
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Agron. , 18 November 2021
Sec. Disease Management
Volume 3 - 2021 | https://doi.org/10.3389/fagro.2021.774863
This article is part of the Research Topic Cocoa Diseases and their Management View all 5 articles
Cacao swollen shoot disease (CSSD) is a damaging disease of Theobroma cacao L. associated with infection by a group of poorly characterized badnaviral species. To establish causality and characterize the symptomatology associated with infection by the badnavirus cacao swollen shoot Ghana M virus (CSSGMV), an infectious clone (1.3-mer) was constructed and used to inoculated cacao “Amelonado” seedlings by biolistic inoculation (BI; n = 18) and agroinoculation (AI; n = 15). Newly expanded leaves of BI (10/18) and AI (12/15) plants developed foliar mosaic and curling symptoms 30-days post inoculation (dpi), with chlorotic mottling and necrotic crinkling being evident by 90 dpi. By 120 dpi, three of 15 AI plants exhibited characteristic stem-swelling. Viral infection was verified by PCR-amplification and sequencing of a 1068 bp fragment of the CSSGMV ORF3 from newly expanding leaves 60 dpi. The PCR results indicated that 14 of 18 and 15 of 15 BI and AI plants, respectively, were systemically infected. The complete CSSGMV genome sequence was determined, by Illumina sequencing, from representative AI and BI plants and shared >99.5% pairwise nucleotide identity with CSSGMV-Nig9 (GenBank Accession No. MH785299). Based on the development of characteristic CSSD symptoms and recovery of partial and complete genome sequences of CSSGMV-Nig9 from systemically infected cacao plants, Koch's postulates have been fulfilled.
The cacao tree (Theobroma cacao L.) is native to the Amazon River Basin in South America and was introduced to West Africa during the 1920s, where it rapidly became a socioeconomically important crop. The West African countries, Cote d'Ivoire, Ghana, Nigeria, and Togo have historically produced ~70–75% of the world's cocoa beans (Ameyaw et al., 2014; FAOSTAT, 2017). To date, at least seven badnavirus species, presumably endemic to West Africa, have been associated with cacao swollen shoot disease (CSSD) in the region (Muller et al., 2018; Ramos-Sobrinho et al., 2020, 2021). Several badnaviruses appear to have re-emerged following an era of reduced CSSD disease incidence and spread, while other species had not been characterized until recently. Together, the CSSD virus complex is responsible for significant losses due to reduced quality and yield, affecting millions of smallholder farmers (Wessel and Quist-Wessel, 2015).
Although the first outbreak of swollen shoot disease occurred in Ghana in 1936 (Steven, 1936), disease symptoms were observed in small cacao plantings as early as 1922 (Paine, 1945). In 1874, T. cacao “Amelonado” was introduced to West Africa due to its production characteristics and suitability to the climate (Muller, 2016). The cacao genotype “Amelonado,” known to be susceptible to CSSD badnaviruses (Thresh et al., 1988), was considered an important factor contributing to the emergence and rapid spread of CSSD badnaviruses in cacao plantations in West Africa (Abrokwah et al., 2016; Muller, 2016; Muller et al., 2018). In 1944, CSSD was reported in Nigeria (Murray, 1945), where native plant species have been implicated as primary virus reservoirs (Abrokwah et al., 2016).
A complex of at least seven badnaviruses (genus Badnavirus, family Caulimoviridae) have been associated with swollen shoot disease, cacao swollen shoot Togo A virus (CSSTAV), cacao swollen shoot Togo B virus (CSSTBV), cacao swollen shoot CD virus (CSSCDV), cacao swollen shoot CE virus (CSSCEV), cacao swollen shoot Ghana M virus (CSSGMV), cacao swollen shoot Ghana N virus (CSSGNV), and Cacao swollen shoot Ghana Q virus (CSSGQV) (Muller and Sackey, 2005; Oro et al., 2012; Chingandu et al., 2017a,b, 2019; Muller et al., 2018; Ramos-Sobrinho et al., 2020). However, Koch's postulates have only been demonstrated for CSSTBV (Hagen et al., 1994). Badnaviruses have circular dsDNA genomes that undergo replication through a greater-than-genome length, terminally redundant, pre-genomic RNA that is used as template for DNA synthesis in the cytoplasm and for translation of the polycistronic viral coding regions (Bhat et al., 2016). Badnaviral genomes range in size from 7.0–9.2 kbp and encode from three to seven open reading frames (ORFs) (King et al., 2011; Bhat et al., 2016). The three canonical ORFs include the virion-associated protein encoded by ORF1 (Cheng et al., 1996), a protein with nucleic acid binding properties encoded by ORF2 (Jacquot et al., 1996), and a polyprotein that is cleaved to release the viral capsid, movement protein, aspartate protease, and the reverse transcriptase and ribonuclease H-like domains involved in genome replication encoded by ORF3 (Medberry et al., 1990; Harper and Hull, 1998). The dsDNA genomes are encapsidated in a 30 × 130 nm bacilliform particle, and are transmitted in a semi-persistent manner by mealybugs (Pseudococcidae) or aphids (Bhat et al., 2016).
Despite the implementation of a number of management practices to mitigate crop losses caused by CSSD, including eradication of virus-infected cacao trees and wild host plants adjacent to plantations, insecticide applications to control the mealybug vectors, and rouging of symptomatic trees (Lister and Thresh, 1957; Thresh, 1958, 1959), virus outbreaks have steadily increased, with the possible emergence of new strains and species (Chingandu et al., 2017a). Disease resistance strategies such as cross protection using a CSSD “mild” strain from Nigeria (Crowdy and Posnette, 1947; Posnette and Todd, 1951, 1955; Ollennu and Owusu, 2003), and planting of CSSD-tolerant/resistant cacao materials, have also been implemented to control the disease (Posnette and Todd, 1951; Dale, 1957; Thresh, 1958; Attafuah and Glendinning, 1965; Thresh et al., 1988). The Upper Amazon species of Theobroma genotypes collected in Peru have been evaluated in cacao breeding programs based on their inherent agronomic traits, resistance to fungal pathogens, vigor, and high yield (Pound, 1938, 1945), but most are susceptible to CSSD badnaviruses (Padi et al., 2018). Although some extent of tolerance has been reported to the severe isolate of CSSTBV from Ghana, known as the New Juaben strain (Crowdy and Posnette, 1947; Legg and Kenten, 1970; Thresh et al., 1988), breeding strategies have been confounded because most producers are smallholders that collect their own seed or plant seed obtained from other sources from which genetically and phenotypically variable trees are produced (Thresh et al., 1988). This has led to a highly heterogenous population of cacao clones where over 70% of the genetic material harbored unknown diversity, resulting from random crossing and potential selection based on variable criteria (Bekele et al., 2001; Jones et al., 2002). Current research is dedicated to discovering and mapping the populations of cultivated cacao (Motamayor et al., 2008; Livingstone et al., 2015, 2017), which can be used to inform breeders of the genetic background of breeding lines. Microsatellite analysis of the earliest cacao collection from the Upper Amazon, sampled from a small area consisting of a few sites located near the Ucayali, Nanay, and Marañon rivers, has shown that although there is high allelic variation, the collection comprises a large number of related family members (Zhang et al., 2009).
The badnavirus CSSGMV is hypothesized to be endemic to Nigeria and is among the most economically important species causing CSSD in cacao in this country (Dongo and Orisajo, 2007; Chingandu et al., 2019). CSSGMV has also been detected in cacao trees in the “living” germplasm collection maintained at the Cacao Research Institute of Ghana (CRIG) (Muller et al., 2018), and on a commercial farm in Cote d'Ivoire (Ramos-Sobrinho et al., 2021). Symptoms associated with CSSGMV, initially named as cacao red vein banding virus, consist of chlorotic mottling, necrotic crinkling, and red vein-banding, which distinguish it from other CSSD species described so far (Chingandu et al., 2019). The CSSD badnavirus from Nigeria was previously reported to cause mild symptoms, relative to the severe symptoms associated with the CSSTBV New Juaben isolate from Ghana. The mild symptom phenotype description was partially based on observations that CSSTBV New Juaben-infected trees frequently developed swollen shoot symptoms, whereas Nigerian isolates did not (Lister and Thresh, 1957; Ollennu and Owusu, 2003; Chingandu et al., 2019). The results of several delivery methods and infectivity for CSSTBV have been reported (Hagen et al., 1994; Jacquot et al., 1999; Friscina et al., 2017). However, Koch's postulates have not been fulfilled for CSSD badnaviruses in Nigeria, and no study has been carried out to compare the specific symptom phenotypes attributable to the plethora of divergent CSSD badnavirus species only recently discovered (Muller and Sackey, 2005; Muller et al., 2018; Ramos-Sobrinho et al., 2020).
Several approaches have been used to construct infectious clones of circular DNA plant viruses like begomoviruses and badnaviruses (Stenger et al., 1991; Buragohain et al., 1994; de Oliveira Ferreira et al., 2008; Nagata and Inoue-Nagata, 2015). An improved method based on polymerase chain reaction (PCR) and “Gibson Assembly,” referred to as PCR-GA (Gibson et al., 2009; Ferro et al., 2019), was used to construct an infectious clone of the cacao-infecting badnavirus CSSGMV isolate Nig9. Infectivity was evaluated by inoculating cacao “Amelonado” seedlings using both biolistic- and Agrobacterium-mediated inoculation. The agroinoculated plants developed virus-like foliar symptoms by 30-days post inoculation (dpi), and stem-swellings were observed in 20% of the infected plants around 120 dpi. The biolistic inoculated plants developed similar leaf symptoms by 30 dpi, but no swelling symptoms were observed by 120 dpi. Partial sequences of CSSGMV-Nig9 were obtained from newly expanded non-inoculated leaves using species-specific PCR primers previously described (Ramos-Sobrinho et al., 2021), and complete viral genome sequences were recovered from representative biolistic and agroinoculated plants by RNA and DNA high throughput-sequencing, respectively. Together, these results provide evidence that the CSSGMV-Nig9 clone is infectious and biologically active in cacao, fulfilling the Koch's postulates.
The two CSSGMV genomic units used for construction of the infectious clone were PCR-amplified from a full-length sequence of CSSGMV-Nig9 (Accession MH785299), previously cloned into the pGreenII vector backbone (Chingandu et al., 2019). The units 1 (2.2 kbp) and 2 (6.9 kbp) were assembled into the pGreenII (EF590266) (Hellens et al., 2000) vector backbone using the PCR-GA protocol (Ferro et al., 2019). Primers were designed based on optimal selection of ~20 nucleotides (nt) that overlapped between the two regions of interest of the cloned CSSGMV genome and selected regions of the plasmid vector (Figure 1). The two CSSGMV-Nig9 units were amplified by PCR using the primer pairs CSSGMV-U1-For/CSSGMV-U1-Rev and CSSGMV-U2-For/CSSGMV-S2-Rev, respectively, and the pGreenII binary plasmid vector was amplified using the pGreenII-noMCS-For/pGreen-noMCS-Rev primers (Table 1). Polymerase chain reaction amplification was carried out using Q5 Hot Start High-Fidelity 2X Master Mix (New England Biolabs, Ipswich, MA USA), in a final volume of 25 : 12.5 μL of 2X Q5 master mix, 0.2 μM of each primer, 1 μL (5 ng) of plasmid DNA (template), and 10.5 μL of nuclease-free water. Cycling parameters were initial denaturation for 2 min at 95°C, followed by 35 cycles of denaturation at 95°C for 20 s, annealing at 58°C for 20 s, and extension at 72°C for 1 min/kbp, and a final extension at 72°C for 10 min. The expected size amplicons were gel-purified using the Gel Band Purification Kit (GE Healthcare, Uppsala, Sweden), treated with the endonuclease DpnI to eliminate potential co-migrating fragments of the template plasmids, and the partial dimer (1.3-mer) of CSSGMV-Nig9 was assembled using Gibson Assembly Cloning Master Mix (New England Biolabs, Ipswich, MA USA) according to the manufacturer's instructions. The PCR-GA assembly product was used to transform competent cells of Escherichia coli DH5α, and clones were confirmed by restriction digestion of the plasmid DNA isolated from several single E. coli colonies with the endonuclease EcoRI, yielding a ~13.6kbp fragment. The clones with expected size inserts were Sanger sequenced at each vector flanking region, and at the junction between units 1 and 2. Further, total plasmid DNA (n = 2) was subjected to Illumina sequencing at the Center for Computational & Integrative Biology (CCIB) of the Massachusetts General Hospital (MGH). The Illumina-based de novo assembled plasmid sequences were compared, in silico, with the CSSGMV-Nig9 genome using the CLUSTALW alignment algorithm (Thompson et al., 2003) implemented in Geneious Prime v.2021.1.1 (www.geneious.com). Finally, the partial dimer construction of CSSGMV-Nig9, herein referred to as pCSSGMV-Nig9-IC, was used to transform electrocompetent cells of Agrobacterium tumefaciens GV3101, previously transformed with the pSoup plasmid (Hellens et al., 2000).
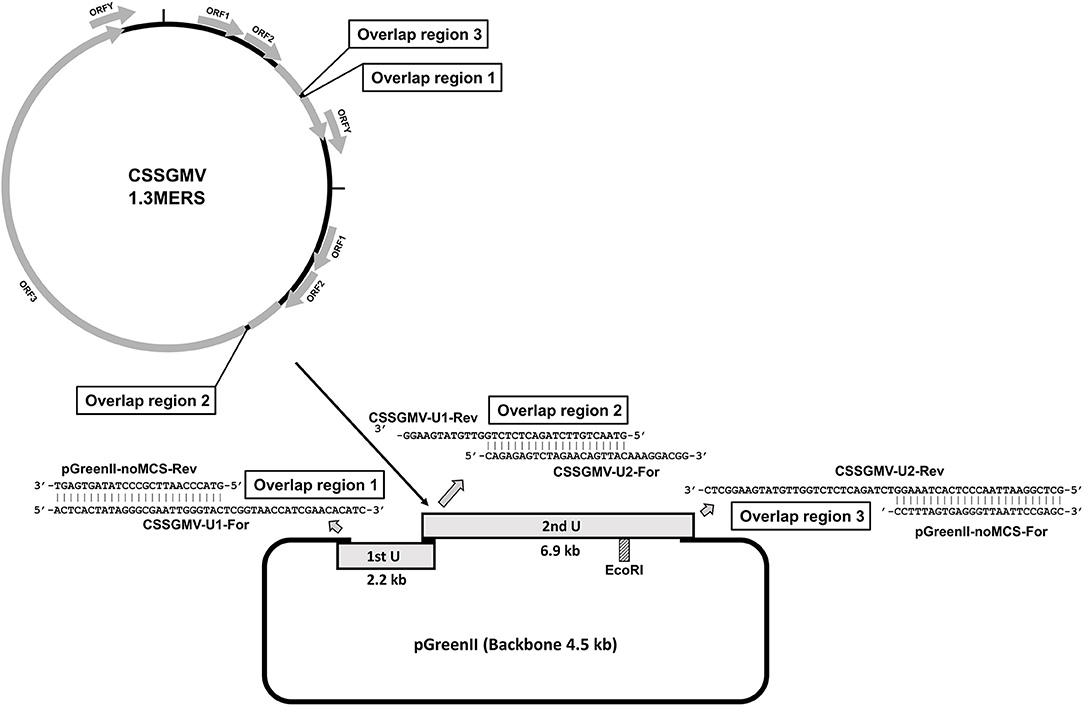
Figure 1. Cacao swollen shoot Ghana M virus (CSSGMV) agroinfectious clone assembly. The first and second viral units are represented by gray rectangles, and the primer-pairs containing overlapping sequences by gray arrows. The EcoRI restriction site (dashed rectangle) was used to confirm 1.3 mers clones by endonuclease digestion.
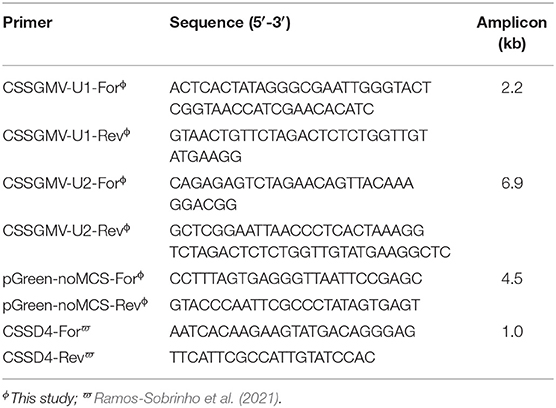
Table 1. Primer sequences used for amplification of the cacao swollen shoot Ghana M virus (CSSGMV) genomic units 1 and 2, pGreenII plasmid vector, and PCR detection of CSSGMV.
Single colonies of A. tumefaciens transformed with pCSSGMV-Nig9-IC were grown, at 28°C while shaking, in liquid Luria Broth-LB media supplemented with kanamycin (50 mg/μL) and rifampicin (50 mg/μL) to an approximate OD600 of 1.0. The cultures were centrifuged at 1,620 rcf for 5 min and resuspended in inoculation buffer [3% sucrose, 0.4% Murashige & Skoog basal salt mixture, 0.5 M 2-N-morpholino-ethanesulfonic acid (MES) pH 5.6, and 37.5 μl of 200 mM acetosyringone, dissolved to a final volume of 50 mL in distilled water and filter sterilized with 0.2 μm nitrocellulose filter], and incubated at 28°C while shaking for 4 h.
A 31-gauge syringe was used to infiltrate 1 mL of the A. tumefaciens-pCSSGMV-Nig9-IC inoculation solution in 30-day-old cacao “Amelonado” seedlings (n = 15) at the leaf blade, midrib, and petiole of the two youngest leaves, and at 0.5 cm of the shoot apical meristem. The control plants were mock-inoculated with A. tumefaciens GV3101 harboring the pGreen/pSoup plasmid, with no viral insert. Agroinoculated (AI) plants were kept in an insect-proof growth chamber at 25 ± 2°C and photoperiod of 16 h for 120-days post agroinoculation (dpa). Plants were observed regularly for symptom development.
Particles were delivered to cacao plants using the Bio-Rad™ Biolistic® PDS-1000/He. The tungsten microparticles (30 mg) were coated with 2.5 μg of pCSSGMV-Nig9-IC plasmid DNA according to the manufacturer's instructions and loaded onto laminated macro-carrier films (0.5 μg of DNA per macro-carrier). The particles were used to inoculate 30-day-old cacao “Amelonado” seedlings (0.5 μg of DNA per plant) using the gene gun at 1,350 psi. The mock control plants were inoculated with particles coated with “empty” pGreen plasmid DNA (no viral insert). The biolistically-inoculated (BI) plants were kept in an insect-proof growth room at 25 ± 2°C and photoperiod of 16 h for 120-days post inoculation (dpi). Plants were observed regularly for symptom development.
Newly expanding cacao leaf samples were collected at 15, 30, 45, and 60 dpi, and total DNA was extracted using the CTAB method (Doyle and Doyle, 1987). The CSSD4 primer set (Ramos-Sobrinho et al., 2021) was used for PCR detection of CSSGMV-Nig9. The PCR reactions were performed in a final volume of 25 μL: 12.5 μL of RedTaq® (Sigma-Aldrich), 0.5 μL of each primer (10 μM), 2 μL of total DNA (template), and 9.5 μL of nuclease-free water. Thermal cycler parameters were 94°C for 2 min as an initial denaturing step, followed by 35 cycles at 94°C for 20 s, 55°C for 15 s, and 72°C for 60 s, with a final extension at 72°C for 10 min. The expected size amplicons (1,068 bp) were cloned in the pGEM-T easy vector (Promega, Madison, WI, USA) according to the manufacturer's protocol and used to transform chemically competent cells of E. coli DH5α. Clones were confirmed by colony PCR amplification using M13 universal primers, and cloned amplicons were bidirectionally Sanger sequenced at Eton Biosciences (San Diego, CA, USA). The nucleotide sequences were assembled in Geneious Prime v2021.1.1 (www.geneious.com) and verified by comparison with the analogous CSSGMV-Nig9 region.
Total RNA and DNA were isolated from 100 mg of fresh leaf tissue collected at 90 dpi from a representative cacao plant that tested positive by PCR using the CSSD4 primers. Total RNA was isolated from a BI plant using a modified silica method (Rott and Jelkmann, 2001), and DNA was isolated from an AI plant according to Doyle and Doyle (1987). The total RNA and DNA preparations were individually subjected to Illumina sequencing at NovoGene Corporation Inc. (Sacramento, CA, USA) using the Illumina MiSeq 3000 platform. Adapters were trimmed from sequences and quality scores below a Phred score of 30 were removed from the data by Novogene. The sequence data was analyzed on FastQC to confirm that reads were trimmed and filtered. The Bowtie2 v2.4.4 program (Langmead and Salzberg, 2012) was used to align host reads against an index built from cacao sequences available in GenBank (Accession Nos. HQ244500, CM001879, CM001888, FR722157, JX065074, and KE132922). The unused reads were then aligned to the CSSGMV-Nig9 genome sequence (Accession MH785299). The consensus genomes from the alignments were extracted in Geneious Prime v2021.1.1 (www.geneious.com). Independently, the raw reads were used for de novo assembly using SPAdes v3.12.0 (Nurk et al., 2013). Finally, single nucleotide variants (SNVs) were characterized in Geneious Prime v2021.1.1 (www.geneious.com). To ensure that the variants were not likely due to sequencing errors or chance, only SNVs represented by ≥ 0.5% of total reads and P values <0.0001% were accepted as reliable predictions (Bansal, 2010).
A partial dimer (1.3-mer) construct of ~13.6 kb in length, which includes the viral units 1 (2.2 kb) and 2 (6.9 kb) and the pGreenII vector backbone (4.5 kb), was obtained for the badnavirus CSSGMV-Nig9 using PCR-GA (Figure 1). The Sanger sequences of the junction regions between vector and units 1 and 2 were 100.0% identical to the CSSGMV-Nig9 genome sequence. Also, results of whole plasmid sequencing indicated that the length of the construct was 13,601 bp, and was 99.9% identical with the CSSGMV-Nig9 genome. Therefore, the accuracy of pCSSGMV-Nig9-IC (1.3-mer) was confirmed by enzyme digestion, Sanger sequencing of the cloned fragment, and whole plasmid sequencing.
The agroinoculated cacao “Amelonado” leaves exhibited mild yellow mosaic symptoms 30 dpa, which developed into yellow chlorotic mottling symptoms 60 dpa (Figure 2A). Only three plants developed characteristic stem-swelling 120 dpa (Figure 2B). The agroinoculated plants declined slowly, with 13 of 15 plants dying by 120 dpa (data not shown). The mock-inoculated (n = 15) and non-inoculated (n = 10) negative control plants were asymptomatic (Figures 2C,D). To assay plants for virus presence or absence, the CSSD4 primer set was used to amplify a partial fragment of the 5' region of CSSGMV ORF3 from total DNA isolated of newly expanded test plants. The expected size amplicon (1068 bp) was detected in 12 of 15 (80%) AI plants 30 dpa, and no amplification product was obtained from the negative control plants (mock- and non-inoculated plants) (Figure 2I; Table 2). At 60 dpa, 15 of 15 (100%) AI plants tested PCR-positive with the CSSD4 primers (Table 2). Also, the expected size amplicon was obtained from total DNA isolated from bark tissue of the two surviving cacao seedlings exhibiting stem-swelling symptoms at 120 dpa (n = 3; data not shown). The cloned amplicons were confirmed as CSSGMV by bi-directional Sanger DNA sequencing. The ORF3 partial sequences (n = 15) shared 99.9–100% nucleotide identity with CSSGMV-Nig9, confirming the cacao plants were systemically infected.
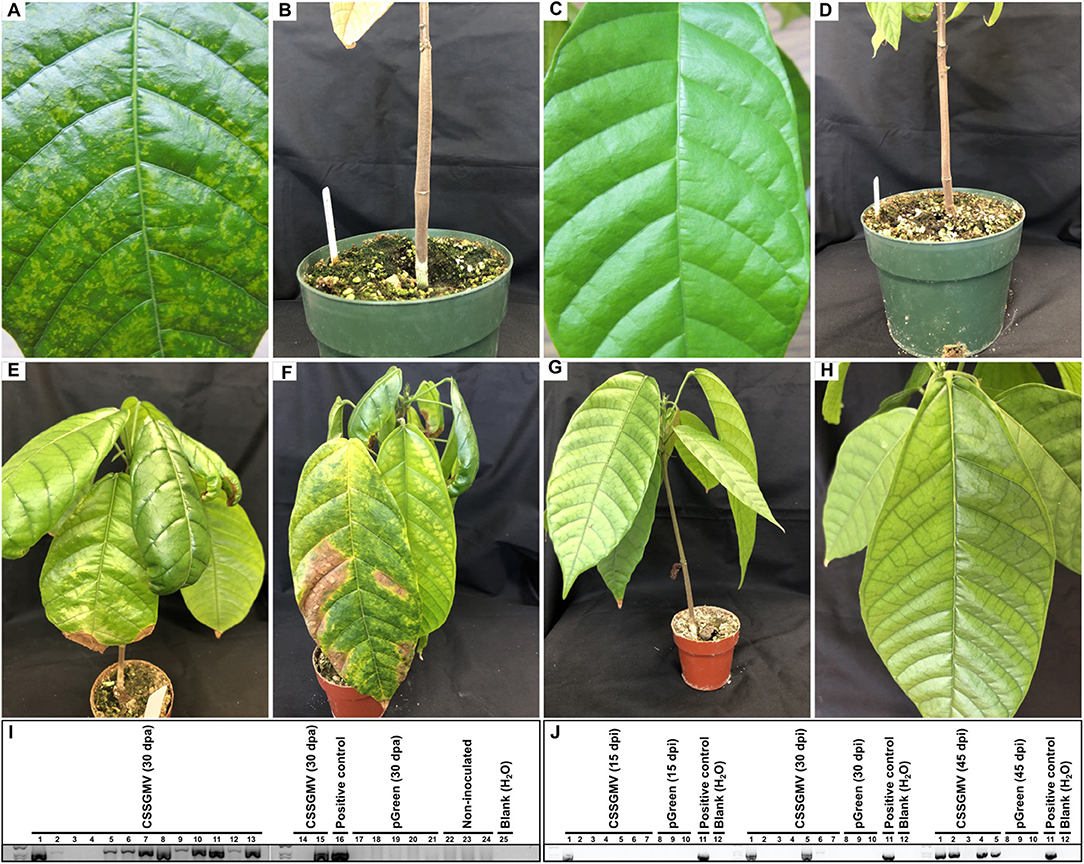
Figure 2. Infectivity assay of the cacao swollen shoot Ghana M virus (CSSGMV) infectious clone in cacao ‘Amelonado’ seedlings. Agroinoculated plants displaying yellow chlorotic mottling (A) and shoot swelling (B) symptoms. No symptoms were observed in mock control plants (C,D). PCR detection of CSSGMV was performed, at 30 days-post-agroinoculation (dpa), using species-specific primers, and expected size amplicons were observed from 12 of 15 agroinoculated plants (lanes 1–15); no amplification fragment was observed from control plants (lanes 17–24) (I). Biolistic-inoculated plants exhibiting symptoms of interveinal chlorosis and necrotic crinkling (E), and interveinal chlorosis, necrotic crinkling, chlorotic mottling, and necrosis spreading from primary veins (F). Symptomless mock control plants (G,H). PCR detection of CSSGMV was performed, at 15, 30, and 45 days-post-inoculation (dpi), using species-specific primers. Expected size amplicons were observed from 1 of 7, 3 of 7, and 4 of 5 plants (lanes 1-7) at 15, 30, and 45 dpi, respectively; no amplification fragment was obtained from mock treatments (lanes 8-10) (J). The partial nucleotide sequences of the cloned amplicons shared 99.9-100% identity with CSSGMV-Nig9.
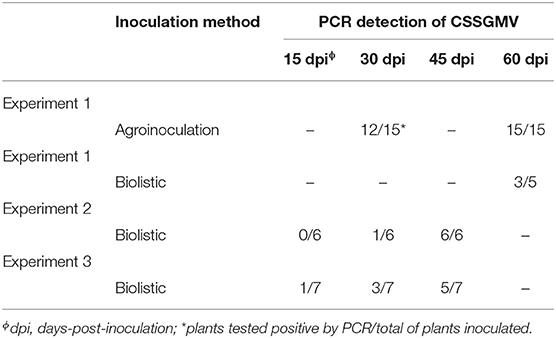
Table 2. Infectivity tests of the cacao swollen shoot Ghana M virus (CSSGMV) infectious clone in cacao “Amelonado” seedlings.
Three replicated experiments were carried out in which cacao “Amelonado” seedlings were biolistically inoculated. The cacao plants developed a similar range of foliar symptoms, but no stem-swelling was observed in any of the BI symptomatic plants. The percentage of infection was lower for the BI experiments, if compared to AI, with 14 of 18 inoculated plants developing interveinal chlorosis, necrotic crinkling, and severe chlorotic mottling. Also, the primary veins extending from the leaf midribs developed necrosis and lesions were observed along the margins of the leaf blade (Figures 2E,F). Similar to the AI experiment, the BI cacao plants declined and most of them died by 120 dpi. No symptoms were observed in negative control plants (Figures 2G,H). The PCR detection results with the CSSD4 primer pair showed that three of five plants (60%) yielded the expected size amplicon by 60 dpi in the first experiment, while the expected size amplicon was obtained for six of six (100%) and five of seven (71%) plants by 45 dpi in the second and third experiments, respectively (Figure 2J; Table 2). In the third experiment, two plants died before 45 dpi, but one of these plants was PCR-positive by 30 dpi (Table 2). Also, one plant (experiment 3) developed symptoms at 15 dpi and was positive by PCR (Figure 2J). No amplification product was obtained from the negative control plants (Figure 2J). The cloned amplicons were confirmed as CSSGMV by bi-directional Sanger DNA sequencing. The ORF3 partial sequences (n = 14) shared 99.9–100% nucleotide identity with CSSGMV-Nig9, confirming the cacao plants were systemically infected.
Total RNA isolated from leaf tissue of a representative BI symptomatic cacao plant, and confirmed positive by PCR amplification, was subjected to Illumina sequencing. Sequencing yielded 1,786 of 96,753,574 total reads (1.8E-3%) that mapped to the CSSGMV-Nig9 genome using Bowtie2. The average depth of coverage was 40x, and the resultant consensus genome sequence shared >99.9% nucleotide identity with CSSGMV-Nig9 (Figure 3). Three nucleotide differences observed in the CSSGMV genome assembled from RNA-Illumina reads were shared with the viral sequence obtained from whole plasmid sequencing, confirming 100% nucleotide identity with pCSSGMV-Nig9-IC. Based on SNVs, five variants with frequencies ranging from 5 to 12% had significant P values between 5.4E-7 and 2.6E-11. The five SNVs were predicted to be transition mutations (e.g., purine to purine; Table 3). These results suggest that the virus was actively undergoing replication as the characterized SNVs had P values below the threshold criteria of predicted sequencing errors (Bansal, 2010; King et al., 2020). The SPAdes de novo assembly recovered the full-length genome of CSSGMV-Nig9 at an average depth of coverage of 17x, and harbored mutations analogous to those from the Bowtie2 guided assembly (Supplementary Table 1).
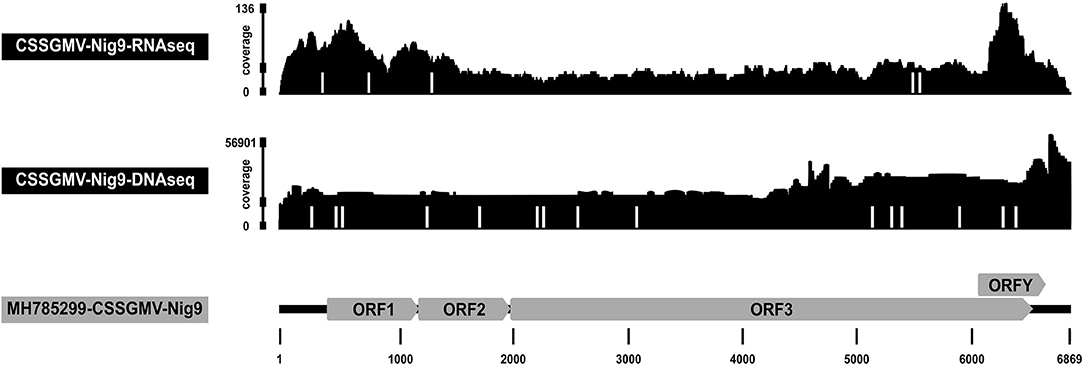
Figure 3. Coverage depth of Illumina reads mapped to the full-length genome of cacao swollen shoot Ghana M virus (CSSGMV)-Nig9. The RNA Illumina sequencing data (top) was obtained from a representative biolistic inoculated cacao plant at 90 days-post-inoculation, while the DNA Illumina reads (middle) were recovered from an agroinoculated plant at 120 days-post-agroinoculation. The positions of single nucleotide variations (SNVs) observed from the RNA and DNA sequencing reads are represented by white bars. The genome organization of CSSGMV-Nig9 is shown at the bottom.
The DNA Illumina results for the AI cacao plant exhibiting stem-swelling symptoms (Figure 2B) yielded 1,036,304 of 371,359,406 total reads (0.28%) that mapped to the CSSGMV-Nig9 genome using Bowtie2, with an average depth of coverage of 22,561x. The resultant consensus sequence shared 100% nucleotide identity with pCSSGMV-Nig9-IC (Figure 3). Based on SNVs, 17 variants with a range of frequencies between 0.5–1.4% had significant P values from 2.3E-80 to 2.8E-264. Two SNVs were predicted as transversion mutations, at frequencies of 0.8% and 1.4%, respectively, while 15 SNVs were identified as transition mutations (Table 3). Because of the high coverage of reads, de novo assembly was not pursued.
Symptoms characteristically reported for CSSD-affected cacao plants are development of swollen areas in shoots, branches, and roots, vein-reddening of newly-expanding leaves, chlorotic veins, deformed and discolored pods, yellow banding along the main vein of leaves, chlorosis and mottling in mature leaves, and tree decline leading to tree death within three to five years (Muller, 2016). The variable nature and range of symptoms observed in discrete geographic areas has been attributed to infection by different suspect badnavirus species (Abrokwah et al., 2016; Muller et al., 2018; Chingandu et al., 2019). Potentially obscuring or confounding differences in CSSD symptoms expected for each distinct badnavirus species is the extensive genomic variation within the diverse T. cacao genetic groups and hybrids (Motamayor et al., 2008; Zhang et al., 2009; Lachenaud and Motamayor, 2017). At least seven CSSD badnaviruses are formally recognized by the International Committee on Taxonomy of Viruses (ICTV) (https://talk.ictvonline.org/ictv-reports/ictv_online_report/reverse-transcribing-dna-and-rna-viruses/w/caulimoviridae/1361/genus-badnavirus), but to date, causality has been demonstrated only for CSSTBV (Hagen et al., 1994; Jacquot et al., 1999; Friscina et al., 2017), where viral inoculation and evaluation of disease symptomatology was carried out in T. cacao “Amelonado.”
Different approaches to construct infectious clones of DNA plant viral genomes have been reported, most often employing conventional restriction endonucleases to produce greater than unit-length or dimeric genome molecules containing two copies of the origin of replication (Buragohain et al., 1994; Jacquot et al., 1999; Nagata and Inoue-Nagata, 2015). The inclusion of two origins of replication has been shown to enhance the likelihood of infectivity and/or to increase the rate of infectivity of infectious clones in experimental inoculation (Stenger et al., 1991; Nagata and Inoue-Nagata, 2015). Additionally, agro-clones of plant pararetroviruses such as badnaviruses require a longer than full-length genome construct to enable production of a pre-genomic RNA with terminally redundant sequences essential for viral replication (Jacquot et al., 1999). In this study, an agroinfectious clone (1.3 partial dimer) of the CSSD-badnavirus CSSGMV isolate Nig9 was constructed using the PCR-GA method (Ferro et al., 2019), and experiments designed to fulfill Koch's postulates were carried out by inoculating seedlings of the CSSD-susceptible cacao “Amelonado” genotype. The chlorotic mottling, necrotic leaf curling, and primary vein necrosis symptoms observed in cacao plants experimentally inoculated with pCSSGMV-Nig9-IC were distinguishable from symptoms observed for cacao plants inoculated with infectious clones of CSSTBV (Hagen et al., 1994; Jacquot et al., 1999; Friscina et al., 2017). Symptom onset, progression, and phenotype also differed between the two species of CSSD badnaviruses. Conditions of light intensity, frequency of watering, and fertilizer nutrition were not reported by Jacquot et al. (1999) or Hagen et al. (1994), precluding a comparison with the results reported here. However, the latter two studies involved the same CSSTBV infectious clone reported similar disease symptomatology, suggesting that laboratory environmental conditions were either similar, or if different, may not have influenced symptom phenotype in cacao. The most notable differences between the symptoms reported for CSSTBV (Hagen et al., 1994; Jacquot et al., 1999; Friscina et al., 2017) and those observed for CSSGMV (this study) are the earlier onset of symptoms that consisted of necrotic crinkling, interveinal chlorosis, chlorotic mottling, and a lower frequency of stem-swelling for CSSGMV-infected plants. For example, the initial mild symptoms of CSSTBV were first apparent 7 weeks post-inoculation (~50 dpi) and swollen shoots and fern-like mosaic were observed around 11 weeks (~77 dpi) (Jacquot et al., 1999). Hagen et al. (1994) inoculated seeds and reported that the symptoms first appeared at the 7-true leaf stage, or about 5–6 weeks after the first leaves matured, while stem swellings were reported at 14 weeks (~98 days). This differs with the results observed here in which foliar symptoms caused by CSSGMV developed by 15 dpi, and regularly around 30 dpi, while swelling symptoms were rarely observed by 120 dpa. Insufficient knowledge about similarities and/or differences in symptom phenotype(s) associated with known CSSD species underscores the need to carry out Koch's postulates and comparative symptom analysis for all known CSSD species.
Many factors have been hypothesized to influence CSSD-badnavirus symptoms and disease severity, including differences in susceptibility among cacao genotypes (Thresh et al., 1988; Wessel and Quist-Wessel, 2015), climate, light intensity (Andres et al., 2018; Domfeh et al., 2021), soil nutrient composition (Andres et al., 2018; Domfeh et al., 2021), and herbivory (Lister and Thresh, 1957; Thresh, 1960a). Here, CSSGMV-inoculated cacao “Amelonado” seedlings were maintained under controlled environment conditions (light and temperature) and regular fertilizer application, in the absence of herbivory. The red vein-banding symptoms previously described in cacao leaves in Nigeria (Thresh, 1960b) has been ascribed to what is now thought to be CSSGMV-infected trees under field conditions, hence the proposed name cacao red vein-banding virus (Chingandu et al., 2019). In this study, red vein-banding symptoms were not observed by 120 dpi in cacao “Amelonado” seedlings inoculated with pCSSGMV-Nig9-IC via either biolistic or agroinoculation. Perhaps the “ephemeral” red vein-banding and other symptoms reported in previous studies can only be recapitulated under field conditions or for differentially susceptible cacao genotypes such as in hybrids that are now commonly planted in West Africa. Further, the red vein-banding symptoms reported in association with CSSGMV in Nigeria (Chingandu et al., 2019) were observed in >5-year-old cacao trees, and the age of the host should be considered as a possible factor, among others that could influence symptomatology. Cacao “Amelonado” plants agroinoculated with a CSSTBV infectious clone exhibited red vein-banding symptoms 60 dpi (Jacquot et al., 1999). Because mixed infections of CSSD badnaviruses and potential co-infection by other plant viruses cannot be ruled out (Chingandu et al., 2019; Ramos-Sobrinho et al., 2020, 2021), badnaviral etiology underlying the previously reported foliar red vein-banding requires further investigation.
Swollen shoot symptoms have been observed infrequently in CSSD-cacao trees in Nigeria, where CSSGMV is believed to be endemic and possibly the most prevalent badnavirus species (Lister and Thresh, 1957; Ollennu and Owusu, 2003; Chingandu et al., 2019). The occasional observation of swollen shoots among the CSSD isolates in Nigeria lead to the categorization of the suspect badnavirus as a “mild” strain when compared to the “severe” New Juaben CSSD-isolate from Ghana, which routinely caused swelling symptoms on infected cacao plants (Lister and Thresh, 1957; Thresh, 1959; Ollennu and Owusu, 2003; Chingandu et al., 2019). Here, 100% (15/15) of the pCSSGMV-Nig9-IC-agroinoculated plants were shown to be systemically infected, with rates of infectivity for BI cacao seedlings ranging from 60–100%. Notably, swelling symptoms on the primary stem (e.g., that will become the trunk of the mature tree) were observed in 20% (3/15) of CSSGMV-agroinoculated plants, while analogous swelling was not observed in the BI plants. Biolistic inoculation of monomers of the Agou1 “severe strain” (accession L14546) from Togo, which appears to be genotypically similar to the New Juaben isolate (accession AJ608931) and recently re-named CSSTBV (Muller et al., 2018), in cacao “Amelonado” resulted in infectivity rates of 65–88%, with at least 30% (9/30) of the seedlings developing stem-swelling symptoms ~120 dpi (Hagen et al., 1994). Agroinoculation of cacao “Amelonado” with an infectious clone (1.2-mers) of CSSTBV-Agou1 resulted in 83% (49/59) and 85% (17/20) of inoculated plants exhibiting swelling symptoms 120 dpa (Jacquot et al., 1999). Based on several reports of experimental inoculation of cacao with an infectious clone of CSSTBV, and results described here, stem-swelling was observed in CSSGMV-infected cacao plants more rarely than has been previously reported for CSSTBV-infected plants, which developed stem-swelling at a higher rate (e.g., 20% for CSSGMV compared to >80% for CSSTBV).
Lastly, notable differences between the inoculation methods used here and by Jacquot et al. (1999) include use of the A. tumefaciens strain GV3101 instead of LBA4404 and the resuspension and incubation of the A. tumefaciens GV3101 in an induction media to induce expression of the Ti plasmid Vir genes. The syringe infiltration inoculation method and plant age were similar, but the percent of infection was higher under the conditions reported here, at 100% compared to 85%. In Hagen et al. (1994), the infectious material was a monomer excised from the plasmid backbone before inoculation, a greater mass of DNA was loaded onto tungsten particles, the pressure of helium pulse was substantially lower, and seeds instead of whole plants were inoculated. However, similar rates of infection were observed, with 100% infection being observed in one of three experiments performed in the present study. Virus detection methods differed in Hagen et al. (1994) and Jacquot et al. (1999), which used ELISA and dot blot hybridization. Also, Hagen et al. (1994) analyzed the test plants at a single time point, compared to the Jacquot et al. (1999) study that analyzed inoculated plants 8- and 12-weeks post-inoculation. The more sensitive detection method used in this study (i.e., PCR amplification) compared to ELISA or dot-blot hybridization may explain the earlier detection of CSSGMV in cacao plants, at 30 and 60 dpi, compared to later time points reported in the previous studies.
The depth of coverage of the CSSGMV-Nig9 genome from both RNA and DNA Illumina sequence reads determined from systemically infected leaves of inoculated cacao plants was considerably high, ranging from 40 to 22,561x. Analysis of the RNA and DNA reads revealed statistically SNVs, indicative of virus population dynamics in an environment conducive to replication, a supposition that is supported by the high number of reads associated with the individual SNVs. In the latter scenario, the SNVs were convincingly attributable to the accumulation of bonafide errors introduced during viral replication, and not to background noise (Bansal, 2010; King et al., 2020).
Evidence for replication of pCSSGMV-Nig9-IC in T. cacao “Amelonado” and systemic infection of CSSGMV-Nig9 was demonstrated in this study. Also, foliar CSSGMV-caused symptoms were distinguishable from CSSTBV-Agou1 in that they developed sooner, and were manifest as necrotic leaf crinkle, primary vein necrosis, and chlorotic mottling. Further, CSSGMV-inoculated plants only rarely developed swollen stems, compared to the high frequency of stem-swelling associated with CSSTBV (Hagen et al., 1994; Jacquot et al., 1999). The absence of the red vein-banding phenotype needs further investigation and could be attributable to different factors, including differences between cacao genotype and environmental effects on symptom development. These results provide ample evidence to suggest that there are discernable symptoms caused by the different species of the CSSD complex, confirming earlier hypotheses (Abrokwah et al., 2016; Muller et al., 2018; Chingandu et al., 2019) and belaboring the need to explore the relationship between symptoms and CSSD-associated badnaviral species. The development of pCSSGMV-Nig9-IC provides a new, tool to pursue studies of plant-virus-vector interactions, characterization of viral genes and gene function, and for screening badnavirus-resistant cacao germplasm. Also, this newly developed CSSGMV infectious clone, together with those of other cacao-infecting badnavirus species, will contribute to a genetically diverse panel of CSSD-badnavirus clones. Such a panel would facilitate identification of badnavirus and host factors through differential gene expression studies and elucidate the genetic basis for differential susceptibilities among CSSD-badnavirus species in the context of cacao genetic group diversity.
The data presented in the study are deposited in the GenBank repository, accession numbers MH785299, and BioProject PRJNA774777.
CK: conceptualization, formal analysis, investigation, data curation, and original draft preparation. RR-S: conceptualization, methodology, investigation, data curation, and review and editing. J-PM: resources, review and editing, and project administration. JB: conceptualization, methodology, data curation, review and editing, and project administration. All authors contributed to the article and approved the submitted version.
The authors thank Mars Wrigley for supporting this research through the UA Sponsored Research agreement number 6038-21000-023-12-T. The funder was not involved in the study design, collection, analysis, interpretation of data, the writing of this article or the decision to submit it for publication.
J-PM was employed by Mars Wrigley.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fagro.2021.774863/full#supplementary-material
Abrokwah, F., Dzahini-Obiatey, H., Galyuon, I., Osae-Awuku, F., and Muller, E. (2016). Geographical distribution of cacao swollen shoot virus molecular variability in Ghana. Plant Dis. 100, 2011–2017. doi: 10.1094/PDIS-01-16-0081-RE
Ameyaw, G. A., Dzahini-Obiatey, H. K., and Domfeh, O. (2014). Perspectives on cocoa swollen shoot virus disease (CSSVD) management in Ghana. Crop Prot. 65, 64–70. doi: 10.1016/j.cropro.2014.07.001
Andres, C., Blaser, W. J., Dzahini-Obiatey, H. K., Ameyaw, G. A., Domfeh, O. K., Awiagah, M. A., et al. (2018). Agroforestry systems can mitigate the severity of cocoa swollen shoot virus disease. Agric. Ecosyst. Environ. 252, 83–92. doi: 10.1016/j.agee.2017.09.031
Attafuah, A., and Glendinning, D. K. (1965). Studies on resistance and tolerance to cocoa viruses in Ghana. Ann. Appl. Biol. 56, 219–225. doi: 10.1111/j.1744-7348.1965.tb01229.x
Bansal, V. (2010). A statistical method for the detection of variants from next-generation resequencing of DNA pools. Bioinformatics 26, i318–i324. doi: 10.1093/bioinformatics/btq214
Bekele, F. L., End, M., and Eskes, A. (2001). Proceedings of the International workshop on new technologies and cocoa breeding. INGENIC
Bhat, A. I., Hohn, T., and Selvarajan, R. (2016). Badnaviruses: the current global scenario. Viruses 8:177. doi: 10.3390/v8060177
Buragohain, A. K., Sung, Y. K., Coffin, R. S., and Coutts, R. H. A. (1994). The infectivity of dimeric potato yellow mosaic geminivirus clones in different hosts. J. Gen. Virol. 75, 2857–2861. doi: 10.1099/0022-1317-75-10-2857
Cheng, C.-P., Lockhart, B. E. L., and Olszewski, N. E. (1996). The ORF I and II proteins ofcommelinayellow mottle virus are virion-associated. Virology 223, 263–271. doi: 10.1006/viro.1996.0478
Chingandu, N., Dongo, L., Gutierrez, O. A., and Brown, J. K. (2019). The previously unidentified, divergent badnavirus species Cacao red vein-banding virus is associated with Cacao swollen shoot disease in Nigeria. Plant Dis. 103, 1302–1308. doi: 10.1094/PDIS-09-18-1561-RE
Chingandu, N., Kouakou, K., Aka, R., Gutierrez, O., and Brown, J. (2017a). Unexpected genome variability at multiple loci suggests Cacao swollen shoot virus comprises multiple, divergent molecular variants. J. Emerg. Virol. Dis. 3, 1846–2473. doi: 10.16966/2473-1846.128
Chingandu, N., Sreenivasan, T. N., Surujdeo-Maharaj, S., Umaharan, P., Gutierrez, O. A., and Brown, J. K. (2017b). Molecular characterization of previously elusive badnaviruses associated with symptomatic cacao in the New World. Arch. Virol. 162, 1363–1371. doi: 10.1007/s00705-017-3235-2
Crowdy, S. H., and Posnette, A. F. (1947). Virus diseases of cacao in West Africa: II. Cross-immunity experiments with viruses 1A, 1B, and 1C. Ann. Appl. Biol. 34, 403–411. doi: 10.1111/j.1744-7348.1947.tb06373.x
Dale, W. T. (1957). “Studies on resistance and tolerance to cacao viruses,” in Proceedings of the Cacao Breeding Conference, Ghana, 3–6.
de Oliveira Ferreira, T., Lemos, T. O., Nagata, T., and Inoue-Nagata, A. K. (2008). One-step cloning approach for construction of agroinfectious begomovirus clones. J. Virol. Methods 147, 351–354. doi: 10.1016/j.jviromet.2007.10.001
Domfeh, O., Ameyaw, G. A., Boakye, A. Y., Opoku-Agyeman, M. O., Arthur, A., Akpertey, A., et al. (2021). Evaluation of mild strain cross protection in cacao–further evidence of the protective potential of cacao swollen shoot virus strain N1 against the New Juabeng (1A). isolate under field conditions. Australas. Plant Pathol. 50, 329–340. doi: 10.1007/s13313-021-00777-1
Dongo, L. N., and Orisajo, S. B. (2007). Status of cocoa swollen shoot virus disease in Nigeria. African J. Biotechnol. 6:2054–2061. doi: 10.5897/AJB2007.000-2317
Doyle, J. J., and Doyle, J. L. (1987). A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 19, 11–15.
FAOSTAT (2017). FAO statistical databases. Available online at: http://www.fao.org/faostat/en/#data/QC/visualize (accessed June 15, 2021).
Ferro, M. M. M., Ramos-Sobrinho, R., Xavier, C. A. D., Zerbini, F. M., Lima, G. S. A., Nagata, T., et al. (2019). New approach for the construction of infectious clones of a circular DNA plant virus using Gibson Assembly. J. Virol. Methods 263, 20–23. doi: 10.1016/j.jviromet.2018.10.017
Friscina, A., Chiappetta, L., Jacquemond, M., and Tepfer, M. (2017). Infection of non-host model plant species with the narrow-host-range Cacao swollen shoot virus. Mol. Plant Pathol. 18, 293–297. doi: 10.1111/mpp.12404
Gibson, D. G., Young, L., Chuang, R.-Y., Venter, J. C., Hutchison, C. A., and Smith, H. O. (2009). Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6, 343–345. doi: 10.1038/nmeth.1318
Hagen, L., Lot, H., Godon, C., Tepfer, M., and Jacquemond, M. (1994). Infection of Theobroma cacao using cloned DNA of cacao swollen shoot virus and particle bombardment. Phytopathology. 84, 1239–1243. doi: 10.1094/Phyto-84-1239
Harper, G., and Hull, R. (1998). Cloning and sequence analysis of banana streak virus DNA. Virus Genes 17, 271–278. doi: 10.1023/A:1008021921849
Hellens, R. P., Edwards, E. A., Leyland, N. R., Bean, S., and Mullineaux, P. M. (2000). pGreen: a versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol. Biol. 42, 819–832. doi: 10.1023/A:1006496308160
Jacquot, E., Hagen, L. S., Jacquemond, M., and Yot, P. (1996). The open reading frame 2 product of cacao swollen shoot badnavirus is a nucleic acid-binding protein. Virology 225, 191–195. doi: 10.1006/viro.1996.0587
Jacquot, E., Hagen, L. S., Michler, P., Rohfritsch, O., Stussi-Garaud, C., Keller, M., et al. (1999). In situ localization of cacao swollen shoot virus in agroinfected Theobroma cacao. Arch. Virol. 144, 259–271. doi: 10.1007/s007050050502
Jones, P. G., Allaway, D., Gilmour, M. D., Harris, C., Rankin, D., Retzel, E. R., et al. (2002). Gene discovery and microarray analysis of cacao (Theobroma cacao L.). varieties. Planta 216, 255–264. doi: 10.1007/s00425-002-0882-6
King, A. M. Q., Lefkowitz, E., Adams, M. J., and Carstens, E. B. (2011). Virus taxonomy: ninth report of the International Committee on Taxonomy of Viruses. Amsterdam: Elsevier
King, D. J., Freimanis, G., Lasecka-Dykes, L., Asfor, A., Ribeca, P., Waters, R., et al. (2020). A systematic evaluation of high-throughput sequencing approaches to identify low-frequency single nucleotide variants in viral populations. Viruses 12:1187. doi: 10.3390/v12101187
Lachenaud, P., and Motamayor, J. C. (2017). The Criollo cacao tree (Theobroma cacao L.): a review. Genet. Resour. Crop Evol. 64, 1807–1820. doi: 10.1007/s10722-017-0563-8
Langmead, B., and Salzberg, S. L. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods. 9:357doi: 10.1038/nmeth.1923
Legg, J. T., and Kenten, R. H. (1970). The resistance and tolerance of different cocoa varieties to cocoa swollen-shoot virus in Ghana. Ann. Appl. Biol. 65, 425–434. doi: 10.1111/j.1744-7348.1970.tb05511.x
Lister, R. M., and Thresh, J. M. (1957). “The history and control of cocoa swollen shoot disease in Nigeria,” in Proceedings of the Cocoa Conference, London. 132–142.
Livingstone, D., Royaert, S., Stack, C., Mockaitis, K., May, G., Farmer, A., et al. (2015). Making a chocolate chip: development and evaluation of a 6K SNP array for Theobroma cacao. DNA Res. 22,279–291. doi: 10.1093/dnares/dsv009
Livingstone, I. I. I. D., Stack, C., Mustiga, G. M., Rodezno, D. C., Suarez, C., Amores, F., et al. (2017). A larger chocolate chip—development of a 15K Theobroma cacao L. SNP array to create high-density linkage maps. Front Plant Sci 8:2008. doi: 10.3389/fpls.2017.02008
Medberry, S. L., Lockhart, B. E. L., and Olszewski, N. E. (1990). Properties of Commelina yellow mottle virus's complete DNA sequence, genomic discontinuities and transcript suggest that it is a pararetrovirus. Nucleic Acids Res. 18, 5505–5513. doi: 10.1093/nar/18.18.5505
Motamayor, J. C., Lachenaud, P., e Mota JW da, S., Loor, R., Kuhn, D. N., Brown, J. S., et al. (2008). Geographic and genetic population differentiation of the Amazonian chocolate tree (Theobroma cacao L). PLoS ONE 3:e3311. doi: 10.1371/journal.pone.0003311
Muller, E. (2016). “Cacao Swollen Shoot Virus (CSSV): history, biology, and genome bt,” in Cacao Diseases: A History of Old Enemies and New Encounters, eds B. A. Bailey, L. W. Meinhardt, (Cham: Springer International Publishing), 337–358.
Muller, E., Ravel, S., Agret, C., Abrokwah, F., Dzahini-Obiatey, H., Galyuon, I., et al. (2018). Next generation sequencing elucidates cacao badnavirus diversity and reveals the existence of more than ten viral species. Virus Res. 244, 235–251. doi: 10.1016/j.virusres.2017.11.019
Muller, E., and Sackey, S. (2005). Molecular variability analysis of five new complete cacao swollen shoot virus genomic sequences. Arch. Virol. 150:53–66. doi: 10.1007/s00705-004-0394-8
Murray, D. F. (1945). First progress report of the Nigeria cocoa survey April 1944-March 1945. First Prog. Rep. Niger. cocoa Surv. 11, 19–45.
Nagata, T., and Inoue-Nagata, A. K. (2015). “Simplified methods for the construction of RNA and DNA virus infectious clones,” in Plant Virology Protocols (New York, NY: Springer), 241–254.
Nurk, S., Bankevich, A., Antipov, D., Gurevich, A., Korobeynikov, A., Lapidus, A., et al. (2013). “Assembling genomes and mini-metagenomes from highly chimeric reads bt,” in Research in Computational Molecular Biology, eds M. Deng, R. Jiang, F. Sun, and X. Zhang (Berlin Heidelberg: Springer), 158–170.
Ollennu, L. A. A., and Owusu, G. K. (2003). Field evaluation of the protective capability of CSSV mild strain N1 against severe strain New Juaben (1A). isolate. Ghana J. Agric. Sci. 36,3–12. doi: 10.4314/gjas.v36i1.2058
Oro, F., Mississo, E., Okassa, M., Guilhaumon, C., Fenouillet, C., Cilas, C., et al. (2012). Geographical differentiation of the molecular diversity of cacao swollen shoot virus in Togo. Arch. Virol. 157, 509–514. doi: 10.1007/s00705-011-1158-x
Padi, F. K., Domfeh, O. K., Arthur, A., and Ofori, A. (2018). “Potential of recurrent selection for developing improved cocoa varieties in Ghana,” in International Symposium on Cocoa Research (ISCR), Lima, Peru, 13-17 November 2017. International Cocoa Organization (ICCO).
Posnette, A. F., and Todd, J. M. (1951). Virus diseases of cacao in West Africa VIII. The search for virus-resistant cacao. Ann. Appl. Biol. 38, 785–800. doi: 10.1111/j.1744-7348.1951.tb07850.x
Posnette, A. F., and Todd, J. M. (1955). Virus diseases of cacao in West Africa IX. strain variation and interference in virus 1A. Ann. Appl. Biol. 43, 433–453. doi: 10.1111/j.1744-7348.1955.tb02493.x
Pound, F. J. (1938). Cacao and witchbroom disease (Marasmius perniciosus). of South America. With notes on other species of Theobroma. Report by Dr. FJ Pound on a visit to Ecuador, the Amazon Valley, and Colombia. April 1937-April 1938. Cacao Witch Dis (Marasmius perniciosus). South Am With notes other species Theobroma Rep by Dr FJ Pound a Visit to Ecuador, Amaz Val Colomb April 1937-April 1938.
Pound, F. J. (1945). A note on the cocoa population of South America. In: Report and Proceedings of the 1945 Cocoa Conference. London. 131–133.
Ramos-Sobrinho, R., Chingandu, N. A., Gutierrez, O., Marelli, J.-P. K., et al. (2020). A complex of badnavirus species infecting cacao reveals mixed infections, extensive genomic variability, and interspecific recombination. Viruses 12:43. doi: 10.3390/v12040443
Ramos-Sobrinho, R., Kouakou, K., Bi, A. B., Keith, C. V., Diby, L., Kouame, C., et al. (2021). Molecular detection of cacao swollen shoot badnavirus species by amplification with four PCR primer pairs, and evidence that Cacao swollen shoot Togo B virus-like isolates are highly prevalent in Côte d'Ivoire. Eur. J. Plant Pathol. 159, 941–947. doi: 10.1007/s10658-021-02203-0
Rott, M. E., and Jelkmann, W. (2001). Characterization and detection of several filamentous viruses of cherry: adaptation of an alternative cloning method (DOP-PCR), and modification of an RNA extraction protocol. Eur. J. Plant Pathol. 107, 411–420. doi: 10.1023/A:1011264400482
Stenger, D. C., Revington, G. N., Stevenson, M. C., and Bisaro, D. M. (1991). Replicational release of geminivirus genomes from tandemly repeated copies: evidence for rolling-circle replication of a plant viral DNA. Proc. Natl. Acad. Sci. 88, 8029–8033. doi: 10.1073/pnas.88.18.8029
Thompson, J. D., Gibson, T. J., and Higgins, D. G. (2003). Multiple sequence alignment using ClustalW and ClustalX. Curr. Protoc. Bioinforma. 203, 2–3. doi: 10.1002/0471250953.bi0203s00
Thresh, J. M. (1958). The control of cacao swollen shoot disease in West Africa: a review of the present situation. Tech. Bull. African Cocoa Res. Inst. 4:36.
Thresh, J. M. (1959). The control of cacao swollen shoot disease in Nigeria. Trop. Agric. 36, 35–44.
Thresh, J. M. (1960a). Capsids as a factor influencing the effect of swollen shoot disease on cacao in Nigeria. Emp. J. Exp. Agric. 28, 193–200.
Thresh, J. M., Owusu, G. K., Boamah, A., and Lockwood, G. (1988). Ghanaian cocoa varieties and swollen shoot virus. Crop Prot. 7, 219–231. doi: 10.1016/0261-2194(88)90043-9
Wessel, M., and Quist-Wessel, P. M. F. (2015). Cocoa production in West Africa, a review and analysis of recent developments. NJAS—Wageningen J. Life Sci. 74, 1–7. doi: 10.1016/j.njas.2015.09.001
Keywords: Caulimoviridae, Badnavirus, emergent disease, tropical tree, seamless cloning, infectious clone
Citation: Keith CV, Ramos-Sobrinho R, Marelli J-P and Brown JK (2021) Construction of an Infectious Clone of the Badnavirus Cacao Swollen Shoot Ghana M Virus and Infectivity by Gene Gun- and Agrobacterium-Mediated Inoculation. Front. Agron. 3:774863. doi: 10.3389/fagro.2021.774863
Received: 13 September 2021; Accepted: 18 October 2021;
Published: 18 November 2021.
Edited by:
David Ian Guest, The University of Sydney, AustraliaReviewed by:
Xinhua Ding, Shandong Agricultural University, ChinaCopyright © 2021 Keith, Ramos-Sobrinho, Marelli and Brown. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Judith K. Brown, amJyb3duQGFnLmFyaXpvbmEuZWR1
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.