
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Agron. , 23 September 2022
Sec. Weed Management
Volume 4 - 2022 | https://doi.org/10.3389/fagro.2022.1010292
This article is part of the Research Topic Women in Agronomy View all 8 articles
Herbicides, such as tembotrione, that inhibit 4-hydroxyphenylpyruvate dioxygenase (HPPD) enzyme are used to control broad spectrum of weeds, primarily in corn, as this crop can metabolize these herbicides via cytochrome P450 activity. In 2018, a Palmer amaranth (Amaranthus palmeri) population, KCTR was found to be resistant to multiple herbicides including, tembotrione in Kansas (KS), USA. However, the mechanism of tembotrione resistance in KCTR is not known. The objective of this study was to characterize the level of tembotrione resistance and investigate the mechanism of resistance to this herbicide in KCTR using a known susceptible Palmer amaranth population (KSS). Tembotrione dose response experiments revealed that KCTR Palmer amaranth is 23 times more resistant to this herbicide, than KSS. No difference in absorption or translocation of [14C] tembotrione between KSS and KCTR was found. However, the time required to metabolize 50% of tembotrione was shorter in KCTR than in KSS. More than 95% of tembotrione was metabolized at 6 hours after treatment (HAT) in the KCTR, compared to only 50% in KSS plants. Application of cytochrome P450-inhibitors (e.g., malathion or piperonyl butoxide), along with tembotrione reversed the resistance in KCTR. Furthermore, the KCTR plants showed 35-fold increase in constitutive expression of CYP81E8 gene compared to KSS. Nonetheless, the HPPD gene expression was not altered in KCTR Palmer amaranth. Our results suggest that enhanced metabolism of tembotrione possibly due to increased expression of CYP81E8 gene contribute to tembotrione resistance in KCTR. Metabolic resistance to herbicides is a challenge for weed management as such resistance predisposes weeds to evolve resistance to unknown herbicides even without selection.
Based on the chemical structure and properties, the 4-hydroxyphenylpyruvate dioxygenase (HPPD)-inhibitors are broadly classified into three chemical families: isoxazoles (e.g., isoxaflutole and pyrasulfotole), pyrazolones (e.g., topramezone), and triketones (e.g., mesotrione and tembotrione) (Lee et al., 1998; Van Almsick, 2009). Tembotrione (2-[2-chloro-4-(methylsulfonyl)-3-[(2,2,2-(trifluoroethoxy)methyl]benzoyl]-1,3-cyclohexanedione) is widely used for the control of wide spectrum of broad-leaved and grass weeds (Beaudegnies et al., 2009; Ahrens et al., 2013). The herbicidal activity of tembotrione is due to the competitive inhibition of HPPD (EC1.13.11.27), a ubiquitous non-heme oxygenase involved in the catabolism of the amino acid tyrosine (Matringe et al., 2005). This enzyme catalyzes the formation of homogentisate, an intermediate in the biosynthesis of α-tocopherols and plastoquinone (Matringe et al., 2005; Ndikuryayo et al., 2017). Plastoquinone serves as a cofactor for the enzyme phytoene desaturase in the carotenoid biosynthetic pathway and acts as an electron carrier between photosystem II (PSII) and the cytochrome b6f complex, hence a key component of photosynthetic machinery (Ksas et al., 2018). When the biosynthesis of this prenylquinone is inhibited, it results in the lack of carotenoids in the newly emerging foliar tissue (Matringe et al., 2005; Schulte and Köcher, 2009; Van Almsick, 2009). Depletion of tocopherols leads to oxidative damage to biological membranes caused by destructive singlet oxygen and eventually plant death (Ksas et al., 2018). In sensitive plants, HPPD-inhibitors exhibit symptoms from yellowing and bleaching to necrosis within 7 to 14 days after application (Lee et al., 1998).
Crops such as corn (Zea mays), rice (Oryza sativa) and wheat (Triticum aestivum) are naturally tolerant to some of the HPPD-inhibitors and hence are registered for use in these crops. While mesotrione and tembotrione are widely used in corn (Mitchell et al., 2001; Ndikuryayo et al., 2017), the pyrazolate and pyrasulfatole are used rice (Matsumoto et al., 2002) and wheat respectively (Hawkes et al., 2019). The natural tolerance of corn was found to be due to an enhanced metabolism of these herbicides by cytochrome P450 (CYP) enzyme activity (Hawkes et al., 2001). Two weed species of Amaranthaceae family, i.e., common waterhemp (Amaranthus tuberculatus) and Palmer amaranth (Amaranthus palmeri) have been found to have evolved resistance to HPPD-inhibitors in the US (Heap, 2022). The first case of HPPD-inhibitor resistance was documented in common waterhemp in Illinois (IL) in 2011 (Hausman et al., 2011). Later, in 2012, a Palmer amaranth population in Stafford County, KS was confirmed with resistance to these herbicides (Nakka et al., 2017) and soon after, another HPPD-inhibitor-resistant Palmer amaranth population was documented in Nebraska (NE) (Sandell et al., 2012; Jhala et al., 2014). More recently, a wild radish (Raphanus raphanistrum) population (Lu et al., 2020) was also reported to have evolved resistance to these herbicides in Australia (Heap, 2022). Similar to corn, all the HPPD-inhibitor-resistant weeds also found to exhibit metabolic resistance to these herbicides (Ma et al., 2013; Gaines et al., 2020).
Palmer amaranth is one of the most troublesome weeds across the US and if uncontrolled can reduce yields in several cropping systems (Ward et al., 2013). Multiple herbicide resistance in Palmer amaranth has become widespread in the US (Heap, 2022). Currently, this weed has evolved resistance to eight herbicide sites of action (SOAs) (Heap, 2022). Recently, we reported a 6-way-resistant Palmer amaranth population, KCTR (Kansas Conservation Tillage Resistant) from KS that survived to 2,4-D, ALS-, PS II-, EPSPS-, PPO- and HPPD- inhibitor herbicides at field recommended rates (Shyam et al., 2021). This population predominantly exhibited metabolic resistance to multiple herbicides (Shyam et al., 2021; Shyam et al., 2022). Metabolism of herbicides is a common non-target site resistance (NTSR) mechanism found in many multiple resistant weed populations (Yuan et al., 2007). Such resistance is primarily mediated by the activity of CYP (Nakka et al., 2017; Pandian et al., 2020; Dimaano and Iwakami, 2021; Han et al., 2021), glutathione S-transferases (GST) (Ma et al., 2013), glycosyl transferases (GTs) (Huang et al., 2021), aryl acylamidases (Hirase and Hoagland, 2006), and ATP-binding cassette (ABC) transporters (Jugulam and Shyam, 2019; Lang et al., 2021). CYPs are one of the important classes of enzymes that are implicated in oxidative metabolism of xenobiotics, including herbicides (Gaines et al., 2020). Plant CYPs belonging to the families 71, 72, 73, 74, 76, 81 have been found to be involved in herbicide detoxification (Han et al., 2021). CYP81 family of enzymes has been identified to metabolize diverse herbicides in corn and weed species such as rigid ryegrass (Lolium rigidum) and late watergrass (Echinochloa phyllopogon) (Dimaano et al., 2020; Dimaano and Iwakami, 2021; Han et al., 2021). CYP81A9 and CYP81A10v7 genes are involved in conferring resistance to mesotrione in corn and rigid ryegrass, respectively (Han et al., 2021; Brazier-hicks et al., 2022).
Increased oxidative metabolism of mesotrione was reported in HPPD-inhibitor resistant common waterhemp (Ma et al., 2013). Similarly, rapid metabolism of mesotrione via 4-hydroxylation has been reported in waterhemp population from NE as well (Kaundun et al., 2017). Enhanced metabolism of mesotrione was also found to confer resistance in Palmer amaranth (Nakka et al., 2017) and wild radish population (Lu et al., 2020). Further, faster 4-hydroxylation followed by glycosylation of tembotrione was reported in tembotrione-resistant Palmer amaranth from NE (Küpper et al., 2018). We previously characterized the mesotrione-resistance in a Palmer amaranth from KS (Nakka et al., 2017). However, the mechanism of resistance to tembotrione in Palmer amaranth populations from KS is not known. The objectives of this study were to a) determine the level of resistance to tembotrione and b) the mechanism of tembotrione resistance in KCTR, using a known tembotrione-susceptible population (KSS) of Palmer amaranth.
The same Palmer amaranth seed used in our previous work (Shyam et al., 2021) was used in this study. Briefly, the KCTR plants that survived repeated applications of 2,4-D treatment were originally collected in summer 2018 and upon testing was found to be resistant to several herbicides including tembotrione. The susceptible population KSS, was collected a nearby field in Riley County, KS. The seed of KSS and KCTR Palmer amaranth were germinated in small trays (25 cm × 15 cm × 2.5 cm) with commercial potting mixture (Miracle Gro). Seedlings at 2–3 cm tall, were transplanted into small pots (6 cm × 6 cm × 6.5 cm) in the greenhouse, maintained at 25/20°C and 15/9 h photoperiod, supplemented with 250 mmol m-2 s-1 of light provided with sodium vapor lamps. When the plants reached 5–6 cm tall, they were transferred to a growth chamber maintained at 32.5/22.5°C, 15/9 h photoperiod. Light in the growth chamber was provided by fluorescent bulbs delivering 550 mmol m-2 s-1 photon flux at plant canopy level. Plants were watered as needed regularly both under greenhouse as well as growth chamber conditions.
The KSS and KCTR Palmer amaranth plants at 8–10 leaf stage and 8–10 cm tall were treated with 0, 11.5, 23, 46, 92*, 184, 368, and 736 g ai ha-1 of tembotrione, Methylated seed Oil and ammonium sulfate (AMS, Liquid N-Pak;Winfield) at 1% v/v and 1% w/v, respectively, were used in all the treatments per label recommendation (* - field recommended dose). Treatments were applied with a bench type track sprayer (Generation III, DeVries Manufacturing, RR 1 Box 184, Hollandale, MN, USA) equipped with a flat-fan nozzle tip (80015LP TeeJet tip, Spraying Systems Co., P.O. Box 7900, Wheaton, IL, USA) delivering 187 L ha-1. Following treatment, plants were returned to the same growth chambers (within 30 min after treatment). Treatments were arranged in a completely randomized design with five replications and the experiment was conducted twice. Treated plants were clipped off at the soil surface 3 weeks after treatment (WAT). Harvested plants were packed in paper bags and oven dried at 60°C (Precision Scientific Thelco Laboratory Oven) for a week and weighed for aboveground dry biomass.
KSS and KCTR Palmer amaranth plants of 8-10 centimeters height were treated with a total of 1.67 kBq of [phenyl-UL-14C]-labeled tembotrione (Bayer AG) with specific activity of 3.38 MBq/mg. Unlabeled tembotrione was added to the radioactive solution to obtain 92 g ai ha-1 tembotrione in a carrier volume of 187 L. Additionally, the adjuvants, methylated seed oil and AMS (Liquid N-Pak;Winfield) were added at 1% v/v and 1% w/v, respectively, to this mixture to enhance droplet-to-leaf surface contact. A total volume of 10 μL containing 100,000 dpm was applied as ten 1 μL droplets on the upper surface of the fourth youngest leaf. The treated plants were returned to the growth chamber within 20 minutes. Plants were harvested at 24, 48, 72 and 96 HAT and separated into treated leaf (TL), leaves above the treated leaf (ATL), and leaves below the treated leaf (BTL). Treated leaves were washed with 5 mL wash solution (10% methanol and 0.05% Tween) twice for 60 s in a 20 mL scintillation vial to remove any unabsorbed herbicide. Radioactivity in the leaf rinsate was measured using liquid scintillation spectrometry (LSS: Tricarb 2100 TR Liquid Scintillation Analyzer; Packard Instrument Co., Meriden, CT, USA). The dissected plant parts were wrapped in a single layer of tissue paper. Plant parts were oven (Precision Scientific Thelco Laboratory Oven) dried at 60°C for 48 h. They were combusted using a biological oxidizer (OX-501, RJ Harvey Instrument). Total radioactivity absorbed into each part was studied by quantification using LSS. Five replications were included in each treatment and the experiment was performed twice. Total [14C] tembotrione absorption was determined as; % absorption = (total radioactivity applied – radioactivity recovered in wash solution) ×100/total radioactivity applied. Herbicide translocation was determined using the following equation
where, 14C al is the amount of 14C measured in the treated leaf, and 14C ot is the amount of 14C detected in other untreated tissues of the plant.
Since corn is known to metabolize tembotrione, we used corn as a positive control in this experiment. Seedlings of KSS and KCTR Palmer amaranth along with corn were grown as described before and moved to the growth chamber to acclimate for 2 − 3 days before applying [phenyl-UL-14C] tembotrione. In order to compare the metabolism profile of KCTR with another susceptible population, we also used another known HPPD-inhibitor susceptible Palmer amaranth seedlings, MSS (Nakka et al., 2017; grown as described above). When seedlings were at 8-10 cm tall, a total of 10 µL of [phenyl-UL-14C] tembotrione consisting of 1.67 kBq was applied on the fourth youngest leaf as 10 droplets of 1 µL each per leaf. At 6, 24 and 48 h after treatment (HAT), treated leaves were harvested and rinsed with 5 mL of wash solution (10% v/v ethanol and 0.5% v/v Tween 20, ThermoFisher Scientific) in a 20 mL scintillation vial for 60 s twice to remove the unabsorbed [phenyl-UL-14C] tembotrione. Washed treated leaves were flash frozen and ground in liquid nitrogen. [phenyl-UL-14C] tembotrione and its metabolites were extracted by incubating in 15 mL of 90% acetone (HPLC grade, Thermo Fisher Scientific) at 4 °C overnight (16 h). Subsequently, the samples were centrifuged at 4 °C at 7000 rpm for 10 min. Supernatant was collected and evaporated using a vacuum concentrator (Centrivap, Labconco) for 2 - 2.5 h at 45 °C. The supernatant was concentrated until a volume of 500 − 1000 µL was reached and centrifuged at 13,000 rpm for 10 min, at room temperature to remove the debris. The total extractable radioactivity [14C] in each sample was measured by liquid scintillation spectrometry and normalized to 3000 dpm/50 µL using acetonitrile (HPLC grade, Thermo Fisher Scientific, Waltham, MA, United States).
Reverse-phase high performance liquid chromatography (HPLC) (Agilent Technologies, 1260 Infinity II) was used to resolve the total extractable radioactivity into parent [14C] tembotrione and its metabolites using Zorbax SB-C18 column (4.6 × 250 mm, 5 µm particle size; Agilent Technologies). Eluent A used was water with 0.1% trifluoroacetic acid (HPLC grade, Thermo Fisher Scientific, Waltham, MA, United States) and eluent B was acetonitrile with 0.1% trifluoroacetic acid (HPLC grade, Thermo Fisher Scientific, Waltham, MA, United States). The elution program used at a flow rate of 1 mL min−1 started at 30% B at 0 min and was in a linear gradient till 11 mins 85% B. Solvent B was at hold 85% till 13 mins and was maintained at 100% from 14-15 mins. %B was again reduced to 70% at 17 mins, 50% at 18 mins and brought back to 30% at 20 mins. Detection was performed with a radioflow detector (EG&G Berthold, LB 509) employed with an admixture flow cell, Z-1000 (Berthold Technologies GmbH) at a flow rate of 1 mL min−1 of scintillation fluid, Ultima-Flo M cocktail (Perkin-Elmer). % Parent [14C] tembotrione remaining in each sample was determined based on the peak areas as a percent of [14C] tembotrione relative to the total extractable radioactivity.
To determine if the involvement of CYPs confers resistance to tembotrione in KCTR, experiments were conducted using two known CYP-inhibitors, malathion and piperonyl butoxide (PBO). The KSS and KCTR plants were grown in the greenhouse (as described previously and under similar growth conditions). Malathion (Spectracide, malathion insect spray concentrate; Spectrum Brands) was applied at 1500 g ai ha-1 along with 0.25% non-ionic surfactant (NIS). PBO (Thermo Fisher Scientific, Waltham, MA, United States) was used at 2100 g ai ha-1 along with 7.5% acetone and 0.25% NIS as adjuvant. Tembotrione was applied at 92 g ai ha-1 with 1% methylated soy oil and 1% ammonium sulphate, 1 h after the inhibitor treatment. Soil drenching of 5 mM malathion at 24 h after primary application at the rate of 50 mL per plant was given as a booster dose in order to retain the efficacy of inhibitor treatment. Non-treated plants were also included in the experiment. Based on our previous work (Shyam et al., 2021) we have selected the time and dose of application of CYP-inhibitors. The plants were clipped off at the soil surface at 3 weeks after treatment (WAT). The same procedure, as mentioned in the earlier tembotrione dose-response assay, was followed for all chemical treatments (malathion, PBO, and tembotrione) and for the collection of aboveground dry biomass.
KSS and KCTR Palmer amaranth plants were grown in the growth chamber (as described above). At 8-10 leaves stage, the young leaves of non-treated as well as 1X tembotrione (92 g ai ha-1)- treated (as described under dose-response assay) plants of KSS and KCTR were harvested at 24 HAT. The leaf samples were flash-frozen immediately in liquid nitrogen and stored at -80°C for isolating RNA. Total RNA was extracted following manufacturer’s protocol of RNA extraction using TRIzol (Ambion, Life Technologies, Carlsbad, California, USA). The quality and quantity of total RNA was determined using agarose gel (1%) electrophoresis and spectrophotometer (NanoDrop 1000, Thermo Scientific), respectively, and the samples were stored at −80°C. Total RNA of 1 μg was treated with DNase I (Thermo Scientific, Waltham, MA, USA) and the DNA free RNA was used for cDNA synthesis using RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific). qPCR reaction was conducted using StepOnePlus™ Real-Time PCR system (Applied Biosystems™, Waltham, MA, USA) for determining the expression of the genes CYP81E8 and HPPD. The primer sequence of CYP81E8 was adopted from Giacomini et al., 2020 and the housekeeping gene, β-tubulin served as an internal control (Godar et al., 2015). CYP81E8 forward (5′-GTACTTTGATTGAACAGTTGCTGGATTTGC-3′); reverse (5′-AGGTTCGTGCTGTGGTTTCTGATC-3′); β-tubulin forward (5′- ATGTGGGATGCCAAGAACATGATGTG-3′) and reverse (5′- TCCACTCCACAAAGTAGGAAGAGTTCT-3′). Primer sequences used for HPPD were forward (5′- CTGTCGAAGTAGAAGACGCAG -3′) and reverse (5′- TACATACCGAAGCACAACATCC-3′); Single product amplification of primers was verified in agarose gel electrophoresis and melting curve analysis. The reaction mixture consisted of 7 μL of PowerUp™ SYBR™ Green Master Mix (Applied Biosystems™), 1 μL each of forward and reverse primers (10 μM) and 1:10 diluted cDNA. Reaction was carried out in triplicates with PCR cycling conditions set as 50°C for 2 min, 95°C for 5 min, and 40 cycles of 95°C for 30 s and 60°C for 1 min as per the manufacturer’s instruction of the master mix (Applied Biosystems™). The relative quantifications were performed using ΔΔCT method. qPCR experiments were carried out with 5-8 biological replicates of KSS and KCTR Palmer amaranth. The fold-changes of CYP81E8 and HPPD gene expression of KCTR was calculated individually relative to the KSS plants.
The genomic DNA from KSS and KCTR plants was isolated using plant Genomic DNA Extraction kit (Thermo Fisher Scientific, Waltham, MA). The primer sets and qPCR conditions used were the same as described above for the CYP81E8 gene expression analysis. The reaction mixture consisted of 30 ng of genomic DNA and the composition of the reaction conditions were same as that used for expression analysis described above. Due to its single copy number, β-tubulin was used as reference gene primer as described in our study (Koo et al., 2018).
All the experiments were conducted in a completely randomized design. Dose-response data (expressed as percentage of the untreated control) were analyzed using ‘drc’ package in R 4.1.2 (Knezevic et al., 2007; Ritz et al., 2015). The four-parameter log-logistic model as shown below was used to show the relationship between herbicide rate and biomass,
where d is asymptotic value of Y at upper limit, b is the slope of the curve around GR50 (the herbicide rate giving response halfway between d and the lower asymptotic limit c), and x is the herbicide rate. The in-built ‘plot’ function in the ‘drc’ package was used to make the dose-response curve. Resistance index (R/S) was calculated as GR50 ratio between KSS and KCTR populations. Absorption and translocation data, expressed as percentage of applied and absorbed, respectively, and metabolism data were analyzed using one-way ANOVA in R 4.1.2 and the means were compared using Tukey’s HSD test. The time course of tembotrione metabolism by KSS and KCTR Palmer amaranth populations was fitted with a three-parameter Weibull regression model. Effect of malathion and PBO on tembotrione metabolism was analyzed using a factorial analysis of variance (ANOVA) in R and contrast comparisons were adjusted by the Tukey’s HSD test results. The data plotted using “ggplot2” package for graphical visualizations. qPCR data was analyzed using Student’s t-test in Origin 2021b software (OriginLab Corp., Northampton, MA, USA). Levene’s test was carried out in R, using the ‘Car’ package to compare the repeated data of the runs for each experiments. If no significance was found between the runs, the data was combined and analyzed together.
The results of tembotrione dose-response assay revealed that the amount of tembotrione required to reduce aboveground dry biomass by 50% (GR50) at 3 WAT was ~378.58 g ai ha-1 for KCTR compared to 16.72 g ai ha-1 for KSS (Figure 1; Table 1). At 48 HAT, the KCTR plants showed injury (bleached) symptoms at the shoot meristem but recovered eventually with all doses of tembotrione (Supplementary Figure S1). However, KCTR showed stunted growth at 3 WAT in response to highest dose of tembotrione (8 X=736 g ai ha-1) treatment (Supplementary Figure S1). KSS plants did not recover from the injury even at low doses of tembotrione application and showed more bleaching symptoms than KCTR (Supplementary Figure S1). KSS population did not show any growth after herbicide treatment and did not survive in the field recommended rate of tembotrione. Based on the GR50, KCTR was found to be 23-fold resistant to tembotrione compared to KSS Palmer amaranth (p< 0.001) (Table 1).
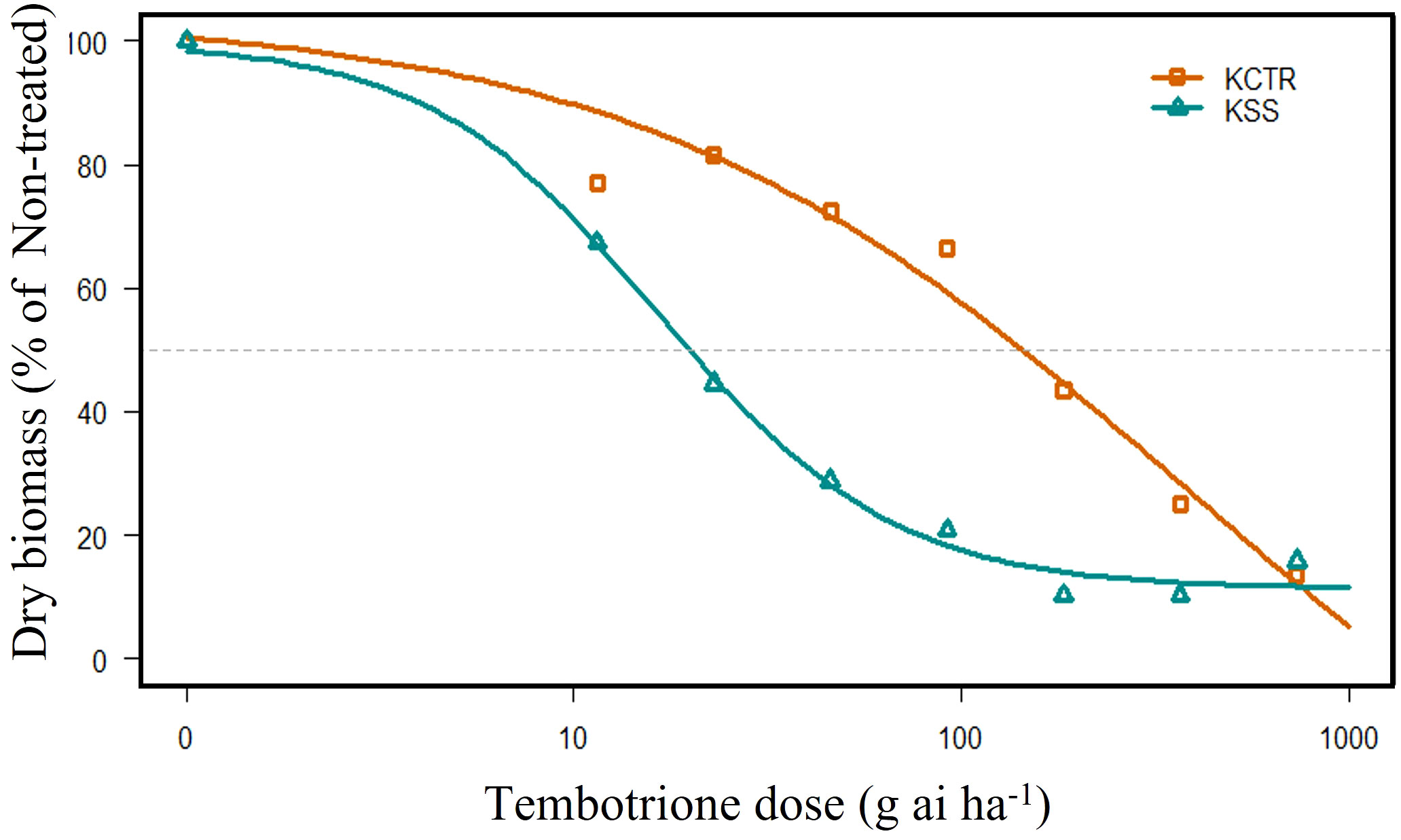
Figure 1 Tembotrione dose-response curves of KSS (susceptible) and KCTR (resistant) palmer amaranth population at 3 weeks after treatment (WAT). Non-linear regression analysis of aboveground dry biomass at different doses of tembotrione fitted using the four-parameter log-logistic model.
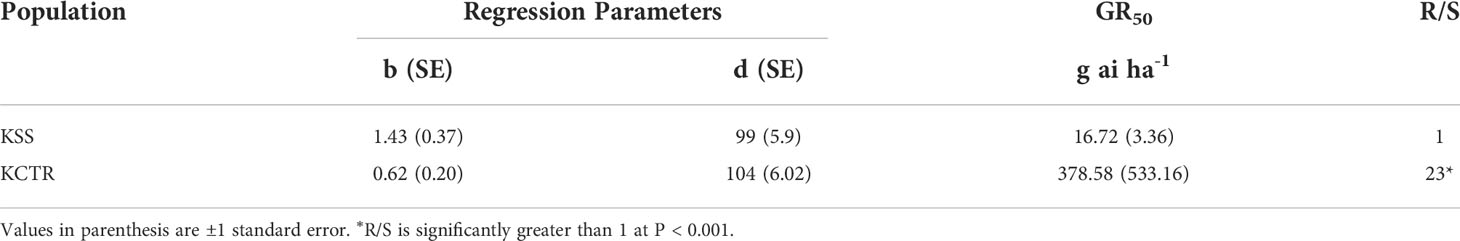
Table 1 Summary of parameters describing the response of KSS (susceptible) and KCTR (resistant) Palmer amaranth aboveground dry biomass to different rates of tembotrione at 3 weeks after treatment (WAT).
To determine if difference in tembotrione absorption or translocation pattern between KSS and KCTR Palmer amaranth population attribute to resistance, we measured the amount of 14C-tembotrione absorbed and translocated in these plants. No difference in the amount of absorbed 14C-tembotrione was found in KSS and KCTR plants. The percentage of total recovery of 14C recovery ranged from 75-80%. The absorption of [14C] tembotrione (% of total applied), in KCTR at 24, 48, 72 and 96 HAT was 94, 94, 94, and 92% respectively, which was not significantly different from the KSS plants which had the absorption of 91, 94, 94 and 93% respectively, at the same time points (Figure 2A). HPPD-inhibitor are systemic herbicides that translocate both via xylem and phloem. While the KCTR plants had showed tembotrione translocation of 7-11% (expressed as % of total [14C] tembotrione absorbed) the KSS plants translocated 12-13% of this herbicide (Figure 2B). Further, there was no difference in the amount of [14C] tembotrione translocated to ATL or BTL between KSS and KCTR plants at all the time points tested, i.e., 24, 48, 72, 96 HAT (Figures 2C–E).
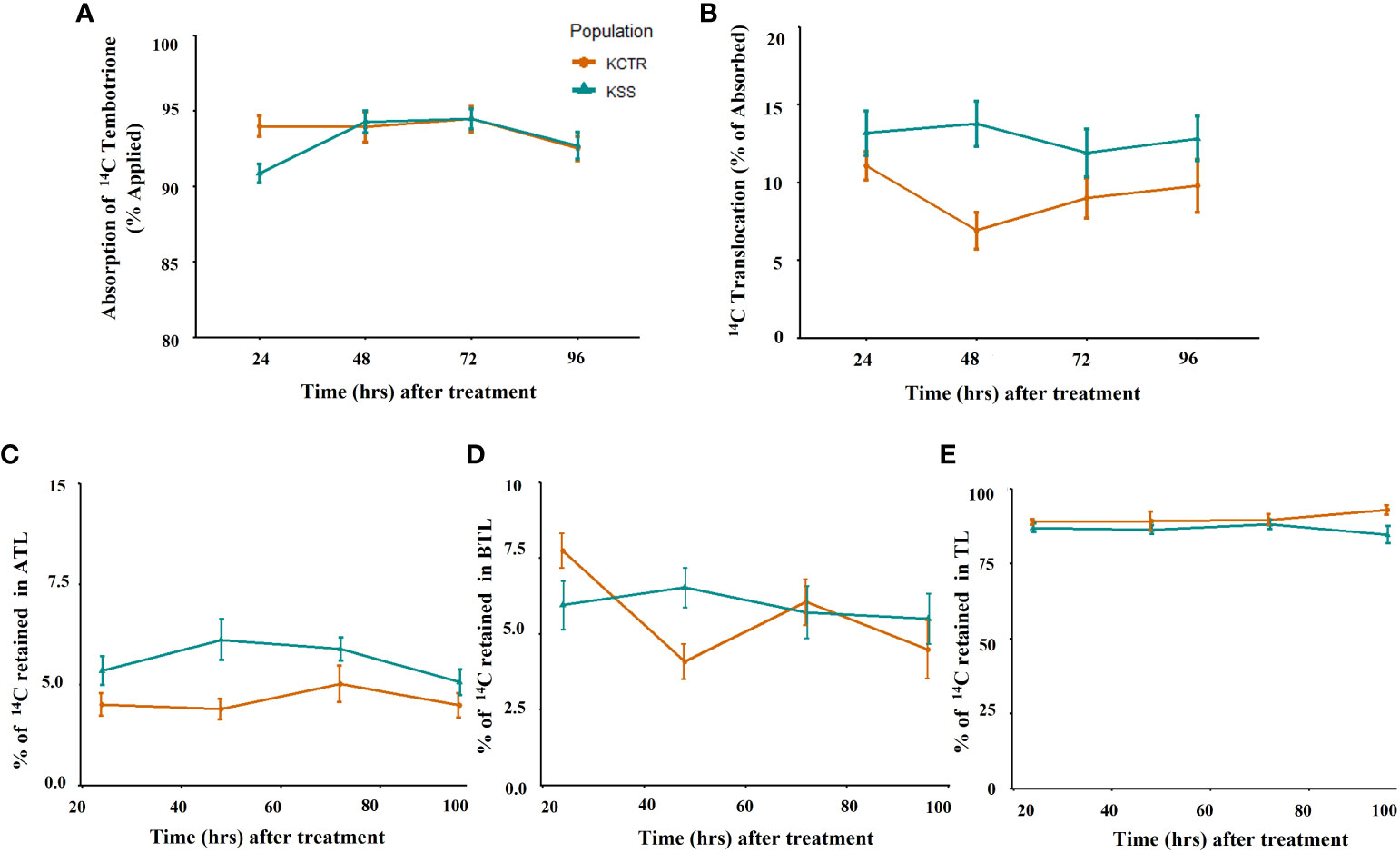
Figure 2 14C tembotrione absorption and translocation in resistant and susceptible Palmer amaranth populations. 14C-tembotrione absorption as % of applied radioactivity to KCTR and KSS plants (A), 14C translocation as % of absorbed radioactivity (B), % radioactivity translocated to the above treated leaf part, ATL (C), % radioactivity translocated to the below treated leaf part, BTL (D), % radioactivity retained in treated leaves (TL) (E). Error bars represent the standard error of means of 6–8 biological replicates.
Metabolism of tembotrione was determined at three time points, i.e., 6, 24, 48 HAT in KSS and KCTR plants. The standard 14C-tembotrione (parent compound) resolved at a peak retention time of 11.4 min, whereas its metabolites were identified at 10 (M1), 9 (M2), 8 (M3), 6.5 (M4) and (3.4) mins. (Figure 3). To determine the percent of applied 14C-tembotrione as parent compound at these time points, we determined the amount of radioactivity at 11.4 min as a fraction of total radioactivity. Our data indicate that the KSS plants had 50% of parent 14C-tembotrione at 6 HAT (Figures 3A and 3I). Conversely, only 5-10% of parent compound was detected in the KCTR plants (p< 0.0001) at this time point (Figures 3B, 3I). However, at 24 HAT, the KCTR plants metabolized 100% of tembotrione (Figures 3D, 3I), while KSS plants showed 5-10% of parent 14C-tembotrione (Figures 3C and 3I). Nonetheless, KSS plants also completely metabolized tembotrione at 48 HAT (Figure 3E). Corn, as expected, showed no parent 14C-tembotrione at 6 or 24 HAT (Figures 3G, H) and two metabolites M1 and M3 were also identified in the 14C-metabolic profile of corn (Figures 3G, H). The metabolic profile of MSS was similar to that of KSS Palmer amaranth with 5% and 65% of parent 14C-tembotrione retained at 6 and 24 HAT respectively (Supplementary Figure S2). The time required for metabolism of 50% of parent tembotrione (T50) was 2.7 and 5.9 h, respectively, in KCTR and KSS plants, suggesting that KCTR plants metabolized tembotrione 2.2 times faster (p< 0.001) than KSS (Figure 4; Table 2).
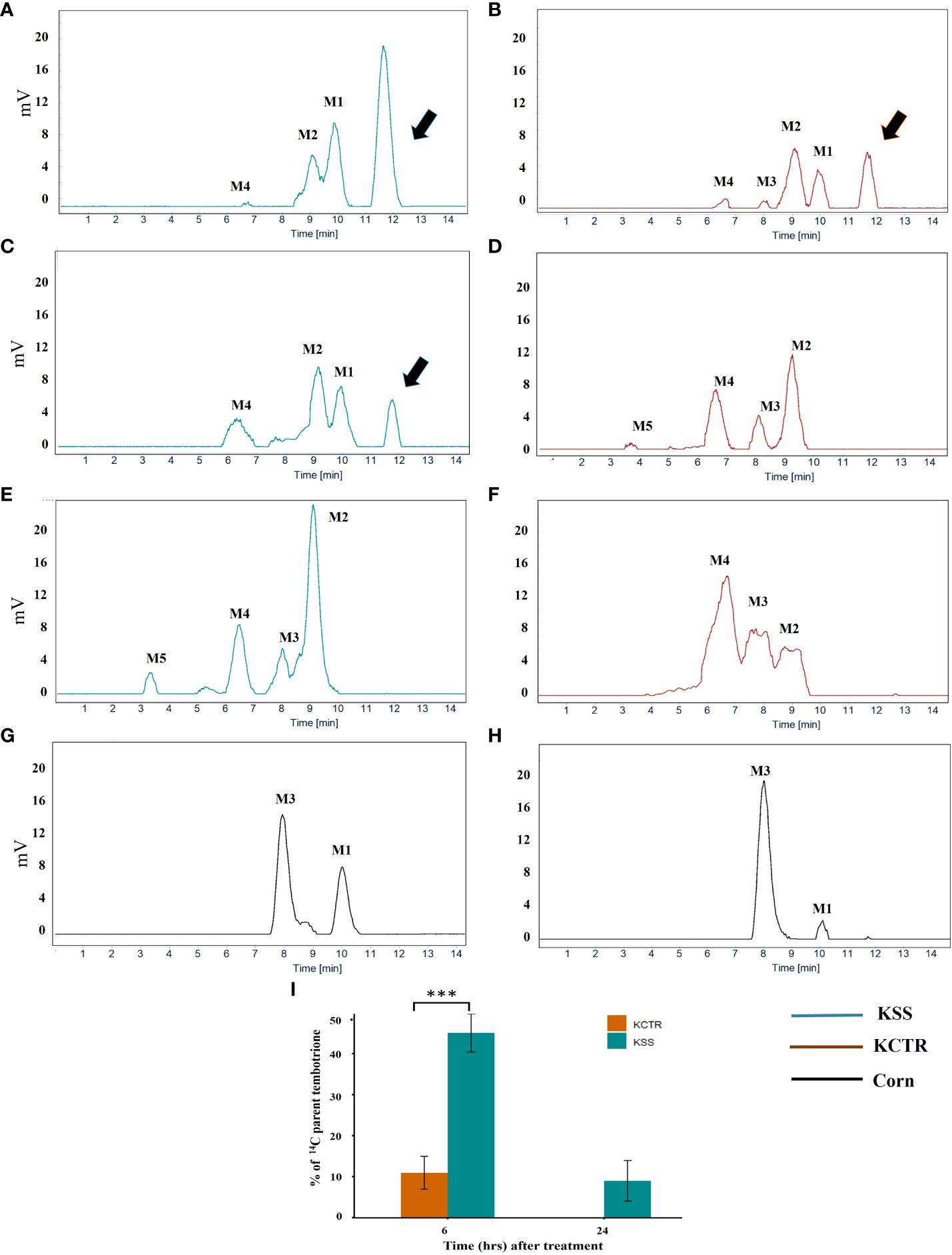
Figure 3 Metabolism of [14C] tembotrione in resistant and susceptible Palmer amaranth populations compared with corn as control. Reverse-phase HPLC chromatograms of samples at different time points, (A, B, G - 6 HAT), (C, D, H– 24 HAT) and (E, F- 48 HAT). Retention time 11.4 min corresponds to [14C] parent tembotrione and is marked with an arrow. (A) KSS; (B) KCTR; (C) KSS; (D) KCTR; (E) KSS; (F) KCTR; (G) Corn; (H) Corn; (I) Represents the amount of [14C] tembotrione remaining as percentage of total in the resistant (KCTR) and susceptible population (KSS). Error bars represent the standard error of mean of 5–7 biological replicates. Asterisks represent significant difference of p < 0.0001.
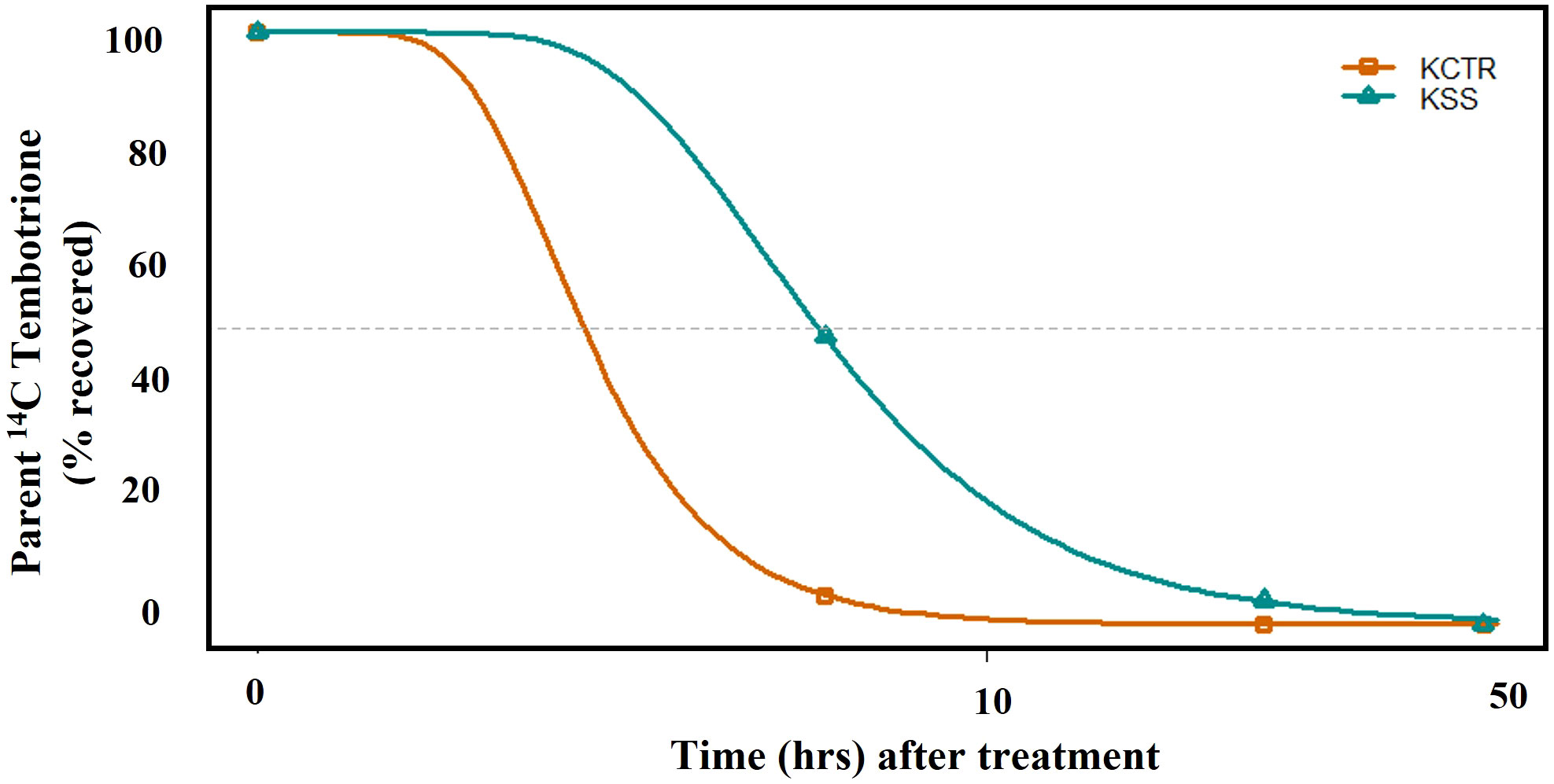
Figure 4 The time course of [14C] tembotrione metabolism (T50) in KSS (susceptible) and KCTR (resistant) Palmer amaranth populations treated with [14C] tembotrione. The data was collected at 6, 24, 48 hours after treatment (HAT).
Upon treatment with the two CYP-inhibitors, i.e., malathion or PBO alone, the KSS or KCTR plants did not show any difference in biomass accumulation (Figure 5). Treatment with tembotrione alone resulted in higher biomass accumulation in KCTR population (65%) compared to KSS plants (8%) relative to their non-treated plants (Figure 5). In response to malathion followed by tembotrione treatments, KCTR exhibited significantly (p< 0.01) lower biomass accumulation (15%) and showed significant growth reduction compared to those treated with tembotrione alone (Figure 5B, Supplementary Figure S3). Similarly, PBO followed by tembotrione treatments caused lesser biomass in KCTR (29%) when compared to tembotrione only treatment (p< 0.1) (Figure 5B). The tembotrione application following inhibitor treatment either killed the KCTR plants or affected its growth drastically (Supplementary Figure S3). In contrast, malathion- or PBO plus tembotrione-treated KSS plants showed no difference in biomass accumulation compared to tembotrione treatment only (Figure 5A).
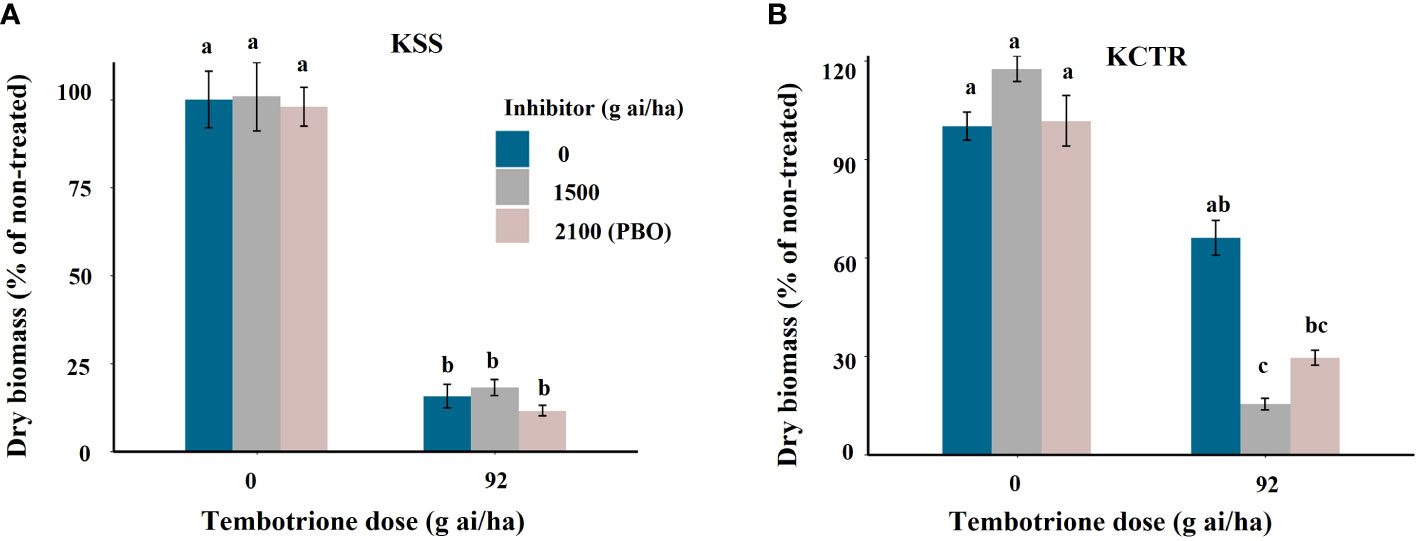
Figure 5 Effect of P450 inhibitors Malathion and PBO on efficacy of tembotrione in Palmer amaranth populations, (A) susceptible (KSS) and (B) resistant (KCTR). Error bars represent standard error of the mean of 5-7 biological replicates. Letters represent significant differences identified by separation of means using Tukey.
CYP81 gene family has been reported to detoxify multiple herbicides including HPPD-inhibitor mesotrione (Dimaano et al., 2020; Han et al., 2021). Since, CYP81E8 is a member of this gene family and has been reported to differentially express in 2,4-D and HPPD-inhibitor-resistant common waterhemp (a close relative of Palmer amaranth) (Giacomini et al., 2020), we investigated whether CYP81E8 gene mRNA transcript abundance accounts for increased metabolism of tembotrione, and resistance to tembotrione in KCTR. We performed qPCR analysis to compare the expression level of CYP81E8 gene. The data indicate that the transcript of CYP81E8 is upregulated in KCTR compared to KSS Palmer amaranth. The constitutive expression level of this gene is 30 to 35-fold higher in KCTR compared to KSS plants (α = 0.05) (Figure 6B). The transcript of this gene in tembotrione-treated KCTR plants was 10 to 14 -fold higher at 24 HAT compared to KSS (α = 0.05) (Figure 6A). This increased transcript levels of KCTR (both constitutive and in response to tembotrione treatment) are likely to play an important role in the rapid metabolism of tembotrione in KCTR population. Further, our data also suggests that the high expression of CYP81E8 is not because of a change in copy number of this gene with reference to β-tubulin gene in KCTR Palmer amaranth (Figure 6C).
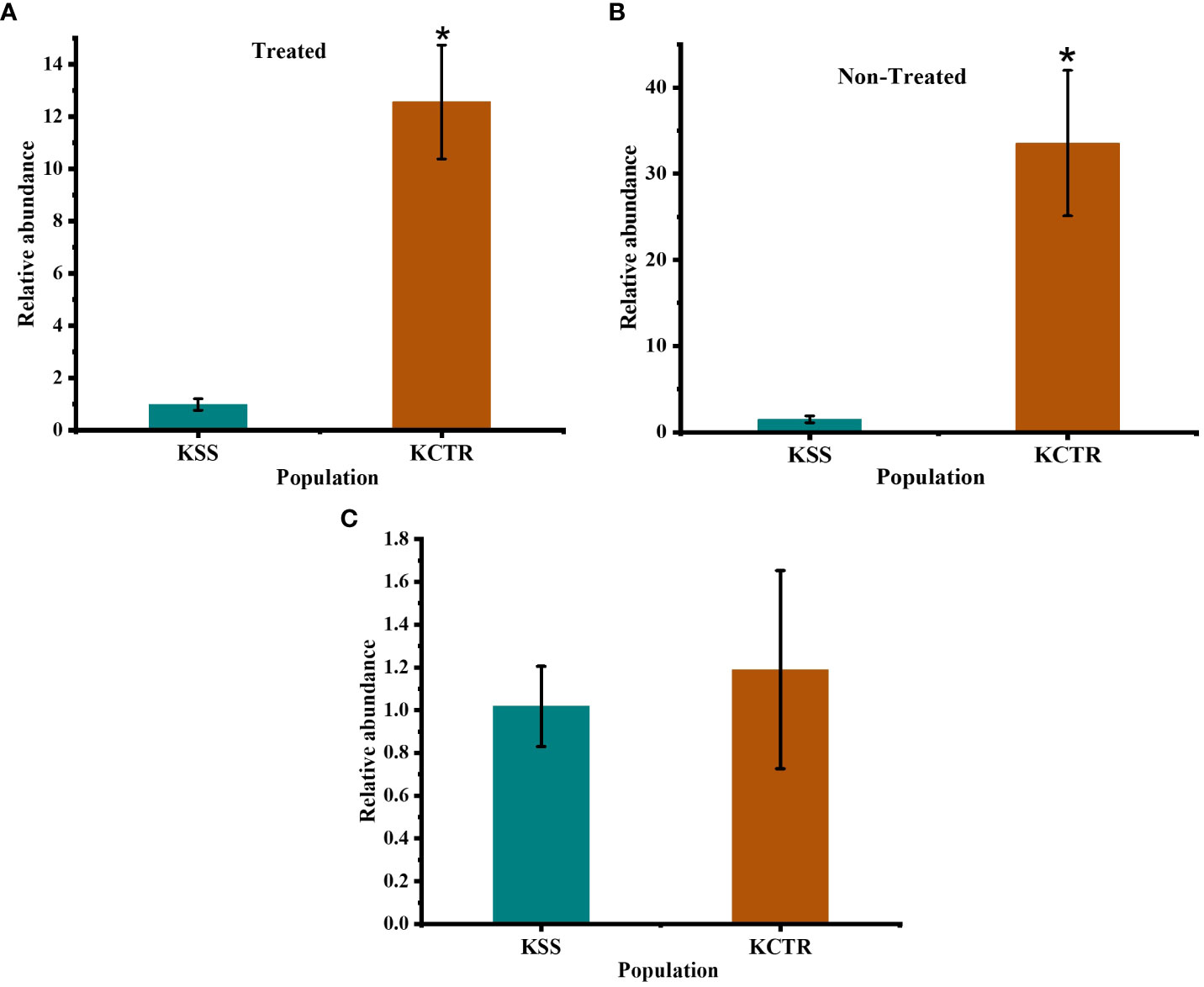
Figure 6 Expression level of CYP81E8 in KCTR relative to KSS population. (A) CYP81E8 in non-treated plants (B) CYP81E8 in tembotrione treated plants collected at 24 HAT (C) copy number analysis of the gene CYP81E8. Bars represent the means ± SE of 8–10 biological replicates. Asterisks above error bars represent significant difference in gene expression compared to susceptible population KSS at α = 0.05.
We tested if increased expression of the HPPD gene, the molecular target of tembotrione, plays a role in conferring resistance to tembotrione in KCTR Palmer amaranth. The results did not show any change in the expression level of HPPD gene either before or at 24 HAT in both KSS and KCTR plants (Supplementary Figure S4).
We recently reported predominance of metabolic resistance in a multiple herbicide resistant Palmer amaranth population KCTR (Shyam et al., 2021; Shyam et al., 2022). This population survived field recommended rates of 2,4-D, mesotrione, tembotrione, atrazine, imazethapyr, imazamox, chlorsulfuron, thifensulfuron, lactofen, fomesafen, glyphosate and metribuzin (Shyam et al., 2021). In this research, we characterized the mechanism of resistance to tembotrione in KCTR population. Resistance to tembotrione by metabolism has also been reported in A. palmeri from Nebraska by Küpper et al. (2018). Till date, only two weed species in the US, i.e., Palmer amaranth and common waterhemp and a wild radish biotype from Australia have known to have evolved resistance to HPPD-inhibitors (Heap, 2022). Previously, we characterized mesotrione resistance in the first case HPPD-inhibitor resistant Palmer amaranth population KSR from KS (Nakka et al., 2017). However, the mechanism of resistance to tembotrione in KCTR Palmer amaranth from KS is unknown. Our tembotrione dose-response assay suggested that the KCTR Palmer amaranth is 23-fold more resistant than the known susceptible population, KSS (Figure 1; Table 1). Previously, the resistance to tembotrione was characterized in a Palmer amaranth population from NE (Küpper et al., 2018) and common waterhemp from IL (Hausman et al., 2011). The resistant common waterhemp population from IL showed ten-fold resistance relative to susceptible population (Hausman et al., 2011) whereas, the Palmer amaranth population from NE had a resistance factor of 3.3 (Küpper et al., 2018). However, the level of resistance in these population is lower than what we found in KCTR. Our data suggest that no changes in tembotrione absorption or translocation that could confer resistance in KCTR (Figure 2).
Triketone herbicides are widely used for weed control in corn as it can rapidly metabolize this herbicide into more polar and less toxic metabolites because of Phase I hydroxylation and dealkylation mediated by CYP activity (Brazier-hicks et al., 2022). In this research, the metabolic profiles of tembotrione showed presence of metabolites M1 and M3 in corn (Figures 3G, H), whereas in Palmer amaranth, M1 and M2 were identified as major metabolites at 6 HAT (Figures 3A, 3C). The more polar metabolites, M3, M4 and M5 were identified as small peaks in KCTR Palmer amaranth at 24 HAT (Figure 3D). Though the metabolite M3 was found in KCTR Palmer amaranth (Figures 3B, 3D and 3F), it was not predominant as that of corn at 24 HAT (Figures 3G, H). Instead, the metabolite M4 was found to be more predominant than M3 in KCTR Palmer amaranth at 24 and 48 HAT (Figures 3D and 3F). Importantly, both corn and KCTR Palmer amaranth had M1 and M3, and the other metabolites identified in Palmer amaranth are different. Küpper et al. (2018) reported that glycosylation and acetylation of tembotrione may result in formation of more polar metabolites. The detoxification pathway scheme for tembotrione has been illustrated by Küpper et al. (2018) and the 14C tembotrione HPLC metabolic profiles in our current study are similar to the one reported previously in Palmer amaranth (Küpper et al., 2018). Further, they also reported that tembotrione reduced at benzoic ring resulting in a non-polar metabolite peak eluting after the retention time of parent herbicide (Küpper et al., 2018). However, the reduced tembotrione metabolite was not found in KCTR Palmer amaranth.
Both mesotrione and tembotrione are members of the benzoylcyclohexane-1,3-dione family of herbicides (Beaudegnies et al., 2009). Mesotrione was found to undergo 4-hydroxylation in corn by CYP activity (Hawkes et al., 2001). Two metabolites, (M3/M4) formed by hydroxylation at cyclohexanedione of tembotrione were reported by Küpper et al. (2018) in Palmer amaranth. In this research, the metabolites M2 and M1 (Figure 3) showed similar HPLC separation as that of hydroxylated tembotrione metabolites, M3/M4 reported by Küpper et al. (2018). These hydroxylated tembotrione metabolites formed by catalytic activity of CYP enzymes could still be toxic unless they are further converted to more polar forms. This is clearly evident from our data at 48 HAT, KSS Palmer amaranth showed the presence of metabolite M2 (Figure 3E) relatively higher than metabolite M4, whereas in KCTR, M4 was relatively higher than M2 and M3 (Figure 3F). In corn, the metabolite M1 was detected as a major metabolite at 6 HAT whereas at 24 HAT, this metabolite was not detected which implies rapid metabolism of tembotrione, soon after the application of this herbicide in corn. However, in KCTR Palmer amaranth the metabolites M1/M2 were detected at both 6 and 24 HAT. Although the metabolic profiles of KCTR and KSS Palmer amaranth are qualitatively similar (Figure 3), the T50 was 2.7 and 5.9 min, respectively, suggesting that KCTR metabolizes this herbicide 2.2 times faster than KSS conferring resistance (Figure 4; Table 2). The T50 of tembotrione was shorter in both KCTR and KSS Palmer amaranth compared to tembotrione-resistant Palmer amaranth population from NE, which also metabolized this herbicide, 2.2 times faster than susceptible population from NE (T50 7.3 and 17.1 h, respectively) (Küpper et al., 2018). Similarly, the mesotrione-resistant Palmer amaranth from Kansas metabolized 14C mesotrione 2.6 times faster than the susceptible population (Nakka et al., 2017). Rapid detoxification of mesotrione by CYPs into 4-OH mesotrione was reported in common waterhemp IL (Ma et al., 2013) and from Nebraska (Kaundun et al., 2017). Furthermore, resistance to syncarpic acid-3, a nonselective, noncommercial HPPD-inhibitor not amenable for metabolism due to the cyclohexanedione ring blocked with alternating methyl and ketone groups was found to be metabolized in common waterhemp through reduction–dehydration–GSH conjugation pathway (Concepcion et al., 2021).
Plants detoxify xenobiotics that are detrimental to their growth and development by enzymatic modification via certain biochemical reactions (Yuan et al., 2007). The initial step of the reaction takes place in Phase I, where activation of the chemical molecule takes place by attachment of certain functional groups, which are exposed to Phase II metabolism (Yuan et al., 2007). This initial step is mediated by the catalytic activity of CYPs and mixed function oxidases (Siminszky, 2006; Yuan et al., 2007). Plant CYP family has known to have more than 300 genes (Hansen et al., 2021), which are grouped into 10 clans and four of them have multiple gene families (Brazier-hicks et al., 2022). Several chemicals that have inhibitory activity on CYPs have been identified, e.g., 1-aminobenzo-triazole (ABT), tetcyclacis (TET), PBO, tridiphane, malathion (organophosphate insecticide), and phorate (Siminszky, 2006). Studies using these chemicals provided indirect evidence for the role of CYPs in metabolizing herbicides as well as other xenobiotics (Siminszky, 2006). Each of these chemicals are known to inhibit a specific class of CYP enzymes and therefore, can provide cues for the involvement of a particular CYP isoform in metabolism of xenobiotics (Oliveira et al., 2018). PBO and malathion are irreversible inhibitors of CYP activity, act as suicide inactivators; substantially reduce the levels of active of CYPs (Hodgson and Levi, 1998). Different CYP inhibitors exhibits varying level of synergistic effect in different weed based on the CYP isoform involved in the metabolism of particular herbicide. For instance, Tardif et al. (1997) have found that PBO inhibited the metabolism of chlorotoluron and simazine in rigid ryegrass, while malathion synergized with chlorsulfuron but not vice versa. In our current study, both PBO and malathion showed the reversal of resistance in KCTR population (Figure 5B), suggesting that the metabolism of tembotrione in KCTR Palmer amaranth is mediated by a particular class (yet to be identified) of CYPs.
Nsf1/Ben1 (gene named for nicosulfuron, Nsf, and bentazon, Ben) identified as the single major locus in corn which corresponded to CYP81A9 was implicated in resistance to nicosulfuron, a sulfonylurea herbicide (Williams and Pataky, 2008; Choe and Williams, 2020). Hitherto, few herbicides metabolizing CYPs have been expressed in heterologous system and characterized in plants (Dimaano et al., 2020; Brazier-hicks et al., 2022). CYP81A9 of corn that was found to metabolize multiple herbicides, including mesotrione, tembotrione and sulcotrione through the oxidation of cyclohexanedione ring (Brazier-hicks et al., 2022). Previous research identified the specific CYP isoforms that are differentially expressed in herbicide-resistant and -susceptible plants were found responsible for metabolism of the herbicides (Giacomini et al., 2020; Han et al., 2021). RNAseq analysis, identified increased expression of CYP81A10v7 in Lolium rigidum conferring resistance to multiple herbicides including the HPPD-inhibitors (Han et al., 2021). CYP81E8 was identified to differentially express in 2,4-D- and HPPD-inhibitor-resistant common waterhemp (Giacomini et al., 2020) and in HPPD-resistant Palmer amaranth population (Kuepper, 2018). This enzyme has also been characterized as isoflavone hydroxylase and was found to be involved in regiospecific hydroxylation of isoflavones in Medicago truncatula (Liu et al., 2003). In this study, KCTR Palmer amaranth was found to have increased expression of the CYP81E8 both before or after tembotrione treatment compared to KSS (Figures 6A, B). The role of other CYPs and differentially expressed genes in conferring tembotrione resistance in KCTR needs further investigation, however our data suggest that this gene could potentially contribute to tembotrione resistance.
Previously we reported in another HPPD-inhibitor-resistant Palmer amaranth, i.e., KSR from Kansas showed both target and non-target site mediated resistance, including high expression of HPPD gene and enhanced metabolism of mesotrione, respectively, confer resistance (Nakka et al., 2017). However, in this research, the KCTR Palmer amaranth did not show any increased expression of HPPD gene (Supplementary Figure S4).
Our results indicate that metabolism of tembotrione contributes to resistance in KCTR Palmer amaranth. Future work identifying the specific tembotrione metabolites using LC/MS will help in predicting the detoxification pathway in KCTR. Our data also suggest that increased expression of CYP81E8 could have a role in imparting resistance in KCTR. Future research using RNAseq analysis will help identify any other candidates involved in metabolism of herbicides in KCTR Palmer amaranth. Regardless, metabolic resistance is a challenge for weed management as the populations are predisposed to evolve resistance to other unknown xenobiotics.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
MJ conceived and supervised the research. TA and CS conducted dose-response, uptake, translocation, metabolism and P450 inhibitor experiments. TA performed qPCR experiment, collected, analyzed the data, and wrote the manuscript. MJ edited and helped to draft the final manuscript. All authors contributed to the article and approved the submitted version.
The financial support from KSU Libraries towards publishing this article is much appreciated.
Authors thank Bayer Crop Science for providing radiolabelled tembotrione. This is contribution number 23-061-J from the Kansas Agricultural Experiment Station.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fagro.2022.1010292/full#supplementary-material
Ahrens H., Lange G., Muller T., Rosinger C., Willms L., Almsick A.v. (2013). 4-hydroxyphenylpyruvate dioxygenase inhibitors in combination with safeners solutions. Angew. Chem. Int. Ed. 52, 9388–9398. doi: 10.1002/anie.201302365
Beaudegnies R., Edmunds A. J. F., Fraser T. E. M., Hall R. G., Hawkes T. R., Mitchell G., et al. (2009). Herbicidal 4-hydroxyphenylpyruvate dioxygenase inhibitors-a review of the triketone chemistry story from a syngenta perspective. Bioorg. Med. Chem. 17, 4134–4152. doi: 10.1016/j.bmc.2009.03.015
Brazier-hicks M., Franco-ortega S., Watson P., Rougemont B., Cohn J., Dale R., et al. (2022). Characterization of cytochrome P450s with key roles in determining herbicide selectivity in maize. ACS Omega 7, 17416–17431. doi: 10.1021/acsomega.2c01705
Choe E., Williams M. M. (2020). Expression and comparison of sweet corn CYP81A9s in relation to nicosulfuron sensitivity. Pest Manage. Sci. 76, 3012–3019. doi: 10.1002/ps.5848
Concepcion J. C. T., Kaundun S. S., Morris J. A., Hutchings S. J., Strom S. A., Lygin A. V., et al. (2021). Resistance to a nonselective 4-hydroxyphenylpyruvate dioxygenase-inhibiting herbicide via novel reduction–dehydration–glutathione conjugation in amaranthus tuberculatus. New Phytol. 232, 2089–2105. doi: 10.1111/nph.17708
Dimaano N. G., Iwakami S. (2021). Cytochrome P450-mediated herbicide metabolism in plants: current understanding and prospects. Pest Manage. Sci. 77, 22–32. doi: 10.1002/ps.6040
Dimaano N. G., Yamaguchi T., Fukunishi K., Tominaga T., Iwakami S. (2020). Functional characterization of cytochrome P450 CYP81A subfamily to disclose the pattern of cross-resistance in echinochloa phyllopogon. Plant Mol. Biol. 102, 403–416. doi: 10.1007/s11103-019-00954-3
Gaines T. A., Duke S. O., Morran S., Rigon C. A. G., Tranel P. J., Küpper A., et al. (2020). Mechanisms of evolved herbicide resistance. J. Biol. Chem. 295, 10307–10330. doi: 10.1074/jbc.REV120.013572
Giacomini D. A., Patterson E. L., Küpper A., Beffa R., Gaines T. A., Tranel P. J. (2020). Coexpression clusters and allele-specific expression in metabolism-based herbicide resistance. Genome Biol. Evol. 12, 2267–2278. doi: 10.1093/GBE/EVAA191
Godar A. S., Varanasi V. K., Nakka S., Prasad P. V. V., Thompson C. R., Mithila J. (2015). Physiological and molecular mechanisms of differential sensitivity of palmer amaranth (Amaranthus palmeri) to mesotrione at varying growth temperatures. PloS One 10, 1–17. doi: 10.1371/journal.pone.0126731
Hansen C. C., Nelson D. R., Møller B. L., Werck-Reichhart D. (2021). Plant cytochrome P450 plasticity and evolution. Mol. Plant 14, 1244–1265. doi: 10.1016/j.molp.2021.06.028
Han H., Yu Q., Beffa R., González S., Maiwald F., Wang J., et al. (2021). Cytochrome P450 CYP81A10v7 in lolium rigidum confers metabolic resistance to herbicides across at least five modes of action. Plant J. 105, 79–92. doi: 10.1111/tpj.15040
Hausman N. E., Singh S., Tranel P. J., Riechers D. E., Kaundun S. S., Polge N. D., et al. (2011). Resistance to HPPD-inhibiting herbicides in a population of waterhemp (Amaranthus tuberculatus) from Illinois, united states. Pest Manage. Sci. 67, 258–261. doi: 10.1002/ps.2100
Hawkes T. R., Holt D. C., Andrews C. J., Thomas P. G., Langford M. P., Hollingworth S., et al. (2001) in The BCPC Conference: Weeds, 2001, Volume 1 and Volume 2. Proceedings of an international conference held at the Brighton Hilton Metropole Hotel, Brighton, UK. 563–568 (Farnham, UK: British Crop Protection Council).
Hawkes T. R., Langford M. P., Viner R., Blain R. E., Callaghan F. M., Mackay E. A., et al. (2019). Characterization of 4-hydroxyphenylpyruvate dioxygenases, inhibition by herbicides and engineering for herbicide tolerance in crops. Pestic. Biochem. Physiol. 156, 9–28. doi: 10.1016/j.pestbp.2019.01.006
Heap I. (2022) International herbicide-resistant weed database. Available at: www.weedscience.org (Accessed June 24, 2022).
Hirase K., Hoagland R. E. (2006). Characterization of aryl acylamidase activity from propanil-resistant barnyardgrass (Echinochloa crus-galli [L.] beauv.). Weed Biol. Manage. 6, 197–203. doi: 10.1111/j.1445-6664.2006.00218.x
Hodgson E., Levi P. E. (1998). 3 - Interactions of Piperonyl Butoxide with Cytochrome P450, in, ed. Jones D. G. B. T.-P. B. (London: Academic Press), 41–53. doi: 10.1016/B978-012286975-4/50005-X
Huang X.-X., Zhao S.-M., Zhang Y.-Y., Li Y.-J., Shen H.-N., Li X., et al. (2021). A novel UDP-glycosyltransferase 91C1 confers specific herbicide resistance through detoxification reaction in arabidopsis. Plant Physiol. Biochem. 159, 226–233. doi: 10.1016/j.plaphy.2020.12.026
Jhala A. J., Sandell L. D., Rana N., Kruger G. R., Knezevic S. Z. (2014). Confirmation and control of triazine and 4-hydroxyphenylpyruvate dioxygenase-inhibiting herbicide-resistant palmer amaranth (Amaranthus palmeri) in Nebraska. Weed Technol. 28, 28–38. doi: 10.1614/wt-d-13-00090.1
Jugulam M., Shyam C. (2019). Non-Target-Site resistance to Herbicides. Plants 8. doi: 10.3390/plants8100417
Kaundun S. S., Hutchings S. J., Dale R. P., Howell A., Morris J. A., Kramer V. C., et al. (2017). Mechanism of resistance to mesotrione in an amaranthus tuberculatus population from Nebraska, USA. PloS One 12. doi: 10.1371/journal.pone.0180095
Knezevic S. Z., Streibig J. C., Ritz C. (2007). Utilizing r software package for dose-response studies: The concept and data analysis. Weed Technol. 21, 840–848. doi: 10.1614/wt-06-161.1
Koo D.-H., Molin W. T., Saski C. A., Jiang J., Putta K., Jugulam M., et al. (2018). Extrachromosomal circular DNA-based amplification and transmission of herbicide resistance in crop weed amaranthus palmeri. Proc. Natl. Acad. Sci. 115, 3332–3337. doi: 10.1073/pnas.1719354115
Ksas B., Légeret B., Ferretti U., Chevalier A., Pospíšil P., Alric J., et al. (2018). The plastoquinone pool outside the thylakoid membrane serves in plant photoprotection as a reservoir of singlet oxygen scavengers. Plant Cell Environ. 41, 2277–2287. doi: 10.1111/pce.13202
Kuepper A. (2018). Molecular genetics of herbicide resistance in palmer amaranth (Amaranthus palmeri): Metabolic tembotrione resistance and geographic origin of glyphosate resistance (Colorado State University, Fort Collins, CO).
Küpper A., Peter F., Zöllner P., Lorentz L., Tranel P. J., Beffa R., et al. (2018). Tembotrione detoxification in 4-hydroxyphenylpyruvate dioxygenase (HPPD) inhibitor-resistant palmer amaranth (Amaranthus palmeri s. wats.). Pest Manage. Sci. 74, 2325–2334. doi: 10.1002/ps.4786
Lang P., Qin Y., Junzhi W., Heping H., Lingfeng M., Alex N., et al. (2021). An ABCC-type transporter endowing glyphosate resistance in plants. Proc. Natl. Acad. Sci. 118, e2100136118. doi: 10.1073/pnas.2100136118
Lee D. L., Knudsen C. G., Michaely W. J., Chin H. L., Nguyen N. H., Carter C. G., et al. (1998). The structure-activity relationships of the triketone class of HPPD herbicides. Pestic. Sci. 54, 377–384. doi: 10.1002/(SICI)1096-9063(199812)54:4<377::AID-PS827>3.0.CO;2-0
Liu C. J., Huhman D., Sumner L. W., Dixon R. A. (2003). Regiospecific hydroxylation of isoflavones by cytochrome P450 81E enzymes from medicago truncatula. Plant J. 36, 471–484. doi: 10.1046/j.1365-313X.2003.01893.x
Lu H., Yu Q., Han H., Owen M. J., Powles S. B. (2020). Evolution of resistance to HPPD-inhibiting herbicides in a wild radish population via enhanced herbicide metabolism. Pest Manage. Sci. 76, 1929–1937. doi: 10.1002/ps.5725
Ma R., Kaundun S. S., Tranel P. J., Riggins C. W., McGinness D. L., Hager A. G., et al. (2013). Distinct detoxification mechanisms confer resistance to mesotrione and atrazine in a population of waterhemp. Plant Physiol. 163, 363–377. doi: 10.1104/pp.113.223156
Matringe M., Sailland A., Pelissier B., Rolland A., Zink O. (2005). P-hydroxyphenylpyruvate dioxygenase inhibitor-resistant plants. Pest Manag. Sci. 61, 269–276. doi: 10.1002/ps.997
Matsumoto H., Mizutani M., Yamaguchi T., Kadotani J. (2002). Herbicide pyrazolate causes cessation of carotenoids synthesis in early watergrass by inhibiting 4-hydroxyphenylpyruvate dioxygenase. Weed Biol. Manage. 2, 39–45. doi: 10.1046/j.1445-6664.2002.00046.x
Mitchell G., Bartlett D. W., Fraser T. E. M., Hawkes T. R., Holt D. C., Townson J. K., et al. (2001). Mesotrione: A new selective herbicide for use in maize. Pest Manage. Sci. 57, 120–128. doi: 10.1002/1526-4998(200102)57:2<120::AID-PS254>3.0.CO;2-E
Nakka S., Godar A. S., Wani P. S., Thompson C. R., Peterson D. E., Roelofs J., et al. (2017). Physiological and molecular characterization of hydroxyphenylpyruvate dioxygenase (HPPD)-inhibitor resistance in palmer amaranth (Amaranthus palmeri S.Wats.). Front. Plant Sci. 8. doi: 10.3389/fpls.2017.00555
Ndikuryayo F., Moosavi B., Yang W.-C., Yang G.-F. (2017). 4−Hydroxyphenylpyruvate dioxygenase inhibitors from chemical biology. J. Agric. Food Chem. 65, 8523–8537. doi: 10.1021/acs.jafc.7b03851
Oliveira M. C., Gaines T. A., Dayan F. E., Patterson E. L., Jhala A. J., Knezevic S. Z. (2018). Reversing resistance to tembotrione in an amaranthus tuberculatus (var. rudis) population from Nebraska, USA with cytochrome P450 inhibitors. Pest Manage. Sci. 74, 2296–2305. doi: 10.1002/ps.4697
Pandian B. A., Sathishraj R., Djanaguiraman M., Prasad P. V. V., Jugulam M. (2020). Role of cytochrome P450 enzymes in plant stress response. Antioxidants 9, 1–15. doi: 10.3390/antiox9050454
Ritz C., Baty F., Streibig J. C., Gerhard D. (2015). Dose-response analysis using r. PloS One 10, 1–13. doi: 10.1371/journal.pone.0146021
Sandell L., Amit J., Kruger G. R. (2012). Evaluation of a putative HPPD-resistant palmer amaranth (Amaranthus palmeri) population in Nebraska. Proc. North Cent. Weed Sci. Soc. 10–1371.
Schulte W., Köcher H. (2009). Tembotrione and combination partner isoxadifen-ethyl – mode of herbicidal action. Bayer CropSci. J 65, 35–52.
Shyam C., Borgato E. A., Peterson D. E., Dille J. A., Jugulam M. (2021). Predominance of metabolic resistance in a six-Way-Resistant palmer amaranth (Amaranthus palmeri) population. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.614618
Shyam C., Peterson D. E., Jugulam M. (2022). Resistance to 2,4-d in palmer amaranth (Amaranthus palmeri) from Kansas is meditated by enhanced metabolism. Weed Sci. 70, 1–32. doi: 10.1017/wsc.2022.29
Siminszky B. (2006). Plant cytochrome P450-mediated herbicide metabolism. Phytochem. Rev. 5, 445–458. doi: 10.1007/s11101-006-9011-7
Tardif F. J., Preston C., Powles S. B. (1997). “Mechanisms of herbicide multiple resistance in lolium rigidum BT - weed and crop resistance to herbicides,”. Eds. De Prado R., Jorrín J., García-Torres L. Weed and Crop Resistance to Herbicides. (Netherlands: Dordrecht: Springer), 117–124. doi: 10.1007/978-94-011-5538-0_12
Van Almsick A. (2009). New HPPD-inhibitors - a proven mode of action as a new hope to solve current weed problems. Outlooks Pest Manage. 20, 27–30. doi: 10.1564/20feb09
Ward S. M., Webster T. M., Steckel L. E. (2013). Palmer amaranth (Amaranthus palmeri): A review. Weed Technol. 27, 12–27. doi: 10.1614/wt-d-12-00113.1
Williams M. M., Pataky J. K. (2008). Genetic basis of sensitivity in sweet corn to tembotrione. Weed Sci. 56, 364–370. doi: 10.1614/ws-07-149.1
Keywords: Amaranthus palmeri, tembotrione resistance, tembotrione metabolism, cytochrome P450, CYP81E8 expression
Citation: Aarthy T, Shyam C and Jugulam M (2022) Rapid metabolism and increased expression of CYP81E8 gene confer high level of resistance to tembotrione in a multiple-resistant Palmer amaranth (Amaranthus palmeri S. Watson). Front. Agron. 4:1010292. doi: 10.3389/fagro.2022.1010292
Received: 02 August 2022; Accepted: 09 September 2022;
Published: 23 September 2022.
Edited by:
Baruch Rubin, Hebrew University of Jerusalem, IsraelReviewed by:
Maor Matzrafi, Volcani Center, IsraelCopyright © 2022 Aarthy, Shyam and Jugulam. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mithila Jugulam, bWl0aGlsYUBrc3UuZWR1
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.