- 1Department of Entomology and Plant Pathology, Auburn University, Auburn, AL, United States
- 2College of Sciences and Mathematics, Auburn University, Auburn, AL, United States
Cotton is a multipurpose crop grown globally, including the United States. Cotton leafroll dwarf virus (CLRDV), a phloem-limited virus (Solemoviridae) transmitted by aphids, causes significant economic losses to cotton cultivation. CLRDV strains (CLRDV-typical and atypical) that were previously prevalent in other countries cause severe symptoms leading to high yield loss. Recently, a new isolate of CLRDV (CLRDV-AL) has been characterized from infected cotton plants in Alabama that are often asymptomatic and difficult to detect, implying a low titer and pathogenicity within the host. Different pathogenicity among certain strains within the same species often correlates with both environmental and molecular factors. Thus, better management and control of the vector-borne disease can be achieved by elucidating host-pathogen interaction, such as host immune response and pathogen counter-response. In this study, we demonstrate the ability of CLRDV-AL to suppress a major host defense response known as RNA silencing and compare the potency of silencing suppression to other strains of the same virus. Also, we discuss the difference in pathogenicity among them by evaluating the observations based on the amino acid variation within the functional domain. Our study provides and suggests a future direction for specifying the strategy to mitigate potential cotton disease severity.
1 Introduction
Cotton is a multipurpose agricultural resource cultivated in most continents, including Africa, Asia, and the Americas. In 2021, 12.8% of the global cotton harvested area was in the United States, and the production was valued at $7.4 billion (reviewed in Tarazi and Vaslin, 2022; Edula et al., 2023). Similarly, like many other crops, cotton is prone to damage from pests and diseases (Xiao et al., 2019; Chohan et al., 2020). Among them, viruses are major pests affecting the yield (Ziegler-Graff, 2020; Tarazi and Vaslin, 2022). Cotton leafroll dwarf virus (CLRDV) is the second most damaging virus to commercial cotton worldwide, reported in all cotton-growing continents (Distéfano et al., 2010; Agrofoglio et al., 2019; Conner et al., 2021). CLRDV is a member of the genus Polerovirus (family Solemoviridae) and the causal agent for the cotton blue disease (Parkash et al., 2021; Sõmera et al., 2021). This disease has caused about 80% yield loss in commercial cotton in South America (Silva et al., 2008). To manage CLRDV and mitigate the loss, the growers’ remedy was increasing the application of pesticides to reduce the population level of the insect vector, Aphis gossypii (Ellis et al., 2016; Galbieri et al., 2017). However, it has not been effective (Galbieri et al., 2017; Mahas et al., 2022). A resistant cotton variety was developed with a dominant resistant gene (Cbd) (Fang et al., 2010). Although this variety showed absolute resistance to CLRDV by restricting systemic virus replication, it was not long before a new strain that broke the resistance was identified in South America (Silva et al., 2008). This resistance-breaking strain was called CLRDV-atypical (CLRDV-at) in contrast to the previous strain CLRDV-typical (CLRDV-ty) (Da Silva et al., 2015). In 2017, CLRDV was reported first time in the United States in Alabama, and the genome sequence analysis identified it as a new strain (CLRDV-AL) (Avelar et al., 2019). Since the first incidence, CLRDV has been reported in many other cotton-growing states (Aboughanem-Sabanadzovic et al., 2019; Alabi et al., 2020; Ali and Mokhtari, 2020; Ali et al., 2020; Faske et al., 2020; Iriarte et al., 2020; Price et al., 2020; Tabassum et al., 2020; Thiessen et al., 2020; Wang et al., 2020; Ramos-Sobrinho et al., 2021; Ferguson and Ali, 2022). All cotton varieties grown in the United States are susceptible to this virus (Edula et al., 2023), and the presence of this pathogen poses a considerable threat. Plants infected in the early season show stunted leaves. During a late season infection, the symptoms include leaf distortion, cupping of leaves, and discoloration of veins (Avelar et al., 2019). However, CLRDV-AL-infected plants are also cryptic as they are often detected in asymptomatic plants, and infected plants do not always test positive for the virus using an amplification-based detection method (Tabassum et al., 2020; Bag et al., 2021; Tabassum et al., 2021).
CLRDV has seven open reading frames (ORFs) that encode proteins necessary for the virus infection cycle (Distéfano et al., 2010; King et al., 2012; Delfosse et al., 2014; Smirnova et al., 2015). Genome analysis has revealed that ORF 0, which encodes a viral suppressor of RNA silencing suppressor (VSR) protein (P0), and is the most divergent region among CLRDV strains (Cascardo et al., 2015; Avelar et al., 2020). Like other plant viruses, certain poleroviral proteins function as VSRs (Almasi et al., 2015; Cascardo et al., 2015; Clavel et al., 2021; Wang et al., 2021; Xiao-Yan et al., 2021). VSRs are virus-encoded proteins to prevent the antiviral mechanism employed by their hosts, which degrades the viral RNA molecules (reviewed in Li and Wang, 2019). Thus, VSRs are considered to play key roles in determining the pathogenicity, virulence, and disease severity of viral pathogens (Sharma and Ikegami, 2010; Wang et al., 2021). Previous studies have shown that CLRDV-ty has a more potent suppression of RNA silencing activity than CLRDV-at and correlated this difference to their symptom severity (Nishiguchi and Kobayashi, 2011; Agrofoglio et al., 2019). Although the difference in amino acid (aa) sequence of CLRDV-AL encoded P0 protein from the other two strains’ P0 proteins was identified (Avelar et al., 2020), its effect on the VSR potency has not yet compared with that of the other strains. In this study, we demonstrated the VSR activity of CLRDV-AL encoded P0 protein via Agrobacterium-mediated transient expression and compared VSR potency to the P0 proteins encoded by two other strains (CLRDV-ty and CLRDV-at).
2 Materials and methods
2.1 Generation of cDNA constructs
The cDNAs of ORF 0 from three different strains of CLRDV were synthesized and cloned into a pUC57mini plasmid (Genscript, Piscataway, NJ, USA) using the reference genome sequences; CLRDV-ty (GenBank accession number: GU167940), CLRDV-at (GenBank accession number: KF359946), and CLRDV-AL (GenBank accession number: MN071395.1). pAI-P0AL, pAI-P0at, and pAI-P0ty, constructs of ORF 0 placed under the CaMV 35S promoter sequence, were generated using these pUC57mini clones as templates. Fragments encompassing the ORF 0 from three different strains of CLRDV were amplified using a Phusion® High-Fidelity DNA polymerase (New England BioLab, Ipswich, MA, USA) by a pair of oligomers, P0-AL.FW.ApaI (5`-ACTAGGGCCCAACAATGTTGAATTTGATCATCTGC-3`) and P0-AL.RV.XbaI (5`-GGACTCTAGATCAACTGCTTTCTTCTTCAC-3`). The amplified product was separated in 0.8% agarose gel by electrophoresis, and the target DNAs were purified from the gel using Zymoclean Gel DNA Recovery Kit (Zymo Research, Irvine, CA, USA). The purified DNAs were digested using ApaI and XbaI restriction endonucleases (New England BioLab) and ligated into the corresponding region of the binary plasmid pAIDEE (pAI; Lin et al., 2014), which was digested with the same restriction endonucleases using T4 DNA ligase (Promega, Madison, WI, USA). Constructs were transformed into E. coli JM109 competent cells (Promega) and the resulting transformants were screened on Luria–Bertani (LB) agar plates containing kanamycin (100 mg/ml). Sequence analysis was performed at Psomagen Inc. (Rockville, MD, USA). pAI-GFP, a construct expressing green fluorescence protein (GFP) within the same vector background, was generated in a similar way using a pair of oligomers GFP.FW.ApaI (5`-ACTAGGGCCCAACAATGGCTAGCAAAGGAGAAG-3`) and GFP.RV.XbaI (5`-GTACTCTAGACTATTTGTAGAGCTCATCC-3`) for the amplification.
2.2 Sequence analysis
Multisequence alignment of amino acid sequence for P0 ORFs of CLRDV-ty, CLRDV-at, and CLRDV-AL was performed using CLUSTAL multiple sequence alignment by MUSCLE (Multiple Sequence Comparison by Log- Expectation at https://www.ebi.ac.uk/Tools/msa/muscle/) (Edgar, 2004). The GenBank accession number for each strain is identified above.
2.3 Agroinfiltration
Agroinfiltration of the constructs was conducted as previously described (Kang et al., 2015). Briefly, Agrobacterium tumefaciens strain GV3101 was transformed with the cloned plasmids by the heat shock method. The resulting transformants were screened on LB agar plates containing kanamycin and rifampicin (100 mg/ml and 50 mg/ml, respectively). A single colony was selected and inoculated in 3 ml liquid LB media containing kanamycin (100 mg/ml). The culture was grown overnight at 28°C, then 50 µl of the culture was transferred into 5 ml of fresh LB media with kanamycin and rifampicin (100 mg/ml and 50 mg/ml, respectively). The cells were grown overnight at 28°C, and the culture was centrifuged at 3,000 rpm for 10 mins. Pelleted cells were resuspended in agroinfiltration buffer (10 mM MES, pH 5.85; 10 mM MgCl2; 150 µM Acetosyringone) at the optical density 1.0 at 600 nm (OD600nm = 1.0). Following the incubation for at least 2 hours without shaking at room temperature, the suspension was infiltrated into the lower surface of fully expanded leaves of six weeks old Nicotiana benthamiana 16c plants using needleless syringes. The infiltrated plants were grown under 16h/8h of light/dark cycle until further analysis.
2.4 Examination of fluorescence in plants
Six weeks old N. benthaminana 16c plants were infiltrated with A. tumefaciens GV3101 cells transformed with pAI-GFP mixed with cells transformed with P0 constructs (pAI-P0AL, pAI-P0at, or pAI-P0ty) or other known VSRs (pPZP-P19 or pPZP-HC-Pro; Qu et al., 2003) or empty pAI vector in equal volumes (v/v, OD600nm = 1.0). The expression of the GFP transgene within this transgenic line (16c) can be silenced when extra copies of the GFP transcript are introduced via a binary vector (Voinnet et al., 1998). However, silencing of GFP induced this way can be suppressed by the simultaneous expression of VSRs (Voinnet et al., 1999). This method has been used to identify VSRs among virus-encoded proteins for decades (Brigneti et al., 1998; Voinnet et al., 1999; Qu et al., 2003; Cui et al., 2005; Li et al., 2014; Dong et al., 2016; Li et al., 2019; Sinha et al., 2021; Wang et al., 2022). Green fluorescence was observed under a long wavelength (365 nm) using a hand-held UV lamp raised 6 inches above the plants at four days post infiltration (dpi) in the dark room. The infiltrated leaves were collected at 4 dpi and examined using an epifluorescence microscope, Echo Revolve (San Diego, CA, USA), with wavelengths specified by FITC cube (EX:470 ± 40 nm and EM:525 ± 50 nm).
2.5 Relative expression levels of GFP mRNA
The relative expression levels of GFP mRNA were evaluated by RT-qPCR using Luna® universal RT-qPCR system (New England Biolabs). Total RNA was extracted from 100 mg of leaf discs of infiltrated N. benthamiana (four discs of 0.9 cm in diameter). The samples were ground in liquid nitrogen using TissueLyser II (Qiagen, Hilden, Germany). The extraction was performed using RNeasy Plant Mini Kit (Qiagen) according to the manufacturer’s protocol. RNA quality and concentration were evaluated using NanoDrop™ Lite Spectrophotometer (ThermoFisher Scientific). One-step RT-qPCR reactions were performed in a CFX Connect™ Real-time PCR detection system (BIO-RAD) with a total volume of 10 μl, using 2 μl of RNA (10 ng/μl). For the amplification of GFP, a pair of oligomers; MFA.Gq-PCR : FW (5`- GATGACGGGAACTACAAGAC -3`) and MFA.Gq-PCR : RV (5`- CGAGTACAACTATAACTCACAC -3`) were used. The reference gene, NbACTIN2, was used as the internal control and was amplified using a pair of NbACTIN2-FW (5`-CAATCCAGACACTGTACTTTCTCTC-3`) and NbACTIN2-RV (5`-AAGCTGCAGGTATCCATGAGACTA-3`) as previously described (Luo et al., 2019). The cycling conditions include the reverse transcription at 55°C for 10 min, incubation at 95°C for 2 min, and 40 cycles of 95°C for 10 s and 60°C for 60 s. Three biological and technical replicates were performed for all treatments. Ct values were averaged for triplicates of each sample prior to calculating relative values using the 2−ΔΔCt method (Livak and Schmittgen, 2001).
3 Results
To verify the ability of the P0 protein encoded by CLRDV-AL (P0-AL) to suppress the RNA silencing, P0-AL was expressed along with a green fluorescent protein (GFP) in Nicotiana benthamiana 16c plants, a transgenic line that constitutively expresses GFP (Figure 1). When there is no suppression, the transiently expressed GFP triggers an RNA silencing response which degrades both endogenous and transient GFP mRNAs, and subsequently, the fluorescence level drops (Figure 1B; EV, top left). However, in the presence of suppressors, such degradation is inhibited, and the level of GFP expression can be visualized under UV range light. In our assay, strong GFP fluorescence was observed in the presence of P19 or HC-Pro at four days post-infiltration (dpi) (Figure 1B; HC-Pro and P19, bottom right and left). P19 and HC-Pro are well-known strong VSRs encoded by Tomato bushy stunt virus and Tobacco etch virus, respectively (Anandalakshmi et al., 1998; Scholthof, 2006). Although P0-AL showed suppression activity of the RNA silencing, the GFP fluorescence level within the region of N. benthamiana 16c leaves co-infiltrated with GFP visualized by UV light was not comparable to the level shown by P19 or HC-Pro co-infiltration (Figure 1B; P0-AL, top right). A similar pattern in the suppression of GFP fluorescence was observed upon the co-expression of GFP along with each VSR or EV in the individual leaves (Figure 1C). The visually assessed relative intensity of the GFP fluorescence suggested that the RNA silencing suppression potency of P0-AL is significantly weaker than that of P19 or HC-Pro because the fluorescence intensity was similar to the basal fluorescence from the non-infiltrated leaves (Figure 1C; P0-AL and Mock). To better evaluate the relative intensity of GFP fluorescence resulting from the suppression of RNA silencing, infiltrated leaf tissue patches were examined using a fluorescence microscope (Figure 1D). The relative fluorescence intensity calculated from more than 15 images per treatment using Region of Interest (ROI) on ImageJ and analyzed by one-way ANOVA test in R (see Materials and Methods) demonstrated significant difference (p < 0.01) in the fluorescence level of P0-AL-infiltrated patches compared to either HC-Pro- or P19-infiltrated patches (Figure 1E; denoted as ‘a’ for HC-Pro or P19, and as ‘b’ for P0-AL). According to the analysis, all three VSRs demonstrated significant (p < 0.01) suppressor activity relative to the empty vector (EV)-infiltrated control (Figure 1E; denoted as ‘c’ for EV control). However, it is noteworthy that the fluorescence level measured from P0-AL-infiltrated patches was not completely distinct from the basal level fluorescence measured from buffer-infiltrated N. benthamiana 16c plants (Figure 1E; denoted as ‘bc’ for 16c control). These results implied that the P0 protein of CLRDV-AL is not a potent silencing suppressor compared to other VSRs, such as P19 and HC-Pro, known for their strong silencing suppression.
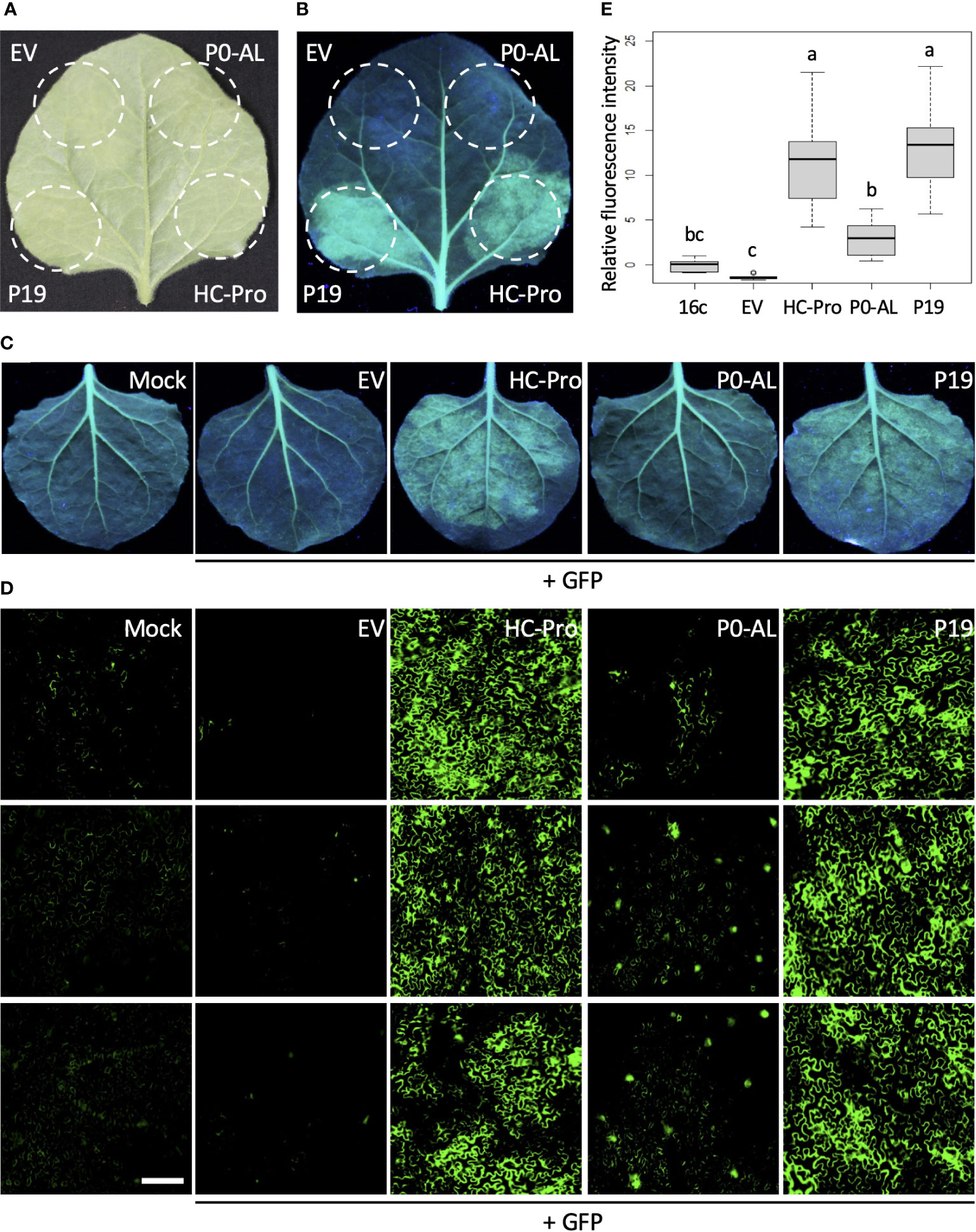
Figure 1 P0 protein of CLRDV-AL is a weak local VSR. Potency of P0 protein of CLRDV-AL as a viral suppressor of RNA silencing (VSR) was compared to the known VSRs, such as P19 or HC-Pro, by co-expressing GFP and each VSR in GFP-transgenic N. benthamiana (16c) plants by Agroinfiltration. Images were taken at 4 days post infiltration (dpi) under the natural light (A) and UV light (B, C). The same leaf is shown for panels (A, B) EV, empty vector. (D) Infiltrated leaf tissue shown in panel (C) was examined using a fluorescence microscope. Three representative images per treatment were shown. Scale bar = 180 µm. (E) The fluorescence intensity was calculated using Region of Interest (ROI) on ImageJ. The relative intensity was analyzed by one-way ANOVA test in R and shown as a box and whisker plot. Each boxplot depicts the interquartile range (middle 50% of the data), the lower and upper edge show the first and third quartile (25th and 75th percentile respectively), median (horizontal line within the box). The whiskers are the contributions within the 1.5 interquartile range; open circles beyond these whiskers are considered as outliers; significant differences, p < 0.01, were denoted by letters.
Previously, the P0 protein encoded by an atypical strain of CLRDV (P0-at) was reported to function as a weaker VSR than the P0 protein encoded by a typical strain of CLRDV (P0-ty) by a similar set of experiments described above (Agrofoglio et al., 2019). To evaluate the VSR potency of P0-AL compared to the P0 proteins from two other strains (P0-ty and P0-at), a similar set of qualitative and quantitative experiments and analysis shown in Figure 1 was performed (Figure 2). Each construct harboring the ORF of P0 from three different strains of CLRDV was co-infiltrated into the leaves of N. benthamiana 16c plants along with the GFP construct via agroinfiltration. When observed under the UV light at four dpi, non-infiltrated leaves showed the basal level of GFP fluorescence generated from their own GFP transgene expression (Figure 2A; Mock). On the contrary, the leaves co-infiltrated by the GFP construct with an empty vector did not show GFP fluorescence and looked rather red due to the autofluorescence from the chloroplasts (Figure 2A; EV). As expected from previous reports, the leaves co-infiltrated with P0-ty or P0-at showed a sharp contrast in fluorescence level (Figure 2A; P0-ty and P0-at). Stronger fluorescence was observed from the leaves co-infiltrated with P0-ty, but less fluorescence was observed from the leaves co-infiltrated with P0-at. Upon the visual observation, the fluorescence level from the leaves co-infiltrated with P0-AL was even lower than the level of P0-at (Figure 2A: P0-AL). The relative intensities of the GFP fluorescence visually evaluated under UV light suggested that the RNA silencing suppression activity of P0-AL is weaker than P0-ty like P0-at. To evaluate the relative potency of the suppression of RNA silencing better, infiltrated leaf tissue patches were examined using a fluorescence microscope (Figure 2A; bottom row, see Supplementary Figure 1 for more representative images), and the relative fluorescence intensity was calculated from more than 15 images per treatment using ROI on ImageJ and analyzed by one-way ANOVA test in R (Figure 2B). The analysis demonstrated a significant difference (p < 0.01) in the fluorescence level from the patches infiltrated with three P0s individually compared to EV-infiltrated patches (Figure 2B; denoted as ‘d’ for EV control). The fluorescence level of the patches infiltrated with P0-ty was also significantly different and distinct from the other two sets of patches infiltrated with the other two P0s (Figure 2B; denoted as ‘a’ for P0-ty). However, the fluorescence level of the patches infiltrated with P0-at that was significantly different from the patches infiltrated with P0-ty (Figure 2B; denoted as ‘b’ for P0-at) was not completely distinct from the patches infiltrated with P0-AL (Figure 2B; denoted as ‘bc’ for P0-AL). Interestingly, according to the analysis, the fluorescence level of P0-AL-infiltrated patches did not show complete distinction from buffer-infiltrated N. benthamiana 16c control (Figure 2B; denoted as ‘cd’ for 16c control). This implied that there was suppression activity for P0-AL, yet the suppression potency was not strong enough. This result is also in accordance with the result shown above (Figure 1E; denoted as ‘b’ and ‘bc’ for P0-AL and 16c control, respectively). Although the fluorescence of GFP under UV light is an indication of the level of silencing or its suppression, this can still be argued because silencing is the mechanism acting on the RNA, yet the analysis based on the observed florescence comes from the translated product of such RNA. To further validate our observation, the relative accumulation of GFP mRNA was analyzed by quantitative real-time PCR (Figure 2C). The GFP mRNA levels in the patches infiltrated with three P0s individually were all significantly higher, p < 0.01, than EV control patches (Figure 2C; significant difference was not noted within the graph). This was consistent with the analysis using the GFP fluorescence shown in Figure 2B. The GFP mRNA level in the patches infiltrated with P0-ty was significantly higher than the levels in the other two sets of patches infiltrated with P0-at or P0-AL. This was also in agreement with the analysis shown in Figure 2B. Remarkably, the GFP mRNA levels between the patches infiltrated with P0-at and P0-AL showed less significant difference (Figure 2C; p < 0.05), which was in line with the result shown in Figure 2B where P0-at and P0-AL were denoted as groups ‘b’ and ‘bc’, respectively. Overall, the analysis of relative GFP mRNA level by RT-qPCR (Figure 2C) supported the quantified GFP fluorescence level analyzed in Figure 2B. These results implied that the silencing suppressor potency of the P0 protein encoded by CLRDV-AL is comparable to the P0 protein encoded by CLRDV-at, which is weaker than the P0 protein encoded by CLRDV-ty.
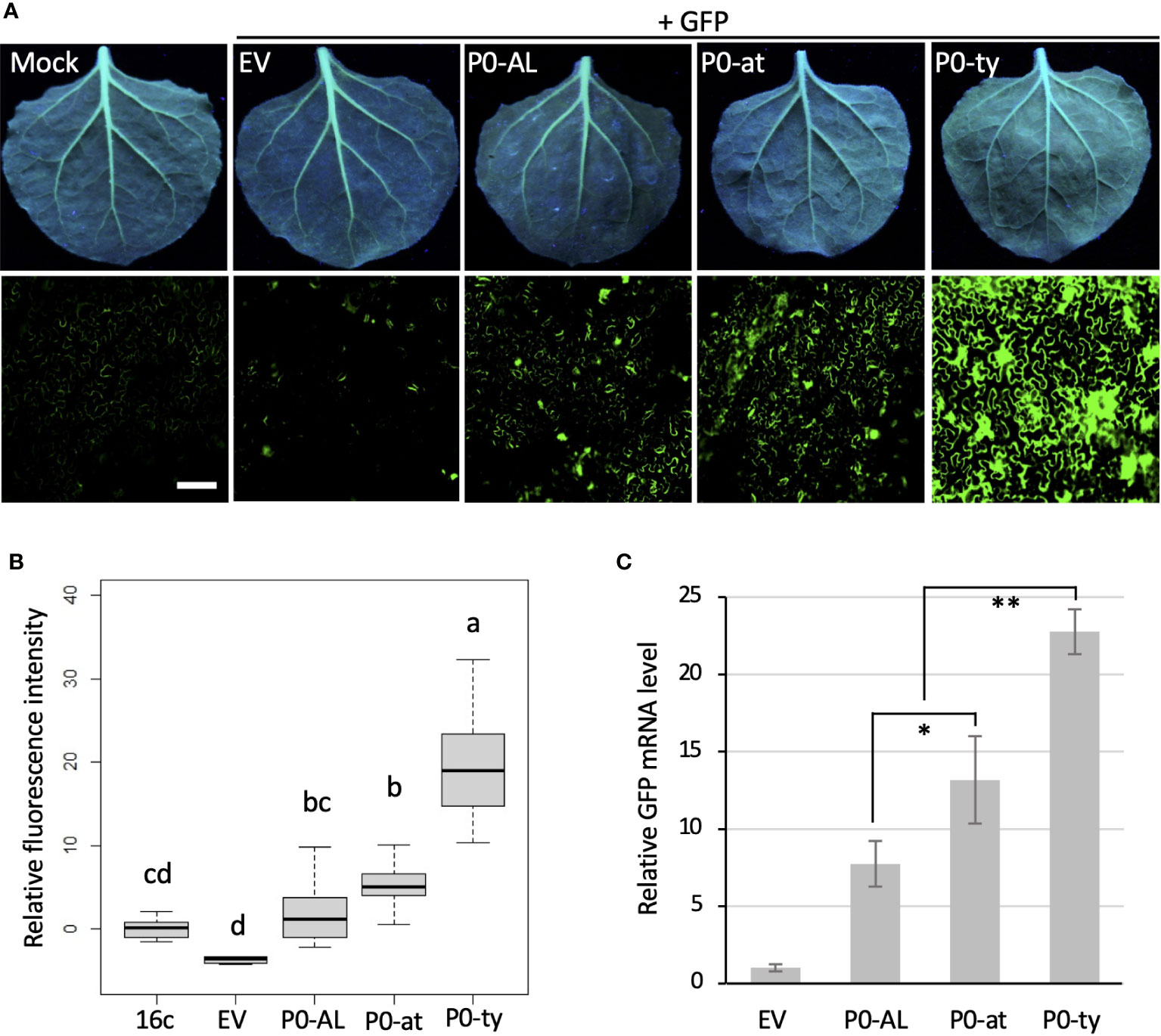
Figure 2 Comparison of VSR potency among the P0s encoded by three CLRDV strains. The suppressor of RNA silencing activity of the P0 proteins from CLRDV-AL, CLRDV-ty, and CLRDV-at was compared by co-expressing GFP and each P0 in GFP-transgenic N. benthamiana (16c) plants by Agroinfiltration. Images were taken at 4 dpi under the UV light (A, top row) and using a fluorescence microscope (A, bottom row). Scale bar = 180 µm. Mock; non-infiltrated N. benthamiana 16c plant, EV; empty vector. See Supplementary Figure 1 for more fluorescence microscopy images. Note that the leaves shown for Mock and EV are the same images used in Figure 1C as this set of Agroinfiltration was performed concurrently. (B) The fluorescence intensity was calculated using Region of Interest (ROI) on ImageJ. The relative intensity was analyzed by one-way ANOVA test in R and shown as a box and whisker plot. Significant differences were denoted by letters. (C) GFP mRNA levels in tissues co-infiltrated with a set shown in (A) were analyzed by RT-qPCR. Error bars indicate the standard deviation of the means of Ct values in three biological repeats using actin2 reference gene and 2-ΔΔCt method. Statistically significant differences determined by Student’s t-test, p < 0.05 or < 0.01, were denoted by asterisks (* or **, respectively). Note that all P0s showed significant difference (p < 0.01) from EV, but not noted.
4 Discussion
CLRDV induces a wide range of various symptoms, such as leaf drooping, wilting, reddening, stunting, and shortened internodes as well as asymptomatic infection which are commonly observed in the US (Agrofoglio et al., 2017; Avelar et al., 2019; Tabassum et al., 2020; Bag et al., 2021). The causes for diverse symptoms are not clearly understood and could be due to myriads of factors, such as abiotic factors, interactions between the virus and host, or a combination of both. Interaction between the virus and host also contributes to the resistance dynamic reported in the cotton-CLRDV pathosystem. Upon the rise of CBD in Brazil, deploying cultivars with a single dominant resistance gene, Rghv1, effectively mitigated the disease (Junior et al., 2008). However, varieties carrying a single dominant resistance gene, Cbd (Fang et al., 2010), became susceptible to CLRDV-at in Argentina (Agrofoglio et al., 2019). Furthermore, the BRS-293 cultivar, which is moderately resistant to typical and atypical CLRDVs (Morello et al., 2010), was susceptible to prevalent CLRDV in the southeastern United States (Brown et al., 2019). Considered a crosstalk between plant immunity such as resistance and viral pathogenicity mediated by RNA silencing and VSR has been proposed (reviewed in Ding, 2010), it is noteworthy to mention that suppression of RNA silencing at variable degrees among different isolates within the same species or among different species within the same genus has been reported from several members of family Solemoviridae (Han et al., 2010; Kozlowska-Makulska et al., 2010; Fusaro et al., 2012; Delfosse et al., 2014; Zhuo et al., 2014; Cascardo et al., 2015; Agrofoglio et al., 2019). Thus, while it is not the sole factor, the characteristics of its VSR does contribute partially to the cryptic pathogenicity of CLRDV-AL.
In this study, we examined the silencing suppression potency of the P0 protein encoded by CLRDV-AL by using GFP co-infiltration assays on N. benthamiana 16c plants and analyzed the measure of suppression both in protein and RNA levels. Our study demonstrated that CLRDV-AL P0 functioned as a local VSR (Figure 1), but the suppression level differed from cognate VSRs from other strains of CLRDV (Figure 2) or VSRs from other viruses (Figure 1). The alignment comparing the aa sequence of the P0-AL to the previously isolated strains from South American countries showed some mutations within the F-box domain known for VSR function (Agrofoglio et al., 2019). Figure 3 summarizes some notably divergent residues within its conserved F-box motif among three CLRDV strains used in this study. P0-AL has a valine (V) substitution which resembles an atypical strain (Figure 3; letter in red), and an arginine (R) substitution which resembles a typical strain (Figure 3; letter in blue). However, P0-AL also carries a unique phenylalanine (F) substitution that is found as either leucine (L) or isoleucine (I) in atypical or typical strains, respectively (Figure 3; letter in green). The collective difference in these residues among the three strains may contribute to our results demonstrating that the P0 protein from CLRDV-AL displays yet another level of silencing suppression potency compared to two other P0s of different strains. Previous study sought the correlation between the resistance-breaking trait of CLRDV-at and its VSR, P0-at, by comparing VSR potency between the P0 proteins from CLRDV-at and CLRDV-ty (Agrofoglio et al., 2019). Agrofoglio et al. (2019) showed that an aa substitution, I to V, present in F-box motif was not sufficient to completely elucidate this question because the revertant P0-at, in which V residue has turned back to I, was still not able to restore the comparable suppression potency demonstrated by P0-ty. Our results with P0-AL demonstrating a similar suppression potency to P0-at can be partially explained by the fact that P0-AL has the same substitution to V in that residue as P0-at. However, other substitutions unique in P0-AL from both P0-at and P0-ty may explain why P0-AL is not precisely at the same degree of VSR potency as P0-at (Figure 2B). Furthermore, it is known that the virus-encoded proteins interact among them during the virus infection within the host to expand their functional landscape and stability (Dao et al., 2020; Wang et al., 2022a; Wang et al., 2022b). As presented study was based on the transient expression of a single viral protein, it is worthy to note that the observed VSR potency could differ in the presence of the other CLRDV proteins under the viral context.

Figure 3 Alignment of the P0s encoded by CLRDV-AL, CLRDV-ty, and CLRDV-at. Amino acid (aa) sequences of the P0 proteins from three CLRDV strains were aligned. Notable divergence in P0-AL, within the F-box motif (shown in a box with black line) linked to RNA silencing suppressor activity, from atypical, typical, or both are noted in blue, red, or green, respectively. Consensus symbols show sequence similarities based on Percent Accepted Mutations (PAM) 250 scoring matrix. ‘*’; indicates aa fully conserved aa residues. ‘:’; indicates conservation between groups of strongly similar properties. ‘.’; indicates conservation between groups of weakly similar properties.
CLRDV is a potentially threatening pest to the cotton industry due to its high mutation rate as a viral pathogen with an RNA genome and the tremendously effective transmission by insect vectors (Jones, 2009; Tarazi and Vaslin, 2022). Considering the economic significance of cotton in the United States, it is imperative to develop predictable strategies to manage potential new strains of CLRDV. Further molecular investigation on P0-AL to understand the cryptic nature of CLRDV-AL and its disease development will provide insight into how different CLRDV strains display a wide range of various disease symptoms and severity. Additionally, identifying host factors playing a role with the P0 protein during the virus infection will help specify the strategy to develop resistant varieties that can mitigate potential loss by CLRDV and sustainably manage resources for cotton growers in the United States and beyond.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Author contributions
MA conducted experiments, analysed the data, and wrote the manuscript. BP conducted experiments and collected the data. KM conceived the study. S-HK conceived the study, designed experiments, and edited the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This research was supported by the research program of the U.S. Department of Agriculture, National Institute of Food and Agriculture, Hatch Project No. AL 15-1-19124 (to S-HK) and the Agricultural Research Service Agreement No. 58-6010-0-011.
Acknowledgments
The authors would like to thank Dr. Feng Qu at the Ohio State University for providing pAIDEE plasmid and other VSRs used in this article. The findings and conclusions in this article have not been formally disseminated by the U. S. Department of Agriculture and should not be construed to represent any agency determination or policy.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fagro.2023.1235168/full#supplementary-material
Supplementary Figure 1 | Infiltrated leaf tissue showing GFP silencing inhibition from Figure 2A was examined using a fluorescence microscope. Representative images used for the analysis of relative fluorescence intensity (Figure 2B) are shown. (A; P0-ty, B; P0-at, C; P0-AL, D; EV). Scale bar = 180 µm.
References
Aboughanem-Sabanadzovic N., Allen T. W., Wilkerson T. H., Conner K. N., Sikora E. J., Nichols R. L., et al. (2019). First Report of Cotton Leafroll Dwarf Virus in upland cotton (Gossypium hirsutum) in Mississippi. Plant Dis. 103, 1798. doi: 10.1094/PDIS-01-19-0017-PDN
Agrofoglio Y. C., Delfosse V. C., Casse M. F., Hopp H. E., Kresic I. B., Distéfano A. J. (2017). Identification of a new cotton disease caused by an atypical Cotton Leafroll Dwarf Virus in Argentina. Phytopathology 107, 369–376. doi: 10.1094/PHYTO-09-16-0349-R
Agrofoglio Y. C., Delfosse Ab V. C., Casse M. F., Hopp H. E., Bonacic Kresic I., Ziegler-Graff V., et al. (2019). P0 protein of Cotton Leafroll Dwarf Virus-atypical isolate is a weak RNA silencing suppressor and the avirulence determinant that breaks the cotton Cbd gene-based resistance. Plant Pathol. 68 (6), 1059–1071. doi: 10.1111/ppa.13031
Alabi O. J., Isakeit T., Vaughn R., Stelly D., Conner K. N., Gaytán B. C., et al. (2020). First report of Cotton Leafroll Dwarf Virus infecting upland cotton (Gossypium hirsutum) in Texas. Plant Dis. 104, 998. doi: 10.1094/PDIS-09-19-2008-PDN
Ali A., Mokhtari S. (2020). First report of Cotton Leafroll Dwarf Virus infecting cotton (Gossypium hirsutum) in Kansas. Plant Dis. 104, 1880. doi: 10.1094/PDIS-12-19-2589-PDN
Ali A., Mokhtari S., Ferguson C. (2020). First report of Cotton Leafroll Dwarf Virus from cotton (Gossypium hirsutum) in Oklahoma. Plant Dis. 104, 2531. doi: 10.1094/PDIS-03-20-0479-PDN
Almasi R., Miller W. A., Ziegler-Graff V. (2015). Mild and severe Cereal Yellow Dwarf Viruses differ in silencing suppressor efficiency of the P0 protein. Virus Res. 208, 199–206. doi: 10.1016/j.virusres.2015.06.020
Anandalakshmi R., Pruss G. J., Ge X., Marathe R., Mallory A. C., Smith T. H., et al. (1998). A viral suppressor of gene silencing in plants. Proc. Natl. Acad. Sci. 95, 13079–13084. doi: 10.1073/pnas.95.22.13079
Avelar S., Ramos-Sobrinho R., Conner K., Nichols R. L., Lawrence K., Brown J. K. (2020). Characterization of the complete genome and P0 protein for a previously unreported genotype of Cotton Leafroll Dwarf Virus, an introduced polerovirus in the United States. Plant Dis. 104, 780–786. doi: 10.1094/PDIS-06-19-1316-RE
Avelar S., Schrimsher D. W., Lawrence K., Brown J. K. (2019). First Report of Cotton Leafroll Dwarf Virus associated with cotton blue disease symptoms in Alabama. Plant Dis. 103, 592. doi: 10.1094/PDIS-09-18-1550-PDN
Bag S., Roberts P. M., Kemerait R. C. (2021). Cotton leafroll dwarf disease: An emerging virus disease on cotton in the U.S. Crops Soils 54, 18–22. doi: 10.1002/crso.20105
Brigneti G., Voinnet O., Li W. X., Ji L. H., Ding S. W., Baulcombe D. C. (1998). Viral pathogenicity determinants are suppressors of transgene silencing in Nicotiana benthamiana. EMBO J. 17, 6739–6746. doi: 10.1093/emboj/17.22.6739
Brown S., Conner K., Hagan A., Jacobson A., Koebernick J., Lawrence K., et al. (2019). Report of a research review and planning meeting on Cotton Leafroll Dwarf Virus (Orange Beach, Alabama: Cotton Incorporated). Available at: https://www.cottoninc.com/cotton-production/ag-research/plant-pathology/cotton-leafroll-dwarf-virus-research/.
Cascardo R. S., Arantes I. L. G., Silva T. F., Sachetto-Martins G., Vaslin M. F. S., Corrêa R. L. (2015). Function and diversity of P0 proteins among cotton leafroll dwarf virus isolates. Virol. J. 12, 123. doi: 10.1186/s12985-015-0356-7
Chohan S., Perveen R., Abid M., Tahir M. N., Sajid M. (2020). “Cotton diseases and their management,” in Cotton production and uses: agronomy, crop protection, and postharvest technologies. Eds. Ahmad S., Hasanuzzaman M. (Singapore: Springer), 239–270.
Clavel M., Lechner E., Incarbone M., Vincent T., Cognat V., Smirnova E., et al. (2021). Atypical molecular features of RNA silencing against the phloem-restricted polerovirus TuYV. Nucleic Acids Res. 49, 11274–11293. doi: 10.1093/nar/gkab802
Conner K., Strayer-Scherer A., Hagan A., Koebernick J., Jacobson A., Bowen K., et al. (2021). “Cotton Leafroll Dwarf Virus,” (Alabama: Alabama Cooperative Extension System). Available at: https://www.aces.edu/blog/topics/crop-production/cotton-leafroll-dwarf-virus/.
Cui X., Li G., Wang D., Hu D., Zhou X. (2005). A begomovirus DNAβ-encoded protein binds DNA, functions as a suppressor of RNA silencing, and targets the cell nucleus. J. Virol. 79, 10764–10775. doi: 10.1128/JVI.79.16.10764-10775.2005
Dao T. N. M., Kang S.-H., Bak A., Folimonova S. Y. (2020). A non-conserved p33 protein of Citrus Tristeza Virus interacts with multiple viral partners. Mol. Plant Microbe Interact. 33, 859–870. doi: 10.1094/MPMI-11-19-0328-FI
Da Silva A. K. F., Romanel E., Silva T., da F., Castilhos Y., Schrago C. G., et al. (2015). Complete genome sequences of two new virus isolates associated with cotton blue disease resistance breaking in Brazil. Arch. Virol. 160, 1371–1374. doi: 10.1007/s00705-015-2380-8
Delfosse V. C., Agrofoglio Y. C., Casse M. F., Kresic I. B., Hopp H. E., Ziegler-Graff V., et al. (2014). The P0 protein encoded by cotton leafroll dwarf virus (CLRDV) inhibits local but not systemic RNA silencing. Virus Res. 180, 70–75. doi: 10.1016/j.virusres.2013.12.018
Ding S.-W. (2010). RNA-based antiviral immunity. Nat. Rev. Immunol. 10, 632–644. doi: 10.1038/nri2824
Distéfano A. J., Bonacic Kresic I., Hopp H. E. (2010). The complete genome sequence of a virus associated with cotton blue disease, cotton leafroll dwarf virus, confirms that it is a new member of the genus Polerovirus. Arch. Virol. 155, 1849–1854. doi: 10.1007/s00705-010-0764-3
Dong K., Wang Y., Zhang Z., Chai L.-X., Tong X., Xu J., et al. (2016). Two amino acids near the N-terminus of Cucumber Mosaic Virus 2b play critical roles in the suppression of RNA silencing and viral infectivity. Mol. Plant Pathol. 17, 173–183. doi: 10.1111/mpp.12270
Edgar R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. doi: 10.1093/nar/gkh340
Edula S. R., Bag S., Milner H., Kumar M., Suassuna N. D., Chee P. W., et al. (2023). Cotton leafroll dwarf disease: An enigmatic viral disease in cotton. Mol. Plant Pathol. 24, 513–526. doi: 10.1111/mpp.13335
Ellis M. H., Stiller W. N., Phongkham T., Tate W. A., Gillespie V. J., Gapare W. J., et al. (2016). Molecular mapping of bunchy top disease resistance in Gossypium hirsutum L. Euphytica 210, 135–142. doi: 10.1007/s10681-016-1713-3
Fang D. D., Xiao J., Canci P. C., Cantrell R. G. (2010). A new SNP haplotype associated with blue disease resistance gene in cotton (Gossypium hirsutum L.). Theor. Appl. Genet. 120, 943–953. doi: 10.1007/s00122-009-1223-y
Faske T. R., Stainton D., Aboughanem-Sabanadzovic N., Allen T. W. (2020). First Report of Cotton Leafroll Dwarf Virus from upland cotton (Gossypium hirsutum) in Arkansas. Plant Dis. 104,2742. doi: 10.1094/PDIS-12-19-2610-PDN
Ferguson C., Ali A. (2022). Complete genome sequence of Cotton Leafroll Dwarf Virus infecting cotton in Oklahoma, USA. Microbiol. Resour Announc 11, e0014722. doi: 10.1128/mra.00147-22
Fusaro A. F., Correa R. L., Nakasugi K., Jackson C., Kawchuk L., Vaslin M. F. S., et al. (2012). The Enamovirus P0 protein is a silencing suppressor which inhibits local and systemic RNA silencing through AGO1 degradation. Virology 426, 178–187. doi: 10.1016/j.virol.2012.01.026
Galbieri R., Boldt A. S., Scoz L. B., Rodrigues S. M., Rabel D. O., Belot J. L., et al. (2017). Cotton blue disease in central-west Brazil: Occurrence, vector (Aphis gossypii) control levels and cultivar reaction. Trop. Plant Pathol. 42, 468–474. doi: 10.1007/s40858-017-0165-1
Han Y.-H., Xiang H.-Y., Wang Q., Li Y.-Y., Wu W.-Q., Han C.-G., et al. (2010). Ring structure amino acids affect the suppressor activity of melon aphid-borne yellows virus P0 protein. Virology 406, 21–27. doi: 10.1016/j.virol.2010.06.045
Iriarte F. B., Dey K. K., Small I. M., Conner K. N., O’Brien G. K., Johnson L., et al. (2020). First report of cotton leafroll dwarf virus in Florida. Plant Dis. 104, 2744. doi: 10.1094/PDIS-10-19-2150-PDN
Jones R. A. C. (2009). Plant virus emergence and evolution: origins, new encounter scenarios, factors driving emergence, effects of changing world conditions, and prospects for control. Virus Res. 141, 113–130. doi: 10.1016/j.virusres.2008.07.028
Junior O., Schuster I., Pinto R., Pires E., Belot J., Silvie P., et al. (2008). Inheritance of resistance to cotton blue disease. Pesquisa Agropecuária Bras. 43, 661–665. doi: 10.1590/S0100-204X2008000500015
Kang S.-H., Bak A., Kim O.-K., Folimonova S. Y. (2015). Membrane association of a nonconserved viral protein confers virus ability to extend its host range. Virology 482, 208–217. doi: 10.1016/j.virol.2015.03.047
King A. M. Q., Adams M. J., Carstens E. B., Lefkowitz E. J. (2012). “Family - Luteoviridae,” in Virus Taxonomy (San Diego: Elsevier), 1045–1053.
Kozlowska-Makulska A., Guilley H., Szyndel M. S., Beuve M., Lemaire O., Herrbach E., et al. (2010). P0 proteins of European beet-infecting poleroviruses display variable RNA silencing suppression activity. J. Gen. Virol. 91, 1082–1091. doi: 10.1099/vir.0.016360-0
Li Z., He Y., Luo T., Zhang X., Wan H., Ur Rehman A., et al. (2019). Identification of key residues required for RNA silencing suppressor activity of p23 protein from a mild strain of Citrus Tristeza Virus. Viruses 11, 782. doi: 10.3390/v11090782
Li F., Huang C., Li Z., Zhou X. (2014). Suppression of RNA silencing by a plant DNA virus satellite requires a host calmodulin-like protein to repress RDR6 expression. PloS Pathog. 10, e1003921. doi: 10.1371/journal.ppat.1003921
Li F., Wang A. (2019). RNA-targeted antiviral immunity: more than just RNA silencing. Trends Microbiol. 27, 792–805. doi: 10.1016/j.tim.2019.05.007
Lin J., Guo J., Finer J., Dorrance A., Redinbaugh M. G., Qu F. (2014). The bean pod mottle virus RNA2-encoded 58K protein is required in cis for RNA2 accumulation. J. Virol. 88, 3213–3222. doi: 10.1128/JVI.03301-13
Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Luo C., Wang Z. Q., Liu X., Zhao L., Zhou X., Xie Y. (2019). Identification and analysis of potential genes regulated by an Alphasatellite (TYLCCNA) that contribute to host resistance against Tomato Yellow Leaf Curl China Virus and its Betasatellite (TYLCCNV/TYLCCNB) Infection in Nicotiana benthamiana. Viruses 11, 442–442. doi: 10.3390/v11050442
Mahas J. W., Hamilton F. B., Roberts P. M., Ray C. H., Miller G. L., Sharman M., et al. (2022). Investigating the effects of planting date and Aphis gossypii management on reducing the final incidence of cotton leafroll dwarf virus. Crop Prot. 158, 106005. doi: 10.1016/j.cropro.2022.106005
Morello C., Suassuna N., Farias F., Lamas F., Pedrosa M., Ribeiro J., et al. (2010). BRS 293: A midseason high-yielding upland cotton cultivar for Brazilian savanna. Crop Breed. Appl. Biotechnol. 10, 180–182. doi: 10.12702/1984-7033.v10n02a13
Nishiguchi M., Kobayashi K. (2011). Attenuated plant viruses: preventing virus diseases and understanding the molecular mechanism. J. Gen. Plant Pathol. 77, 221–229. doi: 10.1007/s10327-011-0318-x
Parkash V., Sharma D. B., Snider J., Bag S., Roberts P., Tabassum A., et al. (2021). Effect of cotton leafroll dwarf virus on physiological processes and yield of individual cotton plants. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.734386
Price T., Valverde R., Singh R., Davis J., Brown S., Jones H. (2020). First report of cotton leafroll dwarf virus in Louisiana. Plant Health Prog. 21, 142–143. doi: 10.1094/PHP-03-20-0019-BR
Qu F., Ren T., Morris T. J. (2003). The Coat Protein of Turnip Crinkle Virus suppresses posttranscriptional gene silencing at an early initiation step. J. Virol. 77, 511–522. doi: 10.1128/JVI.77.1.511-522.2003
Ramos-Sobrinho R., Adegbola R. O., Lawrence K., Schrimsher D. W., Isakeit T., Alabi O. J., et al. (2021). Cotton Leafroll Dwarf Virus US genomes comprise divergent subpopulations and harbor extensive variability. Viruses 13, 2230. doi: 10.3390/v13112230
Scholthof H. (2006). The Tombusvirus-encoded P19: from irrelevance to elegance. Nat. Rev. Microbiol. 4, 405–411. doi: 10.1038/nrmicro1395
Sharma P., Ikegami M. (2010). Tomato leaf curl Java virus V2 protein is a determinant of virulence, hypersensitive response and suppression of posttranscriptional gene silencing. Virology 396, 85–93. doi: 10.1016/j.virol.2009.10.012
Silva T., Corrêa R., Castilho Y., Silvie P., Bélot J.-L., Vaslin M. (2008). Widespread distribution and a new recombinant species of Brazilian virus associated with cotton blue disease. Virol. J. 5, 123. doi: 10.1186/1743-422X-5-123
Sinha K. V., Das S. S., Sanan-Mishra N. (2021). Overexpression of a RNA silencing suppressor, B2 protein encoded by Flock House virus, in tobacco plants results in tolerance to salt stress. Phytoparasitica 49, 299–316. doi: 10.1007/s12600-020-00847-y
Smirnova E., Firth A. E., Miller W. A., Scheidecker D., Brault V., Reinbold C., et al. (2015). Discovery of a small non-AUG-initiated ORF in Poleroviruses and Luteoviruses that is required for long-distance movement. PloS Pathog. 11, e1004868. doi: 10.1371/journal.ppat.1004868
Sõmera M., Fargette D., Hébrard E., Sarmiento C., ICTV report consortium (2021). ICTV virus taxonomy profile: Solemoviridae 2021. J. Gen. Virol. 102, 001707. doi: 10.1099/jgv.0.001707
Tabassum A., Bag S., Suassuna N. D., Conner K. N., Chee P., Kemerait R. C., et al. (2021). Genome analysis of cotton leafroll dwarf virus reveals variability in the silencing suppressor protein, genotypes and genomic recombinants in the USA. PloS One 16, e0252523. doi: 10.1371/journal.pone.0252523
Tabassum A., Roberts P. M., Bag S. (2020). Genome sequence of Cotton Leafroll Dwarf Virus infecting cotton in Georgia, USA. Microbiol. Resour Announc 9, e00812–e00820. doi: 10.1128/MRA.00812-20
Tarazi R., Vaslin M. F. S. (2022). The viral threat in cotton: how new and emerging technologies accelerate virus identification and virus resistance breeding. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.851939
Thiessen L. D., Schappe T., Zaccaron M., Conner K., Koebernick J., Jacobson A., et al. (2020). First report of Cotton Leafroll Dwarf Virus in cotton plants affected by cotton leafroll dwarf disease in North Carolina. Plant Dis. 104, 3275. doi: 10.1094/PDIS-02-20-0335-PDN
Voinnet O., Vain P., Angell S., Baulcombe D.C. (1998). Systemic spread of sequence-specific transgene RNA degradation in plants is initiated by localized introduction of ectopic promoterless DNA. Cell 95, 177–187. doi: 10.1016/S0092-8674(00)81749-3
Voinnet O., Pinto Y. M., Baulcombe D. C. (1999). Suppression of gene silencing: A general strategy used by diverse DNA and RNA viruses of plants. Proc. Natl. Acad. Sci. U.S.A. 96, 14147–14152. doi: 10.1073/pnas.96.24.14147
Wang L., Fan P., Jimenez-Gongora T., Zhang D., Ding X., Medina-Puche L., et al. (2022a). The V2 protein from the Geminivirus Tomato Yellow Leaf Curl Virus largely associates to the endoplasmic reticulum and promotes the accumulation of the viral C4 protein in a silencing suppression-independent manner. Viruses 14, 2804. doi: 10.3390/v14122804
Wang H., Greene J., Mueller J., Conner K., Jacobson A. (2020). First report of Cotton Leafroll Dwarf Virus in cotton fields of South Carolina. Plant Dis. 104, 2532. doi: 10.1094/PDIS-03-20-0635-PDN
Wang Y., Liu H., Wang Z., Guo Y., Hu T., Zhou X. (2022). P25 and P37 proteins encoded by firespike leafroll-associated virus are viral suppressors of RNA silencing. Front. Microbiol. 13, 964156. doi: 10.3389/fmicb.2022.964156
Wang L., Tan H., Medina-Puche L., Wu M., Gomez B. G., Gao M., et al. (2022b). Combinatorial interactions between viral proteins expand the potential functional landscape of the tomato yellow leaf curl virus proteome. PloS Pathog. 18, e1010909. doi: 10.1371/journal.ppat.1010909
Wang L., Tian P., Yang X., Zhou X., Zhang S., Li C., et al. (2021). Key amino acids for Pepper Vein Yellows Virus P0 protein pathogenicity, gene silencing, and subcellular localization. Front. Microbiol. 12, 1653–1653. doi: 10.3389/fmicb.2021.680658
Xiao Q., Li W., Kai Y., Chen P., Zhang J., Wang B. (2019). Occurrence prediction of pests and diseases in cotton on the basis of weather factors by long short term memory network. BMC Bioinf. 20, 688. doi: 10.1186/s12859-019-3262-y
Xiao-Yan Z., Yuan-Yuan L., Wang Y., Jia-Lin Y., Cheng-Gui H. (2021). Comparative analysis of biological characteristics among P0 proteins from different Brassica Yellows Virus genotypes. Biology 10, 1076. doi: 10.3390/biology10111076
Zhuo T., Li Y.-Y., Xiang H.-Y., Wu Z.-Y., Wang X.-B., Wang Y., et al. (2014). Amino acid sequence motifs essential for P0-mediated suppression of RNA silencing in an isolate of potato leafroll virus from Inner Mongolia. Mol. Plant Microbe Interact. 27, 515–527. doi: 10.1094/MPMI-08-13-0231-R
Keywords: plant virus, CLRDV, P0, RNA silencing, cotton, pathogenicity
Citation: Akinyuwa MF, Price BK, Martin KM and Kang S-H (2023) A newly isolated cotton-infecting Polerovirus with cryptic pathogenicity encodes a weak suppressor of RNA silencing. Front. Agron. 5:1235168. doi: 10.3389/fagro.2023.1235168
Received: 05 June 2023; Accepted: 07 July 2023;
Published: 24 July 2023.
Edited by:
Saptarshi Ghosh, University of Florida, United StatesReviewed by:
Ana Julia Distefano, CONICET Institute of Agrobiotechnology and Molecular Biology (IABIMO), ArgentinaSayanta Bera, University of Maryland, College Park, United States
Copyright © 2023 Akinyuwa, Price, Martin and Kang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sung-Hwan Kang, c2hrMDAxNUBhdWJ1cm4uZWR1
 Mary F. Akinyuwa
Mary F. Akinyuwa Bailee K. Price2
Bailee K. Price2 Kathleen M. Martin
Kathleen M. Martin Sung-Hwan Kang
Sung-Hwan Kang