- 1Department of Botany, Indira Gandhi National Tribal University, Amarkantak, India
- 2Department of Plant Sciences, University of Hyderabad, Hyderabad, India
- 3Department of Environmental Science, Indira Gandhi National Tribal University (IGNTU), Amarkantak, India
Synthetic pesticides are extensively used in agriculture to control pests and prevent yield loss. However, excessive use imposes a serious threat to human health, environment, and biodiversity; hence, certain pesticides have been abandoned from agricultural applications. Thus, there is a need to discover potential and eco-friendly pesticides for the effective management of phytopathogens. In current study, Urginea indica bulb extract was evaluated for potential antimicrobials and antioxidant phytochemicals. The methanol and aqueous extracts were prepared from the bulbs of Urginea indica and were evaluated for polyphenol contents, alkaloid, total antioxidant capacity, and iron chelating activity. Aqueous extract exhibited high phenol and flavonoid content, whereas the total antioxidant activity was higher in methanol extract. The iron chelating activity of both methanolic and aqueous extracts was approximately similar. The antioxidant activity of both methanolic and aqueous extracts was expressed in terms of IC50 values for 2,2-diphenyl-1-picrylhydrazyl (DPPH), 2,2′-azino-bis-3-ethylbenzotiazolin-6-sulfonic acid (ABTS), and nitric oxide (NO). The highest IC50 value was observed for DPPH and the lowest for NO in both the extract. Further, fourier transform infrared spectroscopy (FTIR) was performed, which indicated the presence of several functional groups in the extract. In addition, 75 metabolites were recorded through gas chromatography–mass spectrometry (GC-MS), of which 23 were predicted to have antimicrobial activities. Consequently, metabolites were docked with D-alanine-D-alanine ligase A (DdlA) and mitogen-activated protein kinase 1 (MAPK1) of Xanthomonas oryzae pv. oryzae (Xoo) and Magnaporthe oryzae (M.oryzae), respectively, to understand the possible mechanism of interaction between active metabolites and pathogen receptors. Docking study revealed that quinic acid, 3-caffeoyl has highest binding affinity for both DdlA and MAPK1 with respect to reference compound D-cycloserine and Trametinib. Thus, quinic acid, 3-caffeoyl could inhibit both DdlA and MAPK1-mediated signal transduction and, hence, could be used as a promising natural inhibitor of DdlA and MAPK1 receptors. The above results indicate that Urginea indica could be a potential source of bioactive compounds and could be used as a potential source of natural pesticides to suppress phytopathogens.
Introduction
Crops are constantly exposed to various pests like bacteria, fungi, viruses, and insects, which greatly impair their growth, productivity, and quality. To protect crops from these pests, farmers routinely use synthetic chemicals. Although, synthetic chemicals have played vital roles in fulfilling developmental goals such as food security, livelihoods, and economic growth. However, inappropriate and indiscriminate use of pesticides causes undesirable health and environmental damage, and hence, certain chemicals have been abandoned from agriculture applications. In addition, the indelible use of synthetic chemicals directly/indirectly increases soil toxicity, kills beneficial microorganism, accelerates biodiversity loss, and drives the development of resistant pathogens (Nicolopoulou-Stamati et al., 2016; Shabana et al., 2017). Therefore, the damage associated with synthetic pesticides forced to ponder for a sustainable and ecological approach to reform agriculture through the implementation of naturally safe alternative pesticides. Interestingly, plant-derived natural compounds are less toxic, biodegradable, and eco-friendly (Fayaz et al., 2017; Campos et al., 2019). Notably, plants containing various bioactive compounds such as phenolics, flavonoids, coumarins, alkaloids, tannins, and terpenoids have been traditionally used to manage plant, human, and animal pathogens (Valli et al., 2012; Yuan et al., 2016; Anand et al., 2019). Pyrethrum (Tanacetum cinerariifolium), sweet wormwood (Artemisia annua), turmeric (Curcuma longa), eucalyptus (Eucalyptus citriodora), ginger (Zingiber officinale), wild basil (Ocimum gratissimum), lemongrass (Cymbopogon citratus), Neem (Azadirachta indica), etc., have been exploited as a promising source of natural pesticides/insecticides (Nisha et al., 2012; Alzohairy, 2016; Anyanwu and Okoye, 2017).
Rice, the staple food, faces two devastating infections termed as Bacterial Blight (BB) and rice blast and tents significant yield loss globally (Kumar et al., 2020; Jamaloddin et al., 2020). BB is caused by Xanthomonas oryzae pv. oryzae (Xoo), whereas Magrnaporthe oryzae (M. oryzae) is responsible for rice blast. Nowadays, researchers are targeting several receptor proteins of pathogens to understand the molecular mechanism of interaction between natural bioactive compounds and pathogen receptors. In particular, Eicosapentaenoic acid, Phytol, and Salicylic acid methyl ester of Lantana camara have shown greater affinity to peptide deformylase (PDF) of Xoo (Mansoori et al., 2020). Similarly, Copaene from Thespesia lampas has been reported to exhibit strong binding affinity with mitogen-activated protein kinase 1 (MAPK1) of M. oryzae and sucrose hydrolase (SUH)–glucose complex of Xanthomonas axonopodis (Singh et al., 2021a). Recently, Schefflera vinosa molecules including 3-isopropoxy-1,1,1,7,7,7-hexamethyl-3,5,5-tris (trimethylsiloxy) tetrasiloxane and 2-methoxy-5-methylthiophene) have been reported, which targets histone deacetylase and PDF of M. oryzae and Xoo, respectively (Singh et al., 2021b). In current study, D-alanine-D-alanine ligase A (DdlA) and MAPK1 of Xoo and M. oryzae, respectively, were selected for molecular docking studies with bioactive compounds acquired from gas chromatography–mass spectrometry (GC-MS). DdlA catalyzes the biosynthesis of d-alanyl-d-alanine, an essential precursor of peptidoglycan (Pederick et al., 2020). On the other side, MAPK1 regulates cell division through phosphorylation of protein residues (Xu et al., 2017).
Although few plants have been exploited, several plant species are yet to be exploited, and one such species is Urginea indica Kunth (Liliaceae family). It is commonly known as Jangli Pyaj (forest onion) and Bon Pollundu in India (Bashir et al., 2013) and has been used both in the pharmaceutical and agriculture sectors (Aswal et al., 2019). Extract of U. indica has been reported to contain several properties such as antioxidant, anticancer, antidiabetic, antiasthma, antigout, antiallergic, anti-dropsy, cardiac stimulant, wound healing, dyspepsia, and in treatment of various bacterial and plant pathogenic fungal diseases (Shenoy et al., 2006; Pandey and Gupta, 2014). Many phytochemicals like tannins, phenols, alkaloids, and flavonoids were reported in all parts of U. indica, whereas in bulb, steroids, resins, glycosides, carbohydrates, quinones, and saponins were reported (Chittoor et al., 2012; Pandey and Gupta, 2014). Since U. indica contains several medicinal compounds, a deeper investigation is needed for identification of bioactive compounds antagonists of phytopathogens such as Xoo and M. oryzae. Thus, the main objective of the current work was to systematically investigate the secondary metabolites from the crude extract of U. indica bulb and to examine the binding ability of bioactive compounds with Xoo and M. oryzae receptor proteins (DdlA and MAPK1). In addition, phytochemical profiling (qualitative and quantitative) was also performed to understand its antioxidant properties.
Materials and Methods
Reagents
All chemicals and reagents used in these experiments were of analytical grade. Chemicals such as acetic acid (CH3COOH), ammonium hydroxide (NH4OH), methanol, gallic acid monohydrate (C7H6O5•H2O), sodium carbonate (Na2CO3), quercetin dihydrate (C15H14O9), ammonium molybdate (NH4)2MoO4), hydrochloric acid (HCl), chloroform, 2,2-diphenyl-1-picrylhydrazyl (DPPH) and 2,2′-azino-bis-3-ethylbenzotiazolin-6-sulfonic acid (ABTS), L-ascorbic acid, sodium nitroprusside, orthophosphoric acid, napthylene diamine dihydochloride ferric chloride ninhydrin, ammonia, and sodium phosphate (monobasic) were procured from Hi-Media, India. Aluminum chloride (AlCl3), sulfuric acid (H2SO4), and Folin–Ciocalteu reagent were purchased from Central Drug House (P) Ltd., India.
Plant Extract Preparation
Fresh U. indica bulb was collected from Amarkantak, Madhya Pradesh, India (22°40′N 81°45′E/22.67°N 81.75°E/22.67; 81.75). Afterward, samples were cleaned with tap and distilled water. Subsequently, samples were dried at room temperature (RT) and crushed to a fine powder with a mechanical grinder. The powdered materials (10 g) were macerated with 100 ml of 80% methanol and 100 ml of water for three days using a magnetic stirrer. The residues were re-macerated for three times to completely extract the phytoconstituent. The extract was then filtered through the Whatman No. 1 filter paper and concentrated to dry mass at RT. The dried residues were harvested and transferred into airtight bottles and were stored at 4°C in a refrigerator until use. The dried extracts were dissolved in methanol and water as per the requirements of the experiment. The yield of crude extract was calculated and expressed in percentages as: Yield% = weight of the dry extract × 100/weight of the dry plant material (Singh et al., 2021a).
Qualitative Phytochemical Screening
Preliminary phytochemical qualitative screening of U. indica methanolic and aqueous extract (ME and AE) was carried out to determine the existence of biologically active compounds/secondary metabolites like saponins, tannins, flavonoid, and steroids using as method described by Harborne (1998). Similarly, the presence/absence of alkaloids, coumarins, terpenoids, glycosides, phenols, quinones, amino acids, cardiac glycosides, phlobatannins, anthocyanins, and anthraquinones was unveiled using the Roghini and Vijayalakshmi (2018) method.
Quantitative Phytochemical Screening
Total Phenol Content
The total phenol content (TPC) was estimated according to the Folin–Ciocalteu method through spectrophotometer with slight modifications (Singleton et al., 1999). First, methanolic and aqueous solution was made in a concentration of 300 µg/ml and then blended with 10% Folin–Ciocalteu reagent (2.5 ml) followed by 7.5% Na2CO3 (2.5 ml). Whole solution was vortexed for 2 min and dark-incubated at RT for 45 minutes. Subsequently, absorbance was read at 750 nm. Standard calibration curve was made using gallic acid (20–100 μg/ml). The TPC was represented as mg of gallic acid equivalents per gram of dry weight (mg GAE/g dw) and was calculated as mean value ± SD (n = 3).
Total Flavonoids Content (TFC)
The total flavonoids content (TFC) of extract was estimated through the aluminum chloride (AlCl3) colorimetric method (Chang et al., 2002; Kumar et al., 2011). Briefly, reaction solution was made by mixing 2.0 ml of diluted extract/quercetin, AlCl3 (0.1 ml, 10%), and CH3CO2K (0.1 ml, 0.1 mM). Afterward, absorbance was acquired at 415 nm after 2 min of vortexing and 30 min of dark incubation at RT. Quercetin was used to prepare the standard curve (10–60 μg/ml), and, finally, TFC in extract was represented as mg of quercetin equivalents per gram of dry weight (mg QE/g dw) and was calculated as mean value ± SD (n = 3).
Total Antioxidants Activity
The total antioxidants activity (TAA) in ME and AE was evaluated through phosphomolybdenum method (Govindarajan et al., 2003; Kumar et al., 2013) with minor amendments. Altogether, a 0.2-ml extract was amended with 1.8 ml of phosphomolebdenum reagent [0.6M H2SO4, 28mM Na3PO4, and 4mM (NH4)6Mo7O24]. After mixing for 2 min, the solution was incubated for 90 min at 95°C in a water bath. Finally, mixture was cooled at RT, and absorbance was taken at 695 nm through a spectrophotometer. Ascorbic acid (20–100 μg/ml) was taken as the standard, and result was represented as milligrams of ascorbic acid equivalents per gram of dry weight (mg AAE/g dw) and calculated as mean value ± SD (n = 3).
Total Alkaloid Content
The total alkaloid content (TAC) in extract was assessed following the standard method (Harborne, 1973). Plant powder (2.5 gm) was mixed with 200 ml of acetic acid in methanol (10%). Afterward, the mixture was incubated for 4 h at RT. Then, until final precipitation, a few drops of concentrated ammonium hydroxide (NH4OH) were added. The precipitates were then rinsed in 25 ml of 0.1 M NH4OH. Finally, the supernatant was air-dried, and the result was expressed in percentage using the following calculation: Percent alkaloid = weight of alkaloid × 100/weight of sample.
Iron Chelating Activity
The capacity of U. indica crude extract to bind iron (I) was determined using the procedure by Dinis et al. (1994). The presence of ferrous ions is indicated by the development of a red ferrous iron–ferrozine complex, which is evaluated by measuring absorbance at 562 nm. In a separate test tube, 2.0-ml extract and standard (Na2EDTA) at working concentrations of 20, 40, and 100 µg/ml were combined with 400 µl of ferrous sulfate (2.0 mM). The reaction was initiated by the addition of ferrozine into the mixture. Finally, the absorbance of the sample was measured at 562 nm, and the percent inhibition was determined using the following equation: Scavenging effect % = (control absorbance − sample absorbance) ×100/control absorbance.
Nitric Oxide Radical Scavenging Activity
The capacity of the crude plant extract to scavenge the nitric oxide (NO) free radical was assessed through a conventional approach (Sreejayan Rao, 1997) with slight modifications. The reaction was started by adding 200 µl of plant crude extract at various concentrations in separate test tubes, followed by 80 µl of sodium nitroprusside, and then incubated for 15 min under light conditions. After 15 min, 1.0 ml of Griess reagent was added to this combination, and all the tubes were incubated for 45 min at 30°C under light conditions. To halt the process, 40 µl of 0.1% napthylene diamine dihydrochloride in 2.5% H3PO4 was added. Using double-distilled water (DDW), the final volume was increased to 2.0 ml, and the absorbance was measured at λmax = 540 nm. The scavenging activity was determined in terms of NO radicals scavenged using the following equation: Scavenging effect % = (control absorbance − sample absorbance) ×100/control absorbance
2,2-Diphenyl-1-Picrylhydrazyl Radical Scavenging Activity
The extract’s free radical scavenging activity was determined using conventional DPPH tests (Bursal and Gülçin, 2011; Kumar et al., 2011; Kumar et al., 2013). The solution was made by dissolving 4.0 mg of DPPH in 100 ml of methanol, and absorbance was measured at λmax = 516 nm with a spectrophotometer. This solution was combined with 300 µl of standard (Ascorbic acid) or extract at various concentrations (20–220 µg/ml). After shaking, the reaction mixture was incubated in the dark for 15 min at RT. The final absorbance was taken at λmax = 515 nm. The scavenging activity was determined in terms of the DPPH radical scavenged using the following equation: Scavenging effect % = (control absorbance − sample absorbance) ×100/control absorbance.
2,2′-Azino-Bis-3-Ethylbenzotiazolin-6-Sulfonic Acid Radical Scavenging Activity
The capacity of a crude plant extract to scavenge ABTS radicals was assessed through a conventional method (Re et al., 1999). The mechanism of the reaction includes the decolorization of ABTS•+ radicals via the electron donating activity of plant extract antioxidants. ABTS solution (7.0 mM) was produced with DDW and combined in a 1:1 ratio with 2.45 mM K2S2O8 solution. Following that, a 24- to 48-h dark incubation at RT was performed. The ABTS solution was then diluted with methanol, and the absorbance was adjusted at 0.700 ± 0.02 at λmax = 745 nm. A mixture of extract/standard concentrations ranging from 20 to 220 µg/ml (300 µl) was combined with 2.5 ml of solution. After shaking, the entire reaction mixture was incubated in the dark at RT for 15 min. Finally, absorbance was measured at λmax = 745 nm. The scavenging activity was calculated in terms of the percent inhibition of ABTS•+ using the same equation as the percent inhibition of DPPH radical scavenging activity.
Fourier Transform Infrared Spectroscopy Spectral Analysis
The fourier transform infrared spectroscopy (FTIR) spectra of dried U. indica extract were measured at RT, utilizing attenuated total reflectance (ATR) and internal reflection element formed of diamond using ATR equipment Thermo Nicolet iS5 (Thermo Scientific, Madison, WT, USA). This FTIR system is built using a KBr beam splitter and a deuterated triglycine sulfate detector, and it is linked to a Windows-based system running OMNIC software (OMNIC™ Series Software; thermofisher.com). All measurements were collected at a resolution of 4 cm−1 in the spectral band of 4,000 to 500 cm−1. The spectra of the samples were subtracted from the spectra of the air. At each point, the spectra were logged as an absorbance value. During measurement, the ATR crystal surface was cleaned with 70% ethanol on a regular basis.
Gas Chromatography–Mass Spectrometry Analysis
To determine the presence of metabolites, gas chromatography–mass spectrometry (GC-MS) was performed according to Sharma et al. (2021) and Kilambi et al. (2021) (a modified method of Roessner et al., 2000). Fraction of metabolite was prepared by mixing 50 mg of dry powder with 100% chilled methanol (0.95 ml) containing ribitol as internal standard succeed by incubation at 70°C for 15 min with continuous shaking. After incubation, 0.95 ml of chilled water was added and at 2,500g centrifugation was performed for 15 min at 4°C. The supernatant was relocated to a new centrifuge tube and allowed to dry for 120 min. Further, derivatization of obtained residue was done by mixing methoxamine (65 µl) containing pyridine at 37°C for 90 min following the incubation with 65 μl of N-methyl-N-(trimethylsilyl) trifluoroacetamine and FAME mix (20 µl) at 37°C for 30 min. The LECO-PEGASUS GCXGC-TOF-MS system (LECO Corporation, USA) was used to analyze the derivatized sample, which was equipped with a 30-m Rxi-5ms column with 0.25-mm internal diameter and 0.25-μm film thickness (Restek, USA). Sources of injection temperature, interference, and ion temperature were set to 240°C, 225°C, and 200°C, respectively. The following steps were persuaded for sample chromatographic separation: isothermal heating (70°C) for 5 min, succeed by a 5°C min−1 oven temperature ramp to 290°C and 5 min of heating at 290°C. Helium gas was utilized as carrier, and their flow rate was attuned to 1.4 ml/min. Finally, 1 µl of sample was administered in split less mode for investigation, and the range of scan mass was set to 70 to 600 at 2 scans/s.
Metabolite Identifications
Metabolite identification was performed according to Sharma et al. (2021). Data files were generated as netCDF by ChromaTOF software 299 4.50.8.0 chromatography version (LECO Corporation, USA) and examined using MetAlign 3.0 software (www.metalign.nl) (Lommen and Kools, 2012). Several parameters were acquainted such as signal-to-noise ratio of ≥2, baseline correction, noise estimation, and mass peak identification (ion-wise mass alignment). Data reduction and mass extraction of compounds were performed through MSClust analysis (Tikunov et al., 2012). Finally, the files obtained were exported to the NIST MS Search v2.2 program to identify compounds using the NIST (National Institute of Standards and Technology) library and the GOLM metabolome database library (Hummel et al., 2010). All the metabolites were quantified by normalization with ribitol (internal standard).
Molecular Docking
Ligand and Protein Preparation
As mentioned, 66 ligands were acquired from the PubChem database (https://pubchem.ncbi.nlm.nih.gov/) in Protein Data Bank (PDB) format and were converted into PDBQT format for docking analysis. Three-dimensional structure of DDdlA (PDB ID: 4ME6) and MAPK1 (PDB ID: 5z33) was acquired from PDB (https://www.rcsb.org/) in Docking Parameter Format (PDB) format. Heteroatoms including water and others molecules were eliminated using BIOVIA Discovery Studio. Missing atoms of amino acid chain were corrected with Swiss PDB viewer. Active sites of DdlA and MAPK1 were detected from Computed Atlas of Surface Topography of protein (CASTp) online server (Tian et al., 2018) and discovery studio, respectively.
Docking Analysis
Autodock 4.2.6 was used for analysis together with accompanying software, viz., Python 3.9.6 and MGLTools 1.5.6. Protein ligand docking was accomplished by binding one ligand at a time to the protein receptor (Morris et al., 2009). The processed protein was then imported into an Autodock workspace for the insertion of polar hydrogen atoms, as well as Kollman and Gasteiger charges (Ravindranath et al., 2015). Similarly, ligands were incorporated into the same place to define its torsion tree by choosing a root and a rotatable number of bonds. Both the protein and ligands were saved in PDBQT format for further process.
Preparation of Grid Parameter and Autodocking
Grid parameters were defined for both the proteins to navigate the active site for proper interaction of ligands. For DdlA, the center grid box values were set to x = 2.481, y = 12.094, and z = 34.667 and for MAPK1, x = 51.07, y =33.07, and z = 28.2. Grid points for both the receptor proteins in the x, y, and z dimensions were set to 68, 62, and 68, respectively. Autogrid was successfully executed, and then, genetic algorithm was set to the following parameters: 1) numbers of GA runs: 50; 2) population size: 300; 3) energy evaluations number: 2.5 million; and 4) generations number: 27,000. The Lamarckian genetic process was used subsequently, and the output file was stored in DPF. Finally, the program was launched for docking, and output of the final result was saved as docking log file. Binding energy, inhibition constant, and ligand efficiency were calculated, and the complex had lower binding energy that was visualized in discovery studio for 2D and 3D evaluation.
Absorption, Distribution, Metabolism, Elimination, and Toxicity Property Prediction
Each compound which had good binding affinity with proteins was subjected to absorption, distribution, metabolism, elimination, and toxicity (ADMET) property prediction. It is a computer program that calculates the pharmacokinetics and drug-like properties of substances based on their chemical structure. The smiley format was introduced at interface of SwissADME website (http://www.swissadme.ch/), and program was run to generate ADMET properties/parameters.
Statistical Analysis
Both ME and AE were taken in triplicates for all biochemical assays. The results were denoted as mean ± standard deviation (SD) (n = 3). GC-MS results were expressed as an average of five replicates. Two-way analysis of variance (ANOVA) was executed to assess the variation among TPC, TFC, and TAA of ME and AE. The interrelations among DPPH, ABTS, NO radical scavenging assay, and iron chelating activity were studied using the Pearson correlation coefficient test. OriginPro 2016 and GraphPad Prism 8 were used to create the statistical analysis and graph preparation.
Results and Discussion
Preliminary Phytochemical Analysis of Methanolic and Aqueous Extract of U. indica
In both ME and AE, preliminary phytochemical screening discovered the occurrence of alkaloids, steroids, flavonoids, quinones, and coumarins (Table 1). Saponins and terpenoids were found in only AE, whereas amino acids were only detected in ME. Each detected phytochemical has shown several medicinal properties against different microbial diseases. Alkaloids are a diverse group of secondary metabolites that have inspired the development of several antibacterial compounds such as ascididemin, eudistomin, squalamine, and clausenol against Gram-positive and Gram-negative bacteria (Cushnie et al., 2014). Predominantly, flavonoids have been used as an antifungal agent against Candida albicans, Trichophyton rubrum, and Aspergillus flavus (Lee and Chee, 2010; Mohotti et al., 2020; Quiroga et al., 2009; Bitencourt et al., 2013). Terpenoids and their derivatives usually affect oxygen uptake and oxidative phosphorylation, which are essential to microbial survival (Griffin et al., 1999). Plant-derived quinones have been used as an anticancer and antibacterial agent (Goel et al., 2020). Several studies have reported that coumarins exhibited potent antibacterial activity especially against Gram-negative bacteria through their cell membrane damage (Céspedes et al., 2006; Saleem et al., 2010; Yu, 2015). Steroids have been used as cataionic forms for bacterial membrane targets based on specificity (Savage et al., 2002). In addition, there are several reports that suggest that consuming natural food containing phenolics compounds decreases the risk of health disorder due to the antioxidant nature of phenolics (Surh, 2002). Thus, the present investigation suggests that U. indica phytochemicals could be a promising agent, particularly against plant pathogens and in nutraceuticals.

Table 1 Qualitative analysis of phytochemicals presents in methanolic and aqueous extract of U. indica bulb.
Determination of Total Alkaloids, Phenols, Flavonoids, and Antioxidants Contents
TAC was 2.4 ± 0.42% in U. indica powder. TPC, TFC, and TAA calculations were based on the equation obtained from gallic acid, quercetin, and ascorbic acid, respectively (Supplementary Figure 1). The TPC in both ME and AE was slightly similar in terms of mg GAE/g dw. The ME demonstrated 11.73 ± 0.46 mg GAE/g dw, whereas, in aqueous extract, GAE was found to be 12.40 ± 0.20 mg/g dw (Figure 1). The AE exhibited higher amounts of flavonoid content (101.78 ± 0.98 mg QE/g dw) than ME (26.81 ± 0.98 mg/g dw). Further, the TAA was found to be higher in ME (79.09 ± 10.00) as compared to AE (71.03 ± 4.50) in terms of mg AAE/g dw. In the present study, flavonoid content was higher in AE among all tested phytochemicals. In addition, it found that water was a better solvent for the extraction of phenols and flavonoids from U. indica. Both TPC and TAA were similar in ME and AE, suggesting the extraction of polar nature of compounds due to polar nature of phenols (Garcia-Salas et al., 2010). Previous studies also suggest a correlation between total phenol and antioxidants (Kumar et al., 2014). We have also studied the interaction between TPC, TFC, and TAA of ME and AE of U. indica using two-way ANOVA (Figure 1), which demonstrated that TPC and TFC of ME are significantly different at a level of P < 0.05, whereas TAA of ME also exhibited highly dissimilarities with both TPC and TFC at a level of P < 0.0001. However, TPC and TFC of AE have significantly different at a level of P < 0.0001, whereas TAA of AE has variation with TPC at a level of P< 0.0001 and with TFC at a level of P< 0.0001. The above result justifies the significant contribution of flavonoids and phenols to plants’ phosphomolybdate scavenging-related total antioxidant activity. Phenols lose H+ ion to produce phenolate ion, which subsequently reacts with Folin–Ciocalteu reagent to reduce them (Fernandes et al., 2012). As phenolics and many flavonoids contain polar hydroxyl groups, their higher extraction with methanol and water is quite reasonable (Ahmed et al., 2015). High phenolic content in plants makes them a potential source for various therapeutic uses for the treatment of chronic diseases related to microbial infections (Petti and Scully, 2009). Likewise, flavonoid has a wide range of biological functions and is used as an anti-cancer, anti-inflammatory, anti-allergic, anti-thrombotic, anti-tumor inhibitor, and anti-viral agent (Trease and Evans, 2002; Aggarwal and Shishodia, 2006). The presence of phenolics and flavonoids in U. indica makes them a potent antimicrobial agent as it demonstrated microbial inhibitory effects previously (Lim et al., 2007; Jarial et al., 2018; Dzotam et al., 2018). Hence, results obtained demonstrate that U. indica would be of particular interest for further research.
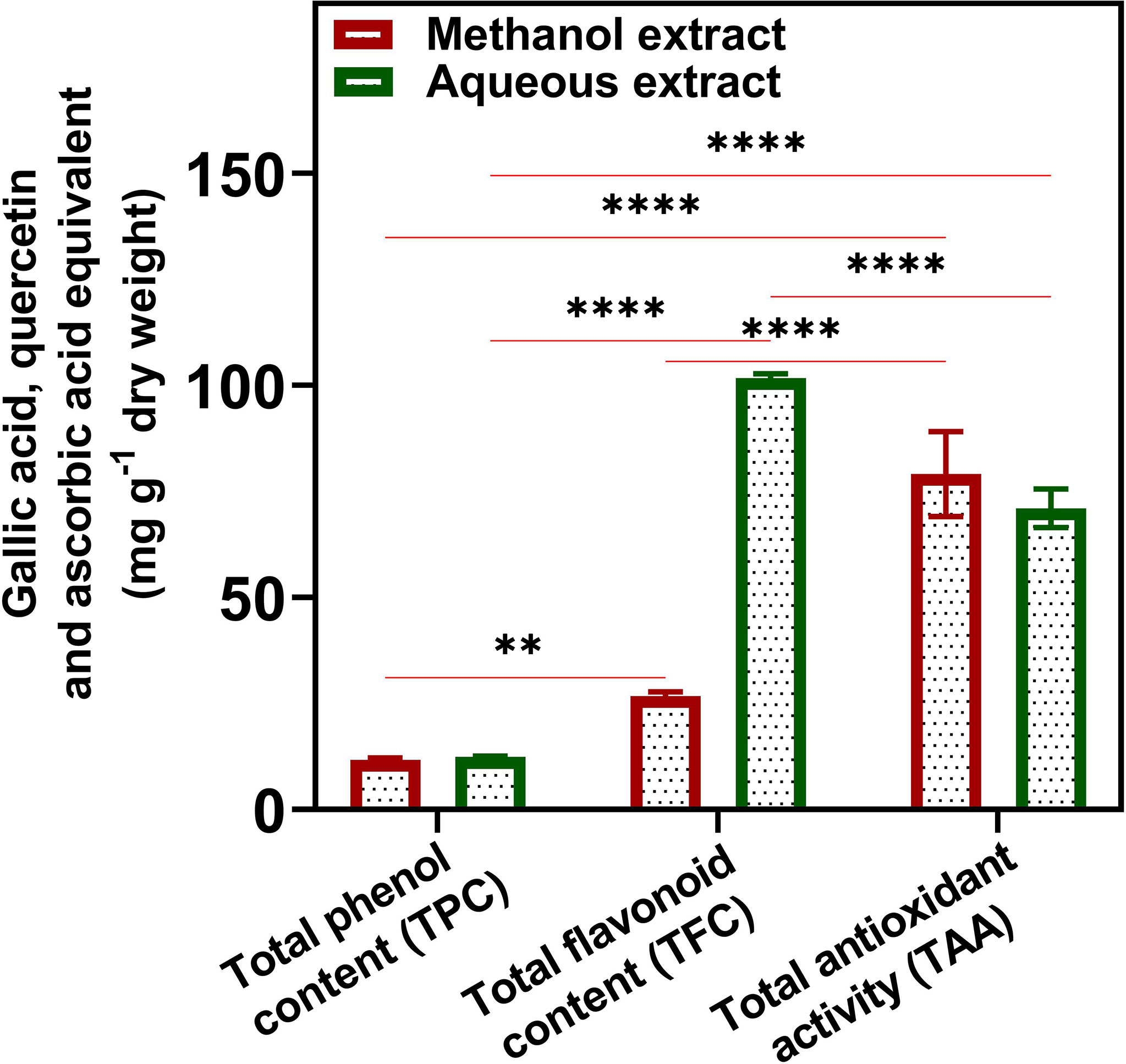
Figure 1 Quantitative total phenol (TPC), flavonoid (TFC), and antioxidant content (TAA) of U. indica methanol and aqueous extract. Each value is the average of three replicates. The figure also represents two-way analysis of variance (ANOVA) among TPC, TFC, and TAA. *, **, and **** represent the significance at p < 0.05, < 0.001, and < 0.0001, respectively.
Iron Chelating Activity Determination
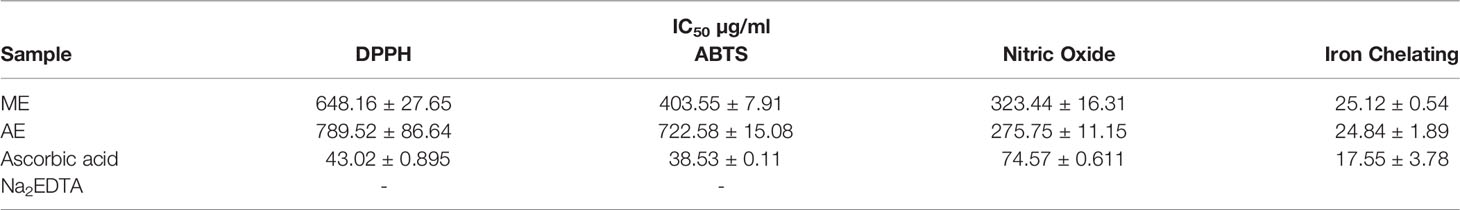
The Fe2+ ion chelating activity of U. indica ME and AE was determined at concentrations of 20, 60, and 100 µg/ml. At a concentration of 100 µg/ml, the result obtained in terms of percent inhibition of Fe2+ ion was determined to be 85.22 ± 0.513 for ME and 85.39 ± 3.69 for AE. As a positive control, Na2EDTA demonstrated chelating activity of 98.68 ± 0.425% at the maximum concentration of 100 µg/ml (Figure 2A). As a result, the chelating activity of standard is greater than that of ME and AE. However, ME’s IC50 value is almost similar to AE’s, indicating that both ME and AE have equal iron chelating potential (Table 2). Previous studies have reported that the iron chelators like caffeic acid act like antioxidants and reactive oxygen species scavengers. Interestingly, iron overload is neutralized by chelation therapy in case of thalassemia and anemia, and synthetic chelators are also used to control oxidative damage, which have shown negative impact on health as well (Ebrahimzadeh et al., 2008). Hence, plant-derived molecules could be used for chelation as an alternative to synthetic chelators.
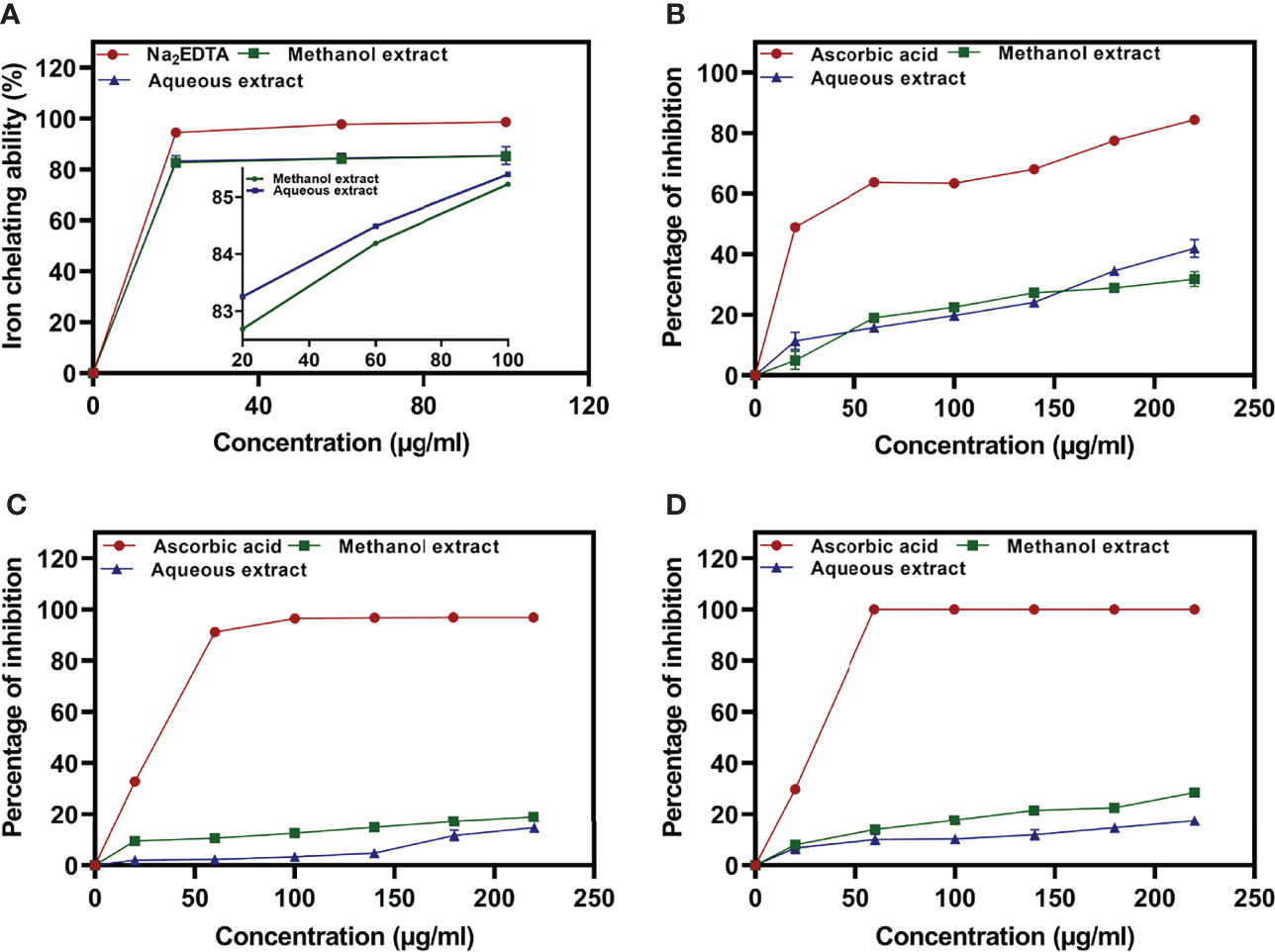
Figure 2 Iron chelating activity of methanolic and aqueous extract of U. indica, along with Na2EDTA standard (A), percentage inhibition of nitric oxide radicals (B), DPPH radicals (C), and ABTS radicals (D) via methanolic and aqueous of U. indica along with ascorbic acid standard.
NO Radical Scavenging Activity Determination
The reactive capability of the NO radical was estimated by measuring the decrease in absorbance at λmax = 546 nm caused by antioxidant activation. The antioxidant activity values of U. indica ME, AE, and standard ascorbic acid at 20–220 µg/ml were 4.96 ± 2.96 to 31.77 ± 2.41, 11.38 ± 2.81 to 41.91 ± 2.96, and 48.95 ± 0.17 to 84.47 ± 0.31, respectively, as shown in Figure 2B. It clearly shows that U. indica has a dose-dependent NO scavenging action, with an IC50 value of 323.44 ± 16.31 µg/ml for ME and 275.75 ± 11.15 µg/ml for AE, whereas ascorbic acid has an IC50 value of 74.57 ± 0.611 µg/ml (Table 2). The Pearson correlation coefficient values of NO radical scavenging activity of plant extract with TAA and iron chelating activity suggest that both are positively correlated with correlation coefficient values of 0.32 and 0.42, respectively. NO overproduction has been related to a variety of chronic diseases, including carcinogenesis and arteriosclerosis. It is poisonous, and it combines with other free radical molecules to increase toxicity (Valko et al., 2007). Because U. indica extract has NO scavenging activity, it may be effective in reducing free radical–related damage and illnesses.
DPPH Radical Scavenging Activity Determination
Concentration-dependent increases in DPPH radical scavenging capacity of U. indica extract are noted as depicted in Figure 2C. The ME and AE of U. indica had the most potent scavenging effect, with an IC50 value of 648.16 ± 27.65 and 789.52 ± 86.64 µg/ml, respectively. When the extract’s scavenging capability was compared to the standard (ascorbic acid), the IC50 value was 43.02 ± 0.895. (Table 2). The methanolic extract’s scavenging activity in terms of percent inhibition varies from 9.55 ± 1.37% to 18.99 ± 1.22%, whereas aqueous extract’s scavenging activity in terms of percent inhibition varies from 1.97 ± 0.12% to 14.72 ± 0.81%, and the standard ranges from 32.82 ± 0.99% to 96.83 ± 0.063% at concentrations ranging from 20 to 220 µg/ml. In terms of low IC50 value, a comparative investigation has shown that ME of U. indica had greater antioxidant activity than AE (Table 2). Pearson correlation result also suggests that DPPH radical scavenging activity has positive correlation with ABTS radical scavenging activity (with correlation coefficient value 0.81) followed by TFC (with correlation coefficient value 0.79) and TPC (with correlation coefficient value 0.55) (Supplementary Figure 2). The DPPH test is an appropriate and widely used technique for determining the free radical scavenging capability of plant extracts. Phenolic compounds contain hydroxyl groups that are responsible for radical scavenging effects due to their redox properties (Evans et al., 1997). A study indicated that the natural antioxidants derived from plants have been reported as an antimicrobial agent with possible mechanisms of microbial enzyme inhibition, substrate deprivation, and protein regulation (Aminov, 2010). Hence, high free radicals’ activity in the current study could be attributed to high flavonoid and phenolic content in plant extract, which may contribute to Xoo and M. oryzae inhibition. Interestingly, extract having both antioxidant and DPPH radical scavenging ability donates hydrogen to free radicals that potentially inhibit the progression of lipid peroxidation (Bamforth et al., 1993).
ABTS Radical Scavenging Activity Determination
The interaction of ABTS and K2S2O8 produces ABTS cation. The hydrogen/electron donating abilities of plant extract were demonstrated by assessing the scavenging potential of ABTS cations using U. indica ME and AE. U. indica crude extract demonstrated concentration-dependent ABTS radical scavenging capacity (20–220 µg/ml), as shown in Figure 2D. The linear response curves were also produced, and the IC50 value for ME and AE was determined to be 403.55 ± 7.91 µg/ml and 722.58 ± 15.08 µg/ml, respectively, and 38.53 ± 0.11 µg/ml for ascorbic acid. ME has a stronger ABTS scavenging activity than AE (Table 2). This experiment also shows that U. indica has free radical scavenging action and, hence, might be employed as an active component in medicines and free radical–related therapies. The correlation of ABTS radical scavenging capacity exhibited a positive correlation with TFC (with correlation coefficient value of 1.0) and TPC (with correlation coefficient value of 0.73) (Supplementary Figure 2). Significant correlation of ABTS with TFC and TPC could be a reason for scavenging potential of U. indica. Actually, the cation radical scavenging activity of ABTS indicates the donating capability of hydrogen. Previous reports suggest that phenolics of high molecular weight possess a greater ability for free radical (ABTS+) quenching (Hagerman et al., 1998). This scavenging feature leads to prevent oxidation of lipids through chain termination reaction and, hence, could be incorporated as an important nutraceutical when consumed with another nutrient as well.
Functional Group Analysis of U. indica Extract by FTIR Spectrum Analysis
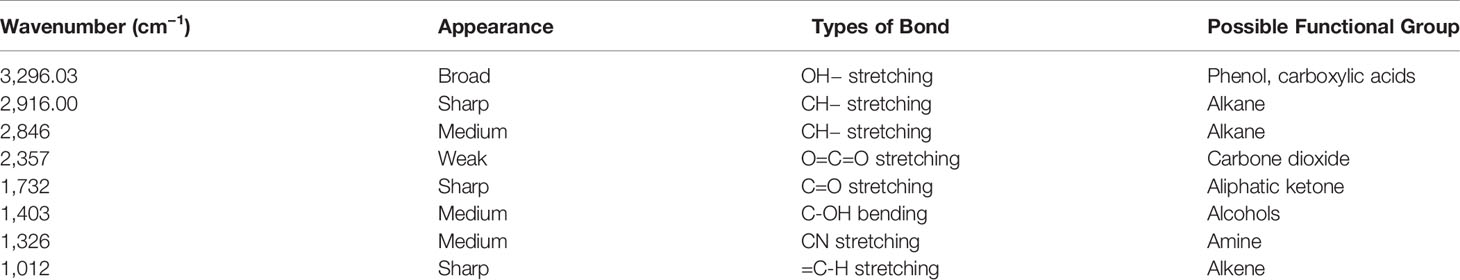
The FTIR study was performed to bring out the fingerprints of the chemical constituents of the U. indica extract (Figure 3). It is a sophisticated approach for detecting functional groups in extracts based on the peak values acquired from the FTIR spectrum. The absorbance bands were analyzed in the region at a resolution between 500 and 4,000 cm−1. Importantly, 3,269 (OH− stretching), 2,916 (CH− stretching), 2,846 (CH− stretching), 2,357 (O=C=O stretching), 1,732 (C=O stretching), 1,403 (C-OH bending), 1,326 (CN components), and 1,021 (=C-H stretching) cm−1 were assigned to several possible functional group such as alkanes, alkenes, amines, aromatic compounds/carboxylic acids, aliphatic ketones, alcohols, and phenol related compounds. These functional groups may be responsible for the crude extract’s antioxidant action as it is an integral part of various secondary metabolites such as phenols and flavonoids (Poojary et al., 2015). Table 3 summarizes the peak value, anticipated functional groups, and type of bond. Previous research indicates that functional group like aldehyde and phenolics participate in different biological functions, such as antiseptics, insecticides, fungicides, and plant immunity (Rao, 2012; Chandra, 2019; Singh et al., 2021b). Hence, it can be assumed that the U. indica antimicrobial property could have been contributed by such a functional group. As a result, its therapeutic usage in Unani medicine appears to be validated. However, it has been reported that FTIR alone could not detect all the functional group of active constituents present in the crude extract, and hence, GC-MS study was also performed.
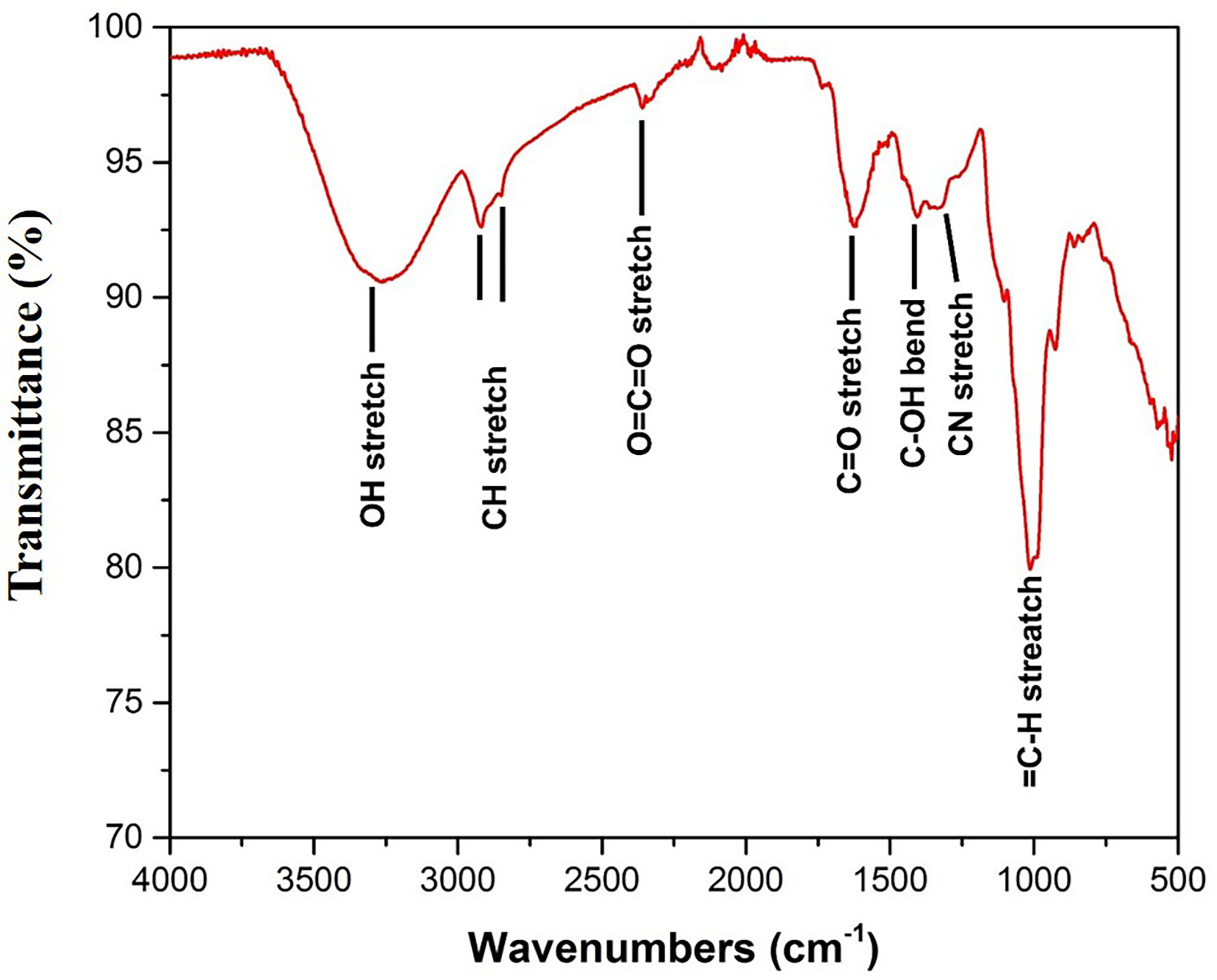
Figure 3 FTIR spectra of U. indica extract. Extract was put on diamond crystal pedestal of ATR and readings were taken from 500 to 4,000 cm−1.
Metabolite Identification in Extract by GC-MS Based Metabolomics
U. indica extract was administered to GC-MS for detection of metabolites. Analysis was performed for five replicates of the sample for accuracy. A total of 75 compounds were detected in GC-MS metabolomics, which was categorized under different groups such as organic acids, amino acids, monoamines, sugars, sugar alcohols, sugar acids, fatty acids, and alcohols (Supplementary Table 1). In terms of relative levels, higher amounts of sugars were detected, viz., sorbose (32.78 ± 3.07), fructose (24.12 ± 1.96), and sucrose (21.80 ± 2.66). followed by organic acids such as malic acid (20.56 ± 1.27) and phosphoric acid (14.26 ± 1.21). Some metabolites were also confined to phenolic compounds, which are quinic acid, 3-caffeoyl quinic acid, tyrosol, pyrogallol, resorcinol, hydroquinone, and caffeic acid. Phenolic compounds detection may confirm the TPC of U. indica extract. Many studies have supported the phenol as an antimicrobial agent both experimentally and theoretically (Alves et al., 2013; Vidhya et al., 2020; Aliye et al., 2021). In silico analysis revealed the potential of phenol including diosmin, morin, and chlorogenic acid, against bacterial urease (Kataria and Khatkar, 2019). In addition, phenolic compounds exhibited Staphylococcus aureus membrane damage under in vitro conditions (Miklasińska-Majdanik et al., 2018). Perez-Castillo et al. (2020) reported that cinnamic acid and benzoic acid act against Candida albicans. Interestingly, 23 compounds identified in U. indica have previously reported antimicrobial activity against different human and plant pathogens (Table 4). Thus, the detection of an array of phytochemicals signifies the endowed medicinal properties of U. indica. These findings may aid in a more rational evaluation of the multipurpose usage of U. indica plant as a potential and natural source of antimicrobial compounds.
Validation of Antimicrobial Property of Plant Extract Through Molecular Docking
To examine binding affinity, molecular docking was performed for 66 compounds (excluded amino acids) against DdlA and MAPK1 of Xoo and M. oryzae, respectively. Virtual screening technologies are often and widely used as a strategy for discovering novel ligands for protein structures. Top three compounds were selected which had maximum binding affinity with both the proteins (Table 5). Binding affinity of compounds with DdlA ranged from −4.45 to −11.7 Kcal/mol (Supplementary Table 2). Highest binding affinity (−11.7 Kcal/mol) was exhibited by quinic acid, 3-caffeoyl with lower inhibition constant (2.64 nM). Next, glucose-1-phosphate (−9.48) and quinic acid (−9.17 Kcal/mol) showed better binding affinity with 112.08 nM constant and 188.24 nM inhibition constant (Ki), respectively, with DdlA protein. Although, it is observed that D-cycloserine (reference compound) with DdlA has low binding affinity (−5.49) compared with the other three compounds that showed highest binding affinity. In case of MAPK1, quinic acid, 3-caffeoyl also exhibited highest binding affinity (−10.45 Kcal/mol) followed by sucrose (−8.46 Kcal/mol) and glucose-1-phosphate (−8.38 Kcal/mol). Ki of quinic acid, 3-caffeoyl (21.87 nM) with MAPK1 was slightly higher than with DdlA. In addition, binding affinity (−8.30 Kcal/mol) of Trametinib (reference compound) was found less than other top three compounds with MAPK1. Further, structural interaction studies for bond formation were visualized in Discovery Studio. Quinic acid, 3-caffeoyl was able to form seven hydrogen bonds with Lys13, Gly47, His107 (two bonds), Leu110, Gly111, and Ser115 residues of DdlA proteins (Figure 4A). Glucose-1-phosphate and quinic acid have formed three and seven hydrogen bonds, respectively (Figures 4B, C). D-cycloserine formed only four hydrogen bonds, which include conventional and carbon hydrogen bonds (Figure 4D). Interaction of MAPK1 and quinic acid, 3-caffeoyl exhibited 10 hydrogen bonds with different amino acid residues, in which Glu105, Gly13, Cys166, and Asp 167 shared one bond each, whereas Lys51, Met107, and Asn154 formed two hydrogen bonds (Figure 4E). However, sucrose and glucose-1-phosphate were able to form five and seven hydrogen bonds, respectively, with MAPK1 (Figures 4F, G). In the case of Trametinib and MAPK1 interaction, major bonds were alkyl and Pi-alkyl with only three hydrogen bonds (Figure 4H). Interestingly, Lys13 and Gly47 of DdlA have shared common bonding with quinic acid, 3-caffeoyl, glucose-1-phosphate, and quinic acid, whereas His107, leu110, and Gly111 formed bonds with both quinic acid, 3-caffeoyl and quinic acid. Similarly, Met107 residue of MAPK1 exhibited common bonding with all three compounds, viz., quinic acid, 3-caffeoyl, sucrose, and glucose-1-phosphate. Common sharing of amino acid with different compounds suggests the key target amino acid residue of both the proteins. The high binding affinity of quinic acid, 3-caffeoyl for both DdlA and MAPK1 protein could be due to presence of caffeoyl group. Previous studies suggest that the increasing number of caffeoyl groups promoted anti-mutagenecity (Yoshimoto et al., 2002), which could be used as a target antimicrobial agent, because bacteria undergo several mutations during growth for survival. In addition, various studies confirmed that quinic acid, 3-caffeoyl inhibited the growth of fungus and bacteria (Karunanidhi et al., 2013; Martínez et al., 2017). So far, several phytochemical-based inhibitors have been predicted through molecular docking analysis against human and plant proteins responsible for disease (Tahir et al., 2019; Basu et al., 2020; Joshi et al., 2021). Recently, Loliolide, Eicosapentaenoic acid, Salicylic Acid Methyl Ester, and Phytol have been reported as antimicrobial agents against MAPK1, PDF, and SUH of M. oryzae and Xoo (Mansoori et al., 2020). Binding energy and Ki were also represented through heat map, which indicates that all ligands have more affinity with DdlA than MAPK1 (Figure 5). However, a linear correlation between binding energy of DdlA and MAPK1 with metabolites showed a positive correlation (R2 = 0.82), which indicates the almost equal potential of ligands for inhibition of target proteins (Supplementary Figure 3). Thus, molecular docking result indicates the higher efficiency of quinic acid, 3-caffeoyl and quinic acid for inhibition of both DdlA and MAPK1 proteins with consideration of their binding affinity and inhibition constant.
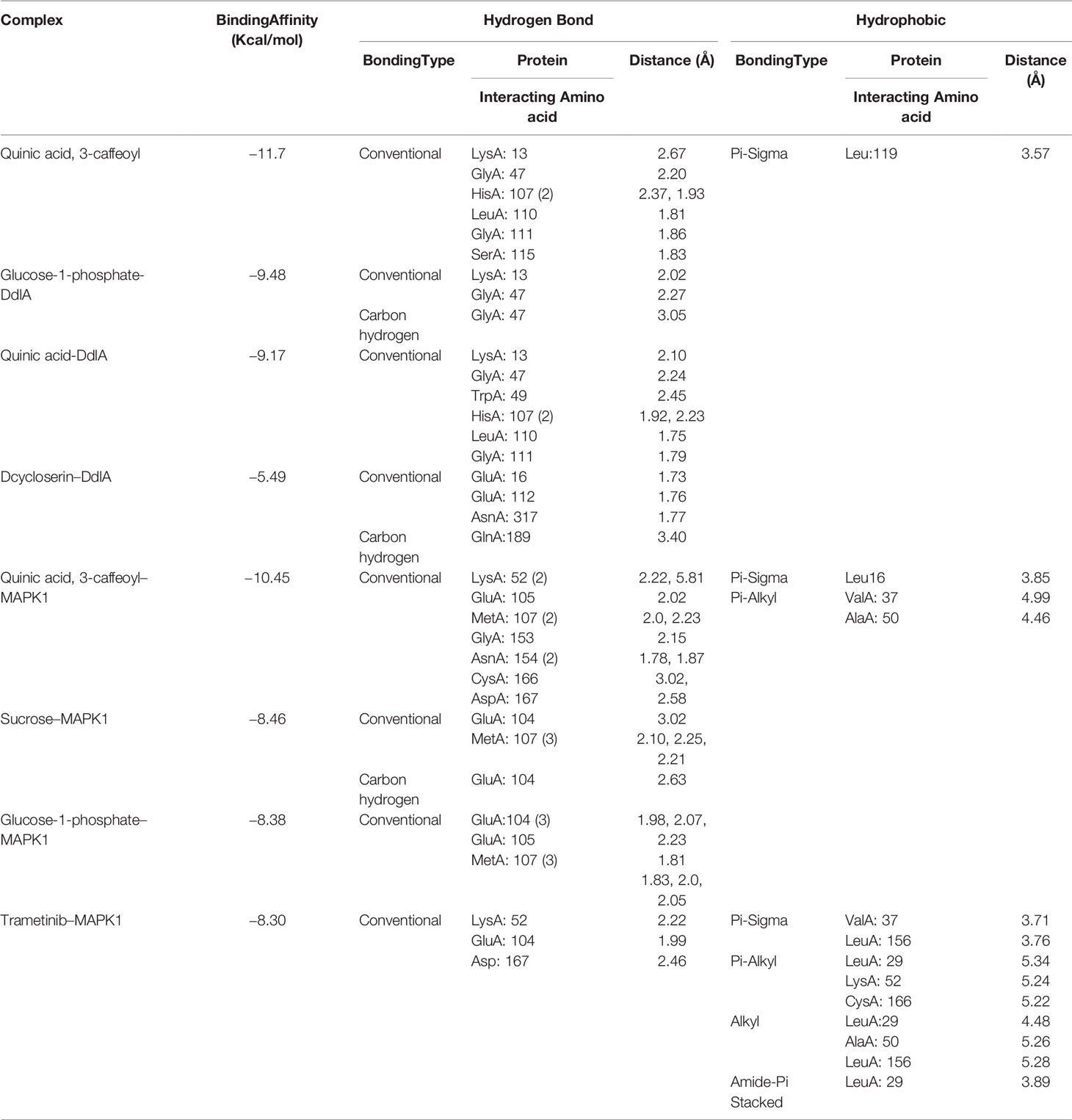
Table 5 Hydrogen bond and hydrophobic interaction of top six compounds with DdlA (Xoo) and MAPK1 (M. oryzae).
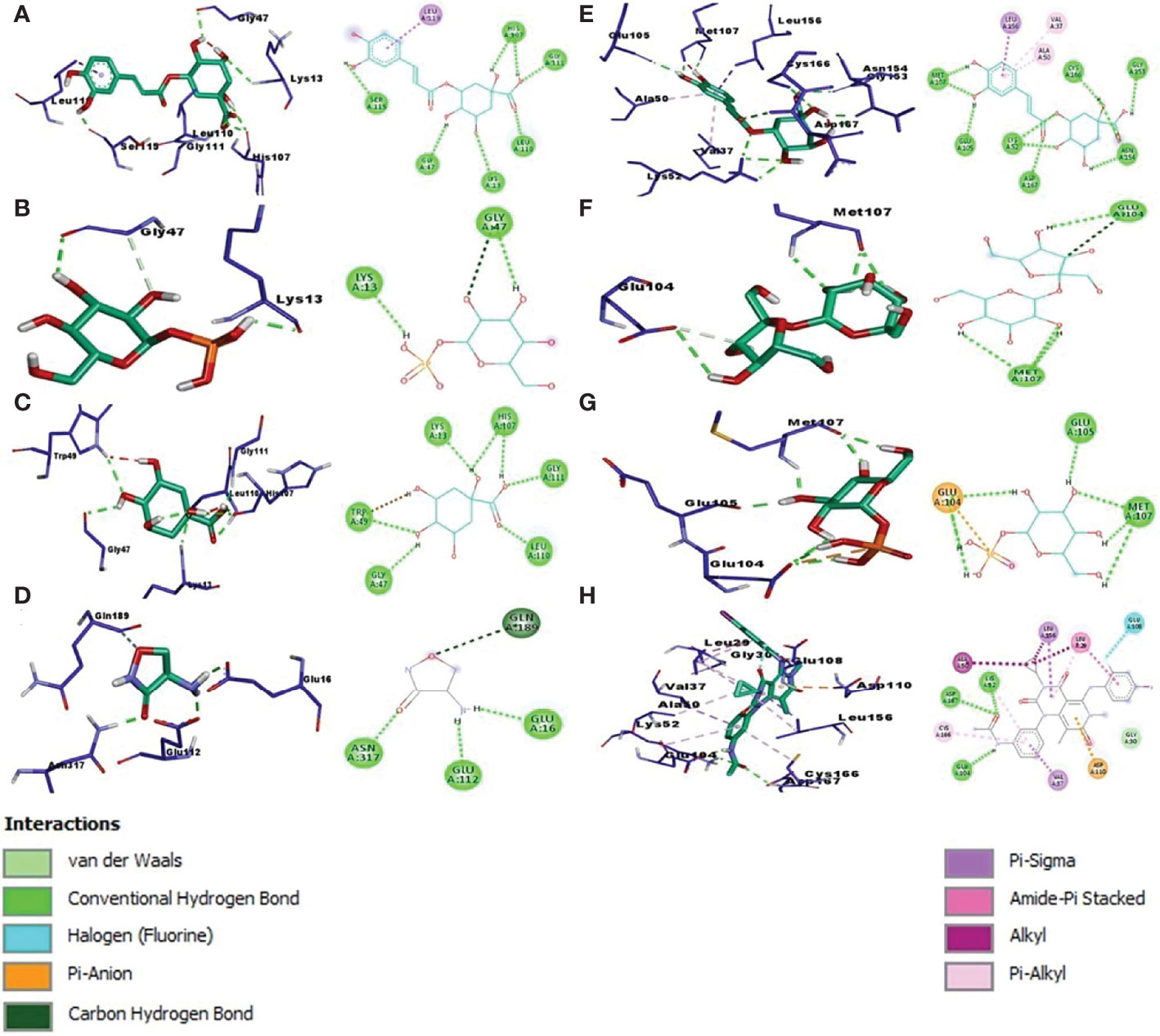
Figure 4 Left panel of each alphabet showing 3D image while right panel indicating the 2D image. Molecular docking interaction and different bond formation of quinic acid, 3-caffeoyl (A), glucose-1-phosphate (B), quinic acid (C), and Dcycloserine (D) with DdlA protein of Xoo. Molecular docking interaction of quinic acid 3 caffeoyl (E), sucrose (F), glucose-1-phosphate (G), and Trametinib (H) with MAPK1 protein of M oryzae.
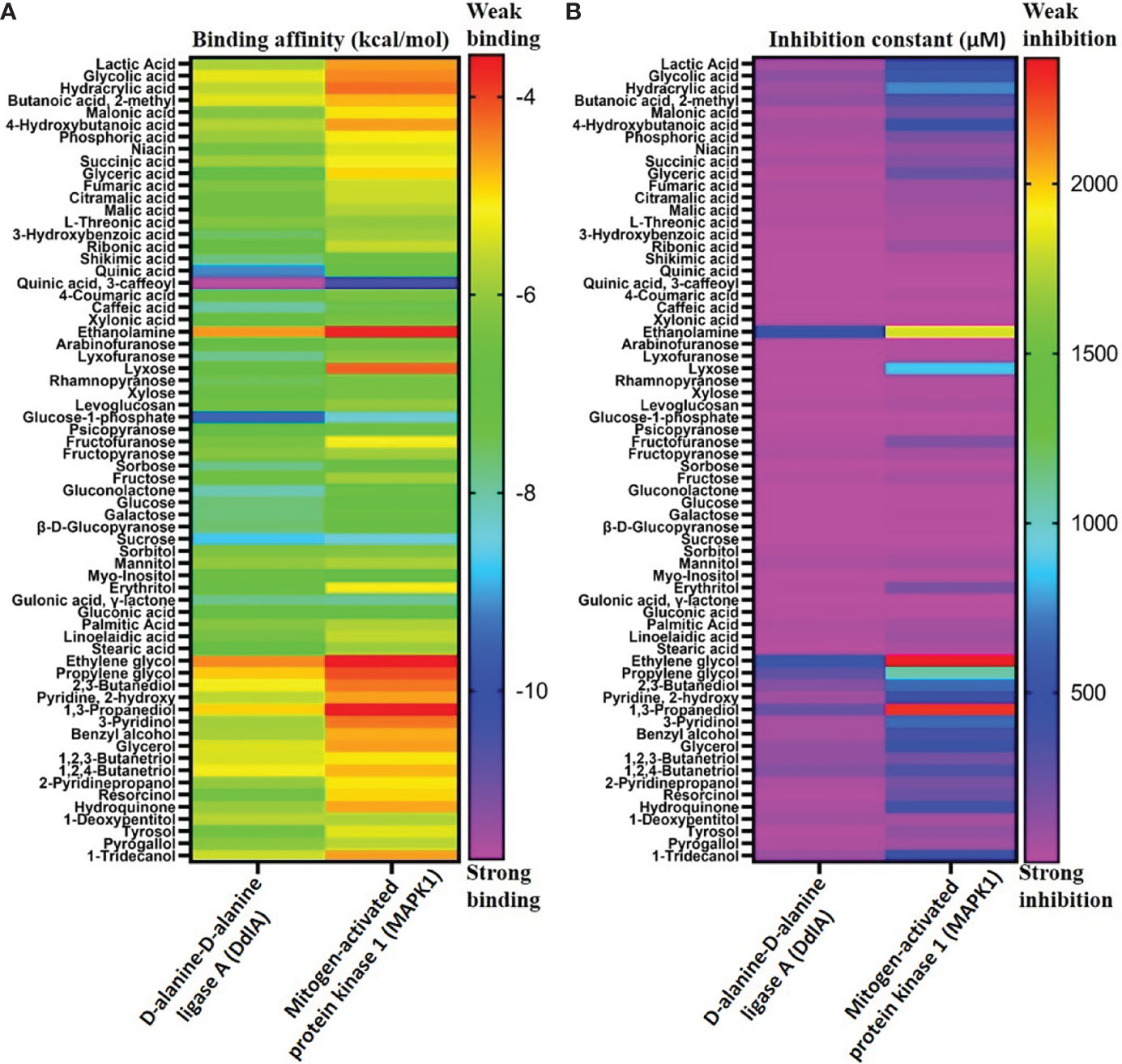
Figure 5 Binding affinity of different metabolites with DdlA (Xoo) and MAPK1 (M oryzae) protein (A). Inhibition constant of metabolites with DdlA (Xoo) and MAPK 1 (M. oryzae) protein (B). Pink and red color representing strong and weak binding affinity, respectively. Heat map represents the strong binding affinity and inhibition constant of metabolites with DdlA (Xoo) than MAPK1 (M. oryzae).
Risk Assessment of Extract Through ADMET Analysis
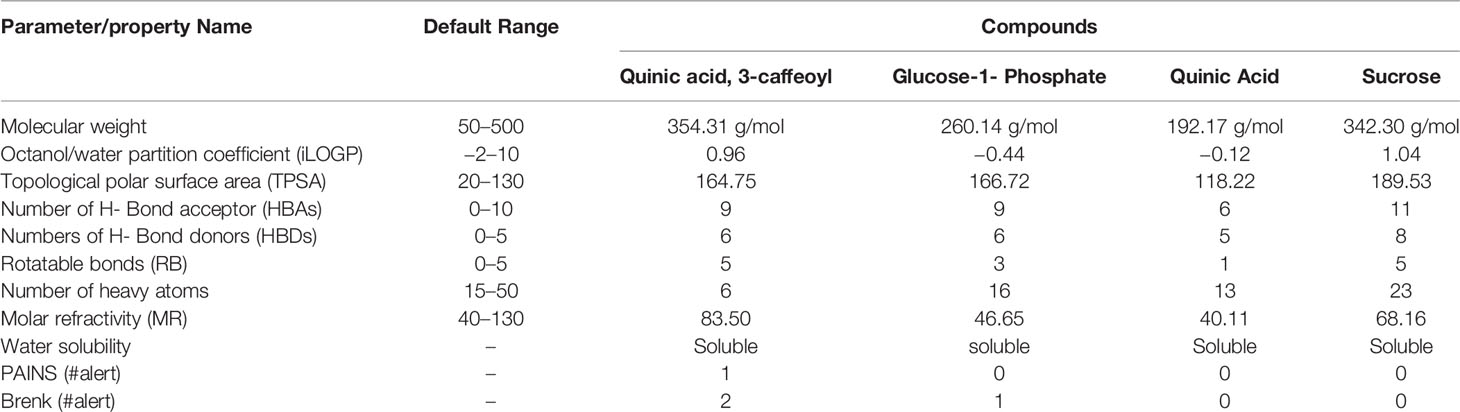
The ADMET study revealed the physicochemical nature of compounds, including the rules of five (MW, iLOGP, HBAs, and HBDs), as well as other parameters like molecular polar surface area (TPSA), rotatable bonds (ROTBs), aromatic heavy atoms numbers, and number of alerts for undesirable substructure (i.e., PAINS #alert AND Brenk #alert). Four compounds were subjected to ADMET analysis and values were compared with the default range for each parameter (Isyaku et al., 2020). It was found that MW, MR, RB, and the number of heavy atoms for each compound were considerable, whereas TPSA was only considerable, for quinic acid (Table 6). In addition, there is no alert for PAINS and Brenk for quinic acid and sucrose, whereas one and two alerts of Brenk for glucose-1-phosphate and quinic acid, 3-caffeoyl respectively. Although, all the compounds are soluble in water, which is necessary for any compounds to be a drug or drug-like. Among all the four compounds, only quinic acid has fitted under default ranges of parameters. Hence, it can be stated that quinic acid could be used as a natural antioxidant and may serve as a safe compound for treatment of BB disease without harming the surrounding flora and fauna.
Conclusion
The current investigation found that both extracts (ME and AE of U. indica) have better and similar antioxidant and free radical scavenging properties. Both ME and AE contain useful phytochemicals such as alkaloids, flavonoids, quinones, coumarin and steroids, saponins, and terpenoids, in which total phenol, alkaloid, flavonoid, and antioxidant confirm its presence through quantitative analysis. The iron chelating activities of both the extracts were also found similar, and dose-dependent scavenging activity of NO and DPPH was also noted. The correlation study indicates that NO radical scavenging activity was positively correlated with TAA and iron chelating activity. Similarly, DPPH radical scavenging activity was also found to be positively correlated with ABTS radical scavenging activity, TFC, and TPC. These results suggest that U. indica could be used to avert free radical–mediated disease occurrence, amelioration, and therapies. The FTIR spectrum analysis indicated the presence of several functional groups that are integral part of various secondary metabolites such as phenol and flavonoids, which could be responsible for the extract’s antimicrobial and antioxidant properties of the extract. The GC-MS investigation for metabolite profiling revealed the presence of several compounds with antimicrobial activity against different human and plant pathogens. Further, molecular docking study showed promising binding affinity of phytochemicals like quinic acid, 3-caffeoyl, glucose-1-phosphate, quinic acid with DdlA protein, and quinic acid, 3-caffeoyl, sucrose, and glucose-1-phosphate with MAPK1 of Xoo and M. oryzae, which could be used to target pathogen receptors to control the rice pathogens. The highest binding affinity of quinic acid, 3-caffeoyl for both DdlA and MAPK1 protein could be due to presence of caffeoyl group. In addition, ADMET analysis revealed the quinic acid as their drug-like behavior. Hence, it can be stated that quinic acid could be used as a natural antioxidant and may serve as a safe compound for treatment of BB disease and rice blast without harming the surrounding flora and fauna. However, more research studies, such as the identification and isolation of reliable specific antimicrobial and antioxidant compounds, are needed and recommended for its broad-spectrum application as well to validate the specific activity against rice pathogens.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
AK, AM, AD, and SKD designed the article. AD, AM, KS, and SKD performed the work and wrote the article. AK and TT corrected the article and finally, all authors read and approved the final article.
Funding
Financial support from MPCST (Endt.No.3879/CST/R&D/BioSci/2018) has been received to conduct the work. AM has received a fellowship from DST INSPIRE (IF 180542).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be constructed as a potential conflict of interest. All have agreed for submission of the manuscript to the Frontiers in Agronomy.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
AK sincerely acknowledges financial supports from MPCST (Endt. No. 3879/CST/R&D/BioSci/2018). AM thanks DST INSPIRE (IF 180542) for the fellowship.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fagro.2022.922306/full#supplementary-material
Supplementary Figure 1 | | Calibration curve of Gallic acid (A), Quercetin (B) and Ascorbic acid (C).
Supplementary Figure 2 | | Pearson’s correlation coefficient (r) test among TPC, TFC, TAA, DPPH, ABTS, nitric oxide radicals scavenging activity, and Iron chelating activity. Correlation coefficients are indicated by the color (Red: positive correlation; blue: negative correlation). TPC = total phneol content, TFC = total flavonoid content, TAA = total antioxidant activity, NO = nitric oxide inhibition, IC = iron chelation.
Supplementary Figure 3 | | Summary of linear correlation between binding affinity of DdlA and MAPK1 in terms of Kcal/mol.
References
Abdel-Rhman S. H., El-Mahdy A. M., El-and Mowafy M. (2015). Effect of Tyrosol and Farnesol on Virulence and Antibiotic Resistance of Clinical Isolates of Pseudomonas Aeruginosa. Biomed. Res. Int. 2015, 456463. doi: 10.1155/2015/456463
Aggarwal B. B., Shishodia S. (2006). Molecular Targets of Dietary Agents for Prevention and Therapy of Cancer. Biochem. Parmacol. 71, 1397–1421. doi: 10.1016/j.bcp.2006.02.009
Ahmed D., Khan M. M., Saeed R. (2015). Comparative Analysis of Phenolics, Flavonoids, and Antioxidant and Antibacterial Potential of Methanolic, Hexanic and Aqueous Extracts From Adiantum Caudatum Leaves. Antioxid. (Basel) 4, 394–409. doi: 10.3390/antiox4020394
Aliye M., Dekebo A., Tesso H., Abdo T., Eswaramoorthy R., Melaku Y. (2021). Molecular Docking Analysis and Evaluation of the Antibacterial and Antioxidant Activities of the Constituents of Ocimum Cufodontii. Sci. Rep. 11, 10101. doi: 10.1038/s41598-021-89557-x
Alves M. J., Ferreira I. C., Froufe H. J., Abreu R. M., Martins A., Pintado M. (2013). Antimicrobial Activity of Phenolic Compounds Identified in Wild Mushrooms, SAR Analysis and Docking Studies. J. Appl. Microbiol. 115, 346–357. doi: 10.1111/jam.12196
Alzohairy M. A. (2016). Therapeutics Role of Azadirachta Indica (Neem) and Their Active Constituents in Diseases Prevention and Treatment. Evid. Based. Complement. Alternat. Med. 2016, 7382506. doi: 10.1155/2016/7382506
Aminov R. I. (2010). A Brief History of the Antibiotic Era: Lessons Learned and Challenges for the Future. Front. Microbio. 1. doi: 10.3389/fmicb.2010.00134
Anand U., Jacobo-Herrera N., Altemimi A., Lakhssassi N. (2019). A Comprehensive Review on Medicinal Plants as Antimicrobial Therapeutics: Potential Avenues of Biocompatible Drug Discovery. Metabolites 9, 258. doi: 10.3390/metabo9110258
Anyanwu M. U., Okoye R. C. (2017). Antimicrobial Activity of Nigerian Medicinal Plants. J. Intercult. Ethnopharmacol. 6, 240–259. doi: 10.5455/jice.20170106073231
Arias-Moliz M. T., Ferrer-Luque C. M., Espigares-Rodríguez E., Liébana-Ureña J., Espigares-García M.. (2008). Bactericidal Activity of Phosphoric Acid, Citric Acid, and EDTA Solutions Against Enterococcus Faecalis. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. Endod. 106 (2), e84–9. doi: 10.1016/j.tripleo.2008.04.002
Aswal S., Kumar A., Semwal R. B., Chauhan A., Kumar A., Lehmann J., et al. (2019). Drimia Indica: A Plant Used in Traditional Medicine and Its Potential for Clinical Uses. Medicina (Kaunas) 55, 255. doi: 10.3390/medicina55060255
Bai J.-R., Wu Y.-P., Elena G., Zhong K., Gao H. (2019). Insight Into the Effect of Quinic Acid on Biofilm Formed by Staphylococcus Aureus. RSC Advances 9 (7), 3938–3945. doi: 10.1039/C8RA09136F
Bamforth C. W., Muller R. E., Walker M. D. (1993). Oxygen and Oxygen Radicals in Malting and Brewing: A Review. J. Am. Soc. Brew Chem. 53, 79–88. doi: 10.1094/ASBCJ-51-0079
Barnes R. H., Karatzas K. A. G. (2020). Investigation Into the Antimicrobial Activity of Fumarate Against Listeria Monocytogenes and Its Mode of Action Under Acidic Conditions. Int. J. Food Microbiol. 324, 108614. doi: 10.1016/j.ijfoodmicro.2020.108614
Bashir S., Abbas S., Gilani A. H., Khan A. (2013). Studies on Bronchodilator and Cardiac Stimulant Activities of Urginea Indica. Bangladesh J. Pharmacol. 8, 249–254. doi: 10.3329/bjp.v8i3.14825
Basu A., Sarkar A., Maulik U. (2020). Molecular Docking Study of Potential Phytochemicals and Their Effects on the Complex of SARS-CoV2 Spike Protein and Human ACE2. Sci. Rep. 10(2020), 17699. doi: 10.1038/s41598-020-74715-4
Bitencourt T. A., Komoto T. T., Massaroto B. G., Miranda C. E. S., Beleboni R. O., Marins M., et al. (2013). Trans-Chalcone and Quercetin Down-Regulate Fatty Acid Synthase Gene Expression and Reduce Ergosterol Content in the Human Pathogenic Dermatophyte. BMC Compl. Altern. Med. 13, 229. doi: 10.1186/1472-6882-13-229
Bursal E., Gülçin İ. (2011). Polyphenol Contents and In Vitro Antioxidant Activities of Lyophilised Aqueous Extract of Kiwifruit (Actinidia Deliciosa). Food Res. Int. 44 (5), 1482–1489. doi: 10.1016/j.foodres.2011.03.031
Campos E. V., Proença P. L., Oliveira J. L., Bakshi M., Abhilash P. C., Fraceto L. F. (2019). Use of Botanical Insecticides for Sustainable Agriculture: Future Perspectives. Ecol. Indica 105, 483–495. doi: 10.1016/j.ecolind.2018.04.038
Céspedes C. L., Avila J. G., Martínez A., Serrato B., Calderón-Mugica J. C., Salgado-Garciglia R. (2006). Antifungal and Antibacterial Activities of Mexican Tarragon (Tagetes Lucida). J. Agric. Food Chem. 54, 3521–3527. doi: 10.1021/jf053071w
Chandra S. (2019). Fourier Transform Infrared (Ft-Ir) Spectroscopic Analysis of Nicotiana Plumbaginifolia (Solanaceae). J. Medicinal Plants 7 (1), 82–85.
Chang C. C., Yang M. H., Wen H. M., Chem J. C. (2002). Estimation of Total Flavonoid Content in Propolis by Two Complementary Colorimetric Methods. J. Food Drug Anal. 10, 178–182. doi: 10.38212/2224-6614.2748
Chong K. P., Rossall S., Atong M. (2009). In Vitro Antimicrobial Activity and Fungitoxicity of Syringic Acid, Caffeic Acid and 4-Hydroxybenzoic Acid Against Ganoderma Boninense. J. Agr. Sci. 1, 15–20. doi: 10.5539/jas.v1n2p15
Chittoor M. S., Binny A. J., Yadlapalli S. K., Cheruku A., Dandu C., Nimmanapalli Y. (2012). Anthelmintic and Antimicrobial Studies of Drimia Indica (Roxb.) Jessop. Bulb Aqueous Extracts. J. Pharm. Res. 5, 3677–3686.
Cushnie T. P., Cushnie B., Lamb A. J. (2014). Alkaloids: An Overview of Their Antibacterial, Antibiotic-Enhancing and Antivirulence Activities. Int. J. Antimicrob. Agents. 44, 377–386. doi: 10.1016/j.ijantimicag.2014.06.001
Dinis T. C. P., Maderia V. M., Almeida L. M. (1994). Action of Phenolic Derivatives (Acetaminophen, Salicylate, and 5-Aminosalicylate) as Inhibitors of Membrane Lipid Peroxidation and as Peroxy Radical Scavengers. Arch. Biochem. Biophys. 315 (1), 161–169. doi: 10.1006/abbi.1994.1485
Dzotam J. K., Simo I. K., Bitchagno G., Celik I., Sandjo L. P., Tane P., et al. (2018). In Vitro Antibacterial and Antibiotic Modifying Activity of Crude Extract, Fractions and 3',4',7-Trihydroxyflavone From Myristica Fragrans Houtt Against MDR Gram-Negative Enteric Bacteria. BMC Complement. Altern. Med. 18, 15. doi: 10.1186/s12906-018-2084-1
Ebrahimzadeh M. A., Pourmorad F., Bekhradnia A. R. (2008). Iron Chelating Activity, Phenol and Flavonoid Content of Some Medicinal Plants From Iran. Afr. J. Biotechnol. 7 (18), 3188–3192.
Evans R. C. A., Miller N. J., Paganga G. (1997). Antioxidant Properties of Phenolic Compounds. Trend. Plant Sci. 4, 152–159. doi: 10.1016/S1360-1385(97)01018-2
Eswaranandam S., Hettiarachchy N. S., Johnson M. G. (2004). Antimicrobial Activity of Citric, Lactic, Malic, or Tartaric Acids and Nisin-Incorporated Soy Protein Film Against Listeria Monocytogenes, Escherichia Coli O157:H7, And Salmonella Gaminara. J. Food Sci. 69, 79–84. doi: 10.1111/j.1365-2621.2004.tb13375.x
Fayaz M., Bhat M. H., Fayaz M., Kumar A., Jain A. K. (2017). Antifungal Activity of Lantana Camara L. Leaf Extracts in Different Solvents Against Some Pathogenic Fungal Strains. Pharmacologia. 8, 105–112. doi: 10.5567/pharmacologia.2017.105.112
Fernandes A. J. D., Ferreira M. R. A., Randau K. P., de Souza T. P., Soares L. A. L. (2012). Total Flavonoids Content in the Raw Material and Aqueous Extractives From Bauhinia Monandra Kurz (Caesalpiniaceae). Sci. World J 2012, 923462. doi: 10.1100/2012/923462
Fernandes S., Gomes I. B., Simões M. (2020). Antimicrobial Activity of Glycolic Acid and Glyoxal Against Bacillus Cereus and Pseudomonas Fluorescens. Food Res. Int. 136, 109346. doi: 10.1016/j.foodres.2020.10934
Garcia-Salas P., Morales-Soto A., Segura-Carretero A., Fernández-Gutiérrez A. (2010). Phenolic-Compound-Extraction Systems for Fruit and Vegetable Samples. Molecules 15, 8813–8826. doi: 10.3390/molecules15128813
Goel S., Parihar P. S., Meshram V. (2020). “Plant-Derived Quinones as a Source of Antibacterial and Anticancer Agents,” in (Eds) Bioactive Natural Products in Drug Discovery. Eds. Singh J., Meshram V., Gupta M. (Singapore: Springer). doi: 10.1007/978-981-15-1394-7_6
Govindarajan R., Rastogi S., Vijayakumar M., Shirwaikar A., Rawat A. K., Mehrotra S., et al. (2003). Studies on the Antioxidant Activities of Desmodium Gangeticum. Biol. Pharm. Bull. 26, 1424–1427. doi: 10.1248/bpb.26.1424
Griffin S. G., Wyllie S. G., Markham J. L., Leach D. N. (1999). The Role of Structure and Molecular Properties of Terpenoids in Determining Their Antimicrobial Activity. Flavour Fragr. J. 14, 322–332. doi: 10.1002/(SICI)1099-1026(199909/10)14:5%3C322::AID-FFJ837%3E3.0.CO;2-4
Hagerman A. E., Riedl K. M., Jones G. A., Sovik K. N., Ritchard N. T., Hartzfeld P. W., et al (1998). High Molecular Weight Plant Polyphenolics (Tannins) as Biological Antioxidants. J. Agric. Food Chem. 46, 1887–92. doi: 10.1021/jf970975b
Harborne J. B. (1973). Phytochemical Methods: A Guide to Modern Techniques of Plant Analysis. 2nd Edn (London; New York, NY: Chapman and Hall Ltd).
Harborne J. B. (1998). Phytochemical Methods: A Guide to Modern Technique of Plant Analysis. 2nd edition (London: Chapman and Hall Ltd), 282.
Hidalgo A. A., Arias Á. J., Fuentes J. A., García P., Mora G. C., Villagra N. A. (2018). Xylose Improves Antibiotic Activity of Chloramphenicol and Tetracycline Against K. Pneumoniae and A. Baumannii in a Murine Model of Skin Infection. Can. J. Infect. Dis. Med. Microbiol., 3467219. doi: 10.1155/2018/3467219
Hummel J., Strehmel N., Selbig J., Walther D., Kopka J. (2010). Decision Tree Supported Substructure Prediction of Metabolites From GC-MS Profiles. Metabolomics 6, 322–333. doi: 10.1007/s11306-010-0198-7
Isyaku Y., Uzairu A., Uba S. (2020). Computational Studies of a Series of 2-Substituted Phenyl-2-Oxo-,2-Hydroxyl- and 2-Acylloxyethylsulfonamides as Potent Anti-Fungal Agents. Heliyon 6 (4), e03724. doi: 10.1016/j.heliyon.2020.e03724
Jamaloddin M., Durga Rani C. V., Swathi G., Anuradha C., Vanisri S., Rajan C., et al. (2020). Marker Assisted Gene Pyramiding (MAGP) for Bacterial Blight and Blast Resistance Into Mega Rice Variety "Tellahamsa". PloS One 15, e0234088. doi: 10.1371/journal.pone.0234088
Jarial R., Thakur S., Sakinah M., Zularisam A. W., Sharad A., Kanwar S. S., et al. (2018). Potent Anticancer, Antioxidant and Antibacterial Activities of Isolated Flavonoids From Asplenium Nidus. J. King Saud Univ. Sci 30, 185–192. doi: 10.1016/j.jksus.2016.11.006
Joray M. B., González M. L., Palacios S. M., Carpinella M. C. (2011). Antibacterial Activity of the Plant-Derived Compounds 23-Methyl-6-O-Desmethylauricepyrone and (Z,Z)-5-(Trideca-4,7-Dienyl)resorcinol and Their Synergy With Antibiotics Against Methicillin-Susceptible and -Resistant Staphylococcus Aureus. J Agric. Food Chem. 59 (21),11534–42. doi: 10.1021/jf2030665
Joshi T., Joshi T., Sharma P., Chandra S., Pande V. (2021). Molecular Docking and Molecular Dynamics Simulation Approach to Screen Natural Compounds for Inhibition of Xanthomonas Oryzae Pv. Oryzae by Targeting Pept. Deformylase. J. Biomol. Struct. Dyn. 39, 823–840. doi: 10.1080/07391102.2020.1719200
Jung K., Miyagawa M., Matsuda A., Amagai Y., Oida K., Okamoto Y., et al (2013). Antifungal Effects of Palmitic Acid Salt and Ultrapure Soft Water on Scedosporium Apiospermum. J. Appl. Microbiol. 115 (3), 711–7. doi: 10.1111/jam.12298
Karunanidhi A., Thomas R., van Belkum A., Neela V. (2013). In Vitro Antibacterial and Antibiofilm Activities of Chlorogenic Acid Against Clinical Isolates of Stenotrophomonas Maltophilia Including the Trimethoprim/Sulfamethoxazole Resistant Strain. Biomed. Res. Int 2013, 392058. doi: 10.1155/2013/392058
Kataria R., Khatkar A. (2019). Molecular Docking of Natural Phenolic Compounds for the Screening of Urease Inhibitors. Curr. Pharm. Biotechnol. 20, 410–421. doi: 10.2174/1389201020666190409110948
Kilambi H. V., Dindu A., Sharma K., Nizampatnam N. R., Gupta N., Thazath N. P., et al. (2021). The New Kid on the Block: A Dominant-Negative Mutation of Phototropin1 Enhances Carotenoid Content in Tomato Fruits. Plant J. 106, 844–861. doi: 10.1111/tpj.15206
Kocaçalişkan I., Talan I., Terzi I. (2006). Antimicrobial Activity of Catechol and Pyrogallol as Allelochemicals. Z Naturforsch C J Biosci. 61 (9–10), 639–42. doi: 10.1515/znc-2006-9-1004
Kumar A., Gul M. Z., Zeeshan A., Bimolata W., Qureshi I. A., Ghazi I. A. (2013). Differential Antioxidative Responses of Three Different Rice Genotypes During Bacterial Blight Infection. Aust. J. Crop Sci. 7, 1893–1900.
Kumar A., John M. M., Gul M. Z., Bimolata W., Ghazi I. A. (2011). Differential Responses of non-Enzymatic Antioxidative System Underwater Deficit Condition in Rice (Oryza sativaL.). Int. Proc. Chem. Biol. Environ. Eng. 9. IACSIT Press, Singapore.
Kumar A., Kumar R., Sengupta D., Das S. N., Pandey M. K., Bohra A., et al. (2020). Deployment of Genetic and Genomic Tools Toward Gaining a Better Understanding of Rice-Xanthomonas Oryzae Pv. Oryzae Interactions for Development of Durable Bacterial Blight Resistant Rice. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.01152
Kumar R., Chandar B., Parani M. (2018). Use of Succinic & Oxalic Acid in Reducing the Dosage of Colistin Against New Delhi Metallo-β-Lactamase-1 Bacteria. Indian J. Med. Res. 147 (1), 97–101. doi: 10.4103/ijmr.IJMR_1407_16
Kumar S., Sandhir R., Ojha S. (2014). Evaluation of Antioxidant Activity and Total Phenol in Different Varieties of Lantana Camara Leaves. BMC Res. Notes 7, 560. doi: 10.1186/1756-0500-7-560
Lee J. A., Chee H. Y. (2010). In Vitro Antifungal Activity of Equol Against Candida Albicans. Mycobiology 38, 328–330. doi: 10.4489/myco.2010.38.4.328
Lim Y. H., Kim I. H., Seo J. J. (2007). In Vitro Activity of Kaempferol Isolated From the Impatiens Balsamina Alone and in Combination With Erythromycin or Clindamycin Against Propionibacterium Acnes. J. Microbiol. 45, 473–477.
Lou Z., Wang H., Shengqi R., Juntao S., Chaoyang M., Jing L. (2012). P-Coumaric Acid Kills Bacteria Through Dual Damage Mechanisms. Food Control 25 (2), 550–54. doi: 10.1016/j.foodcont.2011.11.022
Lou Z., Wang H., Zhu S., Ma C., Wang Z. (2011). Antibacterial Activity and Mechanism of Action of Chlorogenic Acid. J. Food Sci. 76 (6), 398–403. doi: 10.1111/j.1750-3841.2011.02213.x
Lommen A., Kools H. J. (2012). MetAlign 3.0: Performance Enhancement by Efficient Use of Advances in Computer Hardware. Metabolomics 8, 719–726. doi: 10.1007/s11306-011-0369-1
Ma C., He N., Zhao Y., Xia D., Wei J., Kang W. (2019). Antimicrobial Mechanism of Hydroquinone. Appl. Biochem. Biotechnol. 189 (4), 1291–1303. doi: 10.1007/s12010-019-03067-1
Mansoori A., Singh N., Dubey S. K., Thakur T. K., Alkan N., Das S. N., et al. (2020). Phytochemical Characterization and Assessment of Crude Extracts From Lantana Camara L. For Antioxidant and Antimicrobial Activity. Front. Agron. 2. doi: 10.3389/fagro.2020.582268
Martínez G., Regente M., Jacobi S., Del Rio M., Pinedo M., de la Canal L. (2017). Chlorogenic Acid is a Fungicide Active Against Phytopathogenic Fungi. Pestic. Biochem. Physiol. 140, 30–35. doi: 10.1016/j.pestbp.2017.05.012
Miklasińska-Majdanik M., Kępa M., Wojtyczka R. D., Idzik D., Wąsik T. J. (2018). Phenolic Compounds Diminish Antibiotic Resistance of Staphylococcus Aureus Clinical Strains. Int. J. Environ. Res. Public Health 15, 2321. doi: 10.3390/ijerph15102321
Moghayedi M., Ahmadzadeh H., Ghazvini K., Goharshadi E. K.. (2017). Neglected Antibacterial Activity of Ethylene Glycol as a Common Solvent. Microb. Pathog. 107, 457–61. doi: 10.1016/j.micpath.2017.04.022
Mohotti S., Rajendran S., Muhammad T., Strömstedt A. A., Adhikari A., Burman R., et al. (2020). Screening for Bioactive Secondary Metabolites in Sri Lankan Medicinal Plants by Microfractionation and Targeted Isolation of Antimicrobial Flavonoids From Derris Scandens. J. Ethnopharmacol. 246, 112158. doi: 10.1016/j.jep.2019.112158
Morris G. M., Huey R., Lindstrom W., Sanner M. F., Belew R. K., Goodsell D. S., et al. (2009). Autodock4 and AutoDockTools4: Automated Docking With Selective Receptor Flexiblity J. Comput. Chem. 16, 2785–2791. doi: 10.1002/jcc.21256
Nalawade T. M., Bhat K., Sogi S.H. (2015). Bactericidal Activity of Propylene Glycol, Glycerine, Polyethylene Glycol 400, and Polyethylene Glycol 1000 Against Selected Microorganisms. J. Int. Soc. Prev. Community Dent. 5 (2), 114–19. doi: 10.4103/2231-0762.155736
Nicolopoulou-Stamati P., Maipas S., Kotampasi C., Stamatis P., Hens L. (2016). Chemical Pesticides and Human Health: The Urgent Need for a New Concept in Agriculture. Front. Public Health 4. doi: 10.3389/fpubh.2016.00148
Nisha S., Revathi K., Chandrasekaran R., Kirubakaran S., Sathish-Narayanan S., Stout M., et al. (2012). Effect of Plant Compounds on Induced Activities of Defense-Related Enzymes and Pathogenesis Related Protein in Bacterial Blight Disease Susceptible Rice Plant. Physiol. Mol. Plant Pathol., 80:1–9. doi: 10.1016/j.pmpp.2012.07.001
Pandey D., Gupta A. K. (2014). Antimicrobial Activity and Phytochemical Analysis of Urginea Indica From Bastar District of Chhattisgarh. Int. J. Pharm. Sci. Rev. Res. 26, 273–281.
Pederick J. L., Thompson A. P., Bell S. G., Bruning J. B. (2020). D-Alanine-D-Alanine Ligase as a Model for the Activation of ATP-Grasp Enzymes by Monovalent Cations. J. Biol. Chem. 295, 7894–7904. doi: 10.1074/jbc.ra120.012936
Perez-Castillo Y., Lima T. C., Ferreira A. R., Silva C. R., Campos R. S., Neto J. B. A., et al. (2020). Bioactivity and Molecular Docking Studies of Derivatives From Cinnamic and Benzoic Acids. Biomed. Res. Int. 2020, 6345429. doi: 10.1155/2020/6345429
Petti S., Scully C. (2009). Polyphenols, Oral Health and Disease. A Review. J. Dent. 37, 413–423. doi: 10.1016/j.jdent.2009.02.003
Pinho E., Soares G., Henriques M. (2015). Evaluation of antibacterial activity of caffeic acid encapsulated by β-cyclodextrins. J. Microencapsul. 32 (8), 804–10. doi: 10.3109/02652048.2015.1094531
Poojary M. M., Vishnumurthy K. A., Adhikari V (2015). Extraction, Characterization and Biological Studies of Phytochemicals From Mammea Suriga. J. Pharm. Anal. 5, 182–189. doi: 10.1016/j.jpha.2015.01.002
Prado M., Silva E. J., Duque T. M., Zaia A. A., Ferraz C. C., Almeida J. F., et al (2015). Antimicrobial and Cytotoxic Effects of Phosphoric Acid Solution Compared to Other Root Canal Irrigants. J. Appl. Oral Sci. 23 (2), 158–63. doi: 10.1590/1678-775720130691
Quiroga E. N., Sampietro D. A., Sgariglia M. A., Soberón J. R., Vattuone M. A. (2009). Antimycotic Activity of 5′-Prenylisoflavanones of the Plant Geoffroea Decorticans, AgainstSpecies. Int. J. Food Microbiol. 132, 42–46. doi: 10.1016/j.ijfoodmicro.2009.03.013
Rao V. (Ed.) (2012). Phytochemicals: A Global Perspective of Their Role in Nutrition and Health. BoD–Books on Demand. London: Intech Open. doi: 10.5772/1387
Ravindranath P. A., Forli S., Goodsell D. S., Olson A. J., Sanner M. F. (2015). AutoDockFR: Advances in Protein-Ligand Docking With Explicitly Specified Binding Site Flexibility. PloS Comput. Biol. 11, e1004586. doi: 10.1371/journal.pcbi.1004586
Re R., Pellegrinni N., Proteggente A., Pannala A., Yang M., Rice-Evans C. (1999). Antioxidant Activity Applying an Improved ABTS Radical Cation Decolorization Assay. Free Radic. Biol. Med. 26 (9-10), 1231–1237. doi: 10.1016/s0891-5849(98)00315-3
Roessner U., Wagner C., Kopka J., Trethewey R. N., Willmitzer L. (2000). Technical Advance: Simultaneous Analysis of Metabolites in Potato Tuber by Gas Chromatography-Mass Spectrometry. Plant J. 23, 131–142. doi: 10.1046/j.1365-313x.2000.00774.x
Roghini R., Vijayalakshmi K. (2018). Phytochemical Screening, Quantitative Analysis of Flavonoids and Minerals in Ethanolic Extract of Citrus Paradise. Int. J. Pharm. Sci. Res. 9, 4859–4864. doi: 10.19080/JDVS.2019.09.555768
Rosa M. R., Mosqueda-Melgar J., Martín-Belloso O.. (2009). Antimicrobial Activity of Malic Acid Against Listeria Monocytogenes, Salmonella Enteritidis and Escherichia Coli O157:H7 In Apple, Pear and Melon Juices. Food control 20 (2), 105–112. doi: 10.1016/j.foodcont.2008.02.009
Saleem M., Nazir M., Ali M. S., Hussain H., Lee Y. S., Riaz N., et al. (2010). Antimicrobial Natural Products: An Update on Future Antibiotic Drug Candidates. Nat. Prod. Rep. 27, 238–254. doi: 10.1039/b916096e
Savage P. B., Li C., Taotafa U., Ding B., Guan Q. (2002). Antibacterial Properties of Cationic Steroid Antibiotics. FEMS Microbiol. Lett. 217, 1–7. doi: 10.1111/j.1574-6968.2002.tb11448.x
Shabana Y. M., Abdalla M. E., Shahin A. A., El-Sawy M. M., Draz I. S., Youssif A. W. (2017). Efficacy of Plant Extracts in Controlling Wheat Leaf Rust Disease Caused by Puccinia Triticina. Egypt. J. Basic Appl. Sci., 4:67–73. doi: 10.1016/j.ejbas.2016.09.002
Sharipova R. R., Andreeva O. V., Garifullin B. F., Strobykina I. Y., Strobykina A. S., Voloshina A. D., et al (2017). Synthesis and Antimicrobial and Antituberculosis Activity of the First Conjugates of the Diterpenoid Isosteviol and D-Arabinofuranose. Chem. Nat. Compd., 54 (1), 92–7. doi: 10.1007/s10600-018-2267-5
Sharma K., Gupta S., Sarma S., Rai M., Sreelakshmi Y., Sharma R. (2021). Mutations in Tomato 1-Aminocyclopropane Carboxylic Acid Synthase2 Uncover its Role in Development Beside Fruit Ripening. Plant J. 106, 95–112. doi: 10.1111/tpj.15148
Shenoy S. R., Kameshwari M. S., Swaminathan S., Gupta M. N. (2006). Major Antifungal Activity From the Bulbs of Indian Squill Urginea Indica is a Chitinase. Biotechnol. Prog. 22, 631–637. doi: 10.1021/bp050305n
Singh N., Mansoori A., Jiwani G., Solanke A. K. U., Kumar R., Kumar A. (2021b). Evalution of Antioxidant and Antimicrobial Potential of Thespesia Lampas Root Extracts. J. Exp. Biol. Agric. Sci. 9 (1), 87–101. doi: 10.18006/2021.9(1).87.101
Singh N., Mansoori A., Jiwani G., Solanke A. U., Thakur T. K., Kumar R., et al. (2021a). Antioxidant and Antimicrobial Study of Schefflera Vinosa Leaves Crude Extracts Against Rice Pathogens. Arab. J. Chem. 14, 103243. doi: 10.1016/j.arabjc.2021.103243
Singleton V. L., Orthofer R., Lamuela-Raventos R. M. (1999). Analysis of Totalphenols and Other Oxidation Substrates and Antioxidants by Means of Folin-Ciocalteu Reagent. Methods Enzymol. 299, 152–178. doi: 10.1016/S0076-6879(99)99017-1
Sreejayan Rao M. N. (1997). Nitric Oxide Scavenging by Curcuminoids. J. Pharm. Pharmacol. 49 (1), 105–107. doi: 10.1111/j.2042-7158.1997.tb06761.x
Stanojević-Nikolić S., Dimić G., Mojović L., Pejin J., Djukić-Vuković A., Kocić-Tanackov S., et al (2016). Antimicrobial Activity of Lactic Acid against Pathogen and Spoilage Microorganisms. J. Food. Process. Preserv. 40, 990–98. doi: 10.1111/jfpp.12679
Surh Y. J. (2002). Anti-Tumor Promoting Potential of Selected Spice Ingredients With Antioxidative and Anti-Inflammatory Activities: A Short Review. Food Chem. Toxicol. 40, 1091–1097. doi: 10.1016/s0278-6915(02)00037-6
Tahir U. L., Qamar M., Maryam A., Muneer I., Xing F., Ashfaq U. A., et al. (2019). Computational Screening of Medicinal Plant Phytochemicals to Discover Potent Pan-Serotype Inhibitors Against Dengue Virus. Sci. Rep. 9, 1433. doi: 10.1038/s41598-018-38450-1
Tian W., Chen C., Lei X., Zhao J., Liang J. (2018). CASTp 3.0: Computed Atlas of Surface Topography of Proteins. Nucleic Acids Res. 46 (W1), W363–W367. doi: 10.1093/nar/gky473
Tikunov Y. M., Laptenok S., Hall R. D., Bovy A., de Vos R. C. (2012). MSClust: A Tool for Unsupervised Mass Spectra Extraction of Chromatography-Mass Spectrometry Ion-Wise Aligned Data. Metabolomics 8, 714–718. doi: 10.1007/s11306-011-0368-2
Togashi N., Shiraishi A., Nishizaka M., Matsuoka K., Endo K., Hamashima H., et al (2007). Antibacterial Activity of Long-Chain Fatty Alcohols Against Staphylococcus Aureus. Molecules 12 (2), 139–48. doi: 10.3390/12020139
Trease G. E., Evans W. C. (2002). Phytochemicals. In: Pharmacognosy. 15th ed (London: Saunders Publishers), 391–393.
Tripathi P., Rawat G., Yadav S., Saxena R. K. (2015). Shikimic Acid, a Base Compound for the Formulation of Swine/avian Flu Drug: Statistical Optimization, Fed-Batch and Scale Up Studies Along With Its Application as an Antibacterial Agent. Antonie Van Leeuwenhoek 107 (2), 419–31. doi: 10.1007/s10482-014-0340-z
Valko M., Leibfritz D., Moncol J., Mazur M., Telser J. (2007). Free Radicals and Antioxidants Innormal Physiological Functions and Human Disease. Int. J. Biochem. Cell Biol. 39 (1), 44–84. doi: 10.1016/j.biocel.2006.07.001
Valli M., Pivatto M., Danuello A., Castro-Gamboa I., Silva D. H. S., Cavalheiro A. J., et al. (2012). Tropical Biodiversity: Has it Been a Potential Source of Secondary Metabolites Useful for Medicinal Chemistry? Trop. Biodivers. Quím. Nova 35, 2278–2287. doi: 10.1590/S0100-40422012001100036
Vidhya V., Austine A., Arivazhagan M. (2020). Experimental Approach, Theoretical Investigation and Molecular Docking of 2- Chloro-5-Fluoro Phenol Antibacterial Compound. Heliyon 6, e05464. doi: 10.1186/s12906-018-2084-1
Xu C., Liu R., Zhang Q., Chen X., Qian Y., Fang W. (2017). The Diversification of Evolutionarily Conserved MAPK Cascades Correlates With the Evolution of Fungal Species and Development of Lifestyles. Genome Biol. Evol. 9, 311–322. doi: 10.1093/gbe/evw051
Yoshimoto M., Yahara S., Okuno S., Islam M. S., Ishiguro K., Yamakawa O. (2002). Antimutagenicity of Mono-, Di-, and Tricaffeoylquinic Acid Derivatives Isolated From Sweetpotato (Ipomoea Bata Tas L.) Leaf. Biosci. Biotechnol. Biochem. 66, 2336–2341. doi: 10.1271/bbb.66.2336
Yu Y. M. (2015). The Antibacterial Effects and Mechanism of Several Botanical Compounds Against Ralstonia Solanacearum (China: Southwest University). [master’s thesis].
Yuan H., Ma Q., Ye L., Piao G. (2016). The Traditional Medicine and Modern Medicine From Natural Products. Molecules 21, 559. doi: 10.3390/molecules21050559
Keywords: antimicrobial, antioxidant, metabolites, magnaporthe oryzae, pesticides, urginea indica, xanthomonas oryzae
Citation: Mansoori A, Dwivedi A, Sharma K, Dubey SK, Thakur TK and Kumar A (2022) Identification of Potential Inhibitors From Urginea indica Metabolites Against Xanthomonas oryzae pv. oryzae and Magnaporthe oryzae Receptors. Front. Agron. 4:922306. doi: 10.3389/fagro.2022.922306
Received: 17 April 2022; Accepted: 06 June 2022;
Published: 05 July 2022.
Edited by:
Giovanni Bubici, Institute for Sustainable Plant Protection (CNR), ItalyReviewed by:
Sadegh Balotf, University of Tasmania, AustraliaGausiya Bashri, Aligarh Muslim University, India
Copyright © 2022 Mansoori, Dwivedi, Sharma, Dubey, Thakur and Kumar. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Anirudh Kumar, YW5pcnVkaC5rdW1hckBpZ250dS5hYy5pbg==
 Aadil Mansoori
Aadil Mansoori Anurag Dwivedi
Anurag Dwivedi Kapil Sharma
Kapil Sharma Sharad Kumar Dubey
Sharad Kumar Dubey Tarun K. Thakur
Tarun K. Thakur Anirudh Kumar
Anirudh Kumar