- 1State Key Laboratory of Crop Biology, Shandong Agricultural University, Taian, China
- 2College of Agronomy, Shandong Agricultural University, Taian, China
- 3College of Agriculture, Shanxi Agricultural University, Jinzhong, China
- 4College of Life Sciences, Shandong Agricultural University, Taian, China
Sustainable agriculture has become a common trend in countries around the world. This includes returning straw to the field as part of sustainable crop production. However, it is not known whether changing the amount of straw returned to the field will lead to changes in the microbial community during decomposition and subsequent nutrient use by the next season's crop. The design included three treatments: (1) all the summer maize stalks returned to the field (SR); (2) half the summer maize stalks returned to the field (HSR); and (3) no summer maize stalks returned to the field (control: CK), base on all the winter wheat stalks were returned to the field. Compared with CK, the annual yield of SR treatment and HSR treatment increased by 18.8 and 17.3%, respectively. However, there are no significant difference in crop yield between the SR and HSR treatments. Compared with CK, the Proteobacteria increased by 29.9% in SR and 31.2% in HSR, which is the second most common bacterium during the maturity stage of maize season. Different crops season have an important impact on soil microbial community function (ANOSIM, R = 0.5209, P = 0.001). The PCoA analysis of the Faprotax function prediction found that the functional composition in the wheat planting period (wheat jointing and wheat harvest) was more similar, and the functional composition of maize planting period (maize tasseling and maize harvest) was more similar. The HSR can significantly increase the diversity of species in the soil to meet the nutritional demands for crop growth for achieve high and stable crop yield.
Introduction
The relationship between the functional diversity and the species diversity of above-ground plants and underground microbes is an important issue for understanding current ecosystem processes (Andersen et al., 2010). Focusing on this issue will improve our understanding of complex agro-ecosystems which will benefit both the restoration of ecosystem functions and the realization of agricultural functions (McGrady-Steed et al., 1997; Bardgett and Wardle, 2003). In previous research, the above-ground and underground parts of ecosystems have been traditionally isolated for scientific research. There is now a growing recognition of the interactions among these components and the fundamental role of above-ground and underground feedbacks in controlling ecosystem processes and characteristics (Grime, 2001; Van der Putten et al., 2001; Bardgett and Wardle, 2003). There is now a growing recognition of the interactions among these components and the fundamental role of above-ground and underground feedbacks in controlling ecosystem processes and characteristics (Grime, 2001; Van der Putten et al., 2001; Bardgett and Wardle, 2003). The organisms associated with roots and microorganisms have a more direct impact on plants, while at the same time affecting the energy and nutrient flows between plants and microorganisms (Wardle et al., 2004). Studying the interaction between communsities and ecosystems is an area of great interest and requires focus on both the above-ground and underground systems.
There is growing evidence that above-ground plant species have an important impact on the root system and microbial communities in the soil, while changes in underground organisms can also affect the structure and function of above-ground communities. The composition of the soil biota and its adjustment process are greatly affected by different plant species and different amounts of resources returning to the soil. For example, the composition of the microbiome around the roots of different grass species varies, which helps explain why there is different species richness of soil microbes in mixed grasslands (Bardgett et al., 1999). In forests, differences in the secretions produced by co-existing tree species explain the scattered distribution and functionality of soil organisms caused by the “single tree” effect (Saetre and Baath, 2000). In the crop-soil-environment system, the turnover and use of soil nutrients by different crops are mainly achieved by changing the microbial composition and activities (Zhu et al., 2018). At the same time, the process of straw decomposition is also influenced by soil texture, straw quality and climate (Blanco-Canqui and Lal, 2008), and the response of soil microbiome to different stages of straw decomposition is different (Shen et al., 2016).
Agricultural sustainability is a way to make full use of environmental products and services without harming the environment. The return of straw as a means of sustainable agricultural development has been replicated throughout the world (Kesavan and Swaminathan, 2007). As part of the focus of increased environmental protection by the Chinese government in recent years, a series of measures has been introduced to prevent the burning of straw waste to reduce pollution (Ni et al., 2015). Straw application increases the microbial metabolic activity and vinasse amendment causes positive or negative effects on different microbial groups (Pitombo et al., 2015; Suleiman et al., 2018). At the same time, straw incorporation management affects the community structure of soil microorganism, because the incorporation alters their habitat and provides abundant carbon sources (Wang et al., 2014). Straw could improve microbial activity and flora growth, microbial secretions, fungal mycelium which made soil particles cement composite, improved soil structure and promote the formation of soil agglomerates (Zhong et al., 2020). Microbial communities played a critical role in regulating the rate and extent of organic matter decomposition, which strongly determined soil nutrient composition. Hence, the altered soil nutrient status may be the result of modification of microbial communities (Sun et al., 2020).
The Huanghuaihai area of China has a typical winter wheat-summer maize (corn) double-crop planting model, where experiments have been carried out to see if returning all the straw to the field brings diseases or reduces yields. HSR is beneficial to promote the development of summer corn root system and nutrient accumulation through many years of positioning test verification (Gao et al., 2018). However, the relationship between the amount of returned summer maize residues and soil microbes dynamics under double-cropping system is still unclear. The purpose of this study was: (i) to explore the effects of returned summer maize straw on the soil microorganisms across winter wheat-summer maize annual rotation system; (ii) to explore the impact of the diversity of bacterial communities; and (iii) to explore the impact of different amount of straw returned on the bacterial coexistence network. This research could give some additional information to optimize management measures to achieve sustainable agriculture.
Materials and Methods
Experimental Area, Design, and Treatments
This study was conducted from 2012 to 2019 at the State Key Laboratory of Crop Biology and Experimental Farm of Shandong Agricultural University, China (36°11′ N, 117°06′ E, 151 m above sea level). The weather conditions in the maize growing season are shown in Supplementary Figure S1. This region is characterized by brown loam soil and has a temperate continental monsoon climate. The basic physical and chemical properties of the brown loam soil after successive years of straw return in the harvest stage of maize are shown in Supplementary Table S1.
The experiment was arranged as a randomized block design with three replicates, and it included the following three treatments: All the winter wheat straw produced in a season was pulverized mechanically by a harvester and returned to the field using a rotary cultivator, and then maize was sown. The experiment included the following three treatments. (1) all summer maize stalks produced returned to the field (SR), (2) half of the summer maize stalks produced returned to the field (HSR), (3) All summer maize stalks were removed from the experimental area (CK). The amount of stover returned to the field in 2012–2016 can be found in Gao et al. (2020).
Sampling and Measurements
The grain yield (moisture content is approximately 14%) of summer maize at the physiological maturity stage (R6) was estimated from 30 consecutive plants in each row for measure grain yield.
The grain yield (moisture content is ~12%) of winter wheat at maturity was estimated from all the remaining ears within three representative areas of 1 × 1.5 m for measure grain yield.
The Period and Method of Soil Microbial Sampling
In 2017, the soil sampling for each treatment was carried out during the jointing stage (April 7) and the harvesting stage (June 8) of the wheat season, the tasseling stage (August 3) and the maturity stage (October 1) of the maize season (Supplementary Figure S2). The main reasons for choosing these four growth periods are: 1. After entering the jointing of winter wheat, the tillers rapidly differentiate between effective and ineffective polarization, and the boundaries are gradually clear. The leaf area and the length and volume of the stem and ear dozens of times. 2. The harvest period is the key growth period for crop growth. 3. The flowering period is the key period in the process of maize growth and development from vegetative growth to reproductive growth. At the same time, the four growth periods are separated by 2 months, which can better represent the annual microbial dynamics. A 200 g sample of the 0–20 cm soil layer was taken and stored at −80° C (five-point sampling method). The total genomic DNA of 1 g of the soil sample was extracted according to the E.N.Z.A.TM Soil DNA Kit (Omega, USA). The concentration and quality of DNA were measured by NanoDrop 2000 photophotometer (U.S. Thermal Science) and 1% Agarose gel electrophoresis. The V3–V4 hypervariable region of the 16S rRNA gene was amplified using a bacterial universal sequencing primer (338F 5'- ACTCCTACGGGAGGCAGCAG-3'; 806R 5'-GGACTACHVGGGTWTCTAAT-3'). The sequencing platform used the Illumina MiSeq PE250 sequencer. The original sequencing data has been stored in the NCBI Sequence Read Archive (SRA) database under Accession Number PRJNA636222.
Statistical Analysis
The raw data has been analyzed using USEARCH v. 10.0 software (Edgar, 2013). Sequences into OTUs with 97% similarity has clustered through UPARSE-OTU algorithm, and the representative sequence of each OTU of the 16S rRNA gene has classified through the SILVA database (version 123 https://www.arb-silva.de/). The beta diversity of the samples was analyzed using the principal co-ordinates analysis method (PCoA). The alpha diversity indices, including ACE, Chao 1, Shannon, and Simpson, were calculated using Mothur v. 1.34.4. All statistical calculations of 16S rRNA genes in soil samples were performed based on rarefied OTU analysis, with a depth of 26,251 sequences, and bacterial sequences were clustered into 5,376 OTUs (Supplementary Figure S3). The potential microbial functional profiles of bacterial rarefied OTU level data were predicted using FAPROTAX v. 1.1 (Louca et al., 2016). ANOVA and Spearman/Pearson correlation analysis were performed using SPSS (IBM Corporation, USA). Finally, to explore microbial interactions, co-occurrence network analyses were carried out through the Molecular Ecological Network Analyses Pipeline (MENA, http://ieg4.rccc.ou.edu/MENA/main.cgi), and Gephi software was used to make networks diagrams (https://gephi.org/) (Deng et al., 2012).
Results
Yield and Its Composition of Winter Wheat and Summer Maize
SR and HSR can significantly increase yield of winter wheat and summer maize. At the winter wheat season, the yields of the SR treatment and HSR treatment increased by 38.7 and 32.1%, respectively; the harvest ear number increased by 36.3% in SR and by 28.9% in HSR, compared with that in CK. At the summer maize season, the yields of the SR treatment and HSR treatment increased by 5.2, 7.0%, respectively; the number of kernels per ear increased by 31.4% in SR and by 32.0% in HSR, compared with that in CK (Table 1).
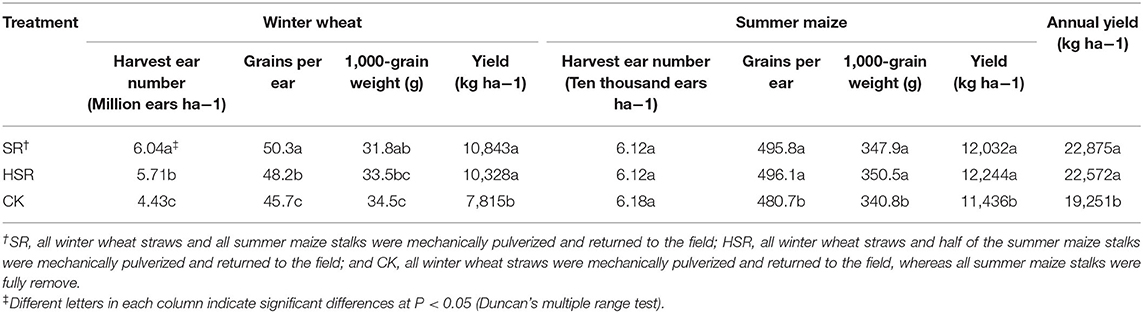
Table 1. Effect of different amount of corn stover returned to the field on yield and its components of summer maize and Winter wheat in 2017.
Soil Bacterial Alpha Diversity and Beta Diversity
In order to focus on species within the community, an analysis of alpha diversity was performed. The α diversity of CK changed relatively little over time, the OTUs and Chao1 showed an upward trend, and the Shannon and Simpson indices showed a downward trend. The shift of α diversity of the returned straw treatment was larger in different periods. The Shannon and Simpson indices of the HSR treatment decreased first, then increased and then decreased. The four indices in the SR treatment showed a trend of increasing first, then decreasing and then increasing (Figure 1).
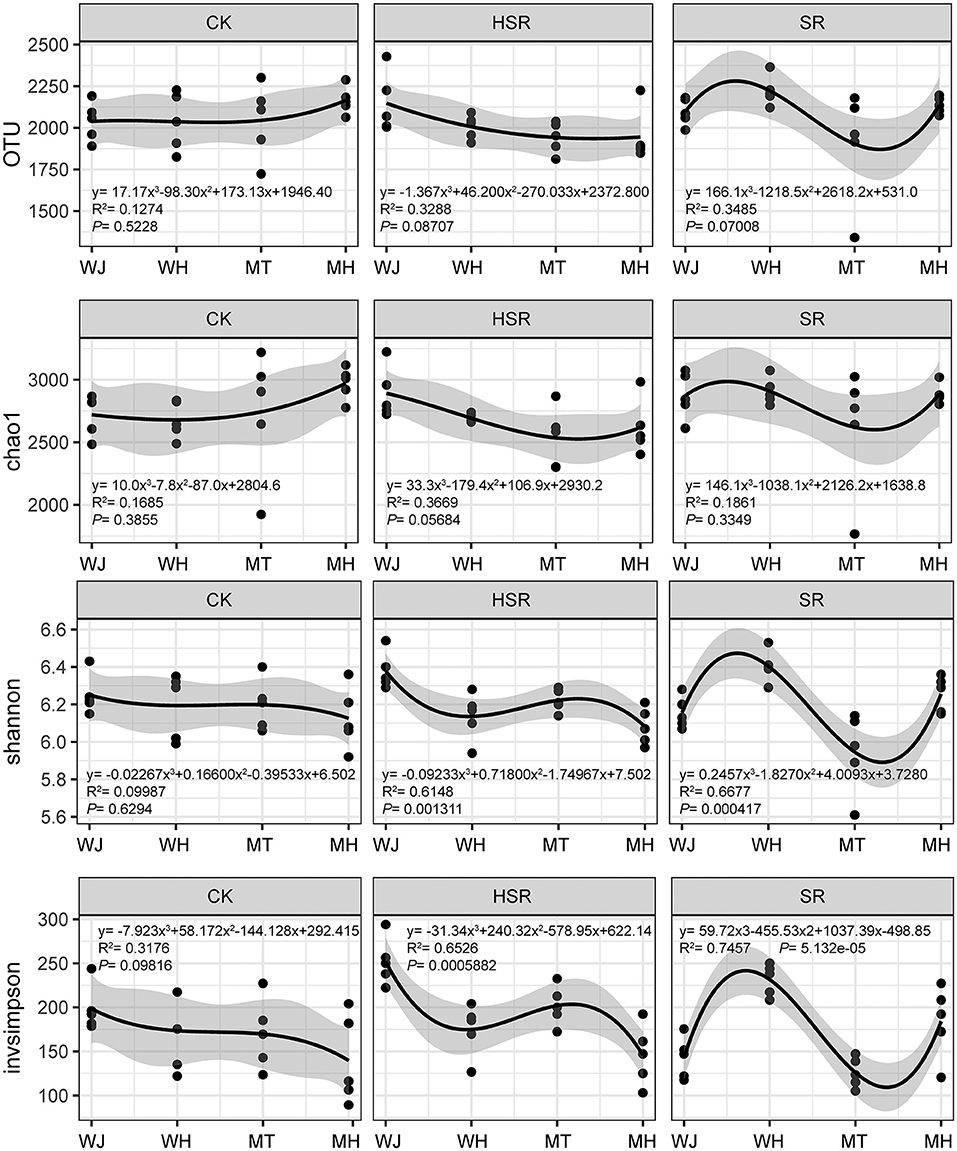
Figure 1. Effect of different amounts of corn straw returned to the field on the alpha diversity of soil bacteria. WJ, the jointing stage of the wheat season; WH, the harvesting stage of the wheat season; MT, the tasseling stage of the corn season; MH, the maturity stage of the corn season.
The effects of different amounts of straw returning on bacterial community structure based on the PCoA and ANOSIM tests. Differences in the amount of straw returned (Figure 2A, ANOSIM, R = 0.2764, P = 0.001) and seasonal differences (ANOSIM, R = 0.2492, P = 0.001) significantly affected the overall bacterial community structure. The bacterial community structure of SR, HSR and CK was significantly different (Figures 2B–E, ANOSIM, P = 0.001). The bacterial community structure was more heterogeneous in the wheat harvest season (Figure 2C, ANOSIM, R = 0.564, P = 0.001) and the maize harvest season (Figure 2E, ANOSIM, R = 0.597, P = 0.001) in the same season.
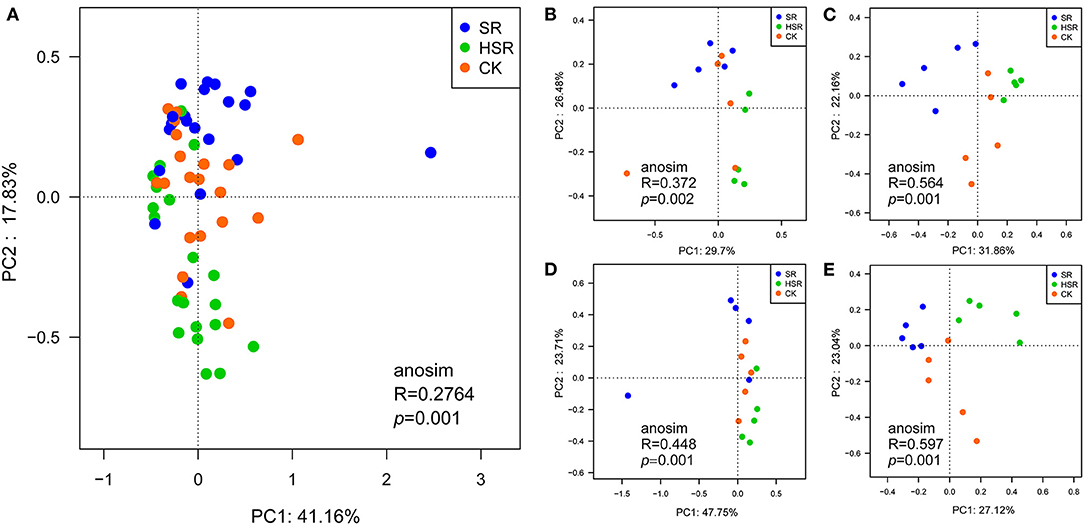
Figure 2. Effect of different amounts of corn straw returned to the field on the beta diversity of soil bacteria. (A) PCoA Analysis of Bacteria Based on Bray-curtis Dissimilarity OTU Matrix. (B) PCoA analysis in the jointing stage of the wheat season. (C) PCoA analysis in the harvesting stage of the wheat season. (D) PCoA analysis in the tasseling stage of the corn season. (E) PCoA analysis in the maturity stage of the corn season.
Bacterial Community Composition and Differences
Ten dominant phyla with relative abundances >1% of the total number of sequences were selected for species composition analysis. Different amounts of returned maize straw had no significant effect on the dominant bacteria at the phylum level. Acidobacteria was the most dominant bacterium (30.1%), followed by Chloroflexi (30.1%), Proteobacteria (11.9%), Actinobacteria (9.7%), Gemmatimonadetes (5.8%), Firmicutes (5.5%), Nitrospirae (3.5%), Saccharibacteria (3.4%), Laterscibacteria (2.8%), and Bacteroidetes (2.3%). Compared with CK, the Proteobacteria increased by 29.9% in SR and 31.2% in HSR, which is the second most common bacterium during the maturity stage of the maize season (Figure 3A). However, different amounts of returned maize straw had no significant effect on the dominant bacteria at the family level. Among these, norank_Acidobacteria was the most dominant bacterium (17.0%), followed by Blastocatellaceae_Subgroup_4 (6.1%), norank_KD4-96 (5.9%), Gemmatimonadaceae (5.4%), Acidobacteriaceae_Subgroup_1 (4.2%), norank_Nitrospira (2.3%), norank_Saccharibacteria (2.1%) (Figure 3B).
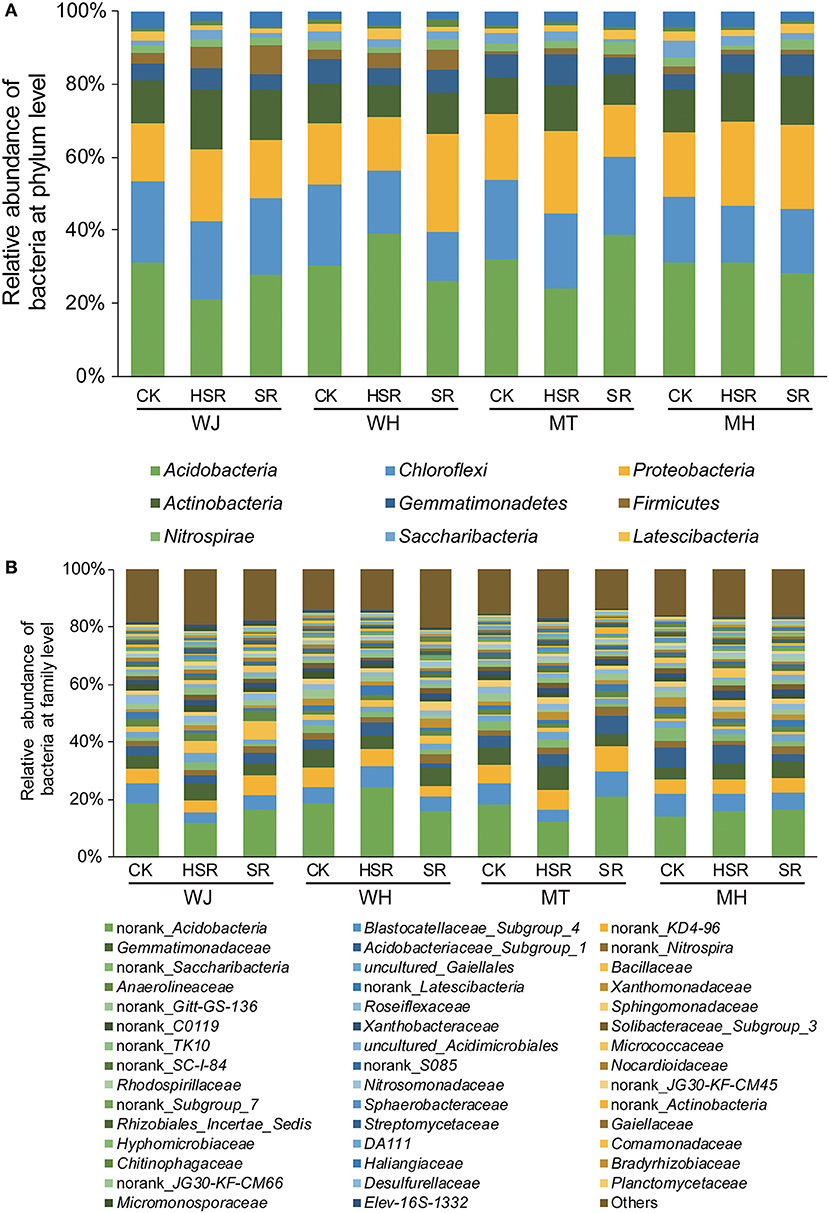
Figure 3. Effect of different amounts of corn straw returned to the field on phylum and family level of bacterial species. J, the jointing stage of the wheat season; H, the harvesting stage of the wheat season; T, the tasseling stage of the corn season; M, the maturity stage of the corn season; SR, all winter wheat straw and all summer maize stalks were pulverized mechanically and returned to the field; HSR, all winter wheat straw and one-half of the summer maize stalks were pulverized mechanically and returned to the field; CK, all winter wheat straw was pulverized mechanically and returned to the field, whereas all summer maize stalks were completely removed.
Welch's t-test was carried out on the microorganisms between each pair of treatments at the genus level. There were 59 different differential microorganisms between the three treatments at the jointing stage of the wheat season, belonging to six phyla (Actinobacteria, Bacteroidetes, Chloroflexi, Firmicutes, Gemmatimonadetes, Proteobacteria), and six differential microorganisms of Rhizobiales (Bradyrhizobium, Rhodoplanes, Nordella, Rhizomicrobium, Roseiarcus, Pseudolabrys), all of which were abundant in HSR (Supplementary Figure S4). There were 83 different differential microorganisms between the three treatments at the harvesting stage of the wheat season, belonging to eight phyla (Actinobacteria, Bacteroidetes, Chloroflexi, Firmicutes, Gemmatimonadetes, Planctomycetes, Proteobacteria, Verrucomicrobia) (Supplementary Figure S5). There were 58 different differential microorganisms between the three treatments at the tasseling stage of the maize season, belonging to eight phyla (Actinobacteria, Bacteroidetes, Chloroflexi, Firmicutes, Gemmatimonadetes, Planctomycetes, Proteobacteria, Verrucomicrobia) (Supplementary Figure S6). There were 77 different differential microorganisms between the three treatments at the maturity stage of the maize season, belonging to 10 phyla (Actinobacteria, Armatimonadetes, Bacteroidetes, Chloroflexi, Cyanobacteria, Firmicutes, Gemmatimonadetes, Planctomycetes, Proteobacteria, Verrucomicrobia) (Supplementary Figure S7), and 13 differential microorganisms of Rhizobium, all of which were abundant in HSR and SR.
At the genus level, 22 dominant genera with an abundance >0.5% were selected. The dominant genera in the four periods changed slightly between Bradyrhizobium, Mesorhizobium, and Rhizomicrobium). These belong to Proteobacteria, Alphaproteobacteria, and Rhizobiales, respectively, and they are involved in nitrogen fixation for ammonia (NH3) or ammonium (). The relative abundance of the three genera was highest in the SR treatment across the whole winter wheat-summer maize season. The relative abundance of Bradyrhizobium in HSR treatment was significantly higher than that in the SR treatment, and the relative abundance of Rhizomicrobium in HSR treatment was significantly higher than that in the SR and the CK treatment, at 0.066 and 0.041, respectively, in the jointing stage of the wheat season. Compared with the CK and SR treatments, the relative abundance of Mesorhizobium in the HSR treatment was the highest at 0.065, but this difference was not significant. In the tasseling stage of the maize season, the relative abundances of Bradyrhizobium and Rhizomicrobium in HSR were significantly higher than those in the CK and SR treatments, at 0.0062 and 0.0062, respectively. The relative abundance of Mesorhizobium of HSR was 0.0042, which was the highest among the three treatments and significantly higher than that of the SR treatment. The presence of Bryobacter of the acid bacillus may be related to CO2 or soil nutrient cycling. The relative abundance of Bryobacter in the HSR treatment was highest in both the returned straw seasons, at 0.012 (significantly higher than the CK treatment in the wheat jointing stage) and 0.006 (significantly higher than the CK and SR treatments in the maize tasseling stage). At the wheat harvest stage, the percentage of Nitrolancea, which was mainly involved in nitrite oxidation, increased in SR and HSR by 125 and 68%, respectively, compared with CK. At the maize harvest stage, compared with CK, Rhodanobacter, which may be involved in denitrification, increased in SR and HSR by 78 and 95%, respectively (Figure 4).
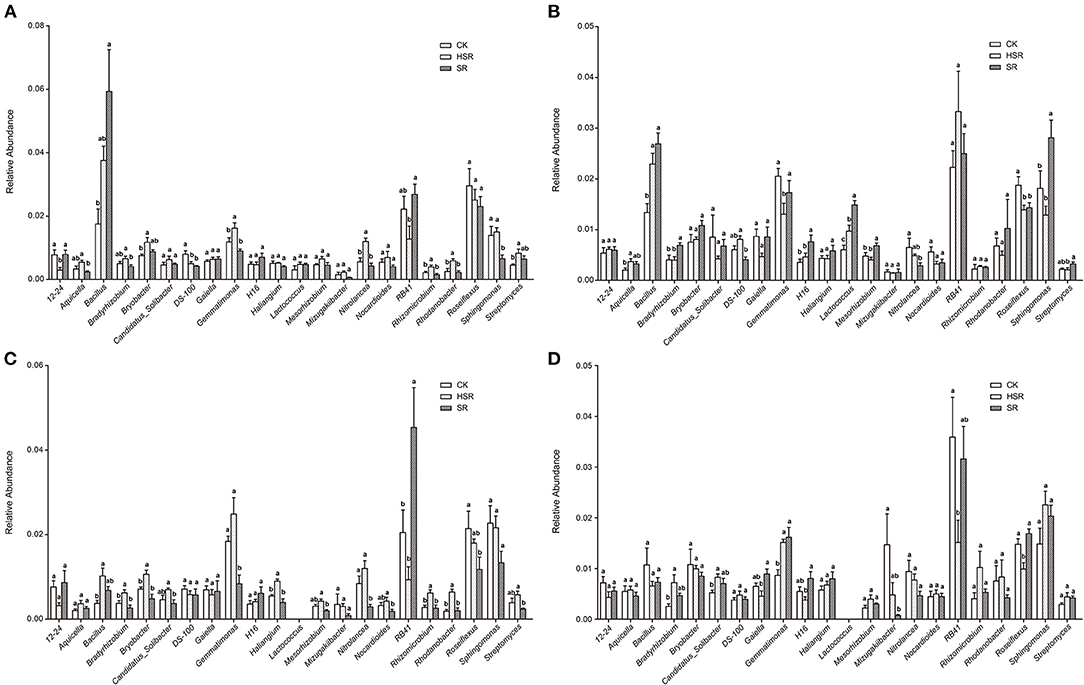
Figure 4. Effect of different amounts of corn straw returned to the field on the genus and difference of dominant bacteria. (A) The jointing stage of the wheat season. (B) The harvesting stage of the wheat season. (C) The tasseling stage of the corn season. (D) The maturity stage of the corn season. Note: different letters in each column indicate significant differences at P < 0.05 (Duncan's multiple range test).
Bacterial Co-occurrence Network Relationships
The Faprotax predictive results showed that the bacteria covered many metabolic functions and were highly enriched in chemoheterotrophy, nitrification, aerobic nitrite oxidation and reduction of nitrate. The PCoA analysis of the Faprotax function prediction found that the functional composition in the wheat planting period (wheat jointing and wheat harvest) was more similar, and the functional composition of maize planting period (maize tasseling and maize harvest) was more similar. Different crops season have an important impact on soil microbial community function (ANOSIM, R = 0.5209, P = 0.001) (Supplementary Figure S8). There was some correlation between different sampling periods [e.g., wheat jointing-maize tasseling (R = 0.441, P = 0.001***)] (Figure 5).
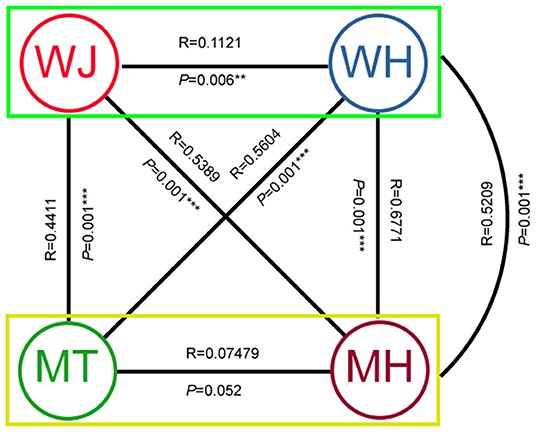
Figure 5. Correlation between different sampling time periods. WJ, the jointing stage of the wheat season; WH, the harvesting stage of the wheat season; MT, the tasseling stage of the corn season; MH, the maturity stage of the corn season.
Network analyses based on the wheat and maize seasons were constructed for finding the potential keystone taxa. The overall topological index showed that the power-law model (power-law R2, with a range of 0.791–0.938) was close to 1, indicating that most nodes in the network linked a small number of edges, while a small number of nodes linked a large number of edges, which is in line with scale-free networks. In the coexisting network, the three treatments (CK, HSR, and SR) had a similar number of nodes. The number of total links and average degree (avgK) of the returned straw treatments (HSR, SR) were significantly higher than those of CK in the wheat season. The number of total links of SR and HSR increased by 43.5 and 52.4%, compared with that of CK, the average degree (avgK) of each node of SR and HSR increased by 33.8 and 31.5%, compared with that of CK, under the wheat season (Figures 6B,C; Supplementary Table S2). In the maize season, the number of total links increased by 2.5% in HSR and 157.7% in SR, compared with that in CK (Figures 6E,F; Supplementary Table S2). This showed that the returned straw treatment increased the interaction between microorganisms, resulting in a more complex network structure.
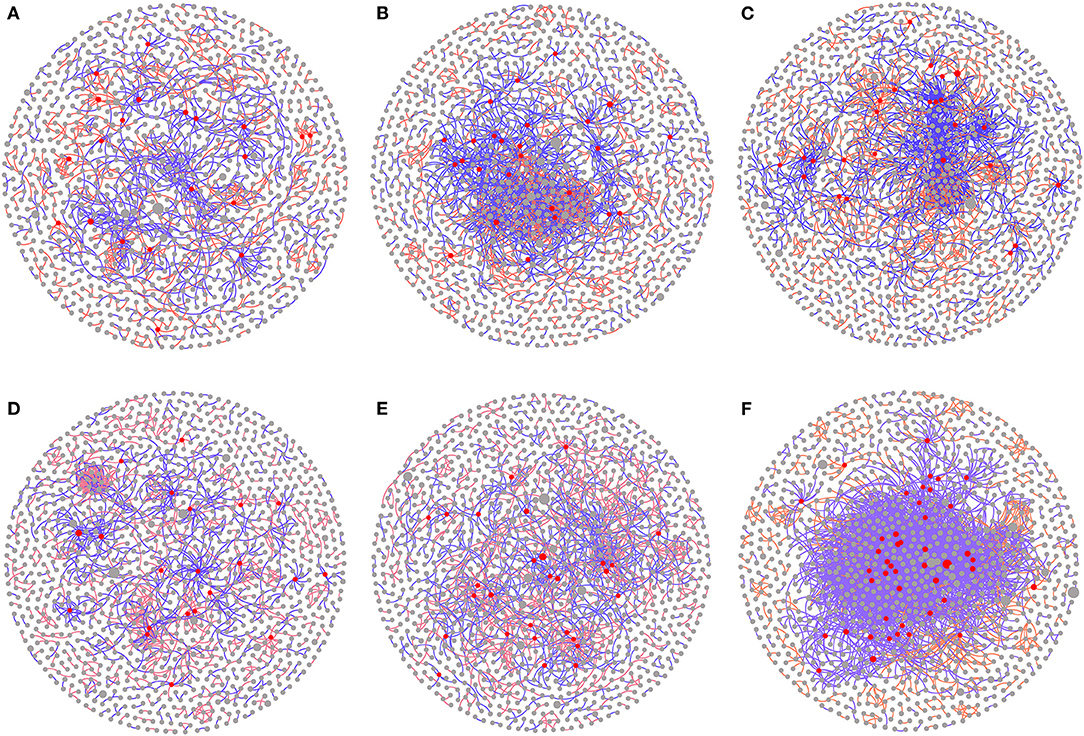
Figure 6. Effect of different amounts of corn straw returned to the field on soil bacterial coexistence network. (A) CK treatment in the wheat season. (B) HSR treatment in the wheat season. (C) SR treatment in the wheat season. (D) CK treatment in the corn season. (E) HSR treatment in the corn season. (F) SR treatment in the corn season. Nodes represent bacterial OTUs. Node colors correspond to their topological role in the network. Red is the core microorganism. The node size corresponds to the relative abundance of the OTU. The blue lines indicate positive correlation, and the red lines indicate negative correlation.
The returned straw treatments significantly increased the number of keystone OTUs in the soil bacterial communities. In the wheat season, there were 21 keystone OTUs in the bacterial community in the CK treatment, which focused on Acidobacteria, Gemmatimonadetes, and Proteobacteria, 26 keystone OTUs in the bacterial community in the HSR treatment, which belong to Acidobacteria, Actinobacteria, Chloroflexi and Proteobacteria, and 25 keystone OTUs of the bacterial community in the SR treatment, which belong to Acidobacteria and Proteobacteria (Figure 7A). The number of keystone OTUs for Actinobacteria and Proteobacteria showed the main increases with returned straw (SR and HSR) in the wheat season. In the maize season, there were 21 keystone OTUs in the bacterial community in the CK treatment, which focused on Acidobacteria, Actinobacteria, and Gemmatimonadetes, 28 keystone OTUs in the bacterial community in the HSR treatment, which focused on Acidobacteria, Actinobacteria, Chloroflexi and Proteobacteria, and 45 keystone OTUs in the bacterial community in the SR treatment which focus on Acidobacteria and Proteobacteria (Figure 7B). The number of keystone OTUs for Chloroflexi and Proteobacteria showed the main increases with returned straw (SR and HSR) in the maize season.
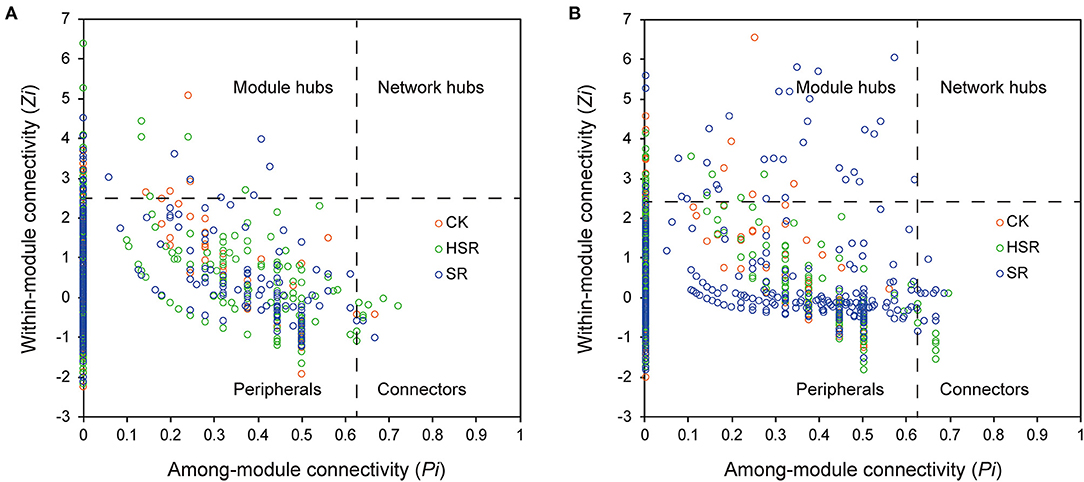
Figure 7. Distribution map of soil bacteria Zi-Pi based on topology. Zi > 2.5 and/or Pi > 0.625 are defined as Keystone OUT. (A) Distribution map of soil bacteria Zi-Pi in the wheat season. (B) Distribution map of soil bacteria Zi-Pi in the wheat season.
Discussion
Straw return to the field is one of the common practices in crop production. However, crop residues are sometimes seen as a serious problem. For example, in the North China Plain, there can be a negative effect if all the straw is returned under double-cropping systems. Crop residue reduction is a promising management to reduce soil degradation and improve the nutrition cycle in agro-ecosystems (Lal, 2012). The decay of straw is restricted by a variety of conditions (e.g., accumulation temperature, water, soil type and crop type), research has focused on finding the correct amount of straw for a specific environment under specific regional conditions, while maintaining the sustainable production capacity of soil. However, the available literature is rarely reported about the effect of changing the amount of returned straw on the soil microorganisms.
First, it is important to understand the temporal response of the soil microbial community after adding straw. Soil microorganisms may cause fluctuations due to the addition of organic matter and return to a stable state after a period of time (Zhang et al., 2017). A considerable number of temporal studies have focused on microbial-driven biogeochemical processes (Meyer et al., 2004) and specific functions in the soil, indirectly showing their activities (Strickland et al., 2009). However, few studies have looked at how the composition and function of general microorganisms cope with agricultural disturbances; in short-term experiments, the microbial community structure of different treatments does not show variability over time (Suleiman et al., 2016). Here, we showed that after many years of experimental treatment, microbiodiversity between different treatments showed different trends (Figure 1), which is inconsistent with the results of Rachid et al. (2016) on the addition of straw.
Second, how much influence does the addition of maize stalks back to the field affect the composition and function of the microbiome? This avoided the possibility that the impact of the amount of returned straw might be masked by comparing two crops over four periods. Through the analysis of beta diversity, we found that there was greater heterogeneity in the harvest season for wheat (Supplementary Figure S5) and maize (Supplementary Figure S7). At the phylum and family level, there were significant differences in the three treatments (Figures 3, 4). In the returned straw treatments, the functions of the more abundant microbial populations related to C-compound degradation, methylotrophic metabolism and to nitrogen metabolism, including nitrogen fixation, denitrification and nitrification. Streptomyces, which plays an important role in the degradation of organic matter (https://microbewiki.kenyon.edu/index.php/streptomyces), had a higher abundance in returned straw treatments.
The returned straw treatments (SR and HSR) also significantly increased the amount of keystone OTUs for Proteobacteria, which mainly played a role in nitrogen fixation (Figure 7; Supplementary Table S2). Rhodanobacter can achieve denitrification, a process of converting nitrate to N2 via microbial enzyme-catalyzed nitrite, nitric acid, and nitrous oxide intermediates (Spain and Krumholz, 2011). Bradyrhizobium bacteria can participate in the nitrogen fixation process, laying the foundation for nutrient absorption of crops and nutrient fixation of soil (Berkum et al., 1985; Stacey, 1995). Sphingomonas can metabolize various carbon sources in low concentrations of nutrients (Kelley et al., 2004). The aforementioned bacterial group has been extensively studied, but we have found a lesser known Gemmatimonadales at this stage. Studies have found that the group activity in Gemmatimonadales increased after rice straw returned to the soil (Whitman et al., 2016; Zhou et al., 2020). In this experiment, the Gemmatimonadales group was highly expressed in the next season of crop cultivation after the straw was returned to the field. The anaerobic reduction of nitrates leads to the decomposition of different physiological abilities (Hesselsoe et al., 2009; Lecomte et al., 2018). The key role of soil microbes in carbon sequestration is well-known (Gougoulias et al., 2014). Previous studies have shown that the nutritional supply of microorganisms is likely to be stable organic matter (Singh et al., 2010; Liang and Balser, 2011). The content of organic carbon in soil is determined by the mineralization and assimilation balance of microorganisms (Moorhead et al., 2012; Xiao et al., 2018), which may depend on the structure and composition of the community (Yang et al., 2011). Bacillales has a high abundance in the processing of returned straw, and members of this phylum are known to grow rapidly in a C-rich environment (Pitombo et al., 2015; Jalme and Gail, 2020), which can resist adverse conditions, laying the foundation for healthy crop growth.
It is also important to know what factors (e.g., crop type or returned straw) have the greatest influence on the functional composition of the microbial community. It is well-known that the composition of microbes is constrained by environmental impacts (Gibbons, 2017). To this end, we chose FAPROTAX for functional prediction, which can analyze earth chemical cycles (Schiff et al., 2017). Different crops have a more significant effect than returned straw on the function of soil microbial communities by the results of this study (Figure 5). In the bacterial coexistence network, returned straw increased the interaction between microorganisms in different crops, which provided strong support for crop growth and development in turn (Figure 6). For microbial keystone OTUs, returned straw increased the number of key OTUs regardless of the crop production conditions. Actinobacteria played a key role in rotting animal and plant remains in the soil in the wheat season (Figure 7A). In previous research found that returned straw significantly increased soil fertility but, with greater amounts of returned straw, the increase in soil fertility ceased (Gao et al., 2020). In the process of improving soil functions, it is important to improve the carbon sequestration capacity of agricultural soils, reduce soil degradation, avoid nutrient depletion, and reduce the impact on carbon dioxide emissions in the agricultural management process (Banerjee et al., 2016). The HSR treatment can significantly reduce the efflux of greenhouse gases and contribute to the global mitigation of greenhouse gas emissions (Gao et al., 2019). The next stage of work is mainly carried out from the following aspects: 1. Purification and utilization of specific functional microbial communities; 2. Research on the relationship between rhizosphere soil microorganisms and crop types; 3. This experimental design will be promoted to achieve stable crop yields while ensuring soil sustainability. These studies provide theoretical basis for the rational use of crop straw resources.
Conclusion
Compared with CK, the annual yield of SR treatment and HSR treatment increased by 18.8, 17.3%, respectively. However, there are no significant difference in crop yield between the SR and HSR treatments. The relative abundance of Bradyrhizobium and Nitrolancea in HSR treatment was significantly higher than that in the CK treatment, which are important bacteria for nitrogen fixation and nitrification-denitrification. This residue management practice provides a foundation by optimizing the microbial community structure in the soil to achieve no significant difference in yield with SR treatment under double-cropping systems.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA636222.
Author Contributions
ZG and JZ contributed to conception and designed the study. JZ organized the database. FG and CZ performed the statistical analysis. FG wrote the first draft of the manuscript. All authors contributed to manuscript revision, read, and approved the submitted version.
Funding
This work was funded by China Agriculture Research System of MOF and MARA (CARS-02-18), Shandong Central Guiding the Local Science and Technology Development (YDZX20203700002548), and Science and Technology Innovation Plan for Higher Education Institutions in Shanxi Province (2021L176).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fagro.2022.855820/full#supplementary-material
References
Andersen, R., Grasset, L., Thormann, M. N., Rochefort, L., and Francez, A. J. (2010). Changes in microbial community structure and function following Sphagnum peatland restoration. Soil Biol. Biochem. 42, 291–301. doi: 10.1016/j.soilbio.2009.11.006
Banerjee, S., Kirkby, C. A., Schmutter, D., Bissett, A., Kirkegaard, J. A., and Richardson, A. E. (2016). Network analysis reveals functional redundancy and keystone taxa amongst bacterial and fungal communities during organic matter decomposition in an arable soil. Soil Biol. Biochem. 97, 188–198. doi: 10.1016/j.soilbio.2016.03.017
Bardgett, R. D., Mawdsley, J. L., Edwards, S., Hobbs, P. J., Rodwell, J. S., and Davies, W. J. (1999). Plant species and nitrogen effects on soil biological properties of temperate upland grasslands. Funct. Ecol. 13, 650–660. doi: 10.1046/j.1365-2435.1999.00362.x
Bardgett, R. D., and Wardle, D. A. (2003). Herbivore-mediated linkages between aboveground and belowground communities. Ecology 84, 2258–2268. doi: 10.1890/02-0274
Berkum, P., Sloger, C., Weber, D. F., Cregan, P. B., and Keyser, H. H. (1985). Relationship between ureide N and N2 fixation, aboveground N accumulation, acetylene reduction, and nodule mass in greenhouse and field studies with Glycine max L. (Merr). Plant Physiol. 77, 53–58. doi: 10.1104/pp.77.1.53
Blanco-Canqui, H., and Lal, R. (2008). Corn stover removal impacts on micro-scale soil physical properties. Geoderma 145, 335–346. doi: 10.1016/j.geoderma.2008.03.016
Deng, Y., Jiang, Y. H., Yang, Y., He, Z., Luo, F., and Zhou, J. (2012). Molecular ecological network analyses. BMC Bioinformat. 13, 1–20. doi: 10.1186/1471-2105-13-113
Edgar, R. C. (2013). UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 10, 996–998. doi: 10.1038/nmeth.2604
Gao, F., Hu, J., Ren, B., Liu, P., Zhao, B., and Zhang, J. (2020). Improving soil properties and grains yield of winter wheat and summer corn under residue management strategies. Agron. J. 112, 4287–4302. doi: 10.1002/agj2.20283
Gao, F., Li, B., Ren, B., Zhao, B., Liu, P., and Zhang, J. (2019). Effects of residue management strategies on greenhouse gases and yield under double cropping of winter wheat and summer maize. Sci. Total Environ. 687, 1138–1146. doi: 10.1016/j.scitotenv.2019.06.146
Gao, F., Zhao, B., Dong, S., Liu, P., and Zhang, J. (2018). Response of maize root growth to residue management strategies. Agron. J. 110, 95–103. doi: 10.2134/agronj2017.06.0307
Gibbons, S. M. (2017). Microbial community ecology: function over phylogeny. Nat. Ecol. Evol. 1, 1–2. doi: 10.1038/s41559-016-0032
Gougoulias, C., Clark, M., and Shaw, J. (2014). The role of soil microbes in the global carbon cycle: tracking the below-ground microbial processing of plant-derived carbon for manipulating carbon dynamics in agricultural systems. J. Sci. Food Agr. 94, 2362–2371. doi: 10.1002/jsfa.6577
Grime, J. P. (2001). Plant Strategies, Vegetation Processes, and Ecosystem Properties, 2nd Edn. Chichester: Wiley.
Hesselsoe, M., Fureder, S., Schloter, M., Bodrossy, L., Iversen, N., Roslev, P., et al. (2009). Isotope array analysis of Rhodocyclales uncovers functional redundancy and versatility in an activated sludge. ISME J. 3, 1349–1364. doi: 10.1038/ismej.2009.78
Jalme, C., and Gail, M. P. (2020). Growing edible mushromms: a conversation between bacteria and fungi. Environ. Microbiol. 22, 858–872. doi: 10.1111/1462-2920.14765
Kelley, S. T., Theisen, U., Angenent, L. T., St. Amand, A., and Pace, N. R. (2004). Molecular analysis of shower curtain biofilm microbes. Appl. and Environ. Microb. 70, 4187–4192. doi: 10.1128/AEM.70.7.4187-4192.2004
Kesavan, P. C., and Swaminathan, M. S. (2007). Strategies and models for agricultural sustainability in developing Asian countries. Philos. T. R. Soc. B 363, 877–891. doi: 10.1098/rstb.2007.2189
Lal, R. (2012). Climate change and soil degradation mitigation by sustainable management of soils and other natural resources. Agric. Res. 1, 199–212. doi: 10.1007/s40003-012-0031-9
Lecomte, S. M., Achouak, W., Abrouk, D., Heulin, T., Nesme, X., and Haichar, F. Z. (2018). Diversifying anaerobic respiration strategies to compete in the rhizosphere. Front. Environ. Sci. 6,139. doi: 10.3389/fenvs.2018.00139
Liang, C., and Balser, T. C. (2011). Microbial production of recalcitrant organic matter in global soils: implications for productivity and climate policy. Nat. Rev. Microbiol. 9, 75–75. doi: 10.1038/nrmicro2386-c1
Louca, S., Parfrey, L. W., and Doebeli, M. (2016). Decoupling function and taxonomy in the global ocean microbiome. Science 353, 1272–1277. doi: 10.1126/science.aaf4507
McGrady-Steed, J., Harris, P., and Morin, P. (1997). Biodiversity regulates ecosystem predictability. Nature 390, 162–165. doi: 10.1038/36561
Meyer, A. F., Lipson, D. A., Martin, A. P., Schadt, C. W., and Schmidt, S. K. (2004). Molecular and metabolic characterization of cold-tolerant alpine soil Pseudomonas sensu stricto. Appl. Environ. Microbiol. 70, 483–489. doi: 10.1128/AEM.70.1.483-489.2004
Moorhead, D. L., Lashermes, G., and Sinsabaugh, R. L. (2012). A theoretical model of c- and n-acquiring exoenzyme activities, which balances microbial demands during decomposition. Soil Biol. Biochem. 53, 133–141. doi: 10.1016/j.soilbio.2012.05.011
Ni, H., Han, Y., Cao, J., Chen, L. W. A., Tian, J., Wang, X., et al. (2015). Emission characteristics of carbonaceous particles and trace gases from open burning of crop residues in China. Atmos. Environ. 123, 399–406. doi: 10.1016/j.atmosenv.2015.05.007
Pitombo, L. M., do Carmo, J. B., de Hollander, M., Rossetto, R., López, M. V., Cantarella, H., et al. (2015). Exploring soil microbial 16S rRNA sequence data to increase carbon yield and nitrogen efficiency of a bioenergy crop. GCB Bioenergy 8, 867–879. doi: 10.1111/gcbb.12284
Rachid, C. T. C. C., Pires, C. A., Leite, D. C. A., Coutinho, H. L. C., Peixoto, R. S., Rosado, A. S., et al. (2016). Sugarcane trash levels in soil affects the fungi but not bacteria in a short-term field experiment. Braz. J. Microbiol. 47, 322–326. doi: 10.1016/j.bjm.2016.01.010
Saetre, P., and Baath, E. (2000). Spatial variation and patterns of soil microbial community structure in a mixed spruce–birch stand. Soil Biol. Biochem. 32, 909–917. doi: 10.1016/S0038-0717(99)00215-1
Schiff, S. L., Tsuji, J. M., Wu, L., Venkiteswaran, J. J., Molot, L. A., Elgood, R. J., et al. (2017). Millions of boreal shield lakes can be used to probe Archaean Ocean biogeochemistry. Sci. Rep. 7, 46708. doi: 10.1038/srep46708
Shen, W., Ni, Y., Gao, N., Bian, B., Zheng, S., Lin, X., et al. (2016). Bacterial community composition is shaped by soil secondary salinization and acidification brought on by high nitrogen fertilization rates. Appl. Soil Ecol. 108, 76–83. doi: 10.1016/j.apsoil.2016.08.005
Singh, B., Bardgett, R., Smith, P., et al. (2010). Microorganisms and climate change: terrestrial feedbacks and mitigation options. Nat. Rev. Microbiol. 8, 779–790. doi: 10.1038/nrmicro2439
Spain, A. M., and Krumholz, L. R. (2011). Nitrate-reducing bacteria at the nitrate and radionuclide contaminated Oak Ridge Integrated Field Research Challenge site: a review. Geomicrobiol. J. 28, 418–429. doi: 10.1080/01490451.2010.507642
Stacey, G., Sanjuan, J., Luka, S., Dockendorff, T., and Carlson, R. W. (1995). Signal exchange in the Bradyrhizobium–soybean symbiosis. Soil Biol. Biochem. 27, 473–483. doi: 10.1016/0038-0717(95)98622-U
Strickland, M. S., Lauber, C., Fierer, N., and Bradford, M. A. (2009). Testing the functional significance of microbial community composition. Ecology 90, 441-−451. doi: 10.1890/08-0296.1
Suleiman, A. K. A., Gonzatto, R., Aita, C., Lupatini, M., Jacques, R. J. S., Kuramae, E. E., et al. (2016). Temporal variability of soil microbial communities after application of dicyandiamide-treated swine slurry and mineral fertilizers. Soil Biol. Biochem. 97, 71–82. doi: 10.1016/j.soilbio.2016.03.002
Suleiman, A. K. A., Lourenço, K. S., Pitombo, L. M., Mendes, L. W., Roesch, L. F. W., Pijl, A., et al. (2018). Recycling organic residues in agriculture impacts soil-borne microbial community structure, function and N2O emissions. Sci. Total Environ. 631, 1089–1099. doi: 10.1016/j.scitotenv.2018.03.116
Sun, K., Han, L., Yang, Y., Xia, X., Yang, Z., Wu, F., et al. (2020). Application of hydrochar altered soil microbial community composition and the molecular structure of native soil organic carbon in a paddy soil. Environ. Sci. Technol. 54, 2715–2725. doi: 10.1021/acs.est.9b05864
Van der Putten, W. H., Vet, L. E., Harvey, J. A., and Wäckers, F. L. (2001). Linking above-and belowground multitrophic interactions of plants, herbivores, pathogens, and their antagonists. Trends Ecol. Evol. 16, 547–554. doi: 10.1016/S0169-5347(01)02265-0
Wang, J. J., Zhang, H. W., Li, X. Y., Su, Z. C., Li, X., and Xu, M. K. (2014). Effects of tillage and residue incorporation on composition and abundance of microbial communities of a fluvo-aquic soil. Eur. J. Soil Biol. 65, 70–78. doi: 10.1016/j.ejsobi.2014.10.003
Wardle, D. A., Bardgett, R. D., Klironomos, J. N., Setäl,ä, H., Van Der Putten, W. H., and Wall, D. H. (2004). Ecological linkages between aboveground and belowground biota. Science 304, 1629–1633. doi: 10.1126/science.1094875
Whitman, T., Pepe-Ranney, C., Enders, A., Koechli, C., Campbell, A., and Buckley, D. H. (2016). Dynamics of microbial community composition and soil organic carbon mineralization in soil following addition of pyrogenic and fresh organic matter. ISME J. 10, 2918–2930. doi: 10.1038/ismej.2016.68
Xiao, H., Li, Z., Chang, X., Nie, X., Liu, C., Dan, L., et al. (2018). The mineralization and sequestration of organic carbon in relation to agricultural soil erosion. Geoderma 329, 73–81. doi: 10.1016/j.geoderma.2018.05.018
Yang, H., Wu, M., Liu, W., Zhang, Z., Zhang, N., and Wan, S. (2011). Community structure and composition in response to climate change in a temperate steppe. Global Change Biol. 17, 452–465. doi: 10.1111/j.1365-2486.2010.02253.x
Zhang, W., Liang, C., Kao-Kniffin, J., He, H., Xie, H., Zhang, X., et al. (2017). Effects of drying and wetting cycles on the transformations of extraneous inorganic N to soil microbial residues. Sci. Rep. 7, 9477. doi: 10.1038/s41598-017-09944-1
Zhong, Y., Liu, J., Jia, X., Shangguan, Z., and Yan, W. (2020). Microbial community assembly and metabolic function during wheat straw decomposition under different nitrogen fertilization treatments. Biol. Fertil. Soils 56, 697–710. doi: 10.1007/s00374-020-01438-z
Zhou, C. B., Heal, K., Tigabu, M., Xia, L., Hu, H., et al. (2020). Biochar addition to forest plantation soil enhances phosphorus availability and soil bacterial community diversity. Forest Ecol. Manag. 455, 117635. doi: 10.1016/j.foreco.2019.117635
Keywords: straw returned, yield, microbes, community structure, double-cropping systems
Citation: Gao F, Zhang C, Gao Z and Zhang J (2022) Response of the Soil Microbe Community to Maize Residue Management Strategies Under Double-Cropping Systems. Front. Agron. 4:855820. doi: 10.3389/fagro.2022.855820
Received: 16 January 2022; Accepted: 27 April 2022;
Published: 16 June 2022.
Edited by:
Pankaj Kumar Verma, Ben-Gurion University of the Negev, IsraelReviewed by:
Jun Zhao, Nanjing Normal University, ChinaParneeta Mishra, National Botanical Research Institute (CSIR), India
Copyright © 2022 Gao, Zhang, Gao and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jiwang Zhang, and6aGFuZ0BzZGF1LmVkdS5jbg==; Zheng Gao, Z2FvemhlbmdAc2RhdS5lZHUuY24=
†ORCID: Fei Gao orcid.org/0000-0003-1637-2071
Zheng Gao orcid.org/0000-0003-1691-0976
‡These authors have contributed equally to this work
 Fei Gao1,2,3†‡
Fei Gao1,2,3†‡ Chao Zhang
Chao Zhang Jiwang Zhang
Jiwang Zhang