- 1Department of Agricultural Biology, Colorado State University, Fort Collins, CO, United States
- 2Department of Microbiology, Immunology, and Pathology, College of Veterinary Medicine and Biomedical Sciences, Colorado State University, Fort Collins, CO, United States
Hemp (Cannabis sativa L.) production has increased significantly in recent years; however, the crop has been understudied in the U.S. since its production declined in the late 1950s. Disease identification and management is an increasing challenge for hemp growers across the country. In 2019, beet curly top virus (BCTV) was first reported in hemp in Colorado. Hence, we were motivated to understand the diversity and prevalence of BCTV strains infecting hemp in Colorado. We detected BCTV at high incidence rate (81%) in leaf samples from 12 counties. Two different strains of BCTV, Worland (Wor) and Colorado (CO) were present as a single or mixed infection in hemp leaf samples. Phylogenetic analysis revealed BCTV sequences from hemp formed a distinct group along with BCTV strains CO and Wor. To determine other potential viral and viroid pathogens in hemp, we performed next generation sequencing (NGS). Virome analysis revealed the presence of both virus and viroid sequences that had high nucleotide sequence identity with GenBank accessions for cannabis cryptic virus, cannabis sativa mitovirus, citrus yellow vein associated virus, opuntia-like virus and hop latent viroid. In contrast, tobacco streak virus sequences were highly variable compared to sequences in GenBank suggesting a possible new genotype of this virus. The data presented here has important implications for the epidemiology and management of the various diseases of hemp and will lead to the development of integrated pest management strategies designed to interrupt transmission cycles and facilitate efficient crop production.
Introduction
Hemp (Cannabis sativa L.) is a multifaceted crop, sourcing communities with food, fiber and medicinal properties (Schluttenhofer and Yuan, 2017). With the passage of the 2014 Farm Bill, hemp is no longer a controlled substance and is now a legal agricultural crop in the U.S. Section 7606 (Legitimacy of Industrial Hemp Research) provided a formal definition of the crop as “the plant Cannabis sativa L. and any part of such plant, whether growing or not, with a delta-9 tetrahydrocannabinol concentration of not more than 0.3% on a dry weight basis.” Currently, 46 U.S. states have passed laws to define the crop and remove barriers to its production. In 2020, hemp was produced on 336,655 acres with 13,475 grower licenses across 34 states, and 46 states with active hemp program and 41 tribes with approved USDA plans, according to “U.S. Hemp Report,” by the organization Vote Hemp (www.votehemp.com). This is a more than 300% increase since 2018 licensed acreage. Colorado, the leading state in hemp production in the new era, increased production from 4,873 licensed acres in 2017 to 36,225 licensed acres in 2020. With the current resurgence of hemp as a crop to be produced within the U.S. there are many challenges with associated pests and diseases that are essentially undescribed, as are the management strategies. Disease identification and management is an increasing challenge for hemp farmers across the country. As production increases, the crop diversifies, and in turn legitimizes, the emergence of viral diseases and their spread is imminent (Fike, 2016).
There are over 100 pathogens that affect hemp with potential to cause economic damage. These include fungal, bacterial, viral, and nematode species that affect hemp during production (Mcpartland, 1994, 1996, 1999; Mcpartland and Cubeta, 1997; Mcpartland et al., 2000). A recent review summarizes important pathogens affecting the cannabis and industrial hemp production in North America (Punja, 2021). The earliest reports of viral syndromes affecting hemp were described as hemp streak virus (HSV) (Röder, 1941) and hemp mosaic virus (HMV) described in 1958 (Ceapoiu, 1958); however, the causal agents of these diseases have yet to be isolated and characterized. Other viruses that are known to infect hemp are alfalfa mosaic virus (AMV), cucumber mosaic virus (CMV), potato virus X (PVX), tomato ringspot virus (TomRSV), potato virus Y (PVY), broad bean wilt virus (BBWV), arabis mosaic virus (ArMV), and raspberry ringspot virus (RpRSV) (Kegler and Spaar, 1997). More recently, cannabis cryptic virus (CanCV) was isolated from hemp plants (Ziegler et al., 2012; Righetti et al., 2018). In addition, hemp was found to be infected with hop latent viroid (HLVd) in California (Bektaş et al., 2019; Warren et al., 2019). A survey of cannabis farms in Israel detected the presence of lettuce chlorosis virus (LCV) in plants showing leaf yellowing, interveinal chlorosis, that are typically associated with general nutrient deficiency (Hadad et al., 2019). Diagnosis of hemp viruses is challenging due to the lack of research that characterizes symptomology and transmission mechanisms of known viruses and novel viruses. Next generation sequencing (NGS) technologies circumvent many of these problems and allows for the characterization of complete genomes from known or novel viruses (Villamor et al., 2019). Indeed a diversity of virus and viroid communities were identified in hemp that were previously unknown using NGS (Nachappa et al., 2020).
In 2019, beet curly top virus (BCTV) was found infecting hemp plants in Colorado (Giladi et al., 2020) and the virus was also detected in Arizona (Hu et al., 2021). Beet curly top virus is a type member of the genus Curtovirus in the family Geminiviridae (Chen and Gilbertson, 2016). Curly top disease is one of the most economically-important diseases for sugar beet production in the western United States; in addition, BCTV has resulted in yield losses in vegetable crops such as tomato, pepper, spinach, cucurbits and common bean (Chen and Gilbertson, 2016). The virus is transmitted only by the beet leafhopper, Circulifer tenellus Baker (Hemiptera: Cicadellidae) in a circulative non-propagative manner (Bennett, 1971) (reviewed in Chen and Gilbertson, 2016). There are 11 strains of BCTV [California/Logan (CA/Logan), Colorado (CO), Kimberly1 (Kim1), Leafhopper 71 (LH71), Mild (Mld), Pepper curly top (PeCT), Pepper yellow dwarf (PeYD), Severe (Svr), Severe pepper (SvrPep), Spinach curly top (SpCT), and Worland (Wor)] and recent isolates sequenced from sugar beet and tomato added the Kim1 and LH71 strains (reviewed in Chen and Gilbertson, 2016; Strausbaugh et al., 2017). The occurrence of the various strains of BCTV varies over time by geographic region and are often observed as co-infections (Strausbaugh et al., 2017; Creamer, 2020).
The objective of this study is to understand diversity and distribution of BCTV, and to analyze the presence of established and emerging virus/viroid communities in hemp in Colorado using NGS analysis. The information obtained in this study will aid in the development of accurate detection methods and effective virus and vector management strategies to minimize disease incidence and spread.
Materials and Methods
Hemp Leaf Tissue and Insect Sample Collection
A total of 78 symptomatic hemp leaf tissue samples were harvested from individual hemp plants from various hemp growers' fields in Delta County, Colorado throughout the 2019 growing season between July and September. In addition, 44 symptomatic leaf tissues were obtained from samples sent to the Plant Diagnostic Clinic at Colorado State University for diagnosis by hemp growers from 12 counties. Approximately 100 mg of leaf tissue was harvested and stored in a 2 mL microcentrifuge tube and placed in −20°C until DNA extraction. To test potential insect vectors of BCTV, leafhopper species were collected in Delta County, Colorado from weeds and surrounding vegetation of several BCTV-infected hemp fields. Approximately 20–30 adult insects were collected using sweep nets. Insects were stored in 90% alcohol and submitted to Dr. Chris Dietrich at the University of Illinois Urbana-Champaign for species identification. Additionally, groups of 3–5 leafhoppers of each species were stored in a 2 mL microcentrifuge tube and placed in −20°C until DNA extraction.
DNA Extraction and PCR Analysis
Plant DNA extraction was performed following the manufacturer's recommendations using the DNeasy Plant Mini Kit (Qiagen Inc., Valencia, CA). All samples were eluted in 100 μL purified water for subsequent PCR analysis. Insect DNA extraction was performed following the manufacturer's protocol using the DNeasy Blood & Tissue Kit (Qiagen). Leafhoppers were separated by species with 3–5 insects per tube and ground using Tissuelyser (Qiagen). All samples were eluted in 100 μL RNase-free water for subsequent PCR analysis. The quantity of leaf and insect DNA was determined using a NanoDrop One spectrophotometer (ThermoFisher Scientific, Waltham, MA) and stored at −20°C until virus detection.
To detect BCTV in hemp leaf tissues and leafhoppers, samples were analyzed by PCR using universal BCTV primers BCTV2-F and BCTV2-R that amplify a 496-bp fragment of the coat protein (CP), a region that is conserved among BCTV strains (Strausbaugh et al., 2017) with GoTaq® Flexi DNA polymerase (Promega, Madison, WI). The amplification cycle consisted of 94°C initial denature for 5 min, 25 cycles of denaturation at 94°C for 1 min, 58°C annealing for 2 min, and 72°C extension for 2 min, followed by a 10-min final extension. All PCR products were visualized on 1% agarose gel. Three different strain-specific primer pairs targeting BCTV-Wor, BCTV-CO, and BCTV-Svr were used and are listed in Table 1. The PCR products were excised from the agarose gels and purified using DNA Clean and Concentrator™-5 (Zymo Research, Irvine, CA). One -two PCR products from each location were randomly selected and submitted for Sanger sequencing at Genewiz Inc. to confirm the virus identity using strain-specific primers (Table 1). The sequences for each BCTV strain were checked for identity against the non-redundant (nr) database using blastn in the NCBI database.
Hemp Leaf Tissue Collection for Virome Analysis
Total RNA was extracted from a composite of five leaves that previously tested positive for BCTV from several locations in Colorado. The samples originated from outdoor hemp production in Delta, Pueblo, Boulder, Rio Blanco and Conejos counties and one indoor production in Larimer County in Colorado. Total RNA was extracted as described above and checked for quantity using a Nanodrop One spectrophotometer (ThermoFisher Scientific) and quality using a Qubit 3.0 fluorometer (ThermoFisher Scientific). Approximately 2 μg of RNA was submitted to the CSU Next Generation Sequencing Facility, where library preparation, quality measurements, and sequencing was performed. Briefly, RNA quality was confirmed using an Agilent Tapestation instrument. Shotgun RNA libraries were constructed using the Kapa Biosystems RNA HyperPrep kit (Roche, IN, USA) according to the manufacturer's instructions. Pooled libraries were sequenced on an Illumina NextSeq 500 instrument to produce single-end 150 nucleotide (nt) reads.
To confirm the presence of low percentage nucleotide (nt) identity viruses (<90%), 1 μg of total RNA was used to synthesize cDNA using Verso cDNA synthesis kit (ThermoFisher Scientific) according to manufacturer's instructions. The RT-PCR was performed using GoTaq® Flexi DNA polymerase (Promega, Madison, WI) with virus-specific primers (Supplementary Table 2). The amplification cycle consisted of 2 min at 95°C, 40 cycles of 30 s at 95°C, 30 s at 55°C and 35 s at 72°C followed by 5 min at 72°C for all viruses except for citrus yellow vein associated virus (CYVaV) which had Tm of 51°C.
Bioinformatic Analyses
Virus and virus-like sequences were identified as previously described (Cross et al., 2018). Analysis scripts are available at https://github.com/stenglein-lab/taxonomy_pipeline. Low quality and adapter sequences were removed using cutadapt software (Martin, 2011) and duplicate reads were collapsed with cd-hit (Li and Godzik, 2006). Host (hemp)-derived reads were removed by bowtie2 alignment (Langmead and Salzberg, 2012) to the hemp reference genome (assembly accession GCF_900626175.1). Remaining non-host reads were assembled into contigs using the Spades assembler (Bankevich et al., 2012). Contigs and non-assembling reads were taxonomically categorized first by nucleotide-level alignment to the NCBI nucleotide (nt) database using blastn, and then by protein-level alignment to the NCBI protein (nr) database using the diamond aligner (Altschul et al., 1990; Buchfink et al., 2015). This produced a comprehensive classification of all non-host reads. Although we focused on viruses, this also constitute a valuable dataset about the entire hemp-associated microbiota (bacteria, fungi, etc.) for future use by us and others. Candidate virus sequences were manually validated by aligning reads to draft genome sequences using bowtie2. Lastly, the raw sequence data was deposited in the NCBI Sequence Read Archive (SRA) repository under NCBI BioProject accession PRJNA762365.
Phylogenetic Analysis
The BCTV-CP sequences from this study were aligned with sequences of BCTV strains categorized by Strausbaugh et al. (2017) and BCTV sequences from hemp deposited in the GenBank database using MUSCLE program (Edgar, 2004) embedded in MEGA X: Molecular Evolutionary Genetics Analysis (Kumar et al., 2018). The evolutionary history was inferred by using the Maximum Likelihood method and Tamura-Nei model (Tamura and Nei, 1993) with 1,000 pseudo-replicates to obtain bootstrap values at each evolutionary node. The complete and partial genomes of virus and viroids from the hemp virome analysis were aligned with corresponding matches in the GenBank database using the same approach to construct phylogenetic trees.
Results
Beet Curly Top Virus Diversity and Distribution in Hemp
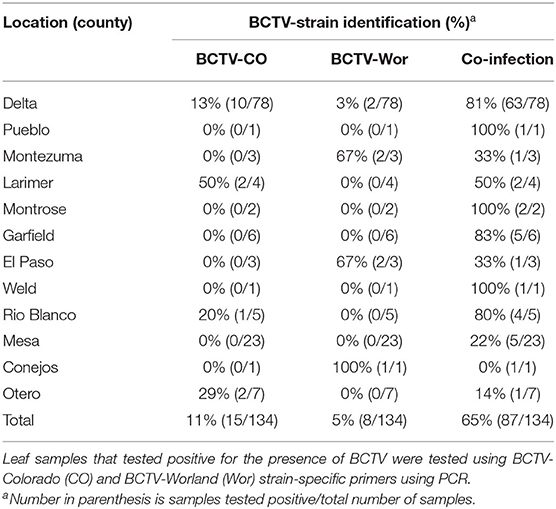
In 2019, we analyzed 134 symptomatic hemp tissue samples from different stages (vegetative and reproductive) of hemp plants from outdoor productions in 12 different counties spanning the predominant hemp-growing regions in Colorado (Figure 1). PCR analyses confirmed BCTV in most hemp leaf samples with 81% (109/134) incidence using the BCTV-universal primer set. Among the samples that tested positive for BCTV, incidence of BCTV-CO was 11% (15/135), BCTV-Wor was 5% (7/134) and coinfection of BCTV-Wor and BCTV-CO was 65% (86/134). In contrast, we did not detect the BCTV-Svr in any of the samples (Table 2). The BCTV-positive plants exhibited various virus symptoms in hemp including yellowing, mottling, curled leaves and stunting (Nachappa et al., 2020). All symptoms observed in hemp were found to be associated with BCTV infection (Supplementary Table 1).

Figure 1. Incidence of beet curly top virus (BCTV) in hemp leaf samples collected during the 2019 field season in Colorado as determined by PCR analysis. Incidence was calculated as number of BCTV-positive hemp leaf tissue samples over total samples tested per county listed in Table 2.
Phylogenetic analysis based on a fragment of the BCTV coat protein (CP) sequences from 20 hemp samples from various locations in Colorado (accessions: MW604759-MW604777) had nt identity to one another between 98.99 and 98.24%. Sequences from this study had 97 to 99% sequence identity with cannabis BCTV sequences from Colorado (isolate BCTV-Can; MK803280) and Arizona (isolate BCTV-Can-AZ; MW182244). The BCTV sequences from the current study had high nt identity with BCTV-Wor strains (97–99%) and lower nt identity with BCTV-CO strains (93–95%) in GenBank. Phylogenetic analysis revealed that BCTV sequences from hemp form a distinct group including both BCTV-Wor and BCTV-CO sequences (Figure 2).
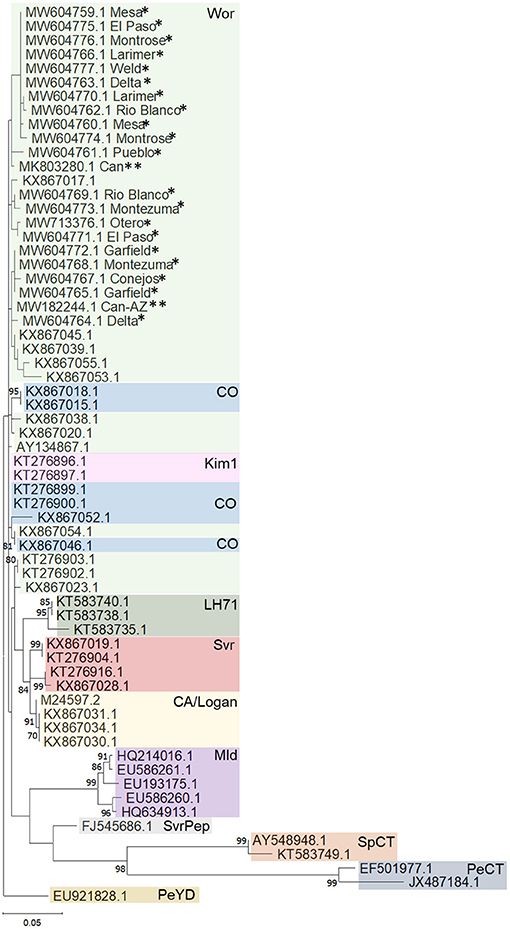
Figure 2. Phylogenetic analysis of partial coat protein sequences of beet curly top virus (BCTV) obtained from hemp samples collected during the 2019 field season in Colorado and other BCTV sequences available in GenBank. Phylogenetic analysis by Maximum Likelihood method was done using MUSCLE in MEGAX. Single asterisks indicate genotypes from Colorado from this study, double asterisk indicate accessions from previously published BCTV-Cannabis genotypes. Bootstrap values are indicated on the nodes. Bootstrap values <70% out of 1,000 replicates are not shown. Scale bar indicates number of substitutions per site. CA/Logan, BCTV strains California/Logan; CO, Colorado; Kim1, Kimberly 1; Mld, Mild; LH71, Leafhopper 71; PeCT, Pepper curly top; PeYD, Pepper yellow dwarf; Svr, Severe; SvrPep, Severe pepper; SpCT, Spinach curly top; Wor, Worland.
Leafhopper species that were collected from weed species around hemp fields in Delta County in Colorado included: C. tenellus, Empoasca sp., Balclutha neglecta, Macrosteles quadrilineatus, Exitianus exitiosus, and Ceratagallia uhleri. Of the six leafhopper species collected, BCTV was only detected in C. tenellus. All nine pooled C. tenellus samples comprising of 3–5 insects per sample were positive for BCTV with 56% (5/9) positive for just BCTV-CO, while the other four samples were coinfected with BCTV-CO and BCTV-Wor 44% (4/9).
Hemp Virome Analysis
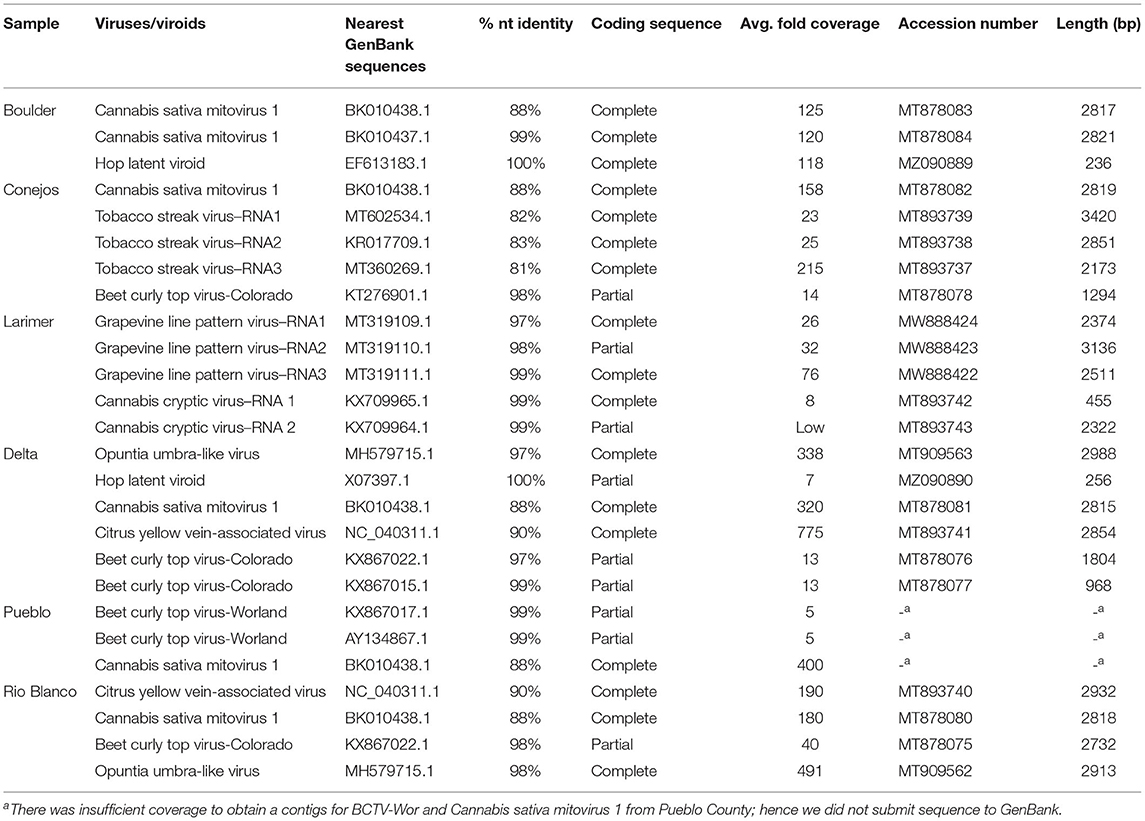
We further explored the hemp virome using NGS of total RNA from leaf samples from Boulder, Conejos, Delta, Pueblo and Rio Blanco counties that were part of the field samples collected in 2019, and leaf samples from indoor hemp cultivation from Larimer County. Datasets contained an average of 12.4 ×106 reads. After removal of low quality and adapter sequences, there was an average of average of 11.6 ×106 sequences remaining per library (94%). Duplicate reads were collapsed leaving an average of 1.4 ×106 unique reads per dataset (12%). Removal of host-derived reads left an average of 7.8 ×104 reads per sample (0.6%) (Supplementary Table 2). Next generation sequence analysis revealed the presence of 7 viruses and one viroid in hemp samples from Colorado (Table 3). We assembled complete or nearly complete genomes of several of the viruses using NGS data (Table 3).
Cannabis sativa mitovirus 1 (CasaMV1) was detected in hemp from all counties with the exception of Larimer County, which was from indoor hemp cultivation (Table 3). Alignment of the complete RNA-dependent RNA polymerase (RdRp) gene of CasaMV1 sequences (MT878080-MT878083) showed high nt similarity with each other (99.89–100%). Four out of five CasaMV sequences had an 88% nt identity to CasaMV1 sequenced from C. sativa (BK010438.1) and one of the sequences from Boulder County had a 99.72% identity match to a CasaMV isolate, also recovered from C. sativa (BK010437.1) (Table 3).
Four of the six counties had BCTV-CO and BCTV-Wor strains (MT878075-MT878078) present which had 97 to 99% nt sequence identity. The sample from Conejos County (MT878078) had 98% nt sequence identity with both a BCTV Cannabis isolate (BCTV-Can-AZ; MW182244) and a sugar beet isolate (CTS14-024; KT276901) determined to be a BCTV-CO strain. The sequences from Delta and Rio Blanco counties were similar to BCTV-CO with nt identity of 96–98% and the sequences from Pueblo matched BCTV-Wor with nt identity of 99% (Table 3). There was insufficient coverage to recover a coding complete assembly for the BCTV-Wor sequences from Pueblo.
Citrus yellow vein-associated virus (CYVaV) was detected in hemp from two counties (Delta and Rio Blanco) and these two sequences (MT893740 and MT893741) were partial sequences of the RdRP of CYVaV gene and shared of 98% nt identity with each other. The sequences from this study had an 90% nt identity with CYVaV identified from citrus (NC_040311) (Figure 3). Opuntia umbra-like viruses (OULV) were also detected in these two locations (MT909563 and MT909562), sharing nt identity of 98% with each other, and a top blastn hit of OULV detected in the barberry fig with 98% nt identity (MH579715). Opuntia umbra-like virus and CYVaV sequences share a 77% nt identity, coming up in one another's NCBI blast searches with shared similarity of the RdRp gene. Citrus yellow vein-associated virus and OULV formed separate groups in phylogenetic analysis (Figure 3).
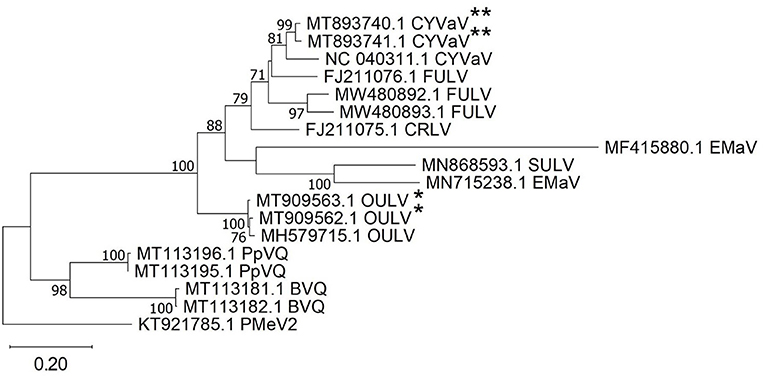
Figure 3. Phylogenetic analysis of RNA-dependent RNA polymerase gene sequences of citrus yellow vein-associated virus (CYVaV) and opuntia umbra-like virus (OULV) were obtained from hemp samples collected during the 2019 field season in Colorado along with all top sequence matches of both detected viruses available in GenBank. Phylogenetic analysis by Maximum Likelihood method was done MUSCLE in MEGAX. Double asterisks indicate CYVaV and single asterisks indicate OULV genotypes from Colorado. Bootstrap values are indicated on the nodes. Scale bar indicates number of substitutions per site. Virus name abbreviations: BVQ, babaco virus Q; CRLV, carrot red leaf luteovirus; FULV, fig umbra-like virus; EMaV, Ethiopia maize-associated virus; PpVQ, papaya virus Q; PMeV2, papaya meleira virus2; SULV, sugarcane umbra-like virus.
Tobacco streak virus (TSV) was detected in hemp virome dataset from Conejos County encoding all three RNAs (MT893737-MT893739). The TSV RNA 1 sequence from the current study (MT893739) had 82% nt identity to a fragment of the replicase gene of TSV sequences from soybean (MT602534). The TSV RNA 2 sequence (MT893738), had 83% nt identity match with the gp1 putative viral polymerase in TSV from Dahlia pinnata (KR017709.1). Lastly, TSV RNA 3 (MT893737) had an 81% identity match to TSV RNA 3 from soybean from Brazil (MT360269.1) which encodes the movement and coat protein gene. These TSV RNA sequences from the current study range from 81 to 83% similar to previously observed TSV detected from various host plants, indicating this as a possible novel genotype of TSV (Figure 4).
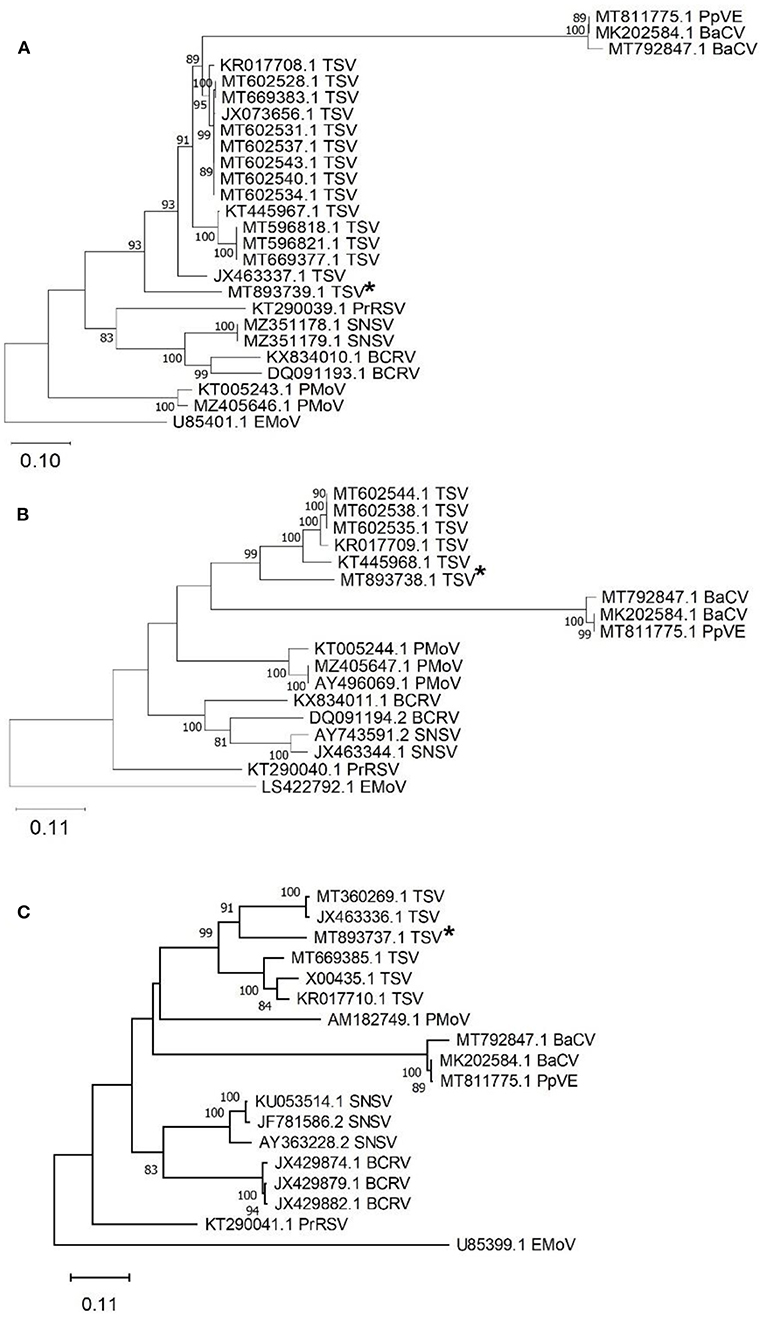
Figure 4. Phylogenetic analysis of tobacco streak virus (TSV) RNA 1-3 obtained from hemp samples collected during the 2019 field season in Colorado and other TSV sequences available in GenBank. (A) TSV RNA 1 encodes the replicase protein gene with elm mottle virus (EMoV) as an outgroup, (B) TSV RNA 2 encodes RNA-dependent RNA polymerase gene with elm mottle virus (EMoV) as an outgroup, and (C) TSV RNA3 encodes the movement protein with parietaria mottle virus (PMoV) as an outgroup. Tobacco streak virus sequences from this study indicated with an asterisk. Phylogenetic analysis by Maximum Likelihood method was done using MUSCLE in MEGAX. Asterisks indicate genotypes from Colorado from this study. Bootstrap values are indicated on the nodes. Bootstrap values <70% out of 1,000 replicates are not shown. Scale bar indicates number of substitutions per site. Virus name abbreviations: BaCV, bean-associated cytorhabdovirus; BCRV, blackberry chlorotic ringspot virus; EMoV, elm mottle virus; PMoV, parietaria mottle virus; PrRSV, privet ringspot virus; PpVE, papaya virus E; SNSV, strawberry necrotic shock virus.
Cannabis cryptic virus (CanCV) corresponding to RNA segments 1 and 2 were assembled (MT893742 and MT893743) from the Larimer County sample. The CanCV RNA 1 (MT893742) sequence matched a partial region of the RdRP gene and shared a 99% nt identity with CanCV RdRp sequences found in C. sativa from Italy and Germany (KX709964 and JN196536). The CanCV RNA 2 segment (MT893743) matched the partial sequence of the CP gene and shared 95–99% nt identity with sequences corresponding to MT893743 and MT893742 sequences from this study.
Grape line pattern virus (GLPV) was detected from Larimer County in three RNA segments. GLPV RNA1 segment (MW888424) had the highest nt identity of 97% to the methyltransferase from a GLPV isolate from grapevine in Hungary (MT319109), followed by the replication protein of Hop yellow virus with a 95% nt match from hops in China (MG727388). GLVP RNA2 segment (MW888423) matched the of GLPV with a nt match of 98% (MT319110) coding for the RdRp gene. GLVP RNA3 (MW888422) had a nt match of 99% to the movement protein of GLPV (MT319111).
Two complete genomes of Hop latent viroid (HLVd) were assembled from Boulder and Delta counties (MZ090889 and MZ090890). The HLVd detected from Delta County (MZ090890) had a 100% shared nt identity to X07397.1, a sequence submitted to GenBank from hops. The second HLVd sequence (MZ090889) from Boulder County, shared a 100% nt identity with HLVd collected from hops growing in a commercial garden in China (EF613183.1).
The majority of the viruses identified through NGS had a high % nt identity (>90%; Table 3). Only three of the viruses, CasaMV1, TSV, and CYVaV were under this threshold. The presence of these low % nt identity (<90%) viruses was confirmed in the respective sample/county with RT-PCR analysis (Supplementary Figure 1). Interestingly, there was a smaller PCR product for CasaMV1 in Boulder County than expected. This is likely due to Boulder County sample having two distinct CasaMV1 isolates (MT878083 and MT878084; Table 3).
Discussion
One of biggest challenges facing the hemp industry is detection and management of viral diseases (Nachappa et al., 2020; Punja, 2021). As hemp acreage increases across the U.S., across the U.S., additional viruses and viroids will likely be found in hemp that can potentially impact the crop, but also serve as a reservoir for disease spread to other economically important crops in the vicinity. In the current study, we assessed the diversity and distribution of BCTV affecting hemp in Colorado using PCR analysis. As a further attempt to identify previously known viruses/viroids in hemp, we also performed NGS analysis.
A survey of symptomatic hemp tissue samples from different stages from outdoor productions across 12 counties revealed high incidence (81%) of BCTV in Colorado. Growers reported disease incidence levels above 90% in some cases and disease severity reaching >50% of leaf area in infected plants. This led to diminished crop yield, including low quality of the flower and overall stunted growth (John House, hemp grower personal communication). The symptoms observed were variable among plants, and disease progression differed across farms (Nachappa et al., 2020). Given the broad host range of the virus and the vector (reviewed in Chen and Gilbertson, 2016), it is possible that curly top disease may become of the most serious disease affecting hemp production. There are 11 BCTV strains and the presence of the strains in a particular location may change over time and are often found as mixed infections (Strausbaugh et al., 2017; Creamer, 2020). For instance, a survey of BCTV strains from Idaho and Oregon showed a reduction in the prevalence of the Svr strain and the number of mixed infections from 2006–2007 (87% Svr and 60% Wor-like; 59% mixed infections) to 2012–2015 (2% Svr and 87% Wor-like; 16% mixed infections) (Strausbaugh et al., 2017). During this time frame a significant shift in crop management occurred when neonicotinoid seed treatments became labeled for use on sugar beet in 2008. Thus, beet leafhoppers coming from the desert in the spring were likely limited in their ability to feed on sugar beet (influence of neonicotinoids lasts at least 77 days) and had to survive on alternate host plants such as common bean and weeds. These alternate hosts may have favored the Wor and CO strains over Svr. Another possibility might have been the addition of strain specific resistance to commercial sugar beet cultivars but the combination of genes in these cultivars is not publicly known. In addition, new strains can emerge likely due to recombination among strains during mixed infections (Strausbaugh et al., 2008; Chen et al., 2010; Bach and Jeske, 2014).
We identified the mild strains, BCTV-CO and BCTV-Wor present as a single or mixed infection in hemp leaf samples in Colorado using strain-specific primers. In contrast, we did not detect the BCTV-Svr in any of the samples. The DNA fragments from Sanger sequencing and whole genomes from NGS revealed high nt similarity to BCTV genotype previously from Colorado (Giladi et al., 2020) and Arizona (Hu et al., 2021). Reports suggest that mild strains (strains that produce mild symptoms such as slight leaf curling, stunting, and vein thickening) are more effective in infecting alternative and weed hosts than severe strains (Chen and Gilbertson, 2009). All the beet leafhoppers collected in the current study also carried BCTV-Wor and CO strains. This complements the findings that CO and Wor are the predominant strains in hemp. The beet leafhopper transmits BCTV in a circulative manner (Bennett, 1971). The leafhopper acquires the virus in as little as 1–2 min, but maximum accumulation occurs after acquisition access period (AAP) of 24–48 h (Soto and Gilbertson, 2003). There is a 4-h latent period before the insect can transmit the virus. The longer the AAP the higher the rate of transmission. The virus does not replicate in the insects (Soto and Gilbertson, 2003). Leafhoppers can transmit the virus by feeding for 15 min and the virus is retained for days to weeks, but there is no transstadial transmission (Bennett, 1971).
There is limited information about curly top epidemiology in Colorado. Beet leafhoppers are most found in the Western Slope of Colorado where outbreaks of this disease have been reported on many crops such as tomato, bean, squash, sugar beets, spinach (Cranshaw, 2020). It is thought that leafhoppers migrate from their overwintering sites in southern states including Arizona and New Mexico to Colorado in spring. The insect survives the winter on various kinds of weedy plants, particularly mustard-family (Bennett, 1971). Hence, the abundance of winter host plants in the southern breeding areas could be an important factor in the number of beet leafhoppers that appear in Colorado in spring. We hypothesize that in winter 2019 moisture conditions were very favorable in the southern breeding areas to support the large population of overwintering plants on which beet leafhopper could develop resulting in large number of migrants that moved into Colorado causing an outbreak in hemp, which is one of the most widely grown crops in the Western slope.
Virome analysis revealed a diversity of viruses and one viroid pathogen infecting hemp in Colorado. The number of identified viruses and viroid in each location ranged from 2 to 5. Beet curly top virus, CasaMV1, and HLVd were commonly present in several locations, whereas other viruses were unique to specific locations. Cannabis sativa mitovirus 1 was detected in hemp from all counties sampled with the exception of Larimer County, where tissues were collected from indoor hemp production. Mitoviruses are capsidless viruses known for their ability to infect eukaryotic mitochondria (Shahi et al., 2019). Several mitovirus sequences have been recovered during the transcriptome analysis of a variety of invertebrates across different phyla (Shi et al., 2016). Complete plant mitovirus genomes were recovered from publicly available transcriptome data of 10 different plant species including hemp, hops and sugar beet (Nibert et al., 2018). These viruses are generally considered to be cryptic viruses and there is no information on the impact on plant hosts.
A new genotype of TSV which is only 81–83% identical to the closest TSV sequence in GenBank was identified from hemp samples from Conejos County. Tobacco streak virus has a wide host range (Brunt et al., 1996) and is transmitted by thrips and pollen (Sdoodee and Teakle, 1988; Sharman et al., 2015) and by seed transmission (Sharman et al., 2009). The virus was first reported to infect hemp in 1971 and had described symptoms of stunting and mosaic patterning (Hartowicz et al., 1971). TSV is a non-enveloped, quasi-spherical virion with tripartite (RNA1, RNA2, and RNA3) segmented linear (+) sense RNA genome and all 3 segments were retrieved in the hemp virome dataset. Further characterization of these sequences is challenging because the leaf samples from number of plants were pooled and/or mixed infection of viruses in these plants.
Cannabis cryptic virus was only detected in the indoor hemp sample from Larimer County and had high nt sequence similarity (99%) to the RdRP gene of previously reported CanCV sequence (Righetti et al., 2018). Historically, interveinal chlorosis and leaf margin wrinkling in hemp was attributed to the so-called hemp streak virus (HSV) (Röder, 1941). In 2012, Ziegler et al. (2012) first identified CanCV accidently while using hemp as a host for a hop latent virus (Carlavirus) and found that the virus was seed-transmissible. More recently, Righetti et al. (2018) tested hemp samples with typical hemp streak syndrome and identified CanCV in all tested samples irrespective of presence and severity of symptoms. This calls into question the role of CanCV in symptomology.
Citrus yellow vein-associated virus and opuntia umbra-like virus were identified from Delta and Rio Blanco counties. Citrus yellow-vein disease (CYVD) was first reported Dr. L. G. Weathers in California in 1957 (Weathers, 1957) resulting in typical yellow vein symptoms. Recently, Kwon et al. (2021) demonstrated that CYVD is associated with a virus-like agent, tentatively named CYVaV and is transmitted via grafting to virtually all citrus varieties. The virus appears to be closely related to unclassified virus-like RNAs in the family Tombusviridae specifically OULV (Kwon et al., 2021). Indeed, phylogenetic analysis of the RdRp and ORF1-3genes placed the CYVaV and OULV from the current study were in a well-supported cluster. Umbraviruses lack a capsid protein (CP) gene, which makes them dependent on a “helper” virus, usually from the Luteoviridae, for replication, encapsidation and cell-to-cell movement (Syller, 2003). However, no helper virus was identified using NGS in this dataset. It is possible that a helper virus too distant to be recognized was missed by the analysis. Indeed, the first report of OULV from Opuntia ficus indica fruit cactus plants with symptoms of pad swelling disease was not associated with Luteoviridae (Felker et al., 2019). The authors hypothesized that as Umbraviruses occur throughout the plant, but Luteoviruses only occur in the phloem, low concentrations of Luteovirus can be expected in the sample. To our knowledge, this is the first report of CYVaV and OULV in hemp.
Hop latent viroid has been previously identified in hemp from symptomatic and asymptomatic plants (Bektaş et al., 2019) and from symptomatic plants (Warren et al., 2019). Viroids are small non-encapsulated infectious pathogens, comprised of closed single stranded RNA molecules and biological resources to drive host specificity (Flores et al., 2005). There was high nt sequence identity (99–100%) between the HLVd genomes retrieved from hemp. Typical symptoms of HLVd include stunting, malformation or chlorosis of leaves, brittle stems, and reduction in yields. Indeed, some of the samples collected from Delta County demonstrated typical HLVd symptoms.
Conclusions and Future Perspectives
Our study identified a diversity of viruses and viroids in hemp in Colorado using NGS. The number of identified viruses and viroid in each location ranged from 2 to 5. Cannabis sativa mitovirus, BCTV and HLVd were commonly present in several locations, whereas other viruses were unique to specific locations. We identified a divergent virus with 81% sequence identity to TSV. Future research should focus on surveying hemp viruses across multiple hemp genotypes, locations, and stages of the crop. Overall, outcomes of this study and future research will result in the identification of unique target sequences for the development of rapid and accurate nucleic acid-based detection tools for viruses and viroids and a rich characterization of hemp-associated pathogens. The viruses/viroids identified in the study were mechanically transmitted, seed-transmitted or insect-transmitted making management of these viruses challenging (Mcpartland et al., 2000; Nachappa et al., 2020; Punja, 2021). Planting certified disease-free materials will be critical to minimize disease spread. Future investigations should explore vector metagenomics to determine potential insect vectors of viruses/viroids associated with hemp with the goal of identifying anticipated threats and developing early prevention tactics. Once the insect vector community is identified, a deeper exploration of timing of insect flights, reproduction and feeding behaviors can be targeted for pest management and interruption of transmission cycles. Lastly, identifying potential sources of resistance to insect pests and viruses is an especially important disease control strategy, as commercial insecticides that are effective against insect vectors in other crops are restricted on hemp.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article.
Author Contributions
PN and JC conceived and designed the experiments and wrote the original draft of the manuscript. JC, KL, AF, and WC collected samples. JC, KL, JM, and TA performed experiments and phylogenetic analyses. MK and MS performed library preparation and bioinformatic analyses. JC, JM, MS, and PN reviewed and edited the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by USDA-NIFA Critical Agricultural Research and Extension (CARE) 2020-04954 and the Foundation for Food and Agriculture Research (FFAR)-Rapid Outcomes from Agricultural Research (ROAR) program.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We would like to thank the hemp growers for sending us samples and for allowing us to survey their fields.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fagro.2021.778433/full#supplementary-material
References
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. doi: 10.1016/S0022-2836(05)80360-2
Bach, J., and Jeske, H. (2014). Defective DNAs of beet curly top virus from long-term survivor sugar beet plants. Virus Res. 183, 89–94. doi: 10.1016/j.virusres.2014.01.028
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. doi: 10.1089/cmb.2012.0021
Bektaş, A., Hardwick, K., Waterman, K., and Kristof, J. (2019). Occurrence of hop latent viroid in cannabis sativa with symptoms of cannabis stunting disease in California. Plant Dis. 103:2699. doi: 10.1094/PDIS-03-19-0459-PDN
Bennett, C. W. (1971). The Curly Top Disease of Sugarbeet and Other Plants. St. Paul, MN: A.P. Society.
Brunt, A., Crabtree, K., Dallwitz, M., Gibbs, A., and Watson, L. (1996). Viruses of Plants. Wallingford: Cab International.
Buchfink, B., Xie, C., and Huson, D. H. (2015). Fast and sensitive protein alignment using DIAMOND. Nat. Methods 12, 59–60. doi: 10.1038/nmeth.3176
Chen, L.-F., Brannigan, K., Clark, R., and Gilbertson, R. L. (2010). Characterization of curtoviruses associated with curly top disease of tomato in California and monitoring for these viruses in beet leafhoppers. Plant Dis. 94, 99–108. doi: 10.1094/PDIS-94-1-0099
Chen, L.-F., and Gilbertson, R. L. (2009). Curtovirus–Cucurbit interaction: acquisition host plays a role in leafhopper transmission in a host-dependent manner. Phytopathology 99, 101–108. doi: 10.1094/PHYTO-99-1-0101
Chen, L.-F., and Gilbertson, R. L. (2016). “CHAPTER 17: Transmission of curtoviruses (Beet curly top virus) by the beet leafhopper (Circulifer tenellus),” in Vector-Mediated Transmission of Plant Pathogens, ed J. K. Brown (St. Paul, MN: The American Phytopathological Society (APS)), 243–262.
Cranshaw, W. (2020). Beet Leafhopper and Beet Curly Top Virus [Online]. Available online at: https://agsci.colostate.edu/hempinsects/ (accessed September 18, 2021).
Creamer, R. (2020). “Beet curly top virus transmission, epidemiology, and management,” in Applied Plant Virology, ed L. P. Awasthi (Amsterdam: Elsevier), 521–527.
Cross, S. T., Kapuscinski, M. L., Perino, J., Maertens, B. L., Weger-Lucarelli, J., Ebel, G. D., et al. (2018). Co-Infection Patterns in Individual Ixodes scapularis Ticks Reveal Associations between Viral, Eukaryotic and Bacterial Microorganisms. Viruses 10:388. doi: 10.3390/v10070388
Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. doi: 10.1093/nar/gkh340
Felker, P., Bunch, R., Russo, G., Preston, K., Tine, J. A., Suter, B., et al. (2019). Biology and chemistry of an Umbravirus like 2989 bp single stranded RNA as a possible causal agent for Opuntia stunting disease (engrosamiento de cladodios)-a review. J. Prof. Assoc. Cactus Dev. 21, 1–31. Available online at: https://www.jpacd.org/jpacd/article/view/3
Fike, J. (2016). Industrial hemp: renewed opportunities for an ancient crop. CRC Crit. Rev. Plant Sci. 35, 406–424. doi: 10.1080/07352689.2016.1257842
Flores, R., Hernández, C., Alba, A. E. M. D., Daròs, J.-A., and Serio, F. D. (2005). Viroids and viroid-host interactions. Annu. Rev. Phytopathol. 43, 117–139. doi: 10.1146/annurev.phyto.43.040204.140243
Giladi, Y., Hadad, L., Luria, N., Cranshaw, W., Lachman, O., and Dumbrovsky, A. (2020). First report of beet curly top virus infecting Cannabis sativa in western Colorado. Plant Disease 104:999. doi: 10.1094/PDIS-08-19-1656-PDN
Hadad, L., Luria, N., Smith, E., Sela, N., Lachman, O., and Dombrovsky, A. (2019). Lettuce chlorosis virus disease: a new threat to cannabis production. Viruses 11:802. doi: 10.3390/v11090802
Hartowicz, L., Knutson, H., Paulsen, A., Eaton, B., and Eshbaugh, E. (1971). “Possible biocontrol of wild hemp,” in North Central Weed Control Conference, Proceedings) (Madison: University of Wisconsin), 2:69.
Hu, J., Masson, R., and Dickey, L. (2021). First report of beet curly top virus infecting industrial hemp (Cannabis sativa) in Arizona. Plant Dis. 105, 1233–1233. doi: 10.1094/PDIS-11-20-2330-PDN
Kegler, H., and Spaar, D. (1997). Zur virusanfälligkeit von hanfsorten (Cannabis sativa L.). Arch. Phytopathol. Plant Protect. 30, 457–464. doi: 10.1080/03235409709383198
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Kwon, S.-J., Bodaghi, S., Dang, T., Gadhave, K. R., Ho, T., Osman, F., et al. (2021). Complete Nucleotide Sequence, Genome Organization, and Comparative Genomic Analyses of Citrus Yellow-Vein Associated Virus (CYVaV). Front. Microbiol. 12:683130. doi: 10.3389/fmicb.2021.683130
Langmead, B., and Salzberg, S. L. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359. doi: 10.1038/nmeth.1923
Li, W., and Godzik, A. (2006). Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22, 1658–1659. doi: 10.1093/bioinformatics/btl158
Martin, M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17:13. doi: 10.14806/ej.17.1.200
Mcpartland, J. (1994). Cannabis pathogens X: phoma, ascochyta and didymella species. Mycologia 86, 870–878. doi: 10.1080/00275514.1994.12026492
Mcpartland, J., and Cubeta, M. A. (1997). New species, combinations, host associations and location records of fungi associated with hemp (Cannabis sativa). Mycol. Res. 101, 853–857. doi: 10.1017/S0953756297003584
Mcpartland, J. M. (1999). A Survey of Hemp Diseases and Pests. Advances in Hemp Research. New York, NY: Food Product Press, 109–128.
Mcpartland, J. M., Clarke, R. C., and Watson, D. P. (2000). Hemp Diseases and Pests: Management and Biological Control: An Advanced Treatise. Wallingford: CABI.
Nachappa, P., Fulladolsa, A. C., and Stenglein, M. (2020). Wild wild west: emerging viruses and viroids of hemp. Outlooks Pest Manag. 31, 175–179. doi: 10.1564/v31_aug_07
Nibert, M. L., Vong, M., Fugate, K. K., and Debat, H. J. (2018). Evidence for contemporary plant mitoviruses. Virology 518, 14–24. doi: 10.1016/j.virol.2018.02.005
Punja, Z. K. (2021). Emerging diseases of Cannabis sativa and sustainable management. Pest Manag. Sci. 77, 3857–3870. doi: 10.1002/ps.6307
Righetti, L., Paris, R., Ratti, C., Calassanzio, M., Onofri, C., Calzolari, D., et al. (2018). Not the one, but the only one: about Cannabis cryptic virus in plants showing ‘hemp streak’disease symptoms. Eur. J. Plant Pathol. 150, 575–588. doi: 10.1007/s10658-017-1301-y
Röder, K. (1941). Einige Untersuchungen über ein an hanf (Cannabis sativa L.) auftretendes virus. Faserforschung 15:77.
Schluttenhofer, C., and Yuan, L. (2017). Challenges towards revitalizing hemp: a multifaceted crop. Trends Plant Sci. 22, 917–929. doi: 10.1016/j.tplants.2017.08.004
Sdoodee, R., and Teakle, D. (1988). Seed and pollen transmission of tobacco streak virus in tomato (Lycopersicon esculentum cv. Grosse Lisse). Aust. J. Agric. Res. 39, 469–474. doi: 10.1071/AR9880469
Shahi, S., Eusebio-Cope, A., Kondo, H., Hillman, B. I., and Suzuki, N. (2019). Investigation of host range of and host defense against a mitochondrially replicating mitovirus. J. Virol. 93, e01503–e01518. doi: 10.1128/JVI.01503-18
Sharman, M., Persley, D. M., and Thomas, J. E. (2009). Distribution in Australia and seed transmission of Tobacco streak virus in Parthenium hysterophorus. Plant Dis. 93, 708–712. doi: 10.1094/PDIS-93-7-0708
Sharman, M., Thomas, J., and Persley, D. (2015). Natural host range, thrips and seed transmission of distinct Tobacco streak virus strains in Queensland, Australia. Ann. Appl. Biol. 167, 197–207. doi: 10.1111/aab.12218
Shi, M., Lin, X.-D., Tian, J.-H., Chen, L.-J., Chen, X., Li, C.-X., et al. (2016). Redefining the invertebrate RNA virosphere. Nature 540, 539–543. doi: 10.1038/nature20167
Soto, M. J., and Gilbertson, R. L. (2003). Distribution and rate of movement of the curtovirus Beet mild curly top virus (family Geminiviridae) in the beet leafhopper. Phytopathology 93, 478–484. doi: 10.1094/PHYTO.2003.93.4.478
Strausbaugh, C., Wintermantel, W., Gillen, A., and Eujayl, I. A. (2008). Curly top survey in the western United States. Phytopathology 98, 1212–1217. doi: 10.1094/PHYTO-98-11-1212
Strausbaugh, C. A., Eujayl, I. A., and Wintermantel, W. M. (2017). Beet curly top virus strains associated with sugar beet in Idaho, Oregon, and a Western US Collection. Plant Dis. 101, 1373–1382. doi: 10.1094/PDIS-03-17-0381-RE
Syller, J. (2003). Molecular and biological features of umbraviruses, the unusual plant viruses lacking genetic information for a capsid protein. Physiol. Mol. Plant Pathol. 63, 35–46. doi: 10.1016/j.pmpp.2003.08.004
Tamura, K., and Nei, M. (1993). Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 10, 512–526.
Villamor, D., Ho, T., Al Rwahnih, M., Martin, R., and Tzanetakis, I. (2019). High throughput sequencing for plant virus detection and discovery. Phytopathology 109, 716–725. doi: 10.1094/PHYTO-07-18-0257-RVW
Warren, J., Mercado, J., and Grace, D. (2019). Occurrence of hop latent viroid causing disease in Cannabis sativa in California. Plant Dis. 103, 2699–2699. doi: 10.1094/PDIS-03-19-0530-PDN
Weathers, L. (1957). A vein yellowing disease of citrus caused by a graft-transmissible virus. Plant Dis. Rept 42, 741–742.
www.votehemp.com (2019). U.S. Hemp License Report [Online]. Washington, DC. Available online at: www.votehemp.com (accessed May 22, 2020).
Keywords: hemp, beet curly top virus, beet leafhopper, virome, next generation sequencing
Citation: Chiginsky J, Langemeier K, MacWilliams J, Albrecht T, Cranshaw W, Fulladolsa AC, Kapuscinski M, Stenglein M and Nachappa P (2021) First Insights Into the Virus and Viroid Communities in Hemp (Cannabis sativa). Front. Agron. 3:778433. doi: 10.3389/fagro.2021.778433
Received: 16 September 2021; Accepted: 08 November 2021;
Published: 15 December 2021.
Edited by:
Zamir Punja, Simon Fraser University, CanadaReviewed by:
Carl Strausbaugh, Northwest Irrigation and Soils Research, Agricultural Research Service, United StatesSudeep Bag, The University of Georgia, United States
Copyright © 2021 Chiginsky, Langemeier, MacWilliams, Albrecht, Cranshaw, Fulladolsa, Kapuscinski, Stenglein and Nachappa. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Punya Nachappa, cHVueWEubmFjaGFwcGFAY29sb3N0YXRlLmVkdQ==
†Present address: Tessa Albrecht, Plant Health Services, LLC, Wellington, CO, United States
 Judith Chiginsky
Judith Chiginsky Kaitlyn Langemeier1
Kaitlyn Langemeier1 Tessa Albrecht
Tessa Albrecht Mark Stenglein
Mark Stenglein Punya Nachappa
Punya Nachappa