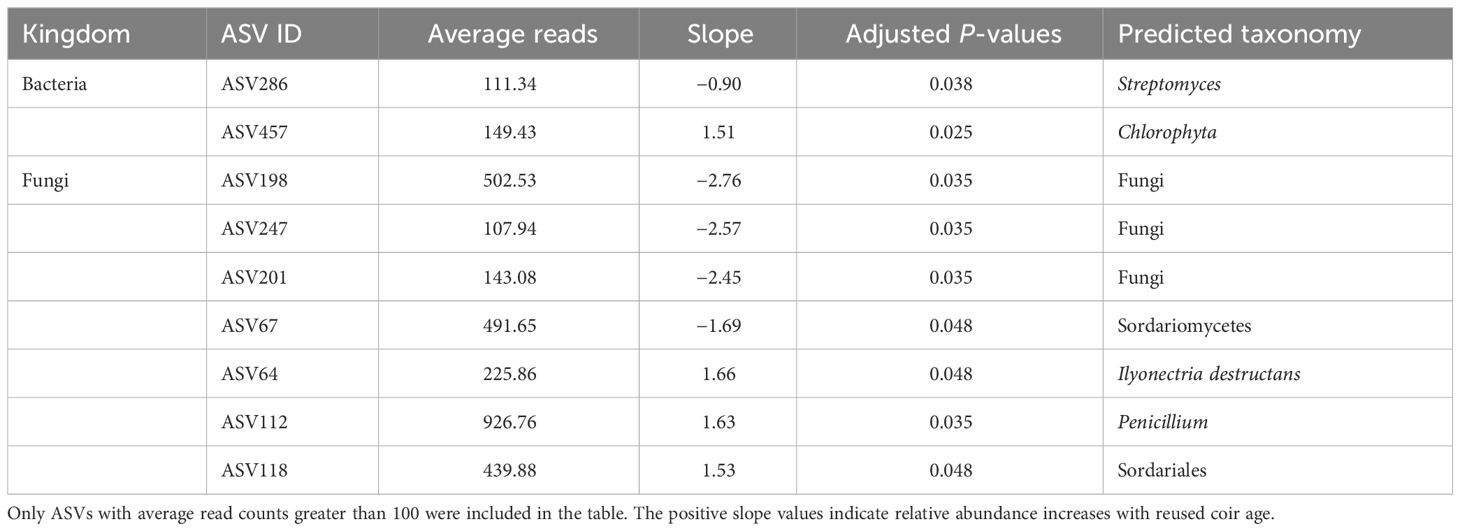
Table 6 Differential analysis summary of bacterial and fungal ASVs in coir that are significantly linearly related to the reused coir age (years).
3.2.4 Fruit yield in relation to microbial ASVs in coir
Fruit yield was not significantly affected by the relative abundance of any of the top 1,526 bacterial or 487 fungal ASVs. Similarly, none of bacterial or fungal PCs were significantly correlated with fruit yield.
4 Discussion
The present study identified many microbial taxa (ASVs), particularly fungal taxa in strawberry roots (rhizoplane and root endophytes), whose relative abundance significantly varied with the age of spent coir. However, the interpretation of these ASVs is made difficult by the fact that most of these ASVs cannot be identified to the taxonomical rank of species. There were only a few ASVs whose increased or reduced relative abundance in reused coir could be interpreted biologically in association with strawberry yield decline in spent coir. Interestingly, neither fungal nor bacterial within-sample diversity was affected by coir age. Moreover, amendment with either oak or beech biochar did not affect the overall microbial composition.
The relative abundance of several bacteria genera in roots was linearly associated with reused coir age, including two Niastella ASVs which increased their abundance with increasing reused coir age. Niastella, as a denitrifier, can mitigate N2O emissions (Nishizawa et al., 2014); its increased abundance may be the result of an increasing level of N fertilizer in the reused coir. Increased abundance was also found for one Actinoplanes ASV, which may have the role of cycling nutrients (Boubekri et al., 2022). Thus, its increased abundance may also be related to increasing amounts of nutrients in reused coir. There were four Streptomyces ASVs, two of which increased their abundance linearly, and the other two decreased their abundance with increasing reused coir age. The Streptomyces genus contains many known plant pathogens (Scholte, 1989; Elphinstone and Wale, 2009) and strains with biological control properties (Law et al., 2017; Mahnkopp-Dirks et al., 2021), making their role in yield decline unclear. Similarly, the decrease in the relative abundance of two genus Kineosporia (Actinomycetes) ASVs is likely unrelated to yield decline since the genus is commonly associated with leaf litter and has not been known to cause plant diseases (Tamura and Suzuki, 2014). For two Novosphingobium ASVs, the relative abundance linearly decreased with increasing reused coir age; Novosphingobium is associated with biodegradation of substrates and is prevalent in environments such as soil and wood (Wang et al., 2018).
The relative abundance of two Ilyonectria ASVs was linearly related to the spent coir age. One ASV (I. destructans, syn. Cylindrocarpon destructans) increased its abundance in strawberry roots with increasing reused coir age; the other (I. liriodendra) decreased. Both species are known pathogens of grapevine, causing root infections (Halleen et al., 2006; Cabral et al., 2012; Reis et al., 2013). The high relative abundance of I. robusta was associated with yield decline in soil-grown strawberry under open-field conditions (Xu et al., 2015). Cylindrocarpon destructans can cause variable degrees of crown and root rot in strawberry (Fang et al., 2011a, b). Similar root diseases in strawberry can also be caused by other non-specific pathogens, including Rhizoctonia (Martin, 2000) and Fusarium oxysporum (Koike et al., 2009), and are commonly referred to as black root rot, a name that is descriptive of the appearance of the roots (Wing et al., 1994). Cylindrocarpon destructans is also implicated in causing ginseng root rot disease and rusty symptoms (Farh et al., 2018). Although Rhizoctonia (Martin, 2000) and Fusarium (Koike et al., 2009) could be strawberry root pathogens, the relative abundance of one Rhizoctonia ASV and one Fusarium ASV in strawberry root decreased with increasing reused coir age. However, it should be noted that only one I. destructans ASV increased its relative abundance in the coir with increasing reused coir age.
Several other ASVs with identification to the species level also showed linear relationships with reused coir ages. The relative abundance of two Pseudorhypophila marina (syn. Zopfiella marina) ASVs, two C. luteo-olivacea ASVs, one C. malorum ASV, and one Ophiostoma stenoceras ASV all significantly decreased with increasing coir age. Cadophora luteo-olivacea is the most prevalent Cadophora species associated with Petri disease and esca of grapevine (Maldonado-González et al., 2020), and a minor post-harvest pathogen of apple (Amaral Carneiro et al., 2022). Pseudorhypophila marina produces zopfinol and the strong antifungal zofimarin (Charria-Girón et al., 2022). Sporothrix stenoceras is a sapwood-colonizing fungus occurring on some coniferous and hardwood hosts (de Beer et al., 2003). Cadophora malorum is present in pear orchard soil and may cause the side rot of pear (Sugar and Spotts, 1992; Sugar, 1993). The relative abundance of one Flagelloscypha minutissima ASV increased with increasing coir age; F. minutissima is a wild mushroom fungus. The relative abundance of three yeast ASVs of Coniochaeta in strawberry root decreased with increasing reused coir age. Coniochaeta endophytes of plants and lichens (Damm et al., 2010; Arnold et al., 2021a, Arnold et al., 2021b) are positively associated with soil health (Huang et al., 2020).
When only microbiota in coir is considered, only a few taxa linearly decreased or increased with reused coir age compared to the root-associated microbiome. This may be partially due to reduced statistical power because fewer samples were available for coir microbiome analysis. Only one Streptomyces ASV had reduced abundance with increasing coir age, and one green alga (Chlorophyta) ASV had increased abundance with increasing age in coir. For fungi, the same I. destructans ASV as in the roots had its abundance linearly increased with increasing spent coir age.
Biochar, as an alternative agricultural practice, may modify and improve soil fertility (Nelson et al., 2011; Prendergast-Miller et al., 2014) when added to peat growing media; biochar enhanced root formation and fruit production in strawberry (De Tender et al., 2016). However, in the present experiments over four seasons, the application of biochar did not lead to any yield benefit (Shuttleworth et al., 2021), agreeing with another strawberry study at East Malling in which biochar application to soil-grown strawberry did not lead to a yield benefit (Jay et al., 2015). Furthermore, the present research also showed that biochar did not significantly affect the root-associated microbiome. Published research generally suggests that amending the substrate or soil with biochar can lead to significant changes in the soil microbiome (Dai et al., 2021; Ren et al., 2023) and sometimes in the rhizosphere microbiome, e.g., rhizobacterial communities in wheat (Li et al., 2023). The lack of biochar-associated effects on the root-associated microbiome may partially result from the application method. By the nature of a compact coir bag, it was not possible to mix biochar with coir well, and thus biochar was only applied to the planting hole. Consequently, the roots may have grown into those areas without biochar.
Of all bacterial and fungal ASVs associated with roots, only one Linnemannia elongata ASV was found to be significantly negatively correlated with fruit yield. Although L. elongata can improve Arabidopsis thaliana foliar growth and seed development (Vandepol et al., 2022; De Tender et al., 2023), its function might be context sensitive and/or strain specific. As explained, we used the number of fruits as a yield indicator, which may not truly reflect the total fruit weight and/or number of marketable fruits.
The reuse of substrate in the UK strawberry production is becoming more frequent for several reasons. Reusing coir significantly reduces the seasonal costs of using virgin coir, carbon footprint related to shipping virgin coir material from South Asia, and also manual labor required for bag disposal and replacement. Reused coir bags are thus cheaper and more sustainable. Our (Shuttleworth et al., 2021) and other independent research (Woznicki et al., 2024) have demonstrated that June-bearing strawberry cultivars such as ‘Malling Centenary’ may be grown in up-to-twice-reused bags with negligible yield and quality penalty. This is likely due to the short cropping season, during which negative effects of substrate microbiome do not manifest in terms of noticeable yield decline. This is, however, our inference since the aforementioned studies did not investigate substrate microbiome. We showed that care needs to be taken when everbearing strawberry cultivars are grown in reused coir, especially if reused for more than two seasons (Shuttleworth et al., 2021). It is likely that detrimental microbial taxa, such as I. destructans identified in this study, accumulate in used coir over several growing seasons and cause a significant yield decrease only when fruit is produced for a prolonged period of time. It may also be the case that the everbearing cultivar used in the study is more susceptible to I. destructans and/or other potentially detrimental microbes that had accumulated in used coir compared to ‘Malling Centenary’. Although directly re-using in everbearing strawberry production may reduce yields, the loss of crop may be outweighed by the reduced input costs and increased sustainability. Using biocontrol and plant growth-promoting microorganisms at planting could mitigate this negative impact in used coir substrate (Lombardi et al., 2020).
In addition to pathogenic and all other components of the root and coir microbiomes, other factors also need to be considered when reusing coir, such as whether pests become established in the spent coir. Physio-chemical properties, including water retention capacity, air porosity, and electrical conductivity, are very important in strawberry production (Diara et al., 2012). These parameters are likely to change with coir reuse (Woznicki et al., 2024), and their optimum may also be variety specific. The reused coir itself can also be converted into biochar; however, care needs to be taken regarding salt and nutrient content, the pH of the biochar produced from it, and the amount added to the media.
In summary, coir age did not alter the alpha diversity of strawberry root and bulk coir microbiomes, but it contributed approximately 10%–20% and 30%–34% of the observed variance in the Bray–Curtis beta diversity indices in strawberry roots and bulk coir microbiomes, respectively. In strawberry roots, the relative abundance of 20 fungal ASVs, including the generalist root pathogen I. destructans, increased with reused coir age, while the abundance of 21 ASVs decreased with increasing coir age. Furthermore, I. destructans abundance increased also in bulk coir. The observed yield decline in strawberry is, therefore, likely to be associated with I. destructans. The contribution of other microbial components as well as the degradation of physiochemical properties of used coir may also contribute to the observed yield decline in reused coir. Adding biochar into planting holes did not cause much changes in strawberry root-associated and coir-associated microbiomes.
Future research is needed to investigate the extent to which I. destructans is responsible for strawberry yield decline in reused coir substrate. Furthermore, other methods of mixing substrate amendments and different amendments that may alleviate yield decline in reused coir need to be investigated—for instance, sterilizing reused coir instead of direct reuse could have a profound effect on microbiomes as well as plant development and fruit production. Amendments of specific beneficial products could then be added post-sterilization before packaging to ensure thorough mixing.
Data availability statement
All raw sequence data are available in the European Nucleotide Archive (ENA) under accession number PRJEB74694.
Author contributions
XX: Conceptualization, Formal analysis, Funding acquisition, Methodology, Project administration, Supervision, Writing – original draft, Writing – review & editing. GD: Formal 4nalysis, Writing – review & editing. JZ: Writing – original draft, Writing – review & editing. TP: Data curation, Investigation, Methodology, Project administration, Resources, Writing – review & editing. MP-R: Conceptualization, Funding acquisition, Investigation, Methodology, Resources, Supervision, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was funded by the Interreg2Seas project Horti-BlueC (www.horti-bluec.eu) Biotechnology and Biological Sciences Research Council (BBSRC) (project number: BB/X011801/1 and BB/R021295/1), and The Director Grant from The East Malling Trust.
Acknowledgments
We thank Jennifer Kingsnorth, Joyce Robinson, Hamish McLean, Josh Weaver, and Georgina Fagg for technical assistance.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fagro.2024.1397974/full#supplementary-material
References
Abad M., Noguera P., PuChades R., Maquieira A., Noguera V. (2002). Physico-chemical and chemical properties of some coconut coir dusts for use as a peat substitute for containerised ornamental plants. Bioresour. Technol. 82, 241–245. doi: 10.1016/S0960-8524(01)00189-4
Alexander P., Bragg N., Meade R., Padelopoulos G., Watts O. (2008). Peat in horticulture and conservation: the UK response to a changing world. Mires Peat 3, 1–10.
Amaral Carneiro G., Walcher M., Baric S. (2022). Cadophora luteo-olivacea isolated from apple (Malus domestica) fruit with post-harvest side rot symptoms in Northern Italy. Eur. J. Plant Pathol. 162, 247–255. doi: 10.1007/s10658-021-02388-4
Amery F., Debode J., Ommeslag S., Visser R., De Tender C., Vandecasteele B. (2021). Biochar for circular horticulture: feedstock related effects in soilless cultivation. Agronomy 11, 629. doi: 10.3390/agronomy11040629
Anders S., Huber W. (2010). Differential expression analysis for sequence count data. Genome Biol. 11, R106–R106. doi: 10.1186/gb-2010-11-10-r106
Arnold A. E., Harrington A. H., Huang Y.-L., Ren J. M., Massimo N. C., Knight-Connoni V., et al. (2021a). Coniochaeta elegans sp. nov., Coniochaeta montana sp. nov. and Coniochaeta nivea sp. nov., three new species of endophytes with distinctive morphology and functional traits. Int. J. System. Evolution. Microbiol. 71. doi: 10.1099/ijsem.0.005003
Arnold A. E., Harrington A. H., U'Ren J. M., Oita S., Inderbitzin P. (2021b). Two new endophytic species enrich the Coniochaeta endophytica, C. prunicola clade: Coniochaeta lutea sp. nov. and C. palaoa sp. nov. Plant Fungal System. 66, 66–78. doi: 10.35535/pfsyst-2021-0006
Benjamin L. R., Aikman D. P. (1995). Predicting growth in stands of mixed species from that in individual species. Ann. Bot. 76, 31–41. doi: 10.1006/anbo.1995.1075
Bodenhausen N., Horton M. W., Bergelson J. (2013). Bacterial communities associated with the leaves and the roots of Arabidopsis thaliana. PloS One 8. doi: 10.1371/journal.pone.0056329
Boubekri K., Soumare A., Mardad I., Lyamlouli K., Ouhdouch Y., Hafidi M., et al. (2022). Multifunctional role of Actinobacteria in agricultural production sustainability: A review. Microbiol. Res. 261, 127059. doi: 10.1016/j.micres.2022.127059
Cabral A., Groenewald J., Rego C., Oliveira H., Crous P. (2012). Cylindrocarpon root rot: multi-gene analysis reveals novel species within the Ilyonectria radicicola species complex. Mycol. Prog. 11, 655–688. doi: 10.1007/s11557-011-0777-7
Charria-Girón E., Surup F., Marin-Felix Y. (2022). Diversity of biologically active secondary metabolites in the ascomycete order Sordariales. Mycol. Prog. 21, 43. doi: 10.1007/s11557-022-01775-3
Chelius M. K., Triplett E. W. (2001). The diversity of archaea and bacteria in association with the roots of Zea mays L. Microbial. Ecol. 41, 252–263. doi: 10.1007/s002480000087
Cole J. R., Wang Q., Fish J. A., Chai B., McGarrell D. M., Sun Y., et al. (2014). Ribosomal Database Project: data and tools for high throughput rRNA analysis. Nucleic Acids Res. 42, D633–D642. doi: 10.1093/nar/gkt1244
Cook C., Magan N., Xu X. M. (2023). Inter-row cropping and rootstock genotype selection in a UK cider orchard to combat apple replant disease. Phytopathol. Res. 5, 28. doi: 10.1186/s42483-023-00184-y
Dai Z., Xiong X., Zhu H., Xu H., Leng P., Li J., et al. (2021). Association of biochar properties with changes in soil bacterial, fungal and fauna communities and nutrient cycling processes. Biochar 3, 239–254. doi: 10.1007/s42773-021-00099-x
Damm U., Fourie P. H., Crous P. W. (2010). Coniochaeta (Lecythophora), Collophora gen. nov. and Phaeomoniella species associated with wood necroses of Prunus trees. Persoonia Mol. Phyl. Evol. Fungi 24, 60–80. doi: 10.3767/003158510X500705
de Beer Z. W., Harrington T. C., Vismer H. F., Wingfield B. D., Wingfield M. J. (2003). Phylogeny of the Ophiostoma stenoceras–Sporothrix schenckii complex. Mycologia 95, 434–441. doi: 10.1080/15572536.2004.11833088
De Tender C., Haegeman A., Vandecasteele B., Clement L., Cremelie P., Dawyndt P., et al. (2016). Dynamics in the strawberry rhizosphere microbiome in response to biochar and Botrytis cinerea leaf infection. Front. Microbiol. 7. doi: 10.3389/fmicb.2016.02062
De Tender C., Vandecasteele M., Ommeslag S., De Zutter N., Vandenbussche E., Haegeman A., et al. (2023). Linnemannia elongata: a key species in chitin-based plant growth promotion. The Phytobiome Journal. doi: 10.1094/PBIOMES-05-23-0031-R
Diara C., Incrocci L., Pardossi A., Minuto A. (2012). Reusing greenhouse growing media. Acta Hortic. 927, 793–800. doi: 10.17660/ActaHortic.2012.927.98
Dixon P. (2003). VEGAN, a package of R functions for community ecology. J. Vege. Sci. 14, 927–930. doi: 10.1111/j.1654-1103.2003.tb02228.x
Edgar R. C. (2013). UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 10, 996–998. doi: 10.1038/nmeth.2604
Edgar R. C., Flyvbjerg H. (2015). Error filtering, pair assembly and error correction for next-generation sequencing reads. Bioinformatics 31, 3476–3482. doi: 10.1093/bioinformatics/btv401
Elphinstone J. G., Thwaites R., Stalham M., Wale S. (2009). Integration of precision irrigation and non-water based measures to suppress common scab of potato. Potato Council. Available from https://chrome-extension://efaidnbmnnnibpcajpcglclefindmkaj/https://projectbluearchive.blob.core.windows.net/media/Default/Research%20Papers/Potatoes/20099%20Final%20Report%20R272.pdf
Fang X., Phillips D., Li H., Sivasithamparam K., Barbetti M. J. (2011a). Comparisons of virulence of pathogens associated with crown and root diseases of strawberry in Western Australia with special reference to the effect of temperature. Scientia Hortic. 131, 39–48. doi: 10.1016/j.scienta.2011.09.025
Fang X. L., Phillips D., Li H., Sivasithamparam K., Barbetti M. J. (2011b). Severity of crown and root diseases of strawberry and associated fungal and oomycete pathogens in Western Australia. Australas. Plant Pathol. 40, 109–119. doi: 10.1007/s13313-010-0019-5
Farh M. E.-A., Kim Y.-J., Kim Y.-J., Yang D.-C. (2018). Cylindrocarpon destructans/Ilyonectria radicicola-species complex: Causative agent of ginseng root-rot disease and rusty symptoms. J. Ginseng Res. 42, 9–15. doi: 10.1016/j.jgr.2017.01.004
Fennimore S. A., Serohijos R., Samtani J. B., Ajwa H. A., Subbarao K. V., Martin F. N., et al. (2024). TIF film, substrates and nonfumigant soil disinfestation maintain fruit yields. California Agric. 67, 139–146. doi: 10.3733/ca.v067n03p139
Gardes M., Bruns T. D. (1993). ITS primers with enhanced specificity for basidiomycetes - application to the identification of mycorrhizae and rusts. Mol. Ecol. 2, 113–118. doi: 10.1111/j.1365-294X.1993.tb00005.x
Halleen F., Schroers H.-J., Groenewald J. Z., Rego C., Oliveira H., Crous P. W. (2006). Neonectria liriodendri sp. nov., the main causal agent of black foot disease of grapevines. Stud. Mycol. 55, 227–234. doi: 10.3114/sim.55.1.227
Hernández-Apaolaza L., Gascó A., Gascó J., Guerrero F. (2005). Reuse of waste materials as growing media for ornamental plants. Biores. Technol. 96, 125–131. doi: 10.1016/j.biortech.2004.02.028
Huang K., Jiang Q., Liu L., Zhang S., Liu C., Chen H., et al. (2020). Exploring the key microbial changes in the rhizosphere that affect the occurrence of tobacco root-knot nematodes. AMB Express 10, 72. doi: 10.1186/s13568-020-01006-6
Jay C. N., Fitzgerald J. D., Hipps N. A., Atkinson C. J. (2015). Why short-term biochar application has no yield benefits: evidence from three field-grown crops. Soil Use Manage. 31, 241–250. doi: 10.1111/sum.12181
Koike S. T., Kirkpatrick S. C., Gordon T. R. (2009). Fusarium wilt of strawberry caused by Fusarium oxysporum in California. Plant Dis. 93, 1077–1077. doi: 10.1094/PDIS-93-10-1077A
Köljalg U., Nilsson R. H., Abarenkov K., Tedersoo L., Taylor A. F. S., Bahram M., et al. (2013). Towards a unified paradigm for sequence-based identification of fungi. Mol. Ecol. 22, 5271–5277. doi: 10.1111/mec.12481
Law J. W.-F., Ser H.-L., Khan T. M., Chuah L.-H., Pusparajah P., Chan K.-G., et al. (2017). The potential of streptomyces as biocontrol agents against the rice blast fungus, Magnaporthe oryzae (Pyricularia oryzae). Front. Microbiol. 8. doi: 10.3389/fmicb.2017.00003
Lehmann J., Rondón M. A. (2006). Bio-char soil management on highly weathered soils in the humid tropics. in “Biological Approaches to Sustainable Soil Systems” ed. Uphoff N.. (Boca Raton: CRC Press), 517–530. doi: 10.1201/9781420017113.ch36
Li W., Hou Y., Long M., Wen X., Han J., Liao Y. (2023). Long-term effects of biochar application on rhizobacteria community and winter wheat growth on the Loess Plateau in China. Geoderma 429, 116250. doi: 10.1016/j.geoderma.2022.116250
Liu Z., Liang T., Kang C. (2023). Molecular bases of strawberry fruit quality traits: Advances, challenges, and opportunities. Plant Physiol. 193, 900–914. doi: 10.1093/plphys/kiad376
Lombardi N., Caira S., Troise A. D., Scaloni A., Vitaglione P., Vinale F., et al. (2020). Trichoderma applications on strawberry plants modulate the physiological processes positively pffecting fruit production and quality. Front. Microbiol. 11. doi: 10.3389/fmicb.2020.01364
Love M. I., Huber W., Anders S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550–550. doi: 10.1186/s13059-014-0550-8
Mahnkopp-Dirks F., Radl V., Kublik S., Gschwendtner S., Schloter M., Winkelmann T. (2021). Molecular barcoding reveals the genus Streptomyces as associated root endophytes of apple (Malus domestica) plants grown in soils affected by apple replant disease. Phytobiomes J. 5, 177–189. doi: 10.1094/PBIOMES-07-20-0053-R/ASSET/IMAGES/LARGE/PBIOMES-07-20-0053-RF3.JPEG
Maldonado-González M. M., del Pilar Martínez-Diz M., Andrés-Sodupe M., Bujanda R., Díaz-Losada E., Gramaje D. (2020). Quantification of Cadophora luteo-olivacea from grapevine nursery stock and vineyard soil using droplet digital PCR. Plant Dis. 104, 2269–2274. doi: 10.1094/PDIS-09-19-2035-RE
Marris E. (2006). Marine natural products: Drugs from the deep. Nature 443, 904–905. doi: 10.1038/443904a
Martin F. N. (2000). Rhizoctonia spp. recovered from strawberry roots in Central Coastal California. Phytopathogy 90, 345–353. doi: 10.1094/PHYTO.2000.90.4.345
Mazzola M., Manici L. M. (2012). Apple replant disease: role of microbial ecology in cause and control. Annu. Rev. Phytopathol. 50, 45–65. doi: 10.1146/annurev-phyto-081211-173005
Meller Harel Y., Elad Y., Rav-David D., Borenstein M., Shulchani R., Lew B., et al. (2012). Biochar mediates systemic response of strawberry to foliar fungal pathogens. Plant Soil 357, 245–257. doi: 10.1007/s11104-012-1129-3
Muyzer G., de Waal E. C., Uitterlinden A. G. (1993). Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59, 695–700. doi: 10.1128/aem.59.3.695-700.1993
Nelson N. O., Agudelo S. C., Yuan W., Gan J. (2011). Nitrogen and phosphorus availability in biochar-amended soils. Soil Sci. 176, 218–226. doi: 10.1097/SS.0b013e3182171eac
Nishizawa T., Quan A., Kai A., Tago K., Ishii S., Shen W., et al. (2014). Inoculation with N2-generating denitrifier strains mitigates N2O emission from agricultural soil fertilized with poultry manure. Biol. Fertil. Soils 50, 1001–1007. doi: 10.1007/s00374-014-0918-7
Norah M., Shumirai Z., Zelma M. L., Upenyu M. (2015). Impacts of untreated sewage discharge on water quality of middle Manyame River: A case of Chinhoyi town, Zimbabwe. Int. J. Environ. Monit. Anal. 3, 133–138. doi: 10.11648/j.ijema.20150303.14
Papp-Rupar M., Karlstrom A., Passey T., Deakin G., Xu X.-M. (2022). The influence of host genotypes on the endophytes in the leaf scar tissues of apple trees and correlation of the endophytes with apple canker (Neonectria ditissima) development. Phytobiome J. 6, 127–138. doi: 10.1094/PBIOMES-10-21-0061-R
Philip L. (2013). Advances in strawberry substrate culture during the last twenty years in the Netherlands and Belgium. Int. J. Fruit Sci. 13, 84–90. doi: 10.1080/15538362.2012.697024
Prendergast-Miller M. T., Duvall M., Sohi S. P. (2014). Biochar–root interactions are mediated by biochar nutrient content and impacts on soil nutrient availability. Eur. J. Soil Sci. 65, 173–185. doi: 10.1111/ejss.12079
Reis P., Cabral A., Nascimento T., Oliveira H., Rego C. (2013). Diversity of Ilyonectria species in a young vineyard affected by black foot disease. Phytopathol. Mediterr. 52, 335–346. doi: 10.14601/Phytopathol_Mediterr-12719
Ren H., Guo H., Shafiqul Islam M., Zaki H. E. M., Wang Z., Wang H., et al. (2023). Improvement effect of biochar on soil microbial community structure and metabolites of decline disease bayberry. Front. Microbiol. 14. doi: 10.3389/fmicb.2023.1154886
Reynolds H. L., Packer A., Bever J. D., Clay K. (2003). Grassroots ecology: Plant-microbe-soil interactions as drivers of plant community structure and dynamics. Ecology 84, 2281–2291. doi: 10.1890/02-0298
Robinson-Boyer L., Feng W., Gulbis N., Hajdu K., Harrison R. J., Jeffries P., et al. (2016). The use of arbuscular mycorrhizal fungi to improve strawberry production in coir substrate. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.01237
Rutigliano F. A., Romano M., Marzaioli R., Baglivo I., Baronti S., Miglietta F., et al. (2014). Effect of biochar addition on soil microbial community in a wheat crop. Eur. J. Soil Biol. 60, 9–15. doi: 10.1016/j.ejsobi.2013.10.007
Scholte K. (1989). The effect of netted scab (Streptomyces spp.) and Verticillium dahliae on growth and yield of potato. Potato Res. 32, 65–73. doi: 10.1007/BF02365818
Sheng M., Gorzsás A., Tuck S. (2016). Fourier transform infrared microspectroscopy for the analysis of the biochemical composition of C. elegans worms. Worm 5, e1132978. doi: 10.1080/21624054.2015.1132978
Shuttleworth L., Papp-Rupar M., Passey T., Xu X.-M. (2021). Extending the lifetime of coconut coir media in strawberry production through reuse and amendment with biochar. Acta Hortic. 1317, 397–402. doi: 10.17660/ActaHortic.2021.1317.46
Sugar D. (1993). The importance of wounds in infection of pear fruit by Phialophora malorum and the role of hydrstatic pressure in spore penetration of wounds. Phytopathology 83, 1083–1086. doi: 10.1094/Phyto-83-1083
Sugar D., Spotts R. A. (1992). Sources of inoculum of Phialophora malorum, causal agent of side rot of pear. Phytopathology 82, 735–738. doi: 10.1094/Phyto-82-735
Tamura T., Suzuki K.-I. (2014). “The Suborder Kineosporiineae,” in The Prokaryotes: Actinobacteria. Eds. Rosenberg E., DeLong E. F., Lory S., Stackebrandt E., Thompson F. (Springer Berlin Heidelberg, Berlin, Heidelberg), 443–453.
Team R. C. D. (2019). R: A Language and environment for statistical computing (Vienna, Austria: R Foundation for Statistical Computing).
Ulrich D., Kecke S., Olbricht K. (2018). What do we know about the chemistry of strawberry aroma? J. Agric. Food Chem. 66, 3291–3301. doi: 10.1021/acs.jafc.8b01115
Vandecasteele B., Similon L., Moelants J., Hofkens M., Visser R., Melis P. (2023). End-of-life stage of renewable growing media with biochar versus spent peat or mineral wool. Nutrient Cycling Agroecosys. doi: 10.1007/s10705-023-10315-8
Vandepol N., Liber J., Yocca A., Matlock J., Edger P., Bonito G. (2022). Linnemannia elongata (Mortierellaceae) stimulates Arabidopsis thaliana aerial growth and responses to auxin, ethylene, and reactive oxygen species. PloS One 17, e0261908. doi: 10.1371/journal.pone.0261908
Vidhana Arachchi L., Somasiri L. (1997). Use of coir dust on the productivity of coconut on sandy soils. Cocas: J. Coconut Res. Instit. Sri Lanka 12, 54–71. doi: 10.4038/cocos.v12i0.2166
Wang J., Wang C., Li J., Bai P., Li Q., Shen M., et al. (2018). Comparative genomics of degradative Novosphingobium strains with special reference to microcystin-degrading Novosphingobium sp. THN1. Front. Microbiol., 9, 2238. doi: 10.3389/fmicb.2018.02238
Wang Y., Zhang Y., Li Z.-Z., Zhao Q., Huang X.-Y., Huang K.-F. (2020). Effect of continuous cropping on the rhizosphere soil and growth of common buckwheat. Plant Product. Sci. 23, 81–90. doi: 10.1080/1343943X.2019.1685895
White T. J., Bruns T. L., Taylor J. W. (1990). “Amplification and direct sequencing of fungal ribosomal RNA genes for phylogentics,” in A Guide to Molecular Methods and Applications. Eds. Innis M. A., Gelfand D. H., Snisky J. J., White T. J. (Academic Press, New York), 315–322.
Wing K. B., Pritts M. P., Wilcox W. F. (1994). Strawberry black root rot: a review. Adv. Strawberry Res. 13, 13–19.
Woznicki T., Kusnierek K., Vandecasteele B., Sønsteby A. (2024). Reuse of coir, peat, and wood fiber in strawberry production. Front. Plant Sci. 14. doi: 10.3389/fpls.2023.1307240
Keywords: coir reuse, strawberry yield decline, Ilyonectria destructans, Cylindrocarpon destructans, biochar amendment
Citation: Xu X, Deakin G, Zhao J, Passey T and Papp-Rupar M (2024) Amplicon-based metagenomics to study the effect of coir age and wood biochar on microbiome in relation to strawberry yield. Front. Agron. 6:1397974. doi: 10.3389/fagro.2024.1397974
Received: 11 March 2024; Accepted: 11 April 2024;
Published: 10 May 2024.
Edited by:
Mohammad Bagher Hassanpouraghdam, University of Maragheh, IranReviewed by:
Federica Caradonia, University of Modena and Reggio Emilia, ItalyAdnan Akhter, University of the Punjab, Pakistan
Copyright © 2024 Xu, Deakin, Zhao, Passey and Papp-Rupar. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiangming Xu, eGlhbmdtaW5nLnh1QG5pYWIuY29t
†Present address: Jingchen Zhao, College of Plant Protection, Northwest A&F University, Yangling, Shaanxi, China
 Xiangming Xu
Xiangming Xu Greg Deakin
Greg Deakin Jingchen Zhao
Jingchen Zhao Tom Passey
Tom Passey Matevz Papp-Rupar
Matevz Papp-Rupar