- 1LSTM, Univ. Montpellier, IRD, CIRAD, INRAE, SupAgro, Montpellier, France
- 2AgroParisTech, Ministère de l'Agriculture et de l'Alimentation, Paris, France
Given the limits of intensive agriculture (pollution, degradation of biodiversity, or soil desertification), it is necessary to develop sustainable alternatives to respond to future agricultural demand. Among these sustainable alternatives is the use of microbial biostimulants. Despite convincing scientific studies on them, their agricultural use remains minor. This can be explained by the lack of efficiency and reliability of their use. This review proposes to develop efficient microbial biostimulants based on the combination of two approaches, namely that of endophytic bacteria from seeds and the Back to the Future approach. Seed endophytic bacteria have a major agroindustrial potential insofar as they stand out from other microbial agents by their resistance, competitiveness, efficiency, and vertical transmission. Contrary to modern cultivars, non-domesticated plants harbor microbiomes which have not been impacted by the processes of domestication and agriculture intensification. The Back to the Future suggests therefore to use interesting microorganisms isolated from non-domesticated plants and to integrate them into modern cultivars. This could result in the rehabilitation of modern microbiomes and lead to make crop cultures more resistant and resilient. The idea consisting in the combination of both approaches aims at obtaining optimized microbiomes. Particular emphasis is placed on integrating these innovative microbiomes into variety breeding programs. Indeed, given the importance of plant-microorganism interactions, particularly from an agronomic point of view, taking the hologenome into account as a unit of selection in breeding programs is essential. This integrative and unprecedented approach to designing breeding programs is promising with a view to reconciling productivity and preservation of agroecosystems.
Introduction
Microbial biofertilizers show limited success in agriculture which can be explained by several factors including (i) competition with microorganisms already present in agricultural soil; (ii) difficult adaptation to the new post-inoculation environment; (iii) an absence of the target crop plant colonization (Wang and Haney, 2020). Therefore, microbial inoculations results are very variable depending on the environment considered and the inoculation's efficiency fluctuates.
It is therefore advisable to proceed with a considered recruitment of microbial strains of interest based on the combination of two approaches:
• The “Back to the Future” approach: the processes of domestication, intensive agriculture, centralized production and processing of seeds have resulted in a depletion of microbial diversity within agroecosystems and the seed microbiome. Therefore, this approach proposes to use microorganisms not affected by modern agriculture by using old varieties or wild species that are phylogenetically close to crop species as resources;
• The approach of seed endophytic bacteria arouses a growing interest: seeds harbor a specific microbiome of agro-industrial interest. Indeed, in addition to being beneficial for plant growth and development, seed endophytic bacteria also have specificities of agronomic interest including their resistance, their competitive advantage, their privileged interactions with the target plant and their vertical transmission. They are also recognized for their efficiency and practicality of use for farmers.
Indeed, it seems judicious to find microbiomes of interest within ancestral varieties or in wild species and to target the seed compartment to isolate the interesting microbiomes in order to benefit from the precious advantages of resistance, competitiveness, and vertical transmission of seed endophytic bacteria.
This methodology would allow new beneficial microorganisms to be inserted and used as selection tools in optimized microbiome design programs. In order to make these inocula efficient and to be part of a sustainability scheme, it is suggested to carry out integrative plant selection programs with the hologenome selection unit including the plant genome and its microbiota.
Recruitment of Seed Endophytic Bacteria
The Seed Endophytic Bacteria Approach
Bomfim et al. (2020) consider that the isolation of seed endophytic bacteria is a promising approach, assuming that seeds harbor a specific microbiome of agro-industrial interest.
Indeed, Shahzad et al. (2018) previously highlighted the particular interest of these bacteria linked to their vertical transmission, their potential for the production of phytohormones, enzymes, antimicrobial compounds, secondary metabolites, and their ability to enhance development and plant yield under (a) biotic stress conditions.
In this context, Bergna et al. (2019) describe seeds as privileged carriers of bacteria beneficial to plants and go so far as to identify the seeds as hotspots for the isolation of beneficial bacteria.
The properties of seed endophytic bacteria are detailed below and are distinguished in two sections: (i) typical PGP properties of seed endophytic bacteria; (ii) elaborate properties of seed endophytic bacteria.
Typical PGP Properties of Seed Endophytic Bacteria
Promotion of Plant Development
Seed endophytic bacteria have the ability to increase crop development and crop yields (Cottyn et al., 2001) by using direct or indirect mechanisms (Santoyo et al., 2016; Shahzad et al., 2017a,b) notingly by eliciting the production of secondary metabolites (Shahzad et al., 2018). They facilitate development by producing phytohormones (Shahzad et al., 2016; Finkel et al., 2020), by improving plant nutrition, or even by improving plants resistance to (a) biotic stresses (Hallmann et al., 1997; Puente et al., 2009b; White et al., 2012, 2018; Maehara et al., 2016; Sülü et al., 2016; Cope Selby et al., 2017).
Numerous studies demonstrate the relevance of seed endophytic bacteria to increase growth and plant yield. These relate to rice (Ruiza et al., 2011; Hardoim et al., 2012; Shahzad et al., 2016, 2017c; Verma et al., 2017; Krishnamoorthy et al., 2020), eucalyptus (Ferreira et al., 2008), maize (Rosenblueth et al., 2012; Liu et al., 2013; Bodhankar et al., 2017; Bomfim et al., 2020), wheat (Díaz Herrera et al., 2016), soybeans (Oehrle et al., 2000), tobacco (Mastretta et al., 2009), Phragimates australis (White et al., 2017), tomato (Xu et al., 2014), chickpea (Mukherjee et al., 2020), peanut (Li L. et al., 2019), or millet (Kumar et al., 2020).
Hormones
Endophytic bacteria help to promote plant development by producing plant growth hormones (Pal et al., 2021). Thus, many studies demonstrate phytohormone production by seed endophytic bacteria isolated respectively from the plant species Tylosema esculentum, rice, tomato, and maize (Ruiza et al., 2011; Liu et al., 2013; Xu et al., 2014; Chimwamurombe et al., 2016; Shahzad et al., 2016).
Among these phytohormones can be cited indole acetic acid, gibberellins and cytokines, whether in combination or not (Pal et al., 2021).
Seed endophytic bacteria producing indole acetic acid in plants have the effect of promoting plant development by promoting root elongation and biomass production, increasing the production of root exudates and improving the resistance of plants (Etesami et al., 2015). In fact, by increasing root surface, produced indole acetic acid improves plant nutrition and water acquisition by plants (Vessey, 2003). Numerous studies show the ability of seed endophytic bacteria to produce indole acetic acid. These studies concern seed endophytic bacteria of diverse plant species detailed in the Table 1 below.
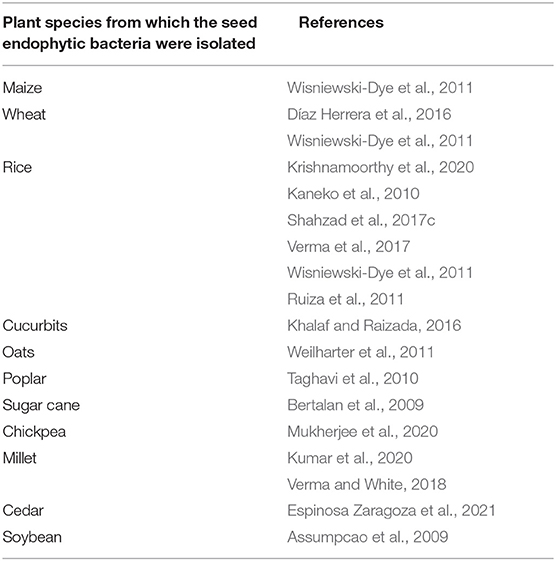
Table 1. Summary table of publications recording the ability of seed endophytic bacteria of several cultivated plant species to produce indole acetic acid.
Gibberellins are growth regulators involved in several physiological processes such as germination or stem growth (Sponsel, 2003). Shahzad et al. (2016) studied the production of gibberellin by endophytic bacteria in rice seeds.
Several studies have also highlighted the role of ACC deaminase (1-aminocyclopropane-1-carboxylate) activity in endophytic bacteria causing a reduction in the ethylene level through the degradation of its precursor to ammonia and alpha-ketoglutarate (Glick et al., 1998). Endophytic bacteria capable of synthesizing ACC deaminase allow plant species with whom they interact to promote their growth while improving their resistance to (a) biotic stresses (Rashid et al., 2012). More specifically, studies on seed endophytic bacteria of cucurbits (Khalaf and Raizada, 2016), rice (Kwak et al., 2012), maize (Johnston-Monje and Raizada, 2011), or poplar (Taghavi et al., 2009) are the instigators of ACC deaminase activity in these seed endophytic bacteria.
Cytokines correspond to a group of growth regulators allowing an improvement in cell divisions and elongations (Salisbury, 1994). Seed endophytic bacteria (Ruiza et al., 2011; Goggin et al., 2015) can produce these growth regulators.
Jasmonic acid is another stress-related growth regulator in plants (Wang et al., 2020). Bacteria can act on this regulator and help protect plants (Sabki et al., 2021) like seed endophytic bacteria from peanut (Li H. et al., 2019).
Nutrition
Endophytic bacteria allow an improvement in plant nutrition through several mechanisms such as nitrogen fixation, phosphate and potassium solubilization, siderophore production, mechanisms allowing several nutrients to be assimilated for plants (N, P, K, Fe, etc.). More particularly, seed endophytic bacteria similarly allow an increase in nutrients directly assimilable by plants, like endophytic bacteria from tobacco seeds (Mastretta et al., 2009) or cacti (Puente et al., 2009a).
Improvement of the Nitrogenous Nutrition of Plants Thanks to the Seed Endophytic Bacteria. Nitrogen plays a crucial role in plant growth and physiology (Leghari et al., 2016). Although it is abundant in the atmosphere, lithosphere and hydrosphere (Greenwood and Earnshaw, 1997), its predominant form being nitrogen, it cannot be directly assimilated by plants. Indeed, it must be reduced to nitrate or ammonium to be used by plants.
Numerous studies have recorded the capacity of certain endophytic bacteria to make dinitrogen assimilable for plants, i.e., in the case of rice and maize (Sturz et al., 2000). More specifically, many studies attest to the ability of seed endophytic bacteria to fix nitrogen in rice (Krause et al., 2006; Yan et al., 2008; Kaneko et al., 2010; Wisniewski-Dye et al., 2011; Annapurna et al., 2018; Krishnamoorthy et al., 2020), sugar cane (Bertalan et al., 2009), wheat (Fouts et al., 2008; Wisniewski-Dye et al., 2011), the cactus (Puente et al., 2009b).
Improving the Phosphate Nutrition of Plants Thanks to Endophytic Seed Bacteria. Phosphorus is a limiting mineral nutrient for plant growth and development. However, it is mainly found in the soil in an insoluble form that cannot be assimilated by plants (Stevenson and Cole, 1999). Endophytic bacteria are able to make insoluble phosphorus in a form that can be assimilated by plants thanks to their enzymatic arsenal (phosphatases) (Walia et al., 2017) or to the secretion of organic acids (Varga et al., 2020). Similarly, numerous studies show the ability of seed endophytic bacteria to solubilize phosphate in the case of the species mentioned below in Table 2.
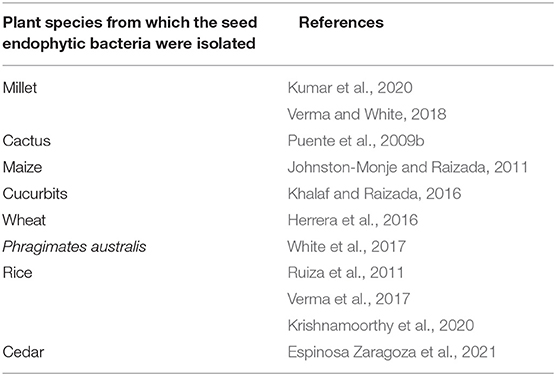
Table 2. Summary table of publications recording the ability of seed endophytic bacteria of several cultivated plant species to solubilize phosphate.
Improving the Iron Nutrition of Plants Thanks to Seed Endophytic Bacteria. Siderophores are low molecular weight compounds secreted by endophytic bacteria (Loaces et al., 2011; Chimwamurombe et al., 2016) and allow iron to be sequestered under limiting conditions. Siderophores have a great affinity for binding ferric ions and transporting them in soluble and assimilable form to plants (Loper and Buyer, 1991). There are several studies showing a siderophore production by endophytic seed bacteria, whether in cucurbits (Khalaf and Raizada, 2016), poplar (Taghavi et al., 2010), Indigofera argentea (Andres-Barrao et al., 2017), olive (Martinez-Garcia et al., 2015), rice (Krishnamoorthy et al., 2020), or millet (Kumar et al., 2020).
Rhizophagy
Rhizophagy corresponds to the ability of some symbiotic or endophytic bacteria to develop in the rhizosphere by capturing available nutrients there and to penetrate into plant tissues by releasing these nutrients (Paungfoo-Lonhienne et al., 2010). As a result, plants directly benefit from nutrients (White et al., 2018), especially in the case of plant-seed endophytic bacteria interactions (White et al., 2018).
Resistance Against (a) Biotic Stresses
Seed endophytic bacteria confer on the plants with which they interact an increased resistance to (a) biotic stresses (Santoyo et al., 2016; Shahzad et al., 2017a). Pal et al. (2021) insist on the ability of seed endophytic bacteria to improve plant resistance to salt stress, drought, heat, frost, heavy metals, or when pests are present. In this regard, studies on this subject are mentioned below and categorized according to the stress considered.
Resistance Against Biotic Stresses
Numerous seed endophytic bacteria have been recorded as potential biocontrol agents (Tayung et al., 2012). These studies involve multiple plant species and are listed in Table 3. Endophytic bacteria can exert a biocontrol action, either:
Directly by inhibiting the pathogen growth i.e., (a) through antimicrobial compounds production (Johnston-Monje and Raizada, 2011; Truyens et al., 2015; Khalaf and Raizada, 2016, 2018; Mitter et al., 2017; Shahzad et al., 2018) including volatile compounds (Mukhopadhyay et al., 1996), lipopeptides (Gagne-Bourgue et al., 2013; Verma and White, 2018), and enzymes with chitinolytic properties (Mukhopadhyay et al., 1996), (b) through the exercise of competition for space and nutrients (Loper and Henkels, 1999);
• Indirectly by improving the resistance of the plant i.e., by triggering a systemic induced resistance (Shahzad et al., 2016).
The potential for biocontrol of seed endophytic bacteria arouses particular interest considering the number of studies relating to this particular property. Indeed, it is possible to find many studies of seed endophytic bacteria including seed endophytic bacteria isolated from.
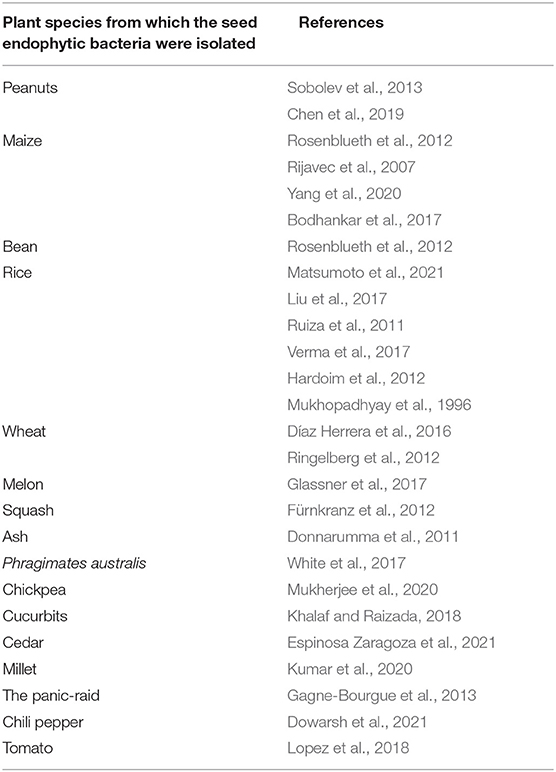
Table 3. Summary table of publications recording the potential of biocontrol of seed endophytic bacteria isolated from several cultivated plant species.
Resistance Against Abiotic Stresses
Several studies have revealed the ability of seed endophytic bacteria to improve plant resistance in the event of abiotic stress i.e., in the event of drought. It is the case of the respective studies by Hardoim et al. (2012) on endophytic rice seed bacteria, Rosenblueth et al. (2012) on endophytic bacteria in corn seeds, Vega et al. (2005) on the endophytic bacteria of coffee seeds, Sziderics et al. (2007) on the endophytic bacteria of pepper, or even Kukkurainen et al. (2005) on seed endophytic bacteria of strawberry.
In addition to all of the beneficial properties mentioned above, seed endophytic bacteria have specific properties giving them agronomic potential.
Elaborate Properties of Seed Endophytic Bacteria
Germination Facilitation and Release of Seed Dormancy
Seed endophytic bacteria play a key role in seedling germination and growth by acting on several parameters including seed viability, germination, and seedling survival (Verma et al., 2019; Rodríguez et al., 2020). The important role of seed endophytic bacteria has been demonstrated by a lack of vigor found in seeds harboring a reduced microbiome (Verma et al., 2017; Escobar Rodríguez et al., 2020). Therefore, this confirms the hypothesis previously emitted by Chee-Sanford et al. (2006), Rodríguez et al. (2017), or even Shearin et al. (2017) who then proposed that these bacteria have the role of promoting the conservation of seeds and the facilitation of seed germination.
Several studies showing this ability to facilitate germination have been carried out. These studies focus in particular on facilitating germination in rice (Mano et al., 2006; Kaga et al., 2009), eucalyptus (Ferreira et al., 2008), cactus (Puente et al., 2009a), or maize (Rijavec et al., 2007).
Competition Ruled Out
Seed endophytic bacteria interact closely with their host (Verma et al., 2017): indeed, their location is ideal (Verma et al., 2017; Shearin et al., 2018) and makes it possible to avoid any competition or concurrence (Truyens et al., 2015; Verma and White, 2018). In this regard, Shahzad et al. (2018) underline the particular ecological potential of seed microbiome that constitutes not only the culmination of the assembly of communities within the seed but also the starting point of the assembly of a new community in the plant organism to become. Gopal and Gupta (2016) consider seeds as the main source of inoculum for crops.
Vectors
Seed endophytic bacteria are currently arousing a real craze in connection with one of their specificity: they can be transmitted vertically and the seeds would then play the role of beneficial bacteria vectors (Truyens et al., 2013; Bergna et al., 2019). Numerous studies underline the importance of this vertical transmission insofar as it allows bacteria to be transmitted from generation to generation, i.e., from seed to seed and ensures the presence of bacteria within subsequent generations (Johnston-Monje et al., 2016; Cope Selby et al., 2017; Frank et al., 2017; Shade et al., 2017; Adam et al., 2018; Nelson et al., 2018; Vannier et al., 2018; Bergna et al., 2019; Hardoim, 2019; White et al., 2019).
Current knowledge on this subject is not extensive and further studies are necessary to better understand vertical transmission of seed endophytic bacteria (Berg and Raaijmakers, 2018; Nelson et al., 2018; Li H. et al., 2019; White et al., 2019).
Proofs and Agribusiness Applications
At first, vertical transmission was only an assumption. Some studies supported this theory without providing certainty based on different arguments, namely:
• Preservation of the microbiome over a long time period: in 2012, Liu et al. identified the presence of a microbiome that would have been preserved over a long period in seed endophytic bacteria from corn, at the same time supporting the theory of vertical transmission (Liu et al., 2012). These results are reinforced by those of Johnston-Monje and Raizada who in 2011 suggested the presence of a microbiome conserved within corn seeds throughout evolution, ethnography and ecology.
• The study of the specific mode of trophic resources acquisition found in invasive plants. In 2013, Rout et al. found another clue to support the vertical transmission theory by studying seed endophytic bacteria of Sorghum halepense (Rout et al., 2013). Indeed, plants developed from seeds whose surface has been disinfected are able to assimilate nitrogen resulting from the reduction of nitrogen by diazotrophic bacteria potentially acquired through the process of vertical transmission.
• The link between genotype and microbiome: Adams and Kloepper evaluated in 2002 the impact of the cotton plants genotype on the populations of endophytic bacteria of seeds, stems and roots (Adams and Kloepper, 2002). They found that different cotton cultivars harbor different bacterial community structures. Another study examining the diversity of endophytic bacteria in corn seeds provided evidence to link the composition of endophytic bacterial communities to the phylogeny of the host plant (Johnston-Monje and Raizada, 2011).
• Analysis of seed endophytic bacteria communities: in 2017, Cope Selby et al. suggested that vertical seed-to-seed transmission is the primary source of Miscanthus endophytes by examining the diversity of seed endophytic bacteria isolated from Miscanthus (Cope Selby et al., 2017).
• Transmission of seed endophytic bacteria to seedlings by GFP labeling: Ferreria analyzed in 2008 the endophytic bacteria of seeds and seedlings of 10 species of Eucalyptus and two hybrids and found that endophytic bacteria are transmitted from seeds to seedlings, in particular by tagging the bacterium Pantoea agglomerans.
• The study of the link between host plant fitness and the microbiome: the study by Bergna et al. (2019) whose goal is to understand the stability and transmission of beneficial bacteria from generation to generation has provided information on vertical transmission. Indeed, Bergna and her team have demonstrated that each biotope and genotype associated with a plant harbors a specific microbiome that is modulated according to the constraints of the plant's ecological niche so as to increase the fitness of the host plant. They also demonstrated that the communities of beneficial bacteria are more selective in the ecological seed niche than in the other compartments of the plant and that this selection is mostly sensitive to the constraints of the biotope. In fact, in the event of the presence of pathogens and absence of nutritional deficit, seeds harbor bacteria adapted for the biocontrol of this pathogen, while an absence of pathogens accompanied by an environment poor in nutrients is the instigator of a predominance of bacteria facilitating plant nutrition. They also identified seeds as primary vector for beneficial bacteria transmission. Kusstascher et al. also suggested this link more recently in 2021. Indeed, by analyzing the respective seed microbiomes over two generations and the rhizosphere of six squash genotypes, they were able to suggest that plants are likely to specifically enrich certain microorganisms in the seeds that can improve fitness, health and growth of daughter plants from seeds.
• The mediation of endophyte transmission by pollen documented in some species (Ambika et al., 2016; Frank et al., 2017).
Since then, recent studies have established the theory of vertical transmission with arguments based on analysis of the microbiome and the location of endophytic seed bacteria.
Studies Related to the Localization of Seed Endophytic Bacteria
In 2019, Li L et al. demonstrated vertical transmission of endophytic peanut bacteria. By using a GFP marker, they showed that the P.g.YMR3 strain can be transmitted vertically from one generation to the next by the ova and gynophores (Li L. et al., 2019).
Faddetta et al. (2021) isolated endophytic bacteria from lemon seeds. Subsequently, they characterized the endophytic bacteria of the different compartments of the next generation lemon plants. They found that the most abundant bacterial genera are identical in lemon leaves and seeds. In addition, they localized these endophytic bacteria in the seeds and lemon leaves revealing colonization of the vascular bundle. They therefore demonstrated the transmission of endophytic bacteria from lemon seeds to plant leaves from different seeds, supporting the theory of vertical transmission.
Studies Related to the Localization of Seed Endophytic Bacteria
In 2020, Liu et al. studied the relationship between the composition and diversity of endophytic bacterial communities of hybrid maize kernels and of the previous generations from which they originated (Liu et al., 2020). It allowed to show a potential relationship of endophytic bacteria between the different genotypes. To this end, they used high-throughput sequencing (HTS) technology to analyze community structures and bacterial diversity in different maize kernels. It has been shown that a large part of seed microbiomes is shared among all the genotypes studied and that the individual female parental line has a greater impact on the microbiome of its hybrid seeds than the male individual parental line. This study provides scientific clues to the endophytes vertical transmission theory over generations of maize.
Abdelfattah et al. (2021) studied the vertical transmission of endophytic bacteria from acorns by developing a culture device to grow acorns in a microbe-free environment while keeping underground and aerial tissues separate. They found that the acorn microbiome is not distributed randomly. Indeed, they found that the phyllosphere microbiome is similar to that of the embryo while the root microbiome is quite distinct. They were thus able to identify a spatial distribution of bacterial communities in both acorns and seedlings indicating heredity, niche differentiation and divergent transmission pathways for the bacterial communities establishment in roots and phyllosphere.
Zhou et al. (2020) analyzed rice grain and rhizosphere samples from two consecutive harvests using the metabarcoding method targeting the 16S rRNA gene region. This analysis allowed to highlight a greater incidence of vertical transmission on the endophytic bacteria communities in rice than horizontal transmission through the rhizosphere (25.5 vs. 10.7%). They also demonstrated that grain moisture and average of annual winter temperature has a significant role on the seed microbiome which may explain the adaptation of the microbiome according to the ecological niche considered.
Tannenbaum et al. (2020) analyzed the ryegrass microbiome over two successive generations by sequencing the V4 region of the 16S rRNA gene. They found that the microbiome of the two ryegrass generations is similar marked by a dominance of Gamma proteobacteria and a presence of Bacilli. The results of this study suggest that the microbiome is transmitted from seed to seed.
Matsumoto et al. (2021) observed that rice plants of the same cultivar are more or less resistant in the presence of a pathogenic organism. Having identified endophytic bacteria from seeds of this cultivar having biocontrol properties toward this pathogen, they integrated high-throughput data, performed gene mutagenesis and molecular interaction tests to better understand this phenomenon. Analysis of these data allowed them to state that the endophytic seed bacteria Sphingomonas melonis is transmitted from generation to generation in resistant rice plants and that it confers resistance to non-resistant plants by producing anthralinic acid.
Mutualistic Coevolution
The transmission from generation to generation of seed endophytic bacteria is likely to preferentially select mutualistic interactions allowing to improve plant host fitness insofar as these endophytes depend on their plant host survival and reproduction (Ewald, 1987; Rudgers et al., 2009). In addition to improving plant host fitness, seed endophytic bacteria also ensure better fitness for the plant host progeny since the latter also benefits from these vertically transmitted endosymbionts (Shade et al., 2017).
Extreme Resistance
Seed ecological niche requires seed endophytic bacteria to be resistant to potentially extreme environmental conditions. Indeed, the seed internal environment:
• Is unstable i.e., in the case of seed maturation process during which starch accumulation and intense dehydration occurs (Mano et al., 2006);
• Induces dehydration and high osmotic pressure (Ebeltagy et al., 2000; Nicholson et al., 2000; Compant et al., 2010; Truyens et al., 2015; Pitzschke, 2016; Lopez et al., 2018);
• Is poor in directly assimilable nutrients (Nicholson et al., 2000; Compant et al., 2010; Truyens et al., 2015);
• Can be frozen i.e., during dormancy (Nelson, 2004; Geisen et al., 2017).
As a result, seed endophytic bacteria have developed the traits of:
• Endospore formation allowing them to survive in an unstable environment (Mano et al., 2006; Compant et al., 2011; Kane, 2011) subject to high osmotic pressures and lack of nutrients (Nicholson et al., 2000; Compant et al., 2010; Truyens et al., 2015);
• Motility allowing them to migrate (Johnston-Monje and Raizada, 2011; Truyens et al., 2015). In this regard, the respective studies of Okunishi et al. (2005) and Ebeltagy et al. (2000) demonstrated the motility trait in the majority of rice seed endophytes;
• Specific mode of nutrition involving the possession of a particular enzymatic arsenal including amylases to use starch (Mano et al., 2006) or even phytases make it possible to use phytate as a source of phosphate (López-López et al., 2010).
The seed endophytic bacteria's particular traits explain their remarkable resistance reported in several studies. Indeed, it has been found that in the case of seed endophytic bacteria isolated from rice, a greater tolerance to high osmotic pressures is present in seed endophytic to those of other compartments (Mano et al., 2006). A similar study on endophytic bacteria of rice seeds also emphasizes this property of tolerance to osmotic pressure (Kaga et al., 2009), just like that of endophytic bacteria of corn seeds tolerating conditions of osmotic pressure simulated by a 40% medium concentration -PEG-6000 (Bodhankar et al., 2017). In the case of seed endophytic bacteria of corn, their tolerance to extreme environments has been demonstrated: 10% salinity or temperatures reaching 60°C (Bodhankar et al., 2017).
Thanks to their extreme resistance, seed endophytic bacteria allow plants to benefit from their specificities even under extremely difficult conditions, both in terms of water and nutrition, and thus make it possible to promote the development of plants such as cacti (Puente et al., 2009a) or quinoa (Pitzschke, 2016) whose ecological niches are also extreme environments.
The Potential of Seed Endophytic Bacteria as Elicitors of Secondary Metabolite Production Due to Their Endophytic Status
Plants are likely to be affected by (a) biotic stresses, notably through the production of a group of low molecular weight compounds called secondary metabolites. Among them, we can mention quinones, anthocyanins, phenolic compounds, flavonoids, alkaloids, terpenoids, and steroids. The latter play a fundamental role in the adaptation of plants in stress situations and in their defense (Wink, 2003).
The elicitation of secondary metabolite production corresponds to the stimulation of the biosynthesis of these chemical compounds following the addition of elicitors (Radman et al., 2003). Ogbe et al. (2020) describe endophytes as elicitors of secondary metabolite production. Indeed, endophytic microorganisms play a role in the production of secondary metabolites (Li et al., 2008; Kusari et al., 2012). These metabolites include: (i) antimicrobial compounds; (ii) phytohormones; (iii) vitamins; (iv) bioprotectants (Singh et al., 2017).
Ogbe et al. (2020) explain that under stress conditions, endophytes assist their host plant to produce secondary metabolites. This assistance comes in different forms: (i) modulation of the production of secondary metabolites present within the endosphere (Lopez-Fernandez et al., 2016; Singh et al., 2017); (ii) production of secondary metabolites by the endophytic microbiome itself (Fu et al., 2017).
Production of Secondary Metabolites Through Endophytes
Only recently have some studies been able to record the potential of endophytic bacteria to increase the production of secondary metabolites to benefit their host plant. Some endophytes have the ability to produce biochemical compounds similar or identical to those produced by plants (Taghinasab and Jajabi, 2020). It is proposed that this ability is due to horizontal gene recombination or to a transfer made during the evolutionary process. For example, the taxol-producing fungus Cladisporium cladosporioides MD2 associated with the host plant Taxus media possesses this trait through the 10-deacetylbaccatin-III-10-O-acetyl transferase gene, which plays a role in taxol biosynthesis and has a 99% similarity to the host plant gene (Zhang et al., 2009). In this regard, some endophytes, such as Colletotrichum gloeosporioides and Piriformospora indica, have been identified for their potential to accumulate certain secondary metabolites i.e., asiaticoside (Gupta and Chaturvedi, 2017). Zhang et al. (2013), on the other hand, were able to record that Mycena sp. endophytes also induce elevated flavonoid and kinsenoside content. Mahmood and Kataoka (2020) corroborate the studies mentioned above. They studied the effects of endophytic bacteria application on cucumber. In particular, they were able to show that such application affects plant metabolomes compared to non-inoculated plants. They recorded an increase in the concentrations of more than half of the secondary metabolites due to the inoculation of the plants. They thus proved that endophytic bacteria are elicitors of secondary metabolites in the endosphere. Similarly, Parthasarathy et al. (2018) showed that sugarcane endophytic bacteria are capable of producing multiple secondary metabolites thanks to secondary analysis shell (antiSMASH4.0) of their genomes.
Modulation of Secondary Metabolite Production by Endophytes
Krishnamoorthy et al. (2020) have highlighted the ability of endophytes to modulate the production of secondary metabolites. Indeed, they studied the modulation of certain secondary metabolites by seed endophytic bacteria of rice. They suggested the following hypothesis: endophytic bacteria would have the potential to regulate Plant Growth Properties by producing secondary metabolites at specific levels. Indeed, they observed differences in secondary metabolite concentrations following seed priming. They suggest that endophytic bacteria play a crucial role in determining the molecular processes that upregulate certain secondary metabolites. Taghinasab and Jajabi (2020) corroborate their postulation. In particular, they mention the role of endophytes in modulating the production of secondary metabolites in cannabis.
Other studies also suggest modulation of secondary metabolite production. This is notably the case of the study by Irmer et al. (1994). They show that alkaloid (pyrrolizidine) biosynthesis in a fabaceae (Crotalaria) is dependent on nodulation by Bradyrhizobium species bacteria, thus showing the control exerted by endophytic bacteria.
This modulation of secondary metabolites production is due to the influence exerted by endophytes on the gene expression of their vegetal host. Indeed, some endophytic microorganisms have the ability to rapidly induce the expression of specific functional genes in their plant host in a specific and selective manner as endophytes and their plant partners co-evolve (Jia et al., 2016; Cui et al., 2017). As a result, they have the potential to regulate their host plant's secondary metabolism to improve its fitness under (a) biotic stresses (Booker et al., 2016; El-Hawary et al., 2016): they have the ability to modify gene expression as well as host plant metabolic pathways through lateral gene transfer, induction, and biotransformation, and as a result have the potential to regulate secondary metabolite concentration (Gluck-Thaler and Slot, 2015). Maggini et al. (2017) investigated the influence of interactions between the plant organism Echinacea purpurea (L.) Moench and its endophyte communities in the production of secondary metabolites. They were able to show different alkamide contents between inoculated and control plants, suggesting a modulation of alkamide biosynthesis following inoculation. To validate this hypothesis, they studied the expression of genes related to alkamide biosynthesis. They found different expression profiles between inoculated and control plants: inoculated plants had higher expression levels. Endophyte-plant interactions thus influence secondary metabolite production, notably through upregulation of gene expression. Cui et al. (2020) corroborate the above mentioned study. They studied the effects of the endophyte Phialocephala fortinii on the regulation of the biosynthesis of secondary metabolites of Rhodiola crenulata. To do so, they used the combination of transcriptome and metabolome to better understand the mechanisms inherent to their interaction. They were able to show that 16,151 genes are differentially regulated, of which 14,706 are up-regulated and 1,445 are down-regulated, as well as the occurrence of 1,432 metabolites including 27 marker metabolites: the endophyte plays an important role in the transcriptomics and metabolic regulation of the host plant R. crenulata. In this way, they were able to highlight the ability of the endophyte to regulate the expression of host plant genes and to rapidly induce an accumulation of secondary metabolites. Ray et al. (2019) also corroborate these results by indicating that inoculation of Papaver somniferum L. with a consortium of bacteria induces an increase in morphine yields via an increase in the expression of the COR gene, a gene essential for morphine biosynthesis.
Singh et al. (2017) and Taghinasab and Jajabi (2020) specify that although endophytes are recorded as fundamental players in the production and modulation of secondary metabolite concentration, the underlying mechanisms are largely unknown.
The choice to favor seed compartment for the isolation of strains of agronomic interest having been explained, it is now necessary to determine which seeds to choose as microbiome resources. In this context, it is proposed to join the approach of seed endophytic bacteria to the “Back to the Future” approach proposed by Berg and Raaijmakers (2018).
The “Back to the Future” Approach
Principle of the Approach
The hypothesis on which this approach is based is that the process of domestication coupled with the practice of intensive agriculture generated loss and depletion of the plant and seed microbiomes (Johnston-Monje and Raizada, 2011; Pérez-Jaramillo et al., 2016; Rybakova et al., 2017), microbiomes that can be regenerated using wild species or old varieties as microbiome resources of interest.
The Practice of Intensive Agriculture and the Loss of Microbial Diversity in the Soil
The practice of intensive agriculture has impacted the agroecosystems microbial diversity, source of beneficial bacteria for plants and at the same time induced the emergence of dysbiotics systems (Weese et al., 2015; Wang and Haney, 2020).
The Strategic Error of the Domestication Process: the Failure to Take Into Account the Holonbiont Component
Plant domestication led to a selection of productive crops, but it has too resulted in indirect repercussions that were not visible. It induced an alteration of the microbial communities associated with the cultures of interest (Leff et al., 2017; Chaluvadi and Bennetzen, 2018; Contreras-Liza, 2021) in connection with a modification of the root architecture, exudation and defense mechanisms (Martínez-Romero et al., 2020). Domestication may have removed resistance and nutrient uptake traits by affecting plant interactions with beneficial microorganisms (Bulgarelli et al., 2013; Chen et al., 2015). Gopal and Gupta (2016) explain this phenomenon by making a comparison with wild plants which evolved by selecting a microbiome of beneficial partners. The development of intensive agriculture and plant domestication have caused this joint evolution to fail, in particular by restricting varietal selection to the plant genome only and not to the hologenome one (Gopal and Gupta, 2016).
A change in the crop varieties microbiomes has been observed in rice, wheat, corn and beans (Peiffer et al., 2013; Perez-Jaramillo et al., 2017; Martínez-Romero et al., 2020). This change has resulted in an impact on the composition and functions of the plant microbiota playing a key role in multiple ecosystem processes and in plant development (Pérez-Jaramillo et al., 2018; Contreras-Liza, 2021). For example, Özkurt et al. (2020) analyzed the bacterial and fungal communities associated with wild wheat (Triticum dicoccoides) and domestic wheat (Triticum aestivum). They showed that vertically transmitted bacteria from the domesticated wheat species (T. aestivum) are less diverse and stable in plants compared to bacteria from the wild wheat species (T. dicoccoides). Shi et al. (2019) also found divergences in terms of microbiota between domesticated and wild species: wild rice and non-domesticated soybeans have a greater abundance of beneficial symbionts than in cultivated species. A trend found in several species consists of a decrease in populations of Bacteriodetes in favor of Proteobacteria in modern cultivars (Germida and Siciliano, 2001; Perez-Jaramillo et al., 2017; Adam et al., 2018).
Depletion of the Seed Microbiome
Berg and Raaijmakers (2018) focused more specifically on the effect of modern agriculture on seed microbiome. Indeed, seed microbiome is affected by several phenomena including (i) treatments carried out on the seeds (disinfection, osmopriming); (ii) seed centralized production leading to a homogenization of plant microbiomes on a global scale and ultimately to an impoverishment of microbiobial diversity within agroecosystems. Chen et al. (2020) also consider that seed microbiome - reservoir of beneficial microorganisms - is susceptible to being impacted by agricultural practices, i.e., in the case of their biological model - Nicotiana tabacum - including the seed microbiome seed that is impacted by the widely used seed coating technique.
The Advisability of Using Non-domesticated Plants
Berg and Raaijmakers (2018) raise the problem posed by this impact on the plant microbiome given its importance for phenotypic and epigenetic plasticity but also plant evolution (Van der Heijden et al., 2016). In order to compensate for a possible loss of the beneficial plant microbiome in modern varieties, Berg and Raaijmakers propose a new approach. The latter consists of the insertion of the beneficial microorganisms missing in modern varieties, microorganisms found in ancestral or wild varieties. The aim of this insertion is to optimize the management of plant resistance to (a) biotic stresses. Pérez-Jaramillo et al. (2016) propose the same approach and highlight the potential of exploring the wild species microbiomes for the isolation of beneficial microorganisms that may have been lost in the domestication process.
According to the Berg and Raaijmakers approach, it would therefore be judicious to use the seed endophytic bacteria microbiome of wild species as a source of growth promoting and biocontrol agents for domesticated species (Wassermann et al., 2017). For instance, the recent study by Roodi et al. (2020) supports the relevance of this approach for agronomic purposes. Roodi et al. isolated endophytic bacteria from the majority of Brassica accessions. They were able to find two strains of Methylobacterium with the ability to promote the development of rapeseed and inhibit the growth of the pathogen Leptosphaeria maculans.
Berg et al. (2017) explain that this methodology is applicable to other crop species and that it resulted in obtaining a new inoculation technology easily incorporated into the seeds of crop species.
Card et al. (2016) underline the commercial interest of this technology in terms of biocontrol and improvement of plant growth both thanks to its efficiency and practicality of use for farmers.
The strains of interest from seeds of old varieties or wild species having the particularity of being transmitted from generation to generation, it is therefore conceivable to integrate them into innovative integrative breeding programs taking into account the essential role of the microbiome.
The Hologenome as a Selection Unit for Innovative and Efficient Breeding Programs
The Combination of the Two Approaches and the Desirability of Integrating Them Into Breeding Programs
Gruber (2017) and Raaijmakers and Mazzola (2016) highlight the perspectives offered by the “Back to the Future” approach and the interest of using microorganisms not affected by the domestication process by using old or ancestral varieties or wild species phylogenetically close to crop species as resources. Indeed, this approach would allow the insertion of new beneficial microorganisms for cultivated plants obtained from non-domesticated plants and use them as selection tools in microbiome design programs (Pérez-Jaramillo et al., 2016).
The approach of seed endophytic bacteria is similarly arousing growing interest. For practical use of bacteria with Plant Growth Promoting traits and having biocontrol properties, seeds are preferred carriers (O'Callaghan, 2016). Given the specificities of interest of seed endophytic bacteria for agricultural purposes including their resistance, competitive advantage and vertical transmission, they arouse commercial interest in agribusiness (Frank et al., 2017; Nelson, 2018). Indeed, Bergna et al. (2019) having demonstrated that seeds are the privileged vectors of beneficial bacteria, the characteristic of vertical transmission of seed endophytic bacteria represents an agro industrial potential and would allow to design seed treatments that would be sustainable insofar as the seed endophytic bacteria are passed from generation to generation. Indeed, they have a particular potential to be integrated into breeding programs, seeds constitute a solution to their selection and their transmission over generations (Berg et al., 2017; Wei and Jousset, 2017).
An Integrative Approach to Variety Selection Programs Taking Into Account the Privileged Role of the Microbiome
The plant phenotype is not only determined in response to an environment but also by its associated microbiota, its own responses to a given environment and the complex interactions between agro ecosystem members (Hardoim et al., 2015). Therefore, Sessitsch and Mitter (2015) consider that varietal selection programs should not be only based on the plant component but also on the importance of the interactions that the latter have with microorganisms. In this context, it is possible to design variety selection programs leading to new phenotypes by adopting an integrative approach, i.e., by modifying genetic information of plants and their associated microbiota. The plant microbial community is considered as the second genome of the plant and plays a key role in plant nutrition and health (Berendsen et al., 2012; Sessitsch and Mitter, 2015). Many perspectives are offered by microorganisms i.e., nitrogen fixation by non-leguminous plants such as wheat, corn (Van Deynze et al., 2018), rice, potato, tomato (Dent and Cocking, 2017). The plant microbiome having significant genetic variability, it is likely to constitute a relevant resource in the breeding strategy (Gopal and Gupta, 2016).
Wang and Haney (2020) consider the plant microbiome as a real reservoir of genes with major agronomic potential: it is a lever for agronomic action and it is possible to optimize it by modifying it in variety selection programs.
Several methods are possible to integrate the microbiome into breeding programs (Bakker et al., 2012; Arif et al., 2020).
Two Levers of Action: the Genomes of Plants and Their Microbiota
Optimized holobiome breeding programs should promote the following objectives:
• Select cultivars receptive to microbial interactions
According to Corbin et al. (2020), it is important to understand what makes a cultivated plant more or less receptive in order to benefit from the advantages of associations with beneficial microorganisms to improve varieties. To this end, it is necessary to take into account plant genes allowing the plant microbiota assembly regulation in future varietal breeding programs. Indeed, given the agricultural practices and intense artificial selection impacts, the downside consisting in the impact on the interactions between cultivated plants and their beneficial microbiota appeared gradually. To tacke the problem, research must develop selection methods that improve these interactions (Porter and Sachs, 2020). Contreras-Liza (2021) supports the production of new cultivated plants phenotypes through the plant selection optimizing the symbiosis with microorganisms.
• Select effective microorganisms
Recent studies stipulating seed endophytic bacteria vertical transmission (Johnston-Monje et al., 2016; Adam et al., 2018; Bergna et al., 2019) arouse interest in particular insofar as it has been shown that plant genotype-specific microbial communities are more represented in the seed compartment than in other plant compartments (Rybakova et al., 2017; Adam et al., 2018; Chen et al., 2020). This represents an opportunity to develop new varietal selection methodologies (Cordovez et al., 2019) since the selection of microbiomes (Mendes et al., 2019) is possible: a transfer of microbiomes from generation to generation has been proven (Berg and Raaijmakers, 2018). Arif et al. (2020) characterize candidate microorganisms based on: (i) their ability to interact with the target plant; (ii) their ability to be transmitted vertically (Mueller and Sachs, 2015; Wei and Jousset, 2017).
Arif et al. (2020) underline the importance of customizing inocula according to the plant species considered.
The Holobiome, a New Unit of Selection
Gopal and Gupta (2016) discuss the holobiome as a unit of selection. The holonbiont is the assembly of the individual plant and the beneficial microorganisms with which it interacts and functions as a unit of biological organization (Bordenstein and Theis, 2015; Theis et al., 2016). This holonbiont having the capacity to replicate and transmit its genetic information is a unit of selection (Zilber-Rosenberg and Rosenberg, 2008; Booth, 2014; Van Opstal and Bordenstein, 2015). The genomic correspondence of the complex symbiotic interactions of the plant holobiont is governed by its holobiome or hologenome including the host and its microbial genome (Guerrero et al., 2013; Bordenstein and Theis, 2015). Arif et al. (2020) consider the prospect of selecting the holonbiont component for varietal selection as promising to improve yields and resilience of agroecosystems. Indeed, beneficial microorganisms provide important ecosystem services (Sankar Ganesh et al., 2017; Syed Ab Rahman et al., 2018; Saied and Chojnacka, 2019). Sahu and Mishra (2021) refer to the holonbiont selection as an emerging discipline integrating the genome and the microbiome. They particularly mention the holonbiont potential to contribute to better resistance of cultivated plants to environmental constraints and to boost agricultural productivity. Corbin et al. (2020) also see the potential to use the holonbiont as a breeding target for plants. Indeed, they underline the strategic error made until then in varietal selection programs that did not consider plants as a unit of selection and not as holonbionts.
Wei and Jousset (2017) propose in this regard to find a way to obtain new plant phenotypes through a combined modification of the genetic information of the plant and its microbiota, made possible by the emergence of a new technology which allows the transmission of the endophytic microbiota from generation to generation.
Methods for Selecting the Hologenome
Modification of the Plant Microbiota Genetic Information
Berg and Raaijmakers (2018) consider the vertical transmission of seed endophytic bacteria as an agronomic opportunity that would allow the development of a new way of designing variety breeding programs. Until then, breeding programs did not take into account the concept of the holobiome and it indeed seems judicious to introduce into the varietal selection criteria new species which would be the most able to constitute an effective microbial community. In this regard, the fact that some studies have shown that the processes of domestication and selection have altered the microbial communities interacting with the plants of interest (Leff et al., 2017; Chaluvadi and Bennetzen, 2018) suggests that the composition microbiome is a trait that can be selected (Wissuwa et al., 2009).
New studies have offered the interesting prospect of combining varietal selection and the introduction of beneficial microorganisms on or in seeds: Berg and Raaijmakers (2018) speak of a “Symbiotic” approach combining the two aspects of treatments pre and pro-biotics. Among these studies of breeding programs based on the composition and functions of the microbiome are those of Adam et al. (2018), Mitter et al. (2017), and Panke-Buisse et al. (2017).
Mitter et al. (2017) succeeded in proving the feasibility of designing new microbiologically improved seeds through community enrichment of endophytic bacteria from wheat grains. They introduced the bacterium Paraburkholderia phytofirmans PsJNT through the inoculation of the flowers. By vertical transmission, the wheat plants grown from the enriched seeds showed both better yield and improved development. This study is interesting as it indicates the feasibility and efficiency of modulating alternative microbiomes to improve agronomic results by targeting seed endophytic bacteria as a microbiological selection tool. Kusstatscher et al. (2021) also stressed the need to adopt an integrative approach to plant breeding taking into account the microbiome parameter. Their study of seed endophytic squash bacteria has helped to support the potential of seed endophytic bacteria for plant microbiome selection.
Adam et al. (2018) studied the squash microbiome of seeds and rhizosphere. They were able to demonstrate that in the case of their biological study model, modulation of the microbiome is possible. Indeed, their study demonstrated a strong impact of the Cucurbita pepo genotype on the seed microbiome composition. They suggested a selection of new cultivars more apt to exploit the indigenous beneficial microorganisms' communities.
In 2017, Panke-Bruise et al. succeeded in selecting microorganisms that influence the flowering of Arabidopsis thaliana. In 10 generations, they have selected the best microcosms based on the phenotypic results of the plant. Plants inoculated with late flowering microbiomes exhibited increased inflorescence (Panke-Buisse et al., 2015).
Modification of the Plant Genetic Information
It is important to select host plants capable of interacting and benefiting from beneficial microorganisms in order to obtain optimum yield improvement. Indeed, Wintermans et al. (2016) demonstrated that plant genotype plays a crucial role in the holonbiont composition: thus, Arabidopsis plants inoculated with the biofertilizer Pseudomonas simiae WCS417r obtained inhomogeneous results in terms of biomass according to the cultivar considered, achieving biomass differences ranging from simple to quadruple. Inoculation with biofertilizers can sometimes be ineffective if the cultivar cannot interact and benefit from the beneficial microorganisms since each cultivar differentially attracts microorganisms and therefore a differentiated construction of the holonbiont takes place (Mitter et al., 2017; Kwak et al., 2018).
A targeted selection of cultivars that can benefit from interactions with beneficial microorganisms seems relevant and feasible insofar as studies on the plant functional genomics have allowed to manipulate plant genomes to attract and maintain beneficial microorganisms (Stringlis et al., 2018).
In this regard, the genotype of non-domesticated species constitutes a precious genetic resource insofar as non-domesticated species have privileged interactions with beneficial microorganisms (Pérez-Jaramillo et al., 2018).
Conclusion
In order to make agriculture productive and sustainable, it would be preferable to combine several approaches linked to the microbiome tool: (i) selection of plants and their genetic modification; (ii) targeted design of specific microbiomes; (iii) customization of the appropriate microbiome in relation to the cultural species of interest and the cultivation methods (Arif et al., 2020).
If studies raise the feasibility of the implementation of both approaches, adjustments remain to be made on (i) the preferred selection method; (ii) stabilization of the microbiomes of interest between successive generations; (iii) methods of configuring optimized microbiomes.
Ultimately, to master hologenome selection programs, it would also be necessary to conduct in-depth studies focused on five priority areas cited by Busby et al. (2017):
• development of holonbionts model for cultivated plants with collections of referenced associated strains and characterized genomes;
• definition of the main microbiomes and metagenomes corresponding to these models;
• definition of the rules governing the assembly of functional microbiomes;
• determining the mechanisms of plant-microorganism interactions;
• characterization of the link between genotype, plant and environment.
Now, the consideration of hologenomes in breeding programs must be taken into account to acquire crops which are both more productive and resilient. However, it is necessary to combine different approaches to obtain efficient holobionts, in particular by adopting agricultural practices that respect agroecosystems and favor the establishment of microbial communities associated with plants (Tosi et al., 2020).
Author Contributions
ML'H: writing—original draft. RD: review and editing. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the funding of the French Institute for the Research and Development (IRD) and the Laboratory of Mediterranean and Tropical Symbioses research unit (LSTM).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We are thankful to IRD for the funding.
References
Abdelfattah, A., Wisniewski, M., Schena, L., and Tack, A. J. M. (2021). Experimental evidence of microbial inheritance in plants and transmission routes from seed to phyllosphere and root. Enviro. Microbiol. 23, 2199–2214. doi: 10.1111/1462-2920.15392
Adam, E., Bernhart, M., Müller, H., Winkler, J., and Berg, G. (2018). The Cucurbita pepo seed microbiome: genotype-specific composition and implications for breeding. Plant Soil 422, 35–49. doi: 10.1007/s11104-016-3113-9
Adams, P. D., and Kloepper, J. W. (2002). Effect of host genotype on indigenous bacterial endophytes of cotton (Gossypium hirsutum L.). Plant Soil 240, 181–189. doi: 10.1023/A:1015840224564
Ambika, M. B., Ratering, S., Rusch, V., Schwiertz, A., Geissler-Plaum, R., Cardinale, M., et al. (2016). Bacterial microbiota associated with flower pollen is influenced by pollination type, and shows a high degree of diversity and species-specificity. Environ. Microbiol. 18, 5161–5174. doi: 10.1111/1462-2920.13524
Andres-Barrao, C., Lafi, F. F., Alam, I., De Zelicourt, A., Eida, A. A., Bokhari, A., et al. (2017). Complete genome sequence analysis of Enterobacter sp. SA187, a plant multi-stress tolerance promoting endophytic bacterium. Front. Microbiol. 8:2023. doi: 10.3389/fmicb.2017.02023
Annapurna, K., Govindasamy, V., Sharma, M., Ghosh, A., and Chikara, S. K. (2018). Whole genome shotgun sequence of Bacillus paralicheniformis strain KMS 80, a rhizobacterial endophyte isolated from rice (Oryza sativa L.). Biotech 8:223. doi: 10.1007/s13205-018-1242-y
Arif, I., Batool, M., and Schenk, P. M. (2020). Plant microbiome engineering: expected benefits for improved crop growh and resilience. Trends Microbiol. 38, 1385–1396. doi: 10.1016/j.tibtech.2020.04.015
Assumpcao, L. D. C., Lacava, P. T., Dias, A. C. F., Azevedo, J. L. D., and Menten, J. O. M. (2009). Diversity and biotechnological potential of endophytic bacterial community of soybean seeds. Pesq. Agrop. Brasileira 44, 503–510. doi: 10.1590/S0100-204X2009000500010
Bakker, M. G., Manter, D. K., Sheflin, A. M., Weir, T. L., and Vivanco, J. M. (2012). Harnessing the rhizosphere microbiome through plant breeding and agricultural management. Plant Soil 360, 1–13. doi: 10.1007/s11104-012-1361-x
Berendsen, R. L., Pieterse, C. M., and Bakker, P. A. (2012). The rhizosphere microbiome and plant health. Trends Plant Sci. 17, 478–486. doi: 10.1016/j.tplants.2012.04.001
Berg, G., Köberl, M., Rybakova, D., Müller, H., Grosch, R., and Smalla, K. (2017). Plant microbial diversity is suggested as the key to future biocontrol and health trends. FEMS Microbiol. Ecol. 93:fix050. doi: 10.1093/femsec/fix050
Berg, G., and Raaijmakers, J. M. (2018). Saving seed microbiomes. ISME J. 12, 1167–1170. doi: 10.1038/s41396-017-0028-2
Bergna, A., Cernava, T., Rändler, M., Grosch, R., Zachow, C., and Berg, G. (2019). Tomato seeds preferably transmit plant beneficial endophytes. Phytobiomes J. 2, 183–193. doi: 10.1094/PBIOMES-06-18-0029-R
Bertalan, M., Albano, R., de Padua, V., Rouws, L., Rojas, C., Hemerly, A., et al. (2009). Complete genome sequence of the sugarcane nitrogen-fixing endophyte Gluconacetobacter diazotrophicus Pal5. BMC Genomics 10:450. doi: 10.1186/1471-2164-10-450
Bodhankar, S., Grover, M., Hemanth, S., Reddy, G., Rasul, S., Yadav, S. K., et al. (2017). Maize seed endophytic bacteria: dominance of antagonistic, lytic enzyme-producing Bacillus spp. Biotech 7:232. doi: 10.1007/s13205-017-0860-0
Bomfim, C. S. G., da Silva, V. B., Cursino, L. H. S., da Silva Mattos, W., Santos, J. C. S., de Souza, L. S. B., et al. (2020). Endophytic bacteria naturally inhabiting commercial maize seeds occupy different niches and are efficient plant growth-promoting agents. Symbiosis 81, 255–269. doi: 10.1007/s13199-020-00701-z
Booker, A., Jalil, B., Frommenwiler, D., Reich, E., Zhai, L. X., Kulic, Z., et al. (2016). The authenticity and quality of Rhodiola rosea products. Phytomedicine 15, 754–762. doi: 10.1016/j.phymed.2015.10.006
Booth, A. (2014). Symbiosis selection and individuality. Biol. Philos. 29, 657–673. doi: 10.1007/s10539-014-9449-8
Bordenstein, S. R., and Theis, K. R. (2015). Host biology in the light of the microbiome: ten principles of holobionts and hologenomes. PLoS Biol. 13:e1002226. doi: 10.1371/journal.pbio.1002226
Bulgarelli, D., Schlaeppi, K., Spaepen, S., Loren Ver van The maat, E., and Schulze- Lefert, P. (2013). Structure and functions of the bacterial microbiota of plants. Ann. Rev. Plant Biol. 64, 807–838. doi: 10.1146/annurev-arplant-050312-120106
Busby, P. E., Soman, C., Wagner, M. R., Friesen, M. L., Kremer, J., Bennett, A., et al. (2017). Research priorities for harnessing plant microbiomes in sustainable agriculture. PLoS Biol. 15:e2001793. doi: 10.1371/journal.pbio.2001793
Card, S., Johnson, L., Teasdale, S., and Caradus, J. (2016). Deciphering endophyte behaviour: the link between endophyte biology and efficacious biological control agents. FEMS Microbiol. Ecol. 92:fiw114. doi: 10.1093/femsec/fiw114
Chaluvadi, S., and Bennetzen, J. L. (2018). Species-associated differences in the below-ground microbiomes of wild and domesticated setaria. Front. Plant Sci. 9:1183. doi: 10.3389/fpls.2018.01183
Chee-Sanford, J. C., Williams, M. M., Davis, A. S., and Sims, G. K. (2006). Do microorganisms influence seed-bank dynamics? Weed Sci. 54, 575–587. doi: 10.1614/WS-05-055R.1
Chen, L., Wu, Y. D., Chong, X. Y., Xin, Q. H., Wang, D. X., and Bian, K. (2019). Seed-borne endophytic Bacillus velezensis LHSB1 mediate the biocontrol of peanut stem rot caused by Sclerotium rolfsii. J. Appl. Microbiol. 128, 803–813. doi: 10.1111/jam.14508
Chen, X., Krug, L., Yang, M., Berg, G., and Cernava, T. (2020). Conventional seed coating reduces prevalence of proteobacterial endophytes in Nicotiana tabacum. Ind. Crops Prod. 155:112784. doi: 10.1016/j.indcrop.2020.112784
Chen, Y. H., Gols, R., and Benrey, B. (2015). Crop domestication and its impact on naturally selected trophic interactions. Ann. Rev. Entomol. 60, 35–58. doi: 10.1146/annurev-ento-010814-020601
Chimwamurombe, P. M., Grönemeyer, J. L., and Reinhold-Hurek, B. (2016). Isolation and characterization of culturable seed-associated bacterial endophytes from gnotobiotically grown Marama bean seedlings. FEMS Microbiol. Ecol. 92:fiw083. doi: 10.1093/femsec/fiw083
Compant, S., Clément, C., and Sessitsch, A. (2010). Plant growth-promoting bacteria in the rhizo-and endosphere of plants: their role, colonization, mechanisms involved and prospects for utilization. Soil Biol. Biochem. 42, 669–678. doi: 10.1016/j.soilbio.2009.11.024
Compant, S., Mitter, B., Colli-Mull, J. G., Gangl, H., and Sessitsch, A. (2011). Endophytes of grapevine flowers, berries, and seeds: identification of cultivable bacteria, comparison with other plant parts, and visualization of niches of colonization. Microb. Ecol. 62, 188–197. doi: 10.1007/s00248-011-9883-y
Contreras-Liza, S. E. (2021). “Plant breeding and microbiome,” in Plant Breeding - Current and Future Views, ed I. Y. Abdurakhmonov (IntechOpen), 107.
Cope Selby, N., Cookson, A., Squance, M., Donnison, I., Flavell, R., and Farrar, K. (2017). Endophytic bacteria in Miscanthus seed: implications for germination, vertical inheritance of endophytes, plant evolution and breeding. GCB Bioenergy 9, 57–77. doi: 10.1111/gcbb.12364
Corbin, K. R., Bolt, B., and Rodriguez Lopez, C. M. (2020). Breeding for beneficial microbial communities using epigenomics. Front. Microbiol. 11:937. doi: 10.3389/fmicb.2020.00937
Cordovez, V., Dini-Andreote, F., Carrión, V. J., and Raaijmakers, J. M. (2019). Ecology and evolution of plant microbiomes. Annu. Rev. Microbiol. 73, 69–88. doi: 10.1146/annurev-micro-090817-062524
Cottyn, B., Regalado, E., Lanoot, B., De Cleene, M., Mew, T., and Swings, J. (2001). Bacterial populations associated with rice seed in the tropical environment. Phytopathology 91, 282–292. doi: 10.1094/PHYTO.2001.91.3.282
Cui, J. L., Gao, X. Y., Vijayakumar, V., Guo, Z. X., Wang, M. L., Wang, J. H., et al. (2020). Regulation by fungal endophyte of Rhodiola crenulata from enzyme genes to metabolites based on combination of transcriptome and metabolome. J. Sci. Food Agric. 100, 4483–4494. doi: 10.1002/jsfa.10489
Cui, J. L., Wang, Y. N., Jiao, J., Gong, Y., Wang, J. H., and Wang, M. L. (2017). Fungal endophyte-induced salidroside and tyrosol biosynthesis combined with signal cross-talk and the mechanism of enzyme gene expression in Rhodiola crenulata. Sci. Rep. 7:12540. doi: 10.1038/s41598-017-12895-2
Dent, D., and Cocking, E. (2017). Establishing symbiotic nitrogen fixation in cereals and other non-legume crops: the greener nitrogen revolution. Agric. Food Secur. 6, 7–9. doi: 10.1186/s40066-016-0084-2
Díaz Herrera, S., Grossi, C., Zawoznik, M., and Groppa, M. D. (2016). Wheat seeds harbour bacterial endophytes with potential as plant growth promoters and biocontrol agents of Fusarium graminearum. Microbiol. Res. 186, 37–43. doi: 10.1016/j.micre.2016.03.002
Donnarumma, F., Capuana, M., Vettori, C., Petrini, G., Giannini, R., Indorato, C., et al. (2011). Isolation and characterisation of bacterial colonies from seeds and in vitro cultures of Fraxinus spp. from Italian sites. Plant Biol. 13, 169–176. doi: 10.1111/j.1438-8677.2010.00334.x
Dowarsh, B., Agarwal, H., Krishnatreya, D. B., Sharma, P. L., Kalita, N., and Agarwala, N. (2021). Evaluation of seed associated endophytic bacteria from tolerant chilli cv. Firingi Jolokia for their biocontrol potential against bacterial wilt disease. Microb. Res. 248:126751. doi: 10.1016/j.micres.2021.126751
Ebeltagy, A., Nishioka, K., Suzuki, H., Sato, T., Sato, Y., Morisaki, H., et al. (2000). Isolation and characterization of endophytic bacteria from wild and traditionally cultivated rice varieties. Soil Sci. Plant Nutr. 46, 617–629. doi: 10.1080/00380768.2000.10409127
El-Hawary, S. S., Mohammed, R., AbouZid, S. F., Bakeer, W., Ebel, R., Sayed, A. M., et al. (2016). Solamargine production by a fungal endophyte of Solanum nigrum. J. Appl. Microbiol. 120, 900–911. doi: 10.1111/jam.13077
Escobar Rodríguez, C., Antonielli, L., Mitter, B., Trognitz, F., and Sessitsch, A. (2020). Heritability and functional importance of the Setaria viridis bacterial seed microbiome. Phytobiomes J. 4, 40–52. doi: 10.1094/PBIOMES-04-19-0023-R
Espinosa Zaragoza, S., Sánchez Cruz, R., Sanzón Gómez, D., Escobar Sandoval, M. C., Yañez Ocampo, G., Morales Constantino, M. A., et al. (2021). Identification of endophytic bacteria of seeds from Cedrela odorata L. (Meliaceae) with biotechnological characteristics. Acta Biol. Colomb. 26, 196–206. doi: 10.15446/abc.v26n2.85325
Etesami, H., Alikhani, H. A., and Hosseini, H. M. (2015). Indole-3-acetic acid (IAA) production trait, a useful screening to select endophytic and rhizosphere competent bacteria for rice growth promoting agents. MethodsX 2, 72–78. doi: 10.1016/j.mex.2015.02.008
Ewald, P. T. W. (1987). Transmission modes and evolution of the parasitism-mutualism continuum. Ann. N. Y. Acad. Sci. 503, 295–306. doi: 10.1111/j.1749-6632.1987.tb40616.x
Faddetta, T., Abbate, L., Alibrandi, P., Arancio, W., Siino, D., Strati, F., et al. (2021). The endophytic microbiota of Citrus limon is transmitted from seed to shoot highlighting differences of bacterial and fungal community structures. Sci. Rep. 11:7078. doi: 10.1038/s41598-021-86399-5
Ferreira, A., Quecine, M. C., Lacava, P. T., Oda, S., Azevedo, J. L., and Araujo, W. L. (2008). Diversity of endophytic bacteria from Eucalyptus species seeds and colonization of seedlings by Pantoea agglomerans. FEMS Microbiol Lett. 287, 8–14. doi: 10.1111/j.1574-6968.2008.01258.x
Finkel, O. M., Castrillo, G., Herrera Paredes, S., Gonzalez, I. S., and Dangl, J. L. (2020). Understanding and exploiting plant beneficial microbes. Curr. Opin. Plant Biol. 38, 155–163. doi: 10.1016/j.pbi.2017.04.018
Fouts, D. E., Tyler, H. L., DeBoy, R. T., Daugherty, S., Ren, Q., Badger, J. H., et al. (2008). Complete genome sequence of the N2-fixing broad host range endophyte Klebsiella pneumoniae 342 and virulence predictions verified in mice. PLoS Genet. 4:e1000141. doi: 10.1371/journal.pgen.1000141
Frank, A., Saldierna Guzman, J., and Shay, J. (2017). Transmission of bacterial endophytes. Microorganisms 5:70. doi: 10.3390/microorganisms5040070
Fu, Y., Yin, Z. H., and Yin, C. Y. (2017). Biotransformation of ginsenoside Rb1 to ginsenoside Rg3 by endophytic bacterium Burkholderia sp. GE 17_7 isolated from Panax ginseng. J. Appl. Microbiol. 122, 1579–1585. doi: 10.1111/jam.13435
Fürnkranz, M., Lukesch, B., Müller, H., Huss, H., Grube, M., and Berg, G. (2012). Microbial diversity inside pumpkins: microhabitat-specific communities display a high antagonistic potential against phytopathogens. Microb. Ecol. 63, 418–428. doi: 10.1007/s00248-011-9942-4
Gagne-Bourgue, F., Aliferis, K., Seguin, P., Rani, M., Samson, R., and Jabaji, S. (2013). Isolation and characterization of indigenous endophytic bacteria associated with leaves of switchgrass (Panicum virgatum L.) cultivars. J. Appl. Microbiol. 114, 836–853. doi: 10.1111/jam.12088
Geisen, S., Kostenko, O., Cnossen, M. C., ten Hooven, F. C., Vreš, B., and van der Putten, W, W. H. (2017). Seed and root endophytic fungi in a range expanding and a related plant species, Front. Microbiol. 8:1645. doi: 10.3389/fmicb.2017.01645
Germida, J., and Siciliano, S. (2001). Taxonomic diversity of bacteria associated with the roots of modern, recent and ancient wheat cultivars. Biol. Fertil. Soils 33, 410–415. doi: 10.1007/s003740100343
Glassner, H., Zchori-Fein, E., Yaron, S., Sessitsch, A., Sauer, U., and Compant, S. (2017). Bacterial niches inside seeds of Cucumis melo L. Plant Soil 2017, 1–3. doi: 10.1007/s11104-017-3175-3
Glick, B. R., Penrose, D. M., and Li, J. (1998). A model for the lowering of plant ethylene concentrations by plant growth-promoting bacteria. J. Theor. Biol. 190, 63–68. doi: 10.1006/jtbi.1997.0532
Gluck-Thaler, E., and Slot, J. C. (2015). Dimensions of horizontal gene transfer in eukaryotic microbial pathogens. PLoS Pathog. 11:e1005156. doi: 10.1371/journal.ppat.1005156
Goggin, E. D., Emery, R. J. N. R., Kurepin, L. V., and Powles, S. B. (2015). A potential role for endogenous microflora in dormancy release, cytokinin metabolim and the response to fluoridone iun Lolium rigidum seeds. Ann. Bot. 115, 293–301. doi: 10.1093/aob/mcu231
Gopal, M., and Gupta, A. (2016). Microbiome selection could spur next-genetation plant breeding strategies. Front. Microbiol. 7:1971. doi: 10.3389/fmicb.2016.01971
Greenwood, N. N., and Earnshaw, A. (1997). Chemistry of the Elements, 2nd Edn. Oxford: Butterworth-Heinemann.
Guerrero, R., Margulis, L., and Berlanga, M. (2013). Symbiogenesis: the holobiont as a unit of evolution. Int. Microbiol. 16, 133–143. doi: 10.2436/20.1501.01.188
Gupta, S., and Chaturvedi, P. (2017). Foliar endophytic diversity of Centella asiatica (L.) Urban in relation to different seasons and leaf-age. Int. J. Curr. Microbiol. Appl. Sci. 6, 468–477. doi: 10.20546/ijcmas.2017.606.054
Hallmann, J., Quadt-Hallmann, A., Mahaffee, W. F., and Kloepper, J. W. (1997). Bacterial endophytes in agricultural crops. Can. J. Microbiol. 43, 895–914. doi: 10.1139/m97-131
Hardoim, P. (2019). “The ecology of seed microbiota,” in Seed Endophytes, eds S. K. Verma and J. F. White, Jr. (Cham: Springer), 103–126. doi: 10.1007/978-3-030-10504-4_6
Hardoim, P. R., Hardoim, C. C., Van Overbeek, L. S., and Van Elsas, J. D. (2012). Dynamics of seed-borne rice endophytes on early plant growth stages. PLoS ONE 7:e30438. doi: 10.1371/journal.pone.0030438
Hardoim, P. R., van Overbeek, L. S., Berg, G., Pirttilä, A. M., Compant, S., Campisano, A., et al. (2015). The hidden world within plants: ecological and evolutionary considerations for defining functioning of microbial endophytes. Microbiol. Mol. Biol. Rev. 79, 294–320. doi: 10.1128/MMBR.00050-14
Herrera, S. D., Grossi, C., Zawoznik, M., and Groppa, M. D. (2016). Wheat seeds harbour bacterial endophytes with potential as plant growth promoters and biocontrol agents of Fusarium graminearum. Microbiol. Res. 186, 37–43. doi: 10.1016/j.micres.2016.03.002
Irmer, S., Podzun, N., Langel, D., Heidemann, F., Kaltenegger, E., Schemmerling, B., et al. (1994). Partial purification and characterization of 1-aminocyclopropane-1-carboxylate deaminase from the plant growth promoting rhizobacterium Pseudomonas putida GR12-2. Can. J. Microbiol. 40, 1019–1025. doi: 10.1139/m94-162
Jia, M., Chen, L., Xin, H. L., Zheng, C. J., Rahman, K., Han, T., et al. (2016). A friendly relationship between endophytic fungi and medicinal plants: a systematic review. Front. Microbiol. 7:906. doi: 10.3389/fmicb.2016.00906
Johnston-Monje, D., Lundberg, D. S., Lazarovits, G., Reis, V. M., and Raizada, M. N. (2016). Bacterial populations in juvenile maize rhizospheres originate from both seed and soil. Plant Soil 405, 337–355. doi: 10.1007/s11104-016-2826-0
Johnston-Monje, D., and Raizada, M. N. (2011). Conservation and diversity of seed associated endophytes in Zea across boundaries of evolution, ethnography and ecology. PLoS ONE 6:e0020396. doi: 10.1371/journal.pone.0020396
Kaga, H., Mano, H., Tanaka, F., Watanabe, A., Kaneko, S., and Morisaki, H. (2009). Rice seeds as sources of endophytic bacteria. Microbes Environ. 24, 154–162. doi: 10.1264/jsme2.ME09113
Kane, K. H. (2011). Effects of endophyte infection on drought stress tolerance of Lolium perenne accessions from the Mediterranean region. Environ. Exp. Bot. 71, 337–344. doi: 10.1016/j.envexpbot.2011.01.002
Kaneko, T., Minamisawa, K., Isawa, T., Nakatsukasa, H., Mitsui, H., Kawaharada, Y., et al. (2010). Complete genomic structure of the cultivated rice endophyte Azospirillum sp. B510. DNA Res. 17, 37–50. doi: 10.1093/dnares/dsp026
Khalaf, E. M., and Raizada, M. N. (2016). Taxonomic and functional diversity of cultured seed associated microbes of the cucurbit family. BMC Microbiol. 16:131. doi: 10.1186/s12866-016-0743-2
Khalaf, E. M., and Raizada, M. N. (2018). Bacterial seed endophytes of domesticated cucurbits antagonize fungal and oomycete pathogens including powdery mildew. Front. Microbiol. 9:42. doi: 10.3389/fmicb.2018.00042
Krause, A., Ramakumar, A., Bartels, D., Battistoni, F., Bekel, T., Boch, J., et al. (2006). Complete genome of the mutualistic, N 2-fixing grass endophyte Azoarcus sp. strain BH72. Nat. Biotechnol. 24:1384. doi: 10.1038/nbt1243
Krishnamoorthy, A., Agarwal, T., Kotamreddy, J. N. R., Bhattacharya, R., Mitra, A., Maiti, T. K., et al. (2020). Impact of seed-transmitted endophytic bacteria on intra- and inter-cultivar plant growth promotion modulated by certain sets of metabolites in rice crop. Microbiol. Res. 241:126581. doi: 10.1016/j.micres.2020.126582
Kukkurainen, S., Leino, A., Vähämiko, S., Kärkkäinen, H. R., Ahanen, K., Sorvari, S., et al. (2005). Occurrence and location of endophytic bacteria in garden and wild strawberry. HortScience 40, 348–352. doi: 10.21273/HORTSCI.40.2.348
Kumar, K., Pal, G., Verma, A., and Verma, S. K. (2020). Seed inhabiting bacteria endophytes on finger millet (Eleusine coracana L.) promote seedling growth an ddevelopment, and protect from fungal disease. South Afr. J. Bot. 134, 91–98. doi: 10.1016/j.sajb.2020.03.032
Kusari, S., Hertweck, C., and Spiteller, M. (2012). Chemical ecology of endophytic fungy: origins of secondary metabolites. Chem. Biol. 19, 792–798. doi: 10.1016/j.chembiol.2012.06.004
Kusstatscher, P., Adam, E., Wicaksono, W. A., Bernhart, M., Olimi, E., Müller, H., et al. (2021). Microbiome-assisted breeding to understand cultivar-dependent assembly in Cucurbita pepo. Front. Plant Sci. 12:642027. doi: 10.3389/fpls.2021.642027
Kwak, M.-J., Song, J. Y., Kim, S.-Y., Jeong, H., Kang, S. G., Kim, B. K., et al. (2012). Complete genome sequence of the endophytic bacterium Burkholderia sp. strain KJ006. J. Bacteriol. 194, 4432–4433. doi: 10.1128/JB.00821-12
Kwak, M. J., Kong, H. G., Choi, K., Kwon, S. K., Song, J. Y., Lee, J., et al. (2018). Rhizosphere microbiome structure alters to enable wilt resistance in tomato. Nat. Biotechnol. 36, 1100–1109. doi: 10.1038/nbt.4232
Leff, J. W., Lynch, R. C., Kane, N. C., and Fierer, N. (2017). Plant domestication and the assembly of bacterial and fungal communities associated with strains of the common sunflower, Helianthus annuus. New Phytol. 214, 412–423. doi: 10.1111/nph.14323
Leghari, S. J., Wahocho, N. A., Laghari, G. M., Hafeez Laghari, A., Mustafa Bhabhan, G., Hussain Talpur, K., et al. (2016). Role of nitrogen for plant growth and development: a review. Adv. Environ. Biol. 10, 209–219.
Li, H., Parmar, S., Sharma, V. K., and White, J. F. (2019). “Seed endophytes and their potential applications,” in Seed Endophytes, eds S. K. Verma and J. F. White, Jr. (Cham: Springer), 35–54. doi: 10.1007/978-3-030-10504-4_3
Li, J., Zhao, G. Z., Chen, H. H., Wang, H. B., Sin, S., Zhu, W. Y., et al. (2008). Antitumour and antimicrobial activities of endophytic streptomycetes from pharmaceutical plants in rainforest. J. Appl. Microbiol. 47, 574–580. doi: 10.1111/j.1472-765X.2008.02470.x
Li, L., Zhang, Z., Pan, S., Li, L., and Li, X. (2019b). Characterization and metabolism effect of seed endophytic bacteria associated with peanut grown in South China. Front. Microbiol. 10:2659. doi: 10.3389/fmicb.2019.02659
Liu, Y., Bai, F., Li, N., Wang, W., and Cheng, C. (2017). Identification of endophytic bacterial strain RSE1 from seeds of super hybrid rice Shenliangyou 5814 (Oryza sativa L.) and evaluation of its antagonistic activity. Plant Growth Regul. 82, 403–408. doi: 10.1007/s10725-017-0265-4
Liu, Y., Yan, H., Zhang, X., Zhang, R., Li, M., Yang, F., et al. (2020). Investigating the endophytic bacterial diversity and community structures in seeds of genetically related maize (Zea mays L.) genotypes, Biotech 10:27. doi: 10.1007/s13205-019-2034-8
Liu, Y., Zuo, S., Xu, L., Zou, Y., and Song, W. (2012). Study on diversity of endophytic bacterial communities in seeds of hybrid maize and their parental lines. Arch. Microbiol. 194, 1001–1012. doi: 10.1007/s00203-012-0836-8
Liu, Y., Zuo, S., Zou, Y., Wang, J., and Song, W. (2013). Investigation on diversity and population succession dynamics of seed endophytic bacteria of maize (Zea mays L., Nongda 108) at different growth stages. Ann. Microbiol. 63, 71–79. doi: 10.1007/s13213-012-0446-3
Loaces, I., Ferrando, L., and Scavino, A. F. (2011). Dynamics, diversity and function of endophytic siderophore-producing bacteria in rice. Microb Ecol. 61, 606–618. doi: 10.1007/s00248-010-9780-9
Loper, J., and Henkels, M. D. (1999). Utilization of heterologous siderophores enhances levels of iron available to Pseudomonas putida in the rhizosphere. Appl. Environ. Microbiol. 65, 5357–5363. doi: 10.1128/AEM.65.12.5357-5363.1999
Loper, J. E., and Buyer, J. S. (1991). Siderophores in microbial interactions on plant surfaces. Mol. Plant Microbe Interact. 41, 5–13. doi: 10.1094/MPMI-4-005
Lopez, S. M. Y., Pastorino, G. N., Franco, M. E. E., Medina, R., Lucentini, C. G., Saparrat, M. C. N., et al. (2018). Microbioal endophytes that live within the seeds of two tomato hybrids cultivated in Argentina, Agronomy 8:136. doi: 10.3390/agronomy8080136
Lopez-Fernandez, S., Compant, S., Vrhovsek, U., Bianchedi, P. L., Sessitsch, A., Pertot, I., et al. (2016). Grapevine colonization by endophytic bacteria shifts secondary metabolism and suggests activation of defense pathways. Plant Soil 405, 155–175. doi: 10.1007/s11104-015-2631-1
López-López, A., Rogel, M. A., Ormeño-Orillo, E., Martínez-Romero, J., and Martínez-Romero, E. (2010). Phaseolus vulgaris seed-borne endophytic community with novel bacterial species such as Rhizobium endophyticum sp. nov. Syst. Appl. Microbiol. 33, 322–327. doi: 10.1016/j.syapm.2010.07.005
Maehara, S., Agusta, A., Kitamura, C., Ohashi, K., and Shibuya, H. (2016). Composition of the endophytic filamentous fungi associated with Cinchona ledgeriana seeds and production of Cinchona alkaloids. J. Nat. Med. 70, 271–275. doi: 10.1007/s11418-015-0954-0
Maggini, V., De Leo, M., Mengoni, A., Gallo, E. G., Miceli, E., Reidel, R. V. B., et al. (2017). Plant-endophytes interaction influences the secondary metabolism in Echinacea purpurea (L.) Moench: an in vitro model. Sci. Rep. 7:16924. doi: 10.1038/s41598-017-17110-w
Mahmood, A., and Kataoka, R. (2020). Metabolite profiling reveals a complex response of plants to application of plant growth-promoting endophytic bacteria. Microbiol. Res. 324:126421. doi: 10.1016/j.micres.2020.126421
Mano, H., Tanaka, F., Watanabe, A., Kaga, H., Okunishi, S., and Morisaki, H. (2006). Culturable surface and endophytic bacterial flora of the maturing seeds of rice plants (Oryza sativa) cultivated in a paddy field. Microbes Environ. 21, 86–100. doi: 10.1264/jsme2.21.86
Martinez-Garcia, P. M., Ruano-Rosa, D., Schiliro, E., Prieto, P., Ramos, C., Rodriguez-Palenzuela, P., et al. (2015). Complete genome sequence of Pseudomonas fluorescens strain PICF7, an indigenous root endophyte from olive (Olea europaea L.) and effective biocontrol agent against Verticillium dahliae. Stand. Genomic Sci. 10:10. doi: 10.1186/1944-3277-10-10
Martínez-Romero, E., Aguirre-Noyola, J. L., Taco-Taype, N., Martínez-Romero, J., and Zuñiga-Dávila, D. (2020). Plant microbiota modified by plant domestication. Syst. Appl. Microbiol. 43:126106. doi: 10.1016/j.syapm.2020.126106
Mastretta, C., Taghavi, S., van der Lelie, D., Mengoni, A., Galardi, F., Gonnelli, C., et al. (2009). Enodphytic Endophytic bacteria from seeds of Nicotiana Tabacum can reduce cadmium phytotoxicity. Int. J. Phytoremed. 11, 251–267. doi: 10.1080/15226510802432678
Matsumoto, H., Fan, X., Wang, Y., Kusstatscher, P., Duan, J., Wu, S., et al. (2021). Bacterial seed endophyte shapes disease resistance in rice. Nat. Plants 7, 60–72. doi: 10.1038/s41477-020-00826-5
Mendes, L. W., de Chaves, M. G., da Fonseca, M. C., Mendes, R., Raaijmakers, J. M., and Tsai, S. M. (2019). Resistance breeding of common bean shapes the physiology of the rhizosphere microbiome. Front. Microbiol. 10:2252. doi: 10.3389/fmicb.2019.02252
Mitter, B., Pfaffenbichler, N., Flavell, R., Compant, S., Antonielli, L., Petric, A., et al. (2017). A new approach to modify plant microbiomes and traits by introducing beneficial bacteria at flowering into progeny seeds. Front. Microbiol. 8:11. doi: 10.3389/fmicb.2017.00011
Mueller, U. G., and Sachs, J. L. (2015). Engineering microbiomes to improve plant and animal health. Trends Microbiol. 23, 606–617. doi: 10.1016/j.tim.2015.07.009
Mukherjee, A., Singh, B. K., and Verma, J. P. (2020). Harnessing chickpea (Cicer arietinum L.) seed endophytes for enhancing plant growth attributes and bio-controllingbio controlling against Fusarium sp. Microbiol. Res. 237:126469. doi: 10.1016/j.micres.2020.126469
Mukhopadhyay, K., Garrison, N. K., Hinton, D. M., Bacon, C. W., Khush, G. S., Peck, H. D., et al. (1996). Identification and characterization of bacterial endophytes of rice. Mycopathologia 134, 151–159. doi: 10.1007/BF00436723
Nelson, E. (2004). Microbial dynamics and interactions in the spermosphere. Annu. Rev. Phytopathol. 42, 271–309. doi: 10.1146/annurev.phyto.42.121603.131041
Nelson, E. B. (2018). The seed microbiome: origins, interactions and impacts. Plant Soil 422, 7–34. doi: 10.1007/s11104-017-3289-7
Nelson, E. B., Simoneau, P., Barret, M., Mitter, B., and Compant, S. (2018). Editorial special issue: the soil, the seed, the microbes and the plant. Plant Soil 422, 1–5. doi: 10.1007/s11104-018-3576-y
Nicholson, W., Munakata, N., Horneck, G., Melosh, H., and Setlow, P. (2000). Resistance of Bacillus endospores to extreme terrestrial and extraterrestrial environments. Microbiol. Mol. Biol. Rev. 64, 548–572. doi: 10.1128/MMBR.64.3.548-572.2000
O'Callaghan, M. (2016). Microbial inoculation of seed for improved crop performance: issues and opportunities. Appl. Microbiol. Biotechnol. 100, 5729–5746. doi: 10.1007/s00253-016-7590-9
Oehrle, N. W., Karr, D. B., Kremer, R. J., and Emerich, D. W. (2000). Enhanced attachment of Bradyrhizobium japonicum to soybean through reduced root colonization of internally seedborne microorganisms. Can. J. Microbiol. 46, 600–606. doi: 10.1139/w00-030
Ogbe, A. E., Finnie, J. F., and Van Staden, J. (2020). The role of endophytes in secondary metabolites accumulation in medicinal plants under abiotic stress. South Afr. J. Bot. 134, 126–134. doi: 10.1016/j.sajb.2020.06.023
Okunishi, S., Sako, K., Mano, H., Imamura, A., and Morisaki, H. (2005). Bacterial flora of endophytes in the maturing seed of cultivated rice (Oryza sativa). Microbes Environ. 20, 168–177. doi: 10.1264/jsme2.20.168
Özkurt, E., Hassani, M. A., Sesiz, U., Künzel, S., Dagan, T., Özkan, H., et al. (2020). Seed-derived microbial colonization of wild emmer and domesticated bread wheat (Triticum dicoccoides and T. aestivum) seedlings shows pronounced differences in overall diversity and composition. mBio. 11:e02637-20. doi: 10.1128/mBio.02637-20
Pal, G., Kumar, K., Verma, A., Kharwar, R. N., and Verma, S. K. (2021). “Seed-inhabiting endophytes,” in New and Future Developments in Microbiol Bitechnology and Bioengineering, eds J. P. Verma, C. A. Macdonald, and A. R. Podil (Cambridge, MA: Elsevier), 35–44. doi: 10.1016/B978-0-444-64325-4.00004-3
Panke-Buisse, K., Lee, S., and Kao-Kniffin, J. (2017). Cultivated sub-populations of soil microbiomes retain early flowering plant trait. Microb. Ecol. 73, 394–403. doi: 10.1007/s00248-016-0846-1
Panke-Buisse, K., Poole, A. C., Goodrich, J. K., Ley, R. E., and Kao-Kniffin, J. (2015). Selection on soil microbiomes reveals reproducible impacts on plant function. ISME J. 9, 980–989. doi: 10.1038/ismej.2014.196
Parthasarathy, A., Gan, H. M., Wong, N. H., Savka, M. A., Steiner, K. K., Henry, K. R., et al. (2018). Isolation and genomic characterization of six endophytic bacteria isolated from Saccharum sp (sugarcane): insights into antibiotic, secondary metabolite and quorum sensing metabolism. J. Genom. 6, 117–121. doi: 10.7150/jgen.28335
Paungfoo-Lonhienne, C., Rentsch, D., Robatzek, S., Webb, R. I., Sagulenko, E., Nösholm, T., et al. (2010). Turning the table: plants consume microbes as source of nutrients. PLoS ONE 5:e11915 doi: 10.1371/journal.pone.0011915
Peiffer, J. A., Spor, A., Koren, O., Jin, Z., Tringe, S. G., Dangl, J. L., et al. (2013). Diversity and heritability of the maize rhizosphere microbiomeunder field conditions. Proc. Natl. Acad. Sci. U. S. A. 110, 6548–6553. doi: 10.1073/pnas.1302837110
Perez-Jaramillo, J. E., Carrion, V. J., Bosse, M., Ferrao, L. F. V., de Hollander, M., Garcia, A. A. F., et al. (2017). Linking rhizosphere microbiome composition of wild and domesticated Phaseolus vulgaris to genotypic and root phenotypic traits. ISME J. 11, 2244–2257. doi: 10.1038/ismej.2017.85
Pérez-Jaramillo, J. E., Carrión, V. J., de Hollander, M., and Raaijmakers, J. M. (2018). The wild side of plant microbiomes. Microbiome 6:143. doi: 10.1186/s40168-018-0519-z
Pérez-Jaramillo, J. E., Mendes, R., and Raaijmakers, J. M. (2016). Impact of plant domestication on rhizosphere microbiome assembly and functions. Plant Mol. Biol. 90, 635–644. doi: 10.1007/s11103-015-0337-7
Pitzschke, A. (2016). Developmental peculiarities and seed-borne endophytes in quinoa: omnipresent, robust bacilli contribute to plant fitness. Front. Microbiol. 7:2. doi: 10.3389/fmicb.2016.00002
Porter, S. S., and Sachs, J. L. (2020). Agriculture and the disruption of plant–microbial Symbiosis. Trends Ecol. Evol. 35, 426–439. doi: 10.1016/j.tree.2020.01.006
Puente, M. E., Li, C. Y., and Bashan, Y. (2009a). Endophytic bacteria in cacti seeds can improve the development of cactus seedlings. Environ. Exp. Bot. 66, 402–408. doi: 10.1016/j.envexpbot.2009.04.007
Puente, M. E., Li, C. Y., and Bashan, Y. (2009b). Rock-degrading endophytic bacteria in cacti. Environ. Exp. Bot. 66, 389–401. doi: 10.1016/j.envexpbot.2009.04.010
Raaijmakers, J. M., and Mazzola, M. (2016). Soil immune responses. Science 352, 1392–1393. doi: 10.1126/science.aaf3252
Radman, R., Saez, T., Bucke, C., and Keshavarz, T. (2003). Elicitation of plants and microbial cell systems. Biotechnol. Appl. Biochem. 37, 91–102. doi: 10.1042/BA20020118
Rashid, S., Charles, T. C., and Glick, B. R. (2012). Isolation and characterization of new plant growth-promoting bacterial endophytes. Agric. Ecosyst. Environ. Appl. Soil Ecol. 61, 217–224. doi: 10.1016/j.apsoil.2011.09.011
Ray, T., Pandey, S. S., Pandey, A., Srivastava, M., Shanker, K., and Kalra, A. (2019). Endophytic consortium with diverse gene-regulating capabilities of Benzylisoquinoline Alkaloids biosynthetic pathway can enhance endogenous Morphine biosynthesis in Papaver somniferum. Front. Microbiol. 10:925. doi: 10.3389/fmicb.2019.00925
Rijavec, T., Lapanje, A., Dermastia, M., and Rupnik, M. (2007). Isolation of bacterial endophytes from germinated maize kernels. Can. J. Microbiol. 53, 802–808. doi: 10.1139/W07-048
Ringelberg, D., Foley, K., and Reynolds, C. M. (2012). Bacterial endophyte communities of two wheatgrass varieties following propagation in different growing media. Can. J. Microbiol. 58, 67–80. doi: 10.1139/w11-122
Rodríguez, C. E., Antonielli, L., Mitter, B., Trognitz, F., and Sessitsch, A. (2020). Heritability and functional importance of the Setaria viridis bacterial seed microbiome. Phytobiomes J. 4, 40–52. doi: 10.1094/PBIOMS-04-19-0023-R
Rodríguez, C. E., Mitter, B., Barret, M., Sessitsch, A., and Compant, S. (2017). Commentary: seed bacterial inhabitants and their routes of colonization. Plant Soil 422, 129–134. doi: 10.1007/s11104-017-3368-9
Roodi, D., Millner, J. P., McGill, C., Johnson, R. D., Jauregui, R., and Card, S. D. (2020). Methylobacterium, a major component of the culturable bacterial endophyte community of wild Brassica seed. PeerJ. 8:e9514. doi: 10.7717/peerj.9514
Rosenblueth, M., López-López, A., Martínez, J., Rogel, M. A., Toledo, I., and Martínez-Romero, E. (2012). Seed bacterial endophytes: common genera, seed to-seed variability and their possible role in plants. Acta Hortic. 938, 39–48. doi: 10.17660/ActaHortic.2012.938.4
Rout, M. E., Chrzanowski, T. H., Westlie, T. K., DeLuca, T. H., Callaway, R. M., and Holben, W. E. (2013). Bacterial endophytes enhance competition by invasive plants. Am. J. Bot. 100, 1726–1737 doi: 10.3732/ajb.1200577
Rudgers, J., Afkhami, M., Rúa, M., Davitt, A., Hammer, S., and Huguet, V. (2009). A fungus among us: broad patterns of endophyte distribution in the grasses. Ecology 90, 1531–1539. doi: 10.1890/08-0116.1
Ruiza, D., Agaras, B., de Werrab, P., Wall, L. G., and Valverde, C. (2011). Characterization and screening of plant probiotic traits of bacteria isolated from rice seeds cultivated in Argentina. J. Microbiol. 49, 902–912. doi: 10.1007/s12275-011-1073-6
Rybakova, D., Mancinelli, R., Wirkstrom, M., Birch-Jensen, F., Postma, J., Ehlers, R. U., et al. (2017). The seed microbiome: cultivar-dependent structure in oilseed rape affects the interaction with symbionts and pathogens. Microbiome 5:104. doi: 10.1186/s40168-017-0310-6
Sabki, M. H., Ong, P. Y., Ibrahim, N., Lee, C. T., Klemes, J. J., Li, C., et al. (2021). A review on abiotic stress tolerance and plant growth metabolite framework by plant growth promoting bacteria for sustainable agriculture. Chem. Eng. Trans. 83, 367–372. doi: 10.3303/CET2183062
Sahu, P. K., and Mishra, S. (2021). Effect of hybridization on endophytes: the endo-microbiome dynamics. Symbiosis. 83. doi: 10.1007/s13199-021-00760-w
Saied, A., and Chojnacka, K. (2019). “Chapter 4 - Fertlizers: Need for New Strategies,” in Global Perspectives and Methods Woodhead Publishing Series in Food Science, Technology and Nutrition, Organic Farming, eds S. Chandran, M. R. Unni, and S. Thomas (Woodhead Publishing), 91–116. doi: 10.1016/B978-0-12-813272-2.00004-5
Salisbury, F. B. (1994). “The role of plant hormones,” in Plant-Environment Interactions, ed R. E. Wilkinson (New York, NY: Marcel Dekker), 39–81.
Sankar Ganesh, K., Sundaramoorthy, P., Nagarajan, M., and Lawrence Xavier, R. (2017). “Role of organic amendments in sustainable agriculture,” in Sustainable Agriculture towards Food Security, ed A. Dhanarajan (Singapore: Springer), 111–124. doi: 10.1007/978-981-10-6647-4_7
Santoyo, G., Moreno-Hagelsieb, G., del Carmen Orozco-Mosqueda, M., and Glick, B. R. (2016). Plant growth-promoting bacterial endophytes. Microbiol. Res. 183, 92–99. doi: 10.1016/j.micres.2015.11.008
Sessitsch, A., and Mitter, B. (2015). 21st-century agriculture: integration of plant microbiomes for improved crop production and food security. Microb. Biotechnol. 8, 32–33. doi: 10.1111/1751-7915.12180
Shade, A., Jacques, M. A., and Barret, M. (2017). Ecological patterns of seed microbiome diversity, transmission, and assembly. Curr. Opin. Microbiol. 37, 15–22. doi: 10.1016/j.mib.2017.03.010
Shahzad, R., Khan, A. L., Bilal, S., Asaf, S., and Lee, I. (2018). What is there in seeds? Vertically transmitted endophytic resources for sustainable improvment in plant growth. Front. Plant Sci. 9:24. doi: 10.3389/fpls.2018.00024
Shahzad, R., Khan, A. L., Bilal, S., Asaf, S., and Lee, I.-J. (2017a). Plant growth promoting endophytic bacteria versus pathogenic infections: an example of Bacillus amyloliquefaciens RWL-1 and Fusarium oxysporum f. sp. Lycopersici in tomato. PeerJ 5:e3107. doi: 10.7717/peerj.3107
Shahzad, R., Khan, A. L., Bilal, S., Waqas, M., Kang, S.-M., and Lee, I.-J. (2017b). Inoculation of abscisic acid-producing endophytic bacteria enhances salinity stress tolerance in Oryza sativa. Environ. Exp. Bot. 136, 68–77. doi: 10.1016/j.envexpbot.2017.01.010
Shahzad, R., Waqas, M., Khan, A. L., Al-Hosni, K., Kang, S.-M., Seo, C.-W., et al. (2017c). Indole acetic acid production and plant growth promoting potential of bacterial endophytes isolated from rice (Oryza sativa L.) seeds. Acta Biol. Hung. 68, 175–186. doi: 10.1556/018.68.2017.2.5
Shahzad, R., Waqas, M., Khan, A. L., Asaf, S., Khan, M. A., Kang, S. M., et al. (2016). Seed-borne endophytic Bacillus amyloliquefaciens RWL-1 produces gibberellins and regulates endogenous phytohormones of Oryza sativa. Plant Physiol. Biochem. 106, 236–243. doi: 10.1016/j.plaphy.2016.05.006
Shearin, Z. R., Filipek, M., Desai, R., Bickford, W. A., Kowalski, K. P., and Clay, K. (2018). Fungal endophytes from seeds of invasive, non-native Phragmites australis and their potential role in germination and seedling growth. Plant Soil 422, 183–194. doi: 10.1007/s11104-017-3241-x
Shearin, Z. R. C., Filipek, M., Desai, R., Bickford, W. A., Kowalski, K. P., and Clay, K. (2017). Fungal endophytes from seeds of invasive, non-native Phragmites australis and their potential role in germination and seedling growth. Plant Soil 422, 183–194. doi: 10.1007/s11104-017-341-x
Shi, S., Chang, J., Tian, L., Nasir, F., Ji, L., Li, X., et al. (2019). Comparative analysis of the rhizomicrobiome of the wild versus cultivated crop: insights from rice and soybean. Arch. Microbiol. 201, 879–888. doi: 10.1007/s00203-019-01638-8
Singh, M., Kumar, A., Singh, R., and Pandey, K. D. (2017). Endophytic bacteria: a new source of bioactive compounds. Biotech 7:315. doi: 10.1007/s13205-017-0942-z
Sobolev, V. S., Orner, V. A., and Arias, R. S. (2013). Distribution of bacterial endophytes in peanut seeds obtained from axenic and control plant material under field conditions. Plant Soil 371, 367–376. doi: 10.1007/s11104-013-1692-2
Sponsel, V. M. (2003). “Gibberellins,” in Encyclopedia of Hormones, Vol. 2, eds H. L. Henry and A.W. Norman (San Diego, CA: Academic), 29–40. doi: 10.1016/B0-12-341103-3/00108-X
Stevenson, F. J., and Cole, M. A. (1999). Cycles of Soils: Carbon, Nitrogen, Phosphorus, Sulfur, Micronutrients. New York, NY: John Wiley and Sons.
Stringlis, I. A., Zhang, H., Pieterse, C. M. J., Bolton, M. D., and de Jjonge, R. (2018). Microbial small molecules – weapons of plant subversion. Nat. Prod. Rep. 35, 410–433. doi: 10.1039/C7NP00062F
Sturz, A. V., Christie, B. R., and Nowak, J. (2000). Bacterial endophytes: potential role in developing sustainable systems of crop production. Crit. Rev. Plant Sci. 19, 1–30. doi: 10.1080/07352680091139169
Sülü, S. M., Bozkurt, I. A., and Soylu, S. (2016). Bitki Büyüme Düzenleyici ve Biyolojik Mücadele Etmeni Olarak Bakteriyel Endofitler. Mustafa Kemal Üniversitesi Ziraat Fakültesi Dergisi 21, 103–111. Available online at: http://dergipark.gov.tr/download/article-file/226513 (accessed June 4, 2021).
Syed Ab Rahman, S. F., Singh, E., Pieterse, C. M. J., and Schenk, P. M. (2018). Emerging microbial biocontrol strategies for plant pathogens. Plant Sci. 267, 102–111. doi: 10.1016/j.plantsci.2017.11.012
Sziderics, A. H., Rasche, F., Trognitz, F., Sessitsch, A., and Wilhelm, E. (2007). Bacterial endophytes contribute to abiotic stress adaptation in pepper plants (Capsicum annuum L.). Can. J. Microbiol. 53, 1195–1202. doi: 10.1139/W07-082
Taghavi, S., Garafola, C., Monchy, S., Newman, L., Hoffman, A., Weyens, N., et al. (2009). Genome survey and characterization of endophytic bacteria exhibiting a beneficial effect on growth and development of poplar trees. Appl. Environ. Microbiol. 75, 748–757. doi: 10.1128/AEM.02239-08
Taghavi, S., Van Der Lelie, D., Hoffman, A., Zhang, Y.-B., Walla, M. D., Vangronsveld, J., et al. (2010). Genome sequence of the plant growth promoting endophytic bacterium Enterobacter sp. 638. PLoS Genet. 6:e1000943. doi: 10.1371/journal.pgen.1000943
Taghinasab, M., and Jajabi, S. (2020). Cannabis microbiome and the role of endophytes in modulating the production of secondary metabolites: an overview. Microorganisms 8:355. doi: 10.3390/microorganisms8030355
Tannenbaum, I., Kaur, J., Mann, R., Sawbridge, T., Rodoni, B., and Spangenberg, G. (2020). Profiling the Lolium perene Microbiome: from seed to seed. Phytobiomes J. 4, 281–289. doi: 10.1094/PBIOMES-03-20-0026-R
Tayung, K., Sarkar, M., and Baruah, P. (2012). Endophytic fungi occurring in Ipomoea carnea tissues and their antimicrobial potentials. Braz. Arch. Biol. Technol. 55, 653–660. doi: 10.1590/S1516-89132012000500003
Theis, K. R., Dheilly, N. M., Klassen, J. L., Brucker, R. M., Baines, J. F., Bosch, T. C., et al. (2016). Getting the hologenome concept right: an eco-evolutionary framework for hosts and their microbiome. mSystems 1:e00028-16. doi: 10.1128/mSystems.00028-16
Tosi, M., Mitter, E. K., Gaiero, J., and Dunfield, K. (2020). It takes three to tango: the importance of microbes, host plant, and soil management to elucidate manipulation strategies for the plant microbiome. Can. J. Microbiol. 66, 413–433. doi: 10.1139/cjm-2020-0085
Truyens, S., Weyens, N., Cuypers, A., and Vangronsveld, J. (2013). Changes in the population of seed bacteria of transgenerationally Cd-exposed Arabidopsis thaliana. Plant Biol. 15, 971–981. doi: 10.1111/j.1438-8677.2012.00711.x
Truyens, S., Weyens, N., Cuypers, A., and Vangronsveld, J. (2015). Bacterial seed endophytes: genera, vertical transmission and interaction with plants. Environ. Microbiol. Rep. 7, 40–50. doi: 10.1111/1758-2229.12181
Van der Heijden, M. G., de Bruin, S., Luckerhoff, L., van Logtestijn, R. S., and Schlaeppi, K. A. (2016). Widespread plant-fungal-bacterial symbiosis promotes plant biodiversity, plant nutrition and seedling recruitment. ISME J. 10, 389–399. doi: 10.1038/ismej.2015.120
Van Deynze, A., Zamora, P., Delaux, P.-M., Heitmann, C., Jayaraman, D., Rajasekar, S., et al. (2018). Nitrogen fixation in a landrace of maize is supported by a mucilage-associated diazotrophic microbiota. PLoS Biol. 16:e2006352. doi: 10.1371/journal.pbio.2006352
Van Opstal, E. J., and Bordenstein, S. R. (2015). Microbione. Rethinking heritability of the microbiome. Science 349, 1172–1173. doi: 10.1126/science.aab3958
Vannier, N., Mony, C., Bittebiere, A. K., Michon-Coudouel, S., Biget, M., and Vandenkoornhuyse, P. (2018). A microorganism's journey between plant generations. Microbiome 6:7979. doi: 10.1186/s40168-018-0459-7
Varga, T., Hixson, K. K., Ahkami, A. H., Sher, A. W., Barnes, M. E., Chu, R. K., et al. (2020). Endophyte-promoted phosphorous solubilization in Populus. Front Plant Sci. 11:567918. doi: 10.3389/fpls.2020.567918
Vega, F. E., Pava-Ripoll, M., Posada, F., and Buyer, J. S. (2005). Endophytic bacteria in Coffea arabica L. J. Basic Microbiol. 45, 371–380. doi: 10.1002/jobm.200410551
Verma, S. K., Kharwar, R. N., and White, J. F. (2019). The role of seed-vectored endophytes in seedling development and establishment. Symbiosis 78, 107–113. doi: 10.1007/s13199-019-00619-1
Verma, S. K., Kingsley, K., Irizarry, I., Bergen, M., Kharwar, R. N., and White, J. F. (2017). Seed vectored endophytic bacteria modulate development of rice seedlings. J. Appl. Microbiol. 122, 1680–1691. doi: 10.1111/jam.13463
Verma, S. K., and White, J. F. (2018). Indigenous endophytic seed bacteria promote seedling development and defend against fungal disease in browntop millet (Urochloa ramosa L.). J. Appl. Microbiol. 124, 764–778. doi: 10.1111/jam.13673
Vessey, J. K. (2003). Plant growth promoting rhizobacteria as biofertilizers. Plant Soil. 255, 571–586. doi: 10.1023/A:1026037216893
Walia, A., Sharla, S. G., Chauhan, A., and Mehta, P. (2017). “Endophytic bacteria: role in phosphate solubilization,” in Endophytes: Crop Productivity and Protection, eds D. Maheshwari and K. Annapurna (Cham: Springer), 61–93. doi: 10.1007/978-3-319-66544-3_4
Wang, J., Song, L., Xu, J., and Li, M. (2020). Funcitons of Jasmonic Acid in plant regulation and response to abiotic stress., Int. J. Mol. Sci. 41:446. doi: 10.3390/ijms21041446
Wang, N. R., and Haney, C. H. (2020). Harnessing the genetic potential of the plant microbiome. Biochem 42, 20–25. doi: 10.1042/BIO20200042
Wassermann, B., Rybakova, D., Müller, C., and Berg, G. (2017). Harnessing the microbiomes of Brassica vegetables for health issues. Sci. Rep. 7:17649. doi: 10.1038/s41598-017-17949-z
Weese, D. J., Heath, K. D., Dentinger, B. T., and Lau, J. A. (2015). Long-term nitrogen addition causes the evolution of less-cooperative mutualists. Evolution 69, 631–642. doi: 10.1111/evo.12594
Wei, Z., and Jousset, A. (2017). Plant breeding goes microbial. Trends Plant Sci. 22, 555–558. doi: 10.1016/j.tplants.2017.05.009
Weilharter, A., Mitter, B., Shin, M. V., Chain, P. S., Nowak, J., and Sessitsch, A. (2011). Complete genome sequence of the plant growth-promoting endophyte Burkholderia phytofirmans strain, PsJN. J. Bacteriol. 193, 3383–3384. doi: 10.1128/JB.05055-11
White, J., Kingsley, K., Verma, S., and Kowalski, K. (2018). Rhizophagy cycle: an oxidative process in plants for nutrient extraction from symbiotic microbes. Microorganisms 6:95. doi: 10.3390/microorganisms6030095
White, J. F., Crawford, H., Torres, M. S., Mattera, R., Irizarry, I., and Bergen, M. (2012). A proposed mechanism for nitrogen acquisition by grass seedlings though oxidation of symbiotic bacteria. Symbiosis 57, 161–171. doi: 10.1007/s13199-012-0189-8
White, J. F., Kingsley, K. I., Kowalski, K. P., Irizarry, I., Micci, A., Soares, M. A., et al. (2017). Disease protection and allelopathic interactions of seed-transmitted endophytic pseudomonads of invasive reed grass (Phragmites australis). Plant Soil 422, 195–208. doi: 10.1007/s11104-016-3169-6
White, J. F., Kingsley, K. L., Butterworth, S., Brindisi, L., Gatei, J. W., Elmore, M. T., et al. (2019). “Seed-vectored microbes: Their roles in improving seedling fitness and competitor plant suppression,” in Seed Endophytes, eds S. K. Verma and J. F. White, Jr. (Cham: Springer), 3–20. doi: 10.1007/978-3-030-10504-4_1
Wink, M. (2003). Evolution of secondary metabolites from an ecological and molecular phylogenetic perspective. Phytochemistry 64, 3–19. doi: 10.1016/S0031-9422(03)00300-5
Wintermans, P. C. A., Bakker, P. A. H. M., and Pieterse, C. M. J. (2016). Natural genetic variation in Arabidopsis for responsiveness to plant growth-promoting rhizobacteria. Plant Mol. Biol. 90, 623–634. doi: 10.1007/s11103-016-0442-2
Wisniewski-Dye, F., Borziak, K., Khalsa-Moyers, G., Alexandre, G., Sukharnikov, L. O., Wuichet, K., et al. (2011). Azospirillum genomes reveal transition of bacteria from aquatic to terrestrial environments. PLoS Genet. 7:1002430. doi: 10.1371/journal.pgen.1002430
Wissuwa, M., Mazzola, M., and Picard, C. (2009). Novel approaches in plant breeding for rhizosphere-related traits. Plant Soil 321, 409–430. doi: 10.1007/s11104-008-9693-2
Xu, M., Sheng, J., Chen, L., Men, Y., Gan, L., Guo, S., et al. (2014). Bacterial community compositions of tomato (Lycopersicum esculentum Mill.) seeds and plant growth promoting activity of ACC deaminase producing Bacillus subtilis (HYT-12-1) on tomato seedlings. World J. Microbiol. Biotechnol. 30, 835–845. doi: 10.1007/s11274-013-1486-y
Yan, Y., Yang, J., Dou, Y., Chen, M., Ping, S., Peng, J., et al. (2008). Nitrogen fixation island and rhizosphere competence traits in the genome of root associated Pseudomonas stutzeri A1501. Proc. Natl. Acad. Sci. U. S. A. 105, 7564–7569. doi: 10.1073/pnas.0801093105
Yang, F., Zhang, R., Wu, X., Xu, T., Ahmad, S., Zhang, X., et al. (2020). An endophytic strain of the genus Bacillus isolated from the seeds of maize has antagonistic activity against maize pathogenic strains. Microb. Pathog. 142:104074. doi: 10.1016/j.micpath.2020.104074
Zhang, F. S., Lv, Y. L., Zhao, Y., and Guo, S. X. (2013). Promoting role of an endophyte on the growth and contents of kinsenosides and flavonoids of Anoectochilus formosanus hayata, a rare and threatened medicinal orchidaceae plant. J. Zhejiang Univ. Sci. B. 14, 785–792. doi: 10.1631/jzus.B1300056
Zhang, P., Zhou, P.-P., and Yu, L. J. (2009). An Endophytic taxol-producing fungus from Taxus media, Cladosporium cladosporioides MD2. Curr. Microbiol. 59:227. doi: 10.1007/s00284-008-9270-1
Zhou, X., Wang, J. T., Zhang, Z. F., Li, W., Chen, W., and Cai, L. (2020). Microbiota in the rhizosphere and seed of rice from China, with reference to their transmission and biogeography. Front. Microbiol. 11:995. doi: 10.3389/fmicb.2020.00995
Keywords: hologenome, holonbionte, seed endophyte, breeding selection, domestication, microbiome, bacteria
Citation: L'Hoir M and Duponnois R (2021) Combining the Seed Endophytic Bacteria and the Back to the Future Approaches for Plant Holonbiont Breeding. Front. Agron. 3:724450. doi: 10.3389/fagro.2021.724450
Received: 13 June 2021; Accepted: 26 July 2021;
Published: 03 September 2021.
Edited by:
Mohamed Farissi, Université Sultan Moulay Slimane, MoroccoReviewed by:
Gausiya Bashri, Aligarh Muslim University, IndiaSaad Farouk, Mansoura Universiy, Egypt
Copyright © 2021 L'Hoir and Duponnois. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Robin Duponnois, cm9iaW4uZHVwb25ub2lzQGlyZC5mcg==
 Maïwenn L'Hoir
Maïwenn L'Hoir Robin Duponnois
Robin Duponnois