- Department of Plant Science and Landscape Architecture, University of Connecticut, Storrs, CT, United States
Oomycetes and fungi were recovered from coconut coir and rockwool substrates where marijuana (Cannabis sativa L. cv. Silver and Citron) plants with root rot and wilt symptoms were grown in a commercial growing facility in Connecticut. The objectives of this study were to identify the isolates collected from these substrates, determine the pathogenicity of the isolates on hemp seedlings in vitro and in vivo, and evaluate the pathogens' sensitivity to mefenoxam. Pythium and Globisporangium isolates were identified by sequencing the mitochondrially-encoded cytochrome oxidase genes (COI and COII) and Fusarium sp. with the translation elongation factor (EF-1α) region and internal transcribed spacer region (ITS4 and ITS5) genes. Three isolates were identified as Globisporangium irregulare (formerly Pythium irregulare), 21 isolates were Pythium myriotylum, and one was Fusarium oxysporum. All the isolates tested were pathogenic to hemp plants in vitro and in vivo, with disease incidence between 6.7 and 100%. Inoculated plants were smaller by 32% or more compared with the non-inoculated control. On average, hemp plants infected with Pythium myriotylum produced the lowest biomass and relative greenness values. None of the Pythium and Globisporangium isolates were resistant to mefenoxam—all were sensitive to ≥5 μg·mL−1 mefenoxam. This is the first report of G. irregulare causing root rot on marijuana and hemp plants. The results of this study provide information about the characteristics of pathogens that can be found potentially in soilless substrates in controlled environment agriculture.
Introduction
The production of cannabis and hemp (Cannabis sativa L.) in controlled environment agriculture (CEA) has increased rapidly in North America and little is known about the diseases that prevail in commercial operations. The production of hemp in the United States increased in production acreage from zero in 2013 to 32,464 in 2018 and to 146,065 in 2019 (Mark et al., 2020). Hemp is grown in controlled environment agriculture for cannabinol (CBD) (Mead, 2017). Many U.S. states and providences in Canada have legalized marijuana production for recreational or medical purposes in controlled environment agriculture (i.e., greenhouses or indoor production facilities). The rapid upsurge in production requires a better understanding of potential plant diseases that might affect the industry.
Cannabis sativa L. is the botanical classification of cultivated strains and varieties of marijuana and hemp (Sawler et al., 2015; Barcaccia et al., 2020). There are genetic and biochemical differences between marijuana and hemp (Sawler et al., 2015). Government agencies use plant tissue concentrations of delta-9 tetrahydrocannabinol (Δ9-THC), a cannabinoid with psychoactive effects on humans, to regulate these two crops. The U.S. Federal and Drug Administration (US-FDA, 2020) classifies marijuana as any strains of C. sativa with >0.3% THC by dry weight (US-FDA, 2020), a Schedule I Controlled Substance. The 2018 Farm Bill defines industrial hemp as plants with <0.3% THC by dry weight (US 115th Congress, 2018). In this project we collected soilless substrate samples from a marijuana production facility and conducted controlled experiments with hemp in our research facility—per local regulation, we could not collect plant tissue from the commercial facility or conduct experiments with marijuana plants.
Pythium and Fusarium spp. are causal agents of damping-off, root rot, and wilt diseases on a wide-range of crops (Agrios, 2004). There are only a few reports about root diseases on marijuana and hemp plants. Punja and Rodriguez (2018) confirmed symptoms of root rot, reduced growth, chlorosis, and wilt caused by P. aphanidermatum, P. dissotocum, P. myriotylum, F. oxysporum, and F. solani in hydroponically-grown marijuana in Canada. We recovered P. myriotylum from necrotic roots of a wilted hemp plant grown in a research greenhouse in Connecticut, USA (McGehee et al., 2019). Root rot and wilt on field-grown C. sativa L. caused by Pythium spp. and Fusarium spp. have been reported in Kentucky, Indiana, Israel, California, Canada, Italy, and North Carolina (Beckerman et al., 2017, 2018; Punja et al., 2018; Sorrentino et al., 2019; Jerushalmi et al., 2020; Thiessen et al., 2020; Pitman et al., 2021). Pythium spp. and Fusarium spp. in greenhouse-grown hemp and marijuana have been reported in Arizona, Canada, and North Carolina (Punja et al., 2019, 2020; Punja, 2020; Thiessen et al., 2020; Hu, 2021). The number of reports on diseases associated with hemp and marijuana production are limited, especially in soilless substrates in indoor production. Therefore, there is a need to identify pathogens associated with these crops in order to develop effective management strategies.
The objectives of this study were to (1) identify fungal and oomycete isolates from soilless substrates obtained from a commercial marijuana growing operation, (2) determine the pathogenicity of the isolates on hemp seedlings, and (3) evaluate the sensitivity of isolates to mefenoxam.
Materials and Methods
Isolate Collection
Soilless substrates (coconut coir and rockwool) with no plant tissue were collected from the root zone of symptomatic plants in a marijuana production facility. Federal and local laws prevented us from collecting plant tissue. The symptoms observed were chlorosis, reduced foliar or root growth compared with surrounding plants of the same stage, and root rot. Coconut coir samples were collected from containers with one C. sativa L. ‘Silver’ plant at a mature stage and from two C. sativa L. ‘Citron’ plants in the early flowering stage. Rockwool was collected from C. sativa L. ‘Citron’ cuttings. The samples were collected from a commercial growing facility on March 19, 2019 in Connecticut, USA.
Dilution plating was conducted using Singleton et al.'s (1992) modified-protocol. Briefly, two grams of each soilless substrate were suspended in 10 mL of sterile-deionized water, followed by three 10-fold serial dilutions. A 100 μL aliquot of each dilution was transferred onto Petri plates with corn meal agar (CMA; Difco Laboratories, Detroit, MI, USA), potato dextrose agar (PDA; Difco Laboratories, Detroit, MI, USA), PARP (CMA amended with pimaricin, ampicillin, rifampicin, and pentachloronitrobenzene; Jeffers and Martin, 1986), or PARP-H (pimaricin, ampicillin, rifampicin, pentachloronitrobenzene, and hymexazol), and then spread evenly with a sterile glass rod. Petri dishes were incubated at room temperature in the dark for 48 h. The hyphal tips of individual colonies were transferred to PARP, PARP-H, PDA, and CMA. Twenty-five isolates were collected during one sampling period.
Morphological Identification
Morphological identification was conducted to identify the organisms at the genus level and guide the selection of genes used for molecular identification. Five mycelial plugs of each isolate were placed on four 2.5 cm fragments of autoclaved tall-fescue leaves in sterilized-deionized water and incubated under constant fluorescent light at room temperature for 4 days (Trigiano et al., 2008). The isolates were identified via microscopy following van der Plaats-Niterink identification key for oomycetes (van der Plaats-Niterink, 1981) and the presence of macro and microconidia, false-heads, and chlamydospores for Fusarium (Leslie and Summerell, 2006).
Molecular Identification
DNA from each isolate was obtained from mycelial mats. Mycelial mats were produced as follows: five 4 mm diameter plugs of 3-day-old cultures grown on CMA were transferred to an empty Petri dish and then filled with 20 mL of V8 broth. The plates were incubated in the dark for 5 days, and then the mycelial mats were rinsed three times with sterile deionized water, and then placed on sterile filter paper to remove excess water. The mycelial mats (80–100 mg) were placed in a 2-mL centrifuge tube with a metal bead, liquid nitrogen was added and lyophilized in a TissueLyzer II machine (Qiagen, Germantown, MD, USA) for 2 min with a frequency at 30 1·s−1 or until a fine powder was formed in the tube. DNA was extracted using the DNeasy Plant Mini Kit (Qiagen, Germantown, MD, USA) and quantified with a NanoDrop Spectrophotometer ND-1000 (Thermo Fisher Scientific, Wilmington, DE, USA).
PCR Amplification
The following genes were selected: mitochondrially-encoded cytochrome oxidase I and II (COI and COII), elongation factor 1-alpha regions (EF1 and EF2), and the internal transcribed spacer regions 4 and 5 (ITS5 and ITS4). Amplification of the COI gene was performed with the OomyCoxI-Levup (5′ TCAWCWMGATGGCTTTTTTCAAC 3′) as the forward primer and OomyCoxI-Levlo (5′ CYTCHGGRTGWCCRAAAAACCAAA 3′) as the reverse primer (Robideau et al., 2011). Amplification of the COII was performed with FM58 (5′ CCACAAATTTCACTACATTGA 3′) as the forward primer and FM66 (5′ TAGGATTTCAAGATCCTGC 3′) as the reverse primer (Martin, 2000). Each 20 μL reaction contained 10 μL GoTaq™ 2x MasterMix (Promega Corp., Madison, WI, USA), 1 μL of 0.4 μM of each primer, and 1 μL of 50 ng of genomic DNA. All amplifications were performed on a T100™ Thermocycler (Bio-Rad, Hercules, CA, USA). COI amplification was conducted with the following program: 2 min at 95°C, 35 cycles of 1 min at 95°C, 1 min at 55°C, and 10 min at 72°C. COII gene amplification was conducted with the following program: 3 min at 96°C, 35 cycles of 1 min at 96°C, 1 min at 55°C, 2 min at 72°C, and 10 min at 72°C. Elongation factors, EF1 (5′ATGGGTAAGGARGACAAGAC 3′), and EF2 (5′ GGARGTACCAGTSATCATG 3′) (O'Donnell et al., 1998), and ITS5 (5′ GGAAGTAAAAGTCGTAACAAGG 3′) and ITS4 (5′ TCCTCCGCTTATTGATATGC 3′) (White et al., 1990) were used for the Fusarium isolate. Amplification of the EF-1α and ITS genes were performed with the following program: 3 min at 95°C, 35 cycles of 30 s at 95°C, 30 s at 55°C, 1 min at 72°C, 7 min at 72°C (Figueroa et al., 2015). Amplicons were separated on a 1.7% agarose gel in 1x Tris-Acetate-EDTA (TAE) buffer at 90 volts for 80 min. Gels were visualized using Quantity One Software (Biorad, Hercules, CA, USA).
Sequence Analysis
All amplicons were purified using ExcelaPure PCR purification kit (Edge BioSystems, San Jose, CA, USA) according to the manufacturer's instructions. Purified products were prepared for sequencing with BigDye™ Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Waltham, MA) and Performa® DTR Ultra 96-Well Plates (Edge Biosystems, San Jose, CA, USA) removed dye terminator using Capillary electrophoresis on a 3730xl DNA Analyzer (Applied Biosystems, Waltham, MA, USA). Sanger sequencing was conducted by DNA Analysis Facility at Yale University and Eurofins Genomics LLC laboratories.
Base pairs were trimmed on each end according to the quality graph. The nucleotide sequences were compared with BLAST nucleotide analysis against publicly available sequences in the GenBank online database (www.ncbi.nlm.nih.gov/BLAST). An isolate was considered homologous to published organisms when there was ≥99% identity match and ≥98% query coverage.
Pathogenicity and Virulence Assays
Pathogenicity was evaluated in vitro and in vivo on hemp cv. Wife. Twenty-five isolates were tested for pathogenicity using an in vitro Petri dish assay based on Broders et al. (2007). The seeds were surface-sterilized with concentrated sulfuric acid (98% H2SO4) for 20 s, rinsed under cold tap water for 20 min, placed on cheese cloth, and tied into a bag. The bag was steeped into 75% ethanol for 2 min, wrung out, steeped in 3% bleach for 20 min, rinsed with sterile deionized water, and imbibed in purified water (Milli-Q®, MilliporeSigma, Burlington, MA, USA) for 48 h. After 48 h, the seed coat was removed using sterile forceps and a scalpel to ensure uniform germination. Five seeds were placed in a 9 cm diameter Petri dish containing water agar and spaced 2 cm from the edge and equidistantly 2 cm from each other. A 2 mm plug from each actively growing isolate cultured on CMA for 4 days was placed in the middle of each Petri dish. The control consisted of hemp seeds placed in a Petri dish with a 2 mm plug of CMA with no isolate present. Petri dishes were maintained at ~21°C in the dark for 7 days. Pathogenicity was evaluated using the following virulence scale: 0 = 100% seed germination with healthy appearance, 1 = 99–70% seed germination, small brown lesions on the radicle or hypocotyl, 2 = 69–30% seed germination, large brown lesions on the hypocotyl and radicle, and 3 = 29–0% germination, coalesced lesions covering the hypocotyl and radicle (Broders et al., 2007; Del Castillo Múnera and Hausbeck, 2016). Seed germination per plate (%) and individual seedling length (cm) were recorded. The experiment was arranged as a complete randomized design with three replicate plates per isolate. The experiment was conducted twice.
An in vivo pathogenicity assay was conducted in the greenhouse. Hemp cv. Wife seeds were sown in 42 mm peat pellets (Jiffy, Lorain, OH, USA), with one seed per peat-pellet. After 2 weeks, one seedling was transferred to a square plastic pot (Dillen® Deep Press Fit, A.M.A. Horticulture, Kingsville, ON, CA) with the following dimensions 10.5 × 10.5 × 12.7 cm filled with a peat-based potting mix (Promix BK25, SunGro, Agawam, MA, USA). Five pots were transferred to a seedling production tray (27.9 × 54.3 × 3.3 cm). A tray with five pots was the experimental unit and received the same treatment. Mycelial mats were used as inoculum (Heungens and Parke, 2000) and were produced as described above. Three mycelial mats of each isolate were placed in a 50 mL centrifuge tube and vortexed at high speed with six sterile silicon beads. The mycelial mats were diluted with 800 mL of sterile deionized water and 50 mL of the suspension was used to inoculate directly into the soilless-substrate in each pot. The negative control consisted of plants treated with 50 mL of sterile deionized water. The trays were maintained in UConn's Plant Science Research greenhouse on benches under high-pressure sodium lights with a photoperiod of 16 h between June and July 2019. Plants were irrigated manually with 15-30-15 at 100 mg·L−1 N. Plants were assessed daily for visual wilting and mortality for 14 consecutive days. The area under disease progress curve (AUDPC) was calculated using the trapezoidal integration method (Campbell and Madden, 1990) from the disease incidence assessments. After 14 days, plant shoots were cut right above the cotyledons, and roots were washed with tap water. Shoots and roots were dried at 21 ± 1°C for 2 weeks and weighed. SPAD 502 Plus chlorophyll meter (Spectrum Technologies, Inc., IL) was used to compare relative greenness as a way to measure the visual chlorosis symptoms. To confirm that the symptoms observed were caused by inoculant, re-isolation of the pathogen from symptomatic plant tissue was achieved by placing 10 pieces of 1 cm triple-washed roots on PARP for the oomycete isolates and PDA for the Fusarium isolate. Colonies isolated from the root tissue were examined morphologically and compared to the originally isolated organism following the methods previously described. The experimental unit consisted of five hemp seedlings in a tray and there were three replicates per treatment per experimental run. The experiment was conducted twice.
Mefenoxam Sensitivity
The sensitivity to mefenoxam of the 25 isolates was evaluated in vitro. Technical grade mefenoxam (99.5% active ingredient; Chem Service Inc., West Chester, PA, USA) was dissolved in dimethyl sulfoxide (DMSO) to a stock concentration of 100 mg·mL−1. Control plates received the same volume of DMSO as fungicide-amended agar. A 2 mm actively growing mycelial plug was transferred to the center of a 9 cm diameter Petri plate containing CMA amended with mefenoxam at 0, 0.5, 5, 10, and 100 μg·mL−1. The plates were incubated for 4 days in the dark at room temperature. The relative growth rate of an isolate at each fungicide concentration was calculated by measuring the hyphal growth in two perpendicular directions and averaging the result of the culture diameter, then dividing it by the radial growth on plates with 0% mefenoxam and multiplying the value by 100 (Olson and Benson, 2011). Isolates were characterized as “sensitive” if the culture diameter had <30% growth compared with the 0% mefenoxam plate, “intermediate resistance” if the culture diameter had between 30 and 50% of growth compared with the 0% mefenoxam plate, or “resistant” if the culture diameter was ≥50% of growth compared with the 0% mefenoxam plate (Olson and Benson, 2011). The experiment was a complete randomized design, with three replicates per isolate at each concentration—where each replicate consisted of one plate. The experiment was conducted twice.
Statistical Analysis
Disease incidence and mefenoxam sensitivity data were analyzed using SAS Version 9.4 (SAS Institute Inc., Cary, NC, USA) to establish effect of isolates on the response variables at α = 0.05. Homogeneity of variance and normality were checked for all measured variables using Kolmogorov-Smirnov test, Cramer-von Mises test, and Kuiper test. Data were analyzed by analysis of variance (ANOVA), and means were separated using Tukey's studentized range HSD (Honestly Significant Difference) separation test using PROC MIXED. Virulence data were analyzed using non-parametric Kruskal-Wallis test to determine differences between medians and Tukey's studentized range HSD on ranked data for separation of groups (Minitab®19, Minitab, LLC, State College, PA, USA).
Phylogenetic Analysis
COI gene regions for oomycete isolates and concatenated ITS and EF-1α regions for Fusarium isolates were aligned with MUSCLE (Edgar, 2004). Phylogenetic and molecular analyses were then conducted on the aligned sequences using Geneious Prime version 2020.2.4 (Biomatters, Ltd., Auckland, NZ) (Kearse et al., 2012). An unweighted pair group method with arithmetic mean (UPGMA) (Michener and Sokal, 1957) was constructed using the Tamura-Nei model (Tamura and Nei, 1993) with bootstrap analysis performed by resampling the data 1,000 times (Felsenstein, 1985). In addition to the sequences obtained in this study, sequences were included from the National Center for Biotechnology Information (NCBI) database for comparison (Tables 1, 2). Phytophthora capsici and Phytophthora nicotianae were used as out-groups for P. myriotylum and G. irregulare phylogenies. F. proliferatum and F. subglutinans were used as out-groups for F. oxysporum phylogeny.
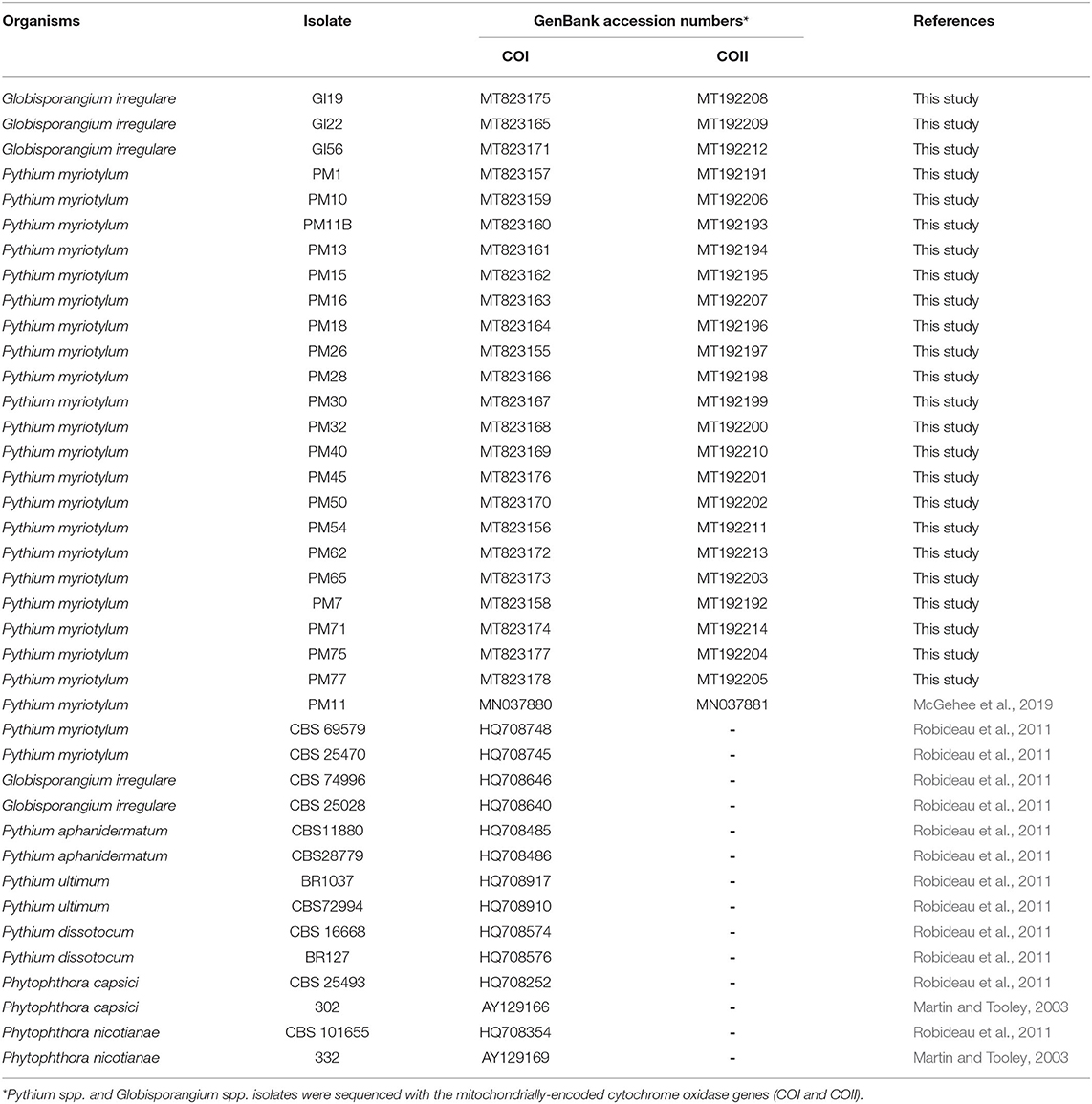
Table 1. Oomycete isolates obtained from coconut coir substrates of symptomatic marijuana plants at a commercial facility in Connecticut and other oomycetes used in the phylogenetic analysis.
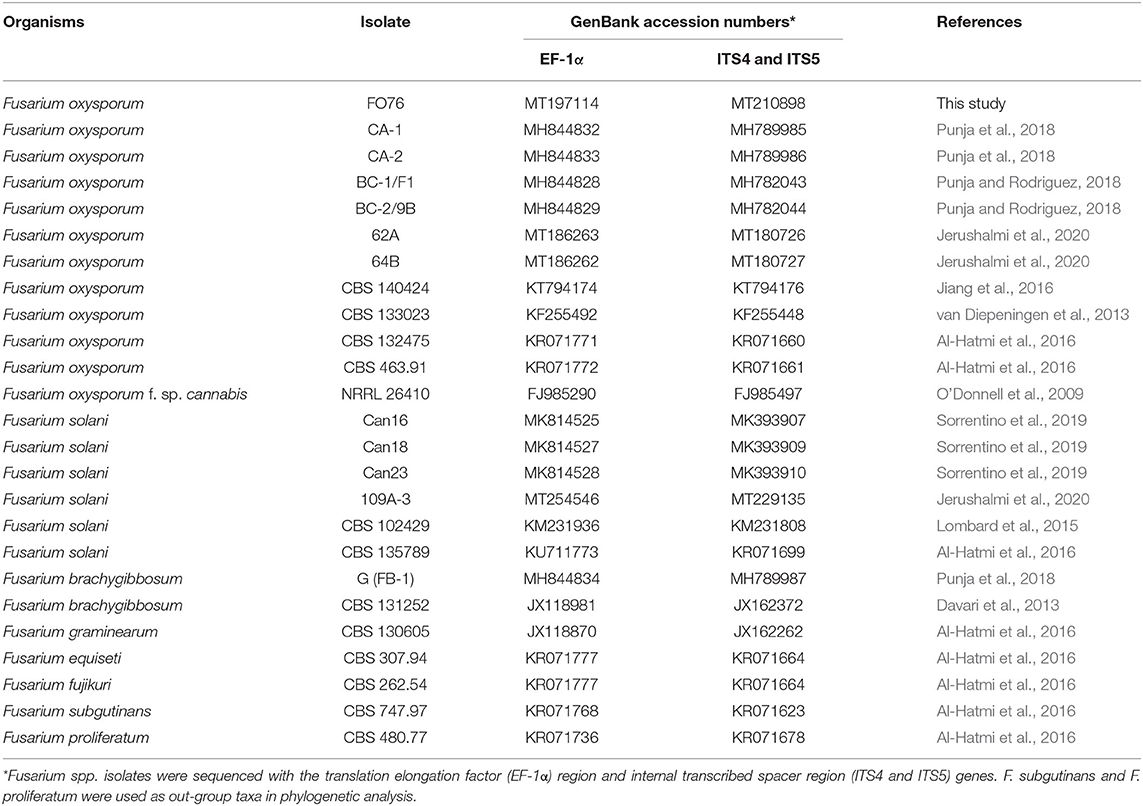
Table 2. Fusarium oxysporum isolate FO76 obtained from the rockwool substrate of a symptomatic marijuana plant at a commercial facility in Connecticut and other Fusarium spp. used in the phylogenetic analysis.
Results
Collection and Identification
Twenty-five isolates were obtained from four soilless-substrate samples from symptomatic marijuana plants collected from an indoor-growing facility. The isolates were identified based on morphological characteristics and then using gene amplification and sequencing.
Morphological Characteristics
The isolates from this study were identified as Globisporangium irregulare (Buisman), Pythium myriotylum (Drechsler), and Fusarium oxysporum (Schltdl). G. irregulare isolates on tall-fescue leaves presented the following characteristics: limoniform hyphal swellings, mostly aplerotic oospores with an average diameter of 18.6 ± 2.8 μm and average wall diameter of 1.3 ± 0.4, intercalary or terminal spherical oogonium (21.6 ± 3.4) with a smooth wall, monoclinous antheridia stalked 1–3 per oogonium with some branches, and spherical sporangia. P. myriotylum had the following characteristics: filamentous-inflated sporangia, globose-shape smooth-surface oogonia with an average diameter of 27.3 ± 2.2 μm, two to six antheridia per oogonium with club-shape and predominately diclinous, apletoric oospores an average diameter of 26 ± 1.9 μm and average wall diameter up to 2.7 ± 1.1 μm thick, and finger-shaped appressoria formed in clusters. F. oxysporum had sickle-shaped microconidia with 3–4 septae. Microconidia were produced in false heads on short monophialides. Chlamydospores were single and terminally positioned.
Identification Based on Nucleotide Sequences
Nucleotide sequences of PCR products from genomic DNA were compared (BLASTn) against publicly available sequences in GenBank®. The isolates were identified as F. oxysporum (n = 1), G. irregulare—also known as P. irregulare (n = 3), and P. myriotylum (n = 21) (Tables 1, 2). Six isolates of P. myriotylum (PM28, PM30, PM40, PM50, PM54, and PM75) and three of G. irregulare (GI19, GI22, and GI56) were isolated from coconut coir samples when the cultivar ‘Citron’ was at the early flowering stage. Six isolates of P. myriotylum (PM16, PM18, PM26, PM45, PM62, and PM65) were isolated from another coconut coir substrate with ‘Citron’ at the early flowering stage, and nine P. myriotylum isolates (PM1, PM7, PM10, PM11, PM13, PM15, PM32, PM71, PM77) were isolated from ‘Silver’ in an 11.4-liter container with coconut coir. One isolate of F. oxysporum (FO76) was collected from rockwool.
Phylogenetic Analysis
Phylogenetic analysis of oomycete isolates based on the rDNA COI gene consisted of 39 taxa including 21 P. myriotylum and three G. irregulare isolates from this study, voucher sequences, and Pythium spp. from Cannabis sativa in published studies obtained from the NCBI database and P. capsici and P. nicotianae as out-group taxa. Distance analysis using the UPGMA algorithm with bootstrap values >75% are displayed in Figure 1. Within the G. irregulare clade two monophyletic groups were formed—one with the isolates from this study (GI19, GI22, and GI56) and another with published sequences (CBS 74996 and CBS 25028) at a 100% bootstrap value. The P. myriotylum clade separated in two groups—PM40 parted from all other isolates. Twenty P. myriotylum isolates from this study separated in 10 subclades with bootstrap values ≥94.3%.
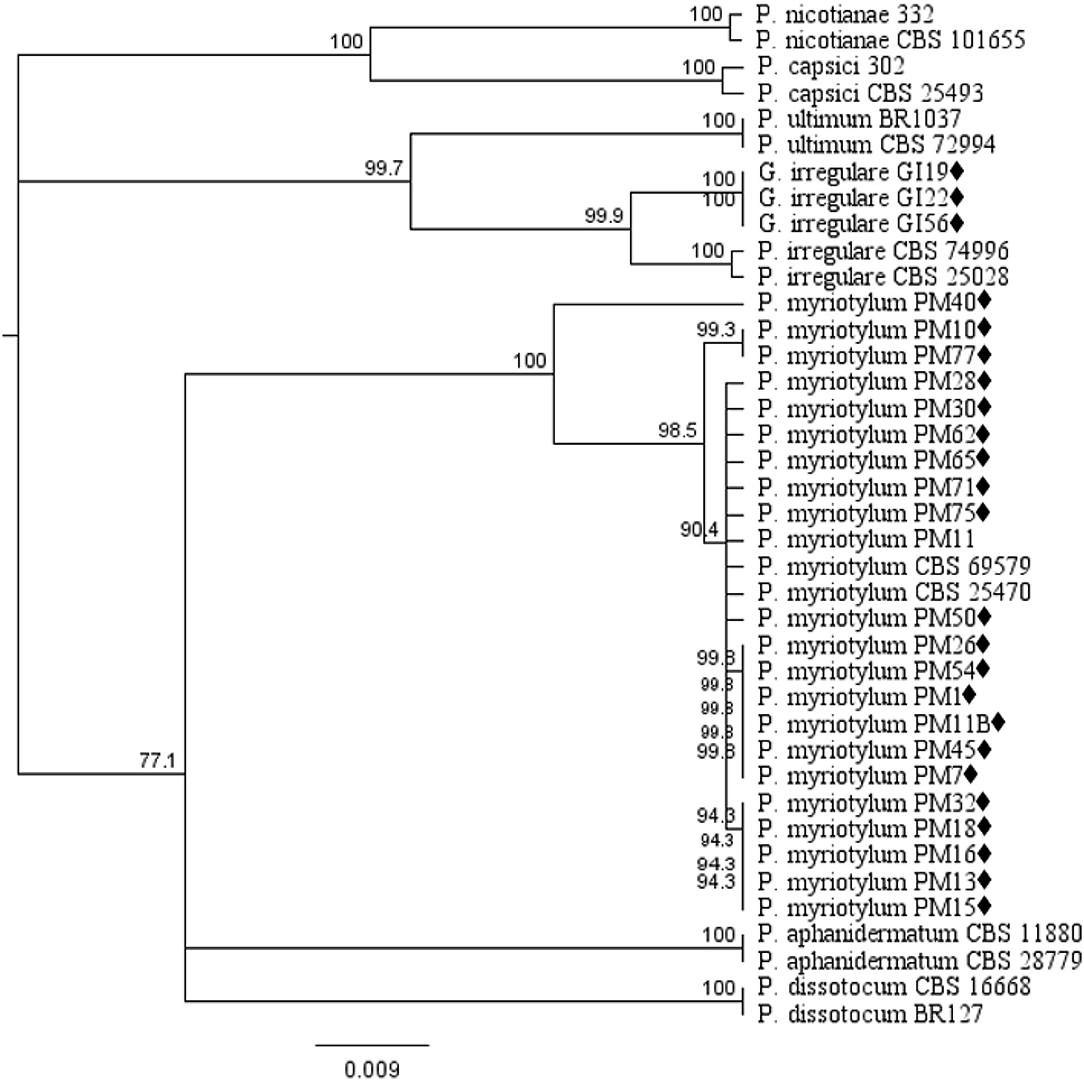
Figure 1. Dendrogram of relationships isolates by among isolates of Pythium and Globisporangium species based on sequence analysis of nuclear rDNA cytochrome oxidase I (COI) genes. The results per isolate were clustered by UPGMA (unweighted pair group method with arithmetic mean) using Tamura-Nei model. Numbers within the tree represent a bootstrap consensus inferred from 1,000 replicates (only values >75% are shown). Phytophthora capsici and Phytophthora nicotianae were used as out-group sequences. ♦ represents the isolates obtained in this study.
The EF-1 α and ITS concatenated tree analyses for Fusarium species consisted of 25 taxa including the F. oxysporum isolate obtained in this study, and 24 taxa obtained from the NCBI database (Figure 2). F. oxysporum FO76 (from this study) clustered with all other F. oxysporum isolates, except F. oxysporum f. sp. cannabis (O'Donnell et al., 2009). F. oxysporum FO76 caused root necrosis, chlorosis, and occasional wilt symptoms in hemp—consistent with the symptoms described by Punja and Rodriguez (2018)—and was >99% identical to the Canadian isolates BC-1 and BC-2 based on a BLASTn analysis. F. oxysporum isolate FO76 was 98 and 95% identical to F. oxysporum isolates CA-1 and CA-2 collected from marijuana roots in California, respectively (Punja et al., 2018).
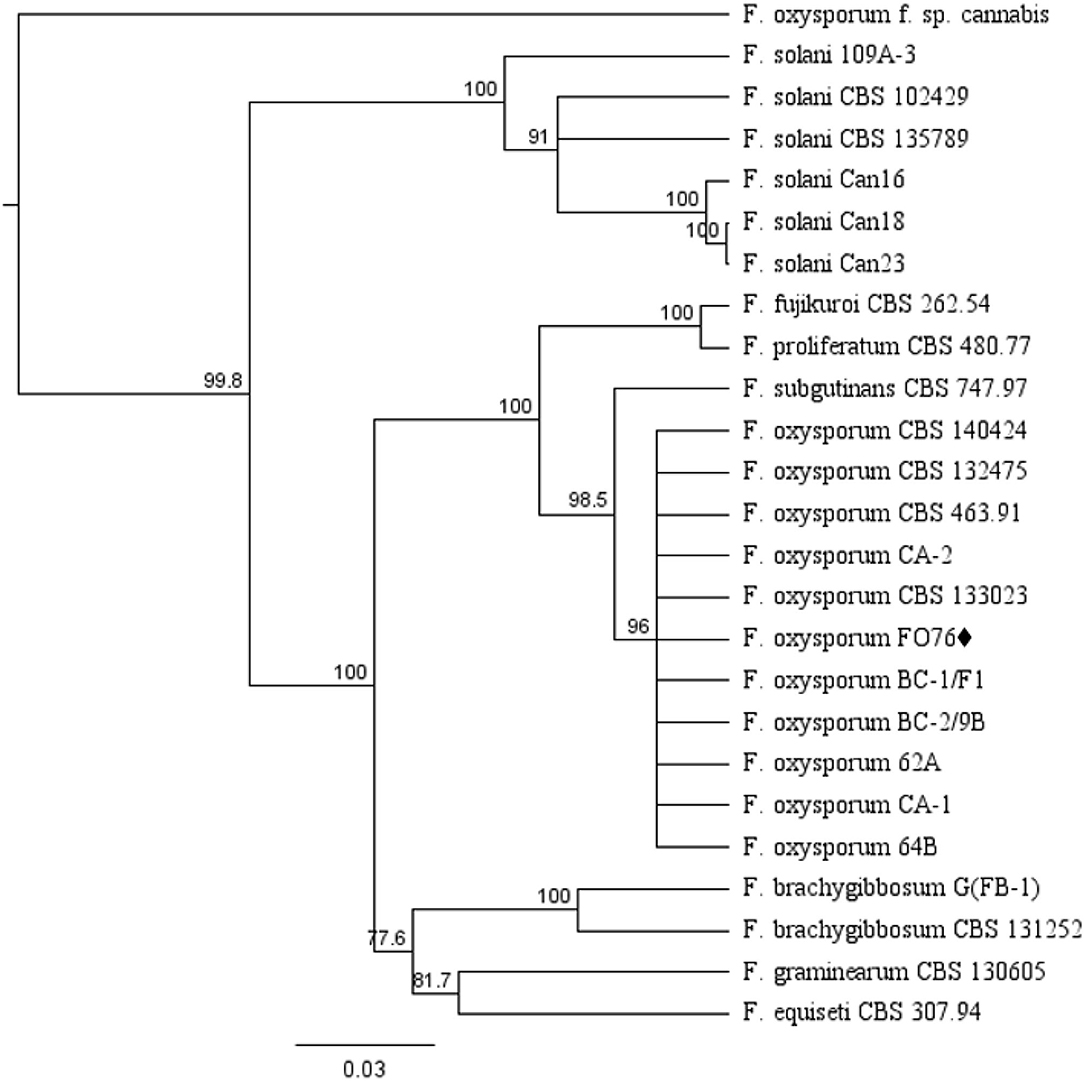
Figure 2. Dendrogram of relationships among isolates of Fusarium species based on sequence analysis of nuclear rDNA elongation factor 1 alpha (EF-1 α) and internal transcribed spacer region (ITS). The results per isolate were clustered by UPGMA (unweighted pair group method with arithmetic mean) using Tamura-Nei model. Numbers within the tree represent a bootstrap consensus inferred from 1,000 replicates (only values >75% are shown). Fusarium subglutinans and Fusarium proliferatum were used as out-group sequences. ♦ represents the isolate obtained in this study.
In vitro Pathogenicity Assays on Hemp Seed
All the isolates, except F. oxysporum FO76, resulted in the highest virulence score (3.0) and were pathogenic to hemp seeds in vitro (Figure 3A). Seed germination was under 30% when inoculated with the Pythium and Globisporangium isolates, whereas the control and F. oxysporum FO76 resulted in 100 and 80% germination, respectively. P. myriotylum PM71 was the only isolate that did not differ from F. oxysporum FO76 in germination percentage (Figure 3B). Seedling length was the longest for hemp seeds in untreated control Petri plates. Seedlings infected with F. oxysporum were ≥60% longer than the ones infected with oomycete isolates, yet all hemp seedlings in contact with isolates were significantly shorter than the negative (non-infected) control (P < 0.0001). Seedling length data is not shown because it presented a similar pattern as germination percentage confirmed with a Pearson correlation (P < 0.001, r = 0.935).
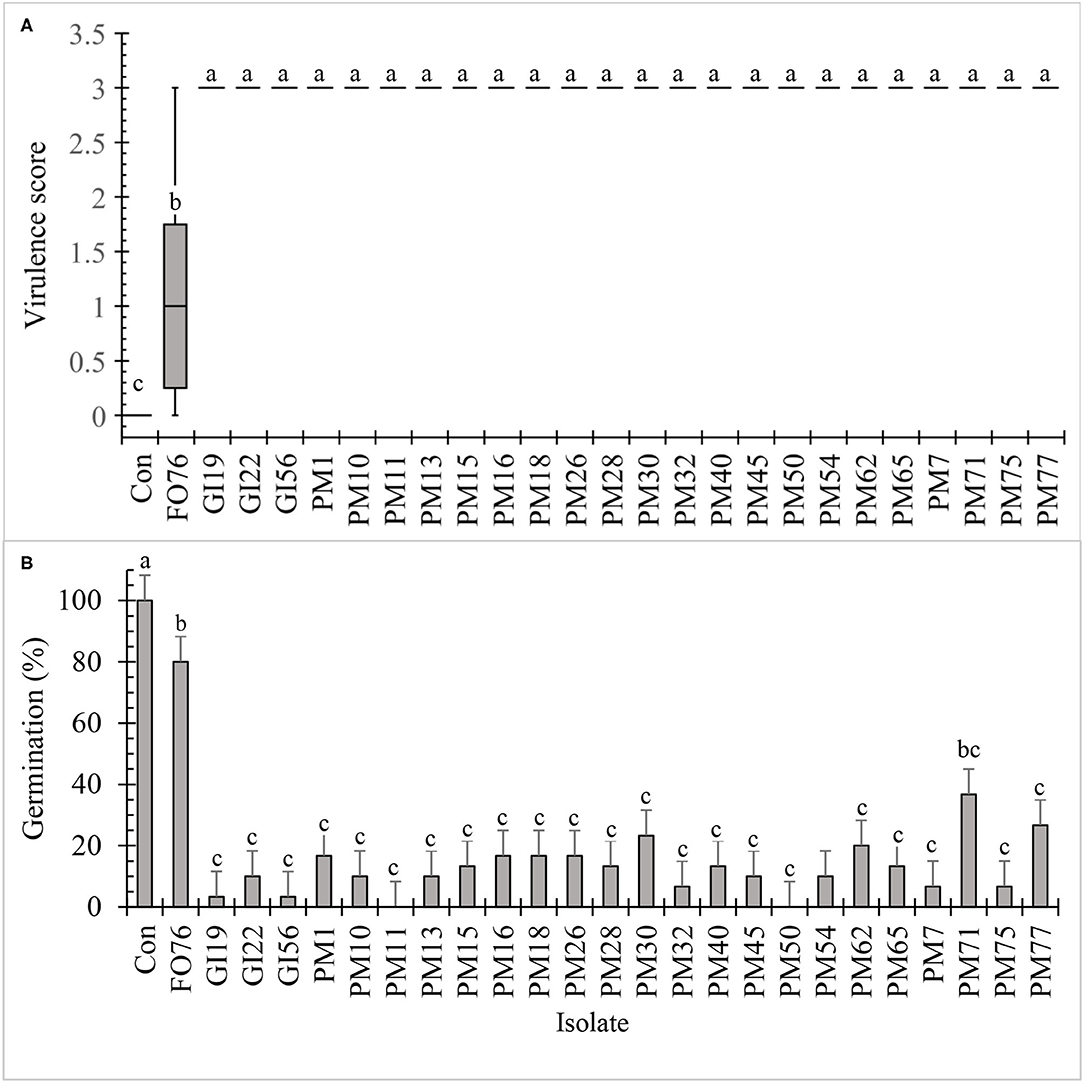
Figure 3. In vitro pathogenicity assay of fungal and oomycete isolates on 7-day-old hemp ‘Wife’ seedlings. FO, Fusarium oxysporum; GI, Globisporangium irregular; PM, Pythium myriotylum; Con, negative control. Boxplot of virulence rating (A) are significantly different according to Krustal-Wallis test (n = 6, α = 0.05). Boxplots followed by the same letter are not different according to Tukey's HSD test on ranked virulence data at α = 0.05. Means of germination data (B) followed by the same letter are not significantly different according to ANOVA and Tukey's HSD test at (n = 6, α = 0.05). Error bars represent standard error.
Pathogenicity Assay in the Greenhouse
Data were analyzed together for both experimental runs due to homogeneity among experimental runs (P > 0.05), except relative greenness and disease incidence represented as the area under the disease progress curve (AUDPC). The control plants did not display any symptoms. In general, plants inoculated with P. myriotylum had the lowest biomass, followed by G. irregulare and F. oxysporum (Figure 4). The control plants had the highest shoot (≥32%) and root (≥48%) biomass compared with all other treatments (Figure 4). Early disease symptoms, wilt and chlorosis, were observed 6 days after plants were inoculated with P. myriotylum isolates. For G. irregulare, disease symptoms occurred at day 8 for isolate GI56 and day 13 for isolate GI22. Disease symptoms were observed for the F. oxysporum isolate FO76 9 days after inoculation. Analysis of variance for the AUDPC values based on disease incidence showed a significant difference among isolates for both experimental runs (Figure 5). P. myriotylum isolates resulted in the highest AUDPC values in both experimental runs (Figure 5). In general, the AUDPC of G. irregulare and F. oxysporum did not differ from the control and P. myriotylum had the highest disease incidence (Figure 5A). The control and G. irregulare isolates resulted in the highest relative greenness (SPAD), indicating no symptoms of chlorosis (Figure 6). Plants inoculated with P. myriotylum isolates had the lowest SPAD readings. F. oxysporum FO76 resulted in lower SPAD values compared to the control, but higher than P. myriotylum (Figure 6). Symptoms for the hemp plants infected with pathogenic isolates included reduced growth, chlorosis, and wilting (Figure 7). Morphological identification of the organisms grown from root fragments cultured on PARP and PDA confirmed that the plants were infected with the organisms originally inoculated (data not shown).
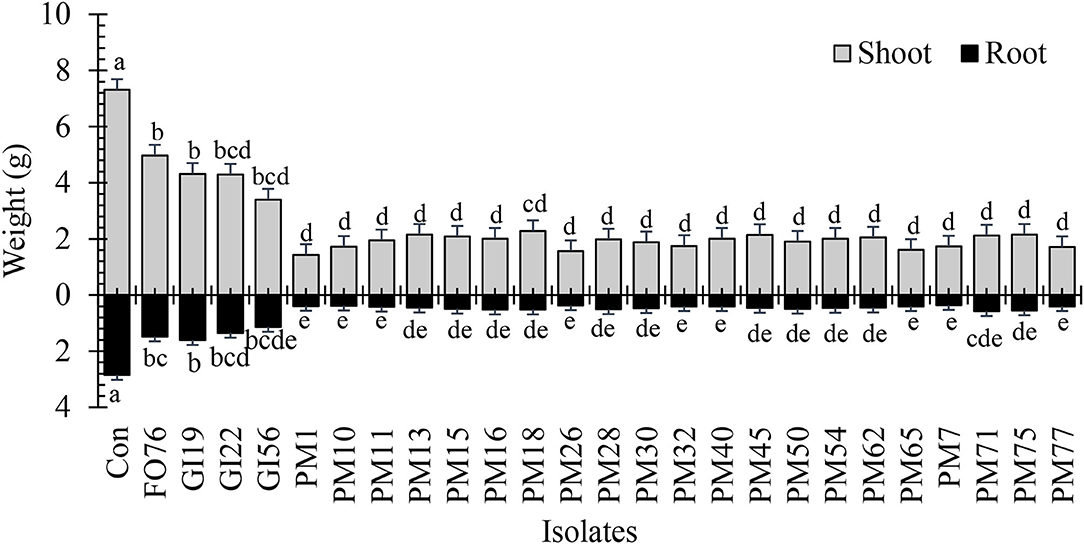
Figure 4. Dry shoot and root weight of 28-day-old hemp ‘Wife’ plants grown in the greenhouse after inoculation with 25 isolates and the control. FO, Fusarium oxysporum; GI, Globisporangium irregular; PM, Pythium myriotylum; Con, negative control. Means (n = 3) within a variable followed by the same letter are not significantly different according to Tukey's HSD test at α = 0.05. Error bars represent standard error.
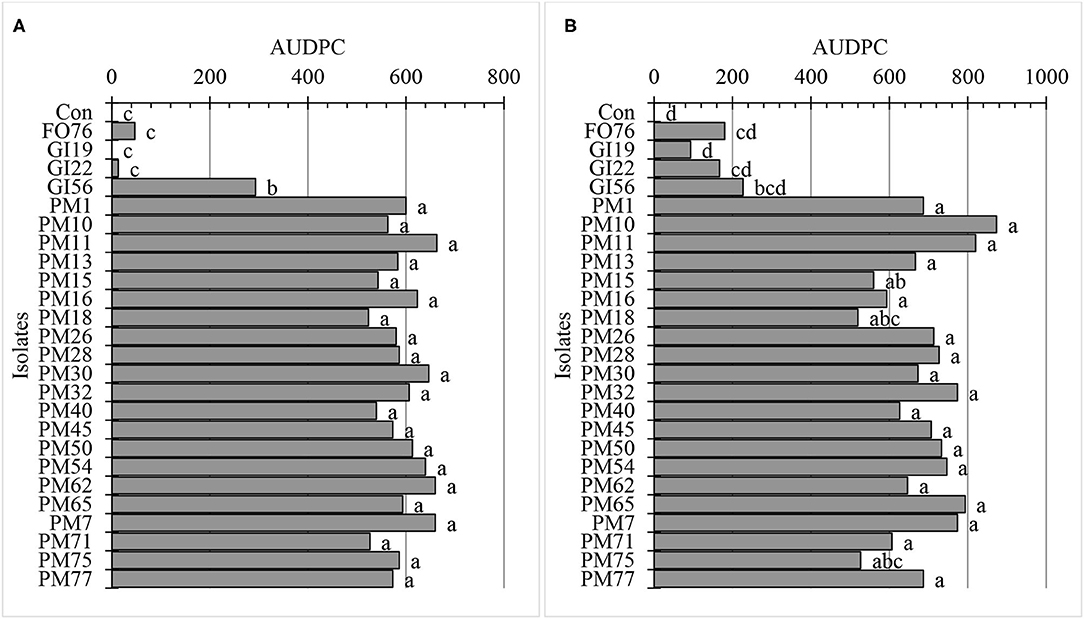
Figure 5. Area under disease progress curve (AUDPC) using disease incidence of hemp ‘Wife’ plants 14 days after inoculation in vivo for (A) the first and (B) second experimental run. FO, Fusarium oxysporum; GI, Globisporangium irregular; PM, Pythium myriotylum; Con, negative control. Means (n = 3) with the same letters are not significantly different according to Tukey's HSD test at α = 0.05. The pooled standard errors were 66 and 42 for runs 1 (A) and run 2 (B), respectively.
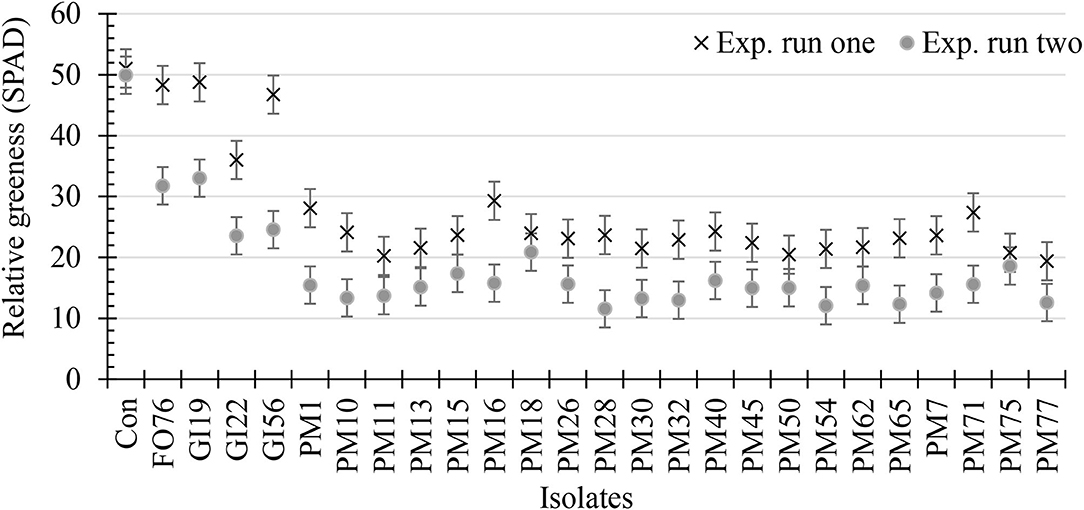
Figure 6. Relative greenness indicating leaf chlorosis or greenness of 28-day-old hemp ‘Wife’ plants 14 days after inoculation with 25 isolates and the control. X's represent the first experimental run, and dots represent the second experimental run (n = 3). FO, Fusarium oxysporum; GI, Globisporangium irregular; PM, Pythium myriotylum; Con, negative control. Error bars represent standard error.

Figure 7. Hemp ‘Wife’ plants 14 days after inoculation with pathogenic isolates in vivo, plus the control. FO, Fusarium oxysporum; GI, Globisporangium irregular; PM, Pythium myriotylum.
Mefanoxam Sensitivity Assay
Dose concentrations, pathogenic isolates, and the interaction between the two were significant (P < 0.0001). The mycelial radial growth rate at 0 μg·mL−1 of mefenoxam resulted in 100% radial growth for all oomycete isolates. G. irregulare isolates had intermediate resistance (>30% mycelial growth) to 0.5 μg·mL−1 mefenoxam (Figure 8). At 5 μg·mL−1, the radial growth ranged from 3.5 to 12.1% and 0 to 12.7% for G. irregulare isolates, and for P. myriotylum, respectively. At 10 μg·mL−1, the radial growth ranged from 2.8 to 10.2%, and 0 to 10% for G. irregulare isolates and P. myriotylum, respectively. P. myriotyum isolates were sensitive (<30% radial growth) at all doses. All oomycete isolates were sensitive to mefenoxam at 5 μg·mL−1 or higher. The oomycetes in this study were not resistant to mefenoxam doses. F. oxysporum was not included in this study.
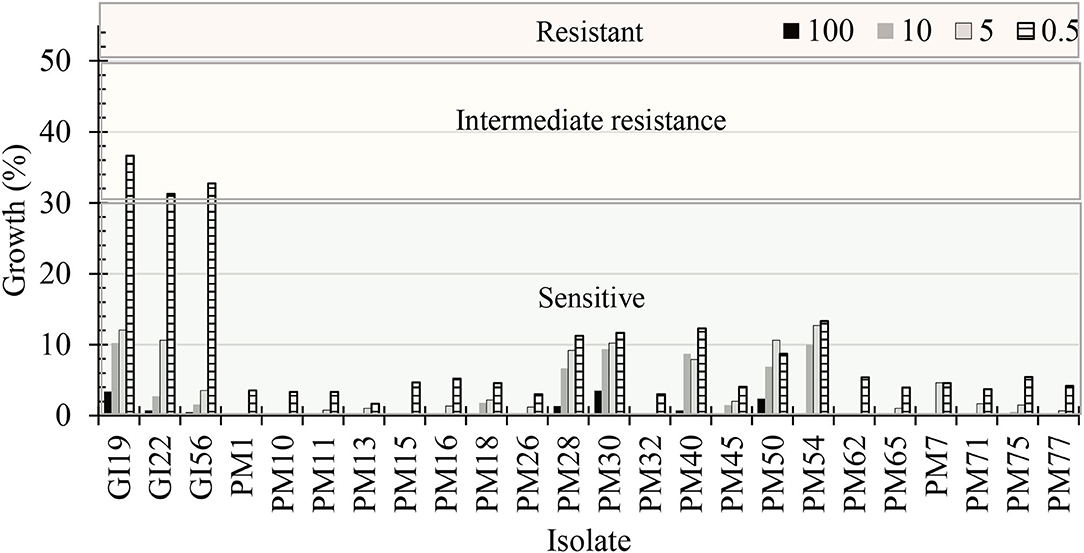
Figure 8. Percentage of radial growth rate on media amended with mefenoxam at 0.5, 5, 10, and 100 μg·mL−1 compared with mycelial radial growth on non-amended (control) plates for oomycete isolates recovered from marijuana substrates (n = 6). GI, Globisporangium irregulare; PM, Pythium myriotylum. Isolates were characterized as “sensitive” if the culture diameter had <30% growth compared with the 0% mefenoxam plate, “intermediate resistance” if the culture diameter had between 30 and 50% of growth compared with the 0% mefenoxam plate, or “resistant” if the culture diameter was ≥50% of growth compared with the 0% mefenoxam plate (Olson and Benson, 2011).
Discussion
This study aimed to investigate if soilborne plant pathogens were present in soilless substrates used at a commercial facility growing marijuana. Our study is limited in scope because we were unable to collect plant tissue from the facilities and we could only collect samples one time. We obtained 21 isolates of P. myriotylum, three of G. irregulare, and one of F. oxysporum and confirmed in two separate experiments that these isolates are pathogenic to hemp ‘Wife’ seeds and seedlings. Plants inoculated with each isolate resulted in varying degrees of growth reduction, chlorosis, and wilting symptoms. P. myriotylum consistently caused the highest virulence (in vitro), lowest germination (in vitro), smallest plants (in vivo), highest AUDPC (in vivo), and most chlorosis (in vivo).
P. myriotylum, the species recovered at the highest frequency in this study, has been previously identified as the causal agent of wilting, stunting, and chlorosis on marijuana and hemp plants grown in soilless systems (Punja and Rodriguez, 2018; McGehee et al., 2019) and G. irregulare had not been reported in hemp or marijuana before. Punja and Rodriguez (2018) found P. dissotocum at the highest frequency (70%). P. aphanidermatum isolates were recovered from diseased cannabis rooted cuttings with symptoms of damping-off, and from flowering plants grown in cocofibre substrate under hydroponic conditions in a commercial greenhouse that were associated with chlorosis of plants and mortality (Punja and Rodriguez, 2018). Thiessen et al. (2020) reported that Pythium root and crown rot caused by P. aphanidermatum, P. myriotylum, and P. ultimum was observed in up to 8% of hemp samples from greenhouses received in the plant diagnostic clinic in North Carolina. These three Pythium species have been associated with other soilless production of leafy greens in hydroponic systems (Stanghellini et al., 1998; Utkhede et al., 2000; McGehee et al., 2018). P. myriotylum has been recovered from other crops grown in soilless substrates including tobacco (Anderson et al., 1997), pepper (Schuerger and Hammer, 2009), tomato (Jenkins and Averre, 1983), and poinsettia (Miyake et al., 2014). These studies suggest that soilless substrates and indoor-production systems might provide favorable conditions for the proliferation of a few Pythium spp. (P. aphanidermatum, P. myriotylum, and P. ultimum). Our study was the first to isolate G. irregulare from any Cannabis sativa. The abundance and diversity of oomycete species in soilless media should be further studied with non-cultured techniques such as high-throughput sequencing metabarcoding.
We observed that P. myriotylum caused more damage to hemp plants than G. irregulare and that G. irregulare exhibited high disease incidence and severity in vitro, but not in vivo—suggesting that G. irregulare isolates may be pre-emergent pathogens. G. irregulare is known to cause severe pre-damping off. For example, 100% seed rot on alfalfa (Berg et al., 2017) and 100% damping-off on carrot seedlings (Howard et al., 1978) have been reported. If G. irregulare does cause pre-emergent damping off, that might explain why G. irregulare has not been previously identified on Cannabis spp.—since most studies studied plants at more advanced crop stages. Alternatively, environmental conditions such as temperature have the potential to influence the interaction of oomycete species with the host, since various Pythium species have temperature-mediated virulence (Matthiesen et al., 2016). In our study, the in vitro experiments were conducted at room temperature (~21°C), while the in vivo assays were conducted in the greenhouse at a higher average temperature of 25 ± 5.9°C. G. irregulare differs from P. myriotylum in that it prefers cooler environments. For instance, a study showed P. aphanidermatum and P. myriotylum caused damping-off on cucumber seedlings at temperatures of 20, 24, 28, and 32°C, whereas G. irregulare caused disease only at 20 and 24°C (Ben-Yephet and Nelson, 1999). Therefore, the variability in virulence seen with G. irregulare isolates may also explain differences in temperature in the two pathogenicity assays.
Phylogenetic analysis verified accurate taxonomic identification for pathogenic isolates collected from this study to reference sequences from curated databases. F. oxysporum isolates recovered from Cannabis sativa L. samples reported in California, Canada, Italy, and Israel were grouped in the same clade as our isolate FO76, indicating phylogenetic similarities. G. irregulare isolates from this study had a high similarity with curated reference sequences (Robideau et al., 2011). However, they formed two separate groups within the G. irregulare clade. Phylogenetic analysis of the P. myriotylum isolates of this study resulted in several subgroups, suggesting some diversity within the isolates and low likelihood that the isolates are a single organism isolated multiple times. These results suggest a diverse population of P. myriotylum within the production facility that was sampled in this project. The morphological characteristics of P. myriotylum isolates recovered from this study were similar to those of PM11 collected from hemp roots in 2019 (McGehee et al., 2019). Both isolates were recovered from the root-zone of Cannabis sativa L. plants in Connecticut, yet the environment and genetic make-up of the hosts differ.
Globisporangium irregulare and P. myriotylum isolates were not resistant to mefenoxam—suggesting low selection pressure of fungicides in the facility. In a previous study, 32 isolates of P. myriotylum recovered from the roots of greenhouse-grown floriculture crops were all sensitive to mefenoxam at 100 μg·mL−1 and 89 out of 127 G. irregulare isolates were resistant (Lookabaugh et al., 2015). Moorman et al. (2002) observed a similar trend with P. myriotylum and G. irregulare presenting resistance to mefenoxam at 100 μg·mL−1. Moorman and Kim (2004) reported 8 out of 13 P. irregulare isolates collected from commercial greenhouses were resistant to 0.078 μg·mL−1 mefenoxam. Another study reported that 43 percent (n = 55) of P. irregulare isolates were resistant to mefenoxam, with 50% of the resistant isolates obtained from greenhouse grown geranium roots (Del Castillo Múnera and Hausbeck, 2016). The State of Connecticut does not allow registration or application of synthetic fungicides for use on marijuana crops grown for human consumption. While we cannot speculate about the exact source of the pathogens in this operation, we presume that it might not come from other agricultural sites where mefenoxam resistance is common.
While the results of this study are limited to a small set of samples from one commercial facility, they provide insights into potential problems and management strategies in cannabis production. The results of our study are also limited by our inability to complete Koch's postulates with the original crop—marijuana—due to legal restrictions. Hemp and marijuana have similar genetics and biosynthetic pathways, but their differences might result in differences in plant disease development. Despite these limitations, our research indicates that isolates in soilless substrates can cause diseases on hemp significantly reducing its potential to grow. Based on this study and previous ones (Punja and Rodriguez, 2018; McGehee et al., 2019) we can also conclude that P. myriotylum is frequently found in the rhizosphere of cannabis plants grown in soilless substrates. Further research should investigate whether some soilless substrates are more or less conducive to harbor specific groups of pathogens, as we observed from our sampling. Except for P. myriotylum (McGehee et al., 2019), the pathogens described here have not been reported to cause root rot and wilt in cannabis plants in Connecticut. Both P. myriotylum and F. oxysporum have been identified causing root rot in Canada (Punja et al., 2018). Further studies and surveys are needed to determine the prevalence of these species and the economic impact these root diseases have on greenhouse cannabis production in other U.S. states.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, MT823175 https://www.ncbi.nlm.nih.gov/genbank/, MT823165 https://www.ncbi.nlm.nih.gov/genbank/, MT823171 https://www.ncbi.nlm.nih.gov/genbank/, MT823157 https://www.ncbi.nlm.nih.gov/genbank/, MT823159 https://www.ncbi.nlm.nih.gov/genbank/, MT823160 https://www.ncbi.nlm.nih.gov/genbank/, MT823161 https://www.ncbi.nlm.nih.gov/genbank/, MT823162 https://www.ncbi.nlm.nih.gov/genbank/, MT823163 https://www.ncbi.nlm.nih.gov/genbank/, MT823164 https://www.ncbi.nlm.nih.gov/genbank/, MT823155 https://www.ncbi.nlm.nih.gov/genbank/, MT823166 https://www.ncbi.nlm.nih.gov/genbank/, MT823167 https://www.ncbi.nlm.nih.gov/genbank/, MT823168 https://www.ncbi.nlm.nih.gov/genbank/, MT823169 https://www.ncbi.nlm.nih.gov/genbank/, MT823176 https://www.ncbi.nlm.nih.gov/genbank/, MT823170 https://www.ncbi.nlm.nih.gov/genbank/, MT823156 https://www.ncbi.nlm.nih.gov/genbank/, MT823172 https://www.ncbi.nlm.nih.gov/genbank/, MT823173 https://www.ncbi.nlm.nih.gov/genbank/, MT823158 https://www.ncbi.nlm.nih.gov/genbank/, MT823174 https://www.ncbi.nlm.nih.gov/genbank/, MT823177 https://www.ncbi.nlm.nih.gov/genbank/, MT823178 https://www.ncbi.nlm.nih.gov/genbank/, MT197114 https://www.ncbi.nlm.nih.gov/genbank/, MT210898.
Author Contributions
CM and RR contributed to the design, implementation, and data analysis of the study. CM conducted the experiments, analyzed the data, and wrote the first draft of the manuscript. RR supervised the findings of this work, verified data analysis, and contributed to the interpretation of results and discussion. All authors contributed to the final manuscript and approved the final version.
Funding
This work was supported by the USDA National Institute of Food and Agriculture, Hatch Multistate project accession number 1020637 and the Floriculture and Nursery Research Initiative.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Al-Hatmi, A. M., van Den Ende, A. H., Stielow, J. B., van Diepeningen, A. D., Seifert, K. A., McCormick, W., et al. (2016). Evaluation of two novel barcodes for species recognition of opportunistic pathogens in Fusarium. Fungal Biol. 120, 231–245. doi: 10.1016/j.funbio.2015.08.006
Anderson, M. G., Fortnum, B. A., and Martin, S. B. (1997). First report on Pythium myriotylum in a tobacco seedling float system in South Carolina. Plant Dis. 81:227. doi: 10.1094/PDIS.1997.81.2.227D
Barcaccia, G., Palumbo, F., Scariolo, F., Vannozzi, A., Borin, M., and Bona, S. (2020). Potentials and challenges of genomics for breeding cannabis cultivars. Front. Plant Sci. 11:573299. doi: 10.3389/fpls.2020.573299
Beckerman, J., Nisonson, H., Albright, N., and Creswell, T. (2017). First report of Pythium aphanidermatum crown and root rot of industrial hemp in the United States. Plant Dis. 101, 1038–1039. doi: 10.1094/PDIS-09-16-1249-PDN
Beckerman, J., Stone, J., Ruhl, G., and Creswell, T. (2018). First report of Pythium ultimum crown and root rot of industrial hemp in the United States. Plant Dis. 102:2045. doi: 10.1094/PDIS-12-17-1999-PDN
Ben-Yephet, Y., and Nelson, E. B. (1999). Differential suppression of damping-off caused by Pythium aphanidermatum, P. irregulare, and P. myriotylum in composts at different temperatures. Plant Dis. 83, 356–360. doi: 10.1094/PDIS.1999.83.4.356
Berg, L. E., Miller, S. S., Dornbusch, M. R., and Samac, D. A. (2017). Seed rot and damping-off of alfalfa in Minnesota caused by Pythium and Fusarium species. Plant Dis. 101, 1860–1867 doi: 10.1094/PDIS-02-17-0185-RE
Broders, K. D., Lipps, P. E., Paul, P. A., and Dorrance, A. E. (2007). Characterization of Pythium spp. associated with corn and soybean seed and seedling disease in Ohio. Plant Dis. 91, 727–735. doi: 10.1094/PDIS-91-6-0727
Campbell, C. L., and Madden, L. V. (1990). Introduction to Plant Disease Epidemiology. New York, NY: John Wiley and Sons.
Davari, M., Wei, S. H., Babay-Ahari, A., Arzanlou, M., Waalwijk, C., van der Lee, T. A. J., et al. (2013). Geographic differences in trichothecene chemotypes of Fusarium graminearum in the northwest and north of Iran. World Mycotoxin J. 6, 137–150. doi: 10.3920/WMJ2012.1493
Del Castillo Múnera, J., and Hausbeck, M. K. (2016). Characterization of Pythium species associated with greenhouse floriculture crops in Michigan. Plant Dis. 100, 569–576. doi: 10.1094/PDIS-03-15-0296-RE
Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. doi: 10.1093/nar/gkh340
Felsenstein, J. (1985). Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39, 783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x
Figueroa, F. R., Isla, L. H., and Hidalgo, O. B. (2015). Molecular diagnosis of Fusarium spp. isolates associated to bud rot of oil palm in Ecuador. Biotecnol. Aplicada 32, 2221–2223.
Heungens, K., and Parke, J. L. (2000). Zoospore homing and infection events: effects of the biocontrol bacterium Burkholderia cepacia AMMDR1 on two oomycete pathogens of pea (Pisum sativum Appl L.). Environ. Microbiol. 66, 5192–5200. doi: 10.1128/AEM.66.12.5192-5200.2000
Howard, R. J., Pratt, R. G., and Williams, P. H. (1978). Pathogenicity to carrots of Pythium species from organic soils of North America. Phytopathology 68, 1293–1296. doi: 10.1094/Phyto-68-1293
Hu, J. (2021). First Report of crown and root rot caused by Pythium myriotylum on hemp (Cannabis sativa) in Arizona. Plant Dis. doi: 10.1094/PDIS-12-20-2712-PDN
Jeffers, S. N., and Martin, S. B. (1986). Comparison of two media selective for Phytophthora and Pythium species. Plant Dis. 70, 1038–1043. doi: 10.1094/PD-70-1038
Jenkins, S. F. Jr, and Averre, C. W. (1983). Root diseases of vegetables in hydroponic culture systems in North Carolina greenhouses. Plant Dis. 67, 968–970. doi: 10.1094/PD-67-968
Jerushalmi, S., Maymon, M., Dombrovsky, A., and Freeman, S. (2020). Fungal pathogens affecting the production and quality of medical Cannabis in Israel. Plants (Basel) 7:882. doi: 10.3390/plants9070882
Jiang, Y., Al-Hatmi, A. M., Xiang, Y., Cao, Y., van den Ende, A. H., Curfs-Breuker, I., et al. (2016). The concept of ecthyma gangrenosum illustrated by a Fusarium oxysporum infection in an immunocompetent individual. Mycopathologia 181, 759–763. doi: 10.1007/s11046-016-0031-6
Kearse, M., Moir, R., Wilson, A., Stones-Havas, S., Cheung, M., Sturrock, S., et al. (2012). Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647–1649. doi: 10.1093/bioinformatics/bts199
Leslie, J. F., and Summerell, B. A. (2006). The Fusarium Laboratory Manual. Ames, IA: Blackwell Publishing. doi: 10.1002/9780470278376
Lombard, L., van der Merwe, N. A., Groenewald, J. Z., and Crous, P. W. (2015). Generic concepts in Nectriaceae. Stud. Mycol. 80, 189–245. doi: 10.1016/j.simyco.2014.12.002
Lookabaugh, E. C., Ivors, K. L., and Shew, B. B. (2015). Mefenoxam sensitivity, aggressiveness, and identification of Pythium species causing root rot on floriculture crops in North Carolina. Plant Dis. 99, 1550–1558. doi: 10.1094/PDIS-02-15-0232-RE
Mark, T. B., Shepherd, J., Olson, D. W., Snell, W., Proper, S., and Thornsbury, S. (2020). Economic Viability of Industrial Hemp in the United States: A Review of State Pilot Programs. USDA- ERS. Economic Information Bulletin No. (EIB-217). U.S. Department of Agriculture, Economic Research Service.
Martin, F. N. (2000). Phylogenetic relationships among some Pythium species inferred from sequence analysis of the mitochondrially encoded cytochrome oxidase II gene. Mycologia 92, 711–727. doi: 10.1080/00275514.2000.12061211
Martin, F. N., and Tooley, P. W. (2003). Phylogenetic relationships among phytophthora species inferred from sequence analysis of mitochondrially encoded cytochrome oxidase I and II genes. Mycologia 95, 269–284. doi: 10.2307/3762038
Matthiesen, R. L., Ahmad, A. A., and Robertson, A. E. (2016). Temperature affects aggressiveness and fungicide sensitivity of four Pythium spp. that cause soybean and corn damping off in Iowa. Plant Dis. 100, 583–591. doi: 10.1094/PDIS-04-15-0487-RE
McGehee, C. S., Apicella, P., Raudales, R., Berkowitz, G., Ma, Y., Durocher, S., et al. (2019). First report of root rot and wilt caused by Pythium myriotylum on hemp (Cannabis sativa L.) in the United States. Plant Dis. 103:3288. doi: 10.1094/PDIS-11-18-2028-PDN
McGehee, C. S., Raudales, R. E., and Elmer, W. H. (2018). First report of Pythium dissotocum causing Pythium root rot on hydroponically grown lettuce in Connecticut. Plant Dis. 102, 2043–2044. doi: 10.1094/PDIS-02-18-0365-PDN
Mead, A. (2017). The legal status of cannabis (marijuana) and cannabidiol (CBD) under U.S. law. Epilepsy Behav. 70, 288–291. doi: 10.1016/j.yebeh.2016.11.021
Michener, C. D., and Sokal, R. R. (1957). A quantitative approach to a problem of classification. Evolution 11, 490–499. doi: 10.1111/j.1558-5646.1957.tb02884.x
Miyake, N., Nagai, H., and Kageyama, K. (2014). Wilt and root rot of poinsettia caused by three high-temperature-tolerant Pythium species in ebb-and-flow irrigation systems. J. Gen. Plant Pathol. 80, 479–489. doi: 10.1007/s10327-014-0542-2
Moorman, G. W., Kang, S., Geiser, D. M., and Kim, S. H. (2002). Identification and characterization of Pythium species associated with greenhouse floral crops in Pennsylvania. Plant Dis. 86, 1227–1231. doi: 10.1094/PDIS.2002.86.11.1227
Moorman, G. W., and Kim, S. H. (2004). Species of Pythium from greenhouses in Pennsylvania exhibit resistance to propamocarb and mefenoxam. Plant Dis. 88, 630–632. doi: 10.1094/PDIS.2004.88.6.630
O'Donnell, K., Gueidan, C., Sink, S., Johnston, P. R., Crous, P. W., Glenn, A., et al. (2009). A two-locus DNA sequence database for typing plant and human pathogens within the Fusarium oxysporum species complex. Fungal Genet. Biol. 46, 936–948. doi: 10.1016/j.fgb.2009.08.006
O'Donnell, K., Kistler, H. C., Cigelnik, E., and Ploetz, R. C. (1998). Multiple evolutionary origins of the fungus causing Panama disease of banana: concordant evidence from nuclear and mitochondrial gene genealogies. Proc. Natl. Acad. Sci. U. S. A. 95, 2044–2049. doi: 10.1073/pnas.95.5.2044
Olson, H. A., and Benson, D. M. (2011). Characterization of Phytophthora spp. on floriculture crops in North Carolina. Plant Dis. 95, 1013–1020. doi: 10.1094/PDIS-09-10-0619
Pitman, T. L., Philbrook, R. N., Vetterli, M. R., and Warren, J. G. (2021). First report of Pythium ultimum causing crown rot in greenhouse-grown Cannabis sativa in California. Plant Dis. 105. doi: 10.1094/PDIS-02-21-0336-PDN
Punja, Z. K. (2020). First report of Fusarium proliferatum causing crown and stem rot, pith necrosis. in cannabis (Cannabis sativa L., marijuana) plants. Can. J. Plant Pathol. doi: 10.1080/07060661.2020.1793222
Punja, Z. K., Collyer, D., Scott, C., Lung, S., Holmes, J., and Sutton, D. (2019). Pathogens and molds affecting production and quality of Cannabis sativa L. Front. Plant Sci. 10:1120. doi: 10.3389/fpls.2019.01120
Punja, Z. K., Ni, L., and Roberts, A. (2020). The Fusarium solani species complex infecting cannabis (Cannabis sativa L., marijuana) plants and a first report of Fusarium (Cylindrocarpon) lichenicola causing root and crown rot. Can. J. Plant Pathol. doi: 10.1080/07060661.2020.1866672
Punja, Z. K., and Rodriguez, G. (2018). Fusarium and Pythium species infecting roots of hydroponically grown marijuana (Cannabis sativa L.) plants. Can. J. Plant Pathol. 40, 498–513. doi: 10.1080/07060661.2018.1535466
Punja, Z. K., Scott, C., and Chen, S. (2018). Root and crown rot pathogens causing wilt symptoms on field-grown marijuana (Cannabis sativa L.) plants. Can. J. Plant Pathol. 40, 528–541. doi: 10.1080/07060661.2018.1535470
Robideau, G. P., de Cock, A. W. A. M., Coffey, M. D., Voglmayr, H., Brouwer, H., Bala, K., et al. (2011). DNA barcoding of oomycetes with cytochrome c oxidase subunit I and internal transcribed spacer. Mol. Ecol. Resour. 11, 1002–1011. doi: 10.1111/j.1755-0998.2011.03041.x
Sawler, J., Stout, J. M., Gardner, K. M., Hudson, D., Vidmar, J., Butlers, L., et al. (2015). The genetic structure of marijuana and hemp. PLoS ONE 10:e0133292. doi: 10.1371/journal.pone.0133292
Schuerger, A. C., and Hammer, W. (2009). Use of cross-flow membrane filtration in a recirculating hydroponic system to suppress root disease in pepper caused by Pythium myriotylum. Phytopathology 99, 597–607. doi: 10.1094/PHYTO-99-5-0597
Singleton, L. L., Mihail, J. D., and Rush, C. M. (1992). Methods for Research on Soilborne Phytopathogenic Fungi. St. Paul, MN: The American Phytopathological Society, 41.
Sorrentino, R., Pergamo, R., Battaglia, V., Raimo, F., Cermola, M., and Lahoz, E. (2019). Characterization and pathogenicity of Fusarium solani causing foot rot on hemp (Cannabis sativa L.) in Southern Italy. J. Plant Dis. Protect. 126, 585–591. doi: 10.1007/s41348-019-00265-1
Stanghellini, M. E., Kim, D. H., Rakocy, J., Gloger, K., and Klinton, H. (1998). First report of root rot of hydroponically grown lettuce caused by Pythium myriotylum in a commercial production facility. Plant Dis. 82:831. doi: 10.1094/PDIS.1998.82.7.831D
Tamura, K., and Nei, M. (1993). Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 10, 512–526.
Thiessen, L. D., Schappe, T., Cochran, S., Hicks, K., and Post, A. R. (2020). Surveying for potential diseases and abiotic disorders of industrial hemp (Cannabis sativa) production. Plant Health Prog. 21, 321–332. doi: 10.1094/PHP-03-20-0017-RS
Trigiano, R. N., Windham, M. T., and Windham, A. S. (2008). Plant Pathology. Concepts and Laboratory Exercises. 2nd Edn. Boca Raton, FL: CRC Press, 209.
US 115th Congress (2018). Agriculture Improvement Act of 2018. Available online at: https://www.agriculture.senate.gov/2018-farm-bill (accessed April 6, 2020).
US-FDA (2020). FDA Regulation of Cannabis and Cannabis Derived Products Including Cannabidiol (CBD). Available online at: https://www.fda.gov/news-events/public-health-focus/fda-regulation-cannabis-and-cannabis-derived-products-including-cannabidiol-cbd (accessed October 29, 2020).
Utkhede, R. S., Lévesque, C. A., and Dinh, D. (2000). Pythium aphanidermatum root rot in hydroponically grown lettuce and the effect of chemical and biological agents on its control. Can. J. Plant Pathol. 22, 138–144. doi: 10.1080/07060660009500487
van Diepeningen, A. D., Feng, P., Ahmed, S., Sudhadham, M., Bunyaratavej, S., and de Hoog, G. S. (2013). Fusarium infections in a tropical dermatologic clinic: a MLST and evidence-based evolution. Mycoses 58, 48–57. doi: 10.1111/myc.12273
White, T. J., Burns, T., Lee, S., and Taylor, J. (1990). “Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics,” in PCR Protocols: A Guide to Methods and Applications, eds M. A. Innis, D. H. Gelfand, J. J. Sninsky, and T. J. White (San Diego, CA: Academic Press, Inc), 315–322. doi: 10.1016/B978-0-12-372180-8.50042-1
Keywords: cannabis, controlled environment agriculture, plant pathogen, substrates, roots
Citation: McGehee CS and Raudales RE (2021) Pathogenicity and Mefenoxam Sensitivity of Pythium, Globisporangium, and Fusarium Isolates From Coconut Coir and Rockwool in Marijuana (Cannabis sativa L.) Production. Front. Agron. 3:706138. doi: 10.3389/fagro.2021.706138
Received: 06 May 2021; Accepted: 06 July 2021;
Published: 06 August 2021.
Edited by:
Zamir Punja, Simon Fraser University, CanadaReviewed by:
Antonio Cellini, University of Bologna, ItalyClaudio Altomare, National Research Council (CNR), Italy
Copyright © 2021 McGehee and Raudales. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rosa E. Raudales, cm9zYS5yYXVkYWxlc0B1Y29ubi5lZHU=
 Cora S. McGehee
Cora S. McGehee Rosa E. Raudales
Rosa E. Raudales