- 1Gulf Coast Research and Education Center, University of Florida, Wimauma, FL, United States
- 2Everglades Research and Education Center, University of Florida, Belle Glade, FL, United States
Corynespora cassiicola (Cc) is a genetically diverse ascomycete found worldwide in tropical and subtropical regions. Cc causes necrotrophic diseases in several plant species, including important crops such as rubber tree, tomato, cotton, and cucumber. Evidence suggests the involvement of one or more Cc host-specific toxins in disease progression. Management of Cc crop diseases is based mainly on pesticide sprays. However, the pathogen’s development of resistance to commonly used fungicides is documented. Resistance breeding is an attractive alternative or supplement to chemical control of Cc crop diseases, but research on this topic is very limited. This review describes the current plant resistance breeding efforts towards Cc resistance and discusses the potential influence of host-specific toxins (HSTs) on such efforts. Although some reports of host resistance exist in a few crops, resistant germplasm and knowledge about resistance mechanisms are limited. Some studies have suggested the involvement of HSTs in disease development, including the upregulation of resistance-related proteins in susceptible reactions and the recessive nature of resistance. In light of this, host resistance may not be achieved through commonly used dominant R-genes.
Introduction
The warm and humid climate of tropical and subtropical regions favors year-round production of many annual crops but also favors the development of numerous plant diseases. Among these are diseases caused by Corynespora cassiicola (Berk. & Curt.) Wei. (Dixon et al., 2009; Farr and Rossman, 2017), an imperfect ascomycete fungus (Schlub et al., 2009) with high intraspecific morphological and genetic diversity (Smith, 2008; Sumabat et al., 2018a) and a very wide trophic habit that includes necrotroph, saprophyte and endophyte (Dixon et al., 2009; Schlub et al., 2009). Corynespora cassiicola infects more than 530 monocot and dicot plant species, including many important crops (Smith, 2008; Chairin et al., 2017; Sumabat et al., 2018b; Qiu et al., 2020). Figure 1A shows a simplified disease cycle from tomato. Beyond plants, the fungus has been isolated from nematode cysts and can cause opportunistic infections in immunocompromised human patients (Carris et al., 1986; Huang et al., 2010; Yan et al., 2016; Arango-Franco et al., 2018). The incidence and severity of C. cassiicola crop diseases have rapidly increased over the last two decades (Pernezny et al., 2005; Schlub et al., 2009; Sumabat et al., 2018a) causing severe yield losses in tomato (Pernezny et al., 1996; Schlub et al., 2009), rubber tree (Silva et al., 2003; Barthe et al., 2007; Qi et al., 2007; Liu et al., 2014), cotton, soybean (Fulmer et al., 2012; Hartman et al., 2016; Sumabat et al., 2018a), and cucumber (Miyamoto et al., 2009).
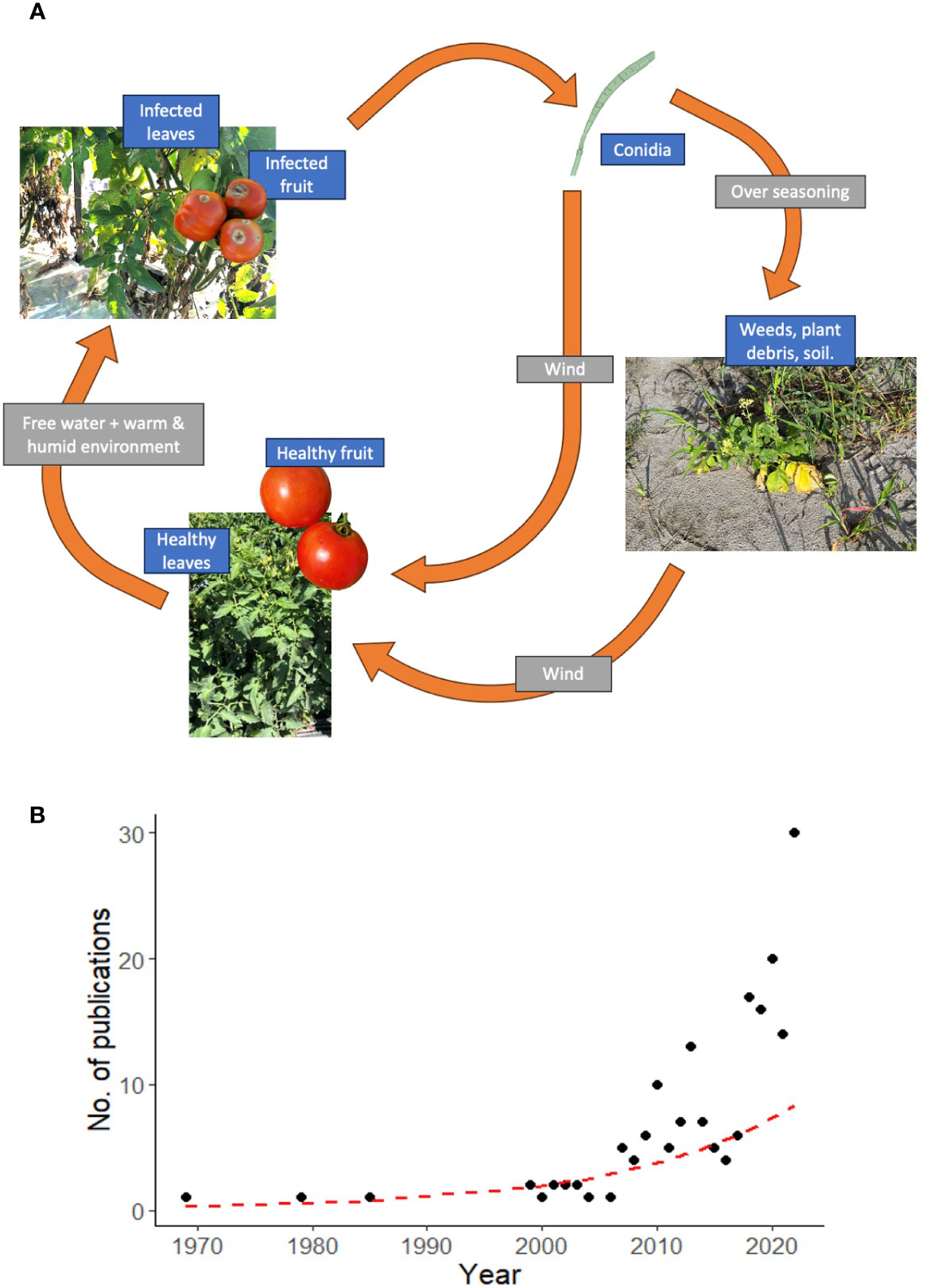
Figure 1 (A) Disease cycle of C. cassiicola in tomato, and (B) Number of scientific publications related to C. cassiicola in the PubMed database from 1969 to 2022.
Control of diseases caused by C. cassiicola can be difficult, especially when the environment is conducive to disease development. Over the years, chemical control has been the primary means of disease management (Blazquez, 1967; Pernezny et al., 2002). However, studies from diverse production regions and several cropping systems have documented increasing issues with fungicide resistance. Resistance to quinone outside inhibitors (QoI), benzimidazoles, succinate dehydrogenase inhibitors (SDHI), and other commonly used fungicides has been detected among C. cassiicola isolates from tomato in Florida (Mackenzie and Vallad, 2017; MacKenzie et al., 2020), from cucumber and tomato in China (Zhu et al., 2018; Duan et al., 2019; Sun et al., 2022), and from cucumber in Japan (Miyamoto et al., 2009). Rondon and Lawrence (2021) cited many other reports of the development of fungicide resistance by C. cassiicola in different crops and regions in addition to those mentioned above. Therefore, current chemical control practices, alone, do not represent a sustainable disease management strategy for C. cassiicola diseases. A search of the PubMed database showed an exponential increase in scientific publications related to C. cassiicola during the last few decades with more than two-thirds of those related to some type of characterization of the fungus or plant diseases caused by it, disease management, or first reports; but only 11 (6%) of the 183 publications between 1969 and 2022 were related to plant resistance (Figure 1B).
Although there has been an increase in disease issues caused by C. cassiicola in the last two decades, few resistance breeding efforts have been reported. The main goal of this review is to address the status of resistance breeding efforts against C. cassiicola, and the potential role of host-specific toxins in such efforts. Reviews discussing the morphology, host range, and diversity of C. cassiicola, as well as the etiology, epidemiology, and management of associated diseases are available (Pernezny and Simone, 1999; MacKenzie et al., 2018; Rondon and Lawrence, 2021; Edwards Molina et al., 2022).
Host-specific toxins and resistance to necrotrophs
The plant immune system consists of two layers: the pathogen-associated molecular patterns (PAMP)-triggered immunity (PTI), and the effector-triggered immunity (ETI) defense systems (Chisholm et al., 2006; Jones and Dangl, 2006; Wang et al., 2014). PTI results in a weak broad-spectrum defense response (Wang et al., 2014). To interfere with PTI, pathogens evolved effectors, which are proteins secreted into the plant cell that target specific sites involved in resistance pathways (Chisholm et al., 2006; Jones and Dangl, 2006). In ETI, pathogen effectors are detected by protein receptors encoded by plant resistance genes (R-genes), triggering a rapid and strong resistance reaction (Chisholm et al., 2006; Jones and Dangl, 2006; Wang et al., 2014). Resistance through either pathway typically behaves as a dominant genetic trait (Joosten and De Wit, 1999; Gomez-Gomez and Boller, 2000 ). ETI often leads to the hypersensitive response (HR), involving the rapid production of reactive oxygen species and programmed cell death at the site of pathogen ingression (Glazebrook, 2005; Wang et al., 2014; Balint-Kurti, 2019).
Resistance to some necrotrophs appears to involve the PTI system, which is triggered when plant pathogen recognition receptors (PRRs) detect PAMPs such as chitin and endopolygalacturonases (Wang et al., 2014). Although ETI is generally effective against biotrophic and hemibiotrophic pathogens which require living cells for successful infection, it is rarely effective against necrotrophic pathogens which actually benefit from the HR (Glazebrook, 2005; Wang et al., 2014). In fact, R-gene-mediated susceptibility to necrotrophs has been demonstrated (Lorang et al., 2007; Faris et al., 2010).
Many necrotrophic pathogens produce host specific toxins (HST) that manipulate the plant ETI system to induce the HR. This is referred to as effector triggered susceptibility (ETS), R-mediated susceptibility, or the inverse gene-for-gene model (Glazebrook, 2005; Lorang et al., 2007; Faris et al., 2010; Oliver et al., 2011; Wang et al., 2014). Resistance to necrotrophic pathogens is thus more commonly achieved in a recessive manner. ETS has been well-studied in a few host-pathogen systems. Southern corn leaf blight, caused by Cochliobolus heterostrophus (or Bipolaris maydis) race T, was a major epidemic in the US in the 1970’s, affecting Texas male-sterile cytoplasm corn (Ullstrup, 1972; Inderbitzin et al., 2010). In this system, the HST T-toxin targets the product of the mitochondrial gene T-urf13, leading to disease (Levings Iii et al., 1995). Wheat-Stagonospora nodorum (causing Stagonospora wheat leaf and glume blotch) and Pyrenophora tritici-repentis (causing tan spot of wheat) are also model systems for ETS (Oliver et al., 2011; Liu et al., 2017). Here, a few specific interactions between HSTs and host susceptibility factors dictate a disease reaction. For instance, the combination of the pathogen HST ToxA and the wheat dominant Tsn1 gene results in susceptibility to tan spot (Oliver et al., 2011). Cochliobolus victoriae (victoria blight of oats) produces the HST victorin which targets the oats R gene Pc2, thereby triggering disease (Navarre and Wolpert, 1999). In each of the host-pathogen systems mentioned above, the absence (or mutation) of the host protein targeted by the pathogen HST led to plant resistance which is a recessive genetic state.
Cassiicolin
Cassiicolin is a necrotrophic toxin produced and diffused by C. cassiicola during infection (Onesirosan et al., 1975; Breton et al., 2000; Barthe et al., 2007; de Lamotte et al., 2007; Déon et al., 2012; Lopez et al., 2018; Ribeiro et al., 2019). Although its exact mode of action remains unknown, a study using a rubber tree isolate suggests that cassiicolin behaves as an HST (Barthe et al., 2007; Déon et al., 2012). To date, eight cassiicolin isoforms have been identified (Cas1 to Cas7, and Cas2.2) (Déon et al., 2014; Lopez et al., 2018; Wu et al., 2018); although, pathogenic isolates with no cassiicolin gene (Cas0), such as tomato isolates, have also been identified (Déon et al., 2012; Déon et al., 2014; Dacones et al., 2022).
Other pathogenicity factors produced by C. cassiicola
The C. cassiicola genome encodes for 2870 putative effectors including CAZymes, lipases, peptidases, secreted proteins, and enzymes associated with secondary metabolism (Lopez et al., 2018). Among those, 92 genes (including 45 secreted effectors) were differentially expressed in susceptible and resistant rubber tree clones, supporting their involvement in disease progression (Lopez et al., 2018). The composition and number of encoded effectors agree with the wide host range and multi-trophic habit (Lopez et al., 2018). Cell wall-degrading enzymes including polygalacturonase, cellulase, neutral xylanase and laccase also play a role in pathogenicity in C. cassiicola (Jia et al., 2021).
Although C. cassiicola isolates infecting tomato in the southeastern US do not code for a cassiicolin isoform, they likely employ other effectors. Interestingly, among other putative necrotrophic effectors, a complete set of the genes necessary to encode a T-toxin-like compound was detected in isolates collected from tomato in the southeastern US (Dacones et al., 2022).
Resistance breeding efforts in crops
Table 1 presents a summary of the current status of breeding efforts in different crops. Although C. cassiicola plant diseases have been reported since the 1960s (Ramakrishnan and Pillay, 1961; Blazquez, 1967), breeding for resistance to C. cassiicola diseases has not been a primary focus (Pernezny and Simone, 1999; Edwards Molina et al., 2022), and disease management has relied on the use of fungicide sprays (Chee, 1990; MacKenzie et al., 2018; de Castro Costa et al., 2023). Disease resistance has been identified in tomato, rubber tree, soybean and cucumber (Bliss et al., 1973; Narayanan and Mydin, 2012; Wen et al., 2015; de Castro Costa et al., 2023). Bliss et al. (1973) reported on resistance in tomato conferred by a single recessive gene in inbred line (PI 120265) derived from S. pimpinellifolium accession PI 112215. New data support the recessive nature of resistance in tomato to C. cassiicola (Sierra-Orozco et al., 2022), which is in line with the ETS resistance model. Unfortunately, PI 120265 demonstrated poor resistance in subsequent disease screenings conducted in 2010-2011 using recently collected C. cassiicola isolates from Florida (G. Vallad, personal communication).
In rubber tree, studies have identified resistant clones but with inconsistent results across studies or even between different experiment settings within the same experiment (i.e., greenhouse and field) (Chee, 1988; Narayanan and Mydin, 2012). Some clones that were previously found resistant were later identified as susceptible; additionally, two pathogenic races were reported (Narayanan and Mydin, 2012). The fact that in both tomato and rubber tree, there have been reports of germplasm which demonstrated resistance in earlier screens but susceptibility in later screens, suggests that the pathogen is evolving new mechanisms of infection or a shift in pathogen population with more aggressive isolates becoming more prevalent.
Resistant soybean cultivars released in the 1950s were associated with negative agronomic traits, and only susceptible soybean cultivars were thereafter used (Edwards Molina et al., 2022). More recently, some soybean varieties have shown resistance or moderate resistance in greenhouse (Patel et al., 2022) or field settings (de Castro Costa et al., 2023). Genetic analysis based on greenhouse inoculations suggested that soybean resistance is quantitative, involving both additive and epistatic effects, and is largely influenced by the environment (Soares and Arias, 2020). In cucumber, earlier work suggested that resistance was conferred by a single dominant gene (Abul-Hayja et al., 1978), but a later study suggests a single recessive gene (Wen et al., 2015). Three different studies have indicated the location of a resistance QTL in chromosome 6, but it is unclear if these have mapped the same or different QTLs (Wen et al., 2015).
The molecular mechanisms of resistance have not been well investigated. Cassiicolin is a primary determinant of pathogenicity and resistance in rubber tree infections (Breton et al., 2000). Pure toxin inoculation of detached leaves demonstrated that resistance in rubber tree appears to be dependent on the concentration of cassiicolin secreted into the plant tissues, with higher cassiicolin concentration producing symptoms in resistant rubber tree clones (Breton et al., 2000). Transcriptome profiling suggested that genes involved in defense and stress response, including disease resistance and leucine-rich repeat genes, were significantly upregulated in the resistant clone GT 1 compared to the susceptible RRII 105 in response to C. cassiicola inoculation (Roy et al., 2019). Fortunato et al. (2017) described a link between disease resistance in soybean and the phenylpropanoid pathway. Histopathology analysis in soybean showed accumulation of phenolic-like compounds in infection sites in resistant cultivars contributing to fungal cell death and conservation of plant cell integrity (Fortunato et al., 2017).
The molecular basis of resistance in cucumber has also been investigated in three separate works. In the first, the ERF transcription factor CsERF004 was involved in resistance to C. cassiicola infection via salicylic acid and ethylene pathways (Liu et al., 2017). In the second, resistance was linked to genes and miRNAs involved in secondary metabolism, especially with the phenylpropanoid pathway (Wang et al., 2018) which agrees with the findings from Fortunato et al. (2017). The third points to a NB-LRR (nucleotide binding site- leucine rich repeat) domain gene on chromosome 6 as a potential candidate for resistance (Wen et al., 2015). Together, these studies point to the involvement of cassiicolin, the phenylpropanoid pathway, resistance genes, and gene transcript reprogramming in the host’s response to infection.
Possible role of HSTs and considerations in resistance breeding efforts
Corynespora cassiicola belongs to the order Pleosporales, which includes many necrotrophic pathogens known (or believed) to produce HSTs (Friesen et al., 2008). As described above, C. cassiicola produces cassiicolin with is an HST. NB-LRR genes are typically involved in plant disease resistance (Chisholm et al., 2006). Interestingly, a susceptible reaction in cucumber was associated with an increased expression of a candidate gene containing a NC-LRR domain (Wen et al., 2015). A recessive gene action of resistance was found which is in line with an ETS model involving HSTs (Wen et al., 2015). Thus evidence suggests that one or more HSTs are likely involved in C. cassiicola pathogenicity. One or more HSTs from C. cassiicola (including cassiicolin) interact with still-unknown host susceptibility factors, thereby triggering susceptibility in a dominant fashion. Because of this, resistance will most likely be achieved through recessive gene action.
The necrotrophic behavior of C. cassiicola, including the involvement of HSTs and thus the potentially recessive action of resistance, presents challenges in finding durable and simple resistance to diseases caused by this pathogen. Additionally, resistance against some necrotrophic pathogens is quantitative (Chaerani et al., 2007; Pogoda et al., 2021), including resistance against C. cassiicola in soybean (Soares and Arias, 2020). Whereas breeding with one or a few recessive genes is feasible, quantitative resistance would be more challenging to adopt using traditional marker assisted selection – especially in vegetable crops which often rely heavily on multiple R-genes for resistance to various pathogens. Instead, genomic selection of resistance may be a better alternative, provided that resistance is not linked to negative marketability traits.
The host-C. cassiicola interaction likely involves toxins. Breton et al. (2000) suggested that host resistance may be based on insensitivity to the toxin driven by a detoxification process. Alternatively, the susceptibility factor targeted by the toxin may be missing in resistant germplasm.
Conclusion
Corynespora cassiicola has become an increasingly relevant pathogen in the last two decades. Formerly, disease management through fungicide sprays was effective, and resistance breeding was not a central focus. However, recent worldwide reports on fungicide resistance in several crops have warned of the ineffectiveness of commonly used fungicides for management of C. cassicola. Additionally, society pressures to move away from reliance on pesticides in crop disease management. Despite this, breeding for resistance against C. cassiicola diseases has been scarce, and active breeding efforts are only reported in rubber tree, soybean and cucumber. Increasing research on resistance breeding is expected to help substantially in the management of diseases caused by this pathogen.
Evidence points the involvement of HSTs from C. cassiicola in disease progression. Because HSTs target susceptibility factors thereby resulting in ETS, and given the necrotrophic nature of C. cassiicola diseases, it is likely that resistance will not be achievable through a typical dominant R-gene. Rather, evidence suggests, that resistance is more likely be achieved in a recessive manner, as is typical for diseases involving HSTs.
Studies focused on finding durable and reliable resistance against C. cassiicola may consider investigating ETS models by identifying host targets of the fungal HSTs. Once identified, those targets may be modified using gene editing tools to develop resistant cultivars. Resistance may also be achieved through traditional breeding techniques by introducing natural mutations in the ETS pathway. In the long term, the adoption of C. cassiicola-resistant crops together with integrated disease management programs, should result in improved crop productivity and quality, and a reduction of pesticide usage.
Author contributions
ES-O: Conceptualization, Investigation, Writing – original draft, Writing – review & editing. GS: Writing – review & editing. SL: Writing – review & editing. GV: Writing – review & editing. SH: Conceptualization, Funding acquisition, Supervision, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported in part by Hatch project FLA-GCR-006019.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abul-Hayja Z., Williams P., Peterson C. (1978). Inheritance of resistance to anthracnose and target leaf spot in cucumbers. Plant Dis. Rep. 62, 43–45.
Arango-Franco C., Moncada-Velez M., Beltran C., Berrio I., Mogollon C., Restrepo A., et al. (2018). Early-onset invasive infection due to Corynespora cassiicola associated with compound heterozygous CARD9 mutations in a Colombian patient. J. Clin. Immunol. 38, 794–803. doi: 10.1007/s10875-018-0549-0
Balint-Kurti P. (2019). The plant hypersensitive response: concepts, control and consequences. Mol. Plant Pathol. 20, 1163–1178. doi: 10.1111/mpp.12821
Barthe P., Pujade-Renaud V., Breton F., Gargani D., Thai R., Roumestand C., et al. (2007). Structural analysis of cassiicolin, a host-selective protein toxin from Corynespora cassiicola. J. Mol. Biol. 367, 89–101. doi: 10.1016/j.jmb.2006.11.086
Blazquez C. H. (1967). Corynespora leaf spot of cucumber. In Proceedings of the Florida State Horticultural Society 80, 177–182.
Bliss F. A., Onesirosan P. T., Arni D. C. (1973). Inheritance of resistance in tomato to target leaf spot. Phytopathology 63, 837–840. doi: 10.1094/Phyto-63-837
Breton F., Sanier C., D’auzac J. (2000). Role of cassiicolin, a host-selective toxin, in pathogenicity of Corynespora cassiicola, causal agent of a leaf fall disease of Hevea. J. Rubber Res. 3, 115–128.
Carris L. M., Glawe D. A., Gray L. E. (1986). Isolation of the soybean pathogens Corynespora cassiicola and Phialophora gregata from cyst of Heterodera glycines in Illinois. Mycologia 78, 503–506. doi: 10.1080/00275514.1986.12025280
Chaerani R., Smulders M. J. M., Van Der Linden C. G., Vosman B., Stam P., Voorrips R. E. (2007). QTL identification for early blight resistance (Alternaria solani) in a Solanum lycopersicum × S. arcanum cross. Theor. Appl. Genet. 114, 439–450. doi: 10.1007/s00122-006-0442-8
Chairin T., Pornsuriya C., Thaochan N., Sunpapao A. (2017). Corynespora cassiicola causes leaf spot disease on lettuce (Lactuca sativa) cultivated in hydroponic systems in Thailand. Australas. Plant Dis. Notes 12, 16. doi: 10.1007/s13314-017-0241-x
Chee K. H. (1988). Studies on sporulation, pathogenicity and epidemiology of Corynespora cassiicola on hevea ruber. J. Nat. Rubb. Res. 3, 21–29.
Chee K. H. (1990). Present status of rubber diseases and their control. Rev. Plant Pathol. 69, 423–430.
Chisholm S. T., Coaker G., Day B., Staskawicz B. J. (2006). Host-microbe interactions: shaping the evolution of the plant immune response. Cell 124, 803–814. doi: 10.1016/j.cell.2006.02.008
Dacones L. S., Kemerait R. C., Brewer M. T. (2022). Comparative genomics of host-specialized populations of Corynespora cassiicola causing target spot epidemics in the southeastern United States. Front. Fungal Biol. 3. doi: 10.3389/ffunb.2022.910232
de Castro Costa É., Dias-Neto J. J., Bizerra L. V. A. S., Tormen N., Paul P. A., Café-Filho A. C. (2023). Varietal response to soybean target spot (Corynespora cassiicola) in the Brazilian Neotropical Savannah. Phytoparasitica. doi: 10.1007/s12600-023-01086-7
de Lamotte F., Duviau M., Sanier C., Thai R., Poncet J., Bieysse D., et al. (2007). Purification and characterization of cassiicolin, the toxin produced by Corynespora cassiicola, causal agent of the leaf fall disease of rubber tree. J. Chromatogr. B-Analytical Technol. Biomed. Life Sci. 849, 357–362. doi: 10.1016/j.jchromb.2006.10.051
Déon M., Bourre J., Gimenez S., Berger A., Bieysse D., De Lamotte F., et al. (2012). Characterization of a cassiicolin-encoding gene from Corynespora cassiicola, pathogen of rubber tree (Hevea brasiliensis). Plant Sci. 185, 227–237. doi: 10.1016/j.plantsci.2011.10.017
Déon M., Fumanal B., Gimenez S., Bieysse D., Oliveira R. R., Shuib S. S., et al. (2014). Diversity of the cassiicolin gene in Corynespora cassiicola and relation with the pathogenicity in Hevea brasiliensis. Fungal Biol. 118, 32–47. doi: 10.1016/j.funbio.2013.10.011
Dixon L., Schlub R., Pernezny K., Datnoff L. (2009). Host specialization and phylogenetic diversity of Corynespora cassiicola. Phytopathology 99, 1015–1027. doi: 10.1094/PHYTO-99-9-1015
Duan Y., Xin W., Lu F., Li T., Li M., Wu J., et al. (2019). Benzimidazole- and QoI-resistance in Corynespora cassiicola populations from greenhouse-cultivated cucumber: An emerging problem in China. Pesticide Biochem. Physiol. 153, 95–105. doi: 10.1016/j.pestbp.2018.11.006
Edwards Molina J. P., Navarro B. L., Allen T. W., Godoy C. V. (2022). Soybean target spot caused by Corynespora cassiicola: a resurgent disease in the Americas. Trop. Plant Pathol. 47, 315–331. doi: 10.1007/s40858-022-00495-z
Faris J. D., Zhang Z., Lu H., Lu S., Reddy L., Cloutier S., et al. (2010). A unique wheat disease resistance-like gene governs effector-triggered susceptibility to necrotrophic pathogens. Proc. Natl. Acad. Sci. 107, 13544. doi: 10.1073/pnas.1004090107
Farr D. F., Rossman A. Y. (2017). Fungal databases, U.S. National fungus collections (United States Department of Agriculture – Agricultural Research Services). Available at: https://nt.ars-grin.gov/fungaldatabases/.
Fortunato A. A., Araujo L., Rodrigues F. Á. (2017). Association of the Production of Phenylpropanoid Compounds at the Infection Sites of Corynespora cassiicola with Soybean Resistance against Target Spot. J. Phytopathol. 165, 131–142. doi: 10.1111/jph.12546
Friesen T. L., Faris J. D., Solomon P. S., Oliver R. P. (2008). Host-specific toxins: effectors of necrotrophic pathogenicity. Cell. Microbiol. 10, 1421–1428. doi: 10.1111/j.1462-5822.2008.01153.x
Fulmer A., Walls J., Dutta B., Parkunan V., Brock J., Kemerait R. (2012). First report of target spot caused by Corynespora cassiicola on cotton in Georgia. Plant Dis. 96, 1066–1066. doi: 10.1094/PDIS-01-12-0035-PDN
Glazebrook J. (2005). Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 43, 205–227. doi: 10.1146/annurev.phyto.43.040204.135923
Gomez-Gomez L., Boller T. (2000). FLS2: An LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol. Cell 5, 1003–1011. doi: 10.1016/S1097-2765(00)80265-8
Hartman G. L., Rupe J. C., Sikora E. J., Domier L. L., Davis J. A., Steffey K. L. eds., (2016). Compendium of soybean diseases and pests. (St. Paul, MN: The American Phytopathological Society), 56–59.
Huang H., Liu C., Liou J., Hsiue H., Hsiao C., Hsueh P. (2010). Subcutaneous infection caused by Corynespora cassiicola, a plant pathogen. J. Infect. 60, 188–190. doi: 10.1016/j.jinf.2009.11.002
Inderbitzin P., Asvarak T., Turgeon B. G. (2010). Six new genes required for production of T-toxin, a polyketide determinant of high virulence of Cochliobolus heterostrophus to maize. Mol. Plant-Microbe Interactions® 23, 458–472. doi: 10.1094/MPMI-23-4-0458
Jia M., Liu X., Zhao H., Ni Y., Liu H., Tian B. (2021). Cell-wall-degrading enzymes produced by sesame leaf spot pathogen Corynespora cassiicola. J. Phytopathol. 169, 186–192. doi: 10.1111/jph.12973
Jones J. D. G., Dangl J. L. (2006). The plant immune system. Nature 444, 323–329. doi: 10.1038/nature05286
Joosten M., De Wit P. (1999). The tomato - Cladosporium fulvum interaction: A versatile experimental system to study plant-pathogen interactions. Annu. Rev. Phytopathol. 37, 335–367. doi: 10.1146/annurev.phyto.37.1.335
Levings Iii C. S., Rhoads D. M., Siedow J. N. (1995). Molecular interactions of Bipolaris maydis T-toxin and maize. Can. J. Bot. 73, 483–489. doi: 10.1139/b95-286
Liu X., Qi Y., Zhang X., Xie Y., Zhang H., Wei Y., et al. (2014). Infection process of Corynespora cassiicola tagged with GFP on Hevea brasiliensis. Australas. Plant Pathol. 43, 523–525. doi: 10.1007/s13313-014-0299-2
Liu D., Xin M., Zhou X., Wang C., Zhang Y., Qin Z. (2017). Expression and functional analysis of the transcription factor-encoding Gene CsERF004 in cucumber during Pseudoperonospora cubensis and Corynespora cassiicola infection. BMC Plant Biol. 17, 96. doi: 10.1186/s12870-017-1049-8
Liu Z., Zurn J. D., Kariyawasam G., Faris J. D., Shi G., Hansen J., et al. (2017). Inverse gene-for-gene interactions contribute additively to tan spot susceptibility in wheat. Theor. Appl. Genet. 130, 1267–1276. doi: 10.1007/s00122-017-2886-4
Lopez D., Ribeiro S., Label P., Fumanal B., Venisse J., Kohler A., et al. (2018). Genome-wide analysis of Corynespora cassiicola leaf fall disease putative effectors. Front. Microbiol. 9. doi: 10.3389/fmicb.2018.00276
Lorang J. M., Sweat T. A., Wolpert T. J. (2007). Plant disease susceptibility conferred by a “resistance” gene. Proc. Natl. Acad. Sci. 104, 14861. doi: 10.1073/pnas.0702572104
MacKenzie K. J., Sumabat L. G., Xavier K. V., Vallad G. E. (2018). A review of Corynespora cassiicola and its increasing relevance to tomato in Florida. Plant Health Prog. 19, 303–309. doi: 10.1094/PHP-05-18-0023-RV
Mackenzie K. J., Vallad G. E. (2017). An evaluation of QoI and SDHI fungicide sensitivity to target spot of tomato in Florida. In: abstracts of presentations at the 2017 southern division meeting. Phytopathology® 107, S3.7–S3.8.
MacKenzie K. J., Xavier K. V., Wen A., Timilsina S., Adkison H. M., Dufault N. S., et al. (2020). Widespread QoI fungicide resistance revealed among Corynespora cassiicola tomato isolates in Florida. Plant Dis. 104, 893–903. doi: 10.1094/PDIS-03-19-0460-RE
Miyamoto T., Ishii H., Seko T., Kobori S., Tomita Y. (2009). Occurrence of Corynespora cassiicola isolates resistant to boscalid on cucumber in Ibaraki Prefecture, Japan. Plant Pathol. 58, 1144–1151. doi: 10.1111/j.1365-3059.2009.02151.x
Narayanan C., Mydin K. (2012). Breeding for disease resistance in Hevea spp. - status, potential threats and possible strategies. Proceedings of the fourth international workshop on the genetics of host-parasite interactions in forestry: Disease and insect resistance in forest trees Eugene, Oregon, July 31- August 5, 2011. Eds. (U.S. Department of Agriculture, Forest Service, Pacific Southwest Research Station), 240–251.
Navarre D. A., Wolpert T. J. (1999). Victorin induction of an apoptotic/senescence-like response in oats. Plant Cell 11, 237–249. doi: 10.1105/tpc.11.2.237
Oliver R. P., Friesen T. L., Faris J. D., Solomon P. S. (2011). Stagonospora nodorum: from pathology to genomics and host resistance. Annu. Rev. Phytopathol. 50, 23–43. doi: 10.1146/annurev-phyto-081211-173019
Onesirosan P., Manubi C. T., Durbin R. D., Morin R. B., Rich D. H., Arni D. C. (1975). Toxin production by Corynespora cassiicola. Physiol. Plant Pathol. 5, 289–295. doi: 10.1016/0048-4059(75)90095-8
Patel S. J., Bowen K. L., Patel J. D., Koebernick J. C. (2022). Evaluating target spot (Corynespora cassiicola) resistance in soybean (Glycine max (L.) Merrill) in a controlled environment. Crop Prot. 159, 106018. doi: 10.1016/j.cropro.2022.106018
Pernezny K., Datnoff L., Mueller T., Collins J. (1996). Losses in fresh-market tomato production in Florida due to target spot and bacterial spot and the benefits of protectant fungicides. Plant Dis. 80, 559–563. doi: 10.1094/PD-80-0559
Pernezny K., Simone G. W. (1999). Target spot of several vegetable crops (Gainesville: University of Florida), 81–88.
Pernezny K., Stoffella P., Collins J., Carroll A., Beaney A. (2002). Control of target spot of tomato with fungicides, systemic acquired resistance activators, and a biocontrol agent. Plant Prot. Sci. 38. doi: 10.17221/4855-PPS
Pernezny K., Stoffella P., Havranek N., Sanchez J., Beany A. (2005). BAM! KICKING CONTROL OF TARGET SPOT UP A NOTCH (Leuven, Belgium: International Society for Horticultural Science (ISHS), 175–180.
Pogoda C. S., Reinert S., Talukder Z. I., Attia Z., Collier-Zans E. C. E., Gulya T. J., et al. (2021). Genetic loci underlying quantitative resistance to necrotrophic pathogens Sclerotinia and Diaporthe (Phomopsis), and correlated resistance to both pathogens. Theor. Appl. Genet. 134, 249–259. doi: 10.1007/s00122-020-03694-x
Qi Y. X., Zhang X., Pu J. J., Xie Y. X., Zhang H. Q., Huang S. L., et al. (2007). Detection of Corynespora cassiicola in Hevea rubber tree from China. Australas. Plant Dis. 2, 153–155. doi: 10.1007/BF03215907
Qiu F., Xu G., Xie C. P., Li X., Zheng F. Q., Miao W. G. (2020). First Report of Corynespora cassiicola Causing Leaf Spot on Avocado (Persea americana) in China. Plant Dis. 104, 1857–1857. doi: 10.1094/PDIS-01-20-0127-PDN
Ramakrishnan T. S., Pillay P. N. R. (1961). Leaf spot of Rubber caused by Corynespora cassiicola (Berk. & Curt.) Wei. Rubb. Bd Bull. 5, 32–35.
Ribeiro S., Tran D., Deon M., Clement-Demange A., Garcia D., Soumahoro M., et al. (2019). Gene deletion of Corynespora cassiicola cassiicolin Cas1 suppresses virulence in the rubber tree. Fungal Genet. Biol. 129, 101–114. doi: 10.1016/j.fgb.2019.05.004
Rondon M. N., Lawrence K. (2021). The fungal pathogen Corynespora cassiicola: A review and insights for target spot management on cotton and soya bean. J. Phytopathol. 169, 329–338. doi: 10.1111/jph.12992
Roy C. B., Liu H., Rajamani A., Saha T. (2019). Transcriptome profiling reveals genetic basis of disease resistance against Corynespora cassiicola in rubber tree (Hevea brasiliensis). Curr. Plant Biol. 17, 2–16. doi: 10.1016/j.cpb.2019.02.002
Schlub R., Smith L. J., Datnoff L., Pernezny K. (2009). An overview of target spot of tomato caused by Corynespora cassiicola. Acta Hortic. 808, 25–28. doi: 10.17660/ActaHortic.2009.808.1
Sierra-Orozco E., Vallad G. E., Hutton S. F. (2022). “Mapping and Characterization of Resistance to Target Spot of Tomato Caused by Corynespora cassiicola from S. galapagense and S. pimpinellifolium.,” in ASHS 2022 Annual Conference, Chicago, Illinois (American Society for Horticultural Science).
Silva W., Karunanayake E., Wijesundera R., Priyanka U. (2003). Genetic variation in Corynespora cassiicola: a possible relationship between host origin and virulence. Mycological Res. 107, 567–571. doi: 10.1017/S0953756203007755
Smith L. J. (2008). Host range, phylogenetic, and pathogenic diversity of Corynespora cassiicola (Berk. & Curt) Wei.Doctor of Philosophy Dissertation (University of Florida).
Soares R., Arias C. (2020). Inheritance of soybean resistance to Corynespora cassiicola. Summa Phytopathologica 46, 85–91. doi: 10.1590/0100-5405/232903
Sumabat L., Kemerait R., Brewer M. (2018a). Phylogenetic diversity and host specialization of Corynespora cassiicola responsible for emerging target spot disease of cotton and other crops in the Southeastern United States. Phytopathology 108, 892–901. doi: 10.1094/PHYTO-12-17-0407-R
Sumabat L., Kemerait R., Kim D., Mehta Y., Brewer M. (2018b). Clonality and geographic structure of host-specialized populations of Corynespora cassiicola causing emerging target spot epidemics in the southeastern United States. PloS One 13, e0205849. doi: 10.1371/journal.pone.0205849
Sun B., Zhu G., Xie X., Chai A., Li L., Fan T., et al. (2022). Rapidly increasing boscalid resistance in Corynespora cassiicola in China. Phytopathology® 112, 1659–1666. doi: 10.1094/PHYTO-12-21-0503-R
Ullstrup A. J. (1972). The impacts of the southern corn leaf blight epidemics of 1970-1971. Annu. Rev. Phytopathol. 10, 37–50. doi: 10.1146/annurev.py.10.090172.000345
Wang X., Jiang N., Liu J., Liu W., Wang G. L. (2014). The role of effectors and host immunity in plant-necrotrophic fungal interactions. Virulence 5, 722–732. doi: 10.4161/viru.29798
Wang X., Zhang D., Cui N., Yu Y., Yu G., Fan H. (2018). Transcriptome and miRNA analyses of the response to Corynespora cassiicola in cucumber. Sci. Rep. 8, 7798. doi: 10.1038/s41598-018-26080-6
Wen C., Mao A., Dong C., Liu H., Yu S., Guo Y.-D., et al. (2015). Fine genetic mapping of target leaf spot resistance gene cca-3 in cucumber, Cucumis sativus L. Theor. Appl. Genet. 128, 2495–2506. doi: 10.1007/s00122-015-2604-z
Wu J., Xie X., Shi Y., Chai A., Wang Q., Li B. (2018). Variation of cassiicolin genes among Chinese isolates of Corynespora cassiicola. J. Microbiol. 56, 634–647. doi: 10.1007/s12275-018-7497-5
Yan X. X., Yu C. P., Fu X. A., Bao F. F., Du D. H., Wang C., et al. (2016). CARD9 mutation linked to Corynespora cassiicola infection in a Chinese patient. Br. J. Dermatol. 174, 176–179. doi: 10.1111/bjd.14082
Keywords: disease, resistance, necrotroph, Corynespora, cassiicola, toxin, pathogen, plant
Citation: Sierra-Orozco E, Sandoya G, Lee S, Vallad G and Hutton S (2023) Need for disease resistance breeding against Corynespora cassiicola in crops. Front. Agron. 5:1275906. doi: 10.3389/fagro.2023.1275906
Received: 10 August 2023; Accepted: 02 October 2023;
Published: 13 November 2023.
Edited by:
Pei Li, Kaili University, ChinaReviewed by:
Zhenchao Wang, Guizhou University, ChinaCopyright © 2023 Sierra-Orozco, Sandoya, Lee, Vallad and Hutton. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Samuel Hutton, c2ZodXR0b25AdWZsLmVkdQ==
 Edgar Sierra-Orozco
Edgar Sierra-Orozco German Sandoya
German Sandoya Seonghee Lee
Seonghee Lee Gary Vallad
Gary Vallad Samuel Hutton
Samuel Hutton