
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Agron. , 18 April 2023
Sec. Weed Management
Volume 5 - 2023 | https://doi.org/10.3389/fagro.2023.1182528
This article is part of the Research Topic Herbicide Efficacy View all 6 articles
Herbicide resistance within Poa annua is widespread in managed turfgrass systems. In 2020, a P. annua collection from a golf course in the southeastern United States was reported to be resistant to indaziflam as well as six other mode-of-action groups. This first report in 2020 suggests that turfgrass managers would benefit from a bioassay to screen other collections with putative indaziflam resistance. A dose-response experiment was conducted with ten concentrations of indaziflam (0, 250, 500, 667, 1000, 1143, 1333, 2000, 4500, and 9000 pM) in Gelrite® culture during 2021 and 2022. An herbicide-susceptible (S1) collection of P. annua, a resistant standard (Site 3A), and a collection with putative resistance to indaziflam (Site 18) were included in this experiment. Petri dishes were filled with 80 mL of Gelrite® (3.75 g L−1) containing technical grade (≥ 98%) indaziflam and rifampicin (1000 µg mL−1). Each plate was sealed with parafilm after placing 15 seeds of a single collection on the Gelrite® surface. At 14 days after seeding (DAS), the length of the radicle (mm) protruding from each seed was recorded with digital calipers. Indaziflam concentrations required to reduce root growth by 70% (EC70) were calculated via non-linear regression. Statistically significant differences were detected among P. annua collections with the EC70 values for the herbicide-susceptible collection measuring 708 pM [95% confidence interval (CI) = 656 to 764 pM] compared to 2130 pM (CI = 1770 to 2644 pM) for Site 3A and 4280 pM (CI = 3464 to 5442) for Site 18. Given that resistant collections exhibited longer root length in the absence of herbicide, confocal microscopy analysis was used to explore differences in root cell count among resistant and susceptible P. annua collections; however, few differences in cell count were detected. Overall, these findings indicate that a discriminatory dose of 708 pM (95% CI = 656 to 764 pM) can be used to differentiate among susceptible and resistant P. annua collections from field sites where poor control is observed following broadcast applications of indaziflam.
Poa annua is an allotetraploid species within the Poaceae family that resulted from a hybridization event between the diploid species Poa supina (male parent) and Poa infirma (female parent) (Huff et al., 2003; Mao and Huff, 2017). P. annua can be found on all seven continents globally (Chwedorzewska, 2008) and is referred to by various common names, including: annual meadowgrass, annual bluegrass, wintergrass, junegrass, suffolkgrass, speargrass, and walkgrass (Gibeault, 1971). Morphological diversity among P. annua is wide-ranging and its characterization as an annual species has been questioned, given that it does not exhibit monocarpic senescence (Carroll et al., 2021a; Carroll et al., 2022).
P. annua is the most troublesome weed of turfgrass (Van Wychen, 2020), commonly infesting golf courses, sports fields, and lawns. A survey conducted by the Golf Course Superintendents Association of America reported that P. annua was the third most abundant species found on United States golf courses in 2015, behind Cynodon spp. and Poa pratensis L. (Gelernter et al., 2017). Sole reliance on herbicides to control P. annua infestations in turfgrass has resulted in extensive selection for herbicide-resistant biotypes (Brosnan et al., 2020a), including herbicidal inhibitors of acetolactate synthase (ALS; Cross et al., 2013; McElroy et al., 2013; Brosnan et al., 2015; Brosnan et al., 2016; Singh et al., 2021), cellular mitosis (Isgrigg et al., 2002; Cutulle et al., 2009; Brosnan et al., 2014; McCullough et al., 2017; Singh et al., 2021), photosystem II (PSII, Kelly et al., 1999; Mengistu et al., 2000; Hutto et al., 2004; Brosnan et al., 2020b), 5-enolpyruvate-shikimate-3-phosphate-synthase (Binkholder et al., 2011; Brosnan et al., 2012; Cross et al., 2015; Breeden et al., 2017), and protoporphyrinogen oxidase (Yu et al., 2018). Multiple resistance in P. annua is an emerging concern to turfgrass managers as these populations become uncontrollable by chemical methods (Brosnan et al., 2020a). Reports of P. annua with resistance to ALS and PS-II inhibitors as well as pre- and postemergence (prodiamine and glyphosate, respectively) herbicides have been documented on golf courses in the United States transition zone (Brosnan et al., 2016; Breeden et al., 2017). Multiple resistance to five modes of action was identified in P. annua from an Australian golf course (Barua et al., 2020). In Texas, Singh et al. (2021) identified a population of P. annua resistant to simazine, amicarbazone, trifloxysulfuron, foramsulfuron, and preemergence applications of pronamide.
The possibility of simultaneous co-evolution of non-target and target-site resistance within the self-pollinating species P. annua has been explored (Laforest et al., 2021). In working with a P. annua collection that contained target site mutations conferring resistance to both PS-II and ALS-inhibiting herbicides, Laforest et al. (2021) confirmed that treatment with an ALS-inhibiting herbicide (trifloxysulfuron) resulted in differential expression of genes associated with non-target site resistance, particularly those associated with oxidative stress. Interestingly, resistant plants also exhibited constitutive expression of genes regulating transmembrane transport prior to treatment. The researchers suggested that continued use of an ALS-inhibitor after evolution of target site resistance may lead to an accumulation of resistance mechanisms affecting other modes of action. For example, P. annua plants characterized by Laforest et al. (2021) were less affected by applications of flumioxazin, indaziflam, and pronamide than herbicide-susceptible controls.
Cellulose biosynthesis inhibiting herbicides (HRAC Group 29) target the cellulose synthase complex (CSC) where cellulose polymers are synthesized and then released to the cell wall (Mutwil et al., 2008; Bashline et al., 2015). Within the CSC, 18 to 24 catalytic cellulose synthase A (CESA) proteins could be targets of cellulose biosynthesis inhibiting herbicides (Jarvis, 2013; Brabham et al., 2014; Tateno et al., 2016). Inhibiting the function of any of these proteins leads to radial root swelling caused by the loss of anisotropic growth in cells undergoing expansion (Brabham et al., 2014). Indaziflam is an alkylazine inhibitor of cellulose biosynthesis with an unknown site-of-action. Brabham et al. (2014) reported that indaziflam increased CESA particle density in the plasma membrane and inhibited polymerization by reducing CESA particle velocity by 65% (Brabham et al., 2014). Indaziflam is also a unique herbicide as it is efficacious on both monocots and dicots, whereas other Group 29 herbicides (e.g., isoxaben) only exhibit dicot activity (Sabba and Vaughn, 1999). Furthermore, plants with target-site resistance to isoxaben are not cross-resistant to indaziflam, indicating a unique site of action (Brabham et al., 2014). When indaziflam is applied to a susceptible plant, radial root swelling and ectopic lignification will occur leading to chlorosis and eventual mortality (Brabham et al., 2014). In Arabidopsis, indaziflam inhibited cellulose production within one hour of treatment in a dose-dependent manner (Brabham et al., 2014).
Indaziflam is a lipophilic herbicide with low water solubility, a dissociation constant (pKa) of 3.5, an octanol/water partition coefficient of 2.8 at pH 7, and residual activity for controlling annual weeds in soil (EPA, 2010; Sebastian et al., 2017). In managed turfgrass, indaziflam (Specticle® FLO. Bayer Environmental Sciences, St. Louis, MO) is used to control problematic grassy weeds such as P. annua, Eleusine indica, and Digitaria spp., as well as annual sedges and broadleaf weeds. Considering that few Group 29 herbicides are used for weed control in turfgrass, indaziflam offered turfgrass managers a new mode of action for managing P. annua populations that have evolved resistance to other modes of action (Brosnan et al., 2014; Brosnan et al., 2015; Brosnan et al., 2017a). For example, a P. annua population with target site resistance to PS-II inhibiting herbicides was controlled ≥ 98% with indaziflam (Brosnan et al., 2017a). Similarly, indaziflam effectively controlled P. annua with resistance to mitotic and ALS-inhibiting herbicides (Brosnan et al., 2014; Brosnan et al., 2015).
The first case of any plant evolving resistance to indaziflam was identified in P. annua collected from a golf course and confirmed via greenhouse and laboratory bioassays (Brosnan et al., 2020c). Plants were identified and collected after escaping preemergence applications of indaziflam (Specticle® FLO. Bayer Environmental Sciences. St. Louis, MO) at maximum label rate (48.7 g ha−1) in the field. P. annua surviving indaziflam treatment (applied early-postemergence to non-tillered plants< 2.5 cm height) in greenhouse and laboratory bioassays also exhibited extensive multiple resistance to several other herbicide mode-of-action groups, including #2 (foramsulfuron), #3 (pronamide), #5 (metribuzin and simazine), #9 (glyphosate), #10 (glufosinate), and #14 (flumioxazin). Inhibitors of cytochrome P450 monooxygenase, including 1-aminobenzotriazole (10 mg L−1), tebuconazole (1510 g ha−1), or malathion (400 g ha−1) could not restore indaziflam susceptibility in these P. annua collections suggesting that metabolism may not be the sole mechanism of resistance in these plants. Interestingly, Brosnan et al. (2020c) demonstrated that concentrations of indaziflam and 1-fluoroethyl triazinediamine (the primary metabolite of indaziflam) were lower in resistant plants compared to susceptible controls and suggested that altered herbicide absorption may be a mechanism of resistance to be further explored in these collections.
Evolution of indaziflam resistance within P. annua is concerning considering that the herbicide helped turfgrass managers control infestations with resistance to other mode of action groups (Brosnan et al., 2014; Brosnan et al., 2015, Brosnan et al., 2017a). There is a need to develop a bioassay to rapidly screen many collections of P. annua for resistance to indaziflam. A gel-based, in vitro, bioassay could help turfgrass managers understand how plants will respond to indaziflam treatment in as few as 14-days compared to the 42 days required for whole-plant testing in a soil medium (Brosnan et al., 2020c). Similar bioassays have been developed for other herbicidal mode of action groups with an array of plant species, including P. annua (Bourgeois et al., 1997; Claude et al., 2004; Cutulle et al., 2009; Kaundun et al., 2011; Kaundun et al., 2014; Brosnan et al., 2017b; Messelhäuser et al., 2021). Moreover, a gel bioassay could identify indaziflam resistance regardless of mechanism (Kaundun, 2021), which is important given that the indaziflam target site is currently unknown. This research aims to develop a bioassay to determine P. annua responses to indaziflam that can be used to discern amongst resistant and susceptible accessions.
Laboratory research was conducted at the University of Tennessee in 2021 and 2022 to develop a bioassay to screen P. annua germplasm response to indaziflam. Herbicide susceptible (S1) and one resistant biotype (Site 3A) used in this experiment were the same as those described by Brosnan et al. (2020c). Collections S1 and Site 3A are from University Park, PA and a golf course fairway in Cummings, GA respectively. Additionally, a collection of P. annua with putative resistance to indaziflam (Site 18) was also included. This collection was submitted to the University of Tennessee Weed Diagnostics Center after escaping treatment with preemergence applications of indaziflam (Specticle® FLO; Bayer Environmental Sciences, St. Louis, MO) on a golf course fairway in Thomasville, GA. Plants were cultured in a glasshouse to produce seed required for research. Plants were placed in greenhouse pots filled with a peat moss growing media (PRO MIX BX General Purpose; Premier Tech Horticulture, Quakertown, PA, USA) and treated with complete fertilizer (20 N–20 P2O5-20 K2O. Peter’s 20–20–20. JR PETERS, Incorporated. Allentown, PA, USA) at 49.5 kg N ha−1 wk−1. Mature seed from each collection were harvested with scissors, dried in a forced-air oven (Laboratory Oven; The Grieve Corporation, Round Lake, IL), and sieved (Standard Test Sieve; Fisher Scientific Company LLC, Hampton, NH) to remove chaff. After processing, seed were stored in glass vials (40 mL Scintillation Vial; Thermo-Fisher Scientific, St. Louis, MO, USA) at −20˚C for a minimum of four weeks to break any potential dormancy (Carroll et al., 2021b).
A natural gelling agent (Gelrite®; Duchefa Biochemie, Haarlem, The Netherlands) was used to culture plants in this research. This agent was prepared by adding 7.5 g Gelrite® to 8.9 g MS Salts & Vitamins (Murashige & Skoog M519 Basal Medium with Vitamins; Phyto Tech Labs, Lenexa, KS) in 1966 mL of distilled H2O in sterilized Erlenmeyer flasks; MS Salts & Vitamins was added to distilled H2O prior to the Gelrite®. The pH of this solution was measured with a pH conductivity meter (Multimeter Model 250; Denver Instrument, Bohemia, NY) and adjusted to 6.5 by adding either hydrochloric acid (HCl) or sodium hydroxide (NaOH) to lower or raise the pH, respectfully. The resultant solution went through a liquid 20 cycle that raised temperature to 121˚C in an autoclave (Amsco Century V120; Steris, Mentor, OH) for 20 minutes. After the autoclave cycle was finished, sealed flasks were placed on a hot plate (FisherBrand Stirring Hot Plate; Thermo Orion Incorporated, Chelmsford, MA) and stirred until cooling to 80˚C. Rifampicin (1000 µg mL−1; Rifamipicin Molecular Biology Reagent; Alfa Aesar, Ward Hill, MA) and technical indaziflam (≥ 98%; Sigma-Aldrich, St. Louis, MO) were added at this temperature to create ten indaziflam concentrations for evaluation: 0, 250, 500, 667, 1000, 1143, 1333, 2000, 4500, 9000 picomolar (pM). These concentrations were selected based results of Brosnan et al. (2020c) who reported that early-postemergence exposure to 633 pM indaziflam in agar culture could differentiate between indaziflam resistant and susceptible collections of P. annua. Technical indaziflam (150 mg) was diluted in 99.5 mL dimethyl sulfoxide with a concentration of 5000 µM. This stock solution was frozen to keep concentrations uniform across all trials. On the day of trial initiation, this bottle was allowed to thaw to facilitate adding the appropriate amount of indaziflam to each flask for each concentration. Serial dilutions using deionized H2O were used to deliver intended pM concentrations of indaziflam to each autoclaved solution.
After adding rifampicin and indaziflam, 80 mL of the resultant solution was added to square petri dishes (120 x 15 mm) within a sterile fume hood (1300 Series A2 Fume Hood; Thermo-Fisher Scientific, St. Louis, MO, USA) using a dispensette (Dispensette S Bottle Top Dispenser; BrandTech Scientific Incorporated, Essex, CT). Plates were left open in the fume hood for 12 min in order to cool before being covered, sealed with parafilm (2 IN All-Purpose Laboratory Film; Amcor, Menasha, WI), and placed in a growth chamber (G1000-Germinator; Conviron, Winnipeg, Manitoba, Canada) set to constant 16˚C temperature and 16 h photoperiod. Plates were unsealed to facilitate adding P. annua seed (15 seeds, one collection per plate) to the Gelrite® surface using sterilized tweezers; seed were pressed into the Gelrite® surface with sterilized spatulas after plating. Each petri dish was then re-sealed with parafilm after seeding and placed in the aforementioned growth chamber at a constant temperature of 16˚C.
Fourteen days after seeding (DAS), responses to indaziflam were evaluated by assessing the presence (or absence) of root or shoot growth (i.e., 1 = yes, 0 = no). Additionally, the length of the radicle (mm) protruding from each seed was measured via digital calipers (CD-6” AX Digimatic Caliper; Mitutoyo Corporation, Kawasaki, Kanagawa, Japan). The experiment was arranged in a completely randomized design with five replications of each indaziflam concentration. Within a given concentration, the 15 seeds per collection were considered sub-samples and used to calculate mean root length values for each P. annua collection that were expressed as a percentage of the non-treated. The experiment was repeated as previously described in January 2022 using five replications of each indaziflam concentration and 15 sub-samples. Data from each experimental run were combined following ANOVA in R (v. 3.6.2) with root length values subjected to non-linear regressions in Prism (Version 9.0.0. GraphPad Prism. La Jolla, CA.) using the ‘EC Anything’ model described below:
In this model, Bottom and Top represent asymptotes that were constrained to 0 and 100 percent, respectively. Y represented root growth as a percentage of the non-treated, and HillSlope represented the steepness of the curve. X represented the indaziflam concentration to reduce root length 70% (EC70). Best-fit parameters for modeled responses were compared using a global sums-of-squares F-test at α = 0.05 with 95% confidence intervals used to compared EC70 concentrations of indaziflam among resistant and susceptible P. annua collections. EC70 values were used to discern amongst potential resistant and susceptible accessions to provide a more stringent filter than EC50 values commonly used in resistance screening (Burgos et al., 2013).
Roots from indaziflam-resistant (Site 3A, Site 18) and -susceptible collections used in Gelrite® assay development were analysed using microscopy to evaluate the cause of apparent increased root length in indaziflam resistant populations. Additional indaziflam-resistant (Site 3B, FL) collections submitted to the University of Tennessee Weed Diagnostics Center were also included, along with an additional indaziflam-susceptible collection (S5) originating from Wake Forest, NC to better support this observation. All seeds were sterilized using 70% ethanol for 5 min, followed by 40 min exposure to a solution of 10% bleach + 0.01% triton X-100, followed by three rinses with distilled water. Plants were grown in vertical, square (10 cm x 10 cm), petri plates, with 1% agar + ½ strength Murashige & Skoog M519 Basal Medium, similar to Brosnan et al. (2020c). Preliminary germination assays showed variability among resistant and susceptible collections. To that end, plates were grown in the dark for three days before being exposed to light (16/8 day/night cycle) in the growth chamber in order to synchronize root emergence.
Five days after light exposure, confocal root tissue images for each P. annua collection were taken using an inverted laser scanning microscope (Nikon-A1RSi. Nikon Instruments. Melville, NY). The microscope was configured using a 20x Plan Apo objective (NA 0.75) and 10X ocular lenses for a total magnification of 200x. A z-series with a step size of 1.9 um was taken for all root images. A 32 μM stock solution of N-(3-triethylammoniumpropyl)-4-(6-(4-(diethylamino) phenyl) hexatrienyl) pyridinium dibromide dye (FM4-64. Invitrogen. Waltham, MA) was applied to root tissue for 3 min and then washed (three times) using distilled water to stain cell membranes. Root tissue from each seedling was placed on a glass slide and seperated from shoot tissue with a scalpel. Post-acquisition image analysis was performed using ImageJ (Schneider et al., 2012) equipped with a cell counter plugin. Images were enhanced using enhance contrast function of Fiji (Schindelin et al., 2012) using a saturated pixels value of 0.35% and the equalize histogram function. The depth of root tissue selected for analysis was determined by drawing five segmented lines of 100 pixels (1 micron = 1.6 pixels, 500 pixels = 312.5-micron length) from the quiescent center of the root tip; the number of cells was counted from quiescent layer to the end of line within the outermost anterior and posterior epidermal layers of each root. Cell count data from each P. annua collection were subjected to a one-way ANOVA using Minitab software (Minitab Statistical Software, 2010) with mean values for each collection compared using a pairwise T-test at α = 0.05.
Significant P. annua collection-by-indaziflam concentration interactions were detected in root length data from each experimental run; therefore, data from both were combined before being subjected to non-linear regression analysis. A non-linear model fit the combined data set with a global sums-of-squares F-test detecting highly significant (F = 117, P< 0.0001) differences among P. annua collections (Figure 1).
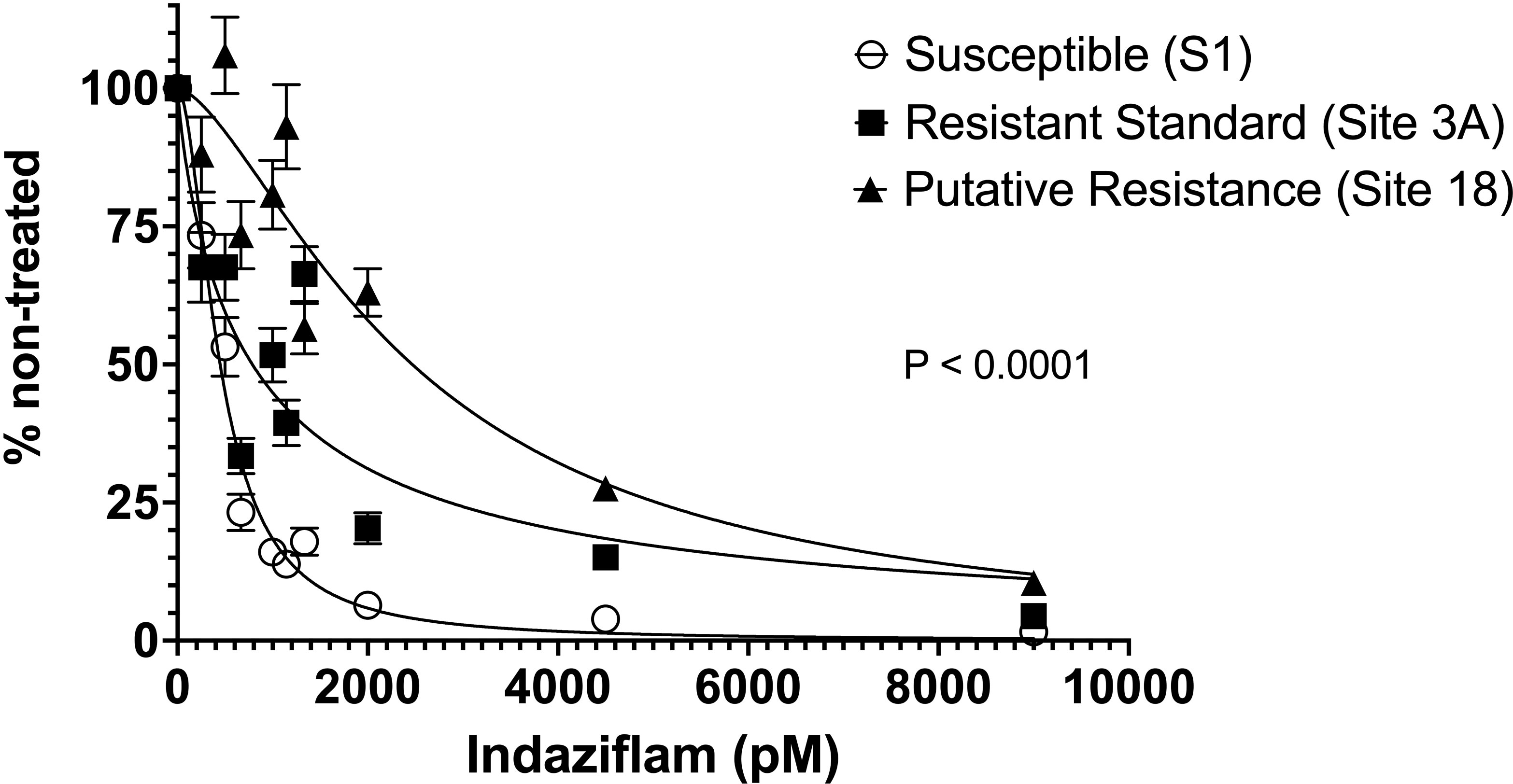
Figure 1 Root length of P. annua collections subjected to increasing concentrations of indaziflam (0, 250, 500, 667, 1000, 1143, 1333, 2000, 4500, and 9000 pM) in Gelrite® culture for 14 days. Root length (mm) measurements were expressed as a percentage of the non-treated (0 pM) for each collection. Error bars presented illustrate the standard error of the mean for each collection (N = 150).
While root length reduced for all P. annua collections as indaziflam concentration increased, there were significant differences in the concentration of indaziflam required to reduce root length 70% (EC70) among P. annua collections in this experiment (Table 1). EC70 values for the herbicide-susceptible collection measured 708 pM [95% confidence interval (CI) = 656 to 764 pM] compared to 2130 pM (CI = 1770 to 2644 pM) for Site 3A and 4280 pM (CI = 3464 to 5442) for Site 18 (Table 1). These values can be used to calculate resistance indices (i.e., R/S ratios) of 3 and 6 for the Site 3A and Site 18 P. annua collections, respectively. Low-level resistance within the P. annua collections evaluated in this study provides further evidence that non-target site mechanisms may be affecting indaziflam efficacy given that non-target site mechanisms are often continuous as opposed to discreet for target-site mutations (Tranel and Wright, 2002; Yu and Powles, 2014).
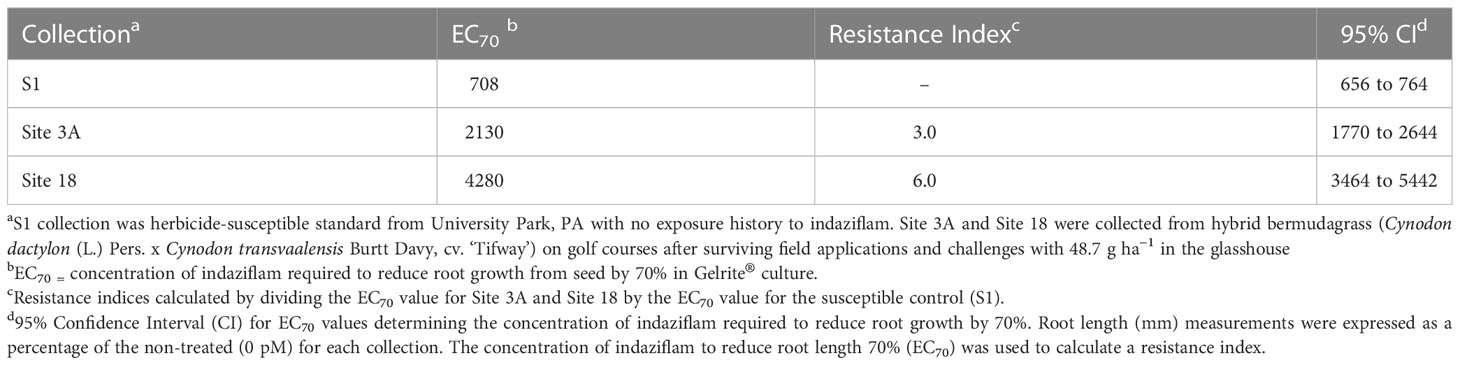
Table 1 P. annua collections subjected to increasing doses of indaziflam (0, 250, 500, 667, 1000, 1143, 1333, 2000, 4500, and 9000 pM) in Gelrite® culture for 14 days.
Root length values for both indaziflam-resistant collections (Site 3A and Site 18) were greater than the susceptible P. annua collection used in this study (Table 2). Microscopy analysis was conducted in order to explore possible differences at the cellular level between the P. annua collections. We hypothesized that enhanced root cell division in indaziflam-resistant collections may be causing the constitutive differences between indaziflam-resistant and indaziflam-susceptible collections in control plates (0 pM indaziflam). Cell count data from indaziflam-resistant and susceptible P. annua collections are presented in Figure 2 with sample microscopy images used for analysis presented in Figure 3. No significant differences were detected in cell number (in the zone of cell division) among the indaziflam-resistant and -susceptible P. annua collections included in this experiment.
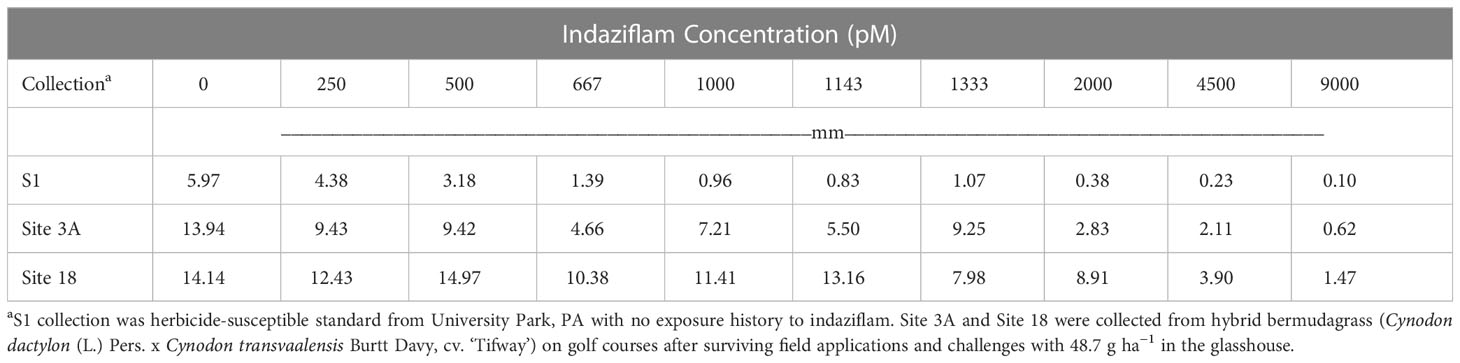
Table 2 Mean root length (mm) data for indaziflam-resistant (Site 3A and Site 18) and -susceptible (S1) collections of P. annua in laboratory assays evaluating root growth in in Gelrite® culture for 14 days.
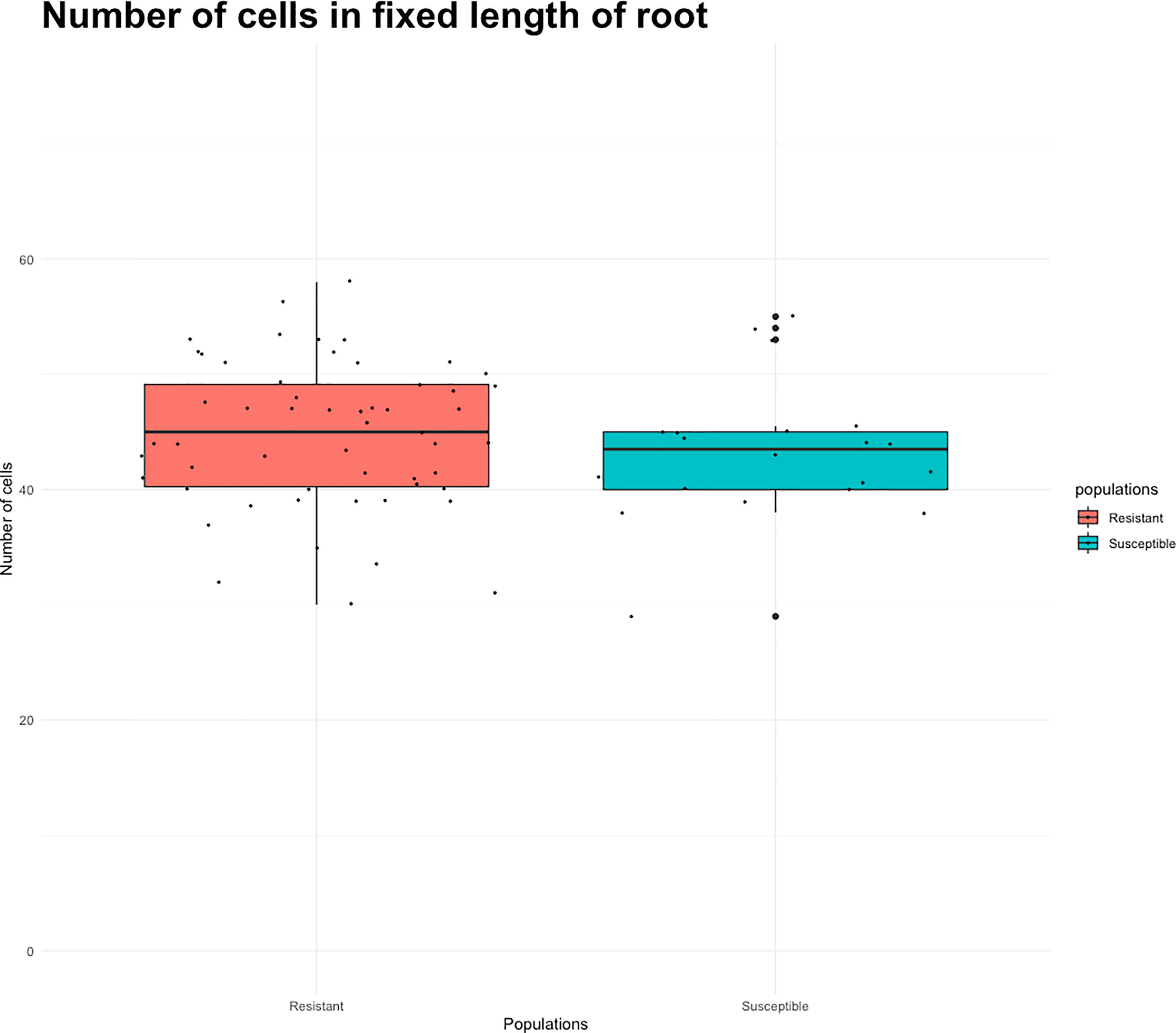
Figure 2 Root cell count data from indaziflam-resistant (Sites 3A, 3B, S18, FL) and indaziflam-susceptible (S1, S5) P. annua collections. Root length was measured at fixed length from the quescient tip of the roots.
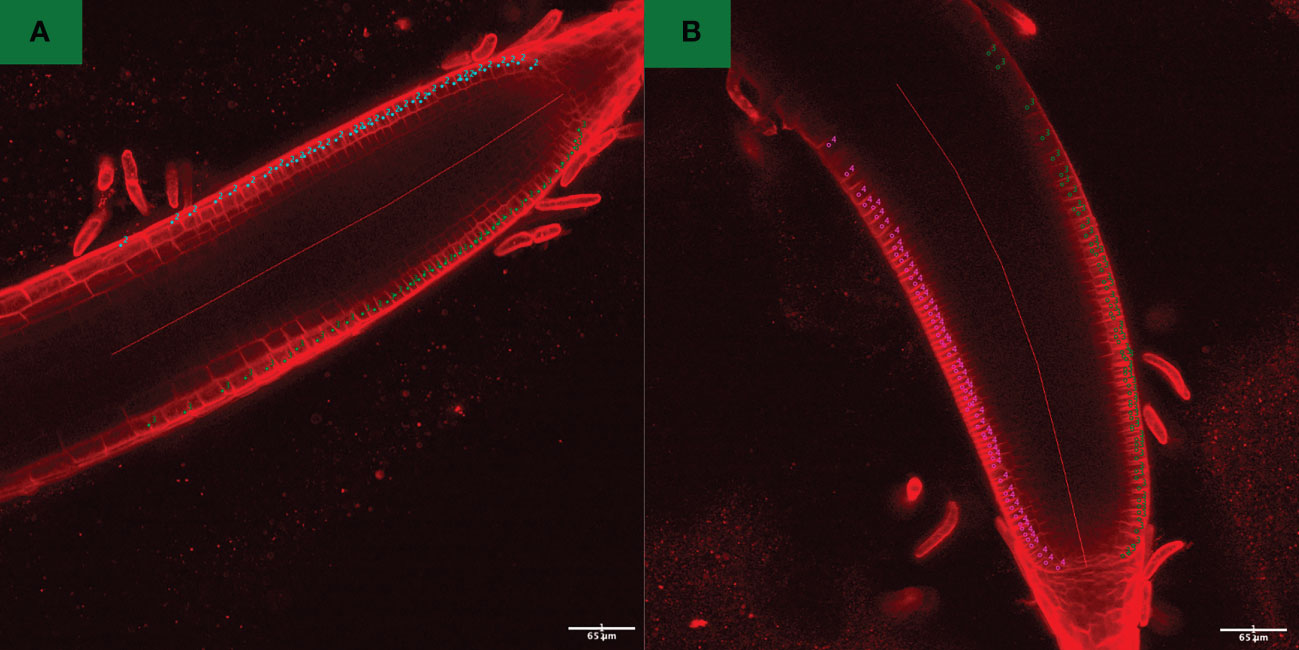
Figure 3 Confocal fluorescence images of (A) indaziflam-resistant (Site 18) and (B) indaziflam-susceptible (S1) P. annua, treated with FM4-64 dye and post-acquisition processed using FIJI function enhance contrast, with equalize histogram enabled. Cell counter plugin was utilized for counting the number of cells in the fixed length (500 pixels) from the quescient center of roots. These count data are presented in Figure 2.
Indaziflam resistance indices in the current study align with previous research using Site 3A and S1 P. annua germplasm (Brosnan et al., 2020c); the researchers determined indaziflam concentrations required to reduce root growth of newly emerged P. annua plants 50% (I50 values) and calculated a resistance index of 6.8 for Site 3A. The difference in resistance factor for collection Site 3A between experiments can most likely be attributed to differences in plant maturity at the time of indaziflam exposure. Brosnan et al. (2020c) calculated a resistance index following early-postemergence indaziflam exposure to emerged seedlings (a labeled use pattern for indaziflam in turfgrass; Anonymous, 2023), whereas preemergence exposure was evaluated in this research. Additionally, EC70 values were used to calculate resistance indices in the current study compared to EC50 values commonly used by other researchers (Cutulle et al., 2009; Kaundun et al., 2014; Brosnan et al., 2020c). Given that there is only a single report of indaziflam resistance in published literature to date (Brosnan et al., 2020c), EC70 values were selected to provide a more stringent filter among putative resistant and susceptible P. annua collections. A limitation of this research is that only a single susceptible collection was included in the dose-response experiments. Future research to refine this assay using additional susceptible P. annua collections is warranted.
A collection of P. annua with putative resistance to indaziflam (Site 18) was included in this research after escaping treatment with preemergence applications of indaziflam on a golf course fairway. P. annua from Site 18 exhibited nearly 2x the level of indaziflam resistance than Site 3A, the resistant standard used in the experiment (Table 1, Figure 1). This represents the highest level of indaziflam resistance reported in the peer-reviewed scientific literature (to date) and a first report of resistance to indaziflam applied preemergence.
Evolution of indaziflam resistance in P. annua is concerning given that many turfgrass managers have effectively used this herbicide to control P. annua biotypes that evolved resistance to other modes of action, including those with multiple resistance. After screening 1,500 plants across ten concentrations, 708 pM indaziflam (95% confidence interval of 656 to 764 pM) consistently separated resistant collections from a susceptible control in gel culture. Sebastian et al. (2017) reported that 251 pM indaziflam reduced root growth of feral rye (Secale cereale L.) in agar culture by 50%. In the current study, 448 pM of indaziflam was required to yield a similar reduction in root growth of herbicide susceptible P. annua in gel culture. The discriminatory concentration identified in this research successfully identified other accessions of P. annua with putative resistance to indaziflam that escaped control in the field (Figure S1). This assay could be used to determine if indaziflam can be a component of an effective herbicide mixture for P. annua control or potentially be expanded to screen response of P. annua accessions to herbicide mixtures. Hulme (2022) explored multiple resistance across an array of weed species and reported that certain mode of action groups cluster together, with a main cluster including ALS and PS-II inhibitors as well as glyphosate. Given widespread reports of resistance to these herbicides in P. annua, challenging resistant plants with indaziflam using this assay would help determine if the herbicide could be a component of an effective mixture for resistance management or falls within the same multiple resistant cluster.
In addition, it is worth noting that root length values for both indaziflam-resistant collections used in this study (Site 3A and Site 18) were significantly greater (>2x) than the susceptible P. annua collection under no indaziflam treatment (Table 2). Microscopy analysis did not show an increased number of cells in the zone of division for these resistant plants suggesting that these differences are not the result of increased cell division. It may be that cells in the resistant plants are elongating more and/or faster than susceptible plants, but this was not explored. Future research exploring this and other growth phenotypes associated with mechanisms of indaziflam resistance in P. annua is warranted as the resistance mechanism may cause or be linked with traits that may be disadvantageous for resistant plants.
A gel bioassay was developed to discern P. annua responses to indaziflam in a 14-day period once seed is available. Comparatively, traditional screening via greenhouse pot studies can require months to complete. This novel bioassay allows turfgrass managers to screen P. annua plants for indaziflam resistance within the same season of management to determine if indaziflam will be an effective component of their P. annua management plans. This bioassay will also aid those challenged with indaziflam resistant P. annua in other cropping systems. For example, indaziflam resistance in P. annua was recently reported in hazelnut (Corylus avellana L.) orchard production (Miranda and Moretti, 2023). One P. annua collection screened using this assay (Site 18) exhibited the highest level of indaziflam resistance reported in the peer-reviewed scientific literature (to date) and serves as a first report of resistance to indaziflam applied preemergence. Future work should be conducted to determine the mechanism of resistance in the indaziflam-resistant P. annua collections described in this research. Doing so would allow for molecular assays to identify indaziflam resistance to be developed that could return results even more rapidly.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Dose-response experiments were designed by JB, EP, and BP and conducted by BP and JV. Analysis of dose-response data and supplemental information was conducted by JB and BP. Root microscopy experiments were designed by EP, conducted by MM, with data analyzed by MM and EP. JB and EP provided insights on overall research effort. BP drafted the original manuscript, which was reviewed, edited, and revised by JB and EP. All authors agreed to the submission.
This research was partially supported by the United States Department of Agriculture – National Institute of Food and Agriculture, Award No. 2018-51181-28436, ‘Research and Extension to Address Herbicide Resistance Epidemic in Annual Bluegrass in Managed Turf Systems’. This work was also supported by Bayer Environmental Sciences, Envu Turf and Ornamentals, and the University of Tennessee Institute of Agriculture Office of AgResearch.
Authors would like to thank Robert Trigiano and Scott Senseman for the insights they provided towards this project. Additionally, authors would like to thank Devon Carroll, Brynn Johnson, and Greg Breeden for their assistance in conducting this research, as well as the University of Tennessee Department of Plant Sciences Graduate Student Association.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fagro.2023.1182528/full#supplementary-material
Anonymous (2023) Specticle FLO herbicide label. Available at: http://www.cdms.net/ldat/ldANH006.pdf (Accessed 29 January 2023).
Barua R., Boutsalis P., Malone J., Gill G., Preston C. (2020). Incidence of multiple herbicide resistance in annual bluegrass (Poa annua) across southeastern Australia. Weed Sci. 68 (4), 340–347. doi: 10.1017/wsc.2020.35
Bashline L., Li S., Zhu X., Gu Y. (2015). The TWD40-2 protein and the AP2 complex cooperate in the clathrin-mediated endocytosis of cellulose synthase to regulate cellulose biosynthesis. Proc. Natl. Acad. Sci. 112 (41), 12870–12875. doi: 10.1073/pnas.1509292112
Binkholder K. M., Fresenburg B. S., Teuton T. C., Xiong X., Smeda R. J. (2011). Selection of glyphosate-resistant annual bluegrass (Poa annua) on a golf course. Weed Sci. 59 (3), 286–289. doi: 10.1614/WS-D-10-00131.1
Bourgeois L., Kenkel N. C., Morrison I. N. (1997). Characterization of cross-resistance patterns in acetyl-CoA carboxylase inhibitor resistant wild oat (Avena fatua). Weed Sci. 45 (6), 750–755. doi: 10.1017/S0043174500088925
Brabham C., Lei L., Gu Y., Stork J., Barrett M., DeBolt S. (2014). Indaziflam herbicidal action: a potent cellulose biosynthesis inhibitor. Plant Physiol. 166 (3), 1177–1185. doi: 10.1104/pp.114.241950
Breeden S. M., Brosnan J. T., Mueller T. C., Breeden G. K., Horvath B. J., Senseman S. A. (2017). Confirmation and control of annual bluegrass (Poa annua) with resistance to prodiamine and glyphosate. Weed Technol. 31 (1), 111–119. doi: 10.1614/WT-D-16-00084.1
Brosnan J. T., Breeden G. K., Mueller T. C. (2012). A glyphosate-resistant biotype of annual bluegrass in Tennessee. Weed Sci. 60 (1), 97–100. doi: 10.1614/WS-D-11-00139.1
Brosnan J. T., Breeden G. K., Vargas J. J., Grier L. (2015). A biotype of annual bluegrass (Poa annua) in Tennessee is resistant to inhibitors of ALS and photosystem II. Weed Sci. 63 (1), 321–328. doi: 10.1614/WS-D-14-00080.1
Brosnan J. T., Breeden G. K., Vargas J. J., Trigiano R. N., Boggess S. L., Staton M. E. (2017a). Confirmation and control of annual bluegrass resistant to photosystem-II-inhibiting herbicides. Int. Turfgrass Soc. Res. J. 13 (1), 675–680. doi: 10.2134/itsrj2016.05.0359
Brosnan J. T., Elmore M. T., Bagavathiannan M. V. (2020a). Herbicide-resistant weeds in turfgrass: current status and emerging threats. Weed Technol. 34 (3), 424–430. doi: 10.1017/wet.2020.29
Brosnan J. T., Reasor E. H., Vargas J. J., Breeden G. K., Kopsell D. A., Cutulle M. A., et al. (2014). A putative prodiamine-resistant annual bluegrass (Poa annua) population is controlled by indaziflam. Weed Sci. 62 (1), 138–144. doi: 10.1614/WS-D-13-00057.1
Brosnan J. T., Vargas J. J., Breeden G. K., Grier L., Aponte R. A., Tresch S., et al. (2016). A new amino acid substitution (Ala-205-Phe) in acetolactate synthase (ALS) confers broad spectrum resistance to ALS-inhibiting herbicides. Planta 243 (1), 149–159. doi: 10.1007/s00425-015-2399-9
Brosnan J. T., Vargas J. J., Breeden G. K., Zobel J. M. (2020b). Herbicide resistance in annual bluegrass on Tennessee golf courses. Crop Forage Turfgrass Manage. 6 (1), e20050. doi: 10.1002/cft2.20050
Brosnan J. T., Vargas J. J., Reasor E. H., Viggiani R., Breeden G. K., Zobel J. M. (2017b). A diagnostic assay to detect herbicide resistance in annual bluegrass (Poa annua). Weed Technol. 31 (4), 609–616. doi: 10.1017/wet.2017.26
Brosnan J. T., Vargas J. J., Spesard B., Netzband D., Zobel J. M., Chen J., et al. (2020c). Annual bluegrass (Poa annua) resistance to indaziflam applied early-postemergence. Pest Manage. Sci. 76 (6), 2049–2057. doi: 10.1002/ps.5740
Burgos N. R., Tranel P. J., Streibig J. C., Davis V. M., Shaner D., Norsworthy J. K., et al. (2013). Confirmation of resistance to herbicides and evaluation of resistance levels. Weed Sci. 61 (1), 4–20. doi: 10.1614/WS-D-12-00032.1
Carroll D. E., Brosnan J. T., McCurdy J. D., De Castro E. B., Patton A. J., Liu W., et al. (2021b). Germinability of annual bluegrass seed during spring in the Eastern united states. Crop Forage Turfgrass Mgmt. 7, e20117. doi: 10.1002/cft2.20117
Carroll D. E., Brosnan J. T., Trigiano R. N., Horvath B. J., Shekoofa A., Mueller T. C. (2021a). Current understanding of the Poa annua life cycle. Crop Sci. 61 (3), 1527–1537. doi: 10.1002/csc2.20441
Carroll D. E., Horvath B. J., Prorock M., Trigiano R. N., Shekoofa A., Mueller T. C., et al. (2022). Poa annua: an annual species? PloS One. doi: 10.1371/journal.pone.0274404
Chwedorzewska K. J. (2008). Poa annua L. in Antarctic: searching for the source of introduction. Polar Biol. 31 (3), 263–268. doi: 10.1007/s00300-007-0353-4
Claude J. P., Didier A., Favier P., Thalinger P. P. (2004). “Development of a European database for the evolution follow-up of resistant black-grass (Alopecurus myosuroides huds.) populations in cereal crops,” in Proc. fourth international weed science congress (Durban, SA Davis: CA: International Weed Science Society), 48.
Cross R. B., McCarty L. B., Tharayil N., McElroy J. S., Chen S., McCullough P. E., et al. (2015). A Pro106 to ala substitution is associated with resistance to glyphosate in annual bluegrass (Poa annua). Weed Sci. 63 (3), 613–622. doi: 10.1614/WS-D-15-00033.1
Cross R. B., McCarty L. B., Tharayil N., Whitwell T., Bridges W. C. (2013). Detecting annual bluegrass (Poa annua) resistance to ALS-inhibiting herbicides using a rapid diagnostic assay. Weed Sci. 61 (3), 384–389. doi: 10.1614/WS-D-12-00172.1
Cutulle M. A., McElroy J. S., Millwood R. W., Sorochan J. C., Stewart C. N. Jr. (2009). Selection of bioassay method influences detection of annual bluegrass resistance to mitotic-inhibiting herbicides. Crop Sci. 49 (3), 1088–1095. doi: 10.2135/cropsci2008.05.0242
EPA. (2010). U. S Environmental protection agency pesticide fact sheet—indaziflam (Washington, DC: U.S. Environmental Protection Agency), p.108.
Gelernter W. D., Stowell L. J., Johnson M. E., Brown C. D. (2017). Documenting trends in land-use characteristics and environmental stewardship programs on US golf courses. Crop Forage Turfgrass Manage. 3 (1), 1–12. doi: 10.2134/cftm2016.10.0066
Gibeault V. A. (1971). Perenniality in Poa annua L.(Doctoral dissertation, Corvallis, OR: Oregon State University).
Huff D. R., Casler M. D., Duncan R. R. (2003). “Annual bluegrass (Poa annua L.),” in Turfgrass biology, genetics, and breeding (Hoboken, NJ: John Wiley & Sons), 39–51.
Hulme P. E. (2022). Hierarchical cluster analysis of herbicide modes of action reveals distinct classes of multiple resistance in weeds. Pest Manage. Sci. 78 (3), 1265–1271. doi: 10.1002/ps.6744
Hutto K. C., Coats G. E., Taylor J. M. (2004). Annual bluegrass (Poa annua) resistance to simazine in Mississippi. Weed Technol. 18 (3), 846–849. doi: 10.1614/WT-03-148R1
Isgrigg J., Yelverton F. H., Brownie C., Warren L. S. (2002). Dinitroaniline resistant annual bluegrass in north Carolina. Weed Sci. 50 (1), 86–90. doi: 10.1614/0043-1745(2002)050[0086:DRABIN]2.0.CO;2
Jarvis M. C. (2013). Cellulose biosynthesis: Counting the chains. Plant Physiol. 163 (4), 1485–1486. doi: 10.1104/pp.113.231092
Kaundun S. S. (2021). Syngenta’s contribution to herbicide resistance research and management. Pest Manage. Sci. 77 (4), 1564–1571. doi: 10.1002/ps.6072
Kaundun S. S., Hutchings S. J., Dale R. P., Bailly G. C., Glanfield P. (2011). Syngenta ‘RISQ’test: a novel in-season method for detecting resistance to post-emergence ACCase and ALS inhibitor herbicides in grass weeds. Weed Res. 51 (3), 284–293. doi: 10.1111/j.1365-3180.2011.00841.x
Kaundun S. S., Hutchings S. J., Harris S. C., Jackson L. V., Shashi-Kiran R., Dale R. P., et al. (2014). A simple in-season bioassay for detecting glyphosate resistance in grass and broadleaf weeds prior to herbicide application in the field. Weed Sci. 62 (4), 597–607. doi: 10.1614/WS-D-14-00046.1
Kelly S. T., Coats G. E., Luthe D. S. (1999). Mode of resistance of triazine-resistant annual bluegrass (Poa annua). Weed Technol. 13 (4), 747–752. doi: 10.1017/S0890037X00042172
Laforest M., Soufiane B., Patterson E. L., Vargas J. J., Boggess S. L., Houston L. C., et al. (2021). Differential expression of genes associated with non-target site resistance in Poa annua with target site resistance to acetolactate synthase inhibitors. Pest Manage. Sci. 77 (11), 4993–5000. doi: 10.1002/ps.6541
Mao Q., Huff D. R. (2017). Characterizing small RNA profiles in allotetraploid Poa annua L. and its diploid parents. Crop Sci. 57 (S1), S-13–S-25. doi: 10.2135/cropsci2016.06.0462
McElroy J. S., Flessner M. L., Wang Z., Dane F., Walker R. H., Wehtje G. R. (2013). A Trp574 to leu amino acid substitution in the ALS gene of annual bluegrass (Poa annua) is associated with resistance to ALS-inhibiting herbicides. Weed Sci. 61 (1), 21–25. doi: 10.1614/WS-D-12-00068.1
McCullough P. E., Yu J., Czarnota M. A. (2017). First report of pronamide-resistant annual bluegrass (Poa annua). Weed Sci. 65 (1), 9–18. doi: 10.1614/WS-D-12-00068.1
Mengistu L. W., Mueller-Warrant G. W., Liston A., Barker R. E. (2000). psbA mutation (valine219 to isoleucine) in Poa annua resistant to metribuzin and diuron. Pest Manage. Science: formerly Pesticide Sci. 56 (3), 209–217. doi: 10.1002/(SICI)1526-4998(200003)56:3<209::AID-PS117>3.0.CO;2-8
Messelhäuser M. H., Linn A. I., Mathes A., Sievernich B., Gerhards R. (2021). Development of an agar bioassay sensitivity test in alopecurus myosuroides for the pre-emergence herbicides cinmethylin and flufenacet. Agronomy 11 (7), 1408. doi: 10.3390/agronomy11071408
Minitab Statistical Software (2010). Computer software (State College, PA: Minitab, Inc). Available at: www.minitab.com.
Miranda J., Moretti M. (2023). Indaziflam resistance in annual bluegrass (Poa annua L.) from hazelnut orchards. Proc. Western Soc Weed Sci.
Mutwil M., Debolt S., Persson S. (2008). Cellulose synthesis: a complex complex. Curr. Opin. Plant Biol. 11 (3), 252–257. doi: 10.1016/j.pbi.2008.03.007
Sabba R. P., Vaughn K. C. (1999). Herbicides that inhibit cellulose biosynthesis. Weed Sci. 47 (6), 757–763. doi: 10.1017/S004317450009144X
Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., et al. (2012). Fiji: An open-source platform for biological-image analysis. Nat. Methods 9 (7), 676–682. doi: 10.1038/nmeth.2019
Schneider C. A., Rasband W. S., Eliceiri K. W. (2012). NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9 (7), 671–675. doi: 10.1038/nmeth.2089
Sebastian D. J., Fleming M. B., Patterson E. L., Sebastian J. R., Nissen S. J. (2017). Indaziflam: a new cellulose-biosynthesis-inhibiting herbicide provides long-term control of invasive winter annual grasses. Pest Manage. Sci. 73 (10), 2149–2162. doi: 10.1002/ps.4594
Singh V., Dos Reis F. C., Reynolds C., Elmore M., Bagavathiannan M. (2021). Cross and multiple herbicide resistance in annual bluegrass (Poa annua) populations from eastern Texas golf courses. Pest Manage. Sci. 77 (4), 1903–1914. doi: 10.1002/ps.6217
Tateno M., Brabham C., DeBolt S. (2016). Cellulose biosynthesis inhibitors – a multifunctional toolbox. J. Exp. Bot. 67 (2), 533–542. doi: 10.1093/jxb/erv489
Tranel P. J., Wright T. R. (2002). Resistance of weeds to ALS-inhibiting herbicides: what have we learned? Weed Sci. 50 (6), 700–712. doi: 10.1614/0043-1745
Van Wychen L. (2020) Survey of the most common and troublesome weeds in grass crops, pasture and turf in the United States and Canada (Weed Science Society of America National Weed Survey Dataset). Available at: https://wssa.net/wp-content/uploads/2020-Weed-Survey_grass-crops.xlsx (Accessed February 27th, 2022).
Yu J., McCullough P. E., Czarnota M. A. (2018). Annual bluegrass (Poa annua) biotypes exhibit differential levels of susceptibility and biochemical responses to protoporphyrinogen oxidase inhibitors. Weed Sci. 66 (5), 574–580. doi: 10.1017/wsc.2018.30
Keywords: herbicide, resistance, cellulose biosynthesis, turfgrass, weeds
Citation: Pritchard BD, Vargas JJ, Mahey M, Brosnan JT and Patterson EL (2023) A bioassay to determine Poa annua responses to indaziflam. Front. Agron. 5:1182528. doi: 10.3389/fagro.2023.1182528
Received: 08 March 2023; Accepted: 05 April 2023;
Published: 18 April 2023.
Edited by:
Baruch Rubin, Hebrew University of Jerusalem, IsraelReviewed by:
Stephen Christopher Marble, University of Florida, United StatesCopyright © 2023 Pritchard, Vargas, Mahey, Brosnan and Patterson. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: James T. Brosnan, amJyb3NuYW5AdXRrLmVkdQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.