
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Agron. , 18 November 2021
Sec. Plant-Soil Interactions
Volume 3 - 2021 | https://doi.org/10.3389/fagro.2021.754646
This article is part of the Research Topic Belowground Interactions and Implications for Nutrient Use Eco-Efficiency in Cropping Systems View all 6 articles
Perennial agroecosystems often seek to optimize productivity by breeding nutrient-efficient, disease-resistant rootstocks. In cacao (Theobroma cacao L.), however, rootstock selection has traditionally relied on locally available open pollinated populations with limited data on performance. Furthermore, rootstock associations with the rhizobiome, or rhizosphere microbiome, have been neglected. Better understanding of rootstock and scion effects on cacao-specific traits, particularly those involved in root-microbe interactions and nutrient acquisition, could contribute to more efficient rootstock selection and breeding. A rootstock-scion interaction study was conducted using three scion genotypes and eight rootstock populations under greenhouse conditions to better understand the relationships among rootstock and scion identities, soil fertility, and rhizobiome composition and the impacts of these factors on plant uptake of macro- and micronutrients. We show that rootstock genotype has a stronger influence than scion on nutrient uptake, bacterial and fungal diversity, and rhizobiome composition, and that the relative contributions of rootstock and scion genotype to foliar nutrient status are dynamic over time. Correlation analysis and stepwise regression revealed complex relationships of soil physicochemical parameters and the rhizobiome to plant nutrition and emphasized strong impacts of microbial diversity and composition on specific nutrients. Linear discriminant analysis effect size estimation identified rootstock-responsive taxa potentially related to plant nutrition. This study highlights the importance of considering root-associated microbial communities as a factor in cacao rootstock breeding and the need for further investigation into mechanisms underlying nutrient acquisition and microbial interactions in grafted plants.
Grafting is a common practice in perennial crops to combine desirable combinations of rootstock and scion traits. Although grafting has been called a “surgical alternative to breeding” for its utility in creating desirable combinations of traits (Albacete et al., 2015), it is better understood as a synergistic counterpart to breeding. Physical attachment of rootstocks bred for disease resistance and nutrient use efficiency to scions bred for fruit quality and quantity enables divergent sets of traits to be combined without incurring linkage drag and yield penalties. However, breeding programs have not yet optimized both rootstocks and scions in many grafted perennial crops, including Theobroma cacao.
Valued for its pods, which are used in chocolate, cocoa butter, and other economically important commodities, cacao is grown in lowland tropical regions such as West Africa, Latin America, and Southeast Asia. Modern cacao breeding programs have only existed since the 1930s, despite the popularity of this crop, which translates to a limited number of generations given the timeframe required for breeding perennials (DuVal et al., 2017). To date, these programs have focused primarily on generating elite scions, leaving the development of nutrient-efficient, stress-tolerant, disease-resistant rootstocks as a significant research gap. One reason may be that at least two of the three primary breeding objectives (bean quality, yield, and disease resistance) are controlled by scions, and soil-borne pathogens that can be controlled with resistant rootstocks are not as significant as foliar and pod pathogens in cacao (Bailey and Meinhardt, 2016). Another reason for the relative disinterest in rootstocks may be the dominance of seedlings over vegetative propagation methods in West Africa, which produces 70% of the world's cacao (Yin, 2004). Difficulties inherent in phenotyping belowground traits may also contribute. Yet rootstock breeding could play a critical role in the future of cacao. Meeting rising consumer demand without conversion of additional land to cacao production will require increasing resource use efficiency, especially at the root-soil interface.
Studies of nutrient use efficiency in cacao rootstocks are rare, but have shown potential that should be exploited to develop rootstocks that effectively capture soil nutrients, even in depleted soils. A comparison of two cacao rootstocks found differences in nitrogen use efficiency and growth response that were related to root dry weight (Ribeiro et al., 2008). Evidence from other tropical perennials such as mango supports a role for rootstock as well as scion identity in leaf nutrient content (Sarkhosh et al., 2021). This can be traced in some cases to soil resource acquisition. In grapevine, for example, contrasting effects on scion vigor were traced to differences in responsiveness to heterogeneous soil nitrogen and in transcriptomic reprogramming of nitrogen-related genes (Cochetel et al., 2017). Further exploration of homologous root and rhizosphere traits in cacao is warranted.
Root associations with the rhizosphere microbiome, or rhizobiome, may also play an important role in soil resource acquisition and nutrient use efficiency in perennial systems but have not been fully characterized in cacao. Grafting is known to affect rhizobiome composition: Small but significant rootstock effects on rhizosphere bacterial communities have been identified in grafted tomato (Poudel et al., 2019) and grapevine (D'Amico et al., 2018; Marasco et al., 2018). These effects may be small [around 3% of variation in tomato rhizosphere bacterial communities, far lower than soil compartment and site effects (Poudel et al., 2019)] but stronger than scion effects (Liu et al., 2018). Furthermore, these root-rhizobiome interactions can affect nutrient uptake (D'Amico et al., 2018). Research in cacao has shown that management practices impact fungal communities (Arévalo-Gardini et al., 2020) and total microbial communities (Buyer et al., 2017), but the effect of rootstock identity on root-soil-microbe interactions affecting plant nutrition has not been examined.
We grafted seedlings of eight rootstock populations with three scion genotypes under greenhouse conditions to test the individual and interactive effects of rootstock and scion identity on cacao seedling nutrition and rhizobiome composition. Furthermore, we investigated the nature of plant-soil-microbe interactions affecting nutrient uptake using a variety of statistical techniques. We hypothesized that (a) rootstock and scion identity would both affect leaf nutrient content; (b) rootstock would have a stronger influence than scion on rhizobiome composition; and (c) soil physicochemical parameters and microbial community composition would interactively affect plant nutrition. Because nutritionally relevant plant-microbe interactions could occur at many scales, we explored potential impacts of individual taxa, the entire rhizobiome, and subsets of bacterial and fungal communities.
Eighteen-month-old T. cacao seedlings representing eight different rootstock populations (BYNC, EQX3348, IMC14, PA107, SCA6, T294, T384, T484) were cleft grafted with budwood from three scions (Criollo 22, Matina 1-6, Pound 7) (Table 1). Seedlings were grafted at this developmental stage because graft take is highest when rootstock diameter is close to the diameter of the scion budwood and the seedling has matured enough to flush. These diverse rootstocks were chosen to represent a range of genetic backgrounds, but detailed information about growth and developmental characteristics was not available due to limited research. Similarly, scions were selected from widely planted elite commercial cultivars representing different cacao genetic groups and origins (Table 1) (Motamayor et al., 2008; Cornejo et al., 2018). The decision to use a diverse range of rootstocks and these specific scions was made to reflect commercial production, where a wide variety of rootstocks (often locally adapted seedlings) are commercially planted and grafted with a smaller pool of elite scions, which are chosen primarily for bean quality and yield. In addition, the larger number of rootstocks was chosen because we hypothesized that rootstock effects would be stronger than scion effects.
Grafted seedlings were maintained under controlled greenhouse conditions in the Mars Breeding Greenhouse at the USDA SHRS in Miami, Florida, USA. Seedlings were maintained in 2 gallon polybags in a substrate of 37% pine bark, 25% perlite, 8% sand, and 30% coir with 6 lbs/yard dolomite and 1.66 lbs/yard micro max. This substrate mixture was autoclaved when first mixed to ensure homogeneous initial conditions, then transferred into polybags. Physical, chemical, and microbiological characteristics were not measured in each polybag at this time because the substrate had been thoroughly mixed and was assumed to be the same in all polybags. At 18 months, the plants were transferred to 7 gallon pots. The seedlings received an application of 15 ml of NUTRICOTE 18-6-18 fertilizer (Arysta LifeScience America Inc., New York, NY, USA) every 6 months and applications of 5 ml of K-Mag 0-0-22 (Mosaic Crop Nutrition, Tampa, FL, USA), which contains 10.8% magnesium and 22% sulfur, at planting and transplanting. Greenhouse temperatures were maintained between 68 and 85°F and humidity was between 30 and 95%. Photosynthetically active radiation ranged from 161 to 485 umol/s/m2 and there was no supplemental lighting.
At 4 and 16 months after grafting, leaf tissue was sampled for analysis to assess how grafting affected scion nutrition soon after graft take and a year after establishment. Five leaves were sampled from recently hardened flushes of each plant. Leaves were dried, ground, and analyzed for nutrient concentrations by Waypoint Analytical (Richmond, VA, USA).
To allow time for rootstock effects on soil and microbial communities to develop, soil was collected at 20 months after grafting for analysis of macro- and micronutrients as well as rhizosphere microbial communities. Soil for nutrient analysis was sieved to pass a 2 mm mesh. pH was measured in a 1:1 soil:water slurry, soil organic matter (SOM) was measured via loss-on-ignition, and plant-available nutrient concentrations were measured with the Mehlich 3 method (Mehlich, 1984). Cation exchange capacity (CEC) was calculated from measured cation concentrations and soil pH.
Genomic DNA was extracted from 0.25 g of rhizosphere soil using DNeasy PowerSoil Pro kits (QIAGEN, Germantown, MD, USA). Bacterial and fungal rhizosphere communities were assessed by sequencing of the V4 and ITS1 regions, respectively, on an Illumina Novaseq platform. The original 515F/806R primer pair was used to amplify the V4 region (Caporaso et al., 2011) and the ITS1F/ITS2 primer pair was used for the ITS1 region (White et al., 1990; Gardes and Bruns, 1993).
Primers and adapters were removed and low-quality reads (length < 60 bp, Q score < 5, or unidentified bases > 10%) were filtered out using fastp software (Chen et al., 2018). Subsequent analyses were done in R software v4.0.3 (R Core Team, 2020). Reads were filtered using the dada2 package v1.18 (Callahan et al., 2016) with a maximum of two expected errors. Paired-end reads were denoised, merged, and used to generate amplicon sequence variant (ASV) tables separately for 16S and ITS sequences, containing 44,150 unique prokaryotic ASVs and 5,153 fungal ASVs. Chimeras were removed for both 16S and ITS sequences. For 16S sequences, products of non-specific priming that fell outside the anticipated 250-256 bp length for the V4 region, chloroplast sequences, and mitochondrial sequences were additionally removed. Taxonomy was assigned using the SILVA reference database v138.1 for prokaryotes (Quast et al., 2013; Yilmaz et al., 2014) and the UNITE database v8.2 for fungi (Abarenkov et al., 2020), and species-level identification was done only in the case of 100% sequence identity. Chloroplasts and mitochondria were removed and subsequent analyses of 16S sequences were restricted to bacteria only.
All analyses were done in R software v4.0.3 (R Core Team, 2020). Leaf nutrient data were scaled to have unit variance and principal components analysis (PCA) was used to ordinate data from each sampling date separately [prcomp function of stats package (R Core Team, 2020)].
Microbial diversity and community composition were investigated separately for bacterial and fungal communities using the vegan package v.2.5-7 (Oksanen et al., 2020). Alpha diversity metrics included richness, Shannon index, and Pielou index (taxonomy-based methods) and Faith's PD (a phylogeny-based method). The effects of rootstock, scion, and the rootstock:scion interaction on microbial alpha diversity metrics were tested with ANOVA followed by post-hoc Tukey tests. Microbial communities were ordinated with principal coordinates analysis (PCoA) based on Bray-Curtis distance matrices of log-transformed count data. Effects of rootstock, scion, and the rootstock:scion interaction on community composition were tested with permutational multivariate analysis of variance (PERMANOVA) with 1,000 permutations (Adonis function of vegan package; Oksanen et al., 2020).
Because the functional relevance of the microbiome for plant nutrition was of interest, we sought to identify individual taxa or groups of taxa associated with variation in leaf nutrients. Kendall correlations between leaf nutrients and microbial ASVs were calculated to test potential direct effects of individual bacterial and fungal taxa on plant nutrition (cor function of stats package). Full Mantel tests were carried out on Bray-Curtis dissimilarity matrices of leaf nutrients, bacterial communities, and fungal communities to assess direct relationships among these datasets (mantel function of vegan package). Partial Mantel tests were used to test correlations between plant and microbial communities when soil nutrients were held constant. Relationships among soil nutrients, microbial communities, and leaf nutrients were then investigated with stepwise regression. Individual models were established for each leaf nutrient from the 16-month sampling point. Predictor variables included all soil nutrients, all measures of microbial diversity discussed previously, and the first two eigenvectors of bacterial and fungal communities. Stepwise regression was implemented and variable importance was calculated for each predictor variable using the leapSeq method in the train function of the caret package v6.0.86 with 10-fold cross-validation and all possible variables (Kuhn, 2020).
Linear discriminant analysis effect size (LEfSe), an ensemble learning approach that identifies features responsible for differentiation among groups, was used to identify rootstock-responsive bacterial and fungal ASVs with potential impacts on plant nutrition. Rootstock-responsive taxa were defined as those that differ significantly between rootstock genotypes. LEfSE identifies microbiome features that may explain differences among groups, tests their significance with Kruskal-Wallis and Wilcoxon rank-sum tests, then uses redundancy analysis to estimate the effect size associated with each feature (Segata et al., 2011). This method was implemented with the lefse function of the microbiomeMarker package v0.0.1.9000 (Cao, 2021). Functionally relevant taxa were prioritized by analyzing the 50 bacterial and fungal ASVs with the highest loadings on the first two eigenvectors, which were shown to be important for plant nutrition in the stepwise regression analysis.
Ranges of leaf nutrient values for grafted seedlings 4 and 16 months after grafting and normal reference ranges are given in Table 2. Four months after grafting, PCA revealed that 46% of variation in leaf nutrients was accounted for by the first two eigenvectors (Figure 1A). Ca, Mg, and Zn had the highest loadings on the first eigenvector, while N, Cu, and P had the highest loadings on the second eigenvector. Significant correlations were also observed between Ca – Mg (p < 0.001), Ca – Zn (p < 0.05), and P – Cu (p < 0.05; Supplementary Figure 1). At 16 months, variation among samples was driven by K, P, Ca, and B in the first eigenvector and Fe, Cu, and Al in the second eigenvector (Figure 1B). Correlations among nutrients were stronger at this point, with three main nutrient groupings that were positively correlated with one another and negatively correlated with other groupings: P-K, S-Ca-Mg-B-Zn-Mn, Fe-Al (Supplementary Figure 2). Some clustering by rootstock was apparent at 4 months, and two groups of rootstocks (BYNC, EQX3348, IMC14, PA107 vs. SCA6, T294, T384, T484) were visible at 16 months (Figure 1).
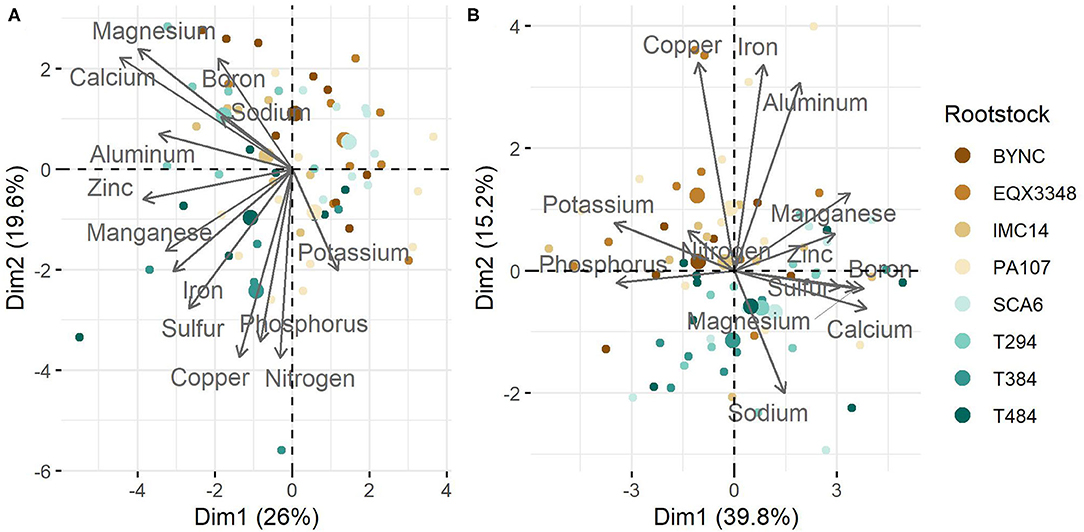
Figure 1. Principal components analysis (PCA) of cacao leaf nutrients at two sampling dates. Values were scaled to have unit variance prior to analysis. (A) At 4 months, 46% of variation in leaf nutrients was accounted for by the first two eigenvectors. Ca, Mg, and Zn had the highest loadings for the first eigenvector (Dim1 above) and N, Cu, and P had the highest loadings for the second eigenvector (Dim2). (B) At 16 months, 55% of variation was accounted for by the first two eigenvectors, which were driven by K, P, Ca, and B (Dim1) and Fe, Cu, and Al (Dim2).
Rootstock, scion, and the rootstock:scion interaction significantly affected leaf nutrients at both sampling dates. At 4 months, the rootstock:scion interaction was significant for B (p < 0.05) and rootstock effects were significant for N (p < 0.001), P (p < 0.01), K (p < 0.05), Mg (p < 0.05), Ca (p < 0.001), Zn (p < 0.001), Mn (p < 0.01), Fe (p < 0.001), Cu (p < 0.05), and Al (p < 0.001). Only Na responded to scion genotype (p < 0.01). At 16 months, fewer nutrients were affected by rootstock or scion identity. B (p < 0.05), Fe (p < 0.001), and Al (p < 0.001) were affected by the rootstock:scion interaction, while only Na (p < 0.01) and Cu (p < 0.001) differed among rootstocks. No nutrients were affected by scion identity at 16 months.
Rootstock identity (but not scion or the rootstock:scion interaction) significantly affected both phylogeny-based and taxonomy-based measures of bacterial alpha diversity (all p < 0.001; Figure 2A). A trend was observed for all measures in which bacterial communities associated with SCA6, T294, T384, and T484 were less diverse than bacterial communities associated with BYNC, EQX3348, IMC14, and PA107, although this distinction was significant only for richness.
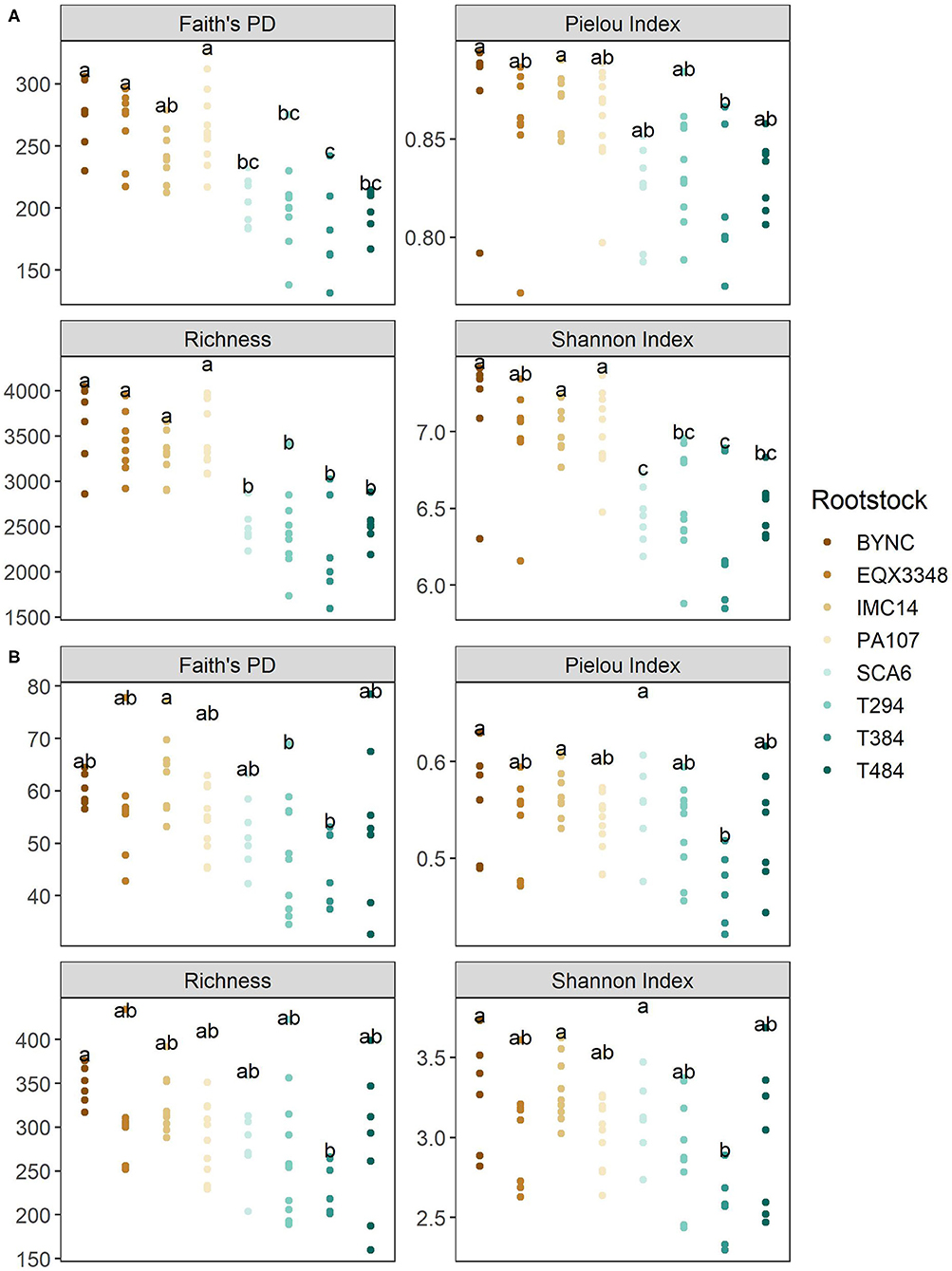
Figure 2. Cacao rootstock effects on microbial alpha diversity. (A) In bacterial communities, Faith's PD (a diversity metric that accounts for phylogeny), the Pielou and Shannon indices (integrating richness and evenness), and taxonomic richness were significantly affected by rootstock (all p < 0.05). Lower diversity was observed in bacterial communities associated with SCA6, T294, T384, and T484. (B) In fungal communities, rootstock identity significantly affected all measures of alpha diversity (all p < 0.05). Fungi associated with T384 showed the lowest alpha diversity.
Rootstock identity (but not scion or rootstock:scion interaction) similarly affected all measures of fungal alpha diversity (richness p < 0.05, Shannon index p < 0.01, Pielou index p < 0.05, Faith's PD p < 0.05; Figure 2B). Unlike for bacteria, however, decreased diversity was observed only in rootstock T384.
PCoA showed clustering of bacterial and fungal communities by two groups of rootstocks (Figure 3). Rhizosphere communities associated with BYNC, EQX3348, IMC14, and PA107 formed a distinct cluster separated from communities associated with SCA6, T294, T384, and T48. PERMANOVA revealed significant effects of rootstock (bacterial R2 = 0.43, fungal R2 = 0.19, both p < 0.001) but not scion or the rootstock:scion interaction on bacterial and fungal communities. The rootstock effect was also observed when the ordination was based on weighted UniFrac distance, which accounts for phylogeny, rather than Bray-Curtis distance (both PERMANOVA p < 0.001, data not shown).
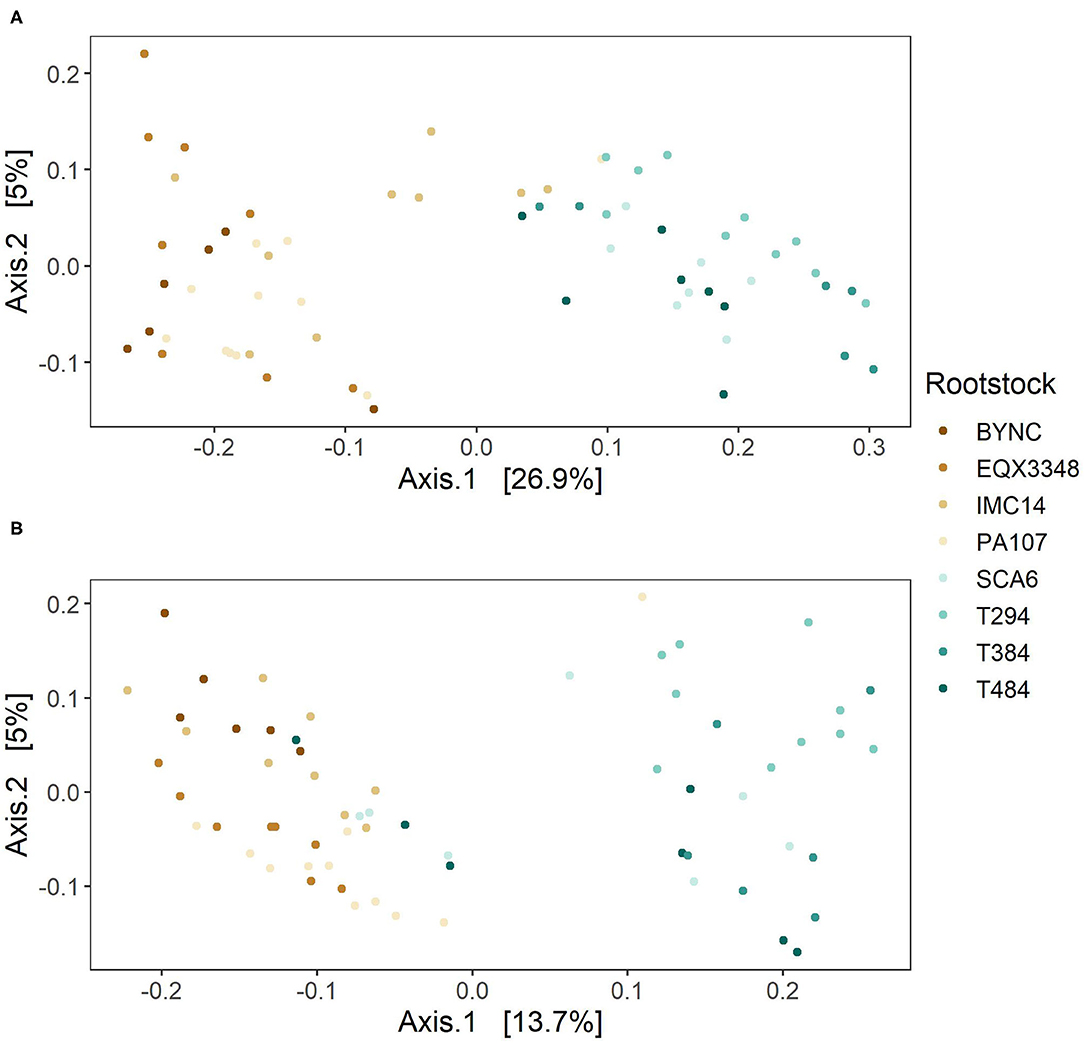
Figure 3. Principal coordinates analysis (PCoA) ordination of rhizobiomes. Separation of (A) bacterial communities and (B) fungal communities by two groups of rootstocks occurred primarily along the first axis, which accounted for 26.9% of variation in bacterial communities and 13.7% of variation in fungal communities. Further clustering by rootstock within each group was evident along the second axis, which accounted for 5% of variation in both bacterial and fungal communities.
High bacterial richness and evenness was observed at the order level, with 303 unique orders represented by at least one ASV (Supplementary Figure 3). The Rhizobiales were the most abundant order with 2,486 ASVs across all samples, followed by the Burkholderiales (1,124) and Chitinophagales (848). Fungal ASVs were distributed across 74 orders, but samples were dominated by fewer taxonomic groups (Supplementary Figure 4). Although over half of fungal sequences were not identifiable to the order level, the Agaricales and Hypocreales were the highest-abundance identifiable orders with 505 and 259 ASVs, respectively.
Correlation analysis did not identify individual bacterial or fungal ASVs whose abundance was strongly and significantly correlated with the concentration of any leaf nutrient (Kendall correlation > 0.75, p < 0.05). Full and partial Mantel tests based on Bray-Curtis dissimilarity matrices were used to test for significant relationships among leaf nutrients, soil nutrients, and bacterial and fungal communities. Leaf nutrients did not vary significantly with either bacterial or fungal communities (full Mantel test, both p > 0.05), even if soil nutrients were held constant (partial Mantel test, both p > 0.05). However, bacterial and fungal communities were correlated with one another (full Mantel test, p < 0.05).
Stepwise regression was implemented for each leaf nutrient individually to investigate the combinations of soil physicochemical properties and microbial parameters that affect foliar nutrition (Supplementary Tables 1–5). Analysis of the relative variable importance in each model showed that measures of microbial diversity and community composition contributed substantially to variation in many leaf nutrients (Figure 4). For example, richness and Faith's PD of bacterial communities were the most important predictor variables in the regression models defined for leaf N, Ca, Zn, and Al, and the second eigenvector of fungal communities (PC2F) was the most important predictor variable for leaf Na. Among soil physicochemical properties, only SOM and Zn had the highest variable importance in multiple models (SOM: leaf Mg, Mn; soil Zn: leaf P, K, and Cu).
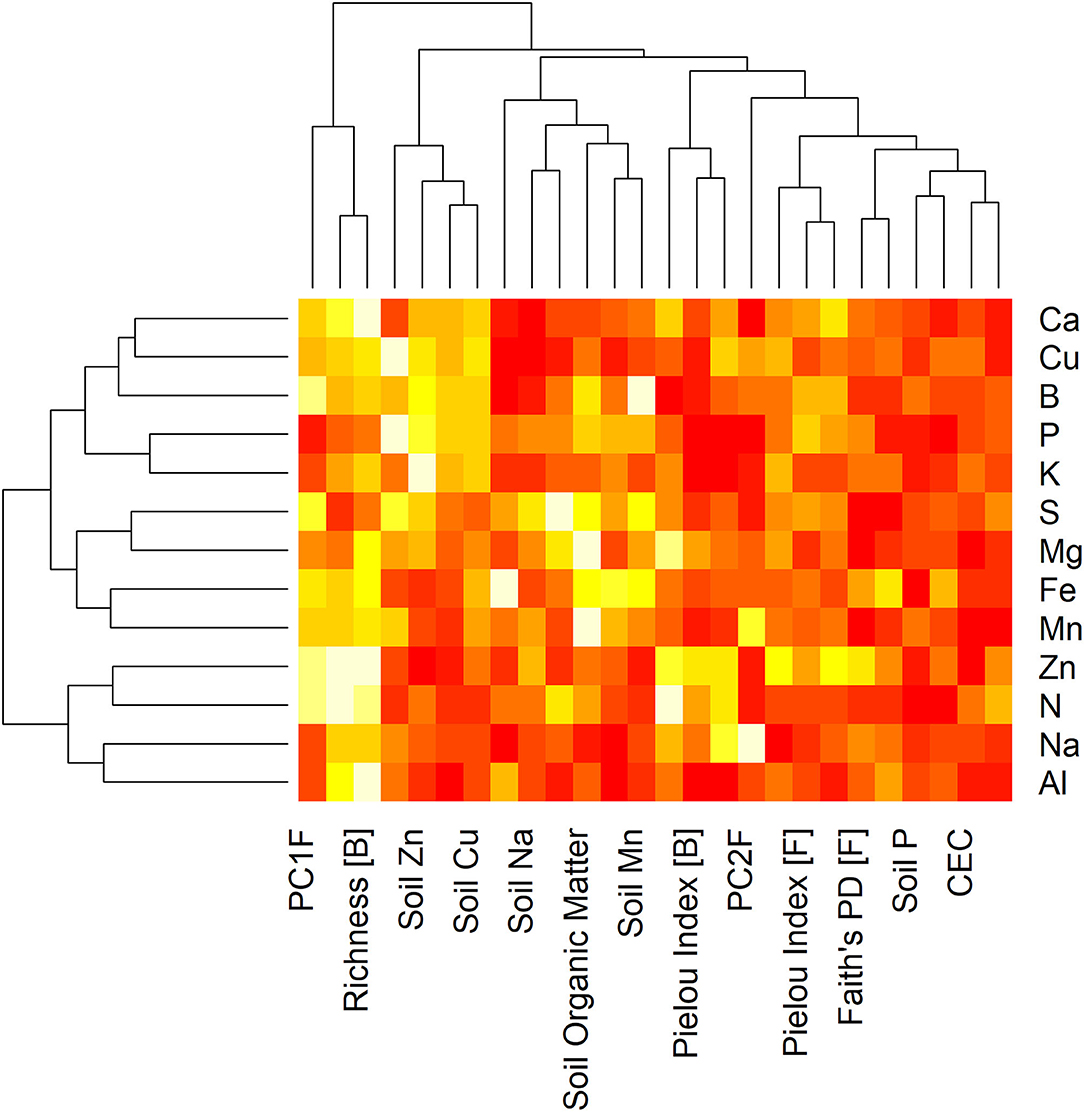
Figure 4. Variable importance heatmap and dendrograms for leaf nutrient models. Stepwise regression analysis identified linear models with the combination of soil physicochemical properties and microbial diversity and composition metrics that best explained the concentrations of each leaf nutrient individually. The heatmap portrays the relative importance of each explanatory variable (columns) in each linear model (rows by response variable). Hierarchical clustering dendrograms along the “x” and “y” axes show similarity among variables and models, respectively. PC1B, PC2B: first and second principal components of bacterial community; PC1F, PC2F: first and second principal components of fungal community; CEC: cation exchange capacity; [B]: bacteria; [F]: fungi.
The Rhizobiales were the most-represented order among rootstock-responsive bacteria potentially related to plant nutrition, and these were enriched in BYNC, EQX3348, and T384 (Figure 5). BYNC, EQX3348, T294, and T384 were all enriched in multiple bacterial orders. In contrast, IMC14 was enriched only in Pseudomonodales, SCA6 in the Bacillales, and PA107 and T484 in groups not identified at the order level.
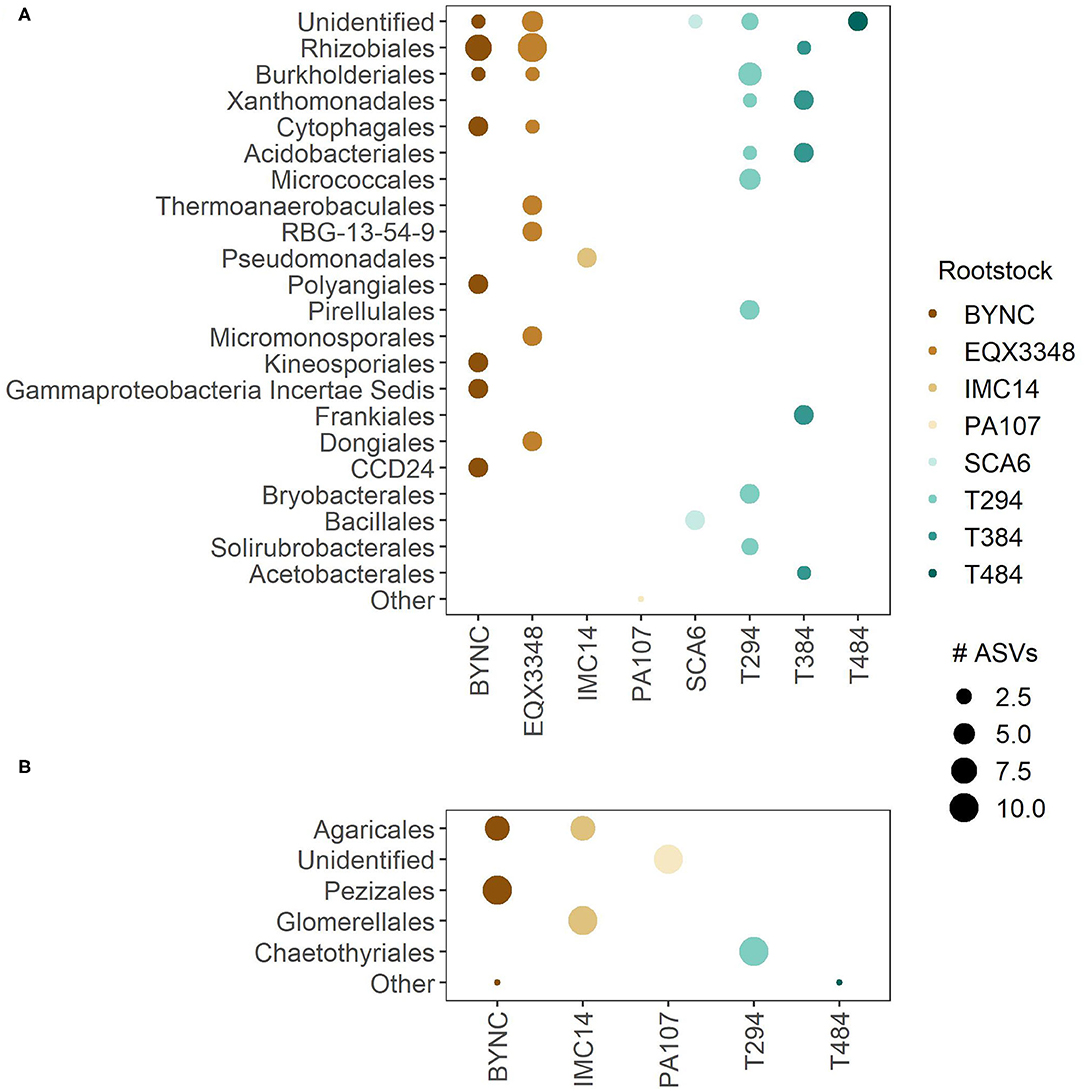
Figure 5. Distribution of rootstock-responsive taxa with potential impacts on plant nutrition. The linear discriminant analysis effect size (LEfSe) method identified (A) bacteria and (B) fungi responsible for differentiation among rootstocks (LDA > 2, p < 0.05). To focus the analysis on taxa potentially relevant for plant nutrition, LEfSe was applied to bacterial and fungal amplicon sequence variants (ASVs) with the highest loadings on the first two eigenvectors used in leaf nutrient models (PC1B, PC2B, PC1F, and PC2F in Figure 4). ASVs are identified to order and classified as “Unidentified” if only higher taxonomic levels were available. “Other” is used to represent orders with fewer than three members. Dots represent the number of ASVs from a given order whose relative abundance was enriched in one rootstock.
BYNC and IMC14 were enriched in the Agaricales, the most-represented order among root-responsive fungi (Figure 5). Other enriched orders were the Pezizales (in BYNC), Glomerellales (in IMC14), and Chaetothyriales (in T294). Enriched fungal groups in PA107 and T484 were not identified at the order level. EQX3348, SCA6, and T384 were not enriched in any rootstock-responsive taxa.
Rootstock-responsive bacteria and fungi were similar to the overall rhizobiome composition (Supplementary Figures 3, 4). The most abundant bacterial orders were consistent between the two datasets, but the composition of fungal communities was more variable. In addition, a large number of fungal sequences could be identified only at the kingdom, phylum, or class levels, making higher-resolution analysis of community composition difficult.
Our findings of dominant rootstock effects on plant nutrition and microbial interactions in cacao suggest that rootstock breeding could be employed to increase nutrient use efficiency. In addition, this strategy would not compromise cacao bean quality, which is under scion control (Prawoto, 1990). Differences in plant nutrition among rootstocks and scions appear to be important primarily for the first few months after grafting. If differences had been observed among rootstocks in nutrient accumulation at both time points, it would have warranted investigation of the genetic basis of differences in nutrient uptake and partitioning as well as rootstock-specific microbial taxa that may have contributed. As the plant matures, plant physiological processes may demand stoichiometric nutrient ratios regardless of plant genotype, an explanation supported by the stronger correlations among nutrients at sixteen as compared to 4 months after grafting (Supplementary Figures 1, 2). Nonetheless, juvenile vigor of rootstocks and young grafts is a relevant trait for field production and a better understanding of the genes responsible for differentiation among rootstocks and scions in this study could help inform cacao breeding programs. A useful next step would be to validate these results by planting the same genotypes in the field under fertile and nutrient-limited conditions.
In accordance with our second hypothesis, rootstock effects outweighed scion effects on root-associated microbes. Bacterial diversity and community composition of both bacteria and fungi followed a similar trend in which rootstocks clustered in two groups: BYNC/EQX3348/IMC14/PA107 and SCA6/T294/T384/T484. Identifying the mechanisms responsible for the higher bacterial diversity in the first group and differences in rhizobiome composition was beyond the scope of this study, but we suggest that genome sequencing and analysis of root exudates from all eight rootstock populations would be useful starting points. The functional implications of reduced bacterial and fungal diversity for nutrient uptake by rootstocks SCA6, T294, T384, and T484 also deserve further investigation. D'Amico et al. (2018) found reduced rhizosphere bacterial diversity in one Vitis vinifera rootstock, including lower abundance of potassium-solubilizing taxa, that was hypothesized to be related to frequent potassium deficiencies in that rootstock.
We used multiple statistical methods to test our hypothesis that soil physicochemical parameters and microbial communities would impact plant nutrition, and to understand the nature of these relationships. The results of stepwise regression show that it may be important to develop new approaches to cacao nutrition that factor in soil biology as well as fertility. However, because this was a greenhouse study, limited variation in soil physicochemical parameters may have led to underestimation of the importance of these variables for plant nutrition. Applying the statistical methods used in this study to field data would be a valuable area for future research. Soil pH, for example, strongly affects nutrient availability and microbial communities in tropical agroecosystems (Juo and Franzluebbers, 2003), but did not show up as particularly important in stepwise regression, likely because of low variability in this study (Figure 4). Interestingly, despite limited variation, soil organic matter (SOM, also a key management concern in tropical soils and particularly in intensive cacao production systems) was the most important variable in multiple plant nutrient models (Figure 4). Increasing SOM in soils under cacao should remain a high priority given its apparent role in plant nutrition as well as significance for microbially-mediated soil health and C sequestration.
We interpret the insignificant nutrient-microbe correlations and Mantel tests and the results of stepwise regression as a call for more complex understanding of plant-soil-microbe interactions at intermediate scales, e.g., between binary associations and entire communities. Identification of functional guilds, statistical methods to identify the most pertinent features of high-dimensionality datasets, and synthetic-community-based studies could prove useful here among other methods (recently reviewed by Gupta et al., 2021). Plant growth promotion or enhanced plant nutrient content mediated by these intermediate-scale groups of microorganisms could result from direct mechanisms (e.g., direct nutrient transfer by symbionts) or indirect mechanisms (e.g., increased bioavailability of soil nutrients), but in either case would be an emergent property of the group rather than attributable to a single organism. Still, we gain insight into potentially beneficial members of the rootstock-responsive taxa identified in this study from single-strain or low-complexity inoculation studies common in the literature.
Many of the rootstock-responsive bacterial taxa identified belonged to orders associated with plant growth promotion, although incomplete taxonomic assignments limit our ability to confirm the presence of plant-growth-promoting rhizobacteria (PGPR) here. For example, the Rhizobiales, Frankiales, and Xanthomonodales, identified as enriched in BYNC, EQX3384, T294, and T384 (Figure 5) include free-living and associative nitrogen-fixing bacteria. Unfortunately, these could be identified to the genus level only in some cases (Pedomicrobium, Devosia, Mycoplana, Hyphomicrobium, Bauldia, Rhodanobacter, Chujaibacter, and Jatrophihabitans), and N fixation is not a conserved trait for all of these genera (Rivas et al., 2002; DeAngelis and Pold, 2020). The Rhizobiales, which include both diazotrophs and pathogens (Carvalho et al., 2010), were the most abundant order across all samples (Supplementary Figure 3). Future metagenomic studies of the cacao rhizobiome could screen for gene clusters involved in N fixation that might become part of biologically-based nutrient management strategies.
The Pseudomonodales and Bacillales, present at low relative abundance in all rhizobiome samples and identified in rootstock-responsive bacteria (Supplementary Figure 3, “Other”), contain taxa capable of increasing plant productivity in many plant hosts by both direct and indirect methods, e.g., producing growth-promoting auxins or suppressing pathogens (Kloepper et al., 2004; Lugtenberg and Kamilova, 2009). Furthermore, strains belonging to the Bacillales have shown promise in cacao. Bacillus subtilis inoculation of germinating seeds increased leaf area by 14%, shoot height by 8%, and graft success rate by 24% (Falcäo et al., 2014), and repeated inoculations of seeds and seedlings increased shoot height by 22% (Leite et al., 2013). Inoculation of 1-month-old cacao seedlings with other Bacillus spp. increased height and biomass by up to 37 and 73%, respectively (Gupta et al., 2011). However, inoculation with Pseudomonas sp. did not promote cacao growth in another study (Guillermo Ramírez et al., 2016).
As with bacteria, incomplete taxonomic assignments limit our ability to clarify the ecological roles of many of the rootstock-responsive fungi. Unfortunately, many of the rootstock-responsive fungi could not be identified by genus or species, the taxonomic levels used for functional guild assignment in fungal trait databases such as FunFun (Zanne et al., 2020), FUNGuild (Nguyen et al., 2016), and FungalTraits (Põlme et al., 2020). Two of the four fungal orders detected, the Glomerellales and the Chaetothyriales, have been found at low abundance (2.0–2.3%) in the cacao phyllosphere, but their roles there were not defined (Wemheuer et al., 2020). The majority of studies investigating fungi as potential plant-growth-promoting agents in cacao have focused on mycorrhizae (Chulan and Ragu, 1986; Cuenca et al., 1990; Chulan and Martin, 1992; Tchameni et al., 2012; Guillermo Ramírez et al., 2016) or Trichoderma sp. (Bae et al., 2009; Tchameni et al., 2011). While 112 ASVs belonging to the genus Trichoderma and 8 belonging to the mycorrhizal order Glomerales were found in fungal rhizobiomes in this study, none were among the rootstock-responsive taxa (Supplementary Figure 4).
The possible presence of pathogenic taxa among rootstock-responsive fungi could have had detrimental impacts on leaf nutrition in diseased plants. The Glomerellales (not to be confused with the Glomerales) contain Colletotrichum sp., cacao pathogens responsible for anthracnose (Nascimento et al., 2019). While two of the rootstock-responsive Glomerellales strains were identified as Musicillium sp., the other two were not identified to the genus level and the possible presence of Colletotrichum sp. could not be ruled out. A member of the Hypocreales enriched in T484 (Figure 5, “Other”) was identified as Fusarium sp. This genus is present at low relative abundance in cacao agroforestry systems (e.g., 1.3–6.0%, Arévalo-Gardini et al., 2020). While it contains pathogens that cause cushion gall, dieback, and other diseases in cacao (Ploetz, 2006; Adu-Acheampong et al., 2012; Rosmana et al., 2013), it also contains strains that may provide biocontrol against Phytophthora palmivora, the causal agent of black pod rot (Hanada et al., 2010).
Despite limited attention to cacao rootstocks and rhizosphere microbial associations to date, our results highlight the potential of coordinated breeding efforts in these areas, combined with informed soil nutrient management, to enhance plant nutrition in cacao. This will be a challenging endeavor for numerous reasons: time and cost required to generate new perennial germplasm, dispersed smallholder production adding logistical difficulties to participatory cacao research and breeding, and remaining knowledge gaps in the genetic basis of cacao-rhizobiome interactions. Nonetheless, significant interest by consumers and industry in sustainable cacao production will help advance the field and translate research into practice.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: NCBI (accession: PRJNA755178).
AD, AP, AT, and JS contributed to the study design and data collection. JS, AD, and TC analyzed the data. JS wrote the manuscript with input from all authors. All authors approved the final version for publication.
JS, AD, AT, and TC are employed by Mars Wrigley.
The remaining author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The authors would like to acknowledge Alana Firl and David Mackill for bioinformatics and project support.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fagro.2021.754646/full#supplementary-material
Abarenkov, K., Zirk, A., Piirmann, T., Pöhönen, R., Ivanov, F., Nilsson, R. H., et al. (2020). UNITE general FASTA Release for Fungi. Version 04.02.2020. UNITE Community. Available online at: https://doi.org/10.15156/BIO/786368 (accessed April 8, 2021).
Adu-Acheampong, R., Archer, S., and Leather, S. (2012). Resistance to dieback disease caused by Fusarium and Lasiodiplodia species in cacao (Theobroma cacao L.) genotypes. Exp. Agric. 48, 85–98. doi: 10.1017/S0014479711000883
Albacete, A., Martínez-Andújar, C., Martínez-Pérez, A., Thompson, A. J., Dodd, I. C., and Pérez-Alfocea, F. (2015). Unravelling rootstock×scion interactions to improve food security. J. Exp. Bot. 66, 2211–2226. doi: 10.1093/jxb/erv027
Arévalo-Gardini, E., Canto, M., Alegre, J., Arévalo-Hernández, C. O., Loli, O., Julca, A., et al. (2020). Cacao agroforestry management systems effects on soil fungi diversity in the Peruvian Amazon. Ecol. Indic. 115:106404. doi: 10.1016/j.ecolind.2020.106404
Bae, H., Sicher, R. C., Kim, M. S., Kim, S.-H., Strem, M. D., Melnick, R. L., et al. (2009). The beneficial endophyte Trichoderma hamatum isolate DIS 219b promotes growth and delays the onset of the drought response in Theobroma cacao. J. Exp. Bot. 60, 3279–3295. doi: 10.1093/jxb/erp165
Bailey, B. A., and Meinhardt, L. W. (2016). Cacao Diseases: A History of Old Enemies and New Encounters. Basel: Springer International Publishing.
Buyer, J. S., Baligar, V. C., He, Z., and Arévalo-Gardini, E. (2017). Soil microbial communities under cacao agroforestry and cover crop systems in Peru. Appl. Soil Ecol. 120, 273–280. doi: 10.1016/j.apsoil.2017.09.009
Callahan, B. J., McMurdie, P. J., Rosen, M. J., Han, A. W., Johnson, A. J. A., and Holmes, S. P. (2016). DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583. doi: 10.1038/nmeth.3869
Caporaso, J. G., Lauber, C. L., Walters, W. A., Berg-Lyons, D., Lozupone, C. A., Turnbaugh, P. J., et al. (2011). Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. U. S. A. 108, 4516–4522. doi: 10.1073/pnas.1000080107
Carvalho, F. M., Souza, R. C., Barcellos, F. G., Hungria, M., and Vasconcelos, A. T. R. (2010). Genomic and evolutionary comparisons of diazotrophic and pathogenic bacteria of the order Rhizobiales. BMC Microbiol. 10, 1–15. doi: 10.1186/1471-2180-10-37
Chen, S., Zhou, Y., Chen, Y., and Gu, J. (2018). fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890. doi: 10.1093/bioinformatics/bty560
Chulan, A., and Ragu, P. (1986). Growth response of Theobroma cacao L. seedlings to inoculation with vesicular-arbuscular mycorrhizal fungi. Plant Soil 285, 279–285. doi: 10.1007/BF02374771
Chulan, H. A., and Martin, K. (1992). The vesicular-arbuscular (VA) mycorrhiza and its effects on growth of vegetatively propagated Theobroma cacao L. Plant Soil 144, 227–233. doi: 10.1007/BF00012879
Cochetel, N., Escudié, F., Cookson, S. J., Dai, Z., Vivin, P., Bert, P. F., et al. (2017). Root transcriptomic responses of grafted grapevines to heterogeneous nitrogen availability depend on rootstock genotype. J. Exp. Bot. 68, 4339–4355. doi: 10.1093/jxb/erx224
Cornejo, O. E., Yee, M.-C., Dominguez, V., Andrews, M., Sockell, A., Strandberg, E., et al. (2018). Population genomic analyses of the chocolate tree, Theobroma cacao L., provide insights into its domestication process. Commun. Biol. 1:167. doi: 10.1038/s42003-018-0168-6
Cuenca, G., Herrera, R., and Meneses, E. (1990). Effects of VA mycorrhiza on the growth of cacao seedlings under nursery conditions in Venezuela. Plant Soil 126, 71–78. doi: 10.1007/BF00041370
D'Amico, F., Candela, M., Turroni, S., Biagi, E., Brigidi, P., Bega, A., et al. (2018). The rootstock regulates microbiome diversity in root and rhizosphere compartments of vitis vinifera cultivar lambrusco. Front. Microbiol. 9:2240. doi: 10.3389/fmicb.2018.02240
DeAngelis, K. M., and Pold, G. (2020). Genome sequences of Frankineae sp. strain MT45 and Jatrophihabitans sp. strain GAS493, two actinobacteria isolated from forest soil. Microbiol. Resour. Announc. 9:e00614-20. doi: 10.1128/MRA.00614-20
DuVal, A., Gezan, S. A., Mustiga, G., Stack, C., Marelli, J. P., Chaparro, J., et al. (2017). Genetic parameters and the impact of off-types for Theobroma cacao L. in a breeding program in Brazil. Front. Plant Sci. 8:2059. doi: 10.3389/fpls.2017.02059
Falcäo, L. L., Silva-Werneck, J. O., Vilarinho, B. R., da Silva, J. P., Pomella, A. W. V., and Marcellino, L. H. (2014). Antimicrobial and plant growth-promoting properties of the cacao endophyte Bacillus subtilis ALB629. J. Appl. Microbiol. 116, 1584–1592. doi: 10.1111/jam.12485
Gardes, M., and Bruns, T. D. (1993). ITS primers with enhanced specificity for basidiomycetes - application to the identification of mycorrhizae and rusts. Mol. Ecol. 2, 113–118. doi: 10.1111/j.1365-294X.1993.tb00005.x
Guillermo Ramírez, J., Osorno, L., and Walter Osorio, N. (2016). Presence of mycorrhizal fungi and a fluorescent Pseudomonas sp. in the rhizosphere of cacao in two agroecosystems and their effects on cacao seedling growth. Agron. Colomb. 34, 385–392. doi: 10.15446/agron.colomb.v34n3.57950
Gupta, A., Gopal, M., and Thomas, G. V. (2011). Efficacy of Rhizospheric Bacillus spp. for Growth Promotion in Theobroma cacao L. seedlings. Available online at: https://www.researchgate.net/publication/237862366 (accessed July 29, 2020).
Gupta, G., Ndiaye, A., and Filteau, M. (2021). Leveraging experimental strategies to capture different dimensions of microbial interactions. Front. Microbiol. 12:700752. doi: 10.3389/fmicb.2021.700752
Hanada, R. E., Pomella, A. W. V., Costa, H. S., Bezerra, J. L., Loguercio, L. L., and Pereira, J. O. (2010). Endophytic fungal diversity in Theobroma cacao (cacao) and T. grandiflorum (cupuaçu) trees and their potential for growth promotion and biocontrol of black-pod disease. Fungal Biol. 114, 901–910. doi: 10.1016/j.funbio.2010.08.006
Juo, A. S. R., and Franzluebbers, K. (2003). Tropical Soils: Properties and Management for Sustainable Agriculture. New York, NY: Oxford University Press. Available online at: https://books.google.com/books?hl=en&lr=&id=dVGgQcuoibAC&oi=fnd&pg=PA3&dq=franzluebbers+tropical+soil+sustainable&ots=I-fie2C3ed&sig=C2BfBRHlUTEqHVI1HCUEC1WVHEc#v=onepage&q=franzluebbers tropical soil sustainable&f=false (accessed June 25, 2021).
Kloepper, J. W., Ryu, C.-M., and Zhang, S. (2004). Induced systemic resistance and promotion of plant growth by Bacillus sp. Phytopathology 94, 1259–1266. doi: 10.1094/PHYTO.2004.94.11.1259
Leite, H. A. C., Silva, A. B., Gomes, F. P., Gramacho, K. P., Faria, J. C., De Souza, J. T., et al. (2013). Bacillus subtilis and Enterobacter cloacae endophytes from healthy Theobroma cacao L. trees can systemically colonize seedlings and promote growth. Appl. Microbiol. Biotechnol. 97, 2639–2651. doi: 10.1007/s00253-012-4574-2
Liu, J., Abdelfattah, A., Norelli, J., Burchard, E., Schena, L., Droby, S., et al. (2018). Apple endophytic microbiota of different rootstock/scion combinations suggests a genotype-specific influence. Microbiome 6:18. doi: 10.1186/s40168-018-0403-x
Lugtenberg, B., and Kamilova, F. (2009). Plant-growth-promoting rhizobacteria. Annu. Rev. Microbiol. 63, 541–556. doi: 10.1146/annurev.micro.62.081307.162918
Marasco, R., Rolli, E., Fusi, M., Michoud, G., and Daffonchio, D. (2018). Grapevine rootstocks shape underground bacterial microbiome and networking but not potential functionality. Microbiome 6:3. doi: 10.1186/s40168-017-0391-2
Mehlich, A. (1984). Mehlich 3 soil test extractant: a modification of mehlich 2 extractant. Commun. Soil Sci. Plant Anal. 15, 1409–1416. doi: 10.1080/00103628409367568
Motamayor, J. C., Lachenaud, P., da Silva E Mota, J. W., Loor, R., Kuhn, D. N., Brown, J. S., et al. (2008). Geographic and genetic population differentiation of the Amazonian chocolate tree (Theobroma cacao L). PLoS ONE 3:e3311. doi: 10.1371/journal.pone.0003311
Nascimento, A. D., Lima, M. O., Feijó, F. M., Júnior, J. H., Sobrinho, R. R., Assunção, I. P., et al. (2019). First report of colletotrichum aeschynomenes causing anthracnose in cacao (Theobroma cacao) in Brazil. Plant Dis. 103, 3284–3284. doi: 10.1094/PDIS-11-18-2047-PDN
Nguyen, N. H., Song, Z., Bates, S. T., Branco, S., Tedersoo, L., Menke, J., et al. (2016). FUNGuild: an open annotation tool for parsing fungal community datasets by ecological guild. Fungal Ecol. 20, 241–248. doi: 10.1016/j.funeco.2015.06.006
Oksanen, J., Blanchet, F. G., Friendly, M., Kindt, R., Legendre, P., Mcglinn, D., et al. (2020). Package “vegan” Title Community Ecology Package Version 2.5-7.
Ploetz, R. C. (2006). Fusarium-induced diseases of tropical perennial crops fusarium-induced diseases of tropical, perennial crops. Phytopathology 96, 648–652. doi: 10.1094/PHYTO-96-0648
Põlme, S., Abarenkov, K., Henrik Nilsson, R., Lindahl, B. D., Clemmensen, K. E., Kauserud, H., et al. (2020). FungalTraits: a user-friendly traits database of fungi and fungus-like stramenopiles. Fungal Divers 105, 1–16. doi: 10.1007/s13225-020-00466-2
Poudel, R., Jumpponen, A., Kennelly, M. M., Rivard, C. L., Gomez-Montano, L., and Garrett, K. A. (2019). Rootstocks shape the rhizobiome: rhizosphere and endosphere bacterial communities in the grafted tomato system. Appl. Environ. Microbiol. 85, e01765-18. doi: 10.1128/AEM.01765-18
Prawoto, A. (1990). A Study on cocoa (Theobroma cacao L.) budding V. The influence of rootstocks on scion-yield quality. Pelita Perkeb. 6, 38–46. doi: 10.22302/iccri.jur.pelitaperkebunan.v6i2.366
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., et al. (2013). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596. doi: 10.1093/nar/gks1219
R Core Team (2020). R: A Language and Environment for Statistical Computing. Available online at: https://www.r-project.org/ (accessed March 30, 2021).
Ribeiro, M. A. Q., Da Silva, J O, Aitken, W. M., Machado, R. C. R., and Baligar, V. C. (2008). Nitrogen use efficiency in cacao genotypes. J. Plant Nutr. 31, 239–249. doi: 10.1080/01904160701853720
Rivas, R., Velázquez, E., Willems, A., Vizcaíno, N., Subba-Rao, N. S., Mateos, P. F., et al. (2002). A new species of Devosia that forms a unique nitrogen-fixing root-nodule symbiosis with the aquatic legume Neptunia natans (L.f.) Druce. Appl. Environ. Microbiol. 68, 5217–5222. doi: 10.1128/AEM.68.11.5217-5222.2002
Rosmana, A., Hikmawati, H., and Asman, A. (2013). Identification of a disease on cocoa caused by Fusarium in Sulawesi. Pelita Perkeb. 29, 210–219. doi: 10.22302/iccri.jur.pelitaperkebunan.v29i3.13
Sarkhosh, A., Shahkoomahally, S., Asis, C., and McConchie, C. (2021). Influence of rootstocks on scion leaf mineral content in mango tree (Mangifera indica L.). Hortic. Environ. Biotechnol. 62, 725–735. doi: 10.1007/s13580-021-00355-w
Segata, N., Izard, J., Waldron, L., Gevers, D., Miropolsky, L., Garrett, W. S., et al. (2011). Metagenomic biomarker discovery and explanation. Genome Biol. 12:R60. doi: 10.1186/gb-2011-12-6-r60
Tchameni, S. N., Ngonkeu, M. E. L., Begoude, B. A. D., Wakam Nana, L., Fokom, R., Owona, A. D., et al. (2011). Effect of Trichoderma asperellum and arbuscular mycorrhizal fungi on cacao growth and resistance against black pod disease. Crop Prot. 30, 1321–1327. doi: 10.1016/j.cropro.2011.05.003
Tchameni, S. N., Nwaga, D., Wakam, L. N., Mangaptche Ngonkeu, E. L., Fokom, R., Kuaté, J., et al. (2012). Growth enhancement, amino acid synthesis and reduction in susceptibility towards Phytophthora megakarya by arbuscular mycorrhizal fungi inoculation in cocoa plants. J. Phytopathol. 160, 220–228. doi: 10.1111/j.1439-0434.2012.01888.x
Wemheuer, F., Berkelmann, D., Wemheuer, B., Daniel, R., Vidal, S., and Daghela, H. B. B. (2020). Agroforestry management systems drive the composition, diversity, and function of fungal and bacterial endophyte communities in theobroma cacao leaves. Microorganisms 8:405. doi: 10.3390/microorganisms8030405
White, T. J., Bruns, T., Lee, S., and Taylor, J. W. (1990). “Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics,” in PCR Protocols: A Guide to Methods and Applications, eds M. A. Innis, D. H. Gelfand, J. J. Sninsky, and T. J. White (New York, NY: Academic Press, Inc.), 315–322.
Yilmaz, P., Parfrey, L. W., Yarza, P., Gerken, J., Pruesse, E., Quast, C., et al. (2014). The SILVA and “all-species Living Tree Project (LTP)” taxonomic frameworks. Nucleic Acids Res. 42, D643–D648. doi: 10.1093/nar/gkt1209
Yin, J. P. T. (2004). Rootstock effects on cocoa in Sabah, Malaysia. Exp. Agric. 40, 445–452. doi: 10.1017/S0014479704002108
Keywords: rootstock, Theobroma cacao, rhizobiome, nutrient uptake, rhizosphere, microbiome, grafting
Citation: Schmidt JE, DuVal A, Puig A, Tempeleu A and Crow T (2021) Interactive and Dynamic Effects of Rootstock and Rhizobiome on Scion Nutrition in Cacao Seedlings. Front. Agron. 3:754646. doi: 10.3389/fagro.2021.754646
Received: 06 August 2021; Accepted: 18 October 2021;
Published: 18 November 2021.
Edited by:
Mohamed Hijri, Université de Montréal, CanadaReviewed by:
Durgesh Kumar Jaiswal, Banaras Hindu University, IndiaCopyright © 2021 Schmidt, DuVal, Puig, Tempeleu and Crow. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jennifer E. Schmidt, amVubmlmZXIuc2NobWlkdEBlZmZlbS5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.