
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Agron., 10 December 2021
Sec. Pest Management
Volume 3 - 2021 | https://doi.org/10.3389/fagro.2021.794312
This article is part of the Research TopicInsights in Pest ManagementView all 4 articles
 Jonathan Willow1,2*
Jonathan Willow1,2* Clauvis Nji Tizi Taning2
Clauvis Nji Tizi Taning2 Samantha M. Cook3
Samantha M. Cook3 Silva Sulg1
Silva Sulg1 Ana I. Silva4
Ana I. Silva4 Guy Smagghe2
Guy Smagghe2 Eve Veromann1
Eve Veromann1The unprecedented target-specificity of double-stranded RNA (dsRNA), due to its sequence-specific mode of action, puts dsRNA at the forefront of biosafe insecticide technology. Since 2007, sensitive target genes have been identified in numerous crop pest insects, with an end goal of applying RNA interference (RNAi) in pest management. Key RNAi targets identified include genes involved in (1) feeding and digestion, (2) production of dsRNases, (3) resistance to insecticides and plant allelochemicals, (4) reproductive fitness, and (5) transmission of plant viruses. Despite the advances, there remain critical knowledge gaps in each of these areas. Particular emphasis must be placed on ensuring RNAi's compatibility with integrated pest management (IPM), via further identification of molecular targets that reduce crop damage while sustaining pest (host) populations for highly specialized biocontrol agents, the latter representing a core pillar of IPM.
In 2007, two studies demonstrated potential for double-stranded RNA (dsRNA) to act as a nucleotide sequence-specific insecticide, as it induced RNA interference (RNAi) in insects consuming dsRNA comprising an inverted repeat of a target gene's coding sequence (Baum et al., 2007; Mao et al., 2007). Baum et al. (2007) demonstrated this concept in western corn rootworm (Diabrotica virgifera) and Colorado potato beetle (Leptinotarsa decemlineata), major pests of maize and potato, respectively. They showed that efficacy varied depending on the gene- and corresponding protein targeted for downregulation. Mao et al. (2007) demonstrated this concept in cotton bollworm (Helicoverpa armigera), through targeting the cytochrome P450 monooxygenase gene CYP6AE14, and showed how its targeted downregulation impairs H. armigera's ability to detoxify the plant allelochemical gossypol; as well as through targeted downregulation of the glutathione-S-transferase gene GST1. Both studies demonstrated that pest control efficacy was based on nucleotide sequence identity, placing RNAi at the forefront of biosafe insecticide technology given its potential for target species-specificity. Since these two landmark studies, researchers have continued to demonstrate potential applications for RNAi technology in crop protection, through targeted downregulation of genes in other agricultural pest insects. While some studies have examined the use of species as screening models for identifying sensitive RNAi targets across crop pest taxa (Knorr et al., 2018, 2021; Mehlhorn et al., 2021), sensitivity of a target gene in one species may not necessarily apply to another species, given the physiological and ecological differences between species, especially between distantly related taxa.
DsRNA's mode of action suggests potential for high target-specificity, as long as dsRNA-expressing transgenes or sprayed dsRNAs are constructed to be target-specific. However, there is always the possibility that non-target organisms could be directly affected by RNAi cultivars or dsRNA sprays, especially if there is sufficient similarity between the target messenger RNA (mRNA) sequence in the target pest species and an mRNA sequence in an exposed non-target species. Nevertheless, rapid improvements in next generation sequencing platforms, and an upward trend in the availability of genome data for non-target species, is improving the exploitation of bioinformatics as a tool for RNAi target selection and risk prediction. This is essential as a first step in the development and risk assessment of both RNAi cultivars and dsRNA spray products.
DsRNA generally induces a lethal phenotype at a slower rate than other insecticides, and dsRNA-resistance has been demonstrated in vivo in both D. virgifera (Khajuria et al., 2018) and L. decemlineata (Mishra et al., 2021). Thus, it is of utmost importance to examine current knowledge regarding RNAi targets that either induce a timelier lethal phenotype, or that are more compatible with integrated pest management (IPM) principles. Here we provide an overview of both advancements and knowledge gaps regarding RNAi targets for potentially achieving sustainable crop protection from insect pests. Key points include targeting molecules involved in feeding and digestion, production of dsRNases (enzymes which degrade dsRNAs), and resistance to chemical compounds (e.g., commercial pesticides, plant allelochemicals); as well as the potential for minimizing RNAi's confliction with agroecological services provided by highly specialized biocontrol agents, which is important due to RNAi's potential for use in integrated pest management (IPM).
Several groups have examined the possibility of targeting insect genes involved in feeding and digestion. Will and Vilcinskas (2015) showed that targeting the salivary sheath protein in pea aphid (Acyrthosiphon pisum) disrupts formation of the salivary sheath (which is needed to facilitate penetration of the aphid's feeding apparatus into plant cell membranes), and interrupts feeding and reduces reproduction (possibly due to disruption of salivary sheath formation and reduced nutrition, respectively). Genes encoding membrane proteins associated with smooth septate junctions, required for functional intestinal barriers, have been shown to be effective RNAi targets in D. virgifera (Hu et al., 2016, 2019), the fruit fly Drosophila melanogaster (Izumi et al., 2019), and L. decemlineata (Petek et al., 2020). Hu et al. (2016, 2019) and Izumi et al. (2019) linked downregulation of these genes with intestinal barrier dysfunction and shortened lifespan. Hu et al. (2016) demonstrated that transgenic maize plants targeting a membrane protein associated with smooth septate junctions were protected from feeding damage. Furthermore, Petek et al. (2020) showed that field spraying of dsRNA targeting a membrane protein associated with smooth septate junctions was equally effective in controlling L. decemlineata as the commercial insecticide spinosad, though its activity was slower (significant mortality at 7 days) compared to that of spinosad (2 days). Salvador et al. (2021) targeted, in both larval and adult stages of cotton boll weevil (Anthonomus grandis), genes coding for production of α-amylase, trehalase and hexokinase, each of these proteins being important for chemical digestion in the midgut of A. grandis. Targeting these molecules resulted in ~60% (α-amylase), 45% (trehalase), and 50% (hexokinase) mortality in A. grandis larvae after 14 days of dsRNA feeding.
Apoptosis, a form of programed cell death in multicellular organisms, is a highly regulated process. While apoptotic events are vital to an insect's longevity (e.g., host control of viral infection), excessive apoptosis results in atrophy. Thus, inhibitors of apoptosis (IAPs) have been examined for suitability as RNAi targets in members of the insect orders Coleoptera (Rodrigues et al., 2017, 2018; Cao et al., 2018; Yoon et al., 2018, 2020; Máximo et al., 2020; Chikami et al., 2021), Diptera (Powell M. et al., 2017), Hemiptera and Lepidoptera (Gurusamy et al., 2020). In dsRNA feeding assays, Máximo et al. (2020) demonstrated that targeting an IAP in L. decemlineata larvae induced greater mortality compared to all other targets examined, but when targeting both the IAP and actin simultaneously, significant inhibition of feeding occurred within 24 h, followed by significant larval mortality within 48 h. Chikami et al. (2021), in a study on the pest ladybeetle Henosepilachna vigitioctopunctata, also demonstrated rapid reduction in feeding within 48 h, followed by rapid reduction in survival of larvae fed dsRNA targeting an IAP. IAPs may be effective RNAi targets in other pest insect taxa, for example in aphids (Figure 1), an important group of crop pests where an evolutionary expansion of IAPs is observed (Lopes et al., 2020). As this expansion has been hypothesized as a contributing factor to stress resilience in aphids (Lopes et al., 2020), targeting IAPs may significantly contribute to enhancing management of this important group of agricultural pests.
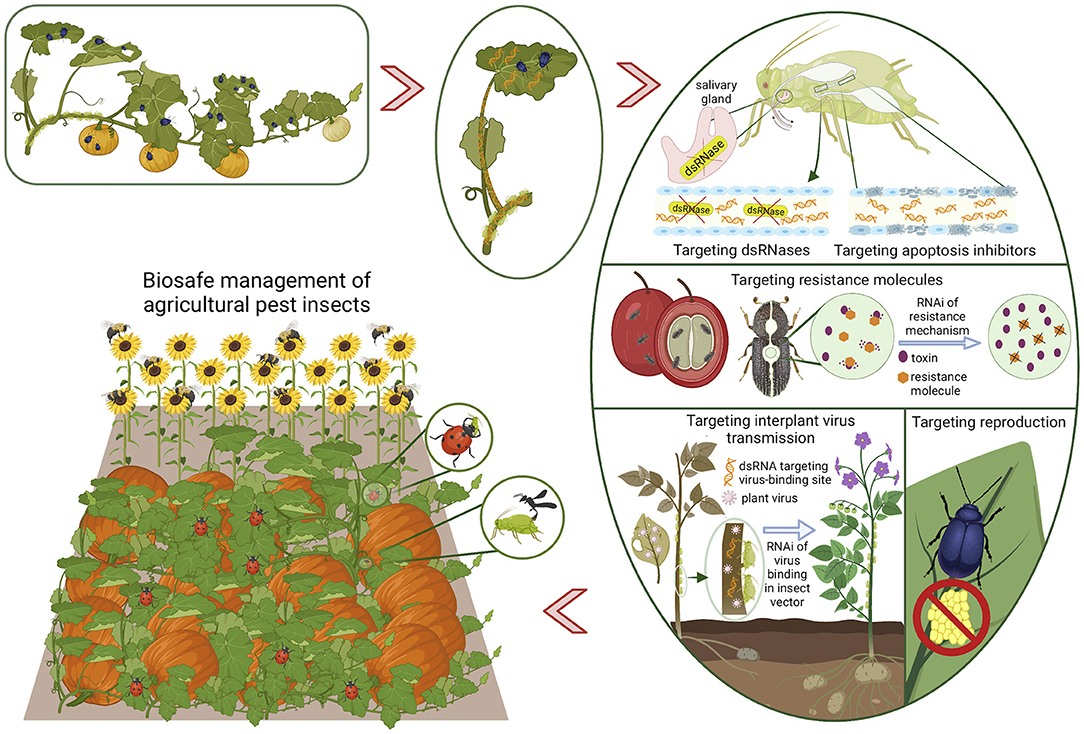
Figure 1. Schematic illustration depicting recent advances in RNAi targets in agricultural pest insects, and the potential utility of targeting these molecules/mechanisms for effective and biosafe crop protection. Crop pest insects take up dsRNA sprayed on- or expressed in plants. Some attractive targets for RNAi include: molecules important in feeding and digestion; enzymes important for detoxification of pesticides or plant allelochemicals; molecules contributing to insect-vectored transmission of plant viruses; and reduction or inhibition of reproduction. The sequence-specificity of dsRNA makes it IPM-compatible, and brings it to the frontier of ecologically sustainable insecticide technology.
DsRNases, especially in the insect midgut or in organs that can influence exogenous dsRNAs in passage to the midgut (e.g., salivary glands), can influence RNAi efficacy (Figure 1). Studies examining stability of exogenous dsRNAs isolated in extracted midgut fluid indicate the role of dsRNases in partial- or complete dsRNA degradation in several pest insects (Wynant et al., 2014; Almeida Garcia et al., 2017; Singh et al., 2017; Peng et al., 2018; Prentice et al., 2019; Tayler et al., 2019; Kaur et al., 2020). However, for the majority of crop pest species, it is unknown what role dsRNases have in inhibiting RNAi post-consumption of exogenous dsRNAs. In vivo co-targeting of dsRNases together with other vital genes has been demonstrated to significantly reduce gene expression and insect performance, compared to targeting only the vital gene, in insects of the orders Coleoptera (Almeida Garcia et al., 2017; Spit et al., 2017; Prentice et al., 2019; Peng et al., 2020a), Hemiptera (Luo et al., 2017; Chung et al., 2018; Kaur et al., 2020; Sharma et al., 2021), and Diptera (Tayler et al., 2019; Giesbrecht et al., 2020). Additional studies also suggest that inadequate oral RNAi is due to the presence of dsRNases in an even greater diversity of insect taxa (Song et al., 2017; Ghodke et al., 2019; Cooper et al., 2020; Fan et al., 2020; Peng et al., 2020b; Yoon et al., 2021).
Interestingly, while a few representatives from some coleopteran families show high sensitivity to oral RNAi (i.e., Chrysomelidae, Coccinellidae), other coleopteran families contain species that are insensitive- or only moderately sensitive to oral RNAi; for example, Nitidulidae (Powell M. E. et al., 2017; Willow et al., 2020, 2021), Curculionidae (Almeida Garcia et al., 2017; Prentice et al., 2019), and Buprestidae (Rodrigues et al., 2017, 2018). As Coleoptera contains a relatively large number of the world's most damaging crop pest species, with relatively few having been examined for RNAi sensitivity, inter- and intrafamily comparative RNAi sensitivity in this diverse order represents a critical avenue of research with many knowledge gaps. Thus, co-targeting endogenous dsRNases may prove vital for RNAi-based management of many coleopteran pest species.
RNAi has been examined as a tool for overcoming insecticide resistance in various insect pests. In Asian citrus psyllid (Diaphorina citri), targeted downregulation of cytochrome P450, an enzyme involved in metabolism of exogenous and endogenous compounds, increased susceptibility to the neonicotinoid insecticide imidacloprid in members of a resistant population (Killiny et al., 2014). Targeting cytochrome P450 in A. grandis without post-treatment of any conventional insecticide was also shown to result in significant mortality (Salvador et al., 2021). Xu et al. (2017) demonstrated that targeted downregulation of seven different nuclear receptor genes in brown planthopper (Nilaparvata lugens) increased susceptibility to the sulfoximine insecticide sulfoxaflor, suggesting a role of these genes in sulfoxaflor detoxification, and thus their potential as RNAi targets for overcoming sulfoxaflor resistance in N. lugens populations. Du et al. (2021) recently demonstrated that targeting a uridine diphosphate-glucuronosyltransferase gene (UGT352A5) in silverleaf whitefly (Bemisia tabaci) reduced resistance to the neonicotinoid insecticide thiamethoxam, as mortality after both 6 and 12 h of thiamethoxam exposure was significantly greater in B. tabaci pre-fed dsUGT352A5 for 48 h, compared to those pre-fed a negative control dsRNA. This finding suggests UGT352A5 as a useful target for overcoming thiamethoxam resistance in B. tabaci populations. While RNAi-based inhibition of broad-spectrum insecticide resistance may be necessary in certain cases, we note that this tactic could also represent an RNAi application that is in conflict with IPM principles.
A recent study by Kang et al. (2021) demonstrated that upregulation of the gene fused (encoding a serine/threonine protein kinase) is involved in diamondback moth's (Plutella xylostella's) resistance to the pesticidal crystal protein Bacillus thuringiensis (Bt)Cry1Ac, and that targeted downregulation of fused results in larval and pupal mortality in BtCry1Ac-resistant strains. Thus, this finding revealed fused as a potential molecular target for overcoming Bt resistance in P. xylostella populations. Guo et al. (2021) also recently provided the first comprehensive mechanistic insight into the mitogen-activated protein kinase (MAPK, another serine/threonine protein kinase) signaling pathway's involvement in insect resistance to Bt toxins. The authors specifically demonstrated the role of MAPK cascades in defending P. xylostella against the insecticidal action of BtCry1Ac. Both of the abovementioned studies provide a platform for investigating the potential for targeting particular genes and/or signaling pathways, in order to suppress Bt resistance in other crop pest species.
Horizontal gene transfer (HGT) events, the non-parental transfer of genetic material between organisms, have given rise to new candidate target genes in crop pest species. For example, the gene MAN1, encoding a mannanase, was transferred from a member of the bacterial Bacillus clade to the genome of coffee berry borer (Hypothenemus hampei), this HGT event preceding H. hampei's invasion from West Africa to Latin America and Asia (Acuña et al., 2012). MAN1 specifically encodes a protein capable of hydrolysing galactomannan, the most abundant polysaccharide in coffee berries. This HGT event would have been essential for H. hampei to adapt to its specialization on host tissues rich in mannans. More recently, evidence emerged that B. tabaci obtained its malonyltransferase gene PMaT1 from an unknown plant species (B. tabaci being highly polyphagous) through an HGT event (Xia et al., 2021). B. tabaci's expression of this gene allows this pest to neutralize allelochemicals that plants use to defend themselves against herbivorous insects, in turn allowing B. tabaci to safely feed on these plants. Xia et al. (2021) demonstrated that targeted downregulation of PMaT1 resulted in almost 100% mortality of B. tabaci fed transgenic tomato plants, after 7 days of feeding. With increasing work being conducted in genomics, it may be found that HGT events in crop pests are far more prevalent than current predictions suggest. It could be supposed that some HGT events represent a prerequisite for certain species to achieve pest status, thereby giving rise to the use of broad-spectrum insecticides, which can in turn affect non-target organisms. These horizontally transferred genes that allow trophic niche occupancy may, in some cases, represent relevant targets for RNAi-based pest management of crop pest insects.
Identifying genetic targets involved in dsRNA resistance represents an important area of research for encouraging RNAi's future in sustainably managing crop pest insects. Khajuria et al. (2018) established a dsRNA-resistant D. virgifera population, demonstrating not only that resistant beetles were insensitive to Snf7-specific dsRNA microinjected into the haemolymph, but also that their resistance was non-specific to dsRNA targeting Snf7, as these beetles also showed resistance to other D. virgifera-specific dsRNAs. This suggested that dsRNA resistance in D. virgifera may involve impaired dsRNA uptake in cell types other than those of the gut, and/or may involve impaired systemic RNAi. More recently, Mishra et al. (2021) demonstrated dsRNA resistance development in L. decemlineata, through continuous rearing of adult survivors that were chronically exposed to L. decemlineata-specific dsRNA. As in Khajuria et al. (2018), this outcome was not specific to a single dsRNA, as L. decemlineata reared for resistance to vATPase-specific dsRNA also showed resistance to another L. decemlineata-specific dsRNA. If precise molecular mechanisms impairing dsRNA uptake were to be identified in a broad variety of pest species, these mechanisms could potentially be co-targeted for downregulation to ameliorate dsRNA resistance in different target species. However, the precise molecular basis of such mechanisms may widely vary between insect taxa, and thus the investigation of this possibility would require a broad level of intertaxon focus.
RNAi as a crop protection measure is growing in prominence due to its potential use in IPM, an essential part of which is the partial reliance on natural enemies of crop pests, in order to avoid pest outbreaks. Thus, RNAi's compatibility with conservation biocontrol strategies represents a prerequisite to the field use of RNAi strategies. Due to the unprecedented target-specificity of dsRNA, RNAi approaches are not expected to directly affect populations of biocontrol agents. Furthermore, as generalist predators may rely on a variety of pest- and non-pest species for acquiring nutrients, considerable reductions in a single crop pest species is unlikely to significantly impact generalist predator populations. However, RNAi's compatibility with IPM becomes especially important when considering potential indirect effects on highly specialized biocontrol agents (i.e., specialist parasitoids) that rely on some availability of the host with which they co-evolved. It is not yet understood how pest-parasitoid interactions may influence adult parasitoid emergence from hosts subjected to RNAi. For example, indirect effects on parasitoid development may depend on whether a parasitoid is idiobiotic (immobilizes host, preventing further host development) vs. koinobiotic (allows host to continue development while being consumed), or whether a parasitoid is endoparasitic (egg laid within host body cavity) vs. ectoparasitic (egg laid on host epicuticle). Despite these critical knowledge gaps, studies have identified RNAi targets potentially compatible with conservation of highly specialized biocontrol agents. Here we focus on these advances.
Targeting genes that are necessary for reproductive fitness (Figure 1), but that are only infrequently- or marginally effective in inducing a lethal phenotype when downregulated, may allow hosts to persist for specialist parasitoids while also controlling the pest population via reductions in next-generation offspring. Targeting vitellogenin receptor [VgR, responsible for endocytic transport of vitellogenin (Vg), precursor to the major yolk protein, into oocytes] and boule (bol, necessary for successful meiosis during spermatogenesis, and for sperm maturation and male fertility), Niu et al. (2017) demonstrated that larval D. virgifera feeding on transgenic maize expressing dsVgR or dsbol transcripts had no significant effect on successful pupation into the adult stage. However, larval feeding on three transgenic maize lines expressing the dsbol transcript resulted in significant reductions in adult oviposition and progeny hatching rate. Targeting D. virgifera VgR significantly reduced adult oviposition in two transgenic maize lines, whereas none affected progeny hatching rate. In another study, Shang et al. (2018) observed shortened reproductive period, slower embryo development and lower fecundity in adult brown citrus aphids (Aphis citricidus) that were fed either dsVg or dsVgR during their fourth-instar nymph stage. The abovementioned studies, while having innate value for managing D. virgifera and A. citricidus populations, can serve as model studies for examining reproduction-targeting RNAi approaches in other crop pests. This may be especially important for those pests that have co-evolved with highly specialized biocontrol agents. While this tactic could affect host availability in the next season, it represents a critical topic of investigation for achieving IPM-compatible RNAi.
An additional study by Will and Vilcinskas (2015) reported significantly reduced reproductive rates in A. pisum after targeting salivary sheath protein in this aphid species. Specifically, the mean number of nymphs born per target-treatment-injected adult was significantly lower than that observed in two different control groups (untreated, and injection of non-A. pisum-specific dsRNA). Interestingly, no effect was observed on survival. The authors suggested that the reduced availability of nutrients, due to the disruption of salivary sheath formation, forced a trade-off in which reproductive fitness was sacrificed to enable survival. It is unknown whether a similar trade-off would occur under natural conditions, as the complexity of physiological and ecological trade-offs should be expected to vary between laboratory and natural conditions.
RNAi may also be used to manage outbreaks of plant viruses vectored by insects, by interfering with plant virus binding sites in the insect (Figure 1). Bahrami Kamangar et al. (2019) demonstrated that, by targeting a cuticular protein in the stylet of the aphid Myzus persicae, transmission rate of Potato virus Y was reduced by 47%. This represents a critical avenue of inquiry for IPM-compatible, RNAi-based management of insect-vectored phytopathogens. Furthermore, this approach to RNAi-based plant protection could serve to prevent interplant virus transmission by other crop pest taxa with similar modes of viral spread (e.g., leafhoppers, planthoppers, and thrips), in addition to having the advantage of minimizing host elimination, representing a potential benefit to both generalist predators and highly specialized biocontrol agents.
Finally, additional RNAi targets that confer no lethal toxicity to the target species may simultaneously facilitate the efficacy of biocontrol agents. For example, reducing the growth rate of a pest species, while allowing it to successfully develop, may lengthen the window of opportunity for biocontrol of the pest population (Benrey and Denno, 1997). Also, since the alarm pheromone is one of the major defenses against predation and parasitism in aphids (Vandermoten et al., 2012), targeting genes involved in alarm pheromone production or their receptors may make aphids more susceptible to biocontrol by natural enemies. These RNAi targets should be examined in mesocosm studies in order to provide evidence for their potential utility in facilitating biocontrol efficacy.
In light of dsRNA representing the frontier of ecologically sustainable insecticide technology, it is encouraging that many advances in genetic target identification have recently been made. Due to the diversity of pest insects responsible for economically-relevant crop yield losses, suitable RNAi target screening models are required in order to speed up the identification of effective molecular targets. However, sensitivity of molecular targets will undoubtedly vary depending on different pest species' vulnerabilities. Disruption of feeding and/or digestion can quicken the pace at which RNAi-induced mortality occurs in the target species. Co-targeting dsRNases can enhance RNAi efficacy in various crop pests. Targeting resistance molecules in crop pests can reduce various pests' ability to detoxify substances either applied for crop protection or produced in planta as allelochemicals. In some cases, IPM compatibility relies on highly specialized biocontrol agents (i.e., specialist parasitoids); and RNAi's compatibility with IPM will require the assurance that RNAi will not interfere with these populations.
JW conceived the manuscript and wrote the original draft. CT made significant contribution to the manuscript structure. AS and JW created the illustrations. All authors made comments/suggestions toward revising the original draft and approved the final version of the manuscript.
The review was funded by the Personal Research Funding project no. PRG1056 of the Estonian Research Council.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
SC acknowledges funding support from the UK Biotechnology and Biological Sciences Research Council (programme BBS/OS/CP/000001—Smart Crop Protection). We acknowledge the Special Research Fund (BOF) of Ghent University and the Research Foundation–Flanders(FWO–Vlaanderen).
Acuña, R., Padilla, B. E., Flórez-Ramos, C. P., Rubio, J. D., Herrera, J. C., Benavides, P., et al. (2012). Adaptive horizontal transfer of a bacterial gene to an invasive insect pest of coffee. Proc. Natl. Acad. Sci. U. S. A. 109, 4197–4202. doi: 10.1073/pnas.1121190109
Almeida Garcia, R., Lima Pepino Macedo, L., Cabral do Nascimento, D., Gillet, F.-X., Moreira-Pinto, C. E., Faheem, M., et al. (2017). Nucleases as a barrier to gene silencing in the cotton boll weevil, Anthonomus grandis. PLoS ONE 12:e0189600. doi: 10.1371/journal.pone.0189600
Bahrami Kamangar, S., Christiaens, O., Taning, C. N. T., De Jonghe, K., and Smagghe, G. (2019). The cuticle protein MPCP2 is involved in Potato virus Y transmission in the green peach aphid Myzus persicae. J. Plant Dis. Prot. 126, 351–357. doi: 10.1007/s41348-019-00232-w
Baum, J. A., Bogaert, T., Clinton, W., Heck, G. R., Feldmann, P., Ilagan, O., et al. (2007). Control of coleopteran insect pests through RNA interference. Nat. Biotechnol. 25, 1322–1326. doi: 10.1038/nbt1359
Benrey, B., and Denno, R. F. (1997). The slow-growth–high-mortality hypothesis: a test using the cabbage butterfly. Ecology 78, 987–999. doi: 10.1890/0012-9658(1997)0780987:TSGHMH2.0.CO
Cao, M., Gatehouse, J. A., and Fitches, E. C. (2018). A systematic study of RNAi effects and dsRNA stability in Tribolium castaneum and Acyrthosiphon pisum, following injection and ingestion of analogous dsRNAs. Int. J. Mol. Sci. 19:1079. doi: 10.3390/ijms19041079
Chikami, Y., Kawaguchi, H., Suzuki, T., Yoshioka, H., Sato, Y., Yaginuma, T., et al. (2021). Oral RNAi of diap1 results in rapid reduction of damage to potatoes in Henosepilachna vigintioctopunctata. J. Pest Sci. 94, 505–515. doi: 10.1007/s10340-020-01276-w
Chung, S. H., Jing, X., Luo, Y., and Douglas, A. E. (2018). Targeting symbiosis-related insect genes by RNAi in the pea aphid-Buchnera symbiosis. Insect Biochem. Mol. Biol. 95, 55–63. doi: 10.1016/j.ibmb.2018.02.004
Cooper, A. M. W., Song, H., Shi, X., Yu, Z., Lorenzen, M., Silver, K., et al. (2020). Molecular characterizations of double-stranded RNA degrading nuclease genes from Ostrinia nubilalis. Insects 11:652. doi: 10.3390/insects11100652
Du, T., Fu, B., Wei, X., Yin, C., Yang, J., Huang, M., et al. (2021). Knockdown of UGT352A5 decreases the thiamethoxam resistance in Bemisia tabaci (Hemiptera: Gennadius). Int. J. Biol. Macromol. 186, 100–108. doi: 10.1016/j.ijbiomac.2021.07.040
Fan, Y.-H., Song, H.-F., Abbas, M., Wang, Y.-L., Li, T., Ma, E.-B., et al. (2020). A dsRNA-degrading nuclease (dsRNase2) limits RNAi efficiency in the Asian corn borer (Ostrinia furnacalis). Insect Sci. 2020:12882. doi: 10.1111/1744-7917.12882
Ghodke, A. B., Good, R. T., Golz, J. F., Russell, D. A., Edwards, O., and Robin, C. (2019). Extracellular endonucleases in the midgut of Myzus persicae may limit the efficacy of orally delivered RNAi. Sci. Rep. 9:11898. doi: 10.1038/s41598-019-47357-4
Giesbrecht, D., Heschuk, D., Wiens, I., Boguski, D., LaChance, P., and Whyard, S. (2020). RNA interference is enhanced by knockdown of double-stranded RNases in the yellow fever mosquito Aedes aegypti. Insects 11:327. doi: 10.3390/insects11060327
Guo, Z., Kang, S., Wu, Q., Wang, S., Crickmore, N., Zhou, X., et al. (2021). The regulation landscape of MAPK signaling cascade for thwarting Bacillus thuringiensis infection in an insect host. PLoS Pathog. 17:e1009917. doi: 10.1371/journal.ppat.1009917
Gurusamy, D., Mogilicherla, K., and Palli, S. R. (2020). Chitosan nanoparticles help double-stranded RNA escape from endosomes and improve RNA interference in the fall armyworm, Spodoptera frugiperda. Arch. Insect Biochem. Physiol. 104:e21677. doi: 10.1002/arch.21677
Hu, X., Richtman, N. M., Zhao, J.-Z., Duncan, K. E., Niu, X., Procyk, L. A., et al. (2016). Discovery of midgut genes for the RNA interference control of corn rootworm. Sci. Rep. 6:30542. doi: 10.1038/srep30542
Hu, X., Steimel, J. P., Kapka-Kitzman, D. M., Davis-Vogel, C., Richtman, N. M., Mathis, J. P., et al. (2019). Molecular characterization of the insecticidal activity of double-stranded RNA targeting the smooth septate junction of western corn rootworm (Diabrotica virgifera virgifera). PLoS ONE 14:e0210491. doi: 10.1371/journal.pone.0210491
Izumi, Y., Furuse, K., and Furuse, M. (2019). Septate junctions regulate gut homeostasis through regulation of stem cell proliferation and enterocyte behavior in Drosophila. J. Cell Sci. 132:232108. doi: 10.1242/jcs.232108
Kang, S., Sun, D., Qin, J., Guo, L., Zhu, L., Bai, Y., et al. (2021). Fused: a promising molecular target for an RNAi-based strategy to manage Bt resistance in Plutella xylostella (L.). J. Pest Sci. 2021:3. doi: 10.1007/s10340-021-01374-3
Kaur, R., Gupta, M., Singh, S., Joshi, N., and Sharma, A. (2020). Enhancing RNAi efficiency to decipher the functional response of potential genes in Bemisia tabaci AsiaII-1 (Gennadius) through dsRNA feeding assays. Front. Physiol. 11:123. doi: 10.3389/fphys.2020.00123
Khajuria, C., Ivashuta, S., Wiggins, E., Flagel, L., Moar, W., Pleau, M., et al. (2018). Development and characterization of the first dsRNA-resistant insect population from western corn rootworm, Diabrotica virgifera virgifera LeConte. PLoS ONE 13:e0197059. doi: 10.1371/journal.pone.0197059
Killiny, N., Hajeri, S., Tiwari, S., Gowda, S., and Stelinski, L. L. (2014). Double-stranded RNA uptake through topical application, mediates silencing of five CYP4 genes and suppresses insecticide resistance in Diaphorina citri. PLoS ONE 9:e110536. doi: 10.1371/journal.pone.0110536
Knorr, E., Billion, A., Fishilevich, E., Tenbusch, L., Frey, M. L. F., Rangasamy, M., et al. (2021). Knockdown of genes involved in transcription and splicing reveals novel RNAi targets for pest control. Front. Agron. 3:64. doi: 10.3389/fagro.2021.715823
Knorr, E., Fishilevich, E., Tenbusch, L., Frey, M. L. F., Rangasamy, M., Billion, A., et al. (2018). Gene silencing in Tribolium castaneum as a tool for the targeted identification of candidate RNAi targets in crop pests. Sci. Rep. 8, 1–15. doi: 10.1038/s41598-018-20416-y
Lopes, M. R., Parisot, N., Gaget, K., Huygens, C., Peignier, S., Duport, G., et al. (2020). Evolutionary novelty in the apoptotic pathway of aphids. Proc. Natl. Acad. Sci. U. S. A. 117, 32545–32556. doi: 10.1073/pnas.2013847117
Luo, Y., Chen, Q., Luan, J., Chung, S. H., Van Eck, J., Turgeon, R., et al. (2017). Towards an understanding of the molecular basis of effective RNAi against a global insect pest, the whitefly Bemisia tabaci. Insect Biochem. Mol. Biol. 88, 21–29. doi: 10.1016/j.ibmb.2017.07.005
Mao, Y.-B., Cai, W.-J., Wang, J.-W., Hong, G.-J., Tao, X.-Y., Wang, L.-J., et al. (2007). Silencing a cotton bollworm P450 monooxygenase gene by plant-mediated RNAi impairs larval tolerance of gossypol. Nat. Biotechnol. 25, 1307–1313. doi: 10.1038/nbt1352
Máximo, W. P. F., Howell, J. L., Mogilicherla, K., Basij, M., Chereddy, S. C. R. R., and Palli, S. R. (2020). Inhibitor of apoptosis is an effective target gene for RNAi-mediated control of Colorado potato beetle, Leptinotarsa decemlineata. Arch. Insect Biochem. Physiol. 104:e21685. doi: 10.1002/arch.21685
Mehlhorn, S., Ulrich, J., Baden, C. U., Buer, B., Maiwald, F., Lueke, B., et al. (2021). The mustard leaf beetle, Phaedon cochleariae, as a screening model for exogenous RNAi-based control of coleopteran pests. Pestic. Biochem. Physiol. 176:104870. doi: 10.1016/j.pestbp.2021.104870
Mishra, S., Dee, J., Moar, W., Dufner-Beattie, J., Baum, J., Dias, N. P., et al. (2021). Selection for high levels of resistance to double-stranded RNA (dsRNA) in Colorado potato beetle (Leptinotarsa decemlineata Say) using non-transgenic foliar delivery. Sci. Rep. 11:6523. doi: 10.1038/s41598-021-85876-1
Niu, X., Kassa, A., Hu, X., Robeson, J., McMahon, M., Richtman, N. M., et al. (2017). Control of western corn rootworm (Diabrotica virgifera virgifera) reproduction through plant-mediated RNA interference. Sci. Rep. 7:12591. doi: 10.1038/s41598-017-12638-3
Peng, Y., Wang, K., Chen, J., Wang, J., Zhang, H., Ze, L., et al. (2020a). Identification of a double-stranded RNA-degrading nuclease influencing both ingestion and injection RNA interference efficiency in the red flour beetle Tribolium castaneum. Insect Biochem. Mol. Biol. 125:103440. doi: 10.1016/j.ibmb.2020.103440
Peng, Y., Wang, K., Fu, W., Sheng, C., and Han, Z. (2018). Biochemical comparison of dsRNA degrading nucleases in four different insects. Front. Physiol. 9:624. doi: 10.3389/fphys.2018.00624
Peng, Y., Wang, K., Zhu, G., Han, Q., Chen, J., Elzaki, M. E. A., et al. (2020b). Identification and characterization of multiple dsRNases from a lepidopteran insect, the tobacco cutworm, Spodoptera litura (Lepidoptera: Noctuidae). Pestic. Biochem. Physiol. 162, 86–95. doi: 10.1016/j.pestbp.2019.09.011
Petek, M., Coll, A., Ferenc, R., Razinger, J., and Gruden, K. (2020). Validating the potential of double-stranded RNA targeting Colorado potato beetle mesh gene in laboratory and field trials. Front. Plant Sci. 11:1250. doi: 10.3389/fpls.2020.01250
Powell, M., Pyati, P., Cao, M., Bell, H., Gatehouse, J. A., and Fitches, E. (2017). Insecticidal effects of dsRNA targeting the Diap1 gene in dipteran pests. Sci. Rep. 7:15147. doi: 10.1038/s41598-017-15534-y
Powell, M. E., Bradish, H. M., Gatehouse, J. A., and Fitches, E. C. (2017). Systemic RNAi in the small hive beetle Aethina tumida Murray (Coleoptera: Nitidulidae), a serious pest of the European honey bee Apis mellifera. Pest Manag. Sci. 73, 53–63. doi: 10.1002/ps.4365
Prentice, K., Smagghe, G., Gheysen, G., and Christiaens, O. (2019). Nuclease activity decreases the RNAi response in the sweetpotato weevil Cylas puncticollis. Insect Biochem. Mol. Biol. 110, 80–89. doi: 10.1016/j.ibmb.2019.04.001
Rodrigues, T. B., Duan, J. J., Palli, S. R., and Rieske, L. K. (2018). Identification of highly effective target genes for RNAi-mediated control of emerald ash borer, Agrilus planipennis. Sci. Rep. 8:5020. doi: 10.1038/s41598-018-23216-6
Rodrigues, T. B., Rieske, L. K. J, Duan, J., Mogilicherla, K., and Palli, S. R. (2017). Development of RNAi method for screening candidate genes to control emerald ash borer, Agrilus planipennis. Sci. Rep. 7:7379. doi: 10.1038/s41598-017-07605-x
Salvador, R., Niz, J. M., Nakaya, P. A., Pedarros, A., and Hopp, H. E. (2021). Midgut genes knockdown by oral dsRNA administration produces a lethal effect on cotton boll weevil. Neotrop. Entomol. 50, 121–128. doi: 10.1007/s13744-020-00819-1
Shang, F., Niu, J.-Z., Ding, B.-Y., Zhang, Q., Ye, C., Zhang, W., et al. (2018). Vitellogenin and its receptor play essential roles in the development and reproduction of the brown citrus aphid, Aphis (Toxoptera) citricidus. Insect Mol. Biol. 27, 221–233. doi: 10.1111/imb.12366
Sharma, R., Taning, C. N. T., Smagghe, G., and Christiaens, O. (2021). Silencing of double-stranded ribonuclease improves oral RNAi efficacy in southern green stinkbug Nezara viridula. Insects 12:115. doi: 10.3390/insects12020115
Singh, I. K., Singh, S., Mogilicherla, K., Shukla, J. N., and Palli, S. R. (2017). Comparative analysis of double-stranded RNA degradation and processing in insects. Sci. Rep. 7:17059. doi: 10.1038/s41598-017-17134-2
Song, H., Zhang, J., Li, D., Cooper, A. M. W., Silver, K., Li, T., et al. (2017). A double-stranded RNA degrading enzyme reduces the efficiency of oral RNA interference in migratory locust. Insect Biochem. Mol. Biol. 86, 68–80. doi: 10.1016/j.ibmb.2017.05.008
Spit, J., Philips, A., Wynant, N., Santos, D., Plaetinck, G., and Vanden Broeck, J. (2017). Knockdown of nuclease activity in the gut enhances RNAi efficiency in the Colorado potato beetle, Leptinotarsa decemlineata, but not in the desert locust, Schistocerca gregaria. Insect Biochem. Mol. Biol. 81, 103–116. doi: 10.1016/j.ibmb.2017.01.004
Tayler, A., Heschuk, D., Giesbrecht, D., Park, J. Y., and Whyard, S. (2019). Efficiency of RNA interference is improved by knockdown of dsRNA nucleases in tephritid fruit flies. Open Biol. 9:190198. doi: 10.1098/rsob.190198
Vandermoten, S., Mescher, M. C., Francis, F., Haubruge, E., and Verheggen, F. J. (2012). Aphid alarm pheromone: an overview of current knowledge on biosynthesis and functions. Insect Biochem. Mol. Biol. 42, 155–163. doi: 10.1016/j.ibmb.2011.11.008
Will, T., and Vilcinskas, A. (2015). The structural sheath protein of aphids is required for phloem feeding. Insect Biochem. Mol. Biol. 57, 34–40. doi: 10.1016/j.ibmb.2014.12.005
Willow, J., Soonvald, L., Sulg, S., Kaasik, R., Silva, A. I., Taning, C. N. T., et al. (2020). First evidence of bud feeding-induced RNAi in a crop pest via exogenous application of dsRNA. Insects 11:769. doi: 10.3390/insects11110769
Willow, J., Soonvald, L., Sulg, S., Kaasik, R., Silva, A. I., Taning, C. N. T., et al. (2021). RNAi efficacy is enhanced by chronic dsRNA feeding in pollen beetle. Commun. Biol. 4, 1–8. doi: 10.1038/s42003-021-01975-9
Wynant, N., Santos, D., Verdonck, R., Spit, J., Van Wielendaele, P., and Vanden Broeck, J. (2014). Identification, functional characterization and phylogenetic analysis of double stranded RNA degrading enzymes present in the gut of the desert locust, Schistocerca gregaria. Insect Biochem. Mol. Biol. 46, 1–8. doi: 10.1016/j.ibmb.2013.12.008
Xia, J., Guo, Z., Yang, Z., Han, H., Wang, S., Xu, H., et al. (2021). Whitefly hijacks a plant detoxification gene that neutralizes plant toxins. Cell 184, 1693–1705.e17. doi: 10.1016/j.cell.2021.02.014
Xu, L., Zhao, C.-Q., Xu, D.-J., Xu, G.-C., Xu, X.-L., Han, Z.-J., et al. (2017). RNAi suppression of nuclear receptor genes results in increased susceptibility to sulfoxaflor in brown planthopper, Nilaparvata lugens. J. Asia-Pac. Entomol. 20, 645–653. doi: 10.1016/j.aspen.2017.03.022
Yoon, J.-S., Ahn, S.-J., Flinn, C. M., and Choi, M.-Y. (2021). Identification and functional analysis of dsRNases in spotted-wing drosophila, Drosophila suzukii. Arch. Insect Biochem. Physiol. 107:e21822. doi: 10.1002/arch.21822
Yoon, J.-S., Koo, J., George, S., and Palli, S. R. (2020). Evaluation of inhibitor of apoptosis genes as targets for RNAi-mediated control of insect pests. Arch. Insect Biochem. Physiol. 104:e21689. doi: 10.1002/arch.21689
Keywords: RNA interference, molecular targets, integrated pest management, crop protection, agronomy, gene silencing, biopesticide, insecticide
Citation: Willow J, Taning CNT, Cook SM, Sulg S, Silva AI, Smagghe G and Veromann E (2021) RNAi Targets in Agricultural Pest Insects: Advancements, Knowledge Gaps, and IPM. Front. Agron. 3:794312. doi: 10.3389/fagro.2021.794312
Received: 13 October 2021; Accepted: 19 November 2021;
Published: 10 December 2021.
Edited by:
Murray B. Isman, University of British Columbia, CanadaReviewed by:
Barbara Manachini, University of Palermo, ItalyCopyright © 2021 Willow, Taning, Cook, Sulg, Silva, Smagghe and Veromann. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jonathan Willow, am9uYXRoYW5AZW11LmVl
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.